Abstract
Post-translational modifications (PTMs) can occur soon after translation or at any stage in the lifecycle of a given protein, and they may help regulate protein folding, stability, cellular localisation, activity, or the interactions proteins have with other proteins or biomolecular species. PTMs are crucial to our functional understanding of biology, and new quantitative mass spectrometry (MS) and bioinformatics workflows are maturing both in labelled multiplexed and label-free techniques, offering increasing coverage and new opportunities to study human health and disease. Techniques such as Data Independent Acquisition (DIA) are emerging as promising approaches due to their re-mining capability. Many bioinformatics tools have been developed to support the analysis of PTMs by mass spectrometry, from prediction and identifying PTM site assignment, open searches enabling better mining of unassigned mass spectra—many of which likely harbour PTMs—through to understanding PTM associations and interactions. The remaining challenge lies in extracting functional information from clinically relevant PTM studies. This review focuses on canvassing the options and progress of PTM analysis for large quantitative studies, from choosing the platform, through to data analysis, with an emphasis on clinically relevant samples such as plasma and other body fluids, and well-established tools and options for data interpretation.
Keywords: post translational modification, quantitative proteomics, body fluids, clinical samples, PTM
1. Introduction
The ability to analyse protein post-translational modifications (PTMs) occurring on a large scale in a biological system yields insight into their roles and relevance to disease states and confers proteomics a unique edge. An immense variety of biological responses occur in this manner—in Bill Bryson’s memorable turn of phrase “depending on mood and metabolic circumstance, (proteins) will allow themselves to be phosphorylated, glycosylated, acetylated, ubiquitinated, farnesylated, sulphated and linked to GPI anchors, among rather a lot else” [1]. The wealth of these changes and their importance in cell signalling and disease led to modified proteins being the focus of clinical and pharmaceutical research as potential drug targets [2,3,4]. Several factors have converged to make the analysis of such modifications possible on a large scale: advances in mass spectrometry (MS) methods including larger multiplexing chemical labelling (Isobaric tag for relative and absolute quantitation—iTRAQ, Tandem Mas Tag—TMT) and other novel label-free quantitation approaches such as DIA/SWATH [5,6], improvements in PTM enrichment strategies [7], the development of more detailed and robust PTM workflows [8,9], and crucially, improvement in bioinformatics tools and databases making the subsequent analysis possible. In generic terms, the ideal goals of all such large scale PTM analyses are easy enough to state: canvass which sites on the proteins are modified, quantify how those modifications change with condition or disease, and determine what the modification achieves in terms of function in the cell.
Recent reviews illustrated the potential of PTMs as disease biomarkers [10], surveyed disease associated PTM changes [11], in cardiovascular disease [12], cancer [13], neurodegenerative disease [14] and diabetes [15], demonstrating a growing interest in characterising PTMs to answer clinical questions [16]. The present review is structured to follow the workflows necessary for undertaking a large scale PTM experiment aiming at the ideal goals described above, in logical order from the sample preparation through to the data analysis, with emphasis on the bioinformatics steps needed. Figure 1 captures the workflow and outlines some of the common approaches used in PTM studies. Our focus is on studies on human plasma and other body fluids and clinically relevant cell lines, and for the sake of clarity we limit our consideration to the five modifications whose study has matured the most: phosphorylation, glycosylation, acetylation, methylation and ubiquitination. The first two sections provide the background in describing the modifications surveyed and their clinical relevance, giving known examples of such studies in relevant body fluids. Section 3 and Section 4 discuss the quantitative mass spectrometry methods amenable to large scale PTM studies, and review the optimum enrichment methods, including some practical aspects and challenges. The remaining sections focus on the bioinformatics and statistical analysis aspects, structured into four basic areas: site localisation (understand which sites are modified), assessments of quantitation and stoichiometry (how quantitation changes with condition or disease and what proportion of peptides is modified), through to available tools for the subsequent analysis and finally network visualisation approaches as well as validation analysis (assign and validate function).
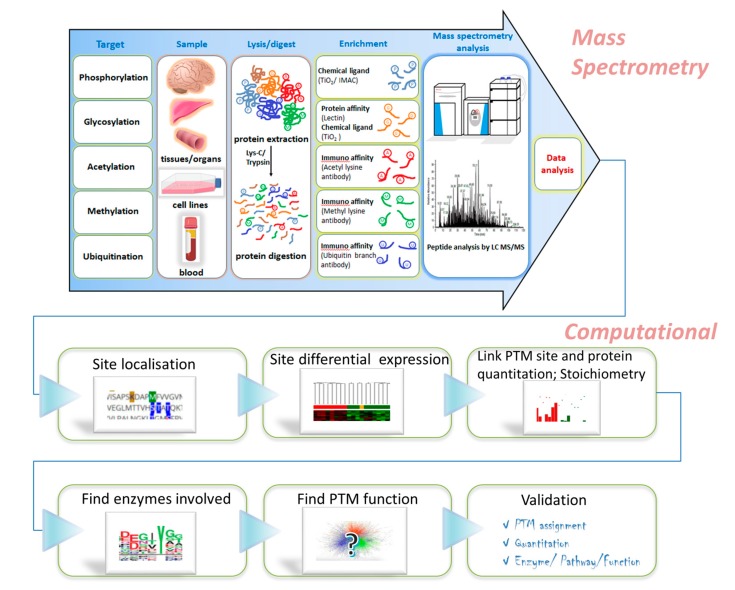
Figure 1.
Schematic PTM Workflow illustrating some of the common steps associated with sample preparation, PTM enrichment, MS and bioinformatics analysis.
2. Modifications with Known Clinical Relevance
The complex phenotype of an organism is attributable to the extensive and diverse phenomenon of protein regulation; this hidden regulation of the protein network is made possible by post translational modifications. A single PTM can re-establish the entire downstream trafficking transforming the protein function and cell fate. Hence PTMs determine the optimal functionalities of several proteins that are involved in an array of physiological and disease states and as such comprises the basis of several drug targeting strategies and diagnostic tests. PTMs determine the protein interactions with other proteins and form the basis of several cellular signalling pathways. These also regulate the cell–cell and cell–matrix interactions and play vital roles in inflammation, host-pathogen interactions, immune modulation, and degenerative and proliferative disorders. According to reports, 100 of the 469 existing PTMs in the UniProt database are found in humans [16]. Although several PTMs have been found to date, transient modifications and low abundances account for some of the many challenges that limit our understanding of PTMs. Here we highlight some examples of known PTMs reported to have clinical relevance in disease pathology and which have considerable information amassed about them in public databases (Figure 2).
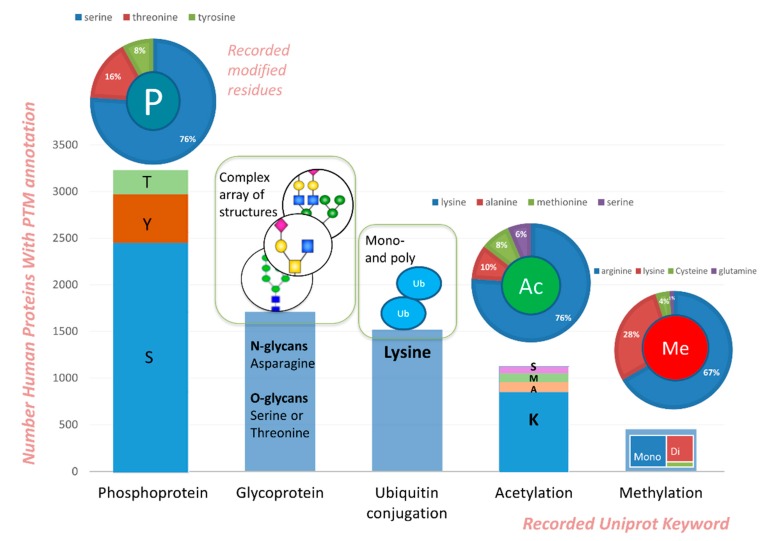
Figure 2.
Human proteins with PTM currently available in the Uniprot database; percentage of modified residues is indicated above the bars, wherever that information is available.
Phosphorylation is one of the most widely studied modifications and can occur in a very dynamic and rapid manner, regulating various signalling pathways in health and disease conditions. The PhosphoSite Plus database records more than 350 proteins with modified PTMs under disease conditions. Phosphopathology is a long-debated concern in neurodegenerative disorders. Early records of hyperphosphorylated neurofibrillary tangles (NFT) in Alzheimer affected brains have been reported as long ago as the 1980s [17,18]. Tau hyperphosphorylation, the major constituent of NFT is another example of disrupted PTM in Alzheimer’s disease (AD) [19]. Detection of Tau phosphorylation in cerebrospinal fluid (CSF) might reflect AD progression and suggest pathological mechanisms underlying the disease [20]. Phosphorylation at S129 of α-synuclein is associated with synucleinopathy lesions like Parkinson’s disease and dementia [21,22]. Atypical phosphorylation patterns of specific proteins are observed in several malignancies as evidenced in non-small cell lung cancer (NSCLC) patient tumour samples [23], serum samples from breast and prostate cancer patients [24], patient-derived acute myeloid leukaemia bone marrow cells (AML) [25], human pancreatic duct tissue of Pancreatic ductal adenocarcinoma patients [26] and renal cell carcinoma tumours from kidney cancer patients [27]. Cardiovascular diseases are also influenced by erroneous protein phosphorylation. A detailed review by Rapundalo et al. has addressed the essential proteins that are negatively modulated by phosphorylation leading to cardiac dysfunction [28].
Glycosylation is another common PTM, which plays an important role in a protein’s structure and function and has been shown to be involved in evolution, development and immunity. Glycoproteomics—the characterisation of the oligosaccharides (glycans) attached to proteins—is an important challenge. Glycans are a chemically diverse structural feature of proteins originating from the enzymatic addition of individual monosaccharide units to asparagine (N-glycans) or serine and/or threonine (O-monosaccharides and O-glycans). The result is a complex array of monosaccharides or glycan structures with different compositions, lengths and linkages.Glycoproteomics is a rapidly emerging field and several modifications and their relevance have been elucidated in recent years. Valero et al. suggested glycosylation of proteins acetylcholinesterase and butyrylcholinesterase in AD patient antemortem tissues as markers of disease progression [29,30,31]. Creutzfeldt-Jakob disease (CJD) is another example where the pathological glycosylation profile of CSF acetylcholinesterase enzyme has been identified [32]. Specific glycosylation patterns of proteins in various brain regions such as the frontal cortex in Parkinson’s disease and temporal lobe samples in Amyotropic Lateral Sclerosis (ALS) have also been reported [33,34]. In breast cancer, glycan changes were shown to mediate disease progression and influence overall survival rates [35]. Supporting these studies, the mRNA and protein levels of core 1 β-1,3-galactosyltransferase (C1GALT1) were observed to be elevated in hepatocellular carcinoma [36]. Similarly, detailed studies reveal genes modifying glycosylation patterns contribute towards disease pathology in different cancer types [10,11,12,13]. Glycated haemoglobin measurement is an important diagnostic test for blood sugar levels over an extended period of time in diabetes [37].
Ubiquitin (Ub) is a small protein composed of 76 amino acids, which can covalently modify other proteins, usually at lysine residues. Ubiquitin is a highly conserved protein often involved in protein degradation, and is implicated in several pathophysiological states including cancer and neurodegeneration. Ubiquitination is a reversible process which involves cleavage of ubiquitin by deubiquitin enzymes (DUBs). Poly ubiquitination is known to target proteins for degradation via 26S proteasome and to recycle ubiquitin. The modification occurs between the C-terminus of Ubiquitin and the amino group of lysine residues, or sometimes the N-terminus of the substrate protein. Ubiquitination can involve mono- or poly ubiquitination and each of the seven lysine residues (Lys-6, Lys-11, Lys-27, Lys-29, Lys-33, Lys-48, and Lys-63) in Ubiquitin itself can serve as potential modification sites for continued poly ubiquitination.
Severe ubiquitin overexpression is observed in lung cancer tissues and a recent study demonstrated the role of E3 ubiquitin ligase NEDD4 in enhancing the EGFR mediated migratory potential of NSCLC cells [38,39]. E3 ubiquitin ligase WWP1 was detected in high levels in bone marrow tissue in acute myeloid leukaemia [40]. Ubiquitin protein ligase E3C (UBE3C) was upregulated in renal cell carcinoma tissues and was reported to be associated with poor survival rate [41]. SPOP mediated ATF2 ubiquitination has been reported in prostate cancer samples [42]. Ubiquitin-conjugating enzyme Ubc13 was found to be upregulated in malignant tissues from breast, pancreas, colon, prostate, and lymphoma [43]. Neurodegenerative disorders are also not exempt from the deleterious effects of ubiquitination as illustrated by the following clinical scenarios. Perry et al have demonstrated the co-existence of ubiquitin in Neurofibrillary tangles (NFT) and neurites associated with senile plaques in AD brain tissues [44]. Lewy bodies in Parkinson’s disease show immunoreactivity to ubiquitin providing insights into the shared PTM feature of several neurodegenerative conditions [45]. Dysregulated ubiquitination was also reported to underlie cardiotoxicity in dilated cardiomyopathy (DCM) and cardiac explants from DCM patients showed hyperubiquitination of proteins with increased mRNA levels of proteins involved in ubiquitin-proteosome pathway [46]. A detailed review on the role of ubiquitination in innate and adaptive immunity highlights the potential implications ubiquitination has on autoimmune disorders as well [47].
Acetylation and methylation are epigenetically relevant PTMs that have well-established roles in disease phenotype by altering replication and transcription regulation. Methylation of arginine by protein methyl transferases (PRMTs) is known to regulate transcription, cell signalling and cell fate. The addition of methyl groups is via covalent attachment to the side chain of arginine and can be either monomethylarginine (monoisotopic, 14.0157 Da), dimethylarginine (symmetric or asymmetric, monoisotopic, 28.0313 Da) or trimethylarginine (monoisotopic 42.0470 Da). Acetylation of the N-terminus (NTA or N-acetylome) by N-terminal acetyltransferases (NATs) plays an important role in protein structure and function. The reversible acetylation of lysine (K-acetylome) by lysine acetyltransferases (KATs) and histone deacetylases (HDACs) plays an important role in histone modification and the regulation gene expression, but the functional changes of reversible acetylation are likely to be far broader than the histones and gene regulation.
Differential expression in the methylation of histone PTM sites of human AD frontal cortex samples contributed towards the disease pathology [48]. Similarly, an increased histone methylation profile was observed in monocytes from type 1 diabetes patients, and a parallel increase in the acetylation pattern was also detectable in these patient samples [49]. Genome-wide expression studies revealed elevated protein arginine methyl transferases 1 and 6 (PRMT1 and PRMT6) expression in lung, pancreatic, breast, and urinary bladder cancers [50,51,52,53]. Chronic obstructive pulmonary disease (COPD) marked the expression of hyperacetylated histone h3.3 rendering resistance to UPS mediated degradation in the airway lumen, alveolar fluid, and plasma samples collected from COPD patients [54]. A significant increase in histone deacetylase 6 levels was evident in oral squamous cell carcinoma with a correlation to tumor aggressiveness suggesting the key roles played by protein deactylation in biochemical processes [55]. Tampered HDAC1 and class II HDAC regulation were evident in human prostate cancer and lung cancer specimens [56,57]. Additionally, rogue acetylation associated with mutant p53 proteins was observed in several tumors [58]. Histone deacetylase inhibitors have been shown to provide neuroprotection to the retinal ganglion cells in experimental model of optic nerve injury [59].
3. Examples of PTM Studies in Plasma or Other Clinically Relevant Fluids
With the rapid advancement in mass spectrometry techniques researchers can gain detail of relevant PTMs in multiple patient samples. Owing to the role played by PTMs in disease, techniques that can analyse PTMs in patient derived biological fluids open up a new horizon for biomarker discovery research. Plasma, serum and other biological fluids are attractive clinical samples for the purpose of discovering disease-relevant biomarkers. The advantage of using such biological fluids for PTM analysis lies in the ease of accessibility and the promise of developing personalised medicine approaches better suited to each individual.
Table 1 highlights several studies carried out in various biological fluids with findings related to the impact of post-translational modifications on disease. The most commonly used samples, not surprisingly, remain plasma and serum. Urine and saliva are other easily accessible samples of general interest; however, information is still lacking on disease-relevant urine or saliva PTMs. More specific body fluids such as cerebrospinal fluid are naturally of high interest for neurological disease studies.
Table 1.
Examples of PTM studies for various clinically relevant biological fluids.
| Body Fluid | PTM | Disease | Description | Reference |
|---|---|---|---|---|
| Plasma | Phosphorylation | Breast cancer | Proteomics study revealed unique phosphopeptide profile of plasma derived extracellular vesicles from breast cancer patients | [60] |
| Phosphorylation | Alzheimer’s disease | Phospho-Tau presence in AD patient plasma samples | [61] | |
| Phosphorylation | Parkinson’s disease | Plasma samples demonstrated higher phosphorylated α-synuclein levels compared to healthy controls | [62] | |
| Acetylation, Ubiquitination | Glioblastoma | LC-MS/MS analysis on plasma from patients with glioblastoma multiform showed decreased acetylated and ubiquitinated peptides | [63] | |
| Ubiquitination | Leukaemia | Unique plasma profile of ubiquitin-proteasome system (UPS) and steep ubiquitin protein levels were detected in AML and ALL patient plasma samples | [64] | |
| Sera | Glycosylation | Alzheimer’s disease | Increased O-GlcNAcylation levels and decreased global glycosylation levels | [65] |
| Glycosylation | Hepatocellular Carcinoma | Increase in the levels of golgi glycoprotein GP73 in HCC patient serum | [66] | |
| Glycosylation | Ovarian cancer | Changes in serum glycome profile of ovarian cancer patients | [67] | |
| Glycosylation | Breast cancer | Abundant fucosylation in metastatic breast cancer patient sera | [68] | |
| Acetylation, Methylation | Leukaemia | Lysine acetylation and arginine mono-methylation as the prevalent PTMs in sera of patients with acute myelogenous leukemia, breast cancer, and non-small-cell lung cancer | [69] | |
| Saliva | Phosphorylation | Control | Large-Scale Phosphoproteomics Analysis of Whole Saliva | [70,71] |
| Glycosylation | Oral ulcer | Proteomic and N-glycoproteomic quantification reveal aberrant changes in the human saliva of oral ulcer patients. | [72] | |
| Glycosylation | Control | Analysis of age and gender associated N-glycoproteome in human whole saliva | [73] | |
| Glycosylation | Control | Identification of N-Linked Glycoproteins in Human Saliva | [74] | |
| Cerebro spinal fluid | Glycosylation | Alzheimer’s disease | Unusually glycosylated acetylcholinesterase in CSF samples of AD patients as a diagnostic molecule | [30] |
| Glycosylation | Schizophrenia | Identification of N-glycosylation changes in the CSF and serum in patients with schizophrenia | [75] | |
| Phosphorylation | Alzheimer’s disease and Parkinson’s disease | CSF of AD and PD patients have also been positive for phospho-Tau and phospho α-Synuclein respectively | [20,76] | |
| Ubiquitination | Alzheimer’s disease and Parkinson’s disease | Accumulation of other PTM, ubiquitin and associated enzymes in the CSF samples of AD and PD patients | [77,78] | |
| Pancreatic fluid | Several | Pancreatitis | Use of pancreatic fluid to outline the PTM profile unique for individuals with chronic pancreatitis | [79] |
| Urine | Glycosylation | Prostate cancer | Characterisation of Glycoproteins from urine samples of prostate cancer patients with different Gleason scores | [80] |
| Glycosylation | Prostate cancer | Investigation of glycoproteome to discriminate prostate cancer (PCa) from benign prostatic hyperplasia (BPH) | [81] | |
| Phosphorylation | Pregnancy | Evaluating the expression of specific phosphoproteins during pregnancy comparison with non-pregnancy. | [82] | |
| Phosphorylation | Bladder cancer | Phosphorylation of urinary tyrosine protein reported for predicting early bladder cancer onset | [83] |
In terms of specific PTMs, protein glycosylation and phosphorylation are the most widely reported PTMs recognised in body fluids relevant to pathological. Aberrant glycation identified in patient serum samples of multiple cancer types indicate the potential of glyco bio-markers in tumor diagnosis, while circulating phosphorylated proteins have been reported extensively in neurodegenerative disorders.
4. Quantitative MS Methods for PTMs Analysis
Mass spectrometry techniques are capable of assessing the modification status of proteins including site localisation and occupancy [84,85,86]. Workflows for PTM analysis often focus on a single modification type—phosphorylation or glycosylation, etc.—and one sample type (blood, plasma, tissue, or cell lines). Substantial progress in PTM analysis has come from cell lines studies, providing the foundations for developing mature PTM workflows, including the establishment of protocols for sample preparation, strategies for PTM enrichment, and sensitive LC-MS/MS detection for PTM quantification. These workflows often make use of carefully selected lysis and digestion protocols, selective purification techniques based on affinity or immunoprecipitation enrichment and quantitative mass spectrometry. It is worth mentioning that these protocols are generally achieved using larger amounts of starting materials (mgs of proteins/peptides) not necessarily ideal for the clinical samples with lower amount of proteins (CSF, tear or saliva). The following section highlights common workflows used for the analysis of phosphorylation, glycosylation, acetylation, methylation, and ubiquitination using quantitative mass spectrometry.
A detailed description of MS methodology including chromatography and mass spectrometry instrumentation is included in the recent phosphoproteomics review of Riley and Coon [87], while data acquisition methods were very recently and comprehensively reviewed in the context of PTM analysis [12], and details on sequence-specific identifications of PTMs including example spectra are provided in the classic review of Choudhary and Mann [88].
4.1. MS General Considerations: Sample Collection And Digestion
Although mass spectrometry has been used extensively to identify proteins and their modifications, large-scale proteome expression profiling in combination with comprehensive PTM site localisation and quantification is a rather ambitious challenge, particularly in the context of spatial and temporal profiling. For clinically relevant studies, stringent SOPs addressing sample collection are required and there is often a need to include reagents and enzymes to inhibit sample degradation after collection and before analysis [10]. For instance, for plasma proteomic studies, it is common practice to use blood collection tubes containing additives, or to introduce inhibitors to avoid undesirable endogenous enzyme activity. As an example, commercially available phosphatase inhibitors are used to avoid undesirable activities of phosphatase enzymes during enrichment of phosphopeptides.
Quantitative assessment of PTMs in the context of clinically relevant studies, represent a very important stepping stone on the path to understand human pathologies. Many quantitative LC-MS/MS approaches make use of the “bottom-up” proteomics strategy, which involves the digestion of proteins using a proteolytic enzyme such as trypsin (or a combination of endopeptidases such as Lys-C and trypsin), then LC-MS/MS analysis and identification of peptides using a protein sequence database and search algorithm. This approach is well suited to analysing PTMs, particularly for the smaller modifications (e.g., phosphorylation, methylation and acetylation), and is now used to identify and quantify different modified and unmodified proteoforms in parallel. Many smaller PTMs introduce a predictable mass shift which can be used to identify the site of modification. However, for larger modifications such as glycosylation, the path to identifying the site and composition of glycans attached to glycoproteins is more challenging, and database search algorithms are not particularly useful in most instances. Instead, various approaches using chemical labelling and cleavage of the glycans are used to identify the site of modification, and more importantly the structure of released glycans.
A major limitation of the bottom-up approach is introduced at the digestion step—the proteoform-origin of each individual peptidoform is lost. For modified peptides, each specific modification site represents a unique peptidoform to be quantified. However, in the context of expression proteomics, unmodified peptides are usually grouped together and differential analysis is used to determine the significance of any change in expression attributed to a change in sample conditions. This approach confounds the parallel quantification of individual peptidoforms arising from a modification and disconnects it from the protein level quantification of each protein/proteoform. It also masks accurate quantification of individual proteoforms arising from sequence mutations or splice variants. Peptide level quantification may yet play an important role in LC-MS/MS-based proteomics.
4.2. MS General Considerations: Stochastic Nature of Acquisition
The MS techniques used for many large-scale proteomic studies rely on Data Dependent Acquisition (DDA). Although modern MS instrumentation allows for increasingly fast data acquisition rates, providing ever increasing numbers of peptide-spectrum matches (PSMs), DDA is stochastic in nature, and missing values in individual DDA experiments lead to a degree of incompleteness in large data sets. Data Independent Acquisition (DIA) approaches such as the Sequential Windowed Acquisition of All Theoretical Fragment Ion Mass Spectra (SWATH-MS) can go some way to overcoming missing values in quantitative studies; however the libraries used to generate reference spectra for identification purposes are currently almost exclusively derived from DDA experiments—so the stochastic element of DDA still remains a challenge. Pooling samples from different groups, treatments or conditions in combination with sample fractionation such as high pH chromatography, can reduces the limitations of stochastic DDA experiments, and this approach has been used to successfully quantify PTMs in SWATH-MS experiments—SWATHProphetPTM [89]. To address missing values and the need for imputation, the inference of peptidoforms (IPF) approach, has been used to target the detection of phosphopetides and multiple PTM peptidoforms in plasma, demonstrating consistent detection and quantification characteristics which are required in large-scale PTM studies [5].
5. Enrichment Considerations
The low abundance of many of the studied PTMs limits the detection of some PTM classes by LC-MS/MS [84], so enrichment of modified peptides is needed to enhance their detection. Key to the detection of large numbers of modified peptides, affinity chromatography and immunoprecipitation techniques are used to substantially enrich PTMs and provide sufficient purification to aid the assessment of modification site-localisation by LC-MS/MS. In most cases the amount of starting material required is significantly larger than that required for expression-based proteomic studies (global proteome profiling), though efforts in the directions of using lower sample amounts have been made [90]. When combined with label and label free MS approaches, site-localisation, PTM occupancy and PTM stoichiometry for individual PTM peptidoforms can be assessed in parallel with global protein expression profiling.
Phosphorylation, glycosylation, methylation, acetylation and ubiquitination each represent distinctly different types of chemical modifications. Consequently, purification strategies have been developed to selectively enrich the different types of modified peptides prior to LC-MS/MS analysis. Although methylation, acetylation and phosphorylation are relatively small chemical modifications, they each impart significant charge-based changes to the amino acid residues they each occur on, often making detection by MS more challenging. Glycosylation and ubiquitination are larger modifications and they can introduce far more complex chemical alterations to proteins, again making MS detection challenging. The unique chemical nature of each type of PTM has led to the development of many highly specific enrichment strategies, with affinity chromatography and immuno-purification being used most often. The following section highlights some of the techniques commonly used to enrichment phosphorylated, glycosylated, methylated, acetylated and ubiquitinated proteins and the MS approaches used for site localisation and PTM quantitation.
5.1. Specific Enrichment Considerations
5.1.1. Phosphorylation
In recent years, phosphopeptide enrichment using immobilised metal affinity chromatography (IMAC), titanium dioxide (TiO2) chromatography, ion exchange chromatography (SCX and SAX), hydrophilic interaction chromatography (HILIC), and immunoprecipitation (IP) have been used very successfully in large-scale phosphoproteomic quantification studies. To achieve a comprehensive assessment of phosphorylation status, larger quantities of protein are generally required compared to amounts needed for proteome only expression profiling. Following extensive refinements of the protocols for phosphopeptide purification using TiO2 and IMAC, enrichment of the phosphopeptide fraction can now approaches 90% in most cases, and studies can now report the identification of tens of thousands of phosphopeptides [91]. LC-MS/MS and LC-MSn approaches using CID, ECD, ETD and photodissociation have proven extremely useful for phospho-site localisation and quantification of phosphopeptides (pS, pT and pY neutral loss of HPO3 79.9663 Da or H3PO4 97.9769).
Although many of the LC/MS-based approaches used in proteomics for large-scale proteome analysis are suitable for phosphopeptide detection and quantitation, some challenges still remain. Firstly, the sub-stoichiometric phosphorylation at any given site is governed by complex biological processes beyond just the addition and remove of phosphate through kinase and phosphates activity. Secondly, the phosphate moiety is relatively labile and the actual site of the modification, and certainly the potential for quantitation, can be hampered during MS analysis. Lastly, phosphopeptides that share the same amino acid sequence will have the same precursor ion mass—this itself can impact the detection of either sequence by conventional DDA methods. For phosphopeptides that share the same amino acid sequence and precursor ion, site localisation can be challenging, particularly if multiple S, T or Y residues are in close proximity within the amino acid sequence, as the number of sequence ions capable of distinguishing individual phosphopeptides from one another reduces.
5.1.2. Glycosylation
Approaches to assess protein glycosylation include the enzymatic release of N-glycans using N-glycosidases such as PNGase F, A or H+, or with various endogylycosidases such as endoglycosidase H, F1, F2, or F3, and the chemical release of O-glycans using reductive β-elimination under alkaline conditions. These approaches are often used to characterise the structural diversity of N- and O-linked glycans. Released glycans can be analysed in their reduced state, but quantitative analysis is complicated by the relatively labile nature of sialic acid. In such cases, derivatisation can be used to enhance quantitative analysis by MS or by incorporating chromophores to optimise quantification by UV or fluorescence detection. Glycoproteins can be enriched using either immunoaffinity and lectin affinity approaches [92], both demonstrating major advantages in glycoprotein analysis. HILIC is well suited to glycopeptide profiling, demonstrating highly specific enrichment of a broad range of glycopeptides.
Some major challenges exist for the site-specific analysis of glycopeptides by MS. The common approach involves enzymatic treatment of the glycoprotein followed by chromatographic separation and subsequent MS analysis of glycopeptides. Large glycopeptides generated using trypsin are poorly enriched using HILIC. Similarly, for MS analysis, some glycopeptides can suffer from poor ionisation efficiency, a high degree of structural heterogeneity among the different glycoforms and a lack of tandem MS spectral features to adequately characterise both the glycan and peptide backbone of the various glycoforms. In addition, identification of the site of glycosylation for N-glycans can be confounded due to the conversion of asparagine to aspartic acid following glycosidase treatment with PNGase F. The detection of aspartic acid could be used for the identification of the site of glycosylation; however, the deamidation of asparagine can result in the false identification of N-glycosites.
5.1.3. Methylation and Acetylation
Early approaches assessing methylation of arginine made use of immunoprecipitation strategies and have more recently been combined with strong cation exchange and HILIC to enhance the recovery and enrichment of methylarginine containing peptides. Given the localisation of methyl groups on arginine residues, Glu-C has been used in place of the commonly used trypsin to yield additional access to complementary fragmentation data for site localisation. Recent studies have identified large numbers of methylation sites using high resolution MS [93], and by using highly specific antibodies against mono and dimethyl arginine and lysine [94].
Acetylation (monoisotopic, Ac, 42.0106 Da) of proteins has been reported at the N-terminus of proteins [95] and at lysine residues. Previous attempts to assess the “acetylome” have made use of immunoaffinity enrichment strategies, which were limited in their ability to sufficiently enrich acetylated peptides. The most in-depth proteomic studies of lysine acetylation make use of immunoprecipitation of acetylated peptides using acetyl lysine antibodies followed by further enrichment and fractionation using strong cation exchange chromatography.
5.1.4. Ubiquitination
Affinity approaches using Ub specific antibodies, Ub remnant (anti diglycyl lysine antibodies), and tandem Ub binding domains have been used for enrichment of Ub proteins. The Ubisite approach, which makes us of a monoclonal antibody specific to ubiquitin and recognising remnant diglycyl lysine has demonstrated substantial enrichment of ubiquitinated proteins and was recently used to identify 63,000 ubiquitination site on 9200 proteins in two human cell lines [96].
Mass spectrometry analysis of ubiquinated peptides can result in the detection of a large number of false positives [97]. The remnant mass of ubiquitination on a target lysine residue is the diglycine sequence (Gly 75-Gly 76) (monoisotopic, 114.0429 Da). Early studies of ubiquitinated peptides on lower-resolution instruments may have resulted in false assignment of ubiquitin as leucine, isoleucine, asparagine and aspartic acid have residual masses within 1Da of the Gly–Gly remnant sequence of ubiquitin. On higher resolving instruments, asparagine (monoisotopic mass, 114.0429 Da) remains an issue when assigning ubiquitin as a PTM by tandem MS. Similarly, non-specific and artifactual alkylation of target peptides with iodoacetamide (carbamidomethylation) can result in over-alkylation (2 × 57.0215 or 114.0429 Da) of free amines at the N-terminus of peptides, or on the side chain of lysine, confound ubiquitination assignment.
5.1.5. Further Considerations: Multiple PTM Studies, Studies without Enrichment
Recent examples demonstrating the combined quantification of multiple PTM types in large-scale studies provide an important stepping-stone for future PTM studies. High throughput quantitative mass spectrometry is becoming increasingly useful at mitigating the issue of sensitivity, enabling screening of several biologically relevant PTMs en masse and with greater ease. PTM crosstalk is outside the scope of this work, but is comprehensively discussed elsewhere [98].
Although measuring both post-translationally modified and unmodified peptides species without the need for prior enrichment appears to be achievable, current mass spectrometry technology is not currently mature enough to handle such stochastic complexity on the fly. Thus, pre-enrichment of post-translationally modified peptides is currently an inevitable step in a large-scale PTM studies. That being said, groups have attempted to circumvent this issue by leveraging the speed and sensitivity of the Orbitrap class of mass spectrometers and achieved comprehensive coverage of the commonly studied PTMs including phosphorylation and N-acetylation without specific enrichment.
6. Pinpointing the Modification—Site Localisation Algorithms
Peptide identification is usually the first step for bioinformatics analysis in bottom-up MS-based proteomics, and the same is true for PTM analysis. In addition to protein identification with standard search engines, unambiguous modification site localisation is an important and challenging part in PTM identification, and several specific algorithms have been proposed for automatic PTM site localisation [99,100,101,102,103].
The challenge of site-localisation stems from two main difficulties. First the database search space will expand in combinatorial fashion for each modification included, thus increasing the computation cost and false positive rate for PTM identification [12]. Second, a peptide with the same amino acid sequence can have multiple same instances of the same amino acid residues and some modifications can modify different amino acid residues within the same sequence, for example, phosphorylation can modify tyrosine (T), serine (S) and threonine (Y) residues when all of them are present in the same peptide [12,103]. Unambiguously identifying a modification site is determined by the presence of one or more fragment ions [103]. For example, a phospho-tyrosine specific immonium ion is commonly used as diagnostic information to determine the presence of a phospho-tyrosine residue [104]. However, very often these ions may be lost or not be detected by mass spectrometers [12,99].
Ascore is one of the earliest automatic site localisation algorithms for phosphorylation [103]. It is a probability-base scoring algorithm based on the presence and intensity of site-determining ions in MS/MS spectra. Site-determining ions are those fragment ions that are exclusive to a site location for a PTM peptide. Ascore is one of the most popular site location algorithms and its performance has been verified in various studies [102,105]. However, although built as a “fit-for-purpose” tool, Ascore can only work for MS/MS and CID fragmentation. With the advent of new types of peptide fragmentation techniques including HCD, ETD and ECD, the fragment ions produced are different from those of the classical CID method, and various algorithms, including Andromeda [100], PhosphoRS [105] and SLoMo [106], have been proposed to cater for these extended fragmentation methods. Mascot Delta Score, a method using the difference between the top two matches of modification sites for the same peptides in the database search, can be used for arbitrary PMT types but is limited to Mascot search results [101,102]. Most peptide search engines have now incorporated one or more PTM site localisation algorithms for PTM identification. For example, ProteinPilot (V5 Sciex) Parogon algorithm has ID focus options, which allow the additional consideration of large sets of biological post-translational modifications [107]; SEQUEST [108] uses the Ascore algorithm; MaxQuant [109] exploits the Andromeda algorithm [100]; and Mascot uses the MD score method [110].
Unlike the direct search approach used in DDA, DIA adopts a mining approach which uses a pre-built spectral library for peptide and protein identification and quantitation [111]. This approach applies to both modified and un-modified peptides. For the PTM peptides to be identified and quantified by DIA or SWATH, both the precursor ions and their fragment ions need to be included in the spectral library [112]. The quality and completeness of the spectral library is a key element to the success of PTM peptide identification. The generation of the peptide library is usually done by combining many runs of DDA experiments [111,112], using DIA pseudo spectra [113] or extending a local spectra library with one or more archived or external libraries [114,115]. Therefore, the traditional site localisation methods for DDA can also apply to DIA.
Besides these DDA-based localisation methods, some recent work has proposed methods of inferring PTM peptides directly from the spectral library [5,89]. Rosenberger et al. proposed the IPF (Inference of PeptideoForms) algorithm which can independently infer modified peptides with specific site-localization using a site-localized or unlocalized spectral library [5]. IPF uses a posterior probability estimation and Bayesian hierarchical model for site localisation. IPF has been integrated into the OpenSwath software [116]. In another similar work, Keller et al proposed a method that infers the putative PTM from the mass shifts exhibited by the precursor and missing fragment ions in the lower-ranking peak groups [89]. This functionality has been added to SWATHProphet [117]. For a more complete review on site localisation, please refer to [12,99].
There are still some limitations to the current PTM identification methods. First, the number of modifications included in each search cannot be too large due to the search space expansion, with an upper limit being about 6–10 modifications [107]. Second, though most search engines can report the estimated confidence of identification on both peptide level or dataset level using an expected score or FDR (False Discovery Rate) based on a target-decoy strategy [118], not many search engines can directly output the site assignment confidence or False Localisation Rate (FLR). Some efforts have been made with either synthetic peptides with known modification sites [102,105] or peptides with only one potential modification sites [119].
7. Understanding the Reproducibility of PTM Quantitation
While site localisation will enable the confident identification of modified peptides via the computational strategies described before, it is their quantification and its change with condition that is crucial to inferring relevance to disease. For this quantitative analysis to be useful, the data collected has to be of sufficient quality to enable it, and the sample size has to be adequately matched to the analysis goals. A useful discussion on sample size in the specific context of large scale PTM analyses can be found in [16]. There are many sources of variation at PTM level in proteomics—from biological, sample preparation through to LC-MS and technical instrument related [10]; these are further enhanced for PTMs, which, through their dynamic nature, can introduce increased variability.
There are two facets to quantitative variability: that of identification, such as determined for instance by the percentage of replicates in which a modification can be measured, and that of quantitation, such as measured by for instance by coefficients of variance. In terms of identification, in the case of PTMs, the overlap between replicates even for cell cultures can be quite low [16]. This low overlap will lead to a high proportion of missing values to be tackled in the subsequent analysis; the issue of missing data is of course not unique to the PTM scenario, where it stems from the stochastic aspect of peptide identification [120,121], with many methods initially developed to handle it in the context of microarrays [122] and in specific proteomics context [123]. The better resolution of current MS instruments and the increased uptake of multiplexed and DIA methods can contribute to lowering the MS variability. As demonstrated in a very recent benchmarking experiment, missing data is less of an issue with TMT though the problem accumulates when a larger number of runs is required, but more so with label free data [6].
Since the label free quantification of the PTM peptides is gaining increasing attention, several measures have been taken to overcome the missing identifications of peptides and PTM peptides. Stochastic effects caused due to sampling issues in DDA are overcome by introducing more biological replicates thus reducing the missing values. Additionally, software such as MaxQuant addresses this issue at the data processing level by performing matching between runs. Through this approach, the software transfers the identification (and thus quantitation) when there is a MS1 feature but not successful MS2 spectra for the corresponding feature. In the DIA space, a SWATH study assessing the quantification of peptidoforms [5] has been used to quantify the presence of PTMs in replicates of a large scale study of plasma from 116 twins, and finds a good median detectability of modifications in the range of 50–100 samples.
Where a large percentage of data is missing across samples, data imputation may be needed, though the validity of imputation is often a hotly debated topic among statisticians; this remains so in the case of PTM analysis. Schwammle et al. [124] recommend against PTM data imputation strategies using methods adopted from proteomics or transcriptomic studies, favouring novel analysis models that handle data absence by different statistical approaches, such as presence-absence models based on Binomial likelihood [125], combining statistical models that assess qualitative differential observation and differential expression [126] or using specific approaches tailored to large scale proteomic studies [127]. However, other established proteomic pipelines make use of data imputation adapted from microarray experiments. For instance the popular impute R package developed for microarray data analysis is part of well-established workflows [6].
Popular packages such as Perseus offer data imputation options including the restriction of values present in a certain percentage of samples, and allow imputing random data from a specific distribution, typically normal. The potential positive effect of matching between runs and imputation with random values strategies is well illustrated in [128]. When choosing whether or how to impute, one should carefully consider several factors: presence of replicates (as imputation will be improved if data can be consolidated across technical replicates), percentage of values imputed (if a very large proportion of the dataset ends up being imputed, then methods of imputing for sparse datasets should be well understood), data distribution after analysis, the impact of imputation (does the imputation modify the resulting data distribution), and, crucially, the stability of the obtained results under imputation. If values sampled from a random distribution are generated to fill in missing data, are the obtained results stable when a different set of random values are imputed, or will a different set of differentially expressed features be obtained for each imputation run? An evaluation of stability can be done by a bootstrapping exercise, or at least simply by repeating the imputation steps more than once and evaluating the effect on differential expression.
A second facet of reproducibility is that of quantitation, typically captured through calculating coefficients of variation or correlations among technical replicates. For clinical samples, understanding coefficients of variance for technical replicates is paramount. In a study demonstrating a new PTM enrichment method combining immunoaffinity purification and LC-MS/MS without depletion, the authors demonstrate reproducibility for plasma technical triplicates in the vicinity of 20% [69]. The large-scale aforementioned twin plasma SWATH study quantifies biological CVs for each modification, with median values in the range of 30%, reporting much lower technical and whole process variability [5]. Alternatively, reproducibility of PTM quantitation can be captured via correlations of technical or biological replicates; for instance phosphopeptide abundance correlations of over 80% were determined for biological replicates using a MaxQuant-based label-free platform [129]. In addition, at a higher level, quantitative reproducibility can be demonstrated by showing conserved processes and motifs [6], and in a more visual manner via multivariate analysis, as we discuss next.
8. Understanding the Changing Levels of Modified Peptides in Context
The extraction of PTM quantitation and possible data imputation is usually followed by normalisation steps, multivariate analysis such as clustering and PCA, and differential expression of the modified peptides. These steps will typically follow the established methodology employed for general shotgun proteomic data generated using the same platform, be it labelled (metabolic or chemical), label free or DIA/SWATH. However, the increased complexity of the PTM multivariate data stems from two aspects: it is carried out at the peptide and even site level, hence yields larger datasets, and it is tightly coupled with the un-modified and protein expression level. We will first briefly discuss the multivariate analysis options, and then in more detail the relationship with protein and un-modified peptide quantitation.
A very recent review [130] describes in visual detail the downstream statistical data analysis options for shotgun proteomics in general, and highlights many of the steps can be carried out in the popular Perseus software suite [109]. Data normalisation is a necessary first step, usually carried out to remove variation stemming from uneven sample loading, with commonly employed methods including total area normalisation, median, quantile-quantile, variance stabilising normalisation, loess and many others. A systematic evaluation of normalisation methods in the context of label free proteomics using public standardised datasets can be found in [131]. The basic statistical tests used for differential expression between conditions remain the standard approaches such as ANOVA or t-tests for differential expression, or their variants such as moderated t-tests accounting for variance shrinkage across the whole dataset [132]. Developed initially in the context of microarrays, these methods are available in the popular R packages limma [133] and SAM [134], with the latter being used in the phosphoproteomics specific context [6].
Because PTMs are quantified at the peptide rather than protein level, the scale of the resulting datasets will be much larger, hence approaches to limit false discoveries when doing repeated tests must be considered. Most commonly used methods include Benjamini and Hochberg (BH) corrections [135], Storey and Tibshirani q-values [136] or more stringent cut-offs if deemed appropriate. For instance, the TMT protocol [8] employed Student t-test BH-corrected p-values 0.001 in addition to fold change requirements of 1.75.
Importantly, a change in abundance in a PTM peptide across conditions of interest could either reflect a change in post translation modification patterns, or a change in abundance of the protein itself [137], thus determining protein abundance changes side-by-side with the PTM changes is crucial. This was demonstrated in a seminal phosphorylation study [138] showing that 25% of the changes in differentially expressed phosphopeptides could be attributed to protein expression changes, thus underscoring the need to account for protein amount normalisation, or at any rate side by side comparison of protein and PTM ratios. However, the process of specific PTM enrichment (if carried out) uncouples the PTM abundance from the protein amount [12], potentially making such normalisation difficult unless the enrichment flow-through is retained.
The stoichiometry—also known as site occupancy—represents the fraction of modified peptides as a percentage of the total protein amount, and provides insight in the down-stream analysis, as a high occupancy coupled with differential regulation of the modification can be a good indicator that the modification is functional [84]. Determining it is a novel part of the PTM data analysis, one not covered by the standard methods for multivariate analysis described above, and one that can be particularly challenging for modifications like phosphorylation, which are both occurring on a rapid time scale, and at low abundance. Stoichiometry is difficult to determine in a standard mass spectrometry PTM analysis, due to the different behaviour of modified and un-modified peptides which makes it hard to compare them directly [139]. If the relative change in protein amounts is captured alongside the relative change in modified peptides, then the relative change in stoichiometry can be understood; however the absolute occupancy will not be directly known [140]. A workflow for determining absolute phosphorylation stoichiometry using stable isotope labelling on a proteome-wide scale has been described in [140], and further improvements have been made to it by using isobaric labelling in a 10-plex TMT setup [141]. In the label-free scenario, Sharma et al describe a computational strategy for determining fractional occupancy of phosphorylated peptides using MaxQuant [129].
9. Online Tools for Subsequent Analysis
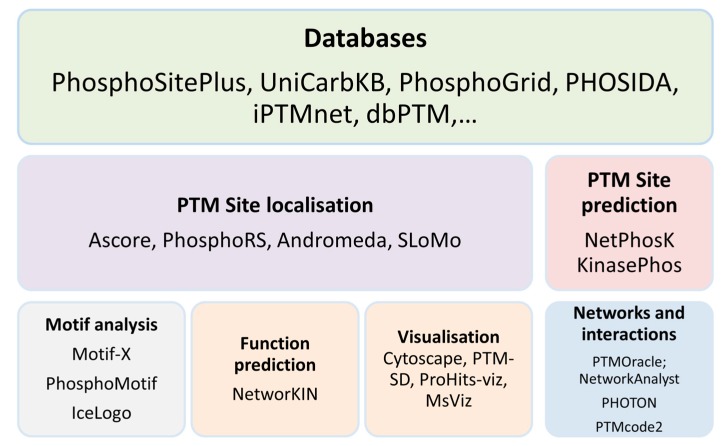
Once PTMs are identified and quantitated, there are numerous tools available for subsequent analysis, and of course numerous databases underpinning the analysis and making prediction possible; PTM databases have been previously reviewed in depth [142]. There are a variety of broad tool categories (databases, tools dealing with site localisation and prediction, motif analysis, function prediction and interaction—Figure 3 captures the main tool categories), and below we highlight some of the more recent additions (Table 2 and Table 3). In addition, due to the more complex challenges of glycan analysis, a vast array of glycoproteomics specific informatics tools have been developed—a recent in-detail book chapter surveys databases and tools in the specific glycomics context [143].
Figure 3.
PTM tool categories, and a few highlighted examples.
Table 2.
PTM annotation databases examples.
| Name | Description | Accessibility | Reference |
|---|---|---|---|
| DEPOD | Manually curated database of human active phosphatases | http://www.depod.bioss.uni-freiburg.de/ | [144] |
| Phospho.ELM | Relational database of in vivo and in vitro phosphorylation data | http://phospho.elm.eu.org/ | [145] |
| UniProt-GOA | Database of gene ontology annotations to UniProt Proteins | https://www.ebi.ac.uk/GOA | [161] |
| PHOSIDA | Database of phosphorylation data from in-house proteomics studies |
https://www.biochem.mpg.de/1144243/Phosida
http://www.phosida.de/ |
[162] |
| PhosphoSitePlus | Manually curated resource of experimentally determined Human and Mouse PTMs | https://www.phosphosite.org/homeAction.action | [163] |
| PhosphoGrid | Database of experimentally determined PTM sites in Saccharomyces cerevisiae | https://phosphogrid.org/ | [150] |
| UniCarbKB | Curated knowledgebase for glycomics and glycobiology research | http://www.unicarbkb.org/ | [151,164] |
| iPTMnet | An integrated database of PTMs in systems biology proteins | https://research.bioinformatics.udel.edu/iptmnet/ | [152] |
| PoGo | Mapping of peptides with PTMs and quantitation to reference genome annotation. | https://www.sanger.ac.uk/science/tools/pogo | [165] |
| PhosPhAt 4.0 | Database and predictor of Arabidopsis thaliana phosphorylation sites based on mass spectrometry experiments. | http://phosphat.uni-hohenheim.de/ | [166] |
| P3DB (Plant Protein Phosphorylation DataBase) | An integrated resource of proteins phosphorylation sites for different plants | http://www.p3db.org/ | [167,168] |
| Human Proteinpedia (Human Protein Reference Database) | An integrated resource of proteins annotations including PTMs derived from different experimental techniques | http://www.humanproteinpedia.org/ | [169] |
| dbPTM | An integrated database of experimentally verified phosphorylation sites encompassing structural and functional analysis along with disease associations | http://dbptm.mbc.nctu.edu.tw/ | [170] |
Table 3.
Examples of recent MS-specific PTM related tools.
| Name | Description | Accessibility | Reference |
|---|---|---|---|
| Specialize (Spectra of complex-PTModified peptides identification tool) | A tool to identify peptides and proteins with PTMs from MS spectra | http://proteomics.ucsd.edu/softwaretools/specialize/ | [171] |
| Protein Prospector | A set of tools to detect and identify the PTMs in through searching for mass shifts in MS spectra | http://prospector.ucsf.edu/prospector/mshome.htm | [172] |
| Byologic, Byonic, Intact Mass, Byomap (Glyco Analysis) | A set of tools to detect and identify N- and O- linked glycans in peptides gathered through MS | https://www.proteinmetrics.com/workflows/ | [173] |
| ScaffoldPTM (ProteomeSoftware) | A tool based on Ascore algorithm to evaluate the assignment of PTM sites on MS-based identified peptides | http://www.proteomesoftware.com/products/ptm/ | [103,174] |
| PhoshoPep 2.0 | Set of tools to study proteins pathways and interactions for MS-derived phosphorylation data from Drosophila melanogaster, Homo sapiens, Caenorhabditis elegans and Saccharomyces cerevisiae. | http://www.unipep.org/phosphopep/index.php | [175] |
| ProteomeScout | An integrated resource of proteins PTMs, experimental data, analysis suite and visualizations. | https://proteomescout.wustl.edu/ | [176] |
| MsViz | An interactive software for manual validation and relative quantitation of PTMs acquired through MS experiments along with spectra visualizations. | http://msviz-public.vital-it.ch/#/about | [160] |
| ProSight Lite | A software tool to align a protein sequence and its PTMs and glycosylation site against MS spectra | http://prosightlite.northwestern.edu/ | [177] |
Many public databases are available for searching for PTM annotation, some are type-specific such as DEPOD [144] and Phospho.ELM [145] and others cover more modification types such as Uniprot [146], PHOSIDA [147], PTMcode [148]. Some only include experimentally verified PTMs such as Phospho.ELM, PhosphoSitePlus [149], PhosphoGrid [150] and UniCarbKB [151], some include known and predicted functional annotations between PTMs such as iPTMnet [152]. Refer to [153,154] for good reviews of PTM databases and the modification type coverage. Building on from the available annotation, PTM site prediction tools leverage the available information in PTM databases, and machine learning algorithms, to predict new PTM sites and binding motifs.
Motif analysis can provide useful sequence pattern information for all identified PTM proteins, and helps identify the significant motifs which, in turn, based on known substrate specificity, can ideally help identify the enzymes involved—though the latter part is still extremely challenging. Motif-x is arguably the most popular tool for identifying the significant motifs in existing PTM peptides [155]. Many studies have used Motif-x in their PTM analysis as a validation of existing motifs or discovery of novel motifs [9,156,157].
Many of the existing PTM analysis tools offer great data visualisation features. For example, Cytoscape with its many visualization apps including PTMOracle and STRING for PPI and networks; Protein data bank and PTM-SD for protein structures [158]; ProHits-viz [159], a suite of web tools for visualizing interaction proteomics data; MsViz [160], a graphical tool for manual validation and quantification of PTMs for small to medium scale experiments.
10. Methods of Functional Analysis
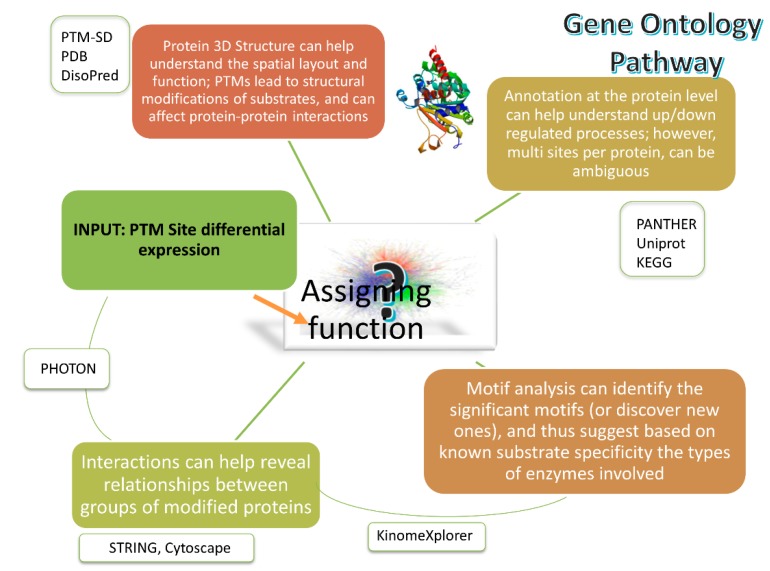
A PTM can either modify protein structures, regulate functions or add a new group [11]. After the quantification analysis of PTM data, and accessing available databases to check whether the PTM/motif is known, we need to understand the complex circuitry of cell signal transmission that the PTM impacts. Functional analysis aims to bring the quantitative PTM analysis into biological contexts based on annotations, protein structures, protein interactions, pathways and networks and interpret the data at the biology level (Figure 4). In this section, we review the general methods currently available for PTM functional analysis.
Figure 4.
Areas relevant to the task of predicting functions of PTM modifications. Several integrative approaches are emerging. Orange arrow indicates the input to the functional analysis tools; green lines indicate the various aspects of PTM functional analysis.
Using the annotations in publicly available databases can help PTM functional analysis from various aspects. Firstly, using ontology categories, such as GeneOntology (GO) and PhosphoSite ontology, can help classify and understand the identified PTMs from the functional levels. For example, the GO molecular function and biological process categories retrieved from PANTHER [178] were used to compare phosphoproteins and total proteins in porcine muscle [157]. A distribution of phosphoprotein types was generated by using the PhosphoSite ontology to classify phosphoproteins of lung cancer cell lines and tumors [23].
Post translational modifications yield structural changes in the substrate, which in turn can affect function and protein-protein interactions—hence 3D structure analysis is important for understanding functional relationships of PTM proteins. The Protein Data Bank [179] is an extensive publicly available data repository of protein 3D structures. PTM-SD [158] provides structurally resolved and experimentally annotated PTMs in protein structures in 3D views. Sharman et al. used DisoPred software [180] to predict disorder signal state for all proteins and mapped to phosphorylated sites and found that protein phosphorylation tends to occur to disordered regions in stimulated cells while independent of structures in unstimulated cells [129].
Protein-protein interaction (PPI), network and pathway analysis leverage PTM functional analysis from single-protein-based to group-of-proteins-based. There are many tools available for protein interaction studies, and some of the most popular ones are listed in Table 4. The group of proteins is usually a set of proteins of biological interest, such as the set of proteins showing a common regulation trend from the quantification analysis. By putting these proteins in the pathway or network context, protein lists are mapped to biological pathways or PPI networks which aids the interpretations and visualisation. There are several tools available for pathway and network analysis for general proteomics data and they vary in the use of function and topological information [181]. However, pathways and networks analysis tools specific for PTMs are still very limited [182]. Pathway and network analysis can offer insight into PTM functions and discover novel pathways or networks; for example, important PPI networks were discovered and different regulatory metabolism mechanisms were revealed for phosphoproteins in porcine muscle proteins by using STRING [9,157]. PTM crosstalk analysis, which aims to identify relationships between different types of PTMs, can also be aided by PPI networks. For example, Grimes et al. integrated three types of PTMs, i.e., phosphorylation, methylation and acetylation, in lung cancer cell lines and outlined lung cancer cell signalling networks [183].
Table 4.
Tools for protein interaction analyses that can also be useful in PTM context.
| Name | Description | Accessibility | Reference |
|---|---|---|---|
| STRING | A database of known and predicted protein–protein interactions from experimental and knowledgebase sources | https://string-db.org/ | [190] |
| NetworkKIN | An approach for motif-based predictions of kinases and phosphoproteins | http://networkin.info/ | [184] |
| Cytoscape | An open source software for molecular interaction networks integration, analysis and visualization, along with gene annotations | https://cytoscape.org/ | [191] |
| ProHits-viz | A suit of tools to perform analysis and visualization of quantitative protein interaction data | https://prohits-viz.lunenfeld.ca/ | [159] |
| PTMOracle | A Cytoscape application for co-visualization and co-analysis of PTMs and protein-protein interactions | http://apps.cytoscape.org/apps/ptmoracle | [192] |
| NetworkAnalyst | An integrated tool to perform gene annotations and protein-protein interaction network analysis along with visualizations | http://www.networkanalyst.ca/ | [193] |
Recent studies combine several of these approaches; for instance, the workflow in [156] starts from the extraction of crosstalk motifs for human PTM peptides downloaded from PhosphoSitePlus using Motif-x and then goes through gene ontology enrichment analysis, kinase analysis with NetworkKIN [184] and network analysis using Cytoscape with the GeneMANIA app [185].
For phosphorylated proteins, kinase analysis can reveal the relationships between phosphorylation sites and the protein kinases. NetworkKIN is a popular tool to model phosphorylation networks by using Kinase-substrate relationships. Qi et al. re-constructed kinase-substrate phosphorylation networks by using predicted site-specific kinase-substrate relation for mouse testis [186]. Rikova et al. characterized tyrosine kinase signalling across non-small cell lung cancer cell lines and tumors and identified known and novel oncogenic kinases [23]. In addition, kinase-substrate enrichment analysis can be used to performed enrichment analysis for phosphorylated substrate groups [187].
Recent integrative approaches have combined several of the steps outlined above, in order to help get closer to extracting meaningful information rather than just providing long lists of PTM sites. The PHOTON tool [188] takes as inputs, sets of differentially quantitated PTMs as arising from quantitative workflows reviewed here, and protein-protein interaction data, and through network-based statistical modeling, generates scores to identify significantly functional signalling proteins. Other developing approaches include modified versions of the single sample gene set enrichment analysis approach [189] tailored to the PTM specific context (PTM-SEA making use of PTMsigDB database). Such tools, when mature and in common use, will speed up the latter, difficult part of PTM functional analysis.
11. Concluding Remarks
The interest in post translational modifications is growing and will continue to be sustained, due not just to increased understanding, but to the practical importance and likely payoff for such studies in the clinic. As quantitative techniques such as TMT and SWATH mature, they will yield increased information in the databases and hence better capacity for the predictive tools, which continue to develop. While keeping in mind the caveats about correct site assignments and some of the other MS related challenges remaining, arguably the part of the workflow ending with generating relative quantitation and relative stoichiometries is getting increasingly reliable and routine. Analysing several PTMs at the same time and PTM crosstalk remains difficult, and for many modifications there is little available data and few usable tools. Function prediction remains a difficult step, though we have seen the beginning of computational tools blending protein interaction data with PTM abundance ratios to generate functional predictions, which constitutes a big step forward. However, validation of the results will still be key, at many levels, from the predicted site assignments, predictive tools performance and all the way through to the predicted enzymes activity.
Acknowledgments
The authors acknowledge the support from the National Health and Medical Research Council (NHMRC) and the Australian Government’s National Collaborative Research Infrastructure Scheme (NCRIS). The authors would like to thank Alison Rodger for the critical reviewing and editing of the manuscript.
Author Contributions
M.M., D.P., J.X.W. and M.J.M. conceptualized the review article. D.P., J.X.W., M.J.M., C.J., Z.N., K.K., Y.W., S.R., V.G. and M.M. contributed in writing, review and editing of the original draft. D.P. compiled the manuscript and M.M. supervised the study.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bryson B., Roberts W. A Short History of Nearly Everything. Volume 33 Broadway Books; New York, NY, USA: 2003. [Google Scholar]
- 2.Cohen P., Tcherpakov M. Will the ubiquitin system furnish as many drug targets as protein kinases? Cell. 2010;143:686–693. doi: 10.1016/j.cell.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 3.Moslehi J.J. Cardiovascular toxic effects of targeted cancer therapies. N. Engl. J. Med. 2016;375:1457–1467. doi: 10.1056/NEJMra1100265. [DOI] [PubMed] [Google Scholar]
- 4.Skaar J.R., Pagan J.K., Pagano M. SCF ubiquitin ligase-targeted therapies. Nat. Rev. Drug Discov. 2014;13:889. doi: 10.1038/nrd4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberger G., Liu Y., Röst H.L., Ludwig C., Buil A., Bensimon A., Soste M., Spector T.D., Dermitzakis E.T., Collins B.C. Inference and quantification of peptidoforms in large sample cohorts by SWATH-MS. Nat. Biotechnol. 2017;35:781. doi: 10.1038/nbt.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hogrebe A., von Stechow L., Bekker-Jensen D.B., Weinert B.T., Kelstrup C.D., Olsen J.V. Benchmarking common quantification strategies for large-scale phosphoproteomics. Nat. Commun. 2018;9:1045. doi: 10.1038/s41467-018-03309-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J., Wang F., Ye M., Zou H. Enrichment and separation techniques for large-scale proteomics analysis of the protein post-translational modifications. J. Chromatogr. A. 2014;1372:1–17. doi: 10.1016/j.chroma.2014.10.107. [DOI] [PubMed] [Google Scholar]
- 8.Navarrete-Perea J., Yu Q., Gygi S.P., Paulo J.A. Streamlined Tandem Mass Tag (SL-TMT) Protocol: An Efficient Strategy for Quantitative (Phospho) proteome Profiling Using Tandem Mass Tag-Synchronous Precursor Selection-MS3. J. Proteome Res. 2018;17:2226–2236. doi: 10.1021/acs.jproteome.8b00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang H., Scheffler T.L., Gerrard D.E., Larsen M.R., Lametsch R. Quantitative proteomics and phosphoproteomics analysis revealed different regulatory mechanisms of Halothane and Rendement Napole genes in porcine muscle metabolism. J. Proteome Res. 2018;17:2834–2849. doi: 10.1021/acs.jproteome.8b00294. [DOI] [PubMed] [Google Scholar]
- 10.Mnatsakanyan R., Shema G., Basik M., Batist G., Borchers C.H., Sickmann A., Zahedi R.P. Detecting post-translational modification signatuRes. as potential biomarkers in clinical mass spectrometry. Expert Rev. Proteom. 2018;15:515–535. doi: 10.1080/14789450.2018.1483340. [DOI] [PubMed] [Google Scholar]
- 11.Thygesen C., Boll I., Finsen B., Modzel M., Larsen M.R. Characterizing disease-associated changes in post-translational modifications by mass spectrometry. Expert Rev. Proteom. 2018;15:245–258. doi: 10.1080/14789450.2018.1433036. [DOI] [PubMed] [Google Scholar]
- 12.Fert-Bober J., Murray C.I., Parker S.J., Van Eyk J.E. Precision profiling of the cardiovascular post-translationally modified proteome: Where there is a will, there is a way. Circ. Res. 2018;122:1221–1237. doi: 10.1161/CIRCRESAHA.118.310966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zavialova M., Zgoda V., Nikolaev E. Analysis of the role of protein phosphorylation in the development of diseases. Biochem. (Mosc.) Suppl. Ser. B Biomed. Chem. 2017;11:203–218. doi: 10.1134/S1990750817030118. [DOI] [PubMed] [Google Scholar]
- 14.Abou-Abbass H., Abou-El-Hassan H., Bahmad H., Zibara K., Zebian A., Youssef R., Ismail J., Zhu R., Zhou S., Dong X., et al. Glycosylation and other PTMs alterations in neurodegenerative diseases: Current status and future role in neurotrauma. Electrophoresis. 2016;37:1549–1561. doi: 10.1002/elps.201500585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wende A.R. Post-translational modifications of the cardiac proteome in diabetes and heart failure. Proteom. Clin. Appl. 2016;10:25–38. doi: 10.1002/prca.201500052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pagel O., Loroch S., Sickmann A., Zahedi R.P. Current strategies and findings in clinically relevant post-translational modification-specific proteomics. Expert Rev. Proteom. 2015;12:235–253. doi: 10.1586/14789450.2015.1042867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cork L.C., Sternberger N.H., Sternberger L.A., Casanova M.F., Struble R.G., Price D.L. Phosphorylated neurofilament antigens in neurofibrillary tangles in Alzheimer’s disease. J. Neuropathol. Exp. Neurol. 1986;45:56–64. doi: 10.1097/00005072-198601000-00005. [DOI] [PubMed] [Google Scholar]
- 18.Sternberger N.H., Sternberger L.A., Ulrich J. Aberrant neurofilament phosphorylation in Alzheimer disease. Proc. Natl. Acad. Sci. USA. 1985;82:4274–4276. doi: 10.1073/pnas.82.12.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grundke-Iqbal I., Iqbal K., Tung Y.C., Quinlan M., Wisniewski H.M., Binder L.I. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc. Natl. Acad. Sci. USA. 1986;83:4913–4917. doi: 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hee-Jin K., Kim S.H., Choi H. Tau phosphorylation at specific site as possible biomarker of clinical severity in alzheimer’s disease. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2014;10:P809. doi: 10.1016/j.jalz.2014.05.1587. [DOI] [Google Scholar]
- 21.Fujiwara H., Hasegawa M., Dohmae N., Kawashima A., Masliah E., Goldberg M.S., Shen J., Takio K., Iwatsubo T. α-Synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 2002;4:160. doi: 10.1038/ncb748. [DOI] [PubMed] [Google Scholar]
- 22.Colom-Cadena M., Pegueroles J., Herrmann A.G., Henstridge C.M., Muñoz L., Querol-Vilaseca M., Martín-Paniello C.S., Luque-Cabecerans J., Clarimon J., Belbin O., et al. Synaptic phosphorylated α-synuclein in dementia with Lewy bodies. Brain. 2017;140:3204–3214. doi: 10.1093/brain/awx275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., et al. Global Survey of Phosphotyrosine Signaling Identifies Oncogenic Kinases in Lung Cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 24.Jagarlamudi K.K., Hansson L.O., Eriksson S. Breast and prostate cancer patients differ significantly in their serum Thymidine kinase 1 (TK1) specific activities compared with those hematological malignancies and blood donors: Implications of using serum TK1 as a biomarker. BMC Cancer. 2015;15:66. doi: 10.1186/s12885-015-1073-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shults K., Flye L., Green L., Daly T., Manro J.R., Lahn M. Patient-derived acute myeloid leukemia (AML) bone marrow cells display distinct intracellular kinase phosphorylation patterns. Cancer Manag. Res. 2009;1:49–59. doi: 10.2147/cmar.s5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C., Wang X., Fang J., Xue J., Xiong X., Huang Y., Hu J., Ling K. EGFR-induced phosphorylation of type Iγ phosphatidylinositol phosphate kinase promotes pancreatic cancer progression. Oncotarget. 2017;8:42621–42637. doi: 10.18632/oncotarget.16730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dushukyan N., Dunn D.M., Sager R.A., Woodford M.R., Loiselle D.R., Daneshvar M., Baker-Williams A.J., Chisholm J.D., Truman A.W., Vaughan C.K., et al. Phosphorylation and Ubiquitination Regulate Protein Phosphatase 5 Activity and Its Prosurvival Role in Kidney Cancer. Cell Rep. 2017;21:1883–1895. doi: 10.1016/j.celrep.2017.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rapundalo S.T. Cardiac protein phosphorylation: Functional and pathophysiological correlates. Cardiovasc. Res. 1998;38:559–588. doi: 10.1016/S0008-6363(98)00063-7. [DOI] [PubMed] [Google Scholar]
- 29.Saez-Valero J., Fodero L.R., Sjogren M., Andreasen N., Amici S., Gallai V., Vanderstichele H., Vanmechelen E., Parnetti L., Blennow K., et al. Glycosylation of acetylcholinesterase and butyrylcholinesterase changes as a function of the duration of Alzheimer’s disease. J. Neurosci. Res. 2003;72:520–526. doi: 10.1002/jnr.10599. [DOI] [PubMed] [Google Scholar]
- 30.Saez-Valero J., Sberna G., McLean C.A., Small D.H. Molecular isoform distribution and glycosylation of acetylcholinesterase are altered in brain and cerebrospinal fluid of patients with Alzheimer’s disease. J. Neurochem. 1999;72:1600–1608. doi: 10.1046/j.1471-4159.1999.721600.x. [DOI] [PubMed] [Google Scholar]
- 31.Saez-Valero J., Barquero M.S., Marcos A., McLean C.A., Small D.H. Altered glycosylation of acetylcholinesterase in lumbar cerebrospinal fluid of patients with Alzheimer’s disease. J. Neurol. Neurosurg. Psychiatry. 2000;69:664–667. doi: 10.1136/jnnp.69.5.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silveyra M.X., Cuadrado-Corrales N., Marcos A., Barquero M.S., Rabano A., Calero M., Saez-Valero J. Altered glycosylation of acetylcholinesterase in Creutzfeldt-Jakob disease. J. Neurochem. 2006;96:97–104. doi: 10.1111/j.1471-4159.2005.03514.x. [DOI] [PubMed] [Google Scholar]
- 33.Moran L.B., Hickey L., Michael G.J., Derkacs M., Christian L.M., Kalaitzakis M.E., Pearce R.K., Graeber M.B. Neuronal pentraxin II is highly upregulated in Parkinson’s disease and a novel component of Lewy bodies. Acta Neuropathol. 2008;115:471–478. doi: 10.1007/s00401-007-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ludemann N., Clement A., Hans V.H., Leschik J., Behl C., Brandt R. O-glycosylation of the tail domain of neurofilament protein M in human neurons and in spinal cord tissue of a rat model of amyotrophic lateral sclerosis (ALS) J. Biol. Chem. 2005;280:31648–31658. doi: 10.1074/jbc.M504395200. [DOI] [PubMed] [Google Scholar]
- 35.Milde-Langosch K., Schütze D., Oliveira-Ferrer L., Wikman H., Müller V., Lebok P., Pantel K., Schröder C., Witzel I., Schumacher U. Relevance of βGal–βGalNAc-containing glycans and the enzymes involved in their synthesis for invasion and survival in breast cancer patients. Breast Cancer Res. Treat. 2015;151:515–528. doi: 10.1007/s10549-015-3425-0. [DOI] [PubMed] [Google Scholar]
- 36.Wu Y.-M., Liu C.-H., Huang M.-J., Lai H.-S., Lee P.-H., Hu R.-H., Huang M.-C. C1GALT1 Enhances Proliferation of Hepatocellular Carcinoma Cells via Modulating MET Glycosylation and Dimerization. Cancer Res. 2013;73:5580–5590. doi: 10.1158/0008-5472.CAN-13-0869. [DOI] [PubMed] [Google Scholar]
- 37.Incani M., Sentinelli F., Perra L., Pani M.G., Porcu M., Lenzi A., Cavallo M.G., Cossu E., Leonetti F., Baroni M.G. Glycated hemoglobin for the diagnosis of diabetes and prediabetes: Diagnostic impact on obese and lean subjects, and phenotypic characterization. J. Diabetes Investig. 2015;6:44–50. doi: 10.1111/jdi.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang Y., Geng Y., Luo J., Shen W., Zhu W., Meng C., Li M., Zhou X., Zhang S., Cao J. Downregulation of ubiquitin inhibits the proliferation and radioresistance of non-small cell lung cancer cells in vitro and in vivo. Sci. Rep. 2015;5:9476. doi: 10.1038/srep09476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao G., Wang R., Sun A., Wei J., Peng K., Dai Q., Yang W., Lin Q. The E3 ubiquitin ligase NEDD4 mediates cell migration signaling of EGFR in lung cancer cells. Mol. Cancer. 2018;17:24. doi: 10.1186/s12943-018-0784-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanarico A.G., Ronchini C., Croce A., Memmi E.M., Cammarata U.A., De Antoni A., Lavorgna S., Divona M., Giacò L., Melloni G.E.M., et al. The E3 ubiquitin ligase WWP1 sustains the growth of acute myeloid leukaemia. Leukemia. 2017;32:911. doi: 10.1038/leu.2017.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen J.L., Wen X.F., Li R.B., Jin Y.C., Wang X.L., Zhou L., Chen H.X. UBE3C promotes growth and metastasis of renal cell carcinoma via activating Wnt/beta-catenin pathway. PLoS ONE. 2015;10:e0115622. doi: 10.1371/journal.pone.0115622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma J., Chang K., Peng J., Shi Q., Gan H., Gao K., Feng K., Xu F., Zhang H., Dai B., et al. SPOP promotes ATF2 ubiquitination and degradation to suppress prostate cancer progression. J. Exp. Clin. Cancer Res. 2018;37:145. doi: 10.1186/s13046-018-0809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu X., Zhang W., Font-Burgada J., Palmer T., Hamil A.S., Biswas S.K., Poidinger M., Borcherding N., Xie Q., Ellies L.G., et al. Ubiquitin-conjugating enzyme Ubc13 controls breast cancer metastasis through a TAK1-p38 MAP kinase cascade. Proc. Natl. Acad. Sci. USA. 2014;111:13870–13875. doi: 10.1073/pnas.1414358111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perry G., Friedman R., Shaw G., Chau V. Ubiquitin is detected in neurofibrillary tangles and senile plaque neurites of Alzheimer disease brains. Proc. Natl. Acad. Sci. USA. 1987;84:3033–3036. doi: 10.1073/pnas.84.9.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galloway P.G., Grundke-Iqbal I., Iqbal K., Perry G. Lewy bodies contain epitopes both shared and distinct from Alzheimer neurofibrillary tangles. J. Neuropathol. Exp. Neurol. 1988;47:654–663. doi: 10.1097/00005072-198811000-00008. [DOI] [PubMed] [Google Scholar]
- 46.Weekes J., Morrison K., Mullen A., Wait R., Barton P., Dunn M.J. Hyperubiquitination of proteins in dilated cardiomyopathy. Proteomics. 2003;3:208–216. doi: 10.1002/pmic.200390029. [DOI] [PubMed] [Google Scholar]
- 47.Hu H., Sun S.-C. Ubiquitin signaling in immune responses. Cell Res. 2016;26:457. doi: 10.1038/cr.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson K.W., Turko I.V. Histone post-translational modifications in frontal cortex from human donors with Alzheimer’s disease. Clin. Proteom. 2015;12:26. doi: 10.1186/s12014-015-9098-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miao F., Chen Z., Zhang L., Liu Z., Wu X., Yuan Y.-C., Natarajan R. Profiles of Epigenetic Histone Post-translational Modifications at Type 1 Diabetes Susceptible Genes. J. Biol. Chem. 2012;287:16335–16345. doi: 10.1074/jbc.M111.330373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kikuchi T., Daigo Y., Katagiri T., Tsunoda T., Okada K., Kakiuchi S., Zembutsu H., Furukawa Y., Kawamura M., Kobayashi K., et al. Expression profiles of non-small cell lung cancers on cDNA microarrays: Identification of genes for prediction of lymph-node metastasis and sensitivity to anti-cancer drugs. Oncogene. 2003;22:2192. doi: 10.1038/sj.onc.1206288. [DOI] [PubMed] [Google Scholar]
- 51.Nakamura T., Furukawa Y., Nakagawa H., Tsunoda T., Ohigashi H., Murata K., Ishikawa O., Ohgaki K., Kashimura N., Miyamoto M., et al. Genome-wide cDNA microarray analysis of gene expression profiles in pancreatic cancers using populations of tumor cells and normal ductal epithelial cells selected for purity by laser microdissection. Oncogene. 2004;23:2385. doi: 10.1038/sj.onc.1207392. [DOI] [PubMed] [Google Scholar]
- 52.Nishidate T., Katagiri T., Lin M.L., Mano Y., Miki Y., Kasumi F., Yoshimoto M., Tsunoda T., Hirata K., Nakamura Y. Genome-wide gene-expression profiles of breast-cancer cells purified with laser microbeam microdissection: Identification of genes associated with progression and metastasis. Int. J. Oncol. 2004;25:797–819. [PubMed] [Google Scholar]
- 53.Yoshimatsu M., Toyokawa G., Hayami S., Unoki M., Tsunoda T., Field H.I., Kelly J.D., Neal D.E., Maehara Y., Ponder B.A.J., et al. Dysregulation of PRMT1 and PRMT6, Type I arginine methyltransferases, is involved in various types of human cancers. Int. J. Cancer. 2010;128:562–573. doi: 10.1002/ijc.25366. [DOI] [PubMed] [Google Scholar]
- 54.Barrero C.A., Perez-Leal O., Aksoy M., Moncada C., Ji R., Lopez Y., Mallilankaraman K., Madesh M., Criner G.J., Kelsen S.G., et al. Histone 3.3 Participates in a Self-Sustaining Cascade of Apoptosis That Contributes to the Progression of Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2013;188:673–683. doi: 10.1164/rccm.201302-0342OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakuma T., Uzawa K., Onda T., Shiiba M., Yokoe H., Shibahara T., Tanzawa H. Aberrant expression of histone deacetylase 6 in oral squamous cell carcinoma. Int. J. Oncol. 2006;29:117–124. doi: 10.3892/ijo.29.1.117. [DOI] [PubMed] [Google Scholar]
- 56.Osada H., Tatematsu Y., Saito H., Yatabe Y., Mitsudomi T., Takahashi T. Reduced expression of class II histone deacetylase genes is associated with poor prognosis in lung cancer patients. Int. J. Cancer. 2004;112:26–32. doi: 10.1002/ijc.20395. [DOI] [PubMed] [Google Scholar]
- 57.Halkidou K., Gaughan L., Cook S., Leung H.Y., Neal D.E., Robson C.N. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate. 2004;59:177–189. doi: 10.1002/pros.20022. [DOI] [PubMed] [Google Scholar]
- 58.Bode A.M., Dong Z. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer. 2004;4:793. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 59.Chindasub P., Lindsey J.D., Duong-Polk K., Leung C.K., Weinreb R.N. Inhibition of Histone Deacetylases 1 and 3 Protects Injured Retinal Ganglion Cells. Investig. Ophthalmol. Vis. Sci. 2013;54:96–102. doi: 10.1167/iovs.12-10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen I.H., Xue L., Hsu C.C., Paez J.S., Pan L., Andaluz H., Wendt M.K., Iliuk A.B., Zhu J.K., Tao W.A. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc. Natl. Acad. Sci. USA. 2017;114:3175–3180. doi: 10.1073/pnas.1618088114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tatebe H., Kasai T., Ohmichi T., Kishi Y., Kakeya T., Waragai M., Kondo M., Allsop D., Tokuda T. Quantification of plasma phosphorylated tau to use as a biomarker for brain Alzheimer pathology: Pilot case-control studies including patients with Alzheimer’s disease and down syndrome. Mol. Neurodegener. 2017;12:63. doi: 10.1186/s13024-017-0206-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Foulds P.G., Diggle P., Mitchell J.D., Parker A., Hasegawa M., Masuda-Suzukake M., Mann D.M.A., Allsop D. A longitudinal study on α-synuclein in blood plasma as a biomarker for Parkinson’s disease. Sci. Rep. 2013;3:2540. doi: 10.1038/srep02540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petushkova N.A., Zgoda V.G., Pyatnitskiy M.A., Larina O.V., Teryaeva N.B., Potapov A.A., Lisitsa A.V. Post-translational modifications of FDA-approved plasma biomarkers in glioblastoma samples. PLoS ONE. 2017;12:e0177427. doi: 10.1371/journal.pone.0177427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma W., Kantarjian H., Zhang X., Wang X., Estrov Z., O’Brien S., Albitar M. Ubiquitin-Proteasome System Profiling in Acute Leukemias and its Clinical Relevance. Leuk. Res. 2011;35:526–533. doi: 10.1016/j.leukres.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frenkel-Pinter M., Shmueli M.D., Raz C., Yanku M., Zilberzwige S., Gazit E., Segal D. Interplay between protein glycosylation pathways in Alzheimer’s disease. Sci. Adv. 2017;3:e1601576. doi: 10.1126/sciadv.1601576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marrero J.A., Romano P.R., Nikolaeva O., Steel L., Mehta A., Fimmel C.J., Comunale M.A., D’Amelio A., Lok A.S., Block T.M. GP73, a resident Golgi glycoprotein, is a novel serum marker for hepatocellular carcinoma. J. Hepatol. 2005;43:1007–1012. doi: 10.1016/j.jhep.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 67.Saldova R., Royle L., Radcliffe C.M., Abd Hamid U.M., Evans R., Arnold J.N., Banks R.E., Hutson R., Harvey D.J., Antrobus R., et al. Ovarian Cancer is Associated with Changes in Glycosylation in Both Acute-Phase Proteins and IgG. Glycobiology. 2007;17:1344–1356. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- 68.Kyselova Z., Mechref Y., Kang P., Goetz J.A., Dobrolecki L.E., Sledge G.W., Schnaper L., Hickey R.J., Malkas L.H., Novotny M.V. Breast Cancer Diagnosis and Prognosis through Quantitative Measurements of Serum Glycan Profiles. Clin. Chem. 2008;54:1166. doi: 10.1373/clinchem.2007.087148. [DOI] [PubMed] [Google Scholar]
- 69.Gu H., Ren J.M., Jia X., Levy T., Rikova K., Yang V., Lee K.A., Stokes M.P., Silva J.C. Quantitative Profiling of Post-translational Modifications by Immunoaffinity Enrichment and LC-MS/MS in Cancer Serum without Immunodepletion. Mol. Cell. Proteom. 2016;15:692–702. doi: 10.1074/mcp.O115.052266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stone M.D., Chen X., McGowan T., Bandhakavi S., Cheng B., Rhodus N.L., Griffin T.J. Large-scale phosphoproteomics analysis of whole saliva reveals a distinct phosphorylation pattern. J. Proteome Res. 2011;10:1728–1736. doi: 10.1021/pr1010247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salih E., Siqueira W.L., Helmerhorst E.J., Oppenheim F.G. Large-scale phosphoproteome of human whole saliva using disulfide–thiol interchange covalent chromatography and mass spectrometry. Anal. Biochem. 2010;407:19–33. doi: 10.1016/j.ab.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y., Wang X., Cui D., Zhu J. Proteomic and N-glycoproteomic quantification reveal aberrant changes in the human saliva of oral ulcer patients. Proteomics. 2016;16:3173–3182. doi: 10.1002/pmic.201600127. [DOI] [PubMed] [Google Scholar]
- 73.Sun S., Zhao F., Wang Q., Zhong Y., Cai T., Wu P., Yang F., Li Z. Analysis of age and gender associated N-glycoproteome in human whole saliva. Clin. Proteom. 2014;11:25. doi: 10.1186/1559-0275-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramachandran P., Boontheung P., Xie Y., Sondej M., Wong D.T., Loo J.A. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J. Proteome Res. 2006;5:1493–1503. doi: 10.1021/pr050492k. [DOI] [PubMed] [Google Scholar]
- 75.Stanta J.L., Saldova R., Struwe W.B., Byrne J.C., Leweke F.M., Rothermund M., Rahmoune H., Levin Y., Guest P.C., Bahn S., et al. Identification of N-Glycosylation Changes in the CSF and Serum in Patients with Schizophrenia. J. Proteome Res. 2010;9:4476–4489. doi: 10.1021/pr1002356. [DOI] [PubMed] [Google Scholar]
- 76.Majbour N.K., Vaikath N.N., van Dijk K.D., Ardah M.T., Varghese S., Vesterager L.B., Montezinho L.P., Poole S., Safieh-Garabedian B., Tokuda T., et al. Oligomeric and phosphorylated alpha-synuclein as potential CSF biomarkers for Parkinson’s disease. Mol. Neurodegener. 2016;11:7. doi: 10.1186/s13024-016-0072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Öhrfelt A., Johansson P., Wallin A., Andreasson U., Zetterberg H., Blennow K., Svensson J. Increased Cerebrospinal Fluid Levels of Ubiquitin Carboxyl-Terminal Hydrolase L1 in Patients with Alzheimer’s Disease. Dement. Geriatr. Cogn. Disord. Extra. 2016;6:283–294. doi: 10.1159/000447239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sjödin S., Hansson O., Öhrfelt A., Brinkmalm G., Zetterberg H., Brinkmalm A., Blennow K. Mass Spectrometric Analysis of Cerebrospinal Fluid Ubiquitin in Alzheimer’s Disease and Parkinsonian Disorders. Proteom. Clin. Appl. 2017;11:1700100. doi: 10.1002/prca.201700100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paulo J.A., Kadiyala V., Brizard S., Banks P.A., Steen H., Conwell D.L. Post-translational Modifications of Pancreatic Fluid Proteins Collected via the Endoscopic Pancreatic Function Test (ePFT) J. Proteom. 2013;92 doi: 10.1016/j.jprot.2013.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jia X., Chen J., Sun S., Yang W., Yang S., Shah P., Hoti N., Veltri B., Zhang H. Detection of aggressive prostate cancer associated glycoproteins in urine using glycoproteomics and mass spectrometry. Proteomics. 2016;16:2989–2996. doi: 10.1002/pmic.201500506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kawahara R., Ortega F., Rosa-Fernandes L., Guimarães V., Quina D., Nahas W., Schwämmle V., Srougi M., Leite K.R., Thaysen-Andersen M. Distinct urinary glycoprotein signatuRes. in prostate cancer patients. Oncotarget. 2018;9:33077. doi: 10.18632/oncotarget.26005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zheng J., Liu L., Wang J., Jin Q. Urinary proteomic and non-prefractionation quantitative phosphoproteomic analysis during pregnancy and non-pregnancy. BMC Genom. 2013;14:777. doi: 10.1186/1471-2164-14-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khadjavi A., Mannu F., Destefanis P., Sacerdote C., Battaglia A., Allasia M., Fontana D., Frea B., Polidoro S., Fiorito G., et al. Early diagnosis of bladder cancer through the detection of urinary tyrosine-phosphorylated proteins. Br. J. Cancer. 2015;113:469. doi: 10.1038/bjc.2015.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Olsen J.V., Mann M. Status of Large-scale Analysis of Post-translational Modifications by Mass Spectrometry. Mol. Cell. Proteom. 2013;12:3444–3452. doi: 10.1074/mcp.O113.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Larsen M.R., Trelle M.B., Thingholm T.E., Jensen O.N. Analysis of posttranslational modifications of proteins by tandem mass spectrometry: Mass Spectrometry For Proteomics Analysis. Biotechniques. 2006;40:790–798. doi: 10.2144/000112201. [DOI] [PubMed] [Google Scholar]
- 86.Doll S., Burlingame A.L. Mass spectrometry-based detection and assignment of protein posttranslational modifications. ACS Chem. Biol. 2014;10:63–71. doi: 10.1021/cb500904b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Riley N.M., Coon J.J. Phosphoproteomics in the Age of Rapid and Deep Proteome Profiling. ACS Chem. Biol. 2016;88:74–94. doi: 10.1021/acs.analchem.5b04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Choudhary C., Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell Biol. 2010;11:427. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 89.Keller A., Bader S.L., Kusebauch U., Shteynberg D., Hood L., Moritz R.L. Opening a SWATH window on posttranslational modifications: Automated pursuit of modified peptides. Mol. Cell. Proteom. 2016;15:1151–1163. doi: 10.1074/mcp.M115.054478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Masuda T., Sugiyama N., Tomita M., Ishihama Y. Microscale phosphoproteome analysis of 10 000 cells from human cancer cell lines. Anal. Chem. 2011;83:7698–7703. doi: 10.1021/ac201093g. [DOI] [PubMed] [Google Scholar]
- 91.Humphrey S.J., Yang G., Yang P., Fazakerley D.J., Stöckli J., Yang J.Y., James D.E. Dynamic Adipocyte Phosphoproteome Reveals that Akt Directly Regulates mTORC2. Cell Metab. 2013;17:1009–1020. doi: 10.1016/j.cmet.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zielinska D.F., Gnad F., Wiśniewski J.R., Mann M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell. 2010;141:897–907. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 93.Larsen S.C., Sylvestersen K.B., Mund A., Lyon D., Mullari M., Madsen M.V., Daniel J.A., Jensen L.J., Nielsen M.L. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal. 2016;9:rs9. doi: 10.1126/scisignal.aaf7329. [DOI] [PubMed] [Google Scholar]
- 94.Guo A., Gu H., Zhou J., Mulhern D., Wang Y., Lee K.A., Yang V., Aguiar M., Kornhauser J., Jia X., et al. Immunoaffinity Enrichment and Mass Spectrometry Analysis of Protein Methylation. Mol. Cell. Proteom. 2014;13:372–387. doi: 10.1074/mcp.O113.027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Drazic A., Myklebust L.M., Ree R., Arnesen T. The world of protein acetylation. Biochim. Biophys. Acta-Proteins Proteom. 2016;1864:1372–1401. doi: 10.1016/j.bbapap.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 96.Akimov V., Barrio-Hernandez I., Hansen S.V., Hallenborg P., Pedersen A.-K., Bekker-Jensen D.B., Puglia M., Christensen S.D., Vanselow J.T., Nielsen M.M. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 2018;25:631. doi: 10.1038/s41594-018-0084-y. [DOI] [PubMed] [Google Scholar]
- 97.Shi Y., Xu P., Qin J. Ubiquitinated proteome: Ready for global? Mol. Cell. Proteom. 2011;10 doi: 10.1074/mcp.R110.006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Venne A.S., Kollipara L., Zahedi R.P. The next level of complexity: Crosstalk of posttranslational modifications. Proteomics. 2014;14:513–524. doi: 10.1002/pmic.201300344. [DOI] [PubMed] [Google Scholar]
- 99.Chalkley R.J., Clauser K.R. Modification site localization scoring: Strategies and performance. Mol. Cell. Proteom. 2012;11 doi: 10.1074/mcp.R111.015305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 101.Lemeer S., Kunold E., Klaeger S., Raabe M., Towers M.W., Claudes E., Arrey T.N., Strupat K., Urlaub H., Kuster B. Phosphorylation site localization in peptides by MALDI MS/MS and the Mascot Delta Score. Anal. Bioanal. Chem. 2012;402:249–260. doi: 10.1007/s00216-011-5469-2. [DOI] [PubMed] [Google Scholar]
- 102.Savitski M.M., Lemeer S., Boesche M., Lang M., Mathieson T., Bantscheff M., Kuster B. Confident phosphorylation site localization using the Mascot Delta Score. Mol. Cell. Proteom. 2011;10 doi: 10.1074/mcp.M110.003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Beausoleil S.A., Villén J., Gerber S.A., Rush J., Gygi S.P. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 104.Boersema P.J., Mohammed S., Heck A.J. Phosphopeptide fragmentation and analysis by mass spectrometry. J. Mass Spectrom. 2009;44:861–878. doi: 10.1002/jms.1599. [DOI] [PubMed] [Google Scholar]
- 105.Taus T., Köcher T., Pichler P., Paschke C., Schmidt A., Henrich C., Mechtler K. Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 2011;10:5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
- 106.Bailey C.M., Sweet S.M., Cunningham D.L., Zeller M., Heath J.K., Cooper H.J. SLoMo: Automated site localization of modifications from ETD/ECD mass spectra. J. Proteome Res. 2009;8:1965–1971. doi: 10.1021/pr800917p. [DOI] [PubMed] [Google Scholar]
- 107.Shilov I.V., Seymour S.L., Patel A.A., Loboda A., Tang W.H., Keating S.P., Hunter C.L., Nuwaysir L.M., Schaeffer D.A. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteom. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 108.Eng J.K., McCormack A.L., Yates J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 109.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 110.Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophor. Int. J. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:183551::AID-ELPS35513.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 111.Rosenberger G., Koh C.C., Guo T., Röst H.L., Kouvonen P., Collins B.C., Heusel M., Liu Y., Caron E., Vichalkovski A. A repository of assays to quantify 10,000 human proteins by SWATH-MS. Sci. Data. 2014;1:140031. doi: 10.1038/sdata.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schubert O.T., Gillet L.C., Collins B.C., Navarro P., Rosenberger G., Wolski W.E., Lam H., Amodei D., Mallick P., MacLean B. Building high-quality assay libraries for targeted analysis of SWATH MS data. Nat. Protoc. 2015;10:426. doi: 10.1038/nprot.2015.015. [DOI] [PubMed] [Google Scholar]
- 113.Tsou C.-C., Avtonomov D., Larsen B., Tucholska M., Choi H., Gingras A.-C., Nesvizhskii A.I. DIA-Umpire: Comprehensive computational framework for data-independent acquisition proteomics. Nat. Methods. 2015;12:258. doi: 10.1038/nmeth.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu J.X., Song X., Pascovici D., Zaw T., Care N., Krisp C., Molloy M.P. SWATH mass spectrometry performance using extended peptide MS/MS assay libraries. Mol. Cell. Proteom. 2016;15 doi: 10.1074/mcp.M115.055558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Noor Z., Wu J.X., Pascovici D., Mohamedali A., Molloy M.P., Baker M.S., Ranganathan S., Wren J. iSwathX: An interactive web-based application for extension of DIA peptide reference libraries. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty660. [DOI] [PubMed] [Google Scholar]
- 116.Röst H.L., Rosenberger G., Navarro P., Gillet L., Miladinović S.M., Schubert O.T., Wolski W., Collins B.C., Malmström J., Malmström L. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 2014;32:219. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- 117.Keller A., Bader S.L., Shteynberg D., Hood L., Moritz R.L. Automated validation of results and removal of fragment ion interferences in targeted analysis of data independent acquisition MS using SWATHProphet. Mol. Cell. Proteom. 2015;14 doi: 10.1074/mcp.O114.044917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Elias J.E., Gygi S.P. Proteome Bioinformatics. Springer; Berlin/Heidelberg, Germany: 2010. Target-decoy search strategy for mass spectrometry-based proteomics; pp. 55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Baker P.R., Trinidad J.C., Chalkley R.J. Modification site localization scoring integrated into a search engine. Mol. Cell. Proteom. 2011;10 doi: 10.1074/mcp.M111.008078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wiśniewski J.R., Mann M. Consecutive proteolytic digestion in an enzyme reactor increases depth of proteomic and phosphoproteomic analysis. Anal. Chem. 2012;84:2631–2637. doi: 10.1021/ac300006b. [DOI] [PubMed] [Google Scholar]
- 121.Gilmore J.M., Kettenbach A.N., Gerber S.A. Increasing phosphoproteomic coverage through sequential digestion by complementary proteases. Anal. Bioanal. Chem. 2012;402:711–720. doi: 10.1007/s00216-011-5466-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Aittokallio T. Dealing with missing values in large-scale studies: Microarray data imputation and beyond. Brief. Bioinform. 2010;11:253–264. doi: 10.1093/bib/bbp059. [DOI] [PubMed] [Google Scholar]
- 123.Lazar C., Gatto L., Ferro M., Bruley C., Burger T. Accounting for the Multiple NatuRes. of Missing Values in Label-Free Quantitative Proteomics Data Sets to Compare Imputation Strategies. J. Proteome Res. 2016;15:1116–1125. doi: 10.1021/acs.jproteome.5b00981. [DOI] [PubMed] [Google Scholar]
- 124.Schwämmle V., Vaudel M. Protein Bioinformatics. Springer; Berlin/Heidelberg, Germany: 2017. Computational and Statistical Methods for High-Throughput Mass Spectrometry-Based PTM Analysis; pp. 437–458. [DOI] [PubMed] [Google Scholar]
- 125.Wang X., Anderson G.A., Smith R.D., Dabney A.R. A hybrid approach to protein differential expression in mass spectrometry-based proteomics. Bioinformatics. 2012;28:1586–1591. doi: 10.1093/bioinformatics/bts193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Webb-Robertson B.-J.M., McCue L.A., Waters K.M., Matzke M.M., Jacobs J.M., Metz T.O., Varnum S.M., Pounds J.G. Combined Statistical Analyses of Peptide Intensities and Peptide Occurrences Improves Identification of Significant Peptides from MS-Based Proteomics Data. J. Proteome Res. 2010;9:5748–5756. doi: 10.1021/pr1005247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ryu S.Y., Qian W.-J., Camp D.G., Smith R.D., Tompkins R.G., Davis R.W., Xiao W. Detecting differential protein expression in large-scale population proteomics. Bioinformatics. 2014;30:2741–2746. doi: 10.1093/bioinformatics/btu341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Keilhauer E.C., Hein M.Y., Mann M. Accurate Protein Complex Retrieval by Affinity Enrichment Mass Spectrometry (AE-MS) Rather than Affinity Purification Mass Spectrometry (AP-MS) Mol. Cell. Proteom. 2015;14:120–135. doi: 10.1074/mcp.M114.041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sharma K., D’Souza R.C., Tyanova S., Schaab C., Wiśniewski J.R., Cox J., Mann M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014;8:1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 130.Sinitcyn P., Rudolph J.D., Cox J. Computational Methods for Understanding Mass Spectrometry–Based Shotgun Proteomics Data. Annu. Rev. Biomed. Data Sci. 2018 doi: 10.1146/annurev-biodatasci-080917-013516. [DOI] [Google Scholar]
- 131.Välikangas T., Suomi T., Elo L.L. A systematic evaluation of normalization methods in quantitative label-free proteomics. Brief. Bioinform. 2018;19:1–11. doi: 10.1093/bib/bbw095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 133.Smyth G.K. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; Berlin/Heidelberg, Germany: 2005. Limma: Linear models for microarray data; pp. 397–420. [Google Scholar]
- 134.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 136.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Solari F.A., Dell’Aica M., Sickmann A., Zahedi R.P. Why phosphoproteomics is still a challenge. Mol. Biosyst. 2015;11:1487–1493. doi: 10.1039/C5MB00024F. [DOI] [PubMed] [Google Scholar]
- 138.Wu R., Dephoure N., Haas W., Huttlin E.L., Zhai B., Sowa M.E., Gygi S.P. Correct interpretation of comprehensive phosphorylation dynamics requiRes. normalization by protein expression changes. Mol. Cell. Proteom. 2011 doi: 10.1074/mcp.M111.009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim M.S., Zhong J., Pandey A. Common errors in mass spectrometry-based analysis of post-translational modifications. Proteomics. 2016;16:700–714. doi: 10.1002/pmic.201500355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wu R., Haas W., Dephoure N., Huttlin E.L., Zhai B., Sowa M.E., Gygi S.P. A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat. Methods. 2011;8:677. doi: 10.1038/nmeth.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lim M.Y., O’Brien J., Paulo J.A., Gygi S.P. Improved Method for Determining Absolute Phosphorylation Stoichiometry Using Bayesian Statistics and Isobaric Labeling. J. Proteome Res. 2017;16:4217–4226. doi: 10.1021/acs.jproteome.7b00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kamath K.S., Vasavada M.S., Srivastava S. Proteomic databases and tools to decipher post-translational modifications. J. Proteom. 2011;75:127–144. doi: 10.1016/j.jprot.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 143.Lisacek F., Mariethoz J., Alocci D., Rudd P.M., Abrahams J.L., Campbell M.P., Packer N.H., Ståhle J., Widmalm G., Mullen E. High-Throughput Glycomics and Glycoproteomics. Springer; Berlin/Heidelberg, Germany: 2017. Databases and associated withols for glycomics and glycoproteomics; pp. 235–264. [DOI] [PubMed] [Google Scholar]
- 144.Duan G., Li X., Köhn M. The human DEPhOsphorylation database DEPOD: A 2015 update. Nucleic Acids Res. 2014;43:D531–D535. doi: 10.1093/nar/gku1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dinkel H., Chica C., Via A., Gould C.M., Jensen L.J., Gibson T.J., Diella F. Phospho. ELM: A database of phosphorylation sites—Update 2011. Nucleic Acids Res. 2010;39:D261–D267. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Consortium U. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gnad F., Ren S., Cox J., Olsen J.V., Macek B., Oroshi M., Mann M. PHOSIDA (phosphorylation site database): Management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Minguez P., Letunic I., Parca L., Bork P. PTMcode: A database of known and predicted functional associations between post-translational modifications in proteins. Nucleic Acids Res. 2012;41:D306–D311. doi: 10.1093/nar/gks1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V., Skrzypek E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2014;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Stark C., Su T.-C., Breitkreutz A., Lourenco P., Dahabieh M., Breitkreutz B.-J., Tyers M., Sadowski I. PhosphoGRID: A database of experimentally verified in vivo protein phosphorylation sites from the budding yeast Saccharomyces cerevisiae. Database. 2010;2010 doi: 10.1093/database/bap026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Campbell M.P., Peterson R., Mariethoz J., Gasteiger E., Akune Y., Aoki-Kinoshita K.F., Lisacek F., Packer N.H. UniCarbKB: Building a knowledge platform for glycoproteomics. Nucleic Acids Res. 2013;42:D215–D221. doi: 10.1093/nar/gkt1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Huang H., Arighi C.N., Ross K.E., Ren J., Li G., Chen S.-C., Wang Q., Cowart J., Vijay-Shanker K., Wu C.H. iPTMnet: An integrated resource for protein post-translational modification network discovery. Nucleic Acids Res. 2018;46:D542–D550. doi: 10.1093/nar/gkx1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chen C., Huang H., Wu C.H. Protein Bioinformatics. Springer; Berlin/Heidelberg, Germany: 2017. Protein bioinformatics databases and resources; pp. 3–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Schwämmle V., Verano-Braga T., Roepstorff P. Computational and statistical methods for high-throughput analysis of post-translational modifications of proteins. J. Proteom. 2015;129:3–15. doi: 10.1016/j.jprot.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 155.Chou M.F., Schwartz D. Biological sequence motif discovery using motif-x. Curr. Protoc. Bioinform. 2011;35:13–15. doi: 10.1002/0471250953.bi1315s35. [DOI] [PubMed] [Google Scholar]
- 156.Peng M., Scholten A., Heck A.J.R., van Breukelen B. Identification of Enriched PTM Crosstalk Motifs from Large-Scale Experimental Data Sets. J. Proteome Res. 2014;13:249–259. doi: 10.1021/pr4005579. [DOI] [PubMed] [Google Scholar]
- 157.Huang H., Larsen M.R., Palmisano G., Dai J., Lametsch R. Quantitative phosphoproteomic analysis of porcine muscle within 24 h postmortem. J. Proteom. 2014;106:125–139. doi: 10.1016/j.jprot.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 158.Craveur P., Rebehmed J., de Brevern A.G. PTM-SD: A database of structurally resolved and annotated posttranslational modifications in proteins. Database. 2014;2014 doi: 10.1093/database/bau041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Knight J.D., Choi H., Gupta G.D., Pelletier L., Raught B., Nesvizhskii A.I., Gingras A.-C. ProHits-viz: A suite of web tools for visualizing interaction proteomics data. Nat. Methods. 2017;14:645–646. doi: 10.1038/nmeth.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Martín-Campos T., Mylonas R., Masselot A., Waridel P., Petricevic T., Xenarios I., Quadroni M. MsViz: A graphical software tool for in-depth manual validation and quantitation of post-translational modifications. J. Proteome Res. 2017;16:3092–3101. doi: 10.1021/acs.jproteome.7b00194. [DOI] [PubMed] [Google Scholar]
- 161.Huntley R.P., Sawford T., Mutowo-Meullenet P., Shypitsyna A., Bonilla C., Martin M.J., O’Donovan C. The GOA database: Gene Ontology annotation updates for 2015. Nucleic Acids Res. 2015;43:D1057–D1063. doi: 10.1093/nar/gku1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gnad F., Gunawardena J., Mann M. PHOSIDA 2011: The posttranslational modification database. Nucleic Acids Res. 2011;39:D253–D260. doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hornbeck P.V., Kornhauser J.M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Campbell M.P., Packer N.H. UniCarbKB: New database featuRes. for integrating glycan structure abundance, compositional glycoproteomics data, and disease associations. Biochim. Biophys. Acta. 2016;1860:1669–1675. doi: 10.1016/j.bbagen.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 165.Schlaffner C.N., Pirklbauer G.J., Bender A., Steen J.A.J., Choudhary J.S. A Fast and Quantitative Method for Post-translational Modification and Variant Enabled Mapping of Peptides to Genomes. J. Vis. Exp. 2018 doi: 10.3791/57633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Durek P., Schmidt R., Heazlewood J.L., Jones A., MacLean D., Nagel A., Kersten B., Schulze W.X. PhosPhAt: The Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 2010;38:D828–D834. doi: 10.1093/nar/gkp810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gao J., Agrawal G.K., Thelen J.J., Xu D. P3DB: A plant protein phosphorylation database. Nucleic Acids Res. 2009;37:D960–D962. doi: 10.1093/nar/gkn733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yao Q., Ge H., Wu S., Zhang N., Chen W., Xu C., Gao J., Thelen J.J., Xu D. P(3)DB 3.0: From plant phosphorylation sites to protein networks. Nucleic Acids Res. 2014;42:D1206–D1213. doi: 10.1093/nar/gkt1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Goel R., Harsha H., Pandey A., Prasad T.K. Human Protein Reference Database and Human Proteinpedia as resources for phosphoproteome analysis. Mol. Biosyst. 2012;8:453–463. doi: 10.1039/C1MB05340J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Huang K.Y., Su M.G., Kao H.J., Hsieh Y.C., Jhong J.H., Cheng K.H., Huang H.D., Lee T.Y. dbPTM 2016: 10-year anniversary of a resource for post-translational modification of proteins. Nucleic Acids Res. 2016;44:D435–D446. doi: 10.1093/nar/gkv1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang J., Anania V.G., Knott J., Rush J., Lill J.R., Bourne P.E., Bandeira N. A turn-key approach for large-scale identification of complex posttranslational modifications. J. Proteome Res. 2014;13:1190–1199. doi: 10.1021/pr400368u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chalkley R.J., Baker P.R., Medzihradszky K.F., Lynn A.J., Burlingame A.L. In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol. Cell. Proteom. 2008;7:2386–2398. doi: 10.1074/mcp.M800021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bern M., Cai Y., Goldberg D. Lookup peaks: A hybrid of de novo sequencing and database search for protein identification by tandem mass spectrometry. Anal. Chem. 2007;79:1393–1400. doi: 10.1021/ac0617013. [DOI] [PubMed] [Google Scholar]
- 174.Vincent-Maloney N., Searle B., Turner M. Probabilistically assigning sites of protein modification with scaffold PTM. J. Biomol. Tech. 2011;22:S36. [Google Scholar]
- 175.Bodenmiller B., Campbell D., Gerrits B., Lam H., Jovanovic M., Picotti P., Schlapbach R., Aebersold R. PhosphoPep—A database of protein phosphorylation sites in model organisms. Nat. Biotechnol. 2008;26:1339–1340. doi: 10.1038/nbt1208-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Matlock M.K., Holehouse A.S., Naegle K.M. ProteomeScout: A repository and analysis resource for post-translational modifications and proteins. Nucleic Acids Res. 2015;43:D521–D530. doi: 10.1093/nar/gku1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fellers R.T., Greer J.B., Early B.P., Yu X., LeDuc R.D., Kelleher N.L., Thomas P.M. ProSight Lite: Graphical software to analyze top-down mass spectrometry data. Proteomics. 2015;15:1235–1238. doi: 10.1002/pmic.201400313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., Thomas P.D. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ward J.J., McGuffin L.J., Bryson K., Buxton B.F., Jones D.T. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20:2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 181.Wu X., Al Hasan M., Chen J.Y. Pathway and network analysis in proteomics. J. Theor. Biol. 2014;362:44–52. doi: 10.1016/j.jtbi.2014.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Minguez P., Letunic I., Parca L., Garcia-Alonso L., Dopazo J., Huerta-Cepas J., Bork P. PTMcode v2: A resource for functional associations of post-translational modifications within and between proteins. Nucleic Acids Res. 2014;43:D494–D502. doi: 10.1093/nar/gku1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Grimes M., Hall B., Foltz L., Levy T., Rikova K., Gaiser J., Cook W., Smirnova E., Wheeler T., Clark N.R. Using Protein Phosphorylation, Acetylation, and Methylation to Outline Lung Cancer Signaling Networks. Sci. Signal. 2018;11 doi: 10.1126/scisignal.aaq1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Linding R., Jensen L.J., Ostheimer G.J., van Vugt M.A., Jørgensen C., Miron I.M., Diella F., Colwill K., Taylor L., Elder K., et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Qi L., Liu Z., Wang J., Cui Y., Guo Y., Zhou T., Zhou Z., Guo X., Xue Y., Sha J. Systematic analysis of the phosphoproteome and kinase-substrate networks in the mouse testis. Mol. Cell. Proteom. 2014 doi: 10.1074/mcp.M114.039073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Casado P., Rodriguez-Prados J.-C., Cosulich S.C., Guichard S., Vanhaesebroeck B., Joel S., Cutillas P.R. Kinase-substrate enrichment analysis provides insights into the heterogeneity of signaling pathway activation in leukemia cells. Sci. Signal. 2013;6:rs6. doi: 10.1126/scisignal.2003573. [DOI] [PubMed] [Google Scholar]
- 188.Rudolph J.D., de Graauw M., van de Water B., Geiger T., Sharan R. Elucidation of Signaling Pathways from Large-Scale Phosphoproteomic Data Using Protein Interaction Networks. Cell Syst. 2016;3:585–593.e583. doi: 10.1016/j.cels.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 189.Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F., Schinzel A.C., Sandy P., Meylan E., Scholl C., et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tay A.P., Pang C.N.I., Winter D.L., Wilkins M.R. PTMOracle: A Cytoscape App for Covisualizing and Coanalyzing Post-Translational Modifications in Protein Interaction Networks. J. Proteome Res. 2017;16:1988–2003. doi: 10.1021/acs.jproteome.6b01052. [DOI] [PubMed] [Google Scholar]
- 193.Xia J., Benner M.J., Hancock R.E. NetworkAnalyst—Integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014;42:W167–W174. doi: 10.1093/nar/gku443. [DOI] [PMC free article] [PubMed] [Google Scholar]