Abstract
Background
The onset of hepatocellular carcinoma (HCC) ranked fifth malignancies all over the world. Increasing evidences showed that the distribution of HCC was related to the incidence of chronic hepatitis B virus (HBV) infection and other factors, such as alcoholism, aflatoxin B1 ingestion and obesity. Recent studies demonstrated that gut dysbiosis plays an important role in liver diseases. However, the researches on gut microbiota of HBV and non-HBV non-HCV related HCC have not been reported. In this study, we investigated the differences between the gut microbiota of HBV related HCC (B-HCC) and non-HBV non-HCV related HCC (NBNC-HCC), finally found some potential bacteria, linking different pathological mechanism of both types of HCCs.
Results
We carried out 16S rRNA analyses in a cohort of 33 healthy controls, 35 individuals with HBV related HCC (B-HCC) and 22 individuals with non-HBV non-HCV (NBNC) related HCC (NBNC-HCC). We found that the species richness of fecal microbiota of B-HCC patients was much higher than other two groups. Interestingly, the feces of NBNC-HCC patients harbored more potential pro-inflammatory bacteria (Escherichia-Shigella, Enterococcus) and reduced levels of Faecalibacterium, Ruminococcus, Ruminoclostridium which results in decrease potential of anti-inflammatory short-chain fatty acids. The feces of NBNC-HCC patients had relatively fewer abundance of multiple biological pathways related to amino acid and glucose metabolism, but high level of transport and secretion in some types. However, the B-HCC patients had opposite results of bacterial composition and associated multiple biological pathways versus NBNC-HCC patients. Meanwhile, we found that aberrant network of gut microbiota occurred differently in B-HCC and NBNC-HCC patients.
Conclusions
Our study indicated that B-HCC and NBNC-HCC patients showed differential abundance of bacteria involved in different functions or biological pathways. We suggested the modification of specific gut microbiota may provide the therapeutic benefit for B-HCC and NBNC-HCC.
Electronic supplementary material
The online version of this article (10.1186/s13099-018-0281-6) contains supplementary material, which is available to authorized users.
Keywords: Gut microbiome, Dysbiosis, Liver cancer, Hepatocellular carcinoma, HBV
Background
Hepatocellular carcinoma (HCC) is the most common form of liver cancer being diagnosed annually [1], which is the fifth global malignancy [2, 3]. With increasing the risk factors such as obesity and fatty liver diseases, the worldwide incidence of HCC also increases [3, 4].
The worldwide distribution of HCC was related to the infection of chronic hepatitis B virus (HBV), especially in China. HBV promote carcinogenesis not only through the direct activation of oncogenic proteins but also indirectly establish chronic inflammation, fibrosis and cirrhosis [5]. Approximately 257 million people are infected with HBV worldwide [6]. For patients with HBV induced cirrhosis, 70–90% of HCC occur against a background of cirrhosis [7]. In addition to HBV, other risk factors include alcoholism, aflatoxin B1 ingestion, obesity, age, diet habits and genetics. In this study, we defined the other elements as non-HBV non-HCV related HCC.
The gut microbiota plays a vital role in physiology, nutrition, immunology and pathogenic processes [8]. The liver interacts with the gut through hepatic portal and bile secretion systems. Increasing evidences have revealed that gut microbiota plays an important role in liver disease formation, pathogenesis and treatment responses [9, 10]. It was reported that microbial translocation, bacteria peptidoglycan and metabolic outcomes can intensify the clinical features of chronic liver diseases [11].
Increasing studies have demonstrated that gut dysbiosis affect liver diseases, such as nonalcoholic alcohol-related liver disease, primary sclerosing cholangitis, fatty liver disease (NAFLD), fibrosis, cirrhosis and HCC [8, 12–16]. Most of these diseases presented a potentially “dysfunctional” gut microbiome. For instance, some cirrhotic patients’ gut microbiota had the high abundance of Enterobacteriaceae, suggesting it an invasion microbiota from the mouth. Meanwhile, some beneficial bacteria declined in the gut of liver diseases, such as Lachnospiraceae [14, 17]. In a recent comprehensive study of gut microbiome in early HCC patients, species diversity decreased in healthy controls compare to cirrhotic patients but increased in early HCC compared with cirrhosis [18]. In addition, butyrate-producing genera declined, however, LPS-producing genera enriched in early HCC patients [18].
However, there are limited comparison researches on gut microbiota of HBV related HCC and non-HBV non-HCV related HCC. The aim of this study was to find the differences in the gut microbiota composition of HBV and non-HBV non-HCV related HCC compared with healthy controls using 16S rRNA sequencing. This would be helpful to find the potential bacteria linking different pathological mechanisms between HBV and non-HBV non-HCV related HCC. It also helps to develop a new non-invasive differential diagnosis and therapeutic procedures for HCC patients with harboring specific gut bacteria.
Results
Cohorts of patients
Volunteers information was collected, including age, gender, weight, height, drinking history, body mass index (BMI) (kg/m2) and blood biochemical indexes (Table 1, Additional file 1). Only two HCC patients were assessed as the Child–Pugh class B, other HCC patients were Child–Pugh class A (Additional file 1). The average MELD score of HCC patients was five (Additional file 1). In total, 2047 operational taxonomic units (OTUs) were obtained from fecal microbiota of three groups for healthy controls, HBV related HCC (B-HCC) patients and non-HBV non-HCV (NBNC) related HCC (NBNC-HCC) patients, averaging 1749, 1285 and 1696, respectively. As shown in Table 1, the coverage values were nearly 1.00 for the sequences in three groups, which indicated that the sequencing depth was enough for the investigation of fecal microbiota of HCC patients or healthy controls.
Table 1.
The basic information of primary data analysis, and species richness indices in the fecal samples
| Characteristic | Healthy controls | NBNC-HCC patients | B-HCC patients |
|---|---|---|---|
| Patients | 33 | 22 | 35 |
| Gender (male/female) | 11/22 | 18/4 | 28/7 |
| Median age | 56.64 ± 9.91 | 61.12 ± 9.99 | 56.83 ± 9.95 |
| Drinking condition | 22/5/3/3 | 6/3/2/11 | 19/10/4/2 |
| BMI (kg/m2) | 30/3 | 13/9 | 22/13 |
| OTUs | 1749 | 1285 | 1696 |
| Coverage | 1.00 | 1.00 | 1.00 |
| Diversity index (median) | |||
| Species | 465 | 466 | 487 |
| ACE | 552.31 | 556.20 | 590.50 |
| CHAO1 | 542.56 | 561.19 | 578.41 |
| Shannon | 5.51 | 5.36 | 5.69 |
| Simpson | 0.93 | 0.90 | 0.94 |
| Beta diversity | 0.39 | 0.42 | 0.40 |
NBNC-HCC non-HBV non-HCV related hepatocellular carcinoma, B-HCC HBV related hepatocellular carcinoma. Drinking condition, none/low level/medium level/high level, BMI Body Mass Index (kg/cm2), normal/overweight, OTU operational taxonomic units, ACE abundance-based coverage estimators
Alterations of gut microbiota composition in HCC patients
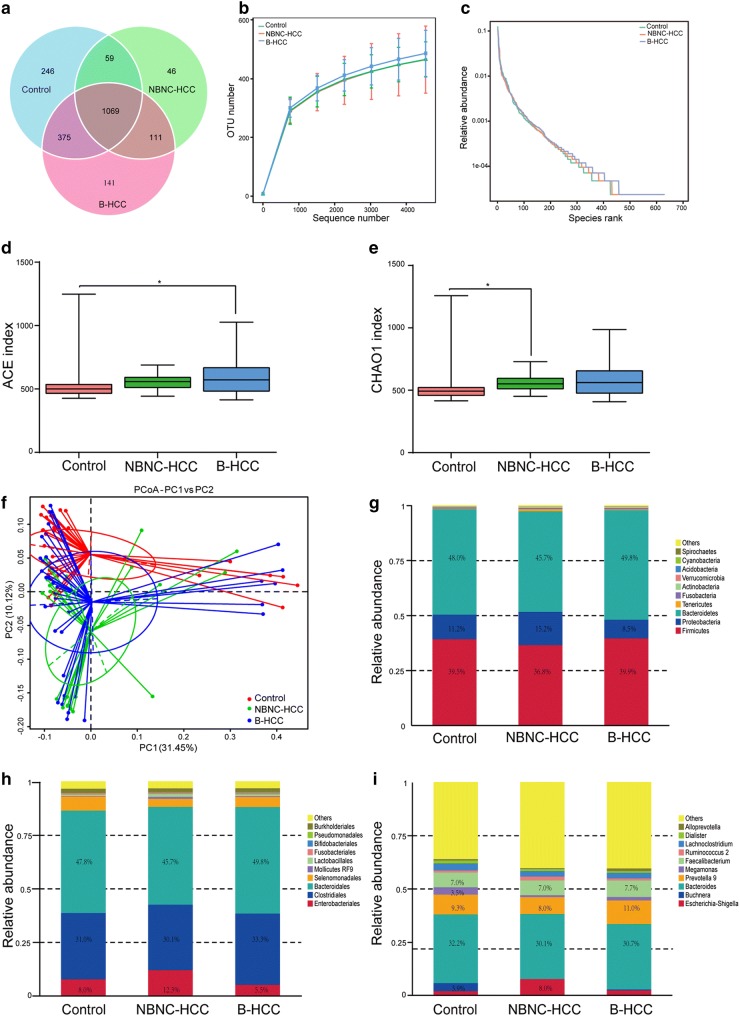
The overlapping OTU of three groups were shown in a Venn diagram (Fig. 1a). These data demonstrated that 246, 46 and 141 OTUs were existed independently in healthy controls, NBNC-HCC and B-HCC patients, respectively. Based on the OTUs analysis, the bacterial communities of B-HCC patients tended to be more heterogeneous, whereas those of the healthy controls and NBNC-HCC patients exhibited similar patterns (Fig. 1b). According to the rank-abundance curves, species richness of B-HCC patients was much higher than other two groups, and all the OTUs were evenly distributed (Fig. 1c).
Fig. 1.
Shift microbiota and diversity in gut microbiota of healthy controls, NBNC-HCC and B-HCC patients. a Venn diagram of OTUs in three groups. b Observed species numbers in three groups. c Rarefaction curve for the comparison of OTUs in three groups. d, e Comparison of the alpha diversity (ACE and CHAO1) based on the OTUs profiles. f Principal Co-ordinates Analysis (PCoA) of bacterial beta diversity based on the unweighted UniFrac distances. Each node represents each sample. Control, NBNC-HCC and B-HCC subjects are colored in red, green and blue, respectively. g–i Relative abundance of the top 10 microbiota at the phylum, order and genus level
Generally, the bacterial alpha diversity indices (such as abundance-based coverage estimators (ACE) and CHAO1 index) in B-HCC patients was the highest (Fig. 1d, e). However, the Shannon and Simpson indices were not significantly different among three groups (P > 0.050, Table 1). The Principal Co-ordinate Analysis (PCoA) of beta diversity calculated on the unweighted UniFrac distances was used for clustering 90 samples into three distinct enterotypes (Fig. 1f).
Distinct differences in bacterial composition were observed among three groups. The microbiome contained 30 phyla, 125 orders and 479 genera in all fecal samples. Bacteroidetes, Firmicutes and Proteobacteria were the most abundant taxonomic groups (Fig. 1g, Additional file 2). The relative abundance of Proteobacteria (15.2%) in NBNC-HCC patients was individually higher than that in other two groups, while the Proteobacteria (8.5%) in B-HCC patients was less. On the contrary, the relative abundance of Firmicutes (36.8%) in NBNC-HCC patients was the lowest among three groups. Based on the order level (Fig. 1h, Additional file 3), Enterobacteriales, Clostridiales, Bacteroidales and Selenomonadales were the most abundant taxonomic groups. For genus level (Fig. 1i, Additional file 4), Escherichia-Shigella, Buchnera, Bacteroides, Prevotella, Megamonas and Faecalibacterium were predominant bacteria. The relative abundance of Escherichia-Shigella was much higher in NBNC-HCC patient (8.0%), however, the abundance of Buchnera and Megamonas were much smaller in NBNC-HCC and B-HCC patients. In B-HCC patients, the abundance of Prevotella was much greater than other two groups. Meanwhile, healthy controls had more Buchnera species.
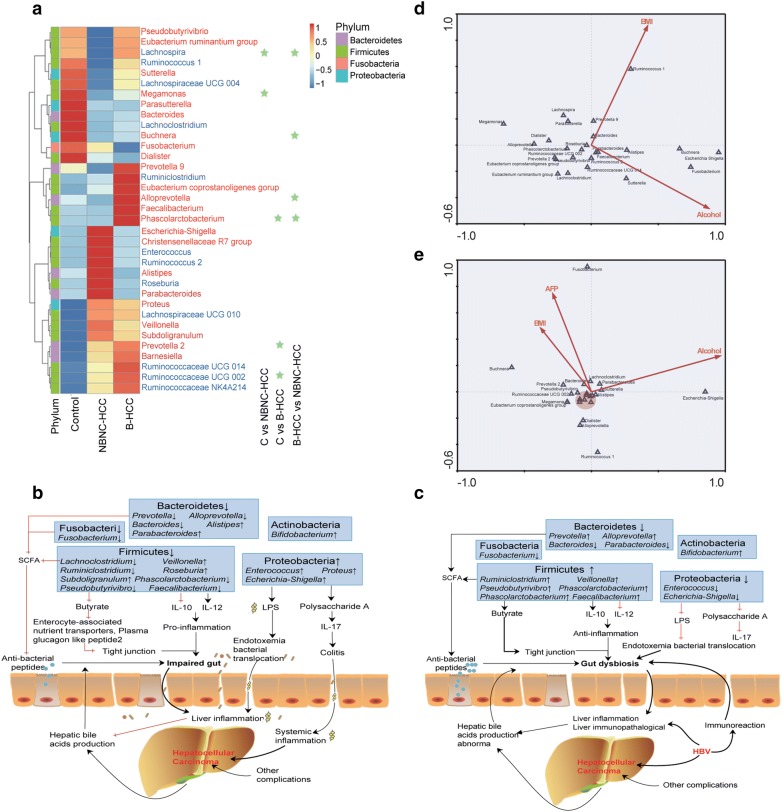
Several similar findings were existed in the top 35 genera heatmap (Fig. 2a, Additional file 5), for example, Proteus, Lachnospiraceae UCG 010, Veillonella, Subdoligranulum, Prevotella 2, Barnesiella and Ruminococcaceae spp., were enriched both in NBNC-HCC and B-HCC patients. However, the differential abundance of bacteria have been found between NBNC-HCC and B-HCC patients displayed the reduced levels of Faecalibacterium, Pseudobutyrivibrio, Lachnoclostridium, Ruminoclostridium, Prevotella 9, Alloprevotella and Phascolarctobacterium (Fig. 2a, b), which may result in the decrease of potential anti-inflammatory short chain fatty acids (SCFAs), especially the butyrate [19, 20]. SCFAs which are intestinal microbial metabolites through dietary fiber play the anti-inflammatory effects on the immune systems [21–23]. Butyrate, the energy for enterocytes, which influences the intestinal barrier through the mucous production and tight junction [24]. In addition, Faecalibacterium inhibits interleukin (IL)-12 secretion and stimulates IL-10 [25] (Fig. 2b). In contrary, potential pro-inflammatory strains including Escherichia-Shigella, Enterococcus, Proteus, Veillonella increased in the gut of NBNC-HCC patients. For instance, Enterococcus can produce polysaccharide A and lipopolysaccharide (LPS), which suppressed IL-17 production, resulting in experimental colitis and promoted LPS translocated into the cell [26, 27].
Fig. 2.
Different genera across three groups. a Heatmap of the top 35 genera. P < 0.050 by Wilcoxon rank sum test with light green star. Red in color represents Gram negative bacteria, blue in color represents Gram positive bacteria. b Changes in intestinal microbiota and the possible relations to intestinal dysfunction, gut dysbiosis and other complications in non-HBV non-HCV related HCC. c Changes in intestinal microbiota and the possible relations to intestinal dysfunction, gut dysbiosis and other complications in HBV related HCC. d The relationship between top 25 genera and body mass index (BMI) and alcohol. e The relationship between top 25 genera and body mass index (BMI), alcohol and alpha fetoprotein (AFP). The red circle shows the genera including Lachnospira, Phascolarctobacterium, Ruminococcus 2, Parasutterella, Prevotella 09, Ruminococcaceae UCG 014, Eubacterium ruminantium group, Faecalibacterium
Interestingly, a group of potential anti-inflammatory bacteria (such as Prevotella, Alloprevotella, Faecalibacterium, Ruminiclostridium) were increased in feces of B-HCC patients (Fig. 2a, c). It is well known that these bacteria are essential for healthy status. For instance, Prevotella is well known to produce propionate in healthy gut [28], and it may be play a protective effect in adult NAFLD patients [29]. Meanwhile, pro-inflammatory bacteria (such as Escherichia-Shigella, Enterococcus) were declined in fecal of B-HCC patients.
The Monte-Carlo tests of canonical correspondence analysis (CCA) revealed the top 25 genera were extremely influenced by alcohol (P = 0.017) and BMI (P = 0.007) (Fig. 2d, Additional file 6). For instance, Escherichia-Shigella was positively associated with alcohol factor. However, the Ruminococcus 2 was also positively associated with BMI factor. Alpha-fetoprotein (AFP) is one of the most useful markers for diagnosing and monitoring HCC [30]. In our study, AFP also had a strong influence on genus assemblages in HCC patients (P = 0.024, Fig. 2e). Due to the gut flora at different stages of liver disease various, so we also predicated the relationship between clinical data and the top 35 genera (Additional files 1 and 7). We calculated correlation of Spearman in all samples. The P value was corrected using the Holm method of R (Version 3.4.4, psych package). The clinical data were mainly focused on the common hepatic function index, which were alanine aminotransferase (ALT), aspartate aminotransferase (AST), glutamyl transpeptidase (GGT), total bilirubin (TBil), albumin and AFP. Several genera (e.g., Enterococcus, Proteus, Tyzzerella 4, Parasutterella, Bifidobacterium) were negatively correlated with GGT, ALT and AST, while Dialister were negatively correlated with albumin. In addition, the index TBil showed positive correction with Parabacteroides.
Identification of potential bacteria biomarkers for HCC
At the genus level, Megamonas, Lachnospira, Eubacterium ventriosum and Lachnospiraceae UCG 001 were significantly decreased in NBNC-HCC patient samples as compared with healthy control samples (P < 0.050, Fig. 3a). In contrast, several genera such as Prevotella, Phascolarctobacterium, Anaerotruncus were particularly enriched in B-HCC patients than that in healthy controls (P < 0.050, Fig. 3b). The proportions of members of Buchnera, Lachnospira, Phascolarctobacterium, Eubacterium ventriosum were increased obviously in B-HCC patient samples compared with NBNC-HCC patients (Fig. 3c). Collectively, these differences revealed the dysbiosis involve in the development of HBV or non-HBV non-HCV related HCC.
Fig. 3.
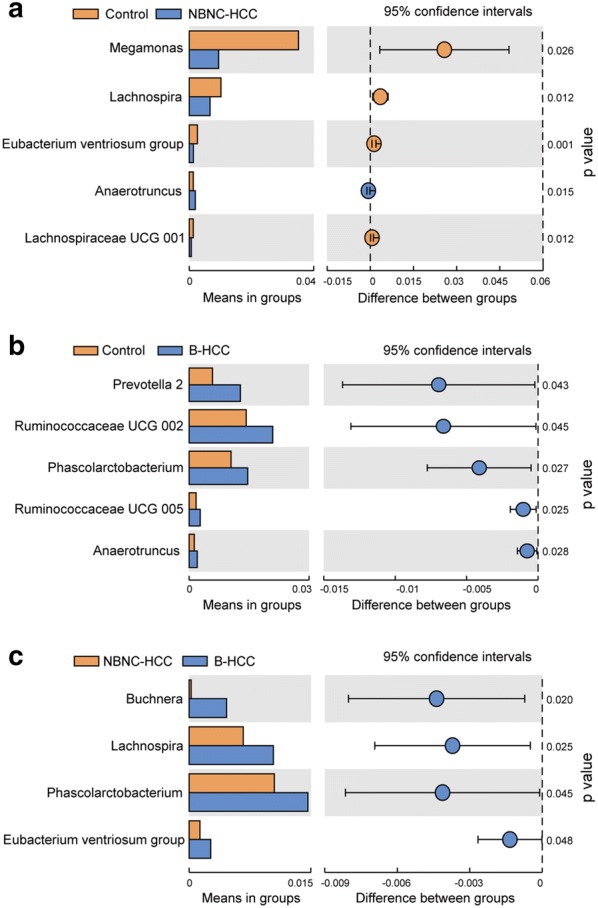
Genera strikingly different in gut microbiota of healthy controls (a), NBNC-HCC (b) and B-HCC (c)
Aberrant ecological networks of microbial communities occurred differently in B-HCC and NBNC-HCC patients.
To explore the relationships among various genera (top 35 and the significantly different genera data, Additional files 5, 8), the ecological networks of three groups were visualized. A striking feature was that taxonomically related genera tend to cluster in healthy controls (Fig. 4a). It was obvious that the within network highly associated connections of healthy controls occupied significant position and the interactions between these nodes were major balance. In NBNC-HCC patients (Fig. 4b), perhaps due to the differences in diet and excessively drinking levels (72.73%, Table 1), NBNC-HCC patients displayed a simpler concurrent network with fewer integrated symbiosis compared to healthy controls. All the interactions of bacteria in NBNC-HCC patients were positively interactions. Nevertheless, patients with B-HCC displayed multifaceted network with lots of genera, and totally grouped into a solo module with association to many other modules (Fig. 4c). Most of the bacteria related to inflammation gathered together. In this small symbiotic network, most of the interactions showed a stronger positive relation, such as Clostridium, Bryobacter, Lachnospiraceae, Buchnera, Burkholderia, Pseudobutyrivibrio. However, fewer interactions were negative, such as Alistipes, Bradyrhizobium and Sutterella, which implicated in competitive relationships for different genera. These observations implicated that the intestinal ecosystem becomes permissive for the development and maintenance of the related taxa in HCC patients.
Fig. 4.
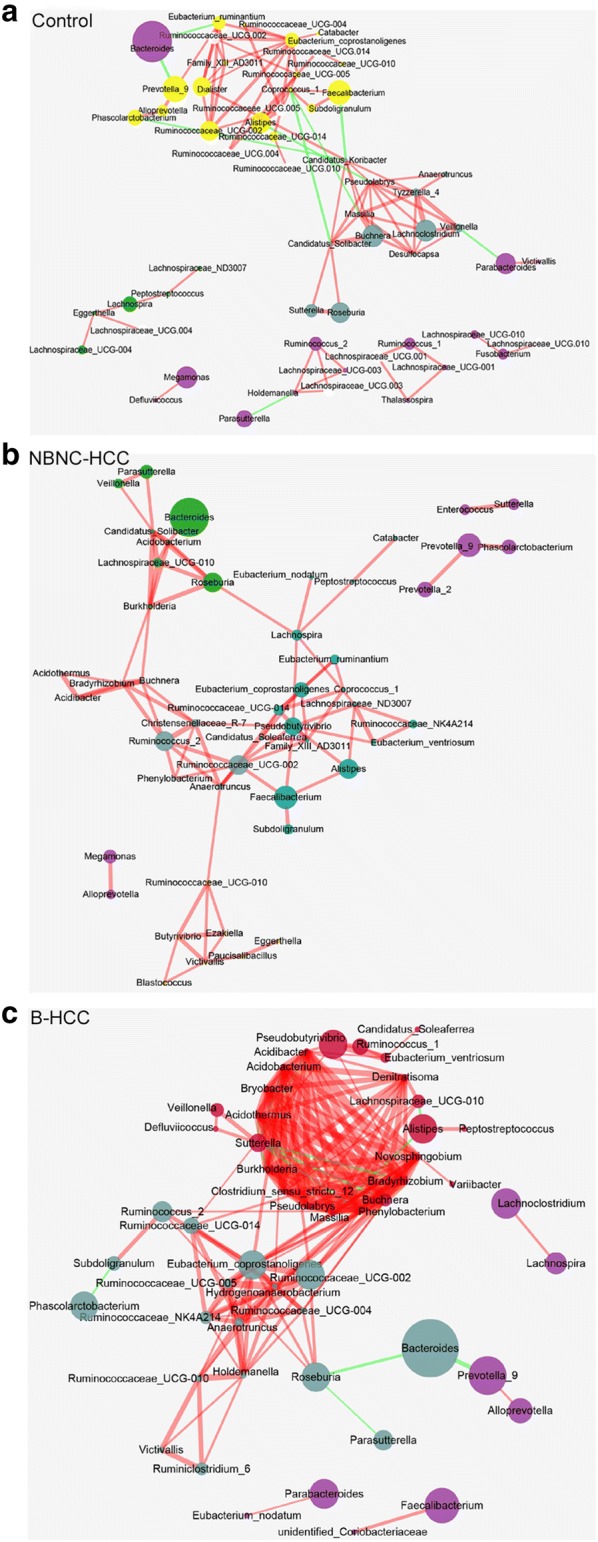
Networks to visualize interactions among different genera in three groups. a healthy controls; b NBNC-HCC patients; c B-HCC patients. Top 35 and significantly different genera are included. The density of the dashed line indicates the Pearson coefficient. Red links are the positive interactions between nodes, green links are the negative interactions. The size of node indicates the relative abundance
The potential multiple biological pathways of NBNC-HCC were different from other two groups.
To further understand the biological functions of genera among HCC patients and healthy controls, we performed Kyoto Encyclopedia of Genes and Genomes database (KEGG) analysis associated with gut microbiota [31]. We identified 15,039 biological pathways in all data. The predicated functions showed unique 109 for healthy controls, 12 for NBNC-HCC patients, and 18 for B-HCC patients (Fig. 5a). The mapped results indicated that multiple biological pathways were divided into seven branches (Fig. 5b). For instance, membrane transport, replication and repair, carbohydrate metabolism and amino acid metabolism were the predominant pathways.
Fig. 5.
Predicated function and multiple biological pathways in three groups. a Venn diagram of the predicated multiple biological pathways. b Annotated of multiple biological pathway distribution in seven major categories. c Heatmap of the top 35 biological pathways across three groups. d The relationship between top 35 biological pathways and genera at the criteria of P < 0.050 by Wilcoxon rank sum test in top 35. +P < 0.050; ++P < 0.010; +++P < 0.001 by Spearman’s correlation analysis. e Heatmap of multiple biological pathways across three groups at the P < 0.100 by Wilcoxon rank sum test. Multiple biological pathways at P < 0.050 by Wilcoxon rank sum test are marked with light green star, P < 0.001 with dark star. Red in color relates with amino acid metabolism, green in color relates to glucose metabolism
Healthy controls and B-HCC patients displayed similar pathways regarding to the top 35 multiple biological pathways (Fig. 5c, Additional files 8, and 9). However, B-HCC patients showed higher abundance of pathways related to chaperones and folding catalysts, general function prediction, DNA replication proteins and chromosome, which further supported that the HBV can destroy the normal function of DNA [5]. In addition, NBNC-HCC patients showed lower abundance of pathways related to amino acid metabolism (such as purine, cysteine and methionine, in red color) and glucose metabolism (such as starch and sucrose, glycolysis/gluconeogenesis, fructose and mannose, in green color). Meanwhile, KEGG analysis showed that microbial functional genes involved in oxidative phosphorylation, amino sugar and nucleotide sugar metabolism were also declined in B-HCC patients. It was also reported that marked depletion of amino acids and nucleotides metabolism in alcohol related cirrhosis patients [32]. In agreement with the study, we noted that some types of transport such as secretion system, transcription factors, other in coupled transporter and ABC transporters are enrichment in the multiple biological pathways of NBNC-HCC patients.
The relationships of multiple biological pathways were predicated to be associated with seven significantly different genera of top 35 genera (Fig. 5d). For instance, Phascolarctobacterium and Alloprevotella involved in the similar potential pathways, such as pyrimidine metabolism, cysteine and methionine metabolism and peptidases. Both genera had a negative relationship to arginine and proline metabolism and pyruvate metabolism. In addition, Ruminococcaceae UCG 002 involved in the potential pathways related to transporter, ribosome, ribosome biogenesis, chromosome, amino acid metabolism (Fig. 5d). Meanwhile, Lachnospira was potentially associated with nucleotide sugar metabolism, amino sugar, fructose and mannose metabolism.
We also identified significant changes in the multiple biological pathways of three groups (Fig. 5e). Five significantly discriminative metabolic pathways (D Arginine and D ornithine metabolism, bisphenol degradation, porphyrin and chlorophyll metabolism, linoleic acid metabolism, fructose and mannose metabolism) between healthy controls and NBNC-HCC patients, and three significantly discriminative pathways (energy metabolism, porphyrin and chlorophyll metabolism, D Arginine and D ornithine metabolism) between healthy controls and B-HCC patients. Due to the taxonomic microbiome, composition was different between two HCC groups, 11 significantly discriminative pathways (biosynthesis of 12, 14 and 16 membered macrolides, mRNA surveillance pathway, indole alkaloid biosynthesis, p53 signaling pathway, small cell lung cancer, toxoplasmosis, betalain biosynthesis, influenza A, viral myocarditis, colorectal cancer, cytochrome P450) between NBNC-HCC and B-HCC patients. The metabolic pathways such as tetracycline biosynthesis and tyrosine metabolism showed divergence enrichment degree among three groups (Additional files 10, 11, 12, 13, 14, 15).
Discussion
Our study focused on comparing the gut microbiota of HCC patients with HBV or without HBV/HCV infection. Like other bacteria associated with hepatic diseases, the bacterial diversity level and composition varied differently between NBNC-HCC and B-HCC patients. Generally, B-HCC patients were found to harbor higher species richness. At the phylum level, there was a decrease in Firmicutes and an increase in Proteobacteria of NBNC-HCC patients. However, Proteobacteria decreased in B-HCC patients. We found that NBNC-HCC patients harbored fewer potential anti-inflammatory bacteria and more pro-inflammatory bacteria. On the contrary, the B-HCC patients harbored more potential anti-inflammatory bacteria. Taken together, our analysis implicated that the gut microbiota plays an important role in the progression of HBV or non-HBV non-HCV related HCC.
It has been reported that Lactobacillus and Bifidobacterium were less in liver diseases [15, 33], but our study found both bacteria were increased in HCC patients and decreased in healthy controls (Additional file 4). Lactobacillus and Bifidobacterium were important probiotics to maintain intestinal microbial homeostasis and gut epithelial barrier [34]. However, recent studies reported that a higher abundance of Bifidobacterium were found be in the tissues of patients with colorectal adenomas or carcinoma [35, 36]. We hypothesized the amount of Bifidobacterium in colorectal adenomas or carcinoma tissue might be related with the tumor differentiation, mucosal barrier, and higher immune response to disease. For instance, Bifidobacterium longum nearly abolished melanoma tumor outgrowth by promoting anti-PD-L1 therapy [37]. By contrast, a recent report showed that commensal Bifidobacterium pseudolongum promoted the development of pancreatic cancer [38]. Thus, the different species of probiotics may have diverging effects in the tumor microenvironment. Taken together, the increasing abundance of Lactobacillus and Bifidobacterium in HCC patients remind us that it should be more attention paid on the probiotic function.
The NBNC-HCC patients harbored fewer anti-inflammatory bacteria and more pro-inflammatory bacteria in our study (Fig. 2a, b). We also noted the higher rate of alcohol consumption in NBNC-HCC patients (72.73%) (Table 1). Heavy drinking of alcohol causes inflammation of numerous organs. The alcohol correlated with pro-inflammatory bacteria, such as Escherichia-Shigella and Enterococcus would enhance leaky gut to the gut dysbiosis [39]. In addition, Escherichia overgrown to interfere the health equilibrium, which enters the liver through blood circulation, leading to the disorder of fatty acid metabolism [26], and becoming component of NAFLD pathogenesis [40]. In agreement with previous studies, our analysis further implicated that the diet and lifestyle habits play a vital role in the development of non-HBV non-HCV related HCC patients.
The B-HCC patients with more potential anti-inflammatory bacteria (such as Prevotella, Faecalibacterium) and fewer pro-inflammatory bacteria (such as Escherichia-Shigella, Enterococcus) were different from previous reports about HBV induced liver diseases [15, 41]. The discrepancy with our findings was probably the progression of liver diseases. The previous studies involved in HBV induced chronic carriers or liver cirrhosis, whereas the subjects of our study were from the HCC. There are essential differences between liver cirrhosis and HCC in accordance with pathogenesis, radiographic measurements, clinical symptoms and signs and other complications. Furthermore, immunoreaction was considered as a significant characteristic in the progression of HCC [3]. HBV uses multiple biological pathways to harness host innate immunity to enhance its replication, which initiates the immunological mechanisms to defense the acute or chronic infection [5]. Therefore, the discrepancy of fecal microbiota between the B-HCC and NBNC-HCC patients in our study perhaps ascribe to the HBV infection. The protective T-cell memory were lack in the chronic HBV infection, and T-cell responses also exhaustion [5, 42]. So, we infer the increased anti-inflammatory bacteria in B-HCC patients may be in response to HBV infection.
Interestingly, Ren et al. [18] indicated the butyrate-producing bacteria declined in early HCC patients, such as Ruminococcus, Feacalibacterium, Clostridium. However, the butyrate-producing bacteria presented heterogeneity in HBV and non-HBV non-HCV related HCC in our study. This further indicated HBV indeed play a role in changes of gut microbiota. Meanwhile, our study involved middle-aged adults (mean year is 56, Table 1, Additional file 1), whereas much older than the previous study [18]. In addition, 30 microbial markers were predicated using to identify early HCC in the previous study [18]. But some bacterial markers were not detected in the present study, such as Gemmiger. The conflict findings are possibly due to the individuals with different regions. The population of our study were all from Jiangsu Province. It has been reported that the diagnostic model of one location may be not used in other location, especially diagnostic efficiency declined with the geographic scale increased [43]. The characteristic changes of gut microbiota had a strongest relationship with the host location [43]. Thus, the diagnosis potential of microbial markers should be considered the geographic differences.
Currently, the treatment of HCC remains a challenge. Thus, it is necessary to develop an effective, life-prolonging strategy in the treatment of HCC patients. Recently, immunotherapy based on chimeric antigen receptor T-cell (CAR-T) [44, 45] or programmed cell death protein 1 (PD-1) [46, 47] has been proved as a promising strategy for cancer treatment. To date, the application of CAR-T cell therapy has some potential values in HCC [48]. In addition, fecal microbiota transplantation (FMT) can improve immune checkpoint inhibitors (ICI) associated colitis, which reconstructed the gut microbiome [49]. Based on the current microbiota analysis of HCC, we also suggest that the direct modification the gut microbiota of HCC patients associated with immunotherapy maybe beneficial for HCC patients (Fig. 6).
Fig. 6.
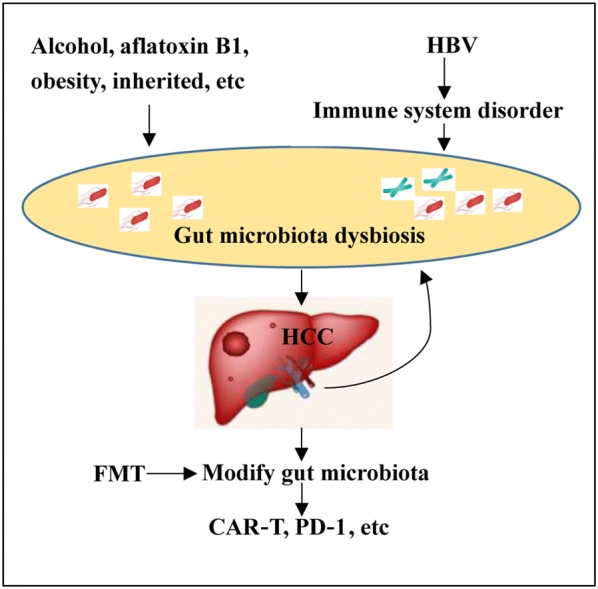
The predicated therapy of HCC patients. FMT, fecal microbiota transplantation; CAR-T, chimeric antigen receptor T-cells; PD-1, cell death protein-1
Conclusions
Our study provides strong evidences that HBV and non-HBV HCV related HCC were associated with different bacteria and displayed the aberrant ecological networks of microbial communities. Our data also provided the additional evidences implicated that the different bacteria play a potential role in the tumorigenesis of both B-HCC and NBNC-HCC patients. We suggest that the gut-liver-axis can be used for monitoring and preventing the progression of liver disease and HCC.
Methods
Study subjects
A total of 57 HCC patients and 33 healthy controls who attended annual physical examination were recruited from September 2016 to May 2017 at Nanjing Medical University Affiliated Cancer Hospital. All participants were provided a written informed consent upon enrolment. This study was approved by the Ethics Committee of Nanjing Medical University.
HCC diagnosis is depending on three factors which includes chronic liver disease background, the positive iconography examination results, or the positive pathological examination. All HCC patients were free from other viral infections, such as Human Immunodeficiency Virus (HIV) [50]. These patients were also free from any other types of liver disease. The HCC patients underwent viral serologic testing (HBsAg and HCVAb). The HCC patients were separated into two groups. Based on the history of HBV or positive HBsAg for more than 6 months, the HBV infection was diagnosed. The patients with HBV were defined as HBV related HCC (B-HCC). While in the other group includes the patients without HBsAg or HCVAb, so-called non-HBV non-HCV related HCC (NBNC-HCC) [51]. The healthy controls were excluded diabetes, metabolic syndrome, hypertension, inflammatory bowel diseases, liver disease and cancers. All of them did not get any antiviral therapy or immunotherapy in the past 6 months.
The physiological characteristics of volunteers including age, weight and drinking condition were investigated (Table 1, Additional file 1). The standard of drinking history was considered as any alcoholic beverage (unit: gram) [52]. Men intake of alcohol less than 9.9 grams daily (or 4.9 g/days for women) was considered as low, while the consumption between 10 and 39.9 g/days (or women 5 and 19.9 g/days) was moderate, more than 40 g/days (or women 20 g/days) was high. Excepting only one HCC patients, 56 HCC patients had the blood test before enrolled, such as AFP, ALT, AST, GGT, TBil, albumin, serum creatinine (SCr) (Additional file 1). Further, the level of cirrhosis was evaluated according to the Child–Pugh and MELD [53, 54].
Fecal sample collection, DNA extraction, PCR amplification, 16S rRNA sequencing
Fecal samples of each participant were obtained at hospital. The time span from sampling to Nanjing Medical University was intended within 24 h. Frozen samples were then stored at − 80 °C until analysis.
The genomic DNA of feces was extracted using kit (#DP328, Tiangen Biotech Co., Ltd., Beijing, China). The DNA concentration was detected using Qubit 2.0 Fluorometer (Thermo Fisher Scientific, USA). PCR was performed to produce V4 regions of the 16S rRNA gene using the conserved primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′), and no template DNA reaction was used as a negative control. PCR products were monitored using the 2% agarose gel. The strips between 400 and 450 bp were purified with GeneJET Gel Extraction Kit (Thermo Fisher Scientific, USA). PCR fragments were sequenced by Novogene Bioinformatics Technology Co., Ltd. (Tianjin, China).
16S rRNA data analysis
Illumina TruSeq DNA PCR-Free Library Preparation Kit (Illumina, USA) was used for generation of sequencing libraries. QIIME software 1.9 package was used to analyze sequences (Quantitative Insights Into Microbial Ecology, http://bio.cug.edu.cn/qiime/). Sequences having ≥ 97% resemblance were categorized as the same operational taxonomic units (OTUs). Alpha and beta diversity were calculated using the relative abundance of OTUs in each sample. The 16S rRNA data were assessed the potential multiple biological pathways of the gut microbiota using PICRUSt [55]. The KEGG ortholog identifiers (KO modules) were used to design the map of metabolic pathways in iPath 2. Datasets are publicly available (Accession number, GSE108847).
Genera interaction in ecological networks of microbial community analysis
To elucidate genera interactions in each group, we constructed three groups of topological overlap networks. The topological overlap of OTU was clustered into modules using WGCNA package of R (Version 3.4.4). The network analysis was visualized using Cytoscape 3.5.1. The threshold was set by Pearson r > 0 P < 0.1, and the topology overlap > 0.01 [56]. The genera including the top 35 and the significantly different genera were used to do network analysis.
Genera canonical correspondence analysis
To examine the distribution of genera associated with personal features (Body Mass Index, alcohol and AFP), canonical correspondence analysis (CCA) were visualized using the software CANOCO 4.5 [57]. Monte-Carlo permutation tests were performed to analyze the personal features played significant influence on the distribution of genera at P < 0.050. To avoid interference of rare species, top 25 genera were included.
Statistical analysis
The alpha diversity index was analyzed suing the QIIME (Version 1.9). PCoA was performed using the ade4 package of R (Version 3.4.4). The comparison of bacterial taxonomic or KO modules was tested by Wilcoxon rank sum test, P value was corrected using the Benjamini–Hochberg method, which named as false discovery rate (FDR) value.
Additional files
Additional file 1. Information of the study cohort used in this study. A total of 90 volunteers consisted of 33 healthy controls, 22 individuals of NBNC-HCC patients and 35 individuals of B-HCC patients were enrolled. The characteristics of participants including physical examination (gender, age, height, weight, BM, drinking condition), blood biochemical indexes (AFP, ALT, AST, GGT, TBil, INR, SCr, total protein, albumin, prothrombin time and activated partial thromboplastin time), Child–Pugh score and class, and MELD score.
Additional file 2. Relative abundance of bacteria at the phylum level.
Additional file 3. Relative abundance of bacteria at the order level.
Additional file 4. Relative abundance of bacteria at the genus level.
Additional file 5. Relative abundance of the top 35 genera.
Additional file 6. Relative abundance of the top 25 genera for canonical correspondence analysis.
Additional file 7. The relationship between six serologic indices (GST, AST, GGT, AFP, TBil, albumin) and top 35 genera is estimated by Spearman’s correlation analysis. *, P < 0.050; **, P < 0.010; ***, P < 0.001.
Additional file 8. Relative abundance of the significantly different genera (P < 0.050 by Wilcoxon rank sum test).
Additional file 9. Relative abundance of the top 35 predicated biological pathways.
Additional file 10. The differences in metabolic pathway between healthy controls and B-HCC patients. Green line is the special metabolism for healthy controls, yellow line is the special metabolism for B-HCC patients, red line for the common metabolism.
Additional file 11. The differences in metabolic pathway between healthy controls and NBNC-HCC patients. Green line is the special metabolism for healthy controls, yellow line is the special metabolism for NBNC-HCC patients, red line for the common metabolism.
Additional file 12. The differences in metabolic pathway between B-HCC and NBNC-HCC patients. Green line is the special metabolism for B-HCC patients, yellow line is the special metabolism for NBNC-HCC patients, red line for the common metabolism.
Additional file 13. The differences in biosynthesis and secondary metabolic pathway between healthy controls and B-HCC patients. Green line is the special metabolism for healthy controls, yellow line is the special metabolism for B-HCC patients, red line for the common metabolism.
Additional file 14. The differences in biosynthesis and secondary metabolic pathway between healthy controls and NBNC-HCC patients. Green line is the special metabolism for healthy controls, yellow line is the special metabolism for NBNC-HCC patients, red line for the common metabolism.
Additional file 15. The differences in biosynthesis and secondary metabolic pathway between B-HCC and NBNC-HCC patients. Green line is the special metabolism for B-HCC, yellow line is the special metabolism for NBNC-HCC patients, red line for the common metabolism.
Authors’ contributions
XL and YZ conceived and designed the study project. QL, FL, YZ, JX, JW, XM and XL did experiments and performed analysis. QL wrote the manuscript. QL, YZ and XL finalized the manuscript. All authors contributed at all stages and critically reviewed the content. All authors read and approved the final manuscript.
Acknowledgements
We greatly thank volunteers that gave informed consent and participated in sample collections. We gratefully thank Dr. Muhammad Haidar Zaman (Key Laboratory of Antibody Technology, National Health and Family Planning Commission, Nanjing Medical University, China) modified the language partly. We appreciate Dr. Kai Mi (China Astronautics Standards Institute, Beijing, China) for helping us perform the network analysis.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The data has been deposited into GEO database (Accession Number GSE108847).
Consent for publication
Not applicable.
Ethics approval and consent to participate
Our study was approved by the Ethics Committee of Nanjing Medical University. All participants provided a written informed consent upon enrolment. The methods were carried out in accordance with the relevant guidelines.
Funding
This work was supported by NSFC grant 81671983 and 81871628 to XL, natural science funding BK20161025, BK20161572 from Jiangsu province to LQ and XL, and starting package from NJMU (2015NJMUZD014) to XL, and 33 Program for High Level Talents of Jiangsu Province (BRA2016508), six talent peaks project in Jiangsu Province (WSW041) and Key Medical Talents of Jiangsu Province (2016KJQWZDRC-03) to YZ.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- AFP
alpha fetoprotein
- ALT
alanine Aminotransferase
- AST
aspartate aminotransferase
- BMI
body mass index
- CAR-T
chimeric antigen receptor T-cell
- CCA
canonical correspondence analysis
- FMT
fecal microbiota transplantation
- GGT
glutamyl transpeptidase
- B-HCC
HBV related hepatocellular carcinoma
- HCC
hepatocellular carcinoma
- INR
International Normalized Ratio
- KEGG
Kyoto Encyclopedia of Genes and Genomes database
- NAFLD
nonalcoholic fatty liver disease
- NBNC-HCC
non-HBV non-HCV related hepatocellular carcinoma
- PD-1
cell death protein-1
- TBil
total bilirubin
Contributor Information
Yewei Zhang, Email: zhangyewei@seu.edu.cn.
Xingyin Liu, Email: xingyinliu@njmu.edu.cn.
References
- 1.Tu T, Budzinska MA, Maczurek AE, Cheng R, Di BA, Warner FJ, et al. Novel aspects of the liver microenvironment in hepatocellular carcinoma pathogenesis and development. Int J Mol Sci. 2014;15:9422–9458. doi: 10.3390/ijms15069422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elserag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu LX, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastro Hepat. 2017;14:527–539. doi: 10.1038/nrgastro.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 5.Tsai KN, Kuo CF, Ou JJ. Mechanisms of hepatitis B virus persistence. Trends Microbiol. 2017;26:33–42. doi: 10.1016/j.tim.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khetrapal SP. Towards ending viral hepatitis as a public health threat: translating new momentum into concrete results in South-East Asia. Gut Pathog. 2018;10:9. doi: 10.1186/s13099-018-0237-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat. 2004;11:97–107. doi: 10.1046/j.1365-2893.2003.00487.x. [DOI] [PubMed] [Google Scholar]
- 8.Betrapally NS, Gillevet PM, Bajaj JS. Gut microbiome and liver disease. Transl Res. 2017;179:49–59. doi: 10.1016/j.trsl.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146:1513–1524. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Wang Y, Zhang X, Liu J, Zhang Q, Zhao Y, et al. Gut microbial dysbiosis is associated with altered hepatic functions and serum metabolites in chronic hepatitis B patients. Front Microbiol. 2017;8:2222. doi: 10.3389/fmicb.2017.02222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodhouse CA, Patel VC, Singanayagam A, Shawcross DL. Review article: the gut microbiome as a therapeutic target in the pathogenesis and treatment of chronic liver disease. Aliment Pharmacol Ther. 2018;47:1–11. doi: 10.1111/apt.14295. [DOI] [PubMed] [Google Scholar]
- 12.Abushanab A, Quigley EMM. The role of the gut microbiota in nonalcoholic fatty liver disease. Nat Rev Gastro Hepat. 2010;7:691–701. doi: 10.1038/nrgastro.2010.172. [DOI] [PubMed] [Google Scholar]
- 13.Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, White MB, et al. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol-Gastr L. 2012;303:675–685. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, et al. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562–572. doi: 10.1002/hep.24423. [DOI] [PubMed] [Google Scholar]
- 15.Lu H, Wu Z, Xu W, Yang J, Chen Y, Li L. Intestinal microbiota was assessed in cirrhotic patients with hepatitis B virus infection. Microb Ecol. 2011;61:693–703. doi: 10.1007/s00248-010-9801-8. [DOI] [PubMed] [Google Scholar]
- 16.Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastro Hepat. 2018;15:397–411. doi: 10.1038/s41575-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764–775. doi: 10.1002/hep.28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H, et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. 2018;1–10. [DOI] [PMC free article] [PubMed]
- 19.Fukui H. Gut microbiome-based therapeutics in liver cirrhosis: basic consideration for the next step. J Clin Transl Hepatol. 2017;5:249–260. doi: 10.14218/JCTH.2017.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin N, Yang F, Li A, Edi P, Chen Y, Shao L, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 21.Clark DP. The fermentation pathways of Escherichia coli. FEMS Microbiol Rev. 1989;5:223. doi: 10.1016/0168-6445(89)90033-8. [DOI] [PubMed] [Google Scholar]
- 22.Corrêaoliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MAR. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunol. 2016;5:e73. doi: 10.1038/cti.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willemsen LE, Koetsier MA, van Deventer SJ, van Tol EA. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E(1) and E(2) production by intestinal myofibroblasts. Gut. 2003;52:1442–1447. doi: 10.1136/gut.52.10.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang HB, Wang P, Wang X, Wan YL, Liu YC. Butyrate enhances intestinal epithelial barrier function via up-regulation of tight junction protein Claudin-1 transcription. Dig Dis Sci. 2012;57:3126–3135. doi: 10.1007/s10620-012-2259-4. [DOI] [PubMed] [Google Scholar]
- 25.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdezhumarán LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J. Ethanol and liver: recent insights into the mechanisms of ethanol-induced fatty liver. World J Gastroenterol. 2014;20:14672. doi: 10.3748/wjg.v20.i40.14672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng J, Wang Q, Liu K, Yang S, Fan X, Liu B, et al. Systemic and splanchnic lipopolysaccharide and endothelin-1 plasma levels in liver cirrhosis before and after transjugular intrahepatic portosystemic shunt. Gastroent Res Pract. 2016;2016:1–5. doi: 10.1155/2016/8341030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 29.Jiang W, Wu N, Wang X, Chi Y, Zhang Y, Qiu X, et al. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci Rep. 2015;5:8096. doi: 10.1038/srep08096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bei R, Mizejewski GJ. Alpha fetoprotein is more than a hepatocellular cancer biomarker: from spontaneous immune response in cancer patients to the development of an AFP-based cancer vaccine. Curr Mol Med. 2011;11:564–581. doi: 10.2174/156652411800615162. [DOI] [PubMed] [Google Scholar]
- 31.Ogata H, Fujibuchi W, Goto S, Kanehisa M. A heuristic graph comparison algorithm and its application to detect functionally related enzyme clusters. Nucleic Acids Res. 2000;28:4021. doi: 10.1093/nar/28.20.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Nan Q, Jing G, Qian G, Fang D, Ding S, et al. Functional gene arrays-based analysis of fecal microbiomes in patients with liver cirrhosis. BMC Genomics. 2014;15:753. doi: 10.1186/1471-2164-15-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu M, Wang B, Fu Y, Chen Y, Yang F, Lu H, et al. Changes of fecal Bifidobacterium species in adult patients with hepatitis B virus-induced chronic liver disease. Microb Ecol. 2012;63:304–313. doi: 10.1007/s00248-011-9925-5. [DOI] [PubMed] [Google Scholar]
- 34.Ashraf R, Shah NP. Immune system stimulation by probiotic microorganisms. Crit Rev Food Sci Nutr. 2014;54:938–956. doi: 10.1080/10408398.2011.619671. [DOI] [PubMed] [Google Scholar]
- 35.Nugent JL, Mccoy AN, Addamo CJ, Wei J, Sandler RS, Keku TO. Altered tissue metabolites correlate with microbial dysbiosis in colorectal adenomas. J Proteome Res. 2014;13:1921–1929. doi: 10.1021/pr4009783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kosumi K, Hamada T, Koh H, Borowsky J, Bullman S, Twombly TS, et al. The amount of bifidobacterium genus in colorectal carcinoma tissue in relation to tumor characteristics and clinical outcome. Am J Pathol. 2018;188:2839–2852. doi: 10.1016/j.ajpath.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayelet S, Leticia C, Nathaniel H, Williams JB, Keston AM, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8(4):403–416. doi: 10.1158/2159-8290.CD-17-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bajaj J, Heuman D, Hylemon P, Sanyal A, White M, Monteith P, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60:940–947. doi: 10.1016/j.jhep.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdou RM, Zhu L, Baker RD, Baker SS. Gut microbiota of nonalcoholic fatty liver disease. Dig Dis Sci. 2016;61:1268–1281. doi: 10.1007/s10620-016-4045-1. [DOI] [PubMed] [Google Scholar]
- 41.Wei X, Yan X, Zou D, Yang Z, Wang X, Liu W, et al. Abnormal fecal microbiota community and functions in patients with hepatitis B liver cirrhosis as revealed by a metagenomic approach. BMC Gastroenterol. 2013;13:1–8. doi: 10.1186/1471-230X-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferrari C. HBV and the immune response. Liver Int. 2015;35:121–128. doi: 10.1111/liv.12749. [DOI] [PubMed] [Google Scholar]
- 43.He Y, Wu W, Zheng H-M, Li P, McDonald D, Sheng H-F, et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat Med. 2018;24:08–27. doi: 10.1038/s41591-018-0164-x. [DOI] [PubMed] [Google Scholar]
- 44.Katz SC, Burga RA, Mccormack E, Wang LJ, Mooring JW, Point G, et al. Phase I hepatic immunotherapy for metastases study of intra-arterial chimeric antigen receptor modified T cell therapy for CEA+ liver metastases. Clin Cancer Res. 2015;21:3149–3159. doi: 10.1158/1078-0432.CCR-14-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Q, Zhang Z, Peng M, Fu S, Xue Z, Zhang R. CAR-T cell therapy in gastrointestinal tumors and hepatic carcinoma: from bench to bedside. Oncoimmunology. 2016;5:e1251539. doi: 10.1080/2162402X.2016.1251539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Routy B, Gopalakrishnan V, Daillère R, Zitvogel L, Wargo JA, Kroemer G. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol. 2018;15:382–396. doi: 10.1038/s41571-018-0006-2. [DOI] [PubMed] [Google Scholar]
- 48.Gao H, Li K, Tu H, Pan X, Jiang H, Shi B, et al. Development of T cells redirected to glypican-3 for the treatment of hepatocellular carcinoma. Clin Cancer Res. 2014;20:6418–6428. doi: 10.1158/1078-0432.CCR-14-1170. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat Med. 2018;24:1804–1808. doi: 10.1038/s41591-018-0238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu J, Zhang T, Cao L, Li A, Zheng K, Zhang N, et al. Toll like receptor7 polymorphisms in relation to disease susceptibility and progression in Chinese patients with chronic HBV infection. Sci Rep. 2017;7:12417. doi: 10.1038/s41598-017-12698-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okamura Y, Sugiura T, Ito T, Yamamoto Y, Ashida R, Uesaka K. The impact of the hepatitis B core antibody status on recurrence in patients with Non-B Non-C hepatocellular carcinoma after curative surgery. Dig Surg. 2017;35:243–251. doi: 10.1159/000479340. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Fan J, Wang B, Lu L, Shi J, Niu J, et al. Guidelines for management of nonalcoholic fatty liver disease: an updated and revised edition. J Clin Hepatol. 2010;15:229–232. [PubMed] [Google Scholar]
- 53.Hoffmann J. Stapler transection of the oesophagus for bleeding oesophageal varices. Scand J Gastroenterol. 1983;18:707–711. doi: 10.3109/00365528309182084. [DOI] [PubMed] [Google Scholar]
- 54.Durand F, Valla D. Assessment of the prognosis of cirrhosis: child-Pugh versus MELD. J Hepatol. 2005;42:S100–S107. doi: 10.1016/j.jhep.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 55.Langille MG, Zaneveld J, Caporaso JG, Mcdonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Braak CJFT, Smilauer P. CANOCO reference manual and canodraw for windows user’s guide: software for canonical community ordination (version 4.5) Ithaca: Microcomputer Power; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Information of the study cohort used in this study. A total of 90 volunteers consisted of 33 healthy controls, 22 individuals of NBNC-HCC patients and 35 individuals of B-HCC patients were enrolled. The characteristics of participants including physical examination (gender, age, height, weight, BM, drinking condition), blood biochemical indexes (AFP, ALT, AST, GGT, TBil, INR, SCr, total protein, albumin, prothrombin time and activated partial thromboplastin time), Child–Pugh score and class, and MELD score.
Additional file 2. Relative abundance of bacteria at the phylum level.
Additional file 3. Relative abundance of bacteria at the order level.
Additional file 4. Relative abundance of bacteria at the genus level.
Additional file 5. Relative abundance of the top 35 genera.
Additional file 6. Relative abundance of the top 25 genera for canonical correspondence analysis.
Additional file 7. The relationship between six serologic indices (GST, AST, GGT, AFP, TBil, albumin) and top 35 genera is estimated by Spearman’s correlation analysis. *, P < 0.050; **, P < 0.010; ***, P < 0.001.
Additional file 8. Relative abundance of the significantly different genera (P < 0.050 by Wilcoxon rank sum test).
Additional file 9. Relative abundance of the top 35 predicated biological pathways.
Additional file 10. The differences in metabolic pathway between healthy controls and B-HCC patients. Green line is the special metabolism for healthy controls, yellow line is the special metabolism for B-HCC patients, red line for the common metabolism.
Additional file 11. The differences in metabolic pathway between healthy controls and NBNC-HCC patients. Green line is the special metabolism for healthy controls, yellow line is the special metabolism for NBNC-HCC patients, red line for the common metabolism.
Additional file 12. The differences in metabolic pathway between B-HCC and NBNC-HCC patients. Green line is the special metabolism for B-HCC patients, yellow line is the special metabolism for NBNC-HCC patients, red line for the common metabolism.
Additional file 13. The differences in biosynthesis and secondary metabolic pathway between healthy controls and B-HCC patients. Green line is the special metabolism for healthy controls, yellow line is the special metabolism for B-HCC patients, red line for the common metabolism.
Additional file 14. The differences in biosynthesis and secondary metabolic pathway between healthy controls and NBNC-HCC patients. Green line is the special metabolism for healthy controls, yellow line is the special metabolism for NBNC-HCC patients, red line for the common metabolism.
Additional file 15. The differences in biosynthesis and secondary metabolic pathway between B-HCC and NBNC-HCC patients. Green line is the special metabolism for B-HCC, yellow line is the special metabolism for NBNC-HCC patients, red line for the common metabolism.
Data Availability Statement
The data has been deposited into GEO database (Accession Number GSE108847).