Abstract
White adipose tissue (WAT) can dynamically expand and remodel through adipocyte hypertrophy and hyperplasia. The relative contribution of these 2 mechanisms to WAT expansion is a critical determinant of WAT function and dysfunction in obesity. However, little is known about the signaling systems that determine the mechanisms of WAT expansion. Here, we show that the GPCR LPA4 selectively activates Gα12/13 proteins in adipocytes and limits continuous remodeling and healthy expansion of WAT. LPA4-KO mice showed enhanced expression of mitochondrial and adipogenesis genes and reduced levels of inhibitory phosphorylation of PPARγ in WAT, along with increased production of adiponectin. Furthermore, LPA4-KO mice showed metabolically healthy obese phenotypes in a diet-induced obesity model, with continuous WAT expansion, as well as protection from WAT inflammation, hepatosteatosis, and insulin resistance. These findings unravel a potentially new signaling system that underlies WAT plasticity and expandability, providing a promising therapeutic approach for obesity-related metabolic disorders.
Keywords: Metabolism
Keywords: Adipose tissue, G-protein coupled receptors, Obesity
LPA4, a Gα12/13-coupling receptor in adipocytes, limits healthy expansion of adipose tissue in obesity.
Introduction
White adipose tissue (WAT) is a highly active metabolic and endocrine organ. Metabolic health depends on the maintenance of normal mass and function of WAT (1). In response to overnutrition, WAT can dynamically expand or remodel through enlargement of existing white adipocytes (hypertrophy) and recruitment of new adipocytes (hyperplasia) (2). The balance between adipocyte hypertrophy and adipocyte hyperplasia in obesity is critical for metabolic health (1). When hypertrophic adipocytes face limits of expansion, the adipocytes die due to WAT inflammation and fibrosis (3). On the other hand, WAT expansion through adipocyte hyperplasia may present a mechanism for healthy fat storage capacity. Indeed, recent studies have determined metabolic benefits from healthy expansion of WAT. Genetic mouse models have shown improvements in their metabolic profile through expansion of adipose tissue by modulating PPARγ activity (4), mitochondrial function (5), or production of adiponectin, an antidiabetic adipokine (6). In addition, the PPARγ-FGF1 axis is essential for continuous expansion of WAT in response to nutritional stress induced by a high-fat diet (HFD) (7). However, the signaling system that determines the mechanisms of WAT expansion remains less understood.
The function of WAT is under the control of multiple cell signaling systems, including that of GPCRs (8). It has been well known that Gαs-coupled GPCR stimulation activates lipolysis, which is counteracted by Gαi-coupled receptors (8). In addition, the Gαq-coupled receptor GPR120 stimulates glucose uptake in adipocytes (9). However, the roles of Gα12/13-coupled receptors in WAT are totally unknown.
Lysophosphatidic acid (LPA) is a pleiotropic lipid mediator acting via specific GPCRs LPA1–6 (10). Importantly, an LPA-producing enzyme, autotaxin (ATX), is abundantly expressed in WAT. Earlier studies have shown the involvement of ATX in the development of obesity and metabolic disorders (11, 12). Among the 6 LPA receptors, LPA1 is involved in adipocyte differentiation and development of obesity (13). In addition, multiple LPA receptors are expressed in immune cells, including macrophages and lymphocytes (14), all of which can affect WAT function (15). Notably, a hierarchical clustering analysis of GPCR mRNA expression revealed that 1 of the LPA receptors, LPA4, is an adipose cluster GPCR (16).
To better understand the roles of GPCRs and LPA in adipose biology, we focused on the adipose cluster GPCR, LPA4. We show that LPA4 selectively activates Gα12/13 proteins in adipocytes. When LPA4-KO mice were fed a standard chow diet, they showed enhanced expression of mitochondrial and adipogenesis genes and a reduced level of inhibitory phosphorylation of PPARγ in WAT. Consistently, the serum adiponectin level was higher in LPA4-KO mice. Moreover, LPA4-KO mice showed metabolically healthy obese (MHO) phenotypes in a diet-induced obesity model, with continuous WAT expansion and protection from WAT inflammation, hepatosteatosis, and insulin resistance.
Results
The adipose cluster GPCR LPA4 selectively activates the Gα12/13 protein in C3H10T1/2- and mouse embryonic fibroblast–derived adipocytes.
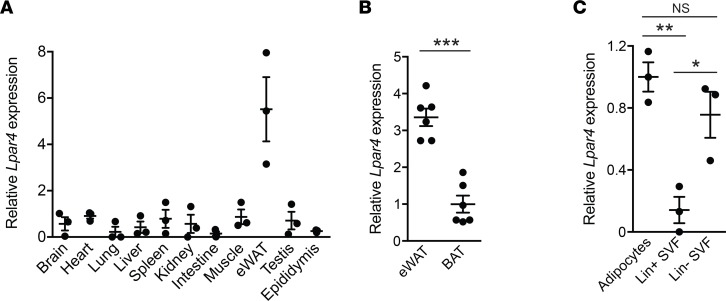
In accordance with a hierarchical clustering analysis of GPCR mRNA expression (16), the expression of mouse Lpar4, the gene encoding LPA4, was the highest in epididymal WAT (eWAT) among the tissues that were examined (Figure 1A). eWAT showed higher expression of Lpar4 than brown adipose tissue (BAT) (Figure 1B). Fractionation of mouse eWAT revealed that Lpar4 expression was high in adipocytes and the lineage-negative (Lin–) population of the stromal vascular fraction (SVF) that contains adipocyte progenitors (Figure 1C).
Figure 1. LPA4 expression levels in various tissues and fractions from adipose tissue.
(A) Mouse Lpar4 mRNA expression in various tissues (n = 3). (B) Comparison of mouse Lpar4 mRNA expression between epididymal white adipose tissue (eWAT) and brown adipose tissue (BAT) (n = 6). (C) Mouse Lpar4 mRNA expression in adipocytes, Lin+ and Lin– populations of the stromal vascular fraction (SVF) isolated from eWAT (n = 3). Each symbol represents an individual mouse. Data are expressed as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, unpaired Student’s t test (B) and 1-way ANOVA with Bonferroni’s post hoc test (C).
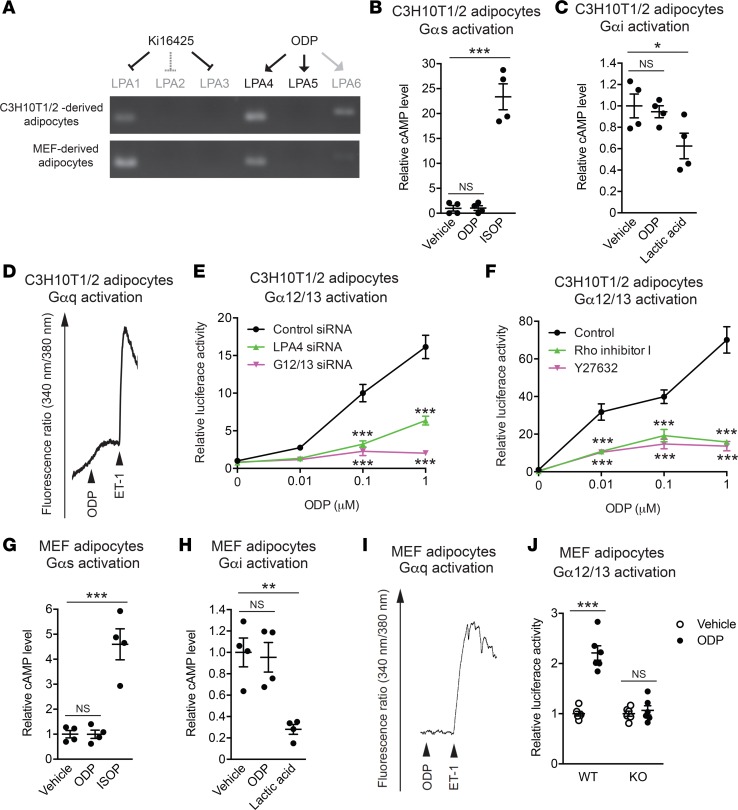
The function of GPCRs is, in part, determined by their coupling Gα proteins, namely Gαs, Gαi, Gαq, and Gα12/13 proteins. To determine the specific Gα proteins that mediate LPA4 signaling in adipocytes, we first used C3H10T1/2-derived adipocytes, which show high Lpar4 mRNA expression (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.97293DS1). One of the difficulties in studying the LPA receptor function is that most cell types express multiple LPA receptor subtypes and no subtype-specific LPA receptor agonists are currently available (10). Indeed, C3H10T1/2-derived adipocytes have mRNA transcripts for LPA1, LPA4, and LPA6 (Figure 2A). To circumvent this problem, we used octadecenyl phosphate (ODP), which is an agonist for LPA4 and LPA5 (Supplemental Figure 2, A and B) (17). Therefore, ODP should predominantly activate LPA4 in C3H10T1/2-derived adipocytes (Figure 2A), although it also partially activates LPA6 (Figure 2A and Supplemental Figure 2C). Furthermore, to inhibit potential weak LPA1 activation by ODP, C3H10T1/2-derived adipocytes were treated with the LPA1–3 antagonist Ki16425 (Figure 2A). Heterologously expressed LPA4 has been shown to activate all classes of Gα proteins, Gαs, Gαi, Gαq, and Gα12/13 proteins (10); however, in contrast to the known Gαs-, Gαi-, and Gαq-coupled adipose GPCR ligands (18, 19), ODP failed to change intracellular cAMP or Ca2+ concentrations in C3H10T1/2-derived adipocytes (Figure 2, B–D). Furthermore, ODP did not affect lipolysis activity, which is regulated by Gαs- or Gαi-coupling GPCR (8), in freshly isolated murine adipocytes (Supplemental Figure 3). Together, these results suggest that the ODP-LPA4 axis does not strongly activate Gαs, Gαi, or Gαq proteins in adipocytes. We next examined the possibility that LPA4 activates Gα12/13 proteins in adipocytes, by using a serum response factor–responsive element (SRF-RE) luciferase reporter assay, which detects signaling downstream of RhoA activation (20). In contrast to the unresponsiveness in the cAMP and Ca2+ assays, ODP induced excessive activation of the SRF-RE reporter gene in a dose-dependent manner (Figure 2E). SRF-RE reporter gene activation by ODP was clearly reversed by siRNA-mediated knockdown of LPA4 or Gα12/13 gene expression (Figure 2E), or by pharmacological inhibition of Rho or ROCK, downstream targets of Gα12/13 proteins, with Rho inhibitor I or Y27632, respectively (Figure 2F). Consistently, ODP treatment induced approximately 3-fold higher RhoA activation in these cells (Supplemental Figure 2D). Next, the predominant coupling of LPA4 to Gα12/13 proteins in adipocytes was further confirmed by a primary cell culture system. We found that Lpar4 expression in isolated Lin– population of the SVF decreased by >10-fold following subculture, which could not be recovered by adipocyte differentiation (Supplemental Figure 1). Therefore, we decided to use mouse embryonic fibroblast–derived (MEF-derived) adipocytes, which maintained substantial expression of Lpar4 mRNA (Supplemental Figure 1). As observed in C3H10T1/2-derived adipocytes, ODP failed to change intracellular cAMP and Ca2+ concentrations in MEF-derived adipocytes from WT mice (Figure 2, G–I). Furthermore, ODP increased SRF-RE reporter gene activity in MEF-derived adipocytes from WT but not from Lpar4-KO mice (Figure 2J). Overall, these results strongly suggest that LPA4 predominantly activates Gα12/13 proteins in adipocytes and prompted us to explore unknown functions of the Gα12/13-coupled receptor in adipocytes, which are most likely distinct from lipolysis and glucose uptake, regulated by Gαs, Gαi, and Gαq protein–coupled receptors (8).
Figure 2. LPA4 selectively activates Gα12/13 protein in C3H10T1/2- and mouse embryonic fibroblast–derived (MEF-derived) adipocytes.
(A) Top, schematic of the assay system for predominant activation of LPA4 with octadecenyl phosphate (ODP) in the presence of Ki16425. Middle and bottom, ethidium bromide–stained agarose gels of reverse transcription PCR (RT-PCR) products of expressed LPA receptors in C3H10T1/2-derived (middle) and MEF-derived (bottom) adipocytes. (B and C) Effects of ODP (2.5 μM) on intracellular cAMP levels in C3H10T1/2-derived adipocytes (n = 4). Isoproterenol (ISOP, 10 μM) and lactic acid (30 mM) were used as positive controls for Gαs (B) and Gαi (C) protein activation, respectively. (D) A representative chart recording of intracellular Ca2+ level in C3H10T1/2-derived adipocytes upon stimulation with ODP (1 μM). Endothelin-1 (ET-1, 100 nM) was used as a positive control for Gαq protein activation. (E) Effects of ODP and siRNA against LPA4 or Gα12/13 on serum response factor–responsive element (SRF-RE) luciferase activity in C3H10T1/2-derived adipocytes (n = 4). (F) Effects of Rho and ROCK inhibition by Rho inhibitor I (2 μg/ml) and Y27632 (10 μM), respectively, on ODP-induced SRF-RE luciferase activity in C3H10T1/2-derived adipocytes (n = 4). (G and H) Effects of ODP (1 μM) on intracellular cAMP levels in MEF-derived adipocytes (n = 4). Isoproterenol (ISOP, 10 μM) and lactic acid (30 mM) were used as positive controls for Gαs (G) and Gαi (H) protein activation, respectively. (I) A representative chart recording of intracellular Ca2+ level in MEF-derived adipocytes upon stimulation with ODP (1 μM). Endothelin-1 (ET-1, 100 nM) was used as a positive control for Gαq protein activation. (J) Effects of ODP (1 μM) on SRF-RE luciferase activity in MEF-derived adipocytes isolated from WT and Lpar4-KO mice (n = 4). Results are representative of at least 3 independent experiments. Data are expressed as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001; 1-way ANOVA with Bonferroni’s (E and F) or Dunnett’s (B,C,G,H) post hoc test, and unpaired Student’s t test (J) were used.
Loss of LPA4 results in enhanced mitochondrial activity in WAT and increased serum adiponectin level on chow diet.
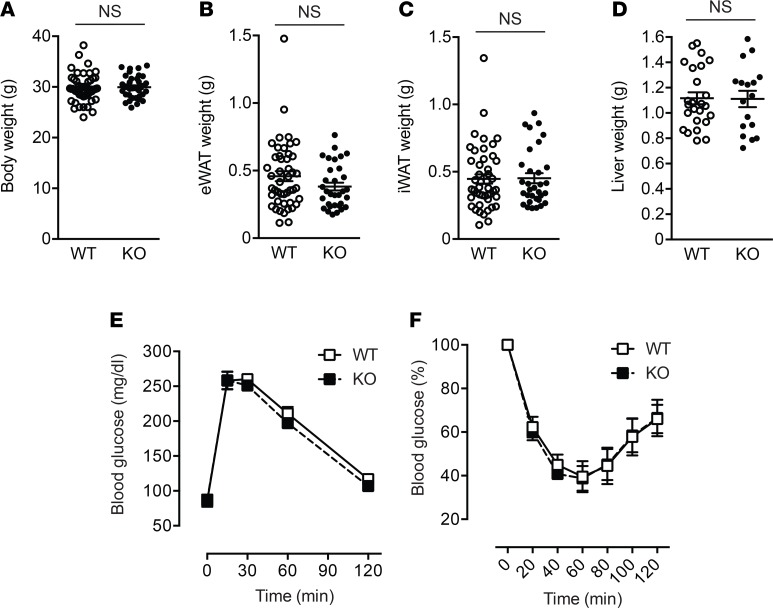
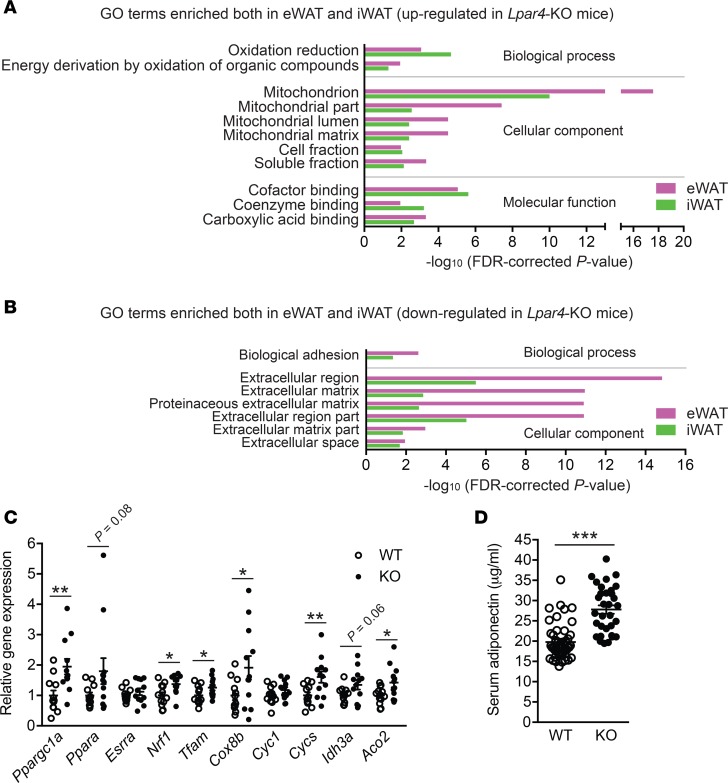
We then analyzed metabolic phenotypes of Lpar4-KO mice. Although ATX is abundantly expressed in mouse WAT, even under standard chow diet conditions (11), we could not detect any obvious phenotypic differences between chow-fed Lpar4-KO and littermate control WT mice in body weight, organ weight, or glucose homeostasis (Figure 3). We next performed microarray expression profiling of WAT transcripts from these mice to identify global changes caused by the deletion of the Lpar4 gene. This analysis revealed significant upregulation of genes involved in mitochondrial functions (Figure 4A and Supplemental Table 1) and downregulation of extracellular matrix–related genes (Figure 4B and Supplemental Table 1) both in eWAT and inguinal WAT (iWAT) of Lpar4-KO mice. We focused on the upregulation of mitochondria-related genes because mitochondrial function in WAT is recognized as a key determinant of whole body metabolism and energy homeostasis (21). In concordance with the microarray analysis, quantitative PCR (qPCR) analysis validated the upregulation of genes comprising the regulators of mitochondrial biogenesis (Ppargc1a and Ppara), their downstream target genes (Nrf1 and Tfam), and other mitochondrial genes (Cox8b, Cycs, Idh3a, and Aco2) in eWAT of chow-fed Lpar4-KO mice (Figure 4C). Recent studies have revealed that adipocyte mitochondrial function is directly linked to the production of adiponectin, an antidiabetic adipokine (21). Consistent with the enhanced expression of genes involved in mitochondrial biogenesis, serum adiponectin levels were higher in Lpar4-KO mice than in WT mice (Figure 4D).
Figure 3. LPA4 ablation has no effects on body weight, organ weight, or glucose homeostasis on chow diet.
(A–D) Whole body (A), epididymal white adipose tissue (eWAT) (B), inguinal WAT (iWAT) (C), and liver (D) weight of chow-fed WT (n = 25–50) and Lpar4-KO mice (n = 17–36). (E) Blood glucose levels of chow-fed WT and Lpar4-KO mice during the oral glucose-tolerance test (n = 7 each). (F) Blood glucose levels of chow-fed WT (n = 10) and Lpar4-KO (n = 6) mice during the insulin-tolerance test. The changes were plotted as a percentage of basal glucose versus time. Each symbol represents an individual mouse. Data are expressed as the mean ± SEM. Unpaired Student’s t test.
Figure 4. Loss of LPA4 results in increased mitochondria-related gene expression and serum adiponectin level on chow diet.
(A and B) Gene Ontology (GO) enrichment analysis of genes upregulated (A) and downregulated (B) in WAT from chow-fed Lpar4-KO mice compared with WT control mice (n = 4 each). Probe sets with FDR < 0.01 were regarded as differentially expressed. GO terms significantly enriched in analyses of both epididymal white adipose tissue (eWAT) and inguinal WAT (iWAT) are shown. (C) Relative expression of mitochondria-related genes in eWAT from chow-fed WT and Lpar4-KO mice. Data are expressed as the mean ± SEM (n = 12 each). (D) Serum adiponectin levels of chow-fed WT (n = 46) and Lpar4-KO (n = 32) mice. Each symbol represents an individual mouse. Data are expressed as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, unpaired Student’s t test.
LPA4 ablation results in enhanced adipose tissue expandability and protection from hepatosteatosis and insulin resistance in a diet-induced obesity model.
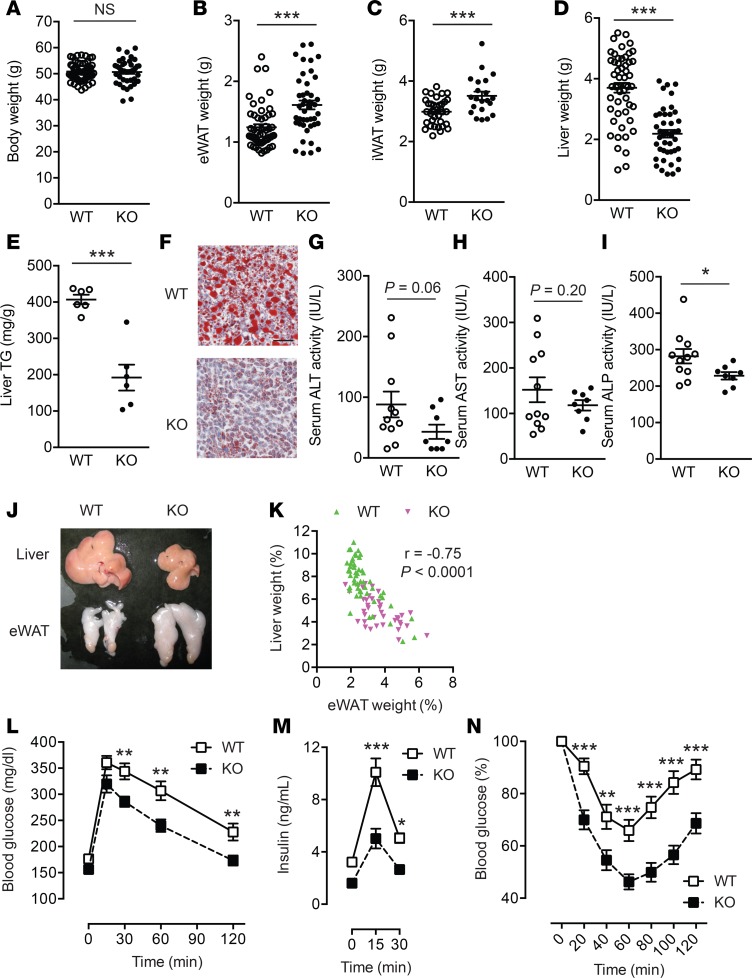
Compromised mitochondrial biogenesis in adipocytes and hypoadiponectinemia are common features of insulin-resistant obesity (21). To further explore the roles of LPA4 signaling pathways in WAT, the metabolic phenotypes of Lpar4-KO mice were examined on HFD. Even in this HFD-induced obesity model, body weight of Lpar4-KO mice was equivalent to that of WT mice (Figure 5A). Furthermore, serum lipid levels were comparable between WT and Lpar4-KO mice (Supplemental Figure 4). However, Lpar4-KO mice had greater WAT mass than WT mice (Figure 5, B and C). In contrast, Lpar4-KO mice were clearly protected from severe hepatomegaly (Figure 5D) and steatosis (Figure 5, E–I). Additionally, eWAT weight showed an inverse correlation with liver weight (Figure 5, J and K), indicating that increased adipose tissue expansion protects Lpar4-KO mice from ectopic fat accumulation in the liver. Consistently, in the glucose-tolerance test, both plasma glucose and insulin levels were significantly lower in Lpar4-KO mice than in WT mice (Figure 5, L and M). In addition, the capacity to regulate blood glucose levels after insulin injection was significantly higher in Lpar4-KO mice than in WT mice (Figure 5N). In accordance with higher Lpar4 expression in WAT than in BAT (Figure 1B), we could not detect any statistically significant differences in BAT weight or marker gene expression, in addition to body temperature or food intake, between WT and Lpar4-KO mice (Supplemental Figure 5).
Figure 5. LPA4 ablation results in enhanced adipose tissue expandability and protection from hepatosteatosis and insulin resistance in a diet-induced obesity model.
(A–D) Whole body (A), epididymal white adipose tissue (eWAT) (B), inguinal WAT (iWAT) (C), and liver (D) weight of high-fat diet–fed (HFD-fed) WT (n = 42–52) and Lpar4-KO mice (n = 25–44). (E) Triglyceride (TG) content of the liver from HFD-fed WT and Lpar4-KO mice (n = 6 each). (F) Representative Oil Red O staining of liver sections from HFD-fed WT and Lpar4-KO mice. Scale bar: 100 μm. (G and I) Serum alanine transaminase (ALT) (G), aspartate transaminase (AST) (H), and alkaline phosphatase (ALP) (I) activities of HFD-fed WT and Lpar4-KO mice (n = 8–10). (J) A representative photograph of livers and eWATs from HFD-fed WT and Lpar4-KO mice. (K) Inverse correlation between eWAT and liver weight from HFD-fed WT (n = 52) and Lpar4-KO (n = 39) mice. (L–N) Blood glucose (L and N) and plasma insulin (M) levels of HFD-fed WT (n = 22–24) and Lpar4-KO (n = 20–21) mice during the oral glucose-tolerance test (L and M) and the insulin-tolerance test (N). Each symbol represents an individual mouse. Data are expressed as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001; unpaired Student’s t test (A–E, G–I) or 1-way ANOVA with Bonferroni’s post hoc test (L–N) were used.
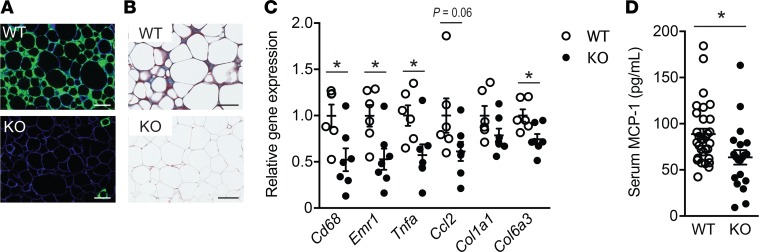
To further investigate the possibility that the metabolic improvements observed in HFD-fed Lpar4-KO mice were associated with increased WAT functions, we performed detailed histological and molecular analyses of eWAT. Mac2-staining of macrophages and Masson’s trichrome staining of collagen in eWAT sections revealed that Lpar4-KO mice were protected from macrophage infiltration and fibrotic change (Figure 6, A and B). Consistently, qPCR analyses showed that the expression of inflammation-related (Cd68, Emr1, Tnfa, and Ccl2) and fibrosis-related (Col1a1 and Col6a3) genes was lower in Lpar4-KO mice than in WT mice (Figure 6C). Reflecting the less inflamed state of eWAT of Lpar4-KO mice, these mice had lower serum MCP-1 levels than WT mice (Figure 6D).
Figure 6. LPA4 ablation inhibits adipose tissue inflammation and fibrosis in a diet-induced obesity model.
(A) Representative fluorescence microscopy images of epididymal white adipose tissue (eWAT) from high-fat diet–fed (HFD-fed) WT and Lpar4-KO mice, costained with anti-Mac2 (green) and anti–caveolin-1 (blue). Scale bar: 100 μm. (B) Representative Masson’s trichrome staining of eWAT sections from HFD-fed WT and Lpar4-KO mice. Scale bar: 100 μm. (C) Relative mRNA expression of inflammation- or fibrosis-related genes in eWAT from HFD-fed WT (n = 6) and Lpar4-KO mice (n = 7). (D) Serum MCP-1 levels of HFD-fed WT (n = 31) and Lpar4-KO (n = 20) mice. Each symbol represents an individual mouse. Data are expressed as the mean ± SEM. *P < 0.05, unpaired Student’s t test.
LPA4 signaling in nonhematopoietic cells exaggerates eWAT inflammation and fibrosis due to diet-induced obesity.
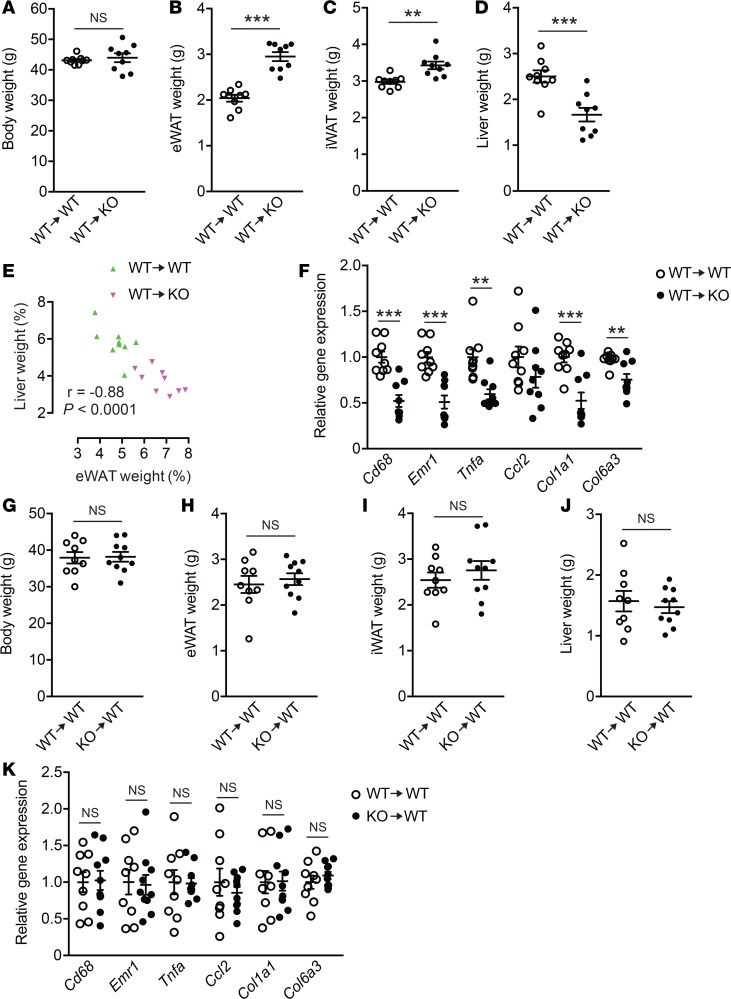
To determine the contribution of LPA4 signaling in nonhematopoietic cells, including adipocytes and adipocyte progenitors, to eWAT inflammation and fibrosis, we reconstituted lethally irradiated WT and Lpar4-KO mice with BM from WT mice and then fed them with HFD (Supplemental Figure 6A). Similar to naive Lpar4-KO mice, WT BM–reconstituted Lpar4-KO mice showed higher WAT mass and protection from liver enlargement (Figure 7, A–E), and lower expression of inflammation- and fibrosis-related genes in eWAT than the WT control mice (Figure 7F). On the other hand, we detected no significant difference between WT BM– and Lpar4-KO BM–reconstituted WT mice (Supplemental Figure 6B) in terms of the inflammation- and fibrosis-related gene expression, as well as the body/organ weights (Figure 7, G–K). Therefore, these results demonstrate that LPA4 signaling in nonhematopoietic cells, likely to be adipocytes or adipocyte progenitors, exaggerates eWAT inflammation and fibrosis due to diet-induced obesity.
Figure 7. WT BM–reconstituted Lpar4-KO mice phenocopy the intact Lpar4-KO mice on high-fat diet (HFD).
(A–D) Whole body (A), epididymal white adipose tissue (eWAT) (B), inguinal WAT (iWAT) (C), and liver (D) weight of HFD-fed WT BM–reconstituted WT (WT→WT) and Lpar4-KO (WT→KO) mice (n = 9 each). (E) Inverse correlation between eWAT and liver weight from HFD-fed WT BM–reconstituted WT (WT→WT) and Lpar4-KO (WT→KO) mice (n = 9 each). (F) Relative mRNA expression of inflammation- or fibrosis-related genes in eWAT from HFD-fed WT BM–reconstituted WT (WT→WT) and Lpar4-KO (WT→KO) mice (n = 9 each). (G–J) Whole body (G), eWAT (H), iWAT (I), and liver (J) weight of HFD-fed WT and Lpar4-KO BM-reconstituted WT mice (WT→WT and KO→WT, respectively, n = 9–10). (K) Relative mRNA expression of inflammation- or fibrosis-related genes in eWAT from HFD-fed WT and Lpar4-KO BM-reconstituted WT mice (WT→WT and KO→WT, respectively, n = 9–10). Each symbol represents an individual mouse. Data are expressed as the mean ± SEM. **P < 0.01, ***P < 0.001, unpaired Student’s t test.
LPA4 signaling inhibits PPARγ-related gene expression and limits continuous remodeling of adipose tissue in a diet-induced obesity model.
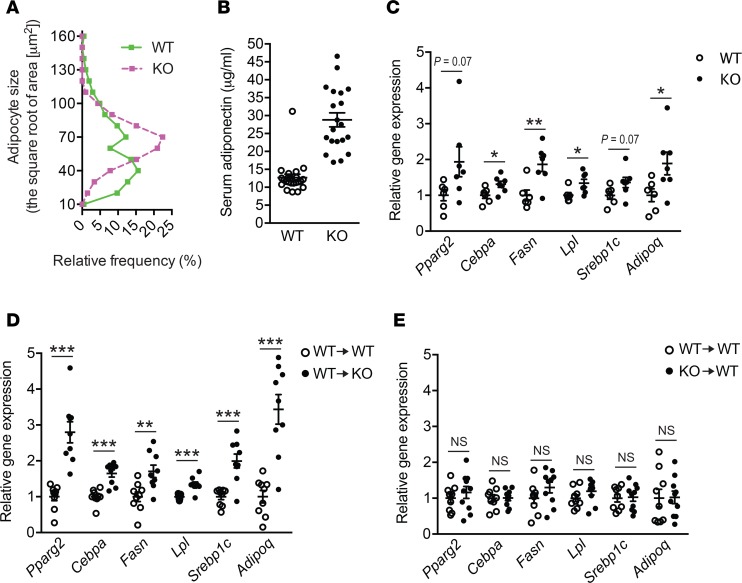
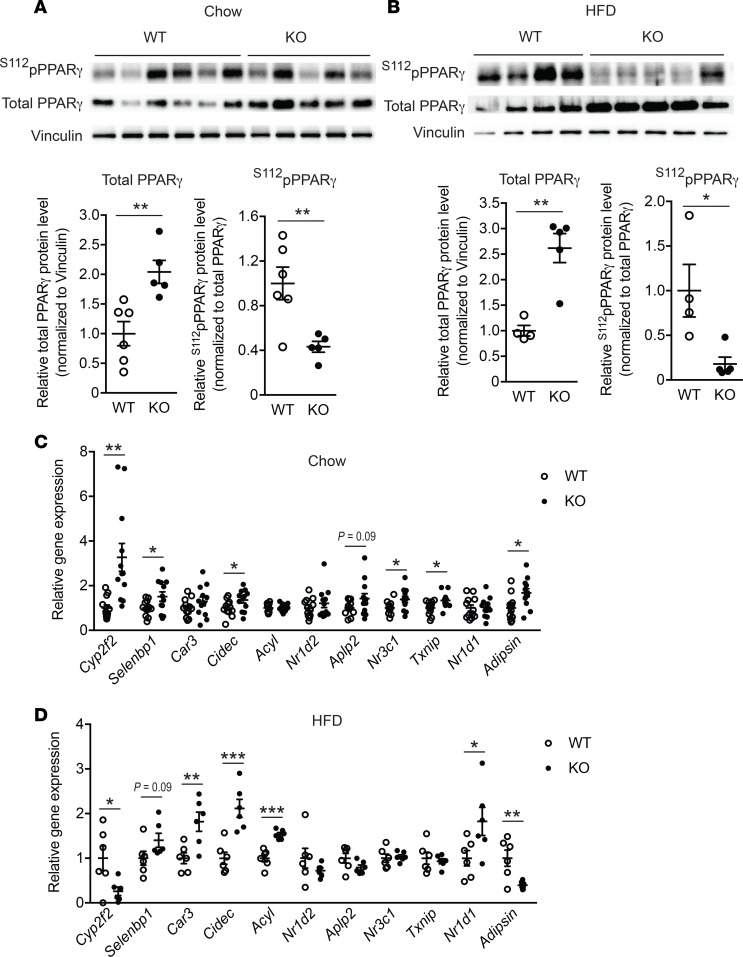
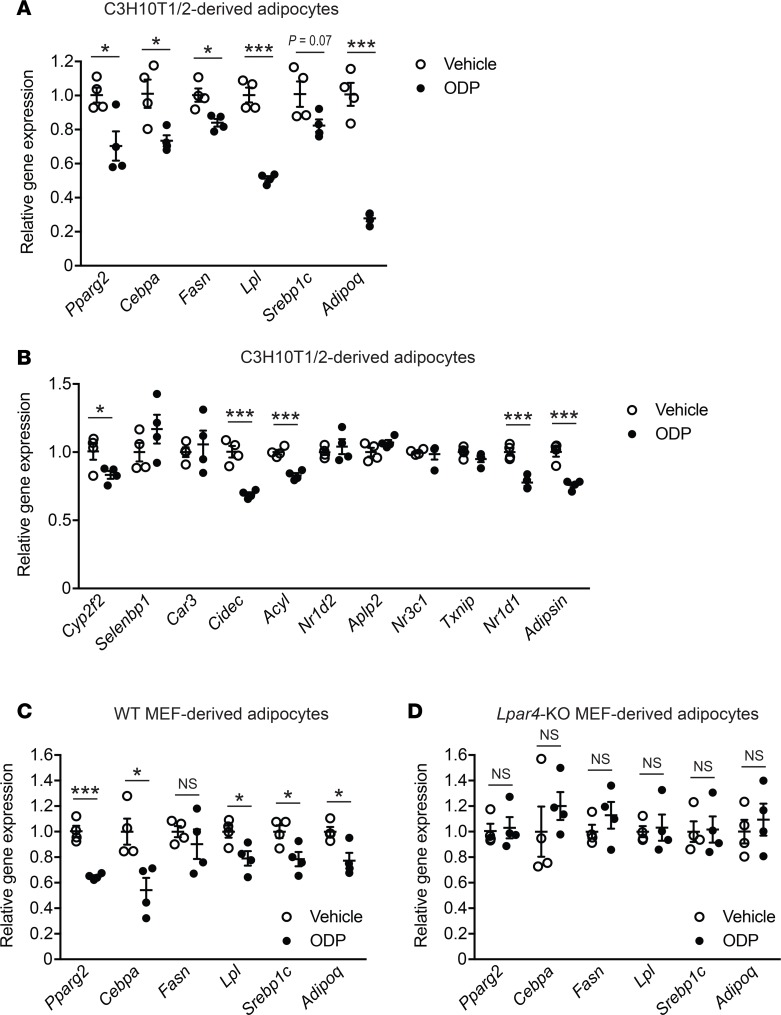
WAT undergoes a continuous remodeling that normally maintains tissue health. However, in the obesity state, the remodeling process may spin out of control, leading to adipocyte death that associates WAT inflammation and fibrosis (3), as shown here in WT mice. Histological analyses revealed that eWAT from Lpar4-KO mice showed a well-organized arrangement of uniform-sized adipocytes, which was distinct from that of WT mice, showing marked heterogeneity in adipocyte size (Figure 6A and Figure 8A). In accordance with the healthy appearance of eWAT in HFD-fed Lpar4-KO mice, these mice had high serum adiponectin levels (Figure 8B) comparable with chow-fed Lpar4-KO mice (Figure 4D). In contrast, HFD-fed WT mice showed lower serum adiponectin levels (Figure 8B) than chow-fed WT mice (Figure 4D). We postulated that LPA4 deficiency improves the remodeling of adipose tissue, with accelerated differentiation of new adipocytes from adipogenic precursor cells. Indeed, the expression of adipogenesis marker genes was considerably higher in eWAT from Lpar4-KO mice than from WT mice (Figure 8C). This difference was completely dependent on the genotype of the nonhematopoietic cells (Figure 8, D and E). Since overall metabolic phenotypes of Lpar4-KO mice seemed similar to those of mice showing higher PPARγ activity (4, 22, 23), we examined the effect of the loss of Lpar4 not only on the expression, but also on the phosphorylation status of PPARγ in vivo. Notably, eWAT from chow- and HFD-fed Lpar4-KO mice showed reduced levels of inhibitory phosphorylation of PPARγ at Ser112 (24) (Figure 9, A and B). Consistently, chow- and HFD-fed Lpar4-KO mice showed global expression changes in genes regulated by phosphorylation of PPARγ (25, 26) (Figure 9, C and D). We then examined the direct effect of LPA4 signaling on adipogenesis in vitro. Consistent with our in vivo data, ODP inhibited the induction of adipogenesis marker gene expression in C3H10T1/2 cells under adipogenic culture conditions (Figure 10A). Furthermore, ODP treatment also changed the expression of genes regulated by phosphorylation of PPARγ in C3H10T1/2-derived adipocytes (Figure 10B). The ODP effects on adipogenesis marker gene expression were confirmed in WT MEF–derived adipocytes (Figure 10C), although little or no significant effect of ODP was detected on the expression of genes regulated by phosphorylation of PPARγ (data not shown). This apparent discrepancy may be due to the lower expression of Lpar4 in MEF-derived adipocytes than in C3H10T1/2-derived adipocytes (Supplemental Figure 1). Because ODP did not affect adipogenesis marker gene expression in MEF-derived adipocytes from Lpar4-KO mice (Figure 10D), the effects of ODP are likely to be LPA4 dependent. Collectively, these results indicate that LPA4 signaling inhibits continuous remodeling of adipose tissues, partly by direct inhibition of the adipogenic program.
Figure 8. LPA4 inhibits continuous remodeling of adipose tissue in a diet-induced obesity model.
(A) Quantification of adipocyte size in epididymal white adipose tissue (eWAT) from high-fat diet–fed (HFD-fed) WT and Lpar4-KO mice. Representative histograms from 3 sections each are shown. (B) Serum adiponectin levels of HFD-fed WT (n = 22) and Lpar4-KO (n = 20) mice. (C) Relative mRNA expression of adipogenesis marker genes in eWAT from HFD-fed WT (n = 6) and Lpar4-KO mice (n = 7). (D) Relative mRNA expression of adipogenesis marker genes in eWAT from HFD-fed WT BM–reconstituted WT (WT→WT) and Lpar4-KO (WT→KO) mice (n = 9 each). (E) Relative mRNA expression of adipogenesis marker genes in eWAT from HFD-fed WT and Lpar4-KO BM-reconstituted WT mice (WT→WT and KO→WT, respectively, n = 9–10). Each symbol represents an individual mouse. Data are expressed as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, unpaired Student’s t test.
Figure 9. LPA4 ablation affects phosphorylation status of PPARγ and expression of genes regulated by PPARγ phosphorylation.
(A and B) Western blotting analysis of total PPARγ and Ser112-phosphorylated PPARγ (S112pPPARγ) in eWAT from chow-fed WT (n = 6) and Lpar4-KO (n = 5) mice (A), as well as high-fat diet–fed (HFD-fed) WT (n = 4) and Lpar4-KO (n = 5) mice (B). (C and D) Relative mRNA expression of genes regulated by phosphorylation of PPARγ in eWAT from chow-fed WT and Lpar4-KO mice (C) (n = 12 each), as well as HFD-fed WT and Lpar4-KO mice (D) (n = 6 each). Each symbol represents an individual mouse. Data are expressed as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, unpaired Student’s t test.
Figure 10. LPA4 activation decreases PPARγ-related gene expression in C3H10T1/2- and mouse embryonic fibroblast–derived (MEF-derived) adipocytes.
(A and B) Effects of octadecenyl phosphate (ODP) on expression of adipogenesis marker genes (A) and genes regulated by phosphorylation of PPARγ (B) in C3H10T1/2-derived adipocytes. C3H10T1/2 cells were differentiated into adipocytes, serum-starved for 24 hours, and treated with 2.5 μM ODP for 24 hours (n = 4). (C and D) Effects of ODP on expression of adipogenesis marker genes in WT-mouse MEF-derived adipocytes (C) and Lpar4-KO MEF-derived adipocytes (D). MEFs were differentiated into adipocytes, serum-starved for 24 hours, and treated with 10 μM ODP for 24 hours (n = 4). Data shown are representative of 2 independent experiments. Data are expressed as the mean ± SEM. *P < 0.05, ***P < 0.001, unpaired Student’s t test.
Discussion
Obesity is closely associated with a cluster of metabolic disorders, including type 2 diabetes; however, there is a subgroup of obese individuals, called MHO, in which these obese individuals do not demonstrate an increased risk of developing obesity-related disorders (27). Recent studies have revealed that the status MHO is a consequence of an increased capacity for storing fat, likely due to improvement in WAT remodeling, mitochondrial function, and adiponectin production (21, 28). Our results reveal that the Lpar4-KO mouse strain is a new animal model of MHO and provides new insights into the mechanisms and factors underlying the MHO phenotype. Furthermore, alterations in the expression or function of LPA4, Gα12/13, and/or Rho/ROCK in WAT may partly explain why some people who are obese can maintain healthy metabolism and do not develop diabetes. Future human studies on LPA4 expression in adipose tissues and LPAR4 single nucleotide polymorphisms may be helpful for understanding the mechanisms of MHO and development of new therapeutic/diagnostic approaches.
The transcriptional regulation of Lpar4 has not been fully elucidated. The abundant expression of Lpar4 in adipose tissue implies that its transcription is regulated by major transcription factors in adipocytes, including PPARγ. Indeed, we observed significant increase of Lpar4 transcripts in C3H10T1/2 cells and MEFs after adipogenic differentiation (Supplemental Figure 1). Notably, Lpar4 expression drastically decreased following culture and expansion of isolated Lin– population of the SVF. Furthermore, the expression of Lpar4 could not be recovered by an adipocyte differentiation procedure. However, the reason for the failure in maintaining Lpar4 expression in the primary adipocyte cultures remains unclear. Further investigations on the factors that repress Lpar4 expression in tissue culture cells and better understanding of Lpar4 transcriptional regulation would yield important information for the future therapeutic/diagnostic application of LPA4.
Earlier studies have shown the involvement of ATX in the development of obesity and metabolic disorders both in mice and humans (12, 29). The adipose tissue abundantly expresses ATX (11, 12, 29) and is considered to be one of the main sources of circulating ATX (12, 29). Adipose tissue–specific ATX-KO mice showed improved systemic metabolism (11, 12); the results of the present study are in agreement with the prodiabetic role of LPA produced by ATX. However, the improvement of systemic metabolism, including protection from steatosis in Lpar4-KO mice shown here, was more drastic than that reported in ATX-KO mice. This discrepancy might reflect complex roles of ATX and LPA in whole body metabolism regulated by multiple LPA receptors. For example, LPA1 is reported to be involved in adipocyte differentiation and development of obesity (13, 29). Furthermore, multiple LPA receptors are expressed in immune cells including macrophages, T cells, and B cells (14), all of which can regulate adipose tissue inflammation and fibrosis. Although ATX is proposed as a drug target for obesity and its related diseases (29), we suppose that targeting LPA4 will provide a more focused and efficient therapeutic strategy for intervention in metabolic disorders.
Because Lpar4-KO mice showed a drastic phenotype under HFD-fed — but not under chow diet-fed — conditions, it is plausible that LPA4 signaling is boosted in diet-induced obesity. However, HFD feeding did not change the mRNA expression of Lpar4 and Lpp3 (encoding LPP3, an LPA degrading enzyme) in WAT (Supplemental Figure 7, A and B). In addition, the Enpp2 (encoding ATX) expression was lower in adipocytes of HFD-fed mice than those of chow-fed mice (Supplemental Figure 7C), which is consistent with that in a previous report (12). Therefore, the drastic phenotype owing to HFD is less likely to be caused by the change in mRNA expression of LPA4 or LPA-producing/degrading enzymes. Notably, LPA is not a single molecular entity, and different LPA species have different agonist activities depending on its receptor subtypes (30). Therefore, LPA4 activity may be affected by the possible difference in the varieties of LPA species between chow and HFD feeding conditions.
Compromised mitochondrial activity in adipocytes, reduced amount/activity of PPARγ, and hypoadiponectinemia are common features of insulin-resistant obesity in both rodents and humans (21, 31, 32). Since all of these are closely related to each other, revealing the primary effect due to Lpar4 deletion is difficult for explaining the whole metabolic phenotypes. In addition, our microarray data suggest that LPA4 signaling directly contributes to fibrotic changes in HFD-fed animals. To identify the primary target of LPA4 action, future time-course studies with conditional deletion of LPA4 or pharmacological blockage of LPA4 signaling are warranted. Analysis of Lpar4-KO mice with various PPARγ ligands, as well as adiponectin-KO or phosphorylation-resistant PPARγ mutant knock-in background, would also be beneficial for elucidating the pathophysiological roles of LPA4 adipocytes in obesity and diabetes.
The metabolic phenotypes of Lpar4-KO mice clearly mimic the insulin-sensitizing effect of thiazolidinediones (TZDs) (22). Although TZDs have been widely used for the treatment of obesity-induced diabetes, they display adverse effects, including fluid retention, which can aggravate congestive heart failure. Since Lpar4-KO mice showed comparable body weight with WT mice, unlike the TZD-treated mice (22), the pharmacological targeting of LPA4 might be a better strategy to improve obesity-induced diabetes and to avoid serious side effects. However, LPA4 is also involved in the embryonic vascular development (33) or hematopoiesis-supporting activity (34), and further studies are necessary to confirm its clinical safety and tolerability.
In summary, we have demonstrated that LPA4 selectively couples to Gα12/13 proteins in adipocytes and limits proper remodeling and healthy expansion of WAT in diet-induced obesity. Although further studies are necessary regarding the role of LPA4 in other aspects of WAT function — including a direct involvement in angiogenesis, hypoxia, and/or fibrosis (3) — our results provide insights to better understand the complex pathophysiology underlying obesity, related disorders, and the obesity paradox (27).
Methods
Mice.
Lpar4-KO mice on a C57BL/6 genetic background were generated as described previously (33). C57BL/6 mice congenic for the Ly5 locus (C57BL/6-Ly5.1 mice) were purchased from Sankyo Labo Service by permission of Hiromitsu Nakauchi (Institute of Medical Science, The University of Tokyo, Tokyo, Japan). Mice were housed on a 12-hour light/dark cycle and fed with standard chow (MF; Oriental Yeast) or HFD (HFD32; CLEA Japan). Mice were fed with standard chow diet for 14–16 weeks or HFD for 14–16 weeks (starting at 6 weeks old). All experiments in this study were conducted on male littermates. Mice were housed under specific pathogen-free conditions in an air-conditioned room in accordance with institutional guidelines.
Cell lines.
Mouse C3H10T1/2 mesenchymal pluripotent cells were obtained from ATCC (catalog CCL-226). RH7777 rat hepatoma cells and B103 rat neuroblastoma cells were originally from Jerold Chun (Scripps Research Institute, La Jolla, California, USA). RH7777 and B103 cells stably expressing HA-tagged LPA receptors were established and maintained as previously described (30, 35, 36). The heterologous expression of LPA receptors in the cells was confirmed by the immunodetection of HA-tag by flow cytometry (35, 36) or IHC conducted essentially as described previously (37). Briefly, the cells were fixed with 2% paraformaldehyde in PBS and incubated with anti-HA (3F10; Roche Diagnostics) for 2 hours. After washing with PBS, the cells were incubated with Alexa Fluor 488–conjugated goat anti–rat IgG for 2 hours. The cells were counterstained with DAPI. The fluorescence microscopy was performed using a laser scanning confocal microscope (LSM510 META, Zeiss).
Chemicals and reagents.
ODP (2.5 mM stock in methanol stored at –80°C) was a gift from Gabor Tigyi (The University of Tennessee, Memphis, Tennessee, USA). 1-Oleoyl LPA (10 mM stock in 50% ethanol stored at –30°C) was from Avanti Polar Lipids. Ki16425 and Y27632 (10 mM stock in DMSO stored at –30°C) were from Calbiochem. Rho inhibitor I (0.1 μg/μl stock in water stored at 4°C) was from Cytoskeleton. siRNAs (ON-TARGETplus SMARTpool) were from Thermo Fisher Scientific.
Adipose tissue fractionation.
eWAT was minced and digested with 0.26 U/ml Liberase TM (Roche Diagnostics) in PBS at 37°C under constant shaking at 125 rpm for 2 hours. During the digestion period, the suspension was triturated several times through a pipet to dissociate the clumps. The suspension was then passed through a 100-μm filter (BD Biosciences) to remove undigested clumps and debris. The effluent was centrifuged at 500 g for 10 minutes, and the floating cells (adipocytes) and pellet (SVF) were collected. Adipocytes were washed twice with PBS containing 0.5% BSA (fatty acid free; MilliporeSigma). Total RNA was collected using RNeasy Lipid Tissue Mini Kit. SVFs were washed twice with PBS containing 0.5% BSA and stained with phycoerythrin-conjugated (PE-conjugated) anti–mouse CD31 (MEC13.3; BioLegend), CD45 (30-F11, BioLegend), and TER119 (BioLegend) antibodies. The cells were further incubated with anti-PE MicroBeads (Miltenyi Biotec) and subjected to magnetic-activated cell sorting (MACS; Miltenyi Biotec) to collect Lin– and Lin+ SVF. MACS was repeated twice to increase the purity.
qPCR.
Total RNA was isolated from the tissues and cells with RNeasy Lipid Tissue Mini Kit (Qiagen), in accordance with the manufacturer’s instructions. cDNA was prepared with SuperScript II, SuperScript III (Invitrogen), or PrimeScript (Takara Bio) reverse transcriptase with random primers (Invitrogen or Takara Bio) and/or oligo-dT primers (Takara Bio). For qPCR, samples were run in a 10-μl reaction using a stepOnePlus real-Time PCR system (Applied Biosystems) with Fast SYBR Green PCR Master Mix (Applied Biosystems) or in a 20-μl reaction using a LightCycler system (Roche Diagnostics) with KAPA SYBR Fast qPCR Kit (Kapa Biosystems) or FastStart DNA Master SYBR Green I (Roche Diagnostics). Relative gene expression levels were calculated after normalization to 36B4 (encoded by Rplp0) or 18S ribosomal RNA (Rn18s) expression level. Primer sequences used are shown in Supplemental Table 2.
Preparation of C3H10T1/2-derived adipocytes.
C3H10T1/2 cells were differentiated into adipocytes as previously described (38). Briefly, the cells were seeded in 100-mm dishes (Corning), grown to be confluent, and kept at confluent for 2 days. Differentiation into adipocytes was induced by treating the confluent cells with 0.5 mM 3-isobutyl-1-methylxanthine (IBMX, MilliporeSigma), 0.25 μM dexamethasone (MilliporeSigma), 10 μg/ml human insulin (MilliporeSigma), and 10 μM pioglitazone (MilliporeSigma) in DMEM (MilliporeSigma) containing 10% FBS (Invitrogen). The medium was changed every 3 days. From day 6 of differentiation, the cells were incubated with DMEM containing 10% FBS and 10 μg/ml human insulin for 2 days. For the experiments, the cells were washed once with PBS and treated with 0.25% trypsin, 2 mM EDTA, and 0.2 U/ml liberase TM at 37°C for 5–10 minutes until most cells were detached from the culture dishes. The cells were collected with DMEM containing 10% FBS, filtered through a 100-μm filter (BD Biosciences) to remove cell aggregates, and reseeded in collagen-coated dishes or multiwell plates (Iwaki).
Preparation of MEF-derived adipocytes.
Primary MEFs were isolated from embryos of WT and Lpar4-KO mice at E12.5. Embryos were removed and further dissected by removing their maternal tissues, yolk sack, heads, and visceral mass. Then, the embryos were sheared through an 18-gauge syringe in DMEM containing 10% FBS, filtered through a 100-μm filter to remove remaining aggregates, and cultured at 37°C. Experiments were performed at passage 3–5. For adipogenesis induction, MEFs were seeded in 100-mm dishes or 24-well plates, grown to be confluent, and kept at confluent for 3 days. Differentiation into adipocytes was induced by treating the confluent cells with 0.5 mM IBMX, 0.25 μM dexamethasone, 10 μg/ml human insulin, and 10 μM pioglitazone DMEM containing 10% FBS. The medium was changed every 2–3 days. From days 4–5 of differentiation, the cells were incubated with DMEM containing 10% FBS and 10 μg/ml human insulin for 4–5 days, with change of medium every 2–3 days. For intracellular Ca2+ or cAMP measurement, the cells were serum starved overnight in DMEM containing 0.1% BSA, washed once with PBS, and treated with 0.25% trypsin, 2 mM EDTA, and 0.2 U/ml Liberase TM at 37°C for 5–10 minutes until most cells were detached from the culture dishes.
LPA receptor expression profile in C3H10T1/2- and MEF-derived adipocytes.
Total RNA and cDNA were prepared as described above. For PCR, samples were run in a 20-μl reaction using ExTaq (Takara Bio) and specific primers for each LPA receptor for 29 cycles. Primer sequences used are shown in Supplemental Table 2. All primers were validated by PCR using cDNAs from tissues or cells that show high expression of individual LPA receptors (data not shown). The PCR products are loaded on a 2% agarose gel.
Measurement of intracellular Ca2+ concentration.
RH7777 cells were serum starved for 5–12 hours in DMEM containing 0.1% BSA and detached using 2 mM EDTA in PBS. C3H10T1/2- and MEF-derived adipocytes were serum starved overnight, detached as described above, and resuspended in DMEM containing 0.1% BSA. The cells were washed once with buffer A (HBSS [MilliporeSigma] containing 25 mM HEPES-NaOH [pH 7.4; Dojindo], 1 mM CaCl2, 1 mM MgCl2, and 0.1% BSA) and loaded with 3 μM Fura-2 AM (Dojindo) and 0.04% pluronic acid (Molecular Probes) at 37°C for 1 hour. The cells were washed once and resuspended in buffer A at a density of 1 × 106 cells/ml. The cell suspension (0.5 ml) was applied to a CAF-110 spectrofluorometer (Jasco) and mixed with 5 μl of ligand solution. The intracellular Ca2+ concentration was measured as the ratio of emission fluorescence at 500 nm in response to excitation at 340 nm and 380 nm. For the experiments with C3H10T1/2- or MEF-derived adipocytes, intracellular Ca2+ concentration was measured in the presence of 10 μM Ki16425.
Impedance measurement.
Real-time impedance traces of ligand-treated B103 cells overexpressing LPA receptors were obtained by the xCELLigence system (Roche Diagnostics) as previously described (36). Briefly, cells were seeded in an E-plate (Roche Diagnostics), followed by a 16-hour culture. The cells were serum starved in DMEM containing 0.1% BSA for 4 hours and stimulated with the indicated ligands. A unitless parameter called cell index (CI) representing impedance of electric flow was continuously monitored every minute. CI after vehicle treatment was subtracted from CI after ligand application to represent the ligand-dependent impedance changes.
Measurement of intracellular cAMP concentration.
C3H10T1/2-derived adipocytes (4 × 104) were reseeded in collagen-coated 96-well plates (Iwaki) as described above, cultured for 24 hours in DMEM containing 10% FBS, and serum starved for 16 hours in DMEM containing 0.1% BSA. The cells were washed with buffer A and incubated in 50 μl of buffer A with 0.5 mM IBMX and 10 μM Ki16425 for 15 minutes at room temperature. The reaction was initiated by adding 50 μl of indicated concentrations of ligands. After a 30-minute incubation at room temperature, the reaction was terminated by adding 10 μl of 10% Tween-20, followed by overnight storage at 4°C. The cAMP concentration in the cell lysate was determined using AlphaScreen cAMP assay kit (PerkinElmer), as recommended by the manufacturer. MEF-derived adipocytes were serum starved overnight, detached as described above, and resuspended in DMEM containing 0.1% BSA. The cells were washed with buffer A, resuspended in buffer A with 0.5 mM IBMX and 10 μM Ki16425, aliquoted to 96-well plate (8 × 104 cells/50 μl), and incubated for 15 minutes at room temperature. The reaction and cAMP concentration measurement were performed as described above.
SRF-RE luciferase assay.
C3H10T1/2- and MEF-derived adipocytes were detached and resuspended in DMEM containing 10% FBS as described above. For the gene knockdown experiments, the resuspended cells (2 × 106) were transfected with ON-TARGETplus SMARTpool mouse LPA4 siRNA (120 pmol), Gα12 and Gα13 siRNA (60 pmol each), or control pool siRNA (120 pmol) using Amaxa Cell Line Nucleofector Kit L (Lonza) with program A-033. The transfected cells were seeded in 24-well collagen-coated plates (Iwaki) at a density of 1 × 105 cells/well, cultured for 16–24 hours in DMEM containing 10% FBS, and subsequently transfected with 4 μg of SRF-RE luciferase-pGL4.35 (Promega) and 1 μg of SV40-Renilla luciferase-pRL (Promega) using xFect transfection reagent (Clontech). After a 4- to 6-hour incubation, the cells were serum starved in DMEM containing 0.1% BSA for 12 hours and stimulated with the indicated concentrations of ODP for 6 hours. Gene knockdown efficiency was >75% as determined by qPCR with primers for Lpar4, Gna12, or Gna13. For the Rho/ROCK inhibition experiments, the resuspended C3H10T1/2-derived adipocytes were seeded in 24-well collagen-coated plates (Iwaki) at a density of 1 × 105 cells/well, cultured for 16–24 hours in DMEM containing 10% FBS, and transfected with reporter genes as described above. After a 16- to 24-hour serum starvation, the cells were treated with either 2 μg/ml Rho inhibitor I for 6 hours or 10 μM Y27632 for 1 hour and stimulated with the indicated concentrations of ODP for 6 hours. For the experiments with MEFs, the resuspended cells were seeded in 24-well collagen-coated plates at a density of 1 × 105 cells/well, cultured for 16–24 hours in DMEM containing 10% FBS, and transfected with reporter genes as described above. After a 24-hour incubation, the cells were serum starved in DMEM containing 0.1% BSA for 12–16 hours and stimulated with the 1 μM of ODP for 6 hour. Firefly and Renilla luciferase activities were measured using Dual luciferase assay kit (Promega) and a MiniLumat LB luminometer (Berthold Technologies). Firefly luciferase values were standardized to Renilla ones.
Rho activity measurement.
C3H10T1/2-derived adipocytes (1 × 105) were reseeded in collagen-coated 35 mm dishes (Iwaki), serum starved in DMEM containing 0.1% BSA for 16 hours, and treated with 2.5 μM ODP for 1 minute in the presence of 10 μM Ki16425. Equal amounts of protein samples were processed for the determination of Rho activation level by G-LISA RhoA activation assay kit (Colorimetric format; Cytoskeleton) with the manufacturer’s instructions.
Lipolysis measurement.
Lipolysis activity of mouse primary adipocytes was determined by using Lipolysis (Adipocyte) kit (MilliporeSigma) according to the manufacturer’s instruction. Briefly, the primary adipocytes were collected from eWAT of WT mice by enzymatic digestion and centrifugation and were stimulated with 10 μM ODP, 100 nM isoproterenol (ISOP), and/or 15 mM lactic acid in the presence of 10 μM Ki16425 for 2 hours. The glycerol concentration was determined by colorimetric detection. All reagents used for adipocyte isolation and lipolysis reaction were components of the kit except for ODP, Ki16425, and lactic acid.
Glucose-tolerance test and insulin-tolerance test.
Glucose tolerance was tested in mice fasted for 16 hours. Lpar4-KO mice and their counterpart WT mice were orally loaded with glucose (1.5 g/kg body weight). Insulin tolerance was tested in mice fasted for 4 hours. Lpar4-KO and their counterpart WT mice were i.p. injected with human insulin (Novo Nordisk Pharma) at a dose of 0.75 U/kg body weight, respectively. In both tests, blood samples were drawn from the tail vein at the times indicated. The glucose levels were measured with an automatic blood glucose meter (Glutest Neo; Sanwa Chemical). For insulin-tolerance test, the changes were plotted as a percentage of basal glucose versus time. Plasma insulin levels after glucose load were determined by ultra–high-sensitivity mouse insulin ELISA kit (Morinaga).
Liver triglyceride content.
Liver triglyceride was collected by Folch’s method and measured by enzymatic reagents (Choletest TG; Sekisui Medical) at Skylight Biotech Inc.
Serum sample assays.
Serum triglyceride and nonesterified fatty acid levels were determined using Triglyceride E-test Wako kit (Wako) and NEFA C-test Wako kit (Wako), respectively. Serum adiponectin and MCP-1 levels were measured using mouse adiponectin (Otsuka Pharmaceutica) and mouse MCP-1 (R&D Systems) ELISA kits, respectively. Serum aspartate transaminase (AST), alanine transaminase (ALT), and alkaline phosphatase (ALP) were measured at Nagahama Life Science Laboratory.
Microarray analysis.
DNA microarray analysis was performed as previously described (39) with minor modifications. In brief, biotinylated amplified RNA was obtained from 100 ng of purified total RNA using GeneChip 3′ IVT Express Kit (Affymetrix). The amplified RNA was then purified, fragmented, and hybridized to an Affymetrix GeneChip mouse Genome 430 2.0 Array. After hybridization at 45°C for 16 hours, the arrays were washed and labeled with PE. Fluorescence signals were scanned using Affymetrix GeneChip System. Affymetrix GeneChip Command Console software was used to reduce the array images to the intensity of each probe (CEL files). The CEL files were quantified with the Factor Analysis for Robust Microarray Summarization (FARMS) method (40) using statistical language R (https://www.r-project.org/) and Bioconductor (http://www.bioconductor.org/) (41). Hierarchical clustering was performed using the pvclust() function (42) in R. To detect the differentially expressed genes (DEGs) between WAT from WT and Lpar4-KO mice, the rank products (RP) method (43) was applied to FARMS quantified data, with the number of permutations set at 1000. Probe sets with a FDR < 0.01 were regarded as differentially expressed. The use of the FARMS preprocessing algorithm followed by the RP selection method has been proposed to be one of the best combinations for accurate detection of differentially expressed transcription (44). Gene annotation enrichment analysis of up- or downregulated probe sets was performed with the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) (45). EASE scores, which are modified Fisher’s exact test P value (46), were used to extract statistically overrepresented Gene Ontology (GO) terms from the DEGs; Benjamini and Hochberg FDR corrections were used to correct the results by multiple testing. GO terms with FDR-corrected P values of <0.05 were regarded as significantly enriched. The data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE66132 (https://www.ncbi.nlm.nih.gov/geo/).
Histological analysis.
Adipose tissues were fixed in 10% formalin overnight at 4°C. For IHC, paraffin-embedded sections were deparaffinized in xylene and rehydrated. The sections were heated at 121°C in 10 mM sodium citrate (pH 6.0) for 10 minutes to retrieve antigens and incubated with 2% goat serum (Invitrogen)/PBS for 30 minutes, followed by incubation with primary or secondary antibody in 2% goat serum/PBS for 1 hour each. The primary antibodies used were rabbit anti-caveolin antibody (1:100; BD Biosciences, 610060) and rat anti-Mac2 antibody (1:100; BioLegend, M3/38); the secondary antibodies used were Alexa Fluor 488 goat anti-rabbit antibody and Alexa Fluor 568 goat anti-rat antibody (Invitrogen), respectively. H&E staining or Masson trichrome staining were performed according to standard procedures. Adipocyte area was manually traced and analyzed with ImageJ (NIH) software. The sizes of 200 or more white adipocytes per mouse were determined in each group.
BM transfer.
BM was isolated from 6-week-old C57BL/6-Ly5.1, WT and Lpar4-KO mice. WT, Lpar4-KO, and C57BL/6-Ly5.1 mice at the age of 6 weeks were irradiated with 8 Gy in 2 split doses with 3-h interval using a cabinet X-ray system CP-160 (Faxitron X-ray Corporation). After 2 hours, 2 × 106 BM cells were injected via the tail vein into the irradiated mice. Five weeks later, recipient mice were fed with HFD32 for 14–16 weeks. The reconstitution efficiencies were evaluated by FACS analysis of peripheral blood.
Western blotting.
eWAT were lysed in ice-cold lysis buffer (150 mM NaCl, 1% Triton-X100, 1% sodium deoxycholate, 50 mM Tris-HCl [pH 7.4], 0.1% SDS, 1 mM EDTA, 1 mM sodium orthovanadate, and 1 mM sodium fluoride) with protease inhibitor cocktail (cOmplete from Roche Diagnostics or SigmaFast from MilliporeSigma). The lysate was centrifuged at 12,000 g for 20 minutes. The resultant supernatants were collected and used for the immunoblot analysis. Equal amounts of protein samples were separated by 8% or 10% SDS-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane, which was then blocked with 5% skim milk and probed with primary antibodies against Ser112-phosphorylated PPARγ (1:3000; MilliporeSigma, MAB3632), total PPARγ (1:5000; Santa Cruz Biotechnology Inc., H-100), and vinculin (1:10,000; MilliporeSigma, V9131). The blots were then probed with horseradish peroxidase–conjugated (HRP–conjugated) secondary antibodies (goat anti–mouse IgG-HRP [Santa Cruz Biotechnology Inc., sc-2005] or goat anti–rabbit IgG-HRP [Santa Cruz Biotechnology Inc., sc-2004]), and the signal was visualized with a ChemiDoc Touch Imaging System (Bio-Rad). Relative band intensities were obtained by densitometric analysis of images using ImageJ software.
Statistics.
Data are expressed as means ± SEM from biological replicates. Statistical analyses were completed with Prism 5.0 or 7.0 software (Graphpad Software). Unpaired Student’s t test (2-tailed) was used for statistical analysis of the difference between 2 groups. Statistical significance among several groups was determined by 1-way ANOVA followed by Bonferroni’s or Dunnett’s post hoc test.
Study approval.
The animal experimental procedures used in this study were approved by Animal Care and Use Committees of The University of Tokyo and Akita University.
Author contributions
KY, NK, TK, SI, and TS designed the research. KY, DK, DY, HI, TON, NA, AS, NS, and SI conducted the experiments. KY, TT, TI, YN, and KA performed microarray analysis. KY, SI, and TS wrote the manuscript.
Supplementary Material
Acknowledgments
We are grateful to H. Shindou, D. Hishikawa, T. Harayama, M.T. Nakano, M. Matsumoto, Y. Takahashi, K. Nagata (National Center for Global Health and Medicine), Y. Kita, S.M. Tokuoka, F. Hamano, K. Yoshida (The University of Tokyo), H. Nakanishi (Akita University), M. Nakamura (Okayama University of Science), and N. Uozumi (Saitama Medical University), in addition to the rest of the Shimizu Lab, for helpful comments, suggestions, and technical supports. We also thank J.I. Miyazaki (Osaka University, Japan) for giving the expression vector pCXN2. This study was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to TS, SI, and KY), The Grants for National Center for Global Health and Medicine 24-001 and 25-201 (to TS), and grants from the Cell Science Research Foundation (to KY), the Takeda Science Foundation (to TS and SI), the Naito Foundation (to SI), the Mochida Memorial Foundation (to SI), and the Uehara Memorial Foundation (to SI). Takao Shimizu, Department of Lipidomics, the University of Tokyo, is financially supported by Shimadzu Corp. and ONO Pharmaceutical Co. Ltd. We would like to thank Editage (www.editage.jp) for English language editing.
Version 1. 12/20/2018
Electronic publication
Footnotes
Conflict of interest: Takao Shimizu is financially supported by Shimadzu Corp. and ONO Pharmaceutical Co. Ltd.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: JCI Insight. 2018;3(24):e97293. https://doi.org/10.1172/jci.insight.97293.
Contributor Information
Keisuke Yanagida, Email: Keisuke.Yanagida@childrens.harvard.edu.
Hidemitsu Igarashi, Email: igrshdmt@med.akita-u.ac.jp.
Daisuke Yasuda, Email: dyasuda@gipc.akita-u.ac.jp.
Daiki Kobayashi, Email: dkobayashi@med.akita-u.ac.jp.
Takayo Ohto-Nakanishi, Email: ohtakayo@alte.jp.
Noriyuki Akahoshi, Email: akahoshi@ac.shoyaku.ac.jp.
Atsushi Sekiba, Email: sekiba@jimu.akita-u.ac.jp.
Tsudoi Toyoda, Email: t.26.toyoda@gmail.com.
Tomoko Ishijima, Email: aishiji@mail.ecc.u-tokyo.ac.jp.
Yuji Nakai, Email: yunakai@hirosaki-u.ac.jp.
Nobuhiro Shojima, Email: nshojima-tky@umin.ac.jp.
Naoto Kubota, Email: nkubota-tky@umin.ac.jp.
Keiko Abe, Email: aka7308@mail.ecc.u-tokyo.ac.jp.
Takashi Kadowaki, Email: kadowaki-3im@h.u-tokyo.ac.jp.
Satoshi Ishii, Email: mame-tky@umin.ac.jp.
Takao Shimizu, Email: tshimizu@ri.ncgm.go.jp.
References
- 1.Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell. 2014;156(1-2):20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berry R, Jeffery E, Rodeheffer MS. Weighing in on adipocyte precursors. Cell Metab. 2014;19(1):8–20. doi: 10.1016/j.cmet.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crewe C, An YA, Scherer PE. The ominous triad of adipose tissue dysfunction: inflammation, fibrosis, and impaired angiogenesis. J Clin Invest. 2017;127(1):74–82. doi: 10.1172/JCI88883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li P, et al. Adipocyte NCoR knockout decreases PPARγ phosphorylation and enhances PPARγ activity and insulin sensitivity. Cell. 2011;147(4):815–826. doi: 10.1016/j.cell.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kusminski CM, et al. MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nat Med. 2012;18(10):1539–1549. doi: 10.1038/nm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JY, et al. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J Clin Invest. 2007;117(9):2621–2637. doi: 10.1172/JCI31021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jonker JW, et al. A PPARγ-FGF1 axis is required for adaptive adipose remodelling and metabolic homeostasis. Nature. 2012;485(7398):391–394. doi: 10.1038/nature10998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Husted AS, Trauelsen M, Rudenko O, Hjorth SA, Schwartz TW. GPCR-Mediated Signaling of Metabolites. Cell Metab. 2017;25(4):777–796. doi: 10.1016/j.cmet.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Oh DY, et al. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell. 2010;142(5):687–698. doi: 10.1016/j.cell.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chun J, Hla T, Lynch KR, Spiegel S, Moolenaar WH. International Union of Basic and Clinical Pharmacology. LXXVIII. Lysophospholipid receptor nomenclature. Pharmacol Rev. 2010;62(4):579–587. doi: 10.1124/pr.110.003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dusaulcy R, et al. Adipose-specific disruption of autotaxin enhances nutritional fattening and reduces plasma lysophosphatidic acid. J Lipid Res. 2011;52(6):1247–1255. doi: 10.1194/jlr.M014985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishimura S, et al. ENPP2 contributes to adipose tissue expansion and insulin resistance in diet-induced obesity. Diabetes. 2014;63(12):4154–4164. doi: 10.2337/db13-1694. [DOI] [PubMed] [Google Scholar]
- 13.Simon MF, et al. Lysophosphatidic acid inhibits adipocyte differentiation via lysophosphatidic acid 1 receptor-dependent down-regulation of peroxisome proliferator-activated receptor gamma2. J Biol Chem. 2005;280(15):14656–14662. doi: 10.1074/jbc.M412585200. [DOI] [PubMed] [Google Scholar]
- 14.Knowlden S, Georas SN. The autotaxin-LPA axis emerges as a novel regulator of lymphocyte homing and inflammation. J Immunol. 2014;192(3):851–857. doi: 10.4049/jimmunol.1302831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zmora N, Bashiardes S, Levy M, Elinav E. The Role of the Immune System in Metabolic Health and Disease. Cell Metab. 2017;25(3):506–521. doi: 10.1016/j.cmet.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Regard JB, Sato IT, Coughlin SR. Anatomical profiling of G protein-coupled receptor expression. Cell. 2008;135(3):561–571. doi: 10.1016/j.cell.2008.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams JR, et al. Unique ligand selectivity of the GPR92/LPA5 lysophosphatidate receptor indicates role in human platelet activation. J Biol Chem. 2009;284(25):17304–17319. doi: 10.1074/jbc.M109.003194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmed K, et al. An autocrine lactate loop mediates insulin-dependent inhibition of lipolysis through GPR81. Cell Metab. 2010;11(4):311–319. doi: 10.1016/j.cmet.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 19.Klepac K, et al. The Gq signalling pathway inhibits brown and beige adipose tissue. Nat Commun. 2016;7:10895. doi: 10.1038/ncomms10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng Z, Garvin D, Paguio A, Stecha P, Wood K, Fan F. Luciferase Reporter Assay System for Deciphering GPCR Pathways. Curr Chem Genomics. 2010;4:84–91. doi: 10.2174/1875397301004010084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kusminski CM, Scherer PE. Mitochondrial dysfunction in white adipose tissue. Trends Endocrinol Metab. 2012;23(9):435–443. doi: 10.1016/j.tem.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 23.Soccio RE, Chen ER, Lazar MA. Thiazolidinediones and the promise of insulin sensitization in type 2 diabetes. Cell Metab. 2014;20(4):573–591. doi: 10.1016/j.cmet.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rangwala SM, et al. Genetic modulation of PPARgamma phosphorylation regulates insulin sensitivity. Dev Cell. 2003;5(4):657–663. doi: 10.1016/S1534-5807(03)00274-0. [DOI] [PubMed] [Google Scholar]
- 25.Choi JH, et al. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature. 2010;466(7305):451–456. doi: 10.1038/nature09291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kroker AJ, Bruning JB. Review of the Structural and Dynamic Mechanisms of PPARγ Partial Agonism. PPAR Res. 2015;2015:816856. doi: 10.1155/2015/816856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phillips CM. Metabolically Healthy Obesity: Personalised and Public Health Implications. Trends Endocrinol Metab. 2016;27(4):189–191. doi: 10.1016/j.tem.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 28.Sun K, Kusminski CM, Scherer PE. Adipose tissue remodeling and obesity. J Clin Invest. 2011;121(6):2094–2101. doi: 10.1172/JCI45887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rancoule C, Dusaulcy R, Tréguer K, Grès S, Attané C, Saulnier-Blache JS. Involvement of autotaxin/lysophosphatidic acid signaling in obesity and impaired glucose homeostasis. Biochimie. 2014;96:140–143. doi: 10.1016/j.biochi.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 30.Yanagida K, Kurikawa Y, Shimizu T, Ishii S. Current progress in non-Edg family LPA receptor research. Biochim Biophys Acta. 2013;1831(1):33–41. doi: 10.1016/j.bbalip.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Asterholm IW, Mundy DI, Weng J, Anderson RG, Scherer PE. Altered mitochondrial function and metabolic inflexibility associated with loss of caveolin-1. Cell Metab. 2012;15(2):171–185. doi: 10.1016/j.cmet.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vernochet C, et al. Adipose tissue mitochondrial dysfunction triggers a lipodystrophic syndrome with insulin resistance, hepatosteatosis, and cardiovascular complications. FASEB J. 2014;28(10):4408–4419. doi: 10.1096/fj.14-253971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sumida H, et al. LPA4 regulates blood and lymphatic vessel formation during mouse embryogenesis. Blood. 2010;116(23):5060–5070. doi: 10.1182/blood-2010-03-272443. [DOI] [PubMed] [Google Scholar]
- 34.Igarashi H, Akahoshi N, Ohto-Nakanishi T, Yasuda D, Ishii S. The lysophosphatidic acid receptor LPA4 regulates hematopoiesis-supporting activity of bone marrow stromal cells. Sci Rep. 2015;5:11410. doi: 10.1038/srep11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanagida K, Ishii S, Hamano F, Noguchi K, Shimizu T. LPA4/p2y9/GPR23 mediates rho-dependent morphological changes in a rat neuronal cell line. J Biol Chem. 2007;282(8):5814–5824. doi: 10.1074/jbc.M610767200. [DOI] [PubMed] [Google Scholar]
- 36.Yanagida K, et al. Identification and characterization of a novel lysophosphatidic acid receptor, p2y5/LPA6. J Biol Chem. 2009;284(26):17731–17741. doi: 10.1074/jbc.M808506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yasuda D, Imura Y, Ishii S, Shimizu T, Nakamura M. The atypical N-glycosylation motif, Asn-Cys-Cys, in human GPR109A is required for normal cell surface expression and intracellular signaling. FASEB J. 2015;29(6):2412–2422. doi: 10.1096/fj.14-267096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eto M, Shindou H, Koeberle A, Harayama T, Yanagida K, Shimizu T. Lysophosphatidylcholine acyltransferase 3 is the key enzyme for incorporating arachidonic acid into glycerophospholipids during adipocyte differentiation. Int J Mol Sci. 2012;13(12):16267–16280. doi: 10.3390/ijms131216267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suyama T, Okada S, Ishijima T, Iida K, Abe K, Nakai Y. High phosphorus diet-induced changes in NaPi-IIb phosphate transporter expression in the rat kidney: DNA microarray analysis. PLoS ONE. 2012;7(1):e29483. doi: 10.1371/journal.pone.0029483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hochreiter S, Clevert DA, Obermayer K. A new summarization method for Affymetrix probe level data. Bioinformatics. 2006;22(8):943–949. doi: 10.1093/bioinformatics/btl033. [DOI] [PubMed] [Google Scholar]
- 41.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22(12):1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 43.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573(1-3):83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 44.Kadota K, Nakai Y, Shimizu K. Ranking differentially expressed genes from Affymetrix gene expression data: methods with reproducibility, sensitivity, and specificity. Algorithms Mol Biol. 2009;4:7. doi: 10.1186/1748-7188-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 46.Hosack DA, Dennis G, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4(10):R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.