Abstract
We present a vision for a Systems Medicine Computing Cloud that draws on progress in the fields of Genomics, Systems Biology, and Biomedical Data Mining. The successful fusion of these areas will create a predictive matrix of biomarkers, genetic variants, and environmental variables that will drastically increase the specificity and timeliness of diagnosis for a wide range of common diseases, while delivering accurate predictions about the efficacy of treatment options. However, the amount of data being generated by each of these areas is staggering, as is the task of managing and analyzing it. Adequate computing infrastructure needs to be developed in order to assemble, manage, and mine the enormous and rapidly growing corpus of ‘omics’ data along with clinical information. We have now arrived at an intersection point between genome technology, cloud computing and biological data mining. This intersection point provides a launch pad for developing a globally applicable cloud computing platform capable of supporting a new paradigm of data intensive, cloud-enabled Systems Medicine.
Technology Drivers
Data intensive science.
The power of computer processors doubles in less than 18 months, as does the capacity of computer storage disks. This exponential growth impacts not only the power of computers, but also the power of scientific instruments and has resulted in an exponentially growing amount of scientific data (Szalay and Gray 2006). In this section, we look at three important changes that have occurred in the last decade regarding data: 1) the explosion in the amount of data produced; 2) a fundamental change in how data is managed and processed; and 3) new algorithms for how data is analyzed using data mining. Together, these changes are beginning to change biology into a data intensive science.
Ubiquitous sequencing; an explosion of data.
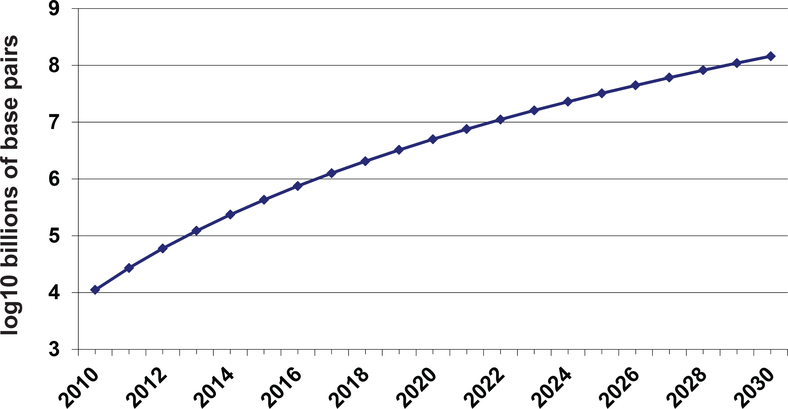
Genomics reveals the ‘parts lists’ and genetic variations that compose each unique human genome. Genome technology has been producing DNA sequence data ever faster, cheaper and in tremendous volumes. The amount of DNA sequence in the public domain has been growing exponentially over the last two decades. Low cost, high throughput DNA sequencing is a disruptive technology that promises to have a major impact on our understanding of biology and our treatment of human diseases. Within a few years, we will be in the era of ubiquitous sequencing. Recently, Stein pointed out that from 1990 until 2004 sequencing output doubled approximately every 19 months, but from 2005 until the present the doubling rate decreased to 5 months due to the development of “NextGen” sequencing technologies (Stein 2010). However, one can also extrapolate more generally based on Genbank data from 1990–2005. In Figure 1 we fit a logarithmic function to these data, and extrapolate the curve into the present day and beyond (this curve was initially calculated in 2007 by KPW for a presentation to the National Science Foundation). The 2011 estimates extrapolated from this curve are approximately 30 Terabases of finished genome that corresponds to 10,000 human genomes. In this sense, the impact of “NextGen” sequencing technology was foreseeable. The total number of complete human genomes sequenced by the end of 2011 worldwide is, in fact, likely to be >10,000, in addition to genome sequencing of other species and “partial genomes” from procedures such as exome capture sequencing, RNA sequencing and Chromatin Immunoprecipitation sequencing (ChIP-seq). Taking this curve as a tentative estimate of future world capacity, we can speculate that by 2015 more than a million human genomes will be sequenced. Storing just the finished sequence would require several Petabytes of disc space.
Fig. 1.
Projected growth of DNA sequence data in the 21st century.
Early in the third decade of the 21st century, we can speculate that the capacity to sequence up to 1 billion people will have been realized. Approximately 3,000 PB of storage will be needed to house 1 billion human genomes. Are these speculations realistic? Perhaps a billion genomes is a bit far-fetched when most of the world’s population does not have access to even basic access to medical professionals. However, with the cost of sequencing likely to drop below $100 per genome in the next decade, and the expected increase in utility of genome information for diagnosing and treating diseases, we can expect genome sequencing to become an important aspect of health care in both developed and developing countries.
Importantly, genome sequencing is simply a baseline. Understanding both normal and disease states requires whole genome expression data at different time points during normal conditions and during disease progression, as well as for different treatment conditions. Similarly, proteomic and metabonomics data monitoring the state of cells is growing increasingly important in identifying disease states and indicating treatments (Nicholson and Lindon 2008; Ponten, Jirstrom et al. 2008; Uhlen, Oksvold et al. 2010; Rubakhin, Romanova et al. 2011). In the future, clinical trials will sample and generate a variety of ‘omics profiles for multiple time points and different conditions (e.g. plus or minus treatment) during normal and diseased states. As the number of temporal points and treatment conditions grows, so too will the amount of data to be sifted. A person’s state of health will be assessed in light of his or her genome sequence, but also the cellular state as measured by hundreds of thousands of analytes. Before these data can be compressed into diagnostic/prognostic panels that are representative of the “whole”, it will be necessary to analyze and mine them in their totality. Because of the enormous scale of data in this new era of ubiquitous sequencing, and concomitant developments of other data-intensive ‘omics technologies, managing and analyzing this data corpus is a formidable challenge.
Cloud computing.
The last ten years or so has seen another important set of advances in how data is managed. A decade ago, an important split occurred between the types of computing systems used by companies providing Internet services, such as Google, and those used by scientists and academics working in high performance computing. By and large, researchers in high performance computing were mainly concerned with writing specialized programs for high-end computers that were used for simulation. To maximize the performance of simulations, the systems are designed to minimize latency; to achieve this, specialized connections were used to connect the various processors and libraries were used to pass messages so that the different processors could exchange relevant information. Writing the code was difficult and managing the (sometimes) large data that simulations produced was an after thought.
In contrast, companies such as Google began collecting very large amounts of data (measured in Petabytes instead of Terabytes) and analyzing it in order to optimize search results as well as the placement of text ads, which provide the bulk of Google’s revenue. To manage and analyze these very large amounts of data (that literally fill a data center), Google developed storage and compute services that scaled to all the computers that filled their data centers and were easy for their software engineers to program (Dean and Ghemawat 2008). To keep costs low, which is by and large not a factor in high performance computing, systems were designed to use commodity computers and to replicate data and redistribute workloads when the components failed (as they do frequently). Instead of designing systems to minimize latency, they designed systems to maximize the rate that data could be processed. This was done by making sure that there were very large numbers of disks and that each disk was matched with a processor, and, later with a core.
In 2006, Amazon introduced another important innovation in computing infrastructure that they called the Amazon Elastic Compute Cloud (EC2). The fundamental idea with EC2 was that a user could use a web portal and obtain one or more virtual machines that they could pay for by the hour. A virtual machine in this context is a process that appears to the user as an actual computer but is in fact one of several such virtual machines that are managed by a single physical computer. This provides two advantages: First, for many data center usage patterns, it was most cost effective to use multiple virtual machines than a single actual physical machine. Second, it is easier to set up and tear down a virtual machine than a physical machine.
With EC2, the cost of one virtual machine for 100 hours is the same as the cost of 100 virtual machines for one hour. With this model, it suddenly became practical for scientists to perform computations on 100 machines for just the time required and let someone else manage the data center infrastructure required to support it. These types of systems developed by Google and Amazon are often referred to as cloud computing systems and promise to have just as large an impact on data intensive biology as they have had on business. In particular, as biology enters the era of large data, cloud computing systems will be one of the tools that will enable biologists to manage and analyze the data produced.
Biomedical data mining.
Biomedical data mining seeks to connect phenotypic data to biomarker profiles and therapeutic treatments, with the goal of creating predictive models of disease detection, progression, and therapeutic response. During the last decade data mining of biological data has become an increasing important technique. Biological data mining includes mining a wide variety of biological data, including 1) mining genomic data (and data from other high throughput technologies such as DNA sequencing, RNA expression, proteomic data, metabolomic data, small molecule screening, etc…), 2) text mining of the biological literature, medical records, etc.; and 3) image mining across a number of modalities, including X-rays, functional MRI, new types of scanning microscopes, etc.
Biomedical Infomics Synthesis
Our thesis is that the explosion of genomic, proteomic, and other ‘omic data, the ability of cloud computing to process and analyze data at the scale of a data center, and new algorithms from data mining and systems biology create what might be called a biomedical-omics-informatics synthesis. We will refer to this more simply as the Biomedical-Infomics Synthesis.
In this section, we discuss three major shifts in biological thinking and approach over the last century: the NeoDarwinian Synthesis, the Molecular Biology Revolution, and Systems Biology. We argue that a Biomedical-Infomics Synthesis is an important emerging component of Systems Biology.
NeoDarwinian Synthesis.
In the middle part of the last century the field of Genetics underwent a transformation known as the NeoDarwinian Synthesis (Dobzhansky 1951). During this period, there was a fusion of the theoretical genetics that explained the inheritance behavior of (mostly) individual genes with simple allelic variants and the evolutionary ideas of Darwin and his intellectual progeny. Remarkably, the NeoDarwinian synthesis happened largely in the absence of the understanding of DNA as the heritable material of all biology and without any but the vaguest notion of the molecular nature of genes. The ramifications of this mid-century synthesis have been immense, leading to improvements in crops and livestock while setting a foundation for interpretation and understanding of the molecular basis of life in the second half of the 20th century as biologists turned to unraveling the basis of DNA and the products it encodes.
Molecular Biology Revolution.
This subsequent Molecular Biology Revolution was catalyzed by the discovery of the double helix in 1953 and subsequently dominated biomedicine and much of biological thinking in general (Watson and Crick 1953; Watson and Crick 1953). Great benefits to society were stimulated by the Molecular Biology Revolution as well, including the development of drugs such as synthetic insulin, humanized antibodies directed against tumors, modification of crops for pest resistance or increased yield, and many other extremely meaningful contributions that relied on the understanding or engineering of one or only a few genes at a time.
However, as geneticists have known for 100 years, most of biology is more complex and involves many genes acting in the context of heterogeneous environments. The molecular biologists for decades chose to ignore this unpleasant reality, going to great lengths to eliminate genetic variability in their model systems, and the geneticists who did not become molecular biologists were largely sidelined academically and in biomedicine, or focused on practical pursuits such as improving agriculture. Only in the last ten to fifteen years has the genetics of complex traits moved into the mainstream of biology, enabled wholly by the advent of genomic technologies that act as the first instrument that can allow scientists to see the totality of variation that contributes to complex traits. The major challenge in all of biology at the beginning of this century is to figure out how complex traits work at a molecular level.
Systems Biology.
With a renewed appreciation for the complexity of biological systems, a modern version of a field known as Systems Biology has affirmed that understanding of complex traits requires their integrated study at the molecular, cellular and organismal levels. While the field has its historical roots largely in metabolic flux analysis, neuronal modeling, and bioengineering, during the last ten years Systems Biology has come to encompass much of modern biology and professes to usher a new era where biological theory and experiment become unified (Ideker, Galitski et al. 2001). It presently connotes two major areas of investigation.
First, systems biologists use genomic scale data to analyze molecular networks, typically by integrating multiple heterogeneous data types that represent different aspects of cellular biology and genetics, to make predictions about network structures and which network substructures (and individual genes or gene products) are crucial for a given phenotype being analyzed. For example, gene expression networks that integrate RNA expression profiling, transcriptional factor binding to the genome, and other data types have now been generated for a vast breadth of traits that range from yeast metabolism (Ideker, Thorsson et al. 2001; Herrgard, Swainston et al. 2008), to embryonic development of fruit flies (Liu, Ghanim et al. 2009; Zinzen, Girardot et al. 2009; Roy, Ernst et al. 2010), to human cancers (e.g.) (Lamb, Crawford et al. 2006; Hua, Kallen et al. 2008), and to dozens of other complex traits.
The second major area of modern Systems Biology echoes its modeling roots, focusing on discrete subsystems where enough data has been gathered to build predictive models that specify non-trivial outcomes of perturbing a given network. This area too has been applied to a wide range of applications that span from modeling the stochastic behavior of microbial chemosensing (Korobkova, Emonet et al. 2004), to the aforementioned transcriptional networks controlling embryonic development in flies (Jaeger, Surkova et al. 2004; Janssens, Hou et al. 2006), to phosphorylation based signaling networks such as those activated by MAP kinases and Receptor Tyrosine Kinases in a wide range of cancers (Jones, Gordus et al. 2006; Ciaccio, Wagner et al. 2010; Morris, Saez-Rodriguez et al. 2011).
But Systems Biology, as it is often formulated, also seeks to understand emergent properties beyond the structures of biological networks and information flow within them. In fact many of the core themes of Systems Biology are identical to the themes identified many years ago by the geneticists that led the NeoDarwinian synthesis, their contemporaries and predecessors. These properties are at the root of biology and include concepts such as emergence of three dimensional structure of cells, tissues and organisms from the simple materials of inheritance, cellular and developmental robustness, modularity of biological systems, group behaviors (such as schooling in fish or swarming in bees), and the process of organic evolution. These concepts, all representing emergent properties of complex systems, are driving much contemporary research in Systems Biology.
Sources of Data for the Biomedical-Infomics Synthesis
Systems Biology provides a natural conceptual framework for launching a Biomedical-Infomics Synthesis. Systems Biology is the modern intellectual home for an integrated view of Biology, undivided into its dozens of subfields and specialties; genomic technologies play a major role; systems biology draws from almost every scientific discipline to address fundamental problems, with particular avidity for computing and engineering. Most importantly for our present thesis, Systems Biology is the site of information integration about biological systems, and the field is extremely active. As mentioned above, data production will grow not only due to genome sequencing but also due to sequencing and otherwise measuring gene products under many different conditions. Two major types of data have proven to yield predictive networks using genomic technology and Systems Biology approaches: perturbations and genetic associations.
Experimental perturbations.
The perturbation approach of hypothesis testing is the basis of modern experimental biology. A system’s output is measured under different experimental or naturally occurring conditions (normal vs. disease, mutant vs. wild type, hormone vs. control treatment, etc…). In the last decade a transition has occurred where investigations have gone from measuring one variable (gene product) at a time to measuring the output of the entire genome – in other words we have transitioned to assaying the state of the entire system. Simultaneously, through miniaturization technologies and development of high throughput screening approaches, perturbation analyses themselves have seen the same scale of expansion whereby the entire contents of genomes are routinely perturbed and then traits (phenotypes) are measured. For example, data matrices with 20,000 perturbations x 10GB of gene expression and corresponding measurements are now foreseeable, potentially generating 10GB of DNA sequence per measurement (200PB). Already it is practical to generate a matrix with hundreds to several thousand perturbations and whole genome measurements. With this transition has been the requirement to implement statistical and computational methods in order to determine which variables are important for a given phenotype being analyzed. More advanced studies have relied on building networks and distilling testable hypotheses from those networks, for example using the types of probabilistic algorithms mentioned above (Ideker, Thorsson et al. 2001; Krogan, Cagney et al. 2006; Amit, Garber et al. 2009; Liu, Ghanim et al. 2009; Costanzo, Baryshnikova et al. 2010; McGary, Park et al. 2010).
Genetic associations.
The second successful experimental approach in Systems Biology has relied on genetic associations. This approach, descended from the same genetic thinking that drove the complex trait geneticists in the 20th century, takes full advantage of genomic technologies. A powerful implementation of this approach is to generate gene expression data alongside genotypic data in order to determine which genetic variants are affecting which genes’ expression. This is known as the expression Quantitative Trait Locus (eQTL) approach (Brem, Yvert et al. 2002; Morley, Molony et al. 2004; Schadt, Lamb et al. 2005; Stranger, Forrest et al. 2007; Stranger, Nica et al. 2007). By comparing populations that are sick vs. healthy, this eQTL approach can identify genes that are hubs in networks that are associated with the trait.
More generally, understanding associations between whole genomic variants and phenotypes across populations will be an important source of data for the Biomedical-Infomics Synthesis.
Evolutionary conservation.
A third approach that is potentially extremely powerful but is just beginning to show its effectiveness in the context of Systems Biology is the evolutionary, or comparative, approach. As an example, perturbation analysis was used in the model organism Drosophila to map a network that controls early developmental pattern formation in the embryo. Using the knowledge that many components of this network are evolutionarily conserved in humans, and involved in diseases such as cancer, the human counterparts were screened for their disease association (Liu, Ghanim et al. 2009). A key conserved factor (predicted by a network centrality metric) was associated with human kidney cancer, and subsequent studies have verified that this gene product can cause cancer in mice and that inhibiting it can kill cancer cells, thus making it a promising drug target for a disease that otherwise is resistant to both chemotherapy and radiation (Li et al., submitted). More generally, using data gleaned from public databases networks in model organisms can be built that map to orthologous networks associated with human diseases. Such networks can help to identify novel candidate genes involved in human disease processes or suggest genes for study in model organisms that could yield human disease insights (McGary, Park et al. 2010).
Medical text.
Today, there are trillions of pages of scholarly text available in the world libraries, and although science-focused text mining is a formidable intellectual challenge, it is beginning to be used to extract new discoveries from this stored text.
The typical stages of text mining include identification of named entities (gene and disease names, organizations and geographic locations etc.), capturing relations among the named entities (such as a reported association between a genetic polymorphism and a disease, or interaction between a protein and a small molecule), and then computational reasoning to construct complex semantic networks from these entities and their relations. Text mining at this scale is challenging due to the sheer volume of the data and to the difficulty extracting what are often quite complex and subtle assertions from the data.
Today, as new experimental data is generated, the process of analyzing the data and deriving text assertions summarizing it, such as identifying the specific genetic polymorphism associated with a disease, is largely manual. Manual processes like these do not scale to the amount of data that will be produced in order to capture millions of variants that must be analyzed from thousands or millions of genomes. Biological data mining is beginning to automate this process, which is essential since deriving these types of text-based assertions from newly generated experimental data provides access to historical context for new data analysis, to previously formulated and supported, untested, or rejected hypotheses, and to legacy observations from multiple research communities.
Electronic Medical Records.
Another important source of data for the Biomedical-Infomics Synthesis are Electronic Medical Records (EMR). Over the next several years, more and more hospitals and medical research centers will be implementing EMR, in part due to financial incentives being offered. Once an EMR system is in place, it is natural to create what is usually called a clinical research data warehouse, so that multiple years of EMRs over entire populations of patients can be analyzed. Patient data for which consent has been provided can be analyzed along with genomic data. This allows phenotype data to be correlated with genomic data for patient populations that sometimes hundreds or thousands in size.
Large scale genomic studies.
Finally, a very important source of data for the Biomedical-Infomics Synthesis are the increasing number of large scale studies. For example, the Encyclopedia of DNA Elements (ENCODE) project is mapping functionality in the human genome (Birney, Stamatoyannopoulos et al. 2007; Myers, Stamatoyannopoulos et al. 2011). The 1,000 Genomes project has mapped already multiple thousands of genomes (http://www.1000genomes.org/; 1000GenomesProject 2010). The Cancer Genome Atlas is identifying the variation associated with more than a dozen types of cancer (http://cancergenome.nih.gov/). Various consortium are sequencing the genomes of patients afflicted with most major complex diseases, nd in addition, a large-scale effort is underway to map the microbial genomes associated with humans (The Human Microbiome Project) (https://commonfund.nih.gov/hmp/). Finally, sequencing of wild and domestic species abounds, for example with the initiation of the 10,000 vertebrates sequencing project (http://genome10k.soe.ucsc.edu/). The end result, in terms of data, is that tens of thousands of human genomes and the genomes of our commensals, our parasites, and even our pets are flooding databases around the world.
A Biomedical Cloud
Although Petabytes of genomic, biological, clinical, text and related data are available for downloading today, there is no way currently to compute over all of this data to make discoveries, and, more importantly, there is no conceptual framework to integrate all this data.
It is important to note that just over a decade ago, the same could have been said about the all the data available from web sites. But during this period, companies such as Google filled data centers with this data, developed software to manage and process this data, and then computed over all of this data to improve algorithms for search, online advertising, and related areas.
We argue that an interconnected network of data center scale facilities (loosely speaking “clouds”) with the appropriate security architecture and a rich set of secure intercloud services is the proper foundation for the Biomedical-Infomics Synthesis. Call this the Biomedical Cloud. It is an example of a community cloud (Mell and Grance 2011). It could be filled with all publicly available data relevant to biology, medicine and health care. Like the data in commercial search engines, such a repository would be accessible to individuals via personal devices, with the compute intensive operations being performed in data centers and associated high performance computing facilities.
Moreover secure private clouds and clinical research data warehouses located at medical centers and hospitals containing Electronic Medical Records and other data with PHI information could be enriched with data from the community Biomedical Cloud.
The following eight requirements seem to be necessary for a Biomedical Cloud:
Appropriate security. A Biomedical Cloud would contain a mixture of data, some of it public, some of it restricted to a collaboration, and some of it restricted because it is human genome data, or contains PHI information. At one extreme, for the most restricted data, specialized secure private clouds would be required where the clouds were designed so that the data remained within the required organization and all appropriate regulations were followed. At the other extreme, public clouds could be used to analyze and distribute publically available data.
Secure communications with private clouds. Secure private clouds and data warehouses containing human data and PHI data will be located at medical research centers and contain data critical to the Biomedical-Infomics Synthesis. With the collections of the necessary consents, approval of suitable protocols, and appropriate secure communications, data in these systems could analyzed along with data in the Biomedical Cloud.
On-demand and scalable storage. A Biomedical Cloud should scale so that it can manage and archive all the data relevant to the Biomedical-Infomics Synthesis.
On-demand and scalable analysis. A Biomedical Cloud should scale so that it could analyze all the data relevant to the Biomedical-Infomics Synthesis without moving the data out of the cloud.
Scalable ingestion of data. A Biomedical Cloud should support the scalable ingestion of biological, medical and health care data, including the ingestion of data from next generation sequencers, genomics databases, and other clouds.
Support data liberation. A Biomedical Cloud should provide both long-term storage for data as well as a mechanism for exporting data so that it can be moved to another cloud or facility.
Peer with other private and community clouds. A Biomedical Cloud should interoperate, preferably via peering, with other clouds so that complex analyses can be done using data that spans multiple clouds. By peering, we mean that the two clouds can exchange data without paying a charge per GB of data transported.
Peer with public clouds. A Biomedical Cloud should interoperate, preferably via peering, with public clouds so that an investigator can analyze data within the genomic cloud or using public clouds.
With a Biomedical Cloud satisfying these requirements, all the Petabytes of available data relevant to the Biomedical-Infomics Synthesis could be collocated in one place, and more importantly, algorithms could be used to integrate and process this data on a continuous basis. With the proper architecture, appropriate security, and scalable algorithms, Terabytes of new data would be added to the Biomedical Cloud each day, processed each night, and available for search each morning. In this way, we would continuously update the connections between phenotypic data, biomarker profiles and therapeutic treatments, with the goal of creating predictive models of disease detection, progression, and therapeutic response. The successful fusion of these areas will create a predictive matrix of biomarkers, genetic variants, and environmental variables that will drastically increase the specificity and timeliness of diagnosis for a wide range of common diseases, while delivering accurate predictions about the efficacy of treatment options. Of course there are many logistical and technical challenges to be surmounted if such a Biomedical Cloud will come into existence. How will personal ‘omics and medical data be protected from being decoded in such an environment? How will physicians make best use of such a powerful resource? However, some version of an organically evolving biomedical infomatics machine is likely to arise in the not too distant future. The ingredients are all in place.
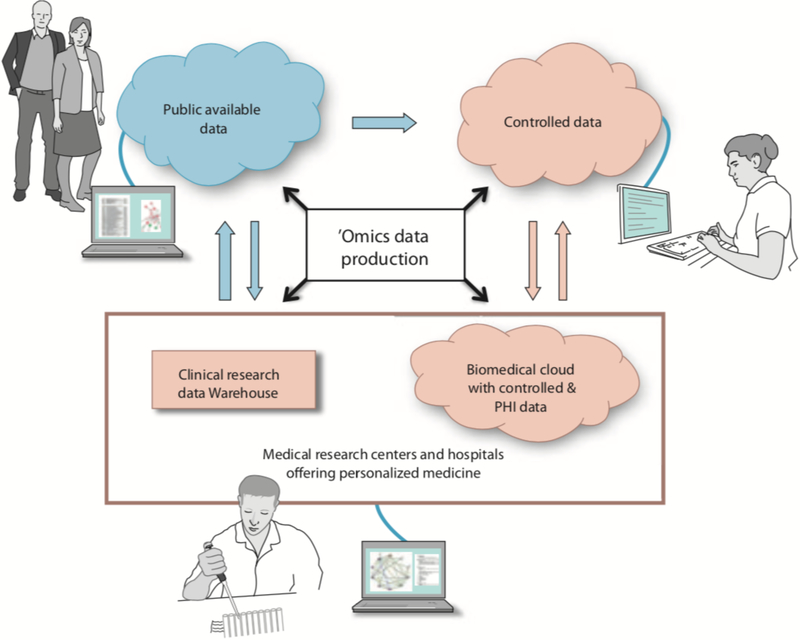
Fig.2.
Biomedical Clouds will interact wit hmedical centres and hospitals providing personalized medicine, as well as researchers and the general public. The blue cloud represents publicly accessible data, whilst pink is used for data that are controlled and have limited access, because of the presence of protected health information, whole human genomes and similar data. Data production may come from many sources, including private and public. There will be multiple Biomedical Clouds associated with medical centers and hospitals that contain controlled data, and these private Biomedical Clouds will be able to ingest data from public Biomedical Clouds. Private clouds may also provide data for research studies via de-identified data sets to public clouds and controlled data to clouds designed to hold such data.
References
- 1000GenomesProject (2010). “A map of human genome variation from population-scale sequencing.” Nature 467(7319): 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit I, Garber M, et al. (2009). “Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses.” Science 326(5950): 257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, et al. (2007). “Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project.” Nature 447(7146): 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem RB, Yvert G, et al. (2002). “Genetic dissection of transcriptional regulation in budding yeast.” Science 296(5568): 752–755. [DOI] [PubMed] [Google Scholar]
- Ciaccio MF, Wagner JP, et al. (2010). “Systems analysis of EGF receptor signaling dynamics with microwestern arrays.” Nature methods 7(2): 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, et al. (2010). “The genetic landscape of a cell.” Science 327(5964): 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean J and Ghemawat S (2008). “MapReduce: simplified data processing on large clusters.” Commun. ACM 51(1): 107–113. [Google Scholar]
- Dobzhansky T (1951). Genetics and the Origin of Species, Columbia University Press. [Google Scholar]
- Herrgard MJ, Swainston N, et al. (2008). “A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology.” Nature biotechnology 26(10): 1155–1160. http://cancergenome.nih.gov/. from http://cancergenome.nih.gov/. http://genome10k.soe.ucsc.edu/. http://www.1000genomes.org/. from http://www.1000genomes.org/. https://commonfund.nih.gov/hmp/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S, Kallen CB, et al. (2008). “Genomic analysis of estrogen cascade reveals histone variant H2A.Z associated with breast cancer progression.” Molecular systems biology 4: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker T, Galitski T, et al. (2001). “A new approach to decoding life: systems biology.” Annual review of genomics and human genetics 2: 343–372. [DOI] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, et al. (2001). “Integrated genomic and proteomic analyses of a systematically perturbed metabolic network.” Science 292(5518): 929–934. [DOI] [PubMed] [Google Scholar]
- Jaeger J, Surkova S, et al. (2004). “Dynamic control of positional information in the early Drosophila embryo.” Nature 430(6997): 368–371. [DOI] [PubMed] [Google Scholar]
- Janssens H, Hou S, et al. (2006). “Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even skipped gene.” Nature genetics 38(10): 1159–1165. [DOI] [PubMed] [Google Scholar]
- Jones RB, Gordus A, et al. (2006). “A quantitative protein interaction network for the ErbB receptors using protein microarrays.” Nature 439(7073): 168–174. [DOI] [PubMed] [Google Scholar]
- Korobkova E, Emonet T, et al. (2004). “From molecular noise to behavioural variability in a single bacterium.” Nature 428(6982): 574–578. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, et al. (2006). “Global landscape of protein complexes in the yeast Saccharomyces cerevisiae.” Nature 440(7084): 637–643. [DOI] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, et al. (2006). “The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease.” Science 313(5795): 1929–1935. [DOI] [PubMed] [Google Scholar]
- Liu J, Ghanim M, et al. (2009). “Analysis of Drosophila segmentation network identifies a JNK pathway factor overexpressed in kidney cancer.” Science 323(5918): 1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGary KL, Park TJ, et al. (2010). “Systematic discovery of nonobvious human disease models through orthologous phenotypes.” Proceedings of the National Academy of Sciences of the United States of America 107(14): 6544–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mell P and Grance T (2011). The NIST Definition of Cloud Computing (Draft): Recommendations of the National Institute of Standards and Technology, National Institute of Standards and Technology. NIST Special Publication; 800–145 (Draft). [Google Scholar]
- Morley M, Molony CM, et al. (2004). “Genetic analysis of genome-wide variation in human gene expression.” Nature 430(7001): 743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MK, Saez-Rodriguez J, et al. (2011). “Training signaling pathway maps to biochemical data with constrained fuzzy logic: quantitative analysis of liver cell responses to inflammatory stimuli.” PLoS computational biology 7(3): e1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RM, Stamatoyannopoulos J, et al. (2011). “A user’s guide to the encyclopedia of DNA elements (ENCODE).” PLoS biology 9(4): e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK and Lindon JC (2008). “Systems biology: Metabonomics.” Nature 455: 1054–1056. [DOI] [PubMed] [Google Scholar]
- Ponten F, Jirstrom K, et al. (2008). “The Human Protein Atlas--a tool for pathology.” The Journal of pathology 216(4): 387–393. [DOI] [PubMed] [Google Scholar]
- Roy S, Ernst J, et al. (2010). “Identification of functional elements and regulatory circuits by Drosophila modENCODE.” Science 330(6012): 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubakhin SS, Romanova EV, et al. (2011). “Profiling metabolites and peptides in single cells.” Nature methods 8(4 Suppl): S20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, et al. (2005). “An integrative genomics approach to infer causal associations between gene expression and disease.” Nature genetics 37(7): 710–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD (2010). “The case for cloud computing in genome informatics.” Genome Biol 11(5): 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, et al. (2007). “Relative impact of nucleotide and copy number variation on gene expression phenotypes.” Science 315(5813): 848–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, et al. (2007). “Population genomics of human gene expression.” Nature genetics 39(10): 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalay A and Gray J (2006). “Science in an exponential world.” Nature 440(7083): 413–414. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, et al. (2010). “Towards a knowledge-based Human Protein Atlas.” Nature biotechnology 28(12): 1248–1250. [DOI] [PubMed] [Google Scholar]
- Watson JD and Crick FH (1953). “Genetical implications of the structure of deoxyribonucleic acid.” Nature 171(4361): 964–967. [DOI] [PubMed] [Google Scholar]
- Watson JD and Crick FH (1953). “Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid.” Nature 171(4356): 737–738. [DOI] [PubMed] [Google Scholar]
- Zinzen RP, Girardot C, et al. (2009). “Combinatorial binding predicts spatiotemporal cis-regulatory activity.” Nature 462(7269): 65–70. [DOI] [PubMed] [Google Scholar]