Abstract
Diverse CRISPR-Cas immune systems protect archaea and bacteria from viruses and other mobile genetic elements. All CRISPR-Cas systems ultimately function by sequence-specific destruction of invading complementary nucleic acids. However, each CRISPR system uses compositionally distinct crRNP [CRISPR (cr) RNA/Cas protein] immune effector complexes to recognize and destroy invasive nucleic acids by unique molecular mechanisms. Previously, we found that Type I-A (Csa) effector crRNPs from Pyrococcus furiosus function in vivo to eliminate invader DNA. Here, we reconstituted functional Type I-A effector crRNPs in vitro with recombinant Csa proteins and synthetic crRNA and characterized properties of crRNP assembly, target DNA recognition and cleavage. Six proteins (Csa 4–1, Cas3″, Cas3′, Cas5a, Csa2, Csa5) are essential for selective target DNA binding and cleavage. Native gel shift analysis and UV-induced RNA–protein crosslinking demonstrate that Cas5a and Csa2 directly interact with crRNA 5′ tag and guide sequences, respectively. Mutational analysis revealed that Cas3″ is the effector nuclease of the complex. Together, our results indicate that DNA cleavage by Type I-A crRNPs requires crRNA-guided and protospacer adjacent motif-dependent target DNA binding to unwind double-stranded DNA and expose single strands for progressive ATP-dependent 3′–5′ cleavage catalyzed by integral Cas3′ helicase and Cas3″ nuclease crRNP components.
Keywords: CRISPR, Cas, Csa, Cas3, Type I-A, Pyrococcus furiosus
Introduction
Many archaea and bacteria harbor RNA-based, CRISPR-Cas (Clustered regularly interspaced short palindromic repeatsCRISPR-associated) adaptive immune systems that provide the organisms with protection against viruses and other inva- sive mobile genetic elements (Barrangou et al. 2007). These immune systems heritably acquire short (30–40 bp) DNA fragments from the invader DNA (called protospacers) and integrate them (as spacer DNAs) into the CRISPR loci of the prokaryotic host genome. CRISPR loci are transcribed to generate precursor CRISPR transcripts. These transcripts are processed to mature CRISPR RNAs (crRNAs). Each mature cRNA contains invader-derived guide sequences flanked by 5′ and/or 3′ CRISPR repeat-derived short sequences (known Communicated by L. Huang. as 5′ and/or 3′ tag elements). Mature crRNAs associate with Cas proteins to form crRNA/Cas protein immune effector complexes (crRNPs) that degrade complementary invader DNA or RNA (depending on the type of CRISPR-Cas system) in a sequence-specific manner (Hille et al. 2018; Jackson and Wiedenheft 2015; Makarova et al. 2015; Terns and Terns 2011).
Type I CRISPR-Cas systems are the most abundant and widespread among the numerous, identified Types I-VI CRISPR-Cas systems (Haft et al. 2005; Makarova et al. 2015; Vestergaard et al. 2014). Type I systems share similarities in Cas protein composition, crRNP structure and mechanism of DNA destruction (Makarova et al. 2011). Typically, Type I immune effector crRNPs contain members of Cas5, Cas7, Cas8 (large subunit) and Cas 11 (small subunit) superfamily proteins (Brendel et al. 2014; Brouns et al. 2008; Hochstrasser et al. 2016; Jackson et al. 2014; Lintner et al. 2011b; Majumdar et al. 2015, 2016; Mulepati et al. 2014; Nam et al. 2012; Plagens et al. 2012; van Duijn et al. 2012; Vestergaard et al. 2014; Wiedenheft et al. 2011; Zhao et al. 2014). However, considerable variability among the protein subunits of Type I effector crRNPs has contributed to partition of Type I CRISPR-Cas systems into several, discrete Type I subtypes: I-A (Csa), I-B (Csh), I-C (Csd), I-D (not applicable), I-E (Cse), I-F (Csy), I-G (Cst) and I-U (unclassified) (Type nomenclature is based on (Makarova et al. 2011, 2015) and three letter designations by Haft et al. (2005) and Vestergaard et al. (2014).
Cas3 superfamily proteins are key defining proteins of Type I systems. Diverse Cas3 proteins share conserved DExH helicase and HD nuclease domains and act as effector nucleases by catalyzing invader DNA cleavage (Brouns et al. 2008; Cady and O’Toole 2011; Huo et al. 2014; Majumadr et al. 2016; Mulepati and Bailey 2013; Plagens et al. 2014; Sinkunas et al. 2013; Westra et al. 2012). Type I-A (Csa) systems (the focus of this study) are unique in that the nuclease and helicase functions are encoded in two separate polypeptides: Cas 3″ (HD nuclease) and Cas3′ (DExH helicase) (Haft et al. 2005; Makarova et al. 2011). Moreover, Cas3″ and Cas3′ are structural components of Type I-A effector crRNP complexes (Majumdar et al. 2015; Plagens et al. 2012, 2014), whereas other Type I crRNPs recruit Cas3 in trans, following DNA recognition and binding (Cady and O’Toole 2011; Mulepati and Bailey 2013; Sinkunas et al. 2013; Xiao et al. 2017, 2018). Type I-A systems are also highly restricted to archaeal lineages, with exceptionally rare occurrences of bacterial I-A systems reported thus far (Makarova et al. 2011, 2015).
All characterized Type I crRNPs recognize and degrade double-stranded DNA targets (in contrast, some CRISPRCas systems recognize and destroy invader RNA [e.g., Type VI (O’Connell 2018)] instead of, or in addition to, DNA [e.g., Type III (Elmore et al. 2015; Estrella et al. 2016; Kazlauskiene et al. 2016)]. For Type I systems, selective binding of target DNAs that are complementary to the crRNA first requires Cas protein-mediated recognition of a short DNA motif (typically 3–5 bases) on the invader DNA called PAM (Protospacer adjacent motif) (Deveau et al. 2008; Hayes et al. 2016; Mojica et al. 2009; Pausch et al. 2017; Rollins et al. 2015; Shah et al. 2013; Wang et al. 2015). Following PAM recognition, Type I crRNPs bind double-stranded DNA targets through base-pairing of crRNA-guide with complementary protospacer sequences of target DNA strand. This leads to R-loop formation and displacement of the non-target DNA strand (Hayes et al. 2016; Sinkunas et al. 2013; Szczelkun et al. 2014; Xiao et al. 2018). Ultimately, Cas3 effector proteins utilize their helicase (DNA unwinding) and DNase activities to progressively nick and destroy target DNAs that are bound by Type I effector crRNPs. However, mechanistic details differ for distinct Type I subtypes (Hochstrasser et al. 2014; Huo et al. 2014; Jiang and Doudna 2015; Jiang et al. 2016; Mulepati and Bailey 2013; Mulepati et al. 2014; Westra et al. 2012).
The hyperthermophilic euryarchaeon, Pyrococcus furiosus (Pfu), has seven CRISPR loci that collectively expressed 200 crRNA species (Terns and Terns 2013). The twentyseven Pfu cas genes are primarily arranged in two gene clusters and encode Cas proteins for: DNA acquisition at CRISPR loci [Cas1, Cas2, Cas4–1, Cas4–2 (Shiimori et al. 2017, 2018)], crRNA biogenesis [Cas6 (Carte et al. 2008, 2010)] and three immune effector crRNPs [Type I-A (Csa), Type I-G (Cst) and Type III-B (Cmr)] Fig. 1a, (Terns and Terns 2013). We have previously identified three distinct native Pfu crRNPs, each comprising either Csa, Cst or Cmr Cas proteins in association with crRNAs from all seven Pfu CRISR loci (Hale et al. 2012; Majumdar et al. 2015). Each of the three co-existing effector crRNPs was demonstrated to function independently in plasmid silencing in vivo (Elmore et al. 2015, 2016). Moreover, we elucidated the structural organization of Type III-B (Cmr) crRNPs (Spilman et al. 2013) and investigated molecular mechanism of crRNAguided target RNA cleavage (Hale et al. 2009, 2012, 2014) as well as crRNA- and transcript-dependent DNA cleavage by the Cmr system (Elmore et al. 2016). Furthermore, we successfully reconstituted active Type I-G (Cst) crRNPs and elucidated important details regarding how the crRNPs assemble and execute DNA recognition and destruction in vitro (Majumdar et al. 2016).
Fig. 1.
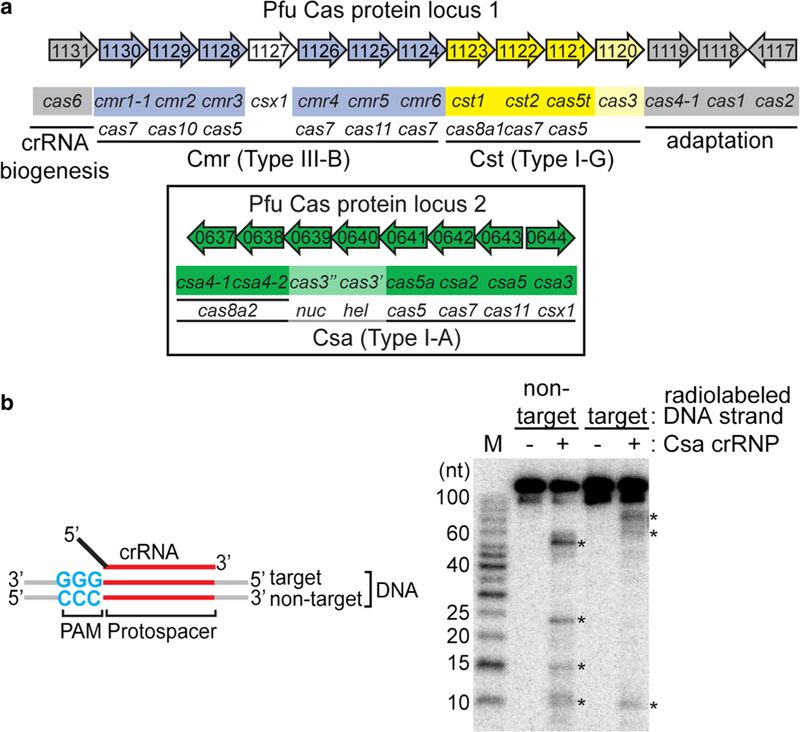
Reconstitution of functional Type I-A crRNPs. a Pyrococcus furiosus (Pfu) cas gene organization and annotation. Genes encoding the crRNA biogenesis protein (cas6, gray), predicted CRISPR DNA integration proteins (cas1, cas2 and cas4, gray) and three effector modules: Type I-A (csa, green and boxed), Type I-G (cst, yellow) and Type III-B (cmr, blue) are shown. The annotated superfamily designation (e.g., cas5, cas7, cas8) is denoted below each gene name. b Schematic of double-stranded DNA substrate used in the assay. Wildtype PAM (blue), protospacer (red), target strand (complementary to crRNA guide) or non-target strand of DNA, 5′ tag (black) and guide (red) of crRNA are labeled. Denaturing PAGE demonstrating cleavage (asterisk) of double-stranded DNA in the absence (−) or presence of Csa crRNP (+) (assembled with six Csa proteins (Csa4–1, Cas3″, Cas3′, Cas5a, Csa2, Csa5) and crRNA). Top label indicates location of 5′ radiolabeling (32P) in either the target strand or non-target strand of double-stranded DNA. 5′ radiolabeled (32P) DNA size-standard (Affymetrix, 10–100 bases) is labeled (M)
In this study, we have characterized Type I-A (Csa) crRNP assembly and function by reconstituting active complexes from recombinant Pfu Csa Cas proteins and synthetic crRNA. The Type I-A crRNP binds and cleaves double-stranded DNA that contains a PAM (5′-NGG-3′ on target strand), in a crRNA-guided manner. We observed that six Csa proteins, Csa4–2, Cas3″, Cas3′, Cas5a, Csa2 and Csa5 (Fig. 1a), are essential minimal protein components of functional Type I-A crRNPs. We have delineated some key molecular features of crRNAs, including the CRISPR repeat-derived, 8-nt 5′ tag sequence and 5′ hydroxyl end group, in Type I-A crRNP assembly. Consistent with findings in other Type I systems, Cas3″ (with conserved HD active site) is the effector nuclease of the Type I-A crRNP that cleaves bound DNA. Based on the analysis of target DNA cleavage profiles, we propose a mechanistic model of invader DNA destruction by the Type I-A crRNP.
Materials and methods
Expression and purification of recombinant Csa proteins
The genes encoding N-terminal 6 × histidine-tagged Pfu Csa Cas proteins were subcloned into either pET24 (Cas3″, Cas5a, Csa2 and Csa5) or pET200 (Csa4–1, Cas3′ and Csa3) expression vectors. Genes encoding a C-terminal 6x histidine-tagged Csa4–2 protein (without stop codon) and untagged Cas3″ and Csa4–1 proteins (with stop codon) were subcloned into a pET101 expression vector. All plasmids were transformed into an E. coli BL21 RIPL strain. Proteins were expressed and purified as described (Majumdar et al. 2016). Briefly, 4 L Luria broth cultures were grown under antibiotic selection to an OD600 of 0.4–0.6 at 37 °C and induced with 1 mM IPTG at room temperature overnight. Cells were resuspended in buffer A (25 mM Tris pH 7.6, 250 mM NaCl) supplemented with 10 mM imidazole and 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and lysed by sonication (Misonix Sonicator 3000). The lysed cells were incubated at 70 °C for 30 min with frequent mixing, centrifuged at 14,000 rpm at 4 °C and the soluble fraction was filtered (0.8 µm filter pore size Millex filter unit, Millipore). His-tagged proteins were purified by Ni2+ affinity column chromatography, washed with buffer A and 25 mM imidazole, and eluted with buffer A at increasing concentrations (50, 100, 200, 500 mM) of imidazole. Peak elution fractions were dialyzed in buffer A to remove imidazole using Slidea-lyzer mini dialysis cassettes (Thermo Fisher), quantified with Qubit 2.0 flurometer (Life Technologies) and stored at 4 °C, prior to use for functional assays.
Preparation of RNA and DNA substrates
Synthetic RNA and DNA oligo sequences are listed in Table S1. 7.01 crRNAs (wild-type 5′ tag and mutant tag) were previously described (Hale et al. 2009, 2012; Majumdar et al. 2016). crRNAs were 3′ radiolabeled (Figs. 7 and S4) with T4 RNA ligase (New England Biolabs) and [α−32P] pCp (3000 Ci/mmol, Perkin Elmer) as previously described (Carte et al. 2008, 2010). Internally labeled 7.01 crRNA was prepared (Fig. 8) by in vitro T7 transcription of repeatspacer unit with [α−32P]-GTP (800 Ci/mmol, 10 mCi/mL, Perkin Elmer) followed by Cas6 digestion as described (Spilman et al. 2013). DNA (MWG, Eurofins) and RNA oligos (Integrated DNA Technologies) were 5′ radiolabeled with T4 polynucleotide kinase (PNK) and [γ−32P] ATP as described (Carte et al. 2008). Radiolabeled double-stranded DNA (Table S2) were made as previously described (Majumdar et al. 2016). Briefly, complementary single-stranded DNA was annealed at a molar ratio of 2:1 of unlabeled to 5′ radiolabeled (32P) in 10 mM Tris (pH 7.5–8.0), 50 mM NaCl, 1 mM EDTA, and incubated at 95 °C for 5 min followed by slow cooling until 23 °C (at the rate of 1 °C per minute). Annealing was confirmed by 8% non-denaturing 0.5 × TBE polyacrylamide gel electrophoresis. Double-stranded DNA substrates were gel purified, extracted with PCI (phenol/ chloroform/isoamyl alcohol) (pH 8.0), ethanol precipitated, resuspended in buffer A and stored at − 20 °C for future use in functional assays.
Fig. 7.
Key crRNA features for Type I-A crRNP assembly. Native gel shift assay of a 3′ radiolabeled (32P) crRNA with no proteins (−), six Csa proteins (Csa4–1, Cas3″, Cas3′, Cas3′, Cas5a, Csa2 and Csa5), five Csa proteins (one Csa protein excluded at a time, denoted with a circled minus) as indicated. b 3′ radiolabeled (32P) crRNAs contain- ing either wild-type 5′ tag (wt., black) or mutated 5′ tag (mut., red) with no protein (−), 2 Csa proteins (Cas5a and Csa2), 6 Csa proteins (same as above). c crRNAs containing 5′-hydroxyl and 3′ phosphate (5′ OH/3′P, wild type in schematics), 5′ and 3′ hydroxyl (5′OH/3′OH), 5′- phosphate, 3′ hydroxyl (5′P/3′OH) with no proteins (−) 2, 6 Csa proteins (same as b). Guide sequence (orange) is wild type in all cases. Unbound crRNA (crRNA), crRNP subcomplexes (asterisk) and Type I-A crRNP complexes (crRNP, bracket) are indicated
Fig. 8.
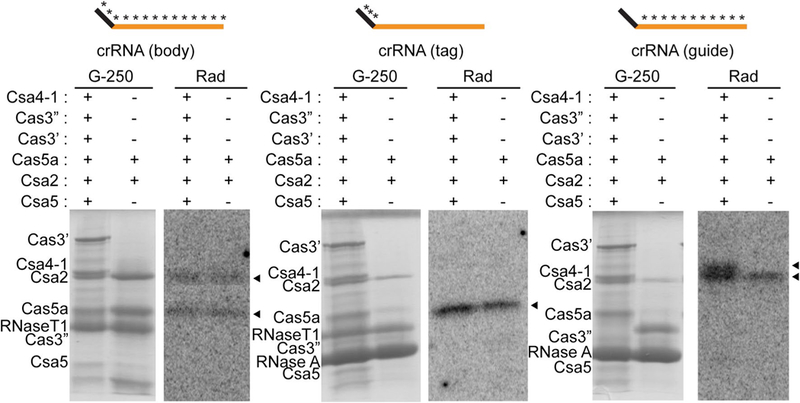
Identification of direct interactions between crRNA and Csa proteins by UV crosslinking. SDS-polyacryladmide gel showing Coomassie blue (G-250) stained Csa proteins and RNase T1 and/or RNase A in each reaction and corresponding autoradiograph images (rad) showing crRNA-interacting Csa proteins (black triangles).Reactions were incubated with either 2 or 6 Csa proteins (same as Fig. 7b, c). The schematic above each panel indicates 32P and 4-thiouridine-labeled crRNAs used in each experiment (5′ tag sequence (black), guide sequence (orange) and radiolabeled regions (asterisks)
Type I-A crRNP reconstitution assays
Csa crRNP complexes were reconstituted as follows: 1 µM of each recombinant Csa Cas protein was incubated with 0.05 pmol of (32P) radiolabeled crRNA (Figs. 7 and S4) in buffer A (25 mM Tris, pH 7.6, 250 mM NaCl), 2 mM ATP, 1.5 mM MgCl2, 1 unit of SUPERase-In ribonuclease inhibitor (Applied Biosystems) and 1 µg of E. coli tRNA (non- specific competitor) at 70 °C for 30 min. Reactions were mixed with loading dye (4% glycerol, 0.05% bromophenol blue and 0.05% xylene cyanol in 1 × TBE) and resolved on 6% native polyacrylamide gels (containing 0.5 × TBE and 8% glycerol) in 0.5 × TBE. The gel was run for 2 h at 110 V, dried and exposed on a phosphor-imaging screen.
DNA binding and cleavage assays
DNA binding and cleavage (Figs. 1, 2, 3, 4, 5, 6 and S1, S2, S3) were assayed as previously described (Majumdar et al. 2016). The Type I-A crRNP complex was reconstituted as mentioned above except with 0.5 pmol of unlabeled crRNA and incubated at 70 °C for 30 min. 0.02–0.05 pmol of 32P radiolabeled DNA substrate and 100 µM NiCl2 were added and incubated at 70 °C for 3 h.
Fig. 2.
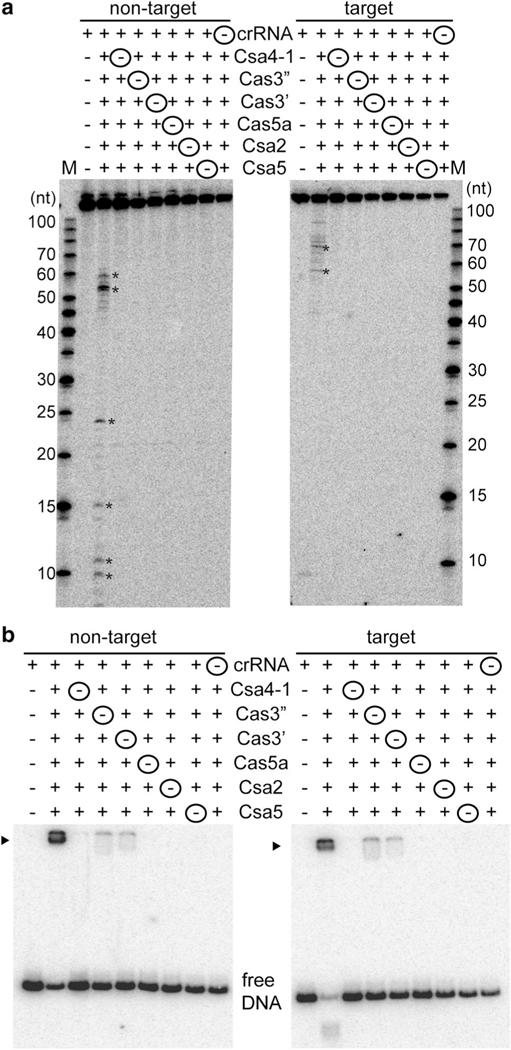
Minimal components of the Type I-A crRNP required for DNA targeting. a Effect on DNA cleavage (major cleavage products are denoted by asterisks) when Csa crRNP complex was reconsti- tuted in the absence of one component at a time. Components that were excluded (− and circled) and included (+) in each Csa crRNP assembly reaction are indicated. 5′ radiolabeled (32P) DNA size- standard (Affymetrix, 10–100 bases) is labeled (M). b Corresponding gel mobility shift assay of samples in a. DNA bound by Csa crRNP (black triangle) and free DNA is indicated. Lane 1 of each gel shows reaction in the absence of any Csa protein and the upper label indi- cates location of radiolabel (32P) in either the non-target or target strand of double-stranded DNA
Fig. 3.
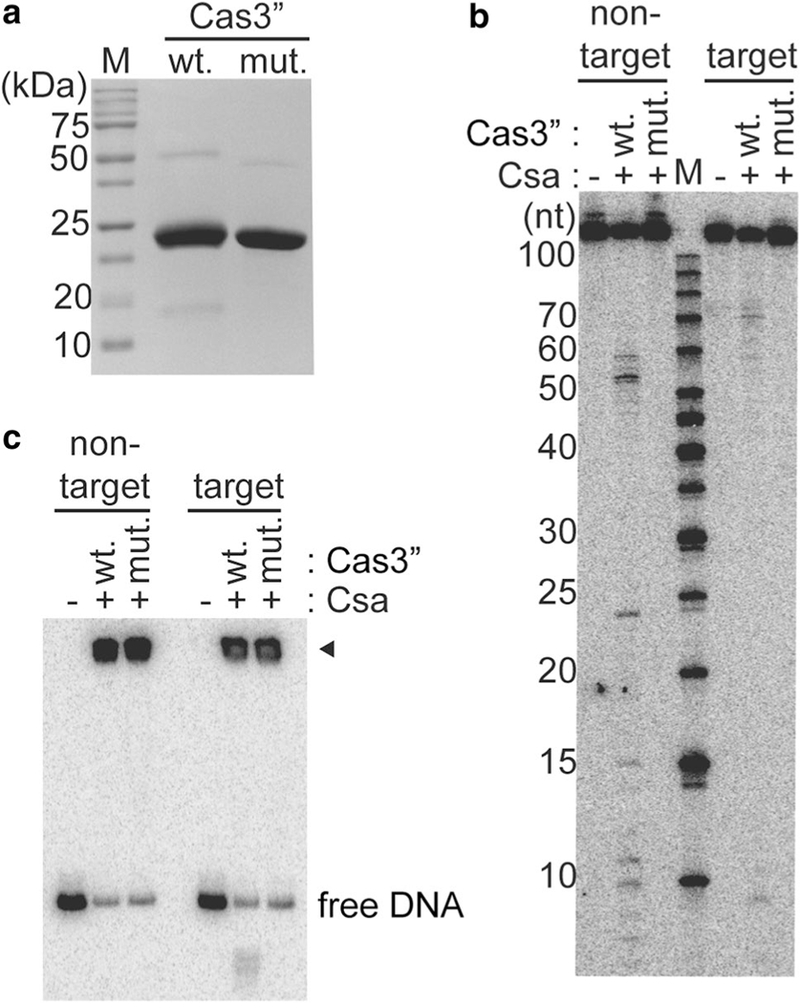
Cas3″ is the effector nuclease of type I-A crRNP. a SDS- PAGE showing recombinant wild-type (wt.) Cas3″ and HD-AA mutant (mut.) Cas3″ proteins. Molecular weight marker proteins (sizes in kDa) are labeled (M). b DNA cleavage with Csa crRNP [+, assembled with crRNA, Csa4–1, Cas3′, Cas5a, Csa2, Csa5 and either Cas3″ (wild-type (wt.) or HD-AA mutant (mut.)]. 5′ radiolabeled (32P) DNA size-standard (Affymetrix, 10–100 bases) is labeled (M). c Gel mobility shift assay of corresponding samples in b. DNA bound by Csa crRNP (black triangle) and free DNA is indicated. b and c Reactions in the absence of any Csa protein are labeled (−) and the upper label indicates location of 5′ radiolabel (32P) in either the nontarget or target strand of double-stranded DNA
Fig. 4.
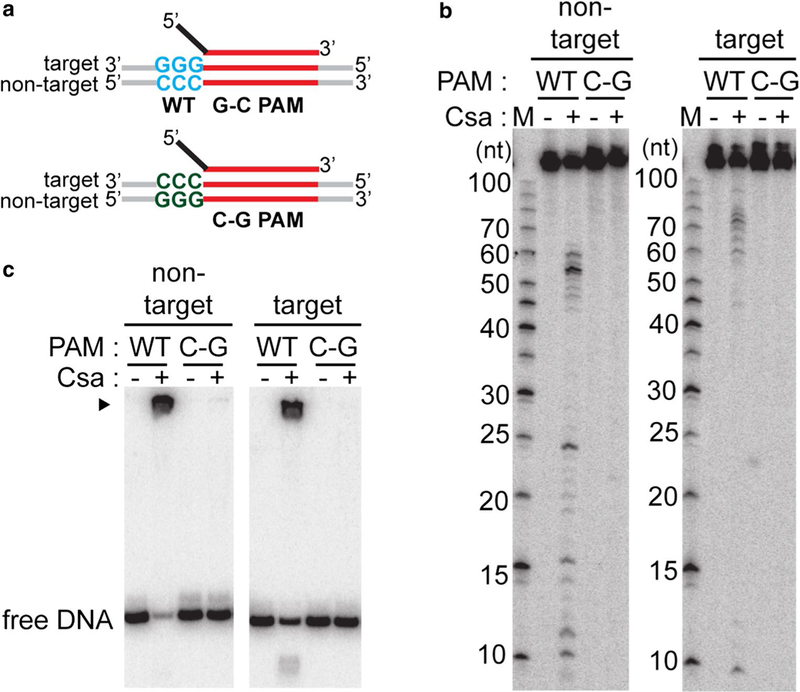
PAM sequence requirements for DNA recognition and cleavage. a Schematic of double-stranded DNA substrates used in assay: crRNA (top) containing 5′ tag (black) and guide (red), double-stranded DNA (target and non-target strands labeled) with wild-type protospacer (red) and PAM [wild-type (blue, WT, G-C PAM) or C-G PAM (green, PAM sequences mutated in both strands)] b DNA cleavage with Csa crRNP (+, assembled with crRNA, Csa4–1, Cas3″, Cas3′, Cas5a, Csa2 and Csa5) and the specified substrates. 5′ radiola-beled (32P) DNA size standard (Affymetrix, 10–100 bases) is labeled (M). c Correspond- ing gel mobility shift assay of samples in b. DNA bound by Csa crRNP (black triangle) and free DNA is indicated. b and c Reaction in the absence of any Csa protein is labeled (−) and top label indicates location of 5′ radiolabel (32P) in either the non-target or target strand of double-stranded DNA
Fig. 5.
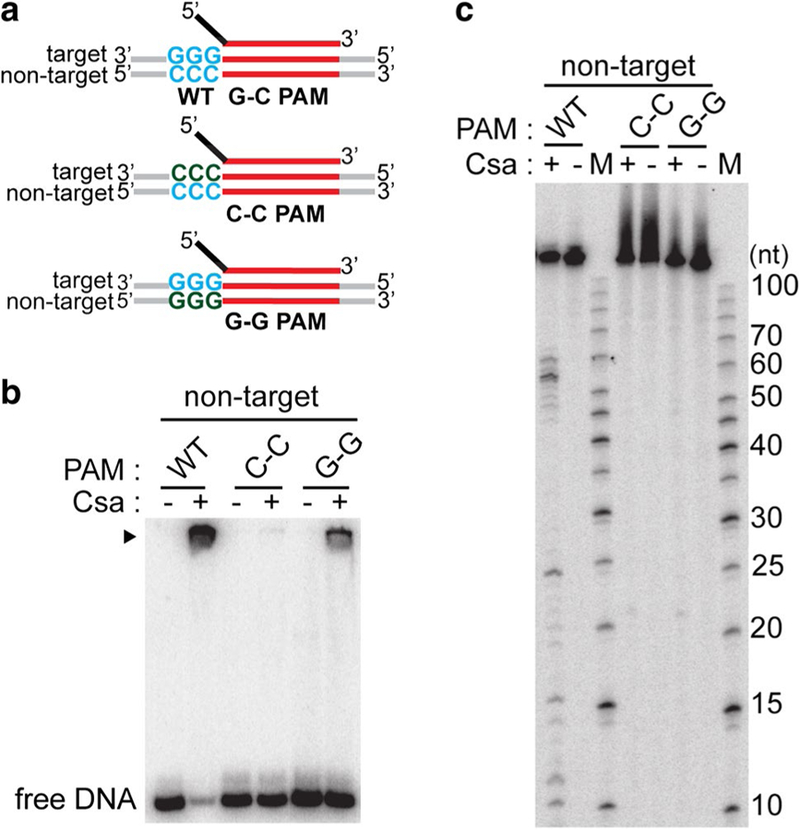
Type I-A crRNPs recognize double-stranded PAM for DNA destruction. a Schematic of double-stranded DNA substrates used in assay: crRNA (top) containing 5′ tag (black) and guide (red), doublestranded DNA (target and non-target strands labeled) with wild-type protospacer (red) and PAM (WT G-C PAM (wild type, blue) or G–G PAM [single-stranded, wild type on target strand (blue) and mutated on non-target strand (Sternberg et al. 2014)] or C–C [single-stranded, wild type on non-target strand (blue) and mutated on target strand (Sternberg et al. 2014)]. b DNA binding and c DNA cleavage with no protein (−) Csa crRNP ((+) assembled with crRNA, Csa4–1, Cas3″, Cas3′, Cas5a, Csa2 and Csa5) and the specified substrates. DNA bound by Csa crRNP (black triangle) and free DNA in b is indicated. 5′ radiolabeled (32P) DNA size standard (Affymetrix, 10–100 bases) (c) is labeled (M). The non-target strand of DNA is 5′-radiolabeled (32P) in all cases
Fig. 6.
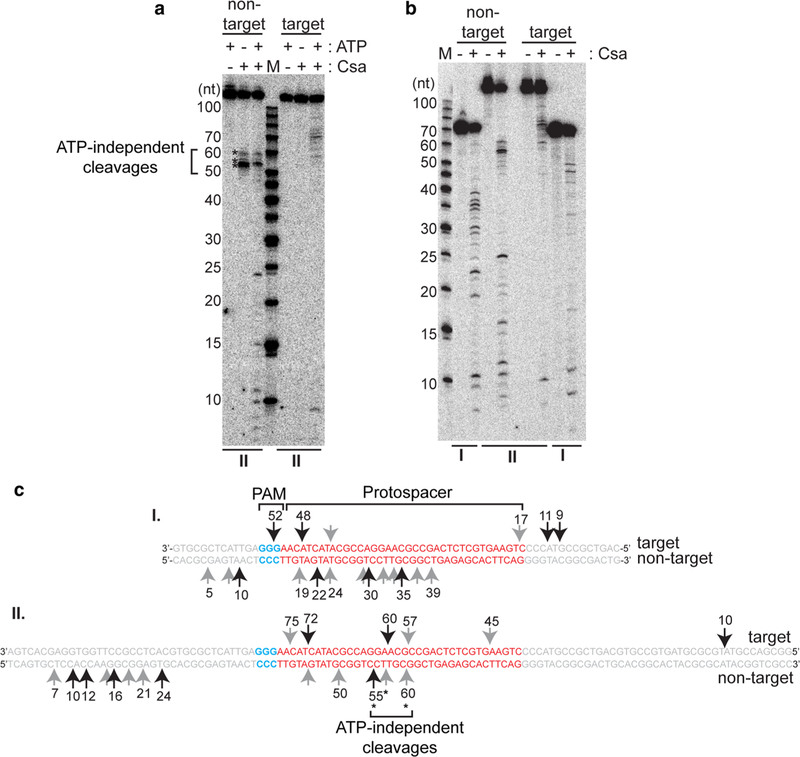
Type I-A crRNP-induced cleavages map to multiple sites in both strands of DNA. a Effect of ATP (∓) on cleavage of double- stranded DNA (117 base pairs, same as Figs. 1, 2, 3, 4, 5 and II in Fig. 6) and b Cleavage profiles of two different sized double-stranded DNA [67 base pairs (I) and 117 base pairs (II)] with no protein (−) or Csa crRNP [(+) assembled with crRNA, Csa4–1, Cas3″, Cas3′, Cas5a, Csa2 and Csa5]. The upper label on each gel indicates location of 5′ radiolabel (32P) on the non-target or target strand of double-stranded DNA. 5′ radiolabeled DNA size-standard (Affymetrix, 10–100 bases) is labeled (M). c Major (long black arrows) and minor (short gray arrows) cleavage products (in a and b) mapped to target and non-target strand sequences of double-stranded DNA substrates (I and II) tested in b. Protospacer (red, labeled), PAM (blue, labeled) and flanking (gray) sequences are indicated. Numbers correspond to size of cleavage products (counting from 5′ end of each strand). ATP- independent cleavages (on non-target strand) are denoted (asterisks and bracket) and labeled (in a and c)
To visualize DNA binding (Figs. 2b, 3c, 4c, 5b and S3), half reactions were mixed with loading dye (4% glycerol, 0.05% bromophenol blue and 0.05% xylene cyanol in 1 × TBE) and separated on 6% native polyacrylamide gels (containing 0.5 × TBE and 8% glycerol) in 0.5 × TBE for 2 h at 110 V. For assaying DNA cleavage, the remain- ing half reactions were treated with 1 µg of Proteinase K (New England Biolabs) at 37 °C for 15 min followed by PCI extraction (pH 8) and ethanol precipitation and resolved on 15% denaturing (7 M urea) polyacrylamide sequencing or mini gels. Gels were dried and exposed to phosphor-imaging screens. 5′ (32P) radiolabeled low- molecular weight single-stranded DNA markers (10–100 bases, Affymetrix) were used to determine the sizes of the observed cleavage products.
Site-directed mutagenesis
To generate the HD-AA mutant of Cas3″, QuikChange site-directed mutagenesis was performed as per manufacture’s guidelines (Agilent Technologies). Primers (from MWG Eurofins) are listed in Table S1. Briefly, the mutants were generated by PCR amplification of wild-type cas3″ (pET24) plasmid followed by DpnI digestion and transformation into an E. coli BL21 RIPL strain. The mutation was confirmed by DNA sequencing. The mutant protein was expressed and purified alongside wild-type Cas3″ (Fig. 3a).
RNA–protein UV crosslinking
crRNA-Csa protein covalent crosslinking was achieved using photoactivable 4-thiouridine and (32P) internally labeled crRNAs as previously described (Majumdar et al. 2016). Briefly, Type I-A crRNP reconstitution reactions were doubled relative to standard crRNP assembly reaction described above and RNasin was omitted. A quarter reaction was assessed for crRNA binding by native gel electrophoresis (Fig. S4b). The remaining reactions were irradiated with 312 nm UV light and the complexes disrupted by addition of 1% SDS treatment and boiling for 10 min, followed by RNase treatment at 37 °C for 1 h. Reactions containing the radiolabeled crRNA and 8 nt 5′ tag-RNA were treated with RNase T1 (1000 U, Thermo Fisher) and guide RNA was treated with RNase A (10 µg, Thermo Fisher). The samples were separated on 12.5% SDS-polyacrylamide gels, visualized by G-250 Coomassie staining, dried, and subjected to phosphor imaging and autoradiography to allow for alignment of protein bands with autoradiographs (Fig. 8).
Protein–protein interaction
1 L culture of N-terminal, 6 × -His-tagged Cas3′ and 500 ml cultures each of untagged Csa4–1, Cas3″ were cultured and induced with IPTG overnight as described above. 2:1:1 cul- tures of Cas3′, Cas3″ and Csa4–1 and 2:1 cultures of Cas3′ and Cas3″ were mixed and the cells were lysed and purified as mentioned before. The samples were affinity purified with Ni–NTA column (Qiagen), eluted with buffer A at increasing concentrations (50, 100, 200, 500 mM) of imidazole and elutions were separated on 12.5% SDS-polyacrylamide gel followed by staining with Coomassie (R-250) staining. The peak elutions are shown (Fig. S5a). The Cas3′ + Cas3″ subcomplex was further purified by gel filtration (Superdex 200, GE Healthcare, provided by Hong Li, Florida State University) (Fig. S5b).
Results
Reconstituted Pfu Type I-A crRNPs cleave double-stranded DNA
Previously, we found that Type I-A (Csa) crRNPs eliminated plasmid DNA in vivo provided that the target DNA contained protospacer sequences complementary to a native crRNA and was immediately flanked by a PAM element (primary consensus: 5′-NGG/CCN-3′) (Elmore et al. 2015). To gain a more detailed understanding of the molecular mechanism of crRNP assembly and target DNA destruction, we sought to reconstitute a functional Type I-A effector crRNP by combining synthetic crRNA and recombinant Csa Cas proteins Csa4–1 and Csa4–2, Cas3″ (predicted HD nuclease), Cas3′ (predicted DExH helicase), Cas5a, Csa2, Csa5 and Csa3 CARF (Cas Rossmann fold) superfamily protein related to Csx1 (Makarova et al. 2014) and putative transcriptional regulator (He et al. 2016; Lintner et al. 2011a; Makarova et al. 2014) (Figs. 1a, S1a). We used a 45 nt crRNA composed of an 8-nt (AUUGAAAG) CRISPR repeat-derived, 5′ tag (black) and entire, 37 nt 7.01 guide sequence (first spacer of CRISPR locus 7, red) (Fig. 1b, schematic). The 7.01 crRNA was previously identified in native Type I-A crRNP complexes (Majumdar et al. 2015) and shown to be capable of guiding function of the Type I-A system in vivo (Elmore et al. 2015). To test for cleavage activity of the reconstituted Type I-A crRNP, we used a double-stranded DNA substrate with 7.01 protospacer (red) sequence, wild-type PAM (5′-GGG/CCC-3′, blue) and random flanking sequences (gray) (Fig. 1b, schematic). The reconstituted Type I-A crRNP cleaved the double-stranded DNA substrate at both the target DNA strand (complementary to crRNA guide) and non-target DNA strand (Fig. 1b).
Six Csa Cas proteins are essential components of functional Type I-A crRNPs
To determine the minimal Cas protein components essential for DNA cleavage, we systematically excluded one Csa pro- tein at a time from the reaction established to assemble functional Type I-A crRNPs (Fig. 2). We observed that Csa4–2 (one of two Csa4 proteins) and Csa3 [a predicted transcrip- tion factor regulating the system (Lintner et al. 2011a)] were dispensable for DNA cleavage activity in vitro (Fig. S1b). In contrast, excluding any one of the six Csa proteins (Csa 4–1, Cas3″, Cas3′ Cas5a, Csa2, Csa5) abolished cleavage of both non-target and target strand of double-stranded DNA (Fig. 2a). The six Csa Cas proteins that form the catalytically active crRNP also bound target DNA as assayed by native gel electrophoretic mobility shift analysis (Fig. 2b). In the absence of any of these six Csa Cas proteins, DNA binding was either greatly reduced (without Cas3″ or Cas3′) or completely abolished (without Csa4–1, Cas5a, Csa2 and Csa5). Furthermore, in the absence of crRNA, all six Csa proteins did not bind or cleave target DNA. Therefore, a crRNA and six Pfu Csa proteins (Csa 4–1, Cas3″, Cas3′ Cas5a, Csa2, Csa5) are essential components of the catalytically active, DNA-cleaving Type I-A effector crRNP. Subsequently, the additional assays described in this study (except Fig. S1b) were performed with crRNPs reconstituted with six Csa proteins and a 45 nt 7.01 synthetic crRNA.
Cas3″ is the Type I-A effector nuclease
Unlike other characterized Type I systems, where nuclease and helicase domains are fused in a single Cas3, Type I-A has two Cas3 proteins. Cas3″ contains the predicted nuclease (HD) domain and Cas3′ contains the (DExH) helicase domain (Haft et al. 2005). Another distinction of Type I-A systems is that Cas3″ and Cas3′ proteins are integral structural components and not recruited in trans (Fig. 2 and Majumdar et al. 2015; Peng et al. 2013; Plagens et al. 2012). However, similar to Cas3 proteins of other Type I systems and consistent with biochemical properties of Cas3″ (Beloglazova et al. 2011; Plagens et al. 2014), site-directed mutagenesis of histidine and aspartate (H60, D61) of Cas3″ to alanines (Fig. 3a) resulted in loss of DNA cleavage by the reconstituted Type I-A crRNP (Fig. 3b) without affecting DNA binding (Fig. 3c).
DNA recognition and cleavage require a functional PAM
Our previous in vivo study highlighted the PAM requirements for plasmid silencing by Type I-A system in Pfu (Elmore et al. 2015). To determine whether the same prin- ciples govern target DNA binding and cleavage by reconsti- tuted Type I-A crRNP, we mutated a functional PAM asso- ciated with Type I-A system (wt. G–C PAM; 5′-GGG-3′ on target strand and 5′-CCC-3′ on the non-target strand) to a mutant variant (C–G PAM; 5′-CCC-3′ on target strand and 5′-GGG-3′ on the non-target strand) (Fig. 4a). The reconsti- tuted Type I-A crRNP only bound and cleaved DNA with wild-type PAM (WT, G–C PAM) but not mutated PAM (C–G PAM) (Figs. 4b, c). Moreover, the Type I-A crRNP did not bind or cleave DNA when the protospacer sequences were mutated such that it lacked complementarity with the 7.01 crRNA guide (data not shown). Furthermore, Csa crRNP complex did not cleave (single-stranded or double- stranded) RNA targets (Fig. S2a). The results indicate that the Type I-A crRNP selectively binds and cleaves DNA that contains a functional PAM and protospacer sequence that is complementary to crRNA.
Type I-A crRNPs recognize double-stranded PAM on DNA targets
To probe whether the Type I-A crRNP recognizes the PAM in single-stranded or double-stranded form, we tested DNAs with hybrid PAMs (changing wild-type G–C PAMs to either a C–C PAM or a G–G PAM, Fig. 5a). We mutated one strand of the PAM (target strand in C–C (green) and non-target strand in G–G (green) (Sternberg et al.) and did not make compensatory mutations in the other strand leaving a region of non-complementarity. Csa crRNPs exhibited weak binding to DNA with single-stranded 5′-GGG-3′ PAM on tar- get strand with reduced efficiency (G–C versus G–G PAM) (Fig. 5b) but failed to cleave the DNA (Fig. 5c). DNA with single-stranded 5′-CCC-3′ PAM on the non-target strand (C–C) was neither bound nor cleaved by Csa complex (C–C PAM, Figs. 5b, c). These results suggest that the Type I-A crRNP recognizes a double-stranded PAM entity for efficient DNA binding and cleavage.
Type I-A crRNP cleaves at multiple sites on target and non-target strands of DNA
We next carried out experiments aimed at gaining a more detailed understanding of how the Type I-A crRNP cleaves target DNA. Given that the Cas3′ subunit of the Type I-A crRNP is a predicted ATP-dependent helicase that likely influences the nuclease activity of the Cas3″ effector DNAse, we tested the effect of ATP on target DNA cleavage by the Type I-A crRNP (Fig. 6a). In the presence of ATP, DNA cleavages were observed at both the target and non-target strands of the DNA substrate (Fig. 6a–c). In contrast, in the absence of ATP, DNA cleavage was restricted to the non-target DNA strand and mapped to a few sites clustered centrally in the protospacer region predicted to become displaced dur- ing R-loop formation (Fig. 6a, c; ATP-independent cleavage sites are indicated by brackets). The Type I-A crRNP bound substrate DNA with similar efficiency with or without ATP (Fig. S3).
We compared ATP-dependent DNA cleavage patterns Type I-A crRNP-mediated for two different sized double- stranded DNA substrates (Figs. 6b, c). Substrate I is 67 bp in length while substrate II is 117 bp long. Both substrate DNAs contain the same PAM and protospacer but have varying 5′ and 3′ flanking sequences. For both DNA substrates, ATP-dependent cleavages on the non-target strand mapped approximately to the middle of the protospacer sequences (red sequences) as well as at sites upstream of the protospacer (Fig. 6b, c). Likewise, target strand cleavages mapped to sites within or upstream of the protospacer (Fig. 6b, c). Additionally, the cleavages map to different sequences in the protospacer region of substrates I and II, although they contain the same protospacer sequence. This indicates that the Cas3″-mediated cleavages are not sequence specific and are influenced by sequences flanking the protospacer, at least in vitro. Collectively, the DNA cleavage profiles indicate that initial cleavages occur within the protospacer and progress in a 3′–5′ direction for each DNA strand.
Csa proteins essential for crRNP formation
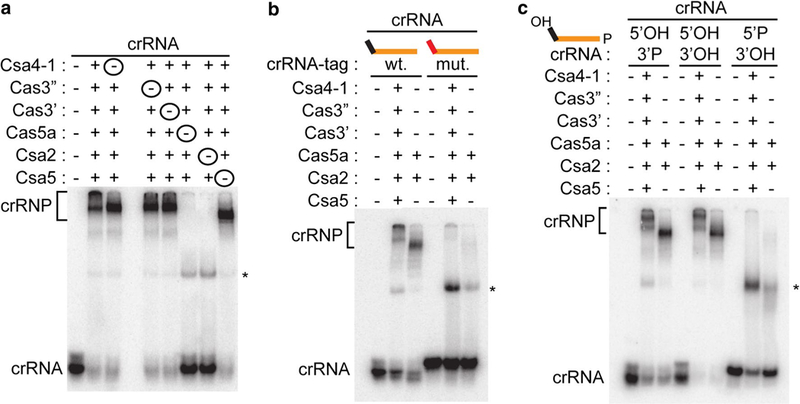
To understand the protein requirements for Type I-A crRNP assembly, we tested the ability of various combinations of Csa proteins to interact with 3′ radiolabeled crRNA using a native gel mobility shift approach (Fig. 7a). Stable, low mobility crRNPs were observed with the same six Csa Cas proteins that were required for DNA binding and cleavage (Fig. 2). While none of the individual Csa proteins bound crRNA, a minimal crRNP core subcomplex was formed with Cas5a and Csa2 (Fig. S4a). Low mobility crRNP complexes were also observed in the absence of either Csa 4–1, Cas3″, Cas3′, or Csa5, indicating the formation of specific crRNACsa protein subcomplexes. The low mobility crRNP complexes did not form if either Cas5a or Csa2 was omitted indicating that the Cas5a and Csa2 proteins are key core components for assembly of intact and functional Type I-A crRNPs (Fig. 7a).
The crRNA 5′ tag and 5′ OH are required for Type I-A crRNP assembly
Next, we examined features of the crRNA required for Type I-A crRNP assembly. During crRNA biogenesis in Pfu, Cas6 cleavage generates mature crRNAs with 5′ termini with an 8-nt CRISPR repeat-derived tag sequence (AUUGAAAG) and a 5′ hydroxyl (OH) end group (Carte et al. 2008, 2010). Previously, we found that the 5′ tag element and 5′ OH of crRNAs were both essential features for assembly of Type III-B (Cmr) and Type I-G (Cst) crRNPs in vitro (Hale et al. 2012; Majumdar et al. 2016). Similarly, we observed that the functional Type I-A crRNP (formed with six Csa proteins and capable of cleaving target DNA) as well as the minimal core crRNP subcomplex (formed with Cas5a and Csa2 but incapable of DNA cleavage) required that the crRNA contains a wild-type 5′ tag sequence (Fig. 7b) and 5′ OH (Fig. 7c). Altering the 3′ end chemical groups of crRNAs (from hydroxyl to phosphate) did not affect formation of either the core Cas5/Cas7 crRNP subcomplex or functional Type I-A crRNPs (Fig. 7c). Therefore, the Cas6-generated, 5′ tag sequence and 5′-OH of the crRNA are critical features for Type I-A crRNP formation.
Cas5a and Csa2 directly interact with 5′ tag and guide sequences of crRNA, respectively
To determine which proteins in Type I-A crRNP complex make direct contacts with crRNA, we performed UV- induced crosslinking of crRNPs assembled with Csa proteins, and internally radiolabeled and 4-thiouridine-containing crRNA (Fig. 8). This approach enables detection of proteins that interact directly with crRNA since UV light covalently crosslinks the 32P-labeled crRNA to the interacting protein(s) which are detected by autoradiography following RNase digestion and SDS-PAGE (Hale et al. 2014; Majumdar et al. 2016). SDS-PAGE and Coomassie Blue (G250) staining was performed to verify the presence of each protein in the UV crosslinking reactions. To visualize interactions at specific crRNA regions (5′ tag or guide), we selectively (32P) radiolabeled the desired region and examined whether the tested proteins became radiolabeled after UV light-induced crRNA–protein crosslinking.
We found that Cas5a, Csa2, and Csa4–1 proteins directly contact crRNA but that Cas3″, Cas3′ or Csa5 do not under the experimental conditions tested (Fig. 8). Cas5a interacts with the 5′ tag and Csa2 and Csa4–1 interact with the guide region of crRNA. Native gel mobility shift assays confirmed that the Cas5a/Csa2 minimal core crRNP as well as the full crRNP (formed by Csa4–1, Cas3″, Cas3′, Cas5a, Csa2 and Csa5 proteins) were formed under the conditions used for RNA-protein UV crosslinking (Fig. S4b).
Discussion
Three significant features distinguish Type I-A (Csa) crRNPs from the many other Type I (I-B, I-C, I-D, I-E, I-F, I-G and I-U) crRNPs (Makarova et al. 2015). First, Type I-A CRISPR systems are highly enriched in archaeal organ- isms, whereas other Type I CRISPR systems are present in both archaea and bacteria (Makarova et al. 2011, 2015). Second, while all Type I systems apparently utilize Cas3 single-stranded DNA nucleases and helicase (DNA unwind- ing) activities for crRNA-guided target DNA destruction, Type I-A systems employ two polypeptides (with separate nuclease (Cas3″) and a helicase (Cas3′) activities), whereas other Type I systems rely on a single Cas3 protein with nuclease and helicase domains. Third, as we show here for Pyrococcus furiousus (Fig. 2 and Majumdar et al. 2015) and as was previously observed in Sulfolobus islandicus (Peng et al. 2013) and Thermoproteus tenax (Plagens et al. 2012, 2014), the Cas3′ and Cas3″ polypeptides of differing Type I-A systems are integral structural components of the effec- tor crRNPs that target destruction of invader DNA. In con- trast, the single Cas3 subunit of other systems acts in trans with the effector crRNP and only becomes recruited and activated for function upon crRNP-target DNA interaction and crRNA-induced R-loop formation of the target DNA (Elmore et al. 2015; Hayes et al. 2016; Jackson et al. 2014; Mulepati et al. 2014; van Erp et al. 2015, 2018; Wang et al. 2016; Xiao et al. 2018; Zhao et al. 2014). In this study, we reconstituted functional Pfu Type I-A crRNPs that cleave double-stranded DNA targets in a PAM (5′-GGG/CCN-3′) and crRNA-dependent manner (Figs. 4, 5). Furthermore, we delineated key crRNA and Cas protein features contributing to Type I-A crRNP assembly and target DNA recognition and cleavage (Figs. 2, 7, 8).
Assembly and architecture of Type I-A crRNPs
We found that six of the eight total Csa proteins encoded at the Pfu Cas protein locus 2 (Fig. 1a), together with a crRNA, were essential for in vitro reconstitution of Type I-A crRNPs capable of specific target DNA recognition and cleavage (Fig. 2, S1b). These proteins include: Csa4–1 (large subunit, Cas8a2 superfamily member), Cas3″ (nuclease), Cas3′ (helicase), Cas5a (Cas5 superfamily member), Csa2 (Cas7 superfamily member) and Csa5 (small subunit, Cas11 superfamily member). [Superfamily Cas nomenclature is according to (Makarova et al. 2015 #136)]. All six Csa proteins found to be required for in vitro activity were also identified as components of the isolated, native Pfu Type I-A crRNPs (Majumdar et al. 2015). While Pfu native complexes contained both Csa4–1 and Csa4–2 (related Cas8a2 proteins), only the Csa4–1 protein was found to be necessary for in vitro activity (Figs. 2, S1b). Typically, Type I-A systems encode just one Csa4 (Cas8a2) subunit and it is unclear if the second Pfu Csa4 protein (Csa4–2) normally plays redundant or distinct roles in the cell or if Csa4–2 is non-functional as suggested by our in vitro results where it could not compensate for loss of Csa4–1 function (data not shown). The Csa3 protein was neither found in native Pfu Type I-A crRNPs (Majumdar et al. 2015) nor was this protein needed for in vitro activity (Fig. S1b). Indeed, there is evidence that Csa3 functions instead to regulate csa gene expression in Sulfolobus (He et al. 2016).
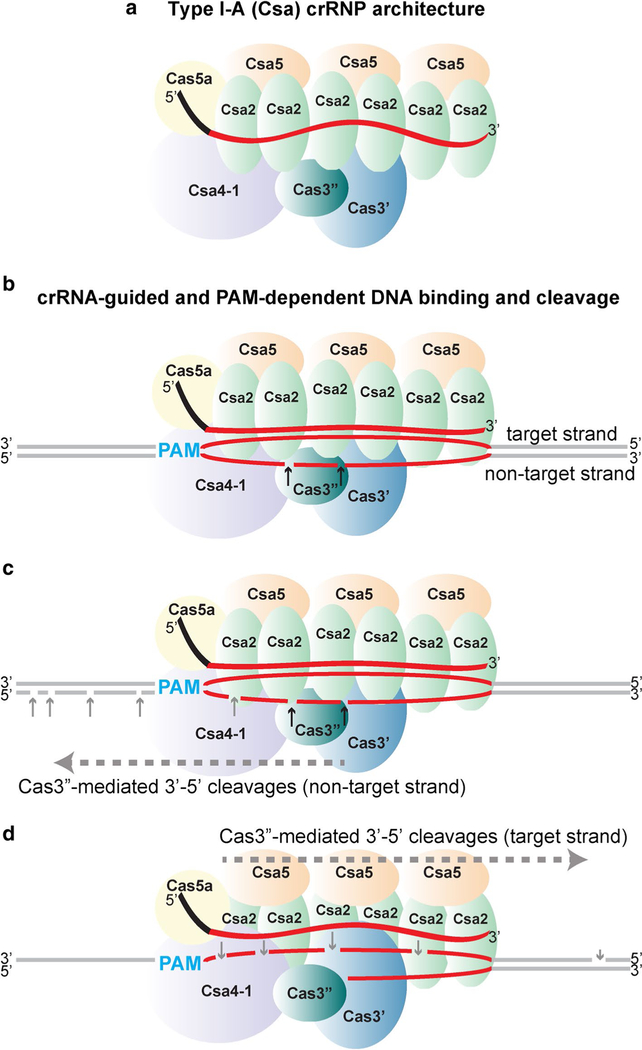
We propose a model for the overall subunit organization of the Pfu Type I-A crRNP (Fig. 9a). This model is based on our specific findings from native gel electrophoresis, UV- induced crRNA-protein crosslinking, and protein–protein interaction analyses (Figs. 7, 8, S5) as well as from principles gained from previous structural studies revealing that the basic organization of protein subunits for diverse Type I and Type III crRNPs is similar (Jackson et al. 2014; Jackson and Wiedenheft 2015; Mulepati et al. 2014; Rouillon et al. 2013; Spilman et al. 2013; Staals et al. 2014; Xiao et al. 2018; Zhao et al. 2014). Similar to other Type I and Type III crRNPs, gel mobility shift analysis indicates that the struc- tural core of the Type I-A crRNP is formed via interactions between Cas5a (Cas5), Csa2 (Cas7) and crRNA (Fig. 7). UV crosslinking revealed that Cas5a (Cas5) directly contacts the crRNA 5′ tag and that Csa2 (Cas7) binds the guide portion of the crRNA (Fig. 8). We show that the nucleotide identity of the 5′ tag and 5′ OH terminus of crRNA are each essential crRNA features for assembly of both the Cas5a/Csa2 core subcomplex as well as the complete (six protein) Type I-A crRNP (Fig. 7b, c). Consistent with structures of Type I-C, Type I-E (Cse), Type III-A (Csm) and Type III-B (Cmr) sys- tems (Hochstrasser et al. 2016; Jackson et al. 2014; Mulepati et al. 2014; Rouillon et al. 2013; Spilman et al. 2013; Staals et al. 2013; Wiedenheft et al. 2011; Xiao et al. 2018; Zhao et al. 2014), we expect that multiple Csa2 (Cas7) proteins bind along length of crRNA guide and 5′ end is capped by binding of a single Cas5a (Cas5) to the repeat-derived 5′ tag (Fig. 9a).
Fig. 9.
Model of Type I-A (Csa) crRNP assembly, DNA recognition and destruction. a Proposed subunit organization for the crRNP. Individual Csa proteins and their positions within the fuctional crRNP are indicated. b DNA recognition, binding and ATP-independent cleavage. Csa4–1 recognizes and interacts with PAM (cyan) enabling the guide sequence of crRNA to base pair with complementary protospacer sequence (red) of target strand of double-stranded DNA, displacing the non-target strand and forming R-loop for- mation. Cas3″ nuclease makes a few nicks in the protospacer region of the displaced non- target strand (black arrows). c, d ATP-dependent, 3′–5′ progressive cleavage in non- target strand and target strands, respectively. Utilizing ATP, the Cas3′ helicase catalyzes unwinding of double-stranded protospacer DNA and Cas3″ nuclease cleaves (gray arrows) progressively in 3′–5′ direction (denoted by dashed gray arrow and labeled
Regarding the relative positions of the other four Csa proteins in the context of the assembled Type I-A crRNP (Fig. 9a), we anticipate that multiple Csa5 (Cas11) proteins join the complex through protein–protein interactions with Csa2 (Cas7) given that Cas11/Cas7 superfamily protein interactions are observed in other characterized Type I and III crRNPs (Jackson et al. 2014; Mulepati et al. 2014; Reeks et al. 2013; Spilman et al. 2013; Staals et al. 2013; Wiedenheft et al. 2011; Xiao et al. 2018; Zhao et al. 2014). Our model places Cas4–1 (Cas8) in the vicinity where the 5′ end of the crRNA is located (Fig. 9) and our UV crosslinking data suggests that Cas4–1 directly contacts the guide RNA at this location (Fig. 8). This positioning is in agreement with the location of other Cas8 orthologs for structurally characterized Type I crRNPs (Hochstrasser et al. 2014; Mulepati et al. 2014; Sashital et al. 2012; Xiao et al. 2018). Finally, the spatial assignment for Cas3′’ (HD nuclease) and Cas3′ (helicase) subunits is in part based on our observed protein–protein interactions between Cas3″, Cas3′ and Cas4–1 (Fig. S5). Furthermore, our model accounts for positioning Cas3″ to the vicinity where initial (ATP- and helicase- independent) cleavages of the non-target DNA strand were observed (Figs. 6, 9b).
DNA binding and cleavage by Type I-A crRNPs
We have determined that the reconstituted Type I-A crRNPs function through PAM- and crRNA-dependent recognition of double-stranded target DNA (Fig. 4), followed by Cas3″ (HD domain) nuclease-mediated, single-strand nicking of non-target and target DNA strands (Figs. 3c, S2b). Based on our results (Figs. 2, 3, 4, 5, 6), and those with distantly related Type I crRNPs, we pro- pose a multi-step model to explain selective recognition and destruction of target DNA by the Type I-A crRNP (Fig. 9b, c, d).
Because Cas3″ (and Cas3 orthologs of other Type I sys- tems) cuts single-stranded DNA rather than double-stranded DNA (Fig. S2b, and Beloglazova et al. 2011; Huo et al. 2014; Sinkunas et al. 2011), an important role of the Type I-A crRNP is to remodel double-stranded target DNA to expose single-strands for Cas3″-mediated cleavage (Fig. 9b). This strand separation step is likely accomplished first by protein-mediated PAM identification mediated by Cas4–1 (Cas8). In other Type I systems, Cas8-PAM interactions trigger local DNA bending or unwinding that triggers crRNA- guided, directional dsDNA unwinding and R-loop formation (Hayes et al. 2016; Hochstrasser et al. 2014; Szczelkun et al. 2014; Xiao et al. 2018) (Fig. 9b). Full R-loop formation generates a crRNA-target DNA strand heteroduplex and an exposed single-stranded, non-target DNA strand that Cas3″ specifically recognizes for initial endonucleolytic cleavage (Fig. 9b).
Our PAM mutational analyses and crRNP/DNA-binding analyses (Fig. 5) suggests a preferential binding of the tar- get strand PAM (5′-GGG’−3′) over non-target strand PAM (5-CCC-3′). However, cleavage activity of the Type I-A crRNP required that the correct sequences be present on both strands of the PAM. These observations are consistent with insights from high-resolution structures of various Type I-E and I-F systems that reveal Cas8 proteins preferentially recognize PAMs in a double-stranded form through major and minor groove interactions (Hayes et al. 2016; Hochstrasser et al. 2014; Huo et al. 2014; Rollins et al. 2015). Other Type I systems rely more heavily of single-stranded (either target or non-target) recognition of PAMs, highlighting variability in mechanism of Cas8/PAM DNA recognition for different Type I crRNPs (Majumdar et al. 2016; Xiao et al. 2017).
We have gained insight into how Cas3″ destroys target DNA following Type I-A crRNP binding. As is the case for other Type I crRNPs which recruit Cas3 nucleases in trans (Hochstrasser et al. 2016; Szczelkun et al. 2014; van Erp et al. 2018; Xiao et al. 2018), we find a strict requirement for PAM recognition and crRNA annealing (R-loop formation) for the activity of Cas3″/Cas3′ DNase-helicase subunits that are integral components of Type I-A crRNPs. This suggests that Type I-A crRNP/DNA interactions and R-loop formation trigger conformational changes that selectively activate the crRNP-associated, Cas3″ nuclease and Cas3′ helicase subunits (Fig. 9b, c). Through mapping DNA cleavages in the absence and presence of ATP (Fig. 6), we view it likely that we have captured information about how Cas3″ cleaves DNA substrates in the absence or presence of contributions from the ATP-dependent Cas3′ helicase partner protein, respectively. Accordingly, in the absence of helicase activity (ATP omission) and likely reflecting the normal initial event, Cas3″ activity becomes constrained such that it carries out endonucleolytic nicks at a couple of closely spaced sites within a centrally located region of the exposed non-target DNA strand of the R-loop structure within the protospacer (Fig. 9b). In contrast, the number and locations of Cas3″-generated cleavages greatly expand when ATP is added to the reaction likely because the Cas3′ helicase activity is fueled by ATP to catalyze progressive 3′–5′ dsDNA unwinding (Fig. 9c, d). As the non-target DNA strand becomes progressively destroyed by processive unwinding and exo and/or endonucleolytic cleavage by Cas3″, this in turn results in progressive unwinding and destruction of the target strand. The end result is that both non-target and target strands become cleaved and destruction of DNA occurs well beyond the borders of the crRNA/ protospacer DNA interaction (Fig. 9c, d).
In summary, we have determined key crRNA features and essential Cas protein components governing assembly of functional Type I-A (Csa) crRNPs and gained important insight into the molecular mechanisms through which Type I-A crRNPs recognize and destroy DNA targets. Our findings provide the foundation for a deeper understanding of Type I-A crRNP DNA recognition and mechanism of action. Of particular value toward this goal would be to obtain highresolution structures of the reconstituted Type I-A crRNP in the presence and absence of bound target DNA. It will be particularly interesting to learn how the activities of the integral Cas3″ and Cas3′ effector nuclease and helicases of this Type I-A crRNP become controlled as the results of target DNA binding and to compare what is learned to that found for other Type I crRNP subtypes that recruit their Cas3 proteins in trans.
Supplementary Material
Acknowledgements
We thank Rebecca Terns for valuable mentorship, discussions and early contributions to the writing of this manuscript. We are also grateful to members of Terns’ lab for their technical input, Claiborne V. C. Glover III for critical reading of the manuscript, and Dr. Hong Li (Florida State University) for contributing purified Cas3″ and Cas3′ proteins shown in Fig. S5b. This work was supported by National Institutes of Health grant R35GM118160 to M.P.T.
References
- Barrangou R et al. (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- Beloglazova N, Petit P, Flick R, Brown G, Savchenko A, Yakunin AF (2011) Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J 30:4616– 4627. 10.1038/emboj.2011.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel J et al. (2014) A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (crispr)-derived rnas (crrnas) in Haloferax volcanii. J Biol Chem 289:7164–7177. 10.1074/jbc.M113.508184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ et al. (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964. 10.1126/science.1159689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady KC, O’Toole GA (2011) Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J Bacteriol 193:3433–3445. 10.1128/JB.01411-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP (2008) Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev 22:3489–3496. 10.1101/gad.1742908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Pfister NT, Compton MM, Terns RM, Terns MP (2010) Binding and cleavage of CRISPR RNA by Cas6. RNA 16:2181–2188. 10.1261/rna.2230110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H et al. (2008) Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bact 190:1390–1400. 10.1128/JB.01412-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore J, Deighan T, Westpheling J, Terns RM, Terns MP (2015) DNA targeting by the type I-G and type I-A CRISPR-Cas systems of Pyrococcus furiosus. Nucleic Acids Res 43:10353–10363. 10.1093/nar/gkv1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore JR, Sheppard NF, Ramia N, Deighan T, Li H, Terns RM, Terns MP (2016) Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR-Cas system. Genes Dev 30:447–459. 10.1101/gad.272153.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrella MA, Kuo FT, Bailey S (2016) RNA-activated DNA cleavage by the Type III-B CRISPR-Cas effector complex. Genes Dev 30:460–470. 10.1101/gad.273722.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE (2005) A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/ Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol 1:e60 10.1371/journal.pcbi.0010060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR et al. (2009) RNA-guided RNA cleavage by a CRISPR RNA- Cas protein complex. Cell 139:945–956. 10.1016/j.cell.2009.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR et al. (2012) Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell 45:292–302. 10.1016/j.molcel.2011.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Cocozaki A, Li H, Terns RM, Terns MP (2014) Target RNA capture and cleavage by the Cmr type III-B CRISPR-Cas effec- tor complex. Genes Dev 28:2432–2443. 10.1101/gad.250712.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes RP et al. (2016) Structural basis for promiscuous PAM recognition in type I-E cascade from E. Coli. Nature 530(7591):499–503. 10.1038/nature16995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Vestergaard G, Peng W, She Q, Peng X (2016) CRISPR-Cas type I-A Cascade complex couples viral infection surveillance to host transcriptional regulation in the dependence of Csa3b. Nucleic Acids Res 45(4):1902–1913. 10.1093/nar/gkw1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille F, Richter H, Wong SP, Bratovic M, Ressel S, Charpentier E (2018) The biology of CRISPR-Cas: backward and forward. Cell 172:1239–1259. 10.1016/j.cell.2017.11.032 [DOI] [PubMed] [Google Scholar]
- Hochstrasser ML, Taylor DW, Bhat P, Guegler CK, Sternberg SH, Nogales E, Doudna JA (2014) CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci USA 111:6618–6623. 10.1073/pnas.1405079111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser ML, Taylor DW, Kornfeld JE, Nogales E, Doudna JA (2016) DNA targeting by a minimal CRISPR RNA-guided cascade. Mol Cell 63:840–851. 10.1016/j.molcel.2016.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y et al. (2014) Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat Struct Mol Biol 21(9):771 10.1038/nsmb.2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Wiedenheft B (2015) A conserved structural chassis for mounting versatile CRISPR RNA-guided immune responses. Mol Cell 58:722–728. 10.1016/j.molcel.2015.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN et al. (2014) Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia Coli. Science 10.1126/science.1256328 [DOI] [PMC free article] [PubMed]
- Jiang F, Doudna JA (2015) The structural biology of CRISPR-Cas systems. Curr Opin Struct Biol 30:100–111. 10.1016/j.sbi.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F et al. (2016) Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351(6275):867–871. 10.1126/science.aad8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskiene M, Tamulaitis G, Kostiuk G, Venclovas C, Siksnys V (2016) Spatiotemporal control of Type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol Cell 62:295–306. 10.1016/j.molcel.2016.03.024 [DOI] [PubMed] [Google Scholar]
- Lintner NG et al. (2011a) The structure of the CRISPR-associated protein Csa3 provides insight into the regulation of the CRISPR/ Cas system. J Mol Biol 405:939–955. 10.1016/j.jmb.2010.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintner NG et al. (2011b) Structural and functional characterization of an archaeal clustered regularly interspaced short palindro- mic repeat (CRISPR)-associated complex for antiviral defense (CASCADE). J Biol Chem 286:21643–21656. 10.1074/jbc.M111.238485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar S et al. (2015) Three CRISPR-Cas immune effector complexes coexist in Pyrococcus furiosus. RNA 21:1147–1158. 10.1261/rna.049130.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar S, Ligon M, Skinner WC, Terns RM, Terns MP (2016) Target DNA recognition and cleavage by a reconstituted Type I-G CRISPR-Cas immune effector complex. Extremophiles: Life Under Extreme Cond 10.1007/s00792-016-0871-5 [DOI] [PMC free article] [PubMed]
- Makarova KS et al. (2011) Evolution and classification of the CRISPR- Cas systems nature reviews. Microbiology 9:467–477. 10.1038/nrmicro2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Anantharaman V, Grishin NV, Koonin EV, Aravind L (2014) CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front Genet 5:102 10.3389/fgene.2014.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS et al. (2015) An updated evolutionary classification of CRISPR-Cas systems nature reviews. Microbiology 13:722–736. 10.1038/nrmicro3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C (2009) Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155:733–740. 10.1099/mic.0.023960-0 [DOI] [PubMed] [Google Scholar]
- Mulepati S, Bailey S (2013) In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional ATP- dependent degradation of DNA target. J Biol Chem 288:22184– 22192. 10.1074/jbc.M113.472233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Heroux A, Bailey S (2014) Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science 10.1126/science.1256996 [DOI] [PMC free article] [PubMed]
- Nam KH, Haitjema C, Liu X, Ding F, Wang H, DeLisa MP, Ke A (2012) Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR-Cas system. Structure 20:1574–1584. 10.1016/j.str.2012.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell M (2018) Molecular Mechanisms of RNA-Targeting by Cas13-containing Type VI CRISPR-Cas Systems. J Mol Biol 10.1016/j.jmb.2018.06.029 [DOI] [PubMed]
- Pausch P, Muller-Esparza H, Gleditzsch D, Altegoer F, Randau L, Bange G (2017) Structural Variation of Type I-F CRISPR RNA Guided DNA Surveillance. Mol Cell 10.1016/j.molcel.2017.06.036 [DOI] [PubMed]
- Peng W, Li H, Hallstrom S, Peng N, Liang YX, She Q (2013) Genetic determinants of PAM-dependent DNA targeting and pre-crRNA processing in Sulfolobus islandicus. RNA Biol 10:738–748. 10.4161/rna.23798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagens A, Tjaden B, Hagemann A, Randau L, Hensel R (2012) Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J Bacteriol 194:2491–2500. 10.1128/JB.00206-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagens A et al. (2014) In vitro assembly and activity of an archaeal CRISPR-Cas type I-A cascade interference complex. Nucleic Acids Res 10.1093/nar/gku120 [DOI] [PMC free article] [PubMed]
- Reeks J, Graham S, Anderson L, Liu H, White MF, Naismith JH (2013) Structure of the archaeal Cascade subunit Csa5: relating the small subunits of CRISPR effector complexes. RNA Biol 10:762–769. 10.4161/rna.23854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins MF, Schuman JT, Paulus K, Bukhari HS, Wiedenheft B (2015) Mechanism of foreign DNA recognition by a CRISPR RNA- guided surveillance complex from Pseudomonas aeruginosa. Nucleic Acids Res 43:2216–2222. 10.1093/nar/gkv094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon C et al. (2013) Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol Cell 52:124–134. 10.1016/j.molcel.2013.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Wiedenheft B, Doudna JA (2012) Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell 46:606–615. 10.1016/j.molcel.2012.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SA, Erdmann S, Mojica FJ, Garrett RA (2013) Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol 10:891–899. 10.4161/rna.23764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiimori M, Garrett SC, Chambers DP, Glover CVC 3rd, Graveley BR, Terns MP (2017) Role of free DNA ends and protospacer adjacent motifs for CRISPR DNA uptake in Pyrococcus furiosus. Nucleic Acids Res 45:11281–11294. 10.1093/nar/gkx839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiimori M, Garrett SC, Graveley BR, Terns MP (2018) Cas4 nucleases define the PAM length, and orientation of dna fragments integrated at CRISPR loci. Mol cell 70(814–824):e816 10.1016/j.molcel.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V (2011) Cas3 is a single-stranded DNA nuclease and ATP- dependent helicase in the CRISPR/Cas immune system. EMBO J 30:1335–1342. 10.1038/emboj.2011.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, Horvath P, Siksnys V (2013) In vitro reconstitution of Cascademediated CRISPR immunity in Streptococcus thermophilus. EMBO J 32:385–394. 10.1038/emboj.2012.352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilman M et al. (2013) Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol Cell 52:146–152. 10.1016/j.molcel.2013.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH et al. (2013) Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell 52:135–145. 10.1016/j.molcel.2013.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH et al. (2014) RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell 56:518–530. 10.1016/j.molcel.2014.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA (2014) DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507(7490):62–67. 10.1038/nature13011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczelkun MD et al. (2014) Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci USA 111(27):9798–9803. 10.1073/pnas.1402597111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns MP, Terns RM (2011) CRISPR-based adaptive immune systems. Curr Opin Microbiol 14:321–327. 10.1016/j.mib.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns RM, Terns MP (2013) The RNA- and DNA-targeting CRISPR- Cas immune systems of Pyrococcus furiosus. Biochem Soc Trans 41:1416–1421. 10.1042/BST20130056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duijn E et al. (2012) Native tandem and ion mobility mass spec- trometry highlight structural and modular similarities in clusteredregularly-interspaced shot-palindromic-repeats (CRISPR)-associ- ated protein complexes from Escherichia coli and Pseudomonas aeruginosa. Mol Cell proteom MCP 112:1430–1441. 10.1074/mcp.M112.020263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp PB, Jackson RN, Carter J, Golden SM, Bailey S, Wiedenheft B (2015) Mechanism of CRISPR-RNA guided recognition of DNA targets in Escherichia coli. Nucleic Acids Res 43:8381–8391. 10.1093/nar/gkv793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp PBG et al. (2018) Conformational Dynamics of DNA binding and Cas3 recruitment by the CRISPR RNA-guided cascade complex. ACS Chem Biol 13:481–490. 10.1021/acschembio.7b00649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard G, Garrett RA, Shah SA (2014) CRISPR adaptive immune systems of archaea. RNA Biol 11:156–167. 10.4161/rna.27990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li J, Zhao H, Sheng G, Wang M, Yin M, Wang Y (2015) Structural and mechanistic basis of PAM-dependent spacer acqui- sition in CRISPR-Cas systems. Cell 163:840–853. 10.1016/j.cell.2015.10.008 [DOI] [PubMed] [Google Scholar]
- Wang X et al. (2016) Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3. Nat Struct Mol Biol 23:868–870. 10.1038/nsmb.3269 [DOI] [PubMed] [Google Scholar]
- Westra ER et al. (2012) CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell 46:595–605. 10.1016/j.molcel.2012.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B et al. (2011) Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477:486–489. 10.1038/nature10402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y et al. (2017) Structure basis for directional r-loop formation and substrate handover mechanisms in Type I CRISPR-Cas system. Cell 170(48–60):e11 10.1016/j.cell.2017.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Luo M, Dolan AE, Liao M, Ke A (2018) Structure basis for RNA-guided DNA degradation by Cascade and Cas3. Science 10.1126/science.aat0839 [DOI] [PMC free article] [PubMed]
- Zhao H et al. (2014) Crystal structure of the RNA-guided immune surveillance cascade complex in Escherichia coli. Nature 10.1038/nature13733 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.