Abstract
Significance: Proline metabolism has complex roles in a variety of biological processes, including cell signaling, stress protection, and energy production. Proline also contributes to the pathogenesis of various disease-causing organisms. Understanding the mechanisms of how pathogens utilize proline is important for developing new strategies against infectious diseases.
Recent Advances: The ability of pathogens to acquire amino acids is critical during infection. Besides protein biosynthesis, some amino acids, such as proline, serve as a carbon, nitrogen, or energy source in bacterial and protozoa pathogens. The role of proline during infection depends on the physiology of the host/pathogen interactions. Some pathogens rely on proline as a critical respiratory substrate, whereas others exploit proline for stress protection.
Critical Issues: Disruption of proline metabolism and uptake has been shown to significantly attenuate virulence of certain pathogens, whereas in other pathogens the importance of proline during infection is not known. Inhibiting proline metabolism and transport may be a useful therapeutic strategy against some pathogens. Developing specific inhibitors to avoid off-target effects in the host, however, will be challenging. Also, potential treatments that target proline metabolism should consider the impact on intracellular levels of Δ1-pyrroline-5-carboxylate, a metabolite intermediate that can have opposing effects on pathogenesis.
Future Directions: Further characterization of how proline metabolism is regulated during infection would provide new insights into the role of proline in pathogenesis. Biochemical and structural characterization of proline metabolic enzymes from different pathogens could lead to new tools for exploring proline metabolism during infection and possibly new therapeutic compounds.
Keywords: proline, pathogenesis, metabolism, PutA, stress
Introduction
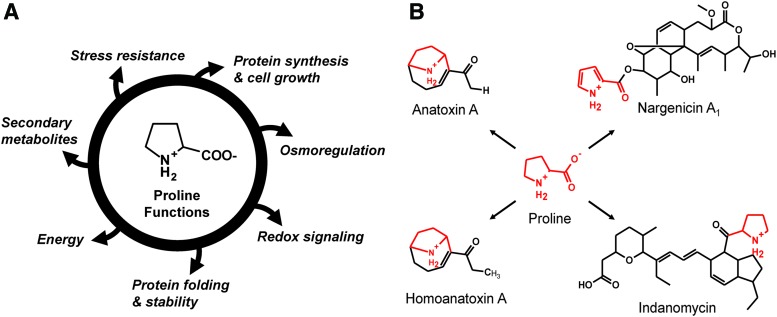
The roles of proline in biology are complex and impact a wide range of cellular processes (Fig. 1). While proline is critical for protein synthesis and cell growth, it also has roles in osmoregulation (29, 53, 106, 155, 247), redox signaling (65, 134, 182, 256, 257), unfolded protein responses (129), protein stability (44, 97, 116, 129, 130, 178), cellular bioenergetics (35, 166, 181, 182, 226), and stress resistance (18, 111, 130, 182, 222, 258) (Fig. 1A). In addition, some organisms use proline directly for the biosynthesis of secondary metabolites with antibacterial or antifungal properties (Fig. 1B) (50, 79, 142, 148, 175, 197, 228).
FIG. 1.
Intracellular roles of proline. (A) Potential biological functions of proline. (B) Examples of proline being directly incorporated into the biosynthesis of antimicrobial compounds. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Proline accumulation and its oxidative metabolism are major mechanisms by which proline affords protective benefits to organisms (130). The properties of proline as an organic osmolyte provide cellular protection against abiotic stress such as drought and osmotic shock (53, 222). The oxidative metabolism of proline enables cells to utilize proline as a carbon and nitrogen source, helping support growth and energy demands (2, 226, 233). Cells must appropriately manage proline levels to support protein biosynthesis and respond to stress and energy needs. Cellular processes such as import, catabolism, biosynthesis, and amino acid recycling coordinate proline flux for optimal growth and stress response. For further details concerning potential mechanisms of proline stress protection see reviews by Liang et al. (130) and Szabados and Savouré (222).
An interesting aspect of proline is its manipulation by pathogenic organisms in response to changes in the environment. There are a growing number of examples of proline facilitating the pathogenicity of different disease-causing organisms. Here, we review the role of proline metabolism in several bacterial and eukaryotic pathogens. Some of the bacterial pathogens highlighted in this review have high priority status for new antibiotics by the World Health Organization such as Helicobacter pylori (clarithromycin resistant), Staphylococcus aureus (methicillin resistant, vancomycin intermediate and resistant), and Mycobacterium tuberculosis. We also examine proline metabolism in entomopathogenic bacteria, which enable soil-dwelling nematodes to infect and multiply within insect larvae. Unfortunately, we were not able review pathogenesis in plants but interested readers are referred to some excellent reviews by Qamar et al. (185), Rizzi et al. (194), and Szabados and Savouré (222). From the literature it is evident that proline affords different benefits to pathogenic organisms according to the pathophysiological environment.
Proline Metabolism
Enzymes of proline metabolism
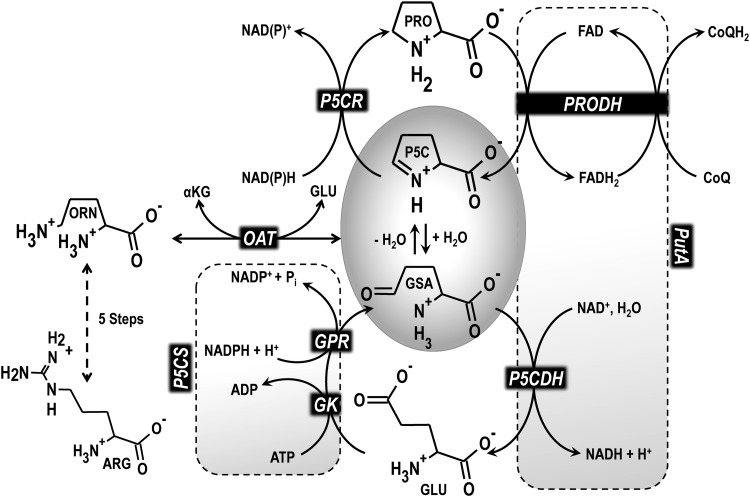
Proline is synthesized from glutamate in a series of steps (Fig. 2) (11, 54, 74). The enzyme γ-glutamyl kinase (GK; EC 2.7.2.11) converts glutamate to γ-glutamyl phosphate, which is next reduced to glutamate-γ-semialdehyde (GSA) by γ-glutamyl phosphate reductase (GPR; EC 1.2.1.41) (54, 74). GSA then cyclizes to form Δ1-pyrroline-5-carboxylate (P5C). In higher organisms such as plants and animals, GK and GPR activities are combined into a bifunctional enzyme known as P5C synthetase (P5CS) (54, 74). In the next step, P5C is reduced to proline by the enzyme P5C reductase (P5CR; EC 1.5.1.2), which utilizes nicotinamide adenine dinucleotide (NADH) and/or nicotinamide adenine dinucleotide phosphate (NADPH) (45, 74). Proline biosynthesis from glutamate is subject to proline feedback inhibition of GK/P5CS thus limiting the intracellular pool of synthesized proline (74, 213, 223).
FIG. 2.
Proline metabolic reactions. Proline biosynthesis begins with phosphorylation of glutamate by GK to form γ-glutamyl phosphate, which is then reduced to GSA by GPR. In higher organisms such as animals and plants, these two steps are catalyzed by the bifunctional enzyme P5CS. GSA cyclizes to P5C followed by NAD(P)H-dependent reduction to proline catalyzed by P5CR. P5C can also be generated from ornithine via OAT. Proline catabolism involves the two-step oxidation of proline to glutamate catalyzed by PRODH and P5CDH. In gram-negative bacteria, the PRODH and P5CDH activities are linked in the PutA bifunctional enzyme. PRODH is a flavin-dependent enzyme that couples the two-electron oxidation of proline to reduction of an electron acceptor in the membrane such as ubiquinone, and thus, proline catabolism feeds reducing equivalents into the respiratory chain. P5CDH pairs the oxidation of GSA to glutamate with reduction of NAD+ to NADH. GK, γ-glutamyl kinase; GPR, γ-glutamyl phosphate reductase; GSA, glutamate-γ-semialdehyde; NAD+, nicotinamide adenine dinucleotide; NADH, nicotinamide adenine dinucleotide; NAD(P)H, reduced nicotinamide adenine dinucleotide (phosphate); OAT, ornithine aminotransferase; P5C, pyrroline-5-carboxylate; P5CDH, P5C dehydrogenase; P5CR, P5C reductase; P5CS, P5C synthetase; PRODH, proline dehydrogenase; PutA, proline utilization A.
P5C can also be provided from ornithine via ornithine aminotransferase (OAT; EC 2.6.1.13), thereby linking proline metabolism to arginine (54, 74). In bacteria, such as M. tuberculosis, arginine serves as a key nitrogen and carbon source for ornithine, proline, and glutamate (91). In organisms that lack GK/GPR, such as Trypanosoma brucei and S. aureus, proline must be scavenged or synthesized via ornithine.
The catabolism of proline to glutamate relies on two enzymatic steps (Fig. 2) (2, 226). The first step involves proline dehydrogenase (PRODH; EC 1.5.99.8), a flavoenzyme that catalyzes the oxidation of proline to P5C with concomitant two-electron reduction of a noncovalently bound flavin adenine dinucleotide (FAD) cofactor (15, 110, 124, 136, 243). This is known as the PRODH reductive half-reaction. The PRODH oxidative half-reaction involves the transfer of electrons from reduced FAD to ubiquinone in the plasma membrane in prokaryotes and inner mitochondrial membrane in eukaryotes, thus linking proline catabolism with the respiratory chain (156, 158, 238). P5C undergoes a nonenzymatic hydrolysis to form GSA, which is then oxidized to glutamate by the nicotinamide adenine dinucleotide (NAD+)-dependent enzyme P5C dehydrogenase (P5CDH; EC 1.5.1.12) (139, 177, 216). P5CDH is also known as l-glutamate-γ-semialdehyde dehydrogenase (GSALDH) (138). Glutamate dehydrogenase or amino acid transaminase converts glutamate to α-ketoglutarate, which feeds into the tricarboxylic acid (TCA) cycle (2, 35).
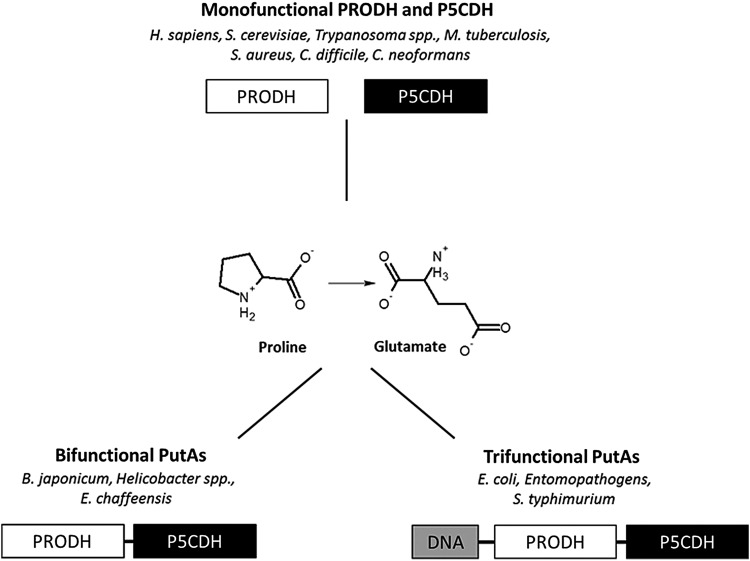
The overall conversion of proline to glutamate by PRODH and P5CDH is conserved across the domains of life, but there are significant differences in the arrangement of enzymes (Fig. 3) (226). In eukaryotes and gram-positive bacteria, the PRODH and P5CDH enzymes are expressed as two discrete monofunctional enzymes, whereas in gram-negative bacteria, the two enzymes are fused onto a single polypeptide called proline utilization A (PutA) (150, 226). Within the PutA family, there are PutAs that have an N-terminal ribbon-helix-helix (RHH) DNA binding domain known as trifunctional PutAs (9, 89, 120, 211, 226). Bacteria with bifunctional PutAs include Helicobacter spp., while those having trifunctional PutAs include Escherichia coli and Salmonella typhimurium (110, 226). Structural details concerning the classification of PutAs can be found in the work by Tanner and coworkers (109, 137, 138, 212, 226).
FIG. 3.
Domain arrangement of proline catabolic enzymes in different organisms. The catabolism of proline to form glutamate is catalyzed by separate PRODH and P5CDH enzymes in eukaryotes and gram-positive bacteria such as Trypanosoma spp., Mycobacterium tuberculosis, Staphylococcus aureus, Clostridium difficile, and Cryptococcus neoformans. In gram-negative bacteria, PRODH and P5CDH are fused onto a single polypeptide called PutA. PutA enzymes can be classified as bifunctional, as in Helicobacter spp. and Ehrlichia chaffeensis or as trifunctional, depending on the presence of an N-terminal DNA binding domain such as in entomopathogenic bacteria, Salmonella typhimurium, and Escherichia coli.
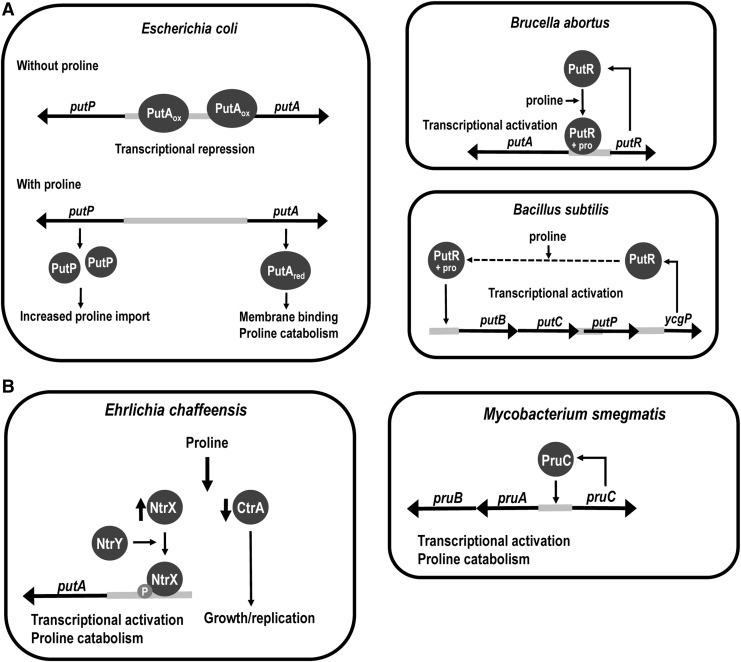
Trifunctional PutAs function as a transcriptional repressor of the put regulon that encodes PutP, a high-affinity proline transporter, and PutA (150, 249, 263). The putP and putA genes are transcribed in opposite directions and are regulated by an intervening put control DNA region (Fig. 4) (87, 150, 264). Characterization of PutA from E. coli has shown that in the absence of proline, PutA binds to specific sites in the put control DNA region, thereby repressing transcription of the put genes (33, 83, 150, 263). In the presence of proline, PutA binds to the membrane rather than DNA, leading to upregulation of put gene expression (32, 172). The mechanism by which PutA regulates put gene expression is dependent on the redox state of the FAD cofactor. When the FAD cofactor is oxidized, that is, in the absence of proline, PutA binds put control DNA. In the presence of proline, the FAD cofactor is reduced, causing a conformational change in PutA that induces tight membrane binding, thereby switching PutA from a DNA binding protein to a membrane-bound proline catabolic enzyme (Fig. 4) (16, 217, 259, 260, 265, 267). To date, no evidence for PutA regulating other genes besides putA and putP has been reported.
FIG. 4.
Regulation of proline metabolism in different organisms. (A) E. coli: trifunctional PutA represses transcription of the putA and putP genes in the absence of proline. When proline becomes available, the FAD cofactor of PutA is reduced and PutA switches to a membrane-bound proline catabolic enzyme thereby relieving repression of the put genes. Brucella abortus: the PutR transcription factor (Lrp-type) is transcribed by the putR gene. PutR DNA binding is activated by proline leading to transcriptional activation of the putA gene. Bacillus subtilis: the PutR transcription factor (PucR-type) is transcribed by the ycgP gene. PutR DNA binding is activated by proline leading to transcriptional activation of putB (PRODH), putC (P5CDH), and putP genes. (B) E. chaffeensis: proline induces increased levels of the transcriptional regulator, NtrX, which on phosphorylation by NtrY binds to the putA promoter region and activates PutA expression. Mycobacterium smegmatis: PruC is transcribed divergently from the pruAB operon. Increased levels of PruC lead to higher expression of pruA (P5CDH) and pruB (PRODH). FAD, flavin adenine dinucleotide; Lrp, leucine-responsive regulatory protein.
Substrate channeling
An important metabolic aspect of PutA enzymes, regardless of being classified as bifunctional or trifunctional, is the channeling of the P5C intermediate between the PRODH and P5CDH active sites. Structural and kinetic data show evidence of substrate channeling for PutAs from five different gram-negative bacteria, such as Bradyrhizobium japonicum and E. coli, suggesting that substrate channeling is a conserved mechanism in the PutA family (10, 138, 157, 210, 211). Evidence for substrate channeling between separate monofunctional PRODH and P5CDH enzymes has been shown for PRODH and P5CDH from the gram-positive bacterium Thermus thermophilus (200). Whether PRODH and P5CDH enzymes in eukaryotes also channel P5C/GSA remains an open question. The benefit of channeling P5C in bacteria is largely to avoid futile proline-P5C cycling and to prevent the buildup of P5C/GSA, which has been shown to form unwanted adducts with other metabolites and exhibit toxic effects (11). These unwanted adducts include pyridoxal phosphate-P5C and acetoacetic acid-P5C, which have been found in human patients (237). P5C/GSA was recently classified as a “Top 30” damage-prone metabolite in a study analyzing the tendency of endogenous metabolites to undergo spontaneous chemical side reactions (125).
Proline and reactive oxygen species
Numerous studies have documented the role of proline catabolism in the protection of cells from oxidative stress and, somewhat counterintuitively, the generation of reactive oxygen species (ROS) (18, 110, 170, 182, 226). PRODH couples proline oxidation with reduction of the ubiquinone pool in the membrane, which can lead to ROS formation and impact intracellular signaling pathways. Proline protection against oxidative stress in mammalian cells was shown to rely on PRODH and downregulation of the forkhead transcription factor class O3 pathway via Akt (166). In Caenorhabditis elegans, PRODH was shown to upregulate the human homologue of Nrf-2, leading to increased antioxidant enzyme levels and life span of the worm (256).
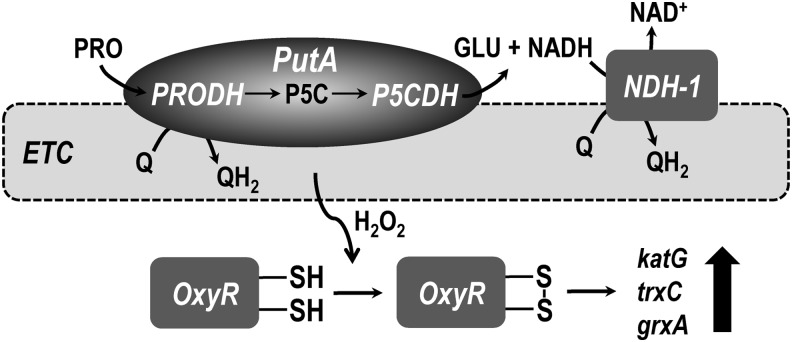
In bacteria, proline oxidative metabolism by PutA has been shown to be important for oxidative stress protection via activation of the OxyR regulon (Fig. 5). Hydrogen peroxide formed by proline catabolism increases important antioxidant enzymes of the OxyR regulon such as catalase, thioredoxin, and glutaredoxin (257). Hydrogen peroxide was hypothesized to be a conserved metabolic by-product of proline catabolism due to proline oxidation being coupled with reduction of respiratory systems in different organisms (257). By increasing antioxidant defenses, proline may provide a preadaptive advantage, which in the context of pathogenesis would prolong cell survival, increase colonization efficiencies, and enhance virulence (134).
FIG. 5.
Production of hydrogen peroxide from proline metabolism in E. coli. Oxidative proline metabolism by PutA leads to reduction of membrane-bound ubiquinone (Q) and formation of NADH, supplying electrons into the electron transport chain via NADH dehydrogenase (NDH-1). Hydrogen peroxide is generated as a by-product during respiratory chain activity to levels sufficient enough to activate the transcriptional regulator OxyR (-S-S-). OxyR subsequently upregulates expression of antioxidant genes such as thioredoxin (trxC), glutaredoxin (grxA), and catalase (katG).
Regulation of proline catabolic genes
Proline utilization in bacteria is dependent on cellular energy status and the nutritional environment. Upregulation of proline catabolic genes relies not only on proline responsive transcription factors but also on global regulatory systems such as carbon catabolite repression (CCR) and nitrogen catabolite repression (NCR). Thus, the full utilization of proline requires the coordination of at least three different regulatory systems depending on the nutrient conditions.
The mechanisms by which proline upregulates expression of proline catabolic genes involve relief of transcriptional repression or activation of transcription depending on the organism. In gram-negative bacteria that contain trifunctional PutA, PutA is an autotranscriptional repressor of the putA and putP genes as described above (Fig. 4). From cell-based transcription assays in E. coli, it appears that PutA more strongly represses the putA gene than putP (262). Therefore, it is likely that putP expression is weakly repressed by PutA, allowing for rapid proline uptake and activation of putA gene expression when extracellular proline becomes available.
In organisms that lack PutA as an autogenous transcriptional repressor (i.e., contain bifunctional PutA), transcription of proline catabolic genes is activated via proline-inducible transcription factors. In gram-negative Rhodobacter capsulatus (107), a transcriptional activator of the leucine-responsive regulatory protein (Lrp/AsnC) family called PutR was shown to activate expression of proline catabolic genes. PutR shares the conserved domains of the Lrp/AsnC family, which are an N-terminal helix-turn-helix DNA binding domain and a C-terminal α/β ligand binding domain (30, 227). PutR has also been demonstrated to activate proline catabolism in gram-negative pathogens, such as Vibrio vulnificus (123) and most recently the putA gene in Brucella abortus (37) (Fig. 4). In the pathogen Pseudomonas aeruginosa PAO1, PruR, a member of the AraC/XylS protein family, was found to be a transcriptional activator of put genes (161). An activator protein of proline catabolism in gram-positive Bacillus subtilis called PutR (ycgP gene) has also been described. PutR in B. subtilis is a PucR-like transcriptional regulator with a C-terminal helix-turn-helix DNA binding domain (17, 95, 155) (Fig. 4).
In yeast Saccharomyces cerevisiae, expression of proline utilization genes PUT1 (PRODH), PUT2 (P5CDH), and PUT4 (proline-specific permease) is activated by a large transcription factor called Put3p (979 amino acids), which contains an N-terminal Zn(II)2Cys6 binuclear cluster DNA binding domain and a C-terminal proline activation domain (126, 208, 220). Proline binding to Put3p induces a conformational change leading to activation of Put3p-DNA binding and expression of the PUT genes (126, 208).
In addition to proline-specific regulation, proline catabolic genes are subject to global carbon and nitrogen regulatory mechanisms. The CCR in bacteria enables pathogens to exploit available carbon sources and also has an important role in the expression of virulence genes. In E. coli, CCR does not inhibit proline metabolism but is required for full induction. The cyclic AMP receptor protein (CRP) functions as an activator by increasing putA and putP promoter activity in poor nutrient environments (41, 163). CRP has two predicted DNA binding sites in the put control region of E. coli and has been shown to bind to put control DNA in response to cAMP (262). In B. subtilis, global CCR is mediated by the catabolite control protein A (CcpA) transcription factor, which under rich glucose conditions, represses genes necessary for metabolism of secondary carbon sources (80, 215). CcpA repression is relieved in glucose-deplete conditions thereby switching on catabolism of alternative carbon sources such as proline (80, 215). Proline catabolic genes in the pathogen S. aureus are also regulated by CcpA (90, 171).
Another global regulator of proline catabolism in gram-positive bacteria is the transcriptional regulator CodY, which is activated by branched-chain amino acids and GTP (215). CodY represses metabolism of poor nutrient sources and regulates virulence genes thereby helping cells adapt to changes in nutrients and the environment (215). CodY repression of proline catabolism is best understood in B. subtilis (12, 17, 31, 153) and is likely a relevant mechanism in other bacteria such as S. aureus (239). Belitsky showed that CodY repression of the proline utilization operon in B. subtilis involves displacement of the proline-specific activator (PutR) from the promoter region (17). As nutrient availability decreases, CodY becomes inactive leading to transcriptional activation of the proline utilization operon (17).
In addition to carbon, cells are faced with decisions of which nitrogen sources to utilize. Ammonium and glutamine are the first choices of nitrogen, whereas proline is of lower preference (215, 245, 246). Proline catabolism is generally responsive to nitrogen status via the global regulatory mechanism known as NCR (19). The put genes in E. coli, Klebsiella aerogenes, and Pseudomonas putida have been shown to be upregulated when cells are grown in limiting nitrogen conditions (41, 235, 251). The nitrogen assimilation control (NAC) regulator mediates the upregulation of put genes (81, 140). NAC, a LysR-type transcription factor, activates σ70-dependent genes to generate ammonium and glutamate for glutamine synthesis (19). In K. aerogenes, putP expression is strongly activated by NAC, while putA expression seems to be activated indirectly by the resulting increase in proline uptake. The overall result of NAC-dependent regulation of put genes is the formation of glutamate for glutamine synthesis (19).
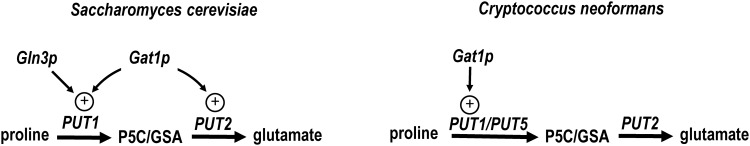
The regulation of proline catabolism by NCR has been studied in fungi as well. In S. cerevisiae, response to changes in nitrogen involves transcriptional regulators, GATA-type zinc finger proteins Gln3p and Gat1p (92) (Fig. 6). In nitrogen-poor medium, Gln3p activates expression of PUT1 and PUT4 (59, 61). Put4p is the yeast transporter specific for proline, but yeast also can import proline through the general amino acid permease (Gap1p) (92). Increased proline transport under nitrogen limitation leads to Put3p activation and further induction of the PUT genes (92). When yeast is grown on rich nitrogen medium, Ure2p interacts with Gln3p, which prevents nuclear localization of Gln3p and transcriptional activation of the PUT genes (114). Ure2p binding to Gln3p is dependent on TOR complex 1 phosphorylation of Gln3p, thus TORC1 has a primary role in the regulation of PUT genes by NCR (14, 126).
FIG. 6.
Regulation of proline catabolic genes in fungi by nitrogen metabolite repression. The GATA transcription factors Gln3p and Gat1p activate proline catabolism in Saccharomyces cerevisiae when grown in poor nitrogen conditions. Gln3p activates PUT1 (PRODH), while Gat1p activates both PUT1 and PUT2 (P5CDH). C. neoformans contains two PRODH genes, PUT1 and PUT5. PUT1 is activated by Gat1p, whereas PUT5 and PUT2 are not regulated by nitrogen metabolite repression.
Proline transport and accumulation
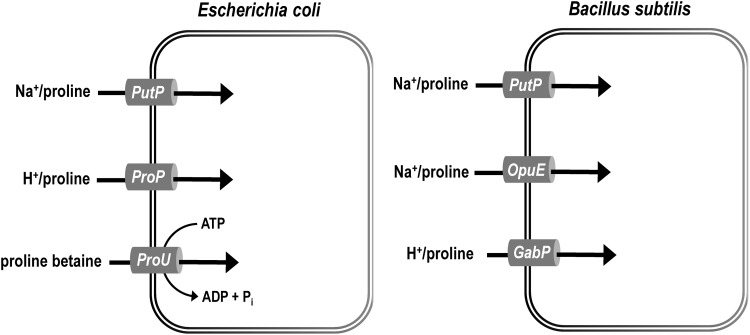
E. coli and Samonella enterica utilize three different transport systems for manipulating intracellular proline levels. These uptake systems include the PutP high-affinity proline/sodium symporter, the ProP proton/osmolyte symporter, and the ABC-type transporter ProU (Fig. 7). PutP displays high specificity and affinity for proline, whereas ProP and ProU exhibit transport activity with a broad range of osmoprotectant compounds (5, 57, 100, 159, 257). ProU comprised membrane-bound ATPase (ProV) and permease (ProW) proteins, and a periplasmic binding protein ProX, which binds glycine betaine and proline betaine (84, 204, 248, 250). Studies of osmoregulatory mechanisms in E. coli have provided a foundation for understanding general prokaryotic osmotic stress responses (5, 51). In response to high external osmotic pressure (e.g., increased NaCl), E. coli accumulate inorganic ions and organic solutes to help maintain cellular hydration and membrane turgor pressure (5, 247). Proline, glutamate, glycine betaine, and polyols such as trehalose are the major organic osmolytes or osmoprotectants (5, 53, 195). E. coli accumulate proline by importing it from the extracellular environment, as proline biosynthesis does not contribute to increased proline levels during osmotic stress (54). The osmolyte solutes transported by ProP and ProU broadly overlap and include proline, glycine betaine, proline betaine, and ectoine (141). ProP exhibits similar affinity for proline and glycine betaine (56), whereas ProU significantly favors glycine betaine over proline (86). Osmotic stress activates expression of the proP gene (40, 132, 149) and the proU operon, encoding the three components of the ProU transport system (proVWX) (5, 60, 104, 149). Wood and coworkers have shown that ProP is also regulated post-translationally (186), with ProP being nearly inactive at low osmolality but becoming maximally activated in a highly osmolality medium (56). The mechanism by which ProP is activated by increased osmolality has been elusive; however, Wood and coworkers recently proposed that the binding of cations (e.g., potassium) to the C-terminal domain of ProP induces a conformational switch that increases transporter activity during osmotic stress (56, 58).
FIG. 7.
Proline transporters in E. coli and B. subtilis. E. coli uptake systems are the high-affinity proline/sodium symporter PutP, the ProP proton/osmolyte symporter, and the ABC-type transporter ProU. B. subtilis contains a PutP homologue of E. coli, OpuE, which is a compatible solute transporter, and the γ-aminobutyrate transporter GabP. The PutP transporter is important for utilizing proline as a nutrient source. ProP and OpuE are the proline transporters most important for osmoregulation and protection against osmotic stress in E. coli and B. subtilis, respectively.
Proline is also a critical osmolyte in gram-positive bacteria such as B. subtilis with intracellular proline concentrations increasing up to 700 mM after osmotic shock (242). B. subtilis contains three main proline transporters: a PutP homologue, OpuE (254) of the Opu family of compatible solute transporters, and the γ-aminobutyrate transporter GabP (255) (Fig. 7). PutP and OpuE share 61% amino acid sequence identity and are high-affinity sodium/proline symporters (155). PutP is critical for use of proline as a nutrient source, while OpuE is important for using proline as an osmostress protectant (236). Bremer and coworkers have shown that OpuE is osmoregulated and responsible for increased proline transport in cells under osmotic stress (236, 254). In contrast to E. coli, B. subtilis also relies on proline biosynthesis to increase intracellular proline levels (28). Intriguingly, B. subtilis has an alternative route for proline biosynthesis involving paralogous enzymes for GK and P5CR (28). This second route is not subject to allosteric feedback inhibition by proline, and thus, B. subtilis is able to increase intracellular proline pools by proline import and biosynthesis (28).
Bacterial Pathogens
Escherichia and Salmonella
E. coli uropathogenic strains are a major cause of bladder infections and, less commonly, kidney infections or pyelonephritis (6, 94). Human urine is a good medium for E. coli bacterial growth with abundant carbon and nitrogen sources from amino acids, citrate, lactate, and ammonia. Identifying metabolic genes that are essential for infection has come from studies of E. coli uropathogens such as E. coli strain CFT073, which was originally isolated from a patient with pyelonephritis (152). Using different metabolic mutants of E. coli strain CFT073 and a mouse urinary tract infection model, Alteri and coworkers showed that mutants of gluconeogenesis and the TCA cycle were significantly defective in colonization, whereas other mutants of central metabolism, such as glycolysis and the pentose phosphate pathway, had no effect on colonization (6). Another study found that TCA and PPP are not critical for E. coli colonization of mouse intestine but that metabolism of gluconate by the Entner–Doudoroff pathway was most important (39). Whether proline oxidation is an important anaplerotic pathway for supplying carbon (i.e., α-ketoglutarate) to the TCA cycle during urinary tract infection is not known.
Besides adapting metabolically, pathogens also need to overcome osmotic challenges to survive and establish infection. Because proline is an osmolyte protectant, its role in osmotic stress protection has been extensively investigated in bacteria, including uropathogens (5, 51). Mutant strains of S. typhimurium that exhibit increased tolerance to osmotic stress were found to have elevated levels of proline (52). In E. coli, proline has been shown to be important for tolerance of high osmotic and urea stress environments such as urine. The importance of proline transport in E. coli pathogenesis has been examined by characterizing the growth and colonization of different proP and proU mutant uropathogenic strains (55). Deletion of proP and proU in the E. coli strain HU734 resulted in slower growth in high-osmolality human urine but did not impact urinary tract colonization in mice (55). Studies of other bacteria showed that the ProU transporter is important for infection of moth larvae by Shigella sonnei (143). Also, ProP was concluded to be critical for survival of S. enterica serovar Typhimurium in desiccated environments such as stainless steel surfaces (75).
Entomopathogenic bacteria
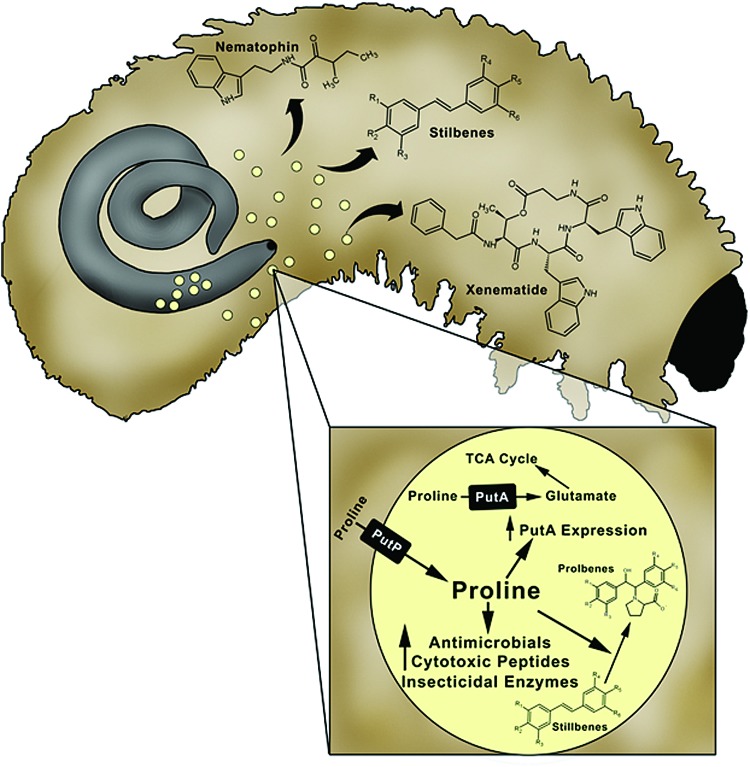
Entomopathogenic bacteria and worms have complex and mutually beneficial relationships (78). Symbiotic relationships involve Xenorhabdus spp. with the Steinernematidae nematode family and Photorhabdus species with nematodes of the Heterorhabditidae family. Soil-dwelling nematodes use nutrient resources from insect hosts for reproduction and propagation (78). To successfully infect insect larvae, the worms utilize enterobacterial symbionts such as Xenorhabdus nematophila and Photorhabdus luminescens (78). Xenorhabdus and Photorhabdus spp. colonize the gut of nematodes in a symbiotic relationship, but transition into a pathogenic lifestyle once inside insect larvae. After being expelled from the worm, the bacteria secrete insecticidal enzymes and cytotoxins (e.g., xenematide) that kill the insect larvae and synthesize antibiotic compounds to ward off competing microbes (22, 23, 50, 66, 119, 175). During the infection process, the bacteria also weaken the innate immune system of the host, thereby facilitating infection, enabling the bacteria to grow, and allowing the worms to reproduce inside the insect carcass (78, 176, 191). A summary of the mutualism between nematodes and Xenorhabdus and Photorhabdus spp is illustrated in Figure 8.
FIG. 8.
Entomopathogenic bacteria and role of proline in insect larvae. After infection of insect larvae, nematodes harboring symbiotic entomopathogens expel the bacteria into the larval host. Cues from the new environment, including increased proline levels, cause the entomopathogens to undergo metabolic shifts, transitioning these bacteria from symbionts of the nematode to pathogens of the worm. This shift in metabolism leads to the generation of cytotoxic peptides, insecticidal enzymes, and antimicrobials that kill the larval host and bacterial competitors. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
This biological cycle in the soil environment is a natural insecticide mechanism that has drawn interest for biocontrol of crop pests (117). Commercial applications of entomopathogenic bacteria and nematodes to soil have been used for natural pest control (69, 117). There are also efforts to identify the toxins and antibiotics produced by Xenorhabdus and Photorhabdus spp. in hopes of finding new molecules for treatment of areas that are often devastated by insects (117). The genome sequencing of X. nematophila and Photorhabdus spp. (e.g., Photorhabdus temperata, P. luminescens, and Photorhabdus asymbiotica) is greatly enhancing these efforts (66).
Insects have an abundant supply of proline in their hemolymph, which has raised curiosity as to the role of proline in the pathogenesis stage of X. nematophila and Photorhabdus spp. (50, 175). Crawford et al. investigated the metabolic changes in entomopathogenic bacteria that are associated with increased production of antimicrobial compounds such as stilbenes, nematophin, and the cyclodepsipeptide xenematide (50). Analysis of insect samples showed that proline concentrations in the hemolymph range from 3 to 73 mM, indicating proline could be a metabolic signal that stimulates antibiotic production (50). Evidence for proline inducing metabolic changes was observed by treating cultures of X. nematophila and P. luminescens with proline (50). Bacterial cultures treated with insect hemolymph or proline (∼5 mM) showed similar increases in the abundance of antimicrobial compounds (50). The role of proline was further tested by disrupting proline transport in P. luminescens, which encodes a homologue of the high-affinity PutP transporter from E. coli. In the ΔputP mutant strain of P. luminescens, proline did not induce a metabolic shift further supporting a key role for proline in the production of antibiotic compounds (50). Proline is also proposed to be part of stilbene detoxification pathway in the host bacterium in a reaction involving stilbene epoxidation and incorporation of l-proline generating less toxic compounds called prolbenes (Fig. 8) (175).
How proline activates antibiotic production in entomopathogenic bacteria is not clear. It may involve Lrp, a nutritional sensor, which could respond to proline and induce Lrp-dependent activation of virulence factors (36, 191). X. nematophila and P. luminescens also encode trifunctional PutAs, indicating that similar to E. coli, putA and putP gene expression would be subject to transcriptional auto-repression in the absence of proline. The PutAs from X. nematophila (1326 residues, Accession No. CBJ89733) and P. luminescens (1326 residues, Accession No. CAE14250) have high-sequence identity (77%) with E. coli PutA (1320 residues, Accession No. AAB59985.1), including residues of the RHH DNA binding domain.
In both genomes of X. nematophila and P. luminescens, the putA and putP genes share a similar arrangement as that found in E. coli with the genes separated by an intervening put control region and the putA and putP genes transcribed in opposite directions. Previously, it was shown that GTTGCA is the consensus DNA binding motif for E. coli PutA (263), with five PutA binding sites in the put control DNA region (263). Analysis of the put control DNA sequences of X. nematophila (Ref Seq: NC_014228.1) and P. luminescens (Ref Seq: NC_005126.1) identified three consensus PutA DNA binding motifs. The fewer PutA binding sites in the put control region of X. nematophila and P. luminescens suggest that transcriptional repression by PutA may be less stringent than that of E. coli. It will be important to characterize the regulation of the putA and putP genes to gain further insights into the DNA binding role of PutA in these bacteria and the proline response mechanism that leads to upregulation of antibiotic production.
Proline is also predicted to serve as an important energy source for these bacteria due to its high abundance in the insect relative to glucose. Proline metabolism was shown to support respiratory activity, which was proposed to be important for the metabolic changes that favor antibiotic production (50). It would also be interesting to know if proline metabolism increases oxidative stress resistance by upregulation of antioxidant enzymes. Analysis of the genomes of X. nematophila and P. luminescens indicates the presence of an OxyR stress response regulator similar to E. coli. These genomes also encode thioredoxin and glutaredoxin proteins, but, interestingly, do not appear to have a katG homologue. A katE gene is predicted in P. luminescens and no catalase is annotated for X. nematophila. In E. coli, katE is not part of the OxyR stress response regulon but instead is upregulated during stationary phase. Activation of the OxyR regulon by proline metabolism in X. nematophila and P. luminescens could potentially facilitate evasion of the innate immune system and colonization of the insect host.
H. pylori
One of the major aspects of pathogenesis is the ability to metabolically adapt to the host or vector environment. An example of a bacterial pathogen that exploits proline as a carbon and energy source is H. pylori, which thrives in microenvironments enriched with proline (160, 162, 193). H. pylori is a microaerophilic gram-negative bacterium that colonizes the lower stomach, is the causative agent of peptic ulcer disease, and is associated with gastric inflammation and gastric cancer (77, 113, 179, 232). Together with l-serine, l-proline has been shown to be a preferred respiratory substrate of Helicobacter and has been found to be significantly elevated in the stomachs of patients infected with H. pylori (160). During infection, H. pylori secretes collagenase, which can breakdown collagen in the extracellular matrix of the stomach epithelium leading to increased proline levels (85, 105). In Helicobacter spp., PutA lacks the DNA binding domain of trifunctional PutAs, and thus, its central biological role is to enable the bacterium to utilize proline as a carbon source by coupling the formation of glutamate with reduction of the respiratory chain (110, 160, 167).
In Helicobacter, the putA and putP genes are arranged in a putAP operon. The putAP operon in H. pylori was shown by Pich et al. to be regulated by the ferric uptake regulator (Fur) (183). Fur represses the expression of iron uptake genes under iron-replete conditions thus preventing the toxic accumulation of metal in bacteria (183). Expression of putA was strongly repressed by Fe2+-Fur indicating that PutA may contribute to pathogenesis by helping with reductive iron acquisition and stress adaptation (183). Other regulators of the putAP operon have not been identified. The genome of H. pylori does not appear to contain homologues for PutR and PruR, which are proline-specific regulators in other gram-negative pathogens as discussed in the Regulation of proline catabolic genes section (110).
Evidence for proline having an important role in Helicobacter infection is from animal studies using H. pylori mutant strains of putA and putP. In a signature tagged mutagenesis screen of 960 mutants of H. pylori, putP was identified along with 46 other genes to be essential for gastric colonization of a Mongolian gerbil oral infection model (105, 193). Similar to E. coli, PutP from H. pylori is an Na+-l-proline symporter with Km of ∼2 μM proline (193). H. pylori is also predicted to have homologues of E. coli ProP and ProU, however, these transporters were not identified as being essential for colonization (229). In another study, a putA mutant strain of H. pylori was unable to colonize mice. The putA mutant strain was defective in flagellar formation and motility, perhaps due to loss of proline respiratory activity, which resulted in lower virulence (162). However, the H. pylori putA mutant also did not import proline from the surrounding media (162). Because Helicobacter putA and putP genes are arranged in a putAP operon, the putA mutant may also have loss of putP expression resulting in a mutant strain deficient in proline import and catabolism (162).
Further insights into the role of PutA in Helicobacter pathogenicity were reported with a putA knockout mutant strain of Helicobacter hepaticus (112). H. hepaticus is a mouse pathogen found in the liver of mice with high incidence of hepatitis and liver tumors. As an enterohepatic Helicobacter spp., H. hepaticus colonizes the lower bowel and progresses to the liver. Knockout of PutA in this murine pathogen did not impact colonization of the liver or cecum, however, inflammation of the liver during disease progression was significantly reduced relative to mice inoculated with the wild-type strain (112). Interestingly, the putA mutant strain exhibited lower catalase expression (katA) relative to wild-type H. hepaticus, indicating that proline oxidation via PutA upregulates katA gene expression and perhaps virulence. Virulence factors, such as catalase, are upregulated in H. pylori to combat oxidative stress from the host neutrophil response, resulting in persistent chronic inflammation (24, 101, 188). PutA metabolism of proline has been shown to increase oxidative stress resistance in E. coli, a potentially conserved feature of proline metabolism that should be further explored in the physiology and pathogenesis of Helicobacter spp. Altogether, these results indicate that proline metabolism plays an integral role in the colonization and pathogenesis of Helicobacter spp.
Ehrlichia chaffeensis
The zoonotic disease of human monocytic ehrlichiosis is caused by infection of white blood cells by the tick-borne bacterial pathogen E. chaffeensis (67). Human monocytic ehrlichiosis is a life-threatening disease, particularly in immunocompromised patients, and is characterized by lethargy, fever, and headache (67). Evidence for a role of proline in this disease was shown recently by Cheng et al., in which E. chaffeensis infection of human monocyte THP-1 cells was decreased on inhibiting proline import by protamine treatment (42). E. chaffeensis, a gram-negative bacterium, has a considerably smaller genome (1.18 Mb) relative to E. coli (4.67 Mb) and is predicted to encode 883 proteins. Interestingly, the E. chaffeenis genome (Ref Seq: CP000236) has a putA gene, but lacks a putP homologue. It appears that E. chaffeensis primarily imports proline via a homologue of the ProP transporter from E. coli. A gene (Accession No. WP_006010458.1) within the E. chaffeensis genome, annotated as a member of the Major Facilitator Superfamily, has 33% amino acid sequence identity with the E. coli osmoregulatory proline/glycine betaine transporter ProP (Accession No. AAC44538.1, 86% query length). Additional analysis of the E. chaffeensis genome shows it lacks proline biosynthesis genes proB (GK) and proA (GPR) but has the proC gene, which encodes P5CR. Therefore, it appears E. chaffeensis is limited in proline biosynthesis and must rely on proline import and proline recycling via an X-Pro aminopeptidase, which is also annotated in the genome.
One of the mechanisms by which proline facilitates E. chaffeensis infection is proposed to involve the utilization of proline as a carbon and energy source (42). The PutA protein from E. chaffeenis (1044 amino acids, Accession No. ABD44729.1) shares 47% amino acid sequence identity with E. coli PutA (78% query length) and was shown to complement a putA mutant E. coli strain, confirming that E. chaffeenis PutA is functional in proline catabolism (42). E. chaffeensis PutA lacks the N-terminal RHH DNA binding of E. coli PutA and is predicted to be a bifunctional PutA (212), and therefore, it is unlikely to have a DNA binding function. The PutA catalyzed oxidation of proline to glutamate followed by conversion of glutamate into α-ketoglutarate by NAD+-dependent glutamate dehydrogenase would funnel carbon into the TCA cycle and support ATP production. The direct coupling of PutA/PRODH activity with the respiratory chain of E. chaffeensis would also provide energy. E. chaffeensis was shown to be deficient in glutamate import, thus supporting a critical role for proline catabolism in supplying intracellular glutamate (42). Glutamine was also shown to enhance E. chaffeensis growth and infection. In addition, expression of glutamine synthetase (GlnA) was shown to have a similar temporal expression pattern and transcriptional regulation as PutA, highlighting an important metabolic link between proline and glutamine (42).
PutA expression is controlled by the two-component regulatory system of nitrogen metabolic regulators NtrY and NtrX (42, 115). NtrY contains an N-terminal sensor domain and C-terminal histidine kinase domain, while NtrX contains an N-terminal phosphorylation domain and C-terminal helix-turn-helix DNA binding domain (115). Cheng et al. showed that NtrX binds to the promoter region of the E. chaffeensis putA gene, with DNA binding affinity significantly increased on phosphorylation of NtrX. These results indicate that putA gene expression is activated by the NtrY/X system (42).
Another potential mechanism by which proline could facilitate infection involves enhanced developmental cycling of E. chaffeensis, which, during infection, involves cycling between small- and large-sized cell bodies (43). The smaller sized aggregates are the more infectious form that enters and exits host cells. On entering a host cell, E. chaffeensis develops into the larger aggregate replicative form. Once reaching stationary phase inside the host, E. chaffeensis transitions back into the smaller sized form and exits the host cell, thereby initiating a new round of infection (43). A key regulatory protein in this process is CtrA, which is highly expressed at stationary phase, downregulates DNA replication, and upregulates genes important for the small-sized aggregate form (42, 43). Interestingly, proline was observed to accelerate the degradation of CtrA in host cells (42). Thus, proline may be a nutrient signal that facilitates the transition of E. chaffeensis to the larger sized replicative form by helping downregulate the CtrA regulon, thereby promoting the developmental cycling of E. chaffeensis during infection.
Figure 4 summarizes the potential effects of proline on CtrA and the regulation of the putA gene, which involves NtrY phosphorylation of NtrX. Exactly how proline enhances CtrA degradation and NtrX phosphorylation by NtrY is not known. Further studies on proline metabolism and regulation in E. chaffeensis seem to be an important area to pursue.
S. aureus
S. aureus is a gram-positive bacterial pathogen that often resides in the nose without signs of illness. However, untreated infection with methicillin-resistant S. aureus can cause a range of illnesses, including potentially life-threatening conditions such as pneumonia and sepsis (206). In S. aureus, proline has an important role in osmoregulation (207). Intracellular levels of proline, along with other osmolytes such as glycine betaine, increase when S. aureus is grown in a high osmotic environment (4, 82, 207). The rise in intracellular proline levels is mediated by at least two transporters: a high-affinity transport system, encoded by the gene putP, and a low-affinity transporter homologous to E. coli ProP (207). The expression of putP increases in tandem with increased osmolarity and decreased intracellular proline levels, indicating that proline import is required for osmotic stress protection (206). In addition, Bae and Miller demonstrated that the low-affinity transport system ProP is activated under periods of high osmolarity, resulting in increased proline accumulation that helps S. aureus tolerate periods of osmotic stress (13).
Proline import has been shown to be critical for infection. A transposon mutation in the putP gene resulted in decreased survival of S. aureus in animal models (205). Accordingly, expression of putP was shown to increase by over 100-fold on S. aureus infection in various mice organs, further demonstrating the importance of rapid proline import during infection (206). Interestingly, the expression of proP also increased, but this increase occurred at a time significantly later than the initial infection (241). These observations have led to a model in which PutP is proposed to be critical for early stages of infection, whereas ProP predominates at later stages of infection after tissue damage has occurred (207).
In addition to proline uptake, S. aureus can also synthesize proline. However, as it lacks proB (GK) and proA (GPR) genes, S. aureus does not synthesize proline from glutamate (231). Instead, this pathogen synthesizes proline from arginine via ornithine and OAT to generate GSA/P5C, which is then reduced to proline by P5CR (proC gene) (Fig. 2) (231). Thus, arginine uptake will lead to increases in intracellular proline (231).
Proline catabolism also supports the growth of S. aureus as demonstrated recently by Fey and coworkers (90). S. aureus PRODH (333 amino acid residues) is encoded by a gene annotated as putA (WP_001196354.1) even though it lacks the P5CDH domain. A second gene called rocA (Accession No. AMV81009) encodes P5CDH. Deletion of putA results in a growth defect in media lacking glucose (90). NMR metabolomics using 13C-labeled proline showed proline is oxidized principally to glutamate with subsequent conversion to α-ketoglutarate by glutamate dehydrogenase thus supplying carbon to the TCA cycle and helping generate ATP (90). These results led Fey and coworkers to propose that proline catabolism is important for the pathophysiology of S. aureus during abscess formation, in which significant tissue remodeling occurs and proline is released from the breakdown of collagen (90). Glucose can become severely limited during infection, whereas amino acids, such as proline, are more abundant and can be utilized as important sources of carbon and energy for survival (90).
Recent immunological studies of S. aureus and Streptococcus pneumoniae discovered P5CDH on the outer surface of the bacteria (131). Whether P5CDH may have another role in virulence unrelated to proline catabolism is unclear, but the association of P5CDH on the bacterial surface has implicated it as a potential component of vaccines against S. aureus (131).
M. tuberculosis
The ability of M. tuberculosis to survive in a highly stressed host environment is a characteristic that makes it such a dangerous human pathogen. By entering a dormant state and undergoing metabolic alterations, M. tuberculosis is able to hide within the human host asymptomatically for long periods of time before reactivating (199). Proline metabolic genes are important for the growth of M. tuberculosis and also appear to have an important role apart from proline utilization as a carbon and energy source (20, 21, 118, 201). M. tuberculosis encodes separate proline catabolic enzymes PRODH and P5CDH, and proline biosynthesis enzymes GK, GPR, and P5CR (118, 209). PRODH couples the oxidation of proline to reduction of menaquinone in the respiratory chain thereby facilitating energy production while P5CDH forms glutamate to supply α-ketoglutarate for the TCA cycle (20, 21, 118, 209).
Insights into the regulation of proline metabolism were gained by Berney et al. who showed that the pruC gene (MSMEG_5120), which is divergently transcribed from the pruAB operon, is a transcriptional activator of the pruA and pruB genes (Fig. 4) (21). PruC is a PucR-type regulatory protein that binds directly to the pruAB promoter and is also membrane associated. Interestingly, PruC activation of pruAB expression was shown to occur independently of exogenous proline, indicating that other regulatory mechanisms are involved (21).
How Mycobacterium regulates proline metabolism in response to nitrogen stress or limiting nitrogen is also important to understand as amino acids are critical for cells to survive in nitrogen-deplete environments. Using global transcriptional analysis, expression levels of genes encoding PRODH, P5CDH, and OAT were found to be significantly induced by nitrogen limitation in Mycobacterium smegmatis consistent with proline being converted to glutamate (244). Using a continuous cell culture system for M. smegmatis, however, Petridis et al. found that these same proline metabolic genes were downregulated under nitrogen limitation (180). A study on M. tuberculosis by Williams et al. reported that OAT expression is significantly upregulated by limiting nitrogen (245). It will be important to continue investigating nitrogen limitation in M. tuberculosis to better understand the metabolic adaptations that enable utilization of different amino acids such as aspartate, which may be an essential nitrogen source for M. tuberculosis infection (70).
Proline catabolism and biosynthesis were identified as essential pathways for optimal growth of Mycobacterium from a large library of transposon insertion mutants (201). Null mutations of the pruA (P5CDH) and pruB (PRODH) genes, which are transcribed from the pruAB operon in saprophytic M. smegmatis, confirmed that PRODH and P5CDH are essential for growth on proline, which is primarily utilized as a growth substrate under low glucose conditions (20, 201). In a transposon site hybridization (TraSH) study of M. tuberculosis, proline metabolic genes were not identified as being essential for survival in macrophages (190). However, a proC (PYCR) mutant of M. tuberculosis, which is auxotrophic for proline, was shown to have severely diminished survival in mouse macrophages and to be avirulent in a mouse infection model (214). The potential of the proC mutant as a vaccine candidate for M. tuberculosis was thus suggested (214). Consistent with these findings, a recent study using a saturating transposon mutagenesis approach of the M. tuberculosis genome reported the proC gene to be essential for in vitro growth (62).
Perhaps related to the essential role of proC is the need of proline for the biosynthesis of the Pro-Glu (PE)/Pro-Pro-Glu (PPE) proteins. These abundant proteins (168 in M. tuberculosis) are found on the bacterial cell surface and have a highly conserved N-terminal domain that contains the PE/PPE motif and a variable C-terminal domain that provides a diverse suite of functions (27). PE/PPE proteins are critical for M. tuberculosis virulence and have multiple roles in pathogenesis, including protection against genotoxic stress and regulation of cytokine production in host cells (27, 164).
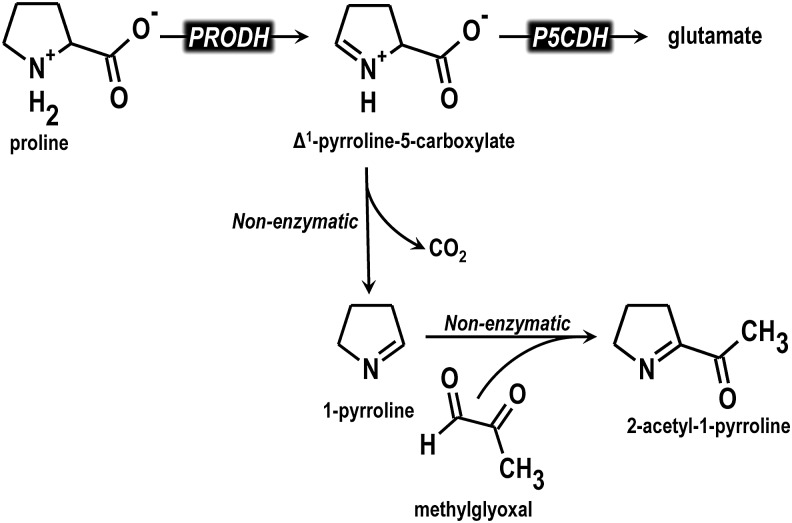
Another key role for proline metabolism in Mycobacterium was shown to involve protection against methylglyoxal, an electrophilic and potentially toxic metabolic intermediate that is formed nonenzymatically from metabolites such as dihydroxyacetone phosphate (Fig. 9) (21). Interestingly, only the first step of proline catabolism, the conversion of proline to P5C, was shown to be necessary for this protection (21). The intermediate of the proline utilization pathway, P5C, is capable of reacting with methylglyoxal to form 2-acetyl-1-pyrroline, which is nontoxic (96). Thus, detoxification of methylglyoxal was proposed to occur directly with P5C, resulting in formation of 2-acetyl-1-pyrroline (Fig. 9) (21). The importance of proline catabolism in protecting against methylglyoxal stress during M. tuberculosis is not yet known (21). However, increased methylglyoxal concentrations have been reported in macrophages infected with M. tuberculosis indicating that proline catabolism, or specifically P5C generation, could serve a protective function by preventing DNA damage and other unwanted modifications by this reactive carbonyl species (187). Whether Mycobaterium PRODH and P5CDH are promising drug targets will require further investigation. Characterization of the M. tuberculosis P5CDH revealed a larger substrate binding site relative to other P5CDH enzymes, suggesting the potential to design a unique molecule that would specifically inhibit proline catabolism in Mycobacteria (118).
FIG. 9.
Role of proline in M. tuberculosis. The intermediate of proline metabolism, P5C, is proposed to undergo nonenzymatic decarboxylation to generate 1-pyrroline. Methylglyoxal reacts with 1-pyrroline to form 2-acetyl-1-pyrroline thus reducing potential cellular damage caused by the buildup of methylglyoxal.
Finally, the significance of osmoregulation in Mycobacterium during growth in macrophages has been tested. An ABC transporter ProXVWZ, predicted to transport glycine betaine, choline, and proline, was shown to be important for the ability of M. tuberculosis to colonize macrophages. ProXVWZ, however, accumulated mainly glycine betaine instead of proline during growth in macrophages (184).
Clostridium difficile
A recent study by the Centers for Disease Control and Prevention estimated that nearly 500,000 Clostridium difficile infections occur in a single year, leading directly to about 15,000 deaths (1). C. difficile is a gram-positive nosocomial pathogen that forms spores and grows anaerobically in the human gut causing diarrhea, inflammation of the colon, and colitis. C. difficile was first isolated from the microflora of newborn infants (88). Certain bacteria from the genus Clostridium use an interesting metabolic pathway for growing on amino acids. The process, called the Strickland reaction or fermentation, was first described in nonpathogenic Clostridium sporogenes and involves the coupling of electron donor amino acids that are oxidatively deaminated with electron acceptor amino acids that are reductively deaminated (25, 169, 218). For example, oxidation of alanine involves deamination and decarboxylation to acetic acid and ammonia coupled to either reductive deamination of glycine to acetyl-phosphate for substrate-level production of ATP or reduction of proline to δ-aminovaleric acid, which is coupled to transmembrane proton translocation (135). In different Clostridium spp., the Strickland reaction has been shown to involve leucine, valine, isoleucine, and alanine as donors and glycine, proline, and hydroxyproline as electron acceptors. In C. difficile, proline is an essential amino acid for growth, along with cysteine, isoleucine, leucine, tryptophan, and valine (103).
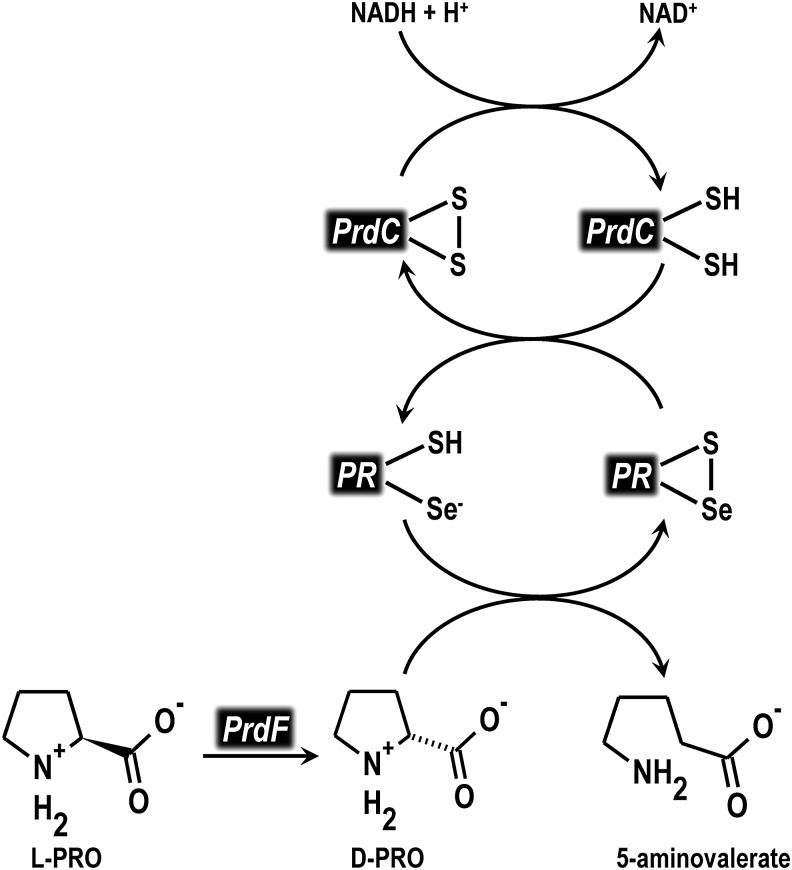
The key enzyme catalyzing the reduction of proline in the Strickland pathway is d-proline reductase (PR), a selenoenzyme that comprised subunits PrdA (45 kDa) and PrdB (27 kDa) with a molecular mass of 280 kDa indicating a higher oligomeric structure (99, 102). PR is specific for d-proline as it does not use l-proline or d- and l-4-hydroxyproline as substrates (8). The catalytic mechanism of PR has been proposed to involve a selenocysteine residue functioning as an active site nucleophile, attacking the α-carbon of d-proline resulting in cleavage of the N-C bond (Fig. 10) (76, 102). Before nucleophilic attack by the selenocysteine, the N-C bond is likely activated by adduct formation between the proline amine and a pyruvoyl group in the active site (102). The selenocysteine substrate adduct would then be resolved by formation of a mixed selenide/sulfide bond with an active site cysteine, releasing δ-aminovalerate (102). The PrdC thiol oxidoreductase, which contains a C-terminal CxxU motif, would then reduce the selenide/sulfide bond of PR to complete the catalytic cycle (76). The physiological electron donor for PrdC is not known but may involve NADH (76).
FIG. 10.
Reduction of d-proline in C. difficile. l-proline is first converted into d-proline by proline racemase. PR then catalyzes the two-electron reduction of d-proline to 5-aminovalerate. The catalytic cycle of PR is completed by the thiol oxidoreductase PrdC, which has been proposed to capture reducing equivalents from NADH. PR, d-proline reductase.
Colonization of the gut by C. difficile involves secretion of glycosylating toxins and production of l-proline aminopeptidase to release trans-4-hydroxy-l-proline and l-proline from host collagen (73, 93). Recently, a trans-4-hydroxy-l-proline dehydratase (t4L-HypD) was identified in the human microbiome from C. difficile (127). The enzyme t4L-HypD is a member of the glycine radical enzyme superfamily and catalyzes the dehydration of trans-4-hydroxy-l-proline to P5C (127). Balskus and coworkers showed that C. difficile enzymes t4L-HypD and P5CR coordinate the conversion of trans-4-hydroxy-l-proline to l-proline, thus providing a mechanism for the bacterium to acquire proline from host and dietary proteins in the gut (127).
Increased levels of proline lead to upregulation of PR in C. difficile to promote growth. Bouillaut and coworkers identified PrdR as the protein responsible for activating transcription of the proline reductase operon that encodes PR subunits (prdA, prdB, and prdD), proline racemase (prdF), and the PrdC thiol oxidoreductase protein (prdC) (25). Therefore, the increased bioavailability of proline during infection leads to upregulation of the proline reductase operon.
The importance of proline during C. difficile infection was examined by Wu and Hurdle using a hamster infection model and a ΔprdF mutant strain made by insertional mutagenesis (253). Because proline racemase is essential for generating d-proline from l-proline, disruption of prdF is expected to block utilization of proline as a Strickland acceptor amino acid. Wu and Hurdle observed no significant difference in the survival between hamsters infected with wild-type C. difficile JIR8094 and the ΔprdF mutant strain (253). These results indicate that proline as a Strickland acceptor amino acid may not be critical for C. difficile pathogenesis. Nevertheless, additional work is needed to fully assess the role of proline metabolism during C. difficile infection, such as testing the impact of PR directly by depleting a PR subunit gene.
Eukaryotic Human Pathogens
Trypanosomatidae family
Trypanosomes are kinetoplastid parasites with life cycles that alternate between infection in humans and insects (196, 234) (Fig. 11). In humans, they are responsible for a number of diseases, including Chagas disease, caused by Trypanosoma cruzi, African trypanosomiasis, also known as African sleeping sickness, caused by T. brucei, and leishmaniasis, caused by various members of the Leishmania genus (196). These parasites adapt to fluctuations in nutrients as they move between human host and insect vector with glucose consumed primarily in mammalian blood and proline utilized as a carbon source in the insect (72, 145, 146, 173, 196, 221, 234).
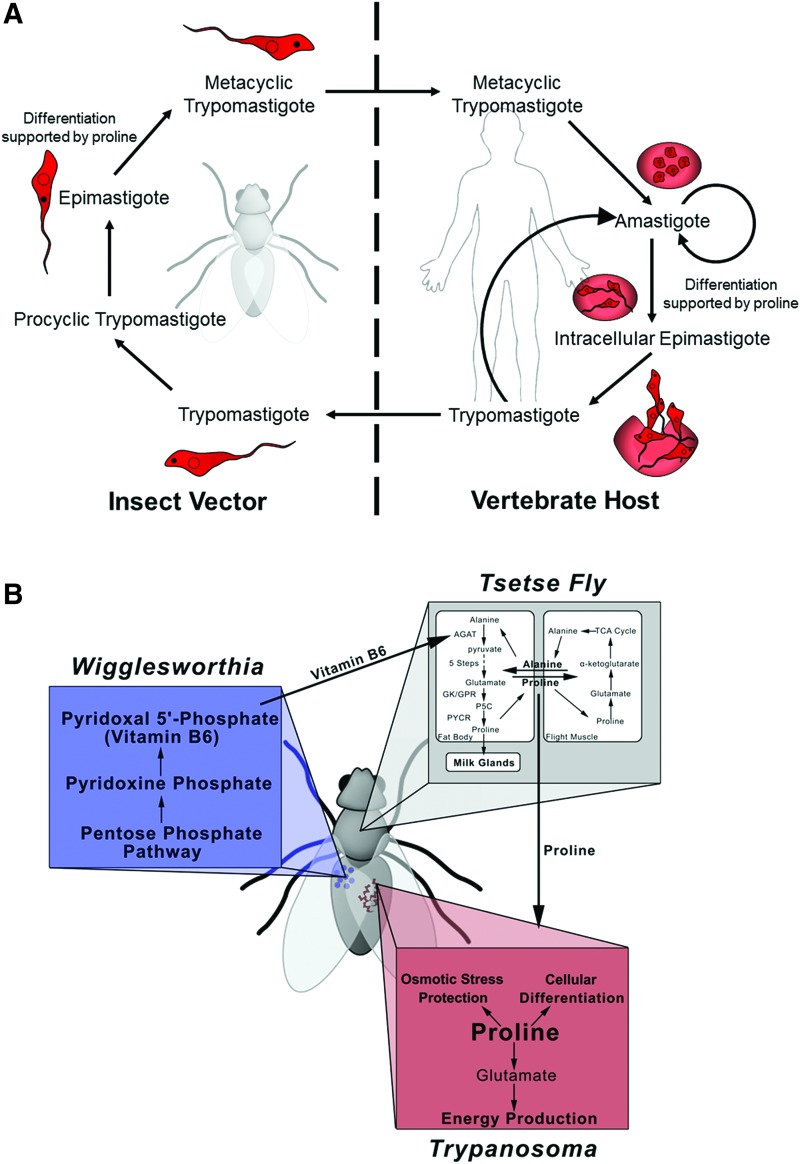
FIG. 11.
Relationship between proline, trypanosomes, and the insect host. (A) Trypanosomes exhibit a complex life cycle, cycling between a vertebrate host and an insect vector. On infection of a vertebrate host, metacyclic trypomastigotes will differentiate into the intracellular amastigote form, a form characterized by no visible flagella. The amastigotes will then reproduce within host cells before differentiating into intracellular epimastigotes, followed by differentiation into trypomastigotes and subsequent bursting of the host cell. The trypomastigotes are then free to infect new cells or be passed into the insect vector. In the insect vector, trypomastigotes differentiate into procyclic trypomastigotes capable of reproduction. The procyclic trypomastigotes will then differentiate into epimastigotes, followed by differentiation into the infective metacyclic trypomastigote form. The trypomastigote and epimastigote forms are primarily distinguished by the location of the kinetoplast (solid, black circle) relative to the nucleus (dashed circle). Proline and its catabolism have been implicated in the differentiation of trypanosomes from the epimastigote to the metacyclic trypomastigote forms and from the amastigote to the intracellular epimastigote forms. (B) In the tsetse fly, proline is utilized as an energy source by flight muscles or during lactation. Alanine generated from proline is then transported to fat bodies for proline regeneration. This conversion of alanine back to proline relies on pyridoxal 5′-phosphate (vitamin B6) generated by the symbiont Wigglesworthia. Trypanosomes actively uptake proline from the insect host for utilization in numerous processes, including energy production, cellular differentiation, and osmotic stress protection. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
The oxidative conversion of proline to glutamate is catalyzed by PRODH and P5CDH in the mitochondrion of trypanosomes (35, 145, 173). Glutamate is subsequently converted to α-ketoglutarate by glutamate dehydrogenase or l-alanine amino acid transferase (35, 145, 173). The flux of α-ketoglutarate into the TCA cycle and formation of key TCA intermediates such as of succinate and malate is required for proline-dependent growth of trypanosomes in the absence of glucose (35, 145, 173). Knockdown of succinate dehydrogenase in proline growth medium was shown to be lethal in T. brucei (48). High glucose levels attenuate proline catabolism, indicating that proline is not the preferred carbon source in glucose-rich environments (48). Recently, transcription levels of PRODH (Tb927.7.210) were found to be regulated by circadian rhythms in the bloodstream form of T. brucei along with other metabolic genes that are associated with ATP production (192).
Evidence for the importance of proline as a carbon and energy source during parasite life cycles and infection has been shown in Trpanosoma spp. (68, 72, 145, 173, 221, 234). When the production of ATP from proline is inhibited in T. cruzi, the ability of this parasite to invade host cells is severely diminished (145, 146). Furthermore, Martins et al. found that proline plays an important role in parasitic translocation across a gastric mucin layer, a process that is vital for host infection by T. cruzi (146). In addition, proline was shown to have a critical role in the differentiation of T. cruzi (38, 203, 230). The differentiation of this parasite between various infective host and replicative vector forms is crucial to disease development and the spreading of disease across a population (196) (Fig. 11A). By using a host cell that requires proline, Tonelli et al. were able to prevent the differentiation of T. cruzi amastigotes (mammalian host form) into trypomastigotes (insect vector form) (230). By adding excess l-proline to the media, differentiation was restored (230).
Interestingly and further highlighting the importance of proline in differentiation, both intracellular proline levels and levels of proline catabolic enzymes in T. cruzi vary as the pathogen's life stage changes. The amastigote form has the highest levels of free proline, whereas PRODH activity is highest in the intracellular epimastigote form and P5CDH activity is elevated in the metacyclic trypomastigote and bloodstream trypomastigote forms (145, 173, 230). Proline racemases also appear to be involved in parasite differentiation. Coutinho et al. have observed that treatment of T. cruzi trypomastigotes with proline racemase antibodies leads to decreased invasion of host cells (49). T. cruzi proline racemase has also been shown to initiate B cell mitogenic activity, but this effect does not appear to be dependent on its enzymatic activity (189).
Trypanosomes have active transport systems for obtaining proline from the environment. In Leishmania major, proline has been shown to be actively transported in promastigote (insect vector residing) and amastigote (mammalian host residing) forms (29). Because proline is abundant in the hemolymph of most insects, including the tsetse fly, trypanosomes can readily acquire proline from the insect host and are not dependent on proline biosynthesis (29). This is significant as not all trypanosomatids are able to synthesize proline from glutamate (29). Trypanosomatids all have P5CR but exhibit genetic differences in GK and GPR. For example, Leishmania spp. encode GK and GPR and T. cruzi encodes a putative P5CS-like protein (TCSYLVIO_005298) with predicted GK and GPR domains; however, T. brucei lacks both GK and GPR. Trypanosomatids also lack OAT, and thus, proline can only be synthesized via GK/GPR.
Proline, alanine, and glutamate are the major amino acids used for osmoregulation by trypanosomes (29, 98). Under hypotonic stress, T. cruzi has been shown to release large amounts of proline and alanine (29, 198). In addition, increased transport and accumulation of proline in T. cruzi have been shown to increase resistance to trypanocidal drugs such as nifurtimox and benznidazole (202). Overexpression of a d, l-proline transporter led to elevated intracellular proline levels and increased resistance to trypanocidal drugs (202). The increased import of proline into the cell also increased resistance to hydrogen peroxide and nitric oxide, suggesting that proline plays a part in multiple stress protection pathways (202).
In the case of trypanosomes, adapting to utilize proline as an energy source involves the addition of a third symbiotic relationship to the already intricate association between the pathogen and insect (Fig. 11B). Proline is a major source of energy during flight in many insects, including the insect vectors of different parasites (35). Tsetse flies, the insect vector of T. brucei, store high amounts of proline in the hemolymph (>100 mM) for ATP production during energetically strenuous activities such as flight and lactation (34, 35, 151). Tsetse flies infected with T. brucei exhibit decreased proline levels in the hemolymph and a concurrent decrease in fecundity indicating an energetic burden due to draining of proline resources (151).
The high levels of proline in the tsetse fly are produced from alanine in an eight-step pathway with the first step catalyzed by alanine-glyoxylate transaminase (AGAT), a pyridoxal 5′-phosphate (PLP)-dependent enzyme (151). Intriguingly, the tsetse fly relies on an obligate symbiont, Wigglesworthia glossinidia, to provide PLP (vitamin B6) for AGAT activity (151). W. glossinidia, a gram-negative bacterium with a genome size of ∼0.7 Mb (3), also provides vitamins necessary for the fertility of female tsetse flies (174). Interestingly, W. glossinidia encodes a trifunctional PutA but lacks the corresponding E. coli PutP homologue and is missing genes for proline biosynthesis (3). The delicate ecosystem whereupon the pathogen and vector are competing for proline as a shared energy source is reliant on a third symbiont that lacks several proline metabolic genes (Fig. 11B) (151).
Cryptococcus
The fungus Cryptococcus, namely the species Cryptococcus neoformans and Cryptococcus gattii, is the causative agent of cryptococcosis in humans (168). Cryptococcosis occurs in immunocompromised individuals due to C. neoformans exposure, whereas C. gattii infects individuals with noncompromised immune systems (168). The spreading of cryptococcal infection into the central nervous system leads to meningoencephalitis. In the pathogenesis of Cryptococcus, the fungus adapts to changes in the nutritional environment and combats oxidative stress defenses of the human immune system. Studies of how proline metabolism contributes to the virulence and pathogenicity of Cryptococcus have implicated P5CDH as having an essential role (122).
Similar to S. cerevisiae, Cryptococcus contains PUT1 and PUT2 genes that encode for PRODH and P5CDH, respectively (122). Intriguingly, C. neoformans encodes a second PRODH gene called PUT5. The biological significance of the PRODH gene duplication is not apparent but suggests that proline catabolism is important for survival. Interestingly, PUT5 was shown to encode the more active PRODH as deletion of PUT1 did not impact the ability of C. neoformans to utilize proline as a nitrogen and carbon source (122). The PUT1, PUT5, and PUT2 transcript levels are upregulated by proline (122). PUT1 is activated by the GATA transcription factor, Gat1p, which regulates genes responsible for nitrogen utilization in C. neoformans (108, 121) (Fig. 6). Unexpectedly, PUT5 and PUT2 expression was found not to be subject to nitrogen metabolite repression, unlike yeast, in which Gat1p activates expression of the PUT1 and PUT2 genes in response to limiting nitrogen availability (121) (Fig. 6). Thus, the regulation of proline catabolism in C. neoformans is dissimilar to that in yeast and will require more investigation, especially in the context of infection.
The role of proline metabolism in the expression of virulence factors and infection was examined by characterizing mutants of PUT1, PUT5, and PUT2 in the C. neoformans strain H99 (122). PRODH genes PUT1 and PUT5 were concluded not to be necessary for expression of cryptococcal virulence factors such as antiphagocytic polysaccharide capsule and melanin, or for infection, as determined using a mouse inhalation infection model (122). In contrast, deletion of PUT2 resulted in significantly lower levels of virulence factors, and using a mouse inhalation infection model, the C. neoformans Δput2 mutant strain was shown to be avirulent with no killing of the infected mice (122). Pathogen virulence was restored on complementation with PUT2, confirming the importance of P5CDH (122). It is suggested that increased superoxide generated from the mitochondria in the Δput2 mutant relative to wild-type C. neoformans results in increased susceptibility to apoptosis or cell death, possibly explaining why P5CDH is critical in C. neoformans virulence (122). Accumulation of P5C has previously been implicated in causing oxidative stress in yeast, suggesting a similar mechanism may be occurring in C. neoformans (165). Higher levels of P5C have been proposed to impact the transfer of reducing equivalents across the mitochondrial membrane via the proline-P5C pathway, thus imposing an oxidative stress burden on the mitochondria (165).
Summary
Proline is crucial for the growth of all the aforementioned pathogens under nutritional stressed conditions that are encountered during infection. In some pathogens, proline is also important for osmoregulation (e.g., E. coli, Trpanosoma spp., S. aureus) and stress resistance (M. tuberculosis, entomopathogens). Infection models using mutant strains of proline import and metabolism have provided insights into the importance of proline during pathogenesis of E. coli (ΔproP, ΔproU), Helicobacter spp. (ΔputA and ΔputP), S. aureus (ΔputP), M. tuberculosis (ΔproC) C. difficile (ΔprdF), and C. neoformans (Δput2 mutant). A summary of the functions of proline in these different pathogens is provided in Table 1.
Table 1.
Summary of Proline Metabolism in Different Pathogens
| Pathogen | Disease | Proline biosynthesis | Proline catabolic enzymes | Proline uptake | Primary functions | References |
|---|---|---|---|---|---|---|
| Ehrlichia chaffeensis | Monocytic ehrlichiosis | P5CR | Bifunctional PutA (PRODH/P5CDH) | H+-symporter (ProP) | Carbon/nitrogen/energy | (42, 67) |
| Entomopathogens | Symbiotic relationship with worms | GK, GPR | Trifunctional PutA (PRODH/P5CDH) | Na+-symporter (PutP) | Carbon/nitrogen/energy | (50, 78, 175) |
| Xenorhabdus nematophila | P5CR | H+-symporter (ProP) | Osmoregulation | |||
| Photorhabdus luminescens | Stress resistance | |||||
| Escherichia coli | Urinary tract infection | GK, GPR | Trifunctional PutA (PRODH/P5CDH) | Na+-symporter (PutP) | Carbon/nitrogen/energy | (152, 227, 248, 249) |
| Osmoregulation | ||||||
| P5CR | H+-symporter (ProP) | Stress resistance | ||||
| Clostridium difficile | Diarrhea, colitis | P5CR | Proline reductase | ABC transporter permease | Energy | (88, 102, 135) |
| Cryptococcus neoformans | Cryptococcosis | GK, GPR | PRODH, P5CDH | Amino acid permease | Carbon/nitrogen/energy | (108, 121, 122, 168) |
| P5CR | ||||||
| Helicobacter spp. | Peptic ulcers, gastric cancer | P5CR | Bifunctional PutA (PRODH/P5CDH) | Na+-symporter (PutP) | Carbon/nitrogen/energy | (105, 110, 112, 160, 162, 193) |
| H+-symporter (ProP) | Stress resistance | |||||
| Mycobacterium tuberculosis | Tuberculosis | GK, GPR | PRODH, P5CDH | Na+-symporter (PutP) | Carbon/nitrogen/energy | (20, 21, 118, 199, 201, 209) |
| P5CR | Stress resistance | |||||
| Staphylococcus aureus | Wide range of infections; skin, blood, endocarditis, osteomyelitis | P5CR | PRODH, P5CDH | Na+-symporter (PutP) | Carbon/nitrogen/energy | (90, 206, 207, 232) |
| H+-symporter | Osmoregulation | |||||
| Stress resistance | ||||||
| Trypanosoma spp. | Chagas disease; African sleeping sickness; leishmaniasis | GK, GPR (varies with species) | PRODH, P5CDH | H+/ATP-dependent transporter | Carbon/nitrogen/energy | (29, 35, 48, 145, 146, 173, 196, 202, 210) |
| Proline/alanine transporter | Osmoregulation | |||||
| Leishmania spp. | P5CR | Stress resistance | ||||
| Proline permease (amino acid/auxin permease) | Cell differentiation |
GK, γ-glutamyl kinase; GPR, γ-glutamyl phosphate reductase; P5CDH, Δ1-pyrroline-5-carboxylate dehydrogenase; P5CR, Δ1-pyrroline-5-carboxylate reductase; PRODH, proline dehydrogenase; PutA, proline utilization A.
Future Directions
Physiology of host/pathogen interactions
Understanding the mechanisms by which pathogens acquire nutrients during infection and survive host innate immunity systems will continue to be an important area of future research (261). Amino acids such as proline are key sources of nitrogen and carbon for pathogens. Some pathogens, such as T. brucei, must import amino acids from the host, including proline (144, 147), whereas others, such as M. tuberculosis, are self-reliant in producing amino acids, including synthesizing their own proline (46, 261). The capacity of a host to limit nutrient availability, which includes amino acids such as proline, is an important defense mechanism (261). The significant attenuation in virulence of the M. tuberculosis proC strain, which is auxotrophic for proline, demonstrates this defense strategy (214).
An intriguing area of research is to explore how pathogens are able to avoid amino acid starvation and in some cases increase host production or transport of amino acids into the host cell (261). In addition, understanding the alternative sources of carbon and nitrogen that pathogens utilize is critical. Although not highlighted in this review, proline has been proposed in Campylobacter jejuni to be a secondary carbon/nitrogen source used after aspartate, asparagine, and serine pools have been drained (252). Interestingly in C. jejuni, a significant upregulation of putA and putP expression at stationary phase was observed to coincide with a large increase in proline utilization (252). Whether proline metabolism is important for C. jejuni survival in the host will need to be determined.
Other factors of pathogenesis that need to be further investigated include examining the effect of proline in the host diet and competition between commensal and pathogenic bacteria. Momose and colleagues have shown that nonpathogenic, proline-consuming strains of E. coli were able to suppress the growth of pathogenic E. coli O157:H7 (154). This suppression was reversed through the addition of proline to the growth media, implying that competition for proline consumption between pathogenic and nonpathogenic strains of E. coli may lead to poor growth and increased elimination of the pathogenic E. coli (154). Thus, dietary proline is a potential environmental factor that should be considered when examining the host microbiota and pathogen colonization (7). This seems especially relevant to the colonization and carcinogenesis of H. pylori, which exhibits considerable pathogenic variability depending on the genetic makeup of the particular strain and human host, and environmental factors such as diet (7).
Also, evidence was reported for the entomopathogen P. asymbiotica being able to infect human cells (47). Different strain isolates of P. asymbiotica from humans have been found, which are able to induce apoptosis in human macrophage cell lines (47). These findings highlight the need to understand more about the biology of entomopathogenic bacteria and the role of proline metabolism to combat these new emerging human pathogens. Clearly, additional investigations are needed to fully define the mechanisms and magnitude by which proline enhances virulence of different pathogenic organisms.
Drug design and targets
Gaining a better understanding of how proline metabolism is involved in the infection and survival of pathogens will be important for improving treatments of infectious diseases.
Targeting unique aspects of proline utilization in prokaryotic and eukaryotic pathogens may enhance selectivity of antibiotics and avoid toxic effects in the host. Structural studies by Tanner's group of PutAs and monofunctional PRODH enzymes strongly indicate that the active site structure and overall fold are similar between bacterial and human enzymes (133, 243). It should be noted, however, that a structure of a eukaryotic PRODH, including human, has not been reported yet. Likewise, bacterial and human P5CDH (or ALDH4A1) share a common fold (133, 216) and are part of the aldehyde dehydrogenase superfamily.
The conservation of active site residues in PRODH and P5CDH enzymes was examined by performing a sequence alignment of the PRODH, P5CDH, and PutA enzymes from the pathogens highlighted in this review. The multiple sequence alignments of the PRODH and P5CDH active sites are shown in Figure 12. For PRODH and P5CDH, key residues in both active sites are identical or highly conserved between pathogenic and human enzymes, denoted in Figure 12 by red arrows. For example, in PRODH, two tandem Arg residues (R556 and R556 in PutA from E. coli), which are required for proline binding in bacterial PutA and PRODH enzymes, are identical in human PRODH (133).
FIG. 12.
Results from multiple sequence alignment of PRODH, P5CDH, and PutA enzymes from pathogenic organisms. Alignment was computed using T-Coffee computing alignment server and the resulting alignment visualized using WebLogo software. The resulting WebLogo images were altered so that residue numbering is according to PutA from E. coli. Shown are the alignments of the PRODH and P5CDH catalytic sites. The GenBank accession numbers of the aligned PutA sequences are E. coli PutA (AAB9985), Helicobacter pylori PutA (WP_001169408.1), E. chaffeensis PutA (WP_011452748.1), Pseudomonas putida PutA (WP_043207299.1), Samonella enterica PutA (WP_050954140.1), Photorhabdus luminescens PutA (WP_036807552.1), Xenorhabdus nematophila PutA (WP_010848062.1), M. tuberculosis PRODH (WP_031710787.1) and P5CDH (EFO75557.1), Trypanosoma cruzi PRODH (ADZ31479.1) and P5CDH (CDG15436.1), Trypanosoma brucei PRODH (XP_845645.1) and P5CDH (XP_822573.1), C. neoformans PRODH (XP_571179.1) and P5CDH (XP_572420.1), and S. aureus PRODH (WP_031824363.1) and P5CDH (AGU62685.1). Red arrows indicate residues that are identical or highly conserved in human PRODH (AAD24775.1) and P5CDH (P30038.3) enzymes. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
In the P5CDH active site, an Arg/Lys-Cys-Ser-Ala motif on the catalytic loop (R916, C917, S918, and A919 in PutA from E. coli) is conserved between pathogenic PutA/P5CDHs and human P5CDH. The Cys residue on this loop is the catalytic nucleophile in the aldehyde reaction and the adjacent Arg (Lys347 in human P5CDH) and Ser residues form water-mediated and direct hydrogen bonds, respectively, to the α-carboxylate of glutamate (216). Thus, it is likely that compounds that target only the PRODH and P5CDH catalytic sites will not specifically inhibit pathogenic enzymes. Illustrating this is that l-tetrahydro-2-furoic acid, a proline analog (124, 266), has been shown to inhibit PutA (158) and human PRODH enzymes (224), and proline metabolism in cell cultures (71, 166, 257). To design specific inhibitors of proline metabolism in pathogenic organisms, properties that are unique to pathogenic enzymes will need to be found by future structural and kinetic studies. For a complete analysis of PRODH and P5CDH structural biology, readers are referred to the review by Tanner in this Forum Issue (225).
A more promising approach, at least for gram-negative pathogens, may be to develop allosteric inhibitors that target the channeling step in PutAs. These types of inhibitors would selectively block the use of proline as a carbon or nitrogen source in gram-negative bacterial pathogens. This approach would be similar to the allosteric inhibition recently reported for tryptophan synthase in M. tuberculosis. A small-molecule inhibitor was discovered to bind to the substrate channel in tryptophan synthase and kill M. tuberculosis (240). Additional work is needed to establish if substrate channeling is present in pathogenic organisms that contain monofunctional PRODH and P5CDH enzymes. If so, perhaps small molecules that block PRODH/P5CDH functional interactions would prevent proline utilization and lead to toxic accumulation of P5C in the pathogenic organism.
Continued studies of proline transport are also needed to guide future drug design. Recently, Jung and colleagues identified residues that are critical for external gate closure of PutP, potentially providing new insights into how the proline transport cycle could be specifically inhibited in bacteria (26). Specific inhibitors of proline transport in trypanosomes would potentially impede the infectious life cycle of these parasites by limiting growth and osmotic stress protection. Another strategy to prevent proline utilization in pathogens would be to use RNA interference or novel antisense synthetic oligomers to silence proline metabolic gene expression (219). Also, the possibility of using proline analogs as antimicrobials needs to be further pursued (57). Deutch and colleagues have shown the viability of this approach using the proline analog, L-selenazolidine-4-carboxylic acid (or, L-selenaproline), which they found inhibits the growth of the uropathogenic E. coli strain CFT073 (63, 64). Selenazolidine compounds, such as L-selenaproline, have been shown not to have toxic effects in mice when supplemented in the diet and were suggested to be viable compounds for cancer therapy and delivery of selenium (128). The potential use of L-selenaproline as an antimicrobial compound against urinary tract infection thus seems worthwhile to pursue.
Acknowledgment
This work was supported, in part, by grants P30GM103335, R01GM061068, and T32 GM008396 from the National Institutes of Health.
Abbreviations Used
- AGAT
alanine-glyoxylate transaminase
- CcpA
catabolite control protein A
- CCR
carbon catabolite repression
- CRP
cyclic AMP receptor protein
- FAD
flavin adenine dinucleotide
- Fur
ferric uptake regulator
- GK
γ-glutamyl kinase
- GPR
γ-glutamyl phosphate reductase
- GSA
glutamate-γ-semialdehyde
- Lrp
leucine-responsive regulatory protein
- NAC
nitrogen assimilation control
- NAD+
nicotinamide adenine dinucleotide
- NADH
nicotinamide adenine dinucleotide
- NCR
nitrogen catabolite repression
- OAT
ornithine aminotransferase
- P5C
Δ1-pyrroline-5-carboxylate
- P5CDH
Δ1-pyrroline-5-carboxylate dehydrogenase
- P5CR
Δ1-pyrroline-5-carboxylate reductase
- P5CS
Δ1-pyrroline-5-carboxylate synthetase
- PE
Pro-Glu
- PLP
pyridoxal 5′-phosphate
- PPE
Pro-Pro-Glu
- PR
d-proline reductase
- PRODH
proline dehydrogenase
- PutA
proline utilization A
- RHH
ribbon-helix-helix
- ROS
reactive oxygen species
- TCA
tricarboxylic acid
References
- 1. Nearly half a million Americans suffered from Clostridium difficile infections in a single year. 2015. www.cdc.gov/media/releases/2015/p0225-clostridium-difficile.html (last accessed January19, 2018)
- 2. Adams E. and Frank L. Metabolism of proline and the hydroxyprolines. Annu Rev Biochem 49: 1005–1061, 1980 [DOI] [PubMed] [Google Scholar]
- 3. Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, and Aksoy S. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet 32: 402–407, 2002 [DOI] [PubMed] [Google Scholar]
- 4. Alreshidi MM, Dunstan RH, Gottfries J, Macdonald MM, Crompton MJ, Ang CS, Williamson NA, and Roberts TK. Changes in the cytoplasmic composition of amino acids and proteins observed in Staphylococcus aureus during growth under variable growth conditions representative of the human wound site. PLoS One 11: e0159662, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Altendorf K, Booth IR, Gralla J, Greie JC, Rosenthal AZ, and Wood JM. Osmotic stress. EcoSal Plus 3: 1–41, 2009 [DOI] [PubMed] [Google Scholar]
- 6. Alteri CJ, Smith SN, and Mobley HL. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog 5: e1000448, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amieva M. and Peek RM., Jr Pathobiology of Helicobacter pylori-induced gastric cancer. Gastroenterology 150: 64–78, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andreesen JR, Wagner M, Sonntag D, Kohlstock M, Harms C, Gursinsky T, Jager J, Parther T, Kabisch U, Grantzdorffer A, Pich A, and Sohling B. Various functions of selenols and thiols in anaerobic gram-positive, amino acids-utilizing bacteria. Biofactors 10: 263–270, 1999 [DOI] [PubMed] [Google Scholar]
- 9. Arentson BW, Hayes EL, Zhu W, Singh H, Tanner JJ, and Becker DF. Engineering a trifunctional proline utilization A chimaera by fusing a DNA-binding domain to a bifunctional PutA. Biosci Rep 36: 1–11, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arentson BW, Luo M, Pemberton TA, Tanner JJ, and Becker DF. Kinetic and structural characterization of tunnel-perturbing mutants in Bradyrhizobium japonicum proline utilization A. Biochemistry 53: 5150–5161, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arentson BW, Sanyal N, and Becker DF. Substrate channeling in proline metabolism. Front Biosci (Landmark Ed) 17: 375–388, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Atkinson MR, Wray LV, Jr, and Fisher SH. Regulation of histidine and proline degradation enzymes by amino acid availability in Bacillus subtilis. J Bacteriol 172: 4758–4765, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bae JH. and Miller KJ. Identification of two proline transport systems in Staphylococcus aureus and their possible roles in osmoregulation. Appl Environ Microbiol 58: 471–475, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beck T. and Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692, 1999 [DOI] [PubMed] [Google Scholar]
- 15. Becker DF. and Thomas EA. Redox properties of the PutA protein from Escherichia coli and the influence of the flavin redox state on PutA-DNA interactions. Biochemistry 40: 4714–4721, 2001 [DOI] [PubMed] [Google Scholar]
- 16. Becker DF, Zhu W, and Moxley MA. Flavin redox switching of protein functions. Antioxid Redox Signal 14: 1079–1091, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Belitsky BR. Indirect repression by Bacillus subtilis CodY via displacement of the activator of the proline utilization operon. J Mol Biol 413: 321–336, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ben Rejeb K, Abdelly C, and Savouré A. How reactive oxygen species and proline face stress together. Plant Physiol Biochem 80: 278–284, 2014 [DOI] [PubMed] [Google Scholar]
- 19. Bender RA. A NAC for regulating metabolism: the nitrogen assimilation control protein (NAC) from Klebsiella pneumoniae. J Bacteriol 192: 4801–4811, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berney M. and Cook GM. Unique flexibility in energy metabolism allows mycobacteria to combat starvation and hypoxia. PLoS One 5: e8614, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Berney M, Weimar MR, Heikal A, and Cook GM. Regulation of proline metabolism in Mycobacteria and its role in carbon metabolism under hypoxia. Mol Microbiol 84: 664–681, 2012 [DOI] [PubMed] [Google Scholar]
- 22. Bode HB. Entomopathogenic bacteria as a source of secondary metabolites. Curr Opin Chem Biol 13: 224–230, 2009 [DOI] [PubMed] [Google Scholar]
- 23. Bode HB, Brachmann AO, Jadhav KB, Seyfarth L, Dauth C, Fuchs SW, Kaiser M, Waterfield NR, Sack H, Heinemann SH, and Arndt HD. Structure elucidation and activity of kolossin A, the d-/l-pentadecapeptide product of a giant nonribosomal peptide synthetase. Angew Chem Int Ed Engl 54: 10352–10355, 2015 [DOI] [PubMed] [Google Scholar]
- 24. Boonjakuakul JK, Canfield DR, and Solnick JV. Comparison of Helicobacter pylori virulence gene expression in vitro and in the Rhesus macaque. Infect Immun 73: 4895–4904, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bouillaut L, Self WT, and Sonenshein AL. Proline-dependent regulation of Clostridium difficile stickland metabolism. J Bacteriol 195: 844–854, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bracher S, Guerin K, Polyhach Y, Jeschke G, Dittmer S, Frey S, Bohm M, and Jung H. Glu-311 in external loop 4 of the sodium/proline transporter PutP is crucial for external gate closure. J Biol Chem 291: 4998–5008, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brennan MJ. The enigmatic PE/PPE multigene family of Mycobacteria and Tuberculosis vaccination. Infect Immun 85: 1–8, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brill J, Hoffmann T, Bleisteiner M, and Bremer E. Osmotically controlled synthesis of the compatible solute proline is critical for cellular defense of Bacillus subtilis against high osmolarity. J Bacteriol 193: 5335–5346, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bringaud F, Barrett MP, and Zilberstein D. Multiple roles of proline transport and metabolism in trypanosomatids. Front Biosci (Landmark Ed) 17: 349–374, 2012 [DOI] [PubMed] [Google Scholar]
- 30. Brinkman AB, Ettema TJ, de Vos WM, and van der Oost J. The Lrp family of transcriptional regulators. Mol Microbiol 48: 287–294, 2003 [DOI] [PubMed] [Google Scholar]
- 31. Brinsmade SR, Alexander EL, Livny J, Stettner AI, Segre D, Rhee KY, and Sonenshein AL. Hierarchical expression of genes controlled by the Bacillus subtilis global regulatory protein CodY. Proc Natl Acad Sci U S A 111: 8227–8232, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brown ED. and Wood JM. Conformational change and membrane association of the PutA protein are coincident with reduction of its FAD cofactor by proline. J Biol Chem 268: 8972–8979, 1993 [PubMed] [Google Scholar]
- 33. Brown ED. and Wood JM. Redesigned purification yields a fully functional PutA protein dimer from Escherichia coli. J Biol Chem 267: 13086–13092, 1992 [PubMed] [Google Scholar]
- 34. Bursell E. Aspects of the metabolism of amino acids in the tsetse fly, Glossina (Diptera). J Insect Physiol 9: 439–452, 1963 [Google Scholar]
- 35. Bursell E. The role of proline in energy metabolism. In: Energy Metabolism in Insects, edited by Downer R, ed. Springer, Boston, MA, 1981, pp. 135–154 [Google Scholar]
- 36. Casanova-Torres AM, Shokal U, Morag N, Eleftherianos I, and Goodrich-Blair H. The global transcription factor Lrp is both essential for and inhibitory to Xenorhabdus nematophila insecticidal activity. Appl Environ Microbiol 83: 1–12, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Caudill MT, Budnick JA, Sheehan LM, Lehman CR, Purwantini E, Mukhopadhyay B, and Caswell CC. Proline utilization system is required for infection by the pathogenic alpha-proteobacterium Brucella abortus. Microbiology 163: 970–979, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chamond N, Goytia M, Coatnoan N, Barale JC, Cosson A, Degrave WM, and Minoprio P. Trypanosoma cruzi proline racemases are involved in parasite differentiation and infectivity. Mol Microbiol 58: 46–60, 2005 [DOI] [PubMed] [Google Scholar]
- 39. Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, Anderson AB, Grissom JE, Laux DC, Cohen PS, and Conway T. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A 101: 7427–7432, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chaulk SG, Smith Frieday MN, Arthur DC, Culham DE, Edwards RA, Soo P, Frost LS, Keates RA, Glover JN, and Wood JM. ProQ is an RNA chaperone that controls ProP levels in Escherichia coli. Biochemistry 50: 3095–3106, 2011 [DOI] [PubMed] [Google Scholar]
- 41. Chen LM. and Maloy S. Regulation of proline utilization in enteric bacteria: cloning and characterization of the Klebsiella put control region. J Bacteriol 173: 783–790, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cheng Z, Lin M, and Rikihisa Y. Ehrlichia chaffeensis proliferation begins with NtrY/NtrX and PutA/GlnA upregulation and CtrA degradation induced by proline and glutamine uptake. MBio 5: e02141, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cheng Z, Miura K, Popov VL, Kumagai Y, and Rikihisa Y. Insights into the CtrA regulon in development of stress resistance in obligatory intracellular pathogen Ehrlichia chaffeensis. Mol Microbiol 82: 1217–1234, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Choudhary S, Kishore N, and Hosur RV. Inhibition of insulin fibrillation by osmolytes: mechanistic insights. Sci Rep 5: 17599, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Christensen EM, Patel SM, Korasick DA, Campbell AC, Krause KL, Becker DF, and Tanner JJ. Resolving the cofactor binding site in the proline biosynthetic enzyme human pyrroline-5-carboxylate reductase 1. J Biol Chem 292: 7233–7243, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, and Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393: 537–544, 1998 [DOI] [PubMed] [Google Scholar]
- 47. Costa SC, Girard PA, Brehelin M, and Zumbihl R. The emerging human pathogen Photorhabdus asymbiotica is a facultative intracellular bacterium and induces apoptosis of macrophage-like cells. Infect Immun 77: 1022–1030, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Coustou V, Biran M, Breton M, Guegan F, Riviere L, Plazolles N, Nolan D, Barrett MP, Franconi JM, and Bringaud F. Glucose-induced remodeling of intermediary and energy metabolism in procyclic Trypanosoma brucei. J Biol Chem 283: 16342–16354, 2008 [DOI] [PubMed] [Google Scholar]
- 49. Coutinho L, Ferreira MA, Cosson A, Batista MM, Batista DG, Minoprio P, Degrave WM, Berneman A, and Soeiro MN. Inhibition of Trypanosoma cruzi proline racemase affects host-parasite interactions and the outcome of in vitro infection. Mem Inst Oswaldo Cruz 104: 1055–1062, 2009 [DOI] [PubMed] [Google Scholar]
- 50. Crawford JM, Kontnik R, and Clardy J. Regulating alternative lifestyles in entomopathogenic bacteria. Curr Biol 20: 69–74, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Csonka LN. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev 53: 121–147, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Csonka LN. Proline over-production results in enhanced osmotolerance in Salmonella typhimurium. Mol Gen Genet 182: 82–86, 1981 [DOI] [PubMed] [Google Scholar]
- 53. Csonka LN. and Hanson AD. Prokaryotic osmoregulation: genetics and physiology. Annu Rev Microbiol 45: 569–606, 1991 [DOI] [PubMed] [Google Scholar]
- 54. Csonka LN. and Leisinger T. Biosynthesis of proline. EcoSal Plus. 2: 1–20, 2007 [DOI] [PubMed] [Google Scholar]
- 55. Culham DE, Dalgado C, Gyles CL, Mamelak D, MacLellan S, and Wood JM. Osmoregulatory transporter ProP influences colonization of the urinary tract by Escherichia coli. Microbiology 144 (Pt 1): 91–102, 1998 [DOI] [PubMed] [Google Scholar]
- 56. Culham DE, Henderson J, Crane RA, and Wood JM. Osmosensor ProP of Escherichia coli responds to the concentration, chemistry, and molecular size of osmolytes in the proteoliposome lumen. Biochemistry 42: 410–420, 2003 [DOI] [PubMed] [Google Scholar]
- 57. Culham DE, Lasby B, Marangoni AG, Milner JL, Steer BA, van Nues RW, and Wood JM. Isolation and sequencing of Escherichia coli gene proP reveals unusual structural features of the osmoregulatory proline/betaine transporter, ProP. J Mol Biol 229: 268–276, 1993 [DOI] [PubMed] [Google Scholar]
- 58. Culham DE, Shkel IA, Record MT, Jr, and Wood JM. Contributions of Coulombic and Hofmeister effects to the osmotic activation of Escherichia coli transporter ProP. Biochemistry 55: 1301–1313, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cunningham TS, Svetlov VV, Rai R, Smart W, and Cooper TG. G1n3p is capable of binding to UAS(NTR) elements and activating transcription in Saccharomyces cerevisiae. J Bacteriol 178: 3470–3479, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dattananda CS, Rajkumari K, and Gowrishankar J. Multiple mechanisms contribute to osmotic inducibility of proU operon expression in Escherichia coli: demonstration of two osmoresponsive promoters and of a negative regulatory element within the first structural gene. J Bacteriol 173: 7481–7490, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Daugherty JR, Rai R, el Berry HM, and Cooper TG. Regulatory circuit for responses of nitrogen catabolic gene expression to the GLN3 and DAL80 proteins and nitrogen catabolite repression in Saccharomyces cerevisiae. J Bacteriol 175: 64–73, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. DeJesus MA, Gerrick ER, Xu W, Park SW, Long JE, Boutte CC, Rubin EJ, Schnappinger D, Ehrt S, Fortune SM, Sassetti CM, and Ioerger TR. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. MBio 8: 1–17, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Deutch CE, Arballo ME, Cooks LN, Gomes JM, Williams TM, Aboul-Fadl T, and Roberts JC. Susceptibility of Escherichia coli to l-selenaproline and other l-proline analogues in laboratory culture media and normal human urine. Lett Appl Microbiol 43: 392–398, 2006 [DOI] [PubMed] [Google Scholar]
- 64. Deutch CE, Spahija I, and Wagner CE. Susceptibility of Escherichia coli to the toxic l-proline analogue l-selenaproline is dependent on two l-cystine transport systems. J Appl Microbiol 117: 1487–1499, 2014 [DOI] [PubMed] [Google Scholar]
- 65. Donald SP, Sun XY, Hu CA, Yu J, Mei JM, Valle D, and Phang JM. Proline oxidase, encoded by p53-induced gene-6, catalyzes the generation of proline-dependent reactive oxygen species. Cancer Res 61: 1810–1815, 2001 [PubMed] [Google Scholar]
- 66. Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, Taourit S, Bocs S, Boursaux-Eude C, Chandler M, Charles JF, Dassa E, Derose R, Derzelle S, Freyssinet G, Gaudriault S, Medigue C, Lanois A, Powell K, Siguier P, Vincent R, Wingate V, Zouine M, Glaser P, Boemare N, Danchin A, and Kunst F. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotechnol 21: 1307–1313, 2003 [DOI] [PubMed] [Google Scholar]
- 67. Dumler JS. and Bakken JS. Ehrlichial diseases of humans: emerging tick-borne infections. Clin Infect Dis 20: 1102–1110, 1995 [DOI] [PubMed] [Google Scholar]
- 68. Ebikeme C, Hubert J, Biran M, Gouspillou G, Morand P, Plazolles N, Guegan F, Diolez P, Franconi JM, Portais JC, and Bringaud F. Ablation of succinate production from glucose metabolism in the procyclic trypanosomes induces metabolic switches to the glycerol 3-phosphate/dihydroxyacetone phosphate shuttle and to proline metabolism. J Biol Chem 285: 32312–32324, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ehlers RU. Mass production of entomopathogenic nematodes for plant protection. Appl Microbiol Biotechnol 56: 623–633, 2001 [DOI] [PubMed] [Google Scholar]
- 70. Ehrt S, Rhee K, and Schnappinger D. Mycobacterial genes essential for the pathogen's survival in the host. Immunol Rev 264: 319–326, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Elia I, Broekaert D, Christen S, Boon R, Radaelli E, Orth MF, Verfaillie C, Grunewald TGP, and Fendt SM. Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells. Nat Commun 8: 15267, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Evans DA. and Brown RC. The utilization of glucose and proline by culture forms of Trypanosoma brucei. J Protozool 19: 686–690, 1972 [DOI] [PubMed] [Google Scholar]
- 73. Fedorko DP. and Williams EC. Use of cycloserine-cefoxitin-fructose agar and l-proline-aminopeptidase (PRO Discs) in the rapid identification of Clostridium difficile. J Clin Microbiol 35: 1258–1259, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fichman Y, Gerdes SY, Kovacs H, Szabados L, Zilberstein A, and Csonka LN. Evolution of proline biosynthesis: enzymology, bioinformatics, genetics, and transcriptional regulation. Biol Rev Camb Philos Soc 90: 1065–1099, 2015 [DOI] [PubMed] [Google Scholar]
- 75. Finn S, Handler K, Condell O, Colgan A, Cooney S, McClure P, Amezquita A, Hinton JC, and Fanning S. ProP is required for the survival of desiccated Salmonella enterica serovar Typhimurium cells on a stainless steel surface. Appl Environ Microbiol 79: 4376–4384, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fonknechten N, Chaussonnerie S, Tricot S, Lajus A, Andreesen JR, Perchat N, Pelletier E, Gouyvenoux M, Barbe V, Salanoubat M, Le Paslier D, Weissenbach J, Cohen GN, and Kreimeyer A. Clostridium sticklandii, a specialist in amino acid degradation: revisiting its metabolism through its genome sequence. BMC Genomics 11: 555, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Forman D, Newell DG, Fullerton F, Yarnell JW, Stacey AR, Wald N, and Sitas F. Association between infection with Helicobacter pylori and risk of gastric cancer: evidence from a prospective investigation. BMJ 302: 1302–1305, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Forst S, Dowds B, Boemare N, and Stackebrandt E. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu Rev Microbiol 51: 47–72, 1997 [DOI] [PubMed] [Google Scholar]
- 79. Garneau-Tsodikova S, Dorrestein PC, Kelleher NL, and Walsh CT. Protein assembly line components in prodigiosin biosynthesis: characterization of PigA,G,H,I,J. J Am Chem Soc 128: 12600–12601, 2006 [DOI] [PubMed] [Google Scholar]
- 80. Gorke B. and Stulke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6: 613–624, 2008 [DOI] [PubMed] [Google Scholar]
- 81. Goss TJ. and Bender RA. The nitrogen assimilation control protein, NAC, is a DNA binding transcription activator in Klebsiella aerogenes. J Bacteriol 177: 3546–3555, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Graham JE. and Wilkinson BJ. Staphylococcus aureus osmoregulation: roles for choline, glycine betaine, proline, and taurine. J Bacteriol 174: 2711–2716, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gu D, Zhou Y, Kallhoff V, Baban B, Tanner JJ, and Becker DF. Identification and characterization of the DNA-binding domain of the multifunctional PutA flavoenzyme. J Biol Chem 279: 31171–31176, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gul N. and Poolman B. Functional reconstitution and osmoregulatory properties of the ProU ABC transporter from Escherichia coli. Mol Membr Biol 30: 138–148, 2013 [DOI] [PubMed] [Google Scholar]
- 85. Guszczyn T. and Sobolewski K. Deregulation of collagen metabolism in human stomach cancer. Pathobiology 71: 308–313, 2004 [DOI] [PubMed] [Google Scholar]
- 86. Haardt M, Kempf B, Faatz E, and Bremer E. The osmoprotectant proline betaine is a major substrate for the binding-protein-dependent transport system ProU of Escherichia coli K-12. Mol Gen Genet 246: 783–786, 1995 [DOI] [PubMed] [Google Scholar]
- 87. Hahn DR, Myers RS, Kent CR, and Maloy SR. Regulation of proline utilization in Salmonella typhimurium: molecular characterization of the put operon, and DNA sequence of the put control region. Mol Gen Genet 213: 125–133, 1988 [DOI] [PubMed] [Google Scholar]
- 88. Hall JC. and O'Toole E. Intestinal flora in new-born infants with a description of a new pathogenic anaerobe, Bacillus difficilis. Am J Dis Child 49: 390–402, 1935 [Google Scholar]
- 89. Halouska S, Zhou Y, Becker DF, and Powers R. Solution structure of the Pseudomonas putida protein PpPutA45 and its DNA complex. Proteins 75: 12–27, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Halsey CR, Lei S, Wax JK, Lehman MK, Nuxoll AS, Steinke L, Sadykov M, Powers R, and Fey PD. Amino acid catabolism in Staphylococcus aureus and the function of carbon catabolite repression. MBio 8: 1–19, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hampel A, Huber C, Geffers R, Spona-Friedl M, Eisenreich W, and Bange FC. Mycobacterium tuberculosis is a natural ornithine aminotransferase (rocD) mutant and depends on Rv2323c for growth on arginine. PLoS One 10: e0136914, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hofman-Bang J. Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol Biotechnol 12: 35–73, 1999 [DOI] [PubMed] [Google Scholar]
- 93. Hryckowian AJ, Pruss KM, and Sonnenburg JL. The emerging metabolic view of Clostridium difficile pathogenesis. Curr Opin Microbiol 35: 42–47, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hu J. and Torres AG. Enteropathogenic Escherichia coli: foe or innocent bystander? Clin Microbiol Infect 21: 729–734, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Huang SC, Lin TH, and Shaw GC. PrcR, a PucR-type transcriptional activator, is essential for proline utilization and mediates proline-responsive expression of the proline utilization operon putBCP in Bacillus subtilis. Microbiology 157: 3370–3377, 2011 [DOI] [PubMed] [Google Scholar]
- 96. Huang TC, Huang YW, Hung HJ, Ho CT, and Wu ML. Delta1-pyrroline-5-carboxylic acid formed by proline dehydrogenase from the Bacillus subtilis ssp. natto expressed in Escherichia coli as a precursor for 2-acetyl-1-pyrroline. J Agric Food Chem 55: 5097–5102, 2007 [DOI] [PubMed] [Google Scholar]
- 97. Ignatova Z. and Gierasch LM. Inhibition of protein aggregation in vitro and in vivo by a natural osmoprotectant. Proc Natl Acad Sci U S A 103: 13357–13361, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Inbar E, Schlisselberg D, Suter Grotemeyer M, Rentsch D, and Zilberstein D. A versatile proline/alanine transporter in the unicellular pathogen Leishmania donovani regulates amino acid homoeostasis and osmotic stress responses. Biochem J 449: 555–566, 2013 [DOI] [PubMed] [Google Scholar]
- 99. Jackson S, Calos M, Myers A, and Self WT. Analysis of proline reduction in the nosocomial pathogen Clostridium difficile. J Bacteriol 188: 8487–8495, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Jung H, Hilger D, and Raba M. The Na(+)/l-proline transporter PutP. Front Biosci (Landmark Ed) 17: 745–759, 2012 [DOI] [PubMed] [Google Scholar]
- 101. Kaakoush NO, Kovach Z, and Mendz GL. Potential role of thiol:disulfide oxidoreductases in the pathogenesis of Helicobacter pylori. FEMS Immunol Med Microbiol 50: 177–183, 2007 [DOI] [PubMed] [Google Scholar]
- 102. Kabisch UC, Grantzdorffer A, Schierhorn A, Rucknagel KP, Andreesen JR, and Pich A. Identification of d-proline reductase from Clostridium sticklandii as a selenoenzyme and indications for a catalytically active pyruvoyl group derived from a cysteine residue by cleavage of a proprotein. J Biol Chem 274: 8445–8454, 1999 [DOI] [PubMed] [Google Scholar]
- 103. Karasawa T, Ikoma S, Yamakawa K, and Nakamura S. A defined growth medium for Clostridium difficile. Microbiology 141 (Pt 2): 371–375, 1995 [DOI] [PubMed] [Google Scholar]
- 104. Kavalchuk K, Madhusudan S, and Schnetz K. RNase III initiates rapid degradation of proU mRNA upon hypo-osmotic stress in Escherichia coli. RNA Biol 9: 98–109, 2012 [DOI] [PubMed] [Google Scholar]
- 105. Kavermann H, Burns BP, Angermuller K, Odenbreit S, Fischer W, Melchers K, and Haas R. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J Exp Med 197: 813–822, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kempf B. and Bremer E. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch Microbiol 170: 319–330, 1998 [DOI] [PubMed] [Google Scholar]
- 107. Keuntje B, Masepohl B, and Klipp W. Expression of the putA gene encoding proline dehydrogenase from Rhodobacter capsulatus is independent of NtrC regulation but requires an Lrp-like activator protein. J Bacteriol 177: 6432–6439, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kmetzsch L, Staats CC, Simon E, Fonseca FL, Oliveira DL, Joffe LS, Rodrigues J, Lourenco RF, Gomes SL, Nimrichter L, Rodrigues ML, Schrank A, and Vainstein MH. The GATA-type transcriptional activator Gat1 regulates nitrogen uptake and metabolism in the human pathogen Cryptococcus neoformans. Fungal Genet Biol 48: 192–199, 2011 [DOI] [PubMed] [Google Scholar]
- 109. Korasick D, Gamage TT, Christgen S, Stiers KM, Beamer LJ, Henzl MT, Becker DF, and Tanner JJ. Structure and characterization of a class 3B proline utilization A: ligand-induced dimerization and importance of the C-terminal domain for catalysis. J Biol Chem 292: 9652–9665, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Krishnan N. and Becker DF. Oxygen reactivity of PutA from Helicobacter species and proline-linked oxidative stress. J Bacteriol 188: 1227–1235, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Krishnan N, Dickman MB, and Becker DF. Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic Biol Med 44: 671–681, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Krishnan N, Doster AR, Duhamel GE, and Becker DF. Characterization of a Helicobacter hepaticus putA mutant strain in host colonization and oxidative stress. Infect Immun 76: 3037–3044, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kuipers E, Thijs JC, and Festen HP. The prevalence of Helicobacter pylori in peptic ulcer disease. Aliment Pharmacol Ther 9: 59–69, 1995 [PubMed] [Google Scholar]
- 114. Kulkarni AA, Abul-Hamd AT, Rai R, El Berry H, and Cooper TG. Gln3p nuclear localization and interaction with Ure2p in Saccharomyces cerevisiae. J Biol Chem 276: 32136–32144, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kumagai Y, Cheng Z, Lin M, and Rikihisa Y. Biochemical activities of three pairs of Ehrlichia chaffeensis two-component regulatory system proteins involved in inhibition of lysosomal fusion. Infect Immun 74: 5014–5022, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Kumar TK, Samuel D, Jayaraman G, Srimathi T, and Yu C. The role of proline in the prevention of aggregation during protein folding in vitro. Biochem Mol Biol Int 46: 509–517, 1998 [DOI] [PubMed] [Google Scholar]
- 117. Kupferschmied P, Maurhofer M, and Keel C. Promise for plant pest control: root-associated pseudomonads with insecticidal activities. Front Plant Sci 4: 287, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Lagautriere T, Bashiri G, Paterson NG, Berney M, Cook GM, and Baker EN. Characterization of the proline-utilization pathway in Mycobacterium tuberculosis through structural and functional studies. Acta Crystallogr D Biol Crystallogr 70: 968–980, 2014 [DOI] [PubMed] [Google Scholar]
- 119. Lang G, Kalvelage T, Peters A, Wiese J, and Imhoff JF. Linear and cyclic peptides from the entomopathogenic bacterium Xenorhabdus nematophilus. J Nat Prod 71: 1074–1077, 2008 [DOI] [PubMed] [Google Scholar]
- 120. Larson JD, Jenkins JL, Schuermann JP, Zhou Y, Becker DF, and Tanner JJ. Crystal structures of the DNA-binding domain of Escherichia coli proline utilization A flavoprotein and analysis of the role of Lys9 in DNA recognition. Protein Sci 15: 2630–2641, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Lee IR, Chow EW, Morrow CA, Djordjevic JT, and Fraser JA. Nitrogen metabolite repression of metabolism and virulence in the human fungal pathogen Cryptococcus neoformans. Genetics 188: 309–323, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Lee IR, Lui EY, Chow EW, Arras SD, Morrow CA, and Fraser JA. Reactive oxygen species homeostasis and virulence of the fungal pathogen Cryptococcus neoformans requires an intact proline catabolism pathway. Genetics 194: 421–433, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lee JH. and Choi SH. Coactivation of Vibrio vulnificus putAP operon by cAMP receptor protein and PutR through cooperative binding to overlapping sites. Mol Microbiol 60: 513–524, 2006 [DOI] [PubMed] [Google Scholar]
- 124. Lee YH, Nadaraia S, Gu D, Becker DF, and Tanner JJ. Structure of the proline dehydrogenase domain of the multifunctional PutA flavoprotein. Nat Struct Biol 10: 109–114, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Lerma-Ortiz C, Jeffryes JG, Cooper AJ, Niehaus TD, Thamm AM, Frelin O, Aunins T, Fiehn O, de Crecy-Lagard V, Henry CS, and Hanson AD. “Nothing of chemistry disappears in biology”: the top 30 damage-prone endogenous metabolites. Biochem Soc Trans 44: 961–971, 2016 [DOI] [PubMed] [Google Scholar]
- 126. Leverentz MK, Campbell RN, Connolly Y, Whetton AD, and Reece RJ. Mutation of a phosphorylatable residue in Put3p affects the magnitude of rapamycin-induced PUT1 activation in a Gat1p-dependent manner. J Biol Chem 284: 24115–24122, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Levin BJ, Huang YY, Peck SC, Wei Y, Martinez-Del Campo A, Marks JA, Franzosa EA, Huttenhower C, and Balskus EP. A prominent glycyl radical enzyme in human gut microbiomes metabolizes trans-4-hydroxy-l-proline. Science 355: 1–9, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Li L, Xie Y, El-Sayed WM, Szakacs JG, and Roberts JC. Characteristics of selenazolidine prodrugs of selenocysteine: toxicity, selenium levels, and glutathione peroxidase induction in A/J mice. Life Sci 75: 447–459, 2004 [DOI] [PubMed] [Google Scholar]
- 129. Liang X, Dickman MB, and Becker DF. Proline biosynthesis is required for endoplasmic reticulum stress tolerance in Saccharomyces cerevisiae. J Biol Chem 289: 27794–27806, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Liang X, Zhang L, Natarajan SK, and Becker DF. Proline mechanisms of stress survival. Antioxid Redox Signal 19: 998–1011, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Lijek RS, Luque SL, Liu Q, Parker D, Bae T, and Weiser JN. Protection from the acquisition of Staphylococcus aureus nasal carriage by cross-reactive antibody to a pneumococcal dehydrogenase. Proc Natl Acad Sci U S A 109: 13823–13828, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Lim B. and Lee K. Stability of the osmoregulated promoter-derived proP mRNA is posttranscriptionally regulated by RNase III in Escherichia coli. J Bacteriol 197: 1297–1305, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Liu LK, Becker DF, and Tanner JJ. Structure, function, and mechanism of proline utilization A (PutA). Arch Biochem Biophys 632: 142–157, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Liu W, Glunde K, Bhujwalla ZM, Raman V, Sharma A, and Phang JM. Proline oxidase promotes tumor cell survival in hypoxic tumor microenvironments. Cancer Res 72: 3677–3686, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lovitt RW, Kell DB, and Morris JG. Proline reduction by Clostridium sporogenes is coupled to vectorial proton ejection. FEMS Microbiol Lett 36: 269–273, 1986 [Google Scholar]
- 136. Luo M, Arentson BW, Srivastava D, Becker DF, and Tanner JJ. Crystal structures and kinetics of monofunctional proline dehydrogenase provide insight into substrate recognition and conformational changes associated with flavin reduction and product release. Biochemistry 51: 10099–10108, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Luo M, Christgen S, Sanyal N, Arentson BW, Becker DF, and Tanner JJ. Evidence that the C-terminal domain of a type B PutA protein contributes to aldehyde dehydrogenase activity and substrate channeling. Biochemistry 53: 5661–5673, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Luo M, Gamage TT, Arentson BW, Schlasner KN, Becker DF, and Tanner JJ. Structures of proline utilization A (PutA) reveal the fold and functions of the aldehyde dehydrogenase superfamily domain of unknown function. J Biol Chem 291: 24065–24075, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Luo M, Singh RK, and Tanner JJ. Structural determinants of oligomerization of delta(1)-pyrroline-5-carboxylate dehydrogenase: identification of a hexamerization hot spot. J Mol Biol 425: 3106–3120, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Macaluso A, Best EA, and Bender RA. Role of the nac gene product in the nitrogen regulation of some NTR-regulated operons of Klebsiella aerogenes. J Bacteriol 172: 7249–7255, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. MacMillan SV, Alexander DA, Culham DE, Kunte HJ, Marshall EV, Rochon D, and Wood JM. The ion coupling and organic substrate specificities of osmoregulatory transporter ProP in Escherichia coli. Biochim Biophys Acta 1420: 30–44, 1999 [DOI] [PubMed] [Google Scholar]
- 142. Maharjan S, Aryal N, Bhattarai S, Koju D, Lamichhane J, and Sohng JK. Biosynthesis of the nargenicin A1 pyrrole moiety from Nocardia sp. CS682. Appl Microbiol Biotechnol 93: 687–696, 2012 [DOI] [PubMed] [Google Scholar]
- 143. Mahmoud RY, Li W, Eldomany RA, Emara M, and Yu J. The Shigella ProU system is required for osmotic tolerance and virulence. Virulence 8: 362–374, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Mantilla BS, Marchese L, Casas-Sanchez A, Dyer NA, Ejeh N, Biran M, Bringaud F, Lehane MJ, Acosta-Serrano A, and Silber AM. Proline metabolism is essential for Trypanosoma brucei brucei survival in the tsetse vector. PLoS Pathog 13: e1006158, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Mantilla BS, Paes LS, Pral EMF, Martil DE, Thiemann OH, Fernandez-Silva P, Bastos EL, and Silber AM. Role of Δ1-pyrroline-5-carboxylate dehydrogenase supports mitochondrial metabolism and host-cell invasion of Trypanosoma cruzi. J Biol Chem 290: 7767–7790, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Martins RM, Covarrubias C, Rojas RG, Silber AM, and Yoshida N. Use of l-proline and ATP production by Trypanosoma cruzi metacyclic forms as requirements for host cell invasion. Infect Immun 84: 3023–3032, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Mathieu C, Macedo JP, Hurlimann D, Wirdnam C, Haindrich AC, Suter Grotemeyer M, Gonzalez-Salgado A, Schmidt RS, Inbar E, Maser P, Butikofer P, Zilberstein D, and Rentsch D. Arginine and lysine transporters are essential for Trypanosoma brucei. PLoS One 12: e0168775, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Mejean A, Mann S, Vassiliadis G, Lombard B, Loew D, and Ploux O. In vitro reconstitution of the first steps of anatoxin-a biosynthesis in Oscillatoria PCC 6506: from free l-proline to acyl carrier protein bound dehydroproline. Biochemistry 49: 103–113, 2010 [DOI] [PubMed] [Google Scholar]
- 149. Mellies J, Wise A, and Villarejo M. Two different Escherichia coli proP promoters respond to osmotic and growth phase signals. J Bacteriol 177: 144–151, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Menzel R. and Roth J. Regulation of the genes for proline utilization in Salmonella typhimurium: autogenous repression by the putA gene product. J Mol Biol 148: 21–44, 1981 [DOI] [PubMed] [Google Scholar]
- 151. Michalkova V, Benoit JB, Weiss BL, Attardo GM, and Aksoy S. Vitamin B6 generated by obligate symbionts is critical for maintaining proline homeostasis and fecundity in tsetse flies. Appl Environ Microbiol 80: 5844–5853, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, and Warren JW. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun 58: 1281–1289, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Molle V, Nakaura Y, Shivers RP, Yamaguchi H, Losick R, Fujita Y, and Sonenshein AL. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J Bacteriol 185: 1911–1922, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Momose Y, Hirayama K, and Itoh K. Competition for proline between indigenous Escherichia coli and E. coli O157:H7 in gnotobiotic mice associated with infant intestinal microbiota and its contribution to the colonization resistance against E. coli O157:H7. Antonie Van Leeuwenhoek 94: 165–171, 2008 [DOI] [PubMed] [Google Scholar]
- 155. Moses S, Sinner T, Zaprasis A, Stoveken N, Hoffmann T, Belitsky BR, Sonenshein AL, and Bremer E. Proline utilization by Bacillus subtilis: uptake and catabolism. J Bacteriol 194: 745–758, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Moxley MA. and Becker DF. Rapid reaction kinetics of proline dehydrogenase in the multifunctional proline utilization A protein. Biochemistry 51: 511–520, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Moxley MA, Sanyal N, Krishnan N, Tanner JJ, and Becker DF. Evidence for hysteretic substrate channeling in the proline dehydrogenase and Delta1-pyrroline-5-carboxylate dehydrogenase coupled reaction of proline utilization A (PutA). J Biol Chem 289: 3639–3651, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Moxley MA, Tanner JJ, and Becker DF. Steady-state kinetic mechanism of the proline:ubiquinone oxidoreductase activity of proline utilization A (PutA) from Escherichia coli. Arch Biochem Biophys 516: 113–120, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Murdock L, Burke T, Coumoundouros C, Culham DE, Deutch CE, Ellinger J, Kerr CH, Plater SM, To E, Wright G, and Wood JM. Analysis of strains lacking known osmolyte accumulation mechanisms reveals contributions of osmolytes and transporters to protection against abiotic stress. Appl Environ Microbiol 80: 5366–5378, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Nagata K, Nagata Y, Sato T, Fujino MA, Nakajima K, and Tamura T. l-Serine, d- and l-proline and alanine as respiratory substrates of Helicobacter pylori: correlation between in vitro and in vivo amino acid levels. Microbiology 149: 2023–2030, 2002 [DOI] [PubMed] [Google Scholar]
- 161. Nakada Y, Nishijyo T, and Itoh Y. Divergent structure and regulatory mechanism of proline catabolic systems: characterization of the putAP proline catabolic operon of Pseudomonas aeruginosa PAO1 and its regulation by PruR, an AraC/XylS family protein. J Bacteriol 184: 5633–5640, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Nakajima K, Inatsu S, Mizote T, Nagata Y, Aoyama K, Fukuda Y, and Nagata K. Possible involvement of put A gene in Helicobacter pylori colonization in the stomach and motility. Biomed Res 29: 9–18, 2008 [DOI] [PubMed] [Google Scholar]
- 163. Nakao T, Yamato I, and Anraku Y. Mapping of the multiple regulatory sites for putP and putA expression in the putC region of Escherichia coli. Mol Gen Genet 214: 379–388, 1988 [DOI] [PubMed] [Google Scholar]
- 164. Namouchi A, Gomez-Munoz M, Frye SA, Moen LV, Rognes T, Tonjum T, and Balasingham SV. The Mycobacterium tuberculosis transcriptional landscape under genotoxic stress. BMC Genomics 17: 791, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Nasuno R, Hirano Y, Itoh T, Hakoshima T, Hibi T, and Takagi H. Structural and functional analysis of the yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism. Proc Natl Acad Sci U S A 110: 11821–11826, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Natarajan SK, Zhu W, Liang X, Zhang L, Demers AJ, Zimmerman MC, Simpson MA, and Becker DF. Proline dehydrogenase is essential for proline protection against hydrogen peroxide-induced cell death. Free Radic Biol Med 53: 1181–1191, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Nedenskov P. Nutritional requirements for growth of Helicobacter pylori. Appl Environ Microbiol 60: 3450–3453, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Ngamskulrungroj P, Chang Y, Roh J, and Kwon-Chung KJ. Differences in nitrogen metabolism between Cryptococcus neoformans and C. gattii, the two etiologic agents of cryptococcosis. PLoS One 7: e34258, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Nisman B. The Stickland reaction. Bacteriol Rev 18: 16–42, 1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Nomura M. and Takagi H. Role of the yeast acetyltransferase Mpr1 in oxidative stress: regulation of oxygen reactive species caused by a toxic proline catablism intermediate. Proc Natl Acad Sci U S A 101: 12616–12621, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Nuxoll AS, Halouska SM, Sadykov MR, Hanke ML, Bayles KW, Kielian T, Powers R, and Fey PD. CcpA regulates arginine biosynthesis in Staphylococcus aureus through repression of proline catabolism. PLoS Pathog 8: e1003033, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Ostrovsky de Spicer P. and Maloy S. PutA protein, a membrane-associated flavin dehydrogenase, acts as a redox-dependent transcriptional regulator. Proc Natl Acad Sci U S A 90: 4295–4298, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Paes LS, Suárez Mantilla B, Zimbres FM, Pral EM, Diogo de Melo P, Tahara EB, Kowaltowski AJ, Elias MC, and Silber AM. Proline dehydrogenase regulates redox state and respiratory metabolism in Trypanosoma cruzi. PLoS One 8: e69419, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Pais R, Lohs C, Wu Y, Wang J, and Aksoy S. The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl Environ Microbiol 74: 5965–5974, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Park HB, Sampathkumar P, Perez CE, Lee JH, Tran J, Bonanno JB, Hallem EA, Almo SC, and Crawford JM. Stilbene epoxidation and detoxification in a Photorhabdus luminescens-nematode symbiosis. J Biol Chem 292: 6680–6694, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Park Y, Herbert EE, Cowles CE, Cowles KN, Menard ML, Orchard SS, and Goodrich-Blair H. Clonal variation in Xenorhabdus nematophila virulence and suppression of Manduca sexta immunity. Cell Microbiol 9: 645–656, 2007 [DOI] [PubMed] [Google Scholar]
- 177. Pemberton TA, Srivastava D, Sanyal N, Henzl MT, Becker DF, and Tanner JJ. Structural studies of yeast delta(1)-pyrroline-5-carboxylate dehydrogenase (ALDH4A1): active site flexibility and oligomeric state. Biochemistry 53: 1350–1359, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Pemberton TA, Still BR, Christensen EM, Singh H, Srivastava D, and Tanner JJ. Proline: Mother Nature's cryoprotectant applied to protein crystallography. Acta Crystallogr D Biol Crystallogr 68: 1010–1018, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Peterson W. Helicobacter pylori and peptic ulcer disease. N Engl J Med 324: 1043–1048, 1991 [DOI] [PubMed] [Google Scholar]
- 180. Petridis M, Benjak A, and Cook GM. Defining the nitrogen regulated transcriptome of Mycobacterium smegmatis using continuous culture. BMC Genomics 16: 821, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Phang JM. The regulatory functions of proline and pyrroline-5-carboxylic acid. Curr Top Cell Regul 25: 91–132, 1985 [DOI] [PubMed] [Google Scholar]
- 182. Phang JM, Liu W, and Zabirnyk O. Proline metabolism and microenvironmental stress. Annu Rev Nutr 30: 441–463, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Pich OQ, Carpenter BM, Gilbreath JJ, and Merrell DS. Detailed analysis of Helicobacter pylori Fur-regulated promoters reveals a Fur box core sequence and novel Fur-regulated genes. Mol Microbiol 84: 921–941, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Price CT, Bukka A, Cynamon M, and Graham JE. Glycine betaine uptake by the ProXVWZ ABC transporter contributes to the ability of Mycobacterium tuberculosis to initiate growth in human macrophages. J Bacteriol 190: 3955–3961, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Qamar A, Mysore KS, and Senthil-Kumar M. Role of proline and pyrroline-5-carboxylate metabolism in plant defense against invading pathogens. Front Plant Sci 6: 503, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Racher KI, Voegele RT, Marshall EV, Culham DE, Wood JM, Jung H, Bacon M, Cairns MT, Ferguson SM, Liang WJ, Henderson PJ, White G, and Hallett FR. Purification and reconstitution of an osmosensor: transporter ProP of Escherichia coli senses and responds to osmotic shifts. Biochemistry 38: 1676–1684, 1999 [DOI] [PubMed] [Google Scholar]
- 187. Rachman H, Kim N, Ulrichs T, Baumann S, Pradl L, Nasser Eddine A, Bild M, Rother M, Kuban RJ, Lee JS, Hurwitz R, Brinkmann V, Kosmiadi GA, and Kaufmann SH. Critical role of methylglyoxal and AGE in Mycobacteria-induced macrophage apoptosis and activation. PLoS One 1: e29, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Rautelin H, Blomberg B, Fredlund H, Jarnerot G, and Danielsson D. Incidence of Helicobacter pylori strains activating neutrophils in patients with peptic ulcer disease. Gut 34: 599–603, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Reina-San-Martin B, Degrave W, Rougeot C, Cosson A, Chamond N, Cordeiro-Da-Silva A, Arala-Chaves M, Coutinho A, and Minoprio P. A B-cell mitogen from a pathogenic trypanosome is a eukaryotic proline racemase. Nat Med 6: 890–897, 2000 [DOI] [PubMed] [Google Scholar]
- 190. Rengarajan J, Bloom BR, and Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci U S A 102: 8327–8332, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Richards GR. and Goodrich-Blair H. Masters of conquest and pillage: Xenorhabdus nematophila global regulators control transitions from virulence to nutrient acquisition. Cell Microbiol 11: 1025–1033, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Rijo-Ferreira F, Pinto-Neves D, Barbosa-Morais NL, Takahashi JS, and Figueiredo LM. Trypanosoma brucei metabolism is under circadian control. Nat Microbiol 2: 17032, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Rivera-Ordaz A, Bracher S, Sarrach S, Li Z, Shi L, Quick M, Hilger D, Haas R, and Jung H. The sodium/proline transporter PutP of Helicobacter pylori. PLoS One 8: e83576, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Rizzi YS, Monteoliva MI, Fabro G, Grosso CL, Larovere LE, and Alvarez ME. P5CDH affects the pathways contributing to Pro synthesis after ProDH activation by biotic and abiotic stress conditions. Front Plant Sci 6: 572, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Roberts MF. Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Systems 1: 5, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Rodrigues JC, Godinho JL, and de Souza W. Biology of human pathogenic trypanosomatids: epidemiology, lifecycle and ultrastructure. Subcell Biochem 74: 1–42, 2014 [DOI] [PubMed] [Google Scholar]
- 197. Roege KE. and Kelly WL. Biosynthetic origins of the ionophore antibiotic indanomycin. Org Lett 11: 297–300, 2009 [DOI] [PubMed] [Google Scholar]
- 198. Rohloff P, Rodrigues CO, and Docampo R. Regulatory volume decrease in Trypanosoma cruzi involves amino acid efflux and changes in intracellular calcium. Mol Biochem Parasitol 126: 219–230, 2003 [DOI] [PubMed] [Google Scholar]
- 199. Rustad TR, Sherrid AM, Minch KJ, and Sherman DR. Hypoxia: a window into Mycobacterium tuberculosis latency. Cell Microbiol 11: 1151–1159, 2009 [DOI] [PubMed] [Google Scholar]
- 200. Sanyal N, Arentson BW, Luo M, Tanner JJ, and Becker DF. First evidence for substrate channeling between proline catabolic enzymes: a validation of domain fusion analysis for predicting protein-protein interactions. J Biol Chem 290: 2225–2234, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Sassetti CM, Boyd DH, and Rubin EJ. Genes requried for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48: 77–84, 2003 [DOI] [PubMed] [Google Scholar]
- 202. Sayé M, Miranda MR, di Girolamo F, de los Milagros Cámara M, and Pereira CA. Proline modulates the Trypanosoma cruzi resistance to reactive oxygen species and drugs through a novel d, l-proline transporter. PLoS One 9: e92028, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Saye M, Miranda MR, Reigada C, and Pereira CA. Trypanosoma cruzi proline transport presents a cell density-dependent regulation. J Eukaryot Microbiol 63: 516–523, 2016 [DOI] [PubMed] [Google Scholar]
- 204. Schiefner A, Breed J, Bosser L, Kneip S, Gade J, Holtmann G, Diederichs K, Welte W, and Bremer E. Cation-pi interactions as determinants for binding of the compatible solutes glycine betaine and proline betaine by the periplasmic ligand-binding protein ProX from Escherichia coli. J Biol Chem 279: 5588–5596, 2004 [DOI] [PubMed] [Google Scholar]
- 205. Schwan WR, Coulter SN, Ng EY, Langhorne MH, Ritchie HD, Brody LL, Westbrock-Wadman S, Bayer AS, Folger KR, and Stover CK. Identification and characterization of the PutP proline permease that contributes to in vivo survival of Staphylococcus aureus in animal models. Infect Immun 66: 567–572, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Schwan WR, Lehmann L, and McCormick J. Transcriptional activation of the Staphylococcus aureus putP gene by low-proline high-osmotic conditions and during infection of murine and human tissues. Infect Immun 74: 399–409, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Schwan WR. and Wetzel KJ. Osmolyte transport in Staphylococcus aureus and the role in pathogenesis. World J Clin Infect Dis 6: 22–27, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Sellick CA. and Reece RJ. Modulation of transcription factor function by an amino acid: activation of Put3p by proline. EMBO J 22: 5147–5153, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Serrano H. and Blanchard JS. Kinetic and isotopic characterization of l-proline dehydrogenase from Mycobacterium tuberculosis. Biochemistry 52: 5009–5015, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Singh H, Arentson BW, Becker DF, and Tanner JJ. Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site. Proc Natl Acad Sci U S A 111: 3389–3394, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Singh RK, Larson JD, Zhu W, Rambo RP, Hura GL, Becker DF, and Tanner JJ. Small-angle X-ray scattering studies of the oligomeric state and quaternary structure of the trifunctional proline utilization A (PutA) flavoprotein from Escherichia coli. J Biol Chem 286: 43144–43153, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Singh RK. and Tanner JJ. Unique structural features and sequence motifs of proline utilization A (PutA). Front Biosci (Landmark Ed) 17: 556–568, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Smith CJ, Deutch AH, and Rushlow KE. Purification and characteristics of a gamma-glutamyl kinase involved in Escherichia coli proline biosynthesis. J Bacteriol 157: 545–551, 1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Smith DA, Parish T, Stoker NG, and Bancroft GJ. Characterization of auxotrophic mutants of Mycobacterium tuberculosis and their potential as vaccine candidates. Infect Immun 69: 1142–1150, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Sonenshein AL. Control of key metabolic intersections in Bacillus subtilis. Nat Rev Microbiol 5: 917–927, 2007 [DOI] [PubMed] [Google Scholar]
- 216. Srivastava D, Singh RK, Moxley MA, Henzl MT, Becker DF, and Tanner JJ. The three-dimensional structural basis of type II hyperprolinemia. J Mol Biol 420: 176–189, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Srivastava D, Zhu W, Johnson WH, Jr, Whitman CP, Becker DF, and Tanner JJ. The structure of the proline utilization a proline dehydrogenase domain inactivated by N-propargylglycine provides insight into conformational changes induced by substrate binding and flavin reduction. Biochemistry 49: 560–569, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Stickland LH. Studies in the metabolism of the strict anaerobes (Genus Clostridium): the reduction of proline by Cl. sporogenes. Biochem J 29: 288–290, 1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Sully EK. and Geller BL. Antisense antimicrobial therapeutics. Curr Opin Microbiol 33: 47–55, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Swaminathan K, Flynn P, Reece RJ, and Marmorstein R. Crystal structure of a PUT3-DNA complex reveals a novel mechanism for DNA recognition by a protein containing a Zn2Cys6 binuclear cluster. Nat Struct Biol 4: 751–759, 1997 [DOI] [PubMed] [Google Scholar]
- 221. Sylvelster D. and Krassner SM. Proline metabolism in Trypanosoma cruzi epimastigotes. Comp Biochem Physiol B 55: 443–447, 1976 [DOI] [PubMed] [Google Scholar]
- 222. Szabados L. and Savouré A. Proline: a multifunctional amino acid. Trends Plant Sci 15: 89–97, 2010 [DOI] [PubMed] [Google Scholar]
- 223. Takagi H. Proline as a stress protectant in yeast: physiological functions, metabolic regulations, and biotechnological applications. Appl Microbiol Biotechnol 81: 211–223, 2008 [DOI] [PubMed] [Google Scholar]
- 224. Tallarita E, Pollegioni L, Servi S, and Molla G. Expression in Escherichia coli of the catalytic domain of human proline oxidase. Protein Expr Purif 82: 345–351, 2012 [DOI] [PubMed] [Google Scholar]
- 225. Tanner JJ. Structural biology of proline catabolic enzymes. Antioxid Redox Signal 30: 650–673, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Tanner JJ. Structural biology of proline catabolism. Amino Acids 35: 719–730, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Thaw P, Sedelnikova SE, Muranova T, Wiese S, Ayora S, Alonso JC, Brinkman AB, Akerboom J, van der Oost J, and Rafferty JB. Structural insight into gene transcriptional regulation and effector binding by the Lrp/AsnC family. Nucleic Acids Res 34: 1439–1449, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Theodore CM, Stamps BW, King JB, Price LS, Powell DR, Stevenson BS, and Cichewicz RH. Genomic and metabolomic insights into the natural product biosynthetic diversity of a feral-hog-associated Brevibacillus laterosporus strain. PLoS One 9: e90124, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, and Venter JC. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388: 539–547, 1997 [DOI] [PubMed] [Google Scholar]
- 230. Tonelli RR, Silber AM, Almeida-de-Faria M, Hirata IY, Colli W, and Alves MJ. l-Proline is essential for the intracellular differentiation of Trypanosoma cruzi. Cell Microbiol 6: 733–741, 2004 [DOI] [PubMed] [Google Scholar]
- 231. Townsend DE, Kaenjak A, Jayaswal RK, and Wilkinson BJ. Proline is biosynthesized from arginine in Staphylococcus aureus. Microbiology 142 (Pt 6): 1491–1497, 1996 [DOI] [PubMed] [Google Scholar]
- 232. Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, and Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med 345: 784–789, 2001 [DOI] [PubMed] [Google Scholar]
- 233. van Amsterdam K. and van der Ende A. Nutrients released by gastric epithelial cells enhance Helicobacter pylori growth. Helicobacter 9: 614–621, 2004 [DOI] [PubMed] [Google Scholar]
- 234. Vickerman K. Developmental cycles and biology of pathogenic trypanosomes. Br Med Bull 41: 105–114, 1985 [DOI] [PubMed] [Google Scholar]
- 235. Vilchez S, Manzanera M, and Ramos JL. Control of expression of divergent Pseudomonas putida put promoters for proline catabolism. Appl Environ Microbiol 66: 5221–5225, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. von Blohn C, Kempf B, Kappes RM, and Bremer E. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol Microbiol 25: 175–187, 1997 [DOI] [PubMed] [Google Scholar]
- 237. Walker V, Mills GA, Mellor JM, Langley GJ, and Farrant RD. A novel pyrroline-5-carboxylic acid and acetoacetic acid adduct in hyperprolinaemia type II. Clin Chim Acta 331: 7–17, 2003 [DOI] [PubMed] [Google Scholar]
- 238. Wanduragala S, Sanyal N, Liang X, and Becker DF. Purification and characterization of Put1p from Saccharomyces cerevisiae. Arch Biochem Biophys 498: 136–142, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Waters NR, Samuels DJ, Behera RK, Livny J, Rhee KY, Sadykov MR, and Brinsmade SR. A spectrum of CodY activities drives metabolic reorganization and virulence gene expression in Staphylococcus aureus. Mol Microbiol 101: 495–514, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Wellington S, Nag PP, Michalska K, Johnston SE, Jedrzejczak RP, Kaushik VK, Clatworthy AE, Siddiqi N, McCarren P, Bajrami B, Maltseva NI, Combs S, Fisher SL, Joachimiak A, Schreiber SL, and Hung DT. A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase. Nat Chem Biol 13: 943–950, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Wetzel KJ, Bjorge D, and Schwan WR. Mutational and transcriptional analyses of the Staphylococcus aureus low-affinity proline transporter OpuD during in vitro growth and infection of murine tissues. FEMS Immunol Med Microbiol 61: 346–355, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Whatmore AM, Chudek JA, and Reed RH. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J Gen Microbiol 136: 2527–2535, 1990 [DOI] [PubMed] [Google Scholar]
- 243. White TA, Krishnan N, Becker DF, and Tanner JJ. Structure and kinetics of monofunctional proline dehydrogenase from Thermus thermophilus. J Biol Chem 282: 14316–14327, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Williams KJ, Bryant WA, Jenkins VA, Barton GR, Witney AA, Pinney JW, and Robertson BD. Deciphering the response of Mycobacterium smegmatis to nitrogen stress using bipartite active modules. BMC Genomics 14: 436, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Williams KJ, Jenkins VA, Barton GR, Bryant WA, Krishnan N, and Robertson BD. Deciphering the metabolic response of Mycobacterium tuberculosis to nitrogen stress. Mol Microbiol 97: 1142–1157, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Wong KH, Hynes MJ, and Davis MA. Recent advances in nitrogen regulation: a comparison between Saccharomyces cerevisiae and filamentous fungi. Eukaryot Cell 7: 917–925, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Wood JM. Bacterial osmoregulation: a paradigm for the study of cellular homeostasis. Annu Rev Microbiol 65: 215–238, 2011 [DOI] [PubMed] [Google Scholar]
- 248. Wood JM. Bacterial osmosensing transporters. Methods Enzymol 428: 77–107, 2007 [DOI] [PubMed] [Google Scholar]
- 249. Wood JM. Genetics of l-proline utilization in Escherichia coli. J Bacteriol 146: 895–901, 1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Wood JM, Bremer E, Csonka LN, Kraemer R, Poolman B, van der Heide T, and Smith LT. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp Biochem Physiol A Mol Integr Physiol 130: 437–460, 2001 [DOI] [PubMed] [Google Scholar]
- 251. Wood JM. and Zadworny D. Characterization of an inducible porter required for l-proline catabolism by Escherichia coli K12. Can J Biochem 57: 1191–1199, 1979 [DOI] [PubMed] [Google Scholar]
- 252. Wright JA, Grant AJ, Hurd D, Harrison M, Guccione EJ, Kelly DJ, and Maskell DJ. Metabolite and transcriptome analysis of Campylobacter jejuni in vitro growth reveals a stationary-phase physiological switch. Microbiology 155: 80–94, 2009 [DOI] [PubMed] [Google Scholar]
- 253. Wu X. and Hurdle JG. The Clostridium difficile proline racemase is not essential for early logarithmic growth and infection. Can J Microbiol 60: 251–254, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Zaprasis A, Bleisteiner M, Kerres A, Hoffmann T, and Bremer E. Uptake of amino acids and their metabolic conversion into the compatible solute proline confers osmoprotection to Bacillus subtilis. Appl Environ Microbiol 81: 250–259, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Zaprasis A, Hoffmann T, Stannek L, Gunka K, Commichau FM, and Bremer E. The gamma-aminobutyrate permease GabP serves as the third proline transporter of Bacillus subtilis. J Bacteriol 196: 515–526, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, Kuhlow D, Guthke R, Platzer M, Kahn CR, and Ristow M. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial l-proline catabolism to induce a transient ROS signal. Cell Metab 15: 451–465, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Zhang L, Alfano JR, and Becker DF. Proline metabolism increases katG expression and oxidative stress resistance in Escherichia coli. J Bacteriol 197: 431–440, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Zhang L. and Becker DF. Connecting proline metabolism and signaling pathways in plant senescence. Front Plant Sci 6: 552, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Zhang W, Zhang M, Zhu W, Zhou Y, Wanduragala S, Rewinkel D, Tanner JJ, and Becker DF. Redox-induced changes in flavin structure and roles of flavin N(5) and the ribityl 2′-OH group in regulating PutA-membrane binding. Biochemistry 46: 483–491, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Zhang W, Zhou Y, and Becker DF. Regulation of PutA-membrane associations by flavin adenine dinucleotide reduction. Biochemistry 43: 13165–13174, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Zhang YJ. and Rubin EJ. Feast or famine: the host-pathogen battle over amino acids. Cell Microbiol 15: 1079–1087, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Zhou Y. DNA Binding of Proline Utilization A. Lincoln: University of Nebraska−Lincoln, 2008 [Google Scholar]
- 263. Zhou Y, Larson JD, Bottoms CA, Arturo EC, Henzl MT, Jenkins JL, Nix JC, Becker DF, and Tanner JJ. Structural basis of the transcriptional regulation of the proline utilization regulon by multifunctional PutA. J Mol Biol 381: 174–188, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Zhou Y, Zhu W, Bellur PS, Rewinkel D, and Becker DF. Direct linking of metabolism and gene expression in the proline utilization A protein from Escherichia coli. Amino Acids 35: 711–718, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Zhu W. and Becker DF. Flavin redox state triggers conformational changes in the PutA protein from Escherichia coli. Biochemistry 42: 5469–5477, 2003 [DOI] [PubMed] [Google Scholar]
- 266. Zhu W, Gincherman Y, Docherty P, Spilling CD, and Becker DF. Effects of proline analog binding on the spectroscopic and redox properties of PutA. Arch Biochem Biophys 408: 131–136, 2002 [DOI] [PubMed] [Google Scholar]
- 267. Zhu W, Haile AM, Singh RK, Larson JD, Smithen D, Chan JY, Tanner JJ, and Becker DF. Involvement of the beta3-alpha3 loop of the proline dehydrogenase domain in allosteric regulation of membrane association of proline utilization A. Biochemistry 52: 4482–4491, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]