Significance
Whole-exome sequencing data from patients with monogenic inborn errors identify thousands of genetic variants in each patient, only a few of which are pathogenic. Identifying pathogenic mutations therefore requires the rigorous filtration of variants that are too common in public databases to cause a disease of the observed incidence. We report that a large proportion of the variants common in patient cohorts are paradoxically absent from public databases. We define these nonpathogenic, cohort-specific common variants that cannot be removed from the analysis as a “blacklist.” We describe these blacklisted variants, demonstrate their usefulness for removing nonpathogenic variants, explain their origin experimentally, and provide a web server and software enabling researchers to automate the creation of their own blacklists.
Keywords: exome, variant, blacklist, WES analysis, WES annotation
Abstract
Computational analyses of human patient exomes aim to filter out as many nonpathogenic genetic variants (NPVs) as possible, without removing the true disease-causing mutations. This involves comparing the patient’s exome with public databases to remove reported variants inconsistent with disease prevalence, mode of inheritance, or clinical penetrance. However, variants frequent in a given exome cohort, but absent or rare in public databases, have also been reported and treated as NPVs, without rigorous exploration. We report the generation of a blacklist of variants frequent within an in-house cohort of 3,104 exomes. This blacklist did not remove known pathogenic mutations from the exomes of 129 patients and decreased the number of NPVs remaining in the 3,104 individual exomes by a median of 62%. We validated this approach by testing three other independent cohorts of 400, 902, and 3,869 exomes. The blacklist generated from any given cohort removed a substantial proportion of NPVs (11–65%). We analyzed the blacklisted variants computationally and experimentally. Most of the blacklisted variants corresponded to false signals generated by incomplete reference genome assembly, location in low-complexity regions, bioinformatic misprocessing, or limitations inherent to cohort-specific private alleles (e.g., due to sequencing kits, and genetic ancestries). Finally, we provide our precalculated blacklists, together with ReFiNE, a program for generating customized blacklists from any medium-sized or large in-house cohort of exome (or other next-generation sequencing) data via a user-friendly public web server. This work demonstrates the power of extracting variant blacklists from private databases as a specific in-house but broadly applicable tool for optimizing exome analysis.
Next-generation sequencing (NGS), particularly whole-exome sequencing (WES) and whole-genome sequencing (WGS), is increasingly being used for the discovery and diagnosis of human genetic disorders (1–3). The number of new disease-causing genetic variants logged by the Human Gene Mutation Database (HGMD) is currently increasing at a rate of ∼10% per annum (4). This increase has coincided with an expansion of the use of WES and WGS (1, 2). The mean number of exonic coding variants per individual relative to the reference human genome is about 20,000 (2, 3), but monogenic disease in any given individual is generally driven by at most two variants. The remaining nonpathogenic variants (NPVs) may be real variants (rare or common, deleterious or neutral), or false/low-quality variants [sequencing artifacts, bioinformatic misprocessing of raw sequencing data, or resulting from limitations to the performance of current quality control (QC) methods]. In practice, analyses of individual exomes aim to generate a short list of high-quality candidate variants by filtering out as many NPVs as possible, while minimizing the risk of false negatives (FNs) due to the removal of true disease-causing mutations. The first step in this process typically involves the use of public databases to identify and remove NPVs through comparisons of their frequency in the general population with the prevalence of the disease considered, its proposed mode of inheritance, and its estimated clinical penetrance. The largest public database currently available is the Genome Aggregation Database (gnomAD), which includes 123,136 exomes and 15,496 genomes from a total of 138,632 individuals (5). For the remaining variants, including those not reported in public databases, various variant-level and gene-level metrics can be used to predict deleteriousness and to select a smaller set of candidate variants for further experimental analysis (6–10).
In studies of rare genetic diseases, public databases are widely used for the initial elimination of common variants [minor allele frequency (MAF) > 0.01] (2, 11). However, some common variants within private databases may be absent from public databases, and most such variants are likely to be NPVs (2, 12). The efficacy with which such variants are identified and used for analyses of exomes from cohorts of patients studied by a particular research group has never been assessed in detail. An approach for detecting false-positive signal (defined as DFS) based on an internal cohort of 118 whole-exome sequences from different individuals generated a shortlist of variants found to be in Hardy–Weinberg (HW) disequilibrium due to excess heterozygosity (the DFS list; 23,389 variants) (13). However, most of these variants (68%) had already been reported in dbSNP (13). Machine learning-based methods for removing false positives (FPs) from sequencing data, such as variant quality score recalibration (VQSR), which uses a clustering score to determine whether a called variant is true (14), can limit the number of NPVs in exome data. However, these methods are subject to several limitations: (i) they are computationally intensive and time-consuming; (ii) they often require a large number of samples; (iii) parameter optimization requires extensive testing; and (iv) the addition of new samples requires reprocessing of the entire cohort. These methods are therefore little used by most researchers, who have small- or medium-sized exome cohorts and may not have access to powerful computing resources. It has been suggested that variants common within a homogeneous cohort and absent from public databases could be filtered out (2), but this approach has not been validated and there are currently no tools for the easy identification and compilation of such variants. In this context, we sought to establish a “blacklist” of variants too frequent in our cohort of 3,104 exomes from patients with severe infectious diseases (15–17).
Results
Determining a Frequency Cutoff for NPVs.
We observed that numerous candidate variant calls (Materials and Methods) (18) predicted to be damaging to the corresponding transcript or protein were present in >1% of our cohort of 3,104 in-house exomes from primary immune deficiency (PID) patients with heterogeneous ancestral backgrounds (19) (i.e., too common to cause PID) but absent from public databases (e.g., 1KG, ExAC, gnomAD). These variants are poor candidates for involvement in rare diseases but are impossible to eliminate by current methods based on variant frequencies in public databases (2). We therefore sought to classify and characterize these variants in a rigorous and comprehensive manner, to enable users to remove them from their WES/WGS analyses. First, we determined a statistical cutoff frequency above which in-house variants should be considered too frequent to cause rare diseases. We found that the MAF of all experimentally validated disease-causing mutations in HGMD followed a Gilbrat distribution (20). We then calculated the 99% Gilbrat distribution confidence interval (CI) for these frequencies and found that the upper boundary of the CI for the frequency of known disease-causing mutations was 0.01 (1%). We therefore used this cutoff as a criterion for the nonpathogenicity of variants (occurring in too many patients in our database to explain a rare monogenic illness). The MAF > 0.01 cutoff used here is an example of the blacklist approach to removing FP variants in studies of rare genetic disorders. The cutoff can be adjusted according to the mode of inheritance and genetic architecture, assumed penetrance, and prevalence of the disease, and the phenotypic homogeneity of the cohort (21). For example, assuming complete penetrance and allelic homogeneity, a rare recessive genetic disorder with a prevalence of 1 in 100,000 could be analyzed with a MAF cutoff of 0.0033, whereas a more common recessive genetic disorder with a prevalence of 1 in 1,000 should be analyzed with a MAF cutoff of 0.033. The assumption of incomplete penetrance may lead to the definition of higher cutoffs, whereas the assumption of allelic/genetic heterogeneity may lead to the use of lower cutoffs.
Generating the Blacklist.
We first designed the reducing FPs in NGS elucidation (ReFiNE) software, an easy-to-use tool for extracting blacklist variants from internal cohorts of WES or WGS data on the basis of a user-defined frequency cutoff (see Materials and Methods for details). ReFiNE creates a blacklist consisting of the full set of variants occurring in >1% (or any user-defined cutoff) of an investigated cohort, which can then be further filtered separately by the user, using MAF cutoffs from a population genetic database of choice. Using ReFiNE, we first collated all variants present at a frequency >1% in our PID WES cohort of 3,104 exomes (Materials and Methods and SI Appendix, Fig. S1) with a depth of coverage (DP) ≥ 5 and mapping quality (MQ) ≥ 30 (Materials and Methods) (5, 22). A large number of multiallelic variants in our cohort were absent from gnomAD for specific chromosomal positions. ReFiNE therefore collapsed all variants at a unique chromosomal position and summed the total number of patients at each of these positions. This generated a list of 780,956 variants, defined as the blacklist. This blacklist is the full list of variants occurring at single chromosomal positions for which >1% of patients had an alternative allele. These variants belonged to two classes: (i) biallelic, with a single alternative allele in our cohort; and (ii) multiallelic, with two or more alternative alleles in our cohort. The blacklist includes variants already reported in public databases, so we needed to extract the subset of variants unique to our method for further analysis. We thus annotated the blacklist with gnomAD, currently the most extensive public population genetics database available (5, 23). We found that 21.4% (167,144) of these 780,956 variants were absent from the gnomAD full exome and genome databases. As these 167,144 variants are not captured by the most extensive public database available, we focused the analysis of our method on this subset of variants, which, for simplicity, we will refer to as blacklist-annotated (BL-A): common in-house variants absent from gnomAD that cannot, therefore, be filtered out of analyses based on gnomAD.
Blacklist Filtering Removes 62% of the NPVs Remaining After Standard Public Database Filtering.
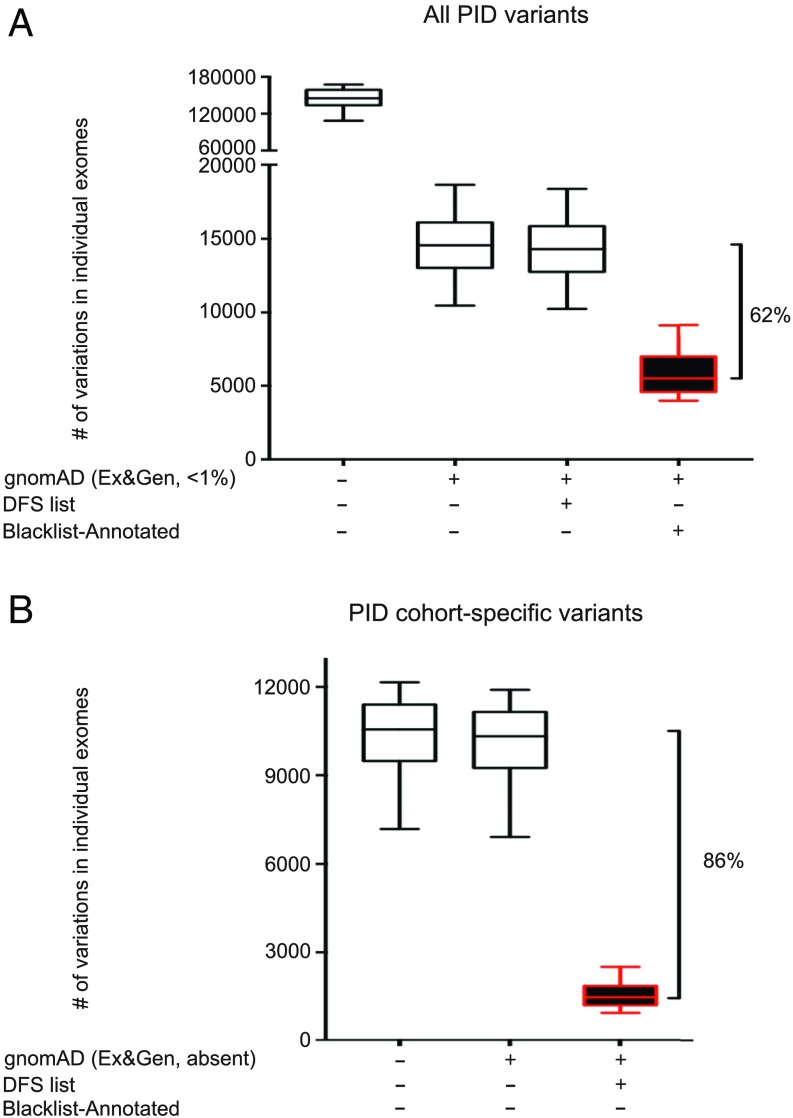
We then assessed the efficacy of BL-A for filtering out NPVs from patient exome data. We first applied the standard procedure for rare genetic disorders, by removing variants with a MAF > 0.01 in gnomAD from our 3,104 exomes (3, 12). This reduced the median number of variants in the patients’ exomes by 90% (Fig. 1A). Subsequent filtering with BL-A removed 62% of the remaining variants that could not be removed by other means (Fig. 1A, a median of 9,056 variants removed per exome). By comparison, the DFS list (13) decreased the median number of these variants by only 1.8% (median of 260 variants removed per exome). BL-A filtering was effective for both coding sequences [coding DNA sequences (CDSs)], including indel, exon-deleted, nonsynonymous, synonymous, and essential splicing variants, and for non-CDS variants, including untranslated region (UTR), nonessential splicing, intronic, downstream, and upstream variants, and for all three exome kits available for our cohort (37, 50, and 71 Mb; SI Appendix, Figs. S2 and S3). We then assessed the performance of BL-A filtering for variants absent from the gnomAD database (i.e., variants private to the PID database), which would be considered among the strongest candidates for a causal role in disease. This approach decreased the number of cohort-private variants potentially associated with PID in each exome by 86%, versus only 2.2% for the DFS list, and was similarly effective for CDS and non-CDS variants (Fig. 1B and SI Appendix, Fig. S4). Thus, when used as a filtering tool, our blacklist was able to remove variants absent from public databases and to decrease the number of candidate variants per exome considerably.
Fig. 1.
Blacklist filtering of 3,104 PID exomes with the PID blacklist. (A) Filtering of all variants in each exome by first removing those common in gnomAD exome and genome databases (MAF greater than 0.01). The remaining variants were subsequently filtered with the blacklist. (B) Filtering of cohort-specific variants in each exome with the blacklist. Filtering with the DFS list is shown for comparison. Error bars represent the 10th to 90th percentiles.
Metric Characteristics of the Variants and Genes Included in the Blacklist.
We then explored whether the QC scores for BL-A variants were similar to those for polymorphic variants (MAF > 0.01) reported in gnomAD. By comparing the median MQ and DP scores for blacklisted variants and polymorphic variants from our cohort (SI Appendix, Fig. S5), we demonstrated that none of these QC metrics could differentiate between these two sets of variants (especially when considering commonly used criteria for hard filtration; see Materials and Methods for further details). We then investigated whether machine learning QC metrics could classify these variants. With VQSR, only 25% of BL-A variants were annotated as “nonpass” (SI Appendix, Table S1). One of the key goals of this approach is providing an efficient tool for researchers who cannot easily perform VQSR. We therefore retained these VQSR nonpass variants in the blacklist. We also assessed the ability of a random forest classifier trained on polymorphic variants from the gnomAD dataset well-characterized by different methods to separate true variants from FP artifacts called by the variant-calling pipeline (5). We then used the same method to construct a new scoring function with the gnomAD dataset. We applied both scoring functions to the blacklist variants and a set of variants present in both the gnomAD dataset and our cohort, with a MAF of more than 1% in each dataset. The score distributions obtained were almost identical (SI Appendix, Fig. S6), demonstrating an inability of this standard classification method to distinguish between the blacklisted variants and true-positive (TP) variants. We then characterized the variants and genes included in BL-A with computational damage prediction metrics. A variant-level analysis revealed that the combined annotation-dependent depletion (CADD) (8) scores for blacklist variants were not significantly different from those for variants not included in the blacklist (SI Appendix, Fig. S7). A gene-level analysis (6) of all genes with blacklist variants (n = 13,665 genes) showed them to have low gene damage index (GDI) values (SI Appendix, Fig. S8). However, some genes with a high GDI have many BL-A variants (e.g., HLA-DRB1, 658 variants; MUC16, 455 variants). Filtration methods based on QC and variant- and gene-level damage prediction metrics would not efficiently detect and remove the blacklist variants absent from gnomAD. These results demonstrate the value of blacklisting as a complementary approach to analyses based on standard public databases, including gnomAD, QC filtering, and damage prediction metrics.
Determining the FN Rate Associated with Blacklist Use.
We estimated the proportion of TP disease-causing mutations removed by the blacklist approach, by screening 129 exomes from patients in our cohort for whom the TP mutations had been validated experimentally. Filtering these exomes with the complete blacklist did not remove any of the known TP mutations (0% FN rate). Even though most variants in any patient are not pathogenic, our analysis indicates that it is very safe to apply the blacklist to patient exomes. We also compared the complete blacklist with the list of 144,641 disease-causing mutations in HGMD and noted an overlap of only 263 variants (0.18% FN rate). These variants are listed as disease-causing in the HGMD dataset, but 47% have a MAF > 0.01 in the gnomAD exome or genome databases, suggesting that are unlikely to be the cause of a rare disorder. These findings indicate that our FN rate is probably lower than the rate of 0.18% for HGMD in the context of rare disorders. Only eight BL-A variants were present in HGMD (0.001% FN rate), indicating that the FN rate for our specific BL-A list was lower than that for gnomAD. Together, these results suggest that the FN rate is very low for this technique (SI Appendix, Table S2). We also screened 3,731,152 somatic cancer-causing or cancer-associated variants available from TCGA (https://cancergenome.nih.gov). We found that 59,151 of these TCGA variants (1.5%) were present in the complete blacklist and 2,471 (0.07%) were present in BL-A. As our blacklist was derived from germline exome data, the presence of these blacklist variants in the TCGA database suggests that they may be FPs that could be removed, as previously reported (24). Together, these data indicate that the blacklist approach results in an extremely low FN rate when applied to patients with rare diseases, and that it is therefore safe to use this approach to remove NPVs from patient exome data.
Practical Application of the Blacklist to the Analysis of Patients’ Exomes.
We assessed the use of blacklisting for practical analyses of patient exomes. We selected a case from our cohort with an autosomal dominant disease-causing mutation described in a previous study (patient D2 from ref. 25). We filtered this patient’s exome with a standard pipeline to identify disease-causing mutations (SI Appendix, Fig. S9). This standard approach reduced the number of candidate variants from 142,473 to 3,526. Taking known mode of inheritance into account and restricting the analysis to CDS variants (excluding synonymous alterations), the number of candidate variants was reduced further, to 231. The inclusion of BL-A in the pipeline decreased the final number of candidate variants to 109 (SI Appendix, Fig. S9), with retention of the known IKZF1 mutation. Overall, this corresponds to a 53% decrease in the number of variants from this patient’s exome to be considered. The remaining variants were high-quality candidates that would probably merit rigorous analysis in exome analyses for patients with diseases of unknown etiology. Thus, blacklisting greatly decreases the number of candidate variants for further study in practice, in exome analyses on individual patients.
Practical Application of Blacklisting to the Analysis of Population Exomes.
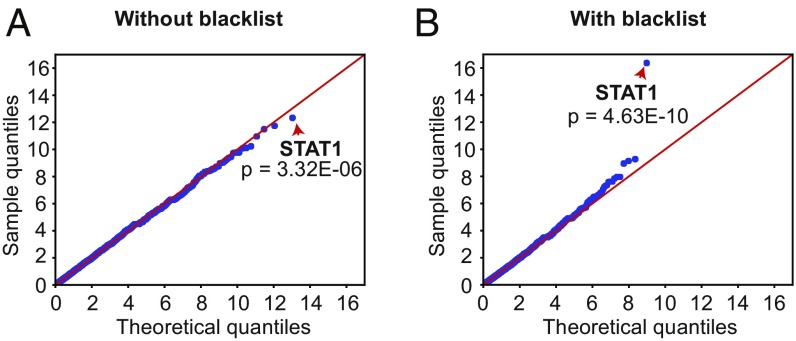
We then explored the use of our blacklist for gene burden analysis for genetic homogeneity at the population level. We compared the number of patients with at least one variant of any given gene between a cohort of 202 patients suffering from chronic mucocutaneous candidiasis (CMC) and 852 phenotypically unrelated controls (26). When standard filtering with public databases was applied in the absence of blacklisting, the enrichment observed for the known disease-causing gene in the CMC cohort, STAT1 (P value = 3.32 × 10−6) was not significant considering the corrected threshold at the genome-wide level (P valuethreshold = 0.05 20554 = 2.43 × 10−6; Fig. 2A). However, following the addition of BL-A to the pipeline, STAT1 was correctly identified as a gene displaying strong and significant genome-wide enrichment in the disease cohort (P value = 4.63 × 10−10; Fig. 2B). In this instance, our blacklist removed two variants present in a large proportion of our PID exomes (both cases and controls) that confounded the statistical comparison between the CMC and control groups. Together with the previous practical example, these analyses demonstrate the power of blacklisting for removing NPVs from patient exomes, both to simplify candidate variant identification in patients and for other large-scale statistical analyses of patient groups.
Fig. 2.
Application of the blacklisting approach to enrichment analysis. Quantile–quantile plots depicting the analysis of genetic homogeneity for a cohort of 202 patients with chronic mucocutaneous candidiasis (CMC) before (A) and after (B) application of the blacklist. The control cohort consisted of 852 unrelated individuals. In each panel, the red arrows indicate STAT1, the known cause of CMC in our cohort, before and after blacklist application.
Characterization of Multiallelic Variants from the Blacklist.
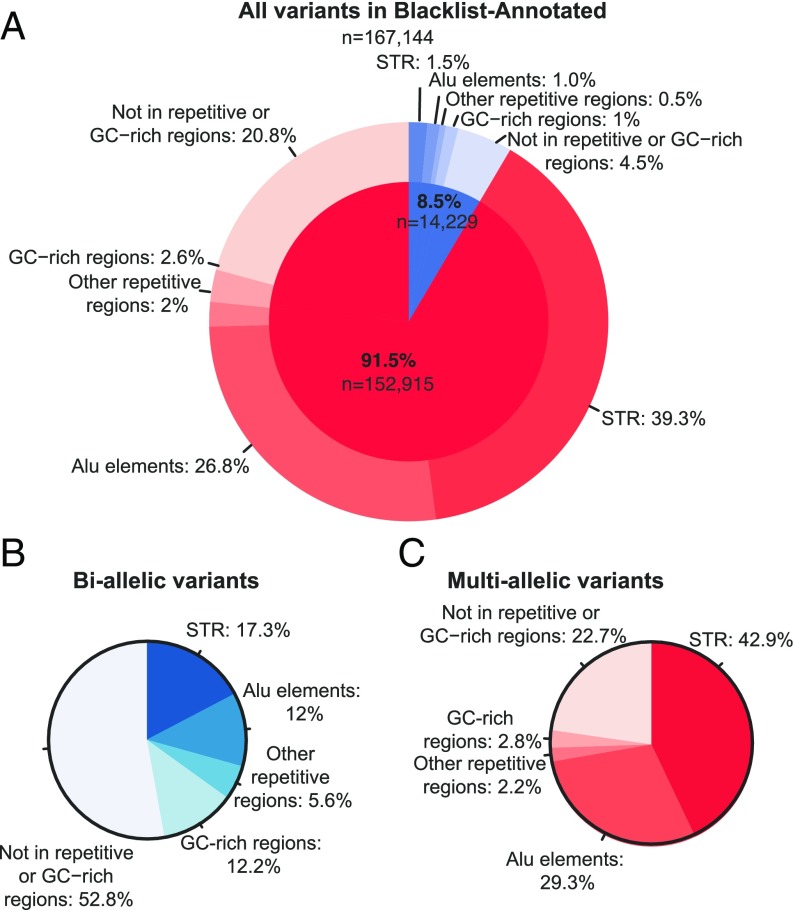
We then characterized the PID cohort BL-A variants (n = 167,144). Most of the variants (91.5%) in the blacklist were multiallelic (SI Appendix, Table S3). The cohort-specific variants present in the blacklist were therefore due to multiallelic sites displaying high levels of variation in our cohort (SI Appendix, Table S4). We began by hypothesizing that the multiallelic variants might lie in low-complexity regions of the human genome, leading to sequencing errors. The annotation of all these variants with RepeatMasker, Simple Repeats, and GC percent tracks from University of California, Santa Cruz (UCSC) Genome confirmed that 118,154 of the 152,915 variants (77.3%) occurred in repetitive or GC-rich regions, and that most (65,646; 56%) were located in short tandem repeat (STR) regions (Fig. 3 and SI Appendix, Table S4).
Fig. 3.
Characterization of the blacklisted biallelic and multiallelic variants in low-complexity regions of the genome. Occurrence of the blacklisted multiallelic (red) and biallelic (blue) variants in repetitive [short tandem repeats (STRs), Alu elements, other repetitive regions] and GC-rich regions; percent relative to the total number of blacklisted variants (A) or the total number of biallelic (B) or multiallelic (C) blacklisted variants.
Characterization of Biallelic Variants from the Blacklist.
We analyzed the biallelic variants, which were also found to be located in repetitive or GC-rich regions, albeit to a lesser extent (6,711; 47.2%) (Fig. 3 and SI Appendix, Table S4). We also characterized these biallelic variants, focusing on those located in CDS regions, in the 1,150 individuals of European origin according to principal-component analysis (PCA) (19), to determine whether these variants were under HW equilibrium. In total, 388 CDS variants were found to be located in repetitive or GC-rich regions; 339 (87.4%) of these variants were in HW equilibrium and 49 (13.6%) were in HW disequilibrium (threshold of P < 10−8; SI Appendix, Table S5). An investigation of the biallelic variants not present in repetitive regions (7,518; 52.8%) yielded a similar distribution, with 209 (89.3%) and 25 (10.7%) of the 234 CDS variants in HW equilibrium and disequilibrium, respectively. Overall, 74 CDS variants were in HW disequilibrium, and in 39 of these variants (52.7%), the cause was an excess of homozygous wild-type (14.9%) or homozygous alternative (37.8%) genotypes (SI Appendix, Table S5). Most of these 39 variants had low coverage (wild-type = 15.6×, alternative = 20.5×; SI Appendix, Table S5), which may have led to miscalls for a homozygous genotype. Most of the variants (35; 47.3%) in HW disequilibrium presented heterozygote excess, with high mean coverage rates of 163× (much higher than the 42.5× coverage of the 548 CDS variants in HW equilibrium), suggesting an excess of reads wrongly mapped to the region (SI Appendix, Table S5). We also studied the 548 biallelic CDS variants in HW equilibrium, to evaluate their distribution across ethnicities. We focused the analysis on the four largest genetic ancestry groups in our cohort (SI Appendix, Fig. S10): European, African, North African, and Middle Eastern, as determined by PCA (19). In total, 200 (36.5%) of these variants were heterogeneously distributed across genetic ancestries (threshold of P < 10−8; SI Appendix, Table S6). The observed heterogeneous distribution was probably due to one specific genetic ancestry in 46 (23%) of the variants (SI Appendix, Table S6). In 20 variants (43.5%), the individual genetic ancestry was Middle Eastern (SI Appendix, Table S6), which is poorly represented in public databases (27), suggesting that these variants are true variants that are more common in this population.
Experimental Investigation of the Blacklisted Variants.
We further investigated the features of BL-A variants. We first focused on biallelic blacklist CDS variants in HW disequilibrium displaying excess heterozygosity and absent from repetitive regions in individuals of European ancestry (n = 35). We found that 48.6% of these variants (n = 17) mapped to four chromosomal regions, in the HLA-DRB1, MUC6, OR8U1, and TAS2R43 genes with consecutive blacklist variants (less than 300 bp) (SI Appendix, Table S7). Most of these regions contain flagged variants annotated in gnomAD (47% in Exome and 65% in Genome, annotated as AC0, RF, and/or InbreedingCoeff; SI Appendix, Table S7). For the remaining variants (referred to as “unique”), we found that the blacklist variants were at the same location (but with different genotypes) as flagged variants annotated in gnomAD, like the consecutive variants (28% in Exome and 50% in Genome, annotated as AC0, RF, and/or InbreedingCoeff). Integrative genomics viewer (IGV) (28) showed that the consecutive variants in these regions belonged to the same reads, suggesting the existence of an “alternative” sequence (referred to as a segmental duplication by gnomAD or as an alternative haplotype; SI Appendix, Figs. S11–S13). These observations strongly suggest that some blacklist biallelic variants define alternative haplotypes belonging to unmapped regions absent from the human reference genome. These variants were probably incorrectly mapped to the region of the reference genome for which the best match was obtained, leading to a mixture of wild-type and alternative alleles in these regions, resulting in higher coverage and a final erroneous heterozygous call. In a second analysis, we focused on multiallelic variants. Most of these variants (77%) were located in low-complexity regions (STRs, Alu elements, GC-rich regions, or other repetitive regions; Fig. 3). IGV analysis of three multiallelic variants absent from these regions and common in our cohort (MAF > 0.9) revealed that they were located in the vicinity of a small stretch of repeated nucleotides (SI Appendix, Figs. S14–S16). Extending the analysis to the 23% of multiallelic variants not previously detected in low-complexity regions (n = 34,761), we found that 83.3% were also located close to mononucleotide repeats (26,165; 75.3%) or to small repetitive stretches (two or more nucleotides; 2,802; 8.1%). Attempts to confirm these variants by Sanger sequencing failed, due to the mononucleotide repeat (SI Appendix, Table S8), strongly suggesting that the WES approach may have been affected by a polymerase artifact similar to that reported in previous studies (29, 30). This exploration of blacklist variants suggests that the multiallelic variants probably resulted from—to a large extent—sequencing/calling errors during WES on low-complexity regions, whereas a proportion of the blacklist biallelic variants, particularly those in HW disequilibrium, were due to mapping errors resulting from the incomplete nature of the GRCh37/GRCh38 genome assembly.
Testing the Blacklist Approach as a General Filtering Method in Three Unrelated Cohorts.
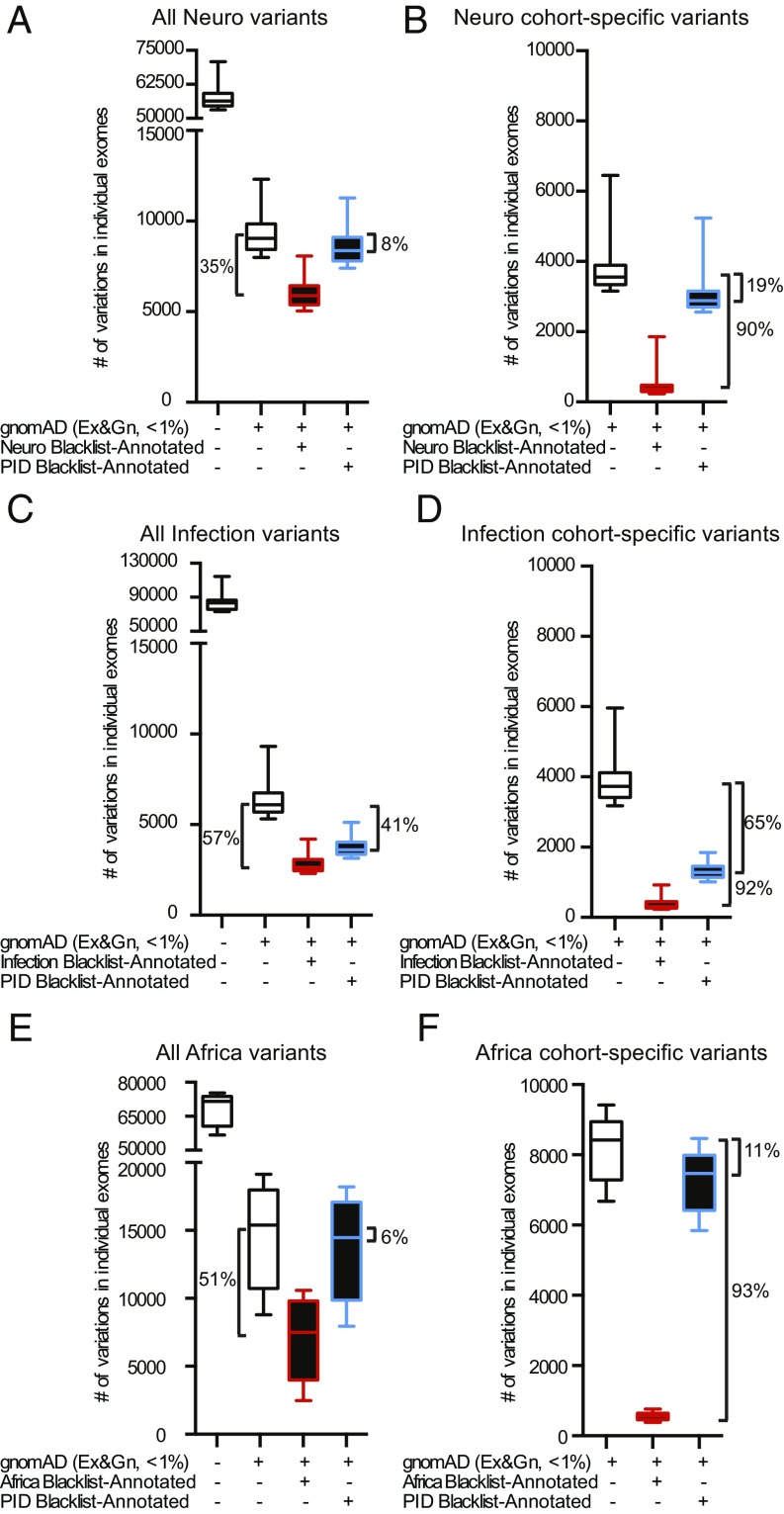
We assessed the suitability of the blacklist approach for filtering in other private databases. We used three unrelated independently processed exome cohorts (from DNA preparation to VCF data): (i) 3,869 exomes from patients suffering from neurological diseases (“Neuro”) (27); (ii) 902 exomes from patients suffering from diseases with an infectious phenotype (“Infection”); and (iii) 400 exomes (100 from Europeans and 300 from Africans) from a study on the demographic history of Central Africans (“Africa”) (31). We first generated separate blacklists for the Neuro, Infection, and Africa cohorts, according to the pipeline described above. After filtering on the basis of MAF > 1% (in the specific cohort) in gnomAD, the application of the cohort-specific blacklists for the Neuro, Infection, and Africa cohorts decreased the number of variants retained by 35%, 57%, and 51%, respectively (a median of 3,160, 3,462, and 7,905 variants per exome, respectively; Fig. 4 A, C, and E). Considering only cohort-private variants (i.e., those appearing in the specific cohort but absent from gnomAD exomes and genomes), applying the cohort-specific blacklists to the Neuro, Infection, and Africa cohorts reduced the number of variants in each exome by 90%, 92%, and 93%, respectively, eliminating a median of 3,195, 3,418, and 7,861 variants per exome, respectively (Fig. 4 B, D, and F). This filtering was effective for both CDS and non-CDS variants (SI Appendix, Fig. S17). A comparison of the four blacklists revealed that a substantial number of variants were unique to each blacklist (SI Appendix, Fig. S18), demonstrating the cohort specificity of the blacklisted variants, particularly for the Africa cohort, probably due to ancestry. Specifically, each blacklist contained 63–91% of the unique biallelic variants (SI Appendix, Fig. S18A and Table S3) and 46–92% of the unique multiallelic variants (SI Appendix, Fig. S18B). A similar pattern was observed when the analysis was restricted to biallelic and multiallelic CDS variants (SI Appendix, Fig. S18 C and D and Table S3). Thus, the efficacy of blacklist filtering in our PID cohort was not due to specific pipeline settings or enrichment within our exomes. Instead, our results suggest that the blacklist method should effectively remove a substantial proportion of the NPVs not already removed by public database analysis from any cohort of exomes considered.
Fig. 4.
Blacklist filtering of unrelated cohort exomes. (A, C, and E) Filtering of all variants in the neurological (A), infectious disease (C), and central African (E) exomes by first removing those common in gnomAD exome and genome databases (MAF greater than 0.01). The remaining variants were subsequently filtered with the Neuro (A), Infection (C), or Africa (E) blacklists (red boxes), or the PID blacklist (blue boxes). (B, D, and F) Filtering of exomes restricted to cohort-specific variants, with the Neuro (B), Infection (D), or Africa (F) blacklists (red boxes), or the PID blacklist (blue boxes). Error bars represent the 10th to 90th percentiles.
Application of the Blacklist to Unrelated Cohorts.
We then assessed whether the originally generated PID blacklist would effectively filter exomes from the unrelated Neuro, Infection, and Africa cohorts used above. We removed variants with a MAF > 0.01 in gnomAD from the Neuro, Infection, and Africa exomes and then applied the PID BL-A. This reduced the median number of remaining variants in the Neuro, Infection, and Africa exomes by 8%, 41%, and 6%, respectively (median of 715, 2,487, and 947 variants per exome, respectively; Fig. 4 A, C, and E, blue box). When the analysis was restricted to cohort-private variants in the Neuro, Infection, and Africa exomes, the PID blacklist decreased the number of variants in individual exomes by 19%, 65%, and 11%, respectively (median of 673, 2,439, and 957 variants per exome, respectively; Fig. 4 B, D, and F, blue box). The superior efficiency of the PID blacklist for the Infection cohort may reflect the library preparation technique (SureSelect) and sequencing technology (HiSeq sequencer) used. Nevertheless, the PID blacklist was shown to be a useful filtering approach in unrelated cohorts in which exomes were captured with different kits and sequencing technologies (SureSelect or Nextera kits and HiSeq 2000 or HiSeq 2500 sequencing, respectively). We also found that filtering our PID exomes with the blacklist from the Neuro cohort did not remove any TP variants from the 129 PID exomes with proven disease-causing mutations. Blacklists are, therefore, effective for filtering exomes other than those with which they were developed and including cohort-private NPVs. However, generating internal blacklists from the cohort under investigation was found to be the most effective approach to removing NPVs.
Determining the Minimum Cohort Size and Saturation Point for the Blacklist.
We sought to determine the minimum sample size appropriate for the generation of a custom blacklist for a cohort of interest. We combined the two largest cohorts studied here—our PID cohort (3,104) and the Neuro cohort (3,869)—and simulated blacklists by randomly sampling various numbers of individuals relative to cohort size, with 30 iterations for each sample size (SI Appendix, Fig. S19). As the Neuro cohort was captured with the 50-Mb kit, which targets CDS, we focused this analysis exclusively on CDS variants. The number of CDS variants in the simulated BL-A increased rapidly with sample size between 10 and 500 individuals, whereas the number of variants increased more slowly when sample size exceeded 500 individuals. We therefore propose the use of samples of at least 500 heterogeneous unrelated individuals, to ensure the reliable capture of common cohort-specific variants. We estimated the saturation point for the blacklist’s CDS variants (less than one new variant added per new individual) at a sample size of ∼2,801 individuals (SI Appendix, Fig. S19). Thus, a blacklist generated with the pipeline described here could be considered “saturated” for the purpose of capturing most of the cohort-specific CDS variants that cannot be removed by public database analysis.
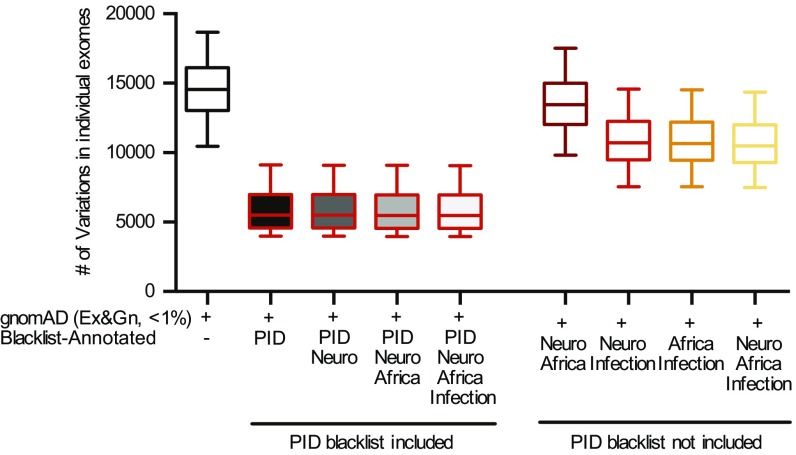
Efficacy of the Combined Blacklist.
Finally, we explored the efficacy of a “universal” blacklist generated by combining the four BL-As presented in this study. We reasoned that the aggregation of blacklists obtained from different cohorts (and different samples/data-processing methods) would result in a “universal blacklist” with the number of filtered variants eventually converging. We tested this hypothesis by aggregating either (i) the four blacklists (PID, Neuro, Infection, and Africa blacklists) into a single “combined blacklist”; or (ii) four combinations from the set of blacklists (Neuro, Infection, Africa) into four combined blacklists (i.e., Neuro–Africa, Neuro–Infection, Africa–Infection, Neuro–Africa–Infection), and applying the combined blacklists obtained in (i) and (ii) to the PID cohort. As the PID blacklist was not included in the four combined blacklists in (ii), we refer to these blacklists as “non–cohort-specific combined blacklists.” These blacklists removed a decreasing number of variants with increasing size of the sets making up the blacklists (Fig. 5). After standard filtering with public databases, the “Neuro–Africa” non–cohort-specific blacklist removed a median of 1,102 (8%) variants, the “Neuro–Infection” non-cohort-specific blacklist removed a median of 3,833 (26%) variants, the “Africa–Infection” non–cohort-specific blacklist removed a median of 3,886 (27%) of variants, and the “Neuro–Africa–Infection” non–cohort-specific blacklist removed a slightly larger number of variants (median of 4,078, or 28% of variants). By contrast, the PID blacklist removed a median of 9,056 variants. The “four combined” blacklist removed a median of 25 (0.45%) additional variants not captured by the PID blacklist alone (Fig. 5). Overall, these findings suggest that the number of variants filtered by the blacklist approach converges with the inclusion of blacklists from additional cohorts, consistent with the results for blacklist saturation. This universal filtering by blacklisting can be effectively applied to other individuals/cohorts. It is most efficient when the sequencing technology used, and the genetic ancestries of the individuals/cohorts under analysis, are similar to the universal blacklist (SI Appendix, Fig. S19). Moreover, the efficiency of a cohort-specific cohort applied to a different cohort (e.g., PID and infection cohorts) was greater for cohorts similar in terms of ethnic background and sequencing procedure (both mostly European and capture with similar kits), consistent with the results in Fig. 4C. Finally, although cohort-specific blacklists maximize the efficiency of this approach, the use of non–cohort-specific combined blacklists is nevertheless a very useful approach for filtering out a large number of unwanted variants, reinforcing the power of blacklist filtering even in the absence of a custom blacklist for the cohort.
Fig. 5.
Efficiency of various combinations of the four blacklists. Filtering of all variants in each PID exome with combinations of the various blacklists, with and without inclusion of the PID blacklist. Error bars represent the 10th to 90th percentiles.
Discussion
An essential step in the analysis of exomes from patients with rare genetic disorders is the removal of NPVs common in public databases (such as gnomAD, Bravo, and TopMed) at frequencies inconsistent with the prevalence, mode of inheritance, and penetrance of the disease (11). In principle, variants found to be common in a private cohort but absent from public databases should also be filtered out. However, only one other previous study has explored the generation of filtering lists based on internal cohorts (13). Moreover, there are currently no tools available for filtering based on allele frequencies in internal cohorts. We report here the identification of in-house variants too common to cause rare monogenic illnesses (typically with a population prevalence of <10−4) in a cohort of 3,104 exomes. We assembled these variants into a blacklist and subsequently explored the use of this blacklist for filtering NPVs from exome sequencing data, using the subset of variants that makes our approach unique (BL-A: those that are absent from public databases). These variants had high-quality metrics and 75% of them would not be captured by the rigorous application of available software, such as VQSR. We further validated this approach in three other independently processed and unrelated cohorts, demonstrating that our blacklist approach is generally, and perhaps universally, effective for filtering variants, and that the generation of blacklists specific to a given cohort significantly increases the number of variants filtered out. We provide a computational tool (ReFiNE) for automatically generating in-house cohort-specific blacklists. We show that our blacklist can be used in synergy with standard public database filtering, to remove variants displaying disproportionate enrichment in an internal cohort.
Public databases such as gnomAD, which represent major population groups (about half of individuals are of European ancestry and the others are a mixture of Admixed Americans, Africans/African Americans, South Asians, East Asians, and Others), are an invaluable resource for estimating the frequency of variants in the general population and in different genetic ancestry groups. However, cohort-specific exomes may contain common variants (e.g., >1%) that are absent from or rare in public databases, partly because they are population-specific variants less represented in gnomAD [as observed for African (31) and Middle Eastern individuals (27)]. Moreover, public databases, such as gnomAD, make considerable efforts to ensure the rigorous removal of FP variants to ensure that they provide high-quality, high-stringency information about variants. However, these public databases do not provide a list of filtered FP variants and their summary statistics for filtration purposes. We demonstrated this with 113 1KG genomes generated by our in-house pipeline, showing that 23% of the variants were absent from the public 1KG database, highlighting discrepancies between the analyzed and released data due to different bioinformatic procedures. Moreover, resources such as dbSNP are difficult to use for FP filtering because their FP variant rate is high (32). Therefore, even when using the latest versions of public databases and gene-level filtration (6, 7), ReFiNE is an effective tool for collecting data independently from external resources.
The technology associated with the NGS analyses (sequencing platform, targeting procedures, and software) is strongly associated with the calling of the variants. We and others have previously observed biases specific for WES and WGS (18) or variant-calling pipelines (33). Differences in technology can therefore lead to the misannotation of variants in a given cohort. The main sources of misannotation are as follows: (i) variants in gnomAD collected by different technologies (PCR for WES and PCR-free plus PCR for WGS) apply rigorous QC cutoffs based on high-quality technologies, resulting in higher proportions of variants from lower-quality technologies being removed; (ii) despite the presence of 15,496 genomes in gnomAD, some genomic regions remain poorly covered or not covered at all, whereas these regions are covered by our cohort and contain variants (2% of our BL-A); (iii) a recent comparative studied revealed strong discrepancies between the variant callers used in NGS analyses (34); these discrepancies have been highlighted by the differences between the gnomAD and ExAC databases (https://macarthurlab.org/2017/02/27/the-genome-aggregation-database-gnomad/); and (iv) the annotation of NGS variants in multiallelic positions is often problematic (35) because current annotation software [SNPeff (36), VEP (37), ANNOVAR (38)] cannot identify these variants efficiently. Indeed, 91.5% of our blacklist variants were located at multiallelic sites according to gnomAD’s genome annotation. Each cohort is unique (in terms of technology, quality, ethnicities). Our blacklisting resource is intended to fill this gap, particularly for researchers without the large exome or genome databases required for filtering with computationally intensive methods, such as VQSR. ReFiNE can, thus, overcome anomalies in sequence alignment or variant-calling processes, such as large indel events (39).
We show here that analyses of variant frequency within internal cohorts constitute an additional method for filtering out variants too common to cause rare disease. The blacklists generated by ReFiNE are easy to use and rapidly identify NPVs that may confound the dissection of patient exomes. As WES and WGS are increasingly widely used for the investigation of genetic disorders in patients, it will be possible to apply the blacklisting approach described here and ReFiNE software to larger cohorts of patients, facilitating the effective identification of NPVs in these cohorts. However, caution is required when generating blacklists with ReFiNE from phenotypically homogeneous cohorts, particularly if of the same underrepresented ethnic origin, as this approach may remove TP variants in such conditions. Finally, such extensive, rapidly generated blacklists (1 h for 3,104 exomes) should increase the efficiency of NPV elimination from exomes and genomes, without the need for the large computer clusters required by current machine-learning algorithms, such as VQSR (a month for 3,104 exomes). As exome capture kits become increasingly efficient, and with the widespread adoption of WGS, the blacklists generated by ReFiNE will facilitate efficient noise reduction in NGS data, independently of the technology used, making it easy to find the needles in increasingly large haystacks of genetic variants in patients.
Materials and Methods
Website Resource.
ReFiNE and precalculated blacklists are available on GitLab (40).
Patient Cohort.
The 3,104 individuals studied here were selected from samples of diverse ancestral origins obtained by our laboratories and recruited with the help of clinicians. This sample was not random, but cohort-specific effects should not have biased the results, as the individuals included had a wide range of different infectious diseases and immune deficiency phenotypes. All study participants provided written informed consent for the use of their DNA in studies aiming to identify genetic risk variants for disease. IRB approval was obtained from The Rockefeller University and Necker Hospital for Sick Children, along with a number of collaborating institutions. The exomes of 3,869 individuals suffering from neurological disease were obtained from the Greater Middle East (GME) Consortium, with recruitment according to a similar protocol (27). The exomes of 902 individuals suffering from severe infectious diseases (Infection cohort) were obtained from patients enrolled in studies coordinated by the laboratory of J.F. at École Polytechnique Fédérale de Lausanne (Lausanne, Switzerland). The exomes of 400 individuals in the Africa cohort were provided by the laboratory of L.Q.-M. at the Pasteur Institute (Paris, France).
WES.
A summary of the technologies and pipelines used for the analysis of the different cohorts is provided in SI Appendix, Table S9.
Rockefeller PID exome sequences.
Genomic DNA from peripheral blood mononuclear cells was extracted and sheared with a Covaris S2 Ultrasonicator. An adaptor-ligated library (Illumina) was generated, and exome capture was performed with SureSelect Human All Exon 37-, 50-, or 71-Mb kits (Agilent Technologies). Massively parallel WES was performed on a HiSeq 2000 or 2500 machine (Illumina), generating 72-, 100-, or 125-base reads. Quality controls were applied at the lane and fastq levels. Specifically, the cutoff used for a successful lane is Pass Filter > 90%, with over 250 M reads for the high-output mode. The fraction of reads in each lane assigned to each sample (no set value) and the fraction of bases with a quality score > Q30 for read 1 and read 2 (above 80% expected for each) were also checked. In addition, the FASTQC tool kit (www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to review base quality distribution, representation of the four nucleotides of particular k-mer sequences (adaptor contamination). We used the Genome Analysis Software Kit (GATK) (version 3.4–46) best-practice pipeline to analyze our WES data (14). Reads were aligned with the human reference genome (hg19), using the maximum exact matches algorithm in Burrows–Wheeler Aligner (BWA) (41). PCR duplicates were removed with Picard tools (picard.sourceforge.net/). The GATK base quality score recalibrator was applied to correct sequencing artifacts. GATK HaplotypeCaller was used to identify variant calls. DP ≥ 5 and MQ ≥ 30 were used as standard hard filtering criteria (22). Variants were annotated with SnpEff (snpeff.sourceforge.net/). Exomes were annotated for PASS and non-PASS variants in gnomAD r2.0.2 (Exome Aggregation Consortium, Broad Institute) and the 1000 Genomes Project Phase 3 (www.internationalgenome.org/) databases. Joint genotyping followed by VQSR filtering was not used because there have been reports of fractions of variants unique to individual samples being missed (https://gatkforums.broadinstitute.org/gatk/discussion/4150/should-i-analyze-my-samples-alone-or-together), rendering this approach unsuitable for our studies. For the purpose of comparison between the blacklist and VQSR approaches, VQSR was calculated with VariantRecalibrator and ApplyRecalibration for both SNPs and indels, with ts_filter_level set to 99.0 and other settings as specified by GATK recommendations. We did not use the InbreedingCoeff as this is discouraged in situations in which the cohort includes members of the same family, as in our cohort. Similarly, we did not include DP among the parameters of the VQSR, as it is not suitable for targeted exome sequencing samples.
GME Consortium neurological exome sequences.
WES for the GME Consortium was performed as previously described (27). Briefly, genomic DNA was extracted from peripheral blood mononuclear cells with Qiagen reagents and captured with the Agilent SureSelect Human All Exome 50-Mb kit. WES was performed on an Illumina HiSeq 2000. The GATK best-practice pipelines were used to analyze WES data (14). BWA was used to align reads with human reference genome NCBI Build 37 (41). The variant-call format files generated were annotated with the Rockefeller pipeline, as described above.
Africa exome sequences.
Whole-exome sequences were obtained for 300 African samples (31), and these data were processed together with those for 100 European individuals (42). All samples were sequenced with the Nextera Rapid Capture Expanded Exome kit, which delivers 62 Mb of genomic content per individual, including exons, UTRs, and microRNAs. Using the GATK Best Practice recommendations (43), we first mapped read-pairs onto the human reference genome (GRCh37) with BWA, version 0.7.7 (41), and reads duplicating the start position of another read were marked as duplicates with Picard Tools, version 1.94 (picard.sourceforge.net/). GATK, version 3.5 (14), was then used for base quality score recalibration (“BaseRecalibrator”), insertion/deletion (indel) realignment (“IndelRealigner”), and SNP and indel discovery for each sample (“Haplotype Caller”).
Infection exome sequences.
WES for the Infection cohort was performed as previously described (44, 45). In brief, genomic DNA was extracted from whole blood with the QIAamp DNA blood kit and captured with the Agilent SureSelect Human All Exome 50-Mb kit (Agilent SureSelect Human all exon V4 or V5) or Illumina Truseq 65-Mb enrichment kit. WES was performed on an Illumina HiSeq 2000 or Illumina HiSeq 2500 machine. BWA-MEM was used to map reads onto the human reference genome hg19 decoy, and GATK, version 3.8 (or an earlier version of this software), was used for data processing and analysis, according to GATK best practice.
Blacklist Creation.
The blacklists used in and provided with this manuscript were created by first collecting unique variants from 3,104 patient exomes and counting the occurrence of each variant (the number of patients reported to have the variant). The QC criteria used to collect these variants were equivalent to those used in gnomAD (MQ ≥ 30). However, we used a lower DP (DP ≥ 5), compatible with research approaches in which investigators want to retain as much information as possible. These criteria correspond to a high degree of QC despite low coverage, but may allow the discovery of true disease-causing variants, as illustrated by the example of the deletion of ISG15, which was initially identified by exome analysis despite a low DP of 4 (46). We did not use the QD value as a QC criterion due to the erroneous calls for some variants (https://gatkforums.broadinstitute.org/gatk/discussion/8912/most-variants-called). We explored the FN rate of the blacklists in the HGMD database and excluded variants that were present in the set of true disease-causing variants in HGMD according to further analyses (47). The measurement of variation at multiallelic sites was rendered more effective by separating variants into biallelic and multiallelic variant groups. Multiallelic variants represent a very specific challenge for the elimination of NPVs from exomes, as variants at multiallelic positions may occur individually in a small number of samples. Collectively, however, these variants may occur in a large proportion of the members of the cohort (i.e., many individuals may contain one of a number of variants at the position). The variants at multiallelic sites are often similar (e.g., G in the reference and an alternative of GA, GAA, GAAA, GAAAA, GAAAAA, etc.) but have remained resistant to removal from exomes by bioinformatic methods. For the capture of these variants, we collapsed all variants at multiallelic sites to a single value by calculating the total number of patients with any variant at the multiallelic position. When this number exceeded 1% of our cohort, all variants at the position concerned were included in the full blacklist. This procedure can thus identify variants present in only a few individuals but nevertheless occurring at positions with a high cumulative burden of variation in a cohort. We then considered biallelic variants. If the number of patients with any individual biallelic variant exceeded 1% of our cohort, the variant concerned was included in the full blacklist. For a schematic diagram of this pipeline, see SI Appendix, Fig. S1.
ReFiNE Generation and Usage.
ReFiNE and subsequent analyses were performed in Python programming language (version 2.7.14; https://www.python.org/) and R, using both default and publicly available libraries. The Python Tkinter module was used to design and implement the graphical interface for ReFiNE. ReFiNE is available as a graphical interface program (including a command-line option) that can be run on a standard laptop and is compatible with comma-separated (CSV) files. ReFiNE can also generate blacklists from WGS data, although this application has yet to be extensively tested. ReFiNE includes an optional parameter for the exclusion of a list of variants from the blacklist regardless of their frequency in the in-house database. This option can be used to remove a small number of known true disease-causing HGMD variants, for example. We also provide precalculated blacklists generated from our cohort of 3,104 PID exomes with cutoffs of 1%, 3%, 5%, and 10%. These blacklists can be used for small cohorts for which it may not be possible to generate custom blacklists. We also provide the PID, Neuro, Infection, Africa, and combined blacklists used in this manuscript, annotated with gnomAD MAFs. Finally, we have constructed a public server (lab.rockefeller.edu/casanova/BL) containing all of the supplemental files, the ReFiNE program, and a user-friendly online tool that can be used to query whether a variant is included in our blacklist or to annotate lists of variants in a similar manner.
Statistics and Figures.
The Scipy library (https://www.scipy.org/) was used for statistical analyses performed in Python. Seaborn (seaborn.pydata.org/) was used to generate figures in Python, together with matplotlib (https://matplotlib.org). Venn diagrams were generated with jvenn software (48). Wordclouds were generated with the WordCloud library (https://github.com/amueller/word_cloud). Prism (GraphPad) was also used for figure generation and statistical analysis.
Simulating minimum sample size and sample size saturation for blacklists.
We determined the minimum number of samples required for the creation of safe blacklists by generating random blacklists based on 10, 50, 100, 250, 500, 1,000, 1,500, 2,000, 2,500, 3,000, 3,500, 4,000, 4,500, 5,000, 5,500, 6,000, or 6,500 individuals from the PID and Neuro cohorts. We weighted the random selection of individuals for the blacklists by project size (i.e., for a sample size of 10, we picked 4 individuals at random from the PID cohort and 6 at random from the Neuro cohort). The selection of individuals for each sample size was repeated 30 times, and full blacklists for each iteration were generated with ReFiNE. The median number of BL-A variants and a 99% CI based on a normal distribution were calculated for each sample size and plotted (SI Appendix, Fig. S18). The number of samples required to reach saturation for blacklist variants was predicted by fitting a logarithmic trendline to the blacklist dataset based on the coefficient of determination (R2). The equation for this line was as follows:
where R2 = 0.7088 (SI Appendix, Fig. S18). We defined saturation as the number of samples for which less than one cohort-specific variant was added to the blacklist per new exome. Based on the best-fit equation, we calculated the saturation point as 2,801 individuals.
Characterization of blacklisted variants by HW equilibrium/disequilibrium, occurrence in low-complexity regions, and allelic distribution across genetic ancestries.
HW disequilibrium was calculated for the blacklisted variants found to be present in the European population (n = 1,150), which constituted the largest population of the PID cohort. χ2 tests were used to assess HW equilibrium. Given the large number of tests performed and the heterogeneity of European origins in our European cohort, a stringent threshold of 10−8 for significance was used for significance. A total of 106 variants with a P value below 10−8 were considered to be in HW disequilibrium and were stratified by excess genotype as follows: excess of heterozygotes (observed no. of heterozygotes > expected no. of heterozygotes, 57 variants), excess wild-type homozygotes (observed no. of wild-type homozygotes > expected no. of wild-type homozygotes, and χ2 for the wild-type homozygote > χ2 for the alternative homozygote, 13 variants), excess alternative homozygotes (observed no. of alternative homozygotes > expected no. of alternative homozygotes, and χ2 for alternative homozygotes > χ2 for wild-type homozygotes, 36 variants).
The occurrence of the variants in low-complexity regions was assessed with the following tracks from the UCSC Genome Browser: RepeatMasker and Simple Repeats (group: Repeats), and GC percent (group: Mapping and Sequencing). RepeatMasker was created from the RepeatMasker program, which screens DNA sequences for interspersed repeats and low-complexity DNA sequences; Simple Repeats reports simple tandem repeats located by Tandem Repeats Finder (TRF), which was designed especially for this purpose. Variants were considered to occur in GC-rich regions in which the G+C content exceeded 80%.
The heterogeneity of ethnicity was assessed in the four largest genetic ancestry groups in our cohort (European, African, North African, and Middle Eastern), for the variants found to be in HW equilibrium in the European population. χ2 tests were used to test the allelic distribution. In total, 203 variants with a P value below 10−8 were considered to be heterogeneous across ancestries. The ancestry driving heterogeneity was unequivocally determined for 67 variants, by testing the allelic distributions of four combinations of three populations from those mentioned above and determining the data for the missing population in the combination from the four that did not reach significance.
Sanger sequencing.
DNA was extracted from 10 SV40-fibroblast cell lines from patients included in our cohort. PCR amplification was performed with Hot-Start Taq Blue DNA Polymerase (Denville Scientific), 85 ng of template genomic DNA, and the primers listed in SI Appendix, Table S10. Sanger sequencing was performed with the BigDye Terminator kit (Perkin-Elmer).
Analysis of variation in patient exomes.
We identified the disease-causing mutation in patient D2 from a previous study (25), using a standard filtration pipeline. In brief, we removed variants with low-quality metrics (DP < 4, MQ < 40, QD < 2) that were common in public databases (variant frequency in gnomAD < 0.0001), variants of high-GDI genes (6), and variants with CADD scores below their gene-specific mutation significance cutoff (9). Gene burden was analyzed in our CMC cohort by first filtering each exome, as described above. We then compared the numbers of individuals with at least one variant for each mutated gene in the patient group between the patient (n = 208) and control (n = 960) groups in a one-tailed Fisher’s exact test. The resulting P values were used to rank genes, to identify those with the highest levels of enrichment in patients.
Supplementary Material
Acknowledgments
D.N.C. and P.D.S. gratefully acknowledge financial support from Qiagen, Inc., through a license agreement with Cardiff University. This work was supported by the National Institutes of Health (Grants P01AI061093, U24AI086037, R18AI048693, T32GM007280, R01AI088364, R01AI095983, and R01AI127564), the French National Research Agency (ANR 14-CE15-0009-01), the Jeffrey Modell Foundation, and the David S. Gottesman Immunology Chair and the Charles Bronfman Institute for Personalized Medicine, Icahn School of Medicine at Mount Sinai.
Footnotes
Conflict of interest statement: A.T. has coauthored multiple papers with J.F. and J.G.G. M.W. coauthored a 2017 paper with J.G.G.
Data deposition: ReFiNE and precalculated blacklists are available at GitLab on https://gitlab.com/pmaffucci/refine.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808403116/-/DCSupplemental.
References
- 1.Casanova JL, Conley ME, Seligman SJ, Abel L, Notarangelo LD. Guidelines for genetic studies in single patients: Lessons from primary immunodeficiencies. J Exp Med. 2014;211:2137–2149. doi: 10.1084/jem.20140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyts I, et al. Exome and genome sequencing for inborn errors of immunity. J Allergy Clin Immunol. 2016;138:957–969. doi: 10.1016/j.jaci.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstein DB, et al. Sequencing studies in human genetics: Design and interpretation. Nat Rev Genet. 2013;14:460–470. doi: 10.1038/nrg3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stenson PD, et al. The Human Gene Mutation Database: Towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet. 2017;136:665–677. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lek M, et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itan Y, et al. The human gene damage index as a gene-level approach to prioritizing exome variants. Proc Natl Acad Sci USA. 2015;112:13615–13620. doi: 10.1073/pnas.1518646112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itan Y, et al. The mutation significance cutoff: Gene-level thresholds for variant predictions. Nat Methods. 2016;13:109–110. doi: 10.1038/nmeth.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itan Y, et al. The human gene connectome as a map of short cuts for morbid allele discovery. Proc Natl Acad Sci USA. 2013;110:5558–5563. doi: 10.1073/pnas.1218167110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao R, et al. Review of current methods, applications, and data management for the bioinformatics analysis of whole exome sequencing. Cancer Inform. 2014;13:67–82. doi: 10.4137/CIN.S13779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacArthur DG, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuentes Fajardo KV, et al. NISC Comparative Sequencing Program Detecting false-positive signals in exome sequencing. Hum Mutat. 2012;33:609–613. doi: 10.1002/humu.22033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alcaïs A, et al. Life-threatening infectious diseases of childhood: Single-gene inborn errors of immunity? Ann N Y Acad Sci. 2010;1214:18–33. doi: 10.1111/j.1749-6632.2010.05834.x. [DOI] [PubMed] [Google Scholar]
- 16.Casanova JL. Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proc Natl Acad Sci USA. 2015;112:E7128–E7137. doi: 10.1073/pnas.1521651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casanova JL. Human genetic basis of interindividual variability in the course of infection. Proc Natl Acad Sci USA. 2015;112:E7118–E7127. doi: 10.1073/pnas.1521644112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belkadi A, et al. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci USA. 2015;112:5473–5478. doi: 10.1073/pnas.1418631112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belkadi A, et al. Exome/Array Consortium Whole-exome sequencing to analyze population structure, parental inbreeding, and familial linkage. Proc Natl Acad Sci USA. 2016;113:6713–6718. doi: 10.1073/pnas.1606460113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones E, Oliphant E, Peterson P. 2001 SciPy: Open source scientific tools for Python, version 1.1.0. Available at https://www.scipy.org/. Accessed December 12, 2018.
- 21.Whiffin N, et al. Using high-resolution variant frequencies to empower clinical genome interpretation. Genet Med. 2017;19:1151–1158. doi: 10.1038/gim.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo Y, Ye F, Sheng Q, Clark T, Samuels DC. Three-stage quality control strategies for DNA re-sequencing data. Brief Bioinform. 2014;15:879–889. doi: 10.1093/bib/bbt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auton A, et al. 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buckley AR, et al. Pan-cancer analysis reveals technical artifacts in TCGA germline variant calls. BMC Genomics. 2017;18:458. doi: 10.1186/s12864-017-3770-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuehn HS, et al. Loss of B cells in patients with heterozygous mutations in IKAROS. N Engl J Med. 2016;374:1032–1043. doi: 10.1056/NEJMoa1512234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toubiana J, et al. International STAT1 Gain-of-Function Study Group Heterozygous STAT1 gain-of-function mutations underlie an unexpectedly broad clinical phenotype. Blood. 2016;127:3154–3164. doi: 10.1182/blood-2015-11-679902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott EM, et al. Greater Middle East Variome Consortium Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat Genet. 2016;48:1071–1076. doi: 10.1038/ng.3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fazekas A, Steeves R, Newmaster S. Improving sequencing quality from PCR products containing long mononucleotide repeats. Biotechniques. 2010;48:277–285. doi: 10.2144/000113369. [DOI] [PubMed] [Google Scholar]
- 30.Clarke LA, Rebelo CS, Gonçalves J, Boavida MG, Jordan P. PCR amplification introduces errors into mononucleotide and dinucleotide repeat sequences. Mol Pathol. 2001;54:351–353. doi: 10.1136/mp.54.5.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez M, et al. The demographic history and mutational load of African hunter-gatherers and farmers. Nat Ecol Evol. 2018;2:721–730. doi: 10.1038/s41559-018-0496-4. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell AA, Zwick ME, Chakravarti A, Cutler DJ. Discrepancies in dbSNP confirmation rates and allele frequency distributions from varying genotyping error rates and patterns. Bioinformatics. 2004;20:1022–1032. doi: 10.1093/bioinformatics/bth034. [DOI] [PubMed] [Google Scholar]
- 33.Hwang S, Kim E, Lee I, Marcotte EM. Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci Rep. 2015;5:17875. doi: 10.1038/srep17875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandmann S, et al. Evaluating variant calling tools for non-matched next-generation sequencing data. Sci Rep. 2017;7:43169. doi: 10.1038/srep43169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campbell IM, et al. Multiallelic positions in the human genome: Challenges for genetic analyses. Hum Mutat. 2016;37:231–234. doi: 10.1002/humu.22944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLaren W, et al. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghoneim DH, Myers JR, Tuttle E, Paciorkowski AR. Comparison of insertion/deletion calling algorithms on human next-generation sequencing data. BMC Res Notes. 2014;7:864. doi: 10.1186/1756-0500-7-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maffucci P, et al. 2018 Data from “Blacklisting variants common in private cohorts but not in public databases optimizes human exome analysis.” GitLab. Available at https://gitlab.com/pmaffucci/refine. Deposited December 19, 2018.
- 41.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quach H, et al. Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell. 2016;167:643–656.e17. doi: 10.1016/j.cell.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asgari S, et al. Severe viral respiratory infections in children with IFIH1 loss-of-function mutations. Proc Natl Acad Sci USA. 2017;114:8342–8347. doi: 10.1073/pnas.1704259114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asgari S, et al. Swiss Pediatric Sepsis Study Exome sequencing reveals primary immunodeficiencies in children with community-acquired Pseudomonas aeruginosa sepsis. Front Immunol. 2016;7:357. doi: 10.3389/fimmu.2016.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bogunovic D, et al. Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science. 2012;337:1684–1688. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stenson PD, et al. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 48.Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. jvenn: An interactive Venn diagram viewer. BMC Bioinformatics. 2014;15:293. doi: 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.