Significance
Scaffold proteins are key regulators of signal transduction that bind multiple proteins of a signaling pathway. This simple tethering model explains how the scaffold colocalizes the components for signal propagation, but it is unclear whether scaffolds ensure signal amplification. Here, we use arrestin-3 and the ASK1-MKK4/7-JNK3 kinase cascade as a model system to explore whether scaffolding amplifies signaling. Our studies suggest a model of scaffold-dependent signal amplification that may be conceptually applicable to other scaffold-dependent signaling pathways.
Keywords: mitogen-activated protein kinase, arrestin, protein scaffold, cell signaling, signal amplification
Abstract
Scaffold proteins tether and orient components of a signaling cascade to facilitate signaling. Although much is known about how scaffolds colocalize signaling proteins, it is unclear whether scaffolds promote signal amplification. Here, we used arrestin-3, a scaffold of the ASK1-MKK4/7-JNK3 cascade, as a model to understand signal amplification by a scaffold protein. We found that arrestin-3 exhibited >15-fold higher affinity for inactive JNK3 than for active JNK3, and this change involved a shift in the binding site following JNK3 activation. We used systems biochemistry modeling and Bayesian inference to evaluate how the activation of upstream kinases contributed to JNK3 phosphorylation. Our combined experimental and computational approach suggested that the catalytic phosphorylation rate of JNK3 at Thr-221 by MKK7 is two orders of magnitude faster than the corresponding phosphorylation of Tyr-223 by MKK4 with or without arrestin-3. Finally, we showed that the release of activated JNK3 was critical for signal amplification. Collectively, our data suggest a “conveyor belt” mechanism for signal amplification by scaffold proteins. This mechanism informs on a long-standing mystery for how few upstream kinase molecules activate numerous downstream kinases to amplify signaling.
Scaffold proteins are critical in the regulation of numerous cellular signaling pathways. The action of protein scaffolds is often attributed to their ability to organize signaling cascades by binding and orienting individual components (1). This tethering model, however, can create a conceptual problem regarding signal amplification: a stable complex would prevent rapid biological response (2).
Among the better studied signaling pathways regulated by scaffolds are mitogen-activated protein kinase (MAPK) cascades (3–6) (SI Appendix, Fig. S1A). MAPK cascades involve three sequentially acting kinases (in general terms, a MAP3K, a MAP2K, and a MAPK). Each kinase becomes competent to phosphorylate downstream substrates when it is both (i) phosphorylated within the activation loop and (ii) liganded with ATP (7). MAPK activity is linked to phosphorylation, so that an “inactive” kinase is unphosphorylated and an “active” kinase has been phosphorylated by upstream kinase(s). The final, activated kinase phosphorylates downstream substrates and yields a biological response. Protein phosphatases terminate signaling by removing the attached phosphates (8). The mechanistic details of MAPK signaling pathways are still contested (3), although spatial and temporal regulators of MAPK proteins play direct roles in signal specificity (5). Mathematical modeling has provided insights into how an initial signal could be amplified through a nonspecific set of MAPK interactions and shown that amplitude and duration of the signal can determine biological outcomes (9, 10).
To elucidate how scaffold proteins modulate MAPK signaling, we focused on arrestin-3 scaffolding of the ASK1-MKK4/7-JNK3 cascade, where biological effects of inappropriate JNK3 activity include neurodegeneration via cellular apoptosis (5, 11). Different names of arrestin proteins are used in the field. We use the systematic names of arrestins, where the number after the dash indicates the order of cloning: arrestin-1 (historic names S-antigen, 48-kDa protein, visual or rod arrestin), arrestin-2 (β-arrestin or β-arrestin1), arrestin-3 (β-arrestin2 or hTHY-ARRX), and arrestin-4 (cone or X-arrestin). Prior studies showed that arrestin-3 scaffolds the JNK3 cascade (4, 12–14) in both a GPCR-dependent (15, 16) and -independent (13, 17–19) fashion. The mechanism of arrestin-mediated MAPK activation, however, remains poorly understood (20). Here we explore the mechanism of receptor-independent, arrestin-3–dependent signal amplification in the JNK3 pathway by combining experimental and computational approaches. To make measurements more experimentally tractable, we used the arrestin-3(1–393) protein, which exhibits a preactivated state in solution, meaning its equilibrium is likely shifted toward the active conformation. This established tool eliminated the need for arrestin activation by GPCRs and nonreceptor activators, and we validated the biological relevance using full-length arrestin-3 activated by the nonreceptor activator IP6. Collectively, our data suggest a “conveyor belt” mechanism that is likely generalizable to other scaffolds.
Results
Purified Arrestin-3 and an Arrestin-Derived T1A Peptide Directly Interact with MKK4, MKK7, and JNK3 in Vitro.
To complement previous reports that arrestin-3 interacts with kinases of the JNK3 cascade in cells, we performed an in vitro pull-down assay. Since this assay primarily detects high-affinity interactions, we focused on the unphosphorylated MAP2Ks and JNK3, which have been suggested as high-affinity arrestin binding partners (13, 21–23). For these assays we fused MBP to either arrestin-3 or the first 25 amino acids of arrestin-3 (a peptide termed T1A that facilitates JNK3 activation in cells) (23). We tested binding in the presence or absence of physiological concentrations of ATP to assess the role of the kinase domain in the arrestin-3 interaction. Unphosphorylated MKK4 exhibited robust binding to both MBP–arrestin-3 and MBP-T1A in the presence of ATP (SI Appendix, Fig. S1B). The quantities of MKK4 that were pulled down in this assay by MBP–arrestin-3 and MBP-T1A did not differ significantly, suggesting that ATP-liganded MKK4 binds largely to the N terminus of arrestin-3. Unphosphorylated MKK7 bound more robustly to MBP–arrestin-3 in the presence of ATP (SI Appendix, Fig. S1C). Its binding to MBP-T1A, although weaker than the full-length protein, also showed ATP dependence. These data suggest that MKK4 and MKK7 share binding sites on the N terminus of arrestin-3 and that ATP-induced conformational changes in the kinase domain facilitate MAP2K binding to arrestin-3.
In the case of unphosphorylated JNK3 (SI Appendix, Fig. S1D) we observed robust binding to full-length arrestin-3, but weak binding to MBP-T1A. This contrasts with the MAP2Ks, where the T1A peptide interacts strongly in each case. The finding that the T1A peptide cannot recapitulate the interaction between unphosphorylated JNK3 and full-length arrestin-3 is consistent with previous reports that JNK3 has multiple sites of interaction on arrestin-3, including contact points outside its N terminus (24). As with the MAP2Ks, unphosphorylated JNK3 binding to arrestin-3 was enhanced by ATP. The binding of doubly phosphorylated JNK3 to either MBP–arrestin-3 or MBP-T1A was not detectable in this assay, suggesting that kinase activation weakens its interaction with arrestin-3.
The Affinities of MKK4/7 and JNK3 for Arrestin-3 Depend on Their Activation State.
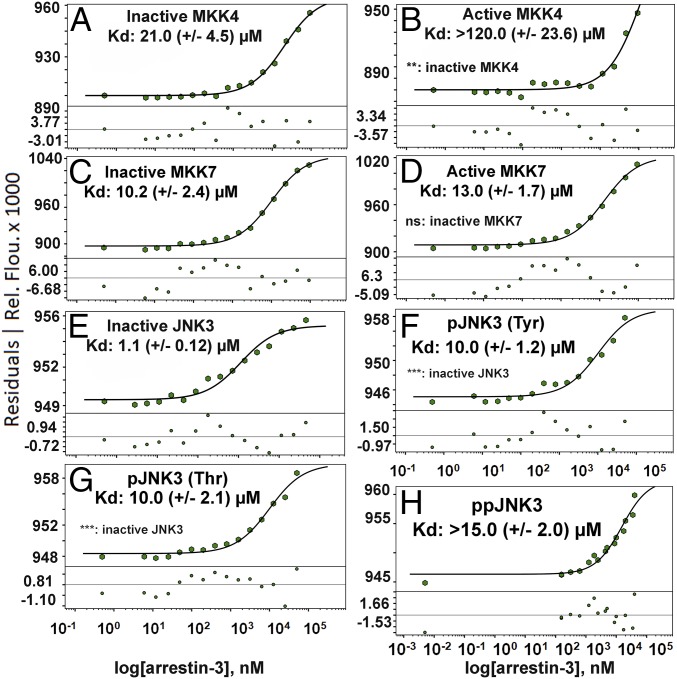
With direct binding of the signaling components established, we next explored how activation altered kinase affinity for arrestin-3. We measured the affinities of MKK4/7 (MAP2Ks) and JNK3 (MAPK) for truncated arrestin-3(1–393). This arrestin-3 construct likely samples the active conformation more readily, which allowed affinity measurements in the absence of activators. The affinity of full-length ASK1 (MAP3K) was not measured as this kinase does not express well enough in Escherichia coli. We measured the affinity of arrestin-3 for all other kinases in different activation states using microscale thermophoresis (MST) (Fig. 1) (25–27) in the presence of ATP and MgCl2.
Fig. 1.
The affinity of arrestin-3 for unphosphorylated and phosphorylated MKK4, MKK7, and JNK3. Average Kd values are shown for each curve (n = 3–6). Binding curves for arrestin-3 with unphosphorylated MKK4 (A), phosphorylated MKK4 (B), unphosphorylated MKK7 (C), phosphorylated MKK7 (D), unphosphorylated JNK3 (E), JNK3 phosphorylated at Tyr-223 (F), JNK3 phosphorylated at Thr-221 (G), and doubly phosphorylated JNK3 (H) in the presence of 1 mM ATP and 2 mM MgCl2 are shown. For the MAP2Ks, MKK4 and MKK7, the preset thermophoresis and T-jump were used to calculate the isotherm, whereas for the MAPK interactions the preset T-jump was used. Points where the fluorescence differed by more than 10% from the average were not included in the curve fit (SI Appendix, Fig. S1). Student’s two-tailed t test was used to assess significance of the differences between inactive (unphosphorylated) and active (phosphorylated) MAP2Ks and one-way ANOVA followed by Dunnett’s post hoc test were used to compare inactive JNK3 to singly or doubly phosphorylated JNK3 (**P < 0.01, ***P < 0.001, ns, not significant).
Activation of the MAP2Ks MKK4/7 occurs when an upstream MAP3K phosphorylates the conserved phosphorylation motif (SXXS or SXXT) in their activation loops (28). The unphosphorylated form of MKK4 exhibited higher affinity for arrestin-3 than the phosphorylated form, with Kd values of 21 (±4.5) µM for the inactive kinase and >120 (±23.6) µM for active MKK4 (Fig. 1 A and B). Importantly, the active kinase measurement suggested only the lower limit of the Kd value due to ligand-induced fluorescent effect at higher concentrations (SI Appendix, Fig. S2). However, the maximum concentrations used in the MST assay exceeded typical arrestin-3 intracellular concentrations (29, 30), and therefore yield a reasonable estimate of relative protein affinities. For MKK7, the unphosphorylated and phosphorylated form did not show statistically significant differences, with Kd values of 10.2 (±2.4) µM for the inactive and 13 (±1.7) µM for active MKK7 (Fig. 1 C and D).
The two MAP2Ks of the cascade, MKK4 and MKK7, work to fully activate the final effector, JNK3, by phosphorylating Tyr-223 and Thr-221, respectively (22, 31). Thus, JNK3 has four potential states: an unphosphorylated (inactive) state, two singly phosphorylated (partially active) states, and one doubly phosphorylated (fully active) state. Affinity measurements between JNK3 and arrestin-3 exhibited the same trend as MKK4, with unphosphorylated JNK3 having a higher affinity than singly or doubly phosphorylated JNK3. Notably, the Kd values of the distinct activation states significantly differed: unphosphorylated JNK3 bound arrestin-3 with high affinity [Kd of 1.1 (±0.12) µM] (Fig. 1E); singly phosphorylated JNK3 exhibited moderate affinities for arrestin-3, with Kd values of 10 (±1.2) µM for JNK3 pThr-221 (Fig. 1F) and 10 (±2.1) µM for JNK3 pTyr-223 (Fig. 1G), whereas doubly phosphorylated JNK3 exhibited lower affinity with a Kd of >15 μM (Fig. 1H). Interestingly, this effect was less pronounced in the absence of ATP (SI Appendix, Fig. S3).
Collectively, the MST affinity measurements indicate that in the presence of ATP, kinase phosphorylation decreases the affinity for arrestin-3. This implies that JNK3 phosphorylation induces its dissociation from arrestin-3, which would vacate the binding site and allow another inactive JNK3 to bind and become phosphorylated.
Kinase Phosphorylation Alters the Binding Site on Arrestin-3.
MAPKs achieve maximum activity when fully phosphorylated on their activation loop (32). While the structures of both unphosphorylated and phosphorylated members of the JNK3 cascade are not available, crystal structures of inactive and active ERK2 (40% identical to JNK3) have been determined (32, 33). Upon full phosphorylation, ERK2 remodels its active site and slightly rotates the N- and C-terminal lobes of the kinase domain (33). These conformational changes induce allosteric shifts in the surface loops that affect the ERK2 interactions with other macromolecules.
We used peptide array analysis to test whether phosphorylation-dependent conformational changes affected the binding position of each kinase on arrestin-3. This technique takes advantage of the fact that arrestins interact with binding partners via peptide-like surface fragments (13, 34–36). Based on our recent finding that the first 25 amino acids of arrestin-3 (T1A) are sufficient to scaffold the JNK3 cascade, we used peptides corresponding to this region to assess whether the binding site on arrestin-3 shifted upon kinase activation (23) (SI Appendix, Fig. S4A). We used the nonhydrolyzable ATP analog AMP-PNP to minimize assay-to-assay variability resulting from ATP hydrolysis during overnight incubation.
Incubation of inactive ASK1 with T1A-derived peptides implicated the first 15 residues of arrestin-3 in ASK1 binding (SI Appendix, Fig. S4 B and C). These residues also participated in binding of the downstream MAP2Ks MKK4/7 (SI Appendix, Fig. S4 D–I). Interestingly, unphosphorylated MKK4 and MKK7 demonstrated similar binding patterns, suggesting that they bind overlapping sites that include the T1A region of arrestin-3; however, phosphorylated MAP2Ks exhibited dissimilar binding. One interpretation of this finding is that the binding sites for these active kinases are optimized to stabilize an ideal orientation for acting on JNK3 (21). Both MAP2Ks exhibited an activation-dependent change in the pattern of peptide binding for T1A-derived peptides (SI Appendix, Fig. S4 E, F, H, and I).
We next assessed binding of final effector JNK3. Unphosphorylated JNK3 exhibited more modest binding to all T1A-derived peptides than did the upstream kinases (SI Appendix, Fig. S4K); however, doubly phosphorylated JNK3 only bound measurably to the T1A1–15 peptide (SI Appendix, Fig. S4L). As the affinity of inactive JNK3 for arrestin-3 in the MST experiments is fairly high, the modest signal observed suggests that JNK3 has additional binding contacts on full-length arrestin-3, in agreement with an earlier report (24) and our pull-down assays (SI Appendix, Fig. S1D). Importantly, JNK3 activation leads to decreased contact with the T1A region of arrestin-3, which is consistent with both the pull-down assays (SI Appendix, Fig. S1) and the lower affinity observed in the MST measurements (Fig. 1 E–H).
Notably, peptide array analysis revealed that unphosphorylated MKK4 and MKK7 bind to overlapping sites within the T1A peptide and that the active forms of MKK4, MKK7, and JNK3 also share binding regions. It is possible that there are additional contact points for each kinase on arrestin-3 that strengthen the interaction. An alternative interpretation is that only kinases that do not share overlapping sites can bind arrestin-3 simultaneously. In this scenario, the data suggest that MKK4 and MKK7 cannot bind to arrestin-3 at the same time, whereas inactive JNK3 can form a complex with arrestin-3 and an upstream kinase.
For comparison, we analyzed the binding of the kinases to the B1A peptide, a homologous peptide to T1A that contains the first 25 amino acids of arrestin-2. We observed that binding to B1A was not significantly affected by the presence of AMP-PNP or phosphorylation, suggesting that kinase contact points on B1A retain similar binding affinities over the course of activation. Because signal amplification requires release of the activated kinase, this difference between T1A and B1A binding may underlie the observed inability of arrestin-2 to facilitate the activation of JNK3 in cells (23).
Computational Modeling of MAP2K Action on JNK3.
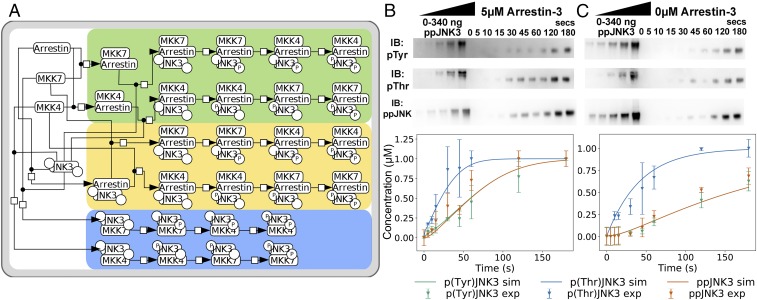
To identify a biochemical reaction mechanism to explain JNK3 activation, we built the JNK3-activation reaction model (JARMv1.0), a Python program that encodes models for the protein interactions relevant to MKK4/7 phosphorylation of JNK3 with and without arrestin-3 (Fig. 2A and SI Appendix, Fig. S5A). JARMv1.0 was encoded using PySB (37) to enable hypothesis testing as we explored plausible mechanisms. We used an in vitro kinase activation assay to give the model a dynamic point of reference for calibration. This assay characterized the JNK3 phosphorylation time course, in the presence or absence of arrestin-3, by MKK7 and MKK4 (Fig. 2 B and C and SI Appendix, Fig. S5B). The data showed 2.2-fold more phosphorylation of Thr-221 than Tyr-223 at 60 s of incubation when arrestin-3 is present. It also showed that ppJNK3 generation in the presence of arrestin-3 displayed a fold change between 1.2 and 1.8 higher than in the absence of arrestin-3.
Fig. 2.
Phosphorylation of inactive JNK3 by upstream kinases MKK4/7. (A) Network of the interactions between MKK4, MKK7, and JNK3 with arrestin-3. Nodes represent the different complexes formed by the interacting species, and the circles in the JNK3 node show the phosphorylation state of the tyrosine (Left) and threonine (Right) sites. The different orders in which JNK3 can be phosphorylated are highlighted by color: arrestin-3:MKK4/7 formation before JNK3 binding (green); arrestin-3:JNK3 complex formation before MKK4/7 binding (yellow); and MKK4/7 binding JNK3 in the absence of arrestin-3 (blue). The convention of Kitano et al. (64) was followed. (B and C) Representative Western blots showing phosphorylation of inactive JNK3 by MKK4 (Top, pTyr), MKK7 (Middle, pThr), or both kinases (Bottom, ppJNK3) at indicated time points upon coincubation of the three kinases in the presence of 5 µM arrestin-3(1–393) (B) or in its absence (C). Standards of ppJNK3 (20–340 ng) were used to calculate the nanograms of singly phosphorylated or doubly phosphorylated JNK3 generated over time (n = 3). JNK3 phosphorylation at Tyr-185, Thr-187, and both residues are shown for arrestin and no-arrestin conditions. Solid lines represent the model simulated trajectories, whereas the symbols show the experimental data used for model calibration.
To populate JARMv1.0 parameters, we used our experimentally determined binding constants (Fig. 1 and SI Appendix, Fig. S6) and values reported in the literature (38). Remaining parameters were estimated using PyDREAM, a Bayesian parameter inference formalism (39). This yielded parameter probability distributions, constrained by our experimental data, rather than single best-fit values (SI Appendix, Fig. S7). Inspecting these distributions, we observed a few that spanned a narrow parameter range, whereas others spanned a broader parameter range. From a statistical perspective, JNK3 activation is less sensitive to parameters with broad distributions and more sensitive to those with a narrow distribution. The calibration results suggest that the system is most sensitive to the catalytic phosphorylation constants of MKK4 and MKK7 (SI Appendix, Fig. S7). This gave us confidence that experimental measurements provided suitable constraints to explore the dynamics of JNK3 phosphorylation even without direct measurements of other parameters.
Calibrated JARMv1.0 showed that the time-dependent concentrations of singly phosphorylated JNK3 in complex (pTyr-JNK3:Arrestin-3 and pThr-JNK3:Arrestin-3) or doubly phosphorylated JNK3 (ppJNK3), exhibit nonlinear dynamics that emerge from multiple simultaneous reactions (Fig. 2 B and C, Top and SI Appendix, Fig. S5E). Our Bayesian calibration estimated that the catalytic constant of JNK3 phosphorylation by MKK7 is two orders of magnitude faster than the one of JNK3 phosphorylation by MKK4 (SI Appendix, Fig. S7). This leads to rapid consumption of unphosphorylated JNK3 and pTyr-JNK3, whereas pThr-JNK3 accumulates in the system over time. In addition, the reaction rate of MKK4 second phosphorylation on the arrestin-3 scaffold is up to one order of magnitude faster than all other second phosphorylation events with or without arrestin-3 (Fig. 2 B and C, Bottom). Taken together, these data suggest that MKK4 phosphorylation is the rate-limiting step in the JNK3 activation pathway, but multiple interactions take place to accomplish this outcome. This is consistent with active MKK4 having a lower affinity for arrestin-3 (Fig. 1 A and B).
JARMv1.0 also showed that the reactions involving arrestin-3 increase the yield of doubly phosphorylated JNK3 threefold at early time points (24 s) (SI Appendix, Fig. S5C) relative to the reaction system without arrestin-3. Previous studies have shown that less than 50% dual phosphorylation over total JNK expression can lead to a robust physiological response (40–42). JARMv1.0 execution places the optimal concentration of arrestin-3 for maximal JNK3 phosphorylation around ∼0.59 µM (SI Appendix, Fig. S5D).
Release of Active JNK3 from Arrestin-3 Is Necessary for Signal Amplification.
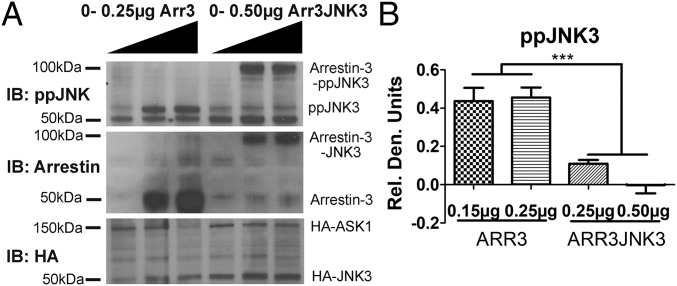
Signal amplification requires activation of multiple downstream signaling molecules following a single upstream stimulus. To test the biological role of the release of activated JNK3 from arrestin-3, we engineered an arrestin-3–JNK3 fusion and expressed it in HEK293 arrestin-2/3 knockout cells (43) (Fig. 3). After 48 h, we assessed the level of JNK3 activation in cells expressing free arrestin-3 or the arrestin-3–JNK3 fusion. We found significantly less JNK3 phosphorylation in cells expressing arrestin-3–JNK3 fusion than in cells expressing free arrestin-3 (Fig. 3B). Importantly, the JNK3 within the arrestin-3–JNK3 fusion was effectively phosphorylated (∼95 kDa band on ppJNK3 blot, Fig. 3A), demonstrating that the fused proteins are functional. These data provide evidence that the release of activated JNK3 increases JNK3 phosphorylation. This supports computational and experimental prediction that once phosphorylated, active JNK3 must dissociate from arrestin-3 to allow an inactive JNK3 molecule to bind, ensuring signal amplification.
Fig. 3.
JNK3 phosphorylation in cells expressing arrestin-3 or arrestin-3-JNK3 fusion. HEK293 arrestin-2/3 knockout cells were transfected with HA-ASK1 (0.1 µg), HA-JNK3 (0.65 µg), and either arrestin-3 (0–0.25 µg) or arrestin-3–JNK3 (0–0.5 µg) DNA for 48 h. (A) The cells were lysed and analyzed by Western using antibodies against ppJNK (9255S, Cell Signaling), arrestin (rabbit polyclonal F431 antibody, ref. 43), and HA (3724, Cell Signaling). QuantityOne software was used to analyze the ppJNK3 bands by densitometry. (B) The quantification of ppJNK3. The background was subtracted for each respective grouping (arrestin-3 and arrestin-3–JNK3) and the intensity of individual bands was divided by the sum total of band intensities (to eliminate the effect of exposure). Statistical analysis was performed using one-way ANOVA followed by Dunnett’s post hoc test with correction for multiple comparisons (***P < 0.001) (n = 5).
Discussion
Since the discovery of the first signaling scaffold (2), scaffold proteins have been the focus of numerous studies as they provide a simple mechanism for information flow in complex intracellular pathways. Prior studies have focused on how scaffold proteins exert their effects through simple tethering and localization of pathway components. There are, however, potential drawbacks to the simple tethering model. Most notable is the sequestration of signaling components leading to a reduction in signal (44). Despite the plethora of literature dedicated to protein scaffolding, the field lacks evidence on how protein scaffolds amplify signals.
It has been long established in the field that arrestin-mediated signaling occurs through both GPCR-dependent (15, 16) and GPCR-independent (13, 17–19) pathways. For example, studies supported the concept that GPCR-bound arrestin-3 increased JNK3 activation, but also demonstrated that the receptor was not obligatory for this function (45, 46). Essentially, in cells overexpressing upstream MAP3K ASK1, arrestin-3 facilitated JNK3 phosphorylation in the absence of receptor activation. Subsequently, an arrestin-3 mutant defective in GPCR binding was found to effectively activate JNK3 (13, 17). In vitro preactivated arrestin-3(1–393) facilitates JNK3 phosphorylation by MKK4 and MKK7 in the absence of GPCRs (47). Finally, a short 25-residue arrestin-3–derived peptide lacking most of receptor-binding elements effectively activated JNK3 in vitro and in cells (23). Thus, arrestin-3 can exist in an active (at least in terms of JNK3 activation) conformation without GPCR binding, likely due to its high flexibility revealed by structural work (48) and molecular dynamics modeling (49).
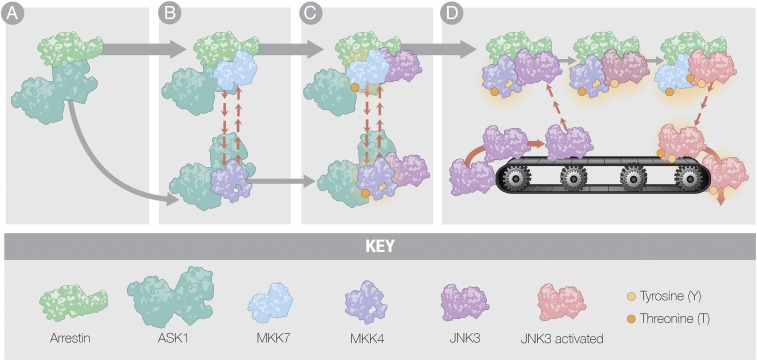
This receptor-independent activation of the JNK3 cascade commences with binding of MAP3K ASK1 to the arrestin scaffold. While ASK1 binds all four isoforms of vertebrate arrestins (13), only arrestin-3 causes an increase in JNK3 phosphorylation (13, 20, 21). Association of phosphorylated ASK1 with arrestin-3 is needed for downstream MAP2K activation. Dual phosphorylation of JNK3 by MKK4/7 is required for full activation of JNK3. Early evidence suggested that only MKK4 associates with the arrestin-3 (50), but later studies showed that both MKK4 and MKK7 bind arrestin-3 (47), albeit with different affinities, which is consistent with reported competition of these kinases for arrestin-3 (21). Therefore, we propose that arrestin-3 promotes the phosphorylation and activation of MKK4/7 by complexing with ASK1 (Fig. 4A). Since our peptide array data show significant overlap of the binding site of inactive MKKs, it is likely that the MKKs exchange on the arrestin-3/ASK1 complex (Fig. 4 B and C).
Fig. 4.
The model of arrestin-3–mediated JNK3 activation. While this model depicts a precise ordering of ASK1 followed by MKK4/7 and finally JNK3, this has yet to be experimentally validated. (A) The upstream MAP3K, apoptosis signal-regulating kinase 1 (ASK1), binds to arrestin-3 in preparation for activation of MKK4/7. (B) MKK4 and MKK7 exchange on arrestin-3 and become activated by ASK1. Both the inactive and active forms of MKK7 have higher affinity for arrestin-3, making it likely that MKK7 is bound more frequently than MKK4, which is supported by our computational modeling (Fig. 2). (C) Inactive JNK3 (purple) binds arrestin-3 so that it may become activated by MKK4 and MKK7. (D) Single phosphorylation of JNK3 on either its threonine or tyrosine residues (partially active JNK3 shown in purple/red) reduces its affinity for arrestin-3; full phosphorylation results in a greater decrease in affinity, allowing JNK3 to dissociate from arrestin-3 to vacate the site where another inactive JNK3 molecule can bind (active JNK3 shown in red).
MKK4/7 phosphorylation of JNK3 on Tyr-223 and Thr-221, respectively, results in its full activation. As previously surmised, MKK4/7 are not expected to bind arrestin-3 simultaneously. Evidence suggests that kinase scaffolding by arrestin-3 could enhance recruitment of additional cascade components via interaction with the arrestin-bound kinases (50); for example, it is well-established that MAPKs form complexes with cognate MAP2Ks and MAP3Ks through docking interactions (51). This type of recruitment would explain how an activated kinase with low affinity for arrestin-3 (i.e., phosphoMKK4) stays associated with the complex and activates its substrate. MST measurements of arrestin-3–JNK3 fusion protein binding to inactive MKK4 (SI Appendix, Fig. S6 A and C) yielded a decrease in affinity compared with arrestin-3 alone (Fig. 1A). Thus, both the MAP2K and MAPK can bind simultaneously to arrestin-3.
Global arrestin concentration changes throughout development: in early neural development it is 7–32 nM, while in late development it is closer to 200 nM (52). For our reported Kd values to be physiologically relevant, local cellular concentrations of arrestin would have to be higher. This is well documented for arrestin in the literature. For example, arrestins are recruited to GPCRs in the plasma membrane (53–55) and to microtubules (18), and it is likely that nonreceptor activators also influence local concentration; indeed, IP6 results in arrestin oligomerization (56, 57). Therefore, the reported Kd values are consistent with this mechanism being physiologically relevant.
To make investigation of this phenomenon experimentally tractable, we employed modified protein tools, including fusion proteins for pull-down assays and His-tagged kinases for the MST measurements. We also used a preactivated arrestin-3(1–393), an established tool in the arrestin field for studying processes affected by activated arrestin (57–62) that eliminates the need for arrestin activation by GPCRs or nonreceptor activators. These modifications did not affect the key conceptual conclusions, as evidenced by the experiments with full-length arrestin-3 in the presence of IP6 (SI Appendix, Fig. S6 B and D). If anything, we might have underestimated the affinity of arrestin-3 for members of the JNK3 cascade, as in-cell experiments (Fig. 3) suggest that the presence of arrestin-3 has a much greater effect on JNK3 activity in the cell than the in vitro data and modeling indicate.
Based on our data, we propose a conveyor belt mechanism of JNK3 activation, where an active JNK3 molecule is exchanged for an inactive JNK3 on an arrestin-3/MKK complex leading to signal amplification (Fig. 4D). This exchange is important biologically; formation of a tight complex would prevent the release of the downstream activated kinase and inhibit physiological response (2). It remains to be elucidated whether this is a unique characteristic of arrestin-3–mediated scaffolding, or whether other scaffolds of JNK activation cascades employ the same mechanism. Our data provide new insight into the mechanism of arrestin-3 scaffolding of the ASK1-MKK4/7-JNK3 cascade; most notably, a >15-fold weaker affinity of doubly phosphorylated JNK3 for arrestin-3 than inactive JNK3, and our model analysis shows a two order of magnitude faster phosphorylation of JNK3 by MKK7 compared with MKK4. Careful comparison of active and inactive JNK3 binding to other purified scaffold proteins is necessary to determine whether this conveyor belt model is applicable to other scaffolds.
Materials and Methods
Detailed procedures for the expression and purification of ASK1, MKK4/7, JNK3, and arrestin, and all biochemical studies are described in SI Appendix, Materials and Methods. The rule-based model described in Fig. 2A was constructed using the PySB python package (37), and model calibration was performed using the python package pyDREAM, which implements an adaptive Markov chain Monte Carlo simulation algorithm. Details of computational modeling are included in SI Appendix (39, 63).
Supplementary Material
Acknowledgments
We thank Dr. Asuka Inoue for the generous gift of the arrestin-2/3 knockout HEK293 cells and Dr. Chad Brautigam for discussions of MST data analysis. This work was supported by American Heart Association Award 16PRE30180007 (to N.A.P.) and Award 18PRE34030017 (to N.A.P.); the Vanderbilt International Scholars Program (O.O.O.); NIH Grants GM077561 and GM109955 (these two grants were replaced by R35 GM122491) (to V.V.G.); GM120569 (to T.M.I.); DA043680 (to T.M.I./V.V.G.); GM123252 (to K.N.D.); and U01-CA215845 (to C.F.L.); a Vanderbilt Discovery Award (to T.M.I./V.V.G.); CPRIT RP140648 (to K.N.D.); Welch F-1390 (to K.N.D.); National Science Foundation MCB 1411482 (to C.F.L.); and Oak Ridge Leadership Computational Facility Department of Energy DE-AC05-00OR22725 (to C.F.L.). This work used facilities supported by Grant P30EY008126 (Vanderbilt Core Grant in Vision Research).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819230116/-/DCSupplemental.
References
- 1.Locasale JW, Shaw AS, Chakraborty AK. Scaffold proteins confer diverse regulatory properties to protein kinase cascades. Proc Natl Acad Sci USA. 2007;104:13307–13312. doi: 10.1073/pnas.0706311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi KY, Satterberg B, Lyons DM, Elion EA. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 3.Brown MD, Sacks DB. Protein scaffolds in MAP kinase signalling. Cell Signal. 2009;21:462–469. doi: 10.1016/j.cellsig.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Good M, Tang G, Singleton J, Reményi A, Lim WA. The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell. 2009;136:1085–1097. doi: 10.1016/j.cell.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayawala K, Gelmi CA, Edwards JS. MAPK cascade possesses decoupled controllability of signal amplification and duration. Biophys J. 2004;87:L01–L02. doi: 10.1529/biophysj.104.051888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witzel F, Maddison L, Blüthgen N. How scaffolds shape MAPK signaling: What we know and opportunities for systems approaches. Front Physiol. 2012;3:475. doi: 10.3389/fphys.2012.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turjanski AG, Hummer G, Gutkind JS. How mitogen-activated protein kinases recognize and phosphorylate their targets: A QM/MM study. J Am Chem Soc. 2009;131:6141–6148. doi: 10.1021/ja8071995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke PR. Signal transduction. Switching off MAP kinases. Curr Biol. 1994;4:647–650. doi: 10.1016/s0960-9822(00)00144-5. [DOI] [PubMed] [Google Scholar]
- 9.Kolch W, Calder M, Gilbert D. When kinases meet mathematics: The systems biology of MAPK signalling. FEBS Lett. 2005;579:1891–1895. doi: 10.1016/j.febslet.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Adler M, Mayo A, Alon U. Logarithmic and power law input-output relations in sensory systems with fold-change detection. PLoS Comput Biol. 2014;10:e1003781. doi: 10.1371/journal.pcbi.1003781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi WS, Klintworth HM, Xia Z. JNK3-mediated apoptotic cell death in primary dopaminergic neurons. Methods Mol Biol. 2011;758:279–292. doi: 10.1007/978-1-61779-170-3_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levchenko A, Bruck J, Sternberg PW. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc Natl Acad Sci USA. 2000;97:5818–5823. doi: 10.1073/pnas.97.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song X, Coffa S, Fu H, Gurevich VV. How does arrestin assemble MAPKs into a signaling complex? J Biol Chem. 2009;284:685–695. doi: 10.1074/jbc.M806124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luttrell LM, Gesty-Palmer D. Beyond desensitization: Physiological relevance of arrestin-dependent signaling. Pharmacol Rev. 2010;62:305–330. doi: 10.1124/pr.109.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eishingdrelo H, et al. ERK and β-arrestin interaction: A converging point of signaling pathways for multiple types of cell surface receptors. J Biomol Screen. 2015;20:341–349. doi: 10.1177/1087057114557233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strungs EG, Luttrell LM. Arrestin-dependent activation of ERK and Src family kinases. In: Gurevich VV, editor. Arrestins - Pharmacology and Therapeutic Potential. Springer; Berlin: 2014. pp. 225–257. [DOI] [PubMed] [Google Scholar]
- 17.Breitman M, et al. Silent scaffolds: Inhibition OF c-Jun N-terminal kinase 3 activity in cell by dominant-negative arrestin-3 mutant. J Biol Chem. 2012;287:19653–19664. doi: 10.1074/jbc.M112.358192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanson SM, et al. Arrestin mobilizes signaling proteins to the cytoskeleton and redirects their activity. J Mol Biol. 2007;368:375–387. doi: 10.1016/j.jmb.2007.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kook S, et al. Caspase-cleaved arrestin-2 and BID cooperatively facilitate cytochrome C release and cell death. Cell Death Differ. 2014;21:172–184. doi: 10.1038/cdd.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rusnak L, Fu H. Regulation of ASK1 signaling by scaffold and adaptor proteins. Adv Biol Regul. 2017;66:23–30. doi: 10.1016/j.jbior.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Kook S, et al. Arrestin-3 binds c-Jun N-terminal kinase 1 (JNK1) and JNK2 and facilitates the activation of these ubiquitous JNK isoforms in cells via scaffolding. J Biol Chem. 2013;288:37332–37342. doi: 10.1074/jbc.M113.510412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhan X, Kaoud TS, Kook S, Dalby KN, Gurevich VV. JNK3 enzyme binding to arrestin-3 differentially affects the recruitment of upstream mitogen-activated protein (MAP) kinase kinases. J Biol Chem. 2013;288:28535–28547. doi: 10.1074/jbc.M113.508085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhan X, et al. Peptide mini-scaffold facilitates JNK3 activation in cells. Sci Rep. 2016;6:21025. doi: 10.1038/srep21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhan X, Perez A, Gimenez LE, Vishnivetskiy SA, Gurevich VV. Arrestin-3 binds the MAP kinase JNK3α2 via multiple sites on both domains. Cell Signal. 2014;26:766–776. doi: 10.1016/j.cellsig.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tso S-C, et al. Using two-site binding models to analyze microscale thermophoresis data. Anal Biochem. 2018;540-541:64–75. doi: 10.1016/j.ab.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 27.Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol. 2011;9:342–353. doi: 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuenda A. Mitogen-activated protein kinase kinase 4 (MKK4) Int J Biochem Cell Biol. 2000;32:581–587. doi: 10.1016/s1357-2725(00)00003-0. [DOI] [PubMed] [Google Scholar]
- 29.Gurevich EV, Benovic JL, Gurevich VV. Arrestin2 and arrestin3 are differentially expressed in the rat brain during postnatal development. Neuroscience. 2002;109:421–436. doi: 10.1016/s0306-4522(01)00511-5. [DOI] [PubMed] [Google Scholar]
- 30.Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci. 2004;25:105–111. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Lawler S, Fleming Y, Goedert M, Cohen P. Synergistic activation of SAPK1/JNK1 by two MAP kinase kinases in vitro. Curr Biol. 1998;8:1387–1390. doi: 10.1016/s0960-9822(98)00019-0. [DOI] [PubMed] [Google Scholar]
- 32.Zhang F, Strand A, Robbins D, Cobb MH, Goldsmith EJ. Atomic structure of the MAP kinase ERK2 at 2.3 A resolution. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 33.Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90:859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 34.ter Haar E, Harrison SC, Kirchhausen T. Peptide-in-groove interactions link target proteins to the beta-propeller of clathrin. Proc Natl Acad Sci USA. 2000;97:1096–1100. doi: 10.1073/pnas.97.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang DS, et al. Structure of an arrestin2-clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking. J Biol Chem. 2009;284:29860–29872. doi: 10.1074/jbc.M109.023366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmed MR, et al. Ubiquitin ligase parkin promotes Mdm2-arrestin interaction but inhibits arrestin ubiquitination. Biochemistry. 2011;50:3749–3763. doi: 10.1021/bi200175q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez CF, Muhlich JL, Bachman JA, Sorger PK. Programming biological models in Python using PySB. Mol Syst Biol. 2013;9:646. doi: 10.1038/msb.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhan X, Kook S, Gurevich EV, Gurevich VV. Arrestin-dependent activation of JNK family kinases. In: Gurevich VV, editor. Arrestins - Pharmacology and Therapeutic Potential. Springer; Berlin: 2014. pp. 259–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shockley EM, Vrugt JA, Lopez CF. PyDREAM: High-dimensional parameter inference for biological models in python. Bioinformatics. 2018;34:695–697. doi: 10.1093/bioinformatics/btx626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muniyappa H, Das KC. Activation of c-Jun N-terminal kinase (JNK) by widely used specific p38 MAPK inhibitors SB202190 and SB203580: A MLK-3-MKK7-dependent mechanism. Cell Signal. 2008;20:675–683. doi: 10.1016/j.cellsig.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lei K, et al. The Bax subfamily of Bcl2-related proteins is essential for apoptotic signal transduction by c-Jun NH(2)-terminal kinase. Mol Cell Biol. 2002;22:4929–4942. doi: 10.1128/MCB.22.13.4929-4942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khalid S, et al. cJun N-terminal kinase (JNK) phosphorylation of serine 36 is critical for p66Shc activation. Sci Rep. 2016;6:20930. doi: 10.1038/srep20930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alvarez-Curto E, et al. Targeted elimination of G proteins and arrestins defines their specific contributions to both intensity and duration of G protein-coupled receptor signaling. J Biol Chem. 2016;291:27147–27159. doi: 10.1074/jbc.M116.754887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Good MC, Zalatan JG, Lim WA. Scaffold proteins: Hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller WE, Lefkowitz RJ. Expanding roles for β-arrestins as scaffolds and adapters in GPCR signaling and trafficking. Curr Opin Cell Biol. 2001;13:139–145. doi: 10.1016/s0955-0674(00)00190-3. [DOI] [PubMed] [Google Scholar]
- 46.Miller WE, et al. Identification of a motif in the carboxyl terminus of β -arrestin2 responsible for activation of JNK3. J Biol Chem. 2001;276:27770–27777. doi: 10.1074/jbc.M102264200. [DOI] [PubMed] [Google Scholar]
- 47.Zhan X, et al. Arrestin-3-dependent activation of c-Jun N-terminal kinases (JNKs) Curr Protoc Pharmacol. 2015;68:2.12.1–12.12.26. doi: 10.1002/0471141755.ph0212s68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhan X, Gimenez LE, Gurevich VV, Spiller BW. Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual subtypes. J Mol Biol. 2011;406:467–478. doi: 10.1016/j.jmb.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sensoy O, Moreira IS, Morra G. Understanding the differential selectivity of arrestins toward the phosphorylation state of the receptor. ACS Chem Neurosci. 2016;7:1212–1224. doi: 10.1021/acschemneuro.6b00073. [DOI] [PubMed] [Google Scholar]
- 50.McDonald PH, et al. Beta-arrestin 2: A receptor-regulated MAPK scaffold for the activation of JNK3. Science. 2000;290:1574–1577. doi: 10.1126/science.290.5496.1574. [DOI] [PubMed] [Google Scholar]
- 51.Tanoue T, Nishida E. Docking interactions in the mitogen-activated protein kinase cascades. Pharmacol Ther. 2002;93:193–202. doi: 10.1016/s0163-7258(02)00188-2. [DOI] [PubMed] [Google Scholar]
- 52.Gurevich EV, Benovic JL, Gurevich VV. Arrestin2 expression selectively increases during neural differentiation. J Neurochem. 2004;91:1404–1416. doi: 10.1111/j.1471-4159.2004.02830.x. [DOI] [PubMed] [Google Scholar]
- 53.Barak LS, Ferguson SSG, Zhang J, Caron MG. A β-arrestin/green fluorescent protein biosensor for detecting G protein-coupled receptor activation. J Biol Chem. 1997;272:27497–27500. doi: 10.1074/jbc.272.44.27497. [DOI] [PubMed] [Google Scholar]
- 54.Namkung Y, et al. Monitoring G protein-coupled receptor and β-arrestin trafficking in live cells using enhanced bystander BRET. Nat Commun. 2016;7:12178. doi: 10.1038/ncomms12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goodman OB, Jr, et al. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383:447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- 56.Milano SK, Kim Y-M, Stefano FP, Benovic JL, Brenner C. Nonvisual arrestin oligomerization and cellular localization are regulated by inositol hexakisphosphate binding. J Biol Chem. 2006;281:9812–9823. doi: 10.1074/jbc.M512703200. [DOI] [PubMed] [Google Scholar]
- 57.Chen Q, et al. Structural basis of arrestin-3 activation and signaling. Nat Commun. 2017;8:1427. doi: 10.1038/s41467-017-01218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Orsini MJ, Benovic JL. Characterization of dominant negative arrestins that inhibit β2-adrenergic receptor internalization by distinct mechanisms. J Biol Chem. 1998;273:34616–34622. doi: 10.1074/jbc.273.51.34616. [DOI] [PubMed] [Google Scholar]
- 59.Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Crystal structure of β-arrestin at 1.9 A: Possible mechanism of receptor binding and membrane translocation. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 60.Shukla AK, et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature. 2014;512:218–222. doi: 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coffa S, et al. The effect of arrestin conformation on the recruitment of c-Raf1, MEK1, and ERK1/2 activation. PLoS One. 2011;6:e28723. doi: 10.1371/journal.pone.0028723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bennett TA, Foutz T, Gurevich V, Sklar L, Prossnitz E. Partial phosphorylation of the N-formyl peptide receptor inhibits G protein association independent of arrestin binding. J Biol Chem. 2002;276:49195–49203. doi: 10.1074/jbc.M106414200. [DOI] [PubMed] [Google Scholar]
- 63.Vrugt JA, Ter Braak CJF. DREAM(D): An adaptive Markov chain Monte Carlo simulation algorithm to solve discrete, noncontinuous, and combinatorial posterior parameter estimation problems. Hydrol Earth Syst Sci. 2011;15:3701–3713. [Google Scholar]
- 64.Kitano H, Funahashi A, Matsuoka Y, Oda K. Using process diagrams for the graphical representation of biological networks. Nat Biotechnol. 2005;23:961–966. doi: 10.1038/nbt1111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.