Abstract
The dramatic increase in antimicrobial resistance for pathogenic bacteria constitutes a key threat to human health. The Centers for Disease Control and Prevention has recently stated that world is on the verge of entering the “post-antibiotic era”, one where more people will die from bacterial infections than from cancer. Recently, nanoparticles (NPs) have emerged as new tools that can be used to combat deadly bacterial infections. Nanoparticle-based strategies can overcome the barriers faced by traditional antimicrobials, including antibiotic resistance. In this Tutorial Review, we have highlighted multiple nanoparticle-based approaches to eliminate bacterial infections providing crucial insight on the design elements that play critical roles in creating antimicrobial nanotherapeutics. In particular, we have focused on the pivotal role played by NP-surface functionality in designing nanomaterials as self-therapeutic agents and delivery vehicles for antimicrobial cargo.
Graphical Abstract
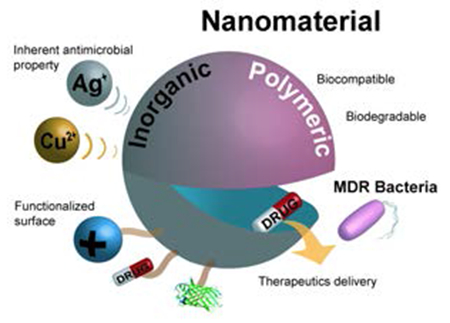
Nanomaterials as self-therapeutic agents and drug-delivery vehicles for antimicrobial therapies.
1. Introduction
The emergence of drug-resistant pathogenic bacteria constitutes one of the dominant challenges in human health. Antibiotic-resistant bacteria cause more than 2 million cases of severe illnesses, including 23,000 annual deaths in the US.1 Recent projections indicate that bacterial infections will result in 10 million annual deaths by 2050, more than that caused by cancer presently.2 Most cases of multi drug-resistant (MDR) infections require prolonged antibiotic therapy with tissue debridement (i.e. surgical removal) in some cases, resulting in low patient compliance and excessive health-care costs. Notably, these infections are responsible for a combined $55 billion per year towards excess health care and societal costs in US.3 Moreover, antibiotic treatment of resistant bacteria further contributes to increased tolerance in surviving bacterial cells. For instance, 40-60% of Staphylococcus aureus strains isolated from US hospitals are resistant to methicillin (MRSA, methicillin-resistant S. aureus) and in some cases even resistant to last-resort antibiotics such as carbapenems and vancomycin.4
Antibiotics act upon bacteria by targeting essential survival processes such as inhibiting cell-wall synthesis and interfering with the synthesis of vital proteins, DNA and RNA. However, bacteria possess the intrinsic ability (derived from aeons of competition) to evolve rapidly through mutations and transfer of DNA (through horizontal gene transfer) to overcome the threat posed by these antibacterials.5 Multiple drug resistance genes from different organisms can be acquired by the same microbe, resulting in emergence of multi-drug resistant (MDR) “superbug”. Studies on resistant-bacteria have found a ‘super-resistance’ gene (NDM-1) that causes enzymatic degradation of β-lactam antibiotics, making bacteria resistant against broad-range of antibiotics.7 Similarly, millions of cases of MDR mycobacterium tuberculosis (MDR-TB) have been reported that are now resistant to current antibiotics.3 Antibiotic resistance in bacteria will increase and the situation will become more dire as the number of MDR strains continues to grow. This rapidly escalating threat has contributed to an urgent need to discover novel antibacterials and new treatment strategies to combat these highly resistant bacteria.
Antimicrobial peptides (AMPs) have emerged as promising therapeutics, demonstrating broad-spectrum activity and reduced acquisition of resistance. These favorable characteristics can be attributed to the unique amphiphilic topology of peptides featuring polycationic headgroups, enabling them to disrupt microbial membrane.6 Building on the structural advantages of both antibiotics and AMPs, nanomaterials provide another potential solution for antimicrobial therapies. For example, nanomaterials can simultaneously disrupt the bacterial membrane and target intracellular components to impede proper functioning of cellular machinery.7 Distinct physio-chemical traits of nanomaterials make them promising candidates to achieve enhanced therapeutic efficacy against resilient MDR infections. Nanomaterials can execute multiple bactericidal pathways, making it difficult for bacteria to adapt against these therapeutics.8 These pathways are dependent upon the inherent core material, shape, size and surface chemistry of nanomaterial scaffolds. Moreover, high therapeutic loading coupled with enhanced ability to penetrate biological membrane make nanomaterials excellent candidates to transport drugs at the site of infection.9 Finally, the ability to modulate nanomaterial interaction with bacterial cellular systems plays a pivotal role in improving therapeutic efficacy of the treatment.10
Nanomaterial-based antimicrobials have demonstrated their efficacy against both planktonic (free floating bacteria) and biofilm (bacterial community) infections.9,11 In this review, we have mainly focused on the nanomaterial-based strategies for combating MDR infections caused by planktonic bacteria. We have divided this review into two main sections based on the role played by nanomaterials in the antibacterial therapy: (1) active therapeutic agent and (2) delivery vehicle for an antimicrobial agent. In both the sections we will focus on the surface functionalization strategies for combating MDR bacteria.
2. Interaction of Bacteria with Nanomaterials
Nanomaterial-bacteria interactions depend upon multiple factors such as electrostatic attraction, hydrophobic and receptor-ligand interaction and Van der Waals forces.7 Fundamental study of interactions between nanomaterials and bacteria provides crucial insight for designing novel antimicrobial agents.
2.1. Bacterial membrane penetration by nanomaterials
Bacteria are mainly classified into Gram-negative and Gram-positive depending upon the structure of their cell wall (Figure 1a). The cell wall of Gram-positive bacteria has a thick peptidoglycan layer (15-100 nm) with polymeric techoic acids and a cytoplasmic membrane underneath. The phosphates present in techoic acid polymeric chains are responsible for bacterial negative charge and serve as binding site for divalent cations in the solution.5 On the other hand, Gram-negative bacteria consists of a cytoplasmic membrane followed by a thin peptidoglycan layer (20-50 nm), further protected by a hydrophobic lipid bilayer consisting of lipopolysaccharides. This additional lipid layer greatly reduces the penetration ability of numerous hydrophobic antibacterial agents such as detergents.12 The bacterial membrane is negatively charged primarily due to presence phosphates and carboxylates as a component of lipopolysaccharides present on Gram-negative bacteria. The structure of bacterial cell wall plays a crucial role in determining the interaction of NPs with the microbes.
Figure 1.
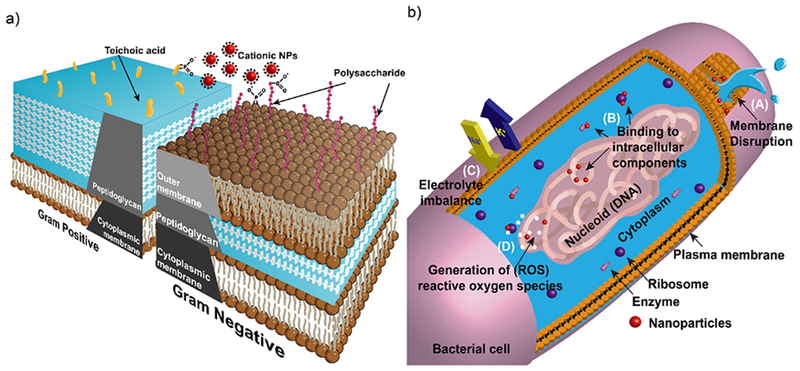
Schematic diagram showing a) cell wall structures of Gram-positive and Gram-negative bacteria. b) antimicrobial mechanism of NPs. i) disruption of cell membrane resulting in cytoplasmic leakage. ii) binding and disruption of intracellular components. iii) disrupting electron transport causing electrolyte imbalance. iv) generation of reactive oxygen species (ROS).
In early studies of nanoparticle-microbe interactions Murphy and coworkers have demonstrated that CTAB coated gold nanorods or nanospheres were homogenously distributed on Gram-positive B. cereus. This phenomenon was attributed to the electrostatic interaction between the positively charged nanomaterial and negatively charged techoic acid moieties on the bacteria.12 Alternatively, mannose substituted gold nanoparticles exhibited binding to pili on Gram-negative E. coli. The pili are hair like structures emanating from bacterial surface that are rich in lectin (sugar-binding proteins) and hence preferentially binding to the mannose coated NPs.13 Building on these observations, Rotello and coworkers demonstrated that cationic NPs possessed toxicity towards bacteria.10 In subsequent studies, the subtle interplay between NPs coverage and membrane structure demonstrated that positively charged hydrophobic AuNPs formed spatial aggregates on the bacterial membrane. Gold nanoparticles (AuNP) of 2 nm core diameter exhibited low toxicity against E. coli (Gram-negative) but rapidly lysed B. subtillis (Gram-positive) bacteria.14 The interaction of specific NP functionality and membrane structure can result in blebbing, tubule formation or other membrane defects.
2.2. Antimicrobial mechanism of NPs
The therapeutic activity of many antibiotics originates from their ability to inhibit cell wall synthesis, interfering with the expression of essential proteins and disrupting DNA replication machinery. However, bacteria have developed the ability to resist each of these mechanisms of action. One fundamental mechanism of bacterial resistance is alteration of the target of the antibiotic.5 For example, modification of cell wall components confers resistance to vancomycin, whereas altered structures of ribosomes resist tetracycline.7 Similarly, bacteria can overexpress enzymes such as β-lactamases and aminoglycosides to degrade antibiotics. Additionally, overexpression of efflux pumps enables bacteria to evade multiple antibiotics simultaneously. Finally, many pathogens such as Chlamydophila pneumonia reside inside the cellular compartments of the host cells to escape from the antibiotics that are mostly confined to extracellular space.4,8
Nanomaterials can overcome the antibiotic-resistance mechanisms owing to their unique physio-chemical properties, enabling nanomaterials to execute multiple novel bactericidal pathways to achieve antimicrobial activity. Nanomaterials can bind and disrupt bacterial membrane causing leakage of cytoplasmic components.9 Upon membrane permeation, nanomaterials can also bind to intracellular components such as DNA, ribosomes and enzymes to disrupt the normal cellular machinery (Figure 1b). Disruption in cellular machinery can lead to oxidative stress, electrolyte imbalance and enzyme inhibition resulting in cell death.7 The bactericidal pathways followed by nanomaterials are inherently dependent upon their core material, shape, size and surface functionalization. In the early studies of nanomaterial-based antimicrobials, researchers varied inherent core materials to generate nanomaterials with different mechanism of action. For example, silver nanoparticle-based antimicrobials utilize free Ag+ ion as active agent. The silver ions disrupt the bacterial membrane and electron transport while simultaneously causing DNA damage.18 Similarly, free Cu2+ions from copper NPs can generate reactive oxygen species (ROS) that disrupts amino acid synthesis and DNA in bacterial cells. On the other hand, ZnO and TiO2 based nanomaterials cause cell membrane damage and generate ROS to kill bacteria.8,11 Different nanomaterial cores can offer a range of antibacterial mechanisms to combat drug-resistant superbugs. However, these non-functionalized nanomaterials often exhibit narrow-spectrum activity against bacterial species. Moreover, they display low therapeutic indices (i.e. selectivity) against healthy mammalian cells, limiting their widespread use in biomedical applications. Surface chemistry of nanomaterials is critical to modulate their interaction with bacteria, improving their broad-spectrum activity while simultaneously reducing their toxicity against mammalian cells.
3. Nanomaterials as active therapeutic agents
Nanomaterials provide a versatile platform to generate novel therapeutic strategies due to their unique physiochemical properties. Nanomaterial are similar in size scale to biomolecular and bacterial cellular systems, enabling additional multivalent interactions as compared to small molecule antibiotics.15 Furthermore, their large surface area enables multivalent interactions with bacteria along with high cargo loading. Additionally, nanomaterials can with appropriate engineering overcome common bacterial drug resistance mechanisms such as overexpression of efflux pumps.23
3.1. Small molecule functionalized NPs
Appropriately functionalized nanomaterials provide a powerful tool to penetrate bacterial cell membrane. In pioneering studies of functionalized nanomaterials-based antimicrobials, Xu’s group generated vancomycin-functionalized gold nanoparticles (AuNPs) to overcome vancomycin-resistant enterococci (VRE).16 In this study, ~5 nm AuNPs were conjugated with bis(vancomycin) cystamide through Au-S bond resulting in ~61 vancomycin molecules per NP (Figure 2). The antimicrobial activity of vancomycin-capped AuNPs (Van@Au) was evaluated by determining their MICs - minimum concentrations to inhibit bacterial growth. Van@Au displayed MIC of 2-4 μg.mL−1, whereas vancomycin alone showed MICs of 64 μg.mL−1 against VRE. Additionally, the functionalized NPs were also effective against E. coli while the antibiotics alone were inactive. In a similar study, aminoglycosidic antibiotics were adsorbed on AuNPs via interaction between Au surface and amine groups of antibiotics.17 Antibiotic capped NPs exhibited high antibacterial efficacy against both Gram-positive and Gram-negative bacterial species. In a non-covalent conjugation approach by Fayaz and coworkers, substantial improvement in broad-spectrum activity of silver nanoparticles (AgNPs) was observed in presence of ampicillin. Hydroxyl and amido groups on ampicillin chelate with AgNPs forming AgNP-ampicillin complex. The ampicillin molecules target the bacterial membrane enabling better penetration of AgNPs which in turn bind with DNA resulting in cell death. The improved antibacterial activity of these formulations can be attributed to enhanced internalization of antibiotics inside bacterial cells coupled with polyvalent effect of concentrated antibiotics present on nanomaterial surface.18
Figure 2.
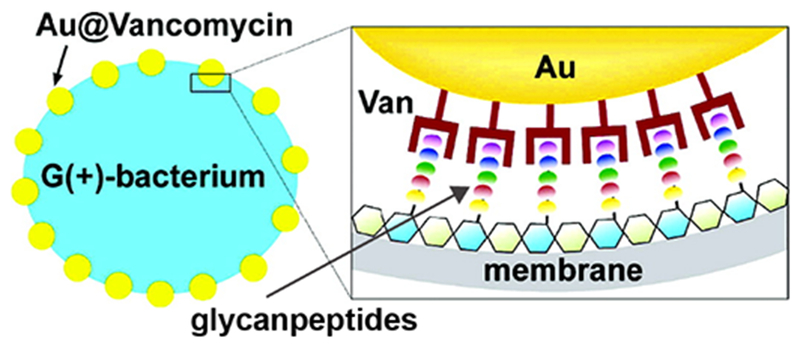
Schematic diagram showing the binding of vancomycin capped gold nanoparticles with vancomycin-resistant enterococci. Reproduced from reference 16 with permission from American Chemical Society, Copyright © 2003.
Another strategy to generate antimicrobial nanoparticles involves functionalizing the nanoparticles with non-antibiotic small molecules. Jiang and coworkers developed 3 nm AuNPs capped with amino-substituted pyrimidines that exhibited antibacterial activities against MDR clinical isolates.19 These positively charged NPs could effectively bind with and disrupt bacterial membrane, resulting in a leakage of bacterial contents such as nucleic acids. Proteomic analysis revealed that internalized NPs could also bind with the DNA and inhibit protein synthesis. Furthermore, these functionalized NPs could also collapse membrane potential and compromise ATPase activities, resulting in cell death. In subsequent studies, Rotello and coworkers utilized ligand engineering to control the surface properties of nanoparticles for combating MDR bacteria, using a library of cationic NPs featuring functional groups with varying chain length, aromatic and non-aromatic characteristics (Figure 3 a, b). The structure-activity relationship revealed that antimicrobial activity of AuNPs could be controlled by varying the hydrophobicity of the ligands on NP surface. Cationic and hydrophobic NP3 could effectively suppress the growth of 11 clinical MDR isolates at concentrations ranging from 8-64 nM. These NPs were able to disrupt bacterial membrane, leaking the cytoplasmic contents and eventually resulting in cell death.20 Significantly, bacteria were unable to develop resistance against these NPs even after 20 passages at sub-MIC concentrations. Notably, these particles showed hemolytic activity (HC50) at 400 nM, providing a therapeutic selectivity (HC50/MIC) of up-to 50-fold. Considering their ability to delay the onset of resistance in bacteria, these NPs were used in combination with antibiotics for the treatment of planktonic MDR bacteria. The results indicated that sub-MIC concentrations of hydrophobic NPs showed synergistic combinations, reducing the antibiotic dosages by 8-16-fold against MDR bacteria.21 The NP-antibiotic combinations showed efficacy against both Gram-negative (E. coli, P. aeruginosa) and Gram-positive (methicillin-resistant S. aureus) strains (Figure 3c). The synergistic response of the combination was attributed to the ability of functionalized NPs to act as efflux pump inhibitor, facilitating accumulation of antibiotics inside the cells (Figure 3d). Furthermore, proteomic analysis of the outer membrane proteins of bacterial cells indicated deregulation of major efflux pump proteins, compromising the detoxification of cells. Notably, NP dosages used in combination therapy were non-toxic against mammalian cells. Combining NPs with antibiotics provides us with a modular approach to combat MDR bacteria while avoiding some of the regulatory issues associated with other nano-antimicrobials.
Figure 3.
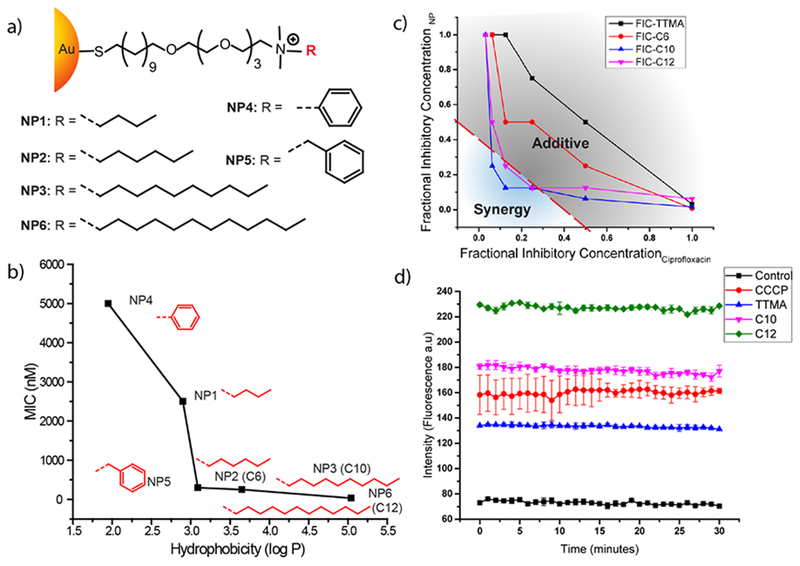
a) Molecular structures of the ligands used to functionalize 2nm AuNPs. b) MIC values of AuNPs with different hydrophobic ligands against E. coli. c) Graphs showing synergistic and additive interactions between the nanoparticles and antibiotic (ciprofloxacin). Data are fractional inhibitory concentration (FICs) of the NPs and antibiotics in combination. Synergy is observed by hydrophobic C10 and C12 NPs. d) Fluorescence kinetics of EtBr accumulation inside E. coli cells, evidencing blocking of efflux pumps by CCCP (efflux pump inhibitor, positive control) and hydrophobic NPs C10 and C12. Reproduced from Reference 20 with permission from American Chemical Society, Copyright © 2014 and Reference 21 with permission from IOP Publishing Ltd, Copyright © 2017.
The surface chemistry of nanoparticles can be further tailored to achieve selectivity between Gram-positive and Gram-negative bacteria. Grzybowski and coworkers developed mixed-charge nanoparticles that exhibit Gram-selective antibacterial activity.22 These NPs were fabricated with different ratios of positively charged (TMA) and negatively charged (MUA) ligands. NPs with ligand ratio (TMA: MUA) 80:20 and 48:52 could selectively kill Gram-negative and Gram-positive bacteria respectively at commensurate rates. Further studies revealed that cationic ligands help the nanoparticles to attach to the bacterial surface and anionic ligands with carboxylate headgroups compete for H-bonding interactions with cell-wall components, disrupting their structural integrity and causing the cell to lyse. In another study, Rotello group further investigated the role surface charge in determining the antimicrobial activity of NPs.23 Researchers fabricated NPs with zwitterionic ligands with different charge orientations, one with positive charge in the outermost layer while the other with positive charge inside the ligand termini. It was determined that NPs with cationic charge in outer layer displayed higher antimicrobial activity as compared to the ones with positive charge inside, with larger particles being more effective.
3.2. Polymer-stabilized Nanomaterials
Polymer stabilized nanomaterials are promising therapeutic agents against multiple antibiotic resistant infectious diseases. For example, Sambhy et al. reported silver bromide nanoparticles stabilized with a cationic polymer i.e., poly(4-vinylpyridine)-co-poly (4-vinyl-N-hexylpyridinium bromide) (AgBr/NPVP), which were found to be very effective to kill both Gram-positive and Gram-negative bacterial strains (Figure 4). The cationic polymer disrupted the cell membrane by binding with the negatively charged membrane of bacteria. Moreover, the biocidal activity of released Ag+ ions was tunable by controlling the size of AgBr NPs.24 Subsequently, Jang’s group reported the design and synthesis of silica–poly (TBAM-co-EGDMA)) core–shell nanoparticles. The polyethylene glycol (PEG) component of the cationic polymer played an important role in the structural stability through crosslinking the polymer shell, and enhanced the antimicrobial activity by reducing bio-adhesion on the nanoparticles. The biocidal efficiency of 17 nm core-shell nanoparticles was found to be 3-fold higher than that of 28 nm sized particles against E. coli, showing the effect of enlarged surface area of nanoparticles on their antimicrobial activity.25 In another example, silica nanoparticles decorated with silver/polyrhodanine composite (SiO2-Ag/PRh) were found to exhibit long-term antimicrobial activity due to contact-active bactericidal effects of polyrhodanine. The combination of positively charged rhodamine on inherently antimicrobial nanoparticles (SiO2-Ag+) showed synergistic effect in killing multiple species of bacteria.26
Figure 4.
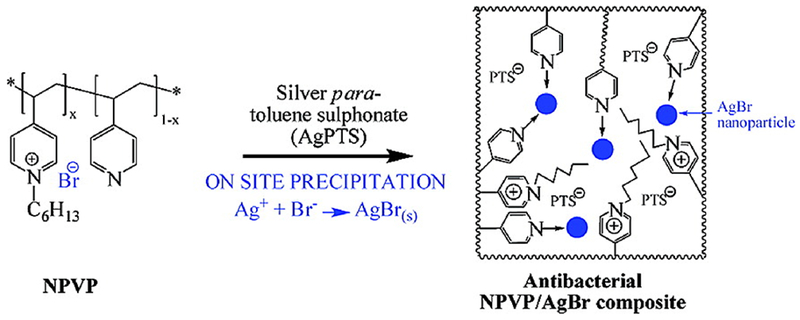
Schematic representation of the synthesis of AgBR/NPVP composite. Reproduced from reference 24 with permission from American Chemical Society, Copyright © 2006.
Magnetic nanoparticles can also be highly efficient recyclable antibiotic therapeutics after surface modification with poly(2-(dimethylamino)ethyl methacrylate. The functionalized nanoparticles maintained 100% killing efficiency against E. coli (105 to 106 E. coli/mg nanoparticles) even after eight cycles (Figure 5). This high bactericidal efficiency arose from the high surface area of nanoparticles enabling high density and stable attachment of antibacterial quaternary ammonium groups on the NP surface.27 In another approach, Kyziol’s group used biocompatible, biodegradable and bactericidal biopolymer chitosan (CS) as reducing and stabilizing agent for the synthesis of AuNPs. The resulting gold nanoparticles stabilized with chitosan of medium molecular weight (~1280 kDa) and highest deacetylation degree (CS-AuNPs, 89 ± 2%) featured the highest antibacterial activity against antibiotic resistant strains S. aureus and P. aeruginosa. The CS-AuNPs showed selective cytotoxicity against bacteria by producing disrupting effect on the microbial membrane, while their internalization was hindered by the eukaryotic cells, showing no cytotoxicity against mammalian somatic and tumoral cells.28 Li’s group also reported the synthesis of polymer-stabilized nontoxic (to mammalian cells) silver nanoparticles (AgNPs@ PDMAEMA-C4) by the reduction of silver nitrate in the presence of cationic polymer made from 2-(dimethylamino)ethyl methacrylate (DMAEMA), displaying strong antimicrobial activity against bacterial infections. The antibacterial activity of resulting AgNPs@ PDMAEMA-C4 was strongly enhanced due to synergistic multivalent and antibacterial mechanisms of the polymer and nanoparticles. These nanoparticles enhanced bacterial cytoplasmic membrane permeability, and subsequently penetrated the cells to strongly inhibit intracellular enzyme activity leading to the cell death. Notably, these nanoparticles promoted the wound healing after eradication of P. aeruginosa and S. aureus infections in an in vivo model.29
Figure 5.
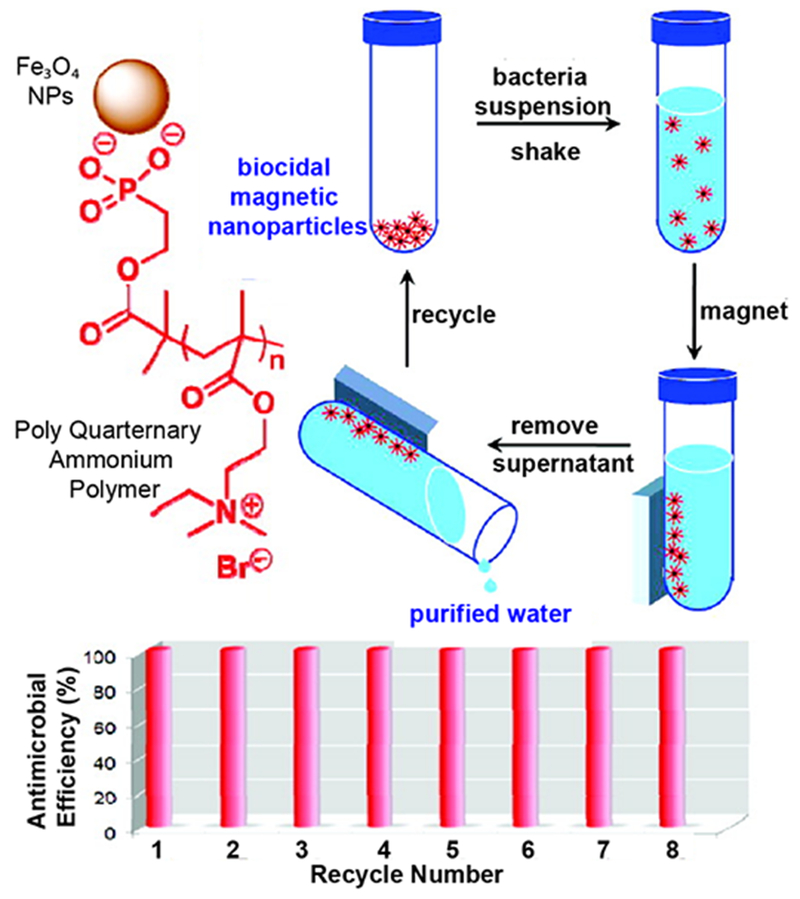
Schematic diagram showing magnetic NP-functionalized with polymer recycled for antibacterial application. Reproduced from reference 27 with permission from American Chemical Society, Copyright © 2011.
Polymeric materials can form self-assembled organic nanoparticles in aqueous solution. The formation of polymeric nanostructures before coming into contact with the cell surface is expected to enable more efficient multivalent interactions with the cell membrane as compared to the individual polymer molecules. Yang and coworkers synthesized biodegradable cationic amphiphilic triblock polycarbonates that self-assembled into micellar nanoparticles upon dissolution in water. The average diameters and zeta potential of these organic nanoparticles could be controlled by varying the molecular weight and length of cationic and hydrophobic poly(carbonate) blocks. These nanoparticles could efficiently disrupt the microbial walls/membrane of Gram-positive bacteria including methicillin-resistant S. aureus (MRSA), inhibiting their growth. Interestingly, the antimicrobial activity of micelle NPs is observed at concentrations above the critical micelle concentration of polymers, indicating the importance of nanostructure formation in determining the bactericidal activity of the polymers. Additionally, these micellar nanoparticles did not induce toxicity to liver and kidney upon intravenous administration in mice at relevant therapeutic concentrations.30
Recently, novel recyclable N-halamine-derivatized cross-linked polymethacrylamide organic nanoparticles were synthesized via a surfactant-free dispersion copolymerization of methacrylamide (MAA) and N, N-methylenebis(acrylamide) (MBAA) as a crosslinker, followed by chlorination using NaOCl. These chloramine-functionalized nanoparticles generated reactive oxygen species (ROS) in organic media, thereby exhibiting remarkable specificity in releasing oxidative chlorine upon association with bacteria.31 Interestingly, these N-halamine NPs specifically encircled the S. aureus membrane, further suggesting a highly specific association between bacteria and NPs.
In a subsequent study, Rotello and coworkers synthesized a library of quaternary ammonium poly(oxanorborneneimides) with different degrees of hydrophobicity and screened their antimicrobial activities (Figure 6 a, b).32 These polymers formed 15nm polymeric NPs (PNPs) in the aqueous solution. Increasing the hydrophobicity of the alkyl chain bridging the cationic headgroup with the backbone, drastically enhanced the antimicrobial activity of PNPs (Figure 6c). The MIC of NPs decreased 1000-fold upon increasing the internal chain length to 11 carbons, while no change in MICs was observed on increasing the hydrophobicity of cationic headgroups. In contrast, cytotoxicity towards mammalian cells increased with the increasing hydrophobicity of the cationic headgroup. The most effective PNP (P5) exhibited minimal hemolysis yielding therapeutic indices (HC50/MIC) as high as 2500, 5-fold higher than previously reported polymers. These PNPs were able to inhibit bacterial growth of 11 different clinical isolates, demonstrating their broad-spectrum activity (Figure 6d). Furthermore, bacteria were unable to develop resistance against these PNPs for up to ~1300 generations, in stark contrast to conventional antibiotics. This study shows that location of hydrophobic and cationic moieties on the polymer structure, determines the antimicrobial activity and therapeutic selectivity of the nanomaterial.
Figure 6.
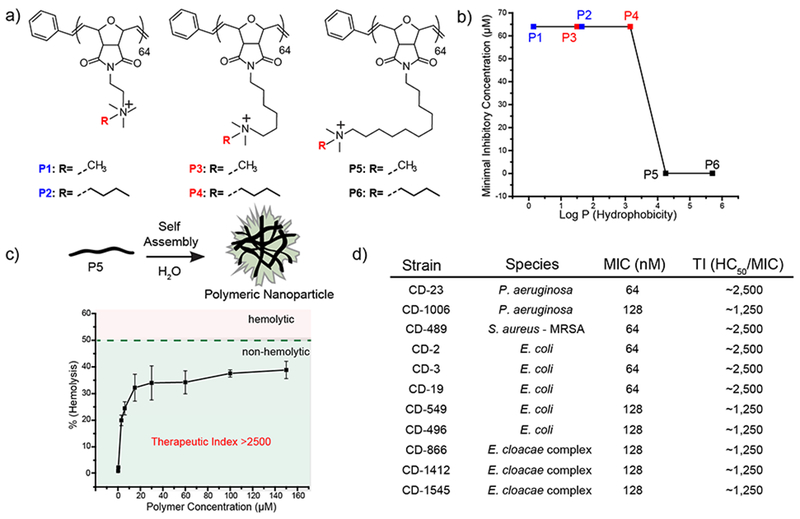
a) Molecular structures of the polymers used in the study. b) Minimal inhibitory Concentrations (MICs) of polymer derivatives plotted against Log P. Log P represents calculated hydrophobicity of each monomer. c) schematic showing self-assembly of polymers and graph showing hemolytic activity against red blood cells. d) Table showing MIC and therapeutic indices of P5 PNPs against clinical isolates. Reproduced from reference 32 with permission from American Chemical Society, Copyright © 2018.
3.3. Biomolecule-functionalized nanomaterials
Biomolecules display specific and strong complementary recognition interactions that can impart biospecificity to nanoparticles. Other important fundamental features of biomolecules include the presence of specific binding domains and the possibility to genetically modify them with specific anchoring groups. Biomolecules such as DNA, proteins and antibodies provide specific attributes to nanoparticles including highly selective and specific recognition properties that are, otherwise, very difficult or nearly impossible to achieve using synthetic materials.15,33 Siamak et al., demonstrated that antibacterial activity of DNA stabilized AgNCs was tunable by varying the oligonucleotide sequence. The sequences tested yielded fluorescent nanoclusters, with antimicrobial activity similar to that of silver nitrate (Figure 7). Researchers prepared a trimeric structure containing the oligonucleotide sequence (Seq-3) with higher contents of cytosine units, that exhibited the highest antimicrobial activity against the growth of Gram-positive and Gram-negative bacteria (S. epidermidis and E. coli) in the sub-micromolar range. Cytosine binds silver better than other nucleobases providing a more structured and stable bioconjugate. Thus, the structure of the DNA used to stabilize the AgNCs plays a key role in the antimicrobial activity, where the most pre-organized systems generates the most efficient antimicrobial agent.34
Figure 7.
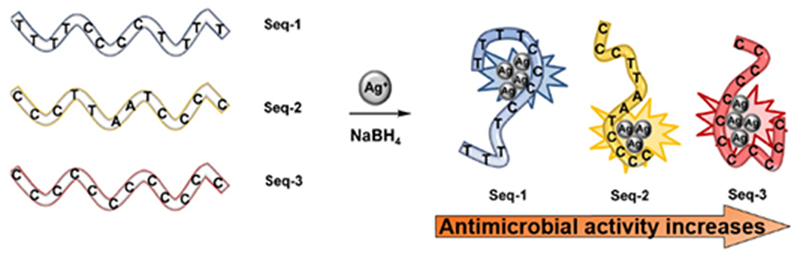
Schematic diagram showing the preparation of DNA-AgNCs. Reproduced from reference 34 with permission from American Chemical Society, Copyright © 2016.
Functionalization of nanoparticles with antimicrobial peptides has also opened up new avenues to combat multi-drug bacterial resistance through synergy arising between the stabilizing agent and nanoparticles.6 Antimicrobial peptides (AMPs) are promising molecules to fight microbial infection owing to their membrane disruption ability. However, compromised antimicrobial activity and decreased stability in presence of serum and proteolytic enzymes have impaired the clinical translation of AMPs. Bi et al. reported that the stability and longevity of AMPs could be enhanced by physically immobilizing peptides on the NP surface. Carbohydrate (phytoglycogen) nanoparticles subjected to β-amylolysis followed by succinate or octenyl succinate substitution were loaded with antimicrobial nisin peptides.35 These peptide-NP assemblies exhibited prolonged bactericidal activity against L. monocytogenes for up to 21h, longer than free peptides. The octenyl succinate substituted NPs showed enhanced stability and activity, owing to stronger electrostatic and hydrophobic interactions between NPs and peptides. In an alternative approach, Ferreira and coworkers covalently conjugated cationic peptide cecropin-melittin with AuNPs exhibiting enhanced antimicrobial activity, even in the presence of proteolytic enzymes at high concentrations. Importantly, cysteine was added at the C-terminus of the cecropin-melittin (CM) to obtain the desired orientation of the peptide on Au surface (CM-S-Au). AMP-conjugated AuNPs showed high loading of ~237 peptides per NP, resulting in permeabilization of bacterial membrane. Moreover, these NP-peptide conjugates eliminated bacterial colonies in an in vivo chronic wound and systemic infection model.36
Protein-nanoparticle conjugates have been used for biological applications due to their increased stability, low size dispersity, high enzyme loading and the potential for targeted delivery. Recently, Hahn and coworkers demonstrated lysozyme conjugated ZnO nanoparticles (L-ZNPs) with enhanced therapeutic activity against S. aureus and E. coli.37 Aminated ZnO NPs were conjugated with lysozyme through glutaraldehyde cross-linking, resulting in positively charged L-ZNP conjugates. Enhanced antimicrobial activity of L-ZNP conjugates has been attributed to the synergy between the action mechanisms of the individual components (ZnONPs and lysozymes). NP-conjugates strongly associate with bacteria causing membrane disruption, followed by ROS mediated oxidative stress caused by released Zn2+ ions. The membrane stress is further aggravated through lysozyme mediated membrane damage to bacterial cells. Furthermore, selective killing of bacteria has been achieved by conjugating antibodies with light absorbing AuNPs by Smeltzer and coworkers. Real-time monitoring of this process was provided by a photothermal (PT) microscope/spectrometer, which showed that laser-generated overheating along with bubble-formation phenomena around clustered gold nanoparticles were the main cause of bacterial damage. This PT strategy was specifically used for selective killing of the Gram-positive S. aureus by targeting the bacterial surface using 10-, 20-, and 40-nm AuNPs conjugated with anti-protein A antibody.38
For bacterial cells, strategies facilitating the thick peptidoglycan layer of the bacterial cell wall as a target are key for the development of useful nanotherapeutics. In mycobacteria, exogenous trehalose is internalized to the mycobacterial cytoplasm by a trehalose transporter system. The important role of trehalose in the cytoplasm in terms of pathogenicity has been used to disrupt trehalose biosynthesis pathways in M. tuberculosis. Following this study, Kalana et al. employed trehalose conjugated nanoparticles to target selectively M. smegmatis over mammalian cells.39
4. Nanomaterials as antimicrobial delivery vehicles
The antimicrobial efficacy of therapeutics can be increased by using delivery vehicles to successfully transport them to the infection site.9,47 Nanoparticle-based drug delivery systems can provide increased drug retention time in blood, reduced nonspecific distribution and targeted delivery of drug at the site of infections. The NP surface chemistry plays a crucial role to ensure NP solubility in the blood stream and to provide a “stealth” invisibility against the body’s natural defense system. The mononuclear phagocytic system can eliminate these nanovehicles from the blood stream unless the vehicles are engineered to escape recognition. Another important biological barrier to nanoparticle-based drug delivery is the process of opsonization. Opsonin proteins in blood rapidly adhere to nanoparticles, facilitating macrophages from the mononuclear phagocytic system (MPS) to bind and remove NPs from circulation.40 Numerous strategies have been used to hide nanoparticles from the MPS to address these limitations. Of these methods, the most preferred is the adsorption or grafting of hydrophilic polymers such as PEG,48 poloxamers (e.g. pluronic-F68),49 polysaccharides like chitosan50 to coat the surface of nanoparticles. These coatings create a ‘cloud’ of uncharged hydrophilic moieties at the particle surface that repel plasma proteins and increase the circulation and retention time in the circulatory system of the body.
4.1. Non-specific cargo release
Delivery of antimicrobial agents using a delivery vehicle can enhance the efficacy of antimicrobial agents as well as circumvent the resistance mechanisms employed by bacteria. For instance, pathogenic bacteria can reside inside mammalian cells to evade from antibiotics. These intracellular infections cannot readily be treated due to limited availability of antibiotics inside the cell. On the other hand, nanomaterials have excellent ability to penetrate inside cells coupled with high drug loading ability. Using the penetration ability of nanomaterials, Biswas and co-workers employed tetracycline encapsulated nanoparticles to effectively treat intracellular S. aureus infections. Chitosan polymers O-substituted with carboxymethyl groups were encapsulated with tetracycline antibiotics, resulting in formation of ~200 nm sized positively charged nanoparticles through ionic gelation method. Cationic NPs showed high uptake inside mammalian cells and subsequently released tetracycline upon binding with the negatively charged bacterial membrane. The survival rate of intracellular S. aureus decreased from 15% to 2.5% upon treatment with nanoparticle encapsulated tetracycline when compared with tetracycline alone.41
Similarly, bacteria utilize their decreased outer-membrane permeability to prevent antibiotics from penetrating inside the cells. Up to 90% of P. aeruginosa isolated from cystic fibrosis patients are highly impermeable to antibiotics including aminoglycosides. However, nanovehicles such as liposomes can easily fuse with bacterial membrane and transport drugs inside the cells, owing to their bacterial membrane-like lipid bilayer composition. Omri and co-workers delivered aminoglycosides to P. aeruginosa using nanoliposome as a vehicle.42 The minimum inhibitory concentrations (MICs) of liposomal aminoglycosides against resistant P. aeruginosa were significantly lower than free aminoglycoside, showing efficient delivery of antibiotics using liposomes. It was observed that liposomes fused easily with membrane of drug-sensitive strain (within 1 hour) as compared to its resistant counterpart that took 6 hours for liposome-bacteria membrane fusion. High dosage of antibiotics is delivered upon membrane-fusion, potentially dominating the bacterial efflux pumps and overcoming the drug-resistance posed by microbe.
In addition to overcoming the bacterial resistance mechanisms, nanomaterial-based delivery vehicles can impart enhanced stability to cargo in physiological media. For example, phytochemicals are plant derived antimicrobials that exhibit broad-spectrum activity against MDR strains. However, low solubility in aqueous solutions limits their widespread application. Recently, Rotello and coworkers have fabricated bactericidal polymer-stabilized nanosponges through self-assembly of synthetically engineered polymers around essential-oil based cores (Figure 8a).43 The polymers (PONI-GMT) serve to stabilize the emulsion and enhance the interaction with bacteria, owing to their positive charge. Further, PONI-GMT possesses maleimide monomers that conjugate with disulfide containing (DTDS) imparting degradability to the nanosponges at five points (Figure 8 b, c). These nanosponges were degradable in presence of endogenous biomolecules such as glutathione and esterase enzymes. The nanosponges (~220 nm) showed high stability in serum and retained their activity even after one year of storage (Figure 8d). These nanosponges disrupted bacterial membrane, eventually causing its death. Moreover, the bacteria were unable to develop resistance against the nanosponges for 20 passages, whereas MIC of conventional antibiotics increased more than a thousand-fold in fewer number of passages (Figure 8e).
Figure 8.
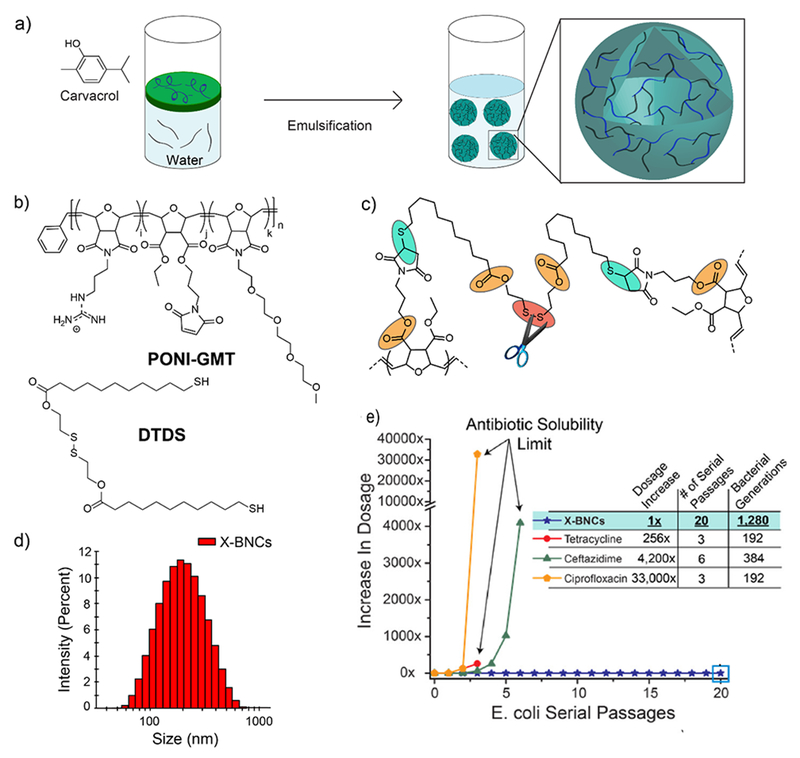
a) Schematic diagram showing the fabrication of biodegradable-polymer stabilized nanosponges. b) Structures of PONI-GMT and DTDS. c) Cross-linked PONI-GMT-DTDS structure showing linkage points reactive to endogenous biomolecules. d) DLS histogram of nanosponges. e) Resistance development assay against E. coli using nanosponges and antibiotics. The y-axis indicates the increase in dosage as compared to the initial bacterial cells (0th passage). Reproduced from reference 43 with permission from American Chemical Society, Copyright © 2018.
Nanomaterial-based delivery vehicles can used to regulate the release of cargo in a sustained and controlled manner. Friedman and coworkers designed nitrite-loaded silane-hydrogel based composite NPs to harness the antimicrobial activity of nitric oxide (NO). These composite NPs were synthesized using tetramethoxysilane (TMOS) as sol-gel matrices, required for thermal conversion of sodium nitrite into NO in presence of glucose. Chitosan and PEG served as additives to control the generation and sustained release of NO upon exposure to moisture. These NPs demonstrated stable release of NO for upto 24 hours and exhibited strong antimicrobial activity against S. aureus. Notably, these NO-releasing NPs were able to treat methicillin-resistant MRSA in a murine wound model.44
Nanoparticles can likewise provide a promising platform for co-delivery of multiple antimicrobials to exhibit synergistic effect against MDR bacteria. Gu and co-workers utilized this strategy by designing mesoporous silica nanoparticles loaded with Ag and levofloxacin (antibiotic).45 Researchers fabricated Ag embedded mesoporous silica nanoparticles, then encapsulated levofloxacin inside the mesopores. The release of Ag+ ions permeabilized the outer membrane of bacteria, making them more susceptible to simultaneously released levofloxacin (Figure 9). Dual-drug loaded NPs showed synergistic effects against MDR E. coli both in vitro and in vivo.
Figure 9.
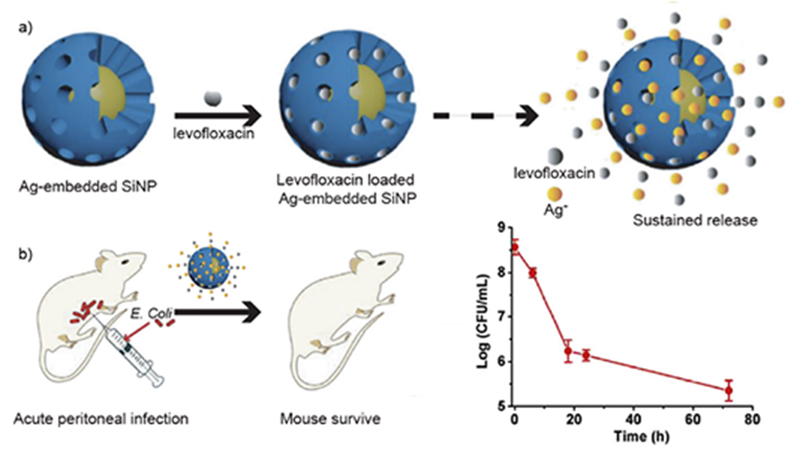
Schematic diagram showing a) fabrication of Ag@MSN loaded with levofloxacin for synergistic treatment of bacteria. b) Schematic illustration showing in vivo infection with graph showing decrease in intraperitoneal colony counts after NP treatment. Reproduced from reference 45 with permission from Elsevier, Copyright © 2016.
4.2. Stimuli-mediated cargo release
Potentially more efficient nanomaterial-based delivery strategies involve on-demand processes that allow the release of pharmaceuticals in a spatio-temporally controlled fashion. This strategy uses stimuli responsive systems that respond to their microenvironment and react in a dynamic way, mimicking the responsiveness of living organisms.40 However, this approach is both complex and sophisticated, requiring biocompatible materials that can undergo structural or chemical changes in response stimuli, e. g. pH changes and excreted enzymes at infection sites.
pH-triggered drug delivery system
Bacterial infections generally feature very low pH (~4.5) values due to their hypoxic nature. The acidic environment of bacterial infectious site can be harnessed in the design of pH sensitive drug delivery systems. Pegylated poly l-histidine can form stable polymeric micellar nanoparticles, however at pH <6.5, histidine residues are protonated resulting in strong interaction with the negatively charged bacterial cell membrane (Figure 10). As a result, the encapsulated vancomycin drug is released at infectious site due to the acidic microenvironment. These nanoparticles prevent non-specific interaction at physiological pH 7.4, thereby increasing the therapeutic activity with declining pH.46 In another study, a charge-adaptive nanomaterial based on polyethylene glycol (PEG), polycaprolactone (PCL) and poly(β-amino ester) (PAE) was used for targeted in vivo vancomycin delivery. This inherently negatively charged nanovehicle (PEG-b-PCL-b-PAE) transitions to overall cationic charge in acidic conditions, increasing the circulation time and facilitating the accumulation of vancomycin in a subcutaneous inflammation model.47
Figure 10.
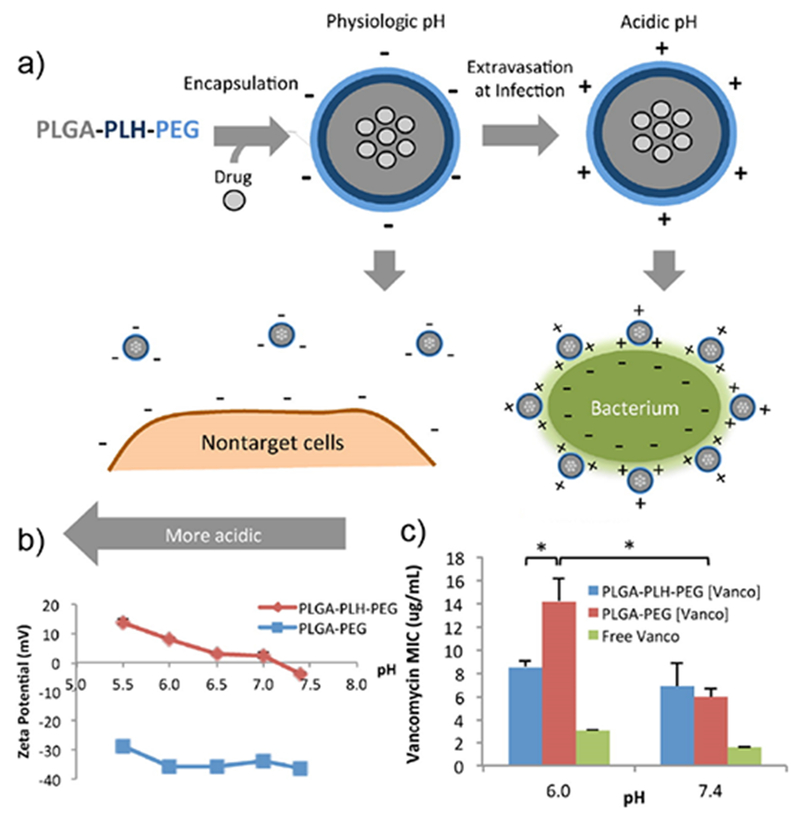
Schematic diagram showing the design of charge switchable NPs for delivery of vancomycin to bacterial cells. b) Zeta potential of NPs, ZP of PLGA-PLH-PEG increases with decreasing pH. c) Minimum inhibitory concentrations (MICs) of vancomycin against S. aureus in different conditions. Significant loss of drug activity observed in PLGA-PEG (Vanco) and free Vanco. Reproduced from reference 46 with permission from American Chemical Society, Copyright © 2012.
Enzyme-sensitive delivery systems
The increase of expression of enzymes at infectious sites including esterases and gelatinases also provide an effective strategy for stimulated delivery of antimicrobials via an enzyme-triggered mechanism. In an earlier study of drug delivery systems, ampicillin antibiotic was mechanically entrapped in polymeric nanocarriers and steadily released on the site of infection due to bio-erosion (enzymatic degradation) of the system.48 Later on, Li et al. reported an “on-demand” delivery system for antibiotics based on supramolecular gelatin nanoparticles (SGNPs) by releasing drugs in the presence of gelatinase at the bacterial infection site. The surface of SGNPs was coated with red blood cell (RBC) membranes (SGNPs@RBC), followed by encapsulation of vancomycin (Van) in gelatin nanoparticles (Van⊂SGNPs@RBC). The coating of RBC membranes imparted biomimetic properties to Van⊂SGNPs@RBC and significantly improved the immune-evading capability of the resulting nanocarriers, which were capable of effectively accumulating at the infection site via enhanced permeability and retention effects. After their delivery at the infection microenvironment, the RBC membranes on the Van⊂SGNPs@RBC acted as detoxifiers to enable further absorption of the exotoxins that were produced by bacteria and relieved their symptoms (Figure 11). Meanwhile, the gelatin core was degraded by overexpressed gelatinase in the infection microenvironment, and the encapsulated vancomycin was released and acted as effective therapeutic agent against pathogenic bacteria.49
Figure 11.
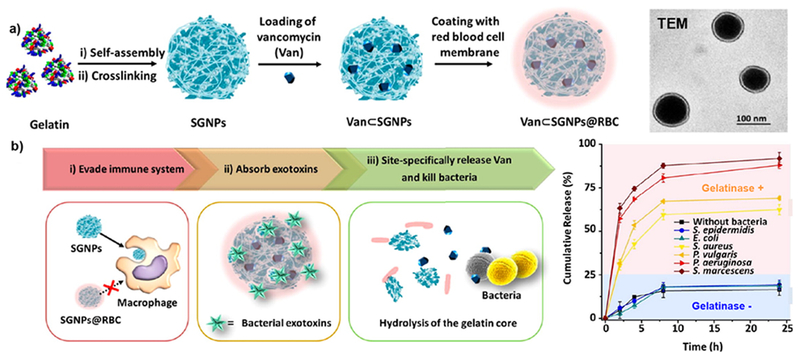
Schematic representation for fabrication of vancomycin encapsulated gelatin nanoparticles with RBC membrane coating layer (Van⊂SGNPs@RBC). (b) Diagram showing the ability of Van⊂SGNPs@RBC to evade macrophage recognition and release drugs in presence of bacterial infection. Graph shows drug releasing profiles of NPs in presence of gelatinase (+/−) bacteria. Reproduced from reference 49 with permission from American Chemical Society, Copyright © 2014.
Bacterial toxin-triggered drug delivery
Due to the abundance of pore-forming bacterial toxins at infected sites, these toxins can be used to trigger selective release of antimicrobials from nanomaterial-based delivery vehicles at the targeted site. This approach provides release of drugs at the infectious sites to kill toxin-secreting bacteria without having harmful side effects on healthy tissues. This strategy was employed to deliver vancomycin to bacterial infections using bacterial toxins (alpha hemolysin) to release the drug from gold nanoparticle-stabilized phospholipid liposomes.50 Attachment of chitosan-decorated gold nanoparticles to liposome surfaces prohibited them from self-fusion with accompanying payload release in normal (non-infection) environments. Once these protected liposomes are near toxin-secreting bacteria these toxins inserted into the liposome membranes, forming pores for releasing the encapsulated therapeutics. The antimicrobial effect are thereby localized to the infection site.
Overall, nanomaterials provide a promising platform for delivery of wide-range of antimicrobial cargo in a site-specific and controlled manner. Nanomaterial-based delivery can address the issues of toxicity and adverse side effects associated with high doses of therapeutic drugs. Similarly, nanomaterial with self-therapeutic activity offers distinctive advantages over conventional antibiotics in terms of overcoming antibiotic resistance and effective treatment. However, the safety profiles of nanomaterials, both acute and long-term toxicity needs to be thoroughly considered for the translation of nanomedicines into clinics.51
5. Summary and Outlook
Tunable surface functionality of nanomaterials provides a versatile platform to design novel antimicrobials to combat multi-drug resistant bacterial infections. In this tutorial, we have discussed multiple strategies that utilize nanomaterials as (i) self-therapeutic agents and (ii) carriers for antimicrobial cargo. Material design can be tailored to achieve high selectivity towards bacteria, overcome drug-resistance pathways and enable synergistic combinations with antibiotics. Nanomaterial-based antimicrobials can be readily used as sterilizers and disinfectants for surfaces in ex vivo applications. With current state of the art, nanomaterials exhibit strong potential to treat topical skin infections including wound biofilms in the near future. However, systemic administration of these nanomaterials still requires multiple aspects to be addressed. Nanomaterial design should be fabricated to retain activity in complex biological media, decrease cytotoxicity and evade recognition by immune system. The structure-activity studies will help in generating strategies to develop effective therapeutic materials. On the biological front, detailed studies of NP-action mechanism should be performed. These studies will enhance the selectivity of these nanomaterials towards specific bacteria, as well as improve their chances to be used as an adjuvant to current antibiotics. Finally, evaluating the pharmacological profiles is essential for the clinical translation of the effective therapeutic materials. Taken together, interdisciplinary research evaluating distinct aspects of nanomaterials based on surface functionality will be able to establish nanomaterials as effective next generation antimicrobials.
Key learning points.
Antibiotic-resistant bacterial infections pose a severe threat to public health.
Nanomaterials have emerged as an alternative tool to combat multi-drug resistant bacteria.
Nanoparticle surface chemistry plays a vital role in determining therapeutic efficacy.
Appropriate surface functionalization can be used to generate self-therapeutic NPs with broad-spectrum activity.
Nanomaterials can also act as delivery vehicles to release antimicrobial agents at the infected site.
Acknowledgements
This research was supported by NIH AI 1347770, GM077173, and EB022641. Authors acknowledge Higher Education Commission Pakistan (HEC) and the US National Academy of Sciences for the support provided through US-Pakistan Science and Technology Cooperation Program.
Footnotes
Conflicts of interest
The authors declare no competing financial interest.
References
- 1.FY15 Detect and Protect Against Antibiotic Resistance Budget Initiative; Centers for Disease Control and Prevention; Atlanta, GA, 2003, http://www.cdc.gov/drugresistance/threat-report-2013/pdf/FY15-DPAR-budget-init.pdf. [Google Scholar]
- 2.Willyard C, Nature, 2017, 543, 15. [DOI] [PubMed] [Google Scholar]
- 3.Prestinaci F; Pezzotti P; Pantosti A Pathog. Glob. Health 2015, 109 (7), 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ventola CL, P T A peer-reviewed J. Formul. Manag, 2015, 40, 277–83. [PMC free article] [PubMed] [Google Scholar]
- 5.Neu HC, Science (80-.)., 1992, 257, 1064–1073. [DOI] [PubMed] [Google Scholar]
- 6.Rajchakit U, U.; Sarojini V Bioconjugate Chem 2017, 28, 2673–2686. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Hu C and Shao L, Int. J. Nanomedicine, 2017, 12, 1227–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hajipour MJ, Fromm KM, Akbar Ashkarran A, Jimenez de Aberasturi D, de Larramendi IR, Rojo T, Serpooshan V, Parak WJ and Mahmoudi M, Trends Biotechnol, 2012, 30, 499–511. [DOI] [PubMed] [Google Scholar]
- 9.Gupta A, Landis RF and Rotello VM, F1000Research, 2016, 5, 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodman CM, McCusker CD, Yilmaz T and Rotello VM, Bioconjug. Chem, 2004, 15, 897–900. [DOI] [PubMed] [Google Scholar]
- 11.Miller KP, Wang L, Benicewicz BC and Decho AW, Chem. Soc. Rev, 2015, 44, 7787–7807. [DOI] [PubMed] [Google Scholar]
- 12.Berry V, Gole A, Kundu S, Murphy CJ and Saraf RF, J. Am. Chem. Soc, 2005, 127, 17600–17601. [DOI] [PubMed] [Google Scholar]
- 13.Lin CC, Yeh YC, Yang CY, Chen CL, Chen GF, Chen CC and Wu YC, J. Am. Chem. Soc, 2002, 124, 3508–3509. [DOI] [PubMed] [Google Scholar]
- 14.Hayden SC, Zhao G, Saha K, Phillips RL, Li X, Miranda OR, Rotello VM, El-Sayed MA, Schmidt-Krey I and Bunz UHF, J. Am. Chem. Soc, 2012, 134, 6920–6923. [DOI] [PubMed] [Google Scholar]
- 15.Jiang Z, Le NDB, Gupta A and Rotello VM, Chem. Soc. Rev, 2015, 44, 4264–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu H, Ho PL, Tong E, Wang L and Xu B, Nano Lett, 2003, 3, 1261–1263. [Google Scholar]
- 17.Nirmala Grace A and Pandian K, Colloids Surfaces A Physicochem. Eng. Asp, 2007, 297, 63–70. [Google Scholar]
- 18.B. K Fayaz AM Girilal M, Yadav R, Kalaichelvan PT, Venketesan R, Nanomedicine, 2010, 6(1), 103–109. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y, Tian Y, Cui Y, Liu W, Ma W and Jiang X, J. Am. Chem. Soc, 2010, 132, 12349–12356. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Robinson SM, Gupta A, Saha K, Jiang Z, Moyano DF, Sahar A, Riley MA and Rotello VM, ACS Nano, 2014, 8, 10682–10686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta A, Saleh NM, Das R, Landis RF, Bigdeli A, Motamedchaboki K, Rosa Campos A, Pomeroy K, Mahmoudi M and Rotello VM, Nano Futur, 2017, 1, 015004. [Google Scholar]
- 22.Pillai PP, Kowalczyk B, Kandere-Grzybowska K, Borkowska M and Grzybowski BA, Angew. Chemie - Int. Ed, 2016, 55, 8610–8614. [DOI] [PubMed] [Google Scholar]
- 23.Huo S, Jiang Y, Gupta A, Jiang Z, Landis RF, Hou S, Liang XJ and Rotello VM, ACS Nano, 2016, 10, 8732–8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambhy V, MacBride MM, Peterson BR and Sen A, J. Am. Chem. Soc, 2006, 128, 9798–9808. [DOI] [PubMed] [Google Scholar]
- 25.Song J, Kong H and Jang J, Chem. Commun, 2009, 5418–5420. [DOI] [PubMed] [Google Scholar]
- 26.Song J, Kim H, Jang Y and Jang J, ACS Appl. Mater. Interfaces, 2013, 5, 11563–11568. [DOI] [PubMed] [Google Scholar]
- 27.Dong H, Huang J, Koepsel RR, Ye P, Russell AJ and Matyjaszewski K, Biomacromolecules, 2011, 12, 1305–1311. [DOI] [PubMed] [Google Scholar]
- 28.Regiel-Futyra A, Kus-Lis̈kiewicz M, Sebastian V, Irusta S, Arruebo M, Stochel G and Kyzioł A, ACS Appl. Mater. Interfaces, 2015, 7, 1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mei L, Lu Z, Zhang X, Li C and Jia Y, ACS Appl. Mater. Interfaces, 2014, 6, 15813–15821. [DOI] [PubMed] [Google Scholar]
- 30.Nederberg F, Zhang Y, Tan JPK, Xu K, Wang H, Yang C, Gao S, Guo XD, Fukushima K, Li L, Hedrick JL and Yang YY, Nat. Chem, 2011, 3, 409–414. [DOI] [PubMed] [Google Scholar]
- 31.Natan M, Gutman O, Lavi R, Margel S and Banin E, ACS Nano, 2015, 9, 1175–1188. [DOI] [PubMed] [Google Scholar]
- 32.Gupta A, Landis RF, Li C-H, Schnurr M, Das R, Lee Y-W, Yazdani M, Liu Y, Kozlova A and Rotello VM, J. Am. Chem. Soc, 2018, 140, 12137–12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katz E and Willner I, Angew. Chemie - Int. Ed, 2004, 43, 6042–6108. [DOI] [PubMed] [Google Scholar]
- 34.Javani S, Lorca R, Latorre A, Flors C, Cortajarena AL and Somoza Á, ACS Appl. Mater. Interfaces, 2016, 8, 10147–10154. [DOI] [PubMed] [Google Scholar]
- 35.Bi L, Yang L, Narsimhan G, Bhunia AK and Yao Y, J. Control. Release, 2011, 150, 150–156. [DOI] [PubMed] [Google Scholar]
- 36.Rai A, Pinto S, Velho TR, Ferreira AF, Moita C, Trivedi U, Evangelista M, Comune M, Rumbaugh KP, Simões PN, Moita L and Ferreira L, Biomaterials, 2016, 85, 99–110. [DOI] [PubMed] [Google Scholar]
- 37.Tripathy N, Ahmad R, Bang SH, Min J and Hahn YB, Chem. Commun, 2014, 50, 9298–9301. [DOI] [PubMed] [Google Scholar]
- 38.Zharov VP, Mercer KE, Galitovskaya EN and Smeltzer MS, Biophys. J, 2006, 90, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jayawardana KW, Jayawardena HSN, Wijesundera SA, De Zoysa T, Sundhoro M and Yan M, Chem. Commun, 2015, 51, 12028–12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torchilin VP, Nat. Rev. Drug Discov, 2014, 13, 813–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maya S; Indulekha S; Sukhithasri V; Smitha KT; Nair SV; Jayakumar R; Biswas R Int. J. Biol. Macromol 2012, 51 (4), 392–399. [DOI] [PubMed] [Google Scholar]
- 42.Mugabe C; Halwani M; Azghani AO; Lafrenie RM; Omri A Antimicrob. Agents Chemother 2006, 50 (6), 2016–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landis RF, Li CH, Gupta A, Lee YW, Yazdani M, Ngernyuang N, Altinbasak I, Mansoor S, Khichi MAS, Sanyal A and Rotello VM, J. Am. Chem. Soc, 2018, 140, 6176–6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pelgrift RY and Friedman AJ, Adv. Drug Deliv. Rev, 2013, 65, 1803–1815. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y; Ding X; Chen Y; Guo M; Zhang Y; Guo X; Gu H Biomaterials 2016, 101, 207–216. [DOI] [PubMed] [Google Scholar]
- 46.Radovic-Moreno AF, Lu TK, Puscasu VA, Yoon CJ, Langer R and Farokhzad OC, ACS Nano, 2012, 6, 4279–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chu L, Gao H, Cheng T, Zhang Y, Liu J, Huang F, Yang C, Shi L and Liu J, Chem. Commun, 2016, 52, 6265–6268. [DOI] [PubMed] [Google Scholar]
- 48.Seijo B, Fattal E, Roblot-Treupel L and Couvreur P, Int. J. Pharm, 1990, 62, 1–7. [Google Scholar]
- 49.Li LL, Xu JH, Bin Qi G, Zhao X, Yu F and Wang H, ACS Nano, 2014, 8, 4975–4983. [DOI] [PubMed] [Google Scholar]
- 50.Pornpattananangkul D, Zhang L, Olson S, Aryal S, Obonyo M, Vecchio K, Huang CM and Zhang L, J. Am. Chem. Soc, 2011, 133, 4132–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elsaesser A and Howard CV, Adv. Drug Deliv. Rev, 2012, 64, 129–137. [DOI] [PubMed] [Google Scholar]


