Abstract
The base excision repair (BER) pathway removes modified nucleobases that can be deleterious to an organism. BER is initiated by a glycosylase, which finds and removes these modified nucleobases. Most of the characterization of glycosylase activity has been conducted in the context of DNA oligomer substrates. However, DNA within eukaryotic organisms exists in a packaged environment with the basic unit of organization being the nucleosome core particle (NCP). The NCP is a complex substrate for repair in which a variety of factors can influence glycosylase activity. In this Review, we focus on the geometric positioning of modified nucleobases in an NCP and the consequences on glycosylase activity and initiating BER.
Keywords: DNA damage, DNA repair, Base excision repair, Glycosylase, Nucleosome, Chromatin
1. Introduction
Genomic DNA provides the code that drives the operation of all living things. Preservation of the genome is crucial for cell and organismal survival, as well as for the faithful transfer of genetic information to the next generation. Despite being the code of life, DNA has a physiochemical composition that renders it susceptible to modification and decomposition [1, 2]. Sources of DNA damage can originate from within the cell or from exogenous DNA damaging agents [3, 4]. Not only are the sources of damage wide ranging, but also there is high diversity in the types of damage including single- and double-strand breaks, inter- and intra-strand crosslinks, abasic sites, and modification of the nucleobases [5].
Fortunately, our cells have a variety of repair processes that are capable of rectifying much of the damage that forms in DNA [6]. One process is the base excision repair (BER) pathway that allows for the removal of modified nucleobases, which we refer to here as lesions. This pathway is initiated by a glycosylase, which cleaves the glycosidic bond that attaches the lesion to the sugar-phosphate backbone and generates an abasic site. Monofunctional glycosylases catalyze only glycosidic bond cleavage, whereas bifunctional glycosylases also catalyze cleavage the backbone 3’ to the abasic site [7]. Subsequent steps in the BER pathway are believed to proceed in an orderly fashion that has been likened to the passing of a baton [8]. AP endonuclease 1 (APE1) incises the backbone to create a nick with 3’-OH and 5’-deoxyribose phosphate (5’-dRP) termini. Polymerase β (pol β) removes the 5’-dRP and catalyzes nucleotide incorporation at the 3’-OH. Lastly, a DNA ligase seals the nick to rejoin the phosphate backbone and complete the repair [5].
Eleven glycosylases have been identified in humans and are categorized based on their structural architecture, which falls into one of six superfamilies [9]. Each glycosylase has specificity for removal of a small number of lesions. Given the average formation of 104 lesions per cell per day a given glycosylase may have to search tens of thousands of nucleobases to identify its target [2, 10]. A majority of glycosylases share the same overall mechanism, despite having different target lesions [9]. Most glycosylases extrude the lesion from the helix via base flipping, plug the resulting space left by the extrahelical lesion with an amino acid residue, and follow an SN1-like mechanism to catalyze glycosidic bond cleavage [7]. It was with the advent of synthetic organic chemistry methods to produce DNA oligomers with a single and site-specifically incorporated lesion that allowed for both these mechanistic studies and substrate specificity of glycosylases to be identified. While these studies provided remarkable advances in our understanding of glycosylase chemistry and biology, they do not account for the packaging of eukaryotic DNA into chromatin. This ubiquitous sequestration of eukaryotic DNA into the DNA-protein complex of chromatin presents a conundrum for BER enzymes, which interact intimately with DNA. Indeed, it has been shown in yeast that within the packaged genome, damage clusters specifically within the packaged environment and glycosylase activity is modulated [11]. In this Review, we focus on efforts to address BER in packaged DNA, with an emphasis on initiating the repair event by glycosylases.
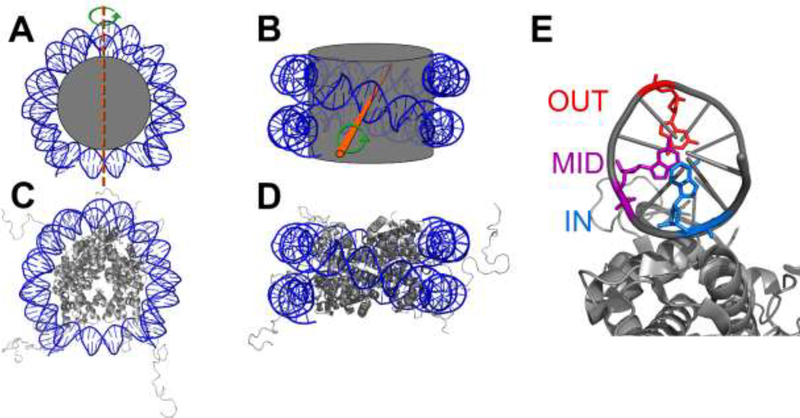
The nucleosome core particle (NCP) is the basic unit of packaging in eukaryotic chromatin. It consists of 145–147 base pairs (bp) of DNA wrapped ~1.7 times in superhelical coils around an octamer core of histone proteins (Fig. 1) [12] with a 2-fold axis of pseudo-symmetry known as the dyad axis. The octamer core is comprised of two copies of each histone protein: H2A, H2B, H3, and H4 [13]. Each histone protein contains a globular domain and an unstructured tail region [12]. The position of each nucleobase in an NCP can be described with respect to the histone core, which can be varied in two ways: (1) rotational position, referring to the helical orientation, i.e. if the nucleobase is facing out towards solution or in towards the histone octamer core; and (2) translational position, referring to the location of the nucleobase relative to the dyad axis. Ensemble [14] and single molecule [15–17] FRET measurements have shown that spontaneous unwrapping of nucleosomal DNA transiently exposes nucleobases that face in towards the histone octamer core (Fig. 2). This dynamic motion ultimately results in increased solution accessibility of nucleobases closer to the ends of DNA compared to bases at the dyad axis. Indeed, restriction enzymes show increased accessibility to recognition sites closer to ends of nucleosomal DNA compared to sites on the dyad axis [18].
Fig. 1.
Representations of the NCP. (A) Top view and (B) side view of the NCP represented as a spool-and-thread cartoon. The DNA is colored blue and the histone octamer core is gray. The dyad axis is indicated by a dashed orange line in (A) and an orange rod coming out of the plane of (B). The pseudosymmetry of the histone octamer about the dyad axis is indicated by the green arrow. In (C) and (D) the histone proteins are shown as ribbon diagrams and images are merged crystal structures of an NCP containing the Widom 601 DNA with a histone octamer containing N-terminal tail regions (Protein Data Bank entries 3lz0 and 1kx5, respectively). (E) Rotational positions of nucleobases relative to the histone octamer core (only one DNA strand is shown for simplicity). Outward (red), midway (purple), and inward (blue) bases are indicated.
Fig. 2.
Representation of an NCP showing an unwrapped state of one end of the DNA from the histone octamer core.
2. Experimental parameters and considerations
There are multiple factors to consider when constructing an NCP model system to reconstitute BER in vitro. Some of these factors include: the histone proteins, the DNA sequence and length, and the location and geometric position of the lesion of interest. It has become increasingly clear that each of these factors can modulate the efficiency of each enzymatic player in the BER pathway. Due to this variety of experimental parameters, one must choose the best option to address the desired question(s). Below we describe some of the most commonly-used systems to assemble NCPs in the laboratory.
2.1. Source of histone proteins
Isolation of histone octamer cores from a biological source.
Many of the first studies on reconstituted NCPs used a biological source of histone octamer cores isolated from chicken erythrocytes [19]. In this procedure, chromatin is isolated and digested with micrococcal nuclease to obtain NCPs containing chicken DNA. The original chicken DNA is then exchanged for the experimental DNA of interest by incremental dialysis from high to low concentrations of salt [19, 20]. Similar experiments have been performed using HeLa cells as a biological source of histone octamer cores [21, 22]. An important consideration of these NCPs assembled using isolated histones is that they are subject to a diverse array of post-translational modifications (PTMs) and represent a heterogeneous population.
Recombinantly expressed histone proteins.
Luger and Richmond reported the first high-resolution crystal structure of an NCP [12]. Crystallization of the NCP required a homogeneous population of NCPs and led to the recombinant expression and purification of individual Xenopus laevis histone proteins in E. coli [12, 23]. These individual histone proteins are then combined to form the histone octamer core. The high conservation of the histone proteins H2A, H2B, H3, and H4 across evolution makes studies using X. laevis histone proteins generalizable to all eukaryotes [24]. Creating NCPs using recombinant histone proteins is an important step if a homogeneous substrate population is required. This homogeneity, however, comes at the expense of histone PTMs which are known to be biologically relevant.
2.2. DNA sequence
While most DNA sequences have affinity for the histone proteins, a small subset (<5%) of sequences have a stronger, sequence-derived affinity that dictates NCP positioning [25]. These DNA are called nucleosome positioning sequences and bind histones in a predictable and reproducible manner with a defined translational and rotational position. The most commonly-used positioning sequences are the 5S rDNA and the Widom 601 sequences.
5S rDNA sequence.
The first demonstration that a DNA duplex could create predictable, positioned nucleosomes used the 5S sequence from the sea urchin L. variegatus [26]. The 5S sequence is appealing to use for in vitro experiments because it is a naturally-occurring sequence that is highly conserved in other organisms [27]. Periodicity of the 5S DNA sequence in an NCP changes relative to the unwrapped version, as revealed by hydroxyl radical footprinting (HRF). It is also known that the periodicity changes within the NCP depending on the translational position relative to the dyad axis [28]. These changes in periodicity based on translational location have also been observed for other DNA sequences [29, 30]. Additionally, the 5S sequence adopts multiple translational positions that are offset by multiples of 10 bp, especially when the length of the DNA surpasses the minimal 145–147 bp required to form an NCP [31–33]. However, incubating the NCPs at elevated temperature allows the DNA to “heat shift” to a single, thermodynamically favored translational position [34].
Widom 601 sequence.
The Widom laboratory recognized the potential benefits of using a positioning sequence and sought to identify the characteristics that define such DNA [35]. Using a SELEX approach they selected from a library of randomized DNA sequences those that had the highest affinity for the histone octamer core. These experiments identified the 601 sequence, a synthetic positioning sequence. Crystal structures of NCPs containing 601 DNA and X. laevis histones serve as useful references for designing experiments to address BER in NCPs [30, 36, 37]. In comparison to the naturally-occurring 5S sequence, the 601 NCP has been shown to be less dynamic and less accessible to digestion by restriction enzymes [18].
2.3. Incorporation of DNA lesion
Single site-specific lesions.
A common experimental set-up utilizes one lesion in a well-defined rotational and translational position. A site of interest for the lesion can be identified, in some cases with reference to a crystal structure, and DNA can be prepared in which the canonical nucleobase is replaced with a lesion. HRF and digestion by exonuclease III can be used to confirm the rotational and translational position of the lesion. A substantial body of literature exists that used site-specific incorporation of a lesion and serves as a major source of our current understanding of BER in NCPs.
Global incorporation of lesions.
Another method of studying BER in NCPs is one that uses global incorporation of lesions. Using either PCR or chemical synthesis techniques a population of DNA sequences can be prepared in which a lesion is globally incorporated across a variety of rotational and translational positions. This technique has been used to create populations of NCPs in which, on a single strand, all the T sites have been replaced by U [38–40], or the G sites have been replaced by 8-oxo-7,8-dihydroguanine (8-oxoG) [41]. While T and G have been globally substituted, each NCP contains only one lesion. By using the entire population of NCPs as a substrate in BER the global profile of repair in packaged DNA can be identified.
3. Glycosylase activity in NCPs
Recognizing that each of the abovementioned parameters may influence enzyme activity, our laboratory set out to develop a model system to compare directly the activity of various glycosylases in an NCP. For these kinetics studies, we sought to eliminate the heterogeneous nature of histone proteins isolated from biological sources and used recombinantly expressed X. laevis histone proteins. While lacking PTMs, we were motivated to use overexpressed histones to assemble reproducibly a homogeneous population of lesion-containing NCP. Similarly, the possibility of multiple translational positions could confound analysis of kinetic data and led us to use the 145 bp 601 DNA sequence rather than 5S DNA. In our work, we have used both site-specific and global incorporation of lesions. Our results are summarized below and then discussed in the context of other work in the field by a number of other research groups.
A minimal kinetic scheme for DNA glycosylases includes the three steps of the enzymatic cycle: (1) binding to the DNA substrate; (2) the chemistry step (kchem), which is cleavage of the glycosidic bond to excise the lesion; and (3) release of the DNA product (krel). For most glycosylases, product release is rate limiting; therefore, kcat measured under steady-state conditions ([glycosylase]<<[DNA]) is a reflection of product release rather than chemistry. By carrying out experiments under single-turnover (STO) conditions ([glycosylase]>>[DNA]), each enzyme has only one chance to perform chemistry before the substrate is consumed and krel does not factor into the kinetic model. When performing STO experiments using duplex substrates, binding to the DNA substrate is known to be fast and kobs = kchem. In NCP substrates however, the kobs values described below represent the slowest step up to and including chemistry and have not been assigned to a specific step. We [42] and others [39] have suggested that kobs reflects conformational dynamics of the NCP.
3.1. Glycosylase activity in the dyad region
We investigated the activity of five glycosylases in the dyad axis region of an NCP under identical reaction conditions: (1) E. coli uracil DNA glycosylase (UDG) acting on U; (2) human 8-oxoguanine glycosylase (hOGG1) on 8-oxoG; (3) E. coli formamidopyrimidine [fapy]-DNA glycosylase (Fpg) on 8-oxoG; (4) human alkyladenine DNA glycosylase (hAAG) on ethenoadenine (εA); and (5) E. coli EndoIII on an oxidized form of U, 5-hydroxyU (5OHU) [42]. These experiments allowed for comparisons between glycosylases based on a variety of factors: structural superfamily, monofunctionality or bifunctionality, or target lesion. Although NCPs are only found in eukaryotes, we used prokaryotic glycosylases in some cases because they are easier to obtain and are better characterized than the human analogs. Additionally, hOGG1 and Fpg both target 8-oxoG, but the eukaryotic and prokaryotic glycosylases are from different superfamilies allowing us to probe the role of structural architecture on activity. For each glycosylase/lesion pair, the rotational position of the lesion was varied as out towards solution, in towards the histone core, or midway between the two.
When comparing the five glycosylases, UDG and hAAG were the most effective at initiating BER at the dyad axis. For UDG acting on an outward facing lesion, product formation is quantitative with kobs of 5.8 min–1, which is less than an order of magnitude slower than the kobs for duplex (35 min–1). Thus in this experimental model with lesions at the dyad axis, UDG activity on an outward facing lesion is not significantly hindered by the histone octamer core. As the solution accessibility of the lesion decreases, the product yield also decreases dramatically, and the kinetics of UDG activity are more complex with two kinetic phases resolved. We [42] and others [39] have proposed that these biphasic kinetics reflect conformational dynamics of the NCP that allow UDG to access otherwise occluded lesions with inward and midway rotational positions.
Similar to the UDG/U system, the degree of activity in hAAG/εA NCP correlates with rotational positioning with the outward facing lesion removed most readily. The kobs are 0.01 and 0.04 min–1 for an outward facing lesion on NCP and duplex substrates, respectively. For all other systems examined including hOGG1/8-oxoG, Fpg/8-oxoG, and EndoIII/5-OHU, glycosylase activity is completely inhibited for lesions positioned at the dyad axis, regardless of rotational position.
Using molecular modeling it was shown that for UDG and hAAG, binding to NCPs at the dyad axis has relatively little steric obstruction from the histone octamer core [42]. In contrast, some residues of Fpg, EndoIII, and hOGG1 are fewer than 5 Å from the histone proteins. These short separation distances suggest that the histone proteins strongly interfere with these glycosylases binding at lesion sites in the dyad region. It is also noteworthy that co-crystal structures of glycosylases and lesion-containing DNA oligomers have revealed that the DNA is bent. UDG and hAAG display relatively small DNA bend angles of 22–45° [43, 44] and 22° [45], respectively. In comparison, for glycosylases that are not active at the dyad axis, larger bend angles have been observed: Endo III (55°) [46], Fpg (66°) [47], and hOGG1 (70°) [48]. While it remains unknown if DNA in an NCP must bend as part of the repair process, it is interesting to consider that the ability to distort DNA would be different in an NCP as compared to free duplex. This concept of torsional flexibility was previously discussed in the context of UDG activity in an NCP relative to pol β [49], where the latter causes a substantial distortion to DNA with a bend angle of ~90° [50]. We suggest torsional flexibility may similarly differentiate activity of glycosylases.
3.2. Glycosylase activity off the dyad
The dyad axis is a region of the NCP where dynamics are limited, as shown by FRET [14–17] and restriction enzyme digestion [18] studies. Using molecular tweezers it has also been observed that DNA unwrapping up to the dyad axis allows for rewrapping and reformation of the NCP [51]. Unwrapping past the dyad axis leads to a dissociation of the DNA from the histones [51]. This ability for transient dissociation of DNA ends may allow for modulation of glycosylase activity by allowing access to otherwise occluded lesions.
We were thus motivated to examine glycosylase activity off the dyad axis. Specifically, we investigated whether the dependence on rotational positioning was upheld when lesions were located in a region where more transient unwrapping occurs. We chose to focus on UDG/U and hOGG1/8-oxoG, since these glycosylase/lesion pairs display and lack activity, respectively, on the dyad. To allow for comparison to results obtained in the dyad region, the same NCP model system was used.
In contrast to the results observed for lesions located in the dyad region, lesions ~20 bp from the end of the DNA were removed by both UDG and hOGG1 irrespective of rotational position, although to varying degrees [52]. Furthermore, biphasic kinetics were observed at all rotational positions. For outward, midway, and inward lesions a fast phase was observed and is within an order of magnitude of kobs for duplex substrates. Specifically, kobs for hOGG1 in NCPs was 0.8–2.0 min−1 depending on rotational position and 2.6 min−1 for duplex. The kobs for UDG was 20.1–42.3 min−1 in NCPs and 12.6 min−1 for duplex. For hOGG1, the second kinetic phase observed in NCPs was ~40-times slower than the first phase; for UDG the second phase was ~130-times slower for outward and inward U and 6-times slower for the midway positioned lesion. These biphasic kinetics suggest that hOGG1 and UDG are able to perform chemistry on a readily accessible sub-population of the NCPs (fast phase) and also on a second sub-population that requires a conformational change to adopt a repairable form (slow phase).
These experiments with lesions positioned off the dyad axis also reflect that solution accessibility is not always predictive of product formation. For hOGG1, near quantitative product formation was observed for the midway lesion, whereas 40–50% product formation was observed for the inward and outward lesions. These results emphasize the existence of distinct microenvironments within an NCP that can have unique modulating effects on glycosylase activity. For example, the histone tails may influence the accessibility of lesions to repair. We observed that acetylation of H2B increased hOGG1 activity for the outward 8-oxoG located off the dyad [52]. Indeed, it has been shown that while the H2B tail associates with the nearby DNA, acetylation causes release of the tail from the DNA which could allow for hOGG1 binding [53]. When considering microenvironments within an NCP another notable feature is that DNA in the dyad region lacks superhelical coils whereas repair in other regions could be confounded by the DNA superhelices.
3.3. Global repair activity of glycosylases in an NCP
Most recently we focused on defining the repair fingerprint of hOGG1 [41]. The repair fingerprint describes the relative excision of 8-oxoG at sites across an NCP with varying rotational and translational positions. To enable comparison to our prior work with site-specifically incorporated lesions, the NCPs were assembled using 601 DNA and X. laevis histones. To prepare 601 DNA with globally-incorporated 8-oxoG we used a mixture of G/8-oxoG phosphoramidites as a reagent during chemical synthesis. The molar ratio of the mixture was chosen to minimize the number of strands with two or more lesions. This strategy yielded a population of oligomers with unbiased distribution of G to 8-oxoG substitutions throughout one strand of the Widom 601 DNA duplex. Incubation of NCPs containing this DNA with hOGG1 revealed the relative activity of the glycosylase at damage locations across the NCP.
One notable observation from the hOGG1 repair fingerprint is that the ~20 bp centered on the dyad axis is a region with relatively low levels of repair. DNA at the dyad axis is known to be relatively straight compared to other regions of an NCP and to adopt an unusual helical periodicity [28, 54], which we suggest modulates hOGG1 activity in this region of packaged DNA. This observation of limited hOGG1 activity at the dyad is consistent with results obtained using site-specific lesions [42, 55]. The repair fingerprint also reveals that hOGG1 efficiently excises most outward lesions outside of the dyad region. Additionally, towards the ends of the DNA some, but not all, lesions that are in or midway can be repaired. Perhaps most importantly, the repair fingerprint demonstrates that no single lesion site can be used to represent repair of packaged DNA but rather that the initiation of BER depends on the location of the lesion.
4. Glycosylase activity in context
4.1. UDG activity in NCPs
A substantial portion of the literature examining glycosylase activity in NCPs has used enzymes from the UDG superfamily: E. coli UDG, human UNG2, and human SMUG1. One of the earliest reports came from the Verreault laboratory, which examined UNG2 and SMUG1 activity in 146 bp 5S NCPs assembled with histone octamer cores isolated from chicken erythrocytes [40]. A reduction in glycosylase activity at U:A mispairs in the NCP compared to duplex was observed. When the translational position of a sites-pecific U was varied between an on and off the dyad location, there was no apparent difference in glycosylase activity for either SMUG1 or UNG2. Using global incorporation, both UNG2 and SMUG1 had the same overall repair pattern on NCPs and duplex, suggesting that the histones do not modulate repair in this system.
The Smerdon group expanded on these observations using NCP substrates with a TG motif positioning sequence flanking a glucocorticoid hormone receptor response element (GRE) and histones from chicken erythrocytes [49]. A correlation between rotational positioning of a U:G mispair and UNG2/APE1 activity was observed. In particular, there was more product formation for outward U than inward sites on the dyad [49, 56]. When the histone tails were removed by trypsin digestion, no change in UNG2/APE1 activity was observed suggesting that the histone tails do not affect these BER enzymes at the dyad axis. In other work, using the TG-GRE-TG NCP [57] and NCPs assembled from 601 DNA and chicken erythrocyte histones [58], they investigated the potential effects of formaldehyde-induced crosslinking of the DNA to the histone octamer core. When formaldehyde-treated NCPs were incubated with either UDG/APE1 or UNG2/APE1, inward sites were not as readily repaired indicating that transient dynamics and DNA breathing facilitate BER. Furthermore, UNG2/APE1 activity was observed in higher-order nucleosome arrays and at rates only 2–3 fold slower than duplex [59]. The Smerdon group also investigated the effect of acetylation of a lysine within H3 (H3K56) and observed decreased pol β activity but no effect on UDG/APE1 activity [60]. This result was observed in both the 601 and 5S sequences and at multiple translational and rotational positions. Most recently, the deletion of a portion of the H2B tail, known as the histone H2B repression domain, was found to enhance UDG/APE1 activity in off dyad regions, which was attributed to enhanced nucleosome dynamics [61].
The Hayes group used a 154 bp 5S sequence derived from Xenopus borealis with chicken erythrocyte histone octamer cores to study U:A mispairs and UDG activity in NCPs [38]. They too found a correlation between glycosylase activity and rotational positioning. In addition, activity was increased when U lesions were located closer to the DNA ends. Using global incorporation, repair was only observed at sites predicted to be facing outward toward solution [38]. A lack of UDG activity at the dyad axis was observed, similar to the result obtained for the repair fingerprint of hOGG1 [41], further indicating that this region of the NCP has a distinct repair profile.
Most recently, the Stivers group studied global incorporation of U:A mispairs in the 601 sequence bound to X. laevis histones and found no correlation between either translational or rotational positioning and UNG2 activity [39]. This result suggests that local DNA-histone environments impede the ability to predict glycosylase activity in the NCP.
4.2. NTH1 and NEIL1 activity in NCPs
The Wallace and Pederson groups have observed endonuclease III-like protein 1 (NTH1) activity on NCP substrates containing site-specific thymine glycol (Tg) lesions [33, 62, 63]. The work used a 184 bp 5S sequence and chicken erythrocyte histone octamer cores. Greater NTH1 activity was seen for outward Tg lesions relative to inward lesions, as well as an increase in activity closer to the ends of the DNA [33]. There was also an increase in glycosylase activity as a function of enzyme concentration, suggesting that an inherent feature of the NCP itself may be rate limiting as opposed to the glycosylase. Specifically, lesion exposure rates, as determined by the DNA unwrapping rates, proved to be rate limiting for NTH1 [63]. This observation was made for NCPs containing both the 601 and 5S sequences, albeit with different unwrapping rates of 0.084 min−1 and 0.18 min−1, respectively. Interestingly, it was recently reported that human cells contain a factor distinct from chromatin remodelers that facilitates NTH1 activity in NCP substrates [64].
A direct comparison of NTH1 and endonuclease VIII-like 1 (NEIL1), which both excise the Tg lesion but belong to different structural superfamilies, was performed [62]. Using biologically relevant concentrations of each glycosylase, NTH1 was more effective at removing occluded sites than NEIL1, while they both displayed similar activity on outward lesions.
4.3. hOGG1 activity in NCPs
The Angelov group reported activity of hOGG1 in the context of both NCPs and dinucleosome substrates assembled with the 601 sequence and X. laevis histones. In the single NCP constructed using a 227 bp 601 DNA sequence, hOGG1 activity was dramatically suppressed relative to duplex when 8-oxoG was near the dyad axis. Activity was recovered in the presence of the SWI/SNF chromatin remodeler and, to a much smaller extent, with the histone variant H2A.Bbd. In the dinucleosome substrate, the 8-oxoG was either near the dyad or in the linker region. Similar to the single NCP substrate, 8-oxoG was not efficiently removed from the dyad axis. However, hOGG1 activity was observed in the linker DNA and the efficiency of 8-oxoG removal was comparable to duplex. When the linker histone H1 was present, hOGG1 activity required a RSC remodeler [55]. These results indicate that chromatin remodelers and histone variants may influence the activity of some glycosylases, particularly for lesions located near the dyad axis.
4.4. MBD4 activity in NCPs
The Ausió group observed Methyl-CpG Binding Domain 4 (MBD4) activity in NCPs using a 164 bp 5S sequence containing a single T:G mispair and chicken erythrocyte histone octamer cores [65]. MBD4 had reduced activity in the NCP compared to duplex. When the T:G mispair was placed in two different off-dyad locations on opposite ends of the wrapped duplex, with the T predicted to be facing outward in both instances, there was no change in activity relative to translational position. However, there was an increase in activity seen in NCPs containing acetylated histones, suggesting that PTMs can modulate glycosylase activity. Histone acetylation has been shown to modulate the activity of other DNA binding proteins and promote unwrapping of the DNA from the histone octamer [66, 67].
5. Conclusion
The NCP is a complex substrate for DNA repair in which a variety of factors can influence glycosylase activity and initiation of BER. While we are beginning to unravel the connections between positioning of a lesion and glycosylase activity, there are many more factors that likely modulate repair and could inform future studies. The contributions of histone PTMs and variants has only begun to be explored. Furthermore, glycosylases do not work in isolation in vivo and have been shown to be influenced by the downstream BER enzymes. The addition of spectator DNA or crowding agents may also aid in creating a more biologically representative analysis of glycosylase activity. The addition of ATPdependent chromatin remodelers and other cellular factors may unwrap and increase accessibility of the DNA for complete BER repair. Finally, the NCP is just the first level of compaction within chromatin. Future studies could include higher-order chromatin structures such as nucleosome arrays.
Acknowledgments
We apologize to those investigators whose research was not cited in the interest of preparing a concise review. Research in the Delaney laboratory has been supported by the National Institute of Environmental Health Sciences (R01ES019296) and the National Science Foundation (MCB-1817417). EEK was supported by the National Institute of General Medical Sciences (T32GM007601). We thank Adam Garlow for preparing the figures and members of the Delaney laboratory for helpful discussions.
ABBREVIATIONS:
- BER
base excision repair
- εA
ethenoadenine
- Fpg
formamidopyrimidine [fapy]-DNA glycosylase
- FRET
fluorescence resonance energy transfer
- hAAG
human alkyladenine DNA glycosylase
- HBR
histone H2B repression domain
- hOGG1
human 8-oxoguanine glycosylase
- HRF
hydroxyl radical footprinting
- 5-OHU
5-hydroxyuracil
- MBD4
Methyl-CpG Binding Domain 4
- NCP
nucleosome core particle
- NEIL1
endonuclease VIII-like 1
- NTH1
endonuclease III-like protein 1
- 8-oxo-G
8-oxo-7,8-dihydroguanine
- PTMs
post-translational modifications
- Tg
thymine glycol
- U
uracil
Footnotes
Conflict of interest
The authors declare no conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Gates KS, An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals, Chem. Res. Toxicol 22 (2009) 1747–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lindahl T, Instability and decay of the primary structure of DNA, Nature 362 (1993) 709–715. [DOI] [PubMed] [Google Scholar]
- [3].Swenberg JA, Lu K, Moeller BC, Gao L, Upton PB, Nakamura J, Starr TB, Endogenous versus exogenous DNA adducts: their role in carcinogenesis, epidemiology, and risk assessment, Toxicol. Sci 120 (2011) S130–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hoeijmakers JH, Genome maintenance mechanisms for preventing cancer, Nature 411 (2001) 366–374. [DOI] [PubMed] [Google Scholar]
- [5].Schermerhorn KM, Delaney S, A kinetic perspective on base excision repair of DNA, Acc. Chem. Res 47 (2014) 1238–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S, Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints, Annu. Rev. Biochem 73 (2004) 39–85. [DOI] [PubMed] [Google Scholar]
- [7].Stivers JT, Jiang YL, A Mechanistic Perspective on the Chemistry of DNA Repair Glycosylases, Chem. Rev 103 (2003) 2729–2760. [DOI] [PubMed] [Google Scholar]
- [8].Prasad R, Shock DD, Beard WA, Wilson SH, Substrate channeling in mammalian base excision repair pathways: passing the baton, J. Biol. Chem 285 (2010) 40479–40488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Brooks SC, Adhikary S, Rubinson EH, Eichman BF, Recent advances in the structural mechanisms of DNA glycosylases, Biochim. Biophys. Acta 1834 (2013) 247271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Friedman JI, Stivers JT, Detection of damaged DNA bases by DNA glycosylase enzymes, Biochemistry 49 (2010) 4957–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mao P, Brown AJ, Malc EP, Mieczkowski PA, Smerdon MJ, Roberts SA, Wyrick JJ, Genome-wide maps of alkylation damage, repair, and mutagenesis in yeast reveal mechanisms of mutational heterogeneity, Genome Res. 27 (2017) 1674–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ, Crystal structure of the nucleosome core particle at 2.8 A resolution, Nature 389 (1997) 251–260. [DOI] [PubMed] [Google Scholar]
- [13].Eickbush TH, Moudrianakis EN, The histone core complex: an octamer assembled by two sets of protein-protein interactions, Biochemistry 17 (1978) 49554964. [DOI] [PubMed] [Google Scholar]
- [14].Yang JG, Narlikar GJ, FRET-based methods to study ATP-dependent changes in chromatin structure, Methods, 41 (2007) 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Buning R, van Noort J, Single-pair FRET experiments on nucleosome conformational dynamics, Biochimie 92 (2010) 1729–1740. [DOI] [PubMed] [Google Scholar]
- [16].Lee JY, Lee J, Yue H, Lee T-H, Dynamics of Nucleosome Assembly and Effects of DNA Methylation, J. Biol. Chem 290 (2014) 4291–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kim J, Wei S, Lee J, Yue H, Lee T-H, Single-Molecule Observation Reveals Spontaneous Protein Dynamics in the Nucleosome, J. Phys. Chem. B 120 (2016) 8925–8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Polach KJ, Widom J, Restriction enzymes as probes of nucleosome stability and dynamics, Methods Enzymol. 304 (1999) 278–298. [DOI] [PubMed] [Google Scholar]
- [19].Hayes JJ, Lee KM, In vitro reconstitution and analysis of mononucleosomes containing defined DNAs and proteins, Methods 12 (1997) 2–9. [DOI] [PubMed] [Google Scholar]
- [20].Libertini LJ, Small EW, Salt induced transitions of chromatin core particles studied by tyrosine fluorescence anisotropy, Nucleic Acids Res. 8 (1980) 3517–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ober M, Lippard SJ, Cisplatin damage overrides the predefined rotational setting of positioned nucleosomes, J. Am. Chem. Soc 129 (2007) 6278–6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mulvihill DJ, Nichol Edamura K, Hagerman KA, Pearson CE, Wang YH, Effect of CAT or AGG interruptions and CpG methylation on nucleosome assembly upon trinucleotide repeats on spinocerebellar ataxia, type 1 and fragile X syndrome, J. Biol. Chem 280 (2005) 4498–4503. [DOI] [PubMed] [Google Scholar]
- [23].Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ, Characterization of nucleosome core particles containing histone proteins made in bacteria, J. Mol. Biol 272 (1997) 301–311. [DOI] [PubMed] [Google Scholar]
- [24].Sullivan SA, Aravind L, Makalowska I, Baxevanis AD, Landsman D, The Histone Database: a comprehensive WWW resource for histones and histone foldcontaining proteins, Nucleic Acids Res. 28 (2000) 320–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lowary PT, Widom J, Nucleosome packaging and nucleosome positioning of genomic DNA, Proc. Natl. Acad. Sci. U.S.A 94 (1997) 1183–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Simpson RT, Stafford DW, Structural features of a phased nucleosome core particle, Proc. Natl. Acad. Sci. U.S.A 80 (1983) 51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Long EO, Dawid IB, Repeated genes in eukaryotes, Annu. Rev. Biochem 49 (1980) 727–764. [DOI] [PubMed] [Google Scholar]
- [28].Hayes JJ, Tullius TD, Wolffe AP, The structure of DNA in a nucleosome, Proc. Natl. Acad. Sci. U.S.A 87 (1990) 7405–7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Prunell A, Periodicity of exonuclease III digestion of chromatin and the pitch of DNA on the nucleosome, Biochemistry 22 (1983) 4887–4894. [DOI] [PubMed] [Google Scholar]
- [30].Vasudevan D, Chua EYD, Davey CA, Crystal Structures of Nucleosome Core Particles Containing the ‘601’ Strong Positioning Sequence, J. Mol. Biol 403 (2010) 1–10. [DOI] [PubMed] [Google Scholar]
- [31].Dong F, Hansen JC, van Holde KE, DNA and protein determinants of nucleosome positioning on sea urchin 5S rRNA gene sequences in vitro, Proc. Natl. Acad. Sci. U.S.A 87 (1990) 5724–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ura K, Hayes JJ, Wolffe AP, A positive role for nucleosome mobility in the transcriptional activity of chromatin templates: restriction by linker histones, EMBO J. 14 (1995) 3752–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Prasad A, Wallace SS, Pederson DS, Initiation of base excision repair of oxidative lesions in nucleosomes by the human, bifunctional DNA glycosylase NTH1, Mol. Cell. Biol 27 (2007) 8442–8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Flaus A, Luger K, Tan S, Richmond TJ, Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals, Proc. Natl. Acad. Sci. U.S.A 93 (1996) 1370–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lowary PT, Widom J, New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning, J. Mol. Biol 276 (1998) 19–42. [DOI] [PubMed] [Google Scholar]
- [36].Bednar J, Garcia-Saez I, Boopathi R, Cutter AR, Papai G, Reymer A, Syed SH, Lone IN, Tonchev O, Crucifix C, Menoni H, Papin C, Skoufias DA, Kurumizaka H, Lavery R, Hamiche A, Hayes JJ, Schultz P, Angelov D, Petosa C, Dimitrov S, Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1, Mol. Cell 66 (2017) 384–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chua EY, Vasudevan D, Davey GE, Wu B, Davey CA, The mechanics behind DNA sequence-dependent properties of the nucleosome, Nucleic Acids Res. 40 (2012) 6338–6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cole HA, Tabor-Godwin JM, Hayes JJ, Uracil DNA Glycosylase Activity on Nucleosomal DNA Depends on Rotational Orientation of Targets, J. Biol. Chem 285 (2010) 2876–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ye Y, Stahley MR, Xu J, Friedman JI, Sun Y, McKnight JN, Gray JJ, Bowman GD, Stivers JT, Enzymatic excision of uracil residues in nucleosomes depends on the local DNA structure and dynamics, Biochemistry 51 (2012) 6028–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nilsen H, Lindahl T, Verreault A, DNA base excision repair of uracil residues in reconstituted nucleosome core particles, EMBO J. 21 (2002) 5943–5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bilotti K, Tarantino ME, Delaney S, hOGG1 removes solution-accessible 8-oxoG lesions from globally-substituted nucleosomes except at the dyad region, Biochemistry, (2018) 1436–1439. [DOI] [PubMed] [Google Scholar]
- [42].Olmon ED, Delaney S, Differential Ability of Five DNA Glycosylases to Recognize and Repair Damage on Nucleosomal DNA, ACS Chem. Biol 12 (2017) 692–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA, Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA, EMBO J. 17 (1998) 5214–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA, A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA, Nature 384 (1996) 87–92. [DOI] [PubMed] [Google Scholar]
- [45].Lau AY, Schärer OD, Samson L, Verdine GL, Ellenberger T, Crystal Structure of a Human Alkylbase-DNA Repair Enzyme Complexed to DNA, Cell 95 (1998) 249–258. [DOI] [PubMed] [Google Scholar]
- [46].Fromme JC, Verdine GL, Structure of a trapped endonuclease III-DNA covalent intermediate, EMBO J. 22 (2003) 3461–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G, Structure of Formamidopyrimidine-DNA Glycosylase Covalently Complexed to DNA, J. Biol. Chem 277 (2002) 19811–19816. [DOI] [PubMed] [Google Scholar]
- [48].Bruner SD, Norman DP, Verdine GL, Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA, Nature 403 (2000) 859–866. [DOI] [PubMed] [Google Scholar]
- [49].Beard BC, Wilson SH, Smerdon MJ, Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes, Proc. Natl. Acad. Sci. U.S.A 100 (2003) 7465–7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Krahn JM, Beard WA, Miller H, Grollman AP, Wilson SH, Structure of DNA Polymerase β with the Mutagenic DNA Lesion 8-Oxodeoxyguanine Reveals Structural Insights into Its Coding Potential, Structure 11 (2003) 121–127. [DOI] [PubMed] [Google Scholar]
- [51].Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD, High-resolution dynamic mapping of histone-DNA interactions in a nucleosome, Nat. Struct. Mol. Biol 16 (2009) 124–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bilotti K, Kennedy EE, Li C, Delaney S, Human OGG1 activity in nucleosomes is facilitated by transient unwrapping of DNA and is influenced by the local histone environment, DNA Repair (Amst) 59 (2017) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Fu I, Cai Y, Geacintov NE, Zhang Y, Broyde S, Nucleosome Histone Tail Conformation and Dynamics: Impacts of Lysine Acetylation and a Nearby Minor Groove Benzo[a]pyrene-Derived Lesion, Biochemistry 56 (2017) 1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Satchwell SC, Drew HR, Travers AA, Sequence periodicities in chicken nucleosome core DNA, J. Mol. Biol 191 (1986) 659–675. [DOI] [PubMed] [Google Scholar]
- [55].Menoni H, Shukla MS, Gerson V, Dimitrov S, Angelov D, Base excision repair of 8-oxoG in dinucleosomes, Nucleic Acids Res. 40 (2012) 692–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Beard BC, Stevenson JJ, Wilson SH, Smerdon MJ, Base excision repair in nucleosomes lacking histone tails, DNA Repair (Amst) 4 (2005) 203–209. [DOI] [PubMed] [Google Scholar]
- [57].Hinz JM, Rodriguez Y, Smerdon MJ, Rotational dynamics of DNA on the nucleosome surface markedly impact accessibility to a DNA repair enzyme, Proc. Natl. Acad. Sci. U.S.A 107 (2010) 4646–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rodriguez Y, Smerdon MJ, The Structural Location of DNA Lesions in Nucleosome Core Particles Determines Accessibility by Base Excision Repair Enzymes, J. Biol. Chem 288 (2013) 13863–13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Nakanishi S, Prasad R, Wilson SH, Smerdon M, Different structural states in oligonucleosomes are required for early versus late steps of base excision repair, Nucleic Acids Res. 35 (2007) 4313–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rodriguez Y, Hinz JM, Laughery MF, Wyrick JJ, Smerdon MJ, Site-specific Acetylation of Histone H3 Decreases Polymerase β Activity on Nucleosome Core Particles in Vitro, J. Biol. Chem 291 (2016) 11434–11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rodriguez Y, Duan M, Wyrick JJ, Smerdon MJ, A cassette of basic amino acids in histone H2B regulates nucleosome dynamics and access to DNA damage, J. Biol. Chem 293 (2018) 7376–7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Odell ID, Newick K, Heintz NH, Wallace SS, Pederson DS, Non-specific DNA binding interferes with the efficient excision of oxidative lesions from chromatin by the human DNA glycosylase, NEIL1, DNA Repair (Amst) 9 (2010) 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Maher RL, Prasad A, Rizvanova O, Wallace SS, Pederson DS, Contribution of DNA unwrapping from histone octamers to the repair of oxidatively damaged DNA in nucleosomes, DNA Repair (Amst) 12 (2013) 964–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Maher RL, Marsden CG, Averill AM, Wallace SS, Sweasy JB, Pederson DS, Human cells contain a factor that facilitates the DNA glycosylase-mediated excision of oxidized bases from occluded sites in nucleosomes, DNA Repair (Amst) 57 (2017) 9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ishibashi T, So K, Cupples CG, Ausió J, MBD4-mediated glycosylase activity on a chromatin template is enhanced by acetylation, Mol. Cell Biol 28 (2008) 4734–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Simon M, North JA, Shimko JC, Forties RA, Ferdinand MB, Manohar M, Zhang M, Fishel R, Ottesen JJ, Poirier MG, Histone fold modifications control nucleosome unwrapping and disassembly, Proc. Natl. Acad. Sci. U.S.A 108 (2011) 12711–12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Manohar M, Mooney AM, North JA, Nakkula RJ, Picking JW, Edon A, Fishel R, Poirier MG, Ottesen JJ, Acetylation of Histone H3 at the Nucleosome Dyad Alters DNA-Histone Binding, J. Biol. Chem 284 (2009) 23312–23321. [DOI] [PMC free article] [PubMed] [Google Scholar]