Abstract
The pyruvate dehydrogenase complex (PDC) is a multienzyme assembly that converts pyruvate to acetyl-CoA. As pyruvate and acetyl-CoA play central roles in cellular metabolism, understanding PDC regulation is pivotal to understanding the larger metabolic network. The activity of mammalian PDC is regulated through reversible phosphorylation governed by at least four isozymes of pyruvate dehydrogenase kinase (PDK). Deciphering which kinase regulates PDC in organisms at specific times or places has been challenging. In this study, we analyzed mouse strains carrying targeted mutations of individual isozymes to explore their role in regulating PDC activity. Analysis of protein content of PDK isozymes in major metabolic tissues revealed that PDK1 and PDK2 were ubiquitously expressed, whereas PDK3 and PDK4 displayed a rather limited tissue distribution. Measurement of kinase activity showed that PDK1 is the principal isozyme regulating hepatic PDC. PDK2 was largely responsible for inactivation of PDC in tissues of muscle origin and brown adipose tissue (BAT). PDK3 was the principal kinase regulating pyruvate dehydrogenase activity in kidney and brain. In a well-fed state, the tissue levels of PDK4 protein were fairly low. In most tissues tested, PDK4 ablation had little effect on the overall rates of inactivation of PDC in kinase reaction. Taken together, these data strongly suggest that the activity of PDC is regulated by different isozymes in different tissues. Furthermore, it appears that the overall flux through PDC in a given tissue largely reflects the properties of the PDK isozyme that is principally responsible for the regulation of PDC activity in that tissue.
Keywords: pyruvate dehydrogenase complex (PDC), pyruvate dehydrogenase kinase (PDC kinase), mitochondria, carbohydrate metabolism, protein phosphorylation, carbohydrate metabolism, mitochondria, oxidative decarboxylation of pyruvate, protein kinase, protein phosphorylation, metabolic flux
Introduction
Pyruvate serves a number of important biological functions. Besides being one of the major sources of carbon for oxidation in the citric acid cycle for energy production, it is also used as a precursor for biosynthesis of fatty acids, glucose, and sterols (1–4). Being an effective scavenger of reactive oxygen species, pyruvate has potent antioxidant and anti-inflammatory properties (5–7). Recent evidence also suggests that pyruvate may act as an inhibitor of histone deacetylases and an inducer of apoptosis in certain tumor cells (8, 9). In mammals, the metabolic fate of pyruvate is largely determined by the activity of mitochondrial pyruvate dehydrogenase complex (PDC)3 due to the physiologically irreversible nature of this reaction (10). Consequently, PDC is critical for maintaining the tissue levels of pyruvate and regulation of intermediary metabolism, oxidative stress, apoptosis, gene transcription, adaptation to food deprivation, and hypoxia (11–16).
PDC is a large multienzyme complex built of multiple copies of three enzymes: pyruvate dehydrogenase (E1), dihydrolipoyl acetyltransferase (E2), and dihydrolipoyl dehydrogenase (E3), which catalyze five consecutive biochemical reactions leading to the synthesis of acetyl-CoA, NADH, and CO2 (17). Activity of the mammalian PDC is regulated through the phosphorylation–dephosphorylation cycle. Phosphorylation and concomitant inactivation of the complex is catalyzed by pyruvate dehydrogenase kinase (PDK), and dephosphorylation and the concomitant reactivation is catalyzed by pyruvate dehydrogenase phosphatase (PDP) (18, 19). The site of this covalent regulation is the pyruvate dehydrogenase component that can be phosphorylated at three sites (sites 1, 2, and 3, respectively). Phosphorylation at site 1 correlates closely with inactivation of the enzyme (18). It is generally believed that reversible phosphorylation of PDC is largely responsible for the adjustments of its activity on a minute-to-minute basis as well as in the long term (e.g. during starvation) (18, 19).
Biochemical and genetic evidence suggests that there are at least four isozymes of kinase (PDK1, PDK2, PDK3, and PDK4) and two isozymes of phosphatase (PDP1 and PDP2), which can contribute to the regulation of PDC activity in mammalian tissues (20–22). PDK isozymes have been implicated in the regulation of intermediary metabolism, oxidative stress, and apoptosis. Their abnormal regulation presumably contributes to diabetes, obesity, cancer, ischemia, and metabolic acidosis (23–27). To understand how this rather broad spectrum of effects comes about, it is necessary to characterize the functionality of native PDK isozymes in various tissues. Analysis of genetically modified mouse models is one of the proven approaches to tackle this issue. Thus, in the present study, we characterized the regulation of PDC in major metabolic tissues of mice using murine strains carrying targeted mutations of individual Pdk genes. Our results provide the first evidence that individual isozymes of kinase play distinctive, only partially overlapping roles in the regulation of PDC activity and, consequently, general metabolism of carbohydrate and lipid fuels.
Results
Expression patterns of PDK isozymes in mouse tissues
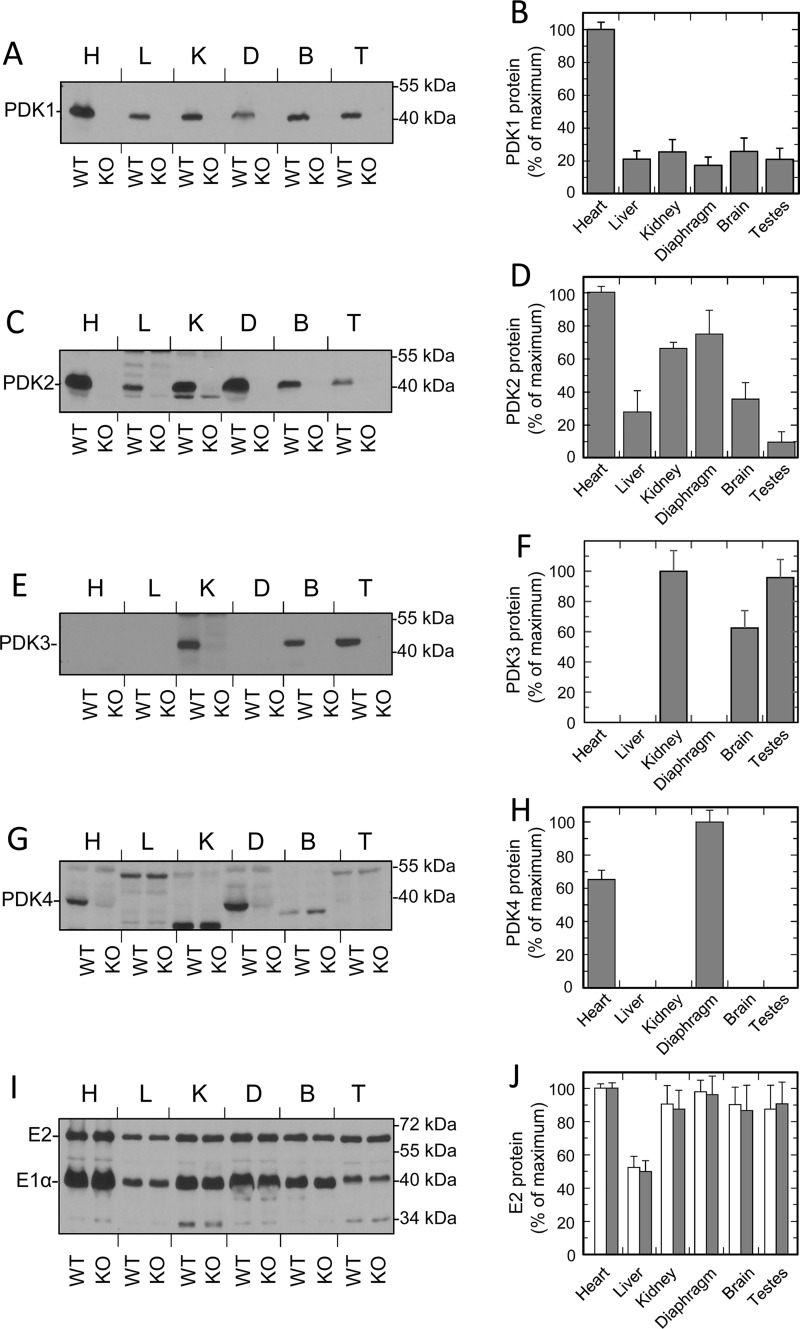
Historically, conclusions about the contributions of different PDK isozymes to the regulation of PDC activity and metabolism, in general, were often based on the analysis of the abundance of their mRNAs by Northern blotting or RT-PCR (28, 29). Although these measurements provide valuable information about the expression patterns of a particular gene, they might be misleading when they are used to predict the activity and/or activity change of the corresponding protein product because, sometimes, the levels of mRNA poorly correlate with the levels of the protein product. For example, PDK4 mRNA tends to show far greater changes than PDK4 protein (30). In this respect, predictions based on the estimates of protein mass by Western blotting or immunohistochemistry appear to be superior. Unfortunately, as far as the isozymes of PDK were concerned, the corresponding estimates were often inconclusive due to poor specificity and cross-reactivity of the available kinase antibodies and the lack of appropriate negative controls. Creation of mouse strains carrying targeted mutations of individual PDK genes allowed for a unique opportunity to investigate expression patterns of the corresponding protein products and, furthermore, to estimate their roles in the regulation of PDC activity through reversible phosphorylation. This paper reports on the expression patterns of the protein products of all four genes encoding PDK isozymes, as well as on their contributions to the overall rate of kinase reaction in the major metabolic tissues obtained from mice maintained in a well-fed state.
General expression patterns of all four PDK proteins in mouse tissues were studied using Western blot analysis (Fig. 1 and Fig. S1). To evaluate the expression pattern of a particular isozyme, each blot contained samples prepared from tissues of WT mice. In addition, each blot contained identically prepared samples made from the corresponding tissues of the littermates carrying a targeted deletion of the gene of interest. This strategy allowed us to identify commercial antibodies for PDK1 that greatly exceeded the sensitivity of our in-house–made antibodies reported previously (30) (Fig. 1A and Fig. S1A). We also found that the previously described antiserum raised against the polypeptide corresponding to the carboxyl end of PDK4 (31) cross-reacted with numerous nonspecific proteins, some of which had electrophoretic mobility virtually identical to that of PDK4 itself (Fig. 1G and Fig. S1G). However, with appropriate negative controls, we were able to reliably identify PDK4 protein in whole-tissue extracts. Systematic application of this approach revealed that in mouse tissues, PDK isozymes display markedly different expression patterns (Fig. 1 and Fig. S1). Some of these patterns differed significantly from the expression patterns identified previously based on the analysis of corresponding mRNAs (28). This was particularly evident for isozyme PDK1 (Fig. 1 (A and B) and Fig. S1 (A and B)). Based on Northern blot analysis, PDK1 appeared to be largely a heart-specific isozyme (28). In agreement with this finding, Western blot analysis showed that the heart samples contained by far the greatest amounts of PDK1 protein. However, Western blotting clearly revealed the presence of PDK1 protein virtually in all tissue samples tested thus far. Although on average the amounts of PDK1 protein in other tissues were significantly lower than in heart, PDK1 was expressed almost ubiquitously. Unlike PDK1, PDK2 was always considered to be a ubiquitous isozyme, playing an essential role in the regulation of PDC activity in the majority of tissues (19, 28). Western blot analysis carried out in this study largely confirmed those earlier observations (Fig. 1 (C and D) and Fig. S1 (C and D)). PDK2 protein had been identified in all tissues tested. The greatest amounts of PDK2 protein were detected in heart, diaphragm, kidney, and red skeletal muscles. Other tissues (i.e. liver, brain, testis, ovaries, and lung) displayed lower levels of PDK2 protein, presumably reflecting the lower numbers of mitochondria present in those tissues and/or lower levels of PDC expression (Fig. 1, I and J).
Figure 1.
Protein levels of PDK isozymes in mouse tissues. A, C, E, G, and I, representative Western blots of PDK1, PDK2, PDK3, PDK4, and E2 and E1α proteins of PDC in mouse tissues (heart (H), liver (L), kidney (K), diaphragm (D), brain (B), and testis (T)). B, D, F, H, and J illustrate the relative abundance of PDK1, PDK2, PDK3, PDK4, and PDC protein in mouse tissues calculated based on the results of scanning densitometry of Western blots. Data are expressed as percentage of the area of the strongest band. Gray and white bars in J correspond to the protein levels of the E2 component in WT and PDK2 knockout mice. Western blotting was carried out as described under “Experimental procedures.” Each lane contains 35 or 17 μg of total whole-tissue protein for kinase or PDC staining, respectively. Data points represent means ± S.D. (error bars) for three mice in each group.
Historically, identification of expression patterns of PDK3 in mammalian tissues was hampered by the extremely low levels of PDK3 mRNA present in major metabolic tissues, making Northern blotting largely unreliable (28). Thus, it was somewhat unexpected that we were able to readily detect PDK3 protein by Western blotting (Fig. 1 (E and F) and Fig. S1 (E and F)). This suggested that PDK3 protein is fairly stable, and small amounts of PDK3 mRNA are sufficient to support the necessary protein levels. In contrast to PDK1 or PDK2 that were expressed ubiquitously, PDK3 was found only in kidney, brain, testis, and lung, whereas the other major metabolic tissues tested did not express appreciable levels of PDK3. This indicates that, unlike PDK1 or PDK2, PDK3 plays a more specialized role, serving metabolic patterns characteristic of kidney, brain, testis, or lung.
Initially, PDK4 was discovered as an inducible isozyme that became overexpressed in starvation and diabetes (21, 30). This was thought to be essential to prevent the aerobic oxidation of carbohydrates and to spare pyruvate and other metabolically related carboxylic acids for gluconeogenesis (30). Over the years, primarily by means of RT-PCR, PDK4 expression had been documented in a variety of tissues and cell lines (19, 29). Here, to avoid variations associated with the effects of fasting, we analyzed the expression patterns of PDK4 protein in a well-fed state under normal feeding conditions. Somewhat unexpectedly, we found that PDK4 displayed a rather limited tissue distribution (i.e. PDK4 was detected only in tissues of muscle origin, such as heart, diaphragm, or skeletal muscle) (Fig. 1 (G and H) and Fig. S1 (G and H)). Importantly, even in tissues of muscle origin, the signal generated by PDK4 tended to be weak, indicating that the protein levels of PDK4 were fairly low. This is consistent with the interpretation that, in a well-fed state, PDK4 plays a rather limited role due to its limited tissue distribution and low tissue abundance. In this respect, brown adipose was the only tissue that consistently displayed elevated levels of PDK4 protein even in a well-fed state (Fig. S1, G and H). The latter indicates that, in a well-fed state, PDK4 might play an important role in the regulation of BAT metabolism and even in the regulation of thermogenesis.
Unfortunately, interpretation of the data obtained by Western blot analysis is limited by the signal saturation, which complicates the direct comparison between samples containing different amounts of antigen. In this context, the comparisons between different samples stained with different antibodies appear to be even less reliable. Thus, to obtain some estimates with respect to the tissue levels of PDK isozymes present in major metabolic tissues, we employed quantitative Western blot analysis. Toward this end, experimental samples made from tissues of WT mice were separated along with identically prepared samples made from tissues of mice that were deficient in isozyme in question. Each of those control samples received various amounts of highly purified recombinant isozyme for quantification purposes. In preliminary experiments, loading conditions were identified where there was a linear relationship between the signals and the amounts of recombinant kinase in standards and where the signals generated by the native kinase in the samples prepared from WT mice were within the linear range of the calibration curve (Figs. S2 and S3). This approach allows one to circumvent, at least to some extent, the issues associated with signal saturation, uneven transfer, nonspecific binding, etc. As shown in Table 1, corresponding analysis revealed that the major metabolic tissues displayed significant variability with respect to the total mass of PDK present in various tissues as well as the relative masses of individual PDK isozymes present in particular tissue. Of all tissues tested, the heart muscle showed the greatest amount of total kinase mass among the major metabolic tissues. It also displayed the greatest disparity in the levels of individual isozymes expressed in one tissue, with PDK2 greatly exceeding the mass of other isozymes (i.e. PDK1 and PDK4) (Table 1). In contrast, the two major kinase isozymes in liver (i.e. PDK1 and PDK2) were present in virtually identical amounts. As discussed above, kidney and brain were identified among just a few tissues expressing isozyme PDK3. Considering that PDK3 is known for its very unusual biochemical characteristics (28), we were interested to see how its tissue amount compares with the tissue amount of isozyme PDK2, which is a “general use” kinase fairly abundant in both tissues. Corresponding analysis showed that PDK3 was relatively abundant in kidney and brain (Table 1). In kidney, PDK3 mass was comparable with that of PDK2. In the brain, the mass of PDK2 exceeded that of PDK3, but, nevertheless, the tissue level of PDK3 in brain was still relatively high. Taken together, these data strongly suggest that, in both tissues, PDK3 should make a significant contribution to the total kinase activity as well as to its responses to various regulatory inputs.
Table 1.
Quantitative Western blot analysis of PDK isozyme expression in select mouse tissues
Protein levels of PDK isozymes in heart, liver, kidney, and brain were quantified as described under “Experimental procedures.” Data points represent means ± S.D. for six mice in each group.
| Tissue | PDK isozyme | PDK protein (ng per 1 mg of total tissue protein) |
|---|---|---|
| Heart | PDK1 | 75 ± 7 |
| PDK2 | 217 ± 11 | |
| PDK4 | 11 ± 2 | |
| Liver | PDK1 | 21 ± 1 |
| PDK2 | 21 ± 3 | |
| Kidney | PDK2 | 34 ± 1 |
| PDK3 | 39 ± 1 | |
| Brain | PDK2 | 44 ± 3 |
| PDK3 | 29 ± 1 |
Regulation of PDC in tissues of muscle origin
Generation of mouse strains carrying targeted mutations of each of the four isozymes of PDK, for the first time, opened up an opportunity to estimate the contributions made by the individual isozymes to the total kinase activity in mouse tissues. Unfortunately, to date, the measurements of kinase activity in crude tissue extract have been complicated by numerous competing activities, such as ATPase, phosphatase, nonspecific kinase, etc. Here, to measure the kinase activity, we employed an indirect method that follows the inactivation of PDC in a kinase reaction (32, 33). To eliminate the interference coming from competing activities, the measurements were conducted utilizing partially purified PDC as described under “Experimental procedures.” In most cases, this approach allowed us to circumvent the majority of known complications. There were two tissues, skeletal muscle and diaphragm, where this approach failed to produce reliable measurements due to the interference from ATPase and phosphatase activities (data not shown). Consequently, to obtain estimates for the kinase activity in skeletal muscle and diaphragm, we took advantage of findings made by the Randle laboratory that demonstrated that PDK can utilize ATPγS as a substrate (34). In general, using ATPγS allows one to avoid the interference coming from ATPases because the majority of ATPases cannot use “nonhydrolyzable” ATPγS as substrate, and, at the same time, it allows one to circumvent the interference coming from phosphatases because phosphatases cannot dephosphorylate thiophosphorylated PDC (34). To evaluate ATPγS as an alternative substrate for PDKs, we carried out model experiments in which inactivation rates of highly purified recombinant PDC by highly purified recombinant isozymes of PDK were estimated with ATP or ATPγS as substrate (Fig. S4, A–E). These experiments showed that all four PDK isozymes could utilize ATPγS. To verify these observations further, we examined the inactivation rates of partially purified PDC from heart muscle that expresses PDK1, PDK2, and PDK4 (Fig. 1). In agreement with the model experiments conducted with recombinant proteins, the native kinases were also capable of utilizing ATPγS (Fig. S4, F–H). It should be pointed out that in all experiments making use of ATPγS, there was an ∼2-fold decrease in the inactivation rates (Fig. S4, E and H). However, the magnitude of this decrease was uniform for all four PDK isozymes and, therefore, did not affect the relative contributions made by the individual isozymes to the total kinase activity. Importantly, employing ATPγS allowed us to reliably estimate the kinase activity in skeletal muscle and diaphragm (Fig. S5, C, D, G, and H) following our standard protocol that was employed to measure the kinase activity in all other tissues with ATP as substrate.
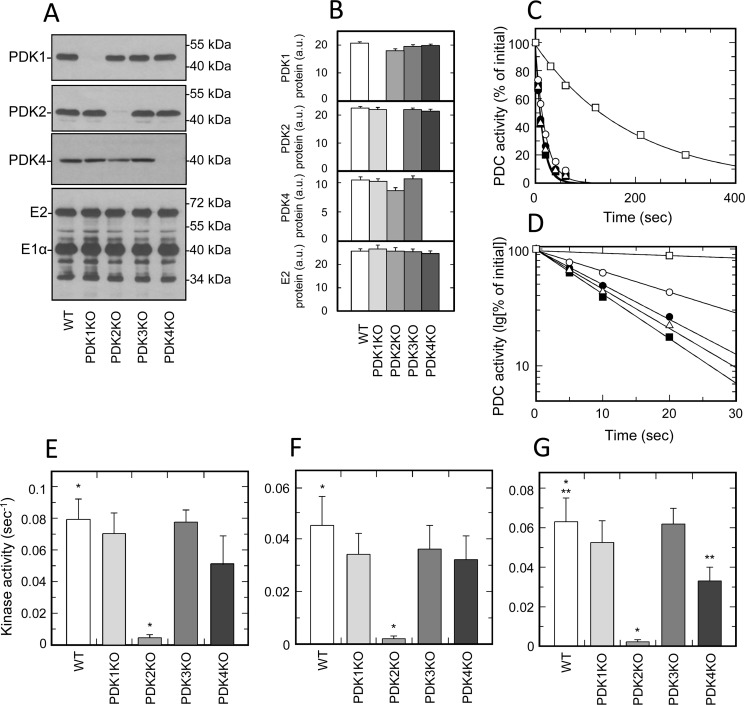
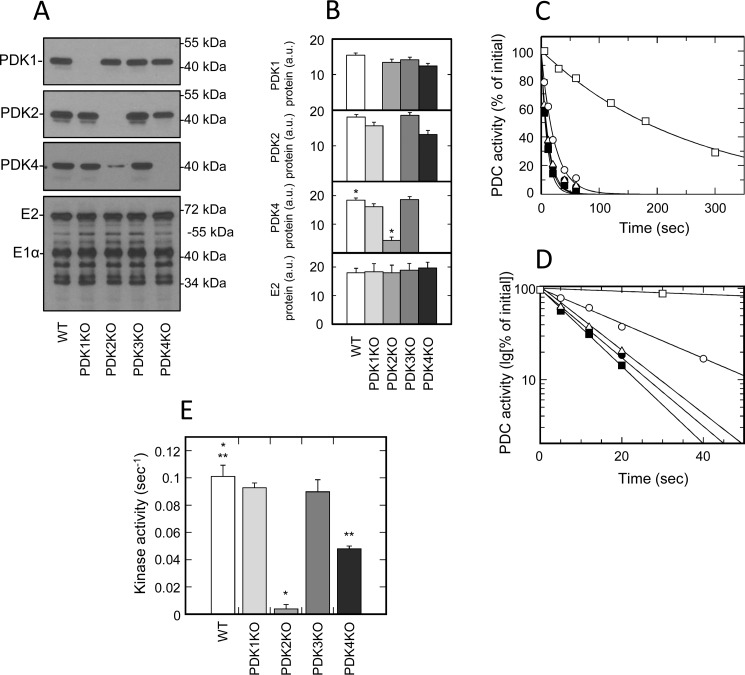
To investigate the contribution of individual isozymes of PDK to the total kinase activity in tissues of muscle origin, we analyzed the inactivation rates of PDC isolated from heart, skeletal muscle, and diaphragm (Fig. 2 and Fig. S5). Based on the results of Western blotting that was carried out on whole-tissue extracts, all muscles of different origin expressed three isozymes of PDK (i.e. PDK1, PDK2, and PDK4) (Fig. 1). To verify that the partially purified preparations of PDC used in activity measurements retained the same isozymic composition, all samples were tested for the presence of individual isozymes by Western blotting. This analysis revealed that, indeed, partially purified preparations of PDC retained an isozymic composition similar to that observed in whole-tissue extracts; PDK1, PDK2, and PDK4 were readily detectable in all samples tested (Fig. 2 (A and B) and Fig. S5 (A, B, E, and F)). Furthermore, none of the samples displayed protein band(s) that could be attributed to isozyme PDK3 (data not shown), which is consistent with the results obtained with whole-tissue extracts. Importantly, we did not observe any compensatory changes in the mass of any isozyme that could stem from ablation of another isozyme. This strongly suggests that the activity measurements conducted with samples prepared from tissues of muscle origin were not affected by reassortment in isozymic composition of PDK. Nevertheless, the results of activity measurements in samples prepared from muscles of different origin that were obtained from the WT and corresponding knockout mice yielded highly unexpected outcomes (Fig. 2 (C–G) and Fig. S5 (C, D, and G)). It was found that PDK1 knockout had little if any effect on the total kinase activity. These outcomes were virtually identical regardless of whether the samples were made from diaphragm or skeletal muscle, where PDK1 is a fairly minor isozyme, or from heart muscle, where PDK1 accounts for a significant proportion of the total kinase mass.
Figure 2.
Isozymic composition of PDK and regulation of activity of PDC isolated from tissues of muscle origin. A, representative Western blots illustrating protein levels of PDK1, PDK2, PDK4, and E2 and E1α proteins of PDC in heart muscle. B, relative abundance of PDK1, PDK2, PDK4, and E2 proteins in heart based on the results of scanning densitometry (data points represent means ± S.D. (error bars) for three mice in each group). White, light gray, gray, dark gray, and black bars, samples obtained from WT, PDK1 knockout, PDK2 knockout, PDK3 knockout, and PDK4 knockout mice, respectively. Isolation of PDC and Western blotting were carried out as described under “Experimental procedures.” Each lane contains 14 or 6 μg of partially purified PDC protein for kinase or PDC staining, respectively. C, representative ATP-dependent inactivation curves of partially purified PDC from heart muscle of WT (dark squares), PDK1 knockout (dark circles), PDK2 knockout (open squares), PDK3 knockout (open triangles), and PDK4 knockout mice (open circles), respectively. An ATP-dependent inactivation assay was carried out as described under “Experimental procedures.” Phosphorylation reactions for heart and diaphragm samples received a partially purified PDC–kinase complex at a final concentration of 1.2 mg/ml. Reactions for samples of skeletal muscles contained PDC–kinase complex at 3.0 mg/ml. D, representative ATP-dependent inactivation curves from C replotted in semilogarithmic coordinates. E, F, and G, kinase activity in preparations of partially purified PDC obtained from heart, skeletal muscle, and diaphragm, respectively. White, light gray, gray, dark gray, and black bars, kinase activity in PDC preparations from WT, PDK1 knockout, PDK2 knockout, PDK3 knockout, and PDK4 knockout mice, respectively. As described under “Experimental procedures,” kinase activities are expressed in terms of pseudo-first-order rate inactivation constants (s−1). Data points represent means ± S.D. for 6–10 mice in each group. p < 0.05 compared with WT group is considered statistically significant. Groups showing statistically significant differences are indicated by single (p ≤ 0.05) and double asterisks (p ≤ 0.01).
A number of studies originating from the Randle laboratory and later confirmed by others demonstrated that the overall inactivation rate of PDC in a kinase reaction correlates with the phosphorylation rate of site 1 located on the α chain of the E1 component (35, 36). Thus, the lack of the effect of PDK1 ablation on the rate of PDC inactivation strongly suggests that the native PDK1, at least in tissues of muscle origin, does not appreciably contribute to the overall inactivation rate of PDC in kinase reaction, presumably due to its inability to compete with other isozyme(s) of PDK for phosphorylation site 1. In agreement with this idea, we have found that the ablation of isozyme PDK2 had a major effect on the overall rate of inactivation of PDC in tissues of muscle origin. This effect was particularly pronounced in the diaphragm of PDK2-null mice, where the overall decrease in inactivation rate reached almost 25-fold, but it was also very strong in skeletal muscles and heart, where it reached 22- and 15-fold, respectively. Taken together, these results suggest that PDK2 is the major isozyme responsible for the inactivation of PDC in tissues of muscle origin. It is also possible that due to its robust activity, PDK2 outcompetes PDK1 for the phosphorylation of site 1, thereby masking PDK1 activity. However, the lack of essential residual activity in the samples prepared from PDK2 knockout mice appears to be in conflict with the latter conclusion and might suggest that PDK1 in tissues of muscle origin does not natively phosphorylate site 1, but rather plays a more specialized role, such as phosphorylation of site 3 (36). Alternatively, there is also a possibility that, natively, PDK1 requires an interaction with PDK2, and, consequently, the ablation of PDK2 down-regulates the PDK1 activity. These and other options are currently under investigation.
Based on Western blot analysis, PDK4 was a rather minor isozyme in the tissues of muscle origin prepared from well-fed mice (Fig. 1 and Table 1). Thus, after studies of PDK2 knockout mice, it appeared to be unlikely that PDK4 could compete with PDK2 for phosphorylation of site 1. Indeed, measurements carried out on samples prepared from heart, skeletal muscle, and diaphragm obtained from PDK4 knockout mice were in general agreement with this idea (Fig. 2, E–G). However, it should be pointed out that all preparations made from tissues of PDK4 knockout mice displayed a somewhat reduced kinase activity. In the samples from heart and skeletal muscle, this decrease in activity never reached sufficient statistical significance. However, in samples prepared from diaphragm, there was a statistically significant almost 2-fold decrease in the overall rate of inactivation of PDC in the kinase reaction. These observations might indicate that PDK4, even in a well-fed state, can contribute to the inactivation rate of PDC, even though its absolute amount is fairly low. The lack of sufficient residual activity in samples prepared from PDK2 knockout mice appears to be in conflict with the latter idea and might suggest that these relationships are more complex and reflect the interactions between different isozymes in the context of a multienzyme complex.
Regulation of PDC in liver
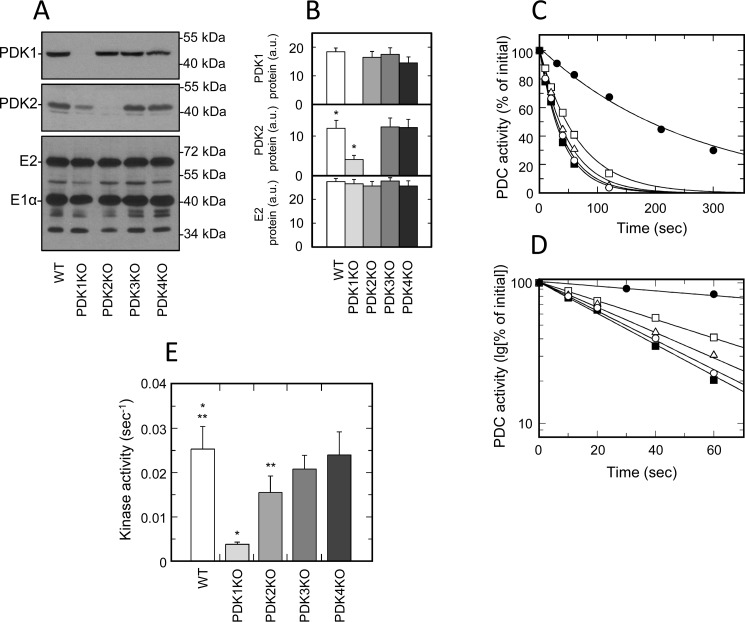
Western blotting carried out on whole-tissue extracts revealed that liver, unlike muscles, expressed only two isozymes of PDK (i.e. PDK1 and PDK2) (Fig. 1). Based on quantitative analysis, both isozymes were present in approximately equal amounts (Table 1). This significantly differed from the expression patterns observed in muscles, where the overall protein mass of PDK2 greatly exceeded the protein masses of other isozymes. Importantly, in liver samples, we were unable to identify either PDK3 or PDK4 proteins. This indicates that these isozymes do not contribute to the regulation of hepatic PDC in a well-fed state. To verify that preparations of hepatic PDC isolated for activity assays retained the isozymic composition of PDK, the corresponding samples were analyzed by Western blotting. As shown in Fig. 3 (A and B), this analysis revealed that the protein levels of PDK1 were virtually identical in samples obtained from WT, PDK2, PDK3, and PDK4 knockout mice. In marked contrast, the protein mass of PDK2 was greatly reduced in PDK1 knockout mice. On average, the protein levels of PDK2 were decreased more than 3-fold, suggesting that, in liver, ablation of PDK1 had a destabilizing effect on PDK2. This response was markedly different than the behavior of PDK isozymes in tissues of muscle origin, where ablation of one individual isozyme did not affect the protein levels of the remaining isozymes. This might suggest that in liver, unlike in muscles, there are much closer interactions between PDK1 and PDK2. Last, in agreement with the results obtained with whole-tissue extracts, analysis of partially purified samples in Western blotting failed to uncover isozymes PDK3 and PDK4 (data not shown). Therefore, it is not surprising that the samples obtained from PDK3 and PDK4 knockout mice did not display any changes with respect to the protein mass of either PDK1 or PDK2.
Figure 3.
Isozymic composition of PDK and regulation of activity of PDC isolated from liver. A, representative Western blots illustrating protein levels of PDK1, PDK2, and E2 and E1α proteins of PDC in liver. B, relative abundance of PDK1, PDK2, and E2 proteins in liver based on the results of scanning densitometry (data points represent means ± S.D. (error bars) for three mice in each group). White, light gray, gray, dark gray, and black bars, samples obtained from WT, PDK1 knockout, PDK2 knockout, PDK3 knockout, and PDK4 knockout mice, respectively. Each lane contains 15 or 6 μg of partially purified PDC protein for kinase or PDC staining, respectively. C, representative ATP-dependent inactivation curves of partially purified PDC from liver of WT (dark squares), PDK1 knockout (dark circles), PDK2 knockout (open squares), PDK3 knockout (open triangles), and PDK4 knockout mice (open circles), respectively. Phosphorylation reactions for liver samples received partially purified PDC–kinase complex at a final concentration of 3.0 mg/ml. D, representative ATP-dependent inactivation curves from C replotted in semilogarithmic coordinates. E, kinase activity in preparation of partially purified PDC obtained from liver. White, light gray, gray, dark gray, and black bars, kinase activity in PDC preparations from WT, PDK1 knockout, PDK2 knockout, PDK3 knockout, and PDK4 knockout mice, respectively. Kinase activities are expressed in terms of pseudo-first-order rate inactivation constants (s−1). Data points represent means ± S.D. for 6–10 mice in each group. p < 0.05 compared with WT group is considered statistically significant. Groups showing statistically significant differences are indicated by single (p ≤ 0.05) and double asterisks (p ≤ 0.01).
Importantly, the marked differences between liver and various muscles in isozymic composition and interactions between the individual isozymes uncovered by Western blotting also translated into a different kinetic behavior assessed following the rates of PDC inactivation in a kinase reaction. Liver samples obtained from mice carrying PDK1 knockout showed a great decrease in the overall inactivation rate of PDC when compared with the corresponding samples isolated from livers of WT littermates (Fig. 3, C–E). On average, the decrease in kinase activity associated with PDK1 ablation reached more than 6-fold. Undoubtedly, at least in part, the decrease in PDC inactivation rate caused by PDK1 ablation could be explained by the down-regulation of PDK2 protein associated with PDK1 knockout (Fig. 3, A and B). However, the sheer magnitude of the effect clearly demonstrates that, in the liver, PDK1 can contribute to the raw inactivation rate of PDC in a kinase reaction that comes about as a result of phosphorylation of site 1. In fact, it appears that PDK1 makes a major contribution to the total inactivation rate of hepatic PDC. This notion is supported by the results of kinetic analysis carried out on samples isolated from livers of PDK2 knockout mice. As shown in Fig. 3 (C–E), samples completely lacking PDK2 retained more than 60% of the total kinase activity. This is in marked contrast to outcomes obtained with samples isolated from tissues of muscle origin, where PDK1 largely did not contribute to the inactivation rate of PDC under similar circumstances. Taken together, these observations suggest that PDK1 is the major isozyme responsible for the inactivation of hepatic PDC in kinase reaction. They might also indicate the existence of multienzyme-specific interactions that are unique for the complexes of different origin.
Regulation of PDC in kidney and brain
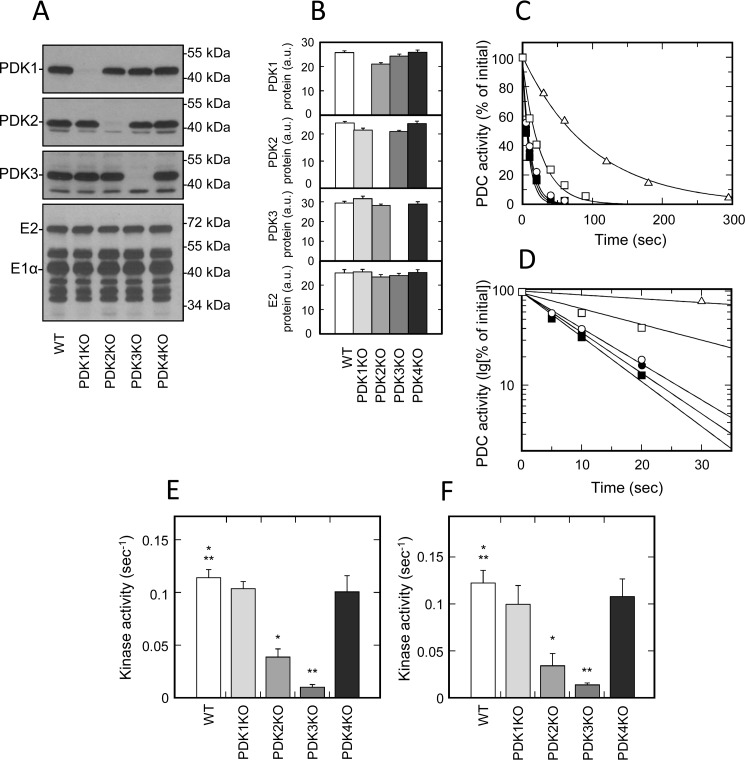
Kidney and brain are two of several tissues that natively express fairly large levels of PDK3 protein (Fig. 1 (E and F) and Fig. S1 (E and F)). Based on the quantitative Western blot analysis, PDK3 protein accounted for almost half of the total kinase mass in kidney (Table 1 and Fig. S3 (G and H)). In mouse brain, the relative protein mass of PDK3 was somewhat lower, but, nevertheless, it still accounted for about 40% of the total kinase mass (Table 1 and Fig. S3 (K and L)). Both tissues expressed PDK1 as well; however, its levels were comparatively low (Figs. 1 (A and B) and 4 (A and B) and Fig. S6 (A and B)). Based on the sheer amount of PDK3 protein expressed in kidney and brain, it would be expected that PDK3 deficiency had a significant impact on the rate of ATP-dependent inactivation of PDC in these tissues. Indeed, in both tissues obtained from PDK3-deficient mice, we observed a great (i.e. more that 10-fold) decrease in the rate of total kinase activity (Fig. 4 (C–F) and Fig. S6 (C and D)). The latter outcome is not particularly surprising, considering that the studies carried out with recombinant kinases established that PDK3 is the most active isozyme of kinase (28, 37). There was just one report showing that PDK3 has a fairly low activity (38), but it is likely that this study was carried out with a catalytically compromised enzyme.
Figure 4.
Isozymic composition of PDK and regulation of activity of PDC isolated from kidney and brain. A, representative Western blots illustrating protein levels of PDK1, PDK2, PDK3, and E2 and E1α proteins of PDC in kidney. B, relative abundance of PDK1, PDK2, PDK3, and E2 proteins in kidney based on the results of scanning densitometry (data points represent means ± S.D. (error bars) for three mice in each group). White, light gray, gray, dark gray, and black bars, samples obtained from WT, PDK1 knockout, PDK2 knockout, PDK3 knockout, and PDK4 knockout mice, respectively. Each lane contains 15 or 6 μg of partially purified PDC protein for kinase or PDC staining, respectively. C, representative ATP-dependent inactivation curves of partially purified PDC from kidney of WT (dark squares), PDK1 knockout (dark circles), PDK2 knockout (open squares), PDK3 knockout (open triangles), and PDK4 knockout mice (open circles), respectively. Phosphorylation reactions for kidney and brain samples received partially purified PDC–kinase complex at a final concentration of 3.0 mg/ml. D, representative ATP-dependent inactivation curves from C replotted in semilogarithmic coordinates. E and F, kinase activity in preparation of partially purified PDC obtained from kidney and brain, respectively. White, light gray, gray, dark gray, and black bars, kinase activity (s−1) in PDC preparations from WT, PDK1 knockout, PDK2 knockout, PDK3 knockout, and PDK4 knockout mice, respectively. Data points represent means ± S.D. for 6–10 mice in each group. p < 0.05 compared with WT group is considered statistically significant. Groups showing statistically significant differences are indicated by single (p ≤ 0.05) and double asterisks (p ≤ 0.01).
In marked contrast to PDK3, the effects of PDK2 deficiency on the total kinase activity in kidney and brain were much more modest (Fig. 4 (C–F) and Fig. S6 (C and D)). On average, PDK2 ablation caused an ∼3.5-fold decrease in kinase activity in brain and an ∼3-fold decrease in kidney. Ablation of either protein did not affect the levels of expression of the remaining isozymes (Fig. 4 (A and B) and Fig. S6 (A and B)), thus suggesting that neither PDK3 nor PDK2 exerts a stabilizing effect on each other. The latter finding is in contrast to the relationships between isozymes PDK1 and PDK2 in liver, where ablation of PDK1 greatly affected the protein levels of PDK2 (Fig. 3, A and B). Taken together, these data suggest that PDK3 is the major kinase isozyme responsible for inactivation of PDC in kidney and brain. This conclusion is in agreement with the earlier studies (39) documenting significant differences in the properties of kinase from kidney versus kinase from heart, where PDK2 is the major isoform (Fig. 2, A–E). The reported differences can be readily explained based on the differences in enzymatic properties of PDK2 and PDK3 that were established during the studies of recombinant kinases (28, 37). In addition to kidney and brain, we observed significant levels of expression of PDK3 protein in testis and lung (Fig. 1 (E and F) and Fig. S1 (E and F)). Thus, it appears that there is a high likelihood that, similarly to brain and kidney, PDK3 is the major isozyme of PDK in those tissues as well.
Regulation of PDC in brown adipose tissue
As discussed above, in a well-fed state, the majority of mouse tissues do not express PDK4 protein in appreciable amounts. By Western blot analysis, we were able to detect PDK4 only in tissues of muscle origin, such as heart, diaphragm, or skeletal muscle (Fig. 1 and Fig. S1). However, even in those tissues, the protein mass of PDK4 under normal feeding conditions was rather low. In this respect, brown adipose was the only tissue that expressed greatly elevated levels of PDK4 (i.e. ∼4 times greater than in heart) (Fig. S1). This suggested that PDK4 might have a significant role in the regulation of PDC in BAT in a well-fed state and, thereby, might have a significant effect on the overall energy metabolism in mice. Here, we examined some of these ideas by analyzing the isozyme composition of PDK in BAT and the effects caused by the ablation of individual isozymes on the total kinase activity (Fig. 5). Based on Western blot analysis, there were three isozymes of PDK expressed in BAT (i.e. PDK1, PDK2, and PDK4) (Fig. 5, A and B). In this respect, the expression pattern of PDK isozymes in BAT was, generally, similar to that observed in tissues of muscle origin. However, in BAT, PDK2 and PDK4 proteins were more abundant, whereas the levels of PDK1 protein tended to be somewhat lower. Importantly, in contrast to tissues of muscle origin, we observed a significant decrease in the tissue levels of PDK4 protein in mice carrying a targeted knockout of PDK2 gene (Fig. 5, A and B). Considering that PDK4 mRNA levels were not affected by the ablation of PDK2 (data not shown), this suggests that, in BAT, PDK2 might have a stabilizing effect on PDK4 protein. This is reminiscent of the relationship between isozymes PDK1 and PDK2 in liver, where ablation of PDK1 had a destabilizing effect on the protein levels of PDK2 protein (Fig. 3, A and B).
Figure 5.
Isozymic composition of PDK and regulation of activity of PDC isolated from BAT. A, representative Western blots illustrating protein levels of PDK1, PDK2, PDK4, and E2 and E1α proteins of PDC in BAT. B, relative abundance of PDK1, PDK2, PDK4, and E2 proteins in BAT based on the results of scanning densitometry (data points represent means ± S.D. (error bars) for three mice in each group). White, light gray, gray, dark gray, and black bars represent samples obtained from WT, PDK1 knockout, PDK2 knockout, PDK3 knockout, and PDK4 knockout mice, respectively. Each lane contains 12 or 3 μg of partially purified PDC protein for kinase or PDC staining, respectively. C, representative ATP-dependent inactivation curves of partially purified PDC from BAT of WT (dark squares), PDK1 knockout (dark circles), PDK2 knockout (open squares), PDK3 knockout (open triangles), and PDK4 knockout mice (open circles), respectively. Phosphorylation reactions for BAT samples received partially purified PDC–kinase complex at a final concentration of 0.6 mg/ml. D, representative ATP-dependent inactivation curves from C replotted in semilogarithmic coordinates. E, kinase activity in preparation of partially purified PDC obtained from BAT. White, light gray, gray, dark gray, and black bars, kinase activity in PDC preparations from WT, PDK1 knockout, PDK2 knockout, PDK3 knockout, and PDK4 knockout mice, respectively. Kinase activities are expressed in terms of pseudo-first-order rate inactivation constants (s−1). Data points represent means ± S.D. for 6–10 mice in each group. p < 0.05 compared with WT group is considered statistically significant. Groups showing statistically significant differences are indicated by single (p ≤ 0.05) and double asterisks (p ≤ 0.01).
Analysis of the total kinase activity in BAT revealed that ablation of PDK2 had the greatest impact on the overall rate of ATP-dependent inactivation of PDC (Fig. 5, C and D). In samples from PDK2-deficient mice, the total kinase activity decreased more than 25-fold. This incredibly strong effect was at least in part due to the 4-fold decrease in the mass of PDK4 protein caused by PDK2 knockout (Fig. 5, A and B). However, it should be emphasized that the majority of this decrease in the total kinase activity undoubtedly reflects the contribution of PDK2 itself. This conclusion is supported by the results showing that the total ablation of PDK4 protein in BAT caused only a 2-fold decrease in the rate of ATP-dependent inactivation of PDC (Fig. 5, C–E). Similarly to the tissues of muscle origin, the ablation of PDK1 had little if any effect on the total kinase activity in BAT, thus suggesting that, in BAT, as in the majority of mouse tissues, excluding liver, PDK1 does not contribute to the raw inactivation rate that reflects the phosphorylation of site 1. In BAT, the overall rate of ATP-dependent inactivation of PDC clearly reflected the activities of isozymes PDK2 and PDK4, with PDK2 making a significantly greater contribution than PDK4. Nevertheless, the contribution made by PDK4 to the inactivation of PDC in BAT appears to be more prominent than the one observed in tissues of muscle origin, such as heart or skeletal muscle. Thus, it is conceivable that certain phenotypic traits associated with PDK4 knockout (31, 40) might, at least in part, stem from the changes in BAT metabolism driven by PDK4 deficiency in addition to the effects caused by PDK4 deficiency in various muscles.
Discussion
During the past 2 decades, the wealth of information on the fine mechanisms responsible for regulation of PDC activity came from the studies of recombinant enzymes (18). The native enzymes in their natural environment did not receive sufficient attention due largely to the very complex structure of this multienzyme assembly. Consequently, a lot of conclusions about the native PDC and its regulation have been drawn based on the known properties of particular recombinant enzymes along with indirect estimates of their abundance in specific model systems under investigation. Creation of mouse strains carrying targeted mutations of all four genes coding for isozymes of PDK gave us a unique opportunity to evaluate the functionality of native isozymes in major metabolic tissues. Even the initial characterization of these strains yielded somewhat unexpected results about the native PDK proteins. Historically, only PDK2 was considered to be a ubiquitous enzyme responsible for the minute-to-minute regulation of PDC, whereas other isozymes were thought to play more specialized roles (19, 28). For example, PDK1 was considered to be a largely heart-specific isozyme (28). However, it turned out that PDK1 protein is readily detectable in all major metabolic tissues. Thus, although the protein levels of PDK1 in heart are significantly higher than in other tissues, its ubiquitous presence cannot be disregarded as far as the regulation of PDC is concerned. Furthermore, historically, there was little information available with respect to the tissue distribution and biological role of isozyme PDK3 (28). Analysis of the corresponding mouse strains carried out in this study revealed that, unlike PDK1 and PDK2, PDK3 displays a rather limited tissue distribution. The large amounts of PDK3 protein were found in kidney, brain, testis, and lung. This fairly narrow expression pattern strongly suggests that PDK3 plays a specialized role in the regulation of general metabolism. At least two of those tissues (i.e. brain and testis) are believed to display an unusual metabolic pattern called “lactate shuttle,” in which some cells, such as astrocytes or Sertoli cells, break down glucose to pyruvate and then secrete it into the surrounding medium in the form of lactate, where it is consumed by other cells, such as neurons or spermatocytes, respectively (41, 42). Biochemical studies of PDK3 provided strong evidence that this isozyme is unique among other PDKs because it does not show any appreciable sensitivity to the inhibitory effects of physiological concentrations of pyruvate (18). This unusual property allows PDK3 to prevent the reactivation of PDC even when there is a buildup of cellular pyruvate; such a unique regulatory property makes PDK3 perfectly suited for the control of PDC activity in lactate producing cells, such as astrocytes or Sertoli cells (41, 42). Kidney, on the other hand, is the major site for gluconeogenesis (43). Renal gluconeogenesis operates even in a well-fed state when kidney breaks down glutamine to control blood pH and recovers its carbon skeleton as glucose through gluconeogenesis (43). For renal gluconeogenesis to operate efficiently, the activity of PDC must be down-regulated. Once again, PDK3 appears to be perfectly suited for this function because its activity is not sensitive to the inhibitory effects of pyruvate and, therefore, it can operate even in nutrition-rich conditions characteristic of the well-fed state. Taken together, these considerations suggest that PDK3 may indeed have a specialized function operating in cell types that display a metabolic pattern(s) that requires down-regulation of PDC activity even in the presence of high concentrations of cellular pyruvate. Last, PDK4 is believed to be an inducible isozyme that becomes overexpressed under conditions such as fasting when there is a need to down-regulate the aerobic oxidation glucose in peripheral tissues to spare pyruvate for gluconeogenesis (19). Consequently, its role in the well-fed state has not been fully assessed. In this study, we found that PDK4 protein is expressed in a well-fed state but displays a fairly narrow expression pattern largely limited to the tissues of muscle origin and BAT. In muscles, PDK4 protein is not particularly abundant. In BAT, on the other hand, it is one of the major isozymes, suggesting that PDK4 might contribute to thermogenesis. One of the intriguing possibilities here would be a PDK-mediated regulation of glyceroneogenesis and lipogenesis in BAT.
Creation of new mouse models of PDK deficiency also, for the first time, allowed us to evaluate the contributions made by individual isozymes to the overall rate of PDC inactivation in kinase reaction. These studies yielded several unexpected results. For example, it turned out that in tissues expressing several isozymes at once, there is no additivity in contributions made by the individual isozymes to the overall inactivation rate of PDC. The lack of additivity strongly suggests the existence of interconnectivity between individual isozymes operating in the context of a multienzyme complex. This interconnectivity might stem, for example, from competition for lipoyl domains, competition for common protein substrate (i.e. E1), direct physical interactions between individual isozymes, or even from cross-phosphorylation of one isozyme by another. In addition, there might be some other, yet to be identified, secondary modifications that selectively affect the activity of a particular isozyme. The latter possibility is supported by studies published by the Randle laboratory (44). Future work will undoubtedly uncover the underlying molecular mechanisms. However, even now it becomes clear that conclusions about the contribution of individual isozymes to the regulation of PDC activity cannot be simply drawn based on the outcomes of Western blot analysis, but should be verified by activity measurements. Another unexpected outcome of this study is the finding that the ubiquitously expressed isozyme PDK1 makes little if any contribution to the overall inactivation rate of PDC in the majority of tissues. This brings about an intriguing possibility that, unlike other isozymes, PDK1 plays a specialized role that regulates PDC activity indirectly. PDK1 is unique among PDK isozymes because of its ability to phosphorylate site 3 (36). A number of studies have shown that phosphorylation of site 3 has a regulatory role because it slows down the reactivation of PDC in the phosphatase reaction (45, 46). Thus, it is feasible that in the majority of tissues, PDK1 regulates the phosphorylation state of PDC through an indirect mechanism (i.e. by down-regulating the reactivation rate of PDC catalyzed by PDPs). Probably the most important outcome of this study is the finding confirming the existence of a tissue-specific regulation of PDC activity by the isozymes of PDK (28). It appears that in every major metabolic tissue, there is one particular isozyme that is largely responsible for the inactivation of PDC in the phosphorylation reaction. In tissues of muscle origin (i.e. heart, skeletal muscle, diaphragm, and BAT), PDK2 activity accounts for the bulk of the phosphorylation rate. In liver, inactivation of PDC in the kinase reaction mainly reflects the activity of PDK1. In kidney and brain, PDK3 appears to be the primary kinase responsible for the inactivation of PDC. In this context, future studies aimed at better understanding of how these native kinases operate are necessary to decipher the molecular mechanisms responsible for the fine-tuning of PDC activity in various tissues.
Experimental procedures
Animals
Agouti C57BL/6N embryonic stem cells carrying the Knockout First allele (47) inserted between exons 1 and 2 of the Pdk1 gene were obtained from the Helmholtz Zentrum Muenchen (Munich, Germany). Embryonic stem cell microinjection into blastocyst embryos was carried out by the Analytical Genomics and Transgenics Core at the University of Alabama at Birmingham. A mouse strain carrying a knockout-targeting mutation of Pdk3 in which exons 7, 8, and 9 were replaced with the LacZ-Neo cassette was obtained from Texas Institute for Genomic Medicine (Houston, TX). Genetic background of both strains was stabilized by backcrossing of heterozygous mice with C57BL/6J WT mice for 10 generations. Both of the newly created strains were fertile and did not demonstrate anatomical abnormalities or metabolic derangements. In our tests, male and female mice displayed similar traits. Further details on these strains will be reported elsewhere.4,5 Previously described PDK2 and PDK4 C57BL/6J knockout mice (31, 48) were obtained as generous gift from Dr. Robert A. Harris (Department of Biochemistry, Indiana University Medical Center).
All experimental protocols were approved by the institutional animal care and use committee of the University of Alabama at Birmingham. Groups of at least 10 WT male mice and 10 PDK knockout male mice (PDK1, PDK2, PDK3, or PDK4 knockout, respectively) were housed under standard conditions of a 12-h dark/12-h light cycle, temperature of 24 ± 2 °C, and relative humidity of 50 ± 10%, with free access to the standard rodent chow (catalogue no. 7071, Harlan) and water. For experiments, animals were sampled in a well-fed state (between 7:00 and 8:00 a.m.). Major metabolic tissues were rapidly removed, freeze-clamped with Wollenberger tongs at the temperature of liquid nitrogen, and stored for analysis at −85 °C.
Western blot analysis
To prepare whole-tissue extracts for Western blot analysis, frozen mouse tissues were pulverized with a mortar and pestle under liquid nitrogen. The resulting tissue samples (20 mg) were homogenized in 4 volumes of buffer containing 50 mm Tris-HCl, pH 6.5, 2% (w/v) SDS, 0.1 m DTT, 1 mm benzamidine, 0.1 mg/ml trypsin inhibitor, 1 μg/ml aprotinin, 0.1 mm N-α-p-tosyl-l-lysine chloromethyl ketone, 1 μm leupeptin, and 1 μm pepstatin A. Homogenates were boiled for 5 min, cooled on ice, and centrifuged at 9,000 × g for 10 min at room temperature. Pellets were extracted the second time using a similar procedure, and the combined supernatants obtained after centrifugation were used for Western blotting.
The overall procedure for Western blot analysis was described previously (30). Briefly, proteins from tissue extracts (17–35 μg of total protein/lane) were separated by SDS-PAGE and transferred to nitrocellulose membrane. Blots were probed with following primary antibodies: rabbit PDC antibodies at a 1:2,000 dilution (30); PDK1 antibodies at a 1:5,000 dilution (catalogue number ADI-KAP-PK112-F, Enzo Life Sciences, Plymouth Meeting, PA); the previously described rabbit PDK2 antibodies (30) diluted 1:3,000; rabbit antipeptide antibodies generated against the C-terminal 19 amino acids of PDK3 (RDASKYKAKQDKIKSNRTF) diluted 1:1,000; and rabbit antipeptide antibodies raised against PDK4 polypeptide (HQENRPSLTPVEAT) diluted 1:3,000. PDK3 and PDK4 antibodies were described previously (31, 40). They were obtained as a generous gift from Dr. Robert A. Harris (Department of Biochemistry, Indiana University Medical Center). Immunoreactive bands were visualized using the Amersham Biosciences ECL immunodetection procedure (GE Healthcare), following the manufacturer's recommendations. Secondary goat anti-rabbit antibodies conjugated with horseradish peroxidase were obtained from Bio-Rad. They were used at a working dilution of 1:10,000. Relative intensities of bands visualized by Western blot analysis were quantified with UN-SCAN-IT Software, version 4.1 (Silk Scientific, Orem, UT). The overall procedure for the analysis of samples containing partially purified PDC was similar, except that the amount of total protein separated by SDS-PAGE was decreased to 6–15 μg of total protein/lane.
In addition to the antibodies listed above, we also tested several antibodies available commercially, such as PDHK1 (C47H1) rabbit mAb (Cell Signaling Technology, Danvers, MA), PDK2 antibody (C-terminal) (catalogue no. AP7039b, ABGENT, San Diego, CA), PDK3 antibody (N-terminal), catalogue no. AP7040a, ABGENT), rabbit polyclonal to PDK4-N-terminal end (catalogue no. 576085, Abcam Inc., Cambridge, MA), and PDK4 (C-16) (catalogue no. sc-14495, Santa Cruz Biotechnology, Inc., Dallas, TX). Unfortunately, with these antibodies, we were unable to obtain reliable results due to low signal strength, multiple nonspecific bands, or both.
The overall Western blotting procedure that was used to quantify the protein levels of PDK isozymes in several mouse tissues was similar to the one described above. For the purpose of quantification, each gel was loaded with tissue samples prepared from WT mice, and, in addition, each gel received 6–7 lanes loaded with different amounts of appropriate recombinant isozyme. To prevent losses of the kinase standards due to nonspecific adsorption during electrophoresis and transfer, as well as to compensate for the nonspecific binding of antibodies, the kinase standards were supplemented with whole-tissue extracts prepared from the corresponding knockout mice. The amount of protein in extracts used for preparation of kinase standards was identical to that of WT extracts used for quantification purposes. In a series of preliminary experiments, the loading conditions were established such that, first, they yielded a linear relationship between the protein amounts of kinase standards loaded and the strengths of signals generated in Western blotting, and, second, they generated a signal from the WT kinase that was well within the linear region of the calibration curve. The blots were quantified using UN-SCAN-IT software. The resulting data were used to construct calibration curves and to calculate the tissue content of each PDK isozyme. Calibration data were fitted to a linear equation (y = a + bx). Fitting was carried out using GraFit software version 5.0 (Erithacus Software Ltd., East Grinstead, UK).
Preparation of partially purified PDC
50–100 mg of powder obtained from frozen tissue pulverized with a mortar and pestle under liquid nitrogen was suspended in 3 volumes of ice-cold Buffer A (30 mm Hepes, 1 mm EDTA, 150 mm KCl, 0.5% (w/v) Triton X-100, 2% (v/v) dialyzed bovine serum, 0.1 mm TPP, 3 mm fresh added DTT, with protease inhibitors 0.1 mm phenylmethylsulfonyl fluoride, 0.01 mm 1-chloro-3-tosylamido-7-amino-2-heptanone, 10 μg/ml trypsin inhibitor, 1 μm leupeptin, pH 7.5 at 4 °C) and homogenized on ice using Potter homogenizer. The homogenate was centrifuged at 9,000 × g for 10 min at 4 °C. The supernatant was transferred into fresh tubes and stored on ice. The pellet was rehomogenized in 2 volumes of Buffer A and centrifuged again under the same conditions. The resulting supernatant was combined with the supernatant obtained after the first centrifugation. The combined extract was precipitated with 1% (w/v) PEG 8000 by incubating at 4 °C for 15 min. The precipitate was removed by centrifugation at 9,000 × g for 10 min at 4 °C. To precipitate PDC, the supernatant was made 4.5% (w/v) with PEG 8000 and incubated for 15 min at 4 °C. The precipitate was recovered by centrifugation as described above. PDC-containing precipitate was resuspended in 2–4 volumes of ice-cold Buffer B (30 mm Hepes, 0.1 mm EDTA, 100 mm KCl, 10 mm MgSO4, 1 mm CaCl2, 0.1% (w/v) Triton X-100, 2% (v/v) dialyzed bovine serum, 5 mm freshly made DTT with protease inhibitors 10 μg/ml trypsin inhibitor, 1 μm leupeptin, pH 7.5, at 4 °C). To reactivate PDC, the extract was incubated at 30 °C for 30 min. After the incubation, the extract was placed on ice for 5–10 min, and then PDC was precipitated with 8% (w/v) PEG 8000 for 15 min at 4 °C. The precipitated protein was collected by centrifugation at 9,000 × g for 10 min at 4 °C. PDC-containing pellet was resuspended in 7–10 volumes of ice-cold Buffer C (20 mm Tris-HCl, pH 7.5, 50 mm KCl, 0.5 mm EDTA, and 5 mm freshly made DTT) and used for the measurement of PDK activity.
Measurement of PDK activity
PDK activity was measured following the ATP-dependent inactivation of PDC as described previously (32). Briefly, phosphorylation reactions were set up in a final volume of 100 μl containing 20 mm Tris-HCl, pH 7.4, 50 mm KCl, 5 mm MgCl2, 5 mm freshly made DTT, 25 mm KF, and 0.6–3 mg/ml of partially purified PDC–kinase complex. After equilibration at 37 °C for 2 min, inactivation reactions were initiated by the addition of ATP or ATPγS to a final concentration of 0.4 mm. At the times indicated in Figs. 2, 3, 4, and 5, 10-μl aliquots of phosphorylation mixture were withdrawn and quenched in 1 ml of PDC reaction mixture. The residual PDC activity was measured spectrophotometrically (33). The resulting PDC activity inactivation data were fitted to a single-exponential decay equation (y = A0e−kt). Fitting of data were carried out with GraFit software version 5.0 (Erithacus Software Ltd.). Kinase activity was expressed in terms of pseudo-first-order rate constants of inactivation reaction (s−1).
Measurement of PDC activity
PDC activity was measured spectrophotometrically following the accumulation of NADH at 340 nm (33). Reactions were set up at 37 °C in a total volume of 1 ml containing 100 mm Tris-HCl, pH 7.8, 0.5 mm EDTA, 1 mm MgCl2, 2 mm DTT, 1 mm NAD+, 0.4 mm CoA, 0.4 mm TPP, 20 μg/ml E3, and 10 μl of PDC inactivation mixture. To prevent the interfering effects of lactate dehydrogenase activity, each reaction received a lactate dehydrogenase inhibitor oxamate added to a final concentration of 25 mm. Reactions were initiated with pyruvate added at 0.8 mm. Accumulation of NADH was monitored using a Cary 100 Bio UV-visible spectrophotometer (Varian Inc., Palo Alto, CA). The rates of the pyruvate dehydrogenase reaction were determined using software provided by the instrument's manufacturer.
Other methods
SDS-PAGE was carried out according to Laemmli (49). Protein concentrations were determined according to Peterson (50) with BSA as a standard. The general procedure for isolation of recombinant, untagged isozymes PDK1, PDK2, and PDK3 using affinity chromatography on lipoyl-bearing domains will be described elsewhere.6 A His6-tagged version of isozyme PDK4 was isolated following the procedure described by Bowker-Kinley et al. (28). A highly purified, recombinant PDC was generated as described (51).
Statistical analysis
All values are presented as means ± S.D. for at least six independent samples. The statistical significance of differences between groups was determined using Student's t test. A p value of ≤0.05 was considered statistically significant.
Author contributions
A. K., A. T., and K. M. P. investigation; A. K., A. T., and K. M. P. methodology; N. Y. K. and K. M. P. conceptualization; N. Y. K. and K. M. P. formal analysis; N. Y. K. and K. M. P. writing-review and editing; K. M. P. resources; K. M. P. data curation; K. M. P. software; K. M. P. supervision; K. M. P. funding acquisition; K. M. P. validation; K. M. P. visualization; K. M. P. writing-original draft; K. M. P. project administration.
Supplementary Material
Acknowledgment
We are grateful to Dr. Robert A. Harris (Department of Biochemistry, Indiana University Medical Center) for PDK2 and PDK4 C57BL/6J knockout mice as well as PDK3- and PDK4-specific antipeptide antibodies.
This work was supported by NIGMS, National Institutes of Health, Grant GM51262. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S6.
A. Klyuyeva, A. Tuganova, N. Y. Kedishvili, and K. M. Popov. Biochemistry and physiology of pyruvate dehydrogenase kinase 3, manuscript in preparation.
A. Klyuyeva, A. Tuganova, N. Y. Kedishvili, and K. M. Popov. Metabolic role of pyruvate dehydrogenase kinase 1, manuscript in preparation.
A. Klyuyeva and K. M. Popov. Isoforms of human pyruvate dehydrogenase kinase, manuscript in preparation.
- PDC
- pyruvate dehydrogenase complex
- PDK
- pyruvate dehydrogenase kinase
- PDK1
- PDK2, PDK3, and PDK4, isozymes 1, 2, 3, and 4 of pyruvate dehydrogenase kinase
- E1
- pyruvate dehydrogenase component of PDC
- E2
- dihydrolipoyl transacetylase component of PDC
- E3
- dihydrolipoyl dehydrogenase component of PDC
- BAT
- brown adipose tissue
- PDP
- pyruvate dehydrogenase phosphatase
- ATPγS
- adenosine 5′-O-(thiotriphosphate).
References
- 1. Pithukpakorn M. (2005) Disorders of pyruvate metabolism and the tricarboxylic acid cycle. Mol. Genet. Metab. 85, 243–246 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 2. Ohlrogge J., Pollard M., Bao X., Focke M., Girke T., Ruuska S., Mekhedov S., and Benning C. (2000) Fatty acid synthesis: from CO2 to functional genomics. Biochem. Soc. Trans. 28, 567–573 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 3. Wahren J., and Ekberg K. (2007) Splanchnic regulation of glucose production. Annu. Rev. Nutr. 27, 329–345 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 4. Story J. A. (1984) Cholesterol synthesis and degradation. Lab. Res. Methods Biol. Med. 10, 217–230 [PubMed] [Google Scholar]
- 5. O'Donnell-Tormey J., Nathan C. F., Lanks K., DeBoer C. J., and de la Harpe J. (1987) Secretion of pyruvate: an antioxidant defense of mammalian cells. J. Exp. Med. 165, 500–514 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mohanty I., Joshi S., Trivedi D., Srivastava S., Tandon R., and Gupta S. K. (2002) Pyruvate modulates antioxidant status of cultured human lens epithelial cells under hypergalactosemic conditions. Mol. Cell. Biochem. 238, 129–135 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 7. Desagher S., Glowinski J., and Prémont J. I. (1997) Pyruvate protects neurons against hydrogen peroxide-induced toxicity. J. Neurosci. 17, 9060–9067 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rajendran P., Williams D. E., Ho E., and Dashwood R. H. (2011) Metabolism as a key to histone deacetylase inhibition. Crit. Rev. Biochem. Mol. Biol. 46, 181–199 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thangaraju M., Carswell K. N., Prasad P. D., and Ganapathy V. (2009) Colon cancer cells maintain low levels of pyruvate to avoid cell death caused by inhibition of HDAC1/HDAC3. Biochem. J. 417, 379–389 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 10. Patel M. S., and Korotchkina L. G. (2006) Regulation of the pyruvate dehydrogenase complex. Biochem. Soc. Trans. 34, 217–222 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 11. Hinoi E., Takarada T., Tsuchihashi Y., Fujimori S., Moriguchi N., Wang L., Uno K., and Yoneda Y. (2006) A molecular mechanism of pyruvate protection against cytotoxicity of reactive oxygen species in osteoblasts. Mol. Pharmacol. 70, 925–935 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 12. Lee Y. J., Kang I. J., Bünger R., and Kang Y. H. (2003) Mechanisms of pyruvate inhibition of oxidant-induced apoptosis in human endothelial cells. Microvasc. Res. 66, 91–101 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 13. Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., and Puigserver P. (2005) Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434, 113–118 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 14. Holness M. J., and Sugden M. C. (1989) Pyruvate dehydrogenase activities during the fed-to-starved transition and on re-feeding after acute or prolonged starvation. Biochem. J. 258, 529–533 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim J. W., Tchernyshyov I., Semenza G. L., and Dang C. V. (2006) HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3, 177–185 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 16. Perrin A., Roudier E., Duborjal H., Bachelet C., Riva-Lavieille C., Leverve X., and Massarelli R. (2002) Pyruvate reverses metabolic effects produced by hypoxia in glioma and hepatoma cell cultures. Biochimie 84, 1003–1011 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 17. Patel M. S., and Roche T. E. (1990) Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J. 4, 3224–3233 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 18. Roche T. E., and Hiromasa Y. (2007) Pyruvate dehydrogenase kinase regulatory mechanisms and inhibition in treating diabetes, heart ischemia, and cancer. Cell. Mol. Life Sci. 64, 830–849 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sugden M. C., and Holness M. J. (2006) Mechanisms underlying regulation of the expression and activities of the mammalian pyruvate dehydrogenase kinases. Arch. Physiol. Biochem. 112, 139–149 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 20. Gudi R., Bowker-Kinley M. M., Kedishvili N. Y., Zhao Y., and Popov K. M. (1995) Diversity of the pyruvate dehydrogenase kinase gene family in humans. J. Biol. Chem. 270, 28989–28994 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 21. Rowles J., Scherer S. W., Xi T., Majer M., Nickle D. C., Rommens J. M., Popov K. M., Harris R. A., Riebow N. L., Xia J., Tsui L. C., Bogardus C., and Prochazka M. (1996) Cloning and characterization of PDK4 on 7q21.3 encoding a fourth pyruvate dehydrogenase kinase isoenzyme in human. J. Biol. Chem. 271, 22376–22382 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 22. Huang B., Gudi R., Wu P., Harris R. A., Hamilton J., and Popov K. M. (1998) Isoenzymes of pyruvate dehydrogenase phosphatase: DNA-derived amino acid sequences, expression, and regulation. J. Biol. Chem. 273, 17680–17688 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 23. Wagman A. S., and Nuss J. M. (2001) Current therapies and emerging targets for the treatment of diabetes. Curr. Pharm. Des. 7, 417–450 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 24. Sugden M. C. (2003) PDK4: a factor in fatness? Obes. Res. 11, 167–169 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 25. Soga T. (2013) Cancer metabolism: key players in metabolic reprogramming. Cancer Sci. 104, 275–281 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martin E., Rosenthal R. E., and Fiskum G. (2005) Pyruvate dehydrogenase complex: metabolic link to ischemic brain injury and target of oxidative stress. J. Neurosci. Res. 79, 240–247 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hollidge-Horvat M. G., Parolin M. L., Wong D., Jones N. L., and Heigenhauser G. J. (1999) Effect of induced metabolic acidosis on human skeletal muscle metabolism during exercise. Am. J. Physiol. 277, E647–E658 [DOI] [PubMed] [Google Scholar]
- 28. Bowker-Kinley M. M., Davis W. I., Wu P., Harris R. A., and Popov K. M. (1998) Evidence for existence of tissue-specific regulation of the mammalian pyruvate dehydrogenase complex. Biochem. J. 329, 191–196 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Woolbright B. L., Choudhary D., Mikhalyuk A., Trammel C., Shanmugam S., Abbott E., Pilbeam C. C., and Taylor J. A. 3rd (2018) The role of pyruvate dehydrogenase kinase-4 (PDK4) in bladder cancer and chemoresistance. Mol. Cancer Ther. 17, 2004–2012 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu P., Sato J., Zhao Y., Jaskiewicz J., Popov K. M., and Harris R. A. (1998) Starvation and diabetes increase the amount of pyruvate dehydrogenase kinase isoenzyme 4 in rat heart. Biochem. J. 329, 197–201 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jeoung N. H., Wu P., Joshi M. A., Jaskiewicz J., Bock C. B., Depaoli-Roach A. A., and Harris R. A. (2006) Role of pyruvate dehydrogenase kinase isoenzyme 4 (PDHK4) in glucose homoeostasis during starvation. Biochem. J. 397, 417–425 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Popov K. M., Kedishvili N. Y., Zhao Y., Gudi R., and Harris R. A. (1994) Molecular cloning of the p45 subunit of pyruvate dehydrogenase kinase. J. Biol. Chem. 269, 29720–29724 [PubMed] [Google Scholar]
- 33. Popov K. M., Shimomura Y., and Harris R. A. (1991) Purification and comparative study of the kinases specific for branched chain α-ketoacid dehydrogenase and pyruvate dehydrogenase. Protein Expr. Purif. 2, 278–286 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 34. Tonks N. K., Kearns A., and Randle P. J. (1982) Pig heart [35S]thiophosphoryl pyruvate dehydrogenase complexes. Eur. J. Biochem. 122, 549–551 [DOI] [PubMed] [Google Scholar]
- 35. Sale G. J., and Randle P. J. (1982) Role of individual phosphorylation sites in inactivation of pyruvate dehydrogenase complex in rat heart mitochondria. Biochem. J. 203, 99–108 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kolobova E., Tuganova A., Boulatnikov I., and Popov K. M. (2001) Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites. Biochem. J. 358, 69–77 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Baker J. C., Yan X., Peng T., Kasten S., and Roche T. E. (2000) Marked differences between two isoforms of human pyruvate dehydrogenase kinase. J. Biol. Chem. 275, 15773–15781 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 38. Kato M., Li J., Chuang J. L., and Chuang D. T. (2007) Distinct structural mechanisms for inhibition of pyruvate dehydrogenase kinase isoforms by AZD7545, dichloroacetate, and radicicol. Structure 15, 992–1004 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Linn T. C., Pettit F. H., Hucho F., and Reed L. J. (1969) α-Keto acid dehydrogenase complexes. XI. Comparative studies of regulatory properties of the pyruvate dehydrogenase complexes from kidney, heart, and liver mitochondria. Proc. Natl. Acad. Sci. U.S.A. 64, 227–234 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jeoung N. H., Rahimi Y., Wu P., Lee W. N., and Harris R. A. (2012) Fasting induces ketoacidosis and hypothermia in PDHK2/PDHK4-double-knockout mice. Biochem. J. 443, 829–839 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chih C. P., and Roberts E. L. Jr. (2003) Energy substrates for neurons during neural activity: a critical review of the astrocyte-neuron lactate shuttle hypothesis. J. Cereb. Blood Flow Metab. 23, 1263–1281 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 42. Boussouar F., and Benahmed M. (2004) Lactate and energy metabolism in male germ cells. Trends Endocrinol. Metab. 15, 345–350 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 43. Gerich J. E., Meyer C., Woerle H. J., and Stumvoll M. (2001) Renal gluconeogenesis: its importance in human glucose homeostasis. Diabetes Care 24, 382–391 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 44. Priestman D. A., Mistry S. C., Kerbey A. L., and Randle P. J. (1992) Purification and partial characterization of rat liver pyruvate dehydrogenase kinase activator protein (free pyruvate dehydrogenase kinase). FEBS Lett. 308, 83–86 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 45. Sugden P. H., Hutson N. J., Kerbey A. L., and Randle P. J. (1978) Phosphorylation of additional sites on pyruvate dehydrogenase inhibits its re-activation by pyruvate dehydrogenase phosphate phosphatase. Biochem. J. 169, 433–435 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Karpova T., Danchuk S., Kolobova E., and Popov K. M. (2003) Characterization of the isozymes of pyruvate dehydrogenase phosphatase: implications for the regulation of pyruvate dehydrogenase activity. Biochim. Biophys. Acta 1652, 126–135 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 47. Skarnes W. C., Rosen B., West A. P., Koutsourakis M., Bushell W., Iyer V., Mujica A. O., Thomas M., Harrow J., Cox T., Jackson D., Severin J., Biggs P., Fu J., Nefedov M., et al. (2011) A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 10.1074/jbc.RA117.001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dunford E. C., Herbst E. A., Jeoung N. H., Gittings W., Inglis J. G., Vandenboom R., LeBlanc P. J., Harris R. A., and Peters S. J. (2011) PDH activation during in vitro muscle contractions in PDH kinase 2 knockout mice: effect of PDH kinase 1 compensation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 300, R1487–R1493 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 49. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 50. Peterson G. L. (1977) A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 83, 346–356 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
- 51. Harris R. A., Bowker-Kinley M. M., Wu P., Jeng J., and Popov K. M. (1997) Dihydrolipoamide dehydrogenase-binding protein of the human pyruvate dehydrogenase complex: DNA-derived amino acid sequence, expression, and reconstitution of the pyruvate dehydrogenase complex. J. Biol. Chem. 272, 19746–19751 10.1074/jbc.RA117.001068 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.