Abstract
Bialaphos resistance (BAR) and phosphinothricin acetyltransferase (PAT) genes, which convey resistance to the broad-spectrum herbicide phosphinothricin (also known as glufosinate) via N-acetylation, have been globally used in basic plant research and genetically engineered crops1–4. Although early in vitro enzyme assays showed that recombinant BAR and PAT exhibit substrate preference toward phosphinothricin over the 20 proteinogenic amino acids1, indirect effects of BAR-containing transgenes in planta, including modified amino acid levels, have been seen but without the identification of their direct causes5,6. Combining metabolomics, plant genetics, and biochemical approaches, we show that transgenic BAR indeed converts two plant endogenous amino acids, aminoadipate and tryptophan, to their respective N-acetylated products in several plant species examined. We report the crystal structures of BAR, and further delineate structural basis for its substrate selectivity and catalytic mechanism. Through structure-guided protein engineering, we generated several BAR variants that display significantly reduced nonspecific activities compared to its wild-type counterpart in vivo. Our results demonstrate that transgenic expression of enzymes can result in unintended off-target metabolism arising from enzyme promiscuity. Understanding of such phenomena at the mechanistic level can facilitate the design of maximally insulated systems featuring heterologously expressed enzymes.
Phosphinothricin is a naturally occurring herbicide derived from the tripeptide antibiotic bialaphos produced by species of Streptomyces soil bacteria. Phosphinothricin is a structural analog of glutamate, and thereby inhibits glutamine synthetase, an essential enzyme for glutamine synthesis and ammonia detoxification in plants, giving rise to its herbicidal activity3. In the 1980s, the bialaphos resistance (BAR) gene and its closely related homolog phosphinothricin acetyltransferase (PAT) gene were isolated from Streptomyces hygroscopicus and Streptomyces viridochromogenes, respectively, and were later broadly used as transgenes to confer herbicide resistance in a variety of major genetically-engineered (GE) crops, including corn, soybean, canola, and cotton7. In addition, BAR and PAT have also gained much utility in basic research as selection markers for generating transgenic plants1. Despite the prevalent use of BAR and PAT in the context of generating herbicide-resistant transgenic plants, whether these bacteria-derived enzymes may possibly interfere with plant endogenous metabolism has not been rigorously investigated.
In research not initially intended to address this issue regarding phosphinothricin-resistance trait, we carried out untargeted metabolomics analysis on senescent leaf extracts prepared from the Arabidopsis thaliana clh2–1 mutant (FLAG_76H05, referred to as FLAG-1 hereafter), which contains a transfer DNA (T-DNA) insertion that abolishes the CHLOROPHYLLASE 2 gene8. This analysis revealed two metabolites that were ectopically accumulated at high levels in clh2–1 compared to wild type (Fig. 1a). Using liquid chromatography-tandem mass spectrometry (LC-MS2), we identified these two metabolites as N-acetyl-aminoadipate and N-acetyl-tryptophan (referred to as acetyl-aminoadipate and acetyl-tryptophan, respectively, hereafter; Fig. 1a and Supplementary Fig. 1). Because the deficiency of CHLOROPHYLLASE 2, a serine esterase8, in clh2–1 does not explain the accumulation of these acetylated metabolites, we hypothesized that the BAR gene present on the T-DNA as a selection marker in clh2–1 might be responsible for their formation. To test this, we extended our metabolomics analysis to additional Arabidopsis T-DNA insertional mutants unrelated to chlorophyll metabolism that carry either BAR (e.g. mutants from the FLAG9 and SAIL10 collections) or alternative antibiotic selection markers (e.g. mutants from the SALK (NTPII, kanamycin resistance)11 and GABI (SULI, sulfadiazine resistance)12 collections (Supplementary Table 1). Senescent leaves of all six T-DNA mutants carrying BAR manifested accumulation of acetyl-aminoadipate and acetyl-tryptophan, while these metabolites were significantly lower or not detected in wild-type plants and T-DNA mutants containing alternative selection markers (Fig. 1b). These results indicate that the ectopic accumulation of these metabolites is likely resulted from the nonspecific N-acetyltransferase activities of transgenic BAR acting upon plant endogenous amino acids.
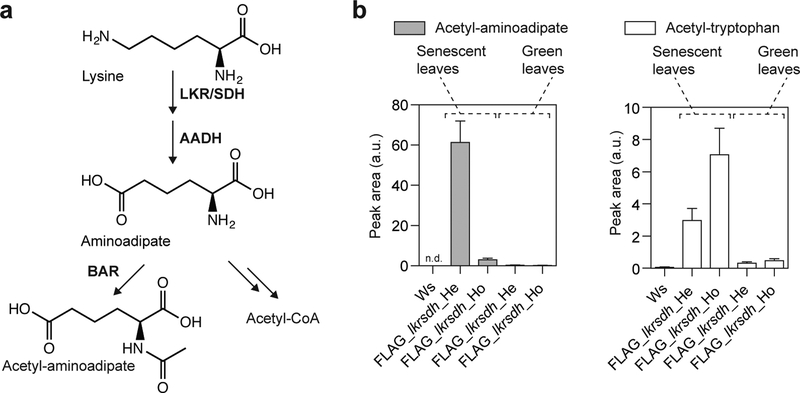
Figure 1 |. Accumulation of acetyl-aminoadipate and acetyl-tryptophan in senescent leaves of Arabidopsis carrying the BAR transgene.
a, Metabolite profiles of senescent leaves from Wassilewskija (Ws) and clh2–1 (FLAG-1), displayed as base peak chromatograms (BPC), reveal the ectopic accumulation of acetyl-aminoadipate (1) and acetyl-tryptophan (2). BPC traces of four biological replicates are displayed. b, Comparative levels of acetyl-aminoadipate and acetyl-tryptophan in Arabidopsis mutants from different insertional mutant collections that contain either BAR (SAIL and FLAG) or alternative selection marker genes (SALK (NTPII, kanamycin resistance) and GABI (SULI, sulfadiazine resistance)). Error bars, mean ± s.d. (n = 3 biological replicates). This experiment was repeated at least three times with similar results. See Supplementary Fig. 2 for absolute quantification. a.u., arbitrary unit; FW, fresh weight; n.d., not detected
We quantified the absolute concentrations of acetyl-aminoadipate and acetyl-tryptophan in senescent leaves of BAR-containing transgenic Arabidopsis to range from 306 to 845 nmole/g and from 14 to 76 nmole/g fresh weight, respectively (Supplementary Fig. 2). While trace level of acetyl-tryptophan can be detected in wild-type Arabidopsis, acetyl-aminoadipate was undetectable in wild-type samples (Supplementary Fig. 2). The ectopic accumulation of acetyl-aminoadipate and acetyl-tryptophan in BAR-containing transgenic Arabidopsis is substantial given that the concentrations of free aminoadipate and tryptophan in senescent leaves of these Arabidopsis lines are in the ranges of 61 to 122 nmole/g and from 1566 to 2663 nmole/g fresh weight, respectively (Supplementary Fig. 2). On the other hand, the concentrations of free amino acids in senescent leaves do not seem to be significantly affected by the expression of BAR, as revealed by the quantification of 21 other amino acids (Supplementary Fig. 2).
To assess whether the nonspecific activities of transgenic BAR also manifest in other plant hosts, we performed metabolic profiling of various tissue samples from phosphinothricin-resistant soybean (Glycine max), canola (Brassica napus), mustard (Brassica juncea) and wheat (Triticum aestivum). Substantially increased accumulation of acetyl-aminoadipate and acetyl-tryptophan was also detected in some tissues of these transgenic lines (Supplementary Fig. 3), indicating that our findings regarding the in vivo nonspecific activities of BAR may apply broadly to a wide range of BAR-containing transgenic plants.
The concentration of free tryptophan is low in photosynthetically active leaves, but increases significantly in senescent leaves13. This is due to enhanced proteolysis during senescence, facilitating remobilization of protein-bound nitrogen and other nutrients to sink organs, such as seeds14. Aminoadipate, an intermediate of lysine degradation, also exhibits a similar accumulation pattern during leaf senescence15. To test whether the BAR-catalyzed production of acetyl-aminoadipate depends on lysine degradation, we analyzed an Arabidopsis mutant from the FLAG collection, FLAG_lkrsdh, in which the BAR-containing T-DNA disrupts At4g33150 encoding the Arabidopsis bifunctional lysine-ketoglutarate reductase/saccharopine dehydrogenase (LKR/SDH, Supplementary Fig. 4)16. LKR/SDH catalyzes the first committed step of lysine degradation, and, together with the subsequent aminoadipate semialdehyde dehydrogenase (AADH), converts lysine to aminoadipate (Fig. 2a). In a segregating population for the FLAG_lkrsdh locus, heterozygous, homozygous and wild-type plants were identified by genotyping, and subjected to LC-MS metabolic profiling after senescence induction (Fig. 2b, Supplementary Fig. 4). Acetyl-aminoadipate occurred at the highest level in the heterozygous mutant, but was greatly reduced in the homozygous mutant, suggesting that the ectopic accumulation of acetyl-aminoadipate in BAR-containing plants is dependent on the activity of LKR/SDH in the lysine degradation pathway in senescent leaves (Fig. 2a). By contrast, the relative abundance of acetyl-tryptophan in the segregating population of FLAG_lkrsdh generally reflected the copy number of the BAR-containing T-DNA transgene, with approximately 2-fold acetyl-tryptophan level observed in the homozygotes compared to the heterozygotes (Fig. 2b). Furthermore, acetyl-aminoadipate and acetyl-tryptophan levels were approximately 10–20 fold higher in senescent leaves than those in green leaves (Fig. 2b), which is likely due to the increased availability of the corresponding free amino acids during senescence. Consistent with these observations in leaves, ectopic accumulation of acetyl-aminoadipate and acetyl-tryptophan was also observed in seeds of multiple BAR-containing T-DNA mutant lines compared to the wild-type controls (Supplementary Fig. 5).
Figure 2 |. BAR-dependent accumulation of acetyl-aminoadipate and acetyl-tryptophan is linked to nitrogen remobilization during senescence.
a, Aminoadipate is derived from the lysine degradation pathway in plants, which can be metabolized by BAR as a nonspecific substrate. b, Comparative levels of acetyl-aminoadipate and acetyl-tryptophan in green and senescent leaves from the heterozygous (He) and homozygous (Ho) FLAG_lkrsdh mutant, harboring a BAR-containing T-DNA that abolishes the Arabidopsis LKR/SDH gene. Error bars, mean ± s.d. (n = 3 biological replicates). a.u., arbitrary unit; n.d., not detected; Ws, Wassilewskija wild-type plants.
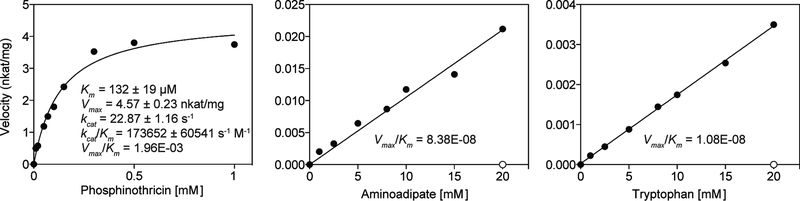
To shed light on the kinetic properties of BAR, we carried out pseudo-first-order enzyme kinetic assays using recombinant BAR against several native and non-native amino acid substrates (Fig. 3 and Supplementary Fig. 6). Similar to published data1,3,17, N-acetylation of phosphinothricin exhibits Michaelis-Menten kinetics with an apparent Km of approximately 132 μM (Fig. 3). Although BAR clearly showed N-acetyltransferase activities toward aminoadipate and tryptophan, Km values for these non-native substrates could not be established, as both substrates reached solubility limit before reaching saturation concentration for BAR. Vmax/Km values of BAR against aminoadipate and tryptophan, which were inferred from Lineweaver-Burk plots, reveals that these two side reactions are less favorable than the acetylation of phosphinothricin. BAR also exhibited relatively higher catalytic activity toward aminoadipate than tryptophan in vitro (Fig. 3).
Figure 3 |. In vitro enzyme kinetic assays of BAR against native and non-native substrates.
An apparent KM value of 132 ± 19.2 μM was obtained for phosphinothricin, similar to previously published data1,3,17. Vmax, kcat, kcat/Km and Vmax/Km values for phosphinothricin are also indicated, as well Vmax/Km values for aminoadipate and tryptophan (estimated from Lineweaver-Burk plots). Aminoadipate and tryptophan are in vitro substrates of BAR but both substrates reached solubility limit before reaching saturation concentration for BAR. Negative controls (open circles) were performed in absence of BAR at the highest substrate concentration tested (20 mM).
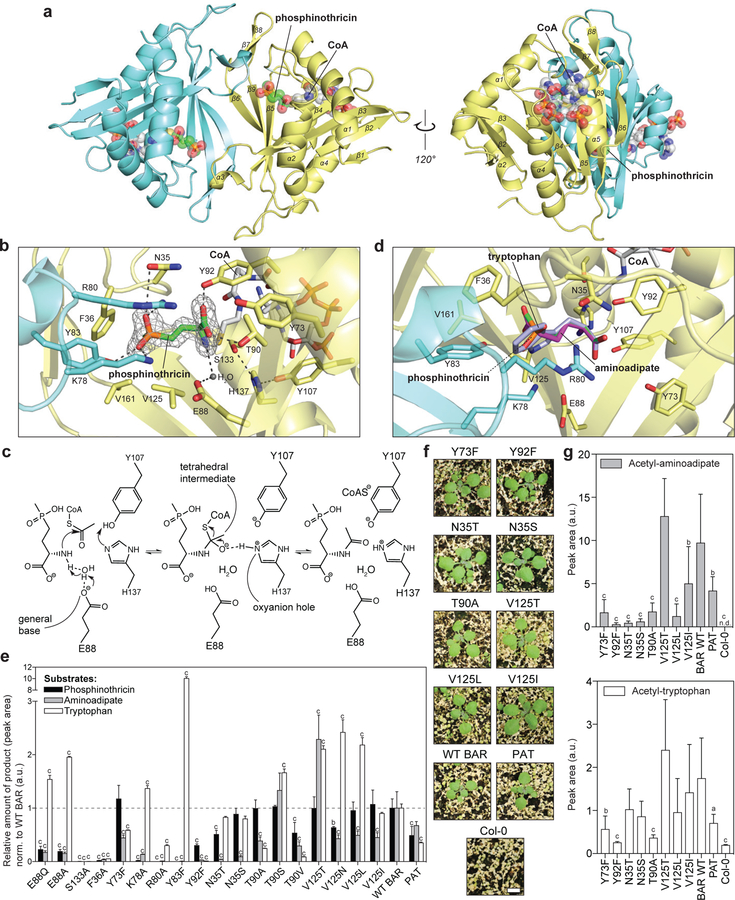
To reveal the structural basis for substrate selectivity and catalytic mechanism of BAR that would enable structure-guided protein engineering, we determined the crystal structures of the BAR/acetyl-CoA holocomplex and the BAR/CoA/phosphinothricin ternary complex (see Supplementary Table 2 for data collection and refinement statistics). Our refined structures revealed that BAR is an αβ protein harboring a globular tertiary structure resembling the previously reported Gcn5-related N-acetyltransferase (GNAT) structures (Supplementary Fig. 7)18–21. BAR crystalizes as a homodimer with two active sites symmetrically distributed around the dimer interface inside a large open cavity (Fig. 4a and Supplementary Fig. 8). The cofactor acetyl-CoA binds to a cleft between α4 and α5 on the opposite side of the dimer interface with the acetyl group pointing toward the catalytic center (Fig. 4a). Close examination of the BAR/acetyl-CoA and BAR/CoA/phosphinothricin structures illuminates the catalytic mechanism of BAR (Fig. 4b, 4c and Supplementary Fig. 9). Similar to other GNATs, BAR utilizes a conserved catalytic Glu88 as a general base to deprotonate the amino group of phosphinothricin through a water molecule as the proton shuttle (Fig. 4b, 4c, and Supplementary Fig. 9)21. The deprotonated amino group then undergoes nucleophilic attack on the carbonyl carbon of acetyl-CoA to produce a tetrahedral intermediate, which is further stabilized by an oxyanion hole formed by a positively charged H137 and its proton donor Y107 (Fig. 4c and Supplementary Fig. 9). Interestingly, the structural feature underlying this oxyanion hole in BAR must have arisen independently from the functionally analogous oxyanion hole previously described in the histone acetyltransferase GCN5, featuring a backbone amide nitrogen instead21. In the final step of the catalytic cycle, coenzyme A is released from the tetrahedral intermediate as a leaving group to produce acetyl-phosphinothricin (Fig. 4c).
Figure 4 |. Structural basis for amino acid N-acetylation catalyzed by BAR and structure-guided engineering of BAR with reduced nonspecific activities.
a, Cartoon representation of BAR homodimer in complex with phosphinothricin and CoA. Two monomers of the dimer are colored in blue and yellow respectively. b, Close-up view of the BAR active site. The |2Fo-Fc| omit electron density map (contoured at 3.0 σ) is shown for phosphinothricin. c, Proposed catalytic mechanism of BAR. d, Docking of tryptophan and aminoadipate within the BAR active site reveals reduced favorable contacts compared to phosphinothricin. e, Enzyme activity assays using purified BAR mutant proteins against phosphinothricin (0.2mM), aminoadipate (1 mM) and tryptophan (1 mM). Wild-type BAR (WT BAR) and PAT from Streptomyces viridochromogenes were also examined as controls. Assays were terminated during the initial linear rate of product formation. The relative amount of product formed by each BAR mutant was normalized to WT BAR for each substrate (value of 1). Error bars, mean ± s.d. (n = 3 technical replicates). f, Photographs of Arabidopsis T1 lines transformed with select BAR mutants 10 days after phosphinothricin treatment (see also Supplementary Fig. 12–14). Scale bar = 0.3 cm. g, Comparative levels of acetyl-aminoadipate and acetyl-tryptophan in phosphinothricin-resistant T2 Arabidopsis plants transformed with selected BAR mutants. Error bars, mean ± s.d. (n = 5–6 biological replicates: Y73F (6), Y92F (6), N35T (5), N35S (5), T90A (6), V125T (5), V125L (6), V125I (6), WT BAR (5), PAT (5)). Significance levels were indicated based on one-way ANOVA with Dunnett’s test for multiple comparisons to WT BAR. a, p-value < 0.1; b, p-value < 0.05; c, p-value < 0.01; a.u., arbitrary unit.
The BAR/CoA/phosphinothricin ternary structure also reveals active-site residues involved in phosphinothricin binding. Within each active site, the methylphosphoryl group of the substrate engages hydrophobic interactions with the surrounding F36, G127, and V161 from the same monomer, whereas the two phosphoryl oxygen atoms are coordinated by K78, R80, and Y83 from the β3-loop-α3 region of the neighboring monomer via a set of hydrogen bonds and electrostatic interactions (Fig. 4b). Furthermore, the amino acid group of phosphinothricin is properly positioned at the catalytic center by a hydrogen-bond network involving the backbone carbonyl group of V125 and the side chains of T90 and Y92 (Fig. 4b). Despite various attempts using co-crystallization and soaking techniques, structures of BAR containing aminoadipate or tryptophan could not be obtained, reflecting the low affinity of these nonspecific substrates to BAR. Simulated docking of these substrates within the active site of the BAR/CoA/phosphinothricin structure reveals fewer favorable interactions as well as potential steric clashes with the surrounding residues compared to phosphinothricin (Fig. 4d).
Site-directed mutagenesis followed by biochemical assays confirmed the roles of many active-site residues predicted by structural analysis (Fig. 4e and Supplementary Fig. 10). Mutating the catalytic E88 to alanine or glutamine greatly reduces the activity of BAR toward phosphinothricin and aminoadipate. Nevertheless, these mutants exhibit higher activity toward tryptophan than that of the wild-type enzyme at the substrate concentration tested (Fig. 4e), suggesting that tryptophan may be deprotonated through an alternative mechanism independent of E88 and/or the first deprotonation step is not rate-limiting for BAR-catalyzed acetyl-tryptophan formation. H137A and Y107F mutants failed to yield sufficient soluble recombinant protein (Supplementary Fig. 10), preventing the role of the oxyanion hole in catalysis to be directly assessed. We thus probed this indirectly by mutating S133, a residue that forms a hydrogen bond with the imidazole ring π-nitrogen of H137 (Fig. 4b). The resulting S133A mutant exhibits completely abolished N-acetyltransferase activity toward the three tested substrates, suggesting an essential role of S133 in catalysis, likely through proper positioning of the imidazoline ring of the histidine within the oxyanion hole (Fig. 4e and Supplementary Fig. 9). Mutants affecting phosphinothricin-binding residues, including F36A, K78A, R80A, Y83F, Y92F, generally show significantly reduced activity toward phosphinothricin and aminoadipate, while K78A and Y83F display increased activity toward the more hydrophobic substrate tryptophan compared to the wild-type enzyme (Fig. 4e).
With the structural information of BAR in hand, we sought to engineer BAR through structure-guided mutagenesis to repress its undesired nonspecific activities toward aminoadipate and tryptophan while maintaining its native activity against phosphinothricin. We selected residue positions N35, Y73, T90, Y92, and V125 for targeted mutagenesis based on structural analysis as well as multiple sequence alignment containing BAR, PAT, and other closely related homologs from bacteria (Fig. 4b, 4d and Supplementary Fig. 11). A set of eleven mutants was first characterized in vitro (Fig. 4e), and eight of them were further tested in transgenic Arabidopsis (Fig. 4f and 4g). All eight BAR mutants confer phosphinothricin resistance in Arabidopsis T1 and T2 generations (Fig. 4f, Supplementary Fig. 12–14). Metabolic profiling of these transgenic lines confirmed that mutations in select active-site residues of BAR can modulate the in vivo nonspecific activities of BAR toward aminoadipate and tryptophan (Fig. 4g). Notably, transgenic Arabidopsis plants containing Y73F, Y92F, N35T, N35D, T90A, V125L, or V125I BAR mutants display significantly reduced levels of acetyl-aminoadipate compared to plants containing wild-type BAR (Fig. 4g). Moreover, plants expressing Y73F, Y92F or T90A BAR mutants exhibit significantly reduced levels of both acetyl-aminoadipate and acetyl-tryptophan compared to plants containing wild-type BAR. These observed differences in acetyl-aminoadipate and acetyl-tryptophan levels are not due to BAR protein levels in transgenic plants (Supplementary Fig. 15), but are consistent with the altered catalytic activities of various BAR mutants measured in vitro (Fig. 4e and Supplementary Fig. 16). Subsequent analysis of N35T and Y92F revealed that both mutants exhibit compromised affinity toward native substrate phosphinothricin in vitro compared to wild-type BAR. However, N35T and Y92F retain largely unaltered catalytic speed in vitro and confer level of resistance to phosphinothricin in planta similar to that of wild-type BAR (Supplementary Fig. 16a and Supplementary Fig. 14). Furthermore, both mutants show more pronounced reduced catalytic activity toward one or both non-native substrates as compared to phosphinothricin (Supplementary Fig. 16).
Transgenic expression of enzymes catalyzing a variety of desirable biochemical reactions in heterologous hosts is a common strategy in both basic biological research and translational biotechnology. Prominent examples include reporter enzymes, such as firefly luciferase and β-glucuronidase, antibiotic/herbicide markers, such as aminoglycoside kinase that confers kanamycin resistance and BAR, and many enzymes used for metabolic engineering purposes in microbes and higher eukaryotes22. Although enzymes are generally considered as perfected catalysts with superior substrate specificity and predictable catalytic mechanism, increasing evidences have raised awareness of the unpredictable behaviors of enzymes and their profound implication in natural and directed evolution of new enzymatic functions23. However, whether and how heterologous expression of a foreign enzyme would interfere with the native metabolic system remains an open question to be addressed on a case-by-case basis.
In this study, we discovered that transgenic expression of the herbicide-resistance enzyme BAR of bacterial origin indeed acetylate two endogenous amino acids, resulting in the ectopic accumulation of acetyl-aminoadipate and acetyl-tryptophan. While acetyl-tryptophan is a naturally occurring metabolite found in numerous plant species, including Arabidopsis, Salsola collina, Glycine max, Solanum lycopersicum, Cocos nucifera, and Ginkgo biloba24,25, to the best of our knowledge, acetyl-aminoadipate has never been reported as an endogenous plant metabolite. Interestingly, in line with our findings, a recent study reported the ectopic accumulation of acetyl-aminoadipate in the flower tissue of a BAR-containing T-DNA mutant of Arabidopsis, which could not be rationalized by the mutated gene26. Despite the widespread use of BAR in GE crops2,27 and the extensive testing and deregulation processes associated with this trait over the past few decades1,3,17,28,29, such phenomenon was not reported elsewhere, probably due to technological limitation in metabolic profiling in the past. Studies have demonstrated indirect effects of BAR-containing transgenes in transgenic lines with high BAR expression, such as reduced fitness and modified amino acid levels, but without identifying their direct causes5,30. However, no negative effect on yield and human health has been reported from the extensive use of phosphinothricin-resistant crops in agriculture over the last two decades.
Our findings suggest that untargeted metabolomics analysis could be a useful methodology for future assessment of GE plants7. This study also provides solutions to reduce the nonspecific activities of BAR through structure-guided enzyme engineering so that its intended herbicide-degrading activity can be maximally insulated from the metabolome of the host.
Materials and Methods
Plant materials
Arabidopsis (Arabidopsis thaliana) Columbia-0 (Col-0) and Wassilewskija (Ws) were used as wild types. T-DNA insertion lines were from the following collections: SALK lines11: SALK_130606 (SALK_1), SALK_051823C (SALK_2), SALK_110649 (SALK_3); SAIL lines10: SAIL_1165_B02 (SAIL_1), SAIL_503_C03 (SAIL_2), SAIL_1235_D10 (SAIL_3); GABI lines12: GABI_453E01 (GABI_1), GABI_833F02 (GABI_2), GABI_453A08 (GABI_3); FLAG lines9: FLAG_076H05 (clh2–18; FLAG_1), FLAG_271B02 (FLAG_2), FLAG_495A09 (FLAG_3), FLAG_271B12 (FLAG_lkrsdh). SALK, SAIL and GABI lines were obtained from the European Arabidopsis Stock Center (http://arabidopsis.info/). The FLAG lines were obtained from the INRA Versailles Arabidopsis Stock Center (http://publiclines.versailles.inra.fr/). Homozygous (and heterozygous for FLAG_lkrsdh) plants were identified by PCR using T-DNA- and gene-specific primers.
Arabidopsis T-DNA lines used for untargeted metabolomics and relative quantification of acetyl-aminoadipate and acetyl-tryptophan were grown on soil under a 12-h-light/12-h-dark photoperiod with fluorescent light of 80 to 120 μmol photons m−2 s−1 at 22°C and 60% relative humidity. For senescence induction, leaves from 5-week-old plants were excised and incubated in permanent darkness on wet filter paper for 8 d at ambient temperature. Transgenic Arabidopsis lines transformed with BAR mutants and Arabidopsis T-DNA lines used for absolute quantification of acetyl-aminoadipate and acetyl-tryptophan were grown on soil under a 16-h-light/8-h-dark photoperiod with fluorescent light of 80 to 120 μmol photons m−2 s−1 at 22°C and 60% relative humidity. For senescence induction, leaves from phosphinothricin-resistant, 4-week-old plants were excised and incubated in permanent darkness on wet filter paper for 6 d at ambient temperature. For measuring the expression of LKR/SDH in FLAG_271B12, seedlings were grown for 7 days on ½ Murashige and Skoog (MS) plates containing 1% sucrose.
Phosphinothricin-resistant Glycine max (Liberty Link trait A2704–12, 283 Morril MC-116, Credenz CZ 3841 LL, Bayer CropScience), wild-type (non-isogenic) Glycine max (Chiba Green; High Mowing Organic Seed), lines were grown on soil under a 16-h-light/8-h-dark photoperiod with fluorescent light of 80 to 120 μmol photons m−2 s−1 at 22°C and 60% relative humidity. Green and senescent leaf samples were collected from 40-days old plants. This experiment was repeated once with similar results.
Phosphinothricin-resistant Brassica napus (Liberty Link trait L252, Bayer CropScience) and wild-type (non-isogenic) Brassica napus (NDC-E12131, NDC-E13285 and NDC-E12027) lines were grown on soil under a 16-h-light/8-h-dark photoperiod with fluorescent light of 80 to 120 μmol photons m−2 s−1 at 22°C and 60% relative humidity. For senescence induction, fully developed cotyledons were excised and incubated in permanent darkness on wet filter paper for 5–7 days at ambient temperature. This experiment was done once.
Wild-type (isogenic) and phosphinothricin-resistant Brassica juncea31 were grown on soil under a 16-h-light/8-h-dark photoperiod with fluorescent light of 80 to 120 μmol photons m−2 s−1 at 22°C and 60% relative humidity. For senescence induction, fully developed cotyledons were excised and incubated in permanent darkness on wet filter paper for 5–7 d at ambient temperature.
Wild-type (isogenic) and phosphinothricin-resistant Triticum aestivum32 were grown on soil under a 16-h-light/8-h-dark photoperiod with fluorescent light of 80 to 120 μmol photons m−2 s−1 at 22°C and 60% relative humidity. For senescence induction, leaves were excised and incubated in permanent darkness on wet filter paper for 5–7 d at ambient temperature.
RNA Isolation and qRT-PCR
Total RNA was extracted using a Qiagen RNeasy Plant Mini Kit according to manufacturer’s instructions (DNase treatment was performed on-column). The concentration and purity of RNA were determined by absorbance at 260/280 nm. First-strand cDNA was synthesized from 1 μg of RNA using SuperScript III Reverse Transcriptase with Oligo dT primers (Thermo Scientific). Reactions were run on a QuantStudio 6 system machine (Thermo Scientific) using Sybr Green Master Mix (Thermo Scientific) using primer listed in Supplementary Fig. 4 and Supplementary Table 3. Gene expression values were calculated using Ct values and normalized using the reference gene At1g1332033.
Metabolite extraction
Arabidopsis and Brassica napus samples were collected in 2 mL Eppendorf tubes containing 500 μL of 1.5 mm glass beads, weighted and snap-frozen in liquid nitrogen. The frozen samples were ground using a MM300 Mixer Mill (Retsch) at 30 Hz for 5 min and stored at −80°C until further processing. Glycine max samples were snap-frozen in liquid nitrogen and ground with a mortar and pestle. Metabolites were extracted using 5–10 (for leaf samples) or 10 −50 volumes (for seed samples; w/v) of ice-cold extraction buffer (80% methanol, 20% water, 0.1% formic acid (v/v/v)). Extracts were homogenized at 30 Hz for 5 min and centrifuged (14,000–16,000 g, 4°C). After re-centrifugation, supernatants were transferred to LC vials and analyzed by LC-MS.
LC-MS analysis of Arabidopsis T-DNA mutants, Brassica juncea and Triticum aestivum (untargeted metabolomics and relative quantification of acetyl-aminoadipate and acetyl-tryptophan)
The LC-MS instrument was composed of an Ultimate 3000 Rapid Separation LC system (Thermo Scientific) coupled to a Bruker Compact ESI-Q-TOF (Bruker Daltonics). The reverse-phase chromatography system consisted of an 150 mm C18 column (ACQUITY UPLC™ BEH, 1.7 μm, 2.1 × 150 mm, Waters), which was developed using LC-MS solvents (Chemie Brunschwig) with a gradient (flow rate of 0.3 mL min−1) of solvent B (acetonitrile with 0.1% (v/v) formic acid) in solvent A (water with 0.1% (v/v) formic acid) as follows (all (v/v)): 5% for 0.5 min, 5% to 100% in 11.5 min, 100% for 4 min, 100% to 5% in 1 min and 5% for 1 min. Electrospray ionization (ESI) source conditions were set as follows: gas temperature, 220°C; drying gas, 9 L min−1; nebulizer, 2.2 BAR; capillary voltage, 4500 V; end plate offset, 500 V. Tuning conditions were set as follows: funnel 1 RF, 250 Vpp; funnel 2 RF, 150 Vpp; isCID energy, 0 eV; hexapole RF, 50 Vpp; quadrupole ion energy, 3.0 eV; quadrupole low mass, 90 m/z; collision cell, 6 eV; pre-pulse storage time, 3 μs. The instrument was set to acquire over the m/z range 50–1300, with an acquisition rate of 4 spectra s−1. Conditions for MS2 of automatically selected precursors (data-dependent MS2) were set as follows: threshold, 1000 counts; active smart exclusion (5×); active exclusion (exclude after 3 spectra, release after 0.2 min, reconsider precursor if current intensity/previous intensity is ≥5); number of precursors, 3; active stepping (basic mode, timing 50%−50%, collision RF from 350 to 450 Vpp, transfer time from 65 to 80 μs, collision energy from 80 to 120%). All data were recalibrated internally using pre-run injection of sodium formate (10 mM sodium hydroxide in 0.2% formic acid, 49.8% water, 50% isopropanol (v/v/v)). After data recalibration using DataAnalysis (version 4.2, Bruker Daltonics) and data conversion to mzXML format using ProteoWizard MSConvert34, metabolite features detected in Ws and FLAG_076H05 (senescent leaves, four replicates) were aligned according to retention time and relatively quantified using XCMS online35 (pairwise comparison using XCMS online pre-set parameters “UPLC/Bruker Q-TOF”). Up-regulated features in FLAG_076H05 were identified at retention times of 2.8 min (labeled as “1” in Fig. 1a, m/z 204.086 (fold change ≥10, p-value ≤0.005, intensity threshold 800,000)) and 6.5 min (labeled as “2” in Fig. 1a, m/z 247.108 (fold change ≥10, p-value ≤0.005, intensity threshold 100,000)) and further characterized as ions derived from N-acetyl-D/L-aminoadipate and N-acetyl-D/L-tryptophan, respectively, by database searches in METLIN36 using MS and MS2 spectra. Relative quantification of acetyl-aminoadipate and acetyl-tryptophan in Arabidopsis mutants from different insertion mutant collections was carried out by QuantAnalysis (version 2.2, Bruker Daltonics) using extracted ion chromatogram (EIC) traces ([M+H]+). Metabolomics data generated in this study have been uploaded to the EBI MetaboLights database (http://www.ebi.ac.uk/metabolights/) with the following accession number (MTBLS553).
Absolute quantification of free amino acids in senescent leaves of Arabidopsis T-DNA mutants
The LC-MS instrument was composed of an Ultimate 3000 Rapid Separation LC system (Thermo Scientific) coupled to a Q-Exactive mass spectrometer (Thermo Scientific). The HILIC chromatography system consisted of SeQuant ZIC-pHILIC Polymeric column (2.1 × 150 mm, 5 μM, EMD Millipore), which was developed using Optima™ LC/MS solvents (Fisher Chemical) with a gradient (flow rate of 0.15 mL min−1) of solvent B (acetonitrile) in solvent A (20 mM ammonium carbonate, 0.1% ammonium hydroxide) as follows (all (v/v)): 80% to 20% in 20 min, 80% to 20% in 0.5 min and 80% for 7.5 min.
The mass spectrometer was operated in full-scan (resolution, 70’0000; AGC target, 1e6; Maximum IT, 20ms) polarity switch mode with the spray voltage set to +/− 3.0 kV, the heated capillary held at 275C, and the HESI probe held at 350C. Seventeen labeled amino acids (MSK-A2–1.2, Cambridge Isotope Laboratories) were added to the extraction solvent (80% methanol, 20% water) and used as internal standards. Standard curves were performed for each 25 amino acids. Acetyl-aminoadipate was synthesized using recombinant BAR as described below and all 24 other amino acids were purchased from Sigma-Aldrich. Data analysis was performed with Xcalibur (Thermo Scientific). Note that values for a few amino acids are shown as relative levels in Supplementary Fig. 2 because their concentrations in some samples were more than 10-fold higher than the highest concentration of the standard.
Absolute quantification of acetyl-aminoadipate and acetyl-tryptophan in seeds of Arabidopsis T-DNA mutants and various tissues of Glycine max and Brassica napus
Metabolites were extracted as described above and then analyzed on an Ultimate 3000 Rapid Separation LC system (Thermo Scientific) coupled to a TSQ Quantum Access MAX triple-quadrupole mass spectrometer (Thermo Scientific). The reverse-phase chromatography system consisted of an 150 mm C18 column (Kinetex 2.6 μm silica core shell C18 100Å pore, Phenomenex) which was developed using Optima™ LC/MS solvents (Fisher Chemical) with a gradient (flow rate of 0.6 mL min−1) of solvent B (acetonitrile with 0.1% (v/v) formic acid) in solvent A (water with 0.1% (v/v) formic acid) as follows (all (v/v)): 2% for 3 min, 2% to 99% in 9 min, 99% for 4 min, 99% to 2% in 1 min and 2% for 1 min. The mass spectrometer was configured to perform two selected-reaction-monitoring scans, each for 0.5 seconds, for acetyl-aminoadipate and acetyl-tryptophan. The m/z resolution of Q1 was set to 0.4 FWHM, the nitrogen collision gas pressure of Q2 was set to 1.5 mTorr, and the Q3 scan width was set to 0.500 m/z in both cases. Selected reaction monitoring for acetyl-aminoadipate was as follows: precursor ion selection at 204.086 m/z on positive ion mode, fragmentation at 10 V, and product ion selection at 144.065 m/z. Selected reaction monitoring for acetyl-tryptophan was as follows: precursor ion selection at 247.107 m/z on positive ion mode, fragmentation at 20 V, and product ion selection at 188.070 m/z. Acetyl-aminoadipate was synthesized using recombinant BAR as described below and used as standard. Pure acetyl-tryptophan was purchased from Sigma-Aldrich.
Heterologous expression of wild-type BAR and activity determination
The BAR coding sequence was amplified by PCR (KaPa HiFi HotStart polymerase; KaPa Biosystems) from genomic DNA extracted from homozygous plants of the SAIL line SAIL_1165_B02 using primers SAIL_BAR_F_pPROEX and SAIL_BAR_R_pPROEX (see Table S3) and then cloned into pProEX Hta (Invitrogen) via EcoRI and HindIII resulting in a 6×His-BAR fusion construct.
6×His-tagged BAR protein was expressed in E. coli BL21(DE3) grown in Terrific Broth medium. At an optical density at 600 nm of 0.6, protein expression was induced with 1.0 mM IPTG and cells were grown at 37°C for 2.5 h. Cells from 1 L culture were harvested by centrifugation and resuspended in 25 mL binding buffer (50 mM Tris-HCl pH 8, 500 mM NaCl, 30 mM imidazole). All the following steps were carried out at 4°C. Cell lysis was performed using a microfluidizer (HC-8000, Microfluidics). The lysate was centrifuged (16,000 g) for 20 min, and the 6×His-tagged BAR protein was purified by metal affinity (5-ml HisTrap HP column, GE Healthcare) and size-exclusion chromatography (HiLoad 16/600 Superdex 200 pg, GE Healthcare) using an ÄKTA Pure FPLC system (GE Healthcare). The 6×His-TEV tag was removed from BAR prior to size-exclusion chromatography by overnight incubation with 1 μg of 6×His-TEV protease37 per 10 μg protein in 50 mM Tris-HCl pH 8, 500 mM NaCl, 1 mM dithiothreitol, followed passage through HisTrap HP column. Purified recombinant BAR was dialyzed in storage buffer (12.5 mM Tris-HCl pH 8, 50 mM NaCl, 2 mM dithiothreitol) and concentrated to 13 mg/mL using an ultra-centrifugal filter (10,000 Da MWCO, Amicon EMD Millipore). The purity of recombinant BAR was assessed by SDS-PAGE (Supplementary Fig. 6a). Purified BAR was aliquoted, snap-frozen in liquid nitrogen and stored at −80°C until further use.
Enzyme assays were carried out in 2 mM Tris-HCl pH 8 and 10 mM acetyl-CoA (Sigma-Aldrich; final volume 25 μl). Before determining the kinetics of BAR with different substrates, time-dependent activity of the purified protein was tested at substrate concentrations of 500 μM L-phosphinothricin (glufosinate ammonium, considered as a 1:1 mixture of L- and D- enantiomers; Sigma-Aldrich) or 1 mM (L-aminoadipate and L-tryptophan; Sigma-Aldrich). Reactions were initiated by the addition of purified BAR at 0.26 μM (assays with L-phosphinothricin) or 150 μM (assays with aminoadipate or tryptophan) and incubated at 25°C for the indicated times (Supplementary Fig.3 b–d). Reactions were stopped by the addition of four volumes of 10% water, 90% acetonitrile (v/v), 5 mM ammonium formate pH 3. Likewise, substrate concentration-dependence was determined by incubating assays for 25 min (assays with L-phosphinothricin), 3 h (assays with aminoadipate) or 7 h (assays with tryptophan; Fig. 3). Stock solutions of aminoadipate and tryptophan at 60 mM were made in 2 mM Tris-HCl pH 8 supplemented with 1 mM N-nonyl β-D-glucopyranoside and substrate concentration-dependence assays employing these two substrates contained 0.33 mM N-nonyl β-D-glucopyranoside. Control assays (Fig. 3) were performed with aminoadipate and tryptophan at 20 mM, but in the absence of BAR.
The assays were analyzed on an Ultimate 3000 Rapid Separation LC system (Thermo Scientific) coupled to a TSQ Quantum Access MAX triple-quadrupole mass spectrometer (Thermo Scientific). Assays on phosphinothricin were analyzed as follows. The normal-phase chromatography system consisted of an 150 mm HILIC column (Kinetex 2.6 μm silica core shell HILIC 100Å pore, Phenomenex), which was developed using Optima™ LC/MS solvents (Fisher Chemical) with a gradient (flow rate of 0.8 mL min−1) of solvent B (50% water, 50% acetonitrile (v/v), 5 mM ammonium formate pH 3) in solvent A (10% water, 90% acetonitrile (v/v), 5 mM ammonium formate pH 3) as follows (all (v/v)): 0% for 2 min, 0% to 70% in 10 min, 70% to 100% in 30 sec, 100% for 90 sec, 100% to 0% in 30 sec and 0% for 3.5 min. The mass spectrometer was configured to perform selected-ion-monitoring scans of 0.5 seconds using Q3 (center mass m/z: 224.068, scan width 1.0 m/z, scan time 0.5 sec). Assays on aminoadipate and tryptophan were analyzed as described above for the absolute quantification of acetyl-aminoadipate and acetyl-tryptophan in planta. Product formation was quantified using standards synthesized using recombinant BAR (acetyl-phosphinothricin and acetyl-aminoadipate) or commercially available (acetyl-tryptophan, Sigma-Aldrich). Km and Vmax value for phosphinothricin were inferred using the Michaelis-Menten kinetics nonlinear regression function under Prism 6 (GraphPad).
X-ray crystallography
Purified BAR protein was incubated with 1 mM acetyl-CoA for >2 hour prior to setting crystal trays. Crystals of BAR were obtained after 3 days at 20 °C in hanging drops containing 1 μL of protein solution (7.5 mg/mL) and 1 μL of reservoir solution (0.18 M calcium acetate, 0.1 M Tris-HCl pH 7, 18% (w/v) PEG 3000, 0.2% (v/v) N-nonyl β-D-glucopyranoside, 1 mM acetyl-CoA). Several crystals were soaked in reservoir solution supplemented with 30 mM L-phosphinothricin for 30–60 min before freezing. Crystals were frozen in reservoir solution supplemented with 15% (v/v) ethylene glycol. Acetylation of phosphinothricin occurred during soaking as no density for the acetyl group of acetyl-CoA was observed in the BAR/CoA/phosphinothricin ternary complex.
X-ray diffraction data were collected on the 24-ID-C beam line of the Structural Biology Center at the Advanced Photon Source (Argonne National Laboratory) equipped with a Pixel Array Detector (Pilatus-6MF). Diffraction intensities were indexed, integrated, and scaled with the iMosflm38 and SCALA39 programs. Initial phases were determined by molecular replacement using Phaser under Phenix40. The search model was an ensemble model generated with Ensembler using 8 protein structures homologous to BAR (PBD codes and % identity to BAR: 2JLM (28%), 3DR8 (35%), 4J3G (31%), 4JXQ (33%), 4MBU (30%), 1VHS (30%), 1YR0 (29%) and 1YVO (35%)). Subsequent structural building and refinements utilized Phenix programs (TSL was used in early rounds of refinement)40. Coot was used for graphical map inspection and manual rebuilding of atomic models41. Crystallographic calculations were performed using Phenix. Molecular graphics were produced with the program PyMol.
Heterologous expression of BAR mutants and activity determination
Single amino acid mutants of BAR were generated using the QuikChange II site-directed mutagenesis kit (Agilent Technologies) and 6×His-BAR in pProEX Hta as template (see Supplementary Table 3 for primer sequences). PAT from Streptomyces viridochromogenes was amplified using primers BAC0327 and BAC0328 from pAG31 vector42 (Addgene 35124) and cloned into BamHI/HindIII-linearized pProEX Hta by Gibson assembly (New England Biolabs). Wild-type 6×His-BAR, 6×His-BAR mutants and 6×His-PAT were expressed in E. coli BL21(DE3) grown in Terrific Broth medium. At an optical density at 600 nm of 0.6, protein expression was induced with 1.0 mM IPTG and cells were grown at 37°C for 2.5 h. Cells from a 150 mL cultures were harvested by centrifugation, lysed using B-PER™ Bacterial Protein Extraction Reagent (Thermo Scientific) and purified by metal affinity using Ni-NTA Agarose (Qiagen). Purified recombinant proteins were concentrated and buffer-exchanged using storage buffer (10 mM Tris-HCl pH 8.0, 0.2 M NaCl, 10% (v/v) glycerol, 1 mM dithiothreitol) and ultra-centrifugal filters (10,000 Da MWCO, Amicon EMD Millipore). The purity of the recombinant proteins was assessed by SDS-PAGE. Final protein concentrations were determined and normalized using a NanoDrop 2000 UV-VIS spectrometer (extinction coefficient: 43430 M−1 cm−1, Thermo Scientific).
Enzyme assays for comparing the relative activity of the purified BAR mutants were carried out in 2 mM Tris-HCl pH 8 and 5 mM acetyl-CoA (Sigma-Aldrich) (final reaction volume 12 μL). Reactions were initiated by the addition of purified recombinant protein at 0.2 μM (assays with L-phosphinothricin at 0.2 mM) or 150 μM (assays with aminoadipate or tryptophan at 1 mM) and incubated at 25°C for 15 min (phosphinothricin), 165 min (aminoadipate), or 330 min (L-tryptophan). Substrate concentration-dependences toward phosphinothricin, aminoadipate and tryptophan were determined for the BAR mutants Y92F and N35T in 2 mM Tris-HCl pH 8 and 10 mM acetyl-CoA (Sigma-Aldrich). Note that assays on aminoadipate and tryptophan were supplemented with 0.33 mM of N-nonyl β-D-glucopyranoside (see also above). Reactions were stopped by the addition of four volumes of 10% water, 90% acetonitrile (v/v), 5 mM ammonium formate pH 3, centrifuged for 2 min (14,000–16,000 g), and transferred to LC vials.
The assays were analyzed on an Ultimate 3000 Rapid Separation LC system (Thermo Scientific) coupled to a TSQ Quantum Access MAX triple-quadrupole mass spectrometer (Thermo Scientific). Assays on phosphinothricin were analyzed as described above. Assays on aminoadipate were analyzed as follows. The reverse-phase chromatography system consisted of an 150 mm C18 column (Kinetex 2.6 μm silica core shell C18 100Å pore, Phenomenex), which was developed using Optima™ LC/MS solvents (Fisher Chemical) with a gradient (flow rate of 0.6 mL min−1) of solvent B (acetonitrile with 0.1% (v/v) formic acid) in solvent A (water with 0.1% (v/v) formic acid) as follows (all v/v): 1% for 2 min, 1% to 30% in 9 min, 30% to 99% in 30 sec, 99% for 30 sec, 99% to 1% in 1 min and 1% for 2 min. The mass spectrometer was configured to perform selected-ion-monitoring scans of 0.5 seconds using Q3 (center mass m/z: 204.086, scan width 0.5 m/z, scan time 0.5 sec). Assays on tryptophan were analyzed as follow: the reverse-phase chromatography system consisted of an 150 mm C18 column (Kinetex 2.6 μm silica core shell C18 100Å pore, Phenomenex) which was developed using Optima™ LC/MS solvents (Fisher Chemical) with a gradient (flow rate of 0.7 mL min−1) of solvent B (acetonitrile with 0.1% (v/v) formic acid) in solvent A (water with 0.1% (v/v) formic acid) as follows (all v/v): 5% for 1 min, 5% to 99% in 9 min, 99% for 2 min, 99% to 5% in 2 min and 5% for 1 min. The mass spectrometer was configured to perform selected-ion-monitoring scans of 0.5 seconds using Q3 (center mass m/z: 247.108, scan width 0.5 m/z, scan time 0.5 sec).
Analysis of BAR mutants in planta
Wild-type BAR from Streptomyces hygroscopicus, selected BAR mutants and wild-type PAT from Streptomyces viridochromogenes were amplified by PCR (Phusion polymerase; New England Biolabs) from pProEX Hta clones (see above) using primers listed in Table S3 and cloned into BpiI-linearized pICH4130843 (Golden Gate entry vector) by Gibson assembly (New England Biolabs). BAR and PAT coding sequences were fused with Agrobacterium tumefaciens mannopine synthase promoter (from pICH85281) and terminator (from pICH77901) into the empty binary vector pICH47732 by Golden Gate assembly43. pICH47732 constructs were transformed into Agrobacterium tumefaciens GV3130 strain by electroporation and transformed into Arabidopsis Col-0 by the floral dip method44. 90 mg of T1 seeds were sown on soil and transformants were selected with Finale® (contains 11.33% glufosinate ammonium; Bayer CropScience) diluted 1:500 in water. Photographs were taken 10 days after herbicide treatment (Fig. 4 and Supplementary Fig. 12). This experiment was repeated once with similar results. T2 seeds from 5 to 6 T1 plants were collected for each BAR mutants, sown on soil and transgenic individuals were selected with Finale® (contains 11.33% glufosinate ammonium; Bayer CropScience) diluted 1:500 in water (Supplementary Fig. 13). This experiment was done once. Metabolites were extracted from dark-incubated leaves collected from T2 phosphinothricin-resistant individuals (senescent leaves from 8–9 individuals were pooled for each T2 population) and then analyzed as described above for the absolute quantification of acetyl-aminoadipate and acetyl-tryptophan in Glycine max and Brassica napus.
To further compare the phosphinothricin tolerance in T2 lines transformed with Y92F, N35T and wild-type BAR, seeds from 5–6 independent lines were germinated on ½ MS medium containing 1% sucrose and 8 μg/mL glufosinate ammonium (45520-Sigma-Aldrich). Seven-days old seedlings were then transformed on soil and further grown for 10 days. Photographs were taken before treatment with four different concentrations of Finale® (0, 0.2X, 1X and 5X; see Supplementary Fig. 14 for further details on the herbicide concentrations). Plants were further grown for 8 days, photographs were taken and the average aerial mass of each T2 populations was measured (average from 8–9 individuals). This experiment was done once.
Protein levels of the BAR mutants in T2 lines were measured as follow. For each protein extraction, equal amounts of aerial tissues from 5–6 transgenic T2 populations were pooled. Total proteins were isolated from frozen samples by homogenization in 5 volumes of ice-cold extraction buffer [50 mM Tris–HCl pH 8, 100 mM NaCl, 0.5% (v/v) TritonX-100, 2mM β-mercaptoethanol] complemented with a protease inhibitor cocktail (Complete; Roche Diagnostics). Samples were centrifuged at 12,000 g for 5 min and protein concentration of the supernatant was determined using the Bradford Assay (Bio-Rad). Proteins were subsequently precipitated with chloroform–methanol and 10 μg were analyzed by SDS-PAGE and immunoblotting as described45. The following antibodies were used for immunoblot analysis: a primary polyclonal antibody against BAR from Streptomyces hygroscopicus produced in rabbit (1:1000; P0374-Sigma-Aldrich) and a polyclonal horseradish peroxidase conjugated goat anti-rabbit IgG as the secondary antibody (1:50000; A0545-Sigma-Aldrich). Substrate detection was performed by chemiluminescence (ECL Western Blotting Substrate™ (Pierce)) and film exposure. This experiment was done once.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. M. Sabatini, G. R. Fink, N. Amrhein and E. Martinoia for helpful discussions. We thank J. M. Cheeseman for providing the phosphinothricin-resistant Glycine max line. We thank J. Varberg for providing the phosphinothricin-resistant Brassica napus line and M. Rahman for providing conventional Brassica napus lines. This work is based on research conducted at the Northeastern Collaborative Access Team (NE-CAT) beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). The Pilatus 6M detector on NE-CAT 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02–06CH11357. This work was supported by the Swiss National Science Foundation (grant 31003A_149389 to S.H. and postdoctoral fellowship P2ZHP3_155258 to B.C.), the EU-funded Plant Fellows program (S.A.), the Pew Scholar Program in the Biomedical Sciences (J.K.W.) and the Searle Scholars Program (J.K.W.).
Footnotes
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
COMPETING FINANCIAL INTERESTS
B.C. and J.K.W. have filed a patent application on BAR and PAT mutants described in this paper that show altered acetyltransferase activity.
REFERENCES
- 1.Wehrmann A, Van Vliet A, Opsomer C, Botterman J & Schulz A The similarities of BAR and PAT gene products make them equally applicable for plant engineers. Nat. Biotechnol 14, 1274–1278 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Duke SO Taking stock of herbicide-resistant crops ten years after introduction. Pest. Manag. Sci 61, 211–218 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Thompson CJ et al. Characterization of the herbicide-resistance gene BAR from Streptomyces hygroscopicus. EMBO J 6, 2519–2523 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wohlleben W et al. Nucleotide sequence of the phosphinothricin N-acetyltransferase gene from Streptomyces viridochromogenes Tü494 and its expression in Nicotiana tabacum. Gene 70, 25–37 (1988). [DOI] [PubMed] [Google Scholar]
- 5.Ren YF et al. A comparative proteomics approach to detect unintended effects in transgenic Arabidopsis. J Genet Genomics 36, 629–639 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Brown RH, Raboy V & Bregitzer P Unintended consequences: high phosphinothricin acetyltransferase activity related to reduced fitness in barley. In Vitro Cellular & Developmental Biology - Plant 49, 240–247 (2013). [Google Scholar]
- 7.The National Academies. Genetically Engineered Crops: Experiences and Prospects (National Academies Press, 2016). [PubMed] [Google Scholar]
- 8.Schenk N et al. The chlorophyllases AtCLH1 and AtCLH2 are not essential for senescence-related chlorophyll breakdown in Arabidopsis thaliana. FEBS Lett 581, 5517–5525 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Samson F et al. FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res 30, 94–97 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sessions A et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alonso JM et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Rosso MG et al. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol 53, 247–259 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Soudry E, Ulitzur S & Gepstein S Accumulation and remobilization of amino acids during senescence of detached and attached leaves: in planta analysis of tryptophan levels by recombinant luminescent bacteria. J. Exp. Bot 56, 695–702 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Hörtensteiner S & Feller U Nitrogen metabolism and remobilization during senescence. J. Exp. Bot 53, 927–937 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Arruda P, Kemper EL, Papes F & Leite A Regulation of lysine catabolism in higher plants. Trends Plant Sci 5, 324–330 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Zhu X, Tang G, Granier F, Bouchez D & Galili G A T-DNA insertion knockout of the bifunctional LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE gene elevates lysine levels in Arabidopsis seeds. Plant Physiol 126, 1539–1545 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vinnemeier J, DrogeLaser W, Pistorius EK & Broer I Purification and partial characterization of the Streptomyces viridochromogenes Tü494 phosphinothricin-N-acetyltransferase mediating resistance to the herbicide phosphinothricin in transgenic plants. Z. Naturforsch. C 50, 796–805 (1995). [Google Scholar]
- 18.Dyda F, Klein DC & Hickman AB GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Bioph. Biom 29, 81–103 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vetting MW et al. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys 433, 212–226 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Srivastava P et al. Structural characterization of a GCN5-related N-acetyltransferase from Staphylococcus aureus. PLoS ONE 9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rojas JR et al. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature 401, 93–98 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Woolston BM, Edgar S & Stephanopoulos G Metabolic engineering: past and future. Annu. Rev. Chem. Biomol. Eng 4, 259–288 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Weng JK & Noel JP The remarkable pliability and promiscuity of specialized metabolism. Cold Spring Harbor symposia on quantitative biology 77, 309–320 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Jin Y-S et al. Chemical and biologically active constituents of Salsola collina. Chem. Nat. Compd 47, 257–260 (2011). [Google Scholar]
- 25.Yu P, Hegeman AD & Cohen JD A facile means for the identification of indolic compounds from plant tissues. Plant J 79, 1065–1075 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Bruckhoff V et al. Functional characterization of CYP94-genes and identification of a novel jasmonate catabolite in flowers. PLoS One 11, e0159875 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green JM & Owen MD Herbicide-resistant crops: utilities and limitations for herbicide-resistant weed management. J. Agric. Food Chem 59, 5819–5829 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herouet C et al. Safety evaluation of the phosphinothricin acetyltransferase proteins encoded by the PAT and BAR sequences that confer tolerance to glufosinate-ammonium herbicide in transgenic plants. Regul. Toxicol. Pharm 41, 134–149 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Dan Y Plant transformation technology revolution in last three decades: historical technology developments (Bentham Science Publishers, 2012). [Google Scholar]
- 30.Brown RH, Raboy V & Bregitzer P Unintended consequences: high phosphinothricin acetyltransferase activity related to reduced fitness in barley. In Vitro Cell Dev-Pl 49, 240–247 (2013). [Google Scholar]
- 31.Song WY, Choi KS, Alexis de A, Martinoia E & Lee Y Brassica juncea plant cadmium resistance 1 protein (BjPCR1) facilitates the radial transport of calcium in the root. Proc. Natl. Acad. Sci. U.S.A 108, 19808–19813, 10.1073/pnas.1104905108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foetzki A et al. Surveying of pollen-mediated crop-to-crop gene flow from a wheat field trial as a biosafety measure. GM crops & food 3, 115–122 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Czechowski T, Stitt M, Altmann T, Udvardi MK & Scheible WR Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant physiology 139, 5–17 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chambers MC et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol 30, 918–920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gowda H et al. Interactive XCMS Online: simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal. Chem 86, 6931–6939 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith CA et al. METLIN: a metabolite mass spectral database. Ther. Drug Monit 27, 747–751 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Tropea JE, Cherry S & Waugh DS Expression and purification of soluble His(6)-tagged TEV protease. Methods Mol. Biol 498, 297–307 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Battye TG, Kontogiannis L, Johnson O, Powell HR & Leslie AG iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D 67, 271–281 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans P Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Goldstein AL & McCusker JH Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541–1553 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Engler C et al. A Golden Gate modular cloning toolbox for plants. ACS Synth. Biol (2014). [DOI] [PubMed] [Google Scholar]
- 44.Clough SJ & Bent AF Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Schelbert S et al. Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21, 767–785 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christ B et al. MES16, a member of the methylesterase protein family, specifically demethylates fluorescent chlorophyll catabolites during chlorophyll breakdown in Arabidopsis. Plant physiology 158, 628–641 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guyer L Characterization of dephytylation and dechelation, two early steps of chlorophyll breakdown in leaves and fruits PhD thesis, Zurich, (2015). [Google Scholar]
- 48.Christ B et al. Cytochrome P450 CYP89A9 is involved in the formation of major chlorophyll catabolites during leaf senescence in Arabidopsis. Plant Cell 25, 1868–1880 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez-Perez JM et al. Functional redundancy and divergence within the Arabidopsis RETICULATA-RELATED gene family. Plant physiology 162, 589–603 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Christ B Chlorophyll breakdown: modifications of colorless chlorophyll catabolites PhD thesis, Zurich, (2013). [Google Scholar]
- 51.Zufferey M et al. The novel chloroplast outer membrane kinase KOC1 is a required component of the plastid protein import machinery. Journal of Biological Chemistry 292, 6952–6964 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pulido P, Llamas E & Rodriguez-Concepcion M Both Hsp70 chaperone and Clp protease plastidial systems are required for protection against oxidative stress. Plant Signal Behav 12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waterhouse AM, Procter JB, Martin DM, Clamp M & Barton GJ Jalview Version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.