Abstract
Objectives:
Finding recent clinical studies that warrant changes in clinical practice (“high impact” clinical studies) in a timely manner is very challenging. We investigated a machine learning approach to find recent studies with high clinical impact to support clinical decision making and literature surveillance.
Methods:
To identify recent studies, we developed our classification model using time-agnostic features that are available as soon as an article is indexed in PubMed®, such as journal impact factor, author count, and study sample size. Using a gold standard of 541 high impact treatment studies referenced in 11 disease management guidelines, we tested the following null hypotheses: 1) the high impact classifier with time-agnostic features (HI-TA) performs equivalently to PubMed’s Best Match sort and a MeSH-based Naïve Bayes classifier; and 2) HI-TA performs equivalently to the high impact classifier with both time-agnostic and time-sensitive features (HI-TS) enabled in a previous study. The primary outcome for both hypotheses was mean top 20 precision.
Results:
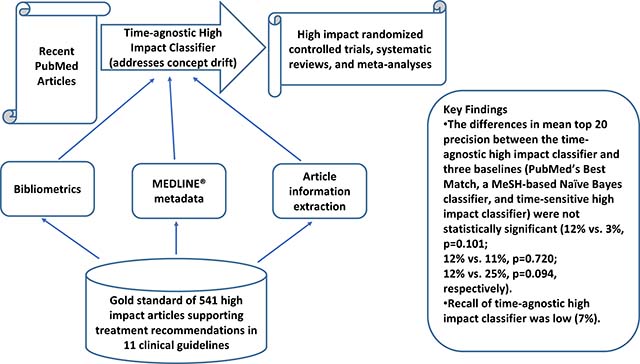
The differences in mean top 20 precision between HI-TA and three baselines (PubMed’s Best Match, a MeSH-based Naïve Bayes classifier, and HI-TS) were not statistically significant (12% vs. 3%, p=0.101; 12% vs. 11%, p=0.720; 12% vs. 25%, p=0.094, respectively). Recall of HI-TA was low (7%).
Conclusion:
HI-TA had equivalent performance to state-of-the-art approaches that depend on time-sensitive features. With the advantage of relying only on time-agnostic features, the proposed approach can be used as an adjunct to help clinicians identify recent high impact clinical studies to support clinical decision-making. However, low recall limits the use of HI-TA for literature surveillance.
Graphical Abstract
1. Introduction
Unmet clinical information needs at the point of care is an important challenge in medical practice [1,2]. Despite substantial advances in information retrieval technology and wide availability of online evidence-based resources, over half of the questions clinicians raise regarding scientific evidence for the care of specific patients are left unanswered [1,2]. These unanswered questions may compromise clinical decision making and patient care quality [3]. Multiple barriers exist that prevent clinicians from pursuing answers to their questions with evidence from the up-to-date primary literature, such as lack of time and doubt that a useful answer exists or can be easily found [2]. As the main resource providing access to the primary literature in health care, PubMed contains answers to most clinical questions [4], but its use at the point of care is still limited due to several challenges [5,6]. One of these challenges is finding reports of studies with a high clinical impact in a timely manner [7]. High impact clinical studies are not only scientifically sound studies but also provide evidence warranting changes in routine clinical practice. Identifying recent scientifically sound and high impact clinical studies is not only a key requirement for literature surveillance efforts [8] but also an essential step for clinicians to adopt evidence-based practice. Literature surveillance is used to update evidence summaries, such as systematic reviews and clinical guidelines [9–11] and could be used to help translate clinical research evidence to routine clinical practice. Also, early prediction of article impact may help clinicians become aware of latest evidence that may warrant changes in clinical practice as soon as it is published, as opposed to having to wait years for relevant accurate predictors to become available.
Previous efforts have investigated methods focused on retrieving reports of scientifically sound studies [12–17]. However, little has been done to identify reports of high impact clinical studies. In a previous study, we developed classification models to automatically identify high impact clinical studies in PubMed using a combination of bibliometrics, social media attention, and citation metadata features [18]. An important limitation of most previous approaches is the reliance on time-sensitive features that take from several months to years after the article publication to be available, such as citation counts and MeSH terms.
In the present study, we proposed a machine learning approach to identify high impact clinical studies right after they are included in PubMed. We build over our previous work using the same gold standard and leveraging several of the features that we previously investigated [18]. However, to classify recently published articles, we only used features that are static or do not change significantly over time, such as journal impact factor, author count, study sample size, funding, and the reputation of the authors’ institutions. We compared this time-agnostic classification model (HI-TA) with prevalent approaches and with our previous classifier that uses time-sensitive features (HI-TS).
2. Background
2.1. Previous related approaches
Previous approaches to retrieving scientifically sound studies fall into three main categories: i) search filters [12,13], ii) citation-based algorithms [14], and iii) supervised machine learning algorithms [15–17]. Search filters include the Clinical Query filters developed by Haynes et al., which are available in PubMed [12,13]. Clinical Queries are Boolean search strategies relying on keywords and MeSH terms that aim to retrieve scientifically sound studies. Filters are available for topics such as treatment, diagnosis, and prognosis; and can be tuned for recall or precision. Citation-based algorithms rely on citation counts and linkage. For example, Bernstam et al. investigated citation count and PageRank methods that rely on the linkage (i.e., citations) analysis between studies [14]. The citation count method only considers one-layer of citation relationships, whereas the PageRank method considers multiple layers. They found that citation-based algorithms outperformed Clinical Queries (precision = 6% versus 0.85%) on retrieving oncology studies included in the Society of Surgical Oncology’s Annotated Bibliography (SSOAB) [14]. Supervised machine learning algorithms include approaches by Aphinyanaphongs et al. and Kilicoglu et al. [15–17]. The former found that a support vector machine classifier with features such as MeSH terms, publication type, and text words outperformed Clinical Queries in retrieving etiology (recall = 0.76 versus 0.28) and treatment (recall = 0.80 versus 0.40) articles contained in the American College of Physicians (ACP) journal club [16]. In addition to the features employed by Aphinyanaphongs et al., Kilicoglu et al. investigated semantic features, such as Unified Medical Language System (UMLS) concepts, UMLS semantic relations, and semantic predications. The classifier with the highest F-measure in retrieving scientifically sound treatment studies from the Clinical Hedges database[19] obtained a precision and recall of 82.5% and 84.3% respectively [15].
Of the previous approaches above, both the Clinical Query filters and the machine learning studies were designed to retrieve scientifically sound studies regardless of clinical impact. On the other hand, the citation-based approach developed by Bernstam et al. can be used to identify high impact clinical studies since it is based on article popularity. In prior work, we investigated machine learning classifiers that combine features used in previous approaches to identify high impact treatment studies. Features included bibliometrics (i.e., journal impact factor and article citation count), social media attention (Altmetric® score [20]), and PubMed metadata (MeSH terms and publication type). Using a gold standard of clinical studies providing evidence to support treatment recommendations in 11 clinical guidelines, our high impact classifier (HI-TS) outperformed Kilicoglu et al.’s Naïve Bayes classifier and PubMed’s relevance sort in terms of top 20 precision (mean = 34% vs. 11% vs. 4% respectively; both p = 0.009). The performance did not decrease significantly after removing proprietary features (i.e., citation count, Altmetric score) (mean top 20 precision = 34% vs. 36%; p = 0.085) [18].
2.2. Time-sensitive features and their limitations
All previous approaches are susceptible to concept drift, a significant and prevalent problem that compromises the model performance due to changes in the model features and/or outcomes over time [21,22]. Concept drift can be addressed by specific techniques, such as periodically updating a prediction model with more recent data or using concept drift learning techniques [23]. However, concept drift is more difficult to address in situations where specific features do not become available for a certain period of time, such as citation counts and MeSH terms.
There are two types of concept drift: sudden and gradual [24]. Methods that depend on MeSH terms and publication type, such as Clinical Query filters [12,13], MeSH-based supervised machine learning algorithms[15–17], and our previous high impact classifier, [18] are susceptible to sudden concept drift, since MeSH terms and publication type are added to PubMed citations 23 to 177 days after the citation is added to PubMed [25]. Once MeSH terms and publication type are added to a citation, its classification output may suddenly change. On the other hand, citation-based algorithms [14] and our previous high impact classifier [18] are susceptible to gradual concept drift because of their dependency on features that change gradually over time such as citation count and journal impact factor. As an article initially classified as “negative” accrues citations, its classification may slowly move towards the “positive” threshold. Citation accrual rate peaks between two to six years after an article is published and then decreases [26]. Besides having to handle a non-linear citation growth pattern, approaches that rely on citation counts may have a substantial time lag until an article starts receiving its first set of citations allowing to compute an accurate citation growth rate. Journal impact factors are based on the number of citations received by the articles published in a particular journal and can be used as a feature for an article’s future citation count. Although impact factors also change over time, most changes are negligible (maximum of ±3.0 between 2013 and 2014 for a group of Journal Citation Reports (JCR) journals [27]) and classification models can use the impact factor at publication time as the classification input for a certain article.
3. Methods
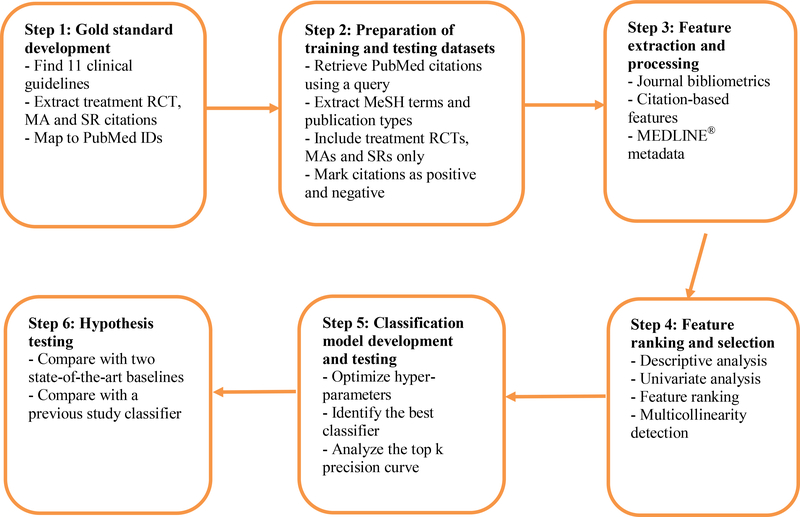
To address the concept drift challenges described above, we investigated an approach (HI-TA) to achieve the most accurate classification model using time-agnostic features that are stable and readily available at article publication time, including author, author affiliation, and references. We also used several features as surrogates for article citation counts, including SJR (SCImago Journal Rank) indicator, SJR journal h-index, and citations per document for the past two years [28]. We developed our study approach in six steps (Figure 1): 1) gold standard development; 2) training and testing datasets preparation; 3) feature extraction and preprocessing; 4) feature ranking and selection; 5) classification model development and testing; and 6) hypothesis testing.
Figure 1 –
Overview of the study method.
3.1. Gold standard development
For positive cases, we used a gold standard developed in a previous study [18], which consists of 629 original clinical studies providing evidence that supports recommendations in 11 clinical guidelines on the treatment of cardiac, autoimmune and respiratory diseases. The study citations and PubMed IDs were extracted from the reference lists of each guideline. Clinical practice guidelines contain evidence-based recommendations for the clinical management of specific conditions. The studies that support clinical guideline recommendations are selected by an expert panel through a systematic literature search, relevance screening and quality appraisal process [29]. Therefore, articles cited in clinical guidelines can be considered the most important in a specific topic and thus are reasonable surrogates for high impact clinical studies. In this research, we focused on treatment studies because most of the questions clinicians raise at the point of care are about treatment [2].
3.2. Preparation of training and testing datasets
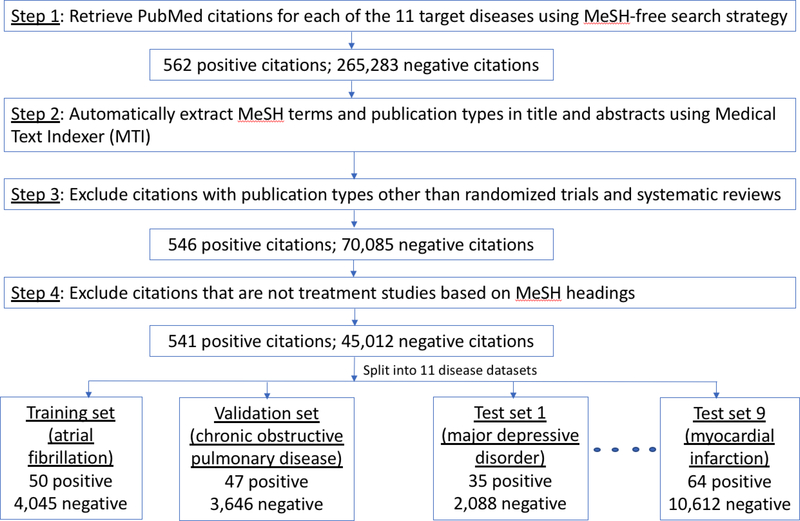
At the time of article publication, time-sensitive features are not available. However, the articles included in the dataset used in this study were published a few years ago, so time-sensitive features, such as MeSH and publication type, are now available for these articles. Since the goal of our study is to find recently published high impact clinical articles, we had to create an experimental environment that reproduces the status of article features soon after their publication. The overall goal is to predict which articles will likely produce a high impact over time before any impact actually happens. The approach consisted of four-steps (Figure 2). First, to find potentially relevant citations for each disease covered in the 11 guidelines, we used PubMed search strategies without time-sensitive terms (i.e., MeSH terms, publication type) that are not readily available once an article is added to PubMed. Each search strategy included three components: a set of keywords representing synonyms for the disease of interest, limited to the article title and abstract; a date filter, matching the time range of the systematic literature search that has been conducted as a part of each guideline development; and other filters, such as citations with an abstract and written in English [30]. Keywords were associated with PubMed’s [Title/Abstract] tag to suppress automatic MeSH term mapping, which is conducted by default in PubMed searches (see example in Box 1; other searches are available in the online supplement, Boxes s1-s10). This search strategy allowed us to stimulate article retrieval before MeSH terms are available. Second, we used the Medical Text Indexer (MTI) to automatically extract MeSH terms and publication types from the titles and abstracts of the retrieved citations. MTI is an automated indexing service for medical text provided by the National Library of Medicine (NLM) [31]. The precision and recall of MTI in 2014 was 60% and 56%, respectively [31]. Third, we selected from the articles retrieved in Step 1, only those with a publication type reflecting high quality scientific methods for clinical research (i.e., randomized controlled trials, systematic reviews) and those with a MeSH subheading related to treatment (i.e., [therapeutic use] and [therapy] MeSH terms along with their hierarchical descendants) [32]. Last, citations in the dataset were marked as “positive” if included in the gold standard and negative otherwise. The resulting dataset consisted of 11 sub datasets with relevant citations for each of the diseases covered in the clinical guidelines.
Figure 2.
Article retrieval process with descriptive statistics of intermediate and final datasets.
Box 1: PubMed search strategy for retrieving relevant citations on the treatment of chronic obstructive pulmonary disease.

3.3. Feature extraction and processing
We used a combination of features investigated in our previous study and a set of new features, including journal bibliometrics, PubMed metadata, and citation-based features. All the features are time-agnostic. Table 1 describes the features used in HI-TA, the steps to obtain each feature, and whether or not the feature is also used in HI-TS. These features were extracted from a variety of resources through a set of Java programs and stored in a local relational database.
Table 1.
Classification features used in HI-TA to identify high impact clinical articles on disease treatment.
| Feature name | Feature description | Data Type | Used in HI-TS |
|---|---|---|---|
| Number of clinically useful sentences | Clinicians prefer sentences that provide patient-specific, actionable recommendations for a particular intervention [33–37]. Clinical useful sentences in the citation title and abstract were identified using a sentence classifier developed in a previous study [38]. | Numeric | No |
| Journal impact factors | Journal impact factors (JIF) are measures of the reputation and Numeric impact of a journal. In general, they are calculated as the ratio between the number of citations received by articles published in the journal and the number of articles published in the journal during a certain period of time (e.g., two years) [39,40]. We retrieved the JIFs for each citation at the time of its publication. We obtained JIFs from the Scimago Journal & Country Rank® (SJR®), developed by Scimago Lab with the data source provided by Scopus® [28]. We obtained three different JIFs from SJR: 1) SJR (SCImago Journal Rank) indicator, which represents the ratio of the weighted citation counts to the documents published in the journal of interest for the past three years [41]; 2) journal h-index, which represents the number of articles in the journal that received more than h citations; and 3) citations per document for the past two years. |
Numeric | Yes |
| Study sample size | Number of participants in the study. High impact clinical studies Numeric often have larger sample sizes [5]. The sample size was extracted using enhanced EasyCIE [42], a rule-based information extraction tool. This tool uses ConText algorithm [43] to identify the numbers within the context of sample size related description in abstracts. Then it applies predefined rules to solve the conflicts if there is any. For instance, if there are multiple numbers that are likely to be the sample size, it will choose the first one. We developed the rules base on 700 training abstracts randomly sampled from PubMed and evaluated on another 100 abstracts. We measured the performance in two metrics: the F1- score and the average numeric difference rate (the average of the normalized difference between the extracted sample sizes and the true sample sizes). The F1-score of the test set is 0.82, and the average numeric difference rate ((Z|Es-Ts|/Ts)/n, Es = extracted sample sizes, Ts = true sample size, n = number of abstracts) is 0.12. Analyses on the extracted sample sizes showed that a sample size of greater than 30000 or smaller than 10 are usually not the actual study sample size. We treated them as missing values of this feature. |
Numeric | Yes |
| Number of grants | Research shows that publications sponsored by grants have higher impact than studies without grant support [44]. We obtained the number of grants supporting a study using the Scopus API [45]. | Numeric | No |
| Number of authors | The number of authors is an independent predictor for the number of citations an article will receive [46]. We obtained this feature using the Scopus API [45]. | Numeric | No |
| Scientific impact of the authors’ institution | The overall scientific impact of the authors’ institution could be a surrogate for the impact of the authors’ work. We collected a snapshot of year 2017 for the following features both for the first author and the corresponding author: 1) total number of citations to publications from the first/corresponding author’s institution; 2) total number of authors from the first/corresponding author’s institution; 3) institution’s average citation count per author. In case an author had multiple affiliations, we used the institution with the highest reputation. We obtained all these features using the Scopus API [45]. | Numeric | No |
| Number of institutions and countries in a study | Multi-center studies are more likely to produce high impact. Collaboration helps better utilize resources and produces higher quality research [47]. In addition, collaborative studies receive more citations [47,48]. Multi-center studies may also have stronger design, such as larger sample sizes and more diverse subjects. We obtained the number of institutions and countries participating in a study using the Scopus API [45]. | Numeric | No |
| Number of bibliographic references | Research shows that article impact (based on citation count) is correlated with the number of bibliographic references included in the article [19,46,49,50]. We obtained this feature using the Scopus API [45]. | Numeric | No |
| Article page count and title word count | Research shows that article impact (based on citation count) is correlated with the article length [46] and title length [51,52]. We used the Scopus API [45] to obtain page count and a Java program to obtain the title word count. | Numeric | No |
| Core clinical journal | Represents whether the journal in which the study is published is part of a subset of core clinical journals. The list was obtained from the union of journals in the MEDLINE Core Clinical journals [53] and the McMaster Premium LiteratUre Service (Plus) journals [54]. Periodic evaluation and updates by experts ensure the quality of these lists [55–57]. | Categorical | Yes |
| Trial registration in ClinicalTrials.gov | Represents whether the study that produced the publication is registered in ClinicalTrials.gov. Registering a clinical trial in national registries, such as ClinicalTrials.gov, is required by funding agencies and by many of the core clinical journals. This feature was extracted from PubMed citation metadata using the eUtils API. | Categorical | Yes |
| Publication in PubMed Central® | Represents whether the article is included in the PubMed Central database. Studies funded by the US National Institutes of Health (NIH) are included in PubMed Central. They tend to be more balanced than commercial funded studies, which is an indication of strong clinical impact [58–60]. This feature was extracted from PubMed citation metadata using the eUtils API. | Categorical | Yes |
3.4. Feature ranking and selection
For feature ranking and selection, we conducted the following steps: 1) Descriptive analysis to obtain mean, standard deviation, and frequency of missing values, which helps us understand the distributions of our data and features. 2) Univariate analyses to determine the relationship between each feature and the outcome variable (i.e., article impact). The Chi-square test was used for categorical independent features and the Mann-Whitney U test was used for numerical features. We only kept the features that were significantly correlated with the outcome variable based on their 99% confidential interval. 3) Feature ranking using the Information Gain attribute evaluator with the Ranker search method in Weka [61]. We only kept the features that had an information gain score greater than zero. 4) Multicollinearity detection among features using variance inflation factors and a threshold of five [62]. In all these steps, we used the citations retrieved for atrial fibrillation (training dataset in classification model development) in the dataset whose number of citations was close to the average across diseases.
3.5. Classification model development and testing
We selected one disease for training (atrial fibrillation), one for validation (chronic obstructive pulmonary disease 2014), and the remainder for evaluation. The size and frequency of positive samples in the training and validation datasets were similar to the overall frequency across all datasets.
To identify an optimal classifier, we evaluated 12 classifiers with the above training and validation datasets: Bayesian Network, Decision Table, J48 (Decision Tree), K-Nearest Neighbor, Logistic Regression, Multilayer Perceptron (Neural Network), Naïve Bayes, Naïve Bayes Multinomial, Random Forest, Simple Logistic Regression, Stochastic Gradient Descent (SGD) and Support Vector Machine (SVM). We selected the optimal hyper-parameters for each classifier following the same method as in our previous study (Table 2) [18]. Overall, we employed an iterative and empirical process to tune the hyperparameters. We started with wide ranges for numeric hyperparameters and several categories for categorical hyperparameters, progressively narrowing the search to focus on the most promising ranges and categories.
Table 2.
- Classification algorithms and the parameters that were varied to identify optimal settings.
| Algorithm | Parameters varied to identify optimal settings |
|---|---|
| Bayesian Network | Search algorithm and estimator algorithm |
| Decision Table | Attribute selection method |
| J48 (decision tree) | Pruning confidence threshold and reduced error pruning |
| K-Nearest Neighbor | Number of nearest neighbors and neighbor weighting methods |
| Logistic Regression | Default parameter setting in Weka |
| Multilayer Perceptron | Learning rate |
| Naive Bayes | Kernel density estimator |
| Naive Bayes Multinomial | Default parameter setting in Weka |
| Random Forest | Number of features, number of trees, and the maximum depth of the trees |
| Simple Logistic | Default parameter setting in Weka |
| Stochastic Gradient Descent Loss function | |
| Support Vector Machine | Kernel type |
Next, we bootstrapped with replacement the validation dataset into 20 datasets. Then we measured the top 20 precision for each classifier using optimal hyperparameter settings and averaged the results to choose the best classification model. If there was a tie between top classifiers, we used the following measurements in descending sequence to break the tie: top 20 mean average precision, top 20 mean reciprocal rank [63], precision, recall, and F-measure.
Using the best classification model, a curve was drawn based on the average over nine datasets for k levels of 10, 20, 50, 100, 200, 300 and 500.
3.6. Hypothesis testing
Hypothesis 1.HI-TA performs equivalently to Kilicoglu et al.’s high quality Naïve Bayes classifier and PubMed’s Best Match sort in terms of top 20 precision.
We relied on the probability score of the Naïve Bayes classifier to rank the citations for Kilicoglu et al’s baseline. PubMed’s Best Match sort is a relevance-based algorithm that uses weighted term frequency and machine learning [64]. We relied on PubMed’s E-utilities service to obtain the ranked citations [65].
Hypothesis 2.HI-TA performs equivalently to HI-TS [18] in terms of top 20 precision.
HI-TS assumes that all time-agnostic and time-sensitive features are available at classification time. We extracted features for HI-TS using the same approach as in our previous study. Next, we ran the same classification model that was prepared and employed in our previous study on the features that were extracted from the first step to get the probability for each citation.
3.6.1. Study outcomes
Top 20 precision was the primary outcome for all hypotheses. We chose this measure because busy clinicians may only have time to read through the first 20 retrieved citations [66] and 20 is the default number of citations per page displayed in PubMed. We also measured top 20 mean reciprocal rank, top 20 mean average precision [63], precision, recall, and F-measure. Since top 20 precision, top 20 mean average precision, and top 20 mean reciprocal are outcomes that depend on a ranked output, we ranked the articles classified as positive according to the optimal classifier’s prediction score/probability for the positive class. For the PubMed baseline, we used PubMed Best Match’s ranked output.
3.6.2. Statistical analysis
We employed the Wilcoxon signed-rank test to assess the significance of the differences between HI-TA and each of the baselines. We used Stata IC 15 for the statistical analyses.
4. Results
The subsections order in this section matches the order in method section except that we removed the gold standard development and feature extraction and processing steps. We re-used the gold standard that was developed in our previous study [18]. The results of feature extraction and processing step is covered in Table 1.
4.1. Retrieval and filtering of candidate citations
A total of 45,553 citations were retrieved with the PubMed search queries, publication type filter and treatment filter for the diseases represented in the 11 guidelines. Among these citations, 541 were high impact clinical studies (recall of 86.0% for the total 629 guideline citations and precision of 1.2% for this initial dataset) (Figure 2).
4.2. Feature ranking and selection
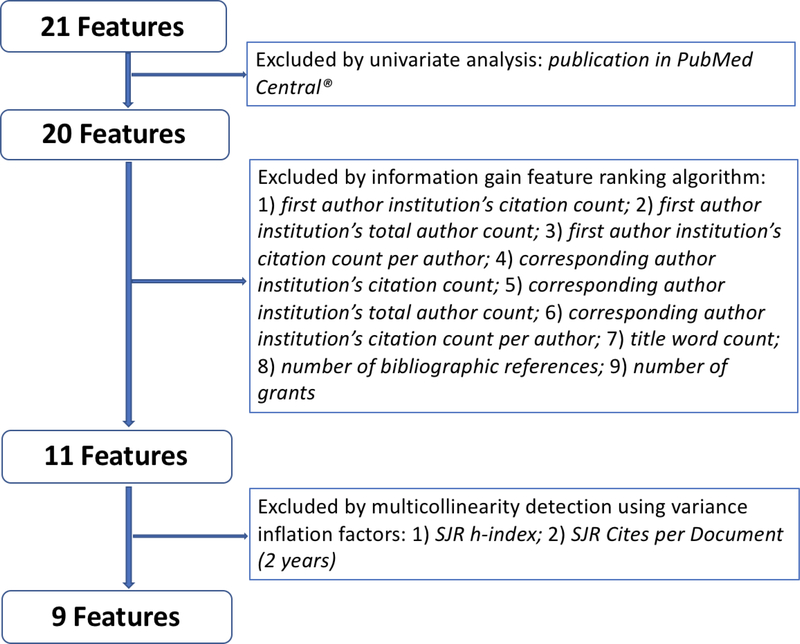
Feature ranking and selection is shown in Figure 3. Detailed results of each step are available in the online supplement, Table s1 - s4. Of the initial set of 21 features, 9 remained in the final set for classifier training (minimal feature set). We found that journal impact factor (SJR) was the top feature followed by core clinical journal, registration in ClinicalTrials.gov, number of authors, study sample size, number of institutions, number of clinically useful sentences, number of countries, and article page count (Table 3).
Figure 3.
Feature ranking and selection.
Table 3.
- Information gain of the final 9 features selected for classifier training
| Rank | Feature | Information gain |
|---|---|---|
| 1 | Journal Impact Factor (SJR) | 0.01794 |
| 2 | Core Clinical Journal | 0.0095 |
| 3 | Registration in ClinicalTrials.gov | 0.00939 |
| 4 | Number of Authors | 0.00799 |
| 5 | Study Sample Size | 0.00423 |
| 6 | Number of Institutions | 0.0034 |
| 7 | Number of Clinically Useful Sentences | 0.0034 |
| 8 | Number of Countries | 0.00338 |
| 9 | Article Page Count | 0.0026 |
We compared the performance between the whole feature set and minimal feature set using multiple classifiers on the pre-allocated training dataset (atrial fibrillation). We found that the performances were equivalent. Therefore, we used the minimal feature set for developing the classification models, optimizing the related hyper-parameters, and testing our two hypotheses.
4.3. Classification model development and testing
J48 classifier with hyper parameters (reduced error pruning = false; confidence factor = 0.2) had the most accurate results compared with all other classifiers (Table 4) and its model was selected for hypothesis testing. Table 4 shows outcomes of the best performance for each classifier with optimized hyper-parameters. The results are the averaged numbers on twenty bootstrapped COPD2014 datasets.
Table 4.
Best performance of each classifier with optimized hyper-parameters.
| Optimized hyper-parameters | Top 20 Precision | Top 20 Mean Average Precision | Top 20 Mean Reciprocal Rank | Precision | Recall | F1 | |
|---|---|---|---|---|---|---|---|
| Bayesian Network | Default | 0.18 | 0.05 | 0.34 | 0.12 | 0.09 | 0.1 |
| Decision Table | Default | 0.02 | 0 | 0.02 | 0 | 0 | 0 |
| J48 (decision tree) | Reduced error pruning = false; Confidence factor =0.2; | 0.24 | 0.11 | 0.46 | 0.39 | 0.09 | 0.14 |
| K-NearestNeighbors | KNN = 4; distance weighting = Weight by 1-distance; | 0.16 | 0.05 | 0.3 | 0 | 0 | 0 |
| Logistic Regression | Default | 0.18 | 0.05 | 0.24 | 0.13 | 0.02 | 0.03 |
| Multilayer Perceptron | Learning rate = 0.6 | 0.22 | 0.08 | 0.28 | 0 | 0 | 0 |
| Naive Bayes | Use kernel estimator = true | 0.01 | 0 | 0.01 | 0.04 | 0.02 | 0.02 |
| Naive Bayes Multinomial | Default | 0.06 | 0.03 | 0.39 | 0 | 0 | 0 |
| Random Forest | Max Depth = 11; Num of Trees = 41; Num of features = 4; | 0.23 | 0.1 | 0.54 | 0.31 | 0.02 | 0.03 |
| Simple Logistic | Default | 0.02 | 0 | 0.02 | 0 | 0 | 0 |
| Stochastic Gradient Descent | Loss function = Log loss (logistic regression) | 0.19 | 0.05 | 0.14 | 0 | 0 | 0 |
| Support Vector Machine | Default | 0.02 | 0 | 0.02 | 0 | 0 | 0 |
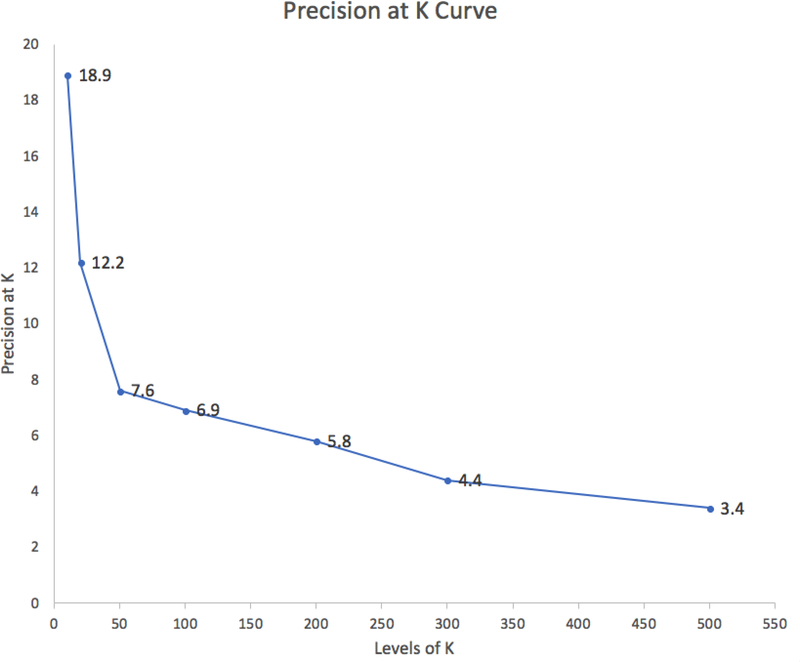
Top k precision curve
The precision among the top 10 to top 50 citations ranged from 18.9% to 7.6% and slowly reduced after the first 50 retrieved results (Figure 4).
Figure 4.
Average precision of HI-TA at different levels of k citations.
4.4. Hypothesis testing
Hypothesis #1: HI-TA performs equivalently to Kilicoglu et al.’s high quality Naïve Bayes classifier and PubMed’s Best Match sort in terms of top 20 precision.
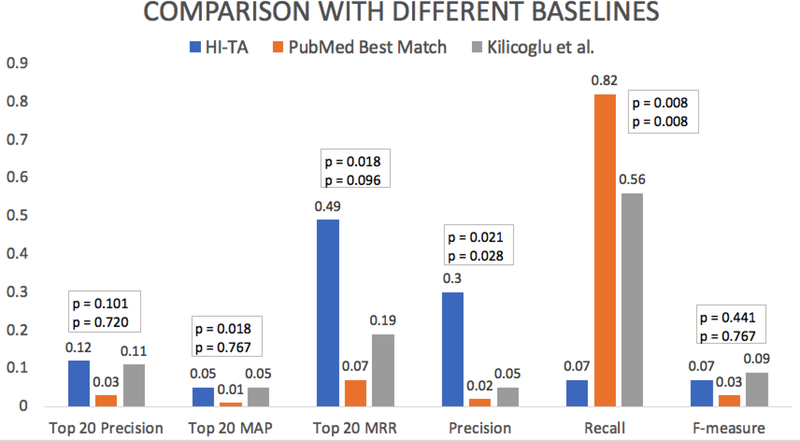
Figure 5 shows the results. The differences between HI-TA and two baselines (Kilicoglu et al.’s classifier and PubMed’s Best Match) were not statistically significant (mean top 20 precision = 12% vs. 11% and 3% respectively; p = 0.720 and p = 0.101). Similar results were found for the secondary outcomes F-measure (mean = 7% vs. 9% and 3% respectively; p = 0.767 and p = 0.441). HI-TA performed equivalently to Kilicoglu et al.’s classifier and outperformed PubMed’s Best Match sort in terms of top 20 mean average precision (mean = 5% vs. 5% and 1% respectively; p = 0.767 and p = 0.018) and top 20 mean reciprocal rank (mean = 49% vs. 19% and 7% respectively; p = 0.096 and p = 0.018). HI-TA outperformed Kilicoglu et al.’s classifier and PubMed’s Best Match sort in terms of precision (mean = 30% vs. 5% and 2% respectively; p = 0.028 and p = 0.021) but performed opposite in terms of recall (mean = 7% vs. 56% and 82% respectively; both p = 0.008).
Figure 5.
Comparison of HI-TA, PubMed’s Best Match sort and Kilicoglu et al.’s high quality Naïve Bayes classifier according to the average top 20 precision, top 20 mean average precision (MAP), top 20 mean reciprocal rank (MRR), precision, recall and F-measure (Experiment #1). The top p value corresponds to the statistical significance of the comparison between HI-TA and PubMed Best Match. The bottom p value corresponds to the statistical significance of the comparison between HI-TA and Kilicoglu et al.’s classifier.
Hypothesis #2: HI-TA performs equivalently to HI-TS in terms of top 20 precision.
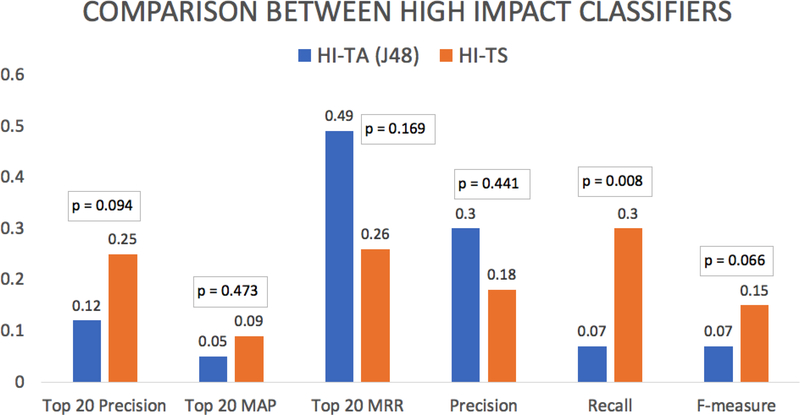
Figure 6 shows the results. There was no significant difference between the two classifiers in terms of mean top 20 precision (mean = 12% vs. 25%; p = 0.094), top 20 mean average precision (mean = 5% vs. 9%; p = 0.473), top 20 mean reciprocal rank (mean = 0.49 vs. 0.26; p = 0.169), precision (mean = 30% vs. 18%; p = 0.441), and F-measure (mean = 7% vs. 15%; p = 0.066). HI-TA had significantly lower recall (mean = 7% vs. 30%; p = 0.008) than HI-TS.
Figure 6.
Comparison between HI-TA versus HI-TS according to the mean top 20 precision, top 20 mean average precision (MAP), top 20 mean reciprocal rank (MRR), precision, recall and F-measure (Experiment #2).
5. Discussion
We investigated a machine learning approach using time-agnostic features to automatically identify recent high impact clinical studies in PubMed. We used a combination of features including citation metadata, bibliometrics, and sample size. To our knowledge, this is the first study that attempted to classify high impact clinical studies addressing the concept drift nature of this problem. We found that the top 20 precision of HI-TA was equivalent to Kilicoglu et al.’s high quality Naïve Bayes classifier (12% vs. 11%; p = 0.720), PubMed’s Best Match sort (12% vs. 3%; p = 0.101), and HI-TS (12% vs. 25%; p = 0.094). Compared with previous approaches [12–18], the main strengths of the proposed method are the use of time-agnostic features and the accessibility of key features such as journal impact factor and article authorship. The main weakness is a low recall (7%), which compromises the use of the classifier for literature surveillance. Our approach could be used as an adjunct to other approaches to help identify high impact studies for clinical decision support.
5.1. Time-agnostic classifier versus time-sensitive baselines
Experiment 1 failed to reject the null hypothesis that HI-TA had equivalent top 20 precision to Kilicoglu et al.’s high quality Naïve Bayes classifier and PubMed’s Best Match ranking. However, HI-TA outperformed PubMed’s Best Match ranking in terms of secondary measurements such as top 20 mean average precision (5% vs. 1%; p = 0.018) and top 20 mean reciprocal rank (49% vs. 7%; p = 0.018). Since both baselines depend on time-sensitive features, HI-TA presents an interesting alternative for retrieving recent articles from PubMed, with equal to better performance than the times-sensitive baselines.
5.2. Time-agnostic versus time-sensitive classifier
Experiment 2 failed to reject the null hypothesis that HI-TA performs equivalently to HITS [18] in terms of top 20 precision (12% vs. 25%; p = 0.094). Still, the absolute difference was relatively large and could be significant with a larger sample size. A couple of factors could have contributed to this finding. HI-TS relies on strong features such as citation counts and the Altmetric® score. The former may not be available until at least a year after publication and the latter is proprietary. Also, the search strategies used to retrieve relevant articles for HI-TS relies on MeSH terms and publication type, which typically take from 23 to 177 days to become available in PubMed citations. Automatic extraction of these features may improve classification accuracy.
5.3. Top k precision curve
The top k precision curve of HI-TA demonstrated among the top 10, 20 and 50 retrieved citations, there were on average 2, 2.5 and 4 high impact clinical articles respectively. Therefore, busy clinicians looking for high impact clinical studies on the treatment of a certain condition would be able to find a couple of those articles within the first set of a default PubMed search results page, which includes 20 citations.
5.4. Potential biases of high impact classification
A number of biases can result from the use of a high impact classifier. First, clinicians could potentially suffer from an automation bias, i.e. overreliance on technology to make decisions [67]. For example, a high impact classifier could bias clinicians to only consider evidence pubished in high impact journals by reserachers with an established track record. This could also lead to confirmation bias, i.e. clinicians developing a narrow view of the evidence that tends to confirm their preexisting viewpoints or beliefs when interpreting new information [68]. Second, articles that are predicted to produce high impact may be more likely to actually succeed in producing impact because the algorithm prediction directs people to read those articles (i.e., a self-fulfilling prophecy). However, due to the fact that our time-agnostic model (HI-TA) does not use any citation features (Table 3), it will be less prone to citation bias, which is another advantage over the time-sensitive model (HI-TS).
5.5. Limitations
An important limitation was that the recall of HI-TA was very low (7%). This has important negative implications, especially for literature surveillance where recall is as critical as precision. Moreover, the relatively small gold standard precluded the use of methods such as deep learning. Future studies should investigate methods to help produce larger gold standards, such as obtaining access to databases that were used to maintain the citations used in systematic review databases (e.g., Cochrane Database of Systematic Reviews [69,70]) and clinical guidelines.
5.6. Future research
Other relevant features, such as scientific productivity or impact of the author team and other free text features, could be explored. The scientific productivity or impact of the author team can be operationalized as the author’s h-index. However, obtaining this feature for each author of every article is computationally expensive (i.e., time). Future studies should investigate approaches to obtain this feature, such as creating a repository of authors with pre-computed h-index. Free-text features such as sample size are available as soon as an article is included in PubMed. Future studies could investigate the utility of free-text features extracted through text mining techniques from the article title and abstract. In addition, a larger gold standard would enable researchers to investigate the effect of other advanced classification models such as deep learning [71] and graphical models such as conditional random fields and hidden Markov models [72–74]. Alternatively, transfer learning methods could be investigated as a part of a deep learning approach. In transfer learning, a model is pre-trained for a similar task in the same domain using larger datasets. The pre-trained model is then “transfered” as input to deep learning training, this time using a small dataset that is the subject of the task of interest [75–77]. Since the pre-trained model captures the underlying relations in data, it helps reduce the required size of the dataset for understanding the new task.
6. Conclusion
We investigated a machine learning approach to identify high impact clinical studies addressing the concept drift challenge faced by previous approaches. We found that a classifier using time-agnostic features (e.g., JIF, authorship, study sample size) performed equivalently to state-of-the-art approaches that depend on time-sensitive features and therefore are susceptible to concept drift. Our classifier could be used as an adjunct to help clinicians identify recent high impact clinical studies in PubMed to support clinical decision making. However, due to low recall, the classifier is less useful for literature surveillance. Time-sensitive features such as citation count still play an important role in identifying high impact clinical studies in PubMed, but only in scenarios where identifying very recent citations is less important.
Supplementary Material
Highlights.
High impact clinical studies provide evidence influencing clinicians’ patient care.
We studied machine learning classifiers to identify high impact studies from PubMed.
To minimize concept drift, we only used time-agnostic features.
The classifier identified 2 to 3 high impact studies out of 20 ranked citations.
Our approach performed equivalently to a baseline with time-sensitive features.
Acknowledgement
This research project was supported by National Library of Medicine grants: R01LM011416 and T15LM007124 and National Cancer Institute Grant U24CA204800. We acknowledge the Richard A. Fay and Carol M. Fay Endowed Graduate Fellowship for funding support. We would like to acknowledge Data Science Librarian Shirley Zhao at University of Utah Eccles Health Sciences Library for her assistance with the Scopus data. We would like to thank Scopus for providing us the data and especially Meshna Koren for providing help on the project and answering specific questions. We would like to acknowledge James G. Mork in the National Library of Medicine (NLM) for processing our initial dataset using NLM Medical Text Indexer (MTI).
Footnotes
Conflict of interest
None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Covell DG, Uman GC, Manning PR, Information needs in office practice: are they being met?, Ann. Intern. Med. 103 (1985) 596–9. http://www.ncbi.nlm.nih.gov/pubmed/4037559 (accessed July 18, 2012). [DOI] [PubMed] [Google Scholar]
- [2].Del Fiol G, Workman TE, Gorman PN, Clinical questions raised by clinicians at the point of care: a systematic review., JAMA Intern. Med 174 (2014) 710–8. doi: 10.1001/jamainternmed.2014.368. [DOI] [PubMed] [Google Scholar]
- [3].Leape LL, Bates DW, Cullen DJ, Cooper J, Demonaco HJ, Gallivan T, Hallisey R, Ives J, Laird N, Laffel G, Systems analysis of adverse drug events. ADE Prevention Study Group., JAMA. 274 (1995) 35–43. http://www.ncbi.nlm.nih.gov/pubmed/7791256 (accessed May 13, 2015). [PubMed] [Google Scholar]
- [4].Maviglia SM, Yoon CS, Bates DW, Kuperman G, KnowledgeLink: impact of context-sensitive information retrieval on clinicians’ information needs., J. Am. Med. Inform. Assoc 13 (2006) 67–73. doi: 10.1197/jamia.M1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ioannidis JPA, Why most published research findings are false., PLoS Med. 2 (2005) e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cook DA, Sorensen KJ, Hersh W, Berger RA, Wilkinson JM, Features of effective medical knowledge resources to support point of care learning: a focus group study., PLoS One. 8 (2013) e80318. doi: 10.1371/journal.pone.0080318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shojania KG, Sampson M, Ansari MT, Ji J, Doucette S, Moher D, How quickly do systematic reviews go out of date? A survival analysis., Ann. Intern. Med 147 (2007) 224–33. http://www.ncbi.nlm.nih.gov/pubmed/17638714 (accessed July 8, 2017). [DOI] [PubMed] [Google Scholar]
- [8].Hertzman-Miller R, FILTERING OUT THE NOISE, Uncovering the “pearls” in clinical literature that have immediate impact on patient care, (2017). https://www.ebsco.com/blog-archives/article/filteringthe-noise.-what-is-systematic-literature-surveillance-and-why-is (accessed March 1, 2018).
- [9].Sampson M, Shojania KG, McGowan J, Daniel R, Rader T, Iansavichene AE, Ji J, Ansari MT, Moher D, Surveillance search techniques identified the need to update systematic reviews, J. Clin. Epidemiol 61 (2008) 755–762. doi: 10.1016/j.jclinepi.2007.10.003. [DOI] [PubMed] [Google Scholar]
- [10].Shojania KG, Sampson M, Ansari MT, Ji J, Garritty C, Rader T, Moher D, Updating Systematic Reviews, Agency for Healthcare Research and Quality (US), 2007. http://www.ncbi.nlm.nih.gov/pubmed/20734512 (accessed February 28, 2018). [PubMed] [Google Scholar]
- [11].Elliott JH, Synnot A, Turner T, Simmonds M, Akl EA, McDonald S, Salanti G, Meerpohl J, MacLehose H, Hilton J, Tovey D, Shemilt I, Thomas J, Living T, Systematic Review Network Hilton J, Perron C, Akl E, Hodder R, Pestridge C, Albrecht L, Horsley T, Platt J, Armstrong R, Nguyen PH, Plovnick R, Arno A, Ivers N, Quinn G, Au A, Johnston R, Rada G, Bagg M, Jones A, Ravaud P, Boden C, Kahale L, Richter B, Boisvert I, Keshavarz H, Ryan R, Brandt L, Kolakowsky-Hayner SA, Salama D, Brazinova A, Nagraj SK, Salanti G, Buchbinder R, Lasserson T, Santaguida L, Champion C, Lawrence R, Santesso N, Chandler J, Les Z, Schünemann HJ, Charidimou A, Leucht S, Shemilt I, Chou R, Low N, Sherifali D, Churchill R, Maas A, Siemieniuk R, Cnossen MC, MacLehose H, Simmonds M, Cossi M-J, Macleod M, Skoetz N, Counotte M, Marshall I, Soares-Weiser K, Craigie S, Marshall R, Srikanth V, Dahm P, Martin N, Sullivan K, Danilkewich A, García LM, Synnot A, Danko K, Mavergames C, Taylor M, Donoghue E, Maxwell LJ, Thayer K, Dressler C, McAuley J, Thomas J, Egan C, McDonald S, Tritton R, Elliott J, McKenzie J, Tsafnat G, Elliott SA, Meerpohl J, Tugwell P, Etxeandia I, Merner B, Turgeon A, Featherstone R, Mondello S, Turner T, Foxlee R, Morley R, van Valkenhoef G, Garner P, Munafo M, Vandvik P, Gerrity M, Munn Z, Wallace B, Glasziou P, Murano M, Wallace SA, Green S, Newman K, Watts C, Grimshaw J, Nieuwlaat R, Weeks L, Gurusamy K, Nikolakopoulou A, Weigl A, Haddaway N, Noel-Storr A, Wells G, Hartling L, O’Connor A, Wiercioch W, Hayden J, Page M, Wolfenden L, Helfand M, Pahwa M, Yepes Nuñez JJ, Higgins J, Pardo JP, Yost J, Hill S, Pearson L, Living systematic review: 1. Introduction-the why, what, when, and how., J. Clin. Epidemiol. 91 (2017) 23–30. doi: 10.1016/j.jclinepi.2017.08.010. [DOI] [PubMed] [Google Scholar]
- [12].Haynes RB, Wilczynski N, McKibbon KA, Walker CJ, Sinclair JC, Developing optimal search strategies for detecting clinically sound studies in MEDLINE., J. Am. Med. Inform. Assoc 1 (1994) 447–58. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=116228&tool=pmcentrez&rendertype=abstract (accessed November 28, 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wilczynski NL, McKibbon KA, Walter SD, Garg AX, Haynes RB, MEDLINE clinical queries are robust when searching in recent publishing years, J. Am. Med. Informatics Assoc 20 (2013) 363–368. doi: 10.1136/amiajnl-2012-001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].V Bernstam E, Herskovic JR, Aphinyanaphongs Y, Aliferis CF, Sriram MG, Hersh WR, Using citation data to improve retrieval from MEDLINE., J. Am. Med. Inform. Assoc 13 (2006) 96–105. doi: 10.1197/jamia.M1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kilicoglu H, Demner-Fushman D, Rindflesch TC, Wilczynski NL, Haynes RB, Towards automatic recognition of scientifically rigorous clinical research evidence, J. Am. Med. Inform. Assoc 16 (2009) 25–31. doi: 10.1197/jamia.M2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Aphinyanaphongs Y, Tsamardinos I, Statnikov A, Hardin D, Aliferis CF, Text categorization models for high-quality article retrieval in internal medicine., J. Am. Med. Inform. Assoc 12 (2005) 207–16. doi: 10.1197/jamia.M1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Aphinyanaphongs Y, Statnikov A, Aliferis CF, A comparison of citation metrics to machine learning filters for the identification of high quality MEDLINE documents., J. Am. Med. Inform. Assoc 13 (2006) 446–55. doi: 10.1197/jamia.M2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bian J, Morid MA, Jonnalagadda S, Luo G, Del Fiol G, Automatic Identification of High Impact Articles in PubMed to Support Clinical Decision Making, J. Biomed. Informatics (In Revis. (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lokker C, McKibbon KA, McKinlay RJ, Wilczynski NL, Haynes RB, Prediction of citation counts for clinical articles at two years using data available within three weeks of publication: retrospective cohort study., BMJ. 336 (2008) 655–7. doi: 10.1136/bmj.39482.526713.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].bout Altmetric and the Altmetric score, (n.d.). https://help.altmetric.com/support/solutions/articles/6000059309-about-altmetric-and-the-altmetric-score (accessed April 4, 2017).
- [21].Žliobaitė I, Learning under Concept Drift: an Overview, (2010). http://arxiv.org/abs/1010.4784 (accessed February 5, 2018).
- [22].Widmer G, Kubat M, Learning in the presence of concept drift and hidden contexts, Mach. Learn. 23 (1996) 69–101. doi: 10.1007/BF00116900. [DOI] [Google Scholar]
- [23].Žliobaitė I, Pechenizkiy M, Gama J, An Overview of Concept Drift Applications, in: Springer, Cham, 2016: pp. 91–114. doi: 10.1007/9783-319-26989-4_4. [DOI] [Google Scholar]
- [24].Tsymbal A, The Problem of Concept Drift: Definitions and Related Work, 2004. citeulike-article-id:2350391.
- [25].Irwin AN, Rackham D, Comparison of the time-to-indexing in PubMed between biomedical journals according to impact factor, discipline, and focus, Res. Soc. Adm. Pharm 13 (2017) 389–393. doi: 10.1016/j.sapharm.2016.04.006. [DOI] [PubMed] [Google Scholar]
- [26].AMIN M, MABE M, Impact Factors: Use and Abuse, Perspect. Publ. (2000). http://www.iuma.ulpgc.es/~nunez/doctorado0910/impactfactor.pdf.
- [27].Kiesslich T, Weineck SB, Koelblinger D, Reasons for Journal Impact Factor Changes: Influence of Changing Source Items, PLoS One. 11 (2016) e0154199. doi: 10.1371/journal.pone.0154199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].SCImago., SJR - SCImago Journal & Country Rank, (2007). http://www.scimagojr.com (accessed February 2, 2018).
- [29].Institute of Medicine (U.S.). Committee on Standards for Developing Trustworthy Clinical Practice Guidelines, Graham R, Clinical practice guidelines we can trust, National Academies Press, 2011. [PubMed] [Google Scholar]
- [30].Demner-Fushman D, Lin J, Answering Clinical Questions with Knowledge-Based and Statistical Techniques, Comput. Linguist. 33 (2007) 63–103. doi: 10.1162/coli.2007.33.1.63. [DOI] [Google Scholar]
- [31].Mork J, Aronson A, Demner-Fushman D, 12 years on – Is the NLM medical text indexer still useful and relevant?, J. Biomed. Semantics 8 (2017) 8. doi: 10.1186/s13326-017-0113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].MeSH Qualifier Hierarchies, (n.d.). https://www.nlm.nih.gov/mesh/subhierarchy.html (accessed February 2, 2018).
- [33].Hoogendam A, Stalenhoef AF, de Vries Robbé PF, Overbeke AJP, Answers to Questions Posed During Daily Patient Care Are More Likely to Be Answered by UpToDate Than PubMed, J. Med. Internet Res. 10 (2008) e29. doi: 10.2196/jmir.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sayyah Ensan L, Faghankhani M, Javanbakht A, Ahmadi S-F, Baradaran HR, To Compare PubMed Clinical Queries and UpToDate in Teaching Information Mastery to Clinical Residents: A Crossover Randomized Controlled Trial, PLoS One. 6 (2011) e23487. doi: 10.1371/journal.pone.0023487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shariff SZ, Bejaimal SAD, Sontrop JM, Iansavichus AV, Weir MA, Haynes RB, Speechley MR, Thind A, Garg AX, Searching for medical information online: a survey of Canadian nephrologists, J. Nephrol 24 (2011) 723–732. doi: 10.5301/JN.2011.6373. [DOI] [PubMed] [Google Scholar]
- [36].Sheets L, Callaghan F, Gavino A, Liu F, Fontelo P, Usability of selected databases for low-resource clinical decision support., Appl. Clin. Inform 3 (2012) 326–33. doi: 10.4338/ACI-2012-06-RA-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Thiele RH, Poiro NC, Scalzo DC, Nemergut EC, Speed, accuracy, and confidence in Google, Ovid, PubMed, and UpToDate: results of a randomised trial, Postgrad. Med. J 86 (2010) 459–465. doi: 10.1136/pgmj.2010.098053. [DOI] [PubMed] [Google Scholar]
- [38].Morid MA, Jonnalagadda S, Fiszman M, Raja K, Del Fiol G, Classification of Clinically Useful Sentences in MEDLINE., AMIA ... Annu. Symp. Proceedings. AMIA Symp 2015 (2015) 2015–24. http://www.ncbi.nlm.nih.gov/pubmed/26958301 (accessed June 28, 2017). [PMC free article] [PubMed] [Google Scholar]
- [39].Garfield E, The history and meaning of the journal impact factor., JAMA. 295 (2006) 90–3. doi: 10.1001/jama.295.1.90. [DOI] [PubMed] [Google Scholar]
- [40].McVeigh ME, Mann SJ, The Journal Impact Factor Denominator, JAMA. 302 (2009) 1107. doi: 10.1001/jama.2009.1301. [DOI] [PubMed] [Google Scholar]
- [41].Detailed description of SJR, (n.d.). http://www.scimagojr.com/SCImagoJournalRank.pdf (accessed February 2, 2018).
- [42].Shi J, Mowery D, Zhang M, Sanders J, Chapman W, Gawron L, Extracting Intrauterine Device Usage from Clinical Texts Using Natural Language Processing, in: 2017 IEEE Int. Conf. Healthc. Informatics, IEEE, 2017: pp. 568–571. doi:10.1109/ICHI.2017.21. [Google Scholar]
- [43].Chapman WW, Hillert D, Velupillai S, Kvist M, Skeppstedt M, Chapman BE, Conway M, Tharp M, Mowery DL, Deleger L, Extending the NegEx lexicon for multiple languages., Stud. Health Technol. Inform 192 (2013) 677–81. http://www.ncbi.nlm.nih.gov/pubmed/23920642 (accessed May 28, 2018). [PMC free article] [PubMed] [Google Scholar]
- [44].Wang J, Shapira P, Is there a relationship between research sponsorship and publication impact? An analysis of funding acknowledgments in nanotechnology papers., PLoS One. 10 (2015) e0117727. doi: 10.1371/journal.pone.0117727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Elsevier Developers - Scopus, (n.d.). https://dev.elsevier.com/scopus.html (accessed February 2, 2018).
- [46].Falagas ME, Zarkali A, Karageorgopoulos DE, Bardakas V, Mavros MN , The impact of article length on the number of future citations: a bibliometric analysis of general medicine journals., PLoS One. 8 (2013) e49476. doi: 10.1371/journal.pone.0049476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hsiehchen D, Espinoza M, Hsieh A, Multinational teams and diseconomies of scale in collaborative research, Sci. Adv 1 (2015) e1500211–e1500211. doi: 10.1126/sciadv.1500211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ahmed A, Adam M, Ghafar NA, Muhammad M, Ebrahim NA, Impact of Article Page Count and Number of Authors on Citations in Disability Related Fields: A Systematic Review Article., Iran. J. Public Health 45 (2016) 1118–1125. http://www.ncbi.nlm.nih.gov/pubmed/27957456 (accessed June 29, 2017). [PMC free article] [PubMed] [Google Scholar]
- [49].Corbyn Z, An easy way to boost a paper’s citations, Nat. News (2010). http://www.nature.com/news/2010/100813/full/news.2010.406.html.
- [50].Vinkler P, Dynamic changes in the chance for citedness, Scientometrics. 54 (2002) 421–434. doi: 10.1023/A:1016086500801. [DOI] [Google Scholar]
- [51].Habibzadeh F, Yadollahie M, Are shorter article titles more attractive for citations? Cross-sectional study of 22 scientific journals., Croat. Med. J 51 (2010) 165–70. http://www.ncbi.nlm.nih.gov/pubmed/20401960 (accessed June 29, 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jacques TS, Sebire NJ, The Impact of Article Titles on Citation Hits: An Analysis of General and Specialist Medical Journals, JRSM Short Rep. 1 (2010) 1–5. doi: 10.1258/shorts.2009.100020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].MEDLINE Abridged Index Medicus (AIM or “Core Clinical”) Journal Titles, (n.d.). https://www.nlm.nih.gov/bsd/aim.html (accessed April 4, 2017).
- [54].McMaster Plus (Premium LiteratUre Service) journals, (n.d.). http://hiru.mcmaster.ca/hiru/journalslist.asp (accessed April 4, 2017). [Google Scholar]
- [55].Journal Selection for Index Medicus®/Medline®, J. Can. Chiropr. Assoc 40 (1996) 47 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2485146/ (accessed March 14, 2016). [Google Scholar]
- [56].Committed selection: abridged index medicus., N. Engl. J. Med 282 (1970) 220–1. doi: 10.1056/NEJM197001222820410. [DOI] [PubMed] [Google Scholar]
- [57].Hemens BJ, Haynes RB, McMaster Premium LiteratUre Service (PLUS) performed well for identifying new studies for updated Cochrane reviews., J. Clin. Epidemiol. 65 (2012) 62–72.e1. doi: 10.1016/j.jclinepi.2011.02.010. [DOI] [PubMed] [Google Scholar]
- [58].Lundh A, Sismondo S, Lexchin J, Busuioc OA, Bero L, Industry sponsorship and research outcome., Cochrane Database Syst. Rev 12 (2012) MR000033. doi: 10.1002/14651858.MR000033.pub2. [DOI] [PubMed] [Google Scholar]
- [59].Sismondo S, Pharmaceutical company funding and its consequences: a qualitative systematic review., Contemp. Clin. Trials 29 (2008) 109–13. doi: 10.1016/j.cct.2007.08.001. [DOI] [PubMed] [Google Scholar]
- [60].Lexchin J, Bero LA, Djulbegovic B, Clark O, Pharmaceutical industry sponsorship and research outcome and quality: systematic review., BMJ. 326 (2003) 1167–70. doi: 10.1136/bmj.326.7400.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH, The WEKA data mining software, ACM SIGKDD Explor. Newsl. 11 (2009) 10. doi: 10.1145/1656274.1656278. [DOI] [Google Scholar]
- [62].O’brien RM, A Caution Regarding Rules of Thumb for Variance Inflation Factors, Qual. Quant 41 (2007) 673–690. doi: 10.1007/s11135-006-9018-6. [DOI] [Google Scholar]
- [63].Manning CD, Raghavan P, Schütze H, Introduction to Information Retrieval, Cambridge University Press, 2008. [Google Scholar]
- [64].Collins M, Updated Algorithm for the PubMed Best Match Sort Order, NLM Tech. Bull; (2017) 414. [Google Scholar]
- [65].Sayers E, E-utilities Quick Start, (2017). https://www.ncbi.nlm.nih.gov/books/NBK25500/ (accessed March 29, 2018).
- [66].Hoogendam A, Stalenhoef AFH, de V PF. Robbé AJ Overbeke PM, Analysis of queries sent to PubMed at the point of care: Observation of search behaviour in a medical teaching hospital, BMC Med. Inform. Decis. Mak 8 (2008) 42. doi: 10.1186/1472-6947-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Goddard K, Roudsari A, Wyatt JC, Automation bias: a systematic review of frequency, effect mediators, and mitigators, J. Am. Med. Informatics Assoc 19 (2011) 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Cook MB, Smallman HS, Human factors of the confirmation bias in intelligence analysis: Decision support from graphical evidence landscapes, Hum. Factors 50 (2008) 745–754. [DOI] [PubMed] [Google Scholar]
- [69].Bastian H, Glasziou P, Chalmers I, Seventy-Five Trials and Eleven Systematic Reviews a Day: How Will We Ever Keep Up?, PLoS Med. 7 (2010) e1000326. doi: 10.1371/journal.pmed.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Starr M, Chalmers I, Clarke M, Oxman AD, The origins, evolution, and future of The Cochrane Database of Systematic Reviews, Int. J. Technol. Assess. Health Care 25 (2009) 182–195. doi: 10.1017/S026646230909062X. [DOI] [PubMed] [Google Scholar]
- [71].Del Fiol G, Michelson M, Iorio A, Cotoi C, Haynes RB, A Deep Learning Method to Automatically Identify Reports of Scientifically Rigorous Clinical Research from the Biomedical Literature: Comparative Analytic Study., J. Med. Internet Res 20 (2018) e10281. doi: 10.2196/10281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Schuler W, AbdelRahman S, Miller T, Schwartz L, Broad-coverage parsing using human-like memory constraints, Comput. Linguist 36 (2010) 1–30. [Google Scholar]
- [73].Velupillai S, Mowery DL, Abdelrahman S, Christensen L, Chapman W, Blulab: Temporal information extraction for the 2015 clinical tempeval challenge, in: Proc. 9th Int. Work. Semant. Eval. (SemEval 2015), 2015: pp. 815–819. [Google Scholar]
- [74].Velupillai S, Mowery DL, Abdelrahman S, Christensen L, Chapman WW, Towards a Generalizable Time Expression Model for Temporal Reasoning in Clinical Notes., AMIA ... Annu. Symp. Proceedings. AMIA Symp. 2015 (2015) 1252–9. http://www.ncbi.nlm.nih.gov/pubmed/26958265 (accessed June 20, 2018). [PMC free article] [PubMed] [Google Scholar]
- [75].Pan SJ, Yang Q, A survey on transfer learning, IEEE Trans. Knowl. Data Eng 22 (2010) 1345–1359. [Google Scholar]
- [76].Zhao W, Research on the deep learning of the small sample data based on transfer learning, in: AIP Conf. Proc., AIP Publishing, 2017: p. 20018. [Google Scholar]
- [77].Ng H-W, Nguyen VD, Vonikakis V, Winkler S, Deep learning for emotion recognition on small datasets using transfer learning, in: Proc. 2015 ACM Int. Conf. Multimodal Interact., ACM, 2015: pp. 443–449. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.