Abstract
Fluorescent markers that bind endogenous target proteins are frequently employed for quantitative live-cell imaging. To visualize the actin cytoskeleton in live cells, several actin-binding probes have been widely used. Among them, Lifeact is the most popular probe with ideal properties, including fast exchangeable binding kinetics. Because of its fast kinetics, Lifeact is generally believed to distribute evenly throughout cellular actin structures. In this study, however, we demonstrate misdistribution of Lifeact toward the rear of lamellipodia where actin filaments continuously move inward along the retrograde flow. Similarly, phalloidin showed biased misdistribution toward the rear of lamellipodia in live cells. We show evidence of convection-induced misdistribution of actin probes by both experimental data and physical models. Our findings warn about the potential error arising from the use of target-binding probes in quantitative live imaging.
Introduction
Quantitative live-cell imaging is becoming increasingly important in the field of biology. Apart from protein probes covalently attached to fluorescent proteins or chemicals, fluorescent reporters that bind endogenous target proteins are frequently employed for quantitative live-cell imaging (1, 2, 3). For proper quantification of target-molecule distribution, two criteria are generally considered to be necessary and sufficient: 1) the fluorescent probe can bind and report all species of targets equally, and 2) it does not perturb dynamics of endogenous targets in vivo.
To visualize the actin cytoskeleton in live cells, fluorescently labeled actin probes including Lifeact, phalloidin, and actin-binding domains from various proteins have been widely used (4). Among them, Lifeact (5) is the most popular probe with ideal properties including fast exchangeable binding kinetics (6). Because of its fast kinetics, Lifeact is generally believed to distribute evenly throughout cellular actin structures. In this study, we demonstrate misdistribution of Lifeact toward the rear of lamellipodia where actin filaments continuously move inward along the retrograde actin flow that is widely observed in adherent cells (7, 8, 9). Similarly, phalloidin, which has been employed as a marker of F-actin in quantitative live-cell imaging (10, 11, 12), showed biased misdistribution toward the rear of lamellipodia in live cells. We show evidence of convection-induced misdistribution of actin probes by both experimental data and physical models. Our data and simulations indicated that the different Kd of the actin probe and retrograde actin flow speeds may give rise to a wide variety of distribution patterns.
Materials and Methods
Plasmids
The expression vectors harboring the defective CMV (cytomegalovirus) promoter (delCMV) for Lifeact-mCherry, Lifeact-EGFP, and EGFP-actin were described (13, 14, 15). miRFP703 cDNA (16) was obtained from Addgene (Cambridge, MA). The expression vector for miRFP703-actin was generated by substituting miRFP703 cDNA for the coding sequence of EGFP in delCMV-EGFP-actin.
Cell culture, transfection and electroporation
Xenopus laevis XTC cells were maintained as described previously (13, 17). The expression constructs were transfected into XTC cells by using polyethyleneimine as described previously (17). Alexa Fluor 546 phalloidin (Thermo Fisher Scientific, Waltham, MA) (1 μM) was electroporated into XTC cells with the Neon transfection system (Invitrogen, Carlsbad, CA) as described previously (15). Keratocytes were isolated from scales of goldfish (Carassius auratus) by culturing between a culture dish and a coverslip, using L-15 Leibovitz medium (Invitrogen) supplemented with 10% fetal bovine serum (Nichirei Biosciences, Tokyo, Japan) and 1% Antibiotic-Antimycotic Mixed Stock Solution (Nacalai Tesque, Kyoto, Japan). Then cells were trypsinized, resuspended with the culture medium, collected by centrifugation, and subjected to electroporation to simultaneously deliver Alexa Fluor 546 phalloidin (0.25 μM) and CF680R-actin (18) as described (15).
Fluorescence microscopy and data analysis
Live-cell imaging was carried out using XTC cells as previously described (17, 19). Briefly, XTC cells were allowed to spread on a poly-L-lysine-coated coverslip in 70% L-15 Leibovitz medium without serum. Keratocytes were allowed to spread on a coverslip in L-15 Leibovitz medium supplemented with 10% fetal bovine serum. Time-lapse imaging of XTC cells and keratocytes was acquired by using a microscope (IX71; Olympus, Tokyo, Japan) equipped with 100-W mercury illumination, a PlanApo 1.40 numerical aperture (NA) 100× oil objective (Olympus), and an electron multiplying charged coupled device (EMCCD) camera (iXon3; Andor Technology, Belfast, Northern Ireland) or a microscope (IX83; Olympus) equipped with a PlanApo 1.40 NA 100× oil objective and an EMCCD camera (Evolve 512; Photometrics, Tucson, AZ).
Immediately after time-lapse imaging, XTC cells and keratocytes were fixed with cytoskeleton buffer (10 mM 2-(N-morpholono)ethane sulfonic acid (pH 6.1), 90 mM KCl, 3 mM MgCl2, 2 mM EGTA) containing 3.7% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 in cytoskeleton buffer for 10 min at room temperature. Alexa 647 phalloidin (Thermo Fisher Scientific), Alexa 546 phalloidin (Thermo Fisher Scientific), or Oregon Green 488 phalloidin (Invitrogen) was used for staining F-actin in XTC cells and keratocytes.
For Lifeact staining in fixed XTC cells, the Lifeact peptide (MGVADLIKKFESISKEE) was synthesized by Sigma-Aldrich (St. Louis, MO), and then the synthesized Lifeact peptide conjugated with an N-terminal Atto 550 fluorophore (Sigma-Aldrich) was used. After the fixation and permeabilization processes described above, the cells were incubated with 1 nM Atto-550 Lifeact in an imaging solution comprising HEPES-buffered solution (10 mM HEPES (pH 7.2), 90 mM KCl, 3 mM MgCl2, 100 μM dithiothreitol [DTT]) with an oxygen-scavenging mix (200 μg/mL glucose oxidase, 35 μg/mL catalase, 4.5 mg/mL glucose, 0.5% 2-mercaptethanol) as described previously (6). Images of Atto-550 Lifeact were acquired using an epifluorescent microscope (IX83) equipped with a PlanApo 1.40 NA 100× oil objective and an EMCCD camera (Evolve 512). Lifeact staining images were generated by integrating images of Atto-550 Lifeact for 150 s (3000 frames). After imaging of Atto-550 Lifeact, the probe was washed out 10 times with the HEPES-buffered solution, and then the cells were stained with rhodamine-phalloidin (Thermo Fisher Scientific).
To measure the retrograde flow speed in cells containing actin probes at a low level, we tracked actin speckles or phalloidin speckles in lamellipodia in a series of time-lapse images as previously described (14, 15). In cells that expressed actin probes at high level, we used kymograph analysis with EGFP-actin to measure the retrograde flow speed.
Fluorescence intensity was measured using the Linescan function in the MetaMorph software (Molecular Devices, San Jose, CA). The background intensity in an area outside of the cell was subtracted from the original image before the measurement. In each cell image, 7–11 lines were placed across the lamellipodium by avoiding condensed F-actin structures. The average fluorescence intensity was shown as a function of distance from the lamellipodium tip for each cell.
Fluorescence anisotropy measurement
The synthesized Lifeact peptide conjugated with an N-terminal Atto 488 fluorophore (Sigma-Aldrich) was used. Rabbit skeletal muscle actin was prepared from acetone powder by one cycle of polymerization and pelleting as described previously (20). Actin was depolymerized in G-buffer (2 mM Tris, 0.2 mM CaCl2, 0.2 mM ATP, 1 mM DTT (pH 8.0)) and then further purified by gel filtration with HiLoad16/60 Superdex 200 (GE Healthcare, Little Chalfont, UK). Cytoplasmic human platelet actin was obtained from Cytoskeleton (Denver, CO). To prepare F-actin, G-actin in G-buffer was polymerized into F-actin by addition of 0.1 volume of 10× polymerizing buffer (200 mM Hepes, 1 M KCl, 20 mM MgCl2, 2 mM DTT, 5 mM ATP, 10 mM EGTA (pH 7.5)) and incubated overnight on ice.
Fluorescence anisotropy measurements were performed using an EnVision plate reader (Perkin Elmer, Waltham, MA) with a polarization filter set (fluorescein isothiocyanate FP label; excitation wavelength 480 nm, emission wavelength 535 nm) at room temperature. Assays with G-actin and F-actin were performed in G-buffer for samples with G-actin and in F-buffer (20 mM Hepes, 0.1 M KCl, 2 mM MgCl2, 0.2 mM DTT, 0.5 mM ATP, 1 mM EGTA (pH 7.5)) for samples with F-actin, respectively. F-actin was fragmented by sonication immediately before mixing with Atto-488-labeled Lifeact (Atto-488 Lifeact). Atto-488 Lifeact (0.1 μM) was mixed with increasing concentrations of G-actin or F-actin in a total volume of 150 μL, and the mixtures were incubated for 15 min at room temperature. Then the horizontal (Ih) and vertical (Iv) components of the fluorescent intensity were measured. The fluorescence anisotropy, r, is calculated using r = (Iv − GIh)/(Iv + 2GIh). The G factor is determined with fluorescein in accordance with the instruction of the manufacturer.
Modeling the distribution of F-actin-binding probe in lamellipodia
In this study, we first considered a model lamellipodium of a single cell moving at a constant speed V (>0) toward the lab frame +X direction. We assumed that the lamellipodium is homogeneous in the Y-direction and the thickness of the lamellipodium in the Z-direction is very thin (100–200 nm). Thus, the model lamellipodium is effectively along one dimension, X, describing a cross section through the lamellipodium with length L. In the cell frame x = X − Vt (0 ≤ x ≤ L), where x = 0 represents the lamellipodial base and x = L indicates the lamellipodial tip, we define Cb(x, t) and Cf(x, t) as the concentrations of F-actin-bound and free actin probe in the lamellipodium at time t. Free actin probes can either bind to F-actin with a rate of kon or diffuse in the cytosol with a diffusion coefficient D. Bound actin probes can unbind from F-actin with a rate of koff or be advected by the actin retrograde flow at a velocity v measured on the cell frame. In this study, we considered two types of F-actin concentration profiles: 1) a uniform distribution (constant) (21, 22) and 2) a linear profile that decreases toward the lamellipodial base, F = ax + b (Fig. 1 C). The spatiotemporal dynamics of Cb and Cf in the cell frame x can be described by the following set of reaction-convection-diffusion equations:
| (1) |
and
| (2) |
To solve Eqs. 1 and 2, the following boundary conditions and conservation law were assigned:
| (3) |
| (4) |
and
| (5) |
The boundary conditions Eqs. 3 and 4 represent no-flux conditions for bound and free probes at the lamellipodial tip, whereas Eq. 5 indicates the conservation law that the total amount of actin probe in the lamellipodia remains constant.
Figure 1.
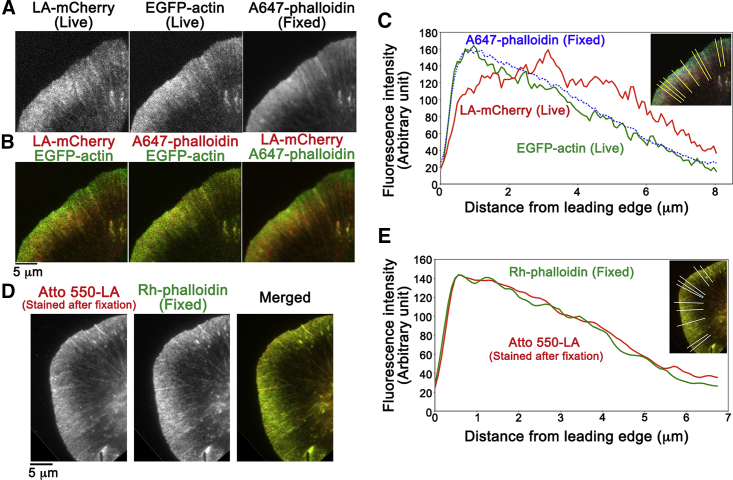
(A) Images of Lifeact-mCherry (LA-mCherry, left) and EGFP-actin (middle) in live Xenopus XTC cells. After fixation, F-actin was stained with Alexa 647 phalloidin (A647-phalloidin, right). Representative data are shown (n = 6 cells). (B) Merged images of (A) showing rear-biased distribution of LA-mCherry in lamellipodia are given. (C) The average fluorescence intensity of the images in (A) along the yellow lines in the inset is shown. (D and E) Similar distribution of Atto-550 Lifeact (Atto-550-LA) and rhodamine-phalloidin (Rh-phalloidin) in fixed XTC cells is shown. Staining with Rh-phalloidin was carried out after washing out Atto-550 Lifeact, which is an exchangeable probe (6). Representative data are shown (n = 3 cells, see Fig. S1).
Equations 1 and 2 were discretized using the second-order finite difference scheme in space (step dx = 0.02 μm) and integrated in time (step dt = 5 μs) by the second-order Runge-Kutta method until they reached the steady-state solution. The initial conditions were set as Cb(x, 0) = 0 and , where represents a uniform random variable ranging from 0 to 1.
Steady-state probe distribution at the uniform F-actin profile
In the case of a uniform F-actin profile (constant), the steady-state solution of Eqs. 1 and 2 can be analytically obtained as
| (6) |
and
| (7) |
where
| (8) |
| (9) |
| (10) |
| (11) |
and
| (12) |
Here, the constant C∗ takes an arbitrary value depending on the total amount of probe in lamellipodia. The fact that ξ2 > 0 means that the dominant term describing the exponential increase of probe concentration toward the lamellipodial base is given by the terms proportional to in Eqs. 6 and 7. The exponential profiles of both free and bound probes greatly differ from the uniform F-actin profile, suggesting that probe distribution no longer reflects the true F-actin distribution.
Values of the model parameters
The model parameters used in this study are summarized in Table S1. The retrograde flow speed v and lamellipodial length L were measured by our experimental data. The diffusion coefficient D of free probe was estimated from that of G-actin in lamellipodia (∼6 μm2 s−1) (23) by using the fact that the diffusion coefficient scales approximately as the inverse of the cube root of the molecular weight (∼29.3 kDa for Lifeact-mCherry and 1.95 kDa for Alexa 647 phalloidin). We calculated the value of the dissociation rate koff of Lifeact-mCherry by dividing ln2 by the half-life of Lifeact-mCherry (23 ms) (6). Because the in vitro value of koff, 2.6 × 10−4 s−1, of phalloidin (24) is much smaller than the inverse of the half-life of actin filaments, the dissociation rate of Alexa 647 phalloidin was estimated by the inverse of averaged halftime of lamellipodial actin filaments (0.08 s−1). As for the probe association rate kon, we used the value of 2.9 × 10−2 μM−1 s−1 for Alexa 647 phalloidin reported in the previous in vitro study (24), whereas we used 2.28 μM−1 s−1 for Lifeact-mCherry based on the relation kon = koff/Kd, where Kd is the equilibrium dissociation constant (13.2 μM for Lifeact-mCherry) (25). The value of uniform F-actin concentration was set as 1000 μM according to the work by Abraham (26). For the case of the linear F-actin profile, the concentration was estimated by using the fluorescence intensity of Alexa647-phalloidin in a fixed cell in our experiment (Fig. 1 C), assuming that the F-actin concentration at the tip of lamellipodia is 1000 μM.
Results
Rear-biased misdistribution of Lifeact and phalloidin in lamellipodia
We first noticed that Lifeact (5) shows rear-biased misdistribution in lamellipodia. We compared distributions of Lifeact and EGFP-actin in lamellipodia of live cells (Fig. 1, A–C), which were then compared to F-actin distribution stained with phalloidin after fixation. Thin lamellipodia contain a dense F-actin meshwork (26). The measured F/G-actin ratio is 6:1 with extraction experiments (13) and 12:1∼20:1 with fluorescent decay after photoactivation (27). Owing to the high F/G-actin ratio in lamellipodia, EGFP-actin showed approximately identical distribution to F-actin (Fig. 1 B, middle). The fluorescence intensities of EGFP-actin and phalloidin were high near the leading edge, with a gradual decrease toward the base of lamellipodia (Fig. 1 C). In marked contrast, Lifeact reached its maximal intensity 2–5 μm away from the leading edge (Fig. 1, B and C). Importantly, when fixed cells were stained with Lifeact, F-actin distribution was identical to that stained with Lifeact (Fig. 1, D and E; Fig. S1), indicating that the rear-biased distribution of Lifeact (Fig. 1 C, also see Fig. 6) occurs specifically in live cells.
Figure 6.
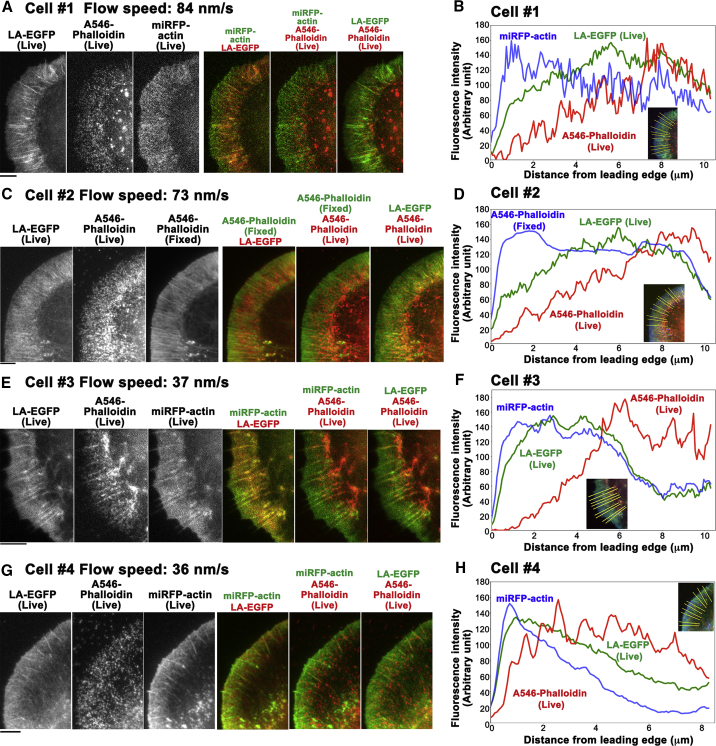
(A, C, E, and G) Images of Lifeact-EGFP (LA-EGFP) and Alexa 546 phalloidin (A546-phalloidin) in four live Xenopus XTC cells with the retrograde flow speeds indicated. The distributions of LA-EGFP and A546-phalloidin are compared to the F-actin distribution visualized by miRFP703-actin in live cells (A, E, and G) or staining with A546-phalloidin after fixation (C). (B, D, F, and H) The average fluorescence intensity of the images in (A, C, and E) or (G) along the yellow lines in the inset is shown. Representative data are shown (n = 11 cells). Bars, 5 μm.
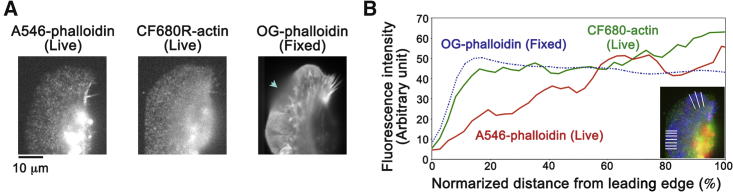
We also found the rear-biased misdistribution of phalloidin by comparing distributions of phalloidin and fluorescently labeled actin (15, 18) in lamellipodia of live fish keratocytes (Fig. 2, A and B; Fig. S2). In live keratocytes, phalloidin was scarcely distributed near the leading edge and gradually increased toward the base of lamellipodia (Fig. 2 A, left, and Fig. 2 B, red line). By contrast, fluorescent dye-labeled actin in live cells (Fig. 2 A, middle, and Fig. 2 B, green line) and F-actin stained with phalloidin after fixation (Fig. 2 A, right, and Fig. 2 B, blue dotted line) distributed almost uniformly in lamellipodia, which was compatible with F-actin distributions in the previous histochemical and electron microscopic studies (21, 22). Similar rear-biased distribution of phalloidin in live cells was found in the previous studies (10, 11, 12, 28). Of note, Vallotton et al. (11) reported a remarkable discrepancy of phalloidin distribution between live-cell and fixed-cell situations by direct comparison in the same cell (11). These observations reveal the apparent misdistribution of both Lifeact and phalloidin toward the base of lamellipodia in living cells.
Figure 2.
(A) Fluorescent speckle images of Alexa 546 phalloidin (A546-phalloidin, left) and CF680R-actin (middle) in a live fish keratocyte. The probes were delivered into keratocytes by electroporation (15). F-actin was stained with Oregon Green phalloidin (OG-phalloidin) after fixation (right). (B) The average fluorescence intensity of the images in (A) along the white lines in the inset is shown. Representative data are shown (n = 4 cells). The region where the lamellipodium detached from the glass was excluded from analysis (the blue arrow in (A)).
Mathematical simulations of convection-induced biased distribution of actin probes
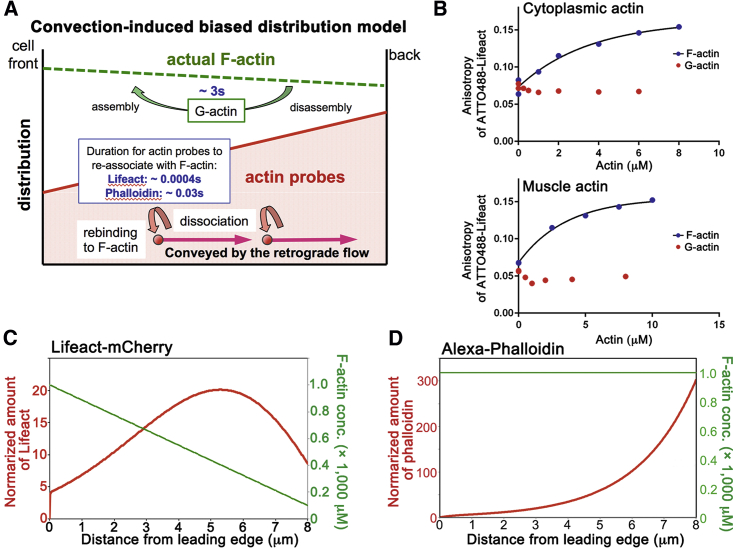
We hypothesized a convection-induced biased distribution mechanism (Fig. 3 A) for the rear-biased misdistribution of actin probes. In lamellipodia, the dense F-actin (∼1000 μM) (26) network is constantly conveyed to the cell center by the retrograde actin flow.
Figure 3.
(A) Convection-induced biased distribution model. Because of fast reassociation and the retrograde actin flow, the probes are biased toward back of lamellipodia. (B) Measurement of Atto-488 Lifeact binding to cytoplasmic (upper) or muscle (lower) G-actin (blue dots) and F-actin (red dots) by fluorescence anisotropy is shown. Atto-488 Lifeact binds to F-actin with the lines showing the best fit to Kd = 3.4 μM for cytoplasmic actin and Kd = 2.3 μM for muscle actin. (C) Calculated distribution of LA-mCherry (red) in a model lamellipodium with a linear decrease in F-actin concentration (green) is shown. (D) Calculated distribution of Alexa phalloidin (red) in a model lamellipodium with a uniform F-actin distribution (green) is shown.
Lifeact has been reported to have a 10- to 30-fold higher affinity for binding to G-actin than to F-actin in vitro (5). We reevaluated the affinities of Lifeact for G- and F-actin by fluorescence anisotropy assay in vitro using Atto-488-labeled Lifeact (Atto 488-Lifeact). F-actin increased fluorescence anisotropy of Atto-488 Lifeact in a concentration-dependent manner (Fig. 3 B). We performed the assays with G-actin in a low-salt condition to prevent actin polymerization. Under the condition, neither muscle nor cytoplasmic G-actin caused an increase in fluorescence anisotropy of Atto-488 Lifeact (Fig. 3 B). We concluded that Lifeact probes shuttle between the F-actin bound state and the free state when introduced in live cells and built a physical model based on this promise.
The association of free Lifeact and phalloidin to F-actin is estimated to occur on the orders of 1 × 10−4 and 1 × 10−2 s, respectively. Thus, rapid reassociation to F-actin might prevent free probes from diffusing back to the front (Fig. 3 A). The convection-induced biased distribution hypothesis requires rigorous testing, especially for the fast exchangeable Lifeact probe (with off rate 30 s−1) (6) because Lifeact travels only ∼1 nm on average per binding event, and diffusion, which depends on the square root of time, may mobilize free probes effectively in a short time. To test this, we developed a physical model to calculate the distributions of Lifeact and phalloidin in lamellipodia. In the model, the dynamics of actin probes in lamellipodia at the position x (cell frame) and time t depends on transport by actin retrograde flow, diffusion in the cytosol, and binding kinetics:
| (13) |
and
| (14) |
where Cf, Cb, and F, respectively, represent concentrations of free, bound actin probes, and F-actin in lamellipodia. v indicates retrograde flow velocity, D represents the diffusion coefficient of free actin probes, and kon and koff are association and dissociation rates, respectively (see Materials and Methods for details of the model). We examined the concentration profile of actin-binding probes in the model lamellipodia at the steady state. Our model produced the concentration profile of total Lifeact (Cf + Cb) as a rear-biased gentle peak (Fig. 3 C), which is similar to our experimental observations (Fig. 1 C). For phalloidin in the lamellipodia of keratocytes, our model predicted an exponential increase of the total phalloidin concentration toward the lamellipodial base (Fig. 3 D), which is also similar to our experimental results (Fig. 2 B).
Several studies suggest the existence of actin oligomers in addition to G- and F-actin in lamellipodia (29, 30, 31). We previously reported that a simulation including actin oligomers provides a good agreement between single-molecule speckle microscopy and FRAP (fluorescence recovery after photobleaching) experiments with EGFP-actin in lamellipodia (29). We also examined the effects of the existence of actin oligomers on the convection-induced distribution of Lifeact using the values we estimated in our previous study (29). The calculated profile of total Lifeact showed that the rear-biased misdistribution of Lifeact remains under the condition, including actin oligomers, whereas the existence of actin oligomers provides slight attenuation of the misdistribution (Fig. S3).
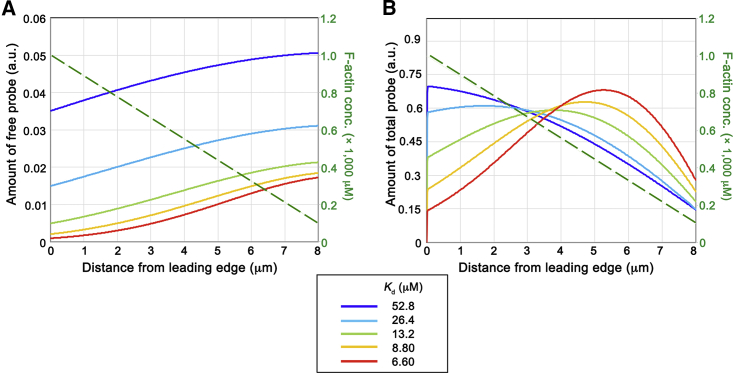
Our model showed that the different Kd of the actin probe gives rise to the rear-biased distribution patterns with various degrees of bias (Fig. 4). Even if the Kd was set to be as large as 53 μM (Fig. 4 B, blue line), the concentration profile clearly deviates from that of F-actin. Therefore, actin probes with faster binding kinetics than fluorescent protein-tagged Lifeact, which binds to F-actin with a Kd of 9–13 μM (25), can still be mislocalized by a convection-induced biased distribution mechanism.
Figure 4.
Simulated concentration profile of free (A) and total (B) actin probes with various Kd. The other parameters, except for kon, are the same as those shown in Fig. 3C and Table S1. F-actin distribution is depicted by a green dotted line. The amount of probe is measured in the same units on both panels.
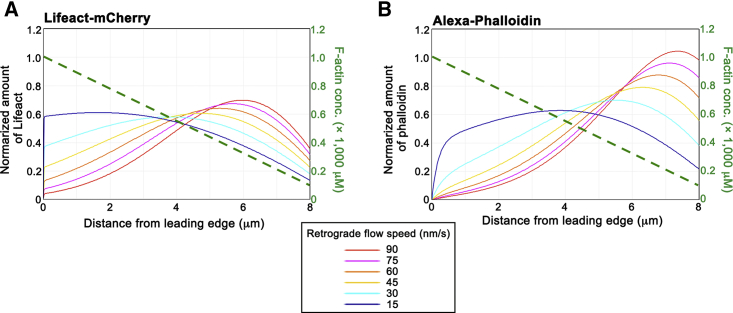
In addition, our physiological model enables us to predict how the retrograde flow speeds affect the convection-induced distribution patterns of the actin probes. Our model showed that the faster retrograde flow speed brings a more rear-biased effect on distributions of the actin probes (Fig. 5).
Figure 5.
Simulated concentration profile of total LA-mCherry (A) and Alexa phalloidin (B) with various retrograde flow speeds. The other parameters, except for retrograde flow speed, are the same as those adopted in Fig. 3C and Table S1. The linear F-actin distribution is depicted by a green dotted line.
To validate these model predictions, we compared distributions of Lifeact (low-affinity F-actin-binding probe) (25), phalloidin (high-affinity F-actin-binding probe), (24) and F-actin simultaneously in lamellipodia with diverse retrograde flow speeds (Fig. 6). We found that the peak of phalloidin intensity was more rear biased than those of Lifeact intensity (Fig. 6, B, D, F, and H. Also see merged images of LA-EGFP and A546-phalloidin in Fig. 6, A, C, E, and G), which is consistent with our model (Fig. 5). The rear-biased misdistribution of Lifeact was more marked in cells with fast retrograde flow speeds (Fig. 6, A–D) than cells with slow retrograde flow speeds (Fig. 6, E–H). These data indicate that the convection-induced misdistribution is highly sensitive to the change in the flow speed of the actin meshwork.
Discussion
In this study, we found a convection-dependent mechanism that may strongly influence the distribution of fluorescent markers. Interruption of F-actin binding of these actin probes by an actin-binding protein cofilin may also affect their distributions. Cofilin competes with phalloidin for binding to F-actin (32) and reduces the affinity of F-actin for Lifeact (25) in vitro. However, cofilin is excluded from the leading edge of keratocytes (33). In XTC cells, cofilin distributes throughout lamellipodia without showing accumulation near the leading edge (34). Therefore, interruption by cofilin cannot explain the rear-biased misdistribution of Lifeact and phalloidin in lamellipodia.
Initially, we started this study to examine whether the retrograde actin flow gives rise to the rear-biased misdistribution of phalloidin that has been used for quantitative live-cell imaging (10, 12). To our surprise, we found that even Lifeact, which exhibits extremely fast exchangeable binding kinetics (5, 6), showed rear-biased misdistribution under the influence of the retrograde actin flow. The convection-induced mechanism presumably influences distributions of actin-binding probes in general (35).
The previous study (36) proposes that the retrograde actin flow increments cellular polarity by convection of associate molecules. Maiuri et al. (36) demonstrated that the speed of retrograde actin flow positively correlates with the cell migration persistence time. The correlation was attributed to the convection of polarity cues by the retrograde flow, which reinforces the asymmetric distribution of polarity cues (36). The authors examined distributions of actin-binding probes, including Lifeact, in live cells with different retrograde flow speeds. In contrast to our observations, Maiuri et al. concluded that increasing the retrograde flow speed did not change the concentration profile of Lifeact (36). Misdistribution of Lifeact might have been overlooked because the distributions of actin-binding probes were not compared to the actual F-actin distribution. In addition, the authors proposed accumulation of a polarity cue to the rear based on the theoretical model including a reaction-diffusion-convection system (36). However, F-actin distribution was not taken into account in the model. Thus, the convection-induced misdistribution of Lifeact was unrecognized in the previous study (36).
Considering that Lifeact travels only ∼1 nm on average per F-actin-binding event, the influence of the retrograde actin flow would seemingly be negligible. However, in this study, both experimental observations and physical models revealed misdistribution of Lifeact toward the rear of lamellipodia. Our observations indicate that the misdistribution of Lifeact is sensitive to the change in the flow speed of the actin meshwork. Our physical model incorporating F-actin distribution and kon and koff values of Lifeact produced the concentration profile of Lifeact as a rear-biased peak, which is not consistent with the F-actin profile. These findings clearly indicate that the retrograde actin flow gives rise to the misdistribution of Lifeact.
Our finding also raises the intriguing possibility that the retrograde actin flow may influence distributions of actin-binding proteins to generate gradient distribution. As described above, cofilin is excluded from the leading edge of keratocytes (33), which may be partly due to the convection-dependent mechanism in lamellipodia.
Our results warn about the potential error arising from the use of target-binding probes in quantitative imaging. Even Lifeact, which has ideal properties as an actin-binding probe, showed noticeable misdistribution under the influence of the retrograde actin flow. Several studies have employed Lifeact or phalloidin for quantitative analyses of the amounts of F-actin (14, 37) and for mapping actin network turnover in lamellipodia (10, 12). We suggest that reevaluation of these studies is necessary for the possible artifact arising from convection-mediated misdistribution of actin probes. A similar problem might occur in live-cell imaging of phenomena accompanied by cytoplasmic flow such as in C. elegans zygotes, Drosophila oocytes, and plant cells. When observing cells with cytoplasmic flow, it is necessary to pay attention to whether target-binding probes report actual localization.
Prompted by our finding of the discrepancy between F-actin and phalloidin distributions, we recently reexamined myosin regulation of actin stability (38). In the previous study (12), phalloidin was employed as an F-actin probe in quantitative fluorescent speckle microscopy in live fish keratocytes. The quantitative fluorescent speckle microscopy study reported that actin-myosin II contraction enhances actin network disassembly (12). However, rear-biased misdistribution of phalloidin may have hindered the accurate monitoring of actin network redistribution. We therefore employed fluorescent DyLight-dye-labeled actin that shows identical distribution to F-actin in lamellipodia. In combination with electroporation-based single-molecule speckle microscopy, which allows direct observation of actin turnover, we revealed that the myosin II inhibitor blebbistatin enhances actin disassembly in lamellipodia of fish keratocytes and lamella of Xenopus XTC cells at an early stage of the inhibition (38). Our data thus provide compelling evidence that actomyosin contraction stabilizes on actin filaments, contrary to the results of earlier studies (12, 39, 40, 41).
Conclusions
In this study, we show evidence of convection-induced misdistribution of actin probes by both experimental data and physical models. Our simulations suggest that a convection-dependent mechanism may strongly influence the distribution of actin-binding probes. Our finding warns about the potential error arising from the use of target-binding probes in quantitative live-cell imaging.
Author Contributions
S.Y., S.T., and T.K. performed experiments and analyzed data. S.Y., S.T., T.K., and N.W. designed experiments. D.T., D.V., and N.W. performed simulations. S.Y., D.T., and N.W. wrote the manuscript. All other authors edited the manuscript.
Acknowledgments
We thank Shu Yamamura for help with line scan analysis.
This work was supported by Japan Society for the Promotion of Science KAKENHI Grant Number JP15K07045 and JP18K06217 (S.Y.), by National Institutes of Health RO1GM114201 (D.V.), by Japan Science and Technology Agency-Core Research for Evolutional Science and Technology Grant Number JPMJCR15G5 (N.W.), and by the Uehara Memorial Foundation (N.W.).
Editor: Kinneret Keren.
Footnotes
Sawako Yamashiro and Daisuke Taniguchi contributed equally to this work.
Supporting Materials and Methods, four figures, and two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)31287-6.
Contributor Information
Sawako Yamashiro, Email: yamashiro.sawako.5c@kyoto-u.ac.jp.
Naoki Watanabe, Email: watanabe.naoki.4v@kyoto-u.ac.jp.
Supporting Material
References
- 1.Gulyani A., Vitriol E., Hahn K.M. A biosensor generated via high-throughput screening quantifies cell edge Src dynamics. Nat. Chem. Biol. 2011;7:437–444. doi: 10.1038/nchembio.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helma J., Schmidthals K., Leonhardt H. Direct and dynamic detection of HIV-1 in living cells. PLoS One. 2012;7:e50026. doi: 10.1371/journal.pone.0050026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamashita S., Tsuboi T., Michiue T. Wide and high resolution tension measurement using FRET in embryo. Sci. Rep. 2016;6:28535. doi: 10.1038/srep28535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melak M., Plessner M., Grosse R. Actin visualization at a glance. J. Cell Sci. 2017;130:525–530. doi: 10.1242/jcs.189068. [DOI] [PubMed] [Google Scholar]
- 5.Riedl J., Crevenna A.H., Wedlich-Soldner R. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiuchi T., Higuchi M., Watanabe N. Multitarget super-resolution microscopy with high-density labeling by exchangeable probes. Nat. Methods. 2015;12:743–746. doi: 10.1038/nmeth.3466. [DOI] [PubMed] [Google Scholar]
- 7.Small J.V. Getting the actin filaments straight: nucleation-release or treadmilling? Trends Cell Biol. 1995;5:52–55. doi: 10.1016/s0962-8924(00)88939-4. [DOI] [PubMed] [Google Scholar]
- 8.Cramer L.P. Molecular mechanism of actin-dependent retrograde flow in lamellipodia of motile cells. Front. Biosci. 1997;2:d260–d270. doi: 10.2741/a189. [DOI] [PubMed] [Google Scholar]
- 9.Yamashiro S., Watanabe N. A new link between the retrograde actin flow and focal adhesions. J. Biochem. 2014;156:239–248. doi: 10.1093/jb/mvu053. [DOI] [PubMed] [Google Scholar]
- 10.Schaub S., Bohnet S., Verkhovsky A.B. Comparative maps of motion and assembly of filamentous actin and myosin II in migrating cells. Mol. Biol. Cell. 2007;18:3723–3732. doi: 10.1091/mbc.E06-09-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vallotton P., Danuser G., Verkhovsky A.B. Tracking retrograde flow in keratocytes: news from the front. Mol. Biol. Cell. 2005;16:1223–1231. doi: 10.1091/mbc.E04-07-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson C.A., Tsuchida M.A., Theriot J.A. Myosin II contributes to cell-scale actin network treadmilling through network disassembly. Nature. 2010;465:373–377. doi: 10.1038/nature08994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe N., Mitchison T.J. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- 14.Ryan G.L., Petroccia H.M., Vavylonis D. Excitable actin dynamics in lamellipodial protrusion and retraction. Biophys. J. 2012;102:1493–1502. doi: 10.1016/j.bpj.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamashiro S., Mizuno H., Watanabe N. New single-molecule speckle microscopy reveals modification of the retrograde actin flow by focal adhesions at nanometer scales. Mol. Biol. Cell. 2014;25:1010–1024. doi: 10.1091/mbc.E13-03-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shcherbakova D.M., Baloban M., Verkhusha V.V. Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat. Commun. 2016;7:12405. doi: 10.1038/ncomms12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe N. Fluorescence single-molecule imaging of actin turnover and regulatory mechanisms. Methods Enzymol. 2012;505:219–232. doi: 10.1016/B978-0-12-388448-0.00020-6. [DOI] [PubMed] [Google Scholar]
- 18.Yamashiro S., Watanabe N. An infrared actin probe for deep-cell electroporation-based single-molecule speckle (eSiMS) microscopy. Sensors (Basel) 2017;17:E1545. doi: 10.3390/s17071545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamashiro S., Mizuno H., Watanabe N. An easy-to-use single-molecule speckle microscopy enabling nanometer-scale flow and wide-range lifetime measurement of cellular actin filaments. Methods Cell Biol. 2015;125:43–59. doi: 10.1016/bs.mcb.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Spudich J.A., Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 21.Svitkina T.M., Verkhovsky A.B., Borisy G.G. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J. Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theriot J.A., Mitchison T.J. Actin microfilament dynamics in locomoting cells. Nature. 1991;352:126–131. doi: 10.1038/352126a0. [DOI] [PubMed] [Google Scholar]
- 23.McGrath J.L., Tardy Y., Hartwig J.H. Simultaneous measurements of actin filament turnover, filament fraction, and monomer diffusion in endothelial cells. Biophys. J. 1998;75:2070–2078. doi: 10.1016/S0006-3495(98)77649-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De La Cruz E.M., Pollard T.D. Kinetics and thermodynamics of phalloidin binding to actin filaments from three divergent species. Biochemistry. 1996;35:14054–14061. doi: 10.1021/bi961047t. [DOI] [PubMed] [Google Scholar]
- 25.Courtemanche N., Pollard T.D., Chen Q. Avoiding artefacts when counting polymerized actin in live cells with LifeAct fused to fluorescent proteins. Nat. Cell Biol. 2016;18:676–683. doi: 10.1038/ncb3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abraham V.C., Krishnamurthi V., Lanni F. The actin-based nanomachine at the leading edge of migrating cells. Biophys. J. 1999;77:1721–1732. doi: 10.1016/S0006-3495(99)77018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiuchi T., Nagai T., Mizuno K. Measurements of spatiotemporal changes in G-actin concentration reveal its effect on stimulus-induced actin assembly and lamellipodium extension. J. Cell Biol. 2011;193:365–380. doi: 10.1083/jcb.201101035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burnette D.T., Schaefer A.W., Forscher P. Filopodial actin bundles are not necessary for microtubule advance into the peripheral domain of Aplysia neuronal growth cones. Nat. Cell Biol. 2007;9:1360–1369. doi: 10.1038/ncb1655. [DOI] [PubMed] [Google Scholar]
- 29.Smith M.B., Kiuchi T., Vavylonis D. Distributed actin turnover in the lamellipodium and FRAP kinetics. Biophys. J. 2013;104:247–257. doi: 10.1016/j.bpj.2012.11.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyoshi T., Tsuji T., Watanabe N. Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: evidence of frequent filament severing. J. Cell Biol. 2006;175:947–955. doi: 10.1083/jcb.200604176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raz-Ben Aroush D., Ofer N., Keren K. Actin turnover in lamellipodial fragments. Curr. Biol. 2017;27:2963–2973.e14. doi: 10.1016/j.cub.2017.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishida E., Iida K., Sakai H. Cofilin is a component of intranuclear and cytoplasmic actin rods induced in cultured cells. Proc. Natl. Acad. Sci. USA. 1987;84:5262–5266. doi: 10.1073/pnas.84.15.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svitkina T.M., Borisy G.G. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuji T., Miyoshi T., Watanabe N. An order of magnitude faster AIP1-associated actin disruption than nucleation by the Arp2/3 complex in lamellipodia. PLoS One. 2009;4:e4921. doi: 10.1371/journal.pone.0004921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belin B.J., Goins L.M., Mullins R.D. Comparative analysis of tools for live cell imaging of actin network architecture. Bioarchitecture. 2014;4:189–202. doi: 10.1080/19490992.2014.1047714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maiuri P., Rupprecht J.F., Voituriez R. Actin flows mediate a universal coupling between cell speed and cell persistence. Cell. 2015;161:374–386. doi: 10.1016/j.cell.2015.01.056. [DOI] [PubMed] [Google Scholar]
- 37.Lee C.W., Vitriol E.A., Zheng J.Q. Dynamic localization of G-actin during membrane protrusion in neuronal motility. Curr. Biol. 2013;23:1046–1056. doi: 10.1016/j.cub.2013.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashiro S., Tanaka S., Watanabe N. Myosin-dependent actin stabilization as revealed by single-molecule imaging of actin turnover. Mol. Biol. Cell. 2018;29:1941–1947. doi: 10.1091/mbc.E18-01-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guha M., Zhou M., Wang Y.L. Cortical actin turnover during cytokinesis requires myosin II. Curr. Biol. 2005;15:732–736. doi: 10.1016/j.cub.2005.03.042. [DOI] [PubMed] [Google Scholar]
- 40.Murthy K., Wadsworth P. Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Curr. Biol. 2005;15:724–731. doi: 10.1016/j.cub.2005.02.055. [DOI] [PubMed] [Google Scholar]
- 41.Haviv L., Gillo D., Bernheim-Groswasser A. A cytoskeletal demolition worker: myosin II acts as an actin depolymerization agent. J. Mol. Biol. 2008;375:325–330. doi: 10.1016/j.jmb.2007.09.066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.