Abstract
Long noncoding RNAs (lncRNAs) transcribed across gene promoters have been detected. These regulate transcription by mechanisms that have not been fully elucidated. We herein show that the chromatin configuration is altered into an accessible state within 290 bp downstream from the initiation site of metabolic-stress-induced lncRNAs (mlonRNAs) in the promoter of the fission yeast fbp1 gene, whose transcription is massively induced upon glucose starvation. Chromatin upstream from fbp1 is progressively altered into an open configuration, as a cascade of transcription of three overlapping mlonRNA species (-a, -b and -c in order) occurs with transcriptional initiation sites progressing 5′ to 3′ upstream of the fbp1 promoter. Initiation of the shortest mlonRNA (mlonRNA-c) induces chromatin remodeling around a transcription factor-binding site and subsequent massive induction of fbp1. We identify the cis-element required for mlonRNA-c initiation, and by changing the distance between mlonRNA-initiation site and the transcription factor-binding site, we show that mlonRNA-initiation effectively induces chromatin remodeling in a limited distance within 290 bp. These results indicate that mlonRNAs are transcribed across the fbp1 promoter as a short-range inducer for local chromatin alterations, and suggest that strict chromatin modulation is archived via stepwise mlonRNA-initiations.
Introduction
Recent transcriptome analyses have revealed that most regions in the human genome are transcribed into RNAs, of which RNAs longer than 200 nucleotides possessing mRNA-like structure (carrying cap-structure and poly-A tail) without protein-coding potential are referred to as long noncoding RNA (lncRNA)1. Various functions of such RNAs have been identified in a range of biological processes2,3. lncRNAs transcribed within gene promoters play a role in the regulation of neighboring genes3. Several lncRNAs interact with polycomb repressive complex 2 (PRC2) and recruit it to target genes, leading to methylation of histone H3K27 following chromatin compaction3,4. Intergenic noncoding transcription at the budding yeast SER3 gene promoter represses the expression of this gene5, and this repressive activity was observed even when >90% of the lncRNA sequence was replaced6. These data indicate that RNA polymerase II (RNAPII) transcription in the regulatory region is sufficient to mediate repression, and that lncRNA itself does not play a direct role. In Schizosaccharomyces pombe (fission yeast), upon glucose starvation, stepwise expression of lncRNAs at the fbp1 gene promoter plays a critical role in chromatin modulation and subsequent gene activation7. This activation is mediated through two distinct mechanisms: (1) the lncRNA itself interacts with Tup1-like corepressors and thereby antagonizes the repressive function of the Tup1-like corepressors and facilitates binding of the Atf1 transcription factor8, and (2) RNAPII-mediated transcription of the lncRNAs mediate chromatin remodeling and further enhance Atf1 transcription-factor binding8,9.
Activation of the fission yeast fbp1 gene as a result of glucose starvation stress is mediated by two transcription factors: Atf1 and Rst210,11. Upon glucose starvation, these transcription factors bind to critical cis-acting binding sequences far upstream from the fbp1 promoter (upstream-activating sequences 1 [UAS1] and 2 [UAS2])10,12,13 (Fig. 1A). In this upstream region, several species of lncRNA, referred to as metabolic stress-induced lncRNAs (mlonRNAs), are transcribed (Fig. 1A, mlonRNA-a, -b and -c in order). Initially, we had defined ‘mRNA type long ncRNA’ as ‘mlonRNA’, when the term ‘lncRNA’ had not been well recognized14. However, after this definition, the term ‘lncRNA’ has been used to mean the ‘mRNA-type long ncRNA’, and thus the definition of mlonRNA was changed to indicate ‘metabolic stress-induced lncRNAs’15. These mlonRNAs are transcribed in a stepwise manner from transcriptional initiation sites located in a 5′ to 3′ direction, leading to chromatin remodeling along their transcribed tract in the upstream region of fbp17,9. RNAPII transcription of mlonRNAs passing across UAS2 is required for histone acetylation, disassembly, and subsequent Rst2 binding7,9. However, the mechanisms underlying the regulation of mlonRNA transcriptional initiation as well as the chromatin remodeling by mlonRNA transcription have not been elucidated. We herein identify the cis-element required for mlonRNA transcriptional initiation and demonstrate that mlonRNA transcription effectively induces chromatin remodeling within 290 bp downstream from the mlonRNA initiation site.
Figure 1.
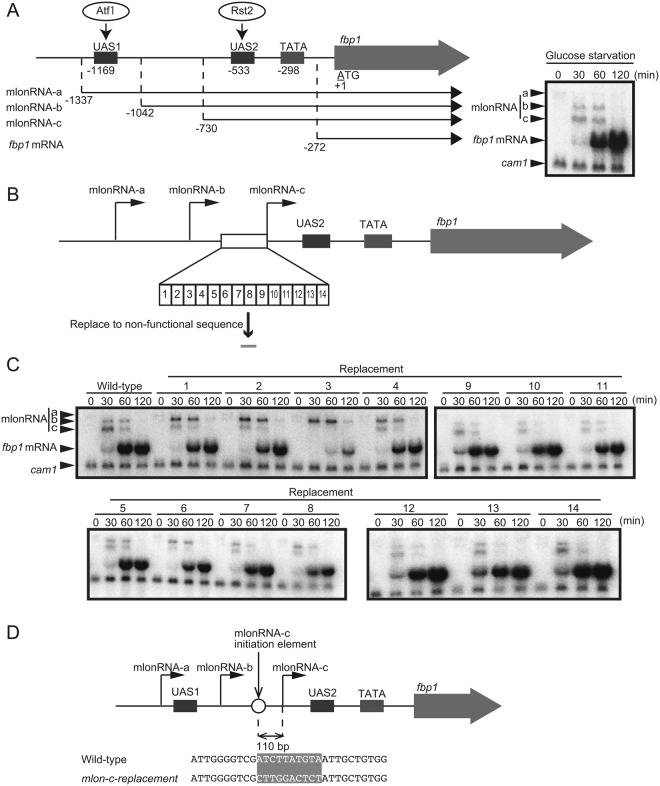
Identification of the cis-acting element required for mlonRNA-c transcriptional initiation. (A) Schematic drawing of the region upstream from fbp1 containing upstream-activating sequences 1 and 2 (UAS 1 and UAS2), the binding sites for transcription factors Atf1 and Rst2, respectively. The mlonRNAs transcribed across the fbp1 upstream region and fbp1 mRNA are presented. The numbers indicate the transcription start-site of the fbp1 transcripts and the distances of UAS1, UAS2, and the TATA box from the first ATG of fbp1 ORF. Representative northern blot image showing expression of the mlonRNAs and fbp1 mRNA during glucose starvation. Wild-type cells were grown to 2.0 × 107 cells/mL in YER medium, then transferred to YED medium. Cells were harvested at the indicated times. cam1 transcript was used as an internal control. (B) Schematic representation of segments covering the region upstream from the mlonRNA-c initiation site. We divided this region into 14 segments, then replaced each segment from the act1 ORF. (C) Northern blot analysis to examine fbp1 transcripts in cells carrying replacement sequences upstream from the mlonRNA-c initiation site. Cells were cultured and harvested as A. (D) The 10 nt sequence replaced in mlon-c-replacement cells is shaded.
Results
Identification of the mlonRNA transcriptional-initiation element
During fbp1 transcriptional activation upon glucose starvation, the chromatin state far upstream from the fbp1 promoter is progressively altered into an open configuration. In this process, several species of mlonRNAs are transcribed in a stepwise manner with transcriptional initiation sites progressing in a 5′ to 3′ direction, thus inducing chromatin remodeling along the same tract7,9. However, how such stepwise mlonRNA transcriptions mediate chromatin remodeling is not well-understood. To investigate the significance of these stepwise mlonRNA transcriptions, we sought to selectively disrupt one of the transcripts in a cascade of transcription of three overlapping mlonRNAs. Since the initiation sites of mlonRNA–a and –b are close to essential cis-element, UAS1, and selective disruption of cis-elements for these mlonRNAs was difficult, we focused our efforts on disrupting the shortest mlonRNA-c transcription. To this end, we searched for the cis-element required for mlonRNA-c initiation by replacing 10–15 bp segments with act1 ORF sequences in the region upstream from the mlonRNA-c transcription initiation site (Fig. 1B). mlonRNA-c transcription is completely lost by replacing a 10 bp segment at 110 bp upstream from the transcription initiation site (Fig. 1C, strain 3). We thus conclude that the cis-acting element required for mlonRNA-c initiation is present in this 10 nt sequence (5′-ATCTTATGTA-3′) (Fig. 1D). Moreover, cells in which this critical sequence had been replaced (mlon-c-replacement cells) exhibit a drastic reduction in fbp1 mRNA transcription (Fig. 1C). These data are consistent with previously appreciated critical role of mlonRNAs in chromatin remodeling and subsequent transcription factor binding at UAS27,9. Given the role played by mlonRNA initiation in chromatin opening, we hypothesized that there is a limitation in distance between the mlonRNA initiation site and the area of chromatin opening, such that the mlonRNA-a and -b initiation sites are too distant to promote chromatin opening at UAS2.
Initiation of mlonRNA-b transcription 190 bp upstream from UAS2 bypasses the requirement for mlonRNA-c transcriptional initiation
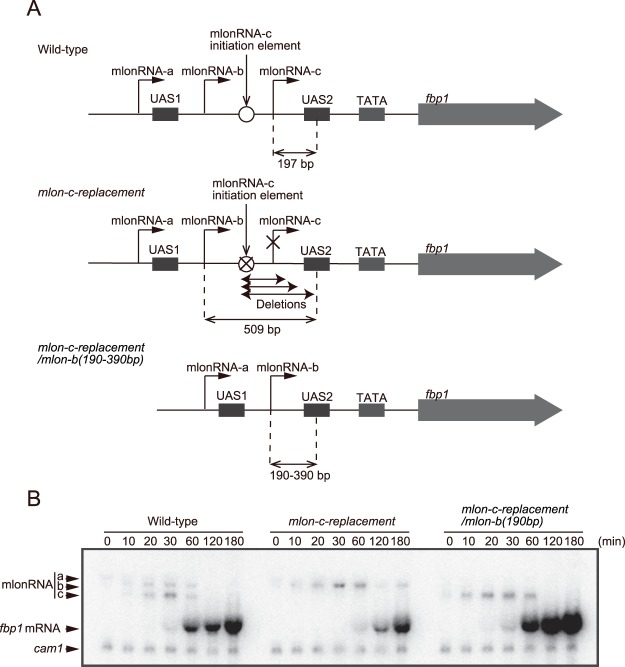
To determine whether mlonRNA initiation affects chromatin remodeling within a limited range, we deleted a 319 bp segment containing the mlonRNA-c initiation element, thus moving the mlonRNA-b initiation site closer to UAS2 (mlon-c-replacement/mlon-b(190 bp); Fig. 2A). In the mlon-c-replacement cells described in Fig. 1, the distance between the mlonRNA-b initiation site and UAS2 is 509 bp, whereas in the mlon-c-replacement/mlon-b(190 bp) cells, the distance of the corresponding region is 190 bp, a distance similar to that of mlonRNA-c and UAS2 in wild-type cells (197 bp) (Fig. 2A). By placing the mlonRNA-b initiation site 190 bp upstream from UAS2, fbp1 induction after glucose starvation is restored (Fig. 2B).
Figure 2.
mlonRNA-b transcriptional initiation 190 bp upstream from UAS2 bypasses the requirement of mlonRNA-c transcriptional initiation for fbp1 induction. (A) Schematic drawing of the region upstream from fbp1 in wild-type, mlon-c-replacement, and mlon-c-replacement/mlon-b(190–390 bp) cells. In mlon-c-replacement/mlon-b(190–390 bp) cells, the distance between the mlonRNA-b initiation site and UAS2 was reduced to 190 bp, 240 bp, 290 bp, 340 bp, and 390 bp. (B) Northern blot analysis in wild-type, mlon-c-replacement, and mlon-c-replacement/mlon-b(190 bp) cells. Cells were cultured and harvested as described in Fig. 1.
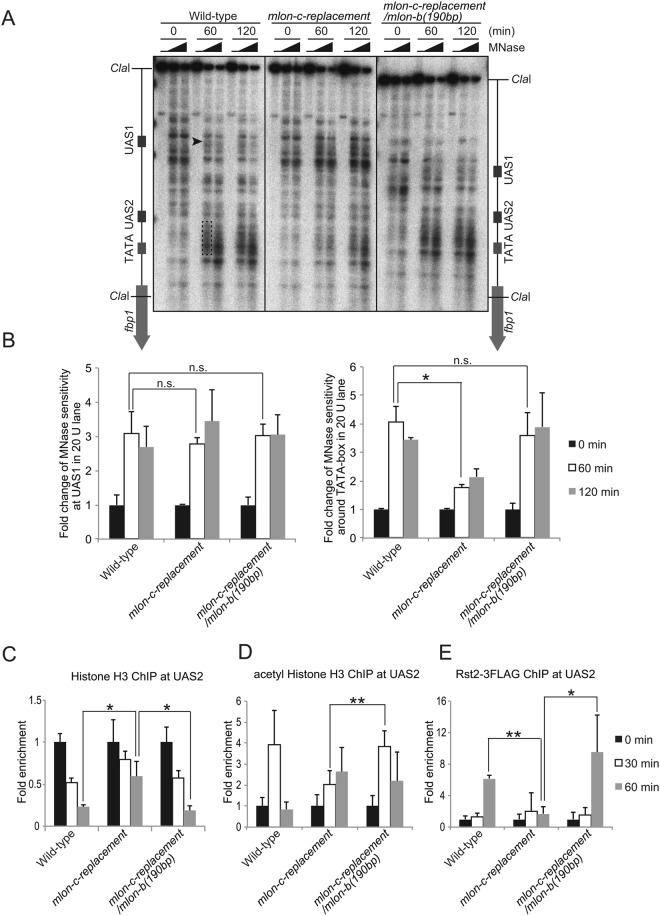
To determine if this recovery is indeed attributable to the restoration of chromatin remodeling at UAS2, we investigated the status of chromatin configuration. To this end, we employed an indirect end-labeling analysis involving partial digestion of chromatin DNA with micrococcal nuclease (MNase) to measure sensitivity to MNase reflecting open chromatin configuration. In wild-type cells, MNase sensitive sites appear near UAS1 followed by sites in the UAS2-TATA box region (Fig. 3A,B). In marked contrast, mlon-c-replacement cells exhibit less prominent MNase sensitive region around UAS2-TATA box (Fig. 3A,B). However, by moving the mlonRNA-b initiation site closer to UAS2 (mlon-c-replacement/mlon-b(190 bp)), MNase sensitivity around UAS2-TATA box region is completely restored to wild-type level (Fig. 3A,B). Consistently, histone binding at UAS2 is progressively reduced in wild-type cells, diminishing to less than 30% of the original state at 60 min after glucose starvation, whereas the mlon-c-replacement cells exhibit impaired histone disassembly, with approximately 60% of histones remaining at 60 min (Fig. 3C). This histone-disassembly defect is also completely restored by moving the mlonRNA-b initiation site closer to UAS2 (mlon-c-replacement/mlon-b(190 bp)) (Fig. 3C).
Figure 3.
mlonRNA-b transcriptional initiation 190 bp upstream from UAS2 bypasses the requirement of mlonRNA-c transcriptional initiation for the efficient chromatin remodeling and transcription-factor binding. (A) Southern blot image showing MNase sensitive sites around the fbp1 promoter in the indicated cells cultured in YED for the indicated time. The isolated chromatin was digested with 0, 20, or 50 units/ml of MNase at 37 °C for 5 min. Purified DNA was digested with ClaI and analyzed by Southern blotting. Black arrowhead indicates region with MNase-sensitive sites at UAS1. Dotted line indicates MNase-sensitive sites around UAS2-TATA box. (B) Histogram shows the quantification of MNase-sensitive bands in the UAS1 (Black arrowhead) and UAS2-TATA box region (Dotted line). The error bars show the standard deviation from at least two independent experiments. (C–E) ChIP analysis to examine histone binding (C), histone acetylation (D) and Rst2 binding (E) at UAS2 in wild-type, mlon-c-replacement, and mlon-c-replacement/mlon-b(190 bp) cells. The relative increase in the ratio at the indicated time after glucose starvation is indicated. Error bars represent standard deviations from at least two independent experiments. p-value was calculated by a Student’s t-test: *p < 0.05, **p < 0.01 and n.s. (not significant, p > 0.05).
We further analyzed the kinetics of histone acetylation and transcription-factor binding at UAS2. At 30 min after glucose starvation, histone acetylation is transiently induced over three fold over the original state in wild-type cells, while mlon-c-replacement cells exhibit delayed and reduced histone acetylation in this region (Fig. 3D). Moreover, Rst2 binding at UAS2 increases > five-fold in the wild-type cells, while the mlon-c-replacement cells exhibit little Rst2 binding after glucose starvation (Fig. 3E). These defects of histone acetylation and Rst2 binding observed in the mlon-c-replacement cells are significantly rescued in mlon-c-replacement/mlon-b(190 bp) cells (p < 0.01 and p < 0.05, respectively) and this cell line shows very similar kinetics to those of the wild-type cells (Fig. 3D,E). These results demonstrate that mlonRNA-b initiation 190 bp upstream from UAS2 efficiently promotes chromatin remodeling at UAS2, and bypasses the requirement of mlonRNA-c transcriptional initiation for fbp1 mRNA induction. We conclude that mlonRNA transcriptional initiation induces chromatin remodeling within a limited distance from the initiation site.
The effective range of mlonRNA transcription-induced chromatin remodeling is approximately 290 bp
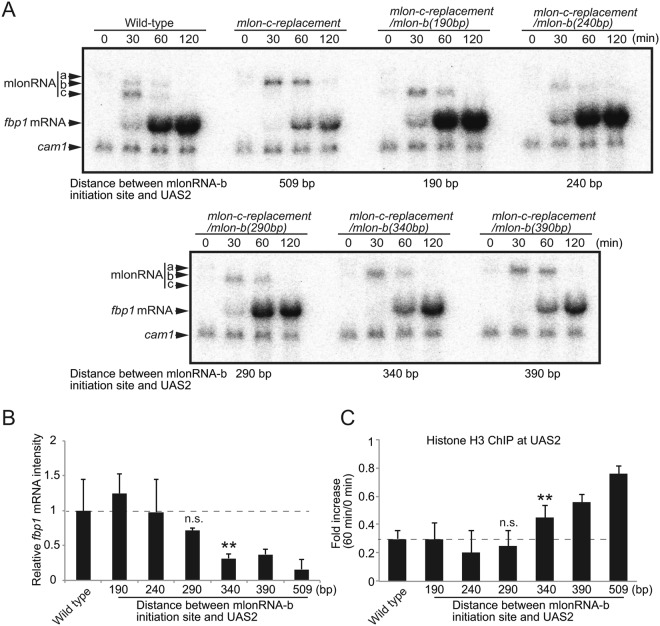
We next sought to identify the limitation in the distance from mlonRNA initiation to induce chromatin remodeling at UAS2. To this end, we examined various distances (240, 290, 340, and 390 bp) between the mlonRNA-b initiation site and UAS2 by generating mlon-c-replacement/mlon-b(240 bp), mlon-c-replacement/mlon-b(290 bp), mlon-c-replacement/mlon-b(340 bp), and mlon-c-replacement/mlon-b(390 bp) cells (Fig. 2A). We found that the induction of fbp1 mRNA lessens as the distance between the mlonRNA-b initiation site and UAS2 increases (Fig. 4A,B). The mlon-c-replacement/mlon-b(190–290 bp) cells exhibit higher or wild-type levels of fbp1 mRNA induction, whereas the mlon-c-replacement/mlon-b(340–509 bp) cells show significantly lower levels of induction compared to wild-type cells (Fig. 4A,B, p < 0.01). The higher level of fbp1 expression in the mlon-c-replacement/mlon-b(190 bp) strain might be attributable to the alteration of genome geometry in this region by sequence deletion, since fbp1 activation is also affected by the local DNA-loop that brings UAS1 and UAS2 into close spatial proximity16. We next evaluated histone disassembly at 60 min after glucose starvation in the cell lines with various segment lengths between the mlonRNA-b initiation site and UAS2. In the wild-type cells, histone H3 binding is reduced to below 30% of the original state. In the mlon-c-replacement cells with 190 bp, 240 bp, and 290 bp segments between the mlonRNA-b initiation site and UAS2, histone disassembly is very similar to that of the wild-type(Fig. 4C, n.s., p > 0.05). In marked contrast, the mlon-c-replacement cells with the 340, 390, and 509 bp segments exhibit histone binding levels of 50%, 60%, and 75%, respectively (Fig. 4C, p < 0.01). These data suggest that mlonRNA initiation efficiently induces histone disassembly within a limited distance of ~290 bp from the initiation site, while chromatin remodeling gradually decreases as the distance from the mlonRNA initiation site increases beyond 290 bp.
Figure 4.
mlonRNA transcriptional initiation promotes chromatin remodeling within an effective range of approximately 290 bp. (A) Northern blot analysis in wild-type, mlon-c-replacement, and mlon-c-replacement/mlon-b(190–390 bp) cells. Cells were cultured and harvested as described in Fig. 1. (B) fbp1 mRNA transcripts were quantified using Image J (https://imagej.nih.gov/ij/) and intensities at 60 min after initiation of glucose starvation relative to those of wild-type cells are presented. Error bars represent standard deviation from three independent experiments (C) Histone binding 60 min after initiation of glucose starvation relative to 0 min (glucose rich condition) was assessed by ChIP analysis. Error bars show the standard deviation from at least two independent experiments. p-value was calculated by a Student’s t-test: *p < 0.05, **p < 0.01 and n.s. (not significant, p > 0.05).
Discussion
Transcriptional activation of the fission yeast fbp1 gene is preceded by RNAPII-mediated transcription of a series of lncRNAs, referred to as mlonRNAs, in a stepwise manner. Transcription of the mlonRNAs induces chromatin remodeling in their transcribed regions, thereby contributing to the access of transcription factors to the fbp1 regulatory elements7. In this study, we identified the cis-acting sequence required for transcriptional initiation of the mlonRNA-c transcript, which is the transcript nearest to the fbp1 promoter. Loss of mlonRNA-c transcription due to inactivating this sequence resulted in a critical defect in chromatin remodeling at the Rst2 transcription-factor binding site located downstream from the mlonRNA-c initiation site. These results indicate the pivotal role played by mlonRNA transcriptional initiation in chromatin remodeling, as the mlonRNA-b transcript overlaps with the mlonRNA-c transcript, showing that transcriptional elongation through this region is not sufficient to promote chromatin remodeling. An interesting possibility is that the RNAPII complex that transcribes mlonRNA might be distinct from the usual RNAPII complex associated with the TATA box, and such mlonRNA transcribing RNAPII may play a role in inducing chromatin remodeling. Should this be true, this mlonRNA-RNAPII would function locally, around the initiation site, since RNAPII accumulates around its transcription-start site and dissociates from the initiation complex after promoter clearance17. This is indeed the case, as we found that mlonRNA initiation efficiently induces chromatin remodeling only within 290 bp of the initiation site (Fig. 4). This range is comparable to the length of two adjacent nucleosomes (294 bp). This is intriguing, since two neighboring nucleosomes tend to be modified in a similar manner18,19. Our current observations further support the notion raised by previous studies that non-canonical transcripts can regulate gene expression20,21, and provides further evidence that lncRNA-transcribing RNAPIIs can act as “pioneers” to rearrange the chromatin array22.
In the analysis to identify the limitation in the distance from mlonRNA initiation to induce chromatin remodeling, we found slightly higher level of fbp1 expression in the mlon-c-replacement/mlon-b(190 bp) strain (Fig. 4B). This higher expression may be attributable to the alteration of the distance between UAS1 and UAS2 by sequence deletion, since fbp1 activation is also affected by the local DNA-loop that brings UAS1 and UAS2 into close spatial proximity16. It is possible that such local DNA-loop structure might be also affected by the mlonRNAs initiations. Since chromatin remodeling is prerequisite for the DNA-loop formation in glucose starvation16, chromatin remodeling defects caused by the mlonRNA-c replacement may indirectly affect on the local DNA-loop structure. Important questions for future studies concern a possible direct contribution of mlonRNA initiation to the formation of such local DNA-loop structure.
We note that the positional relationships between each of the mlonRNA initiation sites and their target elements are very similar, as evidenced by the fact that the distances between (1) the mlonRNA-b initiation site and the cis-element required for mlonRNA-c transcription and (2) the mlonRNA-c initiation site and the cis-element (UAS2) required for fbp1 mRNA are 192 bp and 197 bp, respectively. Stepwise transcriptional activation of several species of mlonRNA may be responsible for converting chromatin into an open configuration upstream from fbp1, thereby contributing to the strict binding-order regulation of transcription factors. The transcription factor Rst2 binding is mediated by a local DNA-loop that brings UAS1 and UAS2 into close spatial proximity16. The formation of an open chromatin configuration upstream from fbp1 is a prerequisite for the formation of this higher-order structure. Thus, the strict regulation of chromatin configuration by a cascade of mlonRNA transcriptions and the subsequent formation of a local DNA-loop structure should cooperatively control fbp1 transcription. Limiting the range in which mlonRNA can effectively induce chromatin remodeling may contribute to this strict chromatin modulation by prohibiting improper chromatin opening. This stepwise regulation via the cascade of mlonRNA transcriptions within the range limitation for each mlonRNA initiation may mediate this strict modulation of gene expression and the appropriate response to glucose starvation stress23.
In the S. pombe core environmental stress response (CESR) gene, the first nucleosome downstream from the transcription-start site (+1 nucleosome) is removed during gene activation. This process is mediated in a histone H3 acetyltransferase, Gcn5 dependent manner24. Gcn5 is recruited through Atf1 binding at UAS1 in the fbp1 gene and is involved in the activation of this gene8,9. It is thus possible that chromatin eviction at UAS2 through mlonRNA-c initiation is mediated by a process involving Atf1 and Gcn5. This hypothesis is supported by the fact that expression of mlonRNA-c and subsequent chromatin remodeling is missing in atf1− cells7. Interestingly, loss of the Tup1-like corepressors in atf1− cells totally rescues the defect in fbp1 induction without recovering mlonRNA-c initiation7. These data suggest a possible counteractive regulation between mlonRNA-initiation-mediated chromatin opening and Tup1-like-corepressor-mediated chromatin compaction. Since mutant fission yeast cells lacking Tup1-like corepressors show inappropriate, nonspecific fbp1 transcription25, this counteractive regulation might be pivotal for precise fbp1 activation resulting from environment stresses.
Methods
Fission-yeast strains, genetic methodology, and cell culture
The fission yeast strains used in this study are listed in Supplementary Table 1. YER medium (yeast extract containing 6% glucose) and YED medium (yeast extract containing 0.1% glucose and 3% glycerol) were used for glucose repression and starvation, respectively26. Transformation was performed using the lithium-acetate method as previously described27.
Primers
The primer sequences used in this study are listed in Supplementary Table S2.
Construction of strains with sequence replacements upstream from the mlonRNA-c initiation site
The fbp1 promoter region including the mlonRNA-c upstream sequence was amplified using a primer set (p1 and p2) and cloned in pCR®-Blunt II-TOPO® (Invitrogen). Replacement of the region upstream from mlonRNA-c with act1 ORF was performed by PCR using the primer sets described in Supplemental Table 2. For replacement numbers 1–14, primer pairs p1 and p3-16, p2 and p17-30 were used respectively. The resulting fragments were purified using QIAquick gel extraction kit (Qiagen). Pairs of fragments were used as templates for PCR amplification using the primer set p1 and p2 to generate replacement constructs for numbers 1–14. Fission yeast cells carrying the ura4 marker gene in the fbp1 promoter were transformed using plasmid carrying the fbp1 upstream sequence, in which a part of the segment upstream from the mlonRNA-c initiation site was replaced with the act1 ORF sequence. To isolate the fbp1 upstream-sequence replacement cells, transformants were selected for uracil auxotrophy using SD plates containing 5-FOA and uracil.
Construction of mlon-c-replacement cells with sequence deletions between the mlonRNA-c initiation element and UAS2
To construct mlon-c-replacement cells with various deletion lengths in the region between the mlonRNA-c initiation element and UAS2, we deleted regions -937 to -619, -669, -719, -769, and -819 (relative to the first A in the ORF as +1) by PCR, using the primer sets p31 and p32 to p36 as described in Supplemental Table 2. We used the fbp1 upstream region cloned in pCR®-Blunt II-TOPO® (Invitrogen) as a template. PCR products were phosphorylated by T4 Polynucleotide Kinase (Takara Bio) and ligated using T4 DNA Ligase (TOYOBO). Fission yeast cells carrying the ura4 marker gene in the fbp1 promoter were transformed with these plasmid constructs and selected for uracil auxotrophy, as above.
Construction of rst2-3flag strains
All rst2-3flag strains were constructed as described previously28.
Northern blot, chromatin immunoprecipitation, and micrococcal nuclease (MNase) digestion assay
Northern blotting and chromatin immunoprecipitation (ChIP) were performed as described previously29. DNA probes for fbp1 and cam1 were amplified by PCR using primer sets p37/p38 and p39/p40, respectively. Anti-histone H3 (Abcam), anti-acetyl histone H3 (millipore), and anti- DYKDDDDK antibody (Wako) were used for the ChIP analysis. DNA concentrations were quantified using Thermal Cycler Dice Real Time (Takara Bio) and THUNDERBIRD® SYBR qPCR Mix (TOYOBO). Primer sets p41/p42 at UAS2 and p43/p44 at the prp3 locus were used for quantitative PCR analysis. MNase digestion assays were performed as described previously16.
Electronic supplementary material
Acknowledgements
We thank all the members of the Hirota laboratory for their help. We acknowledge the Radioisotope Research Center in Tokyo Metropolitan University for support in the use of isotopes. This work was supported in part by JSPS KAKENHI (16K12598, 16H02957, 16H01314 to KH, 17H03711 to KO and 16J02252 to RA) and the Takeda Science Foundation and Yamada Science Foundation (to KH). Correspondence and requests for materials should be addressed to Kouji Hirota (khirota@tmu.ac.jp).
Author Contributions
Conceived and designed the experiments: S.S., R.A. and K.H. Performed the experiments: S.S. and R.A. Wrote the paper: S.S., T.A., C.S.H., K.O. and K.H.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Satoshi Senmatsu and Ryuta Asada contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36049-0.
References
- 1.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang KC, Chang HY. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonasio R, Shiekhattar R. Regulation of Transcription by Long Noncoding RNAs. Annu. Rev. Genet. 2014;48:433–455. doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 6.Martens JA, Wu PYJ, Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005;19:2695–2704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirota K, et al. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–134. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- 8.Takemata N, et al. Local potentiation of stress-responsive genes by upstream noncoding transcription. Nucleic Acids Res. 2016;44:5174–5189. doi: 10.1093/nar/gkw142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adachi A, et al. Interplay between chromatin modulators and histone acetylation regulates the formation of accessible chromatin in the upstream regulatory region of fission yeast fbp1. Genes Genet. Syst. 2017;92:267–276. doi: 10.1266/ggs.17-00018. [DOI] [PubMed] [Google Scholar]
- 10.Neely LA, Hoffman CS. Protein kinase A and mitogen-activated protein kinase pathways antagonistically regulate fission yeast fbp1 transcription by employing different modes of action at two upstream activation sites. Mol. Cell. Biol. 2000;20:6426–6434. doi: 10.1128/MCB.20.17.6426-6434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higuchi T, Watanabe Y, Yamamoto M. Protein Kinase A Regulates Sexual Development and Gluconeogenesis through Phosphorylation of the Zn Finger Transcriptional Activator Rst2p in Fission. Yeast. 2002;22:1–11. doi: 10.1128/MCB.22.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffman CS, Hill C. Glucose sensing via the protein kinase A pathway in Schizosaccharomyces pombe. Biochem. Soc. Trans. 2015;33:257–260. doi: 10.1042/BST0330257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janoo RTK, Neely LA, Braun BR, Whitehall SK, Hoffman CS. Transcriptional regulators of the Schizosaccharomyces pombe fbp1 gene include two redundant Tup1p-like corepressors and the CCAAT binding factor activation complex. Genetics. 2001;157:1205–1215. doi: 10.1093/genetics/157.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirota K, Ohta K. Transcription of mRNA-type long non-coding RNAs (mlonRNAs) disrupts chromatin array. Commun. Integr. Biol. 2009;2:25–26. doi: 10.4161/cib.2.1.7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galipon J, Miki A, Oda A, Inada T, Ohta K. Stress-induced lncRNAs evade nuclear degradation and enter the translational machinery. Genes to Cells. 2013;18:353–368. doi: 10.1111/gtc.12042. [DOI] [PubMed] [Google Scholar]
- 16.Asada R, et al. Recruitment and delivery of the fission yeast Rst2 transcription factor via a local genome structure counteracts repression by Tup1-family corepressors. Nucleic Acids Res. 2017;45:9361–9371. doi: 10.1093/nar/gkx555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Consortium RE, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erdel F. How Communication Between Nucleosomes Enables Spreading and Epigenetic Memory of Histone Modifications. BioEssays. 2017;1700053:1–12. doi: 10.1002/bies.201700053. [DOI] [PubMed] [Google Scholar]
- 20.Schmitt S, Prestel M, Paro R. Intergenic transcription through a Polycomb group response element counteracts silencing. Genes Dev. 2005;19:697–708. doi: 10.1101/gad.326205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.T Sanchez-Elsner, D. Gou., Elisabeth Kremmer, F. S. Noncoding RNAs of Trithorax Response Elements Recruit Drosophila Ash1 to Ultrabithorax. 311, 1118–1123 (2014). [DOI] [PubMed]
- 22.Orphanides G, Reinberg D. RNA polymerase II elongation through chromatin. Nature. 2000;407:471–5. doi: 10.1038/35035000. [DOI] [PubMed] [Google Scholar]
- 23.Chen D, et al. Global Transcriptional Responses of Fission Yeast to Environmental Stress. Mol. Biol. Cell. 2003;14:214–229. doi: 10.1091/mbc.e02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sansó M, et al. Gcn5 facilitates Pol II progression, rather than recruitment to nucleosome-depleted stress promoters, In Schizosaccharomyces pombe. Nucleic Acids Res. 2011;39:6369–6379. doi: 10.1093/nar/gkr255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirota K, et al. Fission yeast global repressors regulate the specificity of chromatin alteration in response to distinct environmental stresses. Nucleic Acids Res. 2004;32:855–862. doi: 10.1093/nar/gkh251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirota K, Hoffman CS, Shibata T, Ohta K. Fission Yeast Tup1-Like Repressors Repress Chromatin Remodeling at the fbp1+ Promoter and the ade6-M26 Recombination Hotspot. Genetics. 2003;165:505–515. doi: 10.1093/genetics/165.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirota K, Tanaka K, Watanabe Y, Yamamoto M. Functional analysis of the C-terminal cytoplasmic region of the M-factor receptor in fission yeast. Genes to Cells. 2001;6:201–214. doi: 10.1046/j.1365-2443.2001.00415.x. [DOI] [PubMed] [Google Scholar]
- 28.Asada R, Takemata N, Hoffman CS, Ohta K, Hirota K. Antagonistic Controls of Chromatin and mRNA Start Site Selection by Tup Family Corepressors and the CCAAT-Binding Factor. Mol. Cell. Biol. 2015;35:847–855. doi: 10.1128/MCB.00924-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirota K, Hoffman CS, Ohta K. Reciprocal nuclear shuttling of two antagonizing Zn finger proteins modulates tup family corepressor function to repress chromatin remodeling. Eukaryot. Cell. 2006;5:1980–1989. doi: 10.1128/EC.00272-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.