Abstract
Little is known about the causes of meningioma. Obesity and obesity-related traits have been reported in several epidemiological observational studies to be risk factors for meningioma. We performed an analysis of genetic variants associated with obesity-related traits to assess the relationship with meningioma risk using Mendelian randomization (MR), an approach unaffected by biases from temporal variability and reverse causation that might have affected earlier investigations. We considered 11 obesity-related traits, identified genetic instruments for these factors, and assessed their association with meningioma risk using data from a genome-wide association study comprising 1,606 meningioma patients and 9,823 controls. To evaluate the causal relationship between the obesity-related traits and meningioma risk, we consider the estimated odds ratio (OR) of meningioma for each genetic instrument. We identified positive associations between body mass index (odds ratio [ORSD] = 1.27, 95% confidence interval [CI] = 1.03–1.56, P = 0.028) and body fat percentage (ORSD = 1.28, 95% CI = 1.01–1.63, P = 0.042) with meningioma risk, albeit non-significant after correction for multiple testing. Associations for basal metabolic rate, diastolic blood pressure, fasting glucose, high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, systolic blood pressure, total cholesterol, triglycerides and waist circumference with risk of meningioma were non-significant. Our analysis provides additional support for obesity being associated with an increased risk of meningioma.
Introduction
Meningioma is the commonest form of brain tumour, affecting approximately one per cent of the adult population1. Although mortality from meningioma is generally low, the disease is associated with substantial morbidity.
Thus far, little is known about the aetiological basis of meningioma with few well-established risk factors for the disease2,3. There is increasing recognition that obesity and obesity-related traits, such as body mass index (BMI), are risk factors for common cancers such as breast, colon, ovarian, renal and pancreatic4. Observational epidemiological studies have however provided inconsistent evidence for obesity and obesity-related traits being risk factors for meningioma5–12. Moreover, in contrast to many cancers, some studies have reported an inverse relationship between hyperglycaemia, type-2 diabetes and meningioma risk13–15. Such observational epidemiological studies can however be influenced by reverse causation, biasing findings.
Mendelian randomisation (MR) uses genetic variants as instrumental variables (IVs) to assess the causal relationship between an exposure, such as a lifestyle-related or environmental risk factor, and an outcome, such as the risk of a disease16. These genetic variants represent unconfounded markers of exposure, provided that these instruments are not associated with disease risk through an alternative mechanism16. As these genetic variants are assigned randomly at conception, they are not influenced by reverse causation, and MR therefore represents an approach orthogonal to traditional observational epidemiological studies. If the assumptions of MR are not violated, then the association between the genetic instruments and the outcome of interest indicates a causal relationship between the exposure and outcome. Parallels have been drawn between MR and randomised controlled trails, in that the random assignment of variants at conception similarly circumvents some of the limitations of observational epidemiological studies17.
Many genetic instruments explain only a small proportion of exposure variation18 and MR frameworks have therefore been developed that consider multiple genetic variants when estimating the causal effect of an exposure on an outcome19. Each additional independent single nucleotide polymorphism (SNP) considered in a multi-SNP approach provides further information on the causal relationship, and combining all valid genetic instruments thereby affords a more precise estimation of the causal effect19.
To further our understanding of meningioma aetiology, we implemented a Generalised Summary-data-based Mendelian Randomisation (GSMR) to evaluate the causal relationship between 11 key obesity-related traits and meningioma risk20. This multi-SNP MR approach is typically more powerful than other summary-data-based MR approaches19. Furthermore, GSMR can account for linkage disequilibrium (LD) between SNPs20. We identified genetic instruments for each obesity-related trait from external genome-wide association studies (GWAS). Using these genetic instruments in conjunction with association statistics from a recent GWAS meta-analysis of meningioma21,22, we estimate the causal effect of each trait on meningioma risk.
Results
We considered 11 obesity-related traits: BMI, basal metabolic rate (BMR), body fat percentage, diastolic blood pressure (DBP), fasting glucose, low-density lipoprotein cholesterol (LDL), high-density lipoprotein cholesterol (HDL), systolic blood pressure (SBP), total cholesterol, triglycerides and waist circumference. SNPs were excluded if there were data missing in either the two meningioma GWAS, or the data sets used to compute LD correlations (see Supplementary Table S1). We performed Heterogeneity in Dependent Instruments (HEIDI) outlier analysis20 to identify those genetic instruments violating MR pleiotropy assumptions, identifying three BMI, three BMR, two body fat percentage and two waist circumference SNPs, which were further excluded from the analyses (see Supplementary Table S1).
The number of SNPs used as IVs for each obesity-related trait, the proportion of variance explained (PVE) by these SNPs, and the mean and standard deviation (SD) for each obesity-related trait in the original discovery study are detailed in Table 1. The PVE was calculated using sample size estimates from each GWAS as well as effect sizes and standard errors for each SNP effect estimate. The SNPs used as IVs for each obesity-related trait, along with the association statistics for these SNPs in respect to disease risk are shown in Supplementary Table S2.
Table 1.
Genetic instruments used to evaluate the causal relationship between obesity-related traits and meningioma risk.
| Obesity-related trait | SNPs* | PVE (%) | Units | Source |
|---|---|---|---|---|
| BMI | 927 | 7.5 | kg/m2 | Yengo et al.43 |
| BMR | 677 | 11.2 | kilojoules | UK Biobank |
| Body fat percentage | 434 | 5.8 | % | UK Biobank |
| DBP | 216 | 3.1 | mmHg | UK Biobank |
| Fasting glucose | 27 | 3.0 | mmol/l | Scott et al.42 |
| HDL | 50 | 5.1 | mg/dl | Willer et al.44 |
| LDL | 19 | 2.2 | mg/dl | Willer et al.44 |
| SBP | 250 | 3.5 | mmHg | UK Biobank |
| Total cholesterol | 29 | 1.1 | mg/dl | Willer et al.44 |
| Triglycerides | 19 | 1.9 | mg/dl | Willer et al.44 |
| Waist circumference | 367 | 4.9 | cm | UK Biobank |
*After removal of SNPs during quality control and LD trimming.
One SNP reported in the original LDL discovery study (rs9411489) has been merged with another SNP (rs635634), and we therefore derived meningioma association statistics using rs635634.
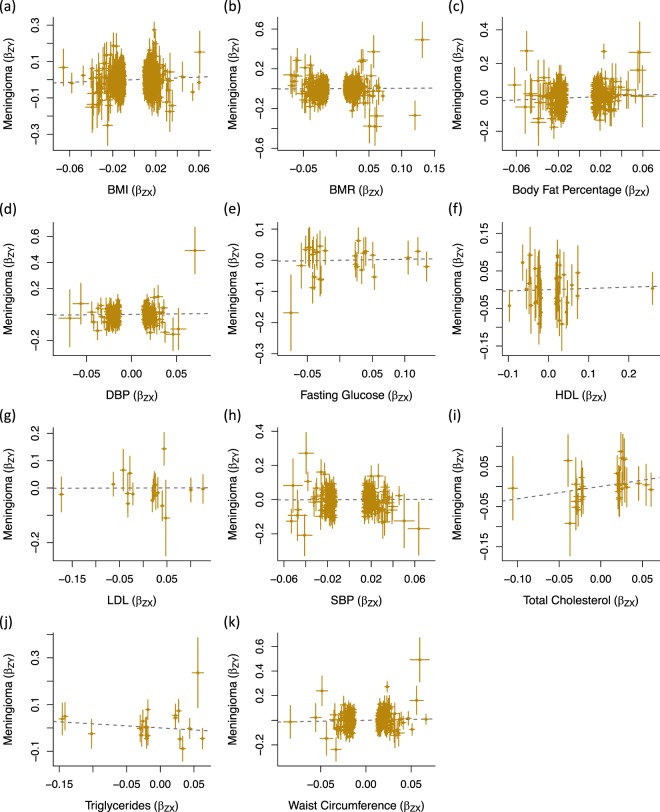
The association of each IV with the respective obesity-related trait and meningioma risk, and the log odds ratios (ORs) estimated by GSMR, are shown in Fig. 1. At a significance threshold of P<0.05, risk of meningioma was not associated with BMR (ORSD = 1.04, 95% confidence interval [CI] = 0.87–1.24, P = 0.67), DBP (ORSD = 1.07, 95% CI = 0.77–1.48, P = 0.68), fasting glucose (ORSD = 1.03, 95% CI = 0.74–1.43, P = 0.85), HDL (ORSD = 1.03, 95% CI = 0.81–1.33, P = 0.79), LDL (ORSD = 1.00, 95% CI = 0.68–1.49, P = 0.98), SBP (ORSD = 1.02, 95% CI = 0.75–1.39, P = 0.89), total cholesterol (ORSD = 1.36, 95% CI = 0.79–2.34, P = 0.26), triglycerides (ORSD = 0.84, 95% CI = 0.54–1.30, P = 0.44) or waist circumference (ORSD = 1.16, 95% CI = 0.89–1.50, P = 0.27) (Table 2). There was however support for a positive relationship between both BMI (ORSD = 1.27, 95% CI = 1.03–1.56, P = 0.028) and body fat percentage (ORSD = 1.28, 95% CI = 1.01–1.63, P = 0.042) with meningioma risk, albeit non-significant after correction for multiple testing.
Figure 1.
Effects of genetic instruments on obesity-related traits and meningioma risk. Shown are the results for (a) BMI, (b) BMR, (c) body fat percentage, (d) DBP, (e) fasting glucose, (f) HDL, (g) LDL, (h) SBP, (i) total cholesterol, (j) triglycerides and (k) waist circumference. For each SNP, the effect for the obesity-related trait is plotted against the effect for meningioma risk. Dashed lines represent GSMR estimates. Error bars represent one standard deviation. BMI: body mass index; BMR; basal metabolic rate; DBP: diastolic blood pressure; GSMR: Generalised Summary-data-based Mendelian Randomisation; HDL: high-density lipoprotein cholesterol; LDL: low-density lipoprotein cholesterol; SNP: single nucleotide polymorphism.
Table 2.
GSMR results for each obesity-related trait and meningioma risk.
| Obesity-related trait | OR (95% CI) | P-value |
|---|---|---|
| BMI | 1.27 (1.03–1.56) | 0.028 |
| BMR | 1.04 (0.87–1.24) | 0.674 |
| Body fat percentage | 1.28 (1.01–1.63) | 0.042 |
| DBP | 1.07 (0.77–1.48) | 0.679 |
| Fasting glucose | 1.03 (0.74–1.43) | 0.853 |
| HDL | 1.03 (0.81–1.33) | 0.792 |
| LDL | 1.00 (0.68–1.49) | 0.980 |
| SBP | 1.02 (0.75–1.39) | 0.887 |
| Total cholesterol | 1.36 (0.79–2.34) | 0.264 |
| Triglycerides | 0.84 (0.54–1.30) | 0.436 |
| Waist circumference | 1.16 (0.89–1.50) | 0.267 |
We used MR-Egger regression to assess whether directional pleiotropy could affect the causal estimates (see Supplementary Fig. S1 and Supplementary Table S3). In none of these analyses was the intercept of the causal effect significantly different from zero (i.e. P < 0.05), thereby providing no evidence that overall unbalanced pleiotropy biased results.
No obesity-related traits were identified by the MR-Egger as being associated with meningioma risk (see Supplementary Table S3). However, whilst a positive relationship between BMI and risk of meningioma was estimated by GSMR (ORSD = 1.27, 95% CI = 1.03–1.56, P = 0.028), a non-significant negative relationship was estimated by MR-Egger regression (ORSD = 0.93, 95% CI = 0.51–1.71, P = 0.83). As MR-Egger regression can be strongly influenced by a single outlying variant23, we conducted leave-one-out analysis to determine whether such a variant could explain this disparity (see Supplementary Table S4). When rs663129 was removed, the OR for BMI and risk of meningioma estimated by MR-Egger regression was 1.06 (95% CI = 0.57–2.00, P = 0.85). Conversely, the removal of rs663129 had little effect on the OR for BMI and risk of meningioma estimated by GSMR (OR = 1.29, 95% CI = 1.05–1.60, P = 0.017). This outlying variant therefore partly explains the difference in causal effects estimated by the two approaches. Single-SNP analyses of each obesity-related trait did not identify any other outlying SNPs (see Supplementary Fig. S2).
Discussion
Twin and family studies have provided strong evidence for large genetic influences on obesity-related traits, with heritability estimates for BMI of between 50% and 90%, leaving the remaining variance attributable to environmental influences24. In our analysis, genetic markers correlated with 11 obesity-related traits were used as proxies for these traits.
As for the common cancers, there is increasing evidence that obesity increases the risk of developing rarer tumours, such as multiple myeloma4,25, inviting speculation that obesity and its related traits provide an environment supportive to tumorigenesis in general. As previously stated, observational epidemiological studies have provided mixed evidence on whether obesity influences the risk of meningioma development5–11. Diabetes and fasting serum glucose levels have been identified as being inversely related to meningioma risk in some studies8,13,14. Anti-diabetic treatment has however paradoxically been reported as being positively related to meningioma risk in one study15. Suggested mechanisms for the association of obesity and obesity-related traits with meningioma risk include chronic inflammation, and increased adipokine-mediated signalling, insulin signalling and insulin-like growth factor (IGF) signalling26,27. Obesity is associated with higher circulating levels of IGF-1, which is known to suppress apoptosis and stimulate tumour growth28. Higher expression of components of the IGF signalling system have also been observed in meningioma, suggesting a role for these proteins in the development of these tumours29. Furthermore, the transcription regulatory factors leptin and leptin receptor have been shown to be important in the proliferation and survival of patient-derived meningioma cell lines30. Leptin hormone concentrations are higher in individuals with high BMI, providing an alternative or complementary mechanism for the association of obesity with meningioma risk30. Although the long-term metabolic consequences of obesity-related traits are highly complex, these mechanisms are compatible with the generic effect of obesity on cancer risk.
Oestrogen pathways also provide a biological explanation for the relationship between obesity and some cancers, including breast and endometrial31. Oestrogen interacts with IGF in the brain32 and such hormonal factors may account for the two-fold increase in meningioma risk observed in women when compared to men33. Both hormone replacement therapy34–37 and oral contraceptive use35–37 have however been inconsistently associated with meningioma risk. In breast cancer, a higher lifetime exposure to oestrogen, as measured using age at menarche and menopause, is associated with increased disease risk38. Reported associations between lifetime oestrogen exposure and meningioma risk have however not been consistent3,39, suggesting a complex relationship between hormone exposure, obesity and risk of meningioma.
Our MR analysis provides evidence for a causal relationship between obesity, specifically higher BMI and body fat percentage, and meningioma risk. An advantage of using MR to identify causal relationships between environmental factors and disease risk is that the random allocation of genetic variants at conception avoids reverse causation, which may have biased causal estimates in previous observational epidemiological studies. MR-Egger analysis of each obesity-related trait did not identify bias caused by directional pleiotropy (see Supplementary Table S3). Therefore, whilst we cannot exclude the possibility that confounders such as type-2 diabetes medication use may be having some effect on the causal estimates, it is unlikely that they are a substantial source of bias. Some potential weak instrument bias is quantified by the low (<10) first-stage F-statistics for BMI, BMR, body fat percentage, DBP, SBP, total cholesterol and waist circumference, with F-statistics of 0.9, 2.0, 1.6, 1.7, 1.6, 4.6 and 1.6 respectively. However, F-statistics for fasting glucose, LDL, HDL and triglycerides indicates that these are strong IVs, with respective values of 13.0, 13.7, 12.3, 11.2, and are hence less likely to be affected by weak instrument bias40.
Our study does however have limited power to detect causal effects for some of the obesity-related traits (Table 3). In particular, our study had less than 80% power to identify ORs of 0.67 or 1.50 for DBP, fasting glucose and total cholesterol. Thus, interpretation of the null results found for these traits in this study are limited, as modest associations between these traits and meningioma risk cannot be excluded.
Table 3.
Odds ratios required for 80% study power.
| Obesity-related trait | OR low | OR high |
|---|---|---|
| BMI | 0.76 | 1.32 |
| BMR | 0.80 | 1.25 |
| Body fat percentage | 0.73 | 1.37 |
| DBP | 0.65 | 1.53 |
| Fasting glucose | 0.65 | 1.54 |
| HDL | 0.78 | 1.28 |
| LDL | 0.70 | 1.43 |
| SBP | 0.67 | 1.49 |
| Total cholesterol | 0.62 | 1.61 |
| Triglycerides | 0.69 | 1.45 |
| Waist circumference | 0.71 | 1.40 |
In conclusion, our analysis provides further evidence that higher BMI and body fat percentage are risk factors for meningioma.
Methods
All data used in this study were retrieved from publicly available summary statistics of published GWAS and we therefore did not seek ethical approval for this specific project. This study used no individual-level data.
Genetic instruments for obesity-related traits
We identified genetic instruments for each obesity-related trait suitable for use in MR analyses by considering the largest GWAS or meta-analysis of each trait conducted to date. Only continuous obesity-related traits were considered, as MR analysis of a binary exposure with a binary outcome using summary data in a two-sample setting can result in inflated causal estimates41. We identified a panel of SNPs associated with each trait at a genome-wide level of significance (P < 5 × 10–8) and minor allele frequency >0.01. Only studies of individuals of European ancestry were considered. Genetic instruments for fasting glucose were retrieved from an analysis by the Meta-Analysis of Glucose and Insulin related traits Consortium (MAGIC)42, instruments for BMI from the Genetic Investigation of Anthropometric Traits (GIANT) consortium43, instruments for HDL, LDL, total cholesterol and triglycerides from the Global Lipids Genetic Consortium (GLGC)44, and instruments for BMR, body fat percentage, DBP, SBP and waist circumference were obtained from GWAS using data from UK Biobank45–47. For each SNP used as a genetic instrument, we obtained the estimated per-allele effect on the obesity-related trait, the standard error of this estimate, and the chromosomal position of the SNP (see Supplementary Table S2). In order to mitigate against co-linearity between SNPs, which can bias causal estimates, we used MR-Base to exclude correlated SNPs at a threshold of r2 > 0.01, retaining the SNPs with the strongest effect on the respective obesity-related trait46,48. Effect size estimates and the standard errors of these estimates were standardized to represent the per-allele effect of the SNP on the exposure in standard deviation units20.
Meningioma association statistics
We examined the association of the respective genetic instruments for each obesity-related trait with risk of meningioma using data from a recent meta-analysis of meningioma GWAS. This GWAS meta-analysis includes 1,606 meningioma patients and 9,823 controls of European descent from two independent GWAS datasets (see Supplementary Table S5)22, and after imputation comprises >9 million genetic variants. Details of the two independent GWAS datasets, including participant recruitment and the quality control procedures conducted, have been reported previously22. We used MR-Base to ensure that the effect of each SNP on the obesity-related trait and meningioma risk corresponded to the same allele46.
Evaluating causal relationships using Mendelian randomisation
For each obesity-related trait, we used GSMR to estimate the OR of meningioma per unit of SD increment of each considered risk factor20. The GSMR R-package contains an implementation of the HEIDI outlier test, which can be used to identify loci that influence multiple phenotypes (i.e. the exposure and outcome considered in an MR analysis)20. LD correlations between SNPs were estimated using data from the 1000 Genomes Project49 and the UK10K50. We used HEIDI outlier tests to identify and exclude SNPs that may have pleiotropic effects on each obesity-related trait and meningioma risk (see Supplementary Table S1).
To derive conclusions concerning which obesity-related traits were causally related to meningioma risk, we considered P < 0.05 as a global significance level. To correct for multiple testing we applied Bonferroni-correction imposing a significance threshold of 0.0045 (i.e. 0.05/11 traits). The power of any MR analysis depends on the PVE captured by the SNPs used as IVs and we therefore estimated the power of each analysis using the method from Burgess et al.51. All statistical analyses were conducted using R (v3.1.2).
We used MR-Egger regression to further assess the violation of MR assumptions caused by directional pleiotropy (see Supplementary Fig. S1 and Supplementary Table S3)52. Significant difference between the slope estimated by MR-Egger regression and the intercept was considered evidence of violation. SNPs failing the HEIDI outlier tests were not included in the MR-Egger regression analyses. Furthermore, we conducted single-SNP MR-Egger analyses and display the results in funnel plots to allow the visual identification of any outlying SNPs (see Supplementary Fig. S2)52. We used leave-one-out analysis, as per Burgess and Thompson23, to assess whether outlying variants could be influencing causal estimates from MR-Egger regression.
Electronic supplementary material
Acknowledgements
H.T. and L.D-H. were supported by Wellcome Trust Summer Student bursaries. A.S. was supported by a Cancer Research UK Clinical Fellowship. Funding in the UK was provided by Cancer Research UK (C1298/A8362), with support from the Bobby Moore Fund.
Author Contributions
Project managed by A.J.C., C.T. and R.S.H. Statistical analyses conducted by H.T., A.J.C., A.S., P.J.L. and L.D.-H. German data analysed by J.S., P.H., M.M.N., K.-H.J. and M.S. USA data analysed by L.C., L.L., H.M.H., I.S., K.M.W., J.M.S., M.B., M.W., J.L.W. and E.B.C. Manuscript written by H.T., A.J.C., A.S., P.J.L. and R.S.H. The final manuscript was reviewed and approved by all authors.
Data Availability
Summary statistics for the obesity-related trait GWAS using UK Biobank data are available from http://www.nealelab.is/uk-biobank/. Genotype data from The Resource for Genetic Epidemiology Research on Aging can be downloaded from The Database of Genotypes and Phenotypes (dbGaP) (accession code phs000674.v2.p2). The 1000 Genomes Project and UK10K imputation panel data are available from the European Genome-phenome Archive (EGA, accession EGAD00001000776). Remaining data are available upon request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hannah Takahashi, Alex J. Cornish and Richard S. Houlston contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36186-6.
References
- 1.Kohler BA, et al. Annual report to the nation on the status of cancer, 1975–2007, featuring tumors of the brain and other nervous system. J Natl Cancer Inst. 2011;103:714–736. doi: 10.1093/jnci/djr077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braganza MZ, et al. Ionizing radiation and the risk of brain and central nervous system tumors: a systematic review. Neuro Oncol. 2012;14:1316–1324. doi: 10.1093/neuonc/nos208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiemels J, Wrensch M, Claus EB. Epidemiology and etiology of meningioma. J Neurooncol. 2010;99:307–314. doi: 10.1007/s11060-010-0386-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kyrgiou M, et al. Adiposity and cancer at major anatomical sites: umbrella review of the literature. The BMJ. 2017;356:j477. doi: 10.1136/bmj.j477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michaud DS, et al. Anthropometric measures, physical activity, and risk of glioma and meningioma in a large prospective cohort study. Cancer Prev Res (Phila) 2011;4:1385–1392. doi: 10.1158/1940-6207.CAPR-11-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiedmann M, et al. Body mass index and the risk of meningioma, glioma and schwannoma in a large prospective cohort study (The HUNT Study) Br J Cancer. 2013;109:289–294. doi: 10.1038/bjc.2013.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niedermaier T, et al. Body mass index, physical activity, and risk of adult meningioma and glioma: A meta-analysis. Neurology. 2015;85:1342–1350. doi: 10.1212/WNL.0000000000002020. [DOI] [PubMed] [Google Scholar]
- 8.Seliger C, et al. Metabolic syndrome in relation to risk of meningioma. Oncotarget. 2017;8:2284–2292. doi: 10.18632/oncotarget.13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiedmann, M. K. H. et al. Overweight, obesity and height as risk factors for meningioma, glioma, pituitary adenoma and nerve sheath tumor: a large population-based prospective cohort study. Acta Oncol, 1–8, 10.1080/0284186X.2017.1330554 (2017). [DOI] [PubMed]
- 10.Claus EB, et al. Exogenous hormone use, reproductive factors, and risk of intracranial meningioma in females. J Neurosurg. 2013;118:649–656. doi: 10.3171/2012.9.JNS12811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schildkraut JM, et al. Endogenous and exogenous hormone exposure and the risk of meningioma in men. J Neurosurg. 2014;120:820–826. doi: 10.3171/2013.12.JNS131170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edlinger M, et al. Blood pressure and other metabolic syndrome factors and risk of brain tumour in the large population-based Me-Can cohort study. J Hypertens. 2012;30:290–296. doi: 10.1097/HJH.0b013e32834e9176. [DOI] [PubMed] [Google Scholar]
- 13.Bernardo BM, et al. Association between prediagnostic glucose, triglycerides, cholesterol and meningioma, and reverse causality. Br J Cancer. 2016;115:108–114. doi: 10.1038/bjc.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claus EB, et al. Family and personal medical history and risk of meningioma. J Neurosurg. 2011;115:1072–1077. doi: 10.3171/2011.6.JNS11129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seliger C, et al. Diabetes, use of metformin, and the risk of meningioma. PLoS One. 2017;12:e0181089. doi: 10.1371/journal.pone.0181089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Human Molecular Genetics. 2014;23:R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nitsch D, et al. Limits to causal inference based on Mendelian randomization: a comparison with randomized controlled trials. Am J Epidemiol. 2006;163:397–403. doi: 10.1093/aje/kwj062. [DOI] [PubMed] [Google Scholar]
- 18.Relton CL, Davey Smith G. Mendelian randomization: applications and limitations in epigenetic studies. Epigenomics. 2015;7:1239–1243. doi: 10.2217/epi.15.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgess S, Butterworth A, Thompson SG. Mendelian Randomization Analysis With Multiple Genetic Variants Using Summarized Data. Genetic Epidemiology. 2013;37:658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Z, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun. 2018;9:224. doi: 10.1038/s41467-017-02317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobbins SE, et al. Common variation at 10p12.31 near MLLT10 influences meningioma risk. Nat Genet. 2011;43:825–827. doi: 10.1038/ng.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Claus EB, et al. Genome-wide association analysis identifies a meningioma risk locus at 11p15.5. Neuro Oncol. 2018 doi: 10.1093/neuonc/noy077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32:377–389. doi: 10.1007/s10654-017-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubois L, et al. Genetic and environmental contributions to weight, height, and BMI from birth to 19 years of age: an international study of over 12,000 twin pairs. PLoS One. 2012;7:e30153. doi: 10.1371/journal.pone.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang TO, et al. Body size in early life and risk of lymphoid malignancies and histological subtypes in adulthood. Int J Cancer. 2016;139:42–49. doi: 10.1002/ijc.30044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Font-Burgada J, Sun B, Karin M. Obesity and Cancer: The Oil that Feeds the Flame. Cell Metabolism. 2016;23:48–62. doi: 10.1016/j.cmet.2015.12.015. [DOI] [PubMed] [Google Scholar]
- 27.Rajaraman P. Hunting for the causes of meningioma–obesity is a suspect. Cancer Prev Res (Phila) 2011;4:1353–1355. doi: 10.1158/1940-6207.CAPR-11-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu H, Rohan T. Role of the insulin-like growth factor family in cancer development and progression. J Natl Cancer Inst. 2000;92:1472–1489. doi: 10.1093/jnci/92.18.1472. [DOI] [PubMed] [Google Scholar]
- 29.Zumkeller W, Westphal M. The IGF/IGFBP system in CNS malignancy. Mol Pathol. 2001;54:227–229. doi: 10.1136/mp.54.4.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du Z, et al. Meningioma transcription factors link cell lineage with systemic metabolic cues. Neuro Oncol. 2018;20:1331–1343. doi: 10.1093/neuonc/noy057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown SB, Hankinson SE. Endogenous estrogens and the risk of breast, endometrial, and ovarian cancers. Steroids. 2015;99:8–10. doi: 10.1016/j.steroids.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 32.Cardona-Gomez GP, Mendez P, DonCarlos LL, Azcoitia I, Garcia-Segura LM. Interactions of estrogens and insulin-like growth factor-I in the brain: implications for neuroprotection. Brain Res Brain Res Rev. 2001;37:320–334. doi: 10.1016/S0165-0173(01)00137-0. [DOI] [PubMed] [Google Scholar]
- 33.Ostrom QT, et al. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008-2012. Neuro Oncol. 2015;17(Suppl 4):iv1–iv62. doi: 10.1093/neuonc/nov189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersen L, et al. Hormone replacement therapy increases the risk of cranial meningioma. Eur J Cancer. 2013;49:3303–3310. doi: 10.1016/j.ejca.2013.05.026. [DOI] [PubMed] [Google Scholar]
- 35.Wigertz A, et al. Risk of brain tumors associated with exposure to exogenous female sex hormones. Am J Epidemiol. 2006;164:629–636. doi: 10.1093/aje/kwj254. [DOI] [PubMed] [Google Scholar]
- 36.Claus EB, et al. Exogenous hormone use and meningioma risk: what do we tell our patients? Cancer. 2007;110:471–476. doi: 10.1002/cncr.22783. [DOI] [PubMed] [Google Scholar]
- 37.Korhonen K, et al. Exogenous sex hormone use and risk of meningioma: a population-based case-control study in Finland. Cancer Causes Control. 2010;21:2149–2156. doi: 10.1007/s10552-010-9634-2. [DOI] [PubMed] [Google Scholar]
- 38.McPherson K, Steel CM, Dixon JM. ABC of breast diseases. Breast cancer-epidemiology, risk factors, and genetics. BMJ. 2000;321:624–628. doi: 10.1136/bmj.321.7261.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jhawar BS, Fuchs CS, Colditz GA, Stampfer MJ. Sex steroid hormone exposures and risk for meningioma. J Neurosurg. 2003;99:848–853. doi: 10.3171/jns.2003.99.5.0848. [DOI] [PubMed] [Google Scholar]
- 40.Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40:740–752. doi: 10.1093/ije/dyq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Disney-Hogg L, et al. Impact of atopy on risk of glioma: a Mendelian randomisation study. BMC Med. 2018;16:42. doi: 10.1186/s12916-018-1027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scott RA, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yengo, L. et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum Mol Genet, 10.1093/hmg/ddy271 (2018). [DOI] [PMC free article] [PubMed]
- 44.Willer CJ, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife7, 10.7554/eLife.34408 (2018). [DOI] [PMC free article] [PubMed]
- 47.Bycroft C, et al. TheUK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palmer TM, et al. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res. 2012;21:223–242. doi: 10.1177/0962280210394459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Consortium GP, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang J, et al. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat Commun. 2015;6:8111. doi: 10.1038/ncomms9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burgess S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int J Epidemiol. 2014;43:922–929. doi: 10.1093/ije/dyu005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics for the obesity-related trait GWAS using UK Biobank data are available from http://www.nealelab.is/uk-biobank/. Genotype data from The Resource for Genetic Epidemiology Research on Aging can be downloaded from The Database of Genotypes and Phenotypes (dbGaP) (accession code phs000674.v2.p2). The 1000 Genomes Project and UK10K imputation panel data are available from the European Genome-phenome Archive (EGA, accession EGAD00001000776). Remaining data are available upon request.