Abstract
Assessment of changes in DNA methylation (DNA-m) has the potential to identify adverse environmental exposures. To examine DNA-m among a subset of participants (n = 369) in the Isle of Wight birth cohort who reported variable near resident traffic frequencies. We used self-reported frequencies of heavy vehicles passing by the homes of study subjects as a proxy measure for TRAP, which were: never, seldom, 10 per day, 1–9 per hour and >10 per hour. Methylation of cytosine-phosphate-guanine (CpG) dinucleotide sequences in the DNA was assessed from blood samples collected at age 18 years (n = 369) in the F1 generation. We conducted an epigenome wide association study to examine CpGs related to the frequency of heavy vehicles passing by subjects’ homes, and employed multiple linear regression models to assess potential associations. We repeated some of these analysis in the F2 generation (n = 140). Thirty-five CpG sites were associated with heavy vehicular traffic. After adjusting for confounders, we found 23 CpGs that were more methylated, and 11 CpGs that were less methylated with increasing heavy vehicular traffic frequency among all subjects. In the F2 generation, 2 of 31 CpGs were associated with traffic frequencies and the direction of the effect was the same as in the F1 subset while differential methylation of 7 of 31 CpG sites correlated with gene expression. Our findings reveal differences in DNA-m in participants who reported higher heavy vehicular traffic frequencies when compared to participants who reported lower frequencies.
Keywords: DNA methylation, air pollution, exposure, traffic, epigenome wide association study
Introduction
Evidence for the health impacts of air pollution has been mounting up for several decades [1–3]. Exposure to ambient air pollutants is associated with both acute and chronic health effects and the impacts are felt on global and local scales [4]. Interestingly, the observed adverse health effects are seen even at very low levels of air pollution exposure, and it is unclear whether any threshold exists (i.e. a concentration below which there are no effects on health) [5]. The concentration of air pollutants can differ in a small geographic area depending on local ambient conditions [6]. Key environmental factors that significantly affect local air quality includes proximity to traffic, wood burning, coal burning, dry cleaning, motor vehicle exhaust and industrial emissions, among others [7–14]. Exposures to such environmental factors are associated with asthma exacerbation [15], although their contribution to the development of the disease is uncertain [16].
For an environmental factor such as traffic, it is often necessary to investigate simple proxies such as distance to roadways and traffic estimates or counts, to help assign individual exposures and account for spatial variability. For instance, there is increasing evidence that living near heavy traffic is associated with increased rates of asthma, cardiovascular disease and dementia [17–21], and chronic air pollution exposure gradients at such small scales are associated with adverse cardiorespiratory effects [22]. In the absence of neighborhood level, air pollution measurements, proximity to traffic, traffic volume, among other methods, can be employed [6, 20, 23–30]. Such substitutes facilitate the characterization of smaller-scale air pollution exposures, and have been operative in some health studies [31–33].
Recent evidence indicates that epigenetics may play an important role in mediating the health effects of air pollution [34]. Indeed, it has been suggested that the extent of epigenetic markers can change progressively and help construct cumulative exposure patterns over time [35]. Interestingly, changes in epigenetic markers can result from exposure to a risk factor such as air pollution, and such changes can potentially serve as predictive biomarkers of susceptibility to adverse health [36]. The epigenetic marker of DNA methylation (DNA-m), which is the addition of a methyl group to cysteine in cytosine-phosphate-guanine (CpG) dinucleotides sequences in the DNA, is reported to be related to air pollution exposures [37, 38], and adverse respiratory health [39], including asthma [40, 41].
Changes in the epigenome and gene expression may be induced by exposure to air pollution [34, 42] and this is relevant to the development of several pathophysiological processes. Difflerential blood DNA-m in response to air pollution exposure from sources such as traffic has been reported [43–45]. We cannot or rarely can directly assess DNA-m in target tissues, such as the lung. However, for many biomarkers, blood changes are considered to constitute a window through which specific processes in other tissues can be assessed. In addition, during development, blood and airways stem from the mesoderm and may represent to have a similar development and susceptibility [46]. For these reasons, the effects of TRAP on epigenome in blood samples represent informative biomarkers of change in the airways.
Given that (i) TRAP exerts its greatest impact on local scales, particularly near roadways [47] and (ii) the mechanistic basis for the effects of TRAP on the epigenome is not well delineated [48], additional studies can provide further evidence and advance the current state of the science [49]. Accordingly, we used the self-reported frequencies of heavy vehicles passing by the homes of study subjects as a proxy measure for TRAP and evaluated their associations with the methylation of CpG sites among 18-year-old participants in the Isle of Wight (IoW) birth cohort, UK (n = 369). Our motivating questions were:
Which specific CpG sites are associated with heavy vehicular traffic in the birth cohort?
Are there any trends in the association between differential DNA-m (both higher and lower) and the frequency of exposure to heavy vehicular traffic?
Results
Characteristics of Study Population
Eighteen percent of the subjects (n = 67) reported never having any heavy vehicles passing by their homes while 82% reported some heavy vehicular traffic outside their homes (Table 1). About 20% had a history of maternal smoking and nearly 50% were exposed to tobacco smoke outside their homes and before age 4 (Table 1). About a quarter of the subjects were current smokers who started smoking at an average of 14.5 (SD 1.5) years. A vast majority of the subjects present a middle class status (72%) with over 90% still living at home with their parents and 70% living in a private residential property. The average body mass index (BMI) was 23.6 (SD 4.3). In this subset with DNA-m, there were more females than males (66% vs. 34%) due to the study design (following until pregnancy) (Table 1).
Table 1:
Comparison of population characteristics of participants in the whole cohort and those with DNA-m at age 18 years
| Whole cohort (n = 1313) | Sample with DNA-m (n = 369) | P-value | |
|---|---|---|---|
| n (%) | n (%) | ||
| Gender | <0.0001 | ||
| Female | 660 (50.27) | 245 (66.4) | |
| Male | 653 (49.73) | 124 (33.6) | |
| Maternal smoking status | 0.24 | ||
| No | 1002 (76.31 | 292 (79.1) | |
| Yes | 305 (23.23) | 75 (20.3) | |
| Unanswered | 6 (0.46) | 2 (0.5) | |
| Frequency of heavy vehicles passing by home | 0.783 | ||
| >10 per hour | 274 (20.87) | 77 (20.9) | |
| 1–9 per hour | 223 (16.98) | 69 (18.7) | |
| 10 per day | 119 ( 9.06) | 36 (9.8) | |
| Seldom | 427 (32.52) | 120 (32.5) | |
| Never | 241 (18.35) | 67 (18.2) | |
| Missing | 29 (2.21) | ||
| Exposure to smoking outside home | 0.41 | ||
| Yes | 598 (45.54) | 180 (48.8) | |
| No | 663 (50.50) | 181 (49.1) | |
| Unanswered | 52 (3.96) | 8 (2.2) | |
| Current smoking status | 0.3 | ||
| No | 910 (69.31) | 270 (73.2) | |
| Yes | 368 (28.03) | 95 (25.8) | |
| Unanswered | 35 (2.67) | 4 (1.1) | |
| Any exposure to environmental tobacco smoke (at 10 years) | 0.23 | ||
| No | 716 (54.5) | 223 ( 60.4) | |
| Yes | 492 (37.5) | 132 (35.8) | |
| Unanswered | 105 (8.0) | 14 (3.8) | |
| Any exposure to environmental tobacco smoke (at birth, 1 year, 2 year or 4 years) | 0.11 | ||
| No | 602 (45.8) | 186 (50.4) | |
| Yes | 708 (53.9) | 182 (49.3) | |
| Unanswered | 3 (0.23) | 1 (0.27) | |
| Socio-economic status | 0.73 | ||
| High | 103 (7.84) | 33 ( 8.9) | |
| Mid | 952 (72.51) | 267 (72.4) | |
| Low | 177 (13.48) | 58 (15.7) | |
| Unanswered | 81 (6.17) | 11 (2.9) | |
| Living with parents | 0.11 | ||
| Yes | 1158 (88.19) | 342 (92.7) | |
| No | 129 (9.82) | 27 (7.3) | |
| Missing | 26 (1.98) | ||
| Type of residential property | 0.78 | ||
| Rented privately | 145 (11.0) | 28 (7.5) | |
| Rented council/housing association | 209 (15.9) | 65 (17.6) | |
| Owned privately | 908 (69.1) | 271 (73.4) | |
| Other | 18 (1.37) | 5 (1.4) | |
| Missing | 33 (2.51) | ||
| Median (p5, p95) | |||
| BMI | 22.1 (18.2, 32.1) | 22.5 (18.7, 32.1) | 0.09 |
| Age subject started smoking | 15 (12, 17) | 15 (12, 17) | 0.88 |
| Time living in present house | 48 (6, 48) | 48 (6, 48) | 0.48 |
Percentage has been rounded up to whole numbers, where applicable.
Which Specific CpG Sites Are Associated with Heavy Vehicular Traffic in the IoW Cohort?
There were a total of 371 CpG sites that were associated with heavy vehicular traffic frequency based on ttscreening results. However, we chose the top CpGs with a cutoff percentage of 70 [m = 70 across 100 total iterations (i = 100)] was used to determine the final pool of potentially important CpG sites (in our case 35 of 371 had a cutoff percentage between 70 and 94). Therefore, a final group of 35 CpGs was selected in step 1 (Tables 2 and 3). The 35 CpG sites are listed in the order of significance based on the epigenome-wide association analysis results. Over 30% of these CpGs were located on Chromosome 1. The identified CpG sites were associated with 34 different genes (two CpG sites—cg11156891 and cg12407057—mapped to one gene ANKRD65). A majority of the CpG sites were located in the body of the identified gene (24 of 35); 4 were 200–1500 bases upstream of the transcriptional start site (TSS), while 2 were 0–200 of the TSS; 3 were within the 5′ untranslated region, and 2 were over 50 kb from the nearest gene (Table 3).
Table 2:
Summary of (a) CpG sites found this is exploratory study, (b) genes associated with the CpGs and (c) list of chemicals documented in the comparative toxicogenomics database that are related to air pollution
| (a) CpG site | (b) Identified gene in this study | (c) Comparative toxicogenomics database chemical (related to air pollution) |
|---|---|---|
| cg25895913 | CDH4 | Benzo(a)pyrene, 7,8-dihydro-7,8-dihydroxybenzo(a)pyrene 9,10-oxide |
| cg11156891 | ANKRD65 | Benzo(a)pyrene |
| cg12407057 | ANKRD65 | Benzo(a)pyrene |
| cg20747739 | FAM132A | Benzo(a)pyrene |
| cg18565510 | ACAP3 | Benzo(a)pyrene |
| cg24843003 | DAZAP1 | Benzo(a)pyrene, air pollutants, occupational, 1-hydroxypyrene |
| cg15730464 | LGI2 | Benzo(a)pyrene, soot, tobacco smoke pollution |
| cg16196077 | RTKN2 | Benzo(a)pyrene, ozone |
| cg02707264 | MYRIP | Benzo(a)pyrene |
| cg03476673 | CRISPLD2 | 7,8-Dihydro-7,8-dihydroxybenzo(a)pyrene 9,10-oxide |
| cg07023532 | ACOT4 | Benzo(a)pyrene |
| cg20255272 | VWA1 | None |
| cg12417992 | SLC6A9 | Benzo(a)pyrene, ozone, dibenzo(a, l)pyrene |
| cg04154465 | WNT2B | None |
| cg12813768 | SYCP1 | None |
| cg14162906 | TMEM222 | 7,8-Dihydro-7,8-dihydroxybenzo(a)pyrene 9,10-oxide |
| cg24361098 | BCL11A | Benzo(a)pyrene |
| cg16147794 | SLC16A10 | Benzo(a)pyrene |
| cg16668397 | JPH3 | None |
| cg26419883 | TRPM5 | Benzo(a)pyrene |
| cg21775675 | TMEM161B | None |
| cg04794690 | PADI3 | Benzo(a)pyrene, particulate matter |
| cg06942649 | FBXO25 | Benzo(a)pyrene |
| cg18459806 | NIN | Benzo(a)pyrene, 7,8-dihydro-7,8-dihydroxybenzo(a)pyrene 9,10-oxide |
| cg20631351 | PALM | Smoke |
| cg00347824 | NSMAF | Benzo(a)pyrene |
| cg17053854 | SEPT9 | Benzo(a)pyrene, 7,8-dihydro-7,8-dihydroxybenzo(a)pyrene 9,10-oxide |
| cg25324786 | RASA3 | Benzo(a)pyrene, 7,8-dihydro-7,8-dihydroxybenzo(a)pyrene 9,10-oxide |
| cg26720961 | TSNARE1 | None |
| cg05575058 | FAM164A | Benzo(a)pyrene |
| cg15742605 | SAMD11 | Smoke |
| cg26185508 | CDCP2 | None |
| cg02378006 | UNC5B | None |
| cg08462127 | MYOM2 | Benzo(a)pyrene |
| cg11017318 | SYT16 | None |
This information was obtained from the UCSC GenomeBroswer (GRCh37/hg19).
Table 3:
Descriptive statistics of the DNA-m of the significant CpG sites in whole blood samples at age 18 (n = 369)
| Raw P-values from tt screening | Chromosome (hg19) | CpG site | Location | Coordinate (hg19) | Minimum | 5th Pctl | Median | 95th Pctl | Maximum |
|---|---|---|---|---|---|---|---|---|---|
| 3.69E–06 | 1 | cg25895913 | Body | 54619445 | –0.804 | –0.414 | 0.025 | 0.366 | 0.612 |
| 2.17E–06 | 1 | cg11156891 | Body | 1373678 | –1.485 | –0.881 | –0.125 | 1.191 | 2.589 |
| 6.84E–06 | 1 | cg12407057 | Body | 44500834 | –0.971 | –0.517 | –0.087 | 0.900 | 2.540 |
| 1.62E–06 | 17 | cg20747739 | TSS1500 | 75463180 | –0.812 | –0.358 | 0.004 | 0.335 | 0.709 |
| 8.53E–06 | 14 | cg18565510 | Body | 51293793 | –0.897 | –0.500 | –0.002 | 0.464 | 0.844 |
| 2.22E–05 | 8 | cg24843003 | Body | 79578035 | –0.904 | –0.449 | 0.003 | 0.416 | 1.093 |
| 2.21E–05 | 2 | cg15730464 | Body | 60748951 | –1.254 | –0.655 | 0.016 | 0.562 | 0.825 |
| 1.48E–05 | 11 | cg16196077 | TSS200 | 2434192 | –1.311 | –0.798 | –0.048 | 0.880 | 1.512 |
| 7.14E–05 | 1 | cg02707264 | 5′UTR | 1366206 | –0.744 | –0.365 | 0.005 | 0.321 | 0.634 |
| 3.00E–05 | 1 | cg03476673 | 5′UTR | 1183257 | –1.149 | –0.598 | –0.015 | 0.608 | 1.208 |
| 2.33E–04 | 3 | cg07023532 | TSS1500 | 39851931 | –0.968 | –0.505 | –0.013 | 0.488 | 1.229 |
| 7.50E–05 | 19 | cg20255272 | Body | 711001 | –1.700 | –1.046 | 0.104 | 0.460 | 0.807 |
| 5.26E–05 | 1 | cg12417992 | Body | 113045271 | –0.572 | –0.347 | –0.025 | 0.396 | 0.716 |
| 8.95E–05 | 1 | cg04154465 | Body | 1238702 | –1.351 | –0.588 | 0.004 | 0.600 | 1.390 |
| 5.58E–05 | 1 | cg12813768 | Body | 115398123 | –1.643 | –0.861 | 0.002 | 0.885 | 1.441 |
| 9.81E–05 | 1 | cg14162906 | TSS1500 | 27647606 | –0.713 | –0.392 | –0.004 | 0.379 | 0.803 |
| 1.22E–04 | 8 | cg24361098 | Body | 143386260 | –1.080 | –0.634 | 0.015 | 0.537 | 0.996 |
| 3.50E–05 | 16 | cg16147794 | Body | 87720712 | –1.217 | –0.578 | –0.022 | 0.660 | 1.565 |
| 1.20E–04 | 5 | cg16668397 | Body | 87441487 | –0.565 | –0.386 | –0.008 | 0.462 | 0.785 |
| 1.16E–04 | 8 | cg26419883 | Body | 2017365 | –0.818 | –0.419 | –0.007 | 0.418 | 0.702 |
| 1.52E–04 | 13 | cg21775675 | ∼50 kb upstream of TMEM161B | 114866221 | –0.987 | –0.460 | –0.005 | 0.518 | 1.296 |
| 1.42E–04 | 19 | cg04794690 | Body | 1409547 | –1.217 | –0.500 | 0.002 | 0.525 | 0.719 |
| 7.64E–05 | 10 | cg06942649 | Body | 64028521 | –1.588 | –0.948 | 0.016 | 0.757 | 1.066 |
| 2.30E–04 | 8 | cg18459806 | 5′UTR | 436813 | –0.610 | –0.359 | 0.002 | 0.373 | 0.807 |
| 2.18E–04 | 8 | cg20631351 | Body | 59571961 | –0.679 | –0.379 | –0.005 | 0.362 | 0.818 |
| 2.01E–04 | 20 | cg00347824 | Body | 60460465 | –1.267 | –0.538 | 0.028 | 0.458 | 1.166 |
| 4.21E–05 | 1 | cg17053854 | Body | 17768059 | –0.474 | –0.289 | 0.001 | 0.277 | 0.682 |
| 2.55E–04 | 1 | cg25324786 | Body | 879383 | –0.935 | –0.568 | 0.021 | 0.466 | 0.985 |
| 3.05E–04 | 14 | cg26720961 | Body | 62279992 | –1.221 | –0.556 | –0.009 | 0.583 | 1.659 |
| 4.63E–04 | 4 | cg05575058 | TSS1500 | 25087441 | –0.907 | –0.455 | –0.029 | 0.459 | 0.804 |
| 3.39E–04 | 6 | cg15742605 | Body | 111405660 | –0.645 | –0.417 | –0.023 | 0.531 | 1.064 |
| 2.17E–04 | 10 | cg26185508 | TSS200 | 73026288 | –0.909 | –0.493 | –0.007 | 0.487 | 0.948 |
| 1.12E–04 | 1 | cg02378006 | Body | 1366274 | –1.212 | –0.576 | –0.015 | 0.595 | 1.240 |
| 2.69E–04 | 16 | cg08462127 | Body | 84870203 | –0.832 | –0.434 | 0.004 | 0.431 | 0.724 |
| 5.65E–04 | 14 | cg11017318 | ∼200 kb upstream of SYT16 | 74057654 | –0.607 | –0.382 | 0.004 | 0.369 | 0.772 |
We also assessed answers to other traffic-related questions such as ‘How often do cars pass your house or on the street less than 100 meters away?’ and ‘How frequently are you annoyed by outdoor air pollution (from traffic industry, etc) in your home if you keep the window open?’. However, these did not have much variability nor did they yield any significant results with the ttScreening package. The CpG by CpG analysis also did not show any statistically significant results for Any versus Never reports of heavy vehicular traffic frequency after adjusting for false discovery rate (FDR; all adjusted P-values were ≥0.4). There was no association between heavy vehicular traffic frequency and cg05575921, located in the aryl hydrocarbon receptor repressor (AHRR) gene. However, there appeared to be an association with self-reported smoking status (among current smokers), tobacco smoke exposure assessed through a questionnaire administered at 10 years and environmental tobacco smoke exposure (Table 4).
Table 4:
Results for linear models for methylation of cg07555921 (AHRR), considered to be a marker for smoking
| Parameter | Estimate | Standard error | P-value |
|---|---|---|---|
| Model 1: Exposure variable only: frequency of heavy vehicular traffic (ref=never), n = 369 | |||
| Heavy vehicular traffic frequency: >10/hour | –0.10 | 0.11 | 0.4 |
| Heavy vehicular traffic frequency: 1–9/hour | 0.13 | 0.11 | 0.2 |
| Heavy vehicular traffic frequency: 10/day | –0.05 | 0.13 | 0.7 |
| Heavy vehicular traffic frequency: Seldom | 0.08 | 0.10 | 0.4 |
| Model 2: Smoking related variables only, n = 342 | |||
| Maternal smoking | –0.07 | 0.09 | 0.4 |
| Current smoking status | 0.77 | 0.08 | <.0001 |
| Smoking outside the home | 0.09 | 0.07 | 0.2 |
| Any environmental tobacco smoke exposure | –0.16 | 0.08 | 0.05 |
| Tobacco smoke exposure at 0–4 years | 0.09 | 0.06 | 0.2 |
| Tobacco smoke exposure at 10 years | 0.15 | 0.09 | 0.09 |
| Model 3: All variables considered apriori among all subjects, n = 329 | |||
| Heavy vehicular traffic frequency: >10/hour | –0.14 | 0.10 | 0.2 |
| Heavy vehicular traffic frequency: 1–9/hour | –0.04 | 0.10 | 0.7 |
| Heavy vehicular traffic frequency: 10/day | –0.08 | 0.13 | 0.5 |
| Heavy vehicular traffic frequency: Seldom | –0.04 | 0.09 | 0.7 |
| Maternal smoking | –0.10 | 0.10 | 0.3 |
| Current smoking status | 0.75 | 0.08 | <.0001 |
| Smoking outside the home | 0.11 | 0.07 | 0.1 |
| Any environmental tobacco smoke exposure | –0.18 | 0.09 | 0.04 |
| Tobacco smoke exposure at 0–4 years | 0.09 | 0.07 | 0.2 |
| Tobacco smoke exposure at 10 years | 0.17 | 0.09 | 0.06 |
| High SES (ref = low SES) | –0.22 | 0.13 | 0.1 |
| Mid SES (ref = low SES) | –0.04 | 0.09 | 0.6 |
| BMI | 0.02 | 0.01 | 0.007 |
| Gender | 0.04 | 0.07 | 0.5 |
| Model 4: All variables considered apriori among current smokers, n = 77 | |||
| Heavy vehicular traffic frequency: >10/hour | –0.13 | 0.30 | 0.7 |
| Heavy vehicular traffic frequency: 1–9/hour | 0.48 | 0.34 | 0.2 |
| Heavy vehicular traffic frequency: 10/day | 0.41 | 0.37 | 0.3 |
| Heavy vehicular traffic frequency: Seldom | 0.20 | 0.31 | 0.5 |
| Maternal smoking | –0.07 | 0.26 | 0.8 |
| Age subject started smoking | 0.20 | 0.06 | 0.002 |
| Smoking outside the home | 0.79 | 0.32 | 0.02 |
| Any environmental tobacco smoke exposure | –0.33 | 0.23 | 0.2 |
| Tobacco smoke exposure at 0–4 years | 0.14 | 0.20 | 0.5 |
| Tobacco smoke exposure at 10 years | 0.32 | 0.25 | 0.2 |
| High SES (ref = low SES) | –0.68 | 0.44 | 0.1 |
| Mid SES (ref = low SES) | –0.04 | 0.24 | 0.9 |
| BMI | 0.03 | 0.02 | 0.1 |
| Gender | 0.04 | 0.21 | 0.9 |
P-values in bold and bold italics denote statistical significance less than or equal to 0.05 and 0.1 respectively.
Gene Set Enrichment Analysis
Using the bioinformatic resource ToppGene Suite [50], we performed a gene enrichment analysis to determine the pathway(s) associated with genes of the significant CpG sites [the respective genes that had the exact CpG coordinates, or if the CpG was located between two genes (i.e. intergenic CpGs), we selected the gene with the closest proximity to the intergenic CpG]. Input parameters for the gene enrichment analysis were as follows: All 34 genes were included in the training set, the hypergeometric probability mass function was used to calculate P-values, and the FDR was controlled at 0.05 using the Benjamini–Hochberg method.
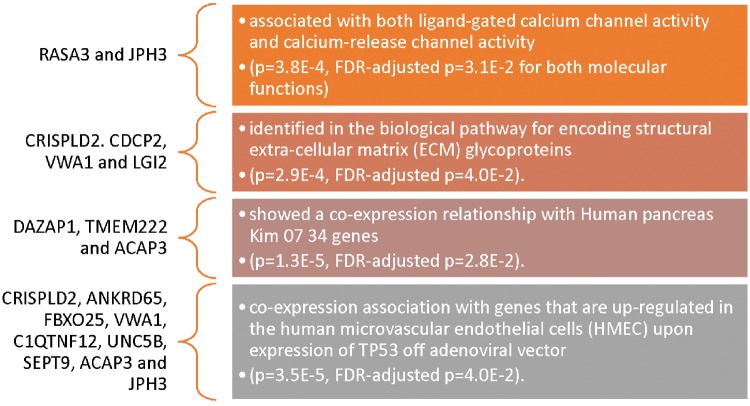
Two genes, RASA3 and JPH3, were associated with both ligand-gated calcium channel activity and calcium-release channel activity (Fig. 1). Four genes (CRISPLD2, CDCP2, VWA1 and LGI2) were identified in the biological pathway for encoding structural extra-cellular matrix (ECM) glycoproteins (all FDR-adjusted P < 0.05, Fig. 1). Additionally, a total of nine genes (CRISPLD2, ANKRD65, FBXO25, VWA1, C1QTNF12, UNC5B, SEPT9, ACAP3 and JPH3) came up as having a coexpression association with genes that are upregulated in the human microvascular endothelial cells.
Figure 1:
Results of gene enrichment analysis identifying potential pathway(s) associated with the genes identified with the significant CpG sites in this study. More details on the gene enrichment analysis can be found in the supplementary material provided
When the analysis is separated into the 23 more methylated and 11 less methylated CpG sites, the latter is identified in the following gene families: zinc fingers (P = 5.3E–3, FDR-adjusted P = 1.7E–2), synaptotagmins (P = 5.6E–3, FDR-adjusted P = 1.7E–2) and A-kinase anchoring proteins (P = 9.9E–3, FDR-adjusted P = 2.0E–2) but no molecular functions are identified. On the other hand, the former is associated with five different molecular functions:
calcium-release channel activity (P = 1.7E–4, FDR-adjusted P = 1.2E–2),
ligand-gated calcium channel activity (P = 1.7E–4, FDR-adjusted P = 1.2E–2),
intracellular ligand-gated ion channel activity (P = 6.E–4, FDR-adjusted P = 2.9E–2),
succinyl-CoA hydrolase activity (P = 1.2E–3, FDR-adjusted P = 4.3E–2) and
metal ion transmembrane transporter activity (P = 1.7E–3, FDR-adjusted P = 4.7E–2).
Additionally, 17 gene families are identified with these 23 more methylated CpGs including CD molecules Type I classical cadherins and Peptidyl arginine deiminases (both: P = 5.2E–3, FDR-adjusted P = 3.4E–2).
Association with Air Pollutants in the Comparative Toxicogenomics Database
The analysis did not reveal any links to air pollutants, as there is currently not enough data on air pollution to factor into biological pathway analyses. However, a search of the description and page index of each gene provided information on reported chemicals related to air pollution in the comparative toxicogenomics database [51]. All but three genes were associated with chemical(s) found in air pollution, e.g. ‘benzo(a)pyrene’, ‘7,8-dihydro-7,8-dihydroxybenzo(a)pyrene 9,10-oxide’, ‘smoke’ and even ‘particulate matter’ (Table 2).
Diseases for Which the Identified Genes Are Enriched
The gene enrichment analysis also identified the following diseases that associated with six of the genes identified with the significant CpG sites in this study
SEPT9 is associated with Orbital separation diminished, hereditary neuralgic amyotrophy (HNA), Brachial Plexus Neuritis, Epiphyses, hemoglobinopathies.
NIN is associated with Orbital separation diminished, Seckel syndrome, HNA, Spondyloepimetaphyseal dysplasia with multiple dislocations, Lumbar scoliosis.
JPH3 is associated with Huntington disease-like 2, Akinetic rigid syndrome, Brachial Plexus Neuropathies
MYOM2 is associated with IgA myeloma, Osteosclerotic Myeloma
BCL11A is associated with Amyotrophy, HNA, hemoglobinopathies, F-cell distribution, fetal hemoglobin levels
PADI3 is associated with Uncombable hair, generalized trichodysplasia
Are There Any Trends in the Association between the Frequencies of Heavy Vehicular Traffic on the IoW and DNA-m?
An initial ANOVA revealed a total of 24 of 35 CpGs with significantly different DNA-m (P < 0.05) when those who reported no heavy vehicles (never) were compared with those who reported any heavy vehicular traffic (Table 5). Further evaluations (using never, seldom, 10 per day, 1–9 per hour or >10 per hour levels) showed all 35 CpGs had significantly different DNA-m for at least one of the five categories of heavy vehicle traffic P ≤ 0.01 (Table 5).
Table 5:
Analysis of variance for unadjusted DNA-M of CpG sites in whole blood samples at age 18 (n = 369)
| CpG site | Any (n = 302) versus no (n = 67) heavy vehicle traffic frequency |
Five categories of heavy vehicle traffic frequency: (1) never (n = 67), (2) seldom (n = 120), (3) 10/day (n = 36), (4) 1–9/hour (n = 69), (5) >10/hour (n = 77) |
||||
|---|---|---|---|---|---|---|
| Mean square | F value (df = 1) | P value | Mean square | F value (df = 4) | P value | |
| cg25895913 | 0.53 | 9.4 | 0.002 | 0.33 | 6.1 | <0.0001 |
| cg11156891 | 4.55 | 9.7 | 0.002 | 2.61 | 5.7 | 0.0002 |
| cg12407057 | 4.55 | 9.7 | 0.002 | 1.37 | 5.2 | 0.0004 |
| cg20747739 | 0.30 | 6.6 | 0.01 | 0.27 | 6.2 | <0.0001 |
| cg18565510 | 0.84 | 9.7 | 0.002 | 0.44 | 5.2 | 0.0005 |
| cg24843003 | 0.97 | 12.3 | 0.0005 | 0.40 | 5.1 | 0.0005 |
| cg15730464 | 1.59 | 13.0 | 0.0004 | 0.78 | 6.6 | <0.0001 |
| cg16196077 | 1.32 | 5.0 | 0.03 | 1.39 | 5.5 | 0.0003 |
| cg02707264 | 0.06 | 1.3 | 0.3 | 0.22 | 5.1 | 0.0006 |
| cg03476673 | 1.38 | 9.5 | 0.002 | 0.71 | 5.0 | 0.0007 |
| cg07023532 | 1.62 | 17.6 | <.0001 | 0.51 | 5.6 | 0.0002 |
| cg20255272 | 0.66 | 3.6 | 0.06 | 0.73 | 4.1 | 0.003 |
| cg12417992 | 0.37 | 8.2 | 0.0046 | 0.20 | 4.5 | 0.001 |
| cg04154465 | 0.48 | 3.4 | 0.07 | 0.56 | 4.0 | 0.003 |
| cg12813768 | 1.08 | 4.3 | 0.04 | 1.13 | 4.7 | 0.001 |
| cg14162906 | 0.49 | 8.9 | 0.003 | 0.26 | 4.7 | 0.001 |
| cg24361098 | 1.38 | 11.1 | 0.0009 | 0.56 | 4.6 | 0.001 |
| cg16147794 | 0.27 | 1.8 | 0.2 | 0.81 | 5.6 | 0.0002 |
| cg16668397 | 0.42 | 6.8 | 0.01 | 0.24 | 3.9 | 0.004 |
| cg26419883 | 0.25 | 4.1 | 0.05 | 0.26 | 4.4 | 0.002 |
| cg21775675 | 0.33 | 3.7 | 0.06 | 0.36 | 4.1 | 0.003 |
| cg04794690 | 1.31 | 14.8 | 0.0001 | 0.49 | 5.5 | 0.0003 |
| cg06942649 | 2.04 | 8.2 | 0.004 | 1.06 | 4.3 | 0.002 |
| cg18459806 | 0.29 | 5.7 | 0.02 | 0.20 | 4.1 | 0.003 |
| cg20631351 | 0.29 | 5.5 | 0.02 | 0.18 | 3.5 | 0.008 |
| cg00347824 | 0.62 | 6.4 | 0.01 | 0.37 | 3.9 | 0.004 |
| cg17053854 | 0.18 | 5.9 | 0.02 | 0.13 | 4.6 | 0.001 |
| cg25324786 | 0.15 | 1.6 | 0.2 | 0.36 | 4.0 | 0.004 |
| cg26720961 | 0.01 | 0.1 | 0.7 | 0.61 | 5.0 | 0.0006 |
| cg05575058 | 0.64 | 8.6 | 0.004 | 0.27 | 3.7 | 0.006 |
| cg15742605 | 0.21 | 2.5 | 0.1 | 0.28 | 3.5 | 0.008 |
| cg26185508 | 0.82 | 9.4 | 0.002 | 0.39 | 4.5 | 0.002 |
| cg02378006 | 0.39 | 3.1 | 0.08 | 0.54 | 4.5 | 0.001 |
| cg08462127 | 0.15 | 2.2 | 0.1 | 0.30 | 4.4 | 0.002 |
| cg11017318 | 0.28 | 5.3 | 0.02 | 0.18 | 3.5 | 0.008 |
P-values in bold and bold italics denote statistical significance less than or equal to 0.05 and 0.1 respectively.
After adjusting for history of maternal smoking, environmental tobacco smoke exposure (0–4 years and/or at 10 years), SES, gender, BMI, current smoking status and/or exposure to smoke outside the home, 34 CpGs remained statistically significant depending on the category of heavy vehicle traffic frequency reported (P ≤ 0.05, range for n = 329–369) (Table 6). We also present results for linear models for the top 35 CpG sites identified with the ttscreening method after adjusting for all confounding factors considered a priori in this study, and the results are similar to Table 6 where associations are still present in 34 of 35 CpG sites for at least one category of the exposure variable (Supplementary Table S1).
Table 6:
Results for multiple linear regression models for CpG sites associated with the frequency of heavy vehicles passing by all subjects’ homes
| CpG | Associated gene | Heavy vehicle frequency (ref = never) | Estimate | Standard error | P value | Significant covariates in final model | Dunnett's test (LSMEAN = never) | Linear trend test (F value, df = 1) P value | Direction of methylation |
|---|---|---|---|---|---|---|---|---|---|
| cg25895913 (n = 355) | CDH4 | >10/hour | 0.15 | 0.04 | 0.0002 | Tobacco smoke exposure (at 10 years); gender | *** | 18.16 | ↑ |
| 1–9/hour | 0.17 | 0.04 | <.0001 | *** | <0.0001 | ||||
| 10/day | 0.06 | 0.05 | 0.2 | ||||||
| Seldom | 0.05 | 0.04 | 0.2 | ||||||
| cg11156891 (n = 329) | ANKRD65 | >10/hour | –0.48 | 0.13 | 0.0002 | Maternal smoking; tobacco smoke exposure (0–4 years and at 10 years); SES; gender; BMI; current smoking status; exposure to smoke outside the home | *** | 14.49 | ↓ |
| 1–9/hour | –0.29 | 0.13 | 0.03 | * | 0.0002 | ||||
| 10/day | –0.19 | 0.15 | 0.2 | ||||||
| Seldom | –0.12 | 0.11 | 0.3 | ||||||
| cg12407057 (n = 329) | ANKRD65 | >10/hour | –0.33 | 0.10 | 0.0006 | Maternal smoking; tobacco smoke exposure (0–4 years and at 10 years); SES; gender; BMI; current smoking status; exposure to smoke outside the home | *** | 15.51 | ↓ |
| 1–9/hour | –0.24 | 0.10 | 0.01 | * | 0.0001 | ||||
| 10/day | –0.08 | 0.12 | 0.5 | ||||||
| Seldom | –0.07 | 0.09 | 0.4 | ||||||
| cg20747739 (n = 362) | FAM132A | >10/hour | 0.14 | 0.04 | <.0001 | Gender; BMI | *** | 19.7 | ↑ |
| 1–9/hour | 0.11 | 0.04 | 0.002 | ** | <0.0001 | ||||
| 10/day | 0.04 | 0.04 | 0.3 | ||||||
| Seldom | 0.03 | 0.03 | 0.4 | ||||||
| cg18565510 (n = 362) | ACAP3 | >10/hour | 0.20 | 0.05 | <.0001 | Gender; BMI | *** | 15.33 | ↑ |
| 1–9/hour | 0.14 | 0.05 | 0.008 | ** | 0.0001 | ||||
| 10/day | 0.09 | 0.06 | 0.1 | ||||||
| Seldom | 0.07 | 0.05 | 0.1 | ||||||
| cg24843003 (n = 369) | DAZAP1 | >10/hour | 0.19 | 0.05 | <.0001 | Gender | *** | 14.73 | ↑ |
| 1–9/hour | 0.15 | 0.05 | 0.002 | ** | 0.0001 | ||||
| 10/day | 0.07 | 0.06 | 0.2 | ||||||
| Seldom | 0.09 | 0.04 | 0.03 | * | |||||
| cg15730464 (n = 348) | LGI2 | >10/hour | 0.28 | 0.06 | <.0001 | Tobacco smoke exposure ( at 10 years); SES; gender | *** | 20.56 | ↓ |
| 1–9/hour | 0.17 | 0.06 | 0.006 | ** | <0.0001 | ||||
| 10/day | 0.03 | 0.07 | 0.7 | ||||||
| Seldom | 0.16 | 0.05 | 0.004 | ** | |||||
| cg16196077 (n = 336) | RTKN2 | >10/hour | –0.27 | 0.09 | 0.003 | Maternal smoking; tobacco smoke exposure (0–4 years and at 10 years); SES; gender; current smoking status; exposure to smoke outside the home | ** | 12.62 | ↓ |
| 1–9/hour | –0.25 | 0.09 | 0.007 | ** | 0.0004 | ||||
| 10/day | –0.14 | 0.11 | 0.2 | ||||||
| Seldom | 0.02 | 0.08 | 0.8 | ||||||
| cg02707264 (n = 329) | MYRIP | >10/hour | –0.11 | 0.04 | 0.005 | Maternal smoking; tobacco smoke exposure (0–4 years and at 10 years); SES; gender; BMI; current smoking status; exposure to smoke outside the home | ** | 14.16 | ↓ |
| 1–9/hour | –0.02 | 0.04 | 0.6 | 0.0002 | |||||
| 10/day | 0.01 | 0.05 | 0.7 | ||||||
| Seldom | 0.02 | 0.03 | 0.5 | ||||||
| cg03476673 (n = 329) | CRISPLD2 | >10/hour | –0.23 | 0.07 | 0.0009 | Maternal smoking; tobacco smoke exposure (0–4 years and at 10 years); SES; gender; BMI; current smoking status; exposure to smoke outside the home | *** ^ | 9.07 | ↓ |
| 1–9/hour | –0.12 | 0.07 | 0.09 | 0.003 | |||||
| 10/day | –0.11 | 0.08 | 0.2 | ||||||
| Seldom | –0.08 | 0.06 | 0.2 | ||||||
| cg07023532 (n = 362) | ACOT4 | >10/hour | 0.18 | 0.05 | 0.0006 | Gender; BMI | *** | 4.39 | ↑ |
| 1–9/hour | 0.23 | 0.05 | <.0001 | *** | 0.04 | ||||
| 10/day | 0.21 | 0.06 | 0.0008 | *** | |||||
| Seldom | 0.14 | 0.05 | 0.003 | ** | |||||
| cg20255272 (n = 348) | VWA1 | >10/hour | 0.24 | 0.07 | 0.0009 | Tobacco smoke exposure (0–4 years only); gender; BMI; SES; current smoking status | ** | 13.83 | ↑ |
| 1–9/hour | 0.13 | 0.07 | 0.08 | 0.0002 | |||||
| 10/day | 0.08 | 0.09 | 0.3 | ||||||
| Seldom | 0.001 | 0.06 | 1.0 | ||||||
| cg12417992 (n = 362) | SLC6A9 | >10/hour | 0.12 | 0.04 | 0.001 | Maternal smoking; tobacco smoke exposure (0–4 years only); SES; gender | ** | 7.91 | ↑ |
| 1–9/hour | 0.11 | 0.04 | 0.003 | * ^ | 0.005 | ||||
| 10/day | 0.10 | 0.04 | 0.02 | ||||||
| Seldom | 0.04 | 0.03 | 0.2 | ||||||
| cg04154465 (n = 351) | WNT2B | >10/hour | 0.23 | 0.06 | 0.0003 | SES; gender; BMI | *** | 16.12 | ↑ |
| 1–9/hour | 0.17 | 0.06 | 0.008 | ** | <0.0001 | ||||
| 10/day | 0.11 | 0.08 | 0.2 | ||||||
| Seldom | 0.02 | 0.06 | 0.7 | ||||||
| cg12813768 (n = 336) | SYCP1 | >10/hour | –0.34 | 0.09 | 0.0001 | Maternal smoking; tobacco smoke exposure (0–4 years and at 10 years); SES; gender; current smoking status; exposure to smoke outside the home | *** | 16.56 | ↓ |
| 1–9/hour | –0.18 | 0.09 | 0.04 | * | <0.0001 | ||||
| 10/day | –0.08 | 0.11 | 0.5 | ||||||
| Seldom | –0.08 | 0.08 | 0.3 | ||||||
| cg14162906 (n = 362) | TMEM222 | >10/hour | 0.11 | 0.04 | 0.004 | BMI; gender | ** | 5.45 | ↑ |
| 1–9/hour | 0.13 | 0.04 | 0.002 | ** | 0.02 | ||||
| 10/day | 0.12 | 0.05 | 0.01 | * | |||||
| Seldom | 0.04 | 0.04 | 0.2 | ||||||
| cg24361098 (n = 362) | BCL11A | >10/hour | 0.25 | 0.06 | <.0001 | BMI; gender | *** | 10.56 | ↑ |
| 1–9/hour | 0.15 | 0.06 | 0.02 | * | 0.001 | ||||
| 10/day | 0.15 | 0.07 | 0.04 | * | |||||
| Seldom | 0.13 | 0.05 | 0.02 | * | |||||
| cg16147794 (n = 329) | SLC16A10 | >10/hour | –0.19 | 0.07 | 0.006 | Maternal smoking; tobacco smoke exposure (0–4 years and at 10 years); SES; gender; BMI; current smoking status; exposure to smoke outside the home | ** | 8.91 | ↓ |
| 1–9/hour | –0.07 | 0.07 | 0.3 | 0.003 | |||||
| 10/day | –0.13 | 0.08 | 0.1 | ||||||
| Seldom | 0.06 | 0.06 | 0.4 | ||||||
| cg16668397 (n = 362) | JPH3 | >10/hour | 0.13 | 0.04 | 0.002 | BMI; gender | ** | 10.56 | ↑ |
| 1–9/hour | 0.13 | 0.04 | 0.003 | ** | 0.001 | ||||
| 10/day | 0.07 | 0.05 | 0.2 | ||||||
| Seldom | 0.03 | 0.04 | 0.4 | ||||||
| cg26419883 (n = 362) | TRPM5 | >10/hour | 0.15 | 0.04 | 0.0003 | BMI; gender | *** | 13.64 | ↑ |
| 1–9/hour | 0.06 | 0.04 | 0.2 | 0.0003 | |||||
| 10/day | 0.05 | 0.05 | 0.3 | ||||||
| Seldom | 0.03 | 0.04 | 0.5 | ||||||
| cg21775675 (n = 329) | TMEM161B | >10/hour | –0.17 | 0.05 | 0.001 | Maternal Smoking; Tobacco Smoke Exposure (0–4 years and at 10 years); SES; gender; BMI; current smoking status; exposure to smoke outside the home | ** | 10.07 | ↓ |
| 1–9/hour | –0.05 | 0.05 | 0.3 | 0.002 | |||||
| 10/day | –0.09 | 0.06 | 0.2 | ||||||
| Seldom | 0.002 | 0.05 | 1.0 | ||||||
| cg04794690 (n = 362) | PADI3 | >10/hour | 0.18 | 0.05 | 0.0005 | BMI; gender | *** | 11.27 | ↑ |
| 1–9/hour | 0.22 | 0.05 | <.0001 | *** | 0.0009 | ||||
| 10/day | 0.08 | 0.06 | 0.2 | ||||||
| Seldom | 0.12 | 0.05 | 0.008 | ** | |||||
| cg06942649 (n = 362) | FBXO25 | >10/hour | 0.26 | 0.08 | 0.002 | BMI; gender | ** | 7.5 | ↑ |
| 1–9/hour | 0.26 | 0.09 | 0.002 | ** | 0.007 | ||||
| 10/day | 0.22 | 0.10 | 0.03 | * | |||||
| Seldom | 0.09 | 0.08 | 0.2 | ||||||
| cg18459806 (n = 329) | NIN | >10/hour | –0.16 | 0.04 | <.0001 | Maternal smoking; tobacco smoke exposure (0–4 years and at 10 years); SES; gender; BMI; current smoking status; exposure to smoke outside the home | *** | 13.46 | ↓ |
| 1–9/hour | –0.07 | 0.04 | 0.07 | * | 0.0003 | ||||
| 10/day | –0.10 | 0.05 | 0.04 | * | |||||
| Seldom | –0.03 | 0.04 | 0.4 | ||||||
| cg20631351 (n = 362) | PALM | >10/hour | 0.12 | 0.04 | 0.001 | BMI; gender | ** | 10.25 | ↑ |
| 1–9/hour | 0.10 | 0.04 | 0.01 | * | 0.002 | ||||
| 10/day | 0.07 | 0.05 | 0.2 | ^ | |||||
| Seldom | 0.03 | 0.04 | 0.3 | ^ | |||||
| cg00347824 (n = 362) | NSMAF | >10/hour | 0.19 | 0.05 | 0.0003 | BMI; gender | *** | 8.63 | ↑ |
| 1–9/hour | 0.09 | 0.05 | 0.09 | 0.004 | |||||
| 10/day | 0.12 | 0.06 | 0.05 | ||||||
| Seldom | 0.06 | 0.05 | 0.2 | ||||||
| cg17053854 (n = 348) | SEPT9 | >10/hour | 0.09 | 0.03 | 0.003 | Tobacco smoke exposure (at 10 years); BMI; gender | ** | 13.15 | ↑ |
| 1–9/hour | 0.09 | 0.03 | 0.003 | ** | 0.0003 | ||||
| 10/day | 0.04 | 0.04 | 0.3 | ||||||
| Seldom | 0.01 | 0.03 | 0.8 | ||||||
| cg25324786 (n = 336) | RASA3 | >10/hour | 0.15 | 0.05 | 0.005 | Maternal smoking; tobacco smoke exposure (0–4 years and at 10 years); SES; gender; current smoking status; exposure to smoke outside the home | ** ^ | 17.15 | ↑ |
| 1–9/hour | 0.09 | 0.05 | 0.09 | <.0001 | |||||
| 10/day | –0.03 | 0.06 | 0.7 | ||||||
| Seldom | –0.03 | 0.05 | 0.5 | ||||||
| cg26720961 (n = 336) | TSNARE1 | >10/hour | 0.12 | 0.06 | 0.05 | Maternal smoking; tobacco smoke exposure (0–4 years and at 10 years); SES; gender; current smoking status; exposure to smoke outside the home | ^ | 9.55 | ↑ |
| 1–9/hour | 0.03 | 0.06 | 0.7 | 0.002 | |||||
| 10/day | –0.01 | 0.08 | 0.9 | ||||||
| Seldom | –0.10 | 0.06 | 0.09 | ||||||
| cg05575058 (n = 329) | FAM164A | >10/hour | –0.16 | 0.05 | 0.001 | Maternal smoking; tobacco smoke exposure (0–4 years and at 10 years); SES; gender; BMI; current smoking status; exposure to smoke outside the home | ** | 7.77 | ↓ |
| 1–9/hour | –0.08 | 0.05 | 0.09 | 0.006 | |||||
| 10/day | –0.10 | 0.06 | 0.1 | ||||||
| Seldom | –0.06 | 0.04 | 0.2 | ||||||
| cg15742605 (n = 329) | SAMD11 | >10/hour | 0.19 | 0.05 | 0.0003 | Maternal smoking; tobacco smoke exposure (0–4 years and at 10 years); SES; gender; BMI; current smoking status; exposure to smoke outside the home | *** | 18.74 | ↑ |
| 1–9/hour | 0.08 | 0.05 | 0.1 | <.0001 | |||||
| 10/day | 0.04 | 0.06 | 0.5 | ||||||
| Seldom | –0.01 | 0.05 | 0.9 | ||||||
| cg26185508 (n = 362) | CDCP2 | >10/hour | 0.15 | 0.05 | 0.002 | BMI; gender | ** | 5.48 | ↑ |
| 1–9/hour | 0.18 | 0.05 | 0.0005 | *** | 0.02 | ||||
| 10/day | 0.17 | 0.06 | 0.005 | ** | |||||
| Seldom | 0.07 | 0.04 | 0.1 | ||||||
| cg02378006 (n = 329) | UNC5B | >10/hour | 0.19 | 0.06 | 0.001 | Maternal smoking; tobacco smoke exposure (0–4 years and at 10 years); SES; gender; BMI; current smoking status; exposure to smoke outside the home | ** | 15.56 | ↑ |
| 1–9/hour | 0.07 | 0.06 | 0.2 | <.0001 | |||||
| 10/day | 0.001 | 0.07 | 1.0 | ||||||
| Seldom | –0.0001 | 0.05 | 1.0 | ||||||
| cg08462127 (n = 329) | MYOM2 | >10/hour | 0.13 | 0.05 | 0.006 | Maternal smoking; tobacco smoke exposure (0–4 years and at 10 years); SES; gender; BMI; current smoking status; exposure to smoke outside the home | ** ^ | 6.48 | ↑ |
| 1–9/hour | 0.04 | 0.05 | 0.4 | 0.01 | |||||
| 10/day | 0.11 | 0.06 | 0.06 | ||||||
| Seldom | –0.02 | 0.04 | 0.7 | ||||||
| cg11017318 (n = 329) | SYT16 | >10/hour | –0.06 | 0.04 | 0.1 | Maternal smoking; tobacco smoke exposure (0–4 years and at 10 years); SES; gender; BMI; current smoking status; exposure to smoke outside the home | 1.37 | NA | |
| 1–9/hour | –0.05 | 0.04 | 0.2 | 0.2 | |||||
| 10/day | –0.08 | 0.05 | 0.1 | ||||||
| Seldom | 0.0001 | 0.04 | 1.0 |
Once the Dunnett’ tests provided statistical evidence of differences in marginal means of the heavy vehicular traffic frequency, a second test for trend is performed to assess a ‘dose–response’ relationship.
P < 0.001;
P < 0.01;
P < 0.05; P < 0.1.
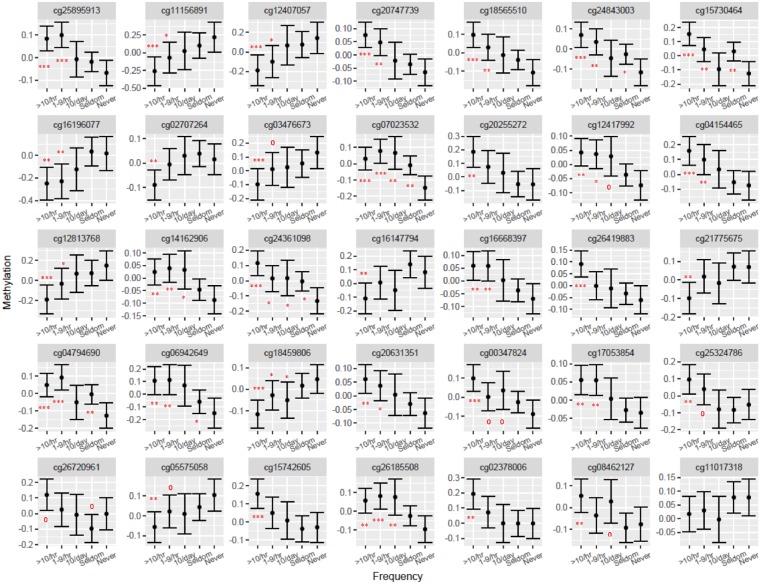
In particular, we found 23 CpGs that were more differentially methylated (Fig. 2). Nineteen of these 23 CpG sites are found in the body of the associated genes while the remaining 4 are located in promoter regions (TSS1500 and TSS200) (Table 3). Conversely of the 11 CpGs that were less methylated with increasing heavy vehicle traffic frequency (Fig. 2), 5 are located in the body of the gene, an additional 5 are found in promoter regions and the last one is ∼50 kb upstream of TMEM161B (Table 3). Among subjects reporting the two highest heavy vehicle traffic frequencies: 1–9 per hour or >10 per hour, statistical significance was consistently reached for the differential methylation observed at these CpG sites (P ≤ 0.05, Table 6).
Figure 2:
Adjusted means (with 95% confidence limits) for DNA-M of 35 significant CpG sites associated with the frequency of heavy vehicles that passed by subjects’ homes for a subset of the IoW birth cohort, F1 generation (range of sample size: 336–369). Adjustments depended on CpG site under consideration and included: Maternal Smoking; Tobacco Smoke Exposure (0–4 years and/or at 10 years); SES; BMI; gender; current smoking status; exposure to smoke outside the home. ***P < 0.001; **P < 0.01; *P < 0.05; P < 0.1. Once the Dunnett’ tests provided statistical evidence of differences in marginal means of the heavy vehicular traffic frequency, a second test for trend is performed to assess a ‘dose–response’ relationship
Stratification by current smoking status, revealed similar trends among smokers and nonsmokers. Although statistically significant differences were only detected for 12 CpGs among smokers and 26 CpGs among nonsmokers, mainly for those reporting >10 heavy vehicles per hour (Supplementary Tables S2 and S3, respectively). Regression results for males only revealed only 10 statistically significant CpG sites with differential methylation: 7 were more methylated and 3 were less methylated (Supplementary Table S4). Results for females indicated 31 significant CpGs with 22 more methylated and 9 less methylated (Supplementary Table S5). The direction of methylation remained the same and the smaller number of significant CpG sites among male subjects is probably due to their smaller sample size in this birth cohort (n = 124).
Results of Replication and Gene Expression Analysis
We replicated the findings for 31 of 35 CpG sites identified in a smaller sample of 140 newborns in the F2 generation. Two CpG sites: cg25895913 (LGI2) and cg00347824 (NSMAF) were associated with traffic frequencies, and the direction of the effect was the same as in the F1 subset. The former CpG site had less methylation, while the latter had more methylation, with increasing vehicular traffic frequency respectively (Table 7). Then, Spearman rank correlation analysis revealed seven CpG sites: cg24843003 (DAZAP1), cg03476673 (CRISPLD2), cg12417992 (SLC6A9), cg04154465 (WNT2B), cg24361098 (BCL11A), cg16668397 (JPH3) and cg17053854 (SEPT9) whose differential methylation was significantly correlated with gene expression (Table 8, partial r ≤ 0.27, P-value ≤ 0.05). For an additional two of these CpG sites: cg14162906 (TMEM222) and cg17053854 (SEPT9), there were marginal correlations with expression data from their associated genes (Table 8, 0.05 > P-value ≤ 0.06).
Table 7:
Results for linear models for CpG sites associated with the frequency of vehicles passing by homes of study subject during pregnancy
| CpG | Associated gene | Heavy vehicle frequency (ref = never) | Estimate | Standard error | P value | Significant covariates in final model | Dunnett's test (LSMEAN = never) | Linear trend test (F value, df = 1) P value | Direction of methylation in F2 generation | Direction of methylation in F1 generation |
|---|---|---|---|---|---|---|---|---|---|---|
| cg15730464 (n = 140) | LGI2 | >10/hour | –0.02 | 0.01 | 0.072 | Gender; birthweight | 3.22 | ↓ | ↓ | |
| 1–9/hour | –0.02 | 0.01 | 0.040 | 0 | 0.07 | |||||
| 10/day | –0.02 | 0.01 | 0.095 | |||||||
| Seldom | –0.02 | 0.01 | 0.118 | |||||||
| cg00347824 (n = 140) | NSMAF | >10/hour | 0.02 | 0.01 | 0.018 | Gender; birthweight | * | 7.36 | ↑ | ↑ |
| 1–9/hour | 0.02 | 0.01 | 0.017 | * | 0.008 | |||||
| 10/day | 0.01 | 0.01 | 0.032 | 0 | ||||||
| Seldom | 0.01 | 0.01 | 0.120 |
Once the Dunnett’ tests provided statistical evidence of differences in marginal means of the heavy vehicular traffic frequency, a second test for trend is performed to assess a ‘dose–response’ relationship.
P < 0.001;
P < 0.01;
P < 0.05; P < 0.1.
Table 8:
Partial Spearman rank correlation coefficients between DNA-m and gene expression in the cord blood of 155 newborns born to a subset of the 369 subjects from the IoW cohort
| CpG | Direction of methylation | Associated gene | Gene expression | Partial correlation coefficient (Spearman) | P value |
|---|---|---|---|---|---|
| cg25895913 | ↑ | CDH4 | A_21_P0009987 | –0.09 | 0.29 |
| cg25895913 | ↑ | CDH4 | A_23_P17593 | 0.01 | 0.92 |
| cg25895913 | ↑ | CDH4 | A_33_P3310976 | –0.07 | 0.40 |
| cg20747739 | ↑ | FAM132A | A_32_P75792 | –0.01 | 0.91 |
| cg18565510 | ↑ | ACAP3 | A_33_P3230290 | 0.01 | 0.93 |
| cg18565510 | ↑ | ACAP3 | A_33_P3398564 | 0.04 | 0.61 |
| cg24843003 | ↑ | DAZAP1 | A_23_P165247 | 0.19 | 0.021 |
| cg24843003 | ↑ | DAZAP1 | A_33_P3359590 | –0.18 | 0.028 |
| cg15730464 | ↓ | LGI2 | A_33_P3358397 | –0.04 | 0.59 |
| cg15730464 | ↓ | LGI2 | A_33_P3393655 | 0.03 | 0.70 |
| cg16196077 | ↓ | RTKN2 | A_21_P0007001 | 0.02 | 0.78 |
| cg16196077 | ↓ | RTKN2 | A_21_P0007002 | –0.09 | 0.30 |
| cg16196077 | ↓ | RTKN2 | A_24_P13041 | 0.06 | 0.45 |
| cg16196077 | ↓ | RTKN2 | A_32_P471485 | –0.04 | 0.61 |
| cg16196077 | ↓ | RTKN2 | A_33_P3219527 | –0.09 | 0.25 |
| cg02707264 | ↓ | MYRIP | A_23_P326760 | 0.05 | 0.58 |
| cg03476673 | ↓ | CRISPLD2 | A_23_P106602 | –0.20 | 0.015 |
| cg12417992 | ↑ | SLC6A9 | A_21_P0001726 | –0.02 | 0.85 |
| cg12417992 | ↑ | SLC6A9 | A_21_P0001727 | –0.04 | 0.61 |
| cg12417992 | ↑ | SLC6A9 | A_23_P11984 | 0.27 | 0.0011 |
| cg12417992 | ↑ | SLC6A9 | A_33_P3402615 | 0.06 | 0.45 |
| cg04154465 | ↑ | WNT2B | A_23_P138352 | 0.16 | 0.0495 |
| cg12813768 | ↓ | SYCP1 | A_23_P722 | –0.02 | 0.83 |
| cg14162906 | ↑ | TMEM222 | A_23_P97442 | 0.09 | 0.30 |
| cg14162906 | ↑ | TMEM222 | A_33_P3259722 | 0.15 | 0.0622 |
| cg24361098 | ↑ | BCL11A | A_21_P0002443 | 0.08 | 0.33 |
| cg24361098 | ↑ | BCL11A | A_21_P0002444 | –0.10 | 0.24 |
| cg24361098 | ↑ | BCL11A | A_21_P0002445 | –0.10 | 0.22 |
| cg24361098 | ↑ | BCL11A | A_24_P402588 | 0.11 | 0.17 |
| cg24361098 | ↑ | BCL11A | A_24_P411186 | 0.26 | 0.0012 |
| cg24361098 | ↑ | BCL11A | A_33_P3249589 | –0.01 | 0.91 |
| cg24361098 | ↑ | BCL11A | A_33_P3249595 | –0.06 | 0.44 |
| cg16147794 | ↓ | SLC16A10 | A_24_P98047 | 0.08 | 0.31 |
| cg16147794 | ↓ | SLC16A10 | A_33_P3308512 | 0.07 | 0.43 |
| cg16668397 | ↑ | JPH3 | A_21_P0008991 | 0.07 | 0.37 |
| cg16668397 | ↑ | JPH3 | A_21_P0008992 | 0.16 | 0.05 |
| cg16668397 | ↑ | JPH3 | A_21_P0008993 | 0.20 | 0.015 |
| cg16668397 | ↑ | JPH3 | A_21_P0008994 | –0.01 | 0.88 |
| cg16668397 | ↑ | JPH3 | A_24_P150791 | 0.16 | 0.048 |
| cg16668397 | ↑ | JPH3 | A_33_P3423721 | –0.20 | 0.016 |
| cg26419883 | TRPM5 | A_23_P87279 | –0.03 | 0.71 | |
| cg21775675 | ↓ | TMEM161B | A_21_P0000876 | –0.02 | 0.83 |
| cg21775675 | ↓ | TMEM161B | A_21_P0004517 | –0.05 | 0.54 |
| cg21775675 | ↓ | TMEM161B | A_23_P156355 | –0.04 | 0.60 |
| cg04794690 | ↑ | PADI3 | A_23_P126869 | 0.00 | 0.97 |
| cg06942649 | ↑ | FBXO25 | A_21_P0005641 | –0.06 | 0.49 |
| cg06942649 | ↑ | FBXO25 | A_21_P0005919 | –0.01 | 0.92 |
| cg06942649 | ↑ | FBXO25 | A_21_P0010595 | 0.07 | 0.40 |
| cg06942649 | ↑ | FBXO25 | A_21_P0012049 | 0.09 | 0.30 |
| cg06942649 | ↑ | FBXO25 | A_21_P0013463 | 0.09 | 0.30 |
| cg06942649 | ↑ | FBXO25 | A_21_P0013464 | –0.01 | 0.93 |
| cg06942649 | ↑ | FBXO25 | A_21_P0013530 | 0.07 | 0.42 |
| cg06942649 | ↑ | FBXO25 | A_21_P0013531 | 0.01 | 0.92 |
| cg06942649 | ↑ | FBXO25 | A_23_P94159 | 0.04 | 0.65 |
| cg06942649 | ↑ | FBXO25 | A_33_P3841368 | 0.04 | 0.60 |
| cg18459806 | ↓ | NIN | A_23_P396353 | 0.06 | 0.47 |
| cg18459806 | ↓ | NIN | A_24_P412512 | 0.02 | 0.84 |
| cg00347824 | ↑ | NSMAF | A_23_P134809 | 0.01 | 0.88 |
| cg17053854 | ↑ | SEPT9- | A_21_P0009276 | –0.11 | 0.17 |
| cg17053854 | ↑ | SEPT9- | A_21_P0009277 | –0.15 | 0.06 |
| cg17053854 | ↑ | SEPT9- | A_21_P0009278 | –0.07 | 0.37 |
| cg17053854 | ↑ | SEPT9- | A_21_P0009279 | –0.20 | 0.014 |
| cg17053854 | ↑ | SEPT9- | A_21_P0009280 | –0.01 | 0.92 |
| cg17053854 | ↑ | SEPT9- | A_21_P0009396 | –0.13 | 0.13 |
| cg25324786 | ↑ | RASA3 | A_21_P0008023 | –0.01 | 0.89 |
| cg25324786 | ↑ | RASA3 | A_33_P3262515 | –0.05 | 0.56 |
| cg26720961 | ↑ | TSNARE1 | A_21_P0005916 | 0.00 | 0.96 |
| cg26720961 | ↑ | TSNARE1 | A_33_P3291567 | –0.06 | 0.44 |
| cg26720961 | ↑ | TSNARE1 | A_33_P3297126 | –0.09 | 0.30 |
| cg15742605 | ↑ | SAMD11 | A_21_P0001250 | –0.06 | 0.45 |
| cg15742605 | ↑ | SAMD11 | A_33_P3818959 | –0.02 | 0.77 |
| cg26185508 | ↑ | CDCP2 | A_33_P3259522 | –0.05 | 0.51 |
| cg02378006 | ↑ | UNC5B | A_23_P52336 | 0.04 | 0.61 |
| cg02378006 | ↑ | UNC5B | A_32_P52153 | 0.13 | 0.11 |
| cg08462127 | ↑ | MYOM2 | A_21_P0005646 | –0.07 | 0.39 |
| cg08462127 | ↑ | MYOM2 | A_23_P258912 | 0.04 | 0.64 |
| cg11017318 | ↓ | SYT16 | A_24_P143324 | 0.02 | 0.85 |
P-values in bold and bold italics denote statistical significance less than or equal to 0.05 and 0.1 respectively.
Discussion
We aimed to answer two questions: (i) Which specific CpG sites are associated with heavy vehicular traffic in the birth cohort? (ii) Are there any trends in the association between differential DNA-m and the frequency of heavy vehicular traffic? Regarding the first question, we found 35 CpG sites to be associated with heavy vehicular traffic. These CpG sites were associated with 34 different genes (two CpG sites—cg11156891 and cg12407057 mapped to the same gene: ANKRD65). Additionally, 31 of these genes have been reported to be associated with air pollution related chemicals such as benzo(a)pyrene in the comparative toxicogenomics database. In adopting an epigenome-wide approach, as opposed to a candidate gene approach, our analysis adds novel information on epigenetic markers for traffic-related air pollution exposure. These exposure-associated changes in the epigenome could be used to identify exposure to air pollutants, particularly those from incomplete combustion of fuels such as diesel which is often used in buses and trucks. With further research, it can also guide the development of effective clinical and public health interventions and reduce the burden of air pollution–related health outcomes.
For the second question on assessing the association between differential DNA-m and traffic-related air pollution, we found 23 CpGs that were more methylated, and 11 CpGs that were less methylated with increasing heavy vehicular traffic frequency for all subjects after adjusting for confounders. These associations between heavy vehicular traffic frequency and DNA-m measurements persisted after stratification by current smoking status for 26 and 12 CpG sites among nonsmokers and smokers, respectively. Among subjects reporting the two highest heavy vehicular frequency levels: 1–9 per hour or >10 per hour, statistical significance was consistently reached for the differential methylation observed at these CpG sites (P ≤ 0.05, Table 6). This exploratory study highlights the fact that epigenetic differences can be observed among subjects exposed to varying frequencies of local traffic.
Our results suggest that exposure to emissions, presumably from the exhaust of heavy vehicles passing by the residences of study subjects, may have an impact on DNA-m. It has been suggested that epigenetic states can convey susceptibility to air pollution, which can lead to biological changes, and ultimately, adverse health [43, 52]. DNA-m profiles can provide insight into aspects of biology such as gene activity and regulation, and our gene enrichment analysis offers examples of how the genes associated with the CpG sites are related to various molecular functions, pathways and some rare diseases. Based on the location of the CpG site such as promoter or body, altered methylation may lead to increased transcription, silencing or altered splicing [53–56]. Hence, a differential transcription level is only one of the consequences of DNA-m. For instance, it has been considered that methylation in promoter regions may lead to changes in gene expression, i.e. gene silencing [57]; and such changes can serve as putative markers or risk factors for altered susceptibility and/or disease states. Additionally, DNA-m can help in identifying CpG sites, and possibly genes, that are more susceptible to environmental exposures [58].
In a replication and gene expression analysis study among 140 newborns from the F2 generation, 6 of the 7 CpG sites that correlated with expression, cg24843003 (DAZAP1), cg12417992 (SLC6A9), cg04154465 (WNT2B), cg24361098 (BCL11A), cg16668397 (JPH3) and cg17053854 (SEPT9), are located in the bodies of the associated genes. The seventh, cg03476673, is found in the 5′UTR region of CRISPLD2. Of the remaining 23 CpG sites with corresponding expression data but no statistically significant correlations, 3 are located in the TSS1500 region including cg14162906 (TMEM222) which achieved a marginal significance. The rest are in the following regions: body of the associated gene (n = 14), TSS200 region (n = 2), 5′UTR region (n = 2), ∼50 kb upstream of TMEM161B (n = 1) and ∼200 kb upstream of SYT16 (n = 1). The association of 31 of 34 genes (identified from CpG sites in this study) to air pollution–related chemicals adds plausibility to potential environment–gene interactions, and can contribute to emerging data that provide a more complete view of environmental exposures. We posit that traffic-related air pollution may be a plausible environmental exposure of interest on the IoW.
With increasing evidence that exposure to air pollution is associated with adverse health outcomes, biologically plausible mechanistic pathways of air pollution’s effects, such as oxidative stress, inflammation, coagulation, endothelial function and hemodynamic response, have been implicated [59]. Exposure to ambient particulate matter, which is known to be emitted from diesel truck traffic, is associated with decreased lung function and increases in respiratory disease and symptoms such as asthma exacerbation [47, 60–62, 72]. Exposure to gaseous air pollutants including nitrogen species (e.g. NO2, NO, NOx) are also associated with deleterious effects such as bronchial reactivity, airway oxidative stress, pulmonary and systemic inflammation [63–66]. Several epidemiologic studies have reported that short-term increases in ambient pollutants such as PM2.5 and nitrogen dioxide (NO2) are associated with increases in airway inflammation in children and adults [67–73].
A recent epigenome-wide meta-analysis by Gruzieva et al. [74] provides evidence on the association between prenatal air pollution exposures and differences in the methylation of several genes in cord blood. In particular, the authors found significant associations between NO2 exposures and DNA-m for CpG sites that mapped to genes in the solute carrier family (SLC), family with sequence similarity (FAM) and transmembrane proteins (TMEM). Five CpG sites [associated gene in square brackets] (cg12417992 [SLC6A9], cg14162906 [TMEM222], cg16147794 [SLC16A10], cg20747739 [FAM132A], cg21775675 [TMEM161B]) related to three gene superfamilies from the Gruzieva et al. meta-analysis were associated with heavy vehicular traffic frequency and DNA-m in our study. The association of one of the CpG sites with FAM132A (codes for an important anti-inflammatory adipokine [75]), strengthens the hypothesis that inflammation may be a possible mechanism though which ambient air pollution affects human health [76].
While underlying molecular alterations of air pollution mediated adverse health remain to be further investigated, another recent study with two European cohorts identified decreasing DNA-m on CpG island shores, shelves and gene bodies with increasing concentrations of nitrogen oxide (NO) species [77]. NO species are currently the best available indicators of spatial variation and mixtures of outdoor urban air pollution such as traffic [78]. Our analysis did not reveal CpG sites associated with the inflammatory genes mentioned in the above study, and to the best of our knowledge, the significant CpG sites reported in our study have not been reported in previous air pollution studies. This may be due to differences in (i) study populations, (ii) exposure assessment and concentrations, (iii) complex multiple biological pathways or (iv) a combination of any of the previous three reasons. These newly identified CpG sites and associated genes are certainly worth exploring in larger cohorts.
In our study, the two CpG sites that were associated with vehicular traffic in both the F1 and F2 generation may be reflective of the effects of TRAP exposures at these two loci. It also suggests possible prenatal exposures to traffic-related air pollutants in the F2 generation. Secondly, correlation between DNA-m and gene expression at 7 of 31 CpG sites (and three marginal correlations) supports the hypothesis that DNA-m is a potential mechanism through which traffic-related air pollutants can affect gene expression. Three of these seven CpG sites are associated with genes previously identified in the literature to be related to inhalation. For instance, CRISPLD2 has been identified as a glucocorticoid responsive gene that modulates cytokine function in airway smooth muscle cells [79]. WNT2B has been reported to be associated with embryonic origins of the lung since the inactivation of WNT2A and WNT2B, resulted in complete absence of lung development [80]. Methylation of JPH3 from sputum samples is a sensitive and specific predictor of chronic mucous hypersecretion in former male smokers [81]. The lack of 100% replication and correlation in our analysis may be due to small sample sizes and exposure misclassification from the use of questionnaire data rather than air pollution data (for instance the questionnaire administered at 18 years specified ‘heavy vehicle’ while the questionnaire during pregnancy only mentioned ‘vehicle’). While our results must be interpreted with caution, there are additional studies that add to the evidence that adverse effects of air pollution that can occur when one is exposed. A recent study, which did not replicate its results in a separate independent cohort, found that living close to major roadways at birth was associated with differential cord blood methylation [82]. Another study, which was also not replicated in an independent cohort found significant associations between long-term air pollution exposure (NO2) and DNA-m for seven CpG sites (Bonferroni corrected threshold P < 1.2E–7) [83].
With continuing indication that exposure to ambient air pollutants may contribute to adverse public health [1, 84], further research is needed to identify the components of air pollution that determine its toxicity and a pristine environment such as the IoW could offer a suitable environment to study ambient air pollutant toxicity. The constituents of the pollution potentially generated by heavy vehicles may need to be identified so that early preventative and possible control strategies can be targeted efficiently. Whether these findings raise the risk for future cellular malfunction and disease is unknown. One main reason for the persistence (or the lack thereof) of such findings could be attributed to small sample sizes. In our case, the nonsmokers were consistently between 248 and 270 while smokers were between 78 and 95 subjects. Another reason could be due to the small magnitude effect sizes that are common with environmental epigenetic research [85]. Profiling of the epigenome over time in this population will help improve understanding of TRAP exposures and how the epigenome responds to this stimuli. Additionally, we found that secondhand smoke exposure is represented by the questions posed to subjects about tobacco smoke exposures since these variables were associated with the methylation of cg07555921 (AHRR), while the exposure variable was not. Therefore, these observed effects of heavy vehicular traffic on DNA-m may be without the contribution of this type of air pollution. Further studies in the future may be needed to examine this in depth.
There are some limitations to this study. First in this study, our exposure variable of interest, heavy vehicular traffic frequency, was ascertained by questionnaire responses from study subjects and we did not attempt to conduct exposure assessment inside or outside their residences, and these analyses were based on current residences (at the time of the blood draw at 18 years old in the F generation) as opposed to conditions in former places of residence. Secondly, the associations observed in this study are informative. However, further analysis may be needed to assess other self-reported exposures such as tobacco exposures, particularly on a cumulative scale. Given that the data in this pilot study are from a birth cohort to which a third generation follow-up has been added, further investigation of the DNA-m of the same subset of this population at earlier time points or in their offspring could address some of these limitations. Thirdly, methylation data were obtained from whole blood but not from specific cell subgroups, due to cost, but while differential methylation may or may not be present in all cell subsets, we believe that important biological insights still may be gained from studying DNA-m in whole blood [86]. Moreover, we did adjust for the cell types in the screening step of the analysis, thereby overcoming this limitation. Additionally, multiple studies have validated the 450K DNA-m array from Illumina [87–89], and this assay is generally accepted in the scientific literature. Hence we did not see a necessity to additionally test the results of specific CpGs from the 450K DNA-m array with methyl-specific qPCR. The use of bisulfite sequencing can be challenging, since it reduces genome complexity and some of the methods may not differentiate between methylcytosine and hydroxymethylcytosine. The incorporation of appropriate controls for bisulfite reactions and careful interpretation of DNA-m level after accounting for cell types can overcome some of these challenges [90]. An overview of major difficulties related to bisulfite sequencing and how to overcome them are presented in the review by Li et al. [91]. Although the correlations between CpG sites and expression data reached statistical significance, the coefficients were weak. One may consider this as a limitation of our study; however, gene expression is influenced by multiple factors and our analysis only focus on the role of DNA-m on gene expression. Future studies with large sample sizes need to further investigate associations between traffic-related DNA-m and gene expression, taking other factors such as genetic polymorphisms and network of related genes, into consideration. Finally, since this is the first study that shows an effect of varying heavy vehicular traffic frequency on DNA-m among residents on the Isle, further replication of these associations in an independent cohort is needed.
Conclusions
Our findings reveal differences in DNA-m in participants who reported higher heavy vehicular traffic frequencies when compared with participants who reported lower frequencies. Such findings may be attributed to TRAP exposure and suggest that further studies are needed.
Materials and Methods
Study Population
Subjects in this study are from a whole population birth cohort established in 1989 on the IoW, UK, to prospectively study the natural history of allergies and asthma. This cohort has been previously described in detail elsewhere [92]. Informed consents and detailed information from questionnaires were obtained from participants at recruitment and at each follow-up year: 1, 2, 4, 10 and 18 years [93]. The questionnaires for the entire birth cohort study are for study-specific objectives, while asthma and allergy symptom questions are from the validated International Study of Asthma and Allergies in Childhood (ISAAC) [92]. Local Research Ethics Committees approved of the parent study, and the Institutional Review Board at the Medical University of South Carolina approved the current study. In this exploratory analysis, we focus on 369 individuals (245 women and 124 men) with DNA-m measurements at age 18 years. Due to the original study question of inheritance via females, we included more females than males at 18 years.
DNA-m Analysis
DNA was extracted from peripheral blood samples and its concentration was determined by Qubit quantitation, as described previously [94]. Genome-wide DNA-m was assessed using the Illumina Infinium Human Methylation 450 beadchip (Illumina, Inc., CA, USA), which interrogates >484 000 CpG sites associated with approximately 24 000 genes. Arrays were processed and imaged using the manufacturer’s recommendations, as described elsewhere [95]. Multiple identical control samples were assigned to each bisulfite conversion batch, and the samples were randomly distributed on microarrays to assess assay variability and to control batch effects respectively.
Methylation levels (β values) were calculated for queried CpG loci using the methylation module of GenomeStudio software [96]. DNA-m levels for each CpG were estimated as the proportion of intensity of methylated (M) over the sum of methylated (M) and unmethylated (U) probes, β = M/[c + M + U] with c being a constant to prevent dividing by zero [97]. DNA-m levels were corrected for batch effect using ‘IMA’ and ‘ComBat’ packages in R [98]. M-values were calculated as log 2 ratio of the intensities of methylated probe versus unmethylated probe, and used in subsequent analysis [99]. The detection P-value for each CpG site was used as a quality control measure of probe performance and CpG sites with: (i) detection P-value > 0.01 in >10% of the samples and (ii) probe single nucleotide polymorphism (SNP) excluded from all analyses.
We estimated the proportion of cell types in adult peripheral blood using the estimateCellCounts() function in minfi package following the Houseman approach [100] using the adult reference panel [101].
Exposure Assessment
The exposure variable of interest, the frequency of heavy vehicular traffic, was determined through questionnaire responses from the subjects to the question: How often do heavy vehicles (e.g. trucks/buses) pass your house or on the street less than 100 meters away? The five-point response included: never, seldom, 10 per day, 1–9 per hour or >10 per hour. We also assessed answers to other air pollution related questions such as ‘How often do cars pass your house or on the street less than 100 meters away?’ and ‘How frequently are you annoyed by outdoor air pollution (from traffic industry, etc) in your home if you keep the window open?’. All subjects were approximately 18 years old when the questionnaire containing these questions was administered. While we have not seen of any study in the literature that has used the same question to assess exposure to TRAP, others have used questionnaire-derived assessments as air pollution exposure variables [102, 103]. Others have used such questions along with proximity to roadways, air pollution measurements, land use regressions together with the validated and widely used International Study of Asthma and Allergies in Childhood (ISAAC) questionnaire to successfully characterize health effects of interest [104–106].
Covariates of Interest
For this exploratory study, the covariates of interest obtained from the subjects’ mothers were as follows: (i) gender; (ii) maternal smoking status during pregnancy obtained from questionnaires at birth of the subject; (iii) tobacco smoke exposure obtained through questionnaires completed at birth and at ages 1, 2, 4 and 10 years. Other covariates were obtained from the questionnaire administered to the subjects at age 18: (iv) socio-economic status (SES) ascertained from the question ‘what is your family’s annual income (estimate)?’; (v) current smoking status, and age subject started to smoke if applicable; (vi) exposure to smoke outside the home ascertained by the question ‘are you regularly exposed to smoking outside the home?’; (vii) BMI calculated from height and weight measurements obtained during the 18-year follow-up, using the following formula: weight (kg)/height (m)^2. In addition, we considered the type of residential property the subjects lived in (rented privately, rented council/housing association, owned privately or other), whether the subjects were still living with their parents, and the duration of living in the present house (obtained in the course of the 4-year follow-up).
Statistical Analysis
Descriptive statistics and chi square tests were used to assess whether the 369 subjects in this study were representative of the total birth cohort. Then, we conducted statistical analyses in two main steps:
Step 1: Epigenome-wide Association Analysis
Screening tool. We employed ttScreening package (an epigenome-wide DNA-m sites screening tool) to examine CpGs that are potentially associated with the frequency of heavy vehicles passing by subjects’ homes at age 18 years. This approach to screen epigenome-wide data was used since it generally performs better and has the potential to control both types I and II errors [107]. Specifically, the ttScreening package conducts surrogate variable analysis, unexplained variation in the data is removed, prior to an iterative training-testing procedure. This training-testing method performs better than methods such as the FDR and the Bonferroni in reducing false-positive and false-negative results. In addition to providing internal validation, the use of training-testing builds more generalized models than those constructed by traditional methods, and can detect additional loci undetectable using traditional methods [107].
The analytical methods implemented in the package employed a screening process that filtered non-informative CpGs through 100 iterations of a training-and-testing (TT) process with robust regressions. We followed the default settings for the ttScreening method: (i) 2 of 3 of the data for training, (ii) the ‘two-step’ method for surrogate variable analysis (sva.method) [108], (iii) 100 iterations for the total number of screenings (iterations), (iv) 50% as the cutoff proportion of those 100 iterations (cv.cutoff) and (v) 0.05 significance level for the training (train.alpha) and testing data (test.alpha). The 100 iterations are recommended by the authors of the ttScreening package to create a balance between computing efficiency and adequate resampling to arrive at true associations. Also 50% is the default for the cutoff proportion since the informative CpGs are usually sparse in comparison to the candidate CpG sites, and the authors’ simulations identified 50% cutoff percentage as suitable for small and large sample sizes [107].
The independent and dependent variables were heavy vehicular traffic frequency and DNA-m, respectively. A CpG was selected as an informative site if it showed statistical significance in at least 70% of iterations. The ttScreening( ) function automatically adjusts for multiple testing using three methods, including FDR, Bonferroni and the TT method [109].
CpG by CpG analysis. As an alternative to the ttscreening method, we also conducted multiple linear regressions with the M values of each CpG while adjusting for all covariates selected apriori and calculated adjusted P-values for the multiple comparisons (p.adjust ( ) command in base R). The exposure variable in this case was classified as ‘Any’ versus ‘No’ heavy vehicular traffic frequency. All procedures in Step 1 were conducted with R (version 3.4.2) [110].
Tobacco smoke exposure. Prior epigenome-wide association studies have shown that the methylation of cg05575921 located in the AHRR gene is a robust indicator of tobacco smoke exposure [111, 112]. Even in different demographics, smoking histories and rates of false-negative self-report of smoking behavior, this CpG site can reliably detect smoking status [113]. Additionally, a recent study revealed that high levels of recent secondhand smoke exposure was inversely associated with DNA-m of cg05575921 in monocytes from nonsmokers, although the effects were weaker when compared with active smokers [114]. Hence we conducted linear regression models with self-reported smoking status and secondhand smoke exposures to examine the relationships between this CpG site and tobacco smoke exposure, as well as our exposure variable: heavy vehicular traffic frequency.
Step 2: Associations between the Frequency of Heavy Vehicular Traffic and DNA-m
To investigate preliminary associations with heavy vehicular traffic frequency, we assessed differences in unadjusted DNA-m of the CpGs identified in the ttscreening method in Step 1 using analysis of variance (ANOVA) on only heavy vehicular frequency. Then, the CpGs were further tested in multiple linear models that included potential confounders to assess their association with the heavy vehicular traffic frequency. A general form of the model is seen in Equation (1):
| (1) |
refers to the DNA-m for the ith subject reporting vth category of heavy vehicular frequency, α is the intercept and ε is the error term. The coefficient is the deviation of grand mean for vth category of heavy vehicle traffic frequency (seldom, 10 per day, 1–9 per hour and >10 per hour) compared to never. The lsmeans statement was used to derive model adjusted means.
Modeling and Variable Selection
For a covariate to be considered a confounder, the estimate of the regression coefficient for heavy vehicle traffic frequency in the reduced model (that excluded the confounder of interest) had to fall outside the range of 10% of the estimate of the full model (the full model includes all covariates considered apriori in this study) [115]. The final models for each CpG site included gender and any identified confounders. Models were assessed for all subjects and then stratified by gender and current smoking status since exposure to tobacco can lead to extensive genome-wide changes in DNA-m [116].
Adjusted DNA-m Means and Trend Test
We performed Dunnett’s tests to compare model adjusted (marginal) means from four heavy vehicle traffic frequency categories (seldom, 10 per day, 1–9 per hour or >10 per hour) against a control group mean (never) to check for statistically significant differences. We also used PROC IML’s ORPOL function in SAS [117] to obtain appropriate coefficients for contrast statements to test for linear trends in increasing heavy vehicular frequency with increasing or decreasing DNA-m measurements, only when marginal means were significantly different from the control mean (never category). When marginal means did not significantly differ from the control category, the results were not provided. P values <0.1 were considered statistically significant for the trend tests. Finally, marginal means for DNA-m were plotted by category of reported heavy vehicle traffic frequency. Step 2 was performed with the SAS statistical package (version 9.4; SAS Institute, Cary, NC, USA). All plots were derived using ‘ggplot’ function in R.
Replication and Gene Expression
Study Population
Thirty-one of 35 significant CpG sites found in the present study for the 369 subjects in the F1 generation were tested in the DNA-m and gene expression data from cord blood in the newborn cohort, the F2 generation (n = 155, born 2006–2013). This step constitutes a replication of the CpGs in a semi-independent cohort. In the F2 generation, there were 76 males and 79 females and the average birthweight was 3459.3 g (standard deviation: 504.6). The median birthweight was 3515 g (n = 148). The exposure variable was obtained from the questionnaire administered to the mothers during pregnancy. The mothers’ answers to this question were used as the exposure (independent) variable of interest: How often do vehicles pass your house or on the street less than 100 meters away? The answers were never, seldom, 10 per day, 1–9 per hour or >10 per hour. When a mother answered the question once instead of three times, this answer was assigned as the frequency of vehicles that passed by the home during the entire pregnancy. If she answered two or three times, the lowest frequency was assumed to be her exposure. This was to be conservative on their exposures since this pregnancy questionnaire did not specify ‘heavy vehicles’, compared to the question posed to them (F1 generation) at age 18. It also allowed for a distribution of responses as follows: never (2), seldom (8), 10 per day (26), 1–9 per hour (39) and >10 per hour (72). Eight mothers did not provide an answer to this question during any of the three trimesters and were excluded from the remaining analysis. Also there were 31 of 35 top CpG sites available for the F2 newborn subset.
Gene Expression Array
At birth, IoW F2 cord blood samples were collected into PAXgene Bone Marrow RNA Tubes and RNA extracted using PAXgene RNA kits (PreAnalytiX GmbH, Switzerland). RNA integrity was verified with the Agilent 2100 Bioanalyzer system. Genome-wide mRNA expression was assessed via one color (Cy3) experiments with the Agilent (Agilent Technologies, Santa Clara, CA) SurePrint G3 Human Gene Expression 8×60k v2 microarray kits. Array content was sourced from RefSeq, Ensembl, UniGene and GenBank databases and provides full coverage of the human transcriptome in 50 599 biological features (including replicate probes and control probes). The oligos were 60mer in length and each transcript was tagged at least once and some had multiple tagging oligos for genes with documented splice variants. Data QC indices and analyses were performed with Agilent GeneSpring software. These data were then percent shift normalized and log2-transformed.
Statistical Analysis
DNA - m data: Linear regression models, Dunnett’s multiple comparison tests and trend tests were used to assess the relationship between the frequency of vehicular traffic and DNA-m, as previously described for the subset from the F1 generation. The models were adjusted for gender and birthweight. Successful replication was defined as having the same direction of differential methylation and a P-value of <0.05.
Gene expression data: We calculated partial Spearman’s rank correlations between the DNA-m at 31 of 35 CpG sites and gene expression data for the associated genes while controlling for cell types (Bcell, CD4T, CD8T, gran, mono, NK and nRBC). Since cord blood includes nucleated red blood cells, we used the cell references provided by Bakulski and colleagues [118, 119].
Supplementary Material
Acknowledgements
The authors are grateful for the participation and cooperation of the children and parents of IoW and the IoW research team in collecting data and technical support. Appreciation also goes to the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust grant reference 090532/Z/09/Z and MRC Hub grant G0900747 91070) for the generation of the methylation data. The authors would also like to acknowledge three anonymous peer reviewers for their valuable input.
Funding
Research reported in this publication was supported by the National Institute Health under Award Numbers R01 AI091905 and R01 HL132321 (PI: Wilfried Karmaus) and R01 AI061471 (PI: Susan Ewart). National Asthma Campaign, UK (Grant No 364) funded 10-year follow-up of this study and NIH/NHLBI R01 HL082925-01 funded the 18-year follow-up (PI: S. Hasan Arshad). The content is the responsibility of the authors and does not represent the official views of the National Institutes of Health.
Conflict of interest statement. None declared.
References
- 1. Kampa M, Castanas E.. Human health effects of air pollution. Environ Pollut 2008;151:362–7. [DOI] [PubMed] [Google Scholar]
- 2. Schwartz J, Bind M-A, Koutrakis P.. Estimating causal effects of local air pollution on daily deaths: effect of low levels. Environ Health Perspect 2017;125:23–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Seaton A, Godden D, MacNee W, Donaldson K.. Particulate air pollution and acute health effects. Lancet 1995;345:176–8. [DOI] [PubMed] [Google Scholar]
- 4. Cohen AJ, Brauer M, Burnett R, Anderson HR, Frostad J, Estep K, Balakrishnan K, Brunekreef B, Dandona L, Dandona R, et al. Estimates and 25-year trends of the global burden of disease attributable to ambient air pollution: an analysis of data from the Global Burden of Diseases Study 2015. Lancet 2017;389:1907–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brunekreef B, Holgate ST.. Air pollution and health. Lancet 2002;360:1233–42. [DOI] [PubMed] [Google Scholar]
- 6. Hoek G, Krishnan RM, Beelen R, Peters A, Ostro B, Brunekreef B, Kaufman JD.. Long-term air pollution exposure and cardio-respiratory mortality: a review. Environ Health 2013;12:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Colvile RN, Hutchinson EJ, Mindell JS, Warren RF.. The transport sector as a source of air pollution. Atmos Environ 2001;35:1537–65. [Google Scholar]
- 8. Hochstetler HA, Yermakov M, Reponen T, Ryan PH, Grinshpun SA.. Aerosol particles generated by diesel-powered school buses at urban schools as a source of children’s exposure. Atmos Environ 2011;45:1444–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim JJ, Smorodinsky S, Lipsett M, Singer BC, Hodgson AT, Ostro B.. Traffic-related air pollution near busy roads: the East Bay Children's. Am J Respir Crit Care Med 2004;170:520–6. [DOI] [PubMed] [Google Scholar]
- 10. Kinney PL, Aggarwal M, Northridge ME, Janssen NA, Shepard P.. Airborne concentrations of PM(2.5) and diesel exhaust particles on Harlem sidewalks: a community-based pilot study. Environ Health Perspect 2000;108:213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumar P, Pirjola L, Ketzel M, Harrison RM.. Nanoparticle emissions from 11 non-vehicle exhaust sources – a review. Atmos Environ 2013;67:252–77. [Google Scholar]
- 12. Schauer JJ, Rogge WF, Hildemann LM, Mazurek MA, Cass GR, Simoneit BR.. Source apportionment of airborne particulate matter using organic compounds as tracers. Atmos Environ 1996;30:3837–55. [Google Scholar]
- 13. Titos G, Lyamani H, Pandolfi M, Alastuey A, Alados-Arboledas L.. Identification of fine (PM 1) and coarse (PM 10-1) sources of particulate matter in an urban environment. Atmos Environ 2014;89:593–602. [Google Scholar]
- 14. Zang-Ho S. Emissions of ozone precursors from a biogenic source and port-related sources in the Largest Port City of Busan, Korea. Asian J Atmos Environ 2015;9:39–47. [Google Scholar]
- 15. Guarnieri M, Balmes JR.. Outdoor air pollution and asthma. Lancet 2014;383:1581–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khreis H, Kelly C, Tate J, Parslow R, Lucas K, Nieuwenhuijsen M.. Exposure to traffic-related air pollution and risk of development of childhood asthma: a systematic review and meta-analysis. Environ Int 2017;100:1–31. [DOI] [PubMed] [Google Scholar]
- 17. Calderón-Garcidueñas L, Villarreal-Ríos R.. Living close to heavy traffic roads, air pollution, and dementia. Lancet 2017;389:675–7. [DOI] [PubMed] [Google Scholar]
- 18. Chen H, Kwong JC, Copes R, Tu K, Villeneuve PJ, van Donkelaar A, Hystad P, Martin RV, Murray BJ, Jessiman B, et al. Living near major roads and the incidence of dementia, Parkinson's disease, and multiple sclerosis: a population-based cohort study. Lancet 2017;389:718–26. [DOI] [PubMed] [Google Scholar]
- 19. Gauderman WJ, Avol E, Lurmann F, Kuenzli N, Gilliland F, Peters J, McConnell R.. Childhood asthma and exposure to traffic and nitrogen dioxide. Epidemiology 2005;16:737–43. [DOI] [PubMed] [Google Scholar]
- 20. Miller KA, Siscovick DS, Sheppard L, Shepherd K, Sullivan JH, Anderson GL, Kaufman JD.. Long-term exposure to air pollution and incidence of cardiovascular events in women. N Engl J Med 2007;356:447–58. [DOI] [PubMed] [Google Scholar]
- 21. Oftedal B, Nystad W, Brunekreef B, Nafstad P.. Long-term traffic-related exposures and asthma onset in schoolchildren in Oslo, Norway. Environ Health Perspect 2009;117:839–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wheeler AJ, Smith-Doiron M, Xu X, Gilbert NL, Brook JR.. Intra-urban variability of air pollution in Windsor, Ontario—measurement and modeling for human exposure assessment. Environ Res 2008;106:7–16. [DOI] [PubMed] [Google Scholar]
- 23. Brauer M, Hoek G, van Vliet P, Meliefste K, Fischer P, Gehring U, Heinrich J, Cyrys J, Bellander T, Lewne M, Brunekreef B.. Estimating long-term average particulate air pollution concentrations: application of traffic indicators and geographic information systems. Epidemiology 2003;14:228–39. [DOI] [PubMed] [Google Scholar]
- 24. Dockery DW, Pope CA, Xu X, Spengler JD, Ware JH, Fay ME, Ferris BG, Speizer FE.. An association between air pollution and mortality in six U.S. cities. N Engl J Med 1993;329:1753–9. [DOI] [PubMed] [Google Scholar]
- 25. Gan WQ, Allen RW, Brauer M, Davies HW, Mancini GJ, Lear SA.. Long-term exposure to traffic-related air pollution and progression of carotid artery atherosclerosis: a prospective cohort study. BMJ Open 2014;4:e004743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jerrett M, Arain A, Kanaroglou P, Beckerman B, Potoglou D, Sahsuvaroglu T, Morrison J, Giovis C.. A review and evaluation of intraurban air pollution exposure models. J Expo Sci Environ Epidemiol 2005;15:185–204. [DOI] [PubMed] [Google Scholar]
- 27. Lipfert F, Wyzga R, Baty J, Miller J.. Traffic density as a surrogate measure of environmental exposures in studies of air pollution health effects: long-term mortality in a cohort of US veterans. Atmos Environ 2006;40:154–69. [Google Scholar]
- 28. Nethery E, Leckie SE, Teschke K, Brauer M.. From measures to models: an evaluation of air pollution exposure assessment for epidemiological studies of pregnant women. Occup Environ Med 2008;65:579–86. [DOI] [PubMed] [Google Scholar]
- 29. Shi L, Zanobetti A, Kloog I, Coull BA, Koutrakis P, Melly SJ, Schwartz JD.. Low-concentration PM2.5 and mortality: estimating acute and chronic effects in a population-based study. Environ Health Perspect 2016;124: 46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zou B, Wilson JG, Zhan FB, Zeng Y.. Air pollution exposure assessment methods utilized in epidemiological studies. J Environ Monit 2009;11:475–90. [DOI] [PubMed] [Google Scholar]
- 31. English P, Neutra R, Scalf R, Sullivan M, Waller L, Zhu L.. Examining associations between childhood asthma and traffic flow using a geographic information system. Environ Health Perspect 1999;107:761–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin S, Munsie JP, Hwang S-A, Fitzgerald E, Cayo MR.. Childhood asthma hospitalization and residential exposure to state route traffic. Environ Res 2002;88:73–81. [DOI] [PubMed] [Google Scholar]
- 33. Zmirou D, Gauvin S, Pin I, Momas I, Sahraoui F, Just J, Le Moullec Y, Brémont F, Cassadou S, Reungoat P, et al. Traffic related air pollution and incidence of childhood asthma: results of the Vesta case-control study. J Epidemiol Commun Health 2004;58:18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hobbs BD, de Jong K, Lamontagne M, Bosse Y, Shrine N, Artigas MS, Wain LV, Hall IP, Jackson VE, Wyss AB, et al. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet 2017;49:426–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Olden K, Lin Y-S., Bussard D.. Epigenome: a biomarker or screening tool to evaluate health impact of cumulative exposure to chemical and non-chemical stressors. Biosensors 2016;6:1–7. [Google Scholar]
- 36. McCullough SD, Dhingra R, Fortin Mc, Diaz-Sanchez D.. Air pollution and the epigenome: a model relationship for the exploration of toxicoepigenetics. Curr Opin Toxicol 2017;6:18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Breton CV, Marutani AN.. Air pollution and epigenetics: recent findings. Curr Environ Health Rep 2014;1:35–45. [Google Scholar]
- 38. Ji H, Hershey GKK.. Genetic and epigenetic influence on the response to environmental particulate matter. J Allergy Clin Immunol 2012;129:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen Z, Salam MT, Eckel SP, Breton CV, Gilliland FD.. Chronic effects of air pollution on respiratory health in Southern California children: findings from the Southern California Children’s Health Study. J Thor Dis 2015;7:46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liang L, Willis-Owen SAG, Laprise C, Wong KCC, Davies GA, Hudson TJ, Binia A, Hopkin JM, Yang IV, Grundberg E, et al. An epigenome-wide association study of total serum immunoglobulin E concentration. Nature 2015;520:670. U188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang IV, Pedersen BS, Liu A, O'Connor GT, Teach SJ, Kattan M, Misiak RT, Gruchalla R, Steinbach SF, Szefler SJ, et al. DNA methylation and childhood asthma in the inner city. J Allergy Clin Immunol 2015;136:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boezen HM. Genome-wide association studies: what do they teach us about asthma and chronic obstructive pulmonary disease? Proc Am Thorac Soc 2009;6:701–3. [DOI] [PubMed] [Google Scholar]
- 43. Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J.. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med 2009;179:572–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bind MA, Lepeule J, Zanobetti A, Gasparrini A, Baccarelli A, Coull BA, Tarantini L, Vokonas PS, Koutrakis P, Schwartz J.. Air pollution and gene-specific methylation in the Normative Aging Study: association, effect modification, and mediation analysis. Epigenetics 2014;9:448–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. De Prins S, Koppen G, Jacobs G, Dons E, Van de Mieroop E, Nelen V, Fierens F, Int Panis L, De Boever P, Cox B, et al. Influence of ambient air pollution on global DNA methylation in healthy adults: a seasonal follow-up. Environ Int 2013;59:418–24. [DOI] [PubMed] [Google Scholar]
- 46. Hogan BL, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CC, Niklason L, Calle E, Le A, Randell SH, et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 2014;15:123–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ji H, Biagini Myers JM, Brandt EB, Brokamp C, Ryan PH, Khurana Hershey GK.. Air pollution, epigenetics, and asthma. Allergy Asthma Clin Immunol 2016;12:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baccarelli A, Bollati V.. Epigenetics and environmental chemicals. Curr Opin Pediatr 2009;21:243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Olden K, Lin Y-S, Gruber D, Sonawane B.. Epigenome: biosensor of cumulative exposure to chemical and nonchemical stressors related to environmental justice. Am J Public Health 2014;104:1816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen J, Bardes EE, Aronow BJ, Jegga AG.. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 2009;37:W305–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Grondin CJ, Davis AP, Wiegers TC, Wiegers JA, Mattingly CJ.. Accessing an expanded exposure science module at the comparative toxicogenomics database. Environ Health Perspect 2018;126:014501–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bind M-A, Baccarelli A, Zanobetti A, Tarantini L, Suh H, Vokonas P, Schwartz J.. Air pollution and markers of coagulation, inflammation and endothelial function: associations and epigene-environment interactions in an elderly cohort. Epidemiology 2012;23:332–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Duncan EJ, Gluckman PD, Dearden PK.. Epigenetics, plasticity, and evolution: how do we link epigenetic change to phenotype? J Exp Zool B Mol Dev Evol 2014;322:208–20. [DOI] [PubMed] [Google Scholar]
- 54. Lev Maor G, Yearim A, Ast G.. The alternative role of DNA methylation in splicing regulation. Trends Genet 2015;31:274–80. [DOI] [PubMed] [Google Scholar]
- 55. Oberdoerffer S. A conserved role for intragenic DNA methylation in alternative pre-mRNA splicing. Transcription 2012;3:106–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shenker N, Flanagan JM.. Intragenic DNA methylation: implications of this epigenetic mechanism for cancer research. Br J Cancer 2012;106:248–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vercelli D. Does epigenetics play a role in human asthma? Allergol Int 2016;65:123–6. [DOI] [PubMed] [Google Scholar]
- 58. Schübeler D. Function and information content of DNA methylation. Nature 2015;517:321–6. [DOI] [PubMed] [Google Scholar]
- 59. Farhi A, Boyko V, Almagor J, Benenson I, Segre E, Rudich Y, Stern E, Lerner-Geva L.. The possible association between exposure to air pollution and the risk for congenital malformations. Environ Res 2014;135:173–80. [DOI] [PubMed] [Google Scholar]
- 60. Alexis NE, Carlsten C.. Interplay of air pollution and asthma immunopathogenesis: a focused review of diesel exhaust and ozone. Int Immunopharmacol 2014;23:347–55. [DOI] [PubMed] [Google Scholar]
- 61. Delfino RJ, Staimer N, Tjoa T, Gillen DL, Schauer JJ, Shafer MM.. Airway inflammation and oxidative potential of air pollutant particles in a pediatric asthma panel. J Expo Sci Environ Epidemiol 2013;23:466–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang JJ, McCreanor JE, Cullinan P, Chung KF, Ohman-Strickland P, Han IK, Jarup L, Nieuwenhuijsen MJ.. Health effects of real-world exposure to diesel exhaust in persons with asthma. Res Rep Health Eff Inst 2009;5–109. discussion 111-23. [PubMed] [Google Scholar]
- 63. Antus B, Kardos Z.. Oxidative stress in COPD: molecular background and clinical monitoring. Curr Med Chem 2015;22:627–50. [DOI] [PubMed] [Google Scholar]
- 64. Ji X, Han M, Yun Y, Li G, Sang N.. Acute nitrogen dioxide (NO2) exposure enhances airway inflammation via modulating Th1/Th2 differentiation and activating JAK-STAT pathway. Chemosphere 2015;120:722–8. [DOI] [PubMed] [Google Scholar]
- 65. Ko FWS, Hui DSC.. Air pollution and chronic obstructive pulmonary disease. Respirology 2012;17:395–401. [DOI] [PubMed] [Google Scholar]
- 66. Patel MM, Chillrud SN, Deepti KC, Ross JM, Kinney PL.. Traffic-related air pollutants and exhaled markers of airway inflammation and oxidative stress in New York City adolescents. Environ Res 2013;121:71–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Adamkiewicz G, Ebelt S, Syring M, Slater J, Speizer F, Schwartz J, Suh H, Gold D.. Association between air pollution exposure and exhaled nitric oxide in an elderly population. Thorax 2004;59:204–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Barraza-Villarreal A, Sunyer J, Hernandez-Cadena L, Escamilla-Nuñez MC, Sienra-Monge JJ, Ramírez-Aguilar M, Cortez-Lugo M, Holguin F, Diaz-Sánchez D, Olin AC, Romieu I.. Air pollution, airway inflammation, and lung function in a cohort study of Mexico City schoolchildren. Environ Health Perspect 2008;116:832–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Delfino RJ, Staimer N, Tjoa T, Arhami M, Polidori A, Gillen DL, George SC, Shafer MM, Schauer JJ, Sioutas C.. Associations of primary and secondary organic aerosols with airway and systemic inflammation in an elderly panel cohort. Epidemiology 2010;21:892–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Koenig JQ, Jansen K, Mar TF, Lumley T, Kaufman J, Trenga CA, Sullivan J, Liu LS, Shapiro G, Larson TV.. Measurement of offline exhaled nitric oxide in a study of community exposure to air pollution. Environ Health Perspect 2003;111:1625–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu L, Poon R, Chen L, Frescura A-M, Montuschi P, Ciabattoni G, Wheeler A, Dales R.. Acute effects of air pollution on pulmonary function, airway inflammation, and oxidative stress in asthmatic children. Environ Health Perspect 2009;117:668–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. McCreanor J, Cullinan P, Nieuwenhuijsen MJ, Stewart-Evans J, Malliarou E, Jarup L, Harrington R, Svartengren M, Han I-K, Ohman-Strickland P, et al. Respiratory effects of exposure to diesel traffic in persons with asthma. N Engl J Med 2007;357:2348–58. [DOI] [PubMed] [Google Scholar]
- 73. Romieu I, Barraza-Villarreal A, Escamilla-Nuñez C, Almstrand A-C, Diaz-Sanchez D, Sly PD, Olin A-C.. Exhaled breath malondialdehyde as a marker of effect of exposure to air pollution in children with asthma. J Allergy Clin Immunol 2008;121:903–9. [DOI] [PubMed] [Google Scholar]
- 74. Gruzieva O, Xu C-J, Breton CV, Annesi-Maesano I, Antó JM, Auffray C, Ballereau S, Bellander T, Bousquet J, Bustamante M, et al. Epigenome-wide meta-analysis of methylation in children related to prenatal NO(2) air pollution exposure. Environ Health Perspect 2017;125:104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Enomoto T, Ohashi K, Shibata R, Higuchi A, Maruyama S, Izumiya Y, Walsh K, Murohara T, Ouchi N.. Adipolin/C1qdc2/CTRP12 protein functions as an adipokine that improves glucose metabolism. J Biol Chem 2011;286:34552–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Esposito S, Tenconi R, Lelii M, Preti V, Nazzari E, Consolo S, Patria MF.. Possible molecular mechanisms linking air pollution and asthma in children. BMC Pulmonary Med 2014;14:31–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Plusquin M, Guida F, Polidoro S, Vermeulen R, Raaschou-Nielsen O, Campanella G, Hoek G, Kyrtopoulos SA, Georgiadis P, Naccarati A, et al. DNA methylation and exposure to ambient air pollution in two prospective cohorts. Environ Int 2017;108:127–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Levy I, Mihele C, Lu G, Narayan J, Brook JR.. Evaluating multipollutant exposure and urban air quality: pollutant interrelationships, neighborhood variability, and nitrogen dioxide as a proxy pollutant. Environ Health Perspect 2014;122:65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Himes BE, Jiang X, Wagner P, Hu R, Wang Q, Klanderman B, Whitaker RM, Duan Q, Lasky-Su J, Nikolos C, et al. RNA-Seq transcriptome profiling identifies CRISPLD2 as a glucocorticoid responsive gene that modulates cytokine function in airway smooth muscle cells. PLoS One 2014;9:e99625–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ornitz DM, Yin Y.. Signaling networks regulating development of the lower respiratory tract. Cold Spring Harb Perspect Biol 2012;4:a008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bruse S, Petersen H, Weissfeld J, Picchi M, Willink R, Do K, Siegfried J, Belinsky SA, Tesfaigzi Y.. Increased methylation of lung cancer-associated genes in sputum DNA of former smokers with chronic mucous hypersecretion. Respir Res 2014;15:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Peng C, den Dekker M, Cardenas A, Rifas-Shiman SL, Gibson H, Agha G, Harris MH, Coull BA, Schwartz J, Litonjua AA, et al. Residential proximity to major roadways at birth, DNA methylation at birth and midchildhood, and childhood cognitive test scores: project viva (Massachusetts, USA). Environ Health Perspect 2018;126:097006–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. de Fc Lichtenfels AJ, van der Plaat DA, de Jong K, van Diemen CC, Postma DS, Nedeljkovic I, van Duijn CM, Amin N, la Bastide-van Gemert S, de Vries M, et al. Long-term air pollution exposure, genome-wide DNA methylation and lung function in the LifeLines Cohort Study. Environ Health Perspect 2018;126:027004-1 to 027004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pope CA III, Dockery DW.. Health effects of fine particulate air pollution: lines that connect. J Air Waste Manage Assoc 2006;56:709–42. [DOI] [PubMed] [Google Scholar]
- 85. Breton CV, Marsit CJ, Faustman E, Nadeau K, Goodrich JM, Dolinoy DC, Herbstman J, Holland N, LaSalle JM, Schmidt R, et al. Small-magnitude effect sizes in epigenetic end points are important in children's environmental health studies: the children's environmental health and disease prevention research center's epigenetics working group. Environ Health Perspect 2017;125:511–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Houseman EA, Kim S, Kelsey KT, Wiencke JK.. DNA methylation in whole blood: uses and challenges. Curr Envir Health Rep 2015;2:145–54. [DOI] [PubMed] [Google Scholar]
- 87. Dedeurwaerder S, Defrance M, Calonne E, Denis H, Sotiriou C, Fuks F.. Evaluation of the infinium methylation 450K technology. Epigenomics 2011;3:771–84. [DOI] [PubMed] [Google Scholar]
- 88. Roessler J, Ammerpohl O, Gutwein J, Hasemeier B, Anwar SL, Kreipe H, Lehmann U.. Quantitative cross-validation and content analysis of the 450k DNA methylation array from Illumina, Inc. BMC Res Notes 2012;5:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yang Y, Sebra R, Pullman BS, Qiao W, Peter I, Desnick RJ, Geyer CR, DeCoteau JF, Scott SA.. Quantitative and multiplexed DNA methylation analysis using long-read single-molecule real-time bisulfite sequencing (SMRT-BS). BMC Genomics 2015;16:350.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kurdyukov S, Bullock M.. DNA methylation analysis: choosing the right method. Biology (Basel) 2016;5:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Li P, Demirci F, Mahalingam G, Demirci C, Nakano M, Meyers BC.. An integrated workflow for DNA methylation analysis. J Genet Genomics 2013;40:249–60. [DOI] [PubMed] [Google Scholar]
- 92. Arshad SH, Holloway JW, Karmaus W, Zhang H, Ewart S, Mansfield L, Matthews S, Hodgekiss C, Roberts G, Kurukulaaratchy R.. Cohort profile: the Isle of Wight Whole Population Birth Cohort (IOWBC). Int J Epidemiol 2018;47:1043–1044i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kurukulaaratchy RJ, Fenn MH, Waterhouse LM, Matthews SM, Holgate ST, Arshad SH.. Characterization of wheezing phenotypes in the first 10 years of life. Clin Exp Allergy 2003;33:573–8. [DOI] [PubMed] [Google Scholar]
- 94. Mukherjee N, Lockett GA, Merid SK, Melén E, Pershagen G, Holloway JW, Arshad SH, Ewart S, Zhang H, Karmaus W.. DNA methylation and genetic polymorphisms of the Leptin gene interact to influence lung function outcomes and asthma at 18 years of age. Int J Mol Epidemiol Genet 2016;7:1–17. [PMC free article] [PubMed] [Google Scholar]
- 95. Bibikova M, J-B Fan. GoldenGate® assay for DNA methylation profiling In: Tost J. (ed.), DNA Methylation: Methods and Protocols. Totowa, NJ: Humana Press, 2009, 149–63. [DOI] [PubMed] [Google Scholar]
- 96. Soto-Ramírez N, Arshad SH, Holloway JW, Zhang H, Schauberger E, Ewart S, Patil V, Karmaus W.. The interaction of genetic variants and DNA methylation of the interleukin-4 receptor gene increase the risk of asthma at age 18 years. Clin Epigenetics 2013;5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kuan PF, Wang S, Zhou X, Chu H.. A statistical framework for Illumina DNA methylation arrays. Bioinformatics 2010;26:2849–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ecker S, Chen L, Pancaldi V, Bagger FO, Fernandez JM, Carrillo de Santa Pau E, Juan D, Mann AL, Watt S, Casale FP, et al. Genome-wide analysis of differential transcriptional and epigenetic variability across human immune cell types. Genome Biol 2017;18:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Du P, Zhang X, Huang C-C, Jafari N, Kibbe WA, Hou L, Lin SM.. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 2010;11:587.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT.. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 2012;13:86–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlen SE, Greco D, Soderhall C, Scheynius A, Kere J.. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One 2012;7:e41361.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lindgren A, Stroh E, Montnemery P, Nihlen U, Jakobsson K, Axmon A.. Traffic-related air pollution associated with prevalence of asthma and COPD/chronic bronchitis. A cross-sectional study in Southern Sweden. Int J Health Geogr 2009;8:2–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Modig L, Forsberg B.. Perceived annoyance and asthmatic symptoms in relation to vehicle exhaust levels outside home: a cross-sectional study. Environ Health 2007;6:29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kim HH, Lee CS, Yu SD, Lee JS, Chang JY, Jeon JM, Son HR, Park CJ, Shin DC, Lim YW.. Near-road exposure and impact of air pollution on allergic diseases in elementary school children: a cross-sectional study. Yonsei Med J 2016;57:698–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Liu W, Cai J, Huang C, Hu Y, Fu Q, Zou Z, Sun C, Shen L, Wang X, Pan J, et al. Associations of gestational and early life exposures to ambient air pollution with childhood atopic eczema in Shanghai, China. Sci Total Environ 2016;572:34–42. [DOI] [PubMed] [Google Scholar]
- 106. Schnass W, Huls A, Vierkotter A, Kramer U, Krutmann J, Schikowski T.. Traffic-related air pollution and eczema in the elderly: findings from the SALIA cohort. Int J Hyg Environ Health 2018;221:861–7. [DOI] [PubMed] [Google Scholar]
- 107. Tong X, Ray MA, Lockett GA, Zhang H, Karmaus WJ.. An efficient approach to screening epigenome-wide data. Biomed Res Int 2016;2016:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Leek JT. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 2007;3:1724–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ray MA, Tong X, Lockett GA, Zhang H, Karmaus WJ.. An efficient approach to screening epigenome-wide data. Biomed Res Int 2016;2016:2615348. 2615348 (1-16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. R Core Team. R (version 3.4.2): A Language and Environment for Statistical Computing https://www.r-project.org/ (3 March 2018, date last accessed), 2016.
- 111. Ladd-Acosta C. Epigenetic signatures as biomarkers of exposure. Curr Environ Health Rep 2015;2:117–25. [DOI] [PubMed] [Google Scholar]
- 112. Philibert R, Hollenbeck N, Andersen E, Osborn T, Gerrard M, Gibbons FX, Wang K.. A quantitative epigenetic approach for the assessment of cigarette consumption. Front Psychol 2015;6:656.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Andersen AM, Philibert RA, Gibbons FX, Simons RL, Long J.. Accuracy and utility of an epigenetic biomarker for smoking in populations with varying rates of false self-report. Am J Med Genet B Neuropsychiatr Genet 2017;174:641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Reynolds LM, Magid HS, Chi GC, Lohman K, Barr RG, Kaufman JD, Hoeschele I, Blaha MJ, Navas-Acien A, Liu Y.. Secondhand tobacco smoke exposure associations with DNA methylation of the aryl hydrocarbon receptor repressor. Nicotine Tob Res 2017;19:442–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Greenland S. Modeling and variable selection in epidemiologic analysis. Am J Public Health 1989;79:340–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zeilinger S, Kuhnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, Weidinger S, Lattka E, Adamski J, Peters A, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One 2013;8:e63812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. SAS Institute Inc. Usage Note 22912: How Can I Obtain Test for Trends (e.g. Linear, Quadratic, Cubic, etc.) Using PROC GLM?http://support.sas.com/kb/22/912.html (3 March 2018, date last accessed), 2003.
- 118. Bakulski KM, Feinberg JI, Andrews SV, Yang J, Brown S, L. McKenney S, Witter F, Walston J, Feinberg AP, Fallin MD.. DNA methylation of cord blood cell types: applications for mixed cell birth studies. Epigenetics 2016;11:354–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Cardenas A, Allard C, Doyon M, Houseman EA, Bakulski KM, Perron P, Bouchard L, Hivert MF.. Validation of a DNA methylation reference panel for the estimation of nucleated cells types in cord blood. Epigenetics 2016;11:773–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kim S, Shen S, Sioutas C, Zhu Y, Hinds WC.. Size distribution and diurnal and seasonal trends of ultrafine particles in source and receptor sites of the Los Angeles basin. J Air Waste Manage Assoc 2002;52:297–307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.