Abstract
High density lipoprotein (HDL) particles are believed to be protective due to their inverse correlation with the prevalence of cardiovascular diseases. However, recent studies show that in some conditions such as heart disease and diabetes, HDL particles can become dysfunctional. Great attention has been directed toward HDL particle composition because the relative abundances of HDL constituents determine HDL’s functional properties. A key factor to consider when studying the structure and composition of plasma particles is the protein glycosylation. Here, we profile the O- and N-linked glycosylation of HDL associated-proteins including the truncated form of Apo CIII and their glycan heterogeneity in a site-specific manner. Apolipoprotein CIII, fetuin A and alpha 1 antitrypsin are glycoproteins associated with lipoproteins and are implicated in many cardiovascular and other disease conditions. A targeted method (UHPLC-QQQ) was used to measure the glycoprotein concentrations and site-specific glycovariations of the proteins in human plasma and compared with HDL particles isolated from the same plasma samples. The proteins found in the plasma are differentially glycosylated compared to those isolated in HDL. The results of this study suggest that glycosylation may play a role in protein partitioning in the blood, with possible functional implications.
Keywords: HDL, Glycosylation, High density lipoprotein, Apo CIII, Fetuin A, Glycopeptides, UPLC, MRM, Tandem mass spectrometry
INTRODUCTION
High density lipoproteins (HDLs) are circulating blood nanoparticles known primarily for their ability to efflux cholesterol from cells, and deliver excess cholesterol to the liver for excretion through bile, a process known as reverse cholesterol transport. In addition to this important function, HDL also plays significant role in blocking the oxidation of low density lipoprotein (LDL), and has additional anti-atherogenic functions including anti-inflammatory and immunomodulatory capacity.1 Interestingly, depending on its composition, HDL may become pro-atherogenic2 by promoting inflammation and enhancing LDL oxidation. For instance, HDL has been found to lose its anti-inflammatory functional characteristics during acute influenza A infection,3 and during chronic inflammatory response.4 Altered HDL composition associated with loss of anti-atherogenic functions has been documented in a variety of disease conditions including diabetes mellitus, oxidative stress, infection, auto-immune diseases, and cardiovascular diseases among others.2
HDLs are composed of subclasses that have unique compositions that enable them to perform specific functions. HDL subclasses in turn possess different kinds of biological molecules, including lipids,5 proteins,6, 7 and enzymes suited for their functions.8 For example, changes in proteins involved in HDL metabolism promote atherosclerosis9 even in elevated plasma HDL.10 Detailed analysis of molecular compositions may unravel the deficiencies associated with altered HDL particles and provide indicators for diseases associated with this loss of functionality.
Glycoproteins are important functional components of HDL.11 Altered HDL glycosylation differentiates between coronary artery disease (CAD) patients with and without atherosclerosis,12 as well as individuals with metabolic syndrome and healthy controls.13 Alterations in glycoprotein composition and site-specific glycosylation differentiate between disease groups and healthy individuals, and they further differentiate between pro- and anti-inflammatory HDL particles.14
Several of the key proteins of HDL are glycosylated and include protease inhibitor alpha 1 antitrypsin (A1AT), human α2-Heremans–Schmid glycoprotein (A2HSG), and apolipoprotein CIII (Apo CIII). A2HSG contains both O- and N- linked glycosylation, Apo CIII contains only O-glycans and A1AT contains only N-glycans. Apo CIII glycosylation has been analyzed by isoelectric focusing (IEF) to diagnose genetic defects in O-glycan biosynthesis in congenital disorders of glycosylation.15 However, the IEF method depends on the availability of sialic acid in the glycan molecule(s) and the technique is not able to distinguish between mono and disialylated or other glycoforms that do not contain sialic acid. Recently, mass spectrometry methods have been used for the analysis of Apo CIII and its isoforms.15–18 The quantitation of intact Apo CIII in the plasma has been carried out in earlier studies using HPLC and a high resolution time of flight instrument where the three most abundant Apo CIII isoforms were analyzed.19
A2HSG, commonly known as fetuin A, is a plasma glycoprotein secreted by the liver and produced in high amounts at early stages of human life.20 Fetuin A in neonates with intrauterine growth restriction (IUGR) lack O-linked sialic acid residues that are usually present in Fetuin A.21 Loss of sialic acid in circulatory glycoproteins affects their half-life, generally leading to faster degradation by liver cells as they bind to asialoglycoprotein receptors (ASGPRs).22 A key function of fetuin A is to protect against vascular calcification by solubilizing calcium and phosphorus in the plasma. The concentration of fetuin A is closely associated with increased HDL cholesterol (HDL-C), triglyceride (TG), and LDL cholesterol (LDL-C) in non-diabetic CAD patients without chronic kidney disease (CKD). Fetuin glycopeptides have been analyzed using mass spectrometry,23–25 and recently, a targeted method for measuring fetuin N-linked glycopeptides in the serum was reported.26 The targeted method we have developed measures both the O- and N-linked fetuin glycopeptides simultaneously. Alpha-1-antitrypsin (A1AT) is one of the serine protease inhibitors (Serpins) mainly produced by hepatocytes and is mainly found in the plasma. As an acute phase protein, A1AT levels increase significantly upon the onset of inflammation or post infection.27
We have developed structural profiling methods for mapping both O- and N-linked glycosylation in a site-specific manner and then measuring their levels in both bulk plasma and isolated HDL particles. The method described here is the first to measure simultaneously the levels of O- and N-linked glycosylation at the site-specific level for HDL-associated glycoproteins. We find that O- and N-linked glycopeptides undergo fragmentation that produces unique oxonium ion responses.28 Multiple reaction monitoring (MRM) is then used to quantitate protein and glycan expression at the site-specific level. The described methods provide a targeted glycan analysis of three key HDL-associated proteins and indicate that the glycosylation of A1AT, A2HSG, and Apo CIII differ between plasma and HDL particles. The variation in glycosylation suggests partitioning toward the particles may be glycan-mediated, and glycoproteins associated with HDL particles represent a distinct subset of the plasma proteome.
EXPERIMENTAL PROCEDURE
Glyco-proteomics analysis
Protein standards for Apo CIII, A1AT, and A2HSG from human plasma were purchased from Sigma-Aldrich (St. Lous, MO). Sequencing grade modified trypsin and dithiothreitol (DTT) were purchased from Promega (Madison, WI). Iodoacetamide (IAA) for capping the reduced disulfide bonds was purchased from Sigma-Aldrich (St. Louis, MO). The 50 mM ammonium bicarbonate (NH4HCO3) buffer for all the trypsin digestion were freshly made just before starting the digestion process. The nanopure water was dissolved in 50 ug of Apo CIII in the container it was purchased in to make 2mg/mL stock solution. Both A1AT and A2HSG solid material were measured using XP26 microbalance (Mettler Toledo, Columbus, OH) and then dissolved in nanopure water to make 5mg/mL stock solution. For the peptide and glycopeptide determination, 20ug of each protein standard was pulled from the stock solution and 50 mM ammonium bicarbonate was added to commence the digestion process.
Each standard solution was reduced using 2 uL of 550 mM DTT at 65oC for 50 min then alkylated using 4 uL of 450 mM IAA in the dark at room temperature for 30 min. The samples were then digested at 37oC overnight using 1ug of trypsin and then the reactions were stopped by placing the samples in −20oC for 1hr. The resulting tryptic digests were injected into the LC QE -Orbitrap instrument for LC-MS/MS data acquisition.
LC−MS/MS Analysis
The digest obtained in the procedure above was subjected to tandem mass spectrometry (MS/MS) analysis using Q-Exactive Orbitrap MS (Thermo Scientific, Waltham, MA). The LC column that was used for the analysis is the Proxeon Easy-nLC II HPLC (Thermo Scientific, Waltham, MA). The peptides were separated using a 75 micron × 150 mm Magic C18 200Å 3U reverse phase column using a 60-min gradient with a flow rate of 3uL/min. The ions were isolated (1.6 m/z window) and fragmented using high energy collision dissociation (HCD) with M/Z scan range of 350–1600. The normalized collision E of 27% was used for the fragmentation.
The tandem MS data was searched and interpreted using Byonic software (Protein Metrics, Inc.).29 For the software analysis, the digestion enzyme was specified as trypsin with full specificity. The precursor and the fragment mass tolerance was set at 10 ppm. Because the alkylation agent IAA was used, the carbamidomethyl modification was fixed. Other modifications like oxidation of methionine and tryptophan, deamination of asparagine and glutamine were set as common modifications. Gln->pyro-Glu at N- terminal of glutamine and glutamic acid were set as rare. The N-glycopeptides were searched against the library of non-sodiated 309 mammalian N-glycans.
The glycopeptides were identified based on the respective spectra quality: the delta mode greater than 10, mass error less than 10 ppm, and log prob of more than 2. The glycopeptides that did not meet these conditions were ignored. Identified glycopeptides were further inspected manually for identity confirmation. Further confirmation was performed by inspecting the presence of unique oxonium fragments that are usually present in the glycopeptides using tandem mass spectrometry. These fragment ions masses include M/Z 204.08 (HexNAc), 366.14 (Hex1HexNAc1), 292.09 (Neu5Ac) and 274.09 (Neu5Ac - H2O).
Biological sample collection
Healthy men and women between 18–65 years of age, with a BMI between 18.5–35 kg/m² were recruited through word-of-mouth, flyers, newspaper advertisements, and postings on university websites to participate in a pilot dietary intervention study. The study was approved by the Institutional Review Board of the University of California at Davis. Subjects who qualified for the study (not smokers, no history of anemia, currently consuming fast food no more than 5 times per week, no documented chronic diseases including diabetes, thyroid disease, metabolic syndrome, cancer (active), or previous cardiovascular events, not currently consuming more than 1 alcoholic drink per day, not participating in any weight loss programs, not following any extreme or unusual exercise or diet patterns, no recent weight fluctuations, not taking prescription lipid medications, and no presence of hyperglycemia) were enrolled in the study and provided written informed consent. The study followed all approved guidelines for the protection of subjects and is registered with ClinicalTrials.gov with NCT # 03205254. In this report, samples from the first 5 subjects recruited to this study, who were following a simplified protocol that was an earlier version of the final approved study described on the ClinicalTrials.gov site, were analyzed and reported.
Following consent, subjects were scheduled for a baseline visit to the Ragle Human Nutrition Research Center located on the UC Davis campus. At the baseline visit, a fasting blood draw and anthropometric measurements were taken. Each participant also completed a health history questionnaire, a physical activity questionnaire, and a 3-day diet record to record their habitual diet. Following the baseline visit, subjects consumed a “healthy” diet which excluded fast food for 7 days, and completed a 3-day diet record during this period. Subjects were instructed to emphasize fruits and vegetables and avoid ‘fast food”. They were given food lists with specific examples of what constituted healthy food choices (e.g. salads, fruits, nuts), and specific examples of foods to avoid (e.g. burgers and fries from a fast food restaurant). At the end of the no fast food diet phase, subjects reported to the Ragle Human Nutrition Research Center again for their first follow-up study visit, which included a fasting blood draw, anthropometric measurements, and the same questionnaires as the baseline study visit.
Following the first follow-up study visit, subjects consumed a fast food diet for 5 days. During this time, subjects were instructed to eat all their meals (i.e. breakfast, lunch, dinner, and snacks) selecting from the sample foods listed on the food list provided, which emphasized convenience foods like burgers and fries from fast food restaurants and avoided “healthy” food items such as salads, whole grain cereal, nuts, and fruits. Subjects ate as much or as little as they wanted throughout the day and at each meal. They were instructed to maintain their normal activity level during the study period and to collect a diet record for each day in the study. Subjects were contacted twice during the dietary period by e-mail or phone and were asked about their general wellbeing. At the end of the 5-day period subjects returned for the second follow-up study visit which also included a fasting blood draw, anthropometric measurements, and the same questionnaires as the baseline study visit.
HDL isolation by ultracentrifugation
HDL was prepared using a modified procedure from a previously report.11 Briefly, freshly prepared potassium bromide (KBr) with densities, 1.063, 1.210 and 1.340 g/mL was used. The densities of KBr were verified using the Densito30PX portable densitometer (Mettler Toledo, Columbus, OH, USA). Briefly, 1.9 mL of plasma was adjusted to d = 1.063 by adding a concentrated KBr solution (d = 1.340). The adjusted plasma was underlain to a KBr density solution of d = 1.063 g/mL for a final volume of 4.7mL in each tube (OptiSeal, Beckman Coulter). Ultracentrifugation was performed for 3 h 10 min at 657,000 x g and 15°C, following which 1.9 mL of the LDL-containing supernatant fraction was collected. The infranate containing the HDL fraction was adjusted to d = 1.210 g/mL. The adjusted low layer fraction was underlain to a KBr density solution of d = 1.210 and then ultracentrifugation was performed for 3 h 20 min at 657,000 x g and 15°C. The top 1.0 mL of solution, which was the HDL fraction at a density range of 1.21 – 1.063 g/mL, was collected and subjected to diafiltration using Amicon ultra-3K centrifugal filter devices. The HDL fractions were desalted with water (Optima LC/MS) by two consecutive steps of centrifugation for 25 min at 4⁰C and 14,000 x g.
Quantification of protein and glycopeptide
Our lab has utilized MRM on the QQQ instrument for effective and efficient serum N-glycopeptide quantitation.30 This method has been utilized to quantify glycoproteins and their glycosylation in the human serum using only 2uL of serum.31 A similar method was recently developed to measure the levels of milk proteins and glycoproteins.32 Most biological samples contain both O- and N-linked glycans, and methods to quantify these glycoforms on a single experimental run are necessary. The method outlined here quantifies both O- and N-linked glycopeptides in the same experimental run for the first time. The parameters were optimized for that unique purpose. The fragmentation patterns and the ionization efficiencies of the O- and N-glycopeptides differ. The molecular and the product ions masses for the glycopeptides were carefully selected from the tandem mass spectrometry data. The developed triple Q (QQQ) method involves ionizing the analytes in the positive ion mode and detecting them at unit resolution.
To quantify the protein levels, known amounts of protein standards were first measured and pooled together. These standards were digested using trypsin and then serially diluted to 6 or more concentration levels with ratios (eg.1.0 : 2.0 : 2.5 : 2.0 : 5.0 : 2.0) to generate calibration curves. These calibration curves were used to estimate the amount of the proteins in the biological samples. All the standards and the samples were analyzed using an Agilent 1290 infinity LC system coupled to an Agilent 6490 triple quadrupole (QqQ) mass spectrometer (Agilent technologies, Santa Clara, CA). The samples were passed through the Agilent eclipse plus C18 pre-column (RRHD 1.8 μm, 2.1 Χ 5 mm) and then separated on the Agilent eclipse plus C18 (RRHD 1.8 μm, 2.1 Χ 100 mm) UHPLC analytical column. The 2 uL trypsin-digested HDL sample solutions were injected into the UHPLC, and the samples were separated at the flow rate of 0.5mL/min. The 14-minute binary gradient used for separation was set as follows: 2% B from 0.0−0.5 min; 2–15% B from 0.5–5 min; 15–44% B from 5−10 min; 44–100% B from 10–12.1 min, and the flush was set at 100% B for 1.1 min and equilibrated for 0.8 min at 2% B before the next run. Solvent B was 90% acetonitrile, 0.1% formic acid while solvent A was composed of 3% acetonitrile, 0.1% formic acid all in nano pure water (v/v).
Each analyte was monitored as it eluted from the LC-ESI using dynamic multiple reaction monitoring (dMRM) mode. This reduced the number of analytes being monitored at any given time during the LC-QQQ run. The collision energy for each glycopeptide was monitored and optimized to achieve the optimum fragmentation and sensitivity. Each glycopeptide transition was composed of the glycopeptide molecular ion of a certain charge as precursor, and the glycopeptide diagnostic oxonium ions such as m/z 204.08 (HexNAc), 366.14 (Hex1HexNAc1), 292.09 (Neu5Ac) and 274.09 (Neu5Ac –H2O) as product ions.
The MRM data was analyzed using Agilent Mass Hunter quantitative analysis software version B-05–02/build 5.2.365.0. The limit of detection and limit of quantitation were determined from the quantitative software signal to noise (S/N) ≥3 and (S/N) ≥6, respectively. The coefficient of variance (CV) was used to obtain the reproducibility of the measurements (shown in table 1). Only a few (15 out of 48) glycopeptides had a CV greater than 10. Nomenclature for each glycopeptide includes the protein name, followed by the nature of the glycan, whether O or N, the site of the glycan attachment, and the glycan composition. The glycan designation is based on the composition (number of hexose (hex) N-acetylhexosamine (HexNAc), fucose (Fuc), and N-acetylneuraminic acid (NeuAc) residues). For example, A2HSG-N-156–5402 denotes the protein fetuin A, N shows it is an N-glycan, 156 is the site, and 5402 indicate 5 hexoses, 4 HexNAc, 0 fucose and 2 NeuAc.
Table 1.
A list of glycopeptides (protein_Glycan type_site_HexnHexNacnFucnSialicAn), the precursor, product, optimized collision energy, the peptide bearing the glycan, the coefficient of variance from three separate measurements, and the two p-values from the mixed model analysis showing differences by sample type (when red, statistically significant after Bonferroni correction), and by time point.
| Protein (mg/mL) ± stdev | A2HSG | A1AT | Apo CIII |
|---|---|---|---|
| HDL | 0.002 ± 0.0004 | 0.093 ± 0.04 | 0.397 ± 0.19 |
| Plasma | 0.108 ± 009 | 0.995 ± 0.24 | 0.037 ± 0.02 |
| Glycopeptide | Precursor Ion | Product Ion | Ret Time (min) | Collision Energy | Peptide | Coefficient of Variation (%) | Group Prob>F | Time point Prob>F |
|---|---|---|---|---|---|---|---|---|
| A1AT_N_70_5402 | 1078.5 | 366.1 | 10.6 | 17 | QLAHQSN70STNIFFSPVSIATAFAMLSLGTK | 4.29 | <.0001 | 0.6652 |
| A1AT_N_70_5412 | 1107.7 | 366.1 | 10.6 | 18 | 23.18 | N/D | N/D | |
| A1AT_N_271_5402 | 991.2 | 366.1 | 8.4 | 15 | YLGN271ATAIFFLPDEGK | 3.16 | <.0001 | 0.6037 |
| A1AT_N_271_5412 | 1027.7 | 366.1 | 6.7 | 16 | 3.47 | N/D | N/D | |
| A1AT_N_271_MC_5402 | 1149.9 | 366.1 | 9.4 | 18 | YLGN271ATAIFFLPDEGKLQHLENELXHDIITK | 6.08 | 0.0171 | 0.7488 |
| A1AT_N_271_MC_5412 | 1179.1 | 366.1 | 7.4 | 19 | 8.36 | N/D | N/D | |
| A1AT_N_107_6513 | 1341.0 | 366.1 | 9.5 | 22 | ADTHDEILEGLNFN107LTEIPEAQIHEGFQELLR | 19.52 | 0.0097 | 0.2198 |
| A1AT_N_107_5411 | 1151.6 | 366.1 | 9.4 | 18 | 3.51 | <.0001 | 0.8388 | |
| A1AT_N_107_6503 | 1311.8 | 366.1 | 9.5 | 21 | 13.35 | 0.13 | 0.0218 | |
| A1AT_N_107_5402 | 1180.6 | 366.1 | 9.4 | 19 | 2.12 | 0.6865 | 0.1956 | |
| A1AT_N_107_5412 | 1209.8 | 366.1 | 9.4 | 19 | 3.11 | 0.0026 | 0.3934 | |
| H2HSG_O_319_1201 | 735.8 | 274.1 | 2.8 | 22 | H319TFMGW325SLGSPSGEVSHPR | 3.75 | N/D | N/D |
| H2HSG_O_319_1102 | 757.8 | 274.1 | 8 | 22 | 27.35 | N/D | N/D | |
| A2HSG_O_346_1101 | 891.4 | 274.1 | 6.5 | 27 | 346TVVQP351SVGAAAGPWPPCPGR | 4.33 | 0.001 | 0.2343 |
| A2HSG_O_346_1102 | 988.5 | 274.1 | 6.3 | 31 | 0.71 | N/D | N/D | |
| A2HSG_O_346_2200 | 916.1 | 366.1 | 7.4 | 28 | 9.37 | 0.0998 | 0.5397 | |
| A2HSG_N_156_5401 | 1229.2 | 366.1 | 7.1 | 20 | VCQDCPLLAPL156NDTR | 0.97 | 0.0018 | 0.7857 |
| A2HSG_N_156_5402 | 994.9 | 366.1 | 7.1 | 16 | 2.82 | <.0001 | 0.7878 | |
| A2HSG_N_156_5421 | 1326.6 | 366.1 | 7.1 | 21 | 0.89 | <.0001 | 0.585 | |
| A2HSG_N_156_6502 | 1086.2 | 366.1 | 7.1 | 17 | 4.11 | 0.0002 | 0.2551 | |
| A2HSG_N_156_6503 | 1159.0 | 366.1 | 7.1 | 18 | 3.29 | <.0001 | 0.8355 | |
| A2HSG_N_156_6510 | 1234.9 | 366.1 | 7.1 | 20 | 16.99 | 0.0064 | 0.7065 | |
| A2HSG_N_156_5412 | 1374.9 | 366.1 | 7.1 | 22 | 18.04 | N/D | N/D | |
| A2HSG_N_156_6513 | 1195.5 | 366.1 | 7.1 | 19 | 2.23 | N/D | N/D | |
| A2HSG_N_176_5401 | 1070.4 | 366.1 | 7.4 | 17 | AALAAFNAQN176NGSNFQLEEISR | 6.57 | N/D | N/D |
| A2HSG_N_176_5402 | 1143.0 | 366.1 | 7.4 | 18 | 4.36 | 0.1947 | 0.9725 | |
| A2HSG_N_176_5412 | 1179.7 | 366.1 | 7.4 | 19 | 5.13 | 0.1102 | 0.1397 | |
| A2HSG_N_176_5431 | 1180.3 | 366.1 | 7.4 | 19 | 7.63 | 0.0494 | 0.7247 | |
| A2HSG_N_176_6502 | 1234.3 | 366.1 | 7.4 | 20 | 4.59 | N/D | N/D | |
| A2HSG_N_176_6512 | 1271.0 | 366.1 | 7.4 | 20 | 4.07 | N/D | N/D | |
| A2HSG_N_176_7600 | 1180.5 | 366.1 | 7.4 | 19 | 8.51 | 0.0169 | 0.8704 | |
| A2HSG_N_176_6501 | 1161.7 | 366.1 | 7.4 | 19 | 13.87 | N/D | N/D | |
| A2HSG_N_176_6503 | 1307.1 | 366.1 | 7.4 | 21 | 11.32 | N/D | N/D | |
| A2HSG_N_176_6513 | 1343.8 | 366.1 | 7.4 | 22 | 17.59 | N/D | N/D | |
| ApoC3_O_74_0300 | 916.1 | 204.1 | 7.4 | 28 | FSEFWDLDPEVRP74TSAVAA | 12.10 | <.0001 | 0.2095 |
| ApoC3_O_74_0310 | 975.4 | 204.1 | 7.9 | 30 | 7.90 | 0.0001 | 0.5369 | |
| ApoC3_O_74_1101 | 931.8 | 274.1 | 8.4 | 29 | 0.62 | <.0001 | 0.9874 | |
| ApoC3_O_74_1102 | 1028.8 | 274.1 | 8.4 | 32 | 3.51 | <.0001 | 0.7864 | |
| ApoC3_O_74_1210 | 951.1 | 366.1 | 8.4 | 29 | 8.40 | 0.0011 | 0.0456 | |
| ApoC3_O_74_1300 | 970.1 | 366.1 | 8.2 | 30 | 11.40 | <.0001 | 0.5914 | |
| ApoC3_O_74_2110 | 937.4 | 366.1 | 8.4 | 29 | 8.40 | <.0001 | 0.6519 | |
| ApoC3_O_74_2212 | 899.6 | 274.1 | 6.5 | 28 | 5.99 | N/D | N/D | |
| ApoC3_O_74_2230 | 1078.8 | 366.1 | 10.7 | 34 | 7.35 | N/D | N/D | |
| ApoC3_O_74_2220 | 1053.8 | 366.1 | 7.3 | 33 | 30.85 | N/D | N/D | |
| ApoC3_O_74_2301 | 1121.2 | 274.1 | 7.8 | 36 | 1.65 | N/D | N/D | |
| ApoC3_O_74_1202 | 1096.5 | 274.1 | 7.7 | 35 | 3.64 | 0.2449 | 0.0601 | |
| ApoC3_O_74_llll | 980.4 | 274.1 | 8 | 30 | 14.80 | N/D | N/D | |
| ApoC3_O_74_2211 | 1102.2 | 274.1 | 8.4 | 35 | 42.21 | N/D | N/D | |
| ApoC3_O_74_2200 | 956.4 | 366.1 | 8 | 30 | 22.17 | N/D | N/D | |
| ApoC3_O_74_1311 | 837.1 | 274.1 | 8.1 | 25 | 35.85 | 0.0011 | 0.2036 | |
| ApoC3_O_74_MC_1101 | 989.1 | 274.1 | 8.3 | 31 | DKFSEFWDLDPEVRP74TSAVAA | 2.93 | 0.7788 | 0.2633 |
| ApoC3_O_74_MC_1102 | 1109.8 | 274.1 | 8.4 | 35 | 4.24 | <.0001 | 0.0863 | |
| ApoC3_O_74_A off_1101 | 908.1 | 274.1 | 8.5 | 28 | FSEFWDLDPEVRP74TSAVA | 5.12 | <.0001 | 0.5105 |
| ApoC3_O_74_A off_1102 | 1005.1 | 274.1 | 8.4 | 31 | 1.96 | 0.0108 | 0.285 |
Statistical analysis
The statistical analyses were performed using JMP (R) Pro13.0.0 64-bit (SAS institute, Cary NC), and Microsoft Office Excel (Microsoft, Seattle WA). The glycopeptides that were detected in less than 70% of the samples were discarded. All the measurements were transformed using log10 transformation to obtain normally distributed values before performing statistical analyses. We aimed to find out whether there were significant glycoform-level differences between sample types (plasma and HDL) and time points (A, B, and C) from the food consumption study. We performed linear mixed model with sample type and time point as the fixed effects and the subject ID (individual samples) as random effects. The p values obtained are shown in the data table (Table 1).
The Bonferroni corrected p-value for significance was calculated to be 0.002 (0.05/33), thus any p-value < 0.002 was considered statistically significant. Principal component analysis was performed to evaluate whether individual groups would cluster together when all the variables are used. The PCA parameters included linear fittings. Canonical plots were extracted with cluster and the constellation plots obtained from the data.
RESULTS AND DISCUSSIONS
Numerous studies have mapped the proteins associated with HDL and even the sub fractions of HDL using several methods6, 18, 33, 34 including intact apolipoproteins,35 and multiple reaction monitoring (MRM) of protein digest.36 Aside from our recent reports on HDL glycosylation, there were few reports where HDL glycosylation was analyzed.11, 18, 37, 38 Our previous studies showed that HDL particles are highly sialylated.11 The present work involves an in-depth characterization of O- and N-linked HDL glycosylation comparing the glycans in plasma with those bound in HDL particles. It is the first quantitative method where both O-and N-linked glycosylation are analyzed in the same experimental run. An LC-QE-orbitrap was used to acquire MS/MS data for peptide and glycopeptide identification. LC-QQQ was used for targeted measurement of the glycopeptides in biological samples using dynamic MRM mode. There were no statistically significant time point effects observed, indicating that the glycoform levels were not affected by the dietary changes the subjects made during the course of this study.
O-linked glycopeptides from Apo CIII.
Apo CIII is a 8.8-kDa protein found in plasma lipoprotein particles including chylomicrons, very low density lipoprotein (VLDLs), low density lipoprotein (LDL) and HDL. Apo CIII is a lipoprotein lipase inhibitor and is involved in triglyceride-rich lipoprotein (TRL) metabolism. It is also an indicator of risk of coronary heart disease and other cardiovascular events.39, 40
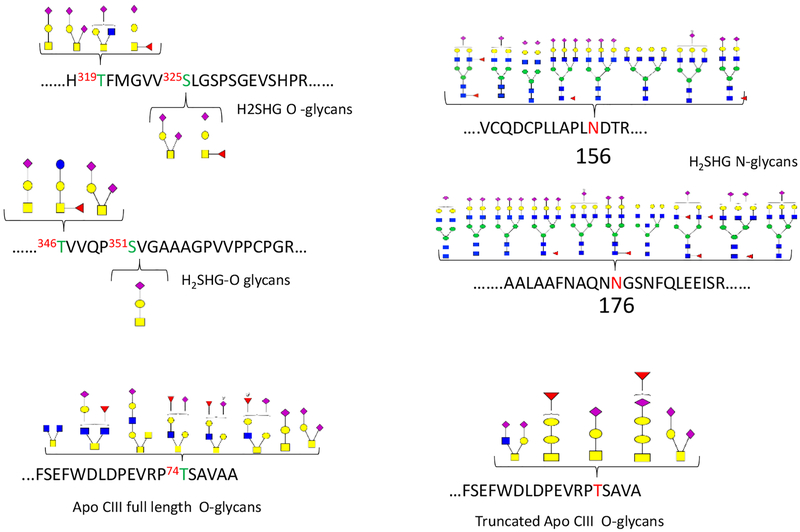
Apo CIII is known to contain only O-linked glycopeptides on a single site.41 The most common Apo CIII glycoforms include asialylated (Apo CIII0), monosialylated (Apo CIII1), and disialiylated (Apo CIII2) forms. We found other less abundant glycoforms from the full-length Apo CIII protein in the tryptic peptide FSEFWDLDPEVRP74TSAVAA near the C-terminus and on the truncated protein (FSEFWDLDPEVRP74TSAVA) where the C terminus alanine residue is removed.
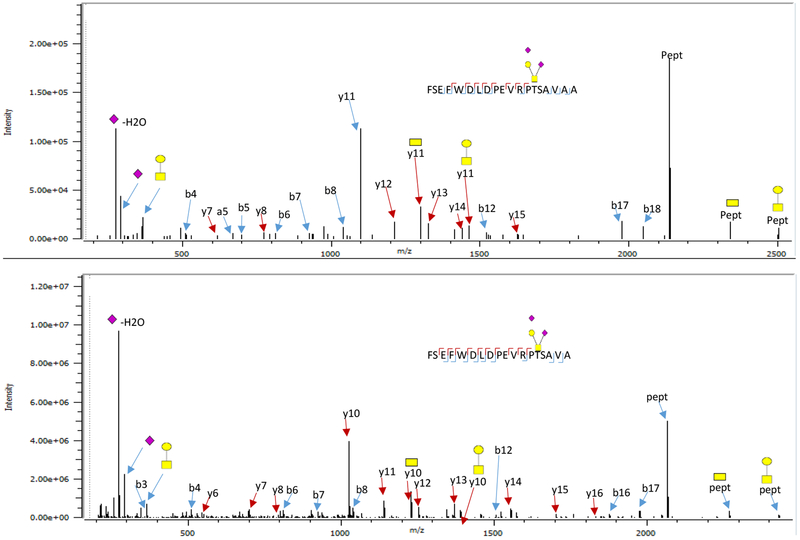
A total of 20 glycoforms were identified in Apo CIII. Figure 1 shows representative tandem mass spectra of Apo CIII O-glycopeptides from a full protein sequence (Figure 1A) and the truncated form (Figure 1B). We obtained extensive structurally informative fragmentation for both the glycan and the peptide that allowed us to unambiguously assign the position of the glycan in the peptide. With the aid of the Byonic software and manual confirmation, we identified the glycopeptide spectra that were observed in the tandem MS data. Apart from the three commonly observed glycoforms, the full sequence Apo CIII contained the following less common glycoforms; 0300, 0310, 1210, 1300, 2110, 2212, 2230, 2220, 2301,1202, 1111, 2200, and 1311 (XXXX represent the number of Hex, HexNac, Fuc, and Sia, respectively). Most of the observed glycoforms were fucosylated and nearly half were sialylated. Other unusual Apo CIII glycoforms have been observed in urine from patients with Schistosoma mansoni, with the observed glycoforms in that study being less sialylated and more fucosylated.17 A number of unusual serum Apo CIII glycoforms have also been observed using top down MALDI –FTICR16 and mass spectrometric immunoassay (MSIA) methods.42
Figure 1.
Tandem mass spectral data of Apo CIII O-glycopeptides obtained from the QE orbitrap: a) Glycan in full sequence protein peptide, and b) the same glycan attached to a truncated (C terminal alanine is removed) form of the protein. Extensive fragmentation able to identify glycan and locate the glycosylation sites is obtained in both glycopeptides
The glycoforms observed in the truncated peptide include 1101, 1102, 1211, 1202, 2110, and 2300. They were lower in abundance and less fucosylated than those observed in the full-length sequence. The non-glycosylated truncated Apo CIII peptide was not detected in our analysis. Truncated Apo CIII has been observed previously and is associated with elevated carboxypeptidase, which hydrolyzes the amino acids at the end of the C-terminus.43 Truncation alters the properties of Apo CIII. Increased carboxypeptidase in the serum has been associated with increased lipase activity in pancreatic cancer patients.43
O- and N-linked glycopeptides for human fetuin A
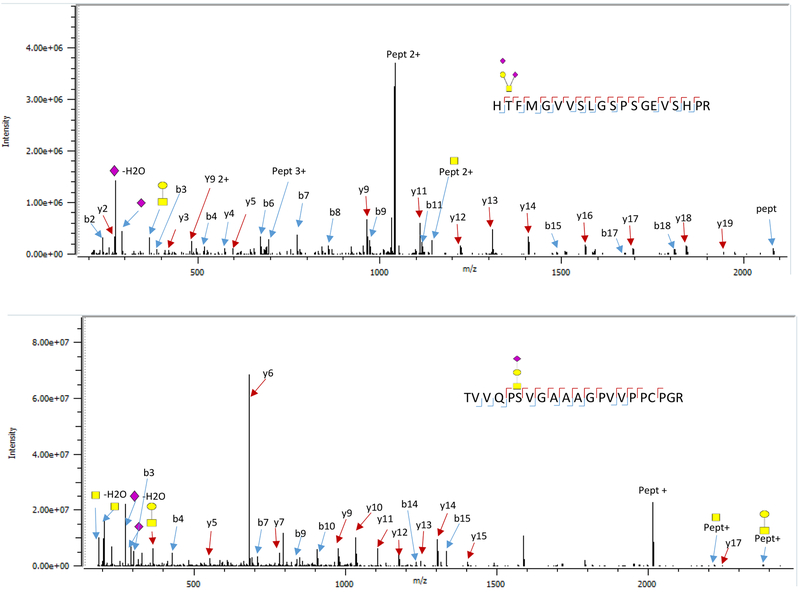
Human fetuin A contains both N- and O-linked glycans. At least 18 N-linked glycoforms (or more correctly glycocompositions) were observed on both sites 156N on peptide, VCQDCPLLAPLNDTR, and 176N the peptide AALAAFNAQNNGSNFQLEEISR. There were 8 glycoforms on site 156 while site 176 was found to contain 10 unique glycoforms (Figure 2). Most of the N-linked glycans in fetuin A are complex bi-antennary and sialylated. Each of these glycopeptide composition is assignable from the peptide and the glycan fragmentation. All the glycopetide spectra contained diagnostic ions (HexHexNac (M/Z 366.14), HexNac (M/Z 204.09) and the sialylated forms had NeuAc-H2O (M/Z 274.09). The diagnostic ions, which are readily available in glycopeptide spectra, are important in the development of a quantitative method. An example of the tandem mass spectrum data for these glycoforms is shown in the supplementary data (Figure S1). As typical of N-linked glycopeptide spectra, the spectrum was dominated with fragments associated with the glycan and a few peptide backbone fragments.
Figure 2.
Fetuin A glycopeptide tandem MS spectra obtained in two sites. a) site 319 and b) site 346. The peptide backbone fragments obtained in both spectra can unambiguously locate the glycan to its site.
O-linked glycans lack a consensus motif like the N-linked glycans, therefore they can be found on any serine (S) and threonine (T) residue. About 12% of human fetuin A amino acids are S and T residues and could potentially be glycosylated. The fetuin O-glycopeptides were observed on four sites (Figure 3). Two glycopeptides contain two sites that are both occupied. The peptide H319TFMGVV325SLGSPSGEVSHPR contains sites 319T and 325S observed with four and two glycans, respectively. The other peptide 346TVVQP351SVGAAAGPVVPPCPGR contains sites 346T and 351S with three glycans and one glycan, respectively. The HCD data provided fragmentation information that was used to unambiguously assign the site of each glycan. Unlike the N-linked glycopeptide spectra, many peptide backbone fragments were observed for the O-glycopeptide spectra. Some peptide ions contained a remnant of the glycan that was attached to the peptide or peptide fragments. For example, the most abundant fragment in Figure 2a was a y11 fragment. This fragment was also found with a GlcNac (Y11+GlcNAc) and with HexHexNac (Y11+HexHexNAc).
Figure 3.
The figure shows the glycomap of both N- and O-linked in Fetuin A and Apo CIII including the truncated form of the Apo CIII protein. The tandem MS data for the glycopeptides resolved the site of the glycans even when the peptide contained two O-linked glycan sites.
Simultaneous quantitation of O- and N-linked glycopeptides in HDL particles
The glycan map of the respective proteins obtained with LC-MS/MS was used to develop a MRM transitions on the triple quadrupole to create a targeted method to quantitate Apo CIII, fetuin A and A1AT and their glycoforms. First, robust peptides for measuring the level of proteins were screened and target peptides were identified. These peptides are AHYDLR, LSITGTYDLK and GWVTDGFSSLK for fetuin A, A1AT and Apo CIII respectively. These tryptic peptides are unique to the respective protein and contain no mis-cleavage sites. The peptide response was tested for repeatability and reproducibility and their linear response was also noted.
The MRM method was first developed and optimized using mixtures of glycoprotein standards. Each transition was optimized individually for the molecular ion mass and the collision energy. The tuning parameters were manually examined to ensure optimal fragmentation and production of the product ion. The final method consisted of a 14-minute separation gradient. Out of the 48 glycopeptide transitions, 21 were O-glycopeptides. The identified O-glycans bound to the peptides were smaller than the N-linked glycopeptides. The O-glycopeptides were fragmented with higher collision energy compared to the N-linked glycopeptides because they appeared more stable than the N-glycans during CID. Equations 1 and 2 show the mathematical expression used to calculate the optimized activation energy for two types of ions. The product ions that produced the most signal in different glycoforms were selected as product ions. The sialylated O-glycoforms produced M/Z 274.09 (NeuAC –H2O) in high signal so this fragment was selected for these types of glycoforms. HexNAc (M/Z 204.09) was the best product ion for other O-linked glycoforms that were not sialylated. The most intense fragment ion for the N-glycopeptides was M/Z 366.14.
N-Linked glycopeptides collision energy equation
| Eq1 |
O-Linked glycopeptides collision energy equation
| Eq 2 |
Glycosylation of protein in plasma versus HDL particles
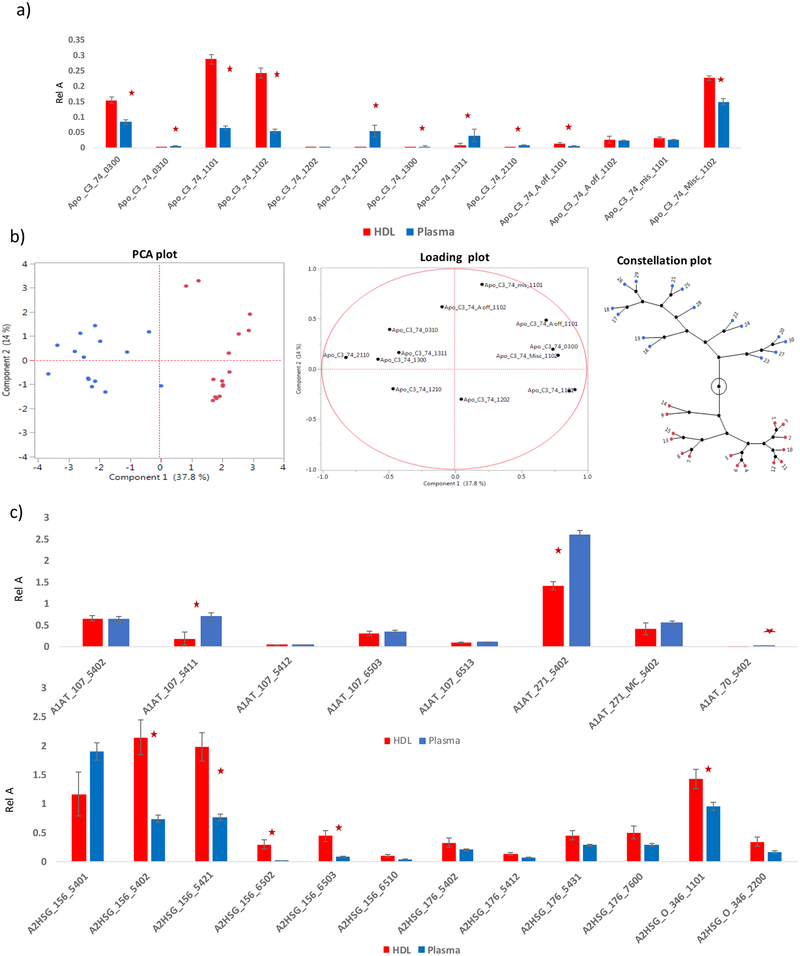
Plasma protein glycosylation has been studied but there has been no reported study comparing HDL glycosylation to the matched plasma.44 Differences in HDL and plasma glycosylation of the same protein would indicate glycan specificity in recruiting proteins into HDL particles. We measured the levels of the glycoproteins and the site-specific glycosylation profiles in plasma and the isolated HDL particles from the five subjects described above. The levels of glycoproteins were estimated using the QQQ response of the peptides from non-glycosylated regions of the protein. The levels for Fetuin A, A1AT and Apo CIII in the HDL were 0.002. 0.093, 0.397 mg/mL, respectively while the plasma levels were 0.108, 0.995, and 0.037 mg/mL, respectively. The level of Apo CIII was higher in HDL compared to the plasma while the levels A1AT and Fetuin A were lower in HDL compared to plasma. This is an indication that Apo CIII protein is enriched in HDL particles (Table 1).
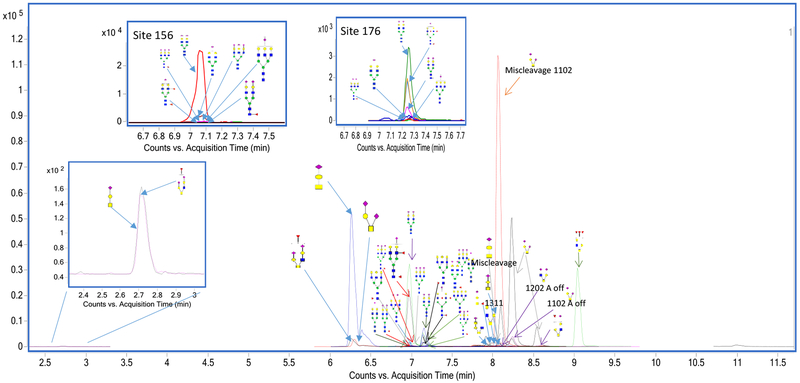
The glycopeptides were quantitated with MRM with the glycopeptide abundance normalized to the protein abundance. Normalizing the glycopeptide abundance decouples it from changes in protein concentration. Shown in Figure 4 is an example of a 14-minute run, with the glycopeptide peaks annotated. Table 1 lists all of the p-values for the mixed model analysis by sample type and time point, indicating that there were no statistically significant differences by time point (no significant effects of diet), but that there were several statistically significant differences by sample type (i.e. plasma vs. HDL) across multiple glycopeptides.
Figure 4.
Extracted ion chromatograms of individual glycoforms as obtained from the UHPLC- QQQ. The glycoforms sharing the same peptide backbone all elute at close retention times because the retention of the glycopeptide in the column is dependent on the peptide backbone properties.
The two most abundant sialylated glycoforms in Apo CIII show unique trends in the full sequence and the truncated forms (Figure 5). Ten out of the 13 Apo CIII glycoforms quantified were found to differ significantly between HDL and plasma (upper panel). The sialylated glycoforms 1101 and 1102 in the full sequence Apo CIII were significantly higher in the isolated HDL particles compared to matched plasma. The fucosylated glycoforms (0310, 1210, 1311, and 2110) were significantly lower in HDL particles compared to plasma. There were no significant differences between plasma and HDL for the disialylated 1102 glycoform in the truncated Apo CIII. The same glycoform shows strong differences in the full Apo CIII protein. The levels of the monosialylated (1101) glycoform in the truncated Apo CIII in plasma and isolated HDL particles were both significant but more pronounced with the full-length protein (Figure 5). Most of the non-sialyated Apo CIII glycoforms were higher in the plasma compared to the levels in HDL particles. HDL extracted from the same individual showed clustering with regard to the glycoform distribution as shown in the constellation map in Figure 5.
Figure 5.
Quantitative data obtained from HDL and matched plasma: a) Apo CIII glycopeptides relative abundances in plasma and HDL. b) Cluster analysis of the Apo CIII data, the PCA analysis shows clear separation between the two groups of samples, the loading plot showing that fucosylated/less sialylated glycans are unique to plasma while sialylated glycoforms are found in higher levels in HDL, c) relative glycopeptide abundances for the A1AT and Fetuin A proteins in plasma and HDL. A * indicate the measurement was significantly different.
Non-glycosylated Apo CIII hinders the breakdown of triglycerides which can lead to hypertriglyceridemia and atherosclerotic diseases.45 Changes in the commonly studied Apo CIII glycoforms have been observed in other diseases like uremia, kidney disease, obesity, and autosomal recessive cutis laxa type 2 patients.13, 46 Sialylation was found to decrease in most of these studies. In type 1 diabetes where Apo CIII promotes Ca2+ dependent B cell death, the levels of sialylated (Apo CIII1 and Apo CIII2) isoforms are significantly elevated while Apo CIII0, which is not glycosylated, is not detectable.47 Apo CIII sialylation is down regulated in many other pathological conditions including cancer.48, 49
Fetuin contains both N- and O- glycosylation. Except the glycoform 6510, all the fetuin glycoforms in in site 156 were significantly elevated in HDL compared to plasma. The N-linked glycoforms on site 176 were not significantly different in HDL compared to plasma. The two O-linked glycoforms on site 346 that were quantified were also found to be higher in HDL compared to plasma. This is an indication that fetuin A that is recruited to HDL has increased glycosylation. Chronic dialysis patients frequently have low A2HSG.50
Fetuin A decreases significantly in response to an infection and/or inflammation and it plays an important role as an anti-inflammatory mediator.51, 52 Glycosylation in fetuin A plays a critical role in which lipids induce insulin resistance. Fetuin A acts as an endogenous ligand of TLR4 to promote lipid-induced insulin resistance.53 More branched and core fucosylated N-linked glycoforms of fetuin A at site 156 have been found to increase significantly in acute pancreatitis and pancreatic cancer patients.54
Eight glycoforms of A1AT were found in both plasma and HDL samples. Most of them had the same levels in both samples. Glycoforms 5411 on site 107 and 5402 on site 271 were higher in HDL compared to plasma samples. Although the levels of these glycoforms do not show strong differences between HDL and plasma, our recent analysis of these glycoforms showed that they are enriched in patients with metabolic syndrome when compared to controls.37 A1AT glycosylation is vital for its secretion by monocytes.55 A1AT glycosylation has been found to differ between serum and hepatocytes,56. Fucosylated biantennary A1AT glycans increase significantly in the serum of hepatocellular carcinoma patients.57 Thus, while glycosylation is important for the secretion of A1AT from cells, it does not appear to be involved in the partitioning of A1AT into HDL. Alternatively, it is possible that most of A1AT that is in plasma is associated with HDL.
CONCLUSION
Site-specific mapping and quantitation add important molecular detail to the analysis of biological samples. This work demonstrates the utility and application of methods for measuring the levels of both N- and O-linked glycosylation in a complex protein mixture in a single experimental run. The additional detail obtained by the site-specificity of the glycosylation pattern of important proteins may provide important information about the functional properties of these proteins, and the HDL particles with which they are associated. This information may be a distinguishing factor of whether the HDL is functional or dysfunctional. In fact, our group recently showed that site-specific differences in the glycosylation of HDL-associated proteins can distinguish between HDL particles that stimulate vs. inhibit cytokine production in stimulated monocytes.14
This pilot study demonstrates that the glycosylation patterns of proteins that associate with HDL particles are distinct from the glycosylation patterns of these same proteins in matched plasma. The results suggest that specific glycoforms are recruited into HDL. Although all glycoforms that are found in HDL are also found in plasma but in different proportions, the data suggest that there are some that are most favored for co-localization with HDL. These results should be confirmed in a larger sample set to demonstrate the generalizability of our observations. Our results demonstrate that a method for the comprehensive analysis of site-specific glycosylation in isolated HDL particles may be useful for the development of lipoprotein-based diagnostics that could improve the diagnosis and management of disease.
Supplementary Material
ACKNOWLEGMENTS
Funding was provided by the National Institutes of Health. Research reported in this publication was supported in part by National Institute of General Medical Sciences under the award number R01GM049077.
BIBLIOGRAPHY
- 1.Eren E; Yilmaz N; Aydin O, High density lipoprotein and it’s dysfunction. The open biochemistry journal. 2012, 6, (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu D; Ji L; Zhang D; Tong X; Pan B; Liu P; Zhang Y; Huang Y; Su J; Willard B; Zheng L, Nonenzymatic glycation of high-density lipoprotein impairs its anti-inflammatory effects in innate immunity. Diabetes Metab Res Rev. 2012, 28, (2), 186–195. [DOI] [PubMed] [Google Scholar]
- 3.Van Lenten BJ; Wagner AC; Nayak DP; Hama S; Navab M; Fogelman AM, High-density lipoprotein loses its anti-inflammatory properties during acute influenza a infection. Circulation. 2001, 103, (18), 2283–2288. [DOI] [PubMed] [Google Scholar]
- 4.Tölle M; Huang T; Schuchardt M; Jankowski V; Prüfer N; Jankowski J; Tietge UJ; Zidek W; van der Giet M, High-density lipoprotein loses its anti-inflammatory capacity by accumulation of pro-inflammatory-serum amyloid A. Cardiovasc. Res 2012, 94, (1), 154–162. [DOI] [PubMed] [Google Scholar]
- 5.Yetukuri L; Söderlund S; Koivuniemi A; Seppänen-Laakso T; Niemelä PS; Hyvönen M; Taskinen M-R; Vattulainen I; Jauhiainen M; Orešič M, Composition and lipid spatial distribution of HDL particles in subjects with low and high HDL-cholesterol. J. Lipid Res. 2010, 51, (8), 2341–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karlsson H; Leanderson P; Tagesson C; Lindahl M, Lipoproteomics II: Mapping of proteins in high-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2005, 5, (5), 1431–1445. [DOI] [PubMed] [Google Scholar]
- 7.Mazur MT; Cardasis HL; Spellman DS; Liaw A; Yates NA; Hendrickson RC, Quantitative analysis of intact apolipoproteins in human HDL by top-down differential mass spectrometry. Proc. Natl. Acad. Sci. USA 2010, 107, (17), 7728–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah AS; Tan L; Lu Long J; Davidson WS, The proteomic diversity of high density lipoproteins: Our emerging understanding of its importance in lipid transport and beyond. J. Lipid Res. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultz JR; Verstuyft JG; Gong EL; Nichols AV; Rubin EM, Protein composition determines the anti-atherogenic properties of HDL in transgenic mice. Nature. 1993, 365, (6448), 762–764. [DOI] [PubMed] [Google Scholar]
- 10.Heinecke JW, The HDL proteome: a marker–and perhaps mediator–of coronary artery disease. J. Lipid Res. 2009, 50, (Supplement), S167–S171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J; Lee H; Zivkovic AM; Smilowitz JT; Rivera N; German JB; Lebrilla CB, Glycomic analysis of high density lipoprotein shows a highly sialylated particle. J. Proteome Res. 2014, 13, (2), 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krishnan S; Huang J; Lee H; Guerrero A; Berglund L; Anuurad E; Lebrilla CB; Zivkovic AM, Combined High-Density Lipoprotein Proteomic and Glycomic Profiles in Patients at Risk for Coronary Artery Disease. J Proteome Res. 2015, 14, (12), 5109–18. [DOI] [PubMed] [Google Scholar]
- 13.Savinova OV; Fillaus K; Jing L; Harris WS; Shearer GC, Reduced Apolipoprotein Glycosylation in Patients with the Metabolic Syndrome. PLoS One. 2014, 9, (8), e104833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krishnan S; Shimoda M; Sacchi R; Kailemia MJ; Luxardi G; Kaysen GA; Parikh AN; Ngassam VN; Johansen K; Chertow GM; Grimes B; Smilowitz JT; Maverakis E; Lebrilla CB; Zivkovic AM, HDL Glycoprotein Composition and Site-Specific Glycosylation Differentiates Between Clinical Groups and Affects IL-6 Secretion in Lipopolysaccharide-Stimulated Monocytes. Sci Rep. 2017, 7, 43728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wada Y; Kadoya M; Okamoto N, Mass spectrometry of apolipoprotein C-III, a simple analytical method for mucin-type O-glycosylation and its application to an autosomal recessive cutis laxa type-2 (ARCL2) patient. Glycobiology. 2012, 22, (8), 1140–1144. [DOI] [PubMed] [Google Scholar]
- 16.Nicolardi S; van der Burgt YEM; Dragan I; Hensbergen PJ; Deelder AM, Identification of New Apolipoprotein-CIII Glycoforms with Ultrahigh Resolution MALDI-FTICR Mass Spectrometry of Human Sera. J. Proteome Res. 2013, 12, (5), 2260–2268. [DOI] [PubMed] [Google Scholar]
- 17.Balog CIA; Mayboroda OA; Wuhrer M; Hokke CH; Deelder AM; Hensbergen PJ, Mass Spectrometric Identification of Aberrantly Glycosylated Human Apolipoprotein C-III Peptides in Urine from Schistosoma mansoni-infected Individuals. Mol. Cell. Proteomics 2010, 9, (4), 667–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savinova OV; Fillaus K; Jing L; Harris WS; Shearer GC, Reduced Apolipoprotein Glycosylation in Patients with the Metabolic Syndrome. PLoS One. 2014, 9, (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jian W; Edom RW; Wang D; Weng N; Zhang S, Relative Quantitation of Glycoisoforms of Intact Apolipoprotein C3 in Human Plasma by Liquid Chromatography–High-Resolution Mass Spectrometry. Anal. Chem 2013, 85, (5), 2867–2874. [DOI] [PubMed] [Google Scholar]
- 20.Reinehr T; Roth CL, Fetuin-A and Its Relation to Metabolic Syndrome and Fatty Liver Disease in Obese Children Before and After Weight Loss. J. Clin. Endocrinol. Metab 2008, 93, (11), 4479–4485. [DOI] [PubMed] [Google Scholar]
- 21.Karamessinis PM; Malamitsi-Puchner A; Boutsikou T; Makridakis M; Vougas K; Fountoulakis M; Vlahou A; Chrousos G, Marked Defects in the Expression and Glycosylation of α2-HS Glycoprotein/Fetuin-A in Plasma from Neonates with Intrauterine Growth Restriction : Proteomics Screening and Potential Clinical Implications. Mol. Cell. Proteomics 2008, 7, (3), 591–599. [DOI] [PubMed] [Google Scholar]
- 22.Hoffmeister KM, The role of lectins and glycans in platelet clearance. Journal of thrombosis and haemostasis : JTH. 2011, 9, (0 1), 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W; Liu H; Li Z, Tandem mass spectrometric characterization of fetuin sialylated glycopeptides enriched by TiO2 microcolumn. Chin. J. Chem . 2011, 29, (11), 2229–2235. [Google Scholar]
- 24.Mayampurath A; Song E; Mathur A; Yu C.-y.; Hammoud Z; Mechref Y; Tang H, Label-free glycopeptide quantification for biomarker discovery in human sera. J. Proteome Res. 2014, 13, (11), 4821–4832. [DOI] [PubMed] [Google Scholar]
- 25.Zauner G; Koeleman CA; Deelder AM; Wuhrer M, Protein glycosylation analysis by HILIC‐LC‐MS of Proteinase K‐generated N‐and O‐glycopeptides. J. Sep. Sci 2010, 33, (6‐7), 903–910. [DOI] [PubMed] [Google Scholar]
- 26.Song E; Pyreddy S; Mechref Y, Quantification of glycopeptides by multiple reaction monitoring liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2012, 26, (17), 1941–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarthy C; Saldova R; Wormald MR; Rudd PM; McElvaney NG; Reeves EP, The Role and Importance of Glycosylation of Acute Phase Proteins with Focus on Alpha-1 Antitrypsin in Acute and Chronic Inflammatory Conditions. J. Proteome Res. 2014, 13, (7), 3131–3143. [DOI] [PubMed] [Google Scholar]
- 28.Eshghi ST; Yang W; Hu Y; Shah P; Sun S; Li X; Zhang H, Classification of Tandem Mass Spectra for Identification of N-and O-linked Glycopeptides. Sci. Rep 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bern M; Kil YJ; Becker C, Byonic: advanced peptide and protein identification software. Curr Protoc Bioinformatics. 2012, 13.20.1–13.20.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong Q; Lebrilla CB; Miyamoto S; Ruhaak LR, Absolute Quantitation of Immunoglobulin G and Its Glycoforms Using Multiple Reaction Monitoring. Anal. Chem 2013, 85, (18), 8585–8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong Q; Ruhaak LR; Stroble C; Parker E; Huang J; Maverakis E; Lebrilla CB, A Method for Comprehensive Glycosite-Mapping and Direct Quantitation of Serum Glycoproteins. J. Proteome Res. 2015, 14, (12), 5179–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang J; Kailemia MJ; Goonatilleke E; Parker EA; Hong Q; Sabia R; Smilowitz JT; German JB; Lebrilla CB, Quantitation of human milk proteins and their glycoforms using multiple reaction monitoring (MRM). Anal. Bioanal. Chem 2017, 409, (2), 589–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reilly MP; Tall AR, HDL proteomics: pot of gold or Pandora’s box? The Journal of Clinical Investigation. 2007, 117, (3), 595–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ronsein GE; Pamir N; von Haller PD; Kim DS; Oda MN; Jarvik GP; Vaisar T; Heinecke JW, Parallel reaction monitoring (PRM) and selected reaction monitoring (SRM) exhibit comparable linearity, dynamic range and precision for targeted quantitative HDL proteomics. J. Proteomics 2015, 113, 388–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazur MT; Cardasis HL; Spellman DS; Liaw A; Yates NA; Hendrickson RC, Quantitative analysis of intact apolipoproteins in human HDL by top-down differential mass spectrometry. Proc. Natl. Acad. Sci. U. S. A 2010, 107, (17), 7728–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoofnagle AN; Becker JO; Oda MN; Cavigiolio G; Mayer P; Vaisar T, Multiple-Reaction Monitoring-Mass Spectrometric Assays Can Accurately Measure the Relative Protein Abundance in Complex Mixtures. Clin. Chem 2012, 58, (4), 777–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krishnan S; Shimoda M; Sacchi R; Kailemia MJ; Luxardi G; Kaysen GA; Parikh AN; Ngassam VN; Johansen K; Chertow GM, HDL Glycoprotein Composition and Site-Specific Glycosylation Differentiates Between Clinical Groups and Affects IL-6 Secretion in Lipopolysaccharide-Stimulated Monocytes. Sci. Rep 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krishnan S; Huang J; Lee H; Guerrero A; Berglund L; Anuurad E; Lebrilla CB; Zivkovic AM, Combined High-Density Lipoprotein Proteomic and Glycomic Profiles in Patients at Risk for Coronary Artery Disease. J. Proteome Res. 2015, 14, (12), 5109–5118. [DOI] [PubMed] [Google Scholar]
- 39.Sacks FM; Furtado JD; Rimm E; Jensen M, Association of levels of HDL-cholesterol apolipoprotein CIII with the risk of coronary heart disease and cardiovascular events. In Google Patents: 2014. [Google Scholar]
- 40.Jensen MK; Rimm EB; Furtado JD; Sacks FM, Apolipoprotein C-III as a Potential Modulator of the Association Between HDL-Cholesterol and Incident Coronary Heart Disease. J Am Heart Assoc. 2012, 1, (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roghani A; Zannis V, Mutagenesis of the glycosylation site of human ApoCIII. O-linked glycosylation is not required for ApoCIII secretion and lipid binding. J. Biol. Chem 1988, 263, (34), 17925–17932. [PubMed] [Google Scholar]
- 42.Trenchevska O; Schaab MR; Nelson RW; Nedelkov D, Development of multiplex mass spectrometric immunoassay for detection and quantification of apolipoproteins CI, C-II, C-III and their proteoforms. Methods. 2015, 81, 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haase R; Menke‐Möllers I; Oette K, Analysis of human apolipoproteins C by isoelectric focusing in immobilized pH gradients. Electrophoresis. 1988, 9, (9), 569–575. [DOI] [PubMed] [Google Scholar]
- 44.Clerc F; Reiding KR; Jansen BC; Kammeijer GS; Bondt A; Wuhrer M, Human plasma protein N. Glycoconjugate J. 2016, 33, (3), 309–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woelter M; Roewer C; Koy C; Reimer T; Rath W; Pecks U; Glocker MO, A proteome signature for intrauterine growth restriction derived from multifactorial analysis of mass spectrometry-based cord blood serum profiling. Electrophoresis. 2012, 33, (12), 1981–1993. [DOI] [PubMed] [Google Scholar]
- 46.Kornak U; Reynders E; Dimopoulou A; van Reeuwijk J; Fischer B; Rajab A; Budde B; Nurnberg P; Foulquier F; Lefeber D; Urban Z; Gruenewald S; Annaert W; Brunner HG; van Bokhoven H; Wevers R; Morava E; Matthijs G; Van Maldergem L; Mundlos S, Impaired glycosylation and cutis laxa caused by mutations in the vesicular H+-ATPase subunit ATP6V0A2. Nat. Genet 2008, 40, (1), 32–34. [DOI] [PubMed] [Google Scholar]
- 47.Juntti-Berggren L; Refai E; Appelskog I; Andersson M; Imreh G; Dekki N; Uhles S; Yu L; Griffiths WJ; Zaitsev S; Leibiger I; Yang S-N; Olivecrona G; Jörnvall H; Berggren P-O, Apolipoprotein CIII promotes Ca2+-dependent β cell death in type 1 diabetes. Proc. Natl. Acad. Sci. U. S. A 2004, 101, (27), 10090–10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harvey SB; Zhang Y; Wilson-Grady J; Monkkonen T; Nelsestuen GL; Kasthuri RS; Verneris MR; Lund TC; Ely EW; Bernard GR; Zeisler H; Homoncik M; Jilma B; Swan T; Kellogg TA, O-Glycoside Biomarker of Apolipoprotein C3: Responsiveness to Obesity, Bariatric Surgery, and Therapy with Metformin, to Chronic or Severe Liver Disease and to Mortality in Severe Sepsis and Graft vs Host Disease. J. Proteome Res. 2009, 8, (2), 603–612. [DOI] [PubMed] [Google Scholar]
- 49.Ueda K; Fukase Y; Katagiri T; Ishikawa N; Irie S; Sato TA; Ito H; Nakayama H; Miyagi Y; Tsuchiya E, Targeted serum glycoproteomics for the discovery of lung cancer‐associated glycosylation disorders using lectin‐coupled ProteinChip arrays. Proteomics. 2009, 9, (8), 2182–2192. [DOI] [PubMed] [Google Scholar]
- 50.Chen H-Y; Chiu Y-L; Hsu S-P; Pai M-F; Lai C-F; Peng Y-S; Kao T-W; Hung K-Y; Tsai T-J; Wu K-D, Association of serum fetuin A with truncal obesity and dyslipidemia in non-diabetic hemodialysis patients. Eur. J. Endocrinol 2009, 160, (5), 777–783. [DOI] [PubMed] [Google Scholar]
- 51.Lebreton J; Joisel F; Raoult J; Lannuzel B; Rogez J; Humbert G, Serum concentration of human alpha 2 HS glycoprotein during the inflammatory process: evidence that alpha 2 HS glycoprotein is a negative acute-phase reactant. J. Clin. Invest 1979, 64, (4), 1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang H; Zhang M; Bianchi M; Sherry B; Sama A; Tracey KJ, Fetuin (α2-HS-glycoprotein) opsonizes cationic macrophagedeactivating molecules. Proc. Natl. Acad. Sci. USA 1998, 95, (24), 14429–14434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pal D; Dasgupta S; Kundu R; Maitra S; Das G; Mukhopadhyay S; Ray S; Majumdar SS; Bhattacharya S, Fetuin-A acts as an endogenous ligand of TLR4 to promote lipid-induced insulin resistance. Nat. Med 2012, 18, (8), 1279–1285. [DOI] [PubMed] [Google Scholar]
- 54.Kontro H; Joenväärä S; Haglund C; Renkonen R, Comparison of sialylated N‐glycopeptide levels in serum of pancreatic cancer patients, acute pancreatitis patients, and healthy controls. Proteomics. 2014, 14, (15), 1713–1723. [DOI] [PubMed] [Google Scholar]
- 55.GROSS V; VOM BERG D; KREUZKAMP J; GANTER U; BAUER J; WÜRTEMBERGER G; SCHULZ-HUOTARI C; BEESER H; GEROK W, Biosynthesis and Secretion of M-and Z-Type α 1-Proteinase Inhibitor by Human Monocytes. Effect of Inhibitors of Glycosylation and of Oligosaccharide Processing on Secretion and Function. Biol. Chem. Hoppe-Seyler. 1990, 371, (1), 231–238. [DOI] [PubMed] [Google Scholar]
- 56.Jeppsson J-O; Larsson C; Eriksson S, Characterization of α1-antitrypsin in the inclusion bodies from the liver in α1-antitrypsin deficiency. New Engl. J. Med 1975, 293, (12), 576–579. [DOI] [PubMed] [Google Scholar]
- 57.Saitoh A; Aoyagi Y; Asakura H, Structural analysis on the sugar chains of human α1-antitrypsin: presence of fucosylated biantennary glycan in hepatocellular carcinoma. Arch. Biochem. Biophys 1993, 303, (2), 281–287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.