Summary
Background
Asthma is a common chronic respiratory disease in children and adults. An important genetic component to asthma susceptibility has long been recognized, most recently through the identification of several genes (e.g., ORMDL3, PDE4D, HLA-DQ, and TLE4) via genome-wide association studies.
Objective
To identify genetic variants associated with asthma affection status using genome-wide association data.
Methods
We describe results from a genome-wide association study on asthma performed in 3855 subjects using a panel of 455 089 single nucleotide polymorphisms (SNPs).
Result
The genome-wide association study resulted in the prioritization of 33 variants for immediate follow-up in a multi-staged replication effort. Of these, a common polymorphism (rs9272346) localizing to within 1 Kb of HLA-DQA1 (chromosome 6p21.3) was associated with asthma in adults (P-value = 2.2E-08) with consistent evidence in the more heterogeneous group of adults and children (P-value = 1.0E-04). Moreover, some genes identified in prior asthma GWAS were nominally associated with asthma in our populations.
Conclusion
Overall, our findings further replicate the HLA-DQ region in the pathogenesis of asthma. HLA-DQA1 is the fourth member of the HLA family found to be associated with asthma, in addition to the previously identified HLA-DRA, HLA-DQB1 and HLA-DQA2.
Introduction
Asthma (MIM 600807) is a syndrome characterized by chronic airway inflammation, airway hyperresponsiveness and intermittent airway obstruction that result in episodic breathlessness, wheeze and cough. Asthma is emblematic of a truly complex disease that develops through the interaction of multiple genetic and environmental factors. The advent of technical and statistical methods for comprehensive genome-wide association studies (GWAS) has helped to identify new loci associated with asthma. The first such study for asthma identified common regulatory variants at and near the ORMDL3/GSDML loci on chromosome 17q21 that were associated with asthma in three populations of European ancestry [1]. These associations have been confirmed in many follow-up replications studies, with the ORMDL3/GSDML representing one of the most consistent asthma associations reported to date [2–4]. A second asthma GWAS identified common intronic variants in phosphodiesterase 4D (PDE4D) to be strongly associated with asthma in Caucasian children and in four of five additional cohorts of diverse ethnicity [5]. A common variant, rs2378383, on chromosome 9q21.31, near transducin-like enhancer of split 4 (TLE4), was associated with childhood asthma in two Mexican cohorts, but not in Caucasians [6]. A GWAS study performed in two independent populations of African ancestry identified three variants associated with asthma in both populations: rs10515807, mapping to alpha-1B-adrenergic receptor (ADRA1B), rs6052761 near prion-related protein (PRNP) and rs1435879 near dipeptidyl peptidase 10 (DPP10), although replication for these associations was not observed in either four additional populations of African descent or three populations of European descent [7]. Other recent asthma GWAS identified other potential asthma variants, RAD50-IL13 [8] and DENND1B, on chromosome 1q31.3 [9]. The GABRIEL project, the largest asthma GWAS to date, consisting of 10 365 asthma cases and 16 110 control individuals, identified genome-wide significant associations between asthma and SNPs in or near IL1RL1/IL18R1, IL33, SMAD3 and IL2RB [10]. The strongest finding was at rs9273349 in the HLADQ region of chromosome 6p21.3 [10]. Prior to this, the HLA region had been identified in several genetic studies as a region associated with asthma and related phenotypes [11–14]. Furthermore, the EVE consortium, a US consortium of similar size to GABRIEL, validated several previously associated regions including loci on 17q21 and genetic variants within the HLA-DQ region. In addition, this study identified and replicated several ethnic-specific associations, including a novel association with asthma in the PYHIN1 gene among subjects of African descent [15].
Although some genetic associations are starting to show consistent, replicable genetic associations, particularly with the larger genetic consortiums, many of the observed genetic associations do not consistently replicate across studies [5, 16–19]. This can be attributed to the heterogeneity of populations studied and differences in statistical power among the studies.
To date, the findings from GWAS have only explained a small percentage of the overall genetic variation for most complex diseases including asthma [20], suggesting that more genetic studies and alternative approaches to gene identification are necessary to localize asthma variants. In addition, the clinical characteristics of asthmatic children and adults are also different; with age, asthmatic symptoms often subside and the nature of the inflammatory response also changes with age. Therefore, the genetic association that is evaluated in children can be described as a causal association with asthma diagnosis, whereas what is evaluated in adults might be described as asthma persistence. It is likely that the genes causing childhood asthma are different from the genes that cause asthma persistence. Additional lines of evidence suggest that the age at onset of asthma may be influenced by genetics, with earlier onset being more likely to implicate a genetic cause for the disease; further suggesting that childhood and adult asthma have important genetic differences [21].
Taken together, it is likely that many additional asthma-susceptibility variants are yet to be discovered. Herein, we describe the results of a GWAS with 3855 subjects, including 1238 asthmatics to further identify and validate genetic variants for asthma.
Methods
Populations
SHARP GWAS cohorts.
The SNP Health Association Resource (SHARe) Asthma Resource Project (SHARP) conducted genome-wide genotyping in adults and children who have participated in the National Heart, Lung, and Blood Institute’s (NHLBI) clinical research trials on asthma. SHARP includes children with asthma who participated in the Childhood Asthma Management Program (CAMP), children who participated in one or more of five clinical trials conducted by the Childhood Asthma Research and Education (CARE) network and adults who participated in one or more of six clinical trials conducted by the Asthma Clinical Research Network (ACRN). For the purposes of these studies, childhood asthma was defined as meeting the diagnostic criteria for a specific study before the age of 18, whereas adult asthma was defined as meeting the diagnostic criteria for a specific study after the age of 18. The ascertainment criteria and study designs for each of the studies conducted by these networks were comparable, although not identical. Because both ACRN and CARE are made up of several substudies, we provide the asthma diagnosis criteria for one representative trial from each study. We also provide the diagnosis criteria for CAMP. Additional explicit details of all studies included in SHARP can be found on the dbGaP website (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000166.v2.p1&phv=71260&phd=1429&pha=&pht=700&phvf=&phdf=&phaf=&phtf=&dssp=1&consent=&temp=1). The primary analysis included here was restricted to Caucasian individuals within SHARP.
CAMP.
Childhood Asthma Management Program was a multi-centre longitudinal clinical trial that followed 1041 asthmatic children for approximately 4 years with inclusion/exclusion criteria that were developed to select individuals with mild to moderate asthma [22]. CAMP was designed to evaluate whether continuous, long-term treatment (over a period of 4–6 years) with either an inhaled corticosteroid (budesonide) or an inhaled non-corticosteroid drug (nedocromil) safely produces an improvement in lung growth as compared with treatment for symptoms only (with albuterol and, if necessary, prednisone, administered as needed). The primary outcome in the study was lung growth, as assessed by the change in forced expiratory volume in 1 s (FEV1, expressed as a percentage of the predicted value) after the administration of a bronchodilator. Secondary outcomes included the degree of airway responsiveness, morbidity, physical growth and psychological development. All recruited children had asthma as defined by having two or more symptoms per week, using an inhaled bronchodilator at least twice weekly or asthma medication daily and airway responsiveness to methacholine ≤ 12.5 mg/mL [23]. Children with severe asthma or other clinically significant conditions were excluded. DNA samples from 2241 probands and their parents underwent genome-wide genotyping as described below.
CARE.
The Asthma Research and Education Network was established in 1999 by the NHLBI. Five clinical centres and a data co-ordinating centre were selected to participate in this research network. As asthma is the most common chronic childhood disease in the United States, the CARE Network was established to evaluate treatments for children with asthma, conducting studies for children with asthma and sharing their findings with the health care community (http://www.asthma-carenet.org/index.html) [24–26]. Asthma diagnosis was similar in all CARE trials. The Pediatric Asthma Controller Trial (PACT) was one of the CARE clinical trials. Asthma diagnosis in PACT was based on the following. First, children had airway responsiveness to methacholine ≤ 12.5 mg/mL. Second, individuals were required to have mild-moderate persistent asthma as defined by either (1) the presence of self-reported symptoms or inhaled bronchodilator (not including pre-exercise) use an average of at least four times per week during the 4 weeks preceding the trial or (2) the presence of diaryreported symptoms or inhaled bronchodilator (not including pre-exercise) use or peak flows in the yellow zone an average of at least four times per week during the 2-week run-in to the clinical trial. DNA samples from 1604 probands and their parents underwent genome-wide genotyping.
ACRN.
Asthma Clinical Research Network was established in 1993 by the NHLBI. The objectives of this multicentre program are to conduct multiple well-designed clinical trials for rapid evaluation of new and existing therapeutic approaches to asthma and to disseminate laboratory and clinical findings to the health care community. From 1993 to 2003, ACRN consisted of six clinical centres and one data co-ordinating centre [26, 27]. Asthma diagnosis was similar in all ACRN trials. In general, the diagnostic criteria for asthma included history of physician diagnosis of asthma and tests for reversible airflow obstruction and/or bronchial hyperresponsiveness. Here, we describe the diagnosis criteria for one representative study, IMProving Asthma Control Trial (IMPACT). Asthma diagnosis was defined in IMPACT by 1) a history of asthma and 2) heightened airway reactivity, shown by reversible airflow obstruction ≥ 12% and ≥ 200 mL improvement in FEV1 following two to four inhalations of albuterol MDI or by methacholine PC20 ≤ 8 mg/mL. DNA samples from 1059 individuals underwent genome-wide genotyping as described below.
Population controls.
We obtained data corresponding to 1432 control individuals from a GWAS of schizophrenia and bipolar disorder [28] that was submitted to dbGaP [http://www.ncbi.nlm.nih.gov/gap]. These individuals did not have a diagnosis of schizophrenia or bipolar disorder and they were not ascertained for the presence or absence of asthma. Therefore, these individuals are considered ‘population controls’ and can be reasonably assumed to have asthma at the rate that is prevalent in the general population.
Genotyping and quality control measures
Affymetrix, Inc. (Santa Clara, CA, USA) performed the genotyping according to manufacturer’s protocol using the Affymetrix Genome-Wide Human SNP Array 6.0. Marker quality control (QC) was performed on all autosomal markers, which were extracted from dbGAP for each of the four cohorts. SNPs were excluded for the following reasons: (1) probe sequence did not map uniquely to the hg18 genome build; (2) genotyping completion rate < 95%; (3) P-value for Hardy-Weinberg equilibrium (HWE) ≤ 1E-06; and (4) > 12 discordances observed among sample replicate pairs. We excluded all SNPs with Mendelian error counts ≥ 4 in either CARE or CAMP trios. Genotypic gender was verified in PLINK [29]. Subject relatedness was verified using rgGRR [30], and by confirming very low rates of parent-child genotype inconsistencies (average within-family rate of 0.14%). Specific information on the QC procedures for individual datasets is available in the Supplemental Data. The data were adjusted for population stratification using Eigenstrat [31] (see Appendix S1).
Statistical analysis
Three primary GWAS analyses were performed: (1) an analysis of all adult asthmatics; (2) an analysis of all childhood asthmatics; and (3) an analysis of the childhood and adult asthmatics combined. We maximized the use of the data by employing both regression-based techniques for case-control data and family-based association tests for family data. Details of the analysis can be found in the Appendix S1.
SNPs were selected for replication using the following approach: First, we verified that the direction of the observed effect was consistent across our primary analyses in children (i.e. CAMP, CARE and Genetic Association Information Network, GAIN), adults (i.e. ACRN and GAIN) and all subjects (i.e. CAMP, CARE, ACRN and GAIN). Second, we identified the SNPs with P-values < 1E-04 from the combined analysis of all subjects. When replication SNPs were not available in replication populations due to differences in genotyping platforms, we identified and evaluated the associations at SNPs that were either in strong linkage disequilibrium with the identified SNP or analysed the imputed SNP data. Joint evidence for association across populations was measured by combining P-values using the sample sizeweighted Liptak method [32]. In combining P-values, hypothesis tests in replication populations had one-sided alternatives (based on the direction of association in the primary SHARP analysis) so that SNPs with association tests in opposite directions would not produce inappropriately small P-values. Overall statistical significance was determined after adjusting for the total number of statistical tests performed based on the total number of genotyped SNPs.
Replication populations
Replication analyses were performed in five cohorts: (1) Children’s Hospital of Philadelphia (CHOP); (2) Children’s Health Study (CHS); (3) Costa Rica; (4) Sepracor/LOCCS/LODO/Illumina; and (5) i2b2 Crimson Asthma Project (iCAP). Details about these cohorts are included in Appendix S1. Each replication study was approved by the Institutional Review Board of the corresponding institution and informed consent was obtained for all study participants.
Results
We performed a GWAS of asthma, combining the three well-characterized asthma cohorts from SHARP and dbGaP population controls genotyped using the Affymetrix 6.0 SNP array. Following exclusion of individuals and markers that did not meet stringent QC criteria, the final dataset consisted of 3855 subjects and 455 089 markers (Tables 1 and 2). In addition to analysing all asthma subjects, we performed separate analyses of childhood and adult asthma. The sample size of the childhood analysis is nearly double that of the adult analysis, both with respect to number of probands/cases and number of total subjects, leading to greater power (~22% more) to detect genetic association in the childhood asthma analysis than in the analysis of adult cohorts (Table 2).
Table 1.
Primary population Characteristics
| CAMP | CARE | ACRN | GAIN | |
|---|---|---|---|---|
| Ascertainment method |
Clinical trail |
Clinical trials |
Clinical trials | GWAS control cohort |
| Number of total individuals |
1280 | 756 | 471 | 1348 |
| Number of probands/cases |
556 | 211 | 471 | 0 |
| Number of parents/ controls |
724 | 545 | 0 | 1348 |
| Age (SD) [min, max] |
8.8 (2.14) [5.2,13.2] |
10.6 (3.45) [2.2,17.8] |
32.5 (10.8) [12.4,65.2] |
50.9 (17.0) [18,90] |
ACRN, Asthma Clinical Research Network; CAMP, Childhood Asthma Management Program; CARE, Childhood Asthma Research and Education.
Table 2.
Summary of the three primary GWA designs
| Childhood asthmatics |
Adult asthmatics |
Combined analysis |
|
|---|---|---|---|
| Total sample size | 3442 | 1819 | 3855 |
| Number of cases/probands |
767 | 471 | 1238 |
| Number of controls |
1348 | 1348 | 1348 |
| Number of parents |
1269 | 0 | 1269 |
| Analysis method |
New screening method |
Logistic regression |
New screening method |
| Power estimate* |
0.86 | 0.64 | 0.98 |
This estimate corresponds to the power to detect that a SNP will be among the top 40 SNPs of the GWAS, assuming an OR = 1.5, a MAF = 0.2, and a disease prevalence of 0.10. The adult analysis had the lowest statistical power. The childhood and combined analyses had adequate statistical power.
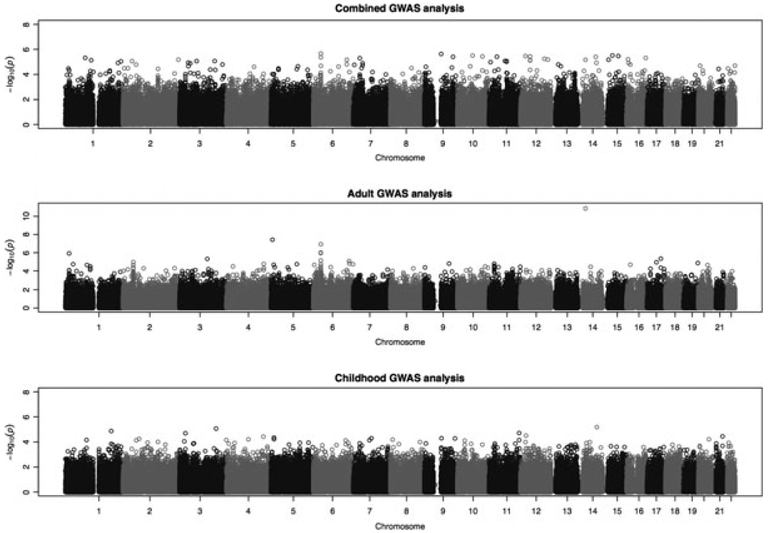
Quantile-quantile plots for all three GWA analyses indicate that the top association P-values are slightly lower than what is expected by chance for the children’s and combined GWAS analysis, in keeping with the conservative nature of the test statistics employed (Figure S1). Despite the conservative nature of the results, the top association P-values obtained via our method are the best candidates for future replication of a study because the analysis methodology maximizes statistical power by combining case-control and family-based data [33]. Deviation from the null distribution was observed among the lowest P-values in the adult analysis, suggesting enrichment for significant associations, with two SNPs (rs17441370 and rs272474) meeting genome-wide significance. SNP rs17441370 resides on chromosome 14q13.1 within an intronic region of AKAP6. SNP rs272474 resides on chromosome 5p15.31 within an intronic region of ubiquitin-conjugating enzyme E2Q family-like 1 UBE2QL1. However, both these SNPs had MAFs just above 5% and our statistical power is limited to rigorously assessing these associations due to the relatively small sample size of the adult cohort. Therefore, these SNPs were not included in the immediate replication efforts. The other top association P-values for the childhood, adult and combined analysis are in the range of 1E-07 and 1E-06, and do not reach statistical significance after correcting for multiple comparisons (Fig. 1). Detailed lists of the top (i.e. < 1E-04) association P-values for each of the primary analyses are included in the Appendix S1 (Tables S1, S2 and S3).
Fig. 1.
Manhattan plots of the three GWA analyses: 1) All asthmatics; 2) Adult asthmatics; 3) Childhood asthmatics. The x-axis represents the chromosomal position of the association test that took place for each SNP. The y-axis represents the -Log10 (p-value) of the association test.
The top 33 SNPs (i.e., P-value < 1E-04) (Table S3) from the combined analysis with consistent effect estimates across the childhood and adult cohorts were selected for replication in four independent populations (CHS, CHOP, Costa Rica, Sepracor/LOCCS/LODO/Illumina). Of the 33 SNPs, we obtained association data in at least one population for 19 SNPs, or SNPs in strong LD to the initial SNP. After taking consistency of effect direction into account, two (i.e. rs9272346, rs1121336) of the 19 SNPs had nominally significant P-values in at least two populations (i.e. the P-value is less than 0.05 before multiple comparison correction) (Table 3). SNP rs9272346 in HLA-DQA1 (6p21.3) achieved genomewide significance in the adult cohort where the initial P-value was 1.2E-07 and the final combined P-value was 2.2E-08 after including two adult replication cohorts (Sepracor/LOCCS/LODO/Illumina P-value = 0.032 and iCAP P-value = 0.042). Further details on this SNP are provided in Table 3, where the association P-values are listed for the childhood, adult and combined cohorts. Although this effect was the strongest in the adult cohort, there is evidence that this association may also exist in children (and hence the combined analysis as well), as the initial P-value was nominally significant (P-value = 0.0053) and the association finding was replicated in CHS Hispanic white subjects (P-value = 0.0019) and the Costa Rica (P-value = 0.039) cohort.
Table 3.
Results for the SNP with strongest evidence of replication across all cohorts
| SNP | MA | SHARP Combined Analysis P-value |
CHS P-value/OR |
Study Type Combined Adult Children |
CHOP (children) P-value/OR |
Costa Rica (children) P-value/OR |
Sepracor/LOCCS/ LODO/Illumina (adult) P-value/OR |
iCAP (adult) P-value/OR |
Gene | Overall Replication P-value |
Combined P-value including SHARP Result |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs9272346 | G | 5.9E-05 | 0.0019/0.76* 0.48/1.00 | Combined | 0.83/1.04† | 0.039/0.85 | 0.032/† | 0.042/0.93 | HLA-DQA1 | 0.027 | 1.0E-04 |
| 5.0E-03 | 0.0019/0.76* 0.48/1.00 | Childhood | 0.83/1.04† | 0.039/0.85 | N/A | N/A | 0.075 | 4.8E-03 | |||
| 1.16E-07 | N/A | Adult | N/A | N/A | 0.032/† | 0.042/0.93 | 0.0067 | 2.2E-08 |
CHS association data for this SNP is based on imputed data and is thus divided into two P-values. Top result corresponds to Hispanic White subjects; bottom result corresponds to Non-Hispanic White subjects. The combined P-values treat the two results as corresponding to two populations of the corresponding size.
Association values for CHOP are for rs1063355 (MA=A), which is in strong LD (r2 = 0.97) with rs9272346.
MA, minor allele; OR, odds ratio.
P-values in replication populations are 1-sided to take differences of effect direction in replication populations relative to primary SHARP analysis into account.
We compared our results to previously published asthma GWAS (Table 4). Of 14 loci representative of previously reported association from asthma GWAS, adequate SNP tagging (r2 ≥ 0.80) by a SNP tested in our study was available for eight SNPs. Nominal association (P < 0.05) was observed for four of the eight SNPs: rs1420101 at the IL1RL1/IL18R1 locus (P-value = 0.01); rs1763231444 tagging rs2416257 at the TSLP/WDR locus (P-value = 0.001); rs992969 tagging rs3939286 at the IL33 locus (P-value = 0.005); and rs4795403 tagging rs7216389 at the ORMDL3/GSDML locus (P-value = 0.039). For these associations, statistical significance is achieved for previously associated SNPs at an alpha level of 0.05 because we are evaluating previously identified genetic associations at individual SNPs. Despite being nominally associated with asthma in our study, none of these variants ranked among the most significant ones.
Table 4.
SHARP analysis association P-values for previously reported associations in asthma GWAS studies in SNP and tagging SNP
| Position | Nearby gene(s) | SNP | Original P-value | Population | Reference | SHARP P-value |
|---|---|---|---|---|---|---|
| 2q12 | IL1RL1/IL18R1 | rs1420101 | 6 × 10−12 | Caucasian, East Asian | [43] | 0.01 |
| 2q12 | IL18R1 | rs3771166 | 4 × 10–12 | Caucasian | [10] | 0.002 |
| 2q12.3 | DPP10 | rs1435879 | 3 × 10–6 | African ancestry | [7] | 0.32 |
| 5q12 | PDE4D | rs1588265 | 4 × 10–7 | Caucasian | [5] | 0.07 |
| 5q22 | WDR/TSLP | rs2416257 | 1 × 10–4 | Caucasian, East Asian | [43] | 0.001 |
| 5q31.1 | RAD50-IL13 | rs2244012 | 3 × 10–7 | Caucasian | [8] | 0.40 |
| 6p21 | HLD-DQ | rs9273349 | 7 × 10–14 | Caucasian | [10] | 0.05 |
| 9q21.31 | TLE4 | rs2378383 | 7 × 10–7 | Hispanic / Mexican | [6] | 0.74 |
| 9q24 | IL33 | rs3939286 | 5 × 10–6 | Caucasian, East Asian | [43] | 0.005 |
| 9q24 | IL33 | rs1342326 | 9 × 10–12 | Caucasian | [10] | 0.09 |
| 15q22 | SMAD3 | rs744910 | 4 × 10–9 | Caucasian | [10] | 0.72 |
| 17q21 | ORMDL3/GSDML | rs7216389 | 9 × 10–11 | Caucasian | [1] | 0.04 |
| 17q21 | GSDMB | rs2305480 | 6 × 10–23 | Caucasian (children) | [10] | 0.14 |
| 17q21 | GSDMA | rs3894194 | 3 × 10–17 | Caucasian (children) | [10] | 0.07 |
| 20q12 | PRNP | rs6052761 | 2 × 10–6 | African ancestry | [7] | 0.65 |
| 22q13 | IL2RB | rs2284033 | 1 × 10–8 | Caucasian | [10] | 0.51 |
rs2786098, rs275358, rs10515807, rs1063355, rs9494145, rs1358786, rs1326772 were also reported in the literature; however there were no adequate tagging SNP in this study to replicate these findings.
Discussion
We performed a large GWAS of asthma and identified several novel asthma candidates that demonstrated strong association in both children and adults. Of these, SNP rs9272346, near HLA-DQA1, achieved genome-wide significance in the adult population. The association finding was also observed in some, but not all, childhood cohorts. The HLA-DQA1 locus is among the MHC class II loci frequently associated with asthma and allergic phenotypes in diverse populations [12, 14, 34–39]. In addition to HLA-DQA1, the HLA-DR/DQ region on chromosome 6p21.3 has also been associated with asthma in several previous GWAS. Specifically, rs9273349 was found to have a genome-wide significant association with asthma in a case-control study with over 10 000 physician-diagnosed asthmatics and over 16 000 controls. An evaluation of the HapMap data suggests that rs9272346 and rs9273349 are in linkage disequilibrium with each other. Therefore, rs9272346 is not likely an independent marker for asthma, but represents the same region that was identified and replicated previously using rs9273349, suggesting that our findings further replicate this HLA-DQ region.
The HLA region of the genome is highly polymorphic, containing over 224 genes that are associated with over 100 different autoimmune and infectious diseases [40]. The literature on HLA and asthma goes back at least 20 years with over 500 articles described in pubmed. In the genomic era, the first HLA gene described to be associated with asthma was HLA-G [41]. Although this study does not identify any polymorphisms within HLA-G, the large number of associations within the HLA region suggests that there are likely multiple genetic variants within this region that influence asthma susceptibility. Recently, investigators have identified the DR/DQ region that has now been replicated in four separate studies [42, 43]. Strong links between HLA and both IL4 and TNF alpha, two of the most replicated candidate genes, emphasize the relationship of HLA to Th2 inflammatory responses. The model suspected for autoimmune disorders is one of HLA specificity and interaction with an environmental antigen simulating an infectious pathogen, either viral or bacterial, via molecular mimicry. In the case of asthma, there appears to be less HLA specificity than is seen in inflammatory bowel disease and celiac disease. Nonetheless, the HLA association results suggest that Th1 (IBD and Celiac Disease) and Th2 (Asthma) autoimmune diseases may be more similar than previously thought.
We evaluated the association of several previously identified associations with asthma and observed significant associations (alpha = 0.05) in half of these SNPs. We note that SNPs near ORMDL3/GSDML, the most consistent and widely replicated region associated with asthma, replicates in our SHARP sample. Although SHARP results support half of previously identified asthma GWAS top hits that were tested, the lack of replication of remaining top hits may be due to: (1) genetic heterogeneity among GWAS populations; (2) insufficient statistical power in SHARP to identify small genetic effects; or (3) agedependent genetic effects.
When performing genetic analyses of phenotypes such as asthma, it is important to consider that agedependent genetic effects may influence the asthma phenotype differently for children and adults [44]. This hypothesis is quite intuitive, as asthmatic children have distinctly different clinical characteristics than asthmatic adults. In addition, adult asthmatics may represent a more severe group of individuals, as many of the milder asthmatic cases resolve by adulthood. In this analysis, we evaluated both the stratified and combined genetic analyses of asthma based on age in an effort to consider the time-dependent nature of asthma. We found that the rs9273349 association finding is notably stronger in the adult asthma cohorts compared to the childhood cohorts. Although the effect of rs9273349 is not completely absent in the childhood cohorts, this association would not have been identified had the childhood cohorts only been evaluated. This suggests that either the childhood cohorts by themselves are underpowered or that age-dependent genetic effects exist for asthma. We selected SNPs for replication based on the combined genetic analysis results to maximize statistical power by utilizing the maximum sample size possible. Further research should be conducted to scrutinize the potential age-dependent genetic effects both in the childhood and adult asthmatic samples by selecting replication SNPs for each sample individually.
The SHARP population is complex because it is made up of several clinical trials. While CAMP is one large trial, CARE and ACRN are comprised of multiple clinical trials. As such, the ascertainment criteria vary throughout SHARP. Some include adults (ACRN) while others include children (CAMP, CARE). Although inclusion criteria varies among the SHARP studies, doctor’s diagnosis of asthma, which is considered the gold standard for asthma diagnosis, is the minimum criterion used. Therefore, although the various clinical trials included in SHARP are heterogeneous in nature, the gold standard for asthma diagnosis is consistently used at a minimum for asthma diagnosis. It is also important to consider how the incorporation of control individuals from the general population affect the analysis results. The control individuals we selected will have asthma at the rate that is observed in the general population (roughly 5%), which will lead to misclassification and hence decreases the overall power of our analysis. The initial manuscript that discusses the analytic method proposed here, evaluated the effect of using unselected controls on the overall power of the analysis and although this reduces the overall power of the analysis, it does not bias the results [33].
In summary, this GWAS identifies a novel sequence variant that is primarily associated with asthma in adults, although there is evidence to suggest that this association also exists among children. Together with the loci described in prior asthma GWAS and with previously validated candidate genes, the common genetic variation underlying asthma is gradually being elucidated.
Supplementary Material
Individual study detail on genotyping and data cleaning.
Complete rosters of the members of the SHARP networks.
Quantile-quantile plot comparing the GWAS analysis p-values to those expected under the null hypothesis. Deviations of measures in the tails indicate that both the Childhood analysis and the combined analysis are conservative. This is not surprising, given that the methodology is slightly conservative; however, the top association p-values remain the most promising for further replication. The adult analysis demonstrates deviations in this tail that may indicate true associations with asthma.
Lists the lowest association P-values for the analysis of asthmatic children. None of these findings meet genome-wide significance.
Lists the top association P-value with the adult asthmatic group.
Shows the top association P-values from the combined analysis of children and adult asthmatics.
Acknowledgement
The single-nucleotide polymorphism health associationasthma resource project (SHARP) was funded by the National Heart Lung and Blood Institute, and was composed of researchers from the Asthma Clinical Research Network (ACRN), the Childhood Asthma Management Program (CAMP) and the Childhood Asthma Research and Education (CARE) network. SHARP genotyping services were provided by Affymetrix, Inc. under U.S. Federal Government contract number N02-HL-6-4278 from the National Heart, Lung, and Blood Institute. Complete rosters of the members of the SHARP networks can be found online as Appendix S2.
Footnotes
Conflict of Interests: Charles Irvin’s work was funded by a grant from the American Lung Association. Stephen Peters has been a member of the NHLBI’s ACRN since its creation in 1993. Homer Boushey received funding from the NIH for his participation in the research of the Inner City Asthma Consortium, from Genentech for research on the airway microbiome of severe asthma, and from GlaxoSmithKline for research on bronchial epithelial cell susceptibility to viral infection. He has served as an ad-hoc consultant or advisory board member for Genentech, GlaxoSmithKline, Kalobios, Pharmaxis, Merck and Johnson & Johnson. Vernon Chinchilli, Blanca Himes, Scott Weiss, Juan Celedon, Barbara Klanderman, Erik Melen, Hakon Hakonarson, Lydiana Avila, Benjamin Raby, Kelan Tantisira, David Mauger, Patrick Sleiman, Fernando Martinez, W. James Gauderman, Frank Gilliland, John Lima, Elliot Israel, Christoph Lange, Jody Senter Sylvia, Manuel Soto Quiros and Jessica Lasky-Su have no conflict of interests to declare
Supporting Information
Additional Supporting Information may be found in the online version of this article:
References
- 1.Moffatt MF, Kabesch M, Liang L et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 2007; 448:470–3. [DOI] [PubMed] [Google Scholar]
- 2.Leung TF, Sy HY, Ng MC et al. Asthma and atopy are associated with chromosome 17q21 markers in Chinese children. Allergy 2009; 64:621–8. [DOI] [PubMed] [Google Scholar]
- 3.Galanter J, Choudhry S, Eng C et al. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med 2008; 177:1194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sleiman PM, Annaiah K, Imielinski M et al. ORMDL3 variants associated with asthma susceptibility in North Americans of European ancestry. J Allergy Clin Immunol 2008; 122:1225–7. [DOI] [PubMed] [Google Scholar]
- 5.Himes BE, Hunninghake GM, Baurley JW et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet 2009; 84:581–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hancock DB, Romieu I, Shi M et al. Genome-wide association study implicates chromosome 9q21.31 as a susceptibility locus for asthma in mexican children. PLoS Genet 2009; 5: e1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathias RA, Grant AV, Rafaels N et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol 2010; 125:336–346 e334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Howard TD, Zheng SL et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol 2010; 125(328-335):e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sleiman PM, Flory J, Imielinski M et al. Variants of DENND1B associated with asthma in children. N Engl J Med 2009; 362:36–44. [DOI] [PubMed] [Google Scholar]
- 10.Moffatt MF, Gut IG, Demenais F et al. A large-scale, consortium-based genomewide association study of asthma. N EnglJ Med 2010; 363:1211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan Z, Randall G, Fan J et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet 2007; 81:829–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munthe-Kaas MC, Carlsen KL, Carlsen KH et al. HLA Dr-Dq haplotypes and the TNFA-308 polymorphism: associations with asthma and allergy. Allergy 2007; 62:991–8. [DOI] [PubMed] [Google Scholar]
- 13.Gao J, Lin Y, Qiu C et al. Association between HLA-DQA1, -DQB1 gene polymorphisms and susceptibility to asthma in northern Chinese subjects. Chin Med J (Engl) 2003; 116:1078–82. [PubMed] [Google Scholar]
- 14.Movahedi M, Moin M, Gharagozlou M et al. Association of HLA class II alleles with childhood asthma and Total IgE levels. Iran J Allergy Asthma Immunol 2008; 7:215–20. [PubMed] [Google Scholar]
- 15.Torgerson DG, Ampleford EJ, Chiu GY et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nature genetics 2011; 43:887–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira M, McRae A, Medland S et al. Association between ORMDL3, IL1RL1 and a deletion on chromosome 17q21 with asthma risk in Australia. Eur J Hum Genet 2011; 19:458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Howard T, Zheng S et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol 2010; 125:328–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeWan A, Triche E, Xu X et al. PDE11A associations with asthma: results of a genome-wide association scan. J Allergy Clin Immunol 2010; 126(871–873):e879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathias R, Grant A, Rafaels N et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol 2010; 125:336–346 e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson G Hints of hidden heritability in GWAS. Nat Genet 2010; 42:558–60. [DOI] [PubMed] [Google Scholar]
- 21.Forno E, Lasky-Su J, Himes B et al. Genome-wide association study of the age of onset of childhood asthma. J Allergy Clin Immunol 2012; 130:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Childhood Asthma Management Program Research Group. The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Control Clin Trials 1999; 20:91–120. [PubMed] [Google Scholar]
- 23.The Childhood Asthma Management Program Research Group. Long-term effects of budesonide or nedocromil in children with asthma. N Engl J Med 2000; 343:1054–63. [DOI] [PubMed] [Google Scholar]
- 24.Lemanske RF Jr, Mauger DT, Sorkness CA et al. Step-up therapy for children with uncontrolled asthma receiving inhaled corticosteroids. N Engl J Med 2010; 362:975–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szefler SJ, Phillips BR, Martinez FD et al. Characterization of within-sub-ject responses to fluticasone and mont-elukast in childhood asthma. J Allergy Clin Immunol 2005; 115:233–42. [DOI] [PubMed] [Google Scholar]
- 26.Sutherland ER, Lehman EB, Teodorescu M et al. Body mass index and phenotype in subjects with mild-to-moderate persistent asthma. J Allergy Clin Immunol 2009; 123(1328–1334):e1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szefler SJ, Martin RJ, King TS et al. Significant variability in response to inhaled corticosteroids for persistent asthma. J Allergy Clin Immunol 2002; 109:410–8. [DOI] [PubMed] [Google Scholar]
- 28.O’Donovan MC, Craddock N, Norton N et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet 2008; 40:1053–5. [DOI] [PubMed] [Google Scholar]
- 29.Purcell S, Neale B, Todd-Brown K et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abecasis GR, Cherny SS, Cookson WO et al. GRR: graphical representation of relationship errors. Bioinformatics 2001; 17:742–3. [DOI] [PubMed] [Google Scholar]
- 31.Price AL, Patterson NJ, Plenge RM et al. Principal components analysis corrects for stratification in genomewide association studies. Nat Genet 2006; 38:904–9. [DOI] [PubMed] [Google Scholar]
- 32.Liptak T On the combination of independent tests. Magyar Tud. Akad. Mat. Kutato’ Int.Ko’zl 1958; 3:171–97. [Google Scholar]
- 33.Lasky-Su J, Won S, Mick E et al. On genome-wide association studies for family-based designs: an integrative analysis approach combining ascertained family samples with unselected controls. Am J Hum Genet 2010; 86:573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bignon JS, Aron Y, Ju LY et al. HLA class II alleles in isocyanate-induced asthma. Am J Respir Crit Care Med 1994; 149:71–5. [DOI] [PubMed] [Google Scholar]
- 35.Gao J, Lin Y, Qiu C et al. Relationship between HLA-DQA1, -DQB1 genes polymorphism and susceptilibity to bronchial asthma among Northern Hans. Zhonghua Yi Xue Za Zhi 2002; 82:379–83. [PubMed] [Google Scholar]
- 36.Guo X, Ni P, Li L. Association between asthma and the polymorphism of HLA-DQ genes. Zhonghua Jie He He Hu Xi Za Zhi 2001; 24:139–41. [PubMed] [Google Scholar]
- 37.Lara-Marquez ML, Yunis JJ, Layrisse Z et al. Immunogenetics of atopic asthma: association of DRB1*1101 DQA1*0501 DQB1*0301 haplotype with Dermatophagoides spp.-sensitive asthma in a sample of the Venezuelan population. Clin Exp Allergy 1999; 29:60–71. [DOI] [PubMed] [Google Scholar]
- 38.Molnar-Gabor E, Endreffy E, Rozsasi A. HLA-DRB1, -DQA1, and -DQB1 genotypes in patients with nasal polyposis. Laryngoscope 2000; 110:422–5. [DOI] [PubMed] [Google Scholar]
- 39.Parapanissiou E, Papastavrou T, Deligi-annidis A et al. HLA antigens in Greek children with allergic bronchial asthma. Tissue Antigens 2005; 65:481–4. [DOI] [PubMed] [Google Scholar]
- 40.Shiina T, Hosomichi K, Inoko H et al. The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet 2009; 54:15–39. [DOI] [PubMed] [Google Scholar]
- 41.Nicolae D, Cox NJ, Lester LA et al. Fine mapping and positional candidate studies identify HLA-G as an asthma susceptibility gene on chromosome 6p21. Am J Hum Genet 2005; 76:349–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knutsen AP, Vijay HM, Kariuki B et al. Association of IL-4RA single nucleotide polymorphisms, HLA-DR and HLA-DQ in children with Alternaria-sensitive moderate-severe asthma. Clin Mol Allergy; 8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shahbazi M, Roshandel D, Omidnyia E et al. Interaction of HLA-DRB1*1501 allele and TNF-alpha −308 G/A single nucleotide polymorphism in the susceptibility to multiple sclerosis. Clin Immunol 2011; 139:277–81. [DOI] [PubMed] [Google Scholar]
- 44.Lasky-Su J, Lyon HN, Emilsson V et al. On the replication of genetic associations: timing can be everything!. Am J Hum Genet 2008; 82:849–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Individual study detail on genotyping and data cleaning.
Complete rosters of the members of the SHARP networks.
Quantile-quantile plot comparing the GWAS analysis p-values to those expected under the null hypothesis. Deviations of measures in the tails indicate that both the Childhood analysis and the combined analysis are conservative. This is not surprising, given that the methodology is slightly conservative; however, the top association p-values remain the most promising for further replication. The adult analysis demonstrates deviations in this tail that may indicate true associations with asthma.
Lists the lowest association P-values for the analysis of asthmatic children. None of these findings meet genome-wide significance.
Lists the top association P-value with the adult asthmatic group.
Shows the top association P-values from the combined analysis of children and adult asthmatics.