Abstract
Background:
Phenols and phthalates are potential endocrine disrupting chemicals (EDCs) that are associated with adverse health outcomes. These EDCs dysregulate a number of biomolecules and pathways, including microRNAs. MicroRNAs can be carried in transport systems called extracellular vesicles (EVs) that are present in most biofluids. EVs in the follicular fluid, which fills the ovarian follicle and influences oocyte developmental competency, carry microRNAs (EV-miRNAs) that have been associated with In Vitro Fertilization (IVF) outcomes. However, it remains unclear whether EDCs affect EV-miRNAs in follicular fluid.
Objectives:
This study sought to determine whether urinary concentrations of phenols and phthalates biomarkers are associated with EV-miRNAs expression in follicular fluid collected from women undergoing IVF treatment.
Methods:
This cross-sectional study included 130 women recruited between January 2014 and August 2016 in a tertiary university-affiliated hospital. Participants provided urine samples during ovarian stimulation and on the day of oocyte retrieval. We assessed urinary concentrations of five phenols, eight phthalate metabolites, and one phthalate alternative metabolite. EV-miRNAs were isolated from follicular fluid and their expression profiles were measured using the TaqMan Open Array® Human microRNA panel. We fitted multivariable linear regression models and principal component analysis to examine associations between individual and molar sums of exposure biomarkers and EV-miRNAs.
Results:
Of 754 miRNAs tested, we detected 133 EV-miRNAs in the microRNA array which expressed in at least 50% of the follicular fluid samples. After adjusting for multiple testing, we identified eight EV-miRNAs associated with individual phenols and phthalate metabolites, as well as molar ΣDEHP that met a q < 0.10 false-discovery rate (FDR) threshold. Hsa-miR-125b, hsa-miR-106b, hsa-miR-374a, and hsa-miR15b was associated with mono(2-ethylhexyl) phthalate concentrations, hsa-let-7c with concentrations mono-2-ethyl-5-hydroxyhexyl phthalate (MEHHP), mono-2-ethyl-5-oxohexyl phthalate (MEOHP), mono-2-ethyl-5-carboxypentyl phthalate (MECPP), and the sum of metabolites of di(2-ethylhexyl) phthalate, hsa-miR-24 with mono-n-butyl phthalate concentrations, hsa-miR-19a with cyclohexane-1,2-dicarboxylic acid monohydroxy isononyl ester (MHiNCH), and hsa-miR-375 with ethyl paraben concentrations. Using Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses, gene targets and pathways of these EV-miRNAs were predicted in silico and 17 KEGG FDR-significant pathways related to follicular development and oocyte competence were identified.
Conclusions:
Our results show that urinary concentrations of select phenol and phthalate metabolites are correlated with altered EV-miRNAs expression in follicular fluid. These findings may provide insight regarding the molecular mechanisms underlying adverse effects of phenol and phthalate exposure on female fertility.
Keywords: Follicular fluid, phthalates, microRNAs, phenols, extracellular vesicles, EDC
1. Introduction:
Phthalates and phenols are two classes of potential endocrine disrupting chemicals (EDCs) present in the environment, food, and everyday consumer products (Baccarelli et al. 2000; Diamanti-Kandarakis et al. 2009). These ubiquitous compounds are associated with adverse health outcomes in humans (Bornehag et al. 2004; Feige et al. 2007; Gore et al. 2015; Miodovnik et al. 2011; Philippat et al. 2012). Human exposure to phthalates and phenols occurs through several routes, including ingestion, inhalation, and dermal contact (Hauser and Calafat 2005). The most common sources of exposure to these chemicals include personal care products (cosmetics, shampoos, perfumes), solvents, medical devices (like IV tubing), thermal receipts, and food packaging materials (Carwile et al. 2011; Guo and Kannan 2013; Hauser and Calafat 2005). Previous epidemiologic studies have shown that some phthalates and phenols are associated with adverse female fertility outcomes (Ehrlich et al. 2012; Hauser et al. 2016b; Jukic et al. 2016; Machtinger et al. 2018a; Meeker and Ferguson 2014; Mínguez-Alarcón et al. 2016; Toft et al. 2012).
There is increasing interest in molecular markers, such as microRNAs, and how they might act as an intermediate between environmental exposures like EDCs and the development of diseases and disorders (Casati et al. 2015; Derghal et al. 2016; Fleisch et al. 2012). MicroRNAs are short, non-coding RNA molecules that can post-transcriptionally regulate gene expression (Li et al. 2007; Lim et al. 2005; Sætrom et al. 2007) and can be free-floating or packaged in extracellular vesicles (EV-miRNAs). Extracellular vesicles (exosomes, micro-vesicles, and other membrane-bound vesicles) have been detected in almost every biofluid, including follicular fluid (Weber et al. 2010), and can act as a vehicle carrying proteins, messenger RNAs (mRNAs), and microRNAs (miRNAs) (Raposo and Stoorvogel 2013). These EV-miRNAs are microRNAs packaged in extracellular vesicles and are stable compared to their free-floating counterparts. They are actively released by viable cells and likely represent an active means of communication between cells and tissues (Machtinger et al. 2016b; Yáñez-Mó et al. 2015). EV-miRNAs expression has been associated with concentrations of phenols and phthalate metabolites, with resultant downstream alterations in gene expression in testicular tissue from zebrafish (Lee et al. 2018), in vitro mouse Sertoli cells (Cho et al. 2010), ovarian tissue from rodents (Liu et al. 2018b), hippocampal tissue from rodents (Luu et al. 2017), and human placental cells (Avissar-Whiting et al. 2010; De Felice et al. 2015b; LaRocca et al. 2016; Meruvu et al. 2016). However, to the best of our knowledge, the relationship between EV-miRNAs expression in follicular fluid and ubiquitous environmental chemicals known to have reproductive toxicity is unstudied.
The ovarian follicle not only houses the oocyte itself, but as the follicle matures, it induces cellular differentiation occurs, creating cellular layers of thecal, granulosa, and cumulus cells. Thecal and granulosa cells make up the membrane of the follicle itself, while cumulus cells surround the encapsulated oocyte (Hennet and Combelles 2012; Rimon-Dahari et al. 2016). The granulosa cells that embed the ovarian follicle secret hyaluronan and chondroitin sulfate that generates an osmotic gradient in the follicle. This gradient pulls in fluid derived from the vasculature of the theca cells that surround the granulosa cells in the ovarian follicle (Rodgers and Irving-Rodgers 2010). The follicular fluid, a critical microenvironment for the development of oocytes (Rodgers and Irving-Rodgers 2010; Zuccotti et al. 2011), contains a mixture of proteins, metabolites, ions, plasma components, numerous other molecules, including EV-miRNAs. Thus, the objective of this study was to determine whether urinary concentrations of phenols and phthalate metabolites are correlated with the expression of EV-miRNAs isolated from follicular fluid in women undergoing in vitro fertilization (IVF). The ability to quantify biomarkers of exposure to phthalates and phenols and how they impact EV-miRNAs in follicular fluid may provide insight into the potential influence of EDCs on female reproduction.
2. Methods
2.1. Ethics
This study was approved by the Sheba Medical Center institutional review board (IRB) in accordance with the Declaration of Helsinki. Authors confirm that all methods were in accordance with the relevant guidelines and regulations. All participants provided written informed consent upon enrollment.
2.2. Study Population:
Between January 2014 and August 2016, women aged 19 to 38 years with six or fewer previous IVF attempts were recruited in a tertiary care university-affiliated hospital in Israel. To increase generalizability, we included in the study both fertile and infertile women. Fertile women were those who had conceived spontaneously in the past and underwent IVF for pre-gestational diagnosis of autosomal recessive diseases. Participants were excluded from the cohort before the EV-miRNAs analysis if they had a diagnosis of polycystic ovarian syndrome (PCOS), endometriosis, were poor responders according to Bologna criteria (Ferraretti et al. 2011), and/or had a male partner with severe male factor infertility. Only women using one regimen (antagonist protocol) were included to avoid potential confounding by the stimulation protocol. All women participated during a single IVF cycle.
2.3. Exposure Assessment:
Study participants provided up to two spot urine samples in a sterile polypropylene cup. Specimens were pooled before further analysis for participants providing more than one urine sample. Collection occurred during stimulation (days 1–7 of gonadotropin injection) and/or on the day of oocyte retrieval. In cases that 2 urine samples were collected, they were pooled before the chemical analysis to save costs. Specific gravity (SG), which is used to correct concentrations for urine dilution, was measured (Comber test strips, Roche, Switzerland), and samples were aliquoted and frozen at −80°C. Frozen samples were shipped to the CDC (Atlanta, GA, USA) for quantification of biomarkers of 11 phenols: 2,4-dichlorophenol, 2,5-dichlorophenol, benzophenone-3, BPA, BPF, BPS, methyl paraben, propyl paraben, ethyl paraben, butyl paraben and triclosan; 17 phthalates metabolites: monoethyl phthalate (MEP), mono-n-butyl phthalate (MnBP), mono-hydroxybutyl phthalate (MHBP), mono-isobutyl phthalate (MiBP), mono-hydroxyisobutyl phthalate (MHiBP), monobenzyl phthalate (MBzP), mono-3-carboxypropyl phthalate (MCPP), mono-2-ethylhexyl phthalate (MEHP), mono-2-ethyl-5-hydroxyhexyl phthalate (MEHHP), mono-2-ethyl-5-oxohexyl phthalate (MEOHP), mono-2-ethyl-5-carboxypentyl phthalate (MECPP), mono-2-ethyl-5-hydroxyhexyl terephthalate (MEHHTP), mono-2-ethyl-5-carboxypentyl terephthalate (MECPTP), mono-isononyl phthalate (MNP), monooxononyl phthalate (MONP), mono(carboxy-isooctyl) phthalate (MCOP), mono(carboxy-isononyl) phthalate (MCNP), and two metabolites of the phthalate althernative DINCH (Cyclohexane-1,2-dicarboxylic acid diisonoyl ether): cyclohexane-1,2-dicarboxylic acid monohydroxy isononyl ester (MHiNCH), and cyclohexane-1,2-dicarboxylic acid monocarboxyisooctyl ester (MCOCH). Due to our smaller sample size, we analyzed metabolites that were detectable in 85% of the participants with the except of ethyl-paraben that was present in (78.5%) but was part of the ∑paraben. Methods for quantifying biomarkers concentrations used online solid phase extraction coupled with high performance liquid chromatography-isotope dilution tandem mass spectrometry following standard quality assurance/quality control procedures as previously described (Silva et al. 2013; Silva et al. 2017; Ye et al. 2005). The biomarkers included in this analysis were Mono-2ethyl-5-carboxypentyl phthalate (MECPP), Mono-2-ethyl-5-hydroxyhexyl phthalate (MEHHP), Mono-2-ethyl-5oxohexyl phthalate (MEOHP), Mono-2-ethylhexyl phthalate (MEHP), Mono-n-butyl phthalate (MBP), Monohydroxybutyl phthalate (MHBP), Mono-isobutyl phthalate (MiBP), Mono-hydroxy-isobutyl phthalate (MHiBP), Cyclohexane-1,2-dicarboxylic acid, monohydroxy isononyl ester (MHiNCH), bisphenol A (BPA), methylparaben, ethyl-paraben, propyl-paraben, and butyl-paraben. Instrumental reading values were used for concentrations below the limit of detection (LOD). Biomarker concentrations were all adjusted for urinary specific gravity using the following formula: Pc= P[(1.014 − 1) / (SG – 1)], where Pc is the SG-corrected biomarker concentration (ng/mL), P is the instrument measured biomarker concentration (ng/mL), and 1.014 is the median SG level in our study population. All analyses used SG-adjusted biomarker concentrations.
2.4. Outcome Assessment:
2.4.1. RNA extraction from follicular fluid:
Follicular fluid (otherwise discarded material) was collected during oocyte retrieval from follicles >18 mm, centrifuged at 1500×g for 15 min. Samples were pre-cleaned using a 0.80 um pore-size polyethersulfone filter (StericupRVP, Merck Millipore) to remove larger proteins and debris and aliquoted into 500 uL for immediate storage at −80°C (Witwer et al. 2013). Only mature (MII) oocytes were examined for RNA analysis. Methods for RNA extraction from biological fluids have been previously described (Pergoli et al. 2017). In short, samples were thawed, centrifuged for 15 min at 1200 × g at room temperature and then centrifuged three times at 1000, 2000, and 3000 × g, respectively, for 15 min at 4°C Following this steps, samples were ultracentrifuged (Beckman Coulter Optima-MAX-XP) at 110,000 × g for 75 min at 4°Cfor the extraction of EV, as ultracentrifugation is considered the standard according to International Society for Extracellular Vesicle recommendations (Gardiner et al. 2016). The pellets obtained were kept at −80°C until use. EV-miRNAs were extracted from the ultracentrifuged pellets using the miRNAeasy Kit and RNeasy CleanUp Kit per the manufacturer (Qiagen, Valencia, CA, USA). The final purified EV-miRNA-enriched RNA was eluted into 20 uL of RNAse-free water and stored at −80°C until further use.
2.4.2. Expression analysis of EV-miRNAs in follicular fluid:
We screened for 754 microRNAs in our EV-miRNA aliquot using the TaqMan Open Array® system. We obtained 758 Crt values for each follicular fluid sample, which included 754 unique miRNAs and four internal controls (ath-miR159a, RNU48, RNU44 and U6). Methods of Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) for screening EV-miRNAs using the microRNA array in biological fluids are published elsewhere (Pergoli et al. 2017). QuantStudio™ 12K Flex is a fixed-content panel containing validated human TaqMan® MicroRNA Assays derived from Sanger miRBase release v.14. All 754 assays have been functionally validated with miRNA artificial templates. The panel is specifically designed to provide specificity for only the mature miRNA targets. TaqMan MicroRNA Assays (spotted in the panel) incorporate a target-specific stem–loop reverse transcription primer allowing to work despite the short length of mature miRNAs (~22 nucleotides) which prohibits conventional design of primers. Briefly, we prepared 3.3 uL of each RNA sample and then reverse-transcribed to cDNA (complementary DNA) and pre-amplified. Pre-amplified samples were mixed with the TaqMan Open Array® Real Time PCR Master Mix (Life Technologies, Foster City, CA) and loaded onto a TaqMan™ OpenArray® Human miRNA panel with the QuantStudio™ AccuFill System Robot (Life Technologies, Foster City, CA). RT-qPCR was performed on the QuantStudio™ 12K Flex Real-Time PCR System with the OpenArray® Platform [QS12KFLEX] (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. Expression levels were calculated in relative cycle threshold values (Crt), estimating the amplification cycle at which the fluorescence levels for each of the analyzed EV-miRNAs exceed the background fluorescence threshold (Enderle et al. 2015).
2.5. Covariate Assessment:
Participant’s height and weight, measured at the beginning of the IVF cycle, were used to calculate body mass index (BMI) (kg/m2). Age, fertility diagnosis [male factor, PGD (pre-gestational diagnosis), unexplained, mechanical, sexual dysfunction, or egg donor], smoking status, number of previous IVF attempts, and number of oocytes retrieved at the start of the IVF cycle were extracted from participants’ medical charts and questionnaire. The batch of EV-miRNAs expression was determined based on the year the samples were sent from Israel to the lab for EV-miRNAs analysis (batch 1 vs batch 2).
2.6. Statistical Analysis:
2.6.1. EV-miRNA-by-EV-miRNA regression analysis
To extract the EV-miRNAs qPCR data, we used Thermo Fisher Cloud Relative Quantification software. To ensure accuracy of in our normalization methods of the EV-miRNAs, as discussed previously (Marabita et al. 2016; Mestdagh et al. 2009; Zeka et al. 2015), we ran algorithms to identify the best normalization strategy. We first applied the NormFinder and geNorm algorithms to select the best normalization strategy among global mean (arithmetic and geometric), RNU48, RNU6, or the average of the four miRNAs with the lowest standard deviation (SD) among subjects. Based on these algorithms, we found that global mean was the best method to normalize the data. EV-miRNAs data was normalized using the global mean (GM) method (ΔCrt_EV-miRNAi = (Crt_EV-miRNAi – Crt_EV-miRNAi_global_mean)) as suggested by Pergoli et al. (Pergoli et al. 2017). All the EV-miRNAs with a Crt value > 28 and/or an amplification score ≤ 1.24 were identified as unexpressed. For the global mean, we coded all those EV-miRNAs that were unexpressed as 28. We calculated the delta Crt based on the global mean across all the miRNAs within that subject and dividing it by the total miRNAs (N=754). All subsequent analyses were performed on only those EV-miRNAs that had expressed values. Standard descriptive statistics were used to explore the characteristics of the study participants and exposure data. Spearman’s correlation coefficients were used to examine correlations between phenols and phthalate metabolites. Adjusted linear regression models were applied to uncover top hit EV-miRNAs. All models were adjusted for a priori covariates: age, body mass index (BMI, calculated from patient height and weight), smoking status, pre-IVF fertility status (fertile vs infertile), and batch number. Sg-adjusted biomarker concentrations were log10 transformed and EV-miRNA outcomes were inverse normally transformed to ensure normality with a standard deviation of one. Regression analyses were further adjusted for any unwanted variation within our high-throughput assay by applying the SVA (surrogate variable analysis) package (Jeffery T. Leek 2017). The SVA package can help identify and remove any batch effects or unwanted sources of variation, seasonal, meteorological, exposure, or technical variables, which are unknown but might be differently distributed in the two batches of samples. It creates surrogate variables, accounting for the unmeasured variation, that act as covariates in our models that would account for any unknown, un-modeled, or other sources of noise (Leek et al. 2012). To account for multiple-testing, we applied the Benjamini-Hochberg FDR “p. adjust” function in R (Benjamini and Hochberg 1995). All statistical analyses were performed in R-version 3.4.0 (R-Core-Team 2017). Statistical significance was set at a p-value < 0.05. For multiple comparisons we chose a less conservative threshold of q-value < 0.10, due to our sample size.
2.6.2. Mixtures of phenols and phthalates:
Molar sums were calculated for metabolites of di 2-(ethyl hexyl) phthalate (ΣDEHP), the dibutyl phthalate metabolites (ΣDBP), and parabens biomarkers (Σparabens) by dividing the concentration of each metabolite by its molecular weight and then summing, e.g. ΣDEHP = [(mEHP × (1/278.34)) + (mEHHP × (1/294.34)) + (mEOHP × (1/292.33)) + (mECPP × (1/308.33))]. We adjusted the molar sums for urinary specific gravity as previously described (Machtinger et al. 2018a) and log10 transformed for the analysis. We ran multivariable linear regression models adjusting for a priori covariates: age, BMI, smoking status, fertility status, and batch number, as well as SVA surrogate variables.
2.7. In silico KEGG Enrichment Pathway Analysis
We performed an in-silico analysis using a web-based tool miRWalk2.0 (http://mirwalk.umm.uni-heidelberg.de/) to investigate gene targets of EV-miRNAs (Dweep et al. 2011). Target predictions included a comparative analysis of seven prediction programs, DIANAmT, miRanda, miRDB, miRWalk, PICTAR5, RNA22, and Targetscan. Only those targets that validated in Target scan and at least five of the other prediction programs were further investigated. The predicted target search examined promoter, 5’-untranslated regions, 3’-untranslated regions, and coding sequences and included a minimum seed length of seven nucleotides (Dweep et al. 2011). Using the genes selected by miRWalk, we ran a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis in the web-based tool WebGestalt (WEB-based GEne SeT AnaLysis Toolkit) (Wang et al. 2017; Zhang et al. 2005) and restricted our results to only those with 10 or more genes listed in the in silico predicted targets and an FDR q-value less than 0.05.
Results:
3.1. Study population
The women in our study were 31 ± 3.7 years of age (mean ± standard deviation), had a BMI of 23.5 ± 4.7 kg/m2, and a mean of 9 ± 5 oocytes retrieved during the IVF cycle (Table 1). Most women were on their first IVF attempt (66%).
Table 1.
Descriptive Statistics of study participants
| Age, years (mean ± SD) | 31 ± 3.7 | |
| BMI, kg/m2 (mean ± SD) | 23.5 ± 4.7 | |
| Number of oocytes retrieved (mean ± SD) | 9 ± 5 | |
| Current Smoking Status (%) | No | 96 (74%) |
| Yes | 34 (26%) | |
| IVF Attempt (%) | First Attempt | 86 (66%) |
| Attempt > 1 | 44 (34%) | |
| Fertility Status (%) | Fertile | 56 (43%) |
| Infertile | 74 (57%) |
Abbreviations: BMI: body mass index, SD: standard deviation, IVF: in vitro fertilization
3.2. Urinary biomarkers
All analyses were run on 130 samples that had exposure biomarkers, demographic, and EV-miRNA data from mature oocytes Supplementary Figure S1. A majority of our participants (73%) provided two spot urine samples (n = 99/136). Most biomarkers were detected in ≥ 85% of the samples, except for ethyl paraben (78.5% > LOD). Specific gravity-adjusted medians, IQRs (interquartile ranges), LOD, and percent of the samples with concentrations above the LOD are shown in Table 2. The correlations between SG-adjusted biomarkers are shown in Supplementary Figure S2.
Table 2.
Exposure distribution of individual urinary metabolites and molar sum for 130 women undergoing IVF treatment
| SG- Adjusteda | |||||||
|---|---|---|---|---|---|---|---|
| Parent Compound | Biomarker | Units | LOD | % Detected | Median | IQR | Max |
| 1,2-Cyclohexane dicarboxylic acid, diisononyl ester (DINCH) | MHINCH | ug/L | 0.4 | 93.80% | 1.18 | (0.75, 2.50) | 74.8 |
| Di(2-ethylhexyl) phthalate (DEHP) | MEHP | ug/L | 0.8 | 91.5% | 3.69 | (2.24, 7.64) | 70.6 |
| MEOHP | ug/L | 0.2 | 100% | 9.71 | (6.48, 16.2) | 145 | |
| MEHHP | ug/L | 0.4 | 100% | 13.49 | (8.72, 22.3) | 215 | |
| MECPP | ug/L | 0.4 | 100% | 20.16 | (13.6, 33.7) | 371 | |
| ∑DEHP | umol/L | -- | -- | 0.16 | (0.11, 0.27) | 2.65 | |
| Di-iso-butyl phthalate (DiBP) | MiBP | ug/L | 0.8 | 99.2% | 24.13 | (16.0, 38.5) | 188 |
| MHiBP | ug/L | 0.4 | 100% | 7.49 | (4.74, 13.2) | 75.0 | |
| Di-n-butyl phthalate (DnBP) | MBP | ug/L | 0.4 | 98.5% | 19.01 | (11.8, 32.2) | 148 |
| MHBP | ug/L | 0.4 | 88.5% | 1.37 | (0.77, 2.39) | 17.1 | |
| ∑DBP | umol/L | -- | -- | 0.24 | (0.15, 0.39) | 1.50 | |
| Bisphenol-A (BPA) | -- | ug/L | 0.2 | 98.5% | 3.20 | (1.96, 5.25) | 31.9 |
| Butyl Paraben | -- | ug/L | 0.1 | 94.6% | 2.73 | (0.44, 14.2) | 297 |
| Ethyl Paraben | -- | ug/L | 1 | 78.5% | 7.14 | (1.45, 14.23 | 441 |
| Methyl Paraben | -- | ug/L | 1 | 98.5% | 99.65 | (20.7, 277) | 1901 |
| Propyl Paraben | -- | ug/L | 0.1 | 100% | 9.10 | (1.68, 43.2) | 581 |
| ∑Parabens | -- | umol/L | -- | -- | 0.86 | (0.19, 2.34) | 19.3 |
LOD = limit of detection, SG = specific gravity, IQR = Inter quartile range
Instrumental reading values were used for biomarker concentrations below the LOD. To adjust for urinary dilution, we used the following formula: Pc =P [(1.014 − 1) / SG −1], where Pc is the SG-corrected biomarker concentration (μg/L), P is the measured biomarker concentration (μg/L), and 1.014 is the mean SG level in our study population.
3.3. Profile of EV-miRNAs in follicular fluid
We screened for EV-miRNAs in a panel of 754 miRNAs and detected 320 in at least one of the 130 samples analyzed. For our analyses, we chose to restrict the EV-miRNAs we analyzed to those that were detected in at least 50% of our samples, resulting in 133 EV-miRNAs. This cut-off was chosen to maximize the number of EV-miRNAs examined while retaining a large enough sample to assess the associations. A list of the selected EV-miRNAs with their detectable levels are provided in Supplemental Table S1. Additionally, a heatmap of the normalized EV-miRNAs used in our analysis for each individual is provided as Supplementary Figure S3.
3.4. Associations between individual biomarker measures and EV-miRNA expression in follicular fluid
To examine the association between the follicular fluid EV-miRNAs and the exposure biomarkers, we ran EV-miRNA-by- EV-miRNA linear regression models for each of our biomarkers. All our models adjusted for age, BMI, smoking status, fertility status, batch, and SVA surrogate variables, which were biomarker specific. The SVA variables accounted for any unmeasured variation within our dataset. After adjusting for multiple testing for the EV-miRNAs, we identified eight EV-miRNAs associated with individual biomarkers concentrations (Table 3; see also Supplemental Tables S2 and S3 for full regression results). We found that a log10 increase (ng/mL) in MBP was associated with a 0.37 [95% CI: −0.57, −0.17] standard deviation decrease of hsa-miR-24 ΔCt, where a lower ΔCt indicates higher relative expression. Additionally, MEHP was significantly associated with hsa-miR-125b (ΔCt effect size: −0.32; 95%CI: −0.50, −0.15), hsa-miR-106b (ΔCt effect size: 0.29; 95%CI: 0.11, 0.46), hsa-miR-374a (ΔCt effect size: 0.30; 95%CI: 0.11, 0.49), and hsa-miR-15b (ΔCt effect size: 0.28; 95%CI: 0.10, 0.47). Ethyl paraben was significantly associated with hsa-miR-375 (ΔCt effect size: 0.28; 95%CI: 0.13, 0.44). Hsa-let-7c was significantly associated with MEOHP (ΔCt effect size: −0.51; 95%CI: −0.79, −0.23), MEHHP (ΔCt effect size: −0.50; 95%CI: −0.79, −0.22), and MECPP (ΔCt effect size: −0.47; 95%CI: −0.74, −0.21). Phthalate alternative MHINCH was significantly associated with hsa-miR-19a (ΔCt effect size: −0.39; 95%CI: −0.60, −0.17) (Table 3).
Table 3.
Number of significant EV-miRNAs for individual exposures and ∑DEHP biomarkers and effect sizes of FDR top hits
| Compound | EV-miRNA name | Percent Expressed in study samples | Beta Coefficient b | 95% CI | Unadjusted p-value | FDR q-valuea | |
|---|---|---|---|---|---|---|---|
| Ethyl-Paraben | hsa-miR-375 | 90% | 0.28 | 0.13 | 0.44 | <0.001 | 0.049 |
| MBP | hsa-miR-24 | 98% | −0.37 | −0.57 | −0.17 | <0.001 | 0.041 |
| MHINCH | hsa-miR-19a | 91% | −0.39 | −0.60 | −0.17 | 0.001 | 0.081 |
| MEHP | hsa-miR-125b | 98% | −0.32 | −0.50 | −0.15 | <0.001 | 0.062 |
| hsa-miR-106b | 100% | 0.29 | 0.11 | 0.46 | 0.001 | 0.099 | |
| hsa-miR-374a | 97% | 0.30 | 0.11 | 0.49 | 0.002 | 0.099 | |
| hsa-miR-15b | 98% | 0.28 | 0.10 | 0.47 | 0.003 | 0.099 | |
| MEOHP | hsa-let-7c | 98% | −0.51 | −0.79 | −0.23 | <0.001 | 0.060 |
| MEHHP | hsa-let-7c | 98% | −0.50 | −0.79 | −0.22 | 0.001 | 0.093 |
| MECPP | hsa-let-7c | 98% | −0.47 | −0.74 | −0.21 | 0.001 | 0.078 |
| ∑DEHP | hsa-let-7c | 98% | −0.55 | −0.85 | −0.26 | <0.001 | 0.035 |
FDR q-value adjusted for multiple testing for the 133 EV-miRNAs but not for the number of biomarkers
For every one unit increase in log10 of exposure, there is an effect size increase/decrease in standard deviation of EV-miRNA ΔCt
3.5. Associations of molar sums of phenol and phthalate biomarkers
We ran separate linear regression models with each of our three molar sums and found that molar ΣDEHP was significantly associated with hsa-let-7c. We found a one unit increase in log10 molar ΣDEHP (umol/L) was associated with a 0.55 (95%CI: −0.85, −0.26) standard deviation decrease in hsa-let-7c ΔCt (Table 3). While we found several significant EV-miRNAs with molar ΣDBP and molar ΣParabens, none were significant after FDR adjustments (Supplemental Table S4).
3.6. In silico KEGG Pathway Analyses
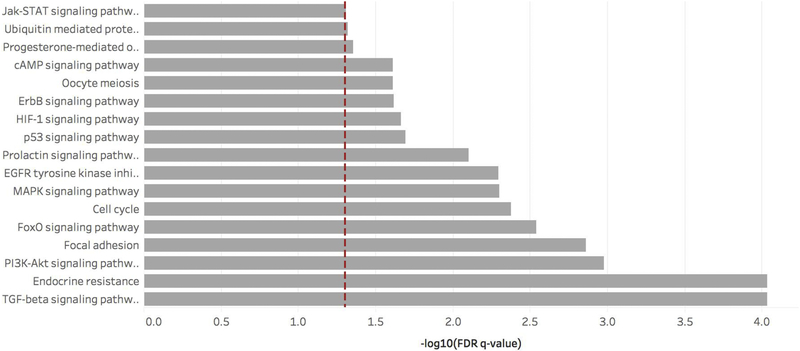
We ran KEGG analyses for the eight FDR-adjusted significant EV-miRNAs, hsa-miR-375, hsa-miR-24, hsamiR-125b, hsa-miR-106b, hsa-miR-374a, hsa-miR-15b, hsa-miR-19a, and hsa-let-7c. The miRWalk tool identified 600 unique genes associated with these EV-miRNAs in the Targetscan database and at least three different EV-miRNA prediction programs. Our in silico analysis using WebGestalt identified 36 enriched KEGG pathways. Each contained at least ten genes identified by the miRWalk tool and had a FDR q-value < 0.05 (Supplemental Table S5). Genes involved with the significant pathways can be identified in Supplemental Table S5 and Supplemental Table S6. Of the 36 KEGG pathways, 17 were associated with follicular development and oocyte maturation and function: TGF-beta signaling, endocrine resistance, PI3K-Akt signaling, focal adhesion, FoxO signaling, cell cycle, MAPK signaling, EGFR tyrosine kinase inhibitor resistance, prolactin signaling, p53 signaling, HIF-1 signaling, ErbB signaling, cAMP signaling, oocyte meiosis, progesterone-mediated oocyte maturation, ubiquitin mediated proteolysis, and Jak-STAT signaling pathways. (Figure 1). The most common of these significant genes, that played a role in identifying these pathways, include mapk1, akt3, map2k1, pik3r3, and raf1 (Supplemental Table S7).
Figure 1. KEGG pathway enrichment.
Enriched KEGG pathways involved in ovary and follicle development, maturation, and fertilization associated with the eight FDR-significant EV-miRNAs. The red dashed line represents the statistically significant FDR threshold (q < 0.05) and the small numbers within each bar indicate number of predicted genes associated with that KEGG pathway.
4. Discussion
In the present study, we identified significant correlations between phenol and phthalate biomarker urinary concentrations and EV-miRNA profiles in follicular fluid. Among these women undergoing IVF, hsa-miR-125b and hsa-miR-15b were positively associated with DEHP, while hsa-miR-106b, and hsa-miR-374a had an inverse association with DEHP (increased DEHP with decreased expression). Concentrations of MBP were positively associated with levels of hsa-miR-24. Hsa-let-7c was positively associated with urinary concentrations of MEOHP, MEHHP, MECPP, and ΣDEHP. Additionally, phthalate alternative MHINCH was positively associated with hsa-miR-19a. Among the phenols examined, increased urinary concentration of ethyl paraben was associated with decreased expression hsa-miR-375. These associations remained significant after FDR adjustment.
To our knowledge, this is the first study to identify correlations between select potential EDCs and EV-miRNA expression in follicular fluid. EV-miRNAs play a role in intra- and inter- cellular communication within the ovarian follicle. Follicular fluid EVs can originate from the oocyte, cumulus or mural granulosa cells in the ovarian follicle. As the field of EVs and reproduction is evolving, the origin of these follicular fluid EVs, their production and their turnover rate remains largely unexplored. Recent studies suggest that EV-miRNAs can impact granulosa cell function and can alter follicular development and oocyte maturation (Di Pietro 2016; Machtinger et al. 2016a) and several of the significant EV-miRNAs that were associated with phthalates and phenols biomarkers in our study play key roles in the ovarian follicle. MiR-375 is expressed in both granulosa cells and oocytes and mediates the effect of genes that regulate follicular growth with downstream proliferation, spread, and apoptosis of cumulus cells (Chen et al. 2017; Liu et al. 2018a). Overexpression of miR-375 blocked the proliferation ability and increased the rate of apoptosis of bovine cumulus cells (Chen et al. 2017; Liu et al. 2018a) and suppressed estradiol production and follicular development in porcine granulosa cells (Yu et al. 2017). In rodents, overexpression of a miR-125b mimic in oocytes and mouse embryonic stem cells could target and block expression of specific genes required for embryo progress beyond the two-cell stage (Kim et al. 2016). In another study, higher levels of miR-24 in bovine culture media was associated with embryos that failed to undergo differentiation. Moreover, when adding more miR-24 to the functional blastocyst containing media, development was significantly altered (Kropp and Khatib 2015). This suggests that over-expression of hsa-miR-375, hsa-miR-24, and hsa-miR-125b in follicular fluid is influenced by environmental exposures and could impact ovarian function through mechanistic pathways yet to be elucidated.
There is growing concern regarding the underlying pathways or mechanisms by which EDCs, such as some phenols and phthalates, impact ovarian reserve, fertility, and pregnancy rates. In rodents, DEHP and its metabolites negatively affect follicular growth and estrogen synthesis (Craig et al. 2014; Gupta et al. 2010). However, in humans, the adverse effects of DEHP on female fertility and pregnancy are not as clear. Epidemiologic studies suggest that higher urinary concentrations of ΣDEHP significantly decreased the antral follicle count among sub-fertile women (Messerlian et al. 2016). Furthermore, an increase in urinary concentrations of DEHP metabolites was associated with a lower number of oocytes retrieved, mature oocytes, fertilized oocytes, and poor embryo quality (Machtinger et al. 2018b) along with lower clinical pregnancy rates and live birth rates following fertility treatment (Hauser et al. 2016a). Higher urinary DEHP metabolite concentrations were also associated with a higher risk of biochemical pregnancy loss (Messerlian et al. 2016). Another study examining the same population as our current study found associations between urinary concentrations of MBP, ΣDEHP, and DEHP metabolites and the number of oocytes (total yield, mature, and/or fertilized) and top quality embryos (Machtinger et al. 2018b). Similarly, in our study, we identified significant associations of EV-miRNAs with urinary concentrations of MBP and ΣDEHP, and individual DEHP metabolites MEHP, MEOHP, MEHHP, and MECPP. The urinary concentration of MHiNCH, a phthalate alternative, was not associated with adverse clinical IVF outcomes (Machtinger et al. 2018b), however, we identified one significant EV-miRNA with MHiNCH in this current study. The agreement between our study and the previous epidemiology studies implies the existence of pathways through which certain phenols and phthalates may affect IVF outcomes. Despite evolving evidence of the negative effects of DEHP on female reproduction, little is known about the possible underlying mechanisms. Our results suggest that exposure to certain phthalates and phenols may affect biological pathways that are associated with follicular development and oocyte maturation by altering the expression of EV-miRNAs in follicular fluid.
We ran KEGG analyses for the eight FDR-adjusted significant EV-miRNAs and identified 17 pathways that were associated with follicular development and oocyte maturation. Additionally, we found five genes that appeared in more than 10 of these KEGG pathways, mapk1, akt3, map2k1, pik3r3, and raf1. MapK1 and map2K1 are mitogen activated protein kinases and are both involved in the meiotic regulation and oocyte maturation, as well as key genes involved in the MAPK signaling pathway (Dupre et al. 2011). Akt3, involved in spindle formation and meiotic maturation (Hoshino and Sato 2008), is also a player in the PI3K-Akt and MAPK pathways. Pik3r3, a gene that is involved in the PI3K-Akt and MAPK pathways, has also been associated with ovarian cancer (Zhang et al. 2007). All five of these genes were identified in the EGFR tyrosine kinase inhibitor, endocrine resistance, ErbB signaling, focal adhesion, FoxO signaling, progesterone-mediated oocyte maturation, prolactin signaling, and PI3K-Akt signaling pathways.
The PI3K-Akt signaling pathway is of great relevance. PI3K-Akt has been associated with recruitment of primordial follicles, a dynamic and tightly controlled process that initiates folliculogenesis (Lopez-Cardona et al. 2017; Sánchez and Smitz 2012), with granulosa proliferation and with ovarian function (Andrade et al. 2017). It is also involved in cell proliferation, apoptosis, DNA repair and protein synthesis. In rodents, exposure to DEHP decreased primordial follicle recruitment by dysregulation of the PI3K signaling pathway (Hannon et al. 2014). In humans, an in vitro study showed that DEHP exposed Hep3B cells induced oxidative stress and oxidative DNA damage (Chen et al. 2013). When a Pl3K-Akt inhibitor was added, reactive oxygen species levels were lower compared to those cells that were exposed to DEHP alone. The oxidative DNA damage persisted, even in the presence of the Pl3K-Akt inhibitor. Additionally, DEHP increased DNA replication rates, but this change was abated by the addition of the Pl3K-Akt inhibitor. The authors concluded that DEHP induced increased cellular proliferation and oxidative damage by activating the Pl3K-Akt pathway (Chen et al. 2013). The Pl3KAkt pathway also activates the mTOR and FoxO pathways, both which are regulated in the oocyte (Makker et al. 2014). In mice, dysregulation of FoxO can lead to stunted oocyte growth and follicle development, as well as luteinization of ruptured follicles (Liu et al. 2007). Deletion of mTOR genes in rodents causes early activation of all primordial follicles and leads to premature ovarian failure (Wang et al. 2014).
While EV-miRNAs have been identified in different tissue types (Ludwig et al. 2016), relatively few studies have examined the associations with phenol and phthalate exposures (Avissar-Whiting et al. 2010; Chou et al. 2017; De Felice et al. 2015a; LaRocca et al. 2016; Liu et al. 2018b; Meruvu et al. 2016; Tilghman et al. 2012). In rodents, exposure to DEHP was associated with increased expression of let-7b, miR-17, miR-181a, and miR-151 (Liu et al. 2018b). In our present study, we did not find similar associations; however, we did observe a positive association between expression miR-151–3p and urinary concentrations of MiBP (p = 0.04). Two studies measured BPA and EV-miRNA in placental tissue and found significant increases in miR-146a (Avissar-Whiting et al. 2010; De Felice et al. 2015a). However, we did not identify a signal with BPA or our parabens. Additionally, an in vitro placental cell line exposed to MEHP increased expression of miR-16 (Meruvu et al. 2016). A recent epidemiology study found decreased expression of free-floating miR-142 in placenta to be associated with urinary phthalate metabolite concentrations (LaRocca et al. 2016). Furthermore, an inverse association was identified between free-floating miR-15a and miR-185 in placenta and urinary phenols (LaRocca et al. 2016). In our current study, we found associations between miR-15a and propyl paraben (p = 0.01), however we did not identify any associations with miR-142 or miR-185. Inconsistencies observed between these prior studies and ours are likely the result of differences in species (human vs rodent) or type of study (in vitro), measurement of microRNAs (free floating vs EV packaged), underlying study populations (USA vs Israel), and exposure type (induced in vitro exposures vs urinary biomarkers capturing environmental exposure). Most of the differences, however, are likely due to differences in the type of tissue used to measure EV-miRNAs. EV-miRNAs are tissue specific and certain EV-miRNAs are present in placental tissues that are not present in follicular fluid (Landgraf et al. 2007).
This study has several limitations. First, our exposure and outcome measurements were not taken in the same tissue type. Although urinary concentrations of phthalates, phenols and DINCH are the gold standard biomarker of exposure, it is unknown how representative urinary concentrations are of concentrations within the follicle. Second, we ran our GO and KEGG pathway analyses on a relatively small number of EV-miRNAs. To minimize that our results may be subject to overfitting of the selected pathways by the software, we set our inclusion of genes to those found by at least six separate miRNA-to-gene prediction softwares. Third, we only quantified EV-miRNA profiles from one follicle and it is unknown whether this represents EV-miRNAs within a woman’s cohort of follicles. Additionally, we present our results as FDR adjustments for the 133 EV-miRNA and not for the FDR adjustments by EV-miRNA and biomarker. Our results, therefore, may be subject to spurious associations and false positives. As it was beyond the scope of our study, we did not collect serum samples in the current studies and could not look adjust to patient hormonal levels as FSH, LH or estradiol. Including only young patients with normal ovarian response, we assume that all of those were in the normal range. We are also limited by the current methods of extracting EV-miRNAs. We can assure we have a pellet enriched in EVs, however, we cannot assume that it is a pure EV fraction. It is possible that miRNAs in the pellet were derived both from necrotic cells, bound to proteins, or packaged in EVs (Endzelins et al. 2017). However, our protocol has been optimized by running thousands of samples from a lab that has a strong experience in large epidemiological studies investigating EV-miRNAs and random samples were checked by Nanosight in order to evaluate EV size distribution. As it was beyond the scope of this study to assess what was the origin of these EV-miRNA in the ovarian follicle, we encourage further studies to test the origin of these EV-miRNA as it may add important information regarding the cell types that are more vulnerable to EDCs. Last, only internal controls were present in replicate reactions and miRNAs were analyzes in a single reaction, as required by the standard protocol of this technique. Addition of technical replicates might help to monitor intra- and inter-assay variability. Despite these limitations, our study has several strengths. This is the first study, to our knowledge, to examine any environmental exposure with EV-miRNA profiles in follicular fluid. In the current study, It is also one of the largest studies to examine EV-miRNAs in follicular fluid, a target sample for IVF outcomes. This study may provide potential mechanistic information which could support previous animal and epidemiological studies on phthalate and phenol exposure and adverse reproductive outcomes such as reduced antral follicle counts, pregnancy loss, and reduced live birth rates.
5. Conclusion:
Our findings suggest that select urinary phenol and phthalate biomarkers are associated with altered expression of eight EV-miRNAs in follicular fluid, hsa-let-7c, hsa-miR-375, hsa-miR-24, hsa-miR-19a, hsa-miR-125b, hsa-miR-106b, hsa-miR-374a, and hsa-miR-15b. Understanding how environmental stimuli might alter the function of EV-miRNAs, especially in follicular fluid, will help elucidate how exposures to these environmental chemicals might affect biological pathways associated with female fertility.
Supplementary Material
HIGHLIGHTS:
Women are exposed to phthalate and phenols before conception.
Exposure to phenols and phthalates are correlated with expression of extracellular vesicle microRNAs (EV-miRNAs) in follicular fluid.
Gene targets and pathways of these EV-miRNAs are related to follicular development and oocyte competence.
Acknowledgements:
The authors gratefully acknowledge Antonia Calafat, Xiaoyun Ye, Manori Silva, Prabha Dwivedi, Ella Samandar, Jim Preau, Xiaoliu Zou, and Tao Jia (CDC, Atlanta, GA) for measuring the urinary concentrations of the environmental biomarkers.
Funding:
This work was supported by the National Institutes of Environmental Health Sciences [Grant R21-ES024236]; Israeli Science Foundation [Grant 1936/12]; the Environmental Health Fund, Israel [Grant 1301]; and Education and Research Center for Occupational Safety and Health CDC/NIOSH [grant award T42/OH008416]
Footnotes
Competing interests
The author(s) declare no competing interests.
Declarations of interest: none
Data Availability
The datasets analyzed during the current study are not publicly available due to protection of participant confidentiality but are available from the corresponding author on reasonable request with assurances and plans in place to protect confidentiality.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- Andrade GM; da Silveira JC; Perrini C; Del Collado M; Gebremedhn S; Tesfaye D; Meirelles FV; Perecin F The role of the PI3K-Akt signaling pathway in the developmental competence of bovine oocytes. PLOS ONE 2017;12:e0185045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avissar-Whiting M; Veiga K; Uhl K; Maccani M; Gagne L; Moen E; Marsit CJ Bisphenol A Exposure Leads to Specific MicroRNA Alterations in Placental Cells. Reproductive toxicology (Elmsford, NY) 2010;29:401–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A; Pesatori AC; Bertazzi PA Occupational and environmental agents as endocrine disruptors: experimental and human evidence. Journal of endocrinological investigation 2000;23:771–781 [DOI] [PubMed] [Google Scholar]
- Benjamini Y; Hochberg Y Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300 [Google Scholar]
- Bornehag CG; Sundell J; Weschler CJ; Sigsgaard T; Lundgren B; Hasselgren M; Hagerhed-Engman L The association between asthma and allergic symptoms in children and phthalates in house dust: a nested case-control study. Environmental health perspectives 2004;112:1393–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carwile JL; Ye X; Zhou X; Calafat AM; Michels KB Canned Soup Consumption and Urinary Bisphenol A: A Randomized Crossover Trial. Jama 2011;306:2218–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati L; Sendra R; Sibilia V; Celotti F Endocrine disrupters: the new players able to affect the epigenome. Frontiers in Cell and Developmental Biology 2015;3:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H; Liu C; Jiang H; Gao Y; Xu M; Wang J; Liu S; Fu Y; Sun X; Xu J; Zhang J; Dai L Regulatory Role of miRNA-375 in Expression of BMP15/GDF9 Receptors and its Effect on Proliferation and Apoptosis of Bovine Cumulus Cells. Cellular Physiology and Biochemistry 2017;41:439–450 [DOI] [PubMed] [Google Scholar]
- Chen X; Qin Q; Zhang W; Zhang Y; Zheng H; Liu C; Yang Y; Xiong W; Yuan J Activation of the PI3K–AKT–mTOR signaling pathway promotes DEHP-induced Hep3B cell proliferation. Food and Chemical Toxicology 2013;59:325–333 [DOI] [PubMed] [Google Scholar]
- Cho H; Kim SJ; Park H-W; Oh M-J; Yu SY; Lee SY; Park C; Han J; Oh J-H; Hwang SY; Yoon S-J A relationship between miRNA and gene expression in the mouse Sertoli cell line after exposure to bisphenol A. BioChip Journal 2010;4:75–81 [Google Scholar]
- Chou WC; Lee PH; Tan YY; Lin HC; Yang CW; Chen KH; Chuang CY An integrative transcriptomic analysis reveals bisphenol A exposure-induced dysregulation of microRNA expression in human endometrial cells. Toxicology in vitro : an international journal published in association with BIBRA 2017;41:133–142 [DOI] [PubMed] [Google Scholar]
- Craig ZR; Singh J; Gupta RK; Flaws JA Co-treatment of mouse antral follicles with 17β-estradiol interferes with mono-2-ethylhexyl phthalate (MEHP)-induced atresia and altered apoptosis gene expression. Reproductive toxicology (Elmsford, NY) 2014;45:45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felice B; Manfellotto F; Palumbo A; Troisi J; Zullo F; Di Carlo C; Sardo ADS; De Stefano N; Ferbo U; Guida M; Guida M Genome–wide microRNA expression profiling in placentas from pregnant women exposed to BPA. BMC Medical Genomics 2015a;8:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felice B; Manfellotto F; Palumbo A; Troisi J; Zullo F; Di Carlo C; Sardo ADS; De Stefano N; Ferbo U; Guida M; Guida M Genome–wide microRNA expression profiling in placentas from pregnant women exposed to BPA. BMC Medical Genomics 2015b;8:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derghal A; Djelloul M; Trouslard J; Mounien L An Emerging Role of micro-RNA in the Effect of the Endocrine Disruptors. Frontiers in Neuroscience 2016;10:318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro C Exosome-mediated communication in the ovarian follicle. Journal of assisted reproduction and genetics 2016;33:303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamanti-Kandarakis E; Bourguignon JP; Giudice LC; Hauser R; Prins GS; Soto AM; Zoeller RT; Gore AC Endocrine-disrupting chemicals: an Endocrine Society scientific statement. Endocrine reviews 2009;30:293–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre A; Haccard O; Jessus C Mos in the oocyte: how to use MAPK independently of growth factors and transcription to control meiotic divisions. Journal of signal transduction 2011;2011:350412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweep H; Sticht C; Pandey P; Gretz N miRWalk – Database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. Journal of Biomedical Informatics 2011;44:839–847 [DOI] [PubMed] [Google Scholar]
- Ehrlich S; Williams PL; Missmer SA; Flaws JA; Berry KF; Calafat AM; Ye X; Petrozza JC; Wright D; Hauser R Urinary Bisphenol A Concentrations and Implantation Failure among Women Undergoing in Vitro Fertilization. Environmental health perspectives 2012;120:978–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enderle D; Spiel A; Coticchia CM; Berghoff E; Mueller R; Schlumpberger M; Sprenger-Haussels M; Shaffer JM; Lader E; Skog J; Noerholm M Characterization of RNA from Exosomes and Other Extracellular Vesicles Isolated by a Novel Spin Column-Based Method. PloS one 2015;10:e0136133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endzelins E; Berger A; Melne V; Bajo-Santos C; Sobolevska K; Abols A; Rodriguez M; Santare D; Rudnickiha A; Lietuvietis V; Llorente A; Line A Detection of circulating miRNAs: comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC cancer 2017;17:730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige JN; Gelman L; Rossi D; Zoete V; Metivier R; Tudor C; Anghel SI; Grosdidier A; Lathion C; Engelborghs Y; Michielin O; Wahli W; Desvergne B The endocrine disruptor monoethyl-hexylphthalate is a selective peroxisome proliferator-activated receptor gamma modulator that promotes adipogenesis. J Biol Chem 2007;282:19152–19166 [DOI] [PubMed] [Google Scholar]
- Ferraretti AP; La Marca A; Fauser BC; Tarlatzis B; Nargund G; Gianaroli L; Definition E w.g.o.P.O.R. ESHRE consensus on the definition of ‘poor response’ to ovarian stimulation for in vitro fertilization: the Bologna criteria. Human reproduction 2011;26:1616–1624 [DOI] [PubMed] [Google Scholar]
- Fleisch AF; Wright RO; Baccarelli AA Environmental Epigenetics: A Role in Endocrine Disease? Journal of molecular endocrinology 2012;49:R61–R67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner C; Di Vizio D; Sahoo S; Thery C; Witwer KW; Wauben M; Hill AF Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. Journal of extracellular vesicles 2016;5:32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore AC; Chappell VA; Fenton SE; Flaws JA; Nadal A; Prins GS; Toppari J; Zoeller RT EDC-2: The Endocrine Society’s Second Scientific Statement on Endocrine-Disrupting Chemicals. Endocr Rev 2015;36:E1–e150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y; Kannan K A Survey of Phthalates and Parabens in Personal Care Products from the United States and Its Implications for Human Exposure. Environmental Science & Technology 2013;47:14442–14449 [DOI] [PubMed] [Google Scholar]
- Gupta RK; Singh JM; Leslie TC; Meachum S; Flaws JA; Yao HHC Di-(2-ethylhexyl) phthalate and Mono-(2-ethylhexyl) phthalate Inhibit Growth and Reduce Estradiol Levels of Antral Follicles In Vitro. Toxicology and applied pharmacology 2010;242:224–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon PR; Peretz J; Flaws JA Daily Exposure to Di(2-ethylhexyl) Phthalate Alters Estrous Cyclicity and Accelerates Primordial Follicle Recruitment Potentially Via Dysregulation of the Phosphatidylinositol 3-Kinase Signaling Pathway in Adult Mice. Biology of Reproduction 2014;90:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser R; Calafat A PHTHALATES AND HUMAN HEALTH. Occupational and Environmental Medicine 2005;62:806–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser R; Gaskins AJ; Souter I; Smith KW; Dodge LE; Ehrlich S; Meeker JD; Calafat AM; Williams PL Urinary Phthalate Metabolite Concentrations and Reproductive Outcomes among Women Undergoing in Vitro Fertilization: Results from the EARTH Study. Environmental health perspectives 2016a;124:831–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser R; Gaskins AJ; Souter I; Smith KW; Dodge LE; Ehrlich S; Meeker JD; Calafat AM; Williams PL; Team ES Urinary Phthalate Metabolite Concentrations and Reproductive Outcomes among Women Undergoing in Vitro Fertilization: Results from the EARTH Study. Environmental health perspectives 2016b;124:831–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennet ML; Combelles CM The antral follicle: a microenvironment for oocyte differentiation. The International journal of developmental biology 2012;56:819–831 [DOI] [PubMed] [Google Scholar]
- Hoshino Y; Sato E Protein kinase B (PKB/Akt) is required for the completion of meiosis in mouse oocytes. Developmental Biology 2008;314:215–223 [DOI] [PubMed] [Google Scholar]
- Leek Jeffery T., W.E. J, Parker Hilary S., Fertig Elana J., Jaffe Andrew E., Storey John D., Zhang Yuqing, and Torres Leonardo Collado. sva: Surrogate Variable Analysis. R; 2017 [Google Scholar]
- Jukic AM; Calafat AM; McConnaughey DR; Longnecker MP; Hoppin JA; Weinberg CR; Wilcox AJ; Baird DD Urinary Concentrations of Phthalate Metabolites and Bisphenol A and Associations with Follicular-Phase Length, Luteal-Phase Length, Fecundability, and Early Pregnancy Loss. Environmental health perspectives 2016;124:321–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K-H; Seo Y-M; Kim E-Y; Lee S-Y; Kwon J; Ko J-J; Lee K-A The miR-125 family is an important regulator of the expression and maintenance of maternal effect genes during preimplantational embryo development. Open Biology 2016;6:160181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropp J; Khatib H Characterization of microRNA in bovine in vitro culture media associated with embryo quality and development. Journal of Dairy Science 2015;98:6552–6563 [DOI] [PubMed] [Google Scholar]
- Landgraf P; Rusu M; Sheridan R; Sewer A; Iovino N; Aravin A; Pfeffer S; Rice A; Kamphorst AO; Landthaler M; Lin C; Socci ND; Hermida L; Fulci V; Chiaretti S; Foà R; Schliwka J; Fuchs U; Novosel A; Müller R-U; Schermer B; Bissels U; Inman J; Phan Q; Chien M; Weir DB; Choksi R; De Vita G; Frezzetti D; Trompeter H-I; Hornung V; Teng G; Hartmann G; Palkovits M; Di Lauro R; Wernet P; Macino G; Rogler CE; Nagle JW; Ju J; Papavasiliou FN; Benzing T; Lichter P; Tam W; Brownstein MJ; Bosio A; Borkhardt A; Russo JJ; Sander C; Zavolan M; Tuschl T A Mammalian microRNA Expression Atlas Based on Small RNA Library Sequencing. Cell 2007;129:1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRocca J; Binder AM; McElrath TF; Michels KB First-Trimester Urine Concentrations of Phthalate Metabolites and Phenols and Placenta miRNA Expression in a Cohort of U.S. Women. Environmental health perspectives 2016;124:380–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J; Kho Y; Kim P-G; Ji K Exposure to bisphenol S alters the expression of microRNA in male zebrafish. Toxicology and Applied Pharmacology 2018;338:191–196 [DOI] [PubMed] [Google Scholar]
- Leek JT; Johnson WE; Parker HS; Jaffe AE; Storey JD The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012;28:882–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SC; Tang P; Lin WC Intronic microRNA: discovery and biological implications. DNA and cell biology 2007;26:195–207 [DOI] [PubMed] [Google Scholar]
- Lim LP; Lau NC; Garrett-Engele P; Grimson A; Schelter JM; Castle J; Bartel DP; Linsley PS; Johnson JM Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005;433:769. [DOI] [PubMed] [Google Scholar]
- Liu C; Yuan B; Chen H; Xu M; Sun X; Xu J; Gao Y; Chen C; Jiang H; Zhang J Effects of MiR-375BMPR2 as a Key Factor Downstream of BMP15/GDF9 on the Smad1/5/8 and Smad2/3 Signaling Pathways. Cellular Physiology and Biochemistry 2018a;46:213–225 [DOI] [PubMed] [Google Scholar]
- Liu J; Wang W; Zhu J; Li Y; Luo L; Huang Y; Zhang W Di(2 ethylhexyl) phthalate (DEHP) influences follicular development in mice between the weaning period and maturity by interfering with ovarian development factors and microRNAs. Environmental Toxicology 2018b;0 [DOI] [PubMed] [Google Scholar]
- Liu L; Rajareddy S; Reddy P; Du C; Jagarlamudi K; Shen Y; Gunnarsson D; Selstam G; Boman K; Liu K Infertility caused by retardation of follicular development in mice with oocyte-specific expression of Foxo3a. Development 2007;134:199. [DOI] [PubMed] [Google Scholar]
- Lopez-Cardona AP; Perez-Cerezales S; Fernandez-Gonzalez R; Laguna-Barraza R; Pericuesta E; Agirregoitia N; Gutierrez-Adan A; Agirregoitia E CB1 cannabinoid receptor drives oocyte maturation and embryo development via PI3K/Akt and MAPK pathways. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 2017;31:3372–3382 [DOI] [PubMed] [Google Scholar]
- Ludwig N; Leidinger P; Becker K; Backes C; Fehlmann T; Pallasch C; Rheinheimer S; Meder B; Stähler C; Meese E; Keller A Distribution of miRNA expression across human tissues. Nucleic Acids Research 2016;44:3865–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu BE; Green SR; Childers CL; Holahan MR; Storey KB The roles of hippocampal microRNAs in response to acute postnatal exposure to di(2-ethylhexyl) phthalate in female and male rats. NeuroToxicology 2017;59:98–104 [DOI] [PubMed] [Google Scholar]
- Machtinger R; Gaskins AJ; Racowsky C; Mansur A; Adir M; Baccarelli AA; Calafat AM; Hauser R Urinary concentrations of biomarkers of phthalates and phthalate alternatives and IVF outcomes. Environment international 2018a;111:23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machtinger R; Gaskins AJ; Racowsky C; Mansur A; Adir M; Baccarelli AA; Calafat AM; Hauser R Urinary concentrations of biomarkers of phthalates and phthalate alternatives and IVF outcomes. Environment international 2018b;111:23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machtinger R; Laurent LC; Baccarelli AA Extracellular vesicles: roles in gamete maturation, fertilization and embryo implantation. Human reproduction update 2016a;22:182–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machtinger R; Laurent LC; Baccarelli AA Extracellular vesicles: roles in gamete maturation, fertilization and embryo implantation. Human Reproduction Update 2016b;22:182–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makker A; Goel MM; Mahdi AA PI3K/PTEN/Akt and TSC/mTOR signaling pathways, ovarian dysfunction, and infertility: an update. Journal of Molecular Endocrinology 2014;53:R103–R118 [DOI] [PubMed] [Google Scholar]
- Marabita F; de Candia P; Torri A; Tegner J; Abrignani S; Rossi RL Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Briefings in bioinformatics 2016;17:204–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker JD; Ferguson KK Urinary Phthalate Metabolites Are Associated With Decreased Serum Testosterone in Men, Women, and Children From NHANES 2011–2012. The Journal of Clinical Endocrinology and Metabolism 2014;99:4346–4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meruvu S; Zhang J; Bedi YS; Choudhury M Mono-(2-ethylhexyl) phthalate induces apoptosis through miR-16 in human first trimester placental cell line HTR-8/SVneo. Toxicology in Vitro 2016;31:35–42 [DOI] [PubMed] [Google Scholar]
- Messerlian C; Souter I; Gaskins AJ; Williams PL; Ford JB; Chiu Y-H; Calafat AM; Hauser R Urinary phthalate metabolites and ovarian reserve among women seeking infertility care. Human reproduction (Oxford, England) 2016;31:75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P; Van Vlierberghe P; De Weer A; Muth D; Westermann F; Speleman F; Vandesompele J A novel and universal method for microRNA RT-qPCR data normalization. Genome Biology 2009;10:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mínguez-Alarcón L; Chiu Y-H; Messerlian C; Williams PL; Sabatini ME; Toth TL; Ford JB; Calafat AM; Hauser R; for the Earth Study, T. Urinary paraben concentrations and in vitro fertilization outcomes among women from a fertility clinic. Fertility and sterility 2016;105:714–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miodovnik A; Engel SM; Zhu C; Ye X; Soorya LV; Silva MJ; Calafat AM; Wolff MS Endocrine Disruptors and Childhood Social Impairment. Neurotoxicology 2011;32:261–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergoli L; Cantone L; Favero C; Angelici L; Iodice S; Pinatel E; Hoxha M; Dioni L; Letizia M; Albetti B; Tarantini L; Rota F; Bertazzi PA; Tirelli AS; Dolo V; Cattaneo A; Vigna L; Battaglia C; Carugno M; Bonzini M; Pesatori AC; Bollati V Extracellular vesicle-packaged miRNA release after short-term exposure to particulate matter is associated with increased coagulation. Particle and Fibre Toxicology 2017;14:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippat C; Mortamais M; Chevrier C; Petit C; Calafat AM; Ye X; Silva MJ; Brambilla C; Pin I; Charles MA; Cordier S; Slama R Exposure to phthalates and phenols during pregnancy and offspring size at birth. Environmental health perspectives 2012;120:464–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R-Core-Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2017 [Google Scholar]
- Raposo G; Stoorvogel W Extracellular vesicles: exosomes, microvesicles, and friends. The Journal of cell biology 2013;200:373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimon-Dahari N; Yerushalmi-Heinemann L; Alyagor L; Dekel N Ovarian Folliculogenesis in: Piprek RP, ed. Molecular Mechanisms of Cell Differentiation in Gonad Development. Cham: Springer: International Publishing; 2016 [DOI] [PubMed] [Google Scholar]
- Rodgers RJ; Irving-Rodgers HF Formation of the ovarian follicular antrum and follicular fluid. Biol Reprod 2010;82:1021–1029 [DOI] [PubMed] [Google Scholar]
- Sætrom P; Snøve O Jr; Rossi JJ Epigenetics and MicroRNAs. Pediatric Research 2007;61:17R. [DOI] [PubMed] [Google Scholar]
- Sánchez F; Smitz J Molecular control of oogenesis. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 2012;1822:1896–1912 [DOI] [PubMed] [Google Scholar]
- Silva MJ; Jia T; Samandar E; Preau JL Jr.; <au, Calafat, AM Environmental exposure to the plasticizer 1,2cyclohexane dicarboxylic acid, diisononyl ester (DINCH) in U.S. adults (2000–2012). Environmental research 2013;126:159–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MJ; Wong LY; Samandar E; Preau JL; Calafat AM; Ye X Exposure to di-2-ethylhexyl terephthalate in a convenience sample of U.S. adults from 2000 to 2016. Archives of toxicology 2017;91:3287–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilghman SL; Bratton MR; Segar HC; Martin EC; Rhodes LV; Li M; McLachlan JA; Wiese TE; Nephew KP; Burow ME Endocrine disruptor regulation of microRNA expression in breast carcinoma cells. PLoS One 2012;7:e32754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toft G; Jonsson BA; Lindh CH; Jensen TK; Hjollund NH; Vested A; Bonde JP Association between pregnancy loss and urinary phthalate levels around the time of conception. Environmental health perspectives 2012;120:458–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J; Vasaikar S; Shi Z; Greer M; Zhang B WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Research 2017;45:W130–W137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-P; Mu X-Y; Guo M; Wang Y-J; Teng Z; Mao G-P; Niu W-B; Feng L-Z; Zhao L-H; Xia G-L Transforming Growth Factor-β Signaling Participates in the Maintenance of the Primordial Follicle Pool in the Mouse Ovary. The Journal of Biological Chemistry 2014;289:8299–8311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JA; Baxter DH; Zhang S; Huang DY; How Huang K; Jen Lee M; Galas DJ; Wang K The MicroRNA Spectrum in 12 Body Fluids. Clinical Chemistry 2010;56:1733–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer KW; Buzas EI; Bemis LT; Bora A; Lasser C; Lotvall J; Nolte-’t Hoen EN; Piper MG; Sivaraman S; Skog J; Thery C; Wauben MH; Hochberg F Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. Journal of extracellular vesicles 2013;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez-Mó M; Siljander PRM; Andreu Z; Bedina Zavec A; Borràs FE; Buzas EI; Buzas K; Casal E; Cappello F; Carvalho J; Colás E; Cordeiro-da Silva A; Fais S; Falcon-Perez JM; Ghobrial IM; Giebel B; Gimona M; Graner M; Gursel I; Gursel M; Heegaard NHH; Hendrix A; Kierulf P; Kokubun K; Kosanovic M; Kralj-Iglic V; Krämer-Albers E-M; Laitinen S; Lässer C; Lener T; Ligeti E; Linē A; Lipps G; Llorente A; Lötvall J; Manček-Keber M; Marcilla A; Mittelbrunn M; Nazarenko I; Nolte-’t Hoen ENM; Nyman TA; O’Driscoll L; Olivan M; Oliveira C; Pállinger É; del Portillo HA; Reventós J; Rigau M; Rohde E; Sammar M; Sánchez-Madrid F; Santarém N; Schallmoser K; Stampe Ostenfeld M; Stoorvogel W; Stukelj R; Van der Grein SG; Helena Vasconcelos M; Wauben MHM; De Wever O Biological properties of extracellular vesicles and their physiological functions. Journal of Extracellular Vesicles 2015;4:27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X; Kuklenyik Z; Needham LL; Calafat AM Automated on-line column-switching HPLC-MS/MS method with peak focusing for the determination of nine environmental phenols in urine. Analytical chemistry 2005;77:5407–5413 [DOI] [PubMed] [Google Scholar]
- Yu C; Li M; Wang Y; Liu Y; Yan C; Pan J; Liu J; Cui S miR-375 mediates CRH signaling pathway in inhibiting E2 synthesis in porcine ovary. Reproduction 2017;153:63–73 [DOI] [PubMed] [Google Scholar]
- Zeka F; Mestdagh P; Vandesompele J RT-qPCR-based quantification of small non-coding RNAs. Methods in molecular biology (Clifton, NJ) 2015;1296:85–102 [DOI] [PubMed] [Google Scholar]
- Zhang B; Kirov S; Snoddy J WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Research 2005;33:W741–W748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L; Huang J; Yang N; Greshock J; Liang S; Hasegawa K; Giannakakis A; Poulos N; O’BrienJenkins A; Katsaros D; Butzow R; Weber BL; Coukos G Integrative genomic analysis of phosphatidylinositol 3’-kinase family identifies PIK3R3 as a potential therapeutic target in epithelial ovarian cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2007;13:5314–5321 [DOI] [PubMed] [Google Scholar]
- Zuccotti M; Merico V; Cecconi S; Redi CA; Garagna S What does it take to make a developmentally competent mammalian egg? Human reproduction update 2011;17:525–540 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.