Abstract
Alzheimer’s disease (AD) is a devastating, progressive neurodegenerative disorder that leads to severe cognitive impairment in elderly patients. Chronic neuroinflammation plays an important role in the AD pathogenesis. Glia maturation factor (GMF), a proinflammatory molecule discovered in our laboratory is significantly upregulated in various regions of AD brains. We have previously reported that GMF is predominantly expressed in the reactive glial cells surrounding the amyloid plaques (APs) in the mouse and human AD brain. Microglia are the major source of proinflammatory cytokines and chemokines including GMF. Recently clustered regularly interspaced short palindromic repeats (CRISPR) based genome editing has been recognized to study the functions of genes that are implicated in various diseases. Here we investigated if CRISPR-Cas9-mediated GMF gene editing leads to inhibition of GMF expression and suppression of microglial activation. Confocal microscopy of murine BV2 microglial cell line transduced with an adeno-associated virus (AAV) co-expressing Staphylococcus aureus (Sa) Cas9 and a GMF specific guide RNA (GMF-sgRNA) revealed few cells expressing SaCas9 while lacking GMF expression, thereby confirming successful GMF gene editing. To further improve GMF gene editing efficiency we developed lentiviral vectors (LVs) expressing either Streptococcus pyogenes (Sp) Cas9 or GMF-sgRNAs. BV2 cells co-transduced with LVs expressing SpCas9 and GMF-sgRNAs revealed reduced GMF expression and the presence of indels in the exons 2 and 3 of the GMF coding sequence. Lipopolysaccharide (LPS) treatment of GMF-edited cells led to reduced microglial activation as shown by reduced p38 MAPK phosphorylation. We believe that targeted in vivo GMF gene editing has a significant potential for developing a unique and novel AD therapy.
Keywords: Adeno-associated virus, Alzheimer’s Disease, CRISPR-Cas9, Glia maturation factor, Lentiviral vectors, Microglia
Introduction
The worldwide incidence of Alzheimer’s disease (AD) is increasing at an alarming pace and is indeed a significant health problem especially due to an increase in the life expectancy and aging population. AD is a progressive neurodegenerative disorder that causes an irreversible cognitive decline in an estimated 5.5 million Americans with $259 billion in healthcare costs in 2017. If the present trend continues, by 2050 the number of AD patients will surpass 16 million with ~$1.1 trillion in healthcare costs (Alzheimer's Association, Chicago, IL) thereby necessitating the development of novel therapeutic strategies to effectively treat AD. Although, a human monoclonal antibody Aducanumab that selectively targets aggregated amyloid-β (Aβ) has shown promising results in the ongoing phase 3 clinical trials, many other clinical trials have witnessed major setbacks including the recent trial of a humanized monoclonal antibody Solanezumab designed to increase the clearance of soluble Aβ peptides from the brain of the AD patients [1-5]. Therefore, towards the fulfillment of the unmet clinical need for an effective therapy to combat AD, our ultimate goal is to develop a robust AD patient-specific personalized precision guided targeted gene editing and stem cell based regenerative therapy.
Recent studies in AD mouse models suggest that Aβ seeding potency peaks during the early phase of AD pathogenesis, thereby highlighting the necessity of directing novel therapeutic strategies at the earliest stages of AD development [6]. Chronic neuroinflammation and neurodegeneration play crucial roles in the pathophysiology of AD [7-12]. Microglia are the key players in neuroinflammation [13-16]. GMF, a proinflammatory molecule, discovered and characterized by our research group has been shown to be highly expressed in AD brain and is associated with neurodegeneration [17-28]. Our prior studies have demonstrated that GMF plays an important role in Parkinson’s disease (PD) pathogenesis and GMF deficiency protects the dopaminergic neuronal loss in the mouse model of Parkinson’s disease [29]. We have also recently shown that GMF-dependent inhibition of mitochondrial PGC1-α triggers oxidative stress-mediated apoptosis in dopaminergic neuronal cells [30]. Although, there are several different approaches including siRNA, shRNA and gene targeting to inhibit or knockdown gene expression, most recently CRISPR-Cas9-mediated gene editing has shown very promising results both in vitro as well as in vivo [31-46]. Here we hypothesized that reducing GMF expression by CRISPR-Cas9-mediated gene editing in microglia represents a novel approach to reduce neuroinflammation and neurodegeneration.
Development of CRISPR based gene editing in microglial cells has proven to be challenging. Here, we have used two different approaches to achieve GMF gene editing in BV2 microglial cells in vitro. In our first approach, we developed a recombinant adeno-associated viral vector AAV-SaCas9-GMF-sgRNA co-expressing Staphylococcus aureus (Sa) Cas9 and GMF sgRNA. Transduction of BV2 cells using AAV-SaCas9-GMF-sgRNA leads to limited GMF gene editing as revealed by confocal microscopy. To further improve GMF gene editing efficiency we developed a dual lentiviral vector system. The first lentiviral vector LV-EF1α-Cas9, expresses Streptococcus pyogenes (Sp) Cas9 and SV40 promoter driven eGFP and Neomycin. The second lentiviral vector expresses a U6 promoter driven GMF sgRNA and SV40 promoter driven mCherry and puromycin. Sequential transduction and selection was performed to generate dual stable BV2 cells simultaneously co-expressing SpCas9 and GMF sgRNA. Our confocal microscopy, flow cytometry, Guide-it Resolvase based mutational analysis and DNA sequencing data validate and confirm successful GMF gene editing in BV2 cells. Our experimental approach and current results lay a strong foundation for the in vivo gene editing as an approach to develop the next generation of precision guided personalized molecular medicine not only for AD but also for other neurodegenerative diseases.
Materials and Methods
Cell Lines:
BV2 [47,48], an immortalized microglial cell line extensively used as a substitute for primary microglia [49] was cultured in DMEM supplemented with 10% FBS, Penicillin (100 U/ml)-streptomycin (100 mg/ml) (Catalog #15140122), 1x Glutamax (Catalog #35050061) and 1x Sodium Pyruvate (catalog #11360070) (all from ThermoFisher Scientific, Waltham, MA ) and 5% CO2 at 37°C. The cells were typsinized using 0.25% Trypsin EDTA (Catalog # 25200-056, ThermoFisher Scientific, Waltham, MA) solution and plated at a density of 1×105 cells per well in a six well plate or 20,000 cells per well of an 8 chamber slide. HEK 293 and HEK 293T used for AAV and lentiviral production were cultured similar to BV2 cells.
Generation of AAV:
A pair of GMF-specific guide RNAs that are compatible with SaCas9 [37] were designed using the Benchling website (https://benchling.com/) and are depicted in Figure 2. The sequences of the GMF Forward and GMF Reverse primers are as follows and were synthesized by Integrated DNA Technologies (IDT: Integrated DNA Technologies Inc, Coralville, IA) :
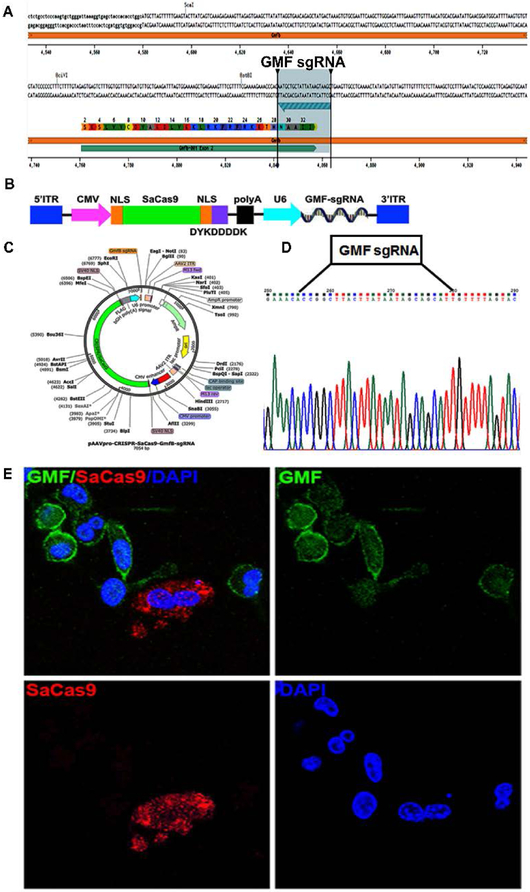
Figure 2: Development of AAV-SaCa9-GMF-sgRNA:
a) Selection of GMF sgRNA: The panel shows the DNA sequence of mouse GMF exon 2 flanked by the introns. The GMF amino acid sequence is indicated by single letter codes. The GMF sgRNA sequence located in the anticlockwise orientation on the lower strand is depicted by the green arrow flanked by the two vertical bars, b) Generation of AAV-SaCas9-GMF-sgRNA: The AAV ITRs flank the expression cassette comprising a CMV promoter driving the expression of a nuclear localized SaCas9 which also harbors the DYKDDDK tag, followed by a polyA signal. The second expression casette bears a U6 promoter driven GMF sgRNA, c) Detailed vector map depicting various elements present in pAAVpro-CRISPR-SaCas9-GMF-sgRNA, d) The DNA sequence of the GMF sgRNA cloned into the AAV vector is depicted flanked by the sequence of the expression cassette, e) Targeted GMF gene editing mediated by AAV-SaCas9-GMF-sgRNA in BV2 cells. The immunostaining for the GMF (green) and the DYKDDDK tag (red) indicates that the non transduced BV2 cells display significant intracellular GMF expression with higher concentrations towards the cell periphery and on the plasma membrane. The GMF non-edited cells do not display any red color and are therefore negative for SaCas9 expression. GMF gene edited cells do not exhibit GMF expression and are higly positive for SaCas9 expression thereby indicating biallelic GMF gene editing mediated by SaCas9 and GMF-sgRNA.
GMF F: ACCGGCTTACTTATAATAGCAGCATT
GMF R: AAACAATGCTGCTATTATAAGTAAGC
The underlined sequences are compatible with the complementary sequences present downstream of the U6 promoter in the prelinearized AAVpro CRISPR/SaCas9 Vector System (Catalog # 632618, Takara Bio USA, Mountain View, CA) that was used for cloning of the in vitro annealed GMF sgRNA. The resulting clones were sequenced to confirm the DNA sequence of the GMF sgRNA. The sequence verified pAAV-SaCas9-GMF-sgRNA clone, pAAV-Helper and the AAV rep-cap plasmid pGG-B1 (Addgene, Cambridge, MA, UMCMTA17-380; kindly provided by Miguel Sena-Esteves, University of Massachusetts, Worcester) were transformed into One Shot Stbl3 chemically competent E. coli (Catalog # C737303, ThermoFisher Scientific, Waltham, MA) as per the manufacturer’s protocol. Large-scale plasmid DNA was isolated using Endofree Plasmid Maxi kit (Catalog # 12362, Qiagen Inc, Germantown, MD). Recombinant AAV-SaCas9-GMF-sgRNA virus was generated by co-transfection of pAAV-SaCas9-GMF-sgRNA, pAAV-Helper and pGG-B1in HEK 293 cells using a calcium phosphate transfection kit (Catalog #631312, Takara Bio USA, Mountain View CA). Briefly, HEK 293 cells in the exponential phase were plated at a cell density of 2.8×107 cells per T225 cm2tissue culture flasks 24 hour prior to transfection. The cells were subjected to fresh medium change 2 hours prior to transfection. For generating recombinant AAV, 50 μg pAAV-SaCas9-GMF-sgRNA, 80 μg pAAV-Helper and 72 μg pGG-B1 were mixed in a sterile 15 ml polypropylene tube and the final volume of 2364 μl was made up with water. Next, 336 μl calcium phosphate solution was added dropwise while vortexing gradually. To this mixture, 2700 μl 2X HEBS was added dropwise while gradually vortexing. The transfection mix was left at room temperature for 15 minutes and added dropwise to the medium in T225 cm2 tissue culture flask. The transfected cells were washed with PBS 12 hours post transfection and replaced with fresh medium. In a modification of the protocol instead of harvesting the transfected cells, the cell supernatant was harvested 72 hours post transfection and centrifuged at 1500 rpm for 15 minutes to remove cellular debris. Carefully the supernatant was transferred to a 50 ml conical tube and subjected to overnight concentration using AAVanced Concentration reagent (System Biosciences, LLC, Palo Alto, CA) as per the manufacturer’s recommendations. The supernatant was centrifuged at 3000 rpm for 30 minutes at 4°C. The concentrated virus was resuspended in Opti-Mem and frozen in 10-20 μl aliquots at −80°C. Infectious virus titer was determined by transduction of HT1080 cells in 12 well plates in triplicates followed by fluorescence microscopy.
Generation of lentiviral vectors:
The lentiviral expression vectors expressing Streptococcus pyogenes Cas9 (CP-LvC9NU-09) and GMF sgRNAs (MCP232778-LvSG03-3-B) or scrambled sgRNA Control (CCPCTR01-LvSG03) were purchased as glycerol stocks (Genecopoeia, Rockville, MD). The nucleotide sequences of the three different GMF sgRNAs as well as scrambled sgRNA used in the present study include: a) GMF sgRNA1: ACTTATAATAGCAGCATTGT, b) GMF sgRNA2: TGACAAGGATGAACGCCTGG, c) GMF sgRNA3: GGTGCTGGATGAGGAGCTCG and Scrambled sgRNA: GCTTCGCGCCGTAGTCTTAG respectively. Large-scale plasmid DNA was isolated using Endofree Plasmid Maxi kit (Catalog # 12362, Qiagen Inc, Germantown, MD). HIV based VSV envelope pseudotyped lentiviral vectors were generated in HEK 293T cells using a calcium phosphate transfection kit (Catalog #631312, Takara Bio USA, Mountain View, CA). Briefly, HEK 293T cells in the exponential phase were plated in T225 cm2 tissue culture flasks 24 hour prior to transfection. The cells were subjected to fresh medium change 2 hours prior to transfection. For generating lentiviral vectors, transfer vector 90 μg (pLV-GMF-sgRNA or pLV-CRISPR-Cas9), 60 μg pMDLg/pRRE (Addgene Plasmid # 78504), 25μg pRSV-Rev (Addgene Plasmid # 12253) and 32 μg pMD2.G (Addgene Plasmid #12251) were mixed in a sterile 15 ml polypropylene tube and the final volume of 2364 μl was made up with water. Next, 336 μl calcium phosphate solution was added dropwise while vortexing gradually. To this mixture, 2700 μl 2X HEBS was added dropwise while gradually vortexing. The transfection mix was left at room temperature for 15 minutes and added dropwise to the medium in T225 cm2 tissue culture flask. The transfected cells were washed 12 hours post transfection and replaced with fresh medium. The lentiviral supernatant was harvested 72 hours post transfection and centrifuged at 1500 rpm for 15 minutes to remove cellular debris. Carefully the supernatant was transferred to a 50 ml conical tube and subjected to overnight concentration using PEG-it Virus Precipitation Solution (System Biosciences, LLC, Palo Alto, CA) as per the manufacturer’s recommendations. The concentrated virus was resuspended in Opti-Mem and frozen in 10-20 μl aliquots at −80°C. Infectious virus titer was determined by transduction of HT1080 cells in 12 well plates in triplicates and by performing fluorescence microscopy.
Generation of BV2 Dual Stable Cell Lines:
Actively proliferating BV2 cells were plated at a cell density of 1×105 cells per well of the six well plate. For AAV and lentiviral transduction, the cells were washed with PBS and maintained in Opti-Mem. Viral transductions were performed at a low multiplicity of infection of 10 in the presence of 8 μg/ml polybrene (Catalog # H9268, Millipore-Sigma, St. Louis, MO) for 8 hours. The cells were washed and maintained in regular growth medium for 72 hours. The lentiviral vector LV-EF1α-SpCas9-eGFP transduced BV2 cells were trypsinized and plated in growth medium containing neomycin (750 μg/ml) for the generation of stable cell line BV2-CRISPR-Cas9. The stable BV2-CRISPR-Cas9 cell line was transduced with the second lentiviral vector LV-GMF-sgRNA-mCherry. A total of three different GMF edited BV2 cell lines using three different GMF-specific sgRNAs were generated.
Immunofluorescence labelling of GMF in BV2 cells:
For the Immunofluorescence staining, BV2 cells grown on the chamber slides were fixed in 4% paraformaldehyde and blocked with 3% bovine serum albumin/PBS plus 0.1% Triton X-100 and incubated with GMF antibody at a dilution of 1:100, (Catalog# ab224322, Abcam, Cambridge, MA) overnight at 4°C. After rinsing with PBS, the slides were incubated with Alexa Fluor 488 conjugated goat anti-rabbit IgG at a 1:500 dilution (Catalog#A-11008, ThermoFisher Scientific, Waltham, MA). For immunostaining of GMF gene edited BV2 cells, we used a 1:100 dilution of primary GMF-β (SP-61) mouse monoclonal antibody (Catalog# sc134347, Santa Cruz Biotechnology, Dallas, TX) followed by incubation with a 1:1000 dilution of Alexa Fluor 647 conjugated donkey anti-mouse IgG (H+L) highly cross-adsorbed secondary antibody (Catalog# A-31571, ThermoFisher Scientific, Waltham, MA). Samples were mounted on Vectashield Antifade Mounting Medium with DAPI solution (Catalog# H-1200, Vector Laboratories, Burlingame, CA). Images were acquired using the Leica TCP SP8 confocal microscope with a 405-nm diode laser and tunable supercontinuum white light laser using either 20x or 63x oil immersion objectives. The following excitation/emission band-pass wavelengths were used: 405/420-480 nm (DAPI), 495/505-550 nm (Alexa Fluor 488), 570/580-630 nm (Alexa Fluor 568) and 647/655-705 nm (Alexa Fluor 647). The images were processed using the Leica Application Suite X software.
Western Blots:
Approximately 2×106 cells were plated in each well of a six well plate for 24 hours and subsequently treated with 1 μg/ml LPS for 30 minutes at 37°C, washed with 1X PBS and lysed using the non denaturing cell lysis buffer as per the manufacturer’s protocol (Catalog #3500-1, Epitomics, Rocklin, CA) supplemented with protease and phosphatase inhibitor cocktail and subjected to 3 cycles of sonication. The cell lystate was centrifuged at 14000 rpm at 4°C for 15 minutes and the clear supernatant was used to determine the protein concentration using BCA. Cell lysates with equal amount of proteins (40 μg) containing the gel loading dye were boiled for 5 minutes and loaded along with Precision Plus Protein Dual Color Standards (Catalog #1610374, Biorad, Hercules, CA) and SeeBlue Plus2 Pre-Stained Standard (Catalog #LC5925, ThermoFisher Scientific, Waltham, MA) on to NuPAGE 4-12% Bis-Tris protein gel (Catalog #NP0335BOX, ThermoFisher Scientific, Waltham, MA) and resolved initially at 100 volts for 10 minutes and 200 volts for approximately 1 hour. The separated proteins were blotted on to PVDF transfer membrane (Catalog #88518, ThermoFisher Scientific, Waltham, MA) at 30 volts for 90 minutes using a wet protein transfer system and NuPage Transfer buffer (Catalog #NP0006-1, ThermoFisher Scientific, Waltham, MA) containing 20% methanol. The membrane was washed briefly and blocked with 5% BSA solution in 1XPBS containing 0.05% Tween 20 for 1 hour. The membrane was sequentially probed with anti-Phospho MAPKp38 and MAPKp38 antibodies (catalog# ab7952 and ab4822, Abcam, Cambridge, MA) at a dilution of 1:800 in 5% BSA containing 1XTBST overnight at 4°C with gentle rocking. The membranes were thoroughly washed 3 times in 1XTBST to remove any unbound antibody and incubated with goat-anti-Rabbit IgG HRP secondary antibody at a dilution of 1:2000 in 5% BSA containing 1XTBST for 1 hour at 4°C with gentle rocking. The membranes were washed thrice with 1XTBST at an interval of 5 minutes each. The specific signal was detected by exposing the membranes with SuperSignal West Pico PLUS Chemiluminescent Substrate (Catalog #34580, ThermoScientific, Rockford, IL) using ChemiDoc-It2 Imaging System and UVP VisionWorksLS Image Acquisition and Analysis Software (UVP LLC, Upland, CA). The blots were stripped using Restore Western Blot Stripping Buffer (Catalog #21059, ThermoScientific, Rockford, IL) as per the manufacturer’s recommendation. β-actin was used as a loading control and was probed using a primary mouse anti-β actin monoclonal antibody (Catalog #A5316, Millipore-Sigma, St. Louis, MO) and a secondary goat anti-mouse IgG HRP antibody (Catalog#sc2005, Santa Cruz Biotechnology, Inc, Dallas, TX). The experiment was performed independently 3 times with similar results.
Flow Cytometry:
Wild type and GMF edited BV2 cells were trypsinized using 0.25% trypsin, resuspended in the growth medium and counted. Approximately 2×106 cells were used for each staining reaction. The cells were washed with PBS, fixed in 4% paraformaldehyde for 15 minutes, washed with PBS and permeabilized using PBS containing 0.01% Tween 20 for 30 minutes. The cells were washed 3 times with PBS and blocked with PBS containing 3% BSA and 0.3M Glycine. Subsequently the cells were subjected to staining with an anti-GMF monoclonal antibody (G2-09) at a 1:100 dilution for 30 minutes. The cells were washed thrice with 1X PBS and stained with Alexa Fluor 647 conjugated secondary antibody (Catalog# A-31571 ThermoFisher Scientific, Waltham, MA) at a dilution of 1:400 for 30 minutes. The cells were washed 3 times to remove any unbound antibodies and also to reduce background fluorescence signal. The cells were analyzed by flow cytometry using BD LSRFortessa X-20. FlowJo software was used to analyze and display the flow cytometry data.
Mutational Analysis and DNA Sequencing of GMF Edited Clones:
The total genomic DNA was isolated from wild type and GMF edited BV2 cells using DNeasy Blood and Tissue Kit (Qiagen Inc, Germantown, MD) as per the manufacturer’s protocol. The DNA concentration and purity were measured by using Nanodrop 2000 Spectrophotometer (ThermoScientific, Rockford, IL). The BV2 genomic DNA sequence flanking the GMF gene edited sequence was PCR amplified using the following PCR primers that were synthesized by Integrated DNA Technologies (IDT: Integrated DNA Technologies Inc, Coralville, IA)
GMF Intron 1F GACTAAAGTGTGCGAATTCAAGCTTGGGAG
GMF Intron 2R CTACGAACACACAATGTGACTTCACATGGC
GMF Intron 2F CATAGACCAAGCTCAGGGTTATCCTTCCC
GMF Intron 3R GAGACACAGGGTGACTGAATGACATAGC
The PCR reaction was performed on a ABI GeneAmp PCR System 9700 using following conditions: an initial DNA denaturation at 98°C for 2 minutes followed by 35 cycles each at 98°C for 10 sec, 60°C for 15 sec, 68°C for 1 minute and finally cooling to 4°C. The PCR fragments were cloned into pCR4TOPO plasmid vector (Catalog #450071, ThermoFisher Scientific, Waltham, MA) and recombinant clones were selected by blue and white colony selection as per the manufacturer’s protocol. The plasmid DNA was isolated using Qiaprep Spin Miniprep Kit (Catalog# 27106, Qiagen Inc, Germantown, MD). The plasmid DNA clones were sequenced at the University of Missouri DNA Core Facility using T3 sequencing primer.
For mutational analysis a heteroduplex DNA hybridization was performed by combining the PCR products from wild type BV2 cells and GMF edited BV2 cells. Briefly 5 μl each of wild type BV2 PCR product and GMF edited BV2 PCR product were mixed together and DNA hybridization was performed a follows: Initial heating at 95°C 5 minutes, followed by 95°C-85°C temperature decrease at 2°C/sec, 85°C-25°C temperature decrease at 0.1°C/sec and final cooling to 4°C. Post hybridization the reaction mixture was treated with 1μl of Guide-it Resolvase and incubated at 37°C for 15 minutes. Entire reaction was run on a 1.8% agarose gel at 125 volts for 45 minutes. The cleavage efficiency was determined using ChemiDoc-It2 Imager (UVP, LLC, Upland, CA).
Results
GMF is a Novel Therapeutic Target for CRISPR-Cas9-Based Gene Editing
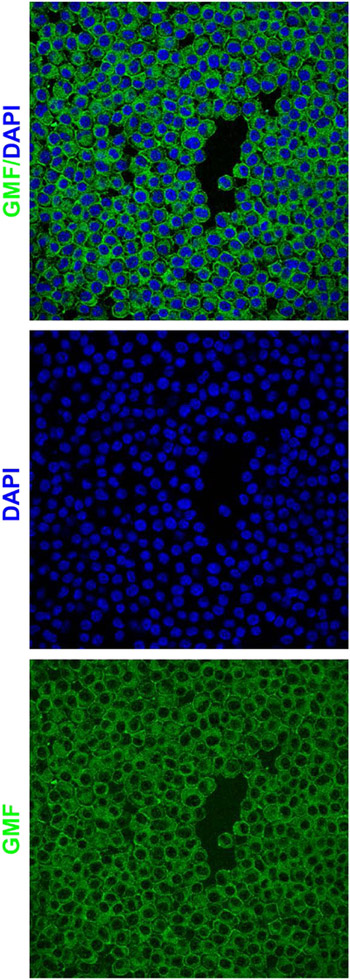
AD is a progressive neurodegenerative disorder in search for a novel cure. Neuroinflammation plays a crucial role during AD progression. We have previously shown that GMF plays a crucial role in the AD pathophysiology including neurodegeneration [17,23,24]. Therefore, we believe that GMF could potentially be an attractive therapeutic target against AD. In our past studies, down-regulation of GMF using conventional shRNA and GMF-specific antibody have been attempted with promising results. In the present communication, we have used the powerful CRISPR/Cas9 gene editing to regulate GMF function. We hypothesized that CRISPR-Cas9-mediated GMF gene editing in microglia is a novel approach to reduce neuroinflammation, neurodegeneration, AD pathophysiology and improve cognitive function. Since microglia play a crucial role in neuroinflammation as well as AD pathophysiology, initially, we performed GMF immunostaining in the BV2 microglial cell line. Our confocal data revealed widespread intracellular as well as cell surface expression of GMF (Figure 1). Our immunofluorescence results suggest that BV2 cells could be potentially useful for studying CRISPR-Cas9-mediated non-homologous end joining to achieve successful GMF gene editing in vitro. However, it is important to note that BV2 cells are relatively resistant to transfection using traditional lipid based transfection reagents. Therefore, we decided to overcome this potential limitation by employing a viral vector based approach to achieve high efficiency GMF gene editing.
Figure 1: GMF is a novel AD therapeutic target for CRISPR-Cas9 based gene editing:
GMF immunostaining in the BV2 microglial cells indicates high-level perinuclear expression of GMF (Green) within the cytoplasm as well as on the cell surface. The nuclei (blue) are stained with DAPI. The images were acquired using the Leica TCP SP8 confocal microscope and processed using Leica Application Suite X software.
AAV-SaCas9-GMF-sgRNA Transduction Leads to Successful GMF Gene Editing
CRISPR-Cas9-mediated gene editing offers an exciting opportunity to specifically ablate GMF gene expression both in vitro as well as in vivo. However, despite significant promise due to the large size (1368 amino acids) of the Streptococcus pyogenes nuclease SpCas9 and due to the AAV packaging limitations it is not possible to simultaneously coexpress SpCas9 as well as GMF specific sgRNA using a single AAV vector. To overcome this limitation we decided to use a smaller (1053 amino acids) CRISPR-Cas9 nuclease derived from Staphylococcus aureus SaCas9 that can be easily packaged along with GMF sgRNA in a single AAV vector. Since the SaCas9 PAM recognition sequence is different from that of SpCas9, we designed a GMF sgRNA as depicted in Figure 2a. The AAV design, the vector map and the cloned GMF sgRNA sequence are depicted in Figure 2b-d. Actively proliferating BV2 microglial cells were cultured at low cell density on poly-D-lysine coated chambered glass slides and transduced with AAV-SaCas9-GMF-sgRNA (Figure 2e) and allowed to proliferate for 72 hours. GMF expression pattern was determined in the non-transduced as well as cells transduced with AAV-CRISPR-GMF-sgRNA. Our confocal data shows a predominantly cytoplasmic GMF expression with higher intensity staining at the cell periphery and plasma membrane in the non-transduced BV2 cells (Figure 1). The BV2 cells transduced with AAV-SaCas9-GMF-sgRNA show high level expression of SaCas9 mostly in the cytoplasm and no detectable GMF expression thereby indicating successful biallelic GMF gene editing (Figure 2e). In contrast the non transduced BV2 cells exhibit widespread GMF expression thereby indicating that they have not been edited for GMF. Despite successful GMF gene editing mediated by AAV-SaCas9-GMF-sgRNA the gene editing efficiency was lower.
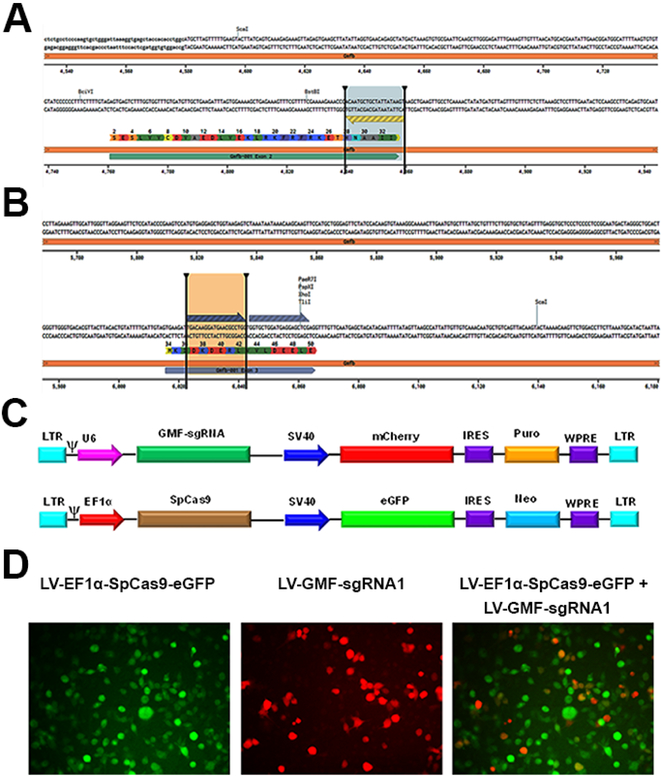
Lentiviral Transduction Leads to High-Level Expression of CRISPR-Cas9 in BV2 Cells and Differential GMF Gene Editing
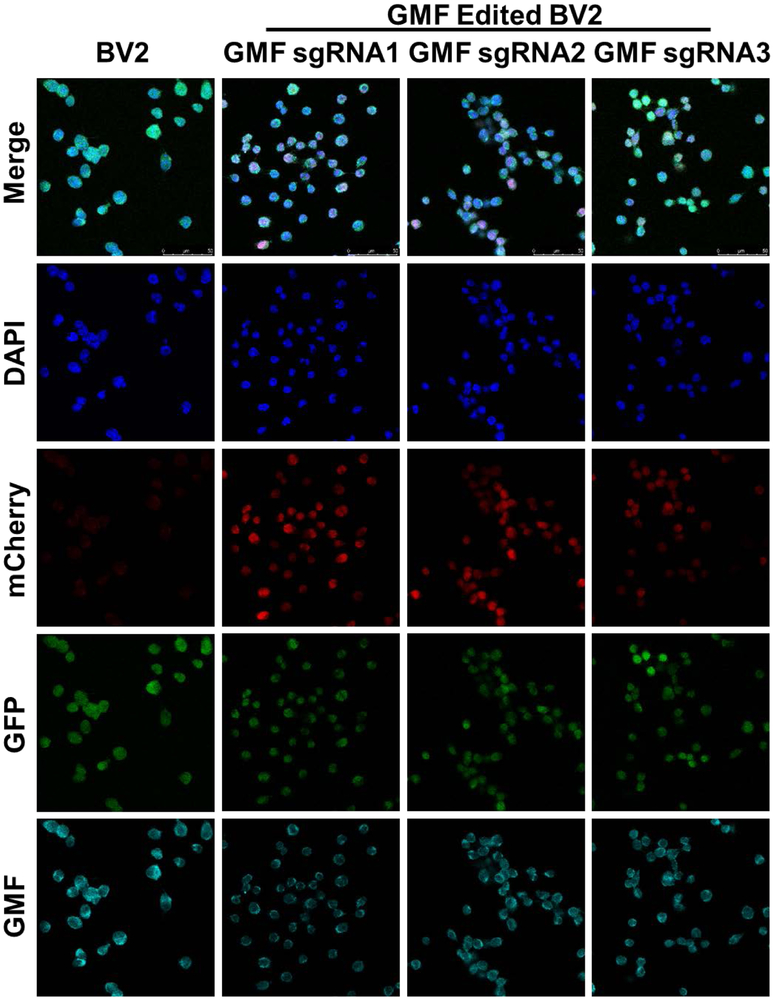
To improve and achieve high level GMF gene editing efficiency in BV2 cells, here we have used a dual lentiviral vector system. We reasoned that GMF gene editing efficiency could potentially be improved by designing new GMF sgRNAs and replacing SaCas9 with more widely used SpCas9. Therefore, first we designed GMF specific 3 separate sgRNAs that are compatible with SpCas9 (Figure 3a, b). In this approach, next we developed a stable BV2 cell line using the lentivirus LV-EF1α-SpCas9-eGFP-Neo (Figure 3c). In LV-EF1α-SpCas9-eGFP-Neo a constitutive EF1α promoter drives the expression of SpCas9 while a SV40 promoter drives the expression of eGFP as well as Neomycin resistance marker. We selected Neomycin resistant BV2 cells stably expressing SpCas9 that were also eGFP positive by fluorescence microscopy (Figure 3d). The morphology of the SpCas9 expressing cells was similar to wild type BV2 cells. The BV2 cells stably expressing SpCas9 were co-transduced with lentiviral vectors expressing either scrambled sgRNA or GMF specific sgRNAs (Figure 3d). Puromycin selection led to the generation of double stable BV2 cell lines simultaneously co-expressing SpCas9 and GMF specific sgRNAs. These cells were immunostained for GMF expression and analyzed for GMF editing by confocal microscopy. The BV2 cells stably expressing SpCas9 alone display a more or less homogenous population of high-level GMF expressing cells that are also positive for GFP expression but negative for mCherry expression (Figure 4, left panel). In comparison, the BV2 cells simultaneously co-expressing SpCas9 as well as GMF specific sgRNAs revealed differential GMF expression and were positive for both GFP as well as mCherry expression (Figure 4, last 3 panels). Unlike AAV-mediated bi-allelic GMF gene editing, surprisingly, the lentiviral transduced cells exhibit differential GMF expression possibly due to mono allelic GMF gene editing despite high-level expression of SpCas9 as well as GMF specific sgRNAs.
Figure 3: Development of LV-EF1α-SpCas9-eGFP and LV-GMF-sgRNAs:
a) Selection of GMF specific sgRNAs: The panel A shows the DNA sequence of mouse GMF exon 2 flanked by the introns. The GMF amino acid sequence is indicated by single letter codes. The GMF sgRNA1 sequence which is slightly different than the GMF sgRNA used in the AAV (due to the differences between SaCas9 and SpCas9 PAM sequences) is also located in the anticlockwise orientation on the lower strand is depicted by the blue arrow flanked by the two vertical bars. b) The panel B shows the GMF sgRNA deisgn of GMF sgRNA2 and GMF sgRNA3 both of which are located in the GMF coding sequence in the exon 3. c) Generation of LV-EF1α-SpCas9-eGFP-Neo and LV-GMF-sgRNAs: A set of 5 different VSV enveloped pseudotyped third generation lentiviral vectors were developed. In the lentiviral vector LV-SpCas9-eGFP-Neo, the EF1α promoter drives the expression of CRISPR-Cas9 and a SV40 promoter drives the expression of eGFP reporter and Neo resistance genes. The lentiviral vector LV-GMF-sgRNA 1, 2 and 3 express GMF sgRNAs under the control of the U6 promoter while the SV40 promoter drives the expression of mCherry reporter and Puromycin resistance marker. In the control lentiviral vector LV-SCRAM-sgRNA a scrambled sgRNA that does not target any known gene was used as a negative gene editing control virus. d) Generation of stable CRISPR-Cas9 expressing cell line: BV2 cells were transduced with the lentiviral vector LV-EF1α-SpCas9-eGFP-Neo to generate a stable cell line using neomycin selection. Co-transduction of BV2-CRISPR-Cas9 cells with LV-GMF-sgRNA 1 shows the presence of mCherry expressing cells and the merged image shows the presence of a small subset of cells showing both eGFP and mCherry expression. Puromycin selection was used to generate stable BV2-CRISPR-Cas9 and GMF-sgRNA expressing cells.
Figure 4: Targeted GMF gene editing using LV-EF1α-SpCas9-eGFP+LV-GMF-sgRNAs:
a) Confocal microscopy of the BV2-CRISPR-Cas9 cells expressing SpCas9 (Green) reveal normal GMF expression (Cyan) with DAPI stained nuclei (Blue) in the far left vertical panel labeled BV2. b) Co-transduction of BV2-CRISPR-Cas9 cells with GMF-specific sgRNA1 leads to a differential GMF editing and partial reduction in GMF expression in the vertical panel labeled as GMF sgRNA1. c) Co-transduction of BV2-CRISPR-Cas9 cells with GMF-specific sgRNA2 leads to a differential GMF editing and partial reduction in GMF expression in the vertical panel labeled GMF sgRNA2. d) Co-transduction of BV2-CRISPR-Cas9 cells with GMF-specific sgRNA3 leads to a differential GMF editing and partial reduction in GMF expression in the vertical panel labeled as GMF sgRNA3. In the panels depicting GMF sgRNAs 1-3, mCherry expression indicates co-expression of GMF sgRNAs. The GMF gene edited BV2 cells with reduced GMF expression are marked with white filled triangles.
SpCas9-Induced Indels in GMF Coding Sequence Predict in vivo Gene Editing Outcomes
We next investigated whether or not GMF gene editing leads to insertions or deletions in the GMF coding sequence. This was achieved first by performing mutational analysis using Guide-it Resolvase on GMF edited and non-edited DNA heteroduplexes. Our mutational analysis revealed a lower level of GMF specific gene editing using all the three GMF specific sgRNAs (Figure 5). The GMF sgRNA2 displayed slightly better cleavage of GMF heteroduplexes thereby suggesting increased GMF editing. In order to investigate the nature of GMF gene editing, we performed DNA sequencing of GMF edited and non-edited DNA clones. Our DNA sequencing results indicate GMF gene editing leads to micro-indels in the exons 2 and 3 as well as mega-deletion of the complete exon 3 of the GMF coding sequence (Figure 6a, b) which in turn are responsible for the loss of GMF expression in the transduced cells. We found the DNA sequence alterations including single and multiple nucleotide changes as well as a single nucleotide deletion in the GMF coding sequence. In the case of GMF sgRNA1, GMF gene editing led to a TG to GT inversion causing the conversion of ATG to AGT (M34S) in one of the clones. In another clone, we found multiple amino acid changes starting after 25th amino acid. These changes include E26A, H28P, N29G, A30C etc. causing a significant alteration in the GMF coding sequence. As compared to GMF sgRNA1, the nucleotide changes were more pronounced with GMF sgRNA2. In the case of GMF sgRNA2, GMF gene editing led to a megadeletion resulting in the complete removal of exon 3 and parts of the flanking introns (nt 5862-6106). Similarly, GMF sgRNA3 also led to GMF gene editing but mostly in the downstream intron (data not shown). There is no similarity or a defined pattern between the GMF gene editing mediated by either of the GMF sgRNAs used in our current study. Based upon our DNA sequencing data we also cannot rule out any potential off target effects. However, our in vitro results do suggest that potentially similar GMF gene editing to occur in vivo. Next, we focused upon monitoring the functional GMF expression post GMF gene editing by flow cytometry. Our flow cytometry data indicates that as compared to the wild type non GMF edited BV2 cells, the GMF expression levels are significantly reduced especially in GMF sgRNA1 and GMF sgRNA2 edited BV2 cells (Figure 7). Surprisingly, GMF gene editing using GMF sgRNA3 did not change GMF expression levels in GMF edited BV2 cells.
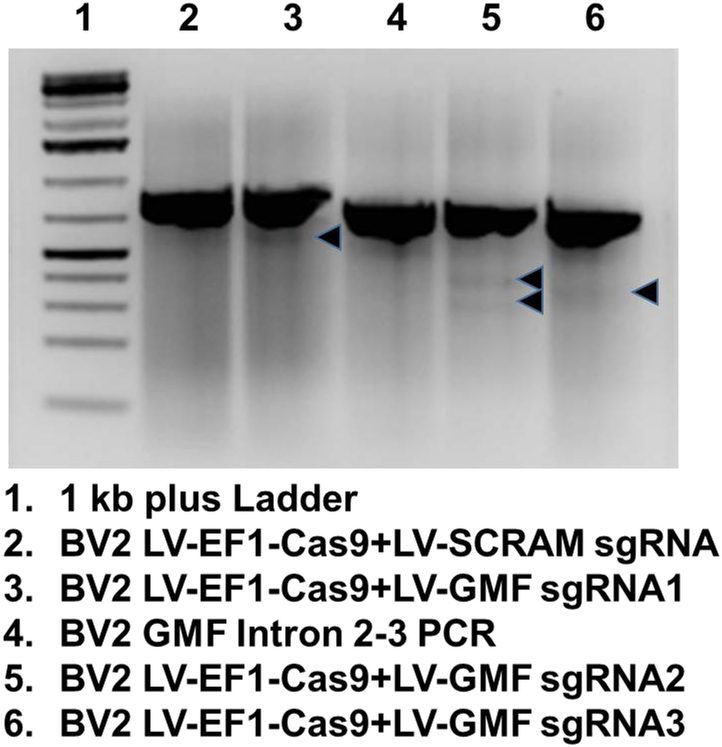
Figure 5: Mutational Analysis reveals GMF gene editing in BV2 cells:
The genomic DNA from the non edited or GMF edited cells was isolated and used for PCR amplification of the GMF edited sequence. The PCR amplified products from the wild type BV2 cells as well as from the GMF edited cells were used for heteroduplex formation and subjected to treatment with Guide-it Resolvase and subsequently analyzed for the cleavage of the mismatched DNA. The DNA mixture was resolved by agarose gel electrophoresis. The DNA in the lanes 2 and 4 does not reveal any cleavage as expected. However, the DNA in the lanes 3, 5 and 6 indicate low levels of DNA cleavage indicating successful GMF gene editing. The faint bands resulting due to Guide-it Resolvase-mediated cleavage are shown as black arrows.
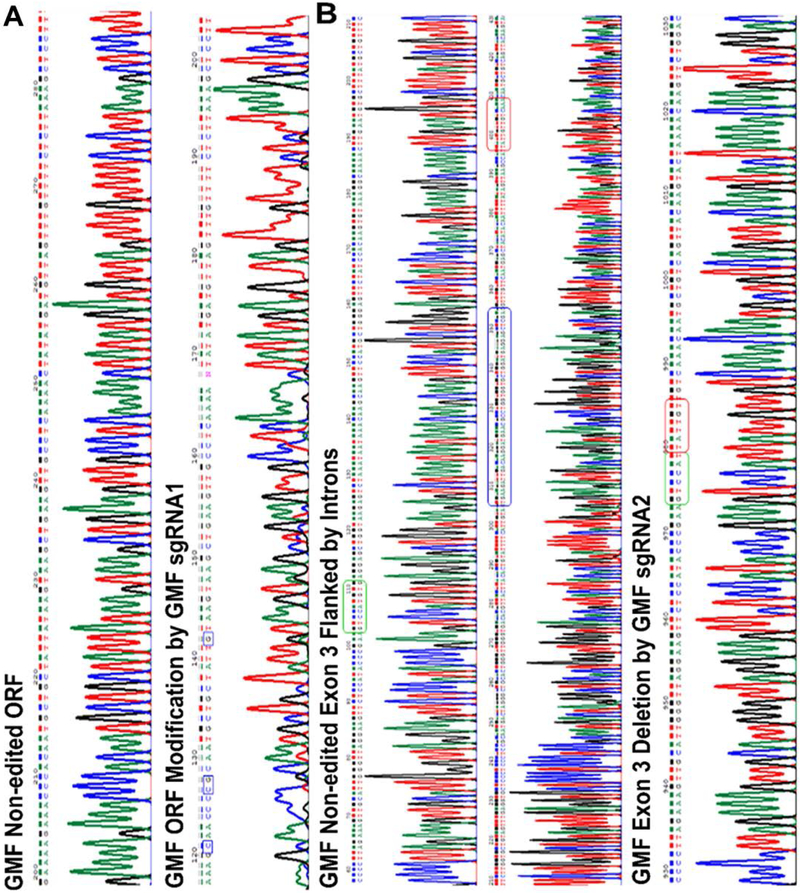
Figure 6A: SpCas9-mediated GMF editing leads to indels in GMF coding sequence:
a) The top panel shows the wild type GMF DNA sequence while the bottom panel shows the GMF edited sequence (using GMF-sgRNA1) from the exon 2 and is flanked by the intron sequences. GMF edited DNA sequence as a result of GMF sgRNA1-mediated editing leads to indels in the GMF coding sequence causing a frameshift in the GMF coding sequence. Specific nucleotide changes are indicated in the blue box, b) The top and the middle panels show the normal non-edited DNA sequence of the GMF exon 3 and the flanking introns. The bottom panel shows the deletion of GMF exon 3 post GMF gene editing. There is a significant change in the nucleotide sequence as a result of GMF gene editing. The green and the red boxes in the top and the middle panels depict the normal GMF DNA sequence. The GMF exon 3 is depicted within the blue box in the middle panel. As a result of GMF gene editing the GMF exon 3 and the flanking partial intronic sequences have been deleted and are represented as green and red boxes in the bottom panel.
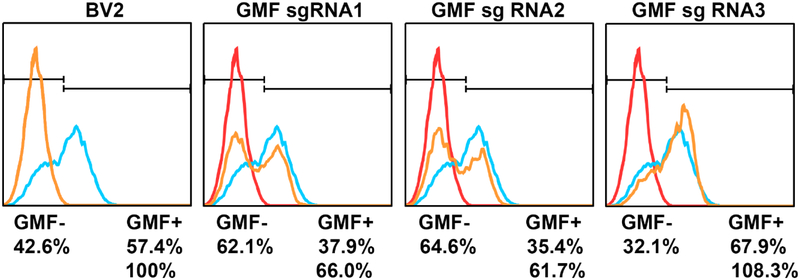
Figure 7: Despite stable expression of SpCas9 and GMF sgRNAs, GMF editing is moderate:
Flow cytometric analysis of GMF gene editing in BV2 cells revealed that as compared to the normal GMF expression in BV2 cells, GMF gene editing leads to significant reduction in GMF expression especially due to GMF sgRNA1 and sgRNA2. Surprisingly there was no significant change in GMF expression due to GMF sgRNA3-mediated GMF gene editing. The changes in GMF expression are indicated in terms of GMF positive and GMF negative cell populations.
GMF Gene Editing Causes Suppression of MAPK Pathway
We have previously shown that overexpression of GMF in astrocytes leads to activation of the p38 MAPK signaling pathway and oxidative toxicity in dopaminergic neurons while inhibition of GMF expression downregulates NFκB signaling pathway to stop progression of neuronal cell death [50,29]. Having established GMF gene editing in BV2 cells, we next investigated whether or not GMF gene editing leads to suppression of MAPK pathway. Wild type BV2 cells as well as GMF edited BV2 cells were treated with 1 μg/ml LPS for 30 minutes and were subsequently analyzed by western blot for phospho p38 MAPK and total p38 MAPK. Our results indicate that the wild type BV2 cells have a basal level of pp38 MAPK and LPS treatment significantly enhances pp38 MAPK expression as compared to p38 MAPK (Figure 8). In contrast, GMF edited BV2 cells have a significantly reduced pp38 MAPK expression at the basal level, which is very slightly enhanced following LPS treatment especially in GMF sgRNA2 edited cells. In contrast, LPS treatment leads to enhanced pp38 MAPK in GMF sgRNA1 and sgRNA3 edited cells similar to non-edited BV2 cells. These functional data validate successful GMF gene editing in BV2 cells and hopefully establish GMF gene editing-mediated inhibition of pp38 MAPK as an effective method to regulate GMF’s proinflammatory action in microglia which could now be explored as a novel therapeutic strategy for the control of neuroinflammatory and neurodegenerative diseases especially AD.
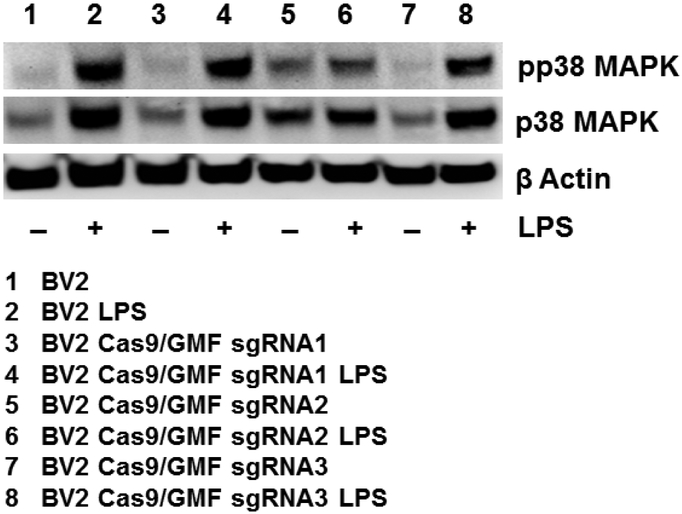
Figure 8: GMF gene editing causes suppression of MAPK pathway activation:
Wild type BV2 cells, and GMF gene edited BV2 cells were treated with 1.0μg/ml LPS for 30 minutes and analyzed for the expression of pp38MAPK and p38 MAPK. The representative western blot data (n=3) indicates that wild type BV2 cells display enhanced pp38 MAPK and p38 MAPK expression in the presence of LPS treatment (lanes 1 and 2). However, GMF sgRNA2-edited BV2 cells exhibit significant reduction in both pp38 MAPK and p38 MAPK expression with and without LPS treatment (lanes 5 and 6). Surprisingly, pp38 MAPK and p38 MAPK expression patterns are very similar between wild type as well as GMF sgRNA1 and sgRNA3-edited BV2 cells as depicted in lanes 3, 4 and 7, 8 respectively.
Discussion
Neuroinflammation and neurodegeneration play significant roles in the development and progression of AD. The activation of the microglial cells initiates a cascade of events that ultimately lead to neurodegeneration and AD pathophysiology. Proinflammatory mediator GMF has been shown to be expressed at elevated levels in AD patients [17,22-25,27]. We therefore believe that GMF represents a novel and an attractive therapeutic target to disrupt glia-neuron interactions and possibly delay and/or halt the progression of AD. Recent advancements in the field of gene editing have made it possible to precisely edit define DNA sequence to ameliorate disease pathology as well as to study disease progression in various disease models [32,51,33,34,37,38,41,43,45].
Here we hypothesized that CRISPR-Cas9-mediated GMF editing in microglial cells leads to inhibition of GMF expression and reduced microglial activation. It is important to note that the GMF gene organization is rather unusual in the sense that the first exon only codes for only one amino acid i.e. the starting codon ATG. Furthermore, the second exon ends with the first nucleotide of the first amino acid of the third exon. Additionally due to lack of canonical SaCas9 PAM sequence entirely within the second exon it was rather challenging to design the much desired GMF sgRNA. As a result, the GMF sgRNA in an anticlockwise orientation targeting the junction of the GMF second exon and the immediate downstream intron was designed. Prior studies have demonstrated that the SaCas9 has an improved on-target profile as compared with SpCas9 due to longer seed sequence requirement, extended PAM sequence and less tolerance for mismatch by SaCas9 [37,51].
We performed a series of experiments to maximize CRISPR-Cas9-mediated GMF gene editing in the BV2 microglial cell line. First, we developed a recombinant AAV simultaneously coexpressing SaCas9 as well as GMF-specific sgRNA. Due to the large size of SpCas9 and the packaging constraints of the AAV system, it is not possible to use a single AAV to package an expression cassette comprising the promoter, SpCas9, polyadenylation sequence as well as a U6 promoter driven GMF sgRNA. Although the split AAV SpCas9 has been used successfully [52,53] and split AAV vector induces higher gene editing efficiency, our major concern was that this strategy will require two recombinant AAVs to simultaneously transduce the less frequent target cells thereby reducing GMF gene editing efficiency. Therefore, our natural choice was to use a single AAV simultaneously co-expressing SaCas9 as well as GMF specific sgRNA. Unlike traditional AAV purification protocols which are often labor intensive, expensive and require multiple rounds of ultracentrifugation and dialysis, our simplified, cost effective and modified AAV purification protocol allows direct concentration of AAV particles secreted into the medium [54,55].
The GMF gene editing efficiency in the transduced BV2 cells was relatively low. One possible explanation could be that our AAV-SaCas9-GMF-sgRNA bears a modified capsid with a higher affinity for the selective transduction of neuronal cells. Previous studies have shown that amongst various AAV serotypes 2, 5, 6, 8 and 9 examined, AAV6 and AAV8 have significantly higher transduction efficiency in microglial cells [56]. Further microglial specificity can be achieved using either novel tropism modified and engineered AAV capsids, robust microglia specific promoter such as TMEM119 [57] and/or by employing neuronal detargeting by incorporating mir-124 target sequences in the AAV 3’UTR [58,59]. Although AAV has been successfully used to achieve genome editing in in vitro primary neuronal cultures as well as postmitotic neurons in vivo [60,61], one of the potential concerns regarding the use of AAV to achieve gene editing in vivo is the cellular immune response. A recent study demonstrated that both AAV9 capsid as well as Cas9 evoke cellular immune response, however, the cellular damage appears to be limited [62]. Most recently, we were informed by the technical support team of Takara that the AAV guide-it vector used in the current study has a 22 bp deletion (nt 2606-2627) in the 5’ITR. Therefore, to improve our gene editing results we decided to modify our approach by using a dual lentiviral transduction approach.
We developed multiple lentiviral vectors expressing either SpCas9, GMF sgRNAs or a scrambled sgRNA. The lentiviral vector LV-EF1α-SpCas9-eGFP-Neo was used to develop a stable BV2 cell line BV2-SpCas9 constitutively expressing SpCas9. This was achieved by using Neomycin selection to eliminate the non-transduced cells. An added advantage of this lentiviral vector is that it co-expresses eGFP for easy visualization of the transduced cells. BV2-CRISPR-Cas9 cells were transduced with three separate lentiviral vectors expressing 3 different GMF sgRNAs to derive three separate BV2 sublines simultaneously co-expressing SpCas9 and either GMF sgRNA1, GMF sgRNA2 or GMF sgRNA3. In addition to mCherry reporter, our GMF sgRNA expressing lentiviral vectors harbor puromycin gene for rapid generation of double stable cell lines.
Our confocal microscopy as well as flow cytometry results confirmed the generation of BV2 cells simultaneously co-expressing SpCas9, eGFP, mCherry, GMF sgRNA, neomycin and puromycin for the ease of selection as well as monitoring the transduction efficiency. Of the 3 BV2 cell lines generated, only subline BV2-SpCas9-GMF-sgRNA1 and BV2-SpCas9-GMF-sgRNA2 revealed appreciable GMF gene editing while BV2-SpCas9-GMF-sgRNA3 had minimal GMF gene editing as revealed by flow cytometry and confocal microscopy. The differences in GMF gene editing between different GMF sgRNAs are expected and are likely to be due to differential accessibility of the SpCas9-sgRNA complexes on the GMF gene. Although, we expected that a vast majority of our cells will exhibit GMF gene editing due to simultaneous coexpression of the SpCas9 as well as the GMF sgRNAs over a prolonged period, this was clearly not the case. It is possible that the GMF editing took place only in one of the GMF alleles with only a very low frequency of biallelic GMF gene editing taking place. This could possibly be due to inefficient translocation of the SpCas9 protein from the cytoplasm to the nucleus. It might also be possible that SpCas9 may have relatively inefficient gene editing efficiency as compared to SaCas9. Most recently, Kichev et al. have reported on the use of plasmid based approach to achieve gene editing of RANK receptor using Cas9 and HDR plasmids in BV2 cells [63]. Although useful for in vitro experiments, this approach clearly lacks in vivo translational potential. Another caveat of the study was lack of confirmation of RANK receptor gene editing by either immunofluorescence, flow cytometry, western blot or T7 Endonuclease I-mediated mutational analysis.
Even though, we observed limited GMF gene editing in BV2 cells, it would be interesting to study GMF gene editing in the primary neurons, astrocytes, and microglia as well as mixed glial-neuronal cultures. It would also be interesting to perform GMF gene editing in the embryonic as well as induced pluripotent stem cells, differentiate them into microglia and perform functional studies [64-66]. Furthermore, it is possible that GMF gene editing will occur at a relatively higher frequency following direct intracranial injections in 5xFAD mice. Moreover, it is possible that the transduced cells will secrete exosomes and or microvesicles containing Cas9 and GMF sgRNAs that will be taken up by the neighboring non-transduced cells. Such a molecular mechanism will significantly enhance in vivo GMF gene editing similar to the bystander effect observed in multiple cancer gene therapy studies. Further It would be interesting to enhance microglial transduction using tropism modified AAV.
Whether GMF gene editing leads to inhibition of inflammatory signaling pathway was confirmed by functional analysis of the MAPK pathway. Our GMF gene editing leads to significant suppression of both phospho p38 MAPK and p38 MAPK in the presence of LPS stimulation. Prior studies have reported that p38 MAPK expression is enhanced in the brains of AD patients [67,68]. Moreover, MAPK kinase 6 and phosphorylated p38 MAPK was found to be upregulated in AD post-mortem brains as well as in the blood of AD patients [69,70]. Our prior studies have shown that GMF overexpression stimulates p38 MAPK activity in C6 and PC12 cells as well as in normal astrocytes [71,50,72-74]. Recently, deficiency of neuronal p38alpha MAPK has been shown to attenuate amyloid pathology in AD mouse and cellular models [75]. Interestingly, mir-744, mir-124a and mir-24 mimics have been shown to suppress both p38 MAPK expression and activation [76]. Similarly, LPS treatment of the hippocampal HT-22 cells has been shown to downregulate mir-132 expression which in turn upregulates the expression of the target gene TRAF6 causing activation of the NFκB and MEK/ERK pathways [77]. MicroRNA-132 deficiencey has been shown to enhance Aβ production and senile plaque deposition in 3xTg-AD mice [78] as well as to aggravate amyloid and tau pathology in human AD patients [79]. Whether, a similar microRNA-mediated molecular mechanism underlies GMF-induced p38 MAPK regulation in vitro as well as in AD patients is presently unknown. In addition, other studies have uncovered the role of microglia in high fat diet induced neuroinflammation [80-83]. We have recently reported that in human AD brain, dysfunctional lysosomal-autophagy pathway triggered and regulated by GMF and the NLRP3 inflammasome along with pro-inflammatory cytokines result in aggregated Aβ and tauopathy [17]. Therefore, we believe that targeted GMF gene editing in microglia could be exploited for halting the progression of neuroinflammation-mediated neurodegeneration in a wide variety of neurodegenerative diseases especially AD, diet-induced metabolic dysfunction including obesity and post-operative cognitive decline. Overall, our data suggest that GMF is a novel therapeutic AD target. An earlier study has reported that IL-4 treatment of BV2 cells lead to a potent M2a response in vitro however bilateral intracranial injections of AAV-IL4 in AD mice led to increased deaths thereby indicating that such an approach may not be appropriate [84].
Generation of stable BV2 microglial cells expressing Cas9 described here will enable CRISPR-based gene editing of multiple AD therapeutic targets simultaneously by multiplexing different target sgRNAs. Multiplexing different target sgRNAs will be very useful to study not only glial development but also to decipher the molecular mechanisms underlying glia-mediated neuroinflammation and neurodegeneration in various CNS disorders. GMF edited BV2 cells will be very useful in studying microglial-neuronal interactions which play a crucial role in AD pathophysiology. To the best of our knowledge, our proof of principle studies on GMF gene editing in the microglial cells is one of the first studies aimed at the future development of personalized precision medicine for the treatment of AD.
Acknowledgements
The authors would like to acknowledge the Veteran Affairs Merit Award I01BX002477 and the National Institutes of Health Grant AG048205 to AZ. The authors would like to thank Prof. Miguel Sena-Esteves, University of Massachusetts, Worcester for providing the AAV plasmid pGG-B1 and Prof. Didier Trono, Ecole Polytechnique Federale de Lausanne (EPFL) for providing the pMDLg/pRRE, pRSV-Rev and pMD2.G lentiviral packaging plasmids. The authors are thankful to Dr. Alexander Jurkevich, Associate Director, University of Missouri Molecular Cytology Core for help with confocal microscopy and Mr. Daniel E. Jackson, University of Missouri Cell and Immunobiology Core for help with Flow Cytometry.
Footnotes
Conflict of Interest
The authors confirm that they have no conflict of interest.
References
- 1.Doody RS, Farlow M, Aisen PS, Alzheimer's Disease Cooperative Study Data A, Publication C (2014) Phase 3 trials of solanezumab and bapineuzumab for Alzheimer's disease. N Engl J Med 370 (15):1460. doi: 10.1056/NEJMc1402193 [DOI] [PubMed] [Google Scholar]
- 2.Doody RS, Thomas RG, Farlow M, Iwatsubo T, Vellas B, Joffe S, Kieburtz K, Raman R, Sun X, Aisen PS, Siemers E, Liu-Seifert H, Mohs R, Alzheimer's Disease Cooperative Study Steering C, Solanezumab Study G (2014) Phase 3 trials of solanezumab for mild-to-moderate Alzheimer's disease. N Engl J Med 370 (4):311–321. doi: 10.1056/NEJMoa1312889 [DOI] [PubMed] [Google Scholar]
- 3.Honig LS, Vellas B, Woodward M, Boada M, Bullock R, Borrie M, Hager K, Andreasen N, Scarpini E, Liu-Seifert H, Case M, Dean RA, Hake A, Sundell K, Poole Hoffmann V, Carlson C, Khanna R, Mintun M, DeMattos R, Selzler KJ, Siemers E (2018) Trial of Solanezumab for Mild Dementia Due to Alzheimer's Disease. N Engl J Med 378 (4):321–330. doi: 10.1056/NEJMoa1705971 [DOI] [PubMed] [Google Scholar]
- 4.Sevigny J, Chiao P, Bussiere T, Weinreb PH, Williams L, Maier M, Dunstan R, Salloway S, Chen T, Ling Y, O'Gorman J, Qian F, Arastu M, Li M, Chollate S, Brennan MS, Quintero-Monzon O, Scannevin RH, Arnold HM, Engber T, Rhodes K, Ferrero J, Hang Y, Mikulskis A, Grimm J, Hock C, Nitsch RM, Sandrock A (2016) The antibody aducanumab reduces Abeta plaques in Alzheimer's disease. Nature 537 (7618):50–56. doi: 10.1038/nature19323 [DOI] [PubMed] [Google Scholar]
- 5.Graham WV, Bonito-Oliva A, Sakmar TP (2017) Update on Alzheimer's Disease Therapy and Prevention Strategies. Annu Rev Med 68:413–430. doi: 10.1146/annurev-med-042915-103753 [DOI] [PubMed] [Google Scholar]
- 6.Ye L, Rasmussen J, Kaeser SA, Marzesco AM, Obermuller U, Mahler J, Schelle J, Odenthal J, Kruger C, Fritschi SK, Walker LC, Staufenbiel M, Baumann F, Jucker M (2017) Abeta seeding potency peaks in the early stages of cerebral beta-amyloidosis. EMBO Rep 18 (9):1536–1544. doi: 10.15252/embr.201744067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerriero F, Sgarlata C, Francis M, Maurizi N, Faragli A, Perna S, Rondanelli M, Rollone M, Ricevuti G (2016) Neuroinflammation, immune system and Alzheimer disease: searching for the missing link. Aging Clin Exp Res. doi: 10.1007/s40520-016-0637-z [DOI] [PubMed] [Google Scholar]
- 8.Kempuraj D, Thangavel R, Selvakumar GP, Zaheer S, Ahmed ME, Raikwar SP, Zahoor H, Saeed D, Natteru PA, Iyer S, Zaheer A (2017) Brain and Peripheral Atypical Inflammatory Mediators Potentiate Neuroinflammation and Neurodegeneration. Front Cell Neurosci 11:216. doi: 10.3389/fncel.2017.00216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ransohoff RM (2016) How neuroinflammation contributes to neurodegeneration. Science 353 (6301):777–783. doi: 10.1126/science.aag2590 [DOI] [PubMed] [Google Scholar]
- 10.Di Benedetto S, Muller L, Wenger E, Duzel S, Pawelec G (2017) Contribution of neuroinflammation and immunity to brain aging and the mitigating effects of physical and cognitive interventions. Neurosci Biobehav Rev 75:114–128. doi: 10.1016/j.neubiorev.2017.01.044 [DOI] [PubMed] [Google Scholar]
- 11.Bronzuoli MR, Iacomino A, Steardo L, Scuderi C (2016) Targeting neuroinflammation in Alzheimer's disease. J Inflamm Res 9:199–208. doi: 10.2147/JIR.S86958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becher B, Spath S, Goverman J (2017) Cytokine networks in neuroinflammation. Nat Rev Immunol 17 (1):49–59. doi: 10.1038/nri.2016.123 [DOI] [PubMed] [Google Scholar]
- 13.Colonna M, Butovsky O (2017) Microglia Function in the Central Nervous System During Health and Neurodegeneration. Annu Rev Immunol 35:441–468. doi: 10.1146/annurev-immunol-051116-052358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornejo F, von Bernhardi R (2016) Age-Dependent Changes in the Activation and Regulation of Microglia. Adv Exp Med Biol 949:205–226. doi: 10.1007/978-3-319-40764-7_10 [DOI] [PubMed] [Google Scholar]
- 15.Crotti A, Ransohoff RM (2016) Microglial Physiology and Pathophysiology: Insights from Genome-wide Transcriptional Profiling. Immunity 44 (3):505–515. doi: 10.1016/j.immuni.2016.02.013 [DOI] [PubMed] [Google Scholar]
- 16.Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, Itzkovitz S, Colonna M, Schwartz M, Amit I (2017) A Unique Microglia Type Associated with Restricting Development of Alzheimer's Disease. Cell 169 (7):1276–1290 e1217. doi: 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- 17.Ahmed ME, Iyer S, Thangavel R, Kempuraj D, Selvakumar GP, Raikwar SP, Zaheer S, Zaheer A (2017) Co-Localization of Glia Maturation Factor with NLRP3 Inflammasome and Autophagosome Markers in Human Alzheimer's Disease Brain. J Alzheimers Dis 60 (3):1143–1160. doi: 10.3233/JAD-170634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan R, Zaheer A, Jaye M, Lim R (1991) Molecular cloning and expression of biologically active human glia maturation factor-beta. J Neurochem 57 (2):483–490 [DOI] [PubMed] [Google Scholar]
- 19.Lim R, Miller JF, Zaheer A (1989) Purification and characterization of glia maturation factor beta: a growth regulator for neurons and glia. Proc Natl Acad Sci U S A 86 (10):3901–3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim R, Zaheer A (1991) Structure and function of glia maturation factor beta. Adv Exp Med Biol 296:161–164 [DOI] [PubMed] [Google Scholar]
- 21.Lim R, Zaheer A, Lane WS (1990) Complete amino acid sequence of bovine glia maturation factor beta. Proc Natl Acad Sci U S A 87 (14):5233–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stolmeier D, Thangavel R, Anantharam P, Khan MM, Kempuraj D, Zaheer A (2013) Glia maturation factor expression in hippocampus of human Alzheimer's disease. Neurochem Res 38 (8):1580–1589. doi: 10.1007/s11064-013-1059-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thangavel R, Kempuraj D, Stolmeier D, Anantharam P, Khan M, Zaheer A (2013) Glia maturation factor expression in entorhinal cortex of Alzheimer's disease brain. Neurochem Res 38 (9):1777–1784. doi: 10.1007/s11064-013-1080-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thangavel R, Kempuraj D, Zaheer S, Raikwar S, Ahmed ME, Selvakumar GP, Iyer SS, Zaheer A (2017) Glia Maturation Factor and Mitochondrial Uncoupling Proteins 2 and 4 Expression in the Temporal Cortex of Alzheimer's Disease Brain. Front Aging Neurosci 9:150. doi: 10.3389/fnagi.2017.00150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thangavel R, Stolmeier D, Yang X, Anantharam P, Zaheer A (2012) Expression of glia maturation factor in neuropathological lesions of Alzheimer's disease. Neuropathol Appl Neurobiol 38 (6):572–581. doi: 10.1111/j.1365-2990.2011.01232.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaheer A, Zaheer S, Thangavel R, Wu Y, Sahu SK, Yang B (2008) Glia maturation factor modulates beta-amyloid-induced glial activation, inflammatory cytokine/chemokine production and neuronal damage. Brain Res 1208:192–203. doi: 10.1016/j.brainres.2008.02.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaheer S, Thangavel R, Sahu SK, Zaheer A (2011) Augmented expression of glia maturation factor in Alzheimer's disease. Neuroscience 194:227–233. doi: 10.1016/j.neuroscience.2011.07.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaheer S, Thangavel R, Wu Y, Khan MM, Kempuraj D, Zaheer A (2013) Enhanced expression of glia maturation factor correlates with glial activation in the brain of triple transgenic Alzheimer's disease mice. Neurochem Res 38 (1):218–225. doi: 10.1007/s11064-012-0913-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan MM, Zaheer S, Nehman J, Zaheer A (2014) Suppression of glia maturation factor expression prevents 1-methyl-4-phenylpyridinium (MPP(+))-induced loss of mesencephalic dopaminergic neurons. Neuroscience 277:196–205. doi: 10.1016/j.neuroscience.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selvakumar GP, Iyer SS, Kempuraj D, Raju M, Thangavel R, Saeed D, Ahmed ME, Zahoor H, Raikwar SP, Zaheer S, Zaheer A (2018) Glia Maturation Factor Dependent Inhibition of Mitochondrial PGC-1alpha Triggers Oxidative Stress-Mediated Apoptosis in N27 Rat Dopaminergic Neuronal Cells. Mol Neurobiol. doi: 10.1007/s12035-018-0882-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callif BL, Maunze B, Krueger NL, Simpson MT, Blackmore MG (2017) The application of CRISPR technology to high content screening in primary neurons. Mol Cell Neurosci 80:170–179. doi: 10.1016/j.mcn.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cyranoski D (2016) CRISPR gene-editing tested in a person for the first time. Nature 539 (7630):479. doi: 10.1038/nature.2016.20988 [DOI] [PubMed] [Google Scholar]
- 33.Ledford H (2017) CRISPR fixes disease gene in viable human embryos. Nature 548 (7665):13–14. doi: 10.1038/nature.2017.22382 [DOI] [PubMed] [Google Scholar]
- 34.Mandal PK, Ferreira LM, Collins R, Meissner TB, Boutwell CL, Friesen M, Vrbanac V, Garrison BS, Stortchevoi A, Bryder D, Musunuru K, Brand H, Tager AM, Allen TM, Talkowski ME, Rossi DJ, Cowan CA (2014) Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell 15 (5):643–652. doi: 10.1016/j.stem.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, Asokan A, Zhang F, Duan D, Gersbach CA (2016) In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351 (6271):403–407. doi: 10.1126/science.aad5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park CY, Kim DH, Son JS, Sung JJ, Lee J, Bae S, Kim JH, Kim DW, Kim JS (2015) Functional Correction of Large Factor VIII Gene Chromosomal Inversions in Hemophilia A Patient-Derived iPSCs Using CRISPR-Cas9. Cell Stem Cell 17 (2):213–220. doi: 10.1016/j.stem.2015.07.001 [DOI] [PubMed] [Google Scholar]
- 37.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520 (7546):186–191. doi: 10.1038/nature14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK, Nieuwenhuis EE, Beekman JM, Clevers H (2013) Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13 (6):653–658. doi: 10.1016/j.stem.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 39.Soldner F, Stelzer Y, Shivalila CS, Abraham BJ, Latourelle JC, Barrasa MI, Goldmann J, Myers RH, Young RA, Jaenisch R (2016) Parkinson-associated risk variant in distal enhancer of alpha-synuclein modulates target gene expression. Nature 533 (7601):95–99. doi: 10.1038/nature17939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staahl BT, Benekareddy M, Coulon-Bainier C, Banfal AA, Floor SN, Sabo JK, Urnes C, Munares GA, Ghosh A, Doudna JA (2017) Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat Biotechnol 35 (5):431–434. doi: 10.1038/nbt.3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, Cong L, Zhang F, Vandenberghe LH, Church GM, Wagers AJ (2016) In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351 (6271):407–411. doi: 10.1126/science.aad5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Traxler EA, Yao Y, Wang YD, Woodard KJ, Kurita R, Nakamura Y, Hughes JR, Hardison RC, Blobel GA, Li C, Weiss MJ (2016) A genome-editing strategy to treat beta-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat Med 22 (9):987–990. doi: 10.1038/nm.4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, Yu H, Xu C, Morizono H, Musunuru K, Batshaw ML, Wilson JM (2016) A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol 34 (3):334–338. doi: 10.1038/nbt.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yin H, Song CQ, Dorkin JR, Zhu LJ, Li Y, Wu Q, Park A, Yang J, Suresh S, Bizhanova A, Gupta A, Bolukbasi MF, Walsh S, Bogorad RL, Gao G, Weng Z, Dong Y, Koteliansky V, Wolfe SA, Langer R, Xue W, Anderson DG (2016) Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol 34 (3):328–333. doi: 10.1038/nbt.3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young CS, Hicks MR, Ermolova NV, Nakano H, Jan M, Younesi S, Karumbayaram S, Kumagai-Cresse C, Wang D, Zack JA, Kohn DB, Nakano A, Nelson SF, Miceli MC, Spencer MJ, Pyle AD (2016) A Single CRISPR-Cas9 Deletion Strategy that Targets the Majority of DMD Patients Restores Dystrophin Function in hiPSC-Derived Muscle Cells. Cell Stem Cell 18 (4):533–540. doi: 10.1016/j.stem.2016.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, Zapatka M, Northcott PA, Schramm K, Belic J, Jones DT, Tschida B, Moriarity B, Largaespada D, Roussel MF, Korshunov A, Reifenberger G, Pfister SM, Lichter P, Kawauchi D, Gronych J (2015) Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun 6:7391. doi: 10.1038/ncomms8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blasi E, Barluzzi R, Bocchini V, Mazzolla R, Bistoni F (1990) Immortalization of murine microglial cells by a v-raf/v-myc carrying retrovirus. J Neuroimmunol 27 (2–3):229–237 [DOI] [PubMed] [Google Scholar]
- 48.Bocchini V, Mazzolla R, Barluzzi R, Blasi E, Sick P, Kettenmann H (1992) An immortalized cell line expresses properties of activated microglial cells. J Neurosci Res 31 (4):616–621. doi: 10.1002/jnr.490310405 [DOI] [PubMed] [Google Scholar]
- 49.Henn A, Lund S, Hedtjarn M, Schrattenholz A, Porzgen P, Leist M (2009) The suitability of BV2 cells as alternative model system for primary microglia cultures or for animal experiments examining brain inflammation. ALTEX 26 (2):83–94 [DOI] [PubMed] [Google Scholar]
- 50.Zaheer A, Yorek MA, Lim R (2001) Effects of glia maturation factor overexpression in primary astrocytes on MAP kinase activation, transcription factor activation, and neurotrophin secretion. Neurochem Res 26 (12):1293–1299 [DOI] [PubMed] [Google Scholar]
- 51.Doudna JA, Charpentier E (2014) Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346 (6213):1258096. doi: 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- 52.Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F (2015) In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol 33 (1):102–106. doi: 10.1038/nbt.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD, Hauschka SD, Chamberlain JR, Chamberlain JS (2017) Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun 8:14454. doi: 10.1038/ncomms14454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doria M, Ferrara A, Auricchio A (2013) AAV2/8 vectors purified from culture medium with a simple and rapid protocol transduce murine liver, muscle, and retina efficiently. Hum Gene Ther Methods 24 (6):392–398. doi: 10.1089/hgtb.2013.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vandenberghe LH, Xiao R, Lock M, Lin J, Korn M, Wilson JM (2010) Efficient serotype-dependent release of functional vector into the culture medium during adeno-associated virus manufacturing. Hum Gene Ther 21 (10):1251–1257. doi: 10.1089/hum.2010.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Su W, Kang J, Sopher B, Gillespie J, Aloi MS, Odom GL, Hopkins S, Case A, Wang DB, Chamberlain JS, Garden GA (2016) Recombinant adeno-associated viral (rAAV) vectors mediate efficient gene transduction in cultured neonatal and adult microglia. J Neurochem 136 Suppl 1:49–62. doi: 10.1111/jnc.13081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, Mulinyawe SB, Bohlen CJ, Adil A, Tucker A, Weissman IL, Chang EF, Li G, Grant GA, Hayden Gephart MG, Barres BA (2016) New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A 113 (12):E1738–1746. doi: 10.1073/pnas.1525528113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosario AM, Cruz PE, Ceballos-Diaz C, Strickland MR, Siemienski Z, Pardo M, Schob KL, Li A, Aslanidi GV, Srivastava A, Golde TE, Chakrabarty P (2016) Microglia-specific targeting by novel capsid-modified AAV6 vectors. Mol Ther Methods Clin Dev 3:16026. doi: 10.1038/mtm.2016.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taschenberger G, Tereshchenko J, Kugler S (2017) A MicroRNA124 Target Sequence Restores Astrocyte Specificity of gfaABC1D-Driven Transgene Expression in AAV-Mediated Gene Transfer. Mol Ther Nucleic Acids 8:13–25. doi: 10.1016/j.omtn.2017.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nishiyama J, Mikuni T, Yasuda R (2017) Virus-Mediated Genome Editing via Homology-Directed Repair in Mitotic and Postmitotic Cells in Mammalian Brain. Neuron 96 (4):755–768 e755. doi: 10.1016/j.neuron.2017.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murlidharan G, Sakamoto K, Rao L, Corriher T, Wang D, Gao G, Sullivan P, Asokan A (2016) CNS-restricted Transduction and CRISPR/Cas9-mediated Gene Deletion with an Engineered AAV Vector. Mol Ther Nucleic Acids 5 (7):e338. doi: 10.1038/mtna.2016.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, Zhu K, Wagers AJ, Church GM (2016) A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods 13 (10):868–874. doi: 10.1038/nmeth.3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kichev A, Eede P, Gressens P, Thornton C, Hagberg H (2017) Implicating Receptor Activator of NF-kappaB (RANK)/RANK Ligand Signalling in Microglial Responses to Toll-Like Receptor Stimuli. Dev Neurosci 39 (1–4):192–206. doi: 10.1159/000464244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beutner C, Roy K, Linnartz B, Napoli I, Neumann H (2010) Generation of microglial cells from mouse embryonic stem cells. Nat Protoc 5 (9):1481–1494. doi: 10.1038/nprot.2010.90 [DOI] [PubMed] [Google Scholar]
- 65.Douvaras P, Sun B, Wang M, Kruglikov I, Lallos G, Zimmer M, Terrenoire C, Zhang B, Gandy S, Schadt E, Freytes DO, Noggle S, Fossati V (2017) Directed Differentiation of Human Pluripotent Stem Cells to Microglia. Stem Cell Reports 8 (6):1516–1524. doi: 10.1016/j.stemcr.2017.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, Bakiasi G, Tsai LH, Aubourg P, Ransohoff RM, Jaenisch R (2016) Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med 22 (11):1358–1367. doi: 10.1038/nm.4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hensley K, Floyd RA, Zheng NY, Nael R, Robinson KA, Nguyen X, Pye QN, Stewart CA, Geddes J, Markesbery WR, Patel E, Johnson GV, Bing G (1999) p38 kinase is activated in the Alzheimer's disease brain. J Neurochem 72 (5):2053–2058 [DOI] [PubMed] [Google Scholar]
- 68.Sun A, Liu M, Nguyen XV, Bing G (2003) P38 MAP kinase is activated at early stages in Alzheimer's disease brain. Exp Neurol 183 (2):394–405 [DOI] [PubMed] [Google Scholar]
- 69.Pei JJ, Braak E, Braak H, Grundke-Iqbal I, Iqbal K, Winblad B, Cowburn RF (2001) Localization of active forms of C-jun kinase (JNK) and p38 kinase in Alzheimer's disease brains at different stages of neurofibrillary degeneration. J Alzheimers Dis 3 (1):41–48 [DOI] [PubMed] [Google Scholar]
- 70.Zhu X, Rottkamp CA, Hartzler A, Sun Z, Takeda A, Boux H, Shimohama S, Perry G, Smith MA (2001) Activation of MKK6, an upstream activator of p38, in Alzheimer's disease. J Neurochem 79 (2):311–318 [DOI] [PubMed] [Google Scholar]
- 71.Zaheer A, Lim R (1998) Overexpression of glia maturation factor (GMF) in PC12 pheochromocytoma cells activates p38 MAP kinase, MAPKAP kinase-2, and tyrosine hydroxylase. Biochem Biophys Res Commun 250 (2):278–282. doi: 10.1006/bbrc.1998.9301 [DOI] [PubMed] [Google Scholar]
- 72.Zaheer A, Zaheer S, Sahu SK, Knight S, Khosravi H, Mathur SN, Lim R (2007) A novel role of glia maturation factor: induction of granulocyte-macrophage colony-stimulating factor and pro-inflammatory cytokines. J Neurochem 101 (2):364–376. doi: 10.1111/j.1471-4159.2006.04385.x [DOI] [PubMed] [Google Scholar]
- 73.Lim R, Zaheer A, Kraakevik JA, Darby CJ, Oberley LW (1998) Overexpression of glia maturation factor in C6 cells promotes differentiation and activates superoxide dismutase. Neurochem Res 23 (11):1445–1451 [DOI] [PubMed] [Google Scholar]
- 74.Lim R, Zaheer A, Yorek MA, Darby CJ, Oberley LW (2000) Activation of nuclear factor-kappaB in C6 rat glioma cells after transfection with glia maturation factor. J Neurochem 74 (2):596–602 [DOI] [PubMed] [Google Scholar]
- 75.Schnoder L, Hao W, Qin Y, Liu S, Tomic I, Liu X, Fassbender K, Liu Y (2016) Deficiency of Neuronal p38alpha MAPK Attenuates Amyloid Pathology in Alzheimer Disease Mouse and Cell Models through Facilitating Lysosomal Degradation of BACE1. J Biol Chem 291 (5):2067–2079. doi: 10.1074/jbc.M115.695916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McCaskill JL, Ressel S, Alber A, Redford J, Power UF, Schwarze J, Dutia BM, Buck AH (2017) Broad-Spectrum Inhibition of Respiratory Virus Infection by MicroRNA Mimics Targeting p38 MAPK Signaling. Mol Ther Nucleic Acids 7:256–266. doi: 10.1016/j.omtn.2017.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ji YF, Wang D, Liu YR, Ma XR, Lu H, Zhang BA (2018) MicroRNA-132 attenuates LPS-induced inflammatory injury by targeting TRAF6 in neuronal cell line HT-22. J Cell Biochem. doi: 10.1002/jcb.26720 [DOI] [PubMed] [Google Scholar]
- 78.Hernandez-Rapp J, Rainone S, Goupil C, Dorval V, Smith PY, Saint-Pierre M, Vallee M, Planel E, Droit A, Calon F, Cicchetti F, Hebert SS (2016) microRNA-132/212 deficiency enhances Abeta production and senile plaque deposition in Alzheimer's disease triple transgenic mice. Sci Rep 6:30953. doi: 10.1038/srep30953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Salta E, Sierksma A, Vanden Eynden E, De Strooper B (2016) miR-132 loss de-represses ITPKB and aggravates amyloid and TAU pathology in Alzheimer's brain. EMBO Mol Med 8 (9):1005–1018. doi: 10.15252/emmm.201606520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Feng X, Valdearcos M, Uchida Y, Lutrin D, Maze M, Koliwad SK (2017) Microglia mediate postoperative hippocampal inflammation and cognitive decline in mice. JCI Insight 2 (7):e91229. doi: 10.1172/jci.insight.91229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tran DQ, Tse EK, Kim MH, Belsham DD (2016) Diet-induced cellular neuroinflammation in the hypothalamus: Mechanistic insights from investigation of neurons and microglia. Mol Cell Endocrinol 438:18–26. doi: 10.1016/j.mce.2016.05.015 [DOI] [PubMed] [Google Scholar]
- 82.Valdearcos M, Douglass JD, Robblee MM, Dorfman MD, Stifler DR, Bennett ML, Gerritse I, Fasnacht R, Barres BA, Thaler JP, Koliwad SK (2017) Microglial Inflammatory Signaling Orchestrates the Hypothalamic Immune Response to Dietary Excess and Mediates Obesity Susceptibility. Cell Metab 26 (1):185–197 e183. doi: 10.1016/j.cmet.2017.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Valdearcos M, Robblee MM, Benjamin DI, Nomura DK, Xu AW, Koliwad SK (2014) Microglia dictate the impact of saturated fat consumption on hypothalamic inflammation and neuronal function. Cell Rep 9 (6):2124–2138. doi: 10.1016/j.celrep.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Latta CH, Sudduth TL, Weekman EM, Brothers HM, Abner EL, Popa GJ, Mendenhall MD, Gonzalez-Oregon F, Braun K, Wilcock DM (2015) Determining the role of IL-4 induced neuroinflammation in microglial activity and amyloid-beta using BV2 microglial cells and APP/PS1 transgenic mice. J Neuroinflammation 12:41. doi: 10.1186/s12974-015-0243-6 [DOI] [PMC free article] [PubMed] [Google Scholar]