Estrogens, often detected in surface waters worldwide, have been classified as endocrine disrupting chemicals and carcinogens. Bacterial degradation is crucial for removing natural estrogens from natural and engineered ecosystems; however, current knowledge regarding the biochemical mechanisms and catabolic enzymes involved in estrogen biodegradation is very limited. Our estrogen metabolite profile and genomic analyses on estrone-degrading bacteria enabled us to characterize the aerobic estrogen degradation pathway. The results greatly expand our understanding of microbial steroid degradation. In addition, the characteristic metabolites, dead-end products, and degradation genes can be used as biomarkers to investigate the fate and biodegradation potential of estrogens in the environment.
KEYWORDS: β-oxidation, 2-oxoacid oxidoreductase, Novosphingobium, Sphingomonas, biodegradation, estrogen, steroid hormones
ABSTRACT
Various bacteria, mainly actinobacteria and proteobacteria, are capable of aerobic estrogen degradation. In a previous study, we used the obligate aerobic alphaproteobacterium Sphingomonas sp. strain KC8 as a model microorganism to identify the initial metabolites involved in the oxygenolytic cleavage of the estrogen A ring: 4-hydroxyestrone, a meta-cleavage product, and a dead-end product pyridinestrone acid. In this study, we identified the downstream metabolites of this aerobic degradation pathway using ultraperformance liquid chromatography–high-resolution mass spectrometry (UPLC-HRMS). 4-Norestrogen-5(10)-en-3-oyl-coenzyme A and its closely related deconjugated (non-coenzyme A [non-CoA]) structure, 4-norestrogenic acid, were detected in the estrone-grown strain KC8 cultures. The structure of 4-norestrogenic acid was elucidated using nuclear magnetic resonance (NMR) spectroscopy. The extracellular distribution and the accumulation of 4-norestrogenic acid in the bacterial cultures indicate that the estrogen-degrading bacteria cannot degrade this deconjugated product. We also observed temporal accumulation and subsequent consumption of a common steroid metabolite, 3aα-H-4α(3′-propanoate)-7aβ-methylhexahydro-1,5-indanedione (HIP), in the bacterial cultures. The metabolite profile and genomic analyses shed light on the biochemical mechanisms involved in the degradation of the A and B rings of natural estrogens. In this proposed aerobic pathway, C-4 of the meta-cleavage product is removed by a 2-oxoacid oxidoreductase through oxidative decarboxylation to produce the 4-norestrogen-5(10)-en-3-oyl-CoA. Subsequently, the B ring is cleaved by hydrolysis. The resulting A/B-ring-cleaved product is transformed into a common steroid metabolite HIP through β-oxidation reactions. Accordingly, the A and B rings of different steroids are degraded through at least three peripheral pathways, which converge at HIP, and HIP is then degraded through a common central pathway.
IMPORTANCE Estrogens, often detected in surface waters worldwide, have been classified as endocrine disrupting chemicals and carcinogens. Bacterial degradation is crucial for removing natural estrogens from natural and engineered ecosystems; however, current knowledge regarding the biochemical mechanisms and catabolic enzymes involved in estrogen biodegradation is very limited. Our estrogen metabolite profile and genomic analyses on estrone-degrading bacteria enabled us to characterize the aerobic estrogen degradation pathway. The results greatly expand our understanding of microbial steroid degradation. In addition, the characteristic metabolites, dead-end products, and degradation genes can be used as biomarkers to investigate the fate and biodegradation potential of estrogens in the environment.
INTRODUCTION
The synthesis and secretion of estrogens occur in most animals, including invertebrates and vertebrates (1, 2). The biosynthetic pathways of estrogens include the elimination of the cholesterol side chain and hydroxylation of the steroid nucleus. In the liver, estrogens undergo structural modifications and are converted into inactive excretion products. The solubility of the resulting products is increased by conjugation with glucuronic acid or sulfate. These conjugates are excreted in urine and feces (3). Primary sources of environmental estrogens thus include human urine and livestock manure (4). In addition, literature and popular media have pointed to 17α-ethinylestradiol, a synthetic estrogen from oral contraceptives, as a major endocrine disrupting chemical (5). Because of the increase in the human population and increasing demands for livestock products, estrogen pollution has become a global concern and challenge (5–7).
Natural and synthetic estrogens have a serious impact on the environment. This has become an increasingly important issue, because exposure to estrogens has adverse physiological and reproductive effects on humans and aquatic animals (8–11). In addition to being endocrine-disrupting chemicals, estrogens have been classified by the World Health Organization as group 1 carcinogens (12). Wastewater treatment plants are crucial for removing estrogens produced by humans and livestock. However, municipal wastewater treatment plants are not able to remove estrogens completely (13, 14). Consequently, estrogens are often detected in surface waters of rivers and coasts near wastewater treatment plants (typically in the nanogram per liter range) (15–19).
Bacterial degradation plays a significant role in the removal of estrogens from the environment (20, 21). Bacteria capable of aerobic estrogen degradation have been isolated from various engineered and natural ecosystems, including activated sludge, seawater, compost, soils, and sandy aquifers; most of these isolates belong to the phyla Actinobacteria and Proteobacteria (22). For example, Fujii et al. (23) reported the isolation and characterization of the first estrogen-degrading sludge bacterium, Novosphingobium tardaugens strain ARI-1. Yoshimoto et al. (24) isolated several estrogen-degrading actinobacteria, identified as Rhodococcus spp.
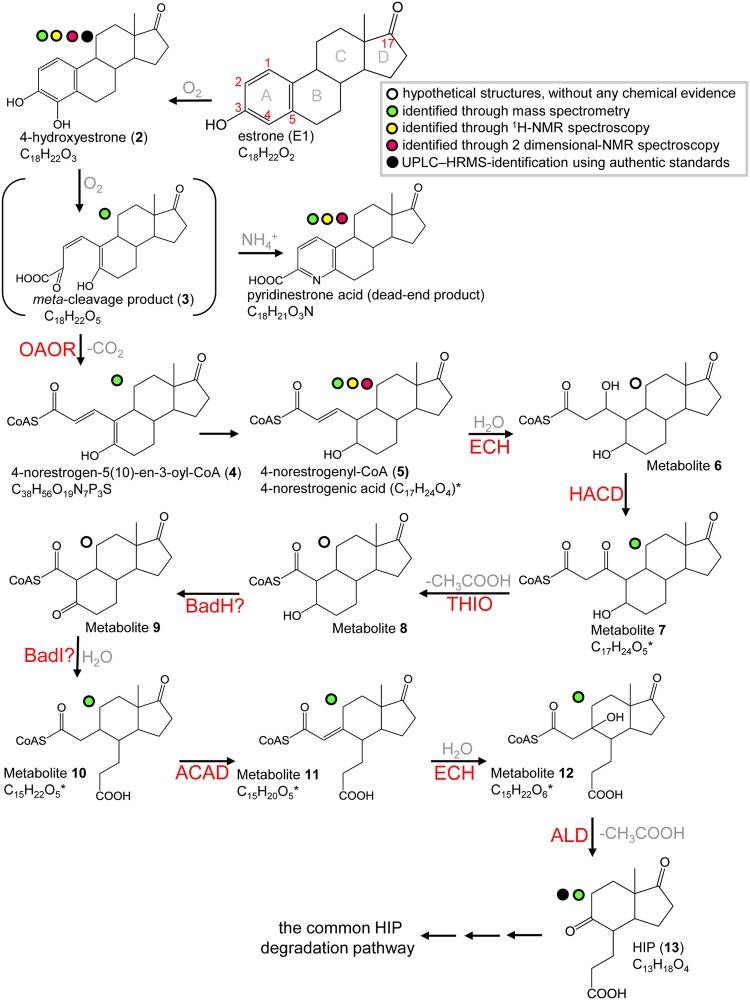
The literature on estrogen degradation pathways is limited. In our recent study (20), we used Sphingomonas sp. strain KC8 (here referred to as strain KC8), a sludge isolate (25), as a model organism to study aerobic estrogen degradation. We identified three initial degradation metabolites: 4-hydroxyestrone (metabolite 2), the meta-cleavage product (metabolite 3), and a dead-end product pyridinestrone acid (see Fig. 1 for the structures of these molecules). We also identified strain KC8 gene clusters likely to be involved in the degradation of estrogen A and B rings (gene clusters I and II) and C and D rings (gene cluster III) (20). Pyridinestrone acid and the key catabolic gene oecC were detected in estrone (E1)-spiked samples collected from wastewater treatment plants and suburban rivers (20, 21), suggesting the prevalence of this aerobic degradation pathway (the 4,5-seco pathway) in the environment. However, the downstream metabolites and corresponding enzymes of this degradation pathway, especially those involved in the A/B ring degradation, remained to be unraveled. In the present study, two estrogen-degrading alphaproteobacteria, strain KC8 (25) and Novosphingobium sp. strain SLCC (21), were used as model microorganisms and incubated with E1. We used ultraperformance liquid chromatography–high-resolution mass spectrometry (UPLC-HRMS) and nuclear magnetic resonance (NMR) spectroscopy to identify the downstream metabolites of this aerobic pathway (Fig. 1). The metabolite profile analysis, together with the genomic analysis, enabled the prediction of biochemical mechanisms operating in the aerobic biodegradation of natural estrogens.
FIG 1.
Proposed bacterial degradation pathway of natural estrogens. *, the deconjugated (non-CoA) metabolites were identified using UPLC-HRMS. The chemical structure of 4-norestrogenic acid, the deconjugated non-CoA structure of metabolite 5, was elucidated using NMR spectroscopy. Abbreviations: ACAD, acyl-CoA dehydrogenase; ALD, aldolase; BadH, 2-hydroxycyclohexanecarboxyl-CoA dehydrogenase; BadI, 2-ketocyclohexanecarboxyl-CoA hydrolase; ECH, enoyl-CoA hydratase; HACD, β-hydroxyacyl-CoA dehydrogenase; OAOR, 2-oxoacid oxidoreductase; THIO, thiolase.
RESULTS AND DISCUSSION
UPLC-HRMS identification of ethyl acetate-extractable estrogen metabolites.
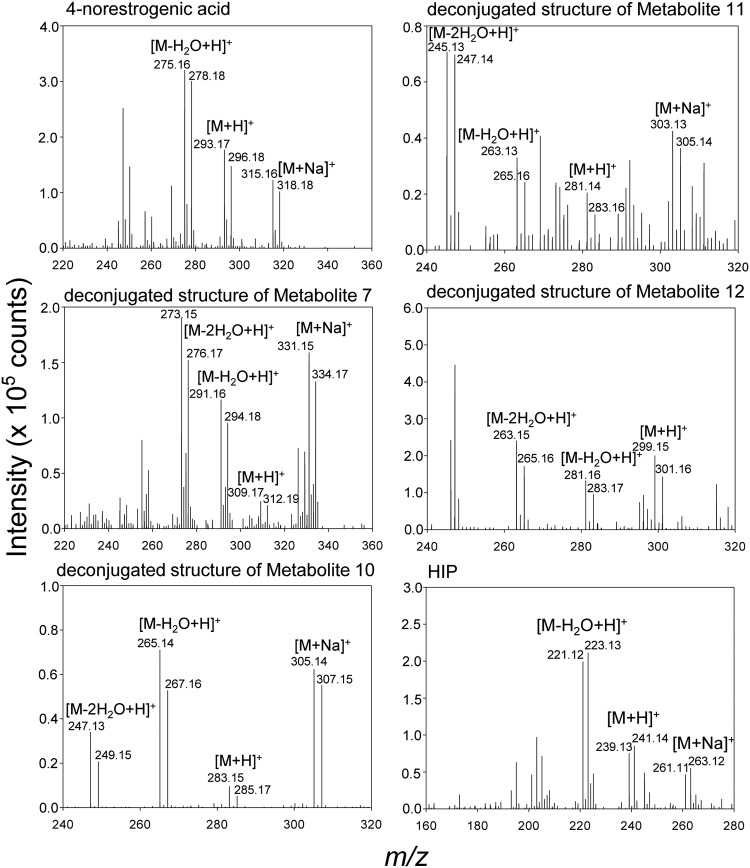
In a previous study (20), we used [3,4C-13C]E1 as a tracer to identify initial metabolites of the 4,5-seco pathway and concluded that bacterial estrogen degradation starts from the A ring. To identify the downstream intermediates of this aerobic pathway, the resting cells of strain KC8 were aerobically incubated with E1 ([2,4,16,16-D4]E1 and unlabeled E1 were mixed at a 1:1 molar ratio), and the hydrophobic estrogen metabolites were extracted using ethyl acetate. Therefore, pairs of adduct ions were observed in the mass spectra of E1-derived metabolites (Fig. 2). We detected 10 E1-derived metabolites in total, including 6 undescribed intermediates (2 C17, 3 C15, and 1 C13 compound) of aerobic estrogen degradation (Table 1). 3aα-H-4α(3′-propanoate)-7aβ-methylhexahydro-1,5-indanedione (HIP), the C13 metabolite, was identified by comparison with the authentic standard (purchased from Sigma-Aldrich) using UPLC-HRMS. The UPLC retention time and HRMS behavior of a detected metabolite were identical to those of the HIP standard (see Fig. S1 in the supplemental material). This result excludes the possibility of detecting structural isomers, which are often observed in chemical analyses relying only on mass spectrometry. A highly similar estrogen metabolite profile was also observed in the resting cell assay of strain SLCC, and the 6 novel estrogen metabolites were also detected (see Table S1).
FIG 2.
ESI-HRMS spectra of the estrogen metabolites of strain KC8 incubated with 1 mM E1. In the resting cell assay, the substrate was composed of [2,4,16,16-D4]E1 and unlabeled E1 (mixed at a 1:1 molar ratio). Only estrogen metabolites newly identified in this study are shown.
TABLE 1.
UPLC-HRMS analysis of metabolites involved in aerobic E1 catabolism by strain KC8
| Compound IDa | UPLC RTb
(min) |
Molecular formula (predicted mol wt)c |
Dominant ion peaks |
Identified product ions |
Observation mode |
|---|---|---|---|---|---|
| E1 | 8.14 | C18H22O2 (270.16) | 253.16 | [M-H2O+H]+ | ESI and APCI |
| 271.17 | [M+H]+ | ESI and APCI | |||
| 4-Hydroxyestrone | 7.41 | C18H22O3 (286.16) | 269.16 | [M-H2O+H]+ | APCI |
| 287.15 | [M+H]+ | ESI and APCI | |||
| 309.15 | [M+Na]+ | ESI | |||
| meta-Cleavage product | 6.85 | C18H22O5 (318.15) | 301.15 | [M-H2O+H]+ | ESI |
| 341.14 | [M+Na]+ | ESI | |||
| Pyridinestrone acid | 4.02 | C18H21O3N (299.15) | 282.17 | [M-H2O+H]+ | ESI |
| 300.16 | [M+H]+ | ESI and APCI | |||
| 322.15 | [M+Na]+ | ESI | |||
| Compound Ie | 5.61 | C16H24O4 (280.17) | 245.15 | [M-2H2O+H]+ | ESI |
| 263.16 | [M-H2O+H]+ | ESI and APCI | |||
| 281.17 | [M+H]+ | ESI and APCI | |||
| 303.16 | [M+Na]+ | ESI | |||
|
4-Norestrogenic acid
(metabolite 5)d |
5.92 | C17H24O4 (292.17) | 257.15 | [M-2H2O+H]+ | ESI |
| 275.16 | [M-H2O+H]+ | ESI and APCI | |||
| 293.17 | [M+H]+ | ESI and APCI | |||
| 315.16 | [M+Na]+ | ESI | |||
| Metabolite 7d | 6.24 | C17H24O5 (308.16) | 273.15 | [M-2H2O+H]+ | ESI and APCI |
| 291.16 | [M-H2O+H]+ | ESI and APCI | |||
| 309.17 | [M+H]+ | ESI and APCI | |||
| 331.15 | [M+Na]+ | ESI | |||
| Metabolite 10d | 5.05 | C15H22O5 (282.15) | 247.13 | [M-2H2O+H]+ | ESI and APCI |
| 265.14 | [M-H2O+H]+ | ESI and APCI | |||
| 283.15 | [M+H]+ | ESI and APCI | |||
| 305.14 | [M+Na]+ | ESI | |||
| Metabolite 11d | 5.47 | C15H20O5 (280.13) | 245.13 | [M-2H2O+H]+ | ESI and APCI |
| 263.13 | [M-H2O+H]+ | ESI and APCI | |||
| 281.14 | [M+H]+ | ESI and APCI | |||
| 303.13 | [M+Na]+ | ESI | |||
| Metabolite 12d | 5.19 | C15H22O6 (298.14) | 263.15 | [M-2H2O+H]+ | ESI and APCI |
| 281.16 | [M-H2O+H]+ | ESI and APCI | |||
| 299.15 | [M+H]+ | ESI | |||
| HIP | 3.78 | C13H18O4 (238.12) | 221.12 | [M-H2O+H]+ | ESI and APCI |
| 239.13 | [M+H]+ | ESI and APCI | |||
| 261.11 | [M+Na]+ | ESI |
ID, identifier. Estrogen metabolites newly identified in this study are in boldface font.
RT, retention time.
The predicated molecular mass was calculated using the atom mass of 12C (12.00), 16O (15.99), and 1H (1.01).
The deconjugated non-CoA structures of the hypothetical CoA ester intermediates were identified using UPLC-HRMS.
Elucidating the structure of a novel estrogen metabolite through NMR analysis.
4-Norestrogenic acid, an unprecedented C17 estrogen metabolite, accumulated in the two E1-degrading strains (Tables 1 and S1). In addition, 4-norestrogenic acid was mostly (approximately 90% produced by bacterial cells) detected in extracellular fractions. To produce a sufficient amount of this compound for NMR analysis, strain SLCC (2 liters) was aerobically incubated with unlabeled E1 (1 mM) for 12 h. The extracellular distribution of 4-norestrogenic acid facilitates the separation of this compound from biomolecules in the cells. 4-Norestrogenic acid was sequentially purified using solid-phase extraction, HPLC, and thin-layer chromatography (TLC). The structure of 4-norestrogenic acid was elucidated using atmospheric pressure chemical ionization (APCI)-HRMS and 2-dimensional NMR spectroscopy.
The TLC-purified compound, obtained as a colorless oil, was assigned the molecular formula of C17H24O4 based on a pseudomolecular ion peak [M+H]+ at m/z 293.17 in the positive mode APCI-HRMS (Fig. 3A). The 1H and 13C NMR signals (Table 2; see also the original NMR spectra in Fig. S2) of this compound were compatible with those of 4-hydroxyestrone (a reference compound) except for the signals on the A and B rings, indicating distinct structural changes in the moiety. Considering the molecular formula, C17H24O4, the double bond equivalence of 4-norestrogenic acid is six, including one ketone at δC 223.7 (C-17), one double bond at δC 145.3 (C-1)/131.7 (C-2), one acid carbonyl at δC 176.0 (C-3), and three rings (Fig. S2B). Comparing the 13C NMR data of the two compounds (the TLC-purified compound and 4-hydroxyestrone), the major differences were the apparent downfield shifts of C-1, C-2, C-3, and C-6, upfield shifts of C-5 and C-10, and disappearance of C-4 (Table 2). These differences suggest that the A ring was cleaved. Mutual 3J correlations of δH 6.26 (H-1)/δH 5.87 (H-2), δH 6.26 (H-1)/δH 1.82 (H-10), δH 1.82 (H-10)/δH 0.93 (H-9), and δH 1.82 (H-10)/δH 3.35 (H-5) were observed in the correlation spectroscopy (COSY) spectrum of the TLC-purified compound (Fig. 3B; see also the original COSY spectrum in Fig. S3A), which were further corroborated by the HMBC correlations of δH 6.26 (H-1)/δC 176.0 (C-3), δH 5.87 (H-2)/δC 56.3 (C-10) and 176.0 (C-3), and δH 1.82 (H-10)/δC 47.0 (C-9) and 73.8 (C-5) (Fig. 3B, see the original heteronuclear multiple bond correlation (HMBC) spectrum in Fig. S3B). The configuration of Δ1(2) was determined to be E form based on the JH-1/H-2 of 15.5 Hz. The TLC-purified compound is named 4-norestrogenic acid, and its structure is shown in Fig. 3B.
FIG 3.
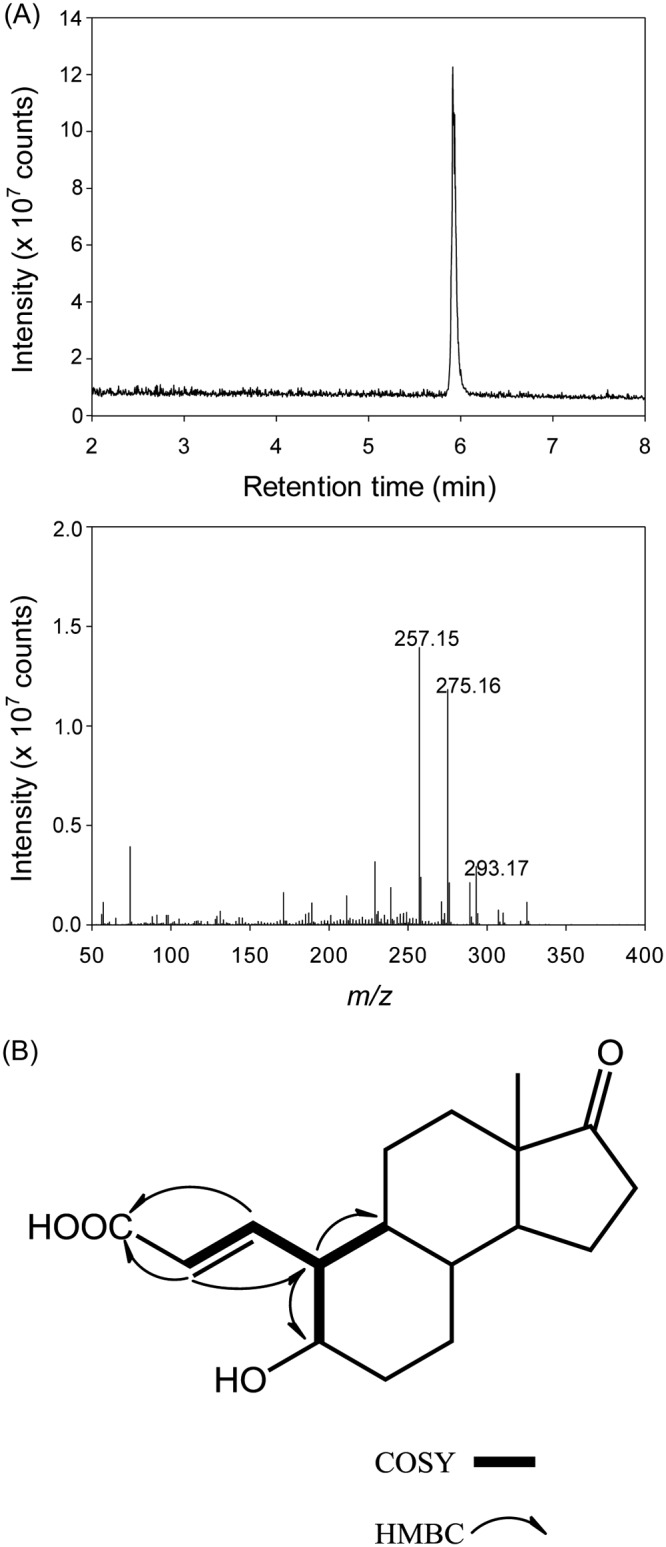
UPLC-APCI-HRMS (A) and key correlations in the 2-dimensional NMR data (B) of the TLC-purified 4-norestrogenic acid. Original NMR spectra are provided in Fig. S2 and S3 in the supplemental material.
TABLE 2.
1H (500 MHz) and 13C NMR (125 MHz) spectral data of 4-norestrogenic acid and 4-hydroxyestrone (a reference)a
| Position | 4-Norestrogenic acid |
4-Hydroxyestrone |
||
|---|---|---|---|---|
| 1H (δ [ppm]) (J [Hz]) | 13C | 1H(δ [ppm]) (J [Hz]) | 13C | |
| 1 | 6.26 dd (9.9, 15.5) | 145.3 | 6.66 d (7.8) | 117.1 |
| 2 | 5.87 d (15.5) | 131.7 | 6.61 d (7.8) | 113.3 |
| 3 | 176.0 | 143.5 | ||
| 4 | 143.5 | |||
| 5 | 3.35 m | 73.8 | 125.0 | |
| 6 | 2.03 m | 35.1 | 2.94 dd (3.8, 12.3) | 24.6 |
| 1.36 m | 2.63 m | |||
| 7 | 1.83 m | 29.2 | 2.38 m | 27.2 |
| 1.10 m | ||||
| 8 | 1.31 m | 40.9 | 1.47 m | 39.4 |
| 9 | 0.93 m | 47.0 | 2.21 m | 45.5 |
| 10 | 1.82 m | 56.3 | 133.0 | |
| 11 | 1.68 m | 28.0 | 2.10 m | 27.4 |
| 1.22 m | 1.37 m | |||
| 12 | 1.68 m | 32.7 | 1.89 m | 32.9 |
| 1.21 m | 1.44 m | |||
| 13 | 49.4 | 49.4 | ||
| 14 | 1.35 m | 51.8 | 1.54 m | 51.7 |
| 15 | 1.95 m | 22.7 | 2.09 m | 22.5 |
| 1.59 m | 1.68 m | |||
| 16 | 2.43 dd (8.8, 19.3) | 36.8 | 2.51 dd (8.3, 18.4) | 36.8 |
| 2.07 dd (8.8, 19.3) | 2.15 dd (8.3, 18.4) | |||
| 17 | 223.7 | 224.0 | ||
| 18 | 0.89 s | 14.4 | 0.93 s | 14.3 |
Measured in methanol-d4.
Role of 4-norestrogenic acid in the estrogen degradation pathway.
At the beginning of the strain KC8 assay, only the substrate E1 was detected. After 4 h of aerobic incubation, two estrogen metabolites, HIP and 4-norestrogenic acid, apparently accumulated. After that, the amount of HIP gradually decreased over time (Fig. 4A). In contrast, 4-norestrogenic acid was not apparently consumed during the aerobic incubation. Similar temporal changes in estrogen metabolites were observed in the resting cell assay of strain SLCC (Fig. 4B). The cell extract of strain KC8 was incubated with 4-norestrogenic acid (0.1 mM), coenzyme A (CoASH; 1 mM), and ATP (5 mM). After 16 h of aerobic incubation, neither substrate consumption nor coenzyme A (CoA) ester production was observed (see Fig. S4). Moreover, strain KC8 cells were incubated with 0.1 mM 4-norestrogenic acid, and the bacterial cells did not import this compound within 6 days (see Fig. S5). These results suggest that transport protein, CoA ligase, or CoA transferase capable of channeling 4-norestrogenic acid back into the estrogen catabolic pathway is absent in strain KC8. It appears that 4-norestrogenic acid cannot be degraded by the estrogen-degrading bacteria. In contrast, the CoA ester of 4-norestrogenic acid, 4-norestrogenyl-CoA (metabolite 5 in Fig. 1), may be a crucial metabolite for aerobic estrogen degradation. The cellular CoA concentration fluctuates depending on the carbon source and growth state (26). CoA is an essential cofactor in numerous biosynthetic and energy-yielding metabolic pathways. When CoA is required in other metabolic pathways, the CoA esters in the estrogen degradation pathway (e.g., 4-norestrogenyl-CoA and metabolites 6 to 12) might be deconjugated by enzymatic activities (e.g., thioesterase or CoA transferase) (26). Alternatively, the abiotic hydrolysis of the CoA esters might occur due to the instability of the thioester structures (27). In this study, we incubated the bacteria with a very high concentration of E1 (1 mM), which may have led to the overproduction of 4-norestrogenic acid and other deconjugated (non-CoA) structures. CoA ester metabolites (28) and their deconjugated (non-CoA) structures (27) were also detected in Sterolibacterium denitrificans DSM 13999 anaerobically grown with a high concentration (millimolar scale) of cholesterol.
FIG 4.
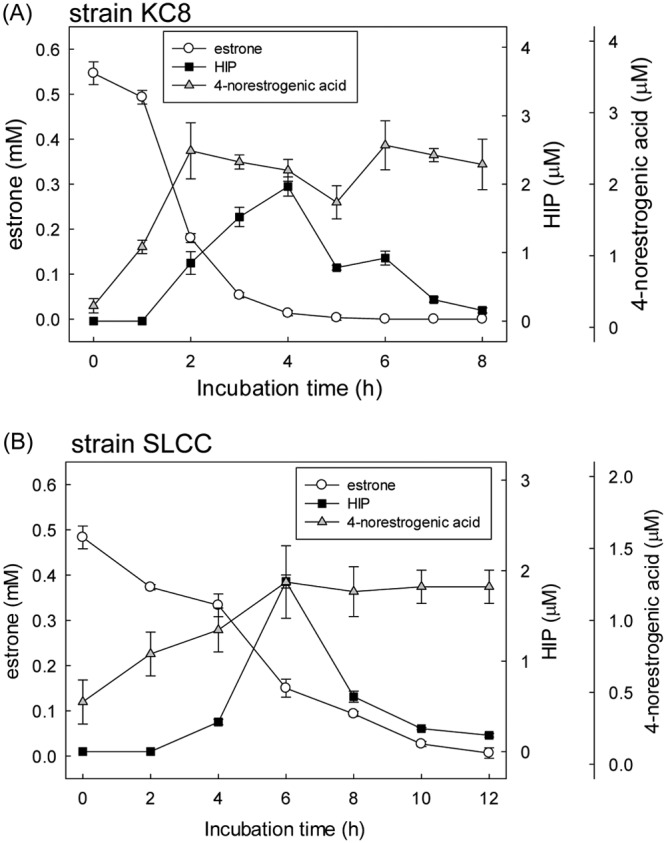
Time course of the E1 consumption as well as the production of 4-norestrogenic acid and HIP in strains KC8 (A) and SLCC (B). The resting cells were aerobically incubated with E1 (1 mM). The [2,4,16,16-D4]E1 and unlabeled E1 were mixed at a 1:1 molar ratio. Quantification of the estrogen metabolites was based on pesudomolecular ion counts corresponding to unlabeled compounds using UPLC-ESI-HRMS. Data shown are the means ± SD from three experimental measurements.
CoA ester intermediates are involved in the aerobic degradation of estrogen A and B rings.
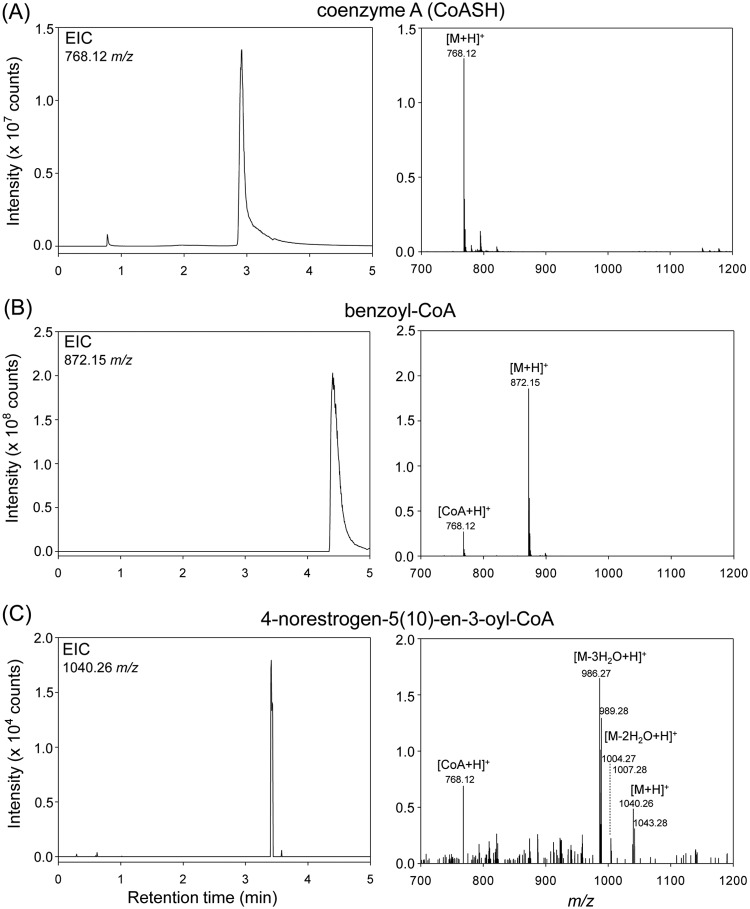
We then identified the hypothetical CoA ester intermediates. Strain KC8 cells in a resting cell assay were incubated with [2,4,16,16-D4]E1 and unlabeled E1 (mixed at a 1:1 molar ratio) for 4 h; therefore, pairs of adduct ion peaks should present in the MS spectra of the E1-derived CoA esters. After incubation, strain KC8 cells were lysed by sonication. The relatively hydrophilic CoA ester intermediates were separated from other bacterial metabolites by solid-phase extraction and subjected to UPLC-electrospray ionization (ESI)-HRMS analysis (Fig. 5). Similar procedures have been applied to extract and identify CoA ester intermediates derived from cholesterol (28) and bile acids (29), suggesting that the method combining solid-phase extraction and UPLC-ESI-HRMS is suitable for metabolite profile analyses of CoA esters. Two authentic standards, CoASH and benzoyl-CoA, were used to confirm the ionization efficiency and investigate the fragmentation patterns of the CoA esters under the UPLC-ESI-HRMS conditions. Pseudomolecular ion [M+H]+ peaks were present in the HRMS spectra of CoASH (retention time, 2.92 min) (Fig. 5A) and benzoyl-CoA (retention time, 4.40 min) (Fig. 5B). Moreover, a fragment ion (m/z 768.12) peak corresponding to the CoA moiety was observed in the HRMS spectrum of benzoyl-CoA (Fig. 5B). Using the solid-phase extraction method, we observed an [M+H]+ peak (m/z 1,040.26; retention time, 3.43 min) corresponding to a hypothetical CoA ester, 4-norestrogen-5(10)-en-3-oyl-CoA (metabolite 4) (Fig. 1), in the UPLC chromatogram of the bacterial extract (Fig. 5C). Moreover, the characteristic fragment ion peak of CoA (m/z 768.12) is also present in the MS spectrum of this compound. Furthermore, pairs of adduct ion peaks, with the m/z difference of 3, were observed in the mass spectrum of 4-norestrogen-5(10)-en-3-oyl-CoA (Fig. 5C), consistent with the loss of an 2H on C-4 of E1 after the decarboxylation step (Fig. 1). We did not detect other hypothetical CoA esters (metabolites 5 to 12). This might indicate that the following reactions leading to HIP are very fast and do not result in the accumulation of the CoA ester intermediates.
FIG 5.
Extracted ion chromatograms (EIC) and MS spectra of two authentic standards, CoASH (A) and benzoyl-CoA (B), and the E1-derived 4-norestrogen-5(10)-en-3-oyl-CoA (metabolite 4) (C). In the resting cell assay of strain KC8, the substrate was composed of [2,4,16,16-D4]E1 and unlabeled E1 (mixed at a 1:1 molar ratio). The fragment ion (m/z 768.12) peak corresponding to the CoA moiety was observed in the MS spectra of benzoyl-CoA and 4-norestrogen-5(10)-en-3-oyl-CoA.
In the aerobic estrogen degradation by Nocardia sp. strain E110, the hydrolytic cleavage of the meta-cleavage product is followed by a decarboxylation reaction, producing a C17 metabolite with cleaved A and B rings (30). However, the enzymes involved were not identified. In the present study, the estrogen metabolite profile analyses of strains KC8 and SLCC also suggest that the B ring of the meta-cleavage product is opened via hydrolysis. The electron deficiency of the carbonyl carbon in the B ring facilitates the hydrolytic cleavage. Moreover, the identification of 4-norestrogen-5(10)-en-3-oyl-CoA, 4-norestrogenic acid, and the B-ring-cleaved metabolites suggests that in the aerobic estrogen degradation by proteobacteria, the C-4 removal through a decarboxylation reaction precedes the B ring cleavage. A similar hydrolytic ring cleavage mechanism was demonstrated in the degradation of cyclohexanecarboxylic acid by an alphaproteobacterium Rhodopseudomonas palustris (31, 32). Comparable estrogen metabolites (e.g., metabolite 10 corresponding to the hydrolytic product pimeloyl-CoA) have been detected in the two E1-degrading strains. However, genes similar (with a deduced amino acid sequence identity >40%) to 2-hydroxycyclohexanecarboxyl-CoA dehydrogenase (BadH) or 2-ketocyclohexanecarboxyl-CoA hydrolase (BadI) were not present in gene cluster II of strain KC8. Nonetheless, further investigation is required to identify the corresponding genes and enzymes involved in the estrogen B ring cleavage.
Gene cluster II of strain KC8 is involved in the degradation of estrogen A and B rings.
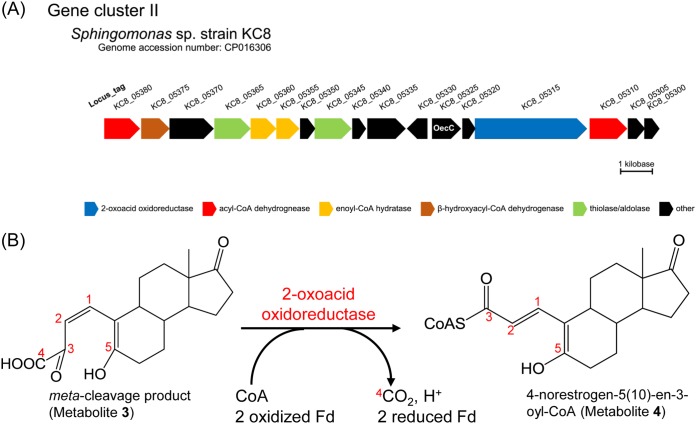
In a previous study (20), we identified a gene cluster (gene cluster II) in the genome of strain KC8, and the genes involved were specifically expressed during aerobic growth on E1. This gene cluster is also present in other estrogen-degrading bacteria. However, the functions of those gene products were unclear except for the 4-hydroxyestrone 4,5-dioxygenase (OecC, the product of KC8_05325). Here, we identified a gene (KC8_05315) encoding a member of the indolepyruvate ferredoxin oxidoreductase (IOR) family (Fig. 6A). Members of this enzyme family contain thiamine diphosphate and [4Fe-4S] clusters and catalyze the decarboxylation of 2-oxoacids [e.g., pyruvate, phenylpyruvate, (indol-3-yl)pyruvate, and 2-oxoglutarate] to form their CoA derivatives (33–35). The C-3 and C-4 of the meta-cleavage product represent a typical 2-oxo acid structure; thus, the meta-cleavage product and a CoA may serve as the substrates for the 2-oxoacid oxidoreductase of strain KC8, producing 4-norestrogen-5(10)-en-3-oyl-CoA through an oxidative decarboxylation (Fig. 6B).
FIG 6.
(A) Identification of genes encoding the β-oxidation enzymes and 2-oxoacid oxidoreductase in gene cluster II of strain KC8. (B) Oxidative decarboxylation of the meta-cleavage product by the 2-oxoacid oxidoreductase (the product of KC8_5315) of strain KC8. Fd, ferredoxin.
In addition, we identified a gene (KC8_05370) encoding a 3-hydroxy-3-methylglutaryl-CoA synthase-like protein; this enzyme appears to use a CoA ester metabolite derived from E1 as the substrate. Genes encoding the β-oxidation enzymes, including acyl-CoA dehydrogenase (KC8_05380 and KC8_05310), enoyl-CoA hydratase (KC8_05355 and KC8_05360), β-hydroxyacyl-CoA dehydrogenase (KC8_05375), and thiolase/aldolase (KC8_05345 and KC8_05365), are present in gene cluster II of strain KC8 (Fig. 6A). Strain KC8 cannot degrade sterols or cholic acid; thus, these β-oxidation genes are not involved in the side chain degradation. Moreover, gene clusters involved in the side chain degradation of steroids (36–40) are not present in the KC8 genome. The potential roles of these β-oxidation enzymes are shown in Fig. 1. Among the two thiolase/aldolase genes, the deduced amino acid sequence of KC8_05345 exhibited higher similarity to those of the aldolases of S. denitrificans (28) (gene product of SDENv1_10308; 45% sequence identity) and Pseudomonas sp. strain Chol1 (40) (gene product of C211_11377; 47% sequence identity), both of which are responsible for the transformation of steroid C22-oyl-CoA to androsta-1,4-diene-3,17-dione; this implies that the product of KC8_05345 might catalyze the aldolytic side chain degradation of the CoA ester of metabolite 12 to produce HIP.
Gene cluster III of strain KC8 is involved in the HIP degradation.
Two pieces of evidence imply that strain KC8 uses the same gene products to degrade the C and D rings of both androgens and estrogens: (i) a conserved gene cluster (gene cluster III) involved in the degradation of steroid C and D rings is present in the strain KC8 genome, and (ii) members of this gene cluster are commonly expressed in the E1- and testosterone-grown cells (20). The gene cluster for C/D ring degradation is widely distributed in testosterone- and cholesterol-degrading bacteria (41, 42), and HIP is a critical substrate for the gene products. The HIP catabolic pathway has been established in Mycobacterium tuberculosis (43); in this pathway, the hydrolytic cleavage of the steroid d ring through EchA20 is followed by the C ring cleavage through another hydrolase, IpdAB. We identified the corresponding strain KC8 genes (KC8_01060 [echA20], KC8_01080 [ipdB], and KC8_01085 [ipdA]). The detection of HIP (this study) and the expression of gene cluster III (20) in E1-grown strain KC8 suggest that the cleavage of the estrogen D ring also precedes the C ring degradation. Consequently, in the aerobic degradation pathway of estrogens, the A, B, D, and C rings of estrogens are sequentially opened (Fig. 1).
Conclusions.
In contrast to the aerobic degradation of the A and B rings of cholesterol (36) and testosterone (44), this study suggests that CoA ester metabolites are largely involved in the aerobic degradation of estrogen A and B rings because of the following: (i) the gene cluster II of strain KC8, which is responsible for the aerobic degradation of estrogen A and B rings, includes at least 7 putative β-oxidation genes, one putative 2-oxoacid oxidoreductase gene, and one putative 3-hydroxy-3-methylglutaryl-CoA synthase gene; (ii) 4-norestrogen-5(10)-en-3-oyl-CoA, the product of the 2-oxoacid oxidoreductase, was detected in the E1-grown cells; and (iii) the biochemical mechanisms involved in estrogen B ring cleavage are highly similar to those involved in the bacterial degradation of cyclohexanecarboxylic acid, and this cyclohexanecarboxylic acid catabolic pathway involves a series of CoA ester intermediates (31, 32).
A highlight of this study was the detection of HIP in the E1-grown bacteria. HIP has been proposed to be a crucial metabolite for the aerobic (36, 44–46) as well as anaerobic (27, 28, 47) degradation of androgens, sterols, and bile acids. Moreover, genes involved in HIP degradation have been identified in the genomes of various aerobes (41, 42) and denitrifying bacteria (47). Accordingly, the A and B rings of different steroids are degraded through several peripheral pathways, including the 9,10-seco pathway (for aerobic degradation of androgens and sterols), the 4,5-seco pathway (for aerobic estrogen degradation), and the 2,3-seco pathway (for anaerobic degradation of androgens and sterols). Nevertheless, these peripheral steroid catabolic pathways finally converge at HIP, and HIP is further degraded through a common central pathway.
The aerobic estrogen degradation pathway was established in bacterial isolates cultivated under laboratory conditions with a very high substrate concentration (1 mM), but our previous studies indicated that the identified metabolites (e.g., the dead-end product pyridinestrone acid) and degradation genes (e.g., oecC) were detectable in the original sludge samples (21) or in various environmental samples treated with a low concentration (0.3 μM) of E1 (20). Moreover, an oecC-containing estrogen degrader, strain SLCC, isolated from the studied wastewater treatment plant was able to degrade E1 with a concentration (3.7 nM) close to environmental levels (21). These results suggest that bacteria may use a highly conserved pathway to degrade natural estrogens, regardless of the substrate concentrations. UPLC–HRMS-based metabolite identification has been successfully applied for monitoring the biodegradation of environmental estrogens (20–22). Compared with intermediates in the degradation pathway, dead-end products such as pyridinestrone acid and 4-norestrogenic acid may serve as more suitable biomarkers for environmental investigations of estrogen biodegradation, because dead-end products are less degradable and thus are more persistent in environmental samples.
MATERIALS AND METHODS
Chemicals and bacterial strains.
Benzoyl-CoA, CoASH, [2,4,16,16-D4]E1 (95 atom % D), 4-hydroxyestrone, and 3aα-H-4α(3′-propanoate)-7aβ-methylhexahydro-1,5-indanedione [HIP; also known as 3-(7a-methyl-1,5-dioxooctahydro-1H-inden-4-yl)propanoic acid] were purchased from Sigma-Aldrich. The other chemicals were of analytical grade and were purchased from Fluka, Mallinckrodt Baker, Merck, and Sigma-Aldrich. Strains KC8 and SLCC were isolated from the municipal wastewater treatment plants as described in our previous studies (21, 25).
Aerobic incubation of bacterial strains with [2,4,16,16-D4]E1.
Strains KC8 and SLCC were used in the resting cell assays. Bacteria were first aerobically grown in R2A medium containing 0.1 mM unlabeled E1 (40 ml in a 250-ml Erlenmeyer flask) at 28°C with shaking (180 rpm). Cells were collected via centrifugation (8,000 × g, 20 min, 15°C) in the exponential growth phase at an optical density at 600 nm (OD600) of 0.5 (optical path, 1 cm). The cell pellet was resuspended in a chemically defined medium described by Chen et al. (20). The cell suspension (OD600 of 2; 10 ml) was fed with 1 mM E1 (unlabeled E1 and [2,4,16,16-D4]E1 mixed at a 1:1 molar ratio) and was aerobically incubated at 28°C with shaking (180 rpm). The cell suspensions of strains KC8 and SLCC were sampled (0.5 ml) every 1 and 2 h, respectively. To facilitate the extraction of carboxylic metabolites of E1, the samples were acidified with 30 μl of 6 N HCl. The resulting samples were extracted twice using equal volumes of ethyl acetate, and the estrogen metabolites were detected by UPLC-ESI-HRMS and UPLC-APCI-HRMS.
UPLC-ESI-HRMS.
Ethyl acetate extractable samples were analyzed by UPLC-ESI-HRMS on a UPLC system coupled to an ESI-mass spectrometer. Separation was achieved on a reversed-phase C18 column (Acquity UPLC BEH C18, 1.7 μm; 100 mm by 2.1 mm; Waters) with a flow rate of 0.4 ml/min at 50°C (column oven temperature). The mobile phase comprised a mixture of two solvents: solvent A (2% [vol/vol] acetonitrile containing 0.1% [vol/vol] formic acid) and solvent B (methanol containing 0.1% [vol/vol] formic acid). Separation was achieved using a linear gradient of solvent B from 5% to 99% over 12 min. Mass spectral data were collected in positive ESI mode in separate runs on a Thermo Fisher Scientific Orbitrap Elite Hybrid Ion Trap-Orbitrap mass spectrometer (Waltham, MA, USA) operated in a scan mode of m/z 50 to 500. The source voltage was set at 3.2 kV, the capillary and source heater temperatures were 360°C and 350°C, respectively, and the sheath, auxiliary, and sweep gas flow rates were 30, 15, and 2 arbitrary units, respectively. The predicted elemental composition of individual intermediates was calculated using Xcalibur mass spectrometry software (Thermo Fisher Scientific).
UPLC-APCI-HRMS.
The ethyl acetate extractable samples or TLC-purified 4-norestrogenic acid were also analyzed using UPLC-APCI-HRMS. The separation conditions for UPLC were the same as those for UPLC-ESI-HRMS. Mass spectral data were obtained using a Thermo Fisher Scientific Orbitrap Elite Hybrid Ion Trap-Orbitrap mass spectrometer (Waltham, MA, USA) equipped with a standard APCI source operating in the positive ion mode. In APCI-HRMS analysis, the capillary and APCI vaporizer temperatures were 120°C and 400°C, respectively; the sheath, auxiliary, and sweep gas flow rates were 40, 5, and 2 arbitrary units, respectively. The source voltage was 6 kV and current was 15 μA. The parent scan was in the range of m/z 50 to 600.
Extraction and identification of CoA ester intermediates derived from E1.
The preparation of the strain KC8 resting cells was the same as described above. The bacterial cells were incubated with unlabeled E1 (0.5 mM) and [2,4,16,16-D4]E1 (0.5 mM) for 4 h. After that, the cells were harvested by centrifugation and stored at −20°C. The frozen cells were gently resuspended in 0.6 ml of double distilled water, and then cells were disrupted by sonication (Bioruptor Pico Sonication System; Diagenode, Denville, NJ, USA) at 4°C for 20 min (20 cycles of 30 s on/30 s off). After sonicating, 30 μl of 6 N HCl was added to the cell lysate, which was vortexed for 5 min and then centrifuged at 13,500 × g for 1 min. The CoA esters were extracted by solid-phase extraction (Bakerbond SPE Octadecyl [C18] disposable extraction column [1 ml] with 100 mg of sorbent) as described (28, 29) with minor modifications. The attached CoA esters were eluted with 1 ml of 30% aqueous methanol (vol/vol) and subjected to UPLC-ESI-HRMS analysis. Two reference compounds, CoASH and benzoyl-CoA, were used to confirm the extraction efficiency of the solid-phase extraction method. The mobile phase for the UPLC separation comprised a mixture of two solvents: solvent A (2% [vol/vol] acetonitrile containing 0.1% [vol/vol] formic acid) and solvent B (acetonitrile containing 0.1% [vol/vol] formic acid); the separation was achieved with a linear gradient of solvent B from 0.1% to 90% within 10.5 min. The ESI-HRMS conditions were the same as described above, except that the parent scan was in the range of m/z 700 to 1,200.
Production of 4-norestrogenic acid by strain SLCC.
Strain SLCC was first aerobically grown in R2A medium containing 0.1 mM unlabeled E1 (totally 4 liters in six 2-liter Erlenmeyer flasks) at 28°C with shaking (180 rpm). Cells were collected by centrifugation (8,000 × g, 20 min, 15°C) in the exponential growth phase at an OD600 of approximately 0.5. The cell pellet was resuspended in 2 liters of a chemically defined medium. The cell suspension was fed 1 mM unlabeled E1 and was aerobically incubated at 28°C with shaking (180 rpm) for 12 h. The supernatant was collected by centrifugation (8,000 × g, 20 min, 15°C). The estrogen metabolites were extracted via solid-phase extraction (Oasis HLB LP extraction cartridges, 6 ml; Waters] with 0.5 g of sorbent). The estrogen metabolites were eluted with 12 ml of methanol, concentrated to 1 ml with a centrifugal evaporator, and purified by HPLC and TLC sequentially. The chemical structure of the TLC-purified metabolite, 4-norestrogenic acid, was elucidated by NMR spectroscopy and UPLC-APCI-HRMS.
Extracellular-intracellular distribution of 4-norestrogenic acid.
The resting cell assays of strains KC8 and SLCC were prepared as described above. The cell suspensions (OD600 of 2; 10 ml) were fed 1 mM unlabeled E1 and then aerobically incubated at 28°C with shaking. The cell suspensions were sampled (1 ml) after 3 h of aerobic incubation with E1. Cells were separated from the medium via centrifugation (13,500 × g, 10 min, 4°C), and the cell pellet was resuspended in 1 ml of double-distilled water. To facilitate the extraction of 4-norestrogenic acid, the samples were acidified using 30 μl of 6 N HCl. The resulting samples were extracted twice using ethyl acetate (1 ml), and the extracted 4-norestrogenic acid was quantified by UPLC-ESI-HRMS.
Incubation of the strain KC8 cell extract with 4-norestrogenic acid and CoASH.
E1-grown strain KC8 cells were harvested by centrifugation. The cell pellet was resuspended in 100 mM HEPES (pH 7.5) containing 10% glycerol and then lysed with a French press (Thermo Fisher Scientific). Soluble proteins were collected by centrifugation (20,000 × g for 30 min, followed by 100,000 × g for 1.5 h) at 4°C. The reaction mixture (1 ml) contained 0.5 ml of soluble protein fraction (20 mg/ml), 0.1 mM 4-norestrogenic acid, 1 mM CoASH, 5 mM ATP, and 10 mM MgSO4. The negative controls were reaction mixtures without soluble proteins or 4-norestrogenic acid. The reaction mixtures were incubated at 30°C for 16 h. The 4-norestrogenic acid and the CoA esters were extracted through liquid-liquid partition and solid-phase extraction, respectively, as described above.
Incubation of strain KC8 cells with 4-norestrogenic acid.
E1-grown strain KC8 cells (OD600 of 0.2; 10 ml) were incubated in a chemically defined medium containing 0.1 mM 4-norestrogenic acid. 17α-Ethinylestradiol (final concentration, 0.05 mM), which cannot be utilized by strain KC8, was added to the bacterial culture to serve as an internal control. After different time intervals of incubation (0, 3, and 6 days), samples (0.5 ml) were withdrawn from the bacterial culture. The bacterial cells were removed via centrifugation (13,500 × g, 20°C, for 10 min). 4-Norestrogenic acid remaining in the supernatant was extracted using ethyl acetate, and the extracted 4-norestrogenic acid was quantified by UPLC-ESI-HRMS.
HPLC.
A reversed-phase (RP) Hitachi HPLC system was used to separate the E1 metabolites. The separation was achieved on an analytical RP C18 column [Luna C18(2), 5 μm, 250 mm by 4.6 mm; Phenomenex, Torrance, CA, USA] with a flow rate of 0.8 ml/min. The separation was performed isocratically at 35°C, with 40% (vol/vol) acetonitrile containing 0.1% (vol/vol) trifluoroacetic acid as an eluent. The estrogen metabolite 4-norestrogenic acid was detected at 210 nm using a photodiode array detector.
TLC.
The estrogen metabolites were separated on silica gel aluminum TLC plates (silica gel 60 F254, 0.2-mm thickness, 20 cm by 20 cm; Merck, Darmstadt, Germany). Ethyl acetate-water-acetic acid (50:1:1 [vol/vol/vol]) was used as the developing solvent system. The estrogen metabolites were visualized by spraying the TLC plates with 30% (vol/vol) H2SO4.
NMR spectroscopy.
1H and 13C NMR spectra were recorded at 27°C by using a Bruker AVIII 500 MHz FT-NMR. Chemical shifts (δ) were recorded and presented in parts per million with deuterated methanol (99.8%) as the solvent and internal reference.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Ministry of Science and Technology of Taiwan (104-2311-B-001-023-MY3 and 107-2311-B-001-021-MY3). We thank the Institute of Plant and Microbial Biology, Academia Sinica, for providing access to the Small Molecule Metabolomics Core Facility (for UPLC-HRMS analyses).
Y.-R.C. designed the research. K.W., Y.-L.C., and Y.-S.W. performed the research. T.-H.L. and K.-H.C. contributed new reagents and analytic tools. P.-H.W., C.-P.Y., and Y.-R.C analyzed the data. Y.-R.C drafted the manuscript. All authors reviewed the manuscript.
The authors declare that they have no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02223-18.
REFERENCES
- 1.Matsumoto T, Osada M, Osawa Y, Mori K. 1997. Gonadal estrogen profile and immunohistochemical localization of steroidogenic enzymes in the oyster and scallop during sexual maturation. Comp Biochem Physiol B Biochem Mol Biol 4:811–817. doi: 10.1016/S0305-0491(97)00233-2. [DOI] [Google Scholar]
- 2.Tarrant AM, Blomquist CH, Lima PH, Atkinson MJ, Atkinson S. 2003. Metabolism of estrogens and androgens by scleractinian corals. Comp Biochem Physiol B Biochem Mol Biol 136:473–485. doi: 10.1016/S1096-4959(03)00253-7. [DOI] [PubMed] [Google Scholar]
- 3.Harvey RA, Ferrier DR. 2011. Biochemistry. Lippincott Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 4.Shore LS, Shemesh M. 2003. Naturally produced steroid hormones and their release into the environment. Pure Appl Chem 75:1859–1871. doi: 10.1351/pac200375111859. [DOI] [Google Scholar]
- 5.Wise A, O'Brien K, Woodruff T. 2011. Are oral contraceptives a significant contributor to the estrogenicity of drinking water? Environ Sci Technol 45:51–60. doi: 10.1021/es1014482. [DOI] [PubMed] [Google Scholar]
- 6.Whitman WB. 2017. Bacteria and the fate of estrogen in the environment. Cell Chem Biol 24:652–653. doi: 10.1016/j.chembiol.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 7.Lange IG, Daxenberger A, Schiffer B, Witters H, Ibarreta D, Meyer HHD. 2002. Sex hormones originating from different livestock production systems: fate and potential disrupting activity in the environment. Anal Chim Acta 473:27–37. doi: 10.1016/S0003-2670(02)00748-1. [DOI] [Google Scholar]
- 8.Teles M, Gravato C, Pacheco M, Santos MA. 2004. Juvenile sea bass biotransformation, genotoxic and endocrine responses to β-naphthoflavone, 4-nonylphenol and 17β-estradiol individual and combined exposures. Chemosphere 57:147–158. doi: 10.1016/j.chemosphere.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 9.Jobling S, Williams R, Johnson A, Taylor A, Gross-Sorokin M, Nolan M, Tyler CR, van Aerle R, Santos E, Brighty G. 2006. Predicted exposures to steroid estrogens in U.K. rivers correlate with widespread sexual disruption in wild fish populations. Environ Health Perspect 114:32–39. doi: 10.1289/ehp.8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinck JE, Blazer VS, Schmitt CJ, Papoulias DM, Tillitt DE. 2009. Widespread occurrence of intersex in black basses (Micropterus spp.) from U.S. rivers, 1995–2004. Aquat Toxicol 95:60–70. doi: 10.1016/j.aquatox.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Lambert MR, Giller GSJ, Barber LB, Fitzgerald KC, Skelly DK. 2015. Suburbanization, estrogen contamination, and sex ratio in wild amphibian populations. Proc Natl Acad Sci U S A 112:11881–11886. doi: 10.1073/pnas.1501065112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.International Agency for Research on Cancer. 2018. Agents classified by the IARC monographs. https://monographs.iarc.fr/list-of-classifications-volumes/.
- 13.Ternes TA, Stumpf M, Mueller J, Haberer K, Wilken RD, Servos M. 1999. Behavior and occurrence of estrogens in municipal sewage treatment plants–I. Investigations in Germany, Canada and Brazil. Sci Total Environ 225:81–90. doi: 10.1016/S0048-9697(98)00334-9. [DOI] [PubMed] [Google Scholar]
- 14.Fan Z, Wu S, Chang H, Hu J. 2011. Behaviors of glucocorticoids, androgens and progestogens in a municipal sewage treatment plant: comparison to estrogens. Environ Sci Technol 45:2725–2733. doi: 10.1021/es103429c. [DOI] [PubMed] [Google Scholar]
- 15.Belfroid AC, Van der Horst A, Vethaak AD, Schäfer AJ, Rijs GBJ, Wegener J, Cofino WP. 1999. Analysis and occurrence of estrogenic hormones and their glucuronides in surface water and waste water in The Netherlands. Sci Total Environ 225:101–108. doi: 10.1016/S0048-9697(98)00336-2. [DOI] [PubMed] [Google Scholar]
- 16.Baronti C, Curini R, D'Ascenzo G, Di Corcia A, Gentili A, Samperi R. 2000. Monitoring natural and synthetic estrogens at activated sludge sewage treatment plants and in a receiving river water. Environ Sci Technol 34:5059–5066. doi: 10.1021/es001359q. [DOI] [Google Scholar]
- 17.Kolodziej EP, Gray JL, Sedlak DL. 2003. Quantification of steroid hormones with pheromonal properties in municipal wastewater effluent. Environ Toxicol Chem 22:2622–2629. doi: 10.1897/03-42. [DOI] [PubMed] [Google Scholar]
- 18.Huang CH, Sedlak DL. 2001. Analysis of estrogenic hormones in municipal wastewater effluent and surface water using enzyme-linked immunosorbent assay and gas chromatography/tandem mass spectrometry. Environ Toxicol Chem 20:133–139. doi: 10.1002/etc.5620200114. [DOI] [PubMed] [Google Scholar]
- 19.Lee YC, Wang LM, Xue YH, Ge NC, Yang XM, Chen GH. 2006. Natural estrogens in the surface water of Shenzhen and the sewage discharge of Hong Kong. Hum Ecol Risk Assess Int J 12:301–312. doi: 10.1080/10807030500533394. [DOI] [Google Scholar]
- 20.Chen YL, Yu CP, Lee TH, Goh KS, Chu KH, Wang PH, Ismail W, Shih CJ, Chiang YR. 2017. Biochemical mechanisms and catabolic enzymes involved in bacterial estrogen degradation pathways. Cell Chem Biol 24:712.e7–724.e7. doi: 10.1016/j.chembiol.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 21.Chen YL, Fu HY, Lee TH, Shih CJ, Huang L, Wang YS, Ismail W, Chiang YR. 2018. Estrogen degraders and estrogen degradation pathway identified in an activated sludge. Appl Environ Microbiol 84:e00001-18. doi: 10.1128/AEM.00001-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu CP, Deeb RA, Chu KH. 2013. Microbial degradation of steroidal estrogens. Chemosphere 91:1225–1235. doi: 10.1016/j.chemosphere.2013.01.112. [DOI] [PubMed] [Google Scholar]
- 23.Fujii K, Kikuchi S, Satomi M, Ushio-Sata N, Morita N. 2002. Degradation of 17β-estradiol by a Gram-negative bacterium isolated from activated sludge in a sewage treatment plant in Tokyo, Japan. Appl Environ Microbiol 68:2057–2060. doi: 10.1128/AEM.68.4.2057-2060.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshimoto T, Nagai F, Fujimoto J, Watanabe K, Mizukoshi H, Makino T, Kimura K, Saino H, Sawada H, Omura H. 2004. Degradation of estrogens by Rhodococcus zopfii and Rhodococcus equi isolates from activated sludge in wastewater treatment plants. Appl Environ Microbiol 70:5283–5289. doi: 10.1128/AEM.70.9.5283-5289.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu CP, Roh H, Chu KH. 2007. 17β-estradiol-degrading bacteria isolated from activated sludge. Environ Sci Technol 41:486–492. doi: 10.1021/es060923f. [DOI] [PubMed] [Google Scholar]
- 26.Takamura Y, Nomura G. 1988. Changes in the intracellular concentration of acetyl-CoA and malonyl-CoA in relation to the carbon and energy metabolism of Escherichia coli K12. J Gen Microbiol 134:2249–2253. doi: 10.1099/00221287-134-8-2249. [DOI] [PubMed] [Google Scholar]
- 27.Lin CW, Wang PH, Ismail W, Tsai YW, El Nayal A, Yang CY, Yang FC, Wang CH, Chiang YR. 2015. Substrate uptake and subcellular compartmentation of anoxic cholesterol catabolism in Sterolibacterium denitrificans. J Biol Chem 290:1155–1169. doi: 10.1074/jbc.M114.603779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warnke M, Jacoby C, Jung T, Agne M, Mergelsberg M, Starke R, Jehmlich N, von Bergen M, Richnow H-H, Brüls T, Boll M. 2017. A patchwork pathway for oxygenase-independent degradation of side chain containing steroids. Environ Microbiol 19:4684–4699. doi: 10.1111/1462-2920.13933. [DOI] [PubMed] [Google Scholar]
- 29.Gan-Schreier H, Okun JG, Kohlmueller D, Langhans CD, Peters V, Ten Brink HJ, Verhoeven NM, Jakobs C, Voelkl A, Hoffmann GF. 2005. Measurement of bile acid CoA esters by high-performance liquid chromatography-electrospray ionisation tandem mass spectrometry (HPLC-ESI-MS/MS). J Mass Spectrom 40:882–889. doi: 10.1002/jms.864. [DOI] [PubMed] [Google Scholar]
- 30.Coombe RG, Tsong YY, Hamilton PB, Sih CJ. 1966. Mechanisms of steroid oxidation by microorganisms X. Oxidative cleavage of estrone. J Biol Chem 241:1587–1595. [PubMed] [Google Scholar]
- 31.Pelletier DA, Harwood CS. 1998. 2-Ketocyclohexanecarboxyl coenzyme A hydrolase, the ring cleavage enzyme required for anaerobic benzoate degradation by Rhodopseudomonas palustris. J Bacteriol 180:2330–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelletier DA, Harwood CS. 2000. 2-Hydroxycyclohexanecarboxyl coenzyme A dehydrogenase, an enzyme characteristic of the anaerobic benzoate degradation pathway used by Rhodopseudomonas palustris. J Bacteriol 182:2753–2760. doi: 10.1128/JB.182.10.2753-2760.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mai X, Adams MW. 1994. Indolepyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. A new enzyme involved in peptide fermentation. J Biol Chem 269:16726–16732. [PubMed] [Google Scholar]
- 34.Tersteegen A, Linder D, Thauer RK, Hedderich R. 1997. Structures and functions of four anabolic 2-oxoacid oxidoreductases in Methanobacterium thermoautotrophicum. Eur J Biochem 244:862–868. doi: 10.1111/j.1432-1033.1997.00862.x. [DOI] [PubMed] [Google Scholar]
- 35.Yan Z, Maruyama A, Arakawa T, Fushinobu S, Wakagi T. 2016. Crystal structures of archaeal 2-oxoacid:ferredoxin oxidoreductases from Sulfolobus tokodaii. Sci Rep 6:33061. doi: 10.1038/srep33061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van der Geize R, Yam K, Heuser T, Wilbrink MH, Hara H, Anderton MC, Sim E, Dijkhuizen L, Davies JE, Mohn WW, Eltis LD. 2007. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc Natl Acad Sci U S A 104:1947–1952. doi: 10.1073/pnas.0605728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capyk JK, Kalscheuer R, Stewart GR, Liu J, Kwon H, Zhao R, Okamoto S, Jacobs WR Jr, Eltis LD, Mohn WW. 2009. Mycobacterial cytochrome P450 125 (Cyp125) catalyzes the terminal hydroxylation of C27 steroids. J Biol Chem 284:35534–35542. doi: 10.1074/jbc.M109.072132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosłoniec KZ, Wilbrink MH, Capyk JK, Mohn WW, Ostendorf M, van der Geize R, Dijkhuizen L, Eltis LD. 2009. Cytochrome P450 125 (CYP125) catalyses C26-hydroxylation to initiate sterol side-chain degradation in Rhodococcus jostii RHA1. Mol Microbiol 74:1031–1043. doi: 10.1111/j.1365-2958.2009.06915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casabon I, Crowe AM, Liu J, Eltis LD. 2013. FadD3 is an acyl-CoA synthetase that initiates catabolism of cholesterol rings C and D in actinobacteria. Mol Microbiol 87:269–283. doi: 10.1111/mmi.12095. [DOI] [PubMed] [Google Scholar]
- 40.Holert J, Kulić Ž, Yücel O, Suvekbala V, Suter MJ, Möller HM, Philipp B. 2013. Degradation of the acyl side chain of the steroid compound cholate in Pseudomonas sp. strain Chol1 proceeds via an aldehyde intermediate. J Bacteriol 195:585–595. doi: 10.1128/JB.01961-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergstrand LH, Cardenas E, Holert J, Hamme JDV, Mohn WW. 2016. Delineation of steroid-degrading microorganisms through comparative genomic analysis. mBio 7:e00166-16. doi: 10.1128/mBio.00166-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holert J, Cardenas E, Bergstrand LH, Zaikova E, Hahn AS, Hallam SJ, Mohn WW. 2018. Metagenomes reveal global distribution of bacterial steroid catabolism in natural, engineered, and host environments. mBio 9:e02345-17. doi: 10.1128/mBio.02345-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crowe AM, Casabon I, Brown KL, Liu J, Lian J, Rogalski JC, Hurst TE, Snieckus V, Foster LJ, Eltis LD. 2017. Catabolism of the last two steroid rings in Mycobacterium tuberculosis and other bacteria. mBio 8:e00321-17. doi: 10.1128/mBio.00321-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horinouchi M, Hayashi T, Kudo T. 2012. Steroid degradation in Comamonas testosteroni. J Steroid Biochem Mol Biol 129:4–14. doi: 10.1016/j.jsbmb.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 45.Lack NA, Yam KC, Lowe ED, Horsman GP, Owen RL, Sim E, Eltis LD. 2010. Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism. J Biol Chem 285:434–443. doi: 10.1074/jbc.M109.058081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sih CJ, Wang KC. 1963. Mechanisms of steroid oxidation by microörganisms.1 II. Isolation and characterization of 3aα-H-4α-[3′-propionic acid]-7aβ-methylhexahydro-1,5-indanedione. J Am Chem Soc 85:2135–2137. doi: 10.1021/ja00897a021. [DOI] [Google Scholar]
- 47.Yang F-C, Chen Y-L, Tang S-L, Yu C-P, Wang P-H, Ismail W, Wang C-H, Ding J-Y, Yang C-Y, Yang C-Y, Chiang Y-R. 2016. Integrated multi-omics analyses reveal the biochemical mechanisms and phylogenetic relevance of anaerobic androgen biodegradation in the environment. ISME J 10:1967–1983. doi: 10.1038/ismej.2015.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.