Abstract
Many nascent long non-coding RNAs (lncRNAs) undergo the same maturation steps as pre-mRNAs of protein-coding genes (PCGs), but they are often poorly spliced. To identify the underlying mechanisms for this phenomenon, we searched for putative splicing inhibitory sequences using the ncRNA-a2 as a model. Genome-wide analyses of intergenic lncRNAs (lincRNAs) revealed that lincRNA splicing efficiency positively correlates with 5′ss strength while no such correlation was identified for PCGs. In addition, efficiently spliced lincRNAs have higher thymidine content in the polypyrimidine tract (PPT) compared to efficiently spliced PCGs. Using model lincRNAs, we provide experimental evidence that strengthening the 5′ss and increasing the T content in PPT significantly enhances lincRNA splicing. We further showed that lincRNA exons contain less putative binding sites for SR proteins. To map binding of SR proteins to lincRNAs, we performed iCLIP with SRSF2, SRSF5 and SRSF6 and analyzed eCLIP data for SRSF1, SRSF7 and SRSF9. All examined SR proteins bind lincRNA exons to a much lower extent than expression-matched PCGs. We propose that lincRNAs lack the cooperative interaction network that enhances splicing, which renders their splicing outcome more dependent on the optimality of splice sites.
INTRODUCTION
The majority of long non-coding RNAs (lncRNAs) were discovered several years ago when it was shown that large parts of the human genome are transcribed (1,2), but only a fraction of these transcripts accounted for protein-coding genes (PCGs) (3). It was suggested that transcripts of non-coding genes (genes with minimal or no protein-coding potential) represent important regulators of PCG expression that control every level of gene expression programs (for reviews see 4-7). In general, lncRNAs are expressed at lower levels (8–10) and are under weaker evolutionary constraints than PCGs (11–14). LncRNAs also display more diverse tissue-specific expression patterns than PCGs (8,12,15) and are modestly enriched in the chromatin and nuclear fractions (8,9,16).
Many nascent lncRNAs contain introns and undergo the same RNA processing steps as pre-mRNAs including capping, splicing, and polyadenylation (17). The biggest similarity with PCGs in terms of genomic structure, gene length etc. has been found for long intergenic non-coding RNAs (lincRNAs) (8,15). It is believed that introns of lncRNAs are spliced by the same splicing machinery as pre-mRNAs (reviewed in 18,19). The 5′ splice site (5′ss) is recognized by the U1 snRNP. The 3′ splice site (3′ss), including the branch point (BP), the polypyrimidine tract (PPT), and the YAG motif at the 3′ end of the intron, is bound by U2 snRNP-associated factors (SF1, U2AF1/2 (U2AF35/65)), which subsequently recruit the U2 snRNP to the BP (reviewed in 19,20). Because cryptic splice sites are relatively abundant throughout the transcribed regions, the recognition of the correct exon boundaries is a crucial step during the splicing process. High fidelity of splice site recognition is mediated throughout a network of interactions that include snRNA base-pairing with sequences around splice sites and the binding of numerous splicing regulatory proteins, e.g. U2 auxiliary factors, SR proteins and hnRNP proteins (reviewed in 20,21). These regulatory factors bind short sequences classified as splicing enhancers or silencers. Moreover, the activities of these regulatory elements are often context-dependent, and they can activate or repress splicing according to their location within the transcript (22–26).
Several bioinformatic studies reported that lncRNAs/lincRNAs, both steady-state and nascent RNAs, are less efficiently spliced than pre-mRNAs of PCGs (10,15,27–29). One possible mechanism explaining the apparent difference in the splicing efficiencies between lincRNAs and PCGs is the absence of proximal RNA Pol II phosphorylation over 5′ss in lincRNA transcripts (10). However, the precise molecular mechanism for this phenomenon has not been elucidated. Here, we combined bioinformatic and experimental approaches to determine cis- and trans-acting factors that are responsible for the poor splicing of lincRNAs.
MATERIALS AND METHODS
Cell culture
HeLa cells were cultured in high glucose (4.5 g/l) DMEM (Sigma) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin (Penicillin/Streptomycin, Gibco) and 10% (v/v) fetal bovine serum (FBS, Gibco) at 37°C and 5% CO2.
Plasmids and transfections
The ncRNA-a2 gene (PCAT6 – ENSG00000228288) was placed under the control of the CMV promoter in the pEGFP-C1 backbone using NheI and HindIII restriction sites, replacing the sequence of the GFP gene with the ncRNA-a2 gene (pEGFP-C1_ncRNA-a2). We introduced Multiplex Identifier barcode sequences (10 nt MID3 and 10 nt MID4, Roche) at the 3′end of the ncRNA-a2 gene to specifically detect transiently expressed ncRNA-a2 transcripts.
The human hemoglobin subunit beta (HBB) gene (ENSG00000244734) was amplified from genomic DNA and cloned between the KpnI and HindIII restriction sites of the pcDNA3 plasmid (pcDNA3_HBB). The ncRNA-a2 gene containing HBB intron 2 was prepared from pEGFP-C1_ncRNA-a2, whereby the HBB intron 2 (pEGFP-C1_ncRNA-a2_HBB-intron2) sequence was amplified from genomic DNA and cloned into the pEGFP-C1_ncRNA-a2 construct by site-directed mutagenesis PCR. The ncRNA-a2 with HBB intron 2 ncRNA-a2 5′ss construct was prepared from pEGFP-C1_ncRNA-a2_HBB-intron2 by site-directed mutagenesis PCR. The HBB gene containing the ncRNA-a2 intron (pcDNA3_HBB_ncRNA-a2-intron) was prepared from pcDNA3_HBB whereby the ncRNA-a2 intron sequence was PCR amplified and cloned into pcDNA3_HBB without HBB intron 2 by site-directed mutagenesis PCR. The HBB with with ncRNA-a2 intron HBB 5′ss construct was prepared from pcDNA3_HBB_ncRNA-a2-intron by site-directed mutagenesis PCR. The plasmid of the ncRNA-a2 gene containing the HBB PPT was cloned in the same way (nucleotides 823–847 of HBB intron 2 were used as the HBB PPT).
NcRNA-a2 deletion mutants (FΔ1-8, RΔ1-7, Δ1-7, ΔPPT, Δ60), ncRNA-a2 mutants with ISE motifs, ncRNA-a2 mutants with modified PPT and 5′ss mutants were prepared by PCR with specific primers using pEGFP-C1_ncRNA-a2 as a template. The FΔ1-8 mutants were prepared by deletion of regions gradually increasing by 20 bp starting 6 bp downstream of the 5′ss. Similarly, RΔ1-7 mutants were prepared by deletion of regions gradually increasing by 20 bp starting 40 bp upstream of the 3′ss. The Δ1-7 mutants were prepared by sequential deletion of 20 bp starting 6 bp downstream of the 5′ss. Mutants with ISE motifs and a negative control with a degenerated motif (30) were introduced 25 bp downstream of the 5′ss (individual ISE motif sequences are listed in the Supplementary Data). In ΔPPT, 4 bp were deleted in the ncRNA-a2 intron sequence (intron positions: 198–201). In Δ60 mutants, 60 bp regions were deleted in the middle of the ncRNA-a2 intron (intron positions: 67–125). In ncRNA-a2 mutants with a modified PPT, nucleotides 181–201 of the ncRNA-a2 intron were modified (T21 – all nucleotides to Ts, CtoT – all Cs to Ts, GAtoT – all Gs and As to Ts). BPs of all lncRNAs used in the study and the HBB intron 2 were predicted by the SVM-BPfinder online tool (31) and sequences between the predicted BP and the 3′ YAG motif were mutated. For further information on the modified sequences, see Supplementary Data.
SNHG8 (ENSG00000269893), BX088651.4 (ENSG00000237357), BX005266.2 (ENSG00000226007), AC005840.2 (ENSG00000256433) and AC116021.1 (ENSG00000254639) genes were amplified from genomic DNA and cloned into the pEGFP-C1 backbone using NheI/AgeI and HindIII/KpnI restriction sites, replacing the sequence of the GFP gene. We introduced Multiplex Identifier barcode sequences (MID5 and MID6, Roche) immediately downstream of the genes to specifically detect ectopically expressed transcripts.
All constructs have been verified by DNA sequencing. Plasmids were transiently transfected into cells using Lipofectamine® 3000 Transfection Reagent (Thermo Fisher Scientific) according to the manufacturer's instructions and incubated for 24 h with a medium change 6 h after the transfection.
RNA isolation, reverse transcription and qPCR
Cells were grown to 90% confluency and RNA was isolated using either the TRIzol reagent (Thermo Fisher Scientific), which allows for simultaneous isolation of RNA and proteins, or the RNAzol reagent (Molecular Research Center). RNA was further precipitated with isopropanol, resuspended in Nuclease-Free Water (Ambion) and treated with Turbo DNase (Ambion) according to the manufacturer's protocol. Reverse transcription was performed with SuperScript III (Thermo Fisher Scientific) using 5 μg of total RNA per 20 μl reaction and either random hexamer primers or primers complementary to barcode sequences downstream of ectopically expressed lincRNAs, respectively. cDNA was analyzed by quantitative PCR using LightCycler 480 (Roche) and LightCycler® 480 SYBR Green I Master (Roche) using the 2−ΔΔCt method [(Ct gene of interest – Ct internal control) sample A – (Ct gene of interest – Ct internal control) sample B]. A list of used primers is provided in the Supplementary Data.
Western blot and antibodies
Proteins were isolated from TRIzol fractions, precipitated with isopropanol and resuspended in NEST-2 buffer (50 mM Tris–HCl pH 6.8, 20 mM EDTA, 5% (w/v) SDS). Proteins were resolved on a 12% (cellular fractions) or 10% (hnRNP H siRNA KD, RIP) SDS-PAGE, blotted onto a nitrocellulose membrane and detected using the indicated antibodies and SuperSignal Femto/Pico West (Thermo Fisher Scientific). For western blot, the following antibodies were used: rabbit α-H3 (Abcam ab1719), rabbit U2B‘‘ (PROGEN 57036), mouse α-tubulin kindly provided by Pavel Draber (Institute of Molecular Genetics of the Czech Academy of Sciences, Prague, Czech Republic), mouse hnRNP F/H (Santa Cruz sc-32310) and mouse U2AF2 (Santa Cruz sc-53942).
Cellular fractionation
Cellular fractionation assays were performed as previously described (32). Cells were grown to 90% confluency, washed with PBS and scraped into PBS/1mM EDTA. The pellet was resuspended in ice-cold NP-40 lysis buffer (10 mM Tris–HCl pH 7.5, 0.15% NP-40, 150 mM NaCl) for 5 min. Then, the lysate was placed on 2.5 volumes of an ice-cold sucrose cushion (24% sucrose in NP-40 lysis buffer) and centrifuged for 10 min at 4°C. The supernatant (cytoplasmic fraction) was stored at 4°C for subsequent RNA isolation. The pellet was washed with ice-cold PBS/1mM EDTA and resuspended in glycerol buffer (50% glycerol, 20 mM Tris–HCl pH 7.9, 75 mM NaCl, 0.5 mM EDTA, 0.85 mM DTT and 0.125 mM PMSF). An equal volume of nuclei lysis buffer (10 mM HEPES pH 7.6, 1 mM DTT, 7.5 mM MgCl2, 0.2 mM EDTA, 0.3 M NaCl, 1 M urea, 1% NP-40) was added, samples were incubated for 2 min on ice and centrifuged for 2 min at 4°C. The supernatant (soluble nuclear fraction) was stored at 4°C for subsequent RNA isolation. The pellet (chromatin fraction) was washed with ice-cold PBS/1 mM EDTA. RNA from all fractions was isolated using the TRIzol reagent as described above.
SiRNA treatment
Pre-annealed siRNA duplexes were obtained from Ambion - hRNP H1 (s6728): 5′ GAAGCAUACUGGUCCAAAUtt 3′, ncRNA-a2: 5′ CCTCCTTACTCTTGGACAAtt 3′, ncRNA-a5: 5′ CCTTGGAGAATAAAGCTTAtt 3′. The negative control # 5 siRNA from Ambion was used as a negative control. SiRNAs were transfected with Oligofectamine (Thermo Fisher Scientific) at a final concentration of 50 nM according to the manufacture's protocol. Cells were incubated for 72 h (hnRNP H) and 48 h (ncRNA-as) and then harvested and analyzed. After siRNA treatment of ncRNAs, the expression of PCGs in their vicinity were evaluated – KDM5B (ENSG00000117139), RABIF (ENSG00000183155), KLHL12 (ENSG00000117153), ADIPOR1 (ENSG00000159346), PQLC3 (ENSG00000162976), ROCK2 (ENSG00000134318), E2F6 (ENSG00000169016).
RNA immunoprecipitation
Cells were grown to 80–90% confluency and 24 hours after transfection with various ncRNA-a2 constructs, cells were washed with PBS and scraped into 2 ml PBS. 2 ml of nuclear isolation buffer (1.28 M sucrose, 40 mM Tris–HCl pH 7.5, 20 mM MgCl2, 4% Triton X-100) and 6 ml of water were added, and cells were incubated 20 min on ice with frequent mixing. Nuclei were pelleted by centrifugation at 2500g for 15 min and resuspended in 1 ml RIP buffer (150 mM KCl, 25 mM Tris–HCl pH 7.4, 5 mM EDTA, 0.5 mM DTT, 0.5% NP-40) with freshly added 100 U/ml RNasin (Promega) and 5 μl Protease Inhibitor Cocktail Set III, EDTA-Free (Calbiochem). Then, nuclei were split into two 500 μl fractions (IP, mock) and mechanically sheared by a dounce homogenizer with three times 20 strokes (0.5 s, 40% amplitude). Nuclear membranes and debris were removed by centrifugation at 13,000 rpm for 10 min. Supernatants were transferred into siliconized tubes, and 10% was frozen and stored at –80°C for RNA/protein isolation (10% inputs). Antibodies were added (IP:2 μg of U2AF2 – Santa Cruz sc-53942, mock: 4 μg of IgG from mouse serum – Sigma I5381) to the remaining supernatants and samples were incubated at 4°C overnight with gentle rotation. Then, 40 μl of Protein G PLUS agarose beads (Santa-Cruz Biotechnology, sc-2002) were added to the lysates and further incubated for 1 h at 4°C with gentle rotation. Beads were pelleted at 2500 rpm for 30 s and washed three times with 500 μl RIP buffer, followed by one additional wash with PBS. Co-precipitated and input RNA and proteins were isolated by resuspending the beads in 1 ml (10% inputs) or 500 μl (IPs) TRIzol reagent (LifeTechnologies). RNA and proteins were isolated and analyzed as described above.
Calculation of splice site strength
The human hg19/GRCh37 genome assembly was used as the reference genome with annotations obtained from GENCODE (Release 19) for both, PCGs and lincRNAs (33). Single-exon genes were filtered out. The remaining list with the coordinates of all intron-containing lincRNAs and PCGs was used to extract positions –3 to +6 and –20 to +3 from each 5′ and 3′ss using a custom python script. The corresponding sequences were retrieved using BEDTools getfasta (34) (v2.17.0), and splice site scores were calculated using the Maximum Entropy Score algorithm (http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html) (35).
Splicing index, exon/intron length and polypyrimidine tract analysis
The splicing index (SI) was computed from RNA-Seq datasets of five different human cell lines (A549, H1-hESC, HepG2, HeLa and MCF7) obtained from the ENCODE project (https://www.encodeproject.org/experiments/) (36). Processed and mapped data files (bam files) were downloaded from the ENCODE website: A549—ENCFF000ELG, H1-hESC—ENCFF000FEH, HepG2—ENCFF074BOV, HeLa—ENCFF485AHM and MCF7— ENCFF000HSJ. To establish a list of expressed genes in each cell line separately, we calculated Reads Per Kilobase of transcript per Million mapped reads (RPKM) values for each gene in each of the cell lines. Only genes with RPKMs higher than 0.01 were considered expressed. The top 10% highly expressed PCGs were filtered out to set an upper expression threshold. All lincRNAs with RPKMs above this threshold were removed from further analysis. The SI was calculated as described previously in (37). Each bam file was divided into two separate files − gapped reads and non-gapped reads. Gapped reads were extracted from the bam files using samtools with an awk command based on CIGAR strings containing the ‘N’-label (gap reads) and mapped to the corresponding 5′ and 3′ss. Non-gapped reads were extracted based on CIGAR strings without ‘N’-label (no gap in read). Alternative exons with overlapping coordinates were excluded from this analysis. To obtain unspliced reads, the last nucleotide in the intron and 25 nucleotides from the exon were extracted at each 3′ss. Reads overlapping the last intronic nucleotide were counted from the extracted non-gapped reads using BEDTools Coverage (v2.17.0) (34) with -split and -s options. Custom R scripts were used to determine the SI of all 3′ss for PCGs and lincRNAs. SIs were computed only for 3′ss that contained both, spliced (gapped) and unspliced reads (overlap with the last nucleotide of the intron) and were calculated as the ratio between the number of spliced and unspliced reads by a custom R script.
Gene and exon co-ordinates were obtained from GENCODE v27. BEDTools Subtract (34) was used to get intronic co-ordinates by substracting the exonic co-ordinates from the gene co-ordinate. Custom R script was used to plot the cumulative distribution for exon and intron lengths.
Obtained SI values were ordered and divided into four groups, representing low, medium, high and highest splicing efficiency. From the grouped introns, 5′ss (3 bp upstream and 6 bp downstream) and a region of 40 bp upstream of the 3′ss including the polypyrimidine tract (PPT) was extracted. Sequence logos were produced using WebLogo (http://weblogo.berkeley.edu/logo.cgi) (38). Percentages of T, G, C and A nucleotides were calculated for each region and mean percentages computed for each group using custom Perl scripts.
Prediction of secondary structures
To evaluate potential secondary structures of lincRNAs and PCGs, we utilized two alternative approaches. First, we randomly selected 5000 introns from PCG or lincRNAs shorter than 2000 bp. Then we applied RNAfold (https://www.tbi.univie.ac.at/RNA/) to predict the base-pairing potential of selected introns using default parameters (39). To calculate the base pairing probability, we divided the number of base-pairs by the total number of intron nucleotides. Alternatively, we randomly selected 5000 introns from PCG or lincRNAs and calculated the minimum free energy of 100 bps around 3′ss and 5′ss using RNAfold (39).
SRSF-binding and splicing silencer motif prediction
SRSF protein-binding motif and splicing silencer consensus motif distributions around splice sites were visualized using the Bioconductor R packages (40). The references for SR binding consensus motifs with IUPAC nucleotide ambiguity codes are listed in the Supplementary Data. Windows of 200 bp (–100 to +100) were extracted around the 5′ss and 3′ss for both, PCGs and lincRNAs. The import.bed function was used from the Rtracklayer Bioconductor R package (41), and the corresponding sequences were extracted using the getSeq function. The getPatternOccurrenceList function of seqPattern R package was used to obtain the position of each consensus pattern occurrence for each sequence. A total number of motifs was counted for each position for the given region. Binding site densities were calculated for each position separately as the number of motifs divided by the number of analyzed sequences. The visualization was done in R (version 3.5.1) with the ggplot2 (https://github.com/hadley/ggplot) (42) package for the final graphical output.
iCLIP library preparation
HeLa cells stably expressing SR proteins tagged with GFP at endogenous levels from bacterial artificial chromosomes (BACs) were grown to 90% confluency and irradiated once with 150 mJ/cm2 UV light (254 nm). Protein G Dynabeads coupled with goat anti-GFP antibodies (D. Drechsel, MPI-CBG, Dresden) were used for immunopurification. Crosslinked, partially digested RNAs to lengths of 60–150 nt were immunopurified using anti-GFP antibodies, reverse transcribed to generate cDNA libraries and subjected to high-throughput sequencing on Illumina HiSeq2000 (single-end 75 nt reads) (43). To normalize for transcript levels, total RNA was isolated from HeLa WT cells (two replicates) and sequenced after rRNA depletion (RiboMinus) on the same Illumina HiSeq2000 machine (single-end 75 nt reads). The raw data were deposited in the GEO database under accession numbers GSE113812-GSE113814.
Analysis of iCLIP and RNA-Seq data
Adapters and barcodes were removed from the iCLIP reads before mapping to the human hg19 genome assembly (Ensembl59 annotation) using Bowtie (version 0.10.1). Uniquely mapped reads were used to extract the crosslink site (first nucleotide of the read) and the statistical significance of binding events (FDR < 0.05) was calculated and compared to randomized co-transcribed regions (44). To obtain comparable numbers of significant binding sites (CLIP tags) replicates that correlated best were pooled.
RNA-Seq reads from HeLa cells (two replicates) were mapped to the hg19 reference using TopHat v2.1.0, where the maximum number of multi mappers (max-multihits) was set to 1. Both mapped replicates were merged using samtools merge option and RPKMs were calculated for each transcript using CoverageBed (34). Transcripts with RKPM values lower than 0.01 were considered as not expressed and were excluded from the analysis. PCGs were filtered to match the expression range of lincRNAs. For this, all lincRNAs were sorted according to their RKPM values into 20 bins. Subsequently, PCGs were randomly selected to match the number and expression levels of lincRNAs in each bin and were further used for analyses as expression-matched PCGs. For each expressed transcript, a window of 200 bp around each 5′ss and 3′ss (–100 to +100) was selected and intersected with the iCLIP data using intersectBed (34). The total number of iCLIP-tags was counted for each region. Mean SR protein binding was determined as the sum of CLIP-tags divided by the number of 200 bp regions used for the analysis. Metagene iCLIP profiles of the 200 bp regions were generated using R/Bioconductor packages (40). First, the input BED file (containing coordinates of all expression-matched transcripts) and the input GFF file (containing genomic reference regions from GENCODE v19) were imported using rtracklayer (45). The GFF file was further processed using GenomicFeatures (45) to extract the intron features (using the GenomicFeatures::intronsByTranscript function). 200 bp around the beginning and the end of introns were selected. iCLIP data coverage was considered as one for each position. The iCLIP counts were converted into a GRanges object using the GenomicRanges (46) and overlapped with the 200 bp regions (using GenomicRanges:: findOverlaps function). Plots were generated using the matplot function in R.
To calculate the cumulative distribution of splicing efficiencies of PTBP1/U2AF2/hnRNP C bound lincRNAs and the sequence compositions of PPTs bound and unbound by U2AF2, the 40 bp regions upstream of 3′ss were intersected with iCLIP data of PTBP1, U2AF2 and hnRNP C (47,48, BioRxiv: https://doi.org/10.1101/179648) using the intersectBed program. SIs were calculated as described above using RNA-Seq data from Xue et al. (47) and Zarnack et al. (48). Cumulative frequencies of SIs for PTBP1/U2AF2/hnRNP C-bound and unbound transcripts at 40bp upstream of 3′ss were plotted using the R script.
Mapping of eCLIP sequence data
For mapping eCLIP sequencing data from HepG2 cell line (BioRxiv: https://doi.org/10.1101/179648) we used GENCODE (GRCh38.p7) genome assembly and the STAR alignment (version 2.4.2a) using the following parameters from ENCODE pipeline: STAR –runThreadN 8 –runMode alignReads –genomeDir GRCh38 Gencode v25 –genomeLoad LoadAndKeep –readFilesIn read1, read2, –readFilesCommand zcat –outSAMunmapped Within –outFilterMultimapNmax 1 –outFilterMultimapScoreRange 1 –outSAMattributes All –outSAMtype BAM Unsorted –outFilterType BySJout –outFilterScoreMin 10 –alignEndsType EndToEnd –outFileNamePrefix outfile as described in Haberman et al. (49), and Chakrabarti et al. (50). All uniquely mapped reads were corrected for over-amplification of PCR duplicates, by using a python script ‘barcode collapse pe.py’ available on GitHub (https://github.com/YeoLab/gscripts/releases/tag/1.0), which is part of the ENCODE eCLIP pipeline (https://www.encodeproject.org/pipelines/ENCPL357ADL/). The visualization part was done in R (version 3.4.1) together with the ggplot2 (https://github.com/hadley/ggplot) (42) and the smoother (https://github.com/config-i1/smooth) package for the final graphical output. Each density graph shows a distribution of raw crosslinking positions of cDNA-starts relative to splice site positions. Gaussian filtering with 10 nt window size was used for the final smoothing of each density line. A set of expression-matched PCGs was created the same way as in the case of iCLIP analysis, but HepG2 RNA-Seq data from ENCODE (HepG2 - ENCFF074BOV) were used.
CRISPR/Cas9-mediated intron knock-out
For designing short-guide RNAs (sgRNAs), online CRISPR Design Tool (51) (http://crispr.mit.edu/) was used. SgRNAs targeting the 5′ and 3′ boundaries of the ncRNA-a2 intron were cloned into pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene plasmid # 42230) (52) using BbsI restriction sites. Guide target sequences for testing guide efficiencies were cloned into pARv-RFP (Red Fluorescent Protein, a gift from Radislav Sedláček, Institute of Molecular Genetics, Czech Academy of Sciences; Addgene plasmid # 60021) (53) using EcoRV and PvuI restriction sites with the introduction of the BamHI restriction site at 5′end of a guide target sequence to allow restriction digest analysis of positive clones. The homology-directed repair (HDR) template including 800bp homology arms flanking the targeted ncRNA-a2 intron was amplified from genomic DNA and cloned into the pBluescript II vector. Before transfections, HDR template was amplified by PCR with specific primers. All sequences were confirmed by DNA sequencing. The sequences of all sgRNAs and guide target sequences used in this study are listed in the Supplementary Data.
Cells were grown to 90% confluency, and an equimolar mixture of plasmids (8 μg of pX330_Cas9_Guide, pARV_GuideTargetSequence, HDR template) was transiently transfected into HeLa cells in Opti-MEMTM I Reduced Serum Medium (Thermo Fisher) using the Lipofectamine LTX Transfection Reagent (Thermo Fisher Scientific) according to the manufacturer's instructions and incubated for 72 h. The culture medium was changed 6 h after transfection and then every 24 h. The inhibitor of non-homologous end-joining (NHEJ) (SCR7, final concentration 1 μM, Xcess Biosciences) was added 16 h prior to transfection. 72 h after transfection, cells were FACS sorted for RFP positivity in single-cell mode. Single cells were grown for 2 weeks until full confluency in a 1:1 fresh/conditioned medium. Positive clones were selected by PCR using primers spanning the deleted sequence (sequences are provided in Supplementary Data). Wild-type and deletion bands were cloned into pGEM-T vectors and confirmed by DNA sequencing.
Isolation of biotin-labeled nascent transcripts
Cells were grown to 80–90% confluency and provided with fresh media containing 500 μM 4-Thiouridine (4-sU; Sigma). Cells were pulsed-labelled for 60 min, and RNA was extracted by TRIzol (Thermo Fisher Scientific) as described before. To biotinylate 4-sU-labeled RNAs, 120 μg total RNA was mixed with 240 μl of 4 mM EZ-Link® HPDP-Biotin (Thermo Fisher Scientific), 120 μl biotinylation buffer (10 mM HEPES pH 7.5, 1 mM EDTA) and 840 μl Nuclease-Free Water (Ambion). The samples were incubated in the dark for 90 min at room temperature. Total RNA including 4sU-Biotin-labeled RNA was extracted by adding 250 μl phenol:chloroform and precipitated overnight with 2.5 vol. 100% ethanol, washed with 70% ethanol and resuspended in 100 μl Nuclease-Free Water (Ambion). 4sU-Biotin-labeled RNA was captured on 100 μl BcMag™ Streptavidin Magnetic Beads (Bioclone Inc), washed twice with washing buffer (0.5 M NaCl, 20 mM Tris–HCl pH 7.5, 1 mM EDTA), eluted with washing buffer containing 0.1 mM DTT (Invitrogen) and precipitated with 2.5 vol. of 100% ethanol. Total RNA that did not bind to magnetic beads served as input. Reverse transcription and quantitative PCR was done as described before.
RESULTS
Intronic sequences determine inefficient splicing of ncRNA-a2
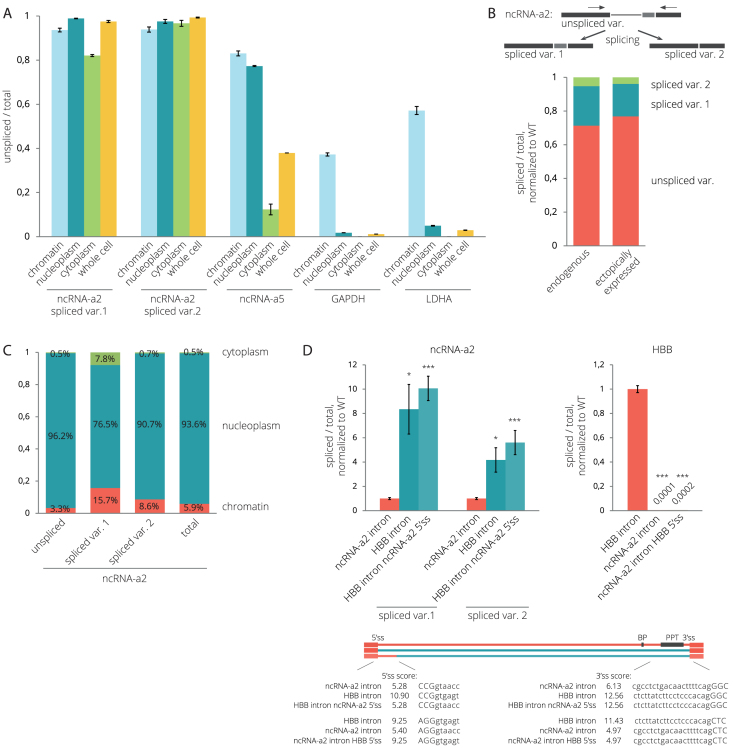
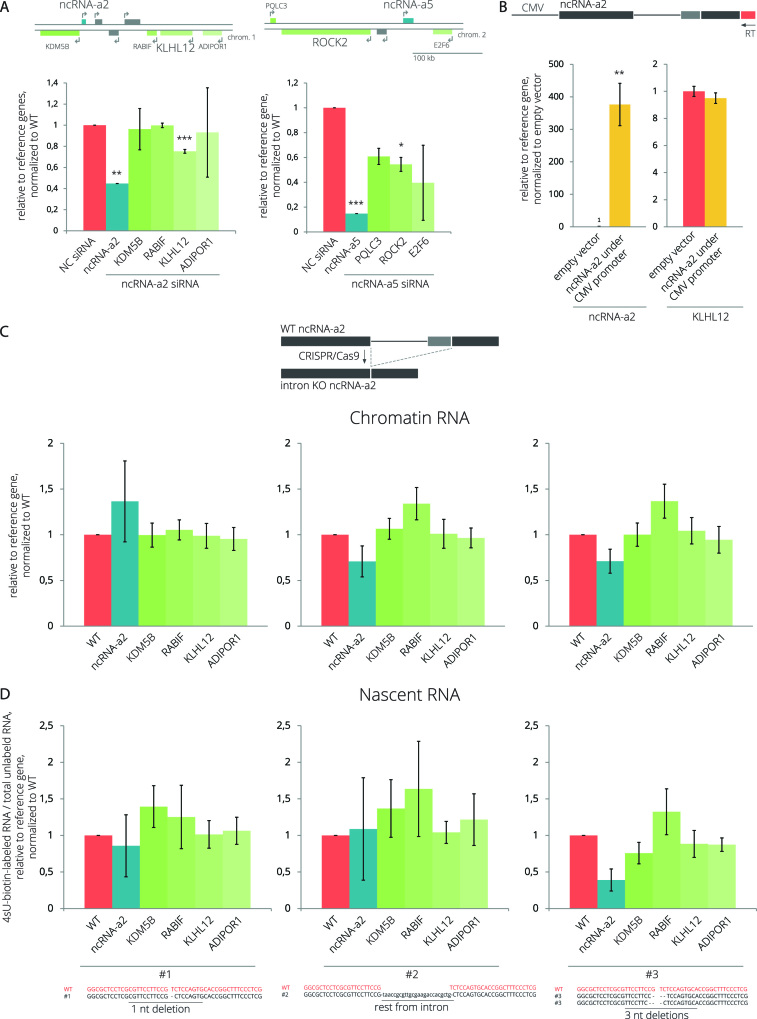
To study the splicing efficiency of lincRNAs, we selected two activating lncRNAs, ncRNA-a2 (PCAT6) and ncRNA-a5 (LINC00570), which stimulate the expression of PCGs located in their genomic vicinity (54). It should be noted that three different transcripts of the ncRNA-a2 (PCAT6) gene and four different transcripts of the ncRNA-a5 (LINC00570) gene are annotated in the Ensembl database (http://www.ensembl.org). However, in HeLa cells, we detected only two variants produced by alternative usage of 3′ss separated by 114 nucleotides producing two ncRNA-a2 transcripts (PCAT6-201, PCAT6-202) (Supplementary Figure S2A). Only one ncRNA-a5 transcript (LINC00570-201) is supported by multiple ESTs in the Ensembl database and annotated in the NCBI Reference Sequence database (www.ncbi.nlm.nih.gov/refseq/). Therefore we focused our analysis on two ncRNA-a2 transcripts (PCAT6-201, PCAT6-202) and the second intron of the LINC00570 transcript (LINC00570-201). To determine their splicing status in different cellular compartments, we fractionated HeLa cells into chromatin, nucleoplasmic and cytoplasmic fractions (Supplementary Figure S1). Using reverse transcription coupled with quantitative PCR (RT-qPCR), we found that nuclear fractions contained predominantly unspliced forms of both lncRNAs (Figure 1A). Strikingly, ∼80% of cytoplasmic ncRNA-a2 retained the intron, compared to only ∼10% for ncRNA-a5 transcripts, revealing large differences in splicing efficiencies between lncRNAs. In contrast, unspliced pre-mRNAs of two PCGs, GAPDH and LDHA, were only detected in the chromatin fraction (Figure 1A). A more detailed analysis of ncRNA-a2 transcripts revealed that the upstream 3′ss was preferentially used, but, in general, splicing at both 3′ss was inefficient (Figure 1B). In addition, ncRNA-a2 seems to reside primarily in the nucleus since ∼76–96% of its transcripts are localized in the nucleoplasm and chromatin fractions (Figure 1C).
Figure 1.
NcRNA-a2 and ncRNA-a5 are less efficiently spliced than PCGs. (A) Splicing efficiencies of lincRNAs (ncRNA-a2 and ncRNA-a5) and PCGs (GAPDH, LDHA) in different cellular fractions. (B) Splicing of endogenous and transiently expressed ncRNA-a2 measured by semi-quantitative RT-PCR. Primers are depicted as arrows above the transcript. (C) The cellular distribution of ncRNA-a2 transcripts. (D) The ncRNA-a2 intron is inefficiently spliced out when inserted into human hemoglobin subunit beta (HBB) pre-mRNA. (A–D) Bar plots show relative RNA levels determined by RT-qPCR. The mean of at least three independent experiments is shown. Error bars indicate SEM; asterisks indicate the statistical significance levels calculated by the two-tailed Student's t-test, *P < 0.05, **P < 0.01, ***P < 0.001.
We selected ncRNA-a2 for further analyses as an example of an inefficiently spliced lincRNA. It has been previously shown that chromatin modifications and promoter sequences can significantly influence the splicing outcome of a PCG (55–60). In order to determine whether promoter or chromatin elements influence ncRNA-a2 splicing, we cloned the whole transcribed ncRNA-a2 sequence into a plasmid containing the CMV promoter, expressed ncRNA-a2 transiently in HeLa cells, and analyzed its splicing using semi-quantitative RT-PCR (Figure 1B and Supplementary Figure S2A). We did not observe any significant differences in the splicing patterns between endogenous and transiently expressed ncRNA-a2. This suggests that the ncRNA-a2 sequence is the dominant factor affecting the efficiency of ncRNA-a2 splicing.
To determine the contribution of exonic or intronic sequences to the observed splicing inefficiency, we swapped introns of ncRNA-a2 and a PCG, human hemoglobin beta subunit (HBB) and transiently expressed chimeric transcripts (Figure 1D). We chose HBB intron 2 because our previous experiment showed its efficient splicing (unpublished data). We observed efficient splicing of the HBB intron when inserted between ncRNA-a2 exons (Figure 1D), and <1% of ncRNA-a2 transcripts with the HBB intron remained unspliced (Supplementary Figure S2B). In contrast, the ncRNA-a2 intron remained largely unspliced when placed between HBB exons (Figure 1D and Supplementary Figure S2B). Since 5′ss sequences extend into the upstream exon, substituting just the intron changes the 5′ss strength of both hybrids. Therefore, we kept the original 5′ss of ncRNA-2 and HBB (8 bp downstream of the exon-intron boundary) and replaced only intronic sequences downstream (constructs ‘ncRNA-a2 with HBB intron ncRNA-a2 5′ss’ and ‘HBB with ncRNA-a2 intron HBB 5′ss’; Figure 1D). In both cases, keeping the original 5′ss sequence increased splicing efficiency of both constructs (1.2× for ‘ncRNA-a2 with HBB intron ncRNA-a2 5′ss’ and 2.1× for ‘HBB with ncRNA-a2 intron HBB 5′ss’). In the case of the ncRNA-a2 construct, the result is surprising because the original ncRNA-a2 5′ss has a much weaker MaxEnt score (MES) (5.28) than the artificial HBB/ncRNA-a2 5′ss (MES 10.90). This suggests that the 5′ss identity does not have a dominant impact on splicing of hybrid RNAs. While some contribution of exonic sequences cannot be ruled out, these results suggest that the ncRNA-a2 intron is largely responsible for the inefficient splicing of the ncRNA-a2 transcript.
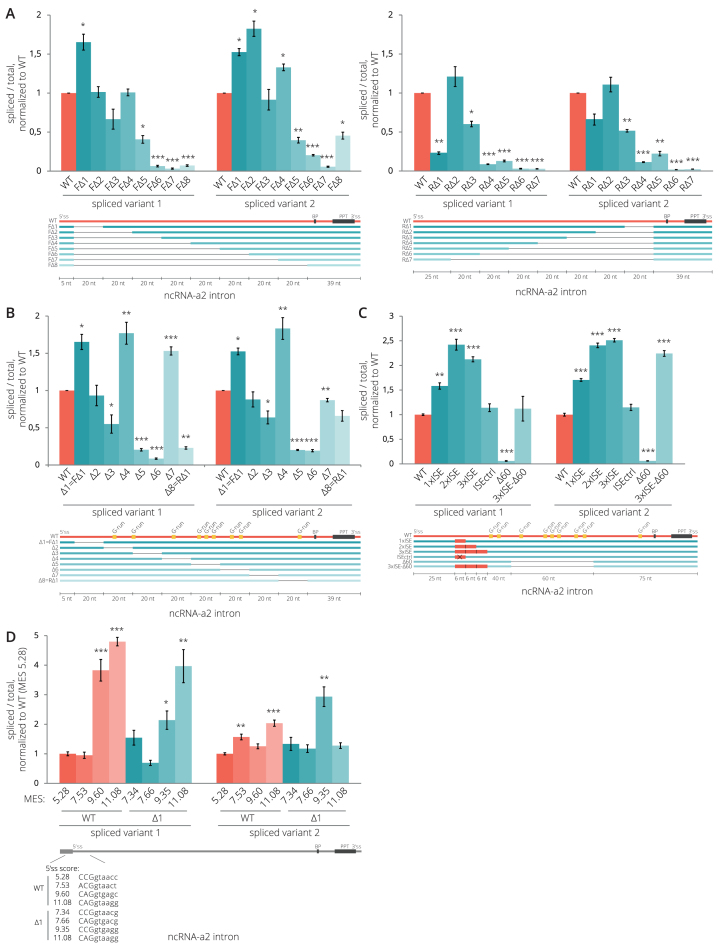
To search for possible splicing regulatory elements, we prepared several deletion mutants of the 204 bp-long ncRNA-a2 intron. We gradually deleted nucleotides either from the 5′ end (F) or the 3′ end (R) of the intron starting 6 nt downstream of the 5′ss and leaving 39 nt upstream of the 3′ss intact (Figure 2A). The deletion of nucleotides 6–25 (mutant FΔ1) partially increased the splicing efficiency, but the majority of deletions reduced splicing efficiencies. We observed a particularly large drop in splicing efficiency when the central intronic region spanning nucleotides 66–125 was deleted (FΔ5 2.5×, FΔ6 15.5×, RΔ5 2.5×, RΔ6 4.9× reduction in splicing efficiency compared to WT). This could be explained either by the fact that the truncated intron is too short to be efficiently recognized by the splicing machinery or that the central intronic sequence contains elements that enhance splicing. To distinguish this, we prepared additional deletion mutants (Δ1-7) and gradually removed a 20 nt sequence window along the intron length (Figure 2B). We observed a partial enhancement of splicing efficiency in mutants Δ1 and Δ4 (nucleotides 6–25 and 66–85) suggesting that these sequences could act as weak splicing silencers. Indeed, predicted binding sites for SR proteins were found in the region 20 nt (nucleotides 6–25) downstream of the 5′ss; a putative binding site for SRSF1 (CTGCCGG), for SRSF2 (CGCTGCCG) and three binding sequences for SRSF6 (CGCGTT, TGCGAA and CGCTGC), which might all act as splicing silencers (61–67).
Figure 2.
Intronic splicing regulatory elements of ncRNA-a2. (A, B) Splicing efficiencies of ncRNA-a2 intron deletion mutants. Deletion of sequences in the center of the intron (mutants Δ5 and Δ6 in (B)) reduces ncRNA-a2 splicing. (C) Insertion of active ISEs (1–3) enhanced splicing while removal of the G-rich splicing enhancer sequence completely abolished splicing (Δ60 mutant). Splicing of the Δ60 mutant was rescued by insertion of 3xISE downstream of 5′ss. A splicing enhancer containing two inactivating point mutations was used as a negative control (ISEctrl). (D) Mutations strengthening the 5′ss improved splicing efficiency of WT as well as Δ1 mutant lacking a putative splicing inhibitory sequence. (A–D) Splicing efficiencies were measured as a fraction of spliced transcripts relative to the total amount of transcripts. Schemes under the charts indicate modifications of the intron sequence of ncRNA-a2 gene. Predicted branch point (BP), the PPT (black), G-run motifs (yellow) and artificial ISEs (red) are indicated. Sequences of all deletion constructs are shown in Supplementary Data. Bar plots show relative RNA levels as determined by RT-qPCR. The mean of at least three independent experiments is shown. Error bars indicate SEM; asterisks indicate the statistical significance levels calculated by two-tailed Student's t-test comparing the individual mutant with WT, *P < 0.05, **P < 0.01, ***P < 0.001.
The splicing efficiency significantly decreased in mutants Δ5 (5×) and Δ6 (11× for spliced variant 1 and 5× for spliced variant 2 compared to WT) indicating that this intronic sequence harbors splicing enhancer(s). This sequence contains several G-rich sequences (G-runs) (Figure 2B), which were previously characterized as splicing enhancers that recruit U1 snRNP and hnRNP F/H proteins when located downstream of 5′ss (68–73). To test whether hnRNP H protein enhances ncRNA-a2 splicing, we knocked it down by RNA interference (RNAi) and observed increased levels of unspliced ncRNAa-2 variants (Supplementary Figure S3). Altogether, this suggests that the ncRNA-a2 contains splicing enhancer(s) in the middle of the intron that is regulated by hnRNP H.
5′ ss sequence is important for lincRNA splicing
Next, we tested whether removal of G-runs in the middle of the intron can be rescued by insertion of intronic splicing enhancers that support recognition of 5′ss. We introduced one, two and three copies of a known intronic splicing enhancer (ISE) motif containing G-runs (30) downstream of the ncRNA-a2 5′ss (Figure 2C). As a control, we introduced a mutated ISE element (ISEctrl). The splicing efficiency significantly increased (1.5–2.5×) in all cases except in the control. Insertion of the ISE element into the ncRNA-a2 lacking the middle G-rich sequence (3xISE-Δ60) rescued splicing to WT level, which is consistent with a model wherein the middle intron G-run sequences promote recognition of the 5′ss.
These results indicate that enhancer sequences that promote 5′ss recognition enhance ncRNA-a2 splicing. To test the role of the 5′ss in ncRNA-a2 splicing more rigorously, we prepared several mutants with increased strengths of the 5′ss. We utilized the WT ncRNA-a2 and the Δ1 mutant lacking the putative inhibitory sequences. Mutations increased the 5′ss MES to 7.53 (WT) and 7.66 (Δ1 mutant), which is approximately one point below the average 5′ss strength of lincRNAs (8.56), 9.60 (WT) and 9.35 (Δ1 mutant), which is similar to the threshold of top 25% 5′ss (9.79 for lincRNAs and 9.80 for PCGs) and to 11.08, which falls into the top 10% of the strongest 5′ss (Figure 2D, for MES distribution see Supplementary Figure S5A). While the average 5′ss strength did not improve splicing efficiency, substitutions leading to strong 5′ss significantly enhanced ncRNA-a2 splicing efficiency (>3.5x), primarily the spliced variant 1. The effect was stronger for WT ncRNA-a2 compared to the Δ1 mutant.
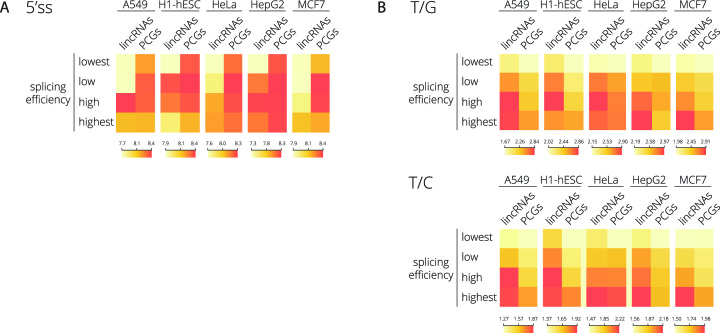
To investigate whether 5′ss strength is a general determinant of lncRNA splicing, we analyzed available RNA-Seq data from five different human cell lines (embryonic stem cells H1-hESC, lung carcinoma A549, cervix carcinoma HeLa, liver carcinoma HepG2 and breast cancer cell line MCF7). To avoid a potential overlap with PCGs, we focused on lincRNAs only. We first selected all PCGs and lincRNAs expressed in a particular cell line (RPKM>0.01) and filtered out the top 10% of highly expressed PCGs as an upper expression threshold to avoid bias from highly expressed genes. All lincRNAs with RPKM values above this threshold were also removed from further analyses to keep lincRNAs and PCGs within the same expression range for each cell line. Splicing indices (SI) were then determined for each individual intron in each cell line separately (Supplementary Figure S4). In total, we analyzed between 1054 and 1770 lincRNA and >77 000 PCGs introns. Although the lincRNA expression is highly cell-specific, we found that lincRNAs were, in general, less efficiently spliced (lower SI) compared to PCGs in all tested cell lines (Supplementary Figure S4).
To evaluate 5′ss strength in differently spliced genes, we categorized lincRNA and PCG introns into four groups based on their splicing efficiencies (increasing SI), and we calculated the mean 5′ss MES for each group (Figure 3A and Supplementary Figure S5). We found a positive correlation between 5′ss strength and splicing efficiency of lincRNAs in four tested cell lines (Pearson's correlation coefficients 0.67–0.94) while no such correlation was found for PCGs (Pearson's correlation coefficients −0.75–0.57) (Supplementary Figure S5). These results suggest that lincRNA splicing is more dependent on 5′ss strength than splicing of PCGs.
Figure 3.
LincRNA splicing efficiency correlates with the strength of 5′ss and PPT. LincRNAs and PCGs were divided into four groups based on their splicing efficiency containing an equal number of transcripts. (A) 5′ss strength or (B) thymidines over guanines (T/G) and thymidines over cytosines (T/C) ratios frequencies in the PPT region was calculated for lincRNAs and PCGs in different cell lines. The color schemes representing the distribution of data are shown under the heat maps. See also Supplementary Figures S5–S8 for further analyses.
Polypyrimidine tract sequence determines the splicing efficiency of lincRNAs
In addition to 5′ss, we also compared sequences at the 3′ end of introns. A detailed analysis of PPT sequences (nucleotides –40 to –1) revealed slightly better conservation of cytidines/thymidines (C/T) nucleotides at position –3 of the YAG sequence in PCGs in all five tested cell lines (Supplementary Figure S6). Interestingly, we found that the stretch of Ts within the PPT of lincRNAs is longer than in PCGs (Supplementary Figure S6). In line with this finding, a higher number of Ts in lincRNA genes versus PCGs was observed in a recent study analyzing lincRNA splice-site strengths (28).
To better understand the role of PPT length and T content, we used the categorized lincRNA and PCG introns based on their splicing efficiencies (see above) and calculated the ratios of Ts over guanidines (T/G) or cytidines (T/C) in the PPT region for the five human cell lines (Figure 3B and Supplementary Figures S7 and S8). The correlation between T content and splicing efficiency was high in both lincRNAs and PCGs (Pearson's correlation coefficients between 0.66 and 0.99; Supplementary Figure S7). However, in all cases, the well-spliced lincRNAs (third and fourth quartiles) contained a higher number of Ts than PCGs (Figure 3B and Supplementary Figure S7). While the average SI of best-spliced lincRNAs reached only 60–76% of PCG SI (dependent on the cell line, Supplementary Figure S5B), the T/C and T/G ratios in this group of lincRNAs were higher by 2–13% and 2–16% respectively (Figure 3B and Supplementary Figure S7) suggesting that the T content in the PPT can play an important role in lincRNA splicing.
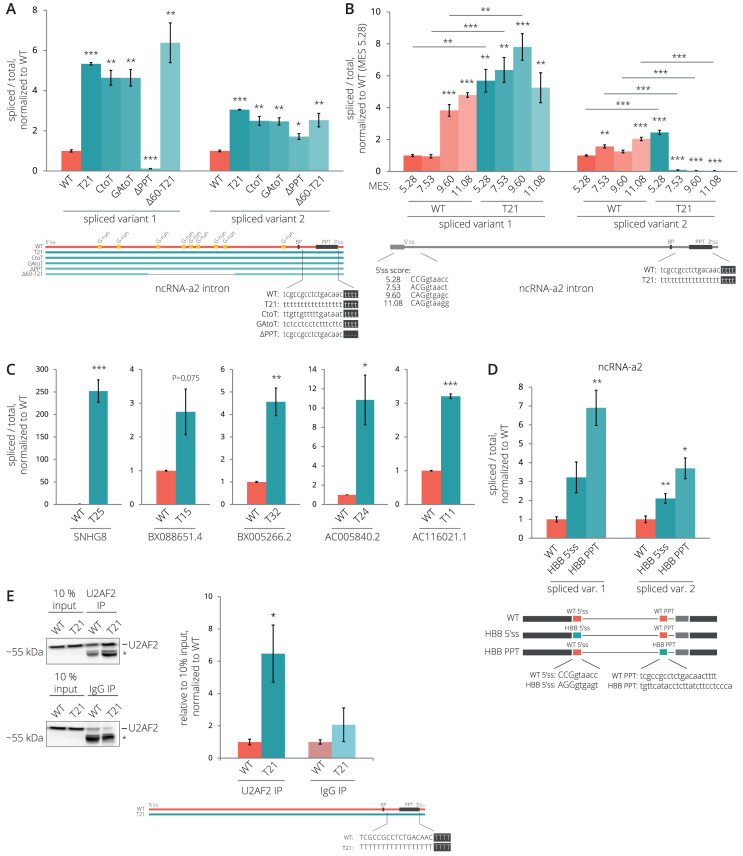
However, comparison of introns that exhibit fast and slow splicing kinetics did not reveal any significant difference in PPT scores (10). Therefore, we decided to experimentally test whether the higher T content in the PPT promotes splicing of lincRNA introns. We utilized the model ncRNA-a2 and increased the number of Ts in its PPT. We either replaced all cytidines (CtoT), all purines (GAtoT), or all nucleotides (T21) with Ts, or deleted a stretch of four Ts upstream of the CAG 3′ss (ΔPPT; Figure 4A). All PPT modifications that increased the T content had a positive effect on the splicing efficiency (4.6–5.3× increase compared to WT), the deletion of Ts inhibited splicing of splicing variant 1 (8.3× reduction with respect to WT) but not splicing variant 2 (1.7× increase with respect to WT). The strong PPT was able to compensate for splicing reduction induced by deletion of the G-run enhancer since the Δ60-T21 construct was spliced 6.4× better than WT ncRNA-a2. Finally, we combined enhancement of 5′ss with the T21 mutation (Figure 4B) and found that the improvement of 5′ss further stimulated splicing of ncRNA-a2 spliced variant 1, but reduced recognition of the downstream 3′ss and the production of spliced variant 2. The only exception was the strongest 5′ss with MES 11.08, which did not enhance splicing when compared with WT PPT containing ncRNA-a2. These results confirm that the 5′ss and the T content in the PPT have a strong and cumulative effect on ncRNA-a2 splicing.
Figure 4.
The PPT is a key determinant of lincRNA splicing efficiency. (A) Splicing efficiencies of ncRNA-a2 after the substitution of Cs (CtoT), Gs (GtoT), all nucleotides (T21) for Ts and deletion of four Ts (ΔPPT) in PPT. (B) Strengthening 5′ss and PPT has a cumulative effect on ncRNA-a2 splicing. Mutations that improve MES of 5′ss (see Figure 2D) were introduced into the T21 mutant, and splicing efficiency was analyzed and compared with WT (the data for WT are identical as in Figure 2D). Asterisks above bars indicate the statistical significance of the individual mutant with respect to WT ncRNA-a2 and asterisks above lines compares WT and T21 constructs with identical 5′ss. (C) Splicing efficiencies of transiently expressed lincRNAs increase after the substitution of nucleotides in PPTs by Ts. (D) The PPT of HBB enhances the splicing of ncRNA-a2. (E) A higher number of Ts in PPT enhances the U2AF2 binding to ncRNA-a2 as determined by RNA immunoprecipitation using the anti-U2AF2 antibody. The position of U2AF2 is shown; the asterisk marks unspecific proteins pulled down in both U2AF2 and control IgG IPs. (A-D) Splicing efficiencies are measured as a fraction of spliced transcripts relative to the total amount of transcripts. (A-E) Bar plots show relative RNA levels as determined by RT-qPCR. The mean of at least three independent experiments is shown. Error bars indicate SEM; asterisks indicate the statistical significance levels calculated by two-tailed Student's t-test comparing the individual mutant with WT, *P < 0.05, **P < 0.01, ***P < 0.001.
To further test the importance of the PPT for lincRNA splicing, we selected five lincRNAs with low SIs and mutated their PPTs. First, we compared splicing efficiencies of endogenous lincRNAs with lincRNAs transiently expressed from CMV-driven plasmid vectors and showed that splicing efficiency is not affected by ectopic expression (Supplementary Figure S9). Then, we converted all nucleotides between the putative BP and the YAG motif into Ts, which significantly enhanced the splicing efficiencies of four out of five tested lincRNAs (Figure 4C). Finally, to test whether a PPT sequence from a PCG that is optimized for splicing can enhance splicing of lincRNA, we replaced the ncRNA-a2 PPT with the PPT sequence from HBB that had higher T/G and T/C ratios than the ncRNA-a2 PPT (Figure 4D). The insertion of the HBB PPT into ncRNA-a2 significantly increased its splicing efficiency, confirming that the ncRNA-a2 PPT is weaker than the HBB PPT. Replacing natural 5′ss ncRNA-a2 sequence with stronger 5′ss from HBB (3 nt upstream and 6 nt downstream of 5′ss) showed partial, but not statistically significant enhancement of spliced variant 1 splicing, which suggests that the PPT sequence is more important than the 5′ss for ncRNA-a2 splicing.
Next, we analyzed how splicing factors, which preferentially bind U-rich sequences in the PPT, interact with lincRNAs and how their binding affect lincRNA splicing. We focused on U2AF2 (U2AF65), hnRNP C and PTBP1, which were all shown to bind to the U-rich sequences in the PPT (44,48,74,75). We utilized publicly available eCLIP (enhanced crosslinking and immunoprecipitation) data from HepG2 cells (BioRxiv: https://doi.org/10.1101/179648) and iCLIP (individual nucleotide–resolution crosslinking and immunoprecipitation) data from Hela cells (47,48) and compared splicing efficiencies of lincRNAs associated/not associated with these proteins. HnRNP C iCLIP data revealed a very low association of hnRNP C with lincRNAs (data not shown), and we included only the hnRNP C eCLIP data analysis. In the U2AF2 data set, lincRNAs bound by U2AF2 tend to have a higher T content in their PPT, longer PPTs and significantly higher splicing efficiencies than lincRNAs that are not bound by U2AF2 (Supplementary Figure S10A–C), in agreement with a recently published analysis (28). In contrast, PTBP1-bound and hnRNP C-bound lincRNAs were spliced as efficiently as unbound lincRNAs (Supplementary Figure S10D and E). These data suggest that U2AF2 binding improves lincRNA splicing efficiency, while PTBP1 and hnRNP C binding do not. To test this prediction experimentally, we transiently expressed ncRNA-a2 WT and its T21 mutant and analyzed their interactions with U2AF2 by RNA immunoprecipitations (RIP) followed by RT-qPCR. We observed that the T21 mutant more efficiently co-precipitated with the U2AF2 protein than the WT transcript (Figure 4E). Altogether these results are consistent with the model that inefficient U2AF2 binding is one of the key factors that reduces splicing efficiencies of lincRNAs.
SR proteins bind less efficiently to lincRNAs
Our data suggest that a strong 5′ss and T-rich PPT together with productive U2AF2 binding are required for efficient lincRNA splicing. To assess whether additional factors may affect lincRNA splicing, we analyzed cumulative lengths of exons and introns and found that lincRNAs contain slightly longer introns and exons than PCGs (Supplementary Figure S11A). Because the length of introns has been associated with splicing efficiency (76–81), longer introns can partially explain lower splicing efficiency of lincRNAs. We also calculated a putative base-pairing and minimum free energy of 5000 randomly selected introns and did not find any relevant differences between lincRNAs and PCGs, which suggest that RNA secondary structure is not the major factor that would determine the splicing difference between PCGs and lincRNAs (Supplementary Figure S11B and C). Finally, we analyzed the presence of known splicing inhibitory sequences 100 bp upstream and downstream of 3′ss. We found only one (out of 12) inhibitory motif (GTAGGT) enriched in lincRNAs over PCGs (Supplementary Figures S12 and S13). Together these results indicate that except for longer introns, lincRNAs do not contain any particular feature that would specifically inhibit their splicing.
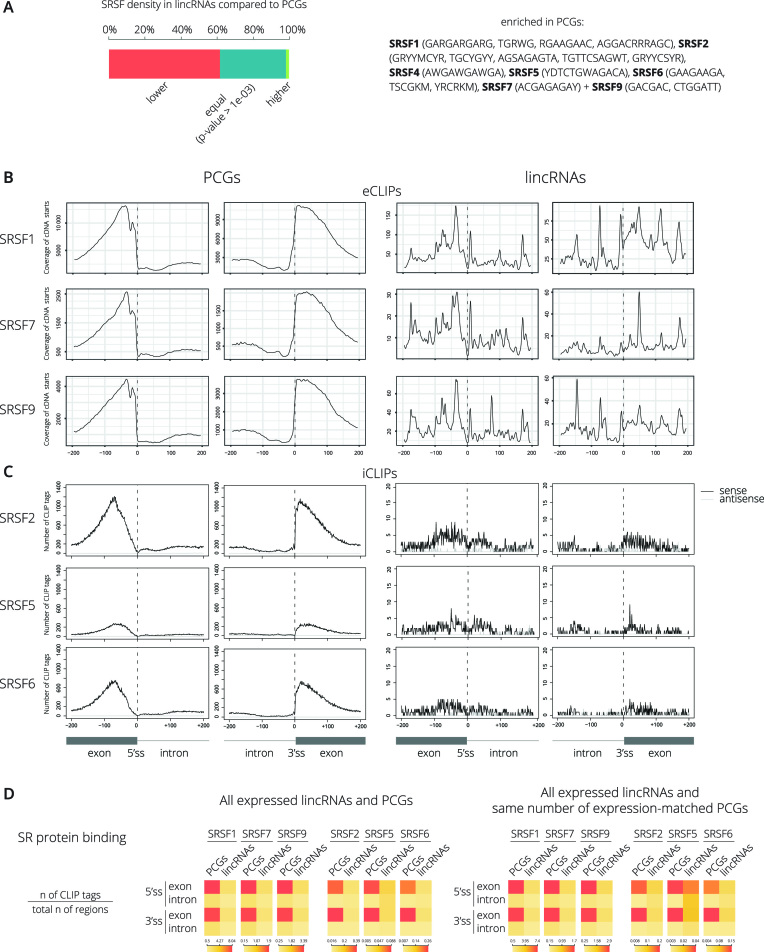
High dependence on strong splice sites could signal that lincRNAs lack additional splicing enhancer sequences that navigate the basic splicing machinery to splice sites. However, previous bioinformatic analyses showed that the global density of exonic splicing enhancers (ESEs) is even slightly higher in lincRNAs than in PCGs (28). In addition, ESEs are conserved in lincRNAs, and no difference in the number of ESEs has been observed between efficiently and inefficiently spliced lincRNAs (13,28,82). To perform a more focused analysis, we searched for the occurrence of ESE motifs that are known to be recognized by SR proteins, general splicing enhancers (83–85). We determined the occurrence of 29 ESE consensus motifs in exons and observed a striking difference in motif densities between lincRNAs and PCGs (Figure 5A and Supplementary Figures S14 and S15). Only one motif (SRSF3 – WCWWC) was significantly enriched in lincRNA exons while the majority of analyzed SR binding motifs were more prevalent in PCGs (Figure 5A).
Figure 5.
SR proteins preferentially bind to exons of PCGs. (A) Combined distribution of 29 SR protein binding motifs within 100 nt regions upstream of 5′ss and downstream of 3′ss between lincRNAs and PCGs. For detail distribution see Supplementary Figures S14 and S15. Motifs enriched in PCG exons are indicated in right. Two SRSF9 binding sites were not detected in lincRNAs. (B) Binding of SRSF1, SRSF7, and SRSF9 within 200 nt regions around 5′ss and 3′ss in lincRNAs and PCGs was analyzed using available eCLIP data (BioRxiv: https://doi.org/10.1101/179648). Protein binding per region 200 bp upstream/downstream of splice sites are shown for PCGs and lincRNAs. (C) Binding of SRSF2, SRSF5, and SRSF6 within 200 nt regions around 5′ss and 3′ss in lincRNAs and PCGs was determined by iCLIP. Protein binding per region 200bp upstream/downstream of splice sites are shown for PCGs and lincRNAs. (D) eCLIP and iCLIP tags (FDR < 0.05) in 200 nt regions upstream and downstream of 5′ss or 3′ss were selected, and the iCLIP tags per nucleotide divided by the total number of expressed regions are shown. Data are shown for all expressed lincRNAs and PCGs (left diagrams), or all expressed lincRNAs and the identical number expression-matched PCGs (right diagrams).
To test whether a smaller number of SR binding motifs in lincRNAs results in a lower interaction with SR proteins, we analyzed available eCLIP data performed with SRSF1, 7 and 9 in HepG2 cells (BioRxiv: https://doi.org/10.1101/179648). All three SR proteins bound efficiently within PCG exons, while their association with lincRNAs was much weaker and we did not detect any significant enrichment over exons (Figure 5B). SR protein binding to lincRNAs was lower compared to the total expressed PCGs (18–26% of binding to PCGs; Figure 5D, left panel). To normalize for the expression level of PCGs and lincRNAs, we created a subset of PCGs that match number and expression level of lincRNAs and repeated the analysis. Similarly, binding of SR proteins to lincRNAs was reduced to 20–30% of expression-matched PCGs (Figure 5D, right panel). To investigate the binding of additional SR proteins not covered by eCLIP, we performed iCLIP in HeLa cell lines stably expressing GFP-tagged SRSF2, SRSF5 or SRSF6 from bacterial artificial chromosomes at near endogenous levels using anti-GFP antibodies as described before (43). iCLIP libraries were prepared in triplicates, submitted to deep sequencing, and significant cross-link events of individual SR proteins were identified (43). Similarly to previous studies (86,87), SR proteins bound preferentially to exonic regions of PCGs (Figure 5C). In agreement with the eCLIP data, binding of all three analyzed SR proteins to lincRNAs was much lower compared to all expressed PCGs (13–30% of binding to PCGs; Figure 5D, left panel) or expression-matched PCGs (56–68% of binding to PCGs; Figure 5D, right panel). Altogether, this confirmed that SR proteins interact poorly with lincRNAs, which is independent of lincRNA expression level.
To test whether residual SR protein binding nevertheless promotes splicing of lincRNAs, we compared splicing efficiencies of bound and unbound lincRNAs (using PCGs as control). Analysis of lincRNA interactions with the three SR proteins identified by iCLIP revealed that binding of SR proteins in lincRNA exons (100 nt downstream of 3′ss) improves their splicing efficiencies (Supplementary Figure S16A). However, no such correlation was found for SR protein interactions with lincRNAs identified by eCLIP (Supplementary Figure S16C), which might be due to the higher noise in eCLIP data compared to iCLIP data (49). In contrast, the stimulatory effect of SR protein binding to exons of PCGs appears stronger in eCLIP than iCLIP data (Supplementary Figure S16B,D). The binding of analyzed SR proteins in intronic regions of PCGs (100 nt upstream of 3′ss) (Supplementary Figure S16B) correlates with lower splicing efficiency, which is consistent with the proposed position-dependent splicing activity of SR proteins (61–67,88).
The role of an intron in the function of ncRNA-a2
NcRNA-a2 and ncRNA-a5 have been suggested to act as transcription enhancers because their depletion by RNAi decreased the expression of some adjacent PCGs (Figure 6A) (54). NcRNA-a2 seems to act in cis because overexpression of ncRNA-a2 from a CMV-driven plasmid did not increase the expression of the target PCG KLHL12 (Figure 6B). Given the ongoing debate about the importance of splicing-associated processes for the function of enhancer-like lncRNAs (89,90, BioRxiv: https://doi.org/10.1101/287706), we tested whether the intron of ncRNA-a2 contributes to its enhancer function. We removed the ncRNA-a2 intron from the endogenous ncRNA-a2 gene locus using CRISPR/Cas9 and isolated three different intron-deleted cell lines (Supplementary Figure S17). To test the importance of ncRNA-a2 splicing on newly transcribed mRNAs, we either isolated RNAs associated with the chromatin fraction or metabolically labelled and isolated nascent RNAs using 4sU-biotin labeling. Expression of neighboring PCGs in three ncRNA-a2 Δintron clonal cell lines was analyzed by RT-qPCR (Figure 6C and D). However, we did not detect any significant difference in transcription of neighboring genes after the deletion of the ncRNA-a2 intron. These results suggest that the intron itself and/or its splicing do not play a significant role in the activating function of ncRNA-a2.
Figure 6.
The ncRNA-a2 intron is not essential for the activation of neighboring gene expression. (A) NcRNA-a2 and ncRNA-a5 were knocked down using siRNAs, and the expression of PCGs located in their genomic vicinity was assayed by RT-qPCR. (B) Expression of the ncRNA-a2 transcript and its target PCG after transient expression of ncRNA-a2. The arrow represents a reverse primer used for RT that is specific for ectopically expressed ncRNA-a2. (C, D) The ncRNA-a2 intron was removed from the genomic locus by CRISPR/Cas9 (top scheme) generating three different clonal cell lines lacking the entire ncRNA-a2 intron. Schemes at the bottom show details of the sequence at the exon/exon boundary after intron deletion in individual cell lines together with WT sequence for comparison. Expression of the ncRNA-a2 gene and PCGs located in its genomic vicinity was analyzed either in the chromatin fraction (C) or in nascent transcripts labeled with 4sU-Biotin (D). (A–D) Bar plots show relative RNA levels as determined by RT-qPCR. The mean of at least three independent experiments is shown. Error bars indicate SEM; asterisks indicate the statistical significance levels calculated by two-tailed Student's t-test comparing the individual mutant with WT, *P < 0.05, **P < 0.01, ***P < 0.001.
DISCUSSION
LincRNAs have been extensively studied in recent years, and previous studies have shown that lncRNAs are less efficiently spliced and polyadenylated in comparison to PCGs (10,15,27–29,91), but the reason for this remained unknown. Here, we calculated SIs for lincRNAs and PCGs expressed in several human cell lines using available ENCODE RNA-Seq data (36) as a proxy for splicing efficiencies of expressed lincRNAs. Our findings show less efficient splicing of lincRNAs in comparison to PCGs in all studied cell lines (Supplementary Figure S4). The bioinformatic results are supported by splicing efficiency analyses of several lincRNAs by quantitative and semi-quantitative RT-PCRs (Figure 1A, Supplementary Figures S2 and S9).
To determine factors affecting lincRNA splicing, we transiently expressed several lincRNAs from a CMV-driven promoter and did not detect any significant changes in their splicing (Figures 1 and Supplementary Figure S9). This result indicates that the promoter and the genomic context do not significantly influence lincRNA splicing profile and that inefficient splicing is an intrinsic property of lincRNA transcripts. To identify potential sequences inhibiting splicing, we created a series of deletion mutants that lack different parts of the ncRNA-a2 intron. We did not find any strong splicing silencers, which is consistent with a bioinformatic analysis that did not reveal any specific accumulation of splicing inhibitory sequences in lincRNAs with respect to PCGs (Figures 2, Supplementary Figures S12 and S13). However, we found that lincRNAs have longer introns and exons than PCGs (Supplementary Figure S11A), which might partially explain their less efficient splicing (76–81). Finally, we analyzed sequences of 5′ and 3′ splice sites and found a positive correlation between the strength of 5′ss and PPT and lincRNA splicing efficiencies (Supplementary Figures S5 and S7). This finding was further supported by experimental evidence showing that increasing the strength of 5′ and 3′ splice sites significantly improved splicing of model lincRNAs (Figures 2 and 4).
The 5′ss and PPT sequences are crucial factors for the splicing efficiency in general, but our data suggest that lincRNA requires stronger 5′ss and PPT containing a high number of Ts to be effectively spliced (Figures 3, Supplementary Figures S5–S8). To understand why lincRNAs are more dependent on basic splice site sequences, we analyzed the presence of known SR protein exonic binding motifs because binding of SR proteins to exons promotes splicing (reviewed in 92,93). We found that the majority of analyzed SR-binding sequences are more abundant in PCG exons while only one motif out of 29 analyzed motifs is enriched in lincRNAs (Figure 5A, Supplementary Figures S14 and S15). Consistently, we show that all analyzed SR proteins exhibit a clear binding preference for PCGs even when we compared lincRNAs with expression-matched PCGs (Figure 5D). This result provides experimental evidence that lincRNAs are unable to secure productive binding of SR proteins. Based on our data we propose a model that lincRNAs lack the cooperative network of positive signals that efficiently navigates the splicing machinery to splice sites. For most lincRNAs, U1 and U2 snRNPs and their auxiliary factors thus have to find splice sites without the help of splicing enhancers, rendering the sequences around exon/intron boundaries more important.
However, it should be noted that the insertion of the ncRNA-a2 intron between PCG exons did not improve splicing efficiency (Figure 1D). This suggests that complete sequence and context is important for correct splicing, which was recently shown for splicing of various 5′ss sequences (94). This is also consistent with studies proposing that the local environment and the continuous sequence of exons and introns information are critical for correct intron definition and removal (15). During evolution, sequences of PCGs were fine-tuned to ensure a robust recognition of intron/exon boundaries and efficient splicing. In contrast to PCGs, where splicing is an essential component of gene expression, this study and the results of Engreitz et al. (90) indicate that introns are not essential for the activating function of lincRNAs. This may result in a lower evolutionary pressure on some lincRNAs to promote efficient splicing. However, it should be noted that the great majority of purifying selection operating on lincRNAs in humans is splicing-related, which suggests that intron presence or its splicing might play some yet unrecognized function in at least some lincRNAs (13).
Evolutionary new transcripts, such as lincRNAs may acquire functional 5′ss and 3′ss over time and splicing may change their functional output. Previous analyses revealed a higher prevalence of cryptic 5′ss over 3′ss, likely reflecting the less complex nature of the 5′ss sequence (95). Indeed, 5′ss can be provided by endogenous retrotransposons (96). Analysis of cryptic 3′ss revealed the importance of the 3′ss sequences and showed that intronic de novo 3′ss arose mainly by AG-creating mutations in existing functional PPTs. In contrast, exonic de novo 3′ss were often induced by mutations improving the PPT, BP sequence or distant auxiliary signals (97). We found the strongest correlation between lincRNA splicing and PPT sequence, which suggests that in the absence of functional splicing enhancers, weak PPT sequence and inefficient U2AF binding represent the major barriers that evolutionary new transcripts have to overcome to become efficiently spliced.
DATA AVAILABILITY
The raw data were deposited in the GEO database under accession number GSE113812-GSE113814.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Josef Pasulka for his help with the bioinformatic analysis, Francois McNicoll for English proofreading and Pavel Draber, David Drechsel, Radek Sedlacek and Petr Kasparek for reagents.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Czech Science Foundation [P305/12/G034]; Czech Academy of Sciences—the DAAD mobility fund [DAAD-17-10]; National Sustainability Program I [LO1419]; institutional support [RVO68378050]; DFG [CEF-MC and SFB902 to M.M.M.]; internal fellowship of the Czech Academy of Sciences [L200521652 to M.K.]; Wellcome Trust grant [106954 to N.H., Boris Lenhard]. Funding for open access charge: Institute of Molecular Genetics.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bertone P., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S. et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004; 306:2242–2246. [DOI] [PubMed] [Google Scholar]
- 2. Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermüller J., Hofacker I.L. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007; 316:1484–1488. [DOI] [PubMed] [Google Scholar]
- 3. Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007; 447:799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mattick J.S., Amaral P.P., Dinger M.E., Mercer T.R., Mehler M.F.. RNA regulation of epigenetic processes. Bioessays. 2009; 31:51–59. [DOI] [PubMed] [Google Scholar]
- 5. Wang K.C., Chang H.Y.. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011; 43:904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patrushev L.I., Kovalenko T.F.. Functions of noncoding sequences in mammalian genomes. Biochem (Mosc.). 2014; 79:1442–1469. [DOI] [PubMed] [Google Scholar]
- 7. Ransohoff J.D., Wei Y., Khavari P.A.. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2017; 19:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012; 22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. et al. Landscape of transcription in human cells. Nature. 2012; 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mukherjee N., Calviello L., Hirsekorn A., de Pretis S., Pelizzola M., Ohler U.. Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat. Struct. Mol. Biol. 2016; 24:86. [DOI] [PubMed] [Google Scholar]
- 11. Wang J., Zhang J., Zheng H., Li J., Liu D., Li H., Samudrala R., Yu J., Wong G.K.-S.. Neutral evolution of ‘non-coding’ complementary DNAs. Nature. 2004; 431:758. [PubMed] [Google Scholar]
- 12. Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L.. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011; 25:1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schüler A., Ghanbarian A.T., Hurst L.D.. Purifying selection on Splice-Related motifs, not expression level nor RNA folding, explains nearly all constraint on human lincRNAs. Mol. Biol. Evol. 2014; 31:3164–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Necsulea A., Soumillon M., Warnefors M., Liechti A., Daish T., Zeller U., Baker J.C., Grutzner F., Kaessmann H.. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014; 505:635–640. [DOI] [PubMed] [Google Scholar]
- 15. Lagarde J., Uszczynska-Ratajczak B., Carbonell S., Pérez-Lluch S., Abad A., Davis C., Gingeras T.R., Frankish A., Harrow J., Guigo R. et al. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat. Genet. 2017; 49:1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cabili M.N., Dunagin M.C., McClanahan P.D., Biaesch A., Padovan-Merhar O., Regev A., Rinn J.L., Raj A.. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015; 16:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Quinn J.J., Chang H.Y.. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016; 17:47–62. [DOI] [PubMed] [Google Scholar]
- 18. Will C.L., Lührmann R.. Spliceosome Structure and Function. Cold Spring Harb. Perspect. Biol. 2011; 3:a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matera A.G., Wang Z.. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014; 15:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. De Conti L., Baralle M., Buratti E.. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscipl. Rev: RNA. 2013; 4:49–60. [DOI] [PubMed] [Google Scholar]
- 21. Sahebi M., Hanafi M.M., van Wijnen A.J., Azizi P., Abiri R., Ashkani S., Taheri S.. Towards understanding pre-mRNA splicing mechanisms and the role of SR proteins. Gene. 2016; 587:107–119. [DOI] [PubMed] [Google Scholar]
- 22. Ule J., Stefani G., Mele A., Ruggiu M., Wang X., Taneri B., Gaasterland T., Blencowe B.J., Darnell R.B.. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006; 444:580–586. [DOI] [PubMed] [Google Scholar]
- 23. Wang Z., Burge C.B.. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA. 2008; 14:802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fu X.-D., Ares M. Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014; 15:689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brillen A.-L., Schöneweis K., Walotka L., Hartmann L., Müller L., Ptok J., Kaisers W., Poschmann G., Stühler K., Buratti E. et al. Succession of splicing regulatory elements determines cryptic 5′ss functionality. Nucleic Acids Res. 2017; 45:4202–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rot G., Wang Z., Huppertz I., Modic M., Lenče T., Hallegger M., Haberman N., Curk T., von Mering C., Ule J.. High-Resolution RNA Maps suggest common principles of splicing and polyadenylation regulation by TDP-43. Cell Rep. 2017; 19:1056–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tilgner H., Knowles D.G., Johnson R., Davis C.A., Chakrabortty S., Djebali S., Curado J., Snyder M., Gingeras T.R., Guigó R.. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012; 22:1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Melé M., Mattioli K., Mallard W., Shechner D.M., Gerhardinger C., Rinn J.L.. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017; 27:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schlackow M., Nojima T., Gomes T., Dhir A., Carmo-Fonseca M., Proudfoot N.J.. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol. Cell. 2017; 65:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Y., Ma M., Xiao X., Wang Z.. Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules. Nat. Struct. Mol. Biol. 2012; 19:1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Corvelo A., Hallegger M., Smith C.W.J., Eyras E.. Genome-Wide association between branch point properties and alternative splicing. PLoS Comp. Biol. 2010; 6:e1001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pandya-Jones A., Black D.L.. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009; 15:1896–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yeo G., Burge C.B.. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004; 11:377–394. [DOI] [PubMed] [Google Scholar]
- 36. Sloan C.A., Chan E.T., Davidson J.M., Malladi V.S., Strattan J.S., Hitz B.C., Gabdank I., Narayanan A.K., Ho M., Lee B.T. et al. ENCODE data at the ENCODE portal. Nucleic Acids Res. 2016; 44:D726–D732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Převorovský M., Hálová M., Abrhámová K., Libus J., Folk P.. Workflow for Genome-Wide determination of Pre-mRNA splicing efficiency from yeast RNA-seq data. Biomed Res Int. 2016; 2016:4783841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lorenz R., Bernhart S.H., Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA package 2.0. Algorith. Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004; 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lawrence M., Gentleman R., Carey V.. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics. 2009; 25:1841–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wickham H. ggplot2: Elegant Graphics for Data Analysis. 2016; NY: Springer-Verlag. [Google Scholar]
- 43. Botti V., McNicoll F., Steiner M.C., Richter F.M., Solovyeva A., Wegener M., Schwich O.D., Poser I., Zarnack K., Wittig I. et al. Cellular differentiation state modulates the mRNA export activity of SR proteins. J. Cell Biol. 2017; 216:1993–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. König J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B., Turner D.J., Luscombe N.M., Ule J.. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010; 17:909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lawrence M., Gentleman R., Carey V.. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics. 2009; 25:1841–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J.. Software for computing and annotating genomic ranges. PLoS Comp. Biol. 2013; 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xue Y., Zhou Y., Wu T., Zhu T., Ji X., Kwon Y.-S., Zhang C., Yeo G., Black D.L., Sun H. et al. Genome-wide Analysis of PTB-RNA Interactions Reveals a Strategy Used by the General Splicing Repressor to Modulate Exon Inclusion or Skipping. Mol. Cell. 2009; 36:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zarnack K., König J., Tajnik M., Martincorena I., Eustermann S., Stévant I., Reyes A., Anders S., Luscombe N.M., Ule J.. Direct Competition between hnRNP C and U2AF65 Protects the Transcriptome from the Exonization of Alu Elements. Cell. 2013; 152:453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Haberman N., Huppertz I., Attig J., König J., Wang Z., Hauer C., Hentze M.W., Kulozik A.E., Le Hir H., Curk T. et al. Insights into the design and interpretation of iCLIP experiments. Genome Biol. 2017; 18:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chakrabarti A.M., Haberman N., Praznik A., Luscombe N.M., Ule J.. Data science issues in studying Protein–RNA interactions with CLIP technologies. Annu. Rev. Biomed. Data Sci. 2018; 1:235–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotech. 2013; 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kasparek P., Krausova M., Haneckova R., Kriz V., Zbodakova O., Korinek V., Sedlacek R.. Efficient gene targeting of the Rosa26 locus in mouse zygotes using TALE nucleases. FEBS Lett. 2014; 588:3982–3988. [DOI] [PubMed] [Google Scholar]
- 54. Ørom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q. et al. Long noncoding RNAs with Enhancer-like function in human cells. Cell. 2010; 143:46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hnilicová J., Hozeifi S., Dusková E., Icha J., Tománková T., Stanek D.. Histone deacetylase activity modulates alternative splicing. PLoS One. 2011; 6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dušková E., Hnilicová J., Staněk D.. CRE promoter sites modulate alternative splicing via p300-mediated histone acetylation. RNA Biol. 2014; 11:865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Salton M., Voss T.C., Misteli T.. Identification by high-throughput imaging of the histone methyltransferase EHMT2 as an epigenetic regulator of VEGFA alternative splicing. Nucleic Acids Res. 2014; 42:13662–13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nieto Moreno N., Giono L.E., Cambindo Botto A.E., Muñoz M.J., Kornblihtt A.R.. Chromatin, DNA structure and alternative splicing. FEBS Lett. 2015; 589:3370–3378. [DOI] [PubMed] [Google Scholar]
- 59. Curado J., Iannone C., Tilgner H., Valcárcel J., Guigó R.. Promoter-like epigenetic signatures in exons displaying cell type-specific splicing. Genome Biol. 2015; 16:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bieberstein N.I., Kozáková E., Huranová M., Thakur P.K., Krchňáková Z., Krausová M., Carrillo Oesterreich F., Staněk D.. TALE-directed local modulation of H3K9 methylation shapes exon recognition. Sci. Rep. 2016; 6:29961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kanopka A., Mühlemann O., Akusjärvi G.. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature. 1996; 381:535. [DOI] [PubMed] [Google Scholar]
- 62. Gallego M.E., Gattoni R., Stévenin J., Marie J., Expert–Bezançon A.. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the β-tropomyosin alternative exon 6A. EMBO J. 1997; 16:1772–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jiang Z.-H., Zhang W.-J., Rao Y., Wu J.Y.. Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:9155–9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. ten Dam G.B., Zilch C.F., Wallace D., Wieringa B., Beverley P.C.L., Poels L.G., Screaton G.R.. Regulation of alternative splicing of CD45 by antagonistic effects of SR protein splicing factors. J. Immunol. 2000; 164:5287–5295. [DOI] [PubMed] [Google Scholar]
- 65. Simard M.J., Chabot B.. SRp30c is a repressor of 3′ splice site utilization. Mol. Cell. Biol. 2002; 22:4001–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang Y., Wang J., Gao L., Lafyatis R., Stamm S., Andreadis A.. Tau exons 2 and 10, which are misregulated in neurodegenerative diseases, are partly regulated by silencers which bind a SRp30c·SRp55 complex that either recruits or antagonizes htra2β1. J. Biol. Chem. 2005; 280:14230–14239. [DOI] [PubMed] [Google Scholar]
- 67. Buratti E., Stuani C., De Prato G., Baralle F.E.. SR protein-mediated inhibition of CFTR exon 9 inclusion: molecular characterization of the intronic splicing silencer. Nucleic Acids Res. 2007; 35:4359–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. McCullough A.J., Berget S.M.. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol. Cell. Biol. 1997; 17:4562–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chou M.-Y., Rooke N., Turck C.W., Black D.L.. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 1999; 19:69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McCullough A.J., Berget S.M.. An intronic splicing enhancer binds U1 snRNPs to enhance splicing and select 5′ splice sites. Mol. Cell. Biol. 2000; 20:9225–9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang E., Dimova N., Cambi F.. PLP/DM20 ratio is regulated by hnRNPH and F and a novel G-rich enhancer in oligodendrocytes. Nucleic Acids Res. 2007; 35:4164–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xiao X., Wang Z., Jang M., Nutiu R., Wang E.T., Burge C.B.. Splice site strength–dependent activity and genetic buffering by poly-G runs. Nat. Struct. Mol. Biol. 2009; 16:1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang E., Mueller W.F., Hertel K.J., Cambi F.. G Run-mediated recognition of proteolipid protein and DM20 5′ splice sites by U1 small nuclear RNA is regulated by context and proximity to the splice site. J. Biol. Chem. 2011; 286:4059–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wagner E.J., Garcia-Blanco M.A.. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 2001; 21:3281–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mulligan G.J., Guo W., Wormsley S., Helfman D.M.. Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of beta-tropomyosin pre-mRNA. J. Biol. Chem. 1992; 267:25480–25487. [PubMed] [Google Scholar]
- 76. Klinz F.-J., Gallwitz D.. Size and position of intervening sequences are critical for the splicing efficiency of pre-mRNA in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1985; 13:3791–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bell M.V., Cowper A.E., Lefranc M.-P., Bell J.I., Screaton G.R.. Influence of intron length on alternative splicing of CD44. Mol. Cell. Biol. 1998; 18:5930–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sterner D.A., Carlo T., Berget S.M.. Architectural limits on split genes. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:15081–15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fox-Walsh K.L., Dou Y., Lam B.J., Hung S.-p., Baldi P.F., Hertel K.J.. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:16176–16181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dewey C.N., Rogozin I.B., Koonin E.V.. Compensatory relationship between splice sites and exonic splicing signals depending on the length of vertebrate introns. Bmc Genomics. 2006; 7:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Louloupi A., Ntini E., Conrad T., Ørom U.A.V.. Transient N-6-Methyladenosine transcriptome sequencing reveals a regulatory role of m6A in splicing efficiency. Cell Rep. 2018; 23:3429–3437. [DOI] [PubMed] [Google Scholar]
- 82. Haerty W., Ponting C.P.. Unexpected selection to retain high GC content and splicing enhancers within exons of multiexonic lncRNA loci. RNA. 2015; 21:333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Paz I., Akerman M., Dror I., Kosti I., Mandel-Gutfreund Y.. SFmap: a web server for motif analysis and prediction of splicing factor binding sites. Nucleic Acids Res. 2010; 38:W281–W285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mueller W.F., Hertel K.J.. Lorkovic Z. The role of SR and SR-related proteins in pre-mRNA splicing. RNA Binding Proteins. 2011; I:NY: Landes Bioscience and Springer Science+Business Media; 1–21. [Google Scholar]
- 85. Müller-McNicoll M., Botti V., de Jesus Domingues A.M., Brandl H., Schwich O.D., Steiner M.C., Curk T., Poser I., Zarnack K., Neugebauer K.M.. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 2016; 30:553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Fairbrother W.G., Holste D., Burge C.B., Sharp P.A.. Single nucleotide Polymorphism–Based validation of exonic splicing enhancers. PLoS Biol. 2004; 2:e268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Xiao X., Wang Z., Jang M., Burge C.B.. Coevolutionary networks of splicing cis-regulatory elements. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:18583–18588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Erkelenz S., Mueller W.F., Evans M.S., Busch A., Schöneweis K., Hertel K.J., Schaal H.. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA. 2013; 19:96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yin Y., Yan P., Lu J., Song G., Zhu Y., Li Z., Zhao Y., Shen B., Huang X., Zhu H. et al. Opposing roles for the lncRNA haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell. 2015; 16:504–516. [DOI] [PubMed] [Google Scholar]
- 90. Engreitz J.M., Haines J.E., Perez E.M., Munson G., Chen J., Kane M., McDonel P.E., Guttman M., Lander E.S.. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016; 539:452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Seidl C.I., Stricker S.H., Barlow D.P.. The imprinted Air ncRNA is an atypical RNAPII transcript that evades splicing and escapes nuclear export. EMBO J. 2006; 25:3565–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Long J.C., Caceres J.F.. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009; 417:15–27. [DOI] [PubMed] [Google Scholar]
- 93. Graveley B.R. Sorting out the complexity of SR protein functions. RNA. 2000; 6:1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wong M.S., Kinney J.B., Krainer A.R.. Quantitative activity profile and context dependence of all human 5′ splice sites. Mol. Cell. 2018; 71:1012–1026.e1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Nakai K., Sakamoto H.. Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene. 1994; 141:171–177. [DOI] [PubMed] [Google Scholar]
- 96. Franke V., Ganesh S., Karlic R., Malik R., Pasulka J., Horvat F., Kuzman M., Fulka H., Cernohorska M., Urbanova J. et al. Long terminal repeats power evolution of genes and gene expression programs in mammalian oocytes and zygotes. Genome Res. 2017; 27:1384–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Královicová J., Christensen M.B., Vorechovský I.. Biased exon/intron distribution of cryptic and de novo 3′ splice sites. Nucleic Acids Res. 2005; 33:4882–4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data were deposited in the GEO database under accession number GSE113812-GSE113814.