Abstract
Alternative pre-mRNA splicing generates functionally distinct transcripts from the same gene and is involved in the control of multiple cellular processes, with its dysregulation being associated with a variety of pathologies. The advent of next-generation sequencing has enabled global studies of alternative splicing in different physiological and disease contexts. However, current bioinformatics tools for alternative splicing analysis from RNA-seq data are not user-friendly, disregard available exon-exon junction quantification or have limited downstream analysis features. To overcome such limitations, we have developed psichomics, an R package with an intuitive graphical interface for alternative splicing quantification and downstream dimensionality reduction, differential splicing and gene expression and survival analyses based on The Cancer Genome Atlas, the Genotype-Tissue Expression project, the Sequence Read Archive project and user-provided data. These integrative analyses can also incorporate clinical and molecular sample-associated features. We successfully used psichomics in a laptop to reveal alternative splicing signatures specific to stage I breast cancer and associated novel putative prognostic factors.
INTRODUCTION
Alternative splicing fosters transcriptome diversity in eukaryotes through the processing of pre-mRNAs from the same gene into distinct transcripts that may encode for proteins with different functions (1,2). Alternative splicing is involved in multiple cellular processes, such as apoptosis and autophagy regulation (1,2), and is especially prevalent in humans, where ∼93% of genes display alternatively spliced transcripts whose regulation may differ across tissues and developmental stages (2–4). Consistently, alternative splicing dysregulation has been linked with cancer, neurodegeneration and other diseases (2,5,6). For instance, splicing alterations mediated by the key regulator SRSF1 may impact multiple hallmarks of cancer, such as resistance to apoptosis and tissue invasion (5).
The relevance of alternative splicing changes in physiological and disease conditions, along with the increasing economic feasibility of next-generation RNA sequencing (RNA-seq), has progressively driven transcriptome-wide alternative splicing studies (3,7–10) and promoted large consortium efforts to assemble publicly accessible splicing data. Such efforts include The Cancer Genome Atlas (TCGA), that catalogues clinical and molecular profiling data from multiple human tumours (11); the Genotype-Tissue Expression (GTEx) project, that focuses on profiling normal human multi-tissue data (12); and the recount2 project, an online resource of processed RNA-seq data for over 2000 studies, mostly from the Sequence Read Archive (SRA) (13). Among the openly available processed data from these projects, counts of RNA-seq reads aligned to exon-exon junctions may be exploited for alternative splicing quantification and further analysis. Indeed, the ability to couple proper differential splicing analysis with, for instance, gene expression, protein domain annotation, clinical information or literature-based evidence enables researchers to extract, from those comprehensive public datasets, valuable insights into the role of alternative splicing in physiological and pathological contexts, as well as putative splicing-associated prognostic factors and therapeutic targets (7–10,14).
Several tools are currently available to quantify, analyse and visualise alternative splicing data. Similarly to psichomics, some analyse alternative splicing based on the commonly-employed and intuitive proportion of reads aligned to splice junctions supporting the inclusion isoform, known as Percent Spliced-In or PSI (3). Examples of such tools are AltAnalyze (15), MISO (16), SpliceSeq (17), VAST-TOOLS (18), rMATS (19), SUPPA (20) and Whippet (21). However, current alternative splicing analysis tools, regardless of their quantification metric, suffer from at least one of the following shortcomings:
Lack of support for imputing pre-processed data (e.g. splice junction read counts), leading to redundant, time-consuming RNA-seq read alignment and exon-exon junction detection, preceding alternative splicing quantification when exon-exon junction quantification is already available (e.g. when analysing TCGA, GTEx or recount2 data).
Limited set of statistical options for differential splicing analysis, mostly relying on median-based non-parametric tests and restricted to pairwise comparisons.
No incorporation of molecular or clinical information enabling analyses that reflect factorial designs or test linear models, for example. This is particularly limiting in the exploration of clinical datasets where, for instance, survival analyses permit assessing the potential prognostic value of alternative splicing events.
No support for transcriptome-wide filtering and sub-setting of events, based on common features or the outcome of statistical analyses, for interactive exploration of individual events of interest.
No user-friendly interactive graphical interface neither support for customisable statistical plots.
The major advantage of exploiting available pre-processed splice junction read counts is that it exempts researchers from storing and processing large FASTQ or BAM files that require expensive computational resources. To our knowledge, no other tool allows the direct performance of transcriptome-wide alternative splicing analysis using splice junction read counts from publicly available RNA-seq datasets (e.g. from TCGA, GTEx and recount2). For instance, jSplice (22) and DIEGO (23) are differential splicing analysis tools that do quantify splicing from junction read counts but it is for the user to convert such counts from the aforementioned projects into the programs’ accepted input formats. Moreover, none of those tools currently incorporates support for survival analysis, exploratory and differential analyses of gene expression, or tests for association between gene expression levels and/or alternative splicing quantifications changes.
To offer a comprehensive pipeline that integrates all the aforementioned features through both a command-line and an easy-to-use graphical interface, we have developed psichomics, an R package to quantify, analyse and visualise alternative splicing and gene expression data using TCGA, GTEx, recount2 and/or user-provided data. Our tool interactively performs dimensionality reduction, differential splicing and gene expression and survival analyses with direct incorporation of molecular and clinical features. We successfully employed psichomics to analyse stage I breast cancer TCGA data and identified alternative splicing events with putative prognostic value. psichomics is freely available in Bioconductor at http://bioconductor.org/packages/psichomics.
MATERIALS AND METHODS
psichomics was developed as an R package with a modular design, allowing to easily modify and extend its components. These include support for multiple file formats and automatic data retrieval from external sources (e.g. TCGA, GTEx and recount2), parsing and standardisation of alternative splicing event identifiers from different programs and annotations and the implementation of a variety of data analysis methodologies.
The program's workflow for alternative splicing analysis begins with the loading of splice junction read count data from the user's computer or external sources, followed by the quantification of alternative splicing (in case no pre-computed quantification is loaded) and subsequent analyses. Alternative splicing quantification is based on RNA-seq reads that align to splice junctions and the genomic coordinates (annotation) of alternative splicing events. The proportion of reads aligned to junctions that support the inclusion isoform, known as the Percent Spliced-In or PSI (3), was the chosen quantification metric.
Exon-exon junction quantification, gene expression and sample-associated data retrieval
Exon-exon junction and gene expression quantifications (obtained from pre-processed RNA-seq data) and clinical data are accessible through FireBrowse's web application program interface (API) for TCGA data retrieval (http://firebrowse.org/api-docs). The FireBrowse API is used in psichomics to automatically download TCGA data according to the user-selected tumour type(s) as tab-delimited files within compressed folders, whose contents are subsequently loaded with minimal user interaction. Data for select SRA projects (including gene expression, exon-exon junction quantification and sample metadata) are also available for automatic retrieval and processing through recount2 (13).
Contrastingly, GTEx does not currently provide any public API for automatic data retrieval, thus requiring the user to manually download exon-exon junction quantification, gene expression and clinical data from the GTEx website (http://gtexportal.org), for instance.
Other SRA projects and user-owned files may also be loaded in appropriate formats, allowing for subsequent alternative splicing analysis from customised data (tutorial on http://rpubs.com/nuno-agostinho/psichomics-custom-data).
Gene expression pre-processing
Gene expression quantifications can be filtered based on user-provided parameters (for instance, to account solely for genes supported by 10 or more reads in 10 or more samples, as performed by default) and normalised by raw library size scaling using function calcNormFactors from R package edgeR (24). Afterwards, counts per million reads (CPM) are computed and log2-transformed (if desired) using the function cpm from edgeR. Log2-transformation is performed by default.
Alternative splicing annotation
Annotations of alternative splicing events are available on-demand in psichomics for the Human hg19 (default) and hg38 genome assemblies. Custom annotation files can also be created by following the appropriate tutorial available at http://rpubs.com/nuno-agostinho/preparing-AS-annotation.
The hg19 annotation of human alternative splicing events was based on files used as input by MISO (16), VAST-TOOLS (18), rMATS (19) and SUPPA (20). Annotation files from MISO and VAST-TOOLS are provided in their respective websites, whereas rMATS and SUPPA identify alternative splicing events and generate such annotation files based on a given isoform-centered transcript annotation. As such, the human transcript annotation was retrieved from the UCSC Table Browser (25) in GTF and TXT formats, so that gene identifiers in the GTF file (misleadingly identical to transcript identifiers) were replaced with proper ones from the TXT version.
The collected hg19 annotation files were non-redundantly merged according to the genomic coordinates and orientation of each alternative splicing event and contain the following event types: skipped exon (SE), mutually exclusive exons (MXE), alternative first exon (AFE), alternative last exon (ALE), alternative 5′ splice site (A5SS), alternative 3′ splice site (A3SS), alternative 5′ UTR length (A5UTR), alternative 3′ UTR length (A3UTR), and intron retention (IR). The resulting hg19 annotation is available as an R annotation package in Bioconductor at http://bioconductor.org/packages/alternativeSplicingEvents.hg19, whereas the hg38 annotation (whose coordinates were converted from those of the hg19 annotation through function liftOver from package rtracklayer (26), based on the hg19 to hg38 chain file from UCSC) is also available as an R annotation package in Bioconductor at http://bioconductor.org/packages/alternativeSplicingEvents.hg38.
Alternative splicing quantification
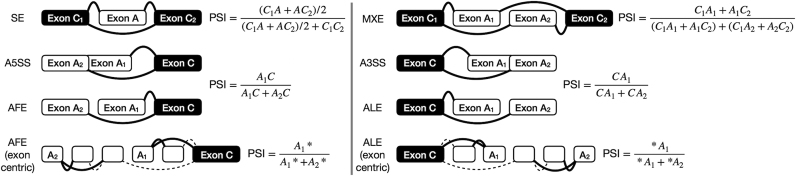
For each alternative splicing event in a given sample, its PSI value is estimated by the proportion of exon–exon junction read counts supporting the inclusion isoform therein (3). The junction reads required for alternative splicing quantification depend on the type of event (Figure 1). Alternative splicing events involving a sum of junction read counts supporting inclusion and exclusion of the alternative sequence below a user-defined threshold (10 by default) are discarded to avoid imprecise quantifications based on insufficient evidence.
Figure 1.
Splice junctions required to quantify alternative splicing based on event type. C1A and AC2 represent read counts supporting junctions between a constitutive (C1 or C2, respectively) and an alternative (A) exon and therefore alternative exon A inclusion, while C1C2 represents read counts supporting the junction between the two constitutive exons and therefore alternative exon A exclusion. A1* and A2* represent the sum of read counts supporting junctions spanning the alternative first (A1) and second (A2) exon, respectively. Legend: skipped exon (SE), mutually exclusive exons (MXE), alternative 5′ splice site (A5SS), alternative 3′ splice site (A3SS), alternative first exon (AFE) and alternative last exon (ALE).
Alternative splicing quantification in psichomics is currently based on exon-exon junction read counts, yet intron retention events require intron-exon junction read counts for their quantification (27), whereas alternative 5′- and 3′-UTR require exon body read counts. psichomics does not currently quantify those types of alternative splicing events.
By default, psichomics quantifies all skipped exon events. However, the user can select to measure other types of alternative splicing events (Figure 1) and may hand in the list of genes whose alternative splicing events are to be specifically quantified. Furthermore, the step of alternative splicing quantification may be avoided if previously performed. psichomics allows the user to save the quantification of alternative splicing in a file to be loaded in a future session.
Data grouping
psichomics allows to group subjects and their samples or genes and their alternative splicing events for subsequent analysis. Subject and sample grouping can be performed based on available phenotypic (e.g. tissue type and histology) and clinical (e.g. disease stage, smoking history and ethnicity) features. Gene and splicing event grouping relies on respective user-provided identifiers. Moreover, the association between subject/sample groups specified by the user and those defined by the outcome of gene expression and alternative splicing analyses or by other clinical categorical variables can be statistically tested with Fisher's exact tests, implemented through function fisher.test from stats (version 3.4.1).
Dimensionality reduction
Dimensionality reduction techniques can be performed on tables containing alternative splicing and gene expression quantifications, with the samples of interest as rows and the selected (if not all) splicing events or genes as columns, after centering and/or scaling the respective distributions (by default, they are only centered).
Principal component analysis (PCA) identifies the combinations of variables that contribute the most to data variance (28) and it is implemented through the singular value decomposition (SVD) algorithm provided by the prcomp function from R package stats (version 3.4.1). The total contribution of each variable (splicing event or gene) towards data variance along selected principal components is measured based on the implementation of fviz_contrib from factoextra (version 1.0.5).
Independent component analysis (ICA), a method used for decomposing data into statistically independent components (29), can also be performed through the fastICA function from the eponymous R package (version 1.2-1), preceded by data centering and/or scaling with the scale function.
As many of the aforementioned functions cannot handle missing data, a user-defined threshold for the accepted number of missing values per alternative splicing event or gene (5%, by default) is used to discard variables before performing dimensionality reduction, whereas the remaining missing values are imputed for each variable as the median from non-missing data samples.
Moreover, samples can be clustered using k-means, partitioning around medoids (PAM) or clustering large applications (CLARA) methods, with the latter being optimised for large datasets and thus preferred by default. The implementation of these methods is based on the kmeans function from stats (version 3.4.1) and pam and clara functions from cluster (version 2.0.6), respectively.
Survival analysis
Kaplan-Meier estimators (and illustrating curves) (30) and proportional hazard (PH) models (31) may be applied to groups of patients defined by the user based on clinical features derived, for instance, from TCGA and user-owned data, with survival distributions being compared using the log-rank test. Survival analyses are implemented in psichomics using functions Surv, survfit, survdiff and coxph from R package survival (32).
To evaluate the prognostic value of a given alternative splicing event, survival analysis can be performed on groups of patients separated based on a given alternative splicing quantification (i.e. PSI) cut-off. Patients with multiple samples are assigned the average PSI value of their respective samples after sample filtering (e.g. when using TCGA data, only tumour samples are used for survival analysis by default). When survival differences are estimated for multiple PSI cut-offs for a single alternative splicing event, psichomics suggests the optimal cut-off that minimises the P-value of the log-rank test used to compare survival distributions, graphically supporting the suggestion with a PSI cut-off versus P-value scatter plot. Survival analysis can also be performed on groups defined by an expression cut-off for a selected gene.
Differential splicing and gene expression analyses
In psichomics, analysis of differential splicing between user-defined groups of samples can be performed on all or selected alternative splicing events. Given the non-normal distribution of PSI values (33,34), median- and variance-based non-parametric tests, such as the Wilcoxon rank-sum (also known as Mann–Whitney U), Kruskal–Wallis rank-sum and Fligner–Killeen tests, are available and recommended (35). Levene's and unpaired t-tests can nonetheless be performed as well. All these tests are available through the stats package (version 3.4.1) with their default settings, except for Levene's test that was implemented based on the leveneTest.default function from the car package (version 2.1-6).
To correct for multiple testing where applicable, P-value adjustment methods for the family-wise error rate (Bonferroni, Holm, Hochberg and Hommel corrections) and the false discovery rate (Benjamini–Hochberg and Benjamini–Yekutieli methods) are available through function p.adjust from package stats (version 3.4.1). By default, multiple testing correction is performed using the Benjamini-Hochberg method.
Although the aforementioned statistical tests are also available to analyse the expression of single genes, genome-wide differential gene expression analysis is implemented based on gene-wise linear model fitting (using lmFit from R package limma (36)) for two selected groups, followed by moderated t-tests and the calculation of log-odds of differential expression, using empirical Bayes moderation of standard errors (function eBayes from limma) and gene-wise variance modelling (limma-trend).
Statistical results can be subsequently explored through density and volcano plots with customisable axes to assist in the identification of the most significant changes when analyzing distributions across single or multiple events, respectively. A corresponding table with the results of all statistical analyses is also available and can be retrieved as a tab-delimited plain text file.
Correlation between gene expression and alternative splicing quantifications
The Pearson product-moment correlation coefficient, Spearman's rho (default) and Kendall's tau, all available with cor.test from stats (version 3.4.1), can be used to correlate gene expression levels with alternative splicing quantifications. Such analyses allow, for instance, to test the association between the expression levels of RNA-binding proteins (RBPs) and PSI levels of interesting splicing events to identify which of these may undergo RBP-mediated regulation. As such, a list of RBPs is provided in-app (37), but the user can also define their own group of genes of interest for the test.
Gene, transcript and protein annotation and literature support
The representational state transfer (REST) web services provided by Ensembl (38), UniProt (39), the Proteins API (40) and PubMed (41) are used in order to annotate genes of interest with relevant biomolecular information (e.g. genomic location, associated transcript isoforms and protein domains, etc.) and related research articles. psichomics also provides the direct link to the cognate entries of relevant external databases, namely Ensembl (42), GeneCards (43), the Human Protein Atlas (44), the UCSC Genome Browser (45), UniProt (39) and VAST-DB (46).
Performance benchmarking
To measure the time taken by psichomics to load data, normalise gene expression, quantify PSIs for skipped exon events and perform global differential expression and splicing analyses between pairs of GTEx tissues and between normal and primary solid tumour samples from multiple TCGA cohorts, the program was run 10 times with the same settings for different combinations of normal human tissues and tumour types in a machine running OS X 10.13.1 with 4 cores and 8GB of RAM, using Safari 11.0.1, RStudio Desktop 1.1.383 and R 3.4.1. The median duration of the 10 runs was used as the performance indicator.
To determine the approximate time complexity of the aforementioned steps in psichomics, gene expression and exon-exon junction quantification datasets were prepared based on approximate distributions obtained from the respective TCGA datasets: negative binomial distributions with a dispersion parameter of 0.25 and 0.2 reads and a mean parameter of 2000 and 100 reads for raw gene expression and exon-exon junction quantification, respectively. Each run was performed on datasets with numbers of samples ranging from 100 to 2500 in intervals of 100 (i.e. 100, 200, 300, …, 2500) and 20 000 genes or 200 000 splice junctions (gene expression or exon-exon junction quantification, respectively). Splice junction identifiers (required for alternative splicing quantification) were randomly retrieved from the TCGA reference annotation. Based on their respective read counts, around 9000 alternative splicing events (i.e. those for which all involved inclusion and exclusion junctions were retrieved) were quantified across selected samples per run. For differential gene expression and splicing analyses, samples were randomly divided into two groups based on the emitted values of a Bernoulli distribution with a probability of success of 50%.
Polynomials of orders 1–6 were fitted to the relation between running time and the number of samples. As the running time is assumed to always increase with an increasing number of analysed samples, fitted polynomials were constrained to be monotone for 0 or more samples, using function monpol from R package MonoPoly (47). The best polynomial fits (Figure 3) were selected based on analyses of variance (ANOVA) between fitted polynomials of consecutive orders, starting with the comparison between polynomials of orders 1 and 2. A polynomial with higher order is only selected if exhibiting a significantly better fit (P-value < 0.05).
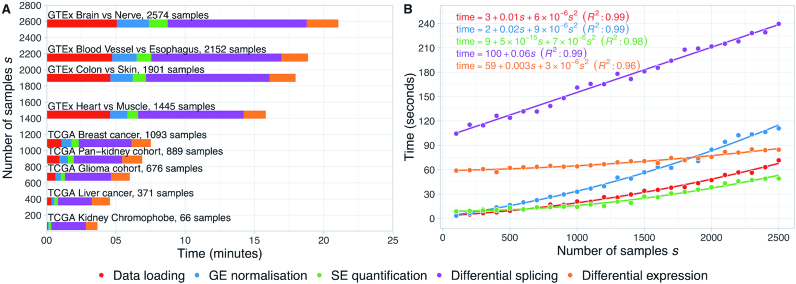
Figure 3.
Performance benchmark for alternative splicing analysis using RNA-seq data from multiple TCGA and GTEx sample types. (A) Median times of 10 runs of data loading, gene expression (GE) normalisation, skipped exon (SE) event quantification and differential expression and splicing analysis (normal versus tumour for TCGA data or pairwise tissue comparison for GTEx data) using psichomics. The default settings were used during the runs. (B) Estimation of the time complexity of each of the aforementioned steps in psichomics. Randomly generated synthetic datasets of different sample size s were used as input. Equations and coefficient of determination (R2) for the best fits are displayed.
Alternative splicing quantification benchmarking
The publicly available RNA-seq data from multiple human, mouse and chicken tissue and cell line samples used in the development of VastDB (46) were aligned with splice-aware STAR (48) against the respective transcript-annotated genomes: UCSC hg19 genome assembly and GENCODE v19 annotation for human, UCSC mm10 genome assembly and GENCODE vM14 annotation for mouse, and Ensembl 70 genome assembly and annotation for chicken. In total, 120/706/34 (human/mouse/chicken) exon skipping events quantified by psichomics (using function quantifySplicing with default settings) were compared with the respective RT-PCR- and VAST-TOOLS-derived PSI values, available from VastDB (46).
Different numbers of junction reads were simulated for different given PSI values to test the impact of read coverage on the accuracy and precision of PSI estimation by psichomics. For each given PSI, junction reads supporting the exon inclusion were simulated as the number of successes obtained from a Bernoulli distribution with the event's junction read coverage (i.e. reads supporting inclusion plus reads supporting exclusion) as the number of observations and the PSI value as the probability of success. Those inclusion reads were then divided by the event's junction read coverage to estimate an ‘observed’ PSI value (as performed by psichomics) that was compared to the given ‘real’ PSI value. These simulations were performed for PSI values from 0 to 1 in 0.1 intervals and event coverages of 10, 20, 50, 100, 500 and 1000 junction read counts, with each combination being tested 10000 times.
TCGASpliceSeq (49) provides pre-computed alternative splicing quantifications across TCGA cohorts. As those quantifications are performed similarly by TCGASpliceSeq and psichomics, PSI estimates for each matching (based on genomic coordinates) alternative splicing event and sample from both tools were correlated across the entire TCGA dataset.
RESULTS
psichomics offers both a graphical and a command-line interface. Although most features are common to both interfaces, we recommend less experienced users to opt for the graphical interface based on the shiny package (version 1.0.5), a web application framework available for R. To start the graphical interface, the user is required to load the psichomics package in R and run function psichomics(), resulting in the automatic launch of the user's default web browser and of the program's graphical interface as a local web app.
Case study: exploration of clinically-relevant, differentially spliced events in breast cancer
Breast cancer is the cancer type with the highest incidence and mortality in women (50) and multiple studies have suggested that transcriptome-wide analyses of alternative splicing changes in breast tumours are able to uncover tumour-specific biomarkers (7,8,14). Given the relevance of early detection of breast cancer to patient survival and in order to pinpoint putative biomarkers that can be exploited from the earlier stages of the disease, we used psichomics to identify novel tumour stage-I-specific molecular signatures based on differentially spliced events.
For the purposes of this case study, default psichomics settings were used unless otherwise stated. The analysis steps summarised below are easily reproducible by following the tutorials on http://rpubs.com/nuno-agostinho/psichomics-tutorial-visual (visual interface) and http://rpubs.com/nuno-agostinho/psichomics-cli-tutorial (command-line interface).
Alternative splicing quantification of the most recent TCGA breast cancer processed RNA-seq data available (28 January 2016) was performed by psichomics for skipped exons, mutually exclusive exons, alternative 5′ and 3′ splice sites and alternative first and last exons.
PCA was performed on alternative splicing and gene expression quantifications. A tumour-stage-independent separation between tumour and normal samples based on alternative splicing is particularly evident (Supplementary Figure S1A-C) and consistent with previous studies (7,8). Some of the events reported as significantly altered by those studies overlap those highlighted in our analysis (Supplementary Figure S1B), including RPS24 alternative exon 6, more excluded in multiple cancer types (8) and considered a potential driver of hepatocellular carcinoma (51).
Nonetheless, this strong tumour-stage-independent separation may be undermining splicing alterations that discriminate the initial stages of tumour progression, i.e. changes that contribute specifically to the separation between normal and tumour stage I samples. Therefore, PCA was performed on the alternative splicing quantification and gene expression data from the 181 tumour stage I and 112 normal breast samples (Figure 2A, B and Supplementary Figure S1D, respectively). Principal component 1, the most explanatory of data variance, separates these two groups for both PCA on alternative splicing quantification and gene expression. Among the top 20 events that most contribute to that separation in principal components 1 and 2, those in genes LRRFIP2, NUMB, EXOC1, MYO18A, FBLN2 and SLMAP (Figure 2B) are described as associated with cancer (8,52–54) and are also among the 12 events that are shared with the top 20 contributors to principal components 1 and 2 separating between all tumour stages and normal samples (Supplementary Figure S1A, B and Tables S1–S2), suggesting that alternative splicing changes discriminating between tumour and normal breast samples can already be identified at the earlier stages of the disease.
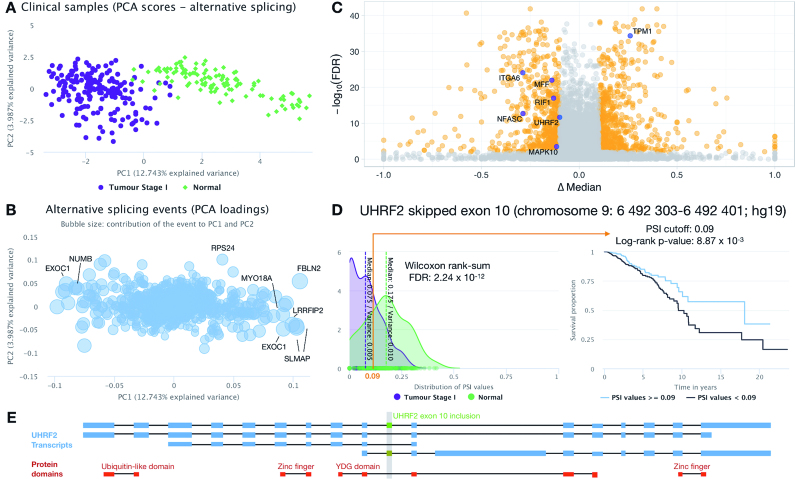
Figure 2.
Alternative splicing analyses on tumour stage I and normal breast cancer samples from TCGA. (A, B) PCA on PSI levels from tumour stage I and normal breast cancer samples; score (A) and loading (B) plots. The loading plot depicts the projection of splicing events on the two first principal components, with selected events labelled with their cognate gene symbol. The bubble size in panel B represents the relative contribution of each alternative splicing event to the selected principal components. (C) Volcano plot of differential splicing analysis performed between tumour stage I and normal breast cancer samples using the Wilcoxon rank-sum test with Benjamini–Hochberg (FDR) adjustment for multiple testing. Significantly differentially spliced events (|Δ median PSI| ≥ 0.1 and FDR ≤ 0.01) are highlighted in orange, with selected events with putative prognostic value depicted in purple. (D) One such event is an UHRF2 skipped exon, whose PSI distributions in tumour stage I and normal samples are depicted in the density plot (left), whereas its prognostic value is illustrated by the Kaplan–Meier survival curves (right; patients separated by a PSI cut-off of 0.09). (E) Protein domain disrupted by UHRF2 exon inclusion. UHRF2 transcripts in blue, UHRF2 exon 10 in green and UniProt domains in red. All images were retrieved from psichomics as is, with the exception of the gene symbol overlay in B and C, the FDR label and the arrow highlighting the PSI cut-off in D and panel E.
Amongst the alternatively spliced genes contributing to the separation between tumour stage I and normal samples, SLMAP encodes a membrane protein suggested to be a mediator of phagocytic signalling of macrophages and a putative biomarker in drug-resistant cancer cells (54). Exon 23 encodes the protein's tail anchor and thus its splicing, which our analyses find altered in breast stage I tumours, determines its subcellular localisation (55).
We also detected alterations in the splicing of exon 12 in NUMB, whose protein is crucial for cell differentiation as a key regulator of the Notch pathway. Of note, the RNA-binding protein QKI has been shown to repress NUMB exon 12 inclusion in lung cancer cells by competing with core splicing factor SF1 for binding to the branchpoint sequence, thereby repressing the Notch signalling pathway, which results in decreased cancer cell proliferation (53). Consistently, when analyzing all TCGA breast normal and tumour samples, we show NUMB exon 12 inclusion is increased in cancer and negatively correlated with QKI expression (Spearman's rho = –0.549, P-value < 0.01; Supplementary Figure S2).
Complementary analyses deemed 1285 events to be differentially spliced between tumour stage I and normal samples (|Δ median PSI| > 0.1 and FDR ≤ 0.01, Benjamini–Hochberg adjustment to Wilcoxon rank-sum test; Figure 2C and Supplementary Table S5) and therefore potential biomarkers for early breast cancer diagnosis. Some of the identified events (for instance, in FBLN2 and AP2B1), have already been described as oncogenic drivers following experimental validation (8). We also looked for alternative splicing alterations between tumour stages I and II, II and III, and III and IV (FDR ≤ 0.05) and across all tumour stages (FDR ≤ 0.05, Benjamini–Hochberg adjustment to Kruskal–Wallis rank sum test). The respective differentially spliced events are listed in Supplementary Tables S6–S9.
Among the differentially spliced events between tumour stage I and normal breast samples, several potentially associated with prognosis were identified in UHRF2, MAPK10, RIF1, MFF, TPM1, ITGA6 and NFASC, based on overall survival analyses stratified by their respective optimal PSI cut-offs (labelled in Figure 2C; survival curves in Figure 2D and Supplementary Figure S3).
Detected alterations in alternative splicing may simply reflect changes in gene expression levels. Therefore, to disentangle these two effects, differential expression analysis between tumour stage I and normal samples was also performed (Supplementary Figure S4). Alternative splicing changes seem to be independent from alterations in the expression of cognate genes for 4 of the 7 prognosis-associated splicing events (labelled points in Supplementary Figure S4).
One of such events is the alternative splicing of UHRF2 exon 10. Cell-cycle regulator UHRF2 promotes cell proliferation and inhibits the expression of tumour suppressors in breast cancer (56). psichomics reveals that higher inclusion of UHRF2 exon 10 is associated with normal samples and better prognosis (Figure 2D), and potentially disrupts UHRF2’s SRA-YDG protein domain, related to the binding affinity to epigenetic marks (Figure 2E). Hence, exon 10 inclusion may suppress UHRF2’s oncogenic role in breast cancer by impairing its activity through the induction of a truncated protein or a non-coding isoform (Figure 2E). Moreover, this hypothesis is independent from gene expression changes, as UHRF2 is not differentially expressed between tumour stage I and normal samples (|log2(fold-change)| < 1; Supplementary Figures S4 and S5A) and there is no significant difference in survival between patient groups stratified by its expression in tumour samples (log-rank P-value = 0.279; Supplementary Figure S5B).
To unveil putative regulators of the splicing of UHRF2 exon 10, its inclusion levels were correlated with the expression of each of ∼1300 RNA-binding proteins (RBPs) identified in a previous study (37) across TCGA breast, all TCGA and GTEx samples (Supplementary Table S10). We found dozens of RBPs with expression significantly correlated with UHRF2 exon 10 inclusion in both the oncological and the physiological contexts and then focused on those for whose knockdowns there are ENCODE RNA-seq data (57) (Supplementary Table S10). Among these, TUFM, whose expression is indeed consistently strongly negatively correlated with UHRF2 exon 10 inclusion levels in TCGA breast samples (Spearman's rho = –0.39, P-value < 10−42; Supplementary Figure S6A), all TCGA tumour types (Spearman's rho = –0.34, P-value < 10−253; Supplementary Figure S6B) and GTEx samples (Spearman's rho = –0.26, P-value < 10−174; Supplementary Figure S6C), is the only one whose knockdown induces a change (in this case, an increase) in UHRF2 exon 10 inclusion (Supplementary Figure S6D). Of note, TUFM (also known as EFTU) is a mitochondrial translation elongation factor (58) and there is indeed an enrichment in genes encoding for mitochondrial ribosomal proteins (e.g. MRPL27, MRPS23, MRPL13, and MRPS7) among those with expression consistently correlated with UHRF2 exon 10 inclusion. Moreover, UHRF2 deletion has been previously associated with mitochondrial dysfunction resulting in cell injury (59). We found TUFM to be overexpressed in multiple cancers (Supplementary Figure S6E) and particularly in breast tumours (Supplementary Figure S6F). Unfortunately, we found neither literature nor data (e.g. binding motifs or RIP/CLIP-seq studies) to hypothesise the direct binding of TUFM to UHRF2 pre-mRNA. Similarly, the absence of available transcriptomes for the knockdowns of other human RBPs, some known to be involved in splicing (e.g. GEMIN6 (60) and SNRNP25 (61)), prevent us from conjecturing on their putative regulatory role on UHRF2 exon 10 splicing. In any case, the reported findings result from analyses performed within psichomics and hint at a potentially overlooked TUFM-mediated exon skipping mechanism that could contribute to further elucidate the role of UHRF2 splicing in cancer.
To our knowledge, the putative prognostic value of UHRF2 exon 10 and its potential regulation by TUFM (or any other RPB topping Supplementary Table S10) have never been described and, together with the finding of both novel and previously validated cancer-specific alternative splicing alterations, demonstrate the potential of psichomics in uncovering alternative splicing-related molecular mechanisms underlying disease and physiological conditions.
Benchmarks
The time required to load, quantify and analyse data from different TCGA and GTEx cohorts was benchmarked. The breast cancer cohort contains the highest number of RNA-seq samples available in TCGA, thus being that for which it takes more time to load, quantify and analyse alternative splicing and gene expression data. Contrastingly, processed data from GTEx come bundled in files containing all tissues. Although only data from specified tissues are loaded, scanning though the large GTEx file still delays data loading. Tissues from GTEx were loaded in pairs for subsequent differential splicing analyses (Figure 3A).
Synthetic datasets for gene expression and exon-exon junction quantification of multiple sample sizes were generated, based on TCGA data distributions, to determine the time complexity of each step in psichomics as a function of the number of input samples s (Figure 3B). Assuming a constant number of genes (20 000 in the benchmark) or exon-exon junctions (200000), the time taken to load data grows quadratically with s. Gene expression normalisation and differential expression are based on commonly-used, time-efficient bioinformatics tools and the times taken for each also grow quadratically with s. Alternative splicing quantification is associated with element-wise operations on matrices of dimensions s by the number of alternative splicing events and takes a runtime approximately proportional to the square of s, for a given number of alternative splicing events (∼9000 for each benchmarked run). Finally, differential splicing is based on multiple, distinct statistical analyses of alternative splicing quantification data and grows linearly with s.
Although jSplice's (22) and DIEGO’s (23) splicing quantifications rely on junction read counts, their alternative splicing module expression and junction usage metrics, respectively, are not directly comparable with psichomics’ PSI values. To evaluate their accuracy in the absence of any known tool with the same input (junction read counts) and output metric (PSI) as psichomics, psichomics-estimated PSI values were compared to those estimated by RT-PCR and using VAST-TOOLS (18) across multiple tissue and cell line samples from human, mouse and chicken (46). VAST-TOOLS follows an analogous, and therefore more directly comparable, procedure for computing PSI values and there is a substantial overlap between the alternative splicing event annotations used by the two tools. psichomics estimates highly correlate with both others, particularly for mouse and human (Supplementary Figure S7), suggesting robustness and reproducibility in alternative splicing quantification by psichomics. Of note, the lower correlation for chicken samples is attributable to a single outlier, as its removal increases the correlation coefficients between psichomics and RT-PCR estimates (Pearson's r = 0.87, P-value < 0.01; Spearman's rho = 0.87, P-value < 0.01) and psichomics and VAST-TOOLS estimates (Pearson's r = 0.93, P-value < 0.01; Spearman's rho = 0.94, P-value < 0.01).
To assess the influence of RNA-seq read coverage on psichomics PSI estimates, different numbers of junction reads per event were simulated for different given PSI values (10000 times for each combination). Supplementary Figure S8 shows that the accuracy of PSI estimation by psichomics is expectedly sensitive to junction read coverage, particularly for intermediate PSI values, with 90% prediction intervals <0.1 for coverage higher than a few hundred reads.
Alternative splicing events annotated by TCGASpliceSeq (49), an online tool that displays pre-computed PSI values across multiple TCGA tumour types, were matched to those from psichomics based on their genomic coordinates. In total, 321 183/757 749 (42%) skipped exon, 70 837/126 725 (56%) alternative 5′ splice site and 90 940/155 799 (58%) alternative 3′ splice site events were successfully matched. When available from both programs, PSI estimates for each of the 482 960 alternative splicing events in each of the 9913 matched samples were compared between TCGASpliceSeq and psichomics, being highly correlated (N = 92 444 302; Pearson's r = 0.97, P-value < 10−15; Spearman's rho = 0.94, P-value < 10−15; Supplementary Figure S9).
DISCUSSION
Alternative splicing is a regulated molecular mechanism involved in multiple cellular processes and its dysregulation has been associated with diverse pathologies (1–3,5). The advent of next-generation sequencing technologies has allowed the investigation of transcriptomes of human biological samples to be expanded to alternative splicing. RNA-seq data, like those yielded by the GTEx and TCGA projects, are indeed playing crucial role in the improvement of our insights into the role of alternative splicing in both physiological and pathological contexts (2,3,6–8).
However, the most commonly used tools for alternative splicing analyses currently do not allow researchers to fully benefit from the wealth of pre-processed RNA-seq data made publicly available by the aforementioned projects. For instance, they lack support for estimating PSIs based on splice junction read counts. Such functionality would allow users to overcome the difficulties caused by the raw RNA-seq data from GTEx and TCGA being under controlled access and, more importantly, their processing requiring computational resources inaccessible to the majority of research labs. psichomics thus exploits pre-processed alternative splicing annotation and exon–exon junction read count data from TCGA and GTEx, two of the richest sources of molecular information on human tissues in physiological and pathological conditions, as well as recount2 and user-owned data, allowing researchers to hasten alternative splicing quantification and subsequent analyses by avoiding the time-consuming alignment of RNA-seq data to a genome or transcriptome of reference followed by splice junction detection.
Together with support for the integration of molecular and sample-associated clinical information, the group creation functionalities featured in psichomics ensure full customisability of data grouping for downstream analyses. Interesting groups to compare in TCGA, for instance, may range from the simple contrast between reformed and current smokers in lung cancer to complex combinations of gender, race, age, country and other subject attributes across multiple cancers. When survival data are available, survival analyses can be performed on samples by PSI or gene expression levels, thereby assessing the putative prognostic value of a respective molecular feature.
The integrative analysis of publicly available TCGA data by psichomics allowed us to identify multiple exons differentially spliced between breast tumour stage I and normal samples, therefore deeming them potential diagnostic biomarkers, and to assess their putative prognostic value. The output of psichomics is validated by identified alternative splicing alterations that have been previously linked to the disease, including events in RPS24, NUMB, FBLN2 and AP2B1. Previously understudied, yet intriguing, events were also identified, such as the skipping of SLMAP exon 23 and UHRF2 exon 10. These may provide novel insights into the early stages of breast cancer development. Indeed, it is of utmost importance to foster alternative splicing analyses of clinical samples as a crucial complement to more conventional research focused on total gene expression.
To ensure researchers with different skills can take the most out of psichomics, users lacking a computational background may feel more comfortable using the intuitive and more accessible graphical interface, whereas advanced users may opt for the command-line view. Should the demanding computational resources for hosting psichomics in a web server become available, we also envisage its web deployment, so that the program's latest version is publicly available on-demand with no installation required, levering the intuitive graphical interface to make alternative splicing analyses more enticing to less computationally-inclined biomedical researchers.
Notwithstanding its merits, a current limitation of psichomics is the current support only for events quantified based on exon–exon junction read counts, as not all types of alternative splicing events can be profiled using splice junction reads alone. For instance, exon–intron junction, exon body and intron body quantifications are vital to confirm intron retention and alternative 5′ and 3′ UTR events over further transcriptional variations (27,62). However, although GTEx (but neither TCGA nor recount2) readily provides intron and exon body read quantification for retrieval, none provides exon–intron junction quantification. As input data may also be user-provided, we are developing support for the missing types of events to be included in a future update.
Another limitation of psichomics is its reliance on existing alternative splicing event annotations and an on the pre-processing of RNA-seq data by third-party pipelines (as is the case for GTEx, TCGA and recount2), depriving the user of the flexibility to identify de novo alternative splicing events. However, as we detail in http://rpubs.com/nuno-agostinho/preparing-AS-annotation, when FASTQ or BAM files are accessible, psichomics supports the loading of alternative splicing annotations generated by different programs that take those files as input, namely rMATS (19), which is able to generate de novo annotations.
Using psichomics, we are able not only to identify novel exons differentially spliced between tumour stage I and normal breast samples but also to pinpoint potentially clinically relevant splicing events by embracing clinical data and evaluating their prognostic value. We expect that fellow researchers and clinicians will be able to intuitively employ psichomics to assist them in uncovering novel splicing-associated prognostic factors and therapeutic targets, as well as in advancing our understanding of how alternative splicing is regulated in physiological and disease contexts.
DATA AVAILABILITY
psichomics is an open-source R package publicly available in Bioconductor at https://bioconductor.org/packages/psichomics, along with graphical and command-line interface tutorials based on the presented case study.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Lina Gallego, Marie Bordone, Maria Teresa Maia, Mariana Ferreira, Ana Carolina Leote and Bernardo de Almeida for testing the program, contributing for its improvement, and all their suggestions on the manuscript. We thank Bárbara Caravela and Susana Vinga for discussions and implementation solutions on variance-based statistical tests. We also thank Ana Rita Grosso and André Falcão for valuable suggestions during the software's development.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Molecular Biology Organization [EMBO Installation Grant 3057 to N.L.B.-M.]; Fundação para a Ciência e a Tecnologia [FCT Investigator Starting Grant IF/00595/2014 to N.L.B.-M., PhD Studentship SFRH/BD/131312/2017 to N.S.-A., project PERSEIDS PTDC/EMS-SIS/0642/2014]; project cofunded by FEDER, via POR Lisboa 2020 — Programa Operacional Regional de Lisboa; PORTUGAL 2020; Fundação para a Ciência e a Tecnologia [LISBOA-01-0145-FEDER-007391]. Funding for open access charge: project cofunded by FEDER, via POR Lisboa 2020 — Programa Operacional Regional de Lisboa; PORTUGAL 2020; Fundação para a Ciência e a Tecnologia [LISBOA-01-0145-FEDER-007391].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kelemen O., Convertini P., Zhang Z., Wen Y., Shen M., Falaleeva M., Stamm S.. Function of alternative splicing. Gene. 2013; 514:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Paronetto M.P., Passacantilli I., Sette C.. Alternative splicing and cell survival: from tissue homeostasis to disease. Cell Death Differ. 2016; 23:1919–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B.. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008; 456:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barbosa-Morais N.L., Irimia M., Pan Q., Xiong H.Y., Gueroussov S., Lee L.J., Slobodeniuc V., Kutter C., Watt S., Colak R. et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012; 338:1587–1593. [DOI] [PubMed] [Google Scholar]
- 5. Oltean S., Bates D.O.. Hallmarks of alternative splicing in cancer. Oncogene. 2014; 33:5311–5318. [DOI] [PubMed] [Google Scholar]
- 6. Gallego-Paez L.M., Bordone M.C., Leote A.C., Saraiva-Agostinho N., Ascensão-Ferreira M., Barbosa-Morais N.L.. Alternative splicing: the pledge, the turn, and the prestige: The key role of alternative splicing in human biological systems. Hum. Genet. 2017; 136:1015–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsai Y.S., Dominguez D., Gomez S.M., Wang Z.. Transcriptome-wide identification and study of cancer-specific splicing events across multiple tumors. Oncotarget. 2015; 6:6825–6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Danan-Gotthold M., Golan-Gerstl R., Eisenberg E., Meir K., Karni R., Levanon E.Y.. Identification of recurrent regulated alternative splicing events across human solid tumors. Nucleic Acids Res. 2015; 43:5130–5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chhibber A., French C.E., Yee S.W., Gamazon E.R., Theusch E., Qin X., Webb A., Papp A.C., Wang A., Simmons C.Q. et al. Transcriptomic variation of pharmacogenes in multiple human tissues and lymphoblastoid cell lines. Pharmacogenomics J. 2017; 17:137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Climente-Gonzalez H., Porta-Pardo E., Godzik A., Eyras E.. The functional impact of alternative splicing in cancer. Cell Rep. 2017; 20:2215–2226. [DOI] [PubMed] [Google Scholar]
- 11. Tomczak K., Czerwińska P., Wiznerowicz M.. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. (Pozn.). 2015; 19:A68–A77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. The GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013; 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Collado-Torres L., Nellore A., Kammers K., Ellis S.E., Taub M.A., Hansen K.D., Jaffe A.E., Langmead B., Leek J.T.. Reproducible RNA-seq analysis using recount2. Nat. Biotechnol. 2017; 35:319–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Anczuków O., Akerman M., Cléry A., Wu J., Shen C., Shirole N.H., Raimer A., Sun S., Jensen M.A., Hua Y. et al. SRSF1-Regulated alternative splicing in breast cancer. Mol. Cell. 2015; 60:105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Emig D., Salomonis N., Baumbach J., Lengauer T., Conklin B.R., Albrecht M.. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res. 2010; 38:W755–W762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Katz Y., Wang E.T., Airoldi E.M., Burge C.B.. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 2010; 7:1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ryan M.C., Cleland J., Kim R., Wong W.C., Weinstein J.N.. SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts. Bioinformatics. 2012; 28:2385–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Irimia M., Weatheritt R.J., Ellis J.D., Parikshak N.N., Gonatopoulos-Pournatzis T., Babor M., Quesnel-Vallières M., Tapial J., Raj B., O’Hanlon D. et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014; 159:1511–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shen S., Park J.W., Lu Z.-X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y.. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alamancos G.P., Pagès A., Trincado J.L., Bellora N., Eyras E.. Leveraging transcript quantification for fast computation of alternative splicing profiles. RNA. 2015; 21:1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sterne-Weiler T., Weatheritt R.J., Best A.J., Ha K.C.H., Blencowe B.J.. Efficient and Accurate Quantitative Profiling of Alternative Splicing Patterns of Any Complexity on a Laptop. Mol. Cell. 2018; doi:10.1016/j.molcel.2018.08.018. [DOI] [PubMed] [Google Scholar]
- 22. Christinat Y., Pawłowski R., Krek W.. jSplice: a high-performance method for accurate prediction of alternative splicing events and its application to large-scale renal cancer transcriptome data. Bioinformatics. 2016; 32:2111–2119. [DOI] [PubMed] [Google Scholar]
- 23. Doose G., Bernhart S.H., Wagener R., Hoffmann S.. DIEGO: detection of differential alternative splicing using Aitchison's geometry. Bioinformatics. 2017; 34:1066–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J.. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004; 32:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lawrence M., Gentleman R., Carey V.. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics. 2009; 25:1841–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Braunschweig U., Barbosa-Morais N.L., Pan Q., Nachman E.N., Alipanahi B., Gonatopoulos-Pournatzis T., Frey B., Irimia M., Blencowe B.J.. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014; 24:1774–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ringnér M. What is principal component analysis. Nat. Biotechnol. 2008; 26:303–304. [DOI] [PubMed] [Google Scholar]
- 29. Hyvärinen A., Oja E.. Independent component analysis: algorithms and applications. Neural Netw. 2000; 13:411–430. [DOI] [PubMed] [Google Scholar]
- 30. Rich J.T., Neely J.G., Paniello R.C., Voelker C.C.J., Nussenbaum B., Wang E.W.. A practical guide to understanding Kaplan-Meier curves. Otolaryngol. Head Neck Surg. 2010; 143:331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Spruance S.L., Reid J.E., Grace M., Samore M.. Hazard ratio in clinical trials. Antimicrob. Agents Chemother. 2004; 48:2787–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Therneau T.M., Grambsch P.M.. Modeling Survival Data: Extending the Cox Model. 2000; 1st ednNY: Springer. [Google Scholar]
- 33. Kakaradov B., Xiong H.Y., Lee L.J., Jojic N., Frey B.J.. Challenges in estimating percent inclusion of alternatively spliced junctions from RNA-seq data. BMC Bioinformatics. 2012; 13(Suppl. 6):S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jia C., Hu Y., Liu Y., Li M.. Mapping Splicing Quantitative Trait Loci in RNA-Seq. Cancer Inform. 2015; 14:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Caravela B.S.R. Alternative splicing analysis for finding novel molecular signatures in renal carcinomas. Master of Science Degree in Biomedical Engineering thesis. 2015; Portugal: Instituto Superior Técnico, Universidade de Lisboa. [Google Scholar]
- 36. Ritchie M.E., Phipson B., Wu Di, Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sebestyén E., Singh B., Miñana B., Pagès A., Mateo F., Pujana M.A., Valcárcel J., Eyras E.. Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res. 2016; 26:732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yates A., Beal K., Keenan S., McLaren W., Pignatelli M., Ritchie G.R.S., Ruffier M., Taylor K., Vullo A., Flicek P.. The ensembl REST API: Ensembl data for any language. Bioinformatics. 2015; 31:143–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu C.H., Apweiler R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R. et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006; 34:D187–D191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nightingale A., Antunes R., Alpi E., Bursteinas B., Gonzales L., Liu W., Luo J., Qi G., Turner E., Martin M.. The Proteins API: accessing key integrated protein and genome information. Nucleic Acids Res. 2017; 45:W539–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roberts R.J. PubMed Central: The GenBank of the published literature. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:381–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cunningham F., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S. et al. Ensembl 2015. Nucleic Acids Res. 2015; 43:D662–D669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fishilevich S., Zimmerman S., Kohn A., Iny Stein T., Olender T., Kolker E., Safran M., Lancet D.. Genic insights from integrated human proteomics in GeneCards. Database (Oxford). 2016; 2016:baw030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. et al. Tissue-based map of the human proteome. Science. 2015; 347:1260419–1260419. [DOI] [PubMed] [Google Scholar]
- 45. Goldman M., Craft B., Swatloski T., Cline M., Morozova O., Diekhans M., Haussler D., Zhu J.. The UCSC Cancer Genomics Browser: update 2015. Nucleic Acids Res. 2015; 43:D812–D817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tapial J., Ha K.C.H., Sterne-Weiler T., Gohr A., Braunschweig U., Hermoso-Pulido A., Quesnel-Vallières M., Permanyer J., Sodaei R., Marquez Y. et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 2017; 27:1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Murray K., Müller S., Turlach B.A.. Fast and flexible methods for monotone polynomial fitting. J. Stat. Comput. Simul. 2016; 86:2946–2966. [Google Scholar]
- 48. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ryan M., Wong W.C., Brown R., Akbani R., Su X., Broom B., Melott J., Weinstein J.. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res. 2016; 44:D1018–D1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A.. Global cancer statistics, 2012. CA Cancer J. Clin. 2015; 65:87–108. [DOI] [PubMed] [Google Scholar]
- 51. Zhang H., Ye J., Weng X., Liu F., He L., Zhou D., Liu Y.. Comparative transcriptome analysis reveals that the extracellular matrix receptor interaction contributes to the venous metastases of hepatocellular carcinoma. Cancer Genet. 2015; 208:482–491. [DOI] [PubMed] [Google Scholar]
- 52. de Miguel F.J., Pajares M.J., Martínez-Terroba E., Ajona D., Morales X., Sharma R.D., Pardo F.J., Rouzaut A., Rubio A., Montuenga L.M. et al. A large-scale analysis of alternative splicing reveals a key role of QKI in lung cancer. Mol. Oncol. 2016; 10:1437–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zong F.-Y., Fu X., Wei W.-J., Luo Y.-G., Heiner M., Cao L.-J., Fang Z., Fang R., Lu D., Ji H. et al. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 2014; 10:e1004289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen K., Yang X., Wu L., Yu M., Li X., Li N., Wang S., Li G.. Pinellia pedatisecta agglutinin targets drug resistant K562/ADR leukemia cells through binding with sarcolemmal membrane associated protein and enhancing macrophage phagocytosis. PLoS ONE. 2013; 8:e74363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Byers J.T., Guzzo R.M., Salih M., Tuana B.S.. Hydrophobic profiles of the tail anchors in SLMAP dictate subcellular targeting. BMC Cell Biol. 2009; 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu J., Liu S., Liu G., Dombkowski A., Abrams J., Martin-Trevino R., Wicha M.S., Ethier S.P., Yang Z.-Q.. Identification and functional analysis of 9p24 amplified genes in human breast cancer. Oncogene. 2012; 31:333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Antonicka H., Sasarman F., Kennaway N.G., Shoubridge E.A.. The molecular basis for tissue specificity of the oxidative phosphorylation deficiencies in patients with mutations in the mitochondrial translation factor EFG1. Hum. Mol. Genet. 2006; 15:1835–1846. [DOI] [PubMed] [Google Scholar]
- 59. Chen X.-R., Sun S.-C., Teng S.-W., Li L., Bie Y.-F., Yu H., Li D.-L., Chen Z.-Y., Wang Y.. Uhrf2 deletion impairs the formation of hippocampus-dependent memory by changing the structure of the dentate gyrus. Brain Struct. Funct. 2018; 223:609–618. [DOI] [PubMed] [Google Scholar]
- 60. Pellizzoni L., Baccon J., Rappsilber J., Mann M., Dreyfuss G.. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J. Biol. Chem. 2002; 277:7540–7545. [DOI] [PubMed] [Google Scholar]
- 61. Will C.L., Schneider C., Hossbach M., Urlaub H., Rauhut R., Elbashir S., Tuschl T., Lührmann R.. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA. 2004; 10:929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dvinge H., Bradley R.K.. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015; 7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
psichomics is an open-source R package publicly available in Bioconductor at https://bioconductor.org/packages/psichomics, along with graphical and command-line interface tutorials based on the presented case study.