Abstract
Cell membranes, despite providing a barrier to protect intracellular constituents, require selective gating for influx of important metabolites including ions, sugars, amino acids, neurotransmitters and efflux of toxins and metabolic end-products. The machinery involved in carrying out this gating process comprises of integral membrane proteins that use ionic electrochemical gradients or ATP hydrolysis, to drive concentrative uptake or efflux. The mechanism through which ion-coupled transporters function is referred to as alternating-access. In the recent past, discrete modes of alternating-access have been described with the elucidation of new transporter structures and their snapshots in altered conformational states. Despite X-ray structures being the primary sources of mechanistic information, other biophysical methods provide information related to the structural dynamics of these transporters. Methods including EPR and smFRET, have extensively helped validate or clarify ion-coupled transport mechanisms, in a near-native environment. This review seeks to highlight the mechanistic details of ion-coupled transport and delve into the biophysical tools and methods that help in understanding these fascinating molecules.
Keywords: Secondary active transport, ion-coupled transport, alternating-access, uniport, symport, antiport
1. Introduction
All cells and cellular organelles are protected by one or more hydrophobic membrane bilayers that segregate the internal constituents from the external environment. This phospholipid bilayer serves as a barrier to most compounds that seek to enter or exit the cell. Cells, however, require a controlled exchange of material including ions, metabolites or metabolic end-products and signaling species to maintain normal physiological processes. Consequently, most cellular and organellar membranes have integral membrane proteins in the form of ion-channels and transporters to facilitate this regulated movement of small molecules, vital for the survival of cells (von Heijne 2006; Gouaux and Mackinnon 2005). While most ion-channels serve as pores for the movement of ions in response to a stimulus, transporters perform the task of gates that move substrates in a controlled fashion into or out of cells (Gouaux and Mackinnon 2005).
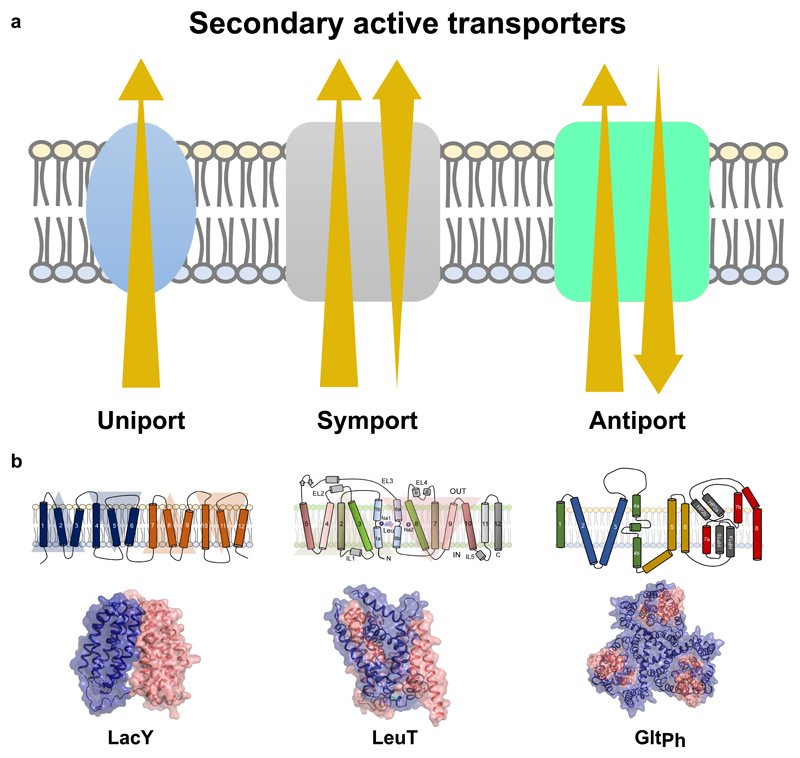
The direction of substrate movement can be in the direction of the concentration gradient or against it (Yan 2015). Transporters that facilitate movement of substrate along the concentration gradient are referred to as uniporters (Yan 2015). Transporters that move substrate(s) against their concentration gradients, couple the process to either ATP hydrolysis or to ionic-electrochemical gradients (Vinothkumar and Henderson 2010). Primary active transporters use ATP hydrolysis to couple substrate movement, whereas most of the secondary active transporters employ ion-coupling to achieve transport. The direction of ion-flow can occur along the direction of substrate movement in case of symporters or against it as observed in antiporters (Fig. 1a)(Shi 2013).
Figure 1.
a, Modes of secondary active transport including uniport (facilitated diffusion), symport and antiport. Direction of ion/substrate flux indicated by arrows. b, Membrane topology and X-ray structures of LacY, LeuT and GltPh. Topologies represent the broad mechanistic classification of transporters.
Ion-coupled transporters in humans are also referred to as solute carriers (SLCs) that form the second largest group of membrane proteins, after GPCRs (Cesar-Razquin et al. 2015). Over 450 SLCs are known in humans and a large subset of them are drug targets (Cesar-Razquin et al. 2015). Together, SLCs are involved in shuttling ions, sugars, neurotransmitters, amino acids/peptides, lipids, and drugs into or out of cells and organelles (Shi 2013).The transporter classification database (TCDB) assigns an enzyme classification style numbering of transport proteins and most of the ion-coupled “porters” are classified under the section 2.A (Saier et al. 2006). Uniporters have very similar structural scaffolds to symporters and antiporters and are generally considered to be secondary active transporters without ion-coupling (Yan 2015). Despite their extensive presence and obvious significance in physiology, secondary active transporters/ solute carriers remain some of the least-studied among integral membrane protein families, prompting calls for enhanced research on these molecules (Cesar-Razquin et al. 2015).
Although quite extensive in their number and substrate-specificity, ion-coupled transporters fall into a small set of structural scaffolds with fewer mechanistic discrepancies (Drew and Boudker 2016). Most secondary active transporters have an inherent symmetry within them and their structural organization reflects this symmetry within the helical repeats (Fig. 1b)(Drew and Boudker 2016). The presence of symmetry allows the transporters to undergo alternating-access, the primary mechanism through which substrates are driven across the membrane bilayer (Forrest et al. 2008; Jardetzky 1966). This review focuses on the mechanistic underpinnings of secondary active transport and discusses recent advances in structural and biophysical methods that have immensely aided in deciphering the structure, dynamics and functional roles of ion-coupled transporters. Although we primarily focus on transporters as a specific case, the methods described here are generally applicable to all integral membrane proteins.
2. Alternating-access
The concept of substrate movement across the membrane was envisaged by Peter Mitchell who proposed a carrier hypothesis for substrate translocation in response to a signal, in the form of phosphorylation. The process of translocation was akin to an enzyme catalyzing a biochemical reaction whereas in case of a transporter, it involves conformational changes to catalyze movement across the membrane (Mitchell 1957).
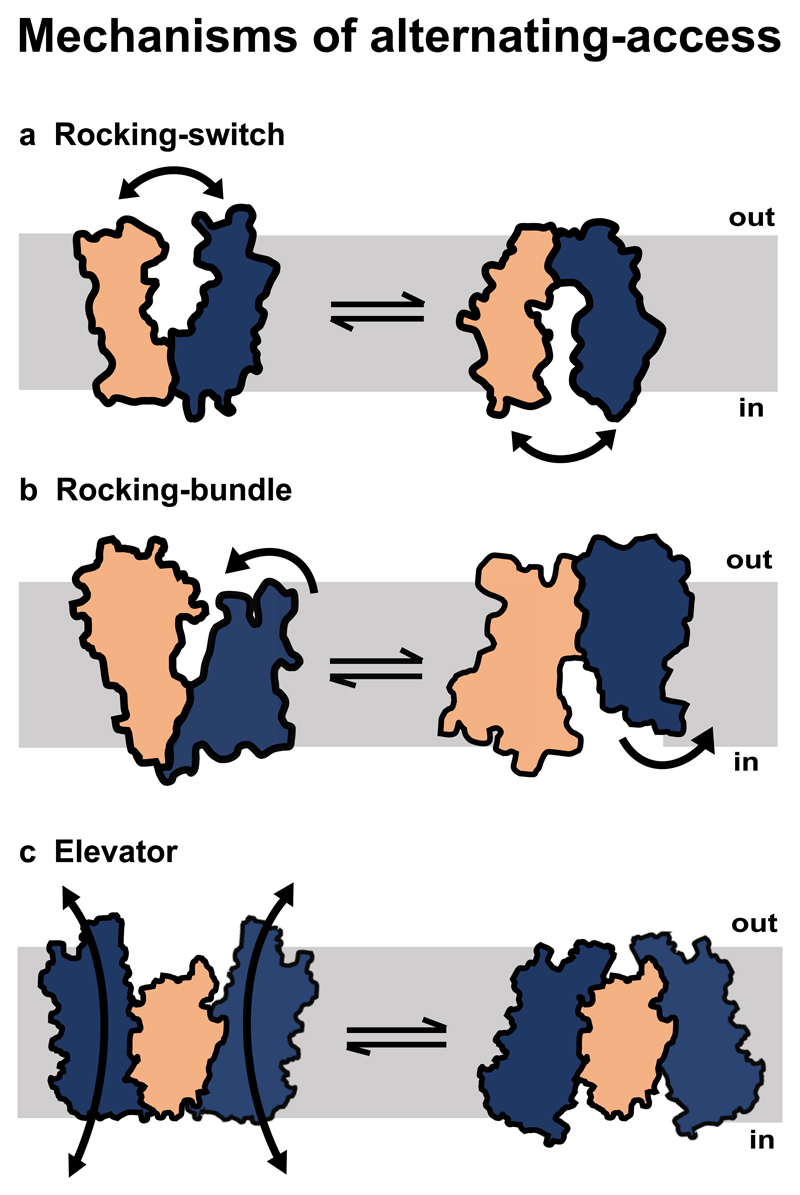
Jardetsky in 1966 put forth a defined role for carriers wherein he proposed that transporters would undergo conformational changes that alternately expose the binding site to either side of the membrane, but never at the same time (Jardetzky 1966). These conformational changes, that occur in response to substrate binding on one side of the compartment followed by its release on the other side, constitutes the alternating-access mechanism (Fig. 2).
Figure 2.
Types of alternating-access mechanisms. Orange and blue colors denote the transporter domains in a, Rocking-switch model, b, Rocking-bundle model. c, In the elevator model, orange region represents the static oligomerization domain whereas the blue regions represent the transport domains.
In the last two decades, high resolution structures of transporters across different families have corroborated the presence of symmetrical halves, that perform alternating-access, albeit with variations to the primary mechanism. Exceptions to alternating-access do occur especially in the chloride channels (CLCs) which transport chloride ions and protons, but given the minimal structural transitions that happen, resemble channel like states (Feng et al. 2010). Despite implying the presence of only two states in alternating-access namely, open-to-in (Oin) and open-to-out (Oout), structures of numerous transporters reveal the presence of asymmetric occluded states including outward-occluded (Oocc) or inward-occluded (Iocc)(Drew and Boudker 2016).
In the recent past, transport was observed to occur in three independent modes while remaining within the bounds of alternating-access. Each mode correlates well with the structural fold of individual molecules (Fig. 2).These are the “rocking-switch” mechanism observed primarily in the major facilitator superfamily (MFS)(Kumar et al. 2014), the multi-drug and toxin extrusion (MATE) family (Radchenko et al. 2015), the SMR (small multi-drug resistance)(Schuldiner 2009) family and the sugars will eventually be exported transporters (SWEETs)(Fig. 2a)(Feng and Frommer 2015). Incidentally, the MFS comprises of the largest set of transporters among all secondary active transporters. The second mechanism is the “rocking-bundle” mechanism prevalent in the proteins with the amino acid, polyamine organo-cation superfamily (ApcT) that includes well-studied members like LeuT(Singh 2008) and Mhp1(Fig. 2b)(Krishnamurthy et al. 2009). A third mechanism common to proteins of the SLC1 family homologues comprising of divalent anion: Na+ coupled transporters is the “elevator” mechanism (Fig. 2c) (Boudker and Verdon 2010).
Proton-coupled transport is also observed in the resistance-nodulation-division (RND) family of proteins that comprise of the AcrA/AcrB/TolC complex in Gram –ve bacteria. The ion-coupled transport, in AcrB, is driven by an asymmetric trimer mechanism by coupling proton gradients to antibacterial efflux. The mechanism is akin to the rotor mechanism of the F1F0 ATPase system and has been reviewed extensively, elsewhere (Pos 2009).
2.1. Rocking-switch mechanism
This is the most prevalent mechanism observed across a wide array of the aforementioned families (Table 1) (Fig. 2a). The mechanism involves conformational changes occurring upon substrate binding on one side of the membrane, triggering a symmetric rigid body reorientation of helices. This allows the bound substrate to gain access to solvent from the cytosolic compartment and subsequently get released. The SWEET transporters represent a minimal structural organization wherein two three-helix bundles (3+3) play a role in the transport process (Tao et al. 2015). The SWEET transporter structure revealed a near-symmetric occluded state which is generally not observed in transporters with greater number of helical domains. An interesting case of rocking switch mechanism is the small multidrug resistance family member EmrE, a four TM helix protein that dimerizes to perform drug efflux (Schuldiner 2009). To add further credence to this, LacY a prototypical member of the MFS, was observed to retain uptake function despite splitting it into two halves (Bibi and Kaback 1990). The ability of these transporters to function as symmetric halves in trans indicates the possibility of gene duplication events resulting in formation of the multi-helix transporters as observed in MFS and MATE families (Reddy et al. 2012).
Table 1. List of transporters with unique structures.
| Mechanism. Clamp and Switch | ||||
|---|---|---|---|---|
| Major Facilitator Superfamily | ||||
| PDB. id | Transporter (TCDB) | Source | Function | Conformations |
| 1PV6, (4OAA) | LacY | E.coli | Lactose: H+ symport | Inward open, outward open |
| 4GBY, (4JA4), (4JA3) | XylE | E.coli | Xylose: H+ symport | Outward open-partly occluded, Inward open, partially occluded inward open |
| 3O7P | FucP | E.coli | Fucose:H+ symport | Outward open |
| 1PW4 | GlpT | E.coli | Glycerol-3-phosphate antiport | Inward open |
| 4APS, (5OXP) | PepTst | S.thermophilus | Peptide:H+ symport | Inward open, occluded |
| 4M64 | MelB | Salmonella typhymurium | Na+: melibiose symport | Outward open- partially occluded, outward open inactive |
| 4LDS | GlcPse | S.epidermidis | Glucose: H+ Symporter | Inward open |
| 4IU8, (4IU9) | NarU | E.coli | Nitrate: nitrite symport or antiport | Inward open-partially occluded, partially inward open |
| 4JR9, (4U4W) | NarK | E.coli | Nitrate: Nitrite antiport | Inward open, Inward open-occluded |
| 5A2N | NRT1.1 | Arabdosis thaliana | Nitrate: H+ symport | Inward open |
| 4J05 | PiPT | Serendipita indica | Phosphate: H+ symport | Inward open-occluded |
| 4IKV | GkPOT | Geobacillus kaustophilus | Peptide: H+ symport | Inward open |
| 2XUT | PepTso | Shewanella oneidensis | Peptide: H+ symport | Inward open-partially occluded |
| 4Q65 | YbgH | E.coli | Peptide: H+ symport | Inward open |
| 4W6V | YePEPT | Yersinia enterocolitica | Peptide: H+ symport | Inward open |
| 5AYO, (5AYM) | BbFPN | Bdellovibrio bacteriovorus | Divalent metal ion uniport | Inward open Outward open |
| 4zp0 | MdfA | E.coli | Multudrug: H+ antiport | Inward open |
| 2GFP | EmrD | E.coli | Multudrug: H+ antiport | Inward open-occluded |
| 3WDO | YajR | E.coli | Inward open | |
| 4PYP | Glut1 | H.sapiens | Glucose uniport | Inward open |
| 4ZWC | Glut3 | H.sapiens | Glucose uniport | Outward open |
| 4YB9, (4YBQ) | Glut5 |
B. taurus Rattus norvegicus |
Glucose uniport | Inward open Outward open |
| MATE family transporters | ||||
| 3MKT | NorM | Vibrio cholerae | Multidrug: Na+/H+ antiporter | Outward open |
| 4LZ6 | DinF | B. halodurans | Multidrug: Na+/H+ antiporter | Outward open |
| 3VVN | PfMATE | Pyrococcus furiosus | Multidrug: Na+/H+ antiporter | Outward open |
| 5YCK | CasMATE | Camelina sativa | Multidrug: Na+/H+ antiporter | Outward open |
| SWEETs/semiSWEETs | ||||
| 5CTH | OsSWEET2B | Oryza sativa | Na+: solute symport | Inward open |
| Mechanism 2. Rocking Bundle | ||||
| LeuT Fold proteins (ApcT superfamily) | ||||
| 2A65 | LeuT | Aquifex aoelicus | Amino acid: Na+ symport | Occluded |
| 4US3 | MhsT | Bacillus halodurans | Na+: L-Tryptophan symport | Inward open-occluded |
| 3GIA | ApcT | M. janaschii | H+: amino acid symport | Inward open |
| 3DH4 | vSGLT | V. cholerae | Na+: sugar symport | |
| 4M48 | dDAT | D. melanogaster | DA: Na+/Cl- symport | Outward open |
| 5I6Z | hSERT | H.sapiens | 5HT: Na+/Cl- symport | Outward open |
| 6C08 | SLC38 | D.rerio | Amino acid: Na+ symport | Inward open |
| 3HFX | CaiT | E.coli | Carnitine: butyrobetaine antiport | Inward open |
| 3WIT | BetP | C.glutaricum | Betaine: Na+ symport | |
| 2JLN, (2JLO) | MHP1 | Microbacterium liquifaciens | Hydantoin: Na+ symport | Outward open, Occluded |
| 3NCY | AdiC | S.typhimurium | Arginine/Agmatine antiport | Outward open |
| Mechanism 3. Elevator | ||||
| 1ZCD | NhaA | E.coli | Na+:H+ antiport | Inward open |
| 5BZ2 | NapA | Thermus thermophilus | Na+:H+ antiport | Inward open |
| 1XFH | GltPh | Pyrococcus horikoshii | Dicarboxylate: Na+ symport | Outward open |
| 5LLM | EAAT1 | H.sapiens | Glutamate: Na+ symport | Outward open |
| 5X9R | CitS | K.pneumoniae | Citrate: Na+ symport | Outward open |
| 5E9S | GltTK | Thermococcus kodakarensis | Dicarboxylate: Na+ symport | |
| 4F35 | VcINDY | Vibrio cholerae | Divalent anion: Na+ symport | Inward open |
| 4R0C | YdaH (AbgT family) | Aclanivorax borkumensis | Drug efflux | Inward open |
The MFS and the MATE transporters have six helix bundles that are symmetrically arranged (6+6)(Yan 2015). Each of the six-helix bundle, has two three-helix bundles that are symmetrically related (Fig. 1b). This results in helices 1, 4, 7, 10 being symmetric equivalents and in proximity to the substrate binding site; helices 2, 5, 8, 11 are long and act as rocker helices and partly line the binding site followed by helices 3, 6, 9 and 12 that make the outer ring, acting as support helices and interacting with the membrane environment (Heng et al. 2015). In some instances, where two additional helices are observed, as in the case of some peptide-oligopeptide transporters (POTs)(Newstead et al. 2011) and DHA2 members of MFS, the additional helices are positioned in the intracellular loop that links the two six helix bundles forming a 6+2+6 arrangement (Reddy et al. 2012). The structures of a few members of MFS have been determined in different conformational states but no snapshots of a single molecule have been captured in all the states of Oout, Oocc, Iocc or Oin (Table 1). We, however, can piece together the transport cycle using snapshots of different molecules in alternate conformational states. For instance, LacY was captured in Oout and Oin conformations (Abramson et al. 2003; Kumar et al. 2014) while XylE was observed in Oocc and Iocc states (Sun et al. 2012). Structures of mammalian GluTs, involved in sugar uniport were also captured in Oout and Oin states (Deng et al. 2014; Nomura et al. 2015). Rocking-switch mode of transport is well observed in case of LacY wherein the two symmetrical halves undergo ~30° movement to alternate from an Oin state to Oout state (Kumar et al. 2014).
In the case of mammalian GluTs, in comparison with occluded state structures of XylE, it was observed that the gating occurs through helices bending to create access to the binding site (Nomura et al. 2015; Sun et al. 2012). Hinge movements in helices TM1, TM7 open the extracellular gate and helices TM4, TM10 allow opening of intracellular gate (Nomura et al. 2015). In most cases, a series of electrostatic interactions are formed and broken during the gating process to allow the transport of the substrate. Given the incidence of hinge movements of helices facilitating transport, as compared to large rigid body motions, the MFS transporters resemble a “gated-pore” with the “rocking-switch” mechanism being revised to a “clamp-and-switch” mechanism (Quistgaard et al. 2016).
2.2. Rocking-bundle mechanism
Members of the amino acid, polyamine, organo-cation transporters (ApcT) superfamily conform to this transport mechanism (Shi 2013). The mechanism is a result of a majority of the TMs, in the transporter, acting as a scaffold and a pair of symmetrical discontinuous helices moving inwards or outwards, to open or close the internal or external gates. While molecules having the rocking-switch mechanism resemble a “V” -shaped architecture, transporters with a rocking-bundle movement have a “K” -shaped architecture (Fig. 2b). This mechanism is best illustrated in case of LeuT, a bacterial amino acid transporter homologous to mammalian neurotransmitter transporters (Table 1)(Krishnamurthy et al. 2009; Krishnamurthy and Gouaux 2012). LeuT and eukaryotic neurotransmitter transporters have a pseudo two-fold symmetry between helices 1-5 and 6-10 with two additional helices TM11 and 12 that are outside these symmetric halves (Fig. 1b)(Yamashita et al. 2005). While a bulk of the helices along TM3 and TM8 form the scaffold domain, helices 1 and 6 which are discontinuous, form the gating helices. TM1b and TM6a together form the extracellular gate whereas TM1a and 6b form the cytosolic gates. The discontinuous region of the gating helices, at the core of the transporter, for the substrate and ion-binding sites. LeuT was solved at a high resolution of 1.9Å with a leucine bound in the binding pocket along with two Na+ ions that are co-transported with the substrate (Yamashita et al. 2005). The conformation of this state resembled an Oocc conformation with solvent having access to the vestibule, but the binding site is secluded from water due to the F253 residue acting as barrier, preventing solvent access to the binding pocket. Co-crystallization of a bulkier amino acid, tryptophan, allowed the outward movement of TM1b (~9°) and TM6a (5.5°)with a hinge-like movement observed in TM2(~8°)(Singh et al. 2008). In the Oin state of the transporter TM1a and TM6b move outward to provide access to the substrate binding pocket (Krishnamurthy and Gouaux 2012). TM1a in particular swings out nearly 45° into the membrane environment to facilitate this access (Krishnamurthy and Gouaux 2012). LeuT was never observed in an Iocc conformational state, although other homologues like ApcT (Shaffer et al. 2009) and MhsT (Malinauskaite et al. 2014) have been observed in this conformation. The gating movements in this family are also associated with the break and formation of salt bridges in the vestibule, much like the rocking-switch mechanism. The movements of the outward gate of LeuT are very similar to movements of dDAT gating helices in its Oocc state, suggesting a conservation of this transport mechanism within this superfamily (K. H. Wang et al. 2015).
2.3. Elevator mechanism
The elevator mechanism is a specialized form of alternating-access that occurs primarily in oligomeric transporters involved in divalent anion:Na+ symporters that have structural similarity to Pyrococcus horikoshii glutamate transporter GltPh (Yernool et al. 2004). The transporters are oligomeric with the elevator mechanism observed in dimeric and/or trimeric molecules (Drew and Boudker 2016). The molecules can distinctly be separated into the transport domain and oligomerization domains (Fig. 2c). While the oligomerization domain remains fixed in its position, the transport domains translate perpendicular to the membrane plane allowing solvent-access to the substrate binding pocket (Reyes et al. 2009). The prototypical member of this set of proteins is GltPh. Structures with similar transport mechanism have recently been elucidated, including EAAT1 (Canul-Tec et al. 2017), VcINDY (Mancusso et al. 2012), CitS (Wohlert et al. 2015) and AbgT (Table 1) (Bolla et al. 2015). Most of these transporters have distinct helical repeats within the oligomerization domain and in the transporter domain (Boudker and Verdon 2010). The distinguishing feature for this fold of proteins is the presence of helical hairpin loops HP1 and HP2 that ensconce the substrate binding site and Na+ binding sites in the vicinity, in close proximity to the discontinuous helix TM7. GltPh in its Oout state resembles a chalice with a concave vestibule formed by the trimer (Yernool et al. 2004). Upon substrate binding, the site between HP1 and HP2 loops moves perpendicular to the plane of the membrane by nearly 19Å and correlated with an 18° twist of the transport domain (Reyes et al. 2009). The vertical movements were also characterized in VcINDY using mutagenesis and in GltPh using smFRET studies. A recent structure of EAAT1 in outward-open state is also likely to follow the same mechanism (Canul-Tec et al. 2017).
Another group of transporters that have the elevator mechanism but a different structural organization are the Na+:H+ antiporters (Lee et al. 2014). These dimeric transporters have transporter domains without hairpin loops but have discontinuous helices that are crossed over to form the substrate binding site(Boudker and Verdon 2010). The proposition of elevator mechanism, in NHA members, was controversial, although in recent past this mechanism is increasingly accepted.
A large fraction of mechanistic information has come from crystallographic studies of transporters in different conformational states making X-ray crystallography the primary tool for understanding transporter structure and function. However, numerous tools help prepare constructs that are suitable for crystallographic studies and retain transport activity. Additional biophysical methods, useful to study transporter dynamics in solution are included in the later sections of the review.
3. Tools to aid in structure determination of transporters
3.1. Pre-crystallization tools
3.1.1. Fluorescence detection size exclusion chromatography (FSEC)
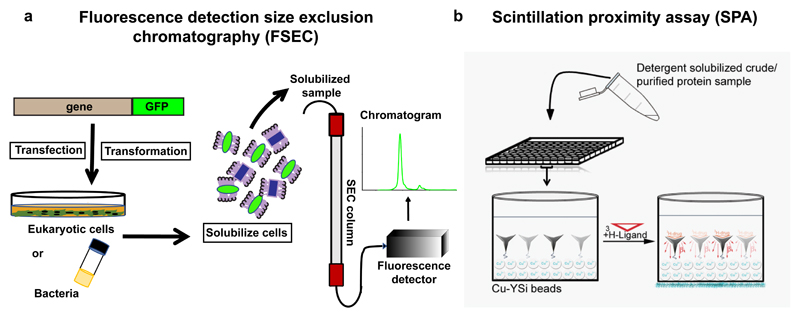
Gouaux and colleagues pioneered the use of tagging a GFP molecule to membrane proteins and monitoring the crude membrane extract on a size exclusion column linked to a fluorimeter (Fig. 3a)(Kawate and Gouaux 2006). The result was the ability to monitor the profile of the membrane protein without using purified material. The profile of the GFP-linked membrane protein provides information on the homogeneity, expression level and aggregation propensity in the background of whole cell extract or solubilized membranes in particular detergent. This simple, yet elegant, screening tool has helped immensely in monitoring well behaved orthologues, optimizing non-ionic detergents, stability measurements and tracking protein behavior upon purification. In recent variations of FSEC, the method was put to use to measure stability of membrane proteins in different detergent/lipid conditions (Hattori et al. 2012) and also to measure formation of heteromeric complexes (Morales-Perez et al. 2016). This method is now a standard tool for any lab working on structures of novel transporter or membrane protein structures.
Figure 3.
a, Schematic representation of fluorescence-detection size exclusion chromatography (FSEC) technique using fluorescent protein-tagged membrane proteins, b, Principle of scintillation proximity assay (SPA) done with Cu-YSi scintillant beads and a radiolabeled inhibitor.
3.1.2. Functional assays to monitor inhibitor binding
Limitations of FSEC lie in its inability to monitor the function of the transporter. Inhibitor binding can effectively be measured in detergent solubilized material using radiolabeled drugs/inhibitors. A binding technique that rapidly provides dissociation constants is the scintillation proximity assay (SPA), that employs copper or biotin coated beads that can interact with either a His-tag or a Strep-II tag (Quick and Javitch 2007). A radiolabeled inhibitor that can bind the transporter with high affinity, emits energy on binding the immobilized transporter, leading to scintillation of the beads, resulting in luminescence that can be quantified as binding (Fig. 3b). The method has extensively been applied to transporters including LeuT (Krishnamurthy et al. 2009), dDAT (Penmatsa et al. 2013) and hSERT (Coleman et al. 2016).In case of the latter two, it was instrumental in optimizing and isolating thermostable mutants (described below). SPA also works for protein reconstituted in nanodiscs, as observed with LeuT reconstituted into nanodiscs (Nasr and Singh 2014), thereby allowing validation of samples meant for cryoEM data collection. FSEC and SPA are powerful complementary tools to perform pre-crystallization screening.
3.1.3. Construct engineering through mutagenesis
Modification of membrane transporters either through single mutants or large-scale mutagenesis has proven to be an effective strategy towards enhancing the stability and crystallization propensity. This was first reported by James Bowie and colleagues for diacylglycerol kinase (DGK), where twenty out 121 residues in a stretch of sequence, were mutated to cysteines. The experiment yielded two cysteine substitutions with greater thermostability than the native DGK. A combination of the two mutants yielded a significantly thermostabilized molecule (Lau et al. 1999). This strategy has since been expanded and employed through alanine or leucine scanning mutagenesis, particularly for the stabilization and structure elucidations of GPCRs (Serrano-Vega et al. 2008) and other integral membrane proteins, including transporters (Green et al. 2015). Examples of thermostabilized GPCRs include β1 adrenergic receptors, neurotensin receptor and adenosine receptor (Vaidehi et al. 2016; Serrano-Vega et al. 2008). The strategy involved creating single alanine mutants through the length of the receptor and identifying mutants that can retain inhibitor binding activity at high temperatures compared to the wild-type constructs. This strategy allowed identification of a small subset of mutants that have a thermostabilizing effect on the receptor and eventually aid in crystallizing them. A similar strategy was employed for the eukaryotic neurotransmitter transporters dDAT and hSERT (Penmatsa et al. 2013; Coleman et al. 2016). Briefly, a subset of the entire sequence, primarily in the TM regions were individually mutated to alanine, leucine and phenylalanine resulting in a single mutant library. Individual mutants were then tested for their ability to retain binding towards an inhibitor, either 3H-nisoxetine (dDAT) or 3H-paroxetine (hSERT), when subjected to high temperatures. Mutants that have a consistently improved binding activity were pooled together, resulting in an additive effect, culminating in thermostabilized constructs. Thermostabilized constructs of dDAT and hSERT retained binding activity at very high temperatures (~60-70°C). Interestingly, in both studies, thermostabilization led to a complete loss of transport activity resulting in a conformation-locked transporter, stabilized in an outward-open inhibitor-bound state (Penmatsa et al. 2013; Coleman et al. 2016).
Besides large-scale scanning mutagenesis, it is quite common to use single or double mutants, towards the purpose of crystallization studies. For instance, LacY, originally determined in the cytosol-open state, carried a single mutation C154G (Abramson et al. 2003). Subsequently, a periplasmic open state of the transporter was created by a combination of G46W/G262W (Kumar et al. 2014). Similarly, a multi-drug efflux pump MdfA that belongs to DHA1 of MFS, could be crystallized using a single mutant Q131R (Heng et al. 2015). Similarly, in GluT1 a combination of N45T and E329Q led to improved crystals of the transporter (Sun et al. 2012). Crystallization of altered conformational states was also made possible by stitching together, mobile domains in transporters. A classic case is the disulfide crosslinking done with GltPh to stabilize the Oin conformation. Mutations K55C(TM2) in the scaffold domain and A364C(HP2) in the transporter domain allowed the formation of a disulfide cross-link that locked the elevator in GltPh, in a cytosol facing conformation (Reyes et al. 2009).
It is generally observed in projects handling crystallization of recalcitrant molecules, that combinations of mutants, stabilizing certain conformations, coupled with the use of crystallization chaperones, significantly enhance the chances of successful crystallization (Hunte and Michel 2002).
3.1.4. Functional analysis for transport activity
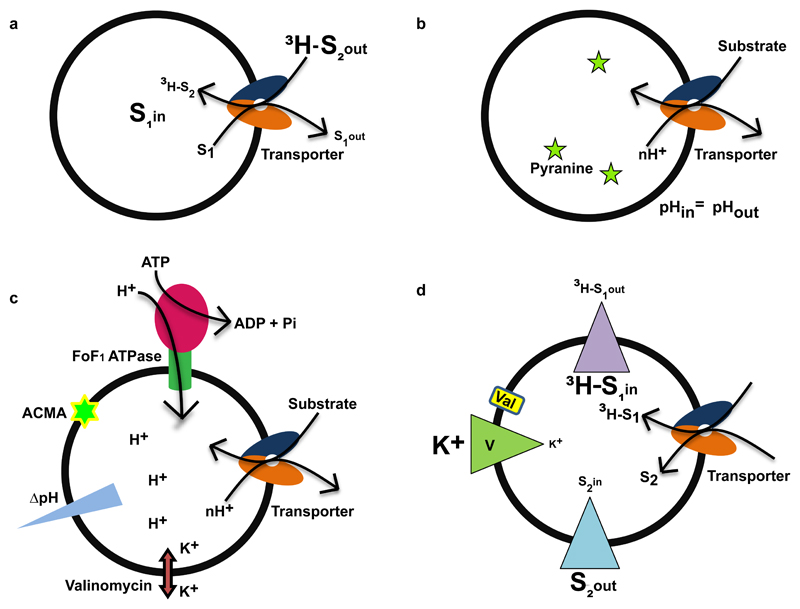
Substrate transport by reconstituted membrane proteins is an evidence for their ability to function independently of other cellular factors. Earlier studies involved whole cell based approaches to study the substrate transport process; conversely, there are some disadvantages as cells have active metabolic mechanisms which can interfere through physical interactions or degradative processes. Transport assays using crude membrane vesicles like spheroplasts or everted vesicles overcome certain drawbacks of doing transport assays in whole cells, while keeping the transporter in its native environment (Futai 1974). However, to understand the process of transport with greater clarity and accuracy, we need to see if the transporter can work in isolation. Reconstitution of integral membrane transporters in an artificial membrane, can clarify whether the molecule maintains its functional integrity, in biochemical isolation. Transporters can function as electrogenic or electroneutral, depending on the substrate charge and the number of co-transported ions, during the transport cycle. Based on the characteristic property of transport, different kinds of proteoliposomal transport assays can be devised to monitor activity (Fig. 4).
Figure 4.
Modes of proteoliposome based transport assays. a, Substrate trapping method used for an antiporter. The unlabelled substrate (S1) is trapped inside the liposome and the radiolabeled substrate (3H-S2) is added externally. The accumulation of radioactive material inside the vesicle is measured. b, Uniform pH is maintained across the membrane, transport of substrate added from outside causes alteration in pH inside the vesicle and detected using altered pyranine fluorescence. c, F1F0 ATPase co-reconstitution based approach is used for proton driven antiporters. Addition of ATP in presence of ATPase creates the required pH gradient needed for transport of the substrate. The change in proton concentration due to active transport process is detected by pH sensitive dye ACMA (9-amino 6-chloro 2-methoxyacridine). Valinomycin in presence of potassium ion helps to keep membrane potential constant. d, Measurement of reversal potential of the membrane at equilibrium is used for electrogenic transporters; radiolabelled substrate (3H-S1) is trapped in high concentration inside the liposome and unlabelled substrate is added in high concentration outside (S2). The transport process is initiated by creating different potentials across the membrane using valinomycin in presence of potassium ions. The alteration of radioactive material is measured along with the reversal potential of the membrane.
For instance, the activity of GadC a GABA/glutamate antiporter from E.coli, was evaluated using a substrate trapping approach wherein GABA was trapped inside the reconstituted vesicles and radiolabeled glutamate was added externally to monitor radiolabel accumulation in the vesicle (Fig. 4a)(Ma et al. 2013). Another approach to monitor H+-driven antiport is to use the transport of substrate, accompanied with the efflux of protons from the proteoliposome. The changes in internal pH are monitored using a ratiometric pH sensitive fluorophore, pyranine that is trapped inside the liposomes (Fig. 4b)(Damiano et al. 1984). A recent study, employed this method to monitor EmrE’s ability to transport guanidinium (Kermani et al. 2018). In an alternate setup, the Na+/H+ antiporter NapA was evaluated through co-reconstitution of F1F0 ATP synthase along with NapA. Addition of external ATP generated the required proton gradient for observing NapA transport and the process was monitored using a pH sensitive dye 9-amino 6-chloro 2-methoxyacridine (ACMA) (Fig. 4c)(Lee et al. 2014). Recently, measuring ion: substrate stoichiometry was made easier with a method developed by Mindell and colleagues wherein, the transporter reversal potential (Erev) is calculated using known values of Δψ and ion-gradients across the membrane (Table 2) (Fig. 4d). The method was applied to measure transport stoichiometry of VcINDY, a bacterial Na+ -coupled succinate transporter and further validated by confirming the known coupling stoichiometry of vSGLT, a bacterial sugar transporter (Mulligan et al. 2016).
Table 2. Transport equation at Erev for transporters with different substrate: ion stoichiometries.
| Process | Transport principle | Stoichiometry |
|---|---|---|
| Uniport | mSain → mSaout | |
| Symport | mSain + nIbin↔mSaout + nIbout |
When* a = -2, b =1 When a = 1, b = 2 When a =0, b = 1 |
| Antiport | mSaout + nIbin↔mSain + nIbout |
When a = -2, b = 2 When a = 1, b = 2 |
R is the universal gas constant, T is the temperature (in °K), F is the Faraday constant, a and b are the substrate and ion charges, respectively, Δψis the voltage difference across the membrane, m and n denotes the number of substrate and ions respectively. I, S denote ion and substrate respectively. At equilibrium with conversion to the base 10 log, and approximating RT/F as 60mV.
3.2. Crystallizations methods
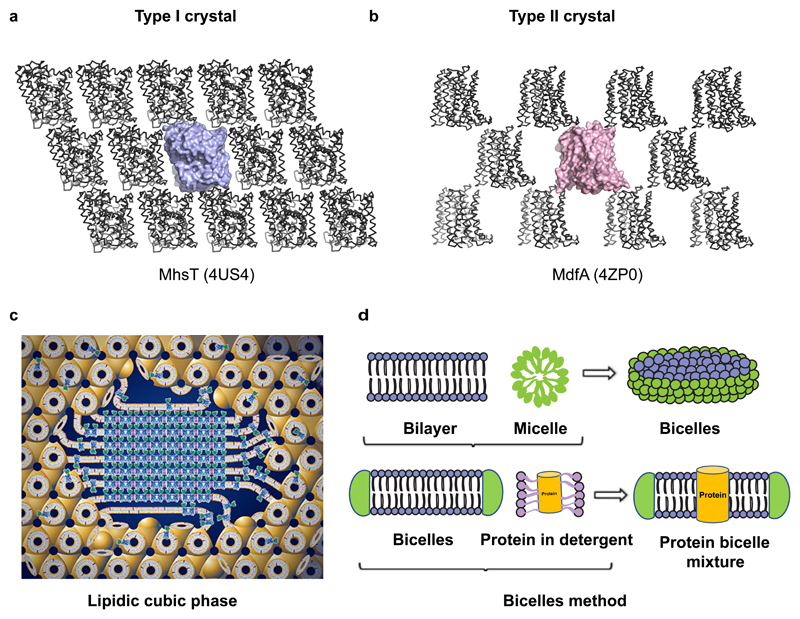
Hartmut Michel classified membrane protein crystals into two forms, type I and type II, based on packing of protein molecules in the crystal (Michel 1983). While the type I crystals of membrane proteins appear as stacks of bilayers, the type II crystal form resembles lattices formed by soluble proteins, with large aqueous channels surrounded by membrane proteins (Fig. 5a,b). Type I crystals might represent a native bilayer like arrangement of membrane proteins and are generally desirable over type II crystals forms. Lipid rich crystallization methods including i. Lipidic cubic phase (LCP) (Fig. 5c) and ii. Bicelle based crystallization (Fig. 5d) have a greater tendency to yield Type I crystals of membrane proteins.
Figure 5.
a, Type I crystal lattice of MhsT (4US4), bacterial sodium driven symporter (NSS family). b, Type II crystal lattice observed in MdfA (4ZP0), a DHA1 multi-drug efflux transporter. c, Schematic representation of Pn3m lipidic cubic phase (LCP) shown with induction of crystal nucleation at the centre (figure used from http://cherezov.usc.edu/resources.htm). d, Pictorial representation of the bicelle crystallisation method.
Landau and Rosenbusch used lipidic cubic phase to obtain crystals of bacteriorhodopsin by mixing defined ratios of protein solution with a host lipid, monoolein (Landau and Rosenbusch 1996). Monoolein or 9.9 monoacyl glycerol(MAG) has a glycerol head group and a single oleic acid with an unsaturation at the 9th carbon. A 3:2 weight ratio mixture of monoolein with water, results in the formation of a clear bicontinuous cubic phase (Pn3m) through self-association (Caffrey 2009). Membrane proteins with or without a crystallization chaperone can be reconstituted into the cubic phase, which upon perturbation by crystallization conditions, results in the formation of lattice contacts leading to crystallization (Fig. 5c). The lattice parameter of the cubic phase and its curvature can be modulated using alternate host lipids with shorter acyl chains or different location of unsaturation which yields cubic phase with reduced curvature and/or altered bilayer thickness. For instance, the bilayer thickness of 7.7 MAG is 41Å, whereas that of monoolein is 51Å. The radius of curvature is larger for 7.7MAG at 56Å whereas monoolein has 52.2Å (Misquitta et al. 2004). The use of LCP is quite extensive with GPCRs and is now the predominant method to crystallize them. Transporters also have successful cases where LCP was helpful in obtaining crystals, although detergent based crystallization has also been quite successful in yielding diffraction quality crystals (Fowler et al. 2015). Crystallization is generally done in glass sandwich plates consuming nanoliter volumes of the cubic phase bolus and reservoir solution. Crystals resulting from LCP tend to have lower solvent content, compared to detergent crystals, resulting in reduced mosaicity. However, the size of the crystals could be small, and prone to radiation damage, requiring data collection and merging of multiple datasets (Liu et al. 2014).
Bicelles, on the other hand, are bilayers that have detergent at the periphery to resemble disc-like micelles (Fig. 5d). Bicelles are made by mixing lipids like DMPC and DPPC with detergents like CHAPSO, at ratios around 2.8:1 (lipid: detergent)(Ujwal and Abramson 2012). Bicelles are characterized by their ability to form gel-like state at room temperature and a solution at 4°C. Membrane proteins incubated with bicelle solution can be used for crystallization screening, similar to detergent micelles. Although a few proteins including LeuT (H. Wang et al. 2012) and VDAC were crystallized in bicelles, it remains a minimally used method. Interestingly, both the lipid rich systems can be doped with other membrane lipids like cholesterol or phospholipids if they are specifically known to improve the behavior of the reconstituted protein.
4. Methods to evaluate dynamic properties of ion-coupled transporters
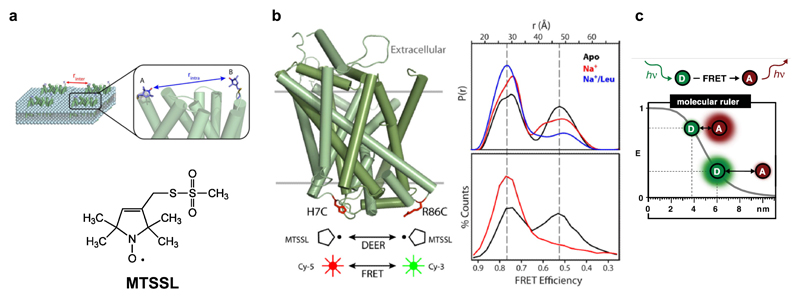
4.1. EPR/DEER measurements
Transporters, as observed in the earlier part of the review, tend to undergo small to large-scale domain movements to affect transport. While crystallographic methods are exceptionally powerful in capturing snapshots of transporters in different states, molecular trajectories of how transporters transition from one state to the other, is not very obvious through crystal structures. In the recent past, electron paramagnetic resonance (EPR) measurements using double electron-electron resonance (DEER), has been used extensively to calculate distance distributions using echo between two spin labels located on mobile elements of a transporter (McHaourab et al. 2011) (Fig. 6a). DEER measurements are capable of long-range distance measurements up to nearly 6-8 nm which is quite capable of observing the modulation between the scaffold domains and gating elements within secondary active transporters (McHaourab et al. 2011). Measurements are performed by chemical crosslinking of spin labels like 1-oxyl-2,2,5,5- tetramethylpyrroline-3-methyl-methanethiosulfonate (MTSSL), with free cysteines strategically introduced in flexible domains, whose dynamics are to be studied. Site-directed spin labeling (SDSL) also requires the removal of excess free cysteines in the protein of interest, to create cys-less, functional constructs. MTSSL labeled integral membrane protein dynamics can be measured in near-native environments, by studying distance distributions of protein reconstituted into liposomes or nanodiscs.
Figure 6.
a, Labeling a transporter at distinct sites using a spin label in case of DEER spectroscopy. b, Distance distribution calculated from spin echos in LeuT and their comparison with smFRET studies performed with fluorophores labeled at the same sites (Figures 6a, b were adapted from Mchaourab, H.S. et al., 2011 after obtaining the publisher’s consent). c, Estimation of dynamic distance measurement for fluorophores in smFRET experiments. Effective distance lies between 1 and 9 nm (Panel adapted from Lerner, E. et al., 2018 after publisher’s consent).
Measurements performed in LeuT by tagging MTSSL spin labels at extracellular elements in EL2, EL4, TM6b, suggested that the extracellular vestibule has improved solvent access in the presence of Na+ and in a leucine-free state (Claxton et al. 2010). Addition of leucine reduces the distance distributions between labels, suggesting the contraction of TM1b, TM6a gates (Fig. 6b). Crystal structure of the substrate free form of LeuT validated this measurement since it was in an Oout state (Krishnamurthy and Gouaux 2012). The high-resolution structure of LeuT in the leucine bound Oout state, reveals a clear inward movement of helices and occlusion of the binding pocket thereby correlating with DEER measurements. On the other hand, DEER measurements at the cytosolic elements of LeuT between the N-terminus and TM3 suggested that movement of TM1a is not as prominent as observed in the crystal structure, in solution (Kazmier et al. 2014).
DEER has also been applied to MFS transporters, LacY a symporter and LmrP an antiporter (Martens et al. 2016). In LacY, measurements at the cytosolic and the periplasmic regions allowed tracking the substrate-induced rocking switch movements in the transporters (Smirnova et al. 2007). In LmrP, DEER measurements aided in demonstrating pH-dependent gating of the transporter and also demonstrated the role of lipid head groups and their interactions with D68 in motif A, that resulted in modulation of drug efflux in LmrP (Masureel et al. 2014). In addition to carrying out measurements in detergent solubilized material, DEER can also be carried out in transporters reconstituted in lipid nanodiscs that allows monitoring the effect of lipids on conformations associated with the transport cycle (Martens et al. 2016).
4.2. smFRET reveals transporter dynamics
smFRET is an analogous method to EPR spectroscopy in providing distance information between FRET pairs, specifically labeled on the transporter. The “molecular-ruler” among FRET pairs extends within a dynamic distance range of 1-9nm allowing monitoring a molecule populating different conformational states (Fig. 6c)(Lerner et al. 2018). Cy3 and Cy5 fluorophores were used to label the cytosolic gate of LeuT at H7 (TM1a) and R86 (IL1). SmFRET between the sites, revealed the presence of two distinct states of the molecule that differ in distance of FRET pair by nearly 13 Å, in the absence of Na+(Zhao et al. 2010). Incremental addition of Na+ resulted in the loss of the second state and populated the Oout state, giving a glimpse into the gating mechanism. SmFRET studies were carried out on GltPh to monitor transport dynamics by labeling the molecule with Cy3 and Cy5 to form donor-acceptor pairs. Sites chosen to label were within a distance to exhibit FRET signal but also undergo significant movements during the transport cycle. I294C at the intracellular loop between HP1-TM7 and N378C at the extracellular surface between loop connecting TM7-HP2 were the sites used to label GltPh. The smFRET studies revealed that the transport domains can function independent of the other protomers and the apo state can cycle between the Oout and Oin state rapidly. Upon binding Na+ and aspartate, the molecule was observed to populate a low FRET Oout state while retaining flickers of high FRET state suggesting, quick shifts to the Oin conformation (Akyuz et al. 2013). A subsequent study also revealed that the humanized version of the transporter with R276S and M395R mutants results in the unlocking of the transport domains allowing the transporter to sample the inward-open states better, thereby allowing the rapid cycling of the glutamate uptake (Akyuz et al. 2015).
5. Monitoring interactions of lipids and transporters
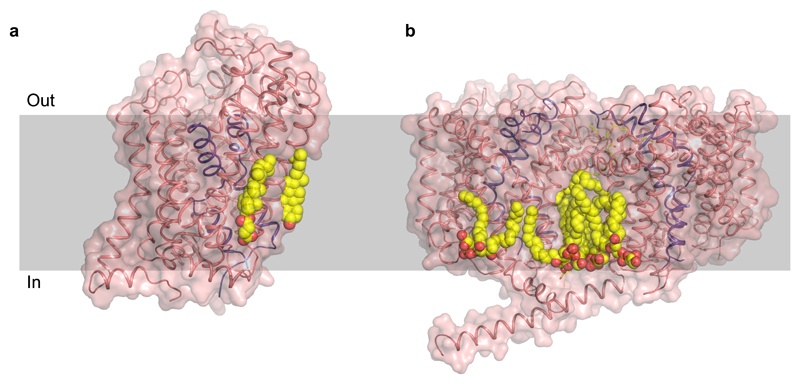
An underlying theme of studying transporter structure and function is the ability of lipids to interact and modulate transport properties. For instance, it was shown through DEER measurements that phosphatidyl ethanol amine (PE) stabilized the inward-open conformation of LmrP, a multi-drug efflux protein, and cardiolipin was involved in the closure of the extracellular gate in LmrP (Martens et al. 2016). In case of BetP, a trimeric betaine transporter with a similar fold as that of LeuT, it was observed to bind POPG lipid in the trimeric interface (Koshy et al. 2013). The lipids were also observed close to the inner leaflet region surrounding TM1 which forms the cytosolic gate in case of the ApcT family transporters. A similar occurrence of a eukaryotic membrane lipid, cholesterol, was observed bound at the interface created by TM1a, TM5 and TM7 in dDAT suggesting a clear role in its ability to allosterically stabilize an inhibitor bound state of the transporter (Penmatsa et al. 2013) (Fig. 7a, b). Cholesterol was also observed in the hSERT structure but away from the TM1 site, in the outer leaflet close to the TM12 region (Coleman et al. 2016). Recent computational studies using coarse grain simulations of hSERT, hDAT, and hNET models have nevertheless, indicated the binding of cholesterol at this site for all three transporters, with varying levels of occupancy (Zeppelin et al. 2018).
Figure 7.
Crystal structures of membrane transporters with bound lipid. a, dDAT crystal structure (4XP4) bound to cholesterol at the interface of TM1a, 5 and 7 and a cholesteryl hemisuccinate bound at TM2, 7 and 11. b, POPG molecules found at the interface of the BetP trimer (4C7R). A third molecule in the trimer was removed for clarity. Head group of the lipid is in close proximity to gating helices.
5.1. Mass spectroscopy of transporters associated with lipid
More often than not, crystal structures do not reveal densities for bound lipid. In the recent past, mass spectroscopy of membrane proteins in gas phase has evolved to obtain signals from intact membrane protein complexes in micelles. This has allowed an unprecedented ability to monitor closely associated lipids that mediate transient or obligate oligomerization in membrane proteins. Carol Robinson and colleagues used ion-mobility mass spectroscopy (IM-MS) to observe that use of non-ionic detergents, above critical micellar concentrations (CMC), allows preservation of the membrane protein complexes, in vacuum using electrospray (Barrera et al. 2008). At higher acceleration voltages, it was observed that the detergent could dissociate from the oligomer resulting in mass spectra corresponding to membrane protein oligomers (Barrera et al. 2008). It was observed in a subsequent study that among non-ionic detergents, C8E4 (tetraethylene glycol monoctyl ether)had lower charge states, thereby, facilitating improved characterization of membrane proteins in gas phase (Laganowsky et al. 2013). This allowed measurement of the stabilizing effects of protein bound lipids, on a wide array of integral membrane proteins. A subsequent study from the same group on integral membrane transporters revealed the role of lipids, particularly cardiolipin, in the dimerization of LeuT. It was observed that one molecule of cardiolipin was bound per protomer of LeuT, in the dimer (Gupta et al. 2017). Cardiolipin was also observed to stabilize sodium: H+ antiporter NapA dimeric interface with the dimer falling apart, when cardiolipin was completely stripped from the protein (Gupta et al. 2017).
These advanced methodologies are some of the approaches through which membrane proteins in general and ion-coupled transporters in specific, have been characterized in the recent past. With the advent of more crystallographic structures and the application of the aforementioned tools, understanding the broad mechanisms and the subtle variations incorporated by individual transporters in the alternating-access mechanism, will increasingly become evident in the near future.
6. Future directions and Conclusions
A major aspect of studying membrane protein dynamics is through computational studies that are not included in this review, since the focus was on experimental methods. Computational methods, including modeling of alternating access (Forrest et al. 2008), all atom molecular dynamics and coarse graining of membrane proteins (Zeppelin et al. 2018) are extensively being used to tease out information ranging from mechanisms of alternating access, role of water and ions in transport process and lipid interactions of membrane proteins. This information is valuable to bolster the findings from crystallographic studies, since a lot of information on dynamics, binding sites, allostery and transport are not apparent, from molecular coordinates.
In addition to computational tools, the advent of cryoEM to elucidate transporter structures is likely to happen, in the near future. Since most ion-coupled transporters are in the size range of 40-60kDa, cryoEM is still in a state where high resolution structural information is difficult to attain. However, novel EM tools like phase plates, and biochemical tools like antibodies that can enhance the size and reduce the conformational heterogeneity, will likely aid in structure determination of transporters through cryoEM in the near future (Vinothkumar and Henderson 2016). Given the important roles that the transporters play in physiology and disease and wide array of methodologies available to characterize them, research in ion-coupled transporters is primed for rapid expansion in the near future.
Supplementary Material
Acknowledgements
The authors would like to thank all members of the Penmatsa lab for their feedback on the manuscript. PM is supported by the IISc-GATE fellowship. AP is an intermediate fellow of the DBT-Wellcome Trust India Alliance (IA/1/15/2/502063) and a recipient of the Innovative Young Biotechnologist Award (IYBA) (BT/09/IYBA/2015/13) from the Dept. of Biotechnology (DBT), India.
References
- Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301(5633):610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- Akyuz N, Altman RB, Blanchard SC, Boudker O. Transport dynamics in a glutamate transporter homologue. Nature. 2013;502(7469):114–118. doi: 10.1038/nature12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akyuz N, Georgieva ER, Zhou Z, Stolzenberg S, Cuendet MA, Khelashvili G, et al. Transport domain unlocking sets the uptake rate of an aspartate transporter. Nature. 2015;518(7537):68–73. doi: 10.1038/nature14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Micelles protect membrane complexes from solution to vacuum. Science. 2008;321(5886):243–246. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- Bibi E, Kaback HR. In vivo expression of the lacY gene in two segments leads to functional lac permease. Proc Natl Acad Sci U S A. 1990;87(11):4325–4329. doi: 10.1073/pnas.87.11.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolla JR, Su CC, Delmar JA, Radhakrishnan A, Kumar N, Chou TH, et al. Crystal structure of the Alcanivorax borkumensis YdaH transporter reveals an unusual topology. Nat Commun. 2015;6 doi: 10.1038/ncomms7874. 6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudker O, Verdon G. Structural perspectives on secondary active transporters. Trends Pharmacol Sci. 2010;31(9):418–426. doi: 10.1016/j.tips.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey M. Crystallizing membrane proteins for structure determination: use of lipidic mesophases. Annu Rev Biophys. 2009;38:29–51. doi: 10.1146/annurev.biophys.050708.133655. [DOI] [PubMed] [Google Scholar]
- Canul-Tec JC, Assal R, Cirri E, Legrand P, Brier S, Chamot-Rooke J, et al. Structure and allosteric inhibition of excitatory amino acid transporter 1. Nature. 2017;544(7651):446–451. doi: 10.1038/nature22064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesar-Razquin A, Snijder B, Frappier-Brinton T, Isserlin R, Gyimesi G, Bai X, et al. A Call for Systematic Research on Solute Carriers. Cell. 2015;162(3):478–487. doi: 10.1016/j.cell.2015.07.022. [DOI] [PubMed] [Google Scholar]
- Claxton DP, Quick M, Shi L, de Carvalho FD, Weinstein H, Javitch JA, et al. Ion/substrate-dependent conformational dynamics of a bacterial homolog of neurotransmitter:sodium symporters. Nat Struct Mol Biol. 2010;17(7):822–829. doi: 10.1038/nsmb.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JA, Green EM, Gouaux E. X-ray structures and mechanism of the human serotonin transporter. Nature. 2016;532(7599):334–339. doi: 10.1038/nature17629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiano E, Bassilana M, Rigaud JL, Leblanc G. Use of the pH sensitive fluorescence probe pyranine to monitor internal pH changes in Escherichia coli membrane vesicles. FEBS Lett. 1984;166(1):120–124. doi: 10.1016/0014-5793(84)80056-3. [DOI] [PubMed] [Google Scholar]
- Deng D, Xu C, Sun P, Wu J, Yan C, Hu M, et al. Crystal structure of the human glucose transporter GLUT1. Nature. 2014;510(7503):121–125. doi: 10.1038/nature13306. [DOI] [PubMed] [Google Scholar]
- Drew D, Boudker O. Shared Molecular Mechanisms of Membrane Transporters. Annu Rev Biochem. 2016;85:543–572. doi: 10.1146/annurev-biochem-060815-014520. [DOI] [PubMed] [Google Scholar]
- Feng L, Campbell EB, Hsiung Y, MacKinnon R. Structure of a eukaryotic CLC transporter defines an intermediate state in the transport cycle. Science. 2010;330(6004):635–641. doi: 10.1126/science.1195230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Frommer WB. Structure and function of SemiSWEET and SWEET sugar transporters. Trends Biochem Sci. 2015;40(8):480–486. doi: 10.1016/j.tibs.2015.05.005. [DOI] [PubMed] [Google Scholar]
- Forrest LR, Zhang YW, Jacobs MT, Gesmonde J, Xie L, Honig BH, et al. Mechanism for alternating access in neurotransmitter transporters. Proc Natl Acad Sci U S A. 2008;105(30):10338–10343. doi: 10.1073/pnas.0804659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler PW, Orwick-Rydmark M, Radestock S, Solcan N, Dijkman PM, Lyons JA, et al. Gating topology of the proton-coupled oligopeptide symporters. Structure. 2015;23(2):290–301. doi: 10.1016/j.str.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futai M. Orientation of membrane vesicles from Escherichia coli prepared by different procedures. J Membr Biol. 1974;15(1):15–28. doi: 10.1007/BF01870079. [DOI] [PubMed] [Google Scholar]
- Gouaux E, Mackinnon R. Principles of selective ion transport in channels and pumps. Science. 2005;310(5753):1461–1465. doi: 10.1126/science.1113666. [DOI] [PubMed] [Google Scholar]
- Green EM, Coleman JA, Gouaux E. Thermostabilization of the Human Serotonin Transporter in an Antidepressant-Bound Conformation. PLoS One. 2015;10(12):e0145688. doi: 10.1371/journal.pone.0145688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K, Donlan JAC, Hopper JTS, Uzdavinys P, Landreh M, Struwe WB, et al. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature. 2017;541(7637):421–424. doi: 10.1038/nature20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M, Hibbs RE, Gouaux E. A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure. 2012;20(8):1293–1299. doi: 10.1016/j.str.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng J, Zhao Y, Liu M, Liu Y, Fan J, Wang X, et al. Substrate-bound structure of the E. coli multidrug resistance transporter MdfA. Cell Res. 2015;25(9):1060–1073. doi: 10.1038/cr.2015.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunte C, Michel H. Crystallisation of membrane proteins mediated by antibody fragments. Curr Opin Struct Biol. 2002;12(4):503–508. doi: 10.1016/s0959-440x(02)00354-8. [DOI] [PubMed] [Google Scholar]
- Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211(5052):969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14(4):673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Kazmier K, Sharma S, Quick M, Islam SM, Roux B, Weinstein H, et al. Conformational dynamics of ligand-dependent alternating access in LeuT. Nat Struct Mol Biol. 2014;21(5):472–479. doi: 10.1038/nsmb.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermani AA, Macdonald CB, Gundepudi R, Stockbridge RB. Guanidinium export is the primal function of SMR family transporters. Proc Natl Acad Sci U S A. 2018;115(12):3060–3065. doi: 10.1073/pnas.1719187115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshy C, Schweikhard ES, Gartner RM, Perez C, Yildiz O, Ziegler C. Structural evidence for functional lipid interactions in the betaine transporter BetP. EMBO J. 2013;32(23):3096–3105. doi: 10.1038/emboj.2013.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy H, Gouaux E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature. 2012;481(7382):469–474. doi: 10.1038/nature10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy H, Piscitelli CL, Gouaux E. Unlocking the molecular secrets of sodium-coupled transporters. Nature. 2009;459(7245):347–355. doi: 10.1038/nature08143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kasho V, Smirnova I, Finer-Moore JS, Kaback HR, Stroud RM. Structure of sugar-bound LacY. Proc Natl Acad Sci U S A. 2014;111(5):1784–1788. doi: 10.1073/pnas.1324141111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganowsky A, Reading E, Hopper JT, Robinson CV. Mass spectrometry of intact membrane protein complexes. Nat Protoc. 2013;8(4):639–651. doi: 10.1038/nprot.2013.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau EM, Rosenbusch JP. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc Natl Acad Sci U S A. 1996;93(25):14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau FW, Nauli S, Zhou Y, Bowie JU. Changing single side-chains can greatly enhance the resistance of a membrane protein to irreversible inactivation. J Mol Biol. 1999;290(2):559–564. doi: 10.1006/jmbi.1999.2905. [DOI] [PubMed] [Google Scholar]
- Lee C, Yashiro S, Dotson DL, Uzdavinys P, Iwata S, Sansom MS, et al. Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights. J Gen Physiol. 2014;144(6):529–544. doi: 10.1085/jgp.201411219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner E, Cordes T, Ingargiola A, Alhadid Y, Chung S, Michalet X, et al. Toward dynamic structural biology: Two decades of single-molecule Forster resonance energy transfer. Science. 2018;359(6373) doi: 10.1126/science.aan1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wacker D, Wang C, Abola E, Cherezov V. Femtosecond crystallography of membrane proteins in the lipidic cubic phase. Philos Trans R Soc Lond B Biol Sci. 2014;369(1647) doi: 10.1098/rstb.2013.0314. 20130314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Lu P, Shi Y. Substrate selectivity of the acid-activated glutamate/gamma-aminobutyric acid (GABA) antiporter GadC from Escherichia coli. J Biol Chem. 2013;288(21):15148–15153. doi: 10.1074/jbc.M113.474502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinauskaite L, Quick M, Reinhard L, Lyons JA, Yano H, Javitch JA, et al. A mechanism for intracellular release of Na+ by neurotransmitter/sodium symporters. Nat Struct Mol Biol. 2014;21(11):1006–1012. doi: 10.1038/nsmb.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancusso R, Gregorio GG, Liu Q, Wang DN. Structure and mechanism of a bacterial sodium-dependent dicarboxylate transporter. Nature. 2012;491(7425):622–626. doi: 10.1038/nature11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens C, Stein RA, Masureel M, Roth A, Mishra S, Dawaliby R, et al. Lipids modulate the conformational dynamics of a secondary multidrug transporter. Nat Struct Mol Biol. 2016;23(8):744–751. doi: 10.1038/nsmb.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masureel M, Martens C, Stein RA, Mishra S, Ruysschaert JM, McHaourab HS, et al. Protonation drives the conformational switch in the multidrug transporter LmrP. Nat Chem Biol. 2014;10(2):149–155. doi: 10.1038/nchembio.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHaourab HS, Steed PR, Kazmier K. Toward the fourth dimension of membrane protein structure: insight into dynamics from spin-labeling EPR spectroscopy. Structure. 2011;19(11):1549–1561. doi: 10.1016/j.str.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel H. Crystallization of membrane proteins. Trends in Biochemical Sciences. 1983;8(2):5. [Review] [Google Scholar]
- Misquitta LV, Misquitta Y, Cherezov V, Slattery O, Mohan JM, Hart D, et al. Membrane protein crystallization in lipidic mesophases with tailored bilayers. Structure. 2004;12(12):2113–2124. doi: 10.1016/j.str.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Mitchell P. A general theory of membrane transport from studies of bacteria. Nature. 1957;180(4577):134–136. doi: 10.1038/180134a0. [DOI] [PubMed] [Google Scholar]
- Morales-Perez CL, Noviello CM, Hibbs RE. Manipulation of Subunit Stoichiometry in Heteromeric Membrane Proteins. Structure. 2016;24(5):797–805. doi: 10.1016/j.str.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan C, Fenollar-Ferrer C, Fitzgerald GA, Vergara-Jaque A, Kaufmann D, Li Y, et al. The bacterial dicarboxylate transporter VcINDY uses a two-domain elevator-type mechanism. Nat Struct Mol Biol. 2016;23(3):256–263. doi: 10.1038/nsmb.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasr ML, Singh SK. Radioligand binding to nanodisc-reconstituted membrane transporters assessed by the scintillation proximity assay. Biochemistry. 2014;53(1):4–6. doi: 10.1021/bi401412e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newstead S, Drew D, Cameron AD, Postis VL, Xia X, Fowler PW, et al. Crystal structure of a prokaryotic homologue of the mammalian oligopeptide-proton symporters, PepT1 and PepT2. EMBO J. 2011;30(2):417–426. doi: 10.1038/emboj.2010.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura N, Verdon G, Kang HJ, Shimamura T, Nomura Y, Sonoda Y, et al. Structure and mechanism of the mammalian fructose transporter GLUT5. Nature. 2015;526(7573):397–401. doi: 10.1038/nature14909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmatsa A, Wang KH, Gouaux E. X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature. 2013;503(7474):85–90. doi: 10.1038/nature12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pos KM. Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta. 2009;1794(5):782–793. doi: 10.1016/j.bbapap.2008.12.015. [DOI] [PubMed] [Google Scholar]
- Quick M, Javitch JA. Monitoring the function of membrane transport proteins in detergent-solubilized form. Proc Natl Acad Sci U S A. 2007;104(9):3603–3608. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quistgaard EM, Low C, Guettou F, Nordlund P. Understanding transport by the major facilitator superfamily (MFS): structures pave the way. Nat Rev Mol Cell Biol. 2016;17(2):123–132. doi: 10.1038/nrm.2015.25. [DOI] [PubMed] [Google Scholar]
- Radchenko M, Symersky J, Nie R, Lu M. Structural basis for the blockade of MATE multidrug efflux pumps. Nat Commun. 2015;6 doi: 10.1038/ncomms8995. 7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VS, Shlykov MA, Castillo R, Sun EI, Saier MH., Jr The major facilitator superfamily (MFS) revisited. FEBS J. 2012;279(11):2022–2035. doi: 10.1111/j.1742-4658.2012.08588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes N, Ginter C, Boudker O. Transport mechanism of a bacterial homologue of glutamate transporters. Nature. 2009;462(7275):880–885. doi: 10.1038/nature08616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Jr, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34(Database issue):D181–186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner S. EmrE, a model for studying evolution and mechanism of ion-coupled transporters. Biochim Biophys Acta. 2009;1794(5):748–762. doi: 10.1016/j.bbapap.2008.12.018. [DOI] [PubMed] [Google Scholar]
- Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Conformational thermostabilization of the beta1-adrenergic receptor in a detergent-resistant form. Proc Natl Acad Sci U S A. 2008;105(3):877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer PL, Goehring A, Shankaranarayanan A, Gouaux E. Structure and mechanism of a Na+-independent amino acid transporter. Science. 2009;325(5943):1010–1014. doi: 10.1126/science.1176088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Common folds and transport mechanisms of secondary active transporters. Annu Rev Biophys. 2013;42:51–72. doi: 10.1146/annurev-biophys-083012-130429. [DOI] [PubMed] [Google Scholar]
- Singh SK. LeuT: a prokaryotic stepping stone on the way to a eukaryotic neurotransmitter transporter structure. Channels (Austin) 2008;2(5):380–389. doi: 10.4161/chan.2.5.6904. [DOI] [PubMed] [Google Scholar]
- Singh SK, Piscitelli CL, Yamashita A, Gouaux E. A competitive inhibitor traps LeuT in an open-to-out conformation. Science. 2008;322(5908):1655–1661. doi: 10.1126/science.1166777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova I, Kasho V, Choe JY, Altenbach C, Hubbell WL, Kaback HR. Sugar binding induces an outward facing conformation of LacY. Proc Natl Acad Sci U S A. 2007;104(42):16504–16509. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zeng X, Yan C, Sun X, Gong X, Rao Y, et al. Crystal structure of a bacterial homologue of glucose transporters GLUT1-4. Nature. 2012;490(7420):361–366. doi: 10.1038/nature11524. [DOI] [PubMed] [Google Scholar]
- Tao Y, Cheung LS, Li S, Eom JS, Chen LQ, Xu Y, et al. Structure of a eukaryotic SWEET transporter in a homotrimeric complex. Nature. 2015;527(7577):259–263. doi: 10.1038/nature15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujwal R, Abramson J. High-throughput crystallization of membrane proteins using the lipidic bicelle method. J Vis Exp. 2012;59:e3383. doi: 10.3791/3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidehi N, Grisshammer R, Tate CG. How Can Mutations Thermostabilize G-Protein-Coupled Receptors? Trends Pharmacol Sci. 2016;37(1):37–46. doi: 10.1016/j.tips.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinothkumar KR, Henderson R. Structures of membrane proteins. Q Rev Biophys. 2010;43(1):65–158. doi: 10.1017/S0033583510000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinothkumar KR, Henderson R. Single particle electron cryomicroscopy: trends, issues and future perspective. Q Rev Biophys. 2016;49:e13. doi: 10.1017/S0033583516000068. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7(12):909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- Wang H, Elferich J, Gouaux E. Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context. Nat Struct Mol Biol. 2012;19(2):212–219. doi: 10.1038/nsmb.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Penmatsa A, Gouaux E. Neurotransmitter and psychostimulant recognition by the dopamine transporter. Nature. 2015;521(7552):322–327. doi: 10.1038/nature14431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlert D, Grotzinger MJ, Kuhlbrandt W, Yildiz O. Mechanism of Na(+)-dependent citrate transport from the structure of an asymmetrical CitS dimer. Elife. 2015;4:e09375. doi: 10.7554/eLife.09375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature. 2005;437(7056):215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- Yan N. Structural Biology of the Major Facilitator Superfamily Transporters. Annu Rev Biophys. 2015;44:257–283. doi: 10.1146/annurev-biophys-060414-033901. [DOI] [PubMed] [Google Scholar]
- Yernool D, Boudker O, Jin Y, Gouaux E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature. 2004;431(7010):811–818. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]
- Zeppelin T, Ladefoged LK, Sinning S, Periole X, Schiott B. A direct interaction of cholesterol with the dopamine transporter prevents its out-to-inward transition. PLoS Comput Biol. 2018;14(1):e1005907. doi: 10.1371/journal.pcbi.1005907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Terry D, Shi L, Weinstein H, Blanchard SC, Javitch JA. Single-molecule dynamics of gating in a neurotransmitter transporter homologue. Nature. 2010;465(7295):188–193. doi: 10.1038/nature09057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.