Abstract
Leaf rust caused by Puccinia triticina is one of the main diseases affecting wheat (Triticum aestivum) production worldwide. Calmodulin (CaM) was found involved in the early stage of signal transduction pathway in response to P. triticina in wheat. To study the function and molecular mechanism of calmodulin (CaM) in signal transduction of wheat against P. triticina, we cloned a putative calmodulin-binding transcription activator (TaCAMTA4), and characterized its molecular structure and functions by using the CaM-encoding gene (TaCaM4-1) as a bait to screen the cDNA library from P. triticina infected wheat leaves. The open reading frame of TaCAMTA4 was 2505 bp encoding a protein of 834 aa, which contained all the four conserved domains of family (CG-1 domain, TIG domain, ANK repeats and CaM-binding domain). TaCaM4-1 bound to TaCAMTA4 by the C-terminal CaM-binding domain in Ca2+-dependent manner in the electrophoretic mobility shift assay (EMSA). Bimolecular fluorescence complementation (BiFC) analysis indicated that the interaction of TaCAMTA4 and TaCaM4-1 took place in the cytoplasm and nucleus of epidermal leaf cells in N. benthamiana. The expression level of TaCAMTA4 genes was down-regulated in incompatible combination after P. triticina infection. Furthermore, virus-induced gene silencing (VIGS)-based knockdown of TaCAMTA4 and disease assays verified that silencing of TaCAMTA4 resulted in enhanced resistance to P. triticina race 165. These results suggested that TaCAMTA4 function as negative regulator of defense response against P. triticina.
Introduction
Plants have developed sophisticated mechanisms to sense and respond to various stresses so as to adapt to their environment. Ca2+ signaling pathway plays an important role in plant defense responses to biotic stresses (e.g., pathogens, hormones, etc.) and abiotic stresses (e.g., cold, hot, drought, etc.)1. Intracellular Ca2+ transient increase is an early signal to induce defense responses in plants2–4. Specific Ca2+ signals could be recognized by Ca2+ receptors and then transferred to other proteins involved in Ca2+ signaling pathways5. Calmodulin (CaM), the primary cellular Ca2+ receptor, recognizes and binds specific Ca2+ signals through its EF-hand domain, and transfers Ca2+ signals by interacting with the calmodulin-binding domains (CaMBD) of target calmodulin-binding proteins (CaMBPs). Many CaMBPs have been identified to play important roles in defense responses to stress in plants4,6–8.
Calmodulin-binding transcription activator (CAMTA), one of the CaMBPs, also known as signaling-responsive protein (SR), is a highly conserved transcription factor family4,6–8 in plants and other multicellular organisms (e.g., mammals, insects, etc.)1. The CAMTA family has four highly conserved domains: the CG-1 DNA-binding domain which specifically recognizes CGCG box and CGTG box in promoter regions of target genes1 and contains a typical bipartite nuclear locating signal (NLS) necessary for nuclear importation; the transcription factor immunoglobin (TIG) domain which is a putative nonspecific DNA-binding domain, a protein-protein interacting domain containing ankyrin (ANK) repeats, and a CaM-binding domain which consists of IQ domain and CaMB domain9.
CAMTAs function in growth and development, as well as stress defenses, in plants10. In A. thaliana genome, six CAMTA genes (AtCAMTA1-6 or AtSR1-6) were identified, and their expressions could be induced by stresses including cold and heat, abscisic acid (ABA), ethylene, methyl jasmonate (MJ), H2O2 or salicylic acid (SA)11, respectively. It has been shown that AtCAMTA1 closely relates with auxin12 and drought signaling13 and AtCAMTA3 plays a role in cold tolerance11. CAMTAs regulate resistance-related genes expression in biotic stress defense processes9,14,15, the DNA-binding domain of AtCAMTA3 could bind the CGCG box in the promoter of EDS1 gene to suppress EDS1 expression, and Ca2+/CaM binding was necessary for AtCAMTA3 to negatively regulate EDS1 expression and plant immunity16.
We have long engaged in the study of the signal transduction mechanism of wheat defense response against P. triticina. Pharmacological tests using Ca2+ signal blockers and activators17–19, subcellular localization of cytoplasmic Ca2+ 20, and cytoplasmic Ca2+ dynamic detection of elicitor stimulated wheat mesophyll protoplasts21 showed that Ca2+ play an important role in the signal transduction pathway in wheat resistance against P. triticina. Real-time quantitative RT-PCR (qRT-PCR) and western blotting showed that, in the early stage of incompatible interactions between wheat and P. triticina, the expression of TaCaM4 was the highest among the eleven CaMs. Furthermore, CAM4-silenced plants showed relatively weak HR (hypersensitive response) signals compared with control. Suppression subtractive hybridization (SSH) indicated that the expression of TaCaM4 increased at 8 hours after P. triticina infection22. All the previous results demonstrated that TaCaMs were involved in the early stage of incompatible interaction processes, and play an important role in the wheat resistance signal transduction pathway against P. triticina. However, the putative molecular mechanism underlying the involvement of CAMs in wheat- P. triticina interaction signal pathway still remain unclear.
In this study, we cloned a putative CAMTA gene by screening cDNA library from wheat leaves infected P. triticina and designated as TaCAMTA4 for its highest homology with AtCAMTA4, and analyzed their gene structures. Furthermore, EMSA was employed to determine the CaM binding motif. We also further verified the interaction by Bimolecular fluorescence complementation (BiFC) analysis in tobacco epidermal leaf cells. The expression profiles of TaCAMTA4 gene were analyzed using quantitative real-time RT-PCR (qRT-PCR). Finally, using VIGS (Virus-induced gene silencing) -based knockdown, we revealed that TaCAMTA4 may negatively regulated wheat basic resistance to P. triticina race 165.
Results
Screening of TaCaM4-1 interacting proteins and amplification of TaCAMTA4 full length cDNA by RACE
To explore the roles of wheat TaCaM4-1 in P. triticina infection, pGBKT7-TaCaM4-1 bait vector was used for screening of wheat yeast two-hybrid cDNA library, resulting in 45 positive clones. To verify that the proteins interact with TaCAM4-1, prey plasmids in the positive clones were extracted and transformed into yeast AH109 together with bait plasmid pGBKT7-TaCaM4-1 in one-on-one method for interaction detection to eliminate false positive clones. Yeast cells with both plasmids (cell number 406, 408, 413, 427, 435, 438 and 439) grew on the selection medium, while the cells with either plasmid was absent, indicating that these proteins interact with TaCAM4-1 in yeast cells (Fig. 1a). Among the 7 candidate genes encoding CaM-binding protein obtained by yeast two-hybrid, an 896-bp sequence (termed Code. 408) was highly homologous to CAMTA genes, with the typical transcription factor characteristics. In order to get the full length cDNA of Code. 408, RACE was conducted to amplify the 5′ end of the cDNA fragment. The resulting PCR amplicon of the full length cDNA was 2704 bp and the open reading frame was 2505 bp (S1 Figure).
Figure 1.

Screening of TaCaM4-1 interacting proteins (a) Interaction tests using yeast two-hybrid assays between TaCAM4-1 and prey proteins. Yeasts harboring TaCAM4-1 and prey proteins were placed in different liquid concentrations on control medium SD/-Trp/-Leu and selection medium SD/-Trp/-Leu/-His/-Ade. For negative controls, pGADT7 without insert TaCAM4-1 was used (pGBKT7-TaCAM4-1 + pGADT7). Experiments were performed three times and a representative result is shown. The full-length blots are presented in Supplementary Fig 1. (b) Phylogenetic analyses of TaCAMTA4 and its homologs from different plant species. The TaCAMTA4 protein sequence was used to perform BLAST searches against the National Center for Biotechnology Information database. TaCAMTA4 and its homologs identified in different organisms were aligned. Gm, Glycine max; Vv, Vitis vinifera; At, Arabidopsis thaliana; Sl, Solanum lycopersicum; Zm, Zea mays L; Bd, Brachypodium distachyon; Ta, Triticum aestivum. (c) TaCAMTA4 conserved domains prediction. CG-1, specific CGCG box-containing DNA sequences; TIG, transcription factor ImmunoGlobin; CaM-binding, calmodulin-binding domain; ANK repeat, ankyrin repeats; Bipartite NLS, nuclear locating signal; aa, amino acids.
Blast analysis on NCBI showed that the Code.408 gene was homologous to CAMTA genes in Glycine max, Vitis vinifera, Solanum lycopersicum, Zea mays and Brachypodium distachyon. Phylogenetic analysis of our gene and six CAMTAs in A. thaliana (AtCAMTA1-6 or AtSR1-6) showed that our gene was highly homologous to AtCAMTA4 (Fig. 1b). As a result, this gene was named TaCAMTA4 and deposited in GenBank (accession No. KC686696). Prediction of conserved domains in TaCAMTA4 indicated that TaCAMTA4 contained all the conserved domains of the CAMTA family: CG-1 domain, TIG domain, ANK repeats, and CaM-binding domain (Fig. 1c). The CG-1 domain is a 130-amino acid highly conserved domain and contains a bipartite NLS necessary for nuclear import. CG-1 domain can bind CGCG box in the promoter region of genes. TIG domain widely exists in endocellular transcription factors and cell surface receptors and functions in interaction with DNAs or proteins. ANK repeats exist in the form of ankyrin tandem repeat but the number of tandems is various in different genes and different species. ANK repeats may function in protein-protein interaction. The probable CaM-binding domain of TaCAMTA4 was found at the C- terminal which could function in CaM recognizing and binding. The bioinformatic analysis suggested that TaCAMTA4 could be a novel member of the wheat CAMTA family.
Interactions between TaCAMTA4 and TaCAM4-1
Previous studies have showed CaM-binding domains of CaMBPs were mostly located on the C-terminal23. Bioinformation analysis revealed that the probably CaM-binding domain of TaCAMTA4 was also on the C-terminal. In order to detect the binding between CaM-binding sites of TaCAMTA4 and TaCaM4-1, two peptide sequences in TaCAMTA4, as AVQAAGRIQATFRVFSLKKKKQKALQNRGS (666–695 aa) and
IRKNVIKIQARFRAHRERNKYKELLQ (725–750 aa) were chosen to conduct the CaM-binding analysis in vitro (Fig. 2a). The two synthetic peptides termed A-S and I-Q were respectively mixed with prokaryotic expressed TaCaM4-1 at peptide/CaM molar ratios of 0, 0.5, 1, 2, 4 and 8 in reaction buffer and spontaneously reacted at room temperature for 1 hour and then were detected by native PAGE. The results showed that, in the presence of 1 mM Ca2+ and in the absence of peptides, TaCaM4-1 appeared as a single band. After A-S or I-Q peptide was added, a second slower mobility band appeared indicating formation of the peptide/CaM complex. And the I-Q/CaM complex band appeared when peptide/CaM molar ratio reached 0.5, whereas A-S/CaM complex band did not appear until peptide/CaM molar ratio reached 1, suggesting the binding characteristics in different regions of the CaM-binding sites. In the presence of 2 mM EGTA, no complex band appeared for either A-S or I-Q (Fig. 2b). The results suggested that TaCAMTA4 could bind to TaCaM4-1 by the C-terminal CaM-binding domain in Ca2+-dependent manner.
Figure 2.
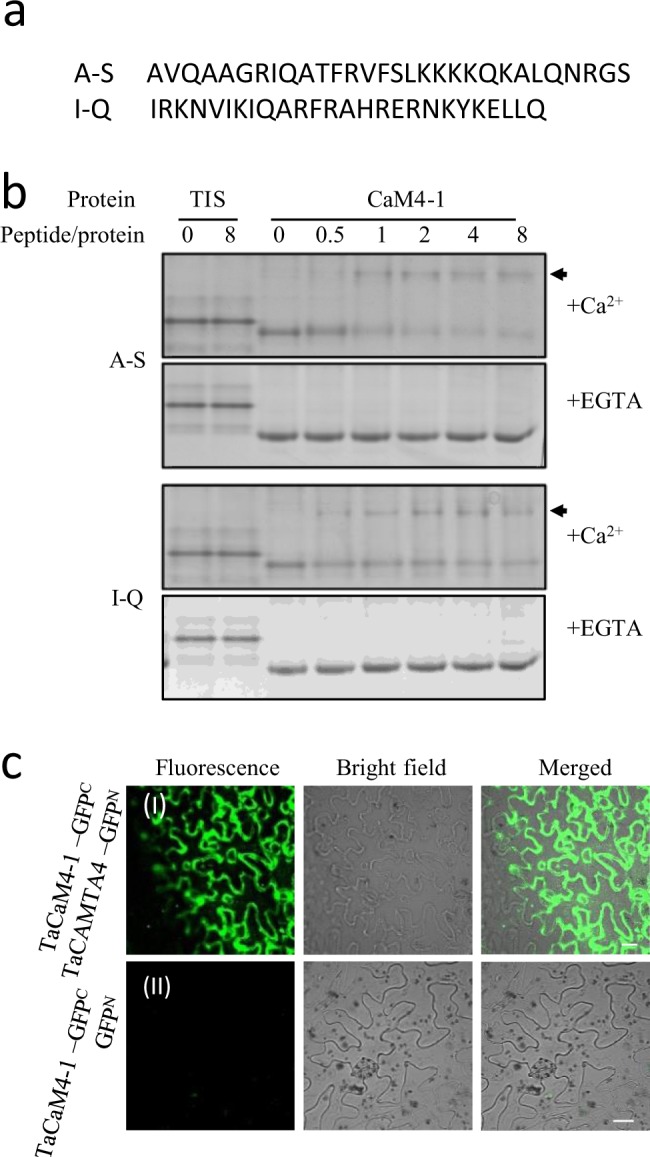
TaCAMTA4 interacted with TaCaM4-1 in EMSA and BiFC assays. (a) Two motif sequences of TaCAMTA4. Two synthetic peptides termed A-S and I-Q. (b) Two motifs of TaCAMTA4 interacted with CaM in vitro with EMSA. A-S peptide and I-Q peptide were mixed with prokaryotic expressed TaCaM4-1 on peptide/protein ratio of 0, 0.5, 1, 2, 4 and 8, respectively, in present of 1 mM Ca2+ or 2 mM EGTA, and detected by native PAGE after reaction 1 hour at room temperature. TIS (trypsin inhibitor) was used as negative control. Arrows indicate the bands of peptide/CaM complex. The full-length gels are presented in Supplementary Figure 2. (c) TaCAMTA4 interacted with TaCAM4-1 by BiFC assays in N. benthamiana. (I). The coding regions of TaCAMTA4 and TaCaM4-1 genes were fused with the N-terminal (GFPN) and C-terminal (GFPC) region of GFP, respectively. The recombinant constructs pSPYCE(MR)- TaCaM4-1/pSPYNE(R)173- TaCAMTA4 were co-transformed into N. benthamiana. (II). The combination of TaCaM4-1 -GFPC and GFPN was used as a negative control. The results were observed 48 h after infiltration, Scale bar, 20 μm.
The interaction between TaCAMTA4 and TaCaM4-1was further confirmed using a BiFC assay, where the N- and C-terminal GFP fragments were fused to TaCAMTA4 and TaCaM4-1, respectively, and co-expressed in N. benthamiana leaves. The fluorescent signals of GFP indicated that interaction between TaCAMTA4 and TaCaM4 co-located in cytoplasm and nucleus (Fig. 2c).
TaCAMTA4 was decreased after P. triticina infection
The involvement of CaM in P. triticina infection has been well confirmed. In order to clarify whether CaM-binding TaCAMTA4 regulates the interaction process, the transcription levels of TaCAMTA4 were detected by qRT-PCR in both incompatible and compatible combinations at different time after P. triticina infection. In the incompatible combination, the expression levels of TaCAMTA4 decreased after P. triticina infection and showed the lowest level at 8 hours after infection, about 40% of the expression level at 0 h. The transcription level rised at 24 h and got back to 0 h level at 48 hours after infection. However, in the compatible combination, the transcription of TaCAMTA4 slightly decreased and the change was not obvious as that in the incompatible combination (Fig. 3). The results indicated that down regulation of TaCAMTA4 expression is required for resistance of wheat against P. triticina.
Figure 3.
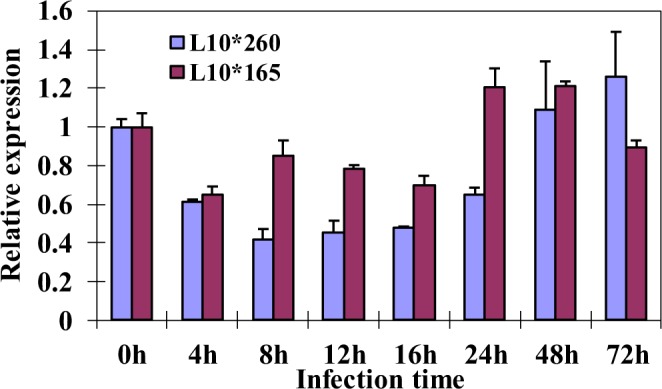
Expression of TaCAMTA4 in different combination plants after P. triticina infection. The transcription levels of wheat TaCAMTA4 were detected by qRT-PCR in both incompatible and compatible combinations at different time points after P. triticina infection. Mock treatment combination was used as control. Constitutively expressed gene EF-1-α was used as a reference. Results are means ± SD with three replicates.
TaCAMTA4 negatively regulate the defense response of wheat against P. triticina
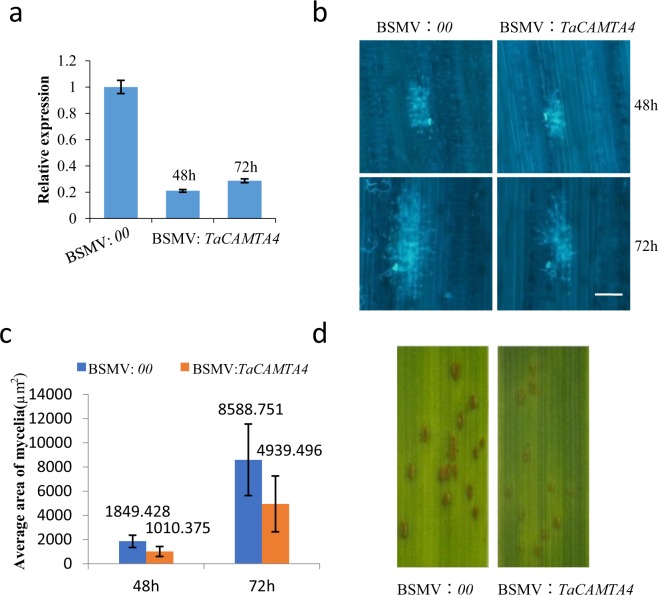
We further hypothesized that silencing TaCAMTA4 may enhance the defense response of wheat Lovrin 10 to rust fungus. A specific fragment of TaCAMTA4 was cloned into the vector of BSMV to silence TaCAMTA4 on the first leaves. BSMV:00 (BSMV empty vector) infiltrated plants were used as control in the VIGS experiment. The newly emerged third leaves were sampled at 48 h and 72 h after race 165 inoculation. To determine the effects of silencing TaCAMTA4 gene and the resistance of wheat to P. triticina, qRT-PCR was used to detect the expression of TaCAMTA4. BSMV: TaCAMTA4-inoculated plants showed reduced TaCAMTA4 expression levels significantly (Fig. 4a), indicating the specific suppression of the target gene in the VIGS plants.
Figure 4.
TaCAMTA4 negatively regulate the defense response of wheat to P. triticina. (a) Expression analysis of TaCAMTA4 in the third leaves inoculated with BSMV: 00 and BSMV: TaCAMTA4 by qRT-PCR. After 12 days BSMV inoculation, the mature urediniospores of P. triticina race 165 were brushed on the newly emerged third leaves. Then the RNA extracted from the third leaves at 48 h and 72 h after P. triticina infection was subjected to qRT-PCR. (b) Biology microscope observations of P. triticina mycelia appearing on leaf pieces at 48 h and 72 h after inoculation. Leaf tissue was stained with 0.1% Florescent Brightener (FB) Bar = 100 μm. (c) The average area of mycelia was statistically analyzed between BSMV: 00(control) and BSMV: TaCAMTA4. Values are means (±SD) of 30 mycelia. The numbers of mycelia per visual field were counted at 48 h and 72 h after P. triticina infection. (d) The phenotype of uredosorus after inoculation with P. triticina. Photos were taken at 13 days after P. triticina infection.
In the meantime, the development of P. triticina hypha was observed at 48 and 72 h (Fig. 4b–d). The average area of mycelia at single infection site was much smaller and the growth rate was slower on TaCAMTA4-silenced plants compared with the control (Fig. 4b). The areas of single sorus at 8 and 13 days after infection were smaller on leaves inoculated with BSMV: TaCAMTA4 than that on the control leaves (Fig. 4d). The chlorosis phenotype on leaves with silenced TaCAMTA4 appeared to be more severe than the control leaves (Fig. 4d). Silencing of TaCAMTA4 resulted in enhanced resistance to P. triticina race 165. These results indicated that TaCAMTA4 regulate the defense response of wheat against P. triticina negatively.
Discussion
Identifying and characterizing CaM target genes (CaMBPs), is crucial for systematically understanding the Ca2+-CaM signal transduction pathway in plant defense responses. A multitude of studies have confirmed that Ca2+-dependent CaM signal transduction pathway plays an important role in plant defense responses17,24–26. In the previous studies, we have preliminarily proved that Ca2+ was involved in the wheat defense response against P. triticina17. In the present study, qRT-PCR results showed CaM play an important role in the wheat defense response against P. triticina. So identification of the CaMBPs is very important for systematically study of the signaling pathway in wheat-P. triticina interaction system. Many CaMBPs have been identified, including the DNA-binding protein family, CAMTAs, by using labeled CaM to screen A. thaliana cDNA library or CaM or CML probes to detect A. thaliana protein chips27–29. However, functions of CAMTAs in plant defense response to pathogen were less reported. CAMTAs can bind to CaM, regulate the transcription of target genes and consequently transfer specific Ca2+ signals30. It has been reported that the six CAMTA genes in A. thaliana were induced by various abiotic stress such as cold, heat, salinity, etc.31. In this work, TaCAMTA4 was identified as a CaM-binding protein in the wheat-P. triticina interaction system, homologous to Arabidopsis CAMTA4 with highly conserved CAMTA domains.
The CaM-binding domain is usually on the C-terminal of CAMTAs. The CaM-binding activity of CAMTAs has already been proved in many studies. Prokaryotic expressed peptide corresponding to the CaM-binding sites of AtSR1 can bind to CaM in the present of Ca2+; but no binding was found32 in the present of EGTA. AtCAMTA1 could bind 35S-CaM in a Ca2+-dependent manner in EMSA33. AtSR1 is Ca2+-dependent CaM-binding protein and the CaM-binding site is on the C-terminal of AtSR1; moreover, all the six SRs in A. thaliana (AtSR1–6) are Ca2+-dependent CaM-binding proteins31. In this study, bioinformatic analysis indicated that the CaM-binding domain was on the C-terminal of TaCAMTA4 and highly homologous to the CaM-binding regions of many other CaM-binding proteins8,25. To further verify the CaM-binding sites of TaCAMTA4, we conducted phylogenetic analysis between CaM-binding domains of TaCAMTA4 and many other CaMBPs and finally chose two peptide sequences (about 30 aa) of TaCAMTA4 for CaM-binding analysis. The results of EMSA showed that the two peptide sequences could interact with TaCaM4-1 in a Ca2+-dependent manner. Meanwhile, the results of BiFC showed that the C-terminal of TaCAMTA4 could interact with TaCaM4-1 in N. benthamiana. Therefore, it can be concluded that TaCAMTA4 could bind TaCaM4-1 by the C-terminal CaM-binding sites in a Ca2+-dependent manner.
Identification of TaCAMTA4 is very important for understanding the function of TaCAMTA4 and studying the Ca2+ signal transduction pathway in the defense response of wheat against P. triticina. In this work, the transcription levels of TaCAMTA4 decreased both in incompatible and compatible combinations in the early stage of infection and decreased more obviously in incompatible combinations. In the incompatible combination, the expression level of TaCAMTA4 reduced to the lowest at 8 h after inoculation. Our previous studies have shown that 8 h after inoculation is the time of substomatal vesicles formation. At 16 h after inoculation the primary hyphae and haustorial mother cells are formed and the haustorial mother cells contact with mesophyll cells to induce hypersensitive reaction(HR) in incompatible combination34. So the genes that were induced to be up-regulated or down-regulated at 8 h might involve in basic resistance. Furthermore, virus-induced gene silencing (VIGS)-based knockdown of TaCAMTA4 assays showed that silencing of TaCAMTA4 resulted in enhanced resistance to P. triticina race 165. The results indicated that TaCAMTA4 may negatively regulate basic resistance of wheat against P. triticina. This work laid a foundation for further studies on TaCAMTA4 to better understand the interaction mechanism between wheat and P. triticina.
Materials and Methods
Plant materials and growth conditions
Wheat (Triticum aesetrum L.) variety Lovrin 10 and P. triticina race 165 made up the compatible combination, while Lovrin 10 and 260 made up the incompatible combination. P.triticina races 165 and 260 were maintained on highly susceptible wheat variety Zhengzhou 5389.
Wheat seeds were sown in 15-cm-diameter pots. Seedlings grew in the greenhouse (20–25°C, 14-h-light/10-h-dark photoperiod, 400 W/m2 light intensity). When seedlings grew to 7 days old with the first leaf fully expanded, the urediniospores of P. triticina were inoculated on the wheat leaves.
Tabacco seeds were sterilized in 2.5% NaClO for 15 min and washed five times with sterile water. Seeds were geminated on the MS medium in the greenhouse (20–25°C, 14-h-light/10-h-dark photoperiod) for 8 days. Then the seedlings were transferred into vermiculite and kept growing for 1 month. N. benthamiana was cultured with Hoagland nutrient solution.
Construction of TaCaM4-1 bait vector and yeast two-hybrid screening
The full coding region of TaCaM4-1 was amplified by PCR from cDNA of P. triticina infected wheat leaves and constructed into pGBKT7 vector in Nde I and EcoR I sites. The primers were designed based on the CaM4-1 gene sequence of wheat variety China Spring.
The recombined vectors pGBKT7-TaCaM4-1 was transferred into yeast AH109 by LiAc method and cDNA library screening was achieved by yeast mating. After yeast mating, the culture was inoculated on SD/-Trp-Leu-His medium and incubated at 28 °C for 7 days. Then the larger colonies were incubated another 7 days on SD/-Trp-Leu-His-Ade medium respectively followed by being transferred onto X-α-gal-based SD/-Trp-Leu-His-Ade medium for coloration detection. The colonies turned blue were regarded as possible positive colonies.
Amplification of full-length cDNA by RACE
Total RNA was extracted from wheat leaves infected by P. triticina. Reverse transcription and RACE PCR were conducted according to the instruction of SMARTTM RACE cDNA Amplification Kit. 5′-RACE primers were designed according to the gene fraction sequence screened by yeast two-hybrid.
Bioinformation analysis
The homologous genes were identified using BLAST (http://blast.ncbi.nlm.nih.gov/Blast). Protein conserved domains prediction and phylogenetic analysis were conducted in InterproScan (http://www.ebi.ac.uk/Tools/InterproScan) and ExpaSy (http://www.expasy.org). PSORT (http://psort.ims.u-tokyo.ac.jp/form/html) was for subcellular location prediction.
CaM-binding analysis by EMSA
The synthesized peptides were dissolved in 10 mM Tris-HCl solution (pH 8.0). Peptide and prokaryotic expressed TaCaM4-1 proteins were mixed respectively in molar ratios of 0, 0.5, 1, 2, 4, and 8 in reaction buffer (10 mM Tris-HCl solution (pH 8.0) with 1 mM CaCl2 or 2 mM EGTA). Total reaction volume was 30 µL. After spontaneously interacting at room temperature for 1 hour, 10 µL of the reaction volume was used for 15% nondenaturing PAGE detection.
Bimolecular fluorescence complementation (BiFC) assay
TaCAMTA4 and TaCaM4-1 CDS sequences were constructed into pSPYNE (R)173 and pSPYCE (MR) respectively. Recombined plasmids were respectively transferred into Arobacterium tumefaciens strain GV3101. The empty vectors pSPYNE (R)173 and pSPYCE (MR), expressing split GFP domains were used as a negative control, Both pSPYNE (R)173- TaCAMTA4 and pSPYCE(MR)- TaCaM4-1 were mixed, and co-transformed into N. benthamiana leaves by pressure injection method. The transformed N. benthamiana were incubated at 25 °C for 40–48 hours before observation. GFP fluorescence was recorded using a confocal laser-scanning microscope (OLYMPUS_FV1000-IX81).
Gene expression analysis by qRT-PCR
Total RNA samples were extracted from infected leaves at 0 h, 4 h, 8 h, 12 h, 16 h, 24 h, 48 h, and 72 h after infection with P. triticina. RNA was purified and reverse transcribed to synthesis the first strand of cDNA. qRT-PCR was conducted using qRT-PCR kit (TaKaRa). Expressions of all genes were assayed for triplicated. Gene expression was calculated using the Delta-Delta cycle threshold method. qRT-PCR was also performed in mock treatment combination used as control. Consecutively expressed gene EF-1-α was used as a reference. Primers for qRT-PCR were designed by Primer 5 and their sequences were listed in S6 Table.
Construction of BSMV-VIGS vector and silencing of TaCAMTA4
A 291 bp specific fragment of TaCAMTA4 (from 72 bp to 362 bp) was cloned to the vector of pSL038-1, to construct BSMV: TaCAMTA4. The primer sequences used to amplify the target gene were as follows: forward, GCGGCCGCCCAGAGAGGTTC; reverse, GCGGCCGCTCATAAGCCGGTTC. The restriction sites of Not I were underlined. In vitro transcription was performed using the mMessage mMachine™ T7 in vitro transcription kit (Ambion) following the manufacturer’s protocol. The tripartite BSMV RNA BSMV: α, BSMV: β, and BSMV: TaCAMTA4 or BSMV: 00 were inoculated on the first fully expended leave of 7–8 days old wheat lovrin 10 by rub inoculation with a gloved finger. The wheat Lovrin 10 infected with BSMV: 00 was as control. For each treatment, ten plants were inoculated.
Virus infection and evaluation of resistance
After 12 days of BSMV inoculation,the mature urediniospores of P. triticina race 165 was brushed on the newly emerged third leaves by using a wet brush. The control wheat was treated the same simultaneously. Then the wheat were placed in a moist dark cabinet for 16 hours. Leaves were collected at 48 h and 72 h after P. triticina infection to detect the expression level of TaCAMTA4 by using qRT-PCR and evaluate the wheat resistance to P. triticina. Primers were designed by Primer 5 and listed in S6 Table. Fluorescent Brightener (FB) was used to dye the leaf pieces to observe the development of hypha. The area of the mycelia was measured. Photos were taken to record the phenotype of leaves and sorus at 8 days and 13 days after P. triticina infection.
Electronic supplementary material
Dataset1, Dataset2, Dataset3, Dataset4, Dataset5, Dataset6.
Acknowledgements
We thank State Key Laboratory Plant Physiology and Biochemistry, China Agriculture University, for friendly providing pSPYNE(R)173 and pSPYCE(MR) vectors. We are grateful to Dr. Steven R. Scofield (Purdue University) for friendly providing BSMV vectors. We are also grateful to professor Longxian Ran and professor Shujun Ge (Hebei Agriculture University) for critical review of this manuscript. This work was financially supported by National Natural Science Foundation of China (30671244; 31171472; 31871548), Specialized Research Found for the Doctoral Program of Higher Education (20111302130001), the Key Basic Research Project of HeBei (12967149D), Natural Science Foundation of HeBei (C2010000787) and Open topic of China Agriculture University (SKLPPBKF1504) .
Author Contributions
Wang D.M., Han S.F. and Hou C.Y. conceived the idea. Wang Y.L. and Wei F.J. planned the work. Wang Y.L. and Zhou H. executed most of the experiments. Wei F.J., Zhang L.F. and Niu X.N. helped in family analysis and data interpretation, Yan C. and Liu N. helped performed gene expression study and biochemical assays. Wang Y.L. and Wei F.J. analyzed the data and wrote the manuscript. All authors read and approved the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yuelin Wang, Fengju Wei and Hui Zhou contributed equally.
Contributor Information
Fengju Wei, Email: weifj98@126.com.
Chunyan Hou, Email: houchunyan@126.com.
Dongmei Wang, Email: dongmeiwang63@126.com.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36385-1.
References
- 1.Reddy ASN, Ali GS, Celesnik H, Day IS. Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell. 2011;23:2010–2032. doi: 10.1105/tpc.111.084988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.White PJ, Broadley MR. Calcium in plants. Ann Bot. 2003;92:487–511. doi: 10.1093/aob/mcg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hetherington AM, Brownlee C. The generation of Ca2+ signals in plants. Annu Rev Plant Biol. 2004;55:401–427. doi: 10.1146/annurev.arplant.55.031903.141624. [DOI] [PubMed] [Google Scholar]
- 4.Cheval C, Aldon D, Galaud JP, Ranty B. Calcium/calmodulin-mediated regulation of plant immunity. Biochim Biophys Acta. 2013;1833:1766–1771. doi: 10.1016/j.bbamcr.2013.01.031. [DOI] [PubMed] [Google Scholar]
- 5.Reddy AS. Calcium: silver bullet in signaling. Plant Science. 2001;160:381–404. doi: 10.1016/S0168-9452(00)00386-1. [DOI] [PubMed] [Google Scholar]
- 6.Du, L., Yang, T., V. Puthanveettil, S. & W. Poovaiah, B. Decoding of calcium signal through calmodulin: calmodulin-binding proteins in plants. 177–233 (2011).
- 7.Choi MS, et al. Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.) J biol chem. 2005;280:40820–40831. doi: 10.1074/jbc.M504616200. [DOI] [PubMed] [Google Scholar]
- 8.Pant P, Iqbal Z, Pandey BK, Sawant SV. Genome-wide comparative and evolutionary analysis of Calmodulin-binding Transcription Activator (CAMTA) family in Gossypium species. Sci Rep. 2018;8:5573. doi: 10.1038/s41598-018-23846-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahman H, Yang J, Xu YP, Munyampundu JP, Cai XZ. Phylogeny of plant CAMTAs and role of AtCAMTAs in nonhost resistance to Xanthomonas oryzae pv. oryzae. Front Plant Sci. 2016;7:177. doi: 10.3389/fpls.2016.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei M, Xu X, Li C. Identification and expression of CAMTA genes in Populus trichocarpa under biotic and abiotic stress. Sci Rep. 2017;7:17910. doi: 10.1038/s41598-017-18219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y, Park S, Gilmour SJ, Thomashow MF. Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J. 2013;75:364–376. doi: 10.1111/tpj.12205. [DOI] [PubMed] [Google Scholar]
- 12.Galon Y, Finkler A, Fromm H. Calcium-regulated transcription in plants. Mol Plant. 2010;3:653–669. doi: 10.1093/mp/ssq019. [DOI] [PubMed] [Google Scholar]
- 13.Pandey N, et al. CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genomics. 2013;14:216. doi: 10.1186/1471-2164-14-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahman, H., Xu, Y. P., Zhang, X. R. & Cai, X. Z. Brassica napus renome possesses extraordinary high Number of CAMTA genes and CAMTA3 contributes to PAMP triggered immunity and resistance to Sclerotinia sclerotiorum. Front Plant Sci. 7 (2016). [DOI] [PMC free article] [PubMed]
- 15.Yue R, et al. Identification and expression profiling analysis of calmodulin-binding transcription activator genes in maize (Zea mays L.) under abiotic and biotic stresses. Front Plant Sci. 2015;6:576. doi: 10.3389/fpls.2015.00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du L, et al. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature. 2009;457:1154–1158. doi: 10.1038/nature07612. [DOI] [PubMed] [Google Scholar]
- 17.Hou C, Wang Z, Wang D. The effects of Ca2+ on the induction of defence responses by wheat leaf rusts. Acta Agriculturae Boreali-Sinica. 2007;22:145–147. [Google Scholar]
- 18.Guan CL, Hou CY, Wang DM. The role of drugs affecting calcium metabolism and calcium channel on HR induced by wheat leaf rust fungus. Journal of Hebei Agricultural University. 2006;29:4–8. [Google Scholar]
- 19.Zhang B, et al. Effect of intracellular calcium stores on hypersensitive reaction induced by wheat leaf rust fungus: effect of intracellular calcium stores on hypersensitive reaction induced by wheat leaf rust fungus. Acta Agronomica Sinica. 2010;36:833–839. doi: 10.3724/SP.J.1006.2010.00833. [DOI] [Google Scholar]
- 20.Liu G, Hou CY, Wang DM. Calcium influx is required for the initiation of the hypersensitive response of Triticum aestivum to Puccinia recondita f.sp. tritici. Physiol Mol Plant P. 2010;74:267–273. doi: 10.1016/j.pmpp.2010.04.002. [DOI] [Google Scholar]
- 21.Liu G, Dang L, Wang D. The influence of elicitor on the distribution pattern of microtubule and the cytosolic calcium in mesophyll protoplast of wheat. Journal of Molecular Cell Biology. 2007;40:205–213. [PubMed] [Google Scholar]
- 22.Yan AH, Zhang LF, Zhang YW, Wang DM. Early stage SSH library construction of wheat near isogenic line TcLr19 under the stress of Puccinia recondita f. sp. Tritici. Frontiers of Agriculture in China. 2009;3:146–151. doi: 10.1007/s11703-009-0045-7. [DOI] [Google Scholar]
- 23.Jauregui E, Du L, Gleason C, Poovaiah BW. Autophosphorylation of calcium/calmodulin-dependent protein kinase (CCaMK) at S343 or S344 generates an intramolecular interaction blocking the CaM-binding. Plant Signal Behav. 2017;12:e1343779. doi: 10.1080/15592324.2017.1343779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma W, Berkowitz GA. Ca2+ conduction by plant cyclic nucleotide gated channels and associated signaling components in pathogen defense signal transduction cascades. New Phytol. 2011;190:566–572. doi: 10.1111/j.1469-8137.2010.03577.x. [DOI] [PubMed] [Google Scholar]
- 25.Kim YS, et al. CAMTA-mediated regulation of salicylic acid immunity pathway genes in Arabidopsis exposed to low temperature and pathogen infection. Plant Cell. 2017;29:2465–2477. doi: 10.1105/tpc.16.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu Y, Xi J, Du L, Suttle JC, Poovaiah BW. Coupling calcium/calmodulin-mediated signaling and herbivore-induced plant response through calmodulin-binding transcription factor AtSR1/CAMTA3. Plant Mol Biol. 2012;79:89–99. doi: 10.1007/s11103-012-9896-z. [DOI] [PubMed] [Google Scholar]
- 27.Reddy VS, Ali GS, Reddy AS. Genes encoding calmodulin-binding proteins in the Arabidopsis genome. J Biol Chem. 2002;277:9840–9852. doi: 10.1074/jbc.M111626200. [DOI] [PubMed] [Google Scholar]
- 28.Reddy AS, Ben-Hur A, Day IS. Experimental and computational approaches for the study of calmodulin interactions. Phytochemistry. 2011;72:1007–1019. doi: 10.1016/j.phytochem.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 29.Popescu SC, et al. Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc Natl Acad Sci USA. 2007;104:4730–4735. doi: 10.1073/pnas.0611615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenzoni G, Liu J, Knight MR. Predicting plant immunity gene expression by identifying the decoding mechanism of calcium signatures. New Phytol. 2018;217:1598–1609. doi: 10.1111/nph.14924. [DOI] [PubMed] [Google Scholar]
- 31.Yang T, Poovaiah BW. A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J Biol Chem. 2002;277:45049–45058. doi: 10.1074/jbc.M207941200. [DOI] [PubMed] [Google Scholar]
- 32.Nie H, et al. SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant physiol. 2012;158:1847–1859. doi: 10.1104/pp.111.192310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouche N, Scharlat A, Snedden W, Bouchez D, Fromm H. A novel family of calmodulin-binding transcription activators in multicellular organisms. J Biol Chem. 2002;277:21851–21861. doi: 10.1074/jbc.M200268200. [DOI] [PubMed] [Google Scholar]
- 34.Qiao M, et al. Changes of nitric oxide and its relationship with H2O2 and Ca2+ in defense interactions between wheat and Puccinia Triticina. PLOS ONE. 2015;10:e0132265. doi: 10.1371/journal.pone.0132265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset1, Dataset2, Dataset3, Dataset4, Dataset5, Dataset6.