Abstract
The last decade has seen an exponential expansion of interest in conjugating multiple enzymes of cascades in close proximity to each other, with the overarching goal being to accelerate the overall reaction rate. However, some evidence has emerged that there is no effect of proximity channeling on the reaction velocity of the popular GOx-HRP cascade, particularly in the presence of a competing enzyme (catalase). Herein, we rationalize these experimental results quantitatively. We show that, in general, proximity channeling can enhance reaction velocity in the presence of competing enzymes, but in steady state a significant enhancement can only be achieved for diffusion-limited reactions or at high concentrations of competing enzymes. We provide simple equations to estimate the effect of channeling quantitatively and demonstrate that proximity can have a more pronounced effect under crowding conditions in vivo, particularly that crowding can enhance the overall rates of channeled cascade reactions.
Introduction
Enzyme-catalyzed reactions are probably the most ubiquitous and elegant reactions on Earth. Typically an enzyme-catalyzed reaction does not occur alone, but instead is a part of a natural metabolic pathway or synthetic cascade. A particular role in enzymatic cascades is played by metabolite or substrate channeling, in which the product of one reaction is directly passed to the active site of the next enzyme in a cascade, either via a physical tunnel in an enzyme-enzyme complex1–4, along an ‘electrostatic highway’3–6, or through proximity of two (or more) enzymes7–10.
Substrate channeling in vivo has also been a subject of yet to be resolved debates11–15. Despite work showing the existence of enzyme-enzyme complexes inside living cells16–21, a consensus has not emerged as to whether, and to what extent, channeling occurs in cells and how it influences reaction velocities. In particular, Poshyvailo et al.22 have argued that direct channeling may slow down the reaction velocity and increase the metabolite pool size, while its main benefit is likely to protect metabolites from degradation or competing side reactions.
In biotechnology, on the other hand, a significant effort has been channeled into developing novel methods to bring and keep enzymes together, hoping to accelerate the reactions by decreasing the diffusion path between the enzymes7–9,21,23–33. Fu et al.26 have assembled glucose oxidase (GOx) and horseradish peroxidase (HRP) enzymes on a DNA origami and reported a 15-fold increase of the overall reaction rate. However, in a recent paper, Zhang et al.34 suggested that the enhanced velocity was due to an increased pH at the origami tile, while the reaction velocity was essentially insensitive to the enzyme proximity under conditions close to the steady state. The same conclusion was also reached in the presence of a competing enzyme (catalase), contrary to expectations.
Herein, we analyse proximity channeling in detail and further clarify if and when enzyme proximity would be beneficial to reaction velocity. To achieve this goal we use a theoretical model, in which the concentration of intermediates was obtained in the presence of two enzymes of a cascade (Fig. 1a). We then solved the appropriate diffusion problem and the reaction rates were computed and compared with the corresponding rates of a non-channeled system (see Methods).
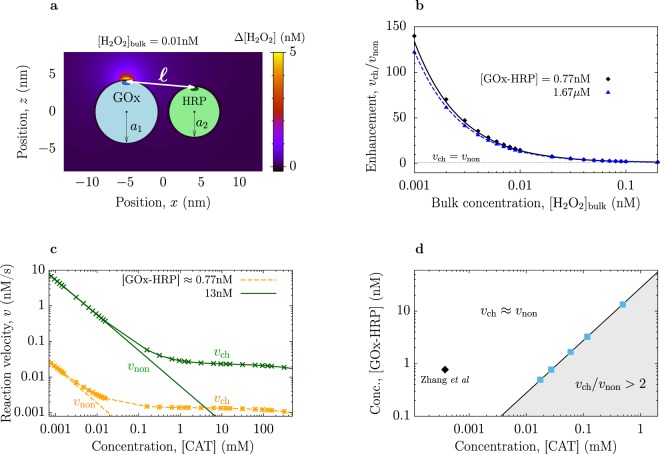
Figure 1.
Effect of proximity channeling on GOx-HRP cascade. (a) A model of a GOx-HRP complex and the distribution of hydrogen peroxide (H2O2, intermediate of the GOx-HRP cascade) shown as a deviation from the bulk concentration, Δ[H2O2] = [H2O2] − [H2O2]bulk. (b) Enhancement vch/vnon due to proximity channeling as a function of the bulk concentration of hydrogen peroxide. Symbols show the results of the full numerical calculations and the lines have been obtained using equation (1). (c) Reaction velocities of the channeled (symbols) and non-channeled (lines) reactions as functions of concentration of catalase (enzyme competing with HRP for hydrogen peroxide). (d) Channeling diagram showing the region where the enzyme proximity can at least double the reaction velocity. The diagram is drawn in the plane of the concentration of enzyme complexes and the concentration of catalase. We have assumed that [GOx-HRP] is the same as [GOx] = [HRP] in the non-channeled system. Squares show the results of the full numerical calculations and the line has been obtained using equation (3). The diamond shows the system with the maximal concentration of catalase studied experimentally by Zhang et al.34.
Before discussing the results of those calculations, however, it is vital to stress, again27,35,36, that, in steady state, enzyme proximity cannot accelerate the reaction velocity in the absence of competing enzymes or degrading intermediates. Indeed, the conservation law dictates that for a tandem reaction (a cascade of two enzyme-catalyzed reactions), the rate of production of intermediates and the rate of their conversion into the final product must be equal in steady state (otherwise the concentration of intermediates would change over time). This means that the reaction velocity is influenced only by the production rate of the first enzyme, which is the same in the channeled and non-channeled system (see equations (6) and (11) in Methods). The conservation-law argument is mathematically rigorous and independent of the position of enzymes, diffusion rates and other parameters. Thus, we believe that enzyme proximity cannot accelerate reactions at low enzyme concentrations or for ‘perfect enzymes’, as suggested in a recent review15. In steady state, an intermediate does not have to diffuse directly between the enzymes for the reaction to occur, since it is already available in the bulk solution; in other words, the diffusion path of an intermediate is determined by the concentration of the second enzyme and by the concentration of intermediates, which are ultimately non-zero in steady state (see equation (10) with kdeg = 0 in Methods).
We decided to consider the effect of proximity before steady-state is achieved. In Fig. 1a we plot the distribution of intermediates (hydrogen peroxide) for the GOx-HRP system of Zhang et al.34 (Supplementry Note S1). For this relatively low non-steady state bulk concentration of the hydrogen peroxide, [H2O2]bulk = 0.01 nM, the enzyme proximity had a dominant contribution on the reaction velocity. Indeed, for the non-channeled system the reaction velocity was vnon = kHRP[HRP][H2O2]bulk = 0.1 pM/s, and vch ≈ 1.5 pM/s for the channeled system, giving the enhancement vch/vnon ≈ 15 (albeit at extremely low reaction rates). At higher bulk concentrations, however, the difference Δ[H2O2] = [H2O2]at−HRP − [H2O2]bulk became small, as compared to [H2O2]bulk, rendering proximity channeling useless. Enzyme proximity accelerated the GOx-HRP cascade at very low H2O2 concentrations, with saturation occurring at a concentration as low as 0.1 nM (Fig. 1b). Assuming [H2O2] = 0 at time t = 0, we estimated that the GOx-HRP cascade would benefit from proximity channeling merely within the first τ1 ≈ 87 ms (see equation (9) in Methods); for other systems τ1 could be larger15,36. However, we shall show that enzyme proximity can accelerate cascade reactions also in steady state, but only in the presence of competing enzymes or if intermediates degrade. A similar conclusion has been reached by Idan and Hess36, who related reaction acceleration to the ratio between the life-time of intermediates and the characteristic time of temporal boost (analogous to our τ1). Here, we provide an alternative clarification and derive simple expressions to estimate rate enhancement quantitatively.
In order to achieve a deeper insight into the proximity-induced enhancement, we solved analytically the diffusion problem for intermediates and the first enzyme of a cascade; then, we estimated the rate enhancement by looking at the excess concentration of intermediates at the location of the active site of the second enzyme. Limiting our considerations to the first term in an infinite series expansion of the exact solution, we have arrived at a simple approximate equation (Supplementry Note S4)
| 1 |
where [I]bulk is the bulk concentration of intermediates (taken the same in the channeled and non-channeled systems), is the distance between the active sites of the two enzymes in a complex (Fig. 1a) and b = (3/4π[E1])−1/3 roughly corresponds to the average distance between the enzymes in the non-channeled system (here [E1] is the enzyme concentration). The reaction constant (in units of s−1) is , assuming , where [S] is the concentration of the first enzyme’s substrate (glucose in the case of GOx) and k(1) and are the turnover number and the Michaelis-Menten constant of the first enzyme, respectively; is the rate due to diffusion over distance (measured in units on nm3/s or, after the appropriate conversion, in units of nM−1s−1), where D is the mutual diffusion coefficient of the first enzyme and intermediates (GOx and hydrogen peroxide for the GOx-HRP system). Figure 1b shows a reasonably good agreement between equation (1) and full numerical calculations.
Physical interpretation of equation (1) is simple. Neglecting the term for simplicity, we found that channeling increased reaction rate when . Under these conditions, the local production of intermediates would be much faster than the rate at which they diffuse away. Clearly, in this case the concentration of intermediates would be enhanced locally at the first enzyme, and hence the reaction velocity could benefit from the enzyme proximity.
Thus, proximity could enhance reaction velocity, but only when the concentration of intermediates in a solution is low, which occurs during the initial stage of reaction, as discussed, or in the presence of competing enzymes. We analysed the effect of competing enzymes by introducing the degradation rate of intermediates kdeg (see Methods); the consumption by competing enzymes was taken into account by setting for low concentrations of intermediates, , where and are the Michaelis-Menten constant and the turnover number, respectively, and [Eco] the concentration of a competing enzyme. The reaction velocities of the channeled and non-channeled GOx-HRP systems, as functions of the concentration of catalase (a competing enzyme that decomposes H2O2 into water and oxygen), showed that the reaction could be accelerated by proximity channeling, but only at high catalase concentrations, with the threshold concentration increasing for increasing concentration of GOx-HRP complexes (Fig. 1c). We compiled our data on a channeling diagram (Fig. 1d), which showed the regimes under which channeling does or does not impact the reaction velocity.
In order to estimate the effect of channeling in the presence of competing enzymes, we replaced the bulk concentration of intermediates in equation (1), [I]bulk, by the steady-state value for a system with non-zero kdeg. This turned out to provide a good approximation for the actual reaction velocity (see below) because the effect of degradation or competing consumption was negligible on the length scales determined by the separation of enzymes in an enzyme-enzyme conjugate. Indeed, in our case the main contribution to enhancement was due to the difference between the bulk and local (at the second enzyme) concentration of intermediates. Straightforward calculations gave a simple expression for the degree of acceleration (Supplementary Note S4B)
| 2 |
where [E] is the concentration of enzyme complexes ([E] ≡ [E12] = [E1] = [E2]) and is the rate constant of the second enzyme (in units of nM−1s−1), assuming .
Equation (2) has two limiting cases. For the proximity-induced rate enhancement is influenced by how slowly the intermediates diffuse on the time scale set by the degradation rate. In this case, straightforward algebra gave a (δv + 1)-fold faster rate due to channeling for the concentrations of enzyme complexes .
In the opposite case, , we have , and the reaction acceleration is determined by how slowly the intermediates diffuse on the time scale set by the reaction rate of the second enzyme. In this diffusion-limited case, fast conversion of intermediates by the second enzyme implies their (locally) low concentration, making it beneficial to keep the two enzymes together in order to increase [I] locally at the second enzyme. Under these conditions, the effect of channeling was independent of the enzyme concentration and degradation rate (but the concentration of intermediates away from an enzyme complex was kept the same as in the non-channeled system).
In a general case, but neglecting the term for simplicity (which is valid for low mM for nm), we obtained a simple linear relation between the concentration of enzyme complexes and the degradation rate of intermediates
| 3 |
This equation gives the concentration of enzyme complexes at which enzyme proximity provides acceleration δv at a given rate of degradation or competing consumption of intermediates. Note that [E]thr is inversely proportional to δv, hence a higher δv will be obtained in a system with the concentration of enzymes below [E]thr. In other words, [E]thr can be viewed as a threshold value below which the proximity of enzymes leads to an acceleration higher than δv. This is shown in Fig. 1d, where equation (3) is represented by the solid line, showing an excellent agreement with the full numerical calculations. Thus, equations (2) and (3) can be convenient estimators of proximity-induced rate enhancement.
Taking now the values of the rate constants from Zhang et al.34, and using , we found for δv = 2 and nm. For [GOx-HRP] =0.77 nM of ref.34, we found the threshold concentration, above which proximity channeling could give a noticeable (more than two-fold) increase in reaction velocity, [CAT]thr ≈ 28 μM. The catalase concentrations in all cases studied in ref.34 were a few orders of magnitude lower than [CAT]thr (see Fig. 5 of ref.34 the highest value from this reference is marked by the diamond in Fig. 1d).
In order to estimate the values of kcat/KM for which channelling could accelerate the overall reaction rate, we took the diffusion constant of substrates D = 0.6 nm2/ns, corresponding to glucose, and the distance between the active sites of two enzymes nm. Using equation (2), we found that enzyme proximity could enhance reaction velocity if at least one of the rate constants, either of the second enzyme of a cascade, , or of a competing enzyme, , would be comparable to or greater than ≈5 nM−1 s−1. While there are enzymes able to catalyze reactions at rates ∼109 M−1 s−1 (e.g. triosephosphate isomerase and carbonic anhydrase), such high rates are rather rare in enzyme kinetics37. Thus, an ‘average’ cascade is unlikely to benefit from enzyme proximity in terms of its reaction velocity. However, our estimate has been done for comparable enzyme concentrations ([E] ≈ [Eco]), increasing the concentration of competing enzymes, [Eco], can make proximity channeling beneficial also for slower enzymes.
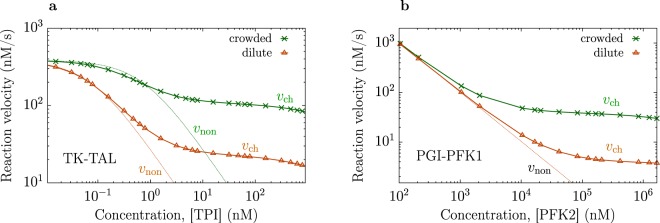
Equation (2) shows that the effect of enzyme proximity becomes more pronounced when the diffusion of intermediates slows down. Indeed, in this case decreases, leading to a higher value of vch/vnon. Reduced rates of diffusion occur under crowding conditions in vivo, where the diffusion coefficients are an order of magnitude lower than in a diluted (typical in vitro) system38–44. Figure 2a shows the effect of crowding on reaction velocities for the transketolase-transaldolase (TK-TAL) system of the pentose phosphate pathway, with glyceraldehyde 3-phosphate (g3p) as an intermediate (Supplementary Note S2). In order to demonstrate this effect, we selected triose-phosphate isomerase (TPI) as a competing enzyme. We found that, perhaps counter-intuitively, crowding increased the overall reaction rate (all considered reactions, except of TPI-catalyzed, were activity-limited, and we assumed their rates crowding-independent; for many enzymes, dependence on crowding is indeed weak45,46, as compared to the effect of channeling). In addition, the regime where channeling enhanced the reaction velocity comparing to the non-channeled system was shifted to higher TPI concentrations. The reason is that TPI is diffusion limited and has a lower efficiency in the crowded system, hence a higher concentration of TPI was required to induce a comparable effect.
Figure 2.
Effect of proximity channeling in dilute and crowded systems. (a) Reaction velocities for channeling of glyceraldehyde 3-phosphate (g3p, intermediate) in the transketolase-transaldolase (TK-TAL) part of the pentose phosphate pathway. A competing enzyme was triose-phosphate isomerase (TPI), which is diffusion limited. (b) Reaction velocities for channeling of fructose 6-phosphate (f6p, intermediate) in the glucose-6-phosphat-isomerase and phosphofructokinase-1 (PGI-PFK1) part of the glycolysis. A competing enzyme was phosphofructokinase-2 (PFK2). All enzymes in (b) are activity limited. The diffusion coefficients of g3p and f6p were taken D = 0.6 nm2/ns in a dilute and D = 0.06 nm2/ns in a crowded system. Dash lines show the reaction velocities for the non-channeled systems. The concentration of enzyme and enzyme conjugates were 26 nM in all systems.
Figure 2b shows the effect of crowding on the glucose-6-phosphate isomerase (PGI) and phosphofructokinase-1 (PFK1) system of the glycolytic pathway (Supplementary Note S3), with PFK2 as a ‘competing enzyme’ and fructose-6-phosphate as an intermediate. In this case all enzymes were activity-limited; here, crowding shifted the region where channeling became beneficial to lower PFK2 concentrations. This is because decreased, while (and hence kdeg, see equation (2)) remained the same, as the system became crowded.
To summarize, we have examined the behaviour of cascade reactions when enzymes are in a close proximity to each other, which is known as proximity channeling. We have shown that the reaction velocity of the popular GOx-HRP cascade is practically independent of channeling for concentrations under typical operating conditions (Fig. 1b–d), thus rationalizing quantitatively the recent experimental results of Zhang et al.34. We also showed that, in general, channeling may lead to orders of magnitude increases in reaction velocities, but a significant enhancement can be obtained only for diffusion-limited reactions (with M−1 s−1) or at high concentrations of competing enzymes. We provide easy-to-use formulae, equations (2, 3), which allow one to estimate the effect of channeling in biotechnological applications and under in vivo conditions prior to experiments. These equations corroborate that channeling may have a more pronounced effect in vivo, where diffusion of enzymes and metabolites is significantly slowed down38–44. In particular, crowding enhances the overall rates of channeled cascade reactions, and can shift the region where proximity becomes more effective towards higher or lower concentrations of competing enzymes, depending on enzyme efficiencies (Fig. 2). Thus, while it is interesting to analyse proximity channeling with in vitro systems, it is vital to consider the effect in vivo in order to reveal the biological and evolutionary role, and the physiological significance of formation of enzyme complexes in living cells15,21,47.
Methods
The intermediate substrates are described by a continuous field C(r, t) ≡ [I] that satisfies the diffusion equation
| 4 |
where kdeg is the degradation rate and/or the rate due to competing side reactions, and D is the mutual diffusion coefficient of intermediates and enzyme-enzyme complexes.
The enzymes are modeled as spherical particles containing small active sites with opening angle α on their surfaces (Fig. 1a and Supplementary Fig. S1). The boundary conditions on the enzyme active sites are
| 5a |
for the first enzyme, and
| 5b |
for the second enzyme of a cascade, and we applied von Neumann boundary conditions () to the rest of the enzyme surfaces. Here is a unit vector normal to the surface.
The total production rate is , where V is the volume of a computational box and dS the surface element. We need to solve only the time independent equation, dC/dt = 0, in order to find the steady-state reaction velocity, of interest in this work.
In steady-state the appropriate boundary condition away from the enzymes is vanishing concentration gradient (von Neumann boundary condition). In this case, and setting kdeg = 0, we obtained by integrating equation (4) once
| 6 |
where A1 is the surface area of the active site of the first enzyme. This equation expresses the conservation law and means that, in steady state and for kdeg = 0, the production rate is determined solely by the production rate of the first enzyme, which is exactly the production rate in the non-channelled system (see equation (11) and below). This argument does not apply to systems that are not in steady state or if kdeg ≠ 0.
The solution to equation (4) has been obtained numerically using F3DM library48. We applied von Neumann boundary condition away from the enzymes to produce all plots, except of Fig. 1a,b (where we kept the concentration of intermediates fixed, as denoted on the plots). The analytical steady-state solution for the system with just the first enzyme allowed us to obtain approximate equations (1, 2); the derivation is presented in Supplementary Note S4.
Similar approaches have been used before and proved to provide correct results6,49.
Homogeneous non-channeled system
We also considered an equivalent homogeneous bulk system, where enzymes do not form complexes and are well mixed. An equation describing the evolution of this system is
| 7 |
where , i = 1, 2, are enzyme’s rate constants, kdeg the degradation rate of intermediates, [I] is the concentration of intermediates and [Ei] the concentration of enzyme Ei. The production rate is .
Equation (7) obeys a simple analytical solution
| 8 |
where is the relaxation or decay constant. Incidentally, this equation gives for the time at which [I](t) = [I]0
| 9 |
The steady-state concentration of intermediates is
| 10 |
and hence the reaction velocity
| 11 |
which becomes for kdeg = 0 if we take the same enzyme concentrations [E] = [E1] = [E2] in order to compare with the channeled system.
To relate enzyme concentrations in the non-channeled system and in the channeled system of Fig. 1a, we considered the computational box as a ‘unit cell’ so that the corresponding enzyme concentration [Ei] = 1/V, where V is the box volume. The rate constants ki and are related by , where Ai is the surface area of the active site of the i’th enzyme. These relations imply that for kdeg = 0 the steady-state velocities of the channeled and non-channeled systems are the same (equations (6) and (11)).
Supplementary information
Reaction parameters and the derivation for the degree of acceleration
Acknowledgements
This work was supported by NCN grant No. 2017/25/B/ST3/02456 to S.K. and by Project FF-30F (No. 0116U001539) to A.K. We are grateful to Martin Cerff for helpful discussions regarding the PGI-PFK system.
Author Contributions
S.K. and E.v.L. initiated the research, A.K. and S.K. derived the equations, A.K., S.C. and S.K. performed numerical calculations, all authors jointly analyzed the results, S.K. and A.J.R. wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37034-3.
References
- 1.Smith NE, Vrielink A, Attwood PV, Corry B. Biological channeling of a reactive intermediate in the bifunctional enzyme dmpfg. Biophys. J. 2012;102:868–877. doi: 10.1016/j.bpj.2012.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith NE, Vrielink A, Attwood PV, Corry B. Binding and channeling of alternative substrates in the enzyme dmpfg: a molecular dynamics study. Biophys. J. 2014;106:1681. doi: 10.1016/j.bpj.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miles EW, Rhee S, Davies DR. The molecular basis of substrate channeling. J. Biol. Chem. 1999;274:12193–12196. doi: 10.1074/jbc.274.18.12193. [DOI] [PubMed] [Google Scholar]
- 4.Huang X, Holden HM, Raushel FM. Channeling of substrates and intermediates in enzyme-catalyzed reactions. Annu. Rev. Biochem. 2001;70:149–180. doi: 10.1146/annurev.biochem.70.1.149. [DOI] [PubMed] [Google Scholar]
- 5.Elcock AH, Huber GA, McCammon JA. Electrostatic channeling of substrates between enzyme active sites: Comparison of simulation and experiment. Biochem. 1997;36:16049–16058. doi: 10.1021/bi971709u. [DOI] [PubMed] [Google Scholar]
- 6.Metzger VT, Eun C, Kekenes-Huskey PM, Huber G, McCammon JAn-drew. Electrostatic channeling in p. falciparum dhfr-ts: Brownian dynamics and smoluchowski modeling. Biophys. J. 2014;107:2394–2402. doi: 10.1016/j.bpj.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y-HP. Substrate channeling and enzyme complexes for biotechnological ap-plications. Biotechnol. Adv. 2011;29:715–725. doi: 10.1016/j.biotechadv.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 8.You C, Myung S, Zhang Y-HP. Facilitated substrate channeling in a self-assembled trifunctional enzyme complex. Angew. Chem. Int. Ed. 2012;51:8787–8790. doi: 10.1002/anie.201202441. [DOI] [PubMed] [Google Scholar]
- 9.You C, Zhang Y-HP. Annexation of a high-activity enzyme in a synthetic three-enzyme complex greatly decreases the degree of substrate channeling. ACS Synth. Biol. 2014;3:380–386. doi: 10.1021/sb4000993. [DOI] [PubMed] [Google Scholar]
- 10.Jeon EY, Baek AH, Bornscheuer UT, Park JB. Enzyme fusion for whole-cell biotransformation of long-chain sec-alcohols into esters. Appl. Microbiol. Biotechnol. 2015;99:6267. doi: 10.1007/s00253-015-6392-9. [DOI] [PubMed] [Google Scholar]
- 11.Wu X-M, Gutfreund H, Lakatos S, Chock PB. Substrate channeling in glycolysis: a phantom phenomenon. Proc. Natl. Acad. Sci. USA. 1991;88:497–501. doi: 10.1073/pnas.88.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornish-Bowden A, Cárdenas ML. Channelling can affect concentrations of metabolic intermediates at constant net flux: artefact or reality? Eur. J. Biochem. 1993;213(87–265):92. doi: 10.1111/j.1432-1033.1993.tb17737.x. [DOI] [PubMed] [Google Scholar]
- 13.Mendes P, Kell DB, Westerhoff HV. Why and when channelling can decrease pool size at constant net flux in a simple dynamic channel. Biochim. et Biophys. Acta. 1996;1289:175–186. doi: 10.1016/0304-4165(95)00152-2. [DOI] [PubMed] [Google Scholar]
- 14.Cornish-Bowden A, Cardenas ML, Letelier J-C, Soto-Andradec J, Abarzuac FG. Understanding the parts in terms of the whole. Biol. Cell. 2004;96(713–271):717. doi: 10.1016/j.biolcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Sweetlove, L. J. & Fernie, A. R. The role of dynamic enzyme assemblies and substrate channelling in metabolic regulation. Nat. Commun.9, 2136 (2018). [DOI] [PMC free article] [PubMed]
- 16.Shearer G, Lee JC, Koo JA, Kohl DH. Quantitative estimation of channeling from early glycolytic intermediates to CO2 in intact Escherichia coli. FEBS Journal. 2005;272:3260–3269. doi: 10.1111/j.1742-4658.2005.04712.x. [DOI] [PubMed] [Google Scholar]
- 17.Srivastava D, et al. Crystal structure of the bifunctional proline utilization a flavoenzyme from bradyrhizobium japonicum. Proc. Natl. Acad. Sci. USA. 2010;107:2878–2883. doi: 10.1073/pnas.0906101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harkewal Singh BW, Arentson DF, Becker, John JT. Structures of the puta peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site. Proc. Natl. Acad. Sci. USA. 2014;111:3389–3394. doi: 10.1073/pnas.1321621111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Proschel M, Detsch R, Boccaccini AR, Sonnewald U. Engineering of metabolic pathways by artificial enzyme channels. Front. Bioeng. Biotechnol. 2015;3:00168. doi: 10.3389/fbioe.2015.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanyal N, Arentson BW, Luo M, Tanner JJ, Becker DF. First evidence for substrate channeling between proline catabolic enzymes. J. Biol. Chem. 2015;290:2225–2234. doi: 10.1074/jbc.M114.625483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt DL, An S. Spatial organization of metabolic enzyme complexes in cells. Biochemistry. 2017;56:3184–3196. doi: 10.1021/acs.biochem.7b00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poshyvailo L, Lieres Evon, Kondrat S. Does metabolite channeling accelerate enzyme-catalyzed cascade reactions? PLoS One. 2017;12:e0172673. doi: 10.1371/journal.pone.0172673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchner A, Tostevin F, Gerland U. Clustering and optimal arrange-ment of enzymes in reaction-diffusion systems. Phys. Rev. Lett. 2013;110:208104. doi: 10.1103/PhysRevLett.110.208104. [DOI] [PubMed] [Google Scholar]
- 24.Jandt U, You C, Zhang Y-HP, Zeng A-P. Compartmentalization and metabolic channeling for multienzymatic biosynthesis: Practical strategies and modeling approaches. Adv. Biochem. Eng. Biotechnol. 2013;137:41–65. doi: 10.1007/10_2013_221. [DOI] [PubMed] [Google Scholar]
- 25.Castellana M, et al. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat. Biotechnol. 2014;32:1011–1018. doi: 10.1038/nbt.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu J, et al. Multi-enzyme complexes on dna scaffolds capable of substrate channelling with an artificial swinging arm. Nat. Nanotech. 2014;9:531–536. doi: 10.1038/nnano.2014.100. [DOI] [PubMed] [Google Scholar]
- 27.Wheeldon I, et al. Substrate channelling as an approach to cascade reactions. Nat. Chemistry. 2016;8:299–309. doi: 10.1038/nchem.2459. [DOI] [PubMed] [Google Scholar]
- 28.Abernathy MH, He L, Tang YJ. Channeling in native microbial pathways: Implications and challenges for metabolic engineering. Biotechnol. Adv. 2017;35:805–814. doi: 10.1016/j.biotechadv.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Rabe KS, Muoller J, Skoupi M, Niemeyer CM. Cascades in com- partments: En route to machine-assisted biotechnology. Angew. Chem. Int. Ed. 2017;56:13574–13589. doi: 10.1002/anie.201703806. [DOI] [PubMed] [Google Scholar]
- 30.Oyetunde T, Bao FS, Chen J-W, Martin HG, Tang Y-jieJ. Leveraging knowledge engineering and machine learning for microbial bio-manufacturing. Biotechnol. Adv. 2018;36:1308–1315. doi: 10.1016/j.biotechadv.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 31.Lee MJ, et al. Engineered synthetic scaffolds for organizing proteins within the bacterial cytoplasm. Nat. Chem. Biol. 2018;14:142–147. doi: 10.1038/nchembio.2535. [DOI] [PubMed] [Google Scholar]
- 32.Wang, T., Qin, X., Liang, C. & Yuan, H. Engineering substrate channel- ing in biosystems for improved efficiency. J. Chem. Technol. Biotechnol.93, 3364–3373, 10.1002/jctb.5731 (2018).
- 33.Ozbakir, H. F., Garcia, K. E. & Banta, S. Creation of a formate: malate oxidore-ductase by fusion of dehydrogenase enzymes with PEGylated cofactor swing arms. Protein Eng. Des. Sel.31, 103–108, 10.1093/protein/gzy005 (2018). [DOI] [PubMed]
- 34.Zhang Y, Tsitkov S, Hess H. Proximity does not contribute to activity enhancement in the glucose oxidase-horseradish peroxidase cascade. Nature Comm. 2016;7:13982. doi: 10.1038/ncomms13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Idan O, Hess H. Engineering enzymatic cascades on nanoscale scaffolds. Curr. Opin. Biotechnol. 2013;24:606–611. doi: 10.1016/j.copbio.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Idan O, Hess H. Origins of activity enhancement in enzyme cascades on scaffolds. ACS Nano. 2013;7:8658–8665. doi: 10.1021/nn402823k. [DOI] [PubMed] [Google Scholar]
- 37.Bar-Even A, et al. The moderately efficient enzyme: Evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry. 2011;50:4402–4410. doi: 10.1021/bi2002289. [DOI] [PubMed] [Google Scholar]
- 38.Rohwer JM, Postma PW, Kholodenko BN, Westerhoff HV. Implications of macromolecular crowding for signal transduction and metabolite channeling. Proc. Natl. Acad. Sci. USA. 1998;95:10547–10552. doi: 10.1073/pnas.95.18.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emmanuel Dauty, Verkman AS. Molecular crowding reduces to a similar extent the diffusion of small solutes and macromolecules: measurement by fluorescence correlation spec- troscopy. J. Mol. Recognit. 2004;17:441–447. doi: 10.1002/jmr.709. [DOI] [PubMed] [Google Scholar]
- 40.Allen P. Minton, Macromolecular crowding. Curr. Biol. 2006;16:R269–R271. doi: 10.1016/j.cub.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 41.McGuffee SR, Elcock AH. Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput. Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kondrat S, Zimmermann O, Wiechert W, Lieres Evon. The effect of composition on diffusion of macromolecules in crowded environment. Phys. Biol. 2015;12:046003. doi: 10.1088/1478-3975/12/4/046003. [DOI] [PubMed] [Google Scholar]
- 43.Blanco PM, Garces JL, Madurga S, Mas F. Macromolecular diffusion in crowded media beyond the hard-sphere model. Soft Matter. 2018;14:3105–3114. doi: 10.1039/C8SM00201K. [DOI] [PubMed] [Google Scholar]
- 44.Kondrat, S. Physics and modelling of intracellular diffusion. In Physics of Life, Vol. 158 Chap. D1, http://arxiv.org/abs/1810.05496v1 (Forschungszentrum Jülich Publishing, 2018).
- 45.Pastor I, et al. Effect of crowding by dextrans in enzymatic reactions. Biophys. Chem. 2014;185:8–13. doi: 10.1016/j.bpc.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 46.David Gomez, Klumpp S. Biochemical reactions in crowded environments: revisiting the effects of volume exclusion with simulations. Front. Phys. 2015;3:45. [Google Scholar]
- 47.Kastritis, P. L. & Gavin, A.-C. Enzymatic complexes across scales, Essays Biochem. EBC20180008, 10.1042/ebc20180008. (2018). [DOI] [PMC free article] [PubMed]
- 48.Kondrat, S. F3DM library and tools, The F3DM library can be downloaded from http://sourceforge.net/projects/f3dm/.
- 49.Song Y, et al. Finite element solution of the steady-state Smoluchowski equation for rate constant calculations. Biophys. J. 2004;86:2017–2029. doi: 10.1016/S0006-3495(04)74263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reaction parameters and the derivation for the degree of acceleration