Abstract
Thioredoxin‐related protein of 14 kDa (TRP14; also named TXNDC17 for thioredoxin domain‐containing protein 17) is a highly conserved and ubiquitously expressed oxidoreductase. It is expressed in parallel with thioredoxin 1 (Trx1, TXN; TXN1), an efficient substrate for the mammalian cytosolic selenoprotein thioredoxin reductase 1 (TrxR1; TXNRD1). However, TRP14, in sharp contrast to Trx1, cannot support the activities of ribonucleotide reductase, peroxiredoxins or methionine sulfoxide reductases, thus is unable to directly support cell proliferation or antioxidant defence through these pathways. However, TRP14 has been shown to efficiently reduce l‐cystine, which thereby indirectly supports glutathione synthesis. TRP14 can also suppress NF‐κB signalling, is functionally linked to STAT3 signalling, and can directly reactivate oxidized protein‐tyrosine phosphatase PTP1B. Furthermore, TRP14 can efficiently reduce persulfidated or nitrosylated cysteine residues in many proteins, thereby having the capacity to modulate signalling through hydrogen sulfide or NO. Additional bioinformatics analyses and observations suggest further roles for TRP14; therefore, further studies of its functions are warranted. Collectively, the results available suggest that TRP14 is a member of the thioredoxin system dedicated to the control of cellular redox signalling pathways.
Linked Articles
This article is part of a themed section on Chemical Biology of Reactive Sulfur Species. To view the other articles in this section visit http://onlinelibrary.wiley.com/doi/10.1111/bph.v176.4/issuetoc
Abbreviations
- BECN1
beclin‐1
- LC8 (DYNLL1)
dynein light chain 1 (light chain 8)
- PTP1B
protein‐tyrosine phosphatase 1B
- RRM2
ribonucleotide reductase subunit M2
- SNO
S‐nitrosylated cysteine residue
- STRING
database of known and predicted protein–protein interactions (string‐db.org)
- TRP14 (TXNDC17)
thioredoxin‐related protein of 14 kDa
- Trx1 (TXN also named TXN1)
thioredoxin 1
- TrxR1 (TXNRD1)
thioredoxin reductase 1
Introduction
Redox control has a critical role in maintaining cellular functions under diverse growth conditions (Trachootham et al., 2008; Holmstrom and Finkel, 2014; Navarro‐Yepes et al., 2014; Ye et al., 2015). Physiological regulation of cellular redox status occurs mainly through the controlled oxidation of reactive cysteine residues in concert with enzymatically‐controlled reduction of such cysteines. The reductive pathways are controlled by two major systems in the cytosol: the glutathione (GSH) and thioredoxin systems.
In the glutathione system, electrons are transferred from NADPH to glutathione disulphide reductase that reduces glutathione disulphide (GSSG). Subsequently, reduced glutathione propels those electrons to numerous enzymes, including the glutathione peroxidases, glutaredoxins and glutathione S‐transferases (Lillig et al., 2008; Toppo et al., 2009; Deponte, 2013; Ye et al., 2015).
The thioredoxin system is composed of different isoforms of thioredoxin and thioredoxin reductase, where the latter use NADPH to reduce the active site disulfide to a dithiol in their thioredoxin substrates. Cytosolic thioredoxin 1 (Trx1), reduced by TrxR1, is an oxidoreductase with a very broad substrate specificity, thus supporting many important functions in cells (Holmgren, 1995; Martin, 1995; Arner and Holmgren, 2000; Gromer et al., 2004). However, it is likely to be impossible for a single protein to recognize and support all reductive pathways in a cell and, in mammals, several auxiliary Trx‐like proteins complement the functions of Trx1, and are presumed to have both overlapping and specific roles in cells (Miranda‐Vizuete et al., 2004; Jimenez et al., 2006; Funato and Miki, 2007; Su et al., 2007; Dammeyer and Arner, 2011). The Trx‐related protein of 14 kDa (TRP14, encoded by TXNDC17) is an interesting member of the Trx system that has, as yet, remained sparsely characterized.
Early on, TRP14, together with Trx1, was found to be expressed in most rat and human tissues when analysed with immunoblotting (Jeong et al., 2004). If searching current databases for its expression, it indeed seems as if TRP14 (TXNDC17) is expressed in most, if not all, human cell types, cell lines and tissues (e.g. see https://www.genecards.org/cgi‐bin/carddisp.pl?gene=TXNDC17&keywords=txndc17#expression, https://www.proteinatlas.org/ENSG00000129235‐TXNDC17/tissue or https://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.408236). At present, it is unclear what role(s) TRP14 carries out in the cells where it is expressed. Recent insights regarding the potential roles of TRP14 in the control of redox signalling events are the main focus of this review.
Potential roles of TRP14 (TXNDC17) in signalling
Identification and characterizations of TRP14 as a regulator of NF‐κB signalling
Mammalian TRP14 has 123 amino acids and was detected in 2000 by a procedure used to search for proteins containing redox‐sensitive cysteine residues with low pKa in rat brains (Kim et al., 2000). Human TRP14 was subsequently discovered in 2004 by Rhee and co‐workers, who characterized TRP14 as a cytosolic protein that is expressed in most cells and tissues, has a low redox potential and, interestingly, is reduced by cytosolic TrxR1 but, in contrast to Trx1, is unable to reduce ribonucleotide reductase subunit M2 (RRM2), peroxiredoxins or methionine sulfoxide reductase, thus suggesting different functions from those of Trx1 (Jeong et al., 2004). The active site motif of human TRP14 is WCPDC (instead of WCGPC as found in Trx1), and in 2004, the Rhee group also solved the crystal structure of TRP14 at 1.8 Å resolution (PDB entry 1WOU), showing that although TRP14 only shares 20% sequence identity with Trx1, the topological structures of the two proteins are very similar. However, in the vicinity of the active site in TRP14, there is an extended loop, an additional α‐helix, and a different distribution of charged residues compared to Trx1, which may explain the differences in substrate specificities between the two oxidoreductases (Woo et al., 2004). It was also shown in 2004, again by Rhee et al., that TRP14 suppresses TNF‐ α‐induced activation of NF‐κB, JNK and p38 MAPKs, while TRP14 in contrast to Trx1 did not regulate the activities of apoptosis signal‐regulating kinase‐1 (Jeong et al., 2004b). The authors later showed that the TRP14‐mediated suppression of NF‐κB activation was uniquely related to its reduction of dynein light chain LC8 (Jung et al., 2008; Jeong et al., 2009).
Further evolutionary insights regarding TRP14 functions
Early on, it was noted that TRP14 orthologues are found in a large number of species, from bacteria, plants, yeast and mammals, and in an alignment of TRP14 sequences from 42 different species the unusual WCPDC active site was completely conserved (Jeong et al., 2004; Jeong et al., 2009). In the decade that has passed since those analyses, many more organisms have been sequenced, and the TRP14 family of sequences has thus grown considerably. When 111 sequences of TRP14 from 89 compiled animal sequences were analysed at the curated TreeFam database of EMBL‐EBI (http://www.treefam.org/family/TF313854) (Li et al., 2006; Ruan et al., 2008), it became evident that TRP14 orthologues are widely distributed throughout the animal kingdom. However, an analysis of the active site motifs of these sequences revealed that the WCPDC active site is not completely conserved. In some of the sequences, it is instead WCPYC (Figure 1); this motif is the sequence of a typical dithiol glutaredoxin active site (Åslund et al., 1997; Holmgren et al., 1998; Fernandes and Holmgren, 2004; Foloppe and Nilsson, 2004; Lillig et al., 2008). At present, it is not clear whether such ‘WCPYC’ active site proteins, otherwise being closely related to TRP14 sequences, should be considered as glutaredoxins or TRP14 variant proteins. An important definition of a glutaredoxin is that it utilizes glutathione during catalysis (Åslund et al., 1997; Holmgren et al., 1998; Fernandes and Holmgren, 2004; Lillig et al., 2008), and those TRP14 variant proteins thereby need to be enzymatically characterized, before they can be fully classified as being either glutaredoxins or TRP14 proteins. Some additional insights into the potential functions of TRP14 can, however, be deduced from a number of biochemical characterizations of TRP14 that have thus far been made with non‐mammalian TRP14 proteins.
Figure 1.

Evolutionary conservation of TRP14 in animals. The accession numbers and amino acid sequences of the region around the active site WCPDC motif are shown for 111 sequences of TRP14 from 89 species, as generated through the curated TreeFam server (see references and details in the text). The human sequence is boxed for reference. Note that the tryptophan in the active site motif is fully conserved throughout these animal species (green). The two active site cysteine residues (yellow) are almost fully conserved, while a few sequences have monothiol active site motifs with only a single Cys residue. The intervening Pro‐Asp amino acids are also conserved in most species (blue), although in some, Asp is substituted by Tyr (grey), thus resulting in a WCPYC active site motif as typically found in glutaredoxins. See text for further details.
In 2007, a TRP14‐encoding gene named AmphiTRP14 was identified in the amphioxus Brahciostom belcheri, which became the first non‐mammalian TRP14 to be characterized (Jiang et al., 2007). AmphiTRP14 shows 56% identity with human TRP14 and is, as in human TRP14, a 123 amino acid protein having a WCPDC active site motif and an estimated molecular weight of 14 kDa. The protein was found to be expressed mainly in hepatic caecum, ovary and hind‐gut of the amphioxus (Jiang et al., 2007). Another TRP14 orthologue was characterized in the clam Venerupis philippinarum, where it was expressed in all tissues but at highest levels in the hepatopancreas and other organs associated with environmental exposure such as gills, hemocytes and mantles (Wang et al., 2011). Interestingly, the expression of that protein was up‐regulated upon exposure to high levels of cadmium or copper, suggesting that it may have a role in antioxidant responses against metal stress in the clam (Wang et al., 2011).
TRP14 was also identified in the fish group Epinephelus coioides. Similar to TRP14 orthologues of other species, it has 123 amino acids with the conserved active site motif WCPDC, and is widely expressed in different organs of the fish, displaying high levels in the liver, muscle, brain, kidney, skin and gill. Interestingly, in that study, it was shown that transcripts for this TRP14 were up‐regulated when the fish was challenged with the pathogenic Singapore grouper iridovirus, again possibly suggesting some protective function of TRP14 (Wei et al., 2013).
Based upon these analyses of TRP14 orthologues in species other than humans, it is reasoned that TRP14 has a role in protection against challenges with oxidative stress, toxic compounds or viral infections. The fact that the protein is so well preserved during evolution points towards some vital but yet unrecognized role of TRP14 in the survival of cells and organisms. The possibility that TRP14 is not a direct antioxidant, but rather an important redox signalling protein that orchestrates protective responses, should not be disregarded, especially considering its lack of direct support of the activities of antioxidant enzymes such as peroxiredoxins or methionine sulfoxide reductases (Jeong et al., 2004). Some recently discovered redox activities of TRP14 should be considered when assessing the possible functional roles of this protein, as shall be discussed next. Several of the activities are also schematically summarized in Figure 2.
Figure 2.
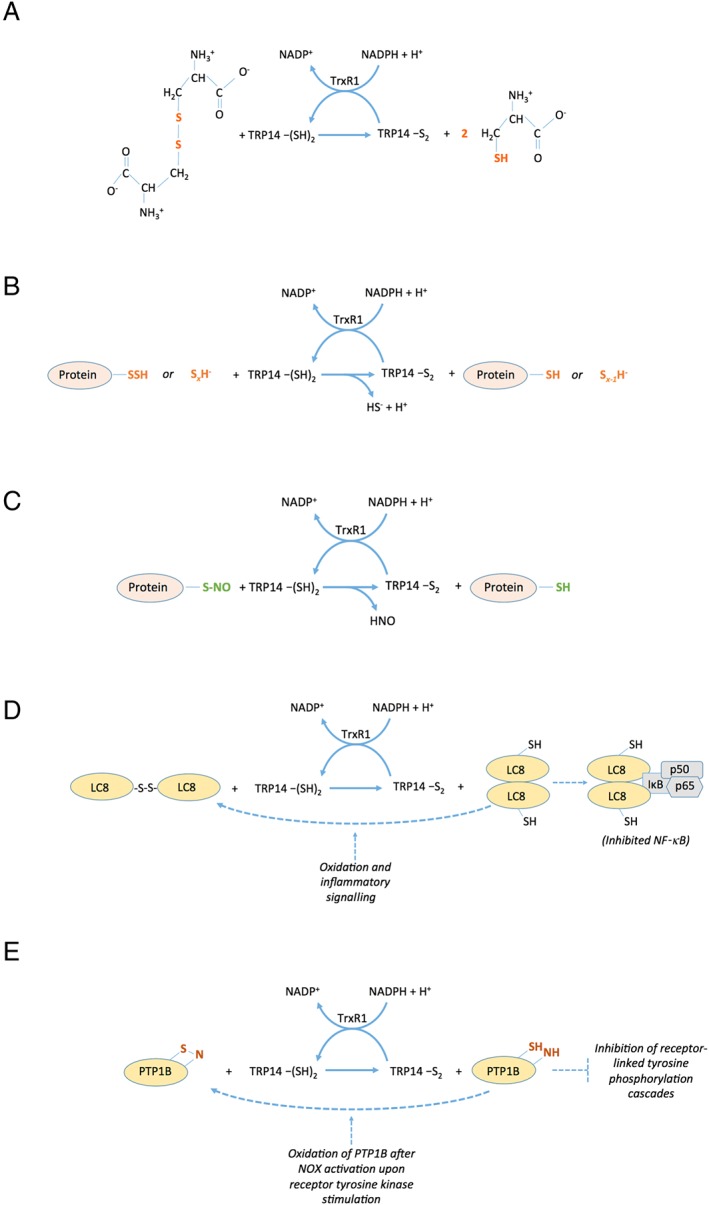
Schematic summary of selected enzymatic reactions catalyzed by TRP14. As is further discussed in the text, TRP14 lacks enzymatic activity with several classic Trx substrates including ribonucleotide reductase, methionine sulfoxide reductases or peroxiredoxins. However, several other enzymatic activities of TRP14, coupled to its own NADPH‐dependent reduction from an active site disulfide to a dithiol by TrxR1, were described in recent years. Some of these are summarized here, with the key features in the different substrates being reduced by TRP14 indicated in colour. The activities include (A) reduction of cystine into two molecules of cysteine, (B) reduction of persulfidated proteins or inorganic polysulfides, (C) reduction of nitrosylated Cys residues (S‐nitrosolyated moieties) in proteins, (D) reduction of a disulfide in LC8 that activates LC8 as an inhibitor of NF‐κB through interactions of reduced LC8 with IκB and (E) reduction of a presumed sulfenylamide motif in oxidized PTP1B that promotes its phosphatase activity, which inhibits receptor‐linked tyrosine phosphorylation cascades.
Cystine reduction by human TRP14
An important antioxidant, protective and cell–cell communicative function of cells is system xC −‐mediated uptake of cystine in exchange with glutamate, with intracellular cystine providing an important source of cysteine for synthesis of glutathione (Conrad and Sato, 2012; Lewerenz et al., 2013). It is in this context that TRP14 could have a potentially important role in that it efficiently reduces the disulfide of cystine, to liberate two cysteine molecules, in a reaction propelled by TrxR1 and NADPH (Pader et al., 2014). Human TRP14 has been shown to be a good substrate for TrxR1, which reduces the active site disulfide of TRP14 to a dithiol using NADPH, while TRP14 is not a substrate for mitochondrial TrxR2 (Jeong et al., 2004; Woo et al., 2004). Interestingly, when coupled to cystine reduction, TRP14 becomes an even better substrate for TrxR1 than Trx1, with the catalytic efficiency (kcat/Km) for TRP14 (2217 min−1·μM−1) being fivefold higher than that for Trx1 (418 min−1·μM−1) (Pader et al., 2014). Moreover, if another substrate of Trx1 other than cystine is present that can be reduced by Trx1 but not by TRP14, then Trx1‐mediated cystine reduction easily becomes blocked, while TRP14 reduction of cystine continues with high efficiency (Pader et al., 2014). These observations possibly suggest that one important role of TRP14, compatible with its involvement in the protection against metal stress or viral infections as implied above, may relate to its catalysis of cystine reduction. Furthermore, several other findings suggest that TRP14 is uniquely positioned to have a role in the control of redox signalling events.
Specific roles of TRP14 in redox signalling
Redox signalling implies the control of cellular functions by enzymatically regulated oxidation or reduction of redox active moieties in key signalling proteins (Winterbourn and Hampton, 2008; Brigelius‐Flohe and Flohe, 2011; Groitl and Jakob, 2014; Sies, 2014; Yoshihara et al., 2014; Sobotta et al., 2015; Winterbourn and Hampton, 2015; Ye et al., 2015). TRP14 can control several distinct steps of redox regulatory pathways, as summarized here.
-
NF‐κB signalling
NF‐κB has critical functions in processes such as inflammation, immunity, cell proliferation, differentiation and survival (Hayden and Ghosh, 2004; Hayden and Ghosh, 2012). TRP14 was initially discovered in 2004 as a suppressor of NF‐κB signalling, as discussed above. Trx1 has also been shown to play different roles in redox regulation of several transcriptional factors, including NF‐κB (Matthews et al., 1992; Mitomo et al., 1994; Heilman and Watson, 2008). However, TRP14 seems to be a more potent regulator of NF‐κB signalling than Trx1, although TRP14 is expressed at lower levels than Trx in cells (Jeong et al., 2004a). In order to try to identify potential cellular substrates of TRP14, a trapping method using an active site monothiol mutant was employed, as typically used for Trx substrates (Motohashi et al., 2001). In that analysis, three potential substrates of TRP14 were identified: LC8, cofilin and ribosomal protein L30, with the reduction of LC8 being suggested to mediate the suppression of NF‐κB signalling by TRP14 (Jeong et al., 2004b; Jung et al., 2008). It was later shown that TRP14 counteracts osteoclast differentiation, which also seems to be an activity that relates to its suppression of NF‐κB activation (Hong et al., 2014). The mode of TRP14‐mediated suppression of NF‐κB is believed to occur through activation of LC8 by reduction of a disulfide in the protein; the reduced LC8 then binds to IκB to prevent its phosphorylation (Jung et al., 2008).
-
STAT3 signalling
By examining the regulation of TRP14 through Chip analyses coupled with a luciferase assay, the transcription factor STAT3 was found to bind the promoter region of the TRP14‐encoding gene TXNDC17 (Zhang et al., 2016). Furthermore, cells became resistant to paclitaxel (Taxol) treatment after either overexpression of TRP14 or stimulation of phosphorylation of STAT3, which in turn correlated with the induction of autophagy (Zhang et al., 2016). These findings suggest that TRP14 is linked to STAT3 signalling, but the full interrelationships between TRP14 and STAT3 signalling are yet unclear. It should be noted that NF‐κB and STAT3 interact and overlap in their signalling pathways (Fan et al., 2013), suggesting that TRP14 can also be indirectly linked to both pathways by being affected, or regulating, either one of these pathways. It is also noteworthy that STAT3 is directly regulated by peroxiredoxin 2 (Sobotta et al., 2015), which in turn is reduced by Trx1 but not by TRP14 (Jeong et al., 2004). These observations suggest that although TRP14 can modulate several pathways of redox signalling, some of these pathways (such as those involving STAT3) can be intricately modulated by many members of the thioredoxin system. The exact roles of TRP14 in this signalling need further scrutiny and may also, noteworthy, be both cell‐, tissue‐ and organism‐specific.
-
Control of autophagy through beclin‐1 (BECN1)
It was found that TRP14 supports the induction of autophagy, as a mechanism of resistance of cancer cells to paclitaxel (Taxol) in a manner dependent upon BECN1. Upon treatment with paclitaxel, an up‐regulation in the expression of both proteins was found. However, when TRP14 expression was lowered using siRNA, the up‐regulation of BECN1 in cells exposed to paclitaxel was prevented, thus suggesting a potential regulation of BECN1 expression by TRP14 (Zhang et al., 2015; Zhang et al., 2016; Zhen et al., 2017). The mechanisms of autophagy as well as BECN1 expression are known to be intimately linked with and affected by oxidative stress and redox signalling pathways (Navarro‐Yepes et al., 2014), but it is yet too early to speculate on exactly how TRP14 directly regulates BECN1 and other processes linked to autophagy.
-
Control of cysteine nitrosylation
One interesting mechanism of redox signalling is that linked to NO and nitrosylation (also called nitrosation) of target cysteine residues in key signalling proteins. Although it may be argued that any reactive cysteine residue can be modified by several different oxidative insults, such as glutathionylation, overoxidation and electrophilic attack, and that nitrosylation may be intrinsically unstable (Wolhuter and Eaton, 2017; Wolhuter et al., 2018), there are nonetheless a vast number of reports suggesting that nitrosylation of cysteine residues may be an important mechanism for control of cell function (Benhar et al., 2009; Wolhuter and Eaton, 2017; Wolhuter et al., 2018). It has long been known that Trx1 can control the nitrosylation state of cysteine residues, both by direct reduction of cysteine S‐nitrosolyated (SNO) groups and by transnitrosylation mechanisms (Benhar et al., 2008; Hashemy and Holmgren, 2008; Benhar et al., 2009; Sengupta and Holmgren, 2013; Benhar, 2015; Benhar, 2016). It is thus interesting to note that TRP14 is also efficient at reducing nitrosylated cysteine residues (Pader et al., 2014). Whether Trx1 and TRP14 have different nitrosylated target proteins, or if one of the two proteins is more dominant than the other in regulating nitrosylation status, is not yet known.
-
Hydrogen sulfide signalling as well as regulation of protein‐tyrosine phosphatase 1B (PTP1B)
Hydrogen sulfide signalling is currently a rapidly expanding research field, with implications in a wide variety of redox signalling systems. There are many mechanisms by which hydrogen sulfide can alter cellular function in a regulated manner. One important mechanism is the formation of persulfidated cysteine residues, that is the addition of one or several sulfur atoms to the sulfur of cysteine through formation of disulfide bonds (Kabil et al., 2014; Nagy, 2015; Kasamatsu et al., 2016; Akaike et al., 2017; Alvarez et al., 2017). It is thus interesting, in relation to the potential functions of TRP14, that TRP14 is efficient at reducing persulfide moieties on cysteine residues in selected target proteins (Doka et al., 2016). Not only can TRP14 reduce persulfidated proteins, as well as inorganic polysulfide compounds in pure systems, but cells with knockdown of TRP14 were also found to display higher levels of endogenous persulfidated proteins, especially after having been challenged with polysulfide (Doka et al., 2016). The reduction of protein persulfides by TRP14 is also interesting in light of the fact that TRP14 can reactivate oxidized forms of protein‐tyrosine phosphatase PTP1B (Dagnell et al., 2013), an important modulator of signalling pathways linked to activation of the tyrosine kinase receptor superfamily (Haj et al., 2003), which has been found to be regulated by persulfidation (Krishnan et al., 2011). It should also be considered that a stable form of PTP1B oxidation is that containing a sulfenylamide motif (Salmeen et al., 2003), which is the form of PTP1B that was believed to be reduced by TRP14 (Dagnell et al., 2013). It should, however, be noted that Trx1, TrxR1 and the glutathione system can also reduce many persulfidated proteins (Doka et al., 2016; Wedmann et al., 2016), as well as regulating PTP1B (Schwertassek et al., 2014; Dagnell et al., 2017). Hence, the importance and specific role(s) of TRP14 in this overall context, if any, remain to be determined.
Additional as yet uncharacterized functions of TRP14?
Less than 20 publications can today be found in PubMed on TRP14 (TXNRD17), and with such few studies published on this protein, it is highly likely that additional unknown functions of TRP14 will be revealed in the coming years. Recently, it was suggested that the clot gene of Drosophila melanogaster, essential for synthesis of the red eye pigment of the fly, encodes for a TRP14 orthologue (Kim, 2018). However, the protein has a typical glutaredoxin active site motif (WCPYC) and indeed exerts high glutaredoxin‐like activities, although it has a predicted overall structure closely similar to TRP14 and also interacts with NF‐κB signalling (Kim, 2018). Thus, the clot protein seems to have features of both glutaredoxins and TRP14. Perhaps it becomes a matter of semantics what protein class it should be considered to belong to.
The fact that TRP14 has such different substrate specificities than Trx1, while both proteins are efficiently supported by cytosolic TrxR1, suggests that TRP14 may have specific but yet unrecognized roles in cells. One such role may perhaps be deduced by analyses of the protein in the STRING database, which was designed with the purpose of identifying possible functions of proteins by analysing co‐expression, fusion genes or physical interactions with other proteins, as reported throughout large database sources (von Mering et al., 2003; Szklarczyk et al., 2017). Performing a STRING analysis for human TRP14 (TXNDC17) indeed gives an interesting result. It is clear that both TRP14 and Trx1 (TXN) are found in an analysis using TRP14 as a query, with the two proteins being linked through TrxR1 (TXNRD1) as a common denominator. More interesting, however, is that the majority of the other proteins predicted to interact with TRP14, or with Trx1, fall into distinct categories rather well separated from each other. For Trx1, the STRING analysis suggests interactions with several proteins involved in the synthesis of deoxyribonucleotides and cell proliferation, with RRM2 being a well‐known and important Trx1 substrate in this context (Arner and Holmgren, 2000; Padovani et al., 2001; Nordlund and Reichard, 2006). It is also interesting to note that Trx1 and TrxR1 are suggested to interact with several enzymes involved in folate metabolism, which may possibly relate to the recently discovered rather strong links between the thioredoxin system and the methionine/homocysteine cycle and thus indirectly folate metabolism (Eriksson et al., 2015; Prigge et al., 2017; Miller et al., 2018). TRP14, however, does not evidently cluster with these systems but seems instead to be predominantly suggested to interact with a number of enzymes that support ubiquitin status and thus proteasome function (Figure 3). Perhaps TRP14 can thereby have functions explaining potential links between TrxR1 activities and proteasome function (Wang et al., 2014; Nagakannan et al., 2016). It should, however, be emphasized that these predictions are primarily based upon database analyses and must be validated experimentally, but could nonetheless suggest lines of forthcoming research further probing the cellular roles of TRP14 in control of redox signalling.
Figure 3.
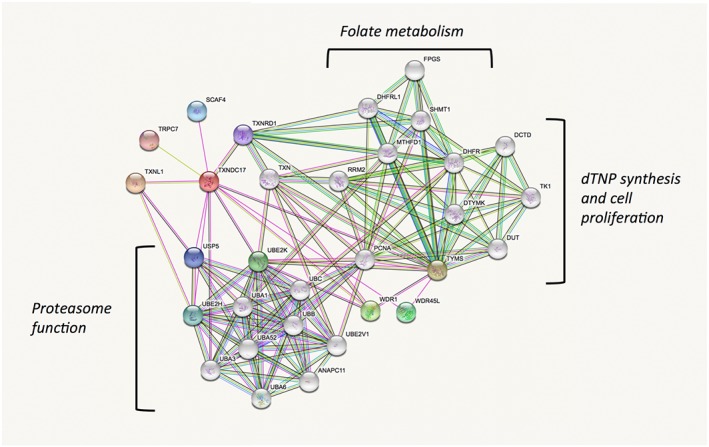
Predicted functional protein partners of TRP14 and its relation to Trx1 according to STRING analysis. The figure summarizes predicted functional protein partners using human TRP14 as query (TXNDC17, red), with the proteins predicted to directly interact with TRP14 shown in coloured nodes and the second shell of interactors shown in non‐coloured nodes. The proposed interactions are those indicated with lines. For full description of the construction of this figure and the prediction basis, please see the text and cited references. Note that TRP14 is correctly predicted to interact with TrxR1 (TXNRD1) and indirectly, through TrxR1, with Trx1 (TXN). The functional overlap between Trx1 and TRP14 is otherwise predicted to be very low, with Trx1 interacting with a cluster of proteins involved in dTNP synthesis and proliferation or folate metabolism, while TRP14 is predicted to possibly interact with ubiquination status and proteasome function, as indicated in the figure. See text for further details.
Conclusions
TRP14 is a ubiquitous cytosolic redox active protein, well‐conserved and present from bacteria to mammals. Its conservation during evolution suggests that this protein should be important for cell or organism survival, but much is still unknown about its roles. Being effectively reduced by TrxR1 but not supporting classic substrates of Trx1 makes TRP14 a perfect candidate for regulation of redox signalling, with unique characteristics complementing those of Trx1. The confirmed activities of TRP14 in NF‐κB regulation, autophagy, persulfide reduction, PTP1B activation, SNO reduction and more implicate TRP14 to be of importance in redox signalling, but at present, little is known about its exact effects and the activities where it may have a predominant role over other redox active proteins. Further studies of TRP14 functions are thus warranted, and forthcoming results on the functions of this protein are likely to be interesting in the context of redox signalling.
Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/18 (Alexander et al., 2017a,b).
Conflict of interest
The authors declare no conflicts of interest.
Acknowledgements
Funding from Karolinska Institutet, The Swedish Cancer Society, The Swedish Research Council and The Knut and Alice Wallenberg Foundation is thankfully acknowledged.
Espinosa, B. , and Arnér, E. S. J. (2019) Thioredoxin‐related protein of 14 kDa as a modulator of redox signalling pathways. British Journal of Pharmacology, 176: 544–553. 10.1111/bph.14479.
References
- Akaike T, Ida T, Wei FY, Nishida M, Kumagai Y, Alam MM et al (2017). Cysteinyl‐tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat Commun 8: 1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Fabbro D, Kelly E, Marrion NV, Peters JA, Faccenda E et al (2017a). The Concise Guide to PHARMACOLOGY 2017/18: Enzymes. Br J Pharmacol 174: S272–S359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Kelly E, Marrion NV, Peters JA, Faccenda E, Harding SD et al (2017b). The Concise Guide to PHARMACOLOGY 2017/18: Overview. Br J Pharmacol 174: S1–S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez L, Bianco CL, Toscano JP, Lin J, Akaike T, Fukuto JM (2017). Chemical biology of hydropersulfides and related species: possible roles in cellular protection and redox signaling. Antioxid Redox Signal 27: 622–633. [DOI] [PubMed] [Google Scholar]
- Arner ES, Holmgren A (2000). Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem 267: 6102–6109. [DOI] [PubMed] [Google Scholar]
- Benhar M (2015). Nitric oxide and the thioredoxin system: a complex interplay in redox regulation. Biochim Biophys Acta 1850: 2476–2484. [DOI] [PubMed] [Google Scholar]
- Benhar M (2016). Emerging roles of protein S‐nitrosylation in macrophages and cancer cells. Curr Med Chem 23: 2602–2617. [DOI] [PubMed] [Google Scholar]
- Benhar M, Forrester MT, Hess DT, Stamler JS (2008). Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science 320: 1050–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhar M, Forrester MT, Stamler JS (2009). Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat Rev Mol Cell Biol 10: 721–732. [DOI] [PubMed] [Google Scholar]
- Brigelius‐Flohe R, Flohe L (2011). Basic principles and emerging concepts in the redox control of transcription factors. Antioxid Redox Signal 15: 2335–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M, Sato H (2012). The oxidative stress‐inducible cystine/glutamate antiporter, system x (c) (−): cystine supplier and beyond. Amino Acids 42: 231–246. [DOI] [PubMed] [Google Scholar]
- Dagnell M, Frijhoff J, Pader I, Augsten M, Boivin B, Xu J et al (2013). Selective activation of oxidized PTP1B by the thioredoxin system modulates PDGF‐β receptor tyrosine kinase signaling. Proc Natl Acad Sci U S A 110: 13398–13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagnell M, Pace PE, Cheng Q, Frijhoff J, Ostman A, Arner ESJ et al (2017). Thioredoxin reductase 1 and NADPH directly protect protein tyrosine phosphatase 1B from inactivation during H2O2 exposure. J Biol Chem 292: 14371–14380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammeyer P, Arner ES (2011). Human protein atlas of redox systems – what can be learnt? Biochim Biophys Acta 1810: 111–138. [DOI] [PubMed] [Google Scholar]
- Deponte M (2013). Glutathione catalysis and the reaction mechanisms of glutathione‐dependent enzymes. Biochimica et Biophysica Acta (BBA) ‐ General Subjects 1830: 3217–3266. [DOI] [PubMed] [Google Scholar]
- Doka E, Pader I, Biro A, Johansson K, Cheng Q, Ballago K et al (2016). A novel persulfide detection method reveals protein persulfide‐ and polysulfide‐reducing functions of thioredoxin and glutathione systems. Sci Adv 2: e1500968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Prigge JR, Talago EA, Arner ESJ, Schmidt EE (2015). Dietary methionine can sustain cytosolic redox homeostasis in the mouse liver. Nat Commun 6: 6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Mao R, Yang J (2013). NF‐κB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell 4: 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes AP, Holmgren A (2004). Glutaredoxins: glutathione‐dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid Redox Signal 6: 63–74. [DOI] [PubMed] [Google Scholar]
- Foloppe N, Nilsson L (2004). The glutaredoxin ‐C‐P‐Y‐C‐ motif: influence of peripheral residues. Structure 12: 289–300. [DOI] [PubMed] [Google Scholar]
- Funato Y, Miki H (2007). Nucleoredoxin, a novel thioredoxin family member involved in cell growth and differentiation. Antioxid Redox Signal 9: 1035–1057. [DOI] [PubMed] [Google Scholar]
- Groitl B, Jakob U (2014). Thiol‐based redox switches. Biochim Biophys Acta 1844: 1335–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromer S, Urig S, Becker K (2004). The thioredoxin system–from science to clinic. Med Res Rev 24: 40–89. [DOI] [PubMed] [Google Scholar]
- Haj FG, Markova B, Klaman LD, Bohmer FD, Neel BG (2003). Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatase‐1B. J Biol Chem 278: 739–744. [DOI] [PubMed] [Google Scholar]
- Harding SD, Sharman JL, Faccenda E, Southan C, Pawson AJ, Ireland S et al (2018). The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucl Acids Res 46: D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemy SI, Holmgren A (2008). Regulation of the catalytic activity and structure of human thioredoxin 1 via oxidation and S‐nitrosylation of cysteine residues. J Biol Chem 283: 21890–21898. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S (2004). Signaling to NF‐kappaB. Genes Dev 18: 2195–2224. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S (2012). NF‐κB, the first quarter‐century: remarkable progress and outstanding questions. Genes Dev 26: 203–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilman J, Watson W (2008). Thioredoxin reductase modulates NFkB signaling at the nuclear compartment. Cancer Res 68: 5268. [Google Scholar]
- Holmgren A (1995). Thioredoxin structure and mechanism: conformational changes on oxidation of the active‐site sulfhydryls to a disulfide. [Review]. Structure 3: 239–243. [DOI] [PubMed] [Google Scholar]
- Holmgren A, Arnér E, Åslund F, Björnstedt M, Liangwei Z, Ljung J et al (1998). Redox regulation by the thioredoxin and glutaredoxin systems In: Montagnier L, Olivier R, Pasquier C. (eds). Oxidative Stress, Cancer, AIDS and Neurodegenerative Diseases. Marcel Dekker, Inc.: New York, pp. 229–246. [Google Scholar]
- Holmstrom KM, Finkel T (2014). Cellular mechanisms and physiological consequences of redox‐dependent signalling. Nat Rev Mol Cell Biol 15: 411–421. [DOI] [PubMed] [Google Scholar]
- Hong S, Huh JE, Lee SY, Shim JK, Rhee SG, Jeong W (2014). TRP14 inhibits osteoclast differentiation via its catalytic activity. Mol Cell Biol 34: 3515–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong W, Chang T‐S, Boja ES, Fales HM, Rhee S (2004a). Roles of TRP14, a thioredoxin‐related protein in tumor necrosis factor‐α signaling pathways. J Biol Chem 279: 3151–3159. [DOI] [PubMed] [Google Scholar]
- Jeong W, Chang TS, Boja ES, Fales HM, Rhee SG (2004b). Roles of TRP14, a thioredoxin‐related protein in tumor necrosis factor‐alpha signaling pathways. J Biol Chem 279: 3151–3159. [DOI] [PubMed] [Google Scholar]
- Jeong W, Jung Y, Kim H, Park SJ, Rhee SG (2009). Thioredoxin‐related protein 14, a new member of the thioredoxin family with disulfide reductase activity: implication in the redox regulation of TNF‐alpha signaling. Free Radic Biol Med 47: 1294–1303. [DOI] [PubMed] [Google Scholar]
- Jeong W, Yoon HW, Lee SR, Rhee SG (2004). Identification and characterization of TRP14, a thioredoxin‐related protein of 14 kDa. New insights into the specificity of thioredoxin function. J Biol Chem 279: 3142–3150. [DOI] [PubMed] [Google Scholar]
- Jiang S, Zhang S, Vuthiphandchai V, Nimrat S (2007). Human TRP14 gene homologue from amphioxus Branchiostoma belcheri: identification, evolution, expression and functional characterization. J Anat 210: 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez A, Pelto‐Huikko M, Gustafsson JA, Miranda‐Vizuete A (2006). Characterization of human thioredoxin‐like‐1: potential involvement in the cellular response against glucose deprivation. FEBS Lett 580: 960–967. [DOI] [PubMed] [Google Scholar]
- Jung Y, Kim H, Min SH, Rhee SG, Jeong W (2008). Dynein light chain LC8 negatively regulates NF‐kappaB through the redox‐dependent interaction with IkappaBalpha. J Biol Chem 283: 23863–23871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabil O, Vitvitsky V, Banerjee R (2014). Sulfur as a signaling nutrient through hydrogen sulfide. Annu Rev Nutr 34: 171–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasamatsu S, Nishimura A, Morita M, Matsunaga T, Abdul Hamid H, Akaike T (2016). Redox signaling regulated by cysteine persulfide and protein polysulfidation. Molecules 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JR, Yoon HW, Kwon KS, Lee SR, Rhee SG (2000). Identification of proteins containing cysteine residues that are sensitive to oxidation by hydrogen peroxide at neutral pH. Anal Biochem 283: 214–221. [DOI] [PubMed] [Google Scholar]
- Kim K (2018). Structure‐based analysis of clot as a thioredoxin‐related protein of 14 kDa in drosophila via experimental and computational approaches. Biotechnol Appl Biochem 65: 338–345. [DOI] [PubMed] [Google Scholar]
- Krishnan N, Fu C, Pappin DJ, Tonks NK (2011). H2S‐induced sulfhydration of the phosphatase PTP1B and its role in the endoplasmic reticulum stress response. Sci Signal 4: ra86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewerenz J, Hewett SJ, Huang Y, Lambros M, Gout PW, Kalivas PW et al (2013). The cystine/glutamate antiporter system x(c)(−) in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxid Redox Signal 18: 522–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Coghlan A, Ruan J, Coin LJ, Heriche JK, Osmotherly L et al (2006). TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res 34: D572–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillig CH, Berndt C, Holmgren A (2008). Glutaredoxin systems. Biochim Biophys Acta 1780: 1304–1317. [DOI] [PubMed] [Google Scholar]
- Martin JL (1995). Thioredoxin‐a fold for all reasons. Structure 3: 245–250. [DOI] [PubMed] [Google Scholar]
- Matthews JR, Wakasugi N, Virelizier JL, Yodoi J, Hay RT (1992). Thioredoxin regulates the DNA binding activity of NF‐kappa B by reduction of a disulphide bond involving cysteine 62. Nucleic Acids Res 20: 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CG, Holmgren A, Arner ESJ, Schmidt EE (2018). NADPH‐dependent and ‐independent disulfide reductase systems. Free Radic Biol Med. 10.1016/j.freeradbiomed.2018.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda‐Vizuete A, Sadek CM, Jimenez A, Krause WJ, Sutovsky P, Oko R (2004). The mammalian testis‐specific thioredoxin system. Antioxid Redox Signal 6: 25–40. [DOI] [PubMed] [Google Scholar]
- Mitomo K, Nakayama K, Fujimoto K, Sun X, Seki S, Yamamoto K (1994). Two different cellular redox systems regulate the DNA‐binding activity of the p50 subunit of NF‐κB in vitro. Gene 145: 197–203. [DOI] [PubMed] [Google Scholar]
- Motohashi K, Kondoh A, Stumpp MT, Hisabori T (2001). Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci 98: 11224–11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagakannan P, Iqbal MA, Yeung A, Thliveris JA, Rastegar M, Ghavami S et al (2016). Perturbation of redox balance after thioredoxin reductase deficiency interrupts autophagy‐lysosomal degradation pathway and enhances cell death in nutritionally stressed SH‐SY5Y cells. Free Radic Biol Med 101: 53–70. [DOI] [PubMed] [Google Scholar]
- Nagy P (2015). Mechanistic chemical perspective of hydrogen sulfide signaling. Methods Enzymol 554: 3–29. [DOI] [PubMed] [Google Scholar]
- Navarro‐Yepes J, Burns M, Anandhan A, Khalimonchuk O, del Razo LM, Quintanilla‐Vega B et al (2014). Oxidative stress, redox signaling, and autophagy: cell death versus survival. Antioxid Redox Signal 21: 66–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund P, Reichard P (2006). Ribonucleotide reductases. Annu Rev Biochem 75: 681–706. [DOI] [PubMed] [Google Scholar]
- Pader I, Sengupta R, Cebula M, Xu J, Lundberg JO, Holmgren A et al (2014). Thioredoxin‐related protein of 14 kDa is an efficient L‐cystine reductase and S‐denitrosylase. Proc Natl Acad Sci U S A 111: 6964–6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padovani D, Mulliez E, Fontecave M (2001). Activation of class III ribonucleotide reductase by thioredoxin. J Biol Chem 276: 9587–9589. [DOI] [PubMed] [Google Scholar]
- Prigge JR, Coppo L, Martin SS, Ogata F, Miller CG, Bruschwein MD et al (2017). Hepatocyte hyperproliferation upon liver‐specific co‐disruption of thioredoxin‐1, thioredoxin reductase‐1, and glutathione reductase. Cell Rep 19: 2771–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Li H, Chen Z, Coghlan A, Coin LJ, Guo Y et al (2008). TreeFam: 2008 update. Nucleic Acids Res 36: D735–D740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK et al (2003). Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl‐amide intermediate. Nature 423: 769–773. [DOI] [PubMed] [Google Scholar]
- Schwertassek U, Haque A, Krishnan N, Greiner R, Weingarten L, Dick TP et al (2014). Reactivation of oxidized PTP1B and PTEN by thioredoxin 1. FEBS J 281: 3545–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta R, Holmgren A (2013). Thioredoxin and thioredoxin reductase in relation to reversible S‐nitrosylation. Antioxid Redox Signal 18: 259–269. [DOI] [PubMed] [Google Scholar]
- Sies H (2014). Role of metabolic H2O2 generation: redox signaling and oxidative stress. J Biol Chem 289: 8735–8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobotta MC, Liou W, Stocker S, Talwar D, Oehler M, Ruppert T et al (2015). Peroxiredoxin‐2 and STAT3 form a redox relay for H2O2 signaling. Nat Chem Biol 11: 64–70. [DOI] [PubMed] [Google Scholar]
- Su D, Berndt C, Fomenko DE, Holmgren A, Gladyshev VN (2007). A conserved cis‐proline precludes metal binding by the active site thiolates in members of the thioredoxin family of proteins. Biochemistry 46: 6903–6910. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M et al (2017). The STRING database in 2017: quality‐controlled protein‐protein association networks, made broadly accessible. Nucleic Acids Res 45: D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toppo S, Flohe L, Ursini F, Vanin S, Maiorino M (2009). Catalytic mechanisms and specificities of glutathione peroxidases: variations of a basic scheme. Biochim Biophys Acta 1790: 1486–1500. [DOI] [PubMed] [Google Scholar]
- Trachootham D, Lu W, Ogasawara MA, Valle N, Huang P (2008). Redox regulation of cell survival. Antioxid Redox Signal 10: 1343–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B (2003). STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 31: 258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Ning X, Chen L, Pei D, Zhao J, Zhang L et al (2011). Responses of thioredoxin 1 and thioredoxin‐related protein 14 mRNAs to cadmium and copper stresses in Venerupis philippinarum. Comp Biochem Physiol C Toxicol Pharmacol 154: 154–160. [DOI] [PubMed] [Google Scholar]
- Wang X, Stafford W, Mazurkiewicz M, Fryknas M, Brjnic S, Zhang X et al (2014). The 19S deubiquitinase inhibitor b‐AP15 is enriched in cells and elicits rapid commitment to cell death. Mol Pharmacol 85: 932–945. [DOI] [PubMed] [Google Scholar]
- Wedmann R, Onderka C, Wei S, Szijarto IA, Miljkovic JL, Mitrovic A et al (2016). Improved tag‐switch method reveals that thioredoxin acts as depersulfidase and controls the intracellular levels of protein persulfidation. Chem Sci 7: 3414–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Ji H, Guo M, Yan Y, Qin Q (2013). Identification and characterization of TRP14, a thioredoxin‐related protein of 14 kDa from orange‐spotted grouper, Epinephelus coioides. Fish Shellfish Immunol 35: 1670–1676. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC, Hampton MB (2008). Thiol chemistry and specificity in redox signaling. Free Radic Biol Med 45: 549–561. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC, Hampton MB (2015). Redox biology: signaling via a peroxiredoxin sensor. Nat Chem Biol 11: 5–6. [DOI] [PubMed] [Google Scholar]
- Wolhuter K, Eaton P (2017). How widespread is stable protein S‐nitrosylation as an end‐effector of protein regulation? Free Radic Biol Med 109: 156–166. [DOI] [PubMed] [Google Scholar]
- Wolhuter K, Whitwell HJ, Switzer CH, Burgoyne JR, Timms JF, Eaton P (2018). Evidence against stable protein S‐nitrosylation as a widespread mechanism of post‐translational regulation. Mol Cell 69: 438–450 e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JR, Kim SJ, Jeong W, Cho YH, Lee SC, Chung YJ et al (2004). Structural basis of cellular redox regulation by human TRP14. J Biol Chem 279: 48120–48125. [DOI] [PubMed] [Google Scholar]
- Ye ZW, Zhang J, Townsend DM, Tew KD (2015). Oxidative stress, redox regulation and diseases of cellular differentiation. Biochim Biophys Acta 1850: 1607–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara E, Masaki S, Matsuo Y, Chen Z, Tian H, Yodoi J (2014). Thioredoxin/Txnip: redoxisome, as a redox switch for the pathogenesis of diseases. Front Immunol 4: 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SF, Wang XY, Fu ZQ, Peng QH, Zhang JY, Ye F et al (2015). TXNDC17 promotes paclitaxel resistance via inducing autophagy in ovarian cancer. Autophagy 11: 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang A, Li H, Zhi H, Lu F (2016). STAT3‐dependent TXNDC17 expression mediates Taxol resistance through inducing autophagy in human colorectal cancer cells. Gene 584: 75–82. [DOI] [PubMed] [Google Scholar]
- Zhen Z, Yang K, Ye L, You Z, Chen R, Liu Y et al (2017). Suberoylanilide hydroxamic acid sensitizes neuroblastoma to paclitaxel by inhibiting thioredoxin‐related protein 14‐mediated autophagy. Cancer Sci 108: 1485–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åslund F, Berndt KD, Holmgren A (1997). Redox potentials of glutaredoxins and other thiol‐disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein‐protein redox equilibria. J Biol Chem 272: 30780–30786. [DOI] [PubMed] [Google Scholar]


