Abstract
The lysosomal degradation pathway of autophagy plays a fundamental role in cellular, tissue and organismal homeostasis and is mediated by evolutionarily conserved autophagy-related (ATG) genes. Definitive etiological links exist between mutations in genes that control autophagy and human disease, especially neurodegenerative, inflammatory disorders and cancer. Autophagy selectively targets dysfunctional organelles, intracellular microbes and pathogenic proteins, and deficiencies in these processes may lead to disease. Moreover, ATG genes have diverse physiologically important roles in other membrane trafficking and signalling pathways. This review discusses the biological functions of autophagy genes from the perspective of understanding – and potentially reversing – the pathophysiology of human disease and aging.
Introduction
A decade has elapsed since our review in 2008 in Cell on “Autophagy in the Pathogenesis of Disease” (Levine and Kroemer, 2008). During this period, more than 33,000 new articles related to autophagy were published; a Nobel prize was awarded for the discovery of the molecular mechanisms of autophagy (Levine and Klionsky, 2017; Mizushima, 2018); and considerable interest has emerged in autophagy modulation as a potential target in clinical medicine (Galluzzi et al., 2017a).
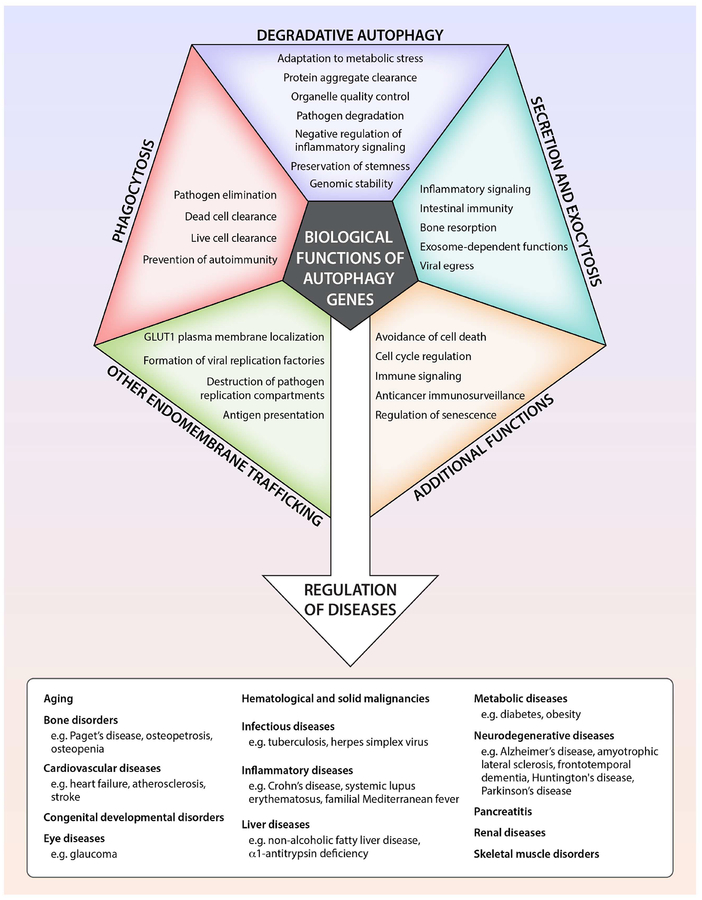
The fundamental concepts discussed in our 2008 review (Levine and Kroemer, 2008) remain unchanged. The lysosomal degradation pathway of macroautophagy (herein referred to as autophagy) plays a crucial role in cellular physiology, including adaptation to metabolic stress, removal of dangerous cargo (e.g. protein aggregates, damaged organelles, intracellular pathogens), renovation during differentiation and development, and prevention of genomic damage. Generally, these and other functions protect against numerous diseases, including infections, cancer, neurodegeneration, cardiovascular disorders, and aging (Mizushima and Komatsu, 2011). Under certain circumstances, autophagy may be detrimental either via its pro-survival effects (such as in cancer progression (Amaravadi et al., 2016)) or via possible cell death-promoting effects (Marino et al., 2014a).
Over the past ten years, significant progress has been made in understanding the molecular mechanisms of autophagy, the regulation of autophagy, and the effects of autophagy on physiology and pathophysiology (Dikic and Elazar, 2018; Galluzzi et al., 2014; Mizushima, 2018). New major conceptual advances underscore the plurality of functions of the autophagic core machinery in various membrane trafficking and signaling events (Cadwell and Debnath, 2018) and delineate the exquisite specificity with which autophagy targets selected cargo for degradation (Gatica et al., 2018). These advances, together with discoveries in human genetics linking ATG gene mutations to specific diseases (Jiang and Mizushima, 2014; van Beek et al., 2018), provide a multidimensional perspective of mechanisms by which ATG gene-dependent pathways protect against mammalian disease.
Herein we review selected highlights of the past decade of research on the biological functions of autophagy genes, primarily from a perspective of understanding and treating human disease.
Autophagy and other Autophagy Gene-Dependent Pathways
The original scientific definition of autophagy (Greek, “self-eating”) is the delivery of cytoplasmic cargo to the lysosome for degradation. There are at least three distinct forms of autophagy — chaperone-mediated autophagy, microautophagy and macroautophagy — which differ in terms of mode of cargo delivery to the lysosome. Macroautophagy is the major catabolic mechanism used by eukaryotic cells to maintain nutrient homeostasis and organellar quality control. It is mediated by a set of evolutionarily conserved genes, the autophagy-related (ATG) genes (Klionsky et al., 2003), originally discovered in yeast genetic screens (Mizushima, 2018). With a few exceptions, all ATG genes are required for the efficient formation of sealed autophagosomes that proceed to fuse with lysosomes.
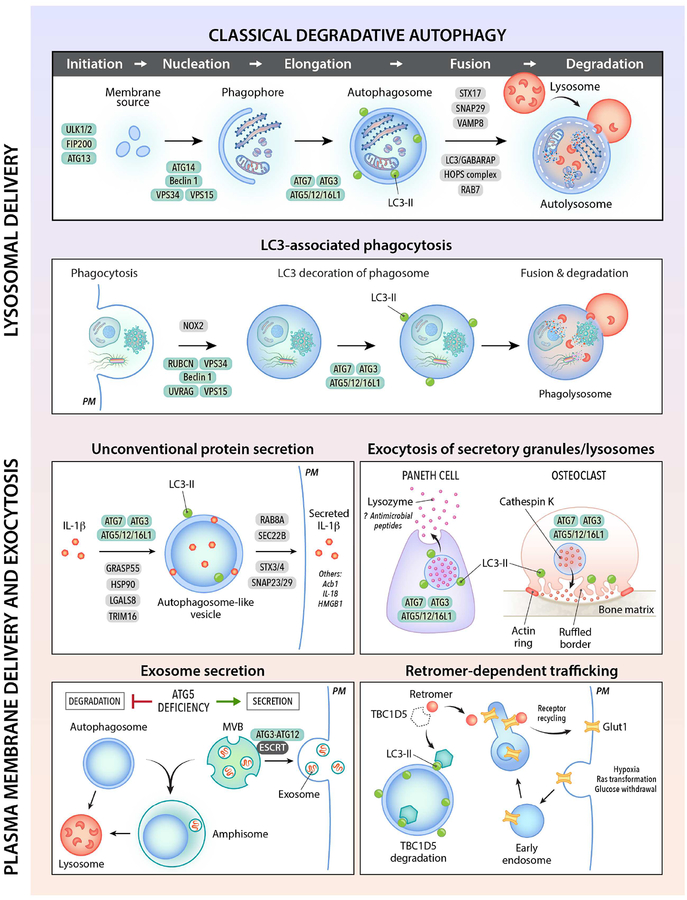
In higher eukaryotes, many ATG genes functionally diversified to facilitate delivery of extracellular cargo to the lysosome, to promote the plasma membrane localization or extracellular release of intracellular cargo, and to coordinate intracellular communication with various cell signaling pathways (Figure 1). These other functions are not, sensu stricto, autophagy and accordingly, will be referred to as ATG gene-dependent pathways. There are broad implications of ATG gene functions in different membrane trafficking and signaling pathways for mammalian cell biology, physiology and disease.
Figure 1. Autophagy gene-dependent membrane trafficking pathways.
Shown are schematic illustrations of different membrane trafficking pathways that involve autophagy (ATG) proteins (green ovals). See text for explanations of each pathway and a discussion of their physiological functions. See Table 1 for examples of genetic mutations that impair autophagy-related pathways which are associated with human disease. The major type of autophagy, macroautophagy, is labeled as “classical degradative autophagy” to distinguish it from other trafficking pathways that utilize overlapping ATG proteins. Due to space limitations, not all ATG proteins, proteins involved in vesicle fusion, or secretary cargo are depicted. PM, plasma membrane. LC3-II (green circle) is the phosphatidyl-ethanolamine-conjugated form of the autophagy protein, LC3.
Degradative Autophagy: The “raison d’être” of Autophagy Genes
The originally discovered function of ATG genes is to orchestrate and mediate the formation of double-membraned structures that deliver intracytoplasmic contents to the lysosome for degradation. This process is conserved in all eukaryotic organisms, occurs at basal levels in nearly all cell types, and is increased by diverse intracellular and extracellular cues. It is essential for cellular homeostasis, cellular protein and organelle quality control, and organismal adaptation to environmental stress. These principles are firmly supported by nearly two decades of studies involving genetic ablation of the autophagy machinery in diverse eukaryotic species (Levine and Kroemer, 2008; Mizushima and Komatsu, 2011).
This lysosomal degradation pathway is usually described as involving a set of ~16–20 core conserved ATG genes. The ATG proteins encoded by these genes are traditionally classified into distinct biochemical and functional groups that act at specific stages of autophagosome initiation or formation. In this scheme (see other recent reviews for details (Dikic and Elazar, 2018; Yu et al., 2018)), the ULK1 serine threonine kinase complex (involving ULK1, FIP200, ATG13 and ATG101) plays a major role in autophagy initiation, phosphorylating multiple downstream factors. Two distinct Beclin 1/class III phosphatidylinositol 3-kinase (PI3KC3) complexes generate phosphatidylinositol 3-phosphate (PI3P) to act in autophagosome nucleation (PI3KC3–C1 involving Beclin 1, VPS34, VPS15 and ATG14) or endolysosomal and autophagolysosomal maturation (PI3KC3–C2 involving Beclin 1, VPS34, VPS15 and UVRAG). Vesicles containing ATG9A, the only transmembrane core ATG protein, supply membrane to autophagosomes. WIPI (WD repeat domain phosphoinositide-interacting) proteins and their binding partners, ATG2A or ATG2B, function in early stages of membrane elongation at the site of PI3P generation. Autophagosome membrane expansion and completion involves two ubiquitin-like protein conjugation systems: the Ub-like ATG12 conjugates with ATG5 and ATL16L1 and the Ub-like LC3 subfamily (ATG8 in yeast) conjugates with membrane-resident phosphatidylethanoloamine (PE). Unlike in yeast, the ubiquitin-like protein conjugation systems are not essential for autophagosomal membrane completion in mammalian cells, although they determine the efficiency of the process (Tsuboyama et al., 2016).
This classification of the ATG proteins has provided a useful framework for studying and understanding autophagy. However, its apparent simplicity is at variance with extensive data indicating a highly complex level of interconnectivity among the ATG proteins and newly described functions of ATG proteins at different stages of autophagy. Based on unbiased proteomic analyses, most ATG proteins interact with other ATG proteins that reside outside of their “classic” functional complex (Behrends et al., 2010). Experimentally, some of these interactions are known to be important for autophagosome formation. For example, FIP200 (a member of the ULK1 kinase complex) interacts with ATG16L1 to properly target it to the isolation membrane (also known as the phagophore) of the nascent autophagosome (Nishimura et al., 2013). ATG14 (a component of the autophagy-specific PI3KC3–C1 complex) also functions in SNARE-driven membrane fusion (Diao et al., 2015). Similarly, Atg13 (a component of the yeast Atg1/mammalian ULK1 kinase complex) interacts with Atg9 to recruit Atg9 vesicles to the pre-autophagosomal structure (Suzuki et al., 2015). The broader interconnectivity and functional multiplicity of core autophagy proteins in autophagosomal biogenesis requires further elucidation. Moreover, as indicated by a recent conditional genetic interactions study using diverse yeast–omics datasets (Kramer et al. 2017), new systems biology approaches will likely identify additional genes required for autophagy, especially those that may function in a stimulus-dependent, cell type-dependent or species-specific manner.
The core ATG proteins, conserved from yeast to humans, are necessary but not sufficient for degradative autophagy. The degradation of autophagosomal cargo cannot proceed without successful fusion to an available and functional lysosome. Research in the past decade has unmasked some of the key factors required for lysosomal biogenesis (Settembre et al., 2013b), autophagolysosomal fusion (Yu et al., 2018), lysosomal function during autophagy (Shen and Mizushima, 2014) and autophagic lysosome reformation (Chen and Yu, 2017). Adenoviral-mediated gene delivery of TFEB, a master transcriptional regulator of lysosomal biogenesis, improves outcomes in various rodent disease models, including Parkinson’s disease, lysosomal storage disorders, tauopathies, α1-antitrypsin deficiency, and hepatic hyperammonemia (Napolitano and Ballabio 2016; Soria et al., 2018). Autophagolysosomal fusion requires changes in lysosomal pH, certain cytoskeleton motor proteins (dynein), tethering factors (the HOPS complex, the Rab GTPase, RAB7), SNARE proteins (the Q-SNARE, syntaxin 17 on autophagosomes which interacts with R-SNARE proteins, SNAP29 and VAMP8 on endosomes/lysosomes), phospholipids, and members of the LC3/GABARAP family (Kriegenburg et al., 2018) that are bridged to tethering factors or SNARES by adaptor proteins. Screens in C. elegans identified novel metazoan-specific genes required for fusion steps in degradative autophagy (Tian et al., 2010). One example relevant to human disease is EPG5, which encodes a RAB7 effector. Autosomal recessive mutation of EPG5 results in Vici syndrome, a neurodevelopmental and multisystem disorder (Table 1). Mutations in genes that regulate lysosomal acidification such as ATP6AP2 and presenilin 1 are associated with X-linked Parkinsonism and Alzheimer’s disease (Table 1). Thus, we must consider regulators of lysosomal biogenesis, the fusion machinery, and determinants of lysosomal function in our efforts to decipher how deficient autophagy leads to disease and how autophagy can be regulated to prevent or treat disease.
Table 1.
Examples of genetic mutations in human disease that impair autophagy
| Gene | Disease | Mechanism | Reference |
|---|---|---|---|
| Mutations in genes required for autophagy and lysosomal function | |||
| ATG16L1 | Crohn’s Disease (CD) | ATGL16L1 T300A is a major risk allele for CD. The T300A polymorphism has a caspase 3 cleavage site that decreases protein levels. T300A knock-in or hypomorphic or intestinal knockout mice show decreased intestinal bacterial clearance; increased cytokine responses; reduced Paneth cell lysozyme secretion and clearance of IRE la protein aggregates during ER stress; enhanced enterocyte TNFa-induced necroptosis; dendritic cell defects in regulatory T cell induction and suppression of mucosal inflammation. | (Jiang and Mizushima, 2014) (Murthy et al., 2014) (Lassen et al., 2014) (Chu et al., 2016) (Bel et al., 2017) (Tschurtschenthaler et al., 2017) (Matsuzawa-Ishimoto et al., 2017) |
| ATG16L2 | Systemic lupus erythematosus (SLE) | ATG16L2 R114W allele is a disease susceptibility gene | (Molineros et al., 2017) |
| ATG5 | Childhood ataxia Systemic sclerosis SLE |
Loss-of-function mutation reduces autophagy and causes ataxia Intronic variants are associated with susceptibility to systemic sclerosis Polymorphisms associated with SLE susceptibility; mouse studies suggest mechanism may involve deficient LC3-associated phagocytosis |
(Kim et al., 2016) (Martinez et al., 2016) |
| ATP6AP2 | X-linked Parkinsonism with spasticity Multisystem disorder |
Exon skipping mutations linked to Parkinsonism Missense mutations associated with immunodeficiency, liver disease, and psychomotor impairment lead to defective lysosomal acidification due to impaired v-ATPase assembly, resulting in defects in autophagy |
(Korvatska et al., 2013) (Rujano et al., 2017) |
| BECN1 | Breast and ovarian cancer | Monoallelic deletion associated with risk and poor prognosis of sporadic breast and ovarian cancer; monoallelic deletion in mice leads to decreased autophagy and increased tumors, including basal-like breast cancer | (Qu et al., 2003) (Yue et al., 2003) (Valente et al., 2014) (Tang et al., 2015) |
| CLEC16A | Diabetes Multiple sclerosis |
CLEC16A variants associated with multiple autoimmune diseases. Mice deficient in Clecl6a have autophagy defects associated with Purkinje degeneration and ataxia, impaired β-cell mitophagy, and autoimmunity | (Soleimanpour et al., 2014) (Schuster et al., 2015) (Bronson et al., 2016) (Redmann et al., 2016) |
| CTNS | Cystinosis | Recessive loss-of-function mutations in CTNS, a gene encoding a proton transporter that exports cysteine from lysosomes associated with renal lysosomal storage disease; gene deletion in mice results in deficient autophagy, altered lysosomal dynamics and accumulation of dysfunctional ROS overproducing mitochondria | (Festa et al., 2018) |
| EPG5 | Vici syndrome | Recessive mutations in EPG5, a gene required for autophagolysosomal fusion results in a neurodevelopmental disorder with multisystem involvement | (Cullup et al., 2013) (Hori et al., 2017) |
| GBA | Gaucher’s disease Parkinson’s disease (PD) | GBA1 encodes the lysosomal enzyme glucocerebrosidase. Homozygous GBA defects cause Gaucher’s disease and heterozygous defects predispose to PD | (Afiaki et al., 2017) |
| GRN | Frontotemporal dementia (heterozygous) or Neuronal ceroid lipofuscinosis (homozygous) | Loss-of-function mutations compromise lysosomal function and autophagic flux | (Chang et al., 2017) |
| LAMP2 | Danon’s cardiomyopathy | X-linked deletion results in vacuolar cardiomyopathy and myopathy; Lamp2 deletion in mice results in autophagosome accumulation and cardiomyopathy | (Nishino et al., 2000) (Tanaka et al., 2000) |
| PIK3R4 (VPSI 5) | Cortical atrophy and epilepsy | Mutations in PIK3R4, a component of the Beclin 1 complex required for endosomal-lysosomal trafficking and autophagy, associated with human neurodevelopmental disease | (Gstrein et al., 2018) |
| SNX14 | Autosomal recessive spinocerebellar ataxia | SNX14 binds lysosomal phosphatidylinositol (3,5)-bisphosphate and is required for autophagosomal clearance | (Akizu et al., 2015) |
|
SPG11, SPG15 (ZFYVE26), SPG49 (TECPR) |
Hereditary spastic paraplegia | SPG15 binds phosphatidylinositol 3-phosphate and SPG49 binds LC3 to function in autophagolysosomal trafficking | (Ebrahimi-Fakhari et al., 2016) |
| WDR45 (WIPI4) | Beta-propeller protein-associated neurodegeneration | WDR45 (WIPI4) binds to phosphoinositide 3-phosphate and interacts with ATG2 and ATG9; disease-associated mutations impair autophagy | (Haack et al., 2012) (Saitsu et al., 2013) (Ebrahimi-Fakhari et al., 2016) |
| Mutations in genes that regulate autophagy and lysosomal function | |||
| APP | Alzheimer’s disease | Mutant amyloid precursor protein expressed in mouse hippocampal neurons inhibits mitophagy and autophagy | (Reddy et al., 2018) |
| AT-1 (SLC33A1) | Spastic paraplegia, Developmental delay Autism spectrum disorders |
AT-1 translocates cytosolic acetyl coA into ER lumen; mutations and duplications associated with a variety of CNS phenotypes in humans; in mice overexpression blocks Atg9a-Faml34b-LC3 interactions, leading to defective ER-phagy and progeria | (Peng et al., 2018) |
| C9orf72 | Amyotrophic lateral sclerosis (ALS) Frontotemporal dementia (FTD) |
Hexanucleotide repeat expansion in C9orf72 gene is most common genetic cause of ALS and FTD. Regulates autophagy and lysosomal homeostasis through interactions with SMCR8, ULK1 and Rab-GTPases | (Nassif et al., 2017) (Corrionero and Horvitz, 2018) |
| ERBB2 | Breast cancer | Amplification of ERBB2 and consequent overexpression of ERBB2 (HER2) interacts with Beclin 1 and inhibits autophagy | (Vega-Rubin-de-Celis et al., 2018) |
| GBA1 | Gaucher’s disese Parkinson’s disease |
Mutations in GBA1 decrease glucocerebrosidase activity, leading to defects in autophagic-lysosomal function and a-synuclein aggregate accumulation | (Schapira, 2015) (Afiaki et al., 2017) |
| GPR65 | Inflammatory bowel disease | GPR65 I231L risk variant of this proton-sensing G protein-coupled receptor impairs lysosomal acidification, decreases intracellular bacterial clearance and alters lipid droplet turnover | (Lassen et al., 2016) |
| HTT (Huntingtin) | Huntington’s disease | PolyQ extension in HTT competitively disrupts interaction between the deubiquitinase ataxin 3 and Beclin 1, leading to enhanced Beclin 1 proteasomal degradation and reduced autophagy | (Ashkenazi et al., 2017) |
| IRGM | Non-alcoholic fatty liver disease (NAFLD) Crohn’s disease Tuberculosis |
IRGM functions in assembly and activation of autophagy machinery; a synonymous variant reduces protein expression leading to reduced autophagy and lipophagy in NAFLD; polymorphisms are associated with risk of Crohn’s disease and tuberculosis | (Jiang and Mizushima, 2014) (Lin et al., 2016) (Chauhan et al., 2016b) |
| LRRK2 | Crohn’s disease (CD) Parkinson’s disease (PD) |
Risk alleles for CD and PD increase kinase activity of leucine-rich repeat kinase 2 and reduce autophagic flux; a protective allele increases flux | (Cooper et al., 2012) (Hui et al., 2018) |
| MeCP2 | Rett syndrome (X-linked neuro-developmental disorder) | Deficiency of methyl-CpG-binding protein-2 (MeCP2), a transcriptional regulator, results in defective autophagy in patient fibroblasts and knockout mouse cerebellum, and mitochondrial retention in erythrocytes | (Sbardella et al., 2017) |
| MTMR3 | Inflammatory Bowel Disease (IBD) | MMTR is a PI3P phosphatase that decreases autophagy. Macrophages from carriers of the risk allele express higher MTMR3 protein levels and have increased pathogen recognition receptor-induced caspase-1 activation and IL-1β secretion | (Lahiri et al., 2015) |
| PLEKHM1 | Osteopetrosis | Disease associated mutants impair binding to RAB7A and secretory lysosome trafficking in osteoclasts | (Stenbeck and Coxon, 2014) |
| RAB7A | Charcot-Marie-Tooth type 2B disease | Disease-associated RAB7A mutants reduce autophagic flux in HeLa cells and patient-derived fibroblasts are autophagy-deficient | (Colecchia et al., 2018) |
| PS1 | Alzheimer’s disease | Mutations in presenilin 1 that disrupt v-ATPase assembly, lysosomal acidification and autophagy cause early onset Alzheimer’s disease | (Lee et al., 2010) |
| PTPN2 | IBD Type 1 Diabetes Juvenile arthritis |
Disease-associated SNP in PTPN2, a gene encoding protein tyrosine phosphatase nonreceptor type 2 causes impaired autophagosome formation and defective bacterial handling in macrophages and intestinal epithelial cells | (Scharl et al., 2012) |
| SMS | Snyder-Robinson syndrome (SRS) | Loss-of-function mutations in spermine synthase (SMS) cause SRS, an X-linked intellectual disability syndrome; deficiency in SMS generates toxic metabolites that impair lysosomal function and autophagic flux | (Li et al., 2017a) |
| TMEM230 | Parkinson’s disease | Transmembrane protein involved in retromer function; loss reduces autophagic cargo degradation and secretory autophagy | (Kim et al., 2017) |
| v-ATPase | Autosomal Recessive Osteoporosis |
Mutations in the α3 subunit encoded by TCIRG1 impair lysosomal acidification at the ruffled border of osteoclasts, leading to defects in bone resorption | (Ochotny et al., 2013) |
| WASP | Wiskott-Aldrich syndrome | Deficiency of the actin cytoskeleton-regulatory WASP protein impairs formation of autophagosomes, resulting in deficient xenophagy and excessive inflammasome activation and pyroptosis | (Lee et al., 2017b) |
| Mutations in genes required for cargo delivery in selective autophagy | |||
| ALFY | Primary microcephaly | Dominant mutation in this autophagy scaffold protein causes human microcephaly | (Kadir et al., 2016) |
| CALC0C02 (NDP52) | Crohn’s disease | Missense mutation of this autophagy adaptor reduces its function and enhances NF-kB activation of inflammatory genes | (Ellinghaus et al., 2013) |
| FAM134B | Hereditary sensory and autonomic neuropathy type II | Mutations disrupt the interaction of this ER protein with LC3 and GABARP to impair ER-phagy | (Khaminets et al., 2015) |
| FANC genes | Fanconi anemia (FA) congenital syndrome Hereditary breast and ovarian cancer Sporadic cancers | FA pathway genes required for clearing damaged mitochondria (mitophagy) and preventing aberrant inflammasome activation | (Sumpter et al., 2016) |
| OPTN1 | Amyotrophic lateral sclerosis (ALS) Primary open angle glaucoma (POAG) Paget’s disease of the bone (PGD) |
Mutations in ALS reduce interaction of this autophagy adaptor with TBK1 and reduce Parkin-dependent mitophagy; mutations in POAG increase interaction with TBK1, activate Bax-dependent apoptosis, and are associated with mitochondrial dysfunction. Truncated protein mutation associated with PGD | (Wong and Holzbaur, 2014) (Li et al., 2016) (Shim et al., 2016) (Silva et al., 2018) |
| PARK2/Parkin | Autosomal recessive and sporadic early onset Parkinson’s disease Colon, lung, and brain cancer |
Parkin is an E3 ligase that functions in mitophagy and xenophagy; mutations are associated with Parkinson’s disease and cancer risk; polymorphisms associated with increased susceptibility to intracellular bacterial infections | (Kitada et al., 1998) (Xu et al., 2014) (Mira et al., 2004) |
| PARK6/PINK1 | Autosomal recessive and sporadic early onset Parkinson’s disease | PINK1 is a serine-threonine kinase that translocates to the outer mitochondrial membrane upon damage, mediating Parkin recruitment and mitophagy | (Jiang and Mizushima, 2014) |
| PEX13 | Zellweger syndrome spectrum disorders | Disease-associated mutations impair mitophagy and patients with mutations have accumulation of abnormal mitochondria | (Lee et al., 2017a) |
| SQSTM1 (p62) | ALS FTD Paget’s disease Distal myopathy |
SQSTM1 is an autophagy adaptor that binds ubiquitin and LC3; mutations in the ubiquitin-binding association domain result in spectrum of multisystem proteinopathies. | (Goode et al., 2014) (Lee et al., 2018) |
| SMURF1 | Ulcerative colitis | SMURF1, a susceptibility gene for ulcerative colitis, encodes an E3 ligase that functions in mitophagy, virophagy and xenophagy of intracellular bacteria | (Franco et al., 2017) |
| TBK1 | ALS Frontotemporal dementia Other neurodegenerative phenotypes POAG |
TBK1 kinase phosphorylates the autophagy receptor OPTN1, increasing its interaction with ATG8 proteins and polyubiquitinated proteins | (Cirulli et al., 2015) (van Beek et al., 2018) |
| TRIM20 | Familial Mediterranean fever |
Disease-associated TRIM20 mutants fail to interact with inflammasome components and target them for autophagic destruction | (Kimura et al., 2015) |
| VPS13D | Ataxia with spasticity | Recessively inherited defects in this ubiquitin-binding protein cause failure in mitophagy and mitochondrial dysfunction | (Seong et al., 2018) |
Beyond Self-Eating: Autophagy Genes Function in Phagocytosis
Several core ATG genes function in a process that shares some similarities with autophagy but involves digestion of unwanted extracellular (rather than intracellular) material. During this process, termed LC3-associated phagocytosis (LAP), single-membraned macroendocytic vacuoles (macropinosomes, phagosomes and entotic vacuoles) engulf extracellular cargo (such as bacteria, dead cells or live cells), become decorated by lipidated LC3, and are directed to the lysosome for degradation (Cadwell and Debnath, 2018; Heckmann et al., 2017). LAP is distinguished from autophagy by four main features: (1) the origin of the vacuolar contents (extracellular versus intracellular), (2) the requirement of cargo engagement of an extracellular receptor for activation, (3) the type of membrane that fuses with the lysosome (single membrane versus double membrane), and (4) the utilization of a subset versus all of the core ATG proteins. LAP requires NADPH-oxidase (NOX2) to generate reactive oxygen species (ROS), certain components of the Beclin 1/VPS34 complexes, PI3P generation, LC3-conjugation to the single membrane of the phagosome, and all components of the LC3 conjugation machinery (Martinez et al., 2011; Martinez et al., 2015; Sanjuan et al., 2007). However, it does not require other core ATG proteins, such as components of the ULK1 complex or the autophagy-specific Beclin 1/VPS34 complex component, ATG14. Somewhat enigmatically, LAP requires Rubicon, an inhibitory component of the autophagy-specific Beclin 1/VPS34 complex. The precise effects of LC3 decoration of phagosomes on their fusion with lysosomes and on lysosomal function are unknown. The presence of LC3 on phagosomes may enhance efficiency of phagolysosomal maturation, perhaps through a mechanism similar to that of LC3/GABARAP family members in autophagolysosomal maturation.
LAP was originally described in murine macrophages during phagocytosis of particles that engage Toll-like receptors (TLRs) (Sanjuan et al., 2007) and is involved in type I interferon (IFN) secretion in response to DNA immune complexes and other TLR9 ligands (Hayashi et al., 2018; Henault et al., 2012). Physiologically important functions of LAP have been identified by comparing phenotypes of mice with myeloid-specific deletion of LAP-specific genes (e.g. Rubicon or NOX2) and autophagy-specific ATG genes (e.g. FIP200 or Atg14). LAP is required for degradation of photoreceptor outer segments by retinal pigment epithelium (RPE), a process necessary for intact vision (Kim et al., 2013b). LAP is induced by the fungus, A. fumigatus, and the intracellular bacterium, L. monocytogenes, and enhances host defense against these pathogens (Gluschko et al., 2018; Martinez et al., 2015). Mice lacking several components of the LAP pathway develop an autoimmune systemic lupus erythematosus (SLE)-like disease, perhaps due to a defect in the clearance of dying cells that triggers enhanced proinflammatory signaling and autoantibody production (Martinez et al., 2016).
Given the crucial roles of receptor-activated phagocytosis in human physiology, it is likely that LAP, like classical autophagy, will emerge as an important pathway in human disease. While the two pathways utilize overlapping genetic machinery, a critical distinction renders them antagonistic. Specifically, Rubicon is required for LAP but suppresses autophagy, and recent studies confirm a mutually inhibitory relationship between LAP and autophagy in photoreceptor degradation in RPE cells (Muniz-Feliciano et al., 2017). It is not clear why these two pathways are counter-regulated; possibly, cells may need to shut off the alternative pathway during stress to avoid competition for overlapping resources (Muniz-Feliciano et al., 2017). At a mechanistic level, it is uncertain how Rubicon functions to promote Beclin 1/VPS34 kinase activity at the phagosome (Martinez et al., 2015), but inhibit it at other organellar sites (Matsunaga et al., 2009). Interestingly, the WD repeat-containing C-terminal domain of ATG16L1 is essential for LC3 recruitment to endolysosomal membranes in LAP, but dispensable for canonical autophagy (Fletcher et al., 2018), illustrating another difference in the molecular roles of an ATG protein in autophagy and LAP.
The genetic overlap and mutual antagonism of LAP and autophagy have practical implications for autophagy-targeted therapies. Theoretically, specificity in autophagy induction might be enhanced by activating the ULK1 complex rather than downstream shared nodes in autophagy and LAP (although the ULK1 kinase complex may have substrates outside of autophagy). The appeal of targeting Rubicon — a negative regulator of autophagy whose knockout in mice has beneficial effects (e.g. improved high-fat diet-induced hepatic steatosis (Tanaka et al., 2016) and increased cardiac protection during lipopolysaccharide-induced sepsis (Zi et al., 2015)) — may be tempered by potential adverse effects of LAP inhibition, such as increased susceptibility to fungal diseases and autoimmunity. Furthermore, treatments that target shared ATG proteins may result in unpredictable effects on each pathway, assuming these proteins are rate-limiting and the two pathways compete for access to these shared core ATG proteins.
The identification of LAP as a lysosomal degradation pathway that utilizes certain core ATG genes requires us to adopt a wider interpretative lens for studies of mice with deletions of these genes. Does deficient LAP versus deficient autophagy contribute to pathological phenotypes in mice with whole body or tissue-specific deficiency of genes such as beclin 1, ATG5, ATG7, or ATG16L1? To what extent does increased autophagy versus decreased LAP contribute to Rubicon knockout phenotypes? Do the GWAS associations between polymorphisms in some of these genes and diseases that involve disordered immune regulation (such as asthma, SLE, and inflammatory bowel disease) (see Table 1) suggest a role for altered LAP in their pathogenesis? The observation that a deficiency of LAP-associated (but not of non-LAP-associated) ATG genes results in a SLE-like syndrome in mice (Martinez et al., 2016) underscores the importance of this question. Specific molecular markers to distinguish LAP from autophagy in both animal models and human disease are needed.
Beyond Lysosomal Degradation: Autophagy Genes Function in Secretion and Exocytosis
ATG genes are used not only for targeting intracellular cargo to the lysosome for degradation, but also for pathways that involve the targeting of intracellular cargo to either the plasma membrane or extracellular environment (Figure 1). Generally, these pathways have been grouped under the umbrella term “secretory autophagy”; however, as the “phagy” part is missing from the process, we prefer the more linguistically precise term of ATG gene-dependent secretion. There are many different types of ATG gene-dependent secretion (reviewed elsewhere from a cell biology perspective [Cadwell and Debnath, 2018]) but the mechanisms governing most of these processes are not well understood. Here, we focus on these pathways as they may relate to mammalian physiology and disease.
Unconventional secretion involves the extracellular release of proteins that lack amino-terminal signal peptide leader sequences and bypass “conventional” transit through the endoplasmic-reticulum (ER)-Golgi apparatus to reach the plasma membrane. A role for ATG proteins (Atg1/5/6/7/8/9/12/17) in this process was first discovered in yeast secretion of the acyl-CoA-binding protein, Acb1 (Duran et al., 2010; Manjithaya et al., 2010). In mammalian cells, unconventional secretion of leaderless proteins, such as the pro-inflammatory cytokines processed by the inflammasome, IL-1β and IL-18, also require the autophagy protein, ATG5 (Dupont et al., 2011). The precise mechanisms underlying ATG gene-dependent unconventional secretion remain unclear. It is not certain whether targets are captured in an autophagosomal lumen (Dupont et al., 2011) and/or the intermembrane space between the double membrane of the autophagosome (Zhang et al., 2015b); nor is it certain how targets are delivered to and released from the plasma membrane. Autophagosome-like vesicles containing IL-1β bypass syntaxin 17-dependent fusion with lysosomes and instead use specific SNAREs and syntaxins involved in vesicle fusion with the plasma membrane for cargo secretion (Kimura et al., 2017).
A function of ATG genes in secretion of pro-inflammatory mediators (and more broadly, other leaderless proteins) could have vast importance for inflammatory disorders and a wide range of other diseases. However, it is currently difficult to assess the physiological importance of ATG gene-dependent secretion of IL-1β and IL-18 in vivo, as macrophage (or hematopoietic cell)-specific deletion of Atg5, Atg16l1, and Atg7 in mice is associated with increased, rather than decreased, levels of IL-1β and IL-18 production (Kimmey et al., 2015; Martinez et al., 2016; Saitoh et al., 2008). These findings may reflect basal functions of ATG genes in the negative control of inflammasome activation (Zhou et al., 2011), whereas the ATG gene-dependent secretion of pro-inflammatory mediators may be unmasked during certain stress conditions, such as inflammasome activation triggered by lysosomal membrane damage (Kimura et al., 2017). The possibility of an autophagy-dependent secretome in vivo warrants further investigation and may lead to the identification of proteomic signatures of autophagy activation as clinically useful serum biomarkers. Theoretically, autophagy-inducing therapies might lead to untoward effects via the unconventional secretion of pro-inflammatory mediators or other pathogenic proteins.
Perhaps the best-established link between ATG gene-dependent secretion and mammalian physiology and disease relates to the exocytosis of secretory granules and lysosomes. Notably, human genome-wide association studies (GWAS) that revealed a polymorphism in a core ATG gene, ATG16L1T300A, as a major risk allele for Crohn’s disease (Barrett et al., 2008) spurred the discovery of a fundamental role for the ATG protein conjugation machinery in secretory granule exocytosis (Cadwell et al., 2008). In mice, hypomorphic expression of Atg16l1, Atg16l1T300A knock-in mutation, Paneth cell-specific deletion of Atg16l1, Atg5, or Atg7, or whole-body deletion of Atg4b results in abnormal granule morphology and a defect in granule exocytosis and lysozyme secretion by Paneth cells (Bel et al., 2017; Cabrera et al., 2013; Cadwell et al., 2008; Lassen et al., 2014), a specialized ileal epithelial cell type that controls the intestinal microbiota by secreting lysozyme and antimicrobial peptides. Similar defects in Paneth cell morphology are observed in patients with (but not those without) the ATG16L1T300A Crohn’s disease risk allele (Cadwell et al., 2008). The precise membrane trafficking mechanisms by which ATG proteins facilitate secretory granule exocytosis in Paneth cells or other cell types remain unknown. However, a recent study indicates that lysozyme is localized in large LC3-positive vesicles in Paneth cells from wild-type but not Atg16l1T300A mice (Bel et al., 2017). Thus, in a manner similar to autophagosome-like vesicles involved in unconventional protein secretion, secretory granules may be earmarked for exocytosis by the presence of LC3 on their surface.
A related, but topologically distinct, link between autophagy and secretory lysosome exocytosis was uncovered in another specialized type of secretory cell, the osteoclast. Osteoclasts resorb bone by a mechanism that involves secretory lysosome fusion with bone-apposed plasma membrane composed of ruffled borders, with the discharge of matrix-degrading molecules into the site of osteal degradation. In mice, the ATG protein conjugation machinery and the Rab GTPase, Rab7, are essential for generating an LC3-labeled ruffled border, cathepsin K release and normal bone resorption (DeSelm et al., 2011), thus indicating a role for ATG genes in mediating polarized secretion of lysosomal contents to the extracellular space. In this scenario, the plasma membrane, not the secretory lysosome, is labeled by LC3. Thus, during secretion, the ATG protein conjugation machinery and resulting lipidated LC3 can function either in the formation of normal secretory granules that properly fuse with the plasma membrane or in the creation of a specialized plasma membrane that fuses with secretory lysosomes.
The predicted clinical outcome of defects in ATG gene-dependent osteoclast functions would be osteopetrosis, a disease marked by abnormally dense bone. Consistent with this prediction, mutations in PLEKHM1, a Rab7 effector, and the v-ATPase α3 subunit involved in lysosomal acidification, are each associated with osteopetrosis in patients (Stenbeck and Coxon, 2014). In contrast, aging, which is associated with reduced autophagy in most cell types (Hansen et al., 2018), is accompanied by osteopenia and osteoporosis in mice and humans. This may reflect the roles of ATG genes in other cell types in the bone that favor bone growth and normal bone density, including protection against endoplasmic reticulum (ER) and oxidative stress in osteoblasts and osteocytes (Li et al., 2018b; Liu et al., 2013a; Onal et al., 2013) and maintenance of the proper function of bone mesenchymal stem cells (Ma et al., 2018). Thus, studies in bone represent an elegant example of how the autophagic machinery can exert different specialized functions in distinct cell types within a given organ – functions that may have opposite effects (such as bone resorption and bone formation) – to orchestrate overall tissue homeostasis. As osteopenia/osteoporosis and associated skeletal fractures are a major cause of morbidity and mortality in aging humans, this area warrants further investigation as a potential clinical target for autophagy upregulation.
Numerous other defects in protein secretion in ATG gene knockout mice have been described, although it is unclear whether they reflect a direct role for ATG genes in autophagy-independent trafficking or more indirect consequences of autophagy deficiency on secretory processes. These include defects in the assembly and secretion of octogonial core proteins which leads to abnormalities in vestibular development (Marino et al., 2010) and defects in pancreatic β-cell insulin granule morphology and secretion (Watada and Fujitani, 2015), melanogenesis and pigmentation (Ganesan et al., 2008), and mucus secretion of airway epithelial cells and intestinal goblet cells (Patel et al., 2013).
Accumulating evidence suggests that ATG proteins also have pleiotropic effects on the cellular release of exosomes, a process that is mediated by fusion of the multivesicular body (MVB) with the plasma membrane (Baixauli et al., 2014; Hessvik and Llorente, 2018). Autophagy induction can prevent – whereas ATG gene silencing or pharmacological inhibition can increase – extracellular release of exosomes, including those containing pathogenic protein cargoes, such as α-synuclein (Fussi et al., 2018), prions (Abdulrahman et al., 2018) and amyloid precursor protein (Miranda et al., 2018). This regulatory mechanism is presumed to involve MVB fusion with autophagosomes, thereby diverting MVB transport away from the plasma membrane. Increased exosome release in the setting of impaired autophagy may function as an alternative quality control pathway to maintain cellular homeostasis and prevent cell death due to proteotoxicity. However, there are also examples in which ATG genes stimulate exosome production. Atg5, but not Atg7, has been shown to decrease late endosome acidification by disrupting the v-ATPase, thereby promoting the production of exosomes that enhance tumor metastasis (Guo et al., 2017). Similarly, the ATG3-ATG12 conjugate which is required for LC3 lipidation during basal (but not starvation) conditions interacts with the ESCRT protein, Alix, and positively controls Alix-dependent exosome biogenesis (Murrow et al., 2015). Given the expanding repertoire of exosome-dependent processes (including neurodegeneration, immune signaling, metabolism, tumor metastasis and viral infection), the effects of the autophagic machinery on the fate of the MVB – lysosomal degradation or exocytosis – may partly underlie the pathophysiological effects of ATG gene mutation.
The ATG machinery modulates retromer function to control the endosome-to-cell-surface recycling pathway (Roy et al., 2017). During metabolic stress, LC3 on autophagic structures binds to the RabGAP protein TBC1D5 to sequester it away from an inhibitory interaction with the retromer complex. This sequestration allows retromers to associate with endosomal membranes and mediate plasma membrane translocation of the glucose transporter, GLUT1, a facilitator of glucose uptake. GLUT1 is required for the low levels of basal glucose uptake required to sustain cellular respiration, and its plasma membrane localization normally increases when cells are exposed to low glucose. Perturbation in ATG protein conjugation may significantly cripple this metabolic homeostatic mechanism involving GLUT1 trafficking and, in addition, affect the cell surface localization of other as-of-yet-unidentified receptors.
Autophagy Genes in Other Dynamic Membrane Events
Non-autophagic functions of ATG genes in membrane trafficking modulate the infection of host cells by viruses, bacteria and other pathogens. These include processes described above such as LAP (which may be partially antagonized by virulent micro-organisms that enter professional phagocytes) and LC3-regulated exocytosis (which is involved in the egress of viruses that either reside inside autophagosomes or whose envelopes become decorated with LC3) (reviewed in [Cadwell and Debnath, 2018]). Many additional ATG gene-dependent membrane reorganization events – or interference with such events – also regulate infection. For example, several ATG genes are required for the formation of intracytoplasmic membrane-associated replication factories of certain medically important RNA viruses, such as hepatitis C virus (Dreux et al., 2009). Similarly, the formation of multi-membranous vacuoles that support replication of the bacterium, B. abortus, involves ATG genes required for class III PI3K activity but not those required for LC3 conjugation (Starr et al., 2012). In contrast, IFN-γ inhibits T. gondii replication by a process involving LC3/GABARAP lipidation and recruitment of IFN-γ-inducible GTPases to the parasitophorous vacuole, where they disrupt the membrane and destroy the parasite’s replicative niche (Choi et al., 2014). Similarly, IFN-γ mediated control of murine norovirus (a model for human epidemics of gastroenteritis) involves labeling membrane-associated viral replication complexes with lipidated LC3 and recruitment of IFN-γ-inducible GTPases (Biering et al., 2017). Thus, marking replication-associated membrane structures by LC3 conjugation may represent a conserved mechanism underlying IFN-γ-mediated control of intracellular pathogen replication. Further understanding of the precise processes by which different subsets of ATG proteins provide or destroy host membranes necessary for different stages of pathogen replication may lead to the development of new anti-infective strategies.
Beyond Membrane Trafficking: Autophagy Proteins Have Other Functions
The autophagy proteins not only help orchestrate the cross-talk of diverse vesicular trafficking pathways, but also interface with multiple other cellular pathways, including (but not limited to) cell death pathways, cell cycle regulation, and innate immune signaling. The interaction of FIP200 with Atg13 is essential for autophagy in vivo and neonatal survival in mice, but the non-autophagic function is sufficient to maintain embryogenesis through a mechanism involving protection against TNFα-induced apoptosis (Chen et al., 2016). Atg7, independently of its E1-like enzymatic activity and function in autophagy, regulates p53-dependent cell cycle arrest and apoptosis, and the neonatal lethality of Atg7 knockout mice is partially rescued by inhibition of the DNA damage response through deletion of the protein kinase Chk2 (Lee et al., 2012). In mice, deletion of Atg9a, but not Atg5, results in a defect in necrosis at the bone surface during developmental morphogenesis (Imagawa et al., 2016). The precise mechanisms underlying these (and additional) functions of individual ATG proteins in cell death and cell cycle regulation are not well understood.
ATG proteins regulate inflammatory and immune signaling both through autophagy-dependent mechanisms (such as by the autophagic removal of damaged mitochondria that produce ROS and activate RIG-I signaling and the NLRP3 inflammasome) and autophagy-independent mechanisms that generally involve ATG protein interactions with immune signaling molecules. For example, the ATG5-ATG12 conjugate inhibits type I IFN signaling in response to viral infection by binding to the CARDs (caspase activation and recruitment domains) of RNA recognition molecules such as RIG-I and MAVs (Jounai et al., 2007). Similarly, the cytosolic DNA sensing innate immunity pathway mediated by cGAS (cyclic GMP-AMP [cGAMP] synthase) and STING (Stimulator of interferon genes) is regulated by autophagy proteins. The generation of cGAMP by cGAS activates ULK1, which phosphorylates and inhibits STING-dependent cytokine production (Konno et al., 2013). As unrestrained STING signaling (either via inherited mutations in the ADAR and ribonuclease H2 complex or gain-of-function mutations in STING) causes human autoinflammatory diseases, ULK1 activating agents have been proposed as potential treatments for such disorders (Konno et al., 2018). Beclin 1 binds cGAS to suppress cGAMP synthesis and halt interferon production (Liang et al., 2014). Atg9a may also function as a negative regulator of innate immune signaling by decreasing the assembly of STING and TBK1 in the presence of double-stranded DNA (Saitoh et al., 2009). Of note, these same RNA-sensing and cytosolic DNA-sensing signaling pathways are activators of autophagy, which is itself an important innate immune effector pathway (Deretic and Levine, 2018). Thus, ATG proteins play a crucial role in both mediating innate immunity and in providing feedback inhibition to fine-tune inflammatory signaling so as to avoid deleterious consequences.
The Selectivity of Autophagy: a Guardian of Cellular Homeostasis
For nearly half a century, the process of macroautophagy was believed to lack cargo specificity. In fact, the morphological identification of an autophagosome required visualizing the simultaneous presence of diverse cytoplasmic contents, such as ER, ribosomes and mitochondria inside a double-membraned vacuole. However, a transformative body of work over the past decade has fully dispelled this notion. We now know that there can be extreme specificity in governing the choice of cargo that is degraded by the autophagosome and an intricate system for earmarking and capturing such cargo. This process, termed selective autophagy, may be more crucial in protection against most mammalian diseases than “bulk autophagy”, which is primarily a homeostatic mechanism during nutrient stress.
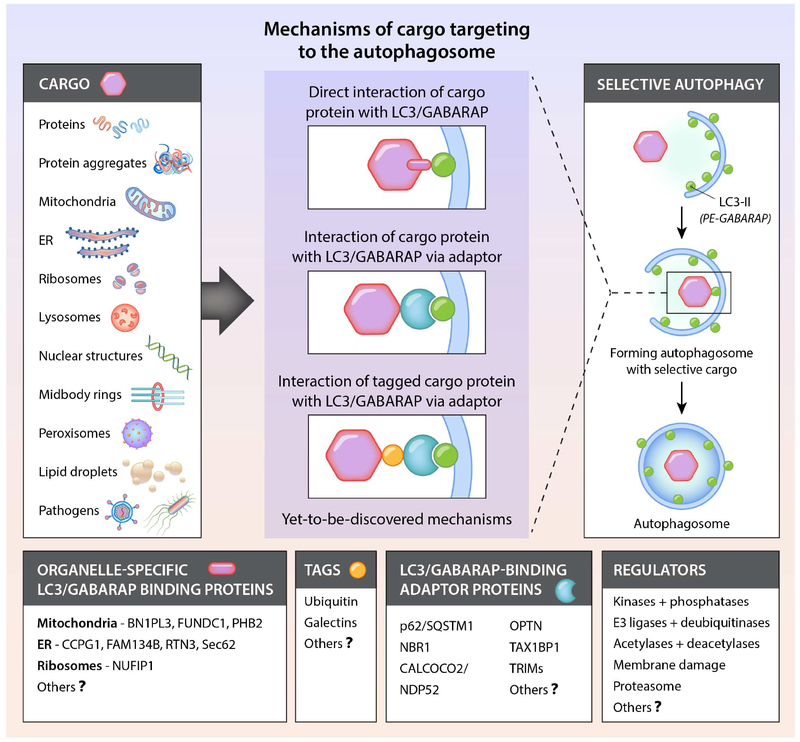
Many parts of the cell can be “selected” for degradation by autophagy (Figure 2 and Table 2). Numerous studies have reported the selective autophagy of various organelles, including mitochondria, ER, peroxisomes, lipid droplets, ribosomes, midbody rings and the nucleus. Autophagy selectively degrades aggregation-prone misfolded proteins such as those involved in the pathogenesis of certain neurodegenerative, skeletal and cardiac muscle, and liver diseases. In addition, it degrades the individual proteins that serve as adaptors to bridge cargo with the nascent phagophore as well as specific inflammatory and immune signaling molecules. Moreover, selective autophagy can target pathogens that reside inside vacuoles or directly inside the cytosol for lysosomal degradation. Once captured, cargo degradation proceeds through a route that involves the same molecular machinery as bulk autophagy. Different forms of selective autophagy are often named by a term comprising a prefix derived from the cargo (e.g. mito-, ER-, ribo-, nucleo-, pexo-, lipo-) and the suffix “phagy”. For selective autophagy of microbial invaders, the term xenophagy is commonly used.
Figure 2. Conceptual overview of selective autophagy.
Shown are the diverse cargoes that are degraded by autophagy and the major known mechanisms by which cargo are attached to LC3 or GABARAP family members on the phagophore membrane. Also listed are currently known organelle-specific LC3/GABARAP-binding proteins, tags that label cargo destined for selective autophagic degradation, LC3/GABRAP-binding adaptor proteins, and factors that regulate the recognition of cargo by adaptors or LC3/GABARAP. Organelle-specific LC3/GABARAP-binding proteins and LC3/GABARAP-binding adaptor proteins interact with LC3/GABARAP via conserved W/F/YxxL/I/V motifs. See Table 2 for information about different types of selective autophagy and their possible roles in physiology and disease.
Table 2.
Types of Selective Autophagy and Possible Roles in Physiology and Disease
| Process (Cargo) | Physiological Function | Possible Pathological Consequences of Defects | References |
|---|---|---|---|
| Proteins | Proteostasis | Aberrant signaling/cellular functions related to effects of increased protein (e.g. p62/SQSTMl inflammatory and pro-tumorigenic signaling; autoinflammatory disorders with defects in TRIM-mediated autophagy of inflammasome components; abnormal iron accumulation and ferroptosis in tissues with defects in ferritin degradation) | (Moscat et al., 2016) (Kimura et al., 2015) (Liu et al., 2016) (Latunde-Dada, 2017) |
| Aggrephagy (Protein aggregates) | Removal of misfolded aggregate-prone proteins labeled by ubiquitin | Enhanced accumulation and detrimental consequences of pathogenic proteins targeted by this mechanism (e.g. β-amyloid, mutant huntingtin, a-synuclein, mutant al-antitrypsin) | (Gatica et al., 2018) (Ueno and Komatsu, 2017) |
| Mitophagy (Mitochondria) | Mitochondrial quality control and homeostasis. Removal of damaged mitochondria, paternal mitochondria during embryogenesis, and mitochondria during erythrocyte differentiation Piecemeal degradation for respiratory chain turnover |
Defective mitophagy may contribute to neurodegenerative diseases, aging, cancer, increased ROS-dependent inflammasome activation and genotoxic stress |
(Rojansky et al., 2016) (Drake et al., 2017) |
| ER-phagy (Endoplasmic reticulum) | Control of ER morphology, turnover, ER luminal proteostasis and recovery from stress | Pathological consequences of defects not defined, but hypothetical role in pathologies associated with abnormal UPR and ER intraluminal proteostasis, including pancreatitis and certain metabolic disorders and aggregopathies. Mutation in ER LC3/GABARAP-binding protein, FAM134B leads to hereditary neuropathy in patients; loss of ER LC3-binding protein CCPG1 leads to injury of exocrine pancreas in mice. Defective ER-phagy in mice (due to AT-1 overexpression) leads to segmental progeria with multiple metabolic and inflammatory phenotypes | (Khaminets et al., 2015) (Grumati et al., 2017) (Smith et al., 2018) (Peng et al., 2018) |
| Ribophagy (Ribosomes) | Required for survival during nutrient starvation, providing source of nucleosides to cell | Not yet known if defects in pathway occur in vivo; if so, would be predicted to disrupt adaptive responses to starvation | (Wyant et al., 2018) |
| Lysophagy (Lysosomes) | Prevents cell destruction and inflammation due to leakage of lyso-somal contents when lysosomal membranes are damaged or ruptured | Defects predicted to be associated with increased cytosol invasion of pathogens, increased lysosomal cell death and inflammation, as well as disruption of lysosomal homeostasis (latter postulated to participate in neurodegeneration); lysosomal damage in autophagy-deficient mice results in acute kidney injury | (Maejima et al., 2013) (Yoshida et al., 2017) (Chauhan et al., 2016a) |
| Nucleophagy (Entire Nucleus) | Nuclear destruction necessary for terminal differentiation of keratinocytes; unknown if required for nuclear removal in red blood cells and lens fiber cells | Perturbations may occur in psoriasis, causing parakeratosis (retention of nuclei in stratum comeum of epidermis) | (Akinduro et al., 2016) |
| Nuclear lamina | Promotes Ras-oncogene-induced senescence | Autophagic degradation of lamin B proposed to be a mechanism of tumor suppression; defects may promote oncogenesis and phenotypes associated with decreased cellular senescence. | (Dou et al., 2015) |
| Micronuclei | Removal of micronuclei generated by mitotic aberration (and cytosolic DNA aggregates that resemble micronuclei) | Defects may contribute to genomic instability associated with autophagy deficiency and pro-inflammatory signaling via cGAS activation; neuroinflammatory autoimmune disorder Aicardi-Goutieres syndrome caused by mutation of DNA repair enzyme RNase H2 resulting in accumulation of micronuclei-like cytosolic DNA aggregates | (Rello-Varona et al., 2012) (Bartsch et al., 2017) |
| Retrotransposon RNA |
Degradation of RNA granules containing retrotransposons may favor genomic stability | Deficiency results in increased retrotransposon insertions into the genome | (Guo et al., 2014) |
| Midbody Rings | Degradation of the midbody, an organelle that contains the remnants of cell division machinery, may regulate cellular fate | Deficiency predicted to alter cellular fate; several mutations affecting midbody proteins cause primary encephalopathies, which is also observed with genetically inherited syndromes associated with defects in autophagic flux | (Kuo et al., 2011) (Mandell et al., 2016) |
| Pexophagy (Peroxisomes) | Perixosomal quality control | Defects may contribute to neurodevelopmental disorders associated with mutations in genes involved in pexophagy, inflammation, aging and age-related diseases, diabetes, cancer and neurodegenerative disorders | (Cipolla and Lodhi, 2017) |
| Lipophagy (Lipid Droplets) | Facilitates transport of lipid droplets to lysosomes for catabolism by lysosomal acid lipase; contributes to lipid homeostasis | Defects are postulated to contribute to pathogenesis of metabolic syndrome, non-alcoholic fatty liver disease and alcoholic fatty liver disease; however, role of defects in lipophagy versus general autophagy pathway remain to be elucidated | (Zechner et al., 2017) (Zhang et al., 2018b) |
| Xenophagy (Intracellular pathogens) | Removal of cytoplasmic bacteria or viruses functions in cell-intrinsic immunity | Mutations in genes required for selective autophagy of pathogens result in enhanced microbial virulence in mouse models of tuberculosis and viral infections. | (Mitchell and Isberg, 2017) (Sumpter et al., 2016) (Franco et al., 2017) |
Major advances have been made in understanding certain aspects of selective autophagy, particularly how cargo binds to the forming phagophore (Figure 2). In most known instances, the cargo either contains an identifiable LC3-interacting region or LIR motif (W/F/Y1×2×3L/I/V4) that directly binds LC3, or it must be labeled with a tag such as ubiquitin, which then binds adaptor proteins that contain both a ubiquitin-binding domain and a LIR motif, thus serving as a bridge between the cargo and the LC3 (or GABARAPs) conjugated to the phagophore membrane. Alternatively, specific proteins (particularly those involved in the inflammasome or IFN signaling) can bind to TRIM (tripartite motif) family members, which serve as adaptors that interact with GABARAPs to target such proteins for autophagic degradation (Kimura et al., 2017). Of note, the proteins we refer to as “adaptors” are often called autophagy “receptors”; however, as these are bridging molecules that are not integral parts of cellular membranes that undergo ligand-dependent activation, the designation as “receptors” can be confusing.
Several layers of control are needed to properly dictate the targeting of cargo for autophagy. In theory, cargo should be disposed of when it is constitutively harmful (e.g. intracellular pathogens), potentially dangerous to the cell (e.g. mitochondrial damage) or obsolete as a result of cellular differentiation (e.g. organelles during erythrocyte maturation). In the scenario where LC3 directly binds to a protein on an organelle containing a LIR motif, there must exist ways to hide or expose the LIR motif in a regulated-fashion. Two mechanisms identified thus far include (1) stimulatory and inhibitory phosphorylations of residues near or in the LIR motif (such as occurs for the mitochondrial outer membrane protein, FUNDC1, that mediates hypoxia-stimulated mitophagy (Lv et al., 2017)) and (2) the exposure of a normally hidden LIR motif (such as occurs when proteasomal-dependent rupture of the outer mitochondrial membrane exposes the inner mitochondrial membrane LC3-binding protein, prohibitin 2 [Wei et al., 2016]). Under circumstances where LC3 binds to an adaptor protein, there must exist ways to recruit the adaptor to the cargo destined for degradation. This process generally involves the concerted action of E3 ligases that ubiquitinate targets (e.g. Parkin, SMURF1), kinases that recruit E3 ligases (e.g. PINK1) or that phosphorylate LIR domains of adaptors (e.g. TBK1), deubiquitinating enzymes (DUBs) that counter E3 ligase activity (e.g. USP30, USP15) (Gatica et al., 2018), and acetylation/deacetylation of mitochondrial and ER target proteins (Peng et al., 2018; Webster et al., 2013).
All mechanisms for earmarking cargo must be tightly coordinated with the formation of autophagosomes to ensure final cargo disposal. Some potential nodes of coordination have recently been described. Certain autophagy adaptors, most notably the TRIM family proteins, bind not only substrate proteins and LC3/GABARAP family members but also assemble the ULK1 and Class III PI3K complexes to initiate autophagosome formation (Kimura et al., 2017). ULK1, a “master kinase” that phosphorylates multiple sites on downstream core autophagy proteins, is recruited to and phosphorylates proteins involved in selective autophagy, such as the LIR domain of the mitochondrial membrane protein, FUNDC1 (Wu et al., 2014). An organelle-specific LC3/GABARAP-binding protein, the ER membrane protein, CCPG1, interacts with a key component of the autophagy-initiating ULK1 complex, FIP200 (Smith et al., 2018). Thus, the machinery involved in selective autophagy substrate recognition may play an active role in autophagy initiation.
For selective autophagy targeting events that involve substrate ubiquitination, the precise mechanisms that dictate the choice between autophagic versus proteasomal degradation are uncertain. In yeast, substrate aggregation and oligmerization of the ubiquitin-binding proteins may favor autophagic degradation (Lu et al., 2017). The lysine residues used for linkage and the length and nature of the ubiquitin chains have also been proposed to contribute to pathway selection (Gatica et al., 2018), but definitive evidence is lacking. Moreover, despite elegant studies characterizing the ubiquitylome of selective autophagy cargo (such as mitochondria and intracellular pathogens) (Grumati and Dikic, 2018), the ubiquitin substrates required for autophagic targeting remain largely undefined.
Selective autophagy seems to involve multiple concurrent targeting mechanisms that act in a cooperative, potentially hierarchical and/or partially redundant manner to ensure proper removal of cargo. This “combinatorial design” may allow specific cell types to more precisely regulate when and how selective autophagy occurs for a given cargo. The partial redundancy also renders it more feasible to study loss-of-function phenotypes of genes required for selective autophagy but dispensable for bulk autophagy (as compared to those required for bulk autophagy), as they are less likely to be lethal to the cell or organism. As selective autophagy genes are partially redundant, this may explain why their loss-of-function mutation seems to be better tolerated in the human population than loss-of-function mutation of core ATG genes.
Indeed, mutations in many of the known molecules involved in selective autophagy are associated with susceptibility to a variety of human diseases (Table 1). Mutations in the genes encoding the adaptor proteins p62/SQSTM and optineurin, the E3 ligase Parkin, and the kinases PINK1 and TBK1, are among the most common causes of familial and early onset neurodegenerative diseases, including Parkinson’s disease, frontotemporal dementia and amyotrophic lateral sclerosis. Hereditary sensory and autonomic neuropathy type II is caused by mutations in an ER-specific LC3 binding protein, FAM134B, required for ER-phagy. Mutations in TRIM20 (also known as pyrin) that impair its ability to target inflammasome components for autophagic degradation result in an inherited autoinflammatory disorder, familial Mediterranean fever. Inflammatory bowel disease-associated genes encode proteins that function in multiple steps of autophagy, including the selective targeting of bacteria by the adaptor, CALCOCO2/NDP52, and the E3 ligase, SMURF1.
The numerous links between mutations in selective autophagy genes and human diseases underscore the likely physiological importance of different forms of selective autophagy (Table 2). However, the precise mechanisms that connect genotype to phenotype remain largely undefined. For example, it is not known why mutations in Parkin and PINK1 are associated with Parkinson’s disease, whereas mutations in optineurin, TBK1, and p62/SQSTM1 are associated with amyotrophic lateral sclerosis and frontotemporal dementia (Table 1). In addition to potential cell non-autonomous effects of mutations in these genes in tissues outside of the brain, cell type-specific differences in various populations of neurons and glia may exist with respect to (1) dependency on subsets of selective autophagy genes; (2) expression and activity of DUBs and other negative feedback mechanisms that regulate selective autophagy; and/or (3) levels and types of stress that mandate different types of selective autophagy responses to maintain homeostasis (e.g. mitophagy or other forms of selective autophagy such as aggrephagy that are relevant to neurodegenerative diseases). Parkin knockout mice (unlike flies lacking Parkin) do not develop spontaneous neurodegeneration, but they do develop dopaminergic neuronal degeneration (resembling that observed in human Parkinson’s disease) when crossed with “mutator” mice with a proof-reading-defective mitochondrial DNA polymerase (PolG) that accumulate mitochondrial mutations (Pickrell et al., 2015). The localization of disease in dopaminergic neurons may be related to increased mitochondrial stress in these cells as compared to other neuronal populations in the brain. Intriguingly, the motor defect and neurodegeneration in Parkin-null/mutator mice can be rescued by deletion of STING, a regulator of Type I IFN responses to cytosolic DNA (Sliter et al. 2018). Thus, aberrant inflammatory signaling as a result of defects in mitophagy may contribute to the pathogenesis of neurodegenerative disease in patients with Parkin or PINK1 mutations.
While most, if not all, forms of selective autophagy are likely to contribute to normal physiology and protection against disease, mitophagy has been the most extensively studied. Mitophagy is an essential component of mammalian developmental and differentiation processes, including elimination of paternal mitochondria from the fertilized egg (Rojansky et al., 2016), removal of mitochondria during red blood cell maturation (Sandoval et al., 2008) and beige-to-white adipocyte differentiation (Lu et al., 2018). In addition to Parkinson’s and other neurodegenerative diseases, defective mitophagy is thought to contribute to organ-specific and systemic inflammatory diseases (Zhao et al., 2018), cancer development and/or progression (Drake et al., 2017), and potentially aging (Lopez-Otin et al., 2016). The removal of damaged mitochondria by mitophagy maintains normal cellular metabolism, reduces mitochondrial generation of ROS that trigger inflammation and genotoxic stress, and prevents mitochondrial release of pro-apoptotic factors. Thus, maintenance of proper mitochondrial function by mitophagy is crucial for cellular and organismal health. Other forms of selective autophagy (including xenophagy) likely operate in a manner analogous manner to mitophagy, in that the mechanisms by which they regulate physiology and disease are a function of the normal “duties” of their substrate and the ensuing pathological consequences of abnormal substrate accumulation (see Table 2).
Our expanding knowledge of the mechanisms and physiological functions of selective autophagy may open up new — albeit unchartered — pathways for drug discovery. Knockdown of the mitochondrial deubiquitinase, USP30, rescues mitophagy defects and disease in flies with pathogenic mutations in Parkin (Bingol et al., 2014), suggesting a potential role for the inhibition of DUBs that target selective autophagy E3 ligases in the treatment of Parkinson’s and other diseases. Indeed, novel highly selective inhibitors of USP30 that accelerate mitophagy have recently been reported (Kluge et al., 2018). As phosphorylation of substrates is also a common mechanism involved in selective autophagic targeting, it may be possible to activate specific kinases to enhance selective autophagy. Potentially, it may also be possible to develop novel strategies to attach high-affinity LIR domains selectively to harmful cargo so that they can be more efficiently captured by an LC3-decorated nascent autophagosome.
Autophagy Regulation: A Nexus for Therapeutics?
Autophagy was originally studied in yeast and mammalian cells as a nutrient stress response pathway. During the past decade, we have dramatically expanded our knowledge of autophagy regulation, particularly the spectrum of physiological and pathophysiological stimuli that control autophagy, the mechanisms that regulate the activity of the core autophagy proteins (Grumati and Dikic, 2018), and the interconnectivity of autophagy with other cellular stress response pathways (Kroemer et al., 2010). These concepts have been reviewed elsewhere; here, we highlight selected aspects relevant to physiology and disease.
Post-translational protein modifications such as phosphorylation, ubiquitination, and acetylation play a central role in coordinating the activity of ATG proteins. In most cases, the upstream kinases/phosphatases, ubiquitin ligases/DUBs, and acetyltransferases simultaneously modify both ATG proteins and proteins involved in other cellular stress-response pathways that are co-regulated with autophagy. As a result, pharmacological targeting of these enzymes will elicit broad-based modulation of multiple intertwined stress-response pathways. Depending on the enzyme and its substrates, such nonspecific targeting may be harmful in some instances, and useful in others.
One important example is the stimulation of AMPK, a low energy-sensing kinase activated by ATP depletion, which phosphorylates multiple proteins to both stimulate catabolic pathways (including autophagy) and restrain anabolic pathways (including mTORC1 signaling), thereby ensuring limitation of ATP consumption and generation of new ATP via breakdown of metabolic products (Herzig and Shaw, 2018). In recent years, AMPK has been shown to not only activate autophagy through inhibition of mTORC1, but also directly phosphorylate several ATG proteins, including ULK1, ATG9A, Beclin 1, and VPS34 (Egan et al., 2011; Kim et al., 2013a). In addition, AMPK promotes mitophagy through effects on ULK1 and stimulates TFEB-dependent activation of the CLEAR (Coordinated Lysosomal Expression and Regulation) network of genes required for autophagy (Herzig and Shaw, 2018). This pro-autophagic activity of AMPK occurs concurrently with its effects on mitochondrial homeostasis and on lipid and glucose metabolism.
AMPK activation may underlie the beneficial effects of metformin, a drug widely prescribed for the treatment of diabetes (Herzig and Shaw, 2018). Metformin activates AMPK indirectly through mitochondrial depletion of ATP, and direct AMPK activators that yield effects similar to metformin are in pre-clinical development. The extent to which autophagy stimulation contributes to beneficial effects of AMPK activation in mice or patients is not known, but it seems likely that autophagy represents a critical part of an AMPK-activated hub that protects against various metabolic diseases, including diabetes, obesity, and non-alcoholic fatty liver disorders, as well as certain cancers and aging-related phenotypes. In Drosophila, deficiency of the Beclin 1 orthologue (ATG6) impairs the ability of metformin to prevent intestinal stem cell aging (Na et al., 2018), and lifespan extension by neuronal AMPK expression requires the fly ULK1 orthologue, ATG1 (Ulgherait et al., 2014). In mice, AMPK upregulation of autophagy is correlated with improved function of aging muscle stem cells (White et al., 2018); additionally, muscle-specific AMPK deficiency results in defective autophagy, fasting-induced hypoglycemia, and aging-associated myopathy (Bujak et al., 2015). In yeast, core ATG genes are required for AMPK-mediated lipid droplet degradation and survival during acute glucose deprivation (Seo et al., 2017).
The lysine acetylation/deacetylation of ATG proteins has emerged as a central node of autophagic control regulated by metabolic sensors involved in lipid, glucose and protein metabolism. Moreover, this control center may function independently from, but intertwined with, AMPK and mTORC1 (Marino et al., 2014b; Su et al., 2017). During acute nutrient depletion, cells undergo a rapid decrease in levels of cytosolic acetyl coenzyme A (AcCoA), which leads to the deacetylation of cellular proteins (Marino et al., 2014b). Sirtuin 1 (which is downstream of AMPK) deacetylates multiple ATGs (e.g. ATG5, ATG7, ATG12, Beclin 1, VPS34, LC3) and thereby promotes autophagy, as does reduced activity of the acetyl transferase EP300 (Madeo et al., 2014; Su et al., 2017). Hence, endogenous activators of sirtuin-1 (e.g. nicotine adenine dinucleotide (NAD+)), endogenous inhibitors of EP300 (e.g. spermidine, a dietary polyamine), and reduced availability of AcCoA (a rate-limiting step for EP300 function) all stimulate autophagy (Madeo et al., 2014).
Compounds that act on these pathways, thereby mimicking the effects of caloric restriction (so-called “caloric restriction mimetics”), are an active area of investigation, and genetic evidence suggests that autophagy is essential for their beneficial effects in vivo. Reservatrol, an indirect sirtuin activator, requires the autophagy machinery for its favorable effects on longevity in nematodes (Morselli et al., 2010). Spermidine-induced autophagy is required for several of its beneficial health effects in model organisms, including lifespan extension in flies, worms, and mice; prevention of cardiac aging in mice; improvement in neuronal function in aging flies; and preservation of myocyte stemness in mice (reviewed in (Madeo et al., 2018)). Moreover, caloric restriction mimetics improve anti-tumor immune surveillance and enhance chemotherapy responses in autophagy-competent, but not autophagy-incompetent, mouse tumor allografts (Pietrocola et al., 2016).
Over the past decade, the lysosome — an organelle traditionally viewed as the downstream “workhorse” for autophagosomal cargo degradation — has been shown to also play a crucial role in the upstream regulation of autophagy (Napolitano and Ballabio, 2016; Shen and Mizushima, 2014). The nutrient-sensing kinase complex, mTORC1, detects both cytosolic and intra-lysosomal amino acids through distinct mechanisms to inhibit autophagy (Saxton and Sabatini, 2017). Amino acids (such as arginine) inside the lysosomal lumen are sensed by the amino acid transporter SLC38A9, which interacts with the lysosomal v-ATPase/Rag/Ragulator complex to activate mTORC1. This both restrains autophagy during baseline conditions and provides feedback inhibition to terminate autophagic responses to acute nutrient depletion. mTORC1 activation in the fed state and/or hyperactivation (as a result of mutations in regulatory signals) switches the cell to a state of anabolic growth and energy storage. Although essential for cell growth and proper metabolic regulation, sustained mTORC1 activation at the organismal level is associated with a variety of pathophysiological consequences, including impaired neonatal gluceoneogenesis and survival (Efeyan et al., 2013); accelerated age-related decline in pancreatic β-cell function (Shigeyama et al., 2008); late-onset muscle atrophy (Castets et al., 2013); altered lipogenesis and adipogenesis (Lee et al., 2016); immune suppression; epileptic seizures and autistic traits; tumorigenesis; and aging (Saxton and Sabatini, 2017). While in some cases, impaired induction of autophagy has been documented in mice with hyperactive mTORC1 signaling and is postulated to contribute to pathological phenotypes (e.g. impaired neonatal gluconeogenesis, late-onset muscle atrophy), the precise role of autophagy inhibition in most diseases associated with mTORC1 signaling remains unknown. There has been some interest in using FDA-approved mTOR inhibitors for the treatment of neurodegenerative disorders that may benefit from autophagy induction (Sarkar, 2013). However, the safety and efficacy of using mTOR inhibitors to induce therapeutic autophagy is uncertain, given the broad range of essential catabolic functions regulated by mTORC1 along with the lack of full specificity of existing agents to target mTORC1 rather than mTORC2 (which functions primarily as an effector of insulin/PI3K signaling).
Both AMPK and mTORC1 participate (in opposite directions) in a signaling axis that links autophagy, the lysosome, and the transcription factor EB (TFEB) and related family members. During the acute response to autophagic stimuli, transcriptional activation is not required, as evidenced by the observation that enucleated cells (cytoplasts) undergo autophagy (Morselli et al., 2011). However, sustained autophagy requires TFEB, a transcription factor that (when inactive) binds to Ragulator at the lysosomal membrane, is phosphorylated by mTORC1, and retained in the cytoplasm by 14–3–3 proteins. Following mTORC1 inhibition, TFEB dephosphorylation releases it from the cytoplasm, allowing its nuclear translocation and subsequent activation of the CLEAR gene network, which includes genes encoding lysosomal hydrolases, lysosomal v-ATPase pumps, lysosomal regulators and autophagy regulators (Puertollano et al., 2018). As noted above, AMPK also activates TFEB-dependent gene expression; this occurs through multiple different mechanisms. In addition, a recent study showed that phosphorylation of acetyl-CoA synthetase 2 (ACSS2) promotes its transport into the nucleus, where it binds to TFEB and favors the acetylation of histone H3 residues within the promoters of TFEB target genes (Li et al., 2017b).
In addition to TFEB, other transcription factors from the same family (such as micropthalmia-associated transcription factor [MITF] and TFE3) (Perera et al., 2015) or from other families (e.g. FOXO3A, HSF1 or TP53) stimulate autophagy (Cai et al., 2018; Kenzelmann Broz et al., 2013). Bromodomain 4 (BRD4), a transcription factor that represses autophagy and lysosomal genes, is displaced from chromatin in response to starvation by a signaling cascade involving an AMPK-SIRT1 axis (Sakamaki et al., 2017). Thus, multiple known (and probably yet-to-be-identified) transcription factors regulate the synthesis of genes required for autophagy (including both the formation of the autophagosome and degradation of its contents by lysosomes). Not surprisingly, the activity of these transcription factors is tightly regulated by numerous signaling factors that also regulate core ATG protein function by post-translational modifications.
Modulation of the activity of TFEB, a master regulator of both lysosomal biogenesis and autophagy, has emerged as a potential therapeutic strategy. Conceptually, this approach is attractive, since limitations in lysosomal numbers and function either occur intrinsically as part of many rare, but devastating, difficult-to-treat, primary diseases (such as lysosomal storage disorders [LSDs]) or are acquired during the progression of diseases associated with the clearance of toxic aggregates progress (such as Huntington’s, Parkinson’s, Alzheimer’s disease and tauopathies). In mice, TFEB overexpression ameliorates several LSDs, neurodegenerative diseases, and α1-antitrypsin deficiency, and it also promotes lipophagy, thereby reducing obesity and associated metabolic syndrome (Napolitano and Ballabio, 2016). The mechanisms by which TFEB overexpression partially corrects lysosomal malfunction in LSDs are not fully understood, but may involve induction of lysosomal exocytosis for the secretion of undigested material.
One potential obstacle to strategies for enhancing TFEB family activity is the risk of tumorigenesis associated with constitutive activation (e.g. renal clear cell carcinoma with TFEB and pancreatic cancer with MITF, TFE3, and TFEB) (Napolitano and Ballabio, 2016). While MITF/TFE3/TFEB-dependent autophagy-lysosomal activation is thought to sustain metabolic reprogramming in pancreatic cancer cells by maintaining intracellular amino acid pools (Perera et al., 2015), further genetic investigations are warranted to confirm that ATG genes are involved in these effects. Moreover, enhanced activity of BRD4, a transcriptional repressor of autophagy, drives another type of cancer, NUT midline carcinoma (Sakamaki et al., 2017), suggesting the effects of transcriptional regulators of autophagy on tumorigenesis may be cell-type specific. It is unclear whether specific subsets of the TFEB-regulated gene network can be induced to avoid genes that contribute to tumorigenesis without losing beneficial effects on the autophagy-lysosomal pathway. An alternative strategy is to activate TFEB on an intermittent basis and/or for limited periods to avoid potential oncogenic effects.
Intriguingly, one of the most widely used medications in the world – aspirin – has been reported to upregulate TFEB in brain cells (via activation of PPARα), induce lysosomal biogenesis, and decrease amyloid plaque pathology in a mouse model of Alzheimer’s-like disease (Chandra et al., 2018). Aspirin (and its active metabolite salicylate) also induces autophagy via inhibition of the acetyltransferase EP300 (Pietrocola et al., 2018) and via AMPK activation/mTORC1 inactivation (Din et al., 2012). However, there is as-of-yet no direct genetic evidence that autophagy contributes to the health benefits of aspirin.
Highly specific activation of autophagy may be possible through strategies that enhance the activity of the upstream components in the core autophagy pathway, i.e. the ULK1 serine/threonine kinase complex and/or Beclin 1/VPS34 lipid kinase complexes. As a key allosteric regulator of VPS34 lipid kinase activity, Beclin 1 activity is tightly regulated by multiple post-translational modifications (ubiquitination, acetylation, phosphorylation) which govern its stability, heterodimeric binding to ATG14 or UVRAG, homodimerization in an inactive form, and/or binding to negative regulators, such as Bcl-2/Bcl-xL (Grumati and Dikic, 2018; Levine et al., 2015). Diverse stress kinases, including AMPK and MAPKAPK2/3, as well as the upstream ATG protein, ULK1, mediate stimulatory phosphorylations of Beclin 1 (Kim et al., 2013a; Park et al., 2018; Wei et al., 2015). The oncogenic kinases, Akt and EGFR, and the EP300 acetylase inhibit the autophagic activity of Beclin 1; mutation of their target post-translational sites in Beclin 1 demonstrates that suppression of Beclin 1-dependent autophagy promotes tumor growth in mouse xenograft models (Sun et al., 2015; Wang et al., 2012; Wei et al., 2013). Enhanced proteasome-mediated degradation of Beclin 1 due to decreased binding of the deubiquitinase, ataxin 3, may contribute to dysregulated autophagy in cells of patients with polyglutamine expansion protein-related diseases, such as Huntington’s and spinocerebellar ataxia type 3 (Ashkenazi et al., 2017).
Disruption of Bcl-2 binding to Beclin 1 represents a central mechanism by which autophagy is activated in response to stress stimuli (such as starvation, exercise, and immune signaling) (He et al., 2012; Wei et al., 2008). This disruption can be triggered by phosphorylation of the BH3 domain of Beclin 1 by DAPK, ubiquitination of the Beclin 1 BH3 domain by the E3 ligase TRAF6, phosphorylation of Bcl-2 by JNK1, or competition by BH3-only proteins (reviewed in [Levine et al., 2015]). Mice containing knock-in non-phosphorylatable mutations in Bcl-2 that prevent disruption of its binding to Beclin 1 are deficient in starvation and exercise-induced autophagy, have decreased exercise endurance, and fail to manifest the beneficial effects of exercise on glucose metabolism (He et al., 2012). Conversely, mice with a knock-in mutation in Beclin 1 that decreases Bcl-2 binding exhibit increased autophagy and extended lifespan and healthspan, including protection against Alzheimer’s-like disease and HER2-mediated breast cancer (Fernandez et al., 2018; Rocchi et al., 2017; Vega-Rubin-de-Celis et al., 2018). Thus, disruption of Beclin 1/Bcl-2 binding may be a safe and effective approach to induce autophagy in vivo; preclinical studies are in progress to develop agents that act through this mechanism (Chiang et al., 2018).
Cell-penetrating peptides (Tat-Beclin 1) derived from a flexible hinge region of Beclin 1 important for VPS34 membrane association and lipid kinase activity (Rostislavleva et al., 2015) are sufficient to induce autophagy in vitro and in vivo (Shoji-Kawata et al., 2013). In mice, Tat-Beclin 1 protects against West Nile virus, chikungunya virus and E. coli bacterial infections; lipopolysaccharide-induced cardiac dysfunction; pressure overload-induced heart failure; hyperammonemia in liver failure and urea cycle disorders, and bone loss in LSDs and in FGF-deficiency (Bartolomeo et al., 2017; Cinque et al., 2015; Shoji-Kawata et al., 2013; Soria et al., 2018; Sun et al., 2018). It also enhances chemotherapeutic effects of murine cancers in immune competent mice (Pietrocola et al., 2016), reduces the growth of human HER2-positive breast cancer xenografts in immune-deficient mice (Vega-Rubin-de-Celis et al., 2018), and acts synergistically with erastin to increase animal survival in an orthotopic pancreatic cancer model (Song et al., 2018). In rats, intrahippocampal injection of Tat-Beclin 1 improves long-term spatial memory (Hylin et al., 2018). In a zebrafish model of human polycystic kidney disease, Tat-Beclin 1 ameliorates renal cyst formation (Zhu et al., 2017). Further studies are needed to examine whether Tat-Beclin 1 induces these effects through autophagy, autophagy-independent effects of Beclin 1, or alternative mechanisms. Moreover, precise definition of its mechanism of action may lead to the development of novel small drug-like molecules that mimic its activity. Recent structural advances elucidating the atomic details of the Beclin 1/VPS34 complexes (reviewed in [Hurley and Young, 2017]) may provide a basis for rational drug design to selectively activate autophagy-specific Beclin 1-associated VP34 lipid kinase activity.
Autophagy in Tissue and Whole-Body Homeostasis
The health of multicellular organisms requires the coordinated regulation of cellular life and death decisions, cell fate determinations, preservation of genomic integrity, immune responses, and metabolic circuitries. The autophagy machinery, via its diverse functions described above (and yet-to-be-discovered mechanisms) plays a crucial role in these processes. Herein, we highlight some recent advances related to the role of autophagy in cell death, preservation of stem cells, tumor suppression, longevity and defense against metabolic diseases.
Autophagy as a Homeostat
During both routine “housekeeping” and responses to acute stress, cells must find ways to maintain adaptive cytoprotective levels of autophagy while simultaneously avoiding potentially maladaptive levels and/or detrimental effects of autophagy. This balance involves self-control of the levels of autophagy, mechanisms of preventing degradation products from becoming toxic to cells, avoidance of degrading essential cargo, and suppression of unwarranted cell death – which likely is a combined function of the aforementioned processes. Cellular self-titration of levels of autophagy involves multiple different inhibitory feedback loops, including feedback regulation of nutrient sensing signals by the generation of amino acids, acetyl-CoA and respiratory substrates (Galluzzi et al., 2014); cytosolic retention of pro-autophagic transcription factors by ATG7 (Simon et al., 2017); and TFEB-mediated activation of mTORC1 (Simon et al., 2017). Cellular toxicity by degradation products may be avoided during autophagy, as evidenced by the observation that the generation of lipid droplets generated by autophagy-dependent dismantling of lipid membranes during starvation-induced autophagy sequesters fatty acids, thereby protecting mitochondria against lipotoxicity and preserving cellular viability (Nguyen et al., 2017). It is not known whether starvation-induced autophagy preferentially induces removal of certain, perhaps aged, structures (and if so, by what mechanisms) or whether it is non-specific. Mitochondria elongate during starvation, which spares them from the autophagic capture that generally occurs after fission (Gomes et al., 2011). Numerous yet-to-be-discovered mechanisms likely protect mitochondria and other organelles from excessive autophagic capture during stress-induced autophagy.
The aforementioned negative feedback loops restrain autophagy to adaptive (rather than maladaptive) levels, allowing this homeostatic pathway to exert cytoprotective effects during stress, and thereby, prevent apoptotic and necroptotic cell death (Marino et al., 2014a). In addition, ATG gene-dependent processes, such as increased plasma membrane localization of the GLUT1 glucose transporter (Roy et al., 2017), may increase the threshold of damage required to kill cells and thereby promote successful organismal adaptation to stress. Promotion of cell survival in vivo during stress-induced autophagy may depend on concurrent antagonism of the Na+,K+-ATPase pump, which normally consumes a large fraction of the ATP available to the cell (Kheloufi et al., 2015). Interestingly, during acute bouts of exercise or nutrient limitation (potent physiological stimuli of autophagy that generally do not result in cell death), endogenous cardiac glycosides that target Na+,K+-ATPase are upregulated 50–500 fold, resulting in decreased cellular ATP consumption (Schoner, 2002).
However, when organisms are pushed beyond physiological limits of energy deprivation, adaptive mechanisms are insufficient to keep cells alive and to prevent tissue damage. In the liver of patients with anorexia nervosa or in neonatal rodents subjected to severe cerebral ischemic injury, a morphologically and genetically distinct form of cell death occurs called autosis, which requires both ATG genes and Na+,K+-ATPase activity (Kheloufi et al., 2015; Liu et al., 2013b). It is not clear how the cell’s major consumer of ATP (i.e. the Na+,K+-ATPase pump) and the cell’s major mobilizer of ATP-generating substrates during stress conditions (i.e. autophagy) interact to regulate life and death decisions of the cell. However, this interaction may represent a fundamental energy homeostatic mechanism that becomes pathologic during different types of ischemic conditions.
Another recently identified bona fide form of autophagic cell death (i.e. demonstrating a genetic requirement for ATG genes) involves GBA1 — the gene encoding the lysosomal enzyme, glucocerebrosidase (GCase) — which metabolizes glucosylceramide (GlcCer) to ceramide and glucose. GBA1 knockdown blocks autophagic cell death in resveratrol-treated lung cancer cells in vitro and developmental midgut death in Drosphila in vivo (Dasari et al., 2017). In humans, homozygous GBA1 mutations lead to Gaucher’s disease, a lysosomal storage disorder, while heterozygous mutations are the most important risk factor for Parkinson’s disease (Schapira, 2015). Interestingly, GBA1 deficiency in the brains of flies and mice leads to an accumulation of GlcCer, impaired autophagic-lysosomal flux, α-synuclein aggregate accumulation, and neurodegeneration; these features are similar to those observed in patients with Parkinson’s disease (Aflaki et al., 2017).
Thus, levels of cellular GCase are crucial for homeostasis; insufficient GCase activity results in a defect in autophagic-lysosomal function and neurodegeneration, whereas excessive GCase activity results in autophagic cell death. It is not yet known how GCase levels are physiologically titrated or how an excess of GCase activity converts adaptive autophagy into lethal autophagy. One possibility is that death occurs as a result of the generation of a metabolic intermediate, sphingosine, which affects lysosomal membrane permeabilization, as lysosome-membrane permeabilization by lysosomal-targeted Bax facilitates autophagic cell death in Bax/Bax knockout murine embryonic fibroblasts (Karch et al., 2017). Future studies are needed to determine the precise mechanisms that convert autophagy from a pro-survival to a cell death pathway, to specifically delineate the role of autophagic cell death pathways in pathophysiology, and to devise therapeutic strategies to block such death in vivo.
Autophagy in Stemness
Accumulating evidence indicates that autophagy is required for stem cell quality control (especially via mitophagy-mediated reduction in ROS levels), energy homeostasis, metabolic reprogramming, and the preservation of fitness. This requirement has multiple disease-related implications, depending on the type of stem cell (e.g. embryonic, adult, cancer) (see [Boya et al., 2018] for detailed recent review). In general, autophagy functions in adult stem cells, including muscle stem cells (satellite cells), hematopoietic stem cells (HSCs), and neural stem cells (NSCs) as a mechanism to prevent exhaustion and aging and to promote quiescence, allowing either self-renewal or differentiation (as needed). Conditional Atg7 deletion leads to a reduction of the muscle satellite cell pool in young mice and hallmarks of premature muscle aging (senescence and DNA damage), and autophagy activation reverses senescence and restores regenerative functions in satellite cells in aged animals (Garcia-Prat et al., 2016). Loss of Atg12 in HSCs impairs maintenance of HSC quiescence and stemness (Ho et al., 2017). Loss of FIP200 results in a progressive loss of neural stem cells (NSCs) and reduced neurogenesis in the adult brain (Wang et al., 2013). Thus, upregulation of autophagy may help stimulate muscle regeneration in sarcopenia, prevent late-onset diseases associated with immune cell senescence, and/or promote adult neurogenesis.
Several studies suggest a role for autophagy in the reprogramming of somatic cells to generate induced pluripotent stem cells (iPSCs) (Boya et al., 2018). The precise mechanisms by which autophagy functions in pluripotency reprogramming are debated, but may involve degradation of transcription factors, ability to sustain glycolytic metabolism (which favors stemness), degradation of mitochondria and other organelles, and mitophagy-mediated limitation of cellular ROS production. While the deletion of core ATG genes or loss of PINK1-dependent mitophagy impairs the reprogramming process (Boya et al., 2018), it is not known if autophagy upregulation improves the efficiency of pluripotent reprogramming for the optimization of stem-cell based therapies.
Additionally, autophagy is important in the origin, differentiation, and survival of cancer stem cells (CSCs), a unique niche which (similar to other stem cell populations) has the potential for self-renewal, but also has the potential for malignancy, initiation of tumor metastasis, and enhanced chemotherapy resistance (Pattabiraman and Weinberg, 2014). CSC formation is enhanced in regions of tumors that are hypoxic, nutrient-depleted and acidic — conditions that favor enhanced autophagy, and CSCs usually have higher rates of basal autophagy than non-cancer stem cells (Boya et al., 2018). Knockdown of core ATG genes in breast CSCs impairs their self-renewal in vitro and impairs their growth when xenografted into mice (Boya et al., 2018). This observation has raised interest in the therapeutic potential of autophagy inhibition of CSCs. However, it is not clear how such cells could be selectively targeted in vivo, and systemic inhibition of autophagy, even in adult mice, is associated with multiple severe toxicities, including multi-organ degeneration and fatal hypoglycemia during fasting (Karsli-Uzunbas et al., 2014). Moreover, this concept is further complicated by the plasticity of CSCs, i.e. the ability to convert to non-cancer stem cells and vice versa. In addition, the depletion of autophagy in HSCs favors the expansion of acute myeloid progenitor cells and development of frank hematological malignancies (Auberger and Puissant, 2017). Given these complexities, it may be premature to consider targeting autophagy in CSCs as an anti-cancer strategy.
Autophagy in Genomic Stability and Tumor Suppression
Besides eliminating ROS-producing dysfunctional mitochondria that are potentially mutagenic, autophagy may promote genomic stability through several additional mechanisms. In the setting of impaired autophagy, the accumulation of the autophagy adaptor and substrate, p62/SQSTM1 (1) inhibits the E3 ligase RNA168 that is essential for histone and chromatin ubiquitination and DNA damage responses (Wang et al., 2016) and (2) activates NRF2 transcription factor of MDM2, which acts through p53-dependent and –independent mechanisms to abrogate normal cell cycle checkpoints (Todoric et al., 2017). Genetic inhibition of autophagosome formation or lysosomal function results in a failure to degrade the small GTPase, RHOA, which leads to cytokinesis failure, multinucleation, and aneuploidy (Belaid et al., 2013). Selective autophagic removal of micronuclei and endogenous retrotransposons may also promote genomic stability (Table 2). Intriguingly, DNA damage repair pathway genes are involved in the selective autophagy of ROS-generating organelles, including pexophagy (e.g. ATM kinase (Zhang et al., 2015a)) and mitophagy (e.g. Fanconi anemia proteins (Sumpter et al., 2016)). Thus, there may be selective pressure for nuclear DNA damage pathways and autophagy proteins to function at multiple levels to protect the genome – both in the cytoplasmic removal of dysfunctional organelles that threaten genomic integrity and, more directly, in the regulation of nuclear events that maintain genomic stability.
The role of autophagy in promoting genomic stability is consistent with its role in tumor suppression. One of the most frequent genetic alterations in sporadic human breast and ovarian cancer is the allelic loss of beclin 1, which is associated with more aggressive cancers and worse patient survival (independently of allelic loss of the nearby tumor suppressor BRCA1) (Liang et al., 1999; Tang et al., 2015; Valente et al., 2014). Mice lacking a copy of beclin 1 develop spontaneous malignancies, demonstrating that it is a haploinsufficient tumor suppressor gene (Cicchini et al., 2014; Qu et al., 2003; Yue et al., 2003), and allelic loss of beclin 1 in immortalized mouse mammary epithelial cells promotes mammary tumorigenesis, DNA damage, and genomic instability in vivo (Karantza-Wadsworth et al., 2007). Similarly, partial autophagy defects in other mouse models (Ambra1+/−, Atg4c−/−, Sh3glb1−/− and mosaic Atg5−/−) are associated with an increased incidence of spontaneous or chemically-induced tumors (Rybstein et al., 2018). In mice, loss of the mitophagy receptor, BNIP3, accelerates progression to metastatic breast cancer (Chourasia et al., 2015), and loss of Parkin results in spontaneous liver tumors, increased radiation-induced lymphoma, enhanced colorectal adenoma development in Apc mutant mice and accelerated KRas-driven pancreatic tumorigenesis (Drake et al., 2017; Li et al., 2018a; Poulogiannis et al., 2010). In patients, PARKIN inactivating mutations are observed in glioblastomas, colorectal carcinoma and other malignancies (Drake et al.; 2017). Numerous oncogenic mutations suppress autophagy through mTORC1 activation (e.g. activating mutations in Akt/class I PI3K, PTEN loss, LKB1 loss), ULK1 and Parkin inhibition (e.g. cytoplasmic accumulation of TP53 mutant proteins) and/or inhibition of the activity of Beclin 1/Class III PI3K complex (e.g. Akt, EGFR, HER2, Bcl-2 amplification or activation) (Rybstein et al., 2018; Vega-Rubin-de-Celis et al., 2018; Levine et al., 2015).
Autophagy has additional cell-autonomous and non-cell-autonomous functions in tumor suppression. At the cell-autonomous level, together with its promotion of genomic instability, autophagy degrades the nuclear lamina (through an interaction with LC3 and lamin B) to promote oncogene-induced senescence (Dou et al., 2015), and it negatively regulates inflammatory signaling which is a strong oncogenic driver (Zhong et al., 2016). It has been proposed that deregulated inflammatory signaling due to defective autophagy may represent a common pro-oncogenic pathway for several cancer risk factors, including obesity, aging, alcohol abuse, chronic infections, and ATG16L1 deficiency/Crohn’ disease (Zhong et al., 2016).
At the cell non-autonomous level, autophagy acts to suppress tumor-promoting inflammatory signaling and to enhance anti-cancer immunity in myeloid cells in the tumor microenvironment. Autophagy in cancer cells is important for cross-presentation as well as facilitating the release of tumor antigens from dying cells and increasing their extracellular availability (Ma et al., 2013). The phenomenon of autophagy-dependent “immunogenic cell death” also leads to the release of ATP and other danger-associated molecular patterns (Michaud et al., 2011), which enhances anti-tumor CTL responses and contributes to the anti-cancer efficacy of chemotherapy and radiation therapy (Galluzzi et al., 2017b). Interestingly, the incidence of KRAS-induced non-small lung cancer is increased by genetic inhibition of autophagy and reduced by autophagy induction through a mechanism that requires T-cell-dependent antitumor immunity (Pietrocola et al., 2016). Taken together, autophagy acts both in cancer cells and myeloid cells to dampen pro-tumorigenic inflammation and to augment adaptive immunity that curtails cancer growth and progression. It is not yet known whether autophagy induction will act synergistically with immune checkpoint inhibitors to boost anti-cancer therapeutic responses.
In parallel with delineation of mechanisms of autophagy in tumor suppression and the promotion of anti-tumor immunity, numerous reports have demonstrated pro-tumorigenic roles of autophagy, primarily in cancers driven by KRAS that require high cellular metabolic activity to sustain survival (Kimmelman and White, 2017). The pro-tumorigenic effects are generally believed to result from the ability of autophagy to sustain tumor cell survival during metabolic stress in a cell-intrinsic fashion and/or in a cell-extrinsic fashion via the provision of nutrients to malignant cells by autophagy in stromal cells in the tumor microenvironment (Katheder et al., 2017; Kimmelman and White, 2017; Yang et al., 2018). These observations have piqued interest in developing autophagy inhibitors for treating certain cancers, a concept that has been explored in mice using two principal approaches: (1) tissue-specific or whole-body-inducible deletion of ATG genes, and (2) lysosomotropic agents such as a chloroquine and hyxdroxychloroquine that inhibit autophagic flux (along with other pharmacological effects). While both approaches reduce KRAS-driven tumor growth in mice (Kimmelman and White, 2017), ATG gene deletion results in inflammation and/or destruction of the organ (i.e. in pancreatic-specific or lung-specific knockouts) or in multi-system degeneration (in whole-body knockouts) (Guo et al., 2013; Karsli-Uzunbas et al., 2014; Rosenfeldt et al., 2013). Furthermore, an extensive examination of the effects of chloroquine on a panel of KRAS mutant tumors failed to show any ATG gene-dependent growth inhibition (Eng et al., 2016). Thus, despite the crucial role of autophagy in malignant cells and stromal cells in promoting tumor growth, conclusive data are lacking to support autophagy inhibition as a viable therapeutic approach, although a recent study of KRAS-driven pancreatic cancer using only mosaic genetic inhibition of autophagy may suggest efficacy with tolerable toxicity (Yang et al., 2018). This issue is further complicated by the aforementioned multiple roles of autophagy in tumor suppression and anti-tumor immunity, along with emerging evidence that autophagy inhibition may promote tumor metastasis (reviewed in [Dower et al., 2018]).
Autophagy in Metabolic Diseases
Growing evidence implicates functional defects in autophagy in various metabolic disorders, including obesity, diabetes, atherosclerosis, and non-alcoholic fatty liver disease (reviewed in [Ueno and Komatsu, 2017; Zhang et al., 2018a]). While these disorders involve genetic and epigenetic factors, excess caloric intake and decreased physical activity are principal driving forces, both of which suppress autophagy. Although there are divergent reports of whether autophagy is enhanced or suppressed in obesity, the preponderance of mouse genetic data indicate that decreased autophagy facilitates the transition from obesity to diabetes and increases the risk of atherosclerosis and non-alcoholic fatty liver disease (NAFLD). Specifically, mice with whole-body partial mutation or tissue-specific (liver or pancreas) deletion of ATG genes or TFEB gain more weight when fed a high-fat diet and have an increased propensity to develop systemic inflammation, diabetes, and hepatic steatosis (Fernandez et al., 2017; Jung et al., 2008; Lim et al., 2014; Settembre et al., 2013a; Singh et al., 2009). In addition, mice with macrophage-specific deletion of Atg5 are more prone to the development of atherosclerotic plaques (Liao et al., 2012; Razani et al., 2012). In humans, genetic variants of IRGM1, a gene required for assembly and activation of the autophagy machinery, are associated with increased risk of NAFLD (Lin et al., 2016). Moreover, patients with NAFLD have elevated hepatic levels of Rubicon, an inhibitor of Beclin 1/VPS34 PI3KC3 activity, and hepatocyte-specific knockout of Rubicon protects mice against high-fat diet-induced impaired autophagy and steatosis (Tanaka et al., 2016). Interestingly, ethanol exposure also inhibits hepatocyte lipophagy by inactivating Rab7 (Schulze et al., 2017), raising the question of whether defective autophagy (lipophagy) may also contribute to alcoholic fatty liver disease.
The precise mechanisms by which deficient autophagy (and ATG gene functions) promote obesity and its metabolic complications are complex and may involve a variety of cell intrinsic effects (e.g. nutrient metabolism; mitochondria, peroxisome, ER and lipid droplet homeostasis), cell extrinsic effects (e.g. release of pro-inflammatory cytokines by aberrant inflammasome activation), and potentially, lack of feedback inhibition of insulin and mTORC1 signaling pathways. The mechanisms by which autophagy is inhibited during obesity are not well understood, but may involve a combination of abnormal lysosomal function in cells with lipid accumulation (Koga et al., 2010) as well as dysregulated endocrine signaling (Zhang et al., 2018a). Autophagy is normally tightly regulated by neuroendocrine signals that are decreased (e.g. insulin and insulin-like growth factors) or increased (e.g. glucagon, fibroblast growth factor 21 [FGF21]) during fasting, and the initiation of and cellular response to these signals are commonly dysregulated in obesity. In mice, deficiency of the fasting hormone FGF21 impairs TFEB activation in hepatocytes, resulting in defective autophagic-lysosomal function and increased lipid accumulation, demonstrating a nutrient-sensing hormonal link between the FGF21-TFEB signaling axis, lysosomal function, and lipid metabolism (Chen et al., 2017). Interestingly, not only do neuroendocrine signals regulate autophagy, but ATG genes may act within subpopulations of neurons to regulate metabolism and feeding behavior. Genetic ablation of certain ATG genes in hypothalamic proopiomelanocortin neurons results in adiposity, glucose intolerance and hyperphagia (Coupe et al., 2012; Malhotra et al., 2015; Quan et al., 2012). Postulated (and potentially non-autophagic) mechanisms involve leptin resistance as well as defects in the unconventional secretion of α-melanocyte-stimulating hormone.
Further studies are needed to determine whether general inducers of autophagy, specific activators of lipophagy, and/or regulators of anti-obesogenic CNS functions of ATG genes may be useful in reducing the morbidity and mortality due to obesity and its associated metabolic disorders. At least in mice, nutritional interventions and physical exercise exert favorable metabolic effects through ATG gene-dependent mechanisms. The benefits of intermittent fasting on high-fat diet-induced loss of pancreatic cells are blocked in autophagy-deficient Lamp2a−/− and Becn1+/− mice (Liu et al., 2017). Moreover, the benefits of prolongation of the intermeal time interval (i.e. feeding mice an isocaloric diet twice-a-day) on adiposity, lipid levels, gluconeogenesis, and age/obesity-associated metabolic defects are impaired in animals with knockout of Atg7 in different organs (proopiomelanocortin neurons, hepatocytes, white adipose tissue, skeletal muscle) (Martinez-Lopez et al., 2017). The ability of chronic exercise to protect against HFD-induced glucose intolerance is compromised in mice that cannot increase autophagy by virtue of a knock-in mutation in Bcl-2 that prevents its disruption from Beclin 1 (He et al., 2012). Physical exercise induces TFEB translocation into muscle fiber nuclei, allowing muscle to adapt by changes in the expression of glucose transporters, glycolytic enzymes and other metabolism-relevant genes (Mansueto et al., 2017). Whether TFEB activation is causally involved in exercise-induced autophagy induction remains to be explored, but its dual role in coordinating insulin sensitivity and glucose homeostasis during exercise and as a master regulator of autophagic-lysosomal function is intriguing. Thus, lifestyle interventions (e.g. dietary and exercise) that are important interventions in preventing metabolic disease may exert such effects, at least in part, through autophagy.
Autophagy in Longevity
Genetic studies using yeasts, worms, flies and mice demonstrate the ATG genes are required for lifespan extension requirement in caloric restriction, loss-of-function insulin signaling and other conserved longevity paradigms (reviewed in (Hansen et al., 2018)). Systemic autophagy induction exerts anti-aging effects in worms, flies and mice (Lopez-Otin et al., 2016), and genetically engineered mice with constitutively increased autophagy have extended lifespan and improved healthspan (e.g. leanness, increased insulin sensitivity, improved muscle function, reduced cardiac and renal fibrosis, and decreased age-related spontaneous tumorigenesis) (Fernandez et al., 2018; Pyo et al., 2013). Interestingly, the offspring of people with exceptional longevity have enhanced activation-induced T cell autophagy and immune function compared to age-matched controls (Raz et al., 2017). Autophagy may prevent aging through improved organellar quality control and homeostasis (e.g. via selective autophagy pathways such as mitophagy, lipophagy, lysophagy, aggrephagy), enhanced insulin sensitivity, maintenance of stemness and promotion of genomic stability. Interestingly, tissue-specific autophagy in certain tissues (e.g. in the muscle, intestine and brain) may also exert favorable effects on longevity, potentially by modulating a range of inter-tissue interactions (Hansen et al., 2018). It is possible that ATG genes may have autophagy-independent effects that promote longevity; for example, their roles in secretion and exocytosis might contribute to inter-tissue effects.
Autophagy gene expression and lysosomal function decline with aging in a range of tissues in worms, flies and mammals (including in human brains), resulting in an age-related decline in autophagic capacity (Hansen et al., 2018). This age-related decline likely contributes both to the aging process itself as well as the development of age-related diseases such as neurodegenerative diseases and cancer. Aside from autophagy inhibition produced by obesity, it is not known what factors contribute to this age-related decline. The molecular mechanisms underlying age-related declines in distinct steps in the autophagic-lysosomal pathway is a fascinating area of future autophagy research.
Concluding Remarks
Autophagy genes function in diverse cell biological pathways, not only in autophagy but also in other processes, to exert widespread physiological functions that protect mammals against aging and a broad range of medically important diseases (Figure 3). Accordingly, a large spectrum of mutations in genes required for autophagy-related pathways have now been implicated in the pathogenesis of human diseases (Table 1).
Figure 3. Diverse biological functions of autophagy genes contribute to their roles in the regulation of mammalian disease.
Shown are the major known biological functions of ATG genes and the broad categories of diseases that they regulate as predicted based on mouse experimental data and human genetic associations. Below major disease categories, some representative specific diseases are noted. Many other examples exist but are not shown due to space limitations.
Some general mechanisms of disease pathogenesis emerge from these links: (1) mutations in genes that regulate mitophagy, such as PARKIN and PINK1, are causally linked to hereditary forms of Parkinson’s disease (and also observed in some cancers) suggesting a crucial role for the consequences of impaired mitophagy (e.g. cellular ROS accumulation, mitochondrial DNA accumulation provoking cGAS/STING dependent inflammatory signaling); (2) mutations in genes encoding autophagy adaptor proteins or their activating kinases (e.g. p62/SQSTM1, optineurin, TBK) contribute to familial forms of amyotrophic lateral sclerosis, frontotemproal dementia and primary open glaucoma, implying a role for deficient selective autophagy in their pathogenesis; (3) mutations in genes that disrupt lysosomal function perturb autophagy and contribute to lysosomal storage disorders (including those with CNS and bone manifestations), Alzheimer’s disease, and Parkinson’s disease; (4) mutations in genes that disrupt autophagolysosomal fusion are often associated with congenital neurodevelopmental disorders; (5) mutations in genes that result in the accumulation of excess protein cargo (e.g polyglutamine expansion proteins, presenilin 1, amyloid precursor protein, αB-crystallin, α1-anti-trypsin) exceed the turnover capacity of autophagy, leading to end-organ proteotoxicity and degeneration; (6) hypomorphic mutations in core autophagy or selective autophagy machinery increase the risk of cancer; (7) dysregulated inflammatory signaling (as a result of defective mitophagy and enhanced inflammasome activation and/or cGAS/STING-mediated interferon signaling) may represent a common downstream event that contributes to diseases that are associated with mutations in the autophagy pathway (e.g. Crohn’s disease, Parkinson’s disease, SLE); (8) specific disorders, such as Crohn’s disease, are associated with several different mutations/polymorphisms in genes (e.g. ATG16L1, CALCOCO2/NDP62, GPR65, IRGM, LRRK2, NOD2) that simultaneously compromise intertwined pathways (e.g. bacterial autophagy, inflammasome regulation, secretion of antimicrobial peptides by Paneth cells) that govern their pathological and clinical manifestations. These genetic links illustrate the wide-ranging impact of autophagy-related pathways on distinct cell types and homeostatic processes.
Therapeutic interventions may aim to restore the wild-type function of the mutated protein, to reverse the specific defects or downstream pathogenetic consequences caused by the gene mutation, and/or to broadly upregulate autophagic activity and lysosomal function. Preclinical studies in animal models with mutations in genes that impair autophagy and lysosomal function will be necessary to determine the optimal therapeutic approaches for diseases due to defects in autophagy-related pathways. Moreover, the intriguing possibility emerges that selectively enhancing or blocking autophagy-related sub-routines might indirectly influence other sub-routines, thus affecting a therapeutically relevant ecosystem of intersecting pathways involving ATG proteins.
ACKOWLEDGMENTS
The authors’ thanks H. Smith for assistance with manuscript preparation and A. Diehl for expert medical illustration. The work in the authors’ laboratories was supported by NIH grants RO1 CA109618 and U19 AI109725 (B.L.), Cancer Prevention Research Institute of Texas (CPRIT) grant RP120718 (B.L), and Fondation Leducq grant 15CBD04 (B.L., G.K.). G.K. was supported by Ligue contre le Cancer (équipe labellisée); Agence National de la Recherche (ANR) – Projets blancs; ANR under the frame of E-Rare-2, the ERA-Net for Research on Rare Diseases; Association pour la recherche sur le cancer (ARC); Cancéropôle Ile-de-France; Chancelerie des universités de Paris (Legs Poix), Fondation pour la Recherche Médicale (FRM); European Research Area Network on Cardiovascular Diseases (ERA-CVD, MINOTAUR); the European Research Council (ERC); Fondation Carrefour; Institut National du Cancer (INCa); Inserm (HTE); Institut Universitaire de France; the LabEx Immuno-Oncology; the RHU Torino Lumière; the Seerave Foundation; the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE); and the SIRIC Cancer Research and Personalized Medicine (CARPEM). We apologize to all those authors’ whose work could not be cited due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST STATEMENT
B.L. is a scientific co-founder of Casma Therapeutics, Inc. G.K. is a scientific co-founder of Samsara Therapeutics, Ltd.
References:
- Abdulrahman BA, Abdelaziz DH, and Schatzl HM (2018). Autophagy regulates exosomal release of prions in neuronal cells. J Biol Chem 293, 8956–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aflaki E, Westbroek W, and Sidransky E (2017). The complicated relationship between Gaucher disease and Parkinsonism: Insights from a rare disease. Neuron 93, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinduro O, Sully K, Patel A, Robinson DJ, Chikh A, McPhail G, Braun KM, Philpott MP, Harwood CA, Byrne C, et al. (2016). Constitutive autophagy and nucleophagy during epidermal differentiation. J Invest Dermatol 136, 1460–1470. [DOI] [PubMed] [Google Scholar]
- Akizu N, Cantagrel V, Zaki MS, Al-Gazali L, Wang X, Rosti RO, Dikoglu E, Gelot AB, Rosti B, Vaux KK, et al. (2015). Biallelic mutations in SNX14 cause a syndromic form of cerebellar atrophy and lysosome-autophagosome dysfunction. Nat Genet 47, 528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi R, Kimmelman AC, and White E (2016). Recent insights into the function of autophagy in cancer. Genes Dev 30, 1913–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazi A, Bento CF, Ricketts T, Vicinanza M, Siddiqi F, Pavel M, Squitieri F, Hardenberg MC, Imarisio S, Menzies FM, et al. (2017). Polyglutamine tracts regulate beclin 1-dependent autophagy. Nature 545, 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auberger P, and Puissant A (2017). Autophagy, a key mechanism of oncogenesis and resistance in leukemia. Blood 129, 547–552. [DOI] [PubMed] [Google Scholar]
- Baixauli F, Lopez-Otin C, and Mittelbrunn M (2014). Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front Immunol 5, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, et al. (2008). Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet 40, 955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomeo R, Cinque L, De Leonibus C, Forrester A, Salzano AC, Monfregola J, De Gennaro E, Nusco E, Azario I, Lanzara C, et al. (2017). mTORC1 hyperactivation arrests bone growth in lysosomal storage disorders by suppressing autophagy. J Clin Invest 127, 3717–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch K, Knittler K, Borowski C, Rudnik S, Damme M, Aden K, Spehlmann ME, Frey N, Saftig P, Chalaris A, et al. (2017). Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy. Hum Mol Genet 26, 3960–3972. [DOI] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, and Harper JW (2010). Network organization of the human autophagy system. Nature 466, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bel S, Pendse M, Wang Y, Li Y, Ruhn KA, Hassell B, Leal T, Winter SE, Xavier RJ, and Hooper LV (2017). Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science 357, 1047–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaid A, Cerezo M, Chargui A, Corcelle-Termeau E, Pedeutour F, Giuliano S, Ilie M, Rubera I, Tauc M, Barale S, et al. (2013). Autophagy plays a critical role in the degradation of active RHOA, the control of cell cytokinesis, and genomic stability. Cancer Res 73, 4311–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biering SB, Choi J, Halstrom RA, Brown HM, Beatty WL, Lee S, McCune BT, Dominici E, Williams LE, Orchard RC, et al. (2017). Viral replication complexes are Targeted by LC3-guided interferon-inducible GTPases. Cell Host Microbe 22, 74–85 e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, Foreman O, Kirkpatrick DS, and Sheng M (2014). The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 510, 370–375. [DOI] [PubMed] [Google Scholar]
- Boya P, Codogno P, and Rodriguez-Muela N (2018). Autophagy in stem cells: repair, remodelling and metabolic reprogramming. Development 145. [DOI] [PubMed] [Google Scholar]
- Bronson PG, Chang D, Bhangale T, Seldin MF, Ortmann W, Ferreira RC, Urcelay E, Pereira LF, Martin J, Plebani A, et al. (2016). Common variants at PVT1, ATG13-AMBRA1, AHI1 and CLEC16A are associated with selective IgA deficiency. Nat Genet 48, 1425–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujak AL, Crane JD, Lally JS, Ford RJ, Kang SJ, Rebalka IA, Green AE, Kemp BE, Hawke TJ, Schertzer JD, et al. (2015). AMPK activation of muscle autophagy prevents fasting-induced hypoglycemia and myopathy during aging. Cell Metab 21, 883–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera S, Fernandez AF, Marino G, Aguirre A, Suarez MF, Espanol Y, Vega JA, Laura R, Fueyo A, Fernandez-Garcia MS, et al. (2013). ATG4B/autophagin-1 regulates intestinal homeostasis and protects mice from experimental colitis. Autophagy 9, 1188–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, and Debnath J (2018). Beyond self-eating: The control of nonautophagic functions and signaling pathways by autophagy-related proteins. J Cell Biol 217, 813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. (2008). A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Li R, Xu X, Zhang L, Lian R, Fang L, Huang Y, Feng X, Liu X, Li X, et al. (2018). CK1alpha suppresses lung tumour growth by stabilizing PTEN and inducing autophagy. Nat Cell Biol 20, 465–478. [DOI] [PubMed] [Google Scholar]
- Castets P, Lin S, Rion N, Di Fulvio S, Romanino K, Guridi M, Frank S, Tintignac LA, Sinnreich M, and Ruegg MA (2013). Sustained activation of mTORC1 in skeletal muscle inhibits constitutive and starvation-induced autophagy and causes a severe, late-onset myopathy. Cell Metab 17, 731–744. [DOI] [PubMed] [Google Scholar]
- Chandra S, Jana M, and Pahan K (2018). Aspirin induces lysosomal biogenesis and attenuates amyloid plaque pathology in a mouse model of Alzheimer’s disease via PPARalpha. J Neurosci 38, 6682–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MC, Srinivasan K, Friedman BA, Suto E, Modrusan Z, Lee WP, Kaminker JS, Hansen DV, and Sheng M (2017). Progranulin deficiency causes impairment of autophagy and TDP-43 accumulation. J Exp Med 214, 2611–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S, Kumar S, Jain A, Ponpuak M, Mudd MH, Kimura T, Choi SW, Peters R, Mandell M, Bruun JA, et al. (2016a). TRIMs and Galectins globally cooperate and TRIM16 and Galectin-3 co-direct autophagy in endomembrane damage homeostasis. Dev Cell 39, 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S, Mandell MA, and Deretic V (2016b). Mechanism of action of the tuberculosis and Crohn disease risk factor IRGM in autophagy. Autophagy 12, 429–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang K, Long A, Jia L, Zhang Y, Deng H, Li Y, Han J, and Wang Y (2017). Fasting-induced hormonal regulation of lysosomal function. Cell Res 27, 748–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang C, Yeo S, Liang CC, Okamoto T, Sun S, Wen J, and Guan JL (2016). Distinct roles of autophagy-dependent and -independent functions of FIP200 revealed by generation and analysis of a mutant knock-in mouse model. Genes Dev 30, 856–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, and Yu L (2017). Recent progress in autophagic lysosome reformation. Traffic 18, 358–361. [DOI] [PubMed] [Google Scholar]
- Chiang WC, Wei Y, Kuo YC, Wei S, Zhou A, Zou Z, Yehl J, Ranaghan MJ, Skepner A, Bittker JA, et al. (2018). High-throughput screens to identify autophagy inducers that function by disrupting Beclin 1/Bcl-2 binding. ACS Chem Biol 13,2247–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Park S, Biering SB, Selleck E, Liu CY, Zhang X, Fujita N, Saitoh T, Akira S, Yoshimori T, et al. (2014). The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity 40, 924–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourasia AH, Tracy K, Frankenberger C, Boland ML, Sharifi MN, Drake LE, Sachleben JR, Asara JM, Locasale JW, Karczmar GS, et al. (2015). Mitophagy defects arising from BNip3 loss promote mammary progression to metastasis. EMBO Rep 16, 1145–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Khosravi A, Kusumawardhani IP, Kwon AH, Vasconcelos AC, Cunha LD, Mayer AE, Shen Y, Wu WL, Kambal A, et al. (2016). Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352, 1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchini M, Chakrabarti R, Kongara S, Price S, Nahar R, Lozy F, Zhong H, Vazquez A, Kang Y, and Karantza V (2014). Autophagy regulator BECN1 suppresses mammary tumorigenesis driven by WNT1 activation and following parity. Autophagy 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinque L, Forrester A, Bartolomeo R, Svelto M, Venditti R, Montefusco S, Polishchuk E, Nusco E, Rossi A, Medina DL, et al. (2015). FGF signalling regulates bone growth through autophagy. Nature 528, 272–275. [DOI] [PubMed] [Google Scholar]
- Cipolla CM, and Lodhi IJ (2017). Peroxisomal dysfunction in age-related diseases. Trends Endocrinol Metab 28, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, Couthouis J, Lu YF, Wang Q, Krueger BJ, et al. (2015). Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 347, 1436–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colecchia D, Stasi M, Leonardi M, Manganelli F, Nolano M, Veneziani BM, Santoro L, Eskelinen EL, Chiariello M, and Bucci C (2018). Alterations of autophagy in the peripheral neuropathy Charcot-Marie-Tooth type 2B. Autophagy, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper O, Seo H, Andrabi S, Guardia-Laguarta C, Graziotto J, Sundberg M, McLean JR, Carrillo-Reid L, Xie Z, Osborn T, et al. (2012). Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson’s disease. Sci Transl Med 4, 141ra190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrionero A, and Horvitz HR (2018). A C9orf72 ALS/FTD ortholog acts in endolysosomal degradation and lysosomal homeostasis. Curr Biol 28, 1522–1535 e1525. [DOI] [PubMed] [Google Scholar]
- Coupe B, Ishii Y, Dietrich MO, Komatsu M, Horvath TL, and Bouret SG (2012). Loss of autophagy in pro-opiomelanocortin neurons perturbs axon growth and causes metabolic dysregulation. Cell Metab 15, 247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullup T, Kho AL, Dionisi-Vici C, Brandmeier B, Smith F, Urry Z, Simpson MA, Yau S, Bertini E, McClelland V, et al. (2013). Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat Genet 45, 83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasari SK, Bialik S, Levin-Zaidman S, Levin-Salomon V, Merrill AH Jr., Futerman AH, and Kimchi A (2017). Signalome-wide RNAi screen identifies GBA1 as a positive mediator of autophagic cell death. Cell Death Differ 24, 1288–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, and Levine B (2018). Autophagy balances inflammation in innate immunity. Autophagy, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSelm CJ, Miller BC, Zou W, Beatty WL, van Meel E, Takahata Y, Klumperman J, Tooze SA, Teitelbaum SL, and Virgin HW (2011). Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell 21, 966–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao J, Liu R, Rong Y, Zhao M, Zhang J, Lai Y, Zhou Q, Wilz LM, Li J, Vivona S, et al. (2015). ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 520, 563–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, and Elazar Z (2018). Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol 19, 349–364. [DOI] [PubMed] [Google Scholar]
- Din FV, Valanciute A, Houde VP, Zibrova D, Green KA, Sakamoto K, Alessi DR, and Dunlop MG (2012). Aspirin inhibits mTOR signaling, activates AMP-activated protein kinase, and induces autophagy in colorectal cancer cells. Gastroenterology 142, 1504–1515 e1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, Xu C, Donahue G, Shimi T, Pan JA, Zhu J, Ivanov A, Capell BC, Drake AM, Shah PP, et al. (2015). Autophagy mediates degradation of nuclear lamina. Nature 527, 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower CM, Wills CA, Frisch SM, and Wang HG (2018). Mechanisms and context underlying the role of autophagy in cancer metastasis. Autophagy 14, 1110–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake LE, Springer MZ, Poole LP, Kim CJ, and Macleod KF (2017). Expanding perspectives on the significance of mitophagy in cancer. Semin Cancer Biol 47, 110–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreux M, Gastaminza P, Wieland SF, and Chisari FV (2009). The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A 106, 14046–14051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, and Deretic V (2011). Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J 30, 4701–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran JM, Anjard C, Stefan C, Loomis WF, and Malhotra V (2010). Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol 188, 527–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi-Fakhari D, Saffari A, Wahlster L, Lu J, Byrne S, Hoffmann GF, Jungbluth H, and Sahin M (2016). Congenital disorders of autophagy: an emerging novel class of inborn errors of neuro-metabolism. Brain 139, 317–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Zoncu R, Chang S, Gumper I, Snitkin H, Wolfson RL, Kirak O, Sabatini DD, and Sabatini DM (2013). Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature 493, 679–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. (2011). Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331, 456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D, Zhang H, Zeissig S, Lipinski S, Till A, Jiang T, Stade B, Bromberg Y, Ellinghaus E, Keller A, et al. (2013). Association between variants of PRDM1 and NDP52 and Crohn’s disease, based on exome sequencing and functional studies. Gastroenterology 145, 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng CH, Wang Z, Tkach D, Toral-Barza L, Ugwonali S, Liu S, Fitzgerald SL, George E, Frias E, Cochran N, et al. (2016). Macroautophagy is dispensable for growth of KRAS mutant tumors and chloroquine efficacy. Proc Natl Acad Sci U S A 113, 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AF, Barcena C, Martinez-Garcia GG, Tamargo-Gomez I, Suarez MF, Pietrocola F, Castoldi F, Esteban L, Sierra-Filardi E, Boya P, et al. (2017). Autophagy couteracts weight gain, lipotoxicity and pancreatic beta-cell death upon hypercaloric pro-diabetic regimens. Cell Death Dis 8, e2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AF, Sebti S, Wei Y, Zou Z, Shi M, McMillan KL, He C, Ting T, Liu Y, Chiang WC, et al. (2018). Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature 558, 136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festa BP, Chen Z, Berquez M, Debaix H, Tokonami N, Prange JA, Hoek GV, Alessio C, Raimondi A, Nevo N, et al. (2018). Impaired autophagy bridges lysosomal storage disease and epithelial dysfunction in the kidney. Nat Commun 9, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher K, Ulferts R, Jacquin E, Veith T, Gammoh N, Arasteh JM, Mayer U, Carding SR, Wileman T, Beale R, et al. (2018). The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J 37, pii: e97840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco LH, Nair VR, Scharn CR, Xavier RJ, Torrealba JR, Shiloh MU, and Levine B (2017). The ubiquitin ligase Smurf1 functions in selective autophagy of Mycobacterium tuberculosis and anti-tuberculous host defense. Cell Host Microbe 21, 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussi N, Hollerhage M, Chakroun T, Nykanen NP, Rosler TW, Koeglsperger T, Wurst W, Behrends C, and Hoglinger GU (2018). Exosomal secretion of alpha-synuclein as protective mechanism after upstream blockage of macroautophagy. Cell Death Dis 9, 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, and Kroemer G (2017a). Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat Rev Drug Discov 16, 487–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Buque A, Kepp O, Zitvogel L, and Kroemer G (2017b). Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol 17, 97–111. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Pietrocola F, Levine B, and Kroemer G (2014). Metabolic control of autophagy. Cell 159, 1263–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan AK, Ho H, Bodemann B, Petersen S, Aruri J, Koshy S, Richardson Z, Le LQ, Krasieva T, Roth MG, et al. (2008). Genome-wide siRNA-based functional genomics of pigmentation identifies novel genes and pathways that impact melanogenesis in human cells. PLoS Genet 4, e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Prat L, Martinez-Vicente M, Perdiguero E, Ortet L, Rodriguez-Ubreva J, Rebollo E, Ruiz-Bonilla V, Gutarra S, Ballestar E, Serrano AL, et al. (2016). Autophagy maintains stemness by preventing senescence. Nature 529, 37–42. [DOI] [PubMed] [Google Scholar]
- Gatica D, Lahiri V, and Klionsky DJ (2018). Cargo recognition and degradation by selective autophagy. Nat Cell Biol 20, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluschko A, Herb M, Wiegmann K, Krut O, Neiss WF, Utermohlen O, Kronke M, and Schramm M (2018). The β2 integrin Mac-1 induces protective LC3-associated phagocytosis of listeria monocytogenes. Cell Host Microbe 23, 324–337 e325. [DOI] [PubMed] [Google Scholar]
- Gomes LC, Di Benedetto G, and Scorrano L (2011). During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol 13, 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode A, Long JE, Shaw B, Ralston SH, Visconti MR, Gianfrancesco F, Esposito T, Gennari L, Merlotti D, Rendina D, et al. (2014). Paget disease of bone-associated UBA domain mutations of SQSTM1 exert distinct effects on protein structure and function. Biochim Biophys Acta 1842, 992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P, and Dikic I (2018). Ubiquitin signaling and autophagy. J Biol Chem 293, 5404–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P, Morozzi G, Holper S, Mari M, Harwardt MI, Yan R, Muller S, Reggiori F, Heilemann M, and Dikic I (2017). Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gstrein T, Edwards A, Pristoupilova A, Leca I, Breuss M, Pilat-Carotta S, Hansen AH, Tripathy R, Traunbauer AK, Hochstoeger T, et al. (2018). Mutations in Vps15 perturb neuronal migration in mice and are associated with neurodevelopmental disease in humans. Nat Neurosci 21, 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Chitiprolu M, Gagnon D, Meng L, Perez-Iratxeta C, Lagace D, and Gibbings D (2014). Autophagy supports genomic stability by degrading retrotransposon RNA. Nat Commun 5, 5276. [DOI] [PubMed] [Google Scholar]
- Guo H, Chitiprolu M, Roncevic L, Javalet C, Hemming FJ, Trung MT, Meng L, Latreille E, Tanese de Souza C, McCulloch D, et al. (2017). Atg5 disassociates the V1V0-ATPase to promote exosome production and tumor metastasis independent of canonical macroautophagy. Dev Cell 43, 716–730 e717. [DOI] [PubMed] [Google Scholar]
- Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, Chen G, Price S, Lu W, Teng X, et al. (2013). Autophagy suppresses progression of K-rasinduced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev 27, 1447–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack TB, Hogarth P, Kruer MC, Gregory A, Wieland T, Schwarzmayr T, Graf E, Sanford L, Meyer E, Kara E, et al. (2012). Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am J Hum Genet 91, 1144–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Rubinsztein DC, and Walker DW (2018). Autophagy as a promoter of longevity: insights from model organisms. Nat Rev Mol Cell Biol. 19, 579–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Taura M, and Iwasaki A (2018). The interaction between IKKalpha and LC3 promotes type I interferon production through the TLR9-containing LAPosome. Sci Signal 11, pii: eaan4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, Sun Q, et al. (2012). Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 481, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann BL, Boada-Romero E, Cunha LD, Magne J, and Green DR (2017). LC3-associated phagocytosis and inflammation. J Mol Biol 429, 3561–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, Sasai M, Latz E, Brinkmann MM, Iwasaki A, et al. (2012). Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity 37, 986–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, and Shaw RJ (2018). AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol 19, 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessvik NP, and Llorente A (2018). Current knowledge on exosome biogenesis and release. Cell Mol Life Sci 75, 193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TT, Warr MR, Adelman ER, Lansinger OM, Flach J, Verovskaya EV, Figueroa ME, and Passegue E (2017). Autophagy maintains the metabolism and function of young and old stem cells. Nature 543, 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori I, Otomo T, Nakashima M, Miya F, Negishi Y, Shiraishi H, Nonoda Y, Magara S, Tohyama J, Okamoto N, et al. (2017). Defects in autophagosome-lysosome fusion underlie Vici syndrome, a neurodevelopmental disorder with multisystem involvement. Sci Rep 7, 3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui KY, Fernandez-Hernandez H, Hu J, Schaffner A, Pankratz N, Hsu NY, Chuang LS, Carmi S, Villaverde N, Li X, et al. (2018). Functional variants in the LRRK2 gene confer shared effects on risk for Crohn’s disease and Parkinson’s disease. Sci Transl Med 10, pii: eaai7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, and Young LN (2017). Mechanisms of autophagy initiation. Annu Rev Biochem 86, 225–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hylin MJ, Zhao J, Tangavelou K, Rozas NS, Hood KN, MacGowan JS, Moore AN, and Dash PK (2018). A role for autophagy in long-term spatial memory formation in male rodents. J Neurosci Res 96, 416–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imagawa Y, Saitoh T, and Tsujimoto Y (2016). Vital staining for cell death identifies Atg9a-dependent necrosis in developmental bone formation in mouse. Nat Commun 7, 13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, and Mizushima N (2014). Autophagy and human diseases. Cell Res 24, 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, et al. (2007). The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A 104, 14050–14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HS, Chung KW, Won Kim J, Kim J, Komatsu M, Tanaka K, Nguyen YH, Kang TM, Yoon KH, Kim JW, et al. (2008). Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab 8, 318–324. [DOI] [PubMed] [Google Scholar]
- Kadir R, Harel T, Markus B, Perez Y, Bakhrat A, Cohen I, Volodarsky M, Feintsein-Linial M, Chervinski E, Zlotogora J, et al. (2016). ALFY-controlled DVL3 autophagy regulates Wnt signaling, determining human brain size. PLoS Genet 12, e1005919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, and White E (2007). Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 21, 1621–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch J, Schips TG, Maliken BD, Brody MJ, Sargent MA, Kanisicak O, and Molkentin JD (2017). Autophagic cell death is dependent on lysosomal membrane permeability through Bax and Bak. Elife 6, pii: e30543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, Kalaany NY, Jacks T, Chan CS, Rabinowitz JD, et al. (2014). Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov 4, 914–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katheder NS, Khezri R, O’Farrell F, Schultz SW, Jain A, Rahman MM, Schink KO, Theodossiou TA, Johansen T, Juhasz G, et al. (2017). Microenvironmental autophagy promotes tumour growth. Nature 541, 417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenzelmann Broz D, Spano Mello S, Bieging KT, Jiang D, Dusek RL, Brady CA, Sidow A, and Attardi LD (2013). Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev 27, 1016–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, et al. (2015). Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522, 354–358. [DOI] [PubMed] [Google Scholar]
- Kheloufi M, Boulanger CM, Codogno P, and Rautou PE (2015). Autosis occurs in the liver of patients with severe anorexia nervosa. Hepatology 62, 657–658. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, and Guan KL (2013a). Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152, 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Zhao H, Martinez J, Doggett TA, Kolesnikov AV, Tang PH, Ablonczy Z, Chan CC, Zhou Z, Green DR, et al. (2013b). Noncanonical autophagy promotes the visual cycle. Cell 154, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Sandford E, Gatica D, Qiu Y, Liu X, Zheng Y, Schulman BA, Xu J, Semple I, Ro SH, et al. (2016). Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. Elife 5, pii: e12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Deng HX, Wong YC, Siddique T, and Krainc D (2017). The Parkinson’s disease-linked protein TMEM230 is required for Rab8a-mediated secretory vesicle trafficking and retromer trafficking. Hum Mol Genet 26, 729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmelman AC, and White E (2017). Autophagy and tumor metabolism. Cell Metab 25, 1037–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmey JM, Huynh JP, Weiss LA, Park S, Kambal A, Debnath J, Virgin HW, and Stallings CL (2015). Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature 528, 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Jain A, Choi SW, Mandell MA, Schroder K, Johansen T, and Deretic V (2015). TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J Cell Biol 210, 973–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Jia J, Kumar S, Choi SW, Gu Y, Mudd M, Dupont N, Jiang S, Peters R, Farzam F, et al. (2017). Dedicated SNAREs and specialized TRIM cargo receptors mediate secretory autophagy. EMBO J 36, 42–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, and Shimizu N (1998). Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn WA Jr., Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, et al. (2003). A unified nomenclature for yeast autophagy-related genes. Dev Cell 5, 539–545. [DOI] [PubMed] [Google Scholar]
- Kluge AF, Lagu BR, Maiti P, Jaleel M, Webb M, Malhotra J, Mallat A, Srinivas PA, and Thompson JE (2018). Novel highly selective inhibitors of ubiquitin specific protease 30 (USP30) accelerate mitophagy. Bioorg Med Chem Lett. 28, 2655–2659. [DOI] [PubMed] [Google Scholar]
- Koga H, Kaushik S, and Cuervo AM (2010). Altered lipid content inhibits autophagic vesicular fusion. FASEB J 24, 3052–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno H, Chinn IK, Hong D, Orange JS, Lupski JR, Mendoza A, Pedroza LA, and Barber GN (2018). Pro-inflammation associated with a gain-of-function mutation (R284S) in the innate immune sensor STING. Cell Rep 23, 1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno H, Konno K, and Barber GN (2013). Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 155, 688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korvatska O, Strand NS, Berndt JD, Strovas T, Chen DH, Leverenz JB, Kiianitsa K, Mata IF, Karakoc E, Greenup JL, et al. (2013). Altered splicing of ATP6AP2 causes X-linked parkinsonism with spasticity (XPDS). Hum Mol Genet 22, 3259–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MH, Farre JC, Mitra K, Yu MK, Ono K, Demchak B, Licon K, Flagg M, Balakrishnan R, Cherry JM, et al. (2017). Active interaction mapping reveals the hierarchical organization of autophagy. Mol Cell 65, 761–774 e765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegenburg F, Ungermann C, and Reggiori F (2018). Coordination of autophagosomelysosome fusion by Atg8 family members. Curr Biol 28, R512–R518. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Marino G, and Levine B (2010). Autophagy and the integrated stress response. Mol Cell 40, 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo TC, Chen CT, Baron D, Onder TT, Loewer S, Almeida S, Weismann CM, Xu P, Houghton JM, Gao FB, et al. (2011). Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat Cell Biol 13, 1214–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri A, Hedl M, and Abraham C (2015). MTMR3 risk allele enhances innate receptorinduced signaling and cytokines by decreasing autophagy and increasing caspase-1 activation. Proc Natl Acad Sci U S A 112, 10461–10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen KG, Kuballa P, Conway KL, Patel KK, Becker CE, Peloquin JM, Villablanca EJ, Norman JM, Liu TC, Heath RJ, et al. (2014). Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci U S A 111, 7741–7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen KG, McKenzie CI, Mari M, Murano T, Begun J, Baxt LA, Goel G, Villablanca EJ, Kuo SY, Huang H, et al. (2016). Genetic coding variant in GPR65 alters lysosomal pH and links lysosomal dysfunction with colitis risk. Immunity 44, 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latunde-Dada GO (2017). Ferroptosis: Role of lipid peroxidation, iron and ferritinophagy. Biochim Biophys Acta 1861, 1893–1900. [DOI] [PubMed] [Google Scholar]
- Lee IH, Kawai Y, Fergusson MM, Rovira II, Bishop AJ, Motoyama N, Cao L, and Finkel T (2012). Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science 336, 225–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, et al. (2010). Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141, 1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MY, Sumpter R Jr., Zou Z, Sirasanagandla S, Wei Y, Mishra P, Rosewich H, Crane DI, and Levine B (2017a). Peroxisomal protein PEX13 functions in selective autophagy. EMBO Rep 18, 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PL, Tang Y, Li H, and Guertin DA (2016). Raptor/mTORC1 loss in adipocytes causes progressive lipodystrophy and fatty liver disease. Mol Metab 5, 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PP, Lobato-Marquez D, Pramanik N, Sirianni A, Daza-Cajigal V, Rivers E, Cavazza A, Bouma G, Moulding D, Hultenby K, et al. (2017b). Wiskott-Aldrich syndrome protein regulates autophagy and inflammasome activity in innate immune cells. Nat Commun 8, 1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Jonson PH, Sarparanta J, Palmio J, Sarkar M, Vihola A, Evila A, Suominen T, Penttila S, Savarese M, et al. (2018). TIA1 variant drives myodegeneration in multisystem proteinopathy with SQSTM1 mutations. J Clin Invest 128, 1164–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, and Klionsky DJ (2017). Autophagy wins the 2016 Nobel Prize in Physiology or Medicine: Breakthroughs in baker’s yeast fuel advances in biomedical research. Proc Natl Acad Sci U S A 114, 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, and Kroemer G (2008). Autophagy in the pathogenesis of disease. Cell 132, 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Liu R, Dong X, and Zhong Q (2015). Beclin orthologs: integrative hubs of cell signaling, membrane trafficking, and physiology. Trends Cell Biol, 25, 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Brazill JM, Liu S, Bello C, Zhu Y, Morimoto M, Cascio L, Pauly R, Diaz-Perez Z, Malicdan MCV, et al. (2017a). Spermine synthase deficiency causes lysosomal dysfunction and oxidative stress in models of Snyder-Robinson syndrome. Nat Commun 8, 1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhang Y, Cheng X, Yuan H, Zhu S, Liu J, Wen Q, Xie Y, Liu J, Kroemer G, et al. (2018a). PINK1 and PARK2 suppress pancreatic tumorigenesis through control of mitochondrial iron-mediated immunometabolism. Dev Cell 46, 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Xie X, Wang Y, Liu J, Cheng X, Guo Y, Gong Y, Hu S, and Pan L (2016). Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins. Nat Commun 7, 12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li D, Ma Z, Qian Z, Kang X, Jin X, Li F, Wang X, Chen Q, Sun H, et al. (2018b). Defective autophagy in osteoblasts induces endoplasmic reticulum stress and causes remarkable bone loss. Autophagy 14, 1726–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yu W, Qian X, Xia Y, Zheng Y, Lee JH, Li W, Lyu J, Rao G, Zhang X, et al. (2017b). Nucleus-translocated ACSS2 promotes gene transcription for lysosomal biogenesis and autophagy. Mol Cell 66, 684–697 e689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Seo GJ, Choi YJ, Kwak MJ, Ge J, Rodgers MA, Shi M, Leslie BJ, Hopfner KP, Ha T, et al. (2014). Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe 15, 228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, and Levine B (1999). Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676. [DOI] [PubMed] [Google Scholar]
- Liao X, Sluimer JC, Wang Y, Subramanian M, Brown K, Pattison JS, Robbins J, Martinez J, and Tabas I (2012). Macrophage autophagy plays a protective role in advanced atherosclerosis. Cell Metab 15, 545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YM, Lim H, Hur KY, Quan W, Lee HY, Cheon H, Ryu D, Koo SH, Kim HL, Kim J, et al. (2014). Systemic autophagy insufficiency compromises adaptation to metabolic stress and facilitates progression from obesity to diabetes. Nat Commun 5, 4934. [DOI] [PubMed] [Google Scholar]
- Lin YC, Chang PF, Lin HF, Liu K, Chang MH, and Ni YH (2016). Variants in the autophagy-related gene IRGM confer susceptibility to non-alcoholic fatty liver disease by modulating lipophagy. J Hepatol 65, 1209–1216. [DOI] [PubMed] [Google Scholar]
- Liu F, Fang F, Yuan H, Yang D, Chen Y, Williams L, Goldstein SA, Krebsbach PH, and Guan JL (2013a). Suppression of autophagy by FIP200 deletion leads to osteopenia in mice through the inhibition of osteoblast terminal differentiation. J Bone Miner Res 28, 2414–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Javaheri A, Godar RJ, Murphy J, Ma X, Rohatgi N, Mahadevan J, Hyrc K, Saftig P, Marshall C, et al. (2017). Intermittent fasting preserves beta-cell mass in obesity-induced diabetes via the autophagy-lysosome pathway. Autophagy 13, 1952–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Tang Q, Liu K, Xie W, Liu X, Wang H, Wang RF, and Cui J (2016). TRIM11 suppresses AIM2 inflammasome by degrading AIM2 via p62-dependent selective autophagy. Cell Rep 16, 1988–2002. [DOI] [PubMed] [Google Scholar]
- Liu Y, Shoji-Kawata S, Sumpter RM Jr., Wei Y, Ginet V, Zhang L, Posner B, Tran KA, Green DR, Xavier RJ, et al. (2013b). Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci U S A 110, 20364–20371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Galluzzi L, Freije JMP, Madeo F, and Kroemer G (2016). Metabolic control of longevity. Cell 166, 802–821. [DOI] [PubMed] [Google Scholar]
- Lu K, den Brave F, and Jentsch S (2017). Receptor oligomerization guides pathway choice between proteasomal and autophagic degradation. Nat Cell Biol 19, 732–739. [DOI] [PubMed] [Google Scholar]
- Lu X, Altshuler-Keylin S, Wang Q, Chen Y, Henrique Sponton C, Ikeda K, Maretich P, Yoneshiro T, and Kajimura S (2018). Mitophagy controls beige adipocyte maintenance through a Parkin-dependent and UCP1-independent mechanism. Sci Signal 11, pii: eaap8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv M, Wang C, Li F, Peng J, Wen B, Gong Q, Shi Y, and Tang Y (2017). Structural insights into the recognition of phosphorylated FUNDC1 by LC3B in mitophagy. Protein Cell 8, 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Galluzzi L, Zitvogel L, and Kroemer G (2013). Autophagy and cellular immune responses. Immunity 39, 211–227. [DOI] [PubMed] [Google Scholar]
- Ma Y, Qi M, An Y, Zhang L, Yang R, Doro DH, Liu W, and Jin Y (2018). Autophagy controls mesenchymal stem cell properties and senescence during bone aging. Aging Cell 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Eisenberg T, Pietrocola F, and Kroemer G (2018). Spermidine in health and disease. Science 359. [DOI] [PubMed] [Google Scholar]
- Madeo F, Pietrocola F, Eisenberg T, and Kroemer G (2014). Caloric restriction mimetics: towards a molecular definition. Nat Rev Drug Discov 13, 727–740. [DOI] [PubMed] [Google Scholar]
- Maejima I, Takahashi A, Omori H, Kimura T, Takabatake Y, Saitoh T, Yamamoto A, Hamasaki M, Noda T, Isaka Y, et al. (2013). Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J 32, 2336–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra R, Warne JP, Salas E, Xu AW, and Debnath J (2015). Loss of Atg12, but not Atg5, in pro-opiomelanocortin neurons exacerbates diet-induced obesity. Autophagy 11, 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell MA, Jain A, Kumar S, Castleman MJ, Anwar T, Eskelinen EL, Johansen T, Prekeris R, and Deretic V (2016). TRIM17 contributes to autophagy of midbodies while actively sparing other targets from degradation. J Cell Sci 129, 3562–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R, Anjard C, Loomis WF, and Subramani S (2010). Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J Cell Biol 188, 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansueto G, Armani A, Viscomi C, D’Orsi L, De Cegli R, Polishchuk EV, Lamperti C, Di Meo I, Romanello V, Marchet S, et al. (2017). Transcription factor EB controls metabolic flexibility during exercise. Cell Metab 25, 182–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Fernandez AF, Cabrera S, Lundberg YW, Cabanillas R, Rodriguez F, Salvador-Montoliu N, Vega JA, Germana A, Fueyo A, et al. (2010). Autophagy is essential for mouse sense of balance. J Clin Invest 120, 2331–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Niso-Santano M, Baehrecke EH, and Kroemer G (2014a). Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 15, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, et al. (2014b). Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell 53, 710–725. [DOI] [PubMed] [Google Scholar]
- Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, and Green DR (2011). Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A 108, 17396–17401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Cunha LD, Park S, Yang M, Lu Q, Orchard R, Li QZ, Yan M, Janke L, Guy C, et al. (2016). Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature 533, 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S, Orchard R, Guan JL, Tan H, Peng J, et al. (2015). Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol 17, 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Martinez-Lopez N, Tarabra E, Toledo M, Garcia-Macia M, Sahu S, Coletto L, Batista-Gonzalez A, Barzilai N, Pessin JE, Schwartz GJ, et al. (2017). System-wide benefits of intermeal fasting by autophagy. Cell Metab 26, 856–871 e855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. (2009). Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 11, 385–396. [DOI] [PubMed] [Google Scholar]
- Matsuzawa-Ishimoto Y, Shono Y, Gomez LE, Hubbard-Lucey VM, Cammer M, Neil J, Dewan MZ, Lieberman SR, Lazrak A, Marinis JM, et al. (2017). Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J Exp Med 214, 3687–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P, Shen S, Kepp O, Scoazec M, Mignot G, et al. (2011). Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 334, 1573–1577. [DOI] [PubMed] [Google Scholar]
- Mira MT, Alcais A, Nguyen VT, Moraes MO, Di Flumeri C, Vu HT, Mai CP, Nguyen TH, Nguyen NB, Pham XK, et al. (2004). Susceptibility to leprosy is associated with PARK2 and PACRG. Nature 427, 636–640. [DOI] [PubMed] [Google Scholar]
- Miranda AM, Lasiecka ZM, Xu Y, Neufeld J, Shahriar S, Simoes S, Chan RB, Oliveira TG, Small SA, and Di Paolo G (2018). Neuronal lysosomal dysfunction releases exosomes harboring APP C-terminal fragments and unique lipid signatures. Nat Commun 9, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell G, and Isberg RR (2017). Innate immunity to intracellular pathogens: Balancing microbial elimination and inflammation. Cell Host Microbe 22, 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N (2018). A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol 20, 521–527. [DOI] [PubMed] [Google Scholar]
- Mizushima N, and Komatsu M (2011). Autophagy: renovation of cells and tissues. Cell 147, 728–741. [DOI] [PubMed] [Google Scholar]
- Molineros JE, Yang W, Zhou XJ, Sun C, Okada Y, Zhang H, Heng Chua K, Lau YL, Kochi Y, Suzuki A, et al. (2017). Confirmation of five novel susceptibility loci for systemic lupus erythematosus (SLE) and integrated network analysis of 82 SLE susceptibility loci. Hum Mol Genet 26, 1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, Criollo A, Galluzzi L, Malik SA, Vitale I, et al. (2010). Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis 1, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Marino G, Bennetzen MV, Eisenberg T, Megalou E, Schroeder S, Cabrera S, Benit P, Rustin P, Criollo A, et al. (2011). Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol 192, 615–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J, Karin M, and Diaz-Meco MT (2016). p62 in cancer: Signaling adaptor beyond autophagy. Cell 167, 606–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz-Feliciano L, Doggett TA, Zhou Z, and Ferguson TA (2017). RUBCN/rubicon and EGFR regulate lysosomal degradative processes in the retinal pigment epithelium (RPE) of the eye. Autophagy 13, 2072–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrow L, Malhotra R, and Debnath J (2015). ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat Cell Biol 17, 300–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy A, Li Y, Peng I, Reichelt M, Katakam AK, Noubade R, Roose-Girma M, DeVoss J, Diehl L, Graham RR, et al. (2014). A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 506, 456–462. [DOI] [PubMed] [Google Scholar]
- Na HJ, Pyo JH, Jeon HJ, Park JS, Chung HY, and Yoo MA (2018). Deficiency of Atg6 impairs beneficial effect of metformin on intestinal stem cell aging in Drosophila. Biochem Biophys Res Commun 498, 18–24. [DOI] [PubMed] [Google Scholar]
- Napolitano G, and Ballabio A (2016). TFEB at a glance. J Cell Sci 129, 2475–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif M, Woehlbier U, and Manque PA (2017). The enigmatic role of C9ORF72 in autophagy. Front Neurosci 11, 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TB, Louie SM, Daniele JR, Tran Q, Dillin A, Zoncu R, Nomura DK, and Olzmann JA (2017). DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev Cell 42, 9–21 e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Kaizuka T, Cadwell K, Sahani MH, Saitoh T, Akira S, Virgin HW, and Mizushima N (2013). FIP200 regulates targeting of Atg16L1 to the isolation membrane. EMBO Rep 14, 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, Mora M, Riggs JE, Oh SJ, Koga Y, et al. (2000). Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature 406, 906–910. [DOI] [PubMed] [Google Scholar]
- Ochotny N, Voronov I, Owen C, Aubin JE, and Manolson MF (2013). The R740S mutation in the V-ATPase a3 subunit results in osteoclast apoptosis and defective early-stage autophagy. J Cell Biochem 114, 2823–2833. [DOI] [PubMed] [Google Scholar]
- Onal M, Piemontese M, Xiong J, Wang Y, Han L, Ye S, Komatsu M, Selig M, Weinstein RS, Zhao H, et al. (2013). Suppression of autophagy in osteocytes mimics skeletal aging. J Biol Chem 288, 17432–17440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Seo M, Jung CH, Grunwald D, Stone M, Otto NM, Toso E, Ahn Y, Kyba M, Griffin TJ, et al. (2018). ULK1 phosphorylates Ser30 of BECN1 in association with ATG14 to stimulate autophagy induction. Autophagy 14, 584–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KK, Miyoshi H, Beatty WL, Head RD, Malvin NP, Cadwell K, Guan JL, Saitoh T, Akira S, Seglen PO, et al. (2013). Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J 32, 3130–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman DR, and Weinberg RA (2014). Tackling the cancer stem cells - what challenges do they pose? Nat Rev Drug Discov 13, 497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Shapiro SL, Banduseela VC, Dieterich IA, Hewitt KJ, Bresnick EH, Kong G, Zhang J, Schueler KL, Keller MP, et al. (2018). Increased transport of acetyl-CoA into the endoplasmic reticulum causes a progeria-like phenotype. Aging Cell, e12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, Lengrand J, Deshpande V, Selig MK, Ferrone CR, et al. (2015). Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature 524, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell AM, Huang CH, Kennedy SR, Ordureau A, Sideris DP, Hoekstra JG, Harper JW, and Youle RJ (2015). Endogenous parkin preserves dopaminergic substantia Nigral neurons following mitochondrial DNA mutagenic stress. Neuron 87, 371–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrocola F, Castoldi F, Markaki M, Lachkar S, Chen G, Enot DP, Durand S, Bossut N, Tong M, Malik SA, et al. (2018). Aspirin recapitulates features of caloric restriction. Cell Rep 22, 2395–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrocola F, Pol J, Vacchelli E, Rao S, Enot DP, Baracco EE, Levesque S, Castoldi F, Jacquelot N, Yamazaki T, et al. (2016). Caloric restriction mimetics enhance anticancer immunosurveillance. Cancer Cell 30, 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulogiannis G, McIntyre RE, Dimitriadi M, Apps JR, Wilson CH, Ichimura K, Luo F, Cantley LC, Wyllie AH, Adams DJ, et al. (2010). PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc Natl Acad Sci U S A 107, 15145–15150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R, Ferguson SM, Brugarolas J, and Ballabio A (2018). The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J 37 pii: e98804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH, Kam TI, Jung S, and Jung YK (2013). Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun 4, 2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. (2003). Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 112, 1809–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan W, Kim HK, Moon EY, Kim SS, Choi CS, Komatsu M, Jeong YT, Lee MK, Kim KW, Kim MS, et al. (2012). Role of hypothalamic proopiomelanocortin neuron autophagy in the control of appetite and leptin response. Endocrinology 153, 1817–1826. [DOI] [PubMed] [Google Scholar]
- Raz Y, Guerrero-Ros I, Maier A, Slagboom PE, Atzmon G, Barzilai N, and Macian F (2017). Activation-induced autophagy is preserved in CD4+ T-cells in familial longevity. J Gerontol A Biol Sci Med Sci 72, 1201–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani B, Feng C, Coleman T, Emanuel R, Wen H, Hwang S, Ting JP, Virgin HW, Kastan MB, and Semenkovich CF (2012). Autophagy links inflammasomes to atherosclerotic progression. Cell Metab 15, 534–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH, Yin X, Manczak M, Kumar S, Jangampalli Adi P, Vijayan M, and Reddy AP (2018). Mutant APP and Amyloid beta-induced defective autophagy, mitophagy, mitochondrial structural and functional changes and synaptic damage in hippocampal neurons from Alzheimer’s disease. Hum Mol Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmann V, Lamb CA, Hwang S, Orchard RC, Kim S, Razi M, Milam A, Park S, Yokoyama CC, Kambal A, et al. (2016). Clec16a is critical for autolysosome function and Purkinje cell survival. Sci Rep 6, 23326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rello-Varona S, Lissa D, Shen S, Niso-Santano M, Senovilla L, Marino G, Vitale I, Jemaa M, Harper F, Pierron G, et al. (2012). Autophagic removal of micronuclei. Cell Cycle 11, 170–176. [DOI] [PubMed] [Google Scholar]
- Rocchi A, Yamamoto S, Ting T, Fan Y, Sadleir K, Wang Y, Zhang W, Huang S, Levine B, Vassar R, et al. (2017). A Becn1 mutation mediates hyperactive autophagic sequestration of amyloid oligomers and improved cognition in Alzheimer’s disease. PLoS Genet 13, e1006962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojansky R, Cha MY, and Chan DC (2016). Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. Elife 5, pii: e17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, Au A, Rai TS, Zheng L, Ridgway R, et al. (2013). p53 status determines the role of autophagy in pancreatic tumour development. Nature 504, 296–300. [DOI] [PubMed] [Google Scholar]
- Rostislavleva K, Soler N, Ohashi Y, Zhang L, Pardon E, Burke JE, Masson GR, Johnson C, Steyaert J, Ktistakis NT, et al. (2015). Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science 350, aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Leidal AM, Ye J, Ronen SM, and Debnath J (2017). Autophagy-dependent shuttling of TBC1D5 controls plasma membrane translocation of GLUT1 and glucose uptake. Mol Cell 67, 84–95 e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujano MA, Cannata Serio M, Panasyuk G, Peanne R, Reunert J, Rymen D, Hauser V, Park JH, Freisinger P, Souche E, et al. (2017). Mutations in the X-linked ATP6AP2 cause a glycosylation disorder with autophagic defects. J Exp Med 214, 3707–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybstein MD, Bravo-San Pedro JM, Kroemer G, and Galluzzi L (2018). The autophagic network and cancer. Nat Cell Biol 20, 243–251. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. (2009). Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A 106, 20842–20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. (2008). Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 456, 264–268. [DOI] [PubMed] [Google Scholar]
- Saitsu H, Nishimura T, Muramatsu K, Kodera H, Kumada S, Sugai K, Kasai-Yoshida E, Sawaura N, Nishida H, Hoshino A, et al. (2013). De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat Genet 45, 445–449, 449e441. [DOI] [PubMed] [Google Scholar]
- Sakamaki JI, Wilkinson S, Hahn M, Tasdemir N, O’Prey J, Clark W, Hedley A, Nixon C, Long JS, New M, et al. (2017). Bromodomain protein BRD4 is a transcriptional repressor of autophagy and lysosomal function. Mol Cell 66, 517–532 e519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, and Wang J (2008). Essential role for Nix in autophagic maturation of erythroid cells. Nature 454, 232–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, et al. (2007). Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257. [DOI] [PubMed] [Google Scholar]
- Sarkar S (2013). Regulation of autophagy by mTOR-dependent and mTOR-independent pathways: autophagy dysfunction in neurodegenerative diseases and therapeutic application of autophagy enhancers. Biochem Soc Trans 41, 1103–1130. [DOI] [PubMed] [Google Scholar]
- Saxton RA, and Sabatini DM (2017). mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371. [DOI] [PubMed] [Google Scholar]
- Sbardella D, Tundo GR, Campagnolo L, Valacchi G, Orlandi A, Curatolo P, Borsellino G, D’Esposito M, Ciaccio C, Cesare SD, et al. (2017). Retention of mitochondria in mature human red blood cells as the result of autophagy impairment in Rett Syndrome. Sci Rep 7, 12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira AH (2015). Glucocerebrosidase and Parkinson disease: Recent advances. Mol Cell Neurosci 66, 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharl M, Wojtal KA, Becker HM, Fischbeck A, Frei P, Arikkat J, Pesch T, Kellermeier S, Boone DL, Weber A, et al. (2012). Protein tyrosine phosphatase nonreceptor type 2 regulates autophagosome formation in human intestinal cells. Inflamm Bowel Dis 18, 1287–1302. [DOI] [PubMed] [Google Scholar]
- Schoner W (2002). Endogenous cardiac glycosides, a new class of steroid hormones. Eur J Biochem 269, 2440–2448. [DOI] [PubMed] [Google Scholar]
- Schulze RJ, Rasineni K, Weller SG, Schott MB, Schroeder B, Casey CA, and McNiven MA (2017). Ethanol exposure inhibits hepatocyte lipophagy by inactivating the small guanosine triphosphatase Rab7. Hepatol Commun 1, 140–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster C, Gerold KD, Schober K, Probst L, Boerner K, Kim MJ, Ruckdeschel A, Serwold T, and Kissler S (2015). The autoimmunity-associated gene CLEC16A modulates thymic epithelial cell autophagy and alters T cell selection. Immunity 42, 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo AY, Lau PW, Feliciano D, Sengupta P, Gros MAL, Cinquin B, Larabell CA, and Lippincott-Schwartz J (2017). AMPK and vacuole-associated Atg14p orchestrate mu-lipophagy for energy production and long-term survival under glucose starvation. Elife 6 pii: e21690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong E, Insolera R, Dulovic M, Kamsteeg EJ, Trinh J, Bruggemann N, Sandford E, Li S, Ozel AB, Li JZ, et al. (2018). Mutations in VPS13D lead to a new recessive ataxia with spasticity and mitochondrial defects. Ann Neurol 83, 1075–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, et al. (2013a). TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 15, 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Medina DL, and Ballabio A (2013b). Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 14, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HM, and Mizushima N (2014). At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem Sci 39, 61–71. [DOI] [PubMed] [Google Scholar]
- Shigeyama Y, Kobayashi T, Kido Y, Hashimoto N, Asahara S, Matsuda T, Takeda A, Inoue T, Shibutani Y, Koyanagi M, et al. (2008). Biphasic response of pancreatic beta-cell mass to ablation of tuberous sclerosis complex 2 in mice. Mol Cell Biol 28, 2971–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim MS, Takihara Y, Kim KY, Iwata T, Yue BY, Inatani M, Weinreb RN, Perkins GA, and Ju WK (2016). Mitochondrial pathogenic mechanism and degradation in optineurin E50K mutation-mediated retinal ganglion cell degeneration. Sci Rep 6, 33830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, et al. (2013). Identification of a candidate therapeutic autophagy-inducing peptide. Nature 494, 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva IAL, Conceicao N, Gagnon E, Caiado H, Brown JP, Gianfrancesco F, Michou L, and Cancela ML (2018). Effect of genetic variants of OPTN in the pathophysiology of Paget’s disease of bone. Biochim Biophys Acta 1864, 143–151. [DOI] [PubMed] [Google Scholar]
- Simon HU, Friis R, Tait SW, and Ryan KM (2017). Retrograde signaling from autophagy modulates stress responses. Sci Signal 10, pii: eaag2791. [DOI] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, and Czaja MJ (2009). Autophagy regulates lipid metabolism. Nature 458, 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Harley ME, Kemp AJ, Wills J, Lee M, Arends M, von Kriegsheim A, Behrends C, and Wilkinson S (2018). CCPG1 is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Dev Cell 44, 217–232 e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimanpour SA, Gupta A, Bakay M, Ferrari AM, Groff DN, Fadista J, Spruce LA, Kushner JA, Groop L, Seeholzer SH, et al. (2014). The diabetes susceptibility gene Clec16a regulates mitophagy. Cell 157, 1577–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Zhu S, Chen P, Hou W, Wen Q, Liu J, Xie Y, Liu J, Klionsky DJ, Kroemer G, et al. (2018). AMPK-mediated BECN1 phosphorylation promotes ferroptosis by directly blocking system Xc(−) activity. Curr Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria LR, Allegri G, Melck D, Pastore N, Annunziata P, Paris D, Polishchuk E, Nusco E, Thony B, Motta A, et al. (2018). Enhancement of hepatic autophagy increases ureagenesis and protects against hyperammonemia. Proc Natl Acad Sci U S A 115, 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliter D, Martinez J, Hao L, Chen X, Sun N, Fischer TD, Burman JL, Li Y, Zhang Z, Narendra DP, Cai H, Borsche M, Klein C, and Youle RJ Parkin and PINK1 mitigate STING-induced inflammation. Nature. 2018; 10.1038/s41586-018-0448-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T, Child R, Wehrly TD, Hansen B, Hwang S, Lopez-Otin C, Virgin HW, and Celli J (2012). Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe 11, 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenbeck G, and Coxon FP (2014). Role of vesicular trafficking in skeletal dynamics. Curr Opin Pharmacol 16, 7–14. [DOI] [PubMed] [Google Scholar]
- Su H, Yang F, Wang Q, Shen Q, Huang J, Peng C, Zhang Y, Wan W, Wong CCL, Sun Q, et al. (2017). VPS34 acetylation controls its lipid kinase activity and the initiation of canonical and non-canonical autophagy. Mol Cell 67, 907–921 e907. [DOI] [PubMed] [Google Scholar]
- Sumpter R Jr., Sirasanagandla S, Fernandez AF, Wei Y, Dong X, Franco L, Zou Z, Marchal C, Lee MY, Clapp DW, et al. (2016). Fanconi anemia proteins function in mitophagy and immunity. Cell 165, 867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Li X, Zhang P, Chen WD, Zhang HL, Li DD, Deng R, Qian XJ, Jiao L, Ji J, et al. (2015). Acetylation of Beclin 1 inhibits autophagosome maturation and promotes tumour growth. Nat Commun 6, 7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Yao X, Zhang QJ, Zhu M, Liu ZP, Ci B, Xie Y, Carlson D, Rothermel BA, Sun Y, et al. (2018). Beclin-1-dependent autophagy protects the heart during sepsis. Circulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki SW, Yamamoto H, Oikawa Y, Kondo-Kakuta C, Kimura Y, Hirano H, and Ohsumi Y (2015). Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proc Natl Acad Sci U S A 112, 3350–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Hikita H, Tatsumi T, Sakamori R, Nozaki Y, Sakane S, Shiode Y, Nakabori T, Saito Y, Hiramatsu N, et al. (2016). Rubicon inhibits autophagy and accelerates hepatocyte apoptosis and lipid accumulation in nonalcoholic fatty liver disease in mice. Hepatology 64, 1994–2014. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lullmann-Rauch R, Janssen PM, Blanz J, von Figura K, and Saftig P (2000). Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406, 902–906. [DOI] [PubMed] [Google Scholar]
- Tang H, Sebti S, Titone R, Zhou Y, Isidoro C, Ross TS, Hibshoosh H, Xiao G, Packer M, Xie Y, et al. (2015). Decreased BECN1 mRNA expression in human breast cancer is associated with estrogen receptor-negative subtypes and poor prognosis. EBioMedicine http://dx.do.org/10.1016/j.ebiom.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todoric J, Antonucci L, Di Caro G, Li N, Wu X, Lytle NK, Dhar D, Banerjee S, Fagman JB, Browne CD, et al. (2017). Stress-activated NRF2-MDM2 cascade controls neoplastic progression in pancreas. Cancer Cell 32, 824–839 e828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschurtschenthaler M, Adolph TE, Ashcroft JW, Niederreiter L, Bharti R, Saveljeva S, Bhattacharyya J, Flak MB, Shih DQ, Fuhler GM, et al. (2017). Defective ATG16L1-mediated removal of IRE1alpha drives Crohn’s disease-like ileitis. J Exp Med 214, 401–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboyama K, Koyama-Honda I, Sakamaki Y, Koike M, Morishita H, and Mizushima N (2016). The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 354, 1036–1041. [DOI] [PubMed] [Google Scholar]
- Ueno T, and Komatsu M (2017). Autophagy in the liver: functions in health and disease. Nat Rev Gastroenterol Hepatol 14, 170–184. [DOI] [PubMed] [Google Scholar]
- Ulgherait M, Rana A, Rera M, Graniel J, and Walker DW (2014). AMPK modulates tissue and organismal aging in a non-cell-autonomous manner. Cell Rep 8, 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente G, Morani F, Nicotra G, Fusco N, Peracchio C, Titone R, Alabiso O, Arisio R, Katsaros D, Benedetto C, et al. (2014). Expression and clinical significance of the autophagy proteins BECLIN 1 and LC3 in ovarian cancer. Biomed Res Int 2014, 462658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beek N, Klionsky DJ, and Reggiori F (2018). Genetic aberrations in macroautophagy genes leading to diseases. Biochim Biophys Acta 1865, 803–816. [DOI] [PubMed] [Google Scholar]
- Vega-Rubin-de-Celis S, Zou Z, Fernandez AF, Ci B, Kim M, Xiao G, Xie Y, and Levine B (2018). Increased autophagy blocks HER2-mediated breast tumorigenesis. Proc Natl Acad Sci U S A 115, 4176–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liang CC, Bian ZC, Zhu Y, and Guan JL (2013). FIP200 is required for maintenance and differentiation of postnatal neural stem cells. Nat Neurosci 16, 532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, and Levine B (2012). Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338, 956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang N, Zhang L, Li R, Fu W, Ma K, Li X, Wang L, Wang J, Zhang H, et al. (2016). Autophagy regulates chromatin ubiquitination in DNA damage response through elimination of SQSTM1/p62. Mol Cell 63, 34–48. [DOI] [PubMed] [Google Scholar]
- Watada H, and Fujitani Y (2015). Minireview: Autophagy in pancreatic beta-cells and its implication in diabetes. Mol Endocrinol 29, 338–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BR, Scott I, Han K, Li JH, Lu Z, Stevens MV, Malide D, Chen Y, Samsel L, Connelly PS, et al. (2013). Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J Cell Sci 126, 4843–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, An Z, Zou Z, Sumpter R, Su M, Zang X, Sinha S, Gaestel M, and Levine B (2015). The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Chiang WC, Sumpter R Jr., Mishra P, and Levine B (2016). Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell 168, 224–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, and Levine B (2008). JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell 30, 678–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, Kinch L, Koduru P, Christudass CS, Veltri RW, et al. (2013). EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell 154, 1269–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JP, Billin AN, Campbell ME, Russell AJ, Huffman KM, and Kraus WE (2018). The AMPK/p27(Kip1) axis regulates autophagy/apoptosis decisions in aged skeletal muscle stem cells. Stem Cell Reports. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC, and Holzbaur EL (2014). Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A 111, E4439–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Tian W, Hu Z, Chen G, Huang L, Li W, Zhang X, Xue P, Zhou C, Liu L, et al. (2014). ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep 15, 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyant GA, Abu-Remaileh M, Frenkel EM, Laqtom NN, Dharamdasani V, Lewis CA, Chan SH, Heinze I, Ori A, and Sabatini DM (2018). NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science 360, 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Lin DC, Yin D, and Koeffler HP (2014). An emerging role of PARK2 in cancer. J Mol Med (Berl) 92, 31–42. [DOI] [PubMed] [Google Scholar]
- Yang A, Herter-Sprie G, Zhang H, Lin EY, Biancur D, Wang X, Deng J, Hai J, Yang S, Wong KK, et al. (2018). Autophagy sustains pancreatic cancer growth through both cell-autonomous and nonautonomous mechanisms. Cancer Discov 8, 276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Yasuda S, Fujita T, Hamasaki M, Murakami A, Kawawaki J, Iwai K, Saeki Y, Yoshimori T, Matsuda N, et al. (2017). Ubiquitination of exposed glycoproteins by SCF(FBXO27) directs damaged lysosomes for autophagy. Proc Natl Acad Sci U S A 114, 8574–8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Chen Y, and Tooze SA (2018). Autophagy pathway: Cellular and molecular mechanisms. Autophagy 14, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, and Heintz N (2003). Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A 100, 15077–15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner R, Madeo F, and Kratky D (2017). Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat Rev Mol Cell Biol 18, 671–684. [DOI] [PubMed] [Google Scholar]
- Zhang J, Tripathi DN, Jing J, Alexander A, Kim J, Powell RT, Dere R, Tait-Mulder J, Lee JH, Paull TT, et al. (2015a). ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol 17, 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Kenny SJ, Ge L, Xu K, and Schekman R (2015b). Translocation of interleukin-1beta into a vesicle intermediate in autophagy-mediated secretion. Elife 4, pii: e11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sowers JR, and Ren J (2018a). Targeting autophagy in obesity: from pathophysiology to management. Nat Rev Endocrinol 14, 356–376. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yao Z, Chen Y, Qian L, Jiang S, Zhou J, Shao J, Chen A, Zhang F, and Zheng S (2018b). Lipophagy and liver disease: New perspectives to better understanding and therapy. Biomed Pharmacother 97, 339–348. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Huang S, Liu J, Wu X, Zhou S, Dai K, and Kou Y (2018). Mitophagy contributes to the pathogenesis of inflammatory diseases. Inflammation 41, 1590–1600. [DOI] [PubMed] [Google Scholar]
- Zhong Z, Sanchez-Lopez E, and Karin M (2016). Autophagy, inflammation, and immunity: A troika governing cancer and its treatment. Cell 166, 288–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yazdi AS, Menu P, and Tschopp J (2011). A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225. [DOI] [PubMed] [Google Scholar]
- Zhu P, Sieben CJ, Xu X, Harris PC, and Lin X (2017). Autophagy activators suppress cystogenesis in an autosomal dominant polycystic kidney disease model. Hum Mol Genet 26, 158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi Z, Song Z, Zhang S, Ye Y, Li C, Xu M, Zou Y, He L, and Zhu H (2015). Rubicon deficiency enhances cardiac autophagy and protects mice from lipopolysaccharide-induced lethality and reduction in stroke volume. J Cardiovasc Pharmacol 65, 252–261. [DOI] [PubMed] [Google Scholar]