Significance
The cellular mechanisms governing tRNA stability in bacteria are not well understood, and in contrast to eukaryotes, a bacterial system for degradation of mature tRNAs has not been described. We investigated the effect of thiolation on tRNA stability in Vibrio cholerae and found that this widespread posttranscriptional tRNA modification modulates the stability of a subset of tRNAs in a stationary phase-specific manner. Additional tRNA modifications were also found to promote tRNA stability. Mechanistic analyses revealed that hypomodified tRNAs can be degraded by the RNA degradosome, a multicomponent RNA degradation complex. Thus, the degradosome serves as a previously unrecognized bacterial tRNA quality control system that mediates clearance of hypomodified tRNAs.
Keywords: tRNA modification, RNA degradosome, thiI, Vibrio cholerae, stationary phase
Abstract
The factors and mechanisms that govern tRNA stability in bacteria are not well understood. Here, we investigated the influence of posttranscriptional modification of bacterial tRNAs (tRNA modification) on tRNA stability. We focused on ThiI-generated 4-thiouridine (s4U), a modification found in bacterial and archaeal tRNAs. Comprehensive quantification of Vibrio cholerae tRNAs revealed that the abundance of some tRNAs is decreased in a ΔthiI strain in a stationary phase-specific manner. Multiple mechanisms, including rapid degradation of a subset of hypomodified tRNAs, account for the reduced abundance of tRNAs in the absence of thiI. Through transposon insertion sequencing, we identified additional tRNA modifications that promote tRNA stability and bacterial viability. Genetic analysis of suppressor mutants as well as biochemical analyses revealed that rapid degradation of hypomodified tRNA is mediated by the RNA degradosome. Elongation factor Tu seems to compete with the RNA degradosome, protecting aminoacyl tRNAs from decay. Together, our observations describe a previously unrecognized bacterial tRNA quality control system in which hypomodification sensitizes tRNAs to decay mediated by the RNA degradosome.
Translation of mRNAs into the proteins that they encode is dependent on tRNAs that function as adapter molecules, coupling the presence of specific mRNA codons to the incorporation of corresponding amino acids into polypeptides. These short noncoding RNA molecules fold into a highly conserved structure that is essential for effective charging by aminoacyl-tRNA synthetases, interaction with the ribosomal translation machinery, and pairing with specific codons in mRNAs (1–3). tRNAs adopt a “clover leaf” secondary structure that generally includes five characteristic elements: the accepter stem, the D-stem loop (D arm), the anticodon stem loop (anticodon arm), the variable loop, and the T-stem loop (T arm) (SI Appendix, Fig. S1A). tRNAs’ characteristic L-shaped tertiary structure results from interactions between these elements (4).
tRNAs are transcribed as longer precursor tRNA molecules, which then undergo endonucleolytic cleavage and exonucleolytic trimming of their 5′ and 3′ ends (5). Additional posttranscriptional modifications are performed by a wide variety of dedicated tRNA-modifying enzymes (6), the activities of which include methylation, pseudouridylation, sulfuration, and coupling of amino acids. Over 100 species of modifications have been discovered across all domains of life (7). Some modifications are introduced in nearly all tRNA species, whereas others are limited to a small subset of tRNA species. Similarly, some modifications are present across all domains of life, while others are limited to a single domain (8). tRNA-modifying enzymes have specificity both for particular tRNA species and for the sites to be modified within them (9). The extent of tRNA modification is not constant; instead, it can vary in response to cellular and environmental factors, such as growth rate and oxygen or nutrient levels (5, 10–14).
Modifications are not uniformly distributed across the length of tRNA molecules (8). Within the anticodon, the first or “wobble” position is particularly subject to modification, and these modifications are often critical for efficient translation (9); consequently, mutations that prevent such modification can result in loss of viability. In contrast, nucleotide modifications within the tRNA core [composed of the T- and D-stem loops and variable loop components that interact within the tertiary structure (4)] and elbow [formed via the interaction of D and T loops (1)] are not generally required for viability, and the absence of a single modification usually does not have an effect on growth (15). Modifications outside of the anticodon region are thought to promote the thermodynamic stability (i.e., structural rigidity) of tRNAs through reinforcing the base interactions in the core or inhibiting unfavorable conformations (8). tRNAs’ highly stable structures are thought to contribute to their intracellular stability; in general, tRNAs exhibit a high melting temperature in vitro and a long half-life within the cell.
Although individual mutations in tRNA modification loci do not generally have detrimental consequences, in yeast the absence of multiple nonessential modifications results in a temperature-sensitive phenotype due to rapid exonuclease-mediated tRNA decay (RTD) of specific tRNA species (16–18). RTD and other mechanisms that degrade hypomodified or mutated mature yeast tRNAs are thought to serve as a surveillance system to eliminate tRNA molecules that have incorrect nucleosides or conformations (19). Bacteria are not known to have a similar surveillance system, and degradation of hypomodified or thermodynamically destabilized mature tRNAs has not been reported. However, mutated thermodynamically unstable precursor tRNA is degraded by PNPase in Escherichia coli (20). In addition, a temperature-sensitive decrease in the abundance of some tRNAs was observed in a tRNA modification mutant in Thermus thermophilus (21). Rapid decreases in the abundance of WT E. coli tRNA after stresses, such as amino acid starvation and oxidative stress, were reported recently (22, 23), but the pathways underlying these decreases have not been identified.
In Vibrio cholerae, the cause of the diarrheal disease cholera, transposon insertion sequencing (TIS) analysis revealed that several tRNA modification loci are required for optimal bacterial growth in an animal model of disease (24). These loci (thiI, miaA, mnmE, and mnmG) were not found to be required for V. cholerae proliferation in nutrient-rich media, although the growth of mutants in culture tubes was not extensively characterized. Consequently, mutations in these loci are thought to impair V. cholerae growth in only a subset of conditions. With the exception of thiI, these genes encode enzymes that synthesize modifications within the anticodon loop and thus, likely directly promote the efficiency or accuracy of translation; however, the product of thiI is thought to modify nucleotides at the junction of the acceptor stem and D stem (SI Appendix, Fig. S1A), which is thought to contribute to maintenance of tRNA structure (25, 26).
The product of thiI is a bifunctional enzyme that catalyzes the synthesis of 4-thiouridine (s4U) and is also required for synthesis of the thiazole moiety of thiamine (27, 28). s4U is a bacteria- and archaea-specific modification that, in E. coli, is present at position 8 in approximately one-half of tRNA species (8, 29). In E. coli tRNA-Tyr, s4U is also found at position 9 (30). The nucleosides modified by ThiI interact with nucleosides in the D-stem loop within the core, and sulfuration of U8 is predicted to promote these interactions, stabilizing tertiary structure (25, 26); consistent with this idea, the lack of s4U was found to decrease the melting temperature of a seryl-tRNA in E. coli (31).
Many of V. cholerae’s tRNAs exhibit strong homology to those of E. coli, and all contain U at position 8, but their thiolation has not been assessed. Here, we investigated the effects of s4U on tRNA homeostasis. We discovered that V. cholerae lacking thiI exhibits stationary phase-specific reductions in the levels of multiple tRNAs that typically contain s4U. A subset of these tRNAs undergoes rapid decay, suggesting that bacteria can utilize a degradative process to eliminate hypomodified tRNAs. Through transposon (Tn) mutagenesis, selection of suppressor mutations, and high-throughput DNA sequencing, we identified additional tRNA modifications that promote tRNA stability and bacteria viability. Genetic and biochemical analyses were used to demonstrate that rapid degradation of hypomodified V. cholerae tRNA is mediated by the RNA degradosome. A system for degradation of mature tRNA has not previously been identified in bacteria.
Results
ThiI Modulates tRNA Abundance in Stationary Phase in a Subset of tRNA Species.
To initiate our studies of V. cholerae thiI and tRNA modification, we confirmed that thiI is associated with the presence of s4U in this organism. Total nucleosides were prepared from total RNA of WT and ΔthiI (VC0894) strains. HPLC analyses enabled us to detect the canonical four nucleosides (C, U, G, A) and s4U, respectively (SI Appendix, Fig. S1B). Nucleosides derived from ΔthiI V. cholerae lacked the s4U detected in the WT sample (SI Appendix, Fig. S1B, red traces), indicating that thiI/VC0894 is required for s4U synthesis in V. cholerae.
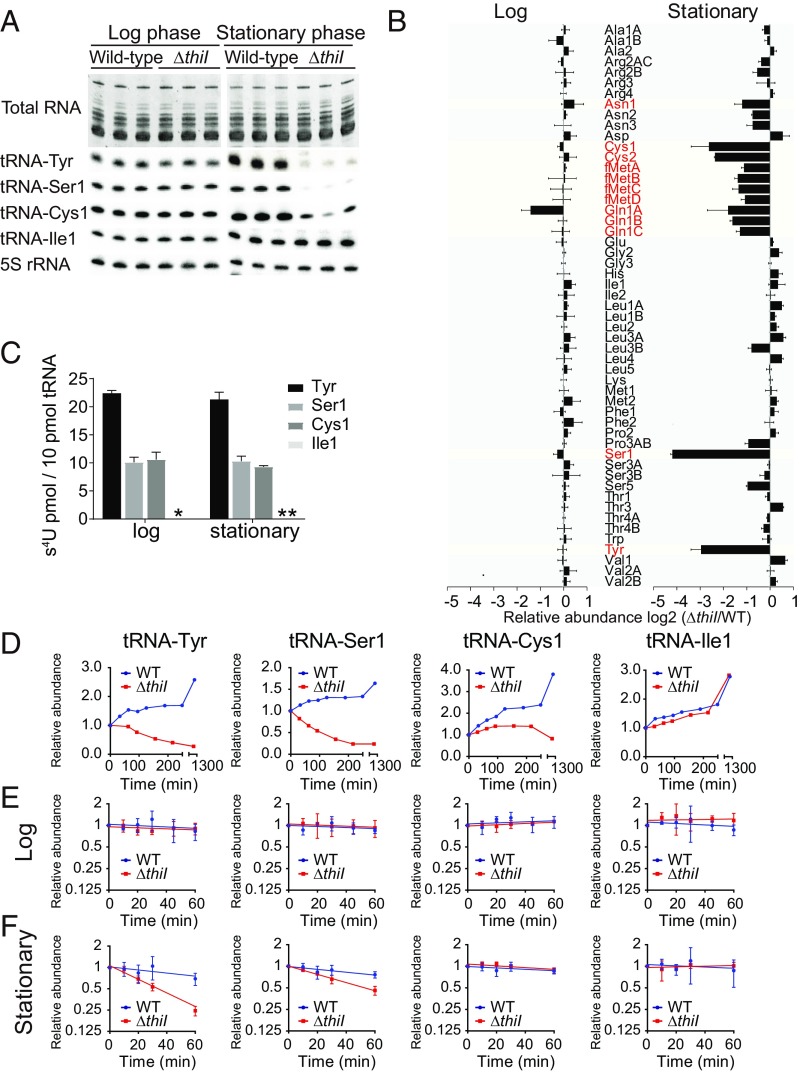
Given previous reports that modifications in tRNAs’ core region can promote the molecules’ thermodynamic stability (8, 31), which could affect their intracellular abundance, we performed a comprehensive comparison of tRNA abundance in WT and ΔthiI cells. Total RNA was extracted from cells harvested at OD600 = 0.5 (late log phase) and 24 h after inoculation and then, electrophoresed on polyacrylamide gels. No difference between the bulk amounts of tRNAs present in the two strains was observed (Fig. 1A). However, Northern blotting, using 53 probes that detect all 55 tRNA species in V. cholerae (two probes detect multiple tRNAs), revealed consistent thiI-dependent differences in tRNA abundance, almost exclusively in stationary phase. We found that 12 tRNA species were significantly less abundant in stationary-phase samples from the ΔthiI strain (fc < 0.5 and P < 0.05) (Fig. 1 A and B and SI Appendix, Fig. S2), with differences reaching as high as 18-fold. tRNA-Tyr, tRNA-Ser1, tRNA-Cys1, and tRNA-Cys2 showed the most profound reductions (Fig. 1 A and B). In log-phase samples, the abundance of only one tRNA (tRNA-Gln1A) was significantly reduced in the ΔthiI strain. These results suggest that ThiI and thus, the presence of s4U modifications modulate tRNA levels in a subset of tRNA species in a stationary phase-specific manner.
Fig. 1.
Some tRNA species lacking s4U exhibit stationary phase-specific tRNA decay. A, Upper shows a polyacryamide gel of total RNA from WT and ΔthiI strains stained with SYBR Gold. A, Lower shows Northern blots probed for the indicated tRNA species. Log-phase cells were harvested at OD600 = 0.5, and stationary-phase cells were harvested 24 h after inoculation. (B) Quantification of the indicated tRNAs from Northern blots in A and SI Appendix, Fig. S2. tRNA species that exhibited a greater than twofold change and a significant difference (P < 0.05 in t test) between WT and ΔthiI samples in stationary phase are colored in red. Bars show the mean values from three independent experiments, with error bars representing SD (calculated from SD of the WT and ΔthiI with error propagation). The list of tRNA sequences with names is shown in Dataset S3. (C) Amount of s4U detected (in 10 pmol) of each tRNA species based on HPLC analysis. Log-phase tRNAs were isolated from cultures with OD600 = 0.4, and stationary-phase tRNAs were isolated from cultures 24 h after inoculation. Each bar represents the mean value of three independent measurements, with error bars representing SD. *s4U was not detected in tRNA-Ile1 from log-phase RNA; **s4U was not analyzed in stationary phase. (D) The relative levels of tRNA-Tyr, tRNA-Ser1, tRNA-Cys1, and tRNA-Ile1 over time based on Northern blotting. The signal at each time point is presented relative to its intensity in cultures at OD600 = 0.5 (time 0) after normalization for RNA loading based on 5S rRNA. WT and ΔthiI curves are colored in blue and red, respectively. (E) Decay curves of tRNAs in log-phase cultures. Rifampicin was added at OD600 = 0.3 followed by sampling for 1 h. Each point represents the mean value of three independent measurements, with error bars representing SD. For each strain background, RNA abundance at each time point is shown relative to abundance in that strain at t = 0. (F) Decay curves of tRNAs in early stationary-phase cultures as shown in E. Rifampicin was added 1 h after cultures reached OD600 = 0.5 (∼OD600 = 1.0) followed by sampling for 1 h.
Almost all of the tRNA species with thiI-dependent reduced abundance in stationary phase are known to harbor s4U in E. coli (29), but the presence of this modification on V. cholerae tRNAs has not been assessed. We isolated four tRNA species (tRNA-Tyr, tRNA-Ser1, tRNA-Cys1, and tRNA-Ile1) from V. cholerae RNA and measured their s4U content by HPLC. tRNA-Tyr contains ∼20 pmol s4U in 10 pmol tRNA in both log and stationary phases (Fig. 1C), suggesting that V. cholerae tRNA-Tyr, like that of E. coli, harbors two s4U(30). tRNA-Ser1 and tRNA-Cys1 each contain ∼10 pmol s4U in 10 pmol tRNA. In contrast, no s4U was detected in tRNA-Ile1, which had been selected to represent tRNAs with abundance that was not altered in the thiI background (Fig. 1C). These data are consistent with the hypothesis that the reduced abundance of some tRNAs in stationary-phase ΔthiI V. cholerae is linked to the tRNAs’ lack of s4U.
s4U Promotes tRNA Stability in Stationary Phase.
We next performed a higher-resolution analysis of tRNA levels over time for tRNAs with abundance that was severely reduced in stationary-phase ΔthiI cells. The level of tRNA-Tyr and tRNA-Ser1 decreased when ΔthiI cultures reached late log phase (Fig. 1D and SI Appendix, Fig. S3A), in contrast to the increase observed in WT cultures. Levels of tRNA-Cys1 did not differ between WT and ΔthiI samples until early stationary phase, when ΔthiI displayed less of an increase than the WT followed by a gradual decrease over late stationary phase. As expected based on its lack of s4U, tRNA-Ile1 levels did not differ between the two strains.
The rapid decrease in tRNA-Ser1 and tRNA-Tyr levels suggests that these tRNAs undergo rapid decay in the ΔthiI background. To test this possibility, we measured the decay of the four tRNAs assessed above in log- and stationary-phase cultures by blocking transcription with rifampicin and using Northern blotting to track levels over time. In early log-phase samples, no decay was observed for any of the tRNA species, consistent with the typical expectation that tRNAs are stable, long-lived species and with our previous observation that the absence of thiI does not generally affect RNA abundance in log phase (Fig. 1E and SI Appendix, Fig. S3B). In contrast, exponential decay of tRNA-Tyr and tRNA-Ser1 was observed in stationary-phase cultures of the ΔthiI strain (Fig. 1F and SI Appendix, Fig. S3C). No decline was observed for tRNA-Cys1 and tRNA-Ile1 from the ΔthiI strain or for any tRNAs from stationary-phase cultures of the WT strain (Fig. 1F). Collectively, these results suggest that the absence of s4U leads to a stationary phase-specific decrease in the abundance of some tRNAs by reducing their intracellular stability but that additional mechanisms also likely contribute to the decline in some tRNA species (e.g., tRNA-Cys1).
Next, we addressed the relationship between thermodynamic instability and rapid tRNA decay. We generated melting curves for tRNA-Tyr and tRNA-Ser1 isolated from WT and ΔthiI strains to test if s4U deficiency affected the stability of tRNA-Tyr and tRNA-Ser1 in vitro (SI Appendix, Fig. S4). At temperatures from ∼40 °C to 80 °C, tRNA-Tyr from ΔthiI cells displayed markedly increased A260 relative to the WT, while tRNA-Ser1 showed a more subtly altered absorbance. Both patterns were associated with a reduced melting temperature of the ΔthiI-derived tRNA, with a melting temperature of 60.1 °C vs. 66.0 °C for tRNA-Tyr and 55.0 °C vs. 61.0 °C for tRNA-Ser1, indicative of the reduced stability of the hypomodified tRNA. This observation is consistent with the idea that rapid decay of hypomodified tRNA could be associated with thermodynamic instability.
Genetic Interactions Between thiI and the Genes Responsible for Other tRNA Modifications.
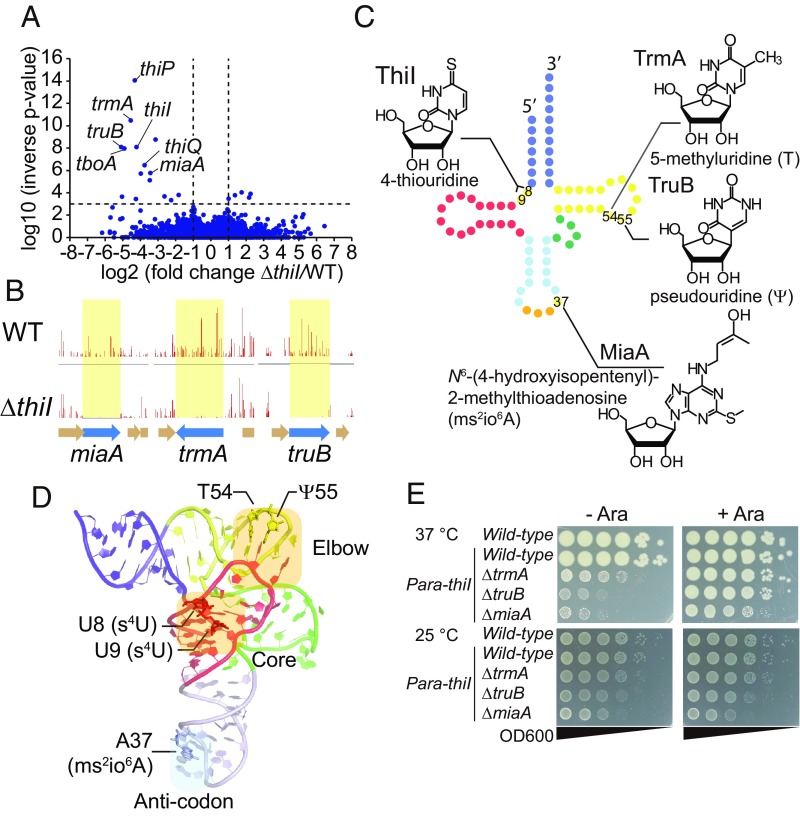
To gain additional insight into the role and impact of s4U, we carried out a TIS screen to define thiI’s genetic interactions. We created a highly saturated Tn insertion library in the ΔthiI strain and determined the frequency with which each potential insertion site was disrupted. The Artist pipeline (32) was used to identify genes in which Tn insertions are underrepresented in the ΔthiI library relative to their frequency in a WT library generated using the same protocol (Dataset S1). Underrepresented loci are expected to be more important for growth and/or viability in the absence of thiI. As expected, the screen identified genes required for thiamine transport (i.e., tboA, thiP, and thiQ) (Fig. 2A and Dataset S1); such genes are necessary for providing cellular thiamine when the biosynthetic pathway is disrupted (e.g., by the absence of thiI). Notably, the set of underrepresented genes also included three genes responsible for tRNA modification: trmA, which enables synthesis of 5-methyluridine at position 54; truB, which enables synthesis of pseudouridine (Ψ) at position 55; and miaA, which enables synthesis of i6A, a precursor of the ms2io6A at position 37 (Fig. 2 A–D). T54 and Ψ55 are located within the T loop and are involved in elbow formation (Fig. 2D); these loci have been found to be modified in all species of E. coli tRNA (29). ms2io6A37 is one of the many modifications generated at this position that enhances tRNA’s decoding capacities (Fig. 2D) (33). Identification of the latter three underrepresented loci suggests that thiI contributes to critical cellular processes in its role in tRNA modification as well as in thiamine biosynthesis.
Fig. 2.
TIS analyses reveal genetic interactions between thiI and other genes that mediate tRNA modifications. (A) Volcano plot of the results from Con-ARTIST analysis comparing WT and ΔthiI Tn libraries (Dataset S1). (B) Artemis plots showing normalized frequencies of Tn insertions in miaA, trmA, and truB. Insertion frequencies per locus are depicted as vertical lines for the WT (Upper) and ΔthiI (Lower) libraries. (C) Schematic secondary structure of tRNA with the chemical structures of the modified nucleosides that are synthesized by the genes identified in the TIS screen. The acceptor stem with 3′ terminus, D arm, anticodon arm, variable loop, T arm, and anticodon are colored in blue, red, light blue, green, yellow, and orange respectively. The names of the modifying enzymes are also shown. (D) Tertiary structure of tRNA-Tyr from T. thermophilus (49) (Protein Data Bank ID code 1H3E). The modified sites identified in the TIS screen are shown. The acceptor stem, D arm, anticodon arm, variable loop, and T arm are colored in blue, red, light blue, green, and yellow, respectively. The elbow and core regions of the tRNA are highlighted in light orange. (E) Growth of double-mutant strains on LB plates with (Right) or without (Left) 0.2% arabinose at 37 °C (Upper) and 25 °C (Lower) for 24 h.
To validate results from this screen, we constructed double mutants in which thiI’s native promoter was replaced by the arabinose-inducible promoter and one additional gene was deleted. While we observed no or only marginal growth defects in all mutants when grown in the presence of arabinose, we observed severe growth defects in all three double mutants in the absence of arabinose (Fig. 2E). Thus, the two modifications work cooperatively to enable optimal growth so that growth is impaired only in the absence of both loci. It is noteworthy that these growth defects were not observed when bacteria were grown at 25 °C rather than 37 °C, perhaps because thermodynamic destabilization of unmodified tRNAs is less pronounced at the lower temperature.
Growth Defects in Double Mutants Are Suppressed by Mutations in tRNA-Tyr and RNase E.
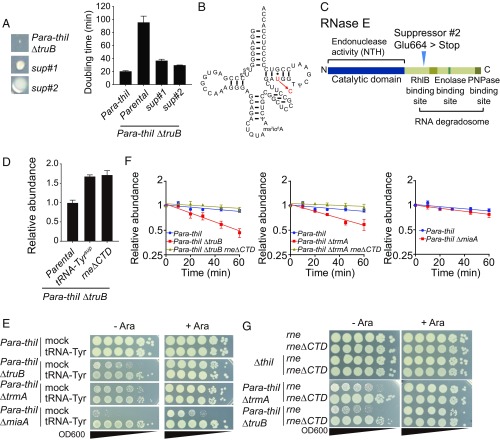
The Para-thiI/ΔtruB strain grown on LB plates without arabinose exhibited heterogeneous colony size (Fig. 3A). The very tiny colonies and two types of larger colonies with distinct morphologies were presumed to be the parental strain and spontaneous suppressor strains, respectively. The slowed growth of the Para-thiI/ΔtruB strain (doubling time = 96 min in the absence of arabinose) was markedly reduced in both suppressor strains (36 and 29 min, respectively) (Fig. 3A). Whole-genome sequencing of DNA from the parental Para-thiI/ΔtruB strain and its derivatives was performed to identify potential suppressor mutations. One suppressor strain was found to have a mutation in a gene encoding tRNA-Tyr that changed U to C at position 51 in the T stem, resulting in a fully matched T stem that is expected to augment tRNA stability (Fig. 3B). The second strain was found to have a stop codon introduced in place of Glu664 at the beginning of the C-terminal domain (CTD) of RNase E (VC2030) (Fig. 3C). The truncated protein retains RNase E’s endonuclease domain, which is involved in rRNA and tRNA precursor processing, but lacks the domain that scaffolds formation of the RNA degradation machinery known as the RNA degradosome (34). The degradosome, which is composed of the RNA helicase RhlB, Enolase, and Polynucleotide phosphorylase PNPase, is thought to degrade structured RNA, but it has not been specifically linked to degradation of mature tRNA. Thus, both suppressor mutations seemed likely to result in tRNA stabilization either of a single species (from the tRNA-Tyr mutation) or more generally (due to absent/defective degradosomes). Northern blotting confirmed that both suppressor mutations increased tRNA-Tyr levels in the Para-thiI/ΔtruB mutant in the absence of arabinose (Fig. 3D).
Fig. 3.
Mutants lacking multiple tRNA modification genes exhibit tRNA decay mediated by the RNA degradosome. (A, Left) Colony morphology of the original Para-thiI/ΔtruB strain and suppressor strains (sup#1 and sup#2) on the same plate. A, Right shows the doubling time in LB medium at 37 °C of the indicated strains. (B) Predicted secondary structure of tRNA-Tyr with the mutation that is found in sup#1. Modifications are predicted based on those previously observed in E. coli. (C) Schematic depiction of the RNase E domain structure with the site of the RNase E Glu664STOP (sup#2) mutation annotated. (D) Relative abundance of tRNA-Tyr in log-phase (OD600 = 0.5) cultures of the indicated strains grown in LB at 37 °C without arabinose. (E) Growth on LB plates with or without 0.2% arabinose of serially diluted double mutants containing either a multicopy vector expressing tRNA-Tyr or an empty vector. Plates were incubated at 37 °C for 24 h. (F) Decay curves of tRNA-Tyr in early log-phase (OD600 = 0.3) cultures of the indicated strains grown in LB medium without arabinose. (G) Growth as in E of serially diluted strains producing either WT RNase E (rne) or RNase E lacking its CTD (rneΔCTD).
Given the profound growth effect of the tRNA-TyrU51C suppressor that should specifically elevate tRNA-Tyr abundance, we also tested whether exogenous expression of tRNA-Tyr countered the growth defect of the Para-thiI/ΔtruB mutant. Introduction of a multicopy vector expressing tRNA-Tyr markedly increased the growth of this strain in the absence of arabinose, rendering it comparable with that of the parental Para-thiI strain (Fig. 3E). In contrast, overproduction of either tRNA-Ser1 or tRNA-Gln1A did not rescue growth of the Para-thiI/ΔtruB mutant (SI Appendix, Fig. S5), suggesting that the growth defect is specifically linked to tRNA-Tyr. The tRNA-Tyr expression vector also increased growth of the Para-thiI/ΔtrmA and Para-thiI/ΔmiaA double mutants, suggesting that the absence of additional tRNA modifications in these strains also reduced tRNA-Tyr abundance and/or activity sufficiently to hamper bacterial growth. These data suggest that V. cholerae is particularly sensitive to tRNA-Tyr depletion. V. cholerae has only one tRNA-Tyr isoacceptor, and therefore, all tRNA-Tyr have s4U. Thus, loss of this tRNA-Tyr is expected to be lethal, and maintenance of proper tRNA-Tyr levels is important for optimal growth.
Rapid Decay of Hypomodified tRNAs Is Mediated by the RNA Degradosome.
The finding that RNase E truncation in the Para-thiI/ΔtruB mutant increased tRNA-Tyr abundance (Fig. 3D) coupled with the observation of the thiI mutant’s rapid tRNA degradation during stationary phase (Fig. 1F) suggested that RNase E might mediate tRNA degradation. To explore this possibility, we measured the decay of tRNA-Tyr in the Para-thiI, Para-thiI/ΔtruB, and RNase E suppressor strains during log-phase growth without arabinose. tRNA-Tyr showed no detectable decay in the single-thiI mutant strain (Fig. 1E); however, it showed exponential decay in the Para-thiI/ΔtruB strain (Fig. 3F and SI Appendix, Fig. S3D). Deletion of the RNase E CTD in the double mutant restored tRNA-Tyr stability to the level observed in the WT (Fig. 1E) or Para-thiI strain (Fig. 3F and SI Appendix, Fig. S3D), providing a possible explanation for this mutant’s increase in tRNA-Tyr levels. Exponential decay of tRNA-Tyr was also observed in the Para-thiI/ΔtrmA mutant, with restoration of its stability by the truncation of RNase E (Fig. 3F and SI Appendix, Fig. S3E). In addition, RNase E truncation increased the growth of both double mutants in the absence of arabinose (Fig. 3G). Collectively, these data suggest that RNA degradosomes can limit the lifespan of hypomodified tRNA species. This is a demonstration of a rapid decay system for hypomodified tRNA in bacteria. It is noteworthy that tRNA decay was not observed in the Para-thiI/ΔmiaA background (Fig. 3F and SI Appendix, Fig. S3F), likely because the anticodon region altered by the miaA mutation is typically less critical for thermodynamic stabilization of the tertiary structure of tRNA; instead, the growth defect in this background is likely attributable to reduced tRNA function.
tRNA-Tyr and tRNA-Ser1 Are Degraded by Distinct Mechanisms.
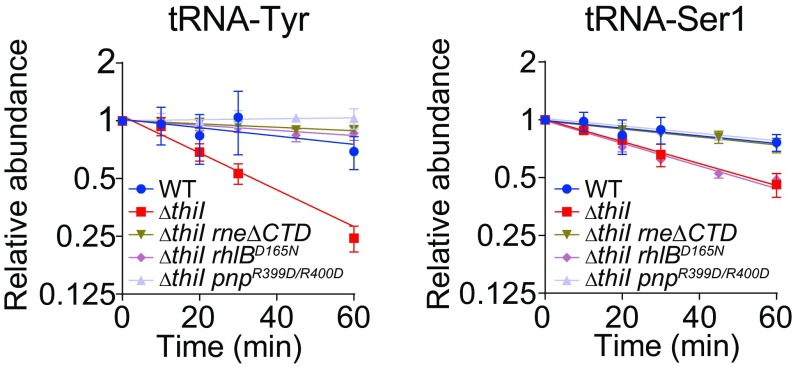
We also assessed whether the rapid decay of some hypomodified tRNAs in stationary-phase cultures of the ΔthiI strain (Fig. 1F) is dependent on the CTD of RNase E. We found that rapid decay of the ΔthiI mutants’ tRNA-Tyr and tRNA-Ser1 was eliminated by deletion of RNase E’s CTD, providing additional support for the involvement of RNA degradosomes in controlling tRNA lifespan (Fig. 4 and SI Appendix, Fig. S3 G and H). As noted above, the degradosome is composed of RNA helicase RhlB, Enolase, and Polynucleotide phosphorylase PNPase in addition to the RNase E CTD. Like RNase E truncation, a catalytic mutation within PNPase (R399D/R400D) (35) suppressed decay of tRNA-Tyr and tRNA-Ser1 in the ΔthiI background; however, a catalytic mutation within RhlB (D165N) (36, 37) suppressed decay of tRNA-Tyr but not of tRNA-Ser1 (Fig. 4). The variability in the requirement for active RhlB suggests that tRNA-specific decay mechanisms may be present.
Fig. 4.
RNA degradosome-mediated tRNA decay in stationary phase in a ΔthiI mutant. Decay curves of tRNA-Tyr and tRNA-Ser1 in early stationary phase (1 h after OD600 = 0.5) in the indicated strains.
RNA Degradosomes Mediate tRNA Decay in Vitro.
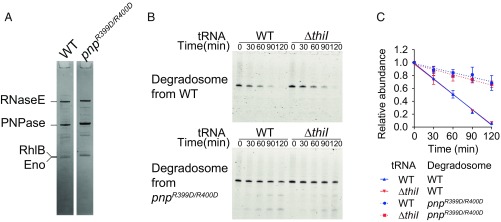
To further support our model that decay of hypomodified tRNAs is mediated by RNA degradosomes, we established an in vitro assay of tRNA decay. An epitope-tagged form of endogenous RNase E was affinity purified from WT and PNPase-deficient (R399D/R400D) strain backgrounds, enabling isolation of associated degradosome proteins as well as RNase E (Fig. 5A). We incubated the purified RNA degradosomes with tRNA-Tyr isolated from the WT or ΔthiI strain and tracked tRNA-Tyr decay. As expected, tRNA-Tyr decay was far more rapid in the presence of degradosomes from WT cells than from the PNPase-deficient strain (Fig. 5 B and C), suggesting that we had largely recapitulated the tRNA decay activity of RNA degradosome in vitro and were not observing degradation catalyzed by RNase E’s endonuclease domain. Surprisingly, we did not see any significant difference in the rate of decay for tRNA-Tyr from WT and ΔthiI cells. These observations suggest that RNA degradosomes do not distinguish hypomodified tRNAs from modified tRNAs via direct recognition of tRNA primary sequence or secondary structure and raise the possibility that additional factors govern degradosome activity and/or alter the susceptibility of hypomodified tRNAs to RNA degradosome-mediated decay in vivo.
Fig. 5.
RNA degradosomes mediate tRNA decay in vitro. (A) Coomasie-stained gel image of Flag-RNase E immunoprecipitated fraction. Indicated bands were identified through peptide mass spectrometry. (B) In vitro tRNA-Tyr decay assay. Time course of tRNA-Tyr resolved in 10% TBE-UREA gel stained with SYBR Gold at the indicated time. Upper and Lower show reactions with degradosomes from the WT and from the PNPase catalytic mutant, respectively. (C) Decay curves of tRNA-Tyr from WT and ΔthiI mutant strains based on data shown in B.
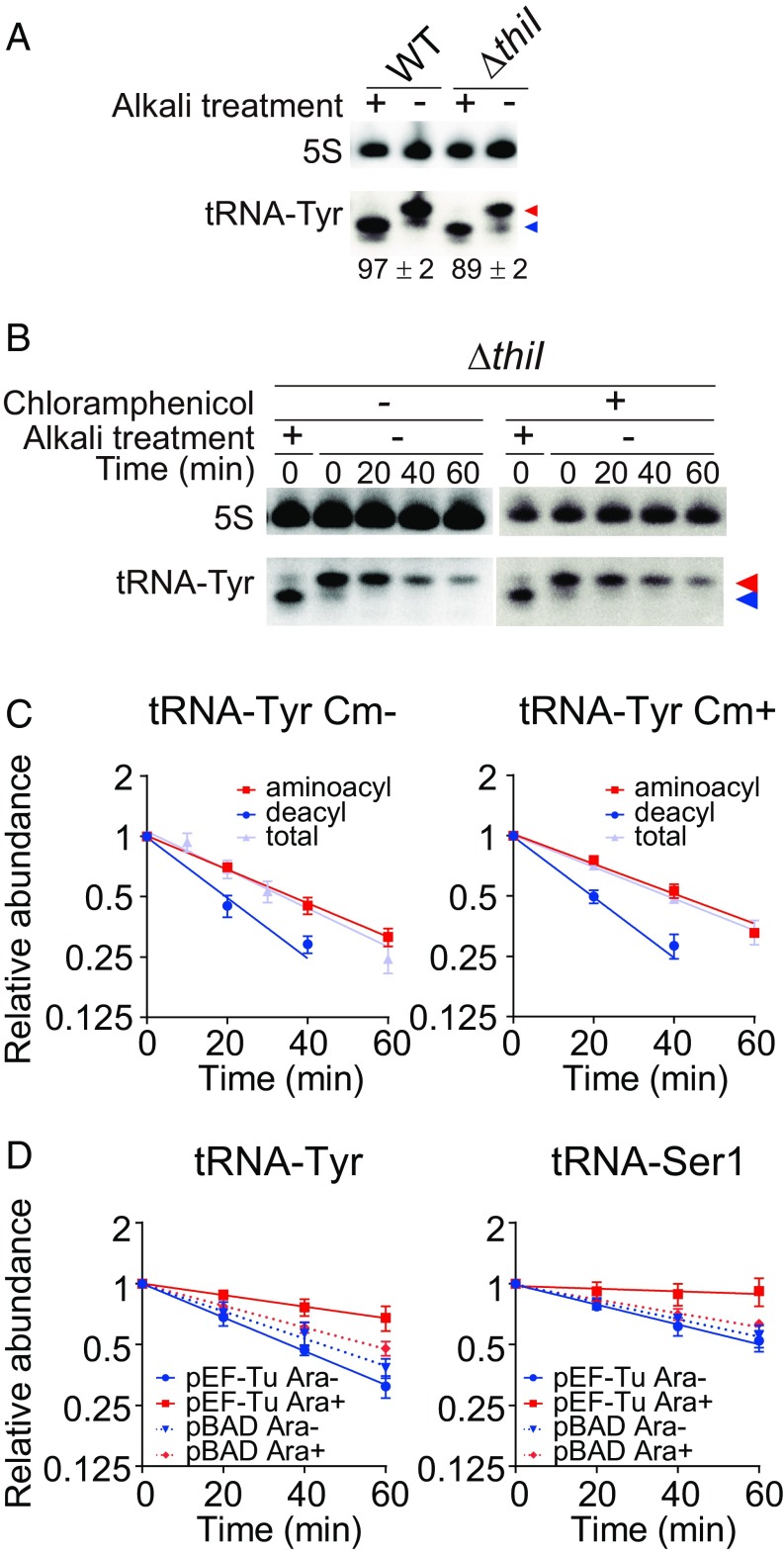
Aminoacylated tRNA-Tyr Undergo RNA Degradosome-Dependent Decay.
PNPase targets the 3′ end of tRNAs, the site of aminoacylation. To investigate whether tRNA aminoacylation status influences RNA degradosome-dependent decay, we first monitored tRNA-Tyr aminoacylation levels in the WT and ΔthiI strains in early stationary phase. Both strains had similar high aminoacylation levels (Fig. 6A), suggesting that the absence of thiI does not drastically affect tRNA-Tyr aminoacylation. Next, we tracked the decay of aminoacylated and deacylated tRNA-Tyr in the ΔthiI strain. Levels of deacylated and aminoacylated tRNAs both decreased after the addition of rifampicin (Fig. 6 B and C and SI Appendix, Fig. S3I), suggesting that both aminoacylated and deacylated tRNAs can be targeted by the RNA degradosome. Furthermore, similar decay patterns of deacylated and aminoacylated tRNAs were observed after addition of chloramphenicol, which blocks translation-mediated conversion of aminoacylated tRNAs to deacylated tRNAs (Fig. 6 B and C), arguing against the possibility that the decrease of aminoacylated tRNA observed above (Fig. 6 B and C) is explained by a shift in the equilibrium between aminoacylated and deacylated tRNA. Taken together, these observations strongly suggest that aminoacylated as well as deacylated tRNAs are targeted by the RNA degradosome for decay.
Fig. 6.
EF-Tu protects aminoacyl-tRNAs from RNA degradosome-mediated decay. (A) Aminoacylation levels of tRNA-Tyr in WT and ΔthiI strains. Total RNAs from early stationary phase (1 h after OD600) were resolved on 6.5% acid gels and analyzed by Northern blotting. Means ± SD of aminoacylation level from three independent experiments are shown. Red and blue arrowheads indicate aminoacylated and deacylated tRNAs, respectively. (B and C) Decay of aminoacylated and deacylated tRNAs in early stationary-phase cultures after rifampicin treatment. Total RNAs were resolved on 6.5% acid gels and analyzed by Northern blotting with or without chloramphenicol (Cm) (B); decay curves of the lower band (deacylated tRNA), upper band (aminoacylated tRNA), and total tRNA-Tyr (calculated from neutral PAGE Northern blotting analysis) are shown in C. Red and blue arrowheads in B indicate aminoacylated and deacylated tRNAs, respectively. (D) Decay curves of tRNA-Tyr and tRNA-Ser1 from the ΔthiI strain containing a vector encoding arabinose-inducible EF-Tu (pEF-Tu) or an empty vector (pBAD) with or without 0.2% arabinose.
Elongation Factor Tu Protects tRNAs from RNA Degradosome-Mediated Decay.
Aminoacylated tRNAs are bound by elongation factor Tu (EF-Tu) in the cytoplasm (38). Since the 3′ end of tRNAs is found within the EF-Tu binding pocket, we hypothesized that this EF protects tRNA from degradosome-mediated decay. To test this hypothesis, an inducible version of EF-Tu was introduced into the ΔthiI strain, and after EF-Tu induction in late log phase, tRNA decay was monitored in early stationary phase. Induction of EF-Tu suppressed the decay of tRNA-Tyr and tRNA-Ser1 (Fig. 6D and SI Appendix, Fig. S3 J and K), suggesting that EF-Tu can protect tRNAs from degradosome-mediated decay.
Discussion
The mechanisms by which bacterial cells govern tRNA stability have been largely unclear. Here, we describe a previously unrecognized tRNA quality control mechanism in bacteria that enables clearance of hypomodified tRNAs (SI Appendix, Fig. S6). V. cholerae lacking ThiI, which generates s4U in a subset of V. cholerae tRNAs, has reduced abundance of tRNAs that contain s4U, particularly in stationary phase. In part, this reduction in tRNA levels reflects rapid degradation of hypomodified tRNA species, a process not previously reported for bacteria. Such rapid degradation is even more pronounced, extending into log-phase growth, in mutants lacking additional tRNA modifications as well as s4U. Our genetic and biochemical analyses indicate that this tRNA degradation is mediated by RNA degradosomes, which seem to be able to target charged as well as noncharged tRNAs. EF-Tu can protect aminoacyl-tRNAs from RNA degradosome-mediated decay. Hypomodified tRNAs subject to degradation exhibited reduced melting temperatures, suggesting that thermodynamic destabilization and loss of typical tRNA structure are likely cues for degradation (SI Appendix, Fig. S4).
The effect of thiI disruption on V. cholerae tRNA abundance is variable, with levels of some tRNAs showing marked changes in stationary phase, while others exhibit minimal or no change. Differences in tRNA abundance do not necessarily reflect the extent of s4U incorporation, since disruption of thiI prompted lesser changes in the abundance of tRNA-Tyr than of tRNA-Ser1, despite tRNA-Tyr containing twice as much s4U. The extent of s4U incorporation in other V. cholerae tRNAs has not been fully determined; however, it is apparent that not all tRNA species include this modification (e.g., tRNA-Ile1 does not), despite the ubiquity of U at position 8 of V. cholerae tRNAs. It is likely that variance at other tRNA positions modulates the need for and benefits of the s4U modification.
Multiple mechanisms likely account for the reduced abundance of some tRNAs in the thiI mutant during stationary phase. A subset of hypomodified tRNAs seems to be subject to rapid degradation that is dependent on RNase E’s C terminus and PNPase catalytic activity, suggesting involvement of RNA degradosomes. Degradation dependent on the RNase E CTD was also observed in log phase for tRNAs lacking multiple modifications. In addition, in vitro analyses confirm that RNA degradosomes can target tRNAs. Collectively, these observations suggest that degradation machinery scaffolded by RNase E’s C terminus underpins a previously unrecognized bacterial tRNA quality control system.
The bacterial tRNA decay system investigated here shares certain features with the yeast rapid tRNA decay system. Both systems can degrade hypomodified aminoacylated tRNAs (16), and our findings suggest that, as in yeast [where elongation factor 1A (EF1A) competes with the decay machinery (39, 40)], EF-Tu (the functional homolog of EF1A) competes with the degradasome. Interestingly, however, the mechanisms of the two degradation machineries differ. The bacterial system relies on PNPase, a 3′ → 5′ exonuclease, whereas the yeast system depends Rat1 and Xrn1, 5′ → 3′ exonucleases (17). We speculate that these systems have convergently evolved to promote tRNA quality control.
Unexpectedly, our results suggest that the RNA degradosome adopts distinct processes to degrade tRNA-Tyr and tRNA-Ser1; the catalytic activity of RhlB facilitates decay of tRNA-Tyr but not that of tRNA-Ser1. Given that RhlB can catalyze unwinding of double-stranded RNAs, RhlB could destroy the secondary and tertiary structures of hypomodified tRNA-Tyr. We hypothesize that disruption of tRNA structure by RhlB might induce dissociation of protein factors associated with tRNA that otherwise protect the molecule and promote its stability. RhlB’s specificity in promoting decay of tRNA-Tyr but not that of other tRNAs (e.g., tRNA-Ser1) could account for the relative rapidity of tRNA-Tyr decay (Fig. 1F). The mechanisms by which RhlB recognizes specific tRNAs remain to be investigated.
The basis for the enhanced susceptibility of hypomodified tRNAs to decay mediated by the RNA degradosome is still unknown. Since rapid degradation was prominent in the thiI trmA and thiI truB double mutants, we speculate that destabilization of the tRNA elbow region, which is critical for generating tRNAs’ canonical L-shaped tertiary structure, is a potent trigger of RNA degradosome-mediated tRNA decay. Formation of the elbow requires interaction of the tRNA T and D arms (Fig. 2 and SI Appendix, Fig. S4), and in solution, such interactions are facilitated by T54 and Ψ55 modifications (generated by TrmA and TruB, respectively) (41). The s4U modification, by stabilizing the tRNA core (Fig. 2D), may also contribute to elbow formation/stability. Thus, the simultaneous absence of s4U and T54 or Ψ55 modifications may markedly reduce the thermodynamic stability of the elbow region. We did not observe preferential degradation of hypomodified tRNAs in vitro, suggesting that RNA degradosomes do not directly recognize alterations in tRNA primary or secondary structure associated with the absence of modification. However, in vivo, it is possible that EF-Tu preferentially protects properly folded tRNAs from degradation if hypomodified tRNAs (with unstable elbow regions) have lower affinity for EF-Tu.
Our findings uncovered another process that distinguishes stationary-phase cell physiology; the thiI mutation had a more marked effect on tRNA abundance in stationary phase than in log phase, and the mutant’s hypomodified tRNAs were only subject to rapid decay during stationary phase. A possible explanation for the latter observation is that stationary-phase tRNAs may have reduced thermodynamic stability, perhaps related to ionic and/or osmotic characteristics of culture media. Additionally, degradosomes may have heightened activity, protective EF-Tu may be less abundant, and/or there may be reduced abundance of alternate degradosome targets during stationary phase.
Finally, our analyses of the role of thiI, its interaction network, and factors that compensate for thiI deficiency were greatly facilitated by high-throughput and genome-wide analyses. Many outstanding questions regarding control of bacterial tRNA metabolism and activity remain to be addressed, and additional applications of similar high-throughput technologies to investigate these issues will be fruitful.
Methods
Strains and Culture Conditions.
Strains and plasmids are listed in SI Appendix, Tables S1 and S2. V. cholerae C6706, a clinical isolate (42), was used in this study. All strains were grown in LB containing 1% NaCl at 37 °C unless otherwise indicated. E. coli SM10 (lambda pir) harboring pSC189 (43) or pCVD442 (44) was cultured in LB plus LB plus carbenicillin (Cb). Antibiotics were used at the following concentrations: 200 μg/mL streptomycin, 50 μg/mL kanamycin, and 50 μg/mL Cb. Arabinose-inducible promoters were induced with 0.2% arabinose.
Strain and Plasmid Construction.
All mutations in C6706 were created using homologous recombination and a derivative of the suicide vector pCVD442 as described (32). Targeting vectors for gene deletions contained 500–1,000 bp of DNA flanking each side of the target gene cloned into pCVD442’s SmaI site using isothermal assembly. The tRNA-Tyr gene was inserted into the BamHI/SalI sites of pACYC184. The tRNA-Gln1A and tRNA-Ser1 genes were inserted into linearized ptRNA-Tyr. The EF-Tu gene was introduced into the SmaI site of pBAD33. For introducing mutations into the chromosomal pnp and rhlB loci, each ORF with flanking regions was initially cloned into pCVD442’s SmaI site, and then, mutations were generated with the PrimeSTAR Mutagenesis Basal Kit (TAKARA-bio); subsequently, allelic exchange was used to introduce these mutant genes into the chromosome. For integration of 3× Flag tag to the 3′ end of the chromosomal rne gene, flag-rne with a flanking region was introduced into pHL100’s SmaI site, transferred to pCVD442′ SmaI site, and delivered into chromosome by allelic exchange.
RNA Extraction.
Total RNA was extracted with TRIzol (Life Technologies) according to the manufacturer’s instructions.
Isolation of Individual tRNAs.
One-liter cultures of log-phase (OD600 = 0.4) and stationary-phase (24-h) V. cholerae cells were harvested, and total RNA was extracted as previously reported (45). Briefly, cells were resuspended in 5 mL buffer [50 mM NaOAc, pH 5.2, 10 mM Mg(OAc)2], mixed with 5 mL water-saturated phenol, and agitated vigorously for 1 h. The aqueous phase was separated by centrifugation, washed with chloroform, and recovered by isopropanol precipitation. RNA was cleaned with the TRIzol reagent, and total RNA was run through a manually packed DEAE column (GE healthcare) to remove contaminants and recovered by isopropanol precipitation. Individual tRNA species were bound to biotinylated DNA probes anchored to high-capacity streptavidin agarose resin (GE Healthcare) in 30 mM Hepes-KOH, pH 7.0, 1.2 M NaCl, 15 mM EDTA, and 1 mM DTT at 68 °C for 30 min with shaking. Beads were washed three times with 15 mM Hepes-KOH, pH 7.0, 0.6 M NaCl, 7.5 mM EDTA, and 1 mM DTT and seven times with 0.5 mM Hepes-KOH, pH 7.0, 20 mM NaCl, 0.25 mM EDTA, and 1 mM DTT. Purified tRNAs were extracted from beads with TRIzol and then gel purified on 10% TBE-UREA gels. Residual DNA probes were eliminated with Turbo DNase (Thermo Fisher Scientific). The probes used in this study are listed in Dataset S2.
Northern Blotting.
In total, 0.1–1 μg RNA was electrophoresed on 10% Novex TBE-UREA gels (Thermofisher) and stained with SYBR Gold (Life Technologies). For evaluation of aminoacylation level, total RNA was electrophoresed on 6.5% urea gels (7 M urea, 100 mM NaOAc, pH 5.0). RNA was transferred to nitrocellulose membranes by semidry blotting and cross-linked twice to membranes with 1,200 μJ UV light. Membranes were incubated in ULTRAhyb-oligo (Life Technologies) at 42 °C for 30 min followed by hybridization overnight at 42 °C with 4 pmol DNA probes radiolabeled using [γ-32P]ATP (PerkinElmer) and T4 Polynucleotide kinase (New England Biolabs). Membranes were washed twice with 2× SSC/0.5% SDS, and then, bound probe was detected using an FLA-5000 phosphoimager (Fuji). The signal intensity of tRNA was normalized to that of 5S rRNA. All DNA oligos were synthesized by Integrated DNA Technology. Probe sequences are listed in Dataset S2.
Propagated errors (σa/b) of relative abundance were calculated by use of the following formula:
a indicates the mean of tRNA abundance in ΔthiI strain, b indicates the mean of tRNA abundance in the WT, σa indicates the SD of tRNA abundance in ΔthiI, σb indicates the SD of tRNA abundance in the WT, and σa/b indicates the propagated SD of relative tRNA abundance.
tRNA Decay Curve Measurement.
LB medium (50 or 100 mL) was inoculated with overnight cultures and then incubated with vigorous shaking at 250 rpm (Innova 2100; New Brunswick Scientific); at selected points (OD600 = 0.3 for log phase and 1 h after OD600 = 0.5 for stationary phase), rifampicin (Fisher BioReagents) was added to cultures at a final concentration of 100 μg/mL. Several time points were characterized per culture; for each, cells in a 1-mL sample were harvested by centrifugation at 21,130 × g for 30 s. Total RNA was extracted using TRIzol reagent, and the abundance of individual tRNAs was quantified by Northern blotting analysis (SI Appendix, Fig. S7) as described above. Average values from three or four biological replicates were fitted to an exponential curve.
Assays of Bacterial Growth.
To generate short-term growth curves, overnight cultures were diluted 1:105 in LB medium and cultured in a Bioscreen C OD reader (Growth Curves USA) with monitoring of OD600 every 15 min for 24 h.
To measure the effect of arabinose on growth of strains with arabinose-inducible thiI, LB medium without arabinose was inoculated with cells grown overnight at 30 °C or 37 °C on LB plates supplemented with 0.2% arabinose. Culture tubes were allowed to stand at 37 °C overnight; cultures were then serially diluted in LB medium and spotted (5 μL) on LB plates with or without arabinose. Plates were photographed 1 d after plating.
Tn Insertion Sequencing.
A Himar1 Tn insertion library in a ΔthiI strain was generated and sequenced as previously described (32). Sequenced reads were mapped onto a V. cholerae reference genome (N16961), and all TA dinucleotides sites (TA sites) were tallied and assigned to annotated genes as described (24). Based on 7,750,593 mapped reads, the library of ∼600,000 colonies contained 134,416 unique Tn insertions corresponding to disruption of 70% of all TA sites. The Con-ARTIST pipeline (24) was used to compare the ΔthiI library with a previously characterized WT library (32) to identify over- and underrepresented genes. Complete results of this analysis are shown in Dataset S1, and raw data files were deposited to the Sequence Read Archive (SRA) database in NCBI (accession number SRP169513) (46).
Whole-Genome Sequencing.
Three colonies of Para-thiI/ΔtruB with distinct sizes and morphologies were cultured in LB overnight, and genomic DNA was extracted with the DNeasy kit (Qiagen). DNA libraries were prepared with the Nextera XT kit (Illumina) according to the manufacturer’s instructions and then sequenced with MiSeq using MiSeq reagent v3 (Illumina). Sequenced reads were mapped to the V. cholerae C6706 reference genome, and SNPs were detected with the CLC Genomic workbench (QIAGEN). Raw data files were deposited to the SRA database in NCBI (accession number SRP169512) (47).
HPLC.
Twenty micrograms total RNA or 35–50 pmol of isolated tRNA was digested with 1.5 μg Nuclease P1 (Sigma Aldrich) and 0.08 U Alkaline phosphatase (Clontech) in 25 mM NH4OAc, pH 7.2, at 37 °C for 1 h. Nucleoside digests of 8 μg total RNA, 10 pmol isolated tRNAs, or s4U standard (MP Biomedical) were subjected to a Ultimate 3000 HPLC system (Dionex) equipped with a reverse-phase HPLC column (Acclaim PolarAdvantageII Dionex). The flow rate was 500 μL/min, and the gradient was made from A buffer (5 mM NH4OAc, pH 5.0) and B buffer (80% Acetonitrile) with the following program: 0% B buffer, 0 min; 35% B buffer, 35 min; 100% B buffer, 40 min; 100% B buffer, 50 min; 0% B buffer, 51 min; and 0% B buffer, 60 min. UV A254 and A334 was monitored for general nucleosides and s4U, respectively. The amount of s4U was calculated from the peak area at 334 nm based on a standard curve made with commercially obtained s4U.
Immunoprecipitation of RNase E.
Immunoprecipitation is done as described in ref. 48. V. cholerae Δrne::rne-flag strains (± pnp mutation) were cultured overnight at 37 °C, diluted by 1,000-fold in 400 mL LB medium, and cultured at 37 °C with vigorous shaking at 200 rpm (Innova 40; New Brunswick Scientific). Cells were harvested at early stationary phase (1 h after OD600 = 0.5) and stored at −80 °C. Cells were resuspended in 10 mL lysis buffer (50 mM Na-Hepes, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 0.5% Triton X-100, 12% Glycerol, 0.5 mM DTT, 1 mM PMSF, 0.2 U/mL DNase I, complete proteinase inhibitor mixture; Roche) and disrupted with EmulsiFlex for 15 min. Five milliliters of cleared supernatant was mixed with 150 μL anti-FLAG M2 affinity gel (Sigma Aldrich) and incubated with gentle rotation at 4 °C overnight. After removal of supernatant, beads were washed twice with wash 1 buffer (50 mM Na-Hepes, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 6% Glycerol, 0.5 mM DTT, 1 mM PMSF) and once with wash 2 buffer (50 mM Na-Hepes, pH 7.5, 150 mM NaCl, 1 mM EDTA, 6% Glycerol, 1 mM PMSF) followed by elution with 200 μL flag elution buffer (25 mM Na-Hepes, pH 7.5, 100 mM NaCl, 0.2 mg/mL 3× FLAG peptide; Sigma Aldrich). Eluent was analyzed by gel electrophoresis, and band identity was confirmed through peptide mass spectrometry.
In Vitro tRNA Degradation Assay.
The amount of degradosomes was normalized with the intensity of the bands of RNase E in the gel stained with Coomassie brilliant blue. Roughly four times higher intensity was observed in degradosomes purified from pnp mutant. Two picomoles tRNA and degradosomes (2.5 μL WT or 0.6 μL pnp) were separately incubated at 37 °C for 15 min in 12.5-μL aliquots containing 20 mM Tris⋅HCl, pH 7.1, 1.5 mM DTT, 5 mM MgCl2, 20 mM NaCl, 10 mM NaHPO4 pH 7.1, and 0.2 U/μL SuperaseIn (Thermo Fisher). The aliquots were mixed to start the assay; then, 4-μL samples were removed at each time point, which were mixed with equal volume of loading solution (9 M Urea, 0.05% Bromophenolblue, 0.05% Xylene cyanol, 2% SDS). tRNAs were resolved by electrophoresis on 10% UREA-TBE PAGE gels and stained with the nucleic acid stain SYBR Gold (Life Technology). Gels were visualized, and the intensity of intact tRNA bands was quantified using FLA-5000.
Melting Curve Measurement.
tRNAs were denatured at 80 °C for 1 min and reannealed by cooling by 1 °C/min in 50 mM Tris⋅HCl, pH 7.6, 100 mM NaCl, and 5 mM MgCl2. Melting curves were generated with a J-815 CD spectropolarimeter with a Peltier temperature controller (JASCO) using 1-mm cuvettes. UV A260 was tracked as temperature was increased from 20 °C to 95 °C by 1 °C/min. The melting temperature of tRNA-Tyr and tRNA-Ser1 from the WT and ΔthiI was determined with the first derivative curve.
Supplementary Material
Acknowledgments
We thank Dr. Brigid Davis for insightful comments on the manuscript and members of the laboratory of M.K.W. for useful discussions. Melting curves was carried out in the Center for Macromolecular Interactions in the Department of Biological Chemistry and Molecular Pharmacology (BCMP) at Harvard Medical School. S.K. was supported by Japan Society for the Promotion of Science Grant 13J09842, and work in the laboratory of M.K.W. is supported by the HHMI and NIH Grant R01-AI-042347.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession nos. SRP169512 and SRP169513).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814130116/-/DCSupplemental.
References
- 1.Zhang J, Ferré-D’Amaré AR. The tRNA elbow in structure, recognition and evolution. Life (Basel) 2016;6:E3. doi: 10.3390/life6010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmeing TM, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 4.Quigley GJ, Rich A. Structural domains of transfer RNA molecules. Science. 1976;194:796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- 5.Shepherd J, Ibba M. Bacterial transfer RNAs. FEMS Microbiol Rev. 2015;39:280–300. doi: 10.1093/femsre/fuv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Machnicka MA, et al. MODOMICS: A database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantara WA, et al. The RNA modification database, RNAMDB: 2011 Update. Nucleic Acids Res. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenz C, Lünse CE, Mörl M. tRNA modifications: Impact on structure and thermal adaptation. Biomolecules. 2017;7:E35. doi: 10.3390/biom7020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Björk GR, Hagervall TG. Transfer RNA modification: Presence, synthesis, and function. Ecosal Plus. 2014;6 doi: 10.1128/ecosalplus.ESP-0007-2013. [DOI] [PubMed] [Google Scholar]
- 10.Sakai Y, Miyauchi K, Kimura S, Suzuki T. Biogenesis and growth phase-dependent alteration of 5-methoxycarbonylmethoxyuridine in tRNA anticodons. Nucleic Acids Res. 2016;44:509–523. doi: 10.1093/nar/gkv1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emilsson V, Näslund AK, Kurland CG. Thiolation of transfer RNA in Escherichia coli varies with growth rate. Nucleic Acids Res. 1992;20:4499–4505. doi: 10.1093/nar/20.17.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vecerek B, Moll I, Bläsi U. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. EMBO J. 2007;26:965–975. doi: 10.1038/sj.emboj.7601553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chionh YH, et al. tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat Commun. 2016;7:13302. doi: 10.1038/ncomms13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buck M, Ames BN. A modified nucleotide in tRNA as a possible regulator of aerobiosis: Synthesis of cis-2-methyl-thioribosylzeatin in the tRNA of Salmonella. Cell. 1984;36:523–531. doi: 10.1016/0092-8674(84)90245-9. [DOI] [PubMed] [Google Scholar]
- 15.Engelke DR, Hopper AK. Modified view of tRNA: Stability amid sequence diversity. Mol Cell. 2006;21:144–145. doi: 10.1016/j.molcel.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Alexandrov A, et al. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 17.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev. 2008;22:1369–1380. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whipple JM, Lane EA, Chernyakov I, D’Silva S, Phizicky EM. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev. 2011;25:1173–1184. doi: 10.1101/gad.2050711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guy MP, et al. Identification of the determinants of tRNA function and susceptibility to rapid tRNA decay by high-throughput in vivo analysis. Genes Dev. 2014;28:1721–1732. doi: 10.1101/gad.245936.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Reimers S, Pandit S, Deutscher MP. RNA quality control: Degradation of defective transfer RNA. EMBO J. 2002;21:1132–1138. doi: 10.1093/emboj/21.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomikawa C, Yokogawa T, Kanai T, Hori H. N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability at high temperatures through a tRNA modification network. Nucleic Acids Res. 2010;38:942–957. doi: 10.1093/nar/gkp1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svenningsen SL, Kongstad M, Stenum TS, Muñoz-Gómez AJ, Sørensen MA. Transfer RNA is highly unstable during early amino acid starvation in Escherichia coli. Nucleic Acids Res. 2017;45:793–804. doi: 10.1093/nar/gkw1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong J, et al. Transfer RNAs mediate the rapid adaptation of Escherichia coli to oxidative stress. PLoS Genet. 2015;11:e1005302. doi: 10.1371/journal.pgen.1005302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pritchard JR, et al. ARTIST: High-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLoS Genet. 2014;10:e1004782. doi: 10.1371/journal.pgen.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neumann P, et al. Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification. Nucleic Acids Res. 2014;42:6673–6685. doi: 10.1093/nar/gku249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar RK, Davis DR. Synthesis and studies on the effect of 2-thiouridine and 4-thiouridine on sugar conformation and RNA duplex stability. Nucleic Acids Res. 1997;25:1272–1280. doi: 10.1093/nar/25.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, et al. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mueller EG, Buck CJ, Palenchar PM, Barnhart LE, Paulson JL. Identification of a gene involved in the generation of 4-thiouridine in tRNA. Nucleic Acids Res. 1998;26:2606–2610. doi: 10.1093/nar/26.11.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jühling F, et al. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffey RH, et al. 15N-labeled tRNA. Identification of 4-thiouridine in Escherichia coli tRNASer1 and tRNATyr2 by 1H-15N two-dimensional NMR spectroscopy. J Biol Chem. 1986;261:12074–12078. [PubMed] [Google Scholar]
- 31.Nomura Y, Ohno S, Nishikawa K, Yokogawa T. Correlation between the stability of tRNA tertiary structure and the catalytic efficiency of a tRNA-modifying enzyme, archaeal tRNA-guanine transglycosylase. Genes Cells. 2016;21:41–52. doi: 10.1111/gtc.12317. [DOI] [PubMed] [Google Scholar]
- 32.Chao MC, et al. A cytosine methyltransferase modulates the cell envelope stress response in the cholera pathogen [corrected] PLoS Genet. 2015;11:e1005666, and erratum (2015) 11:e1005739. doi: 10.1371/journal.pgen.1005666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gustilo EM, Vendeix FA, Agris PF. tRNA’s modifications bring order to gene expression. Curr Opin Microbiol. 2008;11:134–140. doi: 10.1016/j.mib.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aït-Bara S, Carpousis AJ. RNA degradosomes in bacteria and chloroplasts: Classification, distribution and evolution of RNase E homologs. Mol Microbiol. 2015;97:1021–1135. doi: 10.1111/mmi.13095. [DOI] [PubMed] [Google Scholar]
- 35.Jarrige A, Bréchemier-Baey D, Mathy N, Duché O, Portier C. Mutational analysis of polynucleotide phosphorylase from Escherichia coli. J Mol Biol. 2002;321:397–409. doi: 10.1016/s0022-2836(02)00645-9. [DOI] [PubMed] [Google Scholar]
- 36.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: The mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Worrall JA, Howe FS, McKay AR, Robinson CV, Luisi BF. Allosteric activation of the ATPase activity of the Escherichia coli RhlB RNA helicase. J Biol Chem. 2008;283:5567–5576. doi: 10.1074/jbc.M708620200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janiak F, et al. Fluorescence characterization of the interaction of various transfer RNA species with elongation factor Tu.GTP: Evidence for a new functional role for elongation factor Tu in protein biosynthesis. Biochemistry. 1990;29:4268–4277. doi: 10.1021/bi00470a002. [DOI] [PubMed] [Google Scholar]
- 39.Dewe JM, Whipple JM, Chernyakov I, Jaramillo LN, Phizicky EM. The yeast rapid tRNA decay pathway competes with elongation factor 1A for substrate tRNAs and acts on tRNAs lacking one or more of several modifications. RNA. 2012;18:1886–1896. doi: 10.1261/rna.033654.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turowski TW, Karkusiewicz I, Kowal J, Boguta M. Maf1-mediated repression of RNA polymerase III transcription inhibits tRNA degradation via RTD pathway. RNA. 2012;18:1823–1832. doi: 10.1261/rna.033597.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nobles KN, Yarian CS, Liu G, Guenther RH, Agris PF. Highly conserved modified nucleosides influence Mg2+-dependent tRNA folding. Nucleic Acids Res. 2002;30:4751–4760. doi: 10.1093/nar/gkf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Millet YA, et al. Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog. 2014;10:e1004405. doi: 10.1371/journal.ppat.1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiang SL, Rubin EJ. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene. 2002;296:179–185. doi: 10.1016/s0378-1119(02)00856-9. [DOI] [PubMed] [Google Scholar]
- 44.Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimura S, et al. Discovery of the β-barrel-type RNA methyltransferase responsible for N6-methylation of N6-threonylcarbamoyladenosine in tRNAs. Nucleic Acids Res. 2014;42:9350–9365. doi: 10.1093/nar/gku618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kimura S, Waldor MK. 2018 Tnseq of Vibrio cholerae. Sequence Read Archive. Available at https://www.ncbi.nlm.nih.gov/sra/?term=SRP169513. Deposited November 18, 2018.
- 47.Kimura S, Waldor MK. 2018 Whole genome sequencing of Vibrio cholerae. Sequence Read Archive. Available at https://www.ncbi.nlm.nih.gov/sra/?term=SRP169512. Deposited November 18, 2018.
- 48.Gerace E, Moazed D. Affinity pull-down of proteins using anti-FLAG M2 agarose beads. Methods Enzymol. 2015;559:99–110. doi: 10.1016/bs.mie.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yaremchuk A, Kriklivyi I, Tukalo M, Cusack S. Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition. EMBO J. 2002;21:3829–3840. doi: 10.1093/emboj/cdf373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.