Abstract
Infectious diseases claim millions of lives each year. Robust and accurate diagnostics are essential tools for identifying those who are at risk and in need of treatment in low-resource settings. Inorganic complexes and metal-based nanomaterials continue to drive the development of diagnostic platforms and strategies that enable infectious disease detection in low-resource settings. In this review, we highlight works from the past 20 years in which inorganic chemistry and nanotechnology were implemented in each of the core components that make up a diagnostic test. First, we present how inorganic biomarkers and their properties are leveraged for infectious disease detection. In the following section, we detail metal-based technologies that have been employed for sample preparation and biomarker isolation from sample matrices. We then describe how inorganic- and nanomaterial-based probes have been utilized in point-of-care diagnostics for signal generation. The following section discusses instrumentation for signal readout in resource-limited settings. Next, we highlight the detection of nucleic acids at the point of care as an emerging application of inorganic chemistry. Lastly, we consider the challenges that remain for translation of the aforementioned diagnostic platforms to low-resource settings.
1. Introduction
In 1906, William Osler, one of the founders of modern American medicine, stated, “Diagnosis, not drugging, is our chief weapon of offence.”1 Despite the medical advances of the 20th and 21st centuries, infectious diseases remain major global health issues and continue to claim millions of lives each year in low- and middle- income countries (LMICs).2−4 As the global community moves toward control and elimination of infectious disease, it has become evident that there is a pressing need for diagnostic strategies that can be applied in primary health care settings.5−7 This review shines a spotlight on how the applied uses of inorganic chemistry advance the concepts of metals-in-medicine beyond therapeutics and vaccines and into the realm of diagnostics, enabling new tools to meet these global challenges.
1.1. Use Cases
The World Health Organization depicts LMIC health care accessibility and infrastructure as a tiered pyramid structure, in which the best-equipped facilities are the least accessible and facilities with the least amount of resources are most common (Figure 1).8 In this system, a Level 1 facility is a primary care setting with little laboratory infrastructure, trained personnel, or advanced diagnostic technology. In LMICs, both urban and rural Level 1 facilities frequently lack essential resources such as consistent electricity and clean running water. District hospitals (Level 2), regional laboratories (Level 3), and national reference laboratories (Level 4) have more advanced diagnostic technology available with increasing infrastructure; however, these facilities are often inaccessible to most patients in need due to geography, cost, and lack of transportation.8
Figure 1.
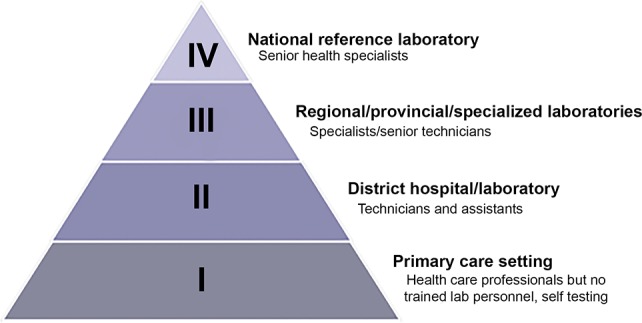
Levels of testing facilities and the types of diagnostic testing and personnel available at each respective level. Adapted with permission from ref (8). Copyright 2018 World Health Organization (https://creativecommons.org/licenses/by-nc-sa/3.0/igo).
Consider the case of early human immunodeficiency virus (HIV) diagnosis in exposed infants. Coulibaly et al. highlight the challenges faced by this population in their 2011 Burkina Faso study.9 HIV-positive mothers were advised to attend a 6-week postnatal appointment at their nearest primary healthcare facility for collection of dried blood spot samples from their exposed infants. Qualified sample collection technicians were often only available once each month, so mothers would have to return to the clinic on that day for testing. Once collected, the dried blood spot samples were sent to district-level hospitals (Level 2), which then sent the samples to a reference laboratory (Level 3). The tertiary laboratory then determined viral load by performing polymerase chain reaction (PCR) to detect HIV genetic material. Test results followed the same circuitous path back to the patients, a process that could take as long as four months. Additionally, in the case of a positive dried blood spot result, the process had to be repeated in order to validate the original results.9 In contrast, the same high-risk newborns in the United States are screened for HIV at birth with additional tests at 2–3 weeks, 1–2 months, and 4–6 months.10 For these patients, results are usually available within 1 or 2 days of sample collection, allowing immediate antiretroviral treatment for HIV-positive children.11 The consequences of diagnostic efficiency versus inefficiency could not be more striking; in the Burkina Faso study, 10% of the HIV-positive infants died before antiretroviral therapy could even be started, while the early treatment initiated in the U.S. improved infant survival by a remarkable 76%.9,12 The stark disparity in response time between these two settings shows just how devastating outcomes can be for patients relying on Level 1 facilities for their healthcare needs.
Disease diagnosis in a Level 1 setting represents a challenge that often requires simple tools that can be used without skilled personnel or significant laboratory or physical infrastructure. The World Health Organization has developed a set of criteria (“ASSURED”) that defines the ideal characteristics for point-of-care (POC) tests in low-resource settings.13 In accordance with these criteria, an ideal test should be affordable to those who are at risk of infection, result in few false-negatives (sensitive) and false-positives (specific), and be user-friendly, rapid and robust, equipment-free, and deliverable to the populations in need of the test. Additionally, the required performance parameters of a test depend on its intended use-case scenario. For individual patient case management, sensitive multiplexed tests are advantageous for determining the source(s) of nonspecific symptoms and selection of appropriate treatment. From an epidemiological standpoint, if the goal is simply to reduce the prevalence of high-intensity infections within a geographic region, the need to quantify disease burden outweighs the need for high analytical sensitivity. If the goal is disease elimination, high analytical sensitivity is one of the most important parameters, since the interruption of local transmission requires the detection of every infection, including extremely low-intensity infections.
1.2. Point-of-Care Device Design
The most sensible approach to designing and developing a POC diagnostic is to examine and optimize each individual component before integrating into a single diagnostic device. In this review, we define the components of a diagnostic to include: (1) the target biomarker, an endogenous indicator of a disease state, which is most often a pathogen or host protein, carbohydrate, or nucleic acid sequence, (2) sample preparation, which allows for biomarker isolation, purification, and/or concentration from complex biological matrices, (3) molecular recognition elements, which specifically capture and detect the target biomarker, (4) signal generation and amplification, and (5) instrumentation for signal read-out. Simple components requiring only a single user step are generally preferable in low-resource Level 1 settings. However, there are few diagnostic technologies that currently can be deployed in these settings, and the lack of appropriate diagnostics in resource-limited settings can lead to tragic outcomes. Fortunately, emerging technologies based on inorganic chemistry and nanomaterials can be exploited as potential solutions. In this review, we focus on how inorganic chemistry and nanomaterials have been utilized in each component of a POC diagnostic for infectious disease detection. Section 2 describes inorganic biomarkers that are associated with infectious diseases and how their properties have been exploited in diagnostic applications. In section 3, we discuss how coordination chemistry has been harnessed for sample preparation and protein biomarker enrichment for POC tests. Section 4 details the critical advancements in inorganic chemistry and nanomaterials for signal generation probes that have allowed the field of POC diagnostics to rapidly progress. In section 5, we provide an overview of field-deployable instrumentation that incorporates the fundamental inorganic chemistries discussed in sections 2–4 for POC diagnosis of infectious disease. Finally, section 6 discusses the emerging trend of using inorganic chemistry and nanomaterials for nucleic acid detection at the point of care.
2. Inorganic Biomarkers of Infectious Diseases
Biomarkers are quantifiable characteristics of a disease state that can be measured accurately and reproducibly. The vast majority of biomarker targets for infectious diseases fall into three main categories: (1) pathogen genetic material, (2) protein and carbohydrate antigens produced by a pathogen, or (3) human host responses to the presence of infection, such as pathogen-specific antibodies. While inorganic markers of infectious disease are far less common than their bioorganic counterparts, metal-based biomarkers have unique properties that can be leveraged for innovative diagnostic strategies, as illustrated in the following two examples.
2.1. Hemozoin
Hemozoin, often called malaria pigment, is a biomineral produced by some parasites that rely on hematophagy as their primary source of nutrients.14 Hemozoin was first described for the malaria parasite Plasmodium falciparum over a century ago and is produced by all Plasmodium species.15−17 In the erythrocytic life stages, malaria parasites digest host hemoglobin in the acidic digestive food vacuole as a source of amino acids. The breakdown of one hemoglobin molecule releases four molecules of heme (ferriprotoporphyrin IX [Fe(III)PPIX]), which are highly toxic to the parasite.17 As Plasmodium spp. lack functional heme oxygenase activity, the parasites remove the free heme by crystallizing it into insoluble, inert hemozoin crystals.17,18 These crystals accumulate in the parasite over the course of its erythrocytic life cycle. After the mature schizont ruptures, hemozoin is released into circulation and rapidly taken up by phagocytes.19 The heme detoxification pathway that produces hemozoin is crucial for malaria parasite survival; therefore, hemozoin indicates the presence of Plasmodia parasites, making it an attractive biomarker for malaria detection.19
Structurally, hemozoin crystals consist of reciprocating Fe(III)PPIX dimers linked through coordination between the central ferric ions and a carboxylate group of one of the propionate moieties on protoporphyrin IV.17,20,21 Dimers assemble into chains through hydrogen bonding of the second propionate porphyrin side chain, and these networks of cross-linked dimers are held together via π–π interactions (Figure 2), resulting in an anisotropic rectangular crystal with unique magnetic, optical, and photoacoustic properties.22
Figure 2.
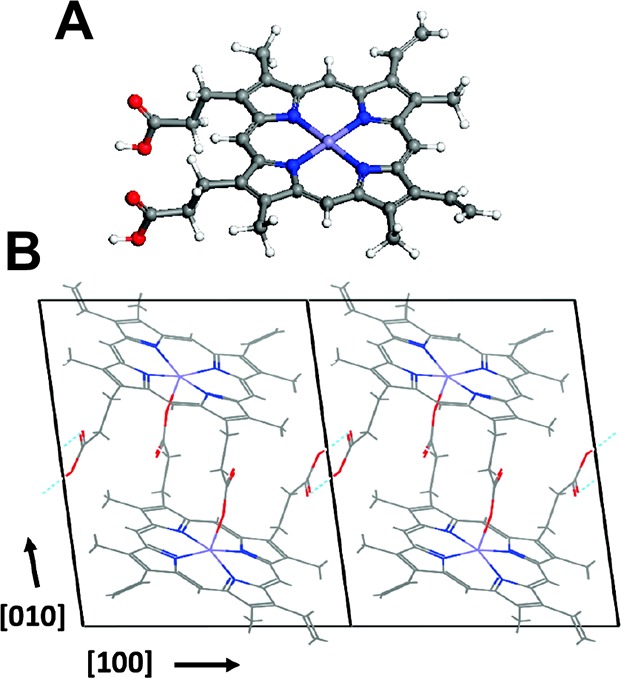
Structure of hemozoin. Adapted from ref (22). Copyright 2011 American Chemical Society.
One such property of hemozoin is its paramagnetism, which is derived from the unpaired electrons on the central Fe(III) ions.23 Like all paramagnetic substances, hemozoin has a magnetic moment and is thus attracted up a magnetic gradient in the presence of an externally applied magnetic field. The paramagnetism of hemozoin has been leveraged for malaria diagnosis in several ways. First, the magnetic properties have been exploited for separation, purification, and concentration of infected red blood cells (iRBCs) from blood samples. With the use of a high-gradient external magnetic field, iRBC separation was first performed by Heidelberger et al.24 in 1946 for the purpose of vaccine development, though at the time the method was described as “applicable only to small blood volumes, [and requiring] apparatus difficult of access.”24 Since then, the availability of high-field permanent magnets and electromagnets has substantially improved, and the time required for magnetic separation of iRBCs has been reduced. Both commercially available systems as well as custom-made microfluidic devices have been used to enrich iRBCs from whole blood samples for analysis by microscopy.25−32
Three groups have explored detection of hemozoin based on the fact that paramagnetic materials decrease nuclear magnetic resonance (NMR) transverse relaxation time (T2) of water, the same phenomenon leveraged for magnetic resonance imaging (MRI) contrast agents. Karl et al.33 first demonstrated that hemozoin could be detected based on decreased T2, though only at parasitemias greater than 10000 parasites/microliter, which is 2 orders of magnitude greater than the detection limits of commercial rapid diagnostic tests. Several years later, Peng et al.34 and Kong et al.35 developed a low-cost (<$2000), homemade micromagnetic resonance device for hemozoin-based malaria detection. They reported detection limits less than 10 parasites/microliter using their method and attributed this drastic improvement to the use of ultrashort echo times and a premeasurement centrifugation step that pelleted red blood cells from whole blood.34 These discordant conclusions led to a reaction by Karl et al.36 and a response by Peng et al.37 Recently, Gossuin et al.38 confirmed the results of Karl et al., indicating that conventional relaxometry alone is insufficient for sensitive malaria detection. These findings led to two hypotheses related to the superior sensitivity measured by Peng et al.: (1) the premeasurement centrifugation of higher-density iRBCs (compared to uninfected RBCs) significantly enriched hemozoin in the portion of the pellet measured, and (2) ultrashort echo times may have allowed for probing of other protons, such as macromolecular protons, which may be more susceptible to the presence of hemozoin.38 A simple, low-resource relaxometry-based detection device remains to be developed, and such a device would ultimately require resources typically not available in primary healthcare settings in LMICs (i.e., consistent availability of electricity and clean water). However, these studies highlight the importance and advantages of sample preparation and enrichment steps in the development of sensitive infectious disease diagnostics.
In addition to its paramagnetic properties, the unique optical properties of hemozoin enable the development of new diagnostic tools for malaria detection. In particular, hemozoin strongly scatters and absorbs light.39 The latter characteristic led to the remarkable development of the Microvascular Microscope (MvM) for needle-free, in vivo detection of hemozoin.40,41 The MvM employed cross-polarized epi-illumination to optically access vessels below the surface of the skin, with green light (λ = 532 nm) illumination for locating vessels and red light (λ = 655 nm) illumination for detecting hemozoin. This method was able to detect parasitemias as low as 0.03% in mouse models, corresponding to approximately 1000 parasites/microliter in humans. Unfortunately, lower parasite density infections were not explored.40 The MvM was able to differentiate between iRBCs and hemozoin-containing white blood cells, which can persist in circulation for weeks after an infection is cleared, based on the diameter and relative intensity of the detected hemozoin.41 Additionally, hemozoin particle velocity could be measured in vivo (Figure 3), potentially allowing for identification of cytoadhesion and sequestration, two phenomena related to disease severity.41,42 Though further sensitivity analysis and field testing is required, the MvM represents a promising, noninvasive tool for malaria detection, particularly if a portable and battery-powered device is developed.
Figure 3.
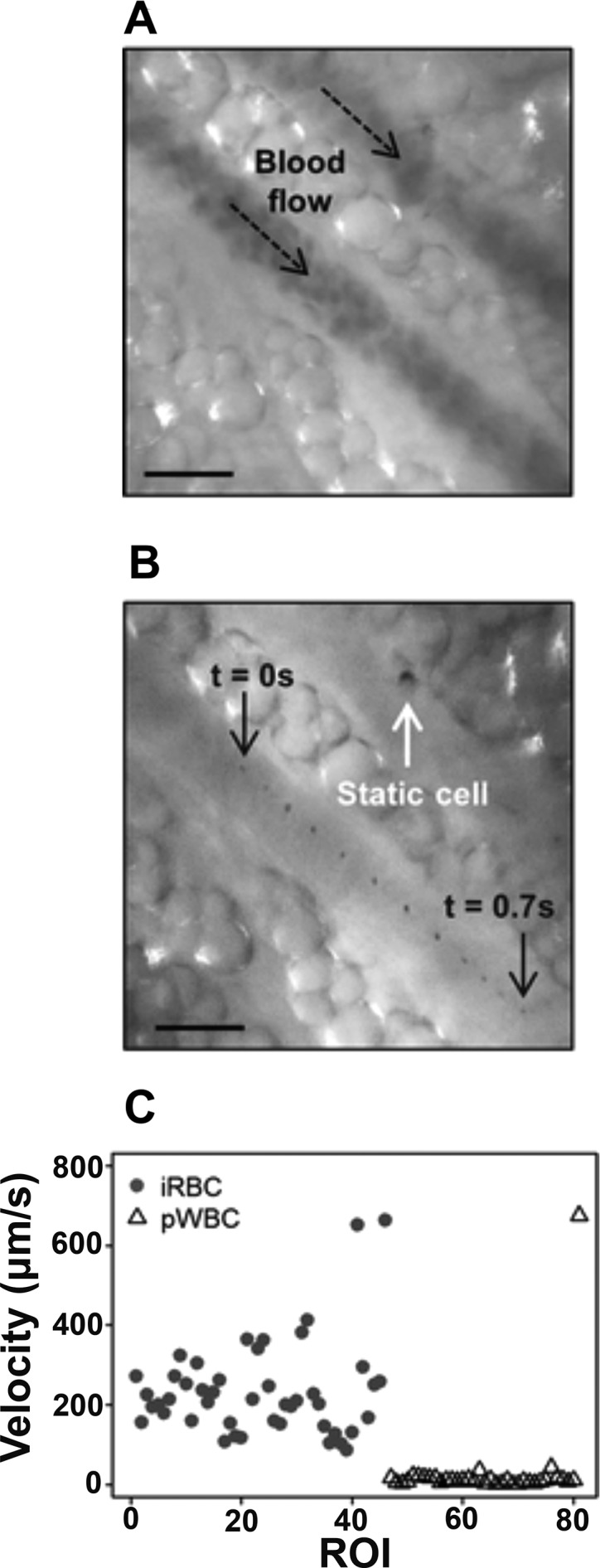
Microvascular Microscope (MvM) measures cell velocity in vivo. (A) Green light illumination detects blood flow through a vessel, and (B) successive frames of red light illumination show a single hemozoin particle moving through the vessel over time. (C) Velocities of iRBCs during acute infection are higher than those of hemozoin-containing white blood cells measured after infection. Adapted with permission from ref (41). Copyright 2017 Burnett et al.; licensee BioMed Central Ltd. (https://creativecommons.org/licenses/by/4.0/legalcode).
Photoacoustic spectroscopy has also demonstrated potential for transdermal malaria detection.43−48 Photoacoustic (PA) hemozoin detection requires excitation with intense light and measurement of the resulting acoustic signals generated by local thermal expansion of the surrounding medium. The utility of in vitro PA detection of hemozoin was first demonstrated by Balasubramanian et al. in 1984.43 Since then, it has been shown that short, intense laser excitation of hemozoin in the parasite can localize heat around the crystal, evaporating the surrounding medium and creating transient, nanosized vapor bubbles. The expansion and collapse of these nanobubbles enhance acoustic detection.44 This technique has allowed for transdermal malaria detection with exceptional sensitivity in mice. This method was also applied to one human patient with high parasite density (8900–65000 parasites/microliter).45−48 Further studies with increased patient sample size will be necessary to determine the true limits of the technology.
Magneto-optical detection exploits both the paramagnetic and optical properties of malarial hemozoin crystals, which have a rectangular shape that affords a high level of both magnetic anisotropy and optical dichroism. In this technique, whole blood (or lysed whole blood) is inserted into a magnetic field, orienting the paramagnetic crystals along the applied field direction. This phenomenon, known as the Cotton-Mouton effect, induces an optical dichroism across the dispersion of crystals, resulting in a detectable change in transmittance of modulated polarized light (λ = 660 nm) (Figure 4).49,50 When applied to patient whole blood samples, the diagnostic sensitivity and specificity of magneto-optical detection were not competitive with rapid diagnostic tests and PCR, though the ease of use is encouraging for POC applications.49,50 In particular, model simulations have suggested that magneto-optic detection of hemozoin could be exploited for the development of a noninvasive probe capable of measuring transmittance of polarized light through a fingertip.51 A similar magneto-optic strategy that employs a fluctuating magnetic field and measures the change in amplitude of transmittance has shown potential in mouse models, but further evaluation of the diagnostic sensitivity and specificity in human samples is needed.52−55 Recently, a commercial magneto-optic device for malaria detection based on hemozoin has demonstrated promise in field evaluations, detecting clinical parasite densities as low as 5 parasites/microliter (Hemex Health).56
Figure 4.
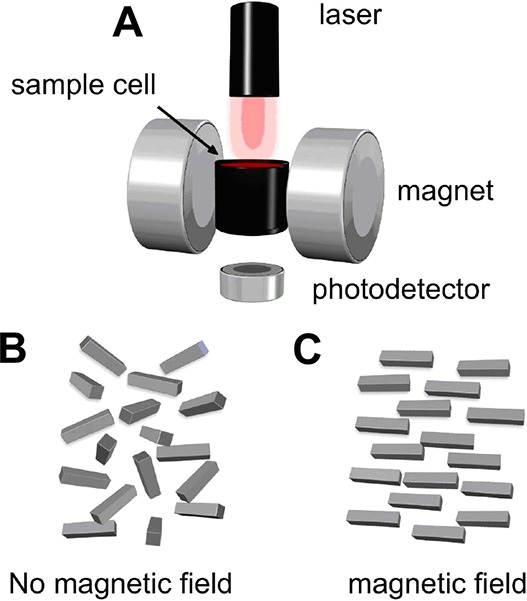
Schematic of magneto-optic device for hemozoin-based diagnosis of malaria. (A) Polarized laser light passes through a sample, and the intensity of the transmitted light is measured over time in the absence and presence of a magnetic field. (B) In the absence of an applied field, hemozoin crystals are randomly aligned throughout the dispersion. (C) Upon application of an external magnetic field, hemozoin aligns along the direction of the field, resulting in a measurable change in transmittance. Adapted with permission from ref (50). Copyright 2010 BioMed Central Ltd. (https://creativecommons.org/licenses/by/2.0/legalcode).
Additional techniques used to detect malarial hemozoin include flow cytometry,57−60 laser-desorption mass spectrometry,61−64 and Raman spectroscopy.65−69 While these strategies are highly sensitive, they require expensive and resource-intensive instrumentation that typically prevent their use in a district hospital setting, local health outpost, or field setting. However, as instruments become more compact, robust, and affordable, similar to some of the technologies detailed in this section, their implementation in field settings could become more feasible.
Importantly, hemozoin is synthesized by other hematophagous parasites that infect humans, including the Schistosoma, Echinostoma, and Opisthorchis trematodes, as well as blood feeding insects, such as Rhodnius and Boophilus.17,70,71 Despite being produced by these additional parasites, hemozoin has not yet been used as a diagnostic biomarker for nonmalarial pathogens. Further, it is important to note that in all of the aforementioned hemozoin-based malarial detection strategies, hemozoin originating from trematodes would produce a positive signal. This is particularly important to consider regarding false positive tests since the geographic distribution of malaria overlaps with Schistosoma.72,73
In addition to confounding signal originating from other hematophagous parasites, hemozoin as a biomarker presents some unique challenges to malaria diagnosis. First, the biocrystal is present at much lower concentrations in the ring stage when compared to the later trophozoite and schizont stages of the erythrocytic cycle.32 Thus, magnetic enrichment or malaria diagnosis strategies based on hemozoin could miss early stages of infection, potentially resulting in false-negatives. Additionally, phagocytic cells ingest iRBCs and hemozoin released into circulation after schizont rupture, and hemozoin-containing white blood cells have been shown to persist in circulation up to 17 days after parasite clearance.74 This persistence can lead to false-positive results in hemozoin-based malarial diagnostics unless an algorithm for distinguishing pigment-containing white blood cells and iRBCs is devised, such as the one developed in the aforementioned work by Burnett et al.41 Despite these challenges, researchers continue to develop innovative techniques for malaria detection using hemozoin as a biomarker. Implementation of these methods will require careful design with cost and equipment reduction in mind in addition to rigorous field evaluation.
2.2. Iron in Schistosome Eggshells
Schistosome eggs are the most common biomarker for schistosomiasis detection. They are produced by adult Schistosoma worm pairs that reside in the intestine and bladder and deposited into the vessels of the gut wall.75 Many eggs become permanently lodged in the intestine, liver, bladder, or urogenital system, leading to chronic inflammation and systemic tissue damage;76 however, some mature eggs are excreted in urine or feces to allow the parasites to continue the transmission cycle.75 Eggs from distinct species are distinguishable by their morphologies. Differences include the overall egg shape (round to oval), spine position, and extent of filamentous coating.77 All of these features are observable by microscopy, which is the gold standard for schistosomiasis diagnosis.
Voided eggs from all Schistosoma species contain mature miracidia within a relatively impermeable eggshell.75 While the dense biopolymer composition of Schistosoma eggshells has been studied since the 1960s,78 the inorganic components of the eggshells were not investigated until 2007, when Jones et al.79 demonstrated the presence of iron in Schistosoma japonicum eggshells (Figure 5A). The group used inductively coupled plasma mass spectrometry (ICP-MS) and energy dispersive spectroscopy (EDS) to provide evidence that iron localized to the eggshell. Although these elemental characterization techniques did not provide specific information about the oxidation state or organization of iron present, the group found that the eggshells contained 121 mg/kg (dry weight) Fe, making up nearly 89% of the iron found in intact eggs.79
Figure 5.
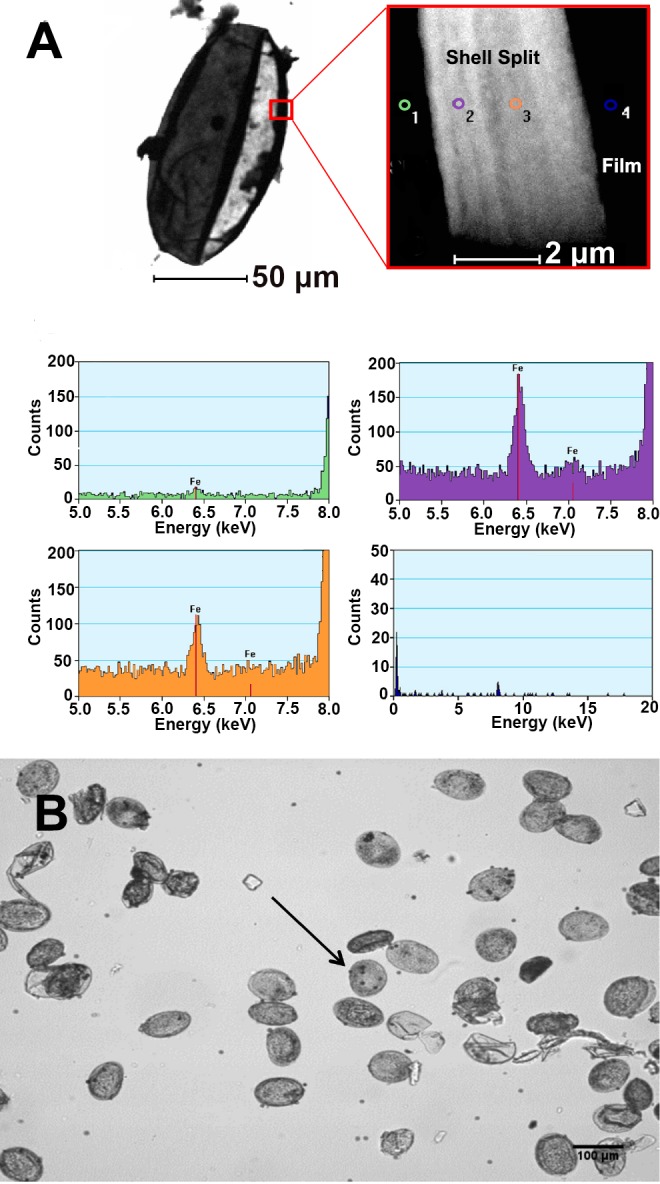
(A) EDS microscopy demonstrates that iron localizes to the schistosome eggshell. Adapted with permission from ref (79). Copyright 2007 Elsevier Ltd. (https://creativecommons.org/licenses/by/4.0/legalcode) (B) Magnetite-coated microparticles bind to S. japonicum eggs after 30 min of incubation. The arrow highlights egg-bound microspheres. Reprinted with permission from ref (84). Copyright 2015 Elsevier.
In the same year as the Jones study, Teixeira et al.80 set out to increase the sensitivity of microscopy for Schistosoma detection using magnetic microparticles to enrich eggs from fecal samples. To accomplish this, stool samples were suspended in water and repeatedly sieved and washed before the sediment was incubated with magnetic particles that bound to the Schistosoma eggs. This technique for egg concentration was dubbed the Helmintex method80 and was demonstrated to be more sensitive than Kato-Katz, the current WHO recommended sample preparation method for schistosome diagnosis in a field setting.81 The initial hypotheses of the Helmintex method were that biotinylated lectins would bind to carbohydrates on the surface of Schistosoma mansoni eggs and that streptavidin-coated magnetic beads could bind the biotin-lectin-egg complexes. Using an external magnet, the eggs could then be isolated and concentrated before detection by microscopy. However, control experiments demonstrated the following: (1) lectins were not necessary for egg binding to magnetic particles, (2) nonmagnetic latex particles did not bind to S. mansoni eggs, and (3) the eggs alone did not migrate to an external magnet (Figure 5B).80 These observations, coupled with the discovery of iron localized to eggshells, led some to hypothesize that, in the presence of an external magnetic field, the paramagnetic beads act as weak bar magnets, enabling attraction of the iron in the shells of the eggs.82
In 2013, Karl et al.83 further probed the iron distribution and magnetic properties of schistosome eggshells to elucidate the mechanism of the Helmintex method. The study found that iron was localized to pores in eggshells, and schistosome eggs demonstrated moderate paramagnetic behavior. However, the eggs did not move in a magnetic field, and the paramagnetic particles bound to parasite eggs in the absence of a magnetic field, suggesting that the mechanism of binding was not magnetic in origin.83 The authors also found that S. japonicum eggs bound more microparticles than S. mansoni, leading to the hypothesis that the filamentous outer structure of the eggs, more prominent in S. japonicum compared to S. mansoni eggs, provided strong, nonmagnetic adhesion based on many relatively weak van der Waals interactions.83 In a separate study, Candido et al.84 also observed that S. japonicum eggs bound more microparticles than S. mansoni. The group used Poisson analysis and found two distinct populations within each species: a high “affinity” group and a low “affinity” group, where affinity was determined by the number of microparticles bound to the visible edge of a single egg. Interestingly, the magnitudes of the high affinity values were similar between the two species, but the fraction of eggs in the high affinity group for S. japonicum was greater than for S. mansoni.84
Collectively, these results suggest that differences in filamentous microspines between the two species do not fully explain the mechanism of egg-microparticle binding. Additionally, the work by Candido et al. opens up many questions, the answers to which could greatly inform further optimization of the Helmintex method. For instance, what are the physiochemical differences between high and low affinity egg groups? Is there a biological difference between the two groups, such as egg maturity or surface antigen expression? Both the Karl et al. and Candido et al. studies were performed on eggs isolated from livers of infected mice, but do these two egg populations appear in the urine or feces of infected humans? If so, should two types of magnetic microspheres be used to ensure the Helmintex method captures all egg types present in a sample?
These questions should be answered carefully and systematically. Given the greater number of microspheres bound to eggs in the high affinity group, the populations should be separable using external magnetic fields of differing strengths. Egg-microparticle binding mechanisms for each population could be interrogated by determining solution properties (e.g., pH, salt, surfactant, chelators, blocking proteins, etc.) required to interrupt the interactions. Additionally, functionalizing microspheres in a way that systematically varies their zeta potential and hydrophobicity could determine the role these physiochemical properties play in binding. These studies will be crucial if the Helmintex method aims to determine infection intensity using quantitative egg enrichment. This is particularly important for schistosomiasis surveillance and morbidity control campaigns, which aim to quantify and reduce the prevalence of high-intensity infections.
The Helmintex method originated with the idea that iron in the eggshells of schistosomes could serve as a handle for magnetically enriching Schistosoma eggs for more sensitive detection in the field setting. While the Helmintex method has already been shown to improve the sensitivity of Kato-Katz, elucidation and optimization of the true binding mechanisms could lead to an even more impactful method for diagnosing low-intensity schistosomiasis infections.
3. Metal-Based Sample Preparation
Metal-based sample preparation techniques that enrich biomarker concentration improve the sensitivity of diagnostics by delivering more biomarker to the test. These strategies can be applied to existing ASSURED diagnostic tests, such as lateral flow assays (LFAs). While the easiest way to deliver more biomarker to the tests would be to add larger sample volumes (100–500 μL), many LFAs cannot accommodate large sample volumes for several reasons:85 first, limited bed volumes in porous paper substrates physically limit the amount of liquid a test can hold. Second, the increase in the number of interfering molecules that results from an increase in sample volume could cause nonspecific cross reaction with the test’s molecular recognition elements (e.g., antibodies). Third, colored biological matrices, such as whole blood, can increase the background signal and decrease the user’s ability to distinguish between positive and negative results. Metal-based sample preparation techniques mitigate these shortcomings, allowing for the concentration of a target from a large volume sample into a smaller, test-compatible volume. Moreover, metal-based sample preparation also removes the target from its original complex biological matrix, decreasing the potential for nonspecific cross-reactivity.85 These low-cost, simple, and robust methods represent a powerful tool for use-case scenarios in which improved sensitivity is needed, such as elimination campaigns. This section will discuss how metal coordination chemistry has been leveraged for simple sample preparation methods applicable to POC diagnostics.
3.1. Metal-Affinity Separation
Coordination chemistry, which is defined as the chemistry of metal atoms or ions that form complexes with one or more ligands,86 can be utilized to coordinate biomarkers with a particular affinity for metal ions. The most significant illustration of this capacity is the isolation of proteins via immobilized metal affinity chromatography (IMAC).87,88 This technique involves the conjugation of various ligands to a solid support matrix such as agarose, cellulose, or silica. These ligands act as Lewis bases, chelating metal ions with a high affinity. The fixed metal ions serve as Lewis acids, coordinating to both the immobilized ligands and specific amino acid residues in a protein biomarker.89 While amino acid affinities toward metal ions vary, histidine demonstrates a particularly high affinity toward metal ions in this format. These interactions can be leveraged to isolate a biomarker from a complex biological matrix. Once separated, the target can be competitively eluted and concentrated into a smaller volume suitable for a POC diagnostic test.87,88
The choice of chelating ligand is critical to IMAC protein separation efficiency. Ligands both fix the metal ion to the solid support and modulate metal-protein binding based on the number and conformation of chelation sites.88,90−92 A multitude of ligands with varying coordination numbers have been employed. Most IMAC platforms contain tridentate or tetradentate chelators, although some bidentate and pentadentate options exist (Figure 6). Tetradentate nitrilotriacetic acid (NTA)93 and tridentate iminodiacetic acid (IDA)94 are two of the most common commercial ligands, but many companies have also developed their own proprietary structures such as TALON95 or Ni-Penta.96 The variety of chelating ligands, many of which possess different coordination numbers, make IMAC tunable, which is beneficial when tailoring the chemistry to different diagnostic applications.87,88,97−100
Figure 6.
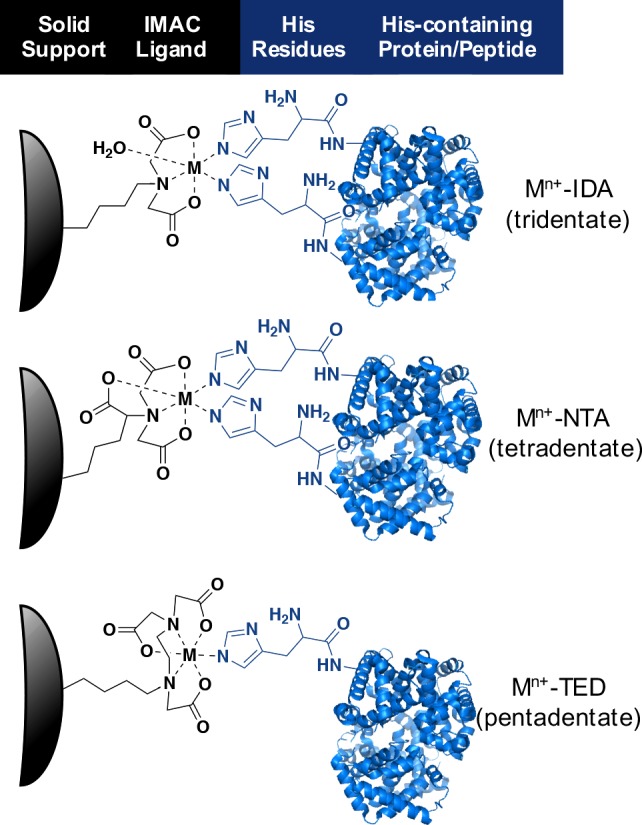
Coordination of IMAC ligands (IDA, NTA, and TED) to a divalent metal for binding histidine residues in proteins. Co2+, Ni2+, Cu2+, and Zn2+ have predominantly been utilized in this application. IDA (iminodiacetic acid) is a tridentate ligand (coordination number = 3), with the remaining three coordination sites in the metal available to bind two histidine residues and a water molecule. NTA (nitrilotriacetic acid) is a tetradentate ligand (coordination number = 4) with two coordination sites for histidine residue binding. TED (tris(carboxymethyl)ethylene diamine) is a pentadentate ligand (coordination number = 5) with one remaining coordination site for protein binding.
Guided by Pearson’s hard–soft acid–base theory, numerous metal ion and ligand combinations have been exploited in IMAC. Borderline acids such as Co2+, Ni2+, Cu2+, and Zn2+ have predominated,86−88 displaying varying levels of affinity and specificity. The determination of which ion will be optimal in a given isolation often depends on the protein in question. Ni(II)-NTA has been one of the most widely utilized metal–ligand combinations in IMAC applications. Until recently, a thorough investigation that compared divalent metals and their molecular recognition capability was lacking.
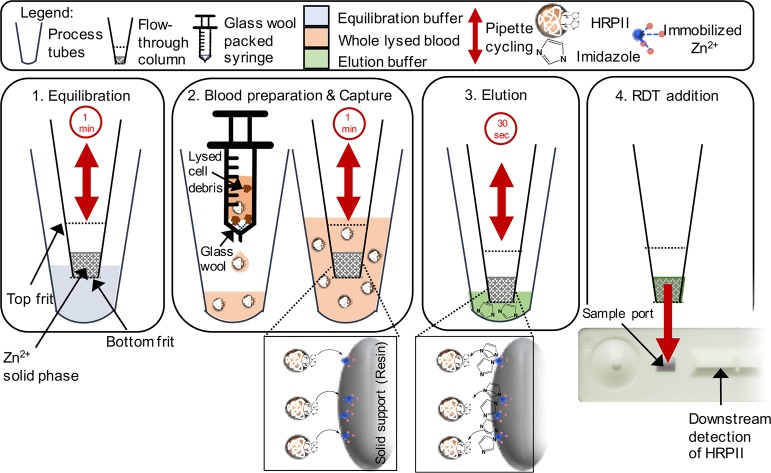
Bauer et al.99 filled this void by conducting a systematic investigation of the affinity of malarial biomarker histidine-rich protein 2 (HRP2) to M2+-functionalized biosensors and solid phase resins (M2+ = Co2+, Ni2+, Cu2+, or Zn2+). HRP2 is the most frequently detected target in malarial rapid diagnostic tests and possesses a unique primary structure containing 35% histidine, making it an ideal histidine-rich target.101 The authors used biolayer interferometry (BLI), an optical biosensor technique, to determine kinetic constants (e.g., kon, KD) for the binding of the various metals to HRP2. The authors found that Co2+ and Zn2+ exhibited the fastest binding kinetics (highest kon) and the highest HRP2 binding affinity (lowest KD) on BLI biosensors.99 When the same IMAC complexes were incorporated into a loose solid phase resin, Zn(II)-NTA resulted in the fastest complete binding of HRP2 per mole of divalent metal while also allowing for the most efficient elution of HRP2 upon the addition of imidazole. Once a metal had been selected, HRP2’s binding and elution behavior was evaluated against a variety of ligands and solid supports. The selected Zn(II)-IDA resin was then integrated into a pipet tip format, enabling direct HRP2 capture from spiked lysed blood samples. This field-deployable sample isolation and concentration tool displayed up to a 4-fold signal enhancement in commercial rapid diagnostic tests (Figure 7). This study99 demonstrated that optimizing the IMAC metal-ligand combination for a particular application is critical for efficient biomarker capture and elution; the initial binding cannot be too tight so as to prevent biomarker elution with a competing ligand (e.g., imidazole) nor too weak so as to attain suboptimal biomarker enrichment.88,99
Figure 7.
Flow-through device for capture of malarial biomarker HRP2 via Zn(II)-IDA chelation to histidine residues. The device incorporated a silica resin solid phase functionalized with Zn(II)-IDA into a pipet tip, which enabled rapid capture of HRP2 from whole blood samples using the principles of IMAC. Subsequent addition of imidazole at a high concentration competitively eluted the HRP2 from the Zn(II)-IDA resin into a smaller volume. This small volume sample was then deposited onto a commercially available rapid diagnostic test which enhanced sensitivity for HRP2 detection in a POC setting. Adapted with permission from ref (99). Copyright 2017 American Institute of Physics.
IMAC has been adapted to a variety of solid support platforms, from agarose gels to rigid silica particles.102,103 Although many of these platforms have become well-established research tools, the emergence of novel materials104−108 has enabled IMAC chemistry in new applications. For example, Wang et al.109 polymerized polyhedral oligomeric silsesquioxanes (POSSs) into micron-sized particles with nanosized pores, which were subsequently functionalized with IDA and charged with Cu2+. This material was then used for specific capture of hemoglobin, which is rich in surface-exposed histidine residues. Incubation of the test solution with the Cu(II)-IDA functionalized material enabled selective capture of hemoglobin versus other proteins without these accessible histidine residues.109 The development of this novel material using the principles of IMAC demonstrates that nanomaterials can be modified for molecular recognition of histidine-rich proteins. These modified nanomaterials have the potential to be incorporated into POC diagnostic frameworks for biomarker capture.
IMAC-functionalized cellulose membranes could provide another excellent option for sample preparation in low-resource settings. Given that these membranes already hold an established place as a laboratory research tool, they could readily be incorporated into field-friendly paper fluidic devices. Opitz et al.110 utilized commercial IDA-functionalized cellulose charged with Zn2+ as a platform for separating influenza virus particles from culture lysate. After an initial metal screening, they saw that Zn2+-charged membranes separated particles with significantly higher affinity the other metals tested. The authors postulated that the influenza hemagglutinin glycoprotein on the particular strain studied (influenza virus A/Puerto Rico/8/34) contained histidine-rich Zn2+-coordinating sequences on each subunit and that the repeated configuration of these subunits as part of the capsid structure contributed to a higher overall binding avidity.110 Although this IMAC-based method for separating virus particles from cell culture was developed to improve the efficiency of laboratory-based research, it is not difficult to imagine extending this concept to a simple sample preparation method for increasing the sensitivity of an influenza rapid test. Applying the developed protocol to a diagnostic test would require feasibility testing on clinical patient samples, ideally, resulting in the design of an integrated device for both sample preparation and virus detection. IMAC-functionalized cellulose membranes present a unique advantage in relation to other materials because of the ease in which they can be incorporated into paper-based diagnostic formats that are usable in low-resource POC settings.
3.2. IMAC on Magnetic Particles
Magnetic particles hold a number of advantages over other solid IMAC supports. The increased surface area of the particles provides increased availability for chemical reactivity and functionalization.111,112 The independent response of paramagnetic particles to an externally applied magnetic field enables active mixing, which has proven especially useful in microfluidic applications. This magnetic susceptibility also allows for easy manipulation of the particles once a targeted biomarker is bound, permitting biomarker isolation and concentration before downstream detection.111,112 Multistep protocols that would otherwise rely heavily on laboratory techniques such as filtration, centrifugation, and column chromatography can easily be performed using magnetic beads and a hand-held external magnet. Aside from chromatographic capabilities, magnetic particles have been employed as signal generation labels that are detected by inductive or magnetoresistive sensors. This application is discussed in detail in sections 4.2.6 and 5.4. Magnetic particles thus represent a promising vehicle for IMAC chemistries in low-resource settings.
The Wright research group recently demonstrated that IMAC-functionalized magnetic beads have the potential to improve commercial POC diagnostic sensitivity for malaria.113 HRP2 was captured from large-volume (100 μL whole blood samples diluted and lysed to 200 μL) P. falciparum-spiked samples via chelation to Ni(II)-NTA on the surface of commercially available magnetic particles. Then, using a specially designed extraction cassette and a hand-held magnet, the HRP2-bound particles were guided through stationary liquid chambers separated by surface-tension valves to wash the sample and remove background. Once purified, the magnetic beads with bound HRP2 were transferred to a final 10 μL elution chamber preloaded with imidazole to release the biomarker. This resulted in a 10-fold theoretical concentration factor for HRP2 due to the transfer from a 100 μL blood sample to a 10 μL elution chamber. After extraction using this self-contained system, the authors added the 10 μL elution volume to a variety of commercial HRP2-based malaria rapid diagnostic tests and found significant signal enhancement across all brands.113,114
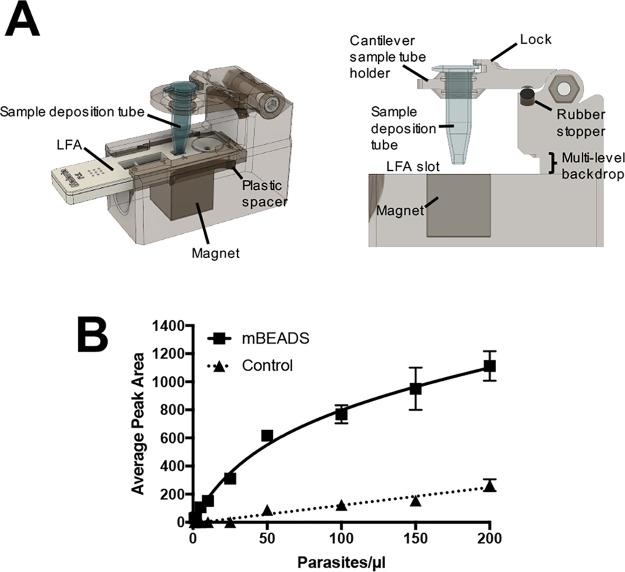
In subsequent studies, the group developed and optimized a hand-held, easy-to-use device85,115 (Figure 8A) in which HRP2-bound, IMAC-functionalized magnetic beads were directly transferred to the sample pad of commercial malaria lateral flow assays. The biomarker was eluted with an imidazole-spiked running buffer. As shown in Figure 8B, this device enabled highly sensitive HRP2 detection with few added user steps, improving the detection limits of commercial malaria tests by over an order of magnitude with minimal added cost per test.85,115 This enhanced sensitivity could make a substantial difference in malaria elimination campaigns, which rely on detecting and treating all infected individuals, including low-density carriers.
Figure 8.
(A) Simple-to-use device for direct transfer of HRP2-bound IMAC-functionalized magnetic beads to a malaria LFA. (B) Improved detection of HRP2 on a commercial LFA using the hand-held device. Adapted with permission from ref (115). Copyright 2017 Royal Society of Chemistry.
One of the primary limitations of IMAC chemistry is its ability to recognize only those proteins with affinities for metal ions. For purification of recombinant proteins from cell culture, this drawback has been overcome via incorporation of short hexahistidine tags (His6-tags) into expressed protein sequences.116,117 Using a similar principle, Bauer et al.117 developed a platform in which target-specific antibodies modified with His6-tags were combined with IMAC magnetic beads to isolate and concentrate biomarkers from biological samples. Thus, the advantages of IMAC for biomarker enrichment were no longer limited to histidine-rich proteins but rather could be extended to any protein biomarker target.
In the context of malaria diagnosis, this expanded capability allows for the detection of the nonhistidine-rich biomarker Plasmodium lactate dehydrogenase (pLDH) alongside HRP2, which is beneficial for several reasons. Plasmodium LDH is essential to parasite survival and is conserved across all species of malaria known to infect humans, whereas HRP2 is produced only by P. falciparum.118 In addition, HRP2-only tests fail to account for parasitic infections containing pfhrp2 gene deletions, which are increasing in prevalence worldwide.72 Finally, HRP2 has been shown to persist in host circulation up to 52 days after successful treatment, but pLDH clears within 24 h after parasite clearance, so a dual assay can distinguish between resolved and active P. falciparum infections.119−122
For these reasons, Bauer et al.117 employed their IMAC-based His6-tagged antibody approach to concentrate both pLDH and HRP2 from large volume samples (200 μL parasite-spiked whole blood) utilizing commercially available IMAC magnetic beads.117 In this dual capture system, HRP2 coordinated directly to the Co(II)-NTA-functionalized surface of the magnetic bead, while pLDH was bound using His6-tagged anti-pLDH antibodies (Figure 9). Following isolation of both biomarkers, HRP2 and the His6-tagged-antibody/pLDH complex were simultaneously eluted with imidazole. When applied to whole blood samples spiked with the P. falciparum parasite, this strategy resulted in a nearly 20-fold improvement in the limit of detection of commercial LFAs.117 This unique platform demonstrates that IMAC-based biomarker enrichment is not limited to histidine-rich proteins and could be expanded to virtually any biomarker for which there is a commercially available antibody.
Figure 9.
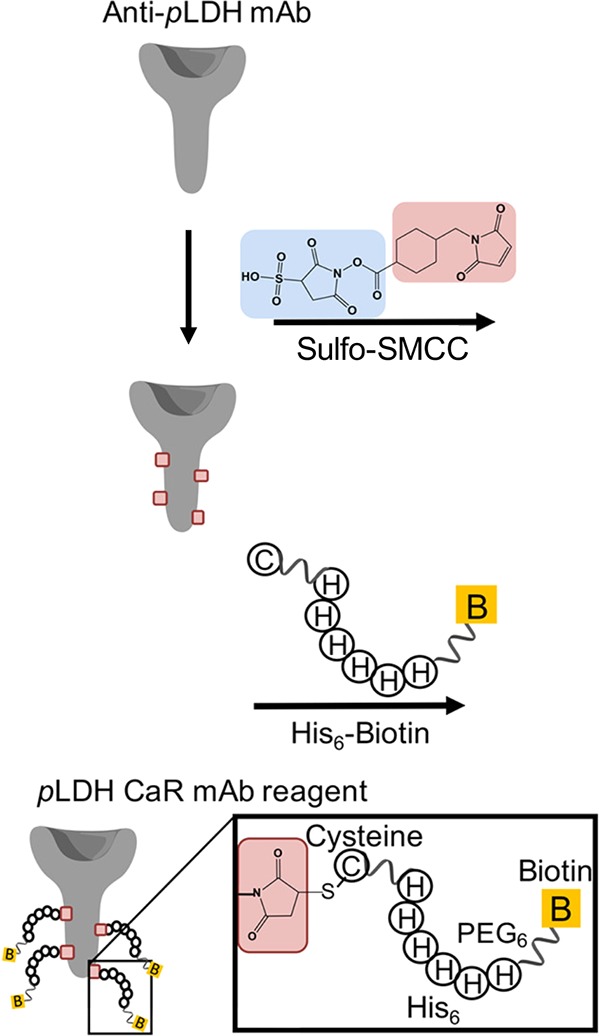
A His6-tag was coupled to an anti-pLDH antibody via Sulfo-SMCC to allow for chelation of the His6-tagged antibody to Co(II)NTA-functionalized magnetic bead. Remaining sites on the bead surface (unoccupied by antibody) permitted coordination of HRP2 directly to the bead surface. Subsequent to biomarker capture, imidazole was added to elute HRP2 and the His-tagged antibody/pLDH complex. Adapted from ref (117). Copyright 2017 American Chemical Society.
One of the challenges associated with employing a His6-tagged antibody/IMAC concentration strategy is that the biomarker remains bound to the antibody during the detection step. The antibody used for sample preparation has the potential to block target binding (via overlapping epitopes or steric effects) to the molecular recognition elements employed in the downstream assay, leading to false-negative results. This challenge can be addressed by designing the diagnostic such that the His6-tagged antibody and downstream detection elements bind to orthogonal epitopes of the target. Furthermore, other His6-tagged molecular recognition elements such as aptamers could be used in place of His6-tagged antibodies. Aptamers are short, single-stranded nucleic acid sequences with secondary structures that afford high (∼nanomolar–picomolar) binding affinities. Compared to antibodies, aptamers are smaller and may target unique epitopes, potentially allowing for this innovative sample preparation strategy to be more universally applied.123−126
Coordination chemistry-based sample preparation technologies have enabled significant improvements in the sensitivities of point-of-care diagnostics. The enhanced sensitivities, which in several examples were improved by a whole order of magnitude, make a strong case for the use of these technologies in elimination campaigns.85,115,117 Previously, biomarker enrichment strategies were not incorporated into low-resource diagnostics because of the restrictive format of the tests. The IMAC-based chemistries described in this section represent innovative yet simple solutions that open the door for the development of POC diagnostics that incorporate sample preparation. While some of the sample preparation methods presented in this section increased the total number of assay steps, these methods were not designed in tandem with the diagnostic devices that were utilized for detection. Moving forward, human-centered device design that integrates sample preparation with detection elements will permit the next generation of POC diagnostics to be developed with minimized user steps and improved sensitivities.
4. Metal-Based Signal Generation
Inorganic metal complexes and nanoparticles have been instrumental in the development of targeted reagents for disease-associated DNA, proteins, or cells. Their biomedical applications include the development of metallodrugs,127,128 enzyme inhibitors,129−131 photothermal132,133 and photodynamic133 therapy agents, and MRI contrast,127,132 in vivo bioimaging,133,134 and protein/cell imaging agents.135,136 Their unique optical, electrochemical, magnetic, and catalytic properties have also enabled their use as signaling modalities in diagnostics. This section focuses on the application of metal-based signal generation probes for the detection of protein biomarkers at the point of care (summarized in Table 1 by biomarker and in Table 2 by signal generation method). (The detection of pathogenic nucleic acids via metal-based signal generation probes is discussed in detail in section 6). First, we describe how inorganic complexes have been utilized in photoluminescent, electrochemical, and electrochemiluminescent applications, respectively. Subsequently, we detail the essential bioconjugation chemistries that permit nanoparticles to be functionalized with molecular recognition elements to generate useful diagnostic reagents. We then discuss how various classes of these functionalized nanoparticles have been used in POC diagnostic assays. Lastly, we consider the catalytic properties of metalloenzymes and metal nanoparticles that have been utilized for signal amplification in POC diagnostics for infectious diseases.
Table 1. Selected Examples of Signal Generation Probes/Methods for Common Infectious Disease Biomarkers.
| signal generation probe/method | biomarker | signal type | equipment | sample matrix | LOD | range | assay time | diagnostic setting |
|---|---|---|---|---|---|---|---|---|
| [Ir(ppy)2(H2O)2]+138 | histidine-rich protein 2 (HRP2) | luminescent | microwell plate and microplate reader | aqueous buffer | 14.5 nM (0.972 μg/mL) | 10–1000 nM | 1.5 h | Level 2–4 |
| ELISA: horseradish peroxidase (HRP) with DAB/H2O2383 | colorimetric | functionalized chromatography paper and cell phone | human serum | 0.32 nM | 0.2–260 nM | ∼2 h | Level 2–4 | |
| ELISA: alkaline phosphatase (ALP) with BCIP/NBT383 | colorimetric | functionalized chromatography paper and cell phone | human serum | 0.15 nM | 0.2–260 nM | ∼2 h | Level 2–4 | |
| ELISA: HRP with DAB/H2O2387 | colorimetric | 2-dimensional paper network (2DPN) | fetal bovine serum | 6.5 ng/mL | 6.25–200 ng/mL | 20 min | Level 1 | |
| [Ir(pbtz)2(dtbpy)]+137 | Interferon-gamma (IFN-γ) | luminescent | luminescence lifetime spectrometer | aqueous buffer | 0.12 nM | 1–300 nM | ∼1 h | Level 2–4 |
| Hemin/G-quadruplex DNAzyme144 | electrochemical | electrochemical workstation | aqueous buffer | 0.1 nM | 0.1–120 nM | ∼1 h | Level 2–4 | |
| [Ir(ppy)2(phen-dione)]+145 | electrochemical | electrochemical workstation | aqueous buffer/5% human serum | 16.3 fM | 50–3000 fM | 3.3 h | Level 2–4 | |
| P-RGO@Au@Ru-SiO2 nanocomposite153 | p24 antigen | ECL | ECL analyzer | aqueous buffer/1% human serum | 1.0 pg/mL | 1.0–10000 pg/mL | 5 h | Level 2–4 |
| AgNPs233 | luminescent | microwell plate and microplate reader | aqueous buffer/human plasma | 8.2 pg/mL | 10–1000 pg/mL | 2 h | Level 2–4 | |
| ELISA: HRP with H2O2 and hydroquinone392 | electrochemical | electrochemical workstation | aqueous buffer/human serum | 8 pg/mL | 0.01–100 ng/mL | 1 h | Level 2–4 | |
| Plasmonic ELISA: Catalase with H2O2 and Au(III)396 | colorimetric | microwell plate and microplate reader | aqueous buffer/human serum | 1.0 ag/mL | 1.0–1000 ag/mL | 6 h | Level 2–4 | |
| Au@Pt core–shell nanozymes435 | colorimetric | lateral flow assay (LFA) | FBS/human serum | 0.8 pg/mL | 1–10000 pg/mL | 20 min | Level 1 |
Table 2. Selected Examples of Inorganic Nanoparticle-Based Signal Generation Probesa.
| signal generation probe/method | biomarkers | signal type | equipment | sample matrix | LOD | range | assay time | diagnostic setting |
|---|---|---|---|---|---|---|---|---|
| AgNPs | YFV NS1, DENV NS1, and ZEBOV glycoprotein229 | colorimetric | lateral flow assay (LFA) | human serum | 150 ng/mL | NR | NR | Level 1 |
| (a) MERS-CoV DNA | colorimetric | microfluidic paper analytical device (μPAD) | aqueous buffer | (a) 1.53 nM | (a) 20–10000 nM | NR | Level 1 | |
| (b) MTB DNA | (b) 1.27 nM | (b) 50–2500 nM | ||||||
| (c) HPV DNA231 | (c) 1.03 nM | (c) 20–2500 nM | ||||||
| CdSe/ZnSQDs | (a) anti-HBV Abs | luminescent | microfluidic device | human serum | (a) 10 pM | 10–10000 pM | ∼1 h | Level 2–4 |
| (b) anti-HCV Abs | (b) 3 pM | |||||||
| (c) anti-gp41 Abs253 | (c) 3 pM | |||||||
| dual (AuNP and QD) readout nanospheres (RNs) | EBOV glycoprotein257 | colorimetric and luminescent | lateral flow assay (LFA) | aqueous buffer/tap water, urine, and plasma | AuNPs: 2 ng/mL | AuNPs: 20−1000 ng/mL | 20 min | Level 1 or Level 2 |
| QDs: 0.18 ng/mL | QDs: 2–1,000 ng/mL | |||||||
| Eu3+ chelate-loaded NPs | hemagglutinin (HA) 1289 | fluorescent | lateral flow assay (LFA) | spiked human nasopharyngeal specimens | 20 HAU/mL | 10–160 HAU/mL | 15 min | Level 2 |
| hepatitis B surface antigen (HBsAg)305 | fluorescent | lateral flow assay (LFA) | Aqueous buffer/Human serum | 30 pg/mL | 50–3130 pg/mL | 30 min | Level 2 | |
| Y2O2S:Yb3+, Er3+ UCPs | circulating anodic antigen (CAA)317 | fluorescent | lateral flow assay (LFA) | human urine | 0.03 pg/mL | 0.03–100 pg/mL | ∼4.5 h | Level 2 |
NR = not reported.
4.1. Inorganic Complexes
The broad array of properties that result when a ligand coordinates to a metal has been leveraged for signal production in a variety of diagnostic formats. Inorganic coordination compounds are valuable as signal generation probes due to their low molecular weight, tunability, and unique catalytic and photophysical properties.130 These compounds have been used in photoluminescent (i.e., fluorescent and phosphorescent),137−142 electrochemical,143−148 and electrochemiluminescent (ECL)149−157 sensing platforms. Each signal generation method has trade-offs with regard to ease-of-use and interpretation, as well as complexity of instrumentation required to measure signal output. When designing a low-resource diagnostic, these trade-offs must be evaluated based on the use-case scenario and testing environment. Collectively, these considerations better inform the developer as to which inorganic complexes are suitable for a particular signal generation method.
4.1.1. Luminescence
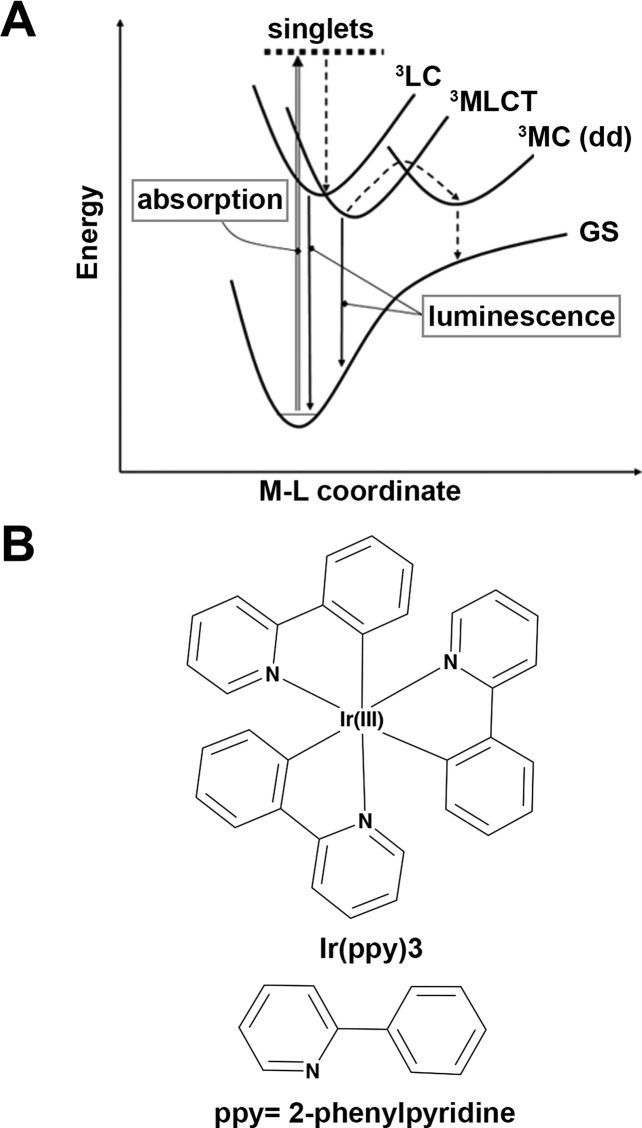
Photoluminescent inorganic complexes possess unique properties and offer several advantages over organic fluorophores as signaling molecules for diagnostics. These advantages include long-lived phosphorescence, significant Stokes shifts, and emission tuneability via ligand composition.137,158,159 Long-lived phosphorescence is beneficial because it is spectrally distinct from the autofluorescent background of biological matrices, leading to improved sensitivity.158 The large Stokes shifts of metal complexes also result in improved sensitivity because self-quenching of signaling molecules is minimized.158 Additionally, the tunable emissions of metal complexes enable their use as signal transducers, whereby ligand exchange reactions that induce changes in emission spectra can be correlated to biomarker concentration.159 To date, cyclometalated d6 complexes of Ir(III) are the most commonly utilized photoluminescent signal generation probes.137−139,141,142,160 Ligands in these cyclometalated Ir(III) complexes (e.g., deprotonated 2-phenylpyridine) demonstrate strong σ-donor effects, which significantly increase the energy of the 1MC (metal-centered) excited state (i.e., the ligand field stabilization energy) that gives rise to nonradiative decay pathways. As a result, excited electrons in these complexes preferentially populate the 1MLCT (metal-to-ligand charge transfer) excited state (as opposed to 1MC). The excited electrons then undergo intersystem crossing and subsequently phosphoresce from a mixture of the 3LC (ligand-centered) and 3MLCT excited states (Figure 10). The high phosphorescent quantum yields with large Stokes shifts make these complexes ideal for signal generation.160,161
Figure 10.
(A) Energy coordinate diagram for cyclometalated complexes of Ir(III). Initial absorption of photons promotes electrons to 1MLCT states, which subsequently undergo intersystem crossing to phosphorescent 3LC and 3MLCT excited states. (B) Structure of [Ir(ppy)3], a common cyclometalated Ir(III) complex. The ligands employed in these complexes (e.g., 2-phenylpyridine) are strong σ-donors that give rise to the luminescent properties of the complex. Adapted with permission from ref (160). Copyright 2007 Springer.
Davis et al.138 designed a “switch-on” Ir(III) detection probe that selectively produced phosphorescent signal in the presence of malarial biomarker HRP2. The cyclometalated Ir(III) complex, [Ir(ppy)2(H2O)2]+, was poorly luminescent in the absence of HRP2; however, when aqua ligands were displaced by HRP2’s histidine residues, a biomarker-dependent 3MLCT/3LC phosphorescent signal was produced (Figure 11). Similar to the principles of IMAC chemistry discussed in section 3, this approach capitalized on the chelation of HRP2’s histidine repeats to the inorganic coordination compound. The assay design aptly showed how the tunable luminescence of Ir(III) complexes could be harnessed for signal transduction, demonstrating that a ligand exchange reaction’s phosphorescent signal could be correlated to biomarker concentration. In addition, the Ir(III) complex displayed a significant Stokes shift (Δλ = 145 nm),138 making it less prone to self-quenching effects and decreases in sensitivity associated with its organic fluorophore counterparts.162
Figure 11.
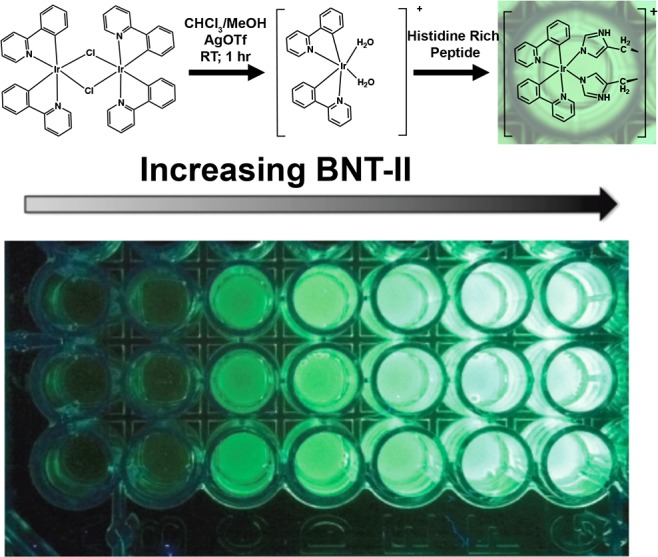
(A) Cyclometalated Ir(III) detection probe for malarial biomarker HRP2. In the absence of HRP2, the diaquabis(2-phenylpyridine)iridium(III) complex demonstrated little to no luminescence. Introduction of a sample containing HRP2 caused displacement of the aqua ligands by the histidine residues in the protein and activated the 3MLCT/3LC phosphorescent signal pathway. (B) Titration of HRP2 biomarker mimic BNT-II, demonstrating increased phosphorescence with increasing biomarker concentration. Adapted with permission from ref (138). Copyright 2014 Elsevier.
This study138 also incorporated the Ir(III) probe into an IMAC magnetic bead-based biomarker isolation and detection assay for recombinant HRP2 which had a detection limit of 14.5 nM. Although less sensitive than an ELISA (enzyme-linked immunosorbent assay), this HRP2 concentration was within the relevant range for clinical diagnosis of malaria. Moreover, the bead-based assay could be completed in less than 2 h, whereas traditional ELISAs require 6–8 h. While the current assay format would only permit its application in better-equipped laboratories, the developed cyclometalated Ir(III) probe could be an exciting detection element in an LFA with HRP2-specific antibodies at the test line. However, this photoluminescence detection mechanism is dependent on existence of a histidine-rich biomarker, so further applications of this particular strategy are limited.
A more generalizable platform using photoluminescent Ir(III) probes was developed by Lin et al.139 for the detection of interferon-gamma (IFN-γ), an inflammatory cytokine biomarker for immunological diseases such as HIV. For molecular recognition and detection of IFN-γ, the assay used two aptamers: one was highly specific for IFN-γ, and the second was a guanine-rich nucleic acid sequence. The two aptamers were designed to be partially complementary and were prehybridized prior to the introduction of a sample containing IFN-γ. In the presence of IFN-γ, the IFN-γ-specific aptamer bound to its target and liberated the guanine-rich sequence. Introduction of K+ ions induced the formation of guanine quadruplexes, which subsequently bound an Ir(III) probe for “switch-on” luminescence that correlated biomarker concentration with phosphorescent signal (Figure 12).
Figure 12.
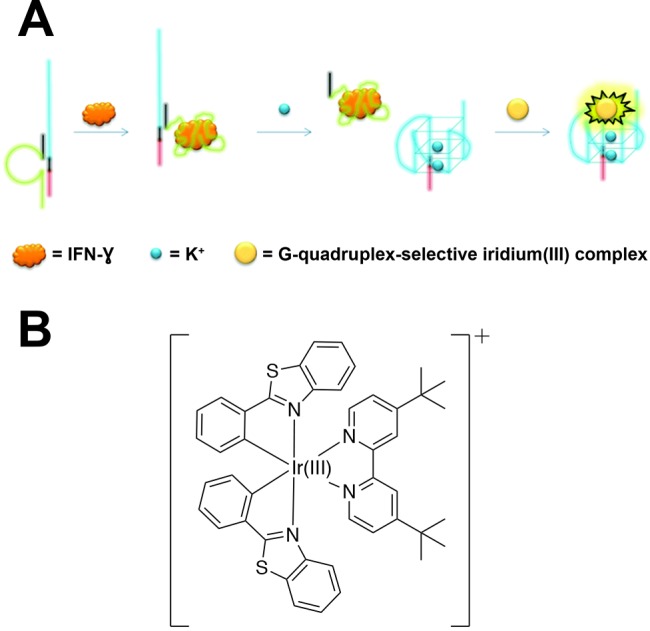
Aptamer-based assay for HIV biomarker IFN-γ using a luminescent Ir(III) detection probe. (A) The assay utilized two aptamers, the first being an IFN-γ-specific aptamer (green) that was prehybridized to a G-quadruplex aptamer (blue). Addition of a sample resulted in the capture of IFN-γ by the first aptamer and liberation of the G-quadruplex aptamer, which underwent a structural switch to form guanine tetrads upon introduction of K+ ions. The G-quadruplexes were specific for a cyclometalated Ir(III) complex that produced phosphorescent signal that was correlated with IFN-γ concentration. (B) Structure of the cyclometalated Ir(III) complex. Adapted with permission from ref (139). Copyright 2015 Royal Society of Chemistry.
This Ir(III) probe possessed a long phosphorescence lifetime (>2.9 μs) and significant Stokes shift (Δλ = 215 nm), thus exhibiting the canonical advantages of inorganic probes over organic fluorophores. The assay demonstrated adequate specificity for IFN-γ versus other proteins commonly found in biological matrices (e.g., human serum albumin and immunoglobulins) and had a limit of detection of 0.12 nM and a dynamic range of 1–300 nM. However, the assay suffered a reduction in sensitivity when conducted in a 0.5% cell extract, and no attempts were made in the complex biological matrices that are pertinent to HIV diagnosis (e.g., blood, urine, saliva, etc.). Nonetheless, the assay established a proof-of-principle for using a photoluminescent cyclometalated Ir(III) complex as a detection probe for an HIV biomarker.139 Moreover, the assay format is generalizable such that it could be applied to virtually any biomarker for which there is a high-affinity aptamer. If combined with one of the sample preparation strategies discussed previously (section 3) or integrated with paper or another field-ready substrate, this Ir(III)-based detection strategy could produce a robust and sensitive assay that is applicable in low-resource diagnostic settings.
4.1.2. Electrochemistry and Electrochemiluminescence (ECL)
By applying a potential (or range of potentials) to a sample containing an electroactive inorganic probe, current or luminescence can be measured as an output for electrochemical or ECL detection, respectively. Electrochemical or ECL-based assays that combine molecular recognition elements, such as antibodies or aptamers, with electroactive inorganic probes have shown promise for infectious disease detection.149,163−165 In particular, inorganic low-spin d6 complexes of Ir(III) and Ru(II), as well as hemin (Ferriprotoporphyrin IX), have been utilized in developing electrochemical- and ECL-based probes.149
Both electrochemistry and ECL are signal generation strategies that are well-suited to the development of POC diagnostics, largely due to (1) capability for miniaturization, (2) low cost, (3) simplicity, (4) rapid time-to-result, and (5) high sensitivity.149,163 This has been aptly demonstrated with the development of paper-based electrochemical diagnostic platforms,166 often termed microfluidic paper-based electrochemical devices (μPEDs)167−169 or electrochemical paper analytical devices (ePADs).170 These tests utilize paper as the assay substrate and have been combined with instrumentation amenable to low-resource settings, such as cell phones, for detection. Incorporation of existing electrochemical- and ECL-based assays into these formats could result in robust and highly sensitive infectious disease diagnostics. This would be impactful for disease surveillance or case management, where robust and sensitive diagnostics are needed.
Electrochemistry
Diagnostic assays with electrochemical detection commonly consist of a three-electrode system: a working electrode (often gold or carbon modified with a molecular recognition element for biomarker capture), a counter electrode (typically platinum), and a reference electrode (commonly a saturated calomel electrode or Ag/AgCl electrode).171 In this format, incubation of a biological sample in an electrochemical cell initially results in capture of the biomarker at the working electrode. Introduction of a biomarker-specific inorganic probe and a supporting electrolyte and subsequent application of an electrochemical method (e.g., cyclic voltammetry, square wave voltammetry, differential pulse voltammetry, amperometry, etc.) results in significant current production at the working electrode. This current can then be correlated to biomarker concentration. With regard to infectious disease biomarker detection, hemin, Ir(III) complexes and Ru(II) complexes have been utilized as electrochemical probes.144−146
Hemin (ferriprotoporphyrin IX) is a Fe3+-containing porphyrin derivative that mimics the enzymatic activity of horseradish peroxidase in catalyzing the reduction of H2O2 (see section 4.3). This enzymatic activity is further enhanced when hemin is incorporated into a G-quadruplex aptamer.143,144,172 Zhang et al.144 developed an electrochemical assay for the detection of HIV-associated IFN-γ that utilized hemin as a key component of the signal generation mechanism. A 3′-thiolated DNA construct was conjugated to a gold working electrode, where the DNA construct contained a G-quadruplex sequence that was specific for hemin and an IFN-γ-specific aptamer. Both the G-quadruplex and the IFN-γ-specific aptamer were prehybridized by an 8-nucleotide hairpin. The authors utilized this functionalized working electrode to perform cyclic voltammetry on samples containing IFN-γ to monitor the reduction of H2O2, which could be correlated to IFN-γ concentration (Figure 13).
Figure 13.
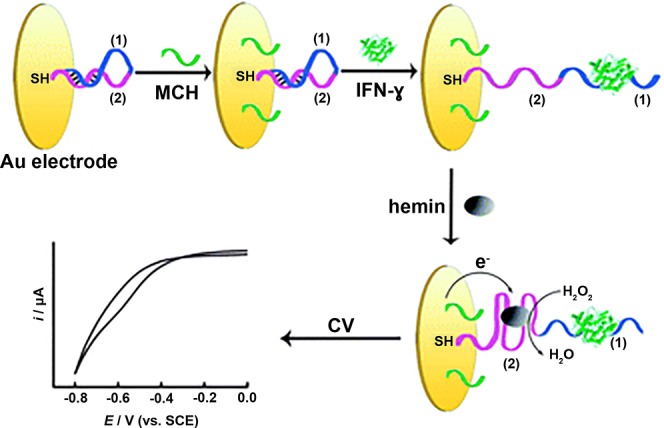
Hemin DNAzyme-based catalytic assay for electrochemical detection of IFN-γ. A thiolated DNA construct was conjugated to a Au working electrode. The DNA construct consisted of a G-quadruplex aptamer specific for hemin (purple) and an IFN-γ-specific aptamer (blue). Both aptamers in the DNA construct were prehybridized by an 8-nucleotide hairpin. In the presence of IFN-γ, the IFN-γ-specific aptamer bound to IFN-γ and opened the hairpin. Hemin was then added, which bound to the G-quadruplex, forming a DNAzyme that could catalyze the reduction of H2O2. This reduction produced a current that was proportional to IFN-γ concentration in the sample. Adapted with permission from ref (144). Copyright 2012 Royal Society of Chemistry.
In samples containing IFN-γ, the IFN-γ-specific aptamer bound to IFN-γ, opening the hairpin and subsequently allowing for hemin to bind to the G-quadruplex. The potential sweep catalyzed the reduction of H2O2 by the hemin/G-quadruplex DNAzyme, producing a cathodic current that was proportional to the concentration of IFN-γ in the sample. The assay yielded a detection limit of 0.1 nM and a dynamic range of 0.1–120 nM144 and thus performed similarly to the luminescent assay developed by Lin et al.139 (section 4.1.1). Though the authors demonstrated the specificity of the assay for IFN-γ versus nontarget proteins BSA and IgG, they did not report whether the assay could be performed in more complex matrices, which can significantly compromise assay performance. More rigorous testing of the assay in biological matrices is therefore needed to demonstrate the robustness of the assay.
Miao and colleagues145 synthesized and implemented an Ir(III) complex-based electrochemical probe for detection of IFN-γ (Figure 14). First, a gold working electrode was functionalized with a molecular beacon (MB) that contained an IFN-γ-specific aptamer as well as a sequence that was partially complementary to a second oligonucleotide strand (H1). In the absence of IFN-γ, the portion of the MB complementary to H1 was hybridized into a hairpin structure. When IFN-γ bound to the aptamer, the H1 sequence was freed and available to hybridize with its complement, H2. The developed Ir(III) probe then bound to the major and minor grooves of the H1–H2 double helix. Oxidation and reduction of the 1,10-phenanthroline-5,6-dione ligand via differential pulse voltammetry (DPV) resulted in IFN-γ concentration-dependent current, thus correlating an electrochemical signal to the presence of target biomarker.
Figure 14.
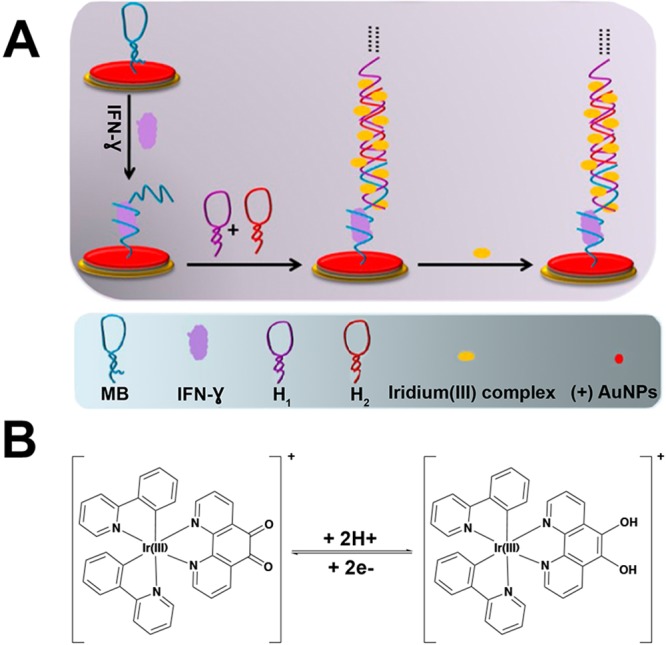
(A) Electrochemical detection assay for HIV biomarker IFN-γ that utilized an Ir(III) redox probe. A gold working electrode was functionalized with a molecular beacon containing an IFN-γ-specific aptamer and an oligonucleotide sequence that was complementary to H1. Hybridization of H1 and H2 formed a double helix to which the Ir(III) probe could bind via the major and minor grooves. (B) The redox reaction of [Ir(ppy)2(phen-dione)]+ was achieved by differential pulse voltammetry, which produced a current that was correlated to the presence of IFN-γ. Adapted with permission from ref (145). Copyright 2017 Nature Publishing Group (https://creativecommons.org/licenses/by/4.0/legalcode).
The electrochemical assay had a detection limit of 16.3 fM and a dynamic range of 50 fM–3.0 pM and could be conducted in samples that were 5.0% human serum. Importantly, this assay yielded an ∼7000-fold improvement in limit of detection compared to the two previously discussed assays for detection of IFN-γ.145 This is largely due to the signal amplification that resulted from the groove-binding of numerous Ir(III) probes to the H1–H2 double helix, which provided multiple signal-generating elements for every one IFN-γ biomarker bound. In its present form, this assay could not be applied in a primary healthcare setting because it utilizes an electrochemical workstation that requires trained personnel and significant laboratory resources. However, a portable electrochemical detection system such as the one described in section 5.5(173) could allow for this highly sensitive assay to be applied in a POC setting.
Electrochemiluminescence
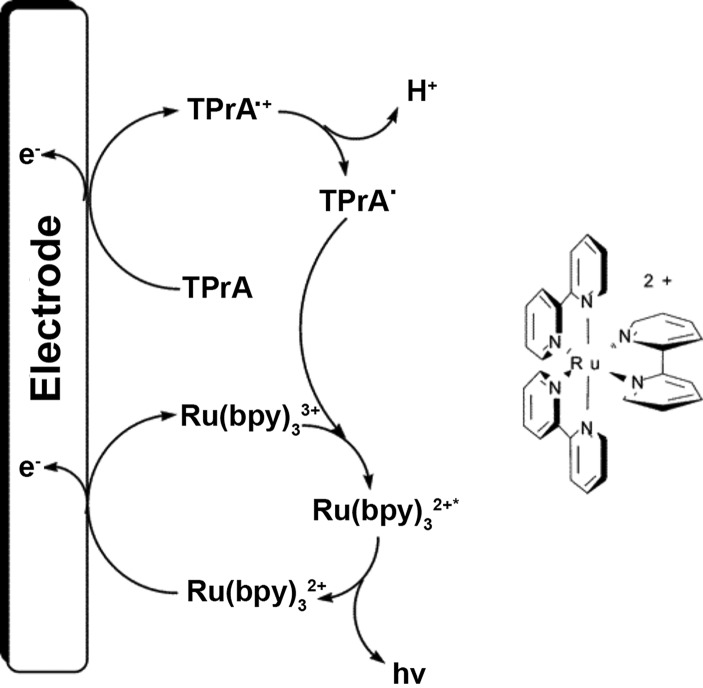
Similar to most electrochemical assays, ECL systems employ a working electrode that is functionalized with a target-specific molecular recognition element. The potential manipulation is similar to voltammetric measurements; however, in ECL, the applied potential also generates an excited state in the ECL probe, which then produces luminescence upon relaxation.149,165 The canonical probe for ECL-based detection is [Ru(bpy)3]2+, a complex (or derivative thereof) which has been used extensively in the development of ECL-based sensors for infectious disease detection.150−154,156 In diagnostic assays, ECL with [Ru(bpy)3]2+ occurs primarily through a coreactant mechanism, where the coreactant is most commonly n-tripropylamine (TPrA) (Figure 15). In coreactant ECL, simultaneous oxidation of [Ru(bpy)3]2+ to [Ru(bpy)3]3+ and TPrA to oxidized TPrA ([TPrA•]+) is followed by deprotonation of [TPrA•]+. An electron is then transferred from TPrA• to [Ru(bpy)3]3+ to form a 3MLCT excited state [Ru(bpy)3]•2+ that is analogous to the 3MLCT state that is populated through absorption of a photon. The triplet excited state then undergoes phosphorescent decay to the ground state, allowing for luminescent detection of target biomarkers.149 Of the various mechanisms for electrogeneration of a luminescent excited state, coreactant ECL has proven to be the most advantageous for the development of diagnostic assays because it permits measurements that can be obtained in aqueous solvents and matrices.
Figure 15.
Coreactant ECL reaction mechanism for [Ru(bpy)3]2+ and n-tripropylamine (TPrA). Oxidation of [Ru(bpy)3]2+ and TPrA at the electrode surface followed by electron transfer from oxidized TPrA to [Ru(bpy)3]2+ forms an excited state [Ru(bpy)3]2+ that then undergoes phosphorescence to decay back to the ground state. Adapted from ref (149). Copyright 2004 American Chemical Society.
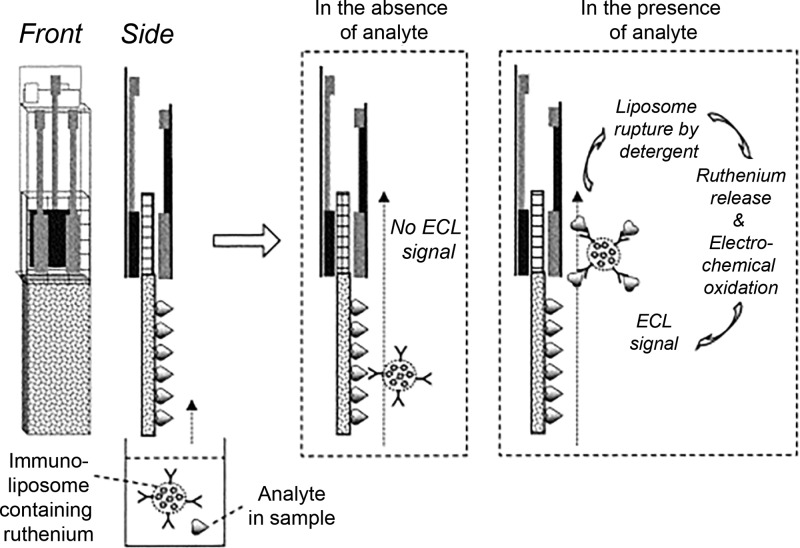
One platform for detection of infectious diseases, pioneered by Yoon et al.,150 used immunoliposomes to encapsulate ECL signal generation probes. Liposomes equipped with a maleimide handle were synthesized by the freeze–thaw method and loaded with [Ru(bpy)3]2+. The maleimide-functionalized liposomes were then conjugated to thiol-activated antibodies specific for the target antigen. The resulting immunoliposomes then served as a target-specific reagent containing an ECL probe that could be released with the application of detergents such as SDS or Triton.
The authors150 incorporated the [Ru(bpy)3]2+-loaded immunoliposome into a membrane strip test for detection of the Legionella antigen. The strip test contained the following components: (1) a nitrocellulose membrane impregnated with Legionella antigen, (2) a glass fiber pad presoaked in SDS, and (3) screen-printed electrodes that could interface with an ECL analyzer. The immunoliposomes were incubated with a buffer solution containing the Legionella antigen, and then the sample solution was allowed to wick up the nitrocellulose strip. Both the antigen in the sample and the antigen embedded in the nitrocellulose competed for binding with the immunoliposome. In positive samples, the immunoliposome was bound by the Legionella antigen in solution and migrated past the nitrocellulose strip to the glass fiber pad. The SDS in the glass fiber pad lysed the immunoliposome, releasing [Ru(bpy)3]2+, which was detected by coreactant ECL and correlated to the antigen concentration (Figure 16).
Figure 16.
Membrane strip test that utilizes ECL probe-loaded liposomes for detection of Legionella antigen. Immunoliposomes functionalized with anti-Legionella antibodies were loaded with the ECL probe [Ru(bpy)3]2+ and incubated with a sample. The sample was then allowed to wick up a nitrocellulose strip that had already been impregnated with the Legionella antigen. In samples containing Legionella antigen, the immunoliposome migrated past the nitrocellulose to a glass fiber pad presoaked with SDS, which lysed the liposome and released [Ru(bpy)3]2+. [Ru(bpy)3]2+ was then detected by coreactant ECL. Adapted with permission from ref (150). Copyright 2003 Elsevier.
The assay could be completed in 20 min and had a limit of detection of 2 ng/mL.150 Compared to a traditional lateral flow assay, the test required additional user manipulation steps (preincubation of the sample and immunoliposome, addition of coreactant, etc.). The assay was also dependent on a benchtop ECL analyzer and, therefore, ill-suited to a Level 1 healthcare setting in its current format. However, this novel platform demonstrated the capabilities of ECL for detecting a target antigen and showed how a [Ru(bpy)3]2+-loaded immunoliposome could be utilized to amplify signal. Variations of this loaded immunoliposome platform have since been applied to the detection of the influenza virus biomarker hemagglutinin.151,152
Recently, Zhou et al.153 developed an ECL-based immunosensor for p24, a HIV biomarker associated with early stage infection and detection of which promotes earlier intervention in HIV cases. The detection probe in this assay was a nanocomposite consisting of gold nanoparticle-decorated graphene (P-RGO@Au) and [Ru(bpy)3]2+-doped silica nanoparticles (Ru-SiO2) (Figure 17). A gold nanoparticle-modified glassy carbon electrode (GCE) was functionalized with an anti-p24 antibody (Ab1) for molecular recognition of p24. A second anti-p24 antibody (Ab2) conjugated to the P-RGO@Au@Ru-SiO2 nanocomposite was used as the detection element. Once the Ab1-p24-Ab2 sandwich was formed on the electrode, coreactant n-tripropylamine (TPA) was added to produce the ECL signal through the mechanism previously discussed.149 The P-RGO@Au@Ru-SiO2 nanocomposite offered several benefits to the system, namely an increased surface area for the ECL reaction and an increased rate of electron transfer. The SiO2 nanoparticles permitted greater [Ru(bpy)3]2+ loading, and the gold nanoparticle-graphene composite increased loading of the [Ru(bpy)3]2+-doped SiO2 nanoparticles. The authors contended that these aspects led to an increased concentration of ECL-probe at the working electrode surface, which ultimately improved the assay’s detection limit (LOD = 1.0 pg/mL) and linear range. Additionally, the authors demonstrated that the p24 antigen could be detected in diluted human serum.153
Figure 17.
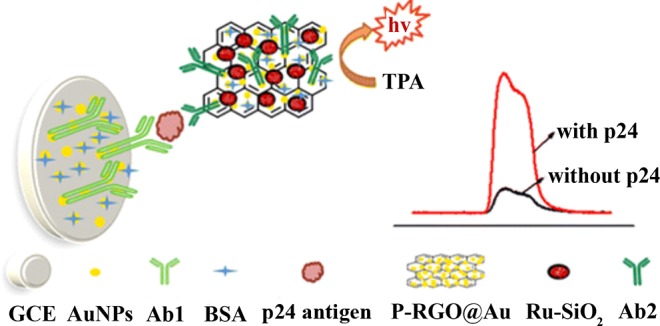
An ECL-based immunosensor for detection of HIV antigen p24. The detection probe was a gold nanoparticle-graphene nanocomposite that was loaded with [Ru(bpy)3]2+-doped SiO2 nanoparticles and generated electrochemiluminescent signal that could be correlated with p24 antigen concentration. Adapted from ref (153). Copyright 2015 American Chemical Society.
A remaining challenge for developing luminescent, electrochemical, and ECL assays that incorporate inorganic complexes is thorough validation in complex biological matrices and clinical samples. The aforementioned luminescent assays138,139 were not rigorously evaluated in biological samples. This is an important consideration in developing luminescent assays because of the high background that can result from the presence of interferents in biological samples. Biofouling of electrodes is a known challenge in developing electrochemical sensors as well.174,175 Although several of the previously mentioned electrochemical and ECL assays were tested in dilute biological matrices,145,153 a more thorough evaluation of how the biological matrix affects the limit of detection is still needed. However, the issue of interference and biofouling could potentially be addressed by integrating these detection platforms with the metal-based sample preparation strategies discussed in section 3, resulting in more robust and sensitive assays.
Inorganic complexes have demonstrated promise for detection of infectious disease biomarkers. The luminescent, electrochemical, and ECL detection platforms discussed in this section all capitalized on the unique properties that result from the interplay between ligands and metal centers. However, inorganic complexes are rarely used as stand-alone probes for biomarker detection. Instead, inorganic nanoparticles have emerged as the predominant probes in infectious disease diagnostics.176,177 Integration of inorganic complexes with nanomaterials represents a potential avenue to developing the next generation of detection probes. One interesting nanomaterial that incorporates inorganic complexes is the metal–organic framework (MOF), which is a nanosized network of organic ligands and metal ion/complex connecting points.178,179 The ligands provide functional handles for the conjugation of molecular recognition elements, while signal-generating metal complexes are installed at the connecting points or encapsulated within the network. Recently, electrochemical aptasensors have been developed that contain aptamer-conjugated MOFs for specific recognition and detection of various protein or small molecule targets.180−182 Moreover, two of the complexes discussed in this section, Ir(ppy)3 and [Ru(bpy)3]2+, have already been incorporated into MOFs for sensing and imaging applications.183,184 By integrating molecular recognition elements into the MOF architecture, the applications of these Ir(III)- and Ru(II)-based MOFs could be further expanded to include luminescent, electrochemical, or ECL detection of infectious disease biomarkers.
4.2. Inorganic Nanoparticles
The recent expansion of nanochemistry has drastically influenced many areas of science. In diagnostics, inorganic nanoparticles have become ubiquitous as detection elements in numerous platforms (e.g., colorimetric, luminescent, electrochemical, thermal, etc.), and each distinct class of nanoparticles possesses unique advantages and caveats.176,177,185−187 The diagnostic setting and use-case scenario determine the applicable detection methods which, in turn, direct which nanoparticles to use. In order to apply inorganic nanoparticles as diagnostic reagents, target-specific molecular recognition elements (i.e., antibodies, aptamers, and oligonucleotide probes) must be functionalized to the nanoparticle surface. This section will first review bioconjugation reactions and nanoparticle characterization methods (section 4.2.1). The list of reactions is not meant to be exhaustive but, rather, to highlight the most essential covalent and affinity-based coupling strategies for nanoparticle bioconjugation. Then, we survey the various types of nanoparticles that have been applied to the detection of infectious diseases with potential applications at the point of care (sections 4.2.2–4.2.6).
4.2.1. Functionalization and Characterization of Nanoparticles
Developing a nanoparticle detection reagent that can be targeted toward a biomarker requires conjugation of molecular recognition elements to its surface. These types of conjugations often require linkers that facilitate attachment to the nanoparticle surface. The chemistries employed for coupling molecular recognition elements to nanoparticle surfaces for diagnostic applications generally rely on a few key bioconjugation reactions and are similar to those used for planar solid supports. However, the smaller size regime of nanoparticles significantly influences the reaction kinetics on nanoparticle surfaces; therefore, bioconjugation reactions must be optimized for a given class and size of nanoparticle.188,189
Covalent Coupling
Given the prevalence of carboxylates and primary amines on nanoparticle surfaces and molecular recognition elements, one of the most common bioconjugation strategies for generating functionalized nanoparticle detection probes is the formation of amide bonds. The conventional reagent for forming amide bonds on nanoparticle surfaces is 1-ethyl-3-(3-(dimethylamino)propyl) carbodiimide (EDC), which produces an activated ester that can subsequently react with a primary amine. However, alone, the EDC-activated ester reacts slowly with amines, often leading to hydrolysis products that inhibit amide coupling. For this reason, EDC is usually paired with sulfo-NHS (N-hydroxysuccinimide) esters to improve the stability of the activated ester intermediate. The sulfo-NHS ester-activated intermediate readily reacts with primary amine nucleophiles to form stable amide linkages (Figure 18A).190,191
Figure 18.
Essential coupling strategies for bioconjugation of inorganic nanoparticles and molecular recognition elements: (A) EDC/Sulfo-NHS condensation of a carboxylic acid and a primary amine. (B) Silica nanoshell formation on a nanoparticle surface using APTES. (C) Dative bonding of thiolated antibody or oligonucleotide to a noble metal nanoparticle. (D) Conjugation of a thiol and a primary amine via heterobifunctional cross-linker Sulfo-SMCC. (E) An example of photochemistry-mediated coupling of a primary amine to a nanoparticle surface using heterobifunctional cross-linker Sulfo-NHS-LC-diazirine. (F) “Click” chemistry [3 + 2] cycloaddition of an azide and an alkyne in the presence of a Cu(I) catalyst.
EDC/Sulfo-NHS coupling is useful in generating both antibody- and nucleic acid-nanoparticle conjugates as diagnostic detection probes. Due to the intrinsic abundance of primary amines in antibodies, they are readily conjugated to carboxylate nanoparticles without the need for initial derivatization.188,192 EDC coupling with imidazole enables conjugation of ethylene diamine to the 5′ phosphate group of nucleic acids, yielding 5′-amine-functionalized oligonucleotides that can be coupled to carboxylated particles.193 Quantum dots, magnetic nanoparticles, and polymeric (latex) particles have found applications in POC diagnostics and are commonly equipped with carboxylates or amines on the surface to enable conjugation via EDC coupling. Quantum dots are conventionally stabilized by dihydrolipoic acid (DHLA) derivatives or various copolymers that install a carboxylate on the surface, and magnetic nanoparticles are coated with carboxylate-containing polymers to enable further functionalization with primary amine-containing biomolecules.188,194−196
Alternatively, both quantum dots and magnetic nanoparticles can be coated with a silica shell derivatized with carboxylate or amine functional groups. Silica nanoshells are formed via the condensation of silica oxide monomers, such as tetraethyl orthosilicate (TEOS) or 3-aminopropyl-triethoxysilane (APTES), yielding surface aminopropyl-silanols (Figure 18B). The surface of the silica can subsequently be coupled to molecular recognition elements via EDC.188 In addition to supplying functional groups for further bioconjugation, the silica encapsulation of other nanoparticles provides biocompatibility and protects the core material from degradation, making it a suitable strategy for the preparation of diagnostic nanoparticle detection probes.197
The other significant reaction class for generating nanoparticle detection probes is the bioconjugation of thiols present either in molecular recognition elements or on nanoparticle surfaces. Thiolated nucleic acids are prepared by the same method as amine-functionalized nucleic acids, except cystamine is used in place of ethylene diamine for the addition of a disulfide that can be reduced with dithiothreitol (DTT) to give a 5′-thiolated oligonucleotide probe.193,198 On the other hand, antibodies generally do not contain free thiols and, therefore, either have to be thiolated (often with Traut’s reagent) or cleaved at the interchain disulfides (via DTT or papain) to generate thiols for subsequent bioconjugation to a nanoparticle surface.192,198
Coupling of thiolated biomolecules to noble metal nanoparticles (referring to gold and silver nanoparticles) is conventionally done via coordinate covalent (dative) bonds, where the lone pairs of the sulfur atoms form stable linkages directly to the nanoparticle surface (Figure 18C).188,191 Alternatively, thiol-containing affinity reagents or nanoparticles can be coupled via a maleimide-derived heterobifunctional cross-linking agent, commonly sulfo-SMCC (succinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate). Thiols react with the maleimide functionality of sulfo-SMCC to form stable thioethers, whereas the NHS-ester component of sulfo-SMCC reacts with primary amines in molecular recognition elements or on nanoparticle surfaces (Figure 18D).191,194,195,197
Photochemical cross-linking and “click chemistry” have also been utilized to produce nanoparticle-based detection probes. Photosensitive functional groups, including phenyl azides and diazirines, generate reactive nitrenes and carbenes, respectively, upon exposure to UV light and allow for insertion into carbon–hydrogen and nitrogen–hydrogen bonds (Figure 18E). Photochemical agents can be integrated into heterobifunctional cross-linkers, such as sulfo-NHS-LC-diazirine, for conjugation of nanoparticles to molecular recognition elements.191,199,200 “Click” chemistry generically refers to bioorthogonal reactions with high yields, minimal side products, and mild conditions. While the most common “click” reaction is the conjugation of an azide to an alkyne in the presence of a Cu(I) catalyst (Figure 18F), there are many classes of “click” reactions that couple nanoparticles and molecular recognition elements.191,201−205
Biotin–Streptavidin
The predominant affinity-based conjugation strategy for nanoparticle functionalization is the biotin–streptavidin system. Biotin is a small molecule that binds with an extraordinarily high affinity (KD = 1.3 × 10–15 M) to the bacterial protein streptavidin.206 Streptavidin is an approximately 60 kDa tetrameric protein, permitting a 4:1 stoichiometry of biotin to streptavidin.206 In general, streptavidin is added to the nanoparticle surface via passive adsorption or a heterobifunctional cross-linker such as sulfo-SMCC. The most common method for biotinylating a molecular recognition element is the coupling of sulfo-NHS-biotin or NHS-(PEG)n-biotin to a primary amine. Although streptavidin–biotin bioconjugation is generally easy, a (PEG)n spacer may be included to optimize for the depth of the streptavidin binding pocket (9 Å) as well as the low relative solubility of biotin.206
Nanoparticle Characterization
Characterization of nanoparticle conjugates is essential for determining the success and extent of particle functionalization, which can drastically affect particle performance in a diagnostic format. There are several characterization techniques applicable to the development of detection probes for diagnostics. UV–visible spectroscopy (UV–vis) is one of the most commonly used methods for biomolecule-functionalized nanoparticles given its simplicity and applicability to the detection of nucleic acids and proteins.207,208 Particle size and distribution can be assessed using dynamic light scattering (DLS) and transmission electron microscopy (TEM),209−211 while functional group characterization on the nanoparticle surface can be evaluated with NMR and/or infrared (IR) spectroscopy.212−215 All of these analytical methods have trade-offs with respect to cost, sensitivity, time of analysis, and complexity of sample preparation. However, because many of the techniques offer complementary information, they can be used in combination to ensure an extensive characterization of inorganic nanoparticle detection probes.
The principal concern associated with antibody modification is decreased antigen-binding activity as a result of overfunctionalization of the variable region of the antibody. Optimization of the bioconjugation reaction minimizes this effect by achieving a balance between sufficient nanoparticle coupling efficiency and retention of antigen binding. The extent of antibody modification can be monitored with functional group-specific chromophores that are detectable by UV–vis, such as Ellman’s reagent for free-thiol detection or HABA (4′-hydroxyazobenzene-2-carboxylic acid) for biotin detection.192,198 Also, biosensor techniques, such as surface plasmon resonance (SPR), quartz crystal microbalance (QCM), and BLI, can be employed to assess the impact of modification on the binding affinity for the antigen.216−218
Decreased target affinity can also originate from suboptimal loading of antibody or nucleic acid on the nanoparticle surface. Insufficient coverage may lead to the nonspecific binding of interferents, whereas overloading the nanoparticle with molecular recognition elements can result in steric effects, decreasing target binding. Simple absorbance-based supernatant assays that measure antibody or nucleic acid concentration both before and after nanoparticle functionalization reactions give an estimation of the element’s surface density and aid in determining the nanoparticle surface saturation.207,208 The straightforward nature of these procedures make them popular strategies for characterizing the extent of functionalization of nanoparticle conjugates for use in infectious disease diagnostics.
The bioconjugation chemistry for attaching a molecular recognition element to a nanoparticle surface is the key component for transforming a nanoparticle with interesting signaling properties into a functional reagent that can be employed in diagnostic assays. The characterization of the nanoparticle postfunctionalization is critical to ensuring optimal signal in downstream diagnostic application. Several classes of functionalized nanoparticles can be utilized in developing POC diagnostics for infectious disease, the choice of which depends on the use-case scenario and desired signal output. Sections 4.2.2–4.2.6 describe the fundamental principles and POC applications for these classes of nanoparticles.
4.2.2. Noble Metal Nanoparticles
Both gold nanoparticles (AuNPs) and silver nanoparticles (AgNPs), collectively referred to as noble metal nanoparticles, possess many unique properties that make them advantageous probes for signal generation in infectious disease diagnostics. Noble metal nanoparticles display a vivid color that is observable with the naked eye due to surface plasmon resonance (SPR). The phenomenon of SPR, illustrated in Figure 19A, occurs when incoming photons strike the nanoparticle surface and generate a dipole that causes electron oscillations (i.e., surface plasmons) with a frequency that resonates with the frequency of the incoming light. Visible light fulfills this resonance condition for noble metal nanoparticles, giving rise to large molar extinction coefficients (on the order of ∼109 M–1cm–1) at visible wavelengths.219 Moreover, a nanoparticle’s color and λmax can be manipulated by controlling its size (Figure 19B).220,221 Noble metal nanoparticles are also easily functionalized with antibodies, aptamers, or oligonucleotide probes to afford specificity to a target biomolecule.177,188 Altogether, these qualities have made noble metal nanoparticles a frequent choice for colorimetric labels in diagnostic assays designed to be interpreted by the naked eye.
Figure 19.
Surface plasmon resonance in noble metal nanoparticles. (A) Incident electromagnetic radiation at visible wavelengths induces electron oscillations (surface plasmons) on the nanoparticle surface that are in resonance with the frequency of the incident light. (B) Effect of nanoparticle size on λmax. Adapted from ref (221). Copyright 2006 American Chemical Society.
It is important to note that we have limited further discussion of AuNPs since their use in diagnostics and sensing applications has been extensively reviewed.177,219,222−226 Given the vast literature that exists for AuNP applications, we have elected to focus on the applications of AgNPs as signal generation probes for infectious disease diagnosis. Similar to AuNPs, localized SPR of AgNPs has been capitalized upon to develop colorimetric assays for infectious disease protein and nucleic acid biomarkers.227−231 Additionally, the conjugation of fluorescent probes (e.g., FAM) to AgNP surfaces, as well as the intrinsic luminescence of some AgNPs, has permitted the development of luminescence-based assays.232,233 The high AgNP conductivity has also been exploited for electrochemical detection of nucleic acids.234−236 Lastly, surface-enhanced Raman spectroscopy (SERS) of AgNPs has been employed for the direct detection of malaria-infected red blood cells because of the significant light scattering of AgNPs and the high specificity of Raman signatures.69,237−240
Colorimetric detection using nanoparticles in paper-based assays has been a widely implemented method for signal readout in primary healthcare settings. In fact, the success of lateral flow assays can be credited in part to the nanoparticle-based detection systems that allow for visual interpretation of results by the end user. The use of functionalized AgNPs as colorimetric detection probes in paper-based diagnostics for infectious disease has been reported for both protein and nucleic acid biomarkers.227,241 For example, Yen et al.229 employed AgNPs in a multiplexed lateral flow assay for detection of dengue, yellow fever, and Ebola virus. The authors synthesized 30-, 41-, and 47 nm AgNPs, which were visually detectable as orange, red, and green, respectively, due to size-dependent SPR shifts. Each nanoparticle of a particular size was specifically functionalized with a distinct target-specific antibody associated with one of the viruses, allowing for detection of all three diseases in a single assay (Figure 20). The detection limit for each biomarker was found to be 150 ng/mL, which is within clinically relevant ranges for each disease but is higher than other published methods.242,243 Such multiplexed diagnostic assays are critical for discriminating between febrile illnesses at the point of care to allow for selection of the appropriate treatment course.
Figure 20.
(A) Silver nanoparticles of different sizes and colors (orange, 30 nm; red, 41 nm; green, 47 nm). (B–D) TEM images of the silver nanoparticles of increasing size. (E) Antibody “sandwiches” that are formed at the LFA test line, where the three different colors of AgNPs were functionalized with three distinct target-specific antibodies. (F) Multiplexed LFAs for detection of yellow fever, Ebola, and dengue virus protein biomarkers on a single test strip. Adapted with permission from ref (229). Copyright 2015 Royal Society of Chemistry.
Colorimetric AgNP aggregation assays also rely on the size-dependence of SPR. When AgNPs aggregate, the decrease in distance between nanoparticles results in interparticle SPR, producing a large red-shift in absorption and distinct color change that is visible to the unaided eye.222,227 These reaction mechanisms are classified as either “cross-linking” or “non-cross-linking.” In cross-linking assays, AgNPs are functionalized with both molecular recognition elements and capping ligands that stabilize the nanoparticles in solution. Addition of a sample of target biomarker results in cross-linking of the functionalized nanoparticles, drastically decreasing the nanoparticle–nanoparticle distance and producing in a spectral shift.244,245 AgNP aggregation assays in this format have been implemented for detection of both infectious disease protein228 and nucleic acid230 biomarkers; however, to the best of our knowledge, cross-linking-based AgNP aggregation has yet to be employed in paper diagnostics or other low-resource platforms.
In non-cross-linking aggregation assays, nanoparticles are either aggregated or stabilized with molecular recognition elements that electrostatically adsorb to the nanoparticle surface. Subsequent addition of a sample containing the diagnostic target then either disrupts or promotes nanoparticle stabilization, producing an observable color change.222 For example, Teengam et al.231 developed a non-cross-linking aggregation assay using AgNPs for multiplexed visual detection of Middle East respiratory syndrome coronavirus, Mycobacterium tuberculosis, and human papillomavirus oligonucleotides in a microfluidic paper analytical device (μPAD). The assay lacked the sensitivity required for detection of pathogenic nucleic acids; however, most sensitive nucleic acid diagnostics incorporate amplification techniques (e.g., PCR) that require extensive resources and infrastructure only available in better-equipped laboratories.246 Nonetheless, the use of a AgNP aggregation-assay in a μPAD enabled multiplexed visual detection of three different nucleic acid biomarkers in a format that could be applicable to a primary healthcare setting.
Taking advantage of the intrinsic fluorescence of some AgNPs, Kurdekar et al.233 developed a fluorescent immunoassay for detection of p24 that utilized a streptavidin-coated AgNP as the detection probe. Using a standard well-plate immunoassay format, adsorbed anti-p24 antibodies captured p24 from samples. A biotinylated anti-p24 detection antibody and the streptavidin-conjugated fluorescent AgNP were subsequently added to generate a fluorescent signal that correlated with p24 concentration. The assay successfully detected p24 standard spiked into plasma as well as the p24 present in HIV-positive patient plasma samples. Moreover, no cross reactivity was observed in samples that were HIV-negative but HBV- or HCV-positive. The limit of detection of the assay was 10 pg/mL, which is an order of magnitude higher than a previously discussed assay for p24 (section 4.1.2).153 This discrepancy could be explained by the lack of signal amplification in the fluorescent AgNP-based immunoassay. In its current format, the fluorescent AgNP immunoassay is not amenable to a resource-limited primary healthcare setting; however, fluorescent AgNPs could be employed in a lateral flow assay format and combined with a portable fluorescent reader, allowing for implementation at the point of care.247
4.2.3. Quantum Dots (QDs)
Quantum dots (QDs) are semiconductor nanocrystals with luminescent and conductive properties that make them advantageous as detection probes in infectious diseases diagnostics. Canonically, QDs consist of combinations of elements from groups II and VI or groups III and V and range in size from 1 to 100 nm.248,249 At these small sizes, QDs absorb photons, generating excited electrons in the conduction band and positively charged holes in the valence band of the semiconductor (Figure 21A). The distance that separates the electron–hole pair is confined to less than the diameter of the nanocrystal, resulting in “particle in a box” behavior known as quantum confinement.249−251 As a result of quantum confinement, QD emission wavelength increases as nanoparticle size increases; thus, QD emission can be tuned via size-controlled synthesis (Figure 21B).252 Because QDs exhibit absorption across a broad spectrum yet have emission spectra with very narrow and tunable bandwidths, they are particularly useful for multiplexed detection of disease biomarkers.194,253 Additionally, QDs demonstrate increased biocompatibility, photostability, quantum yield, and semiconducting properties compared to traditional organic fluorophores, further rendering QDs advantageous as signal generation probes for diagnostics.254−256 QDs have been employed for the detection of biomarkers for Ebola virus,257 tuberculosis,258,259 HIV,253,258,260 malaria,261−264 avian influenza virus,265Escherichia coli,266 and cholera267,268 in fluorescent assays for laboratory and resource-limited settings.
Figure 21.
(A) Quantized energy levels of the valence band and conduction band in a quantum dot (semiconductor nanocrystal). (B) Broad absorption spectra and narrow emission spectra of quantum dots demonstrate the size-dependence of QD absorption and emission. Adapted from ref (252). Copyright 2010 American Chemical Society.
Functionalized QDs or QD-loaded microparticles have been utilized as fluorescent probes for both protein and nucleic acid biomarkers.194,256,269 The surface of QDs can be functionalized with antibodies or antigens for protein detection or oligonucleotide probes for target DNA detection using the bioconjugation strategies discussed in section 4.2.1. For a thorough evaluation of the bioconjugation of QDs, the reader is directed to a number of reviews.194,196,270 Goldman et al.271,272 pioneered the earliest uses of antibody-functionalized QDs as fluorescent detection probes. The authors utilized affinity-based coupling chemistry (i.e., biotin–avidin) as well as electrostatic interactions between charged moieties to functionalize antibodies to the metal surfaces of CdSe/ZnS core–shell quantum dots. The biotin–avidin system was found to be the more simple, reproducible, and robust method for QD bioconjugation of the two, and these functionalized QDs were successfully utilized in a multiplexed fluorescent assay for cholera toxin and staphylococcal enterotoxin B.272 These early works exploited the narrow emission profile and enhanced quantum yield of QDs for antigen detection and laid the groundwork for the development of enhanced, fluorescence-based diagnostics for a variety of targets.
Building upon this previous work, Klostranec et al.253 constructed a multiplexed fluorescence-based microfluidic device for detecting HIV, hepatitis B, and hepatitis C antibodies utilizing antigen-functionalized QD barcode particles. Two types of CdSe/ZnS QDs were synthesized and loaded into polystyrene beads at varying ratios, resulting in three distinct QD barcode particles. Each QD barcode particle was covalently bound to a disease-specific antigen via EDC coupling. The assay employed a “sandwich” format where disease-specific antibodies were captured by the antigen-functionalized QD barcode particles, and a fluorophore-conjugated (AlexaFluor 488) antihuman antibody was used to detect the presence of a target antibody. The narrow peak width of QD emission enabled detection of both the antibody-conjugated fluorophore at ∼520 nm to indicate the presence of target antibody as well as both of the QDs at 570 nm (yellow) or 615 nm (red). The ratio of the red:yellow signal intensity allowed for barcode identification and determination of which of the three target antibodies was present (Figure 22).253
Figure 22.
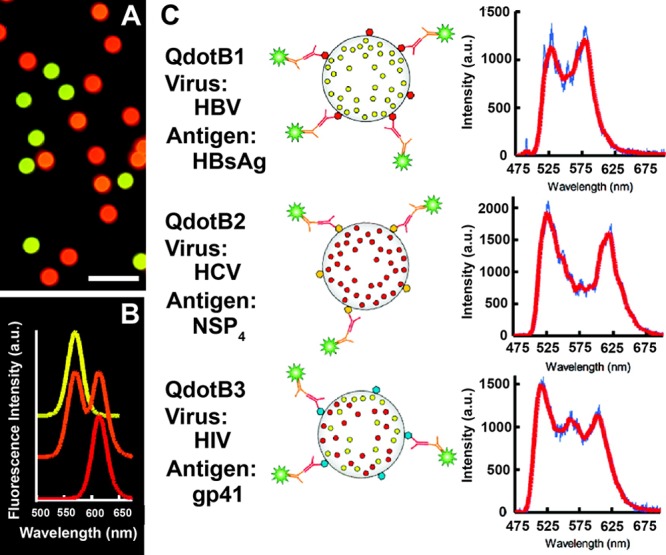
Quantum dot barcodes for fluorescent detection of hepatitis B virus (HBV), hepatitis C virus (HCV), and HIV antibodies. (A) Fluorescent image of the three different QD barcode particles utilized. (B) The unique fluorescent profile for each of the QD barcode particles. (C) Illustrations of each QD barcode particle conjugated to an antigen and their corresponding fluorescence spectra. The peak at 520 nm correlates to the presence of the fluorescent dye, indicating the formation of the antibody sandwich, while the peaks at 570 nm, 615 nm, and the combined peaks at 570 and 615 nm represent the HBV, HCV, and HIV antigen-functionalized QD barcodes, respectively. Reprinted from ref (253). Copyright 2007 American Chemical Society.
The detection limits for antibodies correlating to all three diseases were in the picomolar range, which is a 50-fold improvement compared to commercial ELISAs.253 While the assay enabled multiplexed and sensitive detection of infectious disease biomarkers, the microfluidic platform employed by the authors required extensive user manipulations and complex instrumentation and analysis that would only be viable in higher level healthcare settings. Given the improvements in sensitivity of the assay, adaptation of the technique to a hand-held fluorimeter or cell phone-adapted fluorescence device could potentially allow for deployment in a primary healthcare low-resource setting. The benefit of this strategy is that the biomarker signal and barcode signal are both fluorescence-based, resulting in a multiplexed detection platform that is simplified compared to assays with different detection schemes for biomarkers and barcodes. The authors hypothesized that up to 106 barcode combinations are theoretically possible using this approach due to the narrow and distinct fluorescent peak widths generated by different QD particles, allowing for even greater multiplexing.253 However, such hypotheses should be approached with caution, as highly multiplexed assays can be more susceptible to cross-reactivity, thereby reducing the diagnostic specificity of the individual assays.
QD-loaded particles have also been functionalized with antibodies for the detection of protein biomarkers for infectious disease. Recently, Hu et al.257 developed a lateral flow assay that utilized antibody-coated polymeric particles (260.9 nm) loaded with both QDs (3–6 nm) and AuNPs (20 nm) for detection of Ebola virus (EBOV) glycoprotein. The reporter probes, which the authors dubbed “dual signal readout nanospheres” (RNs), enabled detection of the EBOV glycoprotein either visually via AuNPs or fluorescently via QD emission. Characterization of the RNs revealed that there were dozens of AuNPs adsorbed and hundreds of QDs embedded per every one antibody-coated polymeric particle (Figure 23, panels A and B); thus, for every one biomarker arrested at the test line, there were significantly more signaling moieties to permit more sensitive detection. Additionally, two sets of antibody-functionalized RNs were fabricated and used for each LFA: (1) streptavidin-conjugated and (2) biotin-conjugated RNs. Application of the sample and streptavidin-conjugated RNs to the sample pad resulted in migration of the sample and RNs to the test line and formation of an antibody “sandwich” at the test line. Subsequently, the biotin-conjugated RNs were added and bound to the streptavidin-coated RNs, enhancing the signal based on the high affinity of the biotin–streptavidin interaction.
Figure 23.
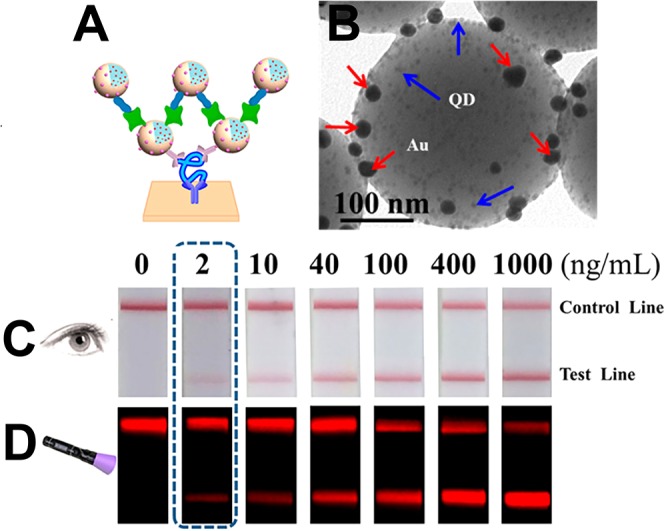
(A) Schematic of the lateral flow assay for detection of EBOV glycoprotein. Polymeric particles containing gold nanoparticles (AuNPs) and quantum dots (QDs), which the authors named dual readout nanospheres (RNs), were utilized as labels for signal readout. (B) TEM image of the dual readout nanosphere, showing AuNPs and QDs adsorbed to the polymeric particle. (C) Titration of EBOV glycoprotein, using the visual detection of AuNPs for signal readout. (D) Titration of EBOV glycoprotein, using the fluorescence of the QDs for signal readout. Adapted from ref (257). Copyright 2017 American Chemical Society.
The authors257 utilized the LFA to detect either the EBOV glycoprotein or the whole virion spiked into samples. The assay demonstrated a broad dynamic range that spanned 3 orders of magnitude for detection of AuNPs with a limit of detection by visual inspection of 2 ng/mL. When the QD fluorescence signal was used, the detection limit was improved to 0.18 ng/mL (Figure 23, panels C and D). The fluorescent detection of this assay was 1–2 orders of magnitude more sensitive than 2 commercial brands of rapid tests for EBOV glycoprotein. Moreover, the fluorescent signal could be quantitated by image processing software that is readily available on a smartphone. Though the authors utilized a benchtop fluorimeter for QD excitation and emission, the use of a smartphone adapted hand-held fluorimeter or dedicated fluorescent lateral flow reader could enable measurements at the point of care.257 The sensitive dual signal readout provides versatility to this platform such that it could be employed in a number of use-case scenarios. If morbidity control were the primary concern, visual detection of AuNPs would suffice. If more sensitive and quantitative measurements were needed for surveillance, intervention management, or an elimination campaign, the fluorescent capabilities of the detection probes could be exploited. However, for a given use-case scenario, likely only one signal readout modality is needed. As a consequence, the additional cost of implementing a dual signal RN, when using a single signal (visual or fluorescent) RN would suffice, needs to be considered. Nonetheless, the assay demonstrated that quantum dots could be incorporated into tests that are readily deployable in primary healthcare settings, though more rigorous field evaluation and validation of the assay would be necessary for clinical use.
There have been significant advances in the use of QD-based detection methods for infectious disease biomarkers. While the complexity of instrumentation required for detecting quantum dots is often still a barrier to their implementation at the point of care, developments in smartphone-based devices are currently enabling the detection of fluorescent labels such as QDs at the point of care.273−275 Fluorescent assays have demonstrated a capacity for multiplexing. Because of the narrow emission profiles demonstrated by QDs, the potential for continued expansion in multiplexing likely lies with QD-based fluorescent assays. Given the importance of detecting comorbidities and coinfections or discriminating between febrile illnesses,276,277 the QD-based technologies represent a platform that has the potential to make a significant impact at the point of care.
4.2.4. Lanthanide Chelate-Doped Nanoparticles
Nanoparticles doped with lanthanide chelates are advantageous as detection labels owing to their large Stokes shifts, sharp emission peaks, stable luminescence, long fluorescence lifetime, and biocompatibility.278,279 In a lateral flow assay format, these particles are often functionalized with target-specific antibodies and then embedded in the conjugate pad of the test. Like QDs, detection of lanthanide chelate-doped nanoparticles requires additional instrumentation for particle excitation and measurement of emission.280,281 Despite this need for additional equipment, these particles are attractive labels for POC diagnostics because they enable highly sensitive biomolecule detection. In the context of elimination or surveillance of a particular infectious disease, the added sensitivity provided by lanthanide chelate-doped nanoparticles could allow for detection of low-intensity infections that would otherwise be missed.
Except lanthanum, all lanthanides are f-block elements; the 4f orbital is gradually filled as the atomic number increases. Because the 4f orbital is largely shielded from the chemical environment by the filled 5s and 5p shells, the lanthanides are very similar in their chemical properties. For example, the most common oxidation state for aqueous lanthanide ions is +3.282,283 Most of these ions are luminescent, though some lanthanides have greater quantum yields than others due to the large energy gaps between the lowest emissive energy level and the highest sublevel of the ground state. As the size of the energy gap increases, lanthanide ions become less prone to lower energy nonradiative decay processes, resulting in greater quantum yields.278 Eu3+ is the most commonly utilized lanthanide for labels in lateral flow assays because of its (1) ideal energy gap between the nondegenerate emissive state of 5D0 and the ground state of 7FJ and (2) convenient emission in the visible region. Consequently, Eu3+ will be the focus of this section.279
Europium(III) itself is not strongly luminescent because most excitations involve forbidden electric dipole f-f transitions, which have low absorption cross sections.278 However, this can be overcome when the ion is coordinated to a sensitizing structure, such as an organic ligand. A simplified Jablonski diagram of the luminescence process for a Eu3+ chelate is depicted in Figure 24. The ligand absorbs the excitation energy and transfers it to a triplet state (T1) via intersystem crossing (ISC). Next, this energy is transferred to one of the 5DJ levels of Eu3+, and, after internal conversion (IET), emission occurs from the 5D0 energy level to any of the 7FJ levels.278,279
Figure 24.
Jablonski diagram of Eu3+ chelate luminescence process. Reprinted with permission from ref (278). Copyright 2014 IOP Publishing Ltd.
In addition to sensitization, chelating ligands also promote luminescence by shielding Eu3+ from aqueous quenching effects and allow for coupling to biomolecules or polymeric materials.278 Chelating ligands typically absorb in the UV (330–370 nm), and the subsequent Eu3+ emission occurs at 580–690 nm.279 Ligands usually contain a chromophore portion (e.g., pyridine, salicylate, phenanthroline, coumarin, pyrazole, triphenylene, and quinolone) and a chelating structure based on multidentate polyacids or macrocycles (e.g., EDTA).278 Tetracycline has also been employed as a ligand for Eu3+.284
These chelate dyes are incorporated into nanoparticles functionalized with target-specific molecular recognition elements for use in lateral flow assays. Most lateral flow assays that employ Eu3+ chelates use commercially available particles, which are often superior due to their monodispersity and quality control standards. The majority of commercially available Eu3+ chelate lateral flow labels are 100–300 nm carboxyl- or streptavidin-decorated polystyrene particles and are readily available from large vendors such as ThermoFisher285 and Expedeon Ltd.286 Eu3+ chelates are easily adsorbed into polymeric (e.g., polystyrene) nanoparticles based on hydrophobic interactions between the polymers and the organic chelating ligands of the complexes. For example, Huhtinen et al.287 found that simply incubating 50 nm polystyrene particles with Eu3+ chelated with dipicolinic acid derivative ligands resulted in 1000–2000 chelates per nanoparticle. This represents a density of approximately 0.02 chelates/nm3. Härmä et al.288 found a similarly high density of approximately 0.05 chelates/nm3 (31000 chelates/particle) in 107 nm commercially available particles. Thus, for each biomolecule bound in a lateral flow format, thousands of associated Eu3+ chelates produce a signal, resulting in a highly sensitive measurement.
The advantages of Eu3+ chelate-loaded polystyrene nanoparticles as labels for lateral flow assays were demonstrated by Yeo et al.,289 who developed a sensitive lateral flow assay for the detection of avian influenza H7 subtype virus. These viruses can be highly pathogenic; for a 2013 H7N9 outbreak in China, a mortality rate of roughly 36% was estimated for hospital patients with laboratory-confirmed infections.290 Currently, laboratory-based molecular testing is required to differentiate between H7 subtype avian influenza and other viral infections. Thus, there is a need for rapid tests that can be performed at the point of care. The authors approached this problem by developing and evaluating a panel of 10 H7 subtype-specific monoclonal antibodies against hemagglutinin 1 of H7N9 influenza. After the best antibody pair was determined, H7 subtype-specific lateral flow assays were constructed with both europium-chelate and gold nanoparticle (AuNP) conjugates. Using a benchtop time-resolved fluorescence reader, the authors found that the detection limits for the Eu3+ chelate-based tests were 25-fold better than those of AuNP-based tests. Additionally, the developed Eu3+-based tests performed better than commercially available tests detecting influenza A nucleoprotein.289 These enhancements over gold particles are impressive, although given the high density of chelates per nanoparticle, it is surprising that larger enhancements were not achieved. One possible explanation could be the efficiency of molecular recognition element adsorption for the two types of nanoparticles.
In addition to avian influenza,289,291 Eu3+ chelate-embedded polymeric nanoparticles have been employed for the detection of E. coli,292 hepatitis B,293 and procalcitonin,281 a host marker for bacterial infections. Several commercial platforms based on these particles have been developed as well, including the Sofia Influenza A+B tests (Quidel, Inc.)294−299 and the aQcare Chlamydia TRF test (Medisensor, Inc.),300,301 which have been extensively evaluated in the literature. The high sensitivity of these assays is derived not only from the high number of Eu3+ chelates embedded in the polymeric nanoparticles but also from the use of time-resolved fluorescence measurements. One of the primary disadvantages of using luminescent probes as labels in lateral flow assays is the high autofluorescence of biological samples and assay components occurring at excitation and emission wavelengths similar to those of the probes.302 This fluorescence can interfere with lateral flow readings, causing high background that leads to low-sensitivity measurements. However, lanthanide chelates overcome these effects because their fluorescence lifetimes are several orders of magnitude longer than conventional fluorescent materials (microseconds or milliseconds compared to nanoseconds).278,279 This allows for the use of time-resolved measurements in which samples are excited with a pulse of UV light, and emission signal is collected after a time delay on the order of 100 ns for a given time period.284 By delaying the collection of emitted signal, the short-lived autofluorescence from biological and assay components is excluded from measurements. While the intensity of the delayed luminescence is slightly decayed from the initial signal output of the chelates, these measurements can be cycled to improve signal-to-noise.303 Of course, the unique instrumentation required for time-resolved measurements makes field-deployment technically challenging, though currently available benchtop devices could allow for measurements in district clinics and laboratories. Recently, Paterson et al.304 developed a promising smartphone-based platform for time-resolved luminescent measurements, which could allow for point-of-care measurements based on Eu3+ chelate particles. However, the lengthy 100 ms time-delay employed in their device is only compatible with the most persistent luminescent nanophosphors such as crystalline SrAl2O4:Eu2+,Dy3+ particles.
While polymeric Eu3+ chelate nanoparticles are the primary materials employed by lanthanide-labeled lateral flow assays, these particles present some disadvantages. Their large size and tendency to swell can cause aggregation in aqueous solutions. Additionally, since they are not covalently bound to the polymers, Eu3+ chelates can leach from the particles.284 Silica particles avoid some of these problems. In one example, Xia et al.305 developed a lateral flow assay for hepatitis B detection using Eu3+ chelate-loaded silica nanoparticles. To synthesize these particles, the authors relied on an iterative method involving silica condensation from TEOS, surface-amination using APTMS, covalent functionalization of chelating ligands to the particles, and loading of Eu3+. This process was repeated five times to achieve a high density of Eu3+ chelates. The resulting 55 nm nanoparticles contained a remarkable 686000 chelates/particle, corresponding to a density of roughly 7.9 Eu3+ chelates/nm3, more than 100 times greater than commercially available polymeric particles.306 These particles were then functionalized with oxidized dextran to serve as a hydrophilic linker, and free amine moieties on antihepatitis B surface antigen (HBsAg)-specific antibodies were coupled to the surface via reductive amination of the aldehyde groups.
After synthesis and characterization, the particles were used as labels in a traditionally formatted lateral flow assay for HBsAg. To measure signal, the strips were illuminated with a UV lamp and imaged using a digital camera. Signal intensity was determined using image analysis in Adobe Photoshop. Unlike the previous example presented, this imaging technique does not take advantage of the long fluorescence lifetime of Eu3+ chelates. However, even without time-resolved measurements, the detection limit of the LFA was found to be 30 pg/mL HBsAg, 100-fold better than a similarly formatted gold-based lateral flow assay.305 The study was then validated by analyzing 286 clinical serum samples using the developed Eu3+ chelate-based lateral flow assay as well as a quantitative ELISA. The two methods were concordant for all samples analyzed and had similar sensitivities.305 Thus, the developed method represents an innovative tool for diagnosis of hepatitis B and provides sufficient sensitivity when compared to clinical methods. In order to implement this assay directly at the point of care, two modifications are necessary: (1) the assay should be optimized for field-friendly whole-blood specimens, and (2) a mobile phone-based detection strategy should be implemented.
Lanthanide chelate-based nanoparticle probes are a promising avenue for developing highly sensitive lateral flow assays. Similar to other fluorescent probes, instrumentation remains the primary barrier to implementation of Eu3+ chelate nanoparticles at the point of care. However, as optical devices are miniaturized and become more affordable, these probes could become commonplace among rapid tests for infectious diseases.
4.2.5. Up-Converting Phosphor Nanoparticles
Up-converting phosphor (UCP) nanoparticles represent a unique and exciting reporter technology for bioassays. Unlike typical fluorescent labels, these ceramic nanoparticles doped with rare earth elements absorb low-energy IR radiation and emit higher-energy visible light.307−309 UCPs are often superior to conventional colorimetric and fluorescent reporters for a number of reasons. First, the high anti-Stokes shift from IR to visible does not occur in nature. Thus, the inherent autofluorescence typically associated with biological samples and assay components does not interfere with UCP measurements. Second, UCP signal is highly stable and does not photobleach, allowing for indefinite storage and repetitive analysis of a single test. This is especially useful for confirming field results in a central laboratory.309 Third, assays are easily multiplexed; distinct particles absorb at the same IR excitation energy and emit wavelengths defined by the dopant. Finally, optical detection of UCP particles offers an inherent boost in analytical sensitivity over visual detection of gold; as few as 10–100 UCP particles can be detected at the test line of a lateral flow strip, while approximately 40000 gold particles are needed for visual detection.310 This increase in sensitivity makes UCP particles attractive labels for diagnostic tests employed in surveillance or elimination campaigns in resource-limited settings, since they would enable the detection of low-intensity or asymptomatic infections. However, similar to QDs and lanthanide-chelate particles discussed in the preceding sections, these benefits of UCP particles must be weighed against the additional cost and resource requirements needed for detection instrumentation.
The primary mechanisms for up-conversion include excited state absorption, energy transfer, and cooperative sensitization. Excited state absorption is the absorption of light by electrons already in an excited state. This requires two photons and equal separation between the ground state, first excited state, and second excited state of a single ion. Energy transfer up-conversion requires two ions, a sensitizer and an activator. In this process, the sensitizer ion is excited and sequentially transfers its energy to the ground state and first excited state of the activator ion.307 Cooperative sensitization is similar to energy transfer, though it involves excitation of two sensitizer ions which simultaneously transfer their energy to the activator ion. Other up-conversion mechanisms include photon avalanche and cross relaxation.311
Energy transfer up-conversion is the most common up-conversion mechanism for bioassay reporter particles, which typically involve a Yb3+ sensitizer (excitation 980 nm) and a rare earth (Er3+, Tm3+, Pr3+, Ho3+, and Gd3+) activator codoped in yttrium fluoride (YF3), yttrium oxide (Y2O3), yttrium oxysulfide (Y2O2S), or sodium yttrium fluoride (NaYF4) crystals.307,309 An illustration of the up-conversion processes for two of these materials are shown in Figure 25.
Figure 25.
Energy transfer from sensitizer Yb3+ to activator (A) Er3+ and (B) Tm3+. The primary green emission peak in Y2O2S:Yb3+, Er3+ requires two resonant energy transfer steps. After phonon relaxation, the double-excited Er3+ ion emits a single photon around 550 nm. The blue emission in Y2O2S:Yb3+, Tm3+ involves three phonon-assisted energy transfer steps.
UCPs have been utilized as labels for both protein and nucleic acid targets in lateral flow assays for a variety of infectious diseases. Recently, Corstjens et al.312−317 developed an ultrasensitive, UCP-based lateral flow assay for schistosomiasis that detects Schistosoma circulating anodic antigen (CAA), a proteoglycan biomarker waste product produced by the parasite. CAA is present in the serum and urine of patients with Schistosoma infections of all known species and has been found to correspond well with worm burden, clearing soon after successful treatment.312,317,318 The developed lateral flow assay employed 400 nm Y2O2S:Yb3+, Er3+ UCPs, which are excited at 980 nm and emit at 550 nm (green) and can detect CAA for a single Schistosoma worm pair in serum.314,317 This assay has been applied to patient samples from endemic areas, and due to its superior sensitivity, it has demonstrated much higher schistosomiasis prevalence than microscopy, serology, and nucleic acid-based tests.315−317,319
The CAA lateral flow assay format is similar to conventional lateral flow assays. CAA-specific antibodies are printed on the test line, and antimouse IgG is printed on the control line. The workflow of the assay in its current form, however, differs from a typical field-ready test. First, a trichloroacetic acid (TCA) extraction is performed on a urine or serum sample, requiring a centrifugation step. The extract supernatant is then combined with running buffer and anti-CAA-functionalized UCP particles and incubated for 1 h on an orbital shaker at 37 °C before the lateral flow strip is added to the solution. The test is allowed to develop and must dry completely (at least 3 h) before scanning and analyzing the strip.312,313 In order to simplify the preparation procedure and decrease the number of steps, a dry reagent kit was later developed in which all buffers and particles were lyophilized.313 This kit was shipped to a tertiary lab in South Africa, where nearly 2000 samples were evaluated by local technicians using both the kit and a CAA ELISA. The researchers found that the UCP-based LFA successfully identified more low-level infections than the CAA ELISA.320
To increase the analytical sensitivity of the assay, Corstjens et al.317 added a spin-filter concentration step to the sample preparation method. This allowed for CAA in urine sample volumes of 0.5–7.5 mL to be concentrated into 20 μL before addition to the lateral flow strip. The resulting detection limits improved as sample volume increased, reaching as low as 0.03 pg/mL for the 7.5 mL assay. To demonstrate clinical applicability, the concentration step was successfully performed on 2 mL patient urine samples from Kenya (high-endemic, S. mansoni) and China (low-endemic, S. japonicum)316,317 It is important to note that, though this sample concentration step improved the sensitivity of the assay, it required laboratory equipment to carry out; all patient samples in these studies were processed in well-equipped laboratories. Further, the additional concentration step increased the cost of this ultrasensitive CAA assay, though sample pooling could make this test more cost-effective and allow for monitoring of worm burdens at the subpopulation level for large-scale surveillance.320 For this UCP-based ultrasensitive assay to be utilized in a field setting, a more robust, field-ready sample preparation method is needed.
Lateral flow assays with UCP reporters have also been developed for detection of protein markers of other infectious diseases, including neurocysticercosis,321 tuberculosis,322,323 leprosy,324−328Yersinia pestis,329,330Brucella,331Vibrio anguillarum,332 hepatitis B,333 respiratory syncytial virus,334 and HIV.335 Additionally, UCP-based multiplexed lateral flow assays have been developed for detection of protein biomarkers for multiple infectious diseases.336,337 These multiplexed diagnostics typically consist of one strip with multiple test lines, each capturing a distinct protein biomarker. In this format, the UCP crystals are the same for each biomarker, though they are functionalized with target-specific antibodies. One disadvantage of highly multiplexed assays on a single strip is the potential increase in cross-reactivity and nonspecific binding that could lead to false-positive results. To mitigate this risk, Hong et al.330 developed a unique, circular cassette with 10 channels for 10 different singleplex lateral flow assays using UCP particles as the reporter labels. One unexplored application of multiplexed detection on a lateral flow assay is the use of UCP particles with identical excitation but differing emission profiles. This could be particularly advantageous for large biomarkers with multiple accessible epitopes and could provide additional clinical information. For example, an assay that captures and detects whole organisms could include a second UCP probe for detecting surface proteins that confer drug resistance, providing both detection and susceptibility results in one assay.
UCP nanoparticles have also been used in nonlateral flow diagnostic formats, including immunohistochemistry,338 microarrays,339 magnetic bead assays,340,341 and plate immunoassays,309 though these platforms are not readily amenable to low-resource, point-of-care settings. Even in the lateral flow format, UCP nanoparticles present interesting challenges for field deployment. Clearly, sample preparation methods, including purification, concentration, and amplification, must be adapted for environments lacking controlled laboratory conditions and should rely on as little electricity as possible. While hand-powered centrifuges342,343 and battery-powered mixers115,344 have been developed, the ideal POC assay will be optimized to preclude these additional resources. Another issue, often unaddressed, is that the lateral flow strips must be completely dry before optical measurements and analysis. This is because water also absorbs in the near-IR, decreasing excitation efficiency of the sensitizer.345 Typically, protocols require at least 3 h of drying time after the lateral flow assay has developed, a long time-to-result for a point-of-care test. Finally, detection of UCP particles relies on optical instrumentation. Although the most sensitive of these devices are benchtop instruments, portable battery-powered devices have been developed, such as the UCP-modified version of Qiagen’s ESEQuant lateral flow reader (“UCP-Quant”).313,314,346
Many of the sample preparation methods described in section 3 can be applied to UCP lateral flow formats. Additionally, innovations in hand-held optical readers discussed in section 5 require only simple adaptations to detect the anti-Stokes shift of UCP particles. There are clear advantages of using UCP nanoparticles as labels in lateral flow assays, including their inherently low background interference and high analytical sensitivity. Innovations in sample preparation methods and the development of portable optical readers will allow for these advantages to be exploited in low-resource, POC settings.
4.2.6. Magnetic Nanoparticles
While most frequently used for sample preparation and biomarker enrichment, magnetic nanoparticles are emerging as promising labels in POC assays. Similar to noble metal nanoparticles, magnetic nanoparticles can be easily functionalized and possess strong optical absorbance, which has led to their use as visual labels on lateral flow assays.347 However, their magnetic properties can also be leveraged for detection, which has advantages over traditional optical detection in a lateral flow format. First, when visual or optical detection of absorbent or fluorescent particles is employed, only signal from the surface (∼10 μm) of the membrane is measurable due to the opacity of the membrane. Typically, nitrocellulose membranes used in LFAs are 100–200 μm thick, so optical detection only allows for signal from the top 10% of the membrane to be measured. In contrast, magnetization measurements can be performed regardless of the opacity of the substrate, taking advantage of the entire volume of the test line on a lateral flow assay.348 In addition, metal-oxide nanoparticles are highly stable, and drying and aggregation do not influence the intensity of magnetic measurements.349 Finally, there are very few biological or assay components that interfere with magnetic measurements.350
The properties of magnetic particles are size- and temperature-dependent.195,351 Iron oxide (typically magnetite or maghemite) particles are the most common magnetic nanoparticles employed in diagnostic applications. In the bulk, these materials are ferri- or ferromagnetic and thus retain magnetization after an external field has been applied. This hysteresis reaches a maximum when particle size is decreased to the point that the material becomes single-domain. As particle size is even further reduced, the hysteresis effect decreases until the particles reach a critical diameter, below which Brownian forces are strong enough to overcome magnetic forces. Thus, at these small sizes, the particles are superparamagnetic; the magnetic moments of the particles are aligned in the presence of an external magnetic field, but they revert back to a nonmagnetic state when the field is removed.351
In the lateral flow format, magnetic nanoparticles are labeled with target-specific molecular recognition elements and employed as conjugates for analyte detection.352 Superparamagnetic behavior is ideal for particles applied as labels for lateral flow assays because the lack of magnetization in the absence of an external field allows for the particles to wick along the strip without aggregating due to magnetic effects.353 Then, the magnetic properties of the particles can be leveraged after labels have bound to the test and control lines. Magnetic nanoparticles are typically detected on lateral flow assays based on induction (Figure 26) or using magnetoresistive sensors.350 In the inductive format, which employs the principles of Faraday’s Law, a magnetic particle-based test is placed in an oscillating magnetic field above a set of induction coils. In the absence of magnetic particles, the net current induced in the coils is zero. However, when particles are present, the direction of their magnetic moments oscillates with the external magnetic field, resulting in a measurable net voltage induced across the coils that is proportional to the total number of particles at the test or control line.354,355 In contrast, magnetoresistive sensors are based on the principle that the electrical resistance of certain materials, which are incorporated in the sensors, can change upon application of an external magnetic field.350 In this format, a magnetic field is applied to para- or superparamagnetic detection elements in order to align their magnetic moments and produce a fringe field. Magnetoresistive sensors are placed close enough to the particles to detect the fringe field they produce based on the change in resistance of the materials within the sensor.356,357 A detailed description of the instrumentation required for these detection methods is provided in section 5.4.
Figure 26.
General format of an inductive sensor for the detection of magnetic nanoparticles on lateral flow assays.
Magnetic nanoparticles have been employed as detection labels in lateral flow assays for protein and whole-cell targets. For example, Handali et al.349 developed two magnetic particle-based lateral flow assays for detection of Taenia solium, a cestode that is prevalent around the world for which swine are intermediate hosts. When contaminated, undercooked pork is ingested, the parasites develop into tapeworms in the human gut (taeniasis). However, if eggs are ingested, the larval stage can infect the human nervous system, potentially forming cysts in the brain. This severe form of the disease, called neurocysticercosis, is a leading cause of epilepsy worldwide.358 The gold standard for diagnosis of taeniasis is observation of eggs in stool by microscopy, though this method is insensitive and labor-intensive. Diagnosis of neurocysticercosis currently requires CT scans of the brain, a technology that is unavailable in low-resource settings.359 As such, there is a pressing need for T. solium rapid diagnostic tests.
Handali et al.349 developed two serological magnetic bead-based lateral flow assays that detected the immune response (i.e., host antibodies) against taeniasis-specific (ES33) and cysticercosis-specific (T24) antigens. For each test, one batch of the recombinant antigen was printed at the test line, and a separate batch was coupled to commercially available carboxylated superparamagnetic particles via EDC/NHS chemistry. The recombinant antigen at the test line and the antigen conjugated to magnetic particles were able to simultaneously bind to host antibodies in the sample, taking advantage of the multivalent nature of anti-ES33 and anti-T24 IgM and IgG antibodies.
The lateral flow assays were evaluated using an induction-based reader and visual reading to detect antibodies in serum samples from endemic areas and nonendemic control regions (Figure 27).349 It was found that the taeniasis-specific ES33 assay had a diagnostic sensitivity and specificity of 95% and 96%, respectively, when evaluated with the magnetic reader. The sensitivity and specificity of the neurocysticercosis T24 test using the magnetic reader were 94% and 99%, respectively, for patients with two or more viable cysts as determined by CT scan or MRI of the brain. For patients with only one viable cyst, the T24 assay only identified 5/15 cases of neurocysticercosis, indicating that the burden of infection was not great enough to elicit an immune response.349 Despite this decrease in sensitivity for single-cyst infections, the developed lateral flow assays are promising alternatives to the current gold standards. Further, because the assays were not species-specific, they could be used to detect porcine cysticercosis as a marker of disease control and transmission. One disadvantage of utilizing superparamagnetic labels is that many magnetic readers are benchtop devices requiring electricity and a laboratory setting. However, in over 70% of the cases, the magnetic nanoparticles could be detected visually.349 While this sensitivity is not ideal, it is possible that initial visual results could be applied in the field with follow-up laboratory-based measurements assessed for the visually negative assays. As hand-held magnetic readers become more commonplace, these assays will become truly impactful.
Figure 27.
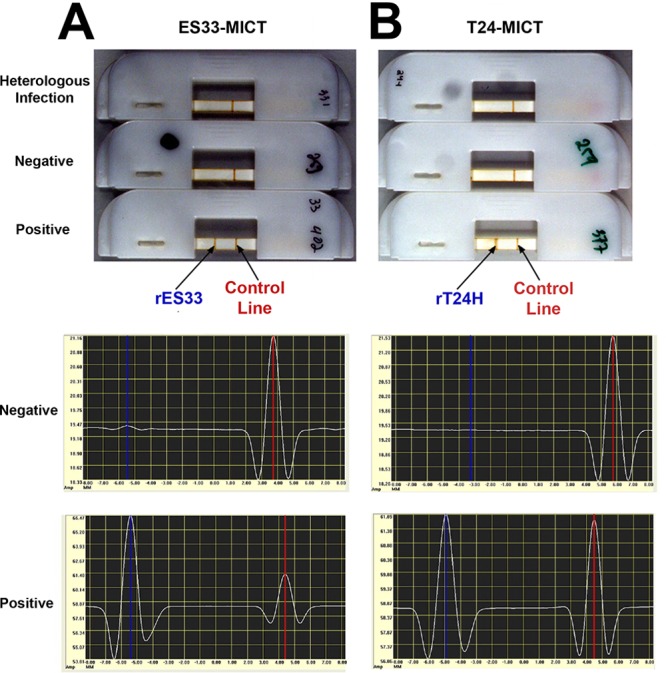
Superparamagnetic lateral flow assay for (A) taeniasis and (B) neurocysticercosis. The top image shows visual signal on lateral flow strips. Below are signal read-outs from negative (middle) and positive (bottom) strips using induction-based magnetic assay reader. Figure adapted with permission from ref (349). Copyright 2010 American Society for Microbiology.
In addition to T. solium, magnetic particles have been employed in protein-detecting lateral flow assays for HIV360,361 and H1N1 influenza.362 Whole cell-detecting magnetic particle-based assays have also been developed for a number of targets, including Vibrio parahemolyticus,363,364Listeria monocytogenes,365 and Bacillus anthracis.366,367 The demonstrated sensitivity of these magnetic particle-based lateral flow assays makes them promising alternatives to traditional assays with colorimetric or fluorescent labels. However, current instrumentation, which will be discussed further in section 5.4, limits their utility at the point of care. One exciting avenue that has yet to be explored for infectious disease detection is the employment of the same superparamagnetic nanoparticles for dual purposes: immunomagnetic biomarker enrichment from large-volume samples and visual labeling for lateral flow assays. Ideally, such a diagnostic would integrate sample preparation and assay into one device. Once developed, the combined effects of target enrichment and magnetic detection could lead to a highly sensitive test capable of detecting low-density infections.
4.3. Metalloenzyme Signal Amplification
One of the most widely utilized techniques for improving the sensitivity of diagnostics is signal amplification, where thousands of signaling molecules are generated for every one biomarker molecule. Though there are now numerous methods for amplifying signal in diagnostic assays, the use of metalloenzymes was one of the first approaches.368,369 Metalloenzymes are metal-containing catalytic proteins in which the presence of particular metals or metal complexes in the tertiary structure is critical to the catalytic turnover of the substrate.370 The predominant metalloenzymes employed in diagnostics are alkaline phosphatase (ALP), horseradish peroxidase (HRP), and catalase. The high catalytic efficiencies of these enzymes enable the rapid conversion of substrate to detectable products.371 Using cross-linking chemistry discussed in section 4.2.1, these metalloenzymes can be conjugated to antibodies or nucleic acids that afford specificity for a target biomarker.192,193
The three primary enzymes used in metalloenzyme detection conjugates rely on different metal ions for catalytic activity. ALP possesses two Zn(II) ions and one Mg(II) ion per enzyme monomer and hydrolyzes phosphate monoesters.372,373 The conventional substrates for ALP in point-of-care diagnostic applications are BCIP (5-bromo-4-chloro-3-indolyl phosphate) and NBT (nitro blue tetrazolium), where dephosphorylation and oxidation of BCIP allows for reduction of yellow NBT dye to a blue/purple formazan precipitate.371 Both HRP and catalase are heme-containing monooxygenases that catalyze reactions with H2O2.374−376 HRP catalyzes the oxidation of various organic substrates with concomitant reduction of H2O2. Conventional HRP substrates in diagnostic assays include DAB (3,3′-diaminobenzidine) and TMB (3,3′,5,5′-tetramethylbenzidine). Catalase, on the other hand, simply catalyzes the disproportionation of H2O2.371,376 Each enzyme has been applied in various diagnostics for infectious disease, canonically for spectrophotometric ELISAs.
4.3.1. ELISAs (Enzyme-Linked Immunosorbent Assays)
Metalloenzyme-antibody conjugates have been widely implemented in ELISAs for the detection of protein biomarkers for disease. This is due to the sensitivity afforded by the use of enzymes for signal amplification as well as the specificity of the antibody–antigen interactions used for molecular recognition.371,377,378 Despite the high sensitivities of ELISAs, their application in POC settings is limited by several factors:378−381 signal readout for ELISAs typically requires a spectrophotometer unavailable in a primary healthcare setting, though there are a growing number of portable alternatives382 (e.g., cellphones or hand-held scanners; see section 5.1.3). ELISAs are time-consuming (∼6–8 h), demand extensive sample handling and solution manipulation, and often require specialized training. Lastly, the lack of thermal and long-term stability of metalloenzymes in ELISAs can lead to suboptimal assay performance, as ELISAs are intrinsically dependent on these metalloenzymes for signal amplification and readout. Thus, ELISAs are usually limited to well-equipped tertiary laboratories. To mitigate these issues, investigators have begun to use paper as the ELISA solid support, increasing the likelihood that these sensitive assays could be used in resource-limited settings. Paper-based ELISAs have been developed for detection of diagnostic markers of HIV and malaria.381,383,384 Long-term stabilization conditions of metalloenzymes on paper have also been investigated to maximize catalytic activity in POC settings.385−387
Paper-based ELISAs seek to address many of the issues that inhibit the use of ELISAs in more resource-limited POC settings. The time to result for paper-based ELISAs is ∼1 h due to the comparatively smaller volumes required when contrasted with conventional benchtop ELISAs. These smaller reagent volumes also significantly reduce the overall cost of ELISAs. Cheng et al.381 first pioneered this method for the detection of serum antibodies against HIV-1 envelope antigen gp41, where an ALP-antibody conjugate was utilized to turn over BCIP/NBT substrates to purple precipitate products. The authors used hydrophilic paper that was patterned with hydrophobic photoresist to fabricate 96 test “wells” that were spatially separated just as in a polystyrene 96-well plate. Samples containing biomarker were added to the paper ELISA card, which was placed on top of a blotting pad to enable wicking of the reagents through the test wells. Though the assay boasted a faster time to result and a lower cost than the analogous conventional ELISA, the sensitivity of the paper-based ELISA was decreased by an order of magnitude compared to the conventional ELISA format.
Lathwal and Sikes383 conducted a systematic investigation of several signal amplification methods for paper-based colorimetric detection of malarial biomarker HRP2. To investigate multiple metalloenzyme–substrate combinations, the same capture antibody was immobilized on aldehyde-modified cellulose for all experiments, and the following detection systems were evaluated: (1) HRP with substrates DAB and H2O2, (2) HRP with substrates TMB and H2O2, (3) ALP with substrates BCIP and NBT, (4) Ag deposition onto AuNPs, and (5) polymerization-based amplification. Because all factors (i.e., capture antibody, capture antibody loading, biomarker, detection antibody) were held constant, the differences observed in each assay were presumed to be the direct result of the signal amplification system used. Positive and negative controls were evaluated at various time points for each signal amplification method to determine an optimal window for visual readout of the signal (Figure 28).383
Figure 28.
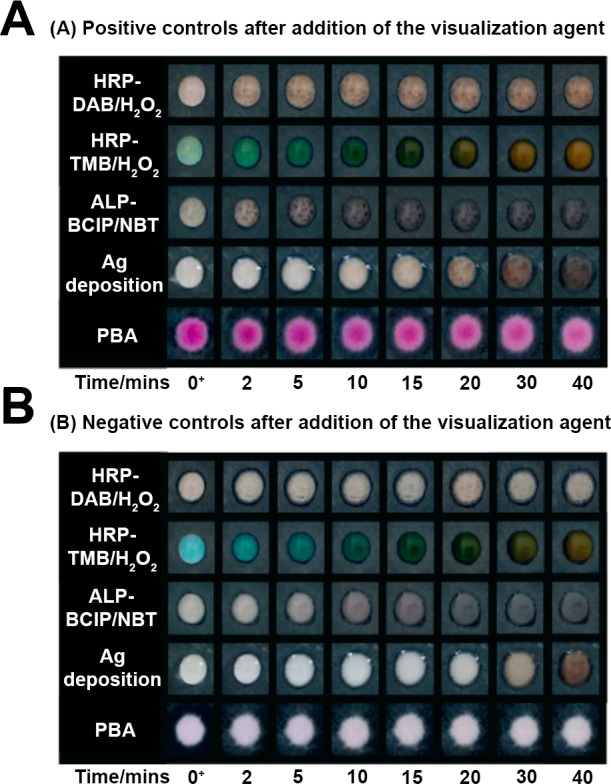
Systematic evaluation of various signal amplification methods for detection of malarial biomarker HRP2. Positive control samples (shown in A) contained HRP2, whereas negative control samples (shown in B) did not. The optimal time window for signal readout could be determined for each method, where the positive controls demonstrated detectable visual signal while the negative controls showed no detectable signal. Adapted with permission from ref (383). Copyright 2016 Royal Society of Chemistry (https://creativecommons.org/licenses/by-nc/3.0/legalcode).
All of the methods had an optimal time point for visual signal readout with the exception of HRP-TMB/H2O2, which produced significant false-positive signals and a color change from blue to brown that was difficult to interpret. Of the 4 remaining methods, the metalloenzyme–substrate combinations (HRP-DAB/H2O2 and ALP-BCIP/NBT) had detection limits an order of magnitude better than Ag deposition and polymerization-based amplification methods when quantified by RGB color intensity.383 Any portable colorimetric reader, even a cell phone, could enable the use of these highly sensitive metalloenzyme conjugates in a primary healthcare setting.
The advantages of paper-based ELISAs are potentially compromised by the instability of metalloenzyme conjugates, since denaturation of metalloenzymes leads to poor turnover of substrate and lower sensitivities.388 For this reason, notable research efforts have been devoted to optimizing long-term storage of metalloenzymes, particularly HRP, for use in microfluidic paper analytical devices (μPADs) or two-dimensional paper networks (2DPNs).385−387 One method involves vacuum drying or freeze-drying enzymes and substrates in the presence of trehalose, a disaccharide often used as a cryoprotectant because of its capacity to stabilize the protein’s interaction with solvents, forming a protective “glass” as it dries.389,390 Ramachandran et al. used a combination of trehalose, bovine serum albumin (BSA), and FeSO4-EDTA for vacuum drying of HRP-conjugated antibodies against malarial biomarker HRP2 into glass fiber pads.387 The stabilized HRP could be stored for 5 months at 45 °C under these conditions without a significant loss in catalytic activity. A glass fiber pad containing all of these reagents was incorporated into a 2DPN for detection of HRP2 with a LOD of 6.5 ng/mL, a value well within clinically relevant concentrations. This assay also demonstrated the utility of metalloenzymes in paper-based POC diagnostics, providing a signal amplification step that is rarely present in conventional paper diagnostics (e.g., lateral flow assays). These advances in enzyme stabilization when combined with simplified paper-based assay formats could potentially allow for ELISA sensitivity to be translated for direct use at the point of care.
4.3.2. Nanoparticle-Assisted Enzymatic Signal Amplification
The integration of noble metal nanoparticles has further augmented the signal amplification capabilities of metalloenzymes in infectious disease diagnostics. Nanoparticles have served as surfaces for the coupling of antibody-metalloenzyme conjugates and have been implemented in electrochemical sensors for detection of infectious disease-associated protein biomarkers.391−393 Zheng et al.392 developed an amperometric immunosensor for detection of HIV antigen p24 that employed a standard “sandwich” format. A glassy carbon electrode was modified with gold nanoparticles to allow for immobilization of anti-p24 capture antibodies. HRP-conjugated anti-p24 antibodies were used for detection, catalyzing the oxidation of substrate hydroquinone in the presence of H2O2. The reaction generated a reductive current at the electrode surface proportional to p24 concentration. The detection limit of the assay was 8 pg/mL, similar to the p24 assays153,233 previously discussed in sections 4.1.2 and 4.2.2. While the assay was robust to human serum samples spiked with p24, the serum samples tested contained a p24 concentration 3 orders of magnitude higher than the LOD.392 The currently required electrochemical workstation would limit the assay’s application to higher level laboratories. Nonetheless, this amperometric method demonstrated the value of integrating noble metal nanoparticles with metalloenzyme conjugates for signal amplification in diagnosis of infectious diseases.
Noble metal nanoparticles have also been utilized as colorimetric signaling probes in ELISAs for detection of protein biomarkers for disease.394−398 So-called “plasmonic” ELISAs utilize the standard “sandwich” assay format with an immobilized capture antibody and a detection antibody that is conjugated to a metalloenzyme. However, as opposed to using conventional metalloenzyme substrates for colorimetric detection, plasmonic ELISAs employ the enzyme as a kinetic tool for nanoparticle nucleation. This causes drastic shifts in SPR and in the absorbance spectra of the nanoparticles that are detectable with the naked eye.399
The plasmonic ELISA pioneered by de la Rica and Stevens396 utilized a catalase-conjugated detection antibody to control the growth of AuNPs in such a manner that was proportional to the concentration of HIV biomarker p24. In the absence of biomarker, bulk Au(III) was reduced by H2O2 to form stable, spherical AuNPs that appeared red in color. When p24 was present in a sample, the catalase-conjugated detection antibody was also present and catalyzed H2O2 disproportionation. The resulting decrease in H2O2 concentration led to kinetically slower nucleation of AuNPs, forming aggregated AuNPs with poorly defined morphology that appeared blue in color (Figure 29). The red-shift in the SPR was highly sensitive toward H2O2 concentration; thus, depletion of H2O2 with catalase that was only present in p24-positive samples allowed for very sensitive detection of p24 with the naked eye. The absorbance was also quantified using a simple spectrophotometric readout, yielding a detection limit of 1.0 ag/mL,396 nearly 6 orders of magnitude more sensitive than the previously discussed detection methods for p24 antigen.153,233,392 The authors also demonstrated that the plasmonic ELISA could detect p24 in HIV-positive serum samples, even identifying p24 in samples from HIV-positive patients with viral loads less than 50 copies/mL.396 The remarkable sensitivity of this assay for naked-eye detection of p24 provides the necessary improvement required for elimination campaigns and active case management. It would permit earlier detection of p24, leading to improved outcomes for HIV patients. The assay could be particularly impactful for the challenges associated with early infant HIV diagnosis in low-resource settings. The plasmonic ELISA presented by the authors still calls for extensive sample handling and user manipulation and is currently unsuitable to a primary healthcare setting. Adaptation of the previously discussed enzyme stabilization measures and integration of the assay to a field-ready format (e.g., paper-based ELISA or 2DPN) could enable its translation to an ultrasensitive field-ready diagnostic for HIV detection.
Figure 29.
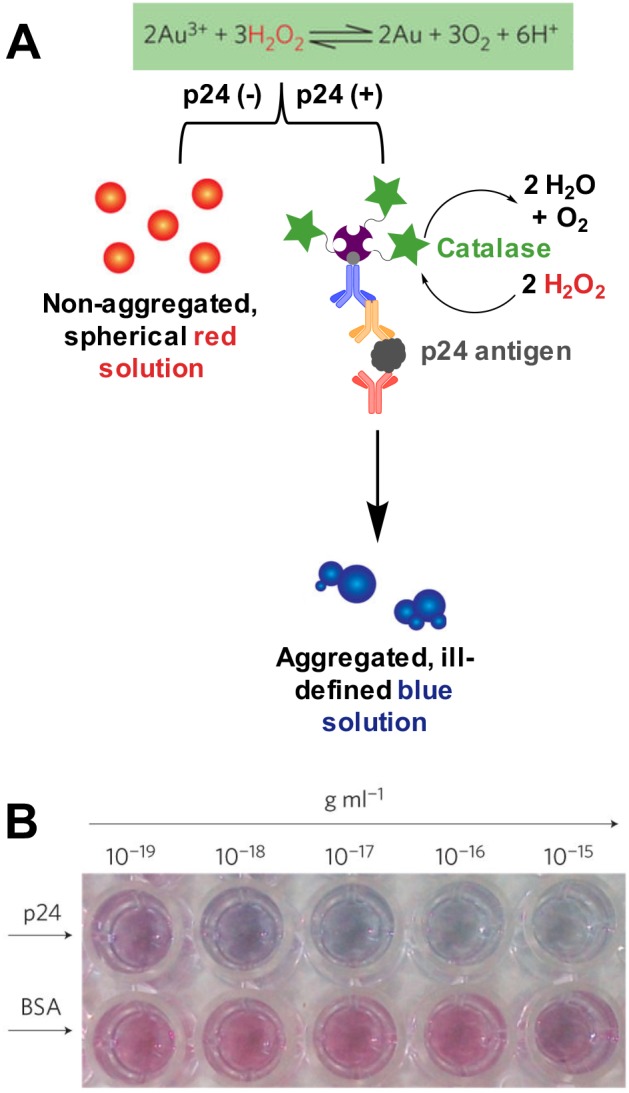
(A) Principle of the plasmonic ELISA. In positive samples, antibody-conjugated catalase depleted H2O2, causing a shift in SPR and absorbance of AuNPs from red to blue that is proportional to p24 concentration. (B) Naked eye detection of p24. The visual data demonstrated that the assay was specific for p24 versus a control protein (BSA). Adapted with permission from ref (396). Copyright 2012 Springer Nature.
Metalloenzymes play a critical role for signal amplification in a number of assays, enabling the sensitive detection of infectious diseases. Several research efforts have been aimed at translating the sensitivity of metalloenzyme-based assays to formats such as paper-based diagnostics that are amenable to resource-limited settings. Although the issue of enzyme storage in paper has been addressed to certain extent,385−387 the long-term stability of enzymes remains a concern for POC diagnostics, especially in climates with elevated temperatures and prevalent infectious disease. There are numerous methods that utilize nonenzymatic means for signal amplification that eliminate the issue of long-term enzyme storage altogether, including nanoparticles that act as enzyme mimics and other metal-based methods. These will be covered in the following section.
4.4. Metal-Based Signal Amplification
While the majority of signal amplification in bioassays is enzyme-based, several interesting metal-based amplification strategies have been developed. These strategies include metal nanoparticle dissolution, nanocrystal ion exchange, enzyme mimics, and reductive nanoparticle enlargement.394,400−403 Compared to enzymes, metal-based signal-amplification has the distinct advantage of long-term storage and thermal stability. However, many of these strategies suffer from the following drawbacks: (1) amplification often requires additional steps to be integrated into point-of-care test workflow, and (2) incorporation into a paper-based format can be technically challenging. However, innovative assay design can overcome these challenges. Integration of signal amplification into point-of-care tests can drastically improve analytical and diagnostic sensitivity, resulting in earlier diagnoses and detection of low-density infections. Therefore, diagnostics which incorporate novel and user-friendly signal amplification steps could fill a need for elimination campaigns and surveillance programs. This section reviews metal-based signal amplification strategies that show promise for application to low-resource infectious disease diagnostics.
4.4.1. Nanoparticle Dissolution and Cation Exchange
At the heart of signal amplification is the principle that, for each biomarker target captured in an assay, many signal-generating elements (particles, molecules, atoms, electrons, photons, etc.) are produced. In a typical nanoparticle-based assay, a single nanoparticle often indicates the presence of one biomolecule. However, a single nanoparticle is a densely packed lattice of thousands to millions of metal atoms, each of which, after nanoparticle dissolution or cation exchange, can be leveraged for signal generation (Figure 30).199,404−406
Figure 30.
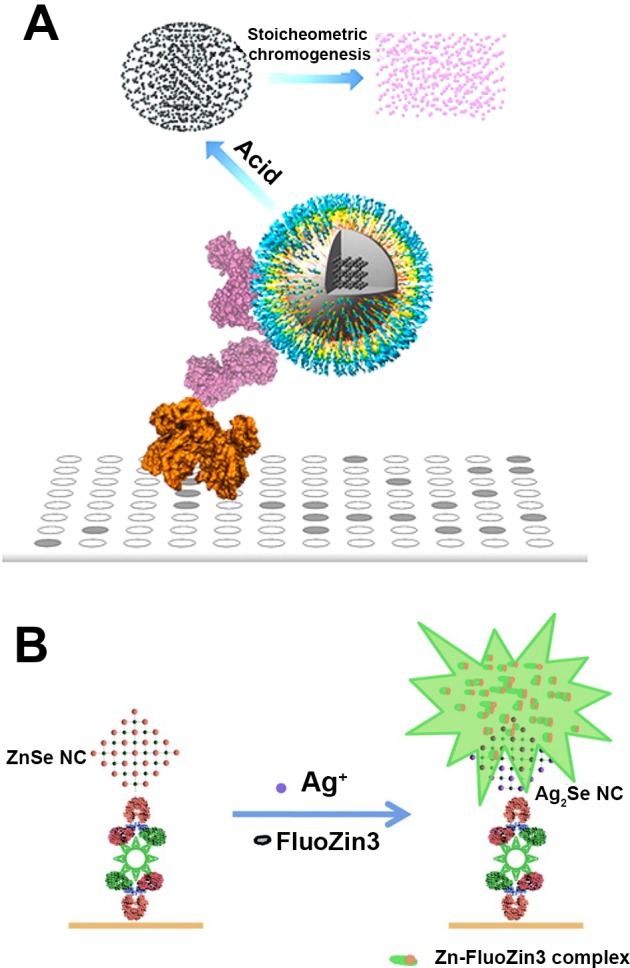
Principles of signal amplification by (A) nanoparticle dissolution and (B) nanocrystal cation exchange. Images adapted from refs (405) and (411). Copyright 2013 and 2011, respectively, American Chemical Society.
Metal ion chelating reagents that result in chromogenic or fluorescent signal have long been used for detection of trace metals for a myriad of applications.407,408 Recently, Tong et al.405 exploited ferrous ion-chelating ferrozine in an iron oxide nanoparticle-linked immunosorbent assay (ILISA). Antibody-functionalized Wüstite (FeO) particles were used as detection elements in direct, competitive, indirect, and sandwich well-plate assays. After functionalized iron-oxide particles bound to the target, the nanocrystals were treated with acid and reducing agents, resulting in stoichiometric conversion to ferrous ions. Thus, thousands of Fe2+ ions were released for each biomarker present. When excess ferrozine was added, solutions changed from transparent to purple upon chelation, with intensities directly proportional to the iron concentration in solution. Proof-of-concept assays detecting mouse IgG had low picomolar detection limits, similar to assays that depend on enzymatic signal amplification. To extend the applicability of their amplification technology, the group also demonstrated its use in a Western blot. By switching the chelating reagent to potassium ferrocyanide (Prussian blue), the reagent could easily be precipitated onto a cellulose membrane when iron oxide particles were used as detection elements.405 While well-plate assays are far from applicable to field settings, the ILISA system could be translated to an aptly designed paper diagnostic format.
Another avenue for signal amplification similar to nanoparticle dissolution is nanocrystal cation exchange, in which the cations within a nanocrystal are place-exchanged with different cations.409 While bulk, solid-phase cation exchange often requires elevated temperatures and several weeks, complete nanoscale exchange can occur within seconds due to increased surface area and lower energy barriers to diffusing ions.410 For cation exchange to be employed in a bioassay, nanocrystals (ZnSe, ZnS, CdSe, or CuS) are first functionalized with target-specific molecular recognition elements, such as antibodies or aptamers, which are utilized as detection probes. Using rapid silver ion exchange, each bound nanocrystal releases thousands of Zn2+, Cd2+, or Cu2+ ions. Zinc and cadmium ions are then detectable using fluorogenic chelating reagents such as FluoZin-3 or Rhod-5N, respectively, and Cu2+ can be detected in a chemiluminescent reaction with luminol and H2O2. These reactions produce an amplified, stoichiometric signal and have been employed in well-plate403,411 and magnetic bead-based412−415 immunoassays for protein403,411,415 and cell412 detection as well as rolling circle amplification for DNA/miRNA413,414 detection. Despite this versatility, the cation exchange method has yet to be applied in paper-based formats. Similar to the ILISA example discussed previously, signal amplification using nanocrystal cation exchange would require a precipitating reagent for metal ion detection as well as clever design of a paper microfluidic device.
4.4.2. Inorganic Nanoparticles as Enzyme Mimics
Enzymatic signal amplification results in high analytical sensitivity and is easily performed in a controlled laboratory environment. However, as discussed in section 4.3, elevated temperatures often lead to decreased catalytic turnover, making it difficult to incorporate enzymatic signal amplification into low-resource infectious disease diagnostics. In recent years, some inorganic nanoparticles, including noble metal, rare earth, and magnetic nanoparticles, have been found to display surprising enzyme-like catalytic activity.416−418 These nanomaterial-based artificial enzymes, originally dubbed “nanozymes” by Manea et al.,419 are highly stable toward a range of temperatures and pHs and cannot be degraded by proteases. Further, catalytic activity of inorganic nanoparticles can be tuned with particle size, shape, coating, modification, and composition. For these reasons, nanozymes have been incorporated into a myriad of sensors for various applications.
In 2007, Gao et al.420 made the surprising discovery that magnetite (Fe3O4) nanoparticles have intrinsic peroxidase-like activity, displaying Michaelis–Menten kinetics consistent with a ping-pong mechanism. Fe3O4 magnetic nanoparticles were found to catalytically turn over several common horseradish peroxidase substrates, including TMB, DAB, and o-phenylenediamine (OPD), with catalytic turnover numbers equal to or improved over horseradish peroxidase. The group demonstrated that the particles could be used in place of horseradish peroxidase in a traditional immunoassay and that the magnetic properties of Fe3O4 could be leveraged further to enrich biomarkers before detection.420 As a result of this seminal work, magnetic nanoparticles have been employed as signal amplification elements in protein and DNA bioassays for a variety of infectious diseases, including Ebola,242 rotavirus,402Mycoplasma pneumonia,421Vibrio cholerae,422Chlamydia trachomatis,401L. monocytogenes,423Enterobacter sakazakii,424 and Salmonella typhimurium.425
Beyond magnetite, several other types of nanoparticles, including gold,426 platinum,427 hybrid particles,428−432 and MOFs,433 have been utilized as enzyme mimics in bioassays for targets such as respiratory syncytial virus,432 avian influenza A,426Salmonella, and E. coli,431 hepatitis C and HIV,428 and Staphylococcus aureus.433 Many of the assays that utilize nanozymes as catalytic turnover reagents for detection are formatted similarly to immunoassays on the surface of well plates.402,421−423 Several groups have translated this technology to the lateral flow format and found that the nanozyme detection element resulted in at least 100-fold improvement in limits of detection when compared to traditional gold nanoparticle-based lateral flow strips.242,424,427,431,434 For example, Loynachan et al.435 developed an ultrasensitive lateral flow assay for the detection of HIV p24 antigen using porous platinum Au@Pt core–shell nanozymes that catalyzed the precipitation of CN/DAB (4-chloro-1-naphthol/3,3′-diaminobenzidine tetrahydrochloride) in the presence of H2O2 (Figure 31). Incorporation of this detection scheme, as well as careful and systematic design of the affinity reagents for p24 capture and detection, resulted in a lateral flow assay with femtomolar detection limits, more sensitive than laboratory-based ELISA methods and nearly 2 orders of magnitude more sensitive than commercially available rapid tests.435 Because visual signal varied linearly with concentration before and after catalytic signal amplification, the test also had an ultrabroad dynamic range. The advantages of the nanoparticle enzyme mimics were directly demonstrated in a stability test in which the activities of lyophilized porous Pt core–shell nanocatalyst conjugate and lyophilized HRP conjugates were measured over time. While the enzyme conjugates lost nearly all activity after 15 days of storage at room temperature, the nanocatalysts maintained 100% of their initial activity while stored at room temperature and nearly 80% of their initial activity while stored at 44 °C for up to 42 days.435 However, it should be noted that the optimal HRP lyophilization and storage conditions387,389 discussed in section 4.3.1 were not employed in this study.
Figure 31.
Format and signal amplification strategy of the ultrasensitive HIV p24 lateral flow assay developed by Loynachan et al. Porous Pt core–shell nanoparticles catalyzed the precipitation of CN/DAB, amplifying visual signal and resulting in improved detection limits and an ultrabroad dynamic range. Figure adapted from ref (435). Copyright 2018 American Chemical Society (https://pubs.acs.org/page/policy/authorchoice_ccby_termsofuse.html).
To achieve such significant improvements in sensitivity using nanozymes, a wash step, a substrate addition step, and a reaction quenching step were inserted into the lateral flow assay workflow after the initial signal development.435 These additional steps make this assay and other similar diagnostics more difficult to perform in the field. Redesigned paper devices or automated lateral flow cassettes could simplify this workflow and allow for application of nanozymes in field settings, providing much-needed signal amplification and improved sensitivity for low-resource infectious disease diagnostics.
4.4.3. Reductive Nanoparticle Enlargement
The use of gold nanoparticles as detection elements in commercially available lateral flow assays is ubiquitous.436 Particles used in these assays are typically 15–50 nm and can be detected either visually (without optical equipment) or using simple hand-held readers.177,219,437 Often, the detection limits of these assays are not low enough to diagnose low-density infections. One simple solution for improving sensitivity is chemically enlarging the particles, making them more visually intense. In this process, a lateral flow assay is run in the typical manner such that, if the antigen is present, a gold nanoparticle-containing sandwich complex is formed at the test line. Next, an enhancement solution consisting of a molecular precursor and reducing agent is added to the test. The gold nanoparticles at the test and control lines serve as nucleation sites for solid metal deposition. The most common format for particle enlargement, silver enhancement, is based on 19th-century photographic techniques and involves the reduction of silver ions (e.g., silver nitrate) to silver metal on the surface of gold particles in an acidic buffered solution containing hydroquinone.438 This process is similar to the seeded-growth nanoparticle synthesis method and can result in particles from hundreds of nanometers to several micrometers in diameter (Figure 32).439
Figure 32.
Silver enhancement of gold nanoparticles at the test line of a lateral flow assay. SEM images show 50 nm gold nanoparticles (left, indicated by arrows) enlarged to 10 μm after reductive deposition of silver (right). This enhancement reaction was performed in an automated cassette developed by Fujifilm. Figure adapted with permission from ref (439). Copyright 2011 Elsevier.
Because silver enhancement found its first biological application in tissue staining in 1935,440 there are many commercially available silver enhancement solutions designated for microscopy applications (Sigma, Thermo, Ted Pella, Nano, etc.). The first application of silver enhancement to paper diagnostics was in 1991 when Horton et al.441 developed a proof-of-concept lateral flow assay for the detection of mouse IgG and demonstrated that silver treatment resulted in a 100-fold improvement in detection limits. Since then, silver enhancement has successfully improved the detection of many infectious diseases by several orders of magnitude, including influenza,439,442 HIV,443 malaria,444−449Y. pestis,450 and E. coli.451
One of the drawbacks to silver enhancement on lateral flow assays is that it must be performed after the test has completed its initial development. Increasing the total number of steps required reduces the likelihood that the test could be applied in the field. For example, one group450 found that silver enhancement improved the detection limits of their Y. pestis test 1000-fold. In order to take advantage of this impressive enhancement, the authors allowed the gold test to develop as usual, washed the strip three times, submerged it into enhancing reagents for 5 min, and then stopped the reaction with sodium thiosulfate. This procedure is feasible in a district hospital or even a local health outpost with minimal resources, and the enhanced assay would be a true asset in these settings should a plague pandemic occur. However, the additional steps required for signal amplification make POC application in a low-resource field setting unlikely.
For this reason, some groups who employ a silver enhancement step in their paper diagnostic assays have moved away from the traditional lateral flow assay format and toward alternative formats that allow for automated delivery of silver enhancement reagents to the test line. Fujifilm439,452 applied their expertise in photo technology to develop a benchtop workstation that automated silver enhancement for influenza rapid diagnostic tests. In this format, a user added the sample to a lateral flow cartridge that contained all reagents necessary for test washing and silver enhancement. The cartridge was inserted into the benchtop workstation, which released the solutions in a timed, automated manner and provided a quantitative readout of the strip after test development was completed. Despite the fact that the instrument greatly simplified the assay protocol for the user, the requirement of a reliable source of electricity greatly inhibits its application to a field setting.
Recently, the Yager and Fu groups have designed 2DPNs that rely on unique paper geometry or embedded paper-fluidic valves to enable programmed, electricity-free delivery of silver enhancement reagents to the assay test line.444−448,453,454 These 2DPNs simply require the addition of sample and buffer to pads that have been preloaded with assay reagents, mirroring the same number of steps required to run a traditional lateral flow assay. The unique physical structures of 2DPNs allow for precisely timed delivery of sample, then gold conjugate, and finally silver enhancement reagents to the test line. Similarly, Cho et al.455 developed a cross-flow chromatography system, in which silver enhancement reagents were embedded into paper and then rehydrated after the lateral flow assay developed. The enhancement reagents flowed perpendicular to the initial immunochromatographic test over the test and control lines. These creative approaches to redesigning the lateral flow assay make silver enhancement the most field-deployable metal-based signal amplification strategy to date, although large-scale manufacturing requirements must be considered to determine the scalability of such test designs.
Finally, it should be noted that while silver enhancement on gold particles is most common, other metals have been used for reductive nanoparticle enlargement for lateral flow enhancement. Gold has been deposited onto gold and silver nanoparticles,443,456−458 and silver onto silver and platinum nanoparticles.459 While there are many other examples of solution and microfluidic assays employing reductive nanoparticle enlargement for visual and electrochemical signal amplification, these formats are less conducive to low-resource disease diagnosis.451,457,460
4.4.4. Bio-Barcodes
The amplification methods discussed thus far increase the number of signaling elements after the initial assay has developed, requiring additional steps post hoc. Another amplification strategy is to generate a greater number of detectable molecules midassay, thereby increasing the downstream signal output of the assay. In 2002, the Mirkin group461 demonstrated that protein biomarker targets could be detected indirectly in a highly sensitive manner by employing gold nanoparticles functionalized with DNA tags. In this strategy, magnetic microparticles functionalized with target-specific antibodies were added to a biological sample. After biomarker capture, the supernatant was removed and intermediate detection elements were added to the particles. These detection elements consisted of gold nanoparticles functionalized with a low density of target-specific antibodies and a very high density of double-stranded barcode DNA. Using an external magnet, the gold nanoparticles that bound the target were separated, and then the barcode DNA was dehybridized and detected downstream (Figure 33).462 Because the number of barcode tags was much higher than the number of captured biomarkers, the resulting signal was amplified when compared to a direct detection method. One powerful advantage of this modular assay format is it could be easily multiplexed, with distinct barcode tags for each target of interest.463 Further, while the biobarcode method was initially applied to the detection of proteins, it has also been applied to nucleic acid detection. Simply replacing the biomarker-specific antibodies with DNA complementary to the nucleic acid target allows for detection of DNA or RNA targets with PCR-like sensitivity.464
Figure 33.
Workflow of biobarcode assay for protein detection. Figure adapted with permission from ref (462). Copyright 2003 American Association for the Advancement of Science.
Since its development, the biobarcode assay format has been utilized for the detection of infectious diseases such as hepatitis B,463 hepatitis C,465,466 variola virus,463 Ebola,463 HIV,463,467,468B. anthracis,464 and Salmonella,469 as well as C-reactive protein470 and a variety of cytokines.471,472 Despite its promise for sensitive detection, the biobarcode assay format suffers from two major drawbacks: (1) the high number of user steps required before the barcode tags are released for detection makes it extremely difficult to adapt to a low-resource format, and (2) low-resource detection of the nucleic acid tags remains a challenge. Initial detection strategies for the biobarcode assay were scanometric; the barcode tags were detected in a sandwich hybridization assay on the surface of a glass slide, with gold nanoparticles as the detection elements. Signal was further enhanced using reductive silver deposition before the slides were evaluated using image analysis.461−464,467,473 Methods such as PCR,466,468 mass spectrometry,474 and electrical detection465 have been employed to detect the barcode tags as well. Nam et al.461,472 developed a colorimetric assay in which the released gold nanoparticle biobarcode tags were introduced to a second solution of gold nanoparticles functionalized with single-stranded DNA complementary to the tags. The hybridization of the biobarcode tags with complementary DNA resulted in aggregation of the two gold nanoparticle-DNA conjugates, yielding a detectable color change. None of these detection methods can be employed in a field setting; however, it is not difficult to imagine developing a hybridization-based lateral flow assay for barcode tag detection or functionalizing the tags themselves with moieties such as biotin or a FLAG peptide that could make them readily detectable in a paper diagnostic format.
As is evidenced by sections 4.3 and 4.4, significant advances have been made in developing metalloenzyme- and metal nanoparticle-based signal amplification strategies for infectious disease diagnosis. However, even with the advancements in metalloenzyme stabilization and highly stable nanoparticle-based approaches, POC assays utilizing signal amplification are still scarce. Most of the assays discussed, even those that employ paper substrates or lateral flow formats, require too many user steps before the signal is generated. Future work that minimizes user steps, for instance, through use of 2DPNs or vertical flow assay formats, will be critical in translating these signal amplification strategies from laboratory tests to assays that can be run at a primary healthcare facility. Moreover, most of the assays discussed in sections 4.3 and 4.4 were also heavily reliant on benchtop, laboratory-based instrumentation for signal readout. The next section will discuss the various types of field-deployable instrumentation that have been developed in an effort to bring quantitative and sensitive assays, such as those discussed in section 4, to low-resource settings.
5. Point-of-Care Instrumentation
Traditionally, point-of-care diagnostics such as lateral flow assays have delivered qualitative visual results. Although this simplicity is advantageous in the field, the diagnostic sensitivity and specificity can suffer from subjective interpretation and user bias errors. Quantitative measurements mitigate these factors, avoiding the data loss associated with qualitative tests and providing insight into important variables such as infection intensities and biomarker expression patterns. In an elimination setting, these parameters allow for robust epidemiological studies into transmission dynamics and intervention efficacy. Novel detection strategies that offer highly sensitive and quantitative results, including many of those outlined in section 4, require instrumentation for signal readout. As a result, the development of appropriate and simple instrumentation will likely become an integral part of the application of diagnostic technologies in low-resource settings.
Incorporation of detection devices into point-of-care diagnostics effectively eliminates the possibility of “equipment-free” tests (E in ASSURED criteria) and reduces the “affordable” nature of simple tests (A in ASSURED criteria).13 However, the potential benefits of instrumentation, including improved sensitivity and reduced bias, will outweigh these disadvantages in many use-case scenarios, and the overall cost of instrumentation per test can be reduced when a device is used to evaluate many tests over time. Additionally, many instruments incorporate common consumer electronic devices, such as mobile phones and smart phones, thereby reducing the equipment burden. This section outlines some of the devices developed for point-of-care infectious disease diagnosis. Table 3 is included at the beginning of section 5 to highlight selected examples of point-of-care instrumentation.
Table 3. Selected Examples of Point-of-Care Instrumentation for Detection of Infectious Diseases.
| technology | device | target | preparation | training | power | associated equipment | projected cost (USD) |
|---|---|---|---|---|---|---|---|
| literature-based prototypes | |||||||
| brightfield/fluorescent (video) microscope | LoaScope/CellScope492,497,498,500 | L. loa, malaria, TB, Helminths | capillaries containing whole blood, stained smears on glass slides | laboratory standard | USB | mobile phone | <∼$30500 |
| brightfield/fluorescent microscope | Global Focus microscope480,485 | TB | stained blood/sputum smears on glass slides | laboratory standard | 2 AA batteries | mobile phone | ∼$240 |
| brightfield/darkfield/fluorescent/lens array microscope | Foldoscope488,489 | Malaria, G. lamblia, L. donovani, T. cruzi, E. coli, S. hematobium eggs | blood and culture films (stained and unstained) on glass slide | laboratory standard | batteries | optional mobile phone or camera | <$1 |
| digital holographic microscope | field-portable pixel super-resolution color microscope (Ozcan group)479 | mammalian cells (cancer) | use-specific commercial kit (ThinPrep) | laboratory standard | not specified | laptop | not specified |
| software-based imaging analysis | mLAB platform512,577 | malaria, HIV | LFA’s required protocol | minimal | mobile device’s rechargeable battery | mobile device with HTML-based browser | not specified |
| QR code-based imaging analysis and assays | beRDT513 | Malaria | custom-designed lateral flow assays | minimal | mobile device’s rechargeable battery | mobile device with iOS app | not specified |
| colorimetric 96-well plate reader | 3D-printed cell phone plate reader (Ozcan group)382 | measles, mumps, HSV1, and HSV2 | ELISA or assay’s required protocol | laboratory standard | Mobile device’s battery with attachment using 6 AAA batteries | mobile device with 3D-printed attachment | not specified |
| spectrophotometer (transmission, reflectance, and intensity) | TRI-analyzer (Cunningham group)516 | fetal fibronectin, phenylalanine | ELISA or assay’s required protocol | laboratory standard | mobile device’s battery with attachment using 3 AA batteries | mobile device with 3D-printed attachment | ∼$550 |
| SPR detection | SPR cell phone sensor (Filippini group)519 | β2 microglobulin control | Biocore CM5 Sensor Chip (GE) protocol or similar | laboratory standard | mobile device’s rechargeable battery | mobile device with optical coupler | not specified |
| fluorescent microscope and cytometer | smartphone-based cytometry tool (Tasoglu group)523 | breast, ovarian, and lung cancer cells | cell staining, mixing in paramagnetic solution, insertion in capillary tube | laboratory standard | mobile device’s Rechargeable battery | mobile device with 3D-printed attachment | ∼$106 |
| electrochemical assay reader | uMed device (Whitesides group)173 | glucose, heavy metals, sodium ions, HRP2 | specific to individual assay | minimal | rechargeable battery | standalone, using phone for data transfer | ∼$25 |
| commercial POC instrumentation | |||||||
| confocal lateral flow reader | ESEQuant (Qiagen, Inc.)346,578 | customizable to any optical signal | LFA’s required protocol | minimal | rechargeable battery, AC power, or car adapter | standalone or with laptop | ∼$1000–10000 (based on quantity and options) |
| CMOS sensor imaging lateral flow reader | RDS-2500 (Detekt Biomedical, LLC)504−506 | visual and fluorescent labels | LFA’s required protocol | minimal | rechargeable battery | standalone or with laptop | (Previous model started at ∼$2000) |
| Raman spectrometer (SERS) | FirstDefender RMX (Thermo Fisher Scientific, Inc.)538,539 | any substance in its chemical library | none; sensor can ID substance surface or in translucent container | minimal | rechargeable battery or 123a disposable batteries | standalone unit | ∼$68000 (Fisher) |
| Raman spectrometer (SERS) | NanoRam (B&W Tek, Inc.)540,541 | any substance in its chemical library | none; sensor can ID substance surface or in translucent container | minimal | rechargeable battery | standalone unit (optional attachments) | ∼$43.500 (VWR) |
| magnetic assay reader | magnetic immuno-chromatographic test (MICT) (MagnaBioSciences, LLC)549,550 | any magnetic lateral flow assay (strip placed in disposable cassette) | LFA’s required protocol | minimal | AC power | standalone unit | ∼$30000 |
5.1. Optical Imaging and Measurements
A vast majority of the metal-based probes and nanoparticles highlighted in section 4 produce optical signals such as absorbance, fluorescence, or phosphorescence. As a result, a diverse array of instrumentation has been developed to facilitate assay readout for imaging and quantitative purposes. This section provides an overview of efforts to develop portable, affordable, and sensitive instruments for optical detection in low-resource settings.
5.1.1. Microscopy
Conventional microscopy remains the gold standard diagnostic technique for many infectious diseases, including malaria, tuberculosis, schistosomiasis, and intestinal protozoa.475 To detect these diseases, a variety of microscopic techniques, such as bright field, dark field, and fluorescence microscopy, are used for direct inspection of stained or unstained smears (blood, sputum, urine, etc.). While microscopy can be a useful diagnostic tool, results depend strongly on infection intensities, sample preparation methods, and the training level of the microscopist.476 These challenges lead to variability that reduces the utility of microscopy-based diagnosis in field settings, especially in low-transmission areas. Additionally, microscope instrumentation can be expensive and bulky. To address these drawbacks, many groups are developing portable, affordable, and easy-to-use microscopy tools suitable for low-resource settings.
One approach to bringing laboratory-based microscopy tools to low-resource settings involves building simple, portable, and/or miniature microscopes from low-cost materials. While this solution does not address some of the primary disadvantages of microscopy as a POC diagnostic (i.e., the need for skilled microscopists and sample preparation requirements), it does allow for conventional microscope platforms to be deployed in difficult settings at low cost. Such devices typically replace high-energy, expensive light sources with LEDs, reducing the expense and power required for illumination in addition to increasing the light source lifetime.477 Structural components of these microscopes are often three-dimensionally (3D) printed,477−480 and affordable plastic or hybrid lenses and fiber optics can replace expensive and delicate glass lenses.477−479,481−484 For example, Miller et al.480,485 developed a portable, low-cost ($240 USD), LED-based fluorescent microscope (Global Focus microscope) weighing just 1 kg and powered by two AA batteries (Figure 34A). The diagnostic utility of the microscope was demonstrated in a small field trial for detection of M. tuberculosis. When samples were evaluated as positive or negative for M. tuberculosis, results from the Global Focus microscope and a laboratory-grade fluorescence microscope were in agreement for 98.4% of cases.485 Many similarly portable and lightweight microscopes have been developed, including tomographic,482 holographic,483 wide-field,484 and fluorescence486 microscopes, some impressively weighing in at less than 200 g.479,482−484 These microscopes have been applied not only to the detection of tuberculosis480,485,486 but also to malaria,481 soil-transmitted helminths,487 schistosomiasis,487 and Hymenolepis nana.482
Figure 34.
Examples of microscopy instrumentation developed for use in low-resource settings. (A) The portable Global Focus fluorescence microscope ($240). Adapted with permission from ref (485). Copyright 2010 Miller et al. (https://creativecommons.org/licenses/by/4.0/legalcode). (B) Layout of an assembled Foldscope. Adapted with permission from ref (488). Copyright 2014 Cybulski et al. in PLOS (https://creativecommons.org/licenses/by/4.0/legalcode). (C) Reverse-lens CellScope mobile phone attachment for diagnosis of schistosomiasis and intestinal protozoa. Adapted with permission from ref (490). Copyright 2016 Coulibaly et al. in PLOS (https://creativecommons.org/licenses/by/4.0/legalcode). (D) Mobile phone-based LoaScope. Adapted with permission from ref (497). Copyright 2015 American Association for the Advancement of Science.
The Foldscope is an example of a low-resource microscope that most embodies the ASSURED criteria.488 Assembled using the principles of origami, the Foldscope is constructed from flat paper, copper tape, a button-cell battery, an LED, polymer filters, and a ball lens (Figure 34B). The entire device costs less than $1, can achieve magnification up to 2180×, and was shown to be capable of detecting Giardia lamblia, Leishmania donovani, Trypanosoma cruzi, E. coli, and Schistosoma hematobium eggs.488 Despite these accomplishments, when the Foldscope was applied to S. hematobium detection in a field setting, the diagnostic sensitivity was just 55.9% when compared to conventional light microscopy.489 This limited sensitivity was likely a result of challenges with physical manipulation of the Foldscope when integrated with a smartphone display. In other words, the Foldscope failed to satisfy the “U” for user-friendly in the ASSURED criteria for this particular use-case. This example demonstrates the importance of rigorous field evaluation of novel diagnostic devices and indicates that improvements to the Foldscope must be made before implementation for case management.
Recently, mobile phone-based microscopy has emerged as a potential alternative to conventional microscopes for infectious disease diagnostics. Phone-based bright-field, fluorescence, and dark-field microscopy devices have been developed, often employing compact, pocket-sized attachments (Figure 34C).490 These devices have been applied to the detection of a myriad of infectious diseases, including malaria,491,492 tuberculosis,492 schistosomiasis,493,494 human papillomavirus (HPV),495 soil-transmitted helminths,496 filarial diseases,497,498 and intestinal protozoa.494,499 Repurposing mobile phones is advantageous due to their widespread adoption and growing connectivity in low-resource settings. Additionally, the acquisition of digital images could allow for image-processing (locally or remotely), automated diagnoses, and near real-time data transmission. These advantages, coupled with disease-specific software applications, could potentially decrease the user variability typically associated with microscopy, improving case management and epidemiological studies.
One example that leverages these advantages is the LoaScope, developed by the Fletcher group (Figure 34D).500,497,498 The LoaScope is a smartphone-based device that combines video microscopy and automated software with onboard quantitative detection based on the movements of filarial parasite Loa loa in whole blood. An application downloaded to the smartphone is used for stage control, video capture, image analysis, and data reporting of quantitative L. loa microfilaria densities. Detection of L. loa is important because high levels of microfilariae are associated with potentially fatal adverse events after treatment with ivermectin, a drug frequently used in mass drug administration efforts against onchocerciasis and lymphatic filariasis (LF). These adverse events have led to the suspension of mass drug administration campaigns and subsequent major setbacks in onchocerciasis and LF elimination efforts. Rapid, point-of-care measurement of L. loa infection intensity could allow for these elimination campaigns to resume.
This device was validated in a field setting and yielded results similar to those of thick smears.497 It was then applied in a large-scale (16,259 participants) test-and-not-treat approach for onchocerciasis in an area with high prevalence that had not participated in mass drug administration campaigns since 1999 due to the prevalence of L. loa.498 Each patient was screened for L. loa burden using the LoaScope, and those with high counts were not given ivermectin but were treated for L. loa with albendazole. This is a striking example of mobile phone-based microscopy in the field; all LoaScope measurements were performed by operators with only one hour of training, and the application-specific software significantly decreased the probability of user error by eliminating the need for subjective interpretation of results. In total, the LoaScope enabled ivermectin delivery to 15522 participants that otherwise would not have received treatment for onchocerciasis, demonstrating the incredible impact mobile phone microscopy can have when applied to detection of infectious diseases.498
Despite the fact that many mobile phone-based microscopes lack the analytical and diagnostic sensitivity of conventional microscopes, the LoaScope is an excellent demonstration that mobile phone microscopy can be highly impactful if applied in an appropriate use-case scenario. Because this particular study focused on identifying patients with very high L. loa burdens, it is unclear to what extent this device could detect low-burden infections. Generalization of this technology to other filarial diseases will also require further study. Additionally, potential interference from other filarial parasites needs to be studied. As work continues in the field of low-resource microscopy, it will be important to develop new tools with specific use-case scenarios in mind. One potential avenue for application-based innovation involves development of low-resource sample preparation methods coupled with novel probes, such as the inorganic complexes and nanoparticles discussed in section 4.
5.1.2. Lateral Flow Analysis
While much progress has been made in the development of rugged, field-applicable microscopy devices, lateral flow assays currently remain the most field-friendly format for infectious disease diagnosis. However, the simple, direct visual readout of lateral flow assays, often cited as one of the primary advantages of this format, is also considered its Achilles heel; difficulties in interpretation resulting from visual impairment and operator bias can result in user-to-user variation. After tests are interpreted, maintaining accurate records and communicating test results to local and national health organizations remain substantial challenges. To overcome these challenges, a number of optical readers have been developed and commercialized to provide rapid, objective, and quantitative measurements of signal on lateral flow assays. The underlying instrumentation within these readers can generally be grouped into two categories: (1) optomechanical scanning and (2) imaging.
For scanning readers, the primary optical components include a light source (LED or laser) and photodiode.501 The specifications of these components are tailored to the specific detection element used in a particular assay. Light source power and wavelength are determined by the absorbance or excitation wavelength of the labels employed in each specific test. The light source is rastered along the lateral flow strip, and a photodiode then detects reflectance or emission, depending on whether colorimetric or fluorescent labels are used. Measurements are typically recorded with millivolt (mV) units, though biomarker concentration can be obtained if a calibration curve is established. When colored particles are present at the control and test lines, they absorb incident light, resulting in a quantifiable decrease in reflectance. For fluorescent labels, the intensity of emitted light increases as the light source passes over the control and test lines.
Optics are configured in either a specular or confocal arrangement.501 In the specular arrangement, which is primarily used for colorimetric labels, the photodiode is positioned such that it collects light that is reflected from the test at the same angle as the incident light from the source. Confocal scanning systems can be used to measure reflectance or fluorescence. In this case, incident light is focused to a point on the lateral flow strip, and the photodiode is placed above this focal point. Light returning from the strip, whether reflected or emitted, passes through an aperture that excludes rays that were not directly from the focal point, eliminating background scattering.502 In the case of fluorescent labels, a dichroic mirror or filter is often placed before the aperture. Both specular and confocal scanning configurations are sensitive to the z position of the lateral flow strip. In the specular arrangement, any small deviation along the z axis can result in the reflected light completely missing the photodiode due to misalignment. Confocal systems are slightly more tolerant to small variations along the z axis, depending on the size of the pinhole aperture. However, even slight variations off the focal plane can drastically decrease the number of photons reaching the photodiode, altering the quantitative output. Another challenge associated with confocal and some specular opto-mechanical scanning strip readers is that a point of incident light only covers 10–20% of the test and control lines.503 This is especially problematic if the concentration of particles varies along the test or control line due to improper flow or manufacturing variations.
Despite these challenges, scanning systems are advantageous due to their low cost and relative simplicity. One of the most widely used commercial systems is the ESEQuant line of lateral flow readers manufactured by Qiagen, Inc. (Figure 35A).346 Qiagen offers customization of optics for colorimetric, fluorescent, chemiluminescent, and upconverting phosphor labels. The user should take into account that these systems often vary in sensitivity. As a general rule, smaller portable systems rarely perform as well as their benchtop counterparts. For example, van Dam et al.313 found that the portable commercial reader, the UCP-Quant (Qiagen), was at least an order of magnitude less sensitive than a custom-modified benchtop reader for evaluating upconverting phosphor-based schistosomiasis lateral flow assays. Consequently, selecting a scanning lateral flow strip reader for a particular application requires consideration of both portability and sensitivity.
Figure 35.
Devices for reading lateral flow assays. (A) The ESEQuant reader scans lateral flow assays with a confocal optics setup. Image adapted with permission from ref (313). Copyright 2013 Elsevier. (B) The Deki ReaderTM for imaging lateral flow assays. Image adapted with permission from ref (507). Copyright 2013 BioMed Central Ltd. (https://creativecommons.org/licenses/by/2.0/legalcode) (C) Barcode-embedded lateral flow assays enable facile data aggregation and communication. Figure adapted with permission from ref (513). Copyright 2017 Royal Society of Chemistry.
Imaging-based lateral flow readers greatly reduce the optical equipment required for quantifying lateral flow assays. These systems produce a picture record of a lateral flow assay using a charge coupled device (CCD) or complementary metal-oxide-semiconductor (CMOS) detector and rely on image processing software to detect and/or quantify the intensity of the test line.503 Benefits of this format include the ability to store test images alongside patient records and a lack of moving parts, making imaging-based readers more robust to field conditions. Further, image analysis software can identify and correct for nonuniform signal, allowing for analysis of the full test line. Imaging-based readers can be used for detection of colorimetric and fluorescent labels. In the latter case, hardware for excitation is required, and filters can be added to improve image quality. Since test position and illumination can affect image analysis, many camera-based lateral flow readers include housing units for run-to-run consistency. Commercial lateral flow imaging devices include the Deki Reader (Fio), AX-2X (Axxin Inc.), SkanSmart (Skannex), and the RDS-2500 PRO (Detekt Biomedical LLC) (Figure 35B).504−506 These devices have been used for quantification of malaria,507−510 dengue,509Burkholderia pseudomallei,509 and Y. pestis(509) lateral flow assays, and some, such as the Deki Reader, have been tested extensively in the field.507−510
Image-based lateral flow quantification translates well to mobile phone technology, especially as smartphone camera quality improves. Mobile phones have been used to quantify lateral flow assays for numerous diseases, including malaria,511−514 HIV,511 and tuberculosis.511 Similar to commercially available imaging-based readers, many of these studies required external devices to ensure consistent illumination and distance from the camera in order to maintain consistent image analysis. Recently, Scherr et al. developed a lateral flow imaging platform using an unmodified mobile phone as the only hardware.512 Relying only on software for image analysis, lateral flow images were captured under ambient lighting conditions. The images were sent to a secure web database (REDCap) and analyzed using a MATLAB-based algorithm. Quantitative results were sent back to REDCap and delivered to the phone in real time. This software-based imaging platform was tolerant to moderate variations in test orientation, distance, and illumination, making data collection as easy as taking a cell phone picture. When compared to the scanning ESEQuant lateral flow reader, this processing platform was found to have improved detection limits. Additionally, the algorithm reliably worked for camera resolutions between 0.5 and 8 megapixels, suggesting that any modern mobile phone camera could capture images with sufficient quality for the image processing software to work.512
As mobile phones are incorporated into low-resource diagnostic workflows, it is important to ask whether the conventional lateral flow format can be redesigned to take full advantage of potential benefits smartphones can offer. In other words, what changes can be made to the diagnostic test format that maintain the simplicity of lateral flow assays but will enable improved quantification, communication, and aggregation of test results? One option is to embed quick response (QR) codes into the lateral flow test. This idea has been approached in a number of ways, including placing stickers on lateral flow tests and adding a transparent overlay in which the test and control lines become part of the QR code.514,515 In these formats, the embedded barcode is either static, providing useful information about the brand, lot, and patient ID, or dynamic, producing a positive/negative read-out. Scherr et al. developed a novel platform in which the control line of the lateral flow assay was replaced with a control grid patterned in the shape of a QR code (Figure 35C).513 The QR code functioned as a flow control, appearing on both positive and negative tests, but not invalid tests. The QR codes also triggered data analysis and enabled corrections for lighting effects. When scanned, manufacturing details were quickly and easily accessed at the same time as the test line was quantified.513 It is not difficult to imagine the impact such a test format could have when combined with the data communication and aggregation abilities of smartphones. Epidemiological and surveillance studies could account for variables that are not even currently considered in most studies, such as manufacturing batch-to-batch variation. Geographic data could also be coupled to specific rapid tests.
As tools for automated reading of lateral flow assays become more prevalent, establishing standards for consistent reporting and data management will be important. Just like with the lateral flow assays themselves, test-reading instrumentation will need to be validated to ensure proper and consistent biomarker quantification across different devices and test brands. Although optical lateral flow readers may detect test lines invisible to the human eye, readers alone will not replace the need for ultrasensitive lateral flow tests.
5.1.3. Other Smartphone-Enabled Optical Measurements
Mobile phones have been adapted for optical measurements beyond lateral flow quantification. One example is the mobile phone-based, hand-held microplate reader developed by Berg et al.382 This device consists of a 3D-printed attachment in which an LED array illuminates each well, and the transmitted light is collected via 96 individual optical fibers. Results are visualized and delivered to the user in one minute within a custom mobile phone app. The device was shown to be more than 98% accurate for immunoassays detecting mumps IgG, measles IgG, and herpes simplex virus IgG (HSV-1 and HSV-2) when compared to an FDA-approved clinical spectrophotometer.382 While microplate-based immunoassays are not feasible in a field setting, this device could make a true impact in resource-limited clinics as an excellent alternative to traditional bulky and expensive spectrophotometers. Additionally, because the device is attached to a mobile phone, it offers the potential to rapidly communicate and aggregate results, further enabling large-scale epidemiological studies.
Mobile phone attachments for other optical detection modalities have been developed as well. In one exciting example, Long et al.516 developed the TRI-Analyzer, a trimodal smartphone attachment capable of measuring transmission, reflection, and emission intensities. This multifunctional device leveraged white light from the smartphone’s rear-facing flash LED or an integrated green laser diode for sample illumination via optical fibers. The optical fibers collected light transmitted through or emitted from the sample or light reflected from a photonic crystal biosensor. This collected light was then collimated and focused before being dispersed by the diffraction grating onto the smartphone CMOS sensor. The intensity at each wavelength was determined by the integration of pixel intensity at the corresponding position along the sensor. This clever detection strategy allowed for full absorbance and emission spectra to be collected with a simple smartphone-based device. In two proof-of-concept assays, the developed device could detect analytes at lower concentrations than those detected using conventional laboratory instrumentation, indicating that the miniaturization of the technology did not result in a trade-off of sensitivity.516 Thus, this trimodal smartphone attachment enables quantitative laboratory-quality optical measurements at one’s fingertips. Similarly formatted mobile phone-based spectrophotometers,517,518 surface plasmon resonance sensors,519−521 and flow cytometers522,523 have also been developed. The miniaturization and decrease in cost associated with these smartphone-enabled tools will make laboratory techniques possible that would otherwise be inaccessible in low-resource settings.
5.2. Thermal Readers
Noble metal nanoparticles have extremely high molar absorptivities, making them ideal visual labels for lateral flow assays. This intense absorbance occurs when particles are excited with light that matches the resonant oscillation of their free electrons. As these electrons relax down to the ground state, they release energy in the form of heat to the surroundings.524 The amount of heat generated is equal to the product of the number of noble metal nanoparticles, the cross-sectional area of the nanoparticles, and the intensity of the incident light.524 Recently, Bischof’s group took advantage of these unique thermophysical properties to develop a novel method for lateral flow quantification based on the thermal contrast of gold nanoparticles.524−526 In this method, a high-powered green laser was used to excite the gold nanoparticles, and an infrared-detecting camera was employed to measure the resulting thermal contrast. An early benchtop prototype of the device demonstrated a 32-fold improvement in detection limits of an FDA approved lateral flow assay for cryptococcal capsular polysaccharide glucuronoxylomannan antigen (CrAg).525 CrAg is a biomarker for cryptococcal infections, which are opportunistic fungal infections that are among the leading causes of death for patients with AIDS and a common cause of adult meningitis in sub-Saharan Africa.525 The thermal contrast-coupled CrAg lateral flow assay was further validated in a second study, displaying 92% accuracy in quantifying CrAg levels in patient samples.527
After successful validation of their prototype device, the group developed a reader that incorporated a green laser, infrared camera, and automated control system into a benchtop box device (Figure 36).526 The study demonstrated the utility and versatility of thermal contrast as a method for quantification of lateral flow assays from a variety of vendors and for multiple diseases. An 8-fold improvement in detection threshold compared to visual inspection was found for lateral flow assays for influenza (BD Veritor), malaria (First Response, SD BIOLINE, and ParaHIT), and Clostridium difficile (RIDAQUICK).526 In a separate study, the group found that this enhancement factor was greater as nanoparticle size increased, suggesting that redesign of lateral flow assays could further improve test sensitivity using a thermal contrast reader.528
Figure 36.
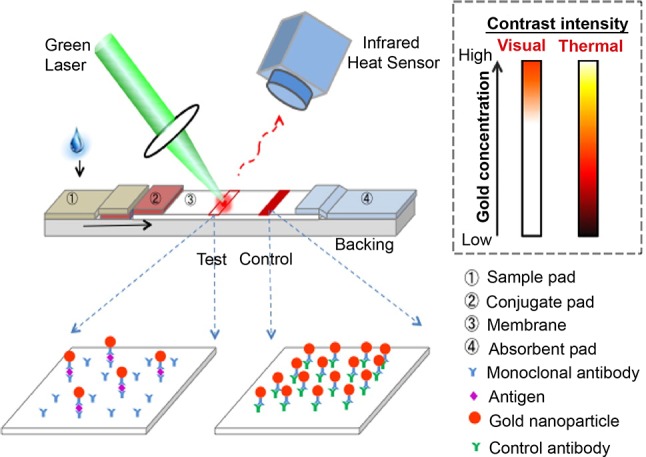
Thermal contrast reader. Figure adapted from ref (526). Copyright 2016 American Chemical Society.
While thermal contrast represents a unique and versatile method for lateral flow quantification, several issues must be addressed before large-scale implementation. These include cost, portability, and background signal reduction from biological interferences such as blood. Additionally, nitrocellulose membranes are highly flammable; while it does not appear that full strip ignition has occurred in the currently published studies, visible burn marks along the nitrocellulose membrane526 suggest that safety studies to determine the limits of the thermal contrast system should be undertaken before widespread implementation. Nevertheless, thermal contrast represents a highly sensitive and promising tool for lateral flow quantification.
5.3. Surface-Enhanced Raman Spectroscopy
Surface-enhanced Raman spectroscopy (SERS) is a highly sensitive vibrational spectroscopy technique for structure-based analyte detection. In this technique, excited localized surface plasmons, typically from noble metal (e.g., gold and silver) substrates with nanoscale features, generate amplified electromagnetic fields, enhancing the inelastic Raman scattering signals of analytes of interest up to 1014 times.529 Instrumentation for SERS detection requires a high-powered laser excitation source, which is typically tuned according to the resonant frequency of the plasmonic substrate. After excitation, filters are used to exclude elastic Rayleigh scattering, allowing only Raman-scattered light to reach the detector.530
The high sensitivity of SERS makes it an attractive detection platform for infectious disease diagnostics, particularly for the detection of low-density infections. Many laboratory-based bioassays for protein, nucleic acid, and whole-cell biomarkers that leverage SERS detection have been developed for a myriad of infectious diseases.68,531−536 These assays are generally either performed on the surface of particles in solution or on a bulk solid substrate. Detection elements typically consist of nanoparticles functionalized with Raman reporter molecules and target-specific molecular recognition elements.529 Once a complex is formed, the nanoparticles provide the plasmonic surface for Raman signal enhancement of the reporter molecule. Nanoparticles are typically gold, silver, or Au@Ag particles and come in a variety of shapes that provide optimized signal enhancement.537 Small-molecule Raman reporters, such as 4-mercaptobenzoic acid, malachite green, and 4-aminothiophenol, are often employed because biomolecule targets typically have complex vibrational signatures and are frequently too far away from the plasmonic surface for sensitive detection and identification.529
The bulky nature and resource requirements of SERS detection remain its primary barrier to implementation in point-of-care settings. Several hand-held Raman spectrometers have been developed and commercialized, including the FirstDefender (ThermoFisher Scientific, Inc.),538,539 NanoRam (B&W Tek),540,541 and Progeny (Rigaku Corp.). These devices are advertised for identification of unknown substances in forensic, environmental, military, and first-response contexts and for quality control in industrial settings. While many studies that utilize SERS-based detection for infectious disease biomarkers suggest that these commercially available portable devices could be utilized for analysis of a SERS-based lateral flow assay, none of the studies have actually implemented a hand-held device.542−547 Accordingly, it is unknown whether such devices have sufficient sensitivity or spatial resolution to use effectively with low-resource diagnostics. These parameters, along with cost analysis, should be studied to determine whether current commercial devices could be implemented.
5.4. Magnetic Detection
Magnetic nanoparticles possess advantages over optical lateral flow labels because their signal is rarely hindered by test opacity or biological interferents. A detailed discussion of these qualities as well as examples of specific assays that employ magnetic nanoparticles are provided in section 4.2.6. Here, we discuss instrumentation required for magnetic particle detection.
There are a variety of methods for detecting paramagnetic and superparamagnetic nanoparticles on lateral flow assays based on either the inductive effect or magnetoresistance. For example, MagnaBioSciences (USA) has developed an induction-based device that has been used extensively in the literature for detection of infectious diseases such as anthrax,366,367T. solium,349 HIV,360,361V. parahemolyticus,364 and L. monocytogenes.365 In this format, the lateral flow assay is placed inside an oscillating magnetic field on a set of induction coils, and the instrument measures the induced electromotive force (V) on the coils. Assays quantified by this technology generally had a greater sensitivity compared to visual detection of iron oxide particles.548 While the current iteration of the MagnaBioSciences reader is a relatively large benchtop device that is unsuitable for use in low-resource settings, a more portable version is being developed.549,550 Other companies, such as Magnasense (Finland) and Magnisense (France), have developed portable, hand-held devices based on inductive detection of paramagnetic particles, though these devices have not been as extensively tested in the literature.
Several groups have developed lateral flow assay readers using magnetoresistive sensors. These devices are based on the principle that the electrical resistance of certain materials, which are incorporated in the sensors, can change upon application of an external magnetic field.350 There are several types of sensors based on magnetoresistance, but the most common are giant magnetoresistive (GMR) sensors, which were once used in the reading heads for hard disk drives.551 When applied to lateral flow assays, GMR sensors are placed in close proximity to the test and control lines, and an external magnetic field is applied to the test. This external field aligns the magnetic moments of paramagnetic or superparamagnetic lateral flow detection reporters. This alignment then produces a fringe field that is detectable by a change in resistance when a constant current is applied across the GMR sensor.551 Magnetoresistive sensors have been applied extensively by researchers to the detection of magnetic particle labels on lateral flow assays; however, most have been proof-of-principle studies using imitative lateral flow assays or tests for biomarkers such as hCG, cardiac troponin 1, and IFN-γ.357,552−555
The high sensitivity of magnetic detection on lateral flow assays makes this technique attractive for infectious disease diagnosis. While magnetic detection necessitates instrumentation for result readout, iron-containing magnetic nanoparticles also have the added advantage of being highly absorbent visual labels.347 Thus, magnetic nanoparticles can potentially be applied to different use-case scenarios with differing sensitivity requirements. In a use-case scenario such as morbidity control, where it is most important to identify high-intensity infections in the field, the visual detection of magnetic particles would be sufficient. When improved sensitivity is desired to detect low-intensity infections for epidemiological or surveillance purposes, analysis using the current instrumentation for reading magnetic nanoparticle labels could be performed in a district hospital or laboratory. However, further work on miniaturization of instrumentation and validation of hand-held readers will be required for field application of magnetic detection on lateral flow assays.
5.5. Electrochemical Instrumentation
As discussed in previous sections, electrochemical detection offers several benefits for POC infectious disease detection, namely that it is inexpensive, sensitive, rapid, and quantitative. The electrochemical detection systems previously discussed were largely proof-of-concept diagnostics for detection of protein or nucleic acid biomarkers; the infrastructure required to carry out these assays would limit their utility in a primary healthcare setting in low- and middle-income countries. Integration of validated and field-ready electrochemical detection systems with developed electrochemical assays represents a viable strategy for implementing the next generation of POC diagnostics.
The blood glucose biosensor is a remarkably successful electrochemical device that is ubiquitously applied at the point of care. Generally, electrochemical glucose meter kits consist of three components: a lancet for sample collection, plastic test strips for wicking of the glucose-containing sample and generation of current, and a device for digital readout of the generated current.556 The test strips house an electrochemical cell that contains a counter-reference electrode, fill-detection electrode, enzyme-coated working electrode, and mediators (Figure 37A). In these devices, the working electrode is coated with the enzyme glucose oxidase (GOx) or glucose dehydrogenase (GDH), and the enzyme oxidizes the glucose present in a blood sample. Ferrocene derivatives or other Group VIII metal complexes act as mediators to facilitate electron transfer from the enzyme redox center to the electrode surface, thereby producing a measurable current.556 Since the development of the first commercial blood glucose biosensor in 1987, the use of this technology has drastically expanded with the current market now offering numerous blood glucose meters that are portable, rapid, sensitive, and quantitative.557 The blood glucose biosensor has had an enormous impact on management of types I and II diabetes where constant monitoring and a rapid time to result are critical to preventing incidences of hyper- and hypoglycemia. Successful adaptation of this technology for field detection of infectious diseases would have a profound impact in disease surveillance and elimination campaigns, where sensitive and quantitative results are desired to best inform healthcare professionals about disease distribution.556,557
Figure 37.
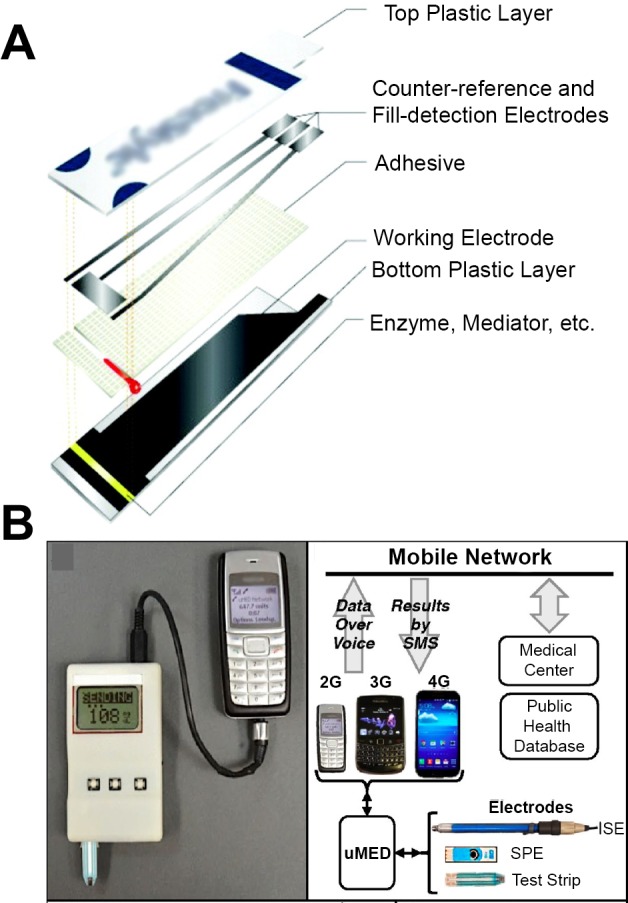
Point-of-care electrochemical diagnostics. (A) Layers comprising a test strip for amperometric measurement of blood glucose using a blood glucose meter. Adapted from ref (556). Copyright 2008 American Chemical Society. (B) Components of the universal mobile electrochemical detector (uMED) that enables point-of-care detection of analytes and subsequent connection to a mobile network. Adapted with permission from ref (173). Copyright 2014 National Academy of Sciences.
Several groups have repurposed commercial blood glucose meters for detection of a variety of targets.167,558−562 Xiang and Lu558 developed a platform that utilized a personal glucose meter for signal output to detect protein (IFN-γ), nucleic acid (adenosine), small molecule (cocaine), or toxic ion (uranium) targets. Their strategy employed a magnetic bead-conjugated aptamer specific for the biomarker of interest. The aptamer was initially prehybridized to a partially complementary DNA strand that was linked to invertase, an enzyme that catalyzes the hydrolysis of sucrose into its fructose and glucose subunits. In biomarker-containing samples, the aptamer bound to the biomarker, releasing the invertase-conjugated complementary DNA into solution. The magnetic beads were then removed from solution using an external magnet, and the addition of sucrose to the supernatant resulted in glucose production that was readily detectable by a personal glucose meter. While this creative application of an extant platform expanded its functionality, it also increased the number of assay steps, potentially limiting its use directly at the point of care. To address this concern, the Lu group562 has since integrated this solution-based assay into a lateral flow strip with dried reagents, eliminating several user steps and further simplifying the platform such that it could be applied at the point of care. This innovative approach is an excellent example of how metal-based sample preparation using magnetic beads can be combined with an existing electrochemical platform for rapid detection of a variety of disease targets.
As the blood glucose meter clearly demonstrates, portable electrochemical instrumentation has enabled minimally trained users to measure analytes at the point of care and rapidly obtain information regarding their health status. Mobile health technologies (i.e., mobile phones and telemedicine) augment these advances in instrumentation, connecting individual users to an entire healthcare infrastructure. Mobile health technologies are useful for tracking patient information and facilitating communication between the patient and the care provider. As these technologies mature, they are becoming increasingly important for modeling diseases and disease trends throughout various populations.563 Integration of portable electrochemical diagnostics with mobile health technologies in so-called “connected” diagnostics has been pursued by interfacing electrochemical devices with mobile phones.173,564 Nemiroski and colleagues173 developed a universal mobile electrochemical detector (uMED) that combined a potentiostat with virtually any mobile phone using audio-based data transmission (Figure 37B). The device could accommodate several electrochemical techniques (amperometry, coulometry, voltammetry, and potentiometry), and the voice system-based data ensured broad compatibility with a variety of mobile phones and networks. The authors employed the uMED to detect blood glucose, heavy metals in water, electrolytes, and malarial biomarker HRP2. Following sample collection and measurement using the potentiostat, the user placed a phone call to a Skype number in a remote location to vocally report the value of the analyte measured. The remote application then extracted the value from the audio data and sent an SMS message back to the user with additional diagnostic information (e.g., “LOW” if the reported blood glucose level was low). Although the uMED was applied to detect an infectious disease biomarker (HRP2), it is limited by the availability of existing compatible testing technology. For instance, HRP2 detection required an offline 96-well plate ELISA to be conducted prior to analysis on the uMED and transmission over a mobile network. As more progress is made in electrochemical diagnostic design, the uMED and other similar devices will be able to reach their full potential as “connected” diagnostics.
As battery-operated and portable devices become more robust, electrochemical detection devices are becoming increasingly viable as POC platforms. The success of the personal glucose meter is a testament to the effective and reliable diagnostic performance that is possible with electrochemical measurements. With the concurrent expansion of information technologies, “connected” electrochemical diagnostics could serve as the link between the chemistries utilized at the point of care and the remote higher level healthcare systems that map disease trends and inform treatment decisions.
5.6. Wearable Diagnostics
In high-income countries, advances in portable electronics and instrumentation have enabled live health tracking at consumers’ fingertips. Wearable commercial health diagnostics such as Apple Watch, Garmin, and FitBit are simple to use and provide real-time updates of heart rate, activities completed, or calories burned. In a similar vein, there are several emerging health-centered tools, such as press-on tattoos,565,566 skin patches,567 and face masks,568 capable of detecting changes in human physiology at the point of use. In addition, the advent of technologies such as Google Glass have allowed for “connected” interpretation of rapid diagnostic tests.515 Overall, wearable instruments combine sample collection, sample preparation, analysis and readout, and data transmission into one device, representing significant progress in diagnosis and health management at the point of care.
Small electrochemical instrumentation has become a focus of innovation, and research in this field has led to a number of skin-based wearable diagnostics. Jia et al.565 developed a noninvasive tattoo biosensor capable of monitoring lactate levels in human sweat, while another tattoo sensor produced by Bandodkar et al.566 detected glucose in the interstitial fluid of the skin (Figure 38A). These technologies are extraordinarily promising for future diagnostic devices, particularly as new biomarkers of infectious diseases are discovered in surface biofluids such as sweat.569
Figure 38.
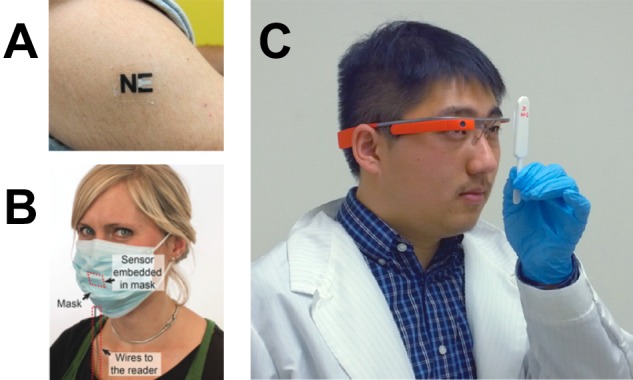
(A) Press-on tattoo sensors for monitoring lactate levels in sweat. Adapted from ref (565). Copyright 2013 American Chemical Society. (B) Paper surgical mask with embedded electrical sensor for monitoring respiration. Adapted with permission from ref (568). Copyright 2016 Wiley-VCH Verlag GmbH & Co. (C) Google Glass used for analysis of an RDT. Adapted from ref (515). Copyright 2014 American Chemical Society.
Recent advances in microprojection570,571 and microneedle572,573 technology have allowed researchers to expand the use of skin-based wearables beneath the surface of skin. In several applications, patches consisting of many microprojections were easily applied to the skin with a gentle press and could draw hundreds of microliters of blood with little-to-no pain.567,574−576 In 2014, Lee et al.567 outlined the design of a multiplexed patch that captured both IgG and malarial biomarker HRP2 from the skin of inoculated mice. The patch consisted of microprojection arrays (MPAs) that were subsequently functionalized with anti-HRP2 antibodies, Fc-specific antimouse IgG antibodies, and antidengue antibodies. The anti-HRP2-functionalized MPAs detected the biomarker, while the anti-IgG-functionalized MPAs acted as a positive control for skin penetration, and the antidengue-functionalized MPA served as a negative control. Though sample collection and preparation were applicable at the point of use, the assay required an ELISA for colorimetric readout of the captured antigen. Further research into integration of this technology with a diagnostic such as a stacked paper-based assay would allow for real-time skin-based monitoring of infectious diseases.
Another forthcoming application for wearable instrumentation is in breath analysis via paper masks. Güder et al.568 developed an electrical sensor to monitor respiration. The sensor consisted of a paper surgical mask printed with graphite electrodes that were attached to a battery and signal amplifier. As a user breathed, the humidity changes from the inhalation and exhalation generated a measurable current across the electrode, which was then detected using a Bluetooth-connected mobile device (Figure 38B). As this technology evolves, it may be possible to adapt it for analysis of breath-based biomarkers, particularly for respiratory infectious diseases such as tuberculosis and influenza. In addition, incorporating the external battery/amplifier pack into the material of the mask may open avenues for broader distribution, particularly to low-resource areas.
Likewise, personal computer displays such as Google Glass have enabled users to harness the power of a technological workstation in a hands-free glasses format. This capacity could significantly improve diagnostic capabilities and effectiveness. In 2014, Feng et al.515 developed a method of lateral flow assay analysis which combined visual examination of a lateral flow test with computerized image analysis and wireless storage capability (Figure 38C). This wearable imaging technique allowed users to collect images of tests and store them in a cloud-based system or locally on the Google Glass headset if Internet access was unavailable. Once the Google Glass headset was connected to a wireless network, the test images could be sent to a secure server to be analyzed, and then the test’s results and analysis were sent back to the viewer’s eyepiece. This method has been demonstrated using rapid diagnostic tests for both HIV antibodies and prostate specific antigen (PSA),515 though it has the potential to be adapted for any lateral flow assay with visual labels. While the use of computer-based imaging may remove some of the inaccuracies that arise from subjective human-based interpretation of results, it must also be noted that there are a number of technological hurdles that should first be addressed. The Google Glass is not capable of autofocusing, and the images were taken under good lighting against white and universally solid, simple backdrops. Thus, this technology is limited in its field application. In addition, test recognition was dependent on preprinted QR codes that were prepared and attached to the tests by researchers before use. In order to more efficiently utilize this technology, the Google Glass application could be adapted to identify test features without the need for a user-supplied QR code and web-based test input. This would simplify the workflow and allow for more high-throughput analysis of diagnostic tests.
Overall, there is significant room for growth and development in the field of wearable instrumentation. Wearable diagnostics, which may have previously seemed more at home in the world of science fiction, are becoming commonplace in health-related research efforts. The development of wearable diagnostic technologies could drastically improve the simplicity of diagnostic measurements and interpretation in regions with little or no laboratory infrastructure. As these technologies continue to be developed for applications in high-resource areas of the world, the scientific community would be remiss not to consider the impact they could have on infectious disease diagnosis in resource-limited settings.
6. Emerging Application: Leveraging Inorganic Chemistry for Nucleic Acid Detection
Thus far, we have highlighted innovative examples of metal-based strategies for improving point-of-care diagnosis of infectious diseases based primarily on protein and inorganic disease biomarkers. In most cases, leveraging inorganic chemistry for sample preparation methods and detection probes would require minimal modification for application at the point of care. It is clear that, despite the potential trade-offs in complexity of workflow and instrumentation, the increased sensitivity offered by these metal-based strategies for biomarker detection could significantly improve morbidity control and elimination efforts.
In many instances, though, sensitive and quantitative detection of the presence or absence of a disease does not paint the full picture needed to implement successful control or elimination campaigns. In some cases, the pathogen’s own genetic material must be detected and extensively studied on a population scale in order to inform strategy. This research provides valuable information on pathogen species and strain distribution and drug resistance, which can inform treatment and diagnostic strategies in a control or elimination campaign.579
The information-rich and highly specific nature of nucleic acid detection makes these biomarkers attractive targets for point-of-care diagnostics. In fact, several groups have begun to apply some of the metal-based detection strategies for proteins and inorganic biomarkers discussed in the preceding sections to the detection of pathogenic DNA or RNA.231,234,235,273,580−587 For instance, many metal-containing nanoparticle probes have been functionalized to detect pathogen-specific nucleic acid sequences in a variety of formats. The size-dependence of SPR for noble-metal nanoparticles has been leveraged in an aggregation assay for multiplexed detection of Middle East respiratory syndrome coronavirus, M. tuberculosis, and HPV in a μPAD format.231 The high conductivity of AgNPs has been exploited for electrochemical detection of M. tuberculosis and HIV.234,235 Luminescent particles (i.e., quantum dots, europium chelate-doped particles, and upconverting phosphors) have been functionalized for the detection of a myriad of diseases, including HIV,273,580−583 HBV,273 HCV,273 HPV,584P. falciparum malaria,585Bacillus cereus,581,582,586,587 and Streptococcus pyogenes.581 Moreover, these nanoparticle-based methods occur in a variety of formats, including lateral flow assays.
Because specific nucleic acid sequences are typically present at much lower concentrations than protein biomarkers in patient samples, a target amplification step is often required before the detection method can be applied. The most common target amplification technique is PCR, a time-consuming enzyme-based method that requires technical expertise, expensive equipment, and significant laboratory infrastructure. These requirements are the primary barriers to sensitive detection of pathogen genetic material in the field. Fortunately, alternative target amplification techniques with potential point-of-care applications have emerged. For example, the biobarcode signal amplification strategy discussed in section 4.4.4 has been adapted for amplification of nucleic acid targets.464 However, in general, isothermal enzymatic amplification techniques have demonstrated the most sensitivity and promise for point-of-care diagnostic applications.
Isothermal amplification techniques do not require any thermal cycling, are simpler to operate, and require fewer resources when compared to traditional PCR. This class of enzymatic amplification techniques includes loop-mediated isothermal amplification (LAMP), helicase-dependent amplification (HDA), rolling circle amplification (RCA), multiple displacement amplification (MDA), recombinase polymerase amplification (RPA), and nucleic acid sequence-based amplification (NASBA).588 Among these strategies, LAMP is one of the most widely validated, having been employed for amplification of nucleic acid targets for a variety of diseases and infections. These include targets for malaria,589−592 HIV,593 HPV,594Bordetella pertussis,595C. trachomatis,595,596Neisserie gonorrheae,595 H1N1 influenza,595,597E. coli,598 and several filarial parasites.599 In addition to malaria diagnosis, LAMP has also been used to detect artemisinin resistance in P. falciparum malaria.600 This technique consists of iterative elongation and recycling of stem-loop DNA structures, usually requiring a constant temperature between 60 and 65 °C.588 The relatively simple workflow of LAMP (when compared to other enzymatic amplification techniques) and its potential for colorimetric visual readout have led to the development of several low-resource devices for nucleic acid detection. One example is the noninstrumented nucleic acid amplification device (NINA), which relies on an exothermal chemical reaction (MgFe powder, saline, and oxygen) coupled to a phase change material for prolonged and precise heating inside a Thermos cup.589,601 Despite its streamlined workflow, the NINA device does not account for sample preparation. The preamplification nucleic acid extraction step still requires laboratory equipment such as a heated water bath and a centrifuge.589 Although the amplification step itself has been modified for use at the point of care, the complete protocol relegates it to the laboratory environment.
In an effort to mitigate these limitations, the Klapperich group594 has developed an inventive method in which nucleic acid extraction, amplification, and detection are fully integrated into a single paper fluidic device. To accomplish this, DNA from clinical samples was precipitated from solution and applied to a paper membrane, which was washed with ethanol to remove impurities. LAMP reaction reagents, which included FITC-conjugated forward and biotinylated reverse loop primers, were then applied to the sample port, which was sealed for a 30 min heating step on a hot plate. The amplified solution was then wicked onto a lateral flow assay equipped with anti-FITC antibodies on the test line and streptavidin-coated AuNPs for detection (Figure 39). In the presence of the target, both primers hybridized to the amplified genetic material, forming a “sandwich” on the lateral flow assay and resulting in a visible test line signal.594
Figure 39.
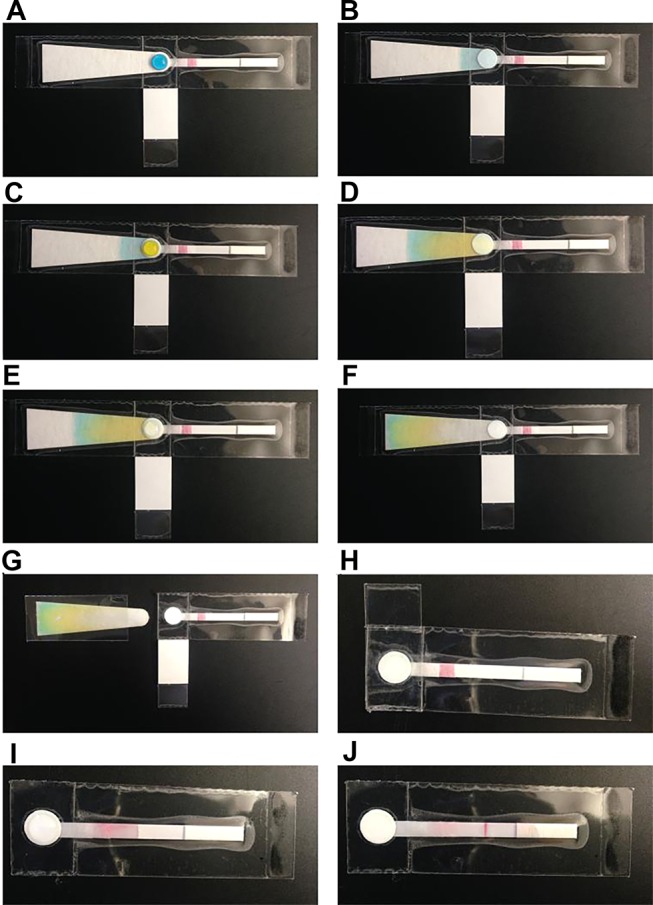
Workflow for detecting nucleic acids on a paper fluidic device. Different dyes were used to illustrate the various components of the assay. (A) The DNA-containing sample (blue dye) is added to the sample port, where it wicks down the test by capillary action (B). The test is then washed twice with ethanol (yellow), which also flows down the test (C–F). (G) The absorbent pad is then removed and discarded. (H) The LAMP reaction mix is then added to the sample port, and the test is folded to protect the sample/LAMP mixture from evaporation. (I) Once the test has been heated on a hot plate for 30 min, the hydrophobic barrier is removed so that the sample port is exposed on top and bottom, allowing it to come into contact with the LFD strip for DNA elution. (J) The sample, once eluted, can then wick down the length of the test strip to generate a signal. Adapted with permission from ref (594). Copyright 2016 Royal Society of Chemistry.
This all-in-one paper-based nucleic acid test is a perfect example of how target extraction and amplification can be innovatively combined with metal-based detection elements. This integration will allow for complex nucleic acid detection protocols to be performed at the point of care that previously were only possible in well-equipped laboratories. Moving forward, several considerations must be addressed, including integration of laboratory-free heating elements, reducing the number of user steps, and large-scale manufacturing feasibility. Nonetheless, combination of the paper-based devices developed by the Klapperich group with simple, integrated heating elements similar to the NINA device could allow for sensitive and information-rich nucleic acid detection in low resource settings. This would enable case management and elimination strategies to be precisely carried out in nearly any healthcare setting.
7. Conclusion
The application of inorganic chemistry and nanomaterials to point-of-care infectious disease diagnosis has enhanced the sensitivity and specificity of diagnostic tests, leading to improved outcomes and reduced transmission. The sample preparation and detection methods presented herein are promising examples of how metal-based strategies can expand the use-case scenarios of diagnostic tests, resulting in access to care that would otherwise be unattainable. However, we have also seen that these same metal-based strategies that improve diagnostic sensitivity and specificity can lead to more complex tests, potentially requiring more user steps, expertise, and time to complete.
These trade-offs of complexity, cost, and speed must be weighed against the potential impact that improved sensitivity and specificity could have in a given use-case scenario. For example, in the case of morbidity control, where the goal is to reduce the prevalence of high-intensity infections, the demand for simple, affordable, and rapid tests likely outweighs the need for highly sensitive tests. Once in the elimination phase, however, where the goal is to completely interrupt local transmission, high sensitivity tests are imperative for detecting and treating low-density infections. Consequently, the need for more sensitive diagnostics likely outweighs small increases in test complexity or cost for elimination campaigns.
In our evaluation of various inorganic complexes and nanomaterials, advantages and caveats became apparent for different classes of materials. The properties of these materials directly affect how they can be applied in diagnostics. Noble metal nanoparticles have frequently been incorporated into paper-based assays, such as lateral flow assays, with visual signal readout because of their stability and superior molar absorptivities.219,221,229,231 Despite the strong optical signal that noble metal nanoparticles afford, these colorimetric assays are often still limited by poor sensitivity. This potentially limits their impact in a use-case scenario such as an elimination campaign.
If better sensitivity is needed, there are essentially three metal-based strategies that we have discussed in this review for further improvement: (1) sample preparation for biomarker enrichment, (2) luminescent detection probes, and (3) signal amplification techniques. The IMAC-functionalized materials used for sample preparation collectively represent one approach for improving assay sensitivity by enriching the biomarker concentration. While some of the sample preparation methods demonstrated increased diagnostic sensitivity, these methods did add to the number of user steps.85,99,113−115,117 Metal-based sample preparation is perhaps the simplest of the three metal-based approaches to increasing sensitivity; however, for it to be truly impactful, it will need to be incorporated into diagnostics that are developed with the end user in mind such that the overall workflow is considered and the assay steps are minimized.
Alternatively, noble metal nanoparticles can be replaced with luminescent detection probes to afford greater sensitivity. Quantum dots, lanthanide chelate-doped nanoparticles, and up-converting phosphor nanoparticles all potentially offer improvements in sensitivity over colorimetric labels, albeit through different mechanisms.257,289,305,309 While luminescent inorganic complexes were incorporated into proof-of-principle assays for the detection of infectious disease biomarkers, they have yet to be utilized in a field-deployable format and lack the sensitivity of the aforementioned nanomaterials.138,139 Quantum dots and up-converting phosphor nanoparticles also have the capability for multiplexing that could be beneficial for case management.252,253,310 Although the various luminescent nanoparticles offer improvements in sensitivity, they also require more instrumentation for signal readout. The increased cost and complexity of signal readout for luminescent nanoparticles must therefore be considered when developing diagnostic strategies to combat infectious diseases.
Lastly, metal-based signal amplification techniques could be incorporated to improve sensitivity. Almost all of the signal amplification techniques discussed in this review (metalloenzyme-based amplification, nanoparticle dissolution, nanoparticle enzyme mimics, reductive nanoparticle enlargement, and biobarcodes) offer colorimetric readouts with improved sensitivity. However, these improvements come at the cost of an increased number of assay steps and thus longer total assay times. While some efforts, such as the p24 LFAs developed by Loynachan et al.,435 demonstrated progress toward more field-friendly signal amplification methods, further improvements in the ease of use and time to result must be made to translate these methods to primary healthcare facilities.
For these metal-based strategies that improve sensitivity, it is clear that simplifying the user interface is a significant challenge that remains. Aside from the user interface, there are several other challenges to translating novel diagnostic strategies that must be considered before implementation. These include: (1) the need for clinical validation, (2) the potential for large-scale manufacturability, (3) the availability of distribution channels, and (4) integration into current health systems. Most of the metal-based strategies for improved infectious disease diagnostics presented in this review have demonstrated some level of feasibility in a small set of patient or mock clinical samples. However, new technology must be clinically validated on a large set of patient samples from endemic regions before implementation. Such studies provide insight into the diagnostic performance and tolerance of day-to-day and interpatient variation in the appropriate clinical context. This rigor can draw attention to challenges and weaknesses that would otherwise go unnoticed in a research laboratory setting.
Production scalability of novel diagnostic technology is another important consideration for successful translation. Even in the research phase of diagnostic development, material choice, device design, and fabrication should be based, in part, on manufacturing scalability in order to streamline implementation. Many of the metal-based strategies presented in this review, particularly the diverse inorganic nanoparticle detection probes used in lateral flow assays, are advantageous in this context because they are easily integrated into existing LFA manufacturing infrastructure.
Beyond the laboratory, it is important to consider distribution channels and how novel tools will be integrated into existing health systems. Region-specific health delivery systems and their respective regulatory approvals are ultimately what define a novel diagnostic device’s implementation. For example, in one region, most rapid testing for infectious diseases may be performed at local health clinics run by a national ministry of health, whereas in another area, privately run dispensaries may be the most common source of primary health care. Understanding the structural differences between such health delivery systems as well as the respective availability of resources could impact device design in the research phase.
Although there are many considerations and barriers to translating novel technologies to the clinical setting, we are optimistic that the continued development of metal-based point-of-care diagnostic strategies will have a profound impact on global health. Returning to the example presented in the introduction, if some of the ultrasensitive p24 detection methods presented in this review396,435 were adapted for use at primary healthcare facilities, the time to result for infant HIV diagnosis at the point of need would be significantly reduced. In areas like Burkina Faso, this would enable earlier initiation of very effective antiretroviral therapies and drastically reduce infant mortality stemming from opportunistic infections.
Finally, the continually increasing connectivity in low- and middle-income countries coupled to sensitive diagnostics will allow for real-time disease surveillance, intervention analysis, and epidemiological studies. “Connected” diagnostics have the potential to accelerate communications between field workers, local health outposts, district hospitals, and national health ministries. The ability to rapidly convey diagnostic results could not only enable active case detection strategies but also would provide faster responses to epidemics such as the 2014–2016 Ebola and 2015–2016 Zika outbreaks. These faster responses could potentially reduce mortality and improve clinical outcomes.
Metal-based strategies for sample preparation and signal generation show great promise for use in the development and improvement of sensitive diagnostic devices for low-resource settings. These devices, coupled with connected, field-deployable instrumentation, will empower rapid response, effective control, and successful elimination of infectious diseases worldwide, ushering in an era of sustainable and improved global health.
Acknowledgments
This work is supported by the National Institutes of Health (R01 AI135937; R21 TW010635-01), NIH/Fogarty International Center (D43 TW009348), the Bill and Melinda Gates Foundation (OPP1123092), and Vanderbilt University through the Laboratories for Innovations in Global Health Technologies. C.F.M. and K.A.R. acknowledge support from the National Science Foundation Graduate Research Fellowship Program (DGE-1145197). We gratefully acknowledge Shellie Richards for her tireless proofreading efforts that sharpened the manuscript.
Biographies
Christine Markwalter earned her B.S. in Chemistry and Mathematics at Agnes Scott College in Decatur, GA, and received her Ph.D. in Chemistry under the direction of Dr. David Wright at Vanderbilt University in Nashville, TN. Her graduate work focused on developing strategies for improving diagnosis of malaria and schistosomiasis in low-resource settings. During her doctoral studies, she was the recipient of a National Science Foundation Graduate Research Fellowship and a NSF Graduate Opportunities Worldwide Fellowship. She is currently a postdoctoral associate with Dr. Myaing Nyunt and Dr. Christopher Plowe at the Duke Global Health Institute working with novel sero-surveillance tools to measure recent and remote malaria exposure in Myanmar.
Andrew Kantor obtained his B.S. in Chemistry from Furman University in Greenville, South Carolina. He is currently pursuing his Ph.D. in Chemistry at Vanderbilt University under the supervision of Dr. David Wright. His research focuses on sample preparation and signal amplification strategies to improve point-of-care diagnostics for malaria.
Carson Moore earned her B.S. in Chemistry from Mercer University and is a graduate student pursuing her Ph.D. in Chemistry at Vanderbilt University under the supervision of Dr. David Wright. Currently, her research focuses on the improvement of low-resource paper diagnostics for malaria and schistosomiasis, emphasizing the interface between chemistry and global health. Her personal research interests include metal-affinity chemistry, surface analysis of affinity ligands, and chemical functionalization of paper.
Kelly Richardson completed her B.S. in Biochemistry at Middle Tennessee State University in 2014. She then joined Prof. David Wright’s group at Vanderbilt University as a Research Assistant and now continues her work in his lab as a National Science Foundation Graduate Research Fellow in Vanderbilt’s Chemistry Ph.D. program. Her research interests focus on the development of low-resource diagnostics and the design of research tools to better understand infectious disease pathology.
David Wright is the Stevenson Professor of Chemistry and serves as Vanderbilt University’s Dean of Sciences in the College of Arts and Science. After graduating magna cum laude from Tulane University with a B.S. in Chemistry and a B.A. in Classics, Wright earned his Ph.D. from the Massachusetts Institute of Technology under Prof. William Orme-Johnson. He completed his education as a Postdoctoral Research Fellow at Boston College under Prof. William H. Armstrong. Wright held appointments at both Boston College and Duquesne University before joining Vanderbilt University in 2001. He has received various research honors, including a National Foundation for Infectious Diseases Young Investigator award (1998), a National Science Foundation CAREER award (2001), and named a Kavli Foundation Fellow (2011) and an American Association for the Advancement of Science Fellow (2014). For the past decade, he has unraveled the mechanism of heme detoxification within the parasite Plasmodium falciparum, the causative agent of malaria. Additionally, he has made important contributions in understanding hemozoin’s immunomodulatory activity in parasite-host interactions, the mechanism of action for antimalarial compounds, and the development of biomarkers for low-resource diagnostics. More recent efforts have focused on the challenges of innovation in low-resource diagnostics. He has focused primarily on malaria biomarkers and new approaches to formatting and improving performance of diagnostics for the detection of asymptomatic patients, drug-resistant infections, and drug-sensitive patients. His lab works in the field in Macha, Zambia; Cape Town, South Africa; and San Marcos, Peru.
Author Present Address
† C.F.M.: Duke Global Health Institute, Durham, NC 27710.
Author Contributions
‡ C.F.M. and A.G.K. contributed equally.
Author Contributions
§ C.P.M. and K.A.R. contributed equally.
The authors declare no competing financial interest.
References
- Osler W.Chauvinism in Medicine. In Aequanimitas, with other Addresses to Medical Students, Nurses and Practitioners of Medicine; P. Blakiston’s Son & Co.: Philadelphia, 1906; p 299. [Google Scholar]
- Armstrong G. L.; Conn L. A.; Pinner R. W. Trends in Infectious Disease Mortality in the United States during the 20th Century. JAMA, J. Am. Med. Assoc. 1999, 281, 61–66. 10.1001/jama.281.1.61. [DOI] [PubMed] [Google Scholar]
- Remais J. V.; Zeng G.; Li G.; Tian L.; Engelgau M. M. Convergence of Non-Communicable and Infectious Diseases in Low- and Middle-Income Countries. Int. J. Epidemiol. 2013, 42, 221–227. 10.1093/ije/dys135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich M. J. Uneven Progress in Preventing and Treating HIV Infections Worldwide. JAMA, J. Am. Med. Assoc. 2017, 317, 687–687. 10.1001/jama.2017.0635. [DOI] [PubMed] [Google Scholar]
- Molyneux D. H.; Hopkins D. R.; Zagaria N. Disease Eradication, Elimination and Control: The Need for Accurate and Consistent Usage. Trends Parasitol. 2004, 20, 347–351. 10.1016/j.pt.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Dowdle W. R. The Principles of Disease Elimination and Eradication. Bull. W. H. O. 1998, 76 (2), 22–25. [PMC free article] [PubMed] [Google Scholar]
- Posthuma-Trumpie G. A.; Korf J.; van Amerongen A. Lateral Flow (Immuno)Assay: Its Strengths, Weaknesses, Opportunities and Threats. A Literature Survey. Anal. Bioanal. Chem. 2009, 393, 569–582. 10.1007/s00216-008-2287-2. [DOI] [PubMed] [Google Scholar]
- Model List of Essential In Vitro Diagnostics, 1st ed.; World Health Organization, 2018. [Google Scholar]
- Coulibaly M.; Meda N.; Yonaba C.; Ouedraogo S.; Congo M.; Barry M.; Thio E.; Siribié I.; Koueta F.; Ye D.; et al. Missed Opportunities for Early Access to Care of HIV-Infected Infants in Burkina Faso. PLoS One 2014, 9, e111240 10.1371/journal.pone.0111240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services . Diagnosis of HIV Infection in Infants and Children. https://aidsinfo.nih.gov/guidelines/html/2/pediatric-arv/55/diagnosis-of-hiv-infection-in-infants-and-children (accessed Feb 19, 2018).
- Violari A.; Cotton M. F.; Gibb D. M.; Babiker A. G.; Steyn J.; Madhi S. A.; Jean-Philippe P.; McIntyre J. A. Early Antiretroviral Therapy and Mortality among HIV-Infected Infants. N. Engl. J. Med. 2008, 359, 2233–2244. 10.1056/NEJMoa0800971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne D. E.; Thompson M.; Brody L. L.; Cotton M.; Draper B.; Laubscher R.; Abdullah M. F.; Myers J. E. Emergence of a Peak in Early Infant Mortality Due to HIV/AIDS in South Africa. AIDS 2009, 23, 101–106. 10.1097/QAD.0b013e32831c54bd. [DOI] [PubMed] [Google Scholar]
- Drain P. K.; Hyle E. P.; Noubary F.; Freedberg K. A.; Wilson D.; Bishai W.; Rodriguez W.; Bassett I. V. Evaluating Diagnostic Point-of-Care Tests in Resource-Limited Settings. Lancet Infect. Dis. 2014, 14, 239–249. 10.1016/S1473-3099(13)70250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S. Q.; Glanfield A.; Gobert G. N.; Jones M. K. Heme and Blood-Feeding Parasites: Friends or Foes?. Parasites Vectors 2010, 3, 108. 10.1186/1756-3305-3-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laveran C. L. A.; Kean B. H.; Mott K. E.; Russell A. J. A Newly Discovered Parasite in the Blood of Patients Suffering from Malaria. Parasitic Etiology of Attacks of Malaria. Rev. Infect. Dis. 1982, 4, 908–911. [DOI] [PubMed] [Google Scholar]
- Noland G. S.; Briones N.; Sullivan D. J. The Shape and Size of Hemozoin Crystals Distinguishes Diverse Plasmodium Species. Mol. Biochem. Parasitol. 2003, 130, 91–99. 10.1016/S0166-6851(03)00163-4. [DOI] [PubMed] [Google Scholar]
- Fong K. Y.; Wright D. W. Hemozoin and Antimalarial Drug Discovery. Future Med. Chem. 2013, 5, 1437–1450. 10.4155/fmc.13.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigala P. A.; Crowley J. R.; Hsieh S.; Henderson J. P.; Goldberg D. E. Direct Tests of Enzymatic Heme Degradation by the Malaria Parasite Plasmodium Falciparum. J. Biol. Chem. 2012, 287, 37793–37807. 10.1074/jbc.M112.414078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronado L. M.; Nadovich C. T.; Spadafora C. Malarial Hemozoin: From Target to Tool. Biochim. Biophys. Acta, Gen. Subj. 2014, 1840, 2032–2041. 10.1016/j.bbagen.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater A. F.; Swiggard W. J.; Orton B. R.; Flitter W. D.; Goldberg D. E.; Cerami A.; Henderson G. B. An Iron-Carboxylate Bond Links the Heme Units of Malaria Pigment. Proc. Natl. Acad. Sci. U. S. A. 1991, 88, 325–329. 10.1073/pnas.88.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagola S.; Stephens P. W.; Bohle D. S.; Kosar A. D.; Madsen S. K. The Structure of Malaria Pigment β-Haematin. Nature 2000, 404, 307–310. 10.1038/35005132. [DOI] [PubMed] [Google Scholar]
- Marom N.; Tkatchenko A.; Kapishnikov S.; Kronik L.; Leiserowitz L. Structure and Formation of Synthetic Hemozoin: Insights From First-Principles Calculations. Cryst. Growth Des. 2011, 11, 3332–3341. 10.1021/cg200409d. [DOI] [Google Scholar]
- Inyushin M.; Kucheryavih Y.; Kucheryavih L.; Rojas L.; Khmelinskii I.; Makarov V. Superparamagnetic Properties of Hemozoin. Sci. Rep. 2016, 6, 26212. 10.1038/srep26212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberger M.; Mayer M.; Demarest C. R. Studies in Human Malaria: I. The Preparation of Vaccines and Suspensions Containing Plasmodia. J. Immunol. 1946, 52, 325–330. [PubMed] [Google Scholar]
- Paul F.; Roath S.; Melville D.; Warhurst D. C.; Osisanya J. O. S. Separation of Malaria-Infected Erythrocytes from Whole Blood: Use of a Selective High-Gradient Magnetic Separation Technique. Lancet 1981, 318, 70–71. 10.1016/S0140-6736(81)90414-1. [DOI] [PubMed] [Google Scholar]
- Trang D. T. X.; Huy N. T.; Kariu T.; Tajima K.; Kamei K. One-Step Concentration of Malarial Parasite-Infected Red Blood Cells and Removal of Contaminating White Blood Cells. Malar. J. 2004, 3, 7. 10.1186/1475-2875-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman P. A.; Thomson J. M.; Fujioka H.; Collins W. E.; Zborowski M. Diagnosis of Malaria by Magnetic Deposition Microscopy. Am. J. Trop. Med. Hyg. 2006, 74, 568–572. 10.4269/ajtmh.2006.74.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. C.; Wilson E. B.; DeRisi J. L. Improved Methods for Magnetic Purification of Malaria Parasites and Haemozoin. Malar. J. 2010, 9, 17. 10.1186/1475-2875-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.; Massoudi M.; Antaki J. F.; Gandini A. Removal of Malaria-Infected Red Blood Cells Using Magnetic Cell Separators: A Computational Study. Appl. Math. Comput. 2012, 218, 6841–6850. 10.1016/j.amc.2011.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J.; Huang H.; Lim H.; Lim C.; Shin S. Magnetic Separation of Malaria-Infected Red Blood Cells in Various Developmental Stages. Anal. Chem. 2013, 85, 7316–7323. 10.1021/ac4012057. [DOI] [PubMed] [Google Scholar]
- Ribaut C.; Berry A.; Chevalley S.; Reybier K.; Morlais I.; Parzy D.; Nepveu F.; Benoit-Vical F.; Valentin A. Concentration and Purification by Magnetic Separation of the Erythrocytic Stages of All Human Plasmodium Species. Malar. J. 2008, 7, 45. 10.1186/1475-2875-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumari D.; Grimberg B. T.; Blankenship D.; Mugasa J.; Mugittu K.; Moore L.; Gwakisa P.; Zborowski M. Application of Magnetic Cytosmear for the Estimation of Plasmodium Falciparum Gametocyte Density and Detection of Asexual Stages in Asymptomatic Children. Malar. J. 2016, 15, 113. 10.1186/s12936-016-1170-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl S.; Gutiérrez L.; House M. J.; Davis T. M. E.; St. Pierre T. G. Nuclear Magnetic Resonance: A Tool for Malaria Diagnosis?. Am. J. Trop. Med. Hyg. 2011, 85, 815–817. 10.4269/ajtmh.2011.11-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W. K.; Kong T. F.; Ng C. S.; Chen L.; Huang Y.; Bhagat A. A. S.; Nguyen N.-T.; Preiser P. R.; Han J. Micromagnetic Resonance Relaxometry for Rapid Label-Free Malaria Diagnosis. Nat. Med. 2014, 20, 1069–1073. 10.1038/nm.3622. [DOI] [PubMed] [Google Scholar]
- Kong T. F.; Ye W.; Peng W. K.; Hou H. W.; Marcos; Preiser P. R.; Nguyen N.-T.; Han J. Enhancing Malaria Diagnosis through Microfluidic Cell Enrichment and Magnetic Resonance Relaxometry Detection. Sci. Rep. 2015, 5, 11425. 10.1038/srep11425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl S.; Mueller I.; St. Pierre T. G. Considerations Regarding the Micromagnetic Resonance Relaxometry Technique for Rapid Label-Free Malaria Diagnosis. Nat. Med. 2015, 21, 1387. 10.1038/nm.3811. [DOI] [PubMed] [Google Scholar]
- Han J.; Peng W. K. Reply to “Considerations Regarding the Micromagnetic Resonance Relaxometry Technique for Rapid Label-Free Malaria Diagnosis.. Nat. Med. 2015, 21, 1387–1389. 10.1038/nm.3959. [DOI] [PubMed] [Google Scholar]
- Gossuin Y.; Ndjolo P. O.; Vuong Q. L.; Duez P. NMR Relaxation Properties of the Synthetic Malaria Pigment β-Hematin. Sci. Rep. 2017, 7, 14557. 10.1038/s41598-017-15238-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson B. K.; Behrend M. R.; Horning M. P.; Hegg M. C. Detection of Malarial Byproduct Hemozoin Utilizing Its Unique Scattering Properties. Opt. Express 2011, 19, 12190–12196. 10.1364/OE.19.012190. [DOI] [PubMed] [Google Scholar]
- Burnett J. L.; Carns J. L.; Richards-Kortum R. In Vivo Microscopy of Hemozoin: Towards a Needle Free Diagnostic for Malaria. Biomed. Opt. Express 2015, 6, 3462–3474. 10.1364/BOE.6.003462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett J. L.; Carns J. L.; Richards-Kortum R. Towards a Needle-Free Diagnosis of Malaria: In Vivo Identification and Classification of Red and White Blood Cells Containing Haemozoin. Malar. J. 2017, 16, 447. 10.1186/s12936-017-2096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp A. M.; Ince C.; Charunwatthana P.; Hanson J.; van Kuijen A.; Faiz M. A.; Rahman M. R.; Hasan M.; Bin Yunus E.; Ghose A.; et al. Direct In Vivo Assessment of Microcirculatory Dysfunction in Severe Falciparum Malaria. J. Infect. Dis. 2008, 197, 79–84. 10.1086/523762. [DOI] [PubMed] [Google Scholar]
- Balasubramanian D.; Rao C. M.; Panijpan B. The Malaria Parasite Monitored by Photoacoustic Spectroscopy. Science 1984, 223, 828–830. 10.1126/science.6695185. [DOI] [PubMed] [Google Scholar]
- Lukianova-Hleb E. Y.; Campbell K. M.; Constantinou P. E.; Braam J.; Olson J. S.; Ware R. E.; Sullivan D. J.; Lapotko D. O. Hemozoin-Generated Vapor Nanobubbles for Transdermal Reagent- and Needle-Free Detection of Malaria. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 900–905. 10.1073/pnas.1316253111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukianova-Hleb E. Y.; Lapotko D. O. Malaria Theranostics Using Hemozoin-Generated Vapor Nanobubbles. Theranostics 2014, 4, 761–769. 10.7150/thno.9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukianova-Hleb E.; Bezek S.; Szigeti R.; Khodarev A.; Kelley T.; Hurrell A.; Berba M.; Kumar N.; D’Alessandro U.; Lapotko D. Transdermal Diagnosis of Malaria Using Vapor Nanobubbles. Emerging Infect. Dis. 2015, 21, 1122–1127. 10.3201/eid2107.150089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C.; Carey K. A.; Nedosekin D. A.; Menyaev Y. A.; Sarimollaoglu M.; Galanzha E. I.; Stumhofer J. S.; Zharov V. P. In Vivo Photoacoustic Flow Cytometry for Early Malaria Diagnosis. Cytometry, Part A 2016, 89, 531–542. 10.1002/cyto.a.22854. [DOI] [PubMed] [Google Scholar]
- Menyaev Y. A.; Carey K. A.; Nedosekin D. A.; Sarimollaoglu M.; Galanzha E. I.; Stumhofer J. S.; Zharov V. P. Preclinical Photoacoustic Models: Application for Ultrasensitive Single Cell Malaria Diagnosis in Large Vein and Artery. Biomed. Opt. Express 2016, 7, 3643–3658. 10.1364/BOE.7.003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D. M.; Heptinstall J.; Matelon R. J.; Savage L.; Wears M. L.; Beddow J.; Cox M.; Schallig H. D. F. H.; Mens P. F. A Magneto-Optic Route toward the In Vivo Diagnosis of Malaria: Preliminary Results and Preclinical Trial Data. Biophys. J. 2008, 95, 994–1000. 10.1529/biophysj.107.128140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mens P. F.; Matelon R. J.; Nour B. Y.; Newman D. M.; Schallig H. D. Laboratory Evaluation on the Sensitivity and Specificity of a Novel and Rapid Detection Method for Malaria Diagnosis Based on Magneto-Optical Technology (MOT). Malar. J. 2010, 9, 207. 10.1186/1475-2875-9-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D. M.; Matelon R. J.; Wears M. L.; Savage L. B. The In Vivo Diagnosis of Malaria: Feasibility Study Into a Magneto-Optic Fingertip Probe. IEEE J. Sel. Top. Quantum Electron. 2010, 16, 573–580. 10.1109/JSTQE.2009.2029068. [DOI] [Google Scholar]
- Butykai A.; Orbán A.; Kocsis V.; Szaller D.; Bordács S.; Tátrai-Szekeres E.; Kiss L. F.; Bóta A.; Vértessy B. G.; Zelles T.; et al. Malaria Pigment Crystals as Magnetic Micro-Rotors: Key for High-Sensitivity Diagnosis. Sci. Rep. 2013, 3, 1431. 10.1038/srep01431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbán Á.; Butykai Á.; Pröhle Z.; Fülöp G.; Zelles T.; Forsyth W.; Hill D.; Schofield L.; Müller I.; Karl S.; et al. Rotating-crystal Malaria Diagnosis: Pre-clinical validation. https://arxiv.org/pdf/1311.4103.pdf (accessed Nov 16, 2017).
- Orbán Á.; Butykai Á.; Molnár A.; Pröhle Z.; Fülöp G.; Zelles T.; Forsyth W.; Hill D.; Müller I.; Schofield L.; et al. Evaluation of a Novel Magneto-Optical Method for the Detection of Malaria Parasites. PLoS One 2014, 9, e96981 10.1371/journal.pone.0096981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbán Á.; Rebelo M.; Molnár P.; Albuquerque I. S.; Butykai A.; Kézsmárki I. Efficient Monitoring of the Blood-Stage Infection in a Malaria Rodent Model by the Rotating-Crystal Magneto-Optical Method. Sci. Rep. 2016, 6, 23218. 10.1038/srep23218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power of the Laboratory At the Point of Need. http://hemexhealth.com/technology/ (accessed July 13, 2018).
- Grobusch M. P.; Hänscheid T.; Krämer B.; Neukammer J.; May J.; Seybold J.; Kun J. F. J.; Suttorp N. Sensitivity of Hemozoin Detection by Automated Flow Cytometry in Non- and Semi-Immune Malaria Patients. Cytometry, Part B 2003, 55, 46–51. 10.1002/cyto.b.10039. [DOI] [PubMed] [Google Scholar]
- Hänscheid T.; Längin M.; Lell B.; Pötschke M.; Oyakhirome S.; Kremsner P. G.; Grobusch M. P. Full Blood Count and Haemozoin-Containing Leukocytes in Children with Malaria: Diagnostic Value and Association with Disease Severity. Malar. J. 2008, 7, 109. 10.1186/1475-2875-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo M.; Shapiro H. M.; Amaral T.; Melo-Cristino J.; Hänscheid T. Haemozoin Detection in Infected Erythrocytes for Plasmodium Falciparum Malaria Diagnosis—Prospects and Limitations. Acta Trop. 2012, 123, 58–61. 10.1016/j.actatropica.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Rebelo M.; Sousa C.; Shapiro H. M.; Mota M. M.; Grobusch M. P.; Hänscheid T. A Novel Flow Cytometric Hemozoin Detection Assay for Real-Time Sensitivity Testing of Plasmodium Falciparum. PLoS One 2013, 8, e61606 10.1371/journal.pone.0061606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirev P. A.; Feldman A. B.; Kongkasuriyachai D.; Scholl P.; Sullivan D.; Kumar N. Detection of Malaria Parasites in Blood by Laser Desorption Mass Spectrometry. Anal. Chem. 2002, 74, 3262–3266. 10.1021/ac025621k. [DOI] [PubMed] [Google Scholar]
- Demirev P. A. Mass Spectrometry for Malaria Diagnosis. Expert Rev. Mol. Diagn. 2004, 4, 821–829. 10.1586/14737159.4.6.821. [DOI] [PubMed] [Google Scholar]
- Scholl P. F.; Kongkasuriyachai D.; Demirev P. A.; Feldman A. B.; Lin J. S.; Jr D. J. S.; Kumar N. Rapid Detection of Malaria Infection in Vivo by Laser Desorption Mass Spectrometry. Am. J. Trop. Med. Hyg. 2004, 71, 546–551. 10.4269/ajtmh.2004.71.546. [DOI] [PubMed] [Google Scholar]
- Nyunt M.; Pisciotta J.; Feldman A. B.; Thuma P.; Scholl P. F.; Demirev P. A.; Lin J. S.; Shi L.; Kumar N.; Sullivan D. J. Detection of Plasmodium Falciparum in Pregnancy by Laser Desorption Mass Spectrometry. Am. J. Trop. Med. Hyg. 2005, 73, 485–490. 10.4269/ajtmh.2005.73.485. [DOI] [PubMed] [Google Scholar]
- Yuen C.; Liu Q. Magnetic Field Enriched Surface Enhanced Resonance Raman Spectroscopy for Early Malaria Diagnosis. J. Biomed. Opt. 2012, 17, 017005. 10.1117/1.JBO.17.1.017005. [DOI] [PubMed] [Google Scholar]
- Hobro A. J.; Konishi A.; Coban C.; Smith N. I. Raman Spectroscopic Analysis of Malaria Disease Progression via Blood and Plasma Samples. Analyst 2013, 138, 3927–3933. 10.1039/c3an00255a. [DOI] [PubMed] [Google Scholar]
- Bilal M.; Saleem M.; Amanat S. T.; Shakoor H. A.; Rashid R.; Mahmood A.; Ahmed M. Optical Diagnosis of Malaria Infection in Human Plasma Using Raman Spectroscopy. J. Biomed. Opt. 2015, 20, 017002. 10.1117/1.JBO.20.1.017002. [DOI] [PubMed] [Google Scholar]
- Garrett N. L.; Sekine R.; Dixon M. W. A.; Tilley L.; Bambery K. R.; Wood B. R. Bio-Sensing with Butterfly Wings: Naturally Occurring Nano-Structures for SERS-Based Malaria Parasite Detection. Phys. Chem. Chem. Phys. 2015, 17, 21164–21168. 10.1039/C4CP04930F. [DOI] [PubMed] [Google Scholar]
- Chen K.; Yuen C.; Aniweh Y.; Preiser P.; Liu Q. Towards Ultrasensitive Malaria Diagnosis Using Surface Enhanced Raman Spectroscopy. Sci. Rep. 2016, 6, 20177. 10.1038/srep20177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. M.; Shi L.; Sullivan D. J. Haemoproteus and Schistosoma Synthesize Heme Polymers Similar to Plasmodium Hemozoin and β-Hematin. Mol. Biochem. Parasitol. 2001, 113, 1–8. 10.1016/S0166-6851(00)00365-0. [DOI] [PubMed] [Google Scholar]
- Pershina A. G.; Saltykova I. V.; Ivanov V. V.; Perina E. A.; Demin A. M.; Shevelev O. B.; Buzueva I. I.; Gutakovskii A. K.; Vtorushin S. V.; Ganebnykh I. N.; et al. Hemozoin “Knobs” in Opisthorchis Felineus Infected Liver. Parasites Vectors 2015, 8, 459. 10.1186/s13071-015-1061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . World Malaria Report 2017; World Health Organization: Geneva, Switzerland, 2017; p 3.
- World Health Organization . Schistosomiasis: Strategic Report 2001–2011 and Strategic Plan 2012–2020; World Health Organization: Geneva, Switzerland, 2013; p 6. [Google Scholar]
- Day N. P.; Pham T. D.; Phan T. L.; Dinh X. S.; Pham P. L.; Ly V. C.; Tran T. H.; Nguyen T. H.; Bethell D. B.; Nguyan H. P.; et al. Clearance Kinetics of Parasites and Pigment-Containing Leukocytes in Severe Malaria. Blood 1996, 88, 4694–4700. [PubMed] [Google Scholar]
- Ashton P. D.; Harrop R.; Shah B.; Wilson R. A. The Schistosome Egg: Development and Secretions. Parasitology 2001, 122, 329–338. 10.1017/S0031182001007351. [DOI] [PubMed] [Google Scholar]
- Colley D. G.; Bustinduy A. L.; Secor W. E.; King C. H. Human Schistosomiasis. Lancet 2014, 383, 2253–2264. 10.1016/S0140-6736(13)61949-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K.; Ishii Y. Fine Structure of Schistosome Eggs as Seen through the Scanning Electron Microscope. Am. J. Trop. Med. Hyg. 1976, 25, 841–844. 10.4269/ajtmh.1976.25.841. [DOI] [PubMed] [Google Scholar]
- Toro-Goyco E.; del Valle M. R. Schistosoma Mansoni. I. Chemical Composition of Eggs. Exp. Parasitol. 1970, 27, 265–272. 10.1016/0014-4894(70)90031-7. [DOI] [PubMed] [Google Scholar]
- Jones M. K.; McManus D. P.; Sivadorai P.; Glanfield A.; Moertel L.; Belli S. I.; Gobert G. N. Tracking the Fate of Iron in Early Development of Human Blood Flukes. Int. J. Biochem. Cell Biol. 2007, 39, 1646–1658. 10.1016/j.biocel.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira C. F.; Neuhauss E.; Ben R.; Romanzini J.; Graeff-Teixeira C. Detection of Schistosoma Mansoni Eggs in Feces through Their Interaction with Paramagnetic Beads in a Magnetic Field. PLoS Neglected Trop. Dis. 2007, 1, e73 10.1371/journal.pntd.0000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldeira K.; Teixeira C. F.; Silveira M. B. da; Fries L. C. C. de; Romanzini J.; Bittencourt H. R.; Graeff-Teixeira C. Comparison of the Kato-Katz and Helmintex Methods for the Diagnosis of Schistosomiasis in a Low-Intensity Transmission Focus in Bandeirantes, Paraná, Southern Brazil. Mem. Inst. Oswaldo Cruz 2012, 107, 690–692. 10.1590/S0074-02762012000500019. [DOI] [PubMed] [Google Scholar]
- Jones M. K.; Balen J. Magnetic Beads for Schistosomiasis Diagnosis. PLoS Neglected Trop. Dis. 2007, 1, e159 10.1371/journal.pntd.0000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl S.; Gutiérrez L.; Lucyk-Maurer R.; Kerr R.; Candido R. R. F.; Toh S. Q.; Saunders M.; Shaw J. A.; Suvorova A.; Hofmann A.; et al. The Iron Distribution and Magnetic Properties of Schistosome Eggshells: Implications for Improved Diagnostics. PLoS Neglected Trop. Dis. 2013, 7, e2219 10.1371/journal.pntd.0002219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candido R. R. F.; Favero V.; Duke M.; Karl S.; Gutiérrez L.; Woodward R. C.; Graeff-Teixeira C.; Jones M. K.; St. Pierre T. G. The Affinity of Magnetic Microspheres for Schistosoma Eggs. Int. J. Parasitol. 2015, 45, 43–50. 10.1016/j.ijpara.2014.08.011. [DOI] [PubMed] [Google Scholar]
- Ricks K. M.; Adams N. M.; Scherr T. F.; Haselton F. R.; Wright D. W. Direct Transfer of HRPII-Magnetic Bead Complexes to Malaria Rapid Diagnostic Tests Significantly Improves Test Sensitivity. Malar. J. 2016, 15, 399. 10.1186/s12936-016-1448-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miessler G. L.; Tarr D. A.. Inorganic Chemistry, 4th ed.; Pearson: Upper Saddle River, NJ, 2010. [Google Scholar]
- Block H.; Maertens B.; Spriestersbach A.; Brinker N.; Kubicek J.; Fabis R.; Labahn J.; Schäfer F.. Immobilized-Metal Affinity Chromatography (IMAC): A Review. In Methods in Enzymology; Burgess R. R., Deutscher M. P., Eds.; Guide to Protein Purification, 2nd ed.; Academic Press, 2009; Vol. 463, Chapter 27, pp 439–473. [DOI] [PubMed] [Google Scholar]
- Porath J. Immobilized Metal Ion Affinity Chromatography. Protein Expression Purif. 1992, 3, 263–281. 10.1016/1046-5928(92)90001-D. [DOI] [PubMed] [Google Scholar]
- Hage D. S.; Anguizola J. A.; Li R.; Matsuda R.; Papastavros E.; Pfaunmiller E.; Sobansky M.; Zheng X.. Chapter 1: Affinity Chromatography. In Liquid Chromatography: Applications; Fanali S., Haddad P. R., Poole C. F., Schoenmakers P., Lloyd D., Eds.; Elsevier: Waltham, MA, 2013; p 11. [Google Scholar]
- Block H.; Maertens B.; Spriestersbach A.; Brinker N.; Kubicek J.; Fabis R.; Labahn J.; Schäfer F.. Immobilized-Metal Affinity Chromatography (IMAC): A Review. In Methods in Enzymology; Burgess R. R., Deutscher M. P., Eds.; Guide to Protein Purification, 2nd ed.; Academic Press, 2009; Vol. 463, Chapter 27, pp 439–473. [DOI] [PubMed] [Google Scholar]
- Arnold F. H. Metal-Affinity Separations: A New Dimension in Protein Processing. Nat. Biotechnol. 1991, 9, 151–156. 10.1038/nbt0291-151. [DOI] [PubMed] [Google Scholar]
- Cheung R. C. F.; Wong J. H.; Ng T. B. Immobilized Metal Ion Affinity Chromatography: A Review on Its Applications. Appl. Microbiol. Biotechnol. 2012, 96, 1411–1420. 10.1007/s00253-012-4507-0. [DOI] [PubMed] [Google Scholar]
- Hochuli E.; Döbeli H.; Schacher A. New Metal Chelate Adsorbent Selective for Proteins and Peptides Containing Neighbouring Histidine Residues. J. Chromatogr. A 1987, 411, 177–184. 10.1016/S0021-9673(00)93969-4. [DOI] [PubMed] [Google Scholar]
- Porath J.; Carlsson J.; Olsson I.; Belfrage G. Metal Chelate Affinity Chromatography, a New Approach to Protein Fractionation. Nature 1975, 258, 598–599. 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]
- Nelson P. S.; Yang T.-T.; Kain S. R.. Method for Purification of Recombinant Proteins. US5962641A, October 5, 1999.
- EDTA-resistant, chemical-tolerant Ni-Penta Agarose-Based Resins. https://marvelgent.com/pages/high-selectivity-ni-penta-agarose-resins-for-secreted-proteins (accessed July 16, 2018).
- Wong J. W.; Albright R. L.; Wang N.-H. L. Immobilized Metal Ion Affinity Chromatography (IMAC) Chemistry and Bioseparation Applications. Sep. Purif. Methods 1991, 20, 49–106. 10.1080/03602549108021408. [DOI] [Google Scholar]
- Yip T. T.; Hutchens T. W.. Immobilized Metal Ion Affinity Chromatography. In Practical Protein Chromatography; Kenney A., Fowell S., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, 1992; Vol. 11, pp 17–31. [DOI] [PubMed] [Google Scholar]
- Bauer W. S.; Richardson K. A.; Adams N. M.; Ricks K. M.; Gasperino D. J.; Ghionea S. J.; Rosen M.; Nichols K. P.; Weigl B. H.; Haselton F. R.; et al. Rapid Concentration and Elution of Malarial Antigen Histidine-Rich Protein II Using Solid Phase Zn(II) Resin in a Simple Flow-through Pipette Tip Format. Biomicrofluidics 2017, 11, 034115. 10.1063/1.4984788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IMAC Resins Selection Guide. https://marvelgent.com/pages/imac-selection-guideline (accessed July 16, 2018). [Google Scholar]
- Panton L. J.; McPhie P.; Lee Maloy W.; Wellems T. E.; Taylor D. W.; Howard R. J. Purification and Partial Characterization of an Unusual Protein of Plasmodium Falciparum: Histidine-Rich Protein II. Mol. Biochem. Parasitol. 1989, 35, 149–160. 10.1016/0166-6851(89)90117-5. [DOI] [PubMed] [Google Scholar]
- Gaberc-Porekar V.; Menart V. Perspectives of Immobilized-Metal Affinity Chromatography. J. Biochem. Biophys. Methods 2001, 49, 335–360. 10.1016/S0165-022X(01)00207-X. [DOI] [PubMed] [Google Scholar]
- Shi Q.-H.; Tian Y.; Dong X.-Y.; Bai S.; Sun Y. Chitosan-Coated Silica Beads as Immobilized Metal Affinity Support for Protein Adsorption. Biochem. Eng. J. 2003, 16, 317–322. 10.1016/S1369-703X(03)00095-0. [DOI] [Google Scholar]
- Xu F.; Wang Y.; Wang X.; Zhang Y.; Tang Y.; Yang P. A Novel Hierarchical Nanozeolite Composite as Sorbent for Protein Separation in Immobilized Metal-Ion Affinity Chromatography. Adv. Mater. 2003, 15, 1751–1753. 10.1002/adma.200305287. [DOI] [Google Scholar]
- Zhao M.; Deng C.; Zhang X. The Design and Synthesis of a Hydrophilic Core–Shell–Shell Structured Magnetic Metal–Organic Framework as a Novel Immobilized Metal Ion Affinity Platform for Phosphoproteome Research. Chem. Commun. 2014, 50, 6228–6231. 10.1039/c4cc01038h. [DOI] [PubMed] [Google Scholar]
- Sun X.; Liu X.; Feng J.; Li Y.; Deng C.; Duan G. Hydrophilic Nb5+-Immobilized Magnetic Core–Shell Microsphere – A Novel Immobilized Metal Ion Affinity Chromatography Material for Highly Selective Enrichment of Phosphopeptides. Anal. Chim. Acta 2015, 880, 67–76. 10.1016/j.aca.2015.04.029. [DOI] [PubMed] [Google Scholar]
- Zheng L.; Dong H.; Hu L. Zirconium-Cation-Immobilized Core/Shell (Fe3O4@Polymer) Microspheres as an IMAC Material for the Selective Enrichment of Phosphopeptides. Ind. Eng. Chem. Res. 2013, 52, 7729–7736. 10.1021/ie4003377. [DOI] [Google Scholar]
- Yan Y.; Zheng Z.; Deng C.; Li Y.; Zhang X.; Yang P. Hydrophilic Polydopamine-Coated Graphene for Metal Ion Immobilization as a Novel Immobilized Metal Ion Affinity Chromatography Platform for Phosphoproteome Analysis. Anal. Chem. 2013, 85, 8483–8487. 10.1021/ac401668e. [DOI] [PubMed] [Google Scholar]
- Wang J.; Zhang R.; Yang X.; Liu X.; Zhang H. Facile Synthesis of Copper(II)-Decorated Functional Mesoporous Material for Specific Adsorption of Histidine-Rich Proteins. Talanta 2018, 176, 308–317. 10.1016/j.talanta.2017.08.016. [DOI] [PubMed] [Google Scholar]
- Opitz L.; Hohlweg J.; Reichl U.; Wolff M. W. Purification of Cell Culture-Derived Influenza Virus A/Puerto Rico/8/34 by Membrane-Based Immobilized Metal Affinity Chromatography. J. Virol. Methods 2009, 161, 312–316. 10.1016/j.jviromet.2009.06.025. [DOI] [PubMed] [Google Scholar]
- Gu H.; Xu K.; Xu C.; Xu B. Biofunctional Magnetic Nanoparticles for Protein Separation and Pathogen Detection. Chem. Commun. 2006, 0, 941–949. 10.1039/b514130c. [DOI] [PubMed] [Google Scholar]
- Colombo M.; Carregal-Romero S.; Casula M. F.; Gutiérrez L.; Morales M. P.; Böhm I. B.; Heverhagen J. T.; Prosperi D.; Parak W. J. Biological Applications of Magnetic Nanoparticles. Chem. Soc. Rev. 2012, 41, 4306–4334. 10.1039/c2cs15337h. [DOI] [PubMed] [Google Scholar]
- Davis K. M.; Swartz J. D.; Haselton F. R.; Wright D. W. Low-Resource Method for Extracting the Malarial Biomarker Histidine-Rich Protein II To Enhance Diagnostic Test Performance. Anal. Chem. 2012, 84, 6136–6142. 10.1021/ac301030m. [DOI] [PubMed] [Google Scholar]
- Davis K. M.; Gibson L. E.; Haselton F. R.; Wright D. W. Simple Sample Processing Enhances Malaria Rapid Diagnostic Test Performance. Analyst 2014, 139, 3026–3031. 10.1039/C4AN00338A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer W. S.; Kimmel D. W.; Adams N. M.; Gibson L. E.; Scherr T.; Richardson K. A.; Conrad J. A.; Matakala H. K.; Haselton F. R.; Wright D. W. Magnetically-Enabled Biomarker Extraction and Delivery System: Towards Integrated ASSURED Diagnostic Tools. Analyst 2017, 142, 1569–1580. 10.1039/C7AN00278E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodelón G.; Mourdikoudis S.; Yate L.; Pastoriza-Santos I.; Pérez-Juste J.; Liz-Marzán L. M. Nickel Nanoparticle-Doped Paper as a Bioactive Scaffold for Targeted and Robust Immobilization of Functional Proteins. ACS Nano 2014, 8, 6221–6231. 10.1021/nn5016665. [DOI] [PubMed] [Google Scholar]
- Bauer W. S.; Gulka C. P.; Silva-Baucage L.; Adams N. M.; Haselton F. R.; Wright D. W. Metal Affinity-Enabled Capture and Release Antibody Reagents Generate a Multiplex Biomarker Enrichment System That Improves Detection Limits of Rapid Diagnostic Tests. Anal. Chem. 2017, 89, 10216–10223. 10.1021/acs.analchem.7b01513. [DOI] [PubMed] [Google Scholar]
- Markwalter C. F.; Davis K. M.; Wright D. W. Immunomagnetic Capture and Colorimetric Detection of Malarial Biomarker Plasmodium Falciparum Lactate Dehydrogenase. Anal. Biochem. 2016, 493, 30–34. 10.1016/j.ab.2015.10.003. [DOI] [PubMed] [Google Scholar]
- Makler M. T.; Palmer C. J.; Ager A. L. A Review of Practical Techniques for the Diagnosis of Malaria. Ann. Trop. Med. Parasitol. 1998, 92, 419–433. 10.1080/00034989859401. [DOI] [PubMed] [Google Scholar]
- Markwalter C. F.; Ricks K. M.; Bitting A. L.; Mudenda L.; Wright D. W. Simultaneous Capture and Sequential Detection of Two Malarial Biomarkers on Magnetic Microparticles. Talanta 2016, 161, 443–449. 10.1016/j.talanta.2016.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson L. E.; Markwalter C. F.; Kimmel D. W.; Mudenda L.; Mbambara S.; Thuma P. E.; Wright D. W. Plasmodium Falciparum HRP2 ELISA for Analysis of Dried Blood Spot Samples in Rural Zambia. Malar. J. 2017, 16, 350. 10.1186/s12936-017-1996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson L. E.; Mudenda L.; Kimmel D. W.; Mbambara S.; Thuma P. E.; Wright D. W.; Markwalter C. F. Characterization of Plasmodium Lactate Dehydrogenase and Histidine-Rich Protein 2 Clearance Patterns via Rapid On-Bead Detection from a Single Dried Blood Spot. Am. J. Trop. Med. Hyg. 2018, 98, 1389–1396. 10.4269/ajtmh.17-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasena S. D. Aptamers: An Emerging Class of Molecules That Rival Antibodies in Diagnostics. Clin. Chem. 1999, 45, 1628–1650. [PubMed] [Google Scholar]
- Toh S. Y.; Citartan M.; Gopinath S. C. B.; Tang T.-H. Aptamers as a Replacement for Antibodies in Enzyme-Linked Immunosorbent Assay. Biosens. Bioelectron. 2015, 64, 392–403. 10.1016/j.bios.2014.09.026. [DOI] [PubMed] [Google Scholar]
- Nery A. A.; Wrenger C.; Ulrich H. Recognition of Biomarkers and Cell-Specific Molecular Signatures: Aptamers as Capture Agents. J. Sep. Sci. 2009, 32, 1523–1530. 10.1002/jssc.200800695. [DOI] [PubMed] [Google Scholar]
- Ellington A. D.; Szostak J. W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Storr T.; Thompson K. H.; Orvig C. Design of Targeting Ligands in Medicinal Inorganic Chemistry. Chem. Soc. Rev. 2006, 35, 534–544. 10.1039/b514859f. [DOI] [PubMed] [Google Scholar]
- Mjos K. D.; Orvig C. Metallodrugs in Medicinal Inorganic Chemistry. Chem. Rev. 2014, 114, 4540–4563. 10.1021/cr400460s. [DOI] [PubMed] [Google Scholar]
- Meggers E. Exploring Biologically Relevant Chemical Space with Metal Complexes. Curr. Opin. Chem. Biol. 2007, 11, 287–292. 10.1016/j.cbpa.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Meggers E. Targeting Proteins with Metal Complexes. Chem. Commun. 2009, 0, 1001–1010. 10.1039/b813568a. [DOI] [PubMed] [Google Scholar]
- Barry N. P. E.; Sadler P. J. Dicarba-Closo-Dodecarborane-Containing Half-Sandwich Complexes of Ruthenium, Osmium, Rhodium and Iridium: Biological Relevance and Synthetic Strategies. Chem. Soc. Rev. 2012, 41, 3264–3279. 10.1039/c2cs15300a. [DOI] [PubMed] [Google Scholar]
- Mornet S.; Vasseur S.; Grasset F.; Duguet E. Magnetic Nanoparticle Design for Medical Diagnosis and Therapy. J. Mater. Chem. 2004, 14, 2161–2175. 10.1039/b402025a. [DOI] [Google Scholar]
- Yao J.; Yang M.; Duan Y. Chemistry, Biology, and Medicine of Fluorescent Nanomaterials and Related Systems: New Insights into Biosensing, Bioimaging, Genomics, Diagnostics, and Therapy. Chem. Rev. 2014, 114, 6130–6178. 10.1021/cr200359p. [DOI] [PubMed] [Google Scholar]
- Smith B. R.; Gambhir S. S. Nanomaterials for In Vivo Imaging. Chem. Rev. 2017, 117, 901–986. 10.1021/acs.chemrev.6b00073. [DOI] [PubMed] [Google Scholar]
- Jain P. K.; Huang X.; El-Sayed I. H.; El-Sayed M. A. Noble Metals on the Nanoscale: Optical and Photothermal Properties and Some Applications in Imaging, Sensing, Biology, and Medicine. Acc. Chem. Res. 2008, 41, 1578–1586. 10.1021/ar7002804. [DOI] [PubMed] [Google Scholar]
- Vellaisamy K.; Li G.; Ko C.-N.; Zhong H.-J.; Fatima S.; Kwan H.-Y.; Wong C.-Y.; Kwong W.-J.; Tan W.; Leung C.-H.; et al. Cell Imaging of Dopamine Receptor Using Agonist Labeling Iridium(III) Complex. Chem. Sci. 2018, 9, 1119–1125. 10.1039/C7SC04798C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D.-L.; Wang M.; Liu C.; Miao X.; Kang T.-S.; Leung C.-H. Metal Complexes for the Detection of Disease-Related Protein Biomarkers. Coord. Chem. Rev. 2016, 324, 90–105. 10.1016/j.ccr.2016.07.010. [DOI] [Google Scholar]
- Davis K. M.; Bitting A. L.; Wright D. W. On-Particle Detection of Plasmodium Falciparum Histidine-Rich Protein II by a “Switch-on” Iridium(III) Probe. Anal. Biochem. 2014, 445, 60–66. 10.1016/j.ab.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Lin S.; He B.; Yang C.; Leung C.-H.; Mergny J.-L.; Ma D.-L. Luminescence Switch-on Assay of Interferon-Gamma Using a G-Quadruplex-Selective Iridium(III) Complex. Chem. Commun. 2015, 51, 16033–16036. 10.1039/C5CC06655G. [DOI] [PubMed] [Google Scholar]
- Liu G.; Zhang K.; Ma K.; Care A.; Hutchinson M. R.; Goldys E. M. Graphene Quantum Dot Based “Switch-on” Nanosensors for Intracellular Cytokine Monitoring. Nanoscale 2017, 9, 4934–4943. 10.1039/C6NR09381G. [DOI] [PubMed] [Google Scholar]
- Liu J.; Vellaisamy K.; Yang G.; Leung C.-H.; Ma D.-L. Luminescent Turn-on Detection of Hg(II) via the Quenching of an Iridium(III) Complex by Hg(II)-Mediated Silver Nanoparticles. Sci. Rep. 2017, 7, 3620. 10.1038/s41598-017-03952-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.; Wang W.; Hu C.; Yang G.; Ko C.-N.; Ren K.; Leung C.-H.; Ma D.-L. The Application of a G-Quadruplex Based Assay with an Iridium(III) Complex to Arsenic Ion Detection and Its Utilization in a Microfluidic Chip. J. Mater. Chem. B 2017, 5, 479–484. 10.1039/C6TB02656G. [DOI] [PubMed] [Google Scholar]
- Pelossof G.; Tel-Vered R.; Elbaz J.; Willner I. Amplified Biosensing Using the Horseradish Peroxidase-Mimicking DNAzyme as an Electrocatalyst. Anal. Chem. 2010, 82, 4396–4402. 10.1021/ac100095u. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Jiang B.; Xiang Y.; Chai Y.; Yuan R. Label-Free and Amplified Electrochemical Detection of Cytokine Based on Hairpin Aptamer and Catalytic DNAzyme. Analyst 2012, 137, 1020–1023. 10.1039/C2AN15962G. [DOI] [PubMed] [Google Scholar]
- Miao X.; Ko C.-N.; Vellaisamy K.; Li Z.; Yang G.; Leung C.-H.; Ma D.-L. A Cyclometalated Iridium(III) Complex Used as a Conductor for the Electrochemical Sensing of IFN-γ. Sci. Rep. 2017, 7, 42740. 10.1038/srep42740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q.; Zhao J.; Li H.; Zhu L.; Li G. Exonuclease III-Based and Gold Nanoparticle-Assisted DNA Detection with Dual Signal Amplification. Biosens. Bioelectron. 2012, 33, 211–215. 10.1016/j.bios.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Huang Y. L.; Gao Z. F.; Luo H. Q.; Li N. B. Sensitive Detection of HIV Gene by Coupling Exonuclease III-Assisted Target Recycling and Guanine Nanowire Amplification. Sens. Actuators, B 2017, 238, 1017–1023. 10.1016/j.snb.2016.07.144. [DOI] [Google Scholar]
- Xiong E.; Li Z.; Zhang X.; Zhou J.; Yan X.; Liu Y.; Chen J. Triple-Helix Molecular Switch Electrochemical Ratiometric Biosensor for Ultrasensitive Detection of Nucleic Acids. Anal. Chem. 2017, 89, 8830–8835. 10.1021/acs.analchem.7b01251. [DOI] [PubMed] [Google Scholar]
- Richter M. M. Electrochemiluminescence (ECL). Chem. Rev. 2004, 104, 3003–3036. 10.1021/cr020373d. [DOI] [PubMed] [Google Scholar]
- Yoon C.-H.; Cho J.-H.; Oh H.-I.; Kim M.-J.; Lee C.-W.; Choi J.-W.; Paek S.-H. Development of a Membrane Strip Immunosensor Utilizing Ruthenium as an Electro-Chemiluminescent Signal Generator. Biosens. Bioelectron. 2003, 19, 289–296. 10.1016/S0956-5663(03)00207-0. [DOI] [PubMed] [Google Scholar]
- Egashira N.; Morita S.; Hifumi E.; Mitoma Y.; Uda T. Attomole Detection of Hemagglutinin Molecule of Influenza Virus by Combining an Electrochemiluminescence Sensor with an Immunoliposome That Encapsulates a Ru Complex. Anal. Chem. 2008, 80, 4020–4025. 10.1021/ac702625d. [DOI] [PubMed] [Google Scholar]
- Katayama Y.; Ohgi T.; Mitoma Y.; Hifumi E.; Egashira N. Detection of Influenza Virus by a Biosensor Based on the Method Combining Electrochemiluminescence on Binary SAMs Modified Au Electrode with an Immunoliposome Encapsulating Ru (II) Complex. Anal. Bioanal. Chem. 2016, 408, 5963–5971. 10.1007/s00216-016-9587-8. [DOI] [PubMed] [Google Scholar]
- Zhou L.; Huang J.; Yu B.; Liu Y.; You T. A Novel Electrochemiluminescence Immunosensor for the Analysis of HIV-1 P24 Antigen Based on P-RGO@Au@Ru-SiO2 Composite. ACS Appl. Mater. Interfaces 2015, 7, 24438–24445. 10.1021/acsami.5b08154. [DOI] [PubMed] [Google Scholar]
- Shao K.; Wang J.; Jiang X.; Shao F.; Li T.; Ye S.; Chen L.; Han H. Stretch–Stowage–Growth Strategy to Fabricate Tunable Triply-Amplified Electrochemiluminescence Immunosensor for Ultrasensitive Detection of Pseudorabies Virus Antibody. Anal. Chem. 2014, 86, 5749–5757. 10.1021/ac500175y. [DOI] [PubMed] [Google Scholar]
- Zhou H.; Chen S.; Gan N.; Li T.; Cao Y.; Jiang Q. Design of Sensitive Biocompatible Quantum-Dots Embedded in Mesoporous Silica Microspheres for the Quantitative Immunoassay of Human Immunodeficiency Virus-1 Antibodies. Electroanalysis 2013, 25, 2384–2393. 10.1002/elan.201300307. [DOI] [Google Scholar]
- Acharya D.; Bastola P.; Le L.; Paul A. M.; Fernandez E.; Diamond M. S.; Miao W.; Bai F. An Ultrasensitive Electrogenerated Chemiluminescence-Based Immunoassay for Specific Detection of Zika Virus. Sci. Rep. 2016, 6, 32227. 10.1038/srep32227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.; Zhu M.; Qiu Y.; Yang P. Novel Electrochemiluminescence-Sensing Platform for the Precise Analysis of Multiple Latent Tuberculosis Infection Markers. ACS Appl. Mater. Interfaces 2017, 9, 18493–18500. 10.1021/acsami.7b03211. [DOI] [PubMed] [Google Scholar]
- Zhao Q.; Li F.; Huang C. Phosphorescent Chemosensors Based on Heavy-Metal Complexes. Chem. Soc. Rev. 2010, 39, 3007–3030. 10.1039/b915340c. [DOI] [PubMed] [Google Scholar]
- Zhao J.; Ji S.; Wu W.; Wu W.; Guo H.; Sun J.; Sun H.; Liu Y.; Li Q.; Huang L. Transition Metal Complexes with Strong Absorption of Visible Light and Long-Lived Triplet Excited States: From Molecular Design to Applications. RSC Adv. 2012, 2, 1712–1728. 10.1039/C1RA00665G. [DOI] [Google Scholar]
- Flamigni L.; Barbieri A.; Sabatini C.; Ventura B.; Barigelletti F.. Photochemistry and Photophysics of Coordination Compounds: Iridium. In Photochemistry and Photophysics of Coordination Compounds II; Balzani V., Campagna S., Eds.; Topics in Current Chemistry; Springer: Berlin, 2007; Vol. 281, pp 143–203. [Google Scholar]
- Wagenknecht P. S.; Ford P. C. Metal Centered Ligand Field Excited States: Their Roles in the Design and Performance of Transition Metal Based Photochemical Molecular Devices. Coord. Chem. Rev. 2011, 255, 591–616. 10.1016/j.ccr.2010.11.016. [DOI] [Google Scholar]
- Lavis L. D.; Raines R. T. Bright Building Blocks for Chemical Biology. ACS Chem. Biol. 2014, 9, 855–866. 10.1021/cb500078u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Electrochemical Biosensors: Towards Point-of-Care Cancer Diagnostics. Biosens. Bioelectron. 2006, 21, 1887–1892. 10.1016/j.bios.2005.10.027. [DOI] [PubMed] [Google Scholar]
- Blackburn G. F.; Shah H. P.; Kenten J. H.; Leland J.; Kamin R. A.; Link J.; Peterman J.; Powell M. J.; Shah A.; Talley D. B. Electrochemiluminescence Detection for Development of Immunoassays and DNA Probe Assays for Clinical Diagnostics. Clin. Chem. 1991, 37, 1534–1539. [PubMed] [Google Scholar]
- Gross E. M.; Durant H. E.; Hipp K. N.; Lai R. Y. Electrochemiluminescence Detection in Paper-Based and Other Inexpensive Microfluidic Devices. ChemElectroChem 2017, 4, 1594–1603. 10.1002/celc.201700426. [DOI] [Google Scholar]
- Chen L.; Zhang C.; Xing D. Paper-Based Bipolar Electrode-Electrochemiluminescence (BPE-ECL) Device with Battery Energy Supply and Smartphone Read-out: A Handheld ECL System for Biochemical Analysis at the Point-of-Care Level. Sens. Actuators, B 2016, 237, 308–317. 10.1016/j.snb.2016.06.105. [DOI] [Google Scholar]
- Nie Z.; Nijhuis C. A.; Gong J.; Chen X.; Kumachev A.; Martinez A. W.; Narovlyansky M.; Whitesides G. M. Electrochemical Sensing in Paper-Based Microfluidic Devices. Lab Chip 2010, 10, 477–483. 10.1039/B917150A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Xiang Y.; Lu Y.; Crooks R. M. Aptamer-Based Origami Paper Analytical Device for Electrochemical Detection of Adenosine. Angew. Chem., Int. Ed. 2012, 51, 6925–6928. 10.1002/anie.201202929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney J. L.; Hogan C. F.; Tian J.; Shen W. Electrogenerated Chemiluminescence Detection in Paper-Based Microfluidic Sensors. Anal. Chem. 2011, 83, 1300–1306. 10.1021/ac102392t. [DOI] [PubMed] [Google Scholar]
- Mettakoonpitak J.; Boehle K.; Nantaphol S.; Teengam P.; Adkins J. A.; Srisa-Art M.; Henry C. S. Electrochemistry on Paper-Based Analytical Devices: A Review. Electroanalysis 2016, 28, 1420–1436. 10.1002/elan.201501143. [DOI] [Google Scholar]
- Harris D. C.Quantitative Chemical Analysis, 8th ed.; W.H. Freeman and Co: New York, 2010. [Google Scholar]
- Travascio P.; Li Y.; Sen D. DNA-Enhanced Peroxidase Activity of a DNA Aptamer-Hemin Complex. Chem. Biol. 1998, 5, 505–517. 10.1016/S1074-5521(98)90006-0. [DOI] [PubMed] [Google Scholar]
- Nemiroski A.; Christodouleas D. C.; Hennek J. W.; Kumar A. A.; Maxwell E. J.; Fernandez-Abedul M. T.; Whitesides G. M. Universal Mobile Electrochemical Detector Designed for Use in Resource-Limited Applications. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 11984–11989. 10.1073/pnas.1405679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouillon R.; Combs Z.; Patel B. A.; O’Hare D. Comparative Study of the Effect of Various Electrode Membranes on Biofouling and Electrochemical Measurements. Electrochem. Commun. 2009, 11, 1409–1413. 10.1016/j.elecom.2009.05.018. [DOI] [Google Scholar]
- Stern E.; Vacic A.; Rajan N. K.; Criscione J. M.; Park J.; Ilic B. R.; Mooney D. J.; Reed M. A.; Fahmy T. M. Label-Free Biomarker Detection from Whole Blood. Nat. Nanotechnol. 2010, 5, 138–142. 10.1038/nnano.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.; Roy I.; Yang C.; Prasad P. N. Nanochemistry and Nanomedicine for Nanoparticle-Based Diagnostics and Therapy. Chem. Rev. 2016, 116, 2826–2885. 10.1021/acs.chemrev.5b00148. [DOI] [PubMed] [Google Scholar]
- Zhou W.; Gao X.; Liu D.; Chen X. Gold Nanoparticles for In Vitro Diagnostics. Chem. Rev. 2015, 115, 10575–10636. 10.1021/acs.chemrev.5b00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.; Lu K.; Poon C.; Lin W. Metal–Organic Frameworks as Sensory Materials and Imaging Agents. Inorg. Chem. 2014, 53, 1916–1924. 10.1021/ic402194c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.-C.; Long J. R.; Yaghi O. M. Introduction to Metal–Organic Frameworks. Chem. Rev. 2012, 112, 673–674. 10.1021/cr300014x. [DOI] [PubMed] [Google Scholar]
- Chen M.; Gan N.; Li T.; Wang Y.; Xu Q.; Chen Y. An Electrochemical Aptasensor for Multiplex Antibiotics Detection Using Y-Shaped DNA-Based Metal Ions Encoded Probes with NMOF Substrate and CSRP Target-Triggered Amplification Strategy. Anal. Chim. Acta 2017, 968, 30–39. 10.1016/j.aca.2017.03.024. [DOI] [PubMed] [Google Scholar]
- Su F.; Zhang S.; Ji H.; Zhao H.; Tian J.-Y.; Liu C.-S.; Zhang Z.; Fang S.; Zhu X.; Du M. Two-Dimensional Zirconium-Based Metal–Organic Framework Nanosheet Composites Embedded with Au Nanoclusters: A Highly Sensitive Electrochemical Aptasensor toward Detecting Cocaine. ACS Sens 2017, 2, 998–1005. 10.1021/acssensors.7b00268. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Dong P.; Sun Z.; Sun C.; Bu H.; Han J.; Chen S.; Xie G. NH 2 -Ni-MOF Electrocatalysts with Tunable Size/Morphology for Ultrasensitive C-Reactive Protein Detection via an Aptamer Binding Induced DNA Walker–Antibody Sandwich Assay. J. Mater. Chem. B 2018, 6, 2426–2431. 10.1039/C8TB00373D. [DOI] [PubMed] [Google Scholar]
- Xie Z.; Ma L.; deKrafft K. E.; Jin A.; Lin W. Porous Phosphorescent Coordination Polymers for Oxygen Sensing. J. Am. Chem. Soc. 2010, 132, 922–923. 10.1021/ja909629f. [DOI] [PubMed] [Google Scholar]
- Liu D.; Huxford R. C.; Lin W. Phosphorescent Nanoscale Coordination Polymers as Contrast Agents for Optical Imaging. Angew. Chem., Int. Ed. 2011, 50, 3696–3700. 10.1002/anie.201008277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ealias A. M.; Saravanakumar M. P. A Review on the Classification, Characterisation, Synthesis of Nanoparticles and Their Application. IOP Conf. Ser.: Mater. Sci. Eng. 2017, 263, 032019. 10.1088/1757-899X/263/3/032019. [DOI] [Google Scholar]
- Syafiuddin A.; Salmiati; Salim M. R.; Kueh A. B. H.; Hadibarata T.; Nur H. A Review of Silver Nanoparticles: Research Trends, Global Consumption, Synthesis, Properties, and Future Challenges. J. Chin. Chem. Soc. 2017, 64, 732–756. 10.1002/jccs.201700067. [DOI] [Google Scholar]
- Jackson T. C.; Patani B. O.; Ekpa D. E. Nanotechnology in Diagnosis: A Review. Adv. Nanopart. 2017, 6, 93–102. 10.4236/anp.2017.63008. [DOI] [Google Scholar]
- Hermanson G. T.Microparticles and Nanoparticles. In Bioconjugate Techniques; Elsevier Inc.: Oxford, U.K., 2013; Chapter 14, pp 549–587. [Google Scholar]
- Aubin-Tam M.-E.Conjugation of Nanoparticles to Proteins. In Nanomaterial Interfaces in Biology; Bergese P., Hamad-Schifferli K., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, 2013; pp 19–27. [DOI] [PubMed] [Google Scholar]
- Hermanson G. T.Zero-Length Crosslinkers. In Bioconjugate Techniques; Elsevier Inc.: Oxford, U.K., 2013; Chapter 4, pp 259–273. [Google Scholar]
- Hermanson G. T.The Reactions of Bioconjugation. In Bioconjugate Techniques; Elsevier Inc.: Oxford, U.K., 2013; Chapter 3, pp 229–258. [Google Scholar]
- Hermanson G. T.Antibody Modification and Conjugation. In Bioconjugate Techniques; Elsevier Inc.: Oxford, U.K., 2013; Chapter 20, pp 867–920. [Google Scholar]
- Hermanson G. T.Nucleic Acid and Oligonucleotide Modification and Conjugation. In Bioconjugate Techniques; Elsevier Inc.: Oxford, U.K., 2013; Chapter 23, pp 959–987. [Google Scholar]
- Medintz I. L.; Uyeda H. T.; Goldman E. R.; Mattoussi H. Quantum Dot Bioconjugates for Imaging, Labelling and Sensing. Nat. Mater. 2005, 4, 435–446. 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- Lu A.-H.; Salabas E. L.; Schüth F. Magnetic Nanoparticles: Synthesis, Protection, Functionalization, and Application. Angew. Chem., Int. Ed. 2007, 46, 1222–1244. 10.1002/anie.200602866. [DOI] [PubMed] [Google Scholar]
- Foubert A.; Beloglazova N. V.; Rajkovic A.; Sas B.; Madder A.; Goryacheva I. Y.; De Saeger S. Bioconjugation of Quantum Dots: Review & Impact on Future Application. TrAC, Trends Anal. Chem. 2016, 83, 31–48. 10.1016/j.trac.2016.07.008. [DOI] [Google Scholar]
- Biju V. Chemical Modifications and Bioconjugate Reactions of Nanomaterials for Sensing, Imaging, Drug Delivery and Therapy. Chem. Soc. Rev. 2014, 43, 744–764. 10.1039/C3CS60273G. [DOI] [PubMed] [Google Scholar]
- Hermanson G. T.Functional Targets for Bioconjugation. In Bioconjugate Techniques; Elsevier Inc.: Oxford, U.K., 2013; Chapter 2, pp 127–228. [Google Scholar]
- Gibson L. E.; Wright D. W. Sensitive Method for Biomolecule Detection Utilizing Signal Amplification with Porphyrin Nanoparticles. Anal. Chem. 2016, 88, 5928–5933. 10.1021/acs.analchem.6b00855. [DOI] [PubMed] [Google Scholar]
- Han Y. D.; Kim H.-S.; Park Y. M.; Chun H. J.; Kim J.-H.; Yoon H. C. Retroreflective Janus Microparticle as a Nonspectroscopic Optical Immunosensing Probe. ACS Appl. Mater. Interfaces 2016, 8, 10767–10774. 10.1021/acsami.6b02014. [DOI] [PubMed] [Google Scholar]
- Iha R. K.; Wooley K. L.; Nyström A. M.; Burke D. J.; Kade M. J.; Hawker C. J. Applications of Orthogonal “Click” Chemistries in the Synthesis of Functional Soft Materials. Chem. Rev. 2009, 109, 5620–5686. 10.1021/cr900138t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canalle L. A.; Vong T.; Adams P. H. H. M.; van Delft F. L.; Raats J. M. H.; Chirivi R. G. S.; van Hest J. C. M. Clickable Enzyme-Linked Immunosorbent Assay. Biomacromolecules 2011, 12, 3692–3697. 10.1021/bm2009137. [DOI] [PubMed] [Google Scholar]
- Haun J. B.; Devaraj N. K.; Hilderbrand S. A.; Lee H.; Weissleder R. Bioorthogonal Chemistry Amplifies Nanoparticle Binding and Enhances the Sensitivity of Cell Detection. Nat. Nanotechnol. 2010, 5, 660–665. 10.1038/nnano.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwe K.; Brechbiel M. W. Growing Applications of “Click Chemistry” for Bioconjugation in Contemporary Biomedical Research. Cancer Biother.Radiopharm. 2009, 24, 289–302. 10.1089/cbr.2008.0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay C. S.; Finn M. G. Click Chemistry in Complex Mixtures: Bioorthogonal Bioconjugation. Chem. Biol. 2014, 21, 1075–1101. 10.1016/j.chembiol.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson G. T.(Strept)Avidin–Biotin Systems. In Bioconjugate Techniques; Elsevier Inc.: Oxford, U.K., 2013; Chapter 11, pp 465–505. [Google Scholar]
- Nwokeoji A. O.; Kilby P. M.; Portwood D. E.; Dickman M. J. Accurate Quantification of Nucleic Acids Using Hypochromicity Measurements in Conjunction with UV Spectrophotometry. Anal. Chem. 2017, 89, 13567–13574. 10.1021/acs.analchem.7b04000. [DOI] [PubMed] [Google Scholar]
- Saha B.; Evers T. H.; Prins M. W. J. How Antibody Surface Coverage on Nanoparticles Determines the Activity and Kinetics of Antigen Capturing for Biosensing. Anal. Chem. 2014, 86, 8158–8166. 10.1021/ac501536z. [DOI] [PubMed] [Google Scholar]
- Lin P.-C.; Lin S.; Wang P. C.; Sridhar R. Techniques for Physicochemical Characterization of Nanomaterials. Biotechnol. Adv. 2014, 32, 711–726. 10.1016/j.biotechadv.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S. DLS and Zeta Potential – What They Are and What They Are Not?. J. Controlled Release 2016, 235, 337–351. 10.1016/j.jconrel.2016.06.017. [DOI] [PubMed] [Google Scholar]
- Casals E.; Pfaller T.; Duschl A.; Oostingh G. J.; Puntes V. Time Evolution of the Nanoparticle Protein Corona. ACS Nano 2010, 4, 3623–3632. 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]
- Lynch I.; Dawson K. A. Protein-Nanoparticle Interactions. Nano Today 2008, 3, 40–47. 10.1016/S1748-0132(08)70014-8. [DOI] [Google Scholar]
- Ingram R. S.; Hostetler M. J.; Murray R. W. Poly-Hetero-ω-Functionalized Alkanethiolate-Stabilized Gold Cluster Compounds. J. Am. Chem. Soc. 1997, 119, 9175–9178. 10.1021/ja971734n. [DOI] [Google Scholar]
- Miller S. A.; Hiatt L. A.; Keil R. G.; Wright D. W.; Cliffel D. E. Multifunctional Nanoparticles as Simulants for a Gravimetric Immunoassay. Anal. Bioanal. Chem. 2011, 399, 1021–1029. 10.1007/s00216-010-4419-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang L.; Wang Y.; Jiang J.; Dong S. PH-Dependent Protein Conformational Changes in Albumin:Gold Nanoparticle Bioconjugates: A Spectroscopic Study. Langmuir 2007, 23, 2714–2721. 10.1021/la062064e. [DOI] [PubMed] [Google Scholar]
- Homola J. Present and Future of Surface Plasmon Resonance Biosensors. Anal. Bioanal. Chem. 2003, 377, 528–539. 10.1007/s00216-003-2101-0. [DOI] [PubMed] [Google Scholar]
- Citartan M.; Gopinath S. C. B.; Tominaga J.; Tang T.-H. Label-Free Methods of Reporting Biomolecular Interactions by Optical Biosensors. Analyst 2013, 138, 3576–3592. 10.1039/c3an36828a. [DOI] [PubMed] [Google Scholar]
- Cooper M. A.; Singleton V. T. A Survey of the 2001 to 2005 Quartz Crystal Microbalance Biosensor Literature: Applications of Acoustic Physics to the Analysis of Biomolecular Interactions. J. Mol. Recognit. 2007, 20, 154–184. 10.1002/jmr.826. [DOI] [PubMed] [Google Scholar]
- Saha K.; Agasti S. S.; Kim C.; Li X.; Rotello V. M. Gold Nanoparticles in Chemical and Biological Sensing. Chem. Rev. 2012, 112, 2739–2779. 10.1021/cr2001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link S.; El-Sayed M. A. Size and Temperature Dependence of the Plasmon Absorption of Colloidal Gold Nanoparticles. J. Phys. Chem. B 1999, 103, 4212–4217. 10.1021/jp984796o. [DOI] [Google Scholar]
- Liz-Marzán L. M. Tailoring Surface Plasmons through the Morphology and Assembly of Metal Nanoparticles. Langmuir 2006, 22, 32–41. 10.1021/la0513353. [DOI] [PubMed] [Google Scholar]
- Song Y.; Wei W.; Qu X. Colorimetric Biosensing Using Smart Materials. Adv. Mater. 2011, 23, 4215–4236. 10.1002/adma.201101853. [DOI] [PubMed] [Google Scholar]
- Wilson R. The Use of Gold Nanoparticles in Diagnostics and Detection. Chem. Soc. Rev. 2008, 37, 2028–2045. 10.1039/b712179m. [DOI] [PubMed] [Google Scholar]
- Boisselier E.; Astruc D. Gold Nanoparticles in Nanomedicine: Preparations, Imaging, Diagnostics, Therapies and Toxicity. Chem. Soc. Rev. 2009, 38, 1759–1782. 10.1039/b806051g. [DOI] [PubMed] [Google Scholar]
- Dykman L.; Khlebtsov N. Gold Nanoparticles in Biomedical Applications: Recent Advances and Perspectives. Chem. Soc. Rev. 2012, 41, 2256–2282. 10.1039/C1CS15166E. [DOI] [PubMed] [Google Scholar]
- de Almeida M. P.; Carabineiro S. A. C. The Role of Nanogold in Human Tropical Diseases: Research, Detection and Therapy. Gold Bull. 2013, 46, 65–79. 10.1007/s13404-013-0086-9. [DOI] [Google Scholar]
- Doria G.; Conde J.; Veigas B.; Giestas L.; Almeida C.; Assunção M.; Rosa J.; Baptista P. V. Noble Metal Nanoparticles for Biosensing Applications. Sensors 2012, 12, 1657–1687. 10.3390/s120201657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz J. D.; Gulka C. P.; Haselton F. R.; Wright D. W. Development of a Histidine-Targeted Spectrophotometric Sensor Using Ni(II)NTA-Functionalized Au and Ag Nanoparticles. Langmuir 2011, 27, 15330–15339. 10.1021/la202937j. [DOI] [PubMed] [Google Scholar]
- Yen C.-W.; de Puig H.; Tam J.; Gómez-Márquez J.; Bosch I.; Hamad-Schifferli K.; Gehrke L. Multicolored Silver Nanoparticles for Multiplexed Disease Diagnostics: Distinguishing Dengue, Yellow Fever, and Ebola Viruses. Lab Chip 2015, 15, 1638–1641. 10.1039/C5LC00055F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso M.; Jiang L.; Cesarman E.; Erickson D. Multiplexed Colorimetric Detection of Kaposi’s Sarcoma Associated Herpesvirus and Bartonella DNA Using Gold and Silver Nanoparticles. Nanoscale 2013, 5, 1678–1686. 10.1039/c3nr33492a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teengam P.; Siangproh W.; Tuantranont A.; Vilaivan T.; Chailapakul O.; Henry C. S. Multiplex Paper-Based Colorimetric DNA Sensor Using Pyrrolidinyl Peptide Nucleic Acid-Induced AgNPs Aggregation for Detecting MERS-CoV, MTB, and HPV Oligonucleotides. Anal. Chem. 2017, 89, 5428–5435. 10.1021/acs.analchem.7b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J.-K.; Ma H.-L.; Cai P.-F.; Wu J.-Y. Highly Selective and Sensitive Nucleic Acid Detection Based on Polysaccharide-Functionalized Silver Nanoparticles. Spectrochim. Acta, Part A 2015, 134, 17–21. 10.1016/j.saa.2014.06.079. [DOI] [PubMed] [Google Scholar]
- Kurdekar A. D.; Chunduri L. A. A.; Chelli S. M.; Haleyurgirisetty M. K.; Bulagonda E. P.; Zheng J.; Hewlett I. K.; Kamisetti V. Fluorescent Silver Nanoparticle Based Highly Sensitive Immunoassay for Early Detection of HIV Infection. RSC Adv. 2017, 7, 19863–19877. 10.1039/C6RA28737A. [DOI] [Google Scholar]
- He Y.; Liu D.; He X.; Cui H. One-Pot Synthesis of Luminol Functionalized Silver Nanoparticles with Chemiluminescence Activity for Ultrasensitive DNA Sensing. Chem. Commun. 2011, 47, 10692–10694. 10.1039/c1cc14389a. [DOI] [PubMed] [Google Scholar]
- Wang R.; Xue C.; Gao M.; Qi H.; Zhang C. Ultratrace Voltammetric Method for the Detection of DNA Sequence Related to Human Immunodeficiency Virus Type 1. Microchim. Acta 2011, 172, 291–297. 10.1007/s00604-010-0490-2. [DOI] [Google Scholar]
- Karaballi R. A.; Nel A.; Krishnan S.; Blackburn J.; Brosseau C. L. Development of an Electrochemical Surface-Enhanced Raman Spectroscopy (EC-SERS) Aptasensor for Direct Detection of DNA Hybridization. Phys. Chem. Chem. Phys. 2015, 17, 21356–21363. 10.1039/C4CP05077K. [DOI] [PubMed] [Google Scholar]
- Anker J. N.; Hall W. P.; Lyandres O.; Shah N. C.; Zhao J.; Van Duyne R. P. Biosensing with Plasmonic Nanosensors. Nat. Mater. 2008, 7, 442–453. 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- Kneipp K.; Kneipp H.; Kneipp J. Surface-Enhanced Raman Scattering in Local Optical Fields of Silver and Gold NanoaggregatesFrom Single-Molecule Raman Spectroscopy to Ultrasensitive Probing in Live Cells. Acc. Chem. Res. 2006, 39, 443–450. 10.1021/ar050107x. [DOI] [PubMed] [Google Scholar]
- Chen F.; Flaherty B. R.; Cohen C. E.; Peterson D. S.; Zhao Y. Direct Detection of Malaria Infected Red Blood Cells by Surface Enhanced Raman Spectroscopy. Nanomedicine 2016, 12, 1445–1451. 10.1016/j.nano.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karadan P.; Aggarwal S.; Anappara A. A.; Narayana C.; Barshilia H. C. Tailored Periodic Si Nanopillar Based Architectures as Highly Sensitive Universal SERS Biosensing Platform. Sens. Actuators, B 2018, 254, 264–271. 10.1016/j.snb.2017.07.088. [DOI] [Google Scholar]
- Lee J.-S.; Lytton-Jean A. K. R.; Hurst S. J.; Mirkin C. A. Silver Nanoparticle–Oligonucleotide Conjugates Based on DNA with Triple Cyclic Disulfide Moieties. Nano Lett. 2007, 7, 2112–2115. 10.1021/nl071108g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D.; Fan K.; Zhang D.; Tan S.; Liang M.; Liu Y.; Zhang J.; Zhang P.; Liu W.; Qiu X.; et al. Nanozyme-Strip for Rapid Local Diagnosis of Ebola. Biosens. Bioelectron. 2015, 74, 134–141. 10.1016/j.bios.2015.05.025. [DOI] [PubMed] [Google Scholar]
- Dias A. C. M. S.; Gomes-Filho S. L. R.; Silva M. M. S.; Dutra R. F. A Sensor Tip Based on Carbon Nanotube-Ink Printed Electrode for the Dengue Virus NS1 Protein. Biosens. Bioelectron. 2013, 44, 216–221. 10.1016/j.bios.2012.12.033. [DOI] [PubMed] [Google Scholar]
- Vilela D.; González M. C.; Escarpa A. Sensing Colorimetric Approaches Based on Gold and Silver Nanoparticles Aggregation: Chemical Creativity behind the Assay. Anal. Chim. Acta 2012, 751, 24–43. 10.1016/j.aca.2012.08.043. [DOI] [PubMed] [Google Scholar]
- Ratnarathorn N.; Chailapakul O.; Henry C. S.; Dungchai W. Simple Silver Nanoparticle Colorimetric Sensing for Copper by Paper-Based Devices. Talanta 2012, 99, 552–557. 10.1016/j.talanta.2012.06.033. [DOI] [PubMed] [Google Scholar]
- Niemz A.; Ferguson T. M.; Boyle D. S. Point-of-Care Nucleic Acid Testing for Infectious Diseases. Trends Biotechnol. 2011, 29, 240–250. 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L.; Wang S.; Song X.; Ge S.; Yu J. 3D Origami-Based Multifunction-Integrated Immunodevice: Low-Cost and Multiplexed Sandwich Chemiluminescence Immunoassay on Microfluidic Paper-Based Analytical Device. Lab Chip 2012, 12, 3150–3158. 10.1039/c2lc40325k. [DOI] [PubMed] [Google Scholar]
- Michalet X. Quantum Dots for Live Cells, in Vivo Imaging, and Diagnostics. Science 2005, 307, 538–544. 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alivisatos A. P. Semiconductor Clusters, Nanocrystals, and Quantum Dots. Science 1996, 271, 933–937. 10.1126/science.271.5251.933. [DOI] [Google Scholar]
- Kippeny T.; Swafford L. A.; Rosenthal S. J. Semiconductor Nanocrystals: A Powerful Visual Aid for Introducing the Particle in a Box. J. Chem. Educ. 2002, 79, 1094–1100. 10.1021/ed079p1094. [DOI] [Google Scholar]
- Brus L. E. Electron–Electron and Electron-hole Interactions in Small Semiconductor Crystallites: The Size Dependence of the Lowest Excited Electronic State. J. Chem. Phys. 1984, 80, 4403–4409. 10.1063/1.447218. [DOI] [Google Scholar]
- Smith A. M.; Nie S. Semiconductor Nanocrystals: Structure, Properties, and Band Gap Engineering. Acc. Chem. Res. 2010, 43, 190–200. 10.1021/ar9001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klostranec J. M.; Xiang Q.; Farcas G. A.; Lee J. A.; Rhee A.; Lafferty E. I.; Perrault S. D.; Kain K. C.; Chan W. C. W. Convergence of Quantum Dot Barcodes with Microfluidics and Signal Processing for Multiplexed High-Throughput Infectious Disease Diagnostics. Nano Lett. 2007, 7, 2812–2818. 10.1021/nl071415m. [DOI] [PubMed] [Google Scholar]
- Bruchez M. Jr. Semiconductor Nanocrystals as Fluorescent Biological Labels. Science 1998, 281, 2013–2016. 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- Chan W. C. W.; Nie S. Quantum Dot Bioconjugates for Ultrasensitive Nonisotopic Detection. Science 1998, 281, 2016–2018. 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- Rosenthal S. J.; Chang J. C.; Kovtun O.; McBride J. R.; Tomlinson I. D. Biocompatible Quantum Dots for Biological Applications. Chem. Biol. 2011, 18, 10–24. 10.1016/j.chembiol.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J.; Jiang Y.-Z.; Wu L.-L.; Wu Z.; Bi Y.; Wong G.; Qiu X.; Chen J.; Pang D.-W.; Zhang Z.-L. Dual-Signal Readout Nanospheres for Rapid Point-of-Care Detection of Ebola Virus Glycoprotein. Anal. Chem. 2017, 89, 13105–13111. 10.1021/acs.analchem.7b02222. [DOI] [PubMed] [Google Scholar]
- Yan Z.; Gan N.; Zhang H.; Wang D.; Qiao L.; Cao Y.; Li T.; Hu F. A Sandwich-Hybridization Assay for Simultaneous Determination of HIV and Tuberculosis DNA Targets Based on Signal Amplification by Quantum Dots-PowerVision Polymer Coding Nanotracers. Biosens. Bioelectron. 2015, 71, 207–213. 10.1016/j.bios.2015.04.010. [DOI] [PubMed] [Google Scholar]
- Cimaglia F.; Aliverti A.; Chiesa M.; Poltronieri P.; De Lorenzis E.; Santino A.; Sechi L. A. Quantum Dots Nanoparticle-Based Lateral Flow Assay for Rapid Detection of Mycobacterium Species Using Anti-FprA Antibodies. Nanotechnol. Dev. 2012, 2, 26–30. 10.4081/nd.2012.e5. [DOI] [Google Scholar]
- Gao Y.; Lam A. W. Y.; Chan W. C. W. Automating Quantum Dot Barcode Assays Using Microfluidics and Magnetism for the Development of a Point-of-Care Device. ACS Appl. Mater. Interfaces 2013, 5, 2853–2860. 10.1021/am302633h. [DOI] [PubMed] [Google Scholar]
- Ku M.-J.; Dossin F. M.; Choi Y.; Moraes C. B.; Ryu J.; Song R.; Freitas-Junior L. H. Quantum Dots: A New Tool for Anti-Malarial Drug Assays. Malar. J. 2011, 10, 118. 10.1186/1475-2875-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.; Chou L. Y. T.; Song F.; Chan W. C. W. Fabrication of Metal Nanoshell Quantum-Dot Barcodes for Biomolecular Detection. Nano Today 2013, 8, 228–234. 10.1016/j.nantod.2013.04.009. [DOI] [Google Scholar]
- Searson P. C.; Castro-Sesquen Y. E.; Kim C.; Gilman R. H.; Sullivan D. J. Nanoparticle-Based Histidine-Rich Protein-2 Assay for the Detection of the Malaria Parasite Plasmodium Falciparum. Am. J. Trop. Med. Hyg. 2016, 95, 354–357. 10.4269/ajtmh.15-0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.; Hoffmann G.; Searson P. C. Integrated Magnetic Bead–Quantum Dot Immunoassay for Malaria Detection. ACS Sens 2017, 2, 766–772. 10.1021/acssensors.7b00119. [DOI] [PubMed] [Google Scholar]
- Li X.; Lu D.; Sheng Z.; Chen K.; Guo X.; Jin M.; Han H. A Fast and Sensitive Immunoassay of Avian Influenza Virus Based on Label-Free Quantum Dot Probe and Lateral Flow Test Strip. Talanta 2012, 100, 1–6. 10.1016/j.talanta.2012.08.041. [DOI] [PubMed] [Google Scholar]
- Zhu H.; Sikora U.; Ozcan A. Quantum Dot Enabled Detection of Escherichia Coli Using a Cell-Phone. Analyst 2012, 137, 2541–2544. 10.1039/c2an35071h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman E. R.; Clapp A. R.; Anderson G. P.; Uyeda H. T.; Mauro J. M.; Medintz I. L.; Mattoussi H. Multiplexed Toxin Analysis Using Four Colors of Quantum Dot Fluororeagents. Anal. Chem. 2004, 76, 684–688. 10.1021/ac035083r. [DOI] [PubMed] [Google Scholar]
- Gharaat M.; Sajedi R. H.; Shanehsaz M.; Jalilian N.; Mirshahi M.; Gholamzad M. A Dextran Mediated Multicolor Immunochromatographic Rapid Test Strip for Visual and Instrumental Simultaneous Detection of Vibrio Cholera O1 (Ogawa) and Clostridium Botulinum Toxin A. Microchim. Acta 2017, 184, 4817–4825. 10.1007/s00604-017-2527-2. [DOI] [Google Scholar]
- Hauck T. S.; Giri S.; Gao Y.; Chan W. C. W. Nanotechnology Diagnostics for Infectious Diseases Prevalent in Developing Countries. Adv. Drug Delivery Rev. 2010, 62, 438–448. 10.1016/j.addr.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Blanco-Canosa J. B.; Wu M.; Susumu K.; Petryayeva E.; Jennings T. L.; Dawson P. E.; Algar W. R.; Medintz I. L. Recent Progress in the Bioconjugation of Quantum Dots. Coord. Chem. Rev. 2014, 263–264, 101–137. 10.1016/j.ccr.2013.08.030. [DOI] [Google Scholar]
- Goldman E. R.; Balighian E. D.; Mattoussi H.; Kuno M. K.; Mauro J. M.; Tran P. T.; Anderson G. P. Avidin: A Natural Bridge for Quantum Dot-Antibody Conjugates. J. Am. Chem. Soc. 2002, 124, 6378–6382. 10.1021/ja0125570. [DOI] [PubMed] [Google Scholar]
- Goldman E. R.; Anderson G. P.; Tran P. T.; Mattoussi H.; Charles P. T.; Mauro J. M. Conjugation of Luminescent Quantum Dots with Antibodies Using an Engineered Adaptor Protein To Provide New Reagents for Fluoroimmunoassays. Anal. Chem. 2002, 74, 841–847. 10.1021/ac010662m. [DOI] [PubMed] [Google Scholar]
- Ming K.; Kim J.; Biondi M. J.; Syed A.; Chen K.; Lam A.; Ostrowski M.; Rebbapragada A.; Feld J. J.; Chan W. C. W. Integrated Quantum Dot Barcode Smartphone Optical Device for Wireless Multiplexed Diagnosis of Infected Patients. ACS Nano 2015, 9, 3060–3074. 10.1021/nn5072792. [DOI] [PubMed] [Google Scholar]
- Petryayeva E.; Algar W. R. Toward Point-of-Care Diagnostics with Consumer Electronic Devices: The Expanding Role of Nanoparticles. RSC Adv. 2015, 5, 22256–22282. 10.1039/C4RA15036H. [DOI] [Google Scholar]
- Petryayeva E.; Algar W. R. Multiplexed Homogeneous Assays of Proteolytic Activity Using a Smartphone and Quantum Dots. Anal. Chem. 2014, 86, 3195–3202. 10.1021/ac500131r. [DOI] [PubMed] [Google Scholar]
- Dhingra B.; Mishra D. Early Diagnosis of Febrile Illness: The Need of the Hour. Indian Pediatr. 2011, 48, 845–849. 10.1007/s13312-011-0134-6. [DOI] [PubMed] [Google Scholar]
- Iroh Tam P.-Y.; Obaro S. K.; Storch G. Challenges in the Etiology and Diagnosis of Acute Febrile Illness in Children in Low- and Middle-Income Countries. J. Pediatric Infect. Dis. Soc. 2016, 5, 190–205. 10.1093/jpids/piw016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuojola J.; Soukka T. Luminescent Lanthanide Reporters: New Concepts for Use in Bioanalytical Applications. Methods Appl. Fluoresc. 2014, 2, 012001. 10.1088/2050-6120/2/1/012001. [DOI] [PubMed] [Google Scholar]
- Syamchand S. S.; Sony G. Europium Enabled Luminescent Nanoparticles for Biomedical Applications. J. Lumin. 2015, 165, 190–215. 10.1016/j.jlumin.2015.04.042. [DOI] [Google Scholar]
- Banerjee R.; Jaiswal A. Recent Advances in Nanoparticle-Based Lateral Flow Immunoassay as a Point-of-Care Diagnostic Tool for Infectious Agents and Diseases. Analyst 2018, 143, 1970–1996. 10.1039/C8AN00307F. [DOI] [PubMed] [Google Scholar]
- Shao X.-Y.; Wang C.-R.; Xie C.-M.; Wang X.-G.; Liang R.-L.; Xu W.-W. Rapid and Sensitive Lateral Flow Immunoassay Method for Procalcitonin (PCT) Based on Time-Resolved Immunochromatography. Sensors 2017, 17, 480. 10.3390/s17030480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Gomez N. C.; Vu H. N.; Skovran E. Lanthanide Chemistry: From Coordination in Chemical Complexes Shaping Our Technology to Coordination in Enzymes Shaping Bacterial Metabolism. Inorg. Chem. 2016, 55, 10083–10089. 10.1021/acs.inorgchem.6b00919. [DOI] [PubMed] [Google Scholar]
- Dong H.; Du S.-R.; Zheng X.-Y.; Lyu G.-M.; Sun L.-D.; Li L.-D.; Zhang P.-Z.; Zhang C.; Yan C.-H. Lanthanide Nanoparticles: From Design toward Bioimaging and Therapy. Chem. Rev. 2015, 115, 10725–10815. 10.1021/acs.chemrev.5b00091. [DOI] [PubMed] [Google Scholar]
- Seydack M. Nanoparticle Labels in Immunosensing Using Optical Detection Methods. Biosens. Bioelectron. 2005, 20, 2454–2469. 10.1016/j.bios.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Thermo Scientific Dyed and Fluorescent Particles Brochure. https://tools.thermofisher.com/content/sfs/brochures/FL_Dyed%20and%20Fluorescent%20Particles_FINAL.pdf (accessed July 18, 2018).
- Time-Resolved Fluorescence. https://www.expedeon.com/resources/applications/time-resolved-fluorescence/ (accessed July 18, 2018).
- Huhtinen P.; Kivelä M.; Kuronen O.; Hagren V.; Takalo H.; Tenhu H.; Lövgren T.; Härmä H. Synthesis, Characterization, and Application of Eu(III), Tb(III), Sm(III), and Dy(III) Lanthanide Chelate Nanoparticle Labels. Anal. Chem. 2005, 77, 2643–2648. 10.1021/ac048360i. [DOI] [PubMed] [Google Scholar]
- Härmä H.; Soukka T.; Lövgren T. Europium Nanoparticles and Time-Resolved Fluorescence for Ultrasensitive Detection of Prostate-Specific Antigen. Clin. Chem. 2001, 47, 561–568. [PubMed] [Google Scholar]
- Yeo S.-J.; Bao D. T.; Seo G.-E.; Bui C. T.; Kim D. T. H.; Anh N. T. V.; Tien T. T. T.; Linh N. T. P.; Sohn H.-J.; Chong C.-K.; et al. Improvement of a Rapid Diagnostic Application of Monoclonal Antibodies against Avian Influenza H7 Subtype Virus Using Europium Nanoparticles. Sci. Rep. 2017, 7, 7933. 10.1038/s41598-017-08328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.; Cowling B. J.; Feng L.; Lau E. H.; Liao Q.; Tsang T. K.; Peng Z.; Wu P.; Liu F.; Fang V. J.; et al. Human Infection with Avian Influenza A H7N9 Virus: An Assessment of Clinical Severity. Lancet 2013, 382, 138–145. 10.1016/S0140-6736(13)61207-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao D. T.; Kim D. T. H.; Park H.; Cuc B. T.; Ngoc N. M.; Linh N. T. P.; Huu N. C.; Tien T. T. T.; Anh N. T. V.; Duy T. D.; et al. Rapid Detection of Avian Influenza Virus by Fluorescent Diagnostic Assay Using an Epitope-Derived Peptide. Theranostics 2017, 7, 1835–1846. 10.7150/thno.18857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing K.-Y.; Peng J.; Liu D.-F.; Hu L.-M.; Wang C.; Li G.-Q.; Zhang G.-G.; Huang Z.; Cheng S.; Zhu F.-F.; et al. Novel Immunochromatographic Assay Based on Eu (III)-Doped Polystyrene Nanoparticle-Linker-Monoclonal Antibody for Sensitive Detection of Escherichia Coli O157:H7. Anal. Chim. Acta 2018, 998, 52–59. 10.1016/j.aca.2017.10.027. [DOI] [PubMed] [Google Scholar]
- Liang R.-L.; Deng Q.-T.; Chen Z.-H.; Xu X.-P.; Zhou J.-W.; Liang J.-Y.; Dong Z.-N.; Liu T.-C.; Wu Y.-S. Europium (III) Chelate Microparticle-Based Lateral Flow Immunoassay Strips for Rapid and Quantitative Detection of Antibody to Hepatitis B Core Antigen. Sci. Rep. 2017, 7, 14093. 10.1038/s41598-017-14427-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. K.; Cho C. H.; Woo M. K.; Nyeck A. E.; Lim C. S.; Kim W. J. Evaluation of Sofia Fluorescent Immunoassay Analyzer for Influenza A/B Virus. J. Clin. Virol. 2012, 55, 239–243. 10.1016/j.jcv.2012.07.008. [DOI] [PubMed] [Google Scholar]
- Lewandrowski K.; Tamerius J.; Menegus M.; Olivo P. D.; Lollar R.; Lee-Lewandrowski E. Detection of Influenza A and B Viruses With the Sofia Analyzer: A Novel, Rapid Immunofluorescence-Based In Vitro Diagnostic Device. Am. J. Clin. Pathol. 2013, 139, 684–689. 10.1309/AJCP7ZTLJCP3LLMA. [DOI] [PubMed] [Google Scholar]
- Arbefeville S. S.; Fickle A. R.; Ferrieri P. Sensitivity of the Quidel Sofia Fluorescent Immunoassay Compared With 2 Nucleic Acid Assays and Viral Culture to Detect Pandemic Influenza A(H1N1)Pdm09. Lab. Med. 2015, 46, 230–234. 10.1309/LMRXBN8P59ITDHQZ. [DOI] [PubMed] [Google Scholar]
- Dunn J. J.; Ginocchio C. C. Point-Counterpoint: Can Newly Developed, Rapid Immunochromatographic Antigen Detection Tests Be Reliably Used for the Laboratory Diagnosis of Influenza Virus Infections?. J. Clin. Microbiol. 2015, 53, 1790–1796. 10.1128/JCM.02739-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelton B.; Nedeljkovic G.; Ratnamohan V. M.; Dwyer D. E.; Kok J. Evaluation of the Sofia Influenza A + B Fluorescent Immunoassay for the Rapid Diagnosis of Influenza A and B. J. Med. Virol. 2015, 87, 35–38. 10.1002/jmv.23976. [DOI] [PubMed] [Google Scholar]
- Gomez S.; Prieto C.; Folgueira L. A Prospective Study to Assess the Diagnostic Performance of the Sofia® Immunoassay for Influenza and RSV Detection. J. Clin. Virol. 2016, 77, 1–4. 10.1016/j.jcv.2016.01.018. [DOI] [PubMed] [Google Scholar]
- Ham J. Y.; Jung J.; Hwang B.-G.; Kim W.-J.; Kim Y.-S.; Kim E.-J.; Cho M.-Y.; Hwang M.-S.; Won D. I.; Suh J. S. Highly Sensitive and Novel Point-of-Care System, AQcare Chlamydia TRF Kit for Detecting Chlamydia Trachomatis by Using Europium (Eu) (III) Chelated Nanoparticles. Ann. Lab. Med. 2015, 35, 50–56. 10.3343/alm.2015.35.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly H.; Coltart C. E. M.; Pai N. P.; Klausner J. D.; Unemo M.; Toskin I.; Peeling R. W. Systematic Reviews of Point-of-Care Tests for the Diagnosis of Urogenital Chlamydia Trachomatis Infections. Sex. Transm. Infect. 2017, 93, S22–S30. 10.1136/sextrans-2016-053067. [DOI] [PubMed] [Google Scholar]
- Croce A. C.; Bottiroli G. Autofluorescence Spectroscopy and Imaging: A Tool for Biomedical Research and Diagnosis. Eur. J. Histochem. 2014, 58, 320–337. 10.4081/ejh.2014.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X.; Knotts M. Time-Resolved Luminescent Lateral Flow Assay Technology. Anal. Chim. Acta 2008, 626, 186–192. 10.1016/j.aca.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Paterson A. S.; Raja B.; Mandadi V.; Townsend B.; Lee M.; Buell A.; Vu B.; Brgoch J.; Willson R. C. A Low-Cost Smartphone-Based Platform for Highly Sensitive Point-of-Care Testing with Persistent Luminescent Phosphors. Lab Chip 2017, 17, 1051–1059. 10.1039/C6LC01167E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X.; Xu Y.; Zhao X.; Li Q. Lateral Flow Immunoassay Using Europium Chelate-Loaded Silica Nanoparticles as Labels. Clin. Chem. 2008, 55, 179–182. 10.1373/clinchem.2008.114561. [DOI] [PubMed] [Google Scholar]
- Xu Y.; Li Q. Multiple Fluorescent Labeling of Silica Nanoparticles with Lanthanide Chelates for Highly Sensitive Time-Resolved Immunofluorometric Assays. Clin. Chem. 2007, 53, 1503–1510. 10.1373/clinchem.2006.078485. [DOI] [PubMed] [Google Scholar]
- Chen J.; Zhao J. X. Upconversion Nanomaterials: Synthesis, Mechanism, and Applications in Sensing. Sensors 2012, 12, 2414–2435. 10.3390/s120302414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S.; Chen G.; Yang C. Sensing Using Rare-Earth-Doped Upconversion Nanoparticles. Theranostics 2013, 3, 331–345. 10.7150/thno.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corstjens P. L. A. M.; Li S.; Zuiderwijk M.; Kardos K.; Abrams W. R.; Niedbala R. S.; Tanke H. J. Infrared Up-Converting Phosphors for Bioassays. IEE Proc.: Nanobiotechnol. 2005, 152, 64–72. 10.1049/ip-nbt:20045014. [DOI] [PubMed] [Google Scholar]
- Niedbala R. S.; Feindt H.; Kardos K.; Vail T.; Burton J.; Bielska B.; Li S.; Milunic D.; Bourdelle P.; Vallejo R. Detection of Analytes by Immunoassay Using Up-Converting Phosphor Technology. Anal. Biochem. 2001, 293, 22–30. 10.1006/abio.2001.5105. [DOI] [PubMed] [Google Scholar]
- Chen G.; Qiu H.; Prasad P. N.; Chen X. Upconversion Nanoparticles: Design, Nanochemistry, and Applications in Theranostics. Chem. Rev. 2014, 114, 5161–5214. 10.1021/cr400425h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corstjens P. L. A. M.; van Lieshout L.; Zuiderwijk M.; Kornelis D.; Tanke H. J.; Deelder A. M.; van Dam G. J. Up-Converting Phosphor Technology-Based Lateral Flow Assay for Detection of Schistosoma Circulating Anodic Antigen in Serum. J. Clin. Microbiol. 2008, 46, 171–176. 10.1128/JCM.00877-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam G. J.; de Dood C. J.; Lewis M.; Deelder A. M.; van Lieshout L.; Tanke H. J.; van Rooyen L. H.; Corstjens P. L. A. M. A Robust Dry Reagent Lateral Flow Assay for Diagnosis of Active Schistosomiasis by Detection of Schistosoma Circulating Anodic Antigen. Exp. Parasitol. 2013, 135, 274–282. 10.1016/j.exppara.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corstjens P. L. A. M.; de Dood C. J.; Kornelis D.; Tjon Kon Fat E. M.; Wilson R. A.; Kariuki T. M.; Nyakundi R. K.; Loverde P. T.; Abrams W. R.; Tanke H. J.; et al. Tools for Diagnosis, Monitoring and Screening of Schistosoma Infections Utilizing Lateral-Flow Based Assays and Upconverting Phosphor Labels. Parasitology 2014, 141, 1841–1855. 10.1017/S0031182014000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopp S.; Corstjens P. L. A. M.; Koukounari A.; Cercamondi C. I.; Ame S. M.; Ali S. M.; de Dood C. J.; Mohammed K. A.; Utzinger J.; Rollinson D.; et al. Sensitivity and Specificity of a Urine Circulating Anodic Antigen Test for the Diagnosis of Schistosoma Haematobium in Low Endemic Settings. PLoS Neglected Trop. Dis. 2015, 9, e0003752 10.1371/journal.pntd.0003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam G. J.; Xu J.; Bergquist R.; de Dood C. J.; Utzinger J.; Qin Z.-Q.; Guan W.; Feng T.; Yu X.-L.; Zhou J.; et al. An Ultra-Sensitive Assay Targeting the Circulating Anodic Antigen for the Diagnosis of Schistosoma Japonicum in a Low-Endemic Area, People’s Republic of China. Acta Trop. 2015, 141, 190–197. 10.1016/j.actatropica.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Corstjens P. L.; Nyakundi R. K.; de Dood C. J.; Kariuki T. M.; Ochola E. A.; Karanja D. M.; Mwinzi P. N.; van Dam G. J. Improved Sensitivity of the Urine CAA Lateral-Flow Assay for Diagnosing Active Schistosoma Infections by Using Larger Sample Volumes. Parasites Vectors 2015, 8, 241. 10.1186/s13071-015-0857-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utzinger J.; Becker S. L.; van Lieshout L.; van Dam G. J.; Knopp S. New Diagnostic Tools in Schistosomiasis. Clin. Microbiol. Infect. 2015, 21, 529–542. 10.1016/j.cmi.2015.03.014. [DOI] [PubMed] [Google Scholar]
- van Dam G. J.; Odermatt P.; Acosta L.; Bergquist R.; de Dood C. J.; Kornelis D.; Muth S.; Utzinger J.; Corstjens P. L. A. M. Evaluation of Banked Urine Samples for the Detection of Circulating Anodic and Cathodic Antigens in Schistosoma Mekongi and S. Japonicum Infections: A Proof-of-Concept Study. Acta Trop. 2015, 141, 198–203. 10.1016/j.actatropica.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Corstjens P. L. A. M.; Hoekstra P. T.; de Dood C. J.; van Dam G. J. Utilizing the Ultrasensitive Schistosoma up-Converting Phosphor Lateral Flow Circulating Anodic Antigen (UCP-LF CAA) Assay for Sample Pooling-Strategies. Infect. Dis. Poverty 2017, 6, 155. 10.1186/s40249-017-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corstjens P. L. A. M.; de Dood C. J.; Priest J. W.; Tanke H. J.; Handali S. Feasibility of a Lateral Flow Test for Neurocysticercosis Using Novel Up-Converting Nanomaterials and a Lightweight Strip Analyzer. PLoS Neglected Trop. Dis. 2014, 8, e2944 10.1371/journal.pntd.0002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corstjens P. L. A. M.; Tjon Kon Fat E. M.; de Dood C. J.; van der Ploeg-van Schip J. J.; Franken K. L. M. C.; Chegou N. N.; Sutherland J. S.; Howe R.; Mihret A.; Kassa D.; et al. Multi-Center Evaluation of a User-Friendly Lateral Flow Assay to Determine IP-10 and CCL4 Levels in Blood of TB and Non-TB Cases in Africa. Clin. Biochem. 2016, 49, 22–31. 10.1016/j.clinbiochem.2015.08.013. [DOI] [PubMed] [Google Scholar]
- Sutherland J. S.; Mendy J.; Gindeh A.; Walzl G.; Togun T.; Owolabi O.; Donkor S.; Ota M. O.; Kon Fat E. T.; Ottenhoff T. H. M.; et al. Use of Lateral Flow Assays to Determine IP-10 and CCL4 Levels in Pleural Effusions and Whole Blood for TB Diagnosis. Tuberculosis 2016, 96, 31–36. 10.1016/j.tube.2015.10.011. [DOI] [PubMed] [Google Scholar]
- Corstjens P. L. A. M.; Zuiderwijk M.; Tanke H. J.; van der Ploeg-van Schip J. J.; Ottenhoff T. H. M.; Geluk A. A User-Friendly, Highly Sensitive Assay to Detect the IFN-γ Secretion by T Cells. Clin. Biochem. 2008, 41, 440–444. 10.1016/j.clinbiochem.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corstjens P. L. A. M.; de Dood C. J.; van der Ploeg-van Schip J. J.; Wiesmeijer K. C.; Riuttamäki T.; van Meijgaarden K. E.; Spencer J. S.; Tanke H. J.; Ottenhoff T. H. M.; Geluk A. Lateral Flow Assay for Simultaneous Detection of Cellular- and Humoral Immune Responses. Clin. Biochem. 2011, 44, 1241–1246. 10.1016/j.clinbiochem.2011.06.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobosha K.; Tjon Kon Fat E. M.; van den Eeden S. J. F.; Bekele Y.; van der Ploeg-van Schip J. J.; de Dood C. J.; Dijkman K.; Franken K. L. M. C.; Wilson L.; Aseffa A.; et al. Field-Evaluation of a New Lateral Flow Assay for Detection of Cellular and Humoral Immunity against Mycobacterium Leprae. PLoS Neglected Trop. Dis. 2014, 8, e2845 10.1371/journal.pntd.0002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corstjens P. L. A. M.; van Hooij A.; Tjon Kon Fat E. M.; van den Eeden S. J. F.; Wilson L.; Geluk A. Field-Friendly Test for Monitoring Multiple Immune Response Markers during Onset and Treatment of Exacerbated Immunity in Leprosy. Clin. Vaccine Immunol. 2016, 23, 515–519. 10.1128/CVI.00033-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hooij A.; Tjon Kon Fat E. M.; van den Eeden S. J. F.; Wilson L.; Batista da Silva M.; Salgado C. G.; Spencer J. S.; Corstjens P. L. A. M.; Geluk A. Field-Friendly Serological Tests for Determination of M. Leprae-Specific Antibodies. Sci. Rep. 2017, 7, 8868. 10.1038/s41598-017-07803-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z.; Zhou L.; Zhao Y.; Wang J.; Huang L.; Hu K.; Liu H.; Wang H.; Guo Z.; Song Y.; et al. Rapid Quantitative Detection of Yersinia Pestis by Lateral-Flow Immunoassay and up-Converting Phosphor Technology-Based Biosensor. Sens. Actuators, B 2006, 119, 656–663. 10.1016/j.snb.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W.; Huang L.; Wang H.; Qu J.; Guo Z.; Xie C.; Zhu Z.; Zhang Y.; Du Z.; Yan Y.; et al. Development of an Up-Converting Phosphor Technology-Based 10-Channel Lateral Flow Assay for Profiling Antibodies against Yersinia Pestis. J. Microbiol. Methods 2010, 83, 133–140. 10.1016/j.mimet.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Qu Q.; Zhu Z.; Wang Y.; Zhong Z.; Zhao J.; Qiao F.; Du X.; Wang Z.; Yang R.; Huang L.; et al. Rapid and Quantitative Detection of Brucella by up-Converting Phosphor Technology-Based Lateral-Flow Assay. J. Microbiol. Methods 2009, 79, 121–123. 10.1016/j.mimet.2009.07.015. [DOI] [PubMed] [Google Scholar]
- Zhao P.; Wu Y.; Zhu Y.; Yang X.; Jiang X.; Xiao J.; Zhang Y.; Li C. Upconversion Fluorescent Strip Sensor for Rapid Determination of Vibrio Anguillarum. Nanoscale 2014, 6, 3804–3809. 10.1039/C3NR06549A. [DOI] [PubMed] [Google Scholar]
- Li L.; Zhou L.; Yu Y.; Zhu Z.; Lin C.; Lu C.; Yang R. Development of Up-Converting Phosphor Technology-Based Lateral-Flow Assay for Rapidly Quantitative Detection of Hepatitis B Surface Antibody. Diagn. Microbiol. Infect. Dis. 2009, 63, 165–172. 10.1016/j.diagmicrobio.2008.10.020. [DOI] [PubMed] [Google Scholar]
- Mokkapati V. K.; Niedbala R. S.; Kardos K.; Perez R. J.; Guo M.; Tanke H. J.; Corstjens P. L. A. M. Evaluation of UPlink–RSV: Prototype Rapid Antigen Test for Detection of Respiratory Syncytial Virus Infection. Ann. N. Y. Acad. Sci. 2007, 1098, 476–485. 10.1196/annals.1384.021. [DOI] [PubMed] [Google Scholar]
- Qiu X.; Thompson J. A.; Chen Z.; Liu C.; Chen D.; Ramprasad S.; Mauk M. G.; Ongagna S.; Barber C.; Abrams W. R.; et al. Finger-Actuated, Self-Contained Immunoassay Cassettes. Biomed. Microdevices 2009, 11, 1175–1186. 10.1007/s10544-009-9334-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corstjens P. L. A. M.; Chen Z.; Zuiderwijk M.; Bau H. H.; Abrams W. R.; Malamud D.; Niedbala R. S.; Tanke H. J. Rapid Assay Format for Multiplex Detection of Humoral Immune Responses to Infectious Disease Pathogens (HIV, HCV, and TB). Ann. N. Y. Acad. Sci. 2007, 1098, 437–445. 10.1196/annals.1384.016. [DOI] [PubMed] [Google Scholar]
- Zhang P.; Liu X.; Wang C.; Zhao Y.; Hua F.; Li C.; Yang R.; Zhou L. Evaluation of Up-Converting Phosphor Technology-Based Lateral Flow Strips for Rapid Detection of Bacillus Anthracis Spore, Brucella Spp., and Yersinia Pestis. PLoS One 2014, 9, e105305 10.1371/journal.pone.0105305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlmans H. J. M. A. A.; Bonnet J.; Burton J.; Kardos K.; Vail T.; Niedbala R. S.; Tanke H. J. Detection of Cell and Tissue Surface Antigens Using Up-Converting Phosphors: A New Reporter Technology. Anal. Biochem. 1999, 267, 30–36. 10.1006/abio.1998.2965. [DOI] [PubMed] [Google Scholar]
- van de Rijke F.; Zijlmans H.; Li S.; Vail T.; Raap A. K.; Niedbala R. S.; Tanke H. J. Up-Converting Phosphor Reporters for Nucleic Acid Microarrays. Nat. Biotechnol. 2001, 19, 273–276. 10.1038/85734. [DOI] [PubMed] [Google Scholar]
- Duan N.; Wu S.; Zhu C.; Ma X.; Wang Z.; Yu Y.; Jiang Y. Dual-Color Upconversion Fluorescence and Aptamer-Functionalized Magnetic Nanoparticles-Based Bioassay for the Simultaneous Detection of Salmonella Typhimurium and Staphylococcus Aureus. Anal. Chim. Acta 2012, 723, 1–6. 10.1016/j.aca.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Wu S.; Duan N.; Shi Z.; Fang C.; Wang Z. Simultaneous Aptasensor for Multiplex Pathogenic Bacteria Detection Based on Multicolor Upconversion Nanoparticles Labels. Anal. Chem. 2014, 86, 3100–3107. 10.1021/ac404205c. [DOI] [PubMed] [Google Scholar]
- Brown J.; Theis L.; Kerr L.; Zakhidova N.; O’Connor K.; Uthman M.; Oden Z. M.; Richards-Kortum R. A Hand-Powered, Portable, Low-Cost Centrifuge for Diagnosing Anemia in Low-Resource Settings. Am. J. Trop. Med. Hyg. 2011, 85, 327–332. 10.4269/ajtmh.2011.10-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhamla M. S.; Benson B.; Chai C.; Katsikis G.; Johri A.; Prakash M. Hand-Powered Ultralow-Cost Paper Centrifuge. Nature Biomedical Engineering 2017, 1, 0009. 10.1038/s41551-016-0009. [DOI] [Google Scholar]
- Scherr T. F.; Ryskoski H. B.; Sivakumar A.; Ricks K. M.; Adams N. M.; Wright D. W.; Haselton F. R. A Handheld Orbital Mixer for Processing Viscous Samples in Low Resource Settings. Anal. Methods 2016, 8, 7347–7353. 10.1039/C6AY01636G. [DOI] [Google Scholar]
- Zheng W.; Huang P.; Tu D.; Ma E.; Zhu H.; Chen X. Lanthanide-Doped Upconversion Nano-Bioprobes: Electronic Structures, Optical Properties, and Biodetection. Chem. Soc. Rev. 2015, 44, 1379–1415. 10.1039/C4CS00178H. [DOI] [PubMed] [Google Scholar]
- Spricigo R.Qiagen Email Communication, 2018.
- Panferov V. G.; Safenkova I. V.; Zherdev A. V.; Dzantiev B. B. Setting up the Cut-off Level of a Sensitive Barcode Lateral Flow Assay with Magnetic Nanoparticles. Talanta 2017, 164, 69–76. 10.1016/j.talanta.2016.11.025. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Xu H.; Wei M.; Gu H.; Xu Q.; Zhu W. Study of Superparamagnetic Nanoparticles as Labels in the Quantitative Lateral Flow Immunoassay. Mater. Sci. Eng., C 2009, 29, 714–718. 10.1016/j.msec.2009.01.011. [DOI] [Google Scholar]
- Handali S.; Klarman M.; Gaspard A. N.; Dong X. F.; LaBorde R.; Noh J.; Lee Y.-M.; Rodriguez S.; Gonzalez A. E.; Garcia H. H.; et al. Development and Evaluation of a Magnetic Immunochromatographic Test To Detect Taenia Solium, Which Causes Taeniasis and Neurocysticercosis in Humans. Clin. Vaccine Immunol. 2010, 17, 631–637. 10.1128/CVI.00511-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G.; Makarov D.; Schmidt O. G. Magnetic Sensing Platform Technologies for Biomedical Applications. Lab Chip 2017, 17, 1884–1912. 10.1039/C7LC00026J. [DOI] [PubMed] [Google Scholar]
- Akbarzadeh A.; Samiei M.; Davaran S. Magnetic Nanoparticles: Preparation, Physical Properties, and Applications in Biomedicine. Nanoscale Res. Lett. 2012, 7, 144. 10.1186/1556-276X-7-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W.-Z.; Cetinel S.; Montemagno C. Application of Biomolecular Recognition via Magnetic Nanoparticle in Nanobiotechnology. J. Nanopart. Res. 2018, 20, 130. 10.1007/s11051-018-4232-4. [DOI] [Google Scholar]
- Haun J. B.; Yoon T.-J.; Lee H.; Weissleder R. Magnetic Nanoparticle Biosensors. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 2010, 2, 291–304. 10.1002/wnan.84. [DOI] [PubMed] [Google Scholar]
- Makiranta J.; Lekkala J. Modeling and Simulation of Magnetic Nanoparticle Sensor. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2006, 2, 1256–1259. 10.1109/IEMBS.2005.1616653. [DOI] [PubMed] [Google Scholar]
- Chen Y.-T.; Kolhatkar A. G.; Zenasni O.; Xu S.; Lee T. R. Biosensing Using Magnetic Particle Detection Techniques. Sensors 2017, 17, 2300. 10.3390/s17102300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Wang Y.; Tu L.; Feng Y.; Klein T.; Wang J.-P. Magnetoresistive Performance and Comparison of Supermagnetic Nanoparticles on Giant Magnetoresistive Sensor-Based Detection System. Sci. Rep. 2015, 4, 5716. 10.1038/srep05716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taton K.; Johnson D.; Guire P.; Lange E.; Tondra M. Lateral Flow Immunoassay Using Magnetoresistive Sensors. J. Magn. Magn. Mater. 2009, 321, 1679–1682. 10.1016/j.jmmm.2009.02.113. [DOI] [Google Scholar]
- World Health Organization. Taeniasis/cysticercosis Fact sheet http://www.who.int/mediacentre/factsheets/fs376/en/ (accessed Dec 16, 2017).
- Del Brutto O. H.; García H. H.. Diagnosis of Taeniasis and Cysticercosis. In Cysticercosis of the Human Nervous System; Springer: Berlin, 2014; pp 87–107. [Google Scholar]
- Workman S.; Wells S. K.; Pau C.-P.; Owen S. M.; Dong X. F.; LaBorde R.; Granade T. C. Rapid Detection of HIV-1 P24 Antigen Using Magnetic Immuno-Chromatography (MICT). J. Virol. Methods 2009, 160, 14–21. 10.1016/j.jviromet.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Granade T. C.; Workman S.; Wells S. K.; Holder A. N.; Owen S. M.; Pau C.-P. Rapid Detection and Differentiation of Antibodies to HIV-1 and HIV-2 Using Multivalent Antigens and Magnetic Immunochromatography Testing. Clin. Vaccine Immunol. 2010, 17, 1034–1039. 10.1128/CVI.00029-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.; Lei X.; Wang W.; Liu Y.; Liang P.; Bao H.; Wang Q.; Guo Y.; Yang J.; Yan Z. Development and Evaluation of a Paramagnetic Nanoparticle Based Immunochromatographic Strip for Specific Detection of 2009 H1N1 Influenza Virus. J. Nanosci. Nanotechnol. 2013, 13, 1684–1690. 10.1166/jnn.2013.7127. [DOI] [PubMed] [Google Scholar]
- Yan J.; Liu Y.; Wang Y.; Xu X.; Lu Y.; Pan Y.; Guo F.; Shi D. Effect of Physiochemical Property of Fe3O4 Particle on Magnetic Lateral Flow Immunochromatographic Assay. Sens. Actuators, B 2014, 197, 129–136. 10.1016/j.snb.2014.02.067. [DOI] [Google Scholar]
- Liu Y.; Zhang Z.; Wang Y.; Zhao Y.; Lu Y.; Xu X.; Yan J.; Pan Y. A Highly Sensitive and Flexible Magnetic Nanoprobe Labeled Immunochromatographic Assay Platform for Pathogen Vibrio Parahaemolyticus. Int. J. Food Microbiol. 2015, 211, 109–116. 10.1016/j.ijfoodmicro.2015.07.005. [DOI] [PubMed] [Google Scholar]
- Shi L.; Wu F.; Wen Y.; Zhao F.; Xiang J.; Ma L. A Novel Method to Detect Listeria Monocytogenes via Superparamagnetic Lateral Flow Immunoassay. Anal. Bioanal. Chem. 2015, 407, 529–535. 10.1007/s00216-014-8276-8. [DOI] [PubMed] [Google Scholar]
- Wang D.-B.; Tian B.; Zhang Z.-P.; Deng J.-Y.; Cui Z.-Q.; Yang R.-F.; Wang X.-Y.; Wei H.-P.; Zhang X.-E. Rapid Detection of Bacillus Anthracis Spores Using a Super-Paramagnetic Lateral-Flow Immunological Detectionsystem. Biosens. Bioelectron. 2013, 42, 661–667. 10.1016/j.bios.2012.10.088. [DOI] [PubMed] [Google Scholar]
- Wang D.-B.; Tian B.; Zhang Z.-P.; Wang X.-Y.; Fleming J.; Bi L.-J.; Yang R.-F.; Zhang X.-E. Detection of Bacillus Anthracis Spores by Super-Paramagnetic Lateral-Flow Immunoassays Based on “Road Closure. Biosens. Bioelectron. 2015, 67, 608–614. 10.1016/j.bios.2014.09.067. [DOI] [PubMed] [Google Scholar]
- Hosseini S.; Vázquez-Villegas P.; Rito-Palomares M.; Martinez-Chapa S. O.. Fundamentals and History of ELISA: The Evolution of the Immunoassays Until Invention of ELISA. In Enzyme-linked Immunosorbent Assay (ELISA); SpringerBriefs in Applied Sciences and Technology; Springer: Singapore, 2018; pp 1–18. [Google Scholar]
- Avrameas S. Coupling of Enzymes to Proteins with Glutaraldehyde: Use of the Conjugates for the Detection of Antigens and Antibodies. Immunochemistry 1969, 6, 43–52. 10.1016/0019-2791(69)90177-3. [DOI] [PubMed] [Google Scholar]
- Valdez C. E.; Smith Q. A.; Nechay M. R.; Alexandrova A. N. Mysteries of Metals in Metalloenzymes. Acc. Chem. Res. 2014, 47, 3110–3117. 10.1021/ar500227u. [DOI] [PubMed] [Google Scholar]
- He J.Practical Guide to ELISA Development. In The Immunoassay Handbook: Theory and Applications of Ligand Binding, ELISA and Related Techniques; Wild D. G., Ed.; Elsevier Inc.: Oxford, U.K., 2013; Chapter 5.1, pp 381–393. [Google Scholar]
- Bertini I.; Luchinat C.. The Reaction Pathways of Zinc Enzymes and Related Biological Catalysts. In Bioinorganic Chemistry; Bertini I., Gray H. B., Stiefel E. I., Valentine J. S., Eds.; University Science Books: Mill Valley, CA, 1994; pp 85–89. [Google Scholar]
- Coleman J. E. Structure and Mechanism of Alkaline Phosphatase. Annu. Rev. Biophys. Biomol. Struct. 1992, 21, 441–483. 10.1146/annurev.bb.21.060192.002301. [DOI] [PubMed] [Google Scholar]
- Gajhede M.; Schuller D. J.; Henriksen A.; Smith A. T.; Poulos T. L. Crystal Structure of Horseradish Peroxidase C at 2.15 Å Resolution. Nat. Struct. Biol. 1997, 4, 1032–1038. 10.1038/nsb1297-1032. [DOI] [PubMed] [Google Scholar]
- Zámocký M.; Koller F. Understanding the Structure and Function of Catalases: Clues from Molecular Evolution and in Vitro Mutagenesis. Prog. Biophys. Mol. Biol. 1999, 72, 19–66. 10.1016/S0079-6107(98)00058-3. [DOI] [PubMed] [Google Scholar]
- Valentine J. S.Dioxygen Reactions. In Bioinorganic Chemistry; Bertini I., Gray H. B., Stiefel E. I., Valentine J. S., Eds.; University Science Books: Mill Valley, CA, 1994; pp 294–297. [Google Scholar]
- Porstmann T.; Kiessig S. T. Enzyme Immunoassay Techniques: An Overview. J. Immunol. Methods 1992, 150, 5–21. 10.1016/0022-1759(92)90061-W. [DOI] [PubMed] [Google Scholar]
- Crowther J. R.The ELISA Guidebook, 2nd ed.; Methods in Molecular Biology; Humana Press: New York, NY, 2009; Vol. 516. [Google Scholar]
- Sajid M.; Kawde A.-N.; Daud M. Designs, Formats and Applications of Lateral Flow Assay: A Literature Review. J. Saudi Chem. Soc. 2015, 19, 689–705. 10.1016/j.jscs.2014.09.001. [DOI] [Google Scholar]
- Kozel T. R.; Burnham-Marusich A. R. Point-of-Care Testing for Infectious Diseases: Past, Present, and Future. J. Clin. Microbiol. 2017, 55, 2313–2320. 10.1128/JCM.00476-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.-M.; Martinez A. W.; Gong J.; Mace C. R.; Phillips S. T.; Carrilho E.; Mirica K. A.; Whitesides G. M. Paper-Based ELISA. Angew. Chem., Int. Ed. 2010, 49, 4771–4774. 10.1002/anie.201001005. [DOI] [PubMed] [Google Scholar]
- Berg B.; Cortazar B.; Tseng D.; Ozkan H.; Feng S.; Wei Q.; Chan R. Y.-L.; Burbano J.; Farooqui Q.; Lewinski M.; et al. Cellphone-Based Hand-Held Microplate Reader for Point-of-Care Testing of Enzyme-Linked Immunosorbent Assays. ACS Nano 2015, 9, 7857–7866. 10.1021/acsnano.5b03203. [DOI] [PubMed] [Google Scholar]
- Lathwal S.; Sikes H. D. Assessment of Colorimetric Amplification Methods in a Paper-Based Immunoassay for Diagnosis of Malaria. Lab Chip 2016, 16, 1374–1382. 10.1039/C6LC00058D. [DOI] [PubMed] [Google Scholar]
- Glavan A. C.; Christodouleas D. C.; Mosadegh B.; Yu H. D.; Smith B. S.; Lessing J.; Fernández-Abedul M. T.; Whitesides G. M. Folding Analytical Devices for Electrochemical ELISA in Hydrophobic RH Paper. Anal. Chem. 2014, 86, 11999–12007. 10.1021/ac5020782. [DOI] [PubMed] [Google Scholar]
- Noh H.; Phillips S. T. Fluidic Timers for Time-Dependent, Point-of-Care Assays on Paper. Anal. Chem. 2010, 82, 8071–8078. 10.1021/ac1005537. [DOI] [PubMed] [Google Scholar]
- Dungchai W.; Chailapakul O.; Henry C. S. Use of Multiple Colorimetric Indicators for Paper-Based Microfluidic Devices. Anal. Chim. Acta 2010, 674, 227–233. 10.1016/j.aca.2010.06.019. [DOI] [PubMed] [Google Scholar]
- Ramachandran S.; Fu E.; Lutz B.; Yager P. Long-Term Dry Storage of an Enzyme-Based Reagent System for ELISA in Point-of-Care Devices. Analyst 2014, 139, 1456–1462. 10.1039/C3AN02296J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel R. M.; Dines M.; Petach H. H. The Denaturation and Degradation of Stable Enzymes at High Temperatures. Biochem. J. 1996, 317, 1–11. 10.1042/bj3170001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelwahed W.; Degobert G.; Stainmesse S.; Fessi H. Freeze-Drying of Nanoparticles: Formulation, Process and Storage Considerations. Adv. Drug Delivery Rev. 2006, 58, 1688–1713. 10.1016/j.addr.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Kaushik J. K.; Bhat R. Why Is Trehalose an Exceptional Protein Stabilizer? An Analysis of the Thermal Stability of Proteins in the Presence of the Compatible Osmolyte Trehalose. J. Biol. Chem. 2003, 278, 26458–26465. 10.1074/jbc.M300815200. [DOI] [PubMed] [Google Scholar]
- de Souza Castilho M.; Laube T.; Yamanaka H.; Alegret S.; Pividori M. I. Magneto Immunoassays for Plasmodium Falciparum Histidine-Rich Protein 2 Related to Malaria Based on Magnetic Nanoparticles. Anal. Chem. 2011, 83, 5570–5577. 10.1021/ac200573s. [DOI] [PubMed] [Google Scholar]
- Zheng L.; Jia L.; Li B.; Situ B.; Liu Q.; Wang Q.; Gan N. A Sandwich HIV P24 Amperometric Immunosensor Based on a Direct Gold Electroplating-Modified Electrode. Molecules 2012, 17, 5988–6000. 10.3390/molecules17055988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Zhang B.; Ye X.; Yan Y.; Huang L.; Jiang Z.; Tan S.; Cai X. Electrochemical Immunosensor for Interferon-γ Based on Disposable ITO Detector and HRP-Antibody-Conjugated Nano Gold as Signal Tag. Mater. Sci. Eng., C 2016, 59, 577–584. 10.1016/j.msec.2015.10.066. [DOI] [PubMed] [Google Scholar]
- de la Rica R.; Aili D.; Stevens M. M. Enzyme-Responsive Nanoparticles for Drug Release and Diagnostics. Adv. Drug Delivery Rev. 2012, 64, 967–978. 10.1016/j.addr.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Lorenzo L.; de la Rica R.; Álvarez-Puebla R. A.; Liz-Marzán L. M.; Stevens M. M. Plasmonic Nanosensors with Inverse Sensitivity by Means of Enzyme-Guided Crystal Growth. Nat. Mater. 2012, 11, 604–607. 10.1038/nmat3337. [DOI] [PubMed] [Google Scholar]
- de la Rica R.; Stevens M. M. Plasmonic ELISA for the Ultrasensitive Detection of Disease Biomarkers with the Naked Eye. Nat. Nanotechnol. 2012, 7, 821–824. 10.1038/nnano.2012.186. [DOI] [PubMed] [Google Scholar]
- Peng M.-P.; Ma W.; Long Y.-T. Alcohol Dehydrogenase-Catalyzed Gold Nanoparticle Seed-Mediated Growth Allows Reliable Detection of Disease Biomarkers with the Naked Eye. Anal. Chem. 2015, 87, 5891–5896. 10.1021/acs.analchem.5b00287. [DOI] [PubMed] [Google Scholar]
- de la Rica R.; Stevens M. M. Plasmonic ELISA for the Detection of Analytes at Ultralow Concentrations with the Naked Eye. Nat. Protoc. 2013, 8, 1759–1764. 10.1038/nprot.2013.085. [DOI] [PubMed] [Google Scholar]
- Satija J.; Punjabi N.; Mishra D.; Mukherji S. Plasmonic-ELISA: Expanding Horizons. RSC Adv. 2016, 6, 85440–85456. 10.1039/C6RA16750K. [DOI] [Google Scholar]
- Lei J.; Ju H. Signal Amplification Using Functional Nanomaterials for Biosensing. Chem. Soc. Rev. 2012, 41, 2122–2134. 10.1039/c1cs15274b. [DOI] [PubMed] [Google Scholar]
- Park K. S.; Kim M. I.; Cho D.-Y.; Park H. G. Label-Free Colorimetric Detection of Nucleic Acids Based on Target-Induced Shielding Against the Peroxidase-Mimicking Activity of Magnetic Nanoparticles. Small 2011, 7, 1521–1525. 10.1002/smll.201001886. [DOI] [PubMed] [Google Scholar]
- Woo M.-A.; Kim M. I.; Jung J. H.; Park K. S.; Seo T. S.; Park H. G. A Novel Colorimetric Immunoassay Utilizing the Peroxidase Mimicking Activity of Magnetic Nanoparticles. Int. J. Mol. Sci. 2013, 14, 9999–10014. 10.3390/ijms14059999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Chen K.; Huang L.; Lu D.; Liang J.; Han H. Sensitive Immunoassay for Porcine Pseudorabies Antibody Based on Fluorescence Signal Amplification Induced by Cation Exchange in CdSe Nanocrystals. Microchim. Acta 2013, 180, 303–310. 10.1007/s00604-012-0934-y. [DOI] [Google Scholar]
- Elzey S.; Grassian V. H. Nanoparticle Dissolution from the Particle Perspective: Insights from Particle Sizing Measurements. Langmuir 2010, 26, 12505–12508. 10.1021/la1019229. [DOI] [PubMed] [Google Scholar]
- Tong S.; Ren B.; Zheng Z.; Shen H.; Bao G. Tiny Grains Give Huge Gains: Nanocrystal-Based Signal Amplification for Biomolecule Detection. ACS Nano 2013, 7, 5142–5150. 10.1021/nn400733t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S.; Fine E. J.; Lin Y.; Cradick T. J.; Bao G. Nanomedicine: Tiny Particles and Machines Give Huge Gains. Ann. Biomed. Eng. 2014, 42, 243–259. 10.1007/s10439-013-0952-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw M. J.; Haddad P. R. The Determination of Trace Metal Pollutants in Environmental Matrices Using Ion Chromatography. Environ. Int. 2004, 30, 403–431. 10.1016/j.envint.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Garrabrants A. C.; Kosson D. S. Use of a Chelating Agent to Determine the Metal Availability for Leaching from Soils and Wastes. Waste Manage. 2000, 20, 155–165. 10.1016/S0956-053X(99)00326-8. [DOI] [Google Scholar]
- Son D. H.; Hughes S. M.; Yin Y.; Alivisatos A. P. Cation Exchange Reactions in Ionic Nanocrystals. Science 2004, 306, 1009–1012. 10.1126/science.1103755. [DOI] [PubMed] [Google Scholar]
- Rivest J. B.; Jain P. K. Cation Exchange on the Nanoscale: An Emerging Technique for New Material Synthesis, Device Fabrication, and Chemical Sensing. Chem. Soc. Rev. 2013, 42, 89–96. 10.1039/C2CS35241A. [DOI] [PubMed] [Google Scholar]
- Yao J.; Schachermeyer S.; Yin Y.; Zhong W. Cation Exchange in ZnSe Nanocrystals for Signal Amplification in Bioassays. Anal. Chem. 2011, 83, 402–408. 10.1021/ac102688s. [DOI] [PubMed] [Google Scholar]
- Sheng Z.; Hu D.; Zhang P.; Gong P.; Gao D.; Liu S.; Cai L. Cation Exchange in Aptamer-Conjugated CdSe Nanoclusters: A Novel Fluorescence Signal Amplification for Cancer Cell Detection. Chem. Commun. 2012, 48, 4202–4204. 10.1039/c2cc00033d. [DOI] [PubMed] [Google Scholar]
- Yao J.; Flack K.; Ding L.; Zhong W. Tagging the Rolling Circle Products with Nanocrystal Clusters for Cascade Signal Increase in the Detection of MiRNA. Analyst 2013, 138, 3121–3125. 10.1039/c3an00398a. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Liu H.; Li R.; Zhang N.; Xiong Y.; Niu S. Chemiluminescence Detection of DNA/MicroRNA Based on Cation-Exchange of CuS Nanoparticles and Rolling Circle Amplification. Chem. Commun. 2015, 51, 6952–6955. 10.1039/C5CC01317H. [DOI] [PubMed] [Google Scholar]
- Xu J.; Zhang Q.; Zhao D.; Liu Y.; Chen P.; Lu G.; Xie H. High Sensitive Detection Method for Protein by Combining the Magnetic Separation with Cation Exchange Based Signal Amplification. Talanta 2017, 168, 91–99. 10.1016/j.talanta.2017.03.018. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Mao Z.; Huang W.; Liu L.; Li J.; Li J.; Wu Q. Redox Enzyme-Mimicking Activities of CeO2 Nanostructures: Intrinsic Influence of Exposed Facets. Sci. Rep. 2016, 6, 35344. 10.1038/srep35344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragg R.; Tahir M. N.; Tremel W. Solids Go Bio: Inorganic Nanoparticles as Enzyme Mimics. Eur. J. Inorg. Chem. 2016, 2016, 1906–1915. 10.1002/ejic.201501237. [DOI] [Google Scholar]
- Wang X.; Guo W.; Hu Y.; Wu J.; Wei H.. Metal-Based Nanomaterials for Nanozymes. In Nanozymes: Next Wave of Artificial Enzymes; SpringerBriefs in Molecular Science; Springer: Berlin, 2016; pp 31–55. [Google Scholar]
- Manea F.; Houillon F. B.; Pasquato L.; Scrimin P. Nanozymes: Gold-Nanoparticle-Based Transphosphorylation Catalysts. Angew. Chem., Int. Ed. 2004, 43, 6165–6169. 10.1002/anie.200460649. [DOI] [PubMed] [Google Scholar]
- Gao L.; Zhuang J.; Nie L.; Zhang J.; Zhang Y.; Gu N.; Wang T.; Feng J.; Yang D.; Perrett S.; et al. Intrinsic Peroxidase-like Activity of Ferromagnetic Nanoparticles. Nat. Nanotechnol. 2007, 2, 577–583. 10.1038/nnano.2007.260. [DOI] [PubMed] [Google Scholar]
- Yang M.; Guan Y.; Yang Y.; Xia T.; Xiong W.; Guo C. A Sensitive and Rapid Immunoassay for Mycoplasma Pneumonia Based on Fe3O4 Nanoparticles. Mater. Lett. 2014, 137, 113–116. 10.1016/j.matlet.2014.08.135. [DOI] [Google Scholar]
- Thiramanas R.; Jangpatarapongsa K.; Tangboriboonrat P.; Polpanich D. Detection of Vibrio Cholerae Using the Intrinsic Catalytic Activity of a Magnetic Polymeric Nanoparticle. Anal. Chem. 2013, 85, 5996–6002. 10.1021/ac400820d. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Huang R.; Liu W.; Liu H.; Zhou X.; Xing D. Rapid and Visual Detection of Listeria Monocytogenes Based on Nanoparticle Cluster Catalyzed Signal Amplification. Biosens. Bioelectron. 2016, 86, 1–7. 10.1016/j.bios.2016.05.100. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Chen Y.; Cheng N.; Xu Y.; Huang K.; Luo Y.; Wang P.; Duan D.; Xu W. Ultrasensitive Detection of Viable Enterobacter Sakazakii by a Continual Cascade Nanozyme Biosensor. Anal. Chem. 2017, 89, 10194–10200. 10.1021/acs.analchem.7b01266. [DOI] [PubMed] [Google Scholar]
- Park J. Y.; Jeong H. Y.; Kim M. I.; Park T. J. Colorimetric Detection System for Salmonella Typhimurium Based on Peroxidase-Like Activity of Magnetic Nanoparticles with DNA Aptamers. J. Nanomater. 2015, 2015, 527126. 10.1155/2015/527126. [DOI] [Google Scholar]
- Ahmed S. R.; Corredor J. C.; Nagy É.; Neethirajan S. Amplified Visual Immunosensor Integrated with Nanozyme for Ultrasensitive Detection of Avian Influenza Virus. Nanotheranostics 2017, 1, 338–345. 10.7150/ntno.20758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.; Kim M. S.; Kweon S. H.; Jeong S.; Kang M. H.; Kim M. I.; Lee J.; Doh J. Simple and Sensitive Point-of-Care Bioassay System Based on Hierarchically Structured Enzyme-Mimetic Nanoparticles. Adv. Healthcare Mater. 2015, 4, 1311–1316. 10.1002/adhm.201500173. [DOI] [PubMed] [Google Scholar]
- Lin X.; Liu Y.; Tao Z.; Gao J.; Deng J.; Yin J.; Wang S. Nanozyme-Based Bio-Barcode Assay for High Sensitive and Logic-Controlled Specific Detection of Multiple DNAs. Biosens. Bioelectron. 2017, 94, 471–477. 10.1016/j.bios.2017.01.008. [DOI] [PubMed] [Google Scholar]
- Ye H.; Yang K.; Tao J.; Liu Y.; Zhang Q.; Habibi S.; Nie Z.; Xia X. An Enzyme-Free Signal Amplification Technique for Ultrasensitive Colorimetric Assay of Disease Biomarkers. ACS Nano 2017, 11, 2052–2059. 10.1021/acsnano.6b08232. [DOI] [PubMed] [Google Scholar]
- Gao Z.; Ye H.; Tang D.; Tao J.; Habibi S.; Minerick A.; Tang D.; Xia X. Platinum-Decorated Gold Nanoparticles with Dual Functionalities for Ultrasensitive Colorimetric in Vitro Diagnostics. Nano Lett. 2017, 17, 5572–5579. 10.1021/acs.nanolett.7b02385. [DOI] [PubMed] [Google Scholar]
- Cheng N.; Song Y.; Zeinhom M. M. A.; Chang Y.-C.; Sheng L.; Li H.; Du D.; Li L.; Zhu M.-J.; Luo Y.; et al. Nanozyme-Mediated Dual Immunoassay Integrated with Smartphone for Use in Simultaneous Detection of Pathogens. ACS Appl. Mater. Interfaces 2017, 9, 40671–40680. 10.1021/acsami.7b12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L.; Li C. M.; Wu W. B.; Huang C. Z. A Colorimetric Immunoassay for Respiratory Syncytial Virus Detection Based on Gold Nanoparticles–Graphene Oxide Hybrids with Mercury-Enhanced Peroxidase-like Activity. Chem. Commun. 2014, 50, 11526–11528. 10.1039/C4CC05155F. [DOI] [PubMed] [Google Scholar]
- Wang S.; Deng W.; Yang L.; Tan Y.; Xie Q.; Yao S. Copper-Based Metal–Organic Framework Nanoparticles with Peroxidase-Like Activity for Sensitive Colorimetric Detection of Staphylococcus Aureus. ACS Appl. Mater. Interfaces 2017, 9, 24440–24445. 10.1021/acsami.7b07307. [DOI] [PubMed] [Google Scholar]
- Kim M. S.; Kweon S. H.; Cho S.; An S. S. A.; Kim M. I.; Doh J.; Lee J. Pt-Decorated Magnetic Nanozymes for Facile and Sensitive Point-of-Care Bioassay. ACS Appl. Mater. Interfaces 2017, 9, 35133–35140. 10.1021/acsami.7b12326. [DOI] [PubMed] [Google Scholar]
- Loynachan C. N.; Thomas M. R.; Gray E. R.; Richards D. A.; Kim J.; Miller B. S.; Brookes J. C.; Agarwal S.; Chudasama V.; McKendry R. A.; et al. Platinum Nanocatalyst Amplification: Redefining the Gold Standard for Lateral Flow Immunoassays with Ultrabroad Dynamic Range. ACS Nano 2018, 12, 279–288. 10.1021/acsnano.7b06229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yetisen A. K.; Akram M. S.; Lowe C. R. Paper-Based Microfluidic Point-of-Care Diagnostic Devices. Lab Chip 2013, 13, 2210. 10.1039/c3lc50169h. [DOI] [PubMed] [Google Scholar]
- Cordeiro M.; Ferreira Carlos F.; Pedrosa P.; Lopez A.; Baptista P. V. Gold Nanoparticles for Diagnostics: Advances towards Points of Care. Diagnostics 2016, 6, 43. 10.3390/diagnostics6040043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker G. W.; Grimelius L.; Danscher G.; Bernatzky G.; Muss W.; Adam H.; Thurner J. Silver Acetate Autometallography: An Alternative Enhancement Technique for Immunogold-Silver Staining (IGSS) and Silver Amplification of Gold, Silver, Mercury and Zinc in Tissues. J. Histotechnol. 1988, 11, 213–221. 10.1179/his.1988.11.4.213. [DOI] [Google Scholar]
- Wada A.; Sakoda Y.; Oyamada T.; Kida H. Development of a Highly Sensitive Immunochromatographic Detection Kit for H5 Influenza Virus Hemagglutinin Using Silver Amplification. J. Virol. Methods 2011, 178, 82–86. 10.1016/j.jviromet.2011.08.017. [DOI] [PubMed] [Google Scholar]
- Oliver C.Use of Immunogold with Silver Enhancement. In Immunocytochemical Methods and Protocols; Oliver C., Jamur M., Eds.; Methods in Molecular Biology; Humana Press, 2010; Vol. 588, pp 311–316. [DOI] [PubMed] [Google Scholar]
- Horton J. K.; Swinburne S.; O’Sullivan M. J. A Novel, Rapid, Single-Step Immunochromatographic Procedure for the Detection of Mouse Immunoglobulin. J. Immunol. Methods 1991, 140, 131–134. 10.1016/0022-1759(91)90135-3. [DOI] [PubMed] [Google Scholar]
- Mitamura K.; Shimizu H.; Yamazaki M.; Ichikawa M.; Nagai K.; Katada J.; Wada A.; Kawakami C.; Sugaya N. Clinical Evaluation of Highly Sensitive Silver Amplification Immunochromatography Systems for Rapid Diagnosis of Influenza. J. Virol. Methods 2013, 194, 123–128. 10.1016/j.jviromet.2013.08.018. [DOI] [PubMed] [Google Scholar]
- Rohrman B. A.; Leautaud V.; Molyneux E.; Richards-Kortum R. R. A Lateral Flow Assay for Quantitative Detection of Amplified HIV-1 RNA. PLoS One 2012, 7, e45611 10.1371/journal.pone.0045611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu E.; Liang T.; Spicar-Mihalic P.; Houghtaling J.; Ramachandran S.; Yager P. Two-Dimensional Paper Network Format That Enables Simple Multistep Assays for Use in Low-Resource Settings in the Context of Malaria Antigen Detection. Anal. Chem. 2012, 84, 4574–4579. 10.1021/ac300689s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz B.; Liang T.; Fu E.; Ramachandran S.; Kauffman P.; Yager P. Dissolvable Fluidic Time Delays for Programming Multi-Step Assays in Instrument-Free Paper Diagnostics. Lab Chip 2013, 13, 2840–2847. 10.1039/c3lc50178g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toley B. J.; McKenzie B.; Liang T.; Buser J. R.; Yager P.; Fu E. Tunable-Delay Shunts for Paper Microfluidic Devices. Anal. Chem. 2013, 85, 11545–11552. 10.1021/ac4030939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridley G. E.; Le H.; Yager P. Highly Sensitive Immunoassay Based on Controlled Rehydration of Patterned Reagents in a 2-Dimensional Paper Network. Anal. Chem. 2014, 86, 6447–6453. 10.1021/ac500872j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toley B. J.; Wang J. A.; Gupta M.; Buser J. R.; Lafleur L.; Lutz B. R.; Fu E.; Yager P. A Versatile Valving Toolkit for Automating Fluidic Operations in Paper Microfluidic Devices. Lab Chip 2015, 15, 1432–1444. 10.1039/C4LC01155D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T.; Robinson R.; Houghtaling J.; Fridley G.; Ramsey S. A.; Fu E. Investigation of Reagent Delivery Formats in a Multivalent Malaria Sandwich Immunoassay and Implications for Assay Performance. Anal. Chem. 2016, 88, 2311–2320. 10.1021/acs.analchem.5b04222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui P.-Y.; Tsai H.-P.; Chiao D.-J.; Liu C.-C.; Shyu R.-H. Rapid Detection of Yersinia Pestis Recombinant Fraction 1 Capsular Antigen. Appl. Microbiol. Biotechnol. 2015, 99, 7781–7789. 10.1007/s00253-015-6663-5. [DOI] [PubMed] [Google Scholar]
- Chen J.; Jackson A. A.; Rotello V. M.; Nugen S. R. Colorimetric Detection of Escherichia Coli Based on the Enzyme-Induced Metallization of Gold Nanorods. Small 2016, 12, 2469–2475. 10.1002/smll.201503682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M.; Katada J.; Chiku H.; Nakamura K.; Oyamada T. Development of Highly Sensitive Immunochromatographic Detection Kit for Seasonal Influenza Virus Using Silver Amplification. Fujifilm Research & Development 2012, 57, 5–10. [Google Scholar]
- Fu E.; Kauffman P.; Lutz B.; Yager P. Chemical Signal Amplification in Two-Dimensional Paper Networks. Sens. Actuators, B 2010, 149, 325–328. 10.1016/j.snb.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu E.; Liang T.; Houghtaling J.; Ramachandran S.; Ramsey S. A.; Lutz B.; Yager P. Enhanced Sensitivity of Lateral Flow Tests Using a Two-Dimensional Paper Network Format. Anal. Chem. 2011, 83, 7941–7946. 10.1021/ac201950g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I.-H.; Seo S.-M.; Paek E.-H.; Paek S.-H. Immunogold–Silver Staining-on-a-Chip Biosensor Based on Cross-Flow Chromatography. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2010, 878, 271–277. 10.1016/j.jchromb.2009.07.016. [DOI] [PubMed] [Google Scholar]
- Cao C.; Gontard L. C.; Tram L. L. T.; Wolff A.; Bang D. D. Dual Enlargement of Gold Nanoparticles: From Mechanism to Scanometric Detection of Pathogenic Bacteria. Small 2011, 7, 1701–1708. 10.1002/smll.201100294. [DOI] [PubMed] [Google Scholar]
- Lei K. F. Quantitative Electrical Detection of Immobilized Protein Using Gold Nanoparticles and Gold Enhancement on a Biochip. Meas. Sci. Technol. 2011, 22, 105802. 10.1088/0957-0233/22/10/105802. [DOI] [Google Scholar]
- Rodríguez M. O.; Covián L. B.; García A. C.; Blanco-López M. C. Silver and Gold Enhancement Methods for Lateral Flow Immunoassays. Talanta 2016, 148, 272–278. 10.1016/j.talanta.2015.10.068. [DOI] [PubMed] [Google Scholar]
- Lin H.-C.; Wang I.-L.; Lin H.-P.; Chang T. C.; Lin Y.-C. Enhancement of an Immunoassay Using Platinum Nanoparticles and an Optical Detection. Sens. Actuators, B 2011, 154, 185–190. 10.1016/j.snb.2010.05.055. [DOI] [Google Scholar]
- Sia S. K.; Linder V.; Parviz B. A.; Siegel A.; Whitesides G. M. An Integrated Approach to a Portable and Low-Cost Immunoassay for Resource-Poor Settings. Angew. Chem., Int. Ed. 2004, 43, 498–502. 10.1002/anie.200353016. [DOI] [PubMed] [Google Scholar]
- Nam J.-M.; Park S.-J.; Mirkin C. A. Bio-Barcodes Based on Oligonucleotide-Modified Nanoparticles. J. Am. Chem. Soc. 2002, 124, 3820–3821. 10.1021/ja0178766. [DOI] [PubMed] [Google Scholar]
- Nam J.-M.; Thaxton C. S.; Mirkin C. A. Nanoparticle-Based Bio-Bar Codes for the Ultrasensitive Detection of Proteins. Science 2003, 301, 1884–1886. 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- Stoeva S. I.; Lee J.-S.; Thaxton C. S.; Mirkin C. A. Multiplexed DNA Detection with Biobarcoded Nanoparticle Probes. Angew. Chem. 2006, 118, 3381–3384. 10.1002/ange.200600124. [DOI] [PubMed] [Google Scholar]
- Nam J.-M.; Stoeva S. I.; Mirkin C. A. Bio-Bar-Code-Based DNA Detection with PCR-like Sensitivity. J. Am. Chem. Soc. 2004, 126, 5932–5933. 10.1021/ja049384+. [DOI] [PubMed] [Google Scholar]
- Chang T.-L.; Tsai C.-Y.; Sun C.-C.; Chen C.-C.; Kuo L.-S.; Chen P.-H. Ultrasensitive Electrical Detection of Protein Using Nanogap Electrodes and Nanoparticle-Based DNA Amplification. Biosens. Bioelectron. 2007, 22, 3139–3145. 10.1016/j.bios.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Yin H.; Ji C.; Yang X.; Wang R.; Yang S.; Zhang H.; Zhang J. An Improved Gold Nanoparticle Probe-Based Assay for HCV Core Antigen Ultrasensitive Detection. J. Virol. Methods 2017, 243, 142–145. 10.1016/j.jviromet.2017.02.007. [DOI] [PubMed] [Google Scholar]
- Tang S.; Zhao J.; Storhoff J. J.; Norris P. J.; Little R. F.; Yarchoan R.; Stramer S. L.; Patno T.; Domanus M.; Dhar A.; et al. Nanoparticle-Based Biobarcode Amplification Assay (BCA) for Sensitive and Early Detection of Human Immunodeficiency Type 1 Capsid (P24) Antigen. JAIDS, J. Acquired Immune Defic. JAIDS, J. Acquired Immune Defic. Syndr. 2007, 46, 231–237. 10.1097/QAI.0b013e31814a554b. [DOI] [PubMed] [Google Scholar]
- Dong H.; Liu J.; Zhu H.; Ou C.-Y.; Xing W.; Qiu M.; Zhang G.; Xiao Y.; Yao J.; Pan P.; et al. Two Types of Nanoparticle-Based Bio-Barcode Amplification Assays to Detect HIV-1 P24 Antigen. Virol. J. 2012, 9, 180. 10.1186/1743-422X-9-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.; Carr D. J.; Alocilja E. C. Fluorescent Bio-Barcode DNA Assay for the Detection of Salmonella Enterica Serovar Enteritidis. Biosens. Bioelectron. 2009, 24, 1377–1381. 10.1016/j.bios.2008.07.081. [DOI] [PubMed] [Google Scholar]
- Broto M.; Galve R.; Marco M.-P. Sandwich NP-Based Biobarcode Assay for Quantification C-Reactive Protein in Plasma Samples. Anal. Chim. Acta 2017, 992, 112–118. 10.1016/j.aca.2017.09.007. [DOI] [PubMed] [Google Scholar]
- Nam J.-M.; Wise A. R.; Groves J. T. Colorimetric Bio-Barcode Amplification Assay for Cytokines. Anal. Chem. 2005, 77, 6985–6988. 10.1021/ac0513764. [DOI] [PubMed] [Google Scholar]
- Nam J.-M.; Jang K.-J.; Groves J. T. Detection of Proteins Using a Colorimetric Bio-Barcode Assay. Nat. Protoc. 2007, 2, 1438–1444. 10.1038/nprot.2007.201. [DOI] [PubMed] [Google Scholar]
- Stoeva S. I.; Lee J.-S.; Smith J. E.; Rosen S. T.; Mirkin C. A. Multiplexed Detection of Protein Cancer Markers with Biobarcoded Nanoparticle Probes. J. Am. Chem. Soc. 2006, 128, 8378–8379. 10.1021/ja0613106. [DOI] [PubMed] [Google Scholar]
- Ahmad R.; Jang H.; Batule B. S.; Park H. G. Barcode DNA-Mediated Signal Amplifying Strategy for Ultrasensitive Biomolecular Detection on Matrix-Assisted Laser Desorption Ionization Time of Flight (MALDI-TOF) Mass Spectrometry. Anal. Chem. 2017, 89, 8966–8973. 10.1021/acs.analchem.7b01535. [DOI] [PubMed] [Google Scholar]
- Hazen K. C.Introduction to Laboratory Diagnosis of Infectious Disease - Infectious Diseases; https://www.merckmanuals.com/professional/infectious-diseases/laboratory-diagnosis-of-infectious-disease/introduction-to-laboratory-diagnosis-of-infectious-disease (accessed July 20, 2018). [Google Scholar]
- Bousema T.; Okell L.; Felger I.; Drakeley C. Asymptomatic Malaria Infections: Detectability, Transmissibility and Public Health Relevance. Nat. Rev. Microbiol. 2014, 12, 833–840. 10.1038/nrmicro3364. [DOI] [PubMed] [Google Scholar]
- Forcucci A.; Pawlowski M. E.; Crannell Z.; Pavlova I.; Richards-Kortum R.; Tkaczyk T. S. All-Plastic Miniature Fluorescence Microscope for Point-of-Care Readout of Bead-Based Bioassays. J. Biomed. Opt. 2015, 20, 105010. 10.1117/1.JBO.20.10.105010. [DOI] [PubMed] [Google Scholar]
- Forcucci A.; Pawlowski M. E.; Majors C.; Richards-Kortum R.; Tkaczyk T. S. All-Plastic, Miniature, Digital Fluorescence Microscope for Three Part White Blood Cell Differential Measurements at the Point of Care. Biomed. Opt. Express 2015, 6, 4433–4446. 10.1364/BOE.6.004433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum A.; Akbari N.; Feizi A.; Luo W.; Ozcan A. Field-Portable Pixel Super-Resolution Colour Microscope. PLoS One 2013, 8, e76475 10.1371/journal.pone.0076475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. R.; Davis G.; Pierce M.; Oden Z. M.; Richards-Kortum R.. Portable, Battery-Operated, Fluorescence Field Microscope for the Developing World. In Proceedings of the Design and Quality for Biomedical Technologies III; International Society for Optics and Photonics: San Francisco, CA, 2010; Vol.7556, p 755608. [Google Scholar]
- Pierce M. C.; Weigum S. E.; Jaslove J. M.; Richards-Kortum R.; Tkaczyk T. S. Optical Systems for Point-of-Care Diagnostic Instrumentation: Analysis of Imaging Performance and Cost. Ann. Biomed. Eng. 2014, 42, 231–240. 10.1007/s10439-013-0918-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isikman S. O.; Bishara W.; Sikora U.; Yaglidere O.; Yeah J.; Ozcan A. Field-Portable Lensfree Tomographic Microscope. Lab Chip 2011, 11, 2222–2230. 10.1039/c1lc20127a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishara W.; Sikora U.; Mudanyali O.; Su T.-W.; Yaglidere O.; Luckhart S.; Ozcan A. Holographic Pixel Super-Resolution in Portable Lensless on-Chip Microscopy Using a Fiber-Optic Array. Lab Chip 2011, 11, 1276–1279. 10.1039/c0lc00684j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum A.; Sikora U.; Ozcan A. Field-Portable Wide-Field Microscopy of Dense Samples Using Multi-Height Pixel Super-Resolution Based Lensfree Imaging. Lab Chip 2012, 12, 1242–1245. 10.1039/c2lc21072j. [DOI] [PubMed] [Google Scholar]
- Miller A. R.; Davis G. L.; Oden Z. M.; Razavi M. R.; Fateh A.; Ghazanfari M.; Abdolrahimi F.; Poorazar S.; Sakhaie F.; Olsen R. J.; et al. Portable, Battery-Operated, Low-Cost, Bright Field and Fluorescence Microscope. PLoS One 2010, 5, e11890 10.1371/journal.pone.0011890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapley A.; Switz N.; Reber C.; Davis J. L.; Miller C.; Matovu J. B.; Worodria W.; Huang L.; Fletcher D. A.; Cattamanchi A. Mobile Digital Fluorescence Microscopy for Diagnosis of Tuberculosis. J. Clin. Microbiol. 2013, 51, 1774–1778. 10.1128/JCM.03432-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmström O.; Linder N.; Ngasala B.; Mårtensson A.; Linder E.; Lundin M.; Moilanen H.; Suutala A.; Diwan V.; Lundin J. Point-of-Care Mobile Digital Microscopy and Deep Learning for the Detection of Soil-Transmitted Helminths and Schistosoma Haematobium. Glob. Health Action 2017, 10 (3), 1337325. 10.1080/16549716.2017.1337325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski J. S.; Clements J.; Prakash M. Foldscope: Origami-Based Paper Microscope. PLoS One 2014, 9, e98781 10.1371/journal.pone.0098781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephraim R. K. D.; Duah E.; Cybulski J. S.; Prakash M.; D’Ambrosio M. V.; Fletcher D. A.; Keiser J.; Andrews J. R.; Bogoch I. I. Diagnosis of Schistosoma Haematobium Infection with a Mobile Phone-Mounted Foldscope and a Reversed-Lens CellScope in Ghana. Am. J. Trop. Med. Hyg. 2015, 92, 1253–1256. 10.4269/ajtmh.14-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Naranjo J. C.; Wei Q.; Ozcan A. Mobile Phone-Based Microscopy, Sensing, and Diagnostics. IEEE J. Sel. Top. Quantum Electron. 2016, 22, 1. 10.1109/JSTQE.2015.2478657. [DOI] [Google Scholar]
- Coulibaly J. T.; Ouattara M.; Keiser J.; Bonfoh B.; N’Goran E. K.; Andrews J. R.; Bogoch I. I. Evaluation of Malaria Diagnoses Using a Handheld Light Microscope in a Community-Based Setting in Rural Côte d’Ivoire. Am. J. Trop. Med. Hyg. 2016, 95, 831–834. 10.4269/ajtmh.16-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslauer D. N.; Maamari R. N.; Switz N. A.; Lam W. A.; Fletcher D. A. Mobile Phone Based Clinical Microscopy for Global Health Applications. PLoS One 2009, 4, e6320 10.1371/journal.pone.0006320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoch I. I.; Koydemir H. C.; Tseng D.; Ephraim R. K. D.; Duah E.; Tee J.; Andrews J. R.; Ozcan A. Evaluation of a Mobile Phone-Based Microscope for Screening of Schistosoma Haematobium Infection in Rural Ghana. Am. J. Trop. Med. Hyg. 2017, 96, 1468–1471. 10.4269/ajtmh.16-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulibaly J. T.; Ouattara M.; D’Ambrosio M. V.; Fletcher D. A.; Keiser J.; Utzinger J.; N’Goran E. K.; Andrews J. R.; Bogoch I. I. Accuracy of Mobile Phone and Handheld Light Microscopy for the Diagnosis of Schistosomiasis and Intestinal Protozoa Infections in Côte d’Ivoire. PLoS Neglected Trop. Dis. 2016, 10, e0004768 10.1371/journal.pntd.0004768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H.; Castro C. M.; Shao H.; Liong M.; Song J.; Pathania D.; Fexon L.; Min C.; Avila-Wallace M.; Zurkiya O.; et al. Digital Diffraction Analysis Enables Low-Cost Molecular Diagnostics on a Smartphone. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 5613–5618. 10.1073/pnas.1501815112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoch I. I.; Andrews J. R.; Speich B.; Utzinger J.; Ame S. M.; Ali S. M.; Keiser J. Mobile Phone Microscopy for the Diagnosis of Soil-Transmitted Helminth Infections: A Proof-of-Concept Study. Am. J. Trop. Med. Hyg. 2013, 88, 626–629. 10.4269/ajtmh.12-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosio M. V.; Bakalar M.; Bennuru S.; Reber C.; Skandarajah A.; Nilsson L.; Switz N.; Kamgno J.; Pion S.; Boussinesq M.; et al. Point-of-Care Quantification of Blood-Borne Filarial Parasites with a Mobile Phone Microscope. Sci. Transl. Med. 2015, 7, 286re4. 10.1126/scitranslmed.aaa3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamgno J.; Pion S. D.; Chesnais C. B.; Bakalar M. H.; D’Ambrosio M. V.; Mackenzie C. D.; Nana-Djeunga H. C.; Gounoue-Kamkumo R.; Njitchouang G.-R.; Nwane P.; et al. A Test-and-Not-Treat Strategy for Onchocerciasis in Loa Loa–Endemic Areas. N. Engl. J. Med. 2017, 377, 2044–2052. 10.1056/NEJMoa1705026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H.; Yaglidere O.; Su T.-W.; Tseng D.; Ozcan A. Cost-Effective and Compact Wide-Field Fluorescent Imaging on a Cell-Phone. Lab Chip 2011, 11, 315–322. 10.1039/C0LC00358A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Switz N. A.; D’Ambrosio M. V.; Fletcher D. A. Low-Cost Mobile Phone Microscopy with a Reversed Mobile Phone Camera Lens. PLoS One 2014, 9, e95330 10.1371/journal.pone.0095330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulstich K.; Gruler R.; Eberhard M.; Lentzsch D.; Haberstroh K.. Handheld and Portable Reader Devices for Lateral Flow Immunoassays. In Lateral Flow Immunoassay; Wong R., Tse H., Eds.; Humana Press, 2009; pp 1–27. [Google Scholar]
- Semwogerere D.; Weeks E. R.. Confocal Microscopy. Encyclopedia of Biomaterials and Biomedical Engineering; Wnek G. E., Bowlin G. L., Eds.; Informa Healthcare Press, 2008; Vol. 1, pp 705–714. [Google Scholar]
- O’Farrell B.Lateral Flow Immunoassay Systems: Evolution from the Current State of the Art to the Next Generation of Highly Sensitive, Quantitative Rapid Assays. In The Immunoassay Handbook; Wild D., Ed.; Elsevier: Oxford, 2013; Chapter 2.4, pp 89–107. [Google Scholar]
- Detekt Biomedical LLC . Lateral Flow Reader Systems > RDS-1500 PRO; http://store.idetekt.com/rds-1500pro.aspx (accessed July 20, 2018).
- Detekt Biomedical LLC . DETEKT RDS-2500 Brochure; 2018.
- Grbavac A.Detekt Biomedical Email Communication; 2018.
- Shekalaghe S.; Cancino M.; Mavere C.; Juma O.; Mohammed A.; Abdulla S.; Ferro S. Clinical Performance of an Automated Reader in Interpreting Malaria Rapid Diagnostic Tests in Tanzania. Malar. J. 2013, 12, 141. 10.1186/1475-2875-12-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera S.; Vallejo A. F.; Quintero J. P.; Arévalo-Herrera M.; Cancino M.; Ferro S. Field Evaluation of an Automated RDT Reader and Data Management Device for Plasmodium Falciparum/Plasmodium Vivax Malaria in Endemic Areas of Colombia. Malar. J. 2014, 13, 87. 10.1186/1475-2875-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansumana R.; Taitt C.; Lamin J. M.; Jacobsen K. H.; Mulvaney S. P.; Leski T.; Bangura U.; Stenger D. Point-of-Need Diagnostics: Biosurveillance with a Device2cloud Capability in Sierra Leone. BMJ. Global Health 2017, 2 (2), A12.2. 10.1136/bmjgh-2016-000260.28. [DOI] [Google Scholar]
- Oyet C.; Roh M. E.; Kiwanuka G. N.; Orikiriza P.; Wade M.; Parikh S.; Mwanga-Amumpaire J.; Boum Y. Evaluation of the Deki Reader, an Automated RDT Reader and Data Management Device, in a Household Survey Setting in Low Malaria Endemic Southwestern Uganda. Malar. J. 2017, 16, 449. 10.1186/s12936-017-2094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudanyali O.; Dimitrov S.; Sikora U.; Padmanabhan S.; Navruz I.; Ozcan A. Integrated Rapid-Diagnostic-Test Reader Platform on a Cellphone. Lab Chip 2012, 12, 2678–2686. 10.1039/c2lc40235a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherr T. F.; Gupta S.; Wright D. W.; Haselton F. R. Mobile Phone Imaging and Cloud-Based Analysis for Standardized Malaria Detection and Reporting. Sci. Rep. 2016, 6, 28645. 10.1038/srep28645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherr T. F.; Gupta S.; Wright D. W.; Haselton F. R. An Embedded Barcode for “Connected” Malaria Rapid Diagnostic Tests. Lab Chip 2017, 17, 1314–1322. 10.1039/C6LC01580H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mthembu C. L.; Sabela M. I.; Mlambo M.; Madikizela L. M.; Kanchi S.; Gumede H.; Mdluli P. Google Analytics and Quick Response for Advancement of Gold Nanoparticle-Based Dual Lateral Flow Immunoassay for Malaria – Plasmodium Lactate Dehydrogenase (PLDH). Anal. Methods 2017, 9, 5943–5951. 10.1039/C7AY01645J. [DOI] [Google Scholar]
- Feng S.; Caire R.; Cortazar B.; Turan M.; Wong A.; Ozcan A. Immunochromatographic Diagnostic Test Analysis Using Google Glass. ACS Nano 2014, 8, 3069–3079. 10.1021/nn500614k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long K. D.; Woodburn E. V.; Le H. M.; Shah U. K.; Lumetta S. S.; Cunningham B. T. Multimode Smartphone Biosensing: The Transmission, Reflection, and Intensity Spectral (TRI)-Analyzer. Lab Chip 2017, 17, 3246–3257. 10.1039/C7LC00633K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.; Tan Y.; Cunningham B. T. Smartphone Fluorescence Spectroscopy. Anal. Chem. 2014, 86, 8805–8813. 10.1021/ac502080t. [DOI] [PubMed] [Google Scholar]
- Gautam S.; Batule B. S.; Kim H. Y.; Park K. S.; Park H. G. Smartphone-Based Portable Wireless Optical System for the Detection of Target Analytes. Biotechnol. J. 2017, 12, 1600581. 10.1002/biot.201600581. [DOI] [PubMed] [Google Scholar]
- Preechaburana P.; Gonzalez M. C.; Suska A.; Filippini D. Surface Plasmon Resonance Chemical Sensing on Cell Phones. Angew. Chem., Int. Ed. 2012, 51, 11585–11588. 10.1002/anie.201206804. [DOI] [PubMed] [Google Scholar]
- Guner H.; Ozgur E.; Kokturk G.; Celik M.; Esen E.; Topal A. E.; Ayas S.; Uludag Y.; Elbuken C.; Dana A. A Smartphone Based Surface Plasmon Resonance Imaging (SPRi) Platform for on-Site Biodetection. Sens. Actuators, B 2017, 239, 571–577. 10.1016/j.snb.2016.08.061. [DOI] [Google Scholar]
- Zhang J.; Khan I.; Zhang Q.; Liu X.; Dostalek J.; Liedberg B.; Wang Y. Lipopolysaccharides Detection on a Grating-Coupled Surface Plasmon Resonance Smartphone Biosensor. Biosens. Bioelectron. 2018, 99, 312–317. 10.1016/j.bios.2017.07.048. [DOI] [PubMed] [Google Scholar]
- Zhu H.; Mavandadi S.; Coskun A. F.; Yaglidere O.; Ozcan A. Optofluidic Fluorescent Imaging Cytometry on a Cell Phone. Anal. Chem. 2011, 83, 6641–6647. 10.1021/ac201587a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton S.; Joshi A.; Syrrist P.; Coskun A. F.; Tasoglu S. 3D-Printed Smartphone-Based Point of Care Tool for Fluorescence- and Magnetophoresis-Based Cytometry. Lab Chip 2017, 17, 2839–2851. 10.1039/C7LC00706J. [DOI] [PubMed] [Google Scholar]
- Qin Z.; Bischof J. C. Thermophysical and Biological Responses of Gold Nanoparticle Laser Heating. Chem. Soc. Rev. 2012, 41, 1191–1217. 10.1039/C1CS15184C. [DOI] [PubMed] [Google Scholar]
- Qin Z.; Chan W. C. W.; Boulware D. R.; Akkin T.; Butler E. K.; Bischof J. C. Significantly Improved Analytical Sensitivity of Lateral Flow Immunoassays by Using Thermal Contrast. Angew. Chem., Int. Ed. 2012, 51, 4358–4361. 10.1002/anie.201200997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Qin Z.; Boulware D. R.; Pritt B. S.; Sloan L. M.; González I. J.; Bell D.; Rees-Channer R. R.; Chiodini P.; Chan W. C. W.; et al. Thermal Contrast Amplification Reader Yielding 8-Fold Analytical Improvement for Disease Detection with Lateral Flow Assays. Anal. Chem. 2016, 88, 11774–11782. 10.1021/acs.analchem.6b03406. [DOI] [PubMed] [Google Scholar]
- Boulware D. R.; Rolfes M. A.; Rajasingham R.; von Hohenberg M.; Qin Z.; Taseera K.; Schutz C.; Kwizera R.; Butler E. K.; Meintjes G.; et al. Multisite Validation of Cryptococcal Antigen Lateral Flow Assay and Quantification by Laser Thermal Contrast. Emerging Infect. Dis. 2014, 20, 45–53. 10.3201/eid2001.130906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L.; Guo S.; Song F.; Gong Y.; Xu F.; Boulware D. R.; McAlpine M. C.; Chan W. C. W.; Bischof J. C. The Role of Nanoparticle Design in Determining Analytical Performance of Lateral Flow Immunoassays. Nano Lett. 2017, 17, 7207–7212. 10.1021/acs.nanolett.7b02302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll A. J.; Harpster M. H.; Johnson P. A. The Development of Surface-Enhanced Raman Scattering as a Detection Modality for Portable in Vitro Diagnostics: Progress and Challenges. Phys. Chem. Chem. Phys. 2013, 15, 20415–20433. 10.1039/c3cp52334a. [DOI] [PubMed] [Google Scholar]
- Sharma B.; Frontiera R. R.; Henry A.-I.; Ringe E.; Van Duyne R. P. SERS: Materials, Applications, and the Future. Mater. Today 2012, 15, 16–25. 10.1016/S1369-7021(12)70017-2. [DOI] [Google Scholar]
- Wang Y.; Ravindranath S.; Irudayaraj J. Separation and Detection of Multiple Pathogens in a Food Matrix by Magnetic SERS Nanoprobes. Anal. Bioanal. Chem. 2011, 399, 1271–1278. 10.1007/s00216-010-4453-6. [DOI] [PubMed] [Google Scholar]
- Ravindranath S. P.; Wang Y.; Irudayaraj J. SERS Driven Cross-Platform Based Multiplex Pathogen Detection. Sens. Actuators, B 2011, 152, 183–190. 10.1016/j.snb.2010.12.005. [DOI] [Google Scholar]
- Tamer U.; Boyacı İ. H.; Temur E.; Zengin A.; Dincer İ.; Elerman Y. Fabrication of Magnetic Gold Nanorod Particles for Immunomagnetic Separation and SERS Application. J. Nanopart. Res. 2011, 13, 3167–3176. 10.1007/s11051-010-0213-y. [DOI] [Google Scholar]
- Guven B.; Basaran-Akgul N.; Temur E.; Tamer U.; Boyacı İ. H. SERS-Based Sandwich Immunoassay Using Antibody Coated Magnetic Nanoparticles for Escherichia Coli Enumeration. Analyst 2011, 136, 740–748. 10.1039/C0AN00473A. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Xu J.; Mi L.; Gong H.; Jiang S.; Yu Q. Multifunctional Magnetic–Plasmonic Nanoparticles for Fast Concentration and Sensitive Detection of Bacteria Using SERS. Biosens. Bioelectron. 2012, 31, 130–136. 10.1016/j.bios.2011.10.006. [DOI] [PubMed] [Google Scholar]
- Yakes B. J.; Lipert R. J.; Bannantine J. P.; Porter M. D. Detection of Mycobacterium Avium Subsp. Paratuberculosis by a Sonicate Immunoassay Based on Surface-Enhanced Raman Scattering. Clin. Vaccine Immunol. 2008, 15, 227–234. 10.1128/CVI.00334-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolsky J.; Kaur S.; Hayashi C.; Batra S. K.; Krasnoslobodtsev A. V. Surface-Enhanced Raman Scattering-Based Immunoassay Technologies for Detection of Disease Biomarkers. Biosensors 2017, 7, 7. 10.3390/bios7010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermo Scientific FirstDefender RMX Handheld Chemical Identification Pricing. https://www.fishersci.com/shop/products/thermo-scientific-firstdefender-rmx-handheld-chemical-identification-5/p-4006497 (accessed July 13, 2018). [Google Scholar]
- FirstDefender RMX Handheld Chemical Identification. https://www.thermofisher.com/order/catalog/product/FIRSTDEFENDERRMX (accessed July 13, 2018). [Google Scholar]
- NanoRam Handheld RAMAN System, B&W Tek. https://us.vwr.com/store/product/13244534/nanoram-handheld-raman-system-b-w-tek (accessed July 13, 2018). [Google Scholar]
- NanoRam® Handheld Raman Spectrometer. http://bwtek.com/products/nanoram/ (accessed July 13, 2018). [Google Scholar]
- Fu X.; Cheng Z.; Yu J.; Choo P.; Chen L.; Choo J. A SERS-Based Lateral Flow Assay Biosensor for Highly Sensitive Detection of HIV-1 DNA. Biosens. Bioelectron. 2016, 78, 530–537. 10.1016/j.bios.2015.11.099. [DOI] [PubMed] [Google Scholar]
- Hwang J.; Lee S.; Choo J. Application of a SERS-Based Lateral Flow Immunoassay Strip for the Rapid and Sensitive Detection of Staphylococcal Enterotoxin B. Nanoscale 2016, 8, 11418–11425. 10.1039/C5NR07243C. [DOI] [PubMed] [Google Scholar]
- Maneeprakorn W.; Bamrungsap S.; Apiwat C.; Wiriyachaiporn N. Surface-Enhanced Raman Scattering Based Lateral Flow Immunochromatographic Assay for Sensitive Influenza Detection. RSC Adv. 2016, 6, 112079–112085. 10.1039/C6RA24418A. [DOI] [Google Scholar]
- Wang X.; Choi N.; Cheng Z.; Ko J.; Chen L.; Choo J. Simultaneous Detection of Dual Nucleic Acids Using a SERS-Based Lateral Flow Assay Biosensor. Anal. Chem. 2017, 89, 1163–1169. 10.1021/acs.analchem.6b03536. [DOI] [PubMed] [Google Scholar]
- Blanco-Covián L.; Montes-García V.; Girard A.; Fernández-Abedul M. T.; Pérez-Juste J.; Pastoriza-Santos I.; Faulds K.; Graham D.; Blanco-López M. C. Au@Ag SERRS Tags Coupled to a Lateral Flow Immunoassay for the Sensitive Detection of Pneumolysin. Nanoscale 2017, 9, 2051–2058. 10.1039/C6NR08432J. [DOI] [PubMed] [Google Scholar]
- Sánchez-Purrà M.; Carré-Camps M.; de Puig H.; Bosch I.; Gehrke L.; Hamad-Schifferli K. Surface-Enhanced Raman Spectroscopy-Based Sandwich Immunoassays for Multiplexed Detection of Zika and Dengue Viral Biomarkers. ACS Infect. Dis. 2017, 3, 767–776. 10.1021/acsinfecdis.7b00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R.Multiplexed Detection with Magnetic Nanoparticles. In Magnetic Nanomaterials; Kumar C. S. S. R., Ed.; Nanomaterials for the Life Sciences; Wiley-VCH: Weinheim, 2009; Vol. 4, pp 55–76. [Google Scholar]
- Wood G.MagnaBioSciences; LLC Email Communication, 2018.
- MagnaBioSciences LLC. MICT® Technology . http://www.magnabiosciences.com/technology.html (accessed July 20, 2018).
- Graham D. L.; Ferreira H. A.; Freitas P. P. Magnetoresistive-Based Biosensors and Biochips. Trends Biotechnol. 2004, 22, 455–462. 10.1016/j.tibtech.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Lei H.; Wang K.; Ji X.; Cui D. Contactless Measurement of Magnetic Nanoparticles on Lateral Flow Strips Using Tunneling Magnetoresistance (TMR) Sensors in Differential Configuration. Sensors 2016, 16, 2130. 10.3390/s16122130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquina C.; de Teresa J. M.; Serrate D.; Marzo J.; Cardoso F. A.; Saurel D.; Cardoso S.; Freitas P. P.; Ibarra M. R. GMR Sensors and Magnetic Nanoparticles for Immuno-Chromatographic Assays. J. Magn. Magn. Mater. 2012, 324, 3495–3498. 10.1016/j.jmmm.2012.02.074. [DOI] [Google Scholar]
- Park J. A Giant Magnetoresistive Reader Platform for Quantitative Lateral Flow Immunoassays. Sens. Actuators, A 2016, 250, 55–59. 10.1016/j.sna.2016.09.016. [DOI] [Google Scholar]
- Ryu Y.; Jin Z.; Kang M. S.; Kim H.-S. Increase in the Detection Sensitivity of a Lateral Flow Assay for a Cardiac Marker by Oriented Immobilization of Antibody. BioChip J. 2011, 5, 193–198. 10.1007/s13206-011-5301-2. [DOI] [Google Scholar]
- Heller A.; Feldman B. Electrochemical Glucose Sensors and Their Applications in Diabetes Management. Chem. Rev. 2008, 108, 2482–2505. 10.1021/cr068069y. [DOI] [PubMed] [Google Scholar]
- Clarke S. F.; Foster J. R. A History of Blood Glucose Meters and Their Role in Self-Monitoring of Diabetes Mellitus. Br. J. Biomed. Sci. 2012, 69, 83–93. 10.1080/09674845.2012.12002443. [DOI] [PubMed] [Google Scholar]
- Xiang Y.; Lu Y. Using Personal Glucose Meters and Functional DNA Sensors to Quantify a Variety of Analytical Targets. Nat. Chem. 2011, 3, 697–703. 10.1038/nchem.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y.; Lu Y. Using Commercially Available Personal Glucose Meters for Portable Quantification of DNA. Anal. Chem. 2012, 84, 1975–1980. 10.1021/ac203014s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y.; Lu Y. Portable and Quantitative Detection of Protein Biomarkers and Small Molecular Toxins Using Antibodies and Ubiquitous Personal Glucose Meters. Anal. Chem. 2012, 84, 4174–4178. 10.1021/ac300517n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan T.; Xiang Y.; Lu Y.. Detection of Protein Biomarker Using a Blood Glucose Meter. In Mobile Health Technologies; Rasooly A., Herold K. E., Eds.; Methods in Molecular Biology; Springer New York: New York, NY, 2015; Vol. 1256, pp 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Shen Z.; Xiang Y.; Lu Y. Integration of Solution-Based Assays onto Lateral Flow Device for One-Step Quantitative Point-of-Care Diagnostics Using Personal Glucose Meter. ACS Sens 2016, 1, 1091–1096. 10.1021/acssensors.6b00270. [DOI] [Google Scholar]
- Dzenowagis J.; Kernen G.. Connecting for Health: Global Vision, Local Insight; World Health Organization: Geneva, 2005. [Google Scholar]
- Lillehoj P. B.; Huang M.-C.; Truong N.; Ho C.-M. Rapid Electrochemical Detection on a Mobile Phone. Lab Chip 2013, 13, 2950–2955. 10.1039/c3lc50306b. [DOI] [PubMed] [Google Scholar]
- Jia W.; Bandodkar A. J.; Valdés-Ramírez G.; Windmiller J. R.; Yang Z.; Ramírez J.; Chan G.; Wang J. Electrochemical Tattoo Biosensors for Real-Time Noninvasive Lactate Monitoring in Human Perspiration. Anal. Chem. 2013, 85, 6553–6560. 10.1021/ac401573r. [DOI] [PubMed] [Google Scholar]
- Bandodkar A. J.; Jia W.; Yardımcı C.; Wang X.; Ramirez J.; Wang J. Tattoo-Based Noninvasive Glucose Monitoring: A Proof-of-Concept Study. Anal. Chem. 2015, 87, 394–398. 10.1021/ac504300n. [DOI] [PubMed] [Google Scholar]
- Lee K. T.; Muller D. A.; Coffey J. W.; Robinson K. J.; McCarthy J. S.; Kendall M. A. F.; Corrie S. R. Capture of the Circulating Plasmodium Falciparum Biomarker HRP2 in a Multiplexed Format, via a Wearable Skin Patch. Anal. Chem. 2014, 86, 10474–10483. 10.1021/ac5031682. [DOI] [PubMed] [Google Scholar]
- Güder F.; Ainla A.; Redston J.; Mosadegh B.; Glavan A.; Martin T. J.; Whitesides G. M. Paper-Based Electrical Respiration Sensor. Angew. Chem., Int. Ed. 2016, 55, 5727–5732. 10.1002/anie.201511805. [DOI] [PubMed] [Google Scholar]
- Adewole O. O.; Erhabor G. E.; Adewole T. O.; Ojo A. O.; Oshokoya H.; Wolfe L. M.; Prenni J. E. Proteomic Profiling of Eccrine Sweat Reveals Its Potential as a Diagnostic Biofluid for Active Tuberculosis. Proteomics: Clin. Appl. 2016, 10, 547–553. 10.1002/prca.201500071. [DOI] [PubMed] [Google Scholar]
- Raphael A. P.; Prow T. W.; Crichton M. L.; Chen X.; Fernando G. J. P.; Kendall M. A. F. Targeted, Needle-Free Vaccinations in Skin Using Multilayered, Densely-Packed Dissolving Microprojection Arrays. Small 2010, 6, 1785–1793. 10.1002/smll.201000326. [DOI] [PubMed] [Google Scholar]
- Bhargav A.; Muller D. A.; Kendall M. A. F.; Corrie S. R. Surface Modifications of Microprojection Arrays for Improved Biomarker Capture in the Skin of Live Mice. ACS Appl. Mater. Interfaces 2012, 4, 2483–2489. 10.1021/am3001727. [DOI] [PubMed] [Google Scholar]
- Park K. Dissolving Microneedle Vaccine Delivery Systems. J. Controlled Release 2016, 225, 314. 10.1016/j.jconrel.2016.02.028. [DOI] [PubMed] [Google Scholar]
- Matsuo K.; Okamoto H.; Kawai Y.; Quan Y.-S.; Kamiyama F.; Hirobe S.; Okada N.; Nakagawa S. Vaccine Efficacy of Transcutaneous Immunization with Amyloid β Using a Dissolving Microneedle Array in a Mouse Model of Alzheimer’s Disease. J. Neuroimmunol. 2014, 266, 1–11. 10.1016/j.jneuroim.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Ng K. W.; Lau W. M.; Williams A. C. Towards Pain-Free Diagnosis of Skin Diseases through Multiplexed Microneedles: Biomarker Extraction and Detection Using a Highly Sensitive Blotting Method. Drug Delivery Transl. Res. 2015, 5, 387–396. 10.1007/s13346-015-0231-5. [DOI] [PubMed] [Google Scholar]
- Coffey J. W.; Meliga S. C.; Corrie S. R.; Kendall M. A. F. Dynamic Application of Microprojection Arrays to Skin Induces Circulating Protein Extravasation for Enhanced Biomarker Capture and Detection. Biomaterials 2016, 84, 130–143. 10.1016/j.biomaterials.2016.01.015. [DOI] [PubMed] [Google Scholar]
- Lee D.-S.; Li C. G.; Ihm C.; Jung H. A Three-Dimensional and Bevel-Angled Ultrahigh Aspect Ratio Microneedle for Minimally Invasive and Painless Blood Sampling. Sens. Actuators, B 2018, 255, 384–390. 10.1016/j.snb.2017.08.030. [DOI] [Google Scholar]
- Schnall R.; Pichon A.; Scherr T.. In-Depth Interviews to Understand the Feasibility of Using the MLab App for Promotion of HIV-Self Testing in Young Men; San Diego, CA, 2018. [Google Scholar]
- Qiagen . ESEQuant Lateral Flow System Technical Flyer 1070062; September 2011.
- Gardy J. L.; Loman N. J. Towards a Genomics-Informed, Real-Time, Global Pathogen Surveillance System. Nat. Rev. Genet. 2017, 19, 9–20. 10.1038/nrg.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.; Hu J. Single Quantum Dot-Based Nanosensor for Multiple DNA Detection. Anal. Chem. 2010, 82, 1921–1927. 10.1021/ac9026675. [DOI] [PubMed] [Google Scholar]
- Abrams W. R.; Barber C. A.; McCann K.; Tong G.; Chen Z.; Mauk M. G.; Wang J.; Volkov A.; Bourdelle P.; Corstjens P. L. a. M.; et al. Development of a Microfluidic Device for Detection of Pathogens in Oral Samples Using Upconverting Phosphor Technology (UPT). Ann. N. Y. Acad. Sci. 2007, 1098, 375–388. 10.1196/annals.1384.020. [DOI] [PubMed] [Google Scholar]
- Chen D.; Mauk M.; Qiu X.; Liu C.; Kim J.; Ramprasad S.; Ongagna S.; Abrams W. R.; Malamud D.; Corstjens P. L. A. M.; et al. An Integrated, Self-Contained Microfluidic Cassette for Isolation, Amplification, and Detection of Nucleic Acids. Biomed. Microdevices 2010, 12, 705–719. 10.1007/s10544-010-9423-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.; Abrams W. R.; Geva E.; de Dood C. J.; González J. M.; Tanke H. J.; Niedbala R. S.; Zhou P.; Malamud D.; Corstjens P. L. A. M.; et al. Development of a Generic Microfluidic Device for Simultaneous Detection of Antibodies and Nucleic Acids in Oral Fluids. BioMed Res. Int. 2013, 2013, 543294. 10.1155/2013/543294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corstjens P.; Zuiderwijk M.; Brink A.; Li S.; Feindt H.; Niedbala R. S.; Tanke H. Use of Up-Converting Phosphor Reporters in Lateral-Flow Assays to Detect Specific Nucleic Acid Sequences: A Rapid, Sensitive DNA Test to Identify Human Papillomavirus Type 16 Infection. Clin. Chem. 2001, 47, 1885–1893. [PubMed] [Google Scholar]
- Ongagna-Yhombi S. Y.; Corstjens P.; Geva E.; Abrams W. R.; Barber C. A.; Malamud D.; Mharakurwa S. Improved Assay to Detect Plasmodium Falciparum Using an Uninterrupted, Semi-Nested PCR and Quantitative Lateral Flow Analysis. Malar. J. 2013, 12, 74. 10.1186/1475-2875-12-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Chen Z.; Corstjens P. L. A. M.; Mauk M. G.; Bau H. H. A Disposable Microfluidic Cassette for DNA Amplification and Detection. Lab Chip 2006, 6, 46–53. 10.1039/B511494B. [DOI] [PubMed] [Google Scholar]
- Qiu X.; Liu C.; Mauk M. G.; Hart R. W.; Chen D.; Qiu J.; Kientz T.; Fiene J.; Bau H. H. A Portable Analyzer for Pouch-Actuated, Immunoassay Cassettes. Sens. Actuators, B 2011, 160, 1529–1535. 10.1016/j.snb.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanoli L. M.; Spoto G. Isothermal Amplification Methods for the Detection of Nucleic Acids in Microfluidic Devices. Biosensors 2013, 3, 18–43. 10.3390/bios3010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sema M.; Alemu A.; Bayih A. G.; Getie S.; Getnet G.; Guelig D.; Burton R.; LaBarre P.; Pillai D. R. Evaluation of Non-Instrumented Nucleic Acid Amplification by Loop-Mediated Isothermal Amplification (NINA-LAMP) for the Diagnosis of Malaria in Northwest Ethiopia. Malar. J. 2015, 14, 44. 10.1186/s12936-015-0559-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton S.; Cheng Q.; Sutherland C. J.; McCarthy J. S. A Simple, High-Throughput, Colourimetric, Field Applicable Loop-Mediated Isothermal Amplification (HtLAMP) Assay for Malaria Elimination. Malar. J. 2015, 14, 335. 10.1186/s12936-015-0848-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchi N. W.; Gaye M.; Diallo M. A.; Goldman I. F.; Ljolje D.; Deme A. B.; Badiane A.; Ndiaye Y. D.; Barnwell J. W.; Udhayakumar V.; et al. Evaluation of the Illumigene Malaria LAMP: A Robust Molecular Diagnostic Tool for Malaria Parasites. Sci. Rep. 2016, 6, 36808. 10.1038/srep36808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohon A. N.; Lee L. D.-Y.; Bayih A. G.; Folefoc A.; Guelig D.; Burton R. A.; LaBarre P.; Chan W.; Meatherall B.; Pillai D. R. NINA-LAMP Compared to Microscopy, RDT, and Nested PCR for the Detection of Imported Malaria. Diagn. Microbiol. Infect. Dis. 2016, 85, 149–153. 10.1016/j.diagmicrobio.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis K. A.; Rudolph D. L.; Morrison D.; Guelig D.; Diesburg S.; McAdams D.; Burton R. A.; LaBarre P.; Owen M. Single-Use, Electricity-Free Amplification Device for Detection of HIV-1. J. Virol. Methods 2016, 237, 132–137. 10.1016/j.jviromet.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez N. M.; Wong W. S.; Liu L.; Dewar R.; Klapperich C. M. A Fully Integrated Paperfluidic Molecular Diagnostic Chip for the Extraction, Amplification, and Detection of Nucleic Acids from Clinical Samples. Lab Chip 2016, 16, 753–763. 10.1039/C5LC01392E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnes J. C.; Rodriguez N. M.; Liu L.; Klapperich C. M. Polyethersulfone Improves Isothermal Nucleic Acid Amplification Compared to Current Paper-Based Diagnostics. Biomed. Microdevices 2016, 18, 30. 10.1007/s10544-016-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnes J. C.; Fan A.; Rodriguez N. M.; Lemieux B.; Kong H.; Klapperich C. M. Paper-Based Molecular Diagnostic for Chlamydia Trachomatis. RSC Adv. 2014, 4, 42245–42251. 10.1039/C4RA07911F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez N. M.; Linnes J. C.; Fan A.; Ellenson C. K.; Pollock N. R.; Klapperich C. M. Paper-Based RNA Extraction, in Situ Isothermal Amplification, and Lateral Flow Detection for Low-Cost, Rapid Diagnosis of Influenza A (H1N1) from Clinical Specimens. Anal. Chem. 2015, 87, 7872–7879. 10.1021/acs.analchem.5b01594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly J. T.; Rolland J. P.; Whitesides G. M. Paper Machine” for Molecular Diagnostics. Anal. Chem. 2015, 87, 7595–7601. 10.1021/acs.analchem.5b00411. [DOI] [PubMed] [Google Scholar]
- Poole C. B.; Li Z.; Alhassan A.; Guelig D.; Diesburg S.; Tanner N. A.; Zhang Y.; Evans T. C.; LaBarre P.; Wanji S.; et al. Colorimetric Tests for Diagnosis of Filarial Infection and Vector Surveillance Using Non-Instrumented Nucleic Acid Loop-Mediated Isothermal Amplification (NINA-LAMP). PLoS One 2017, 12, e0169011 10.1371/journal.pone.0169011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohon A. N.; Menard D.; Alam M. S.; Perera K.; Pillai D. R. A Novel SNP-LAMP Assay for Detection of Artemisinin-Resistant Plasmodium Falciparum Malaria. Open Forum Infect. Dis. 2018, 5, ofy011. 10.1093/ofid/ofy011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigl B. H.; Domingo G.; Gerlach J.; Tang D.; Harvey D.; Talwar N.; Fichtenholz A.; Lew B. van.; LaBarre P.. Noninstrumented Nucleic Acid Amplification Assay. In Microfluidics, BioMEMS, and Medical Microsystems VI; International Society for Optics and Photonics, 2008; Vol. 6886, p 688604. [Google Scholar]