Abstract
The expression of short hairpin RNAs (shRNAs) may result in unwanted activity from the co-processed passenger strand. Recent studies have shown that shortening the stem of conventional shRNAs abolishes passenger strand release. These Dicer-independent shRNAs, expressed from RNA polymerase III (Pol III) promoters, rely on Ago2 processing in resemblance to miR-451. Using strand-specific reporters, we tested two designs, and our results support the loss of passenger strand activity. We demonstrate that artificial primary microRNA (pri-miRNA) transcripts, expressed from Pol II promoters, can potently silence a gene of choice. Among six different scaffolds tested, miR-324 and miR-451 were readily re-targeted to direct efficient knockdown from either a CMV or a U1 snRNA promoter. Importantly, the miR-shRNAs have no passenger strand activity and remain active in Dicer-knockout cells. Our vectors are straightforward to design, as we replace the pre-miR-324 or -451 sequences with a Dicer-independent shRNA mimicking miR-451 with unpaired A-C nucleotides at the base. The use of Pol II promoters allows for controlled expression, while the inclusion of pri-miRNA sequences likely requires Drosha processing and, as such, mimics microRNA biogenesis. Since this improved and tunable system bypasses the requirement for Dicer activity and abolishes passenger strand activity completely, it will likely prove favorable in both research and therapeutic applications in terms of versatility and enhanced safety.
Keywords: RNAi, Dicer-independent shRNA, miR-451, miR-324, agoshRNA, agshRNA, passenger strand activity, Drosha, U1 promoter, Pol-II driven miRNA scaffold
Introduction
RNAi is a process in which small noncoding RNAs silence gene expression in a post-transcriptional manner.1, 2 MicroRNAs (miRNAs) are endogenous effector molecules in mammals, and they exert their repressing function in the RNA-induced silencing complex (RISC) by binding to target mRNA.3, 4 Conventional miRNA biogenesis involves consecutive cleavage of a longer folded primary microRNA (pri-miRNA) precursor transcript, by first Drosha and then Dicer, that results in the formation of a duplex composed of two individual ∼22-nt RNA strands with 3′ overhangs.5, 6, 7, 8 While both miRNA strands may direct silencing, only one strand is preferably loaded into RISC.9
The miRNA pathway can be exploited using several entry points for the targeted downregulation of gene expression, and RNAi remains an attractive method for research or therapeutic purposes.10 One popular entry point is the expression of artificial short hairpin RNAs (shRNAs) that mimic the hairpin structure of the pre-miRNAs produced in the nucleus by the Microprocessor complex Drosha/DGCR8.11, 12, 13 In the cytoplasm, Dicer cleaves of the loop and terminal duplex stability favors preferential loading of the guide strand, which is designed to be fully complementary to the target mRNA.9, 14 However, RISC loading of the passenger strand can occur, and this contributes to the cytotoxicity associated with RNAi, as off-target genes can be repressed by partial complementarity and binding within the seed region.15, 16, 17, 18
Recent discoveries have shown that shortening the stem of expressed shRNAs below 20 bp results in alternative processing that bypasses Dicer.19, 20 These Dicer-independent shRNAs rely on slicing of the 3′ arm by Argonaute-2 (Ago2) between nucleotides 10 and 11 from the 5′ end, which yields a single RNAi-active strand and eliminates unwanted activity of the 3′ arm.19, 20 Further trimming by the poly(A)-specific ribonuclease (PARN) results in shorter RNA species that effectively guide target knockdown.21 Dicer-independent shRNAs mimic the recently described non-canonical biogenesis of miR-451, which also depends on Ago2 and trimming-tailing processes rather than Dicer.22, 23, 24 The Dicer-independent shRNA expression systems reported so far have been limited to RNA polymerase III (Pol III) promoters,25 and tunable systems based on RNA polymerase II (Pol II) promoters are lacking.
In this study, we investigated if Dicer-independent shRNA structures embedded within pri-miRNA transcripts could elicit target knockdown when expressed from Pol II promoters. We demonstrate that both U1 and cytomegalovirus (CMV) promoter-derived miR-324 and miR-451 can be equipped with hairpins that potently target a gene of choice, without any passenger strand activity.
Results
Identification of the Potent Dicer-Independent shRNA agsh12.3
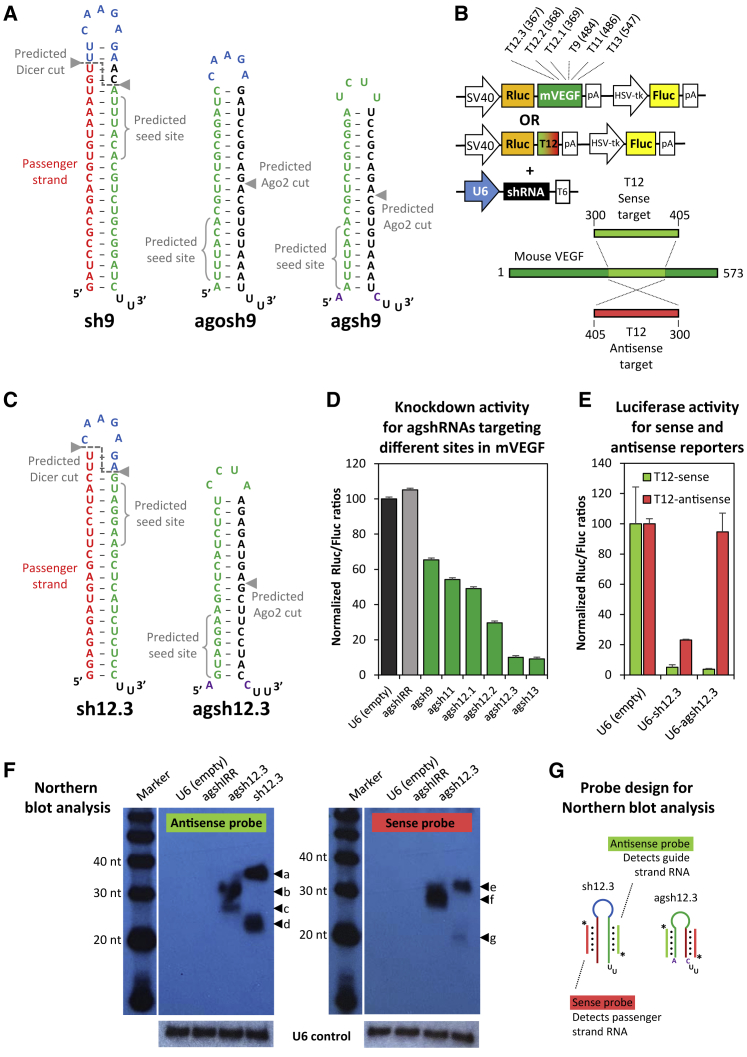
To optimize existing shRNA constructs,26 we set out by re-configuring a potent shRNA from the T.J.C. lab for Dicer-independent processing. This shRNA, named sh9, is targeted toward site 9 in human (h) and murine (m) vascular endothelial growth factor (VEGF), and, expressed from the H1 promoter, it suppresses a transiently expressed reporter by ∼90%.26 As illustrated in Figure 1A, the sh9 design is conventional, with a sense-loop-antisense structure, a 21-bp stem, and a frequently used UUCAAGAGA loop,11 which likely extends the stem further.
Figure 1.
Comparison of Dicer-Independent shRNA Designs and Identification of Potent agshRNAs Targeting VEGF
(A) Predicted secondary structure of a previously reported shRNA targeting site 9 in the VEGF gene,26 based on a conventional sense-loop-antisense design (sh9, left side); Ago2-dependent designs, based on the 19/5 agoshRNA structure, as reported by the B. Berkhout lab20 (agosh9, middle part); or the agshRNA design, as described by the Yen group27 (agsh9, right side). Arrows indicate predicted Dicer and Ago2 cleavage sites, and position 2-7 of the guide RNA (seed site) is shown with braces. The guide and passenger strands are depicted in green and red, respectively. The frequently used Brummelkamp loop11 is shown in blue. The invariant unpaired A-C fork, as also present in miR-451, is highlighted in purple. (B) Schematic diagram of the psiCHECK2-based reporter encoding a Renilla luciferase-mVEGF cDNA fusion transcript (used for screening purposes) and the dedicated T12-sense or T12-antisense reporters (used for testing guide and passenger strand activity, respectively), which are used in combination with the various U6-driven shRNA expression plasmids. Name (T9, target site 9) and location (484, 3′ end of target site in the cDNA) of six chosen targets are indicated at the top. The illustration at the bottom shows the 105-nt-long cDNA region of mVEGF (dubbed T12) encompassing site 12, which was cloned into the psiCHECK2-based reporter shown above in sense or antisense orientation (numbers indicate cDNA position). (C) Predicted secondary structure of the conventional sense-loop-antisense sh12.3 (left side) and the Ago2-dependent agsh12.3 (right side) targeting site 12.3 in VEGF. Arrows and colors as indicated in (A). (D) Knockdown activity of agshRNAs designed to target the mVEGF sites, estimated by co-transfection with the mVEGF-fused reporter, as illustrated in the top part of (B). Renilla luciferase (Rluc) and Firefly luciferase (Fluc) activities were measured in relative units of light (RLU). The Rluc:Fluc ratio was normalized to the empty control (black bar) and plotted as the mean of three replicates plus SDs. An irrelevant agshIRR was included as a non-targeting control (gray bar). (E) Guide and passenger strand activity for agsh12.3 and a conventionally designed shRNA predicted to target site 12.3 using the dedicated T12-sense and T12-antisense reporters, respectively (see B). The Rluc:Fluc ratio was normalized to the empty control and plotted as the mean of three replicates plus SDs. (F) Northern blot analysis of small RNA from transfected HEK293 cells using probes detecting guide strand RNA (antisense probe, autoradiogram on the left side) or passenger strand RNA (sense probe, autoradiogram on the right side). Size of 20-, 30-, and 40-nt bands of the RNA decade marker is indicated, and agsh12.3- and sh12.3-specific bands are marked by arrows and labeled a–g. A cropped image with detection of the native U6 snRNA band (loading control) is shown below. (G) Diagram indicating probe specificity. Antisense probe, light green; sense probe, light red; Fluc, Firefly luciferase; HSV-tk, Herpes simplex virus thymidine kinase promoter; mVEGF, murine vascular endothelial growth factor; pA, polyadenylation signal; Rluc, Renilla luciferase; shRNA, short hairpin RNA; SV40, simian virus 40 promoter; T6, T-rich Pol III termination signal; U6, human U6 snRNA promoter.
Inspired by the work from others,20, 27 we designed two different Dicer-independent shRNAs, entitled agosh9 and agsh9 (see Figure 1A), based on the guide RNA strand from sh9 as predicted by the 5′ counting rule for Dicer processing.28 While agosh9 utilizes the 19/5 design as proposed by Liu and colleagues,20 agsh9 mimics pre-miR-451 with an unpaired basal A-C fork27 (Figure 1A). Dedicated luciferase reporters, sensitive to either guide or passenger strand release (T9-sense and T9-antisense, respectively; Figure S1A), demonstrated high activity of both sh9 strands when expressed from a modified U6 promoter29 (see Figure S1B). As expected, both Dicer-independent shRNA designs eliminated passenger strand activity; however, in the context of VEGF target 9, neither of them retained effective target silencing (Figure S1B). We also tested the activity of agosh9 and agosh9/3, a 19/3 variant with reduced loop size as described by Liu et al.,20 when expressed from the H1 promoter, and we compared it directly to the published pSUPERretro-based sh9 construct (see Figure S1C). While passenger strand activity was abolished from agosh9 and agosh9/3 (data not shown), we again observed only limited activity of the guide strand in our luciferase reporter assay (Figure S1D).
We decided to proceed only with the agsh design,27 due to the simplicity of design and the invariant 5′ A nucleotide, which is ideal in terms of precision and efficacy of transcription when using the Pol III promoter U6.30 Using a Renilla luciferase (Rluc) reporter fused to full-length cDNA sequence of mVEGF for screening knockdown efficiency, we cloned five additional VEGF targeting hairpins based on the agsh design in our pFRT-U6 vector29 (Figure 1B). Two of these agshRNA, entitled agsh12.3 (illustrated in Figure 1C) and agsh13, suppressed the Rluc reporter by approximately 90% (Figure 1D). Given the potent knockdown efficacy, we next verified lack of passenger strand activity of the agsh12.3 construct by using luciferase reporters encompassing ∼100 nt of mVEGF sequence around the 12.3 target site in either sense or antisense orientation (T12-sense and T12-antisense reporter, respectively, as illustrated in Figure 1B). For comparison, we engineered a conventionally designed sh12.3 targeting the same VEGF sequence (illustrated in Figure 1C). As shown in Figure 1E, both designs effectively suppressed the T12-sense luciferase reporter, while the T12-antisense reporter, as expected, demonstrated an absence of passenger strand activity from agsh12.3. Northern blot analysis supported the absence of RNA in the size range of mature miRNA or small interfering RNA (siRNA) for the agsh12.3 construct (Figure 1F).
Using probes complementary to the guide (antisense probe) or the passenger strand (sense probe), as illustrated in Figure 1G, we detected a prominent ∼30-nt band for agsh12.3, which is consistent with Ago2 processing (Figure 1F, bands b and f). The antisense probe also detected a slightly shorter band (labeled c), which is consistent with 3′ end tailing and PARN-directed trimming.21, 25, 31 We note that our 17-nt locked nucleic acid (LNA)-doped sense probe, complementary to the entire agshRNA 3′ stem region, may likely detect the 30-nt Ago2 cleavage product, which encompasses part of the 3′ stem. Shortening the sense probe 4 nt in the 3′ end reduced detection of the ∼30-nt band, although not fully (data not shown). The conventional sh12.3 gave rise to a precursor band of ∼35 nt (Figure 1F, bands a and e), and it showed release of both guide and passenger strand species in the size range consistent with Dicer processing (Figure 1F, bands d and g). Surprisingly, we could not detect the predicted full-length sh12.3 precursor transcript (∼49 nt), and we speculate that this may reflect aberrant processing due to overexpression.
Efficient Target Knockdown by agsh12.3 Embedded in Long Pol II-Expressed pri-miRNA Transcripts
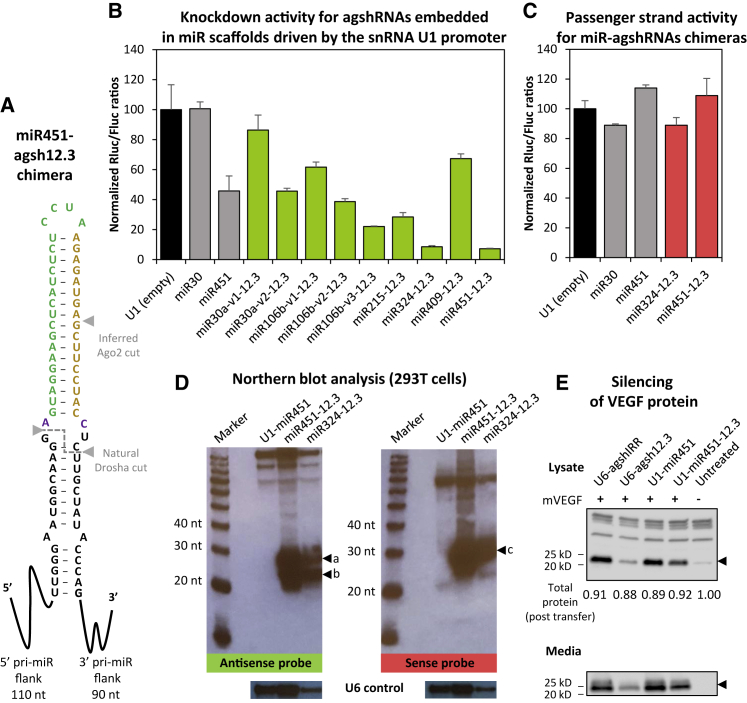
Using the potent agsh12.3-mediated silencing of VEGF as a model for shRNA-encoded Dicer-independent RNAi, we next wanted to explore whether we could transfer the benefits of passenger strand-free knockdown from small Pol III transcripts to larger Pol II-expressed systems. Since the agsh design mimics miR-451 in terms of the stem length and the basal A-C fork, exploiting the flanking sequences of pri-miR-451 was an obvious choice (see Figure 2A). We decided to include five additional miRNAs as potential scaffolds for heterologous expression of agshRNAs (see Figure S2). All stem loop structures and prominent Drosha/Dicer cleavage sites shown in Figure S2 are based on data from miRBase32 and the RNA secondary structure prediction algorithms MFOLD.33 Human miR-215, miR-324, and miR-409 were chosen as they are predicted to have a bulge structure at the native Drosha cleavage site, which makes it convenient to design the pri-miRNA or agshRNA break point. Human miR-30a has often been used to derive potent shRNA-miRNA mimics,34, 35 and we have previously utilized the miR-106b cluster for the expression of shRNAs.36 However, due to the sequence composition of miR-30a, we embedded the agsh12.3 part either to maintain the predicted secondary structure of miR-30a (miR30a-v1) or to mimic the bulge present in miR-451, which allows keeping the unpaired basal A-C fork (miR30a-v2). Similarly, we made a structural miR-106b mimic after using the natural Drosha 5′ cut site as the break point (miR106b-v1) or keeping the A-C fork from agsh12.3 or miR-451 in an asymmetric or symmetric bulge (miR106b-v2 and -v3, respectively). We created two Pol II expression vectors based on the pcDNA3.1 with an identical multiple cloning site (MCS) flanked by either the human U1 small nuclear RNA (snRNA) promoter and a U1 termination signal or the CMV promoter and the bovine growth hormone polyadenylation signal. Synthetic DNA fragments encoding the pri-miRNA-agshRNA chimeras, including ∼90- to 130-nt flanking sequences, were cloned into the U1 or CMV expression vector.
Figure 2.
Potent Knockdown of VEGF Using Dicer-Independent shRNAs Expressed as Pol II-Driven pri-miR-324 or -451 Transcripts
(A) Diagram of the miR451-agsh12.3 chimera showing the sequence and predicted stem loop structure around the Drosha cleavage site. (B) Knockdown activity of U1-expressed agshRNA embedded in various miRNA scaffolds, estimated by co-transfection with psiCHECK-mVEGF-T12-sense. The Rluc:Fluc ratio was normalized to the empty control (black bar) and plotted as the mean of three replicates plus SDs. Endogenous miR-30a and miR-451 were included as non-targeting controls (gray bars). Exact compositions of the miRNA-agshRNA chimeras at the central stem loop structure are shown in Figure S2. (C) Passenger strand activity as measured using the psiCHECK-mVEGF-T12-antisense reporter and plotted as described in (B). (D) Northern blot analysis of small RNA from transfected 293T cells using probes detecting guide strand RNA (antisense probe, autoradiogram on the left side) or passenger strand RNA (sense probe, autoradiogram on the right side). Size of 20-, 30-, and 40-nt bands of the RNA decade marker is indicated, and agsh12.3-specific bands are marked by arrows and labeled a–c. A cropped image with detection of the native U6 snRNA band (loading control) is shown below. (E) Knockdown of intracellular (lysate) and secreted (media) mVEGF by U1-expressed miR451-agsh12.3, estimated by co-transfection with an mVEGF-expressing plasmid and western blot analysis. Total protein levels in each lane after membrane transfer (relative values) are shown below, and lines and arrows indicate protein size markers and the position of the mVEGF band, respectively. Endogenous miR-451 and U6-driven agsh12.3 were included as a non-targeting and a positive control, respectively.
Using the target 12.3 sensitive reporter T12-sense (Figure 1B) in a co-transfection assay, we found potent knockdown from the U1-driven miR324-12.3 and miR451-12.3 vectors and intermediate activity from miR215-12.3 (Figure 2B). Of note, overexpression of the wild-type miRNA transcript in some cases affected the activity of the internal Firefly luciferase (Fluc) control, as, for instance, reflected in the low Rluc:Fluc ratio for miR-451 in Figure 2B. Due to unspecific (and reporter-dependent) effects of miRNA overexpression in our dual luciferase reporter assay, we in all cases in this study estimated and report knockdown efficacy relative to the empty control. Embedding the agsh12.3 hairpin crude (v1) in pri-miR-30a, -106b, or -409 showed limited knockdown of the reporter, although mimicking the symmetric loop at the Drosha cleavage site improved silencing substantially (mir30a-v2 and miR106b-v3). Screening of our panel of 12.3 targeting miRNA-agshRNA chimeras expressed from the CMV promoter showed the same tendency and rank as the U1-driven transcripts (Figure S3A; data not shown).
Based on these results we decided to proceed with the miR-324 and miR-451 scaffolds only. First, we investigated if the miRNA-agshRNA vectors retained a lack of passenger strand activity, as demonstrated for the U6-expressed version. Using the T12-antisense reporter (Figure 1B), both the U1- and CMV-expressed vectors showed no sign of active passenger strand release (Figure 2C; Figure S3B). Northern blot analysis using the sense probe (as used to detect passenger strand release for the U6-driven hairpins) detected a prominent ∼30-nt band for both miRNA scaffolds (Figure 2D, band c). This is consistent with Drosha processing of the primary miRNA transcript and cleavage of an agshRNA-like molecule by Ago2, which in theory would leave eight bases in the 3′ end for (partial) hybridization with our sense probe. Importantly, no miRNA or siRNA-sized bands were detectable with the sense probe. The antisense probe (Figure 2D, left side) detected two specific bands of processed (guide) RNAs, which we estimated to be approximately 29–30 nt and ∼25 nt long (bands a and b, respectively). This is consistent with detection of the Ago2-cleavage product (band a), the 3′-trimming-tailing processes,21, 25, 31 and the generation of shorter RNA species (band b) not detected by the sense probe. However, the resolution does not allow unequivocal distinction between the sizes of band a and band c, and we cannot rule out that these bands have undergone partial 3′ trimming or tailing. Finally, we verified the targeting ability of the U1-miR451-12.3 construct in an independent assay using western blot analysis of cellular and secreted murine VEGF in co-transfected HEK293 cells, which resulted in substantial knockdown (Figure 2E).
Functional knockdown and detection of siRNA-sized species strongly suggested that the miRNA-agshRNA transcripts mature in the nucleus by the Drosha/DGCR8 complex. If the native Drosha cleavage site is faithfully used, a short pre-miR-451- or U6-agshRNA-like hairpin-structured intermediate is released, which may subsequently be exported and processed further in the cytoplasm in accordance with the non-canonical Ago2-dependent pathway (as illustrated in Figure 2A). Hence, we would predict functionality of the miRNA-agshRNA system to be completely independent of Dicer.
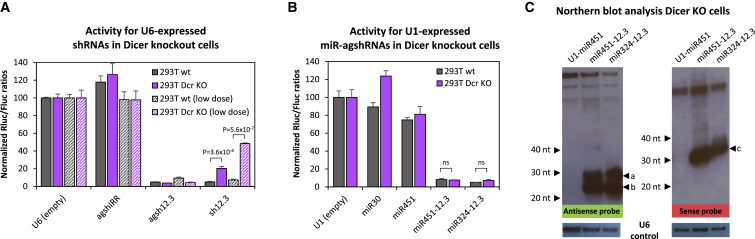
To test this, we took advantage of 293T-derived Dicer-knockout cells37 (NoDice 2-20 and 4-25) for our co-transfection assays. As expected, the U6-expressed agsh12.3 construct silenced the T12-sense reporter just as efficiently in Dicer-deficient cells as the parental 293T cells (Figure 3A; data not shown for NoDice 4-25 cells). In contrast, the conventional sh12.3 lost activity in Dicer-deficient cells, albeit only partially using our standard experimental conditions. The ability of sh12.3 to silence the reporter target in the absence of Dicer could be a result of miR-451-like Ago2-dependent processing, or it may also be a result of sheer overloading and aberrant shRNA processing into antisense molecules acting in a RISC-independent manner. Lowering the shRNA plasmid DNA dose from 34 to 0.5 ng resulted in a more prominent loss of knockdown activity in Dicer-deficient cells (Figure 3A, hatched bars), which tentatively argues that aberrant processing may be in play. As predicted, the Pol II-driven miRNA-agshRNA vectors were equally potent in the absence of cellular Dicer activity (Figure 3B; Figure S3C). Consistent with Dicer-independent processing, we obtained a similar pattern of bands in northern blots of RNA from transfected Dicer-knockout cells as for parental 293T cells (compare Figure 2D with Figure 3C; see Figure S4 for short time exposures).
Figure 3.
Knockdown Activity of miRNA-agshRNAs Is Dicer Independent
(A) Knockdown levels of U6-driven constructs in Dicer-knockout cells37 (NoDice 2-20, purple bars) and the parental 293T control cells (gray bars), as based on psiCHECK-mVEGF-T12-sense reporter activity. The Rluc:Fluc ratio was normalized to the empty control and plotted as the mean of three replicates plus SDs. An irrelevant agshIRR was included as a non-targeting control. For the hatched bars, the agshRNA or shRNA dose was lowered from 34 to 0.5 ng. p values for selected statistical tests are indicated. (B) Knockdown activity of U1-driven miR324- and miR451-agsh12.3 in Dicer-knockout cells (purple bars) and the parental 293T control cells (gray bars) measured and plotted as described in (A). Endogenous miR-30a and miR-451 were included as negative controls. (C) Northern blot analysis of small RNA from transfected DcrKO 293T cells (NoDice 2-20) using probes detecting guide strand RNA (antisense probe, autoradiogram on the left side) or passenger strand RNA (sense probe, autoradiogram on the right side). Arrows indicate RNA size markers for the 20-, 30-, and 40-nt bands, and agsh12.3-specific bands are marked by arrows and labeled a–c. A cropped image with detection of the native U6 snRNA band (loading control) is shown below.
miR324- and miR451-agshRNAs Are Readily Re-targeted to a Gene of Choice
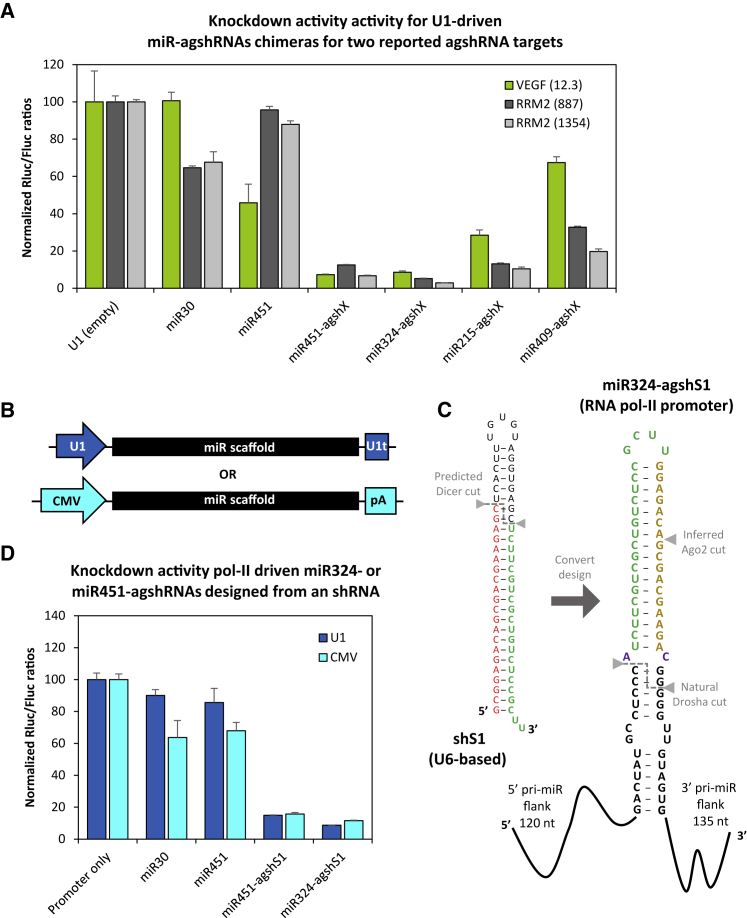
Based on the potent silencing achieved with our VEGF-targeting miRNA-agshRNA vectors, we next wanted to examine whether the miRNA-based platform could be generalized and used as scaffolds for insertion of heterologous (Dicer-independent) shRNAs designed to target other genes. We took advantage of two published and potent hairpins based on the agshRNA design scheme (agsh887 and agsh1354), both targeting the Ribonucleotide reductase regulatory subunit M2 (RRM2) gene.27 As for our VEGF-targeting agsh12.3 hairpin, we simply replaced the endogenous pre-miRNA sequence of miR-324 or miR-451 with the agsh887 and agsh1354 structure, modifying only the 3′ UU tail to either GG or UG, respectively, to maintain base pairing of the distal part of the natural stem. We also included the miR-215 and miR-409 scaffolds, which in the context of VEGF silencing showed moderate or no activity, respectively.
To assess the potency of these constructs, we designed luciferase reporters encompassing a minimal-sized target region for RRM2 target 887 and 1354, and we co-transfected these with the corresponding U1-miRNA-agshRNA constructs. Indeed, miR-324 and miR-451 again showed efficient target silencing. In the context of the RRM2 targets, miR-215 and miR-409 showed improved knockdown performance as compared to silencing of VEGF (Figure 4A). Of note, the RRM2 target 1354, is highly compatible with our Pol II expression system, and it displays a hyper-potent knockdown reminiscent of Pol III-based expression.
Figure 4.
Easy Design and Efficient Targeting of a Gene of Choice
(A) Knockdown activity of U1-driven agshRNA27 targeted against the human RRM2 gene (site 887, dark gray; site 1354, light gray;) when embedded in miR-215, -324, -409, or -451 scaffolds. For comparison, data from Figure 2B have been replicated (VEGF targeting shown in green). The Rluc:Fluc ratio, as measured by dedicated psiCHECK-based reporters, was normalized to the empty control and plotted as the mean of three replicates plus SDs. Endogenous miR-30a and miR-451 were included as negative controls. (B) Schematic diagram of the CMV and U1 Pol II expression cassettes used to generate miRNA-agshRNA transcripts. CMV transcripts terminate using the bovine growth hormone polyadenylation signal (denoted pA), while the U1 snRNA transcripts stop using an U1 terminator box (denoted U1t).66 (C) Diagram depicting the simplicity of reconfiguring an existing conventional shRNA (here illustrated for the HIV-1 site 1-targeting shRNA denoted shS1) into a miR324-agshRNA scaffold for Pol II expression. The 21-mer guide strand for a given target forms the 5′ side and the loop (highlighted in green), while complementary sequences constitute the 3′ side (shown in yellow). The invariant unpaired A-C fork (highlighted in purple), present in the agshRNA design shown in Figure 1A, is simply kept and placed at the natural miR-324 Drosha cleavage site. (D) Knockdown activity of U1- (dark blue) or CMV- (light blue) expressed agshRNA targeted against the HIV-1 site 136, 67 from miR-324 or -451 scaffolds, measured and plotted as described in (A).
To further study the targeting flexibility of the Pol II-expressed miRNA-agshRNA system, we decided to test if a conventionally designed (and Pol III-expressed) shRNA could be directly converted for our U1- or CMV-driven miRNA scaffolds (Figure 4B). We took advantage of a previously published shRNA construct (shS1) that potently targets the HIV-1 tat-rev transcript (site 1).36 As illustrated in Figure 4C, the shS1 has a conventional sense-loop-antisense design and an elongated stem of 27 bp. Based on the predicted guide strand of shS1, we designed miR324- and miR451-agshS1 vectors (Figure 4C). Both miRNA-agshS1 vectors potently suppressed the HIV-1 site 1-fused luciferase reporter, and they showed comparable efficacy when using either the U1 or the CMV promoter (Figure 4D). A knockdown efficacy of approximately 90% is comparable to the potency of the published shRNA construct (shS1) using our experimental conditions (Figure S5).
Discussion
Vector-encoded shRNAs are often used for gene therapy or long-term gene suppression directed by the encoded guide strand. However, unwanted silencing of off-targets remains an important side effect that needs careful attention for any RNAi application. Conventional shRNA design inherently co-delivers a passenger strand with potential off-target-silencing capacity,38 and several strategies to limit this unwanted effect have been reported. These include the following: (1) co-expressing tough decoys to sequester or sponge for the passenger strand,39 (2) adapting structural features such as bulges to favor accurate Dicer processing and asymmetric loading,17, 40 and (3) utilizing Dicer-independent shRNAs.20, 27, 41
In accordance with previous studies,20, 27 we have designed and tested two designs for Pol III-based expression of Dicer-independent shRNAs. Using strand-specific reporters and northern blot analysis, we confirmed the lack of passenger strand activity. In terms of optimal target specificity, it is preferable to have exact processing and, if possible, release of a single guide strand species with high silencing potency. For Ago2-matured shRNA vectors, the 5′ end of the guide strand, which dictates target recognition and seed site match, is dependent on precise transcriptional initiation. The U6 promoter initiates on A or G as compared to the more sloppy H1 promoter.30 Conventional shRNAs, encoding the guide strand on the 3′ arm, in contrast, rely on Dicer cleavage for 5′ end determination, a process that may be imprecise.40, 42 We favor the agshRNA design27 in combination with the U6 promoter, since the invariant 5′ A nucleotide (in the A-C fork) (1) likely favors accurate transcriptional initiation,30, 43 (2) reflects the abundant presence of 5′ As in mature miRNAs44 and may enhance Ago2 binding,45, 46 and (3) imitates the pre-miR-451 structure.
Elimination of the passenger strand may not only alleviate off-target activity but also potentially improve safety by limiting overloading of RISC-Argonaute proteins and Dicer, thereby reducing cell toxicity and competition with endogenous miRNAs.47, 48, 49, 50 Hence, it is crucial to identify hyper-potent target sites and not least control the shRNA dose. Balanced expression using Pol II promoters, combined with tight processing and nuclear export, has been suggested to improve safety as compared to strong Pol III-based systems.51 By incorporating flanking pri-miRNA sequences, we are the first to demonstrate that Dicer-independent shRNAs can be adapted for expression using Pol II promoters. Among six different miRNA scaffolds tested, we demonstrated that miR-324 and -451 can readily be equipped with shRNAs based on the agshRNA design27 and direct potent target suppression. Notably, the miRNA-agshRNA system showed no passenger strand activity, as suggested by northern blot analysis and strand-specific reporters.
The Pol II miRNA-agshRNA transcripts are presumably cleaved by Drosha and Ago2 and trimmed by PARN, in accordance with miR-451 biogenesis and Dicer-independent shRNAs.21, 25, 31 In support of this notion, we found that the miRNA-agshRNA-mediated knockdown and the pattern of RNA processing were unaffected in 293T-knockout cells lacking Dicer. Imprecise Drosha cleavage could potentially result in the release of hairpin structures with a stem length ≤20 bp and, hence, canonical shRNA properties.52, 53 The efficient on-target knockdown, the complete lack of passenger strand activity in reporter assays, and the absence of siRNA-sized species on northern blots when probing for the passenger strand collectively suggest that the Drosha/DGCR8 complex releases an agshRNA-like structure from both the miR-324 and the miR-451 scaffolds. Exact mapping of Drosha cleavage sites is currently ongoing and requires experimental validation, such as deep sequencing. Northern blot analysis using a guide strand-specific probe indicates that both the miR-324- and miR-451-derived agshRNA transcripts give rise to two abundant RNA classes with the approximate sizes of 25 nt and 29–30 nt. This is consistent with the trimming-tailing processes of miR-451 and supported by processing studies of artificial substrates, which led to mainly 23- to 26-mer guide RNA species.21, 27, 31, 54, 55 While structural features and sequence composition in the pre-miRNA region may influence precision and efficiency of Drosha,56, 57, 58, 59 we believe that the miR-324 and -451-based vectors are robust for pre-miRNA replacement, as all the artificial agshRNA structures we tested resulted in efficient target suppression.
We have used the viral CMV promoter and the U1 snRNA promoter to drive miRNA-agshRNA expression. Both promoters display a comparable efficacy in terms of target knockdown. The ability to control Dicer-independent shRNA expression by Pol II promoters evidently allows researchers to use a wide range of application-specific promoters. We have achieved doxycycline-inducible miRNA-agshRNA expression using the CMV-TetO promoter in T-REx-293 cells, and we restricted miRNA-agshRNA-mediated knockdown to retinal pigment epithelium (RPE) cells by utilizing the VMD2 promoter (L.A. and T.J.C., unpublished data).
We believe that our novel Pol II-based miRNA-agshRNA vectors are of general use, as we successfully silenced four independent targets, and may support efficient knockdown of previously identified shRNA targets by simply converting the design. Parameters underlying efficient target suppression by Dicer-independent shRNA are not fully understood, and identifying potent targets sites may be a trial-and-error process. As also noted in the recent review by Herrera-Carrillo and Berkhout,25 current algorithms for predicting efficient siRNAs are not suited for the design of Dicer-independent shRNAs. A very recent paper suggests that synthetic sli-siRNAs (dependent on Ago2 slicing) are more potent than conventional siRNAs but may also be more mismatch tolerant and, hence, display increased off-targeting.54 It remains to be established if this is also the case for vector-encoded Dicer-independent shRNAs and for our Pol II-based miRNA-agshRNA vectors in particular. We speculate that our miRNA-agshRNA vectors may favor RISC incorporation compared to previous Dicer-independent shRNAs, as Drosha cleavage results in a pre-miR-451-like agshRNA structure with a 5′ phosphate group in contrast to the triphosphate group present on Pol III-expressed transcripts.60, 61, 62, 63 Moreover, if Drosha processes our miRNA-agshRNA accurately, well-defined 3′ overhangs may also facilitate handoff to Ago2, as compared to the 3′ end of Pol III transcripts with varying numbers of uracils,61, 64 although the details of RISC loading for this non-canonical pathway remain to be clarified.
In summary, we demonstrate that U1- and CMV-driven miR-324 and miR-451 can be equipped with Dicer-independent shRNAs and elicit efficient target knockdown, without concomitant passenger strand activity. Combining our miRNA-agshRNA system with the plethora of alternative Pol II promoters would facilitate vectorization and allow restricted or controlled expression, which may advance the safe and efficient use of Dicer-independent shRNAs for fundamental research, biotechnology applications, or therapeutic purposes.
Materials and Methods
Plasmid Construction
RNAi targets were cloned into the psiCHECK2.2 vector (Promega, Nacka, Sweden) using the multiple cloning site in the 3′ UTR of the SV40-RLuc gene cassette, using standard cloning procedures. Two ∼100-nt regions of the mouse VEGF gene (cDNA positions 300–405 and 463–563) were PCR amplified using NotI-tagged primers and psiCHECK-mVEGF as template,26 and they were ligated into NotI-linearized psiCHECK.2 plasmid in both orientations to generate sense and antisense reporters with target sites surrounding VEGF site 9 and site 12. The resulting reporter plasmids are abbreviated psiCHECK-mVEGF-T9-sense and -antisense and psiCHECK-mVEGF-T12-sense and antisense, respectively (see Figures 1B and S1B). Reporters for the RRM2 gene were constructed on the basis of the reported sequence of agsh887 and agsh135427 and cloning complementary target sequences into XhoI/NotI-linearized psiCHECK2.2 plasmid. Oligonucleotide cassettes bearing XhoI- and NotI-compatible 4-nt overhangs at each end were generated by annealing two in vitro-phosphorylated partially overlapping oligonucleotides, as described previously.65 psiCHECK.2-HIV-S1 and psiCHECK-mVEGF have been described previously.26, 36
The shRNA expression vector pFRT-U6 harbors the human RNA Pol III promoter U6 modified at positions –2 and –4 to create a BglII site that allows direct oligonucleotide-mediated cloning of shRNAs.29 The shRNA constructs were cloned directionally into BglII/XhoI sites of pFRT-U6 or pSUPER.retro.puro26 as double-stranded oligonucleotide cassettes with 4-nt compatible overhangs, as described previously.65 The pri-miRNA expression vectors pcDNA3-CMV-MCS and pcDNA3-U1-MCS harbor the viral Pol II CMV promoter and the human snRNA promoter U1, respectively. pcDNA3-CMV-MCS was constructed from pcDNA3.1+1 (Invitrogen, Life Technologies, Carlsbad, CA) by cloning an oligonucleotide cassette into the NheI/ApaI sites, creating a new MCS with the following unique restriction sites (5′-3′ order): NheI, XhoI, BamHI, EcoRI, BspEI, AgeI, and ApaI. pcDNA3-U1-MCS was made from pcDNA3.1+1 by substituting the MluI/ApaI fragment holding the CMV promoter with a PCR amplicon harboring the U1 promoter, an MCS, and the U1 terminator, prepared using the pSK-U1-MCS-U1stop plasmid as template (generous gift from the J.J. Rossi lab, City of Hope).
For expression of the endogenous human pri-miR-30a, pri-miR-106b, and pri-miR-451 genes, we amplified and cloned 255-, 281-, and 243-nt regions, respectively, from human genomic DNA using XhoI-tagged forward primer and AgeI-tagged reverse primer for directional cloning into the MCS of pcDNA3-CMV-MCS and pcDNA3-U1-MCS. Chimeric miRNA-agshRNA sequences were purchased as synthetic DNA (GenScript, Piscataway, NJ) in the form of XhoI- and AgeI-flanked inserts in the pUC57 plasmid, and they were subcloned into the MCS of pcDNA3-CMV-MCS and pcDNA3-U1-MCS. We included 80- 120-nt 5′ and 3′ pri-miRNA-flanking sequences similar to the endogenous control genes. All primers were purchased from MWG/Eurofins (Ebersberg, Germany) or TAG Copenhagen (Frederiksberg, Denmark), and oligonucleotide and synthetic DNA sequences are available in Table S1. All vector plasmids have been verified by sequencing of inserts and restriction enzyme digest.
Cell Culturing
HEK293, 293T, or 293T-derived Dcr knockout (KO)37 (NoDice 2-20 or 4-25) cells were cultured at 37°C in 5% (v/v) CO2 and maintained in DMEM supplemented with 10% fetal calf serum, glutamine (2 mM), penicillin (100 U/mL), and streptomycin (0.1 mg/mL), all purchased from Sigma-Aldrich (Milwaukee, WI, USA).
Dual Luciferase Assay
HEK293, 293T, or 293T-derived Dcr KO37 (NoDice 2-20 or 4-25) cells were seeded at a density of 3,000 cells/well in white 96-well plates (Nunclon Delta surface, Thermo Scientific, Denmark). DNA co-transfections were carried out the following day using 34 ng (or 0.5 ng when indicated) shRNA-expressing plasmid and 6 ng psiCHECK.2 reporter plasmid, using X-tremeGENE 9 (Roche, Basel, Switzerland), according to the manufacturer’s protocol. At 2 days after transfection, Rluc and Fluc expression levels were analyzed using the Dual-Glo Luciferase Assay System (Promega, Madison, WI, USA), according to the manufacturer’s protocol. A multisample platereading luminometer (MicroLuminat plus LB 96V, Berthold Technologies, Bad Wild-bad, Germany) was used for measurements of the luminescence. Rluc expression levels were normalized to the Fluc expression levels and presented relative to a negative control. At least three biological replicates were made for all experiments.
VEGF Detection by Western Blot Analysis
HEK293 cells were seeded in 6-well plates at approximately 15% confluency. The next day cells were transfected with 2.1 μg DNA/well (700 ng pT2/CMV-mVEGF164-SV40-neo plasmid26 encoding mVEGF and 1,400 ng plasmid encoding the RNA species) using X-tremeGENE 9 transfection reagent at a DNA-to-transfection reagent ratio of 1:3, according to the manufacturer’s protocol. At 24 hr post-transfection, media were changed to serum-free media. Approximately 48 hr post-transfection, cells and media were harvested. Media were filtered through a 0.20-μM filter (Sarstedt, Nürnbrecht, Germany). Cells were lysed using 150 μL RIPA Lysis and Extraction Buffer/well (Thermo Scientific) supplemented with cOmplete Mini Protease Inhibitor Cocktail (Sigma-Aldrich), according to the manufacturer’s protocol, except that incubation time on ice was 15 min. Total protein concentration of the lysates was determined using Bio-Rad protein assay.
40 μg cell lysates and 32 μL media were electrophoresed on a Mini-PROTEAN TGX 4%–15% stain-free gel (Bio-Rad) in 1× Tris-Glycin-SDS (TGS) buffer (Bio-Rad) and activated for 45 s or two times 45 s using the ChemiDoc MP imaging system (Bio-Rad), before transfer to a polyvinylidene difluoride membrane (Bio-Rad) using the Trans-Blot Turbo Transfer system (Bio-Rad). Total protein was quantified after transfer using the ChemiDoc MP imaging system, and background was subtracted using Image Lab Software (Bio-Rad). The membrane was blocked in 5% w/v skim milk powder (WVR, Radnor, PA, USA) in 1× Tris-buffered saline (TBS) buffer (Thermo Fisher Scientific) with 0.1% v/v Tween20 (Sigma-Aldrich) (TBS-T) for 1 hr at room temperature (RT), and it was incubated with rabbit anti-VEGF antibody (ab46154, Abcam, Cambridge, UK) at a concentration of 1:2,000 overnight at 4°C with constant rocking. The membrane was washed 3 times 5 min in TBS-T before incubation with the secondary horseradish peroxidase (HRP)-conjugated goat-anti-rabbit antibody (Dako, Glostrup, Denmark) for 1 hr at RT. The membrane was washed again, and detection was done using the Clarity Western ECL substrate (Bio-Rad). Imaging was done using the ChemiDoc MP imaging system.
Small RNA Purification and Northern Blot Analysis
For detection of the U6-expressed agsh12.3 and sh12.3, HEK293 cells were transfected with 0.094 pmol inhibitor DNA plasmid and 30 ng GFP DNA plasmid in 6-well plates at approximately 15% confluency. For detection of the U1-expressed transcripts 293T or 293T-derived Dcr KO,37 (2-20 and 4-25) cells were transfected with 26.25 μg DNA in culture flasks (surface area, 75 cm2) at approximately 15% or 20% confluency, respectively, in all cases using X-tremeGENE 9 (Roche Applied Science). Media were replaced after 24 hr.
RNA enriched for small RNAs was isolated 48 hr after transfection using the mirVana miRNA Isolation Kit (Ambion, Austin, TX). For detection of the U1-expressed transcripts, RNA samples were precipitated and resolubilized in 22 μL Gel Loading Buffer II (Ambion), and 10 μL of each sample was electrophoresed in two separate lanes. For the detection of U6-driven siRNA expression, 250 ng small enriched RNA was electrophoresed. Eletrophoresis was done in 10% denaturing polyacrylamide gels (Criterion Precast gel 10% Tris-borate-EDTA [TBE]-urea; Bio-Rad, Copenhagen, Denmark) in 1× TBE buffer for 55–60 min at 150 V, together with a RNA size marker (decade marker RNA; Ambion). The RNA was transferred to a positively charged nitrocellulose membrane, Amersham Hybond-N (GE Healthcare, Little Chalfont, UK) by electroblotting (Trans-blot SD semi-dry transfer cell; Bio-Rad). Ultraviolet-fixed membranes were hybridized overnight in PerfectHyb solution (Sigma-Aldrich, St. Louis, MO) at 42°C with purified 5′ 32P-labeled oligos complementary to the guide strand, the passenger strand, or U6 snRNA. The oligonucleotide probes (10 pmol) were end-labeled with polynucleotide kinase (PNK) (10 units; New England Biolabs) and y32P-ATP (10 μCi) in 10-μL reactions for 1 hr, and they were purified with G-25 Sephadex spin-columns (GE Healthcare). Hybridized membranes with LNA-doped oligonucleotide probes were washed three times with 6× saline-sodium citrate (SSC)/0.2% SDS and then twice with 2× SSC/0.1% SDS, both at 38°C, before exposure to film (CL-XPosure Film, Rockford, IL). Hybridized membranes with DNA oligonucleotide probes were washed twice with 6× SSC/0.2% SDS at 38°C, before exposure to film. DNA probes were purchased from MWG/Eurofins (Ebersberg, Germany), and LNA-doped oligonucleotide probes were purchased from QIAGEN (Copenhagen, Denmark). Probe sequences are available in Table S1.
Statistical Analysis
Data are presented as the mean ± SD. Statistical differences between two groups were evaluated using a two-tailed Student’s t test assuming equal variance. p < 0.05 was considered statistically significant.
Author Contributions
E.K. and L.A. designed the RNAi vectors, and E.K., C.R.C., and S.A. carried out luciferase assays. S.A. performed northern blot and western blot analyses. The Dcr KO cells were a kind gift from Bryan Cullen. The study was conceived by L.A. and supervised by C.K.D., T.J.C., and L.A. The manuscript was written by L.A., and it was reviewed and edited by E.K., S.A., C.K.D., and T.J.C. All authors read and approved the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank Birgit Holm Hansen, Tina Hindkjær, and Izabela Barbara Pytlarz for their technical assistance. The project was financially supported by the Karen Elise Jensen Foundation, the Holger Hjortensbergs Foundation, the Aase and Ejnar Danielsens Foundations, and the Lundbeck Foundation (R126-2012-12456).
Footnotes
Supplemental Information includes five figures and one table and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.11.013.
Supplemental Information
References
- 1.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 5.Denli A.M., Tops B.B., Plasterk R.H., Ketting R.F., Hannon G.J. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 6.Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 7.Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 8.Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 10.Aagaard L., Rossi J.J. RNAi therapeutics: principles, prospects and challenges. Adv. Drug Deliv. Rev. 2007;59:75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 12.Paddison P.J., Caudy A.A., Bernstein E., Hannon G.J., Conklin D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarz D.S., Hutvágner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 15.Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fedorov Y., Anderson E.M., Birmingham A., Reynolds A., Karpilow J., Robinson K., Leake D., Marshall W.S., Khvorova A. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12:1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu S., Jin L., Zhang F., Huang Y., Grimm D., Rossi J.J., Kay M.A. Thermodynamic stability of small hairpin RNAs highly influences the loading process of different mammalian Argonautes. Proc. Natl. Acad. Sci. USA. 2011;108:9208–9213. doi: 10.1073/pnas.1018023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 19.Dallas A., Ilves H., Ge Q., Kumar P., Shorenstein J., Kazakov S.A., Cuellar T.L., McManus M.T., Behlke M.A., Johnston B.H. Right- and left-loop short shRNAs have distinct and unusual mechanisms of gene silencing. Nucleic Acids Res. 2012;40:9255–9271. doi: 10.1093/nar/gks662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y.P., Schopman N.C., Berkhout B. Dicer-independent processing of short hairpin RNAs. Nucleic Acids Res. 2013;41:3723–3733. doi: 10.1093/nar/gkt036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harwig A., Herrera-Carrillo E., Jongejan A., van Kampen A.H., Berkhout B. Deep Sequence Analysis of AgoshRNA Processing Reveals 3′ A Addition and Trimming. Mol. Ther. Nucleic Acids. 2015;4:e247. doi: 10.1038/mtna.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheloufi S., Dos Santos C.O., Chong M.M., Hannon G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cifuentes D., Xue H., Taylor D.W., Patnode H., Mishima Y., Cheloufi S., Ma E., Mane S., Hannon G.J., Lawson N.D. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J.S., Maurin T., Robine N., Rasmussen K.D., Jeffrey K.L., Chandwani R., Papapetrou E.P., Sadelain M., O’Carroll D., Lai E.C. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. USA. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrera-Carrillo E., Berkhout B. Dicer-independent processing of small RNA duplexes: mechanistic insights and applications. Nucleic Acids Res. 2017;45:10369–10379. doi: 10.1093/nar/gkx779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pihlmann M., Askou A.L., Aagaard L., Bruun G.H., Svalgaard J.D., Holm-Nielsen M.H., Dagnaes-Hansen F., Bek T., Mikkelsen J.G., Jensen T.G., Corydon T.J. Adeno-associated virus-delivered polycistronic microRNA-clusters for knockdown of vascular endothelial growth factor in vivo. J. Gene Med. 2012;14:328–338. doi: 10.1002/jgm.2623. [DOI] [PubMed] [Google Scholar]
- 27.Sun G., Yeh S.Y., Yuan C.W., Chiu M.J., Yung B.S., Yen Y. Molecular Properties, Functional Mechanisms, and Applications of Sliced siRNA. Mol. Ther. Nucleic Acids. 2015;4:e221. doi: 10.1038/mtna.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J.E., Heo I., Tian Y., Simanshu D.K., Chang H., Jee D., Patel D.J., Kim V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aagaard L., Bjerregaard B., Kjeldbjerg A.L., Pedersen F.S., Larsson L.I., Rossi J.J. Silencing of endogenous envelope genes in human choriocarcinoma cells shows that envPb1 is involved in heterotypic cell fusions. J. Gen. Virol. 2012;93:1696–1699. doi: 10.1099/vir.0.041764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma H., Wu Y., Dang Y., Choi J.G., Zhang J., Wu H. Pol III Promoters to Express Small RNAs: Delineation of Transcription Initiation. Mol. Ther. Nucleic Acids. 2014;3:e161. doi: 10.1038/mtna.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoda M., Cifuentes D., Izumi N., Sakaguchi Y., Suzuki T., Giraldez A.J., Tomari Y. Poly(A)-specific ribonuclease mediates 3′-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013;5:715–726. doi: 10.1016/j.celrep.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fellmann C., Hoffmann T., Sridhar V., Hopfgartner B., Muhar M., Roth M., Lai D.Y., Barbosa I.A., Kwon J.S., Guan Y. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 35.Zeng Y., Wagner E.J., Cullen B.R. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 36.Aagaard L.A., Zhang J., von Eije K.J., Li H., Saetrom P., Amarzguioui M., Rossi J.J. Engineering and optimization of the miR-106b cluster for ectopic expression of multiplexed anti-HIV RNAs. Gene Ther. 2008;15:1536–1549. doi: 10.1038/gt.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bogerd H.P., Whisnant A.W., Kennedy E.M., Flores O., Cullen B.R. Derivation and characterization of Dicer- and microRNA-deficient human cells. RNA. 2014;20:923–937. doi: 10.1261/rna.044545.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 39.Mockenhaupt S., Grosse S., Rupp D., Bartenschlager R., Grimm D. Alleviation of off-target effects from vector-encoded shRNAs via codelivered RNA decoys. Proc. Natl. Acad. Sci. USA. 2015;112:E4007–E4016. doi: 10.1073/pnas.1510476112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu S., Jin L., Zhang Y., Huang Y., Zhang F., Valdmanis P.N., Kay M.A. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shang R., Zhang F., Xu B., Xi H., Zhang X., Wang W., Wu L. Ribozyme-enhanced single-stranded Ago2-processed interfering RNA triggers efficient gene silencing with fewer off-target effects. Nat. Commun. 2015;6:8430. doi: 10.1038/ncomms9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McIntyre G.J., Yu Y.H., Lomas M., Fanning G.C. The effects of stem length and core placement on shRNA activity. BMC Mol. Biol. 2011;12:34. doi: 10.1186/1471-2199-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herrera-Carrillo E., Gao Z.L., Harwig A., Heemskerk M.T., Berkhout B. The influence of the 5′-terminal nucleotide on AgoshRNA activity and biogenesis: importance of the polymerase III transcription initiation site. Nucleic Acids Res. 2017;45:4036–4050. doi: 10.1093/nar/gkw1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu H.Y., Yan Z., Xu Y., Hu H., Menzel C., Zhou Y.H., Chen W., Khaitovich P. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics. 2009;10:413. doi: 10.1186/1471-2164-10-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frank F., Sonenberg N., Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 46.Harwig A., Kruize Z., Yang Z., Restle T., Berkhout B. Analysis of AgoshRNA maturation and loading into Ago2. PLoS ONE. 2017;12:e0183269. doi: 10.1371/journal.pone.0183269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Birmingham A., Anderson E.M., Reynolds A., Ilsley-Tyree D., Leake D., Fedorov Y., Baskerville S., Maksimova E., Robinson K., Karpilow J. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 48.Castanotto D., Sakurai K., Lingeman R., Li H., Shively L., Aagaard L., Soifer H., Gatignol A., Riggs A., Rossi J.J. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 50.Martin J.N., Wolken N., Brown T., Dauer W.T., Ehrlich M.E., Gonzalez-Alegre P. Lethal toxicity caused by expression of shRNA in the mouse striatum: implications for therapeutic design. Gene Ther. 2011;18:666–673. doi: 10.1038/gt.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boudreau R.L., Martins I., Davidson B.L. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol. Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma H., Wu Y., Choi J.G., Wu H. Lower and upper stem-single-stranded RNA junctions together determine the Drosha cleavage site. Proc. Natl. Acad. Sci. USA. 2013;110:20687–20692. doi: 10.1073/pnas.1311639110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu H., Ye C., Ramirez D., Manjunath N. Alternative processing of primary microRNA transcripts by Drosha generates 5′ end variation of mature microRNA. PLoS ONE. 2009;4:e7566. doi: 10.1371/journal.pone.0007566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun G., Wang J., Huang Y., Yuan C.W., Zhang K., Hu S., Chen L., Lin R.J., Yen Y., Riggs A.D. Differences in silencing of mismatched targets by sliced versus diced siRNAs. Nucleic Acids Res. 2018;46:6806–6822. doi: 10.1093/nar/gky287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang J.S., Maurin T., Lai E.C. Functional parameters of Dicer-independent microRNA biogenesis. RNA. 2012;18:945–957. doi: 10.1261/rna.032938.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Auyeung V.C., Ulitsky I., McGeary S.E., Bartel D.P. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152:844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fang W., Bartel D.P. The Menu of Features that Define Primary MicroRNAs and Enable De Novo Design of MicroRNA Genes. Mol. Cell. 2015;60:131–145. doi: 10.1016/j.molcel.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen T.A., Jo M.H., Choi Y.G., Park J., Kwon S.C., Hohng S., Kim V.N., Woo J.S. Functional Anatomy of the Human Microprocessor. Cell. 2015;161:1374–1387. doi: 10.1016/j.cell.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 59.van den Berg F.T., Rossi J.J., Arbuthnot P., Weinberg M.S. Design of Effective Primary MicroRNA Mimics With Different Basal Stem Conformations. Mol. Ther. Nucleic Acids. 2016;5:e278. doi: 10.1038/mtna.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nykänen A., Haley B., Zamore P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 61.Paul C.P., Good P.D., Winer I., Engelke D.R. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 2002;20:505–508. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- 62.Rossi J.J. Expression strategies for short hairpin RNA interference triggers. Hum. Gene Ther. 2008;19:313–317. doi: 10.1089/hum.2008.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwarz D.S., Hutvágner G., Haley B., Zamore P.D. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell. 2002;10:537–548. doi: 10.1016/s1097-2765(02)00651-2. [DOI] [PubMed] [Google Scholar]
- 64.Good P.D., Krikos A.J., Li S.X., Bertrand E., Lee N.S., Giver L., Ellington A., Zaia J.A., Rossi J.J., Engelke D.R. Expression of small, therapeutic RNAs in human cell nuclei. Gene Ther. 1997;4:45–54. doi: 10.1038/sj.gt.3300354. [DOI] [PubMed] [Google Scholar]
- 65.Aagaard L., Amarzguioui M., Sun G., Santos L.C., Ehsani A., Prydz H., Rossi J.J. A facile lentiviral vector system for expression of doxycycline-inducible shRNAs: knockdown of the pre-miRNA processing enzyme Drosha. Mol. Ther. 2007;15:938–945. doi: 10.1038/sj.mt.6300118. [DOI] [PubMed] [Google Scholar]
- 66.Chung J., Zhang J., Li H., Ouellet D.L., DiGiusto D.L., Rossi J.J. Endogenous MCM7 microRNA cluster as a novel platform to multiplex small interfering and nucleolar RNAs for combinational HIV-1 gene therapy. Hum. Gene Ther. 2012;23:1200–1208. doi: 10.1089/hum.2012.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scherer L.J., Frank R., Rossi J.J. Optimization and characterization of tRNA-shRNA expression constructs. Nucleic Acids Res. 2007;35:2620–2628. doi: 10.1093/nar/gkm103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.