Abstract
Controlled human exposure to the oxidant air pollutant ozone causes decrements in lung function and increased inflammation as evidenced by neutrophil influx into the lung and increased levels of proinflammatory cytokines in the airways. Here we describe a targeted metabolomics evaluation of human bronchioalveolar lavage fluid (BALF) following controlled in vivo exposure to ozone to gain greater insight into its pulmonary effects. In a two-arm cross-over study, each healthy adult human volunteer was randomly exposed to filtered air (FA) and to 0.3 ppm ozone for 2 hr while undergoing intermittent exercise with a minimum of 4 weeks between exposures. Bronchoscopy was performed and BALF obtained at 1 (n = 9) or 24 (n = 23) h post-exposure. Metabolites were detected using ultrahigh performance liquid chromatography-tandem mass spectroscopy. At 1-hour post-exposure, a total of 28 metabolites were differentially expressed (DE) (p < 0.05) following ozone exposure compared to FA-exposure. These changes were associated with increased glycolysis and antioxidant responses, suggesting acute-phase increased energy utilization as part of the cellular response to oxidative stress. At 24-hour post-exposure, 41 metabolites were DE. Many of the changes were in amino acids and linked with enhanced proteolysis. Changes associated with increased lipid membrane turnover were also observed. These later-stage changes were consistent with ongoing repair of airway tissues. Roughly twice as many metabolites were differentially expressed at 24 hour compared to 1-hour post-exposure. The changes at 1 hour reflect responses to oxidative stress while the changes at 24 hour indicate a broader set of responses consistent with tissue repair. These results illustrate the ability of metabolomic analysis to identify mechanistic features of ozone toxicity and aspects of the subsequent tissue response.
Keywords: metabolites, ozone, pulmonary, human
Introduction
Exposure to ozone, a criteria air pollutant, can result in a variety of deleterious responses including mild pulmonary irritation, damage to lung cells, inflammation, exacerbation of lung disease, mortality in susceptible individuals, and at the population level, an increased burden on public health (EPA Integrated Science Assessment of Ozone, 2013). Several key events are recognized components of the progression from exposure to adverse outcome. Inhaled ozone interacts primarily with cells and fluid that line the respiratory airways by reacting with lipid rich components in lung lining fluid as well as cell membranes to form stable lipid peroxides. This results in the induction of oxidative stress, the depletion of antioxidant reservoirs and injury to airway epithelial cells (Bromberg, 2016). Ozonized phospholipids trigger the release of proinflammatory mediators in airway epithelial cells and macrophages, resulting in pulmonary inflammation (Kafoury et al., 1999; Koren et al, 1989; Zemski et al, 2016). Plasma components can migrate across the damaged epithelial barrier and interact with lung lining fluid. Repair processes which include repair of damaged cells, epithelial cell replication and migration to repair damaged areas, and wound closure are typically observed 4 - 72 hr after exposure (Leroy et al., 2015; Nikula et al., 1988.
Transcriptomic and proteomic analyses provide global views of mRNA and protein changes associated with toxicant exposure. These technologies facilitate the investigation of sequences of key events, providing insight into the mechanistic biology linking exposures with adverse outcomes. Recent advances in analytical technology permit the identification of small molecule metabolites that are the final products of the gene expression - protein synthesis sequence. Metabolite profiles reflect alterations in phenotypes and cellular activities at the functional level (Uppal et al., 2016). Metabolites provide insights that are closer to the downstream biological processes as these typically occur at the metabolite level and not the mRNA or protein level. Analysis of metabolites has been widely adopted in medical research and has provided key mechanistic insights in many areas such as quantifying lung injury and cancer biomarker discovery (Cui et al, 2016; Hori et al, 2011; Stringer et al, 2011). To date, however, there have been only a limited number of metabolomic analyses of the effects of environmental exposures on the respiratory tract.
Transcriptomic studies utilizing in vitro and animal models have shown that pathways associated with fatty acid metabolism, cell proliferation, stress response, adhesion molecules, host defense, and metal binding are perturbed by ozone exposure (Holloway et al., 2012). Rodent proteomics studies have associated ozone exposure with oxidative stress, oxidative stress response, and inflammation (Grantziotis et al, 2016; haque et al., 2007; Kierstein et al., 2008; Wattiez et al.,l 2003). Only a small number of metabolomic studies have evaluated the effects of ozone exposure. Air pollutant-induced changes in glucose, lipid, and amino acid metabolism associated with activation of systemic stress response pathways were seen in rodent and human serum samples (Miller et al., 2015; Miller et al., 2016; Ward-Caviness et al., 2016). While serum samples can define systemic perturbations, they are less useful for examination of mechanistic events in the airways that are which are the primary target of ozone. In the current study, we assessed metabolites present in lung lining fluid to characterize the temporal profile of injury, inflammation and repair in the airway from individuals following controlled exposure to both clean air and ozone. This is the first study to report such findings.
Materials and Methods
Study population and experimental design
As was previously described by Devlin et al.(2012), twenty-three non-smoking, healthy adult volunteers participated in the study. Participants were informed of the procedures and potential risk, and each signed an informed consent. The study protocol and consent forms were approved by the University of North Carolina School of Medicine Institutional Review Board on the Protection of the Rights of Human Subjects as well as the US Environmental Protection Agency. Clinical Trial Registration (URL: http://www.clinicaltrials.gov) Unique identifier: NCT01492517.
Each participant was randomly exposed to filtered air (FA) and 0.3 ppm ozone, with the exposures separated by at least four weeks. During the two hour exposures, each participant alternated 15 minute periods of rest with moderate exercise on a cycle ergometer with the appropriate workload set to achieve a minute ventilation of 25 L/min/m2 body surface area. Bronchoalveolar lavage fluid (BALF) was collected from each participant approximately 24 hours after exposure as described previously (Ghio et al., 2000). In brief, a fiber optic bronchoscope was wedged into a segmental bronchus of the right middle lobe, and two 50-ml aliquots of sterile saline were instilled and immediately aspirated. After centrifugation at 1000 g for 10 minutes at 4 °C, cell pellets were removed. The supernatants from the two washes were pooled and stored at −80 °C prior to analysis. Nine of the 23 subjects returned for another randomized, crossover exposure regiment. These volunteers were exposed to FA or 0.3 ppm ozone separated by a minimum of four weeks and BALF was collected one hour after exposure.
Metabolomics analysis
The BALF samples were analyzed for global metabolomic profile by Metabolon Inc. (Durham, NC) with an ultrahigh performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS). Samples were prepared and analyzed as previously described (Evans et al., 2014). In brief, they were extracted in methanol and the recovered small molecules were analyzed using UPLC-MS/MS in four different optimized settings: (1) Acidic positive ion conditions for more hydrophilic compounds were assessed using a C18 column (Waters UPLC BEH C18-2.1×100 mm, 1.7 μm) using water and methanol, containing 0.05% pentafluoropropionic acid (PFP+A) and 0.1% formic acid (FA). (2) Acidic positive ion conditions for more hydrophobic compounds were examined with a C18 column using methanol, acetonitrile, water, 0.05% PFPA and 0.01% FA and was operated at an overall higher organic content. (3) Basic negative ion conditions were optimized with a dedicated C18 column using methanol and water with 6.5 mM Ammonium Bicarbonate at pH 8. (4) The last condition for analysis of negative ions was an HILIC column (Waters UPLC BEH Amide 2.1×150 mm, 1.7 μm) using a gradient consisting of water and acetonitrile with 10mM Ammonium Formate, pH 10.8. The MS analysis alternated between MS and data-dependent MSn scans using dynamic exclusion. The scan range varied slightly between methods but covered 70-1000 m/z. Detailed methods can be found in the supplementary information.
The processes of data extraction, peak identification, and quality control were performed using Metabolon hardware and software. Metabolon’s library for compound identification is based on authenticated standards that contain the retention time/index (RI), mass to charge ratio (m/z), and chromatographic data (including MS/MS spectral data). The identifications are based on the following criteria: retention index within a narrow RI window of the proposed identification, accurate mass match to the library +/− 10 ppm, and the MS/MS forward and reverse scores between the experimental data and authentic standards. The MS/MS scores are based on a comparison of the ions present in the experimental spectrum to the ions present in the library spectrum. While there may be similarities between these molecules based on one of these factors, the use of all three data points can be utilized to distinguish and differentiate metabolites.
Statistical analysis
Metabolites were quantified through assessment of MS/MS area under the curve (AUC). Following log transformation of the AUC data and imputation of missing values as the minimum observed value for each compound, a paired t-test was used to identify metabolites that differed significantly between FA and ozone exposures. MetaboAnalyst (v 3.0, http://www.metaboanalyst.ca/) was used for partial least squares discriminant analysis (PLS-DA) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. PLS-DA was performed for all the metabolites; KEGG pathway analysis was conducted for metabolites with a p < 0.05 for the selected contrasts (ozone 1 hr vs. air 1 hour and ozone 24 hr vs. air 24 hr) and identified with the Human Metabolon Database Compound (HMDB) IDs. The analysis utilized the homo sapiens pathway library with hypergeometric test for over representation analysis and relative-betweeness centrality for pathway topology analysis. For identifying potential upstreaming signaling associations, altered metabolites were submitted for network analysis through Ingenuity Pathway Analysis (IPA, version 36601845, QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis). The Human Metabolon Database Compound (HMDB) IDs, Kyoto Encyclopedia of Genes and Genomes (KEGG) compound IDs, or PubChem IDs were used to map to the IPA database.
Results
Ozone exposure changes the profile of BAL metabolites
A global metabolic analysis was conducted for the BAL samples collected from healthy human volunteers 1- and 24- hours post-air ozone exposure. A total of 134 metabolites were identified by Metabolon. Metabolites present at 1 and 24 hr after ozone exposure with compared with those present at 1 and 24 hr after exposure to clean air. A supervised multivariate statistical analysis was performed using partial least squares-discriminant analysis (PLS-DA). The one-hr data showed a clear separation between the air and ozone groups (Figure 1A). Analysis of the 24 hr data also showed separation, but it was not as pronounced as that seen in the 1 hr data (Figure 1B). Paired t-tests identified 28 and 41 metabolites that were significantly altered (p < 0.05) at 1-hr and 24-hr following ozone exposure, respectively, in comparison with metabolites present following air exposure (Table 1). Additionally, 13 metabolites were altered at both 1 and 24 hr after ozone exposure (Figure 1C). None of these metabolites exceeded the multiple-test corrected cutoff level.
Figure 1.
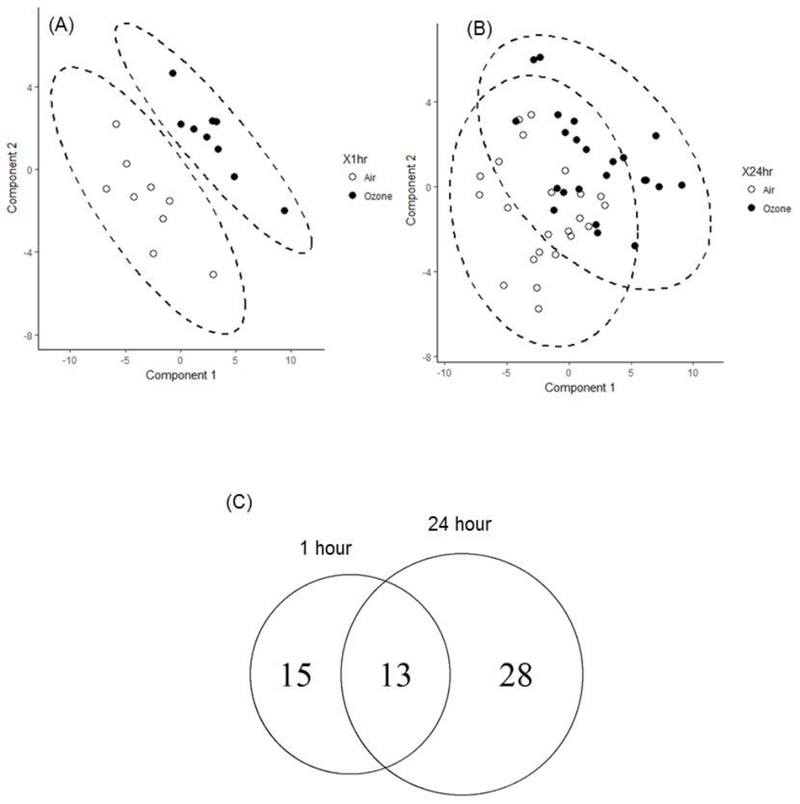
Global metabolomics profiling of human BALF samples after ozone exposure. PLS-DA score plots are calculated from metabolite concentration (A) 1 hour and (B) 24 hour post ozone (●) and clean air (○) exposure with 95 % confidence interval (dashed oval). (C) Venn diagram is plotted for the differentially altered metabolites (p < 0.05) by paired t-test between ozone and air exposure.
Table1.
Significantly altered metabolites in BALF samples. Metabolites that are significantly altered between ozone and air (p < 0.05) are shown with fold change and highlighted in color (yellow for up-regulation and blue for down-regulation) for the 1 hour and 24 hour data. The metabolites are categorized into super- and sub-pathways based on Metabolon’s classification.
| Super Pathway | Sub Pathway | Biochemical Name | FC 1hr | FC 24hr |
|---|---|---|---|---|
| Amino Acid | Glycine, Serine and Threonine Metabolism | glycine |
1.37 | 1.39 |
| serine |
1.39 | 1.69 | ||
| N-acetylserine |
0.98 | 1.20 | ||
| threonine | 1.73 | 1.35 | ||
| Alanine and Aspartate Metabolism | alanine |
1.92 | 1.64 | |
| aspartate |
2.32 | 1.50 | ||
| asparagine | 2.17 | 1.84 | ||
| Glutamate Metabolism | glutamate |
1.75 | 1.40 | |
| glutamine | 1.92 | 1.37 | ||
| Histidine Metabolism | histidine | 1.67 | 1.78 | |
| Lysine Metabolism | lysine | 1.64 | 1.49 | |
| Phenylalanine and Tyrosine Metabolism | phenylalanine |
2.05 | 1.68 | |
| tyrosine |
1.68 | 1.66 | ||
| phenol sulfate |
1.32 | 1.36 | ||
| p-cresol sulfate | 2.14 | 1.32 | ||
| Tryptophan Metabolism | tryptophan | 1.95 | 1.46 | |
| Leucine, Isoleucine and Valine Metabolism | leucine |
1.51 | 1.51 | |
| isoleucine | 1.10 | 1.40 | ||
| Methionine, Cysteine, SAM and Taurine Metabolism | taurine | 1.23 | 1.16 | |
| Urea cycle; Arginine and Proline Metabolism | arginine |
1.72 | 1.48 | |
| ornithine |
1.95 | 1.45 | ||
| proline | 2.22 | 1.48 | ||
| Creatine Metabolism | creatine |
2.25 | 1.17 | |
| creatinine | 1.57 | 1.33 | ||
| Carbohydrate | Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | glucose |
1.93 | 1.47 |
| lactate |
0.78 | 1.52 | ||
| glycerate | 1.19 | 1.54 | ||
| Energy | TCA Cycle | alpha-ketoglutarate | 1.29 | 1.11 |
| Lipid | Carnitine Metabolism | carnitine | 1.86 | 1.35 |
| Inositol Metabolism | myo-inositol | 1.37 | 1.09 | |
| Phospholipid Metabolism | choline phosphate |
1.57 | 1.09 | |
| glycerophosphorylcholine (GPC) |
1.67 | 1.17 | ||
| phosphoethanolamine |
1.38 | 1.10 | ||
| glycerophosphoethanolamine |
2.57 | 1.07 | ||
| 1-palmitoyl-2-oleoyl-GPC (16:0/18:1) |
0.92 | 1.13 | ||
| 1-stearoyl-2-arachidonoyl-GPC (18:0/20:4) |
0.96 | 1.21 | ||
| 1-stearoyl-2-oleoyl-GPC (18:0/18:1) |
0.93 | 1.20 | ||
| 1-stearoyl-2-arachidonoyl-GPI (18:0/20:4) |
1.36 | 1.27 | ||
| 1,2-dilinoleoyl-GPC (18:2/18:2) |
1.01 | 1.17 | ||
| 1-palmitoyl-2-oleoyl-GPS (16:0/18:1) | 0.90 | 1.17 | ||
| Lysolipid | 1-palmitoyl-GPC (16:0) |
1.23 | 1.23 | |
| 1-stearoyl-GPC (18:0) | 1.79 | 1.61 | ||
| Plasmalogen | 1-(1-enyl-stearoyl)-2-linoleoyl-GPE (P-18:0/18:2)* |
0.93 | 1.26 | |
| 1-(1-enyl-stearoyl)-2-arachidonoyl-GPE (P-18:0/20:4)* | 1.07 | 1.14 | ||
| Sphingolipid Metabolism | palmitoyl dihydrosphingomyelin (d18:0/16:0)* |
0.90 | 1.36 | |
| palmitoyl sphingomyelin (d18:1/16:0) |
0.99 | 1.16 | ||
| stearoyl sphingomyelin (d18:1/18:0) |
1.25 | 1.25 | ||
| tricosanoyl sphingomyelin (d18:1/23:0)* |
1.43 | 1.36 | ||
| sphingomyelin (d18:1/20:0, d16:1/22:0)* |
1.04 | 1.17 | ||
| sphingomyelin (d18:1/22:1, d18:2/22:0, d16:1/24:1)* |
1.47 | 1.39 | ||
| sphingomyelin (d18:2/24:1 d18:1/24:2)* | 1.10 | 1.19 | ||
| Nucleotide | Purine Metabolism, (Hypo)Xanthine/Inosine containing | urate |
1.40 | 1.23 |
| allantoin | 2.92 | 1.08 | ||
| Vitamins | Ascorbate and Aldarate Metabolism | threonate | 2.08 | 1.06 |
| Xenobiotics | Drug | N-ethylglycinexylidide |
0.36 | 0.70 |
| salicylate | 1.15 | 1.10 | ||
Ozone exposure primarily increases metabolites associated with amino acids and fatty acids in BAL
Annotation of the metabolites with a p < 0.05 for the ozone-FA paired t-test are listed in Table 1 for each metabolite at 1 and 24 hr post exposure. All but one of the altered metabolites were higher after ozone exposure with an average fold change of 1.77 at 1 hr (range 1.15 ~ 2.92) and of 1.40 at 24-hr (range 1.13 ~ 1.84). The xenobiotic metabolite N-ethylglycinexylidide which was decreased (−2.78) at 1 hr following exposure. Most altered metabolites were amino acids and lipids. Metabolites associated with amino acid metabolism made up 50% of those altered 1 hr after ozone exposure and 49% of those at 24 hrs. Metabolites associated with lipids made up 25% of the differentially expressed metabolites at 1 hr and 44% at 24 hrs. The remaining metabolites were associated with carbohydrate (3.6% at 1 hr and 7.3% at 24 hr), nucleotides (7.1% at 1-hr and 0% at 24-hr), xenobiotics (7.1% at 1-hr and 0% at 24-hr), energy (3.6% at 1-hr and 0% at 24-hr), and vitamins (3.6% at 1-hr and 0% at 24-hr).
Indication of altered energy metabolism
The significant increase in amino acids suggests enhanced protein degradation or synthesis, possibly due to ozone -induced lung injury and inflammation. Both the metabolite and pathway data suggested a significant alteration of arginine, proline, and ornithine metabolism and may be an indication of increased urea cycle activity, which may become active when there is a reliance upon amino acid catabolism (Figure 2A). Increases in arginine and borderline increases in citrulline at both time points also indicate increased nitric oxide production, which may contribute to increased inflammation after ozone exposure. Altered glucose utilization was indicated by elevated levels of glucose and lactate after ozone exposure. The elevation in glucose could be related to blocked mitochondrial utilization or, alternatively, could reflect increased blood levels of glucose in response to the stress of ozone exposure, with leakage of glucose from blood into the lining fluid of the ozone-damaged respiratory tract. The elevations of glutamine and glutamate at both time points and increases of α-ketoglutarate at 24-hr suggest increased use of the mitochondrial carbon pool to support the TCA cycle under impaired glucose utilization. Increased anaerobic metabolism associated with mitochondrial toxicity could also be related to the accumulation of lactate.
Figure 2.
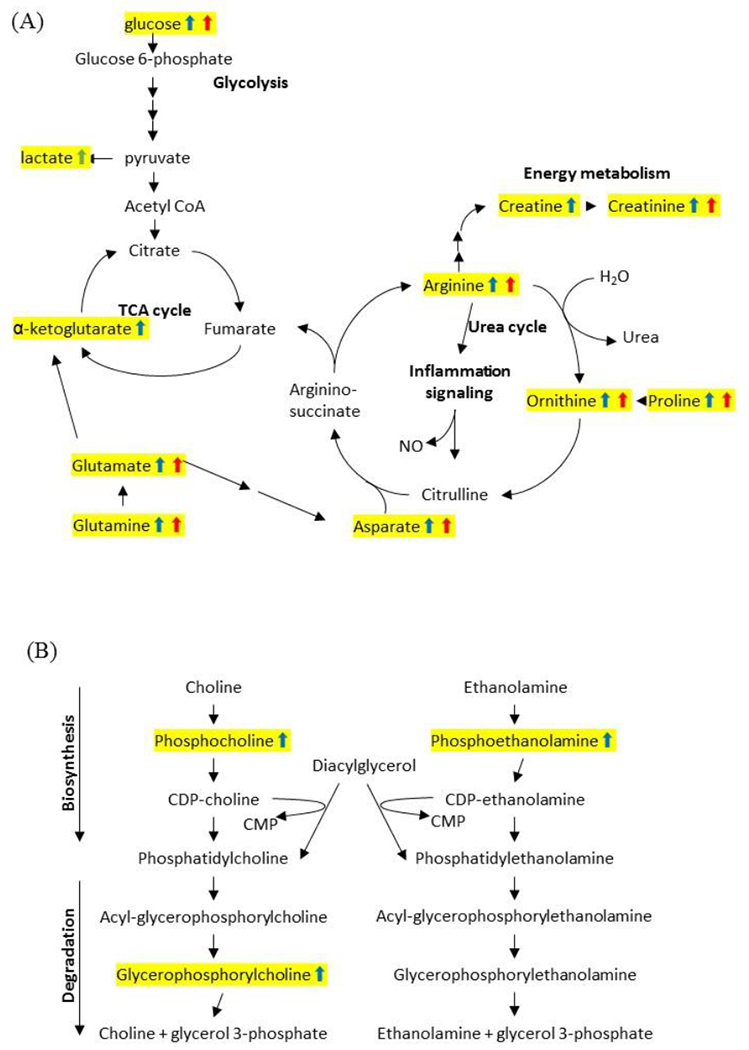
Ozone-induced energy (A) and lipid (B) metabolic pathway alterations. Top altered pathways suggested by the KEGG were plotted along with the significantly altered metabolites. Those metabolites that are altered at 1-hour are marked with blue arrows and those that are altered at 24 hours are marked with red arrows.
Indications of increased lipid membrane turnover in response to ozone exposure
Many ozone-altered metabolites were associated lipid metabolism (25% at 1-hr and 44% at 24-hr). Phospholipid precursors, including significant increases in choline phosphate and phosphoethanolamine as well as the degradation products glycerophosphorylcholine and glycerophosphoethanolamine, were increased 1 hr after ozone exposure (Table 1). These may result from increased lipid membrane turnover following ozone exposure, which is known to attack carbon double bonds on membrane lipids. Similarly, there were also elevated levels of lysophospholipids (1-palmitoyl-GPC and 1-stearoyl-GPC) following ozone exposure, which is consistent with increased lipase activity contributing to an increase in lipid membrane turnover (Table 1 and Figure 2B).
Altered metabolic pathways after ozone exposure
The KEGG database categorized each metabolite based on its functionality (Table 1). However, small molecules function together as an intricate network of interactions where each metabolite is involved in multiple biological processes. In order to rank the significance of altered pathways and overall biological perturbations caused by ozone exposure, we utilized the KEGG database to map altered metabolites in the current study to any associated metabolism pathways and ranked the significance of ozone-altered pathways using the MetaboAnalyst interface with hypergeometric test. Metabolite pathways were divided into three groups: (1) pathways differentially expressed only at 1-hr following exposure, (2) pathways differentially expressed at both time points and, (3) pathways differentially expressed only at 24-hrs after exposure. Using the Human Metabolome Database (HMDB), 40 of the 56 differentially expressed metabolites were mapped to the database. Pathways altered 1 hr after exposure were glycerophospholipid, ether lipid, ascorbate, and aldarate metabolism (Figure 3). The major pathways perturbed at both the 1- and 24- hr time points were aminoacyl-tRNA biosynthesis and amino acid metabolism, including arginine, proline, alanine, aspartate, and glutamate. Pathways perturbed 24-hrs after exposure were aminoacyl-tRNA biosynthesis, amino acid metabolism (cyanoamino acid, glycine, serine, threonine), and lipid metabolism (glycerophospholipid and sphingolipid). The ascorbate and aldarate metabolism alterations at 1 hr suggest possible antioxidant consumption during and after acute ozone exposure. The altered tRNA biosynthesis pathway at both time points suggests increased protein synthesis as part of the repair process after ozone exposure. A detailed KEGG pathway analysis is shown in Supplementary Table 1.
Figure 3.
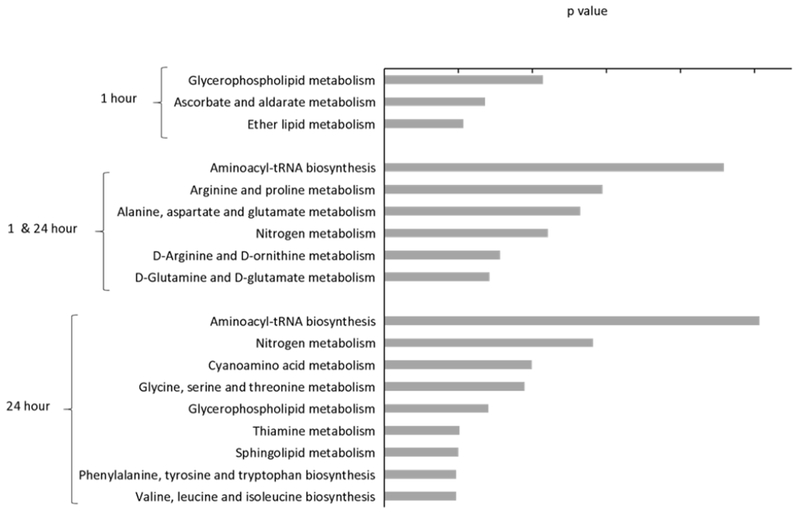
KEGG pathway analysis of the global metabolite alterations between ozone and air. The metabolites were categorized into 3 groups: 1. Only present in the 1-hour data, 2. Present in both the 1- and 24- hour data and, 3. Present in only the 24-hour data. The significantly altered pathways are ranked plotted by the p-value. The significance level for altered KEGG human pathways was set with p < 0.05 and a false discovery rate q < 0.2.
Potential upstream signaling perturbations
Metabolites are the terminal downstream products present following ozone inhalation and reflect the physiological status of the lung. They are linked with changes in upstream gene expression and protein synthesis and function. Therefore, we utilized Ingenuity Pathway Analysis (IPA) to investigate potential upstream signaling network alterations associated with ozone exposure. Metabolites were submitted to IPA with multiple identifiers to optimize metabolite identification. Ninety of the 134 detected metabolites were mapped to the Ingenuity knowledgebase. Network analysis highlighted upstream signaling hubs and connections to measured metabolites for the 1-hr and 24-hr data (Supplementary Figure 1). All the molecules were assessed for the number of connections to other molecules and the 20% most frequently connected molecules were defined as hubs and categorized based on the main biological function (Table 2). Hubs identified in both 1- and 24- hr data were associated with energy metabolism, cell signaling, and inflammation.
Table 2.
IPA Network Analysis for the 20% most frequently connected molecules
| Hub function | Hub name |
|---|---|
| 1 hour | |
| Energy metabolism | Glucose, Insulin, Glutamate, LDL, Gsk3, Creatine, Uric acid |
| Cell signaling | PI3K, Pkc, APP, Akt, ERK, MAPK, NFkB, Mapk, Hsp70 |
| Inflammation | IL-6, TGF-β, Vegf |
| 24 hour | |
| Energy metabolism | Glucose, Insulin, LDL, Glutamate, Albumin, Gsk3, Arginine, Glycine |
| Cell signaling | PI3K, Akt, MAPK, ERK, CDKN1A, Jnk, CDKN1A, PRKCB, CaMKII, NFkB |
| Inflammation | IL-6, EGFR, TGF-β, FCER1A |
Discussion
We conducted a metabolomics analysis of BAL samples from volunteers exposed acutely to ozone, a powerful oxidant, and measured metabolite changes that reflect the associated inflammation and lung injury. The metabolomics analysis identified changes consistent with a sequence of early phase (1-hr) lung damage, altered glucose utilization, activation of oxidative stress responses and subsequent (24-hr) cellular repair, with metabolomic signals of increased energy usage at both time points. The PLS-DA analysis (Figure 1) showed a clear separation of metabolite changes between the 1- and 24-hr timepoints. Amino acids and lipids were the major metabolite alterations associated with ozone exposure (Table 1) and the KEGG metabolism pathway analysis was consistent with initial stress response and subsequent cellular repair (Figure 3).
Several decades of human studies have characterized pulmonary responses to acute ozone exposure. Ozone initially attacks compounds with carbon double bonds, including lipid membranes, resulting in damage to cells and the release of inflammatory mediators such as cytokines, followed by cellular repair (Bromberg, 2016; Devlin et al., 1997; Leikauf et al., 1995). The human airway is lined with an abundance of antioxidants, including urate, ascorbate, and glutathione, that work in a coordinated manner with an inducible antioxidant defense system and that, together, function as a first line of defense against inhaled oxidants such as ozone (Bromberg, 2016). The protective capabilities of this antioxidant defense system have been well characterized. For example, dietary supplementation with ascorbate, carotenoids, and alpha-tocopherol moderated the effects of acute ozone exposure on human pulmonary function (Samet et al., 2001). A controlled human study showed reduced overall antioxidant capacity after acute ozone exposure (Chen et al., 2007). At 1- but not 24-hr, we found significant increases in allantoin (2.92 fold) and threonate (2.08 fold). These are metabolites of the antioxidants urate and ascorbate, respectively. Their early-phase increases presumably reflect ozone-induced oxidative stress. Likewise, their lack of elevation at 24-hr is consistent with the transition from active oxidative stress to the later repair of the oxidative damage. Glutathione was not significantly altered in the BAL samples. Previous human observations showed relatively high concentrations of glutathione (1 ~ 10 mM) relative to the concentrations of urate (200 μM) and ascorbate (50 μM) (van der Vliet et al., 1999) which may result in the lack of glutathione significance in the current study.
Amino acids
Ozone exposure is associated with an initial injury to epithelial cells lining the airways followed by repair or replacement of damaged cells. This is accompanied by an increase in cellular airway inflammation and various biomarkers of inflammation such as interleukins and prostaglandins (Bromberg, 2016). Increased amino acid levels observed in BAL samples were more pronounced at 24-hr, which is suggestive of protein degradation and synthesis, possibly associated with the signaling processes for repair of damaged epithelial cells. The amino acids methionine, cysteine, tryptophan, phenylalanine, and histidine are more sensitive to direct oxidation by ozone (Johnson et al., 1987; Sharma et al., 2010). The elevated levels of tryptophan (2.0 and 1.5 fold), phenylalanine (2.1 and 1.7 fold), and histidine (1.7 and 1.8 fold) seen at 1- and 24-hr respectively in the current study (Table 1) may be associated with the sensitivity as a result of potential protein damage. Branched-chain amino acids (BCAA) are the most hydrophobic amino acids and play an important role in formation of transmembrane proteins. Furthermore, BCAA make up 37% of the amino acids in lung surfactant protein B (Chou et al., 1973). The changes observed for BCAA (leucine 1.5 fold and isoleucine 1.4 fold) at 24-hr suggest an ongoing repair process of lipid membranes and surfactant in the airway. The increases we observed in arginine and ornithine at both 1 and 24 hr suggest increased urea cycle activity (Mori et al., 1998). Nitric oxide is a potent pro-inflammatory mediator produced by alveolar macrophages and has been shown to be reliant on arginine (Wolff et al., 1995; Inoue et al., 2000).
Altered energy-related metabolism
Exposure to air pollutants has been associated with perturbations of energy utilization. For example, epidemiological observation in nondiabetic humans found association between particulate matter exposures and increases in blood glucose ( Peng et al., 2016). Air pollutant exposure is also associated with impaired glucose and lipid metabolism in susceptible subpopulations such as people with diabetes (Yitshak et al., 2016). Animal studies show that acute ozone exposure leads to altered glucose and energy metabolism, global lipid metabolite alteration in circulation, and lung mitochondrial oxidation (Bass et al., 2013; Mustafa et al., 1974). Similar observation was reported where particulate matter exposure leads to glucose dysregulation and altered metabolic syndrome with the implication of diabetes in animal models (Zheng et al., 2013). In this study, we found that ozone perturbed glucose and lactate levels in the BAL samples, with elevations seen at 1- and 24-hr for glucose and 24 hr for lactate.
We observed ozone-induced changes in α-ketoglutarate, glutamate, and glutamine, suggesting impaired mitochondrial function. Prolonged ozone exposure leads to a decline in glucose uptake in ozone-exposed airway epithelial cells (Ahmad et al., 2005), which is consistent with the increased levels of glucose we found in the BAL fluid. Survival of ozone-exposed airway epithelial cells is associated with ATP release from the cells. Impaired glucose utilization and ATP release from ozone-exposed cells, potentially depleting intracellular stores of ATP, could activate alternative pathways for energy generation/metabolism. Glutamine is a conditionally essential amino acid which under catabolic stress scenarios such as injury or sepsis can contribute to the replenishment of TCA cycle through α-ketoglutarate (Altman et al., 2016; Shanware et al., 2014). In fact, proliferating cells with impaired mitochondrial metabolism actively import extracellular glutamine to generate pools of TCA cycle intermediates to support cell growth (Mullen et al., 2011). Glutamine supplementation protected mitochondrial function following oxidant insult in airway epithelial cells (Ahmad et al., 2001). Taken together, the increases in α-ketoglutarate, glutamate, and glutamine seen in our study after ozone exposure suggest that mitochondrial function may be impaired during ozone-induced oxidative stress and that elevated TCA cycle replenishment from alternative source is used to support cellular repair.
Glutathione is an important component of cellular antioxidant defenses and glutathione S-transferase Mu 1 (GSTM1) is a common human polymorphism associated with increased sensitivity to pulmonary effects of ozone (Alexis et al., 2009; Romieu et al., 2004). Among the subjects who participated in this study, 52% were GSTM1 null. Although sample numbers were small, GSTM1 null but not GSTM1 positive subjects had statistically significant elevations (p < 0.05) of glucose, glutamine, and threonate levels in BAL samples after ozone exposure. Threonate is a metabolite of ascorbic acid, and increased levels of threonate are consistent with a shift to ascorbate for oxidant defense given a deficit in the ability use glutathione. Similarly, the elevated levels of glucose and glutamine in GSTM1 null but not in the GSTM1 positive subjects are consistent with increased sensitivity to the oxidative stress caused by ozone ( Peng et al., 2016; Holloway et al., 2012; Zhang et al., 2014) (Supplementary Table 2).
Lipid metabolites
Fatty acids in lung surfactant and epithelial cell membranes are important targets for inhaled ozone. Ozone reacts with fatty acids to form relatively stable lipid peroxides. These can trigger signaling cascades through oxidant-related stress pathways and are also a specific form of cellular oxidative damage. We observed robust increases in phospholipids and their biosynthesis precursors at 1-hr, glycerophosphorylcholines at 24-hr and sphingolipids at 24-hr after ozone exposure. These changes are consistent with previous observations that lipid peroxidation products may act as a secondary messenger that helps to sustain the inflammatory response in the airway. Lung surfactant treated with ozone resulted in oxidized phospholipids which were associated with IL-8 release, impaired mitochondrial activity, and elevated cytotoxicity in airway epithelial and macrophage cells (Uhlson et al., 2002). Furthermore, sphingolipids were identified as important signaling mediators involved in inflammatory regulation associated with stress response and cellular survival in the respiratory airways. Plasmalogen lipids, a type of ether lipid with a vinyl ether linkage at the glycerol backbone, showed high reactivity toward many oxidants (Murphy et al., 2001) and demonstrated reactivity with ozone in lung surfactant (Wynalda et al., 2010). Plasmalogen lipids are associated with respiratory disease, serving as antioxidant and secondary messenger (Braverman et al., 2012), and were elevated at 24-hr in our study. Taken together, the significantly altered glycerophospholipid (1- and 24-hr), ether lipid (1-hr), and sphingolipid (24-hr) pathways suggest surfactant alteration leading to inflammation and potential lipid membrane turnover as a repair mechanism after ozone exposure.
Strength of Metabolomics analysis and
Since altered energy metabolism is one of the major finding derived from the KEGG analysis in the current study, IPA network analysis is consistent with the observed changes in energy-related metabolites. Parallel observations were made for mRNA and protein data with the same samples where many upstream signaling molecules associated with ozone exposure such as PI3K, MAPK, NFkB, and PKC were also observed in the IPA network analysis (data not shown). In future work, an integrated analysis of the mRNA, protein and metabolite data will be conducted. This integrated analysis will provide a more comprehensive understanding of the biological processes perturbed by ozone exposure.
Limitations of this study
A targeted, as opposed to an untargeted, analysis was conducted. While an untargeted analysis can identify thousands of metabolites for biomarker discovery, the targeted analysis employed in this study only quantified the 134 metabolites that could be positively identified. The targeted metabolomics approach may not have the capacity for novel metabolite discovery but instead provides a clearer representation of the altered physiological processes than the untargeted method (Roberts et al., 2012). Also, metabolite identification in the current study may have been limited due to the sample matrix. In comparison with other matrices (e.g. serum, urine, sweat), BAL samples represent relevant physiology for an exposure to ozone. However, sample acquisition involves an invasive procedure and introduction of exogenous fluid, possibly resulting in identification of fewer metabolites (Nobakht et al., 2015). The double-blind crossover design employed in the current study allowed each subject to serve as his or her own control and reduced potential bias. Also, damage to epithelial cells causes disruption in the tight junctions between epithelial cells, resulting in an influx of plasma components into the airways (Koren eet al., 1989). Metabolite changes observed in BAL samples may therefore reflect alterations in both airway and the blood.
Summary
The metabolite changes seen in this study are broadly consistent with the well characterized pulmonary toxicity of inhaled ozone mediated by oxidative stress, the associated stress response, and cellular repair. The clear separation in metabolite changes seen at 1- and 24-hr post exposure indicates the potential for metabolomics studies to support detailed mechanistic investigations of how the airway lining responds to and recovers from the damage caused by ozone exposure. In particular, the ability to track changes in energy-related metabolites such as glucose, glutamine, and α-ketoglutarate illustrates a capability of metabolomic studies that is not provided by upstream transcriptomic and proteomic studies. In future work, we plan to integrate this metabolomics analysis with transcriptomic and proteomic datasets from the same study in support of development of a systems-level quantitative model of ozone toxicity.
Supplementary Material
Acknowledgements
The authors are indebted to Ms. Maryann Bassett, Ms. Heidi Hiers, Ms. Tracey Montilla, Ms. Lisa Dailey, and Ms. Joleen Soukup for their expert medical and technical assistance in the execution of this study.
Funding Information:
This work was supported entirely by EPA intramural funds, since most of the authors are EPA employees.
Footnotes
Publisher's Disclaimer: Disclaimer: The research described in this article has been reviewed by the Environmental Protection Agency and approved for publication. The contents of this article do not necessarily represent Agency policy nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
References
- Ahmad S, Ahmad A, McConville G, Schneider BK, Allen CB, Manzer R, Mason RJ, White CW. Lung epithelial cells release ATP during ozone exposure: signaling for cell survival. Free Radic Biol Med 2005;39:213–226. [DOI] [PubMed] [Google Scholar]
- Ahmad S, White CW, Chang LY, Schneider BK, Allen CB. Glutamine protects mitochondrial structure and function in oxygen toxicity. Am J Physiol Lung Cell Mol Physiol 2001;280:L779–91. [DOI] [PubMed] [Google Scholar]
- Alexis NE, Zhou H, Lay JC, Harris B, Hernandez ML, Lu T-S, Bromberg PA, Diaz-Sanchez D, Devlin RB, Kleeberger SR, Peden DB. The glutathione-S-transferase Mu 1 null genotype modulates ozone-induced airway inflammation in human subjects. J Allergy Clin Immunol 2009;124:1222–1228.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer 2016;16:619–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass V, Gordon CJ, Jarema KA, MacPhail RC, Cascio WE, Phillips PM, Ledbetter AD, Schladweiler MC, Andrews D, Miller D, Doerfler DL, Kodavanti UP. Ozone induces glucose intolerance and systemic metabolic effects in young and aged Brown Norway rats. Toxicol Appl Pharmacol 2013;273:551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman NE, Moser AB. Functions of plasmalogen lipids in health and disease. Biochim Biophys Acta 2012;1822:1442–1452. [DOI] [PubMed] [Google Scholar]
- Bromberg PA. Mechanisms of the acute effects of inhaled ozone in humans. Biochim Biophys Acta 2016;1860:2771–2781. [DOI] [PubMed] [Google Scholar]
- Chen C, Arjomandi M, Balmes J, Tager I, Holland N. Effects of chronic and acute ozone exposure on lipid peroxidation and antioxidant capacity in healthy young adults. Environ Health Perspect 2007;115:1732–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou PY, Fasman GD. Structural and functional role of leucine residues in proteins. J Mol Biol 1973;74:263–281. [DOI] [PubMed] [Google Scholar]
- Cui L, Zheng D, Lee YH, Chan TK, Kumar Y, Ho WE, Chen JZ, Tannenbaum SR, Ong CN. Metabolomics Investigation Reveals Metabolite Mediators Associated with Acute Lung Injury and Repair in a Murine Model of Influenza Pneumonia. Sci Rep 2016;6:26076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin RB, Duncan KE, Jardim M, Schmitt MT, Rappold AG, Diaz-Sanchez D. Controlled exposure of healthy young volunteers to ozone causes cardiovascular effects. Circulation 2012;126:104–111. [DOI] [PubMed] [Google Scholar]
- Devlin RB, Raub JA, Folinsbee LJ. Health effects of ozone. Science and Medicine 1997;4:8–17. [Google Scholar]
- Evans AM, Bridgewater BR, Liu Q, Mitchell MW, Robinson RJ, Dai H, Stewart SJ, DeHaven CD, Miller L. High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics 2014;4:1. [Google Scholar]
- Garantziotis S, Li Z, Potts EN, Kimata K, Zhuo L, Morgan DL, Savani RC, Noble PW, Foster WM, Schwartz DA, Hollingsworth JW. Hyaluronan mediates ozone-induced airway hyperresponsiveness in mice. J Biol Chem 2016;291:19257–19258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghio AJ, Kim C, Devlin RB. Concentrated ambient air particles induce mild pulmonary inflammation in healthy human volunteers. Am J Respir Crit Care Med 2000;162:981–988. [DOI] [PubMed] [Google Scholar]
- Haque R, Umstead TM, Freeman WM, Phelps DS, Floros J. Quantitative Proteomic Profiles of BALF in Wild Type and SP-A KO Mice after Exposure to Ozone. The FASEB Journal 2007;21:A9. [Google Scholar]
- Holloway JW, Savarimuthu Francis S, Fong KM, Yang IA. Genomics and the respiratory effects of air pollution exposure. Respirology 2012;17:590–600. [DOI] [PubMed] [Google Scholar]
- Hori S, Nishiumi S, Kobayashi K, Shinohara M, Hatakeyama Y, Kotani Y, Hatano N, Maniwa Y, Nishio W, Bamba T, Fukusaki E, Azuma T, Takenawa T, Nishimura Y, Yoshida M. A metabolomic approach to lung cancer. Lung Cancer 2011;74:284–292. [DOI] [PubMed] [Google Scholar]
- Inoue H, Aizawa H, Nakano H, Matsumoto K, Kuwano K, Nadel JA, Hara N. Nitric oxide synthase inhibitors attenuate ozone-induced airway inflammation in guinea pigs. Possible role of interleukin-8. Am J Respir Crit Care Med 2000;161:249–256. [DOI] [PubMed] [Google Scholar]
- Johnson DA. Effects of ozone and nitrogen dioxide on human lung proteinase inhibitors. Res Rep Health Eff Inst 1987;5–25.at <https://www.ncbi.nlm.nih.gov/pubmed/3268287>. [PubMed] [Google Scholar]
- Kafoury RM, Pryor WA, Squadrito GL, Salgo MG, Zou X, Friedman M. Induction of inflammatory mediators in human airway epithelial cells by lipid ozonation products. Am J Respir Crit Care Med 1999;160:1934–1942. [DOI] [PubMed] [Google Scholar]
- Kierstein S, Krytska K, Sharma S, Amrani Y, Salmon M, Panettieri RA Jr, Zangrilli J, Haczku A. Ozone inhalation induces exacerbation of eosinophilic airway inflammation and hyperresponsiveness in allergen-sensitized mice. Allergy 2008;63:438–446. [DOI] [PubMed] [Google Scholar]
- Koren HS, Devlin RB, Graham DE, Mann R, McGee MP, Horstman DH, Kozumbo WJ, Becker S, House DE, McDonnell WF. Ozone-induced inflammation in the lower airways of human subjects. Am Rev Respir Dis 1989;139:407–415. [DOI] [PubMed] [Google Scholar]
- Leikauf GD, Simpson LG, Santrock J, Zhao Q, Abbinante-Nissen J, Zhou S, Driscoll KE. Airway epithelial cell responses to ozone injury. Environ Health Perspect 1995;103 Suppl 2:91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy P, Tham A, Wong H, Tenney R, Chen C, Stiner R, Balmes JR, Paquet AC, Arjomandi M. Inflammatory and repair pathways induced in human bronchoalveolar lavage cells with ozone inhalation. PLoS One 2015;10:e0127283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DB, Ghio AJ, Karoly ED, Bell LN, Snow SJ, Madden MC, Soukup J, Cascio WE, Gilmour MI, Kodavanti UP. Ozone Exposure Increases Circulating Stress Hormones and Lipid Metabolites in Humans. Am J Respir Crit Care Med 2016;193:1382–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DB, Karoly ED, Jones JC, Ward WO, Vallanat BD, Andrews DL, Schladweiler MC, Snow SJ, Bass VL, Richards JE, Ghio AJ, Cascio WE, Ledbetter AD, Kodavanti UP. Inhaled ozone (O3)-induces changes in serum metabolomic and liver transcriptomic profiles in rats. Toxicol Appl Pharmacol 2015;286:65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Gotoh T, Nagasaki A, Takiguchi M, Sonoki T. Regulation of the urea cycle enzyme genes in nitric oxide synthesis. J Inherit Metab Dis 1998;21 Suppl 1:59–71. [DOI] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen P-H, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 2011;481:385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RC. Free-radical-induced oxidation of arachidonoyl plasmalogen phospholipids: antioxidant mechanism and precursor pathway for bioactive eicosanoids. Chem Res Toxicol 2001;14:463–472. [DOI] [PubMed] [Google Scholar]
- Mustafa MG, Cross CE. Effects of short-term ozone exposure on lung mitochondrial oxidative and energy metabolism. Arch Biochem Biophys 1974;162:585–594. [DOI] [PubMed] [Google Scholar]
- Nikula KJ, Wilson DW, Giri SN, Plopper CG, Dungworth DL. The response of the rat tracheal epithelium to ozone exposure. Injury, adaptation, and repair. Am J Pathol 1988;131:373–384. [PMC free article] [PubMed] [Google Scholar]
- Nobakht M Gh BF, Aliannejad R, Rezaei-Tavirani M, Taheri S, Oskouie AA. The metabolomics of airway diseases, including COPD, asthma and cystic fibrosis. Biomarkers 2015;20:5–16. [DOI] [PubMed] [Google Scholar]
- Peng C, Bind M-AC, Colicino E, Kloog I, Byun H-M, Cantone L, Trevisi L, Zhong J, Brennan K, Dereix AE, Vokonas PS, Coull BA, Schwartz JD, Baccarelli AA. Particulate Air Pollution and Fasting Blood Glucose in Nondiabetic Individuals: Associations and Epigenetic Mediation in the Normative Aging Study, 2000-2011. Environ Health Perspect 2016;124:1715–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts LD, Souza AL, Gerszten RE, Clish CB. Targeted metabolomics. Curr Protoc Mol Biol 2012;Chapter 30:Unit 3021–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romieu I, Sienra-Monge JJ, Ramírez-Aguilar M, Moreno-Macías H, Reyes-Ruiz NI, Estela del Río-Navarro B, Hernández-Avila M, London SJ. Genetic polymorphism of GSTM1 and antioxidant supplementation influence lung function in relation to ozone exposure in asthmatic children in Mexico City. Thorax 2004;59:8–10. [PMC free article] [PubMed] [Google Scholar]
- Samet JM, Hatch GE, Horstman D, Steck-Scott S, Arab L, Bromberg PA, Levine M, McDonnell WF, Devlin RB. Effect of antioxidant supplementation on ozone-induced lung injury in human subjects. Am J Respir Crit Care Med 2001;164:819–825. [DOI] [PubMed] [Google Scholar]
- Shanware NP, Bray K, Eng CH, Wang F, Follettie M, Myers J, Fantin VR, Abraham RT. Glutamine deprivation stimulates mTOR-JNK-dependent chemokine secretion. Nat Commun 2014;5:4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VK, Graham NJD. Oxidation of Amino Acids, Peptides and Proteins by Ozone: A Review. Ozone: Sci Eng 2010;32:81–90. [Google Scholar]
- Stringer KA, Serkova NJ, Karnovsky A, Guire K, Paine R, Standiford TJ. Metabolic consequences of sepsis-induced acute lung injury revealed by plasma 1H-nuclear magnetic resonance quantitative metabolomics and computational analysis. American Journal of Physiology - Lung Cellular and Molecular Physiology 2011;300:L4–L11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uh S-T, Koo S-M, Jang AS, Park SW, Choi JS, Kim Y-H, Park CS. Proteomic differences with and without ozone-exposure in a smoking-induced emphysema lung model. Korean J Intern Med 2015;30:62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlson C, Harrison K, Allen CB, Ahmad S, White CW, Murphy RC. Oxidized phospholipids derived from ozone-treated lung surfactant extract reduce macrophage and epithelial cell viability. Chem Res Toxicol 2002;15:896–906. [DOI] [PubMed] [Google Scholar]
- Uppal K, Walker DI, Liu K, Li S, Go Y-M, Jones DP. Computational Metabolomics: A Framework for the Million Metabolome. Chem Res Toxicol 2016;29:1956–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US EPA National Center for Environmental Assessment, Research Triangle Park Nc, Environmental Media Assessment Group, Brown J. Integrated Science Assessment (ISA) of Ozone and Related Photochemical Oxidants (Final Report, Feb 2013). 2012;at <https://cfpub.epa.gov/ncea/isa/recordisplay.cfm?deid=247492>.
- van der Vliet A, O’Neill CA, Cross CE, Koostra JM, Volz WG, Halliwell B, Louie S. Determination of low-molecular-mass antioxidant concentrations in human respiratory tract lining fluids. Am J Physiol 1999;276:L289–96. [DOI] [PubMed] [Google Scholar]
- Ward-Caviness CK, Breitner S, Wolf K, Cyrys J, Kastenmüller G, Wang-Sattler R, Schneider A, Peters A. Short-term NO2 exposure is associated with long-chain fatty acids in prospective cohorts from Augsburg, Germany: results from an analysis of 138 metabolites and three exposures. Int J Epidemiol 2016;45:1528–1538. [DOI] [PubMed] [Google Scholar]
- Wattiez R, Noël-Georis I, Cruyt C, Broeckaert F, Bernard A, Falmagne P. Susceptibility to oxidative stress: proteomic analysis of bronchoalveolar lavage from ozone-sensitive and ozone-resistant strains of mice. Proteomics 2003;3:658–665. [DOI] [PubMed] [Google Scholar]
- Wolff DJ, Lubeskie A. Aminoguanidine is an isoform-selective, mechanism-based inactivator of nitric oxide synthase. Arch Biochem Biophys 1995;316:290–301. [DOI] [PubMed] [Google Scholar]
- Wu W, Peden D, Diaz-Sanchez D. Role of GSTM1 in resistance to lung inflammation. Free Radic Biol Med 2012;53:721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynalda KM, Murphy RC. Low-concentration ozone reacts with plasmalogen glycerophosphoethanolamine lipids in lung surfactant. Chem Res Toxicol 2010;23:108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yitshak Sade M, Kloog I, Liberty IF, Schwartz J, Novack V. The Association Between Air Pollution Exposure and Glucose and Lipids Levels. J Clin Endocrinol Metab 2016;101:2460–2467. [DOI] [PubMed] [Google Scholar]
- Zemski Berry KA, Murphy RC. Phospholipid Ozonation Products Activate the 5-Lipoxygenase Pathway in Macrophages. Chem Res Toxicol 2016;29:1355–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wu X, Xiao Y, Chen M, Li Z, Wei X, Tang K. Genetic polymorphisms of glutathione S-transferase M1 and T1, and evaluation of oxidative stress in patients with non-small cell lung cancer. Eur J Med Res 2014;19:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Xu X, Zhang X, Wang A, Zhang C, Hüttemann M, Grossman LI, Chen LC, Rajagopalan S, Sun Q, Zhang K. Exposure to ambient particulate matter induces a NASH-like phenotype and impairs hepatic glucose metabolism in an animal model. J Hepatol 2013;58:148–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


