Abstract
Members of the mint family (Lamiaceae) accumulate a wide variety of industrially and medicinally relevant diterpenes. We recently sequenced leaf transcriptomes from 48 phylogenetically diverse Lamiaceae species. Here, we summarize the available chemotaxonomic and enzyme activity data for diterpene synthases (diTPSs) in the Lamiaceae and leverage the new transcriptomes to explore the diTPS sequence and functional space. Candidate genes were selected with an intent to evenly sample the sequence homology space and to focus on species in which diTPS transcripts were found, yet from which no diterpene structures have been previously reported. We functionally characterized nine class II diTPSs and 10 class I diTPSs from 11 distinct plant species and found five class II activities, including two novel activities, as well as a spectrum of class I activities. Among the class II diTPSs, we identified a neo-cleroda-4(18),13E-dienyl diphosphate synthase from Ajuga reptans, catalyzing the likely first step in the biosynthesis of a variety of insect-antifeedant compounds. Among the class I diTPSs was a palustradiene synthase from Origanum majorana, leading to the discovery of specialized diterpenes in that species. Our results provide insights into the diversification of diterpene biosynthesis in the mint family and establish a comprehensive foundation for continued investigation of diterpene biosynthesis in the Lamiaceae.
Keywords: secondary metabolism, terpenoid, plant biochemistry, transcriptomics, biosynthesis, chemotaxonomy, diterpene synthase, enzymes, Lamiaceae, phytochemistry
Introduction
Diterpenoid specialized metabolites are widespread among plants but are particularly diverse and abundant in the Lamiaceae (mint) family. According to the Dictionary of Natural Products (DNP)4 (version 26.2) (1), more than 13,000 distinct diterpenes have been reported from plants, about 3,000 of those in at least one species from the Lamiaceae. In the Lamiaceae, the majority of diterpenes share a decalin core, characteristic of labdane-related diterpenes. In angiosperms, biosynthesis of labdane-related diterpenes starts with the action of a class II diterpene synthase (diTPS) from the TPS-c subfamily (2, 3), which catalyzes the conversion of the central precursor, geranylgeranyl diphosphate (GGPP), into a bicyclic prenyl diphosphate intermediate (e.g. copalyl diphosphate (CPP)). A class I diTPS from the TPS-e subfamily then acts to remove the diphosphate moiety and form additional rings, double bonds, or hydroxyl groups. TPS-c and TPS-e enzymes are ubiquitous in angiosperms, catalyzing the first two steps in the gibberellin phytohormone biosynthesis pathway, the conversion of GGPP to ent-CPP and of ent-CPP to ent-kaurene, respectively. Labdane-related diterpene-specialized metabolites may arise from alternative decoration of ent-kaurene after the diTPS-catalyzed reactions, from gene duplication and neo-functionalization of ent-CPP and ent-kaurene synthases (4), or from further functional diversification of diTPSs already involved in specialized metabolism. Most class I diTPSs can act on multiple class II products, leading to an increase in the total number of distinct products that can be formed through different combinations of class II and class I enzymes (5). Together, the diTPSs give shape to the diterpene skeleton, which can then undergo further modification by cytochromes P450, acyl transferases, or other enzymes (6–10).
Previous investigations into diterpenoid biosynthetic pathways in the Lamiaceae have focused on medicinal diterpenes, such as the cAMP booster forskolin, from Plectranthus barbatus (syn. Coleus forskohlii); the tanshinones, from Salvia miltiorrhiza, which have many uses in Chinese traditional medicine; the dopaminergic vitexilactone from Vitex agnus-castus; the potential anti-diabetic and vasorelaxant marrubiin, from Marrubium vulgare; and the potent hallucinogen salvinorin A from Salvia divinorum (8, 11–17). Other research was motivated by the industrial value of diterpenes such as sclareol, from Salvia sclarea, which can be used in the semisynthesis of the commodity chemical Ambrox and antioxidant carnosic acid (18–20).
Recently, we made available leaf transcriptomes of 48 species from the Lamiaceae (21). In the present work, we performed a detailed analysis of the available chemotaxonomic and enzyme function data from the Lamiaceae, showing hundreds of diterpene skeletons that could not be accounted for by known enzymes. We therefore saw an opportunity to mine for diTPSs with previously unknown activities. Using homology searches with known diTPSs from mints (Data set S1), we identified a total of 163 candidate diTPSs from the new transcriptomes (Data set S2). By combining and cross-referencing the transcriptome data, chemotaxonomic data, and earlier enzyme data, we narrowed down the list of candidates to select genes with minimal homology to known enzymes, genes from species where the reported diterpenes could not be explained by known activities, and genes from species where no diterpenes have been reported, but where the transcriptome data show an enlargement of the TPS-c or TPS-e gene families.
We report nine class II diTPSs accounting for five diphosphate intermediates and 10 class I diTPSs accounting for a wide variety of additional products. Some of the new enzymes give access to intermediates that were previously difficult or impossible to produce biosynthetically. Specifically, we identified Ajuga reptans ArTPS2 as producing neo-cleroda-4(18),13E-dienyl diphosphate, the likely precursor to a wide variety of bioactive diterpenoids, including antifeedants against insects (22–24). Another class II diTPS, Pogostemon cablin PcTPS1, was found to catalyze the formation of (10R)-labda-8,13E-dienyl diphosphate, a likely precursor to an entire suite of diterpenes from the Lamioideae clade within the Lamiaceae. Further, in Origanum majorana, a culinary herb without reported diterpene accumulation, the characterization of multiple new diTPSs led us to the discovery of some of the corresponding diterpenes in planta. We anticipate that this work will serve as a new foundation for continued discovery of diTPSs in the Lamiaceae. To this end, we have endeavored to make our analyses and results, including gene sequences, raw and processed spectroscopic data, code, and extensive data tables, available in human- and machine-readable formats to be used, adapted, and extended by ourselves and other researchers in the future.
Results
Estimating the diversity of diterpenoids in the Lamiaceae
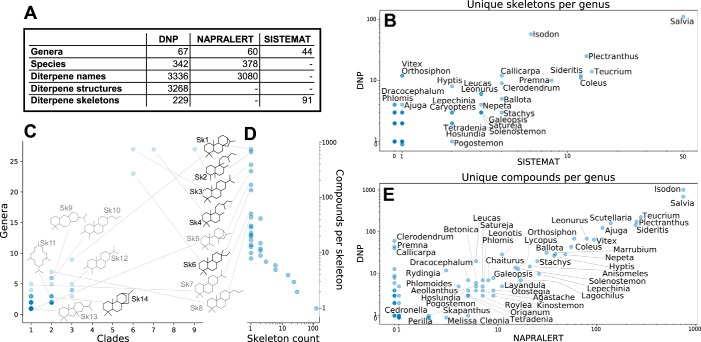
To help determine the most promising species to find previously unknown diTPS activities, it was necessary to compile a data set of diterpene occurrence in Lamiaceae species and a data set of functionally characterized diTPS genes from the Lamiaceae. Information about diterpene occurrence was collected from three sources: SISTEMAT, DNP, and NAPRALERT. SISTEMAT (25) contains Lamiaceae diterpenes reported up to 1997, including 91 unique carbon skeletons (the core alkanes, disregarding all desaturation, acyl-side chains, heteroatoms, and stereochemistry) from 295 species and 51 genera. We were unable to obtain an electronic copy of SISTEMAT, so we reconstructed it based on the figures and tables in the paper.
The DNP (1) includes a wealth of information on diterpenes from the Lamiaceae, including full structures and the species where those structures have been reported. NAPRALERT (26) identifies compounds by their common name rather than their structure or skeleton, associates the compound names to genus and species names, and gives various other information, such as the tissue where the compound was found.
To enable comparison among the databases and cross-referencing with transcriptome and enzyme data, all genus and species names were converted into TaxIDs from the NCBI Taxonomy database (27). To put structure occurrence into clearer evolutionary context, we annotated each genus as a member of one of the 12 primary, monophyletic clades that form the backbone of Lamiaceae, as delineated by Li et al. (28) on the basis of chloroplast genome sequence. In the context of diTPSs, examination of skeletons can be helpful because the skeleton often resembles the diTPS product more obviously than a highly decorated downstream product would. We therefore extracted the skeletons from the DNP structures (Fig. S1 shows a graphical example of skeleton extraction). We assigned each skeleton according its reported presence in the 12 clades, as well as to genera, to help distinguish skeletons that may have arisen early in Lamiaceae evolution versus those that arose more recently. A full tabulation of the skeletons from SISTEMAT and DNP can be found as Data set S3.
The three databases were relatively consistent in their estimations of the diversity and distribution of diterpenes and diterpene skeletons (Fig. 1, A, B, and E). The data are snapshots of reports in the literature and certainly do not comprehensively reflect the true chemical diversity existing in nature. Some genera, such as Salvia, may be overrepresented due to their large number of species or their use in traditional medicine.
Figure 1.
Distribution of diterpenes in the Lamiaceae. A, comparison of different sources for data about Lamiaceae diterpene chemotaxonomy. B, diterpene skeletons per genus according to both the DNP and SISTEMAT. C, distribution of skeletons among the genera and 12 primary clades of Lamiaceae, based on the DNP. A circle in C represents one skeleton, with its vertical position indicating how many genera that skeleton has been reported in and its horizontal position indicating how many of the 12 clades are represented by those genera. Structures are shown for selected skeletons; in black are those where a biosynthetic route is known from the Lamiaceae, and in gray are those for which the pathway remains unknown. D, distribution of compounds among skeletons, based on the DNP. Each circle in D represents a number of skeletons indicated by the horizontal position of the circle, with vertical position indicating the number of compounds reported with each of those skeletons; the data point in the bottom right shows that there are more than 100 skeletons that are represented by only one compound apiece. E, diterpene structures per genus according to both the DNP and NAPRALERT. Data points in B–E are represented by semitransparent circles, so darker spots indicate overlapping data points. Some genus name labels in B and E have been omitted due to space constraints. An exhaustive list of the occurrence of skeletons in genera of Lamiaceae is given in Data set S3, skeleton distribution.
A total of 239 skeletons are represented, with five (the kaurane (Sk1), clerodane (Sk2), abietane (Sk3), labdane (Sk4), and pimarane (Sk6)) being, by far, the most widely distributed and accounting for most of the total structures (Fig. 1, C and D). The clerodane skeleton, for example, has the widest distribution, having been reported in 27 genera representing 9 of the 12 primary clades, absent only in Tectona and two clades from which no diterpenes have yet been reported. The large number of less common, taxonomically restricted skeletons, including over 100 skeletons with only one associated compound (Fig. 1D), suggests that new diterpene synthase activities are continuously and independently arising across the Lamiaceae family tree. Therefore, a search across many species and genera should be a good strategy for finding diterpene synthases with new activities.
Identifying candidate diterpene synthase genes
Through a comprehensive literature search, we built a reference set of known Lamiaceae diTPSs and their activities. 54 functional diTPSs have been reported in this family, corresponding to 30 class II and 24 class I enzymes (Data set S1). Combinations of these diterpene synthases account for 27 distinct products accounting for six different skeletons, the five widely distributed skeletons (Sk1–4 and Sk6) as well as the less common atisane (Sk14) skeleton. This leaves 233 skeletons for which the biosynthetic route remains unknown. It is important to note that a single skeleton can correspond to multiple diTPS products, distinguished through stereochemical configuration, position of double bonds, or functionalization, so there is also a possibility of finding new diTPS activities for skeletons already accounted for by known enzymes.
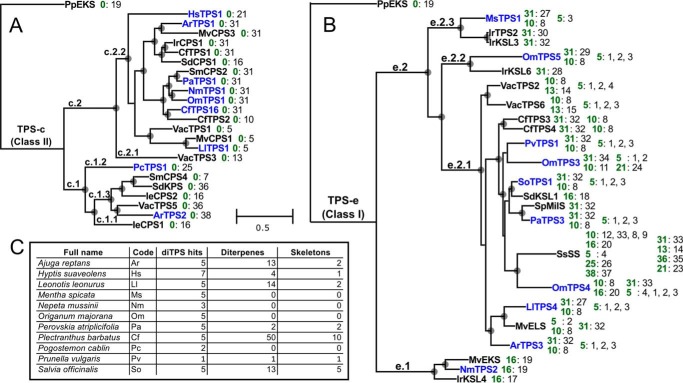
We used BLAST searches (29) with the list of known Lamiaceae diTPSs as query sequences to mine the 48 new Lamiaceae leaf transcriptomes (21) for candidate diTPSs. A total of 163 candidate sequences met our search criteria. The count of diTPS candidates was cross-referenced to the count of diterpenes and diterpene skeletons reported from each species and genus (Fig. 2C and Data set S2). Finally, a phylogenetic tree was generated from the peptide sequences from the reference set, alongside those from the new transcriptome data, including established substrates and products for each enzyme (Fig. 2 (A and B) and Fig. S20). We selected candidate genes from species, such as Mentha spicata and O. majorana, where the transcriptome data showed multiple candidate diTPSs but where few or no diterpene structures have been reported. We also selected genes that had relatively low homology to known enzymes, as judged by visual inspection of the phylogenetic trees, and in this way attempted to evenly cover the sequence homology space. Finally, we chose a few candidates from P. barbatus and Salva officinalis, based on the great diversity of diterpenes that have been reported from their genera.
Figure 2.
Maximum likelihood trees of newly characterized (blue) class II (A), and class I (B) diTPS enzymes in the context of selected previously reported (black) diTPSs from the Lamiaceae. The bifunctional ent-kaurene synthase from P. patens is used as an outgroup. After each enzyme are listed the experimentally verified substrates (green) and their products, numbers corresponding to compound numbers in Fig. 3. The scale bar applies to both trees, and units are substitutions per site. Circles at branch points indicate bootstrap support of at least 75%. C, all of the species we chose to clone diTPSs from, their total number of diTPS candidate sequences, and the number of unique diterpene structures and skeletons for those species, based on the DNP. Abbreviations for species not listed in C are as follows. Ie, Isodon eriocalyx; Ir, Isodon rubescens; Mv, Marrubium vulgare; Sd, Salvia divinorum; Sm, Salvia miltiorrhiza; Sp, Salvia pomifera; Ss, Salvia sclarea; Vac, Vitex agnus-castus.
Characterization of class II diTPSs
Fig. 3 presents a summary of Lamiaceae diTPS activities reported from previous work, together with our newly characterized diTPS activities. Class II activities were established based on comparisons of extracts from Nicotiana benthamiana transiently expressing new genes with those expressing known diTPS combinations. Class II diTPS products retain the diphosphate group from the GGPP substrate. When expressed in vivo, whether in Escherichia coli or N. benthamiana, without a compatible class I diTPS, a diphosphate product degrades to the corresponding alcohol, presumably by the action of nonspecific endogenous phosphatases. Due to difficulties in purifying and structurally characterizing diphosphate class II products, it is customary in the field to instead characterize these derivative alcohols (14, 17), which is the approach we have taken. For clarity, we indicate the alcohol by appending an “a” to the compound number; for example, 16a refers to ent-copalol.
Figure 3.
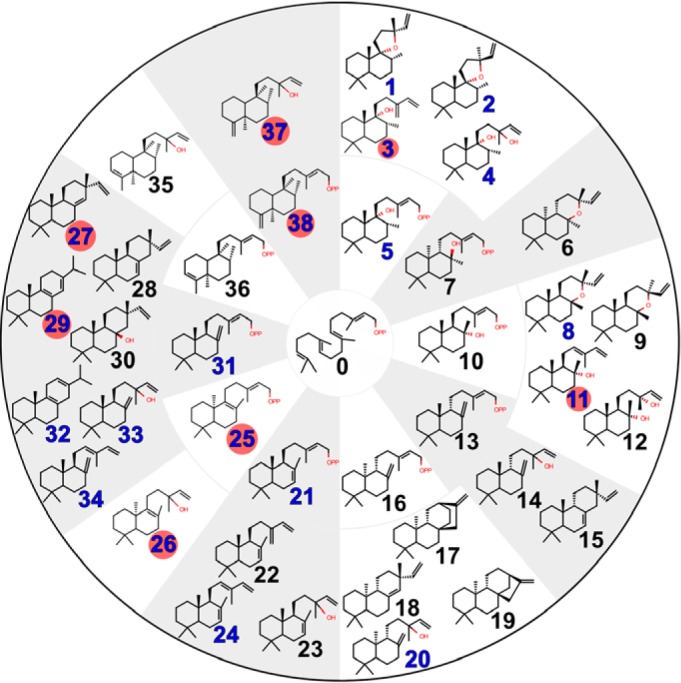
Reported products of diterpene synthases from the Lamiaceae. Blue numbers, compounds experimentally verified to be products of new enzymes from this study. Red circle, compounds that previously were inaccessible biosynthetically or were accessible only as minor components of multiproduct enzymes but are the single product of a newly characterized enzyme. At the center is GGPP, a precursor to all of these diterpenes. Inner ring, class II products; outer ring, class I products derived from the compound in the connected segment of the inner ring.
ArTPS1, PaTPS1, NmTPS1, OmTPS1, and CfTPS16 were identified as (+)-CPP (31) synthases by comparison with products of P. barbatus (syn. C. forskohlii) CfTPS1, and the reference combination of CfTPS1 combined with CfTPS3, yielding miltiradiene (30) (Fig. S7). LlTPS1 was identified as a peregrinol diphosphate (PgPP) (5) synthase based on a comparison of products with M. vulgare MvCPS1 (15), and MvCPS1 combined with M. vulgare 9,13-epoxylabdene synthase (MvELS) (Fig. S5B) and S. sclarea sclareol synthase (SsSS) (Fig. S5D) (31).
HsTPS1 was identified as a (5S,9S,10S)-labda-7,13E-dienyl diphosphate (21) synthase based on comparison with the product of an enzyme from Grindelia robusta, GrTPS2 (32) (Fig. S8B) and by NMR of the alcohol derivative (21a) (Fig. S11). Normal absolute stereochemistry was assigned to the HsTPS1 product based on the optical rotation of 21a, [α]D +8.3° (c. 0.0007, CHCl3) (c.f. lit. [α]D +5°, c. 1.0, CHCl3 (33); [α]D25 +12°, c. 0.69, CHCl3 (34)). When HsTPS1 was expressed in N. benthamiana, we also noticed the formation of labda-7,13(16),14-triene (22) (Fig. S12), which seemed to be enhanced by co-expression with CfTPS3 (Fig. S8B). The combination of HsTPS1 with OmTPS3 produced labda-7,12E,14-triene (24) (Fig. S13) (35), both in N. benthamiana and in vitro (Figs. S8B and S9A), which has previously been accessible only by combinations of bacterial enzymes (36). Labdanes with a double bond at the 7-position have not been reported in Hyptis suaveolens and do not seem to be common in the Lamiaceae. Of nine compounds with the labdane skeleton and a double bond at 7 (Fig. 4), only one was from the same clade as H. suaveolens. (13E)-ent-labda-7,13-dien-15-oic acid, from Isodon scoparius (37), has the opposite absolute stereochemistry to the HsTPS1 product, likely not deriving from a paralog of HsTPS1 because the absolute stereochemistry of labdane skeletons is not known to change after the diTPS steps.
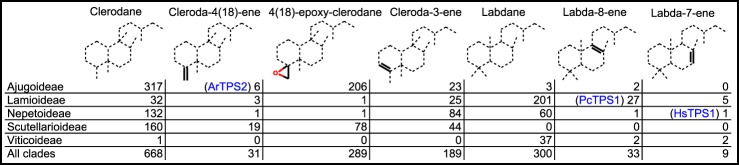
Figure 4.
Distribution among selected Lamiaceae clades of diterpenes with various structural patterns. Blue enzyme names are placed according to the pattern they install and the clade of the species they were cloned from. A solid line indicates that only compounds with the bond type shown at that position are counted. A dashed line indicates that all types of bonds and substituents are counted at that position. Based on data from the DNP (1).
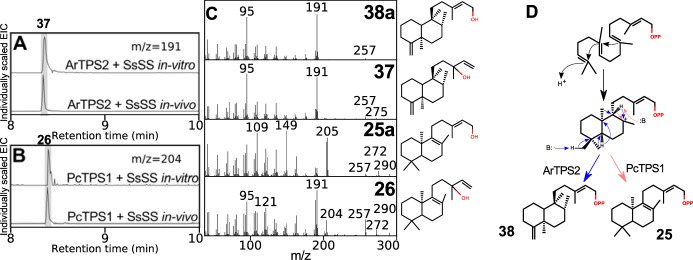
ArTPS2 was identified as a (5R,8R,9S,10R)-neo-cleroda-4(18),13E-dienyl diphosphate (38) synthase. The combination of ArTPS2 and SsSS generated neo-cleroda-4(18),14-dien-13-ol (37) (Fig. 5A). The structures of 37 and 38a were determined by NMR (Figs. S17 and S18), including the comparison of 37 with chelodane (38), which, based on small differences in 13C shifts, may be a stereoisomer at the 13 position, and the comparison of 38a with NMR of its enantiomer (39). Carbon 20 to 19, and 20 to 17 NOE interactions in 37 and 38a (Figs. S17G and S18F) closely resembled those reported for (−)-kolavelol (36a) (17), suggesting (5R,8R,9S,10R) relative stereochemistry. The “neo” absolute configuration was established through optical rotation of 38a, [α]D +30° (c. 0.0025, CHCl3) (c.f. lit. [α]D +20.9°, c. 0.7, CHCl3) (40). Previously reported clerodane diTPSs from the Lamiaceae produce kolavenyl diphosphate (36) (14, 16, 17), which has a double bond at the 3-position. Clerodanes with desaturation at 3 are spread throughout multiple clades but are most common in Nepetoideae (Fig. 4), which includes S. divinorum, one source of a kolavenyl diphosphate synthase. A plausible cyclization mechanism for ArTPS2 can readily be proposed (Fig. 5D). Clerodanes with a double bond at the 4(18) position are rare by comparison, but those with a 4(18)-epoxy moiety make up nearly half of the clerodanes reported in the Lamiaceae, including two-thirds of those reported from the Ajugoideae clade (Fig. 4). One such clerodane is clerodin (41), from which the clerodane skeleton gets its name. Neo-cleroda-4(18),13E-dienyl diphosphate is a logical biosynthetic precursor for the 4(18)-epoxy clerodanes, as we are not aware of any diTPSs that directly produce an epoxide moiety.
Figure 5.
Activities of ArTPS2 and PcTPS1. A and B, GC-MS–extracted ion chromatograms of activity assays for ArTPS2 + SsSS (A) and PcTPS1 + SsSS (B), in vitro with purified protein fed with GGPP and in vivo from N. benthamiana transiently expressing the gene combinations. C, mass spectra for the products of ArTPS2 and PcTPS1, and their combinations with SsSS. D, proposed mechanisms for ArTPS2 (blue) and PcTPS1 (pink).
PcTPS1 was identified as a (10R)-labda-8,13E-dienyl diphosphate (25) synthase. The structure was established by comparison of 13C NMR of 25a with previously reported spectra (34) (Fig. S14). The 10R (ent-) absolute stereochemistry was established by optical rotation of 25a [α]D −64° (c. 0.0008, CHCl3), (c.f. lit. [α]D25 −71.2°, c. 1.11, CHCl3) (42). The combination of PcTPS1 and SsSS, both in vitro, and in N. benthamiana expression produced (10R)-labda-8,14-en-13-ol (26) (Fig. 5B). The structure was determined by comparison of 13C NMR with a published spectrum (43) (Fig. S15G). We can propose a plausible mechanism for PcTPS1 activity (Fig. 5D). The double bond between positions 8 and 9 is present in 33 distinct compounds isolated from the Lamiaceae (Fig. 4). Most occur in the Lamioideae clade, which includes P. cablin, the source of PcTPS1. Absolute stereochemistries of the reported compounds are mixed, with some in the normal configuration (44) and others in the ent-configuration (45). As normal configuration 9-hydroxy labdanes are also abundant in Lamioideae, it is possible that the normal configuration 8(9) desaturated labdanes arise from dehydratase activities downstream of a PgPP synthase (MvCPS1 and its paralogs), whereas those in the ent-configuration arise from paralogs of PcTPS1. Another possibility is that some of the 8(9) desaturated labdanes reported as having normal absolute stereochemistry are actually ent-labdanes that were misassigned, as has occurred in at least one documented case (45).
Characterization of class I diTPSs
Class I diTPS candidates were characterized by transient expression in N. benthamiana in combination with four class II enzymes: CfTPS1, a (+)-CPP (31) synthase; CfTPS2, a labda-13-en-8-ol diphosphate ((+)-8-LPP) (10) synthase (30); LlTPS1, a PgPP (5) synthase; or Zea mays ZmAN2, an ent-CPP (16) synthase (46) (GenBankTM accession number AY562491). Substrates accepted by each enzyme and the products are indicated in Figs. 2B and 6, and GC-MS chromatograms of all combinations tested can be found as Figs. S3–S6.
Figure 6.
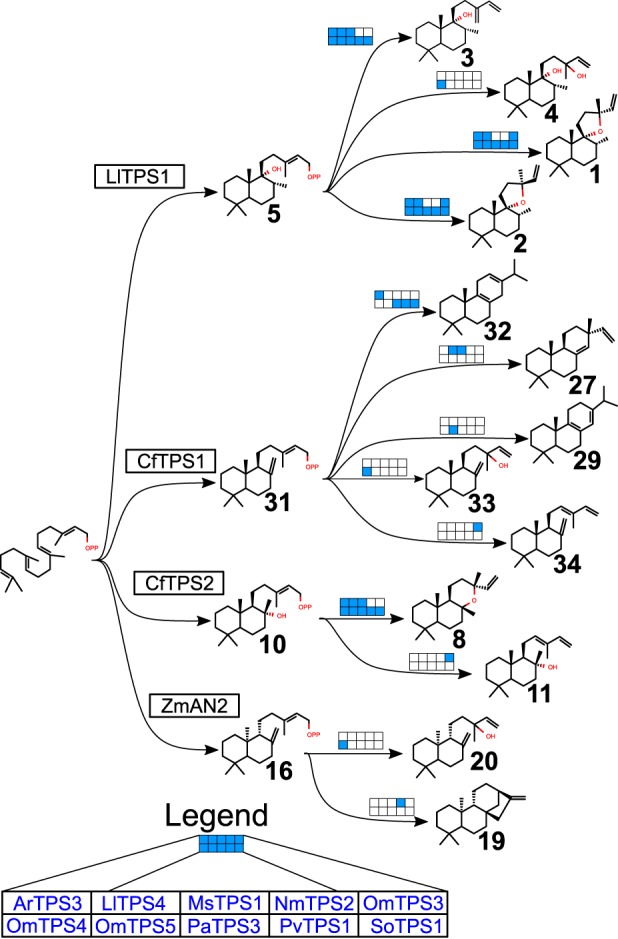
Activities of new class I diTPSs. Filled-in blue boxes indicate which enzymes are capable of each conversion. A diagram including new class II enzymes is available as Fig. S2.
NmTPS2 was identified as an ent-kaurene (19) synthase, converting ent-CPP into ent-kaurene (identified using Physcomitrella patens extract as a standard (47)) but not showing activity with any other substrate. The only other enzyme to show activity with ent-CPP was OmTPS4, which produced ent-manool (20), just as SsSS produces from ent-CPP (31).
PaTPS3, PvTPS1, SoTPS1, ArTPS3, OmTPS4, LlTPS4, OmTPS5, and MsTPS1 converted (+)-8-LPP to 13R-(+)-manoyl oxide (8), verified by comparison with the product of CfTPS2 and CfTPS3 (30). OmTPS3 produced trans-abienol (11), both in vitro and in N. benthamiana (Figs. S4E and S9D). The trans-abienol structure was determined by NMR (Fig. S10), with the configuration of the 12(13) double bond supported by comparison of the NOESY spectrum with that of a commercial standard for cis-abienol (Toronto Research Chemicals, Toronto, Canada). The product from CfTPS2 with OmTPS3 showed clear NOE correlations between positions 16 and 11 (Fig. S10F), whereas the cis-abienol standard showed correlations between 14 and 11 (Fig. S19). Trans-abienol is an alternative precursor to sclareol for semisynthesis of ambroxides, valuable amber odorants in the fragrance industry.
PaTPS3, PvTPS1, SoTPS1, and ArTPS3, LlTPS4, and OmTPS5 converted PgPP to a combination of 1, 2, and 3, with some variation in the ratios between the products. Because perigrinol (5a) spontaneously degrades into 1, 2, and 3 under GC conditions (15), it was difficult to distinguish whether these enzymes have low activity, but specific products, or moderate activity with a mix of products. Nevertheless, differences in relative amounts of the products observed between LlTPS1 alone and in combination with these class I enzymes suggest that they do have some activity on PgPP. OmTPS4 produced 1, 2, 3, and 4. MsTPS1 produced only 3, and OmTPS3 produced only 1 and 2. PgPP products were established by comparison with MvCPS1, MvCPS1 with MvELS (15), and MvCPS1 with SsSS (31).
PaTPS3, PvTPS1, SoTPS1, and ArTPS3 converted (+)-CPP to miltiradiene (32), similarly to CfTPS3. OmTPS4 produced manool (33), as compared with SsSS. LlTPS4 and MsTPS1 produced sandaracopimaradiene (27), by comparison with a product from Euphorbia peplus EpTPS8 (48). OmTPS5 produced palustradiene (29), both in vitro and in N. benthamiana, as compared with a minor product from Abies grandis abietadiene synthase (GenBankTM accession number U50768) (49) (Figs. S3A and S9E). Finally, OmTPS3 produced trans-biformene (34) in vitro and in N. benthamiana (Figs. S3A and S9C), as established by comparison of 13C NMR with the previously reported reference compound (50) (Fig. S16G), with trans-configuration further supported by clear NOE correlations between carbon 16 and 11 and the absence of NOE correlations between 14 and 11 (Fig. S16F).
O. majorana accumulates palustradiene and other diterpenoids
The class I enzymes from O. majorana, OmTPS3, OmTPS4, and OmTPS5, all produced different products from (+)-CPP, which itself is the product of OmTPS1, from the same species. Despite the apparent richness of diterpene synthase activities of enzymes from O. majorana, we did not find any reports of diterpenes from that species either in our database searches (Fig. 2C) or in a subsequent literature search. To determine whether diterpene synthases are active in O. majorana, we looked for the products of the enzyme combinations with extracts from leaf, stem, calyx, corolla, and root. We detected palustradiene (29), the product of OmTPS1 and OmTPS5, in all tissues except roots (Fig. 7). In addition, we detected two diterpene alcohols in the stem, leaf, and calyx. One diterpene alcohol we could not identify, but the other was a close match to a reference spectrum for palustrinol, the 19-hydroxy derivative of palustradiene.
Figure 7.
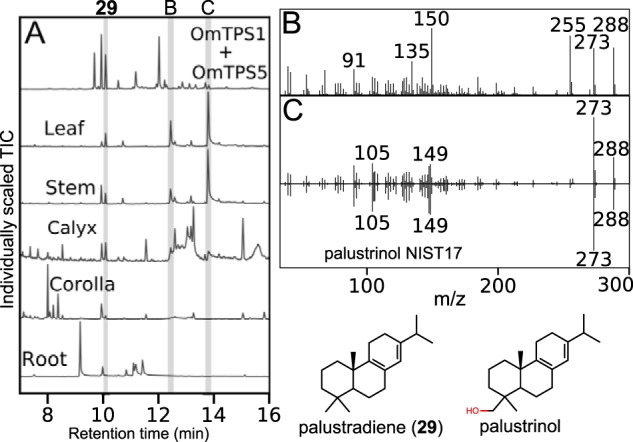
Detection of diterpenoids in O. majorana tissues. A, GC-MS total ion chromatograms of extract from N. benthamiana expressing OmTPS1 and OmTPS5, compared with extracts from various tissues of O. majorana. B, the mass spectrum of peak B, from O. majorana leaf. C, the mass spectrum of peak C from O. majorana leaf compared with reference spectrum for palustrinol from the NIST17 library.
Discussion
Diversification of diterpene synthases in the Lamiaceae
Due to an increase in resolution at the taxonomic level and consistent clustering of enzymes with identical or related function, we propose a hierarchical scheme for classifying TPS genes in the Lamiaceae from the TPS-e and TPS-c subfamilies. TPS-c genes (class II diTPSs) from the Lamiaceae fall broadly into two clades (Fig. 2A and Fig. S20), which we have called c.1 and c.2, further divided into three and two subclades, respectively. The characterized genes from c.1.1 are all ent-CPP (16) synthases, presumably involved in gibberellin biosynthesis. The taxonomic organization among c.1.1 sequences closely resembles the consensus phylogeny generated from 520 genes from each species (21), which, together with the short branch lengths compared with other TPS-c clades, suggests that diTPSs in c.1.1 are highly conserved and evolve slowly. All three of the previously reported enzymes and 14 of 17 new candidates from c.1.1 contained a conserved histidine, which has been implicated in Mg2+-mediated inhibition of TPS-c enzymes involved in primary metabolism (Fig. S21) (51). The conserved histidine is also present in most enzymes in c.1.2 and 1.3, including PcTPS1, and the kolavenyl diphosphate synthases from S. divinorum and V. agnus-castus, demonstrating that it can also be present in enzymes of specialized metabolism (Fig. S21).
The remaining TPS-c clades contain genes involved in specialized metabolism. The only characterized gene from clade c.1.2 is PcTPS1, which makes an ent-labda-8,13E-dienyl diphosphate product (25). Enzymes from clade c.1.3 catalyze the formation of a variety of products, including ent-CPP (52), ent-8-LPP (7) (11), kolavenyl-PP (36), and neo-cleroda-4(18),13E-dienyl diphosphate (38). 36 and 38 are the only products without the labdane (Sk4) skeleton produced by Lamiaceae class II diTPSs. Compounds apparently derived from 36 are widespread among Lamiaceae (Fig. 4), so it is tempting to hypothesize that the progenitor of c.1.3 was a kolavenyl-PP synthase present in an early common ancestor. A histidine residue previously identified as important for coordination of water in the active site of ent-CPP synthases (53), but mutated to an aromatic amino acid in clerodenyl diphosphate synthases (17, 54, 55), was found as a histidine in all c.1.1 enzymes but as phenylalanine in ArTPS2 and PcTPS1 (Fig. S21). In ent-CPP synthases, the water acts as a base to abstract a proton from C-17, leading to the formation of a double bond between C-8 and C-17. The proposed mechanisms for ArTPS2 and PcTPS1 (Fig. 5) require proton abstraction at the C-18 and C-9 positions, respectively, and are consistent with disruption of the coordination of water near C-17 by mutation of the histidine into an aromatic amino acid.
The labdane compounds produced by enzymes in c.1 are all in the ent-configuration. On the other hand, with two exceptions, the known enzymes from clade c.2 all make products with the labdane skeleton in the normal configuration, suggesting that the founder of that clade may have been a normal configuration labdadiene diphosphate synthase. The exceptions are VacTPS3 (14), the only characterized member of c.2.1, which produces syn-CPP (13), and the curious case of SdCPS1 (17), which produces ent-CPP. Consistent with recent results regarding stereocontrol in Lamiaceae TPS-c enzymes (56), most of the enzymes in c.2 feature aromatic amino acids and histidine (Fig. S21), in place of the conserved histidine and arginine, respectively, of c.1.1.
Among TPS-e (class I) genes, all but one of the characterized enzymes from e.1 are ent-kaurene (19) synthases, presumably involved in gibberellin biosynthesis. As with the c.1.1 clade, e.1 reflects the taxonomic distribution among the species. Notable in e.1 are IrKSL4 (57), which is an ent-atiserene synthase, and SmKSL2 (11), which, in addition to ent-kaurene synthase activity, can convert ent-8-LPP 7 into ent-13-epi-manoyl oxide (6). In recent work (48, 58), ent-kaurene synthases from a broad range of species were found to have the ability to convert 7 to 6, so it is likely that this is a general characteristic of all enzymes in the e.1 group. Conserved leucine and isoleucine residues previously implicated in ent-kaurene synthase activities (59, 60) were found in all but three of the new candidate sequences from e.1 (Fig. S22).
Most of the specialized class I diTPSs in the Lamiaceae fall into clade e.2. Enzymes in e.2 have lost the γ domain, present in many diTPSs and located on the opposite end of the peptide from the class I active site (57, 61). Characteristic of enzymes in e.2 is their ability to act on multiple substrates. The extreme example is SsSS (18), which so far has been able to catalyze the dephosphorylation and minor rearrangement of all class II enzyme products that it has been tested with (31, 48). The range of substrates accepted by other enzymes in this group has not been tested systematically, but among the e.2 enzymes characterized in this study, only one (OmTPS4) accepted ent-CPP, and all accepted (+)-CPP (31), (+)-8-LPP (10), and PgPP (5). There is great diversity among the products of e.2 enzymes, with over 20 distinct compounds represented. Most of the enzymes in e.2 convert (+)-CPP to miltiradiene (32) and (+)-8-LPP to 13R-(+)-manoyl oxide (8), with other activities arising sporadically across the clade. Both characterized enzymes in the Nepetoideae-specific e.2.2 have unusual activities; IrKSL6 converts (+)-CPP to isopimara-7,15-diene (28) (57), and OmTPS5 converts (+)-CPP to palustradiene (29). Most of the enzymes in e.2 fall into the e.2.1 clade, which also accounts for most of the known products. Enzymes that we characterized from e.2.1 lent support to emerging functionally consistent subclades. OmTPS4 activity, for three of four substrates tested, mimics that of its nearest homolog (SsSS), notably accepting ent-CPP as a substrate to produce ent-manool (20). LlTPS4 likewise has activities most similar to its closest homolog, MvELS (15), converting PgPP into 9,13(S)-epoxy-labd-14-ene (2) with greater specificity than other enzymes tested, although the products from (+)-CPP are different. From the remaining clade, e.2.3, the three characterized enzymes all come from Nepetoideae and convert (+)-CPP into different products: IrKSL3 produces miltiradiene (57), IrTPS2 produces nezukol (30) (62), and MsTPS1 produces sandaracopimaradiene (27). As noted earlier (63), the known activity-determining residues are almost completely replaced in e.2 compared with e.1. Superficially, there seems to be a correlation between differences in this region and differences in product specificity among e.2 enzymes (Fig. S22), but detailed mutational studies will be needed to assess the importance of individual residue switches.
The existence of two strongly supported subclades of specialized diTPSs within c.1, together with the presence of an ent-atiserene synthase in e.1, suggest that the emergence of specialized diTPSs from the ent-CPP and ent-kaurene synthases of gibberellin biosynthesis is an ongoing process that has occurred multiple times in the Lamiaceae lineage. Whereas it is evident that candidates selected from anywhere in the phylogenetic tree may have novel activities, clades that seem particularly promising and underexplored are c.2.1, c.1.2, and e.2.3. So far, including this work and previous work, diTPSs have been characterized from only four of the 12 major Lamiaceae clades: Ajugoideae, Lamioideae, Nepetoideae, and Viticoideae. Further expanding to the remaining eight Lamiaceae clades may also be a promising strategy for finding new enzyme activities.
The diterpene skeletons of Lamiaceae and how to make them
By considering our newly characterized enzyme activities in the context of chemotaxonomic data and previously described enzymes, we can make some informed speculations about how diverse skeletons arise and what strategies may be used for finding more of the enzymes responsible. All of the six diterpene skeletons with a known biosynthetic route in the Lamiaceae contain a decalin core; Sk2, and Sk4 (Fig. 1, C and D) are skeletons of the direct products of TPS-c enzymes, whereas Sk1, Sk3, Sk6, and Sk14 are skeletons of the products of a TPS-e enzyme acting on a labdadiene diphosphate (Sk4) precursor.
Many diterpene skeletons with an intact decalin core can be plausibly explained by as-yet-undiscovered diTPSs from the TPS-c and TPS-e subfamilies (e.g. through methyl shifts during cyclization). Examples of diTPSs that catalyze methyl shifts are the TPS-c enzymes SdKPS (16, 17) and ArTPS2, which produce the clerodane skeleton (Sk2), and the TPS-e enzyme OmTPS5, which has a product with the abietane skeleton (Sk3). The same mechanisms may form skeletons such as Sk8 and Sk12. Other decalin-containing skeletons, for example the norditerpenes (missing one or more methyl side chains (e.g. Sk7)), are readily explainable by oxidative decarboxylation occurring after the TPS steps. Ring rearrangements catalyzed by TPS-e enzymes also have precedent, for example the generation of ent-kaurene (with skeleton Sk1) or ent-atiserene (with skeleton Sk14) from ent-CPP (with skeleton Sk4) (2, 57), but always preserve the decalin core structure.
Diterpenoids lacking a decalin core are taxonomically restricted within the Lamiaceae, with no single skeleton being reported in more than two clades (Fig. 1C). Many can be explained as modifications occurring after the TPS steps to decalin-containing skeletons. Cytochrome P450–driven ring contraction, akin to that in the gibberellin pathway (64), may play a role in the formation of skeletons such as Sk13 (65). Ring opening and ring expansion may also occur, for example in proposed pathways to compounds with the 6,7-seco-kaurane (Sk5) (66) and icetaxane (Sk9) (67) skeletons, respectively. Skeletons such as cembrane (Sk11) (68, 69), lacking any apparent biosynthetic connection to a decalin core, may arise from diTPSs outside the TPS-c and TPS-e subfamilies. In Euphorbiaceae and Solanaceae, where cembranoid compounds are common, the relevant TPSs come from the TPS-a subfamily (70, 71). Elucidation of pathways to the remaining diterpene skeletons in the Lamiaceae will depend on broadening the search to new genera and species and new TPS subfamilies, eventually moving beyond TPSs to look at cytochromes P450 and other enzyme families.
Implications for biotechnology
Previous work has explored the possibility of producing arrays of compounds by combining class II diTPSs with different class I diTPSs (31, 48). Particularly prolific enzymes for combinatorial biosynthesis have been Cyc2 from the bacterium Streptomyces griseolosporeus (72, 73), which generates alkene moieties on prenyl-diphosphate substrates, and SsSS (18, 20) which installs an alcohol at the 13-position and a double bond at the 14-position; both of these enzymes have demonstrated activity on 12 different class II enzyme products (31). We have found that SsSS is also active on the products of PcTPS1 and ArTPS2. In addition, we have found class I enzymes that provide routes to products that previously were biosynthetically inaccessible or poorly accessible. OmTPS3 is active on class II products with a labdane skeleton and normal absolute configuration, typically generating a 12E-moiety, as in 11, 34, and 24. An enzyme with similar activity, producing 24 and 34, was recently reported from the bacterium Streptomyces cyslabdanicus (36, 74), but was not tested against additional substrates. LlTPS4 produces sandaracopimaradiene (27) from 31, with greater specificity than the earlier enzyme, Euphorbia peplus TPS8 (48). Finally, OmTPS5 enables efficient and specific production of palustradiene (29) from 31. The other known biosynthetic route to 29 is as a minor spontaneous degradation product of 13-hydroxy-8(14)-abietane from Picea abies levopimaradiene/abietadiene synthase (75) and related enzymes.
ArTPS2 is of particular interest for applications in agricultural biotechnology. Neo-clerodane diterpenoids, particularly those with an epoxide moiety at the 4(18) position, such as clerodin, the ajugarins, and the jodrellins (76–78) (Fig. 8), have garnered significant attention for their ability to deter insect herbivores (22–24). The 4(18) desaturated product of ArTPS2, for which no enzyme was previously known, could be used in biosynthetic or semisynthetic routes to these potent insect antifeedants.
Figure 8.
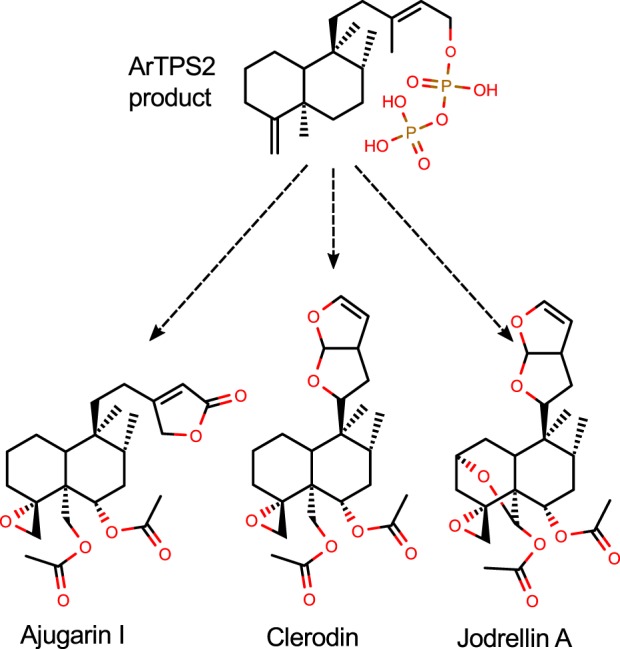
The ArTPS2 product, (5R,8R,9S,10R)-neo-cleroda-4(18),13E-dienyl diphosphate, is the likely precursor to bioactive epoxy-clerodane diterpenoids.
Database-driven phytochemistry
In the traditional plant natural product discovery workflow, compounds are first identified from bulk extractions, and only later, often by decades, are the associated biosynthetic genes identified. In recent years, rapid advances in genome sequencing and transcript profiling have made it much easier and cheaper to obtain detailed genomic data than to obtain comprehensive metabolite data from an organism. A number of strategies have been developed for leveraging genomic data to find novel enzymes, which may or may not be part of pathways to known compounds. Mining of genomes, metagenomes, and metatranscriptomes has led to the identification, from plants, bacteria, and fungi, of natural product pathways that would not have been possible in a metabolomics-first approach (79). In plant terpenoid pathways, genome mining has been used to identify prenyltranferases, cytochromes P450, and TPSs, which have been observed to sometimes occur in genomic clusters (80). In the same spirit as these earlier studies, we took advantage of recently available Lamiaceae leaf transcriptomes to mine for diTPS sequences. Genomics-first approaches may lead back to metabolomics studies, as was the case when we found palustradiene in our O. majorana individual. Alternatively, metabolomics in the source species may be bypassed entirely, with new compounds moved directly to screening for useful biological activities.
In genomics-driven pathway discovery workflows, there is a temptation to not pay much attention to chemotaxonomic data. Part of the reason may be the lack of a comprehensive, user-friendly, open-access database of natural product distribution. Despite this obstacle, in this work we have tried to give equal attention to the decades of accumulated data on natural product distribution as we have to the genomics data. We have found that the chemotaxonomic analysis gives valuable context to enzyme function data (Fig. 4), and its availability helped with interpreting our multispecies diTPS phylogenetic tree and suggesting promising species and genera to target in the future. Aside from its application to terpenoid-related gene discovery, large scale chemotaxonomic analysis has also recently been used to help understand specialty wax biosynthetic pathways across the plant kingdom (81).
Considering that transcriptome data sets for thousands of plant species are already available in public databases, such as the NCBI Sequence Read Archive (82) and Transcriptome Shotgun Assembly (83) archive, and the influx of new data sets is only getting faster, our success in mining the transcriptomes of just 48 species suggests that additional systematic mining of existing transcriptome data could yield many more examples of diverse TPSs and other kinds of enzymes. Systematic, broad transcriptome sampling of additional plant families will help give context to transcriptome mining and improve the chances of finding new enzyme activities. To further enable others to build off our work, we have included detailed data tables of Lamiaceae diTPSs and skeletons as supporting data sets, and, to the extent possible, we have made the data and code used in this study publicly available.
Experimental procedures
Chemotaxonomic database curation
A subset of the NAPRALERT database including all of the occurrences of diterpenoids in mints was obtained as a gift from James Graham (University of Illinois, Chicago, IL). NAPRALERT reports chemical names, but not structures. For the Lamiaceae, the species reported in NAPRALERT largely overlap with those from the DNP, which does include structures, so we made the simplifying assumption that each unique name represents a unique compound, without trying to find structures for the 3,080 Lamiaceae diterpenes in NAPRALERT.
For SISTEMAT, we obtained structure files by redrawing the structures from Vestri Alvarenga et al. (25) into MarvinSketch (ChemAxon, Budapest, Hungary) and the occurrence counts by transcribing the association table into a spreadsheet. A publicly available digital version of SISTEMAT, called SISTEMATX (https://www.sistematx.ufpb.br/),5 exists (84), but there is no option for bulk downloads, limiting our ability to assess its completeness or cross-reference it with other data. We hope that in the future, a publicly available digital chemotaxonomy resource such as SISTEMAT X develops to point where it can be used in gene discovery workflows, but for the present work, the proprietary DNP seemed to be the only viable option for most analyses.
Lamiaceae diterpene structures were obtained from the DNP by searching for them through the DNP web interface (1). Additional compounds were found by searching for individual species names of species for which we had transcriptome data. This additional search step was necessary because some species have been reclassified between families, or their family is not correctly annotated in the DNP. Records for all of the Lamiaceae diterpenes were downloaded and converted into a spreadsheet using a Python script. Species names were extracted from the Biological Source field in a semiautomated method. The DNP contains structural information in the form of InChI strings (85). In most cases, the DNP InChIs do not include stereochemical information, so for consistency, we ignored all stereochemical information. Skeletons were extracted from the structures using the RDKit Python interface. Briefly, all bonds were converted into single bonds, bonds involving at least one noncarbon atom were broken, and the fragment with a carbon count closest to 20 was retained as the skeleton. The resulting skeletons were then manually examined to correct those where the algorithm chose the wrong fragment; for example, a small number of diterpenoids contain acyl side chains of more than 20 carbons, in which case the algorithm would incorrectly select the acyl chain as the skeleton. There are a few cases where sesquiterpenes or other terpenes seem to have been misannotated in the DNP as diterpenes. We chose to leave these in the data set, but their presence or absence does not significantly change any of our analyses.
For all three databases, genus and species names were cross-referenced to TaxIDs from the NCBI Taxonomy database (27), first by automated text comparisons and then by manual inspection of unmatched names. Genus-level TaxID assignments were possible for every entry in NAPRALERT and the DNP, but in some cases, species-level TaxID assignments were not possible.
Candidate gene selection
TransDecoder (version 4.1.0) (86) was used to predict coding sequences from the transcriptome assemblies of the 48 Lamiaceae species. Peptides with 95% or greater identity were merged with CD-HIT (87). The set of known Lamiaceae diTPSs was used as a BLASTP query set against the new peptide sequences. Candidates were filtered to remove sequences with less than 70% coverage or 35% identity to the most similar query peptide. All specific parameters are given in supporting Data set S2, candidate search methods.
To generate phylogenetic trees, peptide sequences were aligned using Clustal Omega (version 1.2.1) (88), and maximum likelihood trees were generated using RAxML (version 8.2.11) (89) using automatic model selection and 1,000 bootstrap iterations. Tree visualizations were generated using ETE3 (90).
Candidates were selected for cloning and characterization based on visual inspection of the phylogenetic trees with the intent to select genes from underrepresented clades. Candidate selection was also influenced by our ability to obtain plant material and by the reported diversity, or lack thereof, of diterpenes in particular genera and species.
Plant material, RNA isolation, and cDNA synthesis
Plants were obtained from different commercial nurseries or botanical gardens (Table S1) and grown in a greenhouse under ambient photoperiod and 24 °C day/17 °C night temperatures. N. benthamiana were grown in a greenhouse under a 16-h light (24 °C) and 8-h dark (17 °C) regime.
Total RNA from leaf tissues of A. reptans, Nepeta mussini, Leonotis leonurus, Perovskia atriplicifolia, and S. officinalis was extracted according to Hamberger et al. (91), whereas total RNA from leaves of Proteus vulgaris, M. spicata, P. cablin, H. suaveolens, and O. majorana was extracted using the Spectrum Plant Total RNA Kit (Sigma-Aldrich). RNA extraction was followed by DNase I digestion using the DNA-freeTM DNA removal kit (Thermo Fisher Scientific). First-strand cDNAs were synthesized from 5 μg of total RNA, with oligo(dT) primer, using the RevertAid First Strand cDNA synthesis kit (Thermo Fisher Scientific). cDNA was diluted 5-fold and used as template for cloning of full-length cDNAs.
Characterization of diTPS genes by transient expression in N. benthamiana
Full-length coding sequences of diTPSs were cloned into pEAQ-HT vector (92) (kindly provided by Prof. G. Lomonossoff, John Innes Centre, UK) using In-Fusion® HD Cloning Plus (Takara Bio). pEAQ-HT vector contains a copy of anti-post-transcriptional gene silencing protein p19 that suppresses the silencing of transgenes (92). Expression vectors carrying full-length coding sequence of candidate diTPS genes were transformed into the LBA4404 Agrobacterium tumefaciens strain by electroporation. DXS and GGPPS are known to be the rate-limiting enzymes in GGPP biosynthesis and have been shown to substantially increase the production of diterpenes in the N. benthamiana system (30, 48). We therefore cloned P. barbatus 1-deoxy-d-xylulose 5-phosphate synthase (CfDXS) (GenBankTM accession number KP889115) and geranylgeranyl diphosphate synthase (CfGGPPS) (GenBankTM accession number KP889114) and created a chimeric polyprotein with an LP4-2A hybrid linker peptide between CfDXS and CfGGPPS. LP4/2A contains the first nine amino acids of LP4 (a linker peptide originating from a natural polyprotein occurring in seeds of Impatiens balsamina) and 20 amino acids of the self-processing FMDV 2A (2A is a peptide from the foot-and-mouth disease virus) (94).
The transformed A. tumefaciens were subsequently transferred to 1 ml of SOC medium and grown for 1 h at 28 °C. 100-μl cultures were transferred to LB-agar solid medium containing 50.0 μg/ml rifampicin and 50.0 μg/ml kanamycin and grown for 2 days. A single-colony PCR-positive clone was transferred to 10 ml of LB medium in a Falcon tube containing 50.0 μg/ml rifampicin and 50.0 μg/ml kanamycin and grown at 28 °C overnight (at 225 rpm). About 1% of the primary culture was transferred to 25 ml of fresh LB medium and grown overnight. Cells were pelleted by centrifugation at 4,000 × g for 15 min and resuspended in 10 ml of water containing 200 μm acetosyringone. Cells were diluted with water-acetosyringone solution to a final A600 of 1.0 and incubated at 28 °C for 2–3 h to increase the infectivity. Equal volumes of culture containing the plasmids with cDNA encoding different diTPS genes were mixed. Each combination of A. tumefaciens culture mixture was infiltrated into independent 4–5-week-old N. benthamiana plants. Plants were grown for 5–7 days in the greenhouse before metabolite extraction. Leaf discs of 2-cm diameter (∼0.1 g fresh weight) were cut from the infiltrated leaves. Diterpenes were extracted in 1 ml of n-hexane with 1 mg/liter 1-eicosene as internal standard at room temperature overnight in an orbital shaker at 200 rpm. Plant material was collected by centrifugation, and the organic phase was transferred to GC vials for analysis.
In vitro enzyme activity assays
To confirm the biosynthetic products obtained in N. benthamiana, diTPS combinations were tested in in vitro assays as described previously (30). TargetP (95) was used for prediction of the plastidial target sequence. Pseudo-mature variant versions of HsTPS1, ArTPS2, PcTPS1, OmTPS3, OmTPS5, SsSS, CfTPS1, CfTPS2, and codon-optimized CfTPS3 (IDT), lacking the predicted plastidial targeting sequences, were cloned in pET-28b(+) (EMD Millipore, Burlington, MA), expressed, and purified from E. coli. pET_diTPS constructs were transformed into chemically competent OverExpressTM C41(DE3) cells (Lucigen, Middleton, WI) and inoculated in a starter culture with Terrific Broth medium and 50 μg/ml kanamycin and grown overnight. About 1% of the starter culture was used to inoculate 50 ml of Terrific Broth medium with 50 μg/ml kanamycin and grown at 37 °C and 200 rpm until A600 reached 0.4. Cultures were grown at 16 °C until A600 of ∼0.6–0.8 was achieved, at which point cultures were induced by 0.2 mm isopropyl 1-thio-β-d-galactopyranoside. Expression was done overnight, and cells were harvested by centrifugation at 5,000 × g at 4 °C for 15 min. Cell pellets were resuspended in lysis buffer containing 20 mm HEPES, pH 7.5, 0.5 m NaCl, 25 mm imidazole, 5% (v/v) glycerol, one protease inhibitor mixture tablet per 100 ml (Sigma-Aldrich), and 0.1 mg/liter lysozyme. The cell pellet was gently shaken for 30 min and subsequently lysed by sonication. Cell lysate was centrifuged for 25 min at 14,000 × g, and the supernatant was subsequently used for purification of the recombinant proteins. Proteins were purified on 1-ml His SpinTrap columns (GE Healthcare) using elution buffer (HEPES, pH 7.5, 0.5 m NaCl, 5% (v/v) glycerol, 350 mm imidazole, and 5 mm DTT) and desalted on PD MiniTrap G-25 columns (GE Healthcare) with a desalting buffer (20 mm HEPES, pH 7.2, 350 mm NaCl, 5 mm DTT, 1 mm MgCl2, 5% (v/v) glycerol). In vitro diTPS assays were performed by adding GGPP to a final concentration of 15 μm and 50–100 μg of purified enzymes in 400 μl of enzyme assay buffer (50 mm HEPES, pH 7.2, 7.5 mm MgCl2, 5% (v/v) glycerol, 5 mm DTT). 500 ml of n-hexane (Fluka GC-MS grade) containing 1 ng/ml 1-eicosene as internal standard were gently added as an overlay onto the reaction mix. Assays were incubated for 60–120 min at 30 °C and ∼75 rpm, and the hexane overlay was subsequently removed by centrifugation at 1,500 × g for 15 min before GC-MS analysis.
Metabolite analysis of O. majorana
20–50 mg of fresh leaf, stem, root, and flowers of O. majorana were harvested. Flowers were further separated with forceps into two parts: the green part (calyx) and the rest of the flower (corolla). Tissues were extracted overnight in 500 μl of methyl tert-butyl ether. Extracts were concentrated to 100 μl and subjected to GC-MS analysis.
GC-MS
All GC-MS analyses were performed on an Agilent 7890A GC with an Agilent VF-5ms column (30 m × 250 μm × 0.25 μm, with 10-m EZ-Guard) and an Agilent 5975C detector. For N. benthamiana and in vitro assays, the inlet was set to 250 °C splitless injection, helium carrier gas with column flow of 1 ml/min. The oven program was 45 °C hold 1 min, 40 °C/min to 230 °C, 7 °C/min to 320 °C, hold 3 min. The detector was activated after a 4-min solvent delay. For analysis of O. majorana extracts, conditions were the same, except that the solvent cutoff was set to 6 min to allow monoterpenes to pass, and the oven program was 45 °C hold 1 min, 40 °C/min to 200 °C, 5 °C/min to 260 °C, 40 °C/min to 320 °C, hold 3 min.
Large-scale production of diterpenes in N. benthamiana for NMR analysis
To produce diterpene levels sufficient for structural analysis by NMR, we made several modifications to the experimental setup of the N. benthamiana system infiltration. A. tumefaciens cultures (500 ml) containing HsTPS1, ArTPS2, CfTPS1, CfTPS2, CfTPS3, PcTPS1, OmTPS3, OmTPS5, and SsSS constructs were separately grown overnight from 20-ml starter cultures and pelleted by centrifugation. Cell pellets were resuspended in water and adjusted to an A600 of 1.0. Resuspended cells were mixed according to the selected combinations together with strains harboring CfDXS/CfGGPPS polyprotein. N. benthamiana plants were submerged in the Agrobacterium suspensions and vacuum-infiltrated at 100 mbar for 30–60 s, similar to a method described previously (48). 15–30 N. benthamiana plants were vacuum-infiltrated with diTPS combinations as well as CfGGPPS and CfDXS. After 5 days, 100–200 g (fresh weight) of leaves were subjected to two rounds of overnight extractions in 500 ml of hexane, which was then concentrated using a rotary evaporator. Compounds were purified on silica gel columns using a mobile phase of hexane with 0–20% ethyl acetate. In some cases, additional rounds of column purification or preparative TLC using a hexane/ethyl acetate or chloroform/methanol mobile phase were necessary to obtain compounds of sufficient purity for structural determination by NMR.
NMR and optical rotation
The NMR spectra for trans-biformene (34) were measured on a Bruker AVANCE 900-MHz spectrometer. All other spectra were measured on an Agilent DirectDrive2 500-MHz spectrometer. All NMR was done in CDCl3 solvent. The CDCl3 peaks were referenced to 7.24 and 77.23 ppm for 1H and 13C spectra, respectively. To aid in the interpretation of NMR spectra, we made extensive use of the NAPROC-13 (96) and Spektraris (97) databases. Reconstruction of 13C spectra from the literature was performed with MestReNova (Mestrelab Research, Santiago de Compostela, Spain). Optical rotation was measured in chloroform at ambient temperature using a PerkinElmer Life Sciences Polarimeter 341 instrument.
Availability of data
GenBankTM numbers for new clones are provided in Data set S1. For code and kinds of data not appropriate for supporting data, and where no appropriate centralized repository exists, we have used the generic database Zenodo. Our Zenodo submission (doi: 10.5281/zenodo.1323366) includes all of the NMR and GC-MS data for this study in multiple formats, as well as the code used to generate the figures. We have also submitted our shift-assigned NMR data for inclusion in the NMRShiftDB (93) database. We are not aware of an open access electron ionization-MS library that accepts submissions from the public, so we have made an AMDIS msl formatted library of background-subtracted reference spectra for compounds observed by GC-MS during this study. The GC-MS library is included in the Zenodo submission and is also available in a Git repository (https://bitbucket.org/seanrjohnson/diterpenoid_databases),5 which we will continue updating with spectra generated in future studies.
Author contributions
S. R. J. and Björn Hamberger conceptualization; S. R. J. resources; S. R. J., W. W. B., and E. M. G. C. data curation; S. R. J. software; S. R. J. and W. W. B. formal analysis; S. R. J., W. W. B., and J. B. validation; S. R. J. visualization; S. R. J. and W. W. B. methodology; S. R. J. writing-original draft; S. R. J., W. W. B., and Björn Hamberger writing-review and editing; W. W. B., J. B., A. T., and Britta Hamberger investigation; Björn Hamberger supervision; Björn Hamberger funding acquisition.
Supplementary Material
Acknowledgments
We thank the Evolutionary Mint Genomics Consortium: Benoît Boachon (Purdue University), C. Robin Buell (Michigan State University), Emily Crisovan (Michigan State University), Natalia Dudareva (Purdue University), Nicolas Garcia (Florida Museum of Natural History), Grant Godden (Michigan State University, Florida Museum of Natural History), Laura Henry (Purdue University), Mohamed O. Kamileen (The John Innes Centre), Heather Rose Kates (Florida Museum of Natural History), Matthew B. Kilgore (Purdue University), Benjamin R. Lichman (The John Innes Centre), Evgeny V. Mavrodiev (Florida Museum of Natural History), Linsey Newton (Michigan State University), Carlos Rodriguez-Lopez (John Innes Centre), Sarah E. O'Connor (John Innes Centre), Douglas Soltis (Florida Museum of Natural History, University of Florida), Pamela Soltis (Florida Museum of Natural History), Brieanne Vaillancourt (Michigan State University), Krystle Wiegert-Rininger (Michigan State University), and Dongyan Zhao (Michigan State University). We thank the facilities at Michigan State University Michigan State University, including the Mass Spectrometry and Metabolomics Core, the Max T. Rogers NMR Facility, and the W. J. Beal Botanical Garden. We thank A. Daniel Jones and Daniel Holmes for essential discussions. We thank James Graham (University of Illinois, Chicago) for providing access to the Lamiaceae diterpenoid data from NAPRALERT. We thank Philip Zerbe (University of California, Davis) for the GrTPS2, MvCPS1, and MvEKS clones. We thank Reuben Peters (Iowa State University) for the ZmAN2 clone. We thank Daniel Moser for technical assistance. We thank the anonymous reviewers for their helpful suggestions.
This work was primarily supported by the Michigan State University Strategic Partnership Grant program (“Evolutionary-Driven Genome Mining of Plant Biosynthetic Pathways”). Bjorn Hamberger, W. W. B., and S. R. J. report a potential conflict of interest: they have filed a patent application (United States Patent Application Serial No. 62/714,216) covering the genes reported in this study. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
This article contains Tables S1–S5, Figs. S1–S22, and Data sets S1–S3.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) MH626616–MH626632.
Code and data types not appropriate for supporting data have been deposited in Zenodo: A database-driven approach finds novel diterpene synthase activities in Lamiaceae (doi: 10.5281/zenodo.1323366).
Data related to diterpenoids, compiled by the Hamberger laboratory at Michigan State University are available at https://bitbucket.org/seanrjohnson/diterpenoid_databases.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- DNP
- Dictionary of Natural Products
- diTPS
- diterpene synthase
- GGPP
- geranylgeranyl diphosphate
- CPP
- copalyl diphosphate
- PgPP
- peregrinol diphosphate
- MvELS
- M. vulgare 9,13-epoxylabdene synthase
- SsSS
- S. sclarea sclareol synthase
- CfDXS
- 1-deoxy-d-xylulose 5-phosphate synthase
- CfGGPS
- geranylgeranyl diphosphate synthase
- LB
- lysogeny broth.
Contributor Information
Benoît Boachon, Purdue University.
C. Robin Buell, Michigan State University.
Emily Crisovan, Michigan State University.
Natalia Dudareva, Purdue University.
Nicolas Garcia, Florida Museum of Natural History.
Grant Godden, Michigan State University, Florida Museum of Natural History.
Laura Henry, Purdue University.
Mohamed O. Kamileen, The John Innes Centre
Heather Rose Kates, Florida Museum of Natural History.
Matthew B. Kilgore, Purdue University
Benjamin R. Lichman, The John Innes Centre
Evgeny V. Mavrodiev, Florida Museum of Natural History
Linsey Newton, Michigan State University.
Carlos Rodriguez-Lopez, John Innes Centre.
Sarah E. O'Connor, John Innes Centre
Douglas Soltis, Florida Museum of Natural History, University of Florida.
Pamela Soltis, Florida Museum of Natural History.
Brieanne Vaillancourt, Michigan State University.
Krystle Wiegert-Rininger, Michigan State University.
Dongyan Zhao, Michigan State University.
Collaborators: Benoît Boachon, C. Robin Buell, Emily Crisovan, Natalia Dudareva, Nicolas Garcia, Grant Godden, Laura Henry, Mohamed O. Kamileen, Heather Rose Kates, Matthew B. Kilgore, Benjamin R. Lichman, Evgeny V. Mavrodiev, Linsey Newton, Carlos Rodriguez-Lopez, Sarah E. O'Connor, Douglas Soltis, Pamela Soltis, Brieanne Vaillancourt, Krystle Wiegert-Rininger, and Dongyan Zhao
References
- 1. Dictionary of Natural Products 26.2. (2018) [online] http://dnp.chemnetbase.com (Accessed January 11, 2018)
- 2. Peters R. J. (2010) Two rings in them all: the labdane-related diterpenoids. Nat. Prod. Rep. 27, 1521–1530 10.1039/c0np00019a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen F., Tholl D., Bohlmann J., and Pichersky E. (2011) The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66, 212–229 10.1111/j.1365-313X.2011.04520.x [DOI] [PubMed] [Google Scholar]
- 4. Zi J., Mafu S., and Peters R. J. (2014) To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. Annu. Rev. Plant Biol. 65, 259–286 10.1146/annurev-arplant-050213-035705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zerbe P., and Bohlmann J. (2015) Plant diterpene synthases: exploring modularity and metabolic diversity for bioengineering. Trends Biotechnol. 33, 419–428 10.1016/j.tibtech.2015.04.006 [DOI] [PubMed] [Google Scholar]
- 6. Hamberger B., and Bak S. (2013) Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120426 10.1098/rstb.2012.0426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Banerjee A., and Hamberger B. (2018) P450s controlling metabolic bifurcations in plant terpene specialized metabolism. Phytochem. Rev. 17, 81–111 10.1007/s11101-017-9530-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pateraki I., Andersen-Ranberg J., Jensen N. B., Wubshet S. G., Heskes A. M., Forman V., Hallström B., Hamberger B., Motawia M. S., Olsen C. E., Staerk D., Hansen J., Møller B. L., and Hamberger B. (2017) Total biosynthesis of the cyclic AMP booster forskolin from Coleus forskohlii. eLife 6, e23001 10.7554/eLife.23001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ondari M. E., and Walker K. D. (2008) The taxol pathway 10-O-acetyltransferase shows regioselective promiscuity with the oxetane hydroxyl of 4-deacetyltaxanes. J. Am. Chem. Soc. 130, 17187–17194 10.1021/ja8067534 [DOI] [PubMed] [Google Scholar]
- 10. Chau M., Walker K., Long R., and Croteau R. (2004) Regioselectivity of taxoid-O-acetyltransferases: heterologous expression and characterization of a new taxadien-5α-ol-O-acetyltransferase. Arch. Biochem. Biophys. 430, 237–246 10.1016/j.abb.2004.07.013 [DOI] [PubMed] [Google Scholar]
- 11. Cui G., Duan L., Jin B., Qian J., Xue Z., Shen G., Snyder J. H., Song J., Chen S., Huang L., Peters R. J., and Qi X. (2015) Functional divergence of diterpene syntheses in the medicinal plant Salvia miltiorrhiza Bunge. Plant Physiol. 169, 1607–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao W., Hillwig M. L., Huang L., Cui G., Wang X., Kong J., Yang B., and Peters R. J. (2009) A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org. Lett. 11, 5170–5173 10.1021/ol902051v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guo J., Zhou Y. J., Hillwig M. L., Shen Y., Yang L., Wang Y., Zhang X., Liu W., Peters R. J., Chen X., Zhao Z. K., and Huang L. (2013) CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc. Natl. Acad. Sci. U.S.A. 110, 12108–12113 10.1073/pnas.1218061110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heskes A. M., Sundram T. C. M., Boughton B. A., Jensen N. B., Hansen N. L., Crocoll C., Cozzi F., Rasmussen S., Hamberger B., Hamberger B., Staerk D., Møller B. L., and Pateraki I. (2018) Biosynthesis of bioactive diterpenoids in the medicinal plant Vitex agnus-castus. Plant J. 93, 943–958 10.1111/tpj.13822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zerbe P., Chiang A., Dullat H., O'Neil-Johnson M., Starks C., Hamberger B., and Bohlmann J. (2014) Diterpene synthases of the biosynthetic system of medicinally active diterpenoids in Marrubium vulgare. Plant J. 79, 914–927 10.1111/tpj.12589 [DOI] [PubMed] [Google Scholar]
- 16. Chen X., Berim A., Dayan F. E., and Gang D. R. (2017) A (−)-kolavenyl diphosphate synthase catalyzes the first step of salvinorin A biosynthesis in Salvia divinorum. J Exp Bot. 68, 1109–1122 10.1093/jxb/erw493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pelot K. A., Mitchell R., Kwon M., Hagelthorn D. M., Wardman J. F., Chiang A., Bohlmann J., Ro D.-K., and Zerbe P. (2017) Biosynthesis of the psychotropic plant diterpene salvinorin A: discovery and characterization of the Salvia divinorum clerodienyl diphosphate synthase. Plant J. 89, 885–897 10.1111/tpj.13427 [DOI] [PubMed] [Google Scholar]
- 18. Caniard A., Zerbe P., Legrand S., Cohade A., Valot N., Magnard J.-L., Bohlmann J., and Legendre L. (2012) Discovery and functional characterization of two diterpene synthases for sclareol biosynthesis in Salvia sclarea(L.) and their relevance for perfume manufacture. BMC Plant Biol. 12, 119 10.1186/1471-2229-12-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Günnewich N., Higashi Y., Feng X., Choi K.-B., Schmidt J., and Kutchan T. M. (2013) A diterpene synthase from the clary sage Salvia sclarea catalyzes the cyclization of geranylgeranyl diphosphate to (8R)-hydroxy-copalyl diphosphate. Phytochemistry 91, 93–99 10.1016/j.phytochem.2012.07.019 [DOI] [PubMed] [Google Scholar]
- 20. Schalk M., Pastore L., Mirata M. A., Khim S., Schouwey M., Deguerry F., Pineda V., Rocci L., and Daviet L. (2012) Toward a biosynthetic route to sclareol and amber odorants. J. Am. Chem. Soc. 134, 18900–18903 10.1021/ja307404u [DOI] [PubMed] [Google Scholar]
- 21. Mint Evolutionary Genomics Consortium (2018) Phylogenomic mining of the mints reveals multiple mechanisms contributing to the evolution of chemical diversity in Lamiaceae. Mol. Plant. 11, 1084–1096 10.1016/j.molp.2018.06.002 [DOI] [PubMed] [Google Scholar]
- 22. Coll J., and Tandrón Y. A. (2008) neo-Clerodane diterpenoids from Ajuga: structural elucidation and biological activity. Phytochem. Rev. 7, 25 [Google Scholar]
- 23. Klein Gebbinck E. A., Jansen B. J. M., and de Groot A. (2002) Insect antifeedant activity of clerodane diterpenes and related model compounds. Phytochemistry 61, 737–770 10.1016/S0031-9422(02)00174-7 [DOI] [PubMed] [Google Scholar]
- 24. Li R., Morris-Natschke S. L., and Lee K.-H. (2016) Clerodane diterpenes: sources, structures, and biological activities. Nat. Prod. Rep. 33, 1166–1226 10.1039/C5NP00137D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vestri Alvarenga S. A., Pierre Gastmans J., do Vale Rodrigues G., Moreno P. R., and de Paulo Emerenciano V. (2001) A computer-assisted approach for chemotaxonomic studies: diterpenes in Lamiaceae. Phytochemistry 56, 583–595 10.1016/S0031-9422(00)00424-6 [DOI] [PubMed] [Google Scholar]
- 26. Loub W. D., Farnsworth N. R., Soejarto D. D., and Quinn M. L. (1985) NAPRALERT: computer handling of natural product research data. J. Chem. Inf. Comput. Sci. 25, 99–103 10.1021/ci00046a009 [DOI] [PubMed] [Google Scholar]
- 27. Federhen S. (2012) The NCBI Taxonomy database. Nucleic Acids Res. 40, D136–D143 10.1093/nar/gkr1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li B., Cantino P. D., Olmstead R. G., Bramley G. L. C., Xiang C.-L., Ma Z.-H., Tan Y.-H., and Zhang D.-X. (2016) A large-scale chloroplast phylogeny of the Lamiaceae sheds new light on its subfamilial classification. Sci. Rep. 6, 34343 10.1038/srep34343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., and Madden T. L. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10, 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pateraki I., Andersen-Ranberg J., Hamberger B., Heskes A. M., Martens H. J., Zerbe P., Bach S. S., Møller B. L., Bohlmann J., and Hamberger B. (2014) Manoyl oxide (13R), the biosynthetic precursor of forskolin, is synthesized in specialized root cork cells in Coleus forskohlii. Plant Physiol. 164, 1222–1236 10.1104/pp.113.228429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jia M., Potter K. C., and Peters R. J. (2016) Extreme promiscuity of a bacterial and a plant diterpene synthase enables combinatorial biosynthesis. Metab. Eng. 37, 24–34 10.1016/j.ymben.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zerbe P., Rodriguez S. M., Mafu S., Chiang A., Sandhu H. K., O'Neil-Johnson M., Starks C. M., and Bohlmann J. (2015) Exploring diterpene metabolism in non-model species: transcriptome-enabled discovery and functional characterization of labda-7,13 E-dienyl diphosphate synthase from Grindelia robusta. Plant J. 83, 783–793 10.1111/tpj.12925 [DOI] [PubMed] [Google Scholar]
- 33. Urones J. G., Marcos I. S., Basabe P., Diez D., Garrido N. M., Alonso C., Oliva I. M., Lithgow A. M., Moro R. F., Sexmero M. J., and Lopez C. (1994) Compounds with the labdane skeleton from Halimium viscosum. Phytochemistry 35, 713–719 10.1016/S0031-9422(00)90593-4 [DOI] [Google Scholar]
- 34. Suzuki H., Noma M., and Kawashima N. (1983) Two labdane diterpenoids from Nicotiana setchellii. Phytochemistry 22, 1294–1295 10.1016/0031-9422(83)80249-0 [DOI] [Google Scholar]
- 35. Roengsumran S., Petsom A., Sommit D., and Vilaivan T. (1999) Labdane diterpenoids from Croton oblongifolius. Phytochemistry 50, 449–453 10.1016/S0031-9422(98)00604-9 [DOI] [PubMed] [Google Scholar]
- 36. Yamada Y., Komatsu M., and Ikeda H. (2016) Chemical diversity of labdane-type bicyclic diterpene biosynthesis in Actinomycetales microorganisms. J. Antibiot. (Tokyo) 69, 515–523 10.1038/ja.2015.147 [DOI] [PubMed] [Google Scholar]
- 37. Xiang W., Li R.-T., Song Q.-S., Na Z., and Sun H.-D. (2004) ent-Clerodanoids from Isodon scoparius. Helv. Chim. Acta 87, 2860–2865 10.1002/hlca.200490257 [DOI] [Google Scholar]
- 38. Rudi A., and Kashman Y. (1992) Chelodane, Barekoxide, and Zaatirin–three new diterpenoids from the marine sponge Chelonaplysilla erecta. J. Nat. Prod. 55, 1408–1414 10.1021/np50088a004 [DOI] [Google Scholar]
- 39. Ohsaki A., Yan L. T., Ito S., Edatsugi H., Iwata D., and Komoda Y. (1994) The isolation and in vivo potent antitumor activity of clerodane diterpenoid from the oleoresin of the Brazilian medicinal plant, Copaifera langsdorfi desfon. Bioorg. Med. Chem. Lett. 4, 2889–2892 10.1016/S0960-894X(01)80834-9 [DOI] [Google Scholar]
- 40. Monaco P., Previtera L., and Mangoni L. (1982) Terpenes from the bled resin of Araucaria hunsteinii. Rendiconto dell'Academia delle scienze fisiche e matematiche 48, 465–470 [Google Scholar]
- 41. Barton D. H. R., Cheung H. T., Cross A. D., Jackman L. M., and Martin-Smith M. (1961) 1003. Diterpenoid bitter principles. Part III. The constitution of clerodin. J. Chem. Soc. 10.1039/JR9610005061 10.1039/JR9610005061 [DOI] [Google Scholar]
- 42. Arima Y., Kinoshita M., and Akita H. (2007) Natural product synthesis from (8aR)- and (8aS)-bicyclofarnesols: synthesis of (+)-wiedendiol A, (+)-norsesterterpene diene ester and (−)-subersic acid. Tetrahedron: Asymmetry 18, 1701–1711 10.1016/j.tetasy.2007.07.010 [DOI] [Google Scholar]
- 43. Wu C.-L., and Lin H. R. (1997) Labdanoids and bis(bibenzyls) from Jungermannia species. Phytochemistry 44, 101–105 10.1016/S0031-9422(96)00519-5 [DOI] [Google Scholar]
- 44. Boalino D. M., McLean S., Reynolds W. F., and Tinto W. F. (2004) Labdane Diterpenes of Leonurus sibiricus. J. Nat. Prod. 67, 714–717 10.1021/np030480i [DOI] [PubMed] [Google Scholar]
- 45. Gray C. A., Rivett D. E. A., and Davies-Coleman M. T. (2003) The absolute stereochemistry of a diterpene from Ballota aucheri. Phytochemistry 63, 409–413 10.1016/S0031-9422(03)00156-0 [DOI] [PubMed] [Google Scholar]
- 46. Harris L. J., Saparno A., Johnston A., Prisic S., Xu M., Allard S., Kathiresan A., Ouellet T., and Peters R. J. (2005) The maize An2 gene is induced by Fusarium attack and encodes an ent-copalyl diphosphate synthase. Plant Mol. Biol. 59, 881–894 10.1007/s11103-005-1674-8 [DOI] [PubMed] [Google Scholar]
- 47. Zhan X., Bach S. S., Hansen N. L., Lunde C., and Simonsen H. T. (2015) Additional diterpenes from Physcomitrella patens synthesized by copalyl diphosphate/kaurene synthase (PpCPS/KS). Plant Physiol. Biochem. 96, 110–114 10.1016/j.plaphy.2015.07.011 [DOI] [PubMed] [Google Scholar]
- 48. Andersen-Ranberg J., Kongstad K. T., Nielsen M. T., Jensen N. B., Pateraki I., Bach S. S., Hamberger B., Zerbe P., Staerk D., Bohlmann J., Møller B. L., and Hamberger B. (2016) Expanding the landscape of diterpene structural diversity through stereochemically controlled combinatorial biosynthesis. Angew. Chem. Int. Ed. Engl. 55, 2142–2146 10.1002/anie.201510650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Peters R. J., Flory J. E., Jetter R., Ravn M. M., Lee H.-J., Coates R. M., and Croteau R. B. (2000) Abietadiene synthase from Grand Fir (Abies grandis): characterization and mechanism of action of the “Pseudomature” recombinant enzyme. Biochemistry 39, 15592–15602 10.1021/bi001997l [DOI] [PubMed] [Google Scholar]
- 50. Bohlmann F., and Czerson H. (1979) Neue labdan- und pimaren-derivate aus Palafoxia rosea. Phytochemistry 18, 115–118 10.1016/S0031-9422(00)90926-9 [DOI] [Google Scholar]
- 51. Mann F. M., Prisic S., Davenport E. K., Determan M. K., Coates R. M., and Peters R. J. (2010) A single residue switch for Mg2+-dependent inhibition characterizes plant class II diterpene cyclases from primary and secondary metabolism. J. Biol. Chem. 285, 20558–20563 10.1074/jbc.M110.123307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li J.-L., Chen Q.-Q., Jin Q.-P., Gao J., Zhao P.-J., Lu S., and Zeng Y. (2012) IeCPS2 is potentially involved in the biosynthesis of pharmacologically active Isodon diterpenoids rather than gibberellin. Phytochemistry 76, 32–39 10.1016/j.phytochem.2011.12.021 [DOI] [PubMed] [Google Scholar]
- 53. Potter K., Criswell J., Zi J., Stubbs A., and Peters R. J. (2014) Novel product chemistry from mechanistic analysis of ent-copalyl diphosphate synthases from plant hormone biosynthesis. Angew. Chem. Int. Ed. Engl. 53, 7198–7202 10.1002/anie.201402911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Potter K. C., Zi J., Hong Y. J., Schulte S., Malchow B., Tantillo D. J., and Peters R. J. (2016) Blocking deprotonation with retention of aromaticity in a plant ent-copalyl diphosphate synthase leads to product rearrangement. Angew. Chem. Int. Ed. Engl. 55, 634–638 10.1002/anie.201509060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hansen N. L., Nissen J. N., and Hamberger B. (2017) Two residues determine the product profile of the class II diterpene synthases TPS14 and TPS21 of Tripterygium wilfordii. Phytochemistry 138, 52–56 10.1016/j.phytochem.2017.02.022 [DOI] [PubMed] [Google Scholar]
- 56. Schulte S., Potter K. C., Lemke C., and Peters R. J. (2018) Catalytic bases and stereocontrol in Lamiaceae class II diterpene cyclases. Biochemistry 57, 3473–3479 10.1021/acs.biochem.8b00193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jin B., Cui G., Guo J., Tang J., Duan L., Lin H., Shen Y., Chen T., Zhang H., and Huang L. (2017) Functional diversification of kaurene synthase-like genes in Isodon rubescens. Plant Physiol. 174, 943–955 10.1104/pp.17.00202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mafu S., Potter K. C., Hillwig M. L., Schulte S., Criswell J., and Peters R. J. (2015) Efficient heterocyclisation by (di)terpene synthases. Chem. Commun. (Camb.) 51, 13485–13487 10.1039/C5CC05754J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xu M., Wilderman P. R., and Peters R. J. (2007) Following evolution's lead to a single residue switch for diterpene synthase product outcome. Proc. Natl. Acad. Sci. U.S.A. 104, 7397–7401 10.1073/pnas.0611454104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jia M., Zhou K., Tufts S., Schulte S., and Peters R. J. (2017) A pair of residues that interactively affect diterpene synthase product outcome. ACS Chem. Biol. 12, 862–867 10.1021/acschembio.6b01075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hillwig M. L., Xu M., Toyomasu T., Tiernan M. S., Wei G., Cui G., Huang L., and Peters R. J. (2011) Domain loss has independently occurred multiple times in plant terpene synthase evolution. Plant J. 68, 1051–1060 10.1111/j.1365-313X.2011.04756.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pelot K. A., Hagelthorn D. M., Addison J. B., and Zerbe P. (2017) Biosynthesis of the oxygenated diterpene nezukol in the medicinal plant Isodon rubescens is catalyzed by a pair of diterpene synthases. PLOS One 12, e0176507 10.1371/journal.pone.0176507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jia M., O'Brien T. E., Zhang Y., Siegel J. B., Tantillo D. J., and Peters R. J. (2018) Changing face: a key residue for the addition of water by sclareol synthase. ACS Catal. 8, 3133–3137 10.1021/acscatal.8b00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Helliwell C. A., Chandler P. M., Poole A., Dennis E. S., and Peacock W. J. (2001) The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. U.S.A. 98, 2065–2070 10.1073/pnas.98.4.2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Han Q.-B., Cheung S., Tai J., Qiao C.-F., Song J.-Z., Tso T.-F., Sun H.-D., and Xu H.-X. (2006) Maoecrystal Z, a cytotoxic diterpene from Isodon eriocalyx with a unique skeleton. Org. Lett. 8, 4727–4730 10.1021/ol061757j [DOI] [PubMed] [Google Scholar]
- 66. Li X.-N., Pu J.-X., Du X., Lou L.-G., Li L.-M., Huang S.-X., Zhao B., Zhang M., He F., Luo X., Xiao W.-L., and Sun H.-D. (2010) Structure and cytotoxicity of diterpenoids from Isodon eriocalyx. J. Nat. Prod. 73, 1803–1809 10.1021/np1004328 [DOI] [PubMed] [Google Scholar]
- 67. González A. G., Andrés L. S., Luis J. G., Brito I., and Rodríguez M. L. (1991) Diterpenes from Salvia mellifera. Phytochemistry 30, 4067–4070 10.1016/0031-9422(91)83468-Z [DOI] [Google Scholar]
- 68. Chen Y.-L., Lan Y.-H., Hsieh P.-W., Wu C.-C., Chen S.-L., Yen C.-T., Chang F.-R., Hung W.-C., and Wu Y.-C. (2008) Bioactive cembrane diterpenoids of Anisomeles indica. J. Nat. Prod. 71, 1207–1212 10.1021/np800147z [DOI] [PubMed] [Google Scholar]
- 69. Li L.-M., Li G.-Y., Pu J.-X., Xiao W.-L., Ding L.-S., and Sun H.-D. (2009) Ent-kaurane and cembrane diterpenoids from Isodon sculponeatus and their cytotoxicity. J. Nat. Prod. 72, 1851–1856 10.1021/np900406c [DOI] [PubMed] [Google Scholar]
- 70. Kirby J., Nishimoto M., Park J. G., Withers S. T., Nowroozi F., Behrendt D., Rutledge E. J. G., Fortman J. L., Johnson H. E., Anderson J. V., and Keasling J. D. (2010) Cloning of casbene and neocembrene synthases from Euphorbiaceae plants and expression in Saccharomyces cerevisiae. Phytochemistry 71, 1466–1473 10.1016/j.phytochem.2010.06.001 [DOI] [PubMed] [Google Scholar]
- 71. Ennajdaoui H., Vachon G., Giacalone C., Besse I., Sallaud C., Herzog M., and Tissier A. (2010) Trichome specific expression of the tobacco (Nicotiana sylvestris) cembratrien-ol synthase genes is controlled by both activating and repressing cis-regions. Plant Mol. Biol. 73, 673–685 10.1007/s11103-010-9648-x [DOI] [PubMed] [Google Scholar]
- 72. Hamano Y., Kuzuyama T., Itoh N., Furihata K., Seto H., and Dairi T. (2002) Functional analysis of eubacterial diterpene cyclases responsible for biosynthesis of a diterpene antibiotic, terpentecin. J. Biol. Chem. 277, 37098–37104 10.1074/jbc.M206382200 [DOI] [PubMed] [Google Scholar]
- 73. Dairi T., Hamano Y., Kuzuyama T., Itoh N., Furihata K., and Seto H. (2001) Eubacterial diterpene cyclase genes essential for production of the isoprenoid antibiotic terpentecin. J. Bacteriol. 183, 6085–6094 10.1128/JB.183.20.6085-6094.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ikeda H., Shin-Ya K., Nagamitsu T., and Tomoda H. (2016) Biosynthesis of mercapturic acid derivative of the labdane-type diterpene, cyslabdan that potentiates imipenem activity against methicillin-resistant Staphylococcus aureus: cyslabdan is generated by mycothiol-mediated xenobiotic detoxification. J. Ind. Microbiol. Biotechnol. 43, 325–342 10.1007/s10295-015-1694-6 [DOI] [PubMed] [Google Scholar]
- 75. Keeling C. I., Madilao L. L., Zerbe P., Dullat H. K., and Bohlmann J. (2011) The primary diterpene synthase products of Picea abies levopimaradiene/abietadiene synthase (PaLAS) are epimers of a thermally unstable diterpenol. J. Biol. Chem. 286, 21145–21153 10.1074/jbc.M111.245951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Geuskens R. B. M., Luteijn J. M., and Schoonhoven L. M. (1983) Antifeedant activity of some ajugarin derivatives in three lepidopterous species. Experientia 39, 403–404 10.1007/BF01963148 [DOI] [Google Scholar]
- 77. Belles X., Camps F., Coll J., and Piulachs M. D. (1985) Insect antifeedant activity of clerodane diterpenoids against larvae of Spodoptera littoralis (Boisd.) (Lepidoptera). J. Chem. Ecol. 11, 1439–1445 10.1007/BF01012143 [DOI] [PubMed] [Google Scholar]
- 78. Anderson J. C., Blaney W. M., Cole M. D., Fellows L. L., Ley S. V., Sheppard R. N., and Simmonds M. S. J. (1989) The structure of two new clerodane diterpenoid potent insect antifeedants from Scutellaria woronowii (Juz); jodrellin A & B. Tetrahedron Lett. 30, 4737–4740 10.1016/S0040-4039(01)80789-9 [DOI] [Google Scholar]
- 79. Challis G. L. (2008) Genome mining for novel natural product discovery. J. Med. Chem. 51, 2618–2628 10.1021/jm700948z [DOI] [PubMed] [Google Scholar]
- 80. Nützmann H.-W., Huang A., and Osbourn A. (2016) Plant metabolic clusters: from genetics to genomics. New Phytol. 211, 771–789 10.1111/nph.13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Busta L., and Jetter R. (2017) Moving beyond the ubiquitous: the diversity and biosynthesis of specialty compounds in plant cuticular waxes. Phytochem. Rev. 17, 1275–1304 10.1007/s11101-017-9542-0 [DOI] [Google Scholar]
- 82. Kodama Y., Shumway M., Leinonen R., and International Nucleotide Sequence Database Collaboration (2012) The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res. 40, D54–D56 10.1093/nar/gkr854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Benson D. A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D. J., Ostell J., and Sayers E. W. (2013) GenBank. Nucleic Acids Res. 41, D36–D42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Scotti M. T., Herrera-Acevedo C., Oliveira T. B., Costa R. P. O., Santos SYKde Rodrigues O. R. P., Scotti L., and Da-Costa F. B. (2018) SistematX, an online web-based cheminformatics tool for data management of secondary metabolites. Molecules 23, 103 10.3390/molecules23010103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Heller S. R., McNaught A., Pletnev I., Stein S., and Tchekhovskoi D. (2015) InChI, the IUPAC International Chemical Identifier. J. Cheminform. 7, 23 10.1186/s13321-015-0068-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., Couger M. B., Eccles D., Li B., Lieber M., MacManes M. D., Ott M., Orvis J., Pochet N., Strozzi F., et al. (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fu L., Niu B., Zhu Z., Wu S., and Li W. (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Huerta-Cepas J., Serra F., and Bork P. (2016) ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 10.1093/molbev/msw046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hamberger B., Ohnishi T., Hamberger B., Séguin A., and Bohlmann J. (2011) Evolution of diterpene metabolism: sitka spruce CYP720B4 catalyzes multiple oxidations in resin acid biosynthesis of conifer defense against insects. Plant Physiol. 157, 1677–1695 10.1104/pp.111.185843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sainsbury F., Thuenemann E. C., and Lomonossoff G. P. (2009) pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7, 682–693 10.1111/j.1467-7652.2009.00434.x [DOI] [PubMed] [Google Scholar]
- 93. Kuhn S., Schlörer N. E., Kolshorn H., and Stoll R. (2012) From chemical shift data through prediction to assignment and NMR LIMS: multiple functionalities of nmrshiftdb2. J. Cheminform. 4, P52 10.1186/1758-2946-4-S1-P52 [DOI] [Google Scholar]
- 94. Sun H., Zhou N., Wang H., Huang D., and Lang Z. (2017) Processing and targeting of proteins derived from polyprotein with 2A and LP4/2A as peptide linkers in a maize expression system. PLoS One 12, e0174804 10.1371/journal.pone.0174804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Emanuelsson O., Nielsen H., Brunak S., and von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 10.1006/jmbi.2000.3903 [DOI] [PubMed] [Google Scholar]
- 96. López-Pérez J. L., Theráon R., del Olmo E., and Díaz D. (2007) NAPROC-13: a database for the dereplication of natural product mixtures in bioassay-guided protocols. Bioinformatics 23, 3256–3257 10.1093/bioinformatics/btm516 [DOI] [PubMed] [Google Scholar]
- 97. Fischedick J. T., Johnson S. R., Ketchum R. E. B., Croteau R. B., and Lange B. M. (2015) NMR spectroscopic search module for Spektraris, an online resource for plant natural product identification: taxane diterpenoids from Taxus × media cell suspension cultures as a case study. Phytochemistry 113, 87–95 10.1016/j.phytochem.2014.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GenBankTM numbers for new clones are provided in Data set S1. For code and kinds of data not appropriate for supporting data, and where no appropriate centralized repository exists, we have used the generic database Zenodo. Our Zenodo submission (doi: 10.5281/zenodo.1323366) includes all of the NMR and GC-MS data for this study in multiple formats, as well as the code used to generate the figures. We have also submitted our shift-assigned NMR data for inclusion in the NMRShiftDB (93) database. We are not aware of an open access electron ionization-MS library that accepts submissions from the public, so we have made an AMDIS msl formatted library of background-subtracted reference spectra for compounds observed by GC-MS during this study. The GC-MS library is included in the Zenodo submission and is also available in a Git repository (https://bitbucket.org/seanrjohnson/diterpenoid_databases),5 which we will continue updating with spectra generated in future studies.