Abstract
Understanding the evolution of mate choice requires dissecting the mechanisms of female preference, particularly how these differ among social contexts and preference phenotypes. Here we study the female neurogenomic response after only 10 minutes of mate exposure in both a sensory component (optic tectum) and a decision-making component (telencephalon) of the brain. By comparing the transcriptional response between females with and without preferences for colorful males, we identified unique neurogenomic elements associated with the female preference phenotype that are not present in females without preference. Network analysis revealed different properties for this response at the sensory-processing and the decision-making levels, and showed that this response is highly centralized in the telencephalon. Furthermore, we identified an additional set of genes that vary in expression across social contexts, beyond mate evaluation. We show that transcription factors among those loci are predicted to regulate the transcriptional response of the genes we found to be associated with female preference.
Introduction
Understanding the evolution of critical animal behaviors requires identifying the underlying mechanisms by which the nervous system produces these behaviors1–5. Many of the most extravagant behaviors in nature are related to mate choice and reproduction. Mate choice has a major effect on organismal fitness, and is therefore subject to powerful natural selection and sexual selection pressures6–8. The steps involved in mating and other behaviors are mediated by changes in neural activity in the brain. Like other input from the external environment to the brain, mating stimuli are translated into neural activity triggered by acute and rapid cascades of gene expression changes. These in turn cause modifications in synaptic activity, metabolic processes or activate further transcriptional pathways1,9,10. We now know that coordinated changes in the expression of many genes (i.e. neurogenomic response11) are the basis of behavioral states9,10, and play a critical role modulating the inherent plasticity that allows our brain to respond appropriately to diverse stimuli12,13.
Studying the gene expression changes that characterize the neurogenomic state behind mating decisions is an important part of dissecting the mechanisms behind mating preferences and mating behavior. Previous studies primarily based on candidate genes and/or whole transcriptomes2,3,9, have identified some key components associated with the neural processes underlying social behaviors and mate preferences3,14–18. Here our goal is to build on this knowledge by characterizing the transcriptional response triggered by different mating contexts, which is key to understanding how the brain coordinates the multitude of behaviors elicited by diverse stimuli and contexts10,19–22. We compared the early transcriptional response in two mating contexts, after exposure to attractive and unattractive males, in females with and without female preference phenotypes. We used the Trinidadian guppy, Poecilia reticulata, a model for studies of sexual selection23–25, in which female preference and male coloration coevolve across natural populations26–28.
Various explanations have been offered for the association between female preference and male color in wild guppies29–31, but recent evidence suggests that the strength of female preference could be linked to brain size and cognitive ability32. Through behavioral tests on selection lines for relative brain size33, we recently showed that replicate small-brained lines have convergently lost their preference for colorful males compared to wild-type and large-brained females32. The variation we found in female preference phenotype in these selection lines32 mirrors variation among natural populations26–28, presenting a unique opportunity to study the neurogenomics of female mating decisions comparatively while controlling for genetic background34.
Previous studies measured whole transcriptome expression changes after 30 minutes of mate exposure35, when the transcriptional response is easily detectable. However, within 10 minutes of mate exposure, guppy females perceive and evaluate males, experience changes in receptivity, and make a decision on whether or not to mate23. Therefore, in order to dissect the early response of the female preference neurogenomic pathway, and understand the transcriptional basis of variation in female preference, we use RNAseq to compare brain gene expression in females from the different selection lines after 10 minutes of exposure to either a colorful (attractive) male, a dull (unattractive) male, or another female (Fig. 1A). We focused on two brain components (Fig. 1C): the optic tectum, because it is involved in sensory processing of visual signals, and the telencephalon, because it integrates those signals to mediate complex decision making, including social and mating decisions36–38.
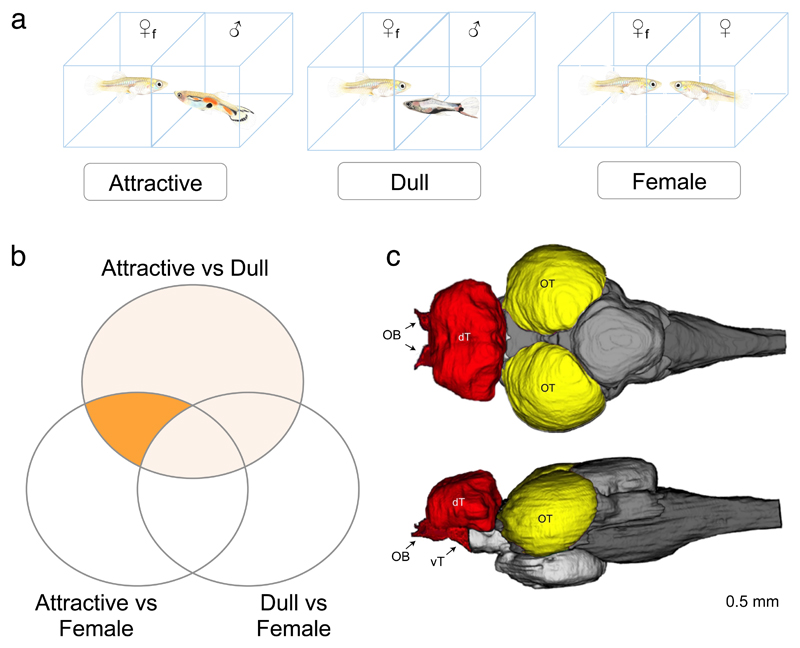
Figure 1. Experimental setup used to find neurogenomic pathways associated with mate preferences.
(A) Diagram of the three treatments: Focal females (♀f) were exposed to either an attractive male (left), a dull male (center) or another female as a control condition (right). Note, guppies are not drawn to scale. (B) Venn diagram illustrating the various pairwise comparisons used to identify differentially expressed genes between treatments. Identification of differentially expressed genes and permutations were performed for each pairwise treatment comparison and separately for Preference and Non-preference lines in both tissues. See Table 1 for results of all comparisons. Area “x” indicates all genes differentially expressed between the attractive and dull treatments and “P” is the final set of Preference DE genes, after filtering to keep only those Attractive vs Dull DE genes that are also differentially expressed in the Attractive vs Female comparison but not in the Dull vs Female (see methods for details). (C) Schematic representation of a top view (top) and lateral view (bottom) of the major regions of the guppy brain. We examined gene expression in the optic tectum (OT, yellow) and the telencephalon (T, red) which included dorsal telencephalon, ventral telencephalon, preoptic area and olfactory bulbs. The latter are less than 2.9% of the mass. The optic tectum samples included the laminated superior area of both hemispheres.
Our results reveal guppy females with clear mate preferences exhibit a distinctive brain transcriptional response following exposure to attractive males. Genes associated with this response are more connected and central in the telencephalon co-expression network, revealing differences in the female mate preference transcriptional cascade in the various components of the brain mediating mating interactions. We also identified genes that vary across different social contexts beyond mate evaluation, and found that these genes exhibit different expression patterns across mating and social encounters. Our results uncover the early components and structure of the genetic networks underlying female mate preferences. These findings have important implications as they provide a foundation to understand the genetics and evolution of mating decisions and mate choice.
Results
Identifying transcriptional response uniquely associated with female preference
We first determined whether there was a transcriptional response uniquely associated with female preference. For this we focused on those genes with significant and concordant differences in expression (DE) between attractive and dull male treatments in Preference females (i.e. females with clear preferences from wild-type and large brain lines32, designated as “X” in Fig. 1B, Fig S1. See methods for details). In order to identify genes associated with the evaluation of an attractive male that fits intrinsic female preference, we filtered these DE genes further, keeping only those that were also differentially expressed between attractive and female treatments, but not between dull and female treatments (area “P” Fig. 1B).
The resulting genes, which are associated with the female preference phenotype in Preference lines, comprised 193 genes in the optic tectum and 106 in the telencephalon (referred to as Preference DE genes, Table 1, Table S1, Supplementary Datasets S1 and S2). Only eight genes were differentially expressed in both tissues. This low overlap is not surprising considering the demonstrated differences in the expression of activity-regulated genes across brain regions in birds and fish10,39. Even though evolutionary models predict sex linkage of female preference genes under the good genes model40,41, we did not observe an enrichment of these candidate genes on the X chromosome (LG12, P>0.05). Instead, we see enrichment of optic tectum and telencephalon Preference DE genes on various autosomes (Table S2). As a species with Y-linked male displays, guppies may be an exception to good genes models41. Importantly, strong female preferences could also evolve from direct selection on sensory system42 or as we hypothesize, on cognitive ability32.
Table 1. Differentially expressed genes.
| OPTIC TECTUM | |||||
| Attractive vs Dull | Attractive vs Female | Dull vs Female | Total (unique genes) |
||
| Preference | Attractive vs Dull genes that pass permutation 5% threshold | 1278 (x) | 1125 | 982 | 2746 |
| Preference DE genes (after filtering§) |
193 (P) | - | - | - | |
| Social DE genes | 357 | ||||
| Non-Preference | Genes that pass permutation 5% threshold | 842 (x) | 1973 | 1449 | 3393 |
| Non-preference DE genes (after filtering§) |
61 (P) | - | - | - | |
| TELENCEPHALON | |||||
| Attractive vs Dull | Attractive vs Female | Dull vs Female | Total (unique genes) |
||
| Preference | Genes that pass permutation 5% threshold | 919 (x) | 746 | 785 | 1999 |
| Preference DE genes (after filtering§) |
106 (P) | - | - | - | |
| Social DE genes | 161 | ||||
| Non-Preference | Genes that pass permutation 5% threshold | 847 (x) | 705 | 677 | 1853 |
| Non-preference DE genes (after filtering§) |
38 (P) | - | - | - | |
Letters in parenthesis refer to Venn diagram sections highlighted in Figure 1.
Genes that were considered differentially expressed between attractive and dull treatments following the permutation 5% cutoff were filtered for concordant expression across all the replicate lines, and for differential expression between attractive vs female and dull vs female keeping only genes in section P of Fig. 1. See text for further details.
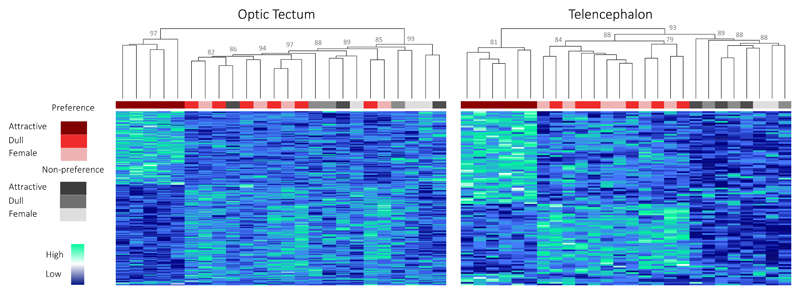
Preference DE genes have a distinct transcriptional signature in Preference females exposed to an attractive male in both tissues, and thus cluster together separately from all the other samples (Fig. 2). However, it is important to note that in the optic tectum, Non-preference samples show differences in the expression of Preference DE genes, similar to those seen in Preference females exposed to a dull male or a female (Fig. 2). There is therefore some activity for Preference DE genes in Non-preference females at the sensory-processing level, suggesting the difference in attractiveness between the two male types is being perceived and processed by Non-preference females. We did not observe this pattern at the decision-making level, in the telencephalon. Here, Non-preference samples group in a third separate cluster, where Preference DE genes do not show any differences in expression. This suggests that Preference DE genes in the telencephalon are not recruited to the decision-making process in Non-preference females. We know these differences are due to the social stimuli as, samples do not follow the same clustering pattern when transcriptome-wide expression is considered (Fig. S2). Moreover, we have previously characterized the genetic differences between large-brained (Preference) and small-brained (Non-preference) lines, and shown that they only differ in the regulation of one locus, Angiopoeitin-134. Expression of this key gene during development influences both the relative brain size and neural density of these fish. We suggest that this developmental difference is indeed the main driver of the variation in brain size between selection lines34.
Figure 2. Hierarchical gene-expression clustering of Preference DE genes.
Hierarchical gene-expression clustering of samples for Preference DE genes differentially expressed between attractive and dull male treatments in the optic tectum (n=193) and telencephalon (n=106). Colors below dendrogram correspond to sample treatment and line as outlined in the legend. Values on top of nodes correspond to bootstrap Approximately Unbiased p-values, computed by multiscale bootstrap resampling90 (all bootstrap values >70%, those <80% not shown for clarity).
We next performed an identical differential expression analysis and filtering in Non-preference females. We found only 61 and 38 loci were differentially expressed between the attractive and dull male treatments in the optic tectum and telencephalon respectively (Non-preference DE genes, Table 1, Supplementary Datasets). Although members of the same gene families were differentially expressed in lines with opposing preference phenotypes (i.e. sodium calcium exchanger proteins, ribosomal proteins among others - Table S1), none of these overlapped with Preference DE genes. Unlike Preference DE genes, Non-preference DE genes do not exhibit a distinct expression signature in Preference females (Fig. S3), and were enriched in different chromosomes as Preference DE genes (Table S2).
Female preference neurogenomic co-expression network attributes and modularity
We next investigated gene relationships in the context of weighted co-expression networks (WGCNA)43,44 for each tissue separately. Co-expression networks allow us to examine the regulatory connections between differentially expressed genes and determine the modular structure of transcriptional responses45. The optic tectum and telencephalon networks retained 6297 genes and 3540 genes respectively (Table S3, Fig. S4; see methods). For subsequent analyses we focus on DE genes remaining in the co-expression networks, as these genes have strong transcriptional connections, a characteristic we might expect for genes at the apex of genetic pathways involved in female preference response. Additionally, we compiled a list of genes previously shown to have roles in social/mating behavior and mate preferences (Table S4), including synaptic plasticity genes (SPG), some of which are immediate early genes (IEG) (Table S5), in order to investigate the network properties of DE genes relative to genes with known roles in social behavior. The context/stimulus dependent plasticity that characterizes the brain, allowing it respond differently to thousands of stimuli, is due in part to the response of genes that alter synaptic connections12,18,46,47.
We found Preference DE genes in the optic tectum and the telencephalon networks have different properties. Our analysis of network attributes reveals Preference DE genes in the optic tectum are distributed throughout the co-expression network with highly variable centrality and connectivity measures (Table 2). In contrast, Preference DE genes are both central and highly connected in the telencephalon network (Table 2, Fig. S4). This suggests the evaluation of males of different qualities causes responses with different characteristics at the sensory-processing and the decision-making levels. The greater centrality and connectivity of Preference DE genes in the telencephalon suggests that we have identified upstream control genes in the decision-making component of the brain, responsible for initiating the transcriptional cascades underlying female preference behaviors. These ultimately lead to the decision to mate, downstream endocrine response and changes in future behavior. Crucially, this pattern was not observed in the telencephalon of Non-preference females in response to an attractive male.
Table 2. Co-expression network centrality and connectivity measures.
| A | n | OPTIC TECTUM | n | TELENCEPHALON | t-test p-value |
|
| Preference DE genes | Degree average1 | 57 | 3.56 (2.83) | 12 | 8.67 (3.64) | 0.02* |
| Clustering Coefficient2 | 0.16 (0.72) | 0.53 (0.53) | <0.001** | |||
| Neighborhood Connectivity3 | 7.84 (3.30) | 21 (3.66) | <0.001** | |||
| Non-preference DE genes | Degree average1 | 31 | 6.48 (3.17) | 6 | 3.83 (2.10) | ns |
| Clustering Coefficient2 | 0.24 (0.56) | 0.49 (0.70) | ns | |||
| Neighborhood Connectivity3 | 11.12 (3.5) | 8.89 (3.56) | ns | |||
| Social affiliation/ female preference genes | Degree average1 | 10 | 13.8 (3.7) | 3 | 1.7 (0.4) | 0.02* |
| Clustering Coefficient2 | 0.34 (0.6) | 0 (0) | <0.01** | |||
| Neighborhood Connectivity3 | 21.3 (3.7) | 2.5 (0.5) | <0.01** | |||
| B | ||||||
| Social affiliation/ female preference genes compared to Preference DE genes | OPTIC TECTUM | TELENCEPHALON | ||||
| Sample sizes | 57/10 | 12/3 | ||||
| Degree average1 | 0.04* | 0.02* | ||||
| Clustering Coefficient2 | <0.01** | <0.001** | ||||
| Neighborhood Connectivity3 | <0.01** | 0.02* | ||||
All p-values correspond to t-tests. Sample sizes in B correspond to Preference DE genes/Social affiliation and female preference genes.
The number of edges, i.e. other genes, each gene is connected to within the network. Central genes in the network will therefore have high degree values as opposed to more peripheral network genes.
The ratio of the number of edges between the neighbors of a gene, and the maximum number of edges that could possibly exist between such neighbors (number between 0 and 1). This is a measure of how connected a gene is relative to how connected it could be given the number of neighbors it has. This value will approach 0 for a loosely connected gene and 1 for a fully connected gene in the center of a network
The average connectivity across all neighbors.
We also find that genes previously associated with mate preference and social and mating behavior3 (Table S4) were significantly more peripheral (i.e. genes with lower gene connectivity at the periphery of the co-expression network) than our Preference DE genes in the telencephalon (Fig. S4). This finding is consistent with the notion that telencephalon Preference DE genes we identified after 10 minutes of treatment exposure are the upstream components of the preference pathway, and induce expression of genes that have been identified by previous work focused on 30 minutes of treatment exposure.
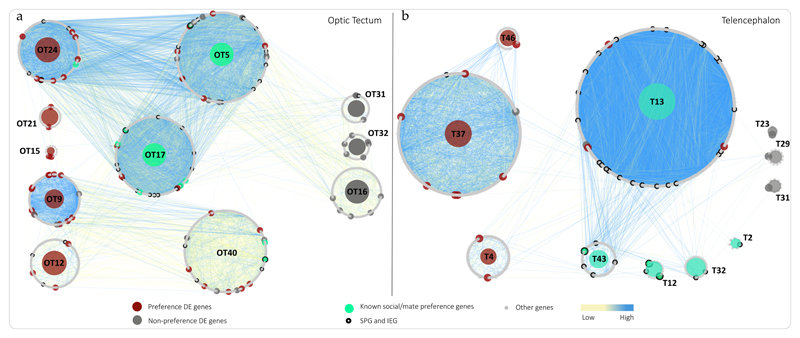
We next identified gene modules in our co-expression network, which represent clusters of genes with highly correlated expression44,48 (Table S3, Fig. S5). Co-expression network modules are a powerful tool in this context, as genes within the same module have been experimentally shown to share functions and/or biological processes45,49. In the optic tectum, five modules (modules OT9, OT12, OT15, OT21 and OT24 - Fig. 3A) are enriched in Preference DE genes. See Table S6 for GO terms associated with these modules.
Figure 3. Optic tectum and telencephalon co-expression networks’ module overview.
Each circle of genes represents a module and the dots forming the module circle represent genes. The size of each module is therefore proportional to the number of genes in that module. The color of each dot refers to its DE category or functional affiliation as shown in the legend. Numbered modules are referred to in text, and correspond to modules after merging (Fig. S5). Modules significantly enriched for Preference DE genes are highlighted in red for Preference lines and grey for Non-preference lines. Modules highlighted in green are significantly enriched in known social behavior/mate preference genes and/or synaptic plasticity genes. Edge connections are highlighted according to weight, with stronger connections, for correlations approaching 1 or -1, shown in blue. Modules with no differentially expressed genes or behavioral genes of interest, as well as edges associated with these modules are hidden for clarity.
Module OT24 is particularly interesting, as it is enriched in Preference DE genes that show strong transcriptional connections to multiple genes known for their role in female preferences in this module and module OT17. Preference DE genes in this subnetwork include gria3, a member of the AMPA glutamate receptor family known to be an important component of the female preference response50. Also scn2a and scn8a, which are known to have molecular functions in brain circuits that mediate specific behaviors51, agap3, involved in signal transduction, syn1, known to be involved in synaptic plasticity and social behavior52, baz2a, which regulates transcription of androgen receptors, and slc24a2, a critical gene in signal transduction53 with known roles in cognition and memory54, and a target of the immediate early gene fosl1. The network structure reveals these genes are connected to other known components of the female preference transcriptional response3,18, including neuroligin-2, neuroligin-3, stmn2a & stmn2b. Such connections, in conjunction with the elevated connectivity and centrality scores, suggest that the Preference DE genes we identified may act to coordinate the transcriptional response behind female preferences documented in previous studies, thus supporting their roles in the initiation of neural and behavioral cascades of female mating decisions.
Once the visual signal travels from the optic tectum into the telencephalon, we see further separation of modules grouping Preference DE genes and modules associated with Non-Preference DE genes. In the telencephalon, modules T4, T37 and T46 are significantly enriched in Preference DE genes while modules T23, T29 and T31 are enriched in Non-Preference DE genes (Fig. 3B). Although not enriched in Preferences DE genes, module T13 is worth noting as it connects three Preference DE genes (out of 12 total) with a very large number of SPG/IEG genes (Fig. 3B). Among the modules enriched in SPG/IEG and social behavior/female preference genes (T2, T12, T13, T32 and T43), modules T12 and T43 group SPG/IEG and genes identified as regulators of female preferences at 30 minutes15,55 that could be activated downstream of the Preference DE genes we identified.
Function and regulation of differentially expressed genes
We found that genes in modules associated with the neurogenomic response of female preference are enriched in pathways underlying neural plasticity13, including ras signaling/long-term potentiation pathways, wnt signaling pathway, neurotrophin signalling pathway and phototransduction (Table S7). Module OT24 in particular, is enriched in GO terms highly relevant to behavior, memory and learning including glutamate receptor signaling pathway (Table S6). We also found that different optic tectum modules are regulated by different sets of transcription factors (TF), and that many of the Preference DE genes are predicted to have TF motifs for immediate early genes egr1, egr2, c-fos and c-jun, as well as neuronal plasticity and long-term memory modulator CREB (Fig. S8).
Telencephalon Preference DE genes include several ribosomal proteins and genes involved in hormone signaling and response, such as eef2 and c2cd5 (Table S6). Promoter analysis shows enrichment for TF motifs for CREB and srf, both part of the CaMK signaling pathway and central regulators of neural plasticity and memory56, as well as pitx2 among others shown in Fig. S8. Aside from ribosomal proteins, all the genes had TF motifs for immediate early genes c-fos and c-jun transcription factors previously associated with activity levels in brain regions mediating various behaviors, including social interactions (Fig. S8).
Preference DE genes in modules OT17 (npr2) and T37 (eef2) have roles in downstream hormone secretion and signaling, being located upstream within the oxytocin signaling pathway, as well as genes in module OT21 (tubb4a and tmem198) in the gonadotropin-releasing hormone receptor pathway, shown to have an important role shaping preferences during interactions with a potential mates57,58 (Table S1). These genes could be responsible for the control of the female physiological changes associated with preparation for mating and reproduction.
Identifying genes that vary in expression in different social interactions
In order to identify genes modulating social interactions beyond mate evaluation, we determined which genes were differentially expressed across all social interactions in all females, independent of their preference phenotype (in Preference and Non-preference lines, Fig. S1). We found 357 such DE genes (denoted Social DE genes) in the optic tectum and 161 in the telencephalon (Table 1, Fig. S6).
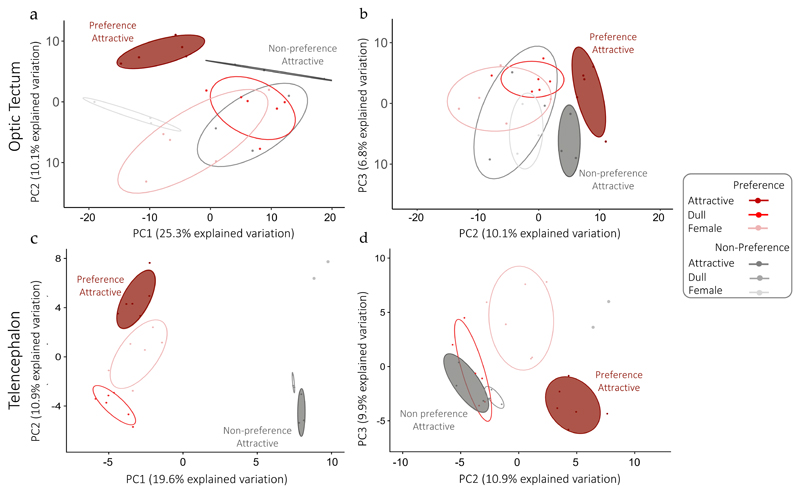
We examined overall differences in the expression patterns of Social DE genes across treatments and lines using principal component analysis (PCA). We found that in both tissues, Preference females exposed to an attractive male exhibit a unique transcriptional signature and cluster as a separate group from the rest of the sample groups based on the first three PCs (Fig. 4). Beyond this, the pattern is different in both tissues. In the optic tectum, except for the attractive treatment in both Preference and Non-preference females, Social DE genes expression in different treatment groups is mostly overlapping (Fig. 4A, 4B). Unlike the optic tectum, PC1 in the telencephalon initially separates samples by preference phenotype (Fig. 4C), however PC2 and PC3 reveal a unique transcriptional pattern in Preference females exposed to an attractive male. Non-preference females lack this unique response to attractive males, so that all male treatments cluster together (Fig. 4D). This suggests that exposure to an attractive male does not trigger a distinct transcriptional response in the telencephalon of Non-preference females.
Figure 4. Differential transcriptional signature of Social DE genes in females exposed to attractive males.
Principal component analysis of Social DE genes in optic tectum (A, n=347) and telencephalon (B, n=161). Points represent samples for each treatment/line group. In graphs on the left the two first principal components are plotted, and in graphs on the right PC2 is plotted against PC3, with the proportion of variance explained by each component printed next to the axes labels.
Social DE genes include genes related to synaptic plasticity, learning, memory and social behavior such as grin1, bdnf, neurod2, fos and egr2b13,16,18,50,59–61. Social DE genes in both tissues are linked in several pathways relevant in behavior such as ras signaling pathway, wnt signaling pathway, GnRH receptor pathway and corticotropin-releasing factor receptor signaling pathway among others (Table S8). Promoter region analysis62 suggests that Preference DE genes in the optic tectum and telencephalon co-expression networks have TF motifs for our Social DE genes (Table S9), indicating that differences in the expression of Social DE genes may trigger distinct transcriptional cascades in the different mating and social contexts of our experiment (Fig. S7, Table S9).
Discussion
Our goal was to characterize the neurogenomic response of female preference by identifying the differences in gene expression triggered by different mating contexts in females with and without a preference for colorful males32. This comparative framework allowed us to investigate which elements of the response differ in females that lack preference for attractive males32, thus identifying the neurogenomic basis of variation in female preferences that are key to sexual selection and sexual conflict. We specifically targeted genes involved in the early female preference neuromolecular response by studying the transcriptional changes after only 10 minutes of mate exposure.
In both the optic tectum and telencephalon, we identified genes that differ in expression in different social contexts (Fig. 4) and found evidence that the transcription factors among these genes likely act as neuromolecular switches triggering distinct neurogenomic states that form the basis of mating decisions and social behaviors. Consistent with this idea, we found multiple genes with unique transcriptional signatures in Preference females exposed to an attractive male, suggesting they are part of the neurogenomic response of female preference (Fig. 2). These Preference DE genes are assembled into discrete genetic modules in the optic tectum and telencephalon, revealing the structure of the transcriptional response uniquely associated with female preference, as well as the connections to other genes known to have regulating roles in social behavior, mate preferences, learning and memory (Fig. 3).
The centrality and connectivity of Preference DE genes in the optic tectum and telencephalon showed that the properties of the response are different in both brain tissues. While we saw a diffuse response associated with female preference at the sensory processing level, with DE genes at all levels of the network, we see a highly centralized response for DE genes in the decision-making telencephalon. In addition to highlighting differences in the properties of the response at the sensory-processing and decision-making levels, a highly centralized response in the telencephalon is exactly what we would expect of the genes that initiate the female preference transcriptional response leading to the alternative mating decisions that follow.
Furthermore, Preference DE genes have similar expression patterns in females with and without preferences in all but the attractive male treatment at the sensory processing level (optic tectum), suggesting that Non-preference females do perceive differences between both types of males. However, at the decision-making level (telencephalon) Preference DE genes are not activated in response to any social interactions in Non-preference females (Fig. 2). These findings, combined with the expression pattern of Social DE genes (PCA, Fig. 4), where we see strong differentiation in telencephalon expression between lines with different preference phenotypes along PC1, suggest there are crucial differences in the neurogenomic response behind social and mating behaviors in the telencephalon. The expression differences seen along PC1 at the decision-making level could be a reflection of the proven differences in cognitive ability between lines33 and consistent with the notion that cognition plays an important role in mating decisions37,46.
Herbert63 originally introduced the idea that limited genetic elements can encode for the multiple behaviors required to appropriately respond to various stimuli in different social and mating contexts, via complex combination of spatial and temporal activation in different brain nuclei. Here, we see evidence for a group of genes that have different expression levels in various mating contexts grouped in several discrete modules associated with female preferences, revealing the modularity of the neurogenomic preference response we observe. We see further evidence of how the brain can flexibly respond to different stimuli in the observation that multiple synaptic plasticity and immediate early genes are present in our Social DE genes, including grin1 (NMDAR), march8, bdnf, thoc6, cant1 and thap6 in the optic tectum and inhba, neurod2, smarcc1, fos, egr2b and thap6 in the telencephalon. Different social behaviors have been shown to be characterized by different patterns of gene activity across the different nodes of the telencephalon forming the social decision-making network76,77, rather than the gene activity of a single node. It would thus be a useful avenue for future research to continue to dissect how the brain mediates its response to mating stimuli by examining detailed patterns of expression of Preference DE genes and Social DE genes across the different nodes of the telencephalon.
The comparative framework we use here allowed the identification of genes and gene modules associated with variation in female preference, and which likely factor in the neurogenomic response behind female mate choice. These findings provide a clear testable hypothesis to investigate the mechanisms behind the repeated and independent evolution of divergent female preference for colorful males across wild guppy populations23,26,64,65. Together, our results reveal the unique transcriptional response related to the earliest stages of female preference behavior, show the modularity of this response, and identify the potential regulatory basis of this transcriptional response. Our approach and results provide a strong comparative framework for studies on the conservation of mate preference transcriptional networks across populations and species.
Materials and Methods
Study system
Guppies used in our experiment are laboratory-raised descendants of Trinidad guppies sampled from the high predation populations of the Quare River (Trinidad). We based our study on guppies from this wild-type population and six selection lines, derived from the wild-type fish, which have been selected on relative brain size. In summary, fish were indirectly selected based on parental brain size achieving a difference of up to 13.6% in relative brain size among three replicate lines selected to have small brains, here denoted small brain lines (SB lines), and three replicate lines selected to have larger brain (LB lines)33,66. All the details on the selection experiment have been previously published33. Brain size in these lines has been shown to carry significant costs and benefits, conferring better cognitive abilities and better response to predators in large brain lines33,66,67. These differences however are not likely due to the accumulation of deleterious alleles in small-brain lines as these were shown to be more fecund33, to have a better immune response68 and faster juvenile growth69. We recently showed females from wild type and selection lines have measurable differences in their female preference for colorful males. While females from LB lines have maintained the clear female preference for colorful males seen in the wild type line, SB females lack this preference32. We demonstrated that this difference in preference phenotype is not due to differences in opsin sequence or expression in the retina, or to variation in color perception across lines32.
For this study, we used virgin females from the fifth generation of selection, all aged approximately 6 months. None of the females used in this experiment were used for other behavioral experiments prior to this study. Fish were raised at a water temperature of 25°C with a 12:12 light:dark schedule, and fed an alternating daily diet of flake food and live Artemia (brine shrimp). After the first onset of sexual maturation, females were placed in 12-liter tanks in groups of 10 fish. All tanks contained gravel, biological filters and Java moss (Vesicularia dubyana). In addition, we allowed visual contact between tanks containing females to enrich the social environment but females never saw a mature male prior the experiment. Experiments were done in accordance with ethical permits approved by Stockholm Ethical Board (Dnr: N173/13, 223/15 and N8/17).
Preference tests
Selection of presentation males
For our study we divided females among three treatments: two treatments represented a male evaluation context, in which females were presented either an attractive male (attractive treatment) or an unattractive male (dull treatment), and a third treatment in which females were exposed to another female representing a general social interaction treatment. Previous studies have demonstrated females are attracted to males with brighter and larger orange areas and longer tails23. Following general methods previously described32, we selected 30 wild-type males from the laboratory population stock for their colorful or dull patterns based on visual inspection. Next, these 30 males were anesthetized with a low dose of benzocaine and photographed on both sides using a Nikon D5300 camera. We scored total coloration, body length, and tail area of each male using the ImageJ software v. 1.4470. Then, we selected the four males with highest and lowest coloration that could be matched by body length. Prior to the trial we made sure that these males were sexually mature by housing them together with females not participant in the experiment and observing their sexual behavior. As color patterns might change over time in young fish, we repeated the whole procedure after 5 days of experiment. In total, we used three sets of colorful-dull males during the experiment. On average, the 12 selected colorful males presented 23% more total coloration, and 16% larger tails than the 12 dull males.
Behavioral treatments
We used a total of 45 wild-type females, 45 large brain females and 45 small brain females divided equally across the three treatments. For the selection lines we used five females each from the three replicates. We allowed each focal female to observe the presented fish for only 10 minutes before ending the experiment based on our findings in a previous female mate choice study in these lines32. This timeframe was chosen based on previous studies32 as an early time point in which differences in female behavior could be observed. This short presentation time also minimizes the possibility of habituation to the experimental setup. Preference tests were carried out in a divided tank (84x40x20 cm), which controlled for the focal female perceiving any chemical or mechanical signals. All fish were netted and transferred to their respective experimental tanks 24h before the start of the experiment for acclimation. We ensured that all females used in gene expression analyses showed sexual interest in the males offered. For this, all trials were followed by an observer through a live broadcast of the experimental setup in a separate room to avoid disturbances. For consistency, all trials were conducted on 15 consecutive days. Focal females belonging to same replicate selection line and the same treatment were presented with different males to avoid uncontrolled male-driven changes in expression. For this, we balanced the number of large-brained, small-brained and wild-type females presented to colorful males, dull males and females respectively per day (nine trials per day). We have previously shown that our selection lines do not significantly differ in any behavior and movement patterns in mating contexts and/or during the preference tests32,71–73. This extensive work showed no evidence for any behavioral differences in perception, activity or swimming behavior that could affect the results.
At the end of each trial, females were euthanized by transfer to ice water. After 45 seconds, and with aid of a Leica S4E microscope, we removed the top of the skull to expose the brain. We cut the olfactory and optic nerves and extracted the following forebrain regions: dorsal telencephalon, ventral telencephalon (harboring the preoptic area) and olfactory bulbs. We severed the telencephalon from the rest of the brain between the ventral telencephalon and thalamus at the “commissura anterioris”, including both the pallium and subpallium regions. The thalamus region was excluded from our samples. As the olfactory bulbs are very small in guppies (typically < 2.9 % of the forebrain mass74), we use “telencephalon” when relating to samples extracted from these forebrain regions. Next, after detachment of the cerebellar region, we dissected out the laminated superior area of the optic tectum (Fig. 1C). Dissection procedure took place in ice water within three minutes. The telencephalon and optic tectum tissue samples were immediately preserved in RNAlater (Ambion) at room temperature for 24 hours and then at -20°C until RNA extraction.
RNA extraction and sequencing
In order to recover sufficient RNA for RNAseq, we pooled tissue from five individuals. For consistency, samples were pooled combining tissue for the same individuals for the optic tectum and telencephalon. This produced three replicate pools per treatment for each the wild-type line, the large-brain line and the small brain lines for optic tectum and telencephalon (three pools per treatment/line = nine pools per line and thus 27 pools in total for each tissue). Each sample pool was homogenized and RNA was extracted using Qiagen’s RNAeasy kits following standard manufacturer’s protocol. Libraries for each sample were prepared and sequenced by the Wellcome Trust Center for Human Genetics at the University of Oxford, UK. All samples were sequenced across 10 lanes on an Illumina HiSeq 4000. We obtained on average 52 million 75bp read pairs per sample (47.1 million read pairs minimum, 72 million maximum).
Assembly construction
Read quality control and trimming
We assessed the quality of reads for each sample using FastQC v.0.11.4. (www.bioinformatics.babraham.ac.uk/projects/fastqc). After verifying initial read quality, reads were trimmed with Trimmomatic v0.3575. We filtered adaptor sequences and trimmed reads if the sliding window average Phred score over four bases was <15 or if the leading/trailing bases had a Phred score <3, removing reads post filtering if either read pair was <33 bases in length. Quality was verified after trimming with FastQC. After trimming we had a total of approximately 537.6 million trimmed read pairs, 44.8 on average per individual (minimum: 36.2 million trimmed read pairs, maximum: 56.2 million trimmed read pairs).
De novo assembly
Because the current guppy genome annotation is incomplete76, we constructed a de novo transcriptome assembly in order to include loci that might be missing from the current annotation. All forward and reverse reads were pooled and assembled de novo with Trinity v2.2 77 using default parameters. We filtered the resulting assembly for non-coding RNA using medaka (Oryzias latipes) and Amazon molly (Poecilia formosa) non-coding RNA sequences as reference in a nucleotide BLAST (Blastn). After eliminating all sequence matching non-coding RNAs we picked the best isoform for each transcript. We defined the best isoform as the one with the highest expression as estimated by mapping the reads to the de novo assembly using RSEM (v1.2.2078). Finally, we used Transdecoder (Transdecoder v3.0.1, http://transdecoder.github.io) with default parameters to filter out all transcripts without an open-reading frame and/or shorter than 150bp (Table S10).
Genome guided assembly
We assembled a genome-guided assembly using the HiSat 2.0.5 - Stringtie v1.3.2 suite79. We based our genome-guided assembly on the published guppy genome assembly (Guppy_female_1.0 + MT, RefSeq accession: GCA_000633615.1, latest release June 2016)76. Samples were individually mapped to the genome and built into transcripts using default parameters but preventing the software from assembling de novo transcripts. The resulting individual assemblies were then merged into a single, non-redundant assembly using the built-in StringTie-merge function. In a similar fashion to the de novo assembly, we filtered out non-coding RNA and chose the best isoform for each transcript based on expression (Table S10).
Reference Transcriptome assembly
We used CD-Hit-Est to obtain a non-redundant reference transcriptome (RefTrans) by fusing the de novo and genome guided assemblies. Transcripts longer than 150bp were clustered if they were >95% similar preserving the longest representative for each cluster.
The resulting reference transcriptome was annotated by performing a BlastX to NCBI’s non-redundant database. The associated gene IDs obtained here were used to search multiple databases in all downstream GO annotations and pathway analysis as detailed below. See Table S10 for details on the final number of transcripts preserved in the reference transcriptome and annotation statistics.
Differential expression
We quantified expression by mapping paired reads for each sample separately to the Reference Transcriptome using RSEM version 1.2.2078, filtering transcripts <2 RPKM (reads per kilobase per million mapped reads), preserving only those transcripts that have expression above this threshold in a least half of the samples for each treatment within a line. After this final filter, a total of 21,131 transcripts were kept for further analysis, 20,396 in the optic tectum and 19,571 in the telencephalon. Using sample correlations in combination with MDS plots based on all expressed transcripts, we determined that out of the 54 samples one optic tectum wild-type attractive male treatment sample, one optic tectum wild-type female treatment and one telencephalon small-brain female treatment sample were significant outliers and were thus excluded from further analysis.
We relied on a random permutation test as described in Ghalambor et al.80. Filtered read counts were normalized using standard function as implemented in DESeq281 (Fig. S1) and used to perform a generalized linear model (GLM) to each transcript, to evaluate the effect of treatment on expression level. Because we were interested in contrasting differences in expression associated with preference, we performed this analysis grouping lines by their preference phenotype, and also carried out the GLM separately for Preference lines (Wild-type and LB lines) and Non-preference lines (SB lines). After grouping samples by the female preference phenotype the analysis was performed with six samples for Preference lines and three samples for Non-preference lines, except for treatments for which we had to remove one outlier (see Table S11 for details on sample sizes). This way we performed GLM to assess the significance of expression differences in pairwise comparisons between attractive and dull treatments, attractive and female treatments and, finally dull and female treatments in Preference and Non-preference lines (Fig. 1B). To control for false positives and determine which transcripts were differentially expressed between treatments we used a random permutations test80. We generated 250 permuted datasets by randomly reassigning the sample names for the entire dataset of each tissue. Then we performed GLM in the exact same way as for the actual data, thus generating an empirical null distribution of 250 p-values for each transcript. A transcript was considered differentially expressed when the statistic for the actual expression data fell below the 5% tail of the permutated data p-value distribution. This method has been shown to better capture the structure of the data and does not assume independence across genes as other multiple test correction methods that can be over-corrective4,82.
Our study relies on the assumption that mRNA levels correlate well with protein levels, which has been well supported in multiple other species 83–86. Here we use a differential expression approach so that the mRNA-to-protein ratio would be the same in all samples and therefore would not impact our results.
Differentially expressed genes involved in the mating decision: comparisons within Preference lines
To determine which genes are involved in the mating decision we focused on the genes we found to be differentially expressed between the attractive and dull treatments in Preference lines. We applied several filters to the initial set of differentially expressed genes that passed the permutation threshold, retaining only those that have a potential role in mate choice based on their expression. We initially filtered out all genes that lack concordant expression (i.e. genes that change in the same direction between pairs of treatments across all replicate samples) between attractive and dull treatments in all Preference lines, and then we retained those genes that are also differentially expressed between attractive and female treatments (Fig. S1). Finally, we excluded genes also differentially expressed in dull male vs female comparisons, keeping only those genes associated with the evaluation of an attractive male (in area P of Fig 1B). Here we assume that any gene important in the evaluation of males of different qualities should also be differentially expressed between the attractive and female treatments, and this way we were able to control for genes that change relative to social interaction alone. We refer to this final set of genes as Preference DE genes (Table 1).
Differentially expressed genes involved social interactions
We initially identified genes involved across the different social interactions we tested, independent of the female preference phenotype and the social context. For this purpose we considered all genes determined to be differentially expressed across all three pairwise treatment comparisons separately within Preferences lines and Non-preference lines. These are genes that are differentially expressed in both mating context and general social interactions. Among these genes we selected only those that are differentially expressed in both Preference and Non-preference females as these are the ones that become differentially expressed in different social context in all the guppies we studied, independent of their selection regime. We refer to these genes as Social DE genes.
Comparative analysis of genes involved in mate evaluation
To address the question of what genes and pathways differ between Preference and Non-preference females, we identified genes that were differentially expressed between attractive and dull treatments in Non-preference lines. We proceeded in the same fashion as described above for Preference DE genes (Non-preference DE genes - Table 1).
Co-expression networks
In order to study the relationship between genes expressed in the optic tectum and telencephalon, we used weighted correlation network analysis, also known as weighted gene co-expression network analysis (WGCNA) using the WGCNA package in R43,44.
We built a weighted co-expression network for each tissue using genes that passed the expression filter described above. This way we avoid using genes with non-significant variance and lowly expressed genes that generally represent transcriptional noise43,44. The input count data used to build co-expression networks was normalized and transformed using the variance-stabilizing transformation as implement in DESeq2 as recommended by WGCNA authors. First, a Similarity matrix of the pairwise correlations between genes was built using log transformed normalized data using a weighted combination of the Pearson correlation and Euclidean distance S = SIGN (corrx) x {|corrx| +[1 - log (distx + 1)]/max[log (distx +1)]/2} as previously described87. We determined the most appropriate soft-threshold to use in order to reduce the number of spurious correlations based on the criterion of approximate scale-free topology44, determined to be six for the telencephalon and four for the optic tectum. We used these soft-thresholds to build the Adjacency matrix and corresponding Topological Overlap matrix (TOM), a matrix of pairwise distance values between genes. Finally, we retained correlations >0.4, based on the correlation value distribution for each tissue, and genes that had >2 connections to other genes in the co-expression networks for all downstream analyses (Fig. S4). Optic tectum and telencephalon network properties are summarized in Table S3.
Module identification
We built a dendrogram of all genes based on the TOM matrix using hierarchical clustering in order to identify the gene modules in each tissue network. We then used the Dynamic Tree Cut method as implemented in WGCNA, using the “tree” method and with a minimum cluster size of 30 genes, to detect the module based on the clustering (Fig. S5). The Dynamic Tree Cut method identified modules whose expression profiles are very similar. We did a further step to merge those modules with highly correlated expression values by estimating module eigengenes as described in43,44 (Fig. S5).
Co-expression network analysis
Final co-expression networks were exported to Cytoscape88 for further network data integration and visualization (Fig. S4). Information on whether a gene was a differentially expressed gene or known to be a gene involved in social interaction and mate preference was attached to the network as metadata so they could be visualized in all downstream network analysis (Figs. 4, S4).
The Network Analyzer tool in Cytoscape was used to calculate network node attributes. These give an indication of how connected and central a gene is in the network. Here we focused on three such attributes89: (1) Degree: the number of edges, i.e. other genes, each gene is connected to within the network. Central genes in the network will therefore have high degree values as opposed to more peripheral network genes. (2) Neighborhood connectivity: defined as the average connectivity, or number of neighbors, for all its neighbors. (3) Clustering coefficient: the ratio of the number of edges between the neighbors of a gene, and the maximum number of edges that could possibly exist between such neighbors (number between 0 and 1). This is a measure of how connected a gene is relative to how connected it could be given the number of neighbors it has. This value will approach 0 for an unconnected gene and 1 for a fully connected gene in the center of a network. We evaluated connectivity and centrality of differentially expressed genes by examining the degree, neighborhood connectivity and clustering coefficient of these genes in the optic tectum and telencephalon networks (Table 2, Fig. S4). We carried out t-tests of log-transformed data to determine whether these attributes differ between optic tectum and telencephalon’s networks for each differentially expressed gene group (attractive vs dull in preference and Non-preference lines) and for gene groups known to be important in mating behavior (lists on tables S4, S5).
We performed enrichment tests to determine whether modules were enriched in differentially enriched genes of any category using one-tail fisher’s exact test (Fig. 3). We carried out similar tests to determine which modules in the network are enriched in gene previously known to be involved in social interactions and or mate preference and in social plasticity genes/immediate early genes (IEG).
Functional analyses
To study the biological functions and pathways associated with differentially expressed genes and gene modules we obtained Gene Ontology (GO) annotations for all expressed genes in the reference transcriptome that had a blast hit to the non-redundant (nr) and Swissprot databases. We performed GO term enrichment tests in TopGO (R package) using the annotated Reference transcriptome we build as background in one-tail Fisher’s exact tests with a p-value threshold of p<0.05 (Table S6).
We determined which known pathways are associated with Preference DE genes within each module using hits to the human database in g:Profiler62. In a similar fashion, we investigated which transcription factors are known to regulate Preference DE genes within each module. This analysis was also based on data for humans, relying on the TransFac transcription factor binding sites database integrated into g:Profiler, as it is far more complete than databases for other species. Although providing a more complete view of the transcription factor motifs associated with Preference DE genes, it is important to keep in mind that some TF motifs are likely to be different in a distant vertebrate like the guppy. Within transcription factor motifs found to be enriched among Preference DE genes we identified those for transcription factors with known roles in mate preference (Table S4) as well as synaptic plasticity and immediate early genes (Table S5). Additionally, we focused on transcription factors belonging to families previously identified in behavioral genetics studies such as zinc finger proteins (znf) or POU domain transcription factors (Fig. S8).
Supplementary Material
Acknowledgements
This work was funded by Marie Sklodowska-Curie Fellowship 654699 and National Science Foundation Postdoctoral Fellowship in Biology 1523669 to N.I.B., by grant agreements 260233 and 680951 from the European Research Council to J.E.M., Swedish Research Council grant 2016-03435 to N.K. and Knut and Alice Wallenberg grant 102 2013.0072 to N.K. We gratefully acknowledge support from a Royal Society Wolfson Merit Award to J.E.M. We wish to thank four anonymous reviewers and our editor for careful reviews and constructive criticism of our manuscript. We especially thank Pedro Almeida, Iulia Darolti, Jake Morris, Vicencio Oostra, Alison Wright and Trevor Price for valuable discussion and help with manuscript preparation. We thank the Oxford Genomics Centre at the Wellcome Centre for Human Genetics (funded by Wellcome Trust grant reference 203141/Z/16/Z) for the generation and initial processing of the sequencing data, and the UCL Legion High Performance Computing Facility (Legion@UCL).
Footnotes
Data accessibility: Normalized counts for all groups of differentially expressed genes as well as all expressed genes are available as Supplementary Datasets. RNA reads have been deposited at the NCBI Sequencing Read Archive, BioProject ID PRJNA413692. Additional data may be requested from the authors.
Author contributions: N.I.B., A.C-L., N.K. and J.E.M. conceived of the study and designed the experiments. A.K. and N.K. created the brain size selection lines. A.K. and S.D.B. performed laboratory work for fish housekeeping. A.C-L. and S.D.B. selected fish for experiments. A.C-L. performed the behavioral tests and dissected brain regions. N.I.B. performed all laboratory RNA work and analyzed data. All authors contributed to writing the manuscript.
Competing interests: The experiment was performed in accordance with ethical applications approved by the Stockholm Ethical Board (Reference number: N173/13, 223/15 and N8/17). These applications are consistent with the Institutional Animal Care and Use Committee guidelines. The authors declare that they have no competing financial interests.
References
- 1.Zayed A, Robinson GE. Understanding the Relationship Between Brain Gene Expression and Social Behavior: Lessons from the Honey Bee. 2012;46:591–615. doi: 10.1146/annurev-genet-110711-155517. [DOI] [PubMed] [Google Scholar]
- 2.O’Connell LA, Hofmann HA. Genes, hormones, and circuits: An integrative approach to study the evolution of social behavior. Frontiers in Neuroendocrinology. 2011;32:320–335. doi: 10.1016/j.yfrne.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Cummings ME. The mate choice mind: studying mate preference, aversion and social cognition in the female poeciliid brain. Anim Behav. 2015;103:249–258. [Google Scholar]
- 4.Ghalambor CK, et al. Non-adaptive plasticity potentiates rapid adaptive evolution of gene expression in nature. Nature. 2015;525:372–375. doi: 10.1038/nature15256. [DOI] [PubMed] [Google Scholar]
- 5.Hitzemann R, et al. Genes, behavior and next‐generation RNA sequencing. Genes, Brain and Behavior. 2013;12:1–12. doi: 10.1111/gbb.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenthal GG. Mate Choice. Princeton University Press; 2017. [Google Scholar]
- 7.Zahavi A. Mate selection—A selection for a handicap. Journal of Theoretical Biology. 1975;53:205–214. doi: 10.1016/0022-5193(75)90111-3. [DOI] [PubMed] [Google Scholar]
- 8.Kokko H, Brooks R, Jennions MD, Morley J. The evolution of mate choice and mating biases. P Roy Soc Lond B. 2003;270:653–664. doi: 10.1098/rspb.2002.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson GE, Fernald RD, Clayton DF. Genes and Social Behavior. Science. 2008;322:896–900. doi: 10.1126/science.1159277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitney O, et al. Core and region-enriched networks of behaviorally regulated genes and the singing genome. Science. 2014;346:1334–+. doi: 10.1126/science.1256780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clayton DF. The Genomic Action Potential. Neurobiology of Learning and Memory. 2000;74:185–216. doi: 10.1006/nlme.2000.3967. [DOI] [PubMed] [Google Scholar]
- 12.Wang SMT, Ramsey ME, Cummings ME. Plasticity of the mate choice mind: courtship evokes choice‐like brain responses in females from a coercive mating system. Genes, Brain and Behavior. 2014;13:365–375. doi: 10.1111/gbb.12124. [DOI] [PubMed] [Google Scholar]
- 13.Cardoso SD, Teles MC, Oliveira RF. Neurogenomic mechanisms of social plasticity. J Exp Biol. 2015;218:140–149. doi: 10.1242/jeb.106997. [DOI] [PubMed] [Google Scholar]
- 14.Cummings ME, et al. Sexual and social stimuli elicit rapid and contrasting genomic responses. P Roy Soc Lond B. 2008;275:393–402. doi: 10.1098/rspb.2007.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch KS, Ramsey ME, Cummings ME. The mate choice brain: comparing gene profiles between female choice and male coercive poeciliids. Genes, Brain and Behavior. 2012;11:222–229. doi: 10.1111/j.1601-183X.2011.00742.x. [DOI] [PubMed] [Google Scholar]
- 16.Ramsey ME, Maginnis TL, Wong RY, Brock C, Cummings ME. Identifying context-specific gene profiles of social, reproductive, and mate preference behavior in a fish species with female mate choice. Frontiers in Neuroscience. 2012;6:62. doi: 10.3389/fnins.2012.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong RY, Oxendine SE, Godwin J. Behavioral and neurogenomic transcriptome changes in wild-derived zebrafish with fluoxetine treatment. BMC Genomics. 2013;14:1. doi: 10.1186/1471-2164-14-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teles MC, Cardoso SD, Oliveira RF. Social Plasticity Relies on Different Neuroplasticity Mechanisms across the Brain Social Decision-Making Network in Zebrafish. Front Behav Neurosci. 2016;10:991–12. doi: 10.3389/fnbeh.2016.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taborsky B, Oliveira RF. Social competence: an evolutionary approach. Trends Ecol Evol. 2012;27:679–688. doi: 10.1016/j.tree.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Weitekamp CA, Hofmann HA. Evolutionary themes in the neurobiology of social cognition. Curr Opin Neurobiol. 2014;28:22–27. doi: 10.1016/j.conb.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Dukas R. Evolutionary Biology of Animal Cognition. 2004;35:347–374. doi: 10.1146/annurev.ecolsys.35.112202.130152. [DOI] [Google Scholar]
- 22.Woolley SC, Doupe AJ. Social Context–Induced Song Variation Affects Female Behavior and Gene Expression. Plos Biol. 2008;6:e62. doi: 10.1371/journal.pbio.0060062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houde AE. Sex, Color, and Mate Choice in Guppies. Princeton University Press; 1997. [Google Scholar]
- 24.Endler JA. Multiple-trait coevolution and environmental gradients in guppies. Trends Ecol Evol. 1995;10:22–29. doi: 10.1016/s0169-5347(00)88956-9. [DOI] [PubMed] [Google Scholar]
- 25.Brooks R. Variation in Female Mate Choice within Guppy Populations: Population Divergence, Multiple Ornaments and the Maintenance of Polymorphism. Genetica. 2002;116:343–358. [PubMed] [Google Scholar]
- 26.Houde AE, Endler JA. Correlated Evolution of Female Mating Preferences and Male Color Patterns in the Guppy Poecilia reticulata. Science. 1990;248:1405–1408. doi: 10.1126/science.248.4961.1405. [DOI] [PubMed] [Google Scholar]
- 27.Endler JA, Houde AE. Geographic variation in female preferences for male traits in Poecilia reticulata. Evolution. 1995;49:456–468. doi: 10.1111/j.1558-5646.1995.tb02278.x. [DOI] [PubMed] [Google Scholar]
- 28.Brooks R, Endler JA. Female guppies agree to differ: phenotypic and genetic variation in mate‐choice behavior and the consequences for sexual selection. Evolution. 2001;55:1644–1655. doi: 10.1111/j.0014-3820.2001.tb00684.x. [DOI] [PubMed] [Google Scholar]
- 29.Sandkam B, Young CM, Breden F. Beauty in the eyes of the beholders: colour vision is tuned to mate preference in the Trinidadian guppy (Poecilia reticulata) Molecular Ecology. 2015;24:596–609. doi: 10.1111/mec.13058. [DOI] [PubMed] [Google Scholar]
- 30.Hughes KA, Houde AE, Price AC, Rodd FH. Mating advantage for rare males in wild guppy populations. Nature. 2013;503:108–110. doi: 10.1038/nature12717. [DOI] [PubMed] [Google Scholar]
- 31.Rodd FH, Hughes KA, Grether GF, Baril CT. A possible non-sexual origin of mate preference: are male guppies mimicking fruit? P Roy Soc Lond B. 2002;269:475–481. doi: 10.1098/rspb.2001.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corral Lopez A, et al. Female brain size affects the assessment of male attractiveness during mate choice. Science Advances. 2017;3:e1601990. doi: 10.1126/sciadv.1601990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotrschal A, et al. Artificial Selection on Relative Brain Size in the Guppy Reveals Costs and Benefits of Evolving a Larger Brain. Curr Biol. 2013;23:168–171. doi: 10.1016/j.cub.2012.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y-C, et al. Expression change in Angiopoietin-1 underlies change in relative brain size in fish. P Roy Soc Lond B. 2015;282 doi: 10.1098/rspb.2015.0872. 20150872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Replogle K, et al. The Songbird Neurogenomics (SoNG) Initiative: Community-based tools and strategies for study of brain gene function and evolution. BMC Genomics. 2008;9:131. doi: 10.1186/1471-2164-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Northcutt RG. Forebrain evolution in bony fishes. Brain Research Bulletin. 2008;75:191–205. doi: 10.1016/j.brainresbull.2007.10.058. [DOI] [PubMed] [Google Scholar]
- 37.Bshary R, Gingins S, Vail AL. Social cognition in fishes. Trends Cogn Sci (Regul Ed) 2014;18:465–471. doi: 10.1016/j.tics.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Salas C, et al. Neuropsychology of learning and memory in teleost fish. Zebrafish. 2006;3:157–171. doi: 10.1089/zeb.2006.3.157. [DOI] [PubMed] [Google Scholar]
- 39.Derycke S, et al. Neurogenomic Profiling Reveals Distinct Gene Expression Profiles Between Brain Parts That Are Consistent in Ophthalmotilapia Cichlids. Frontiers in Neuroscience. 2018;12:e1002962. doi: 10.3389/fnins.2018.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindholm A, Breden F. Sex Chromosomes and Sexual Selection in Poeciliid Fishes. Am Nat. 2010 doi: 10.1086/342898. [DOI] [PubMed] [Google Scholar]
- 41.Kirkpatrick M, Hall DW. Sexual selection and sex linkage. Evolution. 2004;58:683–691. doi: 10.1111/j.0014-3820.2004.tb00401.x. [DOI] [PubMed] [Google Scholar]
- 42.Kirkpatrick M, Ryan MJ. The evolution of mating preferences and the paradox of the lek. Nature. 1991 [Google Scholar]
- 43.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Bin, Horvath S. A General Framework for Weighted Gene Co-Expression Network Analysis. Statistical Applications in Genetics and Molecular Biology. 2005;4 doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 45.Iancu OD, Colville A, Darakjian P, Hitzemann R. Coexpression and cosplicing network approaches for the study of mammalian brain transcriptomes. Int Rev Neurobiol. 2014;116:73–93. doi: 10.1016/B978-0-12-801105-8.00004-7. [DOI] [PubMed] [Google Scholar]
- 46.Cummings ME. Sexual conflict and sexually dimorphic cognition—reviewing their relationship in poeciliid fishes. Behav Ecol Sociobiol. 2018;72:73. [Google Scholar]
- 47.Galizia G, Lledo P-M. Neurosciences - From Molecule to Behavior: a university textbook. Springer Science & Business Media; 2013. [DOI] [Google Scholar]
- 48.Langfelder P, Horvath S. Fast R Functions for Robust Correlations and Hierarchical Clustering. Journal of statistical software. 2012;46 [PMC free article] [PubMed] [Google Scholar]
- 49.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 50.Ramsey ME, Vu W, Cummings ME. Testing synaptic plasticity in dynamic mate choice decisions: N-methyl d-aspartate receptor blockade disrupts female preference. P Roy Soc Lond B. 2014;281:20140047–20140047. doi: 10.1098/rspb.2014.0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krumm N, O’Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37:95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greco B, et al. Autism-related behavioral abnormalities in synapsin knockout mice. Behav Brain Res. 2013;251:65–74. doi: 10.1016/j.bbr.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larhammar D, Nordström K, Larsson TA. Evolution of vertebrate rod and cone phototransduction genes. Phil Trans R Soc Lond B. 2009;364:2867–2880. doi: 10.1098/rstb.2009.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moriguchi S, et al. Reduced CaM Kinase II and CaM Kinase IV Activities Underlie Cognitive Deficits in NCKX2 Heterozygous Mice. Mol Neurobiol. 2017;21:1–12. doi: 10.1007/s12035-017-0596-1. [DOI] [PubMed] [Google Scholar]
- 55.Cummings ME, Ramsey ME. Mate choice as social cognition: predicting female behavioral and neural plasticity as a function of alternative male reproductive tactics. Current Opinion in Behavioral Sciences. 2015;6:125–131. [Google Scholar]
- 56.Wolf C, Linden DEJ. Biological pathways to adaptability--interactions between genome, epigenome, nervous system and environment for adaptive behavior. Genes, Brain and Behavior. 2012;11:3–28. doi: 10.1111/j.1601-183X.2011.00752.x. [DOI] [PubMed] [Google Scholar]
- 57.Cui R, Delclos PJ, Schumer M, Rosenthal GG. Early social learning triggers neurogenomic expression changes in a swordtail fish. P Roy Soc Lond B. 2017;284 doi: 10.1098/rspb.2017.0701. 20170701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okuyama T, et al. A Neural Mechanism Underlying Mating Preferences for Familiar Individuals in Medaka Fish. Science. 2014;343:91–94. doi: 10.1126/science.1244724. [DOI] [PubMed] [Google Scholar]
- 59.Minatohara K, Akiyoshi M, Okuno H. Role of Immediate-Early Genes in Synaptic Plasticity and Neuronal Ensembles Underlying the Memory Trace. Front Mol Neurosci. 2015;8:78. doi: 10.3389/fnmol.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cummings ME. Looking for sexual selection in the female brain. Phil Trans R Soc Lond B. 2012;367:2348–2356. doi: 10.1098/rstb.2012.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kowiański P, et al. BDNF: A Key Factor with Multipotent Impact on Brain Signaling and Synaptic Plasticity. Cell Mol Neurobiol. 2017;358:735–15. doi: 10.1007/s10571-017-0510-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reimand J, et al. g:Profiler-a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44:W83–9. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herbert J. Peptides in the limbic system: neurochemical codes for co-ordinated adaptive responses to behavioural and physiological demand. Prog Neurobiol. 1993;41:723–791. doi: 10.1016/0301-0082(93)90033-o. [DOI] [PubMed] [Google Scholar]
- 64.Alexander HJ, Taylor JS, Wu SST, Breden F. Parallel evolution and vicariance in the guppy (poecilia reticulata) over multiple spatial and temporal scales. Evolution. 2006;60:2352–2369. [PubMed] [Google Scholar]
- 65.Suk HY, Neff BD. Microsatellite genetic differentiation among populations of the Trinidadian guppy. Heredity. 2009;102:425–434. doi: 10.1038/hdy.2009.7. [DOI] [PubMed] [Google Scholar]
- 66.Kotrschal A, Corral Lopez A, Amcoff M, Kolm N. A larger brain confers a benefit in a spatial mate search learning task in male guppies. Behav Ecol. 2015;26:527–532. doi: 10.1093/beheco/aru227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van der Bijl W, Thyselius M, Kotrschal A, Kolm N. Brain size affects the behavioural response to predators in female guppies (Poecilia reticulata) P Roy Soc Lond B. 2015;282 doi: 10.1098/rspb.2015.1132. 20151132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kotrschal A, Kolm N, Penn DJ. Selection for brain size impairs innate, but not adaptive immune responses. P Roy Soc Lond B. 2016;283 doi: 10.1098/rspb.2015.2857. 20152857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kotrschal A, Corral Lopez A, Szidat S, Kolm N. The effect of brain size evolution on feeding propensity, digestive efficiency, and juvenile growth. Evolution. 2015;69:3013–3020. doi: 10.1111/evo.12784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Corral Lopez A, Eckerström-Liedholm S, V Der Bijl W, Kotrschal A, Kolm N. No association between brain size and male sexual behavior in the guppy. Curr Zool. 2015;61:265–273. [Google Scholar]
- 72.Corral Lopez A, Garate-Olaizola M, Buechel SD, Kolm N, Kotrschal A. On the role of body size, brain size, and eye size in visual acuity. Behav Ecol Sociobiol. 2017;71:179. doi: 10.1007/s00265-017-2408-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kotrschal A, et al. Brain size does not impact shoaling dynamics in unfamiliar groups of guppies (Poecilia reticulata) Behavioural Processes. 2018;147:13–20. doi: 10.1016/j.beproc.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 74.Kotrschal A, et al. Evolution of brain region volumes during artificial selection for relative brain size. Evolution. 2017 doi: 10.1111/evo.13373. [DOI] [PubMed] [Google Scholar]
- 75.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Künstner A, et al. The Genome of the Trinidadian Guppy, Poecilia reticulata, and Variation in the Guanapo Population. PLoS ONE. 2016;11:e0169087. doi: 10.1371/journal.pone.0169087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grabherr MG, et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nature biotechnology. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ghalambor CK, et al. Non-adaptive plasticity potentiates rapid adaptive evolution of gene expression in nature. Nature. 2015;525:372–375. doi: 10.1038/nature15256. [DOI] [PubMed] [Google Scholar]
- 81.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Slonim DK. From patterns to pathways: gene expression data analysis comes of age. Nat Genet. 2002;32(Suppl):502–508. doi: 10.1038/ng1033. [DOI] [PubMed] [Google Scholar]
- 83.Dean R, Mank JE. Tissue Specificity and Sex-Specific Regulatory Variation Permit the Evolution of Sex-Biased Gene Expression. Am Nat. 2016;188:E74–E84. doi: 10.1086/687526. [DOI] [PubMed] [Google Scholar]
- 84.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4:117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Futcher B, Latter GI, Monardo P, McLaughlin CS, Garrels JI. A sampling of the yeast proteome. Mol Cell Biol. 1999;19:7357–7368. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lundberg E, et al. Defining the transcriptome and proteome in three functionally different human cell lines. Molecular Systems Biology. 2010;6:450. doi: 10.1038/msb.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Inbar E, et al. The Transcriptome of Leishmania major Developmental Stages in Their Natural Sand Fly Vector. mBio. 2017;8:e00029–17. doi: 10.1128/mBio.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maslov S, Sneppen K. Specificity and Stability in Topology of Protein Networks. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- 90.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.