Abstract
Keloids are benign fibroproliferative skin tumors which can cause disfigurement and disability. While they frequently recur after excision or medical management and can affect 6–16% of African Americans, there is no gold standard therapy. Keloids are challenging to study because there are no animal or in vitro models of this disorder. This makes it very difficult to validate data from treated tissue samples or cells and develop targeted therapies for this disease. In this study, we demonstrate that intralesional 5-FU injection after keloid excision prevents recurrence for 2 years with no reported adverse events. We analyze the expression of treated and untreated biopsies of the same keloids in their native context to capture insights which may be missed by in vitro cell culture models and correct for intra-keloid variability. Random Forest analysis of the microarray data dramatically increased the statistical power of our results; permitting hypothesis-free creation of a gene expression profile of 5-FU treated keloids. Through this analysis, we found a set of genes, including YAP1 and CCL-2, whose expression changes predicts 5-FU therapy status and includes genes which have not previously been associated with keloid biology and of unknown function. We further described keloid heterogeneity for the first time using multidimensional analysis of our microarray results. The methods and tools we developed in this research may overcome some of the challenges in studying keloids and developing effective treatments for this disease.
Introduction:
Keloids are benign fibrino proliferative cutaneous tumors which occur as an extreme of abnormal wound healing [1]. There is no universally accepted therapy for keloids, which often recur [2–4]. This s in part because the molecular mechanisms driving keloid recurrence are unknown. Developing a better understanding of this disease and a treatment protocol which can prevent their recurrence would significantly impact the lives of patients [3].
Studying keloids is complicated by the lack of in vitro and animal models [5]. Microarrays of keloid derived fibroblasts [6–9] and keratinocytes [10, 11] have been used to characterize the cellular response to therapy. This data has produced valuable insights into inflammatory drivers of this disease. However, these analyses have yet to identify definitive disease progression markers. We may improve our understanding of keloids by studying them in their native 3-D context [12]. Acquiring enough clinical biopsies of keloids to permit global gene expression with sufficient statistical power is challenging. Machine learning algorithms, such as Random Forest (RF) analysis, can overcome this difficulty by performing thousands of rounds of analysis on global gene expression arrays to create a predictive gene expression profile with a low percentage of false positives [13].
In this study, we utilize a treatment method of keloid excision and postoperative injection with 5-FU with a 100% response rate and no recurrences after 2 years. We also describe two methods to study keloid response to therapeutics; biopsy of treated and untreated regions for genetic analysis and RF to generate a gene expression profile for treated keloids.
Methods:
5-FU Keloid Treatment and Excision:
All patients received a test injection in the keloid in the clinic. The keloids were then excised to grossly normal tissue and closed with monocryl sutures in the OR. Specimens were taken from the area of injection and a remote area and placed in RNAlater. RNA was extracted from tissue samples with Trizol and the Quiagen RNeasy kit. The RNA was run on an Illumina Human HT-12v4 BeadChip.
Post-operatively, patients were followed weekly and received their first injection into the incision at 2 weeks. 5-FU was injected intradermally at a concentration of 50mg/ml. Patients returned for subsequent injections at 1 to 2 week intervals [14] for 4 rounds of injection. After completion of the 5-FU treatment, patients were scheduled for follow-up at 3, 6, 12 months and then annually for assessment of their scars.
Statistical Analysis:
The microarray data of treated and untreated keloid biopsies from 4 patients were annotated and 26682 coding regions were selected for further analysis based on their statistical significance (P<0.05). This dataset was then analyzed with the RF analysis algorithm to test separation among the treated and untreated samples and identify top contributors to that separation. RF was performed on the genes by generating 50,000 trees [15]. The results of the RF were then visualized using multi-dimensional scaling (MDS). The paired t-test and the fold change were calculated for the top 100 genes from the RF analysis. Hochberg multiple test correction was performed on p values [16].
Results:
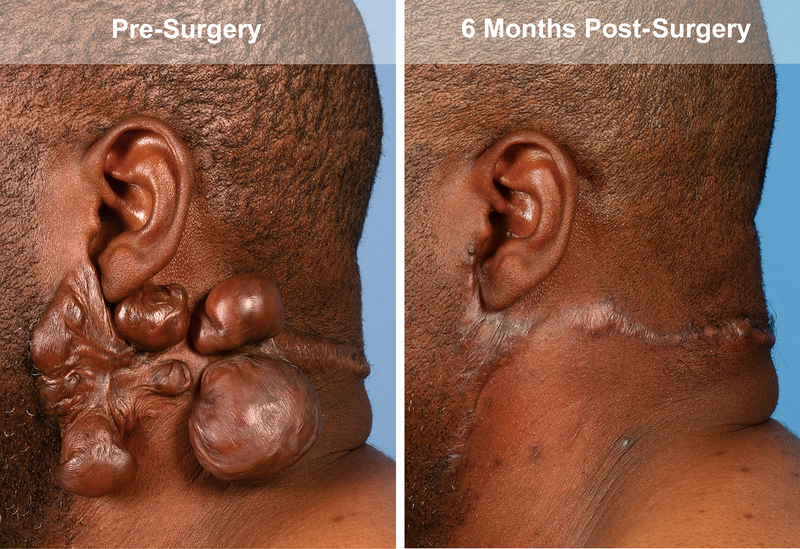
In this study, the keloids of four patients were excised and then the wound bed was treated with 5-FU. These patients did not develop adverse events and showed no recurrence over two years of follow up (Figure 1).
Figure 1).
Representative images taken of the postauricular keloid of patient 1 after excision and treatment with postexisional 5-FU pretreatment and at 6 month follow up.
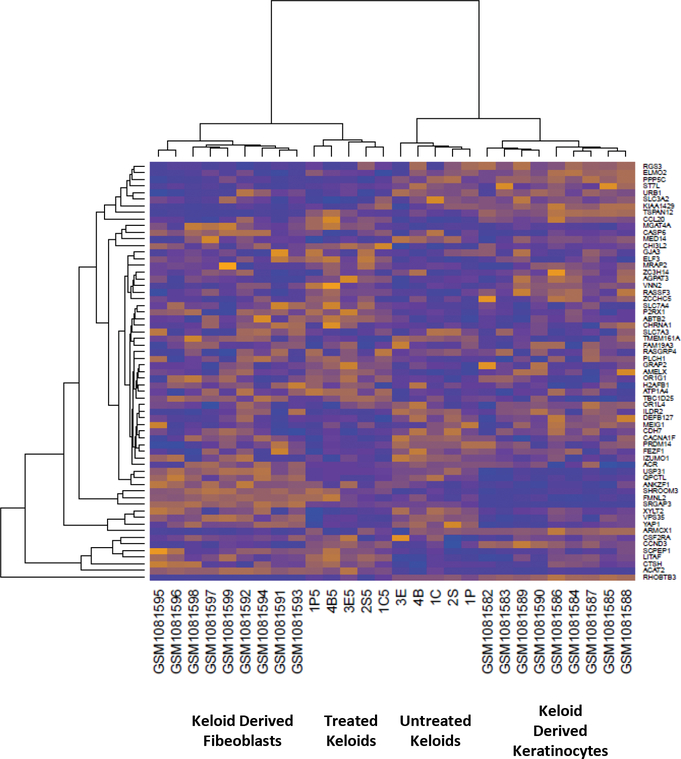
For genetic analysis of the keloid response to 5-FU treatment, we compared microarray results from treated and untreated regions of the same keloid to reduce statistical noise and investigate intra-keloid heterogeneity. Evaluation of the similarity in expression profiles by hierarchical clustering revealed that treated and untreated samples of the same keloid clustered by treatment status. Neither publically available keloid derived fibroblast or keratinocyte microarray data [11] clustered with keloids, highlighting the difference between 2-D culture and the keloid microenvironment (Figure 2). These results suggest that we can detect distinct expression profiles for treated and untreated portions of keloids.
Figure 2).
Heat map comparing keloid derived fibroblasts, keratinocytes, whole untreated keloids, and treated keloids.
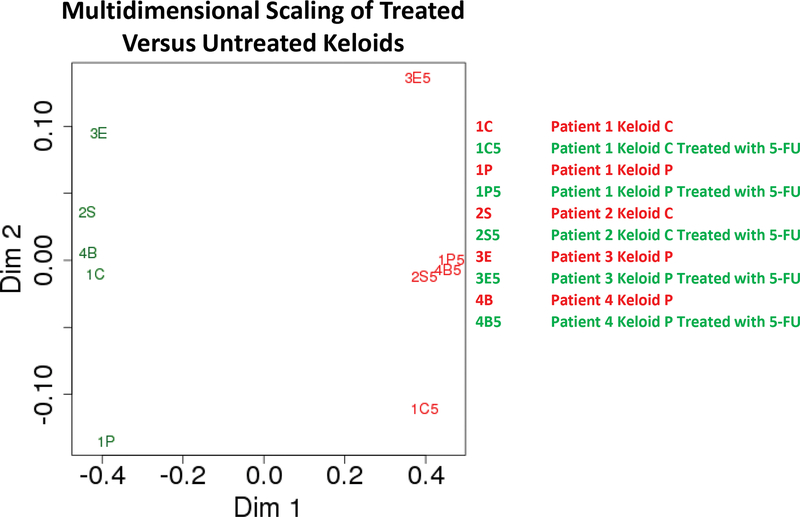
We used RF analysis to determine the 100 genes which were most predictive of keloid treatment. We then calculated the Pearson correlation coefficient (a measure of similarity in the expression network) between each keloid sample relative to every other sample. Multidimensional analysis of this dataset [17] showed separation of keloids into clusters separated by treatment status (Figure 3), visualizing the transcriptional difference between treated and untreated keloids.
Figure 3).
Multidimensional scaling plot showing Euclidean distances of 1-the Pearson coefficient of the 100 most impactful genes from the Random Forest analysis of each keloid sample.
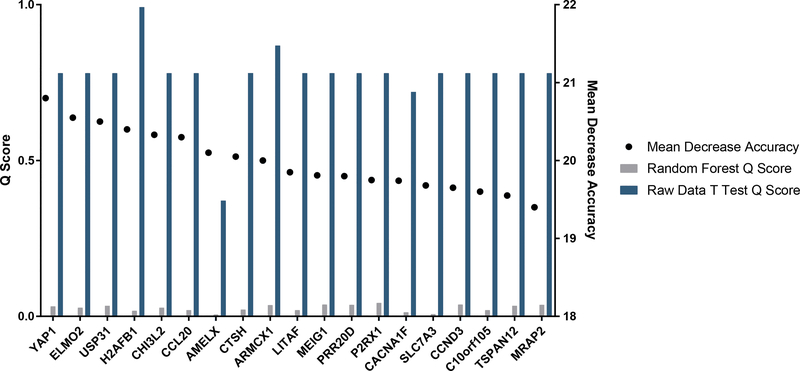
To produce a list of genes which are most likely to influence keloid response to treatment, we selected genes which most influenced the accuracy of the RF model. We also calculated the Q scores (the chance of a result being a false positive) for these genes before and after RF analysis (Figure 4). Repression of YAP1, a pro fibrotic gene associated with skin fibrosis [18–21], and upregulation of CCL2, a regulator of T cell activity downstream of TGF-β [22], was seen in treated keloids. Further, there was no change in the expression of several genes previously linked to keloids: TGF-Beta, PDGF, MMPs, IL-1, IL-6, IL-10. We used Ingenuity Pathway Analysis (IPA) to detect gene networks impacted by 5-FU treatment of whole keloids. Glycosaminoglycan protein linkage region biosynthesis and IL-17A pathways were altered, agreeing with research showing the importance of glycosaminoglycans [23] and IL-17A signaling in keloid biology [24]. These results show that machine learning can be used to create a genetic profile of keloids successfully treated with 5-FU.
Figure 4).
List of the top 20 genes resulting from the Random Forest analysis ranked by how their exclusion from the analysis decreased the accuracy of the model showing the Mean Decrease Accuracy score (the percentage decrease that removal of the gene from analysis reduces the accuracy of the model) and the Q (the percentage chance that a given gene is a false positive) scores for each gene determined by the standard student’s t test before and after Random Forest analysis of the microarray results.
Discussion:
In this study, we evaluated a keloid treatment protocol and two new methods for facilitating the mechanistic study of keloids; taking treated and untreated biopsies from the same tumor and using RF analysis to find significant genes. These methods may help us overcome the lack of in vitro or animal models for keloids by increasing the statistical power we can generate from small numbers of biopsies [25]. Our initial results implicate some drivers of keloid responsiveness to 5-FU treatment. Further study of these and the uncharacterized genes resulting from this study may help us design more targeted strategies for treating and ultimately curing this disfiguring disease.
Acknowledgements:
This work was funded by (TL1TR001104) for RL. This study was supported by NIH 2R01LM009254 and 2R01LM008111 for AKF.
Footnotes
Financial disclosure agreement: None of the authors have any financial arrangements or potential conflicts of interest related to this article.
References
- 1.Nicholas RS, et al. , Patient-related keloid scar assessment and outcome measures. Plast Reconstr Surg, 2012. 129(3): p. 648–56. [DOI] [PubMed] [Google Scholar]
- 2.Alexandrescu D, et al. , Comparative Results in Treatment of Keloids With Intralesional 5-FU/Kenalog, 5-FU/Verapamil, Enalapril Alone,Verapamil Alone, and Laser: A Case Report and Review of the Literature. J Drugs Dermatol, 2016. 15(11): p. 1442–1447. [PubMed] [Google Scholar]
- 3.Mari W, et al. , Novel Insights on Understanding of Keloid Scar: Article Review. J Am Coll Clin Wound Spec, 2015. 7(1–3): p. 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones CD, et al. , The Use of Chemotherapeutics for the Treatment of Keloid Scars. Dermatol Reports, 2015. 7(2): p. 5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luiza Ramos M, Gragnani A, and Masako Ferreira L, Microarray as a new tool to study hypertrophic and keloid scarring. Wounds, 2009. 21(2): p. 57–63. [PubMed] [Google Scholar]
- 6.Wu ZY, et al. , Keloid microRNA expression analysis and the influence of miR-199a-5p on the proliferation of keloid fibroblasts. Genet Mol Res, 2014. 13(2): p. 2727–38. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, et al. , MicroRNA-21 affects proliferation and apoptosis by regulating expression of PTEN in human keloid fibroblasts. Plast Reconstr Surg, 2014. 134(4): p. 561e–73e. [DOI] [PubMed] [Google Scholar]
- 8.Wong VW, et al. , Transcriptional profiling of rapamycin-treated fibroblasts from hypertrophic and keloid scars. Ann Plast Surg, 2014. 72(6): p. 711–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song N, et al. , Enhanced expression of membrane transporter and drug resistance in keloid fibroblasts. Hum Pathol, 2012. 43(11): p. 2024–32. [DOI] [PubMed] [Google Scholar]
- 10.Li M and Wu L, Functional analysis of keratinocyte and fibroblast gene expression in skin and keloid scar tissue based on deviation analysis of dynamic capabilities. Exp Ther Med, 2016. 12(6): p. 3633–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahn JM, et al. , Keloid-derived keratinocytes exhibit an abnormal gene expression profile consistent with a distinct causal role in keloid pathology. Wound Repair Regen, 2013. 21(4): p. 530–44. [DOI] [PubMed] [Google Scholar]
- 12.Marturano-Kruik A, Villasante A, and Vunjak-Novakovic G, Bioengineered Models of Solid Human Tumors for Cancer Research. Methods Mol Biol, 2016. 1502: p. 203–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X and Ishwaran H, Random forests for genomic data analysis. Genomics, 2012. 99(6): p. 323–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta S and Sharma VK, Standard guidelines of care: Keloids and hypertrophic scars. Indian J Dermatol Venereol Leprol, 2011. 77(1): p. 94–100. [DOI] [PubMed] [Google Scholar]
- 15.Wiener LW, M., Classification and Regression by Random Forest. R News, 2002. 2(3): p. 18–12. [Google Scholar]
- 16.Hochberg Y, A Sharper Bonferroni Procedure for Multiple Tests of Significance. Biometrika, 1988. 75(4): p. 800–802. [Google Scholar]
- 17.Rouzier R, et al. , Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res, 2005. 11(16): p. 5678–85. [DOI] [PubMed] [Google Scholar]
- 18.Liu F, et al. , Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am J Physiol Lung Cell Mol Physiol, 2015. 308(4): p. L344–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piersma B, Bank RA, and Boersema M, Signaling in Fibrosis: TGF-beta, WNT, and YAP/TAZ Converge. Front Med (Lausanne), 2015. 2: p. 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee MJ, et al. , YAP and TAZ regulate skin wound healing. J Invest Dermatol, 2014. 134(2): p. 518–25. [DOI] [PubMed] [Google Scholar]
- 21.Low BC, et al. , YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett, 2014. 588(16): p. 2663–70. [DOI] [PubMed] [Google Scholar]
- 22.Brand OJ, et al. , Transforming Growth Factor-beta and Interleukin-1beta Signaling Pathways Converge on the Chemokine CCL20 Promoter. J Biol Chem, 2015. 290(23): p. 14717–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rutkowski D, et al. , An abnormality in glucocorticoid receptor expression differentiates steroid responders from nonresponders in keloid disease. Br J Dermatol, 2015. 173(3): p. 690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cunnusamy K, Chen PW, and Niederkorn JY, IL-17 promotes immune privilege of corneal allografts. J Immunol, 2010. 185(8): p. 4651–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diaz-Uriarte R and Alvarez de Andres S, Gene selection and classification of microarray data using random forest. BMC Bioinformatics, 2006. 7: p. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]