Abstract
We have developed a digital and multiplexed platform for the rapid detection and telemonitoring of infections caused by Ebola and Marburg filoviruses. The system includes a flow cell assay cartridge that captures specific antibodies with microarrayed recombinant antigens from all six species of filovirus, and a smartphone fluorescent reader for high-performance interpretation of test results. Multiplexed viral proteins, which are expandable to include greater numbers of probes, were incorporated to obtain highest confidence results by cross-correlation, and a custom smartphone application was developed for data analysis, interpretation, and communication. The smartphone reader utilizes an opto-electro-mechanical hardware attachment that snaps at the back of a Motorola smartphone and provides a user interface to manage the operation, acquire test results, and communicate with cloud service. The application controls the hardware attachment to turn on LEDs and digitally record the optically enhanced images. Assay processing time is approximately 20 min for microliter amounts of blood, and test results are digitally processed and displayed within 15 s. Furthermore, a secure cloud service was developed for the telemonitoring of test results generated by the smartphone readers in the field. Assay system results were tested with sera from nonhuman primates that received a live attenuated EBOV vaccine. This integrated system will provide a rapid, reliable, and digital solution to prevent the rapid overwhelming of medical systems and resources during EVD or MVD outbreaks. Further, this disease-monitoring system will be useful in resource-limited countries where there is a need for dispersed laboratory analysis of recent or active infections.
Keywords: Ebola, point-of-care, smartphone reader, disease surveillance, telemonitoring
Healthcare decisions that are based on rapid, reliable, and digital data are a high priority measure to prevent the inundation of medical systems during episodes of infectious disease outbreaks. The largest Ebola disease outbreak to date occurred in West Africa (2013–2016) and resulted in more than 11,200 deaths, with approximately 28,000 suspected cases in Guinea, Liberia, and Sierra Leone.1 This recent and unanticipated Ebola epidemic exposed many weaknesses in existing healthcare monitoring and management systems.2 The regional and international responses were compromised by inaccessible, incomplete, and underutilized health information.3 Accessible methods to rapidly monitor biological fluids for detection of suspected infections will result in actionable data for professional healthcare workers. These methods must not only provide accurate diagnosis by untrained personnel, but also overcome the difficulties encountered in the field, and storage of a high volume of confidential test records. However, there are no truly portable diagnostic devices that can be used by care providers with minimal training, especially those designed for detecting Ebola and Marburg virus emergence within the community.4 Polymerase chain reaction (PCR),5,6 and antigen detection tests7−9 are performed and interpreted only in the clinic, and thus rely on movement of patients or specimens to operational laboratories. While PCR methodologies demonstrate excellent sensitivity and specificity, data can only be obtained during the first few days of viremia, using dedicated equipment and highly trained personnel. Similarly, antigen detection tests7−9 are only useful during the first few days of infection, and few assays are commercially available. Enzyme linked immunosorbent assays (ELISA) typically measure antibody against only a single antigen from one species of filovirus, usually Ebolavirus10 (EBOV). Further, ELISAs are time-consuming and require costly instrumentation that is unwieldly and trained personnel to perform, thus restricting use in resource-limited countries.
Smartphones are finding increased use in biosensor applications, especially for optical readers.11 The camera, computing power, and the communication capability to remotely report results make smartphones an attractive choice for integration12−14 into diagnostic and surveillance assays. We previously reported15,16 the development of a filovirus protein microarray that used a desktop laser scanner to characterize antibody responses from human and nonhuman primate survivors of Ebola Viral (EVD) and Marburg Viral Diseases (MVD). Here, we describe a highly portable filovirus disease detection assay that is interfaced with a smartphone reader. The reader system was used to analyze antibody binding to 12 essential antigens from all six species of filoviruses that are arranged in a microarray on a disposable microfluidic chip. Performance of the system prototype was demonstrated with sera from nonhuman primates that were immunized with a recombinant Vesicular Stomatitis Virus–Ebola Virus (rVSV-EBOV) vaccine containing EBOV glycoprotein.17
Experimental Section
Materials and Methods
Recombinant Filovirus Proteins
Full-length genes for nucleoprotein (NP), and the glycoprotein (GP) mucin-like domain fragments for the filovirus species of Reston (RESTV), Bundibugyo (BDBV), Zaire (EBOV), Sudan (SUDV), and Taï Forest ebolavirus (TAFV), and also Marburg marburgvirus (MARV), were cloned into pENTR/TEV/D-TOPO vector (Life Technologies, Grand Island, NY, USA) and sequence verified. All entry vector clones were shuttled into destination bacterial expression vectors via LR reaction (LR Clonase II, Life Technologies, Carlsbad, CA, USA). Specifically, GP mucin open reading frames (ORF) were shuttled into pDESTHisMBP (Addgene plasmid 11085) containing an N-terminal HisMBP tag, while all NP ORFs were shuttled into pDEST17 (Life Technologies) containing an N-terminal His tag. Expression of recombinant GP mucin protein domains were accomplished with BL21-AI Escherichia coli (Life Technologies) transformed with the pDEST17 constructs. Transformed bacterial were cultured in LB supplemented with 0.1% glucose and 100 μg/mL, and induced at 0.4 OD (600 nm) with 0.8% l-arabinose (SIGMA) for 4 h at 30 °C. The bacteria were pelleted by centrifugation (20,000 g, 10 min, 4 °C), and lysed using BPER lysing reagent (Thermo Scientific, Rockford, IL, USA) supplemented with 1 × Halt protease and phosphatase inhibitors cocktail, EDTA-free (Thermo Scientific), 20 mg/mL lysozyme, and 75 U/mL DNase I (Thermo Scientific). The proteins were purified by immobilized metal ion affinity chromatography (IMAC) FPLC using 1 mL HisTrap FF columns (GE Healthcare Lifesciences, Pittsburgh, PA, USA). Binding and washing steps were conducted with 25 mM HEPES, 140 mM NaCl, 25 mM imidazole, pH 7.5, and for protein elution the imidazole concentration was raised to 500 mM. Eluted proteins were examined both by gel-electrophoresis and by mass spectrometry. Expression of recombinant NP proteins were similarly expressed from transformed BL21-AI E. coli bacteria, but due to poor IMAC purification capability of these recombinant proteins, expression conditions were optimized to form inclusion bodies. Induction was done at 0.6 OD (600 nm) with 0.2% l-arabinose (SIGMA-Aldrich, St. Louis, MO, USA) overnight at 18 °C. Pelleting and lysis of the bacteria were performed as described above. Inclusion bodies were washed 2× in 140 mM NaCl, 20 mM Tris-HCl pH 7.5 to remove soluble impurities, at which point >90% of remaining protein was recombinant NP proteins. The proteins were then solubilized in 25 mM HEPES, 140 mM NaCl, 3 M urea, and refolded on the microarray surface. The purified proteins were analyzed by Agilent Bioanalyzer system (Agilent Technologies, Santa Clara, CA, USA). All purified proteins were stored at −20 °C in respective elution buffers, with glycerol added to a final concentration of 25%. For quality control purposes and to validate our assay design, printed microarrays were probed with a panel of purified antibodies directed toward filovirus proteins. The panel included anti-EBOV GP, anti-Sudan GP, anti-RESTV GP antibodies (IBT Biosciences, Gaithersburg, MD, USA). The bound antibodies were detected by Alexa Fluor 647-conjugated goat anti-mouse IgG, and goat anti-rabbit IgG antibodies (Life Technologies). Arrays were scanned with Genepix laser scanner (Molecular Devices, San Jose, CA, USA) and analyzed using Genepix Pro 7 software as described previously.16
Prototype Cartridge
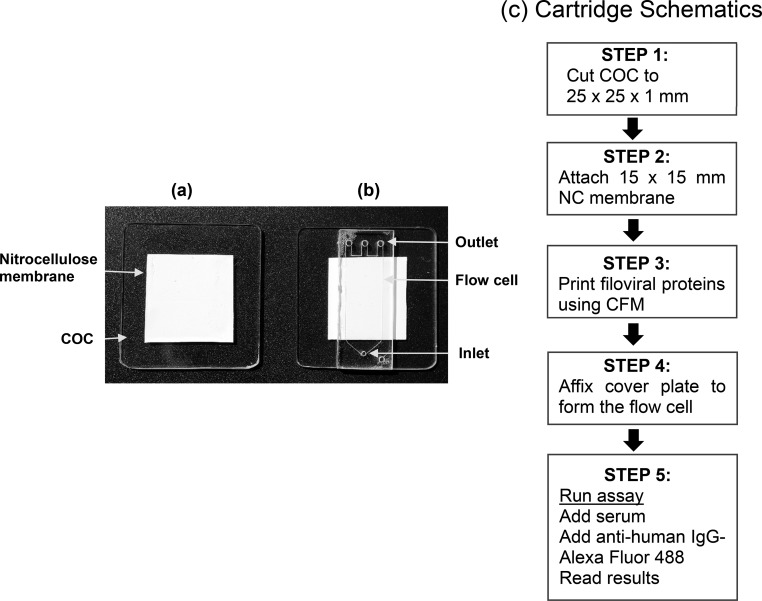
A prototype cartridge (Figure 1) was assembled with the following components. The base substrate was made up of cyclic olefin copolymer (COC) with the following dimensions 25 × 25 × 1 mm3. A polyester backed 0.22 μm nitrocellulose membrane (15 × 15 mm2) was attached (Bio-Rad, Hercules, CA, USA) to the COC substrate (Figure 1a) using polyester double-sided diagnostic microfluidic medical tape, coated with pressure sensitive acrylate adhesive (3M, St. Paul, MN, USA). The recombinant proteins were diluted in PBS at 100 μg/mL concentration and printed on the nitrocellulose membrane in a 4 × 8 array format by continuous flow microprinting (CFM, Carterra, Salt Lake City, UT, USA). The protein samples were delivered to the surface by the printer using 48 microchannels, which allowed the samples to cycle across the nitrocellulose surfaces bidirectionally for 5 min. The spot size of the printed proteins were 400 μm each with a pitch of 500 μm. An adhesive-backed acrylic cover plate (Figure 1b) with a dimension of 7 × 4 × 1 mm3 (Wainamics, Inc., Pleasanton, CA, USA) was pressed over the printed array to form a flow cell with a volume of 100 μL (Figure 1b). The flow cell had one inlet and three outlets to facilitate uniform flow across the membrane.
Figure 1.
Prototype cartridge for the detection of filoviral antibodies. (a) A polyester backed nitrocellulose membrane (0.2 μm) was affixed on a COC slide; (b) an acrylic cover plate with dimensions of 7 × 4 × 1 mm3 was pressed on tightly to form a flow cell with a volume of 100 μL. The cartridge had one inlet and three outlets for uniform flow across the microarray. The schematics (c) show how the cartridge was assembled for the immunoassay.
Smartphone Reader
The smartphone reader consisted of three main components: a Motorola Nexus 6 phone with an opto-electro-mechanical hardware attachment that snaps at the back of the smartphone, a compartment to house the prototype cartridge, and a custom application (App) to run the assay. The hardware attachment was fabricated by 3D printing, using fused deposition modeling and stereolithography technologies (Stratasys Ltd., Eden Prairie, MN, USA). The 3D printed attachment also had a custom-designed tray to hold the prototype cartridge (Figure 2). The Android App provided the user interface to manage the operation, acquire test results, and communicate with cloud service. It also controlled the hardware attachment via flash pulses to turn on specific excitation LEDs and digitally record optically enhanced assay images through the emission filter. The smartphone, which was concealed within the hardware attachment, served as the user interface, digital image acquisition device, and system processer.
Figure 2.
Opto-electro-mechanical device attachment. The 3D printed attachment with the assay chip in panel (a); LEDs for excitation of the fluorophores, the camera aperture with interference filter and external imaging lens in panel (b); the back, front, and side views with the smartphone are shown in panels (c), (d), and (e), respectively.
Detection of Ebola and Marburg Specific Antibodies
The printed microarray chips were blocked for 1 h in blocking buffer (Grace Bio-Laboratories, Bend, OR, USA), probed with serum diluted at 1:150 in phosphate buffered saline (PBS), and washed three times. Bound antibody was detected by incubation (1 h, RT) with Alexa Fluor 647 or 488-conjugated goat anti-human IgG (Invitrogen, Carlsbad, CA, USA) diluted at 1 μg/mL. The microarrays were rinsed with purified water, dried and scanned. A similar process was applied for the prototype cartridge-based flow cell assay. The reagents were injected by pipetting a volume of 100 μL through the inlet using PIPETMAN P200 (Gilson Inc., Middleton, WI, USA) and incubated for 5 min (22 °C). The waste was collected at the outlet with blotting paper.
Results and Discussion
We previously showed that human antibody responses to GP protein were predominantly directed toward the polypeptide region of the highly glycosylated mucin-like domain.15 Hence, in this study only GP mucin proteins were produced for the microarray. The recombinant proteins were physically characterized for molecular size and purity by using a protein chip analyzer (Agilent Technologies). The functional activity of the proteins was confirmed by probing with a panel of purified antibodies directed toward filovirus proteins. The EBOV GP mucin protein was specifically recognized by the rabbit anti-EBOV GP antibodies (Supporting Information Figure S-1a). Similarly, RESTV GP mucin was also detected specifically by mouse anti-RESTV GP antibodies (Figure S-1b). Minor cross-reactivity between MARV NP and BDBV NP was observed for microarrays probed with rabbit anti-SUDV NP (Figure S-1c). Combined, binding data from these control antibodies indicate that the filovirus microarrays were functional and that the protein probes reacted specifically with the correct antibodies.
Alexa Fluor 350, 430, 488, 647, and Quantum Dot 525 fluorophores were evaluated with the smartphone camera to obtain an optimal image that was capable of differentiating between positive and negative samples. The average Stokes shifts of these fluorophores ranged from 17 to 100 nm. Figure S-2 shows the excitation/emission spectra of these fluorophores. Proteins conjugated to the fluorescent tags were printed in a microarray format, and the fluorescent images were scanned by a high-resolution laser scanner (Genepix Scanner 4400a) and the smartphone camera (Figure S-3). Although Alexa Flour 488 had smaller average Stokes shift (30 nm) compared to other fluorophores tested, it produced the strongest signal with the smartphone camera, and was selected for incorporation into the assay.
The proteins were printed on a 15 × 15 mm2 polyester backed nitrocellulose membrane fixed on a plastic (COC) support. Because proteins bind irreversibly to nitrocellulose, no additional chemistry was needed for attachment. An adhesive backed acrylic cover plate was carefully pressed over the COC slide to seal the flow cell. The cover plate had one inlet and three outlets for even flow of reagents across the flow cell and membrane. We examined a cartridge prepared with a microarray containing human IgG, rabbit IgG, and mouse IgGs. Antihuman IgG-Alexa Fluor 488 conjugate was injected, and the fluorescent intensity of the spots were measured by Genepix laser scanner. No fluid leaks were found during the assay. Little or no binding was observed with control IgGs (rabbit and mouse), whereas high binding noted with human IgG (Figure S-4). Keeping the cover plate in place reduced assay signals compared to signals produced without the cover (Figure S-4c). This reduction may be attributed to the change in refractive index created by air trapped between the cover and the microarray. An index matching solution may be injected at the last step to overcome this problem. The results demonstrated that immunoassays could be performed successfully in the prototype cartridge.
An opto-electro-mechanical hardware attachment (Figure 2) that snaps on the back of a Motorola Nexus smartphone device was fabricated by 3D printing (Dimensions Elite (FDM) and Object30 Pro (PolyJet), Stratasys Ltd.) with FDM (Fused Deposition Modeling), and PolyJet technologies. The 3D model of the device was constructed using computer aided design (CAD). The mechanical body isolated the imaging environment and blocked the ambient light entering the imaging area, which was important to obtain reproducible results (Figure 2a). A special tray held the prototype cartridge for imaging by the smartphone camera. Two LEDs (LED Engin, San Jose, CA, USA) on each side of the microarray served as fluorescent dye excitation sources (Figure 2b). The Android App controlled all the steps involved in the operation of the reader device including the acquisition and processing of the image (Figure S-5). The algorithm in the App enhanced the captured images, based on the calibration and cutoff values, and determined if spots present were positive or negative. The results were displayed within 15 s after the image was taken by the smartphone. The use of the dedicated smartphone camera was a critical part of the reader development, but this also limited the ability to manipulate captured images. The CMOS imaging sensors and firmware on smartphones were typically designed and optimized for digital photography purposes. Hence, these devices presented certain limitations for direct use with the microfluidic assay. The digital images collected by the sensors were automatically post-processed and compressed by the phone operating system to provide the best-looking photos. Furthermore, the camera settings had limited preset options that were designed to simplify use. Therefore, total control over the imaging parameters and supporting firmware on smartphone electronic products was not possible. The raw images (i.e., unprocessed data collected by the imaging sensor) could not be acquired on these devices, limiting the ability to control the camera settings (e.g., ISO, aperture, and length of exposure). Similarly, the precision and tolerances were low in the 3D fabrication method used for the opto-electro-mechanical attachment, but were, however, selected for this proof-of-concept study due to ease of production and cost. Injection molding, which requires significant cost and time, will be required for large-scale production.
A Health Insurance Portability and Accountability Act (HIPAA)-compliant cloud service for telemonitoring the test results was also developed. The cloud service was accessed through the SSL (Secure Sockets Layer)-protected web page. The primary function of this service was to report the test results in a secure manner using the smartphone. To this end, we designed the front-end interface with a third-party HIPAA-compliant database provided by TrueVault, a service provider for small businesses. From these readers, each user could register and securely access their own cloud service test results that were stored in the TrueVault database. Users had access to the database through a secure cloud Web site, which displayed the test results as a table and as pins on Google map. The cloud site also enabled study directors and select administrators to upload calibration files to the specific end-user groups in the field.
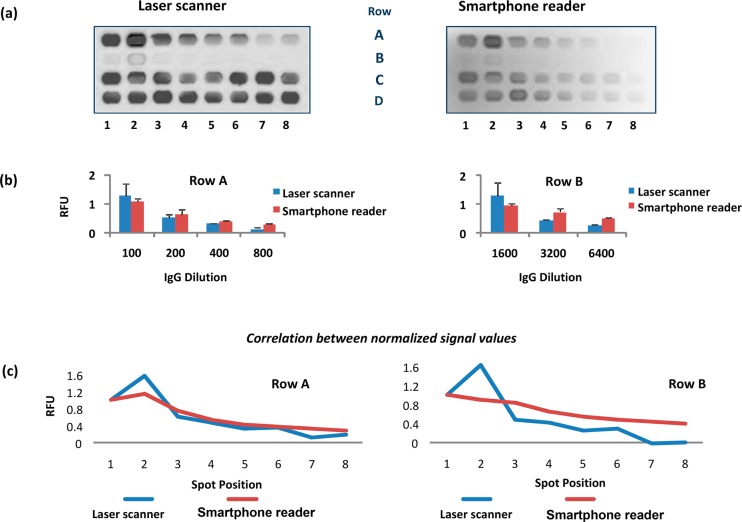
Protein microarrays were printed on the cartridge at various X-Y coordinates to align with the field of view of the smartphone reader. The X-Y coordinates of 7.3 × 13.75 mm2 on the nitrocellulose membrane provided the best results. Figure 3 shows the images of a sample array obtained using with reference Genepix laser scanner as well as smartphone reader. The array (Rows A and B) contained serial dilutions of goat anti-mouse IgG-Alexa Fluor 488 printed in duplicates for measuring linear response. Rows C and D were printed with a fixed concentration of goat anti-mouse IgG-Alexa Fluor 488 and used for illumination correction and calibration. The App utilized Row D elements for the correction of illumination nonuniformity over the chip. Although the laser excitation provided a stronger emission signal than the LED source in the smartphone reader (Figure 3), there was ∼95% correlation between the results.
Figure 3.
Comparison of microarray fluorescent signals produced by the Genepix laser scanner and by the smartphone camera. Serial dilutions of goat anti-mouse IgG-Alexa Fluor 488 in duplicates was printed in Row A and B. Fixed concentration of Goat anti-mouse IgG-Alexa Fluor 488 was printed in Row C and D for image correction and calibration. Panel (a) shows image outputs by the two readers. Panel (b) shows comparison of normalized signal values in relative fluorescence units (RFU), where each row was individually normalized. Panel (c) shows correlation between the normalized RFUs of the two readers.
To test the performance of the integrated assay, we evaluated antibodies from nonhuman primates that were immunized with rVSV-EBOV, a vaccine that protects against EBOV and has undergone human clinical trials.17,18 Sera from Cynomolgus macaque (Macaca fascicularis) that received intramuscular injection of rVSV-EBOV were collected before and 36 days after immunization. Filoviral and control proteins were printed in a 4 × 8 microarray format. Rows A, B, and C contained the filoviral proteins and Row D the calibration spots (Figure 4). The microarray included GP mucin and NP proteins from all six species of filovirus family, human IgG for positive control, buffer control, and goat anti-mouse IgG-Alexa Flour 488 for smartphone reader calibration (spots D1, D3, D6, and D8). The assay was completed in 20 min, and the fluorescent intensities of the spots were measured with the smartphone reader device. We analyzed six different samples that demonstrated quantitative comparisons for vaccinated and prevaccinated NHP sera (spots B3 and B4), as shown in Figure 4a. The relative signal values were calculated based on the local background signal values. Spots B3 and B4 in Figure 4a were printed with EBOV-GP mucin domain proteins. The two NHP prevaccinated sera showed no binding, whereas the vaccinated sera samples produced strong fluorescent signals that indicated recognition of the EBOV GP mucin proteins. The cross-sectional intensity profiles of B3 and B4 spots are shown in Figure 4b. The signal intensities of prevaccinated samples were significantly lower than the vaccinated samples. Thus, the integrated smartphone reader system can clearly distinguish between sera that contains anti-EBOV antibodies and nonvaccinated controls. The microarray contained GP and NP proteins from all six species for broad disease coverage, and high probe specificity was demonstrated with anti-EBOV sera. For this proof-of-concept study, we used sera from NHP vaccinated with a single species (EBOV) of filovirus; however, we have previously used human sera exposed to three different filoviral species and demonstrated the specificity and cross-reactivity.15 By including multiple probes from all six species of filovirus family the specificity can be more precisely determined than by using the single antigens. Antibody levels for filovirus infections are usually described in arbitrary units that do not reference absolute serum concentrations,19,20 making comparisons between methods very difficult. Pathogen-specific IgG concentrations in serum are typically in μM (μg/mL) or higher range and IgM levels, which peak first and are important for disease diagnosis,21 may be a log lower (serum concentration of total IgG 10–16 mg/mL and IgM 1–2 mg/mL). Although IgM was not evaluated here, microarray technology platforms have sensitivities in the low-to-mid picomolar (pM–ng/mL)22 range, and thus should be able to detect relevant antibodies in disease sera. While the system described here detects only filovirus specific IgG, future modifications may allow evaluation of IgM responses to open up the possibility of diagnostic applications.
Figure 4.
Evaluation of the smartphone reader with NHPs vaccinated with rVSV-EBOV vaccine. The microarray was printed with GP mucin proteins from BDBV (A1, A2), MARV (A3, A4), RESTV (A5, A6), SUDV (A7, A8), TAFV (B1, B2), and EBOV (B3, B4); NP proteins from BDBV (B5, B6), MARV (B7, B8), RESTV (C1, C2), SUDV (C3, C4), TAFV (C5, C6), and EBOV (C7, C8). Row D was printed with goat anti-mouse IgG-Alexa Fluor 488 as calibration spots (D1, D3, D6, D8), positive controls of human IgG (D2, D4), and negative controls of buffer (D5, D7). Sera from four vaccinated NHPs and two controls (prevaccinated) were tested in the reader system. Goat anti-human IgG-Alexa Fluor 488 was used for detection of antibodies to microarray. The red windows show the location of EBOV-GP proteins in the array (a). The cross-sectional intensity profiles of B3 and B4 spots are shown in panel (b). The fluorescent signals (RFU) from vaccinated NHP samples are higher than the preimmune NHP controls.
The main objective of this research was to develop a point-of-care platform which will monitor and remotely report data that reveal the emergence and progression of infections caused by Ebola (EVD) and the closely related Marburg (MVD) filoviruses. This goal was accomplished by integrating a microfluidic microarray assay15 with a portable reader that capitalizes on the ease of use and widespread availability of smartphones. The reader capitalizes on widespread availability of smartphones and ease of use. The reader utilizes the computational power of the built-in smartphone and software to guide and monitor proper operation by untrained users especially applicable to resource limited countries. An easy to follow, menu-driven user guide is displayed on the smartphone application, and test results obtained with the device are encrypted and wirelessly transmitted. A HIPAA-compliant secure telemonitoring service provides test results access to care personnel for rapid clinical assessment. The fluorescent-based readout is highly sensitive, and the assay requires only 1 to 2 μL of serum or plasma. The image capturing system employed to maintain image quality, high fluorophore excitation and low background, probe separation limits, and other factors determine the assay multiplexing capacity. The current configuration will accommodate up to 60 individual probes that are printed in 400-μm-diameter spots, and the number of probes can be expanded with further refinements of the detection system. Thus, the multiplexing capacity of the assay can be extended to include additional proteins23−25 from viruses that may co-circulate or co-infect along with filoviruses. For example, yellow fever, Lassa, Rift valley fever, dengue, and other infections can detrimentally affect the survival of EVD and MVD patients. Further, the infection histories from such studies can be used for developing informed diagnosis and understand epidemiology of EVD and MVD. Thus, the platform described here will address important gaps in resources to monitor and remotely report data that reveal the emergence and progression of infections caused by Ebola and Marburg viruses.
Conclusions
We developed a simple, low cost, smartphone-based system for the serological detection of Ebola or Marburg diseases in resource-limited countries. The antibody detection system developed can monitor and report data remotely from infections caused by all six species of virus at the point of use, and will use only a drop of blood, such as that required for a standard glucose analysis by diabetics. The device will also be able to acquire antibody data from both symptomatic and nonsymptomatic cases, and thus enable an additional application and unmet need for disease surveillance. The next generation device in development incorporates a cartridge design for plasma separation to enable direct blood sampling, injection molded opto-electro-mechanical component, and a universal scanner that can be combined with any cell phone.
Acknowledgments
This work was funded by National Institutes of Allergy and Infectious Diseases-Small Business Innovation Research Grant HHSN272201600031C/CELLGEN1608. Research was conducted under an IACUC approved protocol in compliance with the Animal Welfare Act, PHS Policy, and other Federal statutes and regulations relating to animals and experiments involving animals. The facility where this research was conducted is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the United States Army, the National Institutes of Health, or the United States Government.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssensors.8b00842.
Additional figures illustrating the specificity of recombinant proteins, selection of fluorophores, flow cell assay, and smartphone app interface (PDF)
Author Contributions
Production of recombinant proteins, cartridge and assay were developed by USAMRIID team. Smartphone mechanical attachment, App and setup of secure cloud service were developed by NOWDiagnostics team. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Agua-Agum J.; Allegranzi B.; Ariyarajah A.; Aylward R.; Blake I. M.; Barboza P.; Bausch D.; Brennan R. J.; Clement P.; Coffey P.; Cori A.; Donnelly C. A.; Dorigatti I.; Drury P.; Durski K.; Dye C.; Eckmanns T.; Ferguson N. M.; Fraser C.; Garcia E.; Garske T.; Gasasira A.; Gurry C.; Hamblion E.; Hinsley W.; Holden R.; Holmes D.; Hugonnet S.; Jaramillo Gutierrez G.; Jombart T.; Kelley E.; Santhana R.; Mahmoud N.; Mills H. L.; Mohamed Y.; Musa E.; Naidoo D.; Nedjati-Gilani G.; Newton E.; Norton I.; Nouvellet P.; Perkins D.; Perkins M.; Riley S.; Schumacher D.; Shah A.; Tang M.; Varsaneux O.; Van Kerkhove M. D. After Ebola in West Africa–Unpredictable Risks, Preventable Epidemics. N. Engl. J. Med. 2016, 375 (6), 587–96. 10.1056/NEJMsr1513109. [DOI] [PubMed] [Google Scholar]
- Kouadio K. I.; Clement P.; Bolongei J.; Tamba A.; Gasasira A. N.; Warsame A.; Okeibunor J. C.; Ota M. O.; Tamba B.; Gumede N.; Shaba K.; Poy A.; Salla M.; Mihigo R.; Nshimirimana D. Epidemiological and Surveillance Response to Ebola Virus Disease Outbreak in Lofa County, Liberia (March-September, 2014); Lessons Learned. PLoS Curr. 2015, 7, 1. 10.1371/currents.outbreaks.9681514e450dc8d19d47e1724d2553a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancedda C.; Davis S. M.; Dierberg K. L.; Lascher J.; Kelly J. D.; Barrie M. B.; Koroma A. P.; George P.; Kamara A. A.; Marsh R.; Sumbuya M. S.; Nutt C. T.; Scott K. W.; Thomas E.; Bollbach K.; Sesay A.; Barrie A.; Barrera E.; Barron K.; Welch J.; Bhadelia N.; Frankfurter R. G.; Dahl O. M.; Das S.; Rollins R. E.; Eustis B.; Schwartz A.; Pertile P.; Pavlopoulos I.; Mayfield A.; Marsh R. H.; Dibba Y.; Kloepper D.; Hall A.; Huster K.; Grady M.; Spray K.; Walton D. A.; Daboh F.; Nally C.; James S.; Warren G. S.; Chang J.; Drasher M.; Lamin G.; Bangura S.; Miller A. C.; Michaelis A. P.; McBain R.; Broadhurst M. J.; Murray M.; Richardson E. T.; Philip T.; Gottlieb G. L.; Mukherjee J. S.; Farmer P. E. Strengthening Health Systems While Responding to a Health Crisis: Lessons Learned by a Nongovernmental Organization During the Ebola Virus Disease Epidemic in Sierra Leone. J. Infect. Dis. 2016, 214, S153–S163. 10.1093/infdis/jiw345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhurst M. J.; Brooks T. J.; Pollock N. R. Diagnosis of Ebola Virus Disease: Past, Present, and Future. Clin. Microbiol. Rev. 2016, 29 (4), 773–93. 10.1128/CMR.00003-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller S. A.; Bailey D.; Matthews S.; Lumley S.; Sweed A.; Ready D.; Eltringham G.; Richards J.; Vipond R.; Lukaszewski R.; Payne P. M.; Aarons E.; Simpson A. J.; Hutley E. J.; Brooks T. Evaluation of the Biofire FilmArray BioThreat-E Test (v2.5) for Rapid Identification of Ebola Virus Disease in Heat-Treated Blood Samples Obtained in Sierra Leone and the United Kingdom. J. Clin. Microbiol. 2016, 54 (1), 114–9. 10.1128/JCM.02287-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semper A. E.; Broadhurst M. J.; Richards J.; Foster G. M.; Simpson A. J.; Logue C. H.; Kelly J. D.; Miller A.; Brooks T. J.; Murray M.; Pollock N. R. Performance of the GeneXpert Ebola Assay for Diagnosis of Ebola Virus Disease in Sierra Leone: A Field Evaluation Study. PLoS Med. 2016, 13 (3), e1001980 10.1371/journal.pmed.1001980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisen M. L.; Cross R. W.; Hartnett J. N.; Goba A.; Momoh M.; Fullah M.; Gbakie M.; Safa S.; Fonnie M.; Baimba F.; Koroma V. J.; Geisbert J. B.; McCormick S.; Nelson D. K.; Millett M. M.; Oottamasathien D.; Jones A. B.; Pham H.; Brown B. L.; Shaffer J. G.; Schieffelin J. S.; Kargbo B.; Gbetuwa M.; Gevao S. M.; Wilson R. B.; Pitts K. R.; Geisbert T. W.; Branco L. M.; Khan S. H.; Grant D. S.; Garry R. F. Field Validation of the ReEBOV Antigen Rapid Test for Point-of-Care Diagnosis of Ebola Virus Infection. J. Infect. Dis. 2016, 214, S203–S209. 10.1093/infdis/jiw261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisen M. L.; Oottamasathien D.; Jones A. B.; Millett M. M.; Nelson D. S.; Bornholdt Z. A.; Fusco M. L.; Abelson D. M.; Oda S.; Hartnett J. N.; Rowland M. M.; Heinrich M. L.; Akdag M.; Goba A.; Momoh M.; Fullah M.; Baimba F.; Gbakie M.; Safa S.; Fonnie R.; Kanneh L.; Cross R. W.; Geisbert J. B.; Geisbert T. W.; Kulakosky P. C.; Grant D. S.; Shaffer J. G.; Schieffelin J. S.; Wilson R. B.; Saphire E. O.; Branco L. M.; Garry R. F.; Khan S. H.; Pitts K. R.; Viral Hemorrhagic Fever C. Development of Prototype Filovirus Recombinant Antigen Immunoassays. J. Infect. Dis. 2015, 212, S359–67. 10.1093/infdis/jiv353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhurst M. J.; Kelly J. D.; Miller A.; Semper A.; Bailey D.; Groppelli E.; Simpson A.; Brooks T.; Hula S.; Nyoni W.; Sankoh A. B.; Kanu S.; Jalloh A.; Ton Q.; Sarchet N.; George P.; Perkins M. D.; Wonderly B.; Murray M.; Pollock N. R. ReEBOV Antigen Rapid Test kit for point-of-care and laboratory-based testing for Ebola virus disease: a field validation study. Lancet 2015, 386 (9996), 867–74. 10.1016/S0140-6736(15)61042-X. [DOI] [PubMed] [Google Scholar]
- Vu H.; Shulenin S.; Grolla A.; Audet J.; He S.; Kobinger G.; Unfer R. C.; Warfield K. L.; Aman M. J.; Holtsberg F. W. Quantitative serology assays for determination of antibody responses to Ebola virus glycoprotein and matrix protein in nonhuman primates and humans. Antiviral Res. 2016, 126, 55–61. 10.1016/j.antiviral.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada-Gonzalez D.; Merkoci A. Mobile phone-based biosensing: An emerging ″diagnostic and communication″ technology. Biosens. Bioelectron. 2017, 92, 549–562. 10.1016/j.bios.2016.10.062. [DOI] [PubMed] [Google Scholar]
- Lee S.; Kim G.; Moon J. Development of a Smartphone-based reading system for lateral flow immunoassay. J. Nanosci. Nanotechnol. 2014, 14 (11), 8453–7. 10.1166/jnn.2014.9920. [DOI] [PubMed] [Google Scholar]
- Vashist S. K.; Marion Schneider E.; Zengerle R.; von Stetten F.; Luong J. H. Graphene-based rapid and highly-sensitive immunoassay for C-reactive protein using a smartphone-based colorimetric reader. Biosens. Bioelectron. 2015, 66, 169–76. 10.1016/j.bios.2014.11.017. [DOI] [PubMed] [Google Scholar]
- Mudanyali O.; Dimitrov S.; Sikora U.; Padmanabhan S.; Navruz I.; Ozcan A. Integrated rapid-diagnostic-test reader platform on a cellphone. Lab Chip 2012, 12 (15), 2678–86. 10.1039/c2lc40235a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natesan M.; Jensen S. M.; Keasey S. L.; Kamata T.; Kuehne A. I.; Stonier S. W.; Lutwama J. J.; Lobel L.; Dye J. M.; Ulrich R. G. Human Survivors of Disease Outbreaks Caused by Ebola or Marburg Virus Exhibit Cross-Reactive and Long-Lived Antibody Responses. Clin. Vaccine Immunol. 2016, 23 (8), 717–24. 10.1128/CVI.00107-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata T.; Natesan M.; Warfield K.; Aman M. J.; Ulrich R. G. Determination of specific antibody responses to the six species of ebola and Marburg viruses by multiplexed protein microarrays. Clin. Vaccine Immunol. 2014, 21 (12), 1605–12. 10.1128/CVI.00484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Restrepo A. M.; Camacho A.; Longini I. M.; Watson C. H.; Edmunds W. J.; Egger M.; Carroll M. W.; Dean N. E.; Diatta I.; Doumbia M.; Draguez B.; Duraffour S.; Enwere G.; Grais R.; Gunther S.; Gsell P. S.; Hossmann S.; Watle S. V.; Konde M. K.; Keita S.; Kone S.; Kuisma E.; Levine M. M.; Mandal S.; Mauget T.; Norheim G.; Riveros X.; Soumah A.; Trelle S.; Vicari A. S.; Rottingen J. A.; Kieny M. P. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ca Suffit!). Lancet 2017, 389 (10068), 505–518. 10.1016/S0140-6736(16)32621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsell P. S.; Camacho A.; Kucharski A. J.; Watson C. H.; Bagayoko A.; Nadlaou S. D.; Dean N. E.; Diallo A.; Diallo A.; Honora D. A.; Doumbia M.; Enwere G.; Higgs E. S.; Mauget T.; Mory D.; Riveros X.; Oumar F. T.; Fallah M.; Toure A.; Vicari A. S.; Longini I. M.; Edmunds W. J.; Henao-Restrepo A. M.; Kieny M. P.; Keita S. Ring vaccination with rVSV-ZEBOV under expanded access in response to an outbreak of Ebola virus disease in Guinea, 2016: an operational and vaccine safety report. Lancet Infect. Dis. 2017, 17 (12), 1276–1284. 10.1016/S1473-3099(17)30541-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue J.; Tuznik K.; Follmann D.; Grandits G.; Marchand J.; Reilly C.; Sarro Y. D. S.; Pettitt J.; Stavale E. J.; Fallah M.; Olinger G. G.; Bolay F. K.; Hensley L. E. Use of the Filovirus Animal Non-Clinical Group (FANG) Ebola virus immuno-assay requires fewer study participants to power a study than the Alpha Diagnostic International assay. J. Virol. Methods 2018, 255, 84–90. 10.1016/j.jviromet.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower H.; Glynn J. R. A systematic review and meta-analysis of seroprevalence surveys of ebolavirus infection. Sci. Data 2017, 4, 160133. 10.1038/sdata.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macneil A.; Reed Z.; Rollin P. E. Serologic cross-reactivity of human IgM and IgG antibodies to five species of Ebola virus. PLoS Neglected Trop. Dis. 2011, 5 (6), e1175 10.1371/journal.pntd.0001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson J.; Larsson A.; Sjoholm A. G.; Truedsson L.; Jansson B.; Borrebaeck C. A.; Wingren C. Design of recombinant antibody microarrays for serum protein profiling: targeting of complement proteins. J. Proteome Res. 2007, 6 (9), 3527–36. 10.1021/pr070204f. [DOI] [PubMed] [Google Scholar]
- Keasey S.; Pugh C.; Tikhonov A.; Chen G.; Schweitzer B.; Nalca A.; Ulrich R. G. Proteomic basis of the antibody response to monkeypox virus infection examined in cynomolgus macaques and a comparison to human smallpox vaccination. PLoS One 2010, 5 (12), e15547 10.1371/journal.pone.0015547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keasey S. L.; Pugh C. L.; Jensen S. M.; Smith J. L.; Hontz R. D.; Durbin A. P.; Dudley D. M.; O’Connor D. H.; Ulrich R. G. Antibody Responses to Zika Virus Infections in Environments of Flavivirus Endemicity. Clin. Vaccine Immunol. 2017, 24 (4), 1. 10.1128/CVI.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. L.; Pugh C. L.; Cisney E. D.; Keasey S. L.; Guevara C.; Ampuero J. S.; Comach G.; Gomez D.; Ochoa-Diaz M.; Hontz R. D.; Ulrich R. G. Human Antibody Responses to Emerging Mayaro Virus and Cocirculating Alphavirus Infections Examined by Using Structural Proteins from Nine New and Old World Lineages. mSphere 2018, 3 (2), 1. 10.1128/mSphere.00003-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.