Abstract
An extract of Eudistoma sp. provided eudistidine C (1), a heterocyclic alkaloid with a novel molecular framework. Eudistidine C (1) is a racemic natural product composed of a tetracyclic core structure further elaborated with a p-methoxyphenyl group and a phenol-substituted aminoimidazole moiety. This compound presented significant structure elucidation challenges due to the large number of heteroatoms and fully substituted carbons. These issues were mitigated by application of a new NMR pulse sequence (LR-HSQMBC) optimized to detect four- and five-bond heteronuclear correlations and the use of computer-assisted structure elucidation software. Synthesis of eudistidine C (1) was accomplished in high yield by treating eudistidine A (2) with 4(2-amino-1H-imidazol-5-yl)phenol (4) in DMSO. Synthesis of eudistidine C (1) confirmed the proposed structure and provided material for further biological characterization. Treatment of 2 with various nitrogen heterocycles and electron-rich arenes provided a series of analogues (5–10) of eudistidine C. Chiral-phase HPLC resolution of epimeric eudistidine C provided (+)-(R)-eudistidine C (1a) and (−)-(S)-eudistidine C (1b). The absolute configuration of these enantiomers was assigned by ECD analysis. (−)-(S)-Eudistidine C (1b) modestly inhibited interaction between the protein binding domains of HIF-1α and p300. Compounds 1, 2, and 6–10 exhibited significant antimalarial activity against Plasmodium falciparum.
Graphical Abstract
INTRODUCTION
Natural products benefit from “privileged” structures that evolved to interact with essential biopolymers such as proteins and nucleic acids. These secondary metabolites encompass extensive diversity in terms of both their molecular architecture and biological activity and remain a key source of lead compounds and new therapeutic agents.1 Nitrogen-rich natural products represent a broad class of bioactive compounds that interact with important cellular targets, particularly in the context of anticancer and anti-infective agents.2–4 Some recent examples of nitrogenous metabolites include the myxobacterial cytotoxin haprolid,5 the synoxazolidinones, antimicrobial guanidine alkaloids from a marine ascidian,6 the antimalarial β-lactam monamphilectine A,7 and odoamide, a cytotoxic cyclicpeptide from a cyanobacterium.8 There has been significant progress with new genomic tools that enable sequencing-based recognition and annotation of natural products with known biosynthetic machineries, and the structures of encoded secondary metabolites can often be accurately predicted.9–11 However, the identification of completely new molecular scaffolds such as complex polycyclic alkaloids cannot typically be accomplished using genomic methods alone. Such discoveries often depend on approaches utilizing activity-guided fractionation and de novo structural elucidation. In this area, NMR-based structural studies of severely mass-limited samples on the nanomole scale have been facilitated by a new generation of miniaturized cryogenically cooled NMR probes12–14 and the development of sensitivity enhanced NMR pulse sequences.15–18 There has also been dramatic progress in the development of computational approaches that provide predictive structure determination derived from experimental data.19–22 The work described herein illustrates a range of contemporary structural elucidation strategies and techniques, including total synthesis. These were applied in a concerted fashion in the context of the novel heterocyclic alkaloid, eudistidine C (1).
We recently reported the isolation, structural elucidation, and synthesis of eudistidines A (2) and B (3) from an extract of the marine ascidian Eudistoma sp.23 The extract was chemically investigated because it showed activity in a high-throughput screen for inhibitors of the protein–protein interaction between p300 and HIF-1α. We found that 2, but not 3, inhibited this interaction. In the current study, we further interrogated the same extract and identified a related compound that exhibits increased chemical complexity. This compound maintains the tetracyclic core of 2 but is further elaborated at the C-10 position with an unusual aryl-substituted aminoimidazole moiety. Structure elucidation of 1 was complicated by the proton-deficient core of the molecule. The assignment was accomplished using a combination of NMR spectroscopic analyses, including a new pulse sequence specifically optimized for long-range heteronuclear correlations and structure predictive software. Synthetic studies confirmed the structure of eudistidine C (1) and revealed that a variety of electron-rich arenes can react with 2 under mild conditions to provide a series of aryl-substituted analogues. Chiral resolution of racemic 1 provided the two C-10 epimers, and only (−)-(S)-eudistidine C (1b) had significant activity in the p300 and HIF-1α assay. In total, these studies illustrate the utility of applying synergistic technologies in concert to characterize the chemical and biological diversity of new secondary metabolites.
RESULTS AND DISCUSSION
Our initial bioassay-guided fractionation of the Eudistoma sp. extract provided eudistidines A (2) and B (3).23 Further investigation of a side fraction from this effort revealed a 1H NMR signature indicative of a related eudistidine analogue. Repeated chromatographic separations on LH-20 media ultimately provided 10.7 mg of eudistidine C (1).
Eudistidine C (1) was isolated as an optically inactive yellow oil with a molecular formula of C28H21N7O2, as established by HRESI mass spectrometry, which required 22 degrees of unsaturation. The highly aromatic nature of 1 was confirmed by 26 sp2 carbons (δC 167.5, 161.7, 159.5, 159.3, 157.3, 148.0, 147.4, 147.0, 136.5, 131.9, 131.9, 130.9, 130.2, 129.8, 129.8, 129.7, 127.5, 126.1, 126.1, 120.0, 117.6, 115.7, 115.7, 115.2, 115.2, 94.3) and only two sp3 carbons (δC 73.7 and 55.8) in the 13C NMR spectrum (Table 1). 1H NMR (CD3OD) revealed signals attributable to one methoxy group (δH 3.76) and 14 aromatic protons, which constituted four protonated spin systems based on COSY data: a pair of mutually coupled heteroaromatic protons [δH 8.45 (d, J = 5.0 Hz), 6.77 (d, J = 5.3 Hz)], a 1,2-disubstituted benzene ring [δH 8.06 (d, J = 8.0 Hz), 7.81 (t, J = 7.5 Hz), 7.65 (d, J = 8.0 Hz), 7.58 (t, J = 7.5 Hz)], and two para-substituted benzene rings [δH 7.68 (2H d, J = 8.9 Hz), 6.91 (2H d, J = 8.8 Hz) and 6.85 (2H d, J = 8.8 Hz), 6.30 (2H d, J = 8.5 Hz)]. Direct comparison of the 1D and 2D NMR data sets for eudistidine C (1) with those recorded for eudistidines A (2) and B (3) supported the presence of the same tetracylic eudistidine core structure (rings A–D), composed of fused pyrimidine and imidazole rings with embedded guanidine and amidine functionalities (Figure 1 and the Supporting Information). However, the chemical shift of C-10 in 1 (δC 73.7) differed significantly from the C-10 hemiaminal (δC 94.4) in eudistidine A (2) and amide (δC 172.0) in eudistidine B (3). HMBC correlations established that ring E in 1 was a p-methoxybenzene ring that was substituted at C-10 (Figure 1). The remaining para-substituted benzene ring (G) had a resonance at δC 159.1 that was indicative of a hydroxyl substituent.
Table 1.
NMR Spectroscopic Data for Eudistidine C (1) in CD3OD
| position | δC | δNa | δH (mult,J in Hz) | HMBCb |
|---|---|---|---|---|
| 1 | 245.8 | |||
| 2 | 167.5 | 8.45 (d, 5.0) | 1, 3, 3a, 3b, 8c | |
| 3 | 94.3 | 6.77 (d, 5.3) | 1, 2, 3a, 3b, 8b | |
| 3a | 147.4 | |||
| 3b | 117.6 | |||
| 4 | 126.1 | 8.06 (d, 8.0) | 3a, 6, 7a | |
| 5 | 130.2 | 7.58 (t, 7.5) | 3b, 7 | |
| 6 | 136.5 | 7.81 (t, 7.5) | 4, 7a | |
| 7 | 129.7 | 7.65 (d, 8.0) | 3b, 5, 8 | |
| 7a | 147.0 | |||
| 8 | 256.0 | |||
| 8a | 159.5 | |||
| 8b | 169.6 | |||
| 8c | 157.3 | |||
| 9 | n.o.c | |||
| 10 | 73.7 | |||
| 11 | 130.9 | |||
| 12/16 | 129.8 | 7.68 (d, 8.9) | 10, 12/16, 14 | |
| 13/15 | 115.2 | 6.91 (d, 8.8) | 11, 13/15, 14 | |
| 14 | 161.7 | |||
| 14-OMe | 55.8 | 3.76 (s) | 14 | |
| 17 | 126.1 | |||
| 18 | n.o.c | |||
| 19 | 148.0 | |||
| 20 | n.o.c | |||
| 21 | 127.5 | |||
| 22 | 120.0 | |||
| 23/27 | 131.9 | 6.85 (d, 8.8) | 21, 23/27, 25 | |
| 24/26 | 115.7 | 6.30 (d, 8.5) | 22, 24/26, 25 | |
| 25 | 159.3 | |||
| 28 | n.o.c |
15N assignments were based on 1H−15N HMBC correlations. The δN values were not calibrated to an external standard but were referenced to neat NH3 (δ 0.00) using the standard Bruker parameters.
1H−13C (optimized for 8.3 Hz) and 1H−15N (optimized for 8 Hz) HMBC correlations are listed.
Not observed
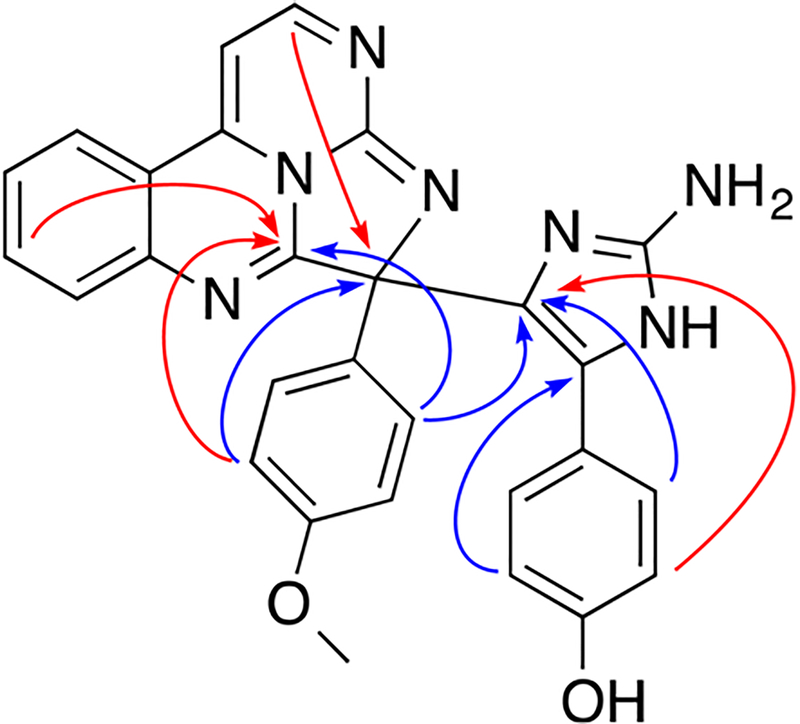
Figure 1.
Selected 1H–13C HMBC (black arrows), 1H–15N (redarrows), and COSY (bold blue lines) correlations for eudistidine C (1)
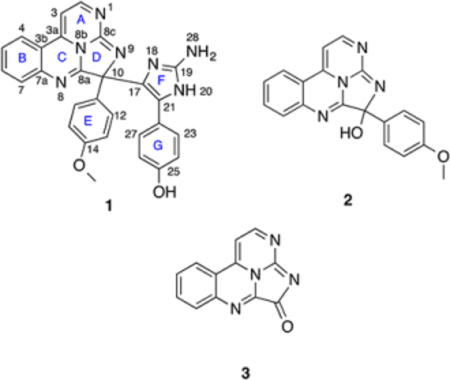
With the tetracyclic eudistidine core and two para-substituted phenyl groups firmly established, molecular formula considerations required the final structure of eudistidine C (1) incorporate an additional C3H3N3 moiety and 3 degrees of unsaturation. The three remaining carbons were quaternary sp2 carbons with chemical shifts of δC 148.0, 127.5, 126.1. The only HMBC correlation to these carbons was a correlation from H-23/27 (δH 6.85) to the carbon at δC 127.5, which revealed that ring G was connected to the C3 fragment at this position. No NH protons were ever observed using a variety of different NMR experiments (1H, 1H–15N HSQC, 1H–15N HMBC) and deuterated solvents (CD3OH, DMSO-d6, DMF-d7). The carbon chemical shifts, in particular the signal at δC 148.0, suggested the remaining fragment was an amino-substituted imidazole ring (ring F) connected via the carbon at δC 126.1 to C-10 and to ring G through the carbon at δC 127.5. Thus, the novel heptacyclic structure of eudistidine C (1) was proposed. However, significant structural ambiguity remained due to the large number of adjacent quaternary carbons and non-protonated nitrogen atoms, along with the lack of observable NH signals. The experimental 13C NMR data recorded with eudistidine C (1) were compared with the DFT-calculated theoretical carbon chemical shifts for structure 1. The computational results were generally in good agreement with the experimentally derived data (Supporting Information), with the largest outliers being carbons, C-11 and C-19, which had Δδ values of 3.8 and 4.5 ppm, respectively. The molecular formula and complete set of NMR data (1H, 13C, COSY, HSQC, HMBC) recorded for 1 were then analyzed using ACD/Structure Elucidator Suite software, which allows for de novo structure elucidation and generates a complete set of structures that fit the correlations observed in the experimental data.21,22 The program generated a total of 419 structural isomers, the 13C NMR data sets for each structure were calculated, and the structures ranked on the basis of cumulative error between the calculated and experimental NMR values. The structure we proposed for eudistidine C (1) was the highest ranked structure identified by the Structure Elucidator software (Supporting Information).
While computer-assisted structure elucidation (CASE) provided a structure consistent with our proposal, we felt that additional evidence was required to complete the assignment due to the proton-deficient nature of 1. The LR-HSQMBC experiment is a recently described NMR pulse sequence optimized to detect very long-range heteronuclear correlations.24 It can be used to detect4,5JCH and4,5JNH correlations, and the utility of these long-range data was clearly demonstrated in the NMR characterization of cervinomycin A2.25 LR-HSQMBC is a high-sensitivity pulse sequence that optimizes responses for very small nJCH/NH couplings (≤0.2 Hz for nJCH), and the resulting data complements experiments such as HMBC for defining structural elements that have few if any detectable protons. The HMBC experiment provides access to2,3J correlations and only occasionally 4J heteronuclear correlations, while LR-HSQMBC data sets are populated with numerous four- and five-bond correlations.26 These long-range correlations can help define structural elements with multiple quaternary carbons and heteroatoms, and link these moieties with protonated portions of the molecule. Crucial 4JCH and 5JCH correlations observed by LR-HSQMBC (Figure 2), including those from H-12/16 (ring E) to C-8a (ring D) and C-17 (ring G) as well as from H-23/27 and H-24/26 (ring F) to C-17, helped confirm the composition and connectivity of the rings in eudistidine C (1). When the LR-HSQMBC correlations were added to the data set used with the ACD/Structure Elucidator, the number of candidate structures was reduced from 419 to 335 and the total processing time was reduced from 247 to 35 s. As before, structure 1 was the highest ranked candidate.
Figure 2.
Key four-bond (blue arrows) and five-bond (red arrows) 1H–13C correlations measured with a 2 Hz optimized LR-HSQMBC experiment digitized with 768 F1 increments
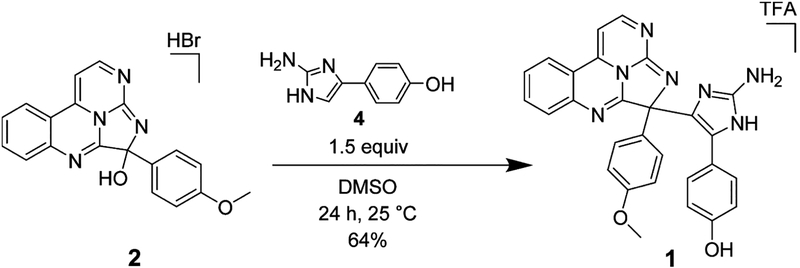
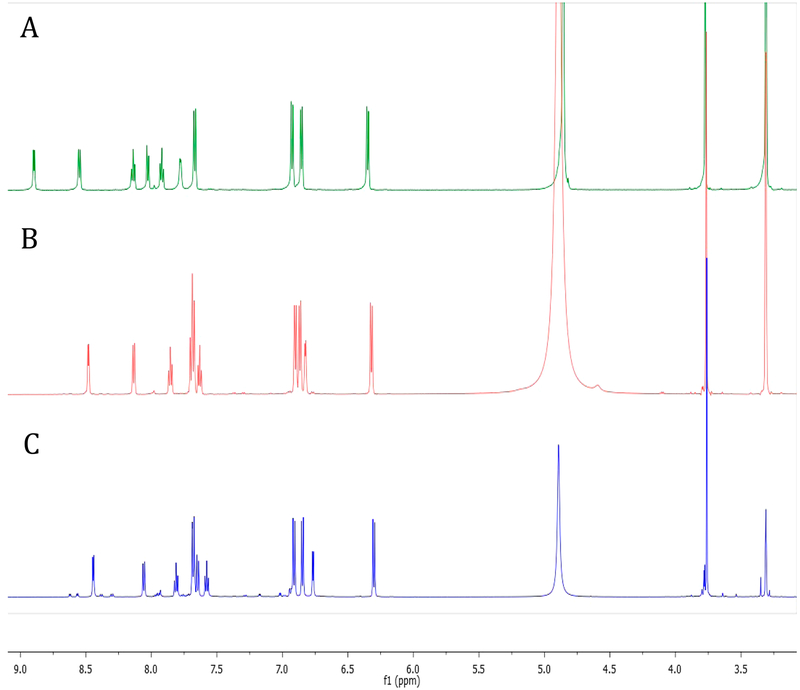
Despite the valuable data obtained by the LR-HSQMBC experiment, no long-range correlations were ever recorded for C-19 or any of the nitrogen atoms associated with ring F. Thus, a portion of eudistidine C (1) remained only partially characterized, with inadequate spectroscopic data to fully define the structure. The synthesis of eudistidine C (1) was viewed as the best way to definitively prove its structure as well as provide access to useful quantities of 1 and its analogues. Inspection of 1 suggested it might arise from a dehydration-driven union of eudistidine A (2) and 3-arylaminoimidazole 4. We recently reported the preparation of 2 using a condensation/cyclization reaction cascade between 4-(2-aminophenyl)pyrimidin-2-amine and 4-(methoxyphenyl)-glyoxal.23 While this synthetic route was concise, isolation of the final product required a scale-limiting preparative HPLC step. We subsequently found that synthetic 2 can be isolated directly from the homogeneous reaction solution by simple addition of 1:1 acetone/hexanes to provide a solid product. This material is the HBr salt of 2 in sufficient purity to use directly in the synthetic studies described below. Alternately, the solid material can be converted to its free base form through preparative HPLC in the presence of NH4OH to give material with NMR spectral features identical to those of natural eudistidine A (2). This four-step, scalable preparation of eudistidine A (2) allowed us to investigate the coupling reaction to form eudistidine C (1) and other analogues. The aminoimidazole coupling partner needed to generate 1 was prepared in one step from the commercially available methyl ether through demethylation with BBr3 to give the desired phenol 4 (Experimental Section). The identity and purity of 4 were verified by NMR analysis (Supporting Information). After a screen of reaction conditions, we found the combination of the HBr salt of 2 and 4 (1.5 equiv) in DMSO at room temperature afforded racemic eudistidine C (1) in 64% yield following preparative HPLC (Figure 3). Notably, the free base of 2 does not undergo this reaction without additional acidic promotor. The synthetic material had NMR spectral features (1H and 13C) identical to those of the natural compound 1 when compared in their free base forms or when both spectra were obtained in CD3OD/0.1% TFA (Supporting Information). A 1:1 mixture of natural and synthetic eudistidine C provided a discrete set of 1H and 13C NMR resonances, which confirmed the identical nature of these materials. Significant spectral differences were observed between the free base and salt forms of these alkaloids (Figure 4). Protonation state, counterion, and sample concentration can all be essential for accurate spectral comparison of natural and synthetic samples of complex heterocycles, such as the eudistidines.
Figure 3.
Synthesis of eudistidine C (1)
Figure 4.
1H NMR spectral characteristics of eudistidine C (1) in CD3OD: (A) synthetic eudistidine C TFA salt, (B) synthetic eudistidine C free base, (C) natural product eudistidine C free base form
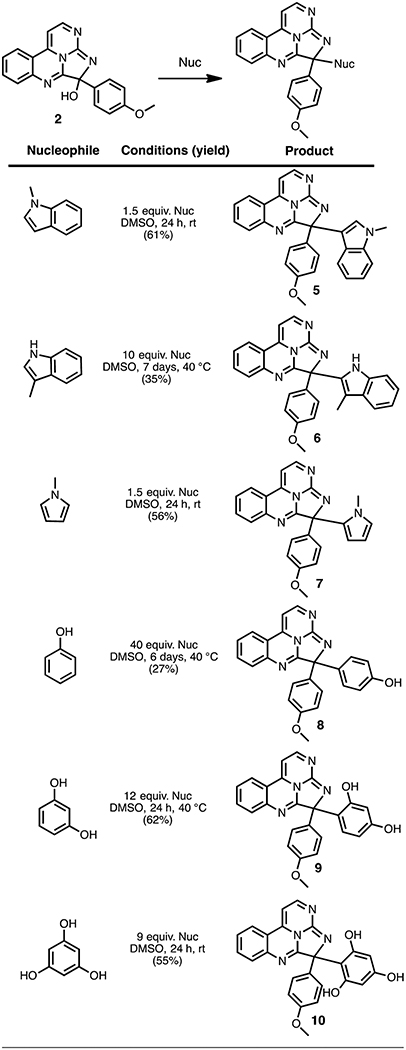
The mild nature of this coupling procedure suggested that alternative nucleophiles might also add to the C-10 position of 2. We first pursued this possibility with a variety of nucleophilic heteroarenes, including N-methylindole, 3-methylindole, and N-methylpyrrole. In all three cases, the corresponding adducts 5–7 could be obtained in useful preparative yield, although the reaction required extended heating at 40 °C in the case of 3-methylindole (Table 2). The connectivity of synthetic adducts was confirmed by 2D NMR, with heterocycle modification occurring at the expected site (C-3 of N-methylindole, C-2 of 3-methylindole, and C-2 of N-methylpyrrole). We further examined the reaction of 2 with phenol, resorcinol, and phloroglucinol. Compounds 8–10 were formed in suitable yields, albeit requiring extended heating in DMSO in the case of phenol and resorcinol. Following these results, the reactivity of 2 with various heteroatom nucleophiles was also evaluated. We found that with several representative alcohols, amines, and thiols, no new adducts could be isolated or observed by LCMS. Thus, it appears the electrophilic reactivity of 2 is confined to carbon nucleophiles. These initial studies generated a series of structural analogues of 1, helped to characterize the reactivity of eudistidine A (2), and will enable future efforts to generate additional analogues of the eudistidine class of natural products.
Table 2.
Reaction of Eudistidine A (2) with Heteroarene and Arene Nucleophiles
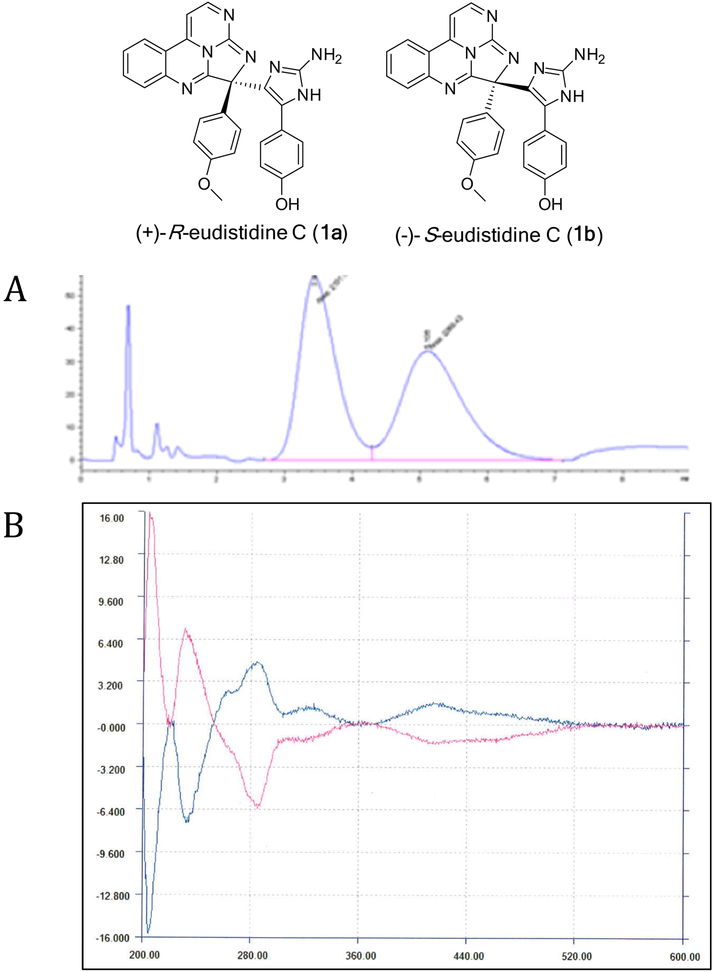
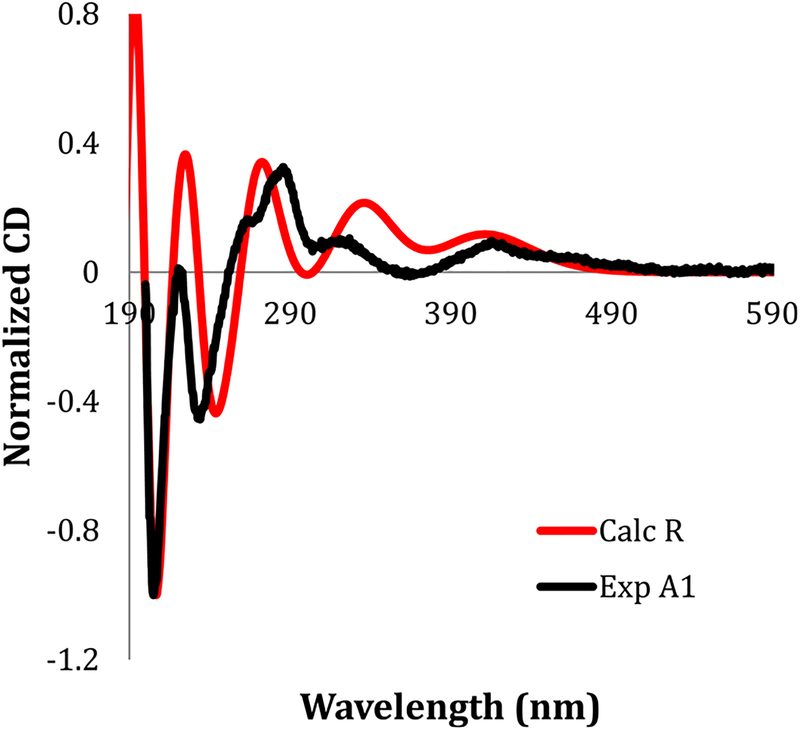
Investigation of the UV absorption properties of eudistidine C (1) in MeOH revealed an absorption maximum at 403 nm. The UV/vis absorption profiles of 1 in aqueous solutions were quite similar to those recorded for eudistidine A (2),23 as they exhibited pronounced pH-dependent spectral shifts. Local absorbance maxima for 1 were observed at 403 and 446 nm, with a hypsochromic shift in more acidic medium and an isosbestic point at approximately 420 nm (Supporting Information). Natural eudistidine C (1) is racemic, as it has no optical rotation [α]D or Cotton effect in its CD spectrum, and the enantiomers are related by inversion at C-10. Eudistidine A (2) natural product is also racemic; however, it has a hemiaminal group at C-10, creating the likelihood that a ring D-opened minor tautomeric form of 2 facilitates rapid racemization at this position. The racemic nature of 1 and its inherent reactivity with nitrogen heteroaromatic compounds under mild conditions suggests that the final aminoimidazole coupling step in the biogenesis of eudistidine C (1) mirrors our synthetic strategy and may not necessarily occur via an enzymatic process. Ring D in 1 cannot open in a manner similar to 2; thus, the two epimers at C-10 likely form during the nucleophilic coupling step and then are configurationally stable. The two enantiomers of the eudistidine C natural product racemate could be resolved by HPLC using a chiral-phase Lux Cellulose-4 analytical column (4.6 × 50 mm). The enantiomers in the natural product occurred as a 1:1 mixture with identical UV spectra but opposite sign Cotton effects when monitored at 240 nm (Supporting Information). Semipreparative separation (10 × 250 mm Lux-4 column) of synthetic eudistidine C provided the two enantiomers in pure form, and they showed equal magnitude but opposite sign Cotton effects in their ECD spectra (Figure 5). The early eluting enantiomer was assigned as (+)-(R)-eudistidine C (1a) by comparing its experimentally measured ECD spectrum with the DFT-calculated spectrum (Figure 6). The late-eluting enantiomer was assigned as (−)-(S)-eudistidine C (1b) in a similar manner (Supporting Information). Eudistidine A (2) could also be resolved by chiral-phase HPLC into its two enantiomers. However, when the eluent of each peak was subsequently dried and reinjected on the HPLC two peaks of equal area were observed, indicating that ring opening and epimerization at C-10 had occurred.
Figure 5.
Chiral-phase HPLC separation of racemic eudistidine C provided (+)-(R)-eudistidine C (1a) and (−)-(S)-eudistidine C (1b). (A) HPLC UV trace monitored at 210 nm of the racemate separated on a Lux Cellulose-4 column (chlorinated cellulose phenylcarbamate) provided two peaks (MH+, m/z 488) of approximately equal area. (B) ECD spectra of the early-eluting HPLC peak [blue; (+)-(R)-eudistidine C (1a)] and late-eluting peak [pink; (−)-(S)-eudistidine C (1b)] had equal magnitude but opposite sign Cotton effects
Figure 6.
Comparison of calculated (red) and experimentally measured (black) ECD spectra for (+)-(R)-eudistidine C (1a).
Racemic eudistidine C (1) was tested in a dose–response version of the screening assay to assess its ability to block binding between the CH1 domain of p300 and the C-TAD domain of HIF-1α. It showed approximately 48% inhibition of the protein–protein interaction at a high test concentration of 500 μM. Testing of the individual enantiomers revealed that (−)-(S)-eudistidine C (1b) had an IC50 of 276 μM, while (+)-(R)-eudistidine C (1a) gave only 27% inhibition at 500 μM. The eudistidine C analogues 5–10 were also evaluated in the HIF-1α/p300 assay. The phloroglucinol adduct (10) was slightly more potent (IC50 = 141 μM), while the N-methylindole (5), 3-methylindole (6), and N-methylpyrrole (7) derivatives had IC50’s of 314, 399, and 356 μM, respectively. The other analogues provided less than 50% inhibition at 500 μM. In an effort to evaluate other biological properties, eudistidines A (2) and C (1) were tested in the NCI-60 panel of human tumor cell lines, but at an initial concentration of 40 μM they were generally noncytotoxic and thus did not undergo further 60-cell testing.27 Since a variety of different alkaloid structural families are known to have antimalarial properties, (+)-(R)-eudistidine C (1a), (−)-(S)-eudistidine C (1b), eudistidine A (2), and analogues 6–10 were also evaluated for inhibitory activity against chloroquine-sensitive (D6) and chloroquine-resistant (W2) strains of the Plasmodium falciparum parasite,28 and all showed significant antimalarial effects (Supporting Information). The most potent were eudistidine A (2), which had IC50 values of 1.4 and 1.1 μM, and the phloroglucinol analogue of eudistidine C (10), which had IC50 values of 1.1 and 0.6 μM against the D6 and W2 strains, respectively.
CONCLUSION
The novel heptacyclic structure of eudistidine C was assigned using a combination of contemporary spectroscopic techniques and CASE analyses. The recently described LR-HSQMBC NMR pulse sequence with its ability to detect four- and five-bond heteronuclear correlations is particularly effective for assigning and verifying structural elements that are distant from protonated portions of a molecule. This technique was most applicable with eudistidine C (1), which has a severely proton-deficient central core region. A hallmark of virtually all natural products studies is the ability to define and verify the structure of the compounds of interest. As such, the continuing adaptation and application of new techniques such as NMR experiments that extend the sensitivity/boundaries for data acquisition are critically important. In addition, CASE programs such as ACD/Structure Elucidator are becoming more powerful and reliable, and they provide important tools that can be effectively utilized in challenging structural elucidation situations. The utility of CASE analysis was highlighted by congruence of the structure obtained from conventional interpretation of the spectroscopic data for eudistidine C with the most favored structural candidate revealed by the software. The new and often synergistic technologies now employed in natural products discovery have confirmed the tremendous untapped potential of this resource and sparked renewed interest in the chemical and biological diversity of secondary metabolites. In spite of recent advances, the architectural complexity and unusual functional group arrays found in some natural scaffolds still present structural elucidation challenges for noncrystalline secondary metabolites. The current study illustrates a range of contemporary structural elucidation strategies and techniques, including total synthesis, that were required to fully address structural questions associated with eudistidine C (1).
The new structural motif of eudistidine C (1) can be accessed in a single step by reaction of eudistidine A (2) with appropriate nucleophiles. The unusual propensity of eudistidine A to react irreversibly with aromatic carbon nucleophiles, but not with nucleophilic heteroatoms such as alkyl alcohols, amines, or thiols, provided a means to generate synthetic eudistidine C and also a variety of heteroarene and phenolic structural analogues (5–10). The effective synthesis of eudistidine C provided proof of its proposed structure and a means to supply an extremely mass-limited natural compound. Preparation of a series of structural analogues of 1 helped delineate the electrophilic reactivity of ring D and the hemiaminal moiety in eudistidine A (2) and also provided additional eudistidine type alkaloids for biological evaluation. Resolution of racemic eudistidine C into its two enantiomers allowed assignment and characterization of (+)-(R)-eudistidine C (1a) and (−)-(S)-eudistidine C (1b).
Initial biological characterization of eudistidine C suggests potential applications in the context of cancer and malaria. (−)-(S)-Eudistidine C (1b) and analogues 5–7 and 10 inhibited interaction of the protein-binding domains of the oncogenic transcription factor HIF-1α and its coactivator p300, albeit only at relatively high concentrations. Small molecule inhibitors of tight protein–protein interactions are notoriously difficult to design or discover. Compounds based on the eudistidine framework represent possible chemical probes into the role of HIF-1α and p300 in the adaptation of cancer cells to low oxygen environments and are leads for the development of cancer therapeutics. Compounds 1a, 1b, 2, and 6–10 also inhibited chloroquine-sensitive and chloroquine-resistant strains of the malaria parasite P. falciparum at low micromolar concentrations. Treatment and prevention of malaria, and especially its drug-resistant variants, is a global health priority, and current drug therapies are inadequate. In total, compound 1 and its readily accessible structural analogues provide a potentially fruitful avenue for discovering novel antimalarial and cancer therapeutics.
EXPERIMENTAL SECTION
General Procedures.
NMR spectra were obtained with an NMR spectrometer equipped with a 3 mm cryogenic probe and operating at 600 MHz for 1H and 150 MHz for 13C. 1H–13C HMBC experiments were optimized for nJ = 8.3 Hz, and 1H–15N HMBC experiments were optimized for nJ = 8 Hz. (+)HRESIMS data for 1 were acquired on a Q-TOF LC/MS. (+)HRESIMS data for 4–10 were conducted on a LTQ-Orbitrap hybrid mass spectrometer system with an API electrospray ion source in positive-ion mode. Preparative reversed-phase HPLC was run on a system using a C18 column (10 μm, 110 Å, 75 × 30 mm) with 0.1% TFA in the aqueous phase or a C18 column (5 μm, 110 Å, 250 × 9.4 mm) with 0.1% formic acid. IR spectra were acquired after MeOH solutions of the sample compounds were applied to a NaCl plate and the solvent was removed under a stream of N2, followed by high vacuum. The three-dimensional structure depicted in the TOC graphic was generated by molecular mechanics minimization using Spartan 14, and the representation was generated in CYL view.29
Animal Material, Isolation, and Purification.
Specimens of the ascidian Eudistoma sp. were collected at a depth range of −3 to −10 m on the Koror side of the collapsed Koror–Babeldaob Bridge in the Koror/Airai Channel, Palau (07°21.64′ N, 134°30.17′ E), in September 1998 by Dr. Patrick Colin, under contract through the Coral Reef Research Foundation for the Natural Products Branch, National Cancer Institute. Taxonomic identification of the ascidian was done by Francoise Monniot, and a voucher specimen (0CDN5649) was deposited at the Smithsonian Institute, Washington, DC.The ascidian specimen (668 g, wet weight) was stored frozen until it was extracted according to the procedures detailed by McCloud30 to give 7.2 g of organic solvent (CH2Cl2–MeOH, 1:1) extract. A portion of the organic extract (812.9 mg) was fractionated on two diol SPE cartridges (2 g) eluting with 9:1 hexane–CH2Cl2 (fraction A), 20:1 CH2Cl2–EtOAc (fraction B), 100% EtOAc (fraction C), 5:1 EtOAc-MeOH (fraction D), and 100% MeOH (fraction E) in a stepwise manner. Repeated size-exclusion LH-20 column chromatography of fraction E (439.5 mg) using MeOH as the eluent yielded eudistidine C (1, 10.7 mg, 1.32%).
Eudistidine C (1):
yellow oil; UV (MeOH) λmax(log ε) 208 (4.36), 219 (4.30), 278 (4.32), 348 (3.40), 439 (3.45) nm; IR λmax 1646, 1626, 1504, 1351, 1246, 1167 cm−1; 1H and 13C NMR data, see Table 1; (+)HRESIMS m/z 488.1848 [M + H]+ (calcd for C28H21N7O2 488.1829).
Synthesis of Eudistidine A (2) HBr Salt.
The reaction to form eudistidine A (2) was run similarly to that reported previously, but the procedure was carried on a larger scale and the isolation procedure was significantly different.23 A solution of 2-bromo-4′-methoxyacetophe-none (620 mg, 2.7 mmol) in DMSO (15 mL) and water (1.5 mL) was heated at 65 °C for 3 h to generate 2-(4-methoxyphenyl)-2-oxoacetaldehyde. The solution was cooled to 4 °C before a solution of 4-(2-aminophenyl)pyrimidin-2-amine (300 mg, 1.8 mmol) in DMSO (2 mL) was added slowly. The resulting solution was heated at 60 °C for 1 h and then cooled back to 4 °C. I2 (450 mg, 1.8 mmol) in DMSO (2 mL) was added slowly, and the brown solution was heated for an additional 1 h at 60 °C. The resulting solution was cooled to room temperature and then added to a stirring solution of 1:1 acetone–hexanes (250 mL). The red solid was filtered, washed (1:1 acetone–hexanes), and then collected and dried (480 mg, 65%). The compound could be subjected to reversed-phase HPLC employing a gradient of 20–60% MeCN/H2O (1% NH4OH) over 10 min to yield the free base form of eudistidine A (2) that by spectroscopic analysis was identical to the natural product.23 The HBr salt of 2 was used in all the synthetic studies described below.
Eudistidine A (2) HBr salt:
1H NMR (DMSO-d6, 600 MHz) δ 9.21 (1H, d, J = 5.8 Hz, H-2), 8.99 (1H, d, J = 8.0 Hz, H-4), 8.40 (1H, d, J = 5.7 Hz, H-3), 8.28 (1H, t, J = 7.8 Hz, H-6), 8.23 (1H, d, J = 8.1 Hz, H-7), 8.17 (1H, br s, 10-OH), 8.10 (1H, t, J = 7.8 Hz, H-5), 7.71(2H, d, J = 8.8 Hz, H-12/16), 7.05 (2H, d, J = 8.8 Hz, H-13/15), 3.79 (3H, s, 14-OCH3); 13C NMR (DMSO-d6, 150 MHz) δ 165.8 (C-2), 160.3 (C-14), 152.2 (C-8c), 151.8 (C-8a), 145.3 (C-3a), 144.9 (C-7a), 137.8 (C-6), 131.3 (C-5), 129.7 (C-7), 128.9 (C-11), 128.3 (C-12/16), 126.9 (C-4), 116.5 (C-3b), 113.8 (C-13/15), 103.2 (C-3), 88.7 (C-10), 55.4 (14-OCH3). Anal. Calcd: C, 55.49; H, 3.68; Br, 19.43; N,13.62. Found: C, 49.39; H, 6.24; Br, 16.45; N. 14.35.31
Synthesis of 4-(2-Amino-1H-imidazol-5-yl)phenol (4).
5-(4-Methoxyphenyl)-1H-imidazol-2-amine (100 mg, 0.53 mmol) was dissolved in CH2Cl2 (5 mL), and the solution was cooled to 0 °C. BBr3 (1 M, 1.1 mL, 1.1 mmol) was added, and the reaction was allowed to warm to room temperature. H2O (2 mL) was added slowly to the resulting heterogeneous mixture. The aqueous layer was loaded directly onto a C18 column and eluted with a gradient of 5–95% MeCN–H2O (no modifier) to provide 4-(2-amino-1H-imidazol-5-yl)phenol, as the HBr salt, as a white solid (100 mg, 70%):32 IR (thin film) λmax 3271, 3142, 3077, 1698, 1674, 1602, 1511, 1352, 1199 cm−1; 1H NMR (DMSO-d6, 400 MHz) δ 12.1 (bs, 2H), 9.70 (s, 1H), 7.44 (d, 2H, J = 8.4 Hz), 7.31 (s, 2H), 7.15 (s, 1H), 6.81 (d, 2H, J = 8.4 Hz); 13C NMR (DMSO-d6, 100 MHz) δ 157.4, 147.2, 127.0, 125.9, 118.7, 115.6, 107.4; (+)HRESIMS m/z 176.0816 [M + H]+ (calc for C9H10N3O 176.0818).
Synthesis of Eudistidine C (1).
Eudistidine A HBr salt (20 mg, 0.048 mmol) was dissolved in DMSO (0.5 mL), and 4-(2-amino-1H-imidazol-5-yl)phenol (4, 15 mg, 0.072 mmol) was added. The solution was allowed to stand for 24 h. The solution was purified by reversed-phase preparative HPLC, employing a gradient of 5–95% MeCN/H2O (0.1% TFA) over 10 min to yield eudistidine C (1, 18 mg, 64%): IR (thin film) λmax 3268, 3100, 1665, 1638, 1525, 1303, 1190 cm−1; 1H NMR (CD3OD, 600 MHz) see the Supporting Information; 13C NMR (CD3OD, 150 MHz) see the Supporting Information; (+)HRESIMS m/z 488.1821 [M + H]+ (calcd for C28H22N7O2 488.1829).
Synthesis of Eudistidine C N-Methylindole Analogue (5).
Eudistidine A HBr salt (20 mg, 0.048 mmol) was dissolved in DMSO (0.5 mL), and N-methylindole (10 mg, 0.072 mmol) was added. The solution was stirred at 22 °C for 24 h and then purified by reversed-phase preparative HPLC, employing a gradient of 5–95% MeCN–H2O (0.1% TFA) over 10 min to yield 5 (16 mg, 61%): IR (thin film) λmax 1664, 1638, 1511, 1256, 1172 cm−1; 1H NMR (CD3OD, 600 MHz) δ 9.02 (1H, d, J = 5.8 Hz, H-2), 8.73 (1H, d, J = 8.0 Hz, H-4), 8.20 (1H, t, J = 7.7 Hz, H-6), 8.15 (1H, d, J = 8.1 Hz, H-7), 8.06 (1H, d, J = 5.6 Hz, H-3), 7.99 (1H, t, J = 7.7 Hz, H-5), 7.60 (2H, d, J = 8.5 Hz, H-12/16), 7.43 (1H, d, J = 8.3 Hz, H-21), 7.21 (2H, m, H-18, H-22), 7.13 (1H, d, J = 7.6 Hz, H-24), 6.98 (3H, m, H-13/15, H-23), 3.81 (3H, s, H-14OCH3), 3.80 (3H, s, H–N19OCH3); 13C NMR (CD3OD, 150 MHz) δ 167.4 (C-2), 161.9 (C-14), 154.5 (C-8c), 154.3 (C-8a), 148.3 (C-3a), 147.0 (C-7a), 139.6 (C-20), 138.7 (C-6), 131.9 (C-5), 131.2 (C-11), 130.9 (C-7), 130.7 (C-18), 129.8 (C-12/16), 127.5 (C-4), 126.2 (C-25), 123.6 (C-22), 121.4 (C-24), 121.1 (C-23), 117.8 (C-3b), 115.3 (C-13/15), 114.0 (C-17), 111.0 (C-21), 103.5 (C-3), 71.5 (C-10), 55.9 (C-14OCH3), 33.1 (C-N19OCH3); (+)HRESIMS m/z 444.1806 [M + H]+ (calcd for C28H22N5O 444.1819).
Synthesis of Eudistidine C 3-Methylindole Analogue (6).
To a solution of eudistidine A HBr salt (5.2 mg, 0.013 mmol) in DMSO (1.0 mL) was added 3-methylindole (17 mg, 0.13 mmol). The orange/brown solution was stirred at 40 °C for 7 days, after which time the reaction was purified directly by preparative reversed-phase HPLC (5–95% MeCN/0.1% TFA/water). The product-containing fractions were concentrated in vacuo to afford 6 (2.5 mg, 35% yield) as a brown solid: IR (thin film) λmax 3179, 1664, 1638, 1512, 1305, 1176 cm−1; 1H NMR (CD3OD, 600 MHz) δ 9.04 (1H, d, J = 5.9 Hz, H-2), 8.76 (1H, d, J = 8.1 Hz, H-4), 8.23 (1H, m, H-6), 8.21 (1H, m, H-7), 8.10 (1H, d, J = 5.8 Hz, H-3), 8.02 (1H, m, H-5), 7.70 (2H, d, J = 8.7 Hz, H-12/16), 7.51 (1H, d, J = 8.1 Hz, H-23), 7.25 (1H, d, J = 8.1 Hz, H-20), 7.13 (1H, t, J = 7.6 Hz, H-21), 7.07 (2H, d, J = 8.7 Hz, H-13/15), 7.04 (1H, t, J = 7.6 Hz, H-22), 3.84 (3H, s, 14-OCH3), 1.91 (3H, s, 25-CH3); 13C NMR (CD3OD, 150 MHz) δ 167.5 (C-2), 162.2 (C-14), 154.4 (C-8c), 153.7 (C-8a), 148.3 (C-3a), 147.0 (C-7a), 138.8 (C-6), 137.5 (C-19), 132.2 (C-5), 131.1 (C-7), 130.6 (C-17), 130.5 (C-11), 130.5 (C-24), 129.7 (C-12/16), 127.5 (C-4), 124.2 (C-21), 120.5 (C-22), 119.8 (C-23), 117.9 (C-3b), 115.6 (C-13/15), 112.7 (C-25), 112.2 (C-20), 103.7 (C-3), 71.6 (C-10), 55.9 (14-OCH3), 9.5 (25-CH3); (+)HRESIMS m/z 444.1808 [M + H]+ (calcd for C28H22N5O 444.1819.
Synthesis of Eudistidine C N-Methylpyrrole Analogue (7).
Eudistidine A HBr salt (20 mg, 0.048 mmol) was dissolved in DMSO (0.5 mL), and N-methylpyrrole (6 mg, 0.072 mmol) was added. The solution was stirred at 22 °C for 24 h and then purified by reversed-phase preparative HPLC, employing a gradient of 5–95% MeCN–H2O (0.1% TFA) over 10 min to yield 7 (13 mg, 56%): IR (thin film) λmax 1663, 1638, 1510, 1301, 1256, 1177 cm−1; 1H NMR (CD3OD, 600 MHz) δ 9.01 (1H, d, J = 5.3 Hz, H-2), 8.74 (1H, d, J = 7.7 Hz, H-4), 8.23 (1H, t, J = 7.9 Hz, H-6), 8.20 (1H, d, J = 8.1 Hz, H-7), 8.06 (1H, d, J = 5.3 Hz, H-3), 8.01 (1H, t, J = 7.8 Hz, H-5), 7.42 (2H, d, J = 8.8 Hz, H-12/16), 7.04 (2H, d, J = 8.8 Hz, H-13/15), 6.82 (1H, br t, J = 2.2 Hz, H-19), 6.26 (1H, m, H-21), 6.09 (1H, br t, J = 3.3 Hz, H-20), 3.82 (3H, s, H-14OCH3), 3.45 (3H, s, H–N18OCH3); 13C NMR (CD3OD, 150 MHz) δ 167.4 (C-2), 162.2 (C-14), 154.3 (C-8c), 153.2 (C-8a), 148.4 (C-3a), 146.7 (C-7a), 138.8 (C-6), 132.2 (C-5), 130.9 (C-7), 130.6 (C-11), 129.6 (C-12/16), 128.9 (C-17), 128.2 (C-19), 127.6 (C-4), 117.8 (C-3b), 115.7 (C-13/15), 112.8 (C-21), 108.1 (C-20), 103.8 (C-3), 71.4 (C-10), 55.9 (C-14OCH3), 36.4 (C-N18OCH3); (+)HRESIMS m/z 394.1656 [M + H]+ (calcd for C24H20N5O 394.1662).
Synthesis of Eudistidine C p-Phenol Analogue (8).
To a solution of eudistidine A HBr salt (6.0 mg, 0.014 mmol) in DMSO (0.3 mL) was added phenol (53 mg, 0.57 mmol). The orange/brown solution was stirred at 40 °C for 6 days, after which time the reaction was purified directly by preparative reversed-phase HPLC employing a gradient of 10–95% MeCN/H2O (0.1% formic acid) over 8 min. The product-containing fractions were concentrated in vacuo to afford 8 (1.8 mg, 27% yield) as a yellow solid: IR (thin film) λmax 3100, 1663, 1639, 1303, 1180 cm−1; 1H NMR (CD3OD, 600 MHz) δ 8.44 (1H, d, J = 5.0 Hz, H-2), 8.22 (1H, d, J = 7.7 Hz, H-4), 7.89 (1H, m, H-6), 7.87 (1H, m, H-7), 7.66 (1H, m, H-5), 7.44 (2H, d, J = 9.4 Hz, H-12/16), 7.34 (2H, d, J = 9.5 Hz, H-18/22), 6.90 (2H, d, J = 8.8 Hz, H-13/15), 6.78 (1H, d, J = 5.1 Hz, H-3), 6.76 (2H, d, J = 8.5 Hz, H-19/21), 3.77 (3H, s, 14-OCH3); 13C NMR (CD3OD, 150 MHz) δ 167.4 (C-2), 162.5 (C-8a), 160.7 (C-14), 158.3 (C-20), 156.3 (C-8c), (C-7a), 147.5 (C-3a), 136.4 (C-6), 135.9 (C-11), 134.6 (C-17), 129.9 (C-7), 129.8 (C-5), 129.8 (C-12/16), 129.8 (C-18/22), 126.3 (C-4), 117.9 (C-3b), 116.1 (C-19/21), 114.7 (C-13/15), 93.7 (C-3), (C-10), 55.7 (14-OCH3); (+)HRESIMS m/z 407.1497 [M + H]+ (calcd for C25H19N4O2 407.1503).
Synthesis of Eudistidine C Resorcinol Analogue (9).
To a solution of eudistidine A HBr salt (10 mg, 0.025 mmol) in DMSO (0.5 mL) was added resorcinol (31 mg, 0.29 mmol). The orange/brown solution was stirred at 40 °C for 24 h, after which time the reaction was purified directly by preparative reversed-phase HPLC employing a gradient of 10–95% MeCN/H2O (0.1% formic acid) over 8 min. The product-containing fractions were concentrated in vacuo to afford 9 (7.2 mg, 62% yield) as a yellow solid: IR (thin film) 3100, 1662, 1527, 1303, 1258, 1185 cm−1; 1H NMR (DMSO-d6 600 MHz) δ 8.42 (1H, d, J = 4.7 Hz, H-2), 8.28 (1H, d, J = 7.8 Hz, H-4), 7.86 (1H, t, J = 7.5 Hz, H-6), 7.78 (1H, d, J = 8.1 Hz, H-7), 7.65 (2H, d, J = Hz, H-12/16), 7.61 (1H, t, J = 7.5 Hz, H-5), 6.96 (2H, d, J = 9.1 Hz, H-13/15), 6.68 (1H, d, J = 8.8 Hz, H-18), 6.79 (1H, d, J = 4.9 Hz, H-3), 6.16 (1H, d, J = 2.3 Hz, H-21), 6.14 (1H, dd, J = 8.7, 2.5 Hz, H-19), 3.74 (3H, s, 14-OCH3); 13C NMR (DMSO-d6, 150 MHz) δ 165.9 (C-2), 162.9 (C-8a), 158.7 (C-14), 158.2 (C-20), 156.9 (C-22), 154.4 (C-8c), 145.8 (C-7a), 145.3 (C-3a), 134.9 (C-6), 133.0 (C-11), 128.8 (C-18), 128.7 (C-12/16), 128.3 (C-7), 128.0 (C-5), 125.3 (C-4), 119.7 (C-17), 116.6 (C-3b), 113.5 (C-13/15), 105.6 (C-19), 103.1 (C-21), 91.7 (C-3), 76.1 (C-10), 55.1 (14-OCH3); (+)HRESIMS m/z 423.1446 [M + H]+ (calcd for C25H19N4O3 423.1452).
Synthesis of Eudistidine C Phloroglucinol Analogue (10).
To a solution of eudistidine A HBr salt (5.6 mg, 0.014 mmol) in DMSO (1.0 mL) was added phloroglucinol (16.4 mg, 0.130 mmol). The orange/brown solution was stirred at 22 °C for 24 h, after which time the reaction was purified directly by preparative reversed-phase HPLC (5–95% MeCN/0.1% TFA/water). The product-containing fractions were concentrated in vacuo to afford 10 (4.2 mg, 55% yield) as a yellow solid: IR (thin film) λmax 3101, 1672, 1627, 1562, 1257, 1183 cm−1; 1H NMR (CD3OD, 600 MHz) δ 8.77 (1H, d, J = 5.2 Hz, H-2), 8.12 (1H, d, J = 8.1 Hz, H-4), 7.63 (1H, t, J = 7.4 Hz, H-6), 7.52 (1H, d, J = 5.3 Hz, H-3), 7.41 (2H, d, J = 8.9 Hz, H-12/16), 7.19 (1H, t, J = 7.8 Hz, H-5), 7.12 (1H, d, J = 8.4 Hz, H-7), 7.01 (2H, d, J = 8.9 Hz, H-13/15), 6.07 (1H, d, J = 1.9 Hz, H-21), 5.85 (1H, d, J = 1.9 Hz, H-19), 3.63 (3H, s, 14-OCH3); 13C NMR (CD3OD, 150 MHz) δ 168.3 (C-2), 163.3 (C-20), 162.0 (C-14), 160.6 (C-18), 157.1 (C-22), 156.5 (C-8c), 149.2 (C-3a), 144.6 (C-7a), 138.4 (C-6), 130.4 (C-12/16), 127.7 (C-4), 126.8 (C-11), 123.1 (C-5), 118.7 (C-7), 115.0 (C-13/15), (C-3b), 104.4 (C-3), 103.3 (C-17), 98.5 (C-21), 90.6 (C-19), (C-10), 55.8 (14-OCH3), not observed (C-8a); (+)HRESIMS m/z 439.1392 [M + H]+ (calcd for C25H19N4O4 439.1401).
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge D. Newman (NCI) for organizing and documenting the collection, the Natural Products Support Group at NCI—Frederick for extraction, T. Bostaph (MTL) and K. Goncharova (MTL) for screening data compilation and analysis, J. Barchi (CBL) for NMR assistance, J. Kelly (CBL) for mass spectrometry analysis, and S. Tarasov and M. Dyba (Biophysics Resource Core, Structural Biophysics Laboratory, CCR) and H. Bokesch (MTL) for assistance with high-resolution mass spectrometry. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This project was also funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E and by the USDA Agricultural Research Service Specific Cooperative Agreement No. 58-6408-1-603. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.joc.6b02380.
1H NMR, 13C NMR, COSY, HSQC, HMBC, and LR-HSQMBC data and UV—vis data for compound 1. 1H NMR and 13C NMR data for compounds 5–10. ACD/Structure Elucidator generation results and table of atomic coordinates for compound 1. ECD spectra for compounds 1a and 1b. p300/HIF-1α and antimalaria testing data for compounds 1a,b, 2, 5–10 (PDF)
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Newman DJ; Cragg GM J. Nat. Prod 2016, 79, 629–661. [DOI] [PubMed] [Google Scholar]
- (2).Ling LL; Schneider T; Peoples AJ; Spoering AL; Engels I; Conlon BP; Mueller A; Schäberle TF; Hughes DE; epstein S; Jones M; Lazarides L; Steadman VA; Cohen DR; Felix CR; Fetterman KA; Millett WP; Nitti AG; Zullo AM; Chen C; Lewis K Nature 2015, 517, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Harvey AL; Edrada-Ebel R; Quinn RJ Nat. Rev. Drug Discovery 2015, 14, 111–129. [DOI] [PubMed] [Google Scholar]
- (4).Newman DJ; Cragg GM Mar. Drugs 2014, 12, 255–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Steinmetz H; Li J; Fu C; Zaburannyi N; Kunze B; Harmrolfs K; Schmitt V; Herrmann J; Reichenbach H; Höfle G; Kalesse M; Müller R Angew. Chem., Int. Ed 2016, 55, 10113–10117. [DOI] [PubMed] [Google Scholar]
- (6).Tadesse M; Strøm MB; Svenson J; Jaspars M; Milne BF; Tørfoss V; Andersen JH; Hansen E; Stensvåg K; Haug T Org. Lett 2010, 12, 4752–4755. [DOI] [PubMed] [Google Scholar]
- (7).Avilés E; Rodríguez AD Org. Lett 2010, 12, 5290–5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Sueyoshi K; Kaneda M; Sumimoto S; Oishi S; Fujii N; Suenaga K; Teruya T Tetrahedron 2016, 72, 5472–5478. [Google Scholar]
- (9).Walsh CT Nat. Chem. Biol 2015, 11, 620–624. [DOI] [PubMed] [Google Scholar]
- (10).Doroghazi JR; Albright JC; Goering AW; Ju K-S; Haines RR; Tchalukov KA; Labeda DP; Kelleher NL; Metcalf WW Nat. Chem. Biol 2014, 10, 963–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Starcevic A; Zucko J; Simunkovic J; Long PF; Cullum J; Hranueli D Nucleic Acids Res 2008, 36, 6882–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Molinski TF Curr. Opin. Biotechnol 2010, 21, 819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Molinski TF Nat. Prod. Rep 2010, 27, 321–329. [DOI] [PubMed] [Google Scholar]
- (14).Hilton BD; Martin GE J. Nat. Prod 2010, 73, 1465–1469. [DOI] [PubMed] [Google Scholar]
- (15).Saurí J; Bermel W; Buevich AV; Sherer EC; Joyce LA; Sharaf MHM; Schiff PL Jr.; Parella T; Williamson RT; Martin GE Angew. ChemInt. Ed 2015, 54, 10160–10164. [DOI] [PubMed] [Google Scholar]
- (16).Paudel L; Adams RW; Király P; Aguilar JA; Foroozandeh M; Cliff MJ; Nilsson M; Sándor P; Waltho JP; Morris GA Angew. Chem., Int. Ed 2013, 52, 11616–11619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Liu Y; Green MD; Marques R; Pereira T; Helmy R; Williamson RT; Bermel W; Martin GE Tetrahedron Lett. 2014, 55, 5450–5453. [Google Scholar]
- (18).Halabalaki M; Vougogiannopoulou K; Mikros E; Skaltsounis AL Curr. Opin. Biotechnol 2014, 25, 1–7. [DOI] [PubMed] [Google Scholar]
- (19).Elyashberg ME; Williams AJ; Martin GE Computer-Assisted Structure Elucidation In Progress in NMR Spectroscopy; Feeney J, Sutcliff L, Eds.; Pergammon: London, 2008; Vol. 53, pp 1–104. [Google Scholar]
- (20).Plainchont B; de Paulo Emerenciano V; Nuzillard J-M Magn. Reson. Chem 2013, 51, 447–453. [DOI] [PubMed] [Google Scholar]
- (21).Moser A; Elyashberg ME; Williams AJ; Blinov KA; DiMartino JC J. Cheminf 2012, 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Naman CB; Li J; Moser A; Hendrycks JM; Benatrehina PA; Chai H; Yuan C; Keller WJ; Kinghorn AD Org. Lett 2015, 17, 2988–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Chan ST; Patel PR; Ransom TR; Henrich CJ; McKee TC; Goey AK; Cook KM; Figg WD; McMahon JB; Schnermann MJ; Gustafson KR J. Am. Chem. Soc 2015, 137, 5569–5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Williamson RT; Buevich AV; Martin GE; Parella T J. Org. Chem 2014, 79, 3887–3894. [DOI] [PubMed] [Google Scholar]
- (25).Blinov KA; Buevich AV; Williamson RT; Martin GE Org. Biomol. Chem 2014, 12, 9505–9509. [DOI] [PubMed] [Google Scholar]
- (26).Lorenc C; Saurí J; Moser A; Buevich AV; Williams AJ; Williamson RT; Martin GE; Peczuh MW ChemistryOpen 2015, 4, 577–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Shoemaker RH Nat. Rev. Cancer 2006, 6, 813–823. [DOI] [PubMed] [Google Scholar]
- (28).Makler MT; Hinrichs DJ Am. J. Trop. Med. Hyg 1993, 48, 205–210. [DOI] [PubMed] [Google Scholar]
- (29).Legault CY CYLview, 1.0b; Université de Sherbrooke, 2009, http://www.cylview.org
- (30).McCloud TG Molecules 2010, 15, 4526–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).The elemental analysis indicates the inclusion of bromine, but not proof of molecular formula, which was conclusively demonstrated in ref 23.
- (32).Nath JP; Mahapatra GN Indian J. Chem. Sect. B 1980, 19B, 536–538 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.