ABSTRACT
Under physiological conditions, PD-1/PD-L1 interactions regulate unwanted over-reactions of immune cells and contribute to maintain peripheral tolerance. However, in tumor microenvironment, this interaction may greatly compromise the immune-mediated anti-tumor activity. PD-1+ NK cells have been detected in high percentage in peripheral blood and ascitic fluid of ovarian carcinoma patients. To acquire information on PD-1 expression and physiology in human NK cells, we analyzed whether PD-1 mRNA and protein are present in resting, surface PD-1−, NK cells from healthy donors. Both different splicing isoforms of PD-1 mRNA and a cytoplasmic pool of PD-1 protein were detected. Similar results were obtained also from both in vitro-activated and tumor-associated NK cells. PD-1 mRNA and protein were higher in CD56dim than in CD56bright NK cells. Confocal microscopy analyses revealed that PD-1 protein is present in virtually all NK cells analyzed. The present findings are compatible with a rapid surface expression of PD-1 in NK cells in response to appropriate, still undefined, stimuli.
KEYWORDS: Natural killer cells, checkpoint inhibitors, PD-1, CD56dim CD56bright, mRNA isoforms, PD-1 cytoplasmic pool
Introduction
NK cells play a major role in innate defenses against viruses and tumors.1,2 In humans, NK cells can be distinguished in two phenotypically and functionally distinct subsets on the basis of CD56 expression levels.3 While CD56dim cells are mainly cytotoxic and are largely prevalent in peripheral blood (PB), CD56bright NK cells display immunoregulatory activity, produce pro-inflammatory cytokines (IFN-γ and TNFα), and are prevalent in tissues and secondary lymphoid organs.4 NK cell function is regulated by an array of surface receptors with activating or inhibitory activity. Natural Cytotoxic Receptors (NCRs), represented by NKp30, NKp44 and NKp46, are some major activating receptors allowing tumor cell recognition and killing.5 Other activating receptors, able to trigger NK cell-mediated killing of tumor cells, are NKG2D and the co-activating receptor/adhesion molecule DNAM-1.6 Important inhibitory receptors, such as KIRs, CD94/NKG2A and LIR-1, regulate NK cell function through the recognition of the classical and non-classical Human Leukocyte Antigen (HLA) class I molecules.7
In recent years, major molecular checkpoints have been shown to control/inactivate different immune cell types, maintain peripheral tolerance and finely regulate the overall immune response. Among these checkpoints, Programmed Cell Death (PD-1) may be expressed on activated T cells regulating their proliferation and antigen-specific responses upon interaction with the PD-ligands 1 (PD-L1) and 2 (PD-L2) expressed by antigen-presenting cells (APC).8-11 However, when PD-L1/PD-L2 are expressed on tumor cells, T cell-mediated anti-tumor activity may be severely compromised.12,13 Recently, PD-1 has been detected in variable proportions also in a subset of fully mature NK cells (CD56dimNKG2A−KIR+CD57+) isolated from some Cytomegalovirus (CMV)-seropositive healthy donors (HDs). In addition, PD-1 was found to be expressed in NK cells derived from a fraction of peripheral blood and, at higher frequency, in ascites of patients with ovarian carcinoma.14 Notably, in all cases, CD56bright cells were consistently negative for PD-1 surface expression. The expression of PD-1 in NK cells is particularly relevant in case of tumors that have lost or down-regulated HLA-I molecules. While these tumors escape T cell control, they become potentially susceptible to NK cell-mediated killing. In this context, the expression of PD-1 on NK cells may acquire a relevant role in NK-mediated anti-tumor response against PDL-1/PDL-2+ tumors.15,16 The recent use of anti-PD-1 or anti-PD-L1 monoclonal antibodies (mAbs) in tumor therapy, resulted in strong improvements of the clinical outcome particularly in melanoma and lung carcinoma patients.17-19 In view of these important findings, it is crucial to acquire information on the expression and on physiological aspects of PD-1 in NK cells. In this study, we analyzed whether PD-1 mRNA and PD-1 protein are detectable in the cytoplasm of resting NK cells of healthy individuals. We show that different splicing isoforms of PD-1 mRNA are present in resting NK cells in all the donors analyzed. In addition, we could consistently detect a cytoplasmic pool of PD-1 protein. Both PD-1 mRNA and protein are present in higher amounts in mature CD56dim, than in CD56bright, immature NK cells. Moreover, PD-1 protein displays a diffuse cytoplasmic distribution and a partial colocalization with Golgi compartments in all NK cells. The presence of a PD-1 cytoplasmic pool is compatible with a rapid surface expression of PD-1 in response to appropriate cellular stimuli.
Results and discussion
Considering both the important role of NK cells in anti-tumor activity and the inhibitory effect exerted by PD-1 in PD-1+ NK cells, we analyzed whether PD-1 expression was dependent on a de novo synthesis or rather a preformed pool of PD-1 mRNA existed in NK cells. In this context, previous studies showed that PD-1 messenger RNA (mRNA) is present, in alternative splicing isoforms, in unfractionated Peripheral Blood Mononuclear Cells (PBMC).20 However, no information was provided on which cell type(s) express such mRNA. The human PD-1 coding sequence was amplified by PCR using a pair of primers allowing the identification of all the splicing isoforms (Figure 1(a)). Freshly isolated PBMC were analyzed for PD-1 surface expression prior to NK purification. A low PD-1 membrane expression, ranging from 0.7% to 2.6% (Figure 1(b)), was observed on fresh PB (“resting”) NK cells in the 14 donors analyzed. Two cell lines NK92 and HEK-293T (hereafter HEK) were used as controls. NK92 is an immortalized clonal cell line derived from a patient with NK-cell lymphoma, which has recently been used in different clinical trials.21 Since, according to the antibody datasheet, HEK were indicated as a positive control, we included this cell line as further control. As shown in Figure 1(c)the full length (FL) form of PD-1 mRNA was the only detectable band in HEK and NK92 cell lines. Analysis of resting NK cells revealed that, although the FL form was the predominant band, other amplification products existed. These bands corresponded to the following isoforms: Δexon3, Δexon2, Δexon2,3 and Δexon2,3,4. Therefore, these data indicate that resting NK cells contain a PD-1 mRNA pool that consists not only of the FL form but also of other splicing variants. On the basis of these results, we further investigated whether also the PD-1 protein, a transmembrane glycoprotein of approximately 50–55 kDa,22 was present in the NK cells. To this end, total protein extracts obtained from resting NK cells of 14 different donors or from the HEK and NK92 cell lines were analyzed. While NK92 cells expressed a 55 kDa protein (upper band), HEK expressed a lighter form of 37 kDa (lower band) (Figure 1(d)) possibly representing the unglycosylated form of the protein. Remarkably, in NK cells from HDs, both PD-1 bands (55 kDa and 37 kDa) were present (Figure 1(d)). To further confirm that the protein detected indeed corresponded to PD-1, we performed experiments for silencing PD-1 expression. For this purpose, NK cells were transfected with validated siRNA specific for PD-1 or with mock siRNA. Reduction in PD-1 mRNA levels was detected only in PD-1-specific siRNA-treated samples (Figure 1(e)). In line with the mRNA data, a parallel reduction in PD-1 protein was also documented (Figure 1(f–g)). Thus, our data indicate that not only PD-1 mRNA but also PD-1 protein are present in resting NK cells from HDs. We further investigated the presence of PD-1 mRNA and protein in in vitro activated NK cells or tumor associated NK cells isolated from malignant pleural effusions (PE) (Figure 2(a–f)). NK cells, isolated from three different HDs, were cultured with IL-2 for 3 weeks. As in resting NK cells, no significant PD-1 surface expression could be detected in such IL-2 treated NK cells (Figure 2(a)), while we could detect the presence of a cytoplasmic pool of both PD-1 mRNA and protein (Figure 2(b,c)). In particular, the RNA analysis for PD-1 isoforms revealed the presence of the FL, Δexon3 and Δexon2,3 splicing variants (Figure 2(b)). On the other hand, neither the Δexon2 nor the Δexon2,3,4 isoforms could be detected in the three different donors analyzed. The PD-1 protein profile was similar to the one detected in resting NK cells, as both the upper and lower bands were present (Figure 2(c)). Of note, in all HDs tested, the upper band appeared to be more expressed as compared to the lower one. Analysis of PD-1 cytoplasmic pool in pathological conditions was performed in NK cells isolated from malignant PE in patients with primary or metastatic tumors (four patients analyzed). Analysis of PD-1 isoforms allowed the detection of all PD-1 splicing variants with the exception of Δexon2,3 (Figure 2(d)). Assessment of PD-1 protein expression revealed the presence of both lower and upper PD-1 bands (Figure 2(e)) in resting and in vitro-activated NK cells. However, in NK cells from PE, the lower band, possibly corresponding to the unmodified protein, was more expressed than the upper band. Thus, both PD-1 cytoplasmic mRNA and protein pool are also present in IL-2-activated NK cells and in tumor-associated NK cells. Remarkably, in addition to PD-1 FL, the Δexon3 isoform was the only one present in all the different NK cell populations analyzed. The Δexon3 isoform is of particular interest because it codifies for a putative soluble PD-1 (sPD-1) protein.20,23 Thus, we investigated whether, in addition to sPD-1 transcript, also a soluble form of PD-1 was released. For this purpose, supernatants collected from resting and in vitro activated NK cells were analyzed by ELISA. The same study was performed also on PE. As shown in Figure 2(f), no sPD-1 was detected in supernatants of resting or IL-2-activated NK cells. These results are in line with previous studies in which no sPD1 was detectable in healthy individuals.20 On the other hand, sPD-1 was detected in PE (Figure 2(f)). These data reveal that a soluble form of PD-1may indeed by released in tumor environment, but they do not prove that it is NK-derived as it may be produced by other cell types present in the pleural cavity.
Figure 1.
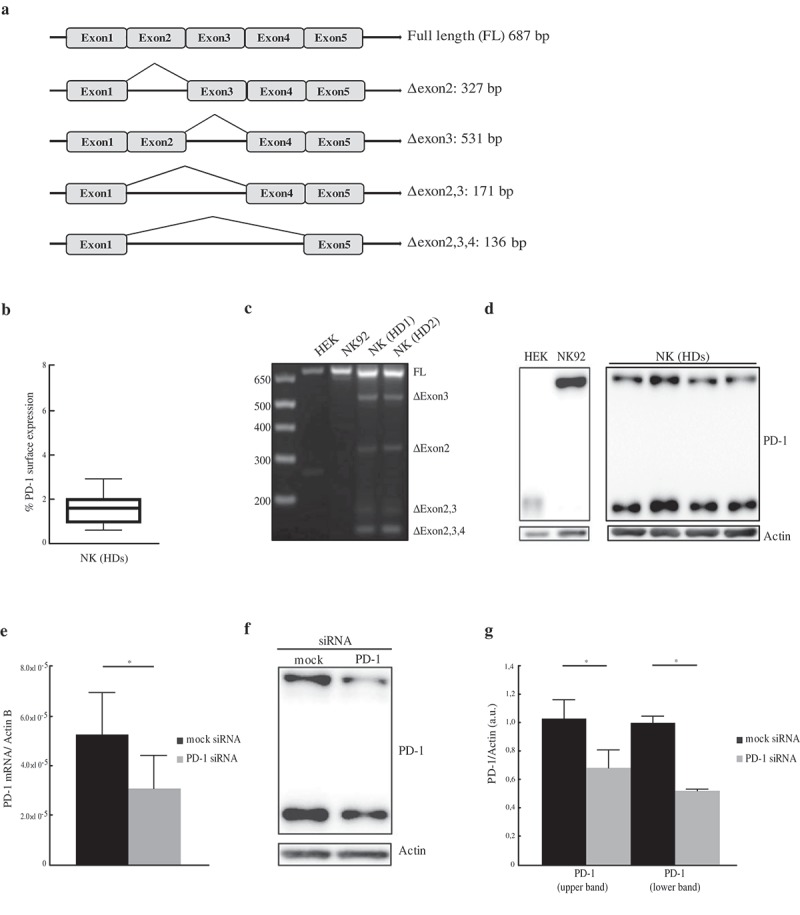
Detection of PD-1 mRNA isoforms and protein. (a) Schematic representation of PD-1 mRNA splicing variants. (b) Percentages of PD-1 surface expression (ranging from 0,7% to 2,6%) in resting NK cells isolated from HDs. (c) Analysis of PD-1 mRNA isoforms on resting NK cells and on both the HEK and NK92 cell lines. NK cells amplification profiles of two donors (HD1 and HD2), out of 14 analyzed, have been reported. (d) Total protein extracts from HEK, NK92 and resting NK cells were analyzed for PD-1 expression. The figure reports the western blot profile of NK cells of four donors out of 14 analyzed. (e–g) PD-1 validation in NK cells transfected with mock or PD-1 siRNA. Samples were collected 72 h after electroporation and analyzed for PD-1 mRNA and protein expression. (e) RT-PCR PD-1 mRNA expression, in mock- (black column) and PD-1 siRNA- (gray column) transfected NK cells. PD-1 expression was normalized over Actin. Values are mean ± SEM. Statistical significance has been determined by paired t-Test. (f) PD-1 protein expression in mock- and PD-1-siRNA transfected NK resting cells. A representative image from three independent experiments has been reported. (g) PD-1 protein quantification of mock- (black columns) and PD-1-siRNA transfected (gray columns) NK cells. PD-1 values, normalized over Actin, have been reported as a fold change calculated as a ratio between PD-1-siRNA and mock samples. Values, relative to three independent experiments, are mean ± SEM. Statistical significance has been determined by paired t-Test.
Figure 2.
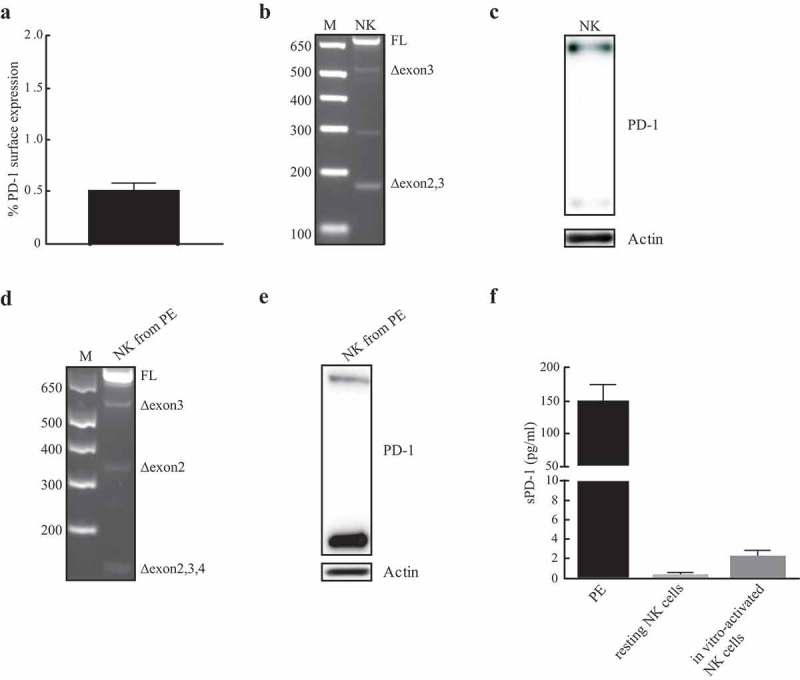
Detection of PD-1 mRNA isoforms and protein in IL-2 activated NK cells and in NK cells isolated from malignant PE. (a) Percentages of PD-1 surface expression in in vitro-activated NK cells isolated from HDs. (b) Analysis of PD-1 mRNA isoforms on NK cells. One representative HD, out of three analyzed, has been reported. (c) Total protein extracts isolated from in vitro-activated NK cells were analyzed for PD-1 expression. (d) Analysis of PD-1 splicing variants in NK from PE. (e) PD-1 protein profile of PE infiltrating NK cells. (f) sPD-1 has been studied in PE or supernatants (SN) of both resting and IL-2-activated NK cells isolated from HDs. Data for PE and HDs are relative to four and three different samples, respectively. sPD-1 concentration (pg/ml) has been calculated by use of a four-point-fit calibration curve of the standard dilutions.
The majority of PB-NK cells are mature cytotoxic CD56dim cells. Therefore, we further analyzed whether both mRNA and protein expression was confined to this NK subset. To clarify this point, we investigated whether also the immature CD56bright cells would express PD-1 mRNA and protein. To this end, CD56dim and CD56bright NK cells subsets were purified by cell sorting (Figure S1) and analyzed separately for both PD-1 mRNA and protein expression. Real-time amplification of the PD-1 coding sequence and western blot analysis showed that also the less mature CD56bright NK cells express both PD-1 mRNA and protein (Figure 3(a–c)). However, a comparative analysis revealed substantial differences in the two cell subsets. Indeed, CD56dim display higher levels of both PD-1 transcript and protein as compared to CD56bright cells. These data clearly indicate that the whole population of PB-NK cells, also including CD56bright cells, contain pools of both PD-1 mRNA and protein. Remarkably, the CD56dim subset, i.e. those cells that may express PD-1 at the cell surface, display higher levels of PD-1. To further assess the type of PD-1 localization in the cytoplasm and to determine whether it was expressed by all NK cells or only by a cell fraction, NK cells were analyzed by immunofluorescence and confocal microscopy. Figure 4(a) shows a diffuse distribution of cytoplasmic PD-1. Notably, cytoplasmic PD-1 was detected in all resting NK cells isolated from PB of different HDs. In a previous study on T cells, PD-1 was found in the proximity of the Golgi compartments.24 Thus, we further investigated whether such localization also occurred in NK cells. To this end, we used GM130 and TGN46 mAbs specific for the cis- and trans-Golgi cisternae, respectively. Confocal microscopy analysis was performed both in resting NK cells and in the NK92 and HEK cell lines. In order to detect possible associations of endogenous PD-1 with the Golgi network, we measured the distance between PD-1 dots close to the cisternae and the Golgi markers (GM130 and TGN46). Resting PB-NK cells showed a substantial colocalization of PD-1 with both Golgi compartments (Figure 4(b,c)). In contrast, in NK92 and HEK cells, the mean distance between PD-1 and Golgi markers was increased (Figures 4(b,c); S2(a,b)). Altogether, these data show that PD-1 is homogeneously expressed in the cytoplasm of all resting NK cells. In addition, it is detectable also in proximity of both the cis- and trans-Golgi cisternae.
Figure 3.
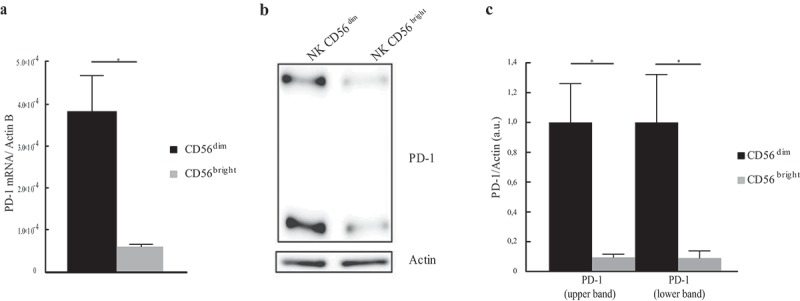
Different PD-1 mRNA and protein expression in CD56dim and CD56bright subsets. (a) PD-1 mRNA expression in sorted CD56dim (black column) and CD56bright (gray column) cells. PD-1 mRNA level was evaluated by RT-PCR amplification and normalized over Actin. Values, relative to three independent experiments are mean ± SEM. Statistical significance has been calculated by unpaired t-Test. (b) PD-1 protein expression in CD56dim and CD56bright NK subsets. A representative image from three independent experiments has been reported. (c) PD-1 protein quantification in CD56bright and CD56dim cells from three independents experiments. PD-1 values, normalized over Actin, are mean ± SEM and represent the PD-1 fold change calculated as a ratio between CD56bright and CD56dim cells. Statistical significance has been determined by unpaired t-Test.
Figure 4.
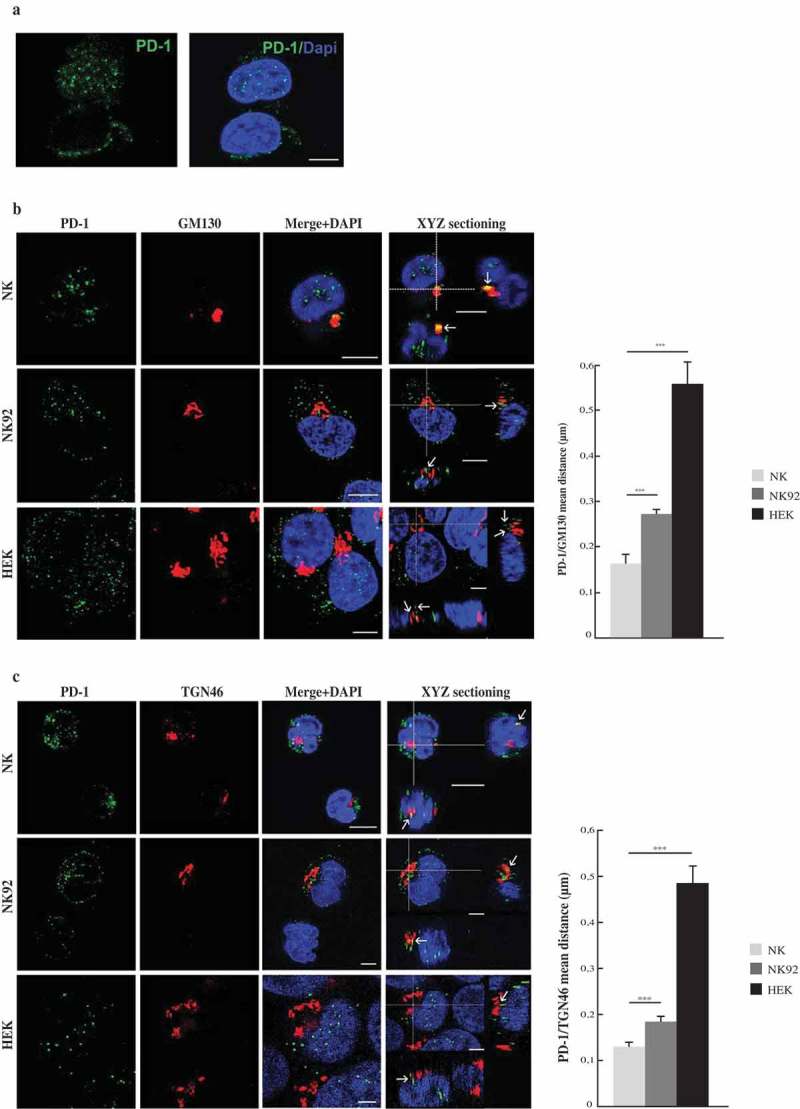
PD-1 cytoplasmic localization. (a) PD-1 immunofluorescence analyses in resting NK cells from different HDs. A representative image from the 14 HDs analyzed has been reported. (b,c) Left panels: confocal microscopy images of PD-1 association with the Cis- (GM130, panel B) and Trans-(TGN46 panel C) Golgi compartments. The analysis was performed on resting NK cells and on both the NK92 and HEK cell lines. Colocalization between PD-1 and Golgi markers is indicated by arrows. A representative image from three independent experiments has been reported. Right panels: quantification of the mean distance between PD-1 and Golgi markers. Values, from three independent experiments, are mean ± SEM. Scale bar 5 µm.
Our present data reveal that, despite the absent/low PD-1 surface expression, a cytoplasmic pool of PD-1 mRNA and protein exists in all resting NK cells. Similarly, PD-1 splicing variant and protein were also detected in IL-2-activated NK cells and in NK cells isolated from malignant PE from patients with primary or metastatic pleural tumors. Different PD-1 mRNA splicing isoforms were detected in all the different NK cell populations analyzed. Of particular interest, the splice variant lacking Exon3 encoding for a putative soluble form of PD-1. Different studies have been performed on a possible role of sPD-1 as an antitumor agent.23 Indeed in mice, delivery at the tumor site of plasmid encoding sPD-1, resulted in increased antitumor immunity.25 Moreover, clinical studies in cancer patients have investigated the presence of sPD-1 and its possible correlation with the overall patients survival.26,27 Our data, in line with published data on PBMC, demonstrate that sPD-1 is not released by resting or in vitro-activated NK cells. Importantly, sPD-1 was detected in malignant PE although its cell source remains to be elucidated. Remarkably, both PD-1 transcript and protein were consistently detectable in both CD56dim and CD56bright resting PB NK cell subsets, although at higher levels in CD56dim NK cells. Of note, intracytoplasmic flow cytometric analysis confirmed that in NK cell populations lacking PD-1 surface expression the presence of PD-1 could be detected in both CD56bright and CD56dim NK cells subsets (data not shown). This is in line with data in T cells in which, despite a strong reduction of PD-1 surface expression in CD4+ cells after exposure to CD28SA, the intracytoplasmic staining revealed high levels of intracellular PD-1.28 In addition, as shown also by confocal microscopy, all resting NK cells analyzed contained a cytoplasmic pool of PD-1, detectable also in proximity of the Golgi compartments. Since surface expression of PD-1 occurs primarily in certain pathological conditions (e.g. tumors/viral infections), it was important to assess whether de novo synthesis was required or rather a preformed cytoplasmic pool was available. Our data revealed the presence of cytoplasmic PD-1 and are compatible with a possible prompt surface expression of PD-1. In addition, the presence of PD-1 mRNA transcripts may allow a rapid protein synthesis. In view of the regulatory role of PD-1 in dampening over-reactive immune cell responses and in maintaining peripheral tolerance, it is conceivable that its expression may be finely tuned by mechanisms ensuring its rapid externalization. These mechanisms have been, in part, identified in T lymphocytes but not in NK cells. It is possible that signaling via NK receptor(s) and/or soluble and cellular factors produced in pathological conditions may lead to PD-1 externalization. Indeed, given the presence of PD-1 vesicles nearby the Golgi compartments, it is conceivable that specific signals are required to promote fusion of PD-1 vesicles within the cytoplasm to the plasma membrane. Preliminary studies in NK cells treated with different cytokines (including IL2, IL15, IL18, IL1β, IL-17, TGF β) or cocultured with some tumor cell lines did not result in PD-1 surface expression. Further studies, including epigenetic analyses, which have been found to correlate with PD-1 expression especially in pathological conditions,29,30 are clearly required to unveil the mechanisms involved in PD-1 surface expression in NK cells.
In conclusion, our study provides further insight into the PD-1 expression in NK cells. Indeed, we show that even resting NK cells are potentially capable of a prompt externalization of PD-1. This notion, together with the possible synthesis of other PD-1 isoforms, may be of relevance in view of the important role of NK cells in killing tumor cells, particularly those that have lost the surface expression of HLA-I molecules, thus escaping cytolytic T lymphocytes-mediated control. Thus, these data may offer clues for further investigations aimed at unveiling the molecular mechanisms that regulate PD-1 protein externalization in NK cells. These notions might allow the design of new therapeutic approaches to improve NK cell-mediated anti-tumor immune responses.
Materials and methods
Samples
This study included 14 buffy coats collected from volunteer blood donors admitted to the blood transfusion center of IRCCS Bambino Gesù Pediatric Hospital after obtaining informed consent. The study was approved by the ethical Committee of IRCCS Bambino Gesù Pediatric Hospital (825/2014). The study on Pleural Effusion was approved by Azienda Sanitaria Locale 3 (ASL, Genova, Italy) Ethics Board (ID 33533184, 29/10/2013).
Cell lines
Human embryonic kidney cells 293T (HEK 293T) were kindly provided by Prof. M.C. Mingari and maintained in Dulbecco’s Modified Eagle Medium (DMEM/F-12, with GlutaMAX) (Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS) from Gibco, 1X Penicillin/streptomycin (Euroclone, Pero, Italy) under 500 µg/ml Geneticin® Selective Antibiotic (G418, Gibco) selection. HEK cell line was initially used as a control for western blot analysis according to the antibody datasheet. NK92 cells were purchased from Leibniz Institute DSMZ–German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany) and cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Euroclone), supplemented with 10% FBS (Gibco), 1% Penicillin/streptomycin (Euroclone), 1% L-Glutamine (Euroclone) and 5 ng/ml of Interleukin-2 (IL-2) (Novartis, Basel, Switzerland).
Isolation of NK cells, flow cytometry analysis and cell sorting
PBMC were obtained from buffy coats after density gradient centrifugation over Ficoll Lympholyte®-H (Cederlane, Burlington, Canada). Highly purified (≥98%) NK cells were subsequently obtained by depletion of non-NK cells using Miltenyi NK cell isolation kit (Miltenyi Biotech, Bergisch Gladbach, Germany), according to the manufacturer’s instruction. To verify the purity of NK cell separation, cells were stained with CD56-Pe-Cy7 (Beckman Coulter, Brea, CA, USA) and CD3-ECD (Beckman Coulter) mAbs for 20 min at 4°C. NK cell samples were acquired using the Cytoflex S (Beckman Coulter) flow cytometer and analyzed with the CytExpert 2.1 software (Beckman Coulter). PD-1 surface staining was performed incubating PBMC with CD56-Pe-Cy7, CD3-ECD, CD19-ECD (Beckman Coulter), CD14-ECD (Beckman Coulter), PD-1 for 30 min at 4°C. The purified anti-PD-1 mAb (PD1.3.1.3 clone, IgG2b) was originally isolated at the Laboratoire Immunologie des Tumeurs, CRCM, Marseille-Luminy (France). IgG2b-PE (Southern Biotech Birmingham, USA) was used as a control. For cell sorting, isolated NK cells were stained with CD56- Pe-Cy7 and CD3-ECD. CD56dim and CD56bright cells were sorted to a purity of ≥98% with the MoFlo Astrios sorter (Beckman Coulter) using the Summit software (Beckman Coulter). The purity of cells separation was controlled by flow cytometry using CD56- Pe-Cy7 and CD3-ECD. After sorting, NK cells were centrifuged at 211 g, for 5 min at 4°C. Pleural Effusion (PE) cells were obtained by centrifugation at 400 g for 10 min. Isolation of NK cells from PE was performed using the Miltenyi NK cell isolation kit (Miltenyi Biotech), according to the manufacturer’s instruction. For protein isolation, NK cell pellets were stored at –80°C; for RNA isolation, cell pellets were resuspended in Trizol (Ambion, Invitrogen Corporation, Applera Norwalk, USA) and then stored at –80°C.
In vitro NK cells activation
To obtain polyclonal activated NK cells, highly purified NK cells obtained from HDs, were cultured with NK macs basal medium (Miltenyi Biotech) supplemented with the NK Macs supplement, 10% FBS (Gibco), 1% L-glutamine (Euroclone), 1% Penicillin/streptomycin (Euroclone), and 600 U/ml of IL-2 (Novartis). Every 3–4 days NK cells were expanded until the right exponential growth phase was reached to perform experiments.
RNA extraction
NK cells pellets were resuspended in Trizol (Ambion). After addition of Chloroform (Sigma-Aldrich, St. Louis, USA) samples were centrifuged at 12,000 g for 15 min at 4°C. The aqueous phase was removed and RNA was then isolated with the Qiagen RNeasy Universal kit (Qiagen, Hilden, Germany) according to manufacturer’s instruction. HEK and NK92 cell pellets were resuspended in Trizol (Applied) and RNA was isolated according to manufacturer’s instruction. After extraction, purity of mRNA was determined by A260/A280 (between 1.2 and 2.2) and A260/A230 (1.7). mRNA samples were then treated with DNase I Amplification Grade (Invitrogen, Carlsbad, CA, USA) to eliminate any genomic DNA contamination. cDNAs were synthetized by oligo dT using the Superspcrit® IV First-Strand Synthesis System (Invitrogen) according to the manufacturer’s instruction.
PCR for PD-1 isoforms
To detect PD-1 mRNA transcripts isoforms, cDNAs were amplified using the EmeralAMP® GT PCR Master Mix (Takara Bio Inc., Kusatsu, Japan). Amplification was carried out with the ProFlex PCR system (Applied Biosystems, Foster City, CA, USA) using the following cycling profile: an initial hold at 95°C for 30 sec, 40 cycles of 94°C for 1 min, 64°C for 1 min and 72°C for 1 min; final extension step of 72°C for 10 min. The following primers were used for the amplification: PD-1 all F: 5’-GCGGCCAGGATGGTTCTTA-3’; PD-1 all R 5’-TACTCCGTCTGCTCAGGGA-3’. PCR products were separated by electrophoresis on a 2% agarose gel. Images were collected using the Uvitec Mini HD9 transilluminator system (Uvitec Ltd. Cambridge).
Real-time PCR
cDNAs from NK cells were synthetized as described above. Quantitative Real-time PCR was performed with the QuantStudio 6 Flex PCR (Applied Biosystems) using the PowerUp SYBR Green Master mix (Applied Biosystems) according to manufacturer’s instruction. Relative quantification of mRNA was determined by the ΔΔCt method. PD-1 mRNA expression was normalized against ActinB expression. Primers for real time are as follows: PD-1-F 5’-CAGGGTGACAGAGAGAAGGG-3’; PD-1-R 5’-CCTGGCTCCTATTGTCCCTC-3’; ActB-F 5’-ACCGCGAGAAGATGACCCAGA-3’; ActB-R 5’-GGATAGCACAGCCTGGATAGCAA-3’
Protein extract and western blot analysis
NK, NK92 and HEK cells pellets were resuspended in RIPA buffer with 1X Halt Protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and incubated on ice for 20 min. After centrifugation for 15 min at 21.130 g at 4°C, supernatants were recovered. Protein concentration was measured with the BCA assay (Perkin Elmer, Waltham, Massachusetts, USA) according to manufacturer’s instruction. Protein extracts were fractionated by SDS-page gel electrophoresis and transferred to a PVDF membrane (Ge Healthcare, Little Chalfont, UK). After incubation in TBST with 5% nonfat dry milk (Cell Signaling Technology, Danvers, Massachusetts, USA) with gentle agitation for 60 min, the membrane was incubated with α-PD-1 1:1000 (Abnova, Taipei, Taiwan) (this mAb was selected based on its good capacity of staining the PD-1 intracytoplasmic form, tested in HEK cell line) or with α-β-Actin 1:10000 (Sigma-Aldrich). After washes, membrane was incubated with the secondary antibody: goat-α-mouse-HRP (Santa Cruz, Dallas, Texas, USA). Signals were developed with the ECL prime system (Ge Healthcare) according to manufacturer’s instruction and detected with the Uvitec Mini HD9 technology (Uvitec Ltd, Rugby, UK). Bands quantification was performed using the Ninealliance© software (Uvitec).
siRNA transfection
NK cells were transfected with Silencer® Negative Control siRNA (mock) (Thermo Fisher Scientific) or with the Silencer® PDCD1 siRNA (Assay ID 5525, Thermo Fisher Scientific) using the Neon electroporation system (Invitrogen), according to manufacturer’s instruction. After transfection NK cells were cultured in RPMI (Euroclone) 10% FBS (Gibco), 1% Penicillin/streptomycin (Euroclone), 1% L-Glutamine (Euroclone) and 600 U/ml of IL-2 (Novartis). NK cells were collected 72 hrs post-transfection, by centrifugation at 211 g at 4°C for 5 min. For subsequent protein isolation, NK cell pellets were immediately stored at −80°C. For RNA isolation, pellets were resuspended in Trizol (Ambion) prior storage at −80°C. Protein and RNA isolations were performed as described above.
sPD-1 ELISA
Soluble PD-1 (sPD-1) release was measured by enzyme-linked immunosorbent assay (ELISA) using the human PD-1 antibody duosets ELISA development kit DY1086 (R&D Systems, Inc. Minneapolis, USA) and the DuoSet ELISA Ancillary Reagent kit 2 DY008 (R&D). Plates were coated overnight with capture antibody (2 ug/ml). The following day, after washes (3 × 300 ul with Wash Buffer 1X) 100 ul of Reagent Diluent (5% BSA final concentration) were added to each well and plate was incubated for 1 hr at 700 rpm. After a washing step as before, 100 ul of sample or standards were added, plate was sealed and incubated for 2 hrs with gentle shaking. Calibration curve consisted of 1:2 dilutions of the standard material ranging from 2,5 ng/ml to 0,0097 ng/ml. The plate was subsequently washed and incubated with 100 ul of Detection Antibody (200 ng/ml) for 2 hrs with gentle shaking. After repeating the washing step, 100 ul of Streptavidin-HRP was added to each well. The plate was incubated in the dark for 40 min, washed and then treated with 100 ul of Substrate Solution for 20 min. After addition of 50 ul of Stop Solution, optical density was immediately measured using the Synergy H1 Reader (Biotek, Winooski, USA). Sample was read at 450 nm and 540 nm wavelengths and to correct optical imperfection reading at 540 nm was subtracted to reading at 450 nm. sPD-1 concentrations (pg/ml) were calculated using the four-point-fit calibration curve of standard dilutions.
Confocal microscopy
Freshly isolated purified blood NK cells were adhered to microscope slides (BD, Franklin Lakes, New Jersey, USA), previously coated with poly-L-lysine (Sigma) for 30 min at room temperature. Cells were fixed with cold Paraformaldehyde 4% (Sigma) for 5 min and then permeabilized with 0.1% Triton X-100 (Sigma Aldrich) for 8 min. After washes with PBS cells were blocked with PBS-BSA3% for 30 min. Cells were then incubated with α-PD-1 1:400 (Abnova) overnight at 4°C. After washes cells were incubated with Alexa Fluor 488 goat-α-mouse 1:800 (Molecular Probe, Eugene, Oregon, USA) for 1 hr at room temperature (RT). Cells were washed in PBS and then incubated with α-GM130 1:200 (Abcam, Cambridge, UK) or α-TGN46 1:200 (Abcam) for 1 hr at RT. Cells were then washed and subsequently incubated with Alexa Fluor 546 goat-α-rabbit 1:400 (Molecular Probe) 1 hr at RT. After washes, cells were mounted with Vectashield® Antifade mounting medium containing 1,5 µg/ml of DAPI (Vectorlab, Burlingame, CA, USA). Samples were stored at 4°C. Confocal microscopy was performed on a Leica TCS-SP8X laser-scanning confocal microscope (Leica Microsystems, Mannheim, Germany) equipped with tunable white light laser (WLL) source, 405 nm diode laser, 3 Internal Spectral Detector Channels (PMT) and 2 Internal Spectral Detector Channels (HyD) GaAsP. Sequential confocal images were acquired using an HCPLAPO 63x oil-immersion objective (1.40 numerical aperture, NA, Leica Microsystems) with a 1024 × 1024 format, scan speed 400 Hz, and z-step size of 0.3 µm. Z-reconstructions were imported into LAS X 3D software (Leica Microsystems) to obtain their 3D surface rendering. Acquisition settings (i.e. lasers’ power, beam splitters, filter settings, pinhole diameters and scan mode) were the same for all examined samples of each staining. The distances between PD-1 spots, closer to the Golgi network, and Golgi cisternae, were manually measured in the focal central plane of the Golgi compartment, using LAS X analysis software (Leica Microsystems) in five digital images randomly selected and acquired for each cell sample. Tables of images are processed using Adobe Photoshop CS4 software (Adobe Systems Inc).
Statistical analysis
Statistical analyses were performed using the GraphPad Prism 6.0 (La Jolla, CA, USA) software. Values were expressed as mean ± SEM. P values less than 0.05 were considered statistically significant. * P˂ 0.05, ** P˂ 0.01 *** P˂ 0.001.
Funding Statement
The present study has been supported by the following grants: Associazione Italiana per la Ricerca sul Cancro (AIRC) [IG 2014 Id. 15283] (LM), [IG 2017 Id. 19920] (LM), [IG 2017 Id. 20312] (AM), Special Project 5 × 1000 no.21147 (LM, AM), Ricerca Corrente Bambino Gesù Children's Hospital [201807P004255] (LM), Progetto Roche per la Ricerca 2017 (SP, EM), and Ministero della Salute [GR-2013-02356568] (PV).
Abbreviations
- mRNA
messanger RNA
- PD-1
Programmed Cell Death protein 1
Acknowledgments
We thank Daniel Olive for providing the anti-PD-1 mAb (PD1.3.1.3 clone). We thank Ezio Giorda and the Flow Cytometry core facility of IRCCS Bambino Gesù Children’s Hospital.
Disclosure of Potential Conflicts of Interest
Moretta A. was a founder and shareholder of Innate-Pharma (Marseille, France). The remaining authors declare that they and their immediate family members have no known conflicts of interest associated with this publication.
Contributions of the authors
Mariotti F.R. designed and performed research, interpreted data and wrote the article; Petrini S performed acquisition and analysis of confocal microscopy images; Ingegnere T. and Tumino N. performed research; Besi F. performed cell sorting; Scordamaglia F. selected clinical samples; Munari E., Pesce S. reviewed the manuscript; Marcenaro E., Moretta, A., Vacca P. and Moretta L. designed research and wrote the paper.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Moretta L, Pietra G, Vacca P, Pende D, Moretta F, Bertaina A, Mingari MC, Locatelli F, Moretta A.. Human NK cells: from surface receptors to clinical applications. Immunol Lett. 2016;178:15–19. doi: 10.1016/j.imlet.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S.. Innate or adaptive immunity: the example of natural killer cells. Science. 2011;331:44–349. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freud AG, Mundy-Bosse BL, Yu J, Caligiuri MA. The broad spectrum of human natural killer cell diversity. Immunity. 2017;47(5):820–833. doi: 10.1016/j.immuni.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper MA, Fehniger TA, Turner SC, Chen KS, Ghaheri BA, Ghayur T, Carson WE, Caligiuri MA. Human natural killer cells: a unique innate immunoregulatory role for the CD56 (bright) subset. Blood. 2001;97(10):3146–3351. doi: 10.1182/blood.V97.10.3146. [DOI] [PubMed] [Google Scholar]
- 5.Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, Biassoni R, Moretta L. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolisis. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 6.Morisaki T, Onishi H, Katano M. Cancer immunotherapy using NKG2D and DNAM-1 systems. Anticancer Res. 2012;32(6):2241–2247. [PubMed] [Google Scholar]
- 7.Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari MC, Moretta L. Receptors for HLA class-I molecule in human natural killer cells. Annu Rev Immunol. 1996;14:619–648. doi: 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- 8.Francisco LM, Sage PT, Sharpe AH. The PD-1 pathway in tolerance and autoimmunity. Immunol Rev. 2010;236:219–242. doi: 10.1111/j.1600-065X.2010.00923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishimura H, Honjo T. PD-1: an inhibitory immunoreceptor involved in peripheral tolerance. Trends Immunol. 2001;22(5):265–268. doi: 10.1016/S1471-4906(01)01888-9. [DOI] [PubMed] [Google Scholar]
- 10.Parry RV, Chemnitz JM, Frauwirth KA, Lanfranco AR, Braunstein I, Kobayashi SV, Linsley PS, Thompson CB, Riley JL. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol Cell Biol. 2005;25(21):9543–9953. doi: 10.1128/MCB.25.21.9543-9553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keir ME, Liang SC, Guleria I, Latchman YE, Qipo A, Albacker LA, Koulmanda M, Freeman GJ, Sayegh MH, Sharpe AH. Tissue expression of PD-L1 mediates peripheral T cell tolerance. J Exp Med. 2006;203(4):883–895. doi: 10.1084/jem.20051776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chinai JM, Janakiram M, Chen F, Chen W, Kaplan M, Zang X. New immunotherapies targeting the PD-1 pathway. Trends Pharmacol Sci. 2015;36(9):587–595. doi: 10.1016/j.tips.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dondero A, Pastorino F, Della Chiesa M, Corrias MV, Morandi F, Pistoia V, Olive D, Bellora F, Locatelli F, Castellano A, et al. PD-L1 expression in metastatic neuroblastoma as an additional mechanism for limiting immune surveillance. Oncoimmunology. 2016;5(1):e1064578. doi: 10.1080/2162402X.2015.1064578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pesce S, Greppi M, Tabellini G, Rampinelli F, Parolini S, Olive D, Moretta L, Moretta A, Marcenaro E. Identification of a subset of human natural killer cells expressing high levels of programmed death 1: a phenotypic and functional characterization. J Allergy Clin Immunol. 2017;139(1):335–346e3. doi: 10.1016/j.jaci.2016.04.025. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Cheng Y, Xu Y, Wang Z, Du X, Li C, Peng J, Gao L, Liang X, Ma C. Increased expression of programmed cell death protein 1 on NK cells inhibits NK-cell-mediated anti-tumor function and indicates poor prognosis in digestive cancers. Oncogene. 2017;36(44):6143–6153. doi: 10.1038/onc.2017.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beldi-Ferchiou A, Lambert M, Dogniaux S, Vely F, Vivier E, Olive D, Dupuy S, Levasseur F, Zucman D, Lebbe C, et al. PD-1 mediates functional exhaustion of activated NK cells in patients with Kaposi sarcoma. Oncotarget. 2016;7(45):72961–72977. doi: 10.18632/oncotarget.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwai Y, Hamanishi J, Chamoto K, Honjo T. Cancer immunotherapies targeting the PD-1 signaling pathway. J Biomed Sci. 2017;24(1):26. doi: 10.1186/s12929-017-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He J, Hu Y, Hu M, Li B. Development of PD-1/PD-L1 pathway in tumor immune microenvironment and treatment for non-small cell lung cancer. Sci Rep. 2015;5:13110. doi: 10.1038/srep13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pitt JM, Vetizou M, Daillere R, Roberti MP, Yamazaki T, Routy B, Lepage P, Boneca IG, Chamaillard M, Kroemer G, et al. Resistance mechanisms to immune-checkpoint blockade in cancer: tumor-intrinsic and -extrinsic factors. Immunity. 2016;44(6):1255–1269. doi: 10.1016/j.immuni.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen C, Ohm-Laursen L, Barington T, Husby S, Lillevang ST. Alternative splice variants of the human PD-1 gene. Cell Immunol. 2005;235(2):109–116. doi: 10.1016/j.cellimm.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Klingemann H, Boissel L, Toneguzzo F. Natural killer cells for immunotherapy – advantages of the NK-92 cell line over blood NK cells. Front Immunol. 2016;7. doi: 10.3389/fimmu.2016.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okazaki T, Honjo T. PD-1 and PD-1 ligands: from discovery to clinical application. Int Immunol. 2007;19(7):813–824. doi: 10.1093/intimm/dxm057. [DOI] [PubMed] [Google Scholar]
- 23.Zhu X, Lang J. Soluble PD-1 and PD-L1: predictive and prognostic significance in cancer. Oncotarget. 2017;8(57):97671–97682. doi: 10.18632/oncotarget.18311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pentcheva-Hoang T, Chen L, Pardoll DM, Allison JP. Programmed death-1 concentration at the immunological synapse is determined by ligand affinity and availability. Proc Natl Acad Sci U S A. 2007;104(45):17765–17770. doi: 10.1073/pnas.0708767104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elhag OA, Hu XJ, Wen-Ying Z, Li X, Yuan YZ, Deng LF, Liu DL, Liu YL, Hui G. Reconstructed adeno-associated virus with the extracellular domain of murine PD-1 induces antitumor immunity. Asian Pac J Cancer Prev. 2012;13(8):4031–4036. [DOI] [PubMed] [Google Scholar]
- 26.Kruger S, Legenstein ML, Rosgen V, Haas M, Modest DP, Westphalen CB, Ormanns S, Kirchner T, Heinemann V, Holdenrieder S, et al. Serum levels of soluble programmed death protein 1 (sPD-1) and soluble programmed death ligand 1 (sPD-L1) in advanced pancreatic cancer. Oncoimmunology. 2017;6(5):e1310358. doi: 10.1080/2162402X.2017.1310358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sorensen SF, Demuth C, Weber B, Sorensen BS, Meldgaard P. Increase in soluble PD-1 is associated with prolonged survival in patients with advanced EGFR-mutated non-small cell lung cancer treated with erlotinib. Lung Cancer. 2016;100:77–84. doi: 10.1016/j.lungcan.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Thaventhiran T, Alhumeed N, Yeang HX, Sethu S, Downey JS, Alghanem AF, Olayanju A, Smith EL, Cross MJ, Webb SD, et al. Failure to upregulate cell surface PD-1 is associated with dysregulated stimulation of T cells by TGN1412-like CD28 superagonist. mAbs. 2014;6(5):1290–1299. doi: 10.4161/mabs.29758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goltz D, Gevensleben H, Dietrich J, Ellinger J, Landsberg J, Kristiansen G, Dietrich D. Promoter methylation of the immune checkpoint receptor PD-1 (PDCD1) is an independent prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncoimmunology. 2016;5(10):e1221555. doi: 10.1080/2162402X.2016.1221555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rover LK, Gevensleben H, Dietrich J, Bootz F, Landsberg J, Goltz D, Dietrich D. PD-1 (PDCD1) Promoter methylation is a prognostic factor in patients with diffuse lower-grade gliomas harboring isocitrate dehydrogenase (IDH) mutations. EBioMedicine. 2018;2:97–104. doi: 10.1016/j.ebiom.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


