Abstract
Low temperature is one of the most important factors affecting plant growth, it causes an stress that directly alters the photosynthetic process and leads to photoinhibition when severe enough. In order to address the photosynthetic acclimation response of Lotus japonicus to cold stress, two ecotypes with contrasting tolerance (MG-1 and MG-20) were studied. Their chloroplast responses were addressed after 7 days under low temperature through different strategies. Proteomic analysis showed changes in photosynthetic and carbon metabolism proteins due to stress, but differentially between ecotypes. In the sensitive MG-1 ecotype acclimation seems to be related to energy dissipation in photosystems, while an increase in photosynthetic carbon assimilation as an electron sink, seems to be preponderant in the tolerant MG-20 ecotype. Chloroplast ROS generation was higher under low temperature conditions only in the MG-1 ecotype. These data are consistent with alterations in the thylakoid membranes in the sensitive ecotype. However, the accumulation of starch granules observed in the tolerant MG-20 ecotype indicates the maintenance of sugar metabolism under cold conditions. Altogether, our data suggest that different acclimation strategies and contrasting chloroplast redox imbalance could account for the differential cold stress response of both L. japonicus ecotypes.
Introduction
Suboptimal temperature is one of the most important abiotic factors limiting plant growth. Frost have a significant impact on agricultural production, depending the extent of damages on factors such as the inherent tolerance of the plant, the severity and duration of the stress, whether it is repeated and whether it occurs in combination with other stressors. When exposed to low but non-freezing temperatures, many plants initiate a cold acclimation process that improves their ability to withstand freezing temperatures1.
Cold acclimation can be defined as the modifications of anatomy, physiology and metabolism that occur in response to sub-optimum temperatures that minimize irreversible freeze-damage and improve plant fitness. This process implies massive changes in gene expression2,3, enzymatic activities4, proteomic changes5 and subcellular reprogramming of metabolism6.
Although the mechanisms behind cold acclimation are not fully understood, it is known that photosynthetic responses to cold but non-freezing temperature can be important. Photosystems, electron transport chain, as well as the Calvin-Benson cycle and other carbon metabolisms reactions, are involved in the production of carbohydrates during photosynthesis. This complex process is finely regulated through different signals, allowing its homeostasis under different environmental conditions. However, its deregulation reduces CO2 fixation capacity and induces photoinhibition7. In general terms, the response to photoinhibition involves different photoprotective mechanisms which avoid a redox imbalance into the electron transport chain, chloroplast ROS generation and protein photodamage8.
Under adverse environmental conditions that restrict the rate of photosynthesis, photon capture can exceed the rate at which the energy can be used, resulting in production of reactive oxygen species (ROS) and cell damage9,10. It has been shown that low temperatures are directly related to ROS increase11,12, and a positive correlation has been observed between detoxification capacity of these molecules and cold stress tolerance13,14. In addition, it has been suggested that an efficient electron sink has the potential to counterbalance the excess of light energy absorption15. In consequence, an increase in carbon assimilation could be strongly relevant in the photosynthetic acclimation capacity and in the avoidance of chloroplast photo-oxidative stress in plants16.
Lotus japonicus is commonly used as a model legume for plant research, specifically concerning plant responses to stress17,18. Recently, we have suggested that a cold-induced redox imbalance is generated in two L. japonicus ecotypes (MG-1 and MG-20), with contrasting photosynthetic adaptability to low temperatures19. The differential response of MG-1 and MG-20 to low temperature conditions has been previously described19. After 7 days of chilling, the tolerant ecotype (MG-20) exhibited a better photosynthetic performance than the sensitive one (MG-1) and higher stomatal conductance, suggesting chloroplast involvement in this proccess19. However, the degree of participation of these organelles in the cold stress response in legumes is poorly studied.
Global proteomic analysis of rice leaves subjected progressively to low temperature20 and chloroplast proteome analysis against drought stress and recovery in tomato21 lead to the conclusion that changes in chloroplast proteins occur in the response and acclimation to abiotic stress. Thus, in the present study we aimed to evaluate the role of chloroplast in the L. japonicus low temperature response, by studying chloroplast stress-responsive proteins under cold stress using a proteomic approach. In addition, confocal microscopy and flow cytometry have been used to identify and quantify chloroplastic ROS generation. We hypothesize that different chloroplast strategies are involved in plant tolerance to cold stress.
Results
Proteomic analysis of L. japonicus chloroplasts under low temperature
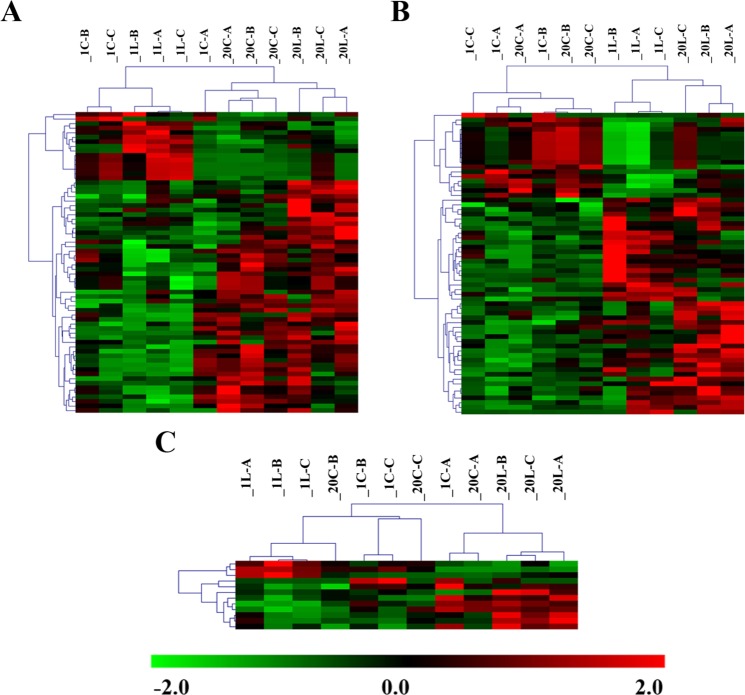
A discriminant analysis of the chloroplast proteome was carried out in the MG-1 and MG-20 L. japonicus ecotypes exposed to low temperature. Hierarchical analysis of the 724 identified proteins allowed us to group the differentially abundant protein in three clusters: 66 proteins differentially abundant only between ecotypes (Fig. 1A), 64 proteins differentially abundant due to low temperature (Fig. 1B) and 12 of them presenting interaction between ecotype and temperature factors (Fig. 1C).
Figure 1.
Hierarchical clusters of significant abundant proteins. The protein abundance is shown as a colour code, from green (lower abundance) to red (higher abundance). (A) Hierarchical cluster based on abundance differences between ecotypes (p < 0.05). (B) Hierarchical cluster based on abundance differences between control and low temperature treatment (p < 0.05). (C) Hierarchical cluster based on abundance differences of proteins with significative interaction between factors (p < 0.05). Three biological replicates are represented for each treatment, in each graph. 1C, MG-1 Control; 1L, MG-1 Low temperature; 20C, MG-20 Control; 20L, MG-20 Low temperature.
The STRING software (version 10.0) was used to estimate functional association networks between the different identified proteins22. Networks were generated according to data from gene co-occurrence, co-expression, curated databases, experimentally determined and textmining, and the differentially abundant proteins between ecotypes and treatments were analysed. When comparing ecotypes, MG-1 chloroplasts showed a greater abundance of proteins related with the “light-reactions” of photosynthesis (in particular, photophosphorylation and antenna proteins) than chloroplast of the MG-20 ecotype (Supplementary Fig. 1A). In contrast, proteins related with nitrogen and carbon metabolism, highlighting saccharose and starch metabolism were more abundant in the MG-20 ecotype. As well, other proteins related with the oxidative stress response, protein folding and translation were also over-represented (Supplementary Fig. 1B).
Proteins differentially abundant depending on temperature are clustered in Supplementary Fig. 2. Under control conditions, a low number of proteins were grouped into the glyoxylate pathway and carbon metabolism (Supplementary Fig. 2A). Nevertheless, a greater functional association network was observed as consequence of low temperature. Proteins related with secondary metabolism, photosynthetic processes, photo-phosphorylation, oxidative stress responses as well as translation, protein folding and degradation were over-represented in these networks (Supplementary Fig. 2B). All proteins used to estimate functional association networks are detailed in Figs 2–4. No functional associations were observed for proteins with significant interactions between ecotypes and the stress treatment (Fig. 1C, Supplementary Table 1).
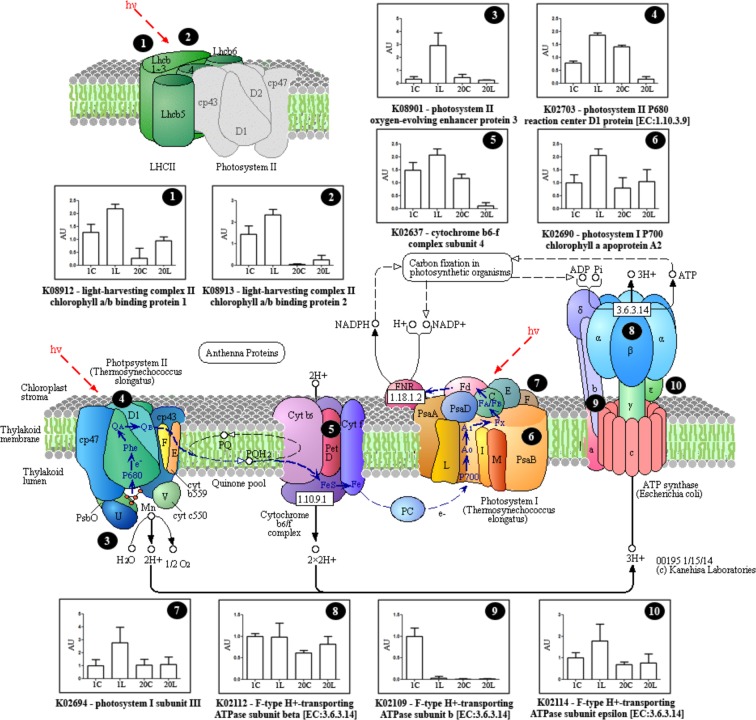
Figure 2.
Differentially abundant proteins related with photosystems and the electron transport chain. Representative scheme showing the different identified proteins and their abundance. Values are the mean ± SD of three biological replicates, and are expressed as arbitrary units (AUs) relative to the MG-1 control treatment. 1C, MG-1 Control; 1L, MG-1 Low temperature; 20C, MG-20 Control; 20L, MG-20 Low temperature. Adapted scheme from the KEGG database (http://www.genome.jp/kegg/pathway/map/map00195.html)63.
Figure 4.

Functional assignation of the differentially abundant proteins in L. japonicus chloroplasts under low temperature. The classification and functional assignation of the significant abundant proteins (p < 0.05) was done using KEGG, LegumeIP and Uniprot databases. Values are the mean ± SD of three biological replicates, and are expressed as arbitrary units (AUs) relative to the MG-1 control treatment. 1C, MG-1 Control; 1L, MG-1 Low temperature; 20C, MG-20 Control; 20L, MG-20 Low temperature.
Differential changes in proteins belonging to photosystems and the electron transport chain
Analysis of the chloroplast proteome showed that the abundance of some proteins related to photosystems and the electron transport chain was different between the MG-1 and the MG-20 ecotypes, what could be related to their response to cold stress. Firstly, the photosystem II (PSII) light-harvesting complex II chlorophyll a/b binding proteins 1 and 2 (Lhcb1 and Lhcb2), increased their abundance as consequence of cold stress in both ecotypes (Fig. 2). However, the levels of these proteins were higher, even under control conditions, in the sensitive MG-1 ecotype (Fig. 2). A contrasting effect was observed in the stress response of the photosystem II (PSII) components, oxygen-evolving protein 3 (OEE3) and D1 protein, and the cytochrome b6f complex subunit 4. While their levels were increased in MG-1 plants as a consequence of cold exposure, they were reduced in the MG-20 tolerant ecotype (Fig. 2). The identified photosystem I (PSI) proteins, P700 chlorophyll a apoprotein A2 and subunit III, raised their abundance as consequence of cold stress in a similar way to the PSII Lhcb proteins, although the abundance of these proteins was almost not affected by stress in the tolerant ecotype (Fig. 2).
Meanwhile, three subunits of the chloroplast ATP synthase [EC:3.6.3.14] (β, ε and b) were also identified (Fig. 2). Protein levels of the subunit β were increased under low temperature in MG-20, while no differences were observed between treatments in MG-1 plants. On the contrary, higher levels of subunit ε were registered under stress in MG-1, but no differences were observed in MG-20 between treatments. Regarding the b subunit, their protein abundance was only reduced in the sensitive MG-1 ecotype under low temperature. No differences between treatments were observed in MG-20, although in both cases its protein levels were lower than those in MG-1.
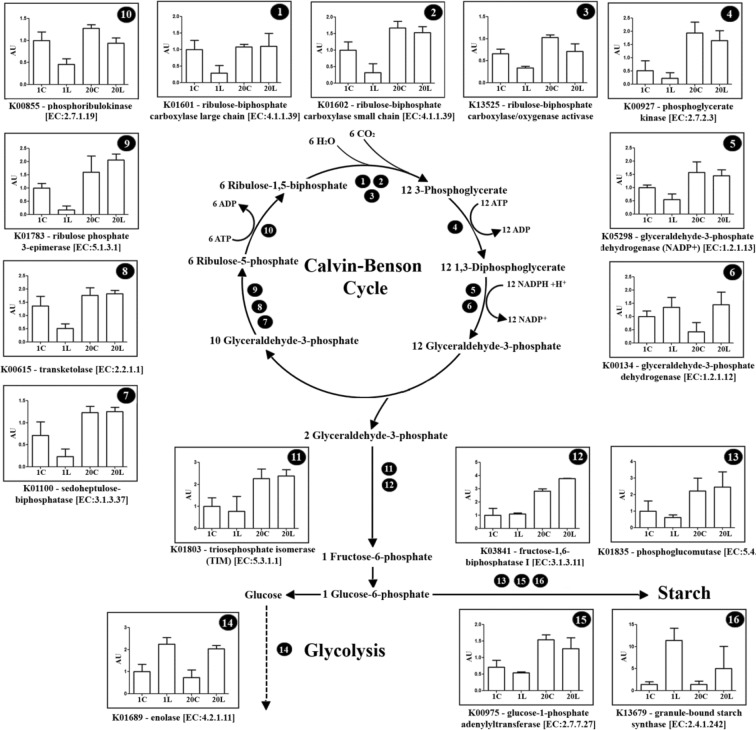
Differentially abundant proteins related with Calvin-Benson cycle, gluconeogenesis and starch biosynthesis
In general, proteins involved in Calvin-Benson cycle were more abundant in the tolerant ecotype MG-20 (Fig. 3). However, as a consequence of low temperature, the level of some of those enzymes was reduced, especially in the MG-1 sensitive ecotype (Fig. 3). Differences to cold response were observed between ecotypes for the following proteins: the large and small subunit of ribulose-1,5-bisphosphate decarboxylase (Rubisco) [EC:4.1.1.39], Rubisco activase, phosphoglycerate kinase [EC:2.7.2.3] and a NADP+ dependent glyceraldehyde-3-phosphate dehydrogenase [EC:1.2.1.13] (Fig. 3). In addition, a reduction in the levels of sedoheptulose 1,7-bisphosphatase [EC:3.1.3.37], transketolase [EC:2.2.1.1] and ribulose-phosphate 3-epimerase [EC:5.1.3.1] were observed under stress in MG-1 (Fig. 3), while no differences were observed between treatments in the MG-20 line. Similar results were obtained for the phosphoribulokinase enzyme [EC:2.7.1.19], although in this case its levels were also reduced in the tolerant ecotype under low temperature (Fig. 3). A NAD+ dependent glyceraldehyde-3-phosphate dehydrogenase isoform [EC:1.2.1.12] was also identified, but this protein increased its abundance under low temperature in both ecotypes.
Figure 3.
Differentially abundant proteins related with the Calvin-Benson cycle and the carbon metabolism. Representative scheme showing the different identified proteins and their abundance. Values are the mean ± SD of three biological replicates, and are expressed as arbitrary units (AUs) relative to the MG-1 control treatment. 1C, MG-1 Control; 1L, MG-1 Low temperature; 20C, MG-20 Control; 20L, MG-20 Low temperature.
Regarding gluconeogenesis proteins, chloroplast isoforms of the triosephosphate isomerase [EC:5.3.1.1] and the fructose-1,6-bisphosphatase were also identified [EC:3.1.3.11] (Fig. 3). The abundance of these proteins was higher in the MG-20 tolerant ecotype, being the fructose-1,6-bisphosphatase levels also increased under stress only in this line. Simultaneously, the levels of enolase [EC:4.2.1.11] increased under low temperature in both ecotypes (Fig. 3).
The abundance of at least three proteins involved in the starch biosynthetic pathway, phosphoglucomutase (PGM, [EC:5.4.2.2]), glucose-1-phosphate adenylyltransferase (AGPase, [EC:2.7.7.27]) and granule-bound starch synthase (GBSS, [EC:2.4.1.242]) was affected by cold (Fig. 3). The PGM and AGPase proteins were more abundant in MG-20 ecotype under both conditions. However, GBSS levels were higher in the sensitive ecotype, only under stress (Fig. 3).
Identification of other proteins participating in the low temperature response
Beyond proteins related to photosynthesis and sugar metabolism, proteomic analysis allowed us to identify a number of proteins whose abundance also differed among ecotypes as a consequence of cold stress (Fig. 4).
The tolerant MG-20 ecotype showed higher levels of proteins involved in antioxidant responses and ROS scavenging, such as superoxide dismutase [EC:1.15.1.1], ascorbate peroxidase [EC:1.11.1.11], peroxidase enzymes [EC:1. 11.1.17], protein thioredoxin 1 and peroxiredoxin [EC:1.11.1.15], and NADH dehydrogenase [EC:1.6.5.3]. In general, the abundance of these proteins was higher in the tolerant ecotype in control and stress conditions, when compared to MG-1 (Fig. 4). On the contrary, thioredoxin reductase [EC:1.8.1.9] was the only protein involved in ROS response which levels increased in the MG-1 ecotype under stress, while no differences were observed in MG-20 between treatments (Fig. 4).
Four heat shock proteins (Hsps) and five chaperones were also identified in the proteomic analysis (Fig. 4). Except for the Hsp 70 kDa and 90 kDa, all other proteins showed an increase in their levels under stress, compared to their abundance in the control condition, in both ecotypes. Similar results were observed for translation related proteins. Elongation factor 1-α (Lj1g3v3690420.1) and the predicted 40S ribosomal protein (Lj3g3v0937870.1) were more abundant in the tolerant ecotype in comparison with MG-1 (Fig. 4). Other proteins as the elongation factor EF-Tu (Lj3g3v3032110.2) and the large ribosomal subunit L9 (Lj3g3v3032110.2), showed no increment in their abundance under low temperature conditions. However, a higher protein level was found in the MG-20 ecotype under this condition (Fig. 4).
Furthermore, the levels of protochlorophyllide reductase [EC:1.3.1.33] and S-adenosylmethionine synthetase [EC:2.5.1.6] were increased under low temperature in both ecotypes, but to a higher extent in MG-20 than in MG-1 (Fig. 4). Similar results were observed for the pyruvate dehydrogenase E1 (β subunit) [EC:1.2.4.1], the dihydrolipoamide dehydrogenase [EC:1.8.1.4] and the myo-inositol-1-phosphate synthase [EC:5.5.1.4] (Fig. 4). On the contrary, a decreased in the abundance of some proteins related with the glyoxylate metabolism was observed under stress condition. In particular, this effect was evidenced for the glutamine synthetase [EC:6.3.1.2] and the glycerate dehydrogenase/hydroxypyruvate reductase [EC:1.1.1.29] (Fig. 4). In the latter case, the reduction was more pronounced in the MG-1 ecotype.
Regarding the nitrogen metabolism, in addition to the glutamine synthetase, the glutamate synthase [EC:1.4.1.7] protein levels were also reduced under low temperature, but only in the sensitive ecotype (Fig. 4). Moreover, no differences in glutamyl-tRNA (Gln) amidotransferase [EC:6.3.5.6] protein levels were observed between treatments in MG-1, while an increased was evidenced in the tolerant ecotype under stress (Fig. 4).
Ultrastructural modifications of L. japonicus chloroplasts under low temperature
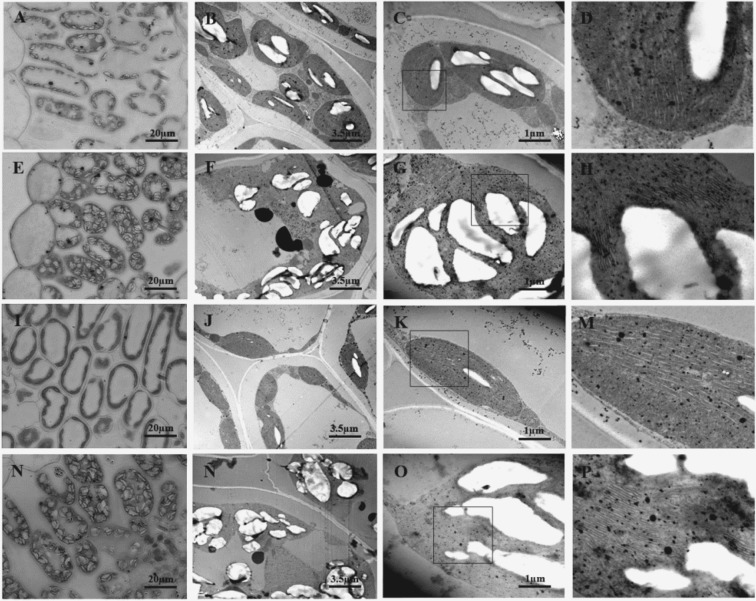
Chloroplasts of MG-1 and MG-20 ecotypes under control and low temperature conditions were observed by transmission electron microscopy. Under cold stress, the chloroplast area and the amount of starch granules increased in both ecotypes, being this increase higher in the MG-20 tolerant line (Fig. 5 and Table 1). Moreover, a higher number of electron-dense particles were registered under low temperature in the chloroplasts of both ecotypes (Fig. 5D,H,M,P). However, their density was higher in the MG-1 ecotype under stress (Fig. 5C,D,G,H,K–M,O,P).
Figure 5.
Micrographs of L. japonicus leaves under 7 d of low temperature and control treatments. Representative images obtained by optical and transmission electron microscopes. (A–D) MG-1 Control. (E–H) MG-1 Low temperature. (I–K,M) MG-20 Control. (N,Ñ,O,P) MG-20 Low temperature. The squares in images (C,G,K,O) delimit the amplified area in (D,H,M,P).
Table 1.
Ultrastructural parameters of L. japonicus chloroplasts.
| Ultrastructural Parameters | MG-1 | MG-20 | ||
|---|---|---|---|---|
| C | L | C | L | |
| Chloroplasts area (µm2) | 9.9 ± 3.5 | 27.2 ± 6.6* | 10.6 ± 3.2 | 38.3 ± 3.6* |
| N° Starch granules | 1.8 ± 1.0 | 5.2 ± 1.3* | 0.8 ± 1.0 | 6.4 ± 2.1* |
| Starch granules area (µm2) | 1.0 ± 0.6 | 2.0 ± 1.1 | 0.4 ± 0.3 | 3.1 ± 1.4* |
Values are the mean ± SD of three biological replicates, for each treatment. The asterisk indicates differences between control and low temperature treatments (Student’s test, p < 0.05), for each ecotype respectively. C, Control; L, Low temperature.
In addition, it was no possible to distinguish chloroplast grana under control conditions, although alterations in the thylakoid membranes were observed in response to stress in both ecotypes. These disruptions were more evident in the MG-1 ecotype (Fig. 5D,H,M,P). Furthermore, a reduction in the vacuole area of the cells was also observed as a consequence of stress (Fig. 5A,E,I,N).
Determination of chloroplast ROS production under low temperature
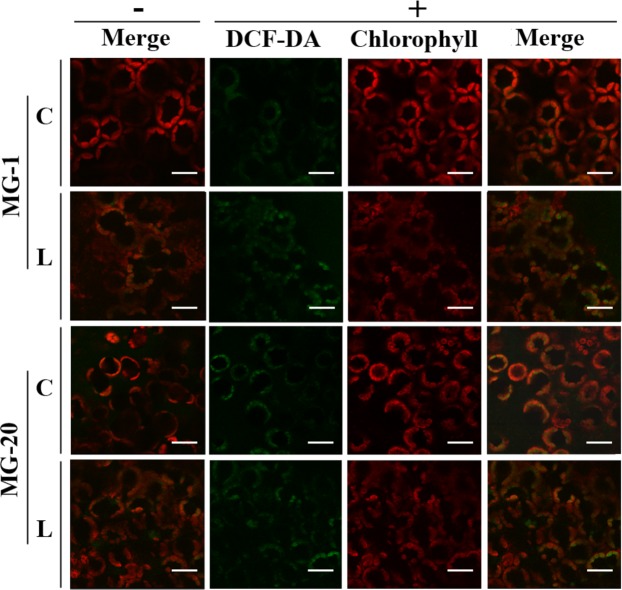
Co-localization of the DCF-DA fluorescence (ROS marker, green) and chlorophyll auto-fluorescence (chloroplasts marker, red) was observed in both ecotypes by confocal microscopy (Fig. 6). No differences due to low temperature were observed in the green fluorescence intensity in MG-20. However, a higher green fluorescence was registered under stress in MG-1 (Fig. 6). When comparing both ecotypes, the tolerant MG-20 ecotype exhibited higher levels of ROS under control conditions than MG-1 plants (Fig. 6). Meanwhile, chlorophyll auto-fluorescence decreased in both ecotypes after cold exposure (Fig. 6). In the absence of DCF-DA, no green fluorescence was registered in L. japonicus leaves observed under the confocal microscope (Supplementary Fig. 3).
Figure 6.
ROS subcellular localization in L. japonicus leaves. Representative images from confocal laser microscopy showing subcellular localization of the DCF-DA fluorescence (green) and chlorophyll autofluorescence (red) in MG-1 and MG-20 ecotypes. C, Control; L, Low temperature; −, DCF-DA incubation without light; +, DCF-DA incubation with light. Scale bars = 30 µm.
In order to quantify the differences observed by confocal microscopy, chloroplast ROS production was determined by flow cytometry in isolated cells. Two cytometric populations (a and b), obtained from L. japonicus leaves, were identified based on differences in their chlorophyll autofluorescence (FL3) and their scattered light (SSC) (Supplementary Fig. 4). Epifluorescence microscopy allowed the identification of chloroplasts and nucleus in a, demonstrating that was composed mainly by whole cells (Supplementary Fig. 4C,D). Due to its cell integrity, flow cytometry was performed on the a population.
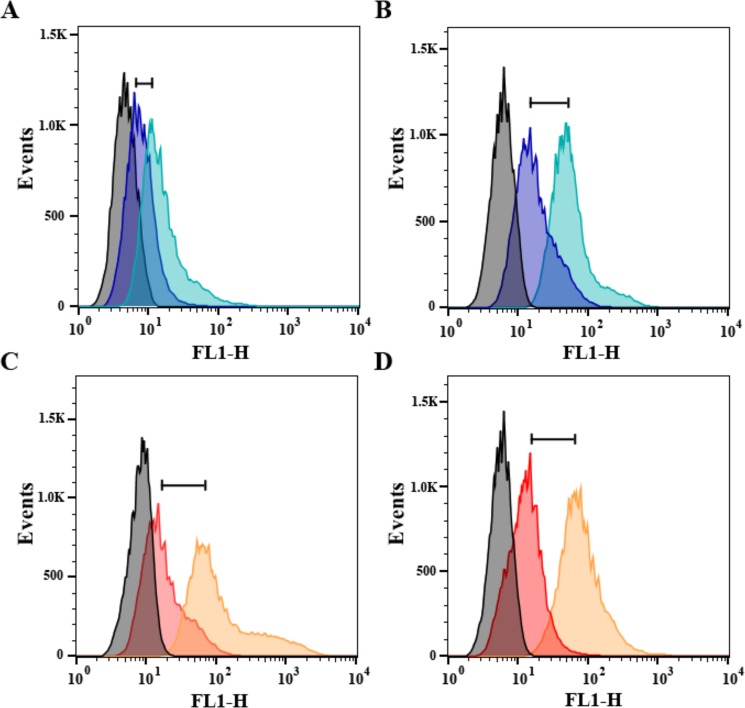
All cells incubated in the absence of DCF-DA showed the same FL1 relative fluorescence (DCF-DA fluorescence) (Fig. 7, and Supplementary Fig. 4). When cells were incubated with DCF-DA but without light exposure, a similar increase in green fluorescence was observed in all populations (Fig. 7, Supplementary Fig. 4). However, treatments with DCF-DA and light exposure showed a higher FL1 fluorescence intensity in cells exposed to cold only in the MG-1 sensitive ecotype (Fig. 7 and Supplementary Fig. 4). In this sense, relative differences between FL1 mode values suggest an increment of almost two-fold in MG-1 ROS levels under stress, compared to its control conditions (Fig. 8). Meanwhile, as observed by confocal microscopy, no significant increment in ROS levels was detected after low temperature treatment in the MG-20 tolerant ecotype. Nevertheless, ROS levels under control conditions were higher in the tolerant ecotype (Figs 7 and 8).
Figure 7.
Representative histograms showing FL1 fluorescence intensity of cells population of L. japonicus leaves under low temperature. (A,B) Histograms of MG-1 cells under control and low temperature treatment, respectively. Blue histogram: incubation with DCF-DA without light. Light blue histogram: incubation with DCF-DA and light. (C,D) Histograms of MG-20 cells under control and low temperature treatment, respectively. Red histogram: incubation with DCF-DA without light. Orange histogram: incubation with DCF-DA and light. Grey histograms: incubation without DCF-DA or light. A total of 40.000 events were analyzed in each case.
Figure 8.
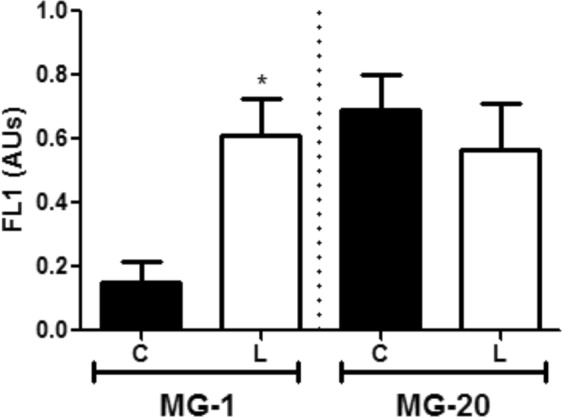
ROS production quantification by flow cytometry. The DFC-DA fluorescence intensity (FL1) was calculated in each case using the formula in Materials and Methods. Values are the mean ± SD of 5 biological replicates and are expressed as arbitrary units (AUs), for each ecotype and treatment. The asterisk indicates differences between control and low temperature treatments (Student’s test, p < 0.05), for each ecotype respectively. C, Control; L, Low temperature.
Discussion
Plants response to different environmental stimuli involves complex transcriptional, protein and metabolic changes23. In particular, chloroplasts participate in stress sensing and signalling through changes in their redox state and their direct implication in photosynthesis and carbon metabolism24. Increasing evidence supports a chloroplastic role in the response and acclimation to abiotic stress in plants25. Moreover, transcriptomic changes produced by cold stress in L. japonicus also suggest chloroplast involvement in the response to low temperature in this species3. In this work, we provide novel insights into the cold stress response in L. japonicus by analysing changes in chloroplastic proteins abundance, which are associated with ROS production and photoinhibition.
Proteomic changes in L. japonicus chloroplast in response to low temperature
We have compared the behaviour of two ecotypes with differential tolerance to low temperature: MG-1 and MG-20. We identified 66 proteins differentially abundant between them (p < 0.05), regardless of the temperature treatment to which they were subjected (Fig. 1A). These differences could be explained due to the genetic variability of the ecotypes26. In addition, approximately 10% of the total identified proteins showed significant changes in their abundance as consequence of low temperature (p < 0.05) (Fig. 1B). Functional assignment and classification of all the differentially abundant proteins allowed the identification of metabolic pathways over-represented in each ecotype (Supplementary Fig. 1), and altered by cold stress (Supplementary Fig. 2). Our results suggest that differences in the chloroplast proteome between MG-1 and MG-20 plants could be related with their differential tolerance to low temperature. Characterization of chloroplast proteome in response to drought stress in tomato21 and in salt-stressed mangrove27 lead to the identification of a number of proteins involved in abiotic stress responses that have been also identified in L. japonicus (Supplementary Figs 1 and 2). Altogether, these results suggest that chloroplast is involved in plant responses to cold stress.
Energy dissipation in the thylakoid membranes
An increase in Lhcb1 and Lhcb2 (LHCII proteins) levels was observed mainly in chloroplasts of the MG-1 plants in response to low temperature (Fig. 2). Beyond solar light absorption, the LHCII complex has also a key role in the regulation, distribution and dissipation of excessive excitation energy under photoinhibitory conditions, avoiding photodamage28. In particular, Lchb1 and Lhcb2 proteins play an important regulatory role in the photoprotective mechanism called Non-Photochemical Quenching (NPQ)29. Other components of the LHCII complex (Lhcb4 and Lhcb6) have also been described as differentially expressed in mangrove chloroplast as a consequence of salt stress27.
In addition, an increase in chloroplast ROS production, ELIP1 protein and the thylakoidal luminal 19 kDa protein levels were observed in MG-1 plants under the low temperature treatment (Figs 2 and 8). ELIP1, a member of the multigenic family of the light harvesting complex, is accumulated under high light and has been related with photoprotection under photo-oxidative conditions30. Meanwhile, although the thylakoid luminal protein 19 kDa has an unknown function, these proteins have also been linked to oxidative stress response in chloroplasts31. Together, these data supports that a cold-induced redox imbalance is generated in L. japonicus chloroplasts, and mainly in the sensitive MG-1 ecotype.
Furthermore, two PSII components were identified in our proteomic study: D1 reaction centre protein and OEE3 (PsbQ, OEC subunit) (Fig. 2). In particular, the D1 protein has been widely characterized as the main PSII photodamage target during photoinhibition32. OEE1 and OEC proteins were also induced in chloroplasts of mangrove under salt stress27 and maize33 and tomato21 under drought stress. Stress conditions produce an excess of excitation energy that damages D1 protein, initiating the PSII damage and repair cycle8. In this process, different chloroplast proteases are involved. Two of them, FtsH and Deg8, increased their levels under low temperature in L. japonicus (Fig. 4). Both proteins participate cooperatively in damaged D1 degradation8, suggesting activation of the PSII damage and repair cycle. These results are consistent with the photoinhibition previously reported under cold stress in both ecotypes19.
The OEE3 subunit has been related with PSII assembly34 and with the PsbO protein, which maintains the manganese cluster stability required for water oxidation35. While OEE3 protein levels were increased under low temperature in the sensitive ecotype MG-1, a reduction was observed in MG-20 plants under the same conditions (Fig. 2). Although these proteins seems to be relevant for PSII function and D1 protein repair cycle, it has been demonstrated that reduced levels of PsbO protein increase abiotic stress tolerance and reduce photoinhibition in transgenic potato plants36. Notwithstanding the photochemical mechanisms related with this tolerance increase are unknown, these results are consistent with the better acclimation of the MG-20 ecotype under cold stress.
Meanwhile, an increment in one subunit of the cytochrome complex b6f and two components of PSI was only observed in the sensitive ecotype under stress (Fig. 2). Both cytochrome b6f and PSI are strongly related to cyclic electron transport (CET)37, which participates in cold acclimation and is induced by an excess of photosynthetic electrons not utilized in carbon assimilation38. This data suggest that CET could be part of the cold-stress responses in Lotus.
Carbon assimilation and starch biosynthesis under cold stress
Higher protein levels of Calvin-Benson cycle and sugar metabolism enzymes were observed in the tolerant ecotype MG-20 compared to MG-1, in response to cold stress (Fig. 3). Accumulation of proteins from these metabolic pathways (in particular Rubisco) has been related to cold stress tolerance in different plant species39,40 and directly with chloroplast responses to abiotic stress21,33. This effect could be explained by a higher electron flux through Calvin-Benson cycle, which could reduce excitation pressure on the electron transport chain components, avoiding photoinhibition16.
We have observed higher levels of sedoheptulose-1,7-bisphosphatase (SBPase) and fructose-1,6-bisphosphatase in the tolerant MG-20 ecotype under cold stress. It has been recently demonstrated the relevance of SBPase and fructose-1,6-bisphosphatase in the abiotic stress response in plants33,41,42. In particular, over-expression of SBPase has been linked to a reduction of the chilling-induce oxidative stress in transgenic tomato lines42. Similar results have been also observed in tobacco plants over-expressing fructose-1,6-bisphosphatase and exposed to saline stress41. Meanwhile, a higher accumulation and activity of the fructose-biphosphate aldolase was linked with cold acclimation in Lolium multiflorum/Festuca arundinacea introgression forms40. Although this enzyme was not identified in our proteomic study, it has been also pointed out the relevance of adjusting photosynthetic carbon assimilation to deal with low temperature stress in the mentioned forage grasses40. Altogether, these data lead to the conclusion that similar cold stress responses could be present in a wide range of species.
Regarding starch metabolism, three enzymes were identified: PGM, AGPase, and GBSS (Fig. 3). The reaction catalyzed by AGPase has been considered as a key step in starch biosynthesis43, although the PGM chloroplast isoform could also be relevant in this process44. Both enzymes showed higher protein levels in the tolerant ecotype (Fig. 3). In addition, an increase in GBSS protein levels (involved in amylose biosynthesis) was observed under cold stress in both ecotypes, suggesting starch composition changes in L. japonicus chloroplasts in response to stress. In fact, an increase in starch granules content was observed in both L. japonicus chloroplasts under the low temperature treatment, being this increment higher in the MG-20 ecotype (Fig. 5 and Table 1).
Differential proteins involved in protein biosynthesis and folding
Complementary to photosynthesis and sugar metabolism, we found a number of proteins responding to cold stress in both ecotypes. Firstly, the DnaK and GroEL chaperones increased their levels in response to low temperature, and to a greater extent in the MG-20 ecotype compared to MG-1 (Fig. 4). These molecules have been related with abiotic stress tolerance in plants21,45. In particular, chloroplast chaperonins have been implicated in Rubisco biosynthesis46, what could suggest their involvement in the photosynthetic acclimation response to stress.
Among the proteins identified, we also found the 1-alpha and Tu elongation factors, which are components of the translation machinery in plants. Both factors have been related with abiotic stress responses, being the last particularly associated with PSII repair under cold stress47,48. Protein levels of both elongation factors were higher in MG-20 under low temperature (Fig. 4). This data could also support the better photosynthetic acclimation response previously reported in the tolerant ecotype19.
ROS as a part of acclimation response to low temperature
Chloroplast ROS generation inhibits protein synthesis49 and could induce or increase the photoinhibitory effect under stress conditions50. Cold stress produces ROS generation and chloroplast redox alterations. The increase in ROS production is associated with a lower plant tolerance to restrictive conditions13,14. Thus, an increase in transcript and protein levels, as well as in the activity of different antioxidant enzymes, have been widely described as part of the cold stress response in plants51, and directly related with chloroplast ROS detoxification in different species13,14. As a consequence, higher protein abundance of different antioxidant enzymes in MG-20 chloroplasts, compared to MG-1, could explain its better acclimation to low temperature stress (Fig. 4). A higher cellular antioxidant capacity has been also related to cold stress tolerance in L. multiflorum/F. arundinacea species40.
Consistently, an increase in chloroplast ROS levels was observed under low temperature conditions in the MG-1 ecotype, while no changes were detected in MG-20 between treatments (Figs 6 and 8). In this sense, we observed alterations in the thylakoid membranes under cold stress (Fig. 5H,P) that could be related with chloroplast redox imbalance52. Disruption of thylakoid membranes was more evident in MG-1, which is consistent with the higher chloroplast ROS accumulation in this ecotype (Figs 6 and 8).
Moreover, the reduction in thioredoxin (Trx), peroxiredoxin and thioredoxin reductase (TrxR) protein levels, observed in both ecotypes under low temperature (Fig. 4), suggests that the Trx-TrxR system could be implicated in the L. japonicus chloroplast response to cold stress. The Trx-TrxR system participates in ROS detoxification, protein redox regulation, as well as in different signalling mechanisms53. In agreement with our results, down-regulation of thioredoxin gene expression by cold stress has been previously reported in plants54. Nevertheless, the fact that Trx-TrxR protein levels were higher in the tolerant MG-20 ecotype, could suggest a higher ROS detoxification capacity in the tolerant ecotype than in the sensitive one.
It is worth mentioning that higher ROS levels were observed in the tolerant ecotype, under control conditions (Figs 6 and 8). Although their cellular toxicity, these molecules have also been widely related with signalling mechanisms in plants54. As a consequence, the observed basal differences in ROS levels could suggest a differential regulation of the redox state and signalling, in response to stress, between ecotypes55. In addition, a regulatory role has been described for the Calvin-Benson cycle and carbon metabolism56,57. These results could imply a tighter regulation of the cold stress response in MG-20 than in MG-1, which would be consistent with its higher tolerance to this constrain condition19.
Concluding remarks
Differences observed in the photosynthetic acclimation process of the L. japonicus MG-1 and MG-20 ecotypes could be explained, at least partially, by a differential chloroplast response. Higher abundance of proteins related with the antioxidant system, heat shock proteins and translation, suggest a better antioxidant response and lower cold-induced protein damage in the MG-20 ecotype. On the other hand, protein changes in the photosynthetic pathways as well as in carbon metabolism, suggest that two different acclimation strategies could be taken place in the chloroplast of the L. japonicus ecotypes studied. While in the MG-1 ecotype these mechanisms could involve mainly energy dissipation processes (such as NPQ and CET), the tolerant ecotype MG-20 would regulate carbon assimilation to cope with photoinhibition. The higher starch accumulation observed in the MG-20 chloroplasts, compared to MG-1, is consistent with these different acclimation strategies. In consequence, only the tolerant ecotype could efficiently balance the absorption and consumption of energy. This would explain the higher ROS levels observed in the MG-1 ecotype and its higher photoinhibition under these conditions. In conclusion, our results emphasize the relevance of the Calvin-Benson cycle and carbon metabolism in the photosynthetic acclimation process of L. japonicus under low temperature.
Materials and Methods
Plant materials, growth conditions and treatments
Seeds of L. japonicus MG-20 and MG-1 ecotypes were incubated for 7 d in a growth chamber with a 16/8 h day/night cycle at 24/21 ± 2 °C and 55/65 ± 5% relative humidity. Light (at an intensity of 250 µmol m−2 s−1) was provided by Grolux F 40 W fluorescent lamps. Seedlings were transferred to 110 mL volume pots containing sterilized sand-perlite (2:1), irrigated with 0.5X Hoagland’s nutrient solution58, and cultivated under the same temperature, relative humidity, and light conditions already described. In order to stress both ecotypes at the same developmental stage, plants with 4–6 fully developed leaves were used for all experiments.
The experimental design was completely randomized, with three biological repetitions per treatment. For both, the control and low temperature treatments, the 3-week old seedlings were placed in a Percival E-30B (Percival Scientific, Perry, IA, USA) growth chamber under a 16/8 photoperiod (day/night). Low temperature treatment was performed at 9/5 °C for 7 d, while control was performed at 24/21 ± 2 °C. Illumination was provided by Philips T8 Standard lamps (F17T8/TL841/ALTO 30PK), with an intensity of 250 µmol m−2 s−1. All the experimental determinations were done at 7 d of control and low temperature treatments, because it was the time when the higher photosynthetically acclimation differences were observed between ecotypes19.
Chloroplast protein isolation
Chloroplasts isolation was performed as described previously59, with modifications. Detached leaves (1 g, approx. 5 plants) were homogenized in a mortar with 10 mL of homogenization buffer (50 mM HEPES pH 7.6, 330 mM sorbitol, 2 mM EDTA, 1 mM MgCl2, 5 mM ascorbic acid, 0.05% m/v BSA, 0.5 PMSF). The homogenate was filtered with gaze and centrifuged at 1,500 g for 7 min at 4 °C. The supernatant was discarded and the precipitate was re-suspended in 1 mL of homogenization buffer. Then, the suspended volume was transferred to a 15 mL Falcon tube with 10 mL of 30% Percoll in washing buffer solution (50 mM HEPES pH 8.0, 330 mM sorbitol, 2 mM EDTA, 1 mM MgCl2, 5 mM ascorbic acid, 0.05% m/v BSA, 0.5 mM PMSF). Samples were centrifuged at 5,000 g for 12 min at 4 °C, and the chloroplast enriched pellet collected.
The pellet was re-suspended in 1 mL of washing buffer, and then centrifuged at 1,500 g for 5 min at 4 °C. The supernatant was discarded and the precipitate was washed again. Finally, samples were re-suspended in 1 mL hypotonic buffer solution (10 mM HEPES/KOH pH 7.9, 4 mM MgCl2, protease inhibitor cocktail -P9599, Sigma, Saint Louis, MO, USA-), centrifuged at 1,500 g for 5 min at 4 °C and re-suspended again in the same buffer. Extracts were shaken for 2 h at 4 °C. Then, chloroplast protein samples were freeze-dried and stored at −80 °C until the proteomic analysis was performed.
LC-MS/MS differential proteomic analysis
Freeze-dried samples were re-suspended in solubilization buffer (50 mM HEPES pH 7.9, 4% CHAPS, protease inhibitor cocktail P9599) and incubated for 1 h at 4 °C, under agitation and centrifuged at 12,000 g for 15 min. This step was repeated twice, and supernatants separated in each sample were used for a label-free differential proteomic analysis. Three independent biological replicates were analyzed per treatment and ecotype.
Samples (40 µg) were loaded into a 12% SDS-PAGE to prepare them for trypsin digestion (Promega), as described60. Then, peptides were loaded onto an analytical column (LC, 3 µm C18-CL, 75 µm × 12 cm, Nikkyo) and analyzed in a mass spectrometer nanoESI qQTOF (5600 TripleTOF, ABSCIEX). Identification of the obtained peaks was carried out with the ProteinPilot software (version 4.5.1, revision 2768; Paragon™ Algorithm 4.5.1.0, 2765; Sciex), using the defaults parameters (trypsin specificity, cys-alkylation, no taxonomy restriction and the search rapid) and the ExPASy database61. Protein identification was performed at 95% confidence.
Peptide and protein areas were calculated using Peak View 1.1 (http://sciex.com/products/software/peakview-software) and the quantitative data obtained analyzed with Marker View 1.3 (http://sciex.com/products/software/markerview-software). Statistical analysis (two factor ANOVA and hierarchical clustering) was performed using MeV (MultiExperiment Viewer) from Tm4 platform62. The experiment was normalized by amount of protein in each sample, and the differentially abundant proteins between ecotypes, stress treatments or the interaction between both factors were identified (p-value < 0.05). The functional analysis was done using the Kyoto Encyclopedia of Genes and Genomes (KEGG)63, UniProt64, LegumeIP65 and Miyakogusa.jp 3.0 (http://www.kazusa.or.jp/lotus/) Functional protein association networks were evaluated with the STRING software (version 10.0)22. Differentially abundant proteins between ecotypes, treatments or with interaction between both factors were analyzed separately. The Arabidopsis thaliana database was used in the analysis, with a required confidence (score) of 0.4. The confidence score is the approximate probability that a predicted link exists between two proteins in the same metabolic map in the KEGG database. The edges in the corresponding graphs showed the predicted functional links between the different proteins, consisting of eight colour lines each one representing one type of evidence. Avalaible databases used for the prediction of functional networks by the STRING software were: gene co-occurrence (blue lines), co-expression (black lines), curated databases (light blue lines), experimentally determined data (violet lines) and textmining (yellow lines).
Chloroplast transmission electron microscopy (TEM)
Ultrastructural analysis of chloroplast was carried out in the second developed leaf of 3 plants per treatment and ecotype. One leaf of 1 plant cultivated individually in a pot was considered as an experimental unit. Leaf sections (1 mm2) were infiltrated and fixed with 2% glutaraldehyde, in sodium phosphate buffer (pH 7.2–7.4), for 2 h at 4 °C. Then, samples were washed three times with the same buffer to remove the excess of fixative, and secondary fixated with 1% osmium tetroxide for 1 h at 4 °C. Dehydration was performed with alcohols, for their inclusion in an epoxy resin. Ultra-thin sections (90 nm) were contrasted with uranyl acetate and lead citrate, and photographed in a transmission electron microscope JEM 1200 EX II (TEM, JEOL, USA).
Image analysis was performed using the ImageJ software (National Health Institute, MD, USA) in a minimum of 20 photographs per treatment and ecotype. Ultrastructural parameters determined were chloroplasts area (µm2) and number and area (µm2) of starch granules. For pair comparisons between temperature treatments, in each ecotype, a Student’s t-test (p < 0.05) was performed with Prism 5 (Graph-pad Software, CA, USA).
Chloroplast ROS detection by confocal microscopy
Confocal microscopy analysis was performed according to Shu et al.66, with some modifications. Briefly, leaves were cut in 1 mm strips and incubated for 10 min with 25 µM 2′,7′-dichlorofluorescein diacetate (DCF-DA), in 20 mM TrisHCl pH 7.8 buffer solution in darkness. Then, samples were washed twice with 20 mM TrisHCl pH 7.0 buffer solution and exposed to a 250 µmol m−2 s−1 light intensity for 1 min, before observation in a confocal microscope LEICA TCS SP5 (Leica, Germany). As fluorescence controls, leaves were incubated with buffer solution without DCF-DA, or with DCF-DA but without light exposition. During the observation, samples were excited at 485 nm (λexc DCF-DA, λem = 535 nm) and 488 nm (λexc chlorophyll autofluorescence, λem = 670–730 nm). Images were processed using the Leica LAS AF LITE software (Leica Microsystems CMS GmbH, Germany). Microscopy was performed on the second developed leaf in 4 plants per treatment and ecotype.
Determination of chloroplast ROS by flow cytometry
Flow cytometer analysis was performed considering an experimental unit as the cell populations isolated from leaves of one plant cultivated individually in a pot. Cells were obtained by partial homogenization of L. japonicus MG-1 and MG-20 leaves. Two hundred mg of leaves were homogenized in 2 mL homogenization buffer solution (50 mM HEPES pH 7.0, 330 mM sorbitol, 2 mM EDTA, 1 mM MgCl2, 5 mM ascorbic acid, 0.05% m/v BSA and 0.5 mM PMSF). The homogenate was filtered with gauze and centrifuged at 1,500 g for 7 min at 4 °C. The supernatant was discarded and the pellet re-suspended in 1 mL of homogenization buffer and diluted 1:8 with the sheath fluid FACSFlowTM (BD Biosciences, CA, USA) for the analysis. Determinations were done using a FACSCalibur flow cytometer (BD Biosciences, Singapore) at the same voltage and at a constant flow rate (<500 events s−1) for all experiments. BD CellQuest software (BD Biosciences, CA, USA) was used for data acquisition. Cytograms analysis was performed using the FlowJo software (TreeStar Inc., CA, USA).
The flow cytometer was equipped with a red Diode laser (λem = 635), which allows acquisition of 6 different parameters simultaneously: 4 colours (FL1, FL2, FL3 and FL4) and two parameters related with light scattering, side scatter (SSC) and the forward scatter. In this study only the FL1, FL3 and SSC parameters were used. FL1 (λexc = 488 nm, λem = 530 ± 30 nm BP) allows the detection of DCF-DA, that emits in green when excited with blue light. The FL3 parameter allows the detection of chlorophyll autofluorescence after excitation with blue light (λexc = 488 nm, λem = 670 nm LP). SSC represents the scattered light by particles at an angle of 90°, and it is related with the internal complexity of the cell and their size.
Cytometric populations obtained from L. japonicus leaves were characterized, physically separated by the flow cytometer and identified. Briefly, populations were filtered through a 0.22 µm polycarbonates filters (Millipore, MA, USA), DAPI stained and observed in an epifluorescence microscope Nikon Eclipse E600 (Nikon Instruments, NY, USA).
Treatments were done incubating samples with 10 µM DCF-DA in the darkness for 10 min, before exposure to 250 µmol m−2 s−1 of light for 1 min. Then, the flow cytometer fluorescence determination was performed. As fluorescence controls, samples were incubated with buffer solution without DCF-DA, or with DCF-DA but without light exposition. To reduce variability between incubation treatments, these were analyzed sequentially in each tube (first without DCF-DA, then with DCF-DA in darkness and finally with DCF-DA and light exposition).
In each case, the FL1 mode was determined for the cytometric population selected (40,000 events), and the difference between the DCF-DAlight and DCF-DAdarkness treatments calculated for this parameter. As the FL1 fluorescence was registered in a logarithmic scale, the difference between the modes in each treatment was calculated as follow:
| 1 |
The obtained results were compared between control and stress treatments for each ecotype. Measurements were performed in 5 plants per treatment and ecotype. The FL1 values are expressed as arbitrary units (UAs). For the statistical analysis, one-way ANOVA was carried out, followed by a Tukey test for multivariate analysis (p < 0.05) using Prism 5 (Graph-pad Software, CA, USA) and InfoStat/L program (Universidad Nacional de Córdoba, Córdoba, Argentina)67. For pair comparisons between temperature treatments, a Student’s t-test (p < 0.05) with Prism 5 (Graph-pad Software, CA, USA) was performed.
One sentence summary
Acclimation responses in two Lotus japonicus ecotypes with different tolerance to cold stress are related to global chloroplast changes that involve its proteome, ultrastructure and ROS levels.
Supplementary information
Acknowledgements
Differential proteomic analysis was carried out at the proteomics division of the S.C.S.I.E (Servei Central de Suport a l’Investigació Experimental) at the University of Valencia (Valencia, Spain). We thank Agustina Ganuza (CIC-INTECH) for technical assistance with flow cytometry. This work was supported by the following grants: PICT of Agencia Nacional de Promoción Científica y Tecnológica (ANPCYT, Argentina) and Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina).
Author Contributions
P.I.C. conceived and performed all the experiments; J.M.V. assisted in the proteomic and flux cytometry experiments; F.J.E. assisted in the proteomic experiment; F.U. assisted in the design and analysis of flux cytometry experiments; P.I.C., O.A.R. and P.C. designed all the experiments and analyzed the data; O.A.R. and P.C. conceived the project; P.I.C., O.A.R. and P.C. wrote the article with contributions of all the authors.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37165-7.
References
- 1.Xin Z, Browse J. Cold comfort farm: the acclimation of plants to freezing temperatures. Plant, Cell & Environment. 2000;23:893–902. doi: 10.1046/j.1365-3040.2000.00611.x. [DOI] [Google Scholar]
- 2.Thomashow MF. Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant physiology. 2010;154:571–577. doi: 10.1104/pp.110.161794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calzadilla P. I., Maiale S. J., Ruiz O. A. & Escaray F. J. Transcriptome response mediated by cold stress in lotus japonicus. Frontiers in Plant Science7 (2016) [DOI] [PMC free article] [PubMed]
- 4.Barrero-Gil J, Salinas J. Post-translational regulation of cold acclimation response. Plant science. 2013;205:48–54. doi: 10.1016/j.plantsci.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Amme S, Matros A, Schlesier B, Mock H-P. Proteome analysis of cold stress response in Arabidopsis thaliana using DIGE-technology. Journal of experimental botany. 2006;57:1537–1546. doi: 10.1093/jxb/erj129. [DOI] [PubMed] [Google Scholar]
- 6.Hoermiller II, et al. Subcellular reprogramming of metabolism during cold acclimation in Arabidopsis thaliana. Plant, cell & environment. 2017;40:602–610. doi: 10.1111/pce.12836. [DOI] [PubMed] [Google Scholar]
- 7.Gururani MA, Venkatesh J, Tran LSP. Regulation of photosynthesis during abiotic stress-induced photoinhibition. Molecular plant. 2015;8:1304–1320. doi: 10.1016/j.molp.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Goh C-H, Ko S-M, Koh S, Kim Y-J, Bae H-J. Photosynthesis and environments: photoinhibition and repair mechanisms in plants. Journal of Plant Biology. 2012;55:93–101. doi: 10.1007/s12374-011-9195-2. [DOI] [Google Scholar]
- 9.Demmig-Adams B, Adams WW., III Photoprotection and other responses of plants to high light stress. Annual review of plant biology. 1992;43:599–626. doi: 10.1146/annurev.pp.43.060192.003123. [DOI] [Google Scholar]
- 10.Tikkanen M, et al. Regulation of the photosynthetic apparatus under fluctuating growth light. Phil. Trans. R. Soc. B. 2012;367:3486–3493. doi: 10.1098/rstb.2012.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ensminger I, Busch F, Huner NPA. Photostasis and cold acclimation: sensing low temperature through photosynthesis. Physiologia Plantarum. 2006;126:28–44. doi: 10.1111/j.1399-3054.2006.00627.x. [DOI] [Google Scholar]
- 12.Grimaud F, et al. Exploring chloroplastic changes related to chilling and freezing tolerance during cold acclimation of pea (Pisum sativum L.) Journal of proteomics. 2013;80:145–59. doi: 10.1016/j.jprot.2012.12.030. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y-P, et al. Enhanced tolerance to oxidative stress in transgenic tobacco plants expressing three antioxidant enzymes in chloroplasts. Plant cell reports. 2007;26:591–598. doi: 10.1007/s00299-006-0253-z. [DOI] [PubMed] [Google Scholar]
- 14.Yoshimura K, et al. Enhancement of stress tolerance in transgenic tobacco plants overexpressing Chlamydomonas glutathione peroxidase in chloroplasts or cytosol. The Plant Journal. 2004;37:21–33. doi: 10.1046/j.1365-313X.2003.01930.x. [DOI] [PubMed] [Google Scholar]
- 15.Murchie EH, Niyogi KK. Manipulation of photoprotection to improve plant photosynthesis. Plant physiology. 2011;155:86–92. doi: 10.1104/pp.110.168831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams WW, III, Muller O, Cohu CM, Demmig-Adams B. May photoinhibition be a consequence, rather than a cause, of limited plant productivity? Photosynthesis research. 2013;117:31–44. doi: 10.1007/s11120-013-9849-7. [DOI] [PubMed] [Google Scholar]
- 17.Signorelli S, Corpas FJ, Borsani O, Barroso JB, Monza J. Water stress induces a differential and spatially distributed nitro-oxidative stress response in roots and leaves of Lotus japonicus. Plant science. 2013;201–202:137–46. doi: 10.1016/j.plantsci.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Campestre MP, et al. The alkaline tolerance in Lotus japonicus is associated with mechanisms of iron acquisition and modification of the architectural pattern of the root. Journal of Plant Physiology. 2016;206:40–48. doi: 10.1016/j.jplph.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Calzadilla PI, et al. Photosynthetic responses mediate the adaptation of two Lotus japonicus ecotypes to low temperature. Plant Science. 2016;250:59–68. doi: 10.1016/j.plantsci.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Cui S, et al. A proteomic analysis of cold stress responses in rice seedlings. Proteomics. 2005;5:3162–3172. doi: 10.1002/pmic.200401148. [DOI] [PubMed] [Google Scholar]
- 21.Tamburino R, et al. Chloroplast proteome response to drought stress and recovery in tomato (Solanum lycopersicum L.) BMC plant biology. 2017;17:40. doi: 10.1186/s12870-017-0971-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D. et al. STRINGv10: protein–protein interaction networks, integrated over the tree of life. Nucleic acids research 43, Database issue D447–D452 (2015). [DOI] [PMC free article] [PubMed]
- 23.Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K. Effects of abiotic stress on plants: a systems biology perspective. BMC plant biology. 2011;11:163. doi: 10.1186/1471-2229-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu J-K. Abiotic Stress Signaling and Responses in Plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ning F. & Wang W. The Response of Chloroplast Proteome to Abiotic Stress. In Drought Stress Tolerance in Plants, Vol 2. pp. 237–249. Springer (2016).
- 26.Hashiguchi M, Tsuruta S, Akashi R. Morphological Traits of Lotus japonicus (Regal) Ecotypes Collected in Japan. Interdiscipinary Bio Central. 2011;3:1–7. [Google Scholar]
- 27.Wang L, et al. Dynamics of chloroplast proteome in salt-stressed mangrove Kandelia candel (L.) Druce. Journal of proteome research. 2013;12:5124–5136. doi: 10.1021/pr4006469. [DOI] [PubMed] [Google Scholar]
- 28.Rinalducci S, Pedersen JZ, Zolla L. Formation of radicals from singlet oxygen produced during photoinhibition of isolated light-harvesting proteins of photosystem II. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 2014;1608:63–73. doi: 10.1016/j.bbabio.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Pietrzykowska M, et al. The light-harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis. The Plant Cell. 2014;26:3646–3660. doi: 10.1105/tpc.114.127373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutin C, et al. Early light-induced proteins protect Arabidopsis from photooxidative stress. Proceedings of the National Academy of Sciences. 2003;100:4921–4926. doi: 10.1073/pnas.0736939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peltier J-B, et al. Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. The Plant Cell. 2002;14:211–236. doi: 10.1105/tpc.010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersson B. & Aro E.-M. Photodamage and D1 protein turnover in photosystem II. In Regulation of photosynthesis. pp. 377–393. Springer (2001).
- 33.Zhao Y. et al. Quantitative Proteomic Analyses Identify ABA-Related Proteins and Signal Pathways in Maize Leaves under Drought Conditions. Frontiers in Plant Science7 (2016). [DOI] [PMC free article] [PubMed]
- 34.Yi X, Hargett SR, Frankel LK, Bricker TM. The PsbQ protein is required in Arabidopsis for photosystem II assembly/stability and photoautotrophy under low light conditions. Journal of Biological Chemistry. 2006;281:26260–26267. doi: 10.1074/jbc.M603582200. [DOI] [PubMed] [Google Scholar]
- 35.Pagliano C, Saracco G, Barber J. Structural, functional and auxiliary proteins of photosystem II. Photosynthesis research. 2013;116:167–188. doi: 10.1007/s11120-013-9803-8. [DOI] [PubMed] [Google Scholar]
- 36.Gururani MA, Upadhyaya CP, Strasser RJ, Yu JW, Park SW. Evaluation of abiotic stress tolerance in transgenic potato plants with reduced expression of PSII manganese stabilizing protein. Plant science. 2013;198:7–16. doi: 10.1016/j.plantsci.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 37.Dumas L, Chazaux M, Peltier G, Johnson X, Alric J. Cytochrome b6f function and localization, phosphorylation state of thylakoid membrane proteins and consequences on cyclic electron flow. Photosynthesis research. 2016;129:307–320. doi: 10.1007/s11120-016-0298-y. [DOI] [PubMed] [Google Scholar]
- 38.Ivanov AG, et al. Implications of alternative electron sinks in increased resistance of PSII and PSI photochemistry to high light stress in cold-acclimated Arabidopsis thaliana. Photosynthesis research. 2012;113:191–206. doi: 10.1007/s11120-012-9769-y. [DOI] [PubMed] [Google Scholar]
- 39.Yamori W, Noguchi KO, Hikosaka K, Terashima I. Phenotypic plasticity in photosynthetic temperature acclimation among crop species with different cold tolerances. Plant Physiology. 2010;152:388–399. doi: 10.1104/pp.109.145862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Augustyniak A, Perlikowski D, Rapacz M, Kościelniak J, Kosmala A. Insight into cellular proteome of Lolium multiflorum/Festuca arundinacea introgression forms to decipher crucial mechanisms of cold acclimation in forage grasses. Plant Science. 2018;272:22–31. doi: 10.1016/j.plantsci.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 41.Chatterjee J, et al. Cloning, characterization and expression of a chloroplastic fructose-1, 6-bisphosphatase from Porteresia coarctata conferring salt-tolerance in transgenic tobacco. Plant Cell, Tissue and Organ Culture (PCTOC) 2013;114:395–409. doi: 10.1007/s11240-013-0334-y. [DOI] [Google Scholar]
- 42.Ding F, Wang M, Zhang S. Overexpression of a Calvin cycle enzyme SBPase improves tolerance to chilling-induced oxidative stress in tomato plants. Scientia Horticulturae. 2017;214:27–33. doi: 10.1016/j.scienta.2016.11.010. [DOI] [Google Scholar]
- 43.Orzechowski S. Starch metabolism in leaves. Acta biochimica polonica. 2008;55:435–445. [PubMed] [Google Scholar]
- 44.Streb S, Egli B, Eicke S, Zeeman SC. The debate on the pathway of starch synthesis: a closer look at low-starch mutants lacking plastidial phosphoglucomutase supports the chloroplast-localized pathway. Plant Physiology. 2009;151:1769–1772. doi: 10.1104/pp.109.144931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim K-H, et al. Overexpression of a chloroplast-localized small heat shock protein OsHSP26 confers enhanced tolerance against oxidative and heat stresses in tall fescue. Biotechnology letters. 2012;34:371–377. doi: 10.1007/s10529-011-0769-3. [DOI] [PubMed] [Google Scholar]
- 46.Gruber AV, Nisemblat S, Azem A, Weiss C. The complexity of chloroplast chaperonins. Trends in plant science. 2013;18:688–694. doi: 10.1016/j.tplants.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Liu X, Rodermel SR, Yu F. A var2 leaf variegation suppressor locus, SUPPRESSOR OF VARIEGATION3, encodes a putative chloroplast translation elongation factor that is important for chloroplast development in the cold. BMC plant biology. 2010;10:287. doi: 10.1186/1471-2229-10-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Momčilović I, et al. Heat-induced accumulation of protein synthesis elongation factor 1A implies an important role in heat tolerance in potato. Planta. 2016;244:671–679. doi: 10.1007/s00425-016-2534-2. [DOI] [PubMed] [Google Scholar]
- 49.Moore M, Gossmann N, Dietz K-J. Redox Regulation of Cytosolic Translation in Plants. Trends in plant science. 2016;21:388–397. doi: 10.1016/j.tplants.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 50.Murata N, Takahashi S, Nishiyama Y, Allakhverdiev SI. Photoinhibition of photosystem II under environmental stress. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 2007;1767:414–421. doi: 10.1016/j.bbabio.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki N, Mittler R. Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Physiologia plantarum. 2006;126:45–51. doi: 10.1111/j.0031-9317.2005.00582.x. [DOI] [Google Scholar]
- 52.Kratsch HA, Wise RR. The ultrastructure of chilling stress. Plant, Cell & Environment. 2000;23:337–350. doi: 10.1046/j.1365-3040.2000.00560.x. [DOI] [Google Scholar]
- 53.Foyer CH, Shigeoka S. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant physiology. 2011;155:93–100. doi: 10.1104/pp.110.166181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zagorchev L, Seal CE, Kranner I, Odjakova M. A central role for thiols in plant tolerance to abiotic stress. International journal of molecular sciences. 2013;14:7405–7432. doi: 10.3390/ijms14047405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mignolet-Spruyt L, et al. Spreading the news: subcellular and organellar reactive oxygen species production and signalling. Journal of experimental botany. 2016;67:3831–44. doi: 10.1093/jxb/erw080. [DOI] [PubMed] [Google Scholar]
- 56.Choudhury FK, Rivero RM, Blumwald E, Mittler R. Reactive oxygen species, abiotic stress and stress combination. The Plant Journal. 2016;90:856–867. doi: 10.1111/tpj.13299. [DOI] [PubMed] [Google Scholar]
- 57.Thormählen I, et al. Thioredoxin f1 and NADPH-dependent thioredoxin reductase C have overlapping functions in regulating photosynthetic metabolism and plant growth in response to varying light conditions. Plant physiology. 2015;169:1766–1786. doi: 10.1104/pp.15.01122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoagland DR, Arnon DI. The water-culture method for growing plants without soil. Circular. California Agricultural Experiment Station. 1950;347:Circular 347. [Google Scholar]
- 59.Phee B-K, et al. Proteomic analysis of the response of Arabidopsis chloroplast proteins to high light stress. Proteomics. 2004;4:3560–3568. doi: 10.1002/pmic.200400982. [DOI] [PubMed] [Google Scholar]
- 60.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Analytical chemistry. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 61.Artimo P, et al. ExPASy: SIB bioinformatics resource portal. Nucleic acids research. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Howe E. et al. Mev: multiexperiment viewer. In Biomedical informatics for cancer research. pp. 267–277. Springer (2010).
- 63.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic acids research. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boutet E. et al. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Plant Bioinformatics: Methods and Protocols, 23–54 (2016). [DOI] [PubMed]
- 65.Li J, Dai X, Liu T, Zhao PX. LegumeIP: an integrative database for comparative genomics and transcriptomics of model legumes. Nucleic acids research. 2012;40:D1221–D1229. doi: 10.1093/nar/gkr939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shu S, et al. The role of putrescine in the regulation of proteins and fatty acids of thylakoid membranes under salt stress. Scientific reports. 2015;5:14390. doi: 10.1038/srep14390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Di Rienzo J. A. et al. InfoStat, versión 2008. Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina (2008).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.