Bacillus subtilis spores are encased in a multiprotein coat that surrounds an underlying peptidoglycan layer, the cortex. How the connection between the two layers is enforced is not well established. Here, we elucidate the role of the peptidoglycan-binding LysM domain, present in two proteins, SafA and SpoVID, that govern the localization of additional proteins to the coat. We found that SafALysM is a protein-protein interaction module during the early stages of coat assembly and a cortex-binding module at late stages in morphogenesis, with the cortex-binding function promoting a tight connection between the cortex and the coat. In contrast, SpoVIDLysM functions only as a protein-protein interaction domain that targets SpoVID to the spore surface at the onset of coat assembly.
KEYWORDS: LysM domain, SpoVID, peptidoglycan-binding protein, spore coat, spore cortex, sporulation
ABSTRACT
At a late stage in spore development in Bacillus subtilis, the mother cell directs synthesis of a layer of peptidoglycan known as the cortex between the two forespore membranes, as well as the assembly of a protective protein coat at the surface of the forespore outer membrane. SafA, the key determinant of inner coat assembly, is first recruited to the surface of the developing spore and then encases the spore under the control of the morphogenetic protein SpoVID. SafA has a LysM peptidoglycan-binding domain, SafALysM, and localizes to the cortex-coat interface in mature spores. SafALysM is followed by a region, A, required for an interaction with SpoVID and encasement. We now show that residues D10 and N30 in SafALysM, while involved in the interaction with peptidoglycan, are also required for the interaction with SpoVID and encasement. We further show that single alanine substitutions on residues S11, L12, and I39 of SafALysM that strongly impair binding to purified cortex peptidoglycan affect a later stage in the localization of SafA that is also dependent on the activity of SpoVE, a transglycosylase required for cortex formation. The assembly of SafA thus involves sequential protein-protein and protein-peptidoglycan interactions, mediated by the LysM domain, which are required first for encasement then for the final localization of the protein in mature spores.
IMPORTANCE Bacillus subtilis spores are encased in a multiprotein coat that surrounds an underlying peptidoglycan layer, the cortex. How the connection between the two layers is enforced is not well established. Here, we elucidate the role of the peptidoglycan-binding LysM domain, present in two proteins, SafA and SpoVID, that govern the localization of additional proteins to the coat. We found that SafALysM is a protein-protein interaction module during the early stages of coat assembly and a cortex-binding module at late stages in morphogenesis, with the cortex-binding function promoting a tight connection between the cortex and the coat. In contrast, SpoVIDLysM functions only as a protein-protein interaction domain that targets SpoVID to the spore surface at the onset of coat assembly.
INTRODUCTION
During endospore development by Bacillus subtilis, a multiprotein coat is assembled around the developing spore. The coat contributes to spore protection but also regulates germination and the interactions of spores with the immediate environment (1–4). Studies on coat assembly have provided important insights onto the mechanisms by which bacterial cells form supramolecular structures or organelles at specific subcellular locations in register with transcriptional cascades and cell morphogenesis (1–4).
Sporulation initiates with the rod-shaped cell dividing close to one of the cell poles to form a larger mother cell and a smaller forespore, the future spore. Following polar division, the mother cell begins a process termed engulfment, with the septal membranes moving toward and eventually fusing at the cell pole, releasing the forespore as a free protoplast surrounded by the mother cell cytoplasm (1–3, 5). A thin layer of peptidoglycan (PG), the germ cell wall, is then formed across the forespore inner membrane and will become the wall of the cell that outgrows from spores during germination. A thicker layer of PG, the spore cortex, is formed across the forespore outer membrane from precursors synthesized in the mother cell. This layer, which occupies the space between the two forespore membranes, is necessary for spore heat resistance (1–3). The coat is assembled around the cortex (1–3, 6, 7). When differentiation is complete, the mature spore is released through lysis of the mother cell. As they are metabolically dormant and resistant under extreme physical and chemical conditions, spores are able to remain viable in the environment for long periods of time (1–4).
Sporulation is controlled by a cascade of RNA polymerase sigma factors that are activated in either the forespore or the mother cell upon the completion of key stages in morphogenesis; σF and σE are activated just after polar division and control gene expression in the forespore and in the mother cell, respectively. Following engulfment completion, σF is replaced by σG, and σK replaces σE. Synthesis of the proteins involved in coat assembly is under the control of σE, σK, and ancillary regulatory proteins that work with these sigma factors and define a transcriptional cascade that produces successive waves of gene expression (1–3, 6, 7).
The coat of B. subtilis is differentiated into a basement layer, an inner lamellar coat, a more external, electron-dense, and striated outer coat, and a crust as the outermost structure (1–3, 6, 7). The formation of each of these layers requires dedicated morphogenetic proteins produced early under σE control and thought to act as hubs that recruit other layer-specific proteins; SpoIVA is essential for the formation of the basal layer, SafA drives assembly of the inner coat, CotE is required for outer coat assembly, and CotZ is required for crust assembly (6, 8–13). Targeting of the layer-specific morphogenetic proteins requires the localization of the SpoIVA ATPase at the onset of engulfment. The localization of SpoIVA involves an interaction with SpoVM, a 26-residue peptide that recognizes the positive curvature of the forespore outer membrane (14–17). SpoIVA recruits SafA, CotE, and CotZ that form an organizational scaffold early in morphogenesis (12, 18, 19) This scaffold is required for a second stage in coat assembly, the encasement, when the layer-specific morphogenetic proteins along with their partners start encircling the forespore. Encasement requires SpoVM and another σE-dependent protein, SpoVID, and occurs in successive waves dependent on the mother cell transcriptional cascade (3, 20).
The encasement protein SpoVID and the inner coat morphogenetic protein SafA both have LysM domains. LysM domains are found in virtually all organisms except the Archaea, and they bind polymers containing N-acetylglucosamine (GlcNAc), which is found in chitin and in PG, or lipooligosaccharides (nodulation factors) (21). SpoVID has an N-terminal domain (N) that resembles a phage coat protein, a middle, possibly extended, domain (M), and a C-terminal targeting signal. The targeting signal consists of a 24-residue stretch called region A and a LysM domain, SpoVIDLysM (8, 12, 22, 23) (Fig. 1A). Both region A and SpoVIDLysM are involved in a direct interaction with SpoIVA that targets SpoVID to the forespore (12, 22). It is not known whether SpoVIDLysM additionally binds to PG. Encasement, in turn, requires a region, termed E, close to the end of the N domain that is required for encasement by all layers of the coat (12, 20, 24, 25).
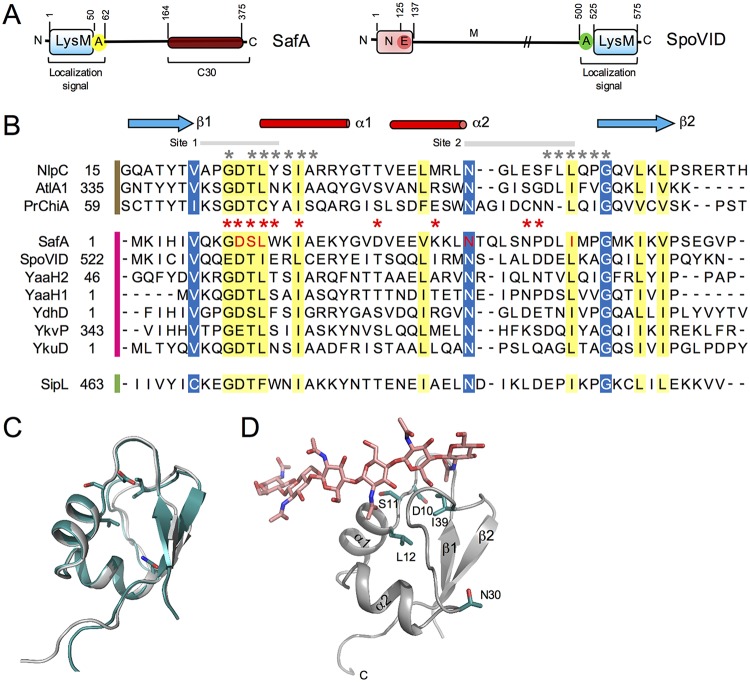
FIG 1.
(A) Diagram of the SafA and SpoVID proteins. SafA (left) has a LysM domain at its N terminus, followed by region A, which together form a localization signal. The SafAC30 region of the full-length protein corresponds to the part that is produced from the safA mRNA by internal translation starting at Met codon 161 or 164. SpoVID (right) is formed by an N domain, followed by the E region (for encasement), a middle domain (M), and a localization signal, formed by region A and a LysM domain. (B) Clustal W alignment of the LysM domain of SafA with the LysM domains of the indicated selected proteins. The LysM sequences are grouped according to the organism of origin and the availability of crystal structures, as follows: B. subtilis, pink bar; C. difficile, green bar; sequences with available structure, brown bar. Residues shaded in yellow represent conserved residues, while residues shaded in blue represent conserved surface-exposed residues. The five residues of SafALysM that were replaced by alanine in this study are shown in red. (C) Superimposition of the homology model generated for SafALysM with that of the template, the N-terminal LysM domain from the putative NlpC/P60 d,l-endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose (PDB ID 4UZ3). SafA, gray; template, blue. (D) Homology model of SafA bound to chitohexose, to locate the substrate binding site. Chitohexose is shown using sticks with carbon atoms colored in pink. To place this molecule, the model structure was superimposed on the template structure containing chitohexose. (C and D) The two α-helices and the two β-strands are labeled in the model, and the positions of the conserved D10, S11, L12, N30, and I39 residues, which were replaced by Ala, are highlighted using sticks and with carbon atoms colored in green.
SafA is produced as a full-length protein of 45 kDa, or SafAFL, and a 30-kDa form, SafAC30, formed through internal translation of the safA mRNA (10, 11, 26). SafAFL has a LysM domain, SafALysM, at its N terminus (Fig. 1A). SafA localizes at the cortex-inner coat interface in mature spores, suggesting that SafALysM might contribute to the localization of the protein (10, 23). It has been suggested that SafALysM might bind to PG in the intermembrane space (10, 23). SafA, however, has no known signals for secretion, and no mechanism is known that could promote its interaction with PG during encasement. Downstream of SafALysM, a region termed A interacts with the N domain of SpoVID and is essential for encasement by SafA (10, 22, 26, 27). SafAC30, which lacks a LysM domain and region A, localizes to the forespore only in the presence of SafAFL, with which it interacts (10, 23). It is not known, however, whether region A is sufficient for encasement or whether SafALysM is also involved.
Here, we have analyzed the effect of single alanine substitutions in SafALysM on the localization of SafA. We show that a class of single Ala substitutions in SafALysM prevents an interaction with SpoVID and encasement by SafA. A second class of mutations that strongly impair the interaction of SafA with purified spore cortex PG, as well as a mutation that blocks the synthesis of a transglycosylase required for spore cortex formation, interfere with a late step in the localization of SafA. We conclude that SafALysM is a protein-protein interaction module during encasement and a cortex PG-binding module that mediates the interaction of SafA with the cortex at late stages in morphogenesis.
RESULTS
Conserved features of the LysM domains of SafA and SpoVID.
SafA has a single LysM domain localized at the N-terminal end of the protein (residues 1 to 50). LysM domains, present in PG- or chitin-binding proteins, share several invariant residues and a common βααβ fold, with the α-helices packed against the antiparallel β-sheets (21, 28, 29) (Fig. 1B and C). We started this investigation by producing a homology model of the LysM domain of SafA to assess whether its overall fold and the positions of conserved amino acids were maintained and to identify candidate surface-exposed residues that could be tested through loss of side-chain mutagenesis for a role in peptidoglycan binding and/or protein localization. We used the structure of the N-terminal LysM domain from the putative NlpC/P60 d,l-endopeptidase from Thermus thermophilus bound to N-acetyl-chitohexaose as our template (30) (PDB ID 4UZ3); not only does it show high sequence identity with the target sequence (44%), but it also is a high-resolution X-ray structure (1.75 Å). The homology model of SafALysM shows the characteristic βααβ fold of the LysM domain (29, 31, 32) (Fig. 1C). Ramachandran plots of the best models located all the residues in the most favored or additional allowed regions (see also Materials and Methods). The binding site for N-acetylglucosamine oligomers (chitin) was determined for the LysM domain of the chitin elicitor receptor kinase 1 of Arabidopsis (AtCERK1) and for one of the two LysM domains of Pteris ryukyuensis chitinase A (PrChi-A) (31, 32) (Fig. 1B, residues marked by the red asterisks). The binding site for PG was determined for the first of the six LysM domains of the AtlA autolysin from Enterococcus faecalis (29) (Fig. 1B, asterisks in gray). These studies have shown that the chitin/PG-binding site is formed mainly by residues located in the loop between β-strand 1 and helix 1 and at the N terminus of helix 1 (site 1), and in the loop between helix 2 and β-strand 2 (site 2) (Fig. 1B). These residues form a long shallow groove at the surface of the LysM domain on one side of the molecule (29, 32). Residues within these two sites are also involved in the recognition of lipochitin-oligosaccharides by cognate Nod-factor receptors (33). Inspection of the model built for SafALysM indicates that the side chains of residues D10, S11 (in site 1), and N30 (site 2) that form the walls of the groove are surface exposed (Fig. 1C and D). The side chains of two other residues, L12 (in site 1) and I39 (in site 2), which form part of the basement of the trench, are also partially exposed (Fig. 1C and D). These residues are also highly conserved among LysM domains (Fig. 1B). To investigate their role in PG binding and in the localization of SafA, variants with single Ala substitutions of D10, S11, L12, N30, and I39 were generated.
Single alanine substitutions in the LysM domain affect the localization of SafA at different stages of morphogenesis.
To examine the localization of SafA, we made use of a previously constructed safA in-frame deletion mutant, ΔsafA (27). We have shown before that a safA-yfp fusion, in which the SafA and yellow fluorescent protein (YFP) moieties are separated by a flexible linker, is functional, as when in single copy at the nonessential amyE locus, it complements the phenotypes caused by the ΔsafA mutation (25). Therefore, we introduced C-terminal yfp fusions to the wild-type (WT) safA gene or to alleles coding for the various single alanine substitutions in SafALysM (D10A, S11A, L12A, N30A, and I39A) (Fig. 1B) at the amyE locus. With respect to engulfment, the early σE-dependent coat proteins can be divided into the following three kinetic classes: class I proteins localize to the surface of the developing spore at the onset of engulfment and track the mother cell membrane so that encasement and engulfment occur simultaneously; class II proteins that localize simultaneously with class I proteins but begin encasement only after engulfment completion from a site, at the mother cell distal (MCD) forespore pole, where fission of the engulfing membranes occurs; and class III proteins that localize simultaneously with class I and class II proteins but begin encasement when phase-dark forespores first appear (20). We have shown before that SafA-YFP behaves as a class II protein; it forms a cap at the mother cell proximal (MCP) forespore pole soon after the onset of engulfment and then a cap at the MCD forespore pole just after engulfment completion, and from this cap, it completes encasement of the forespore (25).
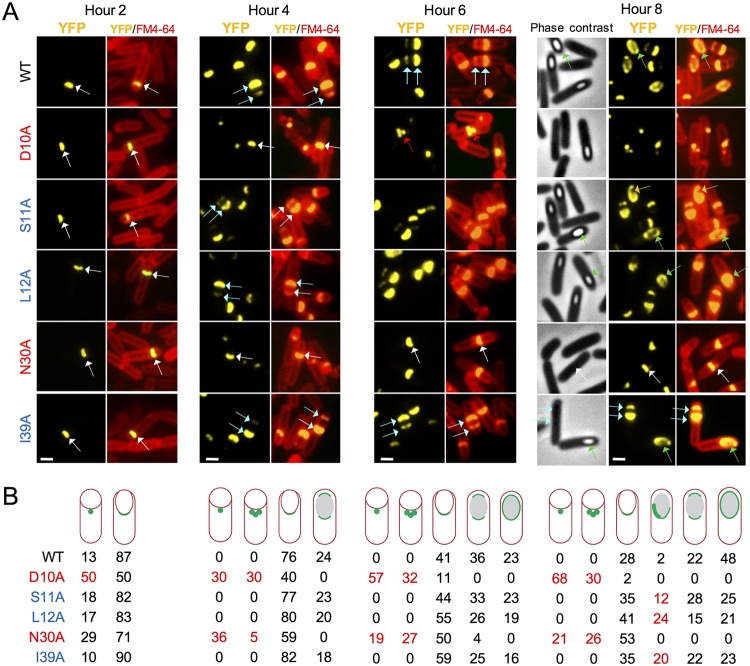
We first examined the localization of SafAWT-YFP after inducing sporulation by the growth and resuspension method (see Materials and Methods). Cells were collected from sporulating cultures 2, 4, 6, and 8 h after the onset of sporulation, stained with the membrane dye FM4-64, and imaged by fluorescence microscopy. In control experiments, we showed that none of the substitutions interfered with the accumulation of SafAFL or SafAC30, as assessed by immunoblotting at different times during sporulation (see Fig. S1 in the supplemental material). At hour 2 of sporulation, a dot of fluorescence was seen at the MCP forespore pole in 13% of the sporangia, but most (87%) showed a cap of fluorescence (Fig. 2A, white arrows; quantification in Fig. 2B). At hour 4, the representation of sporangia with a single cap of fluorescence at the MCP pole pattern decreased to 76% of the sporangia, while caps at the two forespore poles first appeared, representing 24% of the sporangia (Fig. 2A, blue arrows, and Fig. 2B). The two-cap pattern increased to 36% at hour 6, when complete encirclement of the forespore was first noticed for 23% of the sporangia (Fig. 2A, green arrows, and Fig. 2B). Finally, at hour 8, complete encirclement of the forespore by SafAWT-YFP, in 48% of the sporangia, was the prevalent pattern (Fig. 2). The localization pattern of SafA thus fits the description of a class II protein, in agreement with recent results (25). In sharp contrast, for SafAD10A-YFP, 50% of the sporangia scored at hour 2 showed a dot of fluorescence and 50% a single cap (Fig. 2). The single-cap pattern decreased to 40% of the sporangia at hour 4, while the single-dot pattern represented 30% of the sporangia scored, and a new class showing multiple dots dispersed along the engulfing membrane represented 30% of the sporangia (Fig. 2A, red arrows, and Fig. 2B). These two classes combined represented 89% of the sporangia scored at hour 6, while the single-cap pattern decreased to 11% (Fig. 2). In addition, the two-cap pattern was not detected at this or at later times in sporulation (Fig. 2). In an earlier study, we showed that in a spoVID in-frame deletion mutant or in a mutant in which region E was deleted (in both mutants encasement is blocked), the dot pattern of SafA-YFP localization follows a similar evolution, i.e., it increases over time at the expense of the single-cap pattern (25). This suggests that when encasement is blocked, the single cap regresses into one or more dots (25). For SafAN30A-YFP, dots were scored in 29% of the sporangia at hour 2, when 71% of the sporangia showed a single cap (Fig. 2). As for SafAD10A-YFP, the representation of the single-cap pattern decreased to 59% at hour 4, while single dots represented 36% of the sporangia and multiple dots appeared (5% of the sporangia) (Fig. 2). The two-cap pattern was detected only at hour 6, but only in 4% of the sporangia (Fig. 2). Thus, SafAD10A- and SafAN30A-YFP behaved similarly. The SafAS11A-, SafAL12A-, and SafAI39A-YFP fusions formed a different group. Localization of these fusions was similar to the WT up to hour 6 of sporulation, but a distinctive phenotype appeared at hour 8 when sporangia of phase-bright spores appeared, as observed by phase-contrast microscopy (Fig. 2). The appearance of phase-bright spores indicates that a late stage in cortex formation has been reached (2, 27, 34). At hour 8 of sporulation, the percentage of sporangia showing complete encirclement of the forespore was lower than that for the WT (S11A mutant, 25%; L12A mutant, 21%; I39A mutant, 23%) (Fig. 2). Moreover, in a fraction of the sporangia (S11A mutant, 12%; L12A mutant, 24; I39A mutant, 20%) (Fig. 2), the fluorescence signal was asymmetrically distributed around the forespore (Fig. 2A, orange arrows in the S11A mutant).
FIG 2.
Localization of SafA-YFP. (A) Subcellular localization of SafA-YFP in sporulating cells. Strains were induced to sporulate by growth and resuspension. Samples were withdrawn at the indicated times, in hours, after resuspension, defined as the onset of sporulation, stained with the membrane dye FM4-64, and imaged by fluorescence microscopy (phase-contrast images are also shown for the hour 8 sample). The images show the YFP signal or the merge between the YFP and FM4-64 signals. All strains carry an in-frame deletion allele of safA and yfp fusions to the WT gene or to alleles expressing fusion proteins with the indicated single amino acid substitutions, inserted at the nonessential amyE locus. White arrows, single cap; blue arrows, double cap; green arrows, full encirclement; red arrows, multiple dots; orange arrows, asymmetric localization of SafA-YFP around the spore. Scale bar = 1 µm. (B) Scoring of the percentage of sporangia with the represented patterns of SafA-YFP localization at the indicated times after the onset of sporulation. (A and B) Substitutions causing early localization defects are highlighted in red, and those causing late localization defects are in blue. The numbers in red highlight the phenotypes.
We conclude that the D10A and N30A substitutions act mainly by preventing encasement by SafA, while S11A, L12A, and I39A affect localization at a late stage in morphogenesis, when the spore becomes phase bright.
Single alanine substitutions in the LysM domain affect localization of the SafA-dependent YaaH protein.
We also tested whether the D10A and L12A substitutions affected the localization of a SafA-dependent protein, YaaH (20). YaaH is synthesized under the control of σE and requires SafA to encase the spore (20). We have recently shown that YaaH, like SafA, behaves as a kinetic class II protein (25). We scored the localization of a previously constructed YaaH-green fluorescent protein (YaaH-GFP) fusion (20) during sporulation in the WT and in cells producing the D10A and L12A forms of SafA, as representatives of the two main effects, early and late, described above for SafA (Fig. 2). In the WT, YaaH-GFP formed a single cap at the MCP forespore pole in 100% of the sporangia scored at hour 2 (Fig. S2A, white arrows; quantification in Fig. S2B). By hour 4, the most represented pattern were sporangia with two caps of fluorescence (54%); by hour 6, most sporangia (69%) showed a ring of fluorescence, and the representation of this pattern increased to 79% at hour 8, when most sporangia carried phase-bright spores (Fig. S2). Thus, as previously reported, YaaH-GFP behaves as a kinetic class II protein (25). In the safA in-frame deletion mutant, the signal from YaaH-GFP was found dispersed throughout the mother cell cytoplasm in 100% of the sporangia scored at hour 2 (Fig. S2A, red arrows; quantification in Fig. S2B). This pattern persisted at all time points examined (Fig. S2). Thus, in agreement with earlier results, the localization of YaaH-GFP to the forespore surface is completely dependent on safA (20). No dispersed signal was seen for the D10A mutant, with 95% of the sporangia at hour 2 showing a cap of fluorescence at the MCP forespore pole (Fig. S2A, white arrows; Fig. 2B). In 5% of the sporangia, YaaH-GFP seemed to track the engulfing membranes (Fig. S2A, yellow arrows). At hour 4, we found a single cap in 26% of the sporangia, two caps of similar intensity in 15 of the sporangia, and 57% of the sporangia with a cap of reduced fluorescence at the MCD forespore pole (Fig. S2A, blue arrows). These patterns persisted over time (Fig. S2). Additionally, at hour 4, 2% of the sporangia showed complete encasement of the forespore, a number that increased to 18% at hour 6 and to 25% at hour 8 (Fig. S2A, green arrows). In the L12A mutant, 96% of the sporangia at hour 2 showed a cap at the MCP forespore pole (Fig. S2A, white arrow), and in 4%, YaaH-GFP seemed to track the engulfing membranes (yellow arrow). At hour 4, the representation of the MCP cap pattern decreased to 21%, but the double-cap pattern increased to 57% (equal intensity at the two poles) and to 10% (weaker fluorescence at the MCD forespore pole [blue arrows in Fig. S2A]). These two patterns decrease with time, and by hour 8, the double cap that was weaker at the MCD pole was not detected (Fig. S2). By hour 8, complete encasement was seen for 70% of the sporangia, in the same range as for the WT; however, 14% of the sporangia at hour 8 showed asymmetric accumulation of YaaH-GFP around the forespore (Fig. S2A, orange arrow).
For the D10A and L12A mutants, the tracking of the engulfment membranes in a small fraction of the sporangia (5 and 4%, respectively) indicates a behavior characteristic of kinetic class I proteins (20). SpoVID is a class I protein, and it seems possible that in these strains, YaaH-GFP encases the forespore under the direct control of SpoVID. The accumulation of the double-cap pattern with weaker fluorescence at the MCD forespore pole in both the D10A and L12A mutants suggests that encasement of the forespore from this pole is impaired. Finally, as also seen for SafA-YFP, the L12A substitution affects localization of YaaH-GFP at a late stage in morphogenesis, when the cortex is formed and the forespore becomes phase bright.
We conclude that the localization defects imposed upon SafA-YFP by the single amino acid substitutions in the LysM domain translate into deficient assembly of at least one SafA-dependent protein.
Single alanine substitutions in LysM domain affect the distribution of SafA in mature spores.
The deletion of safA leads to the production of spores that are heat resistant but susceptible to lysozyme (10, 11). We note that deletion of safA reduces the titer of lysozyme-resistant spores from about 108 CFU/ml (WT) to over 107 CFU/ml (10, 11). We measured the total viable cell count, as well as the heat-resistant and the lysozyme-resistant cell counts for the WT and the various mutants 48 h after the onset of sporulation. In spite of the localization defects seen for the various forms of SafA, none of the substitutions reduced the titer of heat- or lysozyme-resistant spores (Table S4). Possibly, the various safA alleles herein analyzed retain some functionality, or the LysM is redundant with another region required for SafA localization and function (22). In any event, we proceeded to examine the distribution of SafA in mature spores of the various mutants.
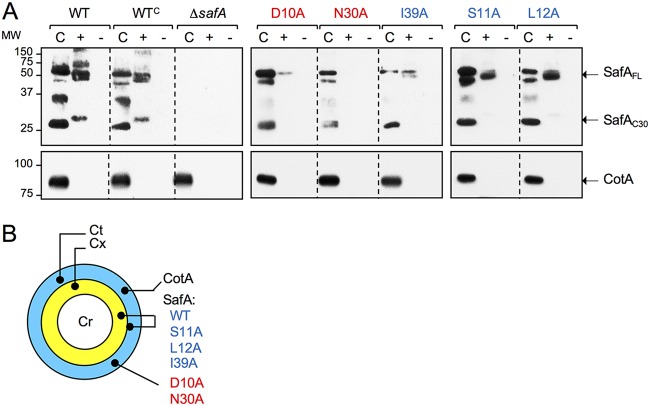
We have shown before that in mature spores, SafA is found both in the coat and in a cortex fraction (25, 27), and we reasoned that the single alanine substitutions in SafALysM could also alter the distribution of the protein in mature spores. We subjected density-gradient-purified spores to a decoating regime to produce a coat fraction, and the decoated spores were reextracted after incubation with lysozyme to produce a “cortex” fraction, or with no treatment, as a control for the effect of the enzyme (27). Proteins in the various fractions were analyzed by SDS-PAGE and immunoblotting with an anti-SafA antibody (10). We examined WT spores and spores of a ΔsafA mutant complemented with the WT safA gene at the amyE locus (WTC) or carrying the different safA alleles at this locus. In agreement with previous results (27), SafAFL and SafAC30 were found in both the cortex and coat fractions of WT and WTC spores, whereas multimeric forms of SafA were detected only in the cortex (Fig. 3A). SafAC30, extracted from the cortex, migrated slower than the form of the protein released from the coat, as also observed before (25, 27) (Fig. 3A). This is most likely because SafAC30 is cross-linked to a small protein or to the peptidoglycan in the cortex fraction (25). Strikingly, no form of SafA was detected in the cortex fraction of N30A spores, and it is highly reduced in D10A spores (Fig. 3A). While SafAFL was detected in the cortex fraction of S11A, L12A, and I39A spores, SafAC30 was detected, albeit at low levels, only in L12A spores (Fig. 3A). As a control for the fractionation experiments, the well-characterized coat protein CotA (35, 36) was found only in the coat fraction (25, 27) (Fig. 3A).
FIG 3.
Localization of SafA in mature spores. (A) Proteins were extracted from density gradient-purified spores of the indicated strains to produce a coat fraction (“C”). The decoated spores were then reextracted following incubation with lysozyme (“+”) or with no lysozyme treatment (“–”). The extracted proteins were resolved by SDS-PAGE and the gels subject to immunoblotting with anti-SafA antibodies or anti-CotA antibodies, as indicated. The positions SafAFL, SafAC30, and CotA are indicated by arrows. The position of molecular weight markers (in kilodaltons) is shown on the left side of the panels. The profile of extractable coat proteins is compared to that of the WT and to an in-frame safA deletion mutant (ΔsafA). The remaining strains carry ΔsafA, and the WT gene (denoted as WTC, for complementation) or alleles expressing proteins with the indicated substitutions, at amyE. (B) Schematic representation of the localization of CotA, SafAWT, and the indicated variants in mature spores. Cr, spore coat; Cx, cortex; Ct, coat. (A and B) The color code for the substitutions is defined in the legend for Fig. 2.
In earlier work, in which the effect of point mutations in region E of spoVID was studied, we suggested that the final localization of SafA in the cortex/coat of mature spores relies on interactions established during engulfment (25). The reduction of SafA in the cortex fraction of D10A and N30A mutants (summarized in Fig. 3B), which show a strong block in encasement, is in line with this suggestion. As for the S11A, L12A, and I39A mutants, the distribution of SafA in mature spores, is closer to that in the WT (Fig. 3B), which is consistent with the quantitatively less pronounced late localization defect.
LysM domain of SafA is required for proper assembly and structure of the coat.
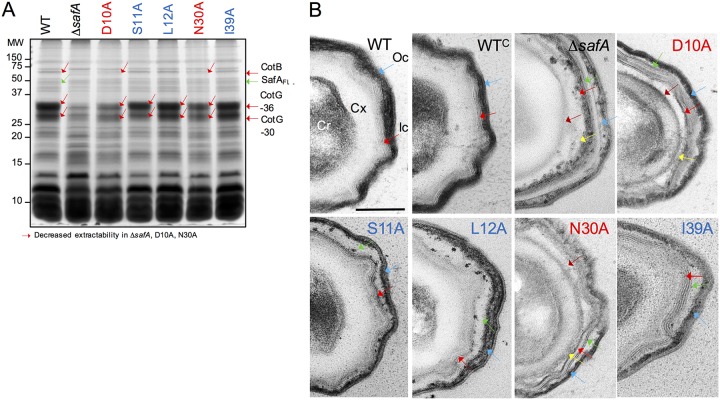
The altered levels and distribution of the various SafA forms in mature spores of the various mutants suggested that the spores could have an altered profile of extractable coat proteins. Density gradient-purified spores were decoated and the collection of extractable coat proteins examined by SDS-PAGE and Coomassie brilliant blue R-250 staining. We found three main species to be absent from or to be less extractable from spores of the safAD10A and safAN30A mutants relative to the WT (Fig. 4A, red arrows). These species include two forms of the abundant protein CotG (CotG-30 and CotG-36 in Fig. 4A), and the 66-kDa form of CotB, whose assembly is known to be CotG dependent (37). Strikingly, these same species also show reduced extractability from ΔsafA mutant spores, as reported before (10, 22) (Fig. 4A). In all, the coat protein profiles for safAD10A and safAN30A spores were very similar to that obtained from ΔsafA spores, while the profiles for safAS11A, safAL12A, and safAI39A spores were close to that of WT spores (Fig. 4A). Thus, the Ala substitutions that affect the localization of SafA-YFP more strongly and early in morphogenesis also cause a more drastic effect on the composition of the coat layers.
FIG 4.
Substitutions in the LysM domain affect the composition and structure of spores. (A) Spores were purified by density gradient centrifugation, and the coat proteins were extracted and resolved by SDS-PAGE. The gel was stained with Coomassie brilliant blue R-250. Spores analyzed were from the WT, a ΔsafA in-frame deletion mutant, and derivatives of the ΔsafA mutant expressing alleles of safA coding for proteins with the indicated substitutions (color code as in Fig. 2) from the amyE locus. The proteins indicated with red arrows show decreased extractability from spores of the ΔsafA, D10A, and N30A mutant strains. The positions of molecular weight (MW) markers, in kilodaltons, are shown on the left side of the panel. (B) The same spores as in panel A were processed for analysis by transmission electron microscopy. Shown are representative specimens for the indicated strains. Cr, spore core; Cx, cortex; Ic, inner coat; Oc, outer coat. Red arrows, inner coat; blue arrows, outer coat; brown arrows, the space between the cortex and inner coat; green arrows, the space between the inner and outer coat; yellow arrows, partially unstructured material that accumulates at the inner edge of the inner coat. Scale bar = 0.2 µm.
The resemblance of the coat protein profiles of safAD10A and safAN30A spores to that of a safA mutant prompted us to examine the ultrastructure of spores by thin-sectioning transmission electron microscopy (TEM). WT spores showed a well-defined lamellar inner coat (Ic) (Fig. 4B, red arrow) in tight connection with the underlying cortex (Cx), and a striated electron-dense outer coat (Oc) closely adherent to the inner coat (Fig. 4B, blue arrow). Spores of a safA insertional mutant show a less electron-dense and thinner outer coat and an additional layer surrounding the outer coat, which we suspect may be a nonadherent crust layer (see Fig. 8 in reference 11). Two other features are important, as follows: first, the cortex is not adjacent to the inner coat, and the inner coat itself is thinner and often did not show the usual lamellar organization (11). Spores of the ΔsafA in-frame mutant share some of these features; both the inner (Fig. 4B, red arrow) and the outer coat (Fig. 4B, blue arrow) appeared thinner and lacked the normal pattern of closely apposed lamellae or striations. Note, for example, that lamellae are seen in the inner coat region, but they are not in close contact (Fig. 4B, red arrows). In addition, neither the outer coat appeared to adhere to the inner coat (Fig. 4B, blue arrow), nor did the inner coat adhere to the cortex (Fig. 4B, brown arrow pointing to a gap between the two layers). Finally, electron-dense unstructured material accumulated in the region between the cortex and the inner coat, associated with the interior edge of the inner coat (Fig. 4B, yellow arrow).
In spores of all of the LysM mutants, the inner and outer coat regions did not adhere (Fig. 4B, green arrows pointing to a gap between the inner and outer coat). Distinctive features of safAD10A and safAN30A spores were the lack of adherence between the cortex and the inner coat (Fig. 4B, gap marked by the brown arrows), with accumulation of electron-dense material at the interior edge of the inner coat, although it was less pronounced than for the ΔsafA mutant spores (yellow arrows). The inner coat showed well-defined lamellae (Fig. 4B, red arrows), but the outer coat appeared thinner (blue arrows), consistent with the reduced representation of the outer coat proteins CotG and CotB from the spore extracts (see above). A distinctive feature of safAS11A, safAL12A, and safAI39A spores was a poorly defined inner coat region (Fig. 4B, red arrows). The inner coat and the cortex, however, were tightly apposed, in contrast to ΔsafA, D10A, and N30A mutant spores (Fig. 4B). The observation that in D10 and N30A spores the cortex and inner coat do not adhere suggests that the presence of SafA in the cortex fraction (see above) is important for this connection and that this localization relies in part on interactions established early during encasement (also see above). The observation that in the S11A, L12A, and I39A changes, which cause a late localization defect, there is a loose inner coat-outer coat connection suggests that a subpopulation of SafA that may reach the spore cortex at a late stage in morphogenesis contributes to this link.
Single alanine substitutions in the LysM domain of SafA differentially affect its interaction with purified spore cortex peptidoglycan.
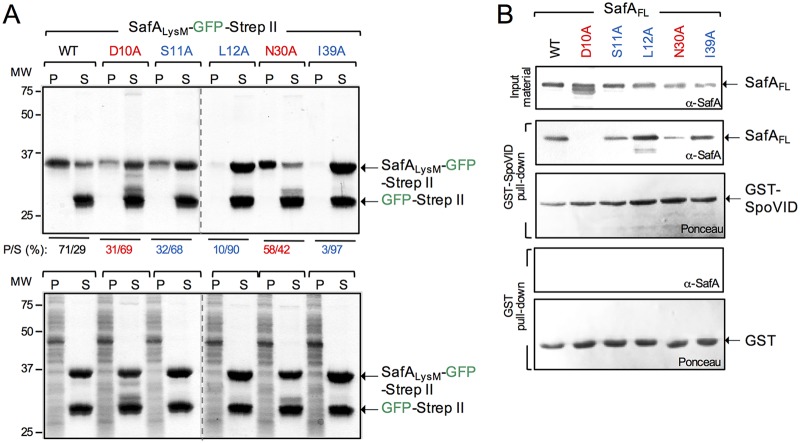
The TEM analysis suggested that at least for the safAD10A and safAN30A mutants, the normal connection between the cortex PG and the inner coat was impaired. Immunogold labeling has shown that SafA localizes at the cortex-inner coat interface in mature spores, and the presence of the LysM domain in the protein suggests that this region of the protein could bind to the cortex (10). Moreover, evidence suggests that it is the C-terminal regions of SafA and SafAC30 that are the main determinants for recruitment of the SafA-dependent proteins. Thus, SafA could act as a molecular staple, linking the cortex and the inner coat (22, 23). If so, the LysM domain could bind to purified cortex, and the D10A and N30A and the other Ala substitutions in the LysM domain herein studied could affect this interaction. To test this idea, we overproduced and partially purified SafALysM-GFP-Strep-tag II (the WT LysM or the domain with the various single Ala substitutions fused to GFP) and GFP-Strep-tag II alone, and we used sedimentation assays to evaluate binding to cortex PG purified from WT spores. SafALysM-GFP-Strep-tag II and GFP-Strep-tag II were mixed together and with purified cortex and incubated, and the mixture was then centrifuged to produce a pellet (P) and a supernatant (S) fraction. The presence of SafALysM-GFP-Strep-tag II in the P or S fraction was revealed by staining the gel with Coomassie brilliant blue R-250 and the P/S ratio determined (see Materials and Methods). In all the assays, GFP-Strep-tag II remained in the supernatant (Fig. 5A). In contrast, 71% of the protein with the WT LysM sequence was found in the pellet (Fig. 5A). The I39A substitution caused the lowest partition into the pellet (3%), followed by the L12A fusion (10%). The remaining fusions showed higher P/S ratios, at 31% for D10A, 32% for S11A, and 58% for N30A (Fig. 5A). None of the fusion proteins were found in the pellet when the incubated mixture was subsequently treated with lysozyme, indicating that their association with the pellet required intact cortex peptidoglycan (Fig. 5A, bottom).
FIG 5.
Substitutions in the LysM domain impair its interaction with peptidoglycan or with SpoVID. (A) SafALysM-GFP-Strep-tag II proteins, WT, and variants with the indicated substitutions were purified along with GFP-Strep-tag II. The SafALysM proteins were mixed with GFP-Strep-tag II included to control for the tags, and with cortex peptidoglycan purified from WT spores. Following incubation, the suspensions were centrifuged and separated into a pellet (P, representing the binding fraction) and a supernatant (S, representing the nonbinding fraction). The gels were stained with Coomassie brilliant blue R-250, and the percentage of SafALysM-GFP-Strep-tag II in the P and S fractions was estimated with ImageJ (shown below the panel). The bottom is a control experiment in which the cortex PG was digested with lysozyme before the proteins were added. The arrows on the right show the position of SafALysM-GFP-Strep-tag II and GFP-Strep-tag II; the positions of molecular weight standards, in kilodaltons, are shown on the left side of the panel. (B) GST-SpoVID pulldown assay. Whole-cell extracts were prepared from E. coli strains producing the forms of SafA with the indicated single amino acid substitutions (color coded as in Fig. 2), GST, or GST-SpoVID. The extracts prepared from the strains producing the various forms of SafA were mixed with the GST-containing (two bottom panels) or GST-SpoVID-containing extracts (two middle panels). Following incubation, proteins were pulled with GST beads. Bound proteins were then eluted and resolved by SDS-PAGE, and the gel was subject to immunoblot analysis with an anti-SafAFL antibody. The membranes were stained with Ponceau red to control for the levels of GST or GST-SpoVID. The presence and levels of the various forms of SafA are shown at the top (input material). The arrows show the positions of SafA, GST, and GST-SpoVID. The positions of molecular weight standards (in kilodaltons) are shown on the left side of the panel.
The I39A and L12A substitutions that caused the largest reduction in the interaction with cortex PG affect the localization of SafA-YFP at a late stage in morphogenesis, whereas D10A and N30A, which arrest encasement, cause the smallest impairment in cortex binding. These results suggest that D10A and N30A affect an interaction required for encasement which does not involve PG binding and that later in morphogenesis, L12A, I39A, and possibly S11A affected an interaction with the spore cortex.
Single alanine substitutions in the LysM domain of SafA differentially affect its ability to interact with SpoVID.
Previous work has shown that region A of SafA, just downstream of the LysM domain, is required for the interaction with SpoVID. Glutathione S-transferase (GST) fusions to progressively shorter fragments of SafA (or internal deletions) were able to pull down SpoVID, as long as the LysM domain and region A were present, but a deletion of region A eliminated the interaction (22). Region A was therefore shown to be essential for the interaction, and a peptide with the sequence of region A interacted with SpoVID (22). It was not tested, however, whether the LysM domain contributed to the interaction in the presence of region A or in the context of SafAFL. Because the interaction of SafA with SpoVID is essential for encasement (22, 25), we wanted to test whether the D10A and N30A substitutions, which block encasement, but not the S11A, L12A, and I39A substitutions, affected the interaction. To test this, we conducted pulldown assays using SafAFL (the WT form or the variants with the various substitution in the LysM domain) overproduced in Escherichia coli and GST-SpoVID. All of the SafAFL forms accumulated in E. coli, as assessed by immunoblotting with anti-SafA antibody (Fig. 5B, top, input material). The WT SafAFL, as well as the L12A and I39A mutants, was pulled down by GST-SpoVID at comparable levels, while the S11A form was pulled down at a slightly lower level (Fig. 5B, middle). In contrast, D10A was not pulled down by GST-SpoVID, whereas the pulldown of N30A was less efficient than for the S11A, L12A, or I39A forms (Fig. 5B, middle). None of the SafAFL forms were pulled down by GST alone, indicating that the interactions detected were specific for the SpoVID moiety of GST-SpoVID (Fig. 5B, bottom).
We infer that the D10A and N30A substitutions impair the interaction of SafAFL with SpoVID, while L12A and I39A, and possibly S11A, do not. Together with the localization phenotype of D10A and N30A mutants (see above), these results establish the role of the LysM domain in an interaction with SpoVID required for encasement. Moreover, results from peptidoglycan-binding assays described above suggest that S11A, L12A, and I39A affect the localization of SafA-YFP at a late stage in morphogenesis, possibly because of an impaired interaction with the spore cortex.
Late localization of SafA is dependent on biogenesis of the cortex.
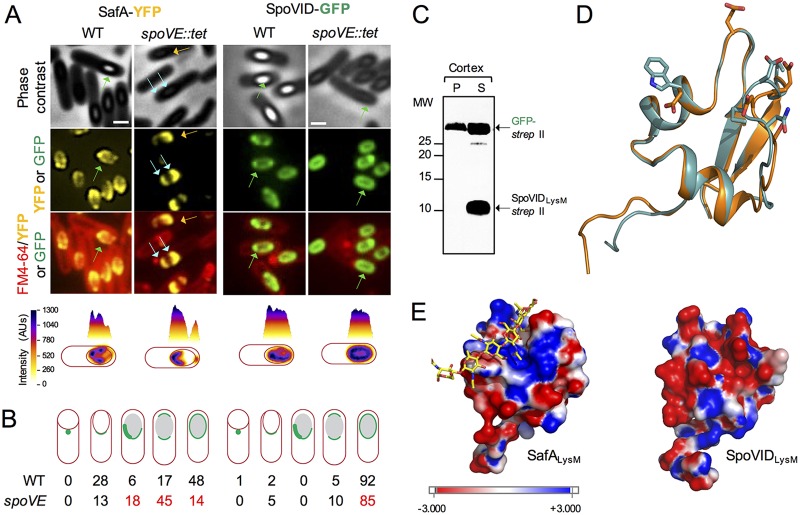
Following engulfment completion, the late mother cell regulator σK is activated. Although several of the key proteins involved in synthesis of the cortex are produced in the mother cell, prior to engulfment completion, under the control of transcription factor σE, it is only under the control of σK that the level of PG precursors reaches a threshold level capable of triggering biogenesis of the cortex (38). Because SafA resides at the cortex-coat interface in mature spores (10) and a fraction of the protein is associated with a cortex fraction (25, 27) (Fig. 3) and because of the mislocalization of the S11A, L12A, and I39A variants of SafA-YFP at a late stage in morphogenesis, when the forespore becomes phase bright, we wanted to test whether the localization of the protein was dependent on assembly of the cortex. We examined the localization of SafA-GFP in cells with a disrupted spoVE gene. spoVE, a σE-controlled gene, codes for a sporulation-specific transglycosylase of the shape, elongation, division, and sporulation (SEDS) family required for synthesis of the cortex (39–43). In spoVE mutants, however, at least some of the coat is still deposited (34, 41). At hour 8 of sporulation in the WT, a time when the phenotype caused by the S11A, L12A, and I39A substitutions is evident (above), SafA-YFP localized as two caps in 17% of the sporangia and completely encircled the forespore in 48% of the sporangia (Fig. 6A and B, left). In spoVE sporangia, however, SafA-YFP encircled the forespore in only 14% of the sporangia, while the two-cap pattern represented 45% of the sporangia (Fig. 6A and B). Moreover, an asymmetric pattern reminiscent of one of the late classes seen for the S11A, L12A, and I39A mutants (Fig. 2) was detected for 18% of the sporangia (only 6% in the WT) (Fig. 6A). Importantly, the localization of SafA-YFP did not differ from the WT during the early hours of sporulation (Fig. S3).
FIG 6.
Localization of SpoVID and SafA in a cortex-less mutant. (A) Localization of SafA-YFP and SpoVID-GFP in WT and spoVE sporangia. Samples were taken from cultures producing SpoVID-GFP or SafA-YFP in either the WT or a congenic spoVE::tet mutant in resuspension medium 8 h after the onset of sporulation (see also Fig. S3 for earlier time points). Cells were stained with FM4-64 and imaged by phase-contrast and fluorescence microscopy. Scale bars = 1 µm. One cell representative of the prevalent localization pattern was chosen to show the distribution of the fluorescence signal (in arbitrary units [AU]) in three-dimensional intensity graphs. Blue arrows, double cap; green arrows, full encirclement; orange arrows, asymmetric localization around the spore. (B) Quantification of SpoVID-GFP or SafA-YFP localization. The percentage of cells showing the localization pattern schematically depicted is shown for the two fusions in the WT and spoVE::tet background. The numbers in red highlight the phenotypes. (C) Binding assay of SpoVIDLysM-Strep-tag II to purified cortex peptidoglycan. SpoVIDLysM-Strep-tag II and GFP-Strep-tag II were partially purified, mixed together, and further mixed with purified cortex peptidoglycan. Following incubation, the mixtures were centrifuged to produce a pellet (P) and a supernatant (S) fraction. Following SDS-PAGE of the P and S samples, the presence of SpoVIDLysM-Strep-tag II or GFP-Strep-tag II in either fraction was assessed by immunoblotting with anti-Strep-tag II antibodies. The position of the relevant species is indicated by arrows, and the positions of molecular weight markers are indicated on the left side of the panels. (D) Comparison of the SpoVID (cyan) and SafA (orange) structures. The model for the LysM domain of SpoVID was based on the crystal structure of the NlpC/P60 d,l-endopeptidase from T. thermophilus (PDB ID 4UZ3) as for SafA (see the legend for Fig. 1). The residues that differ most between the two structures, E9 versus G9, E13 versus W13, D35 versus N35, and D36 versus P36, are highlighted using sticks. (E) Comparison of the electrostatic surface maps of SafA (left) and SpoVID (right). Chitohexose is shown on the SafALysM model, using sticks with carbon atoms colored in yellow, to locate the substrate binding site (Fig. 1).
Thus, the spoVE mutation phenocopied, to some extent, the late localization phenotype of the S11A, L12A, and I39A mutants. Consistent with the PG-binding assays, this suggests that an interaction with the cortex PG involving S11, L12, and I39 is important for the localization of SafA-YFP at a late stage in morphogenesis.
Localization of SpoVID is not dependent on cortex biogenesis, and SpoVIDLysM does not bind to purified cortex.
Since SafA and SpoVID interact during engulfment, and SpoVID also has a LysM domain, we also tested whether SpoVID itself could bind to the spore cortex PG. We investigated this possibility in two ways. First, we examined the localization of SpoVID in WT and spoVE mutant sporangia at hour 8 of sporulation (Fig. 6A). SpoVID-GFP localized as two caps in 5% of the WT sporangia and in 10% of the spoVE mutant sporangia; the fusion protein fully encircled the forespore in 92% of the sporangia in the WT and in 85% of the mutant spoVE sporangia (Fig. 6B). Thus, the localization of SpoVID-GFP was largely independent of formation of the spore cortex.
In addition, we tested whether SpoVIDLysM would bind to purified cortex under the same conditions used in the SafALysM binding assays described above. In these assays, SpoVIDLysM-Strep-tag II and GFP-Strep-tag II fusion proteins were partially purified and mixed with purified cortex, and following incubation, the mixture was centrifuged to produce a pellet and a supernatant fraction. The presence of the proteins in either fraction was monitored with anti-Strep-tag II antibodies. SpoVIDLysM was detected only in the supernatant (Fig. 6C). Thus, under these conditions, we found no interaction of SpoVIDLysM with the cortex. We generated a homology model for SpoVIDLysM using the crystal structure of a LysM domain from the NlpC/P60 d,l-endopeptidase from T. thermophilus, as for SafALysM (30) (see above). As for SafALysM, in Ramachandran plots of the best models, all the residues were located in the most favored regions, supporting the quality of the model (above). Superimposition of the SafALysM and SpoVIDLysM models shows that the overall structure is maintained (Fig. 6D). The surface electrostatic potential maps of SafALysM and SpoVIDLysM, however, show that in SafA, the PG-binding crevice is mainly positively charged (Fig. 6E, left) whereas in SpoVIDLysM, the surface of the cleft is negatively charged (right). This is because residues Gly9, Trp13, and Leu38 in SafA correspond to glutamates in SpoVIDLysM, whereas Pro36 in SafA corresponds to an aspartate in SpoVIDLysM (Fig. 1B). The striking difference in the electrostatic maps of the two proteins may explain why SafA binds PG whereas SpoVIDLysM does not, at least under the conditions used. We note, however, that the replacement of Gly9 and Pro36 may also be relevant, as glycine residues usually confer flexibility, whereas proline imposes secondary structure constraints. In any event, the observation that SpoVIDLysM-Strep-tag II did not bind to PG is consistent with the lack of dependency of SpoVID-GFP localization on cortex biogenesis.
DISCUSSION
SafALysM as a protein-protein and protein-PG interaction module.
Our study unravels the contribution of the LysM domain present in morphogenetic protein SafA to the localization of the protein at different stages of sporulation. We show that SafA has a canonical LysM domain that binds PG. Single Ala substitutions in residues that overlap two sites shown to be involved in chitin/PG/Nod factor binding by LysM domains (29, 31–33) impair the interaction of SafALysM with PG. These residues are D10, S11, and L12 at site 1 and N30 and I39 at site 2 (Fig. 1B), with the S11A, L12A, and I39A substitutions showing the strongest reduction in PG binding. The largest effects on binding of one of the LysM domains from the AtlA autolysin to (GlcN)5 were seen for three residues (T13A, L14A, and I39A) that correspond to S11A, L12A, and I39A in SafALysM, consistent with our results (29) (Fig. 1B).
By analyzing single-amino-acid substitutions in SafALysM, we were able to separate two functions of the protein. SafA is recruited to the spore surface through an interaction with SpoIVA, and at the onset of engulfment, SafA interacts with SpoVID (22). This interaction involves region A in SafA and region E in SpoVID and is essential for encasement by SafA and the SafA-dependent proteins that form the inner coat (22, 25) (Fig. 7A). We now show that at this stage in morphogenesis, SafALysM is also involved in the interaction with SpoVID and that the side chains of D10 and N30 are required for this interaction (Fig. 7A and B). These residues are the ones that showed the smallest contribution for PG binding, suggesting that the PG-binding function of SafALysM may not be required at this stage. SafALysM may function at the onset of engulfment exclusively as a protein-protein interaction module.
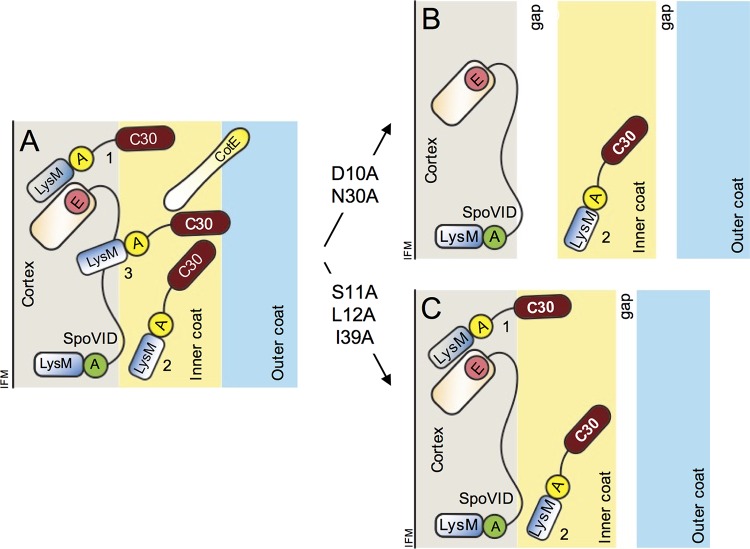
FIG 7.
Role of the LysM domain in spore coat formation. (A) The localization of SafA to the cortex and inner coat relies on its interaction with SpoVID. SafALysM, together with region A (yellow circle), interacts with SpoVID. This interaction, required for encasement, involves residues D10 and N30 in SafALysM and region A. SafA becomes tightly associated with the cortex (1) but also assumes a more peripheral location, close to the edge of the cortex (3). In addition, it is present in the inner coat (2). CotE is found at the inner coat-outer coat interface from where it nucleates assembly of the outer coat. (B) In the D10A and N30 mutants, populations 1 and 3 are missing or greatly reduced and both the cortex-inner coat and inner coat-outer coat interfaces are not properly formed and a gap is seen between the three layers. (C) In the S11A, L12A, or I39A mutants, population 3 is missing. The cortex-inner coat interface is properly formed, but the inner coat-outer coat interface is not. Spores are represented at a late stage in morphogenesis, after synthesis of the spore cortex. At this stage, the forespore outer membrane may no longer exist. SafAC30 is not represented for simplicity, and CotE is omitted from panels B and C also for simplicity. SafAFL, SafAC30, and CotE are thought to polymerize (not represented). The proteins are not drawn to scale. IFM, inner forespore membrane. The diagram is an update of a recent figure (25).
SpoVIDLysM may function only as a protein-protein interaction module.
A LysM domain is also found in the encasement protein SpoVID. Together with a conserved region of 12 residues just upstream, the SpoVIDLysM makes a contribution to an interaction with SpoIVA essential for the targeting of SpoVID to the forespore surface. In fractionation experiments, SpoVID is exclusively detected in the cortex (25). How and when SpoVID reaches the cortex is presently unknown, but it may rely on the interaction with SpoIVA. SipL is an unrelated protein of Clostridium difficile that plays a role analogous to that of SpoVID in coat assembly and carries a LysM domain at its C-terminal end (44). It is not known whether SipLLysM binds to PG, but the domain is involved in an interaction with the C. difficile homolog of SpoIVA (44). Thus, the role of the LysM domain as a protein-protein interaction module is not restricted to SafALysM. Surprisingly, we found that under similar conditions, SpoVIDLysM does not bind to PG. A comparison of the surface charge distributions for SafALysM and SpoVIDLysM suggests that the PG-binding crevice in SpoVIDLysM is more acidic (Fig. 6E), and binding to PG may be influenced by pH, as shown for other LysM domains (21), or by some other factor. Alternatively, SpoVIDLysM may have lost the ability to bind PG and may function exclusively as a protein-protein interaction module.
Four other coat proteins with demonstrated or anticipated roles in cortex degradation during spore germination have LysM domains; they most likely interact with the cortex at least during germination (45–51) (Fig. 1B). YaaH, a SafA-dependent protein (6, 20), carries tandem LysM domains close to its N terminus, and these are sufficient for the coat association of a heterologous protein, β-lactamase (48). Thus, the LysM domain is involved in the localization of other coat proteins. Whether the LysM domain of these proteins functions as a PG- or protein-binding module, or both, is presently unknown. We note, however, that in the D10A and L12A cells scored at the onset of engulfment, YaaH-GFP behaves as a class I protein, of which SpoVID is an example (20) in a small fraction of the sporangia (see Fig. S2 in the supplemental material). It is tempting to speculate that YaaH may bind directly to SpoVID and that the 2 LysM domains at its N terminus may be involved.
SafA as a cortex/inner coat and inner coat/outer coat molecular staple.
We have shown before that the interaction of SafA with SpoVID during engulfment is essential for the localization of SafAFL and SafAC30 to both the cortex and coat (25) (Fig. 7A). In D10A and N30A mutants, SafA is greatly reduced or absent from the cortex fraction but is still detected in the coat (Fig. 3), and in these mutants, the cortex and inner coat fail to adhere (Fig. 4B). This suggests that the localization of SafAFL and SafAC30 to the cortex is important for connecting the cortex and inner coat (Fig. 7A and B, role of SafA population 1). Consistent with this, the cortex and inner coat are tightly adherent in S11A, L12A, and I39A spores, in which at least some SafAFL is still detected in the cortex. Even though these are the substitutions that more strongly affect the interaction with PG, the interaction with SpoVID, unaffected by S11A, L12A, and I39A, seems sufficient to maintain some SafA in the cortex (Fig. 7A and C).
SafA also promotes the connection between the inner and outer coat layers since in spores of all mutants, a gap is between these two layers (Fig. 6 and 7B and C). A role of SafA in outer coat assembly has been noticed since the first articles describing the phenotype of a safA insertional mutant; the abundant CotG protein, a key structural organizer of the outer coat, and the CotG-dependent CotB protein were absent from coat extracts, and TEM images showed a disorganized outer coat (10, 11, 42). Likewise, in D10A and N30A mutant spores, but less so in S11A, L12A, or I39A mutant spores, CotG and CotB are greatly reduced in coat extracts, and the outer coat is disorganized (Fig. 4A; I39A is the exception).
The role of SafA in promoting the inner coat-outer coat connection may be fulfilled by the SafA protein that is associated with the coat. However, the PG-binding ability of SafALysM is important for the localization of SafA at a late stage in sporulation, because (i) the S11A, L12A, and I39A substitutions cause mislocalization of the protein at a late stage in morphogenesis, when the forespore becomes phase bright, a sign that synthesis of the cortex has occurred; (ii) deletion of the gene coding for the SpoVE transglycosylase, essential for synthesis of the spore cortex, largely mimics the effect of the S11A, L12A, and I39A substitutions; (iii) the S11A, L12A, and I39A substitutions are the ones that more strongly impair binding of SafALysM to purified PG. The cortex-associated SafA that is affected by the D10A and N30A substitutions assumes this localization through the interaction with SpoVID (Fig. 7B), whereas the localization defect imposed by S11A, L12A, and I39A may result primarily from an impaired interaction with the cortex. We propose that the D10/N30-dependent population of SafA has a more internal localization than the S11/L12/I39-dependent population (Fig. 7A). Some evidence suggests that the outer forespore membrane is absent in mature spores (52–54), and the more peripheral SafA may reach the cortex when the forespore outer membrane disappears. We speculate that both the coat-associated SafA and the more peripheral S11/L12/I39-dependent cortex-associated SafA allow the protein to come in proximity to the outer coat, contributing to the formation of the inner coat-outer coat interface. By cementing the inner coat-outer coat interface, in turn, SafA indirectly promotes proper assembly of the outer coat and/or maintains the integrity of this layer.
Regardless, our study unravels a new role for SafA, in addition to its well-characterized morphogenetic function as a hub directing assembly of the inner coat proteins, in that SafA promotes the tight adherence of the cortex-inner coat and inner coat-outer coat interfaces.
MATERIALS AND METHODS
Strains and general techniques.
B. subtilis strains used in this study are congenic derivatives of the wild-type strain MB24 (trp2 metC3). Escherichia coli DH5α was used for molecular cloning and for overproduction of Strep-tag II fusion proteins, except for SpoVIDLysM-Strep-tag II, which was expressed in E. coli BL21(DE3). Maintenance and growth experiments for E. coli and B. subtilis were performed in Luria-Bertani medium (LB) with antibiotic selection when needed. Sporulation was induced by resuspension in Sterlini-Mandelstam sporulation medium (SM) (55). All strains used in this study are listed in Table S1 in the supplemental material, and plasmids and oligonucleotide primers required for their construction can be found in Tables S2 and S3, respectively. All the plasmids and strains used in this work are described in the supplemental material.
Fluorescence microscopy and image analysis.
One-milliliter samples were collected from SM cultures at different times. Cells were harvested by centrifugation and resuspended in 0.1 ml of phosphate-buffered saline (PBS), supplemented with 1 µl of a 2-mg/ml solution of the membrane dye FM4-64 (Molecular Probes, Invitrogen). Fluorescence microscopy was performed as previously described (56), and images were analyzed with MetaMorph version 7.7 (Molecular Devices) and ImageJ (http://rsbweb.nih.gov/ij/). For quantification of the subcellular localization, at least 100 sporulating cells were randomly examined and scored. The three-dimensional (3D) graphics of the distribution of the fluorescent signal in sporulating cells were obtained using the ImageJ plug-in Interactive 3D Surface Plot version 2.33.
Accumulation of safA variants during sporulation.
Cultures were grown in defined SM (DSM) at 37°C, and samples were taken at 2, 4, and 6 h after the onset of sporulation. Cells were harvested by centrifugation (7,500 × g, 10 min, at 4°C), resuspended in French press buffer (10 mM Tris [pH 8.0], 10 mM MgCI2, 0.5 mM EDTA, 0.2 M NaCl, 10% glycerol, 0.1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF]), and lysed in a French pressure cell (18,000 lb/in2). The extracts were quantified using the Bio-Rad Mini-PROTEAN system according to the manufacturer’s instructions. Ten micrograms of protein from each sample was prepared for SDS-PAGE, and proteins were resolved, transferred to nitrocellulose membranes, and immunoblotted using anti-SafA antibodies.
Spore purification and fractionation.
Cells were grown in DSM for 24 h after the onset of sporulation, and the produced spores were purified by a two-step gradient of gastrografin (Bayer Schering Pharma) (2, 57). Spore fractioning was performed as described previously (25, 27). Proteins within each spore fraction were resolved by SDS-PAGE and transferred to nitrocellulose membranes for immunoblot analysis with anti-SafA. As a control for the decoating, membranes were stripped with stripping buffer (50 mM Tris [pH 6.8], 2% SDS, 100 mM β-mercaptoethanol) and reprobed with anti-CotA antibodies.
Total coat proteins were extracted by boiling the spores (equivalent to an optical density at 580 nm [OD580] of 2) for 8 min with extraction buffer (10% glycerol, 4% SDS, 10% β-mercaptoethanol, 1 mM DTT, 250 mM Tris [pH 6.8], 0.05% bromophenol blue) and resolved on 15% SDS-PAGE gels (22). The gels were stained with Coomassie brilliant blue R-250.
Cortex isolation and peptidoglycan-binding assays.
Cultures of E. coli producing the various Strep-tag II fusions were grown to mid-log phase, induced with 200 µg/ml anhydrotetracycline, and incubated for 3 to 5 h before harvesting the cells. The pellets corresponding to 50 ml of induced cultures were resuspended in 3 ml of buffer W (100 mM Tris [pH 8.0], 1 mM EDTA) supplemented with 100 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), and cOmplete Mini EDTA-free protease inhibitor cocktail (Roche). Cells were lysed in a French pressure cell (18,000 lb/in2) and the lysates cleared by centrifugation. The Strep-tag II fusion proteins were purified from the lysates using Strep-Tactin–Sepharose columns, according to the manufacturer’s instructions (IBA, GmbH), and finally were quantified by the Bradford method.
For cortex purification, wild-type B. subtilis cells were grown in DSM for 24 h after the onset of sporulation. Spores were purified as described above (2, 57), and the cortex was extracted (58, 59), purified by removal of peptidoglycan-associated polymers, and quantified (60). Peptidoglycan-binding assays were adapted from a study by Yamamoto et al. (60). In short, 1 mg of purified cortex was incubated for 2 h on ice in 40 µl of HEPES buffer, with or without lysozyme (40 µg/ml). Digested and nondigested cortex were washed and resuspended in 30 µl of phosphate-buffered saline with 0.1% Tween 20 (PBS-T). Then, 1 µM purified LysM-(GFP)-Strep-tag II and GFP-Strep-tag II were added. After a 15-min incubation on ice, pellet and supernatant fractions were separated by centrifugation (2 min, 5,000 × g), and the pellets were washed with PBS-T. Proteins within each fraction were resolved by SDS-PAGE and stained with Coomassie brilliant blue R-250. The percentage of SafALysM-GFP-Strep-tag II in the P and S fractions was estimated with ImageJ.
GST pulldown assays.
Cultures of 10 ml (for GST and the SafA variants) or 50 ml (for GST-SpoVID) were grown to an optical density at 600 nm of 0.6 and induced with 1 mM isopropyl-thio-β-d-galactopyranoside (IPTG) for 3 h (for GST and GST-SpoVID) or for 30 min (for SafA variants). Cells were harvested by centrifugation (at 4°C for 10 min at 7,500 × g) and resuspended in 1 ml cold buffer. VPEX-100 buffer (100 mM NaCl, 10 mM Tris [pH 8.0], 1 mM EDTA, 1 mM 2-mercaptoethanol, 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 10% glycerol) (23) was used to resuspend the cells producing GST or GST-SpoVID, while buffer I (PBS containing 0.1% Tween 20 plus 10% glycerol) was used for the resuspension of SafA-producing cells, both supplemented with 1 mM phenylmethylsulfonyl fluoride. Cell lysis was performed in a French pressure cell (18,000 lb/in2). Lysates were cleared by centrifugation (at 4°C for 20 min at 10,000 × g). GST and GST-SpoVID amounts in the soluble fractions were adjusted with VPEX-100 (see above), and the SafA-cleared lysates were adjusted in buffer I. Pulldown assays were performed as previously described (24), except that GST and GST-SpoVID variants were used as baits and SafA was used as prey. Bead fractions were resuspended in SDS protein loading buffer (10% glycerol, 4% SDS, 10% 2-mercaptoethanol, 1 mM DTT, 250 mM Tris [pH 6.8], 0.05% bromophenol blue) and boiled for 5 min, and proteins were resolved on 12.5% SDS-PAGE gels. Gels were transferred to nitrocellulose membranes for immunoblot analysis with anti-SafA antibodies, and the membranes were finally stained with Ponceau S to control for the retention of GST or the GST-SpoVID variants.
Western blot analysis.
Proteins transferred to nitrocellulose membranes were immunoblotted according to the instructions for the SuperSignal West Pico chemiluminescent substrate (Thermo Scientific), using 5% low-fat powder milk in PBS–0.1% Tween 20 (0.001%). Antibodies were used at the following dilutions: anti-SafA (10), 1:15,000; anti-CotA (laboratory stock), 1:1,000; and anti-Strep-tag II (Abcam), 1:1,000. Secondary anti-rabbit and anti-mouse peroxidase-conjugated antibodies (Sigma) were used at the concentrations of 1:5,000 and 1:2,000, respectively.
Transmission electron microscopy.
Spores were collected by centrifugation from cultures 24 h after the onset of sporulation and purified as described above. Spores were fixed and processed for thin-sectioning transmission electron microscopy (TEM), as described before (25).
Homology-based models and calculation of surface electrostatic potential maps.
The structural models of SafA and SpoVIDLysM were generated using the structure of the N-terminal LysM domains from the putative NlpC/P60 d,l-endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose (PDB ID 4UZ3) (30) as the template. The model was built using the software Modeller version 9.6 (61) and setting the refinement degree to slow. The final model corresponds to the one with the lowest value of the objective function out of 20 generated structures. The quality of the models was assessed using PROCHEK (61); Ramachandran plots located all the residues in the most favored or additional allowed regions. Surface electrostatic potential maps of SafALysM and SpoVIDLysM were calculated with the MEAD package (62), where sets of atomic radii and partial charges were taken from the GROMOS 54A7 force field (63).
Supplementary Material
ACKNOWLEDGMENTS
We thank Sérgio Filipe, ITQB NOVA, for helpful discussions and Teresa Costa for the collaboration at the first stages of this work. We acknowledge Erin Tranfield and Ana Sousa of the Electron Microscopy Facility at the Instituto Gulbenkian de Ciência (www.igc.gulbenkian.pt) for technical expertise and sample processing. We thank Ana Henriques for help with the artwork.
This work was financially supported by project LISBOA-01-0145-FEDER-007660 (Microbiologia Molecular, Estrutural e Celular) funded by FEDER funds through COMPETE2020-Programa Operacional Competitividade e Internacionalização (POCI), partially supported by project ONEIDA (LISBOA-01-0145-FEDER-016417) cofunded by Fundos Europeus Estruturais e de Investimento (FEEI) from Programa Operacional Regional Lisboa 2020 and by national funds from the Fundação para a Ciência e a Tecnologia (FCT), and through FCT program IF (IF/00268/2013/CP1173/CT0006) to M.S. We also acknowledge the National Institute of General and Medical Sciences Public Health Services R01 grant GM110000-04 to C.P.M. C.F. (grant SFRH/BD/43200/2008), F.C.P. (grant SFRH/BD/45459/08), and F.N. (grant SFRH/BD/64470/2009) were the recipients of doctoral fellowships from the FCT.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00642-18..
REFERENCES
- 1.Driks A, Eichenberger P. 2016. The spore coat. Microbiol Spectr 4(3):TBS-0023-2016. doi: 10.1128/microbiolspec.TBS-0023-2016. [DOI] [PubMed] [Google Scholar]
- 2.Henriques AO, Moran CP Jr.. 2007. Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol 61:555–588. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- 3.McKenney PT, Driks A, Eichenberger P. 2013. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol 11:33–44. doi: 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Setlow P, Wang S, Li YQ. 2017. Germination of spores of the orders Bacillales and Clostridiales. Annu Rev Microbiol 71:459–477. doi: 10.1146/annurev-micro-090816-093558. [DOI] [PubMed] [Google Scholar]
- 5.Tan IS, Ramamurthi KS. 2014. Spore formation in Bacillus subtilis. Environ Microbiol Rep 6:212–225. doi: 10.1111/1758-2229.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKenney PT, Driks A, Eskandarian HA, Grabowski P, Guberman J, Wang KH, Gitai Z, Eichenberger P. 2010. A distance-weighted interaction map reveals a previously uncharacterized layer of the Bacillus subtilis spore coat. Curr Biol 20:934–938. doi: 10.1016/j.cub.2010.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Popham DL. 2002. Specialized peptidoglycan of the bacterial endospore: the inner wall of the lockbox. Cell Mol Life Sci 59:426–433. doi: 10.1007/s00018-002-8435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beall B, Driks A, Losick R, Moran CP Jr.. 1993. Cloning and characterization of a gene required for assembly of the Bacillus subtilis spore coat. J Bacteriol 175:1705–1716. doi: 10.1128/jb.175.6.1705-1716.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Driks A, Roels S, Beall B, Moran CP Jr, Losick R. 1994. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev 8:234–244. doi: 10.1101/gad.8.2.234. [DOI] [PubMed] [Google Scholar]
- 10.Ozin AJ, Henriques AO, Yi H, Moran CP Jr.. 2000. Morphogenetic proteins SpoVID and SafA form a complex during assembly of the Bacillus subtilis spore coat. J Bacteriol 182:1828–1833. doi: 10.1128/JB.182.7.1828-1833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takamatsu H, Kodama T, Nakayama T, Watabe K. 1999. Characterization of the yrbA gene of Bacillus subtilis, involved in resistance and germination of spores. J Bacteriol 181:4986–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang KH, Isidro AL, Domingues L, Eskandarian HA, McKenney PT, Drew K, Grabowski P, Chua MH, Barry SN, Guan M, Bonneau R, Henriques AO, Eichenberger P. 2009. The coat morphogenetic protein SpoVID is necessary for spore encasement in Bacillus subtilis. Mol Microbiol 74:634–649. doi: 10.1111/j.1365-2958.2009.06886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng LB, Donovan WP, Fitz-James PC, Losick R. 1988. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev 2:1047–1054. doi: 10.1101/gad.2.8.1047. [DOI] [PubMed] [Google Scholar]
- 14.Castaing JP, Nagy A, Anantharaman V, Aravind L, Ramamurthi KS. 2013. ATP hydrolysis by a domain related to translation factor GTPases drives polymerization of a static bacterial morphogenetic protein. Proc Natl Acad Sci U S A 110:E151–E160. doi: 10.1073/pnas.1210554110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramamurthi KS, Clapham KR, Losick R. 2006. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol Microbiol 62:1547–1557. doi: 10.1111/j.1365-2958.2006.05468.x. [DOI] [PubMed] [Google Scholar]
- 16.Ramamurthi KS, Lecuyer S, Stone HA, Losick R. 2009. Geometric cue for protein localization in a bacterium. Science 323:1354–1357. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramamurthi KS, Losick R. 2008. ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol Cell 31:406–414. doi: 10.1016/j.molcel.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullerova D, Krajcikova D, Barak I. 2009. Interactions between Bacillus subtilis early spore coat morphogenetic proteins. FEMS Microbiol Lett 299:74–85. doi: 10.1111/j.1574-6968.2009.01737.x. [DOI] [PubMed] [Google Scholar]
- 19.Qiao H, Krajcikova D, Liu C, Li Y, Wang H, Barak I, Tang J. 2012. The interactions of spore-coat morphogenetic proteins studied by single-molecule recognition force spectroscopy. Chem Asian J 7:725–731. doi: 10.1002/asia.201100795. [DOI] [PubMed] [Google Scholar]
- 20.McKenney PT, Eichenberger P. 2012. Dynamics of spore coat morphogenesis in Bacillus subtilis. Mol Microbiol 83:245–260. doi: 10.1111/j.1365-2958.2011.07936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buist G, Steen A, Kok J, Kuipers OP. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol 68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 22.Costa T, Isidro AL, Moran CP Jr, Henriques AO. 2006. Interaction between coat morphogenetic proteins SafA and SpoVID. J Bacteriol 188:7731–7741. doi: 10.1128/JB.00761-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozin AJ, Samford CS, Henriques AO, Moran CP Jr.. 2001. SpoVID guides SafA to the spore coat in Bacillus subtilis. J Bacteriol 183:3041–3049. doi: 10.1128/JB.183.10.3041-3049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Francesco M, Jacobs JZ, Nunes F, Serrano M, McKenney PT, Chua MH, Henriques AO, Eichenberger P. 2012. Physical interaction between coat morphogenetic proteins SpoVID and CotE is necessary for spore encasement in Bacillus subtilis. J Bacteriol 194:4941–4950. doi: 10.1128/JB.00914-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nunes F, Fernandes C, Freitas C, Marini E, Serrano M, Moran CP Jr, Eichenberger P, Henriques AO. 2018. SpoVID functions as a non-competitive hub that connects the modules for assembly of the inner and outer spore coat layers in Bacillus subtilis. Mol Microbiol doi: 10.1111/mmi.14116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozin AJ, Costa T, Henriques AO, Moran CP Jr.. 2001. Alternative translation initiation produces a short form of a spore coat protein in Bacillus subtilis. J Bacteriol 183:2032–2040. doi: 10.1128/JB.183.6.2032-2040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandes CG, Moran CP Jr, Henriques AO. 2018. Auto-regulation of SafA assembly through recruitment of a protein cross-linking enzyme. J Bacteriol 200:e00066-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bateman A, Bycroft M. 2000. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J Mol Biol 299:1113–1119. doi: 10.1006/jmbi.2000.3778. [DOI] [PubMed] [Google Scholar]
- 29.Mesnage S, Dellarole M, Baxter NJ, Rouget JB, Dimitrov JD, Wang N, Fujimoto Y, Hounslow AM, Lacroix-Desmazes S, Fukase K, Foster SJ, Williamson MP. 2014. Molecular basis for bacterial peptidoglycan recognition by LysM domains. Nat Commun 5:4269. doi: 10.1038/ncomms5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong JE, Midtgaard SR, Gysel K, Thygesen MB, Sorensen KK, Jensen KJ, Stougaard J, Thirup S, Blaise M. 2015. An intermolecular binding mechanism involving multiple LysM domains mediates carbohydrate recognition by an endopeptidase. Acta Crystallogr D Biol Crystallogr 71:592–605. doi: 10.1107/S139900471402793X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu T, Liu Z, Song C, Hu Y, Han Z, She J, Fan F, Wang J, Jin C, Chang J, Zhou JM, Chai J. 2012. Chitin-induced dimerization activates a plant immune receptor. Science 336:1160–1164. doi: 10.1126/science.1218867. [DOI] [PubMed] [Google Scholar]
- 32.Ohnuma T, Onaga S, Murata K, Taira T, Katoh E. 2008. LysM domains from Pteris ryukyuensis chitinase-A: a stability study and characterization of the chitin-binding site. J Biol Chem 283:5178–5187. doi: 10.1074/jbc.M707156200. [DOI] [PubMed] [Google Scholar]
- 33.Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EM, Albrektsen AS, James EK, Thirup S, Stougaard J. 2007. LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J 26:3923–3935. doi: 10.1038/sj.emboj.7601826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piggot PJ, Coote JG. 1976. Genetic aspects of bacterial endospore formation. Bacteriol Rev 40:908–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donovan W, Zheng LB, Sandman K, Losick R. 1987. Genes encoding spore coat polypeptides from Bacillus subtilis. J Mol Biol 196:1–10. doi: 10.1016/0022-2836(87)90506-7. [DOI] [PubMed] [Google Scholar]
- 36.Martins LO, Soares CM, Pereira MM, Teixeira M, Costa T, Jones GH, Henriques AO. 2002. Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J Biol Chem 277:18849–18859. doi: 10.1074/jbc.M200827200. [DOI] [PubMed] [Google Scholar]
- 37.Zilhão R, Serrano M, Isticato R, Ricca E, Moran CP Jr, Henriques AO. 2004. Interactions among CotB, CotG, and CotH during assembly of the Bacillus subtilis spore coat. J Bacteriol 186:1110–1119. doi: 10.1128/JB.186.4.1110-1119.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vasudevan P, Weaver A, Reichert ED, Linnstaedt SD, Popham DL. 2007. Spore cortex formation in Bacillus subtilis is regulated by accumulation of peptidoglycan precursors under the control of sigma K. Mol Microbiol 65:1582–1594. doi: 10.1111/j.1365-2958.2007.05896.x. [DOI] [PubMed] [Google Scholar]
- 39.Daniel RA, Drake S, Buchanan CE, Scholle R, Errington J. 1994. The Bacillus subtilis spoVD gene encodes a mother-cell-specific penicillin-binding protein required for spore morphogenesis. J Mol Biol 235:209–220. doi: 10.1016/S0022-2836(05)80027-0. [DOI] [PubMed] [Google Scholar]
- 40.Fay A, Meyer P, Dworkin J. 2010. Interactions between late-acting proteins required for peptidoglycan synthesis during sporulation. J Mol Biol 399:547–561. doi: 10.1016/j.jmb.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henriques AO, de Lencastre H, Piggot PJ. 1992. A Bacillus subtilis morphogene cluster that includes spoVE is homologous to the mra region of Escherichia coli. Biochimie 74:735–748. doi: 10.1016/0300-9084(92)90146-6. [DOI] [PubMed] [Google Scholar]
- 42.Henriques AO, Glaser P, Piggot PJ, Moran CP Jr.. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol Microbiol 28:235–247. doi: 10.1046/j.1365-2958.1998.00766.x. [DOI] [PubMed] [Google Scholar]
- 43.Real G, Fay A, Eldar A, Pinto SM, Henriques AO, Dworkin J. 2008. Determinants for the subcellular localization and function of a nonessential SEDS protein. J Bacteriol 190:363–376. doi: 10.1128/JB.01482-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Putnam EE, Nock AM, Lawley TD, Shen A. 2013. SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins. J Bacteriol 195:1214–1225. doi: 10.1128/JB.02181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, Ferguson C, Haga K, Sato T, Liu JS, Losick R. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol 2:e328. doi: 10.1371/journal.pbio.0020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eichenberger P, Jensen ST, Conlon EM, van Ooij C, Silvaggi J, Gonzalez-Pastor JE, Fujita M, Ben-Yehuda S, Stragier P, Liu JS, Losick R. 2003. The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis. J Mol Biol 327:945–972. doi: 10.1016/S0022-2836(03)00205-5. [DOI] [PubMed] [Google Scholar]
- 47.Imamura D, Kuwana R, Takamatsu H, Watabe K. 2010. Localization of proteins to different layers and regions of Bacillus subtilis spore coats. J Bacteriol 192:518–524. doi: 10.1128/JB.01103-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kodama T, Takamatsu H, Asai K, Kobayashi K, Ogasawara N, Watabe K. 1999. The Bacillus subtilis yaaH gene is transcribed by SigE RNA polymerase during sporulation, and its product is involved in germination of spores. J Bacteriol 181:4584–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lambert EA, Popham DL. 2008. The Bacillus anthracis SleL (YaaH) protein is an N-acetylglucosaminidase involved in spore cortex depolymerization. J Bacteriol 190:7601–7607. doi: 10.1128/JB.01054-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lambert EA, Sherry N, Popham DL. 2012. In vitro and in vivo analyses of the Bacillus anthracis spore cortex lytic protein SleL. Microbiology 158:1359–1368. doi: 10.1099/mic.0.056630-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steil L, Serrano M, Henriques AO, Volker U. 2005. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology 151:399–420. doi: 10.1099/mic.0.27493-0. [DOI] [PubMed] [Google Scholar]
- 52.Ghosh S, Setlow B, Wahome PG, Cowan AE, Plomp M, Malkin AJ, Setlow P. 2008. Characterization of spores of Bacillus subtilis that lack most coat layers. J Bacteriol 190:6741–6748. doi: 10.1128/JB.00896-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leggett MJ, McDonnell G, Denyer SP, Setlow P, Maillard JY. 2012. Bacterial spore structures and their protective role in biocide resistance. J Appl Microbiol 113:485–498. doi: 10.1111/j.1365-2672.2012.05336.x. [DOI] [PubMed] [Google Scholar]
- 54.Setlow P. 2014. Spore resistance properties. Microbiol Spectrum 2(5):TBS-0003-2012. doi: 10.1128/microbiolspec.TBS-0003-2012. [DOI] [PubMed] [Google Scholar]
- 55.Sterlini JM, Mandelstam J. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J 113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Serrano M, Real G, Santos J, Carneiro J, Moran CP Jr, Henriques AO. 2011. A negative feedback loop that limits the ectopic activation of a cell type-specific sporulation sigma factor of Bacillus subtilis. PLoS Genet 7:e1002220. doi: 10.1371/journal.pgen.1002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seyler RW Jr, Henriques AO, Ozin AJ, Moran CP Jr.. 1997. Assembly and interactions of cotJ-encoded proteins, constituents of the inner layers of the Bacillus subtilis spore coat. Mol Microbiol 25:955–966. doi: 10.1111/j.1365-2958.1997.mmi532.x. [DOI] [PubMed] [Google Scholar]
- 58.Fein JE, Rogers HJ. 1976. Autolytic enzyme-deficient mutants of Bacillus subtilis 168. J Bacteriol 127:1427–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuroda A, Sekiguchi J. 1990. Cloning, sequencing and genetic mapping of a Bacillus subtilis cell wall hydrolase gene. J Gen Microbiol 136:2209–2216. doi: 10.1099/00221287-136-11-2209. [DOI] [PubMed] [Google Scholar]
- 60.Yamamoto H, Miyake Y, Hisaoka M, Kurosawa S, Sekiguchi J. 2008. The major and minor wall teichoic acids prevent the sidewall localization of vegetative dl-endopeptidase LytF in Bacillus subtilis. Mol Microbiol 70:297–310. doi: 10.1111/j.1365-2958.2008.06397.x. [DOI] [PubMed] [Google Scholar]
- 61.Sali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 62.Bashford D. 1997. An object-oriented programming suite for electrostatic effects in biological molecules, p 233–240. In Ishikawa YO, Reynders RR, Tholburn MJVW (ed), Scientific computing in object-oriented parallel environments. Springer, Berlin, Germany. [Google Scholar]
- 63.Schmid N, Eichenberger AP, Choutko A, Riniker S, Winger M, Mark AE, van Gunsteren WF. 2011. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur Biophys J 40:843–856. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.