Abstract
Objective
We describe a stratified sampling design that combines electronic health records (EHRs) and United States Census (USC) data to construct the sampling frame and an algorithm to enrich the sample with individuals belonging to rarer strata.
Materials and Methods
This design was developed for a multi-site survey that sought to examine patient concerns about and barriers to participating in research studies, especially among under-studied populations (eg, minorities, low educational attainment). We defined sampling strata by cross-tabulating several socio-demographic variables obtained from EHR and augmented with census-block-level USC data. We oversampled rarer and historically underrepresented subpopulations.
Results
The sampling strategy, which included USC-supplemented EHR data, led to a far more diverse sample than would have been expected under random sampling (eg, 3-, 8-, 7-, and 12-fold increase in African Americans, Asians, Hispanics and those with less than a high school degree, respectively). We observed that our EHR data tended to misclassify minority races more often than majority races, and that non-majority races, Latino ethnicity, younger adult age, lower education, and urban/suburban living were each associated with lower response rates to the mailed surveys.
Discussion
We observed substantial enrichment from rarer subpopulations. The magnitude of the enrichment depends on the accuracy of the variables that define the sampling strata and the overall response rate.
Conclusion
EHR and USC data may be used to define sampling strata that in turn may be used to enrich the final study sample. This design may be of particular interest for studies of rarer and understudied populations.
Keywords: enrichment sampling, electronic health records, census data
OBJECTIVE
We describe an enrichment-motivated stratified sampling design. We combine electronic health records (EHRs) and United States Census (USC) data to construct sampling strata, and we detail a sampling algorithm that identifies a sample enriched with rarer and underrepresented subpopulations.
BACKGROUND AND SIGNIFICANCE
The United States healthcare system has become more reliant on health information technology and active data collection due in part to the Health Information Technology for Economic and Clinical Health Act of 2009 (HITECH). This Act provides financial incentives to institutions that are implementing and promoting the “meaningful use” of EHR data. As the amount of EHR data proliferates, nationwide efforts (eg, Project HealthDesign) have been initiated to generate novel secondary uses of EHR data to improve public health.1,2 These data are used to reevaluate prior research findings; to develop, assess, and refine predictive models; to aid in the planning of epidemiological and survey studies; and, combined with biorepositories, to understand complex genotype and phenotype relationships.3
To date, research derived from biorepositories is primarily based on individuals of northern European ancestry. To engage more diverse populations in genomic research, surveying under-studied populations is needed to better understand concerns about and barriers to participating in research studies. Such surveys are typically extremely resource intensive, unless one can create a sampling frame with well-defined sampling strata based on demographics and other data.4 Defining such a sampling frame from EHRs is possible since recipients of HITECH funds are required to collect standardized demographic data that may be associated with health disparities.5 The quality of the resulting sampling frame and specifically the variables comprising the sampling strata are dependent on the accuracy and completeness of each institution’s EHR system and may not be sufficient for certain research questions (eg, coarseness of racial/ethnic groups).5–8
In this paper, we describe the study design that we used for the Electronic Medical Records and Genomic (eMERGE) Network’s survey of perspectives on broad consent and data sharing in biomedical research.9 An aim of this multi-site survey was to ensure that under-studied populations were adequately represented (eg, minorities and those from rural areas). Towards this end, we describe how we combined EHR data and USC data to construct sampling strata and the algorithm we used to sample from these strata to maximize diversity of the sample. We also report response rates and the agreement between EHR/USC-defined variables and survey responses.
MATERIALS AND METHODS
Population and data sources
The eMERGE Network was initiated by the National Human Genome Research Institute to develop, disseminate, and apply approaches to research that combines DNA biorepositories with EHR systems for large-scale, high-throughput genetic research.10 The Consent, Education, Regulation and Consultation (CERC) Working Group was commissioned to conduct a broad-based survey on the acceptability of and barriers to broad consent and data sharing for genomics research, especially among those with low socioeconomic status, low education, and rural residence, and younger adults and ethnic and racial minorities.11 Among the eMERGE Network’s 11 U.S. clinical centers, this survey was administered to 7 sites that sampled from their adult patient population, 3 sites that sampled from their pediatric patient population only, and 1 site that sampled from both its adult and pediatric populations. Patients who had an inpatient or outpatient encounter between October 1, 2013, and September 30, 2014, and were not known to be deceased, whose address was geocodable (see “Linking EHR and USC data”), and whose age and gender were available in the EHR were eligible for sampling. Overall, the sampling frame consisted of approximately 2.4 million individuals. Additional details regarding the organizational challenges, including the meeting of human subject guidelines (ie, institution-specific IRB approval), of developing and implementing a national survey have been published previously.9 The completeness of the sociodemographic variables used to define sampling strata within each site’s EHR varied greatly. When EHR data were not available, USC based estimates were used. The following subsections describe the EHR and USC data sources and the process of merging the datasets to create the variables needed to define sampling strata.
EHR data
Table 1 summarizes the EHR data, including percentage of missing data, summarized by population (adult, pediatric) and by site. Within adult sites, the median patient age was 52 years. Fifty-eight percent were female, 87% were white, and 4% were Hispanic/Latino. At pediatric sites, the median age was 8 years, and a majority was male (52%), white (66%), and not Hispanic/Latino (93%). Race and ethnicity were missing from 14% and 16% of adult EHR records, respectively, and from 13% and 12% of pediatric EHR records. We observed substantial site-to-site variability in the availability of race and ethnicity data with values ranging from 67% to 99%.
Table 1.
Marginal distributions of age, gender, race, and ethnicity by population and by site. Age is summarized at the 5th, 25th, 50th (median), 75th, and 95th percentiles, while gender, race, and ethnicity are summarized as the percentages with complete data, along with additional columns summarizing the percentage missing. Age and gender were complete by design
| Age |
Gender |
Race |
Ethnicity |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Q5-Q25-Q50-Q75-Q95 | Female | Missing | White | Black | Asian | AI /AN | NH / PI | Other | Missing | Hispanic/ Latino | |
| Population | ||||||||||||
| Adult | 1787 295 | 22-36-52-65-82 | 58.3 | 14.1 | 87.1 | 5.7 | 2.4 | 0.7 | 0.3 | 3.8 | 15.9* | 4.1 |
| Pediatric | 601 867 | 1-4-8-13-17 | 47.6 | 13.1 | 66.0 | 19.3 | 2.9 | 0.2 | 0.1 | 11.6 | 11.8 | 6.7 |
| Site | ||||||||||||
| Adult | ||||||||||||
| Essentia Institute for Rural Health | 243 092 | 21-35-53-66-84 | 56.7 | 1.0 | 94.8 | 1.1 | 0.4 | 2.1 | 0.1 | 1.5 | 1.3 | 0.9 |
| Kaiser Permanente Washington | 217 959 | 22-35-51-63-78 | 58.3 | 30.1 | 78.3 | 5.5 | 9.7 | 2.0 | 1.3 | 3.1 | 3.0 | 5.3 |
| Geisinger | 356 488 | 22-36-52-66-82 | 58.0 | 3.9 | 96.3 | 2.6 | 0.6 | 0.1 | 0.3 | <0.1 | 7.3 | 2.9 |
| Mayo Clinic | 134 212 | 23-41-57-69-83 | 53.2 | 3.4 | 93.5 | 2.0 | 1.9 | 0.4 | 0.1 | 2.1 | 10.1 | 2.1 |
| Marshfield Clinic | 136 391 | 21-35-52-66-83 | 54.6 | 8.4 | 97.2 | 0.5 | 1.4 | 0.8 | 0.1 | <0.1 | 9.1 | 1.8 |
| Mount Sinai School Medicine | 162 927 | 23-39-54-67-83 | 59.9 | 30.8 | 60.8 | 11.8 | 4.7 | 0.2 | 0.1 | 22.4 | 32.8* | 26.1 |
| Northwestern University | 206 554 | 23-35-47-60-77 | 62.6 | 21.1 | 70.9 | 13.2 | 3.6 | 0.2 | 0.1 | 12.0 | 22.8 | 9.9 |
| Vanderbilt Medical Center | 329 672 | 21-36-52-66-81 | 59.7 | 18.3 | 86.6 | 10.7 | 1.4 | 0.2 | 0.1 | 0.9 | 19.0 | 2.4 |
| Pediatric | ||||||||||||
| Boston Children’s Hospital | 140 304 | 1-4-8-13-17 | 47.4 | 21.5 | 67.5 | 10.0 | 4.2 | 0.2 | 0.1 | 17.9 | 19.9 | 6.8 |
| Cincinnati Children’s Medical Center | 143 994 | 1-3-8-12-16 | 48.4 | 11.3 | 75.8 | 18.5 | 1.7 | 0.1 | 0.1 | 3.9 | 6.2 | 4.5 |
| Children’s Hospital of Philadelphia | 209 755 | 1-4-8-13-17 | 47.6 | 1.0 | 55.3 | 24.7 | 3.1 | 0.1 | 0.1 | 16.7 | 3.0 | 7.0 |
| Vanderbilt Children’s Hospital | 107 814 | 1-3-8-13-16 | 46.6 | 28.1 | 76.0 | 19.1 | 2.5 | 0.3 | 0.1 | 1.9 | 25.6 | 9.6 |
AI/AN = American Indian/Alaska Native, NH/PI = Native Hawaiian/Pacific Islander. *The original file used for sampling at Mt. Sinai contained 70.2% missing ethnicity value, which resulted in the overall adult population having 19.3% missing ethnicity. Sampling was performed using the original data file, while the corrected results are reported in this table.
USC data
Populations with low educational attainment and with rural residences have been understudied in prior research, and data fields that capture these characteristics were not available in any of the EHR systems. For these 2 fields, and for missing values in EHR records, we exploited U.S. Census Bureau data to provide proxy values. For instance, rural residence as determined by the 2010 Census urban areas criteria can be accurately assigned (assuming patients’ addresses in the EHR are accurate), and educational attainment can be estimated by assigning the most-frequent (mode) value from the patients’ census block group. The U.S. Census Bureau administers several surveys each year, in addition to the Decennial Census. This includes the American Community Survey (ACS), an ongoing nationwide program that collects sociodemographic and economic information about the U.S. population.12,13Table 2 describes the USC sources, variable definitions, and transformations used to complete race and ethnicity when missing from the EHR, and for education and rural/(sub)urban living. Household-level USC data were not used (not publicly available), and no attempt was made to individually re-identify USC respondents using patient data.
Table 2.
U.S. Census variables, sources, definitions, and transformations used for imputing missing stratification information
| Stratification Variable | U.S. Census Variable | Source (table, variable names and or numbers) | Description | Variable Transformation |
|---|---|---|---|---|
| Race/Ethnicity | Hispanic or Latino Origin by Race | ACS 2008–2012 5-year summary file (B03002; 001-021) | Number overall and of each race (White alone, Black or African American alone, American Indian / Alaska Native alone, Asian alone, Native Hawaiian / Other Pacific Islander, Some other race alone, Two or more races, White alone not Hispanic or Latino, Hispanic or Latino, Two races including some other race, two races excluding some other race / three or more races) by ethnicity (Hispanic, not Hispanic). | Marginal distributions of race were defined as White (003, 013), Black or African American (004, 014), Asian (006, 016), American Indian/Alaska Native (005, 015), Native Hawaiian/Pacific Islander (007, 017), Other (008, 009, 018, 019). Marginal distributions of ethnicity were defined as: Not Hispanic/Latino (002), Hispanic/Latino (012) |
| Education | Sex by educational attainment for the population 25 years and over | ACS 2008-2012 5-year summary file (B15002; 001-035) | Number of each educational attainment group (no schooling, nursery to fourth grade, 5th and 6th, 7th -8th, 9th, 10th, 11th, 12th with no diploma, HS grad/GED/Alternative, some college less than 1 year, some college one or more years and no degree, associate’s degree, bachelor’s degree, master’s degree, professional school degree, doctorate) by gender for those who are 25 or older. | Marginal distributions of education were defined as: <12 (003–010, 020–027), 12 − <16 (011–014, 028–031), ≥ 16 (015–018, 032–035). |
| Rurality | LSAD10 | 2010 Census urban area criteria | 75=urbanized area (50 000 or more), 76=urban cluster (2500 to 50 000), missing=rural. | 75 or 76 (suburban/urban), missing (rural) |
Linking EHR and USC data
To use USC data, we linked home addresses to USC geographical identifiers. Address processing involved cleaning address fields, such as the primary street address, city, state, and zip code, and applying quality control checks. Processed addresses were geocoded using specialized software, such as ArcGIS and R.14,15 Each of the 11 eMERGE sites participating in the study had its own IRB protocol for the eMERGE CERC survey study, which was approved at each site and included permission to conduct the geocoding. Seven of the 11 sites geocoded their own addresses, and thus personal health information (PHI) for the geocoding did not leave the site and no PHI was shared with the coordinating center (CC). For the 4 centers that required geocoding assistance, a file containing only the physical address fields and an anonymized patient identifier was electronically transmitted from the site to the CC using the site’s preferred secure file transfer system. Geocoded addresses were then linked to USC block group geographical identifiers, which are the most granular identifiers found in USC datasets, using specialized state-specific files and software. The CC managed and curated data obtained from the 2008–2012 ACS summary tables and the 2010 urban areas database and then distributed the data to all sites for merging with the site-specific EHR data. All electronic file transfers from the CC to each center utilized an electronic file transfer system approved by Vanderbilt Medical Center. No PHI, including the physical addresses, was stored at the CC.
Imputing missing EHR data with USC data
To identify the sampling frame, we “filled in” (ie, imputed) missing EHR data (race and ethnicity) using the most-frequent (mode) value from the patient’s census block group. We also used the mode of educational attainment in the census block group as a proxy for individuals’ education. Notably, we imputed data once to define sampling strata, but did not use imputed data (beyond calculating the true sampling weights) for analyses. This is in contrast to more common settings in which multiple imputation serves to fill in missing data used for analysis and to acknowledge uncertainty in the imputed values.
We recognize that defining sampling strata using EHR and USC data may result in misclassifications, and the frequency of these misclassifications is dependent on the accuracy of the EHR and USC data. Misclassification rates may be quantified by comparing the information used to define sampling strata to survey responses using diagnostic summaries such as sensitivity, specificity, and positive and negative predictive values. Even with the sampling strata misclassifications caused by imperfect data sources, we show that including them can result in a far more diverse sample compared to random sampling. Further, by supplementing missing EHR data with USC data, we are able to conduct stratified sampling without removing individuals with missing data (which may result in selection bias).
Sampling scheme
We conducted a disproportionate stratified sampling scheme to identify the sample. Using the combined EHR and USC data, we defined sampling strata at the adult sites based on the cross-classification age (< 35 and ≥35 years), gender, race (white, black or African American, Asian, Native American/Alaska Native, Hawaiian/Pacific Islander, Other), ethnicity (Hispanic or not), educational attainment (less than high school, high school degree or some college, and at least a bachelor’s degree), and rural living (suburban/urban, rural). Patient data from pediatric sites were surrogates for their parents. That is, we sampled the parents from strata defined by the demographics of the child. We defined sampling strata similarly at pediatric sites, except the age variable was categorized as <12 and ≥12 years. These categories were determined using results from an extensive literature review conducted by our team that showed the extent to which some subpopulations are underrepresented in biorepository-derived research and based on the scientific questions of interest.11 The cross-classification of the 6 sampling variables resulted in 288 possible strata, although not all were observed at all sites.
Maximum entropy sampling algorithm
We conducted the sampling design to increase the diversity of those observed compared to the population at each site. Shannon’s entropy, which corresponds to the uncertainty of predicting an individual's sampling stratum, was used to quantify diversity.16 It is defined as H = − where pi denotes the probability of randomly selecting sampling stratum i. For a fixed sample size and for s possible sampling strata, entropy values range from 0 to log2(s) and correspond to the extreme scenarios in which all individuals belong to the same stratum (H = 0) or where individuals are divided equally across all strata [ie, assuming equal numbers of subjects were available from each stratum; H=log2(s)]. We consider only the situation in which the sample size is fixed (eg, due to budgetary constraints), but summarize the relationship between varying sample sizes and entropy in the Supplementary Figure S1. To enrich our final sample with individuals from strata that tended to have small counts, we implemented a maximum entropy sampling (MES) algorithm. The MES algorithm iteratively determines the number of subjects to sample from each stratum so the desired sample size is obtained and the overall entropy is maximized. That is, MES seeks to sample as evenly as possible across strata under the constraints of overall desired sample size and the individual stratum sizes. Once the desired MES stratum counts were calculated, we implemented the sampling procedure with sampling probabilities defined as the ratio of the MES determined sample size for the stratum divided by the stratum size. Within each stratum, sampling preference was given to those with complete (not imputed) stratification information. Finally, we compare this sampling approach to random sampling (RS) to characterize the extent to which the proposed design (which includes both defining the sampling strata using EHR+USC data, and usage of the MES algorithm) improves diversity of those sampled.
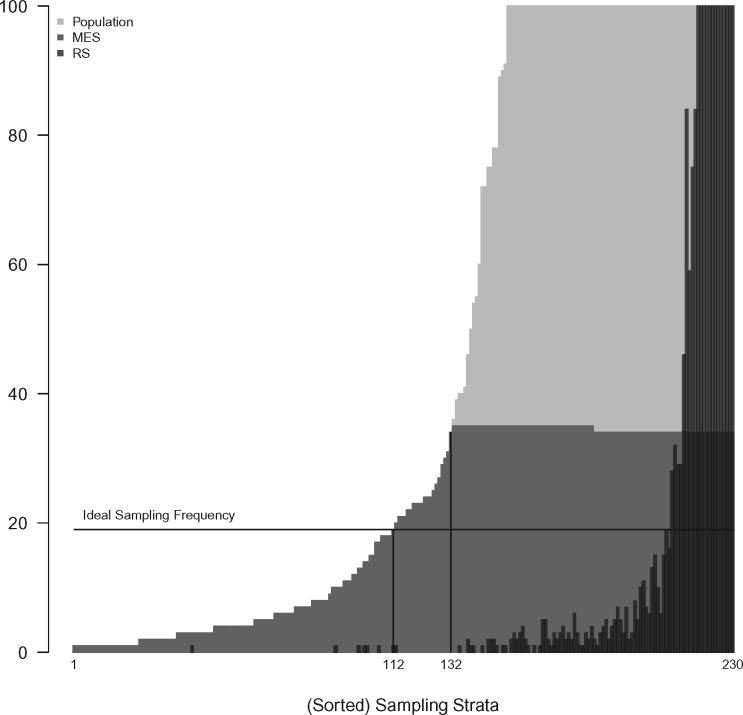
Figure 1 describes the MES algorithm at one of the participating sites, ie, Vanderbilt University Medical Center (VUMC), where 4500 adults were sampled from a population of 329 672. Among the 288 total possible sampling strata, 230 were populated with at least 1 patient. The per stratum frequency in the population is denoted by the light-gray shaded region. Due to the severe skewness of this distribution (ie, stratum sizes ranged from 1 to over 70K with 58% of patients belonging to the top 5 sampling strata), the y-axis of this plot is truncated at 100. When sampling 4500 patients from 230 strata, Shannon entropy is maximized if 19 (4500/230) were sampled from each stratum (see ideal sampling frequency line). However, only 118 strata contained more than 19 patients. To maximize entropy under stratum size constraints, all patients were sampled from the smallest 132 strata, and 34 or 35 patients were sampled from the 98 strata with at least 35 patients. To contrast with MES, the black-shaded region shows the numbers sampled from each stratum in 1 realization of a RS design. As expected, RS results in a far more skewed distribution (ie, with lower Shannon entropy), and those from small strata are unlikely to be included in the sample. In this case, only 41% of the strata would be represented in the sample under this RS design.
Figure 1.
Truncated histograms of the sorted EHR+USC defined sampling strata for the entire Vanderbilt-Adult population and for samples of size 4500 using random (RS) and maximum entropy sampling (MES). The ideal sampling frequency is 19 per stratum with the remaining 130 individuals being randomly selected from available strata. The MES sample is enriched compared to the random sample, especially with individuals from strata with sparse counts (strata 1-112); all individuals belonging to strata 1-132 were included in the final sample. The y-axis is truncated at 100 to highlight the distribution of rare sampling strata (eg, stratum counts ranged from 1 to over 70K with 58% belonging to the top 5 sampling strata).
An R package was written to estimate MES counts for a given vector of stratum counts and an overall sample size. Code, installation instructions, and an example are publicly available at https://github.com/mercaldo/mes.
RESULTS
Enrichment among those sampled
Table 3 summarizes the marginal distributions of the 6 stratification variables using either EHR data only or the combined EHR and USC data from a sample of 90 000 households within the entire eMERGE network. Due to inclusion criteria, age and gender were available on all patients, and so EHR and combined EHR and USC values are identical under RS and MES sampling. At adult sites, the marginal distributions of race and ethnicity remained relatively unchanged after incorporating the USC data, likely due to sampling only 4.9% and 8.9% of participants with imputed race and ethnicity values, respectively. Most individuals lived in census block groups where the mode of the adult educational attainment distribution was between high school and some college (77%) followed by at least a bachelor’s degree (22%). A total of 48% of the sample resided in rural areas. Similar patterns were observed at pediatric sites, though fewer participants (29%) lived in rural areas.
Table 3.
Marginal distributions of stratification variables under random sampling (RS) and maximum entropy sampling (MES) designs when using only EHR data and when using combined EHR and USC data. For a sample on 90 000 households, percentages of non-missing values are reported by population (pediatric, adult)
| Adult Sites |
Pediatric Sites |
|||||
|---|---|---|---|---|---|---|
| EHR Only |
EHR + USC |
EHR Only |
EHR + USC |
|||
| RS | RS | MES | RS | RS | MES | |
| Age | ||||||
| Low age group | 22.9 | 22.9 | 43.8 | 68.7 | 68.7 | 56.7 |
| High age group ears | 77.1 | 77.1 | 56.2 | 31.3 | 31.3 | 43.3 |
| Gender | ||||||
| Female | 58.3 | 58.3 | 52.9 | 47.6 | 47.6 | 49.7 |
| Male | 41.7 | 41.7 | 47.1 | 52.4 | 52.4 | 50.3 |
| Race | ||||||
| White | 87.1 | 87.6 | 34.3 | 66.0 | 69.3 | 33.0 |
| African American | 5.7 | 5.6 | 18.3 | 19.3 | 17.7 | 22.5 |
| Asian | 2.4 | 2.3 | 16.1 | 2.9 | 2.6 | 14.7 |
| AI/AN | 0.7 | 0.6 | 7.1 | 0.2 | 0.1 | 2.5 |
| NH/PI | 0.3 | 0.2 | 4.9 | 0.1 | 0.1 | 1.8 |
| Other | 3.8 | 3.6 | 19.2 | 11.6 | 10.2 | 25.5 |
| Missing | 14.1 | – | – | 13.1 | – | – |
| Ethnicity | ||||||
| Non-Hispanic/Latino | 95.9 | 95.6 | 69.3 | 93.3 | 93.9 | 69.5 |
| Hispanic /Latino | 4.1 | 4.4 | 30.7 | 6.7 | 6.1 | 30.5 |
| Missing | 19.3 | – | – | 11.8 | – | – |
| Education | ||||||
| < HS | – | 1.0 | 11.9 | – | 1.2 | 13.6 |
| HS + some college | – | 76.8 | 54.9 | – | 72.4 | 48.9 |
| ≥ Bachelor’s | – | 22.2 | 33.2 | – | 26.4 | 37.6 |
| Rurality | ||||||
| Suburban/Urban | 52.0 | 62.5 | 70.7 | 63.9 | ||
| Rural | – | 48.0 | 37.5 | – | 29.3 | 36.1 |
Low age group (< 12 in pediatric sites, < 35 in adult sites), high age group (≥ 12 in pediatric sites, ≥ 35 in adult sites), AI/AN = American Indian/Alaska Native, NH/PI = Native Hawaiian/Pacific Islander, HS = high school.
As can be seen from the MES columns in Table 3, the sample identified using the USC-augmented EHR data and the maximum entropy sampling algorithm was enriched with target subpopulations as compared to the sample generated using the same data, but under random sampling. For example, the sample was enriched with all minority races; it was enriched 3-fold for African Americans (18% vs. 6%), 8-fold for Asians (16% vs. 2%), and more than 4-fold (19% vs. 4%) for other races. The sample was also enriched more than 6-fold among those of Hispanic ethnicity (31% vs. 5%), and 12-fold among those without a high school or equivalent degree (12% vs. 1%). However, the survey was administered only in English and written at an 8th-grade literacy level, thus possibly reducing the enrichment of the returned sample.
To further characterize enrichment due to using both EHR+USC derived sampling strata and the MES sampling strategy, entropy values are shown in Table 4. At adult sites, 262 of the 288 possible strata were observed corresponding to a maximum possible entropy of 8.03. The entropy values under random and maximum entropy sampling were 4.39 and 7.35, respectively. Overall, 81% and 72% of the maximum entropy were obtained by merging EHR and USC data and using the MES strategy compared to random sampling in the adult and pediatric sites, respectively.
Table 4.
Sampling frequencies and entropy estimates by sampling method and by population using EHR+USC data. Observed sample (Nsample) and strata frequencies (nstrata) are provided along with maximum entropy (Hmax), entropy under random sampling (RS, HRS), maximum entropy sampling (MES, HMES), and the percentage of maximum entropy accounted for by the MES sample above and beyond that of random sampling
| Nsample | nstrata | Hmax | HRS | HMES | (HMES- HRS) / (Hmax- HRS) | |
|---|---|---|---|---|---|---|
| Population | ||||||
| Adult | 58 500 | 262 | 8.03 | 4.39 | 7.35 | 0.81 |
| Pediatric | 31 500 | 251 | 7.97 | 5.16 | 7.18 | 0.72 |
Survey response rates
The CERC survey sampled 90 000 individuals, and 7761 were excluded due to invalid addresses (n = 7, 504), death/incapacity (n = 168), or previous involvement in the pilot (n = 89). A total of 13 000 surveys were returned, resulting in an overall response rate of 16.7% (N = 9185) at adult sites and 13.9% (N = 3815) at pediatric sites. These response rates, as well as those calculated for each stratification variable, are defined as the number of responses divided by the total number of subjects belonging to the sampling stratum that was defined using EHR+USC data (Table 5). Summarizing the adult sites, participants were less likely to respond if they were young (10.3% if < 35 years and 21.6% if ≥ 35 years), male (16% if male and 17.4% if female), non-white (eg, 13.2% if African American and 20.1% if white), Hispanic or Latino (14.2% if Hispanic and 17.9% if not), reside in low-education census blocks groups (13.6% if education level was <HS and 18.9% if education level was ≥ bachelor’s degree), or residing in non-rural areas (15.5% if urban or suburban and 18.7% if rural). At pediatric sites, response rates corresponded to the parent/guardian of children with the associated demographic characteristic. Like respondents from adult sites, parents or guardians were less likely to respond if their children were young, Hispanic or Latino, and resided in low-education census block groups or non-rural areas. Unlike adult sites, parents or guardians of children that were Asian were more likely to respond than whites (18.5% if Asian and 16.0% if white).
Table 5.
Response rates at adult and pediatric sites. Demographic characteristics are based on sampling strata defined by the combined EHR and USC data
| Adult Sites |
Pediatric Sitesa |
|||
|---|---|---|---|---|
| N | Response Rate | N | Response Rate | |
| Eligible | 54 850 | 16.7 | 27 389 | 13.9 |
| Age | ||||
| Low age group | 23 613 | 10.3 | 15 115 | 13.2 |
| High age group | 31 237 | 21.6 | 12 274 | 14.8 |
| Gender | ||||
| Female | 29 169 | 17.4 | 13 581 | 13.9 |
| Male | 25 681 | 16.0 | 13 808 | 14.0 |
| Race | ||||
| White | 19 099 | 20.1 | 9319 | 16.0 |
| African American | 9703 | 13.2 | 5877 | 10.0 |
| Asian | 8944 | 17.1 | 4171 | 18.5 |
| AI/AN | 3981 | 17.4 | 699 | 13.9 |
| NH/PI | 2711 | 13.4 | 496 | 9.7 |
| Other | 10 502 | 14.2 | 6827 | 12.0 |
| Ethnicity | ||||
| Not Hispanic /Latino | 38 119 | 17.9 | 19 022 | 15.3 |
| Hispanic /Latino | 16 731 | 14.2 | 8367 | 10.8 |
| Education | ||||
| < HS | 6457 | 13.6 | 3414 | 9.0 |
| HS + some college | 30 007 | 16.1 | 13 378 | 12.9 |
| ≥ Bachelor’s | 18 386 | 18.9 | 10 597 | 16.8 |
| Rurality | ||||
| Rural | 20 980 | 18.7 | 10 099 | 15.6 |
| Suburban/Urban | 33 870 | 15.5 | 17 290 | 13.0 |
Low age group (< 12 in pediatric sites, < 35 in adult sites), high age group (≥ 12 in pediatric sites, ≥ 35 in adult sites), AI/AN = American Indian/Alaska Native, NH/PI = Native Hawaiian/Pacific Islander, HS = high school.
Response rates from pediatric sites correspond to the parent/guardian of children with the associated demographic characteristic.
Accuracy of EHR and USC data among respondents
Sensitivity (Se), specificity (Sp), and positive and negative predictive values (PPV, NPV) were used to quantify the accuracy of sampling strata when using data from the EHR only, USC only, and EHR+USC data while using survey response values as the gold standard. Results are summarized in Table 6 for the adult sites because at pediatric sites, the EHR data reflected characteristics of the child while survey responses reflected those of the parent or guardian. EHR age and gender were at least 97% sensitive and specific for the “true” value based on the survey response, and PPV and NPV were also reasonably high even though overall PPV for age < 35 years was only 91%. The low PPV value for age group was largely caused by the following: 1) 73 of the misclassified ages had a self-reported age of 35, which implies that the misclassification was due to aging between the time we sent the survey out and the time the participant sent it back, and 2) 64 of the misclassifications had differences in the EHR and self-reported gender, which may imply that the survey was not completed by the individual to whom the survey was addressed (eg, spouse or caregiver). If these individuals are omitted, then the PPV associated with age group was 96%.
Table 6.
Accuracy measures among CERC survey respondents from the adult sites (N = 9, 185). Frequencies (Nc) and prevalence estimates (P) were computed using self-reported data, and accuracy measures (Se, Sp, PPV, NPV)a were computed by comparing self-reported data (ie, gold-standard) to the EHR and/or USC-derived data. These summaries are also presented by the data source used to define the sampling strata (EHR, USC, or combined)b
| Data Source Used to Define Sampling Strata (Adult Respondents, N = 9185) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
EHR data Only |
USC data Only |
EHR and USC data (combined) |
||||||||||
| Nc | p | Se/Sp | PPV/NPV | Nc | p | Se/Sp | PPV/NPV | Nc | p | Se/Sp | PPV/NPV | |
| Age | ||||||||||||
| < 35 years | 2211 | 24.8 | 99/97 | 91/100 | – | – | – | – | 2211 | 24.8 | 99/97 | 91/100 |
| ≥ 35 years | 6690 | 75.2 | 97/99 | 100/91 | – | – | – | – | 6690 | 75.2 | 97/99 | 100/91 |
| Gender | ||||||||||||
| Female | 5042 | 56.0 | 97/98 | 99/97 | – | – | – | – | 5042 | 56.0 | 97/98 | 99/97 |
| Male | 3969 | 44.0 | 98/97 | 97/99 | – | – | – | – | 3969 | 44.0 | 98/97 | 97/99 |
| Race | ||||||||||||
| White | 4210 | 48.5 | 78/92 | 90/82 | 142 | 53.2 | 62/51 | 59/54 | 4352 | 48.7 | 77/91 | 89/81 |
| African American | 989 | 11.4 | 94/97 | 79/99 | 15 | 5.6 | 60/82 | 17/97 | 1004 | 11.2 | 93/96 | 76/99 |
| Asian | 1440 | 16.6 | 85/96 | 83/97 | 10 | 3.7 | 70/93 | 28/99 | 1450 | 16.2 | 85/96 | 82/97 |
| AI/AN | 240 | 2.8 | 82/94 | 30/100 | 2 | 0.7 | 50/100 | 100/100 | 242 | 2.7 | 82/95 | 30/100 |
| NH/PI | 103 | 1.2 | 71/97 | 21/100 | 0 | 0 | −/100 | −/− | 103 | 1.2 | 71/97 | 21/100 |
| Other | 1692 | 19.5 | 35/88 | 42/85 | 98 | 36.7 | 10/83 | 26/62 | 1790 | 20.0 | 33/88 | 42/84 |
| Ethnicity | ||||||||||||
| Not Hispanic /Latino | 6697 | 80.8 | 90/89 | 97/69 | 495 | 85.5 | 64/73 | 93/26 | 7192 | 81.1 | 89/88 | 97/65 |
| Hispanic /Latino | 1594 | 19.2 | 89/90 | 69/97 | 84 | 14.5 | 73/64 | 26/93 | 1678 | 18.9 | 88/89 | 65/97 |
| Education | ||||||||||||
| < HS | – | – | – | – | 670 | 7.6 | 21/91 | 17/93 | 670 | 7.6 | 21/91 | 17/93 |
| HS + some college | – | – | – | – | 3415 | 38.9 | 66/57 | 49/72 | 3415 | 38.9 | 66/57 | 49/72 |
| ≥ Bachelor’s | – | – | – | – | 4684 | 53.4 | 52/78 | 73/59 | 4684 | 53.4 | 52/78 | 73/59 |
| Ruralityc | ||||||||||||
| Rural | – | – | – | – | 3919 | 42.7 | – | – | 3919 | 42.7 | – | – |
| Suburban/Urban | – | – | – | – | 5266 | 57.3 | – | – | 5266 | 57.3 | – | – |
Nc = number who completed the survey item, Se = sensitivity, Sp = specificity, PPV = positive predicted value, NPV = negative predictive value, AI/AN = American Indian/Alaska Native, NH/PI = Native Hawaiian/Pacific Islander, HS = high school.
Accuracy measures are based on the subset of respondents who provided self-reported data (ie, those who complete the demographic questionnaire item of interest). For example, 284 = 9185—2211—6690 respondents did not complete the survey item related to age.
To define sampling strata, age and gender were based on EHR data for all, as availability of both in the EHR was an inclusion criterion, and education and rurality were based on USC data for all. For race and ethnicity, data presented in the USC Only column correspond to the subset of survey respondents who did not have race and/or ethnicity data recorded in the EHR. For this subset, USC data were used to define their sampling strata.
Accuracy estimates were not calculated for rural living, as the true value is based on the address and not on a participant response.
EHR+USC data showed variable sensitivity for race, ranging from 33% for Other race to 93% for African American race, and the PPV for the smaller minority races was alarmingly low (∼20–40%). Even though EHR+USC data were reasonably sensitive for Hispanic ethnicity (89%), the PPV was only 65%. To gain further insight into the relative contributions to the misclassifications, we provide accuracy estimates for respondents whose stratification data were defined using only EHR data and only USC data, separately. Only 3% (267/8941) and 7% (579/8870) of patients had race and ethnicity values imputed with USC data; however, it is clear that USC data were less accurate than EHR data. This is not surprising, as USC data are based on aggregated, block groups. Utilizing only USC data to determine an individual’s educational attainment resulted in low discriminative and predictive values (eg, <HS: Se = 21%, PPV = 17%).
Overall, the degree of sample enrichment is dependent on the accuracy of the auxiliary data sources (eg, EHR and/or USC data). We observed that using EHR and possibly USC data to identify demographic subgroups may be a reasonable approach for common subgroups (African American race, gender, non-Hispanic ethnicity); however, the smaller subgroups with very low prevalences (American Indian/Alaska Native race, Hispanic ethnicity) are often misclassified, and caution should be taken when using either EHR or USC data for their identification.
The use of USC data to impute missing race and ethnicity represents a tradeoff between sampling frame coverage and enrichment of the observed sample. Creating a sampling frame based only on EHR data yields relatively high accuracy (and therefore higher enrichment) at the cost of a sampling frame that does not cover those with missing EHR data. In contrast, imputing missing EHR data with USC data permits greater sampling frame coverage at the cost of lower accuracy/enrichment. We highlight again that to be included in the sampling frame, we required non-missing age and gender in the EHR data but permitted missing values for race and ethnicity that were imputed with USC data.
DISCUSSION
This paper outlines a complex survey design that utilized both EHR and USC data to define the sampling strata and introduced an algorithm that sought to enrich the final sample with individuals from rare subpopulations. In our sample, we observed substantial enrichment from subpopulations that would not have been observed had a standard random sampling scheme been used. There were several challenges with implementing such a design in this setting, which include: incomplete and inaccurate EHR data, misclassification due to imputing missing EHR data with USC data, the targeting sampling to sparse sampling strata, and, ultimately, induced complexities-associated design-based analyses.
Limitations
The drawbacks of using EHR data for secondary research have been well documented.17,18 As these data are not primarily collected for research purposes, their content and quality may vary by institution. The lack of universally accepted EHR criteria, except for the minimal criteria set by HITECH, may result in these data being insufficient to address certain research questions. In the primary results paper for our study, EHR data were used to define the sampling frame but were not used for primary study analyses.19 Further research is needed to quantify effects of measurement error (or misclassification) on stratification variables, especially because EHR data are seldom complete and are often mismeasured (eg, EHR race, Table 5).20–22
Usage of USC data to impute missing EHR data introduces an additional level of complexity regarding the safeguarding of participants’ personal health information. Census identifiers are not typically stored within an institution’s EHR, and thus physical addresses need to be geocoded to link EHR and USC data. Among the 11 eMERGE sites, 7 geocoded their own addresses, indicating that the infrastructure (eg, software, expertise) to generate these identifiers is available at most academic medical centers.
The overall response rate in this study may have been influenced by the oversampling of subgroups that are less inclined to participate in biomedical research. If sampling strata frequencies are related to willingness to respond, then this enrichment approach may result in a lower than expected response rate (eg, ∼17% in the eMERGE survey). An alternative study design would decrease the number of subjects sampled while increasing resources towards ensuring that those who were sampled answered the survey. However, at the onset of the study, we determined that such a design was impractical across the 11 participating institutions.
Finally, the accuracy of the stratification variables, especially those based solely on USC data (eg, education), may have been affected by the content of the survey and one’s willingness to participate in biomedical research. For example, those individuals who were misclassified as having low education, such as those with having at least a bachelor’s degree, may be more likely to respond resulting in biased accuracy estimates.
CONCLUSION
We have outlined an approach that increases the diversity of a sample by oversampling those subjects who belong to rarer sampling strata. The magnitude of sample enrichment depends on the accuracy of the data used to define the sampling strata as well as the overall response rate. Thus, additional resources may be required to ensure that these variables are correctly enumerated and that sampled subjects complete the questionnaire. This approach may be especially well suited for health disparities research or other endeavors in which it is of interest to elicit information from vulnerable or understudied populations.
FUNDING
This work was supported by the CERC Survey project within the eMERGE Network which was initiated and funded by NHGRI: U01HG006828 (Cincinnati Children’s Hospital Medical Center/Boston Children’s Hospital); U01HG006830 (Children’s Hospital of Philadelphia); U01HG006389 (Essentia Institute of Rural Health, Marshfield Clinic Research Foundation and Pennsylvania State University); U01HG006382 (Geisinger); U01HG006375 (Kaiser Permanente Washington/Group Health Cooperative/University of Washington); 3U01HG006379 (Mayo Clinic); U01HG006380 (Icahn School of Medicine at Mount Sinai); 3U01-HG006388 (Northwestern University); U01HG006378 (Vanderbilt University Medical Center); and 3U01HG006385 (Vanderbilt University Medical Center serving as the Coordinating Center). Additional funding for the CERC Survey project was provided by the NIH Office of the Director through the NIH Common Fund, which is managed by the OD/Office of Strategies Coordination (OSC), award numbers 3U01HG006828-02S1 (Cincinnati Children’s Hospital Medical Center/Boston Children’s Hospital); 3U01HG006830-02S1 (Children’s Hospital of Philadelphia); 3U01HG006389-03S1 (Essentia Institute of Rural Health, Marshfield Clinic Research Foundation and Pennsylvania State University); 3U01HG006382-03S1 (Geisinger); 3U01HG006375-03S1 (Kaiser Permanente Washington/Group Health Cooperative/University of Washington); 3U01HG006379- 03S1 (Mayo Clinic); 3U01HG006380-03S1 (Icahn School of Medicine at Mount Sinai); 3U01HG006388-03S1 (Northwestern University); 3U01HG006378-03S1 (Vanderbilt University Medical Center); and 3U01HG006385-03S1 (Vanderbilt University Medical Center serving as the Coordinating Center). Additional funding for the development of the complex survey design was provided by the National Heart, Lung, and Blood Institute (R01HL094786).
CONTRIBUTORS
Substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data for the work: NDM, KBB, DSC, EWC, JJC, IAH, CRH, GPJ, TK, RL, CAM, JBM, VDM, MFM, JJP, MJS, MES, SCS, JLW, and JSS.
Drafting the work or revising it critically for important intellectual content: NDM, KBB, DSC, EWC, JJC, IAH, CRH, GPJ, TK, RL, CAM, JBM, VDM, MFM, JJP, MJS, MES, SCS, JLW, and JSS.
Final approval of the version to be published: NDM, KBB, DSC, EWC, JJC, IAH, CRH, GPJ, TK, RL, CAM, JBM, VDM, MFM, JJP, MJS, MES, SCS, JLW, and JSS.
Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved: NDM, KBB, DSC, EWC, JJC, IAH, CRH, GPJ, TK, RL, CAM, JBM, VDM, MFM, JJP, MJS, MES, SCS, JLW, and JSS.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
We would like to acknowledge the work of the following CERC Survey workgroup members: Melissa Basford, Mollie Bodin, Diego Campos, Brittany City, Natalia Connolly, Brianne Derveloy, Joe DeWalle, Paul Hitz, Kathryn Jackson, Jebakaran Jebakumar, Mitch Juusola, Dave Kaufman, Brandy Mapes, Keith Marsolo, Rennie Negron, Katherine Nowakowski, Jennifer Pacheco, Peggy Peissig, Lisa Price, Aaron Scrol, Rob Strenn, and Sonja Ziniel. We would also like to thank the referees for their invaluable input that improved the quality of this manuscript.
Conflict of interest statement. None declared.
REFERENCES
- 1. Safran C, Bloomrosen M, Hammond WE, et al. Toward a national framework for the secondary use of health data: an American Medical Informatics Association White Paper. J Am Med Inform Assoc 2007; 14 (1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Casey JA, Schwartz BS, Stewart WF, Adler NE.. Using electronic health records for population health research: a review of methods and applications. Annu Rev Public Health 2016; 37 (1): 61–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008; 84 (3): 362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sudman S, Sirken MG, Cowan CD.. Sampling rare and elusive populations. Science 1988; 240 (4855): 991–6. [DOI] [PubMed] [Google Scholar]
- 5. Douglas MD, Dawes DE, Holden KB, Mack D.. Missed policy opportunities to advance health equity by recording demographic data in electronic health records. Am J Public Health 2015; 105 Suppl 3 (S3): S380–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coorevits P, Sundgren M, Klein GO, et al. Electronic health records: new opportunities for clinical research. J Intern Med 2013; 274 (6): 547–60. [DOI] [PubMed] [Google Scholar]
- 7. Shivade C, Raghavan P, Fosler-Lussier E, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc 2014; 21 (2): 221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Holland AT, Palaniappan LP.. Problems with the collection and interpretation of Asian-American Health Data: omission, aggregation, and extrapolation. Ann Epidemiol 2012; 22 (6): 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith ME, Sanderson SC, Brothers KB, et al. Conducting a large, multi-site survey about patients’ views on broad consent: challenges and solutions. BMC Med Res Methodol 2016; 16 (1): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gottesman O, Kuivaniemi H, Tromp G, et al. The electronic medical records and genomics (eMERGE) network: past, present, and future. Genet Med 2013; 15 (10): 761–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garrison NA, Sathe NA, Antommaria AHM, et al. A systematic literature review of individuals' perspectives on broad consent and data sharing in the United States. Genet Med 2016; 18 (7): 663–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. US Census Bureau. 2008–2012 American Community Survey 5-year estimates. http://www.census.gov/programs-surveys/acs/data/summary-file.html. Accessed June 2014.
- 13. US Census Bureau. 2010 Urban and Rural Classification and Urban Area Criteria. https://www.census.gov/geo/reference/ua/urban-rural-2010.html. Accessed June 2014.
- 14. ESRI. ArcGIS Desktop: Release 10. Redlands, CA: Environmental Systems Research Institute; 2011. [Google Scholar]
- 15. R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2016. https://www.R-project.org/ [Google Scholar]
- 16. Shannon CE. A mathematical theory of communication. Sigmobile Mob Comput Commun Rev 2001; 5 (1): 3–55. [Google Scholar]
- 17. Weiskopf NG, Weng C.. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inform Assoc 2013; 20 (1): 144–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Menachemi N, Collum TH.. Benefits and drawbacks of electronic health record systems. Risk Manag Healthc Policy 2011; 4: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sanderson SC, Brothers KB, Mercaldo ND, et al. Public attitudes toward consent and data sharing in biobank research: a large multi-site experimental survey in the US. Am J Hum Genet 2017; 100 (3): 414–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klinger EV, Carlini SV, Gonzalez I, et al. Accuracy of race, ethnicity, and language preference in an electronic health record. J Gen Intern Med 2015; 30 (6): 719–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grundmeier RW, Song L, Ramos MJ, et al. Imputing missing race/ethnicity in pediatric electronic health records: reducing bias with use of U.S. census location and surname data. Health Serv Res 2015; 50 (4): 946–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fiscella K, Fremont AM.. Use of geocoding and surname analysis to estimate race and ethnicity. Health Serv Res 2006; 41 (4 Pt 1): 1482–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.