Abstract
Breast cancer is the most common cancer among women and the molecular pathways that play main roles in breast cancer regulation are still not completely understood. MicroRNAs (miRNAs) and transcription factors (TFs) are important regulators of gene expression. It is important to unravel the relation of TFs, miRNAs, and their targets within regulatory networks to clarify the processes that cause breast cancer and the progression of it. In this study, mRNA and miRNA expression studies including breast tumors and normal samples were extracted from the GEO microarray database. Two independent mRNA studies and a miRNA study were selected and reanalyzed. Differentially expressed (DE) mRNAs and miRNAs between breast tumor and normal samples were listed by using BRBArray Tools. CircuitsDB2 analysis conducted with DE miRNAs and mRNAs resulted in 3 significant circuits that are SOX10- and hsamiR-301a-dependent. The following significant circuits were characterized and validated bioinformatically by using web-based tools: SOX10→hsa-miR-301a→HOXA3, SOX10→hsa-miR-301a→KIT, and SOX10→hsa-miR-301a→NFIB. It can be concluded that regulatory motifs involving miRNAs and TFs may be useful for understanding breast cancer regulation and for predicting new biomarkers.
Keywords: Breast cancer, miRNA, mRNA, transcription factor, regulatory circuits
1. Introduction
Breast cancer is a complex genetic disorder that is not controlled by a single factor but is rather controlled by many factors (http://www.cancer.org/). Although many factors (genes, microRNAs (miRNAs), transcription factors, etc.) that cause breast cancer and play a role in its development have been identified with the advancement of high-throughput technologies, the molecular mechanisms playing a role in disease regulation have still not been revealed. For this reason, it is important and inevitable to identify the regulatory circuits and networks that will explain the development and progression of breast cancer.
Gene expression regulation is an important mechanism for controlling biological processes in the cell. Transcriptional factors (TFs) are the regulators that function at the transcriptional level, while miRNAs work at the posttranscriptional level. In view of the fact that the transcription of mRNA and miRNAs is controlled by TFs and the expression of TFs is regulated by miRNAs, these two important mechanisms cannot be separated from each other. Therefore, characterization of these combinatorial regulatory mechanisms is important to reveal the biological processes that take part in breast cancer in detail by constructing networks and circuits. miRNAs are small, noncoding RNA molecules that regulate gene expression in posttranscriptional stages (Lagos-Quintana et al., 2001; Lee and Ambros, 2001). miRNAs play important roles in biological processes such as development, cell division, cell differentiation, and programmed cell death. In this context, it is possible that altered miRNA expression may contribute to the development and progression of diseases such as cancer. The presence of abnormal miRNA expression profiling in many diseases, including breast cancer, has been demonstrated and validated by intensifying miRNA studies (Iorio et al., 2005; Lowery et al., 2009; Enerly et al., 2011; Romero-Cordoba et al., 2012) . The finding that miRNA expressions are oeftn dysregulated in cancers has made these molecules important candidates for cancer markers (Calin et al., 2002; Lu et al., 2005) . Therefore, the search for the relationship of miRNAs with target genes (this may be an mRNA or a TF) has great importance for the diagnosis and treatment of breast cancer.
TFs are essential regulatory elements in the transcriptional pathway, which work by binding to target genes that have specific DNA sequences, usually in the promoter region (Latchman, 1997). TFs regulate their targets at the transcription level by inhibiting or enhancing their expression while miRNAs regulate target mRNAs at the posttranscriptional level in the form of inhibition. miRNAs also undergo the transcription process and they regulate their target genes like TFs. When all these reasons are taken into consideration:
· Expression of a miRNA may be regulated by a transcription factor; · Expression of a TF may be regulated by a miRNA; · Similarly, the transcription factor and miRNA may regulate the expression of target genes together.
In recent years, many researchers have been working on TFs, miRNAs, and their regulation mechanism on posttranscriptional and transcriptional levels. With the help of system biology approaches, transcription factor-miRNA-target gene relationships were started to be explored by an increasing number of studies. These studies have mostly used and/or developed bioinformatics and statistical methods and they have reviewed the existing information. The tools that analyze miRNA-TF relationships can be grouped into three subgroups. The tools in the first group are intended to analyze statistically the gene clusters organized by cooperating miRNAs and/ or miRNAs using matched miRNA and mRNA expression profiles (Yoon and De Micheli, 2005; Joung et al., 2007; Tran et al., 2008; Joung and Fei, 2009; Nam et al., 2009) . The tools in the second group again predict gene expression relationships using gene expression data (Naeem et al., 2011) . The tools in the third group provide static circuits, combining statistical data obtained from literature (e.g., CircuitDB) (Friard et al., 2010b) .
In light of the above mentioned information, this work aims to identify breast cancer-specific miRNA-TF-mRNA circuits bioinformatically that could be important in breast cancer development. We have identified regulatory circuits and molecule associations that differ between the normal group and breast cancer patients by using previously performed expression studies.
2. Materials and methods
2.1. Selection of miRNA and mRNA microarray studies
It was aimed to obtain miRNA and mRNA microarray data related to breast cancer in the literature. GEO, a publicly available database at http://www.ncbi.nlm.nih.gov/geo/ (Edgar et al., 2002) , was used for this purpose. “miRNA / mRNA”, “microarray”, and “breast cancer” terms were used together when searching in GEO. The datasets were selected from among the ones that have freshly frozen tumor and normal tissue information. Microarray data obtained from blood samples, formalin-fixed parafinembedded tissues, and cell lines were excluded from the study to prevent biased expression profiles.
As a result, 2 independent mRNA studies and 1 miRNA study including breast tumors and normal samples were selected to be analyzed (Table 1).
Table 1.
The characteristics of the datasets.
2.2. Class comparison analysis
The raw data were obtained from the microarray database (GEO) and normalized by using quantile normalization in BRB-Array Tools, which is an Excel-integrated package for the visualization and statistical analysis of microarray gene expression data. Quantile normalization, one of the most widely adopted methods for analyzing microarray data, was used as the normalization method. Normalization is important to remove any differences that may arise from technical problems and to make the data comparable. Class comparison tests were performed to find out differentially expressed miRNAs and mRNAs between tumor samples and normal samples (P ≤ 0.05, 2-fold change). This test provides powerful methods for ifnding differentially expressed genes when controlling the ratio of false positives. Differentially expressed and common genes (by Venny 2.1.0) were found for tumor vs. normal comparison in GSE3744 and GSE5764 (http:// bioinfogp.cnb.csic.es/tools/venny_old/venny.php). Lastly, differentially expressed miRNAs were found in GSE45666.
2.3. Identification of TFs in DE mRNA lists
Transcription factor lists that have been identified in the human body were accessed by using the Animal Transcription Factor DataBase 2.0 (http://www.bioguo. org/AnimalTFDB/) (Zhang et al., 2015) and the TFs that were in the DE genes were determined by comparison with these lists.
2.4. Circuit analysis
The CircuitsDB2 analysis tool was used to identify the breast cancer-specific circuits as regulatory loops among the members of DE miRNAs, mRNAs, and TFs. This userfriendly tool provides static circuits by combining statistical data obtained from the literature. It contains precompiled transcriptional and posttranscriptional networks, the sets of network motifs, and series of functional and biological information, which are based on JASPAR DB and the ENCODE project. Members of the DE miRNAs, mRNAs, and TFs were searched in microRNA-mediated FFLs in which a TF is the master regulator in CircuitsDB2 (Friard et al., 2010b) .
2.5. Circuit member analysis in diseases, biological processes, and pathways
For the characterization of the members of the significant circuits three different tools, PhenomiR, WebGeshtalt, and DIANA, were used.
The PhenomiR 2.0 database, completely generated by manual curation of experienced annotators, was used to obtain information about differentially regulated miRNA (circuit member miRNA, hsa-miR-301a here) expression in diseases (breast cancer here) (Ruepp et al., 2010) .
WebGestalt is the “WEB-based GEne SeT AnaLysis Toolkit”, in which the functional genomic, proteomic, and large-scale genetic studies from large numbers of gene lists (e.g., DE gene list) are continuously generated. Function analysis of the circuit member genes and TFs was performed with the WebGestalt Protein Interaction Network Module (Wang et al., 2013) .
DIANA-mirPath v.3 was used to search the pathways of circuit member miRNA. It is a miRNA analysis tool that enables users to find targets of mRNAs by using different algorithms and to find pathways in which these genes are significantly enriched (Maragkakis et al., 2009; Vlachos et al., 2015) .
2.6. Validation analysis
For advanced bioinformatics analysis for in silico validation of significant circuits, the mirExTra 2.0 under DIANA was used. Our miRNA and mRNA lists were analyzed using the Central microRNA Discovery Module (CmD) and miRNA-mRNA relationships in the circuits were tried to be validated. This module combines microRNA and mRNA expression data results (names, P-values, fold changes) in order to identify functional microRNAs responsible for changes in mRNA expression by utilizing in silico interactions from DIANA-microT-CDS. In this study, DE miRNA and mRNA lists were used for mirExtra analysis. It enabled us to determine the central regulator miRNAs and their targets and provided us the opportunity to validate our results (Maragkakis et al., 2009; Vlachos et al., 2016) .
3. Results
3.1. The miRNA and mRNA datasets
The miRNA microarray studies and mRNA microarray studies were searched to find differentially expressed miRNAs and mRNAs between breast cancer and normal tissues. Three independent microarray datasets were obtained from GEO (http://www.ncbi.nlm.nih.gov/geo/) (Edgar et al., 2002) .
One of the datasets was a miRNA microarray study performed with 80 breast cancer samples and 15 normal samples (GSE45666 (Lee et al., 2013)). Two of the datasets were mRNA microarray studies (GSE3744 (Richardson et al., 2006) , GSE5764 (Turashvili et al., 2007) ). The total number of breast cancer samples and normal samples were 55 and 22, respectively. The details of the platforms and the sample numbers are given in Table 1.
3.2. Differentially expressed miRNA and mRNA lists
The miRNA microarray study and two mRNA studies (Table 1) were independently analyzed by BRB-Array Tools, which is an integrated package for the visualization and statistical analysis of gene expression data (Simon et al., 2007) . Quantile normalization was used as the normalization method. All studies consisted of 2 groups, which are tumor and normal. Class comparison tests were performed to find out differentially expressed miRNAs and mRNAs between tumor and normal samples. This test provides powerful methods for finding differentially expressed genes when controlling the ratio of false positives. This method is similar to the significance analysis of microarrays method (Tusher et al., 2001) but provides more control of the false discovery rate (Simon et al., 2007) .
As a result of class comparison analysis, miRNA and mRNA lists were obtained. Twenty-three miRNAs (11 upregulated and 12 downregulated; Table S1) were found to be differentially expressed between tumor and normal samples. Since two mRNA studies were performed, the DE mRNAs were listed as common DE genes in both of the studies. A total of 264 mRNAs (187 downregulated and 77 upregulated; Table S1) were found to be differentially expressed between tumor and normal samples and were common for each mRNA study.
3.3. Differentially expressed transcription factors list
To obtain the TFs among DE mRNA lists, which are given in Table S1, AnimalTFDB 2.0 was used (Zhang et al., 2015) . hTis tool has a dataset that consists of 1691 transcription factors in 68 families in humans. When we compared it to the DE list, 18 of the genes in the DE gene list were found to encode proteins functioning as TFs: ATF3, BHLHE41, EHF, FOSB, HOXA3, ID4, IRX1, NFIB, SOX10, MAFF, FOS, NR3C2, STAT1, EGR1, JUN, ZNF662, THRB, and ZBTB16.
3.4. Significant breast cancer-specific circuits
CircuitsDB2 was used to identify important and regulatory circuits that consist of the members of DE miRNA, mRNA, and TF lists. CircuitsDB2 is a web service that includes data as regulatory loops stored in a relational database that can be accessed through a dynamic web interface (Friard et al., 2010b) . With use of this interface it is possible to find different kinds of circuits like feedforward loops (FFLs) in which a TF regulates a miRNA and they both regulate a target gene (Friard et al., 2010a) . Members of the DE miRNA, mRNA, and TF lists were searched in microRNAmediated FFLs in which a TF is the master regulator in CircuitsDB2.
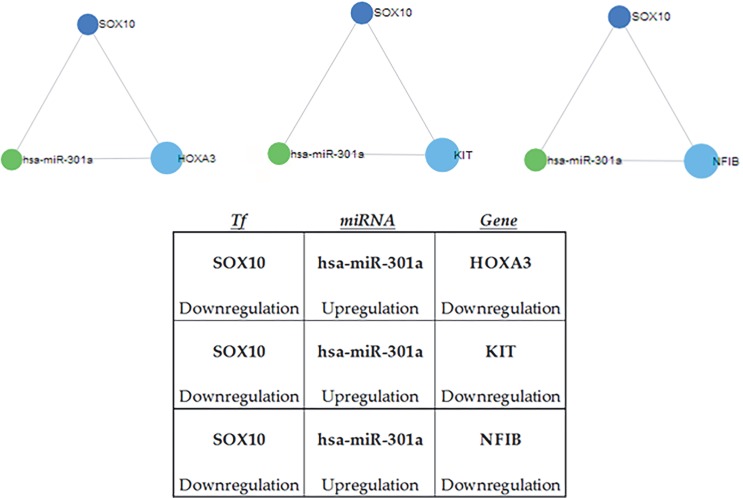
hTree significant circuits that are SOX10- and hsamiR-301a-dependent were identified (Figure 1a) by CircuitsDB2 [5] analysis; in each circuit, hsa-miR-301a was found to be upregulated and its TF target SOX-10 was downregulated. Additionally, the three target genes that were under regulation of both miR-301a and SOX10 were downregulated as expected (Figure 1b).
Figure 1.
Significant circuits, which were results of CircuitsDB2 analysis. a) The triangular shapes are circuit diagrams taken from CircuitsDB2. The dark blue circles represent the TF, the green circles represent the miRNA, and the light blue circles represent the targets. b) Downregulation and upregulation terms indicate the differentiation of expression according to tumor vs. normal.
3.5. Circuit members in diseases, biological processes, and pathways
To characterize the significant circuits in detail, their presence in diseases, biological processes, and signaling pathways were identified in detail by using related webbased tools.
The first step was to search the expression information of miRNA hsa-miR-301a. When breast cancer association data were extracted, concordant with our finding, it was found to be overexpressed in breast tumors. Since the data in PhenomiR (Ruepp et al., 2010) were obtained from independent studies and contain results from both cell lines and tumor samples, our results related to hsa-miR301a could be accepted as robust for breast cancer (Table 2). Given the pathological, histological, and molecular differences (e.g., MCF-7: ER+; SK-BR-3: HER2+; MDAMB-231: triple-negative) of the cell lines in Table 2, hsamiR-301a, which is upregulated in the different cell lines and tumor specimens, may be accepted as a generalized and important marker for breast cancer. Additionally the expression profiles of other members of the circuits, SOX10, HOXA3, KIT, and NFIB, were searched in an independent microarray dataset (GSE17907; 51 tumor and 4 normal breast samples) and compatible to our results found to be significantly downregulated in tumor samples compared to normal ones (SOX10, P-value 1.00E07, 11-fold downregulation; HOXA3, P-value 0.0004616, 4-fold downregulation; KIT, P-value, 3.00E-07, 11-fold downregulation; and NFIB, P-value 0.0012966, 3.7-fold downregulation).
Table 2.
Expression changes of hsa-miR-301a in previous studies in the literature. The table was created with the information obtained from PhenomiR 2.0 (Ruepp et al., 2010).
| No. | ID | miRNA name | Disease | Tissue/cell line | PubMed ID | Regulation | Study design |
|---|---|---|---|---|---|---|---|
| 1 | 447 | hsa-mir-301a | Breast cancer | Breast epithelium | 16754881 | Up | Patient study, phenotype-control |
| 2 | 451 | hsa-mir-301a | Breast cancer | MCF-7 cell | 16192569 | Up | Cell culture study |
| 3 | 366 | hsa-mir-301a | Breast cancer | T-47D cell | 16192569 | Up | Cell culture study |
| 4 | 481 | hsa-mir-301a | Breast cancer | MDA-MB-231 cell | 16192569 | Up | Cell culture study |
| 5 | 377 | hsa-mir-301a | Breast cancer | SK-BR-3 cell | 16192569 | Up | Cell culture study |
| 6 | 482 | hsa-mir-301a | Breast cancer | MDA-MB-361 cell | 16192569 | Up | Cell culture study |
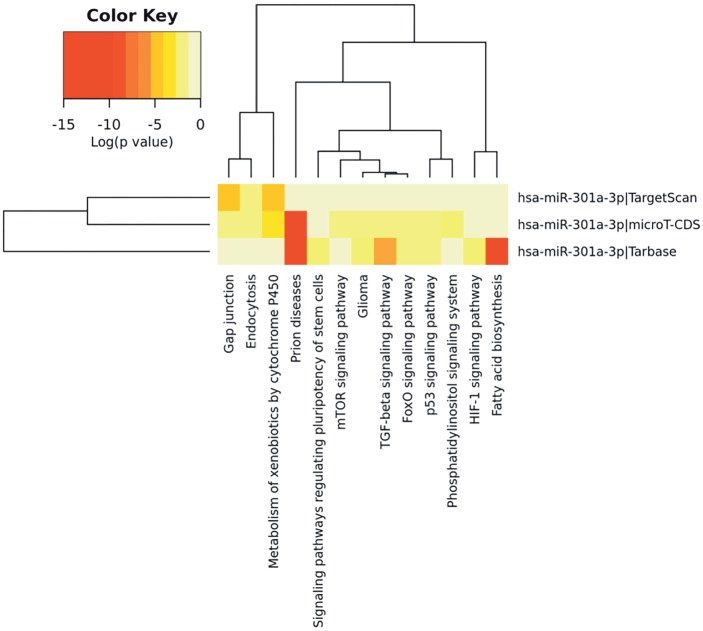
The second step was to find out the pathways in which the targets of hsa-miR-301a were taking part. DIANAmirPath v.3 was used for this purpose (Maragkakis et al., 2009; Vlachos et al., 2015) . This tool aims to find the targets of microRNAs by using 3 different target algorithms (TargetScan, microT-CDS, and Tarbase) and determining the pathways in which these genes are significantly enriched. The members of our circuits, KIT, NFIB, and HOXA3, were among the identified targets (by mirPath) of hsa-miR-301a (data not shown). It was also seen in the HeatMap plotted aeftr the pathway analysis that the targets of this microRNA function in pathways that are known to be important in cancer (Figure 2).
Figure 2.
Pathway enrichment results of hsa-miR-301a targets. Targets of the miRNA and KEGG pathway results were used to construct the heat map, which was obtained from DIANA (Maragkakis et al., 2009; Vlachos et al., 2015). The intensity of color represents the logarithmic P-values.
The third step was function analysis of the genes. For the characterization of the members of the significant circuits, which are SOX10, KIT, NFIB, and HOXA3, function analysis was performed for each of them using WebGestalt protein interaction network module-related function analysis (Table 3) (Wang et al., 2013) .
Table 3.
Function analysis results for the members of the significant circuits. The table was created with the information obtained from the WebGestalt protein interaction network module and related function analysis (Wang et al., 2013).
| Gene name | Biological process | Cellular component | Molecular function |
|---|---|---|---|
| SOX10 | Positive regulation of | Extrinsic to mitochondrial | HMG box domain |
| gliogenesis | outer membrane | binding | |
| P = 4.84e-07 | P = 9.81e-04 | P = 1.91e-11 | |
| KIT | Intracellular signal | Cytosol | Signaling adaptor |
| transduction | activity | ||
| P = 4.67e-10 | P = 3.22e-06 | P = 1.07e-09 | |
| NFIB | Anterior/posterior pattern | Nucleus | Sequence-specific DNA |
| specification | binding | ||
| P = 0e+00 | P = 0e+00 | P = 0e+00 | |
| HOXA3 | Negative regulation of | Perinuclear theca | Biotin-[acetyl-CoA-carboxylase] |
| DNA replication | ligase activity | ||
| P = 1.54e-03 | P = 3.36e-03 | P = 2.03e-03 |
3.6. Advanced bioinformatics analysis for in silico validation
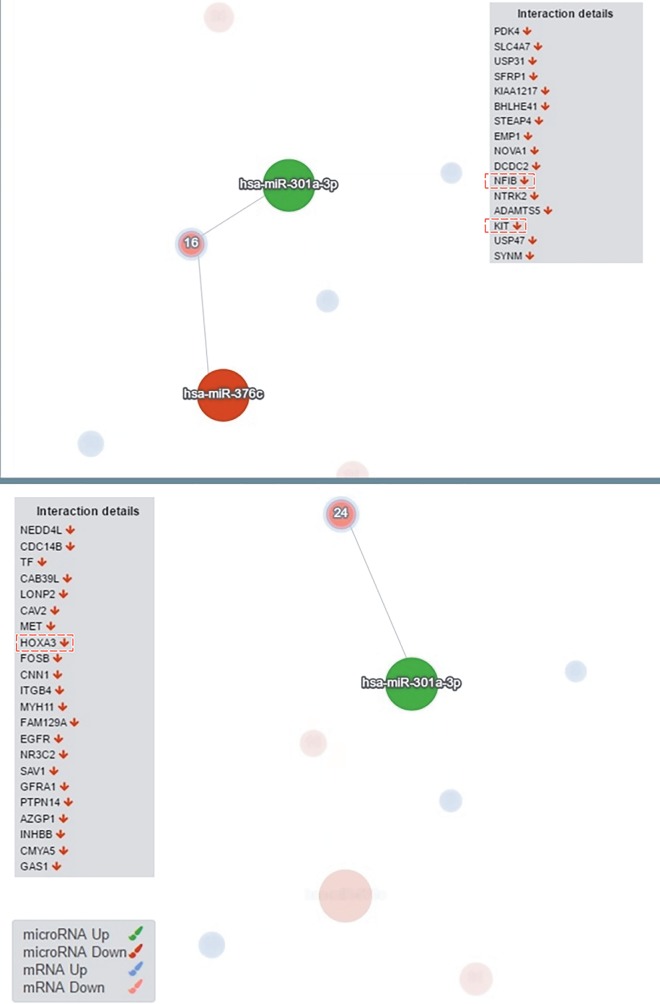
To validate the significant circuits, the DIANA-mirExTra 2.0 tool was used (Maragkakis et al., 2009; Vlachos et al., 2016) . We used DE miRNA and mRNA lists with fold change and P-value information as an input and the algorithm identified which miRNAs were central regulators (interactions based on microT-CDS and threshold type based on fold change with 2-fold change). Analysis showed that the most regulatory upregulated (tumor vs. normal) miRNA was hsa-miR-301a (P = 7.3e03), while the most regulatory downregulated (tumor vs. normal) miRNA was hsa-miR-376c (P = 6.4e-03), which is also the most downregulated DE miRNA in our list. As can be seen in Figure 3, hsa-miR-301a, which is the regulatory miRNA of the 3 circuits (Figure 1), was expressed as one of the central miRNAs, and as can be seen in the interaction details, KIT, NFIB, and HOXA3 are among the regulated target genes (Figure 3). These advanced and independent analysis results confirm and validate our CircuitsDB2 circuit analysis results.
Figure 3.
Validation results of the circuits members. hsa-miR-301a (upregulated) and hsa-miR-376c (downregulated) are the central regulator miRNAs for the tumor vs. normal comparison. KIT and NFIB are targeted by hsa-miR-301a and hsa-miR-376c while HOXA3 is targeted by only hsa-miR-301a. The figure was obtained from DIANA-mirExTra 2.0 (Maragkakis et al., 2009; Vlachos et al., 2016).
4. Discussion
Breast cancer is a complex genetic disorder that is not controlled by a single factor but is controlled by many variables and is still the most common type of cancer in women today (http://www.cancer.org/). Although many scientific studies have been carried out on breast cancer, it is still unclear in many respects. Since breast cancer is a complex disease, to understand the development of breast cancer new biomarkers need to be discovered. In addition to the discovery of new biomarkers, it is also of great importance to investigate their relationships with each other (e.g., miRNA-TF-mRNA circuits). In recent years, systems biology approaches have become more important in the understanding of diseases. The development of the methods that help to understand large molecular data and the molecular mechanisms of breast cancer development is inevitable.
The aim of this study was to analyze the TF-miRNAtarget gene relationships in a global perspective in breast cancer samples and to determine miRNA-TF-mRNA circuits that can play a role in breast cancer.
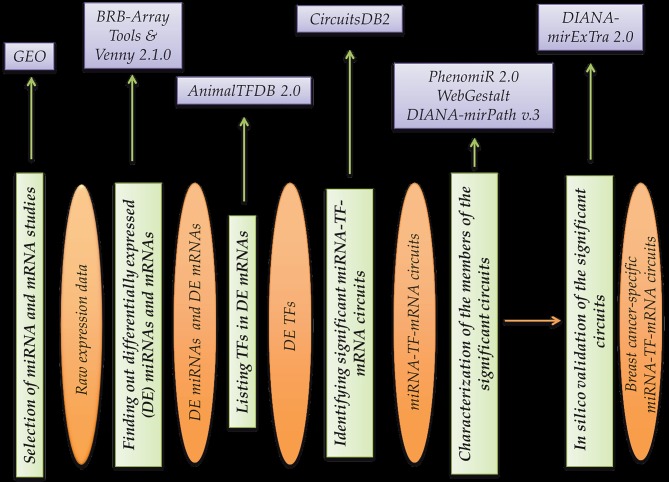
In this study we focused on finding out regulatory miRNA-TF-mRNA circuits specific to breast cancer with the help of various web-based tools and databases. hTrough the workflow designed in the study and the bioinformatics methods used, three independent microarray studies (Table 1) were reanalyzed and were converted to circuits that are important in breast cancer development. The whole process enabled us to make more use of the data in the literature, which was achieved by using a microarray that is a high-throughput and expensive method. The tools used in the workflow are not only used to cross-check the data but also to validate the robustness of the results of this work. Such a work flow can also be applied for other diseases (Figure 4).
Figure 4.
General study design and workflow. Purple boxes represent bioinformatics tools used for the processing of the data, green boxes represent main work steps, and orange ellipses represent input and outputs.
Circuit analysis revealed 3 significant miRNA-TFmRNA circuits that are SOX10- and hsa-miR-301adependent: SOX10→hsa-miR-301a→HOXA3, SOX10→hsamiR-301a→KIT, and SOX10→hsa-miR-301a→NFIB (Figure 1a). hsa-miR-301a, the common member of the circuits, is known to be an important miRNA in breast cancer. It was found to be upregulated in breast tumors for our circuits (Figure 1b). In 2011, it was reported as a novel oncogene in lymph node-negative breast cancer and it was shown to mediate proliferation, migration, and invasion (Shi et al., 2011) . Since then, other studies have been performed regarding the role of miR-301a upregulation in breast cancer (Ma et al., 2014; McDermott et al., 2014) . In 2014, upregulation of that miRNA was found to be associated with poor prognosis in triple-negative breast cancers (Yu et al., 2014) . When we searched the expression information of miRNA in PhenomiR (Ruepp et al., 2010) , it was found to be overexpressed in breast tumors in independent studies (Table 2). Considering the concordance of the literature with our results and the pathological, histological, and molecular variability (e.g., ER+, HER2+, triple-negative) of the samples in Table 2 and in the literature, hsa-miR-301a, which is consistently upregulated in different breast cancer cell lines and tumors, may be accepted as a generalized marker for breast cancer development. The results of DIANA-mirPath together with WebGestalt may strengthen the role of these significant circuits in breast cancer. The pathway enrichment analysis results showed that the target genes and the target TF SOX10 were found to be members of cancer-specific pathways like TGF-beta, mTOR, and p53 signaling pathways. SOX10, which is the common TF in our circuits, was reported to be one of the regulated TFs in breast cancer by Zang et al. Those authors combined seven studies that conducted next-generation sequencing for breast tumors and normal samples and, concordant with our in silico results, SOX10 was found to be downregulated in breast cancers (Zang et al., 2017) .
The two common members of the three circuits in this study are two regulative elements of gene regulation, a miRNA and a TF. The balance of the reciprocal regulation among these two important elements may determine the fate of the cell, which is in our situation cancer progression. In the suggested circuits in this study the regulative effect of miRNA is seen to be dominant and it is consistently upregulated and negatively regulates the TF (SOX10) expression. This tight regulation among miRNATF affected the cancer-related target genes HOXA3 , KIT, and NFIB. The well-known regulation of these target genes in different cancers (Moon et al., 2011; Jachetti et al., 2017; Zhang et al., 2017) and validation of them to be downregulated in breast tumor samples by an independent microarray dataset support the idea of the systems biology approach in cancers originating from different tissues.
In summary, the results of this study show that regulatory motifs involving miRNAs and TFs may be useful for understanding breast cancer regulation and for predicting new biomarkers. Characterization of these combinatorial regulatory mechanisms is important to reveal the biological processes that take part in breast cancer in detail by constructing networks and circuits. Finding out new circuits combining miRNAs, TFs, and mRNAs may provide further insight to improve our knowledge of breast cancer development.
References
- Calin GA , Dumitru CD , Shimizu M , Bichi R , Zupo S , Noch E , Aldler H , Rattan S , Keating M , Rai K et al ( 2002. ). Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia . P Natl Acad Sci USA 99 : 15524 - 15529 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R , Domrachev M , Lash AE ( 2002. ). Gene Expression Omnibus : NCBI gene expression and hybridization array data repository . Nucleic Acids Res 30 : 207 - 210 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enerly E , Steinfeld I , Kleivi K , Leivonen SK , Aure MR , Russnes HG , Ronneberg JA , Johnsen H , Navon R , Rodland E et al ( 2011. ). miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors . PLoS One 6 : e16915 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friard O , Re A , Taverna D , De Bortoli M , Cora D ( 2010a. ). CircuitsDB: a database of mixed microRNA/transcription factor feed-forward regulatory circuits in human and mouse . BMC Bioinformatics 11 : 435 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friard O , Re A , Taverna D , De Bortoli M , Corá D ( 2010b. ). CircuitsDB: a database of mixed microRNA/transcription factor feed-forward regulatory circuits in human and mouse . BMC Bioinformatics 11 : 435 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV , Ferracin M , Liu CG , Veronese A , Spizzo R , Sabbioni S , Magri E , Pedriali M , Fabbri M , Campiglio M et al ( 2005. ). MicroRNA gene expression deregulation in human breast cancer . Cancer Res 65 : 7065 - 7070 . [DOI] [PubMed] [Google Scholar]
- Jachetti E , Rigoni A , Bongiovanni L , Arioli I , Botti L , Parenza M , Cancila V , Chiodoni C , Festinese F , Bellone M et al ( 2017. ). Imatinib spares cKit-expressing prostate neuroendocrine tumors, whereas kills seminal vesicle epithelial-stromal tumors by targeting PDGFR-β . Mol Cancer Ther 16 : 365 - 375 . [DOI] [PubMed] [Google Scholar]
- Joung JG Fei Z Identification of microRNA regulatory modules in Arabidopsis via a probabilistic graphical model. Bioinformatics. 2009;25:387–393. doi: 10.1093/bioinformatics/btn626. [DOI] [PubMed] [Google Scholar]
- Joung JG Hwang KB Nam JW Kim SJ Zhang BT Discovery of microRNA-mRNA modules via population-based probabilistic learning. Bioinformatics. 2007;23:1141–1147. doi: 10.1093/bioinformatics/btm045. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M Rauhut R Lendeckel W Tuschl T Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Latchman S Transcription factors: an overview. Int J Biochem Cell Biol. 1997;2725:1305–1312. doi: 10.1016/s1357-2725(97)00085-x. [DOI] [PubMed] [Google Scholar]
- Lee CH Kuo WH Lin CC Oyang YJ Huang HC Juan HF MicroRNA-regulated protein-protein interaction networks and their functions in breast cancer. Int J Mol Sci. 2013;14:11560–11606. doi: 10.3390/ijms140611560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC Ambros V An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lowery AJ Miller N Devaney A McNeill RE Davoren PA Lemetre C Benes V Schmidt S Blake J Ball G et al 2009. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res 11 R27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J Getz G Miska EA Alvarez-Saavedra E Lamb J Peck D SweetCordero A Ebert BL Mak RH Ferrando AA et al 2005. MicroRNA expression profiles classify human cancers. Nature 435 834 838 [DOI] [PubMed] [Google Scholar]
- Ma F Zhang J Zhong L Wang L Liu Y Wang Y Peng L Guo B Upregulated microRNA-301a in breast cancer promotes tumor metastasis by targeting PTEN and activating Wnt / β -catenin signaling. Gene. 2014;535:191–197. doi: 10.1016/j.gene.2013.11.035. [DOI] [PubMed] [Google Scholar]
- Maragkakis M , Reczko M , Simossis VA , Alexiou P , Papadopoulos GL , Dalamagas T , Giannopoulos G , Goumas G , Koukis E , Kourtis K et al ( 2009. ). DIANA-microT web server: elucidating microRNA functions through target prediction . Nucleic Acids Res 37 : W273 - W276 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott AM , Miller N , Wall D , Martyn LM , Ball G , Sweeney KJ , Kerin MJ ( 2014. ). Identification and validation of oncologic miRNA biomarkers for luminal A-like breast cancer . PLoS One 9 : e87032 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon HG , Hwang KT , Kim JA , Kim HS , Lee MJ , Jung EM , Ko E , Han W , Noh DY ( 2011. ). NFIB is a potential target for estrogen receptor-negative breast cancers . Mol Oncol 5 : 538 - 544 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeem H , Kufner R Zimmer R ( 2011. ). MIRTFnet : Analysis of miRNA regulated transcription factors . PLoS One 6 : e22519 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S , Li M , Choi K , Balch C , Kim S , Nephew KP ( 2009. ). MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression . Nucleic Acids Res 37 : W356 - W362 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AL , Wang ZC , De Nicolo A , Lu X , Brown M , Miron A , Liao X , Iglehart JD , Livingston DM , Ganesan S ( 2006. ). X chromosomal abnormalities in basal-like human breast cancer . Cancer Cell 9 : 121 - 132 . [DOI] [PubMed] [Google Scholar]
- Romero-Cordoba S , Rodriguez-Cuevas S , Rebollar-Vega R , Quintanar-Jurado V , Mafuz-Aziz A , Jimenez-Sanchez G , Bautista-Piña V , Arellano-Llamas R , Hidalgo-Miranda A ( 2012. ). Identification and pathway analysis of microRNAs with no previous involvement in breast cancer . PLoS One 7 : e31904 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A , Kowarsch A , Schmidl D , Buggenthin F , Brauner B , Dunger I , Fobo G , Frishman G , Montrone C , Theis FJ ( 2010. ). PhenomiR: a knowledgebase for microRNA expression in diseases and biological processes . Genome Biol 11 : R6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W , Gerster K , Alajez NM , Tsang J , Waldron L , Pintilie M , Hui AB , Sykes J , P'ng C , Miller N et al ( 2011. ). MicroRNA-301 mediates proliferation and invasion in human breast cancer . Cancer Res 71 : 2926 - 2937 . [DOI] [PubMed] [Google Scholar]
- Simon R , Lam A , Li M , Ngan M , Menenzes S , Zhao Y ( 2007. ). Analysis of gene expression data using BRB-ArrayTools . Cancer Inform 3 : 11 - 17 . [PMC free article] [PubMed] [Google Scholar]
- Tran DH , Satou K , Ho TB ( 2008. ). Finding microRNA regulatory modules in human genome using rule induction . BMC Bioinformatics 9 ( Suppl . 12): S5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turashvili G , Bouchal J , Baumforth K , Wei W , Dziechciarkova M , Ehrmann J , Klein J , Fridman E , Skarda J , Srovnal J et al ( 2007. ). Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis . BMC Cancer 7 : 55 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG , Tibshirani R , Chu G ( 2001. ). Significance analysis of microarrays applied to the ionizing radiation response . P Natl Acad Sci USA 98 : 5116 - 5121 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos IS , Vergoulis T , Paraskevopoulou MD , Lykokanellos F , Georgakilas G , Georgiou P , Chatzopoulos S , Karagkouni D , Christodoulou F , Dalamagas T et al ( 2016. ). DIANA-mirExTra v2. . 0: Uncovering microRNAs and transcription factors with crucial roles in NGS expression data . Nucleic Acids Res 44 : W128 - 134 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos IS , Zagganas K , Paraskevopoulou MD , Georgakilas G , Karagkouni D , Vergoulis T , Dalamagas T , Hatzigeorgiou AG ( 2015. ). DIANA-miRPath v3. . 0: Deciphering microRNA function with experimental support . Nucleic Acids Res 43 : W460 - W466 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J , Duncan D , Shi Z , Zhang B ( 2013. ). WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013 . Nucleic Acids Res 41 : W77 - W83 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S , De Micheli G ( 2005. ). Prediction of regulatory modules comprising microRNAs and target genes . Bioinformatics 21 ( Suppl . 2): ii93 - 100 . [DOI] [PubMed] [Google Scholar]
- Yu H , Li H , Qian H , Jiao X , Zhu X , Jiang X , Dai G , Huang J ( 2014. ). Upregulation of miR-301a correlates with poor prognosis in triple-negative breast cancer . Med Oncol 31 : 283 . [DOI] [PubMed] [Google Scholar]
- Zang H , Li N , Pan Y , Hao J ( 2016. ). Identification of upstream transcription factors (TFs) for expression signature genes in breast cancer . Gynecol Endocrinol 2017. ; 33 : 193 - 198 . [DOI] [PubMed] [Google Scholar]
- Zhang HM , Liu T , Liu CJ , Song S , Zhang X , Liu W , Jia H , Xue Y , Guo AY ( 2015. ). AnimalTFDB 2.0: a resource for expression, prediction and functional study of animal transcription factors . Nucleic Acids Res 43 : D76 - D81 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X , Liu G , Ding L , Jiang T , Shao S , Gao Y , Lu Y ( 2017. ). HOXA3 promotes tumor growth of human colon cancer through activating EGFR/Ras/Raf/MEK/ERK signaling pathway . J Cell Biochem (in press). [DOI] [PubMed]