Abstract
IMPORTANCE
DNA methylation may play an important role in schizophrenia (SZ), either directly as a mechanism of pathogenesis or as a biomarker of risk.
OBJECTIVE
To scan genome-wide DNA methylation data to identify differentially methylated CpGs between SZ cases and controls.
DESIGN, SETTING, AND PARTICIPANTS
Epigenome-wide association study begun in 2008 using DNA methylation levels of 456 513 CpG loci measured on the Infinium HumanMethylation450 array (Illumina) in a consortium of case-control studies for initial discovery and in an independent replication set. Primary analyses used general linear regression, adjusting for age, sex, race/ethnicity, smoking, batch, and cell type heterogeneity. The discovery set contained 689 SZ cases and 645 controls (n = 1334), from 3 multisite consortia: the Consortium on the Genetics of Endophenotypes in Schizophrenia, the Project among African-Americans To Explore Risks for Schizophrenia, and the Multiplex Multigenerational Family Study of Schizophrenia. The replication set contained 247 SZ cases and 250 controls (n = 497) from the Genomic Psychiatry Cohort.
MAIN OUTCOMES AND MEASURES
Identification of differentially methylated positions across the genome in SZ cases compared with controls.
RESULTS
Of the 689 case participants in the discovery set, 477 (69%) were men and 258 (37%) were non–African American; of the 645 controls, 273 (42%) were men and 419 (65%) were non–African American. In our replication set, cases/controls were 76% male and 100% non–African American. We identified SZ-associated methylation differences at 923 CpGs in the discovery set (false discovery rate, <0.2). Of these, 625 showed changes in the same direction including 172 with P < .05 in the replication set. Some replicated differentially methylated positions are located in a top-ranked SZ region from genome-wide association study analyses.
CONCLUSIONS AND RELEVANCE
This analysis identified 172 replicated new associations with SZ after careful correction for cell type heterogeneity and other potential confounders. The overlap with previous genome-wide association study data can provide potential insights into the functional relevance of genetic signals for SZ.
Schizophrenia (SZ) is a neurodevelopmental disorder affecting 1% of the population worldwide, often resulting in lifelong disability.1 Treatments are not efficacious for the most disabling symptoms such as deficits in working memory, attention, and social cognition.2 Understanding the underlying biology of SZ can inform both treatment and prevention strategies. Heritability estimates are high (70%−81%) and some genetic variants have now been identified3 including multiple common variants,4,5 copy number variants,6 and rare mutations.7,8 The Psychiatric Genomics Consortium has identified 108 loci9; despite its size, this study has highlighted modest effect sizes and the importance of regulatory DNA.10 Mechanistic studies in transgenic animals have significant limitations because animal models have different brain structures and cannot fully recapitulate the phenotypic components of SZ.11
Discordance rates between monozygotic twins are 48% to 50%, reflecting a complex genetic architecture and a role for nongenetic factors in the etiology of SZ.12 Environmental risk factors include growing up in lower socioeconomic and urban environments,13 prenatal infections,14 obstetric complications,15 and maternal malnutrition.16 Some of these environmental factors, such as obstetric complications, can interact with genetic risk factors to alter risk or worsen the course of SZ.17
Epigenetic processes, which can be modified by environment but also have a genetic component and regulate genetic signals, have been postulated as links between environmental exposures, genetic risk, and SZ. Epigenetics refers to heritable modifications of DNA or associated factors that have information content beyond the primary DNA sequence. DNA methylation (DNAm), a stable modification of cytosines in CpG dinucleotides, regulates neurobiological processes and is implicated in disorders such as Rett syndrome.18 DNA methylation depends on dietary sources of the essential amino acid methionine, and periods of famine have doubled SZ risk.19,20 The later onset of SZ, its episodic nature, and the induction of psychotic episodes while undergoing treatment with l-methionine21 also suggest a role for epigenetics.
Several case-control studies using blood22–26 and postmortem brain tissues27–30 have revealed differential genome-wide methylation patterns, but the number of CpGs interrogated and sample sizes have been limited. To our knowledge, only 1 large-scale methylome study of SZ of equivalent size to ours has been published. It used a methylated DNA enrichment method (methyl-CpG binding domain protein-enriched genome sequencing) in a Swedish population31 but did not account for cell heterogeneity in blood, which can lead to false-positive results.32,33 Only 1 of the smaller studies in-corporated this critical step in their analysis.26
In this study, we described a comprehensive analysis of genome-scale DNAm in a SZ case-control consortium and its relationship to genetic variation. We profiled genome-wide DNAm in 1334 blood samples using the Illumina HumanMethylation450 BeadChip, accounting for cell type heterogeneity to identify DNAm associated with disease. High-confidence findings were replicated in an independent set of 497 individuals. We established genotypic-methylation relationships and integrated these results with published genome-wide association study data. This study supports the importance of integrating DNAm and quantitative trait analyses into population-based SZ genomic analysis.
Methods
Study Samples
eTable 1 in the Supplement describes the demographic characteristics of the discovery and replication samples. The discovery set consisted of 1334 blood-derived DNA samples (689 SZ cases and 645 controls) from a population of probands and controls derived from the collaborative efforts of 3 ongoing, National Institute of Mental Health–funded, multisite consortia: the Consortium on the Genetics of Endophenotypes in Schizophrenia,34 the Project Among African-Americans to Explore Risks for Schizophrenia,35 and the Multiplex Multigenerational Family Study of Schizophrenia.36 In the replication stage, 497 independent samples (247 SZ cases and 250 controls) from the Genomic Psychiatry Cohort37 were frequency matched with respect to smoking status, age, and sex. Written informed consent was obtained from all individuals, and the study was approved by the Johns Hopkins Medicine Office of Human Subjects Research institutional review board and the Consortium on the Genetics of Endophenotypes in Schizophrenia, the Project Among African-Americans to Explore Risks for Schizophrenia, the Multiplex Multigenerational Family Study of Schizophrenia, and the Genomic Psychiatry Cohort.
Phenotype Assessment
Eligible probands were at least 18 years of age and received SZ diagnosis according to the DSM-IV. Participants from the Project Among African Americans To Explore Risks for Schizophrenia self-identified as African American and had family members available for assessment. Diagnostic assessment was performed using the Diagnostic Interview for Genetic Studies along with medical records. The Multiplex Multigenerational Family Study of Schizophrenia probands were of European American descent from multigenerational families with at least 1 affected family member with SZ or schizoaffective disorder. The Consortium on the Genetics of Endophenotypes in Schizophrenia participants had at least 1 unaffected full sibling. Self-identified race/ethnicity was later confirmed with genetic information derived from the Infinium HumanMethylation450 BeadChip (Illumina) (eFigure 1 in the Supplement).
Data Processing and Statistical Modeling
Microarray Processing and Quality Control
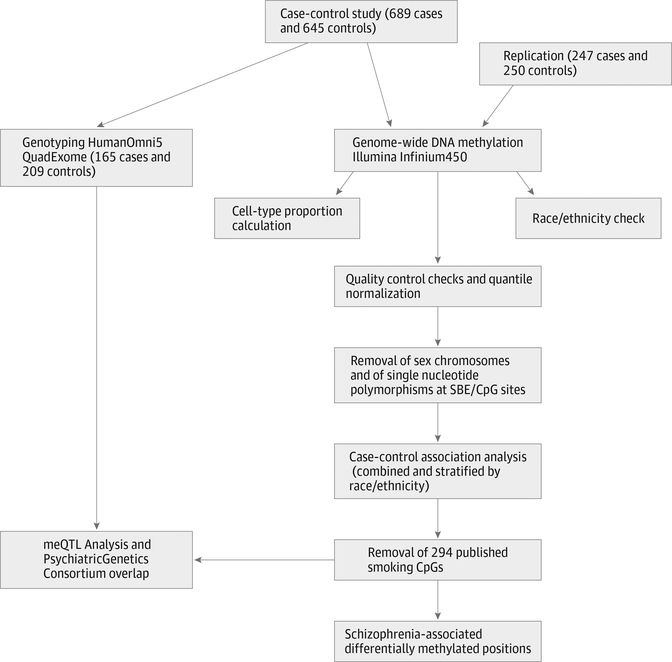
Experimental flow is presented in Figure 1. All analyses were performed in R version 3.1.1 (R Statistics), and raw intensity files were preprocessed and quantile normalized together using the Bioconductor package Minfi, version 1.8.9 (Bioconductor).38 Probes on the sex chromosomes were removed along with probes with annotated single-nucleotide polymorphisms (SNPs) (via the Single Nucleotide Polymorphism database 137 [National Center for Biotechnology Information]) in the CpG site (n = 16 817) or at single base extension sites (n = 7781), which left a total of 456 513 autosomal probes for calculation of beta scale methylation ratios and association analysis. Of the 2891 unique samples in the discovery set (including family members), subsequent samples were dropped according to the following quality control measures: (1) median methylated and median unmethylated signal average less than 10.5 (n = 9); (2) separate clustering in the first principal component of genome-wide DNAm (n = 8); (3) discrepancy between self-reported sex and predicted sex from DNAm and genotypes (n = 18); (4) discrepancy between self-reported race/ethnicity and predicted ancestry from DNAm data (n = 0); and (5) missing age, smoking, or phenotype in the database (n = 84). After removing family members, 1334 samples were left as the discovery arm of the case-control study. For the replication set, the same criteria were applied (1 dropped after principal component analysis and 2 were incongruent for race/ethnicity prediction). The resulting 497 samples constituted the replication set.
Figure 1. Analysis Workflow.
Identification of SZ-Associated Differentially Methylated Positions
To identify differentially methylated positions (DMPs) associated with SZ, we fit a linear regression model using the limma Bioconductor package (Bioconductor).39 We tested the association between proportion methylation values (Illumina “Beta” scale) and SZ diagnosis status at each CpG site, adjusting for sex, age, race/ethnicity (African American vs non–African American), current smoking status, estimated cell proportions for the 6 cell types, and the first 2 principal components of the negative control probes. To adjust for multiple testing, a false discovery rate (FDR) cutoff of less than 0.2 was used for genome-wide significance. We used all of the samples and adjusted for race/ethnicity to improve the power of our study, but also performed 3 additional race/ethnicity- and sex-stratified analyses (African American and non–African American only and male only) for clarification of findings.
The eMethods in the Supplement include a detailed description of genome-wide measurement of DNA methylation, assessment of cell heterogeneity and potential confounders, replication analysis, gene ontology and pathway analyses, Illumina genome-wide genotyping and meQTL analysis, and regression modeling formulas.
Results
Estimating and Correcting for Cell Heterogeneity Across Samples
In epigenetic studies using DNA from heterogeneous tissue, such as blood, it is critical to account for potential confounding by cell type. Each cell type has distinct methylation signatures,40 and differences in relative abundance of these cell populations between SZ cases and controls can create associations that are not causal. We implemented a statistical algorithm that estimates cell proportions based on reference DNAm signatures obtained from cell-specific data.32–37,41 The calculated cell proportions for CD4+ and CD8+ T cells, B cells, monocytes, granulocytes, and natural killer cells for the discovery and replication sets are presented in eFigure 2 in the Supplement. The largest statistically significant differences in cell proportions between cases and controls were observed in CD8+ T cells and monocytes, with SZ showing decreased CD8+ T cells and increased monocytes (CD8+ T cells discovery P value = 3.80E−9, replication P value = 2.95E−5 and monocyte discovery P value = 3.80E−3, replication P value = 8.73E−4). Granulocyte proportions were also higher among SZ cases in the discovery and replication sets (discovery P value = .11 and replication P value = 6.76E−4). CD4+ T cells, natural killer cells, and B cells did not show statistically significant differences in the same direction in both discovery and replication sets.
To our knowledge, only 1 smaller epigenome-wide association study of SZ (N = 105) has accounted for cell heterogeneity.26 Our findings are consistent with the magnitude and direction of their estimated cell proportion differences. We observed no significant association between anti-psychotic use and proportions of CD8+ T-cell counts (P value = .62), monocytes (P value = .58), or granulocytes (P value = .54, eFigure 3 in the Supplement).
Genome-Wide Methylation Differences in Discovery and Replication Sets
Our discovery sample included 1334 unrelated individuals (689 SZ cases and 645 controls) from 3 large NIMH consortia: the Consortium on the Genetics of Endophenotypes in Schizophrenia, the Project Among African-Americans to Explore Risks for Schizophrenia, and the Multiplex Multigenerational Family Study of Schizophrenia (see eMethods and eTable 1 in the Supplement for demographic information).Of the 456 513 CpGs examined, 945 CpGs were associated with case-control status at an FDR of less than 0.20.
We found it critical to account for cell type proportions, even after adjusting for sex, age, race/ethnicity, smoking, and batch effects (removing the first 2 principal components of negative control probes). Cell type correction markedly reduced the number of significant results (eFigure 4 and eFigure 5 in the Supplement). To assess residual confounding after adjusting for cell proportion estimates, we plotted adjusted estimates and residual values against cell proportions and showed that the linear models provided an adequate correction for these factors (eFigure 6 and eFigure 7 in the Supplement). In addition to controlling for smoking status, we further removed 294 CpGs directly associated with smoking.42,43 This eliminated 22 CpGs from the discovery results, leaving 923 SZ - associated DMPs (eTable 2 in the Supplement).
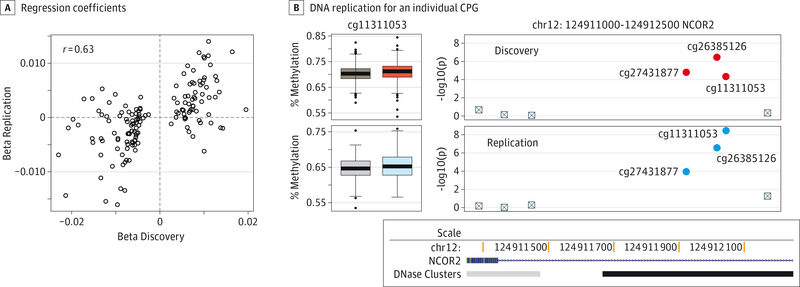
We sought to confirm our results in a replication set of 497 independent samples (247 SZ cases and 250 controls). Of the 923 DMPs from the discovery set, 625 (68%) showed association in the same direction and 172 (19%) of these had a replication P value of less than. 05. Quadrant correspondence (proportion of replication CpG effect estimates in the same direction as the discovery estimate) was even stronger when considering more stringent FDR thresholds among the discovery sample, with 80% correspondence at an FDR of less than 0.05 (Figure 2A).
Figure 2. Changes in DNA Methylation are Concordant Between Discovery and Replication Data Sets.
A, Regression coefficients of the DNA methylation differences at the top-ranked 150 differentially methylated positions in the discovery set are significantly correlated with the coefficients found in the replication set (r = 0.63; P value <.001). B, DNA methylation for an individual CpG in the intron of NCOR2, in the discovery (top) and replication (bottom) data sets, and corresponding P values for neighboring CpGs.
The location, effect size (magnitude of covariate-adjusted methylation change comparing SZ cases with controls), and statistical support for each of the 172 replicated SZ-DMPs are reported in eTable 3 in the Supplement. The first 15 DMPs with the lowest FDR values are presented in the Table. Among the genes near DMPs, those previously associated with SZ include RPS6KA1,44 MAD1L1,5 KLF13,45 SULT4A1,46 NPDC1,47 AUTS2,48 PIK3R1,49 and PNPO.50 We found genes previously reported to play a role in neuronal development and function (S100A2, SULT4A1, HES1, NTM, MARK4, KIFC3, NPDC1, SCAP, DHCR24, HS6ST1, GAS7, CARD10, DYRK2, RPS6KA4, SLC38A10, CUTA, FOXO1, LRFN3, and IRS1) and genes involved in both T-cell development and neuronal-cell fate (ZC3H12D, TCF3, MAP3K1, and IKZF4). Genes with multiple DMPs nearby included 2 genes previously associated with SZ (DDR151 and IFITM152), a component of the mechanistic target of rapamycin signaling pathway that facilitates synaptic plasticity (RPTOR53), and a gene critical in neurogenesis (NCOR2,54 Figure 2B and2C). Genes with SZ-DMPs near them have the same average length as genes on the whole array, but do have more probes per gene.
Table.
Genome-Wide Significant Schizophrenia-Associated DMPsa
| Illumina Gene Annotation | Genes Within 50 kb of Associated CpG | Discovery Set(n = 1334) |
Replication Set(n = 497) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Probe | Chromosome | Position | Δ | P Value | FDR | Δ | P Value | ||
| cg04792777 | 14 | 106322429 | NA | IGHE; IGHG1; IGHD; KIAA0125; MIR4507; MIR4538; MIR4537; MIR4539 | −0.007 | 1.06E–07 | 0.01 | −0.008 | E–<.001 |
| cg13092108 | 1 | 26857284 | RPS6KA1 | RPS6KA1; MIR1976 | −0.008 | 1.16E–07 | 0.01 | −0.009 | 4.38E–05 |
| cg04962621 | 16 | 4714733 | MGRN1 | MGRN1; NUDT16L1; ANKS3 | −0.009 | 1.32E–07 | 0.01 | −0.007 | 6.95E–05 |
| cg13997435 | 1 | 153538406 | S100A2 | S100A6; S100A5; S100A4; S100A3; S100A2; S100A16; S100A14 | −0.007 | 3.26E–07 | 0.01 | −0.005 | 2.77E–02 |
| cg26385126 | 12 | 124912021 | NCOR2 | NCOR2 | 0.006 | 4.99E–07 | 0.01 | 0.010 | 3.91E–07 |
| cg07458272 | 19 | 34744396 | KIAA0355 | LSM14A; KIAA0355 | 0.010 | 5.39E–07 | 0.01 | 0.007 | 2.10E–02 |
| cg13549638 | 17 | 78860076 | RPTOR | RPTOR | 0.013 | 6.70E–07 | 0.01 | 0.008 | 3.72E–02 |
| cg10975863 | 14 | 68830704 | RAD51B | RAD51B | 0.011 | 6.90E–07 | 0.01 | 0.010 | 1.62E–02 |
| cg22891595 | 3 | 193570256 | NA | AK091265 (cDNA) | 0.011 | 6.94E–07 | 0.01 | 0.006 | 4.98E–02 |
| cg06996599 | 6 | 30619232 | C6orf136 | PPP1R10; MRPS18B; ATAT1; DHX16; PPP1R18; NRM; MDC1 | 0.007 | 9.39E–07 | 0.02 | 0.006 | 3.28E–03 |
| cg23387863 | 15 | 77472416 | PEAK1 | PEAK1 | 0.010 | 1.02E–06 | 0.02 | 0.011 | 6.62E–04 |
| cg25323444 | 7 | 2111060 | MAD1L1 | MAD1L1 | 0.011 | 1.07E–06 | 0.02 | 0.011 | 2.32E–04 |
| cg11621113 | 19 | 12776725 | MAN2B1 | ZZNF791; MAN2B1; DHPS; FBXW9; WDR83; TNPO2 | −0.008 | 1.09E–06 | 0.02 | −0.013 | 8.72E–10 |
| cg23009327 | 19 | 1630248 | TCF3 | MBD3; UQCR11; TCF3 | 0.010 | 1.32E–06 | 0.02 | 0.007 | 7.92E–03 |
| cg12939085 | 11 | 67166104 | PPP1CA | POLD4; CLCF1; RAD9A; PPP1CA; TBC1D10C; CARNS1; RPS6KB2; PTPRCAP; CORO1B | 0.008 | 2.45E–06 | 0.02 | 0.006 | 4.65E–03 |
Abbreviations: DMP, differentially methylated positions; FDR, false discovery rate; NA, not available.
Listed are the top 15 replicated results. All replicated DMPs with an FDR of less than 0.20 are listed in eTable 3 in the Supplement.
In male-only analyses, 23 replicated SZ-DMPs were identified (eTable 4 in the Supplement), 16 of which were among the replicated DMPs in the sex-combined analyses. Stratified analysis in non–African American samples revealed similar results, although with less statistical significance given the reduction in sample size (eTable 5 in the Supplement). The African American group showed a greater number of statistically significant DMPs, despite the lower sample size, potentially owing to uncorrected population stratification among samples of African descent because ethnicity adjustment in these data was categorized as African American vs non–African American (eTable 6 in the Supplement). Preliminary post hoc analysis of the influence of antipsychotic medications on DNAm revealed that atypical antipsychotics exerted the biggest effect. We then grouped the SZ cases into 2 subgroups: 1 that received atypical antipsychotics, and 1 that received typical antipsychotics only or no treatment. Four of the 172SZ-DMPs were affected by atypical antipsychotic treatment (P value ≤ .05), with 1 probe inside of the gene MARCH11 and 3 near the promoters of the genes SATB1 and IFITM1 (eFigure 8 in the Supplement).
Pathway and Gene Ontology Analysis
Statistically significant (FDR < 0.2) functional pathway categories among SZ-DMPs included those related to regulation of signaling and signal transduction (eTable 7 in the Supplement). While gene ontology is centered on genes, network analysis focuses on their physical interactions. Consistent with the gene ontology results, the top 3 most relevant regulatory networks from our pathway analysis included (1) amino acid metabolism, energy production, and post-translational modification; (2) hereditary disorder, neurological disease, and psychological disorders (eFigure 9 in the Supplement); and (3) neurological disease, organismal injury and abnormalities, and embryonic development.
Relationship Between SZ-DMPs and SZ-Associated Genetic Loci
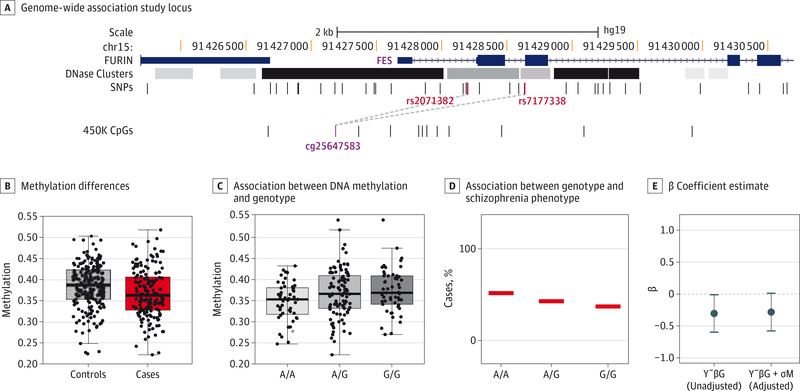
Our replicated SZ-DMPs overlapped 3 genetic regions identified in a 2014 Psychiatric Genomics Consortium mega-analysis9: regions 7, 11, and 37 from that publication list. Enrichment analysis using a Fisher exact test was not statistically significant (P = .47); however, the 450K array covers only 87 of the 105 autosomal PGC regions. In region 11 (chromosome 15), 2 SZ-SNPs (rs7177338 and rs2071382) are associated with methylation at the SZ-DMP (cg25647583), located in an intergenic region and equidistantly located (approximately 500 base pairs) between FURIN and FES genes (Figure 3A-C and eTable 8 in the Supplement). Notably, we observed the same genetic risk association to these SNPs in our data (Figure 3D). We further assessed the potential for DNAm mediation of this genetic risk. We estimated the effect of the SNP genotype G on SZ (SZ affected by G), and the effect of the SNP on SZ, adjusting for methylation (M) at the CpG locus (SZ affected by G + M). Conditioning on M did not significantly reduce the effect of G on SZ (β coefficient estimate for G alone, −0.301; P = .04; β coefficient estimate for G conditioning on M, −0.280; P = .06; Figure 3E), suggesting that the genotype is controlling M and SZ independently.
Figure 3. Genotype-Dependent Differentially Methylated Positions in Psychiatric Genomics Consortium Schizophrenia-Associated Locus.
A, Psychiatric Genomics Consortium Genome-Wide Association Study locus (region 11) with an overlapping schizophrenia (SZ)-associated CpG. Dashed lines indicate single-nucleotide polymorphism–CpG cisregulation between the 2 SZ–single-nucleotide polymorphisms and locus cg25647583. B, Methylation differences between SZ cases and controls at locus cg25647583. C, Association between DNA methylation and genotype. In the Psychiatric Genomics Consortium Genome-Wide Association Study , the A allele is the risk allele (odds ratio = 1.0662). D, Association between genotype and SZ phenotype. The risk allele has increased frequency in our SZ cases. E, β coefficient estimate of the dependence of genotype on SZ phenotype, before and after adjusting for methylation. Error bars represent the 95% CIs for the estimate of β.
Discussion
To our knowledge, our study is the largest epigenome-wide association study of SZ to date, using methods robust to cell-type confounding and with supporting genetic data. We identified and replicated 172 SZ-associated DMPs in 689 cases and 645 controls. The small but significant changes in DNAm are of similar effect size to other methylome studies of complex disorders33,55,56 and highlight the potential usefulness of using blood as a cost-effective surrogate tissue.
The SZ-DMPs discovered in this study include genes important in neuronal function (eg, NCOR2, SULT4A1, and HES1), genes previously associated with SZ (eg, RPS6KA1, MAD1L1, and DDR1), and genes also involved in T-cell development (ZC3H12D, TCF3, and IKZF4). Several of the neuronal genes have been implicated in Alzheimer disease pathology (SORL1,57 HES1,58 MARK4,59 SCAP,60 DHCR24,61 GAS7,62 CARD10,63 CDC37,64 CUTA,65 IRS1,66 and FOXO167), supporting a common network of dysregulation in SZ and Alzheimer disease.68
Schizophrenia cases exhibited statistically significant lower estimated CD8+ T cells and higher monocyte and granulocyte counts than the controls, and these cell proportion differences constitute an important confounder of blood epigenetic studies. The direction of bias can be positive (as is typically presumed), inflating association signals, or negative, reducing ability to see true associations. Cell-specific studies using sorted purified samples are ideal but not always practical in epidemiologic settings, and this adjustment method can alleviate some bias associated with this limitation.
To understand the genetic architecture underlying our results, we integrated methylome-wide data with genotyping information from our samples. The Psychiatric Genomics Consortium association results revealed SZ-SNPs controlling the methylation level of an SZ-DMP (cg25647583), where the risk allele was associated with lower methylation levels in SZ cases. Although our genotyping data set was relatively small (n = 374), we confirmed findings from the large psychiatric consortium meta-analysis.
We attempted to replicate previously published methylome-wide SZ association results. One study reported 450K platform findings among 2 small Japanese study samples (n = 105 discovery and n = 48 replication).26 Although this study adjusted for cell proportions, the samples were not adequately randomized by diagnosis on the arrays, creating batch effects. Attempting to remove this bias using aggressive batch effect correction methods would render the studies non-comparable. We also attempted to replicate the 2014 large (n = 1497) case-control study of SZ samples from Sweden,31 but the samples were assayed using methyl-CpG binding domain protein-enriched genome sequencing, a different technology from the one we used in this study, containing largely non-overlapping CpGs. Only 1 of their 15 replicated methylation blocks overlapped probes available on the 450K array. For their top gene, FAM63B, the closest 450K CpG (cg21149266) lies 126 bp away from their closest validated position (chr15: 59146756). We observed no statistically significant difference between SZ cases and controls at this position in our discovery set (P value = .12)but detected a difference in the same reported direction in our replication se t(P value = .002) (eFigure 10 in the Supplement).
In this emerging area of SZ epigenetics, there is bound to be an initial period of valid and interesting differences of opinion and gaps in our knowledge. Moreover, in the case of SZ, the brain is inaccessible, making blood studies valuable. While the effect sizes are small, that is also the case with genetic variation.69 Other issues will require clarification including the relative causal and risk weight of factors associated with SZ (diet, medications, and stress) and how these factors increase risk or whether they are epiphenomena. The more causal interpretation is supported by the well-known, biologically based brain developmental effects of environmental factors such as the Dutch Hunger Winter and influenza on the developing brains of embryos. A single study cannot answer all of these challenging questions. We will be looking at the relationship of the blood results reported in this study and related brain and neurocognitive data in subsequent studies. Future a priori planned Schizophrenia Psychiatric Genome-Wide Association Study and sequencing of these Consortia cohorts and integration with epigenetic analysis in these same participants will add to the value of both approaches. This study will hopefully be a foundational element raising as many questions as it resolves in an exciting but novel area of SZ research.
Conclusions
We identified novel differential methylation loci between SZ and control individuals from a large, diverse population, using a widely used and cost-effective, high-throughput technology. Using a robust analysis pipeline, with cell proportion correction and sequential replication in independent data sets, we provided candidate loci that contribute to the knowledge of SZ and possibly other psychoses. These findings lay a firm groundwork for the implementation of genome-wide methylation approaches to the understanding of neuropsychiatric disorders.
Supplementary Material
Key Points
Question
Is DNA differentially methylated in schizophrenia?
Findings
This epigenome-wide association study examined differentially methylated positions across the genome in blood-derived DNA samples in a discovery and a replication set. One hundred seventy-two positions showed statistically significant differences in methylation between schizophrenia and control participants in both samples, and a subset of these are located in a region of DNA that is implicated in genome-wide association studies of schizophrenia.
Meaning
DNA is differentially methylated in key regions of genomic DNA in schizophrenia.
Acknowledgments
Funding/Support: This work was supported by the National Institutes of Health grant “A Genome-Wide Methylation Scan for Epigenetic Contributions to Schizophrenia” under the U01 cooperative agreement mechanism: grants 5U01MH085270 (Dr A. P. Feinberg), 5U01MH085264 (Dr R. E. Gur), 5U01MH085269 (Dr Nimgaonkar), 5U01MH085281 (Dr Perry), 5U01MH085265 (Dr Braff), and 5U24MH068457–13 (Jay Tischfield, Rutgers University).
Role of the Funder/Sponsor: The sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Conflict of Interest Disclosures: None reported.
Contributor Information
Carolina Montano, Medical Scientist Training Program and Predoctoral Training Program in Human Genetics, Johns Hopkins University School of Medicine, Baltimore, Maryland; Center for Epigenetics, Johns Hopkins University School of Medicine, Baltimore, Maryland.
Margaret A. Taub, Department of Biostatistics, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland.
Andrew Jaffe, Lieber Institute for Brain Development, Johns Hopkins Medical Campus, Baltimore, Maryland; Department of Mental Health, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland.
Eirikur Briem, Center for Epigenetics, Johns Hopkins University School of Medicine, Baltimore, Maryland.
Jason I. Feinberg, Center for Epigenetics, Johns Hopkins University School of Medicine, Baltimore, Maryland.
Rakel Trygvadottir, Center for Epigenetics, Johns Hopkins University School of Medicine, Baltimore, Maryland.
Adrian Idrizi, Center for Epigenetics, Johns Hopkins University School of Medicine, Baltimore, Maryland.
Arni Runarsson, Center for Epigenetics, Johns Hopkins University School of Medicine, Baltimore, Maryland.
Birna Berndsen, Center for Epigenetics, Johns Hopkins University School of Medicine, Baltimore, Maryland.
Ruben C. Gur, Neuropsychiatry Section, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania.
Tyler M. Moore, Neuropsychiatry Section, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania.
Rodney T. Perry, Department of Epidemiology, University of Alabama at Birmingham, Birmingham.
Doug Fugman, Rutgers University Cell and DNA Repository, Piscataway, New Jersey.
Sarven Sabunciyan, Stanley Division of Developmental Neurovirology, Johns Hopkins, School of Medicine, Baltimore, Maryland.
Robert H. Yolken, Stanley Division of Developmental Neurovirology, Johns Hopkins, School of Medicine, Baltimore, Maryland.
Thomas M. Hyde, Lieber Institute for Brain Development, Johns Hopkins Medical Campus, Baltimore, Maryland.
Joel E. Kleinman, Lieber Institute for Brain Development, Johns Hopkins Medical Campus, Baltimore, Maryland.
Janet L. Sobell, Department of Psychiatry and Behavioral Sciences, Keck School of Medicine of University of Southern California, Los Angeles.
Carlos N. Pato, Department of Psychiatry and Behavioral Sciences, Keck School of Medicine of University of Southern California, Los Angeles.
Michele T. Pato, Department of Psychiatry and Behavioral Sciences, Keck School of Medicine of University of Southern California, Los Angeles.
Rodney C. Go, Department of Epidemiology, University of Alabama at Birmingham, Birmingham.
Vishwajit Nimgaonkar, Department of Psychiatry, University of Pittsburgh, Pittsburgh, Pennsylvania.
Daniel R. Weinberger, Lieber Institute for Brain Development, Johns Hopkins Medical Campus, Baltimore, Maryland.
David Braff, Department of Psychiatry, University of California San Diego School of Medicine, La Jolla; VISN22, Mental Illness Research, Education, and Clinical Center, VA Veterans Affairs San Diego Healthcare System, San Diego, California.
Raquel E. Gur, Neuropsychiatry Section, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania.
Margaret Daniele Fallin, Center for Epigenetics, Johns Hopkins University School of Medicine, Baltimore, Maryland; Department of Mental Health, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland.
Andrew P. Feinberg, Center for Epigenetics, Johns Hopkins University School of Medicine, Baltimore, Maryland; Department of Mental Health, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland; Departments of Medicine and Biomedical Engineering, Johns Hopkins University School of Medicine and Whiting School of Engineering, Baltimore, Maryland.
REFERENCES
- 1.Saha S, Chant D, McGrath J. A systematic review of mortality in schizophrenia: is the differential mortality gap worsening over time? Arch Gen Psychiatry. 2007;64(10):1123–1131. [DOI] [PubMed] [Google Scholar]
- 2.Lieberman JA, Stroup TS, McEvoy JP, et al. ; Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) Investigators. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N Engl J Med. 2005;353(12): 1209–1223. [DOI] [PubMed] [Google Scholar]
- 3.Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13 (8):537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purcell SM, Wray NR, Stone JL, et al. ; International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009; 460(7256):748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ripke S, O’Dushlaine C, Chambert K, et al. ; Multicenter Genetic Studies of Schizophrenia Consortium; Psychosis Endophenotypes International Consortium; Wellcome Trust Case Control Consortium 2. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45(10):1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455(7210): 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Purcell SM, Moran JL, Fromer M, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506(7487):185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girard SL, Gauthier J, Noreau A, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011;43(9):860–863. [DOI] [PubMed] [Google Scholar]
- 9.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarroll SA, Feng G, Hyman SE. Genome-scale neurogenetics: methodology and meaning. Nat Neurosci. 2014;17(6):756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nat Neurosci. 2010;13 (10):1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuang MT, Stone WS, Faraone SV. Genes, environment and schizophrenia. Br J Psychiatry Suppl. 2001;40:s18–s24. [DOI] [PubMed] [Google Scholar]
- 13.Vassos E, Pedersen CB, Murray RM, Collier DA,Lewis CM. Meta-analysis of the association of urbanicity with schizophrenia. Schizophr Bull. 2012; 38(6):1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown AS, Patterson PH. Maternal infection and schizophrenia: implications for prevention. Schizophr Bull. 2011;37(2):284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalman C, Allebeck P, Cullberg J, Grunewald C, Köster M. Obstetric complications and the risk of schizophrenia: a longitudinal study of a national birth cohort. Arch Gen Psychiatry. 1999;56(3): 234–240. [DOI] [PubMed] [Google Scholar]
- 16.Brown AS, Susser ES. Prenatal nutritional deficiency and risk of adult schizophrenia. Schizophr Bull. 2008;34(6):1054–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicodemus KK, Marenco S, Batten AJ, et al. Serious obstetric complications interact with hypoxia-regulated/vascular-expression genes to influence schizophrenia risk. Mol Psychiatry. 2008; 13(9):873–877. [DOI] [PubMed] [Google Scholar]
- 18.Bienvenu T, Chelly J. Molecular genetics of Rett syndrome: when DNA methylation goes unrecognized. Nat Rev Genet. 2006;7(6):415–426. [DOI] [PubMed] [Google Scholar]
- 19.Hoek HW, Brown AS, Susser E. The Dutch famine and schizophrenia spectrum disorders. Soc Psychiatry Psychiatr Epidemiol. 1998;33(8):373–379. [DOI] [PubMed] [Google Scholar]
- 20.St Clair D, Xu M, Wang P, et al. Rates of adult schizophrenia following prenatal exposure to the Chinese famine of 1959–1961. JAMA. 2005;294(5): 557–562. [DOI] [PubMed] [Google Scholar]
- 21.Pollin W, Cardon PV Jr, Kety SS. Effects of amino acid feedings in schizophrenic patients treated with iproniazid. Science. 1961;133(3446):104–105. [DOI] [PubMed] [Google Scholar]
- 22.Dempster EL, Pidsley R, Schalkwyk LC, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20(24): 4786–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishioka M, Bundo M, Koike S, et al. Comprehensive DNA methylation analysis of peripheral blood cells derived from patients with first-episode schizophrenia. J Hum Genet. 2013;58(2):91–97. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Chen J, Ehrlich S, et al. Methylation patterns in whole blood correlate with symptoms in schizophrenia patients. Schizophr Bull. 2014;40(4): 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinoshita M, Numata S, Tajima A, et al. DNA methylation signatures of peripheral leukocytes in schizophrenia. Neuromolecular Med. 2013;15(1): 95–101. [DOI] [PubMed] [Google Scholar]
- 26.Kinoshita M, Numata S, Tajima A, et al. Aberrant DNA methylation of blood in schizophrenia by adjusting for estimated cellular proportions. Neuromolecular Med. 2014;16(4): 697–703. [DOI] [PubMed] [Google Scholar]
- 27.Mill J, Tang T, Kaminsky Z, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82(3):696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Numata S, Ye T, Herman M, Lipska BK. DNA methylation changes in the postmortem dorsolateral prefrontal cortex of patients with schizophrenia. Front Genet. 2014;5:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wockner LF, Noble EP, Lawford BR, et al. Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Transl Psychiatry. 2014;4:e339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pidsley R, Viana J, Hannon E, et al. Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biol. 2014;15(10):483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aberg KA, McClay JL, Nerella S, et al. Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71(3):255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15(2):R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Aryee MJ, Padyukov L, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31(2): 142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calkins ME, Dobie DJ, Cadenhead KS, et al. The Consortium on the Genetics of Endophenotypes in Schizophrenia: model recruitment, assessment, and endophenotyping methods for a multisite collaboration. Schizophr Bull. 2007;33(1):33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aliyu MH, Calkins ME, Swanson CL Jr, et al. ; PAARTNERS Study Group. Project among African-Americans to explore risks for schizophrenia (PAARTNERS): recruitment and assessment methods [published correction appears in Schizophr Res. 2007;90(1–3):369]. Schizophr Res. 2006;87(1–3):32–44. [DOI] [PubMed] [Google Scholar]
- 36.Gur RE, Nimgaonkar VL, Almasy L, et al. Neurocognitive endophenotypes in a multiplex multigenerational family study of schizophrenia. Am J Psychiatry. 2007;164(5):813–819. [DOI] [PubMed] [Google Scholar]
- 37.Pato MT, Sobell JL, Medeiros H, et al. ; Genomic Psychiatry Cohort Consortium. The genomic psychiatry cohort: partners in discovery. Am J Med Genet B Neuropsychiatr Genet. 2013;162B(4):306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30 (10):1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reinius LE, Acevedo N, Joerink M, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One. 2012;7(7):e41361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joubert BR, Håberg SE, Nilsen RM, et al. 450Kepigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012;120(10):1425–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harlid S, Xu Z, Panduri V, Sandler DP, Taylor JA. CpG sites associated with cigarette smoking: analysis of epigenome-wide data from the Sister Study. Environ Health Perspect. 2014;122(7):673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan P, Zhou R, Wang Y, et al. Altered levels of extracellular signal-regulated kinase signaling proteins in postmortem frontal cortex of individuals with mood disorders and schizophrenia. J Affect Disord. 2010;124(1–2):164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stephens SH, Franks A, Berger R, et al. Multiplegenes in the 15q13-q14 chromosomal region are associated with schizophrenia. Psychiatr Genet. 2012;22(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meltzer HY, Brennan MD, Woodward ND, Jayathilake K. Association of Sult4A1 SNPs with psychopathology and cognition in patients with schizophrenia or schizoaffective disorder. Schizophr Res. 2008;106(2–3):258–264. [DOI] [PubMed] [Google Scholar]
- 47.Maccarrone G, Ditzen C, Yassouridis A, et al. Psychiatric patient stratification using biosignatures based on cerebrospinal fluid protein expression clusters. J Psychiatr Res. 2013;47(11):1572–1580. [DOI] [PubMed] [Google Scholar]
- 48.McCarthy SE, Gillis J, Kramer M, et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry. 2014;19(6):652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen C, Zhang C, Cheng L, et al. Correlation between DNA methylation and gene expression in the brains of patients with bipolar disorder and schizophrenia. Bipolar Disord. 2014;16(8):790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song H, Ueno S, Numata S, et al. Association between PNPO and schizophrenia in the Japanese population. Schizophr Res. 2007;97(1–3):264–270. [DOI] [PubMed] [Google Scholar]
- 51.Roig B, Virgos C, Franco N, et al. The discoidin domain receptor 1 as a novel susceptibility gene for schizophrenia. Mol Psychiatry. 2007;12(9):833–841. [DOI] [PubMed] [Google Scholar]
- 52.Arion D, Unger T, Lewis DA, Levitt P, Mirnics K. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol Psychiatry. 2007;62(7):711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lipton JO, Sahin M. The neurology of mTOR. Neuron. 2014;84(2):275–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jepsen K, Solum D, Zhou T, et al. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450(7168):415–419. [DOI] [PubMed] [Google Scholar]
- 55.Huynh JL, Garg P, Thin TH, et al. Epigenome-wide differences in pathology-free regions of multiple sclerosis-affected brains. Nat Neurosci. 2014;17(1):121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lunnon K, Smith R, Hannon E, et al. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat Neurosci. 2014;17(9):1164–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caglayan S, Takagi-Niidome S, Liao F, et al. Lysosomal sorting of amyloid-β by the SORLA receptor is impaired by a familial Alzheimer’s disease mutation. Sci Transl Med. 2014;6(223): 223ra20. [DOI] [PubMed] [Google Scholar]
- 58.Chacón PJ, del Marco Á, Arévalo Á, Domínguez-Giménez P, García-Segura LM, Rodríguez-Tébar A. Cerebellin 4, a synaptic protein, enhances inhibitory activity and resistance of neurons to amyloid-β toxicity. Neurobiol Aging. 2015;36(2):1057–1071. [DOI] [PubMed] [Google Scholar]
- 59.Seshadri S, Fitzpatrick AL, Ikram MA, et al. ; CHARGE Consortium; GERAD1 Consortium; EADI1 Consortium. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010; 303(18):1832–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suzuki R, Ferris HA, Chee MJ, Maratos-Flier E, Kahn CR. Reduction of the cholesterol sensor SCAP in the brains of mice causes impaired synaptic transmission and altered cognitive function. PLoS Biol. 2013;11(4):e1001532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crameri A, Biondi E, Kuehnle K, et al. The role ofseladin-1/DHCR24 in cholesterol biosynthesis, APP processing and Abeta generation in vivo. EMBO J. 2006;25(2):432–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Akiyama H, Gotoh A, Shin RW, et al. A novel role for hGas7b in microtubular maintenance: possible implication in tau-associated pathology in Alzheimer disease. J Biol Chem. 2009;284(47): 32695–32699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nho K, Corneveaux JJ, Kim S, et al. ; Multi-Institutional Research on Alzheimer Genetic Epidemiology (MIRAGE) Study; Add Neuro Med Consortium; Indiana Memory and Aging Study; Alzheimer’s Disease Neuroimaging Initiative (ADNI). Whole-exome sequencing and imaging genetics identify functional variants for rate of change in hippocampal volume in mild cognitive impairment. Mol Psychiatry. 2013;18(7):781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jinwal UK, Abisambra JF, Zhang J, et al. Cdc37/Hsp90 protein complex disruption triggers an autophagic clearance cascade for TDP-43 protein. J Biol Chem. 2012;287(29):24814–24820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hou P, Liu G, Zhao Y, et al. Role of copper andthe copper-related protein CUTA in mediating APP processing and Aβ generation. Neurobiol Aging. 2015;36(3):1310–1315. [DOI] [PubMed] [Google Scholar]
- 66.Kapogiannis D, Boxer A, Schwartz JB, et al. Dysfunctionally phosphorylated type 1 insulin receptor substrate in neural-derived blood exosomes of preclinical Alzheimer’s disease. FASEB J. 2015;29(2):589–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manolopoulos KN, Klotz LO, Korsten P, Bornstein SR, Barthel A. Linking Alzheimer’s disease to insulin resistance: the FoxO response to oxidative stress. Mol Psychiatry. 2010;15(11): 1046–1052. [DOI] [PubMed] [Google Scholar]
- 68.Douaud G, Groves AR, Tamnes CK, et al. A common brain network links development, aging, and vulnerability to disease. Proc Natl Acad Sci U S A. 2014;111(49):17648–17653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43(10):969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.