The mitochondrial genome is a hotspot for divergence in thermal growth differences in yeast.
Abstract
Genetic analysis of phenotypic differences between species is typically limited to interfertile species. Here, we conducted a genome-wide noncomplementation screen to identify genes that contribute to a major difference in thermal growth profile between two reproductively isolated yeast species, Saccharomyces cerevisiae and Saccharomyces uvarum. The screen identified only a single nuclear-encoded gene with a moderate effect on heat tolerance, but, in contrast, revealed a large effect of mitochondrial DNA (mitotype) on both heat and cold tolerance. Recombinant mitotypes indicate that multiple genes contribute to thermal divergence, and we show that protein divergence in COX1 affects both heat and cold tolerance. Our results point to the yeast mitochondrial genome as an evolutionary hotspot for thermal divergence.
INTRODUCTION
The genetic architecture of phenotypic divergence between species is unresolved. There remains considerable uncertainty as to whether evolution occurred through accumulation of numerous small-effect changes (“micromutationism”) or often involves “major genes” of large effect (1). While quantitative trait mapping has been successfully applied to closely related, interfertile species [reviewed in (2)], the results may not be representative of phenotypic divergence in general, because the characters that distinguish sibling species and domesticated organisms evolved over short time scales and potentially favor large-effect loci. However, systematic dissection of divergence between distantly related species has been difficult because of reproductive barriers.
The genus Saccharomyces contains post-zygotically isolated species with substantially diverged genomes, and the ease of genetic manipulation of yeast may allow us to address the genetic architecture of evolution with a systematic approach. While the Saccharomyces species share their preference for fermentative metabolism with many other yeast species (3), they differ dramatically in their thermal growth profile (4, 5). Saccharomyces cerevisiae is the most heat-tolerant species in this lineage, capable of growing at temperatures of 37° to 42°C, while its sister species Saccharomyces paradoxus can grow up to 39°C and the more distantly related Saccharomyces kudriavzevii and Saccharomyces uvarum are more cold-tolerant and only capable of growing at temperatures up to 34° to 35°C (4, 5). Previous studies in yeasts have implicated a small number of genes involved in temperature divergence (4, 6). However, every gene product has the potential to be thermolabile, and only a single systematic screen has been conducted (7), which reported that multiple genes contribute to thermal differences between S. cerevisiae and S. paradoxus, two species with modest differences in heat tolerance.
In the present study, we examined the genetic basis of thermal divergence between S. cerevisiae and S. uvarum, two species that are more divergent at synonymous sites than human and mouse (8, 9). These two species are capable of forming hybrids, but the hybrids cannot produce viable spores. Mechanisms underlying the reproductive isolation could involve mitochondrial-nuclear incompatibilities (10, 11), defects in recombination due to high levels of sequence divergence (12, 13), and chromosomal rearrangements (14, 15). Of relevance, mitochondrial genome variation has been shown to affect high-temperature growth in S. cerevisiae (16, 17) and S. paradoxus (18).
To identify genes involved in the evolution of thermal growth differences, we screened 4792 nonessential genes for noncomplementation and used the reciprocal hemizygosity test (19) to validate genes that came out of the screen. While no single nuclear-encoded genes of large effect were recovered, we did find that mitochondrial DNA (mtDNA) plays a remarkable role in divergence of both heat and cold tolerance across the Saccharomyces species and that multiple mitochondria-encoded genes are involved, including COX1, previously shown to be involved in mitochondrial-nuclear interspecific incompatibilities (11).
RESULTS
A noncomplementation screen for thermosensitive alleles reveals mitochondrial effects
Hybrids of S. cerevisiae and S. uvarum are heat tolerant (Fig. 1A). Thus, deletion of S. cerevisiae heat-tolerant alleles in a hybrid should weaken heat tolerance through noncomplementation. We screened 4792 nonessential genes in the yeast deletion collection for these thermotolerance genes by mating both the MATa (BY4741) and MATα (BY4742) deletion collection to S. uvarum and growing them at high temperature (37°C). For comparison, we also screened the resulting hemizygote collections for two other traits, where the S. cerevisiae phenotype is dominant in the hybrid (Fig. 1A): copper resistance (0.5 mM copper sulfate) and ethanol resistance (10% ethanol at 30°C). We found 80, 13, and 2 hemizygotes that exhibited reduced resistance to heat, copper, and ethanol, respectively, in both the BY4741 and BY4742 hemizygote collections (Fig. 1B). In our initial assessment of these genes, we validated a copper-binding transcription factor, CUP2 (20), for copper resistance through reciprocal hemizygosity analysis (fig. S1).
Fig. 1. A noncomplementation screen identified genes underlying phenotypic divergence between S. cerevisiae and S. uvarum.
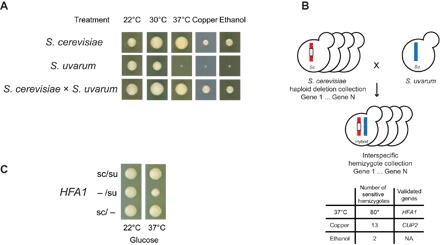
(A) S. cerevisiae and S. uvarum differ in heat (37°C), copper (0.5 mM, 22°C), and ethanol (10%, 30°C) tolerance. The resistant S. cerevisiae alleles are dominant, shown by the hybrid (S. cerevisiae × S. uvarum), compared to S. cerevisiae (diploid, S288C background) and S. uvarum (diploid, CBS7001 background). Growth is after 3 days. (B) S. cerevisiae haploid deletion collection was crossed to S. uvarum to construct an interspecies hemizygote collection. The number of noncomplementing genes is shown for each phenotype; the asterisk indicates that the number includes strains carrying S. uvarum mtDNA. (C) HFA1 hemizygote with only an S. cerevisiae allele (sc/−) shows better 37°C growth than one with only an S. uvarum allele (−/su). Growth is after 5 days. See fig. S1B for quantification. NA, not available.
Nearly all of the heat-sensitive hemizygotes (77 of 80) were from respiration-deficient (“petite”) S. cerevisiae parents. We found that many of these strains carried S. uvarum mtDNA via polymerase chain reaction (PCR) of a mitochondrial marker. Although not extensively tested, other hemizygotes are expected to carry S. cerevisiae mtDNA, a typical outcome of S. cerevisiae × S. uvarum crosses (21). The difference in mtDNA inheritance was likely caused by loss of mtDNA in the S. cerevisiae petite parents. We confirmed one gene (HFA1) by reciprocal hemizygosity analysis (Fig. 1C and fig. S1) that causes a moderate loss of heat tolerance due to the S. uvarum allele in the presence of S. cerevisiae mtDNA. HFA1 encodes a mitochondrial acetyl–coenzyme A (CoA) carboxylase and is involved in mitochondrial fatty acid biosynthesis (22).
The inheritance of S. uvarum mtDNA in heat-sensitive hemizygotes suggested that mtDNA, rather than the deletion, could be the cause. To test whether the species’ mtDNA (“mitotype”) affects heat tolerance, we generated diploid hybrids of wild-type S. cerevisiae and S. uvarum with reciprocal mitotypes and grew them at different temperatures. In comparison to the hybrid with S. cerevisiae mitotype, the hybrid with the S. uvarum mitotype showed reduced fermentative growth (glucose medium) at 37°C compared to 22°C and almost no respiratory growth (glycerol medium) at 37°C (Fig. 2A).
Fig. 2. Mitochondria-encoded genes affect heat and cold tolerance.
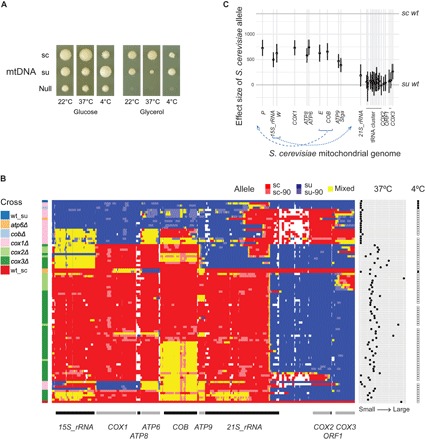
(A) Hybrids with S. cerevisiae (sc) and S. uvarum (su) mtDNA differ in high- and low-temperature growth. Growth is after 5 days (22° and 37°C) or 124 days (4°C). (B) Recombinant strains (rows) derived from mutant crosses (left) are clustered by genotype (middle). Wild-type S. cerevisiae (wt_sc) and S. uvarum (wt_su) mitotypes are at the bottom and top, respectively. Allele identity is shown for 12,574 orthologous single-nucleotide markers (sc and sc-90, S. cerevisiae; su and su-90, S. uvarum; mixed, heterozygous or chimeric; white, no data) in the S. cerevisiae gene order (bottom). Right: 37°C growth is the average size of nonpetite colonies on glycerol plates. Far right: The presence of 4°C glycerol growth is indicated by solid squares. (C) Effect size of S. cerevisiae alleles on 37°C growth on glycerol, with error bars representing 95% confidence intervals. The y axis is scaled to the phenotype of wild-type S. uvarum and S. cerevisiae mitotype (horizontal lines). Transfer RNAs (tRNAs) are labeled by their single-letter amino acid code and a black bar. Blue dashed lines indicate genome positions of S. uvarum genes compared to S. cerevisiae.
S. uvarum is not only known to be heat sensitive but also exhibits enhanced growth at low temperatures relative to S. cerevisiae (4). We thus tested and found that S. uvarum mitotype conferred a growth advantage at 4°C in comparison to S. cerevisiae mitotype (Fig. 2A), suggesting a potential trade-off between the evolution of heat and cold tolerance.
To test whether mtDNA-mediated evolution of temperature tolerance is specific to either the S. cerevisiae or S. uvarum lineages, we generated five additional hybrids with both parental mitotypes using two other Saccharomyces species (fig. S2). In comparison to the 22°C control, we find that both the S. cerevisiae and S. paradoxus nuclear genome conferred heat tolerance to hybrids with S. kudriavzevii and S. uvarum (rhoo comparison), but the S. cerevisiae mitotype conferred heat tolerance in comparison to the S. paradoxus, S. kudriavzevii, and S. uvarum mitotypes on glucose medium. For cold tolerance, we find that the S. uvarum mitotype conferred greater cold tolerance relative to the S. cerevisiae, S. paradoxus, and S. kudriavzevii mitotypes. None of the hybrids was as cold tolerant as S. uvarum on glycerol. Our results suggest that mtDNA has played an important role in divergence of thermal growth profiles among the Saccharomyces species, with heat tolerance evolving primarily on the lineage leading to S. cerevisiae and cold tolerance evolving primarily on the lineage leading to S. uvarum. A related study has shown that these differences have had a direct impact on the domestication of lager-brewing yeast hybrids to low-temperature fermentation (23).
Recombinant analysis identifies contribution of multiple mitochondria-encoded genes
To identify mtDNA genes conferring heat tolerance to S. cerevisiae, we tested whether S. uvarum alleles can rescue the respiratory deficiency of S. cerevisiae mitochondrial gene knockouts at high temperature. We crossed S. uvarum to previously constructed S. cerevisiae mitochondrial knockout strains and plated them on glycerol medium at 37°C. Because heteroplasmy is unstable in yeast, this strategy selects for recombinants between the two mitochondrial genomes: S. uvarum mtDNA is needed to rescue the S. cerevisiae deficiency, and S. cerevisiae mtDNA is needed to grow at high temperature (fig. S3). If the S. uvarum gene required for S. cerevisiae rescue is temperature sensitive, we expect to see no or small colonies on 37°C glycerol plates. Of the six genes tested, COX2 and COX3 deletions were rescued by S. uvarum at high temperature, although the colonies were often smaller than the hybrid with wild-type S. cerevisiae mtDNA. In contrast, COX1 and ATP6 deletions were minimally rescued (Fig. 2B), and COB and ATP8 deletions were not rescued. However, the absence of rescue could also result from a lack of recombination, especially for COB, because its genomic location has moved between the two species.
Using genome sequencing, we mapped breakpoints in 90 recombinants to determine which S. cerevisiae genes are associated with high temperature growth. The recombinants showed hotspots at gene boundaries and within the 21S ribosomal RNA (rRNA) (Fig. 2B). In most cases, the two species’ mtDNA recombine into a circular mitochondrial genome, but sometimes recombination resulted in mitochondrial aneuploidy, particularly for regions where the two species’ mitochondrial genomes are not colinear (see fig. S4B for examples). One complication of measuring mtDNA-dependent heat tolerance is the high rate of mtDNA loss, typically 1% in S. cerevisiae strains, but much higher in the hybrids and variable among recombinants (Supplementary Text and fig. S5). We thus measured the frequency of petites at 22°C and heat tolerance by the size of single colonies at 37°C on glycerol. We found that the petite frequency was associated with the absence of S. cerevisiae ORF1 (F-SceIII (24), a homing endonuclease linked to COX2 (fig. S5, B and C). For heat tolerance, we found a region including four protein-coding genes (COX1, ATP8, ATP6, and COB) with the largest effect (Fig. 2C). The effects associated with these genes are small compared to the total difference between two wild-type mitotypes, suggesting that other regions are required for complete rescue of high-temperature growth. S. cerevisiae COX2 and COX3 showed small but positive effects when the recombinants lacking them were compared to the wild-type S. cerevisiae mitotype (Fig. 2B). The differential heat sensitivity is unlikely to be caused by fitness defects because the recombinants grew normally at 22°C (fig. S4A).
We also found that nearly all mtDNA recombinants did not exhibit 4°C respiratory growth; one strain (S87) derived from the atp6Δ cross (Fig. 2B) was an exception, but another strain with the same mitochondrial genotype did not grow. The 4°C recombinant phenotypes suggest that cold tolerance might require multiple S. uvarum alleles and potentially a different set of genes than those underlying heat tolerance.
COX1 protein divergence affects both thermotolerance and cryotolerance
Because the recombinant strains did not resolve heat tolerance to a single gene, we tested individual genes by replacing S. cerevisiae with S. uvarum alleles via biolistic transformation (fig. S6) (25). We obtained allele replacements for two of the four genes in the region conferring heat tolerance (Fig. 3). For both genes, we used intronless alleles to eliminate incompatibilities in splicing (11).
Fig. 3. COX1 coding alleles affect growth at high and low temperature.
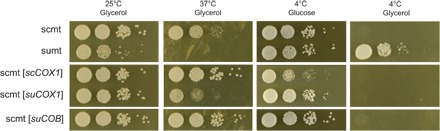
Hybrids carrying allele replacements and two wild-type controls were plated with 1:10 serial dilution and incubated at indicated temperatures. Growth is after 4 days for 25°C and 37°C, 25 days for 4°C on glucose, and 53 days for 4°C on glycerol. sc, S. cerevisiae; su, S. uvarum; mt, mtDNA. Alleles in the brackets were integrated into their endogenous position in S. cerevisiae mtDNA.
We observed a significant difference between S. cerevisiae and S. uvarum COX1 alleles for respiratory growth at 37°C in the hybrid background, with the S. uvarum allele being heat sensitive. The effect was not present at room temperature, and the S. uvarum allele conferred a growth advantage on glucose at 4°C. Thus, divergence in the COX1 coding sequence (CDS) affects both heat and cold tolerance. However, COX1 alleles do not explain the entire difference between the two species’ mitotypes: The strain bearing S. uvarum COX1 had an intermediate level of heat tolerance and did not confer cold tolerance on glycerol, suggesting that other mitochondrial genes are involved. The moderate effect of the COX1 alleles is also consistent with the small effect sizes shown by recombinant analysis (Fig. 2C). Surprisingly, the COX1 allele difference is only seen in the hybrid and not in a diploid S. cerevisiae background (fig. S7), suggesting that the allele difference in the hybrid depends on a dominant interaction with the S. uvarum nuclear genome.
The S. uvarum COB allele replacement rescued respiratory growth at high temperature, demonstrating that the S. uvarum COB protein is not heat sensitive. We were unable to generate the S. cerevisiae intronless COB allele replacement for comparison. Notably, both the intronless S. cerevisiae COX1 and S. uvarum COB allele replacement strains exhibited better growth than wild-type S. cerevisiae mtDNA at 37°C (Fig. 3), implying a dominant-negative role of these introns in the hybrid at high temperature.
DISCUSSION
In Saccharomyces species, the mitochondrial genome is not essential for viability, is large compared to insects and mammals (~86 kb), and is quite variable in intron content (26). While the mitochondrial genome can recombine and introgress between species (18, 24), it also contributes to reproductive isolation through incompatibilities with the nuclear genome (10, 11, 27). Our results show that the mitochondrial genome also makes a significant contribution to one of the most distinct phenotypic differences among the Saccharomyces species: their thermal growth profile. Below, we discuss the implications of our results in relationship to the genetic architecture of species’ phenotypic differences, the role of cytonuclear interactions in phenotypic evolution and reproductive isolation, and mitochondria as a hotspot in the evolution of Saccharomyces species.
Genetic architecture of interspecies differences in thermotolerance
Crosses between closely related, interfertile species have shown that phenotypic divergence can be caused by a few loci of large effect, many loci of small effect, or a mixture of the two (2). In this study, we carried out a genome-wide noncomplementation screen between two diverged yeast species. Of 4792 nonessential genes in our study, we found only one gene (HFA1) that showed a moderate effect on heat tolerance regardless of the mtDNA effect (Fig. 1C). Of relevance, 178 S. cerevisiae deletions are sensitive to 37°C (28), a rate comparable to a subsample we examined in this study (78 of 2251). We can thus conclude that the vast majority of the S. uvarum alleles tested exhibited no detectable loss of function at a temperature they do not experience in their native genome. However, our noncomplementation screen had some limitations. We did not test essential genes and could not detect genes whose effects were masked by mtDNA inheritance or epistasis, which could occur because of the hybrid carrying an otherwise complete complement of both nuclear genomes.
We found that allele differences in HFA1 affect heat tolerance. HFA1 encodes a mitochondrial acetyl-CoA carboxylase and participates in mitochondrial fatty acid synthesis, a process essential to cellular respiration and mitochondrial biogenesis (29). While disruption of HFA1 in S. cerevisiae resulted in a low level of lipoic acid and consequently a temperature-dependent respiratory defect (22, 30), the hemizygote with only the S. uvarum allele showed heat-sensitive growth on glucose but not glycerol (fig. S1C), suggesting that the divergence in heat tolerance of HFA1 might not be directly linked to its role in respiration. Further investigation is needed to elucidate the molecular mechanism by which HFA1 affects thermal divergence.
Although our screen led us to discover a pronounced temperature-dependent effect of mtDNA on respiratory growth and a more subtle effect on fermentative growth, the mtDNA effect explains only a small portion of the large difference in heat tolerance between the two species. The S. cerevisiae × S. uvarum hybrid without mtDNA grows at both 37° and 4°C on glucose (Fig. 2B), indicating that the nuclear genomes carry dominant factors that remain to be identified.
Despite the small number of genes in the mitochondrial genome, our results show that multiple genes within the mitochondrial genome influence heat tolerance. In addition to the large effect of the COX1-COB region, recombinants that inherited S. uvarum COX2 and/or COX3 are considerably more heat sensitive than a hybrid with a complete S. cerevisiae mtDNA genome. Furthermore, while the COX1-linked region showed the largest effect, the COX1 CDS does not explain the entire difference between two species’ mitotypes. Although we ruled out protein-coding changes in S. uvarum COB to be heat sensitive, changes in the other protein-coding sequences and in gene expression remain to be tested.
The cause of mtDNA-mediated differences in cryotolerance is more opaque. At 4°C, only one recombinant with a significant fraction of S. cerevisiae mtDNA grew better than hybrids with an S. cerevisiae mitotype, suggesting that multiple S. uvarum alleles are required for cold tolerance. Although we showed that S. uvarum COX1 increased cold tolerance on glucose, the effect is not seen on glycerol, suggesting that its effect on respiration might depend on the presence of other S. uvarum mitochondrial alleles. However, because the recombinants were all isolated at 37°C, it is possible that they all share some other genetic element or change that facilitates heat tolerance but inhibits 4°C growth.
Cytonuclear interactions in Saccharomyces evolution
In addition to mitochondria-encoded genes, approximately 1000 nuclear genes function in the mitochondria, many of which are involved in expression and regulation of mitochondrial genes and formation of the multisubunit cytochrome b and c complexes (31). Among Saccharomyces species, multiple cytonuclear incompatibilities have been shown to contribute to reproductive isolation. S. uvarum AEP2 cannot regulate the translation of S. cerevisiae ATP9 mRNA (10), while S. cerevisiae MRS1 cannot splice introns of S. paradoxus and S. uvarum COX1 (11). In addition, the S. uvarum RNA binding protein CCM1 has reduced affinity for the S. cerevisiae 15S rRNA (32). While these incompatibilities affect the construction of cybrids, where mtDNA from different species was introduced into S. cerevisiae (27), the phenotypic consequences besides loss of respiration are not known.
Our results show that the mitochondrial genomes of Saccharomyces species influence both heat and cold tolerance and provide multiple lines of evidence for the role of cytonuclear interactions. First, the temperature effects of species’ mitotypes interact with nuclear background (fig. S1). While S. cerevisiae hybrids without mtDNA (rhoo) grow similarly on glucose medium, S. cerevisiae mtDNA confers different levels of heat tolerance in hybrids with S. paradoxus, S. uvarum, and S. kudriavzevii, the latter of which only grows slightly better than the rhoo hybrid.
We also observed interactions between the COX1 allele replacements and their nuclear background. COX1 showed allele differences at high and low temperatures in the hybrid but not in S. cerevisiae. This difference can be explained by a species-specific dominant interaction, as might occur when there are hybrid protein complexes (33). In this scenario, S. uvarum COX1 can function with interacting S. cerevisiae proteins at high temperature but exhibits a loss of function when interacting with temperature-sensitive S. uvarum nuclear factors that are dominant to their S. cerevisiae orthologs. The nuclear factor is unlikely to be the previously reported intron splicing factor MRS1 because our COX1 alleles are intronless.
However, introns might affect temperature sensitivity. The intronless S. cerevisiae COX1 and S. uvarum COB alleles showed better respiratory growth at 37°C than wild-type S. cerevisiae mtDNA, suggesting a dominant negative role of introns in the hybrid. In Saccharomyces, the number and presence of mitochondrial introns is variable between species (34). This contrasts with high conservation of mitochondrial protein-coding sequences, which show over 90% sequence identity between S. cerevisiae and S. uvarum, much higher than the 80% average of nuclear-encoded genes (35). The rapid evolution of introns might require coevolution of splicing factors, such as COX1 and MRS1. The wild-type hybrid with S. cerevisiae mtDNA might be under burden of intron splicing at high temperature caused by dominant negative S. uvarum splicing factors. Nevertheless, many introns self-splice and/or encode maturases or homing endonucleases, which could be temperature sensitive in a nuclear-independent manner.
There is no clear indication that previously reported incompatibilities contribute to the mtDNA temperature phenotypes. The reported cytonuclear incompatibilities are recessive and thus should not contribute to the hybrid phenotypes. For example, although the S. cerevisiae MRS1 is incompatible with S. uvarum COX1, the latter can be correctly spliced by S. uvarum MRS1 in the diploid hybrid, at least at permissive temperatures. One possibility is that S. uvarum MRS1 is heat sensitive, which would explain the heat sensitivity of the S. uvarum mitotype because neither the S. cerevisiae nor S. uvarum MRS1 would splice S. uvarum COX1 at high temperature. Heat sensitivity of S. uvarum MRS1 was tested in our noncomplementation screen, but the result was inconclusive. The S. cerevisiae MRS1 deletion was complemented by the S. uvarum allele in the MATa (BY4741) cross, but its effect was masked by mtDNA inheritance in the MATα (BY4742) cross. In this regard, it is worth noting that S. cerevisiae chromosome 9, which carries MRS1, is duplicated in three of the recombinant strains; in two cases, these strains show increased 37°C growth compared to similar genotypes (table S1).
mtDNA and yeast evolution
It has been proposed that mtDNA plays a disproportionate role in Dobzhansky-Muller incompatibilities. Although it is a small genome, it heavily interacts with nuclear genes and has a high nucleotide substitution rate, leading to coevolution of the mitochondrial and nuclear genomes and multiple interspecific incompatibilities (36). Has adaptation played a role in driving these incompatibilities? Although no direct links are proven, evolution of the mitochondrial genome and mitonuclear epistasis has been linked to multiple phenotypes (21, 37, 38), including 37°C growth (16–18), and deficiencies in mtDNA cause heat sensitivity (39). Here, we show that mtDNA is important for evolution of heat and cold tolerance in distantly related species, caused by the accumulation of multiple small-to-medium effect changes and potentially mitonuclear epistasis. Together, the present and previous findings point to mtDNA as an evolutionary hotspot for yeast speciation and adaptation.
MATERIALS AND METHODS
Strains, growth conditions, and genetic manipulations
Strains used in this study are listed in table S2. S. cerevisiae was maintained on YPD (1% yeast extract, 2% peptone, and 2% dextrose) at 30°C; S. uvarum and S. cerevisiae × S. uvarum hybrids were maintained on YPD at room temperature. Strains were also grown on complete medium (CM; 0.3% yeast nitrogen base with amino acids, 0.5% ammonium sulfate, and 2% dextrose) or dropout medium (CM-xxx; 0.13% dropout powder, 0.17% yeast nitrogen base, 0.5% ammonium sulfate, and 2% dextrose), where xxx represents the missing amino acids when appropriate. SDPSer medium [synthetic dextrose proline d-serine, 2% dextrose, 0.17% yeast nitrogen base without ammonium sulfate or amino acids, l-proline (5 mg/ml), and d-serine (2 mg/ml)] was used to select for dsdAMX4 (40). Antibiotics were added to media when selecting for KanMX, NatMX, and hphMX. YPGly medium (1% yeast extract, 2% peptone, and 3% glycerol) was used to examine respiratory growth.
S. cerevisiae and S. uvarum strains were mated by mixing strains with opposite mating types on YPD at room temperature overnight. Diploid hybrids were obtained by plating the mating mixture to double-selection medium and confirmed by mating-type PCR.
Transformations in this study followed standard lithium acetate methods (41). When transforming S. uvarum or S. cerevisiae × S. uvarum hybrid, we used 37°C for heat shock and room temperature for incubation.
Strains lacking mtDNA (rho0) were generated by overnight incubation with shaking in liquid minimal medium (MM; 0.17% yeast nitrogen base without amino acid and ammonium sulfate, 0.5% ammonium sulfate, and 2% dextrose) containing ethidium bromide (25 μg/ml). Following incubation, the culture was plated to YPD and YPGly to identify nonrespiring colonies.
Interspecific hemizygote collections
trp1 S. uvarum strains YJF2600 and YJF2601 were constructed by replacing TRP1 with hphMX4 in YJF1449 (MATa) and YJF1450 (MATα) in the CBS7001 background (42), respectively. The haploid yeast deletion collections derived from BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) were arrayed in 384-well format using a Singer ROTOR (Singer Instruments, Watchet, UK) and mated to trp1 S. uvarum strains. Diploids were selected on CM-trp-his-leu-lys-ura plates. The resulting two interspecific hybrid collections were hemizygous for 4792 genes.
The hemizygote collections were screened for noncomplementation using the following conditions: (i) YPD at room temperature, 30°C, 35°C, and 37°C; (ii) CM with 0.5 mM copper sulfate at room temperature; and (iii) YPD with 10% ethanol at 30°C. Pictures of plates were taken on the second and fifth day of incubation using a Nikon D3100 camera. Colonies that were visually smaller than wild-type (represented by most of the hemizygotes on the same plate) on day 5 were scored as sensitive, ranging from no growth to slightly sensitive growth. For heat, copper, and ethanol stresses, we found 145, 137, and 26 noncomplemented genes, respectively, from the BY4741 (MATa) cross and 221, 134, and 19 from the BY4742 (MATα) cross, resulting in an intersection of 80, 13, and 2 genes (data file S1).
Respiration-deficient strains (petites) were identified by plating the haploid deletion collection strains on YPGly at 30°C. To estimate the rate of temperature-sensitive deletions, we sampled six plates (~2300 strains) from the haploid deletion collections and assayed their growth on YPD plates at room temperature and 37°C. The rate of heat-sensitive deletions in the subsample was 78 of 2251.
Validation of noncomplementing genes
We first repeated the noncomplementation test in another strain background. We made deletions of candidate genes (HFA1 for heat; TDA1, TDA9, GGC1, TDA4, RPL39, ADD66, YOL075C, CUP2, and CAJ1 for copper) by KanMX in an S. cerevisiae strain YJF173 in the same way as the deletion collection, with the exception that the coding region of HFA1 was defined according to (30). The knockout strains were then crossed to a S. uvarum rho0 strain (YJF2760). Phenotypes of the hemizygotes were assessed at the same conditions as in the screen, and only phenotypes of HFA1 and CUP2 were replicated.
Reciprocal hemizygotes were generated for HFA1 and CUP2. Orthologs of S. cerevisiae HFA1 and CUP2 were knocked out in S. uvarum strain YJF1450 with KanMX. The orthologs were defined according to (42); for HFA1, we included an extra 477 bp (base pairs) upstream of the ATG for the S. uvarum allele, based on translation from a non-AUG start codon at position −372 in S. cerevisiae (30). The S. uvarum deletion strains were then crossed to S. cerevisiae (YJF173), and the resulting hemizygotes were genotyped by PCR and found to carry S. cerevisiae mtDNA. Phenotypes of the two reciprocal hemizygotes were assessed on the same plate, under the same conditions as in the screen.
Interspecific hybrids with reciprocal mitotypes
Interspecific hybrids with reciprocal mitotypes were generated by crossing a rho+ strain from one species to a rho0 strain from another species. Two rho0 colonies from each strain were crossed to control for possible mutagenic effects of the ethidium bromide treatment. Mitotype was confirmed by PCR using primers targeting the transfer RNA (tRNA) clusters in mtDNA (forward, 5′-CCATGTTCAAATCATGGAGAGA-3′; reverse, 5′-CGAACTCGCATTCAATGTTTGG-3′; 95°C for 2 min; 95°C for 30 s, 50°C for 30 s, 72°C for 30 s for 30 cycles; 72°C for 5 min). The expected product sizes are 167 bp for S. cerevisiae, 131 bp for S. paradoxus, 218 bp for S. kudriavzevii, and 100 bp for S. uvarum.
Crosses with mitochondrial knockouts
S. uvarum strain YJF2600 (MATa hoΔ::NatMX trp1Δ::hphMX4) and YJF2601 (MATα hoΔ::NatMX trp1Δ::hphMX4) were crossed to previously constructed S. cerevisiae mitochondrial knockout strains (43–48). S. cerevisiae strains with wild-type mtDNA were crossed in parallel as control. MATa and MATα strains were mixed on YPD and incubated at room temperature overnight. The mating mixtures were either replica-plated (initial trial) or resuspended in sterile water and plated (second trial) onto YPGly. The YPGly plates were incubated at 37°C for 7 to 10 days to select for 37°C-respiring recombinants. The mating mixtures of cox2Δ and cox3Δ crosses were also plated to CM-trp-his-leu-lys-ura at room temperature to select for diploid hybrids, which allowed us to estimate the recombination rate to be about 0.05 to 0.1%. The 37°C-respiring colonies were picked and streaked on YPD at room temperature for single colonies. For the initial trial, the 37°C-respiring cells were streaked on YPD twice. For the cox1Δ and atp6Δ crosses, the plates were left at room temperature for 3 days after 7 days at 37°C incubation, and colonies growing from the recovery period were also picked and streaked. We also tried selecting for recombinants at 33°C and 35°C for the crosses with cobΔ, atp6Δ, and atp8Δ strains, from which we isolated few recombinants at 37°C. However, selection at 35°C did not significantly increase either the number or the size of the recombinant colonies compared to 37°C, and 33°C is too low a temperature to distinguish any heat-tolerant recombinants from nonrecombinant S. uvarum mtDNA; we thus did not sequence colonies from these selections. As a result, 3 + 12, 4 + 48, 3 + 25, 2 + 3, 0 + 7, and 0 + 1 strains (initial trial + second trial) from the cox2Δ, cox3Δ, cox1Δ, cobΔ, atp6Δ, and wild-type D273-10B control crosses, respectively, were generated. A total of 102 strains were subjected to whole-genome sequencing and phenotyping.
Spontaneous mitochondrial recombinants
S. cerevisiae (YJF153, MATa hoΔ::dsdAMX4, YPS163 derivative) and S. uvarum (YJF1450, MATα hoΔ::NatMX, CBS7001 derivative) were mated and streaked onto SDPSer + clonNAT medium to select for diploid hybrids. Three hundred eighty-four colonies on the double-selection plates were picked and arrayed onto one YPD agar plate and subsequently pinned to YPD and YPGly and incubated at room temperature, 37°C, and 4°C. Colony sizes on each plate were scored both manually and quantitatively using ImageJ (49). Strains with recombinant-like temperature phenotypes (r114, r194, r262, r334, r347, and b2), along with two control strains (r21, r23) with typical phenotypes for S. cerevisiae and S. uvarum mitotypes, respectively, were subjected to whole-genome sequencing and phenotyping.
DNA extraction, library preparation, and sequencing
For the unselected putative recombinants and their controls (r21, r23, b2, r334, r114, r194, r262, and r347), DNA was extracted using an mtDNA-enriching protocol (see below). For other strains sequenced in this study, genomic DNA was extracted from 22°C YPD overnight cultures inoculated with cells pregrown on YPGly plates (ZR Fungal/Bacterial DNA MicroPrep kit, Zymo Research).
mtDNA was enriched following a protocol adapted from (50) and (26). YPEG (1% yeast extract, 2% peptone, 2% ethanol, and 2% glycerol) medium (50 ml) was inoculated with overnight YPD starter cultures, shaken at 300 rpm at 22°C. The culture was collected at late-log phase (3000g for 1 min), and the cell pellet was washed twice in 1 ml of sterile distilled water. The cells were then washed in buffer [1.2 M sorbitol, 50 mM tris (pH 7.4), 50 mM EDTA, and 2% β-mercaptoethanol] and centrifuged at 14,000 rpm for 3 min. The cell pellet was weighed, resuspended in solution A [0.5 M sorbitol, 50 mM tris (pH 7.4), 10 mM EDTA, 2% β-mercaptoethanol] containing zymolyase (0.2 mg/ml; Zymo Research) at 7 ml/g of wet weight cells, and incubated at 37°C at 100 rpm for 45 min for osmotic lysis. The suspension was then centrifuged at 4000 rpm for 10 min. The supernatant was decanted to a new tube and centrifuged at 14,000 rpm for 15 min to obtain the crude mitochondrial pellet. The pellet was then incubated in deoxyribonuclease (DNase) treatment solution [0.3 M sucrose, 5 mM MgCl2, 50 mM tris-HCl (pH 8.0), 10 mM CaCl2, RQ1 DNase (100 U/ml; Promega), use 500 μl/g of the initial wet weight] at 37°C at 100 rpm for 30 min to remove nuclear DNA. EDTA (0.5 M) (pH 8.0) was added to a final concentration of 0.2 M to stop the reaction. The mitochondrial pellet was then washed three times by repeated cycles of centrifugation at 15,000 rpm for 10 min and resuspension in 1 ml of solution A to remove DNase and then resuspended in 400 μl of solution B [100 mM NaCl, 10 mM EDTA, and 50 mM tris (pH 8)] and incubated at room temperature for 30 min for lysis. mtDNA was isolated from the solution by phenol-chloroform extraction and ethanol precipitation, followed by a clean-up with a DNA Clean & Concentrator-5 kit (Zymo Research). Alternatively, two samples (r21 and r262) were extracted using the ZR Fungal/Bacterial DNA MicroPrep Kit (Zymo Research) by adding the Fungal/Bacterial DNA binding buffer to the lysed mitochondrial fraction and following the rest of the manufacturer protocol. The yield was typically 10 to 20 ng/g wet weight cells and provided 10- to 100-fold enrichment of mitochondrial reads.
Paired-end libraries were prepared with the Nextera DNA Library Preparation Kit (Illumina) with a modified protocol. Briefly, 3 to 5 ng of DNA were used for each sample, and the tagmentation reaction was performed at a ratio of 0.25 μl of tagmentation enzyme per nanogram of DNA. The tagmented DNA was amplified by KAPA HiFi DNA polymerase for 13 cycles (72°C for 3 min; 98°C for 5 min, 98°C for 10 s, 63°C for 30 s, 72°C for 30 s for 13 cycles; 72°C for 5 min). The PCR was then purified with AMPure beads. Paired-end 2 × 150 Illumina sequencing was performed on a MiniSeq instrument by the DNA Sequencing Innovation Lab in the Center for Genome Sciences and System Biology at Washington University. Ninety-six recombinants generated in the second trial of the mitochondrial mutant crosses were subsequently resequenced on a NextSeq 500 instrument at Duke Center for Genomic and Computational Biology for deeper coverage. The NextSeq and MiniSeq reads were combined in the analysis. The reads were deposited at the Sequence Read Archive under accession no. SRP155764.
Mitochondrial genome assembly
The S. uvarum mitochondrial genome was assembled from high-coverage sequencing of r23. Before assembly, we confirmed that it carried a nonrecombinant S. uvarum mitochondrial genome by mapping the reads to CBS380 (51), a Saccharomyces eubayanus × S. uvarum × S. cerevisiae hybrid that inherited the mitochondria from S. uvarum. To assemble the mitochondrial genome, reads were first cleaned with trimmomatic (52) to remove adapters. They were then assembled using SPAdes assembler (53), included in the wrapper iWGS (54), to produce contigs. Contigs were scaffolded to produce the final assembly through comparison with the output assembly of MITObim (55). The assembly was annotated with MFannot Tool (http://megasun.bch.umontreal.ca/RNAweasel/); ORF1 (F-SceIII) annotation was added manually using Geneious R6 (56). The assembled r23 mitochondrial genome is 64,682 bp and has a total of 5874 gapped bases (GenBank accession no. MH718505). Most gaps are in the intergenic regions, one gap is in VAR1, and three small gaps are in the introns of COB. The r23 mitochondrial genome is 99% identical to CBS380 based on BLAST results.
Read mapping and allele assignment of recombinants
Illumina reads were mapped to a reference that combined the mitochondrial genomes of S. cerevisiae (S288C-R64-2-1) and S. uvarum (r23 mitochondria assembled in this study) using end-to-end alignment in Bowtie 2 (57). Duplicated reads and reads with high secondary alignment scores (XS ≥ AS) or low mapping quality (MQ < 10) were filtered out. With this method, reads from hybrids with nonrecombinant S. cerevisiae or S. uvarum mtDNA were >99.9% correctly mapped to their reference genomes (49,496 of 49,504 for S. cerevisiae and 161,712 of 161,714 for S. uvarum). To characterize aneuploidy and the ratio of mitochondrial to nuclear reads, the reads were remapped to a reference file combining S. cerevisiae (S288C-R64-2-1) and S. uvarum (42) reference genomes using the same method. Coverage of nucleotide positions and chromosomes was generated by samtools depth and samtools idxstats, respectively.
For data visualization and identification of recombination breakpoints, we assigned allele identity for each nucleotide in orthologous regions in the two reference mitochondrial genomes. The total length of orthologous sequences is 16.5 kb (nucmer alignment) and contains mostly coding and tRNA sequences. After removing sites with no coverage in control strains, 12,574 nucleotide positions were subjected to data visualization and allele calling. We called the allele identity of a given nucleotide position based on the ratio of reads that mapped to the S. cerevisiae reference allele to the total number of reads that mapped to the two orthologous alleles (rsc = sc/(sc + su)): rsc = 1 (or no lower than the nonrecombinant S. cerevisiae mtDNA control) was called S. cerevisiae, rsc = 0 (or no higher than the nonrecombinant S. uvarum mtDNA control) was called S. uvarum, and rsc > 0 and rsc < 1 were called mixed. Sites without coverage of either allele were treated as missing data. A relaxed threshold was used in data visualization to account for noise in read mapping (rsc > 0.9 was called S. cerevisiae, labeled as “sc-90”; rsc < 0.1 was called S. uvarum, labeled as “su-90”). With this method, a total of 90 sequenced strains were confirmed to be recombinants.
To quantify the effect size of S. cerevisiae alleles, we counted the number of reads mapped to each protein-coding gene, tRNA, and rRNA by HTSeq count. For each gene, we tested the allele effect across 90 recombinants using a linear model: phenotype ~ allele + petite, where allele is the ratio of S. cerevisiae reads for a given gene and petite is the empirically determined petite rates (see below). Because we used the ratio of S. cerevisiae reads to represent allele identity, the model does not assume dominance; a heterozygous individual (i.e., read ratio = 0.5) should have an intermediate phenotype. P values were extracted from the models and adjusted by the false discovery rate (Benjamini and Hochberg method) to correct for multiple comparisons. While the P value for the petite term is significant in some models, its effect was always estimated to be positive. Because high petite rates should lead to small colonies, we do not consider petite rate to significantly contribute to the phenotype. In addition, aneuploidy and mtDNA copy number variation were present in several recombinants, but the addition of the two variables to the model did not change the effect size and significance of the allele term (phenotype ~ allele + petite + aneuploidy + copy, where aneuploidy is a binary variable indicating the presence or absence of chromosomal duplication and copy is the ratio of mitochondrial to nuclear reads). See data file S2 for all data used in the models.
The unselected putative recombinants were sequenced to high coverage, so we generated contigs and assemblies as in the previous section. The contigs were mapped to S. cerevisiae (r21) and S. uvarum (r23/CBS380/CBS7001) assemblies in Geneious R6 to identify the breakpoints. For the recombinants of lower-quality assemblies (r194, r347, and b2), the contigs were mapped to the best recombinant assembly r114 to improve recombinant construction. Results were confirmed by retaining the Illumina reads from the mitochondrial genome using both reference mitochondrial genomes as baits in HybPiper (58) and mapping them to the reference mitochondrial genomes using Geneious R6.
Recombinant phenotypes
Recombinant strains were first grown on YPGly plates to enrich for respiring cells and then in liquid YPD shaken at room temperature overnight. The overnight culture was diluted 1:105, spread on YPD and YPGly plates, and incubated at 22°, 37°, or 4°C. Pictures of plates were taken on the 5th day for 22° and 37°C YPD plates, on the 6th day for 22° and 37°C YPGly plates, and on the 68th day for 4°C YPD and YPGly plates. Colony sizes on YPGly plates were acquired by the Analyze Particles function in ImageJ (49). Nonsingle colonies were filtered out both by manually marking problematic colonies during analysis and by roundness threshold (roundness > 0.8 for nonpetite colonies). For each strain, sizes of all the nonpetite colonies (colony size > 200 units) were averaged; if no cells were respiring at a given condition, the average of all the (micro)colonies was used instead. Petite rates of the overnight cultures were recorded by counting big/small colonies on 22°C YPD and normal colonies/microcolonies on 22°C YPGly plates, and the two values were averaged. Control strains carrying wild-type S. cerevisiae or S. uvarum mtDNA in the background of D273-10B × CBS7001 were phenotyped in parallel.
Initially, the ~90 strains were phenotyped in three batches. We accounted for the batch effect for the 37°C data by picking three to four strains from each batch and repeating the phenotyping process on the same day at 37°C. Linear models between old data and new data were generated for each batch separately and were used to adjust for an overall batch effect. The 22°C colony sizes were not adjusted.
Mitochondrial allele replacement
Mitochondrial transformation was performed as previously described (fig. S6) (25). Intronless mitochondrial alleles were synthesized by Biomatik. The alleles were Gibson-assembled into an ARG8m-baring pBluescript plasmid such that the mitochondrial allele is flanked by 69- and 1113-bp ARG8m sequences at its 5′ and 3′ end, respectively (fig. S6C). Sequences of the assembled plasmid were confirmed by Sanger sequencing.
Mitochondrial knockout strains were first transformed with PGAL-HO to switch mating types and validated by mating-type PCR. In these strains, the target gene was replaced with ARG8m, so our constructs carrying the allele of interest can integrate into their endogenous loci by homologous recombination with ARG8m (fig. S6C).
We bombarded the mitochondrial plasmid and pRS315 (CEN plasmid carrying LEU2) into S. cerevisiae strain DFS160 (MATα ade2-101 leu2Δ ura3-52 arg8Δ::URA3 kar1-1, rho0) (45) using a biolistic PDS-1000/He particle delivery system (Bio-Rad) and selected for Leu+ colonies on MM plates. The colonies were replica-mated to the mitochondrial knockout strains at 30°C for 2 days. The mating mixtures were replica-plated to YPGly plates and incubated at 30°C. YPGly+ colonies were streaked on YPD, and mating types were determined by PCR. We also isolated the DFS160-derived parent strains that gave rise to the YPGly+ colonies from the master plates. For S. cerevisiae COX1 and COB alleles, the parent strains were remated to the knockout strains for confirmation.
The YPGly+ colonies carry a mitochondrial genome with the allele of interest integrated at their endogenous loci. Because of the kar1-1 mutation in DFS160, we were able to isolate YPGly+ colonies that are diploid, MATa haploid, or MATα haploid. We crossed the MATa transformant (D273-10B background) to an S. uvarum rho0 strain (YJF2760). The hybrid strain and the diploid S. cerevisiae strains directly obtained from the mitochondrial transformation were phenotyped at room temperature, 37°C, and 4°C on YPD and YPGly by spot dilution assays. The allele identity of all the phenotyped strains was confirmed by PCR and restriction digest.
Supplementary Material
Acknowledgments
We thank T. D. Fox, A. Tzagoloff, J. A. del Real Arias, and A. Querol for sharing strains. We thank E. C. P. Baker and members of Fay laboratory for comments and experimental assistance. Funding: This work was supported by the NIH (grant GM080669) to J.C.F. Additional support to C.T.H. was provided by the USDA National Institute of Food and Agriculture (Hatch project 1003258), the National Science Foundation (DEB-1253634), and the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-SC0018409 and DE-FC02-07ER64494 to T. J. Donohue). C.T.H. is a Pew Scholar in the Biomedical Sciences and a Vilas Faculty Early Career Investigator, supported by the Pew Charitable Trusts and the Vilas Trust Estate, respectively. D.P. is a Marie Sklodowska-Curie fellow of the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 747775). Author contributions: Conceptualization: X.C.L. and J.C.F.; methodology: X.C.L., E.A.S., and J.C.F.; investigation: X.C.L.; writing—original draft: X.C.L. and J.C.F.; writing—review and editing: D.P. and C.T.H.; funding acquisition: J.C.F.; resources: D.P., C.T.H., and E.A.S.; supervision: C.T.H. and J.C.F. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The Illumina reads were deposited at the Sequence Read Archive under accession no. SRP155764. The mitochondrial genome assembly of S. uvarum (r23) was deposited at GenBank under accession no. MH718505. Other data, code, and materials are available upon request.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/1/eaav1848/DC1
Supplementary Text
Fig. S1. Reciprocal hemizygosity test of HFA1 and CUP2.
Fig. S2. Fermentative and respiratory growth of interspecific hybrids with reciprocal mitotypes at different temperatures.
Fig. S3. Rescue of S. cerevisiae (sc) mitochondrial knockouts by recombination with S. uvarum (su) mitotypes.
Fig. S4. Recombinant genotypes and examples of recombination breakpoints.
Fig. S5. High petite rate of S. uvarum mitotype and its association with ORF1.
Fig. S6. Procedure for mitochondrial allele replacement.
Fig. S7. Background-dependent allele effects of COX1.
Table S1. Aneuploidy in the recombinants.
Table S2. Strains used in this study.
Data file S1. Results of noncomplementation screen.
Data file S2. Recombinant strain genotypes and phenotypes.
REFERENCES AND NOTES
- 1.Orr H. A., Coyne J. A., The genetics of adaptation: A reassessment. Am. Nat. 140, 725–742 (1992). [DOI] [PubMed] [Google Scholar]
- 2.Orr H. A., The genetics of species differences. Trends Ecol. Evol. 16, 343–350 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Hagman A., Säll T., Compagno C., Piskur J., Yeast “make-accumulate-consume” life strategy evolved as a multi-step process that predates the whole genome duplication. PLOS ONE 8, e68734 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonçalves P., Valério E., Correia C., de Almeida J. M. G. C. F., Sampaio J. P., Evidence for divergent evolution of growth temperature preference in sympatric Saccharomyces species. PLOS ONE 6, e20739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salvadó Z., Arroyo-López F. N., Guillamón J. M., Salazar G., Querol A., Barrio E., Temperature adaptation markedly determines evolution within the genus Saccharomyces. Appl. Environ. Microbiol. 77, 2292–2302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paget C. M., Schwartz J.-M., Delneri D., Environmental systems biology of cold-tolerant phenotype in Saccharomyces species adapted to grow at different temperatures. Mol. Ecol. 23, 5241–5257 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss C. V., Roop J. I., Hackley R. K., Chuong J. N., Grigoriev I. V., Arkin A. P., Skerker J. M., Brem R. B., Genetic dissection of interspecific differences in yeast thermotolerance. Nat. Genet. 50, 1501–1504 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawahara Y., Imanishi T., A genome-wide survey of changes in protein evolutionary rates across four closely related species of Saccharomyces sensu stricto group. BMC Evol. Biol. 7, 9 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mouse Genome Sequencing Consortium , Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Lee H.-Y., Chou J.-Y., Cheong L., Chang N.-H., Yang S.-Y., Leu J.-Y., Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135, 1065–1073 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Chou J.-Y., Hung Y.-S., Lin K.-H., Lee H.-Y., Leu J.-Y., Multiple molecular mechanisms cause reproductive isolation between three yeast species. PLOS Biol. 8, e1000432 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter N., Chambers S. R., Louis E. J., Borts R. H., The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 15, 1726–1733 (1996). [PMC free article] [PubMed] [Google Scholar]
- 13.Liti G., Barton D. B. H., Louis E. J., Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics 174, 839–850 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delneri D., Colson I., Grammenoudi S., Roberts I. N., Louis E. J., Oliver S. G., Engineering evolution to study speciation in yeasts. Nature 422, 68–72 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Fischer G., Neuvéglise C., Durrens P., Gaillardin C., Dujon B., Evolution of gene order in the genomes of two related yeast species. Genome Res. 11, 2009–2019 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Wolters J. F., Charron G., Gaspary A., Landry C. R., Fiumera A. C., Fiumera H. L., Mitochondrial recombination reveals mito–mito epistasis in yeast. Genetics 209, 307–319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paliwal S., Fiumera A. C., Fiumera H. L., Mitochondrial-nuclear epistasis contributes to phenotypic variation and coadaptation in natural isolates of Saccharomyces cerevisiae. Genetics 198, 1251–1265 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leducq J.-B., Henault M., Charron G., Nielly-Thibault L., Terrat Y., Fiumera H. L., Shapiro B. J., Landry C. R., Mitochondrial recombination and introgression during speciation by hybridization. Mol. Biol. Evol. 34, 1947–1959 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinmetz L. M., Sinha H., Richards D. R., Spiegelman J. I., Oefner P. J., McCusker J. H., Davis R. W., Dissecting the architecture of a quantitative trait locus in yeast. Nature 416, 326–330 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Buchman C., Skroch P., Welch J., Fogel S., Karin M., The CUP2 gene product, regulator of yeast metallothionein expression, is a copper-activated DNA-binding protein. Mol. Cell. Biol. 9, 4091–4095 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albertin W., da Silva T., Rigoulet M., Salin B., Masneuf-Pomarede I., de Vienne D., Sicard D., Bely M., Marullo P., The mitochondrial genome impacts respiration but not fermentation in interspecific Saccharomyces hybrids. PLOS ONE 8, e75121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoja U., Marthol S., Hofmann J., Stegner S., Schulz R., Meier S., Greiner E., Schweizer E., HFA1 encoding an organelle-specific acetyl-CoA carboxylase controls mitochondrial fatty acid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 279, 21779–21786 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Baker E. P., Peris D., Moriarty R. V., Li X. C., Fay J. C., Hittinger C. T., Mitochondrial DNA and temperature tolerance in lager yeasts. Sci. Adv. 5, eaav1869 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peris D., Arias A., Orlić S., Belloch C., Pérez-Través L., Querol A., Barrio E., Mitochondrial introgression suggests extensive ancestral hybridization events among Saccharomyces species. Mol. Phylogenet. Evol. 108, 49–60 (2017). [DOI] [PubMed] [Google Scholar]
- 25.N. Bonnefoy, T. D. Fox, Genetic transformation of Saccharomyces cerevisiae mitochondria, in Methods in Cell Biology (Academic Press, 2001), vol. 65, pp. 381–396. [DOI] [PubMed] [Google Scholar]
- 26.Wolters J. F., Chiu K., Fiumera H. L., Population structure of mitochondrial genomes in Saccharomyces cerevisiae. BMC Genomics 16, 451 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Špírek M., Poláková S., Jatzová K., Sulo P., Post-zygotic sterility and cytonuclear compatibility limits in S. cerevisiae xenomitochondrial cybrids. Front. Genet. 5, 454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auesukaree C., Damnernsawad A., Kruatrachue M., Pokethitiyook P., Boonchird C., Kaneko Y., Harashima S., Genome-wide identification of genes involved in tolerance to various environmental stresses in Saccharomyces cerevisiae. J. Appl. Genet. 50, 301–310 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Kastaniotis A. J., Autio K. J., Kerätär J. M., Monteuuis G., Mäkelä A. M., Nair R. R., Pietikäinen L. P., Shvetsova A., Chen Z., Hiltunen J. K., Mitochondrial fatty acid synthesis, fatty acids and mitochondrial physiology. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1862, 39–48 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Suomi F., Menger K. E., Monteuuis G., Naumann U., Kursu V. A. S., Shvetsova A., Kastaniotis A. J., Expression and evolution of the non-canonically translated yeast mitochondrial acetyl-CoA carboxylase Hfa1p. PLOS ONE 9, e114738 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vögtle F.-N., Burkhart J. M., Gonczarowska-Jorge H., Kücükköse C., Taskin A. A., Kopczynski D., Ahrends R., Mossmann D., Sickmann A., Zahedi R. P., Meisinger C., Landscape of submitochondrial protein distribution. Nat. Commun. 8, 290 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jhuang H.-Y., Lee H.-Y., Leu J.-Y., Mitochondrial–nuclear co-evolution leads to hybrid incompatibility through pentatricopeptide repeat proteins. EMBO Rep. 18, 87–101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piatkowska E. M., Naseeb S., Knight D., Delneri D., Chimeric protein complexes in hybrid species generate novel phenotypes. PLOS Genet. 9, e1003836 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sulo P., Szabóová D., Bielik P., Poláková S., Šoltys K., Jatzová K., Szemes T., The evolutionary history of Saccharomyces species inferred from completed mitochondrial genomes and revision in the ‘yeast mitochondrial genetic code’. DNA Res. 24, 571–583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S., Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423, 241–254 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Burton R. S., Barreto F. S., A disproportionate role for mtDNA in Dobzhansky–Muller incompatibilities? Mol. Ecol. 21, 4942–4957 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Solieri L., Antúnez O., Pérez-Ortin J. E., Barrio E., Giudici P., Mitochondrial inheritance and fermentative: Oxidative balance in hybrids between Saccharomyces cerevisiae and Saccharomyces uvarum. Yeast 25, 485–500 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Picazo C., Gamero-Sandemetrio E., Orozco H., Albertin W., Marullo P., Matallana E., Aranda A., Mitochondria inheritance is a key factor for tolerance to dehydration in wine yeast production. Lett. Appl. Microbiol. 60, 217–222 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Zubko E. I., Zubko M. K., Deficiencies in mitochondrial DNA compromise the survival of yeast cells at critically high temperatures. Microbiol. Res. 169, 185–195 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Vorachek-Warren M. K., McCusker J. H., DsdA (D-serine deaminase): A new heterologous MX cassette for gene disruption and selection in Saccharomyces cerevisiae. Yeast 21, 163–171 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A., Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Scannell D. R., Zill O. A., Rokas A., Payen C., Dunham M. J., Eisen M. B., Rine J., Johnston M., Hittinger C. T., The awesome power of yeast evolutionary genetics: New genome sequences and strain resources for the Saccharomyces sensu stricto genus. G3 1, 11–25 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perez-Martinez X., Broadley S. A., Fox T. D., Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 22, 5951–5961 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonnefoy N., Fox T. D., In vivo analysis of mutated initiation codons in the mitochondrial COX2 gene of Saccharomyces cerevisiae fused to the reporter gene ARG8m reveals lack of downstream reinitiation. Mol. Gen. Genet. 262, 1036–1046 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Steele D. F., Butler C. A., Fox T. D., Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc. Natl. Acad. Sci. U.S.A. 93, 5253–5257 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.M. G. Ding, C. A. Butler, S. A. Saracco, T. D. Fox, F. Godard, J. di Rago, B. L. Trumpower, An improved method for introducing point mutations into the mitochondrial cytochrome b gene to facilitate studying the role of cytochrome b in the formation of reactive oxygen species, in Methods in Enzymology (Elsevier, 2009), vol. 456, pp. 491–506. [DOI] [PubMed] [Google Scholar]
- 47.Rak M., Tetaud E., Godard F., Sagot I., Salin B., Duvezin-Caubet S., Slonimski P. P., Rytka J., di Rago J.-P., Yeast cells lacking the mitochondrial gene encoding the ATP synthase subunit 6 exhibit a selective loss of complex IV and unusual mitochondrial morphology. J. Biol. Chem. 282, 10853–10864 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Rak M., Tzagoloff A., F1-dependent translation of mitochondrially encoded Atp6p and Atp8p subunits of yeast ATP synthase. Proc. Natl. Acad. Sci. U.S.A. 106, 18509–18514 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.W. Rasband, ImageJ (U.S. National Institutes of Health, 1997–2016); https://imagej.nih.gov/ij/.
- 50.Fritsch E. S., Chabbert C. D., Klaus B., Steinmetz L. M., A genome-wide map of mitochondrial DNA recombination in yeast. Genetics 198, 755–771 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okuno M., Kajitani R., Ryusui R., Morimoto H., Kodama Y., Itoh T., Next-generation sequencing analysis of lager brewing yeast strains reveals the evolutionary history of interspecies hybridization. DNA Res. 23, 67–80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., Lesin V. M., Nikolenko S. I., Pham S., Prjibelski A. D., Pyshkin A. V., Sirotkin A. V., Vyahhi N., Tesler G., Alekseyev M. A., Pevzner P. A., SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou X., Peris D., Kominek J., Kurtzman C. P., Hittinger C. T., Rokas A., in silico Whole Genome Sequencer and Analyzer (iWGS): A computational pipeline to guide the design and analysis of de novo genome sequencing studies. G3 6, 3655–3662 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hahn C., Bachmann L., Chevreux B., Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—A baiting and iterative mapping approach. Nucleic Acids Res. 41, e129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A., Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson M. G., Gardner E. M., Liu Y., Medina R., Goffinet B., Shaw A. J., Zerega N. J. C., Wickett N. J., HybPiper: Extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Appl. Plant Sci. 4, 1600016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dimitrov L. N., Brem R. B., Kruglyak L., Gottschling D. E., Polymorphisms in multiple genes contribute to the spontaneous mitochondrial genome instability of Saccharomyces cerevisiae S288C strains. Genetics 183, 365–383 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bordonné R., Dirheimer G., Martin R. P., Expression of the oxi1 and maturase-related RF1 genes in yeast mitochondria. Curr. Genet. 13, 227–233 (1988). [DOI] [PubMed] [Google Scholar]
- 61.Jacquier A., Dujon B., An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell 41, 383–394 (1985). [DOI] [PubMed] [Google Scholar]
- 62.Peris D., Moriarty R. V., Alexander W. G., Baker E., Sylvester K., Sardi M., Langdon Q. K., Libkind D., Wang Q.-M., Bai F.-Y., Leducq J.-B., Charron G., Landry C. R., Sampaio J. P., Gonçalves P., Hyma K. E., Fay J. C., Sato T. K., Hittinger C. T., Hybridization and adaptive evolution of diverse Saccharomyces species for cellulosic biofuel production. Biotechnol. Biofuels 10, 78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burt A., Koufopanou V., Homing endonuclease genes: The rise and fall and rise again of a selfish element. Curr. Opin. Genet. Dev. 14, 609–615 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Williams E. H., Butler C. A., Bonnefoy N., Fox T. D., Translation initiation in Saccharomyces cerevisiae mitochondria: Functional interactions among mitochondrial ribosomal protein Rsm28p, initiation factor 2, methionyl-tRNA-formyltransferase and novel protein Rmd9p. Genetics 175, 1117–1126 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lopes C. A., Barrio E., Querol A., Natural hybrids of S. cerevisiae × S. kudriavzevii share alleles with European wild populations of Saccharomyces kudriavzevii. FEMS Yeast Res. 10, 412–421 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/1/eaav1848/DC1
Supplementary Text
Fig. S1. Reciprocal hemizygosity test of HFA1 and CUP2.
Fig. S2. Fermentative and respiratory growth of interspecific hybrids with reciprocal mitotypes at different temperatures.
Fig. S3. Rescue of S. cerevisiae (sc) mitochondrial knockouts by recombination with S. uvarum (su) mitotypes.
Fig. S4. Recombinant genotypes and examples of recombination breakpoints.
Fig. S5. High petite rate of S. uvarum mitotype and its association with ORF1.
Fig. S6. Procedure for mitochondrial allele replacement.
Fig. S7. Background-dependent allele effects of COX1.
Table S1. Aneuploidy in the recombinants.
Table S2. Strains used in this study.
Data file S1. Results of noncomplementation screen.
Data file S2. Recombinant strain genotypes and phenotypes.


