Abstract
Filaggrin (FLG) and corneodesmosin (CDSN) are two key proteins of the human epidermis. FLG loss-of-function mutations are the strongest genetic risk factors for human atopic dermatitis. Studies of the epidermal distribution of canine FLG and CDSN are limited. Our aim was to better characterize the distribution of FLG and CDSN in canine skin. Using immunohistochemistry on beagle skin, we screened a series of monoclonal antibodies (mAbs) specific for human FLG and CDSN. The cross-reactive mAbs were further used using immunoelectron microscopy and Western blotting. The structure of canine CDSN and FLG was determined using publicly available databases. In the epidermis, four anti-FLG mAbs stained keratohyalin granules in the granular keratinocytes and corneocyte matrix of the lower cornified layer. In urea-extracts of dog epidermis, several bands corresponding to proFLG and FLG monomers were detected. One anti-CDSN mAb stained the cytoplasm of granular keratinocytes and cells of both the inner root sheath and medulla of hair follicles. Dog CDSN was located in lamellar bodies, in the extracellular parts of desmosomes and in corneodesmosomes. A protein of 52 kDa was immunodetected. Genomic DNA analysis revealed that the amino acid sequence and structure of canine and human CDSN were highly similar.
Keywords: atopic dermatitis, differentiation, keratinocyte, skin, veterinary medicine
Introduction
Atopic dermatitis (AD) is a common skin disease affecting up to 20% of infants in developed countries.1 Alterations in the epidermal barrier have been identified as critical for the initiation and exacerbation of the disease, and loss of function mutations in the filaggrin (FLG) gene, encoding FLG are the major genetic risk factor in people.1–3 However, the exact sequence of pathophysiological events is still incompletely understood. Animal models of AD are therefore valuable. Mouse models have helped to answer several questions, including deciphering the role of various cytokines,4 but they also have limitations since the mouse disease differs from human AD in many ways, for example, wild-type mice do not spontaneously develop atopic lesions. Human and canine AD share many clinical characteristics, including similar prevalence in the general population, age of onset, affected skin areas, immune characteristics, and so on.5 In particular, young dogs naturally and commonly develop the disease. Thus, canine models of human AD have recently been proposed, including dogs spontaneously developing the disease as well as allergen challenged dogs.5–7 A detailed characterization of epidermal terminal differentiation in dogs is nevertheless necessary to be confident with these models.
FLG and corneodesmosin (CDSN) are key proteins for the epidermal barrier functions in humans and mice.3,8,9 FLG is synthesized in granular keratinocytes as a large precursor, named proFLG, which accumulates in keratohyalin granules. ProFLG consists of multiple FLG units (10 to 12 in humans and 17 in mice) flanked by a short C-terminal domain and a S100-homologous calcium-binding N-terminal domain. Upon cornification, proFLG is dephosphorylated and proteolytically processed to basic FLG monomers with molecular mass of 37 kDa in humans and 25 kDa in mice.10 The basic FLG units interact with and aggregate keratin filaments to yield the intracellular fibrous matrix of cornified cells.11 The following deimination of FLG units results in their detachment from keratins,12 and their degradation into free amino acids in the upper stratum corneum.13 FLG-derived amino acids are necessary for proper hydration of this protective layer.14 In addition, some FLG monomers become cross-linked to the cornified cell envelopes.15,16 The importance of FLG is highlighted by the phenotype of FLG–/– mice and of FLG-depleted skin/epidermis equivalents, which have major alterations of the epidermal barrier.17,18
CDSN is an adhesive glycoprotein secreted by granular keratinocytes and then incorporated into the pre-existing desmosomes.19 It is present in corneodesmosomes, the junctional structures between corneocytes, and plays a key role in stratum corneum cohesion. Its degradation in the upper stratum corneum is necessary for desquamation to occur.20 Nonsense mutations of its gene are responsible for two rare human genodermatoses, namely, peeling skin syndrome type 1 (OMIM#270300) and hypotrichosis simplex of the scalp (OMIM#146520).21,22
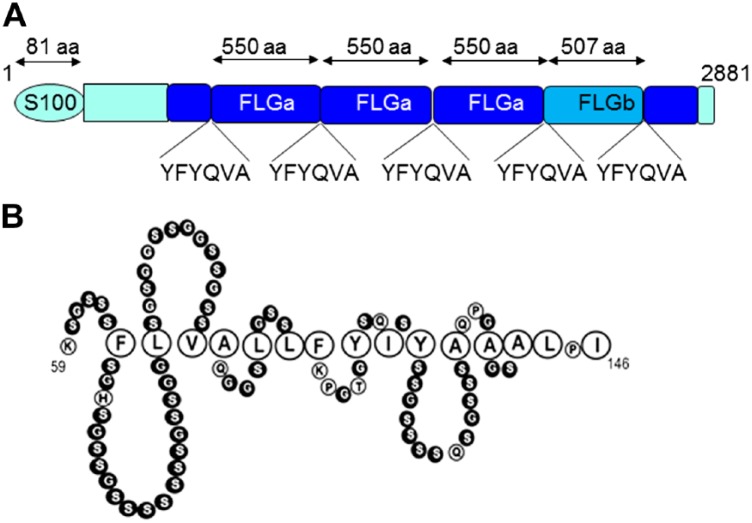
After the recent sequencing of the Canis familiaris genome (canFam3), the sequence of canine FLG, located on chromosome 17, is available. It encodes a 2882 amino acid long precursor composed of, successively (Table 1 and Fig. 1A), a S100-homologous N-terminal domain, a truncated FLG unit, three 549 amino acids long FLG units (referred to FLGa; predicted MW 59 kDa), a shorter 507 amino acids long FLG unit (referred to FLGb; predicted MW 54 kDa), a truncated FLG unit, and a C-terminal unique region.23,24 In prior studies of FLG expression in dog skin, conflicting results have been obtained due to the incomplete characterization and/or lack of specificity of the commercially available or homemade antibodies used.23–26 In addition, none of the previously studied antibodies reacted with ProFLG in Western blotting experiments, therefore the exact processing of the dog FLG precursor is not known. Furthermore, localization of ProFLG and FLG at the ultrastructural level has never been performed.
Table 1.
Sequence of Dog Filaggrin.
| 1– MSTLLENIIAIIDLFHDYSTTDKETDTLSREELKELLEMEFRPILKNPDDPDTADIFMHILDLDHDKKID
FSEFFLMVFKL–81 |
| 82– AQAYYDLTRRQNPQASEQNQKKYAYHTQDEEDDREEDEVEEEKEERIQSSNQAGRNERKGKKTISKSLKG
RRGKEHEQTPDSSRHEGSHDTKGANFGHSDSGSHHYQPSTQTNRSRQTQSRQGKSSSEFRSVKNWESSIN QDSDSEGHTEDSERQYGSGPGNQGESAHGHARNNSRHSESHGGQRKNHGHSESTHRQSDSGTRRRHGSTQ GNSTHTYRQSGSPHRENSSWENTDSFHRHSKSSRRGRQENKQRQSGDGSTHSETGHRKQYSGLRAGTQRG SSTEHTSDNEGHLIDSNTDFTKHHRKSGKTSGKEQRPTQDEVADRSRQPRLHHKKTNKKRQSKSTERDSV STST–435 |
| 436– ERQGRHHQQSQDTSRHTQT GHGSGNSKHRESSVSQASDSEGQSLDSETQSGSVQERSRSSQRRQRGSSAHGSSEHSAS–513 |
| 514–
YFYQVAPQEHFDSAHGQSQSSTRGRQGPRHDQAHDSSRHSGSHEGQAADFGHSESGSRHQQSSTRAQGSR
PSQARQGQSSSEFRPVSNRGSSISQDSDSEGLTEDSERQYGSGSGNQQGSARGHAGDSARHSESQQRHRT NHGQSQSGHGQSDTGTGRRQGSSHGHSLDTSRLPGSHRAGSPSRGHADSVHGRSRSSTGGRRETQHEQST DRSGRSGSEHGPLSSGPRTSRHQESSLSEGHSEDSGREFSTTRGDSGSGARNQRGSTHGQSTDRSTQLGS RQGQTGTHRHSDPAHRDSGSSTRERQGSRHEQSGDRARHAGSRQGQQATRGQPDSAHRDSGSSTRERQGS RHEQSGDRARHAGSRQGQQATRGHPDSAHRDSGSSTRERQGSRHEQSGDRARHAGSRQGQQATRGHPDSA HRDSGSSTRERQGSRHEQSGDRARHTGSQQGQQATRWQPDSAHGDSDLSTVDRQGRHHQQSQDSSRHSRT GHGSGNSKHRESSVSQASDSEGQSLDSETQSGSVQERSRSSQRRQRGSSAHGSSEHSAS–1062 |
| 1063–YFYQVAPQEHFDSAHGQSQSSTRGRQGPRHDQAHDSSRHSGSHEGQAADFGHSESGSRHQQSSTRAQGSR
PSQARQGQSSSEFRPVSNRGSSISQDSDSEGLTEDSERQYGSGSGNQQGSARGHAGDSARHSESQQRHRT NHGQSQSGHGQSDTGTGRRQGSSHGHSLDTSRLPGSHRAGSPSRGHADSVHGRSRSSTGGRRETQHEQST DRSGRSGSEHGPLSSGPRTSRHQESSLSEGHSEDSGREFLTTRGDSGSGARNQRGSTHGQSTDRSTQLGS RQGQTGTHRHSDPAHRDSGSSTRERQGSRHEQSGDRARHAGSHQGQQATRGHPDSAHRDSGSSTRERQGS RHEQSGDRARHAGSHQGQQATRGHPDSAHRDSESSTRERQGSRHEQSGDRARHAGSHQGQQATRGHPDSA HRDSGSSTRERQGSRHEQSGDRARHTGSHQGQQATRWQPDSAHGDSDLSTVDRQGHHHQQSQDSSRHSRT GHGSGNSKHRESSVSQASDSEGQSLDSETQSGSVQERSRSSQRRQRGSSAHGSSEHSAS–1611 |
| 1612–YFYQVAPQEHFDSAHGQSQSSTRGRQGPRHDQAHDSSRHSGSHEGQAADLGHSESGSRHQQSSTRAQGSR
PSQARQGQSSSEFRPVSNRGSSISQDSDSEGLTEDSERQYGSGSGNQQGSARGHAGDSARHSESQQRHRT NHGQSQSGHGQSDTGTGRRQGSSHGHSLDTSRLPGSHRAGSPSRGHADSVHGRSRSSTGGRRETQHEQST DRSGRSGSEHGPLSSGPRTSRHQESSLSEGHSEDSGREFLTTRGDSGSGARNQRGSTHGQSTDRSTQLGS RQGQTGTHRHSDPAHRDSGSRTRERQGSRHEQSGDRARHAGSHQGQQATRGHPDSAHRDSGSSTRERQGS RHEQSGDRARHAGSHQGQQATRGHPDSAHRDSESSTRERQGSRHEQSGDRARHAGSRQGQQATRGQPDSA HRDSGSSTRERQGSRHEQSGDRTRHAGSRQGQQATRGHPDSAHGDSDLSTVDRQGRHHQQSQDSSRHSRT GHGSGNSKHRESSVSQASDSEGQSLDSETQSGSVQERSRSSQRRQRGSSAHGSSEHSAS–2160 |
| 2161–YFYQVAPQEHFDSAHGQSQSSTRGRQGPRHDQAHDSSRHSGSHEGQAADFGHSESGSRHQQSSTRAQGSR
PSQARQGQSSSEFRPVSNRGSSISQDSDSEGLTEDSERQYGSGSGNQQGSARGHAGDSARHSESQQRHRT NHGQSQSGHGQSDTGTGRRQGSSHGHSLDTSRLPGSHRAGSLSRGHADSVHGRSRSSTGGRRETQHEQST DRSGRSGSEHGPLSSGPRTSRHQESSLSEGHSEDSGREFLTTRGDSGSGARNQRGSTHGQSTDRSTQLGS RQGQTGTHRHSDPAHRDSGSSTRERQGSRHEQSGDRARHAGSRQGQQATRGHPDSAHRDSGSSTRERQGS CHEQSGDRARHAGSRQGQQATRGQPDSAHRDSGSSTRERQGSRHEQSGDRARHTGSHQGQQATRWQPDSA ——————————————————————————HGDSDLSTVDRQGHHHQQSQDSSRHSRT GHGSGNSKHRESSVSQASDSEGQSLDSETQSGSVQERSRSSQRRQRGSSAHGSSEHSAS–2667 |
| 2668–YFYQVASQEHFDSAHGQSQSSTRGRQGPRHDQAHDSSRHSGSHEGQAADLGHSESGSRHQQSSTRAQGSR
PSQARQGQSSSDFRPVSNRGSSISQDSDSEGLTEDSERQYGSGSGNQQGSARGHAGDSARHSESQQRHRT NHGQSQSGHGQSDPGIGRRQGSGHGHSLDTSRHPG–2842 |
| 2843–YLQDQRENPGPSDSVFVQSQNGSRSHDSSHVEFKDFCEY–2881 |
Amino acid sequence of dog FLG (NCBI reference XP_022260725.1), showing a proposed structural organization of the protein including the S100A domain (amino acids 1–81), the spacer domain (82–435), truncated FLG subunits (436–513 and 2668–2842), the FLG type-a subunits (514–1062, 1063–1611, and 1612–2160), the FLG type-b subunit (2161–2667), and the COOH-terminal domain (2843–2881). The amino acids are shown using the single letter code. The FLG subunit sequences are identical, except some amino acid changes indicated in red and a 42 amino acids long deletion in the type-b. The underlined YFYQVA sequence corresponds to the putative linker peptide, as compared with FLYQVST in human.27,28 Abbreviation: FLG, Filaggrin.
Figure 1.
Schematic representation of dog profilaggrin (A) and of a tentative structure of dog corneodesmosin region 59–143 (B). Serine and glycine amino acids (in black) are predicted to form the so-called glycine-loops, due to the interactions of the interspersed aromatic and aliphatic residues (enlarged). Abbreviation: FLG, Filaggrin.
The sequence of dog CDSN has not yet been analyzed, and its expression in skin has only been investigated once at the immunomicroscopy level with an incompletely characterized antibody.29
Among a large series of well-characterized murine monoclonal antibodies (mAbs) specific for either human FLG or human CDSN, we identified some antibodies that immunostained normal dog epidermis. These antibodies were used to localize FLG and CDSN on dog skin at the ultrastructural level, and to immunoblot epidermal protein extracts. The purpose of this study was to utilize these mAbs to clarify the location of FLG and CDSN in canine skin and better describe the proteins.
Materials and Methods
Primary Antibodies
The following murine antibodies were used: mAbs to human proFLG and FLG produced after immunization of mice with purified FLG monomers (anti-human filaggrin AHF1 to AHF27),30,31 mAbs to human CDSN (G36-19 and F28-27),19 and mAbs to keratins K10 (RKSE60) and K14 (LL002) (ThermoFisher scientific, Waltham, MA). An isotypic control, MOPC-21, was purchased from Sigma-Aldrich (Saint-Louis, MO). Details are given in Table 2.
Table 2.
Primary Antibodies.
| Antigen | Antibody | Source(Reference) | Dilution/Concentration |
|
|---|---|---|---|---|
| WB | IHa | |||
| Filaggrin | clones AHF1-AHF27 | Home made30,31 | 0.5 µg/ml | 5 µg/ml |
| Corneodesmosin | clones G36-19 and F28-27 | Home made19 | 0.5 µg/ml | 5 µg/ml |
| No specificity | clone MOPC21 | Sigma-Aldrich | 0.5 µg/ml | — |
| Keratin 10 | clone RKSE60 | ThermoFisher | — | 1:100 |
| Keratin 14 | clone LL002 | ThermoFisher | — | 1:100 |
Abbreviations: AHF, anti-human filaggrin; WB, western blotting; IH, immunohistology.
Antigen retrieval was not necessary.
Immunohistology and Immunoelectron Microscopy
All procedures were approved by the Institutional Animal Care and Use Committee of VetAgro Sup (# 1107). Ten beagle dogs, five males and five females, aged between 3 and 5 years, belonging to a research colony were used in the study. Prior to the study, dogs were examined to ensure they were healthy and had no history or evidence of skin lesions. None of the dogs had received systemic or topical therapies for 3 months prior to the start of the study. One punch biopsy (6 mm) of dorsal truncal skin and one punch biopsy of paw pad were taken under general anesthesia and used for immunohistology. Half of each biopsy was fixed in formalin and paraffin embedded; then, 5 µm sections were used for classical hematoxylin and eosin staining and immunohistochemistry (immunoperoxidase staining).32 As the epidermis of paw pads is far thicker than that of hairy skin, paw pad biopsies were used with the aim to improve the visualization of the immunodetected proteins. The other half of each biopsy was fixed in 4% paraformaldehyde/PBS, dehydrated in graded alcohol series with progressive lowering of the temperature (from 4C to −35C), and embedded in Lowicryl K4M resin under UV light at −35C. Ultrathin sections (0.08 µm) collected on Formvar-coated nickel grids were immunolabeled with the mAbs diluted to 5 µg/ml. Immunoreactivities were revealed with a goat antimouse immunogold (10 nm; GE Healthcare, Buckinghamshire, UK). The sections were then counterstained with uranyl acetate and observed with a transmission electron microscope equipped with a Gatan digital camera. A negative control was used with omission of primary antibody and confirmed no immunolabeling occurred. The mAbs were tested in similar conditions on human skin, as positive controls.
Protein Extraction and Immunoblotting
Dorsal skin samples were obtained from two client owned dogs, one beagle and one golden retriever, after agreement of the Ethics Committee. The owners of the animal gave free and informed consent. They received in particular information about the consequences on their dogs and the remedies. One human abdominal skin sample (from a female without any history of skin diseases undergoing plastic surgery) was obtained from Genoskin (Toulouse, France), following informed consent of the donor and as agreed by the French Ministry of Research (#AC-2017-2897). Dermo-epidermal cleavage of the skin, sequential epidermal protein extractions, and protein analysis were performed as previously described.33 In brief, human and dog epidermal proteins were sequentially extracted in Tris-HCl pH 7.4 buffer containing nonidet-P40 detergent (Sigma-Aldrich) and EDTA (TE-NP40 extracts), then in TE buffer containing 8 M urea (TE-U extracts), as previously reported.33 Proteins were separated by polyacrylamide gel electrophoresis in the presence of SDS (SDS-PAGE) on 10% gels and either stained with Coomassie blue or transferred to nitrocellulose membranes for immunoblotting.
Mass Spectrometry
Proteins of the TEU extract obtained from a golden retriever were reduced with 14.5 mM mercaptoethanol, alkylated in 106.5 mM iodoacetamide for 30 min in the dark at room temperature, separated by SDS-PAGE and stained with colloidal Coomassie Blue for 24 hr. Bands corresponding to AHF10-immunodetected 60 and 55 kDa proteins were excised and subjected to in-gel tryptic digestion using sequencing grade modified porcine trypsin (Promega, Charbonnières-les-Bains, France) at 20 ng/ml, as previously described.34 The obtained dried peptide extracts were dissolved in 12 µl of 0.05% trifluoroacetic acid in 2% acetonitrile and analyzed by online nanoLC using an Ultimate 3000 RSLCnano LC system (Thermo Fisher Scientific Dionex, Bremen, Germany) coupled to an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) for data-dependent collision-induced dissociation (CID) fragmentation experiments. Five µl of the peptide extract were loaded on a 300 µm ID × 5 mm PepMap C18 precolumn (ThermoFisher Dionex) at 20 µl/min in 2% acetonitrile, 0.05% trifluoroacetic acid. After 5 min of desalting, peptides were online separated on a 75 µm ID × 50 cm C18 column (in-house packed with Reprosil C18-AQ Pur 3 μm resin; Dr. Maisch Gmbh; Proxeon Biosystems, Odense, Denmark) equilibrated in 95% of buffer A (0.2% formic acid), with a gradient of 5% to 25% of buffer B (80% acetonitrile, 0.2% formic acid) for 80 min then 25% to 50% for 30 min at a flow rate of 300 nL/min. The LTQ Orbitrap Velos was operated in data-dependent acquisition mode with the XCalibur software (version 2.0 SR2; Thermo Fisher Scientific). The survey scan was performed in the Orbitrap on the 350 to 1800 m/z mass range with the resolution set to a value of 60,000. The 20 most intense ions per survey scan were selected with an isolation width of 2 m/z for subsequent data-dependent CID fragmentation and the resulting fragments were analyzed in the linear trap (LTQ). The normalized collision energy was set to 30%. To prevent repetitive selection of the same peptide, the dynamic exclusion duration was set to 60 sec with a 10 ppm tolerance around the selected precursor and its isotopes. Monoisotopic precursor selection was turned on. For internal calibration, the ion at 445.120025 m/z was used as lock mass. The Xcalibur raw files were processed with Proteome Discoverer software (version 2.2.0.388, Thermo Fischer Scientific) for database search with the Mascot search engine (version 2.6.1, Matrix Science, London, UK) combined with the Percolator algorithm (version 2.05) for peptide-spectrum matches search optimization. The following parameters were set for creation of the peak list: parent ions in the mass range 300 to 5000 and no grouping of tandem mass spectrometry (MS/MS) scans. Peak list was searched against the C. familiaris taxonomy (25,539 sequences) of the UniProt protein database implemented with the dog FLG sequence (NCBI protein sequence reference XP_022260725.1). Cysteine carbamidomethylation was set as a fixed modification and methionine oxidation, and protein N-terminal acetylation as variable modifications. Up to three missed trypsin/P cleavages were allowed. Mass tolerances in MS and MS/MS were set to 10 ppm and 0.8 Da, respectively. Mascot results were validated by the target-decoy approach using a reverse database at the same size. The Percolator algorithm was used to calculate a q-value for each peptide-spectrum match; peptides and peptide-spectrum matches were validated based on Percolator q-values at a False Discovery Rate (FDR) set to 1%. Then, the peptide identifications were grouped into proteins according to the law of parsimony and filtered to 1% FDR.
Reconstructed Canine Epidermis
Reconstructed canine epidermis were produced essentially as described previously for reconstructed human epidermis.17,35 Briefly, primary normal canine keratinocytes were obtained from back skin of three different healthy dogs (Labrador) with informed consent of their owners. They were cultured for a maximum of three passages and used, after harvesting using mild trypsinization, to produce reconstructed canine epidermis on polycarbonate culture inserts with 0.4 µm pores (3 × 105 cells seeded per area of 0.63 cm2; Merck Millipore, Billerica, MA). After incubation for 24 hr at 37C in a humidified atmosphere containing 5% CO2, cells were exposed to the air–liquid interface for 10 additional days. The medium was renewed every 2 days.
Results
Filaggrin Expression in Dog Skin
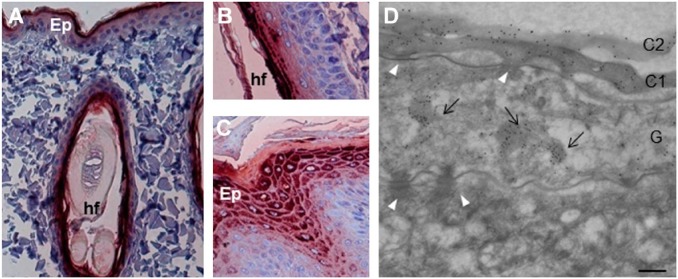
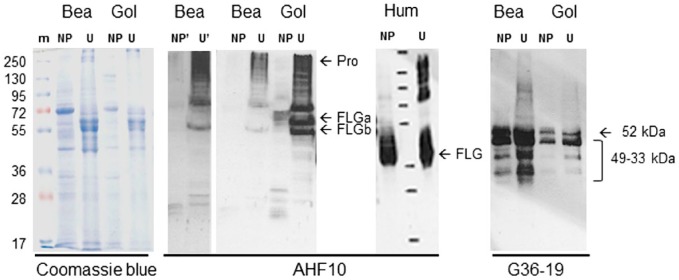
When tested with immunohistochemistry on sections of dorsal skin, four AHF mAbs, namely, AHF10, AHF13, AHF23, and AHF27, directed to human FLG showed a granular cytoplasmic staining of the stratum granulosum and a diffuse staining of the lower stratum corneum in both the inter- and intrafollicular epidermis (Fig. 2A and B and Supplemental Fig. S1A and C). A similar pattern of staining was observed on the skin of paw pad where the granular layer is thicker and therefore more easily visible (Fig. 2C and Supplemental Fig. S1B). The labeling patterns produced by the four mAbs were identical and similar to that previously described on human skin.30 Post-embedding immunoelectron microscopy analysis with AHF10 was then used to localize the protein in the interfollicular epidermis at the ultrastructural level (Fig. 2D). Immunogold particles were clustered over keratohyalin granules in the stratum granulosum and diffusely distributed in the matrix of corneocytes in the lower stratum corneum. No labeling was observed in the spinous and basal layers. Beagle and golden retriever epidermal proteins were then sequentially extracted in the presence of a non-ionic detergent then 8 M urea, and analyzed by western blotting with AHF10, 13, 23, and 27 (Fig. 3 and Fig. S1E). In the urea extract derived from golden retriever dogs, the four antibodies reacted intensely with a very high-molecular mass protein (more than 250 kDa) and three proteins of 55, 60, and 75 kDa. Additional bands of 250 to 100 kDa were less intense. The high-molecular mass protein likely corresponded to canine proFLG, whereas the 60 and 55 kDa may correspond to FLGa and FLGb monomers. The other bands were probably processing intermediates between proFLG and the monomers. The 60 and 55 kDa bands were cut from a SDS-gel and analyzed by mass spectrometry. Confirming they contained FLG, we identified many tryptic peptides derived from this protein (Supplemental Table S1). However, because FLGa and FLGb subunits differ only by a 42 amino acid deletion and by few amino acid substitutions (Table 1), we could not discriminate between the two forms. The same proteins were immunodetected in the urea extracts from a beagle dog except the 60 kDa band. In the TE-NP40 extract, some bands between 60 and 20 kDa were faintly immunodetected, the smaller ones probably corresponded to FLG monomer degradation products. Notably, the patterns differed between the two dog breeds. These reactivities were specific, since the isotypic mAb MOPC-21 did not recognize any proteins (Fig. S1E). The dog FLG detection was similar to that of human proFLG processing (Fig. 3), as previously reported.8,12
Figure 2.
Localization of filaggrin in the skin of healthy dogs. (A) The mAbs of the AHF family were analyzed by immunohistochemistry on sections of dog skin, from back and paw pad. AHF10, 13, 23, and 27 displayed the same pattern of reactivity, but only the data for AHF10 are shown. (A–C) AHF10 stains the upper interfollicular epidermis (A) and the intrafollicular epidermis (B) of the back skin, and the upper epidermis of paw pad (C). (D) The reactivity of AHF10 was further analyzed using immunoelectron microscopy. The mAb labels the first two layers of corneocytes (C1 and C2) and the keratohyalin granules (arrows) in a granular (G) keratinocyte. The spinous (S) keratinocytes as well as desmosomes (white arrowheads) are not labeled. Scale bar = 30 µm (A and C), 25 µm (B) and 0.25 µm (D). Abbreviations: AHF, anti-human filaggrin; Ep, epidermis; hf, hair follicle.
Figure 3.
Western blotting of dog epidermis extracts. Beagle (Bea) and golden (Gol) retriever dog epidermis was sequentially extracted in Tris-EDTA buffer containing either Nonidet-P40 (TE-NP40 extracts, NP) or 8 M urea (TE-U extracts, U). The extracted proteins were separated by SDS-PAGE, Coomassie blue stained, or immunodetected with AHF10 and G36-19 mAbs, as indicated. The immunoblot corresponding to AHF10 reactivity on the extracts of the beagle dog epidermis was exposed for a longer time (NP′ and U′) to highlight the absence of FLGa-corresponding band. The migration of molecular mass markers (m) is indicated on the left in kDa. ProFLG (Pro) and FLG monomers (FLGa and FLGb) are indicated by arrows, as well as the entire CDSN and its proteolytically derived fragments. For comparison, the Western blotting with AHF10 of TE-NP40 and TE-U extracts of human epidermis (Hum) is shown. Abbreviations: AHF, anti-human filaggrin; PAGE, polyacrylamide gel electrophoresis; FLG, Filaggrin; CDSN, corneodesmosin.
Characterization of Canine Corneodesmosin
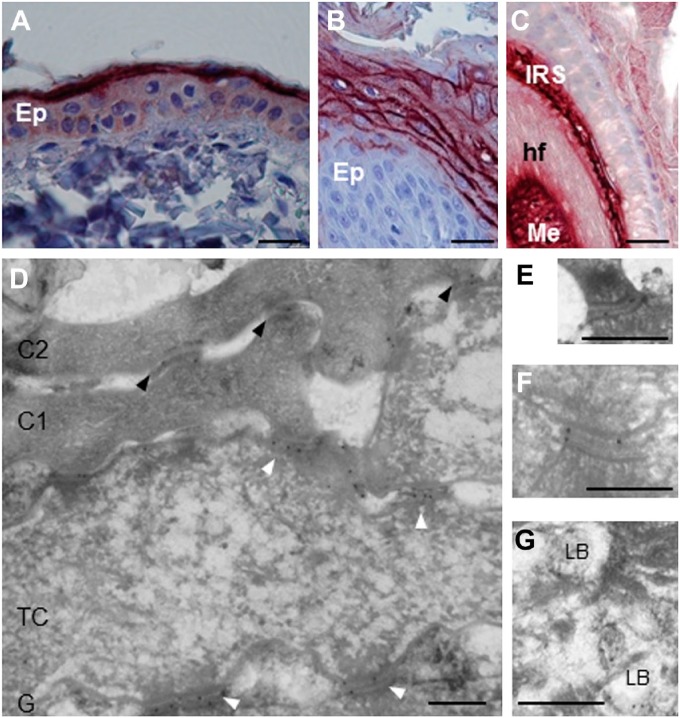
Among the anti-human CDSN mAbs tested on sections of paraffin-embedded skin of beagle dogs, G36-19 showed a cytoplasmic staining of keratinocytes in the lower stratum granulosum and a pericellular staining in the upper stratum granulosum; the immunoperoxidase staining disappeared in the lower stratum corneum (Fig. 4A and B and Fig. S1D). G36-19 also stained the inner root sheath and medulla of hair follicles (Fig. 4C). At the ultrastructural level (Fig. 4D–G), G36-19 labeled corneodesmosomes in the stratum corneum compactum and desmosomes in the transitional and upper granular cells. In the cytoplasm of granular keratinocytes, the mAb labeled lamellar bodies. A similar location of human CDSN has been previously reported.19 When used in western blotting experiments (Fig. 3 and Fig. S1E), G36-19 specifically reacted with several proteins with a molecular mass between 52 and 33 kDa: the entire dog CDSN and its proteolytic fragments. The same proteins were recognized whatever the dog breed. From the open access canine genome database Ensembl (CanFam3.1), the canine CDSN gene (NC_006594.3) was predicted on chromosome 12 at positions 819,488 to 824,033 (4546 bp). It consists of two exons. The first one (132 bp), similar to that of human gene, contains the translation initiation codon. The second one (2391 bp) is separated from the first by a 2023 bp intron and contains the major part of the coding sequence. The deduced coding sequence (UniProtKB-E2QS74) predicts a 498 amino acid–long protein, with a molecular mass of 47,886 Da and 74.1% identity with the human protein (Table 3). In particular, as the human CDSN,36 it contains a high amount of serine (27.3%) and glycine (17.5%) residues, it displays a peptide sequence (position 1–32), a glycine-loop region (position 59–143; Fig. 1B), and the G36-19-recognized epitope (Table 3; YLVP, position 275–278).
Figure 4.
Localization of corneodesmosin in the skin of healthy dogs. (A–C) Using immunohistochemistry, G36-19 mAb stains the upper interfollicular epidermis (Ep) of back skin (A), the upper epidermis of paw pad (B) and both the inner root sheath (IRS) and medulla (Me) of hair follicles (hf, C). (D–G) G36-19 was further used in immunoelectron microscopy. (D) The reactivity is localized in the extracellular part of corneodesmosomes (black arrowheads) in the lower cornified layer, and of desmosomes (white arrowheads) in transitional cells (TC) and granular keratinocytes (G). The first two layers of corneocytes (C1 and C2) are shown. (E, F) Enlargements of desmosomes in transitional and granular cells. (G) lamellar bodies (LB) of granular cells are also labeled. Scale bar = 30 µm (A–C), 0.3 µm (D–G).
Table 3.
Comparison of Human and Dog Corneodesmosin Sequences.
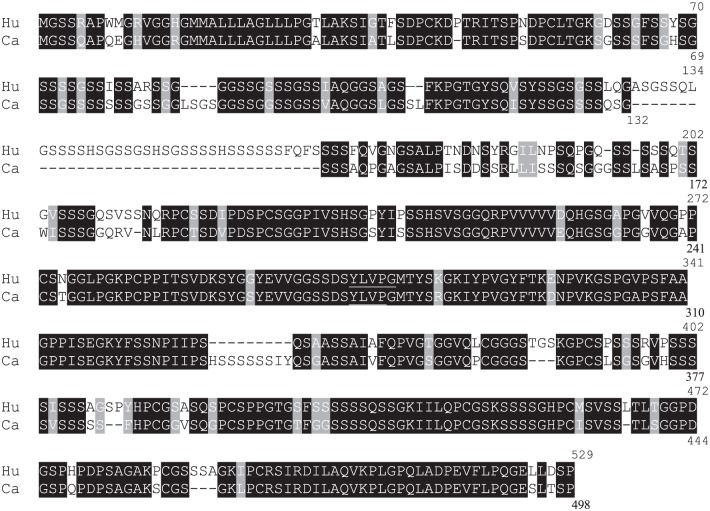
|
Amino acid sequence of human (Hu, #D2IYL2) and dog (Ca, #E2QS74) corneodesmosin, showing high sequence identity. Solid and shaded backgrounds indicate identical (74.1%) or similar (81.1%) amino acids, respectively. Amino acid numbers are indicated on the right. The underlined YLVP sequence corresponds to G36-19 epitope.37 Abbreviation: CDSN, corneodesmosin.
Reconstructed Canine Epidermis
To show the usefulness of these mAbs, we took advantage of them to characterize reconstructed canine epidermis. Indeed, when normal canine primary keratinocytes were seeded on polycarbonate membranes and let differentiate at the air-medium interface for several days, a morphologically differentiated epithelium was obtained, with an organized basal layer, two-to-four layers of spinous keratinocytes, and a granular layer covered by a cornified layer, as shown using Hematoxylin/eosin staining (Fig. 5A). We evidenced that a proper expression of keratins, FLG and CDSN was achieved, using immunohistology (Fig. 5A) and Western blotting (Fig. 5B).
Figure 5.
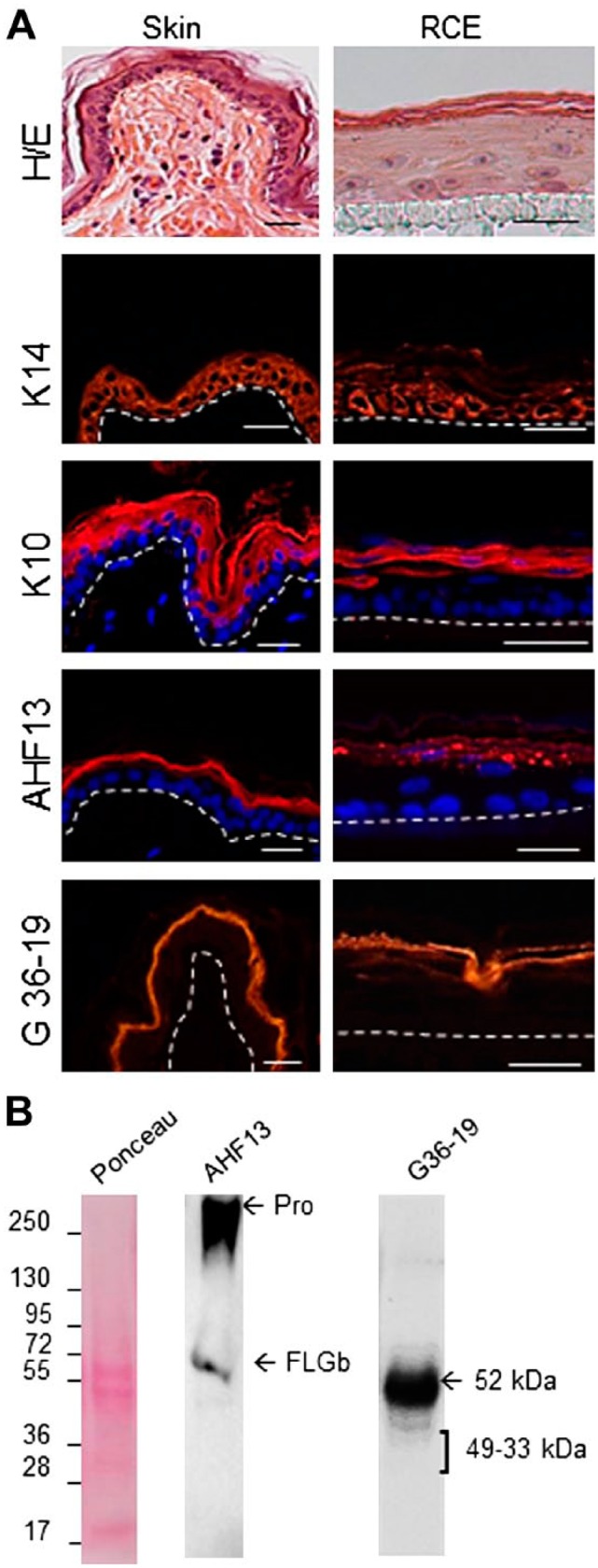
Characterization of reconstructed canine epidermis. (A) Sections of fixed samples of normal dog skin and reconstructed canine epidermis were stained with Hematoxylin and eosin (H/E) and antibodies directed to keratin K14, K10, FLG, and CDSN, as indicated. The dermo-epidermal junction and the polycarbonate membrane/epidermal junction are indicated by a dotted line. Scale bars = 25 µm. In some cases nuclei are labeled with 4′,6′-diamidino-2-phénylindole (Sigma-Aldrich) (blue). (B) Total proteins were extracted from reconstructed canine epidermis, separated by SDS-PAGE, transferred to membranes and Ponceau Red-stained or immunodetected with AHF13 and G36-19 mAbs. The migration of molecular mass markers is indicated on the left in kDa. ProFLG (Pro) and FLGb monomers are indicated by arrows, as well as the entire CDSN and CDSN proteolytically derived fragments. Abbreviation: AHF, anti-human filaggrin; FLG, Filaggrin; CDSN, corneodesmosin; PAGE, polyacrylamide gel electrophoresis.
Discussion
In this study, we identified four mAbs specific for dog FLG and one specific for dog CDSN. Using these mAbs, we demonstrated that proFLG, FLG and CDSN distribution in dog skin was comparable to locations in human and mouse skin.
We are aware of three prior published studies about FLG immunostaining in normal dog skin. In one publication, polyclonal antibodies directed against a synthetic peptide derived from mouse FLG amino acid sequence, but absent from the dog sequence, were used.25 Although the authors suggested a possible cross-reaction with conformational epitopes of the canine protein, the reported results remain questionable. In particular, the antibody stained the nuclei in the basal layer of the epidermis in addition to the expected labeling of the granular layer and the lower stratum corneum. The authors also used an antiserum specific for a synthetic peptide derived from the sequence of the N-terminus of canine ProFLG, but absent from the sequence of FLG monomers.25 Therefore the location of the subunits cannot be deduced from the reported immunofluorescence experiments that revealed labeling of the stratum granulosum and lower stratum corneum.25 In another publication, rabbits were immunized with a peptide GRRESSVTESSDTEND.26 This peptide seems to be derived from the sequence of FLG2 rather than FLG, as shown using a Blast analysis (data not shown). The most reliable data were obtained by Kanda et al.23 who used affinity-purified rabbit antibodies directed to a synthetic peptide with a sequence present in all the canine FLG units (both 59 kDa FLGa and 54 kDa FLGb). On dog skin sections, this antibody stained the cytoplasm of granular keratinocytes with a granular pattern as well as corneocytes. We obtained identical results with four different mAbs, confirming that these are the exact expression sites of canine proFLG and FLG. Moreover, using immunoelectron microscopy, we demonstrated the labeling of keratohyalin granules and the matrix of lower corneocytes. When we analyzed protein extracts of golden retriever epidermis, proFLG, and FLGa and FLGb monomers were immunodetected, as well as processing intermediates between the precursor and the monomers. As previously shown in human and mouse studies, FLG precursor, the intermediates and the basic monomers could be extracted only in the presence of urea, but not a detergent alone. This reflects the fact that proFLG accumulates in keratohyalin granules whereas the monomers associate with keratins. In addition, some bands with a smaller MW were detected in the detergent extracts. These bands probably correspond to FLG monomer-derived fragments, produced during degradation of FLG and the related production of the natural moisturizing factor. For unknown reasons, when Kanda et al. immunoblotted total protein extracts of dog skin, two bands of 59 and 54 kDa were immunodetected but, surprisingly, proFLG was not.23 When reconstructed canine epidermis were treated with pro-inflammatory Th2-type cytokines, a high decrease in the anti-FLG immunolabeling was observed (authors’ unpublished data), as previously observed with human keratinocytes.36 Further validation of the antidog FLG reactivity would be possible using RNA interference technology in the canine epidermis equivalent, as done in reconstructed human epidermis.17
Interestingly, when we analyzed epidermal extracts from one beagle dog, proFLG, FLGb, and intermediates were immunodetected, but not FLGa monomers. Although this is based on results obtained with only two dogs, this may suggest that FLG structure or sequence vary from one dog to the other or from one breed to the other, as it is well known in humans where FLG sequence is highly polymorphic.3 Sequencing of FLG gene in several dogs will be necessary to confirm this finding.
To the best of our knowledge, the distribution of CDSN has only been reported once in the literature.29 The protein has been detected using indirect immunofluorescence in the lowest stratum corneum and, rarely, at the periphery of outermost granular keratinocytes.29 The staining intensity has been reported to decrease after application of house dust-mite allergen-containing patches onto the skin of atopic beagle dogs.29 However, no Western blot analysis has been reported. Our study demonstrated that, just as in humans, canine CDSN is delivered into the extracellular spaces of the stratum granulosum via lamellar bodies, is then readily incorporated into the extracellular parts of desmosomes, and persists in the corneodesmosomes. Dog CDSN is highly similar to the human protein in terms of amino acid sequence and structure; however, it is slightly shorter (498 vs. 529 amino acids). In particular, the region 59 to 143 is formed by 41 serine and 22 glycine residues interspersed by 13 aromatic or aliphatic residues. In human CDSN, the corresponding region forms structural motifs called glycine loops, due to the association of the interspersed aromatic and aliphatic residues, and is responsible for both the oligomerization of the protein and its homophilic adhesive properties.20,38 The entire dog CDSN displays an apparent MW in SDS gels of 52 kDa. Like human CDSN, the canine protein is proteolytically processed, likely in the stratum corneum, to yield 49 to 33 kDa fragments. This probably reflects a progressive loss of its adhesive properties necessary for desquamation to occur, as demonstrated in human.20,38
In conclusion, several antibodies directed against human FLG and CDSN specifically recognized dog proFLG, FLG monomers, and CDSN. The tissue distribution of these proteins is identical in dog and human skin. These antibodies will be useful tools for future studies of various skin diseases of dogs, including ichthyosis due to mutations in the genes encoding enzymes of FLG metabolism.39
Supplemental Material
Supplemental material, DS_10.1369_0022155418798807 for Refined Immunochemical Characterization in Healthy Dog Skin of the Epidermal Cornification Proteins, Filaggrin, and Corneodesmosin by Didier Pin, Valérie Pendaries, Sokhna Keita Alassane, Carine Froment, Nicolas Amalric, Marie-Christine Cadiergues, Guy Serre, Marek Haftek, Emilie Vidémont and Michel Simon in Journal of Histochemistry & Cytochemistry
Acknowledgments
The authors acknowledge the technic help of Carole Pons and members of the histopathology facilities (Toulouse Rio Imagerie, INSERM U1043, Toulouse; Laboratoire de Dermatopathologie, VetAgro Sup, Lyon). Electron microscopy samples were observed at the Centre Technologique des Microstructures (CTµ), Lyon-Bio-Image, University of Lyon. We thank Odile Burlet-Schiltz for access to the Proteome Infrastructure of Toulouse.
Footnotes
Competing Interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: V.P. and N.A. are employed by Synelvia, a private company commercially using reconstructed canine epidermis. M.S. academic research group collaborated with and received grants from Synelvia. The other authors declare that they have no conflict of interest.
Author Contributions: EV, VP, SKA, CF, DP, and MH performed the experiments. DP, MH, and MS designed the conceptual idea for this study and wrote the manuscript. GS has produced the antibodies and wrote the manuscript. NA and M-CC wrote the manuscript. All the authors approved the submission of this manuscript in its final form.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported in part by grants from CNRS, Toulouse University, INSERM, the Région Occitanie, European funds (Fonds Européens de Développement Régional, FEDER), Toulouse Métropole, and the program “Investissement d’Avenir Infrastructures Nationales en Biologie et Santé” (ProFI, Proteomics French Infrastructure project, ANR-10-INBS-08).
ORCID iD: M Simon  https://orcid.org/0000-0003-3655-6329
https://orcid.org/0000-0003-3655-6329
Contributor Information
Didier Pin, University of Lyon, VetAgro Sup, UP Interaction Cellules Environnement, Marcy l’Etoile, France.
Valérie Pendaries, Synelvia, Labège, France.
Sokhna Keita Alassane, UDEAR, INSERM, University of Toulouse, Toulouse, France.
Carine Froment, Institut de Pharmacologie et de Biologie Structurale, University of Toulouse, CNRS, UPS, Toulouse, France.
Nicolas Amalric, Synelvia, Labège, France.
Marie-Christine Cadiergues, UDEAR, INSERM, University of Toulouse, Toulouse, France; Department of Dermatology, Department of Clinical Sciences, National Veterinary School of Toulouse, Toulouse, France.
Guy Serre, UDEAR, INSERM, University of Toulouse, Toulouse, France.
Marek Haftek, LBTIT, CNRS-University of Lyon 1, Lyon, France.
Emilie Vidémont, University of Lyon, VetAgro Sup, UP Interaction Cellules Environnement, Marcy l’Etoile, France.
Michel Simon, UDEAR, INSERM, University of Toulouse, Toulouse, France.
Literature Cited
- 1. Kezic S, Novak N, Jakasa I, Jungersted JM, Simon M, Brandner JM, Middelkamp-Hup MA, Weidinger S. Skin barrier in atopic dermatitis. Front Biosci. 2014;19:542–56. [DOI] [PubMed] [Google Scholar]
- 2. Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, Goudie DR, Sandilands A, Campbell LE, Smith FJ, O’Regan GM, Watson RM, Cecil JE, Bale SJ, Compton JG, DiGiovanna JJ, Fleckman P, Lewis-Jones S, Arseculeratne G, Sergeant A, Munro CS, El Houate B, McElreavey K, Halkjaer LB, Bisgaard H, Mukhopadhyay S, McLean WH. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–6. [DOI] [PubMed] [Google Scholar]
- 3. Le Lamer M, Pellerin L, Reynier M, Cau L, Pendaries V, Leprince C, Méchin MC, Serre G, Paul C, Simon M. Defects of corneocyte structural proteins and epidermal barrier in atopic dermatitis. Biol Chem. 2015;96:1163–79. [DOI] [PubMed] [Google Scholar]
- 4. Oyoshi MK, He R, Kumar L, Yoon J, Geha RS. Cellular and molecular mechanisms in atopic dermatitis. Adv Immunol. 2009;102:135–226. [DOI] [PubMed] [Google Scholar]
- 5. Marsella R, De Benedetto A. Atopic dermatitis in animals and people: an update and comparative review. Vet Sci. 2017;4:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Murray C, Ahrens K, Devalaraja M, Dymond M, Fagura M, Hargreaves A, Holt A, Peers I, Price S, Reens J, Riley R, Marsella R. Use of a canine model of atopic dermatitis to investigate the efficacy of a CCR4 antagonist in allergen-induced skin inflammation in a randomized study. J Invest Dermatol. 2016;136:665–71. [DOI] [PubMed] [Google Scholar]
- 7. Fanton N, Santoro D, Cornegliani L, Marsella R. Increased filaggrin-metabolizing enzyme activity in atopic skin: a pilot study using a canine model of atopic dermatitis. Vet Dermatol. 2017;28:479–e111. [DOI] [PubMed] [Google Scholar]
- 8. Dale BA, Resing KA, Presland RB. Keratohyalin granule proteins. In: Leigh I, Lane B, Watt F, editor. The Keratinocyte Handbook. London: Cambridge University Press; 1994. pp. 323–50. [Google Scholar]
- 9. Sandilands A, Sutherland C, Irvine AD, McLean WH. Filaggrin in the frontline: role in skin barrier function and disease. J Cell Sci. 2009;122:1285–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Resing KA, Walsh KA, Haugen-Scofield J, Dale BA. Identification of proteolytic cleavage sites in the conversion of profilaggrin to filaggrin in mammalian epidermis. J Biol Chem. 1989;264:1837–45. [PubMed] [Google Scholar]
- 11. Mack JW, Steven AC, Steinert PM. The mechanism of interaction of filaggrin with intermediate filaments. The ionic zipper hypothesis. J Mol Biol. 1993;232:50–66. [DOI] [PubMed] [Google Scholar]
- 12. Harding CR, Scott IR. Histidine-rich proteins (filaggrins): structural and functional heterogeneity during epidermal differentiation. J Mol Biol. 1983;170:651–73. [DOI] [PubMed] [Google Scholar]
- 13. Hsu C-Y, Henry J, Raymond A-A, Méchin MC, Pendaries V, Nassar D, Hansmann B, Balica S, Burlet-Schiltz O, Schmitt AM, Takahara H, Paul C, Serre G, Simon M. Deimination of human filaggrin-2 promotes its proteolysis by calpain 1. J Biol Chem. 2011;286:23222–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rawlings AV, Scott IA, Harding CR, Bowser PA. Stratum corneum moisturization at the molecular level. J Invest Dermatol. 1994;103:731–40. [DOI] [PubMed] [Google Scholar]
- 15. Alasdair CS, Steinert PM. Protein composition of cornified cell envelopes of epidermal keratinocytes. J Cell Sci. 1994;107:693–700. [PubMed] [Google Scholar]
- 16. Simon M, Haftek M, Sebbag M, Montézin M, Girbal-Neuhauser E, Schmitt D, Serre G. Evidence that filaggrin is a component of cornified cell envelopes in human plantar epidermis. Biochem J. 1996;317:173–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pendaries V, Malaisse J, Pellerin L, Le Lamer M, Nachat R, Kezic S, Schmitt AM, Paul C, Poumay Y, Serre G, Simon M. Knockdown of filaggrin in a three-dimensional reconstructed human epidermis impairs keratinocyte differentiation. J Invest Dermatol. 2014;134:2938–46. [DOI] [PubMed] [Google Scholar]
- 18. Kawasaki H, Nagao K, Kubo A, Hata T, Shimizu A, Mizuno H, Yamada T, Amagai M. Altered stratum corneum barrier and enhanced percutaneous immune responses in filaggrin-null mice. J Allergy Clin Immunol. 2012;129:1538–46. [DOI] [PubMed] [Google Scholar]
- 19. Serre G, Mils V, Haftek M, Vincent C, Croute F, Réano A, Ouhayoun JP, Bettinger S, Soleilhavoup JP. Identification of late differentiation antigens of human cornified epithelia, expressed in re-organized desmosomes and bound to cross-linked envelope. J Invest Dermatol. 1991;97:1061–72. [DOI] [PubMed] [Google Scholar]
- 20. Jonca N, Leclerc EA, Caubet C, Simon M, Guerrin M, Serre G. Corneodesmosomes and corneodesmosin: from the stratum corneum cohesion to the pathophysiology of genodermatoses. Eur J Dermatol. 2011;21:35–42. [DOI] [PubMed] [Google Scholar]
- 21. Oji V, Eckl KM, Aufenvenne K, Nätebus M, Tarinski T, Ackermann K, Seller N, Metze D, Nürnberg G, Fölster-Holst R, Schäfer-Korting M, Hausser I, Traupe H, Hennies HC. Loss of corneodesmosin leads to severe skin barrier defect, pruritus, and atopy: unraveling the peeling skin disease. Am J Hum Genet. 2010;87:274–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levy-Nissenbaum E, Betz RC, Frydman M, Simon M, Lahat H, Bakhan T, Goldman B, Bygum A, Pierick M, Hillmer AM, Jonca N, Toribio J, Kruse R, Dewald G, Cichon S, Kubisch C, Guerrin M, Serre G, Nöthen MM, Pras E. Hypotrichosis simplex of the scalp is associated with nonsense mutations in CDSN encoding corneodesmosin. Nat Genet. 2003;34:151–3. [DOI] [PubMed] [Google Scholar]
- 23. Kanda S, Sasaki T, Shiohama A, Nishifuji K, Amagai M, Iwasaki T, Kudoh J. Characterization of canine filaggrin: gene structure and protein expression in dog skin. Vet Dermatol. 2013;24:25–31. [DOI] [PubMed] [Google Scholar]
- 24. Suriyaphol G, Theerawatanasirikul S, Suriyaphol P. Filaggrin in canine skin. In: Thyssen JP, Maibach H, editors. Filaggrin. Berlin: Springer-Verlag; 2014. pp.209–21. [Google Scholar]
- 25. Chervet L, Galichet A, McLean WH, Chen H, Suter MM, Roosje PJ, Müller EJ. Missing C-terminal filaggrin expression, NFkappaB activation and hyperproliferation identify the dog as a putative model to study epidermal dysfunction in atopic dermatitis. Exp Dermatol. 2010;19:e343–e346. [DOI] [PubMed] [Google Scholar]
- 26. Santoro D, Marsella R, Ahrens K, Graves TK, Bunick D. Altered mRNA and protein expression of filaggrin in the skin of a canine animal model for atopic dermatitis. Vet Dermatol. 2013;24:329–36. [DOI] [PubMed] [Google Scholar]
- 27. Olivry T, Dunston SM. Expression patterns of superficial epidermal adhesion molecules in an experimental dog model of acute atopic dermatitis skin lesions. Vet Dermatol. 2015;26:53–6. [DOI] [PubMed] [Google Scholar]
- 28. Simon M, Sebbag M, Haftek M, Vincent C, Girbal-Neuhauser E, Rakotoarivony J, Sommé G, Schmitt D, Serre G. Monoclonal antibodies to human epidermal filaggrin, some not recognizing profilaggrin. J Invest Dermatol. 1995;105:432–7. [DOI] [PubMed] [Google Scholar]
- 29. Méchin MC, Enji M, Nachat R, Chavanas S, Charveron M, Ishida-Yamamoto A, Serre G, Takahara H, Simon M. The peptidylarginine deiminases expressed in human epidermis differ in their substrate specificities and subcellular locations. Cell Mol Life Sci. 2005;62:1984–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pin D, Bekrich M, Fantini O, Noel G, Vidémont E. An emulsion restores the skin barrier by decreasing the skin pH and inflammation in a canine experimental model. J Comp Pathol. 2014;151:244–54. [DOI] [PubMed] [Google Scholar]
- 31. Montézin M, Simon M, Guerrin M, Serre G. Corneodesmosin, a corneodesmosome-specific basic protein, is expressed in the cornified epithelia of the pig, guinea pig, rat, and mouse. Exp Cell Res. 1997;231:132–40. [DOI] [PubMed] [Google Scholar]
- 32. Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–8. [DOI] [PubMed] [Google Scholar]
- 33. Frankart A, Malaisse J, De Vuyst E, Minner F, de Rouvroit CL, Poumay Y. Epidermal morphogenesis during progressive in vitro 3D reconstruction at the air-liquid interface. Exp Dermatol. 2012;21:871–75. [DOI] [PubMed] [Google Scholar]
- 34. Pellerin L, Henry J, Hsu CY, Balica S, Jean-Decoster C, Méchin MC, Hansmann B, Rodriguez E, Weindinger S, Schmitt AM, Serre G, Paul C, Simon M. Defects of filaggrin-like proteins in both lesional and nonlesional atopic skin. J Allergy Clin Immunol. 2013;131:1094–102. [DOI] [PubMed] [Google Scholar]
- 35. Jonca N, Guerrin M, Hadjiolova K, Caubet C, Gallinaro H, Simon M, Serre G. Corneodesmosin, a component of epidermal corneocyte desmosomes, displays homophilic adhesive properties. J Biol Chem. 2002;277:5024–9. [DOI] [PubMed] [Google Scholar]
- 36. Bauer A, Waluk DP, Galichet A, Timm K, Jagannathan V, Sayar BS, Wiener DJ, Dietschi E, Müller EJ, Roosje P, Welle MM, Leeb T. A de novo variant in the ASPRV1 gene in a dog with ichthyosis. PLoS Genet. 2017;13:e1006651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McKinley-Grant LJ, Idler WW, Bernstein IA, Parry DA, Cannizzaro L, Croce CM, Huebner K, Lessin SR, Steinert PM. Characterization of a cDNA clone encoding human filaggrin and localization of the gene to chromosome region 1q21. Proc Natl Acad Sci U S A. 1989;86:4848–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thulin CD, Walsh KA. Identification of the amino terminus of human filaggrin using differential LC/MS techniques: implications for profilaggrin processing. Biochemistry. 1995;34:8687–92. [DOI] [PubMed] [Google Scholar]
- 39. Guerrin M, Ishigami A, Méchin M-C, Nachat R, Valmary S, Sebbag M, Simon M, Senshu T, Serre G. cDNA cloning, gene organization and expression analysis of human peptidylarginine deiminase type I. Biochem J. 2003;370:167–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1369_0022155418798807 for Refined Immunochemical Characterization in Healthy Dog Skin of the Epidermal Cornification Proteins, Filaggrin, and Corneodesmosin by Didier Pin, Valérie Pendaries, Sokhna Keita Alassane, Carine Froment, Nicolas Amalric, Marie-Christine Cadiergues, Guy Serre, Marek Haftek, Emilie Vidémont and Michel Simon in Journal of Histochemistry & Cytochemistry