Abstract
Background
A huge outbreak in the men-having-sex-with-men (MSM) has hit Europe during the years 2016–2018. Outbreak control has been hampered by vaccine shortages in many countries, and to minimize their impact, reduction of antigen doses has been implemented. However, these measures may have consequences on the evolution of hepatitis A virus (HAV), leading to the emergence of antigenic variants. Cases in vaccinated MSM patients have been detected in Barcelona, opening the possibility to study HAV evolution under immune pressure.
Methods
We performed deep-sequencing analysis of ten overlapping fragments covering the complete capsid coding region of HAV. A total of 14578255 reads were obtained and used for the analysis of virus evolution in vaccinated versus non-vaccinated patients. We estimated maximum and minimum mutation frequencies, and Shannon entropy in the quasispecies of each patient. Non-synonymous (NSyn) mutations affecting residues exposed in the capsid surface were located, with respect to epitopes, using the recently described crystal structure of HAV, as an indication of its potential role in escaping to the effect of vaccines.
Findings
HAV evolution at the quasispecies level, in non-vaccinated and vaccinated patients, revealed higher diversity in epitope-coding regions of the vaccinated group. Although amino acid replacements occurring in and around the epitopes were observed in both groups, their abundance was significantly higher in the quasispecies of vaccinated patients, indicating ongoing processes of fixation.
Interpretation
Our data suggest positive selection of antigenic variants in some vaccinated patients, raising concerns for new vaccination polices directed to the MSM group.
Keywords: Vaccine escape-mutants, MSM, Quasispecies, Deep-sequencing
Research in context.
Evidence before this study
We searched PubMed using the search terms “hepatitis A” and “MSM”. Since 1998, there has been an increase in outbreaks of hepatitis A in the men-having-sex-with-men (MSM), with a tendency over time to be larger and globally widespread. Hence, it is evident that the MSM group is an endemic at-risk group for hepatitis A. A second search with the terms “hepatitis A” and “vaccine failure” was also done, rendering very few report cases, indicating that the hepatitis A vaccines are highly efficient. However, these failures are always assumed to be due to a lack of seroconversion and never to the emergence of vaccine-escape variants. Nevertheless, a previous study from our group described the occurrence of such variants in an outbreak in the MSM group in Barcelona in 2008.
Lately, there has been a problem of hepatitis A vaccine shortage, which has required Public Health Agencies worldwide to implement changes in the vaccination schedules to reduce the antigenic doses. The advent of the next generation sequencing techniques (NGS) brings the possibility to deep-sequence serum samples from non-vaccinated and vaccinated patients, with the aim to seek for the emergence of antigenic variants.
Added value of this study
Our results represent the first ever-made analysis of the evolution of HAV in vaccinated versus non-vaccinated patients. This study has revealed that, due to the quasispecies dynamics of HAV, the emergence of antigenic variants in and around the main epitopes of the capsid occurs in both, vaccinated and non-vaccinated patients. However, only in vaccinated patients these variants increase in number suggesting their positive selection. Particularly, provided these antigenic variants had a high fitness, their circulation in the MSM group may rapidly increase, and further expand to the general population.
Implications of all the available evidence
Emergence of HAV antigenic variants may become a public health threat and have consequences in the future usefulness of the presently available vaccines. To avoid as much as possible their selection, careful and systematic studies on the impact of new vaccination policies directed to the MSM group are required, to ensure not only a high percent of seroconversion but also a high level of seroconversion. Some measures may include the completion of the vaccination schedule of two doses, the administration of booster doses to vaccinees with low anti-HAV antibodies, and to restrict as much as possible the vaccine post-exposure prophylaxis.
Alt-text: Unlabelled Box
1. Introduction
The men-having-sex-with-men (MSM) group is a relevant target for hepatitis A due to risky practices that favour faecal-oral transmission of high virus doses. Several outbreaks affecting this group have recently occurred in Europe [[1], [2], [3], [4], [5]]. Particularly relevant is the still ongoing outbreak which started in 2016 and has affected over 4000 patients across Europe. Three main circulating strains, belonging to genotype IA, have been detected: VRD_521_2016 [6], RIVM-HAV16-090 [7], and V16-25801 [8]. The control of this outbreak has been hampered by low coverage vaccination in some countries, and by vaccine shortages, which have obliged decision-making administrations to implement some restrictions in the number of vaccine doses administered to patients and close contacts [9]. However, while these measures attempt to avoid as much as possible the spread of the outbreak, and to protect the patients' health, their potential implications on hepatitis A virus (HAV) evolution should be a matter of debate, since the emergence of vaccine escape variants has been previously described [10].
Despite the implementation of vaccine programs since 1999 [11], the 2016–2018 outbreak has affected the MSM group in Barcelona including some vaccinated patients. However, the incidence in Barcelona has been much lower than in other parts of Spain in which no regular vaccination campaigns exist. Next Generation Sequencing (NGS) of the capsid coding region is the best approach for the analysis of the HAV evolution in vaccinated and non-vaccinated patients, setting the ground for measures to avoid as much as possible the emergence of vaccine escape variants.
2. Methods
2.1.1. Study design and participants
The 2016–2018 hepatitis A outbreak in the MSM group in Barcelona affected a total of 159 patients at the end of 2017. Samples from 17% of these patients (n = 27) were used for analysis. Five out of these 27 samples belonged to vaccinated patients. Thirteen samples were chosen for deep-sequencing including five from vaccinated patients, and eight additional samples from non-vaccinated patients. These eight samples were randomly chosen to cover the period of the outbreak.
All data related with the patients, including the clinical parameters and vaccination status, were recorded by the Public Health Agency of Barcelona and analysed anonymously. Data were collected on a routine base, through the hepatitis A surveillance system of Barcelona, which gathers information from physicians, electronic notifications, and laboratory tests. The analyses were carried out retrospectively. Therefore, no informed consent was required because hepatitis A is a mandatory notifiable disease. All data were treated in a strictly confidential manner according to the ethical principles of the Helsinki Declaration of 1964 revised by the World Medical Organization in Edinburgh, 2000 and the Organic Law 15/1999 of Data Protection in Spain.
2.1.2. Molecular procedures
RNA extraction from 150 μl of serum was performed by using the NucleoSpin RNA Virus kit (Macherey-Nagel, Germany), according to the manufacturer's instructions, and eluted in 50 μl. cDNA synthesis was performed using the SuperScript IV Reverse Transcriptase (Invitrogen, ThermoFisher Scientific, MA, USA), using 5 μl of RNA and the primers described in Table 1, Table 2, Table 3 of the web extra material. The cDNA length was of 355 and 1004 nt for genotyping and 3247 nt for deep-sequencing.
Table 1.
Clinical parameters of the thirteen patients analysed by deep-sequencing.
| Patients | Age | ALT U/L (<35)⁎ | Bilirubin mg/dL (<0·3)⁎ | HAV genome copies/mL | Illness duration weeks | HIV status | Vaccination |
|---|---|---|---|---|---|---|---|
| M1 | 31 | 2902 | 8·2 | 2·5 × 105 | 5·7 | + | No |
| M2 | 25 | 1813 | 4·0 | 3·6 × 105 | 3·2 | − | No |
| M4 | 34 | 2722 | NA | 5·6 × 105 | 3·7 | + | No |
| M5 | 48 | 2847 | 6·5 | 5·3 × 104 | 5·7 | − | Yes |
| M6 | 34 | 4852 | 10·8 | 3·2 × 106 | 8·6 | − | Yes |
| M7 | 34 | 2784 | 2·1 | 2·4 × 105 | 7·1 | + | Yes |
| M9 | 28 | 4804 | 6·8 | 1·1 × 107 | 4·0 | − | No |
| M10 | 46 | 9000 | 8·9 | 1·3 × 108 | 4·9 | − | No |
| M17 | NA | 3662 | 13·0 | 9·1 × 104 | 7·1 | + | No |
| M30 | 28 | 528 | 5·9 | 1·2 × 105 | 2·0 | − | No |
| M46 | 24 | NA | 2·5 | 1·7 × 106 | 4·0 | + | Yes |
| M47 | 48 | 3148 | 6·5 | 3·3 × 106 | 4·0 | − | No |
| M48 | NA | 2358 | 6·8 | 1·1 × 106 | 4·3 | + | Yes |
Cuttoff levels.
Table 2.
Genomic characterization of the quasispecies of the complete capsid coding region and part of 2A from vaccinated (n = 5) versus non-vaccinated patients (n = 8). Figures represent the average of all the patients, belonging to a group, for each fragment. Bold figures represent statistically significant differences (p < 0·05) between the non-vaccinated and vaccinated groups. F1 correspond to a non-coding region and F2 includes a very small coding region, hence amino acid-related cells are empty.
| Fragment | Shannon entropy (Mean ± SD) |
Nucleotide Mutation Frequency (Mean ± SD) |
Amino acid Mutation Frequency (Mean ± SD) |
||||
|---|---|---|---|---|---|---|---|
| Nucleotide | Amino acid | Minimum (×10−7) | Maximum (×10−4) | Minimum (×10−7) | Maximum (×10−4) | ||
| VAC- | F1 | 0·03 ± 0·02 | 2·54 ± 3·06 | 2·71 ± 1·98 | |||
| F2 | 0·03 ± 0·02 | 1·17 ± 1·06 | 2·66 ± 2·26 | ||||
| F3 | 0·04 ± 0·02 | 0·02 ± 0·04 | 5·01 ± 4·16 | 2·73 ± 2·16 | 8·88 ± 7·03 | 3·95 ± 2·52 | |
| F4 | 0·04 ± 0·03 | 0·04 ± 0·06 | 4·10 ± 3·67 | 2·32 ± 2·15 | 5·44 ± 5·63 | 2·82 ± 3·79 | |
| F5 | 0·06 ± 0·02 | 0·03 ± 0·01 | 6·94 ± 6·72 | 3·60 ± 1·78 | 8·85 ± 7·49 | 6·55 ± 4·54 | |
| F6 | 0·05 ± 0·02 | 0·02 ± 0·01 | 1·99 ± 2·25 | 3·40 ± 2·67 | 2·82 ± 2·62 | 4·98 ± 6·04 | |
| F7 | 0·04 ± 0·01 | 0·01 ± 0·01 | 2·21 ± 2·17 | 2·64 ± 1·14 | 2·52 ± 3·14 | 4·23 ± 3·96 | |
| F8 | 0·03 ± 0·02 | 0·02 ± 0·02 | 1·72 ± 0·95 | 1·83 ± 1·42 | 3·33 ± 2·54 | 3·14 ± 3·25 | |
| F9 | 0·02 ± 0·01 | 0·01 ± 0·01 | 1·14 ± 0·91 | 1·13 ± 0·65 | 1·63 ± 1·96 | 1·23 ± 1·30 | |
| F10 | 0·05 ± 0·02 | 0·02 ± 0·02 | 1·84 ± 2·18 | 3·28 ± 2·95 | 3·31 ± 3·73 | 7·57 ± 9·50 | |
| VAC+ | F1 | 0·04 ± 0·03 | 4·86 ± 5·73 | 3·27 ± 2·06 | |||
| F2 | 0·03 ± 0·03 | 1·83 ± 1·40 | 2·28 ± 2·63 | ||||
| F3 | 0·04 ± 0·03 | 0·02 ± 0·01 | 3·99 ± 5·57 | 2·98 ± 2·65 | 4·72 ± 2·76 | 4·32 ± 4·24 | |
| F4 | 0·07 ± 0·05 | 0·03 ± 0·03 | 7·55 ± 7·65 | 7·53 ± 6·82 | 10·6 ± 9·14 | 11·5 ± 14·6 | |
| F5 | 0·03 ± 0·01 | 0·01 ± 0·01 | 1·71 ± 1·42 | 2·05 ± 0·88 | 2·41 ± 3·08 | 1·77 ± 2·29 | |
| F6 | 0·04 ± 0·04 | 0·02 ± 0·02 | 1·97 ± 1·28 | 3·11 ± 3·92 | 3·78 ± 3·76 | 3·13 ± 4·22 | |
| F7 | 0·05 ± 0·02 | 0·03 ± 0·01 | 2·29 ± 2·48 | 3·40 ± 2·08 | 3·95 ± 5·28 | 8·15 ± 6·21 | |
| F8 | 0·08 ± 0·03 | 0·05 ± 0·01 | 3·88 ± 4·47 | 7·23 ± 4·46 | 8·97 ± 11·3 | 15·9 ± 12·9 | |
| F9 | 0·06 ± 0·04 | 0·02 ± 0·01 | 4·48 ± 3·30 | 4·33 ± 3·27 | 8·07 ± 8·07 | 5·99 ± 4·72 | |
| F10 | 0·05 ± 0·02 | 0·03 ± 0·02 | 4·05 ± 3·88 | 3·49 ± 1·74 | 10·1 ± 11·5 | 6·54 ± 6·06 | |
Table 3.
Analysis of the NSyn mutations detected in the complete capsid coding region. Data represent the average number of NSyn mutations per patient. In brackets appears the proportion of these mutations present at frequencies above 5%.
| VP2 | VP3 | VP1 | |
|---|---|---|---|
| Non-Vaccinated | 6·13 (0·02) | 6·50 (0·10) | 4·88 (0·03) |
| Vaccinated | 6·20 (0·13) | 3·60 (0·11) | 9·60 (0·20) |
Viral loads were determined as previously described [12], using five μl of the extracted RNA and the RNA UltraSense™ One-Step Quantitative RT-PCR System (Invitrogen, ThermoFisher Scientific), and were expressed as genome copies/ml of serum.
For genotyping, DNA amplification was performed using the FastStart High Fidelity PCR System, dNTPack (Roche Applied Science, Switzerland), with 10 μl of cDNA and using the primers described in Tables 1 and 2 of the web extra material. Consensus sequences were obtained using the ABI Prism® Big Dye™ Terminator Cycle Sequencing Ready Reaction Kit v3·1 (Applied Biosystems, CA, USA) and the ABI Prism 3700 automatic sequencer (Applied Biosystems). The reverse strand was sequenced and all mutations confirmed in the forward strand.
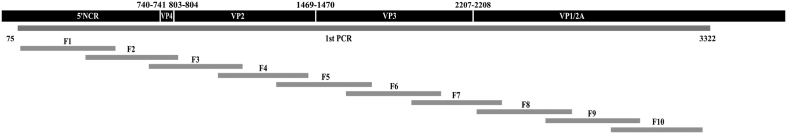
For deep-sequencing, DNA amplification was performed in two steps. In the first PCR, 10 μl of cDNA were amplified using the FastStart High Fidelity PCR System, dNTPack (Roche) and the primers described in Table 3 of the web extra material, to reach a single molecule covering the whole capsid coding region and a fragment of 2A (Fig. 1). Ten nested-PCRs were performed to amplify each of ten fragments, using the same kit but with 5 μl of DNA from the first-round PCR as template. The average fragment size was 437-bp (minimum of 424-bp and maximum of 448-bp) with overlapping regions ranging from 76-bp to 113-bp (Fig. 1). Nested-PCR products were purified from agarose gel bands (QIAquick Gel Extraction Kit, QIAgen, CA, USA), quantified (Qubit™ dsDNA BR Assay Kit, Invitrogene, ThermoFisher), and tested for quality (Bioanalyzer DNA 1000, Agilent, CA, USA). PCR products from a single patient were pooled at a normalized concentration and purified (Agentcourt® AMPure® XP, Beckman Coulter, CA, USA). Another normalization was performed to 1·5 ng/μl, followed by the library preparation protocol using the Kapa HyperPrep Kit (Roche Applied Science, CA, USA), and the SeqCap Adapter Kit A/B (24 Index. Nimblegen, Roche) for the pool indexing. A second cleanup was performed (Kapa Pure Beads, KapaBiosystems, Roche) and a quality control assay was done using the Bioanalyzer. Each pool was normalized to 4 nM, and appropriated volumes of each pool were added to the final library, which was quantified by LightCycler480 (Kapa Library Quantification Kit, KapaBiosystems, Roche). Last dilution of the Library was prepared and mixed with an internal DNA control (PhiX control V3, Illumina, CA, USA) before being sequenced using the MiSeq platform with MiSeq Reagent Kit V3 (Illumina).
Fig. 1.
Diagram of the fragments used for the deep-sequencing analysis covering, the 5′ non-coding-region (5′NCR), the complete capsid region (VP4, VP2, VP3, and VP1), and a fragment of the 2A region.
2.1.3. Phylogenetic analysis
Phylogenetic analyses of a fragment of 255-bp (spanning positions 2958–3230) from the VP1/2A, and a 1004-bp fragment (spanning positions 2208–3212) including the complete VP1 sequence and a short fragment of 2A were performed with the neighbour-joining method (distance calculation by the Kimura-2-parameter correction; pairwise deletion) implemented in the MEGA7 program [13]. Results were validated by 1000 bootstrap replicates. Genotype assignment was based on clustering with reference strains from the sequence database of the European Network [14], and all sequences were deposited to GenBank with accession numbers MH476486 to MH476511 and MK116906 to MK116923.
The same sequences were translated into amino acids and phylogenetic trees based on a sequence of 91 amino acids (spanning positions 251–300 from VP1 and 1–41 from 2A), and on a sequence of 334 amino acids (spanning positions 1–300 from VP1 and 1–35 from 2A) were similarly performed. Amino acid dendograms were constructed using the Neighbour-Joining method with distance calculation using the Poisson correction method.
2.1.4. Ultra-deep sequencing data management
A bioinformatic haplotype-centric procedure was used to exclude full reads that do not meet minimum quality requirements. Briefly, once acquiring raw data from MiSeq (fastq), the first step included the overlap of paired-end reads using FLASH [15,16]. The FLASH parameters were stablished as a minimum overlap between the paired-end reads, R1 and R2, of 20 bp with a maximum of 10% differences. Reads not fulfilling this requirement were discarded. Next, reads were demultipled by specific primers to obtain a fasta file by amplicon and strand. Reads were then collapsed into haplotypes with corresponding frequencies. Haplotypes were aligned with the reference sequence, and haplotypes containing more than two indeterminations, three gaps or 99 differences were also discarded. Accepted indeterminations and gaps were repaired as per the contents of the dominant haplotype. The intersection between forward and reverse haplotypes with abundances not below 0·1% was performed. Finally, all haplotypes with abundances below 0·5% were excluded. All sequences were deposited to GenBank with accession number SRP151545.
The quasispecies diversity was analysed by calculating the minimum and maximum mutation frequencies (synonymous and non-synonymous), and the normalized Shannon entropy (measure of diversity based on the frequency of haplotypes bearing synonymous and non-synonymous mutations), as previously described [17]. All these parameters were calculated for all ten fragments (F1–F10) in the quasispecies of each patient, and expressed as the average of all the patients belonging to the non-vaccinated or the vaccinated groups. The synonymous mutations per synonymous site (Ks), and the non-synonymous mutations per non-synonymous site (Ka) of the haplotypes from each quasispecies were calculated using the DnaSP v6·0 software [18,19]. The average of the Ks and Ka of each haplotype was normalized by its frequency and the Ka/Ks ratio calculated. Finally, the average of the Ka/Ks ratios of all haplotypes from the non-vaccinated and vaccinated groups was figured and used as a measure of positive selection when its value was >1.
2.1.5. Study of the amino acid replacements located on the capsid surface
Analysis of the location of the replacements on the surface of the HAV protomer was performed using the coordinates from the crystal structure [20] (RCSB accession code: 4QPI), and the Discovery Studio software (Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, San Diego: Dassault Systèmes, 2016).
Mutations detected in the protomer surface were classified as located in, close, or far from the epitopes. Mutations directly affecting the epitopes were those coincident with the residues defining monoclonal antibodies (mAbs) escape mutants [21,22], and those included in a mAb footprint area [23]. Additionally, mutations located at a distance of 1–2 residues from the residues identified in mAbs resistant mutants were considered to be closely located to these epitopes.
2.2. Statistical analysis
Statistical differences of the different parameters under analysis between groups of patients were determined using the Student t-test (unpaired) or the Mann-Whitney Rank Sum Test using the SigmaPlot version 11; p values < 0·05 were considered statistically significant.
2.3. Role of the funding source
No role of the funding sources. No conflict of interest to declare. The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
3. Results
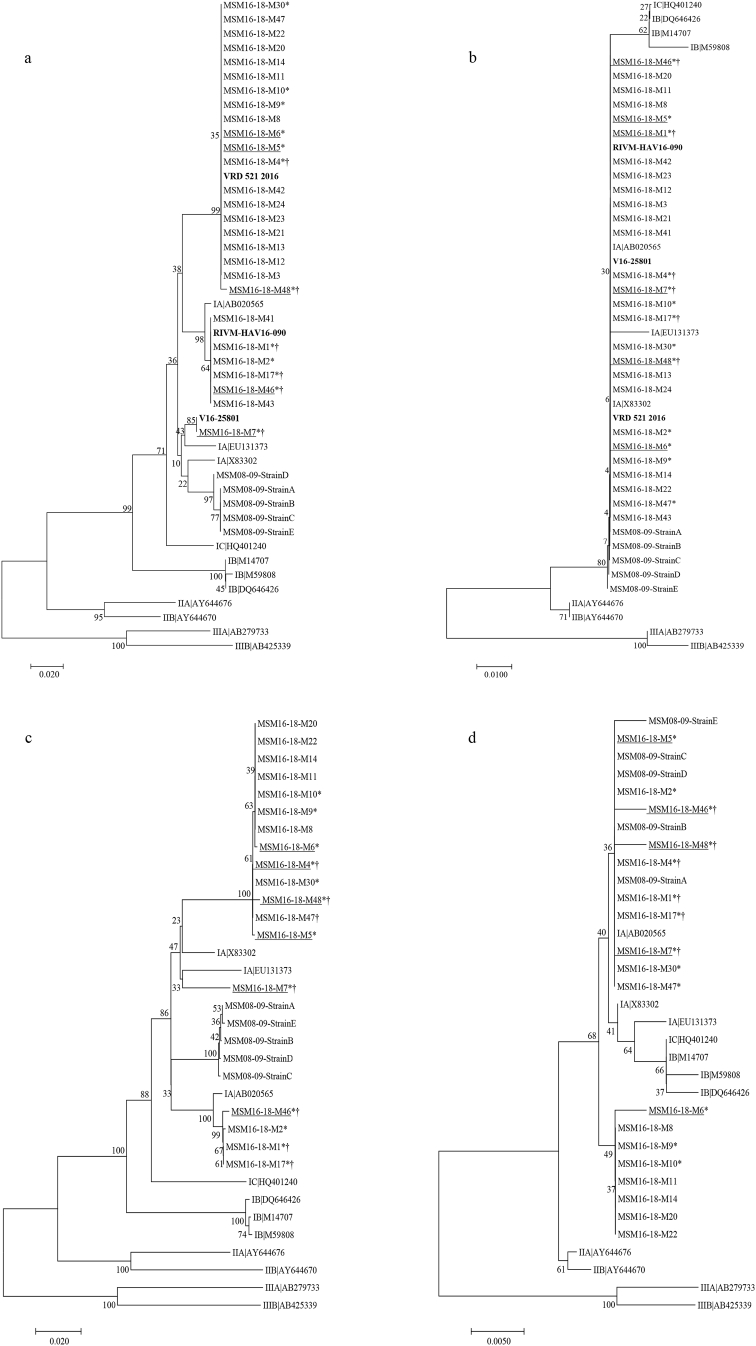
The European hepatitis A outbreak affecting the men-having-sex-with-men (MSM) in the season 2016–2017 affected 159 patients in the city of Barcelona, including six vaccinated patients. The genotype of 27 isolates was determined, proving that 74%, 22%, and 4% of the strains were related with strains VRD_521_2016, RIVM-HAV16-090, and V16-25801, respectively, and were also closely related to the strains isolated in a previous outbreak that occurred in 2008–09 in Barcelona in the MSM group (Fig. 2).
Fig. 2.
Phylogenetic analysis of HAV strains isolated from patients of the men-having-sex-with-men (MSM) 2016–2018 outbreak from Barcelona. (a) Phylogenetic tree based on a 255-bp fragment (spanning positions 2958–3230) from the VP1/2A protein. (b) Phylogenetic tree based on a sequence of 91-amino acids (spanning positions 251–300 from VP1 and 1–41 from 2A). (c) Phylogenetic tree based on a 1004-bp fragment (spanning positions 2208–3212) including the complete VP1 sequence and a short fragment of 2A. (d) Phylogenetic tree based on a sequence of 334-amino acids (spanning positions 1–300 from VP1 and 1–35 from 2A). Nucleotide dendrograms were constructed using the neighbour-joining method with distance calculation by the Kimura-2-parameter, and performing a bootstrap of 1000 replicates. Amino acid dendograms were constructed using the Neighbour-Joining method with distance calculation using the Poisson correction method. Samples from vaccinated individuals (n = 5) are underlined, samples from HIV-infected patients (n = 6) are indicated with the “†” symbol. Samples further analysed by MiSeq Illumina (n = 13) are indicated with “*”. The three main circulating strains in Europe related to this outbreak, VRD_521_20166, RIVM-HAV16-0907, and V16-258018A are included, and labelled in bold, as well as five representative samples from a previous MSM outbreak in Barcelona from 2008 to 09 (MSM08-09). Additionally, eleven reference strains representative of all subgenotypes (subgenotype IA: AB020565, EU13137, X83302; subgenotype IB: M14707, M59808, DQ646426, subgenotype IC: HQ401240; subgenotype IIA: AY644676; subgenotype IIB: AY644670; subgenotype IIIA: AB279733; subgenotype IIIB: AB425339), including the HM175 strain used in the TWINRIX vaccine (subgenotype IB), were also added. All tested samples were obtained in the period 2016–2017.
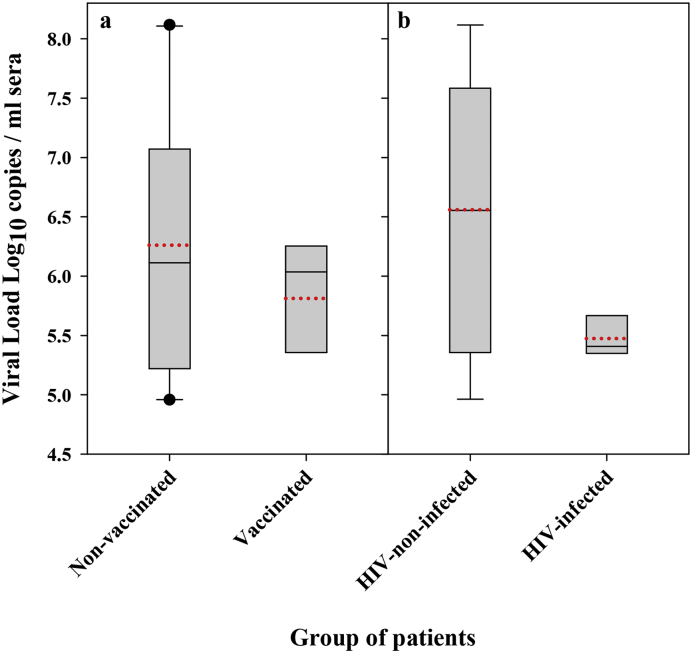
Six out of the 159 patients had been vaccinated (3·77% of all cases), and five of these samples (3·14% of all cases), along with eight additional samples from non-vaccinated patients (5·03% of all samples) were used for the comparative analysis. All sera samples were obtained during the first week after the onset of symptoms. Overall, no significant differences were found in the IgM levels or in the clinical parameters between the non-vaccinated and vaccinated patients (Table 1). Surprisingly, although vaccinated patients tended to show lower HAV viral loads, the difference was not statistically significant (Fig. 3a). Neither HIV-infected patients did show significantly different HAV viral loads than the HIV-non-infected patients, although they tended to be lower (Fig. 3b).
Fig. 3.
HAV viral loads. (a) Boxplot of the average virus load in sera from the non-vaccinated and vaccinated groups. (b) Boxplot of the average virus load in sera from HIV-non-infected and HIV-infected groups. Bottom and upper limits of the boxes represent the 25th and 75th percentiles, respectively. When present (n > 9), bottom and upper whiskers represent 10th and 90th percentiles, respectively. The median is represented by a solid black line and the average as a dotted red line.
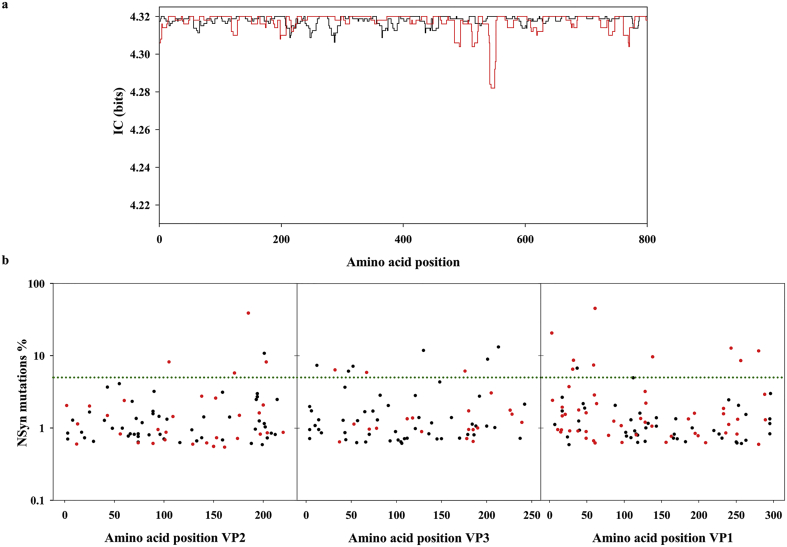
Deep-sequencing analysis was performed on ten fragments (F1–F10) covering the 5′ non-coding-region (5′NCR), the complete capsid region, and a fragment of the 2A region (Fig. 1). A total of 14578255 reads were obtained and used for the analysis of virus evolution in vaccinated versus non-vaccinated patients. The informative content (IC) of each position, obtained using overlapping windows of ten amino acid sliding every three amino acids in a multi-alignment of all the sequences from vaccinated versus non-vaccinated patients, revealed that overall the amino acid variability of the HAV quasispecies is extremely low. The average IC was not different between the non-vaccinated patients and vaccinated patients. However, despite this high degree of conservation, few regions with clear variability differences between the non-vaccinated and vaccinated groups were observed. Additionally, the analysis of the genetic parameters including the entropy and the nucleotide and amino acid mutation frequencies also revealed differences between both groups of patients (Table 2). Integrating the results of the IC and the genetic parameters it was concluded that the region encoded in the fragment F5 (VP3) and the region encoded in fragments F7-F9 (VP1) were more variable in the non-vaccinated patients and vaccinated patients, respectively. These data could suggest the occurrence of positive selection in VP1 and of purifying selection in VP3 in the vaccinated patients.
To look deep inside these possibilities, we plotted the frequency of NSyn mutations detected in the quasispecies of each patient all along the capsid (Fig. 4b). The average number of NSyn mutations in the VP1 coding region in the vaccinated group was clearly above the non-vaccinated group, and yet more remarkably a higher proportion of NSyn mutations at frequencies above 5% (Fig. 4b and Table 3). In contrast, the average number of NSyn mutations in the VP3 coding region in the vaccinated group was clearly below the non-vaccinated group (Fig. 4b and Table 3). Again this suggested purifying selection in VP3, which could be related to some negative epistatic effects increasing the exposure of epitopes, and positive selection in VP1 that was further analysed.
Fig. 4.
Informative content (IC) of each amino acid position along the capsid, and analysis of the frequency of the NSyn mutations. (a) The analysis of the informative content was performed using overlapping windows of 10 amino acids sliding every 3 amino acids. IC values (in bits) range from 0 (maximum variability) to 4·32 (minimum variability). Each point (amino acid position) corresponds to the mean IC from all the sequences of a given group. The black line corresponds to the non-vaccinated group and the red line to the vaccinated group. Values have been normalized by the frequency of each substitution and the number of sequences per group. (b) Frequency (%) of the NSyn mutations detected in the non-vaccinated patients (n = 8; black dots) and vaccinated patients (n = 5; red dots); black dots are more common due to the higher n. The horizontal green line represents the 5% frequency; points over this line represent high-frequency mutations.
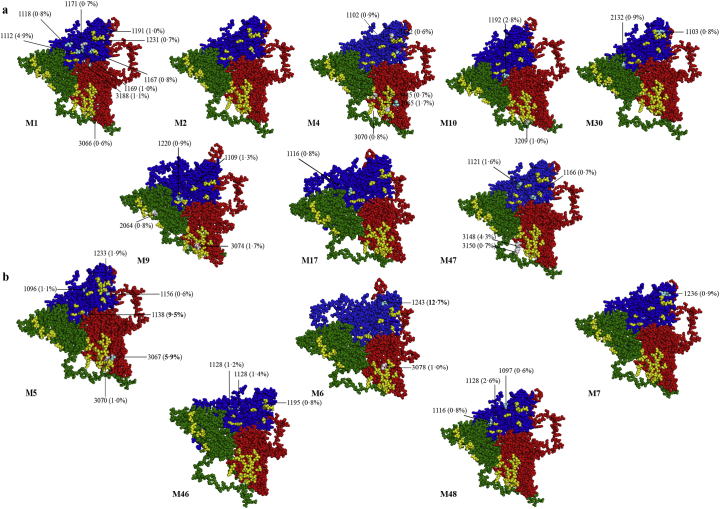
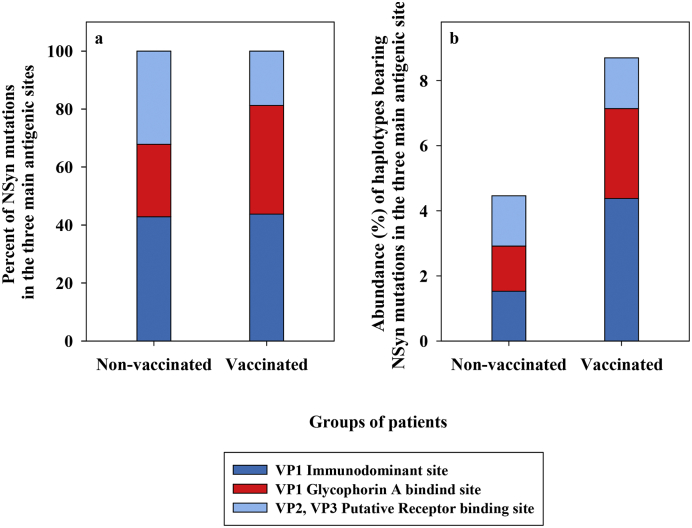
The analysis of NSyn mutations located on the surface of the HAV protomer [20] revealed similar results (Fig. 5). Among the non-vaccinated patients, 11·69% of the NSyn replacements involved residues from antigenic sites, 24·68% involved residues closely located to these sites, and 63·63% were randomly distributed. Similarly, in the vaccinated patients, 4·88% and 36·59% of the replacements were in or closely located to the antigenic sites, respectively, and 58·53% were randomly distributed on the surface of the protomer. Additionally, the average number of NSyn mutations per patient located in or close to the epitopes, was similar in the non-vaccinated and vaccinated groups (Table 4). However, the most striking difference was the abundance of the haplotypes bearing mutations located in or near the antigenic sites in the quasispecies. While in the non-vaccinated patients these variants were found at frequencies below 5%, in the vaccinated patients were much more abundant, again indicating the action of positive selection, particularly in the VP1 epitopes (Fig. 5 and Table 4). Overall, 42·86%, 25·00%, and 32·14% of the NSyn replacements located in and around the antigenic sites in the quasispecies of the non-vaccinated group, and 43·75%, 37·50%, 18·75% in the vaccinated group, were in the VP1 immunodominant site [21], the glycophorin A binding site located in the VP1 protein [22,24], and the putative receptor binding site in the pentamer interface expanding from VP2 to VP3 proteins [23], respectively (Fig. 5, Fig. 6a). More remarkably, the average proportion of haplotypes, from the different patient quasispecies, bearing mutations in and around these sites were of 1·53%, 1·39% and 1·54%, and 4·38%, 2·76% and 1·56% in the non-vaccinated and vaccinated groups, respectively (Fig. 6b). Of note, patient M5 bore the replacements Arg to Ser at position 138 of VP1 (close to the glycophorin A binding site epitope), and Asn to Ile at position 67 of VP3 (close to the VP3 region of the putative receptor binding site), at frequencies of 9·5% and 5·9%, respectively; additionally, patient M6 bore the replacement Leu to Ser at position 243 of VP1 (close to the VP1 immunodominant site) at a frequency of 12·7% (Fig. 5). In contrast, vaccinated patients M7, M46 and M48 showed the same abundance of antigenic variants than non-vaccinated patients.
Fig. 5.
Location of the variants affecting residues in and around the antigenic sites of the HAV capsid. (a) Protomers from non-vaccinated patients (M1, M2, M4, M9, M10, M17, M30, and M47). (b) Protomers from vaccinated patients (M5, M6, M7, M46 and M48). Green protein corresponds to VP2, red to VP3 and blue to VP1. Protomers used to locate the replacements were extracted from the recently resolved crystal structure of HAV [18]. Epitopes (yellow residues) were identified either by mAb escape mutants [19,20] (VP1 and VP3) or mAb footprints [21] (VP2 and VP3). Grey residues represent mutations directly affecting the epitopes. Clear blue residues represent mutations located at a distance of 1–2 residues from the epitopes identified in monoclonal antibodies (mAbs) resistant mutants.
Table 4.
Analysis of the NSyn mutations located in or close to the capsid epitopes. Data represent the number of mutations in the different patients, the average number of mutations, and the proportion of these mutations present at frequencies above 5%, in the non-vaccinated and vaccinated groups.
| Mutations per patient | ||||
|---|---|---|---|---|
| Groups of patients | ||||
| Non-Vaccinated | VP2 | VP3 | VP1 | Total |
| M1 | 0 | 2 | 7 | 9 |
| M2 | 0 | 0 | 0 | 0 |
| M4 | 0 | 3 | 2 | 5 |
| M9 | 1 | 1 | 2 | 4 |
| M10 | 0 | 2 | 0 | 2 |
| M17 | 0 | 0 | 1 | 1 |
| M30 | 1 | 0 | 1 | 2 |
| M47 | 0 | 2 | 3 | 5 |
| Average of mutations per patient | ||||
| 0·25 | 1·25 | 2·00 | 3·50 | |
| Proportion of mutations above 5% | ||||
| 0·00 | 0·00 | 0·00 | 0·00 | |
| Mutations per patient | ||||
| Vaccinated | ||||
| M5 | 0 | 2 | 4 | 6 |
| M6 | 0 | 1 | 1 | 2 |
| M7 | 0 | 0 | 2 | 2 |
| M46 | 0 | 0 | 3 | 3 |
| M48 | 0 | 0 | 3 | 3 |
| Average of mutations per patient | ||||
| 0·00 | 0·60 | 2·60 | 3·20 | |
| Proportion of mutations above 5% | ||||
| 0·00 | 0·33 | 0·15 | 0·19 | |
Fig. 6.
Analysis of NSyn mutations located in and around the main antigenic sites in the non-vaccinated and vaccinated groups. (a) Percent of all NSyn mutations located in or around the antigenic sites distributed in the VP1 immunodominant site (dark blue), the VP1 glycophorin A binding site (red) and the VP2, VP3 putative receptor binding (clear blue). (b) Average of the abundance of haplotypes bearing the NSyn mutations from (a).
Altogether, the analysis of the genomic features and the location of mutations on the protomer surface, suggested positive selection of replacements in the immunodominant site (encoded in fragments F8 and F9) and in the glycophorine A binding site (encoded in fragment F9) in the vaccinated group. In fact, the ratio of NSyn to synonymous variants (Ka/Ks), which is an indication of positive selection when its value is >1, of the F8 and F9 fragments was significantly (p < 0·001) higher in the vaccinated group (Ka/Ks = 1·15 ± 1·13) than in the non-vaccinated group (Ka/Ks = 0·51 ± 0·62). In contrast, no differences were found in the Ka/Ks of the putative receptor binding site (encoded in fragments F3- F6) between the non-vaccinated (Ka/Ks = 0·54 ± 0·60) and vaccinated groups (Ka/Ks = 0·49 ± 0·58), indicating lack of positive selection.
4. Discussion
In the current scenario of hepatitis A vaccine shortage, several countries have implemented as a first measure the reduction of the number of vaccine doses and the antigen content of the doses [9]. Public Health England recommendations for the vaccination of the MSM group in shortage settings, are among the most rational, and based on the following criteria. For immunocompetent adults under 60 years of age, a single dose of vaccine with half the adult antigen content may be enough since its immunogenicity, at one month, is equivalent to the complete antigen content. For patients immunocompromised, with chronic liver disease or aged over 60 years, due to a lower and slower response to vaccine, a higher antigen content dose is required [25]. Recommendations from the Spanish Health System [26] are less comprehensive and based on the administration of a single dose and two doses in the immunocompetent and the immunocompromised patients, respectively. However, despite the need to preserve the vaccine stocks, the implications that the reduction of the overall antigen administered may have on virus evolution and the potential emergence of variants escaping vaccine protection are mostly unknown [27]. Although HAV is an antigenically stable virus due to genetic and structural constraints [28], immune pressure selection may force the emergence of such variants and even of a new serotype.
In this study, we have analysed the evolution of HAV in five cases from vaccinated patients in comparison to the evolution in eight non-vaccinated patients. As expected, mutations located in or close to the capsid antigenic sites were found in both, vaccinated and non-vaccinated patients, and may be explained by the higher tolerance to mutation of the exposed capsid regions, or to the hitchhiking of replacements at antigenic sites belonging to genomes selected for other traits or by genetic drift [22,29]. However, the significantly higher Ka/Ks of the immunodominant and glycophorin A binding sites containing fragments, combined with the higher proportion of variants in these sites in the quasispecies, suggest positive selection of these variants in the vaccinated patients.
The cause of these vaccine failures is unknown. HAV vaccination is estimated to provide immunity up to twenty years in the general population [30], and at least up to ten years in the HIV-infected patients with high seropositivity rates [31]. Two failures were observed in optimally-vaccinated patients. The first patient received two doses of the dual paediatric TWINRIX vaccine during his childhood (M5), was 34-year old and HIV-non-infected, and showed a moderate diversity of antigenic variants but otherwise representing 20% of the virus population, and likely towards fixation of at least two mutations. The time lapse since the vaccination was over twenty years, and this could have caused a reduction of the antibody titres, allowing the virus to replicate in the presence of antibodies, and hence facilitating the selection of escaping variants. Unfortunately, anti-HAV antibody titres are not usually determined in the vaccinated patients. The second patient received two doses of the dual adult TWINRIX vaccine two years before the onset of hepatitis A (M7), was immunocompetent HIV-infected, and showed a moderate diversity of antigenic variants but none of them at a high proportion. The other three failures were in suboptimally-vaccinated patients. The first and second patients received a single dose of the adult dual TWINRIX vaccine 15 months (M46) and 18 months (M48) before the onset of the disease, were both immunocompetent HIV-infected, and again showed moderate diversity of antigenic variants with none at high proportions. Vaccine failures after a single dose have been previously described but data on virus evolution has never been discussed [32]. The third patient, that received two doses of the adult dual TWINRIX vaccine during the incubation period (M6), was 34-year old and HIV-non-infected, and showed a moderate diversity of antigenic variants but with one, affecting the immunodominant site, at a proportion of 12·7%, again suggesting an ongoing process of fixation. Post-exposure vaccination is increasingly being used to prevent cases among contacts, and although is a good preventive measure, failure rates of 4·4%-6·9% have been described [33,34]. HAV replication takes place mostly during the incubation time of the disease and again the low concentration of antibodies would have allowed the virus to replicate in their presence enabling the selection of escaping variants. Regarding the three cases in vaccinated HIV-infected patients, with a repertoire/proportion of HAV antigenic variants similar to that of the non-vaccinated patients, the failure could likely be due to a lack of response to the vaccine as previously described [35]. Alternatively, it could be due to a less prone antigenic variant emergence, since HIV-infected patients showed a tendency to carry a lower HAV genome load. This tendency contrasts with the data available in the literature showing average genome loads of 103–107 and of 107–108 genome copies/ml of sera in HIV-non-infected patients [36,37] and HIV-infected patients [12,38], respectively. Whether this change in the HAV replication capacity observed in HIV-infected patients is related to the recent introduction of new antivirals in the anti-HIV therapy that could act on HAV, remains unknown.
Antigenic variants arising around the immunodominant and the glycophorin A binding sites show higher fitness in the presence of antibodies than the wild-type virus [22]. Consequently, they may represent a public health threat in populations with high anti-HAV seroprevalences, including endemic regions and countries with vaccination programs.
The MSM group is becoming endemic for hepatitis A requiring specific vaccination policies. As expected the number of cases among the vaccinated patients was very low, but the molecular analysis of their quasispecies is essential for the analysis of HAV evolution under immune pressure. Despite the overall low number of samples included in this study, still some recommendations aiming to avoid the emergence of vaccine escape variants can be formulated: vaccination schedule must be completed, booster doses should be administered to those vaccinated long time ago or with low anti-HAV antibodies, and vaccine post-exposure prophylaxis should be reconsidered. These recommendations are particularly relevant for the MSM group, especially in those countries and regions where the virus is endemic.
Acknowledgments
Acknowledgements
We are grateful for the collaboration of the physicians reporting the cases and all the members of the Epidemiology Service, Public Health Agency of Barcelona, particularly to Mireia G Carrasco.
Funding
Spanish Ministry of Economy grants BIO2014-53285-R, BIO2017-83191-R and PI16/00337, all co-financed by the European Regional Development Fund, Spanish Center for the Technological Development grant IDI-20151125, Carlos III-Health Institute Research Network on Hepatic and Digestive Diseases-CIBERehd, and Generalitat de Catalunya Biotechnology Reference Network (XRB).
Declaration of interests
No role of the funding source.
Author contributions
RMP, JQ, and AB conceived the idea and designed the study. AS and DG performed the experimental work. AS, RMP, JG, JQ, AB, TP and SG analysed the data. SM and JAC provided epidemiological data and analysis. RMP, AS, and AB wrote the manuscript. All authors approved the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.11.023.
Contributor Information
Josep Quer, Email: josep.quer@vhir.org.
Rosa M. Pintó, Email: rpinto@ub.edu.
Appendix A. Supplementary data
Supplementary material
References
- 1.Corey L., Holmes K.K. Sexual transmission of hepatitis A in homosexual men: incidence and mechanism. N Engl J Med. 1980;302(8):435–438. doi: 10.1056/NEJM198002213020804. [DOI] [PubMed] [Google Scholar]
- 2.Christenson B., Brostrom C., Bottiger M. An epidemic outbreak of hepatitis A among homosexual men in Stockholm. Hepatitis A, a special hazard for the male homosexual subpopulation in Sweden. Am J Epidemiol. 1982;116(4):599–607. doi: 10.1093/oxfordjournals.aje.a113442. [DOI] [PubMed] [Google Scholar]
- 3.Stene-Johansen K., Tjon G., Schreier E. Molecular epidemiological studies show that hepatitis A virus is endemic among active homosexual men in Europe. J Med Virol. 2007;79(4):356–365. doi: 10.1002/jmv.20781. [DOI] [PubMed] [Google Scholar]
- 4.Urbanus A.T., van Houdt R., van de Laar T.J., Coutinho R.A. Viral hepatitis among men who have sex with men, epidemiology and public health consequences. Euro Surveill. 2009;14(47) doi: 10.2807/ese.14.47.19421-en. [DOI] [PubMed] [Google Scholar]
- 5.Tortajada C., de Olalla P., Diez E. Hepatitis A among men who have sex with men in Barcelona, 1989-2010: insufficient control and need for new approaches. BMC Infect Dis. 2012;12(1):11. doi: 10.1186/1471-2334-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beebeejaun K., Degala S., Balogun K. Outbreak of hepatitis A associated with men who have sex with men (MSM), England, July 2016 to January 2017. Euro Surveill. 2017;22(5) doi: 10.2807/1560-7917.ES.2017.22.5.30454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freidl G.S., Sonder G.J., Bovee L.P. Hepatitis A outbreak among men who have sex with men (MSM) predominantly linked with the EuroPride, the Netherlands, July 2016 to February 2017. Euro Surveill. 2017;22(8) doi: 10.2807/1560-7917.ES.2017.22.8.30468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Werber D., Michaelis K., Hausner M. Ongoing outbreaks of hepatitis A among men who have sex with men (MSM), Berlin, November 2016 to January 2017 - linked to other German cities and European countries. Euro Surveill. 2017;22(5) doi: 10.2807/1560-7917.ES.2017.22.5.30457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ECDC Epidemiological update: hepatitis A outbreak in the EU/EEA mostly affecting men who have sex with men. 2018. https://ecdc.europa.eu/en/news-events/epidemiological-update-hepatitis-outbreak-eueea-mostly-affecting-men-who-have-sex-men-1 (accessed 10/26/2018)
- 10.Perez-Sautu U., Costafreda M.I., Cayla J. Hepatitis A virus vaccine escape variants and potential new serotype emergence. Emerg Infect Dis. 2011;17:734–738. doi: 10.3201/eid1704.101169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godoy P., Carmona G., Manzanares S. Trends and risk factors of hepatitis A in Catalonia after the introduction of a hepatitis A+B vaccination programme. J Viral Hepat. 2018;25(9):1001–1007. doi: 10.1111/jvh.12900. [DOI] [PubMed] [Google Scholar]
- 12.Costafreda M.I., Bosch A., Pintó R.M. Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl Environ Microbiol. 2006;72(6):3846–3855. doi: 10.1128/AEM.02660-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.RIVM HAVNET. 2014. https://www.rivm.nl/en/Topics/H/HAVNET (accessed 10/10/2018)
- 15.Magoc T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregori J., Esteban J.I., Cubero M. Ultra-deep pyrosequencing (UDPS) data treatment to study amplicon HCV minor variants. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0083361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregori J., Perales C., Rodriguez-Frias F., Esteban J.I., Quer J., Domingo E. Viral quasispecies complexity measures. Virology. 2016;493:227–237. doi: 10.1016/j.virol.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 18.Rozas J., Rozas R. DnaSP, DNA sequence polymorphism: an interactive program for estimating population genetics parameters from DNA sequence data. Comput Appl Biosci. 1995;11(6):621–625. doi: 10.1093/bioinformatics/11.6.621. [DOI] [PubMed] [Google Scholar]
- 19.J. Rozas, A. Ferrer-Mata, J.C. Sánchez-Delbarrio, et al., DnaSP, http://www.ub.edu/dnasp/, 2018, (accessed 04/15/2018).
- 20.Wang X., Ren J., Gao Q. Hepatitis A virus and the origins of picornaviruses. Nature. 2015;517(7532):85–88. doi: 10.1038/nature13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ping L.H., Lemon S.M. Antigenic structure of human hepatitis A virus defined by analysis of escape mutants selected against murine monoclonal antibodies. J Virol. 1992;66(4):2208–2216. doi: 10.1128/jvi.66.4.2208-2216.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aragonès L., Bosch A., Pintó R.M. Hepatitis A virus mutant spectra under the selective pressure of monoclonal antibodies: codon usage constraints limit capsid variability. J Virol. 2008;82(4):1688–1700. doi: 10.1128/JVI.01842-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Zhu L., Dang M. Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site. Proc Natl Acad Sci U S A. 2017;114(4):770–775. doi: 10.1073/pnas.1616502114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez G., Aragonès L., Costafreda M.I., Ribes E., Bosch A., Pintó R.M. Capsid region involved in hepatitis A virus binding to glycophorin A of the erythrocyte membrane. J Virol. 2004;78(18):9807–9813. doi: 10.1128/JVI.78.18.9807-9813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.PHE Hepatitis A vaccination in adults - temporary recommendations. 2017. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/727354/Hepatitis_A_vaccine__for_adults_-_temporary_recommendations.pdf (accessed 05/23/2018)
- 26.MSCBS Problemas de suministro de vacunas frente a hepatitis A. Recomendaciones. 2017. https://www.mscbs.gob.es/profesionales/saludPublica/prevPromocion/vacunaciones/docs/Problemas_suministro_HepatitisA.pdf (accessed 05/23/2018)
- 27.Pintó R.M., D'Andrea L., Perez-Rodriguez F.J. Hepatitis A virus evolution and the potential emergence of new variants escaping the presently available vaccines. Future Microbiol. 2012;7(3):331–346. doi: 10.2217/fmb.12.5. [DOI] [PubMed] [Google Scholar]
- 28.Pintó R.M., Pérez-Rodríguez F.-J., D' Andrea L., de Castellarnau M., Guix S., Bosch A. Hepatitis A virus codon usage: implications for translation kinetics and capsid folding. Cold Spring Harb Perspect Med. 2018;8 doi: 10.1101/cshperspect.a031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez G., Bosch A., Gomez-Mariano G., Domingo E., Pintó R.M. Evidence for quasispecies distributions in the human hepatitis A virus genome. Virology. 2003;315(1):34–42. doi: 10.1016/s0042-6822(03)00483-5. [DOI] [PubMed] [Google Scholar]
- 30.Clemens R., Safary A., Hepburn A., Roche C., Stanbury W.J., Andre F.E. Clinical experience with an inactivated hepatitis A vaccine. J Infect Dis. 1995;171(Suppl. 1):S44–S49. doi: 10.1093/infdis/171.supplement_1.s44. [DOI] [PubMed] [Google Scholar]
- 31.Crum-Cianflone N.F., Wilkins K., Lee A.W. Long-term durability of immune responses after hepatitis A vaccination among HIV-infected adults. J Infect Dis. 2011;203(12):1815–1823. doi: 10.1093/infdis/jir180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Innis B.L., Snitbhan R., Kunasol P. Protection against hepatitis A by an inactivated vaccine. JAMA. 1994;271(17):1328–1334. [PubMed] [Google Scholar]
- 33.Victor J.C., Monto A.S., Surdina T.Y. Hepatitis A vaccine versus immune globulin for postexposure prophylaxis. N Engl J Med. 2007;357(17):1685–1694. doi: 10.1056/NEJMoa070546. [DOI] [PubMed] [Google Scholar]
- 34.Whelan J., Sonder G.J., Bovee L., Speksnijder A., van den Hoek A. Evaluation of hepatitis A vaccine in post-exposure prophylaxis, the Netherlands, 2004–2012. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0078914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mor Z., Lurie Y., Katchman E. A case of hepatitis A vaccination failure in an HIV-positive man who had sex with men in Israel. Int J STD AIDS. 2012;23(7):529–530. doi: 10.1258/ijsa.2010.010269. [DOI] [PubMed] [Google Scholar]
- 36.Costa-Mattioli M., Monpoeho S., Nicand E., Aleman M.H., Billaudel S., Ferre V. Quantification and duration of viraemia during hepatitis A infection as determined by real-time RT-PCR. J Viral Hepat. 2002;9(2):101–106. doi: 10.1046/j.1365-2893.2002.00336.x. [DOI] [PubMed] [Google Scholar]
- 37.Normann A., Jung C., Vallbracht A., Flehmig B. Time course of hepatitis A viremia and viral load in the blood of human hepatitis A patients. J Med Virol. 2004;72(1):10–16. doi: 10.1002/jmv.10532. [DOI] [PubMed] [Google Scholar]
- 38.Costa-Mattioli M., Allavena C., Poirier A.S., Billaudel S., Raffi F., Ferre V. Prolonged hepatitis A infection in an HIV-1 seropositive patient. J Med Virol. 2002;68(1):7–11. doi: 10.1002/jmv.10163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material