Abstract
Background
Knowledge of the natural history of colorectal adenomas is limited because these lesions are removed upon detection. The few studies in which small adenomas have been left in situ for a limited period of time, have shown that most lesions remain stable or even completely regress. Specific DNA copy number changes (‘cancer associated events’ or CAEs) are associated with progression of adenomas to cancer. In this study we evaluated whether molecular features of progression correlated with growth of small polyps.
Methods
Small (6–9 mm) colorectal precursor lesions detected on CT-colonography (CTC) were left in situ and re-evaluated with CTC after three years. Based on volumetric change, polyps were classified as either grown, stable or regressed. Surveillance CTC was followed by colonoscopy, during which all lesions were resected. Using DNA isolated from FFPE polyp tissues, low-coverage whole genome sequencing was performed to determine DNA copy number profiles, as well as target enrichment mutation analysis and CpG island methylation phenotype (CIMP) analysis. Expression of DNA mismatch repair (MMR) proteins was determined by immunohistochemistry. Samples were marked as MMR proficient if all MMR proteins were expressed.
Findings
Out of 68 polyps resected at colonoscopy, for 65 (96%) material was available. Of these, 31 (48%) had grown, 27 (41%) remained stable and 7 (11%) regressed. Polyps with at least one CAE had higher growth rates compared to polyps without CAEs (difference 91% growth (95% CI 13–169), p = .023). CAEs were absent in lesions that had partially regressed. Mutations occurred in 94% of the polyps, with higher growth rates being associated with polyps having ≥2 mutations compared to lesions with only 0–1 mutations (difference 99% growth (95% CI 9–189), p = .032). All samples were MMR proficient. No relation between growth and CIMP was observed.
Interpretation
Molecular alterations associated with colorectal cancer, correlated with growth of small polyps and were absent in polyps that regressed. Therefore, this longitudinal study provides in vivo support in the human setting for the functional role of these molecular alterations, that have mostly been identified by cross sectional observations in tissue samples of colorectal adenomas and cancers.
Fund
Alpe d'Huzes- Dutch Cancer Society (project number NKI2013-6338).
Keywords: Colorectal polyps, Natural behaviour, Growth, Molecular profiling
Research in context.
Evidence before this study
We searched Pubmed (from June 1, 2015 up to March 1, 2018) with a combination of relevant key terms including: colonic lesions, polyps, natural history, evolution, growth, progression, follow-up, mutation, copy number changes and molecular profile. Inherent to the fact that colorectal polyps are removed once detected on endoscopy, molecular features of these lesions cannot be correlated to longitudinal observations on phenotype changes. Therefore, the knowledge on the biology of colorectal cancer progression gathered up to now is mainly based on cross-sectional studies. Next to the sequence of mutations classically associated with adenoma to carcinoma progression, we have previously shown that certain DNA copy number alterations are associated with progression of cancer. In one study where polyps were left in situ and followed up in time, mutation burden was correlated with polyp growth. However, comprehensive analysis of DNA alterations (i.e. sequence mutations, promoter methylation, DNA copy number changes and MMR status) in longitudinally observed polyps has never been performed. To enhance our understanding of the natural course of disease in colorectal polyps, we set out to assess the association between DNA alterations and in vivo growth in small colorectal polyps.
Added value of this study
We had access to a unique set of 65 small polyps that had been followed longitudinally, and performed a comprehensive analysis of DNA alterations in these polyps, including DNA copy number, mutation, CIMP and MMR status. For the first time we were able to show that regressed polyps did not harbour DNA copy number alterations or mutations associated with later stages of polyp to cancer progression. Higher growth rates were observed in lesions with cancer associated DNA copy number alterations (cancer associated events or CAEs), as well as in lesions with a higher mutation burden.
Implications of all the available evidence
Since approximately only 5% of all colorectal adenomas eventually progress to cancer, strategies for early detection ideally should focus on identifying only those 5% of lesions that progress and ignore the 95% of polyps that will not harm patients. In this in vivo human longitudinal study we have provided insights in the DNA alterations of small colorectal polyps and their growth over time. These insights can guide the development of new specific and sensitive screening and surveillance methods.
Alt-text: Unlabelled Box
1. Introduction
The development of colorectal cancer (CRC) is a stepwise process, in which normal epithelial cells transform into an adenocarcinoma through a benign intermediate lesion (i.e. mostly adenomas, but also serrated polyps). While the prevalence of adenomas is high, only a minority of approximately 5% eventually progresses into cancer; the remaining lesions do not convert to malignancy or even completely disappear over time [1,2]. Understanding the natural history of disease is at the basis of all strategies for early detection of cancer. In the case of colorectal adenoma to carcinoma progression, the absence of longitudinal observations, like we do have in for instance Barrett's oesophagus and cervical cancer [3], causes an evident blind spot in our knowledge of this disease. Molecular analysis of colorectal polyps that have been left in situ for a few years, even if this concerns small lesions only, provides a unique opportunity to fill some of these gaps.
Cross-sectional studies have shown that adenomas larger than 10 mm have a higher risk of harbouring a focus of cancer (2.1–6.9%), compared to small 6–9 mm lesions (0–0.42%) [4]. During adenoma formation and subsequent malignant progression, several cell signalling pathways get disrupted. An early event is the disruption of the WNT signalling pathway by mutation of the APC gene, followed by mutations in the RAS-MAPK, PI3K-AKT, TGF-ß, and p53 pathways in later stages of progression [5]. The increased rate of new mutations is facilitated by the acquisition of some form of genomic instability, most commonly chromosomal instability (CIN) that is present in about 85% of sporadic CRCs [6]. DNA copy number alterations particularly associated with the transition from adenoma to carcinoma are gains in 8q, 13q and 20q and losses in 8p, 15q, 17p and 18q [[7], [8], [9]]. The presence of two or more of any of these seven CAEs marked adenomas at high risk of progression with high accuracy (78% sensitivity and 78% specificity) [7].
Besides adenomas, also serrated polyps have been recognised as precursors lesions. In serrated polyps a BRAF mutation is typically the initiating event, leading to increased gene promoter hypermethylation (CpG island methylation phenotype or CIMP). When hypermethylation affects the expression of the mismatch repair (MMR) genes, this gives rise to another form of genomic instability, known as microsatellite instability (MSI) [10].
Although the molecular events occurring during CRC development have been widely studied, most observations are from cross-sectional studies as precursor lesions are removed upon detection at colonoscopy. In a few CT-colonography (CTC) studies in which small polyps were left in situ, adenoma growth during follow-up was associated with an advanced adenoma phenotype at resection [2,11]. The aim of the present study was to assess whether polyp growth was related to molecular features of colorectal cancer. For this purpose, we used a unique series of patients with small (6–9 mm) colorectal lesions initially identified by CTC, that were left in situ and ultimately resected after a surveillance interval of three years. This allowed longitudinal assessment of lesion size (i.e. growth) in relation to histological and molecular characteristics at time of resection.
2. Methods
2.1. Study design and participants
In a Dutch multi-centre, randomised controlled screening trial (COlonoscopy or COlonography for Screening (COCOS) trial, 2009–2010, trial number: 2009/03WBO and NTR1829, The Hague, Netherlands) comparing primary colonoscopy to CTC [12], patients in the CTC-arm with one or two small 6–9 mm colorectal lesions were advised to undergo a surveillance CTC after an interval of three years. Patients with more than two 6–9 mm polyps or larger polyps on baseline CTC were referred for colonoscopy directly and therefore not included in the present follow-up study. The 95 small lesions detected on index CTC in this patient subpopulation were thus left in situ and re-measured at follow-up to assess the percentage of volumetric change for each lesion over the entire surveillance interval [2]. Based on volumetric change on CTC over the entire surveillance interval as proportion of baseline volume, lesions were classified as either grown (>30% growth), stable (<30% regression to <30% growth) or regressed (>30% regression). Details of this method and the choice of the 30% cut-off are described elsewhere [2]. Following the surveillance CTC, all patients were offered a colonoscopy for resection of the lesions. Location and size of the polyp on colonoscopy were recorded. The distal colon was defined as rectum and sigmoid. From those lesions for which histopathology was available, formalin fixed paraffin-embedded (FFPE) material was retrieved and reviewed by an expert pathologist (GAM). Based on histopathology and size at colonoscopy, lesions were classified as non-advanced adenoma, advanced adenoma, non-advanced serrated polyp or advanced serrated polyp according to the definitions summarised in Table 1.
Table 1.
Classification of colorectal polyps.
| Lesion type | Abbreviation | Definition |
|---|---|---|
| Non-advanced adenoma | NAA | tubular adenoma (TA) <10 mm with low-grade dysplasia |
| Advanced adenoma | AA | adenoma ≥10 mm and/or with high-grade dysplasia and/or a villous component of ≥25% |
| Non-advanced serrated polyps | NASP | hyperplastic polyp (HP), sessile serrated lesion (SSL), or traditional serrated lesion (TSL) <10 mm without dysplasia |
| Advanced serrated polyp | ASP | hyperplastic polyp (HP), sessile serrated lesion (SSL), or traditional serrated lesion (TSL) ≥10 mm and/or with dysplasia |
Ethics approval from the Dutch Health Council was obtained for COCOS, including surveillance CTC after 3 years. Patients had already given their written informed consent to be contacted for follow-up studies and consented to this study.
2.2. Procedures
2.2.1. DNA isolation
DNA was isolated as previously described [13]. In brief, DNA from FFPE material was isolated following micro-dissection (> 70% tumour cells). A six-day incubation period with proteinase K in lysis buffer (ATL buffer, QIAmp, DNA micro-kit, Qiagen, Venlo, The Netherlands) was performed. Every day, proteinase K (10 μl or 20 ng/μl) was freshly added. DNA was isolated using the QIAmp DNA micro-kit (Qiagen) and concentrations and purity were measured on a Nanodrop ND-1000 spectrophotometer (Isogen, IJsselstein, The Netherlands). Isolated DNA was used as input for copy number analysis, mutation analysis and CIMP analysis, in that order specifically.
2.2.2. DNA copy number analysis
DNA copy number changes were analysed with low-coverage whole genome sequencing (WGS) [14]. Briefly, DNA was fragmented by sonication (Covaris S2, Woburn, MA, USA) and run on the HiSeq 2500 (Illumina, San Diego, CA, USA) on a 65 basepairs single-read modus using the KAPA HyperPrepKit (KAPA Biosystems, KK8504, Wilmington, MA, USA). This yielded a coverage of 0.13× (IQR 0.12–0.14) genome coverage. The WGS reads were analysed with Bioconductor R-package QDNAseq, using a published workflow [15]. For every fixed-sized region of 30 kb on the genome, the relative abundance of sequence reads was used to determine the aberration status, applying corrections for mappability and GC content and removing germ-line specific variations [14]. A wavy pattern seen in copy number plots, ‘genomic waves’, which may be caused by replication timing of proliferating cells [16], were smoothed using NoWaves [17]. This algorithm uses a set of normal samples (in this case of patients with CRC) [18] as reference to correct bins (genomic intervals) which systematically obtain a higher or lower signal. The obtained copy number profiles were segmented into regions of constant log2-read count and aberrations were called as high-level amplification [2], gain [1], normal (0), loss (−1) or homozygous deletion (−2). When the number of called copy number segments was above 200 over the whole genome, while at the same time had a very a high difference between expected and observed noise (>1.5 times the interquartile range of values of all analysed samples), samples were excluded for further analysis. When ≥2 CAEs were present, the lesion was marked as a high-risk adenoma.
2.2.3. Mutation analysis
Samples in which DNA was still available after copy number analysis, were subjected to mutation analysis. DNA libraries were prepared using the KAPA HyperPrep Kit (KAPA Biosystems, Wilmington, MA, USA) as described in the KAPA HyperPrep Kit protocol (KR0961 – v5.16). Target enrichment was performed using a custom 48 gene xGen® Predesigned Gene Capture Pools (Integrated DNA Technologies, San Diego, CA, USA), according to the Rapid Protocol for DNA Probe Hybridisation and Target Capture Using an Illumina TruSeq® Library, Version 2.1, with an extended hybridisation reaction of 24 h. The gene panel consisted of 48 cancer-related genes, including genes most often mutated in colorectal cancer such as APC, KRAS, NRAS, PIK3CA, SMAD4, TP53 and BRAF (Supplementary table 1). Paired-end 65 bp sequencing data were generated with Illumina Hiseq 2500 (Illumina, San Diego, CA, USA), yielding a median of 89× (IQR 55–148) coverage in the target regions, after removal of duplicate reads. The target regions, spanning the exonic sequences of the 48 genes, covered ~3.55 × 105 bp in total.
After adapter trimming, the reads were aligned to the human reference GRCh38 with BWA-MEM [19]. Subsequently base quality scores were recalibrated and the variants were called according to the GATK HaplotypeCaller [20]. Variant effects prediction was performed using SnpEff [21] and external data sources were linked using SnpSift [22]. To exclude DNA polymorphisms present in the normal populations, variants reported in dbSNP as ‘common’ or ‘G5’ were excluded. Furthermore, variants present at ≥1% in the ExAC exome data [23] and variants affecting non-coding sequences were excluded. Variants were required to have a ‘medium’ or ‘high’ effect, according to SnpEff, which led to the exclusion of silent mutations. Mutations were summed per gene and per sample using a representation called Oncoprint, which was created using R Bioconductor, package ComplexHeatmap.
2.2.4. CIMP analysis
Samples in which DNA was still available after copy number and mutation analyses, underwent sodium bisulfite modification (EZ DNA methylation kit, ZYMO research Co., Orange, CA, USA) to determine CIMP status. Nested methylation specific PCR (nested-MSP) for the CIMP marker panel as defined by Weisenberger [24] was performed as described earlier [25]. Ten μl of each MSP reaction was loaded onto a 2% agarose gel, stained with GelStar and visualised using ultraviolet light. Polyps were defined as CIMP-high when ≥3 of the 5 markers (CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1) from the CIMP marker panel were methylated.
2.2.5. MMR-status analysis
Immunohistochemistry (IHC) of the FFPE tumour samples was performed on a BenchMark Ultra autostainer (Ventana Medical Systems, Oro Valley, AZ, USA). Briefly, paraffin sections were cut at 3 μm, heated at 75 °C for 28 min and deparaffinised with EZ prep solution. Heat-induced antigen retrieval was carried out using Cell Conditioning 1 for 32 min at 95 °C (MSH2, MSH6 and PMS2), or 64 min at 95 °C (MLH1). MLH1 was detected using clone M1 (Ready-to-Use, 32 min at 37 °C, Ventana Medical systems), MSH2 using clone G219–1129 (Ready-to-Use, 12 min at 37 °C, Ventana Medical systems), MSH6 using clone EP49 (1/50 dilution, 32 min at 37 °C, Epitomics, Burlingame, CA, USA) and PMS2 using clone EP51 (1/40 dilution, 32 min at 37 °C, Dako).
For PMS2 signal amplification was applied using the Optiview Amplification Kit (4 min, Ventana Medical Systems). Bound antibody was detected using the OptiView DAB Detection Kit and slides were counterstained with Hematoxylin and Bluing Reagent (Ventana Medical Systems, Oro Valley, AZ, USA).
The slides were scored for positivity by an expert pathologist (GAM or PS). In case of positivity of the four MMR genes, the sample was considered MMR proficient. In case expression of one or more MMR genes was lost, the sample was considered MMR deficient.
2.3. Statistics analysis
Supported by the observation that colorectal polyps originating from the same patient differed in morphology, colonic location, histopathology and/or growth, all colorectal lesions were assumed to develop independently (Supplementary table 2). For comparisons of numerical data between two unpaired subgroups, the independent t-test was used. For comparison of categorical data between unpaired subgroups the Chi-square test or Fischer's Exact test was used. For all test, two-sided p ≤ .05 was considered significant. For the comparison of aberration frequencies between two groups, R-package CGHtest was applied, which runs a Chi-square test and a Wilcoxon rank-sum test. A multiple testing correction to the p-values was performed according to the Benjamini and Yekutieli FDR rule, using a cut-off for significance of 0.10 [26].
2.4. Data depository
Sequence data has been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001003284 [27].
3. Results
3.1. Histopathology
Of the 68 polyps resected at the surveillance colonoscopy, material was available for 65 (96%) polyps (Fig. 1). These 65 polyps came from 46 patients (57% male, mean age 66.7 (s.d. 6∙9) years) who had a mean surveillance interval of 3.3 (s.d. 0.29) years. The lesions included 47 (72%) tubular adenomas (TAs) with low grade dysplasia (LGD), 9 (14%) tubulovillous adenomas (TVAs) with LGD, 1 (2%) sessile serrated lesion (SSL) without dysplasia and 8 (12%) hyperplastic polyps (HPs) without dysplasia (Table 2). When using the 30% threshold of volumetric change [2], 48% (31/65) of the lesions had grown, 41% (27/65) remained stable and 11% (7/65) regressed (Table 2).
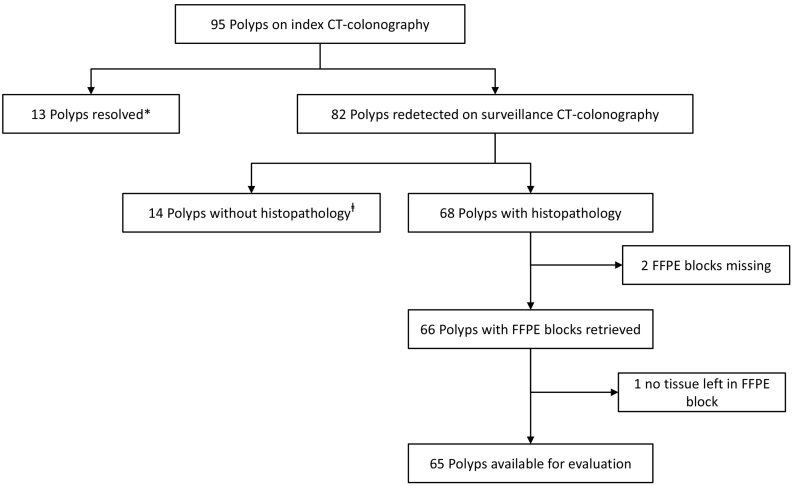
Fig. 1.
Flow-chart of polyps followed longitudinally after initial CT-colonography (CTC) detection. *13 polyps could not be redetected at surveillance CTC, despite good distention and good quality of fecal tagging in the relevant segments for 12 out of the 13 polyps. ⱡ14 polyps had no histopathological diagnosis, because the patient was not referred for colonoscopy (<6 mm polyp at surveillance CTC) (n = 5), the patient refused colonoscopy (n = 5) or polyps were neither detected (n = 3), nor retrieved (n = 1) during colonoscopy.
Table 2.
Clinicopathological characteristics of the 65 longitudinally observed polyps per growth category.
| Grown |
Stable |
Regressed |
|
|---|---|---|---|
| All polyps (n = 65) | n = 31 | n = 27 | n = 7 |
| Location | |||
| Proximal | 16 (52%) | 11 (41%) | 5 (71%) |
| Distal | 15 (48%) | 16 (59%) | 2 (29%) |
| Morphology | |||
| Sessile | 16 (52%) | 14 (52%) | 6 (86%) |
| Pedunculated | 9 (29%) | 11 (41%) | 1 (14%) |
| Flat | 6 (19%) | 2 (7%) | 0 |
| Adenomas (n = 56) | n = 27 | n = 23 | n = 6 |
| Histology | |||
| Tubular | 20 (74%) | 21 (91%) | 6 (100%) |
| Tubullovillous | 7 (26%) | 2 (9%) | 0 |
| Villous | 0 | 0 | 0 |
| Dysplasia | |||
| Low-grade | 27 (100%) | 23 (100%) | 6 (100%) |
| High-grade | 0 | 0 | 0 |
| Serrated polyps (n = 9) | n = 4 | n = 4 | n = 1 |
| Histology | |||
| Sessile serrated | 1 (25%) | 0 | 0 |
| Hyperplastic | 3 (75%) | 4 (100%) | 1 (100%) |
| Dysplasia | |||
| Absent | 4 (100%) | 4 (100%) | 1 (100%) |
| Present | 0 | 0 | 0 |
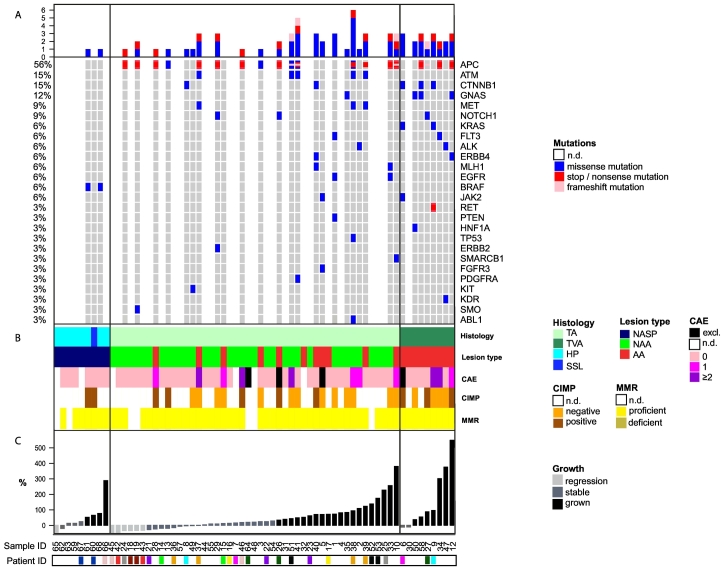
Histopathological features and molecular profiles of the 65 polyps, ranked by growth rate, are summarised in Fig. 2. Of the adenomas, 36% (20/56) were classified as advanced adenomas (Table 1 and Fig. 2B). All nine serrated polyps were classified as non-advanced serrated polyps.
Fig. 2.
Molecular profile and histopathological features ordered by growth rate for hyperplastic polyps and sessile serrated lesions (left), tubulovillous adenomas (middle) and tubular adenomas (right). Each vertical column represents an individual lesion. A. Mutational profile. Only genes that have a mutation in at least one of the samples are shown. The top bars represent the number of mutations per sample. Percentages on the left indicate the prevalence of a specific mutation over all the analysed samples. B. Histopathological features and molecular profile resulting from copy number analysis, CIMP analysis and immunohistochemistry of the mismatch-repair genes. Several samples could not undergo the entire range of molecular analyses due to limited DNA available. Histology: TA = tubular adenoma, TVA = tubulovillous adenoma, HP = hyperplastic polyp, SSL = sessile serrated lesion; Lesion type: NASP = non-advanced serrated polyp, NAA = non-advanced adenoma, AA = advanced adenoma; CAE = cancer associated event, which includes chromosomal gains in 8q, 13q and 20q and losses in 8p, 15q, 17p and 18q. The presence of ≥2 CAEs defines high-risk adenomas. Four samples were excluded from DNA copy number analysis due to poor quality; CIMP = CpG island methylation phenotype; MMR = mismatch repair. C. Growth rates according to CT-colonography measurement during the 3-year surveillance interval. Patient ID is depicted in colour marking only for patients with >1 polyp.
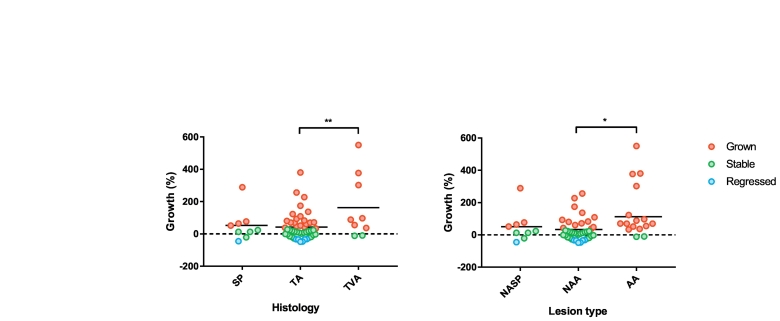
In relation to histopathology, growth rates were significantly higher in TVAs compared to TAs (difference 121% growth (95% CI 42–200), independent t-test, p = .003, Fig. 3A). Also growth rates were significantly higher in polyps that were advanced adenomas at resection compared to non-advanced adenomas (difference 80% growth (95% CI 18–142), independent t-test, p = .012, Fig. 3B).
Fig. 3.
Growth rates for individual polyps classified as A. serrated polyp (SP), tubular adenoma (TA) and tubulovillous adenoma (TVA) with corresponding means (** independent t-test, p < .01) B. non-advanced serrated polyp (NASP), non-advanced adenoma (NAA), and advanced adenoma (AA) (* independent t-test, p < .05). The red, green and blue dots represent lesions that had grown, remained stable or regressed during the 3-year surveillance interval, respectively, according to the threshold of volumetric change.
3.2. DNA copy number analysis
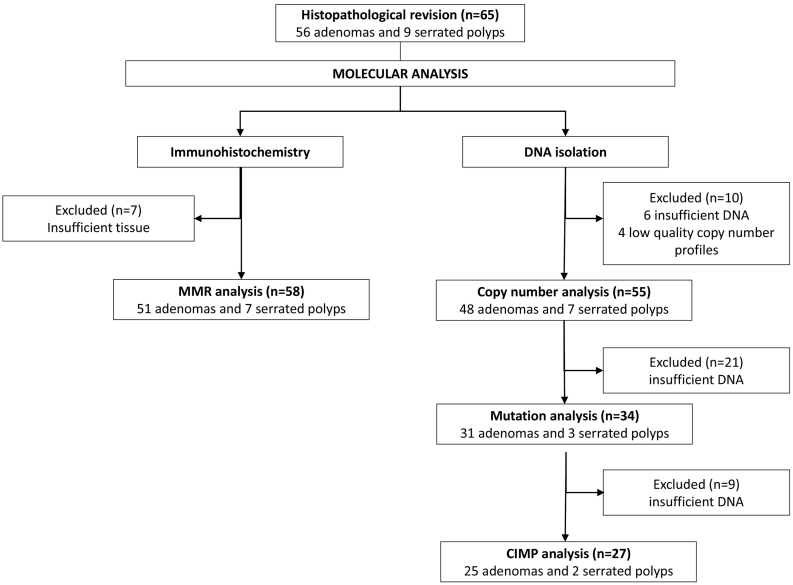
Good quality DNA was obtained from 59 of the 65 retrieved lesions. The copy number profiles of 4/59 samples did not meet the quality criteria, leaving 55 lesions for analysis (Fig. 4). These concerned 48 adenomas and seven serrated polyps (6 HPs and 1 SSL). Twenty percent (11/48) of adenomas, but none of the serrated polyps showed at least one CAE (Fig. 2B). In these adenomas, 13q gain was the most common CAE (91%, 10/11), followed by 20q gain (4/11: 36%), 8q gain (3/11; 27%) and 17p loss (9%; 1/11) (Supplementary table 3).
Fig. 4.
Flow-chart of polyp samples used for molecular analyses. CIMP = CpG island methylation phenotype; MMR = mismatch repair.
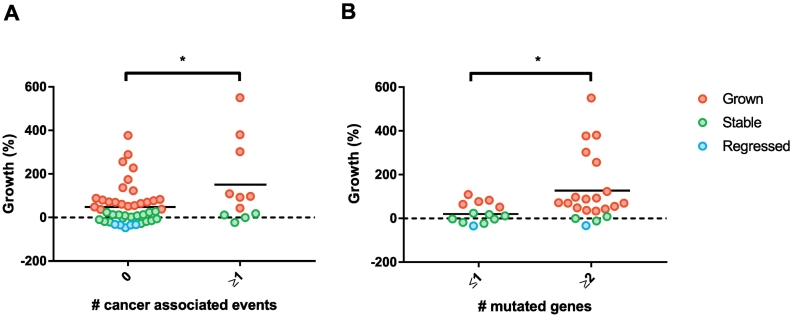
The mean growth rate of lesions with CAEs was significantly higher in lesions with ≥1 CAEs, compared to lesions without CAEs (difference 91% growth (95% CI 13–169), independent t-test, p = .023, Fig. 5A). CAEs were absent in the five lesions that had regressed. Based on the molecular definition of having ≥2 CAEs, 10% (5/48) of the adenomas were classified as being at high risk for progression. Two of these high-risk adenomas were advanced adenomas that had grown, one was an advanced adenoma that had remained stable, one was a non-advanced adenoma that had remained stable and one was a non-advanced adenoma that had grown (Fig. 2B).
Fig. 5.
Growth rates for individual polyps that have A. 0 or ≥ 1 cancer associated events, which include chromosomal gains in 8q, 13q and 20q and losses in 8p, 15q, 17p and 18q (* independent t-test, p < .05) B. ≤1 mutation or ≥ 2 mutations, with corresponding means (* independent t-test, p < .05). The red, green and blue dots represent lesions that had grown, remained stable or regressed during the 3-year surveillance interval, respectively, according to the threshold of volumetric change.
3.3. Mutation analysis
Mutation analysis could be successfully completed for 34 samples (Fig. 4), including 31 adenomas and three serrated polyps (2 HPs and 1 SSL). One or more mutations were observed in 94% (32/34) of the samples (median 2, range 0–5). Mutations of the WNT pathway were found in 74% (23/31) of adenomas, including APC mutation as the most common overall alteration in 61% (19/31) and CTNNB1 mutation in 16% (5/31) (Fig. 2A). Only one adenoma had a mutation in both the APC and CTNNB1 genes; in all other samples these mutations were mutually exclusive. The PI3K-AKT pathway was affected in one adenoma (3%) with a mutation in the PTEN gene. Genetic alterations in the RAS-MAPK pathway occurred in 10% (3/31) of the adenomas, concerning two KRAS mutations (6%; 2/31) and one mutation in ERBB2. No mutations were found in the TGFβ pathway. Sixteen percent (5/31) of adenomas showed mutations in the p53 pathway 16% (5/31), occurring in ATM (16%; 5/31) and/or TP53 (3%; 1/31). In the serrated polyps, BRAF was the only mutation detected (67%; 2/3).
Mutation burden correlated with growth: those lesions with ≥2 mutations had higher growth rates compared to lesions with only 0–1 mutations (difference 99% growth (95% CI 9–189), independent t-test, p = .032, Fig. 5B). APC mutations were present in all growth categories. Six of the 19 (32%) samples with APC mutations also carried one or more mutations in the PI3K, RAS-MAPK, or p53 pathways. Such combinations were not found in lesions that had regressed.
3.4. CIMP analysis
CIMP status was determined on all 27 samples in which DNA was still available (Fig. 4). These were 25 adenomas and two serrated polyps (1 HP and 1 SSL). Eight of 25 adenomas (32%) were CIMP positive (Fig. 2B). Both of the serrated polyps were CIMP positive and located in the distal colon. In the small number of polyps on which CIMP analysis was performed, no statistically significant difference could be observed.
3.5. MMR-status analysis
In all 58 samples (51 adenomas and 7 serrated polyps) that were assessed by IHC (Fig. 4), expression of MLH1, MSH2, MSH6 and PMS2 proteins was present (Supplementary fig. 1), hence these were all classified as MMR proficient (Fig. 2B).
4. Discussion
In the present study, we sought to evaluate whether growing colorectal polyps showed distinct molecular features associated with progression, compared to lesions that regressed or remained stable over time. To this aim, we performed detailed molecular analyses in a set of 65 small (6–9 mm) colorectal polyps that were removed three years after initial identification by CTC.
Higher growth rates were related to presence of non-random DNA copy number alterations associated with colorectal adenoma to carcinoma progression (cancer associated events or CAEs), as well as to increased mutation burden. Regressed lesions did not show CAEs, but did have mutations in genes involved in common CRC pathways, which concerned mostly APC mutations. Because APC inactivation is such an early event in adenoma genesis, it may not prevent lesions from regressing. The only mutation besides APC that was present in one of the regressed lesions was in the SMO gene, a rare mutation occurring in 0.9% of CRCs [28]. The SMO protein is a component of the hedgehog signalling pathway, which has been shown to negatively regulate WNT signalling [29]. In gastric cancers, however, SMO mutations were found not to be associated with altered expression of hedgehog target genes, indicating that these are probably passenger mutations [30].
Our observation that multiple mutations are already present in a substantial proportion of small colorectal polyps is in line with previous observations that detectable mutations occurred at an early stage of polyp development, at a mean size of only 30 ± 35 crypts [31,32]. For perspective; lesions of 10mm3 contain approximately 3 × 105 crypts. Many of the somatic mutations detected in tumours may occur even before morphologically recognisable tumour formation [33,34]. Recent data show that in ~1% of morphologically normal colorectal crypts driver mutations were already present [35]. In approximately one third of the adenomas (6/19; 32%) with APC mutations, also mutations in the PI3K, RAS-MAPK, or p53 pathways occurred. However, only two of these adenomas with additional mutations in the PI3K, RAS-MAPK, or p53 pathways were at high risk of progression based on the criterion of having ≥2 CAEs [7]. Accumulating evidence suggests that whereas mutations in driver genes are already present early in precancerous lesions, chromosomal instability is a late phenomenon during adenoma to carcinoma progression [36]. Therefore, copy number alterations likely play a more critical role in malignant transformation. This is functionally supported by the observation that engineered patient-derived, adenoma organoids with critical driver mutations, only obtained metastatic capacity when CIN was present [37]. The results from our study confirm that mutations, present in regressed, stable and grown lesions, do not reflect polyp risk of progression, whereas CAEs, only present in stable and grown lesions, may likely do so.
Identifying adenomas with a high progression risk is of value for clinical practice. At the current moment, adenomas ≥10 mm, with villous component or high-grade dysplasia are considered high risk - or advanced - adenomas. Yet, this definition gives a suboptimal estimation of the true risk of progression and has been introduced and adopted in literature without much evidence [38]. The presence of more ≥2 CAEs more precisely reflects the natural course of the disease and more specifically identifies adenomas at high risk of progressing to cancer [7]. Previous research has shown that only 25% of advanced adenomas and 3% of non-advanced adenomas presented with ≥2 CAEs [39], suggesting that the majority of advanced adenomas should not be considered high risk, whereas a small proportion of non-advanced adenomas should. The problem associated with the definition of advanced adenoma, is also reflected by studies that investigated the relationship between polyp growth and the risk of progression, taking advanced adenomas as the endpoint [2,11]. In the original description of our cohort [2], the rates of advanced adenomas were 47%, 21% and zero in progressed, stable and regressed lesions, respectively. As 6–9 mm polyps that grow will easily qualify as advanced adenoma, when reaching 10 mm or more, we hypothesised that these rates were an overestimation of the actual risk of progression. Indeed, when focusing on molecular features rather than phenotypical features to define high-risk adenomas, only 13% of the adenomas that had grown, 11% of the stable adenomas and none of the regressed adenomas had ≥2 CAEs. Although numbers are small, these last rates appear to be more consistent with the actual progression risk, as it is estimated that approximately 5% of adenomas eventually progress to cancer [1].
The application of CAEs to identify high risk adenomas could be used in the development of novel diagnostic screening tests. After all, screening programs ideally should aim to detect precursor lesions just before they transform to colorectal cancer, in order to reduce both cancer incidence and mortality. In addition, molecularly-defined high risk adenomas could impact surveillance. According to the current post-polypectomy colonoscopy surveillance guideline, the presence of advanced adenomas shortens the surveillance interval [40]. In our study a total of 18 patients had at least one advanced adenoma based on phenotypical features at follow-up colonoscopy, compared to five patients based on molecular features (of which only three overlapping between the groups), which suggests overdiagnosis is happening with the current strategy. With the technical advancements over the recent years low cost and fast methods for copy number profiling have become available [41]. This makes the use of CAEs for risk stratified surveillance a realistic approach, although first further research is needed to assess the correlation between the presence of molecularly-defined high risk adenomas and the risk of metachronous lesions.
The use of CAEs as progression biomarker only applies to adenomas and not to serrated polyps. In the present cohort, CAEs were absent in serrated polyps irrespective of their growth category. This is not surprising, since progression of this lesion type is associated with the acquisition of MSI instead of CIN and therefore different molecular events characterise high risk serrated polyps. We found that in serrated polyps, BRAF mutations and CIMP positivity were present, but MLH1 was not yet affected. Because no dysplasia was present in any of the serrated polyps studied, this is in line with previous studies showing that MLH1 deficiency coincides with dysplasia in a serrated polyp [42].
For ethical reasons, longitudinal studies leaving polyps in situ for some years can only be done in patients with small polyps, as these are considered low risk. As most of these polyps are still small when removed after follow-up, a practical consequence is that from some polyps only limited amounts of tumour DNA can be obtained for molecular analyses after standard diagnostic procedures. As a result, only a subset (42%) of samples could undergo the entire range of molecular assays. In addition, no paired normal tissue was available, therefore making the analysis dependent on public databases to filter out polymorphisms. Inherent to the fact that histopathology of these small polyps can only be determined at one point in time, being the time of resection of the polyps, evaluating the morphological evolution in relation to the biological evolution of the polyps is not feasible. Despite these limitations, the present study uniquely provides a comprehensive overview of DNA copy number, mutation, CIMP and MSI profiling status of a relatively large series of polyps that were followed longitudinally.
In conclusion, molecular alterations associated with colorectal adenoma to carcinoma progression were related to growth over time, but were absent in regressed lesions. So far, these molecular alterations have been mostly identified by cross-sectional observations in tissue samples from colorectal adenomas and cancers. The present longitudinal study provides in vivo support in the human setting for the functional role of these molecular alterations in this process.
Acknowledgments
Acknowledgements
We thank the Genomic Core Facility at The Netherlands Cancer Institute for their sequencing service. We thank the Core Facility – Molecular Pathology and Biobank for facilitating immunohistochemical staining.
Funding source
MCJvL receives funding from Alpe d'Huzes, Dutch Cancer Society (project number NKI2013-6338). The funders had no role in study design, data collection, analysis and interpretation, decision to publish, or writing of the report. GAM, ED, BC and MCJvL had full access to all the data in the study and GAM had final responsibility to submit for publication.
Declaration of interests
JS is research consultant Robarts Clinical Trials. ED has endoscopic equipment on loan of Olympus and FujiFilm, receives a research grant from FujiFilm, and recieves a honorarium for consultancy from FujiFilm, Tillots and Olympus. GAM has research collaborations with Exact Sciences and Sysmex for other studies regarding early detection of colorectal cancer. The companies provide materials, equipment or (sample) analyses.
Author contributions
ED and GAM designed the present study. ED, EJK and JS designed and conducted the original COCOS trial. MCJvL and BC analysed and interpreted data. MCJvL and MvE did wet lab assays. CR performed bioinformatics analyses. CJTN and JS performed imaging measurements. MCJvL and CJTN were involved in data collection and building the database. GAM and PS were involved with pathology review. MCJvL and CR constructed the figures. MCJvL, BC, ED and GAM wrote the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.12.009.
Appendix A. Supplementary data
Supplementary material
References
- 1.Shinya H., Wolff W.I. Morphology, anatomic distribution and cancer potential of colonic polyps. Ann. Surg. 1979;190:679–683. doi: 10.1097/00000658-197912000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tutein Nolthenius C.J., Boellaard T.N., de Haan M.C. Evolution of screen-detected small (6-9 mm) polyps after a 3-year surveillance interval: assessment of growth with CT colonography compared with Histopathology. Am. J. Gastroenterol. 2015;110:1682–1690. doi: 10.1038/ajg.2015.340. [DOI] [PubMed] [Google Scholar]
- 3.Li X., Galipeau P.C., Paulson T.G. Temporal and spatial evolution of somatic chromosomal alterations: a case-cohort study of Barrett's esophagus. Cancer Prev. Res. (Phila.) 2014;7:114–127. doi: 10.1158/1940-6207.CAPR-13-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vleugels J.L.A., Hazewinkel Y., Fockens P., Dekker E. Natural history of diminutive and small colorectal polyps: a systematic literature review. Gastrointest. Endosc. 2017;85:1169–1176. doi: 10.1016/j.gie.2016.12.014. [e1] [DOI] [PubMed] [Google Scholar]
- 5.Fearon E.R., Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 6.Lengauer C., Kinzler K.W., Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 7.Hermsen M., Postma C., Baak J. Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology. 2002;123:1109–1119. doi: 10.1053/gast.2002.36051. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho B., Postma C., Mongera S. Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut. 2009;58:79–89. doi: 10.1136/gut.2007.143065. [DOI] [PubMed] [Google Scholar]
- 9.Sillars-Hardebol A.H., Carvalho B., Tijssen M. TPX2 and AURKA promote 20q amplicon-driven colorectal adenoma to carcinoma progression. Gut. 2012;61:1568–1575. doi: 10.1136/gutjnl-2011-301153. [DOI] [PubMed] [Google Scholar]
- 10.O'Brien M.J., Yang S., Mack C. Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am. J. Surg. Pathol. 2006;30:1491–1501. doi: 10.1097/01.pas.0000213313.36306.85. [DOI] [PubMed] [Google Scholar]
- 11.Pickhardt P.J., Kim D.H., Pooler B.D. Assessment of volumetric growth rates of small colorectal polyps with CT colonography: a longitudinal study of natural history. Lancet Oncol. 2013;14:711–720. doi: 10.1016/S1470-2045(13)70216-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoop E.M., de Haan M.C., de Wijkerslooth T.R. Participation and yield of colonoscopy versus non-cathartic CT colonography in population-based screening for colorectal cancer: a randomised controlled trial. Lancet Oncol. 2012;13:55–64. doi: 10.1016/S1470-2045(11)70283-2. [DOI] [PubMed] [Google Scholar]
- 13.Voorham QJM, Carvalho B, Spiertz AJ, et al. Chromosome 5q loss in Colorectal Flat Adenomas. Clin. Cancer Res. 18: 4560–9. [DOI] [PubMed]
- 14.Scheinin I., Sie D., Bengtsson H. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014;24:2022–2032. doi: 10.1101/gr.175141.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rausch C., Carvalho B., Fijneman R. QDNAseqFlow: a computational analysis workflow of DNA copy number aberrations from low-coverage whole genome sequencing reads. F1000Research. 2017;6 [Google Scholar]
- 16.Siefert J.C., Georgescu C., Wren J.D., Koren A., Sansam C.L. DNA replication timing during development anticipates transcriptional programs and parallels enhancer activation. Genome Res. 2017;27:1406–1416. doi: 10.1101/gr.218602.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van De Wiel M.A., Brosens R., Eilers P.H.C. vol. 25. 2009. Smoothing Waves in Array CGH Tumor Profiles; pp. 1099–1104. [DOI] [PubMed] [Google Scholar]
- 18.van Dijk E., Biesma H.D., Cordes M. Loss of Chromosome 18q11.2-q12.1 is Predictive for Survival in patients with Metastatic Colorectal Cancer Treated with Bevacizumab. J. Clin. Oncol. 2018;36:2052–2060. doi: 10.1200/JCO.2017.77.1782. [DOI] [PubMed] [Google Scholar]
- 19.Li H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. 2013. http://arxiv.org/abs/1303.3997 published online March 16. (accessed Feb 2, 2018)
- 20.Van der Auwera G.A., Carneiro M.O., Hartl C. Current Protocols in Bioinformatics. John Wiley & Sons, Inc; Hoboken, NJ, USA: 2013. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. 11.10.1-11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cingolani P., Platts A., Wang L.L. A program for annotating and predicting the effects of single nucleotide polymorphisms. SnpEff Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cingolani P., Patel V.M., Coon M. Using drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, snpSift. Front. Genet. 2012;3:35. doi: 10.3389/fgene.2012.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lek M., Karczewski K.J., Minikel E.V. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weisenberger D.J., Siegmund K.D., Campan M. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 25.Derks S., Lentjes M.H.F.M., Hellebrekers D.M.E.I., de Bruïne A.P., Herman J.G., van Engeland M. Methylation-specific PCR unraveled. Cell. Oncol. 2004;26:291–299. doi: 10.1155/2004/370301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini Y., Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
- 27.Lappalainen I., Almeida-King J., Kumanduri V. The European Genome-phenome Archive of human data consented for biomedical research. Nat. Genet. 2015;47:692–695. doi: 10.1038/ng.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jauhri M., Bhatnagar A., Gupta S., Shokeen Y., Minhas S., Aggarwal S. Targeted molecular profiling of rare genetic alterations in colorectal cancer using next-generation sequencing. Med. Oncol. 2016;33:106. doi: 10.1007/s12032-016-0820-2. [DOI] [PubMed] [Google Scholar]
- 29.Akiyoshi T., Nakamura M., Koga K. Gli1, downregulated in colorectal cancers, inhibits proliferation of colon cancer cells involving Wnt signalling activation. Gut. 2006;55:991–999. doi: 10.1136/gut.2005.080333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X.-D., Inzunza H., Chang H. Mutations in the hedgehog pathway genes SMO and PTCH1 in human gastric tumors. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sievers CK, Zou LS, Pickhardt PJ, et al. Subclonal diversity arises early even in small colorectal tumours and contributes to differential growth fates Gut. DOI: 10.1136/gutjnl-2016-312232. [DOI] [PMC free article] [PubMed]
- 32.Gausachs M., Borras E., Chang K. Mutational heterogeneity in APC and KRAS arises at the crypt level and leads to polyclonality in early colorectal tumorigenesis. Clin. Cancer Res. 2017;23:5936–5947. doi: 10.1158/1078-0432.CCR-17-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borras E., San Lucas F.A., Chang K. Genomic Landscape of Colorectal Mucosa and Adenomas. Cancer Prev. Res. (Phila.) 2016;9:417–427. doi: 10.1158/1940-6207.CAPR-16-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomasetti C., Vogelstein B., Parmigiani G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc. Natl. Acad. Sci. 2013;110:1999–2004. doi: 10.1073/pnas.1221068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee-Six H., Ellis P., Osborne R.J. The landscape of somatic mutation in normal colorectal epithelial cells. bioRxiv. 2018:416800. doi: 10.1038/s41586-019-1672-7. [DOI] [PubMed] [Google Scholar]
- 36.Saito T. A temporal shift of the evolutionary principle shaping intratumor heterogeneity in colorectal cancer. Nat. Commun. 2018;9:2884. doi: 10.1038/s41467-018-05226-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matano M., Date S., Shimokawa M. Modeling colorectal cancer using CRISPR-Cas9–mediated engineering of human intestinal organoids. Nat. Med. 2015;21:256. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 38.Winawer S.J., Zauber A.G., Ho M.N. Prevention of colorectal cancer by colonoscopic polypectomy. The National Polyp Study Workgroup. N. Engl. J. Med. 1993;329:1977–1981. doi: 10.1056/NEJM199312303292701. [DOI] [PubMed] [Google Scholar]
- 39.Carvalho B., Diosdado B., Terhaar Sive Droste J.S. Evaluation of Cancer-Associated DNA Copy Number events in Colorectal (Advanced) Adenomas. Cancer Prev. Res. 2018;11:403–412. doi: 10.1158/1940-6207.CAPR-17-0317. [DOI] [PubMed] [Google Scholar]
- 40.Hassan C., Quintero E., Dumonceau J.-M. Post-polypectomy colonoscopy surveillance: European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy. 2013;45:842–851. doi: 10.1055/s-0033-1344548. [DOI] [PubMed] [Google Scholar]
- 41.Abujudeh S, Zeki SS, Lanschot MC van, et al. Low-cost and clinically applicable copy number profiling using repeat DNA. bioRxiv 2018; : 394429. [DOI] [PMC free article] [PubMed]
- 42.Sheridan T.B., Fenton H., Lewin M.R. Sessile serrated adenomas with low- and high-grade dysplasia and early carcinomas. Am. J. Clin. Pathol. 2006;126:564–571. doi: 10.1309/C7JE8BVL8420V5VT. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material