Abstract
The tumor suppressor phosphatase and tensin homolog (PTEN) classically counteracts the PI3K/AKT/mTOR signaling cascade. Germline pathogenic PTEN mutations cause PTEN hamartoma tumor syndrome (PHTS), featuring various benign and malignant tumors, as well as neurodevelopmental disorders such as autism spectrum disorder. Germline and somatic mosaic mutations in genes encoding components of the PI3K/AKT/mTOR pathway downstream of PTEN predispose to syndromes with partially overlapping clinical features, termed the “PTEN-opathies.” Experimental models of PTEN pathway disruption uncover the molecular and cellular processes influencing clinical phenotypic manifestations. Such insights not only teach us about biological mechanisms in states of health and disease, but also enable more accurate gene-informed cancer risk assessment, medical management, and targeted therapeutics. Hence, the PTEN-opathies serve as a prototype for bedside to bench, and back to the bedside, practice of evidence-based precision medicine.
Introduction
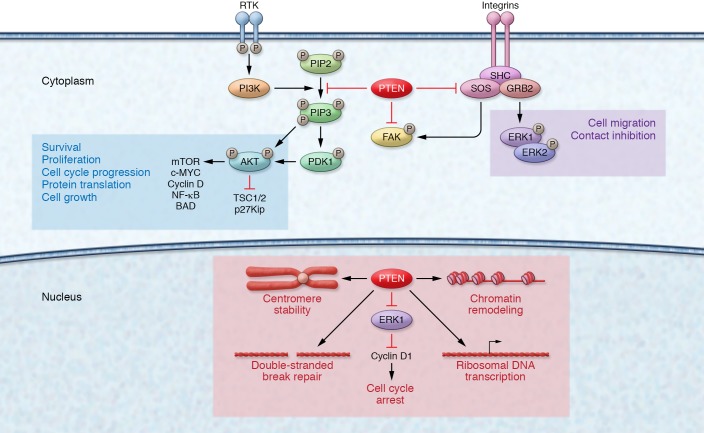
The tumor suppressor gene phosphatase and tensin homolog (PTEN; OMIM 601728) was originally recognized as being mutated somatically in multiple sporadic cancers (1, 2), as well as mutated in the germline of patients with Cowden syndrome (CS; OMIM 158350), a hereditary overgrowth and cancer predisposition disorder (3, 4). PTEN is a dual-specificity phosphatase at two levels. First, PTEN has been shown to dephosphorylate protein substrates on serine/threonine and tyrosine residues, thus acting as a dual-specificity protein phosphatase (5). One example is the tyrosine dephosphorylation of focal adhesion kinase (FAK) to inhibit cell spreading (6). Second, PTEN also dephosphorylates phosphatidylinositol 3,4,5-trisphosphate (PIP3) to phosphatidylinositol 4,5-bisphosphate (PIP2) — hence, PTEN is also a dual-specificity phosphatase in the sense that it dephosphorylates lipid substrates in addition to protein substrates (7). As a lipid phosphatase, PTEN canonically negatively regulates the phosphatidylinositol 3-kinase (PI3K) signaling cascade, thereby dampening downstream protein kinase B (PKB/AKT) signaling (7, 8). Left unchecked, such as through PTEN mutation or inactivation, elevated PIP3 levels cause constitutive activation of AKT with subsequent downstream cascades resulting in, e.g., upregulation of mammalian target of rapamycin (mTOR) signaling (9). This ultimately leads to cell survival, growth, proliferation, and decreased apoptosis (10–12). Notably, AKT represents only one of many PIP3-binding proteins regulated by the PI3K/PTEN axis (13, 14). Although originally believed to be an exclusively cytoplasmic phosphatase, PTEN is now known to also function within the nucleus, contributing to cell cycle regulation, DNA double-strand break repair, genomic stability, and chromatin remodeling (15–20). Therefore, although PTEN exerts much of its function as a lipid phosphatase counteracting the PI3K/AKT/mTOR signaling pathway, PTEN also exerts protein phosphatase–dependent and pan-phosphatase-independent activities within both the cytoplasm and the nucleus (ref. 21 and Figure 1).
Figure 1. Cytoplasmic and nuclear PTEN signaling.
In the cytoplasm, PTEN canonically functions to regulate the PI3K/AKT/mTOR signaling pathway. Under growth factor stimulation, PI3K is activated and catalyzes the phosphorylation of PIP2 to PIP3. PIP3 recruits PDK1 to the plasma membrane, which then contributes to the activation of AKT. AKT regulates a myriad of downstream cellular processes such as cell growth, proliferation, and decreased apoptosis. The lipid phosphatase activity of PTEN counteracts PI3K by dephosphorylating PIP3 to PIP2, thereby dampening AKT activation. In the nucleus, PTEN plays a vital role in maintaining genomic stability, chromosomal architecture, cell cycle control, and the regulation of ribosome biogenesis within nucleoli.
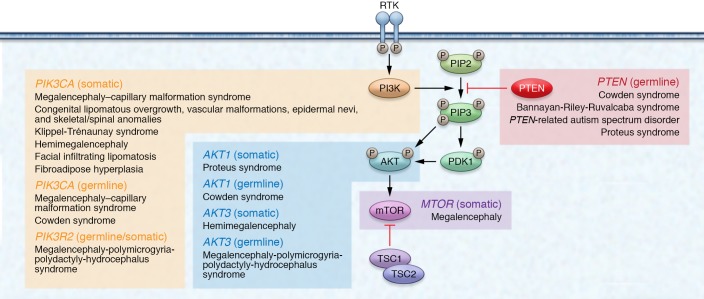
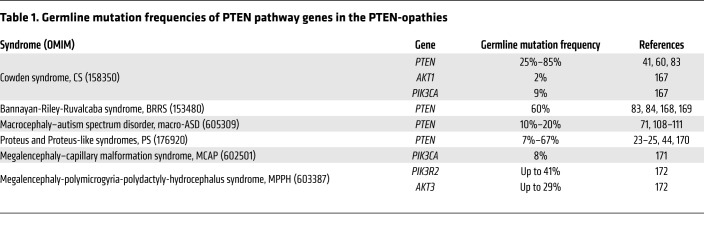
Germline PTEN mutations have been identified in patients with different clinical syndromes, and that subset is termed PTEN hamartoma tumor syndrome (PHTS) (22). Besides PTEN mutation–positive CS, PHTS also encompasses individuals with Bannayan-Riley-Ruvalcaba syndrome (BRRS), Proteus syndrome (PS), and Proteus-like syndrome who have PTEN mutations (22–26). BRRS (OMIM 153480) is a rare congenital disorder classically characterized by macrocephaly in combination with intestinal hamartomatous polyposis, vascular malformations, lipomas, and genital freckling (27, 28). PS (OMIM 176920) is a rare, complex, and highly variable disorder characterized by progressive, postnatal overgrowth of multiple tissues derived from different cell lineages (29). Relatedly, germline and somatic mosaic mutations in other genes encoding components of the PI3K/AKT/mTOR signaling pathway downstream of PTEN predispose patients to partially overlapping sets of clinical manifestations reminiscent of PHTS. These overgrowth syndromes are known as the PTEN-opathies (ref. 30 and Figure 2). A subset of individuals with the PTEN-opathies harbor germline mutations in components of the PTEN signaling cascade (Table 1), predisposing these individuals to overgrowth and/or cancer in different organs. Postzygotic somatic mosaic mutations in PTEN pathway genes cause overgrowth disorders restricted to the tissues where the mutations occurred. One example is PS, in which a somatic mosaic activating AKT1 mutation (p.Glu17Lys) has been identified in more than 90% of individuals meeting clinical diagnostic criteria (31). Finally, somatic mutations in components of the PTEN signaling cascade occurring in postnatal somatic tissue can drive a vast array of sporadic cancers (32–35).
Figure 2. The classic PTEN pathway and associated PTEN-opathies.
The PTEN-opathies encompass a spectrum of disorders with mutations within genes encoding proteins belonging to the PTEN pathway. PIK3CA-related overgrowth spectrum (PROS) includes distinct clinical entities with phenotypic overlap among the different syndromes. These overgrowth disorders are typically associated with postzygotic somatic mosaic PIK3CA mutations in affected tissues and are characterized by segmental overgrowth affecting the body (e.g., CLOVES syndrome, fibroadipose hyperplasia) or the brain (e.g., megalencephaly–capillary malformation syndrome [MCAP], hemimegalencephaly). PIK3CA encodes the catalytic p110α subunit protein of PI3K. Similarly to PTEN dysfunction, PIK3CA activation results in phosphorylation and activation of AKT, ultimately resulting in overgrowth-promoting downstream effects within the PI3K/AKT/mTOR signaling pathway downstream of PTEN. Expectedly, these syndromes show clinical phenotypic overlap with PHTS, including megalencephaly, vascular malformations, overgrowth, and neurocognitive deficits.
Table 1. Germline mutation frequencies of PTEN pathway genes in the PTEN-opathies.
Overgrowth syndromes are important to diagnose, not only for timely disease management, but also because several of these conditions are associated with elevated risks of cancer. Here, we utilize the PTEN-opathies, particularly PHTS, as a model to examine how perturbation of the PTEN signaling pathway leads to a spectrum of heterogeneous clinical phenotypes. We discuss the genetic, functional, and mechanistic insights that put forth why some organs overgrow but never turn malignant while others develop malignancies. Importantly, the elucidation of the underlying mechanisms is of clinical importance since it promotes the implementation of evidence-based medical management and preventative and therapeutic approaches.
PTEN dysfunction and cancer
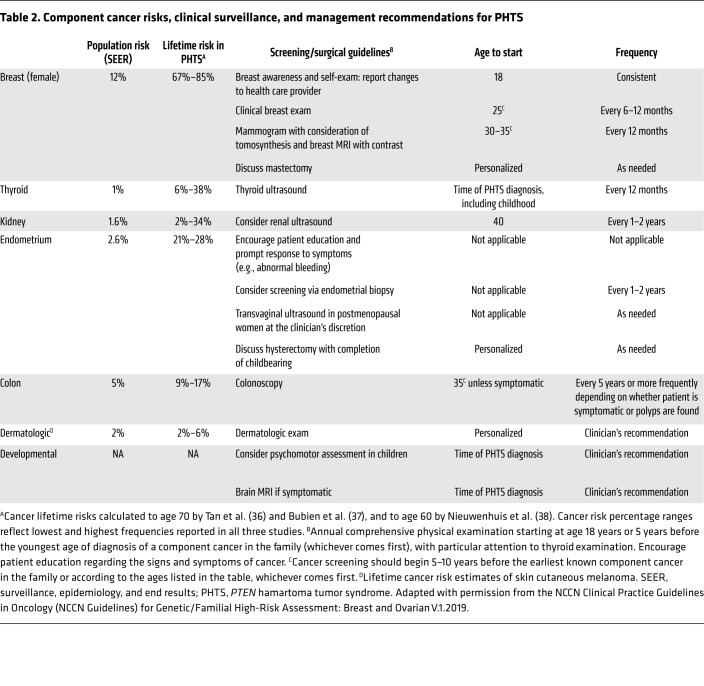
The identification of germline PTEN mutations allowed for the comprehensive elucidation of component cancers and associated lifetime risks (36). Three independent studies revealed elevated risks for breast, thyroid, endometrial, kidney, and colon cancers and melanoma in PHTS (36–38). Similarly to other hereditary cancer syndromes, the risk for bilateral and multifocal cancer is elevated (22). Relatedly, individuals with PHTS have a 7-fold increased risk of developing second malignant primary neoplasms (39). Collectively, these cancer risk assessment studies inform clinical surveillance recommendations and medical management of individuals with germline PTEN mutations (36), with the aim of detecting malignancies at the earliest, most manageable stages (Table 2).
Table 2. Component cancer risks, clinical surveillance, and management recommendations for PHTS.
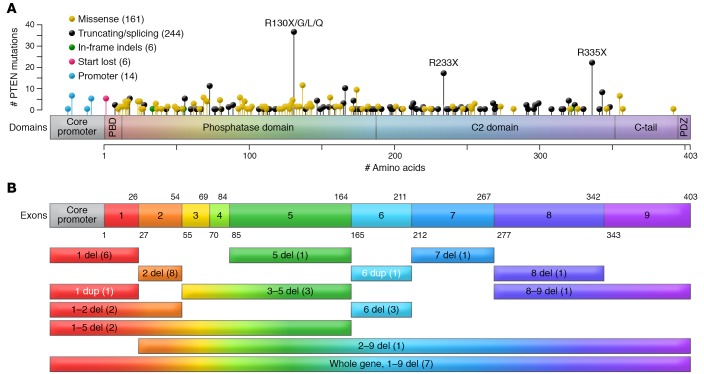
PTEN comprises nine exons canonically encoding a 403–amino acid protein (1, 40). Broadly, PTEN mutations could impact the abundance of PTEN protein, resulting in haploinsufficiency; result in reduced or lost phosphatase activity; act in a dominant-negative manner; and/or result in aberrant localization and function (21). The germline mutation spectrum in PHTS is broad, with mutations affecting all nine exons of PTEN (refs. 36, 39, 41, 42, and Figure 3). Approximately two-thirds of germline PTEN mutations occur in exons 5, 7, and 8 (41). Interestingly, up to 40% of all germline PTEN mutations are located in exon 5, encoding the core catalytic motif, although this exon represents only 20% of the coding sequence (41, 43, 44). Relatedly, two distinct Alu elements have been reported in two unrelated CS patients with identical break points within exon 5, suggesting that this exon is a possible retrotransposition hotspot (45). Mutations within the core catalytic motif typically abrogate pan-phosphatase (lipid and protein) activity, such as mutations affecting p.Cys124, but rarely, mutations such as p.Gly129Glu result in abrogation of lipid phosphatase activity only (4, 46, 47). Interestingly, several PTEN mutations retain partial or even complete catalytic activity (48), suggesting alternative mechanisms for compromised PTEN function. For example, catalytically active mutant PTEN p.Lys289Glu is characterized by a nuclear import defect due to loss of monoubiquitination at p.Lys289 (49). Nuclear PTEN is thought to be protected from polyubiquitination and subsequent proteasome-mediated degradation in the cytoplasm; therefore, it is able to dampen AKT signaling and induce p53-independent apoptosis (49). In support of these observations, nuclear exclusion of PTEN has been associated with more aggressive, advanced-stage cancers (50–54). Relatedly, the N-terminal phosphatase domain contains two ATP-binding motifs, critical for regulating PTEN exit from the nucleus (55). Expectedly, ATP-binding motif mutants (e.g., p.Lys62Arg, p.Tyr65Cys, p.Lys125Glu) do not bind ATP efficiently, resulting in nuclear PTEN mislocalization. This subsequently leads to increased cellular proliferation, reduced/abrogated apoptosis, and increased anchorage-independent growth (56, 57). PTEN has also been shown to be SUMOylated at Lys266 within the C2 domain, which facilitates PTEN binding to the plasma membrane through electrostatic interactions and subsequent suppression of PI3K/AKT signaling, both in vitro and in vivo (58). Additionally, germline PTEN mutations have been observed at Lys254 (Figure 3), a residue that is also SUMOylated to enhance PTEN nuclear import to then function in DNA repair (59). Therefore, mutations at Lys254 result in nuclear exclusion of PTEN and compromised DNA repair mechanisms.
Figure 3. PTEN structure and germline mutation spectrum in PHTS.
(A) PTEN germline mutation spectrum from 431 PHTS patients. PTEN is canonically a 403–amino acid protein. Different types of mutations are depicted in the lollipop plot overlaying the PTEN protein structure. The frequency of mutations correlates with the heights of the vertical lines representing each lollipop. PTEN comprises a PIP2-binding domain (PBD), a phosphatase domain, a C2 domain, and a C-terminal tail including a PDZ-binding domain. The active site is included within amino acid residues 123 and 130. (B) PTEN consists of 9 exons that encode the 403–amino acid protein. The exons are overlaid to match the protein domains in A. Intronic regions are not represented. The colored bars represent large deletions (abbreviated as del) and duplications (abbreviated as dup) annotated by affected exon numbers and the number of affected patients in parentheses. Figure adapted with permission from ref. 174.
Aside from intragenic mutations, approximately 10% of CS patients harbor germline PTEN promoter mutations (60). Pathogenic promoter mutations result in decreased PTEN transcription and translation, the latter due to altered mRNA secondary structure (60, 61). More recently, some unsuspected PTEN intronic variants were shown to result in pathogenic exon skipping, alternative splicing, or the use of cryptic splice sites (62). These splicing changes correlate with significantly lower PTEN protein levels and elevated p-AKT in patients with splicing changes compared with those without aberrant splicing. Finally, large PTEN deletions occur in approximately 3% to 10% of PHTS patients and can be found over the entire coding sequence (41, 42, 60).
Interestingly, PTEN encodes at least two proteins by means of noncanonical translation initiation. The first identified isoform represents a longer PTEN protein, named PTEN-Long (PTEN-L, also known as PTENα), that contains 173 additional amino acids at the amino-terminus due to the usage of an alternative CUG translation initiation site upstream of the canonical AUG sequence (63). Additionally, PTEN-L can be secreted to enter other cells directly, and can be detected in human serum and plasma. PTEN-L has also been shown to interact with canonical PTEN to regulate mitochondrial function and energy production (64). More recently, another N-terminal extended PTEN isoform, named PTENβ, has been identified (65). PTENβ translation is initiated from an AUU codon upstream of the AUG initiation codon for canonical PTEN. This isoform specifically localizes in cell nucleoli, and regulates ribosomal DNA (rDNA) transcription and cellular proliferation. As these newly identified PTEN protein isoforms are characterized by distinct subcellular localizations and biological functions, further studies are warranted to better understand how these isoforms contribute to carcinogenesis. Importantly, since PTEN-L and PTENβ share the canonical PTEN sequence, mutations that impact canonical PTEN would be expected to impact these isoforms as well. However, mutations within the N-terminal extended regions of PTEN-L and PTENβ can have downstream effects independent of canonical PTEN. An intriguing hypothesis is the tissue-specific expression of various PTEN protein isoforms, which could, in turn, predispose PHTS individuals to different phenotypes in a genotype-dependent manner. Indeed, the complex interplay among the PTEN family proteins could partly explain why a wide spectrum of clinical phenotypes are observed in the PTEN-opathies, with implications for the precise clinical management of these disorders.
PTEN dysfunction in PHTS offers important biological insights in the context of common sporadic cancers. Indeed, PTEN is one of the most frequently somatically mutated genes in cancer (66–68). The experimental data, in turn, offer insights regarding how germline PTEN mutations cause the clinical manifestations observed in PHTS. Cell survival, growth, apoptosis, migration, and genomic instability represent processes that influence cell fate and reflect overgrowth and cancer-related phenotypes. In time, it became evident that PTEN is also critical for normal development and physiology (11, 69). These findings help explain the occurrence of neurodevelopmental disorders such as megalencephaly, autism spectrum disorder (ASD), and developmental delay in individuals with PHTS (70–72). Importantly, germline PTEN mutations have been reported in previously undiagnosed individuals with isolated PHTS-related phenotypes, indicating that the syndrome is indeed underdiagnosed (71, 73–79). Certainly, utilizing knowledge about PHTS pathogenesis aids in establishing a molecular diagnosis, itself critical both for understanding the pathomechanisms behind and for subsequent medical management of the observed phenotypes.
Finally, because it is technically challenging to functionally interrogate all germline and somatic PTEN mutations, research efforts have focused on devising high-throughput methods to evaluate pathogenicity. Surprisingly, several residues within the catalytic pocket are shown to be tolerant to mutations, with solvent exposure playing a critical role in dictating tolerance (80). Moreover, several uncharacterized PTEN variants result in decreased PTEN thermodynamic stability and abundance, thus expanding the list of potentially functional variants (81). Collectively, such efforts foster evidence-based, functionally relevant classification of PTEN mutations into more clinically actionable categories. Predictably, meta-analysis of outputs coupled with clinical phenotypic correlations will likely yield more robust classifications. Such analyses culminated in a recently completed effort through ClinGen’s PTEN Variant Curation Expert Panel (82).
Genotype-phenotype correlations and modifiers of cancer risks
As with other inherited cancer syndromes, while it is possible to risk-assess increased organ-specific cancer probabilities, it is still impossible to predict at an individual level who will go on to develop any particular component cancer during his or her lifetime. Hence, multiple studies have attempted to find predictive PTEN genotype-phenotype correlations. Earlier studies revealed an association between PTEN germline mutations and malignant breast disease (83, 84). Missense mutations and mutations within and 5′ of the phosphatase core motif appear to be associated with multiorgan manifestations, serving as a surrogate of disease severity (83). Other groups did not detect such genotype-phenotype correlations (85), likely because their sample size of studied PHTS patients is small (n = 13), compared with the 44 families and 43 probands of the preceding studies (83, 84). More recently, germline PTEN frameshift mutations have been found to be overrepresented, but not absolute, in thyroid cancer (86), nonsense mutations overrepresented in colorectal cancer (36), promoter mutations overrepresented in breast cancer (36), and missense mutations overrepresented in individuals with ASD (87). Interestingly, a theoretical computational approach revealed global 3-dimensional PTEN structural instability and inactive conformation in cancer-associated PTEN mutations, whereas ASD-associated PTEN mutations revealed localized destabilization contributing to partial opening of the active site (88). Such effects cannot be extrapolated from PTEN’s secondary structure alone and indeed provide an important dimension to consider for assessing PTEN genotype–PHTS phenotype associations. Additionally, studies have shown that ASD-associated mutations tend to retain higher PTEN activity relative to non–ASD-associated mutations (80, 89, 90). Whether assayed in vivo in yeast or in vitro in mammalian stable or primary cell lines, partial hypomorphic PTEN lipid phosphatase activity is retained in individuals with ASD, versus total loss of PTEN lipid phosphatase activity in individuals with more severe PHTS-related phenotypes (80, 89–91). Relatedly, it is predicted that PTEN mutations that result in the accumulation of stable inactive PTEN protein would lead to more severe PHTS-related developmental phenotypes and malignancies (90). Finally, imbalances in PTEN subcellular localization could impact PHTS phenotypic manifestations. A murine model of germline-mislocalized cytoplasm-predominant PTEN exhibits macrocephaly and a neurocognitive profile reminiscent of high-functioning ASD (92, 93). Intriguingly, germline-mislocalized nuclear-predominant PTEN can exist in patients with either cancer or ASD (57, 94). In this context, it is tempting to speculate whether mutant nuclear PTEN plays distinct roles in the affected tissues related to the latter disparate phenotypes.
The lack of absolute PTEN genotype-phenotype correlations suggests that additional factors act as phenotypic modifiers in PHTS. A proof-of-principle study showed that approximately 6% of PTEN mutation–positive CS/CS-like individuals also harbor germline variants in genes encoding three of the four subunits of mitochondrial complex II (SDHB, SDHC, SDHD), which were originally discovered as alternative susceptibility genes in PTEN wild-type CS/CS-like patients (95, 96). Individuals carrying SDHx variants show an increased risk of breast and thyroid cancers that surpasses the risks mediated by mutant PTEN alone (95). Interestingly, while individuals with SDHx variants alone show the highest prevalence of thyroid cancer, the coexistence of a PTEN mutation was associated with a 77% snapshot prevalence of breast cancer, as compared with 32% with PTEN mutations alone and 57% with SDHx variants alone. Although the prevalence of thyroid cancer was not significantly elevated in individuals with both PTEN mutations and SDHx variants, the histology was papillary for all tumors versus the notable enrichment in follicular thyroid tumors in individuals with only PTEN mutations. Mechanistically, SDHx variants result in ROS-mediated stabilization of HIF-1α, destabilization and decreased protein expression of p53 due to defective interaction with NQO1, and resistance to apoptosis (95). These data also reveal how mitochondrial dysfunction leads to tumorigenesis subsequent to elevated flavin adenine dinucleotide (FAD) and nicotinamide adenine dinucleotide (NAD+), the cofactor and product of NQO1 enzymatic catalysis, respectively. Subsequent studies showed that SDHD p.G12S and p.H50R variants directly lead to impaired PTEN subcellular localization and function through SRC-induced oxidation, accompanied by apoptosis resistance and induction of cellular migration (97). Importantly, the selective SRC inhibitor bosutinib could rescue these tumorigenic phenotypes only when wild-type PTEN was present. Similarly, SDHD p.G12S and p.H50R variants result in reduced autophagy in a PTEN-dependent manner (98). From a clinical perspective, these data provide mechanistic insights that could explain the increased prevalence of thyroid cancer in CS patients with SDHx variants alone compared with those with PTEN mutations alone, as well as the seemingly paradoxical decreased prevalence of thyroid cancer in the setting of coexisting PTEN mutations and SDHx variants.
A hypothesis-generating pilot study further identified microbiomic differences in fecal samples derived from PTEN mutation–positive patients with and without PHTS component cancers (99). Functional metagenomic analysis revealed enrichment of cancer-relevant biological processes such as folate biosynthesis, genetic information processing, and cell growth/death pathways in fecal samples from PHTS cancer patients compared with those without a cancer diagnosis. These data suggest that gut dysbiosis could also play a role as a cancer risk modifier in PHTS patients. Conceivably, with increased sample sizes and independent replication, we suspect that novel associations will be discovered and expanded beyond cancer, toward phenotypes such as ASD and non-neoplastic overgrowths. Collectively, this knowledge will be impactful for more tailored medical management of PHTS patients.
Germline predisposition — overgrowth versus cancer
The discovery of PTEN as the Cowden syndrome gene paved the way for understanding how its disruption contributes to disease etiology (1, 3, 4). Functional characterization further established PTEN as a bona fide tumor suppressor gene (Figure 1). Studies in Drosophila and mouse models have shown that PTEN and downstream PI3K/AKT/TOR signaling play a central role in regulating cell number and size. Hence, dysfunction of this pathway recapitulates the growth anomalies observed in the PTEN-related human diseases. Drosophila PTEN has been shown to regulate cell number and size when mutated, leading to hyperplastic overgrowth in fruit fly mutant tissue (100). Similarly to mammalian signaling pathways, Drosophila PTEN regulates growth by antagonizing DDP110 (the Drosophila homolog of PI3K), and by acting as a negative regulator of insulin receptor signaling (101–103). With this knowledge of the basic mechanistic principles, what remains elusive, however, is the ability to identify factors that regulate progression from overgrowth to malignancy in a defined set of organs.
The most obvious explanation for organ-specific cancer development could be that the expression of the cancer-related gene, here PTEN, could be limited to the tissues in which malignancies arise. However, PTEN is ubiquitously expressed in all three germ cell layers throughout development, supporting the occurrence of hamartomatous overgrowths and variable multisystem phenotypes in individuals with germline PTEN mutations (11, 69, 104, 105). Homozygous Pten-knockout mice die before birth, further supporting a critical role for PTEN in embryogenesis (11, 105–107). High-level PTEN expression has also been reported during human development in tissues known to be associated with PHTS (104). However, this does not corroborate the tendency of these organs to develop malignancies when PTEN malfunctions. For example, the strongest PTEN protein levels are observed throughout the central and peripheral nervous systems (104), even though brain cancer is not a PHTS component cancer. Nevertheless, neurodevelopmental phenotypes are observed in PHTS, including macrocephaly (about 94% of patients), ASD (108–111), and Lhermitte-Duclos disease (LDD), a pathognomonic hamartomatous overgrowth of the cerebellum (112). Immunohistochemical studies show decreased or absent PTEN expression accompanied by elevated p-AKT in the affected LDD dysplastic gangliocytoma cells (113). Interestingly, murine studies have found that even a subtle reduction in PTEN causes increased tumorigenesis in a tissue-specific manner (114). In humans, reduced PTEN protein dose in CS-derived lymphoblastoid cell lines tends to occur in conjunction with an underlying germline PTEN mutation and to correlate with increasing clinical phenotypic burden (41). Further investigation in CS/CS-like patients with thyroid cancer reveals that low PTEN protein levels from blood-derived lymphoblastoid cells can predict for the presence of a germline PTEN mutation (115). Importantly, low blood PTEN levels correlate with weak or absent PTEN staining in the affected PHTS-derived thyroid tissues. Hence, one possibility is that variable tissue-specific thresholds of PTEN protein dosage could potentially influence particular PHTS-related phenotypes.
Intriguingly, despite the fact that germline PTEN mutations result in component cancers within a restricted set of organs (36), PTEN somatic driver mutations are enriched in multiple sporadic cancer types that are not components of the PHTS spectrum, including prostate cancer, glioblastoma multiforme, and others (1). Moreover, identical germline PTEN mutations often result in apparently disparate phenotypes (e.g., cancer versus nonmalignant overgrowths), including in an intrafamilial manner (116). These observations suggest that additional factors act as overgrowth versus cancer phenotypic modifiers in PHTS. Indeed, while germline PTEN mutations predispose PHTS patients to cancer, it is the landscape of acquired somatic alterations that likely governs cancer initiation and progression. Hence, although the germline PTEN mutations affect all cells of PHTS patients, the tissue-restricted pattern of particular modifying factors could explain the nonrandom progression to malignancy in specific organs. Additionally, the type of germline PTEN mutation could also influence eventual cell fates. For example, germline PTEN mutations such as C-terminal deletions that result in genomic instability could prime tissues that are particularly sensitive to DNA damage for progression to malignancy (117). Finally, the immune system has been recognized as a major determinant of cancer development (118–120). PTEN loss promotes resistance to tumor immune cell infiltration through the production of inhibitory cytokines, hence resulting in immune escape (121). Interestingly, pregnant mice treated with low-dose lipopolysaccharide to induce maternal inflammation produce offspring with brain overgrowth (122). This phenotype is more pronounced in Pten-heterozygous mice compared with wild type, indicating evident crosstalk between genetic susceptibility and the inflamed microenvironment mediated through ROS signaling. Importantly, ROS cause oxidation and subsequent inactivation of PTEN, a mechanism observed in a subset of CS/CS-like patients (97, 123). Hence, the manifestation of a cancer phenotype does represent a complex interplay among predisposing factors, genetic and epigenetic confounders, tissue-specific signaling networks, oncogenic signaling pathways, and microenvironmental context (124).
Molecularly targeted therapeutics
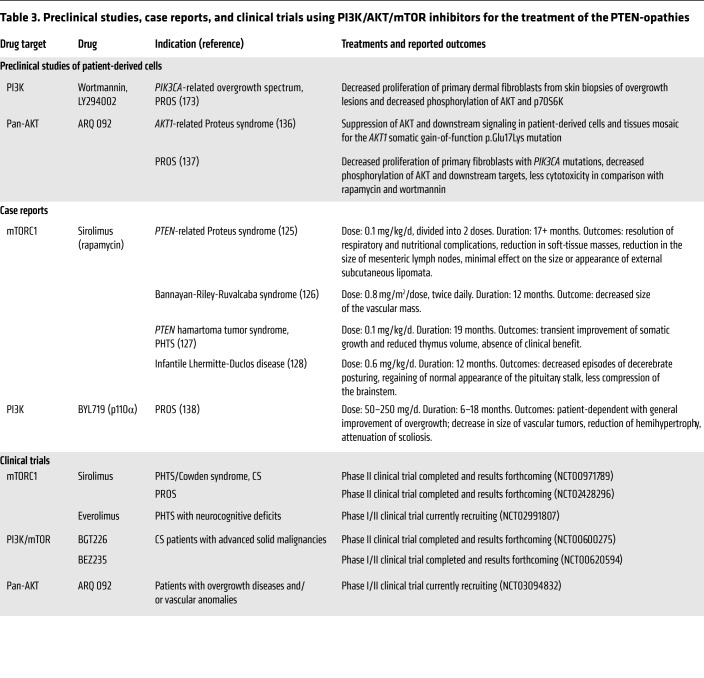
Altered PI3K/AKT/mTOR signaling in the PTEN-opathies implies that PI3K, AKT, and mTOR are germane targets for therapeutic intervention (Table 3). Proof-of-principle case reports demonstrate the use of the mTORC1 inhibitor sirolimus (rapamycin) to alleviate the symptoms and overgrowth manifestations of individuals with PHTS (125–128). Indeed, sirolimus has been used in a phase II open-label clinical trial in individuals with PHTS. Additionally, a double-blind drug-placebo, crossover trial with the mTORC1 inhibitor everolimus is currently accruing PHTS patients with ASD (22). Notably, mTORC1 inhibitors have been used in patients with tuberous sclerosis complex (TSC) (129–132) and Peutz-Jeghers syndrome (PJS) (133). TSC1/2 and STK11/LKB1, the susceptibility genes for TSC and PJS, respectively, are not only upstream of mTOR (9, 134) but are also downstream of PTEN signaling (135).
Table 3. Preclinical studies, case reports, and clinical trials using PI3K/AKT/mTOR inhibitors for the treatment of the PTEN-opathies.
In addition to mTORC1 inhibition, upstream components of the PTEN signaling pathway, such as PI3K and AKT, also serve as candidates for pharmacologic inhibition in the PTEN-opathies (Table 3). As such, AKT and PIK3CA inhibitors have been used in PS and PIK3CA-related overgrowth spectrum (PROS) disorders (136–138). PIK3CA encodes the p110α catalytic subunit protein of PI3K. Preclinical studies using the allosteric pan-AKT inhibitor ARQ 092 revealed suppression of AKT and downstream signaling in cells and tissues from PS patients, mosaic for the AKT1 somatic gain-of-function p.Glu17Lys mutation (136). ARQ 092 treatment of primary fibroblast cells from PROS patients also showed promising results; compared with sirolimus and the PI3K inhibitor wortmannin, ARQ 092 resulted in higher antiproliferative activity and lower cytotoxicity, at least in vitro (137). Relatedly, a recent proof-of-principle study demonstrated the successful usage of the PIK3CA inhibitor BYL719 (alpelisib) in a preclinical murine model of PROS and subsequently for the treatment of 19 patients with severe PROS disorders (138). Importantly, in these PTEN-opathies, all patients harbored somatic mutations; the ultimate goal from treatment is to continuously reduce progrowth signals in affected tissues with minimal toxicity toward normal wild-type cells. However, this becomes more challenging in the germline context, such as PHTS, where a high therapeutic index becomes even more critical since all cells harbor the underlying PTEN mutation. Moreover, constitutional PTEN pathway dysfunction would theoretically necessitate some type of chronic treatment regimen. However, lifelong mTOR and PIK3CA inhibition might not be feasible because of immunosuppressive effects, disruption of systemic glucose homeostasis, and the critical role the PTEN pathway plays in normal tissue and organ development (11, 69, 139, 140). Although isolated case reports and studies (125, 126, 128, 138) show promise for the therapeutic management of the PTEN-opathies, longitudinal studies are necessary to evaluate long-term safety and efficacy.
Another major caveat to molecular targeting of the PI3K/AKT/mTOR pathway is feedback activation of collateral oncogenic signaling pathways, causing resistance. This led to the investigation of combinatorial therapies that would, in theory, effectively target the growth-promoting signals without loss of feedback controls. Indeed, inhibiting mTORC1 has been shown to result in feedback activation of upstream signaling components such as AKT through insulin receptor substrate 1 (IRS1) or through direct phosphorylation at Ser473 by mTORC2 (141). However, experimental studies show promise in that the rebound upregulation of AKT during mTORC1 inhibition can be abrogated by pretreatment or cotreatment with resveratrol, at least in vitro (142). Moreover, PI3K inhibition can result in therapeutic resistance in PIK3CA-mutant cell lines due to a rebound insulin-dependent feedback mechanism (140), or failure to suppress CDK4/6 as evidenced through persistent RB phosphorylation (143). In these contexts, the combination of various PI3K inhibitors with antiglycemic therapies or CDK4/6 inhibitors, respectively, results in the attenuation of the progrowth feedback signaling cascades, hence overcoming resistance. Interestingly, NVP-BEZ235, a dual PI3K/mTOR inhibitor, has been shown to selectively inhibit the growth of a subset of androgen receptor–positive (AR+) breast cancer cell lines (144). AR is positively correlated with PTEN expression in breast cancer, owing to direct PTEN transcription that is mediated by an androgen response element in the PTEN promoter (145, 146). Mechanistically, the beneficial effect of AR activation in combination with PI3K/AKT/mTOR inhibition in AR+/ER+ breast cancers can be explained, at least partially, through PTEN upregulation and MYC suppression (144). Interestingly, the converse phenomenon has been extensively studied in prostate cancer and has been shown to be context-dependent (147–149). As such, PTEN-deficient prostate cancer cells have decreased AR transcription, and PI3K pathway inhibition activates AR signaling by alleviating the feedback inhibition on HER2/3 kinases (148). Therefore, the crosstalk between PTEN and AR signaling will likely be genotype- and context-dependent.
Although most therapeutic strategies are aimed at attenuating downstream oncogenic signaling consequent to PTEN dysfunction, strategies to enhance PTEN levels and/or activity represent promising therapeutic modalities. This is particularly pertinent for the cell-permeable PTEN-L (63) that would theoretically allow the restoration of PTEN levels in the context of PTEN haploinsufficiency. Moreover, PTEN expression and/or activity could also be enhanced through modulating negative and positive regulators of PTEN (e.g., transcription factors, miRNAs, protein ubiquitination machinery, etc.). Certainly, these approaches are context-dependent with respect to baseline endogenous PTEN levels and activity, tissue specificity, and the requisite of establishing long-term effects, among many other factors. Importantly, restoring wild-type PTEN in the context of a stable mutant PTEN protein could worsen the condition owing to dominant-negative effects (150). Another plausible approach is through gene editing of mutant PTEN alleles to restore or even enhance PTEN function (e.g., increased phosphatase activity or recruitment to the plasma membrane) (151). While gene editing poses many challenges, including off-target effects and activation of adaptive immune responses (152, 153), recent advances show promise in mitigating these outcomes (154–156). Undoubtedly, gene editing will be exceptionally challenging in the germline setting where the whole organism is targeted for editing.
Finally, given the broad spectrum and diverse functional consequences of germline PTEN mutations, targeting the PI3K/AKT/mTOR signaling pathway may not be effective in all contexts, e.g., when disease-associated PTEN mutations impact lipid phosphatase–independent functions. One possible approach could be to target the vulnerabilities caused by patient-specific germline PTEN mutations. For example, because PTEN plays a vital role in maintaining genomic integrity in the nucleus, it is possible to therapeutically use PARP inhibitors (157). Moreover, patients with germline PTEN mutations within the C-terminal region could benefit from proteasome inhibitors to mitigate PTEN degradation (158). Indeed, treatment with the proteasome inhibitor MG-132 can restore both nonsense and missense mutant PTEN protein levels in vitro (159). However, restoring missense mutant PTEN levels in the presence of the wild-type allele can also result in dominant-negative effects, and could worsen the condition (150). Predictably, in the era of precision medicine, cancer prevention is key and determining context-specific therapeutic indices will be vital for the effective management of the PTEN-opathies.
Immunotherapy
The PI3K/AKT/mTOR pathway is an important regulator of immunity (160, 161). Since PTEN is a master regulator of this pathway, it is therefore not surprising that PTEN disruption can result in immune dysregulation. The latter is germane in carcinogenesis, whereby immune surveillance, evasion of immune recognition, and a chronically inflamed microenvironment represent major immune hallmarks of cancer (118). Additionally, PI3K/AKT/mTOR pathway activation has been shown to modulate responses to immunotherapy. Loss of PTEN in the sporadic context has been associated with resistance to anti–PD-1 therapy in melanoma (121), a CS component cancer, and more recently in a case study of metastatic uterine leiomyosarcoma (162). Interestingly, activation of the PI3K/AKT/mTOR pathway has been shown to drive expression of PD-1/PD-L1 in a subset of solid tumors, causing immunoresistance (163–165). Indeed, because PTEN seems to be a major immunotherapeutic response predictor, multiple questions arise regarding the promising utility of immunotherapeutic agents in individuals with germline PTEN mutations and cancer. Studies have shown that a subset of individuals with PHTS have autoimmune phenotypes as well as B and T cell–related immune dysfunctions (161, 166). Importantly, reduction in peripheral lymphocyte numbers in comparison with control subjects, including decreased CD4+ cell numbers and hence absolute FOXP3+ Treg numbers, would suggest that these individuals will have a different response to immunotherapy compared with individuals with normal immune systems.
Perspective
The PTEN-opathies represent a paradigm whereby one pathway appears etiologic for a wide spectrum of clinically distinct phenotypes. The recognition and characterization of the PTEN-opathies allow for significant advances in understanding how clinical phenotypic manifestations result from underlying molecular and cellular processes to then guide risk assessment, therapeutics, and preventative strategies. Preclinical studies and clinical trials show promise for the treatment of a subset of the PTEN-opathies. However, this becomes more complex in the germline context, where a high therapeutic index is mandatory, yet exceptionally challenging. Indeed, individuals with germline PTEN mutations have a lifelong predisposition to PHTS-related signs and symptoms, necessitating prolonged treatments that could impact normal growth and development and cause nontargeted cytotoxicity. One of the most serious complications of the PTEN-opathies, particularly PHTS, is the increased lifetime risk for cancer. Although PTEN-enabled organ-specific cancer risk estimates and management guidelines are part of the routine clinical armamentarium of precision care, it remains virtually impossible to absolutely predict which individual (versus a probability) will develop which component malignancy. Nonmalignant component phenotypes of PHTS, such as ASD and severe vascular malformations, can be chronically debilitating and affect quality of life for patients and their families. Intriguingly, identical germline PTEN mutations are observed in patients with these seemingly disparate phenotypes (e.g., cancer versus ASD), indicating that additional factors may act as phenotypic modifiers in PHTS. Hence, future studies elucidating absolute modifiers of disease manifestations and associated signaling networks will be key to define more precise and effective preventative and therapeutic strategies for the individual at risk.
Acknowledgments
We are grateful to our patients and families who have participated in our research studies for over two decades. We are also grateful for our funding sources, including the National Cancer Institute (P01CA124570 and R01CA118989), American Cancer Society (RPG-02-151-01-CCE and Clinical Research Professorship), the National Institute of Neurological Disorders and Stroke (U54NS092090), Breast Cancer Research Foundation, Doris Duke Distinguished Clinical Scientist Award and the Zacconi Center of PTEN Research Excellence (all to CE). LY is an Ambrose Monell Foundation Cancer Genomic Medicine Fellow at the Cleveland Clinic Genomic Medicine Institute. CE is the Sondra J. and Stephen R. Hardis Chair of Cancer Genomic Medicine at the Cleveland Clinic and an American Cancer Society Clinical Research Professor.
Version 1. 01/07/2019
Electronic publication
Version 2. 02/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2019;129(2):452–464.https://doi.org/10.1172/JCI121277.
Contributor Information
Lamis Yehia, Email: yehial@ccf.org.
Joanne Ngeow, Email: jongeow@gmail.com.
Charis Eng, Email: engc@ccf.org.
References
- 1.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 2.Steck PA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15(4):356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 3.Nelen MR, et al. Localization of the gene for Cowden disease to chromosome 10q22-23. Nat Genet. 1996;13(1):114–116. doi: 10.1038/ng0596-114. [DOI] [PubMed] [Google Scholar]
- 4.Liaw D, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16(1):64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 5.Myers MP, et al. P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc Natl Acad Sci U S A. 1997;94(17):9052–9057. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada KM. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science. 1998;280(5369):1614–1617. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 7.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273(22):13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 8.Stambolic V, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95(1):29–39. doi: 10.1016/S0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 9.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4(9):648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 10.Furnari FB, Lin H, Huang HS, Cavenee WK. Growth suppression of glioma cells by PTEN requires a functional phosphatase catalytic domain. Proc Natl Acad Sci U S A. 1997;94(23):12479–12484. doi: 10.1073/pnas.94.23.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19(4):348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 12.Weng LP, et al. PTEN suppresses breast cancer cell growth by phosphatase activity-dependent G1 arrest followed by cell death. Cancer Res. 1999;59(22):5808–5814. [PubMed] [Google Scholar]
- 13.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296(5573):1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 14.Park WS, et al. Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol Cell. 2008;30(3):381–392. doi: 10.1016/j.molcel.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X, Senechal K, Neshat MS, Whang YE, Sawyers CL. The PTEN/MMAC1 tumor suppressor phosphatase functions as a negative regulator of the phosphoinositide 3-kinase/Akt pathway. Proc Natl Acad Sci U S A. 1998;95(26):15587–15591. doi: 10.1073/pnas.95.26.15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weng LP, Brown JL, Eng C. PTEN coordinates G(1) arrest by down-regulating cyclin D1 via its protein phosphatase activity and up-regulating p27 via its lipid phosphatase activity in a breast cancer model. Hum Mol Genet. 2001;10(6):599–604. doi: 10.1093/hmg/10.6.599. [DOI] [PubMed] [Google Scholar]
- 17.Weng LP, Smith WM, Brown JL, Eng C. PTEN inhibits insulin-stimulated MEK/MAPK activation and cell growth by blocking IRS-1 phosphorylation and IRS-1/Grb-2/Sos complex formation in a breast cancer model. Hum Mol Genet. 2001;10(6):605–616. doi: 10.1093/hmg/10.6.605. [DOI] [PubMed] [Google Scholar]
- 18.Shen WH, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128(1):157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 19.Baker SJ. PTEN enters the nuclear age. Cell. 2007;128(1):25–28. doi: 10.1016/j.cell.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 20.Chen ZH, et al. PTEN interacts with histone H1 and controls chromatin condensation. Cell Rep. 2014;8(6):2003–2014. doi: 10.1016/j.celrep.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YR, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat Rev Mol Cell Biol. 2018;19(9):547–562. doi: 10.1038/s41580-018-0015-0. [DOI] [PubMed] [Google Scholar]
- 22. Eng C. PTEN hamartoma tumor syndrome. In: Adam MP, et al., eds. GeneReviews. Seattle, Washington, USA: University of Washington; 2001; pp. 1993–2018. [Google Scholar]
- 23.Zhou X, et al. Association of germline mutation in the PTEN tumour suppressor gene and Proteus and Proteus-like syndromes. Lancet. 2001;358(9277):210–211. doi: 10.1016/S0140-6736(01)05412-5. [DOI] [PubMed] [Google Scholar]
- 24.Smith JM, et al. Germline mutation of the tumour suppressor PTEN in Proteus syndrome. J Med Genet. 2002;39(12):937–940. doi: 10.1136/jmg.39.12.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loffeld A, McLellan NJ, Cole T, Payne SJ, Fricker D, Moss C. Epidermal naevus in Proteus syndrome showing loss of heterozygosity for an inherited PTEN mutation. Br J Dermatol. 2006;154(6):1194–1198. doi: 10.1111/j.1365-2133.2006.07196.x. [DOI] [PubMed] [Google Scholar]
- 26.Kirk EP, Smith JM, Field M, Marshall GM, Marsh DJ. Diagnosis of Proteus syndrome was correct. Am J Med Genet A. 2004;130A(2):214–215; author reply 216. doi: 10.1002/ajmg.a.30337. [DOI] [PubMed] [Google Scholar]
- 27.Gorlin RJ, Cohen MM, Condon LM, Burke BA. Bannayan-Riley-Ruvalcaba syndrome. Am J Med Genet. 1992;44(3):307–314. doi: 10.1002/ajmg.1320440309. [DOI] [PubMed] [Google Scholar]
- 28.Ruvalcaba RH, Myhre S, Smith DW. Sotos syndrome with intestinal polyposis and pigmentary changes of the genitalia. Clin Genet. 1980;18(6):413–416. doi: 10.1111/j.1399-0004.1980.tb01785.x. [DOI] [PubMed] [Google Scholar]
- 29.Cohen MM, Hayden PW. A newly recognized hamartomatous syndrome. Birth Defects Orig Artic Ser. 1979;15(5B):291–296. [PubMed] [Google Scholar]
- 30.Yehia L, Eng C. 65 Years of the double helix: one gene, many endocrine and metabolic syndromes: PTEN-opathies and precision medicine. Endocr Relat Cancer. 2018;25(8):T121–T140. doi: 10.1530/ERC-18-0162. [DOI] [PubMed] [Google Scholar]
- 31.Lindhurst MJ, et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N Engl J Med. 2011;365(7):611–619. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27(41):5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Millis SZ, Ikeda S, Reddy S, Gatalica Z, Kurzrock R. Landscape of phosphatidylinositol-3-kinase pathway alterations across 19 784 diverse solid tumors. JAMA Oncol. 2016;2(12):1565–1573. doi: 10.1001/jamaoncol.2016.0891. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. A pan-cancer proteogenomic atlas of PI3K/AKT/mTOR pathway alterations. Cancer Cell. 2017;31(6):820–832.e3. doi: 10.1016/j.ccell.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan MH, Mester JL, Ngeow J, Rybicki LA, Orloff MS, Eng C. Lifetime cancer risks in individuals with germline PTEN mutations. Clin Cancer Res. 2012;18(2):400–407. doi: 10.1158/1078-0432.CCR-11-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bubien V, et al. High cumulative risks of cancer in patients with PTEN hamartoma tumour syndrome. J Med Genet. 2013;50(4):255–263. doi: 10.1136/jmedgenet-2012-101339. [DOI] [PubMed] [Google Scholar]
- 38.Nieuwenhuis MH, et al. Cancer risk and genotype-phenotype correlations in PTEN hamartoma tumor syndrome. Fam Cancer. 2014;13(1):57–63. doi: 10.1007/s10689-013-9674-3. [DOI] [PubMed] [Google Scholar]
- 39.Ngeow J, Stanuch K, Mester JL, Barnholtz-Sloan JS, Eng C. Second malignant neoplasms in patients with Cowden syndrome with underlying germline PTEN mutations. J Clin Oncol. 2014;32(17):1818–1824. doi: 10.1200/JCO.2013.53.6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li DM, Sun H. TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer Res. 1997;57(11):2124–2129. [PubMed] [Google Scholar]
- 41.Tan MH, et al. A clinical scoring system for selection of patients for PTEN mutation testing is proposed on the basis of a prospective study of 3042 probands. Am J Hum Genet. 2011;88(1):42–56. doi: 10.1016/j.ajhg.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mester J, Eng C. When overgrowth bumps into cancer: the PTEN-opathies. Am J Med Genet C Semin Med Genet. 2013;163C(2):114–121. doi: 10.1002/ajmg.c.31364. [DOI] [PubMed] [Google Scholar]
- 43.Waite KA, Eng C. Protean PTEN: form and function. Am J Hum Genet. 2002;70(4):829–844. doi: 10.1086/340026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eng C. PTEN: one gene, many syndromes. Hum Mutat. 2003;22(3):183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- 45.Crivelli L, et al. Insertion of Alu elements at a PTEN hotspot in Cowden syndrome. Eur J Hum Genet. 2017;25(9):1087–1091. doi: 10.1038/ejhg.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Myers MP, et al. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc Natl Acad Sci U S A. 1998;95(23):13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davidson L, et al. Suppression of cellular proliferation and invasion by the concerted lipid and protein phosphatase activities of PTEN. Oncogene. 2010;29(5):687–697. doi: 10.1038/onc.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han SY, et al. Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer Res. 2000;60(12):3147–3151. [PubMed] [Google Scholar]
- 49.Trotman LC, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128(1):141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tachibana M, et al. Expression and prognostic significance of PTEN product protein in patients with esophageal squamous cell carcinoma. Cancer. 2002;94(7):1955–1960. doi: 10.1002/cncr.0678. [DOI] [PubMed] [Google Scholar]
- 51.Whiteman DC, Zhou XP, Cummings MC, Pavey S, Hayward NK, Eng C. Nuclear PTEN expression and clinicopathologic features in a population-based series of primary cutaneous melanoma. Int J Cancer. 2002;99(1):63–67. doi: 10.1002/ijc.10294. [DOI] [PubMed] [Google Scholar]
- 52.Zhou XP, Gimm O, Hampel H, Niemann T, Walker MJ, Eng C. Epigenetic PTEN silencing in malignant melanomas without PTEN mutation. Am J Pathol. 2000;157(4):1123–1128. doi: 10.1016/S0002-9440(10)64627-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gimm O, et al. Differential nuclear and cytoplasmic expression of PTEN in normal thyroid tissue, and benign and malignant epithelial thyroid tumors. Am J Pathol. 2000;156(5):1693–1700. doi: 10.1016/S0002-9440(10)65040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou XP, et al. PTEN mutational spectra, expression levels, and subcellular localization in microsatellite stable and unstable colorectal cancers. Am J Pathol. 2002;161(2):439–447. doi: 10.1016/S0002-9440(10)64200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lobo GP, Waite KA, Planchon SM, Romigh T, Houghton JA, Eng C. ATP modulates PTEN subcellular localization in multiple cancer cell lines. Hum Mol Genet. 2008;17(18):2877–2885. doi: 10.1093/hmg/ddn185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lobo GP, Waite KA, Planchon SM, Romigh T, Nassif NT, Eng C. Germline and somatic cancer-associated mutations in the ATP-binding motifs of PTEN influence its subcellular localization and tumor suppressive function. Hum Mol Genet. 2009;18(15):2851–2862. doi: 10.1093/hmg/ddp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He X, Ni Y, Wang Y, Romigh T, Eng C. Naturally occurring germline and tumor-associated mutations within the ATP-binding motifs of PTEN lead to oxidative damage of DNA associated with decreased nuclear p53. Hum Mol Genet. 2011;20(1):80–89. doi: 10.1093/hmg/ddq434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang J, et al. SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nat Commun. 2012;3:911. doi: 10.1038/ncomms1919. [DOI] [PubMed] [Google Scholar]
- 59.Bassi C, et al. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science. 2013;341(6144):395–399. doi: 10.1126/science.1236188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou XP, et al. Germline PTEN promoter mutations and deletions in Cowden/Bannayan-Riley-Ruvalcaba syndrome result in aberrant PTEN protein and dysregulation of the phosphoinositol-3-kinase/Akt pathway. Am J Hum Genet. 2003;73(2):404–411. doi: 10.1086/377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teresi RE, Zbuk KM, Pezzolesi MG, Waite KA, Eng C. Cowden syndrome-affected patients with PTEN promoter mutations demonstrate abnormal protein translation. Am J Hum Genet. 2007;81(4):756–767. doi: 10.1086/521051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen HJ, Romigh T, Sesock K, Eng C. Characterization of cryptic splicing in germline PTEN intronic variants in Cowden syndrome. Hum Mutat. 2017;38(10):1372–1377. doi: 10.1002/humu.23288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hopkins BD, et al. A secreted PTEN phosphatase that enters cells to alter signaling and survival. Science. 2013;341(6144):399–402. doi: 10.1126/science.1234907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liang H, et al. PTENα, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab. 2014;19(5):836–848. doi: 10.1016/j.cmet.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liang H, et al. PTENβ is an alternatively translated isoform of PTEN that regulates rDNA transcription. Nat Commun. 2017;8:14771. doi: 10.1038/ncomms14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci U S A. 1999;96(8):4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simpson L, Parsons R. PTEN: life as a tumor suppressor. Exp Cell Res. 2001;264(1):29–41. doi: 10.1006/excr.2000.5130. [DOI] [PubMed] [Google Scholar]
- 68.Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer. 2011;11(4):289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Knobbe CB, Lapin V, Suzuki A, Mak TW. The roles of PTEN in development, physiology and tumorigenesis in mouse models: a tissue-by-tissue survey. Oncogene. 2008;27(41):5398–5415. doi: 10.1038/onc.2008.238. [DOI] [PubMed] [Google Scholar]
- 70.Goffin A, Hoefsloot LH, Bosgoed E, Swillen A, Fryns JP. PTEN mutation in a family with Cowden syndrome and autism. Am J Med Genet. 2001;105(6):521–524. doi: 10.1002/ajmg.1477. [DOI] [PubMed] [Google Scholar]
- 71.Butler MG, et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet. 2005;42(4):318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang L, et al. Brain development and Akt signaling: the crossroads of signaling pathway and neurodevelopmental diseases. J Mol Neurosci. 2017;61(3):379–384. doi: 10.1007/s12031-016-0872-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Won HS, et al. PTEN mutation identified in patient diagnosed with simultaneous multiple cancers. Cancer Res Treat. doi: 10.4143/crt.2017.579. doi: 10.4143/crt.2017.579. [published online ahead of print February 27, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heaney RM, Farrell M, Stokes M, Gorey T, Murray D. Cowden syndrome: serendipitous diagnosis in patients with significant breast disease. Case Series and Literature Review. Breast J. 2017;23(1):90–94. doi: 10.1111/tbj.12691. [DOI] [PubMed] [Google Scholar]
- 75.McBride KL, et al. Confirmation study of PTEN mutations among individuals with autism or developmental delays/mental retardation and macrocephaly. Autism Res. 2010;3(3):137–141. doi: 10.1002/aur.132. [DOI] [PubMed] [Google Scholar]
- 76.Hızarcıoğlu-Gülşen H, Kılıç E, Dominguez-Garrido E, Aydemir Y, Utine GE, Saltık-Temizel İN. Polyposis deserves a perfect physical examination for final diagnosis: Bannayan-Riley-Ruvalcaba syndrome. Turk J Pediatr. 2017;59(1):80–83. doi: 10.24953/turkjped.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 77.Caliskan A, Kohlmann WK, Affolter KE, Downs-Kelly E, Kanth P, Bronner MP. Intramucosal lipomas of the colon implicate Cowden syndrome. Mod Pathol. 2018;31(4):643–651. doi: 10.1038/modpathol.2017.161. [DOI] [PubMed] [Google Scholar]
- 78.Williams AJ, Doherty ES, Hart MH, Grider DJ. Diffuse gastric ganglioneuromatosis: novel presentation of PTEN hamartoma syndrome-case report and review of gastric ganglioneuromatous proliferations and a novel PTEN gene mutation. Case Rep Med. 2018;2018:4319818. doi: 10.1155/2018/4319818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kurek KC, et al. PTEN hamartoma of soft tissue: a distinctive lesion in PTEN syndromes. Am J Surg Pathol. 2012;36(5):671–687. doi: 10.1097/PAS.0b013e31824dd86c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mighell TL, Evans-Dutson S, O’Roak BJ. A saturation mutagenesis approach to understanding PTEN lipid phosphatase activity and genotype-phenotype relationships. Am J Hum Genet. 2018;102(5):943–955. doi: 10.1016/j.ajhg.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matreyek KA, et al. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nat Genet. 2018;50(6):874–882. doi: 10.1038/s41588-018-0122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mester JL, et al. Gene-specific criteria for PTEN variant curation: recommendations from the ClinGen PTEN Expert Panel. Hum Mutat. 2018;39(11):1581–1592. doi: 10.1002/humu.23636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marsh DJ, et al. Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum Mol Genet. 1998;7(3):507–515. doi: 10.1093/hmg/7.3.507. [DOI] [PubMed] [Google Scholar]
- 84.Marsh DJ, et al. PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum Mol Genet. 1999;8(8):1461–1472. doi: 10.1093/hmg/8.8.1461. [DOI] [PubMed] [Google Scholar]
- 85.Nelen MR, et al. Novel PTEN mutations in patients with Cowden disease: absence of clear genotype-phenotype correlations. Eur J Hum Genet. 1999;7(3):267–273. doi: 10.1038/sj.ejhg.5200289. [DOI] [PubMed] [Google Scholar]
- 86.Ngeow J, Mester J, Rybicki LA, Ni Y, Milas M, Eng C. Incidence and clinical characteristics of thyroid cancer in prospective series of individuals with Cowden and Cowden-like syndrome characterized by germline PTEN, SDH, or KLLN alterations. J Clin Endocrinol Metab. 2011;96(12):E2063–E2071. doi: 10.1210/jc.2011-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Frazier TW, Embacher R, Tilot AK, Koenig K, Mester J, Eng C. Molecular and phenotypic abnormalities in individuals with germline heterozygous PTEN mutations and autism. Mol Psychiatry. 2015;20(9):1132–1138. doi: 10.1038/mp.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smith IN, Thacker S, Jaini R, Eng C. Dynamics and structural stability effects of germline PTEN mutations associated with cancer versus autism phenotypes. J Biomol Struct Dyn. doi: 10.1080/07391102.2018.1465854. doi: 10.1080/07391102.2018.1465854. [published online ahead of print May 14, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Spinelli L, Black FM, Berg JN, Eickholt BJ, Leslie NR. Functionally distinct groups of inherited PTEN mutations in autism and tumour syndromes. J Med Genet. 2015;52(2):128–134. doi: 10.1136/jmedgenet-2014-102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leslie NR, Longy M. Inherited PTEN mutations and the prediction of phenotype. Semin Cell Dev Biol. 2016;52:30–38. doi: 10.1016/j.semcdb.2016.01.030. [DOI] [PubMed] [Google Scholar]
- 91.Rodríguez-Escudero I, Oliver MD, Andrés-Pons A, Molina M, Cid VJ, Pulido R. A comprehensive functional analysis of PTEN mutations: implications in tumor- and autism-related syndromes. Hum Mol Genet. 2011;20(21):4132–4142. doi: 10.1093/hmg/ddr337. [DOI] [PubMed] [Google Scholar]
- 92.Tilot AK, et al. Germline disruption of Pten localization causes enhanced sex-dependent social motivation and increased glial production. Hum Mol Genet. 2014;23(12):3212–3227. doi: 10.1093/hmg/ddu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tilot AK, et al. Neural transcriptome of constitutional Pten dysfunction in mice and its relevance to human idiopathic autism spectrum disorder. Mol Psychiatry. 2016;21(1):118–125. doi: 10.1038/mp.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mingo J, et al. A pathogenic role for germline PTEN variants which accumulate into the nucleus. Eur J Hum Genet. 2018;26(8):1180–1187. doi: 10.1038/s41431-018-0155-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ni Y, et al. Germline SDHx variants modify breast and thyroid cancer risks in Cowden and Cowden-like syndrome via FAD/NAD-dependant destabilization of p53. Hum Mol Genet. 2012;21(2):300–310. doi: 10.1093/hmg/ddr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ni Y, et al. Germline mutations and variants in the succinate dehydrogenase genes in Cowden and Cowden-like syndromes. Am J Hum Genet. 2008;83(2):261–268. doi: 10.1016/j.ajhg.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu W, He X, Ni Y, Ngeow J, Eng C. Cowden syndrome-associated germline SDHD variants alter PTEN nuclear translocation through SRC-induced PTEN oxidation. Hum Mol Genet. 2015;24(1):142–153. doi: 10.1093/hmg/ddu425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yu W, Ni Y, Saji M, Ringel MD, Jaini R, Eng C. Cowden syndrome-associated germline succinate dehydrogenase complex subunit D (SDHD) variants cause PTEN-mediated down-regulation of autophagy in thyroid cancer cells. Hum Mol Genet. 2017;26(7):1365–1375. doi: 10.1093/hmg/ddx037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Byrd V, Getz T, Padmanabhan R, Arora H, Eng C. The microbiome in PTEN hamartoma tumor syndrome. Endocr Relat Cancer. 2018;25(3):233–243. doi: 10.1530/ERC-17-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Goberdhan DC, Wilson C. PTEN: tumour suppressor, multifunctional growth regulator and more. Hum Mol Genet. 2003;12(suppl_2):R239–R248. doi: 10.1093/hmg/ddg288. [DOI] [PubMed] [Google Scholar]
- 101.Goberdhan DC, Paricio N, Goodman EC, Mlodzik M, Wilson C. Drosophila tumor suppressor PTEN controls cell size and number by antagonizing the Chico/PI3-kinase signaling pathway. Genes Dev. 1999;13(24):3244–3258. doi: 10.1101/gad.13.24.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang H, et al. PTEN affects cell size, cell proliferation and apoptosis during Drosophila eye development. Development. 1999;126(23):5365–5372. doi: 10.1242/dev.126.23.5365. [DOI] [PubMed] [Google Scholar]
- 103.Oldham S, Stocker H, Laffargue M, Wittwer F, Wymann M, Hafen E. The Drosophila insulin/IGF receptor controls growth and size by modulating PtdInsP(3) levels. Development. 2002;129(17):4103–4109. doi: 10.1242/dev.129.17.4103. [DOI] [PubMed] [Google Scholar]
- 104.Gimm O, Attié-Bitach T, Lees JA, Vekemans M, Eng C. Expression of the PTEN tumour suppressor protein during human development. Hum Mol Genet. 2000;9(11):1633–1639. doi: 10.1093/hmg/9.11.1633. [DOI] [PubMed] [Google Scholar]
- 105.Podsypanina K, et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci U S A. 1999;96(4):1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Suzuki A, et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr Biol. 1998;8(21):1169–1178. doi: 10.1016/S0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- 107.Yamada KM, Araki M. Tumor suppressor PTEN: modulator of cell signaling, growth, migration and apoptosis. J Cell Sci. 2001;114(pt 13):2375–2382. doi: 10.1242/jcs.114.13.2375. [DOI] [PubMed] [Google Scholar]
- 108.Herman GE, Butter E, Enrile B, Pastore M, Prior TW, Sommer A. Increasing knowledge of PTEN germline mutations: two additional patients with autism and macrocephaly. Am J Med Genet A. 2007;143A(6):589–593. doi: 10.1002/ajmg.a.31619. [DOI] [PubMed] [Google Scholar]
- 109.Herman GE, Henninger N, Ratliff-Schaub K, Pastore M, Fitzgerald S, McBride KL. Genetic testing in autism: how much is enough? Genet Med. 2007;9(5):268–274. doi: 10.1097/GIM.0b013e31804d683b. [DOI] [PubMed] [Google Scholar]
- 110.Orrico A, Galli L, Buoni S, Orsi A, Vonella G, Sorrentino V. Novel PTEN mutations in neurodevelopmental disorders and macrocephaly. Clin Genet. 2009;75(2):195–198. doi: 10.1111/j.1399-0004.2008.01074.x. [DOI] [PubMed] [Google Scholar]
- 111.Varga EA, Pastore M, Prior T, Herman GE, McBride KL. The prevalence of PTEN mutations in a clinical pediatric cohort with autism spectrum disorders, developmental delay, and macrocephaly. Genet Med. 2009;11(2):111–117. doi: 10.1097/GIM.0b013e31818fd762. [DOI] [PubMed] [Google Scholar]
- 112.Eng C. Will the real Cowden syndrome please stand up: revised diagnostic criteria. J Med Genet. 2000;37(11):828–830. doi: 10.1136/jmg.37.11.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhou XP, et al. Germline inactivation of PTEN and dysregulation of the phosphoinositol-3-kinase/Akt pathway cause human Lhermitte-Duclos disease in adults. Am J Hum Genet. 2003;73(5):1191–1198. doi: 10.1086/379382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Alimonti A, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010;42(5):454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ngeow J, et al. Utility of PTEN protein dosage in predicting for underlying germline PTEN mutations among patients presenting with thyroid cancer and Cowden-like phenotypes. J Clin Endocrinol Metab. 2012;97(12):E2320–E2327. doi: 10.1210/jc.2012-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Merks JH, et al. PTEN hamartoma tumour syndrome: variability of an entity. J Med Genet. 2003;40(10):e111. doi: 10.1136/jmg.40.10.e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sun Z, et al. PTEN C-terminal deletion causes genomic instability and tumor development. Cell Rep. 2014;6(5):844–854. doi: 10.1016/j.celrep.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cavallo F, De Giovanni C, Nanni P, Forni G, Lollini PL. 2011: the immune hallmarks of cancer. Cancer Immunol Immunother. 2011;60(3):319–326. doi: 10.1007/s00262-010-0968-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Michaud DS, Houseman EA, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. Understanding the role of the immune system in the development of cancer: new opportunities for population-based research. Cancer Epidemiol Biomarkers Prev. 2015;24(12):1811–1819. doi: 10.1158/1055-9965.EPI-15-0681. [DOI] [PubMed] [Google Scholar]
- 120.Singh SK, Dorak MT. Cancer immunoprevention and public health. Front Public Health. 2017;5:101. doi: 10.3389/fpubh.2017.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Peng W, et al. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2016;6(2):202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Le Belle JE, et al. Maternal inflammation contributes to brain overgrowth and autism-associated behaviors through altered redox signaling in stem and progenitor cells. Stem Cell Reports. 2014;3(5):725–734. doi: 10.1016/j.stemcr.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kwon J, et al. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc Natl Acad Sci U S A. 2004;101(47):16419–16424. doi: 10.1073/pnas.0407396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schneider G, Schmidt-Supprian M, Rad R, Saur D. Tissue-specific tumorigenesis: context matters. Nat Rev Cancer. 2017;17(4):239–253. doi: 10.1038/nrc.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marsh DJ, et al. Rapamycin treatment for a child with germline PTEN mutation. Nat Clin Pract Oncol. 2008;5(6):357–361. doi: 10.1038/ncponc1112. [DOI] [PubMed] [Google Scholar]
- 126.Iacobas I, Burrows PE, Adams DM, Sutton VR, Hollier LH, Chintagumpala MM. Oral rapamycin in the treatment of patients with hamartoma syndromes and PTEN mutation. Pediatr Blood Cancer. 2011;57(2):321–323. doi: 10.1002/pbc.23098. [DOI] [PubMed] [Google Scholar]
- 127.Schmid GL, et al. Sirolimus treatment of severe PTEN hamartoma tumor syndrome: case report and in vitro studies. Pediatr Res. 2014;75(4):527–534. doi: 10.1038/pr.2013.246. [DOI] [PubMed] [Google Scholar]
- 128.Zak M, Ledbetter M, Maertens P. Infantile Lhermitte-Duclos disease treated successfully with rapamycin. J Child Neurol. 2017;32(3):322–326. doi: 10.1177/0883073816681340. [DOI] [PubMed] [Google Scholar]
- 129.Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355(13):1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- 130.Yalon M, Ben-Sira L, Constantini S, Toren A. Regression of subependymal giant cell astrocytomas with RAD001 (everolimus) in tuberous sclerosis complex. Childs Nerv Syst. 2011;27(1):179–181. doi: 10.1007/s00381-010-1222-y. [DOI] [PubMed] [Google Scholar]
- 131.Bissler JJ, et al. Everolimus for angiomyolipoma associated with tuberous sclerosis complex or sporadic lymphangioleiomyomatosis (EXIST-2): a multicentre, randomised, double-blind, placebo-controlled trial. Lancet. 2013;381(9869):817–824. doi: 10.1016/S0140-6736(12)61767-X. [DOI] [PubMed] [Google Scholar]
- 132.Krueger DA, Care MM, Agricola K, Tudor C, Mays M, Franz DN. Everolimus long-term safety and efficacy in subependymal giant cell astrocytoma. Neurology. 2013;80(6):574–580. doi: 10.1212/WNL.0b013e3182815428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wei C, et al. Suppression of Peutz-Jeghers polyposis by targeting mammalian target of rapamycin signaling. Clin Cancer Res. 2008;14(4):1167–1171. doi: 10.1158/1078-0432.CCR-07-4007. [DOI] [PubMed] [Google Scholar]
- 134.Gao X, et al. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat Cell Biol. 2002;4(9):699–704. doi: 10.1038/ncb847. [DOI] [PubMed] [Google Scholar]
- 135.Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412(2):179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lindhurst MJ, Yourick MR, Yu Y, Savage RE, Ferrari D, Biesecker LG. Repression of AKT signaling by ARQ 092 in cells and tissues from patients with Proteus syndrome. Sci Rep. 2015;5:17162. doi: 10.1038/srep17162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ranieri C, et al. In vitro efficacy of ARQ 092, an allosteric AKT inhibitor, on primary fibroblast cells derived from patients with PIK3CA-related overgrowth spectrum (PROS) Neurogenetics. 2018;19(2):77–91. doi: 10.1007/s10048-018-0540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Venot Q, et al. Targeted therapy in patients with PIK3CA-related overgrowth syndrome. Nature. 2018;558(7711):540–546. doi: 10.1038/s41586-018-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dumont FJ, Su Q. Mechanism of action of the immunosuppressant rapamycin. Life Sci. 1996;58(5):373–395. doi: 10.1016/0024-3205(95)02233-3. [DOI] [PubMed] [Google Scholar]
- 140.Hopkins BD, et al. Suppression of insulin feedback enhances the efficacy of PI3K inhibitors. Nature. 2018;560(7719):499–503. doi: 10.1038/s41586-018-0343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mahalingam D, Sankhala K, Mita A, Giles FJ, Mita MM. Targeting the mTOR pathway using deforolimus in cancer therapy. Future Oncol. 2009;5(3):291–303. doi: 10.2217/fon.09.9. [DOI] [PubMed] [Google Scholar]
- 142.He X, Wang Y, Zhu J, Orloff M, Eng C. Resveratrol enhances the anti-tumor activity of the mTOR inhibitor rapamycin in multiple breast cancer cell lines mainly by suppressing rapamycin-induced AKT signaling. Cancer Lett. 2011;301(2):168–176. doi: 10.1016/j.canlet.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 143.Vora SR, et al. CDK 4/6 inhibitors sensitize PIK3CA mutant breast cancer to PI3K inhibitors. Cancer Cell. 2014;26(1):136–149. doi: 10.1016/j.ccr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang Y, Yu Q, He X, Romigh T, Altemus J, Eng C. Activation of AR sensitizes breast carcinomas to NVP-BEZ235’s therapeutic effect mediated by PTEN and KLLN upregulation. Mol Cancer Ther. 2014;13(2):517–527. doi: 10.1158/1535-7163.MCT-13-0655. [DOI] [PubMed] [Google Scholar]
- 145.Wang Y, et al. Differential regulation of PTEN expression by androgen receptor in prostate and breast cancers. Oncogene. 2011;30(42):4327–4338. doi: 10.1038/onc.2011.144. [DOI] [PubMed] [Google Scholar]
- 146.Wang Y, He X, Ngeow J, Eng C. GATA2 negatively regulates PTEN by preventing nuclear translocation of androgen receptor and by androgen-independent suppression of PTEN transcription in breast cancer. Hum Mol Genet. 2012;21(3):569–576. doi: 10.1093/hmg/ddr491. [DOI] [PubMed] [Google Scholar]
- 147.Lin HK, Hu YC, Lee DK, Chang C. Regulation of androgen receptor signaling by PTEN (phosphatase and tensin homolog deleted on chromosome 10) tumor suppressor through distinct mechanisms in prostate cancer cells. Mol Endocrinol. 2004;18(10):2409–2423. doi: 10.1210/me.2004-0117. [DOI] [PubMed] [Google Scholar]
- 148.Carver BS, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19(5):575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee SH, Johnson D, Luong R, Sun Z. Crosstalking between androgen and PI3K/AKT signaling pathways in prostate cancer cells. J Biol Chem. 2015;290(5):2759–2768. doi: 10.1074/jbc.M114.607846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Papa A, et al. Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell. 2014;157(3):595–610. doi: 10.1016/j.cell.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nguyen HN, et al. Engineering ePTEN, an enhanced PTEN with increased tumor suppressor activities. Proc Natl Acad Sci U S A. 2014;111(26):E2684–E2693. doi: 10.1073/pnas.1409433111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Charlesworth CT, et al. Identification of pre-existing adaptive immunity to Cas9 proteins in humans. bioRxiv. doi: 10.1101/243345. doi: 10.1101/243345. [published online ahead of print January 5, 2018]. [DOI] [Google Scholar]
- 154.Ricciardi AS, et al. In utero nanoparticle delivery for site-specific genome editing. Nat Commun. 2018;9(1):2481. doi: 10.1038/s41467-018-04894-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ma H, et al. Correction of a pathogenic gene mutation in human embryos. Nature. 2017;548(7668):413–419. doi: 10.1038/nature23305. [DOI] [PubMed] [Google Scholar]
- 156.Collins FS, Gottlieb S. The next phase of human gene-therapy oversight. N Engl J Med. 2018;379(15):1393–1395. doi: 10.1056/NEJMp1810628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dillon LM, Miller TW. Therapeutic targeting of cancers with loss of PTEN function. Curr Drug Targets. 2014;15(1):65–79. doi: 10.2174/1389450114666140106100909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Georgescu MM, Kirsch KH, Akagi T, Shishido T, Hanafusa H. The tumor-suppressor activity of PTEN is regulated by its carboxyl-terminal region. Proc Natl Acad Sci U S A. 1999;96(18):10182–10187. doi: 10.1073/pnas.96.18.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.He X, Arrotta N, Radhakrishnan D, Wang Y, Romigh T, Eng C. Cowden syndrome-related mutations in PTEN associate with enhanced proteasome activity. Cancer Res. 2013;73(10):3029–3040. doi: 10.1158/0008-5472.CAN-12-3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Okkenhaug K. Signaling by the phosphoinositide 3-kinase family in immune cells. Annu Rev Immunol. 2013;31:675–704. doi: 10.1146/annurev-immunol-032712-095946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen HH, et al. Immune dysregulation in patients with PTEN hamartoma tumor syndrome: analysis of FOXP3 regulatory T cells. J Allergy Clin Immunol. 2017;139(2):607–620.e15. doi: 10.1016/j.jaci.2016.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.George S, et al. Loss of PTEN is associated with resistance to anti-PD-1 checkpoint blockade therapy in metastatic uterine leiomyosarcoma. Immunity. 2017;46(2):197–204. doi: 10.1016/j.immuni.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Parsa AT, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med. 2007;13(1):84–88. doi: 10.1038/nm1517. [DOI] [PubMed] [Google Scholar]
- 164.Crane CA, et al. PI(3) kinase is associated with a mechanism of immunoresistance in breast and prostate cancer. Oncogene. 2009;28(2):306–312. doi: 10.1038/onc.2008.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lastwika KJ, et al. Control of PD-L1 Expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res. 2016;76(2):227–238. doi: 10.1158/0008-5472.CAN-14-3362. [DOI] [PubMed] [Google Scholar]
- 166.Heindl M, et al. Autoimmunity, intestinal lymphoid hyperplasia, and defects in mucosal B-cell homeostasis in patients with PTEN hamartoma tumor syndrome. Gastroenterology. 2012;142(5):1093–1096.e6. doi: 10.1053/j.gastro.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 167.Orloff MS, et al. Germline PIK3CA and AKT1 mutations in Cowden and Cowden-like syndromes. Am J Hum Genet. 2013;92(1):76–80. doi: 10.1016/j.ajhg.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Marsh DJ, et al. Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat Genet. 1997;16(4):333–334. doi: 10.1038/ng0897-333. [DOI] [PubMed] [Google Scholar]
- 169.Longy M, et al. Mutations of PTEN in patients with Bannayan-Riley-Ruvalcaba phenotype. J Med Genet. 1998;35(11):886–889. doi: 10.1136/jmg.35.11.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Orloff MS, Eng C. Genetic and phenotypic heterogeneity in the PTEN hamartoma tumour syndrome. Oncogene. 2008;27(41):5387–5397. doi: 10.1038/onc.2008.237. [DOI] [PubMed] [Google Scholar]
- 171. Mirzaa G, et al. PIK3CA-related segmental overgrowth. In: Adam MP, et al., eds. GeneReviews. Seattle, Washington, USA: University of Washington; 2013; pp. 1993–2018. [Google Scholar]
- 172. Mirzaa G. MPPH syndrome. In: Adam MP, et al., eds. GeneReviews. Seattle, Washington, USA: University of Washington; 2016; pp. 1993–2018. [Google Scholar]
- 173.Loconte DC, et al. Molecular and functional characterization of three different postzygotic mutations in PIK3CA-related overgrowth spectrum (PROS) patients: effects on PI3K/AKT/mTOR signaling and sensitivity to PIK3 inhibitors. PLoS One. 2015;10(4):e0123092. doi: 10.1371/journal.pone.0123092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ngeow J, Sesock K, Eng C. Clinical implications for germline PTEN spectrum disorders. Endocrinol Metab Clin North Am. 2017;46(2):503–517. doi: 10.1016/j.ecl.2017.01.013. [DOI] [PubMed] [Google Scholar]