Abstract
The genome is packaged and organized nonrandomly within the 3D space of the nucleus to promote efficient gene expression and to faithfully maintain silencing of heterochromatin. The genome is enclosed within the nucleus by the nuclear envelope membrane, which contains a set of proteins that actively participate in chromatin organization and gene regulation. Technological advances are providing views of genome organization at unprecedented resolution and are beginning to reveal the ways that cells co-opt the structures of the nuclear periphery for nuclear organization and gene regulation. These genome regulatory roles of proteins of the nuclear periphery have important influences on development, disease and ageing.
Early cytological experiments determined that the cell nucleus — the largest, most readily observed subcellular structure — contains the genetic determinants that direct the development of functional tissues1. We now know that the eukaryotic nucleus contains the majority of the genetic information of the cell in the form of discrete chromosomes2, which occupy distinct, nonrandomly positioned territories within the nucleus3. Recent innovations in sequencing-based methods have enabled the visualization of the 3D organization of whole genomes. Chromosome conformation capture (3C) and related technologies such as HiC have made it possible to infer genome folding by detecting contact frequencies throughout the genome4. In general, studies using sequencing-based methods agree with those using microscopy-based approaches that intrachromosomal contacts are more frequent than interchromosomal contacts, supporting the existence of chromosome territories, and that gene-rich chromatin domains interact with each other more frequently than with gene-poor chromatin domains5,6. In fact, gene-rich, transcriptionally active and gene-poor, transcriptionally repressed chromatin form distinct megabase-scale compartments, designated A and B compartments, respectively5.
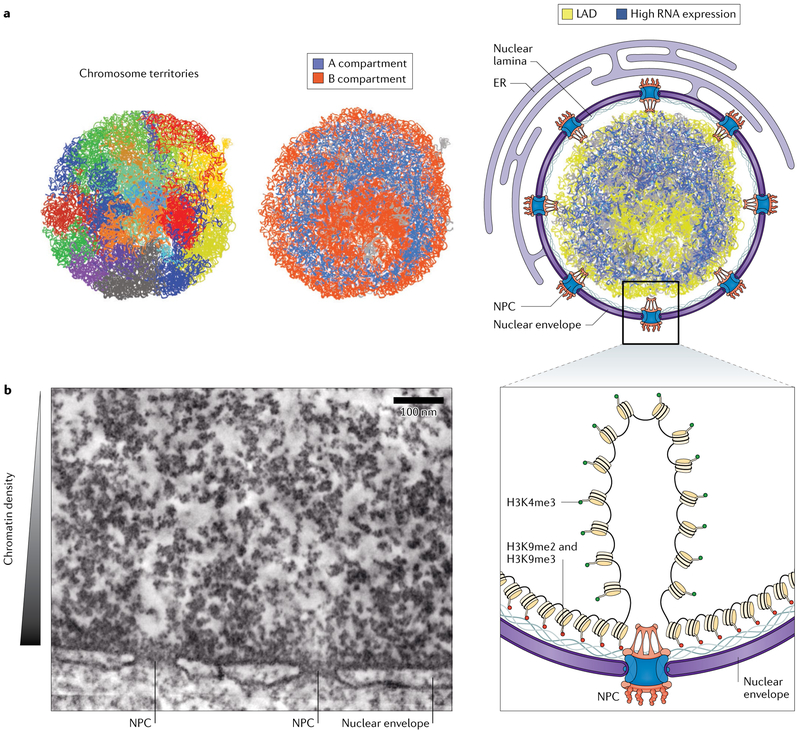
Chromatin compartments adopt specific positions relative to a major structural landmark of the nucleus, the nuclear periphery (FIG. 1a). The nucleus is separated from the cytoplasm by the nuclear envelope, a double membrane bilayer that, although contiguous with the endoplasmic reticulum membrane network, is enriched for a unique set of proteins on the basis of their affinity for other structures, including chromatin and the nuclear lamina7. The nuclear lamina, which underlies the inner membrane of the nuclear envelope, is composed of a meshwork of filamentous proteins, the nuclear lamins, and acts as a structural scaffold for the nucleus8. Nuclear pore complexes (NPCs) are large multiprotein assemblies that perforate the nuclear envelope membrane and control the passage of material between the nucleus and cytoplasm (reviewed in REF.9). HiC analyses indicate that the B compartment preferentially occupies the nuclear periphery, whereas the A compartment is more centrally located within the nucleus10. As visualized by electron microscopy, heterochromatin is prominently clustered underneath the nuclear lamina in most cell types11 and is interspersed with regions of less tightly packed euchromatin underneath the NPCs (FIG. 1b). Accordingly, the nuclear periphery is generally a repressive environment where gene activity is limited, with the prominent exception of genomic regions that are closely associated with NPCs12.
Fig. 1 ∣. Chromatin organization within the nucleus.
a ∣ Cross-sections through 3D models of genome structures generated from single cell Hi-C, coloured according to individual chromosomes (left panel), whether the sequence is in the A or B compartment (middle) and whether the sequence is part of a lamina-associated domain (LAD) (yellow) or contains highly expressed genes (blue) (right). b ∣ ChromEMT (chromatin electron microscopy tomography) depicts the organization of chromatin within the nucleus. Chromatin density is greatest at the nuclear periphery and decreases within the nuclear interior, with the prominent exception of decondensed euchromatin underneath nuclear pore complexes (NPCs). Gene-rich euchromatin that is transcriptionally active and enriched in histone 3 lysine 4 trimethylation (H3K4me3) marks (green) is located within the nucleoplasm, and gene-poor heterochromatin that is transcriptionally repressed and enriched for H3K9 dimethylation (H3K9me2) and H3K9me3 marks (red) is located at the nuclear periphery. ER, endoplasmic reticulum. Part a adapted from REF.10, Springer Nature Limited. EM image in part b adapted with permission from REF.11, Ou, H. D. et al. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357, eaag0025 (2017) https://doi.org/10.1126/science.aag0025. Reprinted with permission from AAAS.
Ongoing work is illuminating how the organization of the genome within the nucleus guides efficient gene activation and gene repression. In recent years, roles have emerged for the proteins and structures of the nuclear periphery in establishing nonrandom genome organization, maintaining nuclear organization over time and regulating gene expression. Here, from the perspective of the nuclear periphery, we follow the life cycle of the nucleus from assembly, to the supporting roles that the nuclear periphery plays in adapting the genome to cell type-specific functions, to the decline in nuclear organization during ageing. Although we focus on recent advances in mammalian systems, where relevant, we also highlight select findings from other model eukaryotes.
Building a nucleus
Cell type specificity of nuclear organization
In multicellular organisms, cells within different tissues position their chromosomes in characteristic ways. In a specific cell type, these positions are maintained in relation to other chromosomes as well as to landmarks such as the nuclear periphery13. This nonrandom positioning has important effects on genome activity and function. For example, in female mammals, X chromosome inactivation is accompanied by positioning of the inactive X close to the nuclear lamina14, and repression of genes encoded by the inactive X requires the expression of the lamina-associated protein lamin B receptor (LBR), which interacts with the Xist RNA15. Cell type-specific positioning of specific gene loci has also been observed. For example, inactive immunoglobulin loci are positioned near the nuclear periphery in haematopoietic progenitor cells but within the nuclear interior in differentiated B cell progeny. This movement into the nuclear interior is accompanied by gene activation16. These examples highlight the relationship between genome organization and cellular function and indicate that cellular differentiation is often accompanied by genome reorganization17. Intriguingly, to revert differentiated cells to a pluripotent state, genome reorganization must also occur (BOX 1).
Box 1 ∣. Nuclear organization in pluripotency and reprogramming.
When compared with differentiated cells, the genome of pluripotent cells exists in a comparatively malleable state with low levels of heterochromatin152 and frequent long-range interactions153. Chromatin is relaxed and deformable, with genomic loci capable of exploring large volumes within the nucleus154.
By contrast, differentiated cell types follow lineage-restricted gene expression programmes that are promoted by cell type-specific chromatin modifications and more stable chromatin organization. This organization must be deconstructed in order to effectively reprogramme a differentiated cell to pluripotency. Reprogramming is extremely inefficient, suggesting that formidable barriers exist155. Such chromatin barriers seem to include DNA methylation156, histone modifications157, and positioning and composition of nucleosomes158.
The composition of major nuclear structures including the nuclear pore complex (NPC)45, the nuclear lamina82 and the nuclear envelope7 is also distinct in pluripotent cells, and the need to remodel nuclear structures may also limit reprogramming efficiency. For example, lamin A expression is abnormally high in non-viable embryos generated from nuclear reprogramming159. Reprogramming involves cell division and allows a differentiated cell to be transformed into almost any other cell type by passage through a pluripotent state. By contrast, transdifferentiation involves the direct transformation of one cell type into another; for example, fibroblasts can be converted into neuronal141,160 and muscular161 cell types. Because this process does not involve cell division, it is likely that nuclear reorganization is more limited. Further investigation into the effects of reprogramming and transdifferentiation on nuclear organization is likely to yield further insight into the mechanisms that control genome activity, as well as the means to more effectively manipulate cell identity.
Nuclear organization after mitosis
In order for cells to maintain nonrandom nuclear organization, mechanisms must exist to re-establish organization after cell division. The nuclear periphery disassembles in mitotic prophase and reassembles during anaphase, telophase and early G1 phase (reviewed in detail in REF.18). Meanwhile, chromatin compacts into discrete mitotic chromosomes to enable the faithful separation of a complete genome into each daughter nucleus. Nevertheless, cell type-specific features of gene positioning and gene expression are maintained through cell division19, implying that mechanisms for ‘remembering’ chromatin organization through cell division exist.
In recent years, some of the supporting roles that proteins of the nuclear periphery play in re-establishing this organization have been illuminated. For example, in anaphase, the DNA crosslinking protein barrier-to-auto-integration factor (BAF) coats and binds chromosomes into a single chromatin mass that serves as a surface for nuclear envelope assembly20. Affinity for BAF recruits nuclear envelope proteins21 and proteins of the nuclear lamina to the chromatin surface (reviewed in REF18). In parallel, several components of the NPC, the nucleoporin proteins (NUPs), bind to histones and act as seeding sites for future NPC assembly22,23. The transcriptional repressor heterochromatin protein 1 (HP1) and associated proteins, such as proline-rich protein 14 (PRR14)24 and LBR25, bridge chromatin interactions with the nascent nuclear lamina.
It remains unclear whether structures of the nuclear periphery exhibit any selectivity in binding to specific genomic regions during nuclear assembly. However, the same genomic regions tend to re-establish contact with the nuclear periphery after mitosis26. Furthermore, single-cell HiC analyses indicate that chromatin decondenses anisotropically as cells exit mitosis. The euchromatic A compartment decondenses to a greater extent than the gene-poor, heterochromatic B compartment, and the A compartment adopts a central position whereas the B compartment moves towards the nuclear periphery and perinucleolar regions27. Chromatin mobility progressively decreases as cells proceed towards interphase28. Repressive chromatin marks, in particular histone 3 lysine 9 dimethylation and trimethylation (H3K9me2 and H3K9me3, respectively), are specifically enriched beneath the nuclear lamina by the localized actions of histone deacetylases and methylases, which are in turn recruited to the nuclear lamina through protein–protein interactions29,30. These types of protein–chromatin and chromatin–chromatin interactions may help to maintain the legacy of cell type-specific gene expression through cell divisions or over long periods of time in non-dividing cells.
Temporal and physical constraints
Reorganizing the genome is usually not a rapid process. Specific DNA sequences have been identified that possess an intrinsic ability to localize to particular nuclear regions and can direct the positioning of other genomic loci when introduced in trans. However, this re-positioning requires passage through mitosis26,31. Similarly, it may be that haematopoietic progenitor cells can undergo rapid and dramatic changes to genome organization as they differentiate into B cells because of their rapid proliferation rate16. However, there are examples of chromatin reorganization in the absence of cell division. The terminal differentiation of rod photoreceptor cells in the mouse retina involves complete inversion of the relative positions of euchromatic and heterochromatic domains over the course of several weeks as peripheral tethers for heterochromatin are lost32. On a faster timescale and smaller physical scale, damaged telomeres can explore large volumes of the nucleus over minutes to hours33, although whether DNA lesions elsewhere in the genome also exhibit increased mobility remains controversial34,35. Movement of a chromatin locus is profoundly affected by the level of compaction of its immediate chromatin environment36, and loci embedded within heterochromatin are generally less mobile than loci positioned within euchromatin37. In fact, drug-induced or transcription-induced decompaction of chromatin can promote the movement of genomic loci within the nucleus38. Decatenation of chromatin by topoisomerases may help to resolve tangled chromatin and promote movement or reorganization of chromatin domains39. As we discuss in subsequent sections, association of chromatin with the nuclear periphery generally serves as a stable anchor for genome organization rather than as a platform for the dynamic movement of loci.
Gene regulation by the nuclear periphery
The proteins and structures of the nuclear periphery actively participate in facilitating the organization of the genome within the nucleus. The following sections of this Review focus on the roles that the NPCs and the nuclear lamina play in regulating the genome.
Nucleoporins regulate gene expression
NPC structure and function.
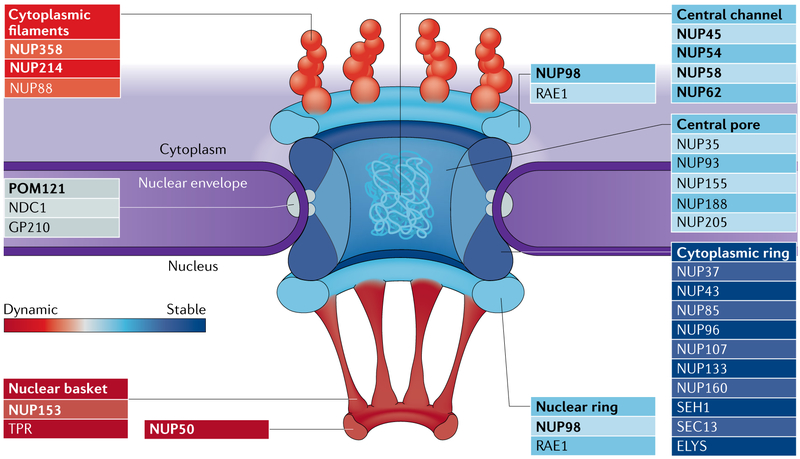
The NPC is the major gateway for transport between the nucleus and the cytoplasm (FIG. 2). The mammalian NPC is a modular assembly of ~30 distinct NUPs that form ~120 MDa structures (reviewed in REF.9). NUPs can be functionally divided into two classes (FIG. 2). The first class of NUPs represents the structural framework of the NPC and is not directly involved in nuclear transport. Many of these structural NUPs are very stable within the NPC assembly40, and a subset of these essentially do not turn over for the entire lifespan of a cell41,42. The second class of NUPs contain unstructured domains rich in phenylalanine and glycine amino acids (FG repeats) that function directly in nuclear transport by binding soluble transport receptors. The conformational flexibility of the FG-repeat NUPs is key to their role in nuclear transport43 but may also allow these proteins to conform to a wide array of binding partners and mediate other functions. Importantly, many of these NUPs shuttle dynamically on and off the NPC40.
Fig. 2 ∣. NPC structure and dynamics.
The nuclear pore complex (NPC) is the major gateway for transport between the nucleus and the cytoplasm. Transport through NPCs involves the nucleoporin proteins (NUPs), the GTPase Ran, importin and exportin transport receptors and specialized factors that promote transport of Large protein and RNA complexes (not shown). Whereas core structural elements of the NPC are very stable, other NUPs shuttle dynamically on and off the NPC40. Average NUP dynamics are represented for each subcomplex on the basis of their residence times at NPCs49; subcomplexes range from highly dynamic (red) to fairly stable (blue). Individual NUPs within the NPC are Labelled, and NUPs in bold contain repeat domains rich in phenylalanine and glycine amino acids (FG repeats). GP210, nuclear pore membrane glycoprotein 210; POM121, nuclear envelope pore membrane protein POM121; RAE1, ribonucleic acid export 1.
Evolution and specialization of the NPC.
Although the overall structure of the NPC is highly conserved, the physical size of the NPC has roughly doubled from yeast to humans9, and NUP genes are among the fastest-evolving genes in the genome44. Recent work suggests that the functional repertoire of the NPC has expanded in mammals, as in many cases the depletion of specific NUPs does not have an impact on nucleocytoplasmic transport but does have tissue-specific effects on development and differentiation. For example, the NPC protein NUP133 is dispensable for embryonic stem (ES) cell proliferation but is essential for differentiation into neural lineages45. By contrast, high expression of NUP153 in ES cells46 and in neural progenitor cells47 promotes pluripotency, and NUP153 downregulation accompanies differentiation. Furthermore, NUP50 (REF.48) and NUP210 (REF.49) are required for muscle differentiation but are dispensable in proliferating myoblasts. These and other examples of tissue-specific requirements for NUPs are suggestive of transport-independent functions. In this section, we review recent work that indicates that NUPs also have the ability to regulate the genome.
NPC–genome contacts regulate gene expression at the nuclear periphery.
Early studies in yeast demonstrated that NPCs bind to the genome50,51 and that gene tethering to the NPC promotes both the activation of genes in response to stimulus52,53 and the stable repression of ribosomal and subtelomeric genes54. Elegant work in yeast has determined that Nup100, which is homologous to human NUP98, is required for tethering activated genes to the NPC55. Transcription factors and chromatin-modifying enzymes also cooperate to promote tethering of activated genes to the yeast NPC56. Work in metazoan systems also implicates NPCs in mediating both gene activation and repression. For example, in pluripotent cells, NUP153 binds to both transcription start sites46,47 and termination sites47, promoting silencing of some targets and activation of others. During muscle differentiation, NUP210 scaffolds the transcription factor myocyte-specific enhancer factor 2C (MEF2C) at the NPC, promoting activation of its target genes57.
Furthermore, NPCs contribute to higher-order chromatin organization in metazoans. As mentioned above, dense heterochromatin is a prominent feature of the nuclear periphery, whereas regions of less-compact euchromatin underlie NPCs12 (FIG. 1). The formation of these heterochromatin exclusion zones beneath NPCs has been shown to depend on the NPC component nucleoprotein TPR58, suggesting that NPCs actively maintain a domain of euchromatin at the nuclear periphery. In Drosophila melanogaster, NUP98 mediates enhancer–promoter looping of poised inducible genes at the NPC59. This arrangement potentiates transcriptional activation in response to stimulus by the hormone ecdysone59. In human cells, NPCs bind to super-enhancers, which are regulatory structures that drive the expression of key genes that specify cell identity60. These NUP-associated super-enhancers localize to the nuclear periphery, and ablation of NPC tethering results in super-enhancer disruption and transcriptional dysregulation. Super-enhancers bound and regulated by NPCs vary across cell types, but in each context they regulate groups of genes involved in defining cellular identity60, suggesting that NPC association with these regions could be important for maintaining cellular fate.
It remains unclear what factors dictate whether NPC–genome interactions activate or repress transcription, but analyses of genome binding by chromatin immunoprecipitation followed by sequencing (ChIP–seq) and DNA adenine methyltransferase identification (DamID) both indicate that NUPs show affinity for distinct regions of chromatin50. This observation suggests that mechanisms exist to specialize the interaction of individual NUPs and/or NPCs with the genome. Given that most NUPs have no demonstrated ability to bind directly to DNA or to chromatin, NUP-associated transcription factors47,56,57, chromatin-modifying complexes146 and other unidentified factors likely mediate the distinct chromatin-binding preferences of NUPs.
Nucleoporins regulate gene expression within the nucleus.
In higher eukaryotes, individual NUPs have acquired spatially and functionally distinct roles in different nuclear domains (FIG. 3). In principle, any NUP that associates dynamically with the NPC40 could take on additional roles within the nucleoplasm. Many NUP-bound loci are found within the nuclear volume when visualized by fluorescence in situ hybridization (FISH), indicating that these interactions can occur independently of the NPC. For example, NUP98 binds genes both at the NPC and within the nucleoplasm61,62, and association within the nucleoplasm correlates with high levels of target gene activation62. Similarly, NUP153 associates with genes both within the nucleus and at the NPC46,60, although whether gene binding has distinct effects on gene activity in each of these domains is less clear. Other NPC components, including NUP50, NUP88, SEC13 and TPR, also bind to the genome within the nuclear interior48,61,63,64. These interactions regulate the expression of genes involved in differentiation, development, cell cycle progression and the antiviral response61,63–65. Recent advances in our understanding of the mechanics of NUP-mediated transcriptional regulation within the nucleus have particularly focused on NUP98.
Fig. 3 ∣. NPC-mediated gene regulation.
a ∣ Nuclear pore complexes (NPCs) bind enhancers at the nuclear periphery to facilitate enhancer–promoter looping for gene activation. NPCs also recruit specific transcription factors (TFs) to the nuclear periphery to control gene profiles involved in cell fate. b ∣ NPCs recruit chromatin modifiers to the nuclear periphery to mediate gene repression at specific genomic loci, including ribosomal, subtelomeric and early differentiation genes. c ∣ Nucleoporin proteins (NUPs) bind the genome within the nucleus and interact with an array of proteins, including histone modifiers and RNA helicases, to regulate gene expression. d ∣ NUPs can regulate gene expression through post-transcriptional processes, such as mRNA splicing and stability. ATP-dependent RNA helicase A (DHX9) has been shown to facilitate NUP-mediated mRNA splicing, but other proteins involved in these processes still remain to be identified. m7G, 7-methylguanosine.
NUP98 binding on the genome coincides with marks of transcriptional activity, including RNA polymerase II association, nuclease accessibility and H3K4me3 (REFS59,61,62,66). NUP98 binding is enriched at transcription start sites, at times to an extent comparable to known transcription factors66, and the depletion of NUP98 potently downregulates target gene expression61,66, suggesting that NUP98 functions as a transcriptional co-activator. NUP98 can also maintain previously expressed genes in a poised state in order to amplify expression in response to future stimuli (such as a nutrient or hormone), a phenomenon known as transcriptional memory. This transcriptional memory has been observed both at the NPC59 and within the nucleoplasm67.
However, NUP98 has no demonstrated ability to bind to DNA68, implying that another factor (or factors) recruits it to the genes that it regulates. In D. melanogaster, enzymes that catalyse transcription-promoting chromatin modifications have been implicated in recruiting NUP98 to chromatin. Specifically, the binding of NUP98 to chromatin requires the Non-Specific Lethal (NSL) complex, which mediates H4K16 acetylation69, a chromatin mark that antagonizes chromatin compaction70. Chromatin-bound NUP98 can then promote the expression of genes targeted by the H3K4 histone-lysine N-methyltransferase trithorax69. Similarly, in mammalian cells, NUP98 binds to the histone-lysine N-methyltransferases SETD1A and SETD1B and promotes the deposition of H3K4me3 (REF.66). This interaction is subverted in leukaemias caused by translocations of the NUP98 gene71. These fusions lack the NPC-targeting carboxyterminal domain of NUP98 but retain the ability to interact with chromatin modifiers72. Emerging evidence indicates that NUP98 fusions drive leukaemia by hyperactivating genes that promote sustained proliferation of haematopoietic progenitors73. These findings highlight broad interdependencies between NUP98, chromatin modifiers and gene activation that are only just beginning to be understood.
The roles of NUPs in gene regulation have further expanded with the recent evolution of a hominoid-specific NUP variant, soluble nuclear envelope pore membrane protein POM121 (sPOM121), which cooperates with NUP98 to bind the genome in human cells74. Importantly, sPOM121 lacks an NPC-anchoring domain and is instead found exclusively within the nucleoplasm. sPOM121 recruits the NUP107–160 complex and potentially other factors to NUP98 genome binding sites. As sPOM121 expression is readily detectable in every human tissue, in many cases at levels surpassing full-length POM121 (REF.74), sPOM121 could have a major role in NUP-mediated gene regulation.
Post-transcriptional regulation of gene expression by NUPs.
Gene expression is initially driven by transcription, yet there are many regulatory processes between the transcription of nascent RNA and mRNA translation that can modulate gene expression75. Evidence that transcription, splicing and RNA export are coupled at the NPC provided early credence to the idea that NUPs also regulate gene expression after transcription17,76. Roles for NUP98 have recently been uncovered in mRNA splicing and stability. For example, NUP98 interacts with the ATP-dependent RNA helicase A (DHX9) and co-binds to specific mRNA transcripts to regulate their splicing77. Moreover, NUP98 seems to protect specific p53-induced transcripts from degradation by the exosome78. It remains unclear whether NUP98 can bind directly to mRNAs for post-transcriptional gene regulation or whether NUP98 works in concert with other proteins to regulate this process. However, these discoveries highlight the increased functionality of NUPs beyond transport and transcription regulation.
Although several NUPs have been functionally linked to gene regulation, many of these NUPs exist as stable subcomplexes within the context of the NPC (FIG. 2), and it remains unclear which cohort of binding partners specific NUPs might be associated with when binding the genome. For example, NUP153 binds to NUP50 and the transport factor importin-α to function in nuclear transport79, but interdependencies between NUP50 and NUP153 in gene regulation have not been defined. Similarly, NUP98 and ribonucleic acid export 1 (RAE1) function together to promote mRNA export80, but it remains unclear whether they cooperate in gene regulation. The ability of additional NUPs to dynamically dissociate from the NPC and associate with chromatin remains to be explored. One potentially interesting candidate is ELYS, which may be unique among NUPs in its ability to bind directly to nucleosomal DNA in vitro22. Overall, many open questions about the extent and mechanism of gene regulation by NPCs and by individual nucleoporins remain.
Gene regulation by lamins
The nuclear lamina, which underlies the nuclear envelope and is interspersed with NPCs (FIG. 1), is composed of four proteins in mammalian somatic cells: the A-type lamins, lamin A and lamin C, and the B-type lamins, lamin B1 and lamin B2 (REF.8). Lamins assemble into nonpolar, bundled filaments that provide support against strain and that scaffold the genome8,81. A-type lamins are expressed at low levels in pluripotent cells and at higher levels in differentiated tissues, whereas at least one B-type lamin is expressed in every cell type82,83. The nuclear lamina functions in close concert with a cohort of associated proteins, including proteins of the nuclear envelope and non-membrane-bound nuclear proteins. Many nuclear lamina-associated proteins are expressed only in selected tissues, perhaps contributing to specialized functions of this structure7. The nuclear lamina is essential for organogenesis, as Lmnb1-null and Lmnb2-null mice each die at birth with defects in brain, lung and diaphragm development84,85, whereas Lmna-null mice exhibit cardiac and muscular defects and die within days of birth86. Mutations to the lamin genes, most prominently to LMNA, cause at least 15 distinct diseases, which are known as laminopathies (reviewed in REF.87). This Review focuses on the functions of the nuclear lamina in gene regulation.
Lamina association confers gene repression.
A dense layer of heterochromatin underneath the nuclear lamina is a prominent feature of the nuclear periphery in most cells11 (FIG. 1). DamID88 with lamin B1 fusion proteins has been extensively employed to identify the regions of the genome that are in close proximity to the nuclear lamina26,89,90. These lamina-associated domains (LADs), which exhibit a median size of 500 kb (REF.86), exhibit heterochromatic features, including low gene density, low transcriptional activity and late replication timing89. In mammalian cells, up to one-third of the genome and 10% of coding genes are found within LADs91,92. They are enriched with repressive histone modifications, including H3K9me2, H3K9me3 (REFS29,89) and H3K27me3 (REFS89,93), and are sparsely populated with active chromatin marks such as H3K4me89. In addition to gene-poor LADs that are consistent across tissues, a subset of LADs known as variable LADs (vLADs) contain lineage-specific genes90. Genes found in vLADs are often irrelevant to the functions of the cell type in which they are found91,94, suggesting that vLADs maintain cell identity by restricting gene expression.
Do LADs represent an intrinsically repressive nuclear domain, or do they result from gene inactivity? When a reporter gene is inserted within a LAD, its expression is lower than the same reporter residing in the nuclear interior95,96. Sequences derived from LADs can home to the lamina and undergo transcriptional repression when randomly integrated into the genome26,93, suggesting that this process is actively mediated. Finally, changes to genome–lamina association usually correlate with changes in gene expression during differentiation91. Taken together, these findings indicate that the residence of genomic regions within LADs correlates with lower gene activity. By analysing the features of genomic regions in close proximity to the lamina, the mechanisms through which LADs form and mediate gene silencing have begun to emerge.
Cooperation of chromatin readers, writers and tethers at the nuclear lamina.
Much of what we know about LADs has been defined by analysing the genomic regions in close proximity to lamin B1 (REFS89,91,97). How do different lamin isoforms contribute to LAD organization? Lamin isoforms generally exhibit very similar genome binding profiles98, although the A-type lamins also interact with euchromatin within the nucleoplasm99. By moving between binding sites within the nucleus and at the nuclear lamina, A-type lamins could potentially modulate LAD organization100. Intriguingly, depletion of A-type lamins is sufficient to disrupt LAD tethering in differentiated cells, although these cells also express B-type lamins93,101. It remains unclear whether A-type lamins can directly mediate LAD interactions or whether the disruption of A-type lamins displaces associated proteins that also participate in LAD tethering.
Although there is some evidence that lamins can bind directly to chromatin102,103, nuclear lamina-associated proteins also contribute to LAD assembly. For example, LBR, which is localized to the inner membrane of the nuclear envelope, associates with lamin B1 (REF.104) and exhibits a very similar genome binding profile to lamin B1 (REF.60). Given that LBR also binds to H3K9me2-modified and H3K9me3-modified chromatin via HP1 (REF.105) and to H4K20me2-modified chromatin through its Tudor domain106, this protein could function as a chromatin ‘reader’ at the nuclear lamina. The ability of LBR to bridge interactions between the lamina and chromatin has clear functional importance as it is required to tether heterochromatin to the nuclear periphery in developing tissues25. LBR and lamin A/C trade this role through development107,108; developing tissues rely on LBR for peripheral heterochromatin tethering, whereas differentiated tissues seem to rely on lamin A/C25. It may be that a lamin A/C-associated protein mediates heterochromatin tethering in differentiated tissues, but such a functional partner remains to be identified.
Another group of proteins that participate in LAD organization are the LEM-domain family of nuclear envelope proteins, named for the founding members lamina-associated polypeptide 2 (LAP2), emerin, and MAN1 inner nuclear membrane protein109. LEM domains interact with the DNA cross-bridging protein BAF109, which brings these nuclear envelope proteins into close contact with chromatin. Emerin tethers muscle differentiation genes to the nuclear periphery, helping to maintain muscle progenitors110,111, whereas LAP2β limits the differentiation capacity of cardiac muscle progenitors by a similar mechanism26. A number of other, less well-characterized nuclear envelope proteins, such as nuclear envelope transmembrane protein 23 (NET23), NET39, transmembrane protein 38A (TMEM38A) and wolframin (WFS1), seem to have supporting roles in genome organization across differentiated tissues112,113.
Chromatin reading and editing enzymes cooperate with nuclear envelope protein tethers to maintain LADs. LAP2β and emerin recruit the histone deacetylase HDAC3 to the nuclear lamina to mediate gene repression in progenitor cells30,114. Disruption of histone-lysine N-methyltransferases that deposit H3K9me2 (EHMT2; also known as G9a)115, H3K9me3 (SUV39H1)116 or H3K27me3 (EZH2)93 moderately affects LAD gene positioning and activity. Furthermore, pharmacological inhibition of histone deacetylation, H3K9me2 or H3K27me3 causes striking decompaction of LADs101, suggesting that chromatin state promotes the tight clustering of repressed LAD domains in relation to each other as well as their close association to the lamina. When spread over the kilobase to megabase scales of an entire LAD, these interactions may cooperatively promote chromatin compaction and reinforce gene repression. Consistent with this idea, LAD regions of a chromosome cluster tightly together at the nuclear periphery of individual cells, whereas non-LAD regions on the same chromosome are more diffuse101. This compaction may limit transcription factor access and decrease the likelihood that a LAD resident gene will be expressed100 (FIG. 4). The recently described ability of heterochromatin domains to exist in a separate phase suggests an alternative means for LAD regions to coalesce and limit transcription factor access117,118. Overall, nuclear lamina anchoring, reinforcement of repressive chromatin marks and compaction of LAD regions cooperate to stably maintain cell type-specific gene expression profiles.
Fig. 4 ∣. Nuclear lamina-mediated genome organization.
The nuclear lamina underlies the inner nuclear membrane (INM) and is connected to the genome through lamin-associated proteins. Lamin-associated domains (LADs) are large, gene-poor regions of heterochromatin that interact with the lamina at the nuclear periphery and are often enriched in repressive histone modifications, such as histone 3 lysine 9 methylation (H3K9me) and histone 3 lysine 27 acetylation (H3K27ac) (red). Histone modifiers and transcriptional regulators have been suggested to cooperate with INM proteins to maintain LADs. A-type lamins, in association with lamina-associated polypeptide 2α (LAP2α), can also interact with regions of euchromatin within the nucleus that are enriched in active histone modifications, including H3K4me and H3K9ac (green). BAF, barrier-to-autointegration factor; H3K9me2, H3K9 dimethylation; H3K9me3, H3K9 trimethylation; HP1, heterochromatin protein 1; LBR, lamin B receptor.
The reach of lamins extends within the nucleus.
Although the nuclear lamina is a prominent feature of the nuclear periphery, the A-type lamins are also found within the nuclear volume99. Whereas the B-type lamins preferentially associate with heterochromatin at the nuclear periphery, the A-type lamins bind to both heterochromatic and euchromatic regions99,119. The abundance of A-type lamins within the nucleoplasm is tuned by a variety of factors, including the expression levels of a nucleoplasmic binding partner, LAP2α99,120, and phosphorylation events121. Depleting cells of LAP2α, and thus diminishing the levels of nucleoplasmic lamin A, allows derepression of some lamin-bound genes99. Nucleoplasmic lamins seem to limit the mobility of chromatin within the nuclear interior122 and exert anti-proliferative and pro-differentiation effects120,123.
Some disease-causative mutations deplete the nucleoplasmic pool of lamin A. For example, a dominant negative progeria-linked mutation causes lamin A to be constitutively lipid modified and associated with the nuclear envelope, which depletes A-type lamins from the nucleoplasm124. This nucleoplasmic depletion of A type lamins is accompanied by global disorganization of heterochromatin marks125,126 and derepression of some genomic regions, including satellite DNA125 and ribosomal DNA127. Because ribosomal DNA transcription controls the morphology, activity and composition of the nucleolus128, deregulation upon lamin A disruption also affects the function and organization of this nuclear compartment127. Overall, nucleoplasmic lamins seem to perform important functions in regulating the activity of the genome and the organization of the nucleus that remain incompletely understood.
The ageing nucleus
Once nuclear organization is established, how is it maintained over time as cells age? The fundamental defining feature of ageing is a decline in the functional capacity of organs, which in turn results from incremental impairments in cellular homeostasis. Several hallmarks of ageing converge on the nucleus, and mutations to nuclear proteins cause premature ageing disorders129, highlighting the nucleus as a weak point in cellular ageing. Nuclear ageing includes changes to the genome itself as well as to the protein structures that enclose and organize it. The ageing genome is subject to telomere shortening, disruptions to epigenetic modifications, increased transcriptional noise and accumulation of DNA damage129. Separately, declining protein homeostasis contributes both generally to cellular ageing and specifically to functional declines in nuclear structures such as the NPC130. The lifetime of the nucleus depends on the extent of cellular turnover in a tissue, and nuclei encounter distinct age-associated disruptions in rapidly proliferating cells (FIG. 5a) versus long-lived cells (FIG. 5b).
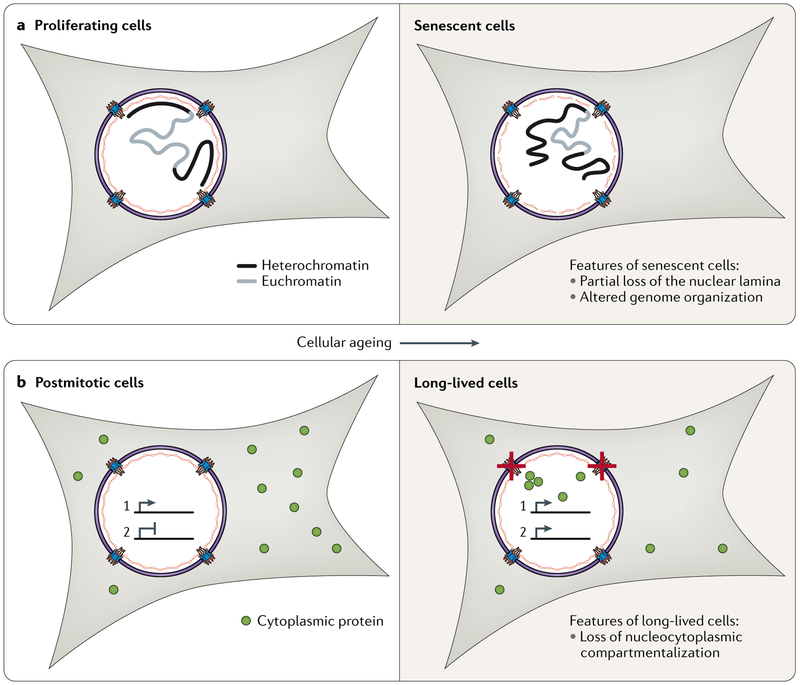
Fig. 5 ∣. Nuclear decline over cellular ageing.
a ∣ Proliferating cells eventually exit the cell cycle and senesce as cellular ageing progresses. These senescent cells exhibit a disrupted lamina meshwork and altered chromatin organization. b ∣ Postmitotic cells do not divide and must maintain their function over cellular ageing. However, the identification of proteins that undergo limited turnover in non-dividing cells suggests that these proteins accumulate damage that leads to nuclear decline. Two prominent characteristics of long-lived postmitotic cells are the loss of nucleocytoplasmic compartmentalization and increased transcriptional noise.
Nuclear remodelling in senescent cells
The function of many epithelial tissues depends on regular renewal. Nuclei in such proliferative tissues encounter challenges including progressive shortening of telomeres and accumulation of DNA damage and mutations129, all of which may push cells towards the irreversible cell cycle arrest known as senescence. Although senescence is an important brake against tumorigenesis, senescent cells accumulate in ageing tissues and exert negative paracrine effects by secreting pro-inflammatory factors131. Senescence onset is accompanied by widespread changes to nuclear organization (FIG. 5a). Lamin B1 is degraded by autophagy132,133, and peripheral lamin-associated heterochromatin becomes much less prominent134,135. Senescent cells exhibit dysregulated gene expression and derepression of repetitive elements136. Depletion of lamin B1 can drive cells into senescence prematurely132, which suggests that remodelling of genome–lamina contacts is an important event in senescence onset. Strikingly, the lamina can be so weakened in senescent cells that it no longer adequately protects the genome; naked chromatin fragments appear in the cytosol of senescent cells137, where they promote the pro-inflammatory signalling that is characteristic of senescent cells by diverting innate immune response factors138.
Nuclear decline in long-lived cell types
Although the cells of many tissues in the body are periodically renewed, organs including the heart139 and brain140 contain substantial numbers of cells that are born in early development and face a demand for lifelong performance. In such long-lived cell types, nuclear organization must be maintained for decades. Maintenance of the nuclear permeability barrier declines as neurons age, allowing cytoplasmic proteins such as tubulin to enter and accumulate within the nucleus49,141 (FIG. 5b). Furthermore, nuclear proteins represent over half of the small cohort of proteins that are stable in neurons for years42,142. Over the lifetime of a cell, the accumulation of aberrant modifications to proteins can perturb critical processes. For example, eye lens crystallin is a long-lived protein that misfolds as a result of protein damage over time, causing cataracts130. It is similarly possible that long-lived histones, NUPs and lamins are vulnerable to damage, misfolding or aggregation and thus may underlie declines in nuclear integrity with age.
Recent reports indicate that declining nuclear integrity is accelerated in neurodegenerative disorders, including Huntington disease143 and amyotrophic lateral sclerosis (ALS)144. Neurodegeneration-linked polypeptides seem to exert toxic effects on nucleocytoplasmic transport by sequestering the FG-repeat NUPs, including NUP98, that line the NPC channel145,146. Given the important roles of NUP98 in mediating gene activation, it is possible that expression of NUP98 target genes is also impaired when NUP98 is sequestered by toxic aggregates. Whether functional declines in other long-lived nuclear structures such as nucleosomes or the lamina also contribute to degenerative and age-associated diseases remains unexplored. Nonetheless, indications of nuclear integrity decline in ageing and age-linked disease raise the possibility that the functional decline of long-lived nuclear structures promotes both cellular ageing and, beyond some threshold, neurodegeneration.
Conclusions and perspectives
Over the past decade, our view of the nuclear membrane has evolved from a passive membrane barrier to a dynamic interface that controls key regulatory aspects of cell division, differentiation and disease. Comparative genomics studies suggest an evolutionary trend towards increased complexity and multifunctionality at the nuclear periphery. The NPC is a remarkably ancient protein complex that first arose at least 1.5 billion years ago147. Lamins are evolutionarily younger, although they still represent the founding members of the intermediate filament protein family147. The lamina has increased in complexity through recent metazoan evolution, with developmentally controlled A-type lamins first appearing in D. melanogaster148. The expansion of lamin genes and splice isoforms has likely allowed for the diversification of lamin functions within distinct nuclear domains and across tissues, which remain to be fully elucidated. Surprisingly, nucleoporin genes are among the fastest-evolving genes147, defying the expectation that proteins involved in critical cellular processes should be constrained by negative selection. The recently evolved ability of individual NUPs to associate with the genome within the nucleus has likely greatly extended the influence of the NPC in gene regulation and might reflect an evolutionary trend towards more complex patterns of gene expression and genome organization149. In particular, it is tempting to speculate that the recently evolved hominoid-specific soluble NUP variant sPOM121 may have contributed to a diversification in function through its ability to interact with both NUP98 and the nonameric NUP107–160 complex, which could nucleate distinct chromatin-associated NUP complexes within the nucleoplasm74.
The nuclear lamina and the NPC both contribute to the organization of the genome and to transcription regulation. Whereas the net effect of lamina association is almost always gene repression100, association with NPCs can either activate or repress genes. Association of a locus with the lamina does not induce its association with nearby NPCs95, underscoring the independence of these modes of tethering. The factors that dictate whether NPC–genome interactions activate or repress loci remain to be determined. Moreover, it remains unclear how individual lamin isoforms and lamina-associated proteins contribute to scaffolding LADs. However, it is evident that lamina association is only one of several strategies for repressing gene activity, as of the thousands of genes that decrease in expression during ES cell differentiation, only hundreds become LAD residents91.
It may be that nuclear lamina-directed genome organization is more important in differentiated cell types, as pluripotent cells lacking B-type lamins exhibit surprisingly modest defects in genome organization85,150,151. Compared with LADs, NUP-binding profiles are considerably narrower and cover a smaller and more variable portion of the genome. NUPs often bind at the level of individual regulatory elements associated with a specific gene, including enhancers, promoters and transcription start sites47,59,60,66. Unlike LADs, NUP interactions can be rapidly induced or disrupted by environmental stimuli59,67.
Clearly, cells balance several strategies for organizing genomes, which may be further illuminated in the future by applying emerging approaches to additional systems. Single-cell HiC27, DamID92 and FISH6,101 can reveal profound changes in the organization of individual cells that may be masked in population-wide analyses. These approaches will continue to advance our understanding of consistent versus variable properties of genome organization. Similarly, evaluating nuclear organization in primary cells, in 3D cultures and within tissues is likely to further define context-specific nuclear organization.
HiC.
A technique used to study genome organization by identifying chromosomal interactions both in cis and in trans throughout the entire genome.
Nuclear periphery.
The outermost region of the nucleus, which includes the nuclear envelope and associated proteins, the nuclear lamina and the nuclear pore complexes.
X chromosome inactivation.
Transcriptional silencing of one X chromosome at random in XX female cells for dosage compensation between XX females and XY males.
Telomeres.
Repetitive sequences found at the ends of chromosomes for maintenance of genomic integrity.
Chromatin immunoprecipitation followed by sequencing.
(ChIP–seq). An assay that combines chromatin immunoprecipitation with DNA sequencing to identify protein–DNA interactions within the genome.
DNA adenine methyltransferase identification.
(DamID). An assay that fuses Escherichia coli DNA adenine methyltransferase with a protein of interest to induce adenine methylation in proximity to the fusion protein. Adenine-methylated DNA fragments represent protein-binding sites within the genome.
Fluorescence in situ hybridization.
(FISH). A fluorescence microscopy technique used to visualize specific genomic regions within the nucleus using fluorescently tagged DNA probes designed to hybridize to the region of interest.
Transcriptional memory.
The state in which genes are poised for rapid transcriptional reactivation after an initial stimulus.
Satellite DNA.
Highly repetitive non-coding sequences within heterochromatic regions of the genome.
Nucleolus.
A region within the nucleus where ribosomal RNA transcription, processing and assembly occur.
Senescence.
The cellular state in which cells permanently exit the cell cycle but do not undergo cell death.
Autophagy.
A process in which cellular material is recycled following degradation by the lysosome or vacuole.
Acknowledgements
The authors thank members of the Hetzer laboratory for providing insightful critiques on this article and apologize to those whose work could not be cited owing to space restrictions. This work was supported by US National Institutes of Health grant R01NS096786, the Nomis Foundation and the Glenn Center for Aging Research.
Footnotes
Competing interests
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer information
Nature Reviews Genetics thanks J. H. Brickner, R. Wozniak and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
References
- 1.Laubichler MD & Davidson EH Boveri’s long experiment: sea urchin merogones and the establishment of the role of nuclear chromosomes in development. Dev. Biol 314, 1–11 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cremer T & Cremer C Rise, fall and resurrection of chromosome territories: a historical perspective. Part I. The rise of chromosome territories. Eur. J. Histochem 50, 161–176 (2006). [PubMed] [Google Scholar]
- 3.Croft JA et al. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol 145, 1119–1131 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sati S & Cavalli G Chromosome conformation capture technologies and their impact in understanding genome function. Chromosoma 126, 33–44 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Lieberman-Aiden E et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article provides the first demonstration of the HiC method for probing genome conformation indicating global properties of chromatin folding — that chromosomes occupy distinct territories and that chromatin separates into megabase-scale A and B compartments on the basis of chromatin activity and gene density.
- 6.Wang S et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science 353, 598–602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uses iterative FISH to demonstrate that regions of single chromosomes that partition into different compartments defined by genomic analyses also pack into distinct domains within single cells.
- 7.Wong X, Luperchio TR & Reddy KL NET gains and losses: the role of changing nuclear envelope proteomes in genome regulation. Curr. Opin. Cell Biol 28, 105–120 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Burke B & Stewart CL The nuclear lamins: flexibility in function. Nat. Rev. Mol. Cell. Biol 14, 13–24 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Beck M & Hurt E The nuclear pore complex: understanding its function through structural insight. Nat. Rev. Mol. Cell. Biol 18, 73–89 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Stevens TJ et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ou HD et al. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357, eaag0025 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capelson M & Hetzer MW The role of nuclear pores in gene regulation, development and disease. EMBO Rep 10, 697–705 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parada LA, McQueen PG & Misteli T Tissue-specific spatial organization of genomes. Genome Biol. 5, R44 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rego A, Sinclair PB, Tao W, Kireev I & Belmont AS The facultative heterochromatin of the inactive X chromosome has a distinctive condensed ultrastructure. J. Cell Sci 121, 1119–1127 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Chen C-K et al. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354, 468–472 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Kosak ST et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296, 158–162 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Blobel G Gene gating: a hypothesis. Proc. Natl Acad. Sci 82, 8527–8529 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wandke C & Kutay U Enclosing chromatin: reassembly of the nucleus after open mitosis. Cell 152, 1222–1225 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Gerlich D et al. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell 112, 751–764 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Samwer M et al. DNA cross-bridging shapes a single nucleus from a set of mitotic chromosomes. Cell 170, 956–972.e23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson DJ, Vargas JD, Hsiao JP & Hetzer MW Recruitment of functionally distinct membrane proteins to chromatin mediates nuclear envelope formation in vivo. J. Cell Biol 186, 183–191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zierhut C, Jenness C, Kimura H & Funabiki H Nucleosomal regulation of chromatin composition and nuclear assembly revealed by histone depletion. Nat. Struct. Mol. Biol 21, 617–625 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vagnarelli P et al. Repo-man coordinates chromosomal reorganization with nuclear envelope reassembly during mitotic exit. Dev. Cell 21, 328–342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poleshko A et al. The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell Rep. 5, 292–301 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solovei I et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152, 584–598 (2013). [DOI] [PubMed] [Google Scholar]; By comparing lamina composition with heterochromatin positioning across species and tissues, this study indicates that either LBR or lamin A/C is required in mammals to tether heterochromatin to the nuclear periphery.
- 26.Zullo JM et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell 149, 1474–1487 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Nagano T et al. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 547, 61–67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walter J, Schermelleh L, Cremer M, Tashiro S & Cremer T Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J. Cell Biol 160, 685–697 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Towbin BD et al. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150, 934–947 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Poleshko A et al. Genome-nuclear lamina interactions regulate cardiac stem cell lineage restriction. Cell 171, 573–587 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article presents a beautiful demonstration of a tissue context in which lamin-directed gene repression is essential for differentiation into a functional tissue.
- 31.Kumaran RI & Spector DL A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol 180, 51–65 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solovei I et al. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137, 356–368 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Lottersberger F, Karssemeijer RA, Dimitrova N & de Lange T 53BP1 and the LINC complex promote microtubule-dependent DSB mobility and DNA repair. Cell 163, 880–893 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soutoglou E et al. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol 9, 675–682 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dion V & Gasser SM Chromatin movement in the maintenance of genome stability. Cell 152, 1355–1364 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Lucas JS, Zhang Y, Dudko OK & Murre C 3D trajectories adopted by coding and regulatory DNA elements: first-passage times for genomic interactions. Cell 158, 339–352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chubb JR, Boyle S, Perry P & Bickmore WA Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol 12, 439–445 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Therizols P et al. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science 346, 1238–1242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Canela A et al. Genome organization drives chromosome fragility. Cell 170, 507–521.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rabut G, Doye V & Ellenberg J Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol 6, 1114–1121 (2004). [DOI] [PubMed] [Google Scholar]
- 41.D’Angelo MA, Raices M, Panowski SH & Hetzer MW Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 136, 284–295 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toyama BH et al. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 154, 971–982 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; This proteomic identification of long-lived proteins identifies several nuclear structures as being extremely long lived.
- 43.Lemke EA The multiple faces of disordered nucleoporins. J. Mol. Biol 428, 2011–2024 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang S & Presgraves DC Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science 323, 779–782 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lupu F, Alves A, Anderson K, Doye V & Lacy E Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev. Cell 14, 831–842 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacinto FV, Benner C & Hetzer MW The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev. 29, 1224–1238 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toda T et al. Nup153 interacts with Sox2 to enable bimodal gene regulation and maintenance of neural progenitor cells. Cell Stem Cell 21, 618–634.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Buchwalter AL, Liang Y & Hetzer MW Nup50 is required for cell differentiation and exhibits transcription-dependent dynamics. Mol. Biol. Cell 25, 2472–2484 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D’Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH & Hetzer MW A change in nuclear pore complex composition regulates cell differentiation. Dev. Cell 22, 446–458 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casolari JM et al. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117, 427–439 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Schmid M et al. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell 21, 379–391 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Brickner JH & Walter P Gene recruitment of the activated INO1 locus to the nuclear membrane. PLOS Biol. 2, e342 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taddei A et al. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441, 774–778 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Van de Vosse DW et al. A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell 152, 969–983 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Light WH, Brickner DG, Brand VR & Brickner JH Interaction of a DNA zip code with the nuclear pore complex promotes H2A. Z incorporation and INO1 transcriptional memory. Mol. Cell 40, 112–125 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brickner J Genetic and epigenetic control of the spatial organization of the genome. Mol. Biol. Cell 28, 364–369 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raices M et al. Nuclear pores regulate muscle development and maintenance by assembling a localized Mef2C complex. Dev. Cell 41, 540–554 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krull S et al. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 29, 1659–1673 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pascual-Garcia P et al. Metazoan nuclear pores provide a scaffold for poised genes and mediate induced enhancer-promoter contacts. Mol. Cell 66, 63–76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ibarra A, Benner C, Tyagi S, Cool J & Hetzer MW Nucleoporin-mediated regulation of cell identity genes. Genes Dev. 30, 2253–2258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that NPCs bind and regulate cell type-specific super-enhancers, which are important regulatory structures in the human genome.
- 61.Capelson M et al. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140, 372–383 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides the first evidence that NUPs regulate genes independently of the NPC in metazoans.
- 62.Liang Y, Franks TM, Marchetto MC, Gage FH & Hetzer MW Dynamic association of NUP98 with the human genome. PLOS Genet. 9, e1003308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vaquerizas JM et al. Nuclear pore proteins Nup153 and megator define transcriptionally active regions in the Drosophila genome. PLOS Genet. 6, e1000846 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalverda B, Pickersgill H, Shloma VV & Fornerod M Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140, 360–371 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Panda D et al. Nup98 promotes antiviral gene expression to restrict RNA viral infection in Drosophila. Proc. Natl Acad. Sci 111, E3890–E3899 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Franks TM et al. Nup98 recruits the Wdr82-Set1A/ COMPASS complex to promoters to regulate H3K4 trimethylation in hematopoietic progenitor cells. Genes Dev. 31, 2222–2234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Light WH et al. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLOS Biol. 11, e1001524 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kasper LH et al. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol. Cell. Biol 19, 764–776 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pascual-Garcia P, Jeong J & Capelson M Nucleoporin Nup98 associates with Trx/MLL and NSL histone-modifying complexes and regulates Hox gene expression. Cell Rep. 9, 433–442 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Akhtar A & Becker PB Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 5, 367–375 (2000). [DOI] [PubMed] [Google Scholar]
- 71.Gough SM, Slape CI & Aplan PD NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood 118, 6247–6257 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Franks TM & Hetzer MW The role of Nup98 in transcription regulation in healthy and diseased cells. Trends Cell Biol. 23, 112–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu H et al. NUP98 fusion proteins interact with the NSL and MLL1 complexes to drive leukemogenesis. Cancer Cell 30, 863–878 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Franks TM et al. Evolution of a transcriptional regulator from a transmembrane nucleoporin. Genes Dev. 30, 1155–1171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper defines a key step in the evolution of a NUP gene — losing functions related to nuclear transport and taking on functions in gene regulation.
- 75.Schoenberg DR & Maquat LE Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet 13, 246–259 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.García-Oliver E, García-Molinero V & Rodriguez-Navarro S mRNA export and gene expression: the SAGA-TREX-2 connection. Biochim. Biophys. Acta 1819, 555–565 (2012). [DOI] [PubMed] [Google Scholar]
- 77.Capitanio JS, Ben Montpetit & Wozniak RW Human Nup98 regulates the localization and activity of DExH/D-box helicase DHX9. eLife 6, e18825 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Singer S et al. Nuclear pore component Nup98 is a potential tumor suppressor and regulates posttranscriptional expression of select p53 target genes. Mol. Cell 48, 799–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Makise M et al. The Nup153-Nup50 protein interface and its role in nuclear import. J. Biol. Chem 287, 38515–38522 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ren Y, Seo H-S, Blobel G & Hoelz A Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1. Proc. Natl Acad. Sci 107, 10406–10411 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kirby TJ & Lammerding J Emerging views of the nucleus as a cellular mechanosensor. Nat. Cell Biol 103, 1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eckersley-Maslin MA, Bergmann JH, Lazar Z & Spector DL Lamin A/C is expressed in pluripotent mouse embryonic stem cells. Nucleus 4, 53–60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rober RA, Weber K & Osborn M Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development 105, 365–378 (1989). [DOI] [PubMed] [Google Scholar]
- 84.Coffinier C et al. Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol. Biol. Cell 22, 4683–4693 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim Y et al. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science 334, 1706–1710 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kubben N et al. Post-natal myogenic and adipogenic developmental: defects and metabolic impairment upon loss of A-type lamins. Nucleus 2, 195–207 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schreiber KH & Kennedy BK When lamins go bad: nuclear structure and disease. Cell 152, 1365–1375 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van Steensel B, Delrow J & Henikoff S Chromatin profiling using targeted DNA adenine methyltransferase. Nat. Genet 27, 304–308 (2001). [DOI] [PubMed] [Google Scholar]
- 89.Guelen L et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 (2008). [DOI] [PubMed] [Google Scholar]
- 90.Meuleman W et al. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 23, 270–280 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peric-Hupkes D et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell 38, 603–613 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kind J et al. Genome-wide maps of nuclear lamina interactions in single human cells. Cell 163, 134–147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; This first analysis of genome association with the lamina in single cells indicates some consistent and some variable features of lamina association.
- 93.Harr JC et al. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J. Cell Biol 208, 33–52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shevelyov YY et al. The B-type lamin is required for somatic repression of testis-specific gene clusters. Proc. Natl Acad. Sci. USA 106, 3282–3287 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reddy KL, Zullo JM, Bertolino E & Singh H Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452, 243–247 (2008). [DOI] [PubMed] [Google Scholar]
- 96.Akhtar W et al. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell 154, 914–927 (2013). [DOI] [PubMed] [Google Scholar]
- 97.Pickersgill H et al. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet 38, 1005–1014 (2006). [DOI] [PubMed] [Google Scholar]
- 98.Kind J & van Steensel B Stochastic genome-nuclear lamina interactions: modulating roles of Lamin A and BAF. Nucleus 5, 124–130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gesson K et al. A-type lamins bind both hetero- and euchromatin, the latter being regulated by lamina-associated polypeptide 2 alpha. Genome Res. 26, 462–473 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van Steensel B & Belmont AS Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169, 780–791 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Luperchio TR et al. Chromosome conformation paints reveal the role of lamina association in genome organization and regulation. Preprint at bioRxiv 10.1101/122226 (2017). [DOI] [Google Scholar]; Using a modified FISH technique, this study provides striking evidence that lamina-associated regions of chromatin are densely compacted at the lamina and that disrupting lamins or chromatin modifiers disrupts compaction and lamina tethering in single cells.
- 102.Yuan J, Simos G, Blobel G & Georgatos SD Binding of lamin A to polynucleosomes. J. Biol. Chem 266, 9211–9215 (1991). [PubMed] [Google Scholar]
- 103.Taniura H, Glass C & Gerace L A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J. Cell Biol 131, 33–44 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ye Q & Worman HJ Primary structure analysis and lamin B and DNA binding of human LBR, an integral protein of the nuclear envelope inner membrane. J. Biol. Chem 269, 11306–11311 (1994). [PubMed] [Google Scholar]
- 105.Ye Q & Worman HJ Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J. Biol. Chem 271, 14653–14656 (1996). [DOI] [PubMed] [Google Scholar]
- 106.Hirano Y et al. Lamin B receptor recognizes specific modifications of histone H4 in heterochromatin formation. J. Biol. Chem 287, 42654–42663 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Clowney EJ et al. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell 151, 724–737 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Oldenburg A et al. A lipodystrophy-causing lamin A mutant alters conformation and epigenetic regulation of the anti-adipogenic MIR335 locus. J. Cell Biol 216, 2731–2743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brachner A & Foisner R Evolvement of LEM proteins as chromatin tethers at the nuclear periphery. Biochem. Soc. Trans 39, 1735–1741 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Frock RL et al. Lamin A/C and emerin are critical for skeletal muscle satellite cell differentiation. Genes Dev 20, 486–500 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Demmerle J, Koch AJ & Holaska JM Emerin and histone deacetylase 3 (HDAC3) cooperatively regulate expression and nuclear positions of MyoD, Myf5, and Pax7 genes during myogenesis. Chromosome Res. 21, 765–779 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Malik P et al. NET23/STING promotes chromatin compaction from the nuclear envelope. PLOS ONE 9, e111851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Robson MI et al. Tissue-specific gene repositioning by muscle nuclear membrane proteins enhances repression of critical developmental genes during myogenesis. Mol. Cell 62, 834–847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Somech R et al. The nuclear-envelope protein and transcriptional repressor LAP2beta interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J. Cell Sci 118, 4017–4025 (2005). [DOI] [PubMed] [Google Scholar]
- 115.Yokochi T et al. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc. Natl Acad. Sci 106, 19363–19368 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bian Q, Khanna N, Alvikas J & Belmont AS β-Globin cis-elements determine differential nuclear targeting through epigenetic modifications. J. Cell Biol 203, 767–783 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Strom AR et al. Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Larson AG et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lund EG, Duband-Goulet I, Oldenburg A, Buendia B & Collas P Distinct features of lamin A-interacting chromatin domains mapped by ChIP-sequencing from sonicated or micrococcal nuclease-digested chromatin. Nucleus 6, 30–39 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Naetar N et al. Loss of nucleoplasmic LAP2alpha-lamin A complexes causes erythroid and epidermal progenitor hyperproliferation. Nat. Cell Biol 10, 1341–1348 (2008). [DOI] [PubMed] [Google Scholar]
- 121.Kochin V et al. Interphase phosphorylation of lamin A. J. Cell Sci 127, 2683–2696 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bronshtein I et al. Loss of lamin A function increases chromatin dynamics in the nuclear interior. Nat. Commun 6, 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Markiewicz E Remodelling of the nuclear lamina and nucleoskeleton is required for skeletal muscle differentiation in vitro. J. Cell Sci 118, 409–420 (2005). [DOI] [PubMed] [Google Scholar]
- 124.Vidak S, Kubben N, Dechat T & Foisner R Proliferation of progeria cells is enhanced by lamina-associated polypeptide 2α (LAP2α) through expression of extracellular matrix proteins. Genes Dev. 29, 2022–2036 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shumaker DK & Goldman RD Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl Acad. Sci 103, 8703–8708 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.McCord RP et al. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res. 23, 260–269 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Buchwalter A & Hetzer MW Nucleolar expansion and elevated protein translation in premature aging. Nat. Commun 8, 328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McStay B & Grummt I The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol 24, 131–157 (2008). [DOI] [PubMed] [Google Scholar]
- 129.López-Otín C, Blasco MA, Partridge L, Serrano M & Kroemer G The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Toyama BH & Hetzer MW Protein homeostasis: live long, won’t prosper. Nat. Rev. Mol. Cell. Biol 14, 55–61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rodier F & Campisi J Four faces of cellular senescence. J. Cell Biol 192, 547–556 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shimi T et al. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 25, 2579–2593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dou Z et al. Autophagy mediates degradation of nuclear lamina. Nature 527, 105–109 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shah PP et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 27, 1787–1799 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lenain C et al. Massive reshaping of genome-nuclear lamina interactions during oncogene-induced senescence. Genome Res. 27, 1634–1644 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.De Cecco M et al. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 12, 247–256 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ivanov A et al. Lysosome-mediated processing of chromatin in senescence. J. Cell Biol 202, 129–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dou Z et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 166, 822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study indicates a link between weakening of the nuclear periphery, pro-inflammatory signalling and senescence that implicates declining nuclear function in ageing and cancer.
- 139.Bergmann O et al. Dynamics of cell generation and turnover in the human heart. Cell 161, 1566–1575 (2015). [DOI] [PubMed] [Google Scholar]
- 140.Spalding KL, Bhardwaj RD, Buchholz BA, Druid H & Frisén J Retrospective birth dating of cells in humans. Cell 122, 133–143 (2005). [DOI] [PubMed] [Google Scholar]
- 141.Mertens J et al. Directly reprogrammed human neurons retain aging-associated transcriptomic signatures and reveal age-related nucleocytoplasmic defects. Cell Stem Cell 17, 705–718 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Savas JN, Toyama BH, Xu T, Yates JR & Hetzer MW Extremely long-lived nuclear pore proteins in the rat brain. Science 335, 942–942 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gasset-Rosa F et al. Polyglutamine-expanded huntingtin exacerbates age-related disruption of nuclear integrity and nucleocytoplasmic transport. Neuron 94, 48–57.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang K et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article presents the first indication that NPCs are disrupted by toxic peptides that can cause neurodegenerative disease.
- 145.Lee K-H et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167, 774–788 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shi KY et al. Toxic PRnpoly-dipeptides encoded by theC9orf72repeat expansion block nuclear import and export. Proc. Natl Acad. Sci. USA 114, E1111–E1117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Devos DP, Gräf R & Field MC Evolution of the nucleus. Curr. Opin. Cell Biol 28, 8–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK & Spann TP Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 16, 533–547 (2002). [DOI] [PubMed] [Google Scholar]
- 149.Madhani HD The frustrated gene: origins of eukaryotic gene expression. Cell 155, 744–749 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Amendola M & van Steensel B Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells. EMBO Rep. 16, 610–617 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zheng X, Kim Y & Zheng Y Identification of lamin B-regulated chromatin regions based on chromatin landscapes. Mol. Biol. Cell 26, 2685–2697 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wen B, Wu H, Shinkai Y, Irizarry RA & Feinberg AP Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet 41, 246–250 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Du Z et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547, 232–235 (2017). [DOI] [PubMed] [Google Scholar]
- 154.Arai R et al. Reduction in chromosome mobility accompanies nuclear organization during early embryogenesis in Caenorhabditis elegans. Sci. Rep 7, 3631 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pasque V, Miyamoto K & Gurdon JB Efficiencies and mechanisms of nuclear reprogramming. Cold Spring Harb. Symp. Quant. Biol 75, 189–200 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Simonsson S & Gurdon J DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat. Cell Biol 6, 984–990 (2004). [DOI] [PubMed] [Google Scholar]
- 157.Sridharan R et al. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1 γ in reprogramming to pluripotency. Nat. Cell Biol 15, 872–882 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pasque V et al. Histone variant macroH2A marks embryonic differentiation in vivo and acts as an epigenetic barrier to induced pluripotency. J. Cell Sci 125, 6094–6104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Moreira PN, Robl JM & Collas P Architectural defects in pronuclei of mouse nuclear transplant embryos. J. Cell Sci 116, 3713–3720 (2003). [DOI] [PubMed] [Google Scholar]
- 160.Abernathy DG et al. MicroRNAs induce a permissive chromatin environment that enables neuronal subtype-specific reprogramming of adult human fibroblasts. Cell Stem Cell 21, 332–348.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ieda M et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]